
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Endocrinol., 19 February 2019
Sec. Neuroendocrine Science
Volume 10 - 2019 | https://doi.org/10.3389/fendo.2019.00064
This article is part of the Research TopicNeuroendocrine Control of Energy Homeostasis in Non-mammalian Vertebrates and InvertebratesView all 19 articles
Numerous neuropeptide systems have been implicated to coordinately control energy homeostasis, both centrally and peripherally. However, the vertebrate neuropeptide Y (NPY) system has emerged as the best described one regarding this biological process. The protostomian ortholog of NPY is neuropeptide F, characterized by an RXRF(Y)amide carboxyterminal motif. A second neuropeptide system is short NPF, characterized by an M/T/L/FRF(W)amide carboxyterminal motif. Although both short and long NPF neuropeptide systems display carboxyterminal sequence similarities, they are evolutionary distant and likely already arose as separate signaling systems in the common ancestor of deuterostomes and protostomes, indicating the functional importance of both. Both NPF and short-NPF systems seem to have roles in the coordination of feeding across bilaterian species, but during chordate evolution, the short NPF system appears to have been lost or evolved into the prolactin releasing peptide signaling system, which regulates feeding and has been suggested to be orthologous to sNPF. Here we review the roles of both NPF and sNPF systems in the regulation of feeding and metabolism in invertebrates.
The nomenclature of the two distinct neuropeptide families, short neuropeptide F (sNPF) on the one hand, and neuropeptide F (NPF) on the other hand, has led to confusing data and conclusions in literature. Whereas, NPF shares a common bilaterian ancestor with vertebrate neuropeptide Y (NPY), the sNPF system appears to be protostomian-specific according to (1). In this review, we summarize work on the regulation of feeding and metabolism by NPF and sNPF in invertebrates and attempt to clarify annotation ambiguities that have become apparent upon large-scale phylogenomic analyses of NPF and sNPF systems across bilaterians (1, 2).
NPY is part of a large neuropeptide family that besides NPY, includes peptide YY (PYY) and the pancreatic polypeptide (PP) (3). The 36 amino acid NPY was isolated in 1982 from pig brain extracts (4) and was found to be widely distributed in the CNS of vertebrates (5, 6). Whereas, NPY is found at all levels of the brain-gut axis, PYY, and PP seem to be predominantly expressed by endocrine cells of the digestive system (7). The three peptides share a characteristic secondary structure called the pancreatic polypeptide-fold (PP-fold) (8) that is fundamental for the full activation of the NPY G protein coupled receptors (NPYRs). These GPCRs comprise the Y1, Y2, Y4, Y5, and Y6 subtypes (9, 10). The Y1, Y2, and Y5 receptor isoforms preferentially bind NPY and PYY (11), while Y4 is specific for PP (12). The Y6 receptor exists as a truncated inactive protein in most mammals, and is only a functionally active receptor in mice and rabbits (13). While NPY centrally promotes feeding and reduces energy expenditure, PYY and PP mediate satiety. In addition, NPY family neuropeptides have various other functions that go beyond the regulation of feeding and appetite (14).
The first invertebrate NPY-like peptide was discovered in 1991 in the tapeworm Moniezia expansa using a C-terminally directed pancreatic polypeptide (PP) antiserum (15). HPLC purification of the PP-immunoreactive (IR) extract followed by automated Edman degradation sequencing identified a 39 amino acid tapeworm peptide that displays sequence similarities with the 36 amino acid vertebrate NPY (15). Based on its C-terminal sequence that ends with a phenylalanine (F) instead of a tyrosine (Y), this peptide was named neuropeptide F instead of neuropeptide Y. After this discovery, NPF-like peptides have been identified in other flatworms and in molluscs (16–20), all of which typically display an RPRF-amide C-terminal sequence and a length ranging from 36 to 40 amino acids.
The first sNPF peptides were discovered in insects, including the Colorado potato beetle Leptinotarsa decemlineata and the desert locust Schistocerca gregaria (21, 22) using antisera raised against M. expansa “long” NPF (23). These insect peptides consist of only 8 to 10 amino acids instead of 36 to 40 amino acids as typical for vertebrate NPY and flatworm or mollusc NPF. Based on their carboxyterminal RLRFamide sequence, which is similar to the RPRFamide motif of the “long” NPFs from flatworms and molluscs (15), they were designated “short” NPFs or sNPFs (24).
NPF receptors (NPFRs) were initially cloned from the brain of the pond snail Lymnaea stagnalis (25) and subsequently from Drosophila larvae (26, 27). Both receptors retained the typical features of vertebrate NPYRs and they showed the highest homology to the mammalian NPYR 2 isoform (28). Upon expression of these NPFRs in Chinese Hamster Ovary (CHO) cells, it was found that the respective NPF peptides inhibit forskolin-stimulated adenylyl cyclase activity, in accordance with vertebrate NPYRs signaling through Gi/o small proteins (25, 26, 29). In addition, the Drosophila NPF receptor can be activated by mammalian NPY-type neuropeptides when expressed in Xenopus oocytes (27).
The first sNPF receptor (sNPFR) was cloned from D. melanogaster (30) and later on from the fire ant Solenopsis invicta (31) and the mosquito Anopheles gambiae (32). Different Drosophila sNPF variants elicit a calcium response in CHO cells (30, 33) or in Xenopus oocytes (26, 34) when these are transformed to express the Drosophila sNPFR.
After the cloning of a long 36 amino acid NPF neuropeptide precursor in Drosophila melanogaster (35) and the sequencing of the Drosophila genome (36), it became evident that not all NPF/NPY-immunoreactive peptides that were designated as NPF were actually long NPFs such as those isolated from M. expansa and D. melanogaster. This led to the introduction of the terms long NPF (or simply NPF) and short NPF (sNPF) (24, 37). Alignment studies of neuropeptide precursors from both vertebrates and invertebrates pointed out the diversity in the consensus sequences for sNPF and NPF (38, 39), and suggested that short NPFs are restricted to protostomian phyla, while long NPFs are conserved across bilaterians.
In the past, several studies in insects have demonstrated overlapping functions of NPF and sNPF signaling with respect to feeding and metabolism, suggesting a common evolutionary origin of sNPF and NPF neuropeptides and receptors. This hypothesis was also supported by structural similarities between both invertebrate sNPF and NPF receptors and vertebrate NPY receptors (NPYR) (in particular with vertebrate NPY2R) (28, 39). Recent phylogenomic analyses of neuropeptide receptor families in bilaterians, however, show that sNPF and NFP receptors share only a distant common ancestor, explaining the structural similarities between both.
Both NPF and sNPF signaling systems in invertebrates have been implicated in the regulation of a diverse array of biological processes including reproduction, growth, nociception, circadian clock, learning, feeding and metabolism and they function mainly as neuromodulators or neurohormones [For an extensive review see (39)]. Here we will review past research on NPF and sNPF in the regulation of feeding-related behaviors and metabolism in protostomes. For each discussed phylum, we will briefly introduce the initial characterization of the sNPF and NPF signaling systems, followed by the current knowledge on their function(s) in feeding and metabolism. We will make a clear distinction between NPF and sNPF and will point to annotation ambiguities where appropriate. Clarifying annotation errors is not a goal here but will be crucial to further understand the evolution of the functions of sNPF and NPF neuropeptides signaling systems. Although the current picture is still not entirely clear due to insufficient genome information from all phyla, we now have increased insight in the evolutionary history of sNPF and NPF systems (Figures 1, 2). It is clear that NPF and sNPF are distinct neuropeptides systems, both involved in the regulation of feeding and metabolism. Both systems branched off from their common ancestor early in evolution, prior to the split of the deuterostome and protostome lineages. Therefore, NPF and sNPF signaling systems are a prime example to ask questions on the sustained evolutionary selection for both systems in protostomes in contrast to the deuterostomian chordate lineage where only the NPF orthologs (NPYs) have been conserved and even have been duplicated multiple times, but where the sNPF system seems to have been lost or evolved into the prolactin-releasing peptide system.
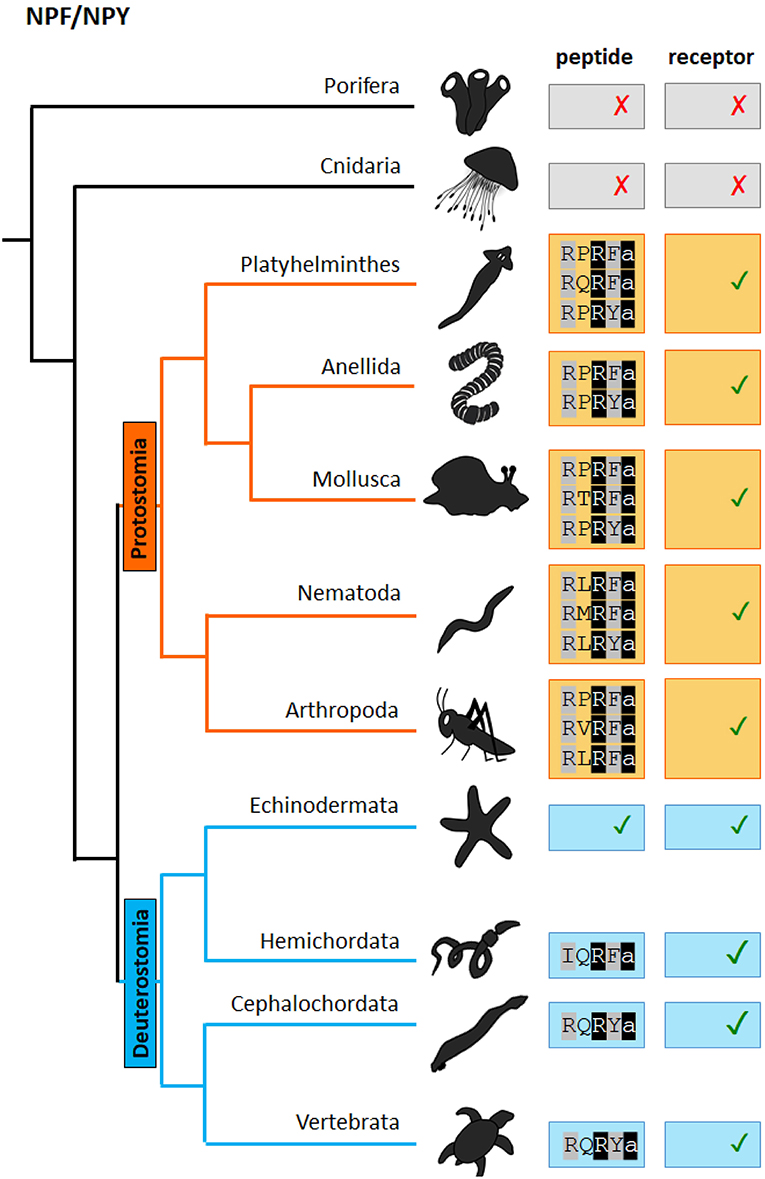
Figure 1. Scheme of the presence of NPF/NPY neuropeptides and receptors in distinct phyla of the metazoan evolutionary tree. The NPF(Y) C-terminal sequence motif is indicated. “✓” and “✗,” respectively represent the presence or absence of the peptide/receptor in the corresponding phylum. Evidence for the presence of both NPY/F peptides and receptors in cephalochordates and hemichordates can be found in Elphick et al. (40).
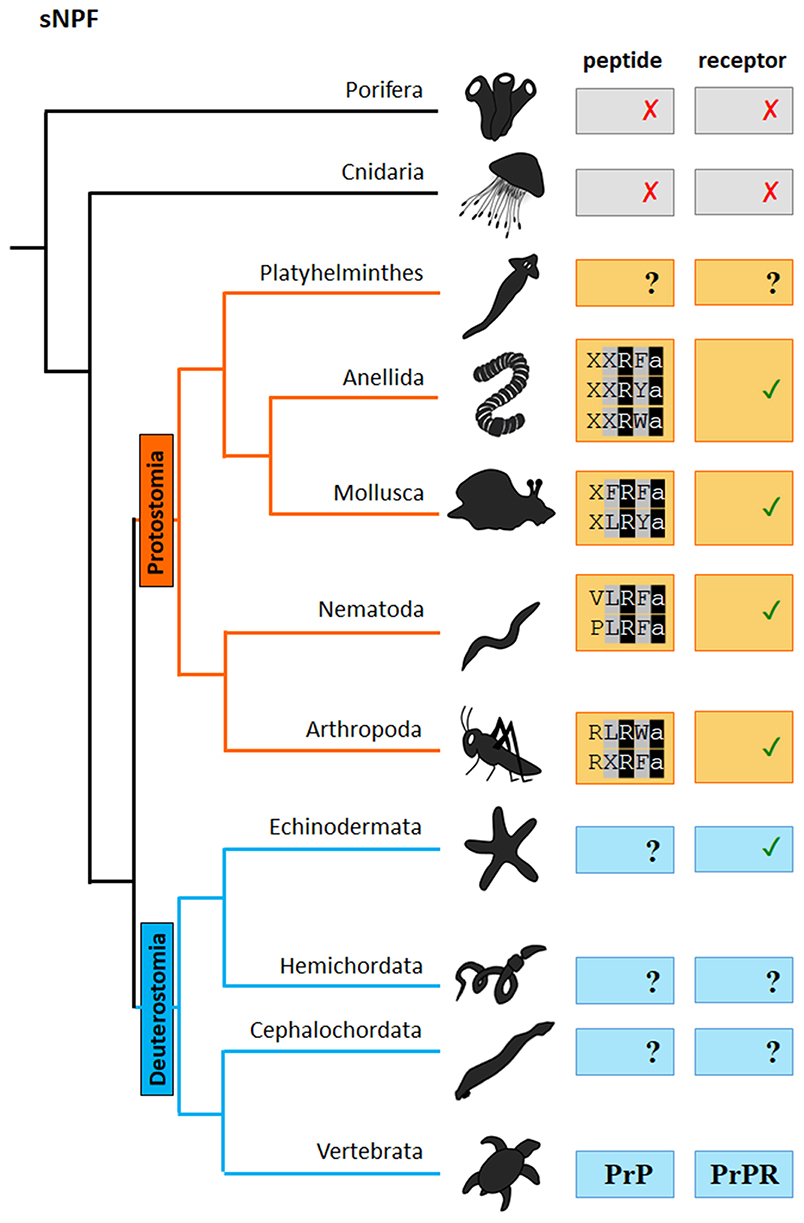
Figure 2. Scheme of the presence of sNPF neuropeptides and receptors in distinct phyla of the metazoan evolutionary tree. “✓” and “✗,” respectively represent the presence or absence of the peptide/receptor in the corresponding phylum. “?” indicates that the presence of the peptide/receptor is not clear or has so far not been investigated. PrP: prolactin. PrPR: prolactin receptor. The sNPF sequence motif is indicated. The first X in the Annelida peptide motif represents a hydrophobic amino acid, while the second X indicates a Phe, Leu or Met amino acid residue. The X in the Mollusca peptide motif represents a hydrophobic amino acid. The X in the Arthropoda peptide motif represents a Leu or Ile amino acid residue.
Figure 3 shows an alignment of representative NPF neuropeptides that have been biochemically isolated or identified by genome sequencing (for a broader overview of protostomian sNPF and NPF peptides, see Supplementary Tables 1, 2). NPFs generally consist of more than 28 amino acids, apart from the shorter predicted C. elegans orthologs and some truncated insect NPFs, and share the common RXRF/Yamide carboxyterminal motif (Figure 1).

Figure 3. Amino acid sequence alignment of representative NPFs from different invertebrate phyla. Genus and species abbreviations used in the alignment are: Schme, Schmidtea mediterranea; Drome, Drosophila melanogaster; Aplca, Aplysia californica; Caeel, Caenorhabditis elegans; Ampfi, Amphiura filiformis; Ophvi, Ophionotus victoriae; Ophar, Ophiopsila aranea. Identical residues are highlighted in black and conserved residues in gray.
Most studies on the regulation of feeding and metabolism by NPF were conducted in the genetically tractable model organism, D. melanogaster. In 1992, cloning and functional expression of two distinct D. melanogaster NPY-like receptors revealed that they could be activated by mammalian NPY and peptide YY (27). The first insect NPY-like peptide was also identified in D. melanogaster, and consisted of 36 amino acids with a characteristic RVRFa carboxyterminal sequence (35). The C-terminal F residue, instead of the vertebrate NPY-defining Y residue, prompted the name conversion from NPY to NPF, previously adopted in other protostomes as well (15). In 2002, Drosophila NPF was shown to dose-dependently activate the Drosophila NPY-like receptor NPFR when expressed in CHO cells (26). By means of immunocytochemistry and in situ hybridization, NPF was localized in the midgut and brain of D. melanogaster, suggesting a role in feeding, digestion and/or metabolism (35).
The first experimental evidence for a role of Drosophila NPF in the regulation of metabolism was provided by analysis of npf transcription levels following sugar exposure. A sugar-rich diet fed to D. melanogaster larvae evoked npf expression in two distinct neurons of the suboesophageal ganglion. Additional experiments with mutant flies deficient in sugar sensing, highlighted that not sugar ingestion, but taste perception of sugar was essential for npf expression (41). Subsequent studies showed that npf expression is high in young, foraging larvae and low in older larvae that display food aversion and burrowing. Experimentally induced overexpression and downregulation of npf transcript levels shifted these stage-specific feeding-related phenotypes (42).
In-depth characterization of the NPF signaling pathway revealed that NPF functions downstream of insulin signaling to regulate feeding in Drosophila larvae. NPF does not specifically influence total food intake, but may rather regulate food choice behavior (43, 44). NPF neurons are hypothesized to modulate the reward circuit to acquire lower-quality foods upon food deprivation (43). NPF signaling through its NPFR receptor promotes the intake of noxious food in starved flies and inhibits the aversive response that is normally elicited. In satiated flies, however, activation of the insulin signaling pathway results in the inhibition of the NPF-induced feeding response toward noxious food (44). NPF thus regulates a feeding response, that integrates both food attractiveness and hunger state (45). Together, these studies show that lower quality food or noxious food can evoke an NPF-mediated feeding response in starved flies, while in metabolically satiated flies, NPF is inhibited resulting in the intake of higher quality food (44, 45).
NPF signaling is also required at the intersection of feeding and stress, namely for the regulation of cold-resistant feeding behavior (46). NPFR1 is expressed in fructose-responsive sensory neurons in the thorax, suggesting that NPF may modulate these neurons directly (47). NPF could modulate the activity of the transient receptor potential ion channel A (TRPA) called PAIN and inhibits the regular avoidance response to aversive stimuli.
NPFR1 colocalizes with the majority of dopaminergic neurons in the larval D. melanogaster CNS, suggesting extensive interplay between these two signaling pathways (48, 49). Firstly, NPF expression in the brain of D. melanogaster may represent the food-deprived state, in which dopaminergic neurons in the mushroom bodies (MBs) promote appetitive memory formation (48). Secondly, NPF signaling modulates dopaminergic transmission in D. melanogaster in appetitive olfaction. NPFR1-expressing dopaminergic neurons display projections toward DL2-lateral horn (LH) neurons that receive olfactory inputs, and modulation of these neurons by NPF is instrumental to stimulate odor-induced appetitive feeding (49). NPF thus can stimulate (MBs) or inhibit (DL2-LH neurons) dopaminergic signaling in functionally distinct neurons even if it does not appear to be implicated in octopaminergic regulation of feeding motivation (50). In relation to this, NPF has recently been proposed to modulate odor-aroused appetitive behavior through a newly characterized DA/NPF-mediated circuit (51). When an appetitive odorant is perceived, the information is integrated in the DL2 neurons and transmitted to Dop1R1 neurons that express NPF. The dorsomedial pair of NPF neurons are essential for the proper manifestation of odor-aroused appetitive behavior (51). NPF signaling also seems to be implicated in odorant detection and is required for sensitization and correct odorant perception by a subset of olfactory neurons named antennal basiconics (ab)3A that besides NPFR also express the olfactory receptor (OR)22a, which responds to different fruity odorants (52). Related to motivation, sucralose can cause stimulation of a gustatory receptor that signals via dopamine and octopamine to activate a reward pathway in which NPF is also implicated. Supplementation of sucralose to the fly's diet caused an imbalance between energy and food sweetness that is signaled by the sweet taste receptor Gr64a. A mechanistic analysis of this sucralose response identified the NPF system as a critical downstream component in this pathway, confirming a conserved role for NPF in sucralose-sweetened food intake stimulation (53).
Since NPF is involved in the regulation of feeding and metabolism on multiple levels in D. melanogaster, its effect on obesity has also been studied. Leptin is a neuropeptide involved in the regulation of food attraction, food intake and body weight and its Drosophila homolog is Upd-1. Interestingly, the Upd-1 receptor Domeless is expressed in the brain's NPF-neurons. The odor-activated food response that is normally elicited by NPF and regulated by internal metabolic status (45), is perturbed in flies lacking Upd-1 in the NPF neurons. When the upstream regulation by Upd-1 and Domeless is absent, NPF-neurons do not register the satiety status of the flies and always display odor responses on the same level as starved flies, leading to overconsumption of food and obesity (54). The enzymatic cofactor tetrahydrobiopterin (BH4) is another compound that can affect the activity of NPF neurons in satiety. BH4, synthesized by the adipose tissue, inhibits NPF signaling by blocking its release and thus induces satiety (55).
In Drosophila, NPF modulates both food intake and wakefulness (56). In particular, NPF signaling plays a fundamental role in wake extension during deprived feeding conditions, in order to facilitate the search of new food sources. However, the neuron clusters for wake and feeding phenotypes were completely unrelated, suggesting an independent regulation of the two behaviors (56).
Confusion between NPF and sNPF partly results from insect research where a clear distinction between both neuropeptide systems remained difficult for a long time, due to the identification of so-called “NPY-like” neuropeptides that differed in length from vertebrate NPY. Studies in the insects, Locusta migratoria, Schistocerca gregaria (57), and Helicoverpa zea (58), in which only a C-terminal fragment of the “full-length” NPF was isolated and identified by mass spectrometry, contributed to this confusion. Identification of the respective neuropeptide precursor sequences by RNAseq and genome sequencing has clarified this issue. Comparison of the coding sequences of these peptides shows that they are truncated forms of larger NPF neuropeptides and that the shorter C-terminal fragments may thus either be artifacts from the extraction procedure or may be created by extensive posttranslational processing in vivo (57, 58).
The earliest indications of NPF being involved in feeding and metabolic regulation in various insect species were based on immunocytochemical localization studies. Its expression in the digestive system or its temporal regulation of expression in response to food intake suggested a regulatory role of NPF in the control of feeding and metabolism. In the yellow fever mosquito, Aedes aegypti, NPF-IR was detected in the midgut and in the suboesophageal ganglion where the regulation of food intake resides. Analysis and sequencing of the immunoreactive peptides from head and midgut extracts confirmed the presence of NPF. A. aegypti NPF inhibits transepithelial ion transport in the anterior stomach, suggesting a function in the digestive system (59). The titer of A. aegypti NPF in the haemolymph is influenced by feeding and decreases drastically after a blood meal (60). Recent studies indicate that genetic and pharmacological disruption of the mosquito NPF pathway results in abnormal host-seeking behavior and blood-feeding (61).
In S. gregaria, however, injection of NPF increases food intake, while RNA interference of npf transcripts decreases food intake. These treatments resulted in weight gain for the peptide-injected group and stunted weight gain for the knockdown group, which implies a stimulatory role of NPF in feeding (62). S. gregaria NPF transcription is spatiotemporally regulated in response to feeding, with high levels in starved animals that drop in the brain, optic lobes, and midgut upon feeding, but significantly rise in the suboesophageal ganglion (62). Also in Bombyx mori, knockdown of NPFR resulted in a reduction of food intake and growth, pointing toward a role for NPF as a positive regulator of feeding (63).
A typical long NPF as well as its truncated form have been found in Rhodnius prolixus (64). NPF-like-IR is present in the stomatogastric nervous system of Rhodnius, which regulates feeding. In addition, radioimmunoassay quantification showed a decrease in the intensity of NPF-like IR material in the cell bodies and axons after feeding, suggesting a release into the haemolymph (65).
In the honey bee Apis mellifera (66) and in the wasp Nasonia vitripennis (67), the NPF sequence does not retain the canonical RPRF/Yamide C-terminal sequence, displaying instead a C-terminal KARYamide motif. Hymenopteran NPF protein precursor sequences show nevertheless clear similarities with NPF/Y precursors of other invertebrate phyla, having YY residues in position 30–31 as well as other conserved amino acids. Interestingly, genomes of hymenopteran species do not seem to encode a clear ortholog of the NPF receptor. It is therefore not unthinkable that evolutionary pressure drastically shaped the so far unknown receptor in hymenopterans into a GPCR with unrecognizable orthology and that receptor-neuropeptide co-evolution resulted in a modification of its ligand.
In A. mellifera, NPF levels differ according to the age and tasks of the worker bees. Younger workers providing brood care display low NPF intensities in in situ hybridization and qPCR analysis, while older workers that are responsible for foraging display higher expression levels (68). This may suggest a stimulatory role for NPF in food searching and foraging behavior of worker bees, but experiments providing a causal relationship are currently lacking.
Information about NPF orthologs in arthropods outside the insect orders is currently increasing. Bioinformatic analysis of genomes and transcriptomes revealed the presence of NPF and NPF receptors in chelicerates, including spiders, scorpions and mites (69). An in silico analysis using the D. melanogaster npf transcript as a query against the crustacean expressed sequence tags (ESTs) database revealed putative hits for the shrimp Marsupenaeus japonicus and the water flea Daphnia magna (70). The predicted pro-peptides, respectively, encode a 32 and 38 amino acid NPF peptide, which both display the typical RPRFamide carboxy terminus. A similar analysis also revealed an NPF ortholog in Daphnia pulex (71). In another study, npf transcripts have been isolated from mixed eyestalk ganglia of Litopenaeus vannamei and from the brains of Melicertus marginatus (72). Both penaeid shrimp species have two identical transcript sequences and one transcript differs from the other by an in-frame 37-amino acid insertion in the middle of the coding sequence of NPF. Both transcripts are broadly distributed in the nervous system of the animals and the short one is also expressed in some of the midgut samples. A diet supplemented with the shorter NPF induced a significant increase in food intake and growth in juvenile L. vannamei suggesting an orexigenic action for the NPF (72).
While a neuropeptide from the crab Pugettia productav has been suggested to resemble vertebrate NPY because of its SQRYamide carboxyterminal sequence (73), recent phylogenetic analysis indicates that the RYamide neuropeptide family, which is widespread in arthropods (74, 75), is evolutionarily distinct from the NPF/NPY peptide family (1).
In nematodes, neuropeptide signaling systems suggested to be orthologous to NPY/NPF were initially discovered in C. elegans (76, 77). Neuropeptide receptor 1 (NPR-1) was cloned from a solitary wild-type strain and firstly assigned to the NPYR family based on its sequence similarity with the vertebrate NPYR groups (78). In a following whole-genome analysis study, using the L. stagnalis lymnokinin GPCR as a query, C. elegans NPR-1, NPR-2, NPR-3, NPR-4, NPR-5, NPR-6, NPR-7, NPR-8, NPR-10, NPR-11, NPR-12, and NPR-13 were all annotated as NPY-like receptors (76, 79). Two of these NPRs, NPR-11, and NPR-12 cluster closest with the Drosophila NPFR as shown by phylogenetic tree analysis (76) and thus appear to be true NPF homologs in C. elegans. These NPFR orthologs are also encoded in the genomes of all other nematodes investigated (76, 80).
A specific role in feeding or metabolism has not been investigated for the nematode NPF system. The only functional study that has been carried out is on the NPFR ortholog, NPR-11, which is involved in local search behavior of C. elegans when the animal is removed from its bacterial food source (81). In this work, the neuropeptide-like protein 1 (NLP-1), although not containing a peptide with a typical NPF-motif, was reported to act upon NPR-11 in the AIA interneuron to modulate local search behavior.
The first evidence for NPY orthologs in platyhelminths was found in the central and peripheral nervous system of the cestode M. expansa, from which a 39 amino acid peptide was identified by plasma desorption mass spectrometry (PDMS) upon isolation monitored by pancreatic polypeptide (PP) antiserum (15). The primary structure of this M. expansa NPF neuropeptide displays strong sequence similarities with vertebrate neuropeptide Y (15). Subsequently, immunochemical analysis using antibodies against M. expansa NPF and against vertebrate PP, Peptide tyrosine tyrosine (PYY) and Substance P (SP) revealed the presence of immunoreactive substances in other cestodes (82–85) and other flatworm classes, including turbellarians (16, 86, 87), monogeneans (88–91), and trematodes (17, 92–94). So far, the immunopositive material has only been identified in Arthurdendyus triangulates (16), Schistosoma mansoni and in Schistosoma japonicum (17). All three platyhelminth peptides have a length between 36 and 39 amino acids and display the NPF-typical RPRF carboxyterminal sequence. In addition, phylogenetic tree analysis identified several GPCRs similar to NPF/Y receptors in the S. mansoni GPCR repertoire (95).
Studies on a putative role of NPF in feeding and metabolism have as yet not been performed in platyhelminths, but PP-, PYY-, and M. expansa NPF-IR is present in neurons innervating the oral and ventral suckers of Schistosoma mansoni (94), in the pharynx musculature of Procerodes littoralis (87) and in fibers and cells of the intestinal wall of Microstomum lineare (86), suggesting a possible role in the modulation of food intake and digestion. However, these results should be interpreted with caution as cross immunoreactivity with other neuropeptides cannot be excluded.
One year after the identification of NPF in platyhelminths, several independent studies demonstrated the presence of NPF-like peptides in different species of molluscs. Following the detection of PP-IR in circumoesophageal ganglia extracts of the garden snail Helix aspersa (18), PMDS and automated Edman degradation led to the identification of an of 39 amino acid peptide that displays pronounced sequence similarities with NPF (18). Two additional NPF orthologs were discovered in the sea slug Aplysia californica (19) and in the cephalopod Loligo vulgaris (20). All display the C-terminal RPRF motif that typifies invertebrate NPF (19). Interestingly, gel permeation chromatography of the squid extract resolved two peaks of PP-IR with different molecular weights. One was suggested to contain (long) NPF and the second one contained a nine amino acid peptide harboring an N-terminal tyrosine and the NPF-typical RPRF C-terminus, but lacking the PP-fold structure (PYF) (20). This short peptide is either the result of an extraction artifact or the product from another as yet unknown neuropeptide precursor.
The only NPY/NPF receptor homolog known in molluscs to date has been cloned in the snail L. stagnalis (25). It is broadly expressed in the CNS. The 39-amino acid L. stagnalis NPF, which displays the typical RPRF C-terminal sequence and the N-terminal proline functionally activates this receptor, when expressed Chinese Hamster Ovary (CHO) cells (25).
A role for NPF in feeding and metabolism has been reported in various molluscan species. Injection of A. californica NPY (Aplca-NPY or better Aplca-NPF) into the hemocoel of the animal resulted in a dose-dependent reduction of food intake (96). In A. californica, the feeding process consists of an initial ingestion program triggered by the presence of food and initiated by the cerebral ganglion that is progressively converted to an egestion program when the esophagus and the gut signal satiation. The change in feeding state depends on the activity of the feeding central pattern generator (CPG) that receives signals derived from cerebral and buccal ganglia. In particular, the cerebral-buccal interneuron 2, located in the cerebral ganglion, initiates the ingestion program promoting the activity of the CPG interneuron B40. When the animal approaches satiation, the release of the Aplca-NPF enhances the egestion program by reducing the activity of B40 and promoting the activation of the egestion-promoting neuron B20, also located in the CPG. Aplca-NPF thus acts as a satiety signal that balances the activity of the antagonistic CPG interneurons B40 and B20 to potentiate the egestion program (96). Although the role of NPY as an orexigenic agent has been widely demonstrated in vertebrates (97–99) and in D. melanogaster (35, 41), the A. californica ortholog seems to display the opposite function. In humans and rodents, the gut released peptide YY3−36 isoform (PYY3−36) has been reported to inhibit food intake (100). It is produced postprandially by the intestinal L-cells in proportion to the calories ingested and, upon release in the blood circulation, it reaches the arcuate nucleus of the hypothalamus (101). Here, PYY3−36 acts on NPY2R, which is an inhibitory presynaptic receptor expressed in NPY neurons, modulating the activity of the NPY orexigenic pathway and leading to decrease in appetite (100, 101). Therefore, the localization and the functional activity of Aplca-NPF seems to be more closely related to the vertebrate PYY3−36 than to NPY (96). However, administration of L. stagnalis NPY (Lymst-NPY or better Lymst-NPF) in the snail led to a reduction in growth and reproduction without clear short-term effects on food intake (102). Thus, NPF regulation of energy flows appears to be conserved in L. stagnalis, while the regulation of food intake seems to be controlled by a leptin-like factor named L. stagnalis storage feedback factor (Lymst-SFF). This peptide is released from the glycogen cells of the mantle edge lining the shell of the animal that represents the only energy reserve in L. stagnalis.
A recent study on filter feeding has reported a “vertebrate-like” action of Ruditapes philippinarum NPF (Rudph-NPF) (103). Bivalves actively control food uptake by adjusting the filtration rate according to internal metabolic signaling. In accordance with this, injection of Rudph-NPF in the hemocoel resulted in a dose-dependent increase of 23% of filtration rate. In addition, a qPCR analysis demonstrated a large increase of Rudph-NPF mRNA levels in the visceral ganglion during starvation that rapidly declined after feeding, resembling the NPY expression pattern in vertebrates (104–107). Rudph-NPF injection was also associated with an increase in insulin and monoamine (serotonin and dopamine) levels, suggesting a coordinated regulation of filter feeding by multiple internal signaling pathways. In conclusion, a controversial role of mollusc NPF in the control of feeding has emerged from the works reviewed here: Aplca-NPF has an anorexigenic effect and seems to be functionally related to the vertebrate PYY3−36 isoform, while the Rudph-NPF showed an orexigenic effect more related to the vertebrate NPY. More interestingly, the Lymst-NPF does not exhibit any clear regulation of food intake, but modulates growth and reproduction. A possible explanation for this diversity could be related to the differential tissue expression of NPF receptors that are present in the different mollusc species. Since only the NPF receptor of L. stagnalis has been characterized so far, additional receptor identification studies are needed to elucidate the role of NPF in the regulation of feeding in molluscs.
The presence of NPF(Y) orthologs in annelids has been assessed by bioinformatics in the genomes of the earthworm Lumbricus rubellus, the polychaete worms Alvinella pompejana and Capitella telata, and the leech Helobdella robustaas (108, 109). In all species the predicted NPFs range from 29 to 43 amino acids in length and in the oligo-and polychaetes they display the NPF-typical RPRFamide carboxy terminus. In addition, a large transcriptome analysis of Platynereis dumerilii identified four NPF paralogous named NPY-1 to 4 of 38 to 48 amino acids in length (110). All the four genes show the typical RPRFamide carboxyterminal sequence and a partial sequence of the NPY-4 pro-neuropeptides has been confirmed by mass spectrometry (110). Although the NPY-4 receptor 1 does not seem to cluster with vertebrate NPY receptors, screening of P. dumerilii orphan GPCRs against Platynereis peptide mixtures revealed that this receptor is activated by three of NPF paralogues (NPY-1, NPY-3 and NPY-4, all displaying the NPF-typical RPRFamide carboxyterminal motif) in a calcium-based cellular receptor assay (111).
NPF encoding genes seem to have expanded in annelids. In Capitella, three predicted paralogous genes encode NPF (109), and in Helobdella five paralogues have been predicted, of which one is probably a pseudogene. Interestingly, three of these five paralogous sequences contain an NPY-typical RPRYamide C-terminal motif instead of the canonical invertebrate RPRFamide sequence (109). It has been shown that leech salivary gland neuropeptides, including NPY, aid in suppressing inflammation in their hosts from which they suck blood (112) and therefore, some of the diversified NPF(Y)s in leeches might be the result of convergent evolution because of their ectoparasitic lifestyle. An alternative explanation resides in the genome organization, gene structure, and functional content of the Spiralia (annelids and molluscs), which appear to be more similar to those of some invertebrate deuterostome genomes (Amphioxus and sea urchin) as well as non-bilaterian metazoans (such as cnidarians, sponges, and placozoans) than to those of ecdysozoan an platyhelminth protostomes that have been sequenced to date (113, 114). On the other hand, similarity-based clustering for neuropeptides and GPCRs of metazoans revealed that the predicted NPF peptides and receptors of annelids strongly cluster with the ones of arthropods, platyhelminths, and molluscs (2). Evidently, for a comprehensive genomic understanding of the metazoan radiation, a far larger sampling of genomes will be needed. Since only bioinformatic and phylogenetic analyses have been performed on annelids NPFs, their function remains elusive as yet.
One of the first investigations on the presence of evolutionarily conserved neuropeptides in echinoderms has been carried out in the starfish Marthasterias glacialis (115). Among the tested antisera, porcine PYY and human PP-like IR was found to be present in the endocrine cells and the basoepithelial plexus of the digestive tract. Analysis of the sea urchin Strongylocentrotus purpuratus genome revealed the existence of NPF(Y) receptors (116, 117). In ambulacrarians that comprise both echinoderms and hemichordates both NPY receptors and NPY neuropeptides (in Saccoglossus kowalevskii) have been predicted (2) (1, 118). In addition, a bioinformatic study focussing on echinoderms has recently shown the presence of NPY orthologs in Ophiopsila aranea, Asterias rubens and Amphiura filiformis (119). The aligned peptides lack the RQRYamide canonical consensus sequence of vertebrate NPYs but do contain a conserved RYamide carboxy terminus as well as other key amino acids (Figure 3). Functional studies on echinoderm NPY are currently lacking.
Short NPFs are short neuropeptides of 8 to12 amino acids in length and display the typical C-terminal consensus sequence M/T/L/FRFa (Figures 2, 4). As mentioned above short NPF and long NPF (NPY) are evolutionarily distant from one another. It has long been assumed that sNPF neuropeptides are confined to arthropods (http://www.neurostresspep.eu), but it has now become clear that both sNPF and NPF systems already originated in the common ancestor of protostomes and deuterostomes. Whereas, the long NPF (NPY) signaling system has been retained in both lineages, the short NPF signaling system appears to have been highly conserved across protostomes, and possibly in echinoderms (deuterostomes) as well. This raises important questions as to whether the regulatory functions of sNPF and NPF systems can be correlated with the differential lifestyles and environments of respective species. Although the sNPF system seems to have been lost in vertebrates, sNPF receptors have been shown to cluster with vertebrate prolactin-releasing peptide receptors (2), which also have a prominent role in the regulation of feeding behavior. Further research is needed to clarify this issue.

Figure 4. Amino acid sequence alignment of representatives of sNPFs from different invertebrate phyla. Genus and species abbreviations used in the alignment are: Caeel, Caenorhabditis elegans; Cragi, Crassostrea gigas; Aplca, Aplysia californica; Drome, Drosophila melanogaster; Aedae, Aedes aegypti; Capte, Capitella telata; Plasp, Platynereis species. Identical residues are highlighted in black and conserved residues in gray.
In Drosophila, four sNPF neuropeptides were predicted from the genome sequence (120), and later on identified by mass spectrometry-based peptidomics (121). Their cognate receptor was identified by means of a calcium-based receptor assay in CHO cells (30).
The use of snpf mutant flies showed that sNPF is involved in the regulation of food intake and body size in D. melanogaster. sNPF increases food intake in larval and adult flies, yet does not prolong the feeding period in larvae or modulate food preferences, as opposed to NPF (122). Pathway analysis of the sNPF signaling system revealed an interaction with insulin signaling regulating growth. sNPF activates extracellular-activated receptor kinases (ERKs) in insulin-producing cells (IPCs), which in turn modulate the expression of insulin (123, 124). In addition, insulin signaling is implicated in a negative feedback loop controlling sNPF expression and inhibiting food intake (124–127). In starved flies where insulin levels are low, sNPFR1 expression is upregulated resulting in facilitation of food search behavior (125). A microarray study revealed additional interaction partners of the sNPF system in D. melanogaster. The most pronounced and confirmed upregulated gene after sNPF administration is mnb, a Mnb/Dyrk1a kinase that activates the FOXO transcription factor through Sir2/Sirt1 deacetylase action (126). FOXO then in turn activates snpf transcription, providing a positive feedback loop. Mnb/Dyrk1a was localized in sNPFR1-expressing neurons, further evidencing an interaction with the sNPF signaling system (126). Genetic experiments using a combination of RNAi and overexpression lines of sNPFR1 and putative interaction partners revealed that activation of mnb transcription is attained through Gαs, PKA and CREB (126). In addition, there is a positive feedback loop from CREB to sNPF in the regulation of energy homeostasis. CREB can dimerize with cAMP-regulated transcription coactivator (Crtc) to stimulate the expression of sNPF, resulting in an attenuation of the immune response and an increased starvation resistance. When this CREB/Crtc-dependent activation of sNPF is absent, the immune response is stimulated while depleting energy reserves (128).
MicroRNA (miRNA) regulation was also reported to be involved in the sNPF/Dilp signaling pathway in D. melanogaster IPCs. The conserved miRNAmiR-9a is able to bind to the 3′-UTR of sNPFR-1 mRNA and downregulates translation leading to a decreased production of sNPFR-1 in the IPCs (129). This inhibition of the sNPF signaling system results in a growth reduction. The NPF and sNPF system in D. melanogaster thus modulate feeding behavior in distinct manners but are both clearly essential for proper control of food intake and metabolism. The importance of the influence of both these systems on this delicate balance was demonstrated in a study investigating resistance to amino acid starvation. Reducing or increasing the expression of either npf or snpf drastically decreases the resistance to amino acid starvation and reduces lifespan on culture media deprived of amino acids (130).
Four peptides, termed “head peptides” were isolated from A. aegyptii head extracts using an FMRFamide-directed antiserum (131). These head peptides have long been assumed to be the sNPFs of A. aegyptii. However, they could not be detected in the completed A. aegypti genome by bioinformatics, nor in any tissue by mass spectrometric analyses. In contrast, the mature peptides encoded by the snpf gene of A. aegypti were found to be abundantly present in different parts of the CNS and also in the gut (132). Furthermore, A. aegypti NPYR1 is activated by sNPF-3, which indicates that this receptor is an sNPF receptor, and not an NPY receptor ortholog as stated in the study (133).
The intricate involvement of sNPF in feeding and metabolism has been demonstrated in many insects but depending on the species sNPF can act as a stimulating or inhibiting factor.
In A. aegypti, multiple sNPFs inhibit both serotonin-induced peristaltic contractions and ion transport of the anterior stomach using in vitro preparations, thus showing a negative modulation of serotonin-induced digestive action (59). Aedes sNPF receptor expression is significantly upregulated for 3 days post blood-feeding with a peak 48 h after the blood meal (133). In contrast to sNPFR expression, the amount of sNPF significantly drops in the antennal lobes following a blood meal. This drop in sNPF coincides with an inhibition of odor-mediated host-seeking behavior. Injection of sNPF is sufficient to mimic this inhibition in host-seeking behavior (134). In the mosquito Culex quinquefasciatus, sNPF precursor and receptor expression drops 27 h post sugar feeding, while a significant increase in sNPFR could be observed 27 h post blood-feeding (135). Thus, there seems to be a specific difference in the regulation of sNPF signaling according to the meal type. Taken together, these studies show that tissue-specific changes in components of the sNPF signaling pathway are instrumental in subtle modulation of feeding, food choice and food searching-related behaviors in mosquitoes.
In S. gregaria, sNPF transcription is inhibited in starved animals, while there is a temporal increase of sNPF transcription immediately after feeding (136). sNPF injection inhibits food intake and knockdown of sNPFR or sNPF precursor significantly increases food intake in S. gregaria (136, 137). These results prompted the idea that sNPF functions as a satiety factor that inhibits food intake in S. gregaria. Further characterization of the sNPF signaling pathway in S. gregaria revealed that the nutrient content in the haemolymph regulates sNPF transcription through the insulin signaling pathway (138).
In the cockroach Periplaneta americana, an upregulation of sNPF-IR cells in the midgut was shown upon starvation, while expression of digestive enzymes was drastically downregulated. In addition, refeeding significantly decreased sNPF IR within 3 h, suggesting an inhibitory function of sNPF on digestion (139). However, the possibility that the sNPF antiserum also recognizes NPF cannot be dismissed. Ex vivo incubation experiments of P. americana showed that sNPF directly inhibits the release of proteases, amylases, and lipases when co-incubated with midguts actively producing these digestive enzymes (139). sNPF injection in fed cockroaches increases locomotion to a level comparable to that of starved animals (140). This suggests that sNPF modulates locomotion in starved animals and that increasing circulating sNPF levels in fed animals override satiety and evoke food searching behavior.
In the silkworm B. mori, starvation caused decreased transcriptional levels of sNPFR, which again links sNPF to a crucial role in fed animals. In addition, mass spectrometric analysis revealed that sNPF levels in the brain decrease during starvation and increase upon refeeding (141). Injection of Bommo-sNPF-2 reduced the latency to feed (142). All these observations suggest that sNPF positively regulates food searching and feeding in this insect species.
A differential peptidomics study in the Colorado potato beetle, Leptinotarsa decemlineata, reveals that sNPF is absent in diapausing adults, but present in active beetles (143). This might indicate that sNPF is important in actively feeding animals having increased metabolic activity, while unnecessary in diapausing animals that are metabolically inactive.
In the honey bee, A. mellifera, food deprivation causes a significant upregulation of sNPF receptor transcription, pointing to a role of sNPF in starvation-resistance or the stimulation of foraging (68, 125). In another hymenopteran species, S. invicta, a downregulation of sNPFR transcription was observed in mated queens that were starved, compared to well-fed congeners (31), again suggesting the importance of sNPF signaling during feeding or metabolically active states.
Although sNPF has several other described functions, it seems to be mainly involved in the regulation of feeding.
The first crustacean sNPF was discovered in the giant freshwater prawn Macrobrachium rosenbergii two decades ago (144). A recent in silico study in this shrimp revealed the presence in the eyestalk and CNS of sNPF transcripts encoding four sNPF peptides (145). Another transcriptome study in ice krill Euphausia crystallorophias lead to the discovery of two sNPF precursors that cleave into several active peptides (146). A unique crustacean sNPF, containing an Asp residue in position 2, was found in Daphnia pulex. This makes Dappu-sNPF more similar to insect sNPFs than other crustacean sNPFs (147). Bioinformatic analysis of genomes and transcriptomes revealed the presence of sNPF and sNPF receptors in chelicerates (69). So far, no functional studies on crustacean or chelicerate sNPFs have been performed.
C. elegans sNPF neuropeptides display more sequence variation compared to other invertebrate sNPFs, which may be attributed to the extensive expansion and diversification of the sNPF signaling system in nematodes. Figure 4 shows that peptides derived from three distinct C. elegans sNPF neuropeptide precursors, FLP 15, FLP-18 and FLP-21, display the canonical motif XLRFa in accordance with the XXR(F/Y/W)amide C-terminal of sNPF in other protostomes.
A large scale phylogenetic analysis on bilaterian neuropeptide receptors indicated that also sNPF receptors underwent a large expansion in nematodes (1). Phylogenetic analysis showed that several C. elegans NPRs cluster with the Drosophila sNPF receptor, with NPR-6 being the closest (76). Also, other C. elegans receptors, including NPR-1,2,3,4,5, NPR-10, and NPR-13 cluster with the Drosophila sNPF receptor in agreement with the large expansion of sNPF receptors as postulated by Mirabeau and Joly (1). FLP-21 derived sNPFs have been shown to activate two candidate sNPF receptors, NPR-1, and NPR-2 in cell-based receptor assays (148–150). Similarly, FLP-18 derived peptides have been identified as a ligand for the NPR-1, NPR-4, and NPR-5 candidate sNPF receptors. FLP-15 has been shown to interact with the NPR-3 sNPF receptor (148, 149, 151, 152).
The role of nematode sNPF receptors in feeding has been examined for NPR-1, NPR-4, NPR-5, and their ligands. Although above-mentioned candidate sNPF receptors have been found in genomes of other nematodes as well, all the feeding-related functional data regarding sNPF in nematodes have been almost exclusively obtained in C. elegans (76, 80).
The sNPF receptor NPR-1 is a suppressor of food-dependent aggregation behavior in C. elegans (78). When food is present, some wild-type C. elegans strains, including the standard laboratory strain N2 Bristol, slow down their movement and disperse as solitary animals across a bacterial lawn. Other strains move faster and aggregate at the border of the food lawn (78). The difference between solitary and aggregation behavior comes down to a single amino acid change in NPR-1. Solitary worms have a gain-of-function allele of the neuropeptide receptor NPR-1, NPR-1 215V (valine at position 215), whereas aggregating animals have the natural isoform NPR-1 215F (phenylalanine at position 215). Worms with disturbed npr-1 expression display solitary behavior (78). Bacterial odor influences the aggregation of npr-1 animals, as does population density, although not when food is absent (153). Aggregation behavior is mainly driven by ambient oxygen levels. C. elegans escapes atmospheric levels of 21% oxygen, which signals exposure at the surface, by aggregating in groups of animals at the border of a bacterial lawn, where local oxygen levels are reduced.
The functions of NPR-1 in food-dependent aggregation behavior are mediated by two short NPF encoding genes, flp-18 and flp-21 (149). Deletion of flp-21 increases the food-dependent aggregation behavior in NPR-1 215V and 215F worms, yet not to the level of the npr-1 null mutant (149).
Mutants of flp-18 have an altered metabolism, higher fat accumulation in the intestine and reduced oxygen consumption. Both npr-4 and npr-5 mutants display the same phenotypes suggesting that flp-18-mediated fat accumulation is executed by both receptors (151). Another effect of FLP-18 was observed in local search behavior. When wild type worms are removed from their food source, they increase their turning and reversing movements and explore the local area. When this withdrawal from food continues for a longer period, they start to search for food in bigger areas by inhibiting their turning and reversing behavior. Animals lacking flp-18 fail to make this behavioral switch (151). This switch is regulated by AIY interneurons via NPR-4 signaling (154–156). AIY release of FLP-18 is also involved in dauer formation, a transition state aiding in survival when food is scarce. This effect of FLP-18 is controlled by NPR-5 in ASJ neurons (151).
Deletion of npr-2 and npr-7 have been associated with an increase in the intestinal fat storage, but the underlying mechanism seems different from the one observed in npr-4 and npr-5 mutants, since FLP-18, the ligand acting upon these last two receptors, is not active on NPR-2 and NPR-7 (151). The ligand of NPR-2 has been identified as FLP-21. NPR-2, along with NPR-1, seems to increase the adaptation to noxious stimuli in the absence of food. However, neither FLP-21 or FLP-18 are involved in this process (150). Although the same peptide is involved in the regulation of aggregation behavior, the action of FLP-21 upon NPR-2 in the modulation of fat storage has never been assessed.
A recent study in the Pacific oyster Crassostrea gigas pointed toward the homology of C. gigas LFRFamide neuropeptides and arthropod sNPFs (157). Yet, it is important to note that LFRFamide peptides lack the Arg residue at the fourth position from the C-terminus and have a Phe instead of a Leu residue at the third position from the C-termunus, which typified all known sNPFs at that time (158). Using in silico techniques, (157) found a Cragi-sNPFR-like receptor and showed that three Cragi-LFRFamide peptides from the same precursor activate this sNPF receptor in a dose-dependent manner (157). They further evidenced that the receptor is differentially expressed in males and females and is upregulated in starved oysters. These results may suggest a role in energy metabolism and reproduction (157) and make the system a convincing functional sNPF ortholog.
Even though the Cragi-sNPF-like system is the first one that was functionally characterized in molluscs, it is not the first LFRFamide (or sNPF) neuropeptide that was discovered in this phylum. Initially, LFRFamides or sNPFs were discovered in gastropods (159) and since then also in cephalopods and oysters (160, 161). In A. californica, LFRFamide peptides or sNPFs have an inhibitory effect in buccal neurons (162). In L. stagnalis, the sNPF-encoding gene is upregulated in response to Trichobilharzia ocellata infection, indicating a role in energy metabolism and reproduction (163). Another example is the involvement of sNPF peptides in feeding along with learning and memory in the cuttlefish Sepia officinalis (164, 165). For a detailed review see Bigot et al. (157) and Zatylny-Gaudin and Favrel (161).
A bioinformatic study on C. telata predicts the presence of an LFRWamide neuropeptide encoding gene reported to be closely related to the mollusk gene (109) that encodes sNPF receptor activating LFRFamides or sNPFs (157). The genome of C. telata also encodes an sNPF receptor for which the activating ligand is currently unknown (114). In Platynereis the RYamide gene (110), encoding LFRWamides and XXRYamides, displays high sequence similarities with the LFRFamide (sNPF) precursor of mollusks (Figure 4). The Platynereis NKY receptor appears to be a candidate sNPF receptor. It was found to be activated in vitro (although at a high EC50 of 120 nM) by KAFWQPMMGGPLPVETRLASFGSRIEPDRTEPGSGPNGIKAMRYamide (111). This neuropeptide does, however, not belong to the NPF, nor the sNPF family. In vivo studies will be needed to demonstrate the cognate ligand of the annelid sNPF receptor. Knowledge on the function of sNPF signaling in annelids remains so far elusive.
Information of sNPF in echinoderms is almost non-existing. Only in A. rubens, a GPCR that resembles an sNPF-type receptor rather than an NPF receptor has recently been identified (166).
Despite their early evolutionary origin and subsequent evolutionary separation, NPF and sNPF neuropeptidergic signaling systems both control similar feeding aspects. In the species investigated, both of them converge to up- or downregulation of insulin signaling depending on the internal feeding state of the animal. It is remarkable to conclude that almost all invertebrate phyla retained both systems, even if their function in feeding is similar. It is, however, clear from the protostomian species investigated that both systems are needed for optimal regulation of feeding. This may suggest that both systems are probably controlling slightly different pathways underlying feeding behaviors. In vertebrates, sNPF signaling seems to have been lost during evolution, or may have evolved into the prolactin releasing peptide signaling system, which also regulates feeding and has been suggested to be orthologous to sNPF. Vertebrate long NPFs such as NPY, PPY and PP neuropeptide genes show an evolutionary expansion, either to compensate for the possible loss of sNPF, or to adapt to vertebrate-specific life styles and feeding.
LS and SZ conceptually designed the study. MF and IH equally contributed in writing the first draft of the manuscript. LS, SZ, LT, and IB wrote sections of the manuscript and critically revised it. All authors contributed to the final draft, have read and approved the submitted version.
This study was supported by the European Research Council, grant nr 340318. IB and SZ were fellows of the Research Foundation Flanders (FWO).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All authors acknowledge the valuable input they received from Olivier Mirabeau. The authors acknowledge funding from ERC Advanced grant 340318.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fendo.2019.00064/full#supplementary-material
1. Mirabeau O, Joly J-S. Molecular evolution of peptidergic signaling systems in bilaterians. Proc Natl Acad Sci USA. (2013) 110:E2028–37. doi: 10.1073/pnas.1219956110
2. Jekely G. Global view of the evolution and diversity of metazoan neuropeptide signaling. Proc Natl Acad Sci USA. (2013) 110:8702–7. doi: 10.1073/pnas.1221833110
3. Larhammar D, Blomqvist AG, Söderberg C. Evolution of neuropeptide Y and its related peptides. Comp Biochem Physiol C. (1993) 106:743–52.
4. Tatemoto K, Carlquist M, Mutt V. Neuropeptide Y—a novel brain peptide with structural similarities to peptide YY and pancreatic polypeptide. Nature (1982) 296:659–60. doi: 10.1038/296659a0
5. Allen YS, Adrian TE, Allen JM, Tatemoto K, Crow TJ, Bloom SR, et al. Neuropeptide Y distribution in the rat brain. Science (1983) 221:877–9.
6. De Quidt ME, Emson PC. Distribution of neuropeptide Y-like immunoreactivity in the rat central nervous system–II. Immunohistochemical analysis. Neuroscience (1986) 18:545–618.
7. Greeley GH, Hill FLC, Spannagel A, Thompson JC. Distribution of peptide YY in the gastrointestinal tract of the rat, dog, and monkey. Regul Pept. (1987) 19:365–72.
8. Fuhlendorff J, Johansen NL, Melberg SG, Thøgersen H, Schwartz TW. The antiparallel pancreatic polypeptide fold in the binding of neuropeptide Y to Y1 and Y2 receptors. J Biol Chem. (1990) 265:11706–12.
10. Michel MC, Beck-Sickinger A, Cox H, Doods HN, Herzog H, Larhammar D, et al. XVI. International Union of Pharmacology recommendations for the nomenclature of neuropeptide Y, peptide YY, and pancreatic polypeptide receptors. Pharmacol Rev. (1998) 50:143–50.
11. Larhammar D, Söderberg C, Lundell I. Evolution of the neuropeptide Y family and its receptors. Ann N Y Acad Sci. (1998) 839:35–40.
12. Gregor P, Feng Y, DeCarr LB, Cornfield LJ, McCaleb ML. Molecular characterization of a second mouse pancreatic polypeptide receptor and its inactivated human homologue. J Biol Chem. (1996) 271:27776–81.
13. Matsumoto M, Nomura T, Momose K, Ikeda Y, Kondou Y, Akiho H, et al. Inactivation of a novel neuropeptide Y/peptide YY receptor gene in primate species. J Biol Chem. (1996) 271:27217–20.
14. Loh K, Herzog H, Shi Y-C. Regulation of energy homeostasis by the NPY system. Trends Endocrinol Metab. (2015) 26:125–35. doi: 10.1016/j.tem.2015.01.003
15. Maule AG, Halton DW, Fairweather I, Johnston CF, Buchanan KD, Shaw C, et al. Neuropeptide F: a novel parasitic flatworm regulatory peptide from Moniezia expansa (Cestoda: Cyclophyllidea). Parasitology (1991) 102:309–16. doi: 10.1017/S0031182000062648
16. Curry WJ, Shaw C, Johnston CF, Thim L, Buchanan KD. Neuropeptide F: Primary structure from the tubellarian, Artioposthia triangulata. Comp Biochem Physiol Part C Comp. (1992) 101:269–74. doi: 10.1016/0742-8413(92)90272-9
17. Humphries JE, Kimber MJ, Barton YW, Hsu W, Marks NJ, Greer B, et al. Structure and bioactivity of neuropeptide F from the human parasites Schistosoma mansoni and Schistosoma japonicum. J Biol Chem. (2004) 279:39880–5. doi: 10.1074/jbc.M405624200
18. Leung PS, Shaw C, Maule AG, Thim L, Johnston CF, Irvine GB. The primary structure of neuropeptide F (NPF) from the garden snail, Helix aspersa. Regul Pept. (1992) 41:71–81. doi: 10.1016/0167-0115(92)90515-V
19. Rajpara SM, Garcia PD, Roberts R, Eliassen JC, Owens DF, Maltby D, et al. Identification and molecular cloning of a Neuropeptide Y homolog that produces prolonged inhibition in Aplysia neurons. Neuron (1992) 9:505–13. doi: 10.1016/0896-6273(92)90188-J
20. Smart D, Shaw C, Johnston C, Thim L, Halton D, Buchanan K. Peptide tyrosine phenylalanine: a novel neuropeptide F-related nonapeptide from the brain of the squid, Loligo vulgaris. Biochem Biophys Res Commun. (1992) 186:1616–23. doi: 10.1016/S0006-291X(05)81593-1
21. Schoofs L, Clynen E, Cerstiaens A, Baggerman G, Wei Z, Vercammen T, et al. Newly discovered functions for some myotropic neuropeptides in locusts. Peptides (2001) 22:219–27. doi: 10.1016/S0196-9781(00)00385-5
22. Spittaels K, Verhaert P, Shaw C, Johnston RN, Devreese B, Van Beeumen J, et al. Insect neuropeptide F (NPF)-related peptides: Isolation from colorado potato beetle (Leptinotarsa decemlineata) brain. Insect Biochem Mol Biol. (1996) 26:375–82. doi: 10.1016/0965-1748(95)00104-2
23. Maule AG, Shaw C, Halton DW, Brennan GP, Johnston CF, Moore S. Neuropeptide F (Moniezia expansa): localization and characterization using specific antisera. Parasitology (1992) 105:505. doi: 10.1017/S0031182000074680
24. Vanden Broeck J. Neuropeptides and their precursors in the fruitfly, Drosophila melanogaster. Peptides (2001) 22:241–54. doi: 10.1016/S0196-9781(00)00376-4
25. Tensen CP, Cox KJA, Burke JF, Leurs R, Van Der Schors RC, Geraerts WPM, et al. Molecular cloning and characterization of an invertebrate homologue of a neuropeptide Y receptor. Eur J Neurosci. (1998) 10:3409–16. doi: 10.1046/j.1460-9568.1998.00350.x
26. Garczynski SF, Brown MR, Shen P, Murray TF, Crim JW. Characterization of a functional neuropeptide F receptor from Drosophila melanogaster. Peptides (2002) 23:773–80. doi: 10.1046/j.1460-9568.2003.02719.x
27. Li XJ, Wu YN, North RA, Forte M. Cloning, functional expression, and developmental regulation of a neuropeptide Y receptor from Drosophila melanogaster. J Biol Chem. (1992). 267:9–12.
28. Larhammar D, Salaneck E. Molecular evolution of NPY receptor subtypes. Neuropeptides (2004) 38:141–51. doi: 10.1016/j.npep.2004.06.002
29. Lundberg JM, Beck-Sickinger A, Cox H, Doods HN, Herzog H, Larhammar D, et al. Pharmacology of cotransmission in the autonomic nervous system: integrative aspects on amines, neuropeptides, adenosine triphosphate, amino acids and nitric oxide. Pharmacol Rev. (1996) 48:113–78.
30. Mertens I, Meeusen T, Huybrechts R, De Loof A, Schoofs L. Characterization of the short neuropeptide F receptor from Drosophila melanogaster. Biochem Biophys Res Commun. (2002) 297:1140–8. doi: 10.1016/S0006-291X(02)02351-3
31. Chen M-E, Pietrantonio PV. The short neuropeptide F-like receptor from the red imported fire ant, Solenopsis invicta Buren (Hymenoptera: Formicidae). Arch Insect Biochem Physiol. (2006) 61:195–208. doi: 10.1002/arch.20103
32. Garczynski SF, Crim JW, Brown MR. Characterization and expression of the short neuropeptide F receptor in the African malaria mosquito, Anopheles gambiae. Peptides (2007) 28:109–18. doi: 10.1016/J.PEPTIDES.2006.09.019
33. Garczynski SF, Brown MR, Crim JW. Structural studies of Drosophila short neuropeptide F: occurrence and receptor binding activity. Peptides (2006) 27:575–82. doi: 10.1016/J.PEPTIDES.2005.06.029
34. Reale V, Chatwin HM, Evans PD. The activation of G-protein gated inwardly rectifying K+ channels by a cloned Drosophila melanogaster neuropeptide F-like receptor. Eur J Neurosci. (2004) 19:570–6. doi: 10.1111/j.0953-816X.2003.03141.x
35. Brown MR, Crim JW, Arata RC, Cai HN, Chun C, Shen P. Identification of a Drosophila brain-gut peptide related to the neuropeptide Y family. Peptides (1999) 20:1035–42. doi: 10.1016/S0196-9781(99)00097-2
36. Adams MD. The Genome Sequence of Drosophila melanogaster. Science (2000) 287:2185–95. doi: 10.1126/science.287.5461.2185.
37. De Loof A, Baggerman G, Breuer M, Claeys U, Cerstiaens A, Clynen E, et al. Gonadotropins in insects: an overview. Arch Insect Biochem Physiol. (2001) 47:129–38. doi: 10.1002/arch.1044
38. McVeigh P, Kimber M, Novozhilova E, Day T. Neuropeptide signalling systems in flatworms. Parasitology (2006) 131:S41. doi: 10.1017/S0031182005008851
39. Nässel DR, Wegener C. A comparative review of short and long neuropeptide F signaling in invertebrates: any similarities to vertebrate neuropeptide Y signaling? Peptides (2011) 32:1335–55. doi: 10.1016/J.PEPTIDES.2011.03.013
40. Elphick MR, Mirabeau O, Larhammar D. Evolution of neuropeptide signalling systems. J Exp Biol. (2018) 221:jeb151092. doi: 10.1242/jeb.151092
41. Shen P, Cai HN. Drosophila neuropeptide F mediates integration of chemosensory stimulation and conditioning of the nervous system by food. J Neurobiol. (2001) 47:16–25. doi: 10.1002/neu.1012
42. Wu Q, Wen T, Lee G, Park JH, Cai HN, Shen P. Developmental control of foraging and social behavior by the Drosophila neuropeptide Y-like system. Neuron (2003) 39:147–61. doi: 10.1016/S0896-6273(03)00396-9
43. Wu Q, Zhang Y, Xu J, Shen P. Regulation of hunger-driven behaviors by neural ribosomal S6 kinase in Drosophila. Proc Natl Acad Sci USA. (2005) 102:13289–94. doi: 10.1073/pnas.0501914102
44. Wu Q, Zhao Z, Shen P. Regulation of aversion to noxious food by Drosophila neuropeptide Y- and insulin-like systems. Nat Neurosci. (2005) 8:1350–5. doi: 10.1038/nn1540
45. Beshel J, Zhong Y. Graded encoding of food odor value in the Drosophila brain. J Neurosci. (2013) 33:15693–704. doi: 10.1523/JNEUROSCI.2605-13.2013
46. Lingo PR, Zhao Z, Shen P. Co-regulation of Cold-Resistant Food Acquisition by Insulin- and Neuropeptide Y-like Systems in Drosophila melanogaster. Neuroscience (2007) 148:371–4. doi: 10.1016/j.neuroscience.2007.06.010.Co-regulation
47. Xu J, Sornborger AT, Lee JK, Shen P. Drosophila TRPA channel modulates sugar-stimulated neural excitation, avoidance and social response. Nat Neurosci. (2008) 11:676–82. doi: 10.1038/nn.2119
48. Krashes MJ, DasGupta S, Vreede A, White B, Armstrong JD, Waddell S. A neural circuit mechanism integrating motivational state with memory expression in Drosophila. Cell (2009) 139:416–27. doi: 10.1016/j.cell.2009.08.035
49. Wang Y, Pu Y, Shen P. Neuropeptide-gated perception of appetitive olfactory inputs in Drosophila larvae. Cell Rep. (2013) 3:820–30. doi: 10.1016/j.celrep.2013.02.003
50. Zhang T, Branch A, Shen P. Octopamine-mediated circuit mechanism underlying controlled appetite for palatable food in Drosophila. Proc Natl Acad Sci USA. (2013) 110:15431–6. doi: 10.1073/pnas.1308816110
51. Pu Y, Zhang Y, Zhang Y, Shen P. Two Drosophila Neuropeptide Y-like Neurons define a reward module for transforming appetitive odor representations to motivation. Sci Rep. (2018) 8:11658. doi: 10.1038/s41598-018-30113-5
52. Lee S, Kim Y-J, Jones WD. Central peptidergic modulation of peripheral olfactory responses. BMC Biol. (2017) 15:35. doi: 10.1186/s12915-017-0374-6
53. Wang QP, Lin YQ, Zhang L, Wilson YA, Oyston LJ, Cotterell J, et al. Sucralose promotes food intake through NPY and a neuronal fasting response. Cell Metab. (2016) 24:75–90. doi: 10.1016/j.cmet.2016.06.010
54. Beshel J, Dubnau J, Zhong Y. A Leptin analog locally produced in the brain acts via a conserved neural circuit to modulate obesity-linked behaviors in Drosophila. Cell Metab. (2017) 25:208–17. doi: 10.1016/j.cmet.2016.12.013
55. Kim DH, Shin M, Jung SH, Kim YJ, Jones WD. A fat-derived metabolite regulates a peptidergic feeding circuit in Drosophila. PLoS Biol. (2017) 15:1–24. doi: 10.1371/journal.pbio.2000532
56. Chung BY, Ro J, Hutter SA, Miller KM, Guduguntla LS, Kondo S, et al. Drosophila Neuropeptide F signaling independently regulates feeding and sleep-wake behavior. Cell Rep. (2017) 19:2441–50. doi: 10.1016/j.celrep.2017.05.085
57. Clynen E, Schoofs L. Peptidomic survey of the locust neuroendocrine system. Insect Biochem Mol Biol. (2009) 39:491–507. doi: 10.1016/j.ibmb.2009.06.001
58. Huang Y, Crim JW, Nuss AB, Brown MR. Neuropeptide F and the corn earworm, Helicoverpa zea: a midgut peptide revisited. Peptides (2011) 483–92. doi: 10.1016/j.peptides.2010.09.014
59. Onken H, Moffett SB, Moffett DF. The anterior stomach of larval mosquitoes (Aedes aegypti): effects of neuropeptides on transepithelial ion transport and muscular motility. J Exp Biol. (2004) 207:3731–9. doi: 10.1242/jeb.01208
60. Stanek DM, Pohl J, Crim JW, Brown MR. Neuropeptide F and its expression in the yellow fever mosquito, Aedes aegypti. Peptides (2002) 23:1367–78. doi: 10.1016/S0196-9781(02)00074-8
61. Duvall LB, Ramos-Espiritu L, Barsoum KE, Glickman JF, Vosshall LB. Novel small molecule agonists of an Aedes aegypti neuropeptide Y receptor block mosquito biting behavior. bioRxiv (2018) 393793. doi: 10.1101/393793
62. Van Wielendaele P, Dillen S, Zels S, Badisco L, Vanden Broeck J. Regulation of feeding by Neuropeptide F in the desert locust, Schistocerca gregaria. Insect Biochem Mol Biol. (2013) 43:102–14. doi: 10.1016/j.ibmb.2012.10.002
63. Deng X, Yang H, He X, Liao Y, Zheng C, Zhou Q, et al. Activation of Bombyx neuropeptide G protein-coupled receptor A4 via a Gαi-dependent signaling pathway by direct interaction with neuropeptide F from silkworm, Bombyx mori. Insect Biochem Mol Biol. (2014) 45:77–88. doi: 10.1016/j.ibmb.2013.12.007
64. Ons S, Richter F, Urlaub H, Pomar RR. The neuropeptidome of Rhodnius prolixus brain. Proteomics (2009) 9:788–92. doi: 10.1002/pmic.200800499
65. Gonzalez R, Orchard I. Characterization of neuropeptide F-like immunoreactivity in the blood-feeding hemipteran, Rhodnius prolixus. Peptides (2008) 29:545–58. doi: 10.1016/j.peptides.2007.11.023
66. Hummon AB, Richmond TA, Verleyen P, Baggerman G, Huybrechts J, Ewing MA, et al. From the genome to the proteome: uncovering peptides in the Apis brain. Science (2006) 314:647–9. doi: 10.1126/science.1124128
67. Hauser F, Neupert S, Williamson M, Predel R, Tanaka Y, Grimmelikhuijzen CJP. Genomics and peptidomics of neuropeptides and protein hormones present in the parasitic wasp Nasonia vitripennis. J Proteome Res. (2010) 9:5296–310. doi: 10.1021/pr100570j
68. Ament SA, Velarde RA, Kolodkin MH, Moyse D, Robinson GE. Neuropeptide Y-like signalling and nutritionally mediated gene expression and behaviour in the honey bee. Insect Mol Biol. (2011) 20:335–45. doi: 10.1111/j.1365-2583.2011.01068.x
69. Veenstra JA. Neuropeptide evolution: Chelicerate neurohormone and neuropeptide genes may reflect one or more whole genome duplications. Gen Comp Endocrinol. (2016) 229:41–55. doi: 10.1016/J.YGCEN.2015.11.019
70. Christie AE, Cashman CR, Brennan HR, Ma M, Sousa GL, Li L, et al. Identification of putative crustacean neuropeptides using in silico analyses of publicly accessible expressed sequence tags. Gen Comp Endocrinol. (2008) 156:246–64. doi: 10.1016/j.ygcen.2008.01.018
71. Gard AL, Lenz PH, Shaw JR, Christie AE. Identification of putative peptide paracrines/hormones in the water flea Daphnia pulex (Crustacea; Branchiopoda; Cladocera) using transcriptomics and immunohistochemistry. Gen Comp Endocrinol. (2009) 160:271–87. doi: 10.1016/j.ygcen.2008.12.014
72. Christie AE, Chapline MC, Jackson JM, Dowda JK, Hartline N, Malecha SR, et al. Identification, tissue distribution and orexigenic activity of neuropeptide F (NPF) in penaeid shrimp. J Exp Biol. (2011) 214:1386–96. doi: 10.1242/jeb.053173
73. Stemmler EA, Bruns EA, Gardner NP, Dickinson PS, Christie AE. Mass spectrometric identification of pEGFYSQRYamide: a crustacean peptide hormone possessing a vertebrate neuropeptide Y (NPY)-like carboxy-terminus. Gen Comp Endocrinol. (2007) 152:1–7. doi: 10.1016/j.ygcen.2007.02.025
74. Mekata T, Kono T, Satoh J, Yoshida M, Mori K, Sato T, et al. Purification and characterization of bioactive peptides RYamide and CCHamide in the kuruma shrimp Marsupenaeus japonicus. Gen Comp Endocrinol. (2017) 246:321–30. doi: 10.1016/j.ygcen.2017.01.008
75. Veenstra JA, Khammassi H. Rudimentary expression of RYamide in Drosophila melanogaster relative to other Drosophila species points to a functional decline of this neuropeptide gene. Insect Biochem Mol Biol. (2017) 83:68–79. doi: 10.1016/j.ibmb.2017.03.001
76. Cardoso JCR, Félix RC, Fonseca VG, Power DM. Feeding and the rhodopsin family G-protein coupled receptors in nematodes and arthropods. Front Endocrinol. (2012) 3:1–22. doi: 10.3389/fendo.2012.00157
77. Hobert O. The Neuronal Genome Of Caenorhabditis elegans. In: The C. elegans Research Community, editor. WormBook. p. 1-106. Available online at: http://www.wormbook.org
78. De Bono M, Bargmann CI. Natural variation in a neuropeptide Y receptor homolog modifies social behavior and food response in cel. Cell (1998) 94:679–89.
79. Keating CD, Kriek N, Daniels M, Ashcroft NR, Hopper NA, Siney EJ, et al. Whole-genome analysis of 60 G protein-coupled receptors in Caenorhabditis elegans by gene knockout with RNAi. Curr Biol. (2003) 13:1715–20. doi: 10.1016/j.cub.2003.09.003
80. McCoy CJ, Atkinson LE, Zamanian M, McVeigh P, Day TA, Kimber MJ, et al. New insights into the FLPergic complements of parasitic nematodes: informing deorphanisation approaches. EuPA Open Proteomics (2014) 3:262–72. doi: 10.1016/j.euprot.2014.04.002
81. Chalasani SH, Kato S, Albrecht DR, Nakagawa T, Abbott LF, Bargmann CI. Neuropeptide feedback modifies odor-evoked dynamics in Caenorhabditis elegans olfactory neurons. Nat Neurosci. (2010) 13:615–21. doi: 10.1038/nn.2526
82. Fairweather I, Macartney GA, Johnston CF, Halton DW, Buchanan KD. Immunocytochemical demonstration of 5-hydroxytryptamine (serotonin) and vertebrate neuropeptides in the nervous system of excysted cysticercoid larvae of the rat tapeworm, Hymenolepis diminuta (Cestoda, Cyclophyllidea). Parasitol Res. (1988) 74:371–9. doi: 10.1007/BF00539460
83. Gustafsson MKS, Fagerholm HP, Halton DW, Hanzelova V, Maule AG, Reuter M, et al. Neuropeptides and serotonin in the cestode, Proteocephalus exiguus: an immunocytochemical study. Int J Parasitol. (1995) 25:673–82. doi: 10.1016/0020-7519(94)00169-O
84. Hrckova G, Halton DW, Maule AG, Brennan GP, Shaw C, Johnston CF. Neuropeptide F-immunoreactivity in the tetrathyridium of Mesocestoides corti (Cestoda: Cyclophyllidea). Parasitol Res. (1993) 79:690–5. doi: 10.1007/BF00932512
85. Marks NJ, Maule AG, Halton DW, Shaw C, Johnston CF. Distribution and immunochemical characteristics of neuropeptide F (NPF) (Moniezia expansa)-immunoreactivity in Proteocephalus pollanicola (Cestoda: proteocephalidea). Comp Biochem Physiol Part C, Comp. (1993) 104:381–6. doi: 10.1016/0742-8413(93)90004-5
86. Reuter M, Maule AG, Halton DW, Gustafsson MKS, Shaw C. The organization of the nervous system in Platyhelminthes. The neuropeptide F-immunoreactive pattern in Catenulida, Macrostomida, Proseriata. Zoomorphology (1995) 115:83–97. doi: 10.1007/BF00403257
87. Reuter M, Gustafsson MKS, Sahlgren C, Halton DW, Maule AG, Shaw C. The nervous system of Tricladida. I Neuroanatomy of Procerodes littoralis (Maricola, Procerodidae): an immunocytochemical study. Invertebr Neurosci. (1995) 1:113–22. doi: 10.1007/BF02331909
88. Brennan GP, Ramasamy P. Ultrastructure of the surface structures and electron immunogold labeling of peptide immunoreactivity in the nervous system of Pseudothoracocotyla indica (Polyopisthocotylea: Monogenea). Parasitol Res. (1996) 82:638–46. doi: 10.1007/s004360050178
89. Maule AG, Shaw C, Halton DW, Johnston CF, Fairweather I. Localization, quantification, and characterization of pancreatic polypeptide immunoreactivity in the parasitic flatworm Diclidophora merlangi and its fish host (Merlangius merlangus). Gen Comp Endocrinol. (1989) 74:50–6. doi: 10.1016/0016-6480(89)90113-5
90. Maule AG, Halton DW, Johnston CF, Fairweather I, Shaw C. Immunocytochemical demonstration of neuropeptides in the fish-gill parasite, Diclidophora merlangi (Monogenoidea). Int J Parasitol. (1989) 19:307–16. doi: 10.1016/0020-7519(89)90142-2
91. Maule AG, Brennan GP, Halton DW, Shaw C, Johnston CF, Moore S. Neuropeptide F-immunoreactivity in the monogenean parasite Diclidophora merlangi. Parasitol Res. (1992) 78:655–60. doi: 10.1007/BF00931516
92. Halton DW, Halton DW, Maule AG, Brennan GP, Shaw C, Johnston CF. Comparative analyses of the neuropeptide F (NPF)- and FMRFamide-related peptide (FaRP)-immunoreactivities in Fasciola hepatica and Schistosoma spp. Parasitology (1995) 110:371–81. doi: 10.1017/S0031182000064714
93. Magee RM, Fairweather I, Halton DW, Johnston CF, Shaw C. Immunocytochemical demonstration of neuropeptides in the nervous system of the liver fluke, Fasciola hepatica (trematoda, digenea). Parasitology (1989) 98:227–38. doi: 10.1017/S0031182000062132
94. Skuce PJ, Johnston CF, Fairweather I, Halton DW, Shaw C, Buchanan KD. Immunoreactivity to the pancreatic polypeptide family in the nervous system of the adult human blood fluke, Schistosoma mansoni. Cell Tissue Res. (1990) 261:573–81. doi: 10.1007/BF00313537
95. Hahnel S, Wheeler N, Lu Z, Wangwiwatsin A, McVeigh P, Maule A, et al. Tissue-specific transcriptome analyses provide new insights into GPCR signalling in adult Schistosoma mansoni. PLOS Pathog. (2018) 14:e1006718. doi: 10.1371/journal.ppat.1006718
96. Jing J, Vilim FS, Horn CC, Alexeeva V, Hatcher NG, Sasaki K, et al. From hunger to satiety: reconfiguration of a feeding network by Aplysia neuropeptide Y. J Neurosci. (2007) 27:3490–502. doi: 10.1523/JNEUROSCI.0334-07.2007
97. Clark JT, Kalra PS, Kalra SP. Neuropeptide Y stimulates feeding but inhibits sexual behavior in rats*. Endocrinology (1985) 117:2435–42. doi: 10.1210/endo-117-6-2435
98. Levine AS, Morley JE. Neuropeptide Y: a potent inducer of consummatory behavior in rats. Peptides (1984) 5:1025–9. doi: 10.1016/0196-9781(84)90165-7
99. Mercer RE, Chee MJS, Colmers WF. The role of NPY in hypothalamic mediated food intake. Front Neuroendocrinol. (2011) 32:398–415. doi: 10.1016/j.yfrne.2011.06.001
100. Batterham RL, Cowley MA, Small CJ, Herzog H, Cohen MA, Dakin CL, et al. Gut hormone PYY3-36 physiologically inhibits food intake. Nature (2002) 418:650–4. doi: 10.1038/nature00887
101. le Roux CW, Bloom SR. Peptide YY, appetite and food intake. Proc Nutr Soc. (2005) 64:213–6. doi: 10.1079/PNS2005427
102. De Jong-Brink M, Ter Maat A, Tensen CP. NPY in invertebrates: molecular answers to altered functions during evolution. Peptides (2001) 22:309–15. doi: 10.1016/S0196-9781(01)00332-1
103. Wang X, Miao J, Liu P, Pan L. Role of neuropeptide F in regulating filter feeding of Manila clam, Ruditapes philippinarum. Comp Biochem Physiol Part B Biochem Mol Biol. (2017) 205:30–8. doi: 10.1016/j.cbpb.2016.12.001
104. Ji W, Ping HC, Wei KJ, Zhang GR, Shi ZC, Yang R, et al. Ghrelin, neuropeptide Y (NPY) and cholecystokinin (CCK) in blunt snout bream (Megalobrama amblycephala): CDNA cloning, tissue distribution and mRNA expression changes responding to fasting and refeeding. Gen Comp Endocrinol. (2015) 223:108–19. doi: 10.1016/j.ygcen.2015.08.009
105. Marks JL, Li M, Schwartz M, Porte D, Baskin DG. Effect of fasting on regional levels of neuropeptide Y mRNA and insulin receptors in the rat hypothalamus: an autoradiographic study. Mol Cell Neurosci. (1992) 3:199–205. doi: 10.1016/1044-7431(92)90039-5
106. Silverstein JT, Breininger J, Baskin DG, Plisetskaya EM. Neuropeptide Y-like gene expression in the salmon brain increases with fasting. Gen Comp Endocrinol. (1998) 110:157–65. doi: 10.1006/gcen.1998.7058
107. Yokobori E, Azuma M, Nishiguchi R, Kang KS, Kamijo M, Uchiyama M, et al. Neuropeptide Y stimulates food intake in the zebrafish, Danio rerio. J Neuroendocrinol. (2012) 24:766–73. doi: 10.1111/j.1365-2826.2012.02281.x
108. Nuss AB, Forschler BT, Crim JW, TeBrugge V, Pohl J, Brown MR. Molecular characterization of neuropeptide F from the eastern subterranean termite Reticulitermes flavipes (Kollar) (Isoptera: Rhinotermitidae). Peptides (2010) 31:419–28. doi: 10.1016/j.peptides.2009.09.001
109. Veenstra JA. Neuropeptide evolution: Neurohormones and neuropeptides predicted from the genomes of Capitella teleta and Helobdella robusta. Gen Comp Endocrinol. (2011) 171:160–75. doi: 10.1016/j.ygcen.2011.01.005
110. Conzelmann M, Williams EA, Krug K, Franz-Wachtel M, Macek B, Jékely G. The neuropeptide complement of the marine annelid Platynereis dumerilii. BMC Genomics (2013) 14:906. doi: 10.1186/1471-2164-14-906
111. Bauknecht P, Jékely G. Large-scale combinatorial deorphanization of Platynereis neuropeptide GPCRs. CellReports (2015) 12:684–93. doi: 10.1016/j.celrep.2015.06.052
112. Liu WH, Chen Y, Bai XW, Yao HM, Zhang XG, Yan XW, et al. Identification and characterization of a novel neuropeptide (neuropeptide Y-HS) from leech salivary gland of Haemadipsa sylvestris. Chin J Nat Med. (2016) 14:677–82. doi: 10.1016/S1875-5364(16)30080-2
113. Raible F, Tessmar-Raible K, Osoegawa K, Wincker P, Jubin C, Balavoine G, et al. Evolution: vertebrate-type intron-rich genes in the marine annelid Platynereis dumerilii. Science (2005) 310:1325–6. doi: 10.1126/science.1119089
114. Simakov O, Marletaz F, Cho S-J, Edsinger-Gonzales E, Havlak P, Hellsten U, et al. Insights into bilaterian evolution from three spiralian genomes. Nature (2013) 493:526–31. doi: 10.1038/nature11696
115. Martínez A, López J, Montuenga LM, Sesma P. Regulatory peptides in gut endocrine cells and nerves in the starfish Marthasterias glacialis. Cell Tissue Res. (1993) 271:375–80. doi: 10.1007/BF00318624
116. Burke RD, Angerer LM, Elphick MR, Humphrey GW, Yaguchi S, Kiyama T, et al. A genomic view of the sea urchin nervous system. Dev. Biol. (2006) 300:434–60. doi: 10.1016/j.ydbio.2006.08.007
117. Sodergren E, Weinstock GM, Davidson EH, Cameron RA, Gibbs RA, Angerer RC, et al. The genome of the sea urchin Strongylocentrotus purpuratus. Science (2006) 314:941–52. doi: 10.1126/science.1133609
118. Elphick MR, Mirabeau O. The evolution and variety of RFamide-type neuropeptides: insights from deuterostomian invertebrates. Front Endocrinol. (Lausanne) (2014). 5:93. doi: 10.3389/fendo.2014.00093
119. Zandawala M, Moghul I, Guerra LAY, Delroisse J, Abylkassimova N, Hugall AF, et al. Discovery of novel representatives of bilaterian neuropeptide families and reconstruction of neuropeptide precursor evolution in ophiuroid echinoderms. Open Biol. (2017) 7:170129. doi: 10.1098/rsob.170129
120. Hewes RS, Taghert PH. Neuropeptides and neuropeptide receptors in the Drosophila melanogaster genome. Genome Res. (2001) 11:1126–42. doi: 10.1101/gr.169901
121. Baggerman G, Cerstiaens A, De Loof A, Schoofs L. Peptidomics of the larval Drosophila melanogaster central nervous system. J Biol Chem. (2002) 277:40368–74. doi: 10.1074/jbc.M206257200
122. Lee KS, You KH, Choo JK, Han YM, Yu K. Drosophila short neuropeptide F regulates food intake and body size. J Biol Chem. (2004) 279:50781–9. doi: 10.1074/jbc.M407842200
123. Kapan N, Lushchak OV, Luo J, Nässel DR. Identified peptidergic neurons in the Drosophila brain regulate insulin-producing cells, stress responses and metabolism by coexpressed short neuropeptide F and corazonin. Cell Mol Life Sci. (2012) 69:4051–66. doi: 10.1007/s00018-012-1097-z
124. Lee KS, Kwon OY, Lee JH, Kwon K, Min KJ, Jung SA, et al. Drosophila short neuropeptide F signalling regulates growth by ERK-mediated insulin signalling. Nat Cell Biol. (2008) 10:468–75. doi: 10.1038/ncb1710
125. Root CM, Ko KI, Jafari A, Wang JW. Presynaptic facilitation by neuropeptide signaling mediates odor-driven food search. Cell (2011) 145:133–44. doi: 10.1016/j.cell.2011.02.008
126. Hong SH, Lee KS, Kwak SJ, Kim AK, Bai H, Jung MS, et al. Minibrain/Dyrk1a regulates food intake through the Sir2-FOXO-sNPF/NPY pathway in Drosophila and mammals. PLoS Genet. (2012) 8:1–15. doi: 10.1371/journal.pgen.1002857
127. Ko KI, Root CM, Lindsay SA, Zaninovich OA, Shepherd AK, Wasserman SA, et al. Starvation promotes concerted modulation of appetitive olfactory behavior via parallel neuromodulatory circuits. Elife (2015) 4:e08298. doi: 10.7554/eLife.08298
128. Shen R, Wang B, Giribaldi MG, Ayres J, Thomas JB, Montminy M. Neuronal energy-sensing pathway promotes energy balance by modulating disease tolerance. Proc Natl Acad Sci USA. (2016) 113:E3307–14. doi: 10.1073/pnas.1606106113
129. Suh YS, Bhat S, Hong SH, Shin M, Bahk S, Cho KS, et al. Genome-wide microRNA screening reveals that the evolutionary conserved miR-9a regulates body growth by targeting sNPFR1/NPYR. Nat Commun. (2015) 6:1–11. doi: 10.1038/ncomms8693
130. Slade JD, Staveley BE. Manipulation of components that control feeding behavior in Drosophila melanogaster increases sensitivity to amino acid starvation. Genet Mol Res. (2016) 15:1–12. doi: 10.4238/gmr.15017489
131. Matsumoto S, Brown MR, Crim JW, Vigna SR, Lea AO. Isolation and primary structure of neuropeptides from the mosquito, Aedes aegypti, immunoreactive to FMRFamide antiserum. Insect Biochem. (1989) 19:277–83. doi: 10.1016/00i20-1790(89)90073-5
132. Predel R, Neupert S, Garczynski SF, Crim JW, Brown MR, Russell WK, et al. Neuropeptidomics of the mosquito Aedes aegypti. J Proteome Res. (2010) 9:2006–15. doi: 10.1021/pr901187p
133. Liesch J, Bellani LL, Vosshall LB. Functional and genetic characterization of neuropeptide Y-like receptors in Aedes aegypti. PLoS Negl Trop Dis. (2013) 7.(10):e2486 doi: 10.1371/journal.pntd.0002486
134. Christ P, Reifenrath A, Kahnt J, Hauser F, Hill SR, Schachtner J, et al. Feeding-induced changes in allatostatin-A and short neuropeptide F in the antennal lobes affect odor-mediated host seeking in the yellow fever mosquito, Aedes aegypti. PLoS ONE (2017) 12:1–15. doi: 10.1371/journal.pone.0188243
135. Christ P, Hill SR, Schachtner J, Hauser F, Ignell R. Functional characterization of mosquito short neuropeptide F receptors. Peptides (2018) 103:31–9. doi: 10.1016/j.peptides.2018.03.009
136. Dillen S, Verdonck R, Zels S, Van Wielendaele P, Vanden Broeck J. Identification of the short neuropeptide F precursor in the desert locust: evidence for an inhibitory role of sNPF in the control of feeding. Peptides (2014) 53:134–9. doi: 10.1016/j.peptides.2013.09.018
137. Dillen S, Zels S, Verlinden H, Spit J, van Wielendaele P, Vanden Broeck J. Functional characterization of the short neuropeptide F receptor in the desert locust, Schistocerca gregaria. PLoS ONE (2013) 8:e53604. doi: 10.1371/journal.pone.0053604
138. Dillen S, Chen Z, Vanden Broeck J. Nutrient-dependent control of short neuropeptide F transcript levels via components of the insulin/IGF signaling pathway in the desert locust, Schistocerca gregaria. Insect Biochem Mol Biol. (2016) 68:64–0. doi: 10.1016/j.ibmb.2015.11.007
139. Mikani A, Wang QS, Takeda M. Brain-midgut short neuropeptide F mechanism that inhibits digestive activity of the American cockroach, Periplaneta americana upon starvation. Peptides (2012) 34:135–44. doi: 10.1016/j.peptides.2011.10.028
140. Mikani A, Watari Y, Takeda M. Brain-midgut cross-talk and autocrine metabolastat via the sNPF/CCAP negative feed-back loop in the American cockroach, Periplaneta americana. Cell Tissue Res. (2015) 362:481–96. doi: 10.1007/s00441-015-2242-4
141. Nagata S, Matsumoto S, Nakane T, Ohara A, Morooka N, Konuma T, et al. Effects of starvation on brain short neuropeptide F-1,−2, and−3 levels and short neuropeptide F receptor expression levels of the silkworm, Bombyx mori. Front Endocrinol. (2012) 3:1–8. doi: 10.3389/fendo.2012.00003
142. Nagata S, Morooka N, Matsumoto S, Kawai T, Nagasawa H. Effects of neuropeptides on feeding initiation in larvae of the silkworm, Bombyx mori. Gen Comp Endocrinol. (2011) 172:90–5. doi: 10.1016/j.ygcen.2011.03.004
143. Huybrechts J, De Loof A, Schoofs L. Diapausing Colorado potato beetles are devoid of short neuropeptide F I and II. Biochem Biophys Res Commun. (2004) 317:909–16. doi: 10.1016/j.bbrc.2004.03.136
144. Sithigorngul P, Saraithongkum W, Jaideechoey S, Longyant S, Sithigorngul W. Novel FMRFamide-like neuropeptides from the eyestalk of the giant freshwater prawn Macrobrachium rosenbergii. Comp Biochem Physiol B Biochem Mol Biol. (1998) 120:587–95. doi: 10.1016/S0305-0491(98)10051-2.
145. Suwansa-Ard S, Thongbuakaew T, Wang T, Zhao M, Elizur A, Hanna PJ, et al. In silico neuropeptidome of female Macrobrachium rosenbergii based on transcriptome and peptide mining of eyestalk, central nervous system and ovary. PLoS ONE (2015) 10:e123848. doi: 10.1371/journal.pone.0123848
146. Toullec JY, Corre E, Bernay B, Thorne MAS, Cascella K, Ollivaux C, et al. Transcriptome and peptidome characterisation of the main neuropeptides and peptidic hormones of a Euphausiid: the ice krill, Euphausia crystallorophias. PLoS ONE (2013) 8:e71609. doi: 10.1371/journal.pone.0071609
147. Dircksen H, Neupert S, Predel R, Verleyen P, Huybrechts J, Strauss J, et al. Genomics, transcriptomics, and peptidomics of Daphnia pulex neuropeptides and protein hormones. J. Proteome Res. (2011) 10:4478–504. doi: 10.1021/pr200284e
148. Kubiak TM, Larsen MJ, Zantello MR, Bowman JW, Nulf SC, Lowery DE. Functional annotation of the putative orphan Caenorhabditis elegans G-protein-coupled receptor C10C6.2 as a FLP15 Peptide Receptor. J Biol Chem. (2003) 278:42115–20. doi: 10.1074/jbc.M304056200
149. Rogers C, Reale V, Kim K, Chatwin H, Li C, Evans P, et al. Inhibition of Caenorhabditis elegans social feeding by FMRFamide-related peptide activation of NPR-1. Nat Neurosci. (2003) 6:1178–85. doi: 10.1038/nn1140
150. Ezcurra M, Walker DS, Beets I, Swoboda P, Schafer WR. Neuropeptidergic signaling and active feeding state inhibit nociception in Caenorhabditis elegans. J Neurosci. (2016) 36:3157–69. doi: 10.1523/JNEUROSCI.1128-15.2016
151. Cohen M, Reale V, Olofsson B, Knights A, Evans P, de Bono M. Coordinated regulation of foraging and metabolism in C. elegans by RFamide neuropeptide signaling. Cell Metab. (2009) 9:375–85. doi: 10.1016/j.cmet.2009.02.003
152. Kubiak TM, Larsen MJ, Bowman JW, Geary TG, Lowery DE. FMRFamide-like peptides encoded on the flp-18 precursor gene activate two isoforms of the orphan Caenorhabditis elegans G-protein-coupled receptor Y58G8A.4 heterologously expressed in mammalian cells. Biopolymers (2007) 90:339–48. doi: 10.1002/bip.20850
153. De Bono M, Tobin, DM, Davis, MW, Avery, L, Bargmann CI. Social feeding in Caenorhabditis elegans is induced by neurons that detect aversive stimuli. Nature (2002) 419:899–903. doi: 10.1038/nature01169
154. Gray JM, Hill JJ, Bargmann CI. A circuit for navigation in Caenorhabditis elegans. Proc Natl Acad Sci USA. (2005) 102:3184–91. doi: 10.1073/pnas.0409009101
155. Tsalik EL, Hobert O. Functional mapping of neurons that control locomotory behavior in Caenorhabditis elegans. J Neurobiol. (2003) 56:178–97. doi: 10.1002/neu.10245
156. Wakabayashi T, Kitagawa I, Shingai R. Neurons regulating the duration of forward locomotion in Caenorhabditis elegans. Neurosci Res. (2004) 50:103–11. doi: 10.1016/j.neures.2004.06.005
157. Bigot L, Beets I, Dubos M-P, Boudry P, Schoofs L, Favrel P. Functional characterization of a short neuropeptide F-related receptor in a lophotrochozoan, the mollusk Crassostrea gigas. J Exp Biol. (2014) 217:2974–82. doi: 10.1242/jeb.104067
158. Nässel DR, Enell LE, Santos JG, Wegener C, Johard HAD. A large population of diverse neurons in the Drosophila central nervous system expresses short neuropeptide F, suggesting multiple distributed peptide functions. BMC Neurosci (2008) 9:90. doi: 10.1186/1471-2202-9-90
159. Kuroki Y, Kanda T, Kubota I, Ikeda T, Fujisawa Y, Minakata H, et al. FMRFamide-related peptides isolated from the prosobranch mollusc Fusinus ferrugineus. Acta Biol Hung. (1993) 44:41–4.
160. Stewart MJ, Favrel P, Rotgans BA, Wang T, Zhao M, Sohail M, et al. Neuropeptides encoded by the genomes of the Akoya pearl oyster Pinctata fucata and Pacific oyster Crassostrea gigas: a bioinformatic and peptidomic survey. BMC Genomics (2014) 15:840. doi: 10.1186/1471-2164-15-840
161. Zatylny-Gaudin C, Favrel P. Diversity of the RFamide peptide family in mollusks. Front Endocrinol. (2014) 5:1–14. doi: 10.3389/fendo.2014.00178
162. Cropper EC, Brezina V, Vilim FS, Harish O, Price DA, Rosen S, et al. FRF peptides in the ARC neuromuscular system of Aplysia: purification and physiological actions. J Neurophysiol. (1994) 72:2181–95. doi: 10.1152/jn.1994.72.5.2181
163. Hoek RM, van Kesteren RE, Smit AB, de Jong-Brink M, Geraerts WP. Altered gene expression in the host brain caused by a trematode parasite: neuropeptide genes are preferentially affected during parasitosis. Proc Natl Acad Sci USA. (1997) 94:14072–6. doi: 10.1073/pnas.94.25.14072
164. Zhang Z, Goodwin E, Loi PK, Tublitz NJ. Molecular analysis of a novel FMRFamide-related peptide gene (SOFaRP2) and its expression pattern in the brain of the European cuttlefish Sepia officinalis. Peptides (2012) 34:114–9. doi: 10.1016/j.peptides.2011.07.011
165. Zhang Z, Tublitz NJ. Expression of the SOFaRP2 gene in the central nervous system of the adult cuttlefish Sepia officinalis. Neuropeptides (2013) 47:149–55. doi: 10.1016/j.npep.2013.01.003
166. Zhong X, Yañez Guerra LA, Zampronio C, Jones A, Mirabeau O, Butts T, et al. (2018). Discovery of a Neuropeptide Y/short Neuropeptide F Related Signalling System in Echinoderms. CECE2018, abstract book 4A-O1. Available online at: http://cece2018.org/projects/
167. Husson SJ, Mertens I, Janssen T, Lindemans M, Schoofs L. Neuropeptidergic signaling in the nematode Caenorhabditis elegans. Prog Neurobiol. (2007) 82:33–55. doi: 10.1016/J.PNEUROBIO.2007.01.006
168. Zoephel J, Reiher W, Rexer K-H, Kahnt J, Wegener C. Peptidomics of the agriculturally damaging larval stage of the cabbage root fly Delia radicum (Diptera: Anthomyiidae). PLoS ONE (2012) 7:e41543. doi: 10.1371/journal.pone.0041543
169. Kimber MJ, Fleming CC, Bjourson AJ, Halton DW, Maule AG. FMRFamide-related peptides in potato cyst nematodes. Mol Biochem Parasitol. (2001) 116:199–208. doi: 10.1016/S0166-6851(01)00323-1
170. Huang Y, Brown MR, Lee TD, Crim JW. RF-amide peptides isolated from the midgut of the corn earworm, Helicoverpa zea, resemble pancreatic polypeptide. Insect Biochem Mol Biol. (1998) 28:345–56. doi: 10.1016/S0965-1748(98)00007-1
171. Vilim FS, Sasaki K, Rybak J, Alexeeva V, Cropper EC, Jing J, et al. Distinct mechanisms produce functionally complementary actions of neuropeptides that are structurally related but derived from different precursors. J Neurosci. (2010) 30:131–47. doi: 10.1523/JNEUROSCI.3282-09.2010
172. Veenstra JA. Neurohormones and neuropeptides encoded by the genome of Lottia gigantea, with reference to other mollusks and insects. Gen Comp Endocrinol. (2010) 167:86–103. doi: 10.1016/j.ygcen.2010.02.010
173. Li C, Kim K. Family of FLP peptides in Caenorhabditis elegans and related nematodes. Front. Endocrinol. (2014) 5:150. doi: 10.3389/fendo.2014.00150
174. Husson SJ, Clynen E, Baggerman G, Janssen T, Schoofs L. Defective processing of neuropeptide precursors in Caenorhabditis elegans lacking proprotein convertase 2 (KPC-2/EGL-3): mutant analysis by mass spectrometry. J Neurochem. (2006) 98:1999–2012. doi: 10.1111/j.1471-4159.2006.04014.x
175. Veenstra JA, Lambrou G. Isolation of a novel RFamide peptide from the midgut of the American cockroach, Periplaneta americana. Biochem Biophys Res Commun. (1995) 213:519–24. doi: 10.1006/bbrc.1995.2162
176. Cowden C, Stretton AOW. Eight novel FMRFamide-like neuropeptides isolated from the nematode Ascaris suum. Peptides (1995) 16:491–500. doi: 10.1016/0196-9781(94)00211-N
177. Li C, Kim K. Neuropeptide gene families in Caenorhabditis elegans, in Neuropeptide Systems as Targets for Parasite and Pest Control. Boston, MA: Springer (2010). p. 98–137. doi: 10.1007/978-1-4419-6902-6_6
178. Pandit AA, Ragionieri L, Marley R, Yeoh JGC, Inward DJG, Davies S-A, et al. Coordinated RNA-Seq and peptidomics identify neuropeptides and G-protein coupled receptors (GPCRs) in the large pine weevil Hylobius abietis, a major forestry pest. Insect Biochem Mol Biol. (2018) 101:94–107. doi: 10.1016/j.ibmb.2018.08.003
179. McVeigh P, Leech S, Mair GR, Marks NJ, Geary TG, Maule AG. Analysis of FMRFamide-like peptide (FLP) diversity in phylum Nematoda. Int J Parasitol. (2005) 35:1043–60. doi: 10.1016/j.ijpara.2005.05.010
180. Husson SJ, Landuyt B, Nys T, Baggerman G, Boonen K, Clynen E, et al. Comparative peptidomics of Caenorhabditis elegans versus C. briggsae by LC-MALDI-TOF MS. Peptides (2009) 30:449–57. doi: 10.1016/j.peptides.2008.07.021
181. Huybrechts J, Bonhomme J, Minoli S, Prunier-Leterme N, Dombrovsky A, Abdel-Latief M, et al. Neuropeptide and neurohormone precursors in the pea aphid, Acyrthosiphon pisum. Insect Mol Biol. (2010) 19:87–95. doi: 10.1111/j.1365-2583.2009.00951.x
182. Clynen E, Husson SJ, Schoofs L. Identification of New Members of the (Short) Neuropeptide F family in locusts and Caenorhabditis elegans. Ann N Y Acad Sci. (2009) 1163:60–74. doi: 10.1111/j.1749-6632.2008.03624.x
183. Husson SJ, Clynen E, Baggerman G, De Loof A, Schoofs L. Discovering neuropeptides in Caenorhabditis elegans by two dimensional liquid chromatography and mass spectrometry. Biochem Biophys Res Commun. (2005) 335:76–86. doi: 10.1016/j.bbrc.2005.07.044
184. Zhang G, Fang X, Guo X, Li L, Luo R, Xu F, et al. The oyster genome reveals stress adaptation and complexity of shell formation. Nature (2012) 490:49–54. doi: 10.1038/nature11413
185. Husson SJ, Reumer A, Temmerman L, De Haes W, Schoofs L, Mertens I, et al. Worm peptidomics. EuPA Open Proteomics (2014) 3:280–90. doi: 10.1016/j.euprot.2014.04.005
186. Zatylny-Gaudin C, Bernay B, Zanuttini B, Leprince J, Vaudry H, Henry J. Characterization of a novel LFRFamide neuropeptide in the cephalopod Sepia officinalis. Peptides (2010) 31:207–14. doi: 10.1016/j.peptides.2009.11.021
187. Marks NJ, Shaw C, Halton DW, Thompson DP, Geary TG, Li C, et al. Isolation and preliminary biological assessment of AADGAPLIRFamide and SVPGVLRFamide from Caenorhabditis elegans. Biochem Biophys Res Commun. (2001) 286:1170–6. doi: 10.1006/bbrc.2001.5524
188. Hoek RM, Li KW, Van Minnen J, Lodder JC, De Jong-Brink M, Smit AB, et al. LFRFamides: a novel family of parasitation-induced -RFamide neuropeptides that inhibit the activity of neuroendocrine cells in Lymnaea stagnalis. J Neurochem. (2005) 92:1073–80. doi: 10.1111/j.1471-4159.2004.02927.x
189. Peymen K, Watteyne J, Frooninckx L, Schoofs L, Beets I. The FMRFamide-like peptide family in nematodes. Front Endocrinol. (2014) 5:90. doi: 10.3389/fendo.2014.00090
190. Husson SJ, Janssen T, Baggerman G, Bogert B, Kahn-Kirby AH, Ashrafi K, et al. Impaired processing of FLP and NLP peptides in carboxypeptidase E (EGL-21)-deficient Caenorhabditis elegans as analyzed by mass spectrometry. J Neurochem. (2007) 102:246–60. doi: 10.1111/j.1471-4159.2007.04474.x
191. Lee YS, Nakahara K, Pham JW, Kim K, He Z, Sontheimer EJ, et al. Distinct roles for Drosophila Dicer-1 and Dicer-2 in the siRNA/miRNA silencing pathways. Cell (2004) 117:69–81. doi: 10.1016/S0092-8674(04)00261-2
192. Caers J, Boonen K, Van Den Abbeele J, Van Rompay L, Schoofs L, Van Hiel MB. Peptidomics of neuropeptidergic tissues of the tsetse Fly Glossina morsitans morsitans. J Am Soc Mass Spectrom. (2015) 26:2024–38. doi: 10.1007/s13361-015-1248-1
Keywords: neuropeptide F, neuropeptide Y, short neuropeptide F, feeding behavior, neuropeptide evolution, G protein coupled receptor, protostomes, neuromodulation
Citation: Fadda M, Hasakiogullari I, Temmerman L, Beets I, Zels S and Schoofs L (2019) Regulation of Feeding and Metabolism by Neuropeptide F and Short Neuropeptide F in Invertebrates. Front. Endocrinol. 10:64. doi: 10.3389/fendo.2019.00064
Received: 01 October 2018; Accepted: 23 January 2019;
Published: 19 February 2019.
Edited by:
Ian Orchard, University of Toronto Mississauga, CanadaReviewed by:
Meet Zandawala, Brown University, United StatesCopyright © 2019 Fadda, Hasakiogullari, Temmerman, Beets, Zels and Schoofs. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Liliane Schoofs, bGlsaWFuZS5zY2hvb2ZzQGt1bGV1dmVuLmJl
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.