
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Endocrinol. , 06 December 2018
Sec. Cellular Endocrinology
Volume 9 - 2018 | https://doi.org/10.3389/fendo.2018.00748
This article is part of the Research Topic SIRT family in Endocrinology View all 19 articles
The mammalian Sirtuins (SIRT1-7) are an evolutionarily conserved family of NAD+-dependent deacylase and mono-ADP-ribosyltransferase. Sirtuins display distinct subcellular localizations and functions and are involved in cell survival, senescence, metabolism and genome stability. Among the mammalian Sirtuins, SIRT1 and SIRT6 have been thoroughly investigated and have prominent metabolic regulatory roles. Moreover, SIRT1 and SIRT6 have been implicated in obesity, insulin resistance, type 2 diabetes mellitus (T2DM), fatty liver disease and cardiovascular diseases. However, the roles of other Sirtuins are not fully understood. Recent studies have shown that these Sirtuins also play important roles in inflammation, mitochondrial dysfunction, and energy metabolism. Insulin resistance is the critical pathological trait of obesity and metabolic syndrome as well as the core defect in T2DM. Accumulating clinical and experimental animal evidence suggests the potential roles of the remaining Sirtuins in the regulation of insulin resistance through diverse biological mechanisms. In this review, we summarize recent advances in the understanding of the functions of Sirtuins in various insulin resistance-associated physiological processes, including inflammation, mitochondrial dysfunction, the insulin signaling pathway, glucose, and lipid metabolism. In addition, we highlight the important gaps that must be addressed in this field.
The increasing prevalence of obesity and associated metabolic syndrome (including type 2 diabetes mellitus [T2DM], nonalcoholic fatty liver disease [NAFLD], atherosclerosis and atherosclerotic heart disease) is an increasingly severe challenge in public health (1). Insulin resistance is the critical, universal pathological feature of these diseases, especially T2DM. Insulin is a major hormone secreted by pancreatic β cells after nutrient stimulation and plays a critical role in reducing blood glucose concentration by facilitating glucose uptake by skeletal muscle and adipose tissue and inhibiting endogenous glucose production in the liver (2–4). Insulin resistance occurs when cells are incapable of efficiently responding to a normal dose of insulin (2–4). The development of insulin resistance is a multistep, complex process influenced by genetics and environments (3). Although the precise pathogenesis of insulin resistance remains incompletely understood, several mechanisms are proposed to be involved, including defects in the insulin signaling pathway, ectopic lipid accumulation, systemic inflammation, mitochondrial dysfunction, oxidative stress, and endoplasmic reticulum (ER) stress, as reviewed elsewhere (1, 5, 6).
The mammalian Sirtuins are a family of NAD+-dependent deacetylases (7). This family consists of seven members (SIRT1-SIRT7), which share the conserved Sirtuin domain conferring NAD+-dependent deacetylase activity but have variable amino- and carboxy-terminal extensions and display distinct subcellular localization and biological functions (8, 9). SIRT1 is mainly localized to the nucleus (10), but it shuttles between the nucleus and cytoplasm during development and in response to physiological and pathological stress (11). In contrast to SIRT1, mammalian SIRT2 is mainly localized to the cytoplasm (10, 12) but is also found in the nucleus (13). SIRT3, SIRT4, and SIRT5 are localized to mitochondria (10, 14), whereas SIRT6 and SIRT7 are found in the nucleus (10). SIRT6 is a chromatin-associated protein, and SIRT7 resides in the nucleolus (15, 16). Deacetylase activity was initially reported as conserved in mammalian Sirtuins, but different Sirtuins exhibit different acyl group preferences (17). Among the seven Sirtuins, SIRT4-7 exhibit weak or undetectable deacetylation activity in vitro (10, 18). SIRT2 reportedly possesses efficient demyristoylase activity (18). SIRT5 is reportedly an efficient NAD+-dependent protein, lysine desuccinylase and demalonylase (19), while SIRT4 and SIRT6 reportedly possess ADP-ribosyltransferase activity (20, 21).
Mammalian Sirtuins regulate a wide variety of cellular processes, including metabolism, mitochondrial homeostasis, oxidative stress, inflammation, autophagy and apoptosis (22). Sirtuins also play important roles in aging and aging-related diseases, such as obesity, T2DM, cardiovascular disease, cancer, neurodegenerative diseases (23). SIRT1 and SIRT6 are the most extensively characterized Sirtuins. A large body of literature indicates that these two Sirtuins play an important role in metabolism, and they have also recently attracted increased attention with regard to their protective roles in maintaining insulin sensitivity, as reviewed elsewhere (24–26). Compared to the metabolic roles of SIRT1 and SIRT6, the metabolic roles of other Sirtuins remain poorly understood. Here, we review the recent advances in the understanding of the roles of Sirtuins in inflammation, mitochondrial dysfunction, and oxidative stress and discuss their possible roles in insulin resistance.
Obesity-induced chronic, low-grade inflammation is one of the most important contributors to the pathogenesis of insulin resistance (27–29). Adipose tissue is not only an insulin-targeting organ for lipid metabolism but also an endocrine organ that secretes hormones, cytokines, and chemokines to influence insulin sensitivity. For instance, adipocytes secrete adipokines, such as leptin and adiponectin, to promote insulin sensitivity (30, 31), and resistin and retinol-binding protein 4 (RBP4) to impair insulin sensitivity (32, 33). Importantly, adipose tissue is a critical initiator of the inflammatory response to obesity (28). In obesity, metabolism and gene expression of adipocytes change, resulting in increased lipolysis of adipocytes, the release of free fatty acids and proinflammatory cytokines and activation of M1 macrophages (27–29). M1 macrophages produce a large number of proinflammatory mediators, such as TNF-α, IL-1β, and resistin, that act on adipocytes to induce an insulin-resistant state and activate inflammatory pathways in insulin-targeting cells. Ultimately, ectopic lipid deposition and increased expression of inflammatory mediators in the liver and skeletal muscle lead to impaired insulin signaling and exacerbate systemic insulin resistance (29). Signals from all the proinflammatory mediators converge on inflammatory signaling pathways, including jun-n-terminal kinase (JNK) and inhibitor of nuclear factor κB (NF-κB) kinase (IKK) (27, 34, 35). Inhibition of insulin receptor downstream signaling is the primary mechanism for inflammation-induced insulin resistance (27). Activated JNK or IKK can phosphorylate the insulin receptor (IR) and insulin receptor substrate (IRS) proteins and decrease their tyrosine phosphorylation, thus leading to decreased activation of PI3-kinase and Akt and resistance to the metabolic actions of insulin (34, 36). In addition, activation of the JNK and IKK pathways can induce the production of inflammatory mediators, while the Sirtuin family plays essential roles in inflammation, which comprehensively contributes to insulin resistance.
NF-κB is a key transcriptional factor that mediates the expression of multiple inflammatory factors, including TNF-α, IL-1β, and IL-6. The acetylation of NF-κB promotes its nuclear translocation and activation. SIRT1 has been demonstrated to repress inflammation in multiple tissues and cells (37–40). In particular, SIRT1 suppresses inflammation in both adipocytes (39, 41) and macrophages (42), which leads to a reduction of adipose tissue inflammation. SIRT1 deacetylates p65 subunit of NF-κB at lysine 310 (K310) and inhibits the transcriptional activity of NF-κB (43). Moreover, SIRT1 interacts with transducing-like enhancer of split 1 (TLE1), a co-repressor of NF-κB, to inhibit NF-κB-mediated transcription (44). In addition, SIRT1 deacetylates activator protein-1 (AP-1) to reduce the expression of COX-2 in macrophages and deacetylates p53 to repress macrophage activation (45, 46). Similar to SIRT1, SIRT2 also binds to NF-κB and mediates the deacetylation of NF-κB subunit p65 at K310, which leads to the inhibition of the expression of NF-κB target inflammatory genes in fibroblasts, macrophages and microglial cells (47–49). SIRT2-mediated inhibition of NF-κB and inflammation contributes to its anti-inflammatory function in an experimental colitis mouse model (49), neuroinflammation (48, 50, 51), collagen-induced arthritis (52), and microvascular inflammation in ob/ob septic mice (53). In addition to SIRT2, SIRT4 can regulate the activation of NF-κB. SIRT4 has been shown to negatively regulate cigarette smoke extract (CSE)-induced NF-κB activation by inhibiting the degradation of IκBα and inhibiting NF-κB target gene expression, including the proinflammatory cytokines IL-1β, TNF-α, and IL-6, resulting in inhibition of CSE-induced mononuclear cell adhesion to human pulmonary microvascular endothelial cells (54). SIRT4 can prevent NF-κB nuclear translocation as well as the transcriptional activity of NF-κB, thereby suppressing inflammation in human umbilical vein endothelial cells (55). Interestingly, the role of SIRT5 in inflammation is controversial. Recently, Qin and colleagues showed that SIRT5 deficiency decreased toll-like receptor (TLR)-triggered inflammation in both acute and immunosuppressive phases of sepsis (56). Mechanistically, SIRT5 competes with SIRT2 to interact with NF-κB p65 in a deacetylase activity-independent manner and thus blocks the deacetylation of p65 by SIRT2, which consequently leads to the activation of the NF-κB pathway and induction of its downstream cytokines in macrophages (56). However, Wang and colleagues found that SIRT5 desuccinylates and actives pyruvate kinase isoform M2 (PKM2) by promoting its dimerization and nuclear accumulation, thereby decreasing proinflammatory cytokine IL-1β production in LPS-activated macrophages (57). As hyperproduction of IL-1β contributes to increased susceptibility to inflammatory bowel disease, Sirt5-deficient mice are more susceptible to dextran sulfate sodium (DSS)-induced colitis (57). Interestingly, Sirt6 deficient mice display increased expression of NF-κB-dependent genes in multiple tissues (58). Sirt6 deletion increases inflammation in the mice adipose tissue and promotes HFD-induced insulin resistance (59, 60). Mechanistically, SIRT6 binds to the NF-κB subunit RelA and deacetylates histone H3 lysine 9 (H3K9) at NF-κB target gene promoters, which leads to a reduction of NF-κB target gene expression (58). These findings suggest that the Sirtuins target inflammatory transcriptional factors (e.g., NF-κB and AP1) directly or indirectly to contribute to insulin resistance comprehensively.
The anti-inflammatory role of Sirtuins involves other mechanisms. The Nod-like receptor family, pyrin domain-containing 3 (NLRP3) inflammasome is a multiprotein complex that orchestrates the innate immune responses of macrophages by controlling the activation of caspase-1 and the release of the proinflammatory cytokines IL-1β and IL-18 (61–63). Obesity-related inflammation is partly mediated by the NLRP3 inflammasome, and NLRP3 activation exacerbates obesity-linked diseases (64, 65). Resveratrol, a SIRT1 activator, inhibits ionizing irradiation-induced inflammation in mesenchymal stem cells via suppressing NLRP3 inflammasome activation (66). In a murine model of sepsis, Sirt1 deletion results in increasing lung inflammasome activation and inflammatory lung injury (67). A recent study demonstrated that silybin prevents NLRP3 inflammasome activation during NAFLD through SIRT2 (68). However, further studies are needed to clarify the mechanism underlying SIRT2-mediated regulation of NLRP3 inflammasome activity. In a human fasting/refeeding study, Traba and colleagues observed that fasting leads to a reduction in NLRP3 inflammasome activation (69). SIRT3 deletion in a human macrophage line increases NLRP3 inflammasome activation, accompanied by excessive mitochondrial ROS production (69). Pharmacologic and genetic SIRT3 activation enhances mitochondrial function and suppresses NLRP3 activity in THP-1 monocyte cells and in leukocytes extracted from healthy volunteers and from refeeding individuals (69). The authors concluded that nutrient levels regulate the NLRP3 inflammasome partly through SIRT3-mediated mitochondrial homeostatic control. Similarly, Chen et al. reported that trimethylamine-N-oxide (TMAO) increases ROS production by inhibiting the SIRT3-SOD2-mitochondrial signaling pathway, which leads to NLRP3 inflammasome activation and consequently promotes vascular inflammation (70). Defective autophagy in monocytes or macrophages might result in NLRP3 inflammasome activation and cause vascular metabolic inflammation (71–73). Acetylation of ATG5, an autophagy-related protein, inhibits autophagosome maturation and induces NLRP3 inflammasome activation (74). Recently, Liu and colleagues demonstrated that SIRT3 binds with ATG5 and deacetylates it, while SIRT3-deficient macrophages display impaired autophagy, leading to accelerated NLRP3 inflammasome activation and endothelial dysfunction (73). These studies suggest that SIRT3 may inhibit NLRP3 inflammasome activation by regulating mitochondrial function, ROS production, and autophagy. As SIRT2 has also been shown to regulate NLRP3 inflammasome activation (68), the potential synergistic effect on regulation of NLRP3 inflammasome activation between SIRT3 and SIRT2 needs further study. Previous studies have highlighted the anti-inflammatory role of SIRT3 in obesity-related diseases, including insulin resistance. The function of SIRT3 in inflammasome regulation largely depends on SIRT3-mediated activation of MnSOD and suppression of oxidative stress. These findings implicate that the SIRT1-SIRT3 indirectly regulate the activation of the NLRP3 inflammasome, which may be involved in the modulation of insulin resistance. However, whether Sirtuins directly regulate inflammasome remains unknown.
The roles of Sirtuins in inflammation significantly contribute to their functions during insulin resistance. For instance, activation of SIRT1 leads to the repression of JNK and IKK inflammatory pathways greatly and subsequently improves glucose tolerance, reduced hyperinsulinemia, and enhanced systemic insulin sensitivity (40). SIRT1 also controls the inflammatory status of macrophages and T lymphocytes to regulate the metabolism (insulin sensitivity) and inflammation of adipose tissues in obese mice (41, 75–77). In addition, SIRT6 is important for macrophage activation and TNFα production (78). Myeloid Sirt6 deficiency causes insulin resistance in HFD–fed mice by eliciting macrophage polarization toward an M1 phenotype (79), and facilitates the development of HFD-induced atherosclerosis (80). Deletion of Sirt6 in T cells or myeloid-derived cells is sufficient to induce liver inflammation and fibrosis (81). Interestingly, SIRT1 and SIRT6 can coordinate a switch from glucose to fatty acid oxidation during the acute inflammatory response (82). Therefore, Sirtuins not only regulate the inflammatory pathways within the target cells (e.g., hepatocytes, skeletal muscle cells, adipocytes) to affect their insulin sensitivity but also regulate inflammatory cells infiltrated in the organs, where the Sirtuins respond to inflammatory and metabolic insults and subsequently regulate insulin sensitivity and disease by targeting inflammatory cell activation and differentiation. Notably, the Sirtuins may also cooperate in diverse types of cells or within the same type of cell during inflammation-associated insulin resistance.
Mitochondria are the primary site for ATP generation and ROS production. Mitochondrial dysfunction results in decreased ATP production, increased ROS production and accumulated mitochondrial DNA damage, which contribute to insulin resistance (5). Cells eliminate ROS by expressing endogenous antioxidant enzymes, including manganese superoxide dismutase (MnSOD), catalase, glutathione peroxidase (GPX) and glutathione reductase (GRx) (83, 84). An imbalance between the production of ROS and antioxidant enzymes leads to oxidative stress, which has been implicated in the pathogenesis of insulin resistance, obesity, and diabetes (1, 85).
There are fewer mitochondria in muscles of T2DM patients than those of insulin-sensitive individuals (86). Marked reduction of oxidative phosphorylation in the mitochondria can be detected in the liver and skeletal muscle of T2DM patients and insulin-resistant individuals (87, 88). Peroxisome proliferator-activated receptor gamma coactivator 1-alpha (PGC-1α) is a transcriptional coactivator that regulates mitochondrial biogenesis and respiration (89, 90). SIRT1 has been shown to deacetylate PGC-1α at several lysine residues and regulates PGC-1α activity, which activates the transcription of genes involved in mitochondrial biogenesis (91). SIRT1 activator resveratrol promotes PGC-1α activity and increases the number of mitochondria in muscle cells, which improves mitochondrial function and protects mice against diet-induced obesity and insulin resistance (92). In addition to mitochondria biogenesis, SIRT1 regulates mitochondrial function through clearance of damaged mitochondria (93). Mechanistically, SIRT1 binds to and deacetylates autophagy regulators (including ATG5, ATG7, and ATG8) to induce mitochondria autophagy or mitophagy (93). SIRT1-mediated deacetylation of FoxO1 and FoxO3a is also known to induce the expression of autophagy pathway components (94, 95). Accumulating evidence has shown that SIRT1 safeguards cells from oxidative stress. SIRT1 reduces ros production by deacetylating and activating FoxO3a to upregulate expression of MnSOD and catalase (95, 96). SIRT1 promotes the transcriptional activity of Nuclear factor (erythroid-derived 2)-like 2 (NRF2) by deacetylating it and upregulates the expression of NRF2 target antioxidant genes, including MnSOD, catalase, glutathione, and heme oxygenase-1 (HO-1) (97).
Oxidative stress increases SIRT2 expression in vivo and in vitro (98, 99). SIRT2 can bind to FoxO3a and deacetylate it, leading to an increase in FoxO3a transcriptional activity, upregulation of the expression of FoxO target genes such as MnSOD, Bim, and p27Kip1, and a consequent decrease in ROS generation (98). Nicotinamide adenine dinucleotide phosphate (NADPH) is a functionally important metabolite that is required to generate the reduced form of glutathione (GSH) to maintain cellular redox potential. In response to oxidative stimuli, SIRT2 deacetylates and activates glucose 6-phosphate dehydrogenase (G6PD), a key enzyme in the pentose phosphate pathway (PPP), resulting in increased cytosolic NADPH to attenuate oxidative damage (100). Similarly, oxidative stress increases the glycolytic enzyme phosphoglycerate mutase (PGAM)-SIRT2 interaction, leading to deacetylation and activation of PGAM, which increases the cellular NADPH level to counteract oxidative damage (101). Lysine 4-oxononanoylation (4-ONylation) is a newly discovered histone posttranslational modification that disrupts the interaction between histone H3 and DNA, thereby preventing nucleosome assembly under oxidative stress (102). SIRT2 was reported to remove the 4-oxononanoyl (4-ONyl) lysine groups on histones and attenuate the negative impact of protein 4-ONylation caused by oxidative stress (103). This study provides novel evidence that SIRT2 may exert antioxidation effects through epigenetic modification. Oxidative stress is not completely caused by mitochondria, and whether SIRT2 can influence oxidative stress by regulating mitochondrial function is unclear.
Interestingly, a recent work by Liu and colleagues observed that SIRT2 can localize to the inner mitochondrial membrane of mouse central nervous system cells and that the acetylation of several metabolic mitochondrial proteins is altered in Sirt2-deficient mice. In mice, deletion of Sirt2 causes mitochondrial morphological changes, increases oxidative stress and decreases ATP production in MEFs and brain tissues (104), indicating that SIRT2 may deacetylate antioxidant enzymes in the mitochondria directly.
SIRT3, a central mitochondrial deacetylase, deacetylates, and activates mitochondrial enzymes to regulate mitochondrial metabolism, oxidative stress, cell survival, and longevity (105). SIRT3 has been shown to play a pivotal role in maintaining mitochondrial function and ROS homeostasis. For example, SIRT3 deacetylates complex I and complex II of the electron transport chain to promote electron transport and regulate energy homeostasis (106, 107). SIRT3 deacetylates cyclophilin D (CypD), the regulatory component of the mitochondrial permeability transition pore (mPTP), preventing opening of the mPTP and mitochondrial dysfunction (108). SIRT3 binds to and deacetylates 8-oxoguanine-DNA glycosylase 1 (OGG1), a DNA repair enzyme that excises 7,8-dihydro-8-oxoguanine from the damaged genome, resulting in the repair of mitochondrial DNA (mtDNA) damage, protection of mitochondrial integrity, defense against mitochondrial dysfunction and prevention of stress-induced cellular apoptosis (109). In contrast, Sirt3 deficiency has been linked to increased ROS production (110, 111). The antioxidant action of SIRT3 involves MnSOD and mitochondrial isocitrate dehydrogenase 2 (IDH2). SIRT3 has been shown to block cardiac hypertrophy by deacetylating FOXO3a and upregulating the expression of FOXO3a target genes such as MnSOD and catalase, decreasing ROS generation (112). Notably, the antioxidative action of SIRT3 may also be attributed to its direct deacetylation and promotion of the enzyme activity of MnSOD. SIRT3 deacetylates two critical lysine residues (K53 and K89) of MnSOD that promote MnSOD antioxidation activity, thereby reducing cellular ROS (113). Direct deacetylation of lysine residues, including K68 and K122 by SIRT3, also increases the enzyme activity of MnSOD and decreases ROS production (114, 115). In addition to MnSOD, SIRT3 directly deacetylates and activates mitochondrial IDH2, which results in increased NADPH levels and an increased ratio of reduced to oxidized GSH in mitochondria, thus protecting cells from oxidative stress-induced damage (110, 116). These studies suggest that SIRT3 activity is necessary to prevent mitochondrial dysfunction and reduce oxidative stress.
SIRT4 functions as an efficient mitochondrial ADP-ribosyl transferase that negatively impacts gene expression and various metabolic processes in mitochondria. Our previous work demonstrated that SIRT4 promotes angiotensin II-induced development of cardiac hypertrophy by inhibiting the interaction of SIRT3 and MnSOD, which increases MnSOD acetylation levels, decreases its activity and leads to elevated ROS accumulation (117). SIRT4 may play a role different from that of SIRT3 in regulating mitochondrial function and oxidative stress. Sirt4 deficiency in vivo and in vitro increases the expression of genes involved in fatty acid β-oxidation and oxidative phosphorylation, thus enhancing fatty acid oxidation and mitochondrial respiration in liver and muscle (118, 119). SIRT4 increases stress-induced mitochondrial ROS production and interacts with the long form of GTPase optic atrophy 1 (L-OPA1) to promote mitochondrial fusion, thereby inhibiting mitophagy and decreasing the removal of dysfunctional mitochondria (120).
SIRT5 functions to deacetylate, demalonylate, and desuccinylate multiple proteins in mitochondria (19). SIRT5 is involved in the regulation of mitochondrial fatty acid β-oxidation, the urea cycle, and cellular respiration (84, 121). SIRT5 binds to, desuccinylates and activates copper-zinc superoxide dismutase 1 (SOD1) to eliminate ROS (122). Moreover, SIRT5 desuccinylates IDH2 and deglutarylates G6PD, thus activating both NADPH-producing enzymes to scavenge ROS (123). SIRT5 also binds, desuccinylates and inhibits the activity of the glycolysis enzyme PKM2, which facilitates the diversion of glucose metabolites into the pentose phosphate shunt and then produces sufficient NADPH to eliminate ROS (123). Interestingly, a very recent study showed that SIRT5 is present in peroxisomes and can bind, desuccinylate and inhibit ACOX1, the first and rate-limiting enzyme in fatty acid β-oxidation and a major producer of H2O2, attenuating peroxisome-induced oxidative stress (124).
SIRT6 plays an important role in DNA repair, genomic stability, and cellular senescence, however, the role of SIRT6 in oxidative stress has not been well clarified. According to recent studies, SIRT6 is believed to protect cells against oxidative stress. Pan et al. (125) found that SIRT6 serves as an NRF2 coactivator by interacting with NRF2 and RNA polymerase II to transactivate NRF2-regulated antioxidant genes, including HO-1. SIRT6 activates AMPK-FoxO3a axis to initiate expression of MnSOD and catalase and protects cardiomyocytes against ischemia/reperfusion-induced injury (126).
Sirt7 deficiency in mice induces multisystemic mitochondrial dysfunction (127). SIRT7 deacetylates GABPβ1, a master regulator of nuclear-encoded mitochondrial genes, enables it to form the transcriptionally active GABPa/GABPβ heterotetramer, and promotes mitochondria function (127). Additionally, the mitochondrial unfolded protein response [UPR(mt)] is mediated by the interplay of SIRT7 and NRF1 and is coupled to cellular energy metabolism and proliferation (128).
Under the condition of insulin resistance, normal pancreatic β cells increase the production of insulin to maintain blood glucose levels. However, this compensatory response fails, and relative insulin insufficiency develops. Then, glucose tolerance is impaired, and T2DM eventually occurs. SIRT1, SIRT4, and SIRT6 reportedly regulate pancreatic β cell function (Figure 1) (24, 129, 130). According to accumulating evidence, SIRT1 and SIRT6 repress pancreatic β cell dysfunction, attenuating the development of T2DM. β cell-specific SIRT1 transgenic mice exhibit enhanced insulin secretion and improved glucose tolerance to high glucose stimulation (131). Mechanistically, through repressing UCP2 expression, SIRT1 enhances ATP production in pancreatic β cells, to shut down the potassium channel, resulting in the influx of calcium and finally the secretion of insulin (131, 132). SIRT1 can induce NeuroD and MafA expression via deacetylating and activating FoxO1, which protects pancreatic β cells against oxidative damage and preserves pancreatic β cells function (133). In contrast to SIRT1 and SIRT6, SIRT4 functions as a negative regulator of insulin secretion in β cells. Glutamate dehydrogenase (GDH) promotes the metabolism of glutamate and glutamine, generating ATP to further promote insulin secretion. In pancreatic β cells, SIRT4 represses the activity of GDH by ADP-ribosylation, thereby downregulating insulin secretion in response to amino acids under calorie-sufficient conditions (20). SIRT4 also controls leucine oxidation to regulate insulin secretion (134). Given that SIRT3 deacetylases GDH and increases its activity in hepatocytes (135), SIRT3 may function in β cell mitochondria to promote insulin secretion. Recent studies have provided evidence to support this notion. Caton et al. observed that SIRT3 expression markedly decreases in islets isolated from T2DM patients, as well as in mouse islets or INS1 cells (136). Sirt3 knockdown in INS1 cells results in increased production of cellular ROS and IL-1β, increased β cell apoptosis and reduced insulin secretion (136). SIRT3 deficiency predisposes pancreatic β cells to oxidative stress-induced dysfunction and reduces glucose-induced insulin secretion (137). By contrast, SIRT3 overexpression inhibits ER stress and attenuates palmitate-induced pancreatic β cell dysfunction (138, 139). Therefore, SIRT3 and SIRT4 play opposing roles in regulating insulin secretion in pancreatic β cells. In addition, insulin secretion impairment is observed in Sirt6 knockout pancreatic β cells, which is mediated by suppression of the FoxO1-Pdx1-Glut2 pathway (140). Sirt6 deletion in pancreatic β cells also reduces ATP production and increases mitochondrial damage which induces cell apoptosis and impairs glucose-stimulated insulin secretion (129, 141). β cell-specific Sirt6-ko mice are glucose intolerance and are defective in glucose-stimulated insulin secretion, in spite do not show abnormality in endocrine morphology, pancreatic β cell mass or insulin production (130). Sirt6 deficiency also results in aberrant upregulation of thioredoxin-interacting protein (TXNIP) in pancreatic β cells, which inhibits insulin secretion (130).
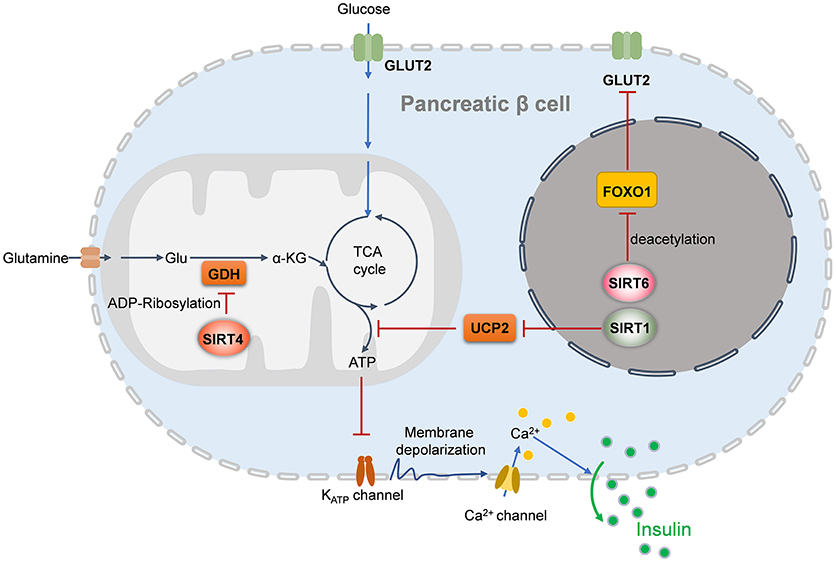
Figure 1. Sirtuins regulates insulin secretion of pancreatic beta cells. In the nucleus, SIRT1 induces insulin secretion through the reduction of UCP2 expression and the enhancement of depolarization in pancreatic β cells, while SIRT6 deacetylates FOXO1 and promotes the expression of GLUT2, which facilitates glucose uptake and insulin secretion. In the mitochondria, SIRT4 promotes the ADP-ribosylation and inactivation of GDH, leading the repression of ATP generation and inhibition of insulin secretion. UCP2, uncoupling protein 2; Glu, glutamate; GDH, glutamate dehydrogenase; GLUT2, glucose transporter 2; α-KG, alpha-ketoglutarate.
Insulin resistance, the inability of cells to efficiently respond to a normal dose of insulin, is caused by impaired insulin signaling and postreceptor intracellular defects (4). Insulin binding to its receptor results in IR phosphorylating itself and several intracellular substrates. The phosphorylated substrates interact with intracellular effectors, leading to the activation of the PI3K-Akt pathway, which is responsible for most of the metabolic actions of insulin, and the Ras-MAPK pathway, which controls cell growth and differentiation (4, 142). Impaired Akt activation is a key factor in metabolic disorders involving insulin resistance. Accumulating evidence suggests that Sirtuins participate in insulin signaling in target cells (Figure 2).
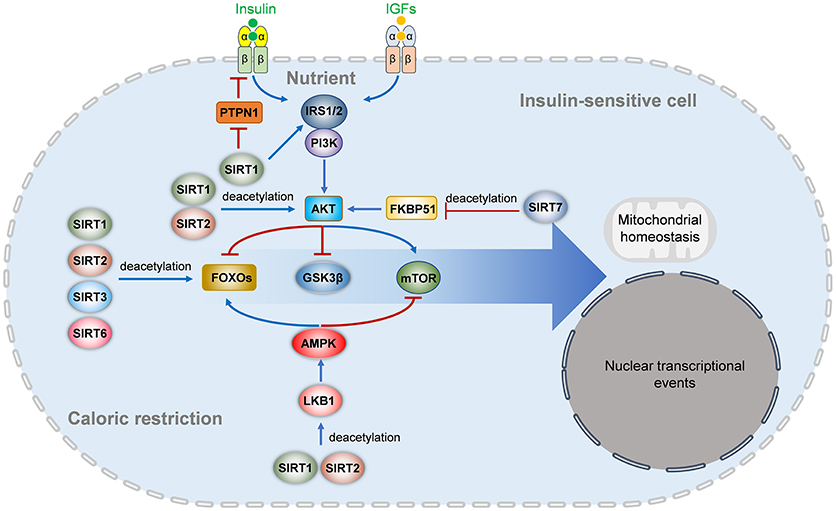
Figure 2. Sirtuins regulates insulin signaling pathways. In nutrient enough conditions, insulin and insulin-like growth factors activate the IRS-PI3K-AKT signaling and downstream FOXO and mTOR signaling to regulate multiple aspects of metabolism, survival, mitochondrial homeostasis, nuclear transcriptional events, and other cellular behaviors. The insulin receptor activation is inhibited by PTPN1, which is repressed by SIRT1. SIRT1 also deacetylates IRS2 and represses IRS1 phosphorylation and PI3K-AKT activation. SIRT1 and SIRT2 also deacetylate AKT to activate its activation directly, while SIRT7 indirectly inhibits AKT activation by deacetylating FKBP51. Under energetic stress or caloric restriction, multiple members of the Sirtuins family are activated. SIRT1, SIRT2, SIRT3, and SIRT6 can directly deacetylate FOXOs (FOXO1 and FOXO3a), while SIRT1 and SIRT2 activate the LKB1-AMPK signaling to activate FOXO and inhibit mTOR signaling. IGF, insulin-like growth factor; IRS, insulin receptor substrate; PTPN1, tyrosine-protein phosphatase non-receptor type 1; PI3K, phosphatidylinositol-4,5-bisphosphate 3-kinase; mTOR, mammalian target of rapamycin; AMPK, AMP-activated protein kinase; LKB1, liver kinase B1; FKBP51, FK506-binding protein 51; GSK3β, glycogen synthase kinase-3 beta.
SIRT1 positively regulates insulin signaling and Akt activation at multiple levels. SIRT1 represses transcription of PTPN1, a negative regulator of the insulin signal transduction cascade, at the chromatin level and improves insulin sensitivity (3). Knockdown of Sirt1 in 3T3-L1 adipocytes increases phosphorylation of JNK, as well as serine phosphorylation of insulin receptor substrate 1 (IRS-1), which leads to decrease tyrosine phosphorylation of IRS-1, and then inhibit phosphorylation of Akt (39). Inhibition of SIRT1 activity reduces insulin-induced IRS-2 deacetylation, which prevents insulin-induced IRS-2 tyrosine phosphorylation (143). SIRT1 mediates deacetylation of Akt regulates binding of Akt to phosphatidylinositol 3,4,5-trisphosphate (PIP3) which is necessary for Akt membrane localization and activation (144).
SIRT2 can directly regulate the insulin signaling pathway, but its role is controversial. SIRT2 can deacetylate and activate Akt through the Akt/glycogen synthase kinase-3β (GSK3β)/β-catenin signaling pathway, finally resulting in aberrant proliferation and survival of myeloid leukemia cells and epithelial-mesenchymal transition of HCC (145, 146). Interestingly, Ramakrishnan and colleagues showed that SIRT2 is a novel Akt interactor and is required for optimal Akt activation under normal conditions (147). Pharmacological or genetic inhibition of SIRT2 decreases Akt activation in 3T3-L1 preadipocytes and HeLa cells, whereas SIRT2 overexpression enhances the activation of Akt and its downstream targets, such as GSK3 and p70-S6-kinase (147). Insulin-induced Akt activation requires Akt binding to inositol 1,4,5-trisphosphate, which leads to Akt conformational changes and facilitates its phosphorylation by PDK1 and mTORC1 (147). Acetylation at Lys20 blocks Akt activation by restricting the binding of Akt to inositol 1,4,5-trisphosphate (144). However, the authors were unable to detect Akt acetylation in the experiment, and they could not determine whether the effects of SIRT2 on Akt are dependent on changing the acetylation level of Akt (147). However, the opposite results have also been reported. Arora et al. reported that SIRT2 is upregulated in insulin-resistant skeletal muscle cells, and inhibition of SIRT2 by pharmacological or genetic means improves phosphorylation of Akt and GSK3β and increases insulin-stimulated glucose uptake (148). Similarly, in insulin-resistant neuro-2a cells, inhibition of SIRT2 by pharmacological or genetic means also enhances the activity of Akt and increases insulin-stimulated glucose uptake (149). Therefore, SIRT2 may regulate Akt in both direct (activity-dependent) and indirect (activity-independent) manners, which may largely rely on the metabolic status of the cells. SIRT2 can deacetylate and regulate the function of FOxO transcription factors, which are direct Akt targets (98, 150). Interestingly, the Akt-independent pathway also contributes to the function of SIRT2 in the regulation of insulin sensitivity. TUG acetylation modulates its interaction with Golgi matrix proteins and enhances its function to trap GLUT4 storage vesicles in intracellular (151). Insulin mobilizes the exocytic translocation of GLUT4 glucose transporters by triggering TUG proteolysis to accelerate glucose uptake in fat and muscle. SIRT2-mediated TUG deacetylation controls insulin sensitivity in vivo and in vitro (151).
The role of SIRT6 in the insulin signaling pathway is controversial. Sirt6-deficient mice die about 4 weeks of age, exhibiting severe metabolic defects, including low insulin and hypoglycemia (15). Xiao et al. found that Sirt6 deficiency increases Akt phosphorylation through modulating insulin signaling upstream of Akt, including insulin receptor, IRS1, IRS2, and enhances insulin signaling, leading to hypoglycemia (152). On the contrary, Sirt6 transgenic mice show increased insulin sensitivity in skeletal muscle and liver and exhibit enhanced insulin-induced Akt activation in gastrocnemius (153).
In addition, SIRT7 can regulate Akt signaling. SIRT7 regulates the acetylation of FKBP51, which then regulates Akt activation. Acetylated FKBP51 enhances Akt activity by blocking its interaction with PHLPP-Akt. SIRT7 deacetylates FKBP51 at two major lysine residues. SIRT7 suppresses Akt activation and modulates cell sensitivity to genotoxic agents (154). The inhibitory effects of SIRT7 on Akt activation were also observed in murine hearts.
Adipose tissue, liver, and muscle are the primary insulin-responsive organs. Insulin regulates blood glucose concentrations by suppressing hepatic glucose output and stimulating glucose uptake by muscle and adipose tissue. In addition, insulin promotes energy storage in adipose tissue, liver, and muscle by stimulating lipogenesis, glycogen, and protein synthesis but inhibiting lipolysis, glycogenolysis and protein catabolism (155).
The impaired lipogenic/adipogenic capacity of adipose tissue leads to increased body fat mass and adverse metabolic consequences (4, 156). Adipose tissue is not only an excessive energy storage pot but also a highly active endocrine organ that secretes proteins, adipokines, cytokines and chemokines to influence insulin sensitivity. Circulating FFAs derived from adipocytes are involved in the accumulation of triglycerides and fatty acid-derived metabolites in muscle and liver, which is a contributing factor to insulin resistance (155). In addition, adipose tissue is an important initiator of the inflammatory response to obesity (28), and FFAs from adipose tissue are important ER stress-triggering factors (28).
The liver, as the central organ responsible for maintaining lipid and glucose hemostasis in the body, plays a crucial role in insulin sensitivity and metabolic diseases. During prolonged fasting or starvation, the liver converts lipids to available energy through fatty acid oxidation and provides glucose to maintain normal blood glucose, initially by glycogenolysis and then by switching to gluconeogenesis (157, 158). Under energy abundance conditions, the liver promotes glycogenesis and lipogenesis to store energy. Insulin stimulates glycogen accumulation and blocks gluconeogenesis and glycogenolysis in the liver to suppress hepatic glucose output (155, 158, 159). In the condition of insulin resistance, suppression of hepatic glucose output is impaired, while increased FFA from adipocytes leads to ectopic lipid accumulation in the liver, which exacerbates insulin resistance (29).
Skeletal muscle is the major site for insulin-stimulated glucose disposal in vivo as well as the main energy consumer of lipid catabolism that strongly influences whole-body lipid metabolism (155, 160). The ability to switch between glucose and lipid oxidation is crucial for skeletal muscle to maintain physiological function and metabolic hemostasis (161). Intramuscular fatty acid metabolite accumulation may cause insulin resistance (162).
The functions of Sirtuins in regulating glucose and lipid metabolism as well as insulin sensitivity have been widely investigated in adipose tissue, liver and skeletal muscles (Figure 3).
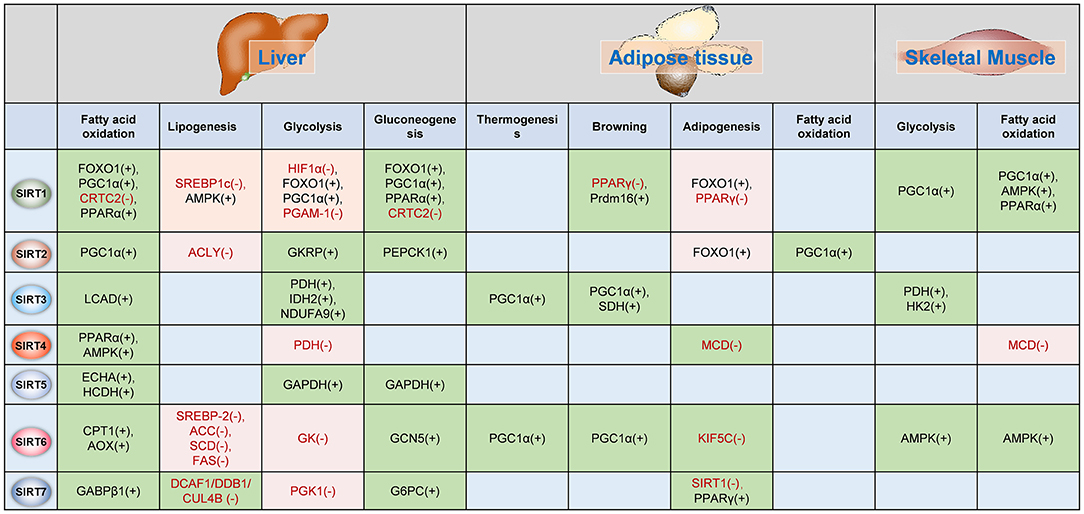
Figure 3. Sirtuins regulates metabolism in insulin-target organs. The functions of Sirtuins in the regulation of glucose and fatty acid metabolism in the liver, adipose tissue, and skeletal muscle. The (–) indicates Sirtuin represses the activation/expression of this target, whereas (+) indicates Sirtuin promotes the activation/expression of the target. The green background indicates Sirtuin promotes the biological process whereas the pink background indicates the Sirtuin represses the biological process. PGC-1α, peroxisome proliferator-activated receptor gamma coactivator 1-alpha; TORC2, CREB regulated transcription coactivator 2; PPARα, peroxisome proliferator-activated receptor alpha; SREBP1c, Sterol response element-binding protein 1c; AMPK, AMP-activated protein kinase; HIF1α, hypoxia-inducible factor 1 alpha; PGAM-1, phosphoglycerate mutase 1; CRTC2, CREB regulated transcription coactivator 2; PPARγ, peroxisome proliferator-activated receptor gamma; Prdm16, PR domain containing 16; PEPCK1, phosphoenolpyruvate carboxykinase 1; LCAD, long-chain acyl-CoA dehydrogenase; PDH, pyruvate dehydrogenase; IDH2, isocitrate dehydrogenase; NDUFA9, NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 9; SDH, succinate dehydrogenase; HK2, hexokinase 2; MCD, malonyl-CoA decarboxylase; ECHA, trifunctional enzyme subunit alpha; HCDH, hydroxyacyl-Coenzyme A dehydrogenase; GAPDH, Glyceraldehyde 3-phosphate dehydrogenase; SREBP-2, sterol regulatory element-binding protein 2; ACC, acetyl-CoA carboxylase; SCD, stearoyl-CoA desaturase; FAS, fatty acid synthase; GK, glucokinase; GCN5, general control non-repressed protein 5; KIF5C, kinesin heavy chain isoform 5C; GABPβ1, GA binding protein β1; DCAF1/DDB1/GUL4B, DDB1-CUL4-associated factor 1 (DCAF1)/damage-specific DNA binding protein 1 (DDB1)/cullin 4B (CUL4B) complex; PGK1, phosphoglycerate kinase 1; G6PC, glucose-6-phosphatase, catalytic subunit; GKRP, glucokinase regulatory protein.
The role of SIRT1 in regulating hepatic gluconeogenesis is controversial under the condition of calorie restriction. Resveratrol has been shown to improve glucose homeostasis in insulin-resistant mice by reducing hepatic gluconeogenesis and increase insulin sensitivity in adipose tissue, skeletal muscle and liver (163). On the contrary, resveratrol causes nuclear translocation of FoxO1 in hepatocytes via SIRT1-dependent deacetylation, which leads to activation of gluconeogenesis and increased hepatic glucose output (164). Pyruvate induces SIRT1 protein in the liver during fasting, and once SIRT1 is induced, which can increase gluconeogenic genes expression and promote hepatic glucose output through interacting and deacetylating PGC-1α (165). Conversely, during prolonged fasting, SIRT1 deacetylates CREB regulated transcription coactivator 2 (CRTC2) and promotes its ubiquitin-dependent degradation to inhibit gluconeogenic gene expression, leading to decreased hepatic glucose output (166). On the other hand, upregulation of FOXO1 and PGC-1α activity by SIRT1 leads to activation of gluconeogenic gene expression in hepatic cells and increase hepatic glucose production (157, 166). In addition, SIRT1 regulates the activity of PGC-1α, and glycolytic enzyme phosphoglycerate mutase-1 (PGAM1), inducing repression of glycolytic genes in response to fasting (165, 167). These studies suggest an important role of SIRT1 in maintaining energy balance under fasting. Liver-specific Sirt1 ko mice develop hepatic steatosis when underwent fasting and obesity when fed with HFD in insulin-dependent and independent manners (38, 168). Sirt1 transgenic mice show better glucose tolerance and insulin sensitivity and are almost entirely protected from hepatic steatosis with HFD treatment (37, 169). On one hand, SIRT1 deacetylates and activates PGC-1α, increasing fatty acid β-oxidation in the liver (38). On the other hand, SIRT1 plays an important role in inhibiting lipogenesis in the liver. For example, resveratrol can increase the levels of sterol regulatory element-binding protein (SREBP), a critical regulator of lipid and sterol homeostasis in eukaryotes, in livers of alcohol-treated mice and alleviate alcoholic fatty liver (170). SRT1720, a SIRT1 activator, ameliorates fatty liver through suppressing the expression of lipogenic enzymes, including SREBP-1c, acetyl-CoA carboxylase, and fatty acid synthase, in obesity and insulin resistant mice (171). SIRT1 can directly deacetylate SREBP, and control SREBP protein stability via SREBP ubiquitination, leading to attenuating SREBP target lipogenic gene expression and inhibiting lipid synthesis and fat storage (172). Otherwise, SIRT1 activation by polyphenols acts as an upstream regulator in the LKB1/AMPK signaling axis, resulting in repression expression of acetyl-CoA carboxylase and fatty acid synthase and reduction of lipid accumulation in hepatocytes (173).
PPARγ and CCAAT/enhancer-binding protein α (C/EBPα) are master regulators of adipogenesis (156, 174). SIRT1 overexpression inhibits the expression of PPARγ and C/EBPα in 3T3-L1 adipocytes (175). SIRT1 represses PPARγ by docking with its cofactors nuclear receptor co-repressor (NCoR) and silencing mediator of retinoid and thyroid hormone receptors (SMRT) and suppresses adipogenesis (175). In differentiated fat cells, upregulation of SIRT1 by resveratrol triggers lipolysis and loss of fat, but SIRT1 inhibitor nicotinamide reduces the release of free fatty acid (175).
There is a strong correlation between the presence of intramyocellular lipid in skeletal muscle and liver and progression of T2DM (176, 177). Fasting induces PGC-1α deacetylation by SIRT1 in skeletal muscle, and that is required for activation of mitochondrial fatty acid oxidation genes (178). SIRT1 overexpression protects C2C12 myotubes against fatty acid-induced insulin resistance through transcriptional repression of PTP1B (3). In skeletal muscle, SIRT1 acts downstream of AMPK signaling, deacetylates and modulates the activity of PGC-1α, FOXO1, and FOXO3a to inhibit lipogenesis and promote energy consumption (179). SIRT1 overexpression or resveratrol treatment increases insulin-induced Akt phosphorylation and activation via interacting with the PI3K adapter subunit p85 (180). These studies suggest SIRT1 plays a positive role in ameliorating insulin sensitivity in skeletal muscle. However, several studies have demonstrated that skeletal muscle-specific overexpression of SIRT1 does not enhance whole-body energy expenditure or skeletal muscle insulin sensitivity under normal or overfeeding conditions (181, 182). These controversial results suggest SIRT1 in other metabolic tissues, such as adipose tissue, liver or intestinal tissue, may play a role in the metabolic benefits of SIRT1 activation (183). Notably, high-intensity interval training increases SIRT1 activity in human skeletal muscle and mice with muscle-specific inactivation of the SIRT1 deacetylase domain displayed reduced myofiber size, impaired muscle regeneration, and derepression of muscle developmental genes (184, 185). Therefore, SIRT1-mediated metabolic balance is important for skeletal muscle homeostasis and regeneration.
SIRT2 is widely distributed and has been detected in a wide range of metabolic tissues, including the brain, muscle, liver, pancreas and adipose tissue. SIRT2 expression is regulated by metabolic status. For instance, the expression of SIRT2 is elevated in the white adipose tissue (WAT) of cr mice (98). SIRT2 gene expression increased in the peripheral blood mononuclear cells of obese subjects following an 8-week hypocaloric diet (186). By contrast, SIRT2 protein expression in visceral WAT from human obese subjects and a mouse model of diet-induced obesity is downregulated compared with that in WAT from lean controls (187). SIRT2 gene expression is significantly lower in peripheral blood mononuclear cells of obese children with insulin resistance than in those without insulin resistance (188). A growing body of literature has indicated that SIRT2 is involved in regulating various metabolic processes, including adipocyte differentiation, hepatic gluconeogenesis, and insulin action. SIRT2 mRNA is more abundant than other Sirtuins in adipose tissue in vivo and preadipocytes in culture (150), implicating the possible important role of SIRT2 in adipose tissue. FOXO1 acts as an adipogenesis inhibitor (189). In adipose tissue, FOXO1 can interact with PPARγ and negatively regulate its transcriptional activity (190) or bind to the PPARγ promoter region and suppress its expression (191). In mouse 3T3-L1 preadipocytes, SIRT2 interacts with and deacetylates FOXO1, which antagonizes FOXO1 phosphorylation and promotes nuclear retention of FOXO1, leading to repression of the expression of PPARγ and C/EBPα as well as genes marking terminal adipocyte differentiation, such as Glut4, aP2, and fatty acid synthase (150). SIRT2 also suppresses adipogenesis by deacetylating FOXO1 to promote the binding of FOXO1 to PPARγ and subsequent repression of PPARγ transcriptional activity (192). Increased de novo lipogenesis is an important contributor to increased adipose mass (193). ATP-citrate lyase (ACLY) is the building block for de novo lipid synthesis, which converts glucose-derived citrate into acetyl-CoA (194). ACLY is acetylated on multiple lysine residues in response to high glucose and promotes lipogenesis, while SIRT2 deacetylates and destabilizes ACLY, leading to reduced lipogenesis (195). Transcriptional regulators such as PPARs and the coactivator PGC-1α play key roles in the process of fatty acid β-oxidation, which determines whole-body energy expenditure (196). Through transcriptional repression of SIRT2, hypoxia-inducible factor 1α (HIF1α) decreases deacetylation of PGC-1α and further diminishes fatty acid β-oxidation in WAT. Adipocyte-specific HIF1α inactivation leads to increased expression of SIRT2 and attenuates dietary-driven obesity in mice (187). These studies suggest that SIRT2 contributes to the control of adipose tissue mass by inhibiting adipogenesis and lipogenesis but promoting fatty acid β-oxidation.
SIRT2 deacetylates and subsequently increases the stability of PEPCK1, the gluconeogenic rate-limiting enzyme under conditions of glucose deprivation, leading to increased gluconeogenesis (197, 198). FOXO1 and PGC-1α reportedly activate the process of gluconeogenesis in the liver by increasing the transcription of gluconeogenic enzyme genes and are considered negative regulators of insulin sensitivity in the liver (199–201). Insulin suppresses gluconeogenesis by regulating the FOXO1-PGC-1α interaction (199). SIRT2 deacetylates and activates FOXO1/PGC-1α in adipocytes (150, 187, 192), which implies that SIRT2 may enhance gluconeogenesis through the FOXO1-PGC-1α pathway. However, whether the role of SIRT2 in gluconeogenesis depends on different nutrient conditions must be elucidated. In addition, the roles of SIRT2 in metabolic diseases are largely unknown.
Human SIRT3 is expressed in a variety of metabolically active tissues, including muscle, liver, kidney, heart, brain, and BAT (202–204). The expression of SIRT3 in the liver and adipose tissue of mice increases during cr (205–208). A single nucleotide polymorphism in the human SIRT3 gene has been correlated with the reduced enzymatic efficiency of SIRT3 and the development of metabolic syndrome (209). Compared with WT mice, Sirt3-KO mice fed an HFD show accelerated obesity, insulin resistance, hyperlipidemia, and hepatic steatosis (209).
High levels of SIRT3 occur in the BAT. Cold exposure upregulates SIRT3 expression in the BAT. Increasing the expression of PGC-1α and uncoupling protein 1 (UCP1) by sustained expression of SIRT3 in brown adipocytes leads to increased thermogenesis (7, 206). Fatty acid β-oxidation in the BAT of Sirt3-KO mice is significantly reduced (210). Although SIRT3 maintains a low level in WAT (206), several studies refer to the role of SIRT3 in regulating WAT lipid metabolism. cr activates SIRT3 expression in both white and brown adipose tissue; SIRT3 expression decreases in the BAT of several lines of genetically obese mice (206). In a human study, SIRT3 gene expression was decreased in VAT from morbid subjects (211) and WAT from children with obesity (212). Although the role of SIRT3 in WAT lipid metabolism is intricate and unclear, these studies suggest that SIRT3 may play a protective role in obesity.
Sirt3-deficient mice show higher levels of fatty acid β-oxidation intermediate products and triglycerides in the liver during fasting and develop hepatic steatosis (210). Metabolomic analyses of fasted Sirt3-deficient mice revealed that SIRT3 is involved in fatty acid β-oxidation and modulates fatty acid β-oxidation at multiple points, such as short-chain L-3-hydroxy acyl-CoA dehydrogenase (SCHAD), very-long-chain acyl-CoA dehydrogenase (VLCAD) and 3-ketoacyl-CoA thiolase, in addition to LCAD (208). LCAD is a key enzyme in mitochondrial fatty acid β-oxidation, and LCAD deficiency causes hepatic steatosis and hepatic insulin resistance (213, 214). SIRT3 promotes hepatic fatty acid β-oxidation through deacetylation and activation of LCAD (210). However, hepatocyte-specific Sirt3-KO mice do not show any obvious metabolic phenotype under either chow or HFD conditions, despite a marked global hyperacetylation of mitochondrial proteins (215). These conflicting findings from global Sirt3-KO mice and tissue-specific KO mice suggest that the roles of SIRT3 in other cell types may be important for SIRT3-mediated metabolic effects in the liver.
SIRT3 expression decreases in the skeletal muscle of diabetic and HFD-fed mice (36, 216). CR and exercise upregulate SIRT3 expression in mouse skeletal muscle (162). These findings suggest that SIRT3 is involved in skeletal muscle metabolism. Although muscle-specific Sirt3 KO in mice shows no obvious effects on global metabolic hemostasis under normal conditions (215), striking results have been shown in global Sirt3-KO mice. Global Sirt3-KO mice exhibit decreased oxygen consumption and enhanced oxidative stress in skeletal muscle that leads to impaired insulin signaling (36). The deletion of Sirt3 in vivo and in vitro induces hyperacetylation of the pyruvate dehydrogenase (PDH) E1α subunit and leads to decreased PDH enzymatic activity (161). Inhibition of PDH activity reduces glucose oxidation and results in a switch to fatty acid β-oxidation, thus leading to a loss of skeletal muscle metabolic flexibility (161). In addition, HFD-fed Sirt3-KO mice exhibit increased insulin resistance due to defects in skeletal muscle glucose uptake (217). These studies suggest that SIRT3 may protect insulin sensitivity in skeletal muscle.
The expression of SIRT4 is upregulated in the liver and adipose tissues in rodents fed an HFD (218, 219). SIRT4 deacetylates and inhibits malonyl CoA decarboxylase (MCD), an enzyme producing acetyl-CoA from malonyl CoA, consequently repressing fatty acid oxidation but promoting lipogenesis in WAT and skeletal muscle under nutrient abundance conditions. Sirt4-KO mice display increased exercise tolerance and protection against diet-induced obesity (220).
SIRT4 inhibition in mouse primary hepatocytes increases fatty acid oxidation gene expression, leading to increased fat oxidative capacity in liver (118). The same result is obtained in muscle (118). Similarly, primary hepatocytes from Sirt4-KO mice exhibit higher rates of fatty acid oxidation. SIRT4 suppresses PPARα activity and inhibits hepatic fatty acid oxidation by modulating SIRT1 activity (221). Livers from NAFLD patients exhibit increased SIRT4 and lipogenic gene expression (222). These results support the notion that SIRT4 is likely to inhibit fatty acid oxidation and potentiate ectopic lipid storage in liver and skeletal muscle.
SIRT5 is highly expressed in metabolic tissues, including the heart, skeletal muscle, brain, liver, and kidney (121). Using a label-free quantitative proteomic approach, Rardin et al. characterized the lysine succinylome in liver mitochondria and revealed a major role for SIRT5 in regulating many metabolic pathways, including β-oxidation and ketogenesis (223). Park et al. revealed that SIRT5 desuccinylates a set of metabolic enzymes in mitochondria that are involved in amino acid degradation, the TCA cycle and fatty acid metabolism (224). In contrast to the other two mitochondrial Sirtuins, SIRT5 protein levels do not change during CR (121, 207). However, similar to Sirt3-KO and Sirt4-KO mice, Sirt5-KO mice do not show any overt metabolic abnormalities under either normal chow or HFD conditions (225). Sirt5 deficiency does not protect or sensitize mice to the development of HFD-induced obesity, hypertension, and insulin resistance (225). The results from Sirt5-KO mice suggest that SIRT5 is not dispensable for cellular metabolism, at least under normal conditions. Subsequent studies have shown promising results. In humans, SIRT5 gene expression decreases in the liver of NAFLD patients (222), and the expression of SIRT5 in adipose tissue is positively correlated with insulin sensitivity (226). Using affinity enrichment and label-free quantitative proteomics, Nishida et al. characterized the SIRT5-regulated lysine malonylome (227). Pathway analysis identified gluconeogenesis and glycolysis as the pathways most enriched in SIRT5-regulated malonylated proteins (227). SIRT5 regulates glyceraldehyde phosphate dehydrogenase (GAPDH), a glycolytic enzyme, through demalonylation of lysine 184 (227). According to these results, SIRT5 may play a critical role in regulating glucose and lipid metabolism and preserving insulin sensitivity. Mitochondria-specific Sirtuin knockout mice show no obvious metabolic abnormalities, indicating that mitochondrial Sirtuins serve as nutrient sensors to maintain energy homeostasis.
SIRT6 expression is decreased in adipose tissue of db/db mice but increased in adipose tissue of human individuals with weight loss (228, 229), suggesting that SIRT6 plays a role in adipose tissue. Chen et al. (230) demonstrated that SIRT6 is required for mitotic clonal expansion during adipogenesis by inhibiting expression of kinesin family member 5C (KIF5C) and subsequent increasing CK2 kinase activity. Sirt6 transgenic mice exhibit resistance to HFD-induced obesity and insulin resistance (231). Conversely, fat-specific Sirt6 knockout increases blood glucose levels and hepatic steatosis, and sensitizes mice to HFD-induced obesity and insulin resistance (59, 60, 232). SIRT6 overexpression downregulates a set of PPARγ target genes that are involved in lipid metabolism, lipid transport and adipogenesis (231). Especially, SIRT6 decreases expressions of ANGPTL4, a negative regulator of lipoprotein lipase, and diglyceride acyltransferase 1 (DGAT1), a key enzyme in triglycerides synthesis, leading to the increased serum triglyceride clearance and reducing triglyceride synthesis in adipose tissues (231). Sirt6 deletion decreases FoxO1 transcriptional activity by increasing its acetylation and phosphorylation and reduces expression of adipose triglyceride lipase (ATGL), a key lipolytic enzyme, reducing lipolysis (59). Fat-specific Sirt6 knockout not only induces obesity and insulin resistance but also impairs the thermogenic function of brown adipocytes (232). Yao et al. (232) found Sirt6 deletion decreases ATF2 binding to the PGC-1α promoter, leading to reducing the expression of PGC-1α and PGC-1α target thermogenic genes.
The hepatic SIRT6 level is reduced in obese/diabetic mice and gluconeogenic genes were higher in Sirt6-deficient livers whereas ectopic re-expression of SIRT6 suppressed gluconeogenesis and normalizes glycemia (228, 233). Mechanistically, SIRT6 interacts with and increases the activity of general control non-repressed protein 5 (GCN5), an acetyltransferase, which, in turn, catalyzes the acetylation of PGC-1α, suppressing gluconeogenic gene expression such as phosphoenolpyruvate carboxykinase C (PEPCK-C) and glucose 6-phosphatase, and resulting in repression of hepatic glucose output (228). p53 directly activates expression of SIRT6, which subsequently interacts with and deacetylates FoxO1, leading to FoxO1 export to the cytoplasm, and finally, reduce the expression of gluconeogenetic genes such as glucose 6-phosphatase alpha and phosphoenolpyruvate carboxykinase 1 (234). Human fatty liver samples exhibited significantly lower levels of SIRT6 than normal controls and liver-specific deletion of Sirt6 in mice causes increased glycolysis, triglyceride synthesis, reduced β-oxidation, and leads to liver steatosis (235). Rosiglitazone, an agonist of PPARγ, increases the expression of SIRT6, PGC-1α, and FoxO1, and AMPK phosphorylation in rat liver and ameliorates hepatic lipid accumulation (236). Sirt6 knockdown abolished the effects of rosiglitazone (236), suggesting Sirt6 at least partly mediates the metabolic effects of rosiglitazone. Altogether, those evidence suggest that SIRT6 significantly participates in glucose and lipid metabolism in the liver.
SIRT6 also regulates metabolic homeostasis in the skeletal muscle. Sirt6 transgenic mice show enhanced insulin sensitivity in skeletal muscle and exhibit enhanced insulin-induced activation of Akt in the gastrocnemius (153). By contrast, skeletal muscle-specific Sirt6 ko mice exhibit impaired glucose homeostasis and insulin sensitivity, attenuating whole-body energy expenditure (237). Mechanistically, Sirt6 deletion decreases AMPK activity and subsequently decreases the expression of genes involved in glucose and lipid uptake, fatty acid oxidation, and mitochondrial oxidative phosphorylation (237). Further studies are needed to elucidate the direct mechanism underlying SIRT6 function in skeletal muscle.
SIRT7 is the least characterized Sirtuin of the seven mammalian Sirtuins. SIRT7 protein levels are high in the liver, spleen, and testis, whereas are low in the muscle, heart, and brain of mice (16). In human, Sirt7 mRNA is expressed in various tissues (10). Recent reports clarify the important roles of SIRT7 in a variety of biological processes including DNA repair, chromatin assembly, and aging. However, the role of SIRT7 in metabolism remains largely unknown. The expression of Sirt7 mRNA level is upregulated in adipose tissues of obese patients (238). In HFD-fed mice, Sirt7 knockout decreased the expression of the fatty acid transporter CD36 in WAT (239). In addition, Sirt7 knockout led to an increase of thermogenesis along with increased expression of UCP1 and DIO2 in BAT (239). These results suggest SIRT7 regulates lipid metabolism in adipocytes. Recently, Fang al et. found that SIRT7 restricts SIRT1 activity by preventing SIRT1 auto-deacetylation, and increasing SIRT1 activity in Sirt7-KO mice blocks PPARγ and adipocyte differentiation, thereby decreases the accumulation of white fat (240, 241). Together, these findings implicate the important role of SIRT7 in the regulation of fatty acid metabolism.
Up to now, there are three studies linking SIRT7 to the liver lipid metabolism using independently generated mouse models. Shin et al. reported that Sirt7-KO mice developed steatosis resembling human fatty liver disease (242). Selectively overexpression of SIRT7 in the liver of Sirt7-KO mice via adeno-associated virus 8 (AAV8)-mediated gene transfer prevents the development of fatty liver (242). The authors found expressions of inflammatory markers and lipogenic genes are increased in Sirt7-deficient livers, and they clarified the underlying mechanism as SIRT7 repressing the expression of ribosomal proteins through decreasing Myc activity and further suppressing ER stress (242). Ryu et al. generated a different Sirt7-KO mouse by deleting exons 6-9, and observed more general metabolic defects including hepatic microvesicular steatosis, increased blood lactate levels, reduced exercise performance, cardiac dysfunction and age-related hearing loss induced by multisystemic mitochondrial dysfunction (127). Mechanistically, SIRT7 deacetylates GABPβ1, thereby enables it to form the transcriptionally active GABPα/GABPβ heterotetramer, and then promotes mitochondria function (127). Another study has the opposite result. Yoshizawa et al. reported that Sirt7-KO mice, deleting exons 4-9, are resistant to HFD induced fatty liver, obesity, and glucose intolerance (239). TR4 is a nuclear receptor involved in lipid metabolism and its target genes increase fatty acid uptake and triglyceride synthesis and storage (243). Hepatic SIRT7 was reported to increase TR4 expression through binding with DCAF1/DDB1/CUL4B E3 ubiquitin ligase complex and inhibiting TR4 degradation (239). It is difficult to explain the divergence of three Sirt7-KO mouse models with different genetic background. Liver-specific knockout or Sirt7 transgene mouse model may be helpful to clarify the role of SIRT7 in liver lipid metabolism (239, 242). In addition to lipid metabolism, SIRT7 is involved in glucose metabolism. SIRT7 regulates acetylation at the K323 site of phosphoglycerate kinase 1 (PGK1), an important enzyme in glycolysis, decreases PGK1 enzyme activity and inhibits glycolysis in liver cancer cells (244). Yoshizawa et al. Found that Sirt7-KO mice show decreased expression of the hepatic glucose-6-phosphatase catalytic subunit (G6PC), a key gluconeogenic enzyme, and resistance to glucose intolerance (239). Mechanistically, glucose deprivation stimulates SIRT7 binding to the promoter of G6PC, and deacetylating H3K18 in the G6PC promoter, which results in elevated G6PC expression and promotion of hepatic gluconeogenesis (245).
Aging is a complex process accompanied by the declines in basal metabolic rate and physical activity. Aging is one of the major risk factors contributing to the development of insulin resistance, obesity, T2DM and metabolic syndrome (246). During the aging process, chronic inflammation and mitochondria dysfunction in pancreatic β cells and insulin-sensitive organs have been demonstrated to be major mechanisms linking aging and insulin resistance (247–250). As mentioned above, Sirtuins play important roles in regulating inflammation and mitochondria function. Sirtuins are critically involved in lifespan and healthspan. Deficiency of Sirtuins (SIRT1, SIRT6, and SIRT7) is associated with shortened lifespan and metabolic diseases (251). Our recent evidence also demonstrated that SIRT2 deficiency also facilitated the aging-related development of cardiac dysfunction, including hypertrophy and fibrosis (252). By contrast, germline or cell-specific overexpression of SIRT1 or SIRT6 were reported to expand lifespan and defense metabolic diseases in insulin-dependent and independent manners (253–255).
Sirtuin-targeted strategies show promising in repressing aging-related insulin resistance and metabolic diseases. For instance, the SIRT1 activator SRT1720 extends lifespan and improves the health of mice fed a standard diet (256, 163). It is well established that caloric restriction (CR), the Sirtuin activator, is an effective and reliable means to defense against aging and extend the lifespan and healthspan of mammals, including monkeys (257). Activation of vascular SIRT1 by CR leads to the repressing of aging-related metabolic vascular diseases, including atherosclerosis and aortic aneurysm (258–260). In human studies, CR also can reduce insulin resistance significantly and delay the onset of metabolic diseases (261, 262). Although the mechanisms by which CR extend lifespan are not fully understood, Sirtuins have been implicated to mediate beneficial effects of CR on aging (263). Notably, the CR mimetics (metformin, resveratrol, rapamycin) could expand lifespan and repress diseases related to insulin resistance in rodents partially through activation of Sirtuins (257, 264). Our data showed that SIRT2 contributes to the effects of metformin on aging-related diseases, including cardiac remodeling (252). Currently, clinical trials investigating the anti-aging effects of metformin is undergoing (ClinicalTrials.gov Identifier: NCT02432287).
Importantly, the Sirtuins do not function in individual metabolic organs or cell types alone during aging. Instead, the Sirtuins orchestrate the crosstalk between different organs or between different cell types within the local microenvironmental niche to maintain metabolic homeostasis and prevent against insulin resistance. The SIRT1 activator SRT3025 provides atheroprotection in Apoe−/− mice by reducing hepatic Pcsk9 secretion and enhancing Ldlr expression (265). Resveratrol activates duodenal SIRT1 to initiate a gut-brain-liver neuronal axis that improves hypothalamic insulin sensitivity in rats (183). SIRT1 in intestinal stem cells also contributes to the protection roles of caloric restriction on aging (266). In addition, SIRT3 activation by nitrite and metformin improves insulin sensitivity in skeletal muscle and normalizes pulmonary hypertension associated with heart failure with preserved ejection fraction (267). Sirtuins also regulate inflammatory cells within the local microenvironmental niches to regulate insulin resistance in an autocrine or paracrine manner (41, 79, 80).
Therefore, targeting Sirtuins could be a promising strategy for improvement of insulin sensitivity and metabolic status of the whole body. However, activation of Sirtuins alone may not archive the biggest benefits because of the exhaustion of the endogenous NAD. Sirtuin activator in supplement with NAD precursor may represent a better therapeutic strategy for repressing aging-related insulin resistance and metabolic diseases.
Insulin resistance is a critical pathological feature of obesity and metabolic syndrome and plays a key role in the pathogenesis of T2DM and attendant cardiovascular complications. Moreover, insulin resistance provides a therapeutic strategy to prevent, delay or treat T2DM, obesity, and metabolic syndrome by improving insulin sensitivity. Although insulin resistance is a complex metabolic disorder that has remained poorly understood, Sirtuin family members are involved in the potential cellular mechanisms of the pathogenesis of insulin resistance. According to accumulating evidence in the past decades, Sirtuin family members have emerged as a nutrient sensor to maintain energy homeostasis. Cellular and animal studies have demonstrated that Sirtuins play an important role in regulating glucose and lipids by modulating crucial enzymes in metabolic pathways and interfering with inflammation, oxidative stress, mitochondrial dysfunction, ER stress, and the insulin signaling pathway.
Sirtuins respond to environmental (diet and lifestyle) or metabolic (obesity, fasting, and diabetes) insults at mRNA and protein levels in insulin-sensing organs (268, 269). The roles of Sirtuins in regulating glucose and lipid metabolism as well as insulin resistance in liver, adipose tissue, and skeletal muscle make their importance in regulating metabolic diseases, including T2DM and diabetic complications (269).
Nevertheless, many additional studies are needed.
1. Different Sirtuins may have the same downstream targets, such as FoxO3a, FoxO1, PGC-1α, and GDH, and there is cross-talk among Sirtuin family members (117, 56, 270). How do different Sirtuin members coordinate to regulate the same downstream targets?
2. The Sirtuin family comprises NAD+-dependent histone deacetylases; however, recent results have revealed that Sirtuin members can act in a deacetylase-independent manner (117). How can we determine the functions and activities of Sirtuins in addition to their deacetylation function in insulin resistance?
3. In addition to Sirtuins, there are other epigenetic modification enzymes, including SUV39H1 and EZH2 are involved in insulin resistance and T2DM (271–276). There is an interaction between SUV39H1 and Sirtuins, including SIRT1, SIRT3, and SIRT7 (277–281). EZH2 is reportedly the deacetylating substrate of SIRT1 (282, 283). SIRT2 negatively regulates JMJD2A expression in human non-small cell lung cancer tissues (284). Is JMJD2A involved in insulin resistance? In the condition of insulin resistance, how do these epigenetic modification enzymes influence each other and consequently act on insulin sensitivity?
4. Obesity, insulin resistance, and T2DM are aging-related abnormalities. Sirtuins, especially SIRT1, SIRT2, and SIRT6, are characterized as protectors of aging and aging-related diseases. Whether the promotive effect of aging by the decline of Sirtuin activity is involved in insulin resistance deserves further investigation. In addition, cell senescence-induced organ dysfunction and aging (senescaging) are common during physiological and pathological aging processes (268). Selective elimination of senescent cells, or senolysis, was reported to delay aging and aging-related metabolic diseases including congestive decline, atherosclerosis, cardiac diseases, and osteoarthritis (285–291). However, it remains to elucidate that what are the physiological and pathological functions of cellular senescence in organs during aging and that whether Sirtuins regulate senescaging in insulin resistance and healthy conditions.
All authors listed have made a substantial, direct and intellectual contribution to the work. SZ is responsible for literature collection and article draft. XT designed the Figures and revised the manuscript. H-ZC is the leading principal investigator who directed the study and data analysis, and prepared the manuscript. All authors approved publication of this work.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
This work was supported by the National Natural Science Foundation of China (Grant Nos. 81600038, 31571193 and 81800273). H-ZC is also supported by the Youth Top-notch Talent Support Program and the Youth Yangtze River Scholar Program in China. XT is also supported by the Postdoctoral Innovative Talents Support Program (BX20180206), the China Postdoctoral Science Foundation (2018M631084) and Full-time Postdoctoral Research and Development Fund of Sichuan University (2018SCU12010). We apologize to those scientists whose work we could not highlight owing to space limitations. We thank Miss. Xiao-Feng Chen for reading the manuscript and helpful suggestions.
1. Samuel VT, Shulman GI. Mechanisms for insulin resistance: common threads and missing links. Cell (2012) 148:852–71. doi: 10.1016/j.cell.2012.02.017
2. Saltiel AR. New perspectives into the molecular pathogenesis and treatment of type 2 diabetes. Cell (2001) 104:517–29. doi: 10.1016/S0092-8674(01)00239-2
3. Sun C, Zhang F, Ge X, Yan T, Chen X, Shi X, et al. SIRT1 improves insulin sensitivity under insulin-resistant conditions by repressing PTP1B. Cell Metab. (2007) 6:307–19. doi: 10.1016/j.cmet.2007.08.014
4. Gomes P, Fleming Outeiro T, Cavadas C. Emerging role of sirtuin 2 in the regulation of mammalian metabolism. Trends Pharmacol Sci. (2015) 36:756–68. doi: 10.1016/j.tips.2015.08.001
5. Kim JA, Wei Y, Sowers JR. Role of mitochondrial dysfunction in insulin resistance. Circul Res. (2008) 102:401–14. doi: 10.1161/CIRCRESAHA.107.165472
6. Muoio DM, Newgard CB. Mechanisms of disease: molecular and metabolic mechanisms of insulin resistance and beta-cell failure in type 2 diabetes. Nat Rev. (2008) 9:193–205. doi: 10.1038/nrm2327
7. Nogueiras R, Habegger KM, Chaudhary N, Finan B, Banks AS, Dietrich MO, et al. Sirtuin 1 and sirtuin 3: physiological modulators of metabolism. Physiol Rev. (2012) 92:1479–514. doi: 10.1152/physrev.00022.2011
8. Frye RA. Characterization of five human cDNAs with homology to the yeast SIR2 gene: Sir2-like proteins (sirtuins) metabolize NAD and may have protein ADP-ribosyltransferase activity. Biochem Biophys Res Commun. (1999) 260:273–9. doi: 10.1006/bbrc.1999.0897
9. North BJ, Verdin E. Sirtuins: Sir2-related NAD-dependent protein deacetylases. Genome Biol. (2004) 5:224. doi: 10.1186/gb-2004-5-5-224
10. Michishita E, Park JY, Burneskis JM, Barrett JC, Horikawa I. Evolutionarily conserved and nonconserved cellular localizations and functions of human SIRT proteins. Mol Biol Cell (2005) 16:4623–35. doi: 10.1091/mbc.e05-01-0033
11. Tanno M, Sakamoto J, Miura T, Shimamoto K, Horio Y. Nucleocytoplasmic shuttling of the NAD+-dependent histone deacetylase SIRT1. J Biol Chem. (2007) 282:6823–32. doi: 10.1074/jbc.M609554200
12. North BJ, Marshall BL, Borra MT, Denu JM, Verdin E. The human Sir2 ortholog, SIRT2, is an NAD+-dependent tubulin deacetylase. Mol Cell (2003) 11:437–44. doi: 10.1016/S1097-2765(03)00038-8
13. Vaquero A, Scher MB, Lee DH, Sutton A, Cheng HL, Alt FW, et al. SIRT2 is a histone deacetylase with preference for histone H4 Lys 16 during mitosis. Genes Dev. (2006) 20:1256–61. doi: 10.1101/gad.1412706
14. Huang JY, Hirschey MD, Shimazu T, Ho L, Verdin E. Mitochondrial sirtuins. Biochim Biophys Acta (2010) 1804:1645–51. doi: 10.1016/j.bbapap.2009.12.021
15. Mostoslavsky R, Chua KF, Lombard DB, Pang WW, Fischer MR, Gellon L, et al. Genomic instability and aging-like phenotype in the absence of mammalian SIRT6. Cell (2006) 124:315–29. doi: 10.1016/j.cell.2005.11.044
16. Ford E, Voit R, Liszt G, Magin C, Grummt I, Guarente L. Mammalian Sir2 homolog SIRT7 is an activator of RNA polymerase I transcription. Genes Dev. (2006) 20:1075–80. doi: 10.1101/gad.1399706
17. Rauh D, Fischer F, Gertz M, Lakshminarasimhan M, Bergbrede T, Aladini F, et al. An acetylome peptide microarray reveals specificities and deacetylation substrates for all human sirtuin isoforms. Nat Commun. (2013) 4:2327. doi: 10.1038/ncomms3327
18. Teng YB, Jing H, Aramsangtienchai P, He B, Khan S, Hu J, et al. Efficient demyristoylase activity of SIRT2 revealed by kinetic and structural studies. Sci Rep. (2015) 5:8529. doi: 10.1038/srep08529
19. Du J, Zhou Y, Su X, Yu JJ, Khan S, Jiang H, et al. SIRT5 is a NAD-dependent protein lysine demalonylase and desuccinylase. Science (2011) 334:806–9. doi: 10.1126/science.1207861
20. Haigis MC, Mostoslavsky R, Haigis KM, Fahie K, Christodoulou DC, Murphy AJ, et al. SIRT4 inhibits glutamate dehydrogenase and opposes the effects of calorie restriction in pancreatic beta cells. Cell (2006) 126:941–54. doi: 10.1016/j.cell.2006.06.057
21. Liszt G, Ford E, Kurtev M, Guarente L. Mouse Sir2 homolog SIRT6 is a nuclear ADP-ribosyltransferase. J Biol Chem. (2005) 280:21313–20. doi: 10.1074/jbc.M413296200
22. Guarente L, Franklin H. Epstein lecture: sirtuins, aging, and medicine. N Engl J Med. (2011) 364:2235–44. doi: 10.1056/NEJMra1100831
23. Morris BJ. Seven sirtuins for seven deadly diseases of aging. Free Radic Biol Med. (2013) 56:133–71. doi: 10.1016/j.freeradbiomed.2012.10.525
24. Liang F, Kume S, Koya D. SIRT1 and insulin resistance. Nat Rev. Endocrinol. (2009) 5:367–73. doi: 10.1038/nrendo.2009.101
25. Cao Y, Jiang X, Ma H, Wang Y, Xue P, Liu Y. SIRT1 and insulin resistance. J Diabetes Compl. (2016) 30:178–83. doi: 10.1016/j.jdiacomp.2015.08.022
26. Kuang J, Chen L, Tang Q, Zhang J, Li Y, He J. The role of SIRT6 in obesity and diabetes. Front Physiol. (2018) 9:135. doi: 10.3389/fphys.2018.00135
27. Wellen KE, Hotamisligil GS. Inflammation, stress, and diabetes. J Clin Investig. (2005) 115:1111–9. doi: 10.1172/JCI25102
28. Olefsky JM, Glass CK. Macrophages, inflammation, and insulin resistance. Annu Rev Physiol. (2010) 72:219–46. doi: 10.1146/annurev-physiol-021909-135846
29. Lumeng CN, Saltiel AR. Inflammatory links between obesity and metabolic disease. J Clin Investig. (2011) 121:2111–7. doi: 10.1172/JCI57132
30. Scherer PE. Adipose tissue: from lipid storage compartment to endocrine organ. Diabetes (2006) 55:1537–45. doi: 10.2337/db06-0263
31. Wang P, Mariman E, Renes J, Keijer J. The secretory function of adipocytes in the physiology of white adipose tissue. J Cell Physiol. (2008) 216:3–13. doi: 10.1002/jcp.21386
32. Steppan CM, Bailey ST, Bhat S, Brown EJ, Banerjee RR, Wright CM, et al. The hormone resistin links obesity to diabetes. Nature (2001) 409:307–12. doi: 10.1038/35053000
33. Yang Q, Graham TE, Mody N, Preitner F, Peroni OD, Zabolotny JM, et al. Serum retinol binding protein 4 contributes to insulin resistance in obesity and type 2 diabetes. Nature (2005) 436:356–62. doi: 10.1038/nature03711
34. Hirosumi J, Tuncman G, Chang L, Gorgun CZ, Uysal KT, Maeda K, et al. A central role for JNK in obesity and insulin resistance. Nature (2002) 420:333–6. doi: 10.1038/nature01137
35. Zick Y. Role of Ser/Thr kinases in the uncoupling of insulin signaling. Int J Obes Related Metab Disord. (2003) 27(Suppl. 3):S56–60. doi: 10.1038/sj.ijo.0802503
36. Jing E, Emanuelli B, Hirschey MD, Boucher J, Lee KY, Lombard D, et al. Sirtuin-3 (SIRT3) regulates skeletal muscle metabolism and insulin signaling via altered mitochondrial oxidation and reactive oxygen species production. Proc Natl Acad Sci USA (2011) 108:14608–13. doi: 10.1073/pnas.1111308108
37. Pfluger PT, Herranz D, Velasco-Miguel S, Serrano M, Tschop MH. SIRT1 protects against high-fat diet-induced metabolic damage. Proc Natl Acad Sci USA (2008) 105:9793–8. doi: 10.1073/pnas.0802917105
38. Purushotham A, Schug TT, Xu Q, Surapureddi S, Guo X, Li X. Hepatocyte-specific deletion of SIRT1 alters fatty acid metabolism and results in hepatic steatosis and inflammation. Cell Metab. (2009) 9:327–38. doi: 10.1016/j.cmet.2009.02.006
39. Yoshizaki T, Milne JC, Imamura T, Schenk S, Sonoda N, Babendure JL, et al. SIRT1 exerts anti-inflammatory effects and improves insulin sensitivity in adipocytes. Mol Cell Biol. (2009) 29:1363–74. doi: 10.1128/MCB.00705-08
40. Yoshizaki T, Schenk S, Imamura T, Babendure JL, Sonoda N, Bae EJ, et al. SIRT1 inhibits inflammatory pathways in macrophages and modulates insulin sensitivity. Am J Physiol. Endocrinol Metab. (2010) 298:E419–428. doi: 10.1152/ajpendo.00417.2009
41. Gillum MP, Kotas ME, Erion DM, Kursawe R, Chatterjee P, Nead KT, et al. SIRT1 regulates adipose tissue inflammation. Diabetes (2011) 60:3235–45. doi: 10.2337/db11-0616
42. Hui X, Zhang M, Gu P, Li K, Gao Y, Wu D, et al. Adipocyte SIRT1 controls systemic insulin sensitivity by modulating macrophages in adipose tissue. EMBO Rep. (2017) 18:645–57. doi: 10.15252/embr.201643184
43. Yeung F, Hoberg JE, Ramsey CS, Keller MD, Jones DR, Frye RA, et al. Modulation of NF-kappaB-dependent transcription and cell survival by the SIRT1 deacetylase. EMBO J. (2004) 23:2369–80. doi: 10.1038/sj.emboj.7600244
44. Ghosh HS, Spencer JV, Ng B, McBurney MW, Robbins PD. SIRT1 interacts with transducin-like enhancer of split-1 to inhibit nuclear factor kappaB-mediated transcription. Biochem J. (2007) 408:105–111. doi: 10.1042/BJ20070817
45. Zhang R, Chen HZ, Liu JJ, Jia YY, Zhang ZQ, Yang RF, et al. SIRT1 suppresses activator protein-1 transcriptional activity and cyclooxygenase-2 expression in macrophages. J Biol Chem. (2010) 285:7097–110. doi: 10.1074/jbc.M109.038604
46. Nakamura K, Zhang M, Kageyama S, Ke B, Fujii T, Sosa RA, et al. Macrophage heme oxygenase-1-SIRT1-p53 axis regulates sterile inflammation in liver ischemia-reperfusion injury. J Hepatol. (2017) 67:1232–42. doi: 10.1016/j.jhep.2017.08.010
47. Rothgiesser KM, Erener S, Waibel S, Luscher B, Hottiger MO. SIRT2 regulates NF-kappaB dependent gene expression through deacetylation of p65 Lys310. J Cell Sci. (2010) 123:4251–8. doi: 10.1242/jcs.073783
48. Pais TF, Szego EM, Marques O, Miller-Fleming L, Antas P, Guerreiro P, et al. The NAD-dependent deacetylase sirtuin 2 is a suppressor of microglial activation and brain inflammation. EMBO J. (2013) 32:2603–16. doi: 10.1038/emboj.2013.200
49. Lo Sasso G, Menzies KJ, Mottis A, Piersigilli A, Perino A, Yamamoto H, et al. SIRT2 deficiency modulates macrophage polarization and susceptibility to experimental colitis. PLoS ONE (2014) 9:e103573. doi: 10.1371/journal.pone.0103573
50. Zhang Y, Chi D. Overexpression of SIRT2 alleviates neuropathic pain and neuroinflammation through deacetylation of transcription factor nuclear factor-kappa B. Inflammation (2018) 41:569–78. doi: 10.1007/s10753-017-0713-3
51. Yuan F, Xu ZM, Lu LY, Nie H, Ding J, Ying WH et al. SIRT2 inhibition exacerbates neuroinflammation and blood–brain barrier disruption in experimental traumatic brain injury by enhancing NF-κB p65 acetylation and activation. J Neurochem. (2016) 136:581–93. doi: 10.1111/jnc.13423
52. Lin J, Sun B, Jiang C, Hong H, Zheng Y. SIRT2 suppresses inflammatory responses in collagen-induced arthritis. Biochem Biophys Res Commun. (2013) 441:897–903. doi: 10.1016/j.bbrc.2013.10.153
53. Wang X, Buechler NL, Martin A, Wells J, Yoza B, McCall CE, et al. Sirtuin-2 regulates sepsis inflammation in ob/ob mice. PLoS ONE (2016) 11:e0160431. doi: 10.1371/journal.pone.0160431
54. Chen Y, Wang H, Luo G, Dai X. SIRT4 inhibits cigarette smoke extracts-induced mononuclear cell adhesion to human pulmonary microvascular endothelial cells via regulating NF-kappaB activity. Toxicol Lett. (2014) 226:320–7. doi: 10.1016/j.toxlet.2014.02.022
55. Tao Y, Huang C, Huang Y, Hong L, Wang H, Zhou Z, et al. SIRT4 suppresses inflammatory responses in human umbilical vein endothelial cells. Cardiovasc Toxicol. (2015) 15:217–23. doi: 10.1007/s12012-014-9287-6
56. Qin K, Han C, Zhang H, Li T, Li N, Cao X. NAD(+) dependent deacetylase Sirtuin 5 rescues the innate inflammatory response of endotoxin tolerant macrophages by promoting acetylation of p65. J Autoimmun. (2017) 81:120–9. doi: 10.1016/j.jaut.2017.04.006
57. Wang F, Wang K, Xu W, Zhao S, Ye D, Wang Y, et al. SIRT5 desuccinylates and activates pyruvate kinase M2 to block macrophage IL-1beta production and to prevent DSS-induced colitis in mice. Cell Rep. (2017) 19:2331–44. doi: 10.1016/j.celrep.2017.05.065
58. Kawahara TL, Michishita E, Adler AS, Damian M, Berber E, Lin M, et al. SIRT6 links histone H3 lysine 9 deacetylation to NF-kappaB-dependent gene expression and organismal life span. Cell (2009) 136:62–74. doi: 10.1016/j.cell.2008.10.052
59. Kuang J, Zhang Y, Liu Q, Shen J, Pu S, Cheng S, et al. Fat-specific SIRT6 ablation sensitizes mice to high-fat diet-induced obesity and insulin resistance by inhibiting lipolysis. Diabetes (2017) 66:1159–71. doi: 10.2337/db16-1225
60. Xiong X, Zhang C, Zhang Y, Fan R, Qian X, Dong XC. Fabp4-Cre-mediated SIRT6 deletion impairs adipose tissue function and metabolic homeostasis in mice. J Endocrinol. (2017) 233:307–14. doi: 10.1530/JOE-17-0033
61. Franchi L, Eigenbrod T, Munoz-Planillo R, Nunez G. The inflammasome: a caspase-1-activation platform that regulates immune responses and disease pathogenesis. Nat Immunol. (2009) 10:241–7. doi: 10.1038/ni.1703
62. Lamkanfi M, Dixit VM. Mechanisms and functions of inflammasomes. Cell (2014) 157:1013–22. doi: 10.1016/j.cell.2014.04.007
63. Youm YH, Nguyen KY, Grant RW, Goldberg EL, Bodogai M, Kim D, et al. The ketone metabolite beta-hydroxybutyrate blocks NLRP3 inflammasome-mediated inflammatory disease. Nat Med. (2015) 21:263–9. doi: 10.1038/nm.3804
64. Vandanmagsar B, Youm YH, Ravussin A, Galgani JE, Stadler K, Mynatt RL, et al. The NLRP3 inflammasome instigates obesity-induced inflammation and insulin resistance. Nat Med. (2011) 17:179–88. doi: 10.1038/nm.2279
65. Lee HM, Kim JJ, Kim HJ, Shong M, Ku BJ, Jo EK. Upregulated NLRP3 inflammasome activation in patients with type 2 diabetes. Diabetes (2013) 62:194–204. doi: 10.2337/db12-0420
66. Fu Y, Wang Y, Du L, Xu C, Cao J, Fan T, et al. Resveratrol inhibits ionising irradiation-induced inflammation in MSCs by activating SIRT1 and limiting NLRP-3 inflammasome activation. Int J Mol Sci. (2013) 14:14105–18. doi: 10.3390/ijms140714105
67. Gao R, Ma Z, Hu Y, Chen J, Shetty S, Fu J. SIRT1 restrains lung inflammasome activation in a murine model of sepsis. Am J Physiol. (2015) 308:L847–853. doi: 10.1152/ajplung.00274.2014
68. Zhang B, Xu D, She L, Wang Z, Yang N, Sun R, et al. Silybin inhibits NLRP3 inflammasome assembly through the NAD(+)/SIRT2 pathway in mice with nonalcoholic fatty liver disease. FASEB J. (2018) 32:757–67. doi: 10.1096/fj.201700602R
69. Traba J, Kwarteng-Siaw M, Okoli TC, Li J, Huffstutler RD, Bray A, et al. Fasting and refeeding differentially regulate NLRP3 inflammasome activation in human subjects. J Clin Investig. (2015) 125:4592–600. doi: 10.1172/JCI83260
70. Chen ML, Zhu XH, Lang HD, Mi MT. Trimethylamine-N-oxide induces vascular inflammation by activating the NLRP3 inflammasome through the SIRT3-SOD2-mtROS signaling pathway. J Am Heart Assoc. (2017) 6:e006347. doi: 10.1161/JAHA.117.006347
71. Harris J, Hartman M, Roche C, Zeng SG, O'Shea A, Sharp FA, et al. Autophagy controls IL-1beta secretion by targeting pro-IL-1beta for degradation. J Biol Chem. (2011) 286:9587–97. doi: 10.1074/jbc.M110.202911
72. Chen Y, Li X, Boini KM, Pitzer AL, Gulbins E, Zhang Y, et al. Endothelial Nlrp3 inflammasome activation associated with lysosomal destabilization during coronary arteritis. Biochim Biophys Acta (2015) 1853:396–408. doi: 10.1016/j.bbamcr.2014.11.012
73. Liu P, Huang G, Wei T, Gao J, Huang C, Sun M, et al. Sirtuin 3-induced macrophage autophagy in regulating NLRP3 inflammasome activation. Biochim Biophys Acta (2018) 1864:764–77. doi: 10.1016/j.bbadis.2017.12.027
74. Nakahira K, Haspel JA, Rathinam VA, Lee SJ, Dolinay T, Lam HC, et al. Autophagy proteins regulate innate immune responses by inhibiting the release of mitochondrial DNA mediated by the NALP3 inflammasome. Nat Immunol. (2011) 12:222–230. doi: 10.1038/ni.1980
75. Cho KW, Lumeng CN. SIRT1: a guardian at the gates of adipose tissue inflammation. Diabetes (2011) 60:3100–2. doi: 10.2337/db11-1308
76. Zhang J, Lee SM, Shannon S, Gao B, Chen W, Chen A, et al. The type III histone deacetylase SIRT1 is essential for maintenance of T cell tolerance in mice. J Clin Invest. (2009) 119:3048–58. doi: 10.1172/JCI38902
77. Liu Z, Gan L, Liu G, Chen Y, Wu T, Feng F, et al. SIRT1 decreased adipose inflammation by interacting with Akt2 and inhibiting mTOR/S6K1 pathway in mice. J Lipid Res. (2016) 57:1373–81. doi: 10.1194/jlr.M063537
78. Jiang H, Khan S, Wang Y, Charron G, He B, Sebastian C, et al. SIRT6 regulates TNF-alpha secretion through hydrolysis of long-chain fatty acyl lysine. Nature (2013) 496:110–3. doi: 10.1038/nature12038
79. Lee Y, Ka SO, Cha HN, Chae YN, Kim MK, Park SY, et al. Myeloid sirtuin 6 deficiency causes insulin resistance in high-fat diet–fed mice by eliciting macrophage polarization toward an m1 phenotype. Diabetes (2017) 66:2659–68. doi: 10.2337/db16-1446
80. Zhang ZQ, Ren SC, Tan Y, Li ZZ, Tang X, Wang TT, et al. Epigenetic regulation of NKG2D ligands is involved in exacerbated atherosclerosis development in SIRT6 heterozygous mice. Sci Rep. (2016) 6:23912. doi: 10.1038/srep23912
81. Xiao C, Wang RH, Lahusen TJ, Park O, Bertola A, Maruyama T, et al. Progression of chronic liver inflammation and fibrosis driven by activation of c-JUN signaling in SIRT6 mutant mice. J Biol Chem. (2012) 287:41903–13. doi: 10.1074/jbc.M112.415182
82. Liu TF, Vachharajani VT, Yoza BK, McCall CE. NAD+-dependent sirtuin 1 and 6 proteins coordinate a switch from glucose to fatty acid oxidation during the acute inflammatory response. J Biol Chem. (2012) 287:25758–69. doi: 10.1074/jbc.M112.362343
83. Pham-Huy LA, He H, Pham-Huy C. Free radicals, antioxidants in disease and health. Int J Biomed Sci. (2008) 4:89–96.
84. Singh CK, Chhabra G, Ndiaye MA, Garcia-Peterson LM, Mack NJ, Ahmad N. The role of sirtuins in antioxidant and redox signaling. Antiox Redox Signal. (2018) 28:643–61. doi: 10.1089/ars.2017.7290
85. Maiese K. New insights for oxidative stress and diabetes mellitus. Oxidat Med Cell Long. (2015) 2015:875961. doi: 10.1155/2015/875961
86. Kelley DE, He J, Menshikova EV, Ritov VB. Dysfunction of mitochondria in human skeletal muscle in type 2 diabetes. Diabetes (2002) 51:2944–50. doi: 10.2337/diabetes.51.10.2944
87. Petersen KF, Dufour S, Befroy D, Garcia R, Shulman GI. Impaired mitochondrial activity in the insulin-resistant offspring of patients with type 2 diabetes. N Engl J Med. (2004) 350:664–71. doi: 10.1056/NEJMoa031314
88. Lowell BB, Shulman GI. Mitochondrial dysfunction and type 2 diabetes. Science (2005) 307:384–7. doi: 10.1126/science.1104343
89. Wu Z, Puigserver P, Andersson U, Zhang C, Adelmant G, Mootha V, et al. Mechanisms controlling mitochondrial biogenesis and respiration through the thermogenic coactivator PGC-1. Cell (1999) 98:115–24. doi: 10.1016/S0092-8674(00)80611-X
90. Finck BN, Kelly DP. PGC-1 coactivators: inducible regulators of energy metabolism in health and disease. J Clin Investig. (2006) 116:615–22. doi: 10.1172/JCI27794
91. Nemoto S, Fergusson MM, Finkel T. SIRT1 functionally interacts with the metabolic regulator and transcriptional coactivator PGC-1{alpha}. J Biol Chem. (2005) 280:16456–60. doi: 10.1074/jbc.M501485200
92. Lagouge M, Argmann C, Gerhart-Hines Z, Meziane H, Lerin C, Daussin F, et al. Resveratrol improves mitochondrial function and protects against metabolic disease by activating SIRT1 and PGC-1alpha. Cell (2006) 127:1109–22. doi: 10.1016/j.cell.2006.11.013
93. Lee IH, Cao L, Mostoslavsky R, Lombard DB, Liu J, Bruns NE, et al. A role for the NAD-dependent deacetylase SIRT1 in the regulation of autophagy. Proc Natl Acad Sci USA (2008) 105:3374–9. doi: 10.1073/pnas.0712145105
94. Hariharan N, Maejima Y, Nakae J, Paik J, Depinho RA, Sadoshima J. Deacetylation of FoxO by SIRT1 Plays an Essential Role in Mediating Starvation-Induced Autophagy in Cardiac Myocytes, Circ Res. (2010) 107:1470–82. doi: 10.1161/CIRCRESAHA.110.227371
95. Kume S, Uzu T, Horiike K, Chin-Kanasaki M, Isshiki K, Araki S, et al. Calorie restriction enhances cell adaptation to hypoxia through SIRT1-dependent mitochondrial autophagy in mouse aged kidney. J Clin Investig. (2010) 120:1043–55. doi: 10.1172/JCI41376
96. Brunet A, Sweeney LB, Sturgill JF, Chua KF, Greer PL, Lin Y, et al. Stress-dependent regulation of FOXO transcription factors by the SIRT1 deacetylase. Science (2004) 303:2011–15. doi: 10.1126/science.1094637
97. Zhang B, Zhai M, Li B, Liu Z, Li K, Jiang L, et al. Honokiol ameliorates myocardial ischemia/reperfusion injury in type 1 diabetic rats by reducing oxidative stress and apoptosis through activating the SIRT1-Nrf2 signaling pathway. Oxidat Med Cell Long. (2018) 2018:3159801. doi: 10.1155/2018/3159801
98. Wang F, Nguyen M, Qin FX, Tong Q. SIRT2 deacetylates FOXO3a in response to oxidative stress and caloric restriction. Aging Cell (2007) 6:505–14. doi: 10.1111/j.1474-9726.2007.00304.x
99. Lynn EG, McLeod CJ, Gordon JP, Bao J, Sack MN. SIRT2 is a negative regulator of anoxia-reoxygenation tolerance via regulation of 14-3-3 zeta and BAD in H9c2 cells. FEBS Lett. (2008) 582:2857–62. doi: 10.1016/j.febslet.2008.07.016
100. Wang YP, Zhou LS, Zhao YZ, Wang SW, Chen LL, Liu LX, et al. Regulation of G6PD acetylation by SIRT2 and KAT9 modulates NADPH homeostasis and cell survival during oxidative stress. EMBO J. (2014) 33:1304–20. doi: 10.1002/embj.201387224
101. Xu Y, Li F, Lv L, Li T, Zhou X, Deng CX, et al. Oxidative stress activates SIRT2 to deacetylate and stimulate phosphoglycerate mutase. Cancer Res. (2014) 74:3630–42. doi: 10.1158/0008-5472.CAN-13-3615
102. Galligan JJ, Rose KL, Beavers WN, Hill S, Tallman KA, Tansey WP, et al. Stable histone adduction by 4-oxo-2-nonenal: a potential link between oxidative stress and epigenetics. J Am Chem Soc. (2014) 136:11864–6. doi: 10.1021/ja503604t
103. Jin J, He B, Zhang X, Lin H, Wang Y. SIRT2 reverses 4-oxononanoyl lysine modification on histones. J Am Chem Soc. (2016) 138:12304–7. doi: 10.1021/jacs.6b04977
104. Liu G, Park SH, Imbesi M, Nathan WJ, Zou X, Zhu Y, et al. Loss of NAD-dependent protein deacetylase sirtuin-2 alters mitochondrial protein acetylation and dysregulates mitophagy. Antioxid Redox Signal. (2017) 26:849–63. doi: 10.1089/ars.2016.6662
105. Weir HJ, Lane JD, Balthasar N. SIRT3: a central regulator of mitochondrial adaptation in health and disease. Genes Cancer (2013) 4:118–24. doi: 10.1177/1947601913476949
106. Ahn BH, Kim HS, Song S, Lee IH, Liu J, Vassilopoulos A, et al. A role for the mitochondrial deacetylase SIRT3 in regulating energy homeostasis. Proc Natl Acad Sci USA (2008) 105:14447–52. doi: 10.1073/pnas.0803790105
107. Cimen H, Han MJ, Yang Y, Tong Q, Koc H, Koc EC. Regulation of succinate dehydrogenase activity by SIRT3 in mammalian mitochondria. Biochemistry (2010) 49:304–11. doi: 10.1021/bi901627u
108. Hafner AV, Dai J, Gomes AP, Xiao CY, Palmeira CM, Rosenzweig A, et al. Regulation of the mPTP by SIRT3-mediated deacetylation of CypD at lysine 166 suppresses age-related cardiac hypertrophy. Aging (2010) 2:914–23. doi: 10.18632/aging.100252
109. Cheng Y, Ren X, Gowda AS, Shan Y, Zhang L, Yuan YS, et al. Interaction of SIRT3 with OGG1 contributes to repair of mitochondrial DNA and protects from apoptotic cell death under oxidative stress. Cell Death Dis. (2013) 4:e731. doi: 10.1038/cddis.2013.254
110. Someya S, Yu W, Hallows WC, Xu J, Vann JM, Leeuwenburgh C, et al. SIRT3 mediates reduction of oxidative damage and prevention of age-related hearing loss under caloric restriction. Cell (2010) 143:802–12. doi: 10.1016/j.cell.2010.10.002
111. Finley LW, Carracedo A, Lee J, Souza A, Egia A, Zhang J, et al. SIRT3 opposes reprogramming of cancer cell metabolism through HIF1alpha destabilization. Cancer Cell (2011) 19:416–28. doi: 10.1016/j.ccr.2011.02.014
112. Sundaresan NR, Gupta M, Kim G, Rajamohan SB, Isbatan A, Gupta MP. SIRT3 blocks the cardiac hypertrophic response by augmenting Foxo3a-dependent antioxidant defense mechanisms in mice. J Clin Investig. (2009) 119:2758–71. doi: 10.1172/JCI39162
113. Qiu X, Brown K, Hirschey MD, Verdin E, Chen D. Calorie restriction reduces oxidative stress by SIRT3-mediated SOD2 activation. Cell Metab. (2010) 12:662–7. doi: 10.1016/j.cmet.2010.11.015
114. Chen Y, Zhang J, Lin Y, Lei Q, Guan KL, Zhao S, et al. Tumour suppressor SIRT3 deacetylates and activates manganese superoxide dismutase to scavenge ROS. EMBO Rep. (2011) 12:534–41. doi: 10.1038/embor.2011.65
115. Tao R, Coleman MC, Pennington JD, Ozden O, Park SH, Jiang H, et al. SIRT3-mediated deacetylation of evolutionarily conserved lysine 122 regulates MnSOD activity in response to stress. Mol Cell (2010) 40:893–904. doi: 10.1016/j.molcel.2010.12.013
116. Yu W, Dittenhafer-Reed KE, Denu JM. SIRT3 protein deacetylates isocitrate dehydrogenase 2 (IDH2) and regulates mitochondrial redox status. J Biol Chem. (2012) 287:14078–86. doi: 10.1074/jbc.M112.355206
117. Luo YX, Tang X, An XZ, Xie XM, Chen XF, Zhao X, et al. SIRT4 accelerates Ang II-induced pathological cardiac hypertrophy by inhibiting manganese superoxide dismutase activity. Eur Heart J. (2017) 38:1389–98. doi: 10.1093/eurheartj/ehw138
118. Nasrin N, Wu X, Fortier E, Feng Y, Bare OC, Chen S, et al. SIRT4 regulates fatty acid oxidation and mitochondrial gene expression in liver and muscle cells. J Biol Chem. (2010) 285:31995–2002. doi: 10.1074/jbc.M110.124164
119. Ho L, Titus AS, Banerjee KK, George S, Lin W, Deota S, et al. SIRT4 regulates ATP homeostasis and mediates a retrograde signaling via AMPK. Aging (2013) 5:835–49. doi: 10.18632/aging.100616
120. Lang A, Anand R, Altinoluk-Hambuchen S, Ezzahoini H, Stefanski A, Iram A, et al. SIRT4 interacts with OPA1 and regulates mitochondrial quality control and mitophagy. Aging (2017) 9:2163–89. doi: 10.18632/aging.101307
121. Nakagawa T, Lomb DJ, Haigis MC, Guarente L. SIRT5 Deacetylates carbamoyl phosphate synthetase 1 and regulates the urea cycle. Cell (2009) 137:560–70. doi: 10.1016/j.cell.2009.02.026
122. Lin ZF, Xu HB, Wang JY, Lin Q, Ruan Z, Liu FB, et al. SIRT5 desuccinylates and activates SOD1 to eliminate ROS. Biochem Biophys Res Commun. (2013) 441:191–5. doi: 10.1016/j.bbrc.2013.10.033
123. Zhou L, Wang F, Sun R, Chen X, Zhang M, Xu Q, et al. SIRT5 promotes IDH2 desuccinylation and G6PD deglutarylation to enhance cellular antioxidant defense. EMBO Rep. (2016) 17:811–22. doi: 10.15252/embr.201541643
124. Chen XF, Tian MX, Sun RQ, Zhang ML, Zhou LS, Jin L, et al. SIRT5 inhibits peroxisomal ACOX1 to prevent oxidative damage and is downregulated in liver cancer. EMBO Rep. (2018) 19:e45124. doi: 10.15252/embr.201745124
125. Pan H, Guan D, Liu X, Li J, Wang L, Wu J, et al. SIRT6 safeguards human mesenchymal stem cells from oxidative stress by coactivating NRF2. Cell Res. (2016) 26:190–205. doi: 10.1038/cr.2016.4
126. Wang XX, Wang XL, Tong MM, Gan L, Chen H, Wu SS, et al. SIRT6 protects cardiomyocytes against ischemia/reperfusion injury by augmenting FoxO3alpha-dependent antioxidant defense mechanisms. Basic Res Cardiol. (2016) 111:13. doi: 10.1007/s00395-016-0531-z
127. Ryu D, Jo YS, Lo Sasso G, Stein S, Zhang H, Perino A, et al. A SIRT7-dependent acetylation switch of GABPbeta1 controls mitochondrial function. Cell Metab. (2014) 20:856–69. doi: 10.1016/j.cmet.2014.08.001
128. Mohrin M, Shin J, Liu Y, Brown K, Luo H, Xi Y, et al. Stem cell aging. A mitochondrial UPR-mediated metabolic checkpoint regulates hematopoietic stem cell aging. Science (2015) 347:1374–7. doi: 10.1126/science.aaa2361
129. Xiong X, Sun X, Wang Q, Qian X, Zhang Y, Pan X, et al. SIRT6 protects against palmitate-induced pancreatic beta-cell dysfunction and apoptosis. J Endocrinol. (2016) 231:159–65. doi: 10.1530/JOE-16-0317
130. Qin K, Zhang N, Zhang Z, Nipper M, Zhu Z, Leighton J, et al. SIRT6-mediated transcriptional suppression of Txnip is critical for pancreatic beta cell function and survival in mice. Diabetologia (2018) 61:906–18. doi: 10.1007/s00125-017-4542-6
131. Moynihan KA, Grimm AA, Plueger MM, Bernal-Mizrachi E, Ford E, Cras-Meneur C, et al. Increased dosage of mammalian Sir2 in pancreatic beta cells enhances glucose-stimulated insulin secretion in mice. Cell Metab. (2005) 2:105–17. doi: 10.1016/j.cmet.2005.07.001
132. Bordone L, Motta MC, Picard F, Robinson A, Jhala US, Apfeld J, et al. SIRT1 regulates insulin secretion by repressing UCP2 in pancreatic beta cells. PLoS Biol. (2006) 4:e31. doi: 10.1371/journal.pbio.0040031
133. Kitamura YI, Kitamura T, Kruse JP, Raum JC, Stein R, Gu W, et al. FoxO1 protects against pancreatic beta cell failure through NeuroD and MafA induction. Cell Metab. (2005) 2:153–63. doi: 10.1016/j.cmet.2005.08.004
134. Anderson KA, Huynh FK, Fisher-Wellman K, Stuart JD, Peterson BS, Douros JD, et al. SIRT4 is a lysine deacylase that controls leucine metabolism and insulin secretion. Cell Metab. (2017) 25:838-855.e815. doi: 10.1016/j.cmet.2017.03.003
135. Lombard DB, Alt FW, Cheng HL, Bunkenborg J, Streeper RS, Mostoslavsky R, et al. Mammalian Sir2 homolog SIRT3 regulates global mitochondrial lysine acetylation. Mol Cell Biol. (2007) 27:8807–14. doi: 10.1128/MCB.01636-07
136. Caton PW, Richardson SJ, Kieswich J, Bugliani M, Holland ML, Marchetti P, et al. Sirtuin 3 regulates mouse pancreatic beta cell function and is suppressed in pancreatic islets isolated from human type 2 diabetic patients. Diabetologia (2013) 56:1068–77. doi: 10.1007/s00125-013-2851-y
137. Zhou Y, Chung ACK, Fan R, Lee HM, Xu G, Tomlinson B, et al. SIRT3 deficiency increased the vulnerability of pancreatic beta cells to oxidative stress-induced dysfunction. Antiox Redox Signal. (2017) 27:962–76. doi: 10.1089/ars.2016.6859
138. Kim M, Lee JS, Oh JE, Nan J, Lee H, Jung HS, et al. SIRT3 Overexpression attenuates palmitate-induced pancreatic beta-cell dysfunction. PLoS ONE (2015) 10:e0124744. doi: 10.1371/journal.pone.0124744
139. Zhang HH, Ma XJ, Wu LN, Zhao YY, Zhang PY, Zhang YH, et al. Sirtuin-3 (SIRT3) protects pancreatic beta-cells from endoplasmic reticulum (ER) stress-induced apoptosis and dysfunction. Mol Cell Biochem. (2016) 420:95–106. doi: 10.1007/s11010-016-2771-5
140. Song MY, Wang J, Ka SO, Bae EJ, Park BH. Insulin secretion impairment in SIRT6 knockout pancreatic beta cells is mediated by suppression of the FoxO1-Pdx1-Glut2 pathway. Sci Rep. (2016) 6:30321. doi: 10.1038/srep30321
141. Xiong X, Wang G, Tao R, Wu P, Kono T, Li K, et al. Sirtuin 6 regulates glucose-stimulated insulin secretion in mouse pancreatic beta cells. Diabetologia (2016) 59:151–60. doi: 10.1007/s00125-015-3778-2
142. Taniguchi CM, Emanuelli B, Kahn CR. Critical nodes in signalling pathways: insights into insulin action. Nat Rev Mol Cell Biol. (2006) 7:85–96. doi: 10.1038/nrm1837
143. Zhang J. The direct involvement of SIRT1 in insulin-induced insulin receptor substrate-2 tyrosine phosphorylation. J Biol Chem. (2007) 282:34356–364. doi: 10.1074/jbc.M706644200
144. Sundaresan NR, Pillai VB, Wolfgeher D, Samant S, Vasudevan P, Parekh V, et al. The deacetylase SIRT1 promotes membrane localization and activation of Akt and PDK1 during tumorigenesis and cardiac hypertrophy. Sci Signal. (2011) 4:ra46. doi: 10.1126/scisignal.2001465
145. Dan L, Klimenkova O, Klimiankou M, Klusman JH, van den Heuvel-Eibrink MM, Reinhardt D, et al. The role of sirtuin 2 activation by nicotinamide phosphoribosyltransferase in the aberrant proliferation and survival of myeloid leukemia cells. Haematologica (2012) 97:551–9. doi: 10.3324/haematol.2011.055236
146. Chen J, Chan AW, To KF, Chen W, Zhang Z, Ren J, et al. SIRT2 overexpression in hepatocellular carcinoma mediates epithelial to mesenchymal transition by protein kinase B/glycogen synthase kinase-3beta/beta-catenin signaling. Hepatology (2013) 57:2287–98. doi: 10.1002/hep.26278
147. Ramakrishnan G, Davaakhuu G, Kaplun L, Chung WC, Rana A, Atfi A, et al. SIRT2 deacetylase is a novel AKT binding partner critical for AKT activation by insulin. J Biol Chem. (2014) 289:6054–66. doi: 10.1074/jbc.M113.537266
148. Arora A, Dey CS. SIRT2 negatively regulates insulin resistance in C2C12 skeletal muscle cells. Biochim Biophys Acta (2014) 1842:1372–8. doi: 10.1016/j.bbadis.2014.04.027
149. Arora A, Dey CS. SIRT2 regulates insulin sensitivity in insulin resistant neuronal cells. Biochem Biophys Res Commun. (2016) 474:747–52. doi: 10.1016/j.bbrc.2016.05.029
150. Jing E, Gesta S, Kahn CR. SIRT2 regulates adipocyte differentiation through FoxO1 acetylation/deacetylation. Cell Metab. (2007) 6:105–14. doi: 10.1016/j.cmet.2007.07.003
151. Belman JP, Bian RR, Habtemichael EN, Li DT, Jurczak MJ, Alcazar-Roman A, et al. Acetylation of TUG protein promotes the accumulation of GLUT4 glucose transporters in an insulin-responsive intracellular compartment. J Biol Chem. (2015) 290:4447–63. doi: 10.1074/jbc.M114.603977
152. Xiao C, Kim HS, Lahusen T, Wang RH, Xu X, Gavrilova O, et al. SIRT6 deficiency results in severe hypoglycemia by enhancing both basal and insulin-stimulated glucose uptake in mice. J Biol Chem. (2010) 285:36776–84. doi: 10.1074/jbc.M110.168039
153. Anderson JG, Ramadori G, Ioris RM, Galie M, Berglund ED, Coate KC, et al. Enhanced insulin sensitivity in skeletal muscle and liver by physiological overexpression of SIRT6. Mol Metab. (2015) 4:846–56. doi: 10.1016/j.molmet.2015.09.003
154. Yu J, Qin B, Wu F, Qin S, Nowsheen S, Shan S, et al. Regulation of serine-threonine kinase Akt Activation by NAD+-dependent deacetylase SIRT7. Cell Rep. (2017) 18:1229–40. doi: 10.1016/j.celrep.2017.01.009
155. Saltiel AR, Kahn CR. Insulin signalling and the regulation of glucose and lipid metabolism. Nature (2001) 414:799–806. doi: 10.1038/414799a
156. Farmer SR. Transcriptional control of adipocyte formation. Cell Metab. (2006) 4:263–273. doi: 10.1016/j.cmet.2006.07.001
157. Puigserver P, Rhee J, Donovan J, Walkey CJ, Yoon JC, Oriente F, et al. Insulin-regulated hepatic gluconeogenesis through FOXO1-PGC-1alpha interaction. Nature (2003) 423:550–5. doi: 10.1038/nature01667
158. Bouche C, Serdy S, Kahn CR, Goldfine AB. The cellular fate of glucose and its relevance in type 2 diabetes. Endocrine Rev. (2004) 25:807–30. doi: 10.1210/er.2003-0026
159. Weyer C, Bogardus C, Mott DM, Pratley RE. The natural history of insulin secretory dysfunction and insulin resistance in the pathogenesis of type 2 diabetes mellitus. J Clin Investigat. (1999) 104:787–94. doi: 10.1172/JCI7231
160. Smith AG, Muscat GE. Skeletal muscle and nuclear hormone receptors: implications for cardiovascular and metabolic disease. Int J Biochem Cell Biol. (2005) 37:2047–63. doi: 10.1016/j.biocel.2005.03.002
161. Jing E, O'Neill BT, Rardin MJ, Kleinridders A, Ilkeyeva OR, Ussar S, et al. SIRT3 regulates metabolic flexibility of skeletal muscle through reversible enzymatic deacetylation. Diabetes (2013) 62:3404–17. doi: 10.2337/db12-1650
162. Palacios OM, Carmona JJ, Michan S, Chen KY, Manabe Y, Ward JL, et al. Diet and exercise signals regulate SIRT3 and activate AMPK and PGC-1alpha in skeletal muscle. Aging (2009) 1:771–783. doi: 10.18632/aging.100075
163. Milne JC, Lambert PD, Schenk S, Carney DP, Smith JJ, Gagne DJ, et al. Small molecule activators of SIRT1 as therapeutics for the treatment of type 2 diabetes. Nature (2007) 450:712–6. doi: 10.1038/nature06261
164. Frescas D, Valenti L, Accili D. Nuclear trapping of the forkhead transcription factor FoxO1 via SIRT-dependent deacetylation promotes expression of glucogenetic genes. J Biol Chem. (2005) 280:20589–95. doi: 10.1074/jbc.M412357200
165. Rodgers JT, Lerin C, Haas W, Gygi SP, Spiegelman BM, Puigserver P. Nutrient control of glucose homeostasis through a complex of PGC-1alpha and SIRT1. Nature (2005) 434:113–8. doi: 10.1038/nature03354
166. Liu Y, Dentin R, Chen D, Hedrick S, Ravnskjaer K, Schenk S, et al. A fasting inducible switch modulates gluconeogenesis via activator/coactivator exchange. Nature (2008) 456:269–73. doi: 10.1038/nature07349
167. Hallows WC, Yu W, Denu JM. Regulation of glycolytic enzyme phosphoglycerate mutase-1 by SIRT1 protein-mediated deacetylation. J Biol Chem. (2012) 287:3850–8. doi: 10.1074/jbc.M111.317404
168. Li Y, Wong K, Giles A, Jiang J, Lee JW, Adams AC, et al. Hepatic SIRT1 attenuates hepatic steatosis and controls energy balance in mice by inducing fibroblast growth factor 21. Gastroenterology (2014) 146:539–549.e537. doi: 10.1053/j.gastro.2013.10.059
169. Li Y, Xu S, Giles A, Nakamura K, Lee JW, Hou X, et al. Hepatic overexpression of SIRT1 in mice attenuates endoplasmic reticulum stress and insulin resistance in the liver. FASEB J. (2011) 25:1664–79. doi: 10.1096/fj.10-173492
170. Ajmo JM, Liang X, Rogers CQ, Pennock B, You M. Resveratrol alleviates alcoholic fatty liver in mice. Am J Physiol. (2008) 295:G833–842. doi: 10.1152/ajpgi.90358.2008
171. Yamazaki Y, Usui I, Kanatani Y, Matsuya Y, Tsuneyama K, Fujisaka S, et al. Treatment with SRT1720, a SIRT1 activator, ameliorates fatty liver with reduced expression of lipogenic enzymes in MSG mice. Am J Physiol. (2009) 297:E1179–1186. doi: 10.1152/ajpendo.90997.2008
172. Walker AK, Yang F, Jiang K, Ji JY, Watts JL, Purushotham A, et al. Conserved role of SIRT1 orthologs in fasting-dependent inhibition of the lipid/cholesterol regulator SREBP. Genes Dev. (2010) 24:1403–17. doi: 10.1101/gad.1901210
173. Hou X, Xu S, Maitland-Toolan KA, Sato K, Jiang B, Ido Y, et al. SIRT1 regulates hepatocyte lipid metabolism through activating AMP-activated protein kinase. J Biol Chem. (2008) 283:20015–26. doi: 10.1074/jbc.M802187200
174. Gesta S, Tseng YH, Kahn CR. Developmental origin of fat: tracking obesity to its source. Cell (2007) 131:242–56. doi: 10.1016/j.cell.2007.10.004
175. Picard F, Kurtev M, Chung N, Topark-Ngarm A, Senawong T, Machado De Oliveira R, et al. SIRT1 promotes fat mobilization in white adipocytes by repressing PPAR-gamma. Nature (2004) 429:771–6. doi: 10.1038/nature02583
176. Guan HP, Li Y, Jensen MV, Newgard CB, Steppan CM, Lazar MA. A futile metabolic cycle activated in adipocytes by antidiabetic agents. Nat Med. (2002) 8:1122–8. doi: 10.1038/nm780
177. Kelley DE, Goodpaster BH, Storlien L. Muscle triglyceride and insulin resistance. Annu Rev Nutr. (2002) 22:325–46. doi: 10.1146/annurev.nutr.22.010402.102912
178. Gerhart-Hines Z, Rodgers JT, Bare O, Lerin C, Kim SH, Mostoslavsky R, et al. Metabolic control of muscle mitochondrial function and fatty acid oxidation through SIRT1/PGC-1alpha. EMBO J. (2007) 26:1913–23. doi: 10.1038/sj.emboj.7601633
179. Canto C, Gerhart-Hines Z, Feige JN, Lagouge M, Noriega L, Milne JC, et al. AMPK regulates energy expenditure by modulating NAD+ metabolism and SIRT1 activity. Nature (2009) 458:1056–60. doi: 10.1038/nature07813
180. Frojdo S, Durand C, Molin L, Carey AL, El-Osta A, Kingwell BA, et al. Phosphoinositide 3-kinase as a novel functional target for the regulation of the insulin signaling pathway by SIRT1. Mol Cell Endocrinol. (2011) 335:166–76. doi: 10.1016/j.mce.2011.01.008
181. White AT, McCurdy CE, Philp A, Hamilton DL, Johnson CD, Schenk S. Skeletal muscle-specific overexpression of SIRT1 does not enhance whole-body energy expenditure or insulin sensitivity in young mice. Diabetologia (2013) 56:1629–37. doi: 10.1007/s00125-013-2912-2
182. White AT, Philp A, Fridolfsson HN, Schilling JM, Murphy AN, Hamilton DL, et al. High-fat diet-induced impairment of skeletal muscle insulin sensitivity is not prevented by SIRT1 overexpression. Am J Physiol. (2014) 307:E764–772. doi: 10.1152/ajpendo.00001.2014
183. Côté CD, Rasmussen BA, Duca FA, Zadeh-Tahmasebi M, Baur JA, Daljeet M, et al. Resveratrol activates duodenal SIRT1 to reverse insulin resistance in rats through a neuronal network. Nat Med. (2015) 21:498. doi: 10.1038/nm.3821
184. James Ryall G, Dell'Orso S, Derfoul A, Juan A, Zare H, Feng X, et al. The NAD+-dependent SIRT1 deacetylase translates a metabolic switch into regulatory epigenetics in skeletal muscle stem cells. Cell Stem Cell (2015) 16:171–83. doi: 10.1016/j.stem.2014.12.004
185. Gurd BJ, Perry CGR, Heigenhauser GJF, Spriet LL, Bonen A. High-intensity interval training increases SIRT1 activity in human skeletal muscle. Appl Physiol Nutr Metab. (2010) 35:350–7. doi: 10.1139/H10-030
186. Crujeiras AB, Parra D, Goyenechea E, Martinez JA. Sirtuin gene expression in human mononuclear cells is modulated by caloric restriction. Eur J Clin Investig. (2008) 38:672–8. doi: 10.1111/j.1365-2362.2008.01998.x
187. Krishnan J, Danzer C, Simka T, Ukropec J, Walter KM, Kumpf S, et al. Dietary obesity-associated Hif1alpha activation in adipocytes restricts fatty acid oxidation and energy expenditure via suppression of the SIRT2-NAD+ system. Genes Dev. (2012) 26:259–70. doi: 10.1101/gad.180406.111
188. Arab Sadeghabadi Z, Nourbakhsh M, Pasalar P, Emamgholipour S, Golestani A, Larijani B, et al. Reduced gene expression of sirtuins and active AMPK levels in children and adolescents with obesity and insulin resistance. Obes Res Clin Pract. (2018) 12:167–73. doi: 10.1016/j.orcp.2017.10.004
189. Nakae J, Kitamura T, Kitamura Y, Biggs WH 3rd, Arden KC, Accili D. The forkhead transcription factor Foxo1 regulates adipocyte differentiation. Dev Cell (2003) 4:119–29. doi: 10.1016/S1534-5807(02)00401-X
190. Dowell P, Otto TC, Adi S, Lane MD. Convergence of peroxisome proliferator-activated receptor gamma and Foxo1 signaling pathways. J Biol Chem. (2003) 278:45485–91. doi: 10.1074/jbc.M309069200
191. Armoni M, Harel C, Karni S, Chen H, Bar-Yoseph F, Ver MR, et al. FOXO1 represses peroxisome proliferator-activated receptor-gamma1 and -gamma2 gene promoters in primary adipocytes. A novel paradigm to increase insulin sensitivity. J Biol Chem. (2006) 281:19881–91. doi: 10.1074/jbc.M600320200
192. Wang F, Tong Q. SIRT2 suppresses adipocyte differentiation by deacetylating FOXO1 and enhancing FOXO1's repressive interaction with PPARgamma. Mol Biol Cell (2009) 20:801–8. doi: 10.1091/mbc.e08-06-0647
193. Strable MS, Ntambi JM. Genetic control of de novo lipogenesis: role in diet-induced obesity. Crit Rev Biochem Mol Biol. (2010) 45:199–214. doi: 10.3109/10409231003667500
194. Wellen KE, Hatzivassiliou G, Sachdeva UM, Bui TV, Cross JR, Thompson CB. ATP-citrate lyase links cellular metabolism to histone acetylation. Science (2009) 324:1076–80. doi: 10.1126/science.1164097
195. Lin R, Tao R, Gao X, Li T, Zhou X, Guan KL, et al. Acetylation stabilizes ATP-citrate lyase to promote lipid biosynthesis and tumor growth. Mol Cell (2013) 51:506–18. doi: 10.1016/j.molcel.2013.07.002
196. Vega RB, Huss JM, Kelly DP. The coactivator PGC-1 cooperates with peroxisome proliferator-activated receptor alpha in transcriptional control of nuclear genes encoding mitochondrial fatty acid oxidation enzymes. Mol Cell Biol. (2000) 20:1868–76. doi: 10.1128/MCB.20.5.1868-1876.2000
197. Zhao S, Xu W, Jiang W, Yu W, Lin Y, Zhang T, et al. Regulation of cellular metabolism by protein lysine acetylation. Science (2010) 327:1000–4. doi: 10.1126/science.1179689
198. Jiang W, Wang S, Xiao M, Lin Y, Zhou L, Lei Q, et al. Acetylation regulates gluconeogenesis by promoting PEPCK1 degradation via recruiting the UBR5 ubiquitin ligase. Mol Cell (2011) 43:33–44. doi: 10.1016/j.molcel.2011.04.028
199. Herzig S, Long F, Jhala US, Hedrick S, Quinn R, Bauer A, et al. CREB regulates hepatic gluconeogenesis through the coactivator PGC-1. Nature (2001) 413:179–83. doi: 10.1038/35093131
200. Nakae J, Biggs WH 3rd, Kitamura T, Cavenee WK, Wright CV, Arden KC, et al. Regulation of insulin action and pancreatic beta-cell function by mutated alleles of the gene encoding forkhead transcription factor Foxo1. Nat Genet. (2002) 32:245–53. doi: 10.1038/ng890
201. Yoon JC, Puigserver P, Chen G, Donovan J, Wu Z, Rhee J, et al. Control of hepatic gluconeogenesis through the transcriptional coactivator PGC-1. Nature (2001) 413:131–8. doi: 10.1038/35093050
202. Onyango P, Celic I, McCaffery JM, Boeke JD, Feinberg AP. SIRT3, a human SIR2 homologue, is an NAD-dependent deacetylase localized to mitochondria. Proc Natl Acad Sci USA (2002) 99:13653–8. doi: 10.1073/pnas.222538099
203. Su AI, Wiltshire T, Batalov S, Lapp H, Ching KA, Block D, et al. A gene atlas of the mouse and human protein-encoding transcriptomes. Proc Natl Acad Sci USA (2004) 101:6062–7. doi: 10.1073/pnas.0400782101
204. Wu C, Orozco C, Boyer J, Leglise M, Goodale J, Batalov S, et al. BioGPS: an extensible and customizable portal for querying and organizing gene annotation resources. Genome Biol. (2009) 10:R130. doi: 10.1186/gb-2009-10-11-r130
205. Han E, Hilsenbeck SG, Richardson A, Nelson JF. cDNA expression arrays reveal incomplete reversal of age-related changes in gene expression by calorie restriction. Mech Ageing Dev. (2000) 115:157–74. doi: 10.1016/S0047-6374(00)00119-6
206. Shi T, Wang F, Stieren E, Tong Q. SIRT3, a mitochondrial sirtuin deacetylase, regulates mitochondrial function and thermogenesis in brown adipocytes. J Biol Chem. (2005) 280:13560–7. doi: 10.1074/jbc.M414670200
207. Schwer B, Eckersdorff M, Li Y, Silva JC, Fermin D, Kurtev MV, et al. Calorie restriction alters mitochondrial protein acetylation. Aging Cell (2009) 8:604–6. doi: 10.1111/j.1474-9726.2009.00503.x
208. Hallows WC, Yu W, Smith BC, Devries MK, Ellinger JJ, Someya S, et al. SIRT3 promotes the urea cycle and fatty acid oxidation during dietary restriction. Mol Cell (2011) 41:139–49. doi: 10.1016/j.molcel.2011.01.002
209. Hirschey MD, Shimazu T, Jing E, Grueter CA, Collins AM, Aouizerat B, et al. SIRT3 deficiency and mitochondrial protein hyperacetylation accelerate the development of the metabolic syndrome. Mol Cell (2011) 44:177–90. doi: 10.1016/j.molcel.2011.07.019
210. Hirschey MD, Shimazu T, Goetzman E, Jing E, Schwer B, Lombard DB, et al. SIRT3 regulates mitochondrial fatty-acid oxidation by reversible enzyme deacetylation. Nature (2010) 464:121–5. doi: 10.1038/nature08778
211. Garcia-Fuentes E, Santiago-Fernandez C, Gutierrez-Repiso C, Mayas MD, Oliva-Olivera W, Coin-Araguez L, et al. Hypoxia is associated with a lower expression of genes involved in lipogenesis in visceral adipose tissue. J Transl Med. (2015) 13:373. doi: 10.1186/s12967-015-0732-5
212. Zamora-Mendoza R, Rosas-Vargas H, Ramos-Cervantes MT, Garcia-Zuniga P, Perez-Lorenzana H, Mendoza-Lorenzo P, et al. Dysregulation of mitochondrial function and biogenesis modulators in adipose tissue of obese children. Int J Obes. (2018) 42:618–24. doi: 10.1038/ijo.2017.274
213. Kurtz DM, Rinaldo P, Rhead WJ, Tian L, Millington DS, Vockley J, et al. Targeted disruption of mouse long-chain acyl-CoA dehydrogenase gene reveals crucial roles for fatty acid oxidation. Proc Natl Acad Sci USA (1998) 95:15592–7. doi: 10.1073/pnas.95.26.15592
214. Zhang D, Liu ZX, Choi CS, Tian L, Kibbey R, Dong J, et al. Shulman GI, Mitochondrial dysfunction due to long-chain Acyl-CoA dehydrogenase deficiency causes hepatic steatosis and hepatic insulin resistance. Proc Natl Acad Sci USA (2007) 104:17075–80. doi: 10.1073/pnas.0707060104
215. Fernandez-Marcos PJ, Jeninga EH, Canto C, Harach T, de Boer VC, Andreux P, et al. Muscle or liver-specific SIRT3 deficiency induces hyperacetylation of mitochondrial proteins without affecting global metabolic homeostasis. Sci Rep. (2012) 2:425. doi: 10.1038/srep00425
216. Yechoor VK, Patti ME, Ueki K, Laustsen PG, Saccone R, Rauniyar R, et al. Distinct pathways of insulin-regulated versus diabetes-regulated gene expression: an in vivo analysis in MIRKO mice. Proc Natl Acad Sci USA (2004) 101:16525–30. doi: 10.1073/pnas.0407574101
217. Lantier L, Williams AS, Williams IM, Yang KK, Bracy DP, Goelzer M, et al. SIRT3 is crucial for maintaining skeletal muscle insulin action and protects against severe insulin resistance in high-fat-fed mice. Diabetes (2015) 64:3081–92. doi: 10.2337/db14-1810
218. Chen YR, Fang SR, Fu YC, Zhou XH, Xu MY, Xu WC. Calorie restriction on insulin resistance and expression of SIRT1 and SIRT4 in rats. Biochem Cell Biol. (2010) 88:715–22. doi: 10.1139/O10-010
219. de Pinho L, Andrade JM, Paraiso A, Filho AB, Feltenberger JD, Guimaraes AL, et al. Diet composition modulates expression of sirtuins and renin-angiotensin system components in adipose tissue. Obesity (2013) 21:1830–5. doi: 10.1002/oby.20305
220. Laurent G, German NJ, Saha AK, de Boer VC, Davies M, Koves TR, et al. SIRT4 coordinates the balance between lipid synthesis and catabolism by repressing malonyl CoA decarboxylase. Mol Cell (2013) 50:686–98. doi: 10.1016/j.molcel.2013.05.012
221. Laurent G, de Boer VC, Finley LW, Sweeney M, Lu H, Schug TT, et al. SIRT4 represses peroxisome proliferator-activated receptor alpha activity to suppress hepatic fat oxidation. Mol Cell Biol. (2013) 33:4552–61. doi: 10.1128/MCB.00087-13
222. Wu T, Liu YH, Fu YC, Liu XM, Zhou XH. Direct evidence of sirtuin downregulation in the liver of non-alcoholic fatty liver disease patients. Ann Clin Labor Sci. (2014) 44:410–8.
223. Rardin MJ, He W, Nishida Y, Newman JC, Carrico C, Danielson SR, et al. SIRT5 regulates the mitochondrial lysine succinylome and metabolic networks. Cell Metab. (2013) 18:920–33. doi: 10.1016/j.cmet.2013.11.013
224. Park J, Chen Y, Tishkoff DX, Peng C, Tan M, Dai L, et al. SIRT5-mediated lysine desuccinylation impacts diverse metabolic pathways. Mol Cell (2013) 50:919–30. doi: 10.1016/j.molcel.2013.06.001
225. Yu J, Sadhukhan S, Noriega LG, Moullan N, He B, Weiss RS, et al. Metabolic characterization of a SIRT5 deficient mouse model. Sci Rep. (2013) 3:2806. doi: 10.1038/srep02806
226. Jukarainen S, Heinonen S, Ramo JT, Rinnankoski-Tuikka R, Rappou E, Tummers M, et al. Obesity is associated with low NAD(+)/SIRT pathway expression in adipose tissue of BMI-discordant monozygotic twins. J Clin Endocrinol Metab. (2016) 101:275–83. doi: 10.1210/jc.2015-3095
227. Nishida Y, Rardin MJ, Carrico C, He W, Sahu AK, Gut P, et al. SIRT5 Regulates both cytosolic and mitochondrial protein malonylation with glycolysis as a major target. Mol Cell (2015) 59:321–32. doi: 10.1016/j.molcel.2015.05.022
228. Dominy JE Jr, Lee Y, Jedrychowski MP, Chim H, Jurczak MJ, Camporez JP, et al. The deacetylase SIRT6 activates the acetyltransferase GCN5 and suppresses hepatic gluconeogenesis. Mol Cell (2012) 48:900–13. doi: 10.1016/j.molcel.2012.09.030
229. Moschen AR, Wieser V, Gerner RR, Bichler A, Enrich B, Moser P, et al. Adipose tissue and liver expression of SIRT1, 3, and 6 increase after extensive weight loss in morbid obesity. J Hepatol. (2013) 59:1315–22. doi: 10.1016/j.jhep.2013.07.027
230. Chen Q, Hao W, Xiao C, Wang R, Xu X, Lu H, et al. SIRT6 is essential for adipocyte differentiation by regulating mitotic clonal expansion. Cell Rep. (2017) 18:3155–66. doi: 10.1016/j.celrep.2017.03.006
231. Kanfi Y, Peshti V, Gil R, Naiman S, Nahum L, Levin E, et al. SIRT6 protects against pathological damage caused by diet-induced obesity. Aging Cell (2010) 9:162–73. doi: 10.1111/j.1474-9726.2009.00544.x
232. Yao L, Cui X, Chen Q, Yang X, Fang F, Zhang J, et al. Cold-inducible SIRT6 regulates thermogenesis of brown and beige fat. Cell Rep. (2017) 20:641–54. doi: 10.1016/j.celrep.2017.06.069
233. Zhong L, D'Urso A, Toiber D, Sebastian C, Henry RE, Vadysirisack DD, et al. The histone deacetylase SIRT6 regulates glucose homeostasis via Hif1alpha. Cell (2010) 140:280–93. doi: 10.1016/j.cell.2009.12.041
234. Zhang P, Tu B, Wang H, Cao Z, Tang M, Zhang C, et al. Tumor suppressor p53 cooperates with SIRT6 to regulate gluconeogenesis by promoting FoxO1 nuclear exclusion. Proc Natl Acad Sci USA (2014) 111:10684–9. doi: 10.1073/pnas.1411026111
235. Kim HS, Xiao C, Wang RH, Lahusen T, Xu X, Vassilopoulos A, et al. Hepatic-specific disruption of SIRT6 in mice results in fatty liver formation due to enhanced glycolysis and triglyceride synthesis. Cell Metab. (2010) 12:224–36. doi: 10.1016/j.cmet.2010.06.009
236. Yang SJ, Choi JM, Chae SW, Kim WJ, Park SE, Rhee EJ, et al. Activation of peroxisome proliferator-activated receptor gamma by rosiglitazone increases SIRT6 expression and ameliorates hepatic steatosis in rats. PLoS ONE (2011) 6:e17057. doi: 10.1371/journal.pone.0017057
237. Cui X, Yao L, Yang X, Gao Y, Fang F, Zhang J, et al. SIRT6 regulates metabolic homeostasis in skeletal muscle through activation of AMPK. Am J Physiol Endocrinol Metab. (2017) 313:E493–E505. doi: 10.1152/ajpendo.00122.2017
238. Kurylowicz A, Owczarz M, Polosak J, Jonas MI, Lisik W, Jonas M, et al. SIRT1 and SIRT7 expression in adipose tissues of obese and normal-weight individuals is regulated by microRNAs but not by methylation status. Int J Obes. (2016) 40:1635–42. doi: 10.1038/ijo.2016.131
239. Yoshizawa T, Karim MF, Sato Y, Senokuchi T, Miyata K, Fukuda T, et al. SIRT7 controls hepatic lipid metabolism by regulating the ubiquitin-proteasome pathway. Cell Metab. (2014) 19:712–21. doi: 10.1016/j.cmet.2014.03.006
240. Fang J, Ianni A, Smolka C, Vakhrusheva O, Nolte H, Kruger M, et al. SIRT7 promotes adipogenesis in the mouse by inhibiting autocatalytic activation of SIRT1. Proc Natl Acad Sci USA (2017) 114:E8352–E8361. doi: 10.1073/pnas.1706945114
241. Cioffi M, Vallespinos-Serrano M, Sara Trabulo M, Pablo Fernandez-Marcos J, Ashley Firment N, Berta Vazquez N, et al. MiR-93 Controls Adiposity via Inhibition of SIRT7 and Tbx3. Cell Rep. (2015) 12:1594–605. doi: 10.1016/j.celrep.2015.08.006
242. Shin J, He M, Liu Y, Paredes S, Villanova L, Brown K, et al. SIRT7 represses Myc activity to suppress ER stress and prevent fatty liver disease. Cell Rep. (2013) 5:654–65. doi: 10.1016/j.celrep.2013.10.007
243. Kang HS, Okamoto K, Kim YS, Takeda Y, Bortner CD, Dang H, et al. Nuclear orphan receptor TAK1/TR4-deficient mice are protected against obesity-linked inflammation, hepatic steatosis, and insulin resistance. Diabetes (2011) 60:177–88. doi: 10.2337/db10-0628
244. Hu H, Zhu W, Qin J, Chen M, Gong L, Li L, et al. Acetylation of PGK1 promotes liver cancer cell proliferation and tumorigenesis. Hepatology (2017) 65:515–28. doi: 10.1002/hep.28887
245. Jiang L, Xiong J, Zhan J, Yuan F, Tang M, Zhang C, et al. Ubiquitin-specific peptidase 7 (USP7)-mediated deubiquitination of the histone deacetylase SIRT7 regulates gluconeogenesis. J Biol Chem. (2017) 292:13296–311. doi: 10.1074/jbc.M117.780130
246. Hall JA, Dominy JE, Lee Y, Puigserver P. The sirtuin family's role in aging and age-associated pathologies. J Clin Investig. (2013) 123:973–9. doi: 10.1172/JCI64094
247. Petersen KF, Befroy D, Dufour S, Dziura J, Ariyan C, Rothman DL, et al. Mitochondrial dysfunction in the elderly: possible role in insulin resistance. Science (2003) 300:1140–2. doi: 10.1126/science.1082889
248. Park MH, Kim DH, Lee EK, Kim ND, Im DS, Lee J, et al. Age-related inflammation and insulin resistance: a review of their intricate interdependency. Arch Pharm Res. (2014) 37:1507–14. doi: 10.1007/s12272-014-0474-6
249. Fabbri E, Chia CW, Spencer RG, Fishbein KW, Reiter DA, Cameron D, et al. Insulin resistance is associated with reduced mitochondrial oxidative capacity measured by 31P-magnetic resonance spectroscopy in participants without diabetes from the baltimore longitudinal study of aging. Diabetes (2017) 66:170–6. doi: 10.2337/db16-0754
250. Wu H, Ballantyne CM. Skeletal muscle inflammation and insulin resistance in obesity. J Clin Investig. (2017) 127:43–54. doi: 10.1172/JCI88880
251. Finkel T, Deng CX. Mostoslavsky R, Recent progress in the biology and physiology of sirtuins. Nature (2009) 460:587–91. doi: 10.1038/nature08197
252. Tang X, Chen XF, Wang NY, Wang XM, Liang ST, Zheng W, et al. SIRT2 acts as a cardioprotective deacetylase in pathological cardiac hypertrophy. Circulation (2017) 136:2051–67. doi: 10.1161/CIRCULATIONAHA.117.028728
253. Kanfi Y, Naiman S, Amir G, Peshti V, Zinman G, Nahum L, et al. The sirtuin SIRT6 regulates lifespan in male mice. Nature (2012) 483:218. doi: 10.1038/nature10815
254. Sundaresan NR, Vasudevan P, Zhong L, Kim G, Samant S, Parekh V, et al. The sirtuin SIRT6 blocks IGF-Akt signaling and development of cardiac hypertrophy by targeting c-Jun. Nat Med. (2012) 18:1643. doi: 10.1038/nm.2961
255. Satoh A, Brace CS, Rensing N, Cliften P, Wozniak DF, Herzog ED, et al. SIRT1 extends life span and delays aging in mice through the regulation of Nk2 homeobox 1 in the DMH and LH. Cell Metab. (2013) 18:416–30. doi: 10.1016/j.cmet.2013.07.013
256. Sarah Mitchell J, Martin-Montalvo A, Evi Mercken M, Hector Palacios H, Theresa Ward M, Abulwerdi G, et al. The SIRT1 activator SRT1720 extends lifespan and improves health of mice fed a standard diet. Cell Rep. (2014) 6:836–43. doi: 10.1016/j.celrep.2014.01.031
257. Madeo F, Pietrocola F, Eisenberg T, Kroemer G. Caloric restriction mimetics: towards a molecular definition. Nat Rev Drug Discov. (2014) 13:727–40. doi: 10.1038/nrd4391
258. Chen HZ, Wang F, Gao P, Pei JF, Liu Y, Xu TT, et al. Age-associated sirtuin 1 reduction in vascular smooth muscle links vascular senescence and inflammation to abdominal aortic aneurysm. Circ Res. (2016) 119:1076–88. doi: 10.1161/CIRCRESAHA.116.308895
259. Liu Y, Wang TT, Zhang R, Fu WY, Wang X, Wang F, et al. Calorie restriction protects against experimental abdominal aortic aneurysms in mice. J Exp Med. (2016) 213:2473–88. doi: 10.1084/jem.20151794
260. Winnik S, Auwerx J, Sinclair DA, Matter CM. Protective effects of sirtuins in cardiovascular diseases: from bench to bedside. Eur Heart J. (2015) 36:3404–12. doi: 10.1093/eurheartj/ehv290
261. Weiss EP, Racette SB, Villareal DT, Fontana L, Steger-May K, Schechtman KB, et al. Washington University School of Medicine, Improvements in glucose tolerance and insulin action induced by increasing energy expenditure or decreasing energy intake: a randomized controlled trial. Am J Clin Nutr. (2006) 84:1033–42. doi: 10.1093/ajcn/84.5.1033
262. Meehan CA, Cochran E, Mattingly M, Gorden P, Brown RJ. Mild caloric restriction decreases insulin requirements in patients with type 2 diabetes and severe insulin resistance. Medicine (2015) 94:e1160. doi: 10.1097/MD.0000000000001160
263. Houtkooper RH, Pirinen E, Auwerx J. Sirtuins as regulators of metabolism and healthspan. Nat Rev Mol Cell Biol. (2012) 13:225–38. doi: 10.1038/nrm3293
264. Martin-Montalvo A, Mercken EM, Mitchell SJ, Palacios HH, Mote PL, Scheibye-Knudsen M, et al. improves healthspan and lifespan in mice. Nat Commun. (2013) 4:2192. doi: 10.1038/ncomms3192
265. Miranda MX, van Tits LJ, Lohmann C, Arsiwala T, Winnik S, Tailleux A, et al. The SIRT1 activator SRT3025 provides atheroprotection in Apoe-/- mice by reducing hepatic Pcsk9 secretion and enhancing Ldlr expression. Eur Heart J. (2015) 36:51–9. doi: 10.1093/eurheartj/ehu095
266. Igarashi M, Guarente L. mTORC1 and SIRT1 cooperate to foster expansion of gut adult stem cells during calorie restriction. Cell (2016) 166:436–50. doi: 10.1016/j.cell.2016.05.044
267. Lai YC, Tabima DM, Dube JJ, Hughan KS, Vanderpool RR, Goncharov DA, et al. SIRT3–AMP-activated protein kinase activation by nitrite and metformin improves hyperglycemia and normalizes pulmonary hypertension associated with heart failure with preserved ejection fraction. Circulation (2016) 133:717–31. doi: 10.1161/CIRCULATIONAHA.115.018935
268. Ding YN, Tang X, Chen HZ, Liu DP. Epigenetic regulation of vascular aging and age-related vascular diseases, In: Wang Z, editor. Aging and Aging-Related Diseases: Mechanisms and Interventions. Singapore: Springer (2018). p. 55–75. doi: 10.1007/978-981-13-1117-8_4
269. Bonkowski MS, Sinclair DA. Slowing ageing by design: the rise of NAD+ and sirtuin-activating compounds. Nat Rev Mol Cell Biol. (2016) 17:679. doi: 10.1038/nrm.2016.93
270. Masri S, Rigor P, Cervantes M, Ceglia N, Sebastian C, Xiao C, et al. Partitioning circadian transcription by SIRT6 leads to segregated control of cellular metabolism. Cell (2014) 158:659–72. doi: 10.1016/j.cell.2014.06.050
271. Villeneuve LM, Reddy MA, Lanting LL, Wang M, Meng L, Natarajan R. Epigenetic histone H3 lysine 9 methylation in metabolic memory and inflammatory phenotype of vascular smooth muscle cells in diabetes. Proc Natl Acad Sci USA (2008) 105:9047–52. doi: 10.1073/pnas.0803623105
272. Raychaudhuri N, Thamotharan S, Srinivasan M, Mahmood S, Patel MS, Devaskar SU. Postnatal exposure to a high-carbohydrate diet interferes epigenetically with thyroid hormone receptor induction of the adult male rat skeletal muscle glucose transporter isoform 4 expression. J Nutr Biochem. (2014) 25:1066–76. doi: 10.1016/j.jnutbio.2014.05.011
273. Yang B, Yang J, Bai J, Pu P, Liu J, Wang F, et al. SUV39H1 protects from myocardial ischemia-reperfusion injury in diabetic rats. Cell Physiol Biochem. (2014) 33:1176–85. doi: 10.1159/000358686
274. Jiang Z, Shi D, Tu Y, Tian J, Zhang W, Xing B, et al. Human proislet peptide promotes pancreatic progenitor cells to ameliorate diabetes through FOXO1/menin-mediated epigenetic regulation. Diabetes (2018) 67:1345–55. doi: 10.2337/db17-0885
275. Tian Y, Wong VW, Wong GL, Yang W, Sun H, Shen J, et al. Histone deacetylase HDAC8 promotes insulin resistance and beta-catenin activation in NAFLD-associated hepatocellular carcinoma. Cancer Res. (2015) 75:4803–16. doi: 10.1158/0008-5472.CAN-14-3786
276. Shi H, Munk A, Nielsen TS, Daughtry MR, Larsson L, Li S, et al. Skeletal muscle O-GlcNAc transferase is important for muscle energy homeostasis and whole-body insulin sensitivity. Mol Metab. (2018) 11:160–77. doi: 10.1016/j.molmet.2018.02.010
277. Vaquero A, Scher M, Erdjument-Bromage H, Tempst P, Serrano L, Reinberg D. SIRT1 regulates the histone methyl-transferase SUV39H1 during heterochromatin formation. Nature (2007) 450:440–4. doi: 10.1038/nature06268
278. Bosch-Presegue L, Raurell-Vila H, Marazuela-Duque A, Kane-Goldsmith N, Valle A, Oliver J, et al. Stabilization of SUV39H1 by SIRT1 is part of oxidative stress response and ensures genome protection. Mol Cell (2011) 42:210–23. doi: 10.1016/j.molcel.2011.02.034
279. Yang G, Zhang X, Weng X, Liang P, Dai X, Zeng S, et al. SUV39H1 mediated SIRT1 trans-repression contributes to cardiac ischemia-reperfusion injury. Basic Res Cardiol. (2017) 112:22. doi: 10.1007/s00395-017-0608-3
280. Ren JH, Hu JL, Cheng ST, Yu HB, Wong VKW, Law BYK, et al. SIRT3 restricts HBV transcription and replication via epigenetic regulation of cccDNA involving SUV39H1 and SETD1A histone methyltransferases. Hepatology (2018) 68:1260–76. doi: 10.1002/hep.29912
281. Kumari P, Popescu D, Yue S, Bober E, Ianni A, Braun T. SIRT7 inhibits SIRT1-mediated activation of SUV39H1. Cell Cycle (2018) 17:1403–12. doi: 10.1080/15384101.2018.1486166
282. Shu YN, Dong LH, Li H, Pei QQ, Miao SB, Zhang F, et al. CKII-SIRT1-SM22alpha loop evokes a self-limited inflammatory response in vascular smooth muscle cells. Cardiovasc Res. (2017) 113:1198–207. doi: 10.1093/cvr/cvx048
283. Li M, Hong W, Hao C, Li L, Wu D, Shen A, et al. SIRT1 antagonizes liver fibrosis by blocking hepatic stellate cell activation in mice. FASEB J. (2018) 32:500–11. doi: 10.1096/fj.201700612R
284. Xu W, Jiang K, Shen M, Qian Y, Peng Y. SIRT2 suppresses non-small cell lung cancer growth by targeting JMJD2A. Biol Chem. (2015) 396:929–36. doi: 10.1515/hsz-2014-0284
285. Baker DJ, Childs BG, Durik M, Wijers ME, Sieben CJ, Zhong J, et al. Naturally occurring p16 Ink4a-positive cells shorten healthy lifespan. Nature (2016) 530:184. doi: 10.1038/nature16932
286. Childs BG, Baker DJ, Wijshake T, Conover CA, Campisi J, van Deursen JM. Senescent intimal foam cells are deleterious at all stages of atherosclerosis. Science (2016) 354:472–7. doi: 10.1126/science.aaf6659
287. Childs BG, Li H, van Deursen JM. Senescent cells: a therapeutic target for cardiovascular disease. J Clin Invest. (2018) 128:1217–28. doi: 10.1172/JCI95146
288. Jeon OH, Kim C, Laberge RM, Demaria M, Rathod S, Vasserot AP, et al. Local clearance of senescent cells attenuates the development of post-traumatic osteoarthritis and creates a pro-regenerative environment. Nat Med. (2017) 23:775–81. doi: 10.1038/nm.4324
289. Jeon OH, David N, Campisi J, Elisseeff JH. Senescent cells and osteoarthritis: a painful connection. J Clin Invest. (2018) 128:1229–37. doi: 10.1172/JCI95147
290. Bussian TJ, Aziz A, Meyer CF, Swenson BL, van Deursen JM, Baker DJ. Clearance of senescent glial cells prevents tau-dependent pathology and cognitive decline. Nature (2018) 562:578–82. doi: 10.1038/s41586-018-0543-y
Keywords: sirtuins, insulin resistance, senescaging, inflammation, mitochondrial dysfunction
Citation: Zhou S, Tang X and Chen H-Z (2018) Sirtuins and Insulin Resistance. Front. Endocrinol. 9:748. doi: 10.3389/fendo.2018.00748
Received: 30 August 2018; Accepted: 26 November 2018;
Published: 06 December 2018.
Edited by:
Yang Yang, Northwest University, ChinaReviewed by:
Pieter de Lange, Università degli Studi della Campania “Luigi Vanvitelli” Caserta, ItalyCopyright © 2018 Zhou, Tang and Chen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiaoqiang Tang, dGFuZ3hpYW9xaWFuZ0BzY3UuZWR1LmNu
Hou-Zao Chen, Y2hlbmhvdXphb0BpYm1zLmNhbXMuY24=; aG91emFvQGdtYWlsLmNvbQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.