- 1Beijing Key Laboratory of Reproductive Endocrinology and Assisted Reproductive Technology and Key Laboratory of Assisted Reproduction, Ministry of Education, Department of Obstetrics and Gynecology, Center for Reproductive Medicine, Peking University Third Hospital, Beijing, China
- 2Peking-Tsinghua Center for Life Sciences, Academy for Advanced Interdisciplinary Studies, Peking University, Beijing, China
Sirtuins comprise a family of nicotinamide adenine dinucleotide (NAD+)-dependent lysine deacylases that regulate the life span, aging, and metabolism. Seven sirtuin family members (SIRT1-7) have been identified in mammals, including humans. Despite the indispensable role of mitochondrial sirtuin 4 (SIRT4) in metabolic regulation, the primary enzymatic activity of SIRT4 remains enigmatic. SIRT4 possesses ADP-ribosyltransferase, lipoamidase and deacylase activities. Interestingly, the enzymatic activities and substrates of SIRT4 vary in different tissues and cells. SIRT4 inhibits insulin secretion in pancreatic β cells and regulates insulin sensitivity as a deacylase in the pancreas. SIRT4 represses fatty acid oxidation (FAO) in muscle and liver cells differently. SIRT4 has also been identified as a mitochondrial-localized tumor suppressor. A comprehensive understanding of the enzymology of SIRT4 in metabolism is essential for developing novel therapeutic agents for human metabolic diseases. This review will update the roles of SIRT4 in cellular and organismal metabolic homeostasis.
Introduction
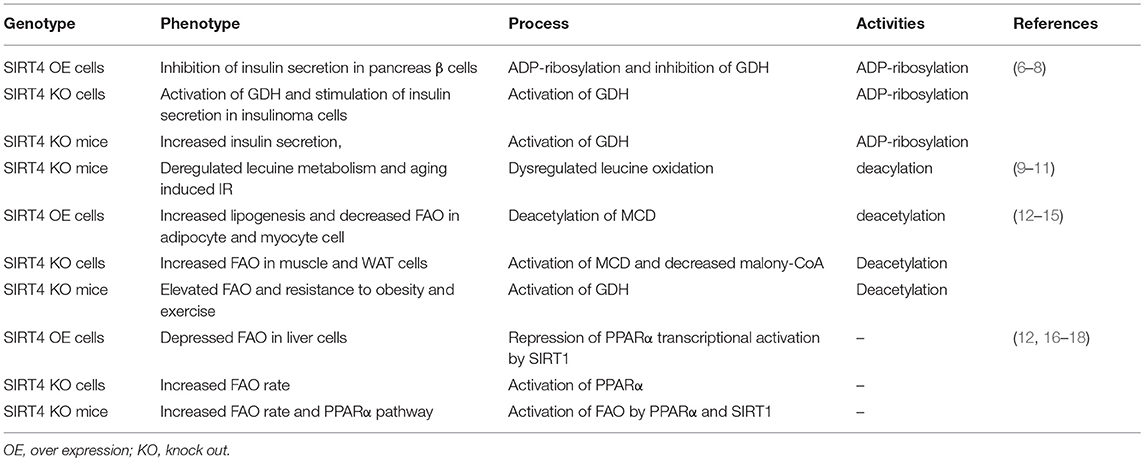
Sirtuins include β-NAD+-dependent deacylases and ADP-ribosyltransferases involved with metabolism and aging (1, 2). Sirtuins (SIRT1-7) can be divided into nuclear (SIRT1, SIRT6, and SIRT7), mitochondrial (SIRT3, SIRT4, and SIRT5) and cytosolic (SIRT2) forms, and some sirtuins are found in more than one compartment (Figure 1B). The mitochondrial sirtuins are involved in metabolic regulation and antioxidative defense. In contrast to other members of SIRT, the enzymatic activities of SIRT4 have remained unclear (3, 4). SIRT4 is critical for cellular metabolism and DNA damage responses in mitochondria. SIRT4 contains an N-terminal mitochondrial signal sequence that directs of its localization to mitochondria (4). SIRT4 has the requisite amino acids to participate in deacylase reactions (5), namely, a homologous sirtuin deacylase domain, a conserved catalytic histidine, and a Rossmann fold/NAD+-binding motif (aa 62–82, 143–146, 260–262, and 286–288) (Figure 1A). Previous genetic studies divide human sirtuins into four classes. SIRT4 belongs to the class II human sirtuins (Figure 1C). SIRT4 mRNA and protein are detected in human muscle, kidney, testis, and liver cells (6). This expression pattern is consistent with the functions of SIRT4. In this review, we summarize the roles of SIRT4 in cellular metabolic homeostasis, including the regulation of insulin secretion and fatty acid oxidation and the effect on tumor cells of different tissues (Table 1).
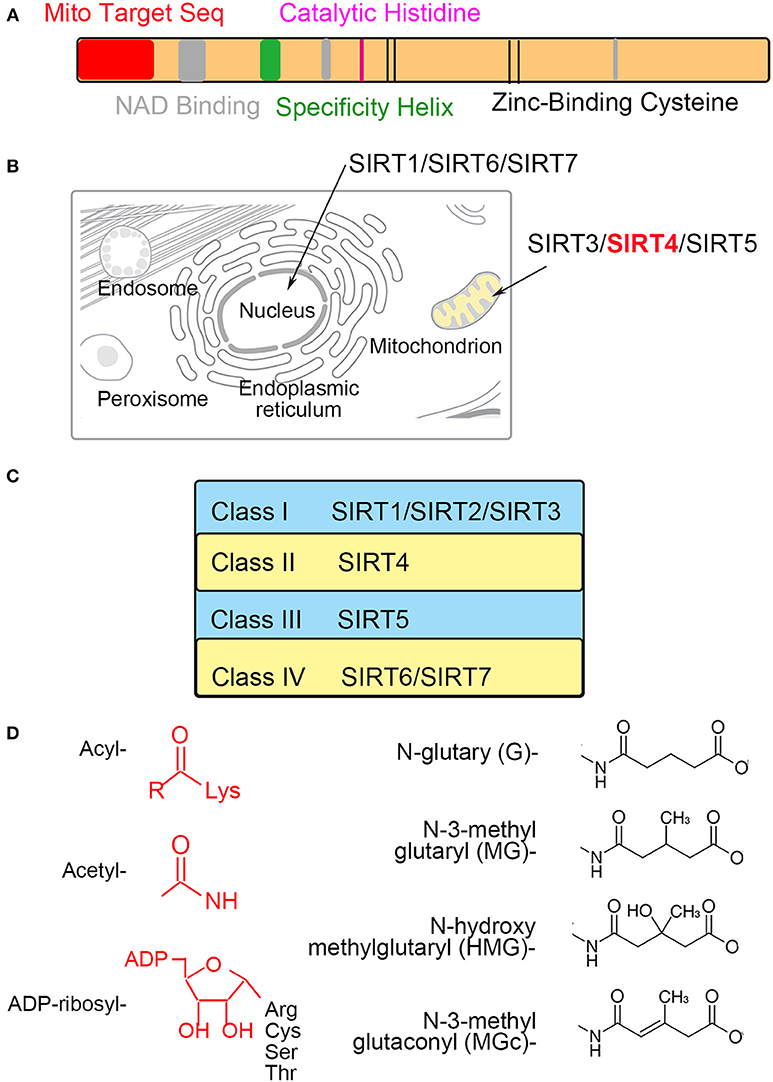
Figure 1. The characteristics of mitochondrial SIRT4 (A). Sequence analyses show the amino acid sequence of the mitochondrial localization, NAD+-binding, substrate specificity, catalytic and Zn2+-binding domains (B). SIRT4 is localized in the mitochondrion (C). SIRT4 is belong to a class II human sirtuins (D). Structure of groups catalyzed by SIRT4.
SIRT4 Inhibits Insulin Secretion in Pancreatic β Cells
SIRT4 was initially found to reduce glutamate dehydrogenase (GDH) activity, thereby inhibiting insulin secretion in pancreatic β cells (7, 9). GDH is known to facilitate glutamine metabolism and ATP production, thus inducing insulin secretion (19, 20). GDH is ADP-ribosylated and inhibited by SIRT4, subsequently repressing leucine-mediated insulin secretion (7, 21). The depletion of SIRT4 in insulinoma cells could activate GDH, thus increasing amino acid-stimulated insulin secretion. Pancreatic β cells derived from SIRT4 knockout mice and those from mice on calorie restriction (CR) as a dietary regimen show a similar effect of insulin secretion (8, 22). Phosphodiesterase (PDE) can be used as a probe for ADP-ribosylation because it cleaves the ADP-ribose and can relieve inhibition. GDH from SIRT4-deficient mice is insensitive to (PDE), and incubation with PDE increases the GDH activity of wild-type lysates to the KO samples. It indicates the absence of the enzymatic cleavage of ADP-ribose would decrease ADP-ribosylation of GDH (6, 8).
A recent study demonstrated that SIRT4 catalyzed the removal of novel lysine modifications: methylglutaryl (MG)-lysine, hydroxymethylglutaryl (HMG)-lysine, and 3-methylglutaconyl (MGc)-lysine (10, 11). SIRT4 participates in leucine oxidation by removing these modifications sequentially. These modifications were discovered and characterized through phylogenetic and structural analysis (Figure 1D). The α-helical region containing the catalytic pocket of SIRT4 was associated with an interaction with negatively charged acyl- modifications (23, 24). In a complementary study, a recombinant SIRT4 protein could remove glutaryl-, MG-, HMG-, and MGc-lysine modifications. Further, SIRT4 overexpression in cells resulted in decreased glutaryl-, MG-, and HMG-lysine expression levels. The methylcrotonyl-CoA carboxylase complex (MCCC) associated with leucine catabolism and interacting with SIRT4 was marked with these new modifications in vivo (10). The absence of SIRT4 increased and destabilized MCCC acylation, leading to decreased leucine flux. The level of a key allosteric GDH activator, leucine, was changed due to the repression of GDH activity and glutamine metabolism by SIRT4 (25, 26). Knowledge about these new modifications provided new insights into SIRT4 function in further metabolism studies (Figure 2).
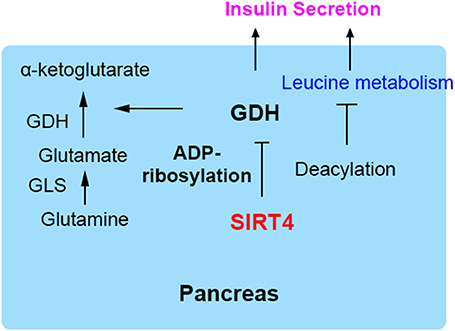
Figure 2. SIRT4 regulates insulin secretion in pancreatic β cells. GDH is ADP-ribosylated and inhibited by SIRT4, repressing insulin secretion.
SIRT4 Inhibits Fatty Acid Oxidation in Muscle and Fat Cells
The repressive effect of SIRT4 against FAO in muscle cells is regulated by the deacetylation and inhibition of the activity of mitochondrial malonyl-CoA decarboxylase (MCD), leading to an increase in malonyl-CoA (12, 13, 27). Malonyl CoA is a key metabolite that inhibits fat catabolism and promotes fat synthesis (28). There are two enzymes that regulate cellular malonyl-CoA levels. MCD catalyzes the conversion of malonyl-CoA to acetyl-CoA, while acetyl-CoA carboxylase (ACC) converts acetyl-CoA back to malonyl-CoA via a reaction regulated by the phosphorylation of AMPK (14, 29). Cytosolic fatty acids transported into the mitochondrial matrix cross the outer and inner mitochondrial membranes for β-oxidation, and this key step is catalyzed by carnitine palmitoyltransferase (CPT1), which is inhibited by accumulated malonyl-CoA (30, 31). During the nutritional rich state (Fed), when the metabolic intermediates funnel into fat synthesis and energy storage, the steady state is increased malonyl-CoA. This suppresses entry of fatty acids into mitochondria and weakens FAO sequentially (15). Conversely, in the fasted state, the malonyl-CoA is decreased and the FAO is evaluated for energy production (Figure 3). Therefore, SIRT4 regulates malonyl-CoA levels of muscle and WAT of mice in the fed and fasted state. In addition, as a unit of fatty acid synthesis, the accumulated malonyl-CoA promotes and provides a carbon skeleton for fatty acid biosynthesis in white adipose tissue (WAT) (32, 33). A previous study reported that SIRT4 overexpression increased lipogenesis and decreased fatty acid oxidation, while SIRT4 knockdown showed the opposite effect on lipid synthesis and catabolism in mouse adipocyte and myocyte cell lines (3, 34). Furthermore, the regulation pattern was confirmed in SIRT4 knockout mice, which showed elevated FAO associated with a resistance to diet-induced obesity and an increased exercise tolerance (24, 35, 36). Additionally, the interaction of MCD and SIRT4 in the mitochondrial matrix was demonstrated by coimmunoprecipitation. SIRT4 lacked detectable histone deacetylase activity, but it deacetylated MCD in a substrate-specific manner (37, 38). Further, mitochondrial located SIRT3 was confirmed and excluded the possibility of deacetylation activity, because interaction between SIRT3 and SIRT4. SIRT3 showed no detectable interaction with MCD indicating the specific function of SIRT4. SIRT4 coordinated lipid homeostasis by promoting lipid anabolism and repressing lipid catabolism (39, 40).
Figure 3. SIRT4 mediates fatty acid oxidation in muscle and adipose cells. In fed state, SIRT4 decreases the activity of the mitochondrial malonyl-CoA decarboxylase (MCD), which can increase malonyl-CoA levels, thus inhibiting FAO. In fasted state, the level malonyl-CoA of is decreased conversely.
SIRT4 Represses Hepatic FAO in Liver Cells
Liver plays a role in energy homeostasis by balancing lipid metabolism and energetic demands in organisms (12, 16). Under nutrient-rich conditions, hepatic lipogenesis, and lipoprotein secretion are activated. In contrast, during fasting, hepatic FAO is stimulated to provide the organism with ketone bodies as cellular energy fuel for the brain. One of the key mediators of the hepatic response to fasting is nuclear receptor peroxisome proliferation-activated receptor α (PPARα). SIRT4 negatively mediated fatty acid oxidation in liver cells by suppressing the transcriptional activity of PPARα (17, 41). More specifically, the interaction of SIRT1 and PPARα was disrupted by SIRT4, thus attenuating the activation of PPARα transcriptional activity via SIRT1 to inhibit FAO (18). In normal liver cells, by interacting with PPARα, SIRT1 is recruited to the PPARα response element (PPRE), which catalyzes the Nε-acetyl-lysine deacetylation of the transcriptional coactivator peroxisome proliferator-activated receptor gamma coactivator 1-alpha (PGC-1α) to promote FAO (42). Additionally, SIRT4 competes with other sirtuins including SIRT1 for β-NAD+, leading to decreased SIRT1 activity and a reduced effect of SIRT1 on both the transcriptional activity of PPARα and FAO (43, 44). During fasting, FAO is promoted as a source of cellular energy. The decreased SIRT4 levels in liver cells demonstrate the inhibitory effect of SIRT4 on liver FAO (45).
The roles of SIRT4 in liver cells were supported by knockout and overexpression experiments in cellular and animal models. In SIRT4 knockdown and knockout primary hepatocytes, the expression of mitochondrial and fatty acid metabolism enzyme genes was increased significantly (17). Consistent with increased FAO gene expression, SIRT4 knockdown hepatocytes exhibited higher rates of FAO than wild-type cells. The increase in FAO gene expression in SIRT4 knockout mice was consistent with the results in primary hepatocytes (12). Additionally, SIRT1 mRNA and protein levels were also evaluated both in vitro and in vivo by the knockdown of SIRT4. In SIRT4/SIRT1 double-knockdown hepatocytes, there was not an increase in FAO compared with that induced by SIRT4 knockdown only (17). These results suggested that SIRT4-modulated FAO was dependent on SIRT1 in primary hepatocytes (22, 46). Moreover, SIRT4 overexpression repressed SIRT1 activation of PPARα and consequently inhibited hepatic FAO (Figure 4).
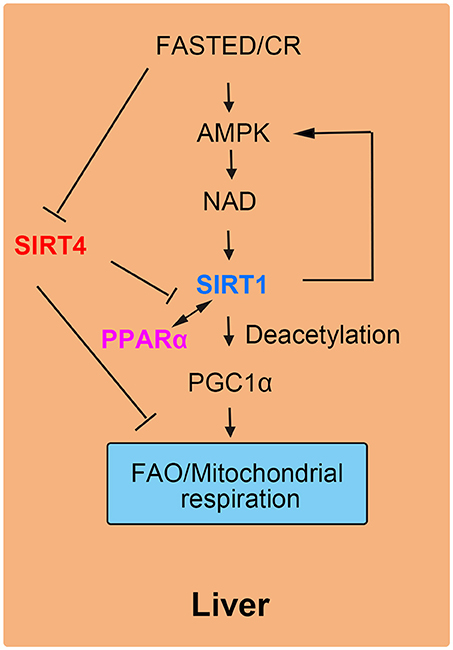
Figure 4. SIRT4 mediates fatty acid oxidation in liver cells. The interaction of SIRT1 and PPARα is blocked by SIRT4, leading to a decrease in FAO.
SIRT4 Participates in Cellular ATP Homeostasis
In addition to regulating FAO, SIRT4 was shown to contribute to cellular ATP homeostasis and mitochondrial biogenesis. Deletion of SIRT4 decreased ATP levels, and overexpression of SIRT4 increased ATP levels (3, 47). ATP/ADP translocase 2 (ANT2a, transmembrane protein spanning the inner mitochondrial membrane) helped SIRT4 mediate cellular ATP homeostasis. Mechanistically, ANT Nε-acylation facilitated mitochondrial respiration by enhancing mitochondrial uncoupling and decreasing ATP production (48). Therefore, SIRT4 catalyzed the Nε-acyl-lysine deacylation of ANT2 to reduce mitochondrial uncoupling, leading to enhanced ATP production. In the mitochondrial signaling pathway, the depletion of SIRT4 activated AMPK to decrease ATP levels. Activated AMPK phosphorylates and inhibits cytosolic ACC, leading to a reduction in MCA and the promotion of FAO as mentioned above.
SIRT4 Acts as a TUMOR Suppressor
SIRT4 is necessary for the regulation of FAO in normal cells but is also identified as a mitochondrial-localized tumor suppressor (49) (Table 2). SIRT4 has been recognized to possess tumor-suppressive effects due to the crucial regulatory role of mitochondrial metabolism in tumourigenesis (34, 50).
SIRT4 plays an important role in the response to DNA damage by regulating the DNA damage-induced inhibition of glutamine catabolism (4, 57, 59). DNA damage increases the flux through the pentose phosphate pathway and decreases glutamine uptake and the levels of TCA cycle intermediates. Mitochondrial glutaminase can catabolize glutamine to form glutamate via mitochondrial GDH and aspartate aminotransferase activity (60). Previous research showed that SIRT4 ADP-ribosylated and inhibited GDH and was thus involved in DNA damage-induced inhibition of glutamine metabolism and anaplerosis (33, 39).
SIRT4 possesses a tumor suppressive effect because of its inhibitory effect on glutamine catabolism and its antiproliferative effect on cells with DNA damage (61, 62). Indeed, SIRT4 expression is upregulated by DNA damage and is downregulated in many types of human tumor tissues and cells (44). SIRT4 depletion leads to both elevated glutamine-dependent proliferation and stress-induced genomic instability, resulting in tumourigenic phenotypes. SIRT4 inhibits glutamine catabolism, especially replenishment into the citric acid cycle, creating a cellular state promoting DNA damage repair (63). In the absence of SIRT4, DNA damage results in delayed DNA repair and increased chromosomal aneuploidies, suggesting that SIRT4 could protect cells from spontaneous damage. Furthermore, SIRT4 null mice spontaneously develop lung tumors (19, 64).
SIRT4 can specifically repress the growth of B cells induced by the transcriptional factor c-Myc through inhibiting glutamine metabolism induced by the abnormal activation of c-Myc in c-Myc-dependent cancers (52, 65). In human Burkitt lymphoma cells, the overexpression of SIRT4 repressed glutamine metabolism and glutamine-dependent cell proliferation, as observed in cells treated with glycolysis inhibitors, promoting cell death (66). Consistent with these findings, the depletion of SIRT4 resulted in increased glutamine-absorbing and GDH enzymic activity in a mouse model of Burkitt lymphoma induced by c-Myc dysregulation. Additionally, SIRT4 suppressed Burkitt lymphoma driven by c-Myc, independent of c-Myc activity. Furthermore, SIRT4 was found to be overexpressed in colorectal cancer cell lines and to increase E-cadherin expression, which resulted in the suppression of cell proliferation and invasion (59). During the development of colorectal cancer invasion, SIRT4 expression was decreased. The suppressive role of SIRT4 is also mediated by inhibiting glutamine metabolism in colorectal cancer (54). Moreover, SIRT4 could enhance the sensitivity of colorectal cancer cells to the chemical drug 5-fluorouracil by inhibiting the cell cycle, thus showing the antiproliferative effect of SIRT4 overexpression (55). Recent reports have demonstrated that mammalian target of rapamycin complex1 (mTORC1) is correlated with increased nutrient uptake and metabolism. mTORC1 promotes glutamine supplement by activating GDH required transcriptional repression of SIRT4. Specifically mTORC1 suppresses SIRT4 by destabilizing of cAMP-responsive element binding 2 (CREB2) (67). Overexpression of SIRT4 decreases cell growth, transformation and development of tumor. Furthermore, leucine is an important positive regulator of mTORC1, which activity may be regulated by SIRT4 by reducing intracellular leucine levels (Figure 5). Therefore, as a glutamine steward, SIRT4 acts as a necessary component of the DNA damage response pathway that manages the metabolic blockade of glutamine metabolism, the cell cycle, and tumor suppression.
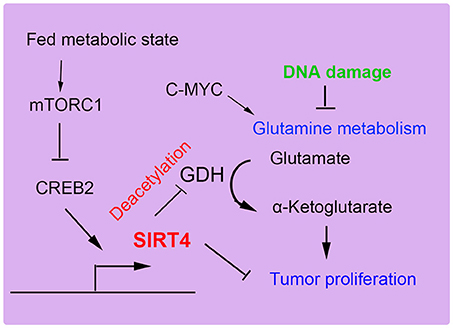
Figure 5. SIRT4 plays a role in tumor suppression. SIRT4 suppresses tumor proliferation by inhibiting glutamine metabolism and DNA damage repair.
Conclusion
In this review, we summarize different roles of SIRT4 in terms of enzymatic activities and functions in specific tissues and cells. SIRT4 possesses ADP-ribosyltransferase, lipoamidase and deacylase activities (46, 68, 69). Importantly, the enzymatic activities, and substrates of SIRT4 vary in different tissues and cells (Figure 6). SIRT4 inhibits insulin secretion as an ADP-ribosyltransferase and regulates insulin sensitivity as a deacylase in the pancreas. SIRT4 represses fatty acid oxidation (FAO) in the muscles and livers differently. SIRT4 has also been identified as a mitochondrial-localized tumor suppressor. SIRT4 is a less well-understood mammalian sirtuin, its cellular functions require further discovery. Knowledge of roles SIRT4 in cellular metabolism is helpful to provide new insights in the potential development of therapeutic agents for human diseases by targeting enzymatic activities (36).
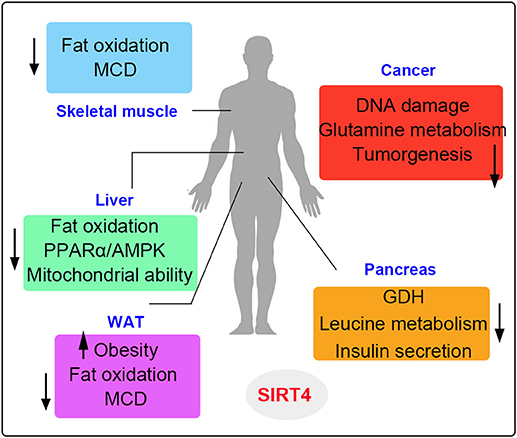
Figure 6. The metabolic roles of SIRT4 in various organs. SIRT4 is mainly involved in the metabolism of muscle, liver, fat, pancreatic, and cancer cells. The up arrows indicate the increased regulation pathways and the down arrows indicate the decreased regulation pathways induced by SIRT4.
Author Contributions
ZM and JG draft the original manuscript. YY proofed and edited the finalized manuscript.
Funding
This work was supported in part by the National Key R&D Program of China (2016YFC1000601) and the National Natural Science Funds for the general program (81571400, 81771580).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
1. Finkel T, Deng CX, Mostoslavsky R. Recent progress in the biology and physiology of sirtuins. Nature (2009) 460:587–91. doi: 10.1038/nature08197
2. Tan Y, Xu Z, Tao J, Ni J, Zhao W, Lu J, et al. A SIRT4-like auto ADP-ribosyltransferase is essential for the environmental growth of Mycobacterium smegmatis. Acta Biochim Biophys Sin (Shanghai). (2016) 48:145–52. doi: 10.1093/abbs/gmv121
3. Li Y, Zhou Y, Wang F, Chen X, Wang C, Wang J, et al. SIRT4 is the last puzzle of mitochondrial sirtuins. Bioorg Med Chem. (2018) 26:3861–5. doi: 10.1016/j.bmc.2018.07.031
4. van de Ven RAH, Santos D, Haigis MC. Mitochondrial sirtuins and molecular mechanisms of aging. Trends Mol Med. (2017) 23:320–331. doi: 10.1016/j.molmed.2017.02.005
5. Min J, Landry J, Sternglanz R, Xu R-M. Crystal structure of a SIR2 homolog–NAD complex. Cell (2001) 105:269–79. doi: 10.1016/S0092-8674(01)00317-8
6. Ahuja N, Schwer B, Carobbio S, Waltregny D, North BJ, Castronovo V, et al. Regulation of insulin secretion by SIRT4, a mitochondrial ADP-ribosyltransferase. J Biol Chem. (2007) 282:33583–92. doi: 10.1074/jbc.M705488200
7. Argmann C, Auwerx J. Insulin secretion: SIRT4 gets in on the act. Cell (2006) 126:837–9. doi: 10.1016/j.cell.2006.08.031
8. Haigis MC, Mostoslavsky R, Haigis KM, Fahie K, Christodoulou DC, Murphy AJ, et al. SIRT4 inhibits glutamate dehydrogenase and opposes the effects of calorie restriction in pancreatic beta cells. Cell (2006) 126:941–54. doi: 10.1016/j.cell.2006.06.057
9. Zaganjor E, Vyas S, Haigis MC. SIRT4 is a regulator of insulin secretion. Cell Chem Biol. (2017) 24:656–8. doi: 10.1016/j.chembiol.2017.06.002
10. Anderson KA, Huynh FK, Fisher-Wellman K, Stuart JD, Peterson BS, Douros JD, et al. SIRT4 is a lysine deacylase that controls leucine metabolism and insulin secretion. Cell Metab. (2017) 25:838–55 e815. doi: 10.1016/j.cmet.2017.03.003
11. Kumar S, Lombard DB. For certain, SIRT4 activities! Trends Biochem Sci. (2017) 42:499–501. doi: 10.1016/j.tibs.2017.05.008
12. Nasrin N, Wu X, Fortier E, Feng Y, Bare OC, Chen S, et al. SIRT4 regulates fatty acid oxidation and mitochondrial gene expression in liver and muscle cells. J Biol Chem. (2010) 285:31995–2002. doi: 10.1074/jbc.M110.124164
13. Acs Z, Bori Z, Takeda M, Osvath P, Berkes I, Taylor AW, et al. High altitude exposure alters gene expression levels of DNA repair enzymes, and modulates fatty acid metabolism by SIRT4 induction in human skeletal muscle. Respir Physiol Neurobiol. (2014) 196:33–7. doi: 10.1016/j.resp.2014.02.006
14. Koves TR, Ussher JR, Noland RC, Slentz D, Mosedale M, Ilkayeva O, et al. Mitochondrial overload and incomplete fatty acid oxidation contribute to skeletal muscle insulin resistance. Cell Metab. (2008) 7:45–56. doi: 10.1016/j.cmet.2007.10.013
15. Laurent G, German NJ, Saha AK, de Boer VCJ, Davies M, Koves TR, et al. SIRT4 coordinates the balance between lipid synthesis and catabolism by repressing malonyl CoA decarboxylase. Molecular Cell (2013) 50:686–98. doi: 10.1016/j.molcel.2013.05.012
16. Schirmer H, Pereira TC, Rico EP, Rosemberg DB, Bonan CD, Bogo MR, et al. Modulatory effect of resveratrol on SIRT1, SIRT3, SIRT4, PGC1alpha, and NAMPT gene expression profiles in wild-type adult zebrafish liver. Mol Biol Rep. (2012) 39:3281–9. doi: 10.1007/s11033-011-1096-4
17. Laurent G, de Boer VC, Finley LW, Sweeney M, Lu H, Schug TT, et al. SIRT4 represses peroxisome proliferator-activated receptor alpha activity to suppress hepatic fat oxidation. Mol Cell Biol. (2013) 33:4552–61. doi: 10.1128/MCB.00087-13
18. Lu M, Wan M, Leavens KF, Chu Q, Monks BR, Fernandez S, et al. Insulin regulates liver metabolism in vivo in the absence of hepatic Akt and Foxo1. Nat Med. (2012) 18:388–95. doi: 10.1038/nm.2686
19. Kim EA, Yang SJ, Choi SY, Lee WJ, Cho SW. Inhibition of glutamate dehydrogenase and insulin secretion by KHG26377 does not involve ADP-ribosylation by SIRT4 or deacetylation by SIRT3. BMB Rep. (2012) 45:458–63. doi: 10.5483/BMBRep.2012.45.8.040
20. Matsumoto M, Han S, Kitamura T, Accili D. Dual role of transcription factor FoxO1 in controlling hepatic insulin sensitivity and lipid metabolism. J Clin Invest. (2006) 116:2464–72. doi: 10.1172/JCI27047
21. Chen YR, Fang SR, Fu YC, Zhou XH, Xu MY, Xu WC. Calorie restriction on insulin resistance and expression of SIRT1 and SIRT4 in rats. Biochem Cell Biol. (2010) 88:715–22. doi: 10.1139/O10-010
22. Carrico C, Meyer JG, He W, Gibson BW, Verdin E. The mitochondrial acylome emerges: proteomics, regulation by sirtuins, and metabolic and disease implications. Cell Metab. (2018) 27:497–512. doi: 10.1016/j.cmet.2018.01.016
23. Pannek M, Simic Z, Fuszard M, Meleshin M, Rotili D, Mai A, et al. Crystal structures of the mitochondrial deacylase Sirtuin 4 reveal isoform-specific acyl recognition and regulation features. Nat Commun. (2017) 8:1513. doi: 10.1038/s41467-017-01701-2
24. Choubey SK, Prabhu D, Nachiappan M, Biswal J, Jeyakanthan J. Molecular modeling, dynamics studies and density functional theory approaches to identify potential inhibitors of SIRT4 protein from Homo sapiens : a novel target for the treatment of type 2 diabetes. J Biomol Struct Dyn. (2017) 35:3316–29. doi: 10.1080/07391102.2016.1254117
25. Newgard CB, An J, Bain JR, Muehlbauer MJ, Stevens RD, Lien LF, et al. A Branched-Chain amino acid-related metabolic signature that differentiates obese and lean humans and contributes to insulin resistance. Cell Metab. (2009) 9:565–6. doi: 10.1016/j.cmet.2009.05.001
26. Mathias RA, Greco TM, Cristea IM. Identification of sirtuin4 (SIRT4) protein interactions: uncovering candidate acyl-modified mitochondrial substrates and enzymatic regulators. Methods Mol Biol. (2016) 1436:213–39. doi: 10.1007/978-1-4939-3667-0_15
27. Zeng G, Liu H, Wang H. Amelioration of myocardial ischemia-reperfusion injury by SIRT4 involves mitochondrial protection and reduced apoptosis. Biochem Biophys Res Commun. (2018) 502:15–21. doi: 10.1016/j.bbrc.2018.05.113
28. Saha AK, Ruderman NB. Malonyl-CoA and AMP-activated protein kinase: an expanding partnership. Mol Cell Biochem. (2003) 253:65–70. doi: 10.1023/A:1026053302036
29. Liu B, Che W, Xue J, Zheng C, Tang K, Zhang J, et al. SIRT4 prevents hypoxia-induced apoptosis in H9c2 cardiomyoblast cells. Cell Physiol Biochem. (2013) 32:655–62. doi: 10.1159/000354469
30. Bugger H, Witt CN, Bode C. Mitochondrial sirtuins in the heart. Heart Fail Rev. (2016) 21:519–28. doi: 10.1007/s10741-016-9570-7
31. Luo YX, Tang X, An XZ, Xie XM, Chen XF, Zhao X, et al. SIRT4 accelerates Ang II-induced pathological cardiac hypertrophy by inhibiting manganese superoxide dismutase activity. Eur Heart J. (2017) 38:1389–98. doi: 10.1093/eurheartj/ehw138
32. Boulange CL, Claus SP, Chou CJ, Collino S, Montoliu I, Kochhar S, et al. Early metabolic adaptation in C57BL/6 mice resistant to high fat diet induced weight gain involves an activation of mitochondrial oxidative pathways. J Proteome Res. (2013) 12:1956–68. doi: 10.1021/pr400051s
33. Jokinen R, Pirnes-Karhu S, Pietilainen KH, Pirinen E. Adipose tissue NAD(+)-homeostasis, sirtuins and poly(ADP-ribose) polymerases -important players in mitochondrial metabolism and metabolic health. Redox Biol. (2017) 12:246–63. doi: 10.1016/j.redox.2017.02.011
34. Parik S, Tewary S, Ayyub C, Kolthur-Seetharam U. Loss of mitochondrial SIRT4 shortens lifespan and leads to a decline in physical activity. J Biosci. (2018) 43:243–7. doi: 10.1007/s12038-018-9754-5
35. Elkhwanky MS, Hakkola J. Extranuclear sirtuins and metabolic stress Antioxid Redox Signal. (2017) 28:662–76. doi: 10.1089/ars.2017.7270
36. George J, Ahmad N. Mitochondrial sirtuins in cancer: emerging roles and therapeutic potential Cancer Res. (2016) 76:2500–6. doi: 10.1158/0008-5472.CAN-15-2733
37. Wood JG, Schwer B, Wickremesinghe PC, Hartnett DA, Burhenn L, Garcia M, et al. Sirt4 is a mitochondrial regulator of metabolism and lifespan in Drosophila melanogaster. Proc Natl Acad Sci USA. (2018) 115:1564–9. doi: 10.1073/pnas.1720673115
38. Saltiel AR, Kahn CR. Insulin signalling and the regulation of glucose and lipid metabolism. Nature (2001) 414:799–806. doi: 10.1038/414799a
39. Hershberger KA, Martin AS, Hirschey MD. Role of NAD(+) and mitochondrial sirtuins in cardiac and renal diseases. Nat Rev Nephrol. (2017) 13:213–25. doi: 10.1038/nrneph.2017.5
40. Dolinsky VW. The role of sirtuins in mitochondrial function and doxorubicin-induced cardiac dysfunction. Biol Chem. (2017) 398:955–74. doi: 10.1515/hsz-2016-0316
41. Langlet F, Haeusler RA, Linden D, Ericson E, Norris T, Johansson A, et al. Selective inhibition of FOXO1 activator/repressor balance modulates hepatic glucose handling. Cell (2017) 171:824–35 e818. doi: 10.1016/j.cell.2017.09.045
42. Osborne B, Bentley NL, Montgomery MK, Turner N. The role of mitochondrial sirtuins in health and disease. Free Radic Biol Med. (2016) 100:164–74. doi: 10.1016/j.freeradbiomed.2016.04.197
43. Kulkarni SS, Canto C. Mitochondrial post-translational modifications and metabolic control: sirtuins and beyond. Curr Diabetes Rev. (2017) 13:338–51. doi: 10.2174/1573399812666160217122413
44. Shukla S, Sharma A, Pandey VK, Raisuddin S, Kakkar P. Concurrent acetylation of FoxO1/3a and p53 due to sirtuins inhibition elicit Bim/PUMA mediated mitochondrial dysfunction and apoptosis in berberine-treated HepG2 cells. Toxicol Appl Pharmacol. (2016) 291:70–83. doi: 10.1016/j.taap.2015.12.006
45. Banks AS, Kim-Muller JY, Mastracci TL, Kofler NM, Qiang L, Haeusler RA, et al. Dissociation of the glucose and lipid regulatory functions of FoxO1 by targeted knockin of acetylation-defective alleles in mice. Cell Metab. (2011) 14:587–97. doi: 10.1016/j.cmet.2011.09.012
46. Gertz M, Steegborn C. Using mitochondrial sirtuins as drug targets: disease implications and available compounds. Cell Mol Life Sci. (2016) 73:2871–96. doi: 10.1007/s00018-016-2180-7
47. Ho L, Titus AS, Banerjee KK, George S, Lin W, Deota S, et al. SIRT4 regulates ATP homeostasis and mediates a retrograde signaling via AMPK. Aging (Albany NY) (2013) 5:835–49. doi: 10.18632/aging.100616
48. Zeng J, Jiang M, Wu X, Diao F, Qiu D, Hou X, et al. SIRT4 is essential for metabolic control and meiotic structure during mouse oocyte maturation. Aging Cell (2018) 29:e12789. doi: 10.1111/acel.12789
49. Sun H, Huang D, Liu G, Jian F, Zhu J, Zhang L. SIRT4 acts as a tumor suppressor in gastric cancer by inhibiting cell proliferation, migration, and invasion. Onco Targets Ther. (2018) 11:3959–68. doi: 10.2147/OTT.S156143
50. Huang G, Zhu G. Sirtuin-4 (SIRT4), a therapeutic target with oncogenic and tumor-suppressive activity in cancer. Onco Targets Ther. (2018) 11:3395–400. doi: 10.2147/OTT.S157724
51. Bradbury CA, Khanim FL, Hayden R, Bunce CM, White DA, Drayson MT, et al. Histone deacetylases in acute myeloid leukaemia show a distinctive pattern of expression that changes selectively in response to deacetylase inhibitors. Leukemia (2005) 19:1751–9. doi: 10.1038/sj.leu.2403910
52. Jeong SM, Lee A, Lee J, Haigis MC. SIRT4 protein suppresses tumor formation in genetic models of Myc-induced B cell lymphoma. J Biol Chem. (2014) 289:4135–44. doi: 10.1074/jbc.M113.525949
53. Roth M, Chen WY. Sorting out functions of sirtuins in cancer. Oncogene (2014) 33:1609–20. doi: 10.1038/onc.2013.120
54. Miyo M, Yamamoto H, Konno M, Colvin H, Nishida N, Koseki J, et al. Tumour-suppressive function of SIRT4 in human colorectal cancer. Br J Cancer (2015) 113, 492–9. doi: 10.1038/bjc.2015.226
55. Zhu Y, Wang G, Li X, Wang T, Weng M, Zhang Y. Knockout of SIRT4 decreases chemosensitivity to 5-FU in colorectal cancer cells. Oncol Lett. (2018) 16:1675–81. doi: 10.3892/ol.2018.8850
56. Huang G, Cui F, Yu F, Lu H, Zhang M, Tang H, et al. Sirtuin-4 (SIRT4) is downregulated and associated with some clinicopathological features in gastric adenocarcinoma. Biomed Pharmacother. (2015) 72:135–9. doi: 10.1016/j.biopha.2015.04.013
57. Fu L, Dong Q, He J, Wang X, Xing J, Wang E, et al. SIRT4 inhibits malignancy progression of NSCLCs, through mitochondrial dynamics mediated by the ERK-Drp1 pathway. Oncogene (2017) 36:2724–36. doi: 10.1038/onc.2016.425
58. Garber ME, Troyanskaya OG, Schluens K, Petersen S, Thaesler Z, Pacyna-Gengelbach M, et al. Diversity of gene expression in adenocarcinoma of the lung. Proc Natl Acad Sci USA. (2001) 98:13784–9. doi: 10.1073/pnas.241500798
59. Wang L, Zhou H, Wang Y, Cui G, Di LJ. CtBP maintains cancer cell growth and metabolic homeostasis via regulating SIRT4. Cell Death Dis. (2015) 6:e1620. doi: 10.1038/cddis.2014.587
60. Komlos D, Mann KD, Zhuo Y, Ricupero CL, Hart RP, Liu AY, et al. Glutamate dehydrogenase 1 and SIRT4 regulate glial development. Glia (2013) 61:394–408. doi: 10.1002/glia.22442
61. Jeong SM, Xiao C, Finley LW, Lahusen T, Souza AL, Pierce K, et al. SIRT4 has tumor-suppressive activity and regulates the cellular metabolic response to DNA damage by inhibiting mitochondrial glutamine metabolism. Cancer Cell (2013) 23:450–63. doi: 10.1016/j.ccr.2013.02.024
62. Potthast AB, Heuer T, Warneke SJ, Das AM. Alterations of sirtuins in mitochondrial cytochrome c-oxidase deficiency. PLoS ONE (2017) 12:e0186517. doi: 10.1371/journal.pone.0186517
63. Fernandez-Marcos PJ, Serrano M. Sirt4: the glutamine gatekeeper. Cancer Cell (2013) 23:427–8. doi: 10.1016/j.ccr.2013.04.003
64. Germain D. Sirtuins and the estrogen receptor as regulators of the mammalian mitochondrial UPR in cancer and aging. Adv Cancer Res. (2016) 130:211–56. doi: 10.1016/bs.acr.2016.01.004
65. Gao P, Tchernyshyov I, Chang TC, Lee YS, Kita K, Ochi T, et al. c-Myc suppression of miR-23a/b enhances mitochondrial glutaminase expression and glutamine metabolism. Nature (2009) 458:762–5. doi: 10.1038/nature07823
66. de Moura MB, Uppala R, Zhang YX, Van Houten B, Goetzman ES. Overexpression of mitochondrial sirtuins alters glycolysis and mitochondrial function in HEK293 cells. PLoS ONE (2014) 9:e106028. doi: 10.1371/journal.pone.0106028
67. Csibi A, Fendt SM, Li C, Poulogiannis G, Choo AY, Chapski DJ, et al. The mTORC1 pathway stimulates glutamine metabolism and cell proliferation by repressing SIRT4. Cell (2013) 153:840–54. doi: 10.1016/j.cell.2013.04.023
68. Celichowski P, Jopek K, Szyszka M, Tyczewska M, Malendowicz LK, Rucinski M. Mitochondrial sirtuins in the rat adrenal gland: location within the glands of males and females, hormonal and developmental regulation of gene expressions. Folia Histochem Cytobiol. (2017) 55:190–202. doi: 10.5603/FHC.a2017.0020
Keywords: SIRT4, mitochondrial, enzymatic activities, insulin secretion, fatty acid metabolism, tumor suppressor
Citation: Min Z, Gao J and Yu Y (2019) The Roles of Mitochondrial SIRT4 in Cellular Metabolism. Front. Endocrinol. 9:783. doi: 10.3389/fendo.2018.00783
Received: 26 August 2018; Accepted: 12 December 2018;
Published: 07 January 2019.
Edited by:
Yang Yang, Northwest University, ChinaReviewed by:
Brenna Osborne, University of Copenhagen, DenmarkShingo Kajimura, University of California, San Francisco, United States
Copyright © 2019 Min, Gao and Yu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yang Yu, eXV5YW5nNTAxMkBob3RtYWlsLmNvbQ==
†These authors have contributed equally to this work
 Zheying Min
Zheying Min Jiangman Gao1†
Jiangman Gao1† Yang Yu
Yang Yu