
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Cell. Infect. Microbiol., 19 October 2020
Sec. Bacteria and Host
Volume 10 - 2020 | https://doi.org/10.3389/fcimb.2020.584751
This article is part of the Research TopicBacterial Effectors as Drivers of Human Disease: Models, Methods, MechanismsView all 16 articles
The type VI secretion system (T6SS) is a transmembrane multiprotein nanomachine employed by many Gram-negative bacterial species to translocate, in a contact-dependent manner, effector proteins into adjacent prokaryotic or eukaryotic cells. Typically, the T6SS gene cluster encodes at least 13 conserved core components for the apparatus assembly and other less conserved accessory proteins and effectors. It functions as a contractile tail machine comprising a TssB/C sheath and an expelled puncturing device consisting of an Hcp tube topped by a spike complex of VgrG and PAAR proteins. Contraction of the sheath propels the tube out of the bacterial cell into a target cell and leads to the injection of toxic proteins. Different bacteria use the T6SS for specific roles according to the niche and versatility of the organism. Effectors are present both as cargo (by non-covalent interactions with one of the core components) or specialized domains (fused to structural components). Although several anti-prokaryotic effectors T6SSs have been studied, recent studies have led to a substantial increase in the number of characterized anti-eukaryotic effectors. Against eukaryotic cells, the T6SS is involved in modifying and manipulating diverse cellular processes that allows bacteria to colonize, survive and disseminate, including adhesion modification, stimulating internalization, cytoskeletal rearrangements and evasion of host innate immune responses.
Gram-negative bacteria depend on specific secretion systems, numbered Type I through Type VII, to transport proteins outside the cell for survival and fitness. It has been estimated that >25% of pathogenic and non-pathogenic proteobacteria encode between one and six Type VI secretion systems (T6SS) (Bingle et al., 2008; Boyer et al., 2009). The T6SS is a dynamic contractile protein nanomachine, evolutionarily related to bacteriophage tails, which delivers protein effectors in a contact-dependent manner into diverse cellular types, including other bacteria, fungi, and host eukaryotic cells. As reviewed elsewhere (Records, 2011; Basler, 2015; Cianfanelli et al., 2016b; Nguyen et al., 2018; Cherrak et al., 2019; Navarro-Garcia et al., 2019; Hernandez et al., 2020), the T6SS gene cluster encodes 13 core components for apparatus assembly. The system can be divided in three substructures, (i) a membrane complex (TssJLM) anchored to the inner membrane and associated to the outer membrane (Aschtgen et al., 2008; Ma et al., 2009b; Durand et al., 2015; Logger et al., 2016; Rapisarda et al., 2019; Yin et al., 2019), (ii) a baseplate complex assembled by a wedge (TssEFGK) (Brunet et al., 2015; Cherrak et al., 2018; Nazarov et al., 2018) and a spike (VgrG and, in some cases PAAR proteins) (Shneider et al., 2013; Brunet et al., 2015; Renault et al., 2018) and (iii) the dynamic tail complex that comprises the inner tube (Hcp) (Ballister et al., 2008; Brunet et al., 2014; Douzi et al., 2014) and the contractile sheath (TssBC) that wraps around the Hcp tube and propels the spike (Bonemann et al., 2009; Basler et al., 2012; Broms et al., 2013; Zhang et al., 2013; Kube et al., 2014).
The T6SS can translocate effector proteins in two modular ways: binding of an additional protein domain to structural components of the needle, Hcp, PAAR, or VgrG (specialized or evolved effectors) or by non-covalent direct or indirect interactions, via adaptor proteins, with any of the components of the needle (cargo effectors) (Shneider et al., 2013; Durand et al., 2014; Whitney et al., 2014; Alcoforado Diniz et al., 2015; Ma et al., 2017; Pissaridou et al., 2018). There are T6SS effector chaperone (TEC), or adaptor (Tap-1), proteins that are essential for toxin loading and delivery through binding to VgrG and effector proteins (Liang et al., 2015; Bondage et al., 2016; Flaugnatti et al., 2016; Jana and Salomon, 2019). TEC and Tap-1 proteins share a highly conserved domain of unknown function (DUF4123) and are not secreted; they exhibit a low pI values and are often genetically encoded upstream of their cognate effector genes or downstream of vgrG genes (Liang et al., 2015; Unterweger et al., 2015). Proteins containing DUF2169 domains are commonly found downstream of vgrG and upstream of DUF4150-containing effector genes and also serve as adaptor or chaperone in binding the N-terminal PAAR or PAAR-like domains of its cognate effector to the tip for translocation (Bondage et al., 2016; Santos et al., 2019). The DUF1795 containing proteins, namely Eag proteins, bind and stabilize the N-terminal PAAR-containing domains of their cognate effectors. Eag chaperone family members are frequently encoded adjacent to putative effectors with predicted transmembrane domains (Cianfanelli et al., 2016a; Quentin et al., 2018).
It has been reported that the T6SS mainly functions as a device for inter-bacterial competition to inject toxic antibacterial proteins into rival bacterial cells, thus modulating polymicrobial communities. More recently, the range of known functions of the T6SS has extended, including action against microbial fungi, biofilm formation and transport of ions. The T6SS also functions as a classical virulence factor by delivering toxins that allow bacteria to manipulate and subvert eukaryotic cells.
The T6SS toxins targeting eukaryotic cells are varied in biological and biochemical functions (Hachani et al., 2016). In general, different bacterial species use and adapt their T6SS for specific roles according to the host, niche or survival strategy of the organism and there is also considerable diversity in effector portfolio. In this review, we discuss and summarize the activity, target and mode of delivery of eukaryotic cell-targeting T6SS toxins important in pathogenicity, which interact and manipulate different components of the host cell. The effectors below revised are categorized accordingly to the bacterial species that encodes them.
Table 1 and Figure 1 give a general overview of functionalities of the eukaryotic T6SS effectors described in the text.
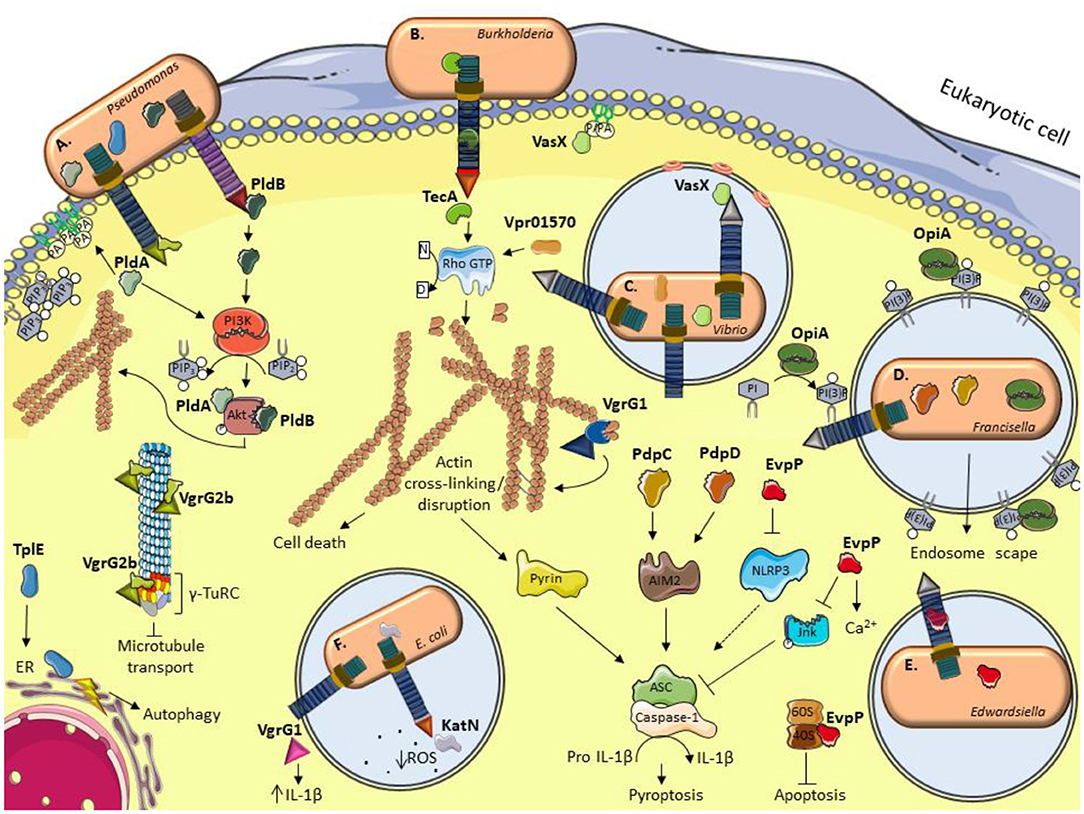
Figure 1. Schematic illustration of current models for the role of some anti-eukaryotic effectors. (A) P. aeruginosa delivers PldA and PldB which bind Akt to allow bacterial internalization via the induction of the PI3K pathway. VgrG2b enables bacterial internalization by interacting with members of the microtubule γ-TuRC complex. TplE targets the endoplasmic reticulum (ER) and induces stress and autophagy. (B) B. cenocepacia TecA is a deamidase that disrupts actin cytoskeleton by deamidating Rho GTPases and activates the Pyrin inflammasome. (C) The V. cholerae evolved VgrG1 interacts with and cross-link actin, leading to an accumulation of toxic actin oligomers and altering host cell morphology. VasX binds the lipid membrane phosphatidic acid (PA) and is thought to form pores in lipid bilayers. V. proteolyticus Vpr01570 contains a deamidase domain that activates Rho GTPases. (D) F. tularensis OpiA is a kinase able to phosphorylate phosphatidylinositol (PI) and binding to phosphatidylinositol trisphosphate [PI(3)P] for its recruitment to endosomal membranes. PdpC and PdpD activate AIM2 inflammasome. (E) E. tarda translocates EvpP which modifies calcium flux and has an inhibitory role in NLRP3 inflammasome by reducing Jnk phosphorylation and ASC oligomerization. E. ictalurid EvpP interacts with ribosomal protein S5 to negatively regulate apoptosis. (F) The EHEC effector KatN is a catalase that contributes to the survival in macrophages by hydrolyzing and decreasing the levels of reactive oxygen species (ROS). VgrG1 is secreted by ExPEC that alters the IL-1β levels. This figure was prepared using free templates on the Servier medical art website (https://smart.servier.com/).
Vibrio cholerae is a natural free-living bacterium widely distributed in aquatic environments and also the environment within human hosts. V. cholerae is a non-invasive intestinal pathogen; O1 and O139 serogroup cause the diarrheal disease cholera. The first reported T6SS toxin targeting eukaryotic cells was VgrG-1. Mutants unable to produce this protein lack the ability to secrete Hcp or to infect amoebae and mammalian macrophages, suggesting that rather in addition to be an essential component of the T6SS apparatus, VgrG-1 is a genuine effector (Pukatzki et al., 2007; Zheng et al., 2011). VgrG-1 carries a large (395 amino acids) C-terminal extension with homology to the actin cross-linking domain (ACD) of the RtxA toxin, a member of the MARTX family (Durand et al., 2012). VgrG-1 catalyzes in vitro the covalent cross-linking of two G-actin monomers in a Mg2+/Mn2+-ATP dependent manner and in vivo induced massive cross-linking of cytosolic actin in macrophages and from harvested intestines in an infant mice model of infection (Pukatzki et al., 2007; Ma and Mekalanos, 2010; Durand et al., 2012). Actin oligomers disrupt the normal inter-subunit interface in the actin filament and prevent polymerization (Satchell, 2009; Heisler et al., 2015). Bacterial internalization by endocytosis is needed for VgrG-1 ACD domain translocation into phagocytic cells to impair their function and cause cell death, preventing bacterial clearance from the gut (Ma et al., 2009a). VgrG-1 also possesses an actin binding motif (ABM) on the surface of the ACD similar to WH2 domain. Actin nucleation is inhibited by this ACD-ABM because the motif can bind and sequester actin monomers; this binding domain is also indispensable for ACD mediated actin cross-linking (Dutta et al., 2019). VgrG-1 forms homotrimeric and heterotrimeric complexes by interacting with VgrG-2 and VgrG-3 (Pukatzki et al., 2007). The crystal structure of VgrG-1-ACD (PDB 4DTD) reveals a V-shaped structure formed of 12 β-strands and 9 α-helices and an active site composed of 5 residues; one of these, Glu-16, is the critical residue for the cross-linking activity (Durand et al., 2012).
Another noteworthy T6SS V. cholerae protein is the virulence-associated secretion protein X (VasX or VCA0020), encoded in the T6SS gene cluster downstream of hcp and vgrG-2. The 121-kDa protein VasX requires the T6SS transcriptional activator VasH for expression, and a functional T6SS apparatus for secretion with the VgrG spike as carrier for its delivery. VasX plays a role in T6SS mediated virulence, killing amoebae by a mechanism that depends on actin cross-linking (Zheng et al., 2011; Dong et al., 2013). VasX carries an N-terminal Pleckstrin-homology (PH) domain that binds to membrane lipids including phosphatidic acid (PA) and each of the phosphatidylinositol phosphates (PIP). Since inositol phosphates are rarely found in bacteria the PH domain of VasX may have a role in binding to host membrane lipids and is thought to form pores in lipid bilayers (Miyata et al., 2011). VasX also carries a motif named MIX (marker for type six effectors) and a C-terminal colicin domain important for its secretion and T6SS assembly (Salomon et al., 2014; Liang et al., 2019).
Vibrio proteolyticus (Vpr) is a marine bacterium that has been previously isolated from corals with yellow band disease. Three T6SS effectors with putative anti-eukaryotic activities were identified by analyzing the Vpr secretome. Vpr01570 contains an N-terminal MIX V domain and a C-terminal CNF1 (cytotoxic necrotizing factor 1) deamidase domain that targets and activates Rho GTPases. Vpr01570 exogenously expressed in macrophages induces actin cytoskeleton rearrangements, including assembly of contractile actin stress fibers and ruffles at the top of the cells in a T6SS-dependent manner. Vpr01570 induces toxicity when expressed in yeast and these effects depend on the CNF1 domain (Ray et al., 2017). Vpr01580 is encoded next to the Vpr01570 encoding gene and also contains a MIX V domain; its homologous proteins are cytotoxic and contain Rhs repeats. Vpr00400 is homologous to the C-terminal domain of the toxic protein Txp40 which has insecticidal activity. Additional studies are required to elucidate the role of Vpr01580 and Vpr00400 (Ray et al., 2017).
Enterohemorrhagic Escherichia coli (EHEC) is a human intestinal pathogen responsible for outbreaks of bloody diarrhea and hemolytic uremic syndrome worldwide. KatN is 84% identical to the Mn2+-containing catalase KatN of Salmonella enterica and the specific activity of KatN is 268.3 U/mg protein (Wan et al., 2017). KatN contributes to the EHEC response to oxidative stress in vitro; OxyR and RpoS are involved in katN transcription activation and H-NS, a global regulator, in its repression. After phagocytosis, EHEC induces the expression of T6SS, and translocated KatN contributes to the survival of intracellular bacteria in macrophages by hydrolyzing and decreasing the levels of reactive oxygen species (ROS) providing an ideal niche for bacterial growth and further infection (Wan et al., 2017).
Extra-intestinal pathogenic Escherichia coli (ExPEC) strains can cause urinary tract, bloodstream, prostate, and other infections at non-intestinal sites, leading to disease in humans and other animals. They are a serious threat to human public health and high risk for food safety. Porcine ExPEC causes meningitis, pneumonia, arthritis, and septicemia and is multidrug-resistant. The VgrG protein, a core component and a T6SS effector, performs diverse functions as an effector in addition to its structural component role. ExPEC VgrG1 plays a role in bacterial adherence, multiplication, and also a main role in evasion of innate immune response. In the absence of VgrG1, the serum level of IL-1β in mice is significantly reduced (Zong et al., 2019).
One of the most virulent opportunistic pathogens is Pseudomonas aeruginosa, commonly found in soil and water as well as in plants and humans. P. aeruginosa is metabolically versatile and can cause a wide range of severe opportunistic infections in patients with cancer, cystic fibrosis and burns. The P. aeruginosa genome encodes three evolutionary distinct T6SS clusters, the H1–3-T6SSs, which are expressed simultaneously, each secreting a variable set of toxins. The H1-T6SS targets bacteria, while H2-3-T6SS targets bacteria and are also involved in internalization into eukaryotic cells (Mougous et al., 2006; Sana et al., 2012, 2016).
Phospholipases D (PLDs) are found in only a very limited number of prokaryotic organisms but, when present, they often play a role in bacterial pathogenesis. The 122-kDa protein PldA (Tle5a) from P. aeruginosa has high homology with eukaryotic PLDs; the protein is secreted via H2-T6SS and delivered as a cargo effector via their cognate VgrG4b. PldA possesses two HXKXXXXD catalytic motifs and it has phospholipase calcium-regulated activity in vitro. PldA enzymatic activity resulting in phosphatidylcholine hydrolysis depends on a catalytic histidine residue (H855) (Wilderman et al., 2001; Russell et al., 2013; Wettstadt et al., 2019). PldA can induce cell death through PA accumulation via PLD activity, primarily aimed against phosphatidylethanolamine (Russell et al., 2013; Jiang et al., 2014).
The 83-kDa protein PldB (Tle5b) is a P. aeruginosa H2- and H3-T6SS-dependent PLD effector delivered via their cognate VgrG5 and is able to translocate into human epithelial cells. PldB possesses two HXKXXXXD catalytic motifs that play a crucial role in toxicity. PldA and PldB do not share homology, suggesting that they have developed similar functions by convergent evolution (Jiang et al., 2014; Wettstadt et al., 2019). A study deciphering the prevalence of genes encoding T6SS effectors in clinical isolates found that the prevalence of pldA was increased in isolates responsible for severe acute pulmonary infection and septicemia. In contrast, pldB prevalence was high in all isolates (Boulant et al., 2018). PldA and PldB are not involved in bacterial adhesion but promote intracellular invasion of host eukaryotic cells by activation of the phosphatidylinositol 3-kinase (PI3K)/Akt signaling pathway that is crucial for cell growth, proliferation, and programmed cell death. After injection into epithelial cells, PldA and PldB directly interact with Akt1 and/or Akt2 kinase, resulting in activation of the PI3K-Akt pathway. Indeed, Akt phosphorylation at serine 473 promotes remodeling of the apical membrane in which protrusions enriched in phosphatidylinositol-3,4,5-triphosphate (PIP3) and actin enables bacterial entry (Bleves et al., 2014; Jiang et al., 2014).
VgrG2b is conserved in all P. aeruginosa strains present in the Pseudomonas genome database. VgrG2b is a 113-kDa protein that contains the conserved VgrG domain homologous to gp27 and gp5 phage-tail proteins followed by a domain of unknown function, DUF2345, and a C-terminal extension with a Zn2+-dependent metallopeptidase domain (LFIHEMTHVW). It is an evolved VgrG with double function as a structural component of the secretion machinery and a true effector translocated via the H2-T6SS required for invasion of host cells. VgrG2b injection precedes internalization; its C-terminal domain interacts with α- and β-tubulin complexes and with the γ-tubulin complexes, such as the γ-tubulin small complex (γTuSC) and the γ-tubulin ring complex (γ-TuRC) involved in microtubule nucleation. This interaction allows bacterial uptake into epithelial cells to be mediated by actin cytoskeletal rearrangement (Sana et al., 2015; Wood et al., 2019). The crystal structure of Vgr2b C-terminal encompassing residues 833-1019 (PDB 6H56) presents a metallopeptidase fold (Wood et al., 2019).
Sana et al. (2016) proposed a working model for the interplay of T6SS effectors PldA, PldB and VgrG2b in P. aeruginosa internalization. First, VgrG2b is translocated via H2-T6SS, causing the polarization of epithelial cells by targeting the microtubule network, promoting microtubule nucleation at the membrane by interacting with γ-TuRC. These novel sites of non-radial microtubule nucleation interfere with the transport of microtubule-dependent cargoes in the cell, like PI3K. Simultaneously, PldA and PldB are translocated by the different H2 and H3-T6SSs, activating Akt which allows actin-dependent membrane protrusion that enables bacterial internalization into the epithelial cells.
Another effector, TplE, contains a eukaryotic PGAP1 (post-glycosylphosphatidylinositol attachment to proteins 1)-like domain. TplE is translocated into epithelial cells in an H2-T6SS-dependent manner and localizes to host endoplasmic reticulum (ER), causing a contraction of the ER surrounding the nuclear periphery. TplE phospholipase activity is not involved in localization but is required for disruption of ER structure. TplE induces the upregulation of Bip and CHOP chaperones that are biomarkers for ER stress and induces the splicing of XBP1 mRNA, suggesting that the TplE-induced unfolded protein response is dependent on the IRE1α-XBP1 signaling pathway. It was also reported that autophagic flux is induced by TplE delivery into human epithelial cells (Jiang et al., 2016).
Klebsiella pneumoniae is a ubiquitous species in nature, a gut commensal, and an opportunistic pathogen in humans. As a prominent nosocomial pathogen, it can cause a wide range of infections, including urinary tract, respiratory tract or blood infections, bacteremia and liver abscesses. Due to the regular occurrence of multiple antibiotic-resistant isolates, K. pneumoniae is considered a global public health concern. In K. pneumoniae three different T6SS loci were defined, and a gene encoding a PLD family protein Pld1 is located within a type VI secretion system locus (Sarris et al., 2011; Lery et al., 2014). Pld1 is a Tle5 homolog, has two conserved HXKXXXXD motif and is expressed during K. pneumoniae virulence in a mouse model of pneumonia. The pld1 phospholipase mutant was strongly attenuated in vivo, suggesting an effect on lipid metabolism in K. pneumoniae pathogenesis (Lery et al., 2014).
VgrG4 encodes a C-terminal domain of unknown function DUF2345. VgrG4 is needed for bacteria-induced killing of the fungal pathogen Candida albicans and Saccharomyces cerevisiae, implicating the T6SS in intoxication of fungal cells. The DUF2345 domain is sufficient for the anti-eukaryotic activity (Storey et al., 2020).
Francisella tularensis is one of the most infectious intracellular pathogens known. After entering the body via the skin, mucous membranes, or respiratory or gastrointestinal tracts, it causes tularemia, a necrotizing bronchopneumonia that leads to sepsis and death. The T6SS encoded by the Francisella pathogenicity island (FPI) is critical for the virulence of this bacterium. In contrast F. tularensis subsp. novicida (F. novicida) has low virulence in humans, but is highly virulent in mice and thus often used as a laboratory model for tularemia (Eshraghi et al., 2016). PdpC (pathogenicity determinant protein C) is a 156-kDa protein encoded within the FPI that contributes to phagosomal escape, trafficking to lysosomes and intramacrophage replication. PdpC plays a role in virulence in the mouse model, as demonstrated by the ΔpdpC mutant causing significantly lower mortality in mice with a corresponding reduction in bacterial burden in organs. PdpC is required to activate the AIM2 inflammasome and ΔpdpC induces lower levels of type I interferon production (Lindgren et al., 2013a,b; Long et al., 2013; Uda et al., 2014; Eshraghi et al., 2016; Ozanic et al., 2016; Brodmann et al., 2017).
PdpD is a protein encoded within the FPI; its export requires VgrG and PdpA. This effector contributes to intramacrophage growth and phagosome rupture. PdpD is also required to activate the AIM2 inflammasome (Eshraghi et al., 2016; Brodmann et al., 2017).
OpiA and OpiB are encoded by open reading frames located outside of the FPI and recently identified as T6SS substrates. They contribute to intracellular growth. There are no homologs of OpiA found outside of Francisella, and in silico analyses were unable to identify characterized domains or motifs within the protein. The OpiB C-terminus is homologous to the ankyrin repeat domains mediating protein-protein interactions that are normally found in eukaryotic proteins. The OpiB N-terminus constitutes an evolutionarily plastic cysteine protease (Eshraghi et al., 2016). OpiA belongs to a family of wortmannin-resistant bacterial PI3K enzymes with members found in a wide range of intracellular pathogens. OpiA can phosphorylate PI but not PIP2. OpiA binds phosphatidylinositol 3-phosphate [PI(3)P] in a selective and high-affinity manner serving as a mechanism for the specific recruitment of OpiA to endosomal membranes. OpiA acts on the Francisella-containing phagosome, leading to efficient bacterial escape from late endosomes into the cytoplasm of infected cells (Ledvina et al., 2018). The protein is translocated into phagocytic cells and reduces the levels of TNF-α, a pro-inflammatory cytokine from monocytes required to block intracellular replication. OpiA contributes to the pathogenesis of F. tularensis, as demonstrated using a chicken embryo infection model (Cantlay et al., 2020).
The protein IglE (intracellular growth locus E) is translocated into macrophages (Broms et al., 2012). The ΔiglE mutant has a slower intracellular growth rate in human macrophages, suggesting a role for this protein in intracellular replication. IglE interacts with β-tubulin, pericentrin and with microtubule organizing centers. It inhibits the dynein- based intracellular trafficking in host cells, allowing F. novicida to escape from fusion with lysosomes (Shimizu et al., 2019).
Edwardsiella tarda infects a wide range of hosts including fish, birds, reptiles and humans. In humans, it causes both intestinal and extra-intestinal infections, mainly in individuals with impaired immune systems. Edwardsiellosis in fish is a devastating disease predominant in worldwide aquaculture industries, making it of particular importance to the fishing industry (Zheng and Leung, 2007). EvpP (E. tarda virulence protein P) transcription is iron-dependent. EvpP is a 20-kDa protein that is not conserved in other bacteria and contains no conserved domains or motifs. It is secreted via T6SS and the EvpP C-terminus interacts with EvpV (Hcp homolog) (Zheng and Leung, 2007; Hu et al., 2014). In an in vivo fish model, EvpP plays a role in proliferation and infection. This toxin also mediates hemolytic activity in sheep erythrocytes and contributes to mucus adhesion and serum resistance of Japanese flounder. EvpP is important for internalization into epithelial papilloma of carp cells (Wang et al., 2009). The protein localizes in the membrane after injection and has an inhibitory role in NLRP3 inflammasome activation by reducing Jnk phosphorylation and ASC oligomerization. It was reported that ΔevpP induced higher intracellular calcium flux than wildtype E. tarda indicating that EvpP-mediated manipulation of the Jnk-ASC could be traced upstream to intracellular Ca2+ signaling (Chen et al., 2017).
Edwardsiella ictaluri causes enteric septicemia of catfish and is the most important endemic infectious disease in catfish aquaculture industry. EvpP toxin is involved in adhesion and internalization of E. ictaluri in catfish ovary cells. EvpP plays a role in growth regulation in the phagolysosome where oxidative stress and limited nutrients are present, and also favors survival and increases apoptosis and necrosis in catfish anterior kidney macrophages (Kalindamar et al., 2020).
Edwardsiella piscicida is abundant in water and causes food and waterborne infections in fish, animals and humans (Leung et al., 2019). Using an in vivo zebrafish larvae infection model EvpP inhibits immune cells recruitment via Jnk-MAPK signaling cascades. EvpP reduces the expression of cxcl8a (chemokine ligand 8) and mmp13 (matrix metallopeptidase 13) transcripts, indicating that EvpP plays a role in inhibiting the recruitment of neutrophils. Meanwhile, EvpP also inhibits the Jnk-caspy inflammasome and IL-1β expression to suppress neutrophil recruitment, thereby promoting bacterial colonization (Tan et al., 2019). EvpP is also able to reduce Annexin V binding and activation of cleaved caspase-3 involved in apoptosis. This effector interacts with ribosomal protein S5 (RPS5), most likely resulting in downregulation of apoptosis-associated pathways in macrophages (Qin et al., 2020).
Burkholderia cenocepacia is widespread in the environment, particularly within the rhizosphere. B. cenocepacia is also an opportunistic pathogen causing chronic lung infections in patients with cystic fibrosis as well as in other immunocompromised patients (Loutet and Valvano, 2010). The 17-kDa protein TecA is a non-VgrG T6SS effector responsible for actin disruption in vivo. TecA and other bacterial homologs bear a cysteine protease-like catalytic triad, which inactivates Rho GTPases by deamidating a conserved asparagine in the GTPase switch-I region. RhoA deamidation induces Pyrin inflammasome activation (Aubert et al., 2016).
Burkholderia thailandensis is a soil saprophyte of low virulence. Burkholderia pseudomallei is the causative agent of melioidosis, a serious and often fatal human infection. These species, referred as the Bptm group, encode several T6SSs but the type VI secretion system 5 (T6SS-5) is the one required for virulence in mammalian infection models. VgrG-5 is a substrate of T6SS-5 and is translocated into macrophages. VgrG-5 C-terminal domain is involved in mediating multinucleated giant cell formation, membrane fusion and virulence in mice (Schwarz et al., 2014; Toesca et al., 2014).
Serratia marcescens occurs naturally in soil and water. It is associated with urinary and respiratory infections, endocarditis, osteomyelitis, septicemia, wound infections, eye infections, and meningitis. Tfe1 (T6SS antifungal effector 1) is an antifungal small T6SS toxin (20 kDa), deletion of Tfe1 encoding gene resulted in a four-fold increase in recovery of viable Candida albicans target cells compared with the wild type bacteria. Tfe1 causes cell distortion and lysis in both the budding and filamentous forms of C. albicans. Tfe1 inhibits growth of S. cerevisiae and induces abnormally large vacuoles and cell lysis, confirming the fungicidal role of this effector. Tfe1 intoxication results in membrane depolarization by loss of membrane potential, which is not due to pore formation but can lead to a loss of membrane integrity and cell death (Trunk et al., 2018, 2019). Removal of Tfe2 (T6SS antifungal effector 2) encoding gene, resulted in almost complete loss of activity against S. cerevisiae or Candida glabrata and reduced activity against C. albicans. Tfe2 is a small protein (26 kDa) which, when expressed in S. cerevisiae, is able to inhibit its growth. Tfe2 intoxication disrupts nutrient uptake and amino acid metabolism and causes autophagy. Tfe1 and Tfe2 act on different cellular targets in fungal cells (Trunk et al., 2018, 2019).
Aeromonas hydrophila is common in freshwater environments and causes disease in fish, reptiles, amphibians, and humans. It causes a broad spectrum of infections (including septicemia, meningitis, endocarditis) in humans and severe motile septicemia in warmwater fishes. The 103-kDa protein VgrG1 is translocated by the T6SS. VgrG1 contains a vegetative insecticidal protein domain at its C-terminus with actin ADP-ribosyltransferase activity. This effector alters the actin cytoskeleton and induces apoptosis in epithelial cells (Suarez et al., 2010).
Yersinia pseudotuberculosis is an enteric pathogen, which usually grows in the environment and can be transmitted to mammalian hosts through ingestion of contaminated food or water. It typically causes a broad range of gastrointestinal diseases, from enteritis to mesenteric lymphadenitis (Yang et al., 2018). Y. pseudotuberculosis contains four T6SS clusters. The T6SS-4 secreted substrate YezP (Yersinia extracellular zinc-binding protein) is a Zn2+-binding protein that has the ability to rescue the sensitivity to oxidative stress exhibited by T6SS mutants when added to extracellular milieu. YezP plays a role in virulence for mice but its contribution to the infection process requires additional investigation (Wang et al., 2015).
Bacterial pathogens employ many strategies to invade mammalian hosts, damage tissues, organelles and prevent the immune system from responding. One strategy is the secretion of proteins (effectors) across membranes. As we described in this review, these toxins are secreted and injected into host cells via the T6SS and exist both as evolved VgrGs and cargo effectors. Translocated effectors can play many roles in eukaryotic cells, which promote bacterial virulence ranging from attachment to directly intoxicating target cells and disrupting their functions to finally establishing a replicative niche and successful colonization.
The clearance of pathogens depends on the host innate immune responses that take place at early stages of infection and in which macrophages and neutrophils are the essential players. Once inside the macrophage, intracellular bacteria can reside in vacuoles or in the cytosol, depending on their effector repertoire which help them to evade host defense and continue the infection cycle and replicate. Here, we described 27 T6SS effectors employed by several bacterial species to promote virulence in eukaryotic cells. These effectors can display similar or complementary functions into host cells and modulate the same central pathway of the host cell (e.g., inflammasome) or having different roles. Moreover, a pathogen may secrete several proteins to produce the same outcome (e.g., PldA and PldB).
Vibrio, Pseudomonas, Burkholderia and Aeromonas species translocate toxins in a T6SS-dependent manner leading to resistance to phagocytosis, inflammasome activation, as well as bacterial internalization by manipulating the actin cytoskeleton. Vibrio, Pseudomonas, and Burkholderia in particular, disrupt the host cell cytoskeleton, targeting actin, although Pseudomonas and Francisella effectors target the microtubules. The interference with immunity pathways is a hallmark function achieved by T6SS-dependent effectors. Burkholderia, Vibrio, Francisella and Edwardsiella inject toxins involved in the activation of the different inflammasomes that lead to the secretion of proinflammatory cytokines. In this context, activation of the inflammasome can be important for the clearance of the pathogen, suggesting the possibility that the T6SS effectors may also have a role as anti-virulence factors. This notion is supported from results using TecA deficient mutants in experimental mice infection whereby the mutant bacteria were able to kill infected mice while the parental strain was cleared. This clearance effect was abolished in infections using Pyrin inflammasome-defective mice (Aubert et al., 2016).
Another important mechanism of host defense is the generation of reactive oxygen species to eradicate intracellular bacteria. E. coli and Yersinia T6SSs deliver effectors with the ability to modulate the oxidative stress and protect the pathogen from ROS and allowing growth. Finally, Serratia delivers effectors into fungal cells, causing depolarization of the plasma membrane and metabolism disrupted, leading to cell death.
In recent years, remarkable progress has been made toward elucidating the function of eukaryotic effectors of the T6SS, which has contributed to better understand several aspects of bacterial pathogenesis. However, our understanding of the molecular mechanism of many T6SS-secreted toxins awaits detailed functional analysis, including biochemical, biophysical, immunological and structural studies. The kinetics of effector delivery is also an open question since very little is known on whether their translocation is regulated in a temporal and spatial manner and the signals that triggers their secretion.
JM and MV: wrote the manuscript.
MV acknowledges funding from the Medical Research Council (MR/P022480/1).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The authors would like to thank Dr. Keren Turton for critical review of this manuscript.
Alcoforado Diniz, J., Liu, Y. C., and Coulthurst, S. J. (2015). Molecular weaponry: diverse effectors delivered by the Type VI secretion system. Cell. Microbiol. 17, 1742–1751. doi: 10.1111/cmi.12532
Aschtgen, M. S., Bernard, C. S., De Bentzmann, S., Lloubes, R., and Cascales, E. (2008). SciN is an outer membrane lipoprotein required for type VI secretion in enteroaggregative Escherichia coli. J. Bacteriol. 190, 7523–7531. doi: 10.1128/JB.00945-08
Aubert, D. F., Xu, H., Yang, J., Shi, X., Gao, W., Li, L., et al. (2016). A burkholderia type VI effector deamidates Rho GTPases to activate the pyrin inflammasome and trigger inflammation. Cell Host Microbe 19, 664–674. doi: 10.1016/j.chom.2016.04.004
Ballister, E. R., Lai, A. H., Zuckermann, R. N., Cheng, Y., and Mougous, J. D. (2008). In vitro self-assembly of tailorable nanotubes from a simple protein building block. Proc. Natl. Acad. Sci. U.S.A. 105, 3733–3738. doi: 10.1073/pnas.0712247105
Basler, M. (2015). Type VI secretion system: secretion by a contractile nanomachine. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 370. doi: 10.1098/rstb.2015.0021
Basler, M., Pilhofer, M., Henderson, G. P., Jensen, G. J., and Mekalanos, J. J. (2012). Type VI secretion requires a dynamic contractile phage tail-like structure. Nature 483, 182–186. doi: 10.1038/nature10846
Bingle, L. E., Bailey, C. M., and Pallen, M. J. (2008). Type VI secretion: a beginner's guide. Curr. Opin. Microbiol. 11, 3–8. doi: 10.1016/j.mib.2008.01.006
Bleves, S., Sana, T. G., and Voulhoux, R. (2014). The target cell genus does not matter. Trends Microbiol. 22, 304–306. doi: 10.1016/j.tim.2014.04.011
Bondage, D. D., Lin, J. S., Ma, L. S., Kuo, C. H., and Lai, E. M. (2016). VgrG C terminus confers the type VI effector transport specificity and is required for binding with PAAR and adaptor-effector complex. Proc. Natl. Acad. Sci. U.S.A. 113, E3931–E3940. doi: 10.1073/pnas.1600428113
Bonemann, G., Pietrosiuk, A., Diemand, A., Zentgraf, H., and Mogk, A. (2009). Remodelling of VipA/VipB tubules by ClpV-mediated threading is crucial for type VI protein secretion. EMBO J. 28, 315–325. doi: 10.1038/emboj.2008.269
Boulant, T., Boudehen, Y. M., Filloux, A., Plesiat, P., Naas, T., and Dortet, L. (2018). Higher prevalence of PldA, a Pseudomonas aeruginosa trans-kingdom H2-type VI secretion system effector, in clinical isolates responsible for acute infections and in multidrug resistant strains. Front. Microbiol 9:2578. doi: 10.3389/fmicb.2018.02578
Boyer, F., Fichant, G., Berthod, J., Vandenbrouck, Y., and Attree, I. (2009). Dissecting the bacterial type VI secretion system by a genome wide in silico analysis: what can be learned from available microbial genomic resources? BMC Genomics 10:104. doi: 10.1186/1471-2164-10-104
Brodmann, M., Dreier, R. F., Broz, P., and Basler, M. (2017). Francisella requires dynamic type VI secretion system and ClpB to deliver effectors for phagosomal escape. Nat. Commun 8:15853. doi: 10.1038/ncomms15853
Broms, J. E., Ishikawa, T., Wai, S. N., and Sjostedt, A. (2013). A functional VipA-VipB interaction is required for the type VI secretion system activity of Vibrio cholerae O1 strain A1552. BMC Microbiol. 13:96. doi: 10.1186/1471-2180-13-96
Broms, J. E., Meyer, L., Sun, K., Lavander, M., and Sjostedt, A. (2012). Unique substrates secreted by the type VI secretion system of Francisella tularensis during intramacrophage infection. PLoS ONE 7:e50473. doi: 10.1371/journal.pone.0050473
Brunet, Y. R., Henin, J., Celia, H., and Cascales, E. (2014). Type VI secretion and bacteriophage tail tubes share a common assembly pathway. EMBO Rep. 15, 315–321. doi: 10.1002/embr.201337936
Brunet, Y. R., Zoued, A., Boyer, F., Douzi, B., and Cascales, E. (2015). The type VI secretion TssEFGK-VgrG phage-like baseplate is recruited to the TssJLM membrane complex via multiple contacts and serves as assembly platform for tail tube/sheath polymerization. PLoS Genet. 11:e1005545. doi: 10.1371/journal.pgen.1005545
Cantlay, S., Haggerty, K., and Horzempa, J. (2020). OpiA, a type six secretion system substrate, localizes to the cell pole and plays a role in bacterial growth and viability in francisella tularensis LVS. J. Bacteriol. 202:e00048-20. doi: 10.1128/JB.00048-20
Chen, H., Yang, D., Han, F., Tan, J., Zhang, L., Xiao, J., et al. (2017). The bacterial T6SS effector EvpP prevents NLRP3 inflammasome activation by inhibiting the Ca(2+)-dependent MAPK-Jnk pathway. Cell Host Microbe 21, 47–58. doi: 10.1016/j.chom.2016.12.004
Cherrak, Y., Flaugnatti, N., Durand, E., Journet, L., and Cascales, E. (2019). “Structure and activity of the type VI secretion system,” in Protein Secretion in Bacteria, eds M. Sandkvist, E. Cascales, and P. J. Christie. 7. doi: 10.1128/9781683670285.ch26
Cherrak, Y., Rapisarda, C., Pellarin, R., Bouvier, G., Bardiaux, B., Allain, F., et al. (2018). Biogenesis and structure of a type VI secretion baseplate. Nat. Microbiol. 3, 1404–1416. doi: 10.1038/s41564-018-0260-1
Cianfanelli, F. R., Alcoforado Diniz, J., Guo, M., de Cesare, V., Trost, M., and Coulthurst, S. J. (2016a). VgrG and PAAR proteins define distinct versions of a functional type VI secretion system. PLoS Pathog. 12:e1005735. doi: 10.1371/journal.ppat.1005735
Cianfanelli, F. R., Monlezun, L., and Coulthurst, S. J. (2016b). Aim, load, fire: the type VI secretion system, a bacterial nanoweapon. Trends Microbiol. 24, 51–62. doi: 10.1016/j.tim.2015.10.005
Dong, T. G., Ho, B. T., Yoder-Himes, D. R., and Mekalanos, J. J. (2013). Identification of T6SS-dependent effector and immunity proteins by Tn-seq in Vibrio cholerae. Proc. Natl. Acad. Sci. U.S.A. 110, 2623–2628. doi: 10.1073/pnas.1222783110
Douzi, B., Spinelli, S., Blangy, S., Roussel, A., Durand, E., Brunet, Y. R., et al. (2014). Crystal structure and self-interaction of the type VI secretion tail-tube protein from enteroaggregative Escherichia coli. PLoS ONE 9:e86918. doi: 10.1371/journal.pone.0086918
Durand, E., Cambillau, C., Cascales, E., and Journet, L. (2014). VgrG, Tae, Tle, and beyond: the versatile arsenal of Type VI secretion effectors. Trends Microbiol. 22, 498–507. doi: 10.1016/j.tim.2014.06.004
Durand, E., Derrez, E., Audoly, G., Spinelli, S., Ortiz-Lombardia, M., Raoult, D., et al. (2012). Crystal structure of the VgrG1 actin cross-linking domain of the Vibrio cholerae type VI secretion system. J. Biol. Chem. 287, 38190–38199. doi: 10.1074/jbc.M112.390153
Durand, E., Nguyen, V. S., Zoued, A., Logger, L., Pehau-Arnaudet, G., Aschtgen, M. S., et al. (2015). Biogenesis and structure of a type VI secretion membrane core complex. Nature 523, 555–560. doi: 10.1038/nature14667
Dutta, P., Jijumon, A. S., Mazumder, M., Dileep, D., Mukhopadhyay, A. K., Gourinath, S., et al. (2019). Presence of actin binding motif in VgrG-1 toxin of Vibrio cholerae reveals the molecular mechanism of actin cross-linking. Int. J. Biol. Macromol. 133, 775–785. doi: 10.1016/j.ijbiomac.2019.04.026
Eshraghi, A., Kim, J., Walls, A. C., Ledvina, H. E., Miller, C. N., Ramsey, K. M., et al. (2016). Secreted effectors encoded within and outside of the Francisella pathogenicity island promote intramacrophage growth. Cell Host Microbe 20, 573–583. doi: 10.1016/j.chom.2016.10.008
Flaugnatti, N., Le, T. T., Canaan, S., Aschtgen, M. S., Nguyen, V. S., Blangy, S., et al. (2016). A phospholipase A1 antibacterial Type VI secretion effector interacts directly with the C-terminal domain of the VgrG spike protein for delivery. Mol. Microbiol. 99, 1099–1118. doi: 10.1111/mmi.13292
Hachani, A., Wood, T. E., and Filloux, A. (2016). Type VI secretion and anti-host effectors. Curr. Opin. Microbiol. 29, 81–93. doi: 10.1016/j.mib.2015.11.006
Heisler, D. B., Kudryashova, E., Grinevich, D. O., Suarez, C., Winkelman, J. D., Birukov, K. G., et al. (2015). Actin-directed toxin. ACD toxin-produced actin oligomers poison formin-controlled actin polymerization. Science 349, 535–539. doi: 10.1126/science.aab4090
Hernandez, R. E., Gallegos-Monterrosa, R., and Coulthurst, S. J. (2020). Type VI secretion system effector proteins: effective weapons for bacterial competitiveness. Cell. Microbiol. 22, e13241. doi: 10.1111/cmi.13241
Hu, W., Anand, G., Sivaraman, J., Leung, K. Y., and Mok, Y. K. (2014). A disordered region in the EvpP protein from the type VI secretion system of Edwardsiella tarda is essential for EvpC binding. PLoS ONE 9:e110810. doi: 10.1371/journal.pone.0110810
Jana, B., and Salomon, D. (2019). Type VI secretion system: a modular toolkit for bacterial dominance. Future Microbiol. 14, 1451–1463. doi: 10.2217/fmb-2019-0194
Jiang, F., Wang, X., Wang, B., Chen, L., Zhao, Z., Waterfield, N. R., et al. (2016). The Pseudomonas aeruginosa type VI secretion PGAP1-like effector induces host autophagy by activating endoplasmic reticulum stress. Cell Rep. 16, 1502–1509. doi: 10.1016/j.celrep.2016.07.012
Jiang, F., Waterfield, N. R., Yang, J., Yang, G., and Jin, Q. (2014). A Pseudomonas aeruginosa type VI secretion phospholipase D effector targets both prokaryotic and eukaryotic cells. Cell Host Microbe 15, 600–610. doi: 10.1016/j.chom.2014.04.010
Kalindamar, S., Kordon, A. O., Abdelhamed, H., Tan, W., Pinchuk, L. M., and Karsi, A. (2020). Edwardsiella ictaluri evpP is required for colonisation of channel catfish ovary cells and necrosis in anterior kidney macrophages. Cell. Microbiol. 22:e13135. doi: 10.1111/cmi.13135
Kube, S., Kapitein, N., Zimniak, T., Herzog, F., Mogk, A., and Wendler, P. (2014). Structure of the VipA/B type VI secretion complex suggests a contraction-state-specific recycling mechanism. Cell Rep. 8, 20–30. doi: 10.1016/j.celrep.2014.05.034
Ledvina, H. E., Kelly, K. A., Eshraghi, A., Plemel, R. L., Peterson, S. B., Lee, B., et al. (2018). A phosphatidylinositol 3-kinase effector alters phagosomal maturation to promote intracellular growth of francisella. Cell Host Microbe 24, 285–295.e288. doi: 10.1016/j.chom.2018.07.003
Lery, L. M., Frangeul, L., Tomas, A., Passet, V., Almeida, A. S., Bialek-Davenet, S., et al. (2014). Comparative analysis of Klebsiella pneumoniae genomes identifies a phospholipase D family protein as a novel virulence factor. BMC Biol 12:41. doi: 10.1186/1741-7007-12-41
Leung, K. Y., Wang, Q., Yang, Z., and Siame, B. A. (2019). Edwardsiella piscicida: a versatile emerging pathogen of fish. Virulence 10, 555–567. doi: 10.1080/21505594.2019.1621648
Liang, X., Kamal, F., Pei, T. T., Xu, P., Mekalanos, J. J., and Dong, T. G. (2019). An onboard checking mechanism ensures effector delivery of the type VI secretion system in Vibrio cholerae. Proc. Natl. Acad. Sci. U. S. A. 116, 23292–23298. doi: 10.1073/pnas.1914202116
Liang, X., Moore, R., Wilton, M., Wong, M. J., Lam, L., and Dong, T. G. (2015). Identification of divergent type VI secretion effectors using a conserved chaperone domain. Proc. Natl. Acad. Sci. U.S.A. 112, 9106–9111. doi: 10.1073/pnas.1505317112
Lindgren, M., Broms, J. E., Meyer, L., Golovliov, I., and Sjostedt, A. (2013a). The Francisella tularensis LVS DeltapdpC mutant exhibits a unique phenotype during intracellular infection. BMC Microbiol 13:20. doi: 10.1186/1471-2180-13-20
Lindgren, M., Eneslatt, K., Broms, J. E., and Sjostedt, A. (2013b). Importance of PdpC, IglC, IglI, and IglG for modulation of a host cell death pathway induced by Francisella tularensis. Infect. Immun. 81, 2076–2084. doi: 10.1128/IAI.00275-13
Logger, L., Aschtgen, M. S., Guerin, M., Cascales, E., and Durand, E. (2016). Molecular dissection of the interface between the type VI secretion TssM cytoplasmic domain and the TssG baseplate component. J. Mol. Biol. 428, 4424–4437. doi: 10.1016/j.jmb.2016.08.032
Long, M. E., Lindemann, S. R., Rasmussen, J. A., Jones, B. D., and Allen, L. A. (2013). Disruption of Francisella tularensis Schu S4 iglI, iglJ, and pdpC genes results in attenuation for growth in human macrophages and in vivo virulence in mice and reveals a unique phenotype for pdpC. Infect. Immun. 81, 850–861. doi: 10.1128/IAI.00822-12
Loutet, S. A., and Valvano, M. A. (2010). A decade of Burkholderia cenocepacia virulence determinant research. Infect. Immun. 78, 4088–4100. doi: 10.1128/IAI.00212-10
Ludu, J. S., de Bruin, O. M., Duplantis, B. N., Schmerk, C. L., Chou, A. Y., Elkins, K. L., et al. (2008). The Francisella pathogenicity island protein PdpD is required for full virulence and associates with homologues of the type VI secretion system. J. Bacteriol. 190, 4584–4595. doi: 10.1128/JB.00198-08
Ma, A. T., McAuley, S., Pukatzki, S., and Mekalanos, J. J. (2009a). Translocation of a Vibrio cholerae type VI secretion effector requires bacterial endocytosis by host cells. Cell Host Microbe 5, 234–243. doi: 10.1016/j.chom.2009.02.005
Ma, A. T., and Mekalanos, J. J. (2010). In vivo actin cross-linking induced by Vibrio cholerae type VI secretion system is associated with intestinal inflammation. Proc. Natl. Acad. Sci. U.S.A. 107, 4365–4370. doi: 10.1073/pnas.0915156107
Ma, J., Pan, Z., Huang, J., Sun, M., Lu, C., and Yao, H. (2017). The Hcp proteins fused with diverse extended-toxin domains represent a novel pattern of antibacterial effectors in type VI secretion systems. Virulence 8, 1189–1202. doi: 10.1080/21505594.2017.1279374
Ma, L. S., Lin, J. S., and Lai, E. M. (2009b). An IcmF family protein, ImpLM, is an integral inner membrane protein interacting with ImpKL, and its walker a motif is required for type VI secretion system-mediated Hcp secretion in Agrobacterium tumefaciens. J. Bacteriol. 191, 4316–4329. doi: 10.1128/JB.00029-09
Miyata, S. T., Kitaoka, M., Brooks, T. M., McAuley, S. B., and Pukatzki, S. (2011). Vibrio cholerae requires the type VI secretion system virulence factor VasX to kill dictyostelium discoideum. Infect. Immun. 79, 2941–2949. doi: 10.1128/IAI.01266-10
Mougous, J. D., Cuff, M. E., Raunser, S., Shen, A., Zhou, M., Gifford, C. A., et al. (2006). A virulence locus of Pseudomonas aeruginosa encodes a protein secretion apparatus. Science 312, 1526–1530. doi: 10.1126/science.1128393
Navarro-Garcia, F., Ruiz-Perez, F., Cataldi, A., and Larzabal, M. (2019). Type VI secretion system in pathogenic Escherichia coli: structure, role in virulence, and acquisition. Front. Microbiol. 10:1965. doi: 10.3389/fmicb.2019.01965
Nazarov, S., Schneider, J. P., Brackmann, M., Goldie, K. N., Stahlberg, H., and Basler, M. (2018). Cryo-EM reconstruction of type VI secretion system baseplate and sheath distal end. EMBO J. 37:e97103. doi: 10.15252/embj.201797103
Nguyen, V. S., Douzi, B., Durand, E., Roussel, A., Cascales, E., and Cambillau, C. (2018). Towards a complete structural deciphering of Type VI secretion system. Curr. Opin. Struct. Biol. 49, 77–84. doi: 10.1016/j.sbi.2018.01.007
Ozanic, M., Marecic, V., Lindgren, M., Sjostedt, A., and Santic, M. (2016). Phenotypic characterization of the Francisella tularensis DeltapdpC and DeltaiglG mutants. Microbes Infect. 18, 768–776. doi: 10.1016/j.micinf.2016.07.006
Pissaridou, P., Allsopp, L. P., Wettstadt, S., Howard, S. A., Mavridou, D. A. I., and Filloux, A. (2018). The Pseudomonas aeruginosa T6SS-VgrG1b spike is topped by a PAAR protein eliciting DNA damage to bacterial competitors. Proc. Natl. Acad. Sci. U.S.A. 115, 12519–12524. doi: 10.1073/pnas.1814181115
Pukatzki, S., Ma, A. T., Revel, A. T., Sturtevant, D., and Mekalanos, J. J. (2007). Type VI secretion system translocates a phage tail spike-like protein into target cells where it cross-links actin. Proc. Natl. Acad. Sci. U.S.A. 104, 15508–15513. doi: 10.1073/pnas.0706532104
Qin, L., Wang, X., Gao, Y., Bi, K., and Wang, W. (2020). Roles of EvpP in Edwardsiella piscicida-macrophage interactions. Front. Cell. Infect. Microbiol. 10:53. doi: 10.3389/fcimb.2020.00053
Quentin, D., Ahmad, S., Shanthamoorthy, P., Mougous, J. D., Whitney, J. C., and Raunser, S. (2018). Mechanism of loading and translocation of type VI secretion system effector Tse6. Nat. Microbiol. 3, 1142–1152. doi: 10.1038/s41564-018-0238-z
Rapisarda, C., Cherrak, Y., Kooger, R., Schmidt, V., Pellarin, R., Logger, L., et al. (2019). In situ and high-resolution cryo-EM structure of a bacterial type VI secretion system membrane complex. EMBO J. 38:e100886. doi: 10.15252/embj.2018100886
Ray, A., Schwartz, N., de Souza Santos, M., Zhang, J., Orth, K., and Salomon, D. (2017). Type VI secretion system MIX-effectors carry both antibacterial and anti-eukaryotic activities. EMBO Rep. 18, 1978–1990. doi: 10.15252/embr.201744226
Records, A. R. (2011). The type VI secretion system: a multipurpose delivery system with a phage-like machinery. Mol. Plant Microbe Interact. 24, 751–757. doi: 10.1094/MPMI-11-10-0262
Renault, M. G., Zamarreno Beas, J., Douzi, B., Chabalier, M., Zoued, A., Brunet, Y. R., et al. (2018). The gp27-like hub of VgrG serves as adaptor to promote hcp tube assembly. J. Mol. Biol. 430, 3143–3156. doi: 10.1016/j.jmb.2018.07.018
Russell, A. B., LeRoux, M., Hathazi, K., Agnello, D. M., Ishikawa, T., Wiggins, P. A., et al. (2013). Diverse type VI secretion phospholipases are functionally plastic antibacterial effectors. Nature 496, 508–512. doi: 10.1038/nature12074
Salomon, D., Kinch, L. N., Trudgian, D. C., Guo, X., Klimko, J. A., Grishin, N. V., et al. (2014). Marker for type VI secretion system effectors. Proc. Natl. Acad. Sci. U.S.A. 111, 9271–9276. doi: 10.1073/pnas.1406110111
Sana, T. G., Baumann, C., Merdes, A., Soscia, C., Rattei, T., Hachani, A., et al. (2015). Internalization of Pseudomonas aeruginosa strain PAO1 into epithelial cells is promoted by interaction of a T6SS effector with the microtubule network. MBio 6:e00712-15. doi: 10.1128/mBio.00712-15
Sana, T. G., Berni, B., and Bleves, S. (2016). The T6SSs of Pseudomonas aeruginosa strain PAO1 and their effectors: beyond bacterial-cell targeting. Front. Cell. Infect. Microbiol. 6:61. doi: 10.3389/fcimb.2016.00061
Sana, T. G., Hachani, A., Bucior, I., Soscia, C., Garvis, S., Termine, E., et al. (2012). The second type VI secretion system of Pseudomonas aeruginosa strain PAO1 is regulated by quorum sensing and Fur and modulates internalization in epithelial cells. J. Biol. Chem. 287, 27095–27105. doi: 10.1074/jbc.M112.376368
Santos, M. N. M., Cho, S. T., Wu, C. F., Chang, C. J., Kuo, C. H., and Lai, E. M. (2019). Redundancy and specificity of type VI secretion vgrG Loci in antibacterial activity of Agrobacterium tumefaciens 1D1609 strain. Front. Microbiol. 10:3004. doi: 10.1101/740209
Sarris, P. F., Zoumadakis, C., Panopoulos, N. J., and Scoulica, E. V. (2011). Distribution of the putative type VI secretion system core genes in Klebsiella spp. Infect. Genet. Evol. 11, 157–166. doi: 10.1016/j.meegid.2010.09.006
Satchell, K. J. (2009). Actin crosslinking toxins of gram-negative bacteria. Toxins 1, 123–133. doi: 10.3390/toxins1020123
Schwarz, S., Singh, P., Robertson, J. D., LeRoux, M., Skerrett, S. J., Goodlett, D. R., et al. (2014). VgrG-5 is a Burkholderia type VI secretion system-exported protein required for multinucleated giant cell formation and virulence. Infect. Immun. 82, 1445–1452. doi: 10.1128/IAI.01368-13
Shimizu, T., Otonari, S., Suzuki, J., Uda, A., Watanabe, K., and Watarai, M. (2019). Expression of Francisella pathogenicity island protein intracellular growth locus E (IglE) in mammalian cells is involved in intracellular trafficking, possibly through microtubule organizing center. Microbiologyopen 8:e00684. doi: 10.1002/mbo3.684
Shneider, M. M., Buth, S. A., Ho, B. T., Basler, M., Mekalanos, J. J., and Leiman, P. G. (2013). PAAR-repeat proteins sharpen and diversify the type VI secretion system spike. Nature 500, 350–353. doi: 10.1038/nature12453
Storey, D., McNally, A., Astrand, M., Sa-Pessoa Graca Santos, J., Rodriguez-Escudero, I., Elmore, B., et al. (2020). Klebsiella pneumoniae type VI secretion system-mediated microbial competition is PhoPQ controlled and reactive oxygen species dependent. PLoS Pathog. 16:e1007969. doi: 10.1371/journal.ppat.1007969
Suarez, G., Sierra, J. C., Erova, T. E., Sha, J., Horneman, A. J., and Chopra, A. K. (2010). A type VI secretion system effector protein, VgrG1, from Aeromonas hydrophila that induces host cell toxicity by ADP ribosylation of actin. J. Bacteriol. 192, 155–168. doi: 10.1128/JB.01260-09
Tan, J., Yang, D., Wang, Z., Zheng, X., Zhang, Y., and Liu, Q. (2019). EvpP inhibits neutrophils recruitment via Jnk-caspy inflammasome signaling in vivo. Fish Shellfish Immunol. 92, 851–860. doi: 10.1016/j.fsi.2019.05.051
Toesca, I. J., French, C. T., and Miller, J. F. (2014). The Type VI secretion system spike protein VgrG5 mediates membrane fusion during intercellular spread by pseudomallei group Burkholderia species. Infect. Immun. 82, 1436–1444. doi: 10.1128/IAI.01367-13
Trunk, K., Coulthurst, S. J., and Quinn, J. (2019). A new front in microbial warfare-delivery of antifungal effectors by the type VI secretion system. J. Fungi. 5:50. doi: 10.3390/jof5020050
Trunk, K., Peltier, J., Liu, Y. C., Dill, B. D., Walker, L., Gow, N. A. R., et al. (2018). The type VI secretion system deploys antifungal effectors against microbial competitors. Nat. Microbiol. 3, 920–931. doi: 10.1038/s41564-018-0191-x
Uda, A., Sekizuka, T., Tanabayashi, K., Fujita, O., Kuroda, M., Hotta, A., et al. (2014). Role of pathogenicity determinant protein C (PdpC) in determining the virulence of the Francisella tularensis subspecies tularensis SCHU. PLoS ONE 9:e89075. doi: 10.1371/journal.pone.0089075
Unterweger, D., Kostiuk, B., Otjengerdes, R., Wilton, A., Diaz-Satizabal, L., and Pukatzki, S. (2015). Chimeric adaptor proteins translocate diverse type VI secretion system effectors in Vibrio cholerae. EMBO J. 34, 2198–2210. doi: 10.15252/embj.201591163
Wan, B., Zhang, Q., Ni, J., Li, S., Wen, D., Li, J., et al. (2017). Type VI secretion system contributes to Enterohemorrhagic Escherichia coli virulence by secreting catalase against host reactive oxygen species (ROS). PLoS Pathog. 13:e1006246. doi: 10.1371/journal.ppat.1006246
Wang, T., Si, M., Song, Y., Zhu, W., Gao, F., Wang, Y., et al. (2015). Type VI secretion system transports Zn2+ to combat multiple stresses and host immunity. PLoS Pathog. 11:e1005020. doi: 10.1371/journal.ppat.1005020
Wang, X., Wang, Q., Xiao, J., Liu, Q., Wu, H., Xu, L., et al. (2009). Edwardsiella tarda T6SS component evpP is regulated by esrB and iron, and plays essential roles in the invasion of fish. Fish Shellfish Immunol. 27, 469–477. doi: 10.1016/j.fsi.2009.06.013
Wettstadt, S., Wood, T. E., Fecht, S., and Filloux, A. (2019). Delivery of the Pseudomonas aeruginosa phospholipase effectors PldA and PldB in a VgrG- and H2-T6SS-dependent manner. Front. Microbiol. 10:1718. doi: 10.3389/fmicb.2019.01718
Whitney, J. C., Beck, C. M., Goo, Y. A., Russell, A. B., Harding, B. N., De Leon, J. A., et al. (2014). Genetically distinct pathways guide effector export through the type VI secretion system. Mol. Microbiol. 92, 529–542. doi: 10.1111/mmi.12571
Wilderman, P. J., Vasil, A. I., Johnson, Z., and Vasil, M. L. (2001). Genetic and biochemical analyses of a eukaryotic-like phospholipase D of Pseudomonas aeruginosa suggest horizontal acquisition and a role for persistence in a chronic pulmonary infection model. Mol. Microbiol. 39, 291–303. doi: 10.1046/j.1365-2958.2001.02282.x
Wood, T. E., Howard, S. A., Forster, A., Nolan, L. M., Manoli, E., Bullen, N. P., et al. (2019). The Pseudomonas aeruginosa T6SS delivers a periplasmic toxin that disrupts bacterial cell morphology. Cell Rep. 29, 187–201.e187. doi: 10.1016/j.celrep.2019.08.094
Yang, X., Pan, J., Wang, Y., and Shen, X. (2018). Type VI secretion systems present new insights on pathogenic yersinia. Front. Cell. Infect. Microbiol. 8:260. doi: 10.3389/fcimb.2018.00260
Yin, M., Yan, Z., and Li, X. (2019). Architecture of type VI secretion system membrane core complex. Cell Res. 29, 251–253. doi: 10.1038/s41422-018-0130-7
Zhang, X. Y., Brunet, Y. R., Logger, L., Douzi, B., Cambillau, C., Journet, L., et al. (2013). Dissection of the TssB-TssC interface during type VI secretion sheath complex formation. PLoS ONE 8:e81074. doi: 10.1371/journal.pone.0081074
Zheng, J., Ho, B., and Mekalanos, J. J. (2011). Genetic analysis of anti-amoebae and anti-bacterial activities of the type VI secretion system in Vibrio cholerae. PLoS ONE 6:e23876. doi: 10.1371/journal.pone.0023876
Zheng, J., and Leung, K. Y. (2007). Dissection of a type VI secretion system in Edwardsiella tarda. Mol. Microbiol. 66, 1192–1206. doi: 10.1111/j.1365-2958.2007.05993.x
Zong, B., Zhang, Y., Wang, X., Liu, M., Zhang, T., Zhu, Y., et al. (2019). Characterization of multiple type-VI secretion system (T6SS) VgrG proteins in the pathogenicity and antibacterial activity of porcine extra-intestinal pathogenic Escherichia coli. Virulence 10, 118–132. doi: 10.1080/21505594.2019.1573491
Keywords: bacterial pathogenesis, secretion system, T6SS, host manipulation, effector
Citation: Monjarás Feria J and Valvano MA (2020) An Overview of Anti-Eukaryotic T6SS Effectors. Front. Cell. Infect. Microbiol. 10:584751. doi: 10.3389/fcimb.2020.584751
Received: 18 July 2020; Accepted: 22 September 2020;
Published: 19 October 2020.
Edited by:
Teresa Thurston, Imperial College London, United KingdomReviewed by:
Rajagopal Kammara, Central Food Technological Research Institute (CSIR), IndiaCopyright © 2020 Monjarás Feria and Valvano. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Miguel A. Valvano, bS52YWx2YW5vQHF1Yi5hYy51aw==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.