- 1Centre for Chronobiology, University of Basel, Basel, Switzerland
- 2Transfaculty Research Platform Molecular and Cognitive Neurosciences, University of Basel, Basel, Switzerland
- 3Psychiatry and Clinical Psychobiology, Division of Neuroscience, Scientific Institute and University Vita-Salute San Raffaele, Milan, Italy
In healthy humans, seasonality has been documented in psychological variables, chronotype, sleep, feeding, metabolic and autonomic function, thermoregulation, neurotransmission, and hormonal response to stimulation, thus representing a relevant factor to account for, especially when considering the individual susceptibility to disease. Mood is largely recognized as one of the central aspects of human behavior influenced by seasonal variations. This historical notion, already mentioned in ancient medical reports, has been recently confirmed by fMRI findings, which showed that seasonality in human cognitive brain functions may influence affective control with annual variations. Thus, seasonality plays a major role in mood disorders, affecting psychopathology, and representing the behavioral correlate of a heightened sensitivity to factors influencing circannual rhythms in patients. Although the genetic basis of seasonality and seasonal affective disorder (SAD) has not been established so far, there is growing evidence that factors affecting the biological clock, such as gene polymorphisms of the core clock machinery and seasonal changes of the light-dark cycle, exert a marked influence on the behavior of patients affected by mood disorders. Here we review recent findings about the effects of individual gene variants on seasonality, mood, and psychopathological characteristics.
Introduction
Seasonality is a central aspect of environmental variability, which has strongly influenced life on Earth by driving the development of biodiversity among living organisms and the evolution of extreme physiological adaptations and behaviors, such as migration and hibernation. In most species, periodic variations of environmental conditions, particularly those related to the light-dark cycle and depending on latitude, season, and time of day, require that internal timing mechanisms induce the adaption of behavioral or physiological functions to such changes (1).
Biological rhythms with an approximate 24-h period, close to the daily light-dark cycle, are known as circadian rhythms and defined by three fundamental properties: persistence of an ~24-h rhythm, entrainability, and temperature compensation (2). The observation that these endogenous processes are also present among organisms such as cyanobacteria, which represent one of the earliest and most primitive species, suggests that circadian rhythms implicated a clear evolutionary advantage (1).
Clock Genes and Mood Regulation
At the cellular level, circadian rhythms are generated by a core molecular clock consisting of multiple transcriptional/translational feedback loops (3). The transcription factors circadian locomotor output cycles kaput (CLOCK) and brain and muscle Arnt-like (ARNTL), or neuronal pas domain protein 2 (NPAS2) proteins, dimerize and initiate the expression of the clock proteins PERIOD (PER1, PER2, PER3), and CRYPTOCHROME (CRY1, CRY2). With rising accumulation, PER1-3 and CRY1/2 inhibit CLOCK:ARNTL (or CLOCK:NPAS2) activity and therefore block their own expression (3). An additional feedback loop is generated by CLOCK:ARNTL (or CLOCK:NPAS2) mediated transcription of REV-ERB and RORs, which in turn also regulate ARNTL transcription (see Figure 1).
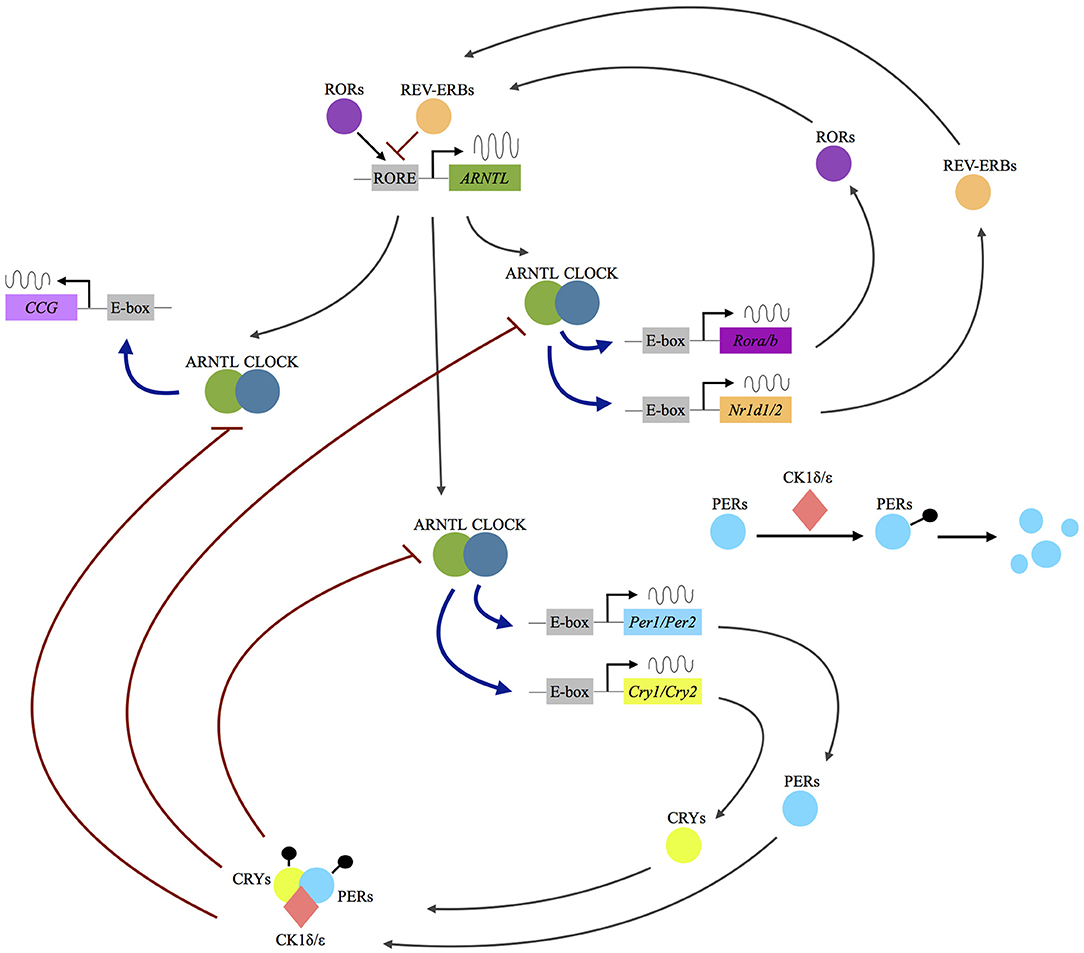
Figure 1. Molecular mechanisms of the circadian clockwork. Following the dimerization of the transcription factors circadian locomotor output cycles kaput (CLOCK) and brain and muscle Arnt-like (ARNTL) or neuronal pas domain protein 2 (NPAS2) proteins, the expression of the clock proteins period (PER1, PER2, PER3) and cryptochrome (CRY1, CRY2) is initiated. The PER and CRY proteins interact with the serine/threonine kinases casein kinase 1 δ/ε, (CK1 δ/ε) and form a complex allowing nuclear translocation. In the nucleus they act as inhibitors of CLOCK:ARNTL (or CLOCK:NPAS2) activity and therefore block their own expression. An additional feedback loop is generated by CLOCK:ARNTL (or CLOCK:NPAS2) mediated transcription of REV-ERB and rar-related orphan receptor A/B (RORA/B), which in turn also regulate ARNTL transcription. Up to 10% of the human genome is under the influence of the molecular clock (clock-controlled genes, CCG). RORE: ROR response element.
As recently reviewed by Albrecht, there is already solid scientific evidence showing that the above-mentioned proteins “not only self-promote their own temporally fluctuating transcription, but also regulate the transcription of a large number of clock-controlled genes (CCGs) and/or modulate key molecular pathways via protein–protein interactions, such as the monoaminergic system, the HPA axis or neurogenic pathways” [(4), p. 1]. Several cellular processes in the brain are under the control of the circadian clock, including “differentiation, growth, motility and apoptosis, immune functions and neuroinflammation, neurogenesis, and neuroplasticity” [(5), p. 236]. A desynchronization of the circadian gene network and disruption of its downstream mechanisms has therefore widespread potential implications for a vast array of physiological processes.
Hampp et al. demonstrated that the functional triade of PER2, ARNTL, and NPAS2 and their encoded proteins, directly regulate the activation of the monoamine oxidase A gene (Maoa). In fact, the transcription and activity of the MAOA enzyme in the mesolimbic neurons is decreased in mice carrying a genetic deletion of the Per2 gene, causing an increase of the dopamine levels and an altered neuronal activity in the striatum, as well as behavioral changes (6, 7).
Dopamine is an important neurotransmitter in the reward system, and its levels in the nucleus accumbens show a circadian rhythmicity (6, 8). Considering that many other brain areas of the reward system, including the ventral tegmental area, prefrontal cortex, and amygdala, are also involved in both mood regulation and clock genes expression, this suggests that the entire reward circuit may be under the influence of the circadian clock, via dopamine metabolism (5).
Cryptochromes (CRY2 and CRY1) are key components of the molecular clock, which drive several functions of the circadian pacemaker (9) and are necessary for the development of intercellular networks in the suprachiasmatic nucleus (10). CRY2 and CRY1 proteins are functionally repressors of the transcription-translation loops, and inhibitors of the cyclic adenosine monophosphate signal pathway (11–14). Due to these important molecular properties at the circadian clock level, it has been suggested that CRY2 and CRY1 may play a major role in the metabolism of glucose and lipids (15, 16) and contribute to mood regulation on daily basis, as well as to seasonal variations in mood and behavior (17).
Finally, PER3 is one of the most robustly rhythmic genes in humans and animals, playing a significant role in the temporal organization of peripheral tissues and being associated with diurnal preference, mental disorders, non-visual responses to light, as well as brain and cognitive responses to sleep loss/circadian misalignment (18). Some genetic variants are supposed to interfere with the stabilizing effect of PER3 on PERIOD1/2 proteins, which play critical roles in circadian timing. These findings suggest that PER3 may represent an important element of the missing molecular linkage between sleep and mood regulation by adapting these processes to seasonal changes (19).
Clock Genes in Mood Disorders
Several human population genetic studies have identified specific single nucleotide polymorphisms (SNPs) or variable number of tandem repeats (VNTR, see Supplementary Table 1) of different circadian clock genes that are associated with mood disorders (20, 21). These associations remain controversial, since most findings could either not be replicated or hold up to correction for multiple testing (22). From a pathophysiological point of view, recent experimental work, and mathematical models suggest that changes in period length and/or decreased amplitude of the circadian oscillation may depend on the impact of specific polymorphisms on the overall function in terms of structure and stability of a given clock protein (23). By as of yet poorly understood processes, the resulting functional changes of the clock-machinery and misalignment between clock-regulated functions and the environment can influence core psychopathological features of mood disorders, including the timing of onset and recurrence of illness episodes, individual symptomatology, and response to treatments (5).
Depressive Disorder
In depressive disorder (DD) (7), two TIMELESS polymorphisms have been found to be associated with excessive daytime fatigue among women, as well as a two-way interaction of TIMELESS and ARNTL (rs1868049) with early-morning awakening among men (24). Lavebratt et al. demonstrated that RORA, PER2, and NPAS2 are associated with DD and the onset of depression within 3 years independently from financial strain (25). Both an increased or decreased PER3 transcriptional activity may implicate a higher risk for MDD. In particular, Shi and colleagues identified a missense mutation in hPER3 (hPER3-P856A), which slightly lengthens the circadian period and is related to MDD in females, by likely driving changes in clock-controlled genes as opposed to SCN timing. Moreover, the authors describe other sex-dependent associations of common polymorphisms with a CLOCK variant protective of MDD in males and NPAS2 polymorphisms with association of MDD especially in females (26). NPAS2 and CRY1 were also linked with DD in a study by Soria et al. (27), with the latter finding replicated by Hua et al. (28), who, instead, did not find any association of CRY2 (rs10838524) with major depressive disorder (MDD). However, Kovanen et al. suggested that CRY2 and the protein kinase C delta binding protein (PRKCDBP, or CAVIN3) variants may represent risk factors for MDD (29). Finally, the best association between a SNP and MDD based on genome-wide association studies has been found for NR1D1 (30).
Bipolar Disorder
The observation that patients with bipolar disorder (BD) show alterations in circadian rhythms, and recurrent fluctuations of mood and sleep disturbances (31) has suggested a possible dysfunction of the biological clock in the pathogenesis of BD (32). Moreover, since heritability in BD is estimated to be as high as 85% (33), an increasing interest in identifying genetic risk factors has supported different association studies looking at the link between BD and some core clock genes (7).
Significant SNPs associations with bipolar 1 disorder were found for TIMELESS and ARNTL (34), as well as for NPAS2, RORB 9, and CRY2 (35). Gonzalez et al. performed a family-based association study of circadian genes and BD in a Latino population, reporting nominal associations between SNPs of CSNK1E, ARNTL, CSNK1D, CLOCK, as well as statistically significant associations between CSNK1E and ARNTL haplotypes and BD, with either increased susceptibility or protective effect against the development of the disorder respectively (36). Shi et al. demonstrated the three-way interaction of BHLHE40, TMEM165 (transmembrane protein 165), and CSNK1E with bipolar disorder (37), while McGrath et al. focusing their analysis on the RORA and RORB genes, found that 4 RORB SNPs were associated with bipolar 1 disorder (38). Etain et al. indicated a significant association of TIMELESS and of RORA with BD (39), while Lee et al. found CLOCK 3111T/C to have significant allelic and genotypic associations with the disease (40). GSK3beta was associated with bipolar type 2 disorder in women (41). General associations of NR1D1 (42) and of VIP (27) with BD were also reported. In genome-wide association studies, the associations of ARNTL, GSK3beta, RORB, and CRY 2 gene variants with BD have gained further support (30, 43).
Circadian Genes and Phenotypic Characteristics in Bipolar Disorder
Genetic polymorphisms influencing clock genes functions have shown major effects on the phenotypic clinical features of disease (44). A SNP in CLOCK gene, which is known to influence diurnal preference in healthy subjects (45), also impacts on bipolar patients, leading to worsening of insomnia, higher evening activity and delayed sleep onset. Carriers of the allelic C variant also showed a higher episode recurrence rate and different neuropsychological performance (46–48), while the G allele of the same polymorphism has been linked with symptoms of appetite disturbances in females (49). A correlation with violent suicide attempts was shown for other SNPs in CLOCK and TIMELESS, while the latter is also associated with the lifetime number of suicide attempts and a positive family history of suicide (50). A VNTR of PER3 gene was shown to influence the general age of onset, as well as a postpartum depressive onset of the disorder (51, 52). PER3 was also linked to an increased preference for the evening hours in daily activity among BD patients (42). Maciukiewicz et al. observed further associations between SNPs of ARNTL variants with sleep, appetite and depressive dimensions in BD (49).
A functional SNP in the promoter region of the GSK3beta gene (nt −171 to +29), which also shows a general association to impulsivity and suicide risk among patients with bipolar disease, was found to influence the age at onset of BD, as well as the response to treatment with antidepressant, lithium salts and chronotherapeutics (53–55). This polymorphism was recently shown to also influence white matter microstructure of bipolar patients under ongoing lithium treatment (56) and gray matter volumes in areas critical for the generation and control of affect implicated in BD pathophysiology (57).
Other polymorphisms influencing treatment response, such as the mood stabilizer effect of lithium salts (variant in the promoter of NR1D1) and a general association with positive treatment response (CRY1) have been described (58). Finally, Sjöholm et al. identified two risk haplotypes and one protective haplotype in the CRY2 gene associated with rapid cycling in BD (59) (see Supplementary Table 1).
Genetics of Seasonality and Seasonal Affective Disorder
The interplay between mood variations and seasonal rhythms in humans has received renewed interest since the diagnosis of Seasonal Affective Disorder (SAD) was proposed by Rosenthal in 1984, as “a condition characterized by recurrent depressive episodes that occur annually at the same time each year” [(60), p. 72]. The observation that many adults experience a “subsyndromal SAD”, with milder vegetative symptoms in the fall/winter months (61, 62), suggested that “seasonality may be a dimensional process rather than a discrete syndrome” [(63), p. 315].
Serotonergic Genes
Although the genetic basis of seasonality and SAD has not yet been completely identified, several studies suggest that both conditions have an inherited component (64–66). From a pathophysiological point of view, the typical symptoms of SAD, such as overeating, carbohydrate craving, weight gain, and oversleeping, point to a dysfunction of the serotonergic system (66). Moreover, the serotonin level in the human hypothalamus shows seasonal variations, with a general decrease during the winter season (67). The serotonin hypothesis is also supported by the large therapeutic evidence that selective serotonin reuptake inhibitors (SSRIs) and bright light therapy are effective in winter SAD (68–71), with reversion of this effect by rapid tryptophan depletion (70, 72).
Therefore, the first pioneer genetic studies focused on the molecular components of the serotonergic system (73). Rosenthal et al. showed that the short (s), as opposed to the long (l), allele of the 5-HT transporter linked polymorphism (5-HTTLPR) contributes to the trait of seasonality and is a risk factor for SAD (74). First reports showing an association of this variant with general susceptibility and several features of the clinical course among patients with SAD (75–77) could not be corroborated by a meta-analysis by Johansson et al., but the authors concluded that the polymorphism may have an effect on seasonal behavioral traits (78, 79).
Recent Positron Emission Tomography (PET) studies showed a significantly higher activity of serotonin transporter binding potential in several brain regions, during fall and winter, compared to spring and summer, in healthy volunteers (80, 81). Furthermore, “the first [11C]DASB PET longitudinal study investigating whole-brain seasonal 5-HTT fluctuations in both patients with SAD and in healthy individuals reported that a whole-brain seasonal change in 5-HTT predicted symptom severity in patients with SAD, an effect primarily driven by females with the short 5-HTTLPR genotype (S' carriers)” [(82), p. 2], (83). These findings were later confirmed by other groups (83, 84).
The serotonin 5-HT2A receptor gene has also been proposed as major candidate gene in association studies of seasonality and SAD (85, 86). In particular, it has been suggested that “downregulation of 5-HT2A receptors may underlie the therapeutic effects of SSRIs” [(64), p. 656], (87) and the effectiveness of light therapy in the treatment of SAD has also been linked to an alteration of the sensitivity of 5-HT2A receptors (76). Moreover, specific sequence polymorphisms in the coding region of the serotonin 5-HT2A receptor gene have been found to be associated with the clinical features and course of depressive disorder or directly with seasonality and SAD (64, 86, 88–90).
Circadian Genes
Apart from an extensive connection between SAD and the serotoninergic system, genes of the core clock family have also been implicated in the disease. After a first report of a SNP in NPAS2 being linked to SAD (91), Partonen et al. found further SNPs of PER2, ARNTL, and NPAS2 to be associated with seasonality and SAD (92, 93).
Kim et al. also reported an association of NPAS2 and ARNTL, especially with the metabolic components of seasonality (body weight and appetite). In addition, they found increased seasonal variations of mood and behavior among individuals carrying a CLOCK polymorphism previously implicated in bipolar disorder (40, 46–48, 94). These recent findings are in contrast with a previous work from the same group, showing that the same SNP of CLOCK is not associated with seasonal fluctuations in a sample of Korean college students (95).
Furthermore, another recent investigation highlighted the impact of two rare genetic variants of the PERIOD3 gene (PER3) on a circadian phenotype and a seasonal mood trait, which may be especially critical under conditions of short photoperiod (e.g., during the winter season) (19).
Other Genetic Findings
Environmental light detection in humans is mediated by melanopsin containing intrinsically photosensitive retinal ganglion cells (ipRGCs), which are located in the inner retina (96–98). Some polymorphisms of the melanopsin gene may be linked to a greater sensitivity to light, thus determining functional variations in ipRGC activity. During shortened photoperiods (e.g., during the winter months) this may contribute to inter-individual differences in sleep and alertness (99, 100). A missense variant (P10L) in the melanopsin (OPN4) gene, which has also been found in SAD patients, has been proposed to contribute to changes in melanopsin sensitivity (99). Reduced retinal light sensitivity, especially during the winter months, as a pathophysiological hypothesis of SAD (101–103) recently gained first supporting evidence. A study by Roecklein et al. found a reduced post-illumination pupil response (PIPR) in SAD patients, compared with controls, in winter but not in summer (104).
A study by Delavest et al. investigating the rs2072621 polymorphism of the X-linked GPR50 gene, a member of the G protein-coupled melatonin receptor subfamily, found an association with SAD in females, thus providing the first potential gender-specific molecular link between the hormone melatonin and SAD (105).
Yang et al. studied the relationship between ST8SIA2 and NCAM1, two genes forming the polysialic acid neural cell adhesion molecule (NCAM) complex in the SCN, and circadian preferences, as well as seasonality, in healthy adult Korean subjects. The association of 8 SNPs of ST8SIA2 and 2 SNPs of NCAM1 with seasonality remained significant after correction for multiple testing (106).
Another study by Nam et al. found that the GNB3 (G-protein β3 subunit) C825T polymorphism, which is associated with various medical conditions (107, 108) and psychiatric disorders, including recurrent winter depression or SAD (109, 110), also plays a role in seasonal variations in mood, body weight, energy level, and appetite, particularly in females.
Conclusions
Gene polymorphisms of the core clock machinery and seasonal changes of the light-dark cycle substantially impact on the behavior of patients with mood disorders. The relationship between biological clock and behavior suggests a specific sensibility of these patients to psychobiological factors that can modify the circadian timing system, such as environmental synchronizers (light phase and seasonal photoperiod changes), and conditions directly perturbing the clock (sleep deprivation, or phase advance/delay). These factors can trigger or worsen the severity of mood disorders, but also be successfully exploited to treat manic and depressive episodes (111).
Current models of circadian homeostasis suggest that the hierarchical control exerted by the SCN on circadian rhythms of behavior, physiological functions, and on peripheral clocks (112), interacts with homeostatic mechanisms that also contribute to these phenomena. In rodents, a similar dependence of behavior on clock gene mutations occurs in the absence of other regulators of circadian rhythmicity, such as melatonin, and is abolished when these homeostatic components are restored (113). Therefore, we suggest that the high sensitivity of mood-disordered patients to clock gene variants is underpinned by a deficit in homeostatic mechanisms regulating the circadian timing system. Recent discoveries in humans of yet unknown circulating substances affecting the circadian phenotype and overcoming the timing of the clock gene machinery (114, 115), lead to hypothesize that a systematic investigation of these mechanisms will shed new light on the nature of circadian disruption in mood disorders.
Author Contributions
Both authors certify that they have participated sufficiently in the work to take public responsibility for the content, including participation in the concept, design, writing, or revision of the manuscript. In particular: CG conceived, designed and drafted the manuscript. FB drafted and critically reviewed the manuscript. CG and FB approved the final version of the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors would like to acknowledge Sandra Hackethal, MD for her precious contribution in reviewing the final version of the manuscript and drafting the graphic contents.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fendo.2018.00481/full#supplementary-material
References
1. Hut RA, Beersma DGM. Evolution of time-keeping mechanisms: early emergence and adaptation to photoperiod. Philos Trans R Soc Lond B Biol Sci. (2011) 366:2141–54. doi: 10.1098/rstb.2010.0409
3. Albrecht U. Timing to perfection: the biology of central and peripheral circadian clocks. Neuron (2012) 74:246–60. doi: 10.1016/j.neuron.2012.04.006
4. Albrecht U. Molecular mechanisms in mood regulation involving the circadian clock. Front Neurol. (2017) 8:30. doi: 10.3389/fneur.2017.00030
5. Benedetti F, Terman M. Much ado about…a moody clock. Biol Psychiatry (2013) 74:236–7. doi: 10.1016/j.biopsych.2013.05.037
6. Hampp G, Ripperger JA, Houben T, Schmutz I, Blex C, Perreau-Lenz S, et al. Regulation of monoamine oxidase a by circadian-clock components implies clock influence on mood. Curr Biol. (2008) 18:678–83. doi: 10.1016/j.cub.2008.04.012
7. Partonen T. Circadian clock genes and mood disorders. In: López-Muñoz F, Srinivasan V, de Berardis D, Álamo C, Kato TA, editors. Melatonin, Neuroprotective Agents and Antidepressant Therapy. New Delhi: Springer (2016). p. 319–34.
8. Hood S, Cassidy P, Cossette MP, Weigl Y, Verwey M, Robinson B, et al. Endogenous dopamine regulates the rhythm of expression of the clock protein PER2 in the rat dorsal striatum via daily activation of D2 dopamine receptors. J Neurosci. (2010) 30:14046–58. doi: 10.1523/JNEUROSCI.2128-10.2010
9. Lamia KA, Papp SJ, Yu RT, Barish GD, Uhlenhaut NH, Jonker JW, et al. Cryptochromes mediate rhythmic repression of the glucocorticoid receptor. Nature (2011) 480:552–6. doi: 10.1038/nature10700
10. Ono D, Honma S, Honma K. Cryptochromes are critical for the development of coherent circadian rhythms in the mouse suprachiasmatic nucleus. Nat Commun. (2013) 4:1666. doi: 10.1038/ncomms2670
11. Dardente H, Fortier EE, Martineau V, Cermakian N. Cryptochromes impair phosphorylation of transcriptional activators in the clock: a general mechanism for circadian repression. Biochem J. (2007) 402:525–36. doi: 10.1042/BJ20060827
12. Oztürk N, Song SH, Ozgür S, Selby CP, Morrison L, Partch C, et al. Structure and function of animal cryptochromes. Cold Spring Harb Symp Quant Biol. (2007) 72:119–31. doi: 10.1101/sqb.2007.72.015
13. Ye R, Selby CP, Chiou YY, Ozkan-Dagliyan I, Gaddameedhi S, Sancar A. Dual modes of CLOCK:BMAL1 inhibition mediated by cryptochrome and period proteins in the mammalian circadian clock. Genes Dev. (2014) 28:1989–98. doi: 10.1101/gad.249417.114
14. Ye R, Selby CP, Ozturk N, Annayev Y, Sancar A. Biochemical analysis of the canonical model for the mammalian circadian clock. J Biol Chem. (2011) 286:25891–902. doi: 10.1074/jbc.M111.254680
15. Kelly MA, Rees SD, Hydrie MZ, Shera AS, Bellary S, O'Hare JP, et al. Circadian gene variants and susceptibility to type 2 diabetes: a pilot study. PLoS ONE (2012) 7:e32670. doi: 10.1371/journal.pone.0032670
16. Machicao F, Peter A, Machann J, Königsrainer I, Böhm A, Lutz SZ, et al. Glucose-raising polymorphisms in the human clock gene cryptochrome 2 (CRY2) affect hepatic lipid content. PLoS ONE (2016) 11:e0145563. doi: 10.1371/journal.pone.0145563
17. Kovanen L, Donner K, Kaunisto M, Partonen T. CRY1 and CRY2 genetic variants in seasonality: A longitudinal and cross-sectional study. Psychiatry Res. (2016) 242:101–10. doi: 10.1016/j.psychres.2016.05.044
18. Archer SN, Schmidt C, Vandewalle G, Dijk DJ. Phenotyping of PER3 variants reveals widespread effects on circadian preference, sleep regulation, and health. Sleep Med Rev. (2017) 40:109–26. doi: 10.1016/j.smrv.2017.10.008
19. Zhang L, Hirano A, Hsu PK, Jones CR, Sakai N, Okuro M, et al. A PERIOD3 variant causes a circadian phenotype and is associated with a seasonal mood trait. Proc Natl Acad Sci USA. (2016) 113:E1536–44. doi: 10.1073/pnas.1600039113
20. Etain B, Milhiet V, Bellivier F, Leboyer M. Genetics of circadian rhythms and mood spectrum disorders. Eur Neuropsychopharmacol. (2011) 21(Suppl. 4) S676–82. doi: 10.1016/j.euroneuro.2011.07.007
21. Partonen T. Clock gene variants in mood and anxiety disorders. J Neural Transm. (2012) 119:1133–45. doi: 10.1007/s00702-012-0810-2
22. Vadnie CA, McClung CA. Circadian Rhythm disturbances in mood disorders: insights into the role of the suprachiasmatic nucleus. Neural Plast. (2017) 2017:1504507. doi: 10.1155/2017/1504507
23. Liberman AR, Halitjaha L, Ay A, Ingram KK. Modeling strengthens molecular link between circadian polymorphisms and major mood disorders. J Biol Rhythms (2018) 33:318–36. doi: 10.1177/0748730418764540
24. Utge SJ, Soronen P, Loukola A, Kronholm E, Ollila HM, Pirkola S, et al. Systematic analysis of circadian genes in a population-based sample reveals association of TIMELESS with depression and sleep disturbance. PLoS ONE (2010) 5:e9259. doi: 10.1371/journal.pone.0009259
25. Lavebratt C, Sjöholm LK, Partonen T, Schalling M, Forsell Y. PER2 variation is associated with depression vulnerability. Am J Med Genet B Neuropsychiatr Genet. (2010) 153B:570–81. doi: 10.1002/ajmg.b.31021
26. Shi SQ, White MJ, Borsetti HM, Pendergast JS, Hida A, Ciarleglio CM, et al. Molecular analyses of circadian gene variants reveal sex-dependent links between depression and clocks. Trans Psychiatry (2016) 6:e748. doi: 10.1038/tp.2016.9
27. Soria V, Martínez-Amorós E, Escaramís G, Valero J, Pérez-Egea R, García C, et al. Differential association of circadian genes with mood disorders: CRY1 and NPAS2 are associated with unipolar major depression and CLOCK and VIP with bipolar disorder. Neuropsychopharmacology (2010) 35:1279–89. doi: 10.1038/npp.2009.230
28. Hua P, Liu W, Chen D, Zhao Y, Chen L, Zhang N, et al. Cry1 and Tef gene polymorphisms are associated with major depressive disorder in the Chinese population. J Affect Disord. (2014) 157:100–3. doi: 10.1016/j.jad.2013.11.019
29. Kovanen L, Donner K, Kaunisto M, Partonen T. PRKCDBP (CAVIN3) and CRY2 associate with major depressive disorder. J Affect Disord. (2017) 207:136–40. doi: 10.1016/j.jad.2016.09.034
30. Byrne EM, Heath AC, Madden PA, Pergadia ML, Hickie IB, Montgomery GW, et al. Testing the role of circadian genes in conferring risk for psychiatric disorders. Am J Med Genet B Neuropsychiatr Genet. (2014) 165B:254–60. doi: 10.1002/ajmg.b.32230
31. Riemann D, Voderholzer U, Berger M. Sleep and sleep-wake manipulations in bipolar depression. Neuropsychobiology (2002) 45(Suppl. 1):7–12. doi: 10.1159/000049255
32. Dallaspezia S, Benedetti F. Chronobiology of bipolar disorder: therapeutic implication. Curr Psychiatry Rep. (2015) 17:606. doi: 10.1007/s11920-015-0606-9
33. McGuffin P, Rijsdijk F, Andrew M, Sham P, Katz R, Cardno A. The heritability of bipolar affective disorder and the genetic relationship to unipolar depression. Arch Gen Psychiatry (2003) 60:497–502. doi: 10.1001/archpsyc.60.5.497
34. Mansour HA, Wood J, Logue T, Chowdari KV, Dayal M, Kupfer DJ, et al. Association study of eight circadian genes with bipolar I disorder, schizoaffective disorder and schizophrenia. Genes Brain Behav. (2006) 5:150–7. doi: 10.1111/j.1601-183X.2005.00147.x
35. Mansour HA, Talkowski ME, Wood J, Chowdari KV, McClain L, Prasad K, et al. Association study of 21 circadian genes with bipolar I disorder, schizoaffective disorder, and schizophrenia. Bipolar Disord. (2009) 11:701–10. doi: 10.1111/j.1399-5618.2009.00756.x
36. Gonzalez R, Gonzalez S, Villa E, Ramirez M, Zavala J, Armas R, et al. Identification of circadian gene variants in bipolar disorder in Latino populations. J Affect Disord. (2015) 186:367–75. doi: 10.1016/j.jad.2015.07.014
37. Shi J, Wittke-Thompson JK, Badner JA, Hattori E, Potash JB, Willour VL, et al. Clock genes may influence bipolar disorder susceptibility and dysfunctional circadian rhythm. Am J Med Genet B Neuropsychiatr Genet. (2008) 147B:1047–55. doi: 10.1002/ajmg.b.30714
38. McGrath CL, Glatt SJ, Sklar P, Le-Niculescu H, Kuczenski R, Doyle AE, et al. Evidence for genetic association of RORB with bipolar disorder. BMC Psychiatry (2009) 9:70. doi: 10.1186/1471-244X-9-70
39. Etain B, Jamain S, Milhiet V, Lajnef M, Boudebesse C, Dumaine A, et al. Association between circadian genes, bipolar disorders and chronotypes. Chronobiol Int. (2014) 31:807–14. doi: 10.3109/07420528.2014.906445
40. Lee KY, Song JY, Kim SH, Kim SC, Joo EJ, Ahn YM, et al. Association between CLOCK 3111T/C and preferred circadian phase in Korean patients with bipolar disorder. Prog Neuropsychopharmacol Biol Psychiatry (2010) 34:1196–201. doi: 10.1016/j.pnpbp.2010.06.010
41. Szczepankiewicz A, Skibinska M, Hauser J, Slopien A, Leszczynska-Rodziewicz A, Kapelski P, et al. Association analysis of the GSK-3β T-50C gene polymorphism with schizophrenia and bipolar disorder. Neuropsychobiology (2006) 53:51–6. doi: 10.1159/000090704
42. Kripke DF, Nievergelt CM, Joo E, Shekhtman T, Kelsoe JR. Circadian polymorphisms associated with affective disorders. J Circadian Rhythm (2009) 7:2. doi: 10.1186/1740-3391-7-2
43. Le-Niculescu H, Patel SD, Bhat M, Kuczenski R, Faraone SV, Tsuang MT, et al. Convergent functional genomics of genome-wide association data for bipolar disorder: comprehensive identification of candidate genes, pathways and mechanisms. Am J Med Genet B Neuropsychiatr Genet. (2009) 150B:155–81. doi: 10.1002/ajmg.b.30887
44. Dallaspezia S, Benedetti F. Melatonin, circadian rhythms, and the clock genes in bipolar disorder. Curr Psychiatry Rep. (2009) 11:488–93. doi: 10.1007/s11920-009-0074-1
45. Katzenberg D, Young T, Finn L, Lin L, King DP, Takahashi JS, et al. A CLOCK polymorphism associated with human diurnal preference. Sleep (1998) 21:569–76. doi: 10.1093/sleep/21.6.569
46. Serretti A, Cusin C, Benedetti F, Mandelli L, Pirovano A, Zanardi R, et al. Insomnia improvement during antidepressant treatment and CLOCK gene polymorphism. Am J Med Genet B Neuropsychiatr Genet. (2005) 137B:36–9. doi: 10.1002/ajmg.b.30130
47. Benedetti F, Dallaspezia S, Fulgosi MC, Lorenzi C, Serretti A, Barbini B, et al. Actimetric evidence that CLOCK 3111 T/C SNP influences sleep and activity patterns in patients affected by bipolar depression. Am J Med Genet B Neuropsychiatr Genet. (2007) 144B:631–5. doi: 10.1002/ajmg.b.30475
48. Benedetti F, Serretti A, Colombo C, Barbini B, Lorenzi C, Campori E, et al. Influence of CLOCK gene polymorphism on circadian mood fluctuation and illness recurrence in bipolar depression. Am J Med Genet. (2003) 123B:23–6. doi: 10.1002/ajmg.b.20038
49. Maciukiewicz M, Dmitrzak-Weglarz M, Pawlak J, Leszczynska-Rodziewicz A, Zaremba D, Skibinska M, et al. Analysis of genetic association and epistasis interactions between circadian clock genes and symptom dimensions of bipolar affective disorder. Chronobiol Int. (2014) 31:770–8. doi: 10.3109/07420528.2014.899244
50. Pawlak J, Dmitrzak-Weglarz M, Maciukiewicz M, Wilkosc M, Leszczynska-Rodziewicz A, Zaremba D, et al. Suicidal behavior in the context of disrupted rhythmicity in bipolar disorder-Data from an association study of suicide attempts with clock genes. Psychiatry Res. (2015) 226:517–20. doi: 10.1016/j.psychres.2015.01.010
51. Benedetti F, Dallaspezia S, Colombo C, Pirovano A, Marino E, Smeraldi E. A length polymorphism in the circadian clock gene Per3 influences age at onset of bipolar disorder. Neurosci Lett. (2008) 445:184–7. doi: 10.1016/j.neulet.2008.09.002
52. Dallaspezia S, Lorenzi C, Pirovano A, Colombo C, Smeraldi E, Benedetti F. Circadian clock gene Per3 variants influence the postpartum onset of bipolar disorder. Eur Psychiatry (2011) 26:138–40. doi: 10.1016/j.eurpsy.2010.11.009
53. Benedetti F, Bernasconi A, Lorenzi C, Pontiggia A, Serretti A, Colombo C, et al. A single nucleotide polymorphism in glycogen synthase kinase 3-beta promoter gene influences onset of illness in patients affected by bipolar disorder. Neurosci Lett. (2004) 355:37–40. doi: 10.1016/j.neulet.2003.10.021
54. Benedetti F, Serretti A, Colombo C, Lorenzi C, Tubazio V, Smeraldi E. A glycogen synthase kinase 3-beta promoter gene single nucleotide polymorphism is associated with age at onset and response to total sleep deprivation in bipolar depression. Neurosci Lett. (2004) 368:123–6. doi: 10.1016/j.neulet.2004.06.050
55. Benedetti F, Serretti A, Pontiggia A, Bernasconi A, Lorenzi C, Colombo C, et al. Long-term response to lithium salts in bipolar illness is influenced by the glycogen synthase kinase 3-beta −50 T/C SNP. Neurosci Lett. (2005) 376:51–5. doi: 10.1016/j.neulet.2004.11.022
56. Benedetti F, Bollettini I, Barberi I, Radaelli D, Poletti S, Locatelli C, et al. Lithium and GSK3-beta promoter gene variants influence white matter microstructure in bipolar disorder. Neuropsychopharmacology (2013) 38:313–27. doi: 10.1038/npp.2012.172
57. Benedetti F, Poletti S, Radaelli D, Locatelli C, Pirovano A, Lorenzi C, et al. Lithium and GSK-3beta promoter gene variants influence cortical gray matter volumes in bipolar disorder. Psychopharmacology (2015) 232:1325–36. doi: 10.1007/s00213-014-3770-4
58. McCarthy MJ, Nievergelt CM, Shekhtman T, Kripke DF, Welsh DK, Kelsoe JR. Functional genetic variation in the Rev-Erbalpha pathway and lithium response in the treatment of bipolar disorder. Genes Brain Behav. (2011) 10:852–61. doi: 10.1111/j.1601-183X.2011.00725.x
59. Sjöholm LK, Backlund L, Cheteh EH, Ek IR, Frisén L, Schalling M, et al. CRY2 is associated with rapid cycling in bipolar disorder patients. PLoS ONE (2010) 5:e12632. doi: 10.1371/journal.pone.0012632
60. Rosenthal NE, Sack DA, Gillin JC, Lewy AJ, Goodwin FK, Davenport Y, et al. Seasonal affective disorder: a description of the syndrome and preliminary findings with light therapy. Arch Gen Psychiatry (1984) 41:72–80. doi: 10.1001/archpsyc.1984.01790120076010
61. Kasper S, Wehr TA, Bartko JJ, Gaist PA, Rosenthal NE. Epidemiological findings of seasonal changes in mood and behavior. a telephone survey of Montgomery County, Maryland. Arch Gen Psychiatry (1989) 46:823–33. doi: 10.1001/archpsyc.1989.01810090065010
62. Magnusson A. An overview of epidemiological studies on seasonal affective disorder. Acta Psychiatr Scand. (2000) 101:176–84. doi: 10.1034/j.1600-0447.2000.101003176.x
63. Levitan RD. The chronobiology and neurobiology of winter seasonal affective disorder. Dialogues Clin Neurosci. (2007) 9:315–24.
64. Molnar E, Lazary J, Benko A, Gonda X, Pap D, Mekli K, et al. Seasonality and winter-type seasonal depression are associated with the rs731779 polymorphism of the serotonin-2A receptor gene. Eur Neuropsychopharmacol. (2010) 20:655–62. doi: 10.1016/j.euroneuro.2010.04.009
65. Madden PA, Heath AC, Rosenthal NE, Martin NG. Seasonal changes in mood and behavior. The role of genetic factors. Arch Gen Psychiatry (1996) 53:47–55. doi: 10.1001/archpsyc.1996.01830010049008
66. Sher L, Goldman D, Ozaki N, Rosenthal NE. The role of genetic factors in the etiology of seasonal affective disorder and seasonality. J Affect Disord. (1999) 53:203–10. doi: 10.1016/S0165-0327(98)00194-3
67. Carlsson A, Svennerholm L, Winblad B. Seasonal and circadian monoamine variations in human brains examined post mortem. Acta Psychiatr Scand. (1980) 61:75–85. doi: 10.1111/acps.1980.61.s280.75
68. Lee TM, Blashko CA, Janzen HL, Paterson JG, Chan CC. Pathophysiological mechanism of seasonal affective disorder. J Affect Disord. (1997) 46:25–38. doi: 10.1016/S0165-0327(97)00076-1
69. Moscovitch A, Blashko CA, Eagles JM, Darcourt G, Thompson C, Kasper S, et al. A placebo-controlled study of sertraline in the treatment of outpatients with seasonal affective disorder. Psychopharmacology (2004) 171:390–7. doi: 10.1007/s00213-003-1594-8
70. Lam RW, Levitt AJ, Levitan RD, Enns MW, Morehouse R, Michalak EE, et al. The Can-SAD study: a randomized controlled trial of the effectiveness of light therapy and fluoxetine in patients with winter seasonal affective disorder. Am J Psychiatry (2006) 163:805–12. doi: 10.1176/ajp.2006.163.5.805
71. Pae CU, Masand PS, Peindl K, Mannelli P, Han C, Marks DM, et al. An open-label, rater-blinded, flexible-dose, 8-week trial of escitalopram in patients with major depressive disorder with atypical features. Prim Care Companion J Clin Psychiatry (2008) 10:205–10. doi: 10.4088/PCC.v10n0305
72. Spillmann MK, Van der Does AJ, Rankin MA, Vuolo RD, Alpert JE, Nierenberg AA, et al. Tryptophan depletion in SSRI-recovered depressed outpatients. Psychopharmacology (2001) 155:123–7. doi: 10.1007/s002130000669
73. Partonen T. Chapter 7: Circadian systems biology in seasonal affective disorder. In: Timo Partonen, Pandi-Perumal SR, editors. Seasonal Affective Disorder: Practice and Research. 2nd ed. Oxford: Oxford University Press (2009).
74. Rosenthal NE, Mazzanti CM, Barnett RL, Hardin TA, Turner EH, Lam GK, et al. Role of serotonin transporter promoter repeat length polymorphism (5-HTTLPR) in seasonality and seasonal affective disorder. Mol Psychiatry (1998) 3:175–7. doi: 10.1038/sj.mp.4000360
75. Praschak-Rieder N, Willeit M, Winkler D, Neumeister A, Hilger E, Zill P, et al. Role of family history and 5-HTTLPR polymorphism in female seasonal affective disorder patients with and without premenstrual dysphoric disorder. Eur Neuropsychopharmacol. (2002) 12:129–34. doi: 10.1016/S0924-977X(01)00146-8
76. Willeit M, Praschak-Rieder N, Neumeister A, Zill P, Leisch F, Stastny J, et al. A polymorphism (5-HTTLPR) in the serotonin transporter promoter gene is associated with DSM-IV depression subtypes in seasonal affective disorder. Mol Psychiatry (2003) 8:942–6. doi: 10.1038/sj.mp.4001392
77. Thierry N, Willeit M, Praschak-Rieder N, Zill P, Hornik K, Neumeister A, et al. Serotonin transporter promoter gene polymorphic region (5-HTTLPR) and personality in female patients with seasonal affective disorder and in healthy controls. Eur Neuropsychopharmacol. (2004) 14:53–8. doi: 10.1016/S0924-977X(03)00064-6
78. Johansson C, Willeit M, Levitan R, Partonen T, Smedh C, Del Favero J, et al. The serotonin transporter promoter repeat length polymorphism, seasonal affective disorder and seasonality. Psychol Med. (2003) 33:785–92. doi: 10.1017/S0033291703007372
79. Sher L, Greenberg BD, Murphy DL, Rosenthal NE, Sirota LA, Hamer DH. Pleiotropy of the serotonin transporter gene for seasonality and neuroticism. Psychiatr Genet. (2000) 10:125–30. doi: 10.1097/00041444-200010030-00004
80. Praschak-Rieder N, Willeit M, Wilson AA, Houle S, Meyer JH. Seasonal variation in human brain serotonin transporter binding. Arch Gen Psychiatry (2008) 65:1072–8. doi: 10.1001/archpsyc.65.9.1072
81. Buchert R, Schulze O, Wilke F, Berding G, Thomasius R, Petersen K, et al. Is correction for age necessary in SPECT or PET of the central serotonin transporter in young, healthy adults? J Nucl Med. (2006) 47:38–42.
82. Mc Mahon B, Andersen SB, Madsen MK, Hjordt LV, Hageman I, Dam H, et al. Seasonal difference in brain serotonin transporter binding predicts symptom severity in patients with seasonal affective disorder. Brain (2016) 139(Pt 5):1605–14. doi: 10.1093/brain/aww043
83. Nørgaard M, Ganz M, Svarer C, Fisher PM, Churchill NW, Beliveau V, et al. Brain networks implicated in seasonal affective disorder: a neuroimaging PET study of the serotonin transporter. Front Neurosci. (2017) 11:614. doi: 10.3389/fnins.2017.00614
84. Tyrer AE, Levitan RD, Houle S, Wilson AA, Nobrega JN, Meyer JH. Increased seasonal variation in serotonin transporter binding in seasonal affective disorder. Neuropsychopharmacology (2016) 41:2447–54. doi: 10.1038/npp.2016.54
85. Yatham LN, Lam RW, Zis AP. Growth hormone response to sumatriptan (5-HT1D agonist) challenge in seasonal affective disorder: effects of light therapy. Biol Psychiatry (1997) 42:24–9. doi: 10.1016/S0006-3223(96)00243-0
86. Lee HJ, Sung SM, Lim SW, Paik JW, Leen Kim. Seasonality associated with the serotonin 2A receptor−1438 A/G polymorphism. J Affect Disord. (2006) 95:145–8. doi: 10.1016/j.jad.2006.05.001
87. Maj J, Bijak M, Dziedzicka-Wasylewska M, Rogoz R, Rogóz Z, Skuza G, et al. The effects of paroxetine given repeatedly on the 5-HT receptor subpopulations in the rat brain. Psychopharmacology (1996) 127:73–82. doi: 10.1007/BF02805977
88. Enoch MA, Goldman D, Barnett R, Sher L, Mazzanti CM, Rosenthal NE. Association between seasonal affective disorder and the 5-HT2A promoter polymorphism, −1438G/A. Mol Psychiatry (1999) 4:89–92. doi: 10.1038/sj.mp.4000439
89. Arias B, Gutiérrez B, Pintor L, Gastó C, Fa-anás L. Variability in the 5-HT(2A) receptor gene is associated with seasonal pattern in major depression. Mol Psychiatry (2001) 6:239–42. doi: 10.1038/sj.mp.4000818
90. Levitan RD, Masellis M, Basile VS, Lam RW, Jain U, Kaplan AS, et al. Polymorphism of the serotonin-2A receptor gene (HTR2A) associated with childhood attention deficit hyperactivity disorder (ADHD) in adult women with seasonal affective disorder. J Affect Disord. (2002) 71:229–33. doi: 10.1016/S0165-0327(01)00372-X
91. Johansson C, Willeit M, Smedh C, Ekholm J, Paunio T, Kieseppä T, et al. Circadian clock-related polymorphisms in seasonal affective disorder and their relevance to diurnal preference. Neuropsychopharmacology (2003) 28:734–9. doi: 10.1038/sj.npp.1300121
92. Partonen T, Treutlein J, Alpman A, Frank J, Johansson C, Depner M, et al. Three circadian clock genes Per2, Arntl, and Npas2 contribute to winter depression. Ann Med. (2007) 39:229–38. doi: 10.1080/07853890701278795
93. Kovanen L, Saarikoski ST, Aromaa A, Lönnqvist J, Partonen T. ARNTL (BMAL1) and NPAS2 gene variants contribute to fertility and seasonality. PLoS ONE (2010) 5:e10007. doi: 10.1371/journal.pone.0010007
94. Kim HI, Lee HJ, Cho CH, Kang SG, Yoon HK, Park YM, et al. Association of CLOCK, ARNTL, and NPAS2 gene polymorphisms and seasonal variations in mood and behavior. Chronobiol Int. (2015) 32:785–91. doi: 10.3109/07420528.2015.1049613
95. Paik JW, Lee HJ, Kang SG, Lim SW, Lee MS, Kim L. CLOCK gene 3111C/T polymorphism is not associated with seasonal variations in mood and behavior in Korean college students. Psychiatry Clin Neurosci. (2007) 61:124–6. doi: 10.1111/j.1440-1819.2007.01621.x
96. Berson DM, Dunn FA, Takao M. Phototransduction by retinal ganglion cells that set the circadian clock. Science (2002) 295:1070–3. doi: 10.1126/science.1067262
97. Hattar S, Liao H, Takao M, Berson D, Yau K. Melanopsin-containing retinal ganglion cells: architecture, projections, and intrinsic photosensitivity. Science (2002) 295:1065–70. doi: 10.1126/science.1069609
98. Ruby NF, Brennan TJ, Xie X, Cao V, Franken P, Heller HC, et al. Role of melanopsin in circadian responses to light. Science (2002) 298:2211–3. doi: 10.1126/science.1076701
99. Roecklein KA, Rohan KJ, Duncan WC, Rollag MD, Rosenthal NE, Lipsky RH, et al. A missense variant (P10L) of the melanopsin (OPN4) gene in seasonal affective disorder. J Affect Disord. (2009) 114:279–85. doi: 10.1016/j.jad.2008.08.005
100. Roecklein KA, Wong PM, Franzen PL, Hasler BP, Wood-Vasey WM, Nimgaonkar VL, et al. Melanopsin gene variations interact with season to predict sleep onset and chronotype. Chronobiol Int. (2012) 29:1036–47. doi: 10.3109/07420528.2012.706766
101. Terman M, Reme C, Rafferty B, Gallin P, Terman J. Bright light therapy for winter depression: potential ocular effects and theoretical implications. Photochem Photobiol. (1990) 51:781–92. doi: 10.1111/php.1990.51.6.781
102. Hébert M, Beattie CW, Tam EM, Yatham LN, Lam RW. Electroretinography in patients with winter seasonal affective disorder. Psychiatry Res. (2004) 127:27–34. doi: 10.1016/j.psychres.2004.03.006
103. Gagné A-M, Hébert M. Atypical pattern of rod electroretinogram modulation by recent light history: a possible biomarker of seasonal affective disorder. Psychiatry Res. (2011) 187:370–4. doi: 10.1016/j.psychres.2010.08.010
104. Roecklein K, Miller M, Donofry S, Dupont C, Hasler B, Franzen P, et al. Melanopsin driven pupillary reflexes in seasonal affective disorder. Meeting Abstract, Society for Light Treatment and Biological Rhythms. Neuropsychobiology (2016) 74:248. doi: 10.1159/000477426
105. Delavest M, Even C, Benjemaa N, Poirier MF, Jockers R, Krebs MO. Association of the intronic rs2072621 polymorphism of the X-linked GPR50 gene with affective disorder with seasonal pattern. Eur Psychiatry (2012) 27:369–71. doi: 10.1016/j.eurpsy.2011.02.011
106. Yang SY, Baek JH, Cho Y, Cho EY, Choi Y, Kim Y, et al. Effects of genetic variants of ST8SIA2 and NCAM1 genes on seasonal mood changes and circadian preference in the general population. Chronobiol Int. (2017) 35:1–11. doi: 10.1080/07420528.2017.1410827
107. Holtmann G, Siffert W, Haag S, Mueller N, Langkafel M, Senf W, et al. G-protein beta 3 subunit 825 CC genotype is associated with unexplained (functional) dyspepsia. Gastroenterology (2004) 126:971–79. doi: 10.1053/j.gastro.2004.01.006
108. Jacobi J, Hilgers KF, Schlaich MP, Siffert W, Schmieder RE. 825T allele of the G-protein beta3 subunit gene (GNB3) is associated with impaired left ventricular diastolic filling in essential hypertension. J Hypertens (1999) 17:1457–62. doi: 10.1097/00004872-199917100-00014
109. Willeit M, Praschak-Rieder N, Zill P, Neumeister A, Ackenheil M, Kasper S, et al. C825T polymorphism in the G protein beta3-subunit gene is associated with seasonal affective disorder. Biol Psychiatry (2003) 54:682–6. doi: 10.1016/S0006-3223(03)00169-0
110. Johansson C, Willeit M, Aron L, Smedh C, Ekholm J, Paunio T, et al. Seasonal affective disorder and the G-protein beta-3-subunit C825T polymorphism. Biol Psychiatry (2004) 55:317–9. doi: 10.1016/S0006-3223(03)00640-1
111. Wirz-Justice A, Benedetti F, Terman M editors. Chronotherapeutics for affective disorders. In: A Clinician's Manual for Light and Wake Therapy. 2nd ed. Basel: Karger (2013). p. 32.
112. Takahashi JS, Hong HK, Ko CH, McDearmon EL. The genetics of mammalian circadian order and disorder: implications for physiology and disease. Nat Rev Genet. (2008) 9:764–75. doi: 10.1038/nrg2430
113. Shimomura K, Lowrey PL, Vitaterna MH, Buhr ED, Kumar V, Hanna P, et al. Genetic suppression of the circadian Clock mutation by the melatonin biosynthesis pathway. Proc Natl Acad Sci USA. (2010) 107:8399–403. doi: 10.1073/pnas.1004368107
114. Brown SA, Kunz D, Dumas A, Westermark PO, Vanselow K, Tilmann-Wahnschaffe A, et al. Molecular insights into human daily behavior. Proc Natl Acad Sci USA. (2008) 105:1602–7. doi: 10.1073/pnas.0707772105
Keywords: seasonality, mood disorders, clock genes, circadian rhythm, seasonal affective disorder
Citation: Garbazza C and Benedetti F (2018) Genetic Factors Affecting Seasonality, Mood, and the Circadian Clock. Front. Endocrinol. 9:481. doi: 10.3389/fendo.2018.00481
Received: 02 June 2018; Accepted: 03 August 2018;
Published: 23 August 2018.
Edited by:
Mario Caba, Universidad Veracruzana, MexicoReviewed by:
Axel Steiger, Max-Planck-Institut für Psychiatrie, GermanyTimo Partonen, National Institute for Health and Welfare, Finland
Copyright © 2018 Garbazza and Benedetti. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Corrado Garbazza, Y29ycmFkby5nYXJiYXp6YUB1cGticy5jaA==
 Corrado Garbazza
Corrado Garbazza Francesco Benedetti
Francesco Benedetti