- 1Division of Cardiovascular Medicine, Department of Medicine, University of Massachusetts Medical School, Worcester, MA, United States
- 2Department of Biochemistry and Molecular Biology, Clark University, Worcester, MA, United States
- 3Department of Quantitative Health Sciences, University of Massachusetts Medical School, Worcester, MA, United States
Introduction: Epicardial adipose tissue (EAT) has been linked to incidence and recurrence of atrial fibrillation (AF), but the underlying mechanisms that mediate this association remain unclear. Circulating microRNAs (miRNAs) contribute to the regulation of gene expression in cardiovascular diseases, including AF. Thus, we sought to test the hypothesis that circulating miRNAs relate to burden of EAT.
Methods: We examined the plasma miRNA profiles of 91 participants from the miRhythm study, an ongoing study examining associations between miRNA and AF. We quantified plasma expression of 86 unique miRNAs commonly expressed in cardiomyocytes using quantitative reverse transcriptase polymerase chain reaction (qPCR). From computed tomography, we used validated methods to quantify the EAT area surrounding the left atrium (LA) and indexed it to body surface area (BSA) to calculate indexed LA EAT (iLAEAT). Participants were divided into tertiles of iLAEAT to identify associations with unique miRNAs. We performed logistic regression analyses adjusting for factors associated with AF to examine relations between iLAEAT and miRNA. We performed further bioinformatics analysis of miRNA predicted target genes to identify potential molecular pathways are regulated by the miRNAs.
Results: The mean age of the participants was 59 ± 9, 35% were women, and 97% were Caucasian. Participants in the highest tertile of iLAEAT were more likely to have hypertension, heart failure, and thick posterior walls. In regression analyses, we found that miRNAs 155-5p (p < 0.001) and 302a-3p (p < 0.001) were significantly associated with iLAEAT in patients with AF. The predicted targets of the miRNAs identified were implicated in the regulation of cardiac hypertrophy, adipogenesis, interleukin-8 (IL-8), and nerve growth factor (NGF) signaling.
Conclusion: miRNA as well as EAT have previously been linked to AF. Our finding that iLAEAT and miRNAs 155-5p and 302a-3p are associated suggest a possible direct link to between these entities in the development and maintenance of AF. Further research is needed to study causal relationships between these biomarkers.
Introduction
Atrial fibrillation (AF) is the most prevalent heart rhythm disorder in the world, with nearly 7 million Americans and 34 million individuals globally affected by it (1–4). It is associated with heart failure, stroke, dementia, and poor quality of life (1). Clinical risk prediction scores, including CHA2DS2VASC, perform modestly well in predicting outcomes of AF but explain little about the mechanisms behind disease onset and progression and only explain a small proportion of observed risk. As such, it is important to examine the molecular processes that drive AF, as it may afford new biomarkers and therapeutic avenues.
MicroRNA (miRNA) is a class of small non-coding RNA that are endogenously produced and have important regulatory function. miRNAs have been implicated in structural and electrical remodeling that are central to the development of AF (5–9). In addition to their regulatory role, they serve as biomarkers of disease states, as they exist in the plasma with remarkable stability (10). Examining the plasma miRome has provided unique insight into the molecular processes that underlie the pathogenesis of AF.
Epicardial adipose tissue (EAT) is a layer of metabolically active adipocytes that lie between the visceral pericardium and the myocardium without fascial boundaries. Due to its close proximity, this adipose depot has paracrine and vasocrine effects on the myocardium and plays a critical role in the development and maintenance of AF (11–13). Several groups, including ours, have shown correlation between EAT and onset as well as severity of AF. Our group has previously shown that an indexed measure of EAT surrounding the left atrium, called iLAEAT, is independently associated with the severity and recurrence of AF after catheter ablation (14).
Although much is known about the interaction of these biomarkers individually and their effects on the pathogenesis of AF, the relationship between EAT and plasma miRNAs has not been studied before. Thus, we hypothesized that increasing levels of EAT measured on computed cardiac tomography are associated with expression of circulating plasma miRNAs and tested this using data from a prospectively recruiting, contemporary cohort of AF patients.
Methods
Study Population
As part of the miRhythm study examining links between circulating miRNAs and AF, 584 participants were recruited at the University of Massachusetts Medical Center (UMMC) between April 2011 to July 2017 (15). Study protocol was approved by the University of Massachusetts Institutional Review Board (IRB #14875). Written consent was obtained from all participants to analyze pre-ablation miRNA expression. Of these, 91 participants underwent cardiac CT scan for evaluation of pulmonary venous anatomy prior to catheter ablation. These participants were included in our analysis. Participant information, including demographic, clinical, and baselines laboratory data, were abstracted from the UMMC AF Treatment Registry and hospital medical records by trained staff.
EAT Measurements
EAT was measured using validated methods (16). CT scans were performed by technicians at UMMC with the Siemens Somatom Definition Flash 128 dual source CT scanner. Unenhanced CT scans were taken with a standardized FLASH protocol. Enhanced scans were performed by injecting contrast dye and scanning using sequential acquisition. Calculation of contrast timing was done by giving the patient a 15 mL contrast test bolus followed by a 50 mL saline injection at 5–6 mL/s. Acquisition times were held constant between 1 and 2 s. EAT was measured by scanning the heart in a four-chamber view, with the bottom of the heart in front view. Locations of EAT around the right and left ventricles and the left atrium were traced by identifying regions of CT scans within a threshold of−43 Hounsfield units (HU) for unenhanced CT scans and−15 HU for enhanced CT scans. The inter and intra-observer reproducibility for EAT measurement was (r = 0.89 and 0.95), respectively. EAT was indexed to body surface area and reported as iLAEAT.
Echocardiographic Measurements
Complete 2D echocardiograms were performed during hospitalization. Linear dimensions and 2D volumes were measured according to ASE guidelines (17). We quantified left atrial (LA) volume), left ventricular end-diastolic (LVIDd) and end-systolic (LVIDs) dimensions, posterior wall thickness (PWT), interventricular septum thickness at end–diastole (IVSTd), left ventricular (LV) mass and left atrial function index (LAFI). LV mass was calculated by LV mass = 0.8 (1.04 [LVID + PWTd + SWTd]3 – [LVID]3) + 0.6 g (18). LAFI was calculated by the previously validated formula: LAFI = LA emptying fraction*LVOT-VTI / LAESVI (19).
MiRNA Identification and Profiling
Ten cc of venous blood was obtained for study purposes after routine femoral venous sheath placement for catheter ablation of AF. This blood was processed to isolate plasma and stored as previously described (20). Briefly, blood was collected into blood collection tubes with solution containing sodium citrate. Samples were then centrifuged at 2,500 g for 22 min at 4°C. Plasma was separated from the cells and frozen at 80°C within 90 min of draw.
For this study, we selected 86 plasma miRNAs based on our RNAseq experiments done on 20 participants with cardiovascular disease and 20 without cardiovascular disease from the Framingham Heart Study Offspring 2 cohort (8th visit). We selected top expressed miRNAs from these 40 individuals for study in our RT-qPCR experiments (20). For all experiment, RNA was extracted from 200 μL of plasma. All methods used for cDNA synthesis, pre-amplification, and qPCR were performed according to Qiagen miScript Microfluidics Handbook by using Qiagen miScript Assays by the High-Throughput Gene Expression & Biomarker Core Laboratory at the University of Massachusetts Medical School (21). RNAs were isolated, and RT reactions performed by miScript II RT kit (Cat. No: 218161, Qiagen, Fredrick, MD, USA). miScript Microfluidics PreAMP Kit (Cat. No: 331455, Qiagen, Fredrick, MD, USA) was used for preamplification reactions. qPCR reactions were run using on BioMark System. BioMark System can detect single miRNA copy at 26–27 Cq compare to conventional qPCR platforms 36–37 (22).
We normalized the volume of plasma did not employ further methods of normalization in our dataset such as global mean normalization. Global mean is not ideal with our modest sample size and there are no reliable “housekeeper” gene to normalize miRNAs in the plasma (20). We believe that our Ct values are representative of relative concentrations. Differentially expressed miRNAs were analyzed using miRDB, an online database that captures miRNA and gene target interactions (23, 24). Network and functional analyses were generated through the use of Qiagen's Ingenuity Pathway Analysis (25).
Statistical Analyses
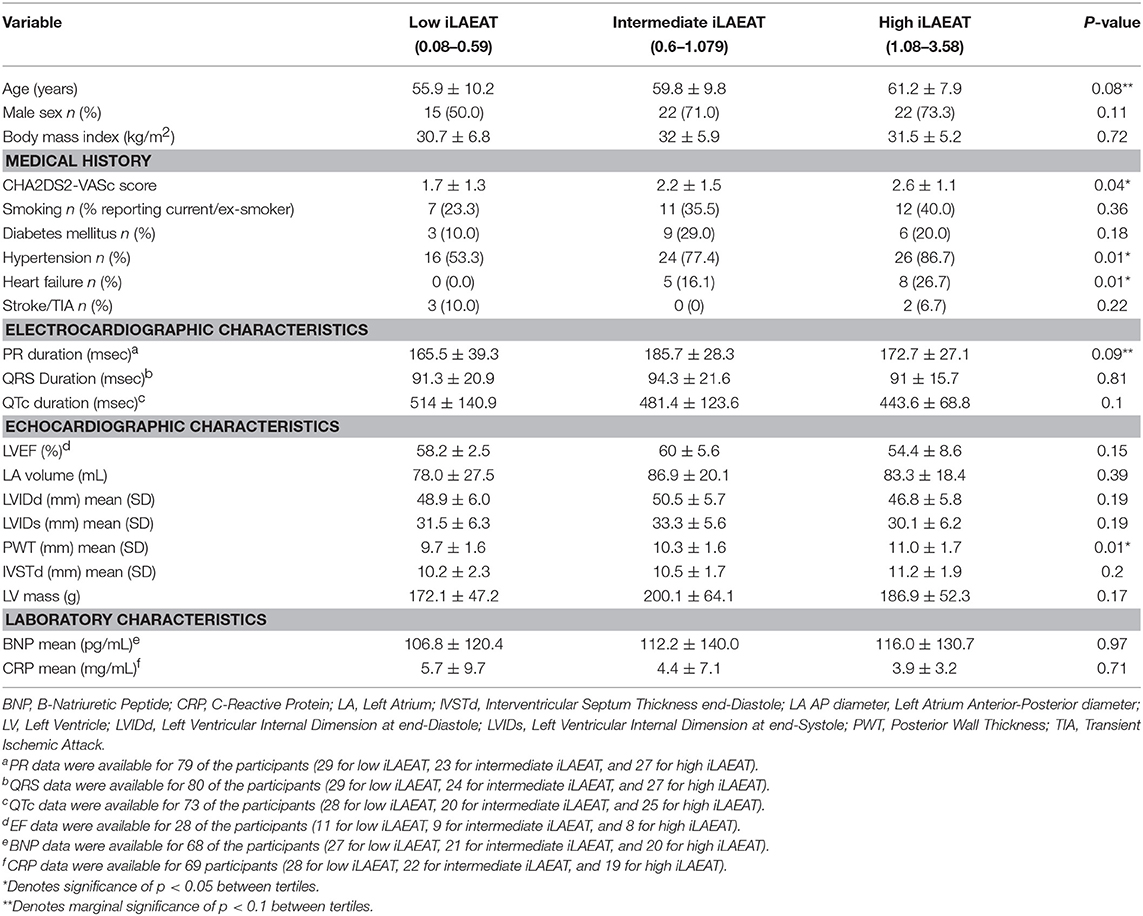
Participants were divided into tertiles based upon iLAEAT values, placing them into either low (iLAEAT 0.08–0.59, n = 30), intermediate (iLAEAT 0.6–1.079, n = 31), or high (iLAEAT 1.08–3.58, n = 30) groups. Tertiles were created to identify participants at the greatest clinical risk for AF burden and recurrence. We examined the relationship between demographic, clinical, and echocardiographic variables and tertiles of iLAEAT using either χ2-squared tests for categorical variables or ANOVAs for continuous variables. A p-value of 0.05 was used as the standard significance threshold for statistical tests.
Pre-ablation miRNA expression was compared to iLAEAT using a series of linear regression models. Covariates included in the models were identified based upon characteristics that were determined to be significantly different between iLAEAT tertiles and included age, hypertension, heart failure, and posterior wall thickness. Bonferroni correction was also applied to account for multiple tests.
Results
We present participant characteristics based upon iLAEAT tertiles in Table 1. Participants almost exclusively identified as Caucasian (97%), and the majority were male (65%). Participants in the highest tertile of iLAEAT tended to be older, more likely to have a history of hypertension (86.7%, p = 0.01) and heart failure (26.7%, p = 0.01) and higher CHA2DS2-VASc scores (p = 0.04), thicker posterior walls (p = 0.01), and longer PR durations than did participants in the lower two tertiles. Participants with persistent AF have higher iLAEAT (Figure 1). As previously observed, participants who have higher iLAEAT have a higher likelihood of having persistent AF (14). Furthermore, there is a non-significant association in decreasing LAFI with increasing iLAEAT tertile (Figure 1).
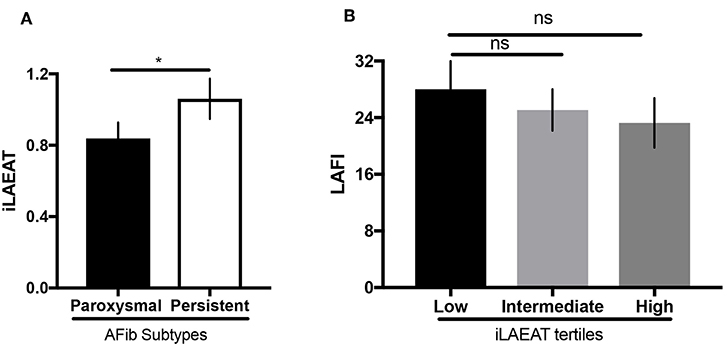
Figure 1. iLAEAT and left atrial function. (A) iLAEAT is higher in participants with persistent AF. (B) LAFI tended to be lower in patients with higher iLAEAT tertile. iLAEAT and LAFI data were available for 71 participants in our cohort (26 for low iLAEAT, 23 for intermediate iLAEAT, and 22 for high iLAEAT). Statistical analysis was performed using unpaired student t-test for AF subtypes (*p < 0.05) and one-way ANOVA analysis for iLAEAT tertiles. Error bars represent SEM. Panel (A) is adapted from work by Sanghai et al. (14).
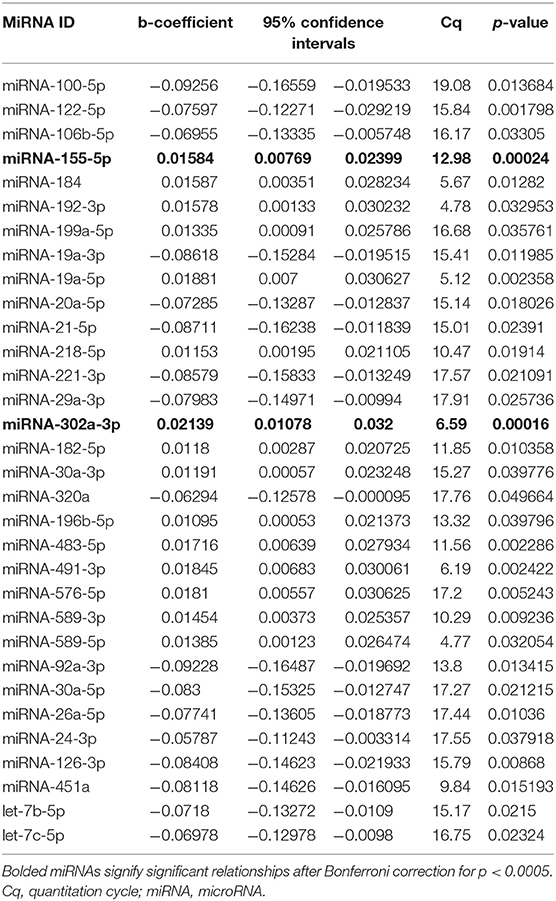
Results from qPCR analysis of 86 miRNAs are included in Supplemental Table 1. Analysis of miRNA expression level as well as regression models run between iLAEAT and Cq are illustrated in Table 2. After statistical correction for multiple tests, two miRNAs (miR-155-5p and−302a-3p) remained significantly associated with higher tertile of iLAEAT. Although, we do not know the source, potential targets or the mechanisms by which they are transported in the plasma, we believe that plasma miRNAs are derived from cellular sources. Therefore, analysis of miRNA downstream targets and pathways may reveal information regarding the molecular pathways that are activated.
Thus, miRNAs downstream targets were analyzed using miRDB, an online database that captures miRNA and gene target interactions (23, 24). We do not have experimental data to guide selection of relevant downstream targets, and thus we included all predicted targets of miR-155-5p and−302a-3p in our analysis. As miRNA are known to act in concert, we used the combined targets of miR-155-5p and−302a-3p to perform further analysis (26). Ingenuity Pathway Analysis (IPA) was utilized to identify the molecular network and cellular toxicity pathways regulated by predicted targets. Canonical pathways were mapped to allow for visualization of the shared biological pathways through the common genes. The pathways identified and their associated genes are shown in Supplementary Table 2 and the top 10 significant pathways are displayed in Figure 2, including adipogenesis, cardiac hypertrophy, nerve growth factor (NGF), and IL-8 signaling.
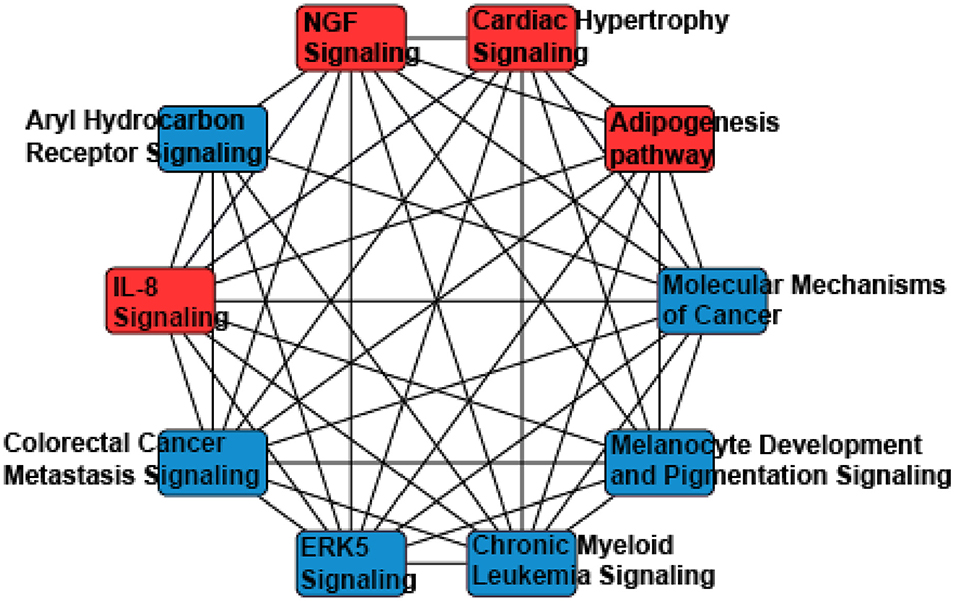
Figure 2. A network analysis of predicted targets of miR-155-5p and miR-302a-3p as performed by Ingenuity Pathway Analysis. Nodes represents signaling pathways, and lines are protein targets that are common between nodes. IL-8, Interleukin 8; NGF, Nerve Growth Factor; ERK5, Extracellular Related Kinase 5.
Discussion
Due to its proximity to the autonomic ganglia, myocardium, and coronary arteries, epicardial adipocytes can influence neighboring cells via both paracrine and vasocrine signaling (27). EAT has been implicated in the pathogenies of AF. We and others demonstrated previously that EAT is associated with incident and severity of AF (14, 28). miRNAs regulate the molecular processes that trigger the structural and electrophysiological remodeling key to the development of AF (6–9, 21). They are associated with incident and recurrent AF, and their dynamic nature after catheter ablation suggests that they are related to the pathogenesis of AF (5, 15, 21). In this work, we explore the relationship between iLAEAT and plasma miRNA and hypothesize their potential role in creating a substrate for AF.
We found two circulating miRNAs, miRNA 155-5p and 302a-3p, significantly associated with iLAEAT. Our pathway analysis of the downstream targets of miRNAs 155-5p and 302a-3p implicated them in the regulation of adipogenesis, cardiac hypertrophy, IL-8, and NGF signaling. In our cohort, increased iLAEAT is associated with increased posterior wall thickness. It is possible that miRNAs 155-5p and 302a-3p regulate pathways to increase formation of adipocytes and myocyte hypertrophy, as evident by echocardiographic phenotypes. It is unclear to us why in our dataset there is a significant association between iLAEAT and PWT and not IVSTd or LV mass. Perhaps, iLAEAT is in close proximity to the posterior wall and thus is able to exert mechanical stress or paracrine effects to cause local hypertrophy. Alternatively, we may be under-powered to examine the differences in IVSTd or LV mass in different tertiles of iLAEAT.
EAT has been shown to secrete adipokines that influence electrical remodeling (29). Our analysis predicts that miRNA 155-5p and 302a-3p regulate NGF signaling. There is much evidence suggesting that autonomic remodeling plays an important role in the pathogenesis of AF (30–32). The ganglionated plexi (GP), consisting of autonomic ganglia, are located on the epicardial surface of the heart and are typically surrounded by EAT (Figure 3) (33). Adipocytes have been shown to secrete NGF (34). NGF is up-regulated in the presence of AF and can induce the hyperactivity of GP (32, 35). In animal models, Yang et al. found that high plasma NGF levels create atrial substrate for AF and increase the incidence of inducible AF and its duration (36). Our analysis of miRNAs 155-5p and 302a-3p suggests a mechanism in which adipose tissue upregulates NGF to potentiate AF.
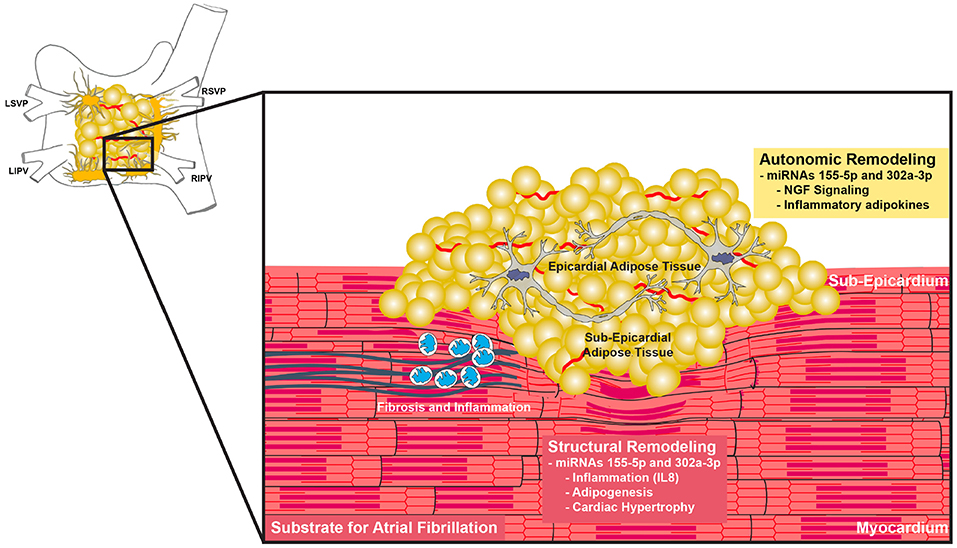
Figure 3. Proposed functions of miR-155-5p and miR-302a-3p. Increase EAT with concurrent upregulation of miR-155-5p and miR-302a-3p cause changes in adipokine secretion such as IL-8 and NGF. These changes lead to autonomic dysregulation and increased inflammation, creating a substrate for atrial fibrillation.
Several studies from the Sata laboratory have shown that EAT adipokine imbalance is a strongly linked of the development of atherosclerosis in men (37–39). Similarly, adipokines released from EAT may alter immune profiles the myocardium and surrounding tissue to increase inflammation and vulnerability to rhythm disturbances (40). Our results further support the hypothesis that EAT is an inflammatory mediator that is involved in adverse cardiac remodeling. EAT potentially contributes to inflammation and adverse structural remodeling via secretion of inflammatory adipokines such as IL-8 (40, 41). Circulating levels of IL-8 are associated to obesity-related factors such as BMI, waist circumference, C-reactive protein, interleukin (IL) 6, and tumor necrosis factor α (TNF-α) (42–44). IL-8, IL-6, IL-10, and TNF-α concentrations have been shown to be independently associated with AF (45). Patients with post-operative AF after coronary artery bypass graft surgery have higher serum IL-8 concentrations, indicating a role for inflammation in the development of AF post-surgery (46). miRNAs 155-5p and 302a-3p are implicated in the regulation of IL-8 and are associated with increased EAT, suggesting that secretion of IL-8 may be one of the ways in which EAT promotes inflammation to create a substrate for AF.
Strengths and Limitations
We leveraged data from a prospectively enrolled contemporary cohort of participants with AF undergoing miRNA profiling, echo phenotyping as well as EAT quantification using cardiac CT. We analyzed plasma concentration levels of commonly expressed miRNAs in cardiomyocytes of a cohort of 91 patients undergoing CA and CT scans at UMMC.
Despite these study strengths, there are notable study limitations. Notably, we do not know the sources of the miRNAs, the mechanism by which they are found in the blood or if they are indeed involved in a signaling cascade. These biological questions are of great importance and will provide mechanistic insight. Unfortunately, they are beyond the scope of our study. Our modest sample size leaves for the possibility of falsely negative associations. This may explain why the negative association between iLAEAT and LAFI was not statistically significant. There is also a lack of diversity in patients sampled inhibits us from making generalizable claims. Furthermore, the associations made in this study between iLAEAT and miRNA expression are purely correlational. We have not tested a causal relationship between miRNAs 155-5p, 302a-3p and iLAEAT. Further experimentation at the bench is needed to elucidate the exact relationship between these miRNAs and EAT.
Conclusion
In this study, we assessed the relationship between relative EAT area and plasma miRNA expression in patients with AF. We observed a significant, positive association between relative EAT area and miRNAs 155-5p and 302a-3p. Future validation and mechanistic studies are needed to improve our understanding of the pathological role of EAT in AF development.
Data Availability
The datasets analyzed in this manuscript are not publicly available. Requests to access the datasets should be directed to ZGF2aWQubWNtYW51cyYjeDAwMDQwO3VtYXNzbWVkLmVkdQ==.
Author Contributions
DM, TF, and JF supervised this work. DM, TF, K-VT, SS, JM, and MS contributed to hypothesis generation, conceptual design, and data analysis. KT, DL, and K-VT conducted experiments and analyzed data. All contributed to manuscript preparation.
Funding
K-VT's time was supported by 5T32HL120823, 1U01HL105268 from the National Heart, Lung and Blood Institute. DM's time was supported by 1R15HL121761, R01HL126911, R01HL137734, R01HL137794, R01HL13660 from the National Heart, Lung and Blood Institute. JF's time was supported by UH3TR000921 from the NIH Common Fund and 16SFRN31740000 from the American Heart Association.
Conflict of Interest Statement
DM has received research grant funding from Bristol-Myers Squibb, Boeringher-Ingelheim, Pfizer, Samsung, Philips Healthcare, Biotronik, has received consultancy fees from Bristol-Myers Squibb, Pfizer, Flexcon, Boston Biomedical Associates, and has inventor equity in Mobile Sense Technologies, Inc. (CT).
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcvm.2019.00115/full#supplementary-material
References
1. Pellman J, Sheikh F. Atrial fibrillation: mechanisms, therapeutics, and future directions. Compr Physiol. (2015) 5:649–65. doi: 10.1002/cphy.c140047
2. Czick ME, Shapter CL, Silverman DI. Atrial fibrillation: the science behind its defiance. Aging Dis. (2016) 7:635–56. doi: 10.14336/AD.2016.0211
3. Schnabel RB, Yin X, Gona P, Larson MG, Beiser AS, McManus DD, et al. 50 year trends in atrial fibrillation prevalence, incidence, risk factors, and mortality in the Framingham Heart Study: a cohort study. Lancet. (2015) 386:154–62. doi: 10.1016/S0140-6736(14)61774-8
4. Benjamin EJ, Virani SS, Callaway CW, Chamberlain AM, Chang AR, Cheng S, et al. Heart disease and stroke statistics-2018 update: a report from the American heart association. Circulation. (2018) 137:e67–492. doi: 10.1161/CIR.0000000000000573
5. McManus DD, Lin H, Tanriverdi K, Quercio M, Yin X, Larson MG, et al. Relations between circulating microRNAs and atrial fibrillation: data from the Framingham Offspring Study. Heart Rhythm. (2014) 11:663–9. doi: 10.1016/j.hrthm.2014.01.018
6. Creemers EE, Tijsen AJ, Pinto YM. Circulating microRNAs: novel biomarkers and extracellular communicators in cardiovascular disease? Circ Res. (2012) 110:483–95. doi: 10.1161/CIRCRESAHA.111.247452
7. Quiat D, Olson EN. MicroRNAs in cardiovascular disease: from pathogenesis to prevention and treatment. J Clin Invest. (2013) 123:11–8. doi: 10.1172/JCI62876
8. Gupta SK, Bang C, Thum T. Circulating microRNAs as biomarkers and potential paracrine mediators of cardiovascular disease. Circ Cardiovasc Genet. (2010) 3:484–8. doi: 10.1161/CIRCGENETICS.110.958363
9. Cardin S, Guasch E, Luo X, Naud P, Le Quang K, Shi Y, et al. Role for MicroRNA-21 in atrial profibrillatory fibrotic remodeling associated with experimental postinfarction heart failure. Circ Arrhythm Electrophysiol. (2012) 5:1027–35. doi: 10.1161/CIRCEP.112.973214
10. Shah R, Patel T, Freedman JE. Circulating extracellular vesicles in human disease. N Engl J Med. (2018) 379:2180–1. doi: 10.1056/NEJMc1813170
11. Iacobellis G. Local and systemic effects of the multifaceted epicardial adipose tissue depot. Nat Rev Endocrinol. (2015) 11:363–71. doi: 10.1038/nrendo.2015.58
12. Wong CX, Ganesan AN, Selvanayagam JB. Epicardial fat and atrial fibrillation: current evidence, potential mechanisms, clinical implications, and future directions. Eur Heart J. (2017) 38:1294–302. doi: 10.1093/eurheartj/ehw045
13. Batal O, Schoenhagen P, Shao M, Ayyad AE, Van Wagoner DR, Halliburton SS, et al. Left atrial epicardial adiposity and atrial fibrillation. Circ Arrhythm Electrophysiol. (2010) 3:230–6. doi: 10.1161/CIRCEP.110.957241
14. Sanghai SR, Sardana M, Hansra B, Lessard DM, Dahlberg ST, Aurigemma GP, et al. Indexed left atrial adipose tissue area is associated with severity of atrial fibrillation and atrial fibrillation recurrence among patients undergoing catheter ablation. Front Cardiovasc Med. (2018) 5:76. doi: 10.3389/fcvm.2018.00076
15. Vaze A, Donahue K, Spring M, Sardana M, Tanriverdi K, Freedman JE, et al. Plasma MicroRNAs relate to atrial fibrillation recurrence after catheter ablation: longitudinal findings from the MiRhythm study. J Clin Exp Cardiol. (2017) 8:502. doi: 10.4172/2155-9880.1000502
16. Marwan M, Achenbach S. Quantification of epicardial fat by computed tomography: why, when and how? J Cardiovasc Comput Tomogr. (2013) 7:3–10. doi: 10.1016/j.jcct.2013.01.002
17. Lang RM, Badano LP, Mor-Avi V, Afilalo J, Armstrong A, Ernande L, et al. Recommendations for cardiac chamber quantification by echocardiography in adults: an update from the American society of echocardiography and the European association of cardiovascular imaging. Eur Heart J Cardiovasc Imaging. (2015) 16:233–70. doi: 10.1093/ehjci/jev014
18. Devereux RB, Alonso DR, Lutas EM, Gottlieb GJ, Campo E, Sachs I, et al. Echocardiographic assessment of left ventricular hypertrophy: comparison to necropsy findings. Am J Cardiol. (1986) 57:450–8. doi: 10.1016/0002-9149(86)90771-X
19. Sardana M, Ogunsua AA, Spring M, Shaikh A, Asamoah O, Stokken G, et al. Association of left atrial function index with late atrial fibrillation recurrence after catheter ablation. J Cardiovasc Electrophysiol. (2016) 27:1411–9. doi: 10.1111/jce.13086
20. Freedman JE, Gerstein M, Mick E, Rozowsky J, Levy D, Kitchen R, et al. Diverse human extracellular RNAs are widely detected in human plasma. Nat Commun. (2016) 7:11106. doi: 10.1038/ncomms11902
21. McManus DD, Tanriverdi K, Lin H, Esa N, Kinno M, Mandapati D, et al. Plasma microRNAs are associated with atrial fibrillation and change after catheter ablation (the miRhythm study). Heart Rhythm. (2015) 12:3–10. doi: 10.1016/j.hrthm.2014.09.050
22. Jang JS, Simon VA, Feddersen RM, Rakhshan F, Schultz DA, Zschunke MA, et al. Quantitative miRNA expression analysis using fluidigm microfluidics dynamic arrays. BMC Genomics. (2011) 12:144. doi: 10.1186/1471-2164-12-144
23. Wong N, Wang X. miRDB: an online resource for microRNA target prediction and functional annotations. Nucleic Acids Res. (2015) 43(Database issue):D146–52. doi: 10.1093/nar/gku1104
24. Wang X. Improving microRNA target prediction by modeling with unambiguously identified microRNA-target pairs from CLIP-ligation studies. Bioinformatics. (2016) 32:1316–22. doi: 10.1093/bioinformatics/btw002
25. Krämer A, Green J, Pollard J, Tugendreich S. Causal analysis approaches in ingenuity pathway analysis. Bioinformatics. (2014) 30:523–30. doi: 10.1093/bioinformatics/btt703
27. Antonopoulos AS, Antoniades C. The role of epicardial adipose tissue in cardiac biology: classic concepts and emerging roles. J Physiol. (2017) 595:3907–17. doi: 10.1113/JP273049
28. Nakanishi K, Fukuda S, Tanaka A, Otsuka K, Sakamoto M, Taguchi H, et al. Peri-atrial epicardial adipose tissue is associated with new-onset nonvalvular atrial fibrillation. Circ J. (2012) 76:2748–54. doi: 10.1253/circj.CJ-12-0637
29. Lin YK, Chen YC, Chen JH, Chen SA, Chen YJ. Adipocytes modulate the electrophysiology of atrial myocytes: implications in obesity-induced atrial fibrillation. Basic Res Cardiol. (2012) 107:293. doi: 10.1007/s00395-012-0293-1
30. Chen PS, Tan AY. Autonomic nerve activity and atrial fibrillation. Heart Rhythm. (2007) 4(3 Suppl.):S61–4. doi: 10.1016/j.hrthm.2006.12.006
31. Stavrakis S, Nakagawa H, Po SS, Scherlag BJ, Lazzara R, Jackman WM. The role of the autonomic ganglia in atrial fibrillation. JACC Clin Electrophysiol. (2015) 1:1–13. doi: 10.1016/j.jacep.2015.01.005
32. Qi B, Wei Y, Chen S, Zhou G, Li H, Xu J, et al. Nav1.8 channels in ganglionated plexi modulate atrial fibrillation inducibility. Cardiovasc Res. (2014) 102:480–6. doi: 10.1093/cvr/cvu005
33. Hou Y, Scherlag BJ, Lin J, Zhou J, Song J, Zhang Y, et al. Interactive atrial neural network: determining the connections between ganglionated plexi. Heart Rhythm. (2007) 4:56–63. doi: 10.1016/j.hrthm.2006.09.020
34. Peeraully MR, Jenkins JR, Trayhurn P. NGF gene expression and secretion in white adipose tissue: regulation in 3T3-L1 adipocytes by hormones and inflammatory cytokines. Am J Physiol Endocrinol Metab. (2004) 287:E331–9. doi: 10.1152/ajpendo.00076.2004
35. Cai L, Liu S. Nerve growth factor mediates the vious cycle between hyperactivity of ganglionated plexus and atrial fibrillation. Clin Pharmacol Biopharm. (2018) 7:183. doi: 10.4172/2167-065X.1000183
36. Yang M, Zhang S, Liang J, Tang Y, Wang X, Huang C, et al. Different effects of norepinephrine and nerve growth factor on atrial fibrillation vulnerability. J Cardiol. (2019). doi: 10.1016/j.jjcc.2019.04.009. [Epub ahead of print].
37. Shimabukuro M, Hirata Y, Tabata M, Dagvasumberel M, Sato H, Kurobe H, et al. Epicardial adipose tissue volume and adipocytokine imbalance are strongly linked to human coronary atherosclerosis. Arterioscler Thromb Vasc Biol. (2013) 33:1077–84. doi: 10.1161/ATVBAHA.112.300829
38. Dagvasumberel M, Shimabukuro M, Nishiuchi T, Ueno J, Takao S, Fukuda D, et al. Gender disparities in the association between epicardial adipose tissue volume and coronary atherosclerosis: a 3-dimensional cardiac computed tomography imaging study in Japanese subjects. Cardiovasc Diabetol. (2012) 11:106. doi: 10.1186/1475-2840-11-106
39. Hirata Y, Tabata M, Kurobe H, Motoki T, Akaike M, Nishio C, et al. Coronary atherosclerosis is associated with macrophage polarization in epicardial adipose tissue. J Am Coll Cardiol. (2011) 58:248–55. doi: 10.1016/j.jacc.2011.01.048
40. Soeki T, Sata M. Role of epicardial adipose tissue in atrial fibrillation. Circ J. (2012) 76:2738–9. doi: 10.1253/circj.CJ-12-1283
41. Bruun JM, Pedersen SB, Richelsen B. Regulation of interleukin 8 production and gene expression in human adipose tissue in vitro. J Clin Endocrinol Metab. (2001) 86:1267–73. doi: 10.1210/jc.86.3.1267
42. Straczkowski M, Dzienis-Straczkowska S, Stêpieñ A, Kowalska I, Szelachowska M, Kinalska I. Plasma interleukin-8 concentrations are increased in obese subjects and related to fat mass and tumor necrosis factor-alpha system. J Clin Endocrinol Metab. (2002) 87:4602–6. doi: 10.1210/jc.2002-020135
43. Hadi HA, Alsheikh-Ali AA, Mahmeed WA, Suwaidi JM. Inflammatory cytokines and atrial fibrillation: current and prospective views. J Inflamm Res. (2010) 3:75–97. doi: 10.2147/JIR.S10095
44. Fitzgibbons TP, Czech MP. Epicardial and perivascular adipose tissues and their influence on cardiovascular disease: basic mechanisms and clinical associations. J Am Heart Assoc. (2014) 3:e000582. doi: 10.1161/JAHA.113.000582
45. Li J, Solus J, Chen Q, Rho YH, Milne G, Stein CM, et al. Role of inflammation and oxidative stress in atrial fibrillation. Heart Rhythm. (2010) 7:438–44. doi: 10.1016/j.hrthm.2009.12.009
Keywords: epicardial adipose tissue, microRNA, atrial fibrillati, cardiac remodeling, inflammation
Citation: Tran K-V, Majka J, Sanghai S, Sardana M, Lessard D, Milstone Z, Tanriverdi K, Freedman JE, Fitzgibbons TP and McManus D (2019) Micro-RNAs Are Related to Epicardial Adipose Tissue in Participants With Atrial Fibrillation: Data From the MiRhythm Study. Front. Cardiovasc. Med. 6:115. doi: 10.3389/fcvm.2019.00115
Received: 12 June 2019; Accepted: 26 July 2019;
Published: 14 August 2019.
Edited by:
Fabio Martelli, Policlinico San Donato (IRCCS), ItalyReviewed by:
Yuri D'Alessandra, Centro Cardiologico Monzino (IRCCS), ItalyMasataka Sata, Tokushima University, Japan
Copyright © 2019 Tran, Majka, Sanghai, Sardana, Lessard, Milstone, Tanriverdi, Freedman, Fitzgibbons and McManus. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: David McManus, ZGF2aWQubWNtYW51cyYjeDAwMDQwO3VtYXNzbWVkLmVkdQ==
†Co-first authors
 Khanh-Van Tran
Khanh-Van Tran Jordan Majka
Jordan Majka Saket Sanghai1
Saket Sanghai1 Darleen Lessard
Darleen Lessard