- The Wellcome-Wolfson Building, Centre for Experimental Medicine, Queen's University Belfast, Belfast, United Kingdom
Cardiovascular disease (CVD), despite the advances of the medical field, remains one of the leading causes of mortality worldwide. Discovering novel treatments based on cell therapy or drugs is critical, and induced pluripotent stem cells (iPS Cells) technology has made it possible to design extensive disease-specific in vitro models. Elucidating the differentiation process challenged our previous knowledge of cell plasticity and capabilities and allows the concept of cell reprogramming technology to be established, which has inspired the creation of both in vitro and in vivo techniques. Patient-specific cell lines provide the opportunity of studying their pathophysiology in vitro, which can lead to novel drug development. At the same time, in vivo models have been designed where in situ transdifferentiation of cell populations into cardiomyocytes or endothelial cells (ECs) give hope toward effective cell therapies. Unfortunately, the efficiency as well as the concerns about the safety of all these methods make it exceedingly difficult to pass to the clinical trial phase. It is our opinion that creating an ex vivo model out of patient-specific cells will be one of the most important goals in the future to help surpass all these hindrances. Thus, in this review we aim to present the current state of research in reprogramming toward the cardiovascular system's regeneration, and showcase how the development and study of a multicellular 3D ex vivo model will improve our fighting chances.
Introduction
The cardiovascular or circulatory system (CVS) consists of the heart, the blood vessels and almost 5 l of blood that continuously gets pumped throughout the body transferring everything that is needed to maintain homeostasis of nutrients, wastes and gases. A severely damaged CVS is incompatible with life making its successful treatment, especially at an early stage, crucial as each year passes. When one or more of its components are malfunctioning the end-result is complicated diseases—some of them extremely difficult to diagnose.
Stem cell technology represents a big hope for treating unmet clinical needs, including in the context of cardiovascular disease. The ability to self-renew indefinitely and to differentiate in all the three germ layers makes them an attractive candidate both for drug development and personalized cell therapies. Using a variety of source cells, we can now generate endothelial cells (ECs), cardiomyocytes (CMs), vascular smooth muscle cells (VSMCs), and pericytes (PCs) or even progenitor cells to be used for transplantation and to create engineered organs. At the same time, we are able to further study developmental vasculogenesis and angiogenesis in vitro and identify possible mechanisms of pathogenesis by comparing models created by patient cells.
Not to be carried away, we note the limitations and challenges currently present in the use of the ESC—and iPS—derived cell lines both in vitro and in vivo. Issues with tumorigenesis are present with the vast majority of the cell lines due to the genetic stability of the clones. All iPS cell lines are genetically screened and subsequently characterized in vivo with tumorigenesis assays with the successful establishment giving a positive result; in contrast, the iPS-derived cell lines ought to present a negative result. Still, the high levels of proliferation of the cells in their early passages cause concerns when it comes to their clinical application; it is worth mentioning that Mandai et al—who just last year were the first to succeed in transplanting a sheet of retinal pigment epithelial (RPE) cells differentiated from iPS Cells in a patient with neovascular age-related macular degeneration—excluded their second patient due to detecting copy-number alterations in the iPS Cells they derived from them (1). Similarly, the high variability between different lines in respect to both maturity and subtype needs to be addressed. It is well-established that iPS Cells carry the identical genetic anomalies related to the source donor—a fact which makes them ideal for disease modeling. Several types of CVDs have already been modeled including: Hypertrophic cardiomyopathy (HCM), Dilated cardiomyopathy (DCM), Barth syndrome (BTHS), Long-QT (LQT), Catecholaminergic polymorphic ventricular tachycardia (CPVT) and Arrhythmogenic right ventricular cardiomyopathy (ARVC) but, as it will be discussed further on, the models are incomplete (2–4). To address these problematics in the last few years, teams from all over the world come up with new ideas every day: genetic manipulation using the CRISPR/Cas9 technology, direct reprogramming of somatic cells bypassing the pluripotent state, creation of small molecule cocktails for in situ direct reprogramming of local cell populations to name a few.
In this review, we discuss what the current state of the stem cell field is and how close or far away we are from designing a potential strategy for clinical cardiovascular therapies that combines successfully a multicellular model.
Pluripotency Reprograming
In 1981, Evans, Kaufman and Martin reported the establishment of the first mouse embryonic stem cells (ESCs) in culture (5, 6), even though it took 17 years until Thompson et al. developed the first human ESCs lines in 1998 (7). Being able to study the differentiation of cells in vitro creates, for the first time, the opportunity to extensively look at the underlying mechanisms, as well as the opportunity to develop new and advanced treatments.
During those decades it was universally acknowledged that specialized cells reach a point when they cannot differentiate or de-differentiate any more making the process terminal. In 1987, Davis et al. transfected fibroblasts with the cDNA of MyoD and it gave rise to a population of myocytes (8). That was the first challenge of the irreversibility of differentiation and 19 years later the field of stem cells was revolutionized by Yamanaka, Takahashi et al. with the establishment of the first mouse (9) and human (10) induced pluripotent stem cells (iPS Cells) in 2006 and 2007, respectively. Subsequently, the iPS Cells were incorporated into high quality research with teams differentiating them into neurons, cardiomyocytes, hepatocytes endothelial cells etc. Strategies for furthering the field of personalized medicine started developing as the clinical significance of patient specific iPS cell lines is undeniable.
The original protocol developed by Yamanaka utilizing a retroviral vector transduction of the four reprogramming factors Oct4, Sox2, Klf4, and C-myc (OSKM) has been modified since aiming to increases in efficiency of reprogrammed cells and/or the generation of footprint-free iPS cell lines that lack integration of any viral vector sequences into their genomes (Figure 1). C-Myc as a known oncogene was substituted with Wnt3 improving the efficiency of the generation of mouse iPS Cells (miPS Cells) colonies (11). Another group reported the addition of Lin28 and Nanog with the OSKLN derived iPS Cells appearing similar to both Embryonic Stem Cells (ESCs) and OSKM-derived iPS Cells (12). Other delivery methods were also applied: Non integrating viruses like adenovirus (13) and Sendai virus (14) were developed for the reprogramming of human fibroblasts or blood cells into iPS Cells but the efficiencies of the reprogramming were 0.0002 and ~1% respectively. Traditional molecular manipulation methods have also been used successfully among them Cre/LoxP (15) and piggyback (16, 17). Others have established mRNA (18), miRNA (19–21), proteins (22), episomal plasmid transfections (12), or minicircle vectors with a varied combination of genetic modulation (23). Since the CRISPR/Cas9 technology was established in eukaryotic cells (24, 25) steps were taken into combining the two revolutionary technologies and creating a new more versatile approach to the genetic editing of human iPS Cells (26, 27). The ability to simultaneously differentiate cells and genetically modify them—as first described in Howden et al. (28)—raises hope for the disease remodeling field.
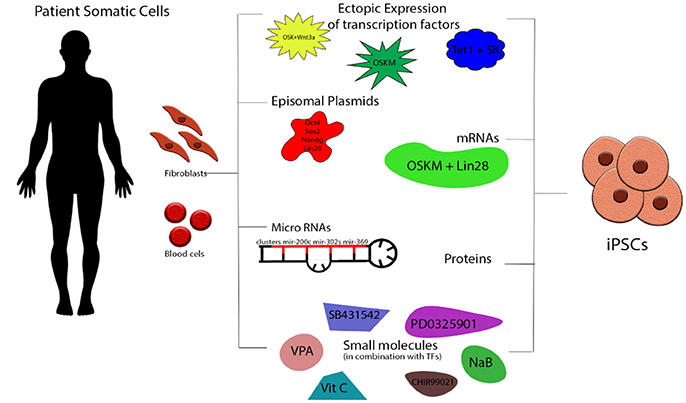
Figure 1. Schematic presentation of iPS Cells generation. Somatic cells (fibroblasts, peripheral blood cells etc) can be isolated from healthy donors and patients and be directly reprogrammed into iPS Cells by the ectopic expression of transcription factors via retro-/lenti-/adeno/Sendai viral transduction. Most commonly used are the combinations of OSKM, OSKNL, OSKL, and TSK. Other strategies involved episomal plasmids (OSLN), mRNAs (OSKML), miRNAs (variety from cluster 200s, 300s, 367/9s), proteins and small molecules including histone modifiers, metabolic modulators, and signaling pathway inhibitors. O, Oct4; S, Sox2; K, Klf4; C, c-Myc; N, NANOG; L, Lin28; T, TET1.
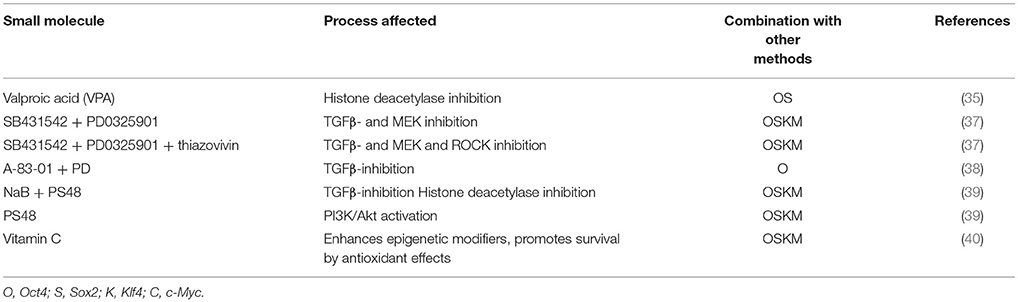
Soon, it became apparent that even when following the same protocol, both the efficiency of the reprogramming and the stability varied between iPS cell lines, presenting quite a challenge, especially with patient specific lines. The cause of that variability is most probably due to the parental line or even disease-specific mutations, therefore, making it clear that the genetic background is crucial to the differentiation potential of a donor-line; curiously surpassing even that of the source-specific variation (29, 30). Large scale screenings and extensive study of the pathways involved in pluripotency led to the discovery of both different transcription factors (31–33) that can be combined and also small molecules that enhance the reprogramming efficiency (34). Epigenetic modifiers like inhibition of histone demethylation (35), inhibition of transforming growth factor-β (TGF-β), MEK and ROCK signaling pathways as well as metabolic modulation and induction of glycolysis (36) were shown to improve the iPS Cells induction (Table 1).
No one can deny that the establishment of patient specific iPS Cells technology gives breath to novel ideas for drug discovery by making in vitro screening of side effects as well as new drug development. The question is; is this enough? In respect to the cardiovascular field, as it will be further discussed, the answer is edging toward no.
Cardiomyocyte Reprogramming
Adult CMs have a very low regenerative ability, mainly coming from the differentiation of cardiac progenitor cells instead of the replacement of the damaged ones via cell division as it was showcased in genetic-fate mapping projects in 2007 (41, 42). Extensive damage leads to scar formation from the activated fibroblasts causing cardiac remodeling and heart failure (HF). Heart transplantation which has been the standard treatment for patients with end-stage HF is still plagued by several issues such as donor shortage, major post-surgery complications such as stroke, bleeding and infection due to chronic immunosuppression (43). The question, as a result, remains. How would we be able to overcome these and regenerate the heart?
The last decade or so, several potential strategies based on stem cells and cell reprogramming have been proposed as an answer to that question. Cell transplantation of ESCs—or iPS Cells-derived CMs (iPS Cells-CMs) is a very promising tool for “remusculising” a failing heart, as showcased in several studies both in small rodents and in non-human primates (44, 45). Alternately, generation of CMs from endogenous sources in situ through differentiation of resident cardiac progenitors or the transdifferentiation of local populations like cardiac fibroblasts or pericytes (46, 47) is another promising approach. Last but not least, cardiac tissue engineering has been evolving rapidly in parallel trying to create fully functional 3-dimensional (3D) biometric constructs from cells derived from iPS to replace the damaged myocardium (48, 49).
Cardiac cells were some of the first cells that were derived from mouse ESCs back in 1985 (50) and subsequently, with the road to pluripotency open, multiple teams in the last decade have been able to differentiate iPS Cells into cardiac progenitors and CMs. The first murine iPS Cells-CMs were derived in 2008 by three groups using the embryonic bodies (EBs) method. Specifically, Mauritz et al. compared the differentiation of an established ESC line and that of an iPSC line toward CMs and they reported a successful conversion, albeit with a much lower efficiency (51). Schenke-Layland et al. exposed EBs to collagen type IV (CollV) and selected Fetal Liver Kinase+ (Flk+) cells through magnetic separation, which, in turn, were differentiated into functional CMs, SMCs, ECs, and hematopoietic cells (52). Narazaki et al. also, modified their protocol and cultured their Flk+ cells on OP9 stroma cells inducing self-beating CMs (53). For human cells, the first iPS Cells-CMs were reported by Zhang et al. in 2009 when they used OSK and Lin28 to generate iPS Cells and then differentiated them using the EB method (54). Many techniques have been described since, and the cells generated have all the advantages that come by that type of differentiation: they can be patient-specific and compatible, making them ideal candidates both for disease study, remodeling and cell therapy (55). Apart from the EB method, differentiation in a monolayer has also been described with high efficiencies by multiple groups. The last 5 years iPS Cells-CMs in varied stages of maturity are produced in larger scales with the help of bioreactors. The murine myocardial infraction model (56) has been used widely to confirm the derived cells' capability of heart tissue regeneration and reduction of scarring. As the technology evolved, rodent models were gradually replaced by non-human primates (44) and pigs (57), and in 2015 Menasché et al. reported the first human ESC-derived cardiac progenitors transplant to patients with advanced ischemic heart failure (58, 59). Even though complications like ventricular arrhythmias may occur post-transplant, their success is considerable. So if we take into consideration the similarities between ESCs and iPS Cells, the baseline of a patient-specific robust cell therapy strategy is set.
In parallel, the concept of transdifferentiating cardiac fibroblasts or other non-myocytes that localize in the heart tissue into CMs also attracts a lot of attention. The first attempts to reprogram cells in vivo started in 2009, when Takeuchi and Bruneau demonstrated that overexpression of Gata4, Tbx5, and the interacting chromatin remodeling protein, Baf60c, converts non-cardiogenic mesoderm into beating CMs in the embryo by a mechanism involving the induction of Nkx2-5 by Gata4 and Baf60c (60). The exogenous production of CMs was revolutionized a year later when Ieda et al. reported the discovery of a 3-factor cocktail, Gata4, Mef2c, and Tbx5 (GMT), successfully reprogramming murine cardiac fibroblasts (mCFs) into induced CM-like cells in vitro (61). Shortly thereafter, three independent studies proved that the non-myocyte pool in the adult mouse heart consisting mainly of CFs can be transdifferentiated in vivo via injecting directly the GMT cocktail into the mouse heart with (GMHT) or without Hand2, and reprogram in vivo CFs into adult induced-CMs (62–64). This resulted in the regeneration of the myocardium and the improvement of cardiac function.
Unfortunately, the low efficiency of most of the in vitro reprogramming protocols—especially when using human cells—as well as concerns for the integration of viral DNA into the host, paved the way of constant modifications, additions and alternations in the GMHT protocol (65). Transcription factors were added (66–68), replaced (69–71), supplemented with small molecules (67, 72) and miRNAs (73) and finally omitted in favor of miRNAs (74), which do not incorporate into the host chromosome, presenting a much safer future clinical application. More recently, Huang et al. proposed a chemically-induced reprogramming in vivo with the combination of CHIR99021; RepSox; Forskolin; VPA; Parnate; TTNPB and Rolipram, successfully inducing CMs from CFs in adult mice and resulting into a reduction of the fibrotic tissue after myocardial infraction (75) (Figure 2). The mechanisms surrounding reprogramming are still left to be elucidated but all these changes contribute little by little to increasing our understanding and, as will be discussed later, to the design of a strategy to combat CVD.
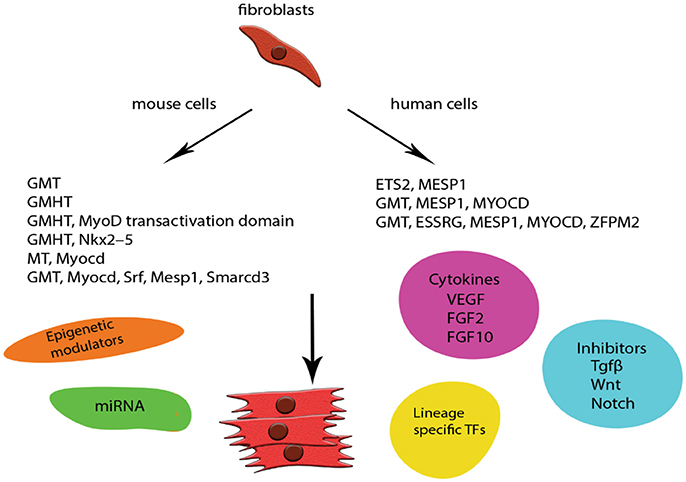
Figure 2. Schematic representation of examples of direct in vitro reprogramming to cardiomyocytes. CMs were one of the first cell lines to be transdifferentiated from mouse and human fibroblasts. The low efficiency of the original GMT protocol especially in human cells was addressed by modifications and addition of more lineage specific factors. Safety concerns were addressed by incorporating new techniques like delivery of miRNA and episomal plasmids coding specific TFs. Small molecules are also widely used with the addition of cytokines and TGFβ inhibition enhancing the final populations and giving rise to robust protocols. G, Gata4; M, Mef2c; T, Tbx5; H, Hand2.
Endothelial Cell Reprogramming
The vascular endothelium controls vascular function and structure, mainly via nitric oxide (NO) production and play a pivotal role in the CVS. EC dysfunction is attributed to be the cause of severe complications, many times proven to be fatal. As the years pass, a growing list of pathological conditions and diseases including hypertension, hypercholesterolemia, diabetes mellitus, congestive heart failure, hyperhomocysteinemia, and even the aging process itself, are associated with EC dysfunction (76).
Differentiation of ECs is governed by several factors, including the immediate microenvironment, interactions with surrounding cells, and the local release of cytokines and growth factors. In the early stages of embryonic development, angiogenesis occurs through the expansion of the vascular plexus with vessel sprouting. The primitive vascular plexus remodels into a highly organized vascular network in which larger vessels ramify into smaller ones and become surrounded by mural cells, which stabilize the newly formed vessels and provide strength to control blood flow and blood pressure (77). In adults with pathological conditions such as cancer or ischemia, or even wound healing, the ECs have the ability to reactivate the angiogenic process making them invaluable for survival.
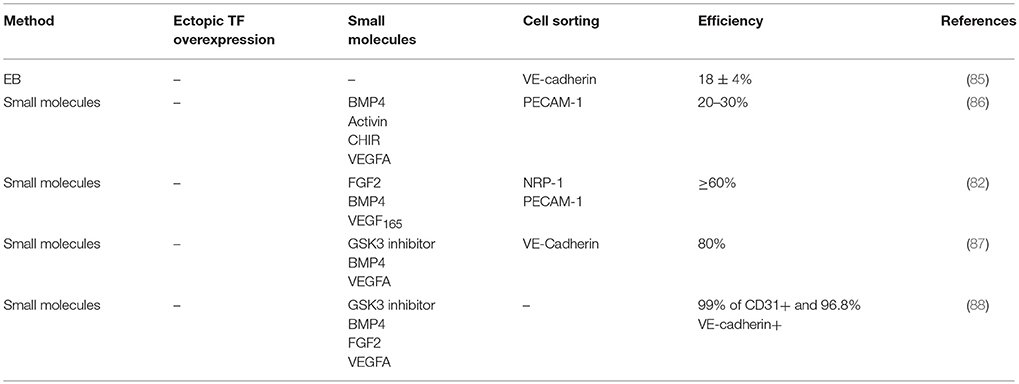
ESC-derived ECs were one of the first to be developed after the establishment of the first colonies with the traditional methods: formation of EBs from the mesoderm germ layer, from which both hematopoietic cells and ECs emerge, and their subsequent exposure to a variety of growth factors, which enhances their differentiation inside the EBs (78, 79). Two-dimensional (2D) cultures under feeder or feeder-free conditions are also very popular and, even though the efficiencies leave something to be desired, advances in the phenotypic stability are being made every day. ESCs gave their place to iPS Cells while linage specific additions to the OSKM factors gave rise to a variety of protocols generating EC progenitors or mature ECs. Especially in the case of endothelial progenitor cells (EPCs), despite the invaluable therapeutic potential they present, the many subtypes that exist create a tricky unit to work with (80–82). A similar challenge is presented with the more mature ECs with different subtypes presenting differences in proliferation and functionality (83, 84). Cell sorting for endothelial progenitor markers like CD34, CD105, Neurophilin1 (NRP1), Vascular Endothelial Growth Factor Receptor 2 (VEGFR2), and PECAM1 (CD31) as well as more mature ones such as VE-Cadherin (CD144) are used to enhance the populations and ensure high purities of the target population (Table 2). Severe limitations like the length of time it takes in generating iPS Cells from the source cells and, in turn, differentiating them into new cell types, challenged the scientific groups. The idea of bypassing the pluripotency state and going down the road of direct reprogramming through epigenetic and linage-specific modulations was reported in 2012 by Margariti et al. when the OSKM factors were transferred to human fibroblasts for 4 days and generated a population of partial-iPS which did not form tumors in vivo and then differentiated them into functional ECs able to revascularize tissue engineered vessels (89). Li et al. used only two of the Yamanaka factors—Oct4 and Klf4—to transdifferentiate human fibroblasts to endothelial-like cells capable of expressing CD31, von Willebrand Factor (vWF) and CD144 that were functional in vivo as well (90). Wong et al. lays emphasis on the importance of epigenetics and describes the potential of using miRNAs (91). Different miRNAs are described to enhance endothelial differentiation including miR-99b,−181a, and−181b (92),−199b (93),−21 (94) of which the overexpression is reported to increase endothelial marker expression and functionality. Apart from fibroblasts, other cell sources have been identified during the last 10 years like mature amniotic cells (95), blood (96), SMCs (97). In addition, genes that have been dubbed as “master key regulators” like Quaking (98–100) and ETV2 (101–104) due to their invaluable role in endothelial function hold a potential to be useful in developing new direct reprogramming strategies.
Direct reprogramming via small molecules and chemical compounds alone has been reported in many cell lines including neuronal cells, glial cells, neural stem cells, brown adipocytes, hepatocytes, CMs, somatic progenitor cells by the regulation of cell signaling pathways and/or histone modification (105). ECs have not been successfully derived yet but further elucidation of the pathways involved both in cell signaling and, in their metabolism, can lead the way. Already scientific groups are turning to single-cell RNA-seq to delve deeper into the heterogeneity of the iPS-derived cell populations. They aim to assess the protocols in use as well as to further study the differentiation process; with Paik et al. publishing a large scale screening of iPS-ECs earlier this year (106).
Smooth Muscle Cells
Smooth muscle cells (SMCs) are highly specialized cells whose major function when matured is the contraction and regulation of blood vessel tone-diameter, blood pressure, and blood flow distribution. Since the late 70s, SMCs have been widely accepted as the main contributors in the pathogenesis of atherosclerosis (107, 108). More specifically, the theory suggested that, in response to vascular injury, SMCs migrate from the media into the intima, where they turn into foam cells and produce extracellular matrix. Almost 30 years later that view was challenged when many scientific teams presented evidence that SMC progenitor cells and hematopoietic stem cells differentiate into SMCs in the intima (109–112). That, in combination with the widely discussed heterogeneity of origin [as different developmental stage SMCs appear with different phenotypes and different source populations (113, 114)], made them the center of attention. Stem cell technology provided the ideal way of studying the different mechanisms of their derivation as well as an opportunity of further understanding the way both mature SMCs and progenitors contribute to the pathophysiology of CVDs.
Again, the EB formation method was used for ES-derived SMCs with Haller et al. reporting that the exposure of the EBs in retinoic acid and dibutyrylcydic adenosine mono-phosphate (db-cAMP) induced differentiation of spontaneously contracting cell clusters in 67% compared with 10% of untreated controls (115). Huang et al. also reported their success in differentiating ESCs into SMCs by adding trans retinoic acid in a monolayer culture with 93% of them expressing SM α-actin and SM-MHC (116). Further studies into cell signaling during the process of differentiation of ESCs and MSCs to SMCs showed that both TGF-β and the Notch pathway, as well as the Bone Morphogenic Proteins (BMPs) (117), are important for the expression of the vascular SMC markers (118, 119). Histone deacetylation plays a main role in vascular homeostasis as well as neuronal, controlling the migration proliferation and differentiation toward SMCs (120–122).
With the introduction of iPS Cells technology, different protocols have been applied into directing the iPS Cells toward SMCs depending on the desired linage (Figure 3). More recently, Steinbach et al. in 2016 described the stepwise administration of key members of the TGF-β superfamily to generate lateral plate-derived vascular SMCs (VSMCs) from human iPS Cells (127). Yang et al. used a combination of Fibroblast Growth Factor (FGF), VEGF and TGFβ to generate VSMCs reporting as well a diversity in their endpoint culture (128).
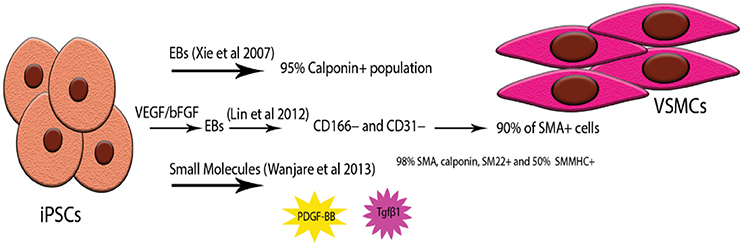
Figure 3. Examples of highly pure iPSC-VSMCs. Due to the heterogeneity of origin as well as the many subtypes of VSMCs acquiring highly pure populations has proven to be challenging. Three types of protocols have been used successfully; simple embryonic body formation and selection of the Calponin positive population (123, 124), combination of EBs and monolayer culture by pre-treating the hiPS Cells with VEGF (125), forming the EBs and then expanding the CD166- /CD31- population in a monolayer again, small molecule treatment of iPS Cells monolayers with PDGF-BB and Tgfβ1 (126).
Direct differentiation protocols have also been applied: most notably, Karamariti et al. used PiPS Cells and 4 days later, the PiPS-SMCs were expressing a full panel of VSMC markers including calponin, SMA-α and SM22α as well as elastin and collagen, characteristically seen in the VSMCs of large arteries (129).
The existence of different VSMCs lineages, occurring as a result of germ layer formation during embryological development, further adds to the complexity of iPS Cells differentiation techniques, as mentioned before. The current studies have provided only a glimpse to a very complicated system but the potential is there. Even though most of the CVD models or tissue engraftments are based on induced-CMs or induced-ECs, we believe VSMCs are an important part of the cardiovascular physiology that should not be bypassed.
Pericytes and their Potential
Perivascular pericytes, or mural cells, envelop the vascular tube surface and are integral in the formation of blood vessels. They are multipotent cells that are heterogeneous in their origin, function, morphology and surface markers. Thus, many controversies have been sparked in respect of their characterization. They are integral for the regulation of blood flow, the stability and permeability of vascular structure and angiogenesis (130–132). Blood vessels lacking pericytes become hyperdilated and haemorrhagic, leading to pathological changes ranging from diabetic retinopathy to embryonic death. They are known to have high level of plasticity and differentiate into other cell types. They also have tissue- specific properties which have been extensively reviewed (132–134).
Dar et al. used EB formation and demonstrated the isolation of CD105+CD90+CD73+CD31− multipotent clonogenic mesodermal precursors. After expansion, the cells expressed markers, like CD146, Neural/glial antigen 2 (NG2), and Beta-Type Platelet-Derived Growth Factor Receptor (PDGFRβ) (135). Orlova et al. described in 2014 the generation of iPS Cells-derived ECs that were expressing a plethora of markers and were functional in vivo; at the same time, using TGFβ3 and BMP4, they differentiated the CD31- fraction of their selection toward pericytes (86).
Another tool in their application into regeneration of the vasculature is cell therapy using pericytes isolated from patients; as they are abundant in various sites on the human body. In 2013, Katare used the mouse myocardial infraction model and demonstrated that transplantation of pericytes, expanded from redundant human leg veins, relocated around the vessels of the peri-infarct zone and released a variety of transcription factors. These enhanced ECs and CMs survival and proliferation (VEGF, angiopoietin) and others inhibited cardiac hypertrophy and fibrosis while promoting angiogenesis (MiR-132 inhibits Ras-GAP, angiotensin 1 receptor (AT1R) and MeCP2 (136).
ESCs and iPS Cells have been used as a source of pericytes (135) but two main concerns have been raised that have yet to be addressed in their entirety. The first is that the efficiencies of the protocols described are usually really low and the second is the lack of distinct pericyte specific markers due to the large heterogeneity of the populations. Most commonly used and widely accepted are PDGFR-β, CD44, CD90, NG2, and α-SMA but they are all strongly present in other cell types so functional assays and extensive expression profiling is needed to complete a characterization. The fact that there is no standardized way of getting a homogenous population of cells is a hindrance to the design of novel therapies however possible epigenetic and secretome screening may help us study the mechanisms of pathogenesis they are involved in.
Discussion
It is commonly acknowledged that, thus far, disease models are incomplete. In vitro co-culture systems are difficult to maintain and although they are very informative, they cannot accurately model the complex and structured in vivo environment. At the same time, animal models are a very useful tool in research for cell therapies and drug development but the different species to species physiology creates major barriers in the clinic. The ideal for many is a 3D in vitro model that will allow for safe testing of therapies and will be modeled with human cells. Naturally, the most promising cell types to be used are human iPS Cells or reprogrammed cells.
With respect to the cardiovascular field, analyzing the three points of structure is crucial to planning future strategies. First and foremost, EC dysfunction is widely acknowledged as one of the leading causes of complications for patients suffering from diabetes and other vascular diseases. Secondly, CM damage combined with the heart's low regeneration ability has proved to be one of the most difficult points to address during the study of the pathophysiology of a disease both in vitro and in/ex vivo as well as in the development of therapies (137). Thirdly, SMCs are deeply integrated into the pathogenesis of the atherosclerotic plaque but the knowledge of the mechanism behind their differentiation during different stages of the disease is lacking (114). Last but not least, pericytes may have been relatively overlooked—possibly due to their heterogeneity—but are integral to the preservation of vascular rheology and homeostasis (134). Keeping these in mind, it is obvious to see that an attempt of excluding a component may lead into not being able to complete the puzzle. Tissue engineering technology is advancing rapidly and experimentation of new biomaterials and re-vascularization strategies is a fact. Engineering 3D cardiac tissue with a physiologically relevant microenviroment is quite challenging. Most promising are the re-vascularization strategies of the bioengineered graft and the maturity of the cells that will be used, since the reprogrammed cells—especially the CMs—are usually more immature types, and not what we would see in a functional human heart. As presented extensively by Costa- Almeida et al. constant vascularization is critical based on cellular strategies combining EC transplantation with support cells, which will produce growth factors, cytokines, hormones and other bioactive molecules essential to the stability of the scaffold (138).
If we take a step back we will see that currently we are getting closer but we still have a long way to go. In vitro culture (and co-culture) models are very useful for studying different cell type interactions but we are still missing the complexity of the cell signaling interplay in tissue. Future research should be focusing not only in getting novel insights into the process of angiogenesis but in combining our knowledge of the interaction of heterotypic cells to develop ex vivo models of the CVS. Considering the difficulty of acquiring mature cells from patients, iPS Cell-derived or reprogrammed cells are ideal candidates for modeling these. Concerns about the phenotypical stability of the differentiated cells should also be addressed by further studying the epigenetic process in which we erase the cell memory and direct them in a different path. The advantages are significant for personalized and regenerative medicine as well as drug development and testing, revealing a potential role of these models for their manipulation into patient-specific scaffolds for heart and vessel damage (Figure 4).
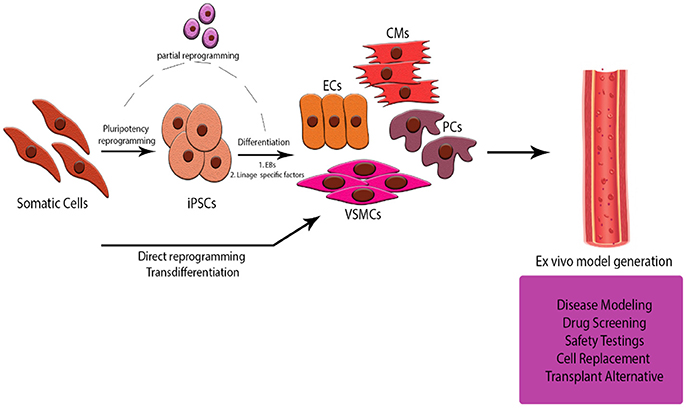
Figure 4. Schematic Representation of the abstract. Generation of an ex-vivo tissue model for studying the cell to cell interaction in a 3D CVS model is offering multiple advantages over current animal models and co-culture in vitro systems. Using cells derived from iPS Cells or somatic cells of donors will help us look deeper into the pathogenesis of diseases, customize and safely test novel drug treatments and cell therapies for patients, even develop strategies for alternative sources of tissue for transplants.
Author Contributions
MT conception and design, manuscript writing. SK design, final approval of manuscript. RC final approval of manuscript. CY final approval of manuscript. AM conception and design, financial support, final approval of manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The reviewer KP and handling Editor declared their shared affiliation.
Acknowledgments
This work was supported by Grants from BBSRC and the British Heart Foundation.
References
1. Mandai M, Watanabe A, Kurimoto Y, Hirami Y, Morinaga C, Daimon T, et al. Autologous induced stem-cell–derived retinal cells for macular degeneration. N Engl J Med. (2017) 376:1038–46. doi: 10.1056/NEJMoa1608368
2. Yang C, Al-Aama J, Stojkovic M, Keavney B, Trafford A, Lako M, et al. Concise review: cardiac disease modeling using induced pluripotent stem cells. Stem Cells (2015) 33:2643–51. doi: 10.1002/stem.2070
3. Dell'Era P, Benzoni P, Crescini E, Valle M, Xia E, Consiglio A, et al. Cardiac disease modeling using induced pluripotent stem cell-derived human cardiomyocytes. World J Stem Cells (2015) 7:329–42. doi: 10.4252/wjsc.v7.i2.329
4. Tanaka A, Yuasa S, Node K, Fukuda K. Cardiovascular disease modeling using patient-specific induced pluripotent stem cells. Int J Mol Sci. (2015) 16:18894–922. doi: 10.3390/ijms160818894
5. Evans MJ, Kaufman MH. Establishment in culture of pluripotential cells from mouse embryos. Nature (1981) 292:154–6.
6. Martin GR. Isolation of a pluripotent cell line from early mouse embryos cultured in medium conditioned by teratocarcinoma stem cells. Proc Natl Acad Sci USA. (1981) 78:7634–8.
7. Thomson JA, Itskovitz-Eldor J, Shapiro SS, Waknitz MA, Swiergiel JJ, Marshall VS, et al. Embryonic stem cell lines derived from human blastocysts. Science (1998) 282:1145–7.
8. Davis RL, Weintraub H, Lassar AB. Expression of a single transfected cDNA converts fibroblasts to myoblasts. Cell (1987) 51:987–1000.
9. Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell (2006) 126:663–76. doi: 10.1016/j.cell.2006.07.024
10. Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell (2007) 131:861–72. doi: 10.1016/j.cell.2007.11.019
11. Marson A, Foreman R, Chevalier B, Bilodeau S, Kahn M, Young RA, et al. Wnt signaling promotes reprogramming of somatic cells to pluripotency. Cell Stem Cell (2008) 3:132–5. doi: 10.1016/j.stem.2008.06.019
12. Yu J, Vodyanik MA, Smuga-Otto K, Antosiewicz-Bourget J, Frane JL, Tian S, et al. Induced pluripotent stem cell lines derived from human somatic cells. Science (2007) 318:1917–20. doi: 10.1126/science.1151526
13. Zhou W, Freed CR. Adenoviral gene delivery can reprogram human fibroblasts to induced pluripotent stem cells. Stem Cells (2009) 27:2667–74. doi: 10.1002/stem.201
14. Fusaki N, Ban H, Nishiyama A, Saeki K, Hasegawa M. Efficient induction of transgene-free human pluripotent stem cells using a vector based on Sendai virus, an RNA virus that does not integrate into the host genome. Proc Jpn Acad Ser B Phys Biol Sci. (2009) 85:348–62. doi: 10.2183/pjab.85.348
15. Soldner F, Hockemeyer D, Beard C, Gao Q, Bell GW, Cook EG, et al. Parkinson's disease patient-derived induced pluripotent stem cells free of viral reprogramming factors. Cell (2009) 136:964–77. doi: 10.1016/j.cell.2009.02.013
16. Woltjen K, Michael IP, Mohseni P, Desai R, Mileikovsky M, Hämäläinen R, et al. piggyBac transposition reprograms fibroblasts to induced pluripotent stem cells. Nature (2009) 458:766–70.doi: 10.1038/nature07863
17. Kaji K, Norrby K, Paca A, Mileikovsky M, Mohseni P, Woltjen K. Virus-free induction of pluripotency and subsequent excision of reprogramming factors. Nature (2009) 458:771–5. doi: 10.1038/nature07864
18. Warren L, Manos PD, Ahfeldt T, Loh Y-H, Li H, Lau F, et al. Highly efficient reprogramming to pluripotency and directed differentiation of human cells with synthetic modified mRNA. Cell Stem Cell (2010) 7:618–30. doi: 10.1016/j.stem.2010.08.012
19. Anokye-Danso F, Trivedi CM, Juhr D, Gupta M, Cui Z, Tian Y, et al. Highly efficient miRNA-mediated reprogramming of mouse and human somatic cells to pluripotency. Cell Stem Cell (2011) 8:376–88. doi: 10.1016/j.stem.2011.03.001
20. Subramanyam D, Lamouille S, Judson RL, Liu JY, Bucay N, Derynck R, et al. Multiple targets of miR-302 and miR-372 promote reprogramming of human fibroblasts to induced pluripotent stem cells. Nat Biotechnol. (2011) 29:443–8. doi: 10.1038/nbt.1862
21. Poleganov MA, Eminli S, Beissert T, Herz S, Moon J-I, Goldmann J, et al. Efficient reprogramming of human fibroblasts and blood-derived endothelial progenitor cells using nonmodified RNA for reprogramming and immune evasion. Hum Gene Ther. (2015) 26:751–66. doi: 10.1089/hum.2015.045
22. Kim D, Kim C-H, Moon J-I, Chung Y-G, Chang M-Y, Han B-S, et al. Generation of human induced pluripotent stem cells by direct delivery of reprogramming proteins. Cell Stem Cell (2009) 4:472–6. doi: 10.1016/j.stem.2009.05.005
23. Malik N, Rao MS. A review of the methods for human iPSC derivation. Methods Mol Biol. (2013) 339:823–6. doi: 10.1007/978-1-62703-348-0_3
24. Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE, et al. RNA-guided human genome engineering via Cas9. Science (2013) 339:823–6. doi: 10.1126/science.1232033
25. Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, et al. Multiplex genome engineering using CRISPR/Cas systems. Science (2013) 339:819–23. doi: 10.1126/science.1231143
26. Smith C, Gore A, Yan W, Abalde-Atristain L, Li Z, He C, et al. Whole-genome sequencing analysis reveals high specificity of CRISPR/Cas9 and TALEN-based genome editing in human iPSCs. Cell Stem Cell (2014) 15:12–3.doi: 10.1016/j.stem.2014.06.011
27. Hockemeyer D, Jaenisch R. Induced pluripotent stem cells meet genome editing. Cell Stem Cell (2016) 18:573–86.doi: 10.1016/j.stem.2016.04.0136
28. Howden SE, Maufort JP, Duffin BM, Elefanty AG, Stanley EG, Thomson JA. Simultaneous reprogramming and gene correction of patient fibroblasts. Stem Cell Rep. (2015) 5:1109–18. doi: 10.1016/j.stemcr.2015.10.009
29. Rouhani F, Kumasaka N, de Brito MC, Bradley A, Vallier L, Gaffney D. Genetic background drives transcriptional variation in human induced pluripotent stem cells. PLoS Genet. (2014) 10:e1004432. doi: 10.1371/journal.pgen.1004432
30. Kyttälä A, Moraghebi R, Valensisi C, Kettunen J, Andrus C, Pasumarthy KK, et al. Genetic variability overrides the impact of parental cell type and determines iPSC differentiation potential. Stem Cell Rep. (2016) 6:200–12. doi: 10.1016/j.stemcr.2015.12.009
31. Boyer LA, Lee TI, Cole MF, Johnstone SE, Levine SS, Zucker JP, et al. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell (2005) 122:947–56. doi: 10.1016/j.cell.2005.08.020
32. Loh Y-H, Wu Q, Chew J-L, Vega VB, Zhang W, Chen X, et al. The Oct4 and Nanog transcription network regulates pluripotency in mouse embryonic stem cells. Nat Genet. (2006) 38:431–40.doi: 10.1038/ng1760
33. Zhou X, Meng G, Nardini C, Mei H. Systemic evaluation of cellular reprogramming processes exploiting a novel R-tool: eegc. Bioinformatics (2017) 33:2532–8. doi: 10.1093/bioinformatics/btx205
34. Lin T, Ambasudhan R, Yuan X, Li W, Hilcove S, Abujarour R, et al. A chemical platform for improved induction of human iPSCs. Nat Methods (2009) 6:805–8. doi: 10.1038/nmeth.1393
35. Huangfu D, Osafune K, Maehr R, Guo W, Eijkelenboom A, Chen S, et al. Induction of pluripotent stem cells from primary human fibroblasts with only Oct4 and Sox2. Nat Biotechnol. (2008) 26:1269–75. doi: 10.1038/nbt.1502
36. Yoshida Y, Takahashi K, Okita K, Ichisaka T, Yamanaka S. Hypoxia enhances the generation of induced pluripotent stem cells. Cell Stem Cell (2009) 5:237–41. doi: 10.1016/j.stem.2009.08.001
37. Noggle S, Fung H-L, Gore A, Martinez H, Satriani KC, Prosser R, et al. Human oocytes reprogram somatic cells to a pluripotent state. Nature (2011) 478:70–5. doi: 10.1038/nature10397
38. Zhu S, Li W, Zhou H, Wei W, Ambasudhan R, Lin T, et al. Reprogramming of human primary somatic cells by OCT4 and chemical compounds. Cell Stem Cell (2010) 7:651–5.doi: 10.1016/j.stem.2010.11.015
39. Ichida JK, Blanchard J, Lam K, Son EY, Chung JE, Egli D, et al. A small-molecule inhibitor of Tgf-β signaling replaces Sox2 in reprogramming by inducing Nanog. Cell Stem Cell (2009) 5:491–503. doi: 10.1016/j.stem.2009.09.012
40. Esteban MA, Wang T, Qin B, Yang J, Qin D, Cai J, et al. Vitamin C enhances the generation of mouse and human induced pluripotent stem cells. Cell Stem Cell (2010) 6:71–9. doi: 10.1016/j.stem.2009.12.001
41. Quaini F, Urbanek K, Graiani G, Lagrasta C, Maestri R, Monica M, et al. The regenerative potential of the human heart. Int J Cardiol. (2004) 95(Suppl. 1):S26–8. doi: 10.1016/S0167-5273(04)90008-3
42. Hsieh PCH, Segers VFM, Davis ME, MacGillivray C, Gannon J, Molkentin JD, et al. Evidence from a genetic fate-mapping study that stem cells refresh adult mammalian cardiomyocytes after injury. Nat Med. (2007) 13:970–4. doi: 10.1038/nm1618
43. Toyoda Y, Guy TS, Kashem A. Present status and future perspectives of heart transplantation. Circ J. (2013) 77:1097–110. doi: 10.1253/circj.CJ-13-0296
44. Chong JJH, Yang X, Don CW, Minami E, Liu Y-W, Weyers JJ, et al. Human embryonic-stem-cell-derived cardiomyocytes regenerate non-human primate hearts. Nature (2014) 510:273–7. doi: 10.1038/nature13233
45. Shiba Y, Fernandes S, Zhu W-Z, Filice D, Muskheli V, Kim J, et al. Human ES-cell-derived cardiomyocytes electrically couple and suppress arrhythmias in injured hearts. Nature (2012) 489:322–5. doi: 10.1038/nature11317
46. Senyo SE, Steinhauser ML, Pizzimenti CL, Yang VK, Cai L, Wang M, et al. Mammalian heart renewal by pre-existing cardiomyocytes. Nature (2012) 493:433–6. doi: 10.1038/nature11682
47. Beltrami AP, Barlucchi L, Torella D, Baker M, Limana F, Chimenti S, et al. Adult cardiac stem cells are multipotent and support myocardial regeneration. Cell (2003) 114:763–76. doi: 10.1016/S0092-8674(03)00687-1
48. Chiu LLY, Iyer RK, Reis LA, Nunes SS, Radisic M. Cardiac tissue engineering: current state and perspectives. Front Biosci. (2012) 17:1533–50. doi: 10.2741/4002
49. Jafarkhani M, Salehi Z, Kowsari-Esfahan R, Shokrgozar MA, Rezaa Mohammadi M, Rajadas J, et al. Strategies for directing cells into building functional hearts and parts. Biomater Sci. (2018) 6:1664–90. doi: 10.1039/c7bm01176h
50. Doetschman TC, Eistetter H, Katz M, Schmidt W, Kemler R. The in vitro development of blastocyst-derived embryonic stem cell lines: formation of visceral yolk sac, blood islands and myocardium. J Embryol Exp Morphol. (1985) 87:27–45.
51. Mauritz C, Schwanke K, Reppel M, Neef S, Katsirntaki K, Maier LS, et al. Generation of functional murine cardiac myocytes from induced pluripotent stem cells. Circulation (2008) 118:507–17. doi: 10.1161/CIRCULATIONAHA.108.778795
52. Schenke-Layland K, Rhodes KE, Angelis E, Butylkova Y, Heydarkhan-Hagvall S, Gekas C, et al. Reprogrammed mouse fibroblasts differentiate into cells of the cardiovascular and hematopoietic lineages. Stem Cells (2008) 26:1537–46. doi: 10.1634/stemcells.2008-0033
53. Narazaki G, Uosaki H, Teranishi M, Okita K, Kim B, Matsuoka S, et al. Directed and systematic differentiation of cardiovascular cells from mouse induced pluripotent stem cells. Circulation (2008) 118:498–506. doi: 10.1161/CIRCULATIONAHA.108.769562
54. Zhang J, Wilson GF, Soerens AG, Koonce CH, Yu J, Palecek SP, et al. Functional cardiomyocytes derived from human induced pluripotent stem cells. Circ Res. (2009) 104:e30–41. doi: 10.1161/CIRCRESAHA.108.192237
55. Yoshida Y, Yamanaka S. Induced pluripotent stem cells 10 years later: for cardiac applications. Circ Res. (2017) 120:1958–68. doi: 10.1161/CIRCRESAHA.117.311080
56. Kumar M, Kasala ER, Bodduluru LN, Dahiya V, Sharma D, Kumar V, et al. Animal models of myocardial infarction: mainstay in clinical translation. Regul Toxicol Pharmacol. (2016) 76:221–30. doi: 10.1016/j.yrtph.2016.03.005
57. Kawamura M, Miyagawa S, Fukushima S, Saito A, Miki K, Ito E, et al. Enhanced survival of transplanted human induced pluripotent stem cell-derived cardiomyocytes by the combination of cell sheets with the pedicled omental flap technique in a porcine heart. Circulation (2013) 128(11 Suppl. 1):S87–94. doi: 10.1161/CIRCULATIONAHA.112.000366
58. Menasché P, Vanneaux V, Fabreguettes J-R, Bel A, Tosca L, Garcia S, et al. Towards a clinical use of human embryonic stem cell-derived cardiac progenitors: a translational experience. Eur Heart J. (2015) 36:743–50. doi: 10.1093/eurheartj/ehu192
59. Menasché P, Vanneaux V, Hagège A, Bel A, Cholley B, Cacciapuoti I, et al. Human embryonic stem cell-derived cardiac progenitors for severe heart failure treatment: first clinical case report: Figure 1. Eur Heart J. (2015) 36:2011–7. doi: 10.1093/eurheartj/ehv189
60. Takeuchi JK, Bruneau BG. Directed transdifferentiation of mouse mesoderm to heart tissue by defined factors. Nature (2009) 459:708–11. doi: 10.1038/nature08039
61. Ieda M, Fu J-D, Delgado-Olguin P, Vedantham V, Hayashi Y, Bruneau BG, et al. Direct reprogramming of fibroblasts into functional cardiomyocytes by defined factors. Cell (2010) 142:375–86. doi: 10.1016/j.cell.2010.07.002
62. Qian L, Huang Y, Spencer CI, Foley A, Vedantham V, Liu L, et al. In vivo reprogramming of murine cardiac fibroblasts into induced cardiomyocytes. Nature (2012) 485:593–8.doi: 10.1038/nature11044
63. Song K, Nam Y-J, Luo X, Qi X, Tan W, Huang GN, et al. Heart repair by reprogramming non-myocytes with cardiac transcription factors. Nature (2012) 485:599–604. doi: 10.1038/nature11139
64. Inagawa K, Miyamoto K, Yamakawa H, Muraoka N, Sadahiro T, Umei T, et al. Induction of cardiomyocyte-like cells in infarct hearts by gene transfer of Gata4, Mef2c, and Tbx5. Circ Res. (2012) 111:1147–56. doi: 10.1161/CIRCRESAHA.112.271148
65. Wang L, Liu Z, Yin C, Asfour H, Chen O, Li Y, et al. Stoichiometry of Gata4, Mef2c, and Tbx5 influences the efficiency and quality of induced cardiac myocyte reprogramming. Circ Res. (2015) 116:237–44. doi: 10.1161/CIRCRESAHA.116.305547
66. Hirai H, Katoku-Kikyo N, Keirstead SA, Kikyo N. Accelerated direct reprogramming of fibroblasts into cardiomyocyte-like cells with the MyoD transactivation domain. Cardiovasc Res. (2013) 100:105–13. doi: 10.1093/cvr/cvt167
67. Cao N, Huang Y, Zheng J, Spencer CI, Zhang Y, Fu J-D, et al. Conversion of human fibroblasts into functional cardiomyocytes by small molecules. Science (2016) 352:1216–20. doi: 10.1126/science.aaf1502
68. Fu J-D, Stone NR, Liu L, Spencer CI, Qian L, Hayashi Y, et al. Direct reprogramming of human fibroblasts toward a cardiomyocyte-like state. Stem Cell Rep. (2013) 1:235–47. doi: 10.1016/j.stemcr.2013.07.005
69. Christoforou N, Chellappan M, Adler AF, Kirkton RD, Wu T, Addis RC, et al. Transcription factors MYOCD, SRF, Mesp1 and SMARCD3 enhance the cardio-inducing effect of GATA4, TBX5, and MEF2C during direct cellular reprogramming. PLoS ONE (2013)8:e63577. doi: 10.1371/journal.pone.0063577
70. Nam Y-J, Song K, Luo X, Daniel E, Lambeth K, West K, et al. Reprogramming of human fibroblasts toward a cardiac fate. Proc Natl Acad Sci USA. (2013) 110:5588–93. doi: 10.1073/pnas.1301019110
71. Islas JF, Liu Y, Weng K-C, Robertson MJ, Zhang S, Prejusa A, et al. Transcription factors ETS2 and MESP1 transdifferentiate human dermal fibroblasts into cardiac progenitors. Proc Natl Acad Sci USA. (2012) 109:13016–21. doi: 10.1073/pnas.1120299109
72. Wang H, Cao N, Spencer CI, Nie B, Ma T, Xu T, et al. Small molecules enable cardiac reprogramming of mouse fibroblasts with a single factor. Cell Rep. (2014) 6:951–60. doi: 10.1016/j.celrep.2014.01.038
73. Muraoka N, Yamakawa H, Miyamoto K, Sadahiro T, Umei T, Isomi M, et al. MiR-133 promotes cardiac reprogramming by directly repressing Snai1 and silencing fibroblast signatures. EMBO J. (2014) 33:1565–81. doi: 10.15252/embj.201387605
74. Jayawardena TM, Egemnazarov B, Finch EA, Zhang L, Payne JA, Pandya K, et al. MicroRNA-mediated in vitro and in vivo direct reprogramming of cardiac fibroblasts to cardiomyocytes. Circ Res. (2012) 110:1465–73. doi: 10.1161/CIRCRESAHA.112.269035
75. Huang C, Tu W, Fu Y, Wang J, Xie X. Chemical-induced cardiac reprogramming in vivo. Cell Res. (2018) 28:686–9.doi: 10.1038/s41422-018-0036-4
76. Tousoulis D, Simopoulou C, Papageorgiou N, Oikonomou E, Hatzis G, Siasos G, et al. Endothelial dysfunction in conduit arteries and in microcirculation. novel therapeutic approaches. Pharmacol Ther. (2014) 144:253–67. doi: 10.1016/j.pharmthera.2014.06.003
77. Carmeliet P. Angiogenesis in life, disease and medicine. Nature (2005) 438:932–6.doi: 10.1038/nature04478
78. Yoder MC. Differentiation of pluripotent stem cells into endothelial cells. Curr Opin Hematol. (2015) 22:252–7. doi: 10.1097/MOH.0000000000000140
79. Wilson HK, Canfield SG, Shusta EV, Palecek SP. Concise review: tissue-specific microvascular endothelial cells derived from human pluripotent stem cells. Stem Cells (2014) 32:3037–45. doi: 10.1002/stem.1797
80. Medina RJ, O'Neill CL, Humphreys MW, Gardiner TA, Stitt AW. Outgrowth endothelial cells: characterization and their potential for reversing ischemic retinopathy. Investig Opthalmol. Vis Sci. (2010) 51:5906. doi: 10.1167/iovs.09-4951
81. Moubarik C, Guillet B, Youssef B, Codaccioni J-L, Piercecchi M-D, Sabatier F, et al. Transplanted late outgrowth endothelial progenitor cells as cell therapy product for stroke. Stem Cell Rev Rep. (2011) 7:208–20. doi: 10.1007/s12015-010-9157-y
82. Prasain N, Lee MR, Vemula S, Meador JL, Yoshimoto M, Ferkowicz MJ, et al. Differentiation of human pluripotent stem cells to cells similar to cord-blood endothelial colony–forming cells. Nat Biotechnol. (2014) 32:1151–7. doi: 10.1038/nbt.3048
83. Turner RR, Beckstead JH, Warnke RA, Wood GS. Endothelial cell phenotypic diversity: in situ demonstration of immunologic and enzymatic heterogeneity that correlates with specific morphologic subtypes. Am J Clin Pathol. (1987) 87:569–75.
84. Marcelo KL, Goldie LC, Hirschi KK. Regulation of endothelial cell differentiation and specification. Circ Res. (2013) 112:1272–87. doi: 10.1161/CIRCRESAHA.113.300506
85. Adams WJ, Zhang Y, Cloutier J, Kuchimanchi P, Newton G, Sehrawat S, et al. Functional vascular endothelium derived from human induced pluripotent stem cells. Stem Cell Rep. (2013) 1:105–13. doi: 10.1016/j.stemcr.2013.06.007
86. Orlova V V, van den Hil FE, Petrus-Reurer S, Drabsch Y, ten Dijke P, Mummery CL. Generation, expansion and functional analysis of endothelial cells and pericytes derived from human pluripotent stem cells. Nat Protoc. (2014) 9:1514–31. doi: 10.1038/nprot.2014.102
87. Patsch C, Challet-Meylan L, Thoma EC, Urich E, Heckel T, O'Sullivan JF, et al. Generation of vascular endothelial and smooth muscle cells from human pluripotent stem cells. Nat Cell Biol. (2015) 17:994–1003. doi: 10.1038/ncb3205
88. Harding A, Cortez-Toledo E, Magner NL, Beegle JR, Coleal-Bergum DP, Hao D, et al. Highly efficient differentiation of endothelial cells from pluripotent stem cells requires the MAPK and the PI3K pathways. Stem Cells (2017) 35:909–19. doi: 10.1002/stem.2577
89. Margariti A, Winkler B, Karamariti E, Zampetaki A, Tsai T, Baban D, et al. Direct reprogramming of fibroblasts into endothelial cells capable of angiogenesis and reendothelialization in tissue-engineered vessels. Proc Natl Acad Sci USA. (2012) 109:13793–8.doi: 10.1073/pnas.1205526109
90. Li J, Huang NF, Zou J, Laurent TJ, Lee JC, Okogbaa J, et al. Conversion of human fibroblasts to functional endothelial cells by defined factors. Arterioscler Thromb Vasc Biol. (2013) 33:1366–75. doi: 10.1161/ATVBAHA.112.301167
91. Wong WT, Huang NF, Botham CM, Sayed N, Cooke JP. Endothelial cells derived from nuclear reprogramming. Circ Res. (2012) 111:1363–75. doi: 10.1161/CIRCRESAHA.111.247213
92. Kane NM, Howard L, Descamps B, Meloni M, McClure J, Lu R, et al. Role of microRNAs 99b, 181a, and 181b in the differentiation of human embryonic stem cells to vascular endothelial cells. Stem Cells (2012) 30:643–54. doi: 10.1002/stem.1026
93. Chen T, Margariti A, Kelaini S, Cochrane A, Guha ST, Hu Y, et al. MicroRNA-199b modulates vascular cell fate during iPS cell differentiation by targeting the notch ligand Jagged1 and enhancing VEGF signaling. Stem Cells (2015) 33:1405–18. doi: 10.1002/stem.1930
94. Di Bernardini E, Campagnolo P, Margariti A, Zampetaki A, Karamariti E, Hu Y, et al. Endothelial lineage differentiation from induced pluripotent stem cells is regulated by microRNA-21 and transforming growth factor β2 (TGF-β2) pathways. J Biol Chem. (2014) 289:3383–93. doi: 10.1074/jbc.M113.495531
95. Ginsberg M, Schachterle W, Shido K, Rafii S. Direct conversion of human amniotic cells into endothelial cells without transitioning through a pluripotent state. Nat Protoc. (2015) 10:1975–85. doi: 10.1038/nprot.2015.126
96. Yeh ETH, Zhang S, Wu HD, Körbling M, Willerson JT, Estrov Z. Transdifferentiation of human peripheral blood CD34+-enriched cell population into cardiomyocytes, endothelial cells, and smooth muscle cells in vivo. Circulation (2003) 108:2070–3.doi: 10.1161/01.CIR.0000099501.52718.70
97. Hong X, Margariti A, Le Bras A, Jacquet L, Kong W, Hu Y, et al. Transdifferentiated human vascular smooth muscle cells are a new potential cell source for endothelial regeneration. Sci Rep. (2017) 7:5590.doi: 10.1038/s41598-017-05665-7
98. Bohnsack BL, Lai L, Northrop JL, Justice MJ, Hirschi KK. Visceral endoderm function is regulated byquaking and required for vascular development. Genesis (2006) 44:93–104.doi: 10.1002/gene.20189
99. de Bruin RG, van der Veer EP, Prins J, Lee DH, Dane MJC, Zhang H, et al. The RNA-binding protein quaking maintains endothelial barrier function and affects VE-cadherin and β-catenin protein expression. Sci Rep. (2016) 6:21643. doi: 10.1038/srep21643
100. Cochrane A, Kelaini S, Tsifaki M, Bojdo J, Vilà-González M, Drehmer D, et al. Quaking is a key regulator of endothelial cell differentiation, neovascularization, and angiogenesis. Stem Cells (2017) 35:952–66. doi: 10.1002/stem.2594
101. Tanaka T, Izawa K, Maniwa Y, Okamura M, Okada A, Yamaguchi T, et al. ETV2-TET1/TET2 complexes induce endothelial cell-specific Robo4 expression via promoter demethylation. Sci Rep. (2018) 8:5653. doi: 10.1038/s41598-018-23937-8
102. Morita R, Suzuki M, Kasahara H, Shimizu N, Shichita T, Sekiya T, et al. ETS transcription factor ETV2 directly converts human fibroblasts into functional endothelial cells. Proc Natl Acad Sci USA. (2015) 112:160–5.doi: 10.1073/pnas.1413234112
103. Oh S-Y, Kim JY, Park C. The ETS Factor, ETV2: a master regulator for vascular endothelial cell development. Mol Cells (2015) 38:1029–36. doi: 10.14348/molcells.2015.0331
104. Van Pham P, Vu NB, Nguyen HT, Huynh OT, Truong MT-H. Significant improvement of direct reprogramming efficacy of fibroblasts into progenitor endothelial cells by ETV2 and hypoxia. Stem Cell Res Ther. (2016) 7:104. doi: 10.1186/s13287-016-0368-2
105. Takeda Y, Harada Y, Yoshikawa T, Dai P. Chemical compound-based direct reprogramming for future clinical applications. Biosci Rep. (2018) 38:BSR20171650. doi: 10.1042/BSR20171650
106. Paik DT, Tian L, Lee J, Sayed N, Chen IY, Rhee S, et al. Large-scale single-cell RNA-seq reveals molecular signatures of heterogeneous populations of human induced pluripotent stem cell-derived endothelial cells. Circ Res. (2018) 123:443–50. doi: 10.1161/CIRCRESAHA.118.312913
107. Ross R, Glomset JA. Atherosclerosis and the arterial smooth muscle cell: proliferation of smooth muscle is a key event in the genesis of the lesions of atherosclerosis. Science (1973) 180:1332–9.
109. Sata M, Saiura A, Kunisato A, Tojo A, Okada S, Tokuhisa T, et al. Hematopoietic stem cells differentiate into vascular cells that participate in the pathogenesis of atherosclerosis. Nat Med. (2002) 8:403–9.doi: 10.1038/nm0402-403
110. Daniel J-M, Sedding DG. Circulating smooth muscle progenitor cells in arterial remodeling. J Mol Cell Cardiol. (2011) 50:273–9. doi: 10.1016/j.yjmcc.2010.10.030
111. Han C, Campbell GR, Campbell JH. Circulating bone marrow cells can contribute to neointimal formation. J Vasc Res. (2001) 38:113–9.doi: 10.1159/000051038
112. Hillebrands JL, Klatter FA, van den Hurk BM, Popa ER, Nieuwenhuis P, Rozing J. Origin of neointimal endothelium and alpha-actin-positive smooth muscle cells in transplant arteriosclerosis. J Clin Invest. (2001) 107:1411–22. doi: 10.1172/JCI10233
113. Wang G, Jacquet L, Karamariti E, Xu Q. Origin and differentiation of vascular smooth muscle cells. J Physiol. (2015) 593:3013–30. doi: 10.1113/JP270033
114. Margariti A, Zeng L, Xu Q. Stem cells, vascular smooth muscle cells and atherosclerosis. Histol Histopathol. (2006) 21:979–85. doi: 10.14670/HH-21.979
115. Haller H, Bychkov R, Erdmann B, Lindschau C, Haase H, Morano I, et al. From totipotent embryonic stem cells to spontaneously contracting smooth muscle cells: a retinoic acid and db-cAMP in vitro differentiation model. FASEB J. (1997) 11:905–15.
116. Huang H, Zhao X, Chen L, Xu C, Yao X, Lu Y, et al. Differentiation of human embryonic stem cells into smooth muscle cells in adherent monolayer culture. Biochem Biophys Res Commun. (2006) 351:321–7.doi: 10.1016/j.bbrc.2006.09.171
117. Zhang P, Li J, Tan Z, Wang C, Liu T, Chen L, et al. Short-term BMP-4 treatment initiates mesoderm induction in human embryonic stem cells. Blood (2008) 111:1933–41.doi: 10.1182/blood-2007-02-074120
118. Sinha S, Hoofnagle MH, Kingston PA, McCanna ME, Owens GK. Transforming growth factor-β1 signaling contributes to development of smooth muscle cells from embryonic stem cells. Am J Physiol Physiol. (2004) 287:C1560–8.doi: 10.1152/ajpcell.00221.2004
119. Kurpinski K, Lam H, Chu J, Wang A, Kim A, Tsay E, et al. Transforming growth factor-β and notch signaling mediate stem cell differentiation into smooth muscle cells. Stem Cells (2010) 28:734–42. doi: 10.1002/stem.319
120. Yan Z-Q, Yao Q-P, Zhang M-L, Qi Y-X, Guo Z-Y, Shen B-R, et al. Histone deacetylases modulate vascular smooth muscle cell migration induced by cyclic mechanical strain. J Biomech. (2009) 42:945–8. doi: 10.1016/j.jbiomech.2009.01.012
121. Zhou B, Margariti A, Zeng L, Xu Q. Role of histone deacetylases in vascular cell homeostasis and arteriosclerosis. Cardiovasc Res. (2011) 90:413–20. doi: 10.1093/cvr/cvr003
122. Xu SS, Alam S, Margariti A. Epigenetics in vascular disease - therapeutic potential of new agents. Curr Vasc Pharmacol. (2014) 12:77–86.
123. Xie C-Q, Zhang J, Villacorta L, Cui T, Huang H, Chen YE. A highly efficient method to differentiate smooth muscle cells from human embryonic stem cells. Arterioscler Thromb Vasc Biol. (2007) 27:e311–2.doi: 10.1161/ATVBAHA.107.154260
124. Ge X, Ren Y, Bartulos O, Lee MY, Yue Z, Kim K-Y, et al. Modeling supravalvular aortic stenosis syndrome with human induced pluripotent stem cells. Circulation (2012) 126:1695–704. doi: 10.1161/CIRCULATIONAHA.112.116996
125. Lin B, Kim J, Li Y, Pan H, Carvajal-Vergara X, Salama G, et al. High-purity enrichment of functional cardiovascular cells from human iPS cells. Cardiovasc Res. (2012) 95:327–35. doi: 10.1093/cvr/cvs185
126. Wanjare M, Kuo F, Gerecht S. Derivation and maturation of synthetic and contractile vascular smooth muscle cells from human pluripotent stem cells. Cardiovasc Res. (2013) 97:321–30. doi: 10.1093/cvr/cvs315
127. Steinbach SK, Husain M. Vascular smooth muscle cell differentiation from human stem/progenitor cells. Methods (2016) 101:85–92. doi: 10.1016/j.ymeth.2015.12.004
128. Yang L, Geng Z, Nickel T, Johnson C, Gao L, Dutton J, et al. Differentiation of human induced-pluripotent stem cells into smooth-muscle cells: two novel protocols. PLoS ONE (2016) 11:e0147155. doi: 10.1371/journal.pone.0147155
129. Karamariti E, Margariti A, Winkler B, Wang X, Hong X, Baban D, et al. Smooth muscle cells differentiated from reprogrammed embryonic lung fibroblasts through DKK3 signaling are potent for tissue engineering of vascular grafts. Circ Res. (2013) 112:1433–43. doi: 10.1161/CIRCRESAHA.111.300415
130. Pallone TL, Silldorff EP. Pericyte regulation of renal medullary blood flow. Exp Nephrol. (2001) 9:165–70.doi: 10.1159/000052608
131. Hellström M, Gerhardt H, Kalén M, Li X, Eriksson U, Wolburg H, et al. Lack of pericytes leads to endothelial hyperplasia and abnormal vascular morphogenesis. J Cell Biol. (2001) 153:543–53. doi: 10.1083/jcb.153.3.543
132. Ahmed TA, El-Badri N. Pericytes: The role of multipotent stem cells in vascular maintenance and regenerative medicine. Adv Exp Med Biol. (2017) 7:452–64. doi: 10.1007/5584_2017_138
133. Bergers G, Song S. The role of pericytes in blood-vessel formation and maintenance. Neuro Oncol. (2005) 7:452–64.doi: 10.1215/S1152851705000232
134. Ferland-McCollough D, Slater S, Richard J, Reni C, Mangialardi G. Pericytes, an overlooked player in vascular pathobiology. Pharmacol Ther. (2017) 171:30–42. doi: 10.1016/j.pharmthera.2016.11.008
135. Dar A, Domev H, Ben-Yosef O, Tzukerman M, Zeevi-Levin N, Novak A, et al. Multipotent vasculogenic pericytes from human pluripotent stem cells promote recovery of murine ischemic limb. Circulation (2012) 125:87–99. doi: 10.1161/CIRCULATIONAHA.111.048264
136. Katare RG, Madeddu P. Pericytes from human veins for treatment of myocardial ischemia. Trends Cardiovasc Med. (2013) 23:66–70. doi: 10.1016/j.tcm.2012.09.002
137. Sadahiro T, Yamanaka S, Ieda M. Direct cardiac reprogramming: progress and challenges in basic biology and clinical applications. Circ Res. (2015) 116:1378–91. doi: 10.1161/CIRCRESAHA.116.305374
Keywords: reprogramming, vascular cells, regeneration, cardiovascular system, induced pluripotent stem cells (iPS Cells)
Citation: Tsifaki M, Kelaini S, Caines R, Yang C and Margariti A (2018) Regenerating the Cardiovascular System Through Cell Reprogramming; Current Approaches and a Look Into the Future. Front. Cardiovasc. Med. 5:109. doi: 10.3389/fcvm.2018.00109
Received: 04 June 2018; Accepted: 24 July 2018;
Published: 20 August 2018.
Edited by:
Gabor Foldes, Imperial College London, United KingdomReviewed by:
Koralia Paschalaki, Imperial College London, United KingdomElisa Avolio, University of Bristol, United Kingdom
Copyright © 2018 Tsifaki, Kelaini, Caines, Yang and Margariti. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Andriana Margariti, YS5tYXJnYXJpdGlAcXViLmFjLnVr
 Marianna Tsifaki
Marianna Tsifaki Sophia Kelaini
Sophia Kelaini Rachel Caines
Rachel Caines Chunbo Yang
Chunbo Yang Andriana Margariti
Andriana Margariti