
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
PERSPECTIVE article
Front. Earth Sci. , 04 September 2019
Sec. Biogeoscience
Volume 7 - 2019 | https://doi.org/10.3389/feart.2019.00225
This article is part of the Research Topic Deep Carbon Science View all 17 articles
The deep marine biosphere is one of the largest, and yet least explored, microbial habitats on the planet. Quantifying the extent, diversity, and activity of subsurface microbial communities is a crucial part of understanding their role in global biogeochemical cycles. Even though deep biosphere habitats can vary widely in chemistry, temperature, turnover rates, and energy sources, all subsurface microbes inherently experience high pressures. While not all subsurface microbes require elevated pressures, for many high pressures are essential to their cellular function and metabolism. Thus, when targeting this elusive portion of the biosphere, it is critical to maintain in situ pressure while sampling and cultivating subsurface microorganisms. In this perspective paper we highlight the sampling and cultivation technologies available to study these communities under in situ conditions. Maintaining elevated pressures throughout sampling, transfer, cultivation, and isolation is challenging, and more often than not samples are decompressed at some point during sample handling, potentially leading to biases in both community diversity and isolate physiology. The development of devices that maintain in situ pressures during sampling and allow for sample transfer without decompression have begun to address this challenge (like the PUSH – Pressurized Underwater Sample Handler). Such vessels can be used for both retrieval and enrichment of deep subsurface samples, as well as high-pressure growth and physiology experiments, thus expanding possibilities for deep biosphere exploration. Finally, we discuss the significant need to develop and share high-pressure facilities across the deep biosphere community, in order to expand the opportunities to discover novel piezophiles from the deep subsurface.
In this communication we consider the deep biosphere to include all environments at and above 10 MPa in the water column (10 MPa/km), subseafloor sediments (15 MPa/km) and all the potential reservoirs in the continental and oceanic crust (25 MPa/km) (Jannasch and Taylor, 1984; Oger and Jebbar, 2010; Picard and Daniel, 2013), though we focus primarily on marine environments. The deep-sea, defined as waters and sediments beneath 1000 m depth, is the Earth’s largest ecosystem, representing 65% of the planet’s surface and encompassing 88% of the global biosphere (Jannasch and Taylor, 1984). The ocean has an average depth of 3800 m and an average pressure of 38 MPa (Herring and Clarke, 1971), and even the deepest waters (Mariana Trench at 11 km, 110 MPa) are known to host life (Glud et al., 2013; Nunoura et al., 2015; Tarn et al., 2016). The habitable zones of the continental crust are similarly vast and underexplored, and also host unique microbial communities (Stevens and McKinley, 1995; Takai et al., 2001; Moser et al., 2005; Lollar et al., 2006; Lavalleur and Colwell, 2013; Borgonie et al., 2015). This remote and dark biosphere hosts a range of subsurface habitats typically characterized by extreme conditions (Jørgensen and Boetius, 2007). They are mostly nutrient-poor – excluding the hot spots (e.g., hydrothermal vents) – and have slow or transient energy fluxes supplied by water-rock reactions and circulating crustal fluids (Orcutt et al., 2011). Despite harsh conditions, these ecosystems define the largest microbial habitat on Earth (Whitman et al., 1998; Kallmeyer et al., 2012; Bar-On et al., 2018). The subsurface is estimated to hold ∼15% of the total terrestrial biomass (Bar-On et al., 2018), and resident microbial communities potentially play an important role in mediating global biogeochemical cycles (D’Hondt et al., 2004; D’Hondt et al., 2009). However, only 5% of the oceans have been investigated using remote instruments and less than 0.01% has been sampled and studied (Ramirez-Llodra et al., 2010). Despite being such a large fraction of Earth’s biomass the microbial diversity, activity and distribution, metabolic pathways, and energy fluxes of the subsurface biosphere are poorly understood because access is limited by technical and economic challenges. Over the past decade, international drilling programs (IODP – International Ocean Discovery Program, ICDP – International Continental Scientific Drilling Program), as well as national submersible facilities have expanded subsurface microbiology research to better understand how intra-terrestrial life inhabiting the deep crust and deep oceans plays a role within the deep biosphere. Subsurface technologies and recent advances in this field are well documented in Schrenk et al. (2010), Edwards et al. (2011), and Edwards et al. (2012).
A majority of prokaryotes live under high-pressure conditions, and thus pressure is a governing factor for the distribution of life (Picard and Daniel, 2013), and subsurface microorganisms are classified by their physiological responses (e.g., growth rates, cell yields) to pressure (Abe, 2007; Fang et al., 2010). Usually, well-adapted surface microorganisms are negatively impacted by high-pressure conditions (piezosensitive), even though some can grow at elevated pressures (piezotolerant). Likewise, microorganisms isolated from the subsurface often grow optimally at high-pressure (piezophiles), and obligate piezophiles are unable to grow at ambient (surface) pressures. In the case of the deep marine biosphere, evidence suggests that decompression can deleteriously impact cellular viability, including morphological changes (Chastain and Yayanos, 1991), membrane cell rupture (Park and Clark, 2002), and piezophile inactivation (Yayanos and Dietz, 1982; Yayanos and Dietz, 1983). Furthermore, significant shifts in community composition and gene expression have been noted as a result of sample decompression (La Cono et al., 2015; Edgcomb et al., 2016). To date, the number of facultative and obligate piezophiles is limited to ∼56 (Figure 1A and Supplementary Table S2) (Picard and Daniel, 2013; Jebbar et al., 2015). Among the obligate piezophiles, Colwellia marinimaniae has the highest optimum growth pressure at 120 MPa (Kusube et al., 2017). This psychrophile and the hyperthermophilic Pyrococcus yayanosii (Zeng et al., 2009), currently claim the highest pressure limits of growth (140 and 120 MPa, respectively). Although the majority of currently known piezophiles were isolated following decompression, these isolates may be notable exceptions, representing only the fraction of the subsurface biosphere tolerant of decompression. Indeed a much broader microbial community has been identified in the subsurface through alternative methods, including genetic and molecular approaches (Orsi et al., 2013), in situ metabolic rate measurements (Jørgensen et al., 1992; Kallmeyer and Boetius, 2004; Kallmeyer et al., 2012), intact polar lipid analysis (Zink et al., 2003; Lipp et al., 2008), stable isotope enrichments (Morono et al., 2011), cell counts (Parkes et al., 1994), and growth of viable cultures (D’Hondt et al., 2004; Smith et al., 2011). It should be noted that many of these observations were made on decompressed samples. While such results of ambient pressure experiments might extend our view of the deep biosphere, they are nonetheless biased toward surface pressures and likely do not reflect microbial processes at in situ pressure conditions.
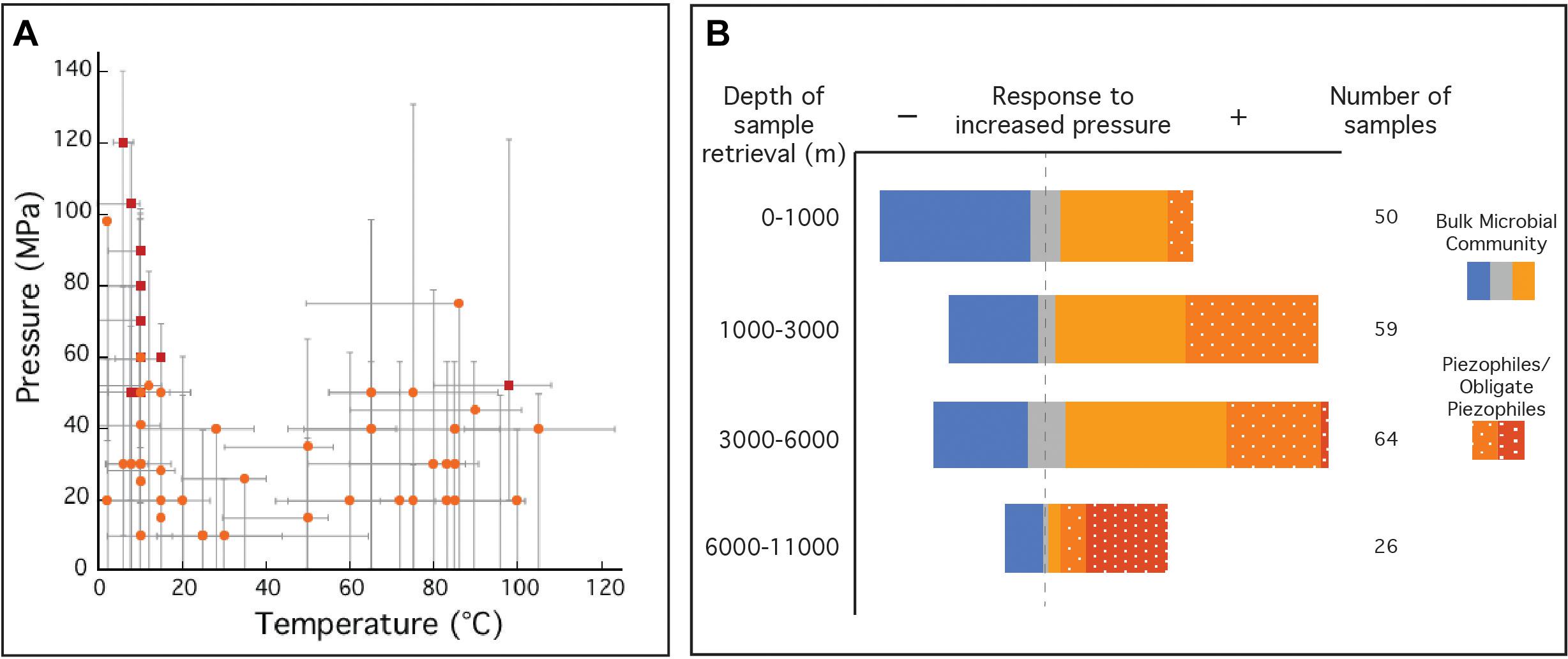
Figure 1. (A) Optimal temperature and pressure growth conditions for known piezophiles (light orange circles) and obligate piezophiles (red squares). Gray bars indicate the reported temperature and pressure growth ranges for each isolate. (B) Collection of natural samples and isolates with sample origins at various depths and their response to elevated pressure compared to ambient pressure (0.1 MPa). Natural samples (solid), where metabolic activity was monitored, was reported to have either a negative (solid blue), neutral (solid gray) or positive (solid orange) response to increased pressure compared to growth at 0.1 MPa. Isolated piezophiles (dotted orange) are described here as having optimum growth at pressures above 0.1 MPa and obligate piezophiles (dotted red) are those that require elevated pressures for growth and cannot grow at 0.1 MPa. This figure is not an exhaustive list of all natural samples taken from the deep biosphere with reported pressure tolerances and does not include piezotolerant organisms (optimum growth pressure at 0.1 MPa). Temperature and pressure data for isolates in A,B were collected from references listed in Supplementary Table S2. Data for natural samples were extracted from Picard and Daniel (2013).
The limited number of piezophilic isolates also hinders our ability to investigate the effects of high-pressure on metabolism and physiology (Tamburini, 2006). Piezophiles exhibit different strategies to cope with elevated pressure and maintain cell integrity over a wide pressure range (Bartlett, 2002; Abe, 2007; Oger and Jebbar, 2010) including (i) cell wall and lipid membrane biochemistry (DeLong and Yayanos, 1985; Allen and Bartlett, 2002; Cario et al., 2015); (ii) intracellular salt content and osmolyte regulation (Martin et al., 2002; Cario et al., 2016b); (iii) specific high-pressure gene expressions (Kato and Qureshi, 1999; Campanaro et al., 2005; Simonato et al., 2006; Vannier et al., 2015; Michoud and Jebbar, 2016); and (iv) macromolecule structural modulation (Kawano et al., 2004; Rosenbaum et al., 2012). These unique piezophilic strategies have been identified via a limited number of high-pressure cultivation experiments, but there are diverse, uncultured species within the deep biosphere that undoubtedly have unknown metabolic and physiological strategies. In the absence of additional isolates, these unknown functions have been probed with high-pressure experiments conducted on bulk samples from the deep biosphere (e.g., sediments, fluids, enrichment cultures). Among these >200 varied experiments one general trend that has emerged is that retrieval depth is broadly correlated with a positive response to elevated pressure (Figure 1B). These results suggest that decompression and/or ambient pressure conditions are a challenge for subsurface organisms, and that incorporating elevated pressure conditions into sampling and experimental protocols could greatly improve experimental outcomes. Below we review the sampling and cultivation techniques commonly adopted in marine subsurface biosphere research, the limitations inherent to these approaches, and strategies to improve subsurface exploration through technology development and new collaborative models (Figure 2).
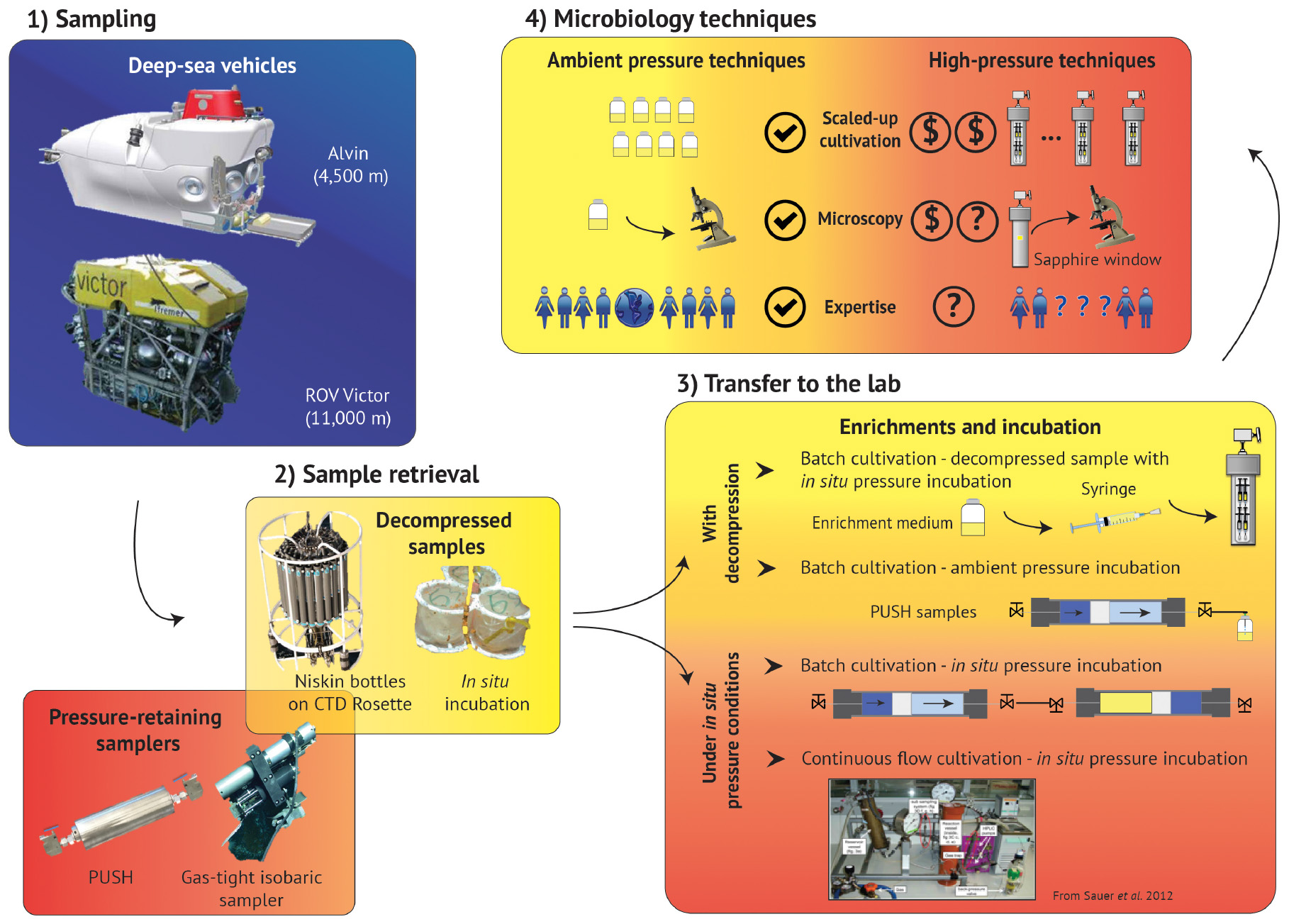
Figure 2. Summary schematic of the deep-sea exploration workflow, from sampling to laboratory experiments. Access to deep marine environments is enabled with submersibles (1 – Sampling), along with the various devices that support 2 – Sample retrieval. 3 – Transfer to the lab and subsequent cultivation techniques often include batch cultivation, but can also accommodate continuous flow methods, though many of these techniques require sample decompression. 4 – Microbiology techniques are much more robust for ambient pressure conditions, and high-pressure analogs of these methods require technology development, high-pressure expertise, and can also increase the costs, and thus decrease the accessibility, of many of these protocols. (Box 1: Alvin picture is reprinted from Woods Hole Oceanographic Institution website (link: https://www.whoi.edu/oceanus/feature/building-the-next-generation-alvin-submersible/) 2019 Copyright WHOI, ROV Victor picture is reprinted from Ifremer website (link: https://wwz.ifremer.fr/grands_fonds/Les-moyens/Les-engins/Les-robots/Robots-Ifremer/Le-Victor-6000) 2019 Copyright IFREMER; Box 2: PUSH is a personal picture, Isobaric sampler picture is reprinted from Woods Hole Oceanographic Institution website (link: https://www.whoi.edu/press-room/news-release/surprisingly-productive-microbes-fuel-deep-sea-ecosystem/) 2019 Copyright WHOI, CTD rosette picture is Reprinted from NOAA website (link: https://oceanexplorer.noaa.gov) 2019 Copyright NOAA, in situ incubation is reprinted with permission from Rassa et al. (2009). Copyright 2019 Taylor & Francis Group; Box 3: Reprint with permission from Sauer et al. (2012). © 2012 Sauer, Glombitza, and Kallmeyer (CC BY-NC 3.0). 2019.)
Since the discovery of deep-sea and subsurface microbial life (Zobell, 1952; Corliss et al., 1979; Parkes et al., 1994), the development of advanced instrumentation for observing, retrieving, and manipulating samples has illuminated some of the key characteristics of the deep geosphere and biosphere (Tengberg et al., 1995; Ramirez-Llodra et al., 2010). International collaboration and national efforts are the hallmarks of subsurface exploration, including drilling and submersibles. Similarly large international programs support the research communities that explore the deep biosphere and improve our knowledge of the subsurface microbiome (cf. ICOMM – International Census of Marine Microbes; C-DEBI – Center for Dark Energy Biosphere Investigations; and DCO – Deep Carbon Observatory). Only three human expeditions have explored the deepest part of the ocean (Mariana Trench, 11 km) (Oppenheimer and Zobell, 1952; Gallo et al., 2015; Fitzherbert, 2019), leaving 95% of the ocean unexplored directly (Ramirez-Llodra et al., 2010), even though other HOVs (human occupied vehicles), like the Shinkai 6500 and Nautile that reach up to 6500 m, allow for indirect exploration of nearly 98% of the ocean floor. Moreover, a veritable army of landers, ROVs (remotely operated vehicles), and AUVs (autonomous underwater vehicles) enable much of today’s deep ocean exploration (Supplementary Table S1) (Tengberg et al., 1995; Reysenbach and Götz, 2001; Yayanos, 2001; Tamburini, 2006), while a new class of Internet operated vehicles (IOVs) and mobile platforms provide extended monitoring of both the seafloor and the water column (Aguzzi et al., 2019).
While drilling and exploration vehicles remain few, instruments to obtain subsurface samples are more numerous (Supplementary Table S1). The paltry list of isolated piezophiles reflects the difficulty of both retrieving and cultivating piezophiles from subsurface samples. Because decompression affects cellular viability, and because most samples are decompressed during sample handling, it remains possible that most piezophilic strains elude current sampling and culturing techniques. Therefore, several pressure-retaining devices have been developed to enable sampling without decompression (Jannasch et al., 1973; Yayanos, 1977; Bianchi et al., 1999; Tamburini et al., 2013). Recently, Tamburini’s team used a pressure-retaining sampler in the bathypelagic zone to measure prokaryote activity under in situ pressure conditions, and demonstrated that decompression suppresses both piezophilic metabolisms and water column community composition (Garel et al., 2019). A similar pressure-retaining sampler was developed as part of DCO’s PRIME (Piezophile Research Instrumentation for Microbial Exploration) Facility. The Pressurized Underwater Sample Handler System (PUSH) is similar to Bianchi’s sampler (Bianchi et al., 1999), and uses a floating piston system to enable sampling, transport, transfer, and cultivation of high-pressure fluids without decompression (Rogers et al., 2016b).
Accessing the deep marine biosphere and retrieving samples at in situ pressures has proven a significant technological challenge for deep biosphere exploration, but these improvements can be amplified if subsequent laboratory studies, particularly cultivation experiments, maintain high-pressure conditions. It is particularly important to maintain in situ pressure conditions during sampling, transfer to the laboratory, and throughout cultivation and storage, in order to avoid decompression bias. When targeting novel deep biosphere microbes, replicating in situ environmental conditions (i.e., pressure, temperature, pH, volatile composition, low nutrients concentrations) and subsampling without decompression are crucial aspects of the cultivation process. Since traditional laboratory culturing techniques are not easily adapted to high-pressure, customized devices and protocols have been developed. Yayanos and colleagues, the pioneers of high-pressure microbiology, developed early high-pressure cultivation apparatus (Yayanos and Pollard, 1969; Yayanos et al., 1979; Yayanos et al., 1982; Yayanos et al., 1984; Yayanos, 1986; DeLong et al., 1997), and these have since been expanded (Taylor and Jannasch, 1976; Bernhardt et al., 1987; Nauhaus et al., 2002; Takai et al., 2008). Cultivating piezophiles requires expertise in both high-pressure equipment (e.g., hydrostatic pumps, high-pressure vessels) (Kato et al., 2008; Kato, 2011) as well as manipulating high-pressure cultures. Most of these high-pressure devices are batch reactors, using a compressible reaction container in static high-pressure vessels. Typically, a stoppered syringe containing inoculated growth medium will be pressurized in a hydrostatic pressure vessel (e.g., Takai et al., 2008). Unfortunately, this setup requires decompression during subsampling, potentially suppressing growth rates and cell viability (Cario et al., 2016a; Rogers et al., 2016a, Oliver, 2019). Variable-volume reactors, like the PUSH and others (Seyfried et al., 1979; Bianchi et al., 1999; Cario et al., 2016a; Rogers et al., 2016b; Garel et al., 2019; Oliver, 2019; Supplementary Table S1), can overcome this limitation, and have recently been used for batch cultivation without decompression, identifying two pressure-tolerant strains previously thought to be pressure-sensitive (Cario et al., 2016a; Oliver, 2019). This technology has been extended to allow for high-pressure transfer (Oliver, 2019), a crucial step in replicating traditional liquid culture microbial cultivation and expanding the possibilities for high-pressure isolation from enrichment cultures. A further improvement that better replicates natural geological environments are continuous flow-through systems. Jannasch and colleagues first developed a high-pressure (71 MPa maximum) continuous culture system for low temperatures (Jannasch et al., 1996). Sauer et al. designed a flow-through bioreactor (120°C and 60 MPa) that allows for volatile equilibration of the culture medium, subsampling without decompression, and the inclusion of a solid phase (Sauer et al., 2012). Recently, in the Rogers’ lab, a similar continuous flow reactor was built that extended this pressure range to 100 MPa and incorporated a module for sample fixation prior to decompression (Supplementary Table S1).
Deep ocean sediments can also host a significant portion of the subsurface biosphere (Jørgensen and Boetius, 2007; Kallmeyer et al., 2012; Lloyd et al., 2013); However, retrieving deep ocean sediments and maintaining in situ pressures during cultivation and transfer of sediment present additional challenges. High-pressure sediments have been retrieved during drilling with pressure-coring systems (Reed et al., 2002; Kubo et al., 2014), but only the DeepIsoBUG system (Parkes et al., 2009) can transfer sediment under pressure for subsequent cultivation. Otherwise, most methods developed for high-pressure incubation and enrichment of sediments require decompression. Nonetheless, these devices can accommodate general batch cultivation, enhanced volatile concentrations, and flow-through conditions, and have confirmed that microbial metabolic rates are faster at elevated pressures in the presence of increased volatile substrate concentrations (Nauhaus et al., 2002; Bowles et al., 2011; Sauer et al., 2012). Given the extent of the subsurface microbiome contained in deep-sea sediments, the deleterious impact of decompression, and the documented importance of elevated pressure on growth and metabolism, expansion and improvement of high-pressure sediment manipulation remains ripe for high-impact technological developments.
In the modern era of microbiology, the capabilities of Earth’s extremophiles have far outpaced our ability to conceive of the bounds of habitability. For each perceived limit – temperature, pressure, pH, energy flux, etc. – a novel and hard-to-imagine extremophile has surpassed expectations and extended the reach of potential habitable environments. In contrast, our technological capabilities have not kept pace with this ever-expanding habitable envelope. Even with the current known temperature and pressure limits of life, we have not yet explored most of the habitable subsurface. The primary limitations are the expense of drilling and oceanographic expeditions, together with technological limitations for sampling and sample manipulation. In the deepest parts of the ocean, pressures can reach 110 MPa, and access to these environments is limited to only a few ROVs and landers. Recently, Bartlett and colleagues developed a modular lander capable of sampling these hadal trenches at depths greater than 6000 m (Peoples et al., 2019). While spatially limited, these regions nonetheless represent 45% of the depth range of the ocean (Jamieson, 2015). Sampling at these pressures also poses technological challenges, as most samplers are designed to access meso- and bathypelagic zones (200–1000 and 1000–4000 m, respectively), and only a few are designed to reach deeper zones. Access to subsurface crustal and sedimentary ecosystems is limited by drilling technology, and high-pressure retrieval of solid-phase samples is rare. To our knowledge only fluid samples have been collected from deep crustal aquifers at in situ pressures (Hatanpää et al., 2005; Hallbeck and Pedersen, 2008) and high-pressure sediment retrieval is pressure-limited (Kubo et al., 2014). These latter examples highlight one of the more daunting challenges to understanding the deep biosphere, which is the scarcity of samples. Subsurface sampling is not only sparse, but often targets specific kinds of environments (e.g., hydrothermal systems, trenches, etc.), resulting in a lack of coverage for the “average” deep subsurface. Therefore, it is difficult to extrapolate across the entire subsurface biosphere, leaving large errors on our estimates for subsurface biomass, as well as metabolic, physiologic and genetic diversity.
The last few decades have witnessed a dramatic increase in subsurface exploration and research, extending our access to and understanding of the deep biosphere, nonetheless, sampling and analysis challenges remain. Here we focus on four areas of opportunity for innovation (Figure 2): (i) sampling at in situ pressure conditions; (ii) high-pressure cultivation techniques; (iii) dissemination of high-pressure skills among the community; and (iv) increasing participation and/or incorporation of high-pressure sampling into deep-sea and drilling expeditions. We propose that community priorities going forward focus on deepening our knowledge of the deep biosphere and its role in global biogeochemical cycles, particularly by addressing technology development and implementing novel management and collaborative models to increase community access to high-pressure samples and devices.
Despite the ubiquity of prokaryotes within the deep biosphere, only a limited number have been cultivated (e.g., 0.1%) (D’Hondt et al., 2004). Illustrative of this point is the distribution of the isolated piezophiles (Figure 1A), with pressure maxima among the psychrophiles of the hadal zones and vent hyperthermophiles. The moderate temperature and pressure regime (e.g., ∼20 –60°C and ∼40–80 MPa) lacks piezophilic isolates despite the prevalence of such conditions within deep-sea sediments (Picard and Daniel, 2013). Similarly, the moderate pressure extent of the isolated hyperthermophilic piezophiles (Popt < 80 MPa) reflects the conditions of the most accessible fluids from mid-ocean ridge high-temperature vents. Ecosystems at higher pressures and more moderate temperatures (e.g., subduction zones, crust off-axis of spreading centers) are among the least accessed habitats of the subsurface. Exploring the full breadth of the piezosphere will require new drilling and sampling techniques targeting high-pressure retrieval and manipulations of rock and sediment samples. More broadly across the piezosphere, better cultivation methods that mimic in situ environmental conditions (i.e., temperature, pressure, chemical composition), could improve enrichment and isolation of novel piezophiles. Targeting several metabolic and physiologic niches requires scaling up high-pressure cultivation either by vastly increasing the number of pressure vessels or developing new technologies for multiplexing techniques (Kallmeyer et al., 2003). The deep biosphere also hosts microbial communities with slow metabolic rates surviving on minimal energy sources (D’Hondt et al., 2002). Traditional enrichment and isolation techniques favor fast growth under high-energy conditions, and are less likely to capture the slow pace of the deep biosphere microbiome. In situ or long-term incubations could help target the deep slow biosphere, but these require innovations for long-term incubations and high-pressure retrieval. While it is clear that every microbiological and molecular tool should be brought to bear to understand the deep biosphere, many of these need to be adapted to accommodate the elevated pressures of the subsurface. For example, monitoring metabolic rates during controlled incubations often relies on radiolabeled substrates (Frank et al., 2015), but adding radioactive material to pressure vessels introduces significant shipboard and laboratory safety concerns. Electrochemical monitoring of in situ chemistry during laboratory cultivation is also quite common in benchtop microbiology, but a more significant technological challenge when incubations are conducted at elevated pressures. Visualizing cells and microbial communities with high-pressure microscopy can be accomplished via specialized equipment (e.g., DACs), but is hardly as routine as it is in the traditional microbiology workflow. Even the most basic methods like dilution-to-extinction isolation protocols, proteomic analyses, or compound-specific stable isotope measurements, require either large numbers or larger volumes of high-pressure devices, limiting wide-spread use.
Molecular techniques, which are often used to give a more robust picture of microbial diversity compared to cultivation-based approaches, have indeed revealed that subsurface microbial diversity is broader than that represented by the cultivated piezophiles (Orsi et al., 2013). However, recent work suggests that even molecular techniques can suffer from decompression bias (Garel et al., 2019). Nonetheless, continued application of next-generation –omics techniques could further elucidate the diversity and activity of the subsurface microbiome (Nunoura et al., 2010), and be used to improve culturing efforts. For example, GeoChip-based high-throughput metagenomics have highlighted metabolic functions within dynamic microbial communities (He et al., 2007; Wang et al., 2009). Furthermore, the combination of stable-isotope probing (Webster et al., 2010; Lin et al., 2013; Fortunato and Huber, 2016), FISH-SIMS (Behrens et al., 2008) and single-cell sorting (Lasken, 2007) could further elucidate the links between microbial diversity, environmental conditions, and ecosystem processes, and develop more robust models of the deep biosphere and its connection to long-term biogeochemical processes.
Finally, we believe that sharing high-pressure expertise and equipment within the deep biosphere community, and expanding the scope of high-pressure microbial research, will advance our understanding of the deep biosphere. The number of facilities that can sample and manipulate subsurface samples at in situ pressures is considerably smaller than the number of research groups that regularly sample and study the deep subsurface. The barrier to entry (expense and expertise) into high-pressure techniques is high, but better coordination and new collaborative models can change this paradigm. The DCO, via the PRIME facility, supported the purchase of several PUSH samplers to share within the community, with the goal of isolating and characterizing more piezophilic microorganisms. However, equipment sharing will only be successful if it is accompanied by the requisite personnel and expertise. Therefore, new collaborative models that include equipment, personnel, and funding need to be developed to keep community-wide facilities successful and sustainable.
The Earth’s deep biosphere not only represents a crucial segment of the global biosphere and modern biogeochemical cycles, but it also serves as a window into Earth’s habitable environments throughout its history. Furthermore, as the exploration for life beyond Earth shifts to other solar system bodies, including the subsurface of Mars (Stamenković et al., 2019), or the subsurface ocean worlds of Europa, Enceladus and other moons, understanding how these environments can foster life becomes crucial to this exploration. Therefore, the technology developed to explore Earth’s modern deep biosphere will eventually become a key component of missions to these bodies to search for signatures of extant extraterrestrial life or its remnant biosignatures.
The datasets generated for this study are available on request to the corresponding author.
AC, GO, and KR conceived the study. AC wrote the original draft. KR and GO wrote sections of the manuscript. GO and AC created the tables and figures. All authors contributed to the revisions of the manuscript, tables, and figures, and approved the submitted version.
Funding for this work was provided by NASA (NNX13AP2G9 and 80NSSC17K0252 to KR), the Deep Carbon Observatory (Subawards: 10371-07, 10561-01, and 10311-11 to KR), an NSF Graduate Fellowship (FAIN 1247271 and 1744655 to GO), and a GSA Research Grant to GO. Additional support was provided by startup funds from Rensselaer Polytechnic Institute to KR.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The development of the PUSH devices and PRIME facility originated at the DCO Sandpit Workshop on Bioreactors, convened by Isabelle Daniel and sponsored by the Deep Carbon Observatory. We appreciate the contributions from the workshop participants and the entire DCO community in support of this initiative. We further thank Isabelle Daniel, Hervé Cardon, Bruce Watson, and Chris Hoff for technical support. Finally, we thank the two reviewers for helpful comments.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/feart.2019.00225/full#supplementary-material
Abe, F. (2007). Exploration of the effects of high hydrostatic pressure on microbial growth, physiology and survival: perspectives from piezophysiology. Biosci. Biotechnol. Biochem. 71, 2347–2357. doi: 10.1271/bbb.70015
Aguzzi, J., Chatzievangelou, D., Marini, S., Fanelli, E., Danovaro, R., Flögel, S., et al. (2019). New high-tech flexible networks for the monitoring of deep-sea ecosystems. Environ. Sci. Technol. 53, 6616–6631. doi: 10.1021/acs.est.9b00409
Allen, E. E., and Bartlett, D. H. (2002). Structure and regulation of the omega-3 polyunsaturated fatty acid synthase genes from the deep-sea bacterium Photobacterium profundum strain SS9. Microbiology 148, 1903–1913. doi: 10.1099/00221287-148-6-1903
Bar-On, Y. M., Phillips, R., and Milo, R. (2018). The biomass distribution on Earth. Proc. Natl. Acad. Sci. U.S.A. 115, 6506–6511. doi: 10.1073/pnas.1711842115
Bartlett, D. H. (2002). Pressure effects on in vivo microbial processes. Biochim. Biophys. Acta 1595, 367–381. doi: 10.1016/s0167-4838(01)00357-0
Behrens, S., Lösekann, T., Pett-Ridge, J., Weber, P. K., Ng, W.-O., Stevenson, B. S., et al. (2008). Linking microbial phylogeny to metabolic activity at the single-cell level by using enhanced element labeling-catalyzed reporter deposition fluorescence in situ hybridization (EL-FISH) and NanoSIMS. Appl. Environ. Microbiol. 74, 3143–3150. doi: 10.1128/AEM.00191-08
Bernhardt, G., Jaenicke, R., and Lüdemann, H.-D. (1987). High-pressure equipment for growing methanogenic microorganisms on gaseous substrates at high temperature. Appl. Environ. Microbiol. 53, 1876–1879.
Bianchi, A., Garcin, J., and Tholosan, O. (1999). A high-pressure serial sampler to measure microbial activity in the deep sea. Deep Sea Res. I Oceanogr. Res. Papers 46, 2129–2142. doi: 10.1016/s0967-0637(99)00039-4
Borgonie, G., Linage-Alvarez, B., Ojo, A., Mundle, S., Freese, L. B., Van Rooyen, C., et al. (2015). Eukaryotic opportunists dominate the deep-subsurface biosphere in South Africa. Nat. Commun. 6:8952. doi: 10.1038/ncomms9952
Bowles, M. W., Samarkin, V. A., and Joye, S. B. (2011). Improved measurement of microbial activity in deep-sea sediments at in situ pressure and methane concentration. Limnol. Oceanogr. Methods 9, 499–506. doi: 10.4319/lom.2011.9.499
Campanaro, S., Vezzi, A., Vitulo, N., Lauro, F., D’Angelo, M., Simonato, F., et al. (2005). Laterally transferred elements and high pressure adaptation in Photobacterium profundum strains. BMC Genomics 6:122.
Cario, A., Daniel, I., and Rogers, K. L. (2016a). “Increased pressure tolerance of a marine bacterium when cultivated under high-pressure conditions without subsampling decompression,” in Proceedings of the 11th International Congress on Extremophiles 2016, Kyoto.
Cario, A., Jebbar, M., Thiel, A., Kervarec, N., and Oger, P. M. (2016b). Molecular chaperone accumulation as a function of stress evidences adaptation to high hydrostatic pressure in the piezophilic archaeon Thermococcus barophilus. Sci. Rep. 6:29483. doi: 10.1038/srep29483
Cario, A., Grossi, V., Schaeffer, P., and Oger, P. M. (2015). Membrane homeoviscous adaptation in the piezo-hyperthermophilic archaeon Thermococcus barophilus. Front. Microbiol. 6:1152. doi: 10.3389/fmicb.2015.01152
Chastain, R. A., and Yayanos, A. A. (1991). Ultrastructural changes in an obligately barophilic marine bacterium after decompression. Appl. Environ. Microbiol. 57, 1489–1497.
Corliss, J. B., Dymond, J., Gordon, L. I., Edmond, J. M., von Herzen, R. P., Ballard, R. D., et al. (1979). Submarine thermal springs on the Galapagos Rift. Science 203, 1073–1083. doi: 10.1126/science.203.4385.1073
DeLong, E., Franks, D., and Yayanos, A. (1997). Evolutionary relationships of cultivated psychrophilic and barophilic deep-sea bacteria. Appl. Environ. Microbiol. 63, 2105–2108.
DeLong, E. F., and Yayanos, A. (1985). Adaptation of the membrane lipids of a deep-sea bacterium to changes in hydrostatic pressure. Science 228, 1101–1103. doi: 10.1126/science.3992247
D’Hondt, S., Jørgensen, B. B., Miller, D. J., Batzke, A., Blake, R., Cragg, B. A., et al. (2004). Distributions of microbial activities in deep subseafloor sediments. Science 306, 2216–2221. doi: 10.1126/science.1101155
D’Hondt, S., Rutherford, S., and Spivack, A. J. (2002). Metabolic activity of subsurface life in deep-sea sediments. Science 295, 2067–2070. doi: 10.1126/science.1064878
D’Hondt, S., Spivack, A. J., Pockalny, R., Ferdelman, T. G., Fischer, J. P., Kallmeyer, J., et al. (2009). Subseafloor sedimentary life in the South Pacific Gyre. Proc. Natl. Acad. Sci. 106, 11651–11656. doi: 10.1073/pnas.0811793106
Edgcomb, V. P., Taylor, C., Pachiadaki, M. G., Honjo, S., Engstrom, I., and Yakimov, M. (2016). Comparison of Niskin vs. in situ approaches for analysis of gene expression in deep Mediterranean Sea water samples. Deep Sea Res. II Top. Stud. Oceanogr. 129, 213–222. doi: 10.1016/j.dsr2.2014.10.020
Edwards, K., Fisher, A., and Wheat, C. G. (2012). The deep subsurface biosphere in igneous ocean crust: frontier habitats for microbiological exploration. Front. Microbiol. 3:8. doi: 10.3389/fmicb.2012.00008
Edwards, K. J., Wheat, C. G., and Sylvan, J. B. (2011). Under the sea: microbial life in volcanic oceanic crust. Nat. Rev. Microbiol. 9, 703–712. doi: 10.1038/nrmicro2647
Fang, J., Zhang, L., and Bazylinski, D. A. (2010). Deep-sea piezosphere and piezophiles: geomicrobiology and biogeochemistry. Trends Microbiol. 18, 413–422. doi: 10.1016/j.tim.2010.06.006
Fitzherbert, S. (2019). Deepest Submarine Dive in History, Five Deeps Expedition Conquers Challenger Deep. Dallas, TX: FiveDeepsPress, 1–7.
Fortunato, C. S., and Huber, J. A. (2016). Coupled RNA-SIP and metatranscriptomics of active chemolithoautotrophic communities at a deep-sea hydrothermal vent. ISME J. 10, 1925–1938. doi: 10.1038/ismej.2015.258
Frank, K. L., Rogers, K. L., Rogers, D. R., Johnston, D. T., and Girguis, P. R. (2015). Key factors influencing rates of heterotrophic sulfate reduction in active seafloor hydrothermal massive sulfide deposits. Front. Microbiol. 6:1449. doi: 10.3389/fmicb.2015.01449
Gallo, N. D., Cameron, J., Hardy, K., Fryer, P., Bartlett, D. H., and Levin, L. A. (2015). Submersible-and lander-observed community patterns in the Mariana and New Britain trenches: influence of productivity and depth on epibenthic and scavenging communities. Deep Sea Res. I Oceanogr. Res. Papers 99, 119–133. doi: 10.1016/j.dsr.2014.12.012
Garel, M., Bonin, P., Martini, S., Guasco, S., Roumagnac, M., Bhairy, N., et al. (2019). Pressure-retaining sampler and high-pressure systems to study deep-sea microbes under in situ conditions. Front. Microbiol. 10:453. doi: 10.3389/fmicb.2019.00453
Glud, R. N., Wenzhöfer, F., Middelboe, M., Oguri, K., Turnewitsch, R., Canfield, D. E., et al. (2013). High rates of microbial carbon turnover in sediments in the deepest oceanic trench on Earth. Nat. Geosci. 6, 284–288. doi: 10.1038/ngeo1773
Hallbeck, L., and Pedersen, K. (2008). Characterization of microbial processes in deep aquifers of the Fennoscandian Shield. Appl. Geochem. 23, 1796–1819. doi: 10.1016/j.apgeochem.2008.02.012
Hatanpää, E., Manninen, P., and Apilo, S. (2005). Representativity of Gas Samples Taken with the Pressurized Water Sampling System (PAVE) 1995-2004, Vol. 44. Eurajoki: Posiva Oy, 123.
He, Z., Gentry, T. J., Schadt, C. W., Wu, L., Liebich, J., Chong, S. C., et al. (2007). GeoChip: a comprehensive microarray for investigating biogeochemical, ecological and environmental processes. ISME J. 1, 67–77. doi: 10.1038/ismej.2007.2
Jamieson, A. (2015). The Hadal Zone: Life in the Deepest Oceans. Cambridge: Cambridge University Press.
Jannasch, H. W., and Taylor, C. D. (1984). Deep-sea microbiology. Annu. Rev. Microbiol. 38, 487–487.
Jannasch, H. W., Wirsen, C. O., and Doherty, K. W. (1996). A pressurized chemostat for the study of marine barophilic and oligotrophic bacteria. Appl. Environ. Microbiol. 62, 1593–1596.
Jannasch, H. W., Wirsen, C. O., and Winget, C. L. (1973). A bacteriological pressure-retaining deep-sea sampler and culture vessel. Deep Sea Res. Oceanogr. Abstracts 20, 661–664. doi: 10.1016/0011-7471(73)90033-8
Jebbar, M., Franzetti, B., Girard, E., and Oger, P. (2015). Microbial diversity and adaptation to high hydrostatic pressure in deep-sea hydrothermal vents prokaryotes. Extremophiles 19, 721–740. doi: 10.1007/s00792-015-0760-3
Jørgensen, B. B., and Boetius, A. (2007). Feast and famine—microbial life in the deep-sea bed. Nat. Rev. Microbiol. 5, 770–781. doi: 10.1038/nrmicro1745
Jørgensen, B. B., Isaksen, M. F., and Jannasch, H. W. (1992). Bacterial sulfate reduction above 100°C in deep-sea hydrothermal vent sediments. Science 258, 1756–1757. doi: 10.1126/science.258.5089.1756
Kallmeyer, J., and Boetius, A. (2004). Effects of temperature and pressure on sulfate reduction and anaerobic oxidation of methane in hydrothermal sediments of Guaymas Basin. Appl. Environ. Microbiol. 70, 1231–1233. doi: 10.1128/aem.70.2.1231-1233.2004
Kallmeyer, J., Ferdelman, T. G., Jansen, K.-H., and Jørgensen, B. B. (2003). A high-pressure thermal gradient block for investigating microbial activity in multiple deep-sea samples. J. Microbiol. Methods 55, 165–172. doi: 10.1016/s0167-7012(03)00138-6
Kallmeyer, J., Pockalny, R., Adhikari, R. R., Smith, D. C., and D’Hondt, S. (2012). Global distribution of microbial abundance and biomass in subseafloor sediment. Proc. Natl. Acad. Sci. 109, 16213–16216. doi: 10.1073/pnas.1203849109
Kato, C. (2011). “Cultivation methods for piezophiles,” in Extremophiles Handbook, ed. K. Horikoshi (Tokyo: Springer), 719–726. doi: 10.1007/978-4-431-53898-1_34
Kato, C., Nogi, Y., and Arakawa, S. (2008). “Isolation, cultivation, and diversity of deep-sea piezophiles,” in High-Pressure Microbiology, eds C. Michiels, D. Bartlett, and A. Aersten (Washington, DC: ASM Press), 203–217. doi: 10.1128/9781555815646.ch12
Kato, C., and Qureshi, M. H. (1999). Pressure response in deep-sea piezophilic bacteria. J. Mol. Microbiol. Biotechnol. 1, 87–92.
Kawano, H., Nakasone, K., Matsumoto, M., Yoshida, Y., Usami, R., Kato, C., et al. (2004). Differential pressure resistance in the activity of RNA polymerase isolated from Shewanella violacea and Escherichia coli. Extremophiles 8, 367–375. doi: 10.1007/s00792-004-0397-0
Kubo, Y., Mizuguchi, Y., Inagaki, F., Yamamoto, K., Schultheiss, P., Grigar, K., et al. (2014). new hybrid pressure-coring system for the drilling vessel Chikyu. Sci. Drill. 17, 37–43. doi: 10.5194/sd-17-37-2014
Kusube, M., Kyaw, T. S., Tanikawa, K., Chastain, R. A., Hardy, K. M., Cameron, J., et al. (2017). Colwellia marinimaniae sp. nov., a hyperpiezophilic species isolated from an amphipod within the Challenger Deep, Mariana Trench. Int. J. Syst. Evol. Microbiol. 67, 824–831. doi: 10.1099/ijsem.0.001671
La Cono, V., Smedile, F., La Spada, G., Arcadi, E., Genovese, M., Ruggeri, G., et al. (2015). Shifts in the meso-and bathypelagic archaea communities composition during recovery and short-term handling of decompressed deep-sea samples. Environ. Microbiol. Rep. 7, 450–459. doi: 10.1111/1758-2229.12272
Lasken, R. S. (2007). Single-cell genomic sequencing using multiple displacement amplification. Curr. Opin. Microbiol. 10, 510–516. doi: 10.1016/j.mib.2007.08.005
Lavalleur, H. J., and Colwell, F. S. (2013). Microbial characterization of basalt formation waters targeted for geological carbon sequestration. FEMS Microbiol. Ecol. 85, 62–73. doi: 10.1111/1574-6941.12098
Lin, Y. S., Lipp, J. S., Elvert, M., Holler, T., and Hinrichs, K. U. (2013). Assessing production of the ubiquitous archaeal diglycosyl tetraether lipids in marine subsurface sediment using intramolecular stable isotope probing. Environ. Microbiol. 15, 1634–1646. doi: 10.1111/j.1462-2920.2012.02888.x
Lipp, J. S., Morono, Y., Inagaki, F., and Hinrichs, K.-U. (2008). Significant contribution of Archaea to extant biomass in marine subsurface sediments. Nature 454, 991–994. doi: 10.1038/nature07174
Lloyd, K. G., May, M. K., Kevorkian, R. T., and Steen, A. D. (2013). Meta-analysis of quantification methods shows that archaea and bacteria have similar abundances in the subseafloor. Appl. Environ. Microbiol. 79, 7790–7799. doi: 10.1128/AEM.02090-13
Lollar, B. S., Lacrampe-Couloume, G., Slater, G., Ward, J., Moser, D., Gihring, T., et al. (2006). Unravelling abiogenic and biogenic sources of methane in the Earth’s deep subsurface. Chem. Geol. 226, 328–339. doi: 10.1016/j.chemgeo.2005.09.027
Martin, D., Bartlett, D. H., and Roberts, M. F. (2002). Solute accumulation in the deep-sea bacterium Photobacterium profundum. Extremophiles 6, 507–514. doi: 10.1007/s00792-002-0288-1
Michoud, G., and Jebbar, M. (2016). High hydrostatic pressure adaptive strategies in an obligate piezophile Pyrococcus yayanosii. Sci. Rep. 6:27289. doi: 10.1038/srep27289
Morono, Y., Terada, T., Nishizawa, M., Ito, M., Hillion, F., Takahata, N., et al. (2011). Carbon and nitrogen assimilation in deep subseafloor microbial cells. Proc. Natl. Acad. Sci. 108, 18295–18300. doi: 10.1073/pnas.1107763108
Moser, D. P., Gihring, T. M., Brockman, F. J., Fredrickson, J. K., Balkwill, D. L., Dollhopf, M. E., et al. (2005). Desulfotomaculum and Methanobacterium spp. dominate a 4-to 5-kilometer-deep fault. Appl. Environ. Microbiol. 71, 8773–8783. doi: 10.1128/aem.71.12.8773-8783.2005
Nauhaus, K., Boetius, A., Krüger, M., and Widdel, F. (2002). In vitro demonstration of anaerobic oxidation of methane coupled to sulphate reduction in sediment from a marine gas hydrate area. Environ. Microbiol. 4, 296–305. doi: 10.1046/j.1462-2920.2002.00299.x
Nunoura, T., Takaki, Y., Hirai, M., Shimamura, S., Makabe, A., Koide, O., et al. (2015). Hadal biosphere: insight into the microbial ecosystem in the deepest ocean on Earth. Proc. Natl. Acad. Sci. 112, E1230–E1236. doi: 10.1073/pnas.1421816112
Nunoura, T., Takaki, Y., Kakuta, J., Nishi, S., Sugahara, J., Kazama, H., et al. (2010). Insights into the evolution of Archaea and eukaryotic protein modifier systems revealed by the genome of a novel archaeal group. Nucleic Acids Res. 39, 3204–3223. doi: 10.1093/nar/gkq1228
Oger, P., and Jebbar, M. (2010). The many ways of coping with pressure. Res. Microbiol. 161, 799–809. doi: 10.1016/j.resmic.2010.09.017
Oliver, G. C. (2019). Exploring Microbial Growth of a Model Extremophile, Archaeoglobus Fulgidus, at Elevated Pressures. Ph.D. dissertation, Rensselaer Polytechnic Institute, Troy, NY, 142.
Oppenheimer, C. H., and Zobell, C. E. (1952). The growth and viability of 63 species of marine bacteria as influenced by hydrostatic pressure. J. Mar. Res. 11, 10–18.
Orcutt, B. N., Sylvan, J. B., Knab, N. J., and Edwards, K. J. (2011). Microbial ecology of the dark ocean above, at, and below the seafloor. Microbiol. Mol. Biol. Rev. 75, 361–422. doi: 10.1128/MMBR.00039-10
Orsi, W. D., Edgcomb, V. P., Christman, G. D., and Biddle, J. F. (2013). Gene expression in the deep biosphere. Nature 499, 205–208. doi: 10.1038/nature12230
Park, C. B., and Clark, D. S. (2002). Rupture of the cell envelope by decompression of the deep-sea methanogen Methanococcus jannaschii. Appl. Environ. Microbiol. 68, 1458–1463. doi: 10.1128/aem.68.3.1458-1463.2002
Parkes, R. J., Cragg, B. A., Bale, S. J., Getlifff, J. M., Goodman, K., Rochelle, P. A., et al. (1994). Deep bacterial biosphere in Pacific Ocean sediments. Nature 371, 410–413. doi: 10.1038/371410a0
Parkes, R. J., Sellek, G., Webster, G., Martin, D., Anders, E., Weightman, A. J., et al. (2009). Culturable prokaryotic diversity of deep, gas hydrate sediments: first use of a continuous high-pressure, anaerobic, enrichment and isolation system for subseafloor sediments (DeepIsoBUG). Environ. Microbiol. 11, 3140–3153. doi: 10.1111/j.1462-2920.2009.02018.x
Peoples, L. M., Norenberg, M., Price, D., McGoldrick, M., Novotny, M., Bochdansky, A., et al. (2019). A full-ocean-depth rated modular lander and pressure-retaining sampler capable of collecting hadal-endemic microbes under in situ conditions. Deep Sea Res. I Oceanogr. Res. Papers 143, 50–57. doi: 10.1016/j.dsr.2018.11.010
Picard, A., and Daniel, I. (2013). Pressure as an environmental parameter for microbial life—a review. Biophys. Chem. 183, 30–41. doi: 10.1016/j.bpc.2013.06.019
Ramirez-Llodra, E., Brandt, A., Danovaro, R., De Mol, B., Escobar, E., German, C., et al. (2010). Deep, diverse and definitely different: unique attributes of the world’s largest ecosystem. Biogeosciences 7, 2851–2899. doi: 10.5194/bg-7-2851-2010
Rassa, A. C., Mcallister, S., Safran, S., and Moyer, C. (2009). Zeta-Proteobacteria dominate the colonization and formation of microbial mats in low-temperature hydrothermal vents at Loihi Seamount, Hawaii. Geomicrobiol. J. 26, 623–638. doi: 10.1080/01490450903263350
Reed, D. W., Fujita, Y., Delwiche, M. E., Blackwelder, D. B., Sheridan, P. P., Uchida, T., et al. (2002). Microbial communities from methane hydrate-bearing deep marine sediments in a forearc basin. Appl. Environ. Microbiol. 68, 3759–3770. doi: 10.1128/aem.68.8.3759-3770.2002
Reysenbach, A.-L., and Götz, D. (2001). Methods for the study of hydrothermal vent microbes. Methods Microbiol. 30, 639–656. doi: 10.1016/s0580-9517(01)30066-1
Rogers, K. L., Cario, A., Cardon, H., and Daniel, I. (2016a). “Experimental high-pressure cultivation with the PUSH50: increased pressure tolerance of a marine bacterium without subsampling decompression,” in Proceedings of the 5th International Workshop on Deep-Sea Microbiology, Kyoto, 10.
Rogers, K. L., Cario, A., Cardon, H., and Daniel, I. (2016b). “Introducing the PUSH50: A New Instrument to Extend Microbial Exploration within the Deep Biosphere,” in Proceedings of the 11th International Congress on Extremophiles, Kyoto.
Rosenbaum, E., Gabel, F., Durá, M. A., Finet, S., Cléry-Barraud, C., Masson, P., et al. (2012). Effects of hydrostatic pressure on the quaternary structure and enzymatic activity of a large peptidase complex from Pyrococcus horikoshii. Arch. Biochem. Biophys. 517, 104–110. doi: 10.1016/j.abb.2011.07.017
Sauer, P., Glombitza, C., and Kallmeyer, J. (2012). A system for incubations at high gas partial pressure. Front. Microbiol. 3:25. doi: 10.3389/fmicb.2012.00025
Schrenk, M. O., Huber, J. A., and Edwards, K. J. (2010). Microbial provinces in the subseafloor. Annu. Rev. Mar. Sci. 2, 279–304. doi: 10.1146/annurev-marine-120308-081000
Seyfried, W., Gordon, P., and Dickson, F. (1979). A new reaction cell for hydrothermal solution equipment. Am. Mineral. 64, 646–649.
Simonato, F., Campanaro, S., Lauro, F., Vezzi, A., D’Angelo, M., Vitulo, N., et al. (2006). Piezophilic adaptation: a genomic point of view. J. Biotechnol. 126, 11–25. doi: 10.1016/j.jbiotec.2006.03.038
Smith, A., Popa, R., Fisk, M., Nielsen, M., Wheat, C. G., Jannasch, H. W., et al. (2011). In situ enrichment of ocean crust microbes on igneous minerals and glasses using an osmotic flow-through device. Geochem. Geophys. Geosyst. 12:Q06007.
Stamenković, V., Beegle, L., Zacny, K., Arumugam, D., Baglioni, P., Barba, N., et al. (2019). The next frontier for planetary and human exploration. Nat. Astronomy 3, 116–120.
Stevens, T. O., and McKinley, J. P. (1995). Lithoautotrophic microbial ecosystems in deep basalt aquifers. Science 270, 450–455.
Takai, K., Komatsu, T., Inagaki, F., and Horikoshi, K. (2001). Distribution of archaea in a black smoker chimney structure. Appl. Environ. Microbiol. 67, 3618–3629. doi: 10.1128/aem.67.8.3618-3629.2001
Takai, K., Nakamura, K., Toki, T., Tsunogai, U., Miyazaki, M., Miyazaki, J., et al. (2008). Cell proliferation at 122°C and isotopically heavy CH4 production by a hyperthermophilic methanogen under high-pressure cultivation. Proc. Natl. Acad. Sci.U.S.A. 105, 10949–10954. doi: 10.1073/pnas.0712334105
Tamburini, C. (2006). “Life under pressure. Deep-sea microbial ecology,” in Life as We Know it Series: Cellular Origin and Life in Extreme Habitats and Astrobiology, ed. J. Seckbach (Berlin: Springer), 1–17.
Tamburini, C., Boutrif, M., Garel, M., Colwell, R. R., and Deming, J. W. (2013). Prokaryotic responses to hydrostatic pressure in the ocean–a review. Environ. Microbiol. 15, 1262–1274. doi: 10.1111/1462-2920.12084
Tarn, J., Peoples, L. M., Hardy, K., Cameron, J., and Bartlett, D. H. (2016). Identification of free-living and particle-associated microbial communities present in hadal regions of the Mariana Trench. Front. Microbiol. 7:665. doi: 10.3389/fmicb.2016.00665
Taylor, C. D., and Jannasch, H. W. (1976). Subsampling technique for measuring growth of bacterial cultures under high hydrostatic pressure. Appl. Environ. Microbiol. 32, 355–359.
Tengberg, A., De Bovee, F., Hall, P., Berelson, W., Chadwick, D., Ciceri, G., et al. (1995). Benthic chamber and profiling landers in oceanography—a review of design, technical solutions and functioning. Prog. Oceanogr. 35, 253–294. doi: 10.1016/0079-6611(95)00009-6
Vannier, P., Michoud, G., Oger, P., þór Marteinsson, V., and Jebbar, M. (2015). Genome expression of Thermococcus barophilus and Thermococcus kodakarensis in response to different hydrostatic pressure conditions. Res. Microbiol. 166, 717–725. doi: 10.1016/j.resmic.2015.07.006
Wang, F., Zhou, H., Meng, J., Peng, X., Jiang, L., Sun, P., et al. (2009). GeoChip-based analysis of metabolic diversity of microbial communities at the Juan de Fuca Ridge hydrothermal vent. Proc. Natl. Acad. Sci. 106, 4840–4845. doi: 10.1073/pnas.0810418106
Webster, G., Rinna, J., Roussel, E. G., Fry, J. C., Weightman, A. J., and Parkes, R. J. (2010). Prokaryotic functional diversity in different biogeochemical depth zones in tidal sediments of the Severn Estuary, UK, revealed by stable-isotope probing. FEMS Microbiol. Ecol. 72, 179–197. doi: 10.1111/j.1574-6941.2010.00848.x
Whitman, W. B., Coleman, D. C., and Wiebe, W. J. (1998). Prokaryotes: the unseen majority. Proc. Natl. Acad. Sci. 95, 6578–6583. doi: 10.1073/pnas.95.12.6578
Yayanos, A., and Pollard, E. (1969). A study of the effects of hydrostatic pressure on macromolecular synthesis in Escherichia coli. Biophysics 9, 1464–1482. doi: 10.1016/s0006-3495(69)86466-0
Yayanos, A. A. (1977). Simply actuated closure for a pressure vessel: design for use to trap deep-sea animals. Rev. Sci. Instr. 48, 786–789. doi: 10.1063/1.1135150
Yayanos, A. A. (1986). Evolutional and ecological implications of the properties of deep-sea barophilic bacteria. Proc. Natl. Acad. Sci. 83, 9542–9546. doi: 10.1073/pnas.83.24.9542
Yayanos, A. A. (2001). Deep-sea piezophilic bacteria. Methods Microbiol. 30, 615–637. doi: 10.1016/s0580-9517(01)30065-x
Yayanos, A. A., and Dietz, A. S. (1982). Thermal inactivation of a deep-sea barophilic bacterium, isolate CNPT-3. Appl. Environ. Microbiol. 43, 1481–1489.
Yayanos, A. A., and Dietz, A. S. (1983). Death of a hadal deep-sea bacterium after decompression. Science 220, 497–498. doi: 10.1126/science.220.4596.497
Yayanos, A. A., Dietz, A. S., and Vanboxtel, R. (1979). Isolation of a deep-sea barophilic bacterium and some of its growth characteristics. Science 205, 808–810. doi: 10.1126/science.205.4408.808
Yayanos, A. A., Dietz, A. S., and Vanboxtel, R. (1982). Dependence of reproduction rate on pressure as a hallmark of deep-sea bacteria. Appl. Environ. Microbiol. 44, 1356–1361.
Yayanos, A. A., Vanboxtel, R., and Dietz, A. S. (1984). High-pressure-temperature gradient instrument - Use for determining the temperature and pressure limits of bacterial growth. Appl. Environ. Microbiol. 48, 771–776.
Zeng, S., Birrien, J., Fouquet, Y., Cherkashov, G., Jebbar, M., Querellou, J., et al. (2009). Pyrococcus CH1, an obligate piezophilic hyperthermophile: extending the upper pressure-temperature limits for life. ISME J. 3, 873–876. doi: 10.1038/ismej.2009.21
Zink, K.-G., Wilkes, H., Disko, U., Elvert, M., and Horsfield, B. (2003). Intact phospholipids—microbial “life markers” in marine deep subsurface sediments. Organic Geochem. 34, 755–769. doi: 10.1016/s0146-6380(03)00041-x
Keywords: deep biosphere, high-pressure microbiology, decompression, sampling, cultivation
Citation: Cario A, Oliver GC and Rogers KL (2019) Exploring the Deep Marine Biosphere: Challenges, Innovations, and Opportunities. Front. Earth Sci. 7:225. doi: 10.3389/feart.2019.00225
Received: 31 May 2019; Accepted: 16 August 2019;
Published: 04 September 2019.
Edited by:
Sabin Zahirovic, University of Sydney, AustraliaReviewed by:
Bernhard Wehrli, ETH Zürich, SwitzerlandCopyright © 2019 Cario, Oliver and Rogers. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Anaïs Cario, YW5haXMuY2FyaW9AY25ycy5mcg==; Karyn L. Rogers, cm9nZXJrNUBycGkuZWR1
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.