- 1Henan Key Laboratory of Ion-Beam Bioengineering, School of Agricultural Sciences, Zhengzhou University, Zhengzhou, China
- 2The Third Affiliated Hospital Xinxiang Medical University, Xinxiang, China
- 3School of Food Science and Technology, Henan University of Technology, Zhengzhou, China
- 4School of Food and Biological Engineering, Henan University of Animal Husbandry and Economy, Zhengzhou, China
We investigated the in vitro effects of Lactobacillus casei ZX633 on gut microorganism composition in infants with diarrhea. For this purpose, 103 feces samples from healthy infants (healthy group) and 300 diarrhea samples from infants (diarrhea group) were collected, and diarrhea feces were treated with L. casei ZX633, which was previously isolated from healthy infant feces (treatment group). We used microbial dilution plate methods, high performance liquid chromatography (HPLC) and high-throughput sequencing approaches to analyze viable main microorganism counts, short chain fatty acid (SCFA) concentrations, and intestinal microbiota composition in feces, respectively. Our data showed that L. casei ZX633 supplementation increased the numbers of Escherichia coli, yeasts, lactic acid bacteria (LAB) and aerobic-bacteria, raised propionic acid levels but reduced four other SCFAs, which are close to the healthy group. Alpha diversity results indicated that microbial diversity and richness decreased in treatment group. Bacterial community analyses revealed that microbial structures of the treatment group tended toward the healthy group; i.e., Escherichia-Shigella and Clostridioides abundance increased, and there was a reduction in the abundance of Streptococcus, Bacteroides, Enterococcus and Veillonella. In conclusion, L. casei ZX633 isolated from healthy infant feces, may be effective in improving infant diarrhea microbiota, potentially providing a new probiotic strain to reduce the incidence of diarrhea associated with bacterial disease in infants.
Introduction
Diarrhea is defined as the passage of three or more loose liquid stools per day, whereas a diarrheal duration of 14 days constitutes acute or persistent diarrhea (Walker et al., 2013). This condition is an enormous health problem for children under 5 years old, and globally, is related to significant morbidity and mortality rates (Wen et al., 2018). Annually, there are approximately 1.7 billon cases of childhood diarrhea disease, with 760,000 deaths, thus representing the second leading cause of death in children (Gallardo et al., 2017).
Diarrhea arises from infectious diarrhea due to bacterial, viral and parasitic organisms, and non-infectious diarrhea, derived from malnutrition and food allergies (Shin and Shin, 2018). Escherichia coli, Salmonella enteritidis, Shigella, and Staphylococcus aureus are the main common pathogens that cause bacterial diarrhea (Wang et al., 2018). Generally, the bacterial condition is treated with antibiotics, however, antibiotic use is considered a double-edged sword, as adverse effects can become evident with antibiotic usage, especially antibiotic resistance. Previous study has shown that antibiotic misuse is a severe problem, causing compositional and richness changes in intestinal microbiota, potentially detrimental to host health (Pilla et al., 2019).
Increasingly, interest has turned toward natural alternatives, as a result of antibiotic resistant effects and restrictions on the uncontrolled use of antibiotics. One alternative treatment for bacterial diarrhea are probiotics, natural and live microorganisms which when administered in adequate amounts confer a health benefit on the host (Hill et al., 2014). They improve the intestinal microbiota balance by competitive inhibition of the colonization of pathogenic bacteria in the intestinal tract (Bubnov et al., 2015; Bautista-Gallego et al., 2019; Wongsen et al., 2019). In addition, they also have beneficial effects on the gut immune system (Bubnov et al., 2015). Current evidence suggests that some probiotics such as Lactobacillus and Bifidobacteria modulate gut microflora homeostasis, and may exert protective effects against diarrhea (Kwok et al., 2014, 2015). Lactobacillus rhamnosus GG notably reduced diarrheal duration in children upon receiving a dosage no <1010 colony-forming units (CFU) per day (Li et al., 2019), whereas Lactobacillus casei increased Bifidobacteria and Lactobacillus counts in stool and improved diarrhea in infants (Lai et al., 2019). In addition, lactic acid bacteria (LAB) from fecal origins have been used as potential probiotics for diarrhea, and are considered promising since these bacteria have long time associations with humans (Gheziel et al., 2019). Therefore, a LAB strain was isolated from healthy infant feces and used as a reagent in this study.
The main study aim was to investigate whether L. casei ZX633 supplementation improved diarrhea in infants. Using in vitro approaches, we assessed differences in fecal microflora in healthy, diarrhea and diarrhea feces supplemented with L. casei ZX633. Our data may provide a new probiotic strain for treating diarrhea associated with bacterial disease.
Materials and Methods
Sample Collection
All experimental procedures in this study were approved by the Institutional Ethics Committee of Zhengzhou University. Prior to the collection of the children's stool and data, consent was given by their parents. A total of 403 fecal samples (300 diarrhea fecal samples and 103 healthy fecal samples) were collected from infants <5 years old at the Third Affiliated Hospital of Xinxiang Medical University, China. Before the study, we selected diarrhea fecal samples with high leukocyte counts which reflect a bacterial infection. Samples were kept at 4°C, and transported under the same conditions to Zhengzhou University, where they were stored at −80°C until required.
Bacterial Strains and Treatment of Diarrhea Stool Samples
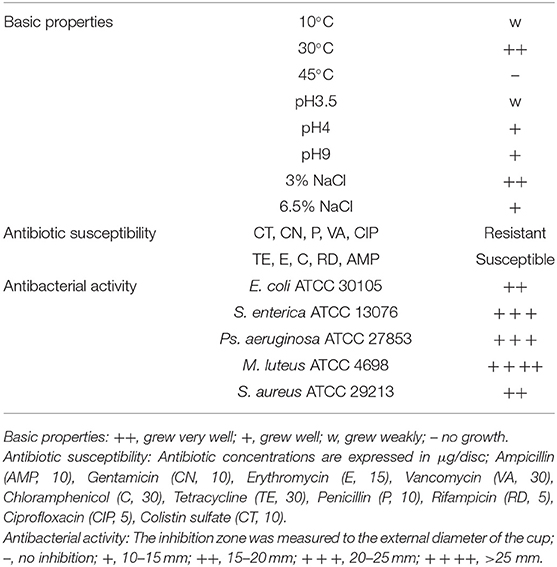
L. casei ZX633 was isolated from the feces of a healthy infant with strong antimicrobial activities, which had reported in our previous research (Wang et al., 2020) (Table 1). The strain was cultured on Man Rogosa Sharpe agar (MRS, Merck, Darmstadt, Germany) at 30°C, under anaerobic conditions for 48 h. Diarrhea samples (1 g) were homogenized in 200 μl L. casei ZX633 fermentation broth (previously cultured in MRS broth at 30°C for ~8 h), using a vortex mixer to reach a LAB viable cell concentration of approximately 105 CFU/g. The mixture was then incubated at 37°C for 48 h (Treatment).
Isolation and Identification of LAB Strains
Samples were homogenized in sterile water using a vortex mixer. After this, 10-fold, 103-fold, and 105-fold dilutions were made and plated onto MRS agar, then grown for 48 h at 30°C under anaerobic conditions. One representative colony of each morphology type (different size, shape and/or color) was picked and streaked onto fresh MRS agar. Pure cultures, with catalase negative activity, were identified by genetic analysis using PCR and 16S rRNA sequencing. The universal PCR primers, 27F (5′-AGAGTTTGATCCTGGCTCAG-3′) and 1492R (5′-GGTTACCTTGTTACGACTT-3′) (Zhang et al., 2016) were used to amplify the 16S rRNA gene. PCR parameters consisted of an initial heating at 95°C for 3 min; followed by 35 cycles of 15 s at 95°C, 15 s at 60°C and 90 s at 72°C, and a final extension at 72°C for 5 min. PCR products were sequenced by the Huada Biotech Company (Zhengzhou, China), and compared to strains in the GenBank database using BLAST, on the NCBI website (https://blast.ncbi.nlm.nih.gov/Blast.cgi).
Microbial Counting
Microbial communities were counted by the dilution plate procedure, according to Zhang et al. (2017). Samples (0.1 g) were suspended in 900 μl sterile water and mixed. Ten-fold and serial dilutions (10−2 to 10−5) were then prepared in sterile water. Then, 20 μl of each dilution dilutions was spread onto various selection agar plates.
Bacterial colony numbers were counted, based on different selection media and colony appearance. LAB colonies were counted on MRS agar after incubation at 30°C for 48 h, under anaerobic conditions. Aerobic bacteria, yeasts and E. coli were cultured on nutrient agar (NA), potato dextrose agar (PDA) with 10% dihydroxysuccinic acid solution (final concentration 1.5%), and eosin-methylene blue medium (EMB) at 37°C for 48 h, respectively. Separately, 10−1 and 10−2 dilutions were spread onto NA and Clostridium difficile agar (CLO) to ascertain Bacillus subtilis (37°C for 48 h) and C. difficile (37°C for 48 h under anaerobic conditions) growth, after dilutions were incubated for 15 min at 75°C. Colonies were counted, and the logarithmic numbers of viable colony-forming units in fecal samples (log CFU/g) were calculated. Three replicates were analyzed.
Quantification of Short Chain Fatty Acid (SCFA) in Fecal Samples
SCFAs including lactic, formic, acetic, propanoic and butyric acid were measured by high performance liquid chromatography (HPLC), using a Waters Alliance series 2695 (Zhang et al., 2018). Fecal samples were centrifuged (6,000 rpm for 10 min) and supernatants were filtered through a 0.22 μm pore-size filter (Jin Teng, Zhengzhou, China) prior to analysis. HPLC conditions were: a Carbomix H-NP10 column (7.8 × 300 mm); mobile phase, 2.5 mmol/l H2SO4; flow rate, 0.6 ml/min; temperature: 55°C. SCFA levels were determined from the area under the peak, detected at 240 nm using a calibration curve.
DNA Extraction and PCR Amplification
Total DNA was extracted from fecal samples using the bacterial DNA Kit (D3350-02, Omega Biotek, Norcross, GA, USA). After extraction, DNA integrity and quality was checked by 1% agarose gel electrophoresis and a NanoDrop 2000 UV-Vis spectrophotometer (Thermo Scientific, Wilmington, USA). PCR reaction were performed in 20 μl volumes, containing 4 μl Fast pfu buffer, 2 μl 2.5 mM dNTPs, 0.8 μl each primer (5 μM), 0.4 μl Fast pfu polymerase, 0.2 μl bovine serum albumin (BSA) and 10 ng template DNA. The V3–V4 regions of bacterial 16S rDNA was amplified with using the primers, 338F (5′-ACTCCTACGGGAGGCAGCAG-3′) and 806R (5′-GGACTACHVGGGTWTCTAAT-3′) (Yang et al., 2019). PCR parameters were 3 min denaturation at 95°C, 29 cycles of 30 s at 95°C, 30 s annealing at 53°C, 45 s elongation at 72°C and a final extension at 72°C for 10 min.
Illumina MiSeq Sequencing
PCR products were sequenced on an Illumina MiSeq PE300 platform (Shanghai Majorbio Bio-pharm Technology Co. Ltd., China), which measured diversity and bacterial composition in infant feces.
Statistical Analysis
LAB and microbial communities, and SCFA levels were analyzed using IBM SPSS Statistics 25.0 and Origin 2017. A one-way analysis of variance (ANOVA) was used to compare the means with a significant difference at p < 0.05. Data from high throughput sequencing were analyzed on the free online platform, Majorbio Cloud Platform (http://www.majorbio.com). Sequences were de-multiplexed and quality-filtered using the QIIME (version 1.9.1 http://qiime.org/install/index.html), and the obtained high quality reads were clustered into Operational taxonomic units (OTUs) using UPARSE (version 7.0 http://www.drive5.com/uparse/), at 97% similarity. Chao, Ace, Shannon, Simpson index and Good's coverage were computed using Mothur (version 1.30.2 https://www.mothur.org/wiki/Download_mothur) for analysis of alpha-diversities. The similarity and differences between samples were compared using the shared and unique OTUs of the Venn diagram. The taxonomy of each 16S rRNA gene sequence was analyzed by Ribosome Database Project (RDP) Classifier (version 2.11 http://rdp.cme.msu.edu/) against the Silva (SSU 132) 16S rRNA database. Spearman correlations were used to analyze the SCFAs and bacterial relationships using the pheatmap package.
Results
LAB Strain Differences Between Diarrhea and Healthy Fecal Samples
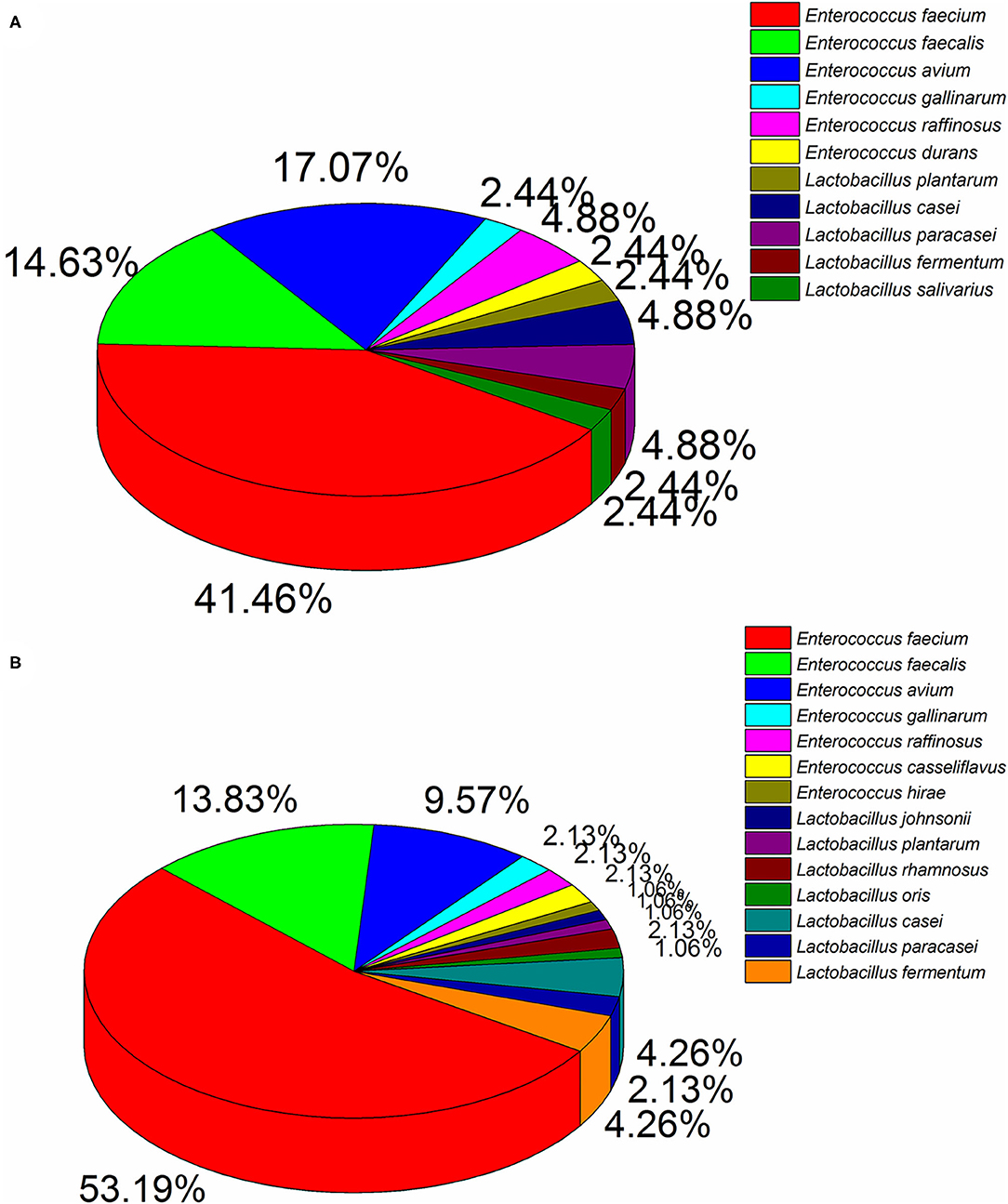
Ninety four and 41 LAB strains were isolated from diarrhea and healthy infant fecal samples, respectively. According to 16S rDNA sequencing analyses (Figure 1), Enterococcus was the predomination LAB. Healthy feces comprised mainly E. faecium (41.46%), E. avium (17.07%), E. faecalis (14.63%), and E. raffinosus (4.88%), whereas diarrhea feces comprised mostly E. faecium (53.19%), E. faecalis (13.83%), and E. avium (9.57%). Similarly, members of the Lactobacillus genera were also detected, of which L. casei (4.88%), L. paracasei (4.88%), L. fermentum (2.44%), and L. plantarum (2.44%) were primarily found in healthy feces, and L. casei (4.26%), L. fermentum (4.26%), L. paracasei (2.13%), and L. rhamnosus (2.13%) were most common in diarrhea feces.
Microbial Counts
Common microbial communities, based on dilution plating are shown (Figure 2). Aerobic-bacteria and LAB strains in diarrhea feces were observably lower than healthy feces (P < 0.05), however, cell viability increased after treatment with L. casei ZX633, and showed no significant differences with healthy infant feces. E. coli and yeast were not remarkably different between diarrhea and healthy feces, but these numbers were lower in diarrhea infant feces. However, these numbers increased and approached the same levels as healthy feces after treatment with L. casei ZX633.
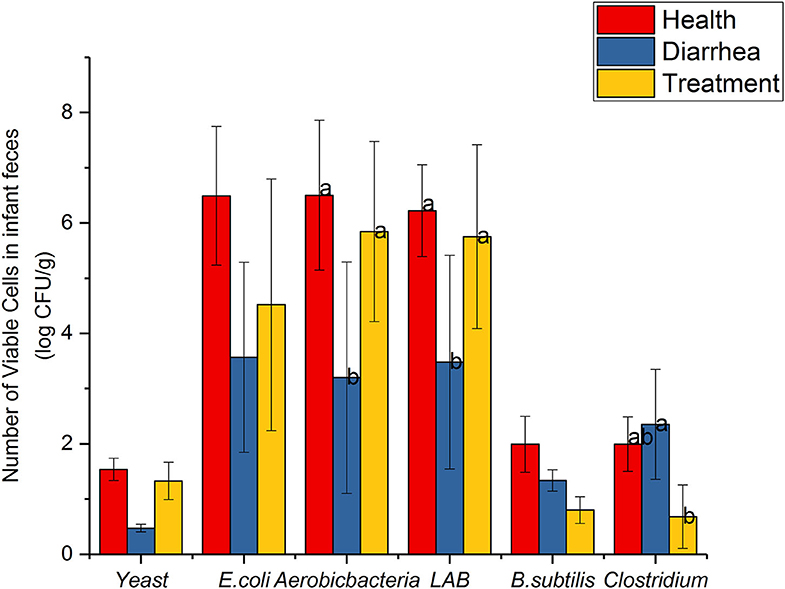
Figure 2. Common viable microbes in infant feces. Different letters above the bars denote statistically significant differences among groups (P < 0.05).
SCFA Levels
SCFA level assessment by HPLC is shown (Figure 3). When compared with healthy feces, lactic, formic, acetic and butyric acid levels were higher in diarrhea feces, but were reduced after treatment with L. casei ZX633. In contrast, propionic acid levels were significantly (P < 0.05) lower in diarrhea feces than healthy feces, but these levels increased after treatment.
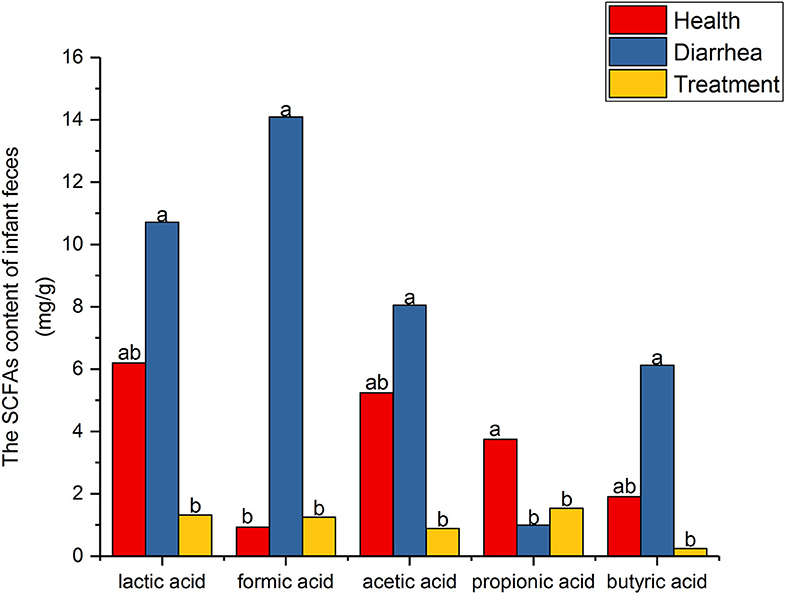
Figure 3. SCFA levels in infant feces. Different letters above the bars denote statistically significant differences among groups (P < 0.05).
Microbial Diversity in Infant Feces
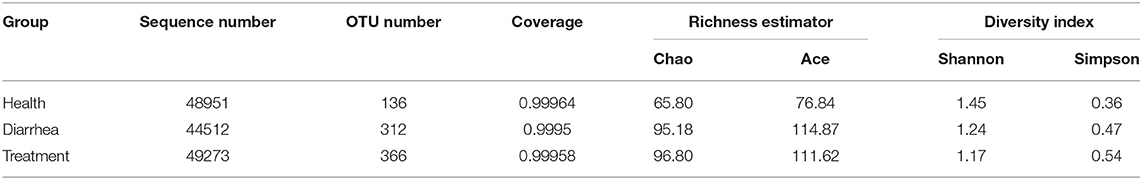
High throughput sequence analyses of the V3–V4 region of 16S rDNA were used to assess bacterial diversity of infant feces. At shown (Table 2), the mean valid sequences from healthy, diarrhea and treated samples were 48,951, 44,512, and 49,273, respectively. These reads were clustered into 488 OTUs based on 97% sequence identity. The average Good's coverage for samples was >99%, indicating that sequencing depth was adequate for robust sequence analyses.
Microbial community richness was evaluated based on alpha diversity (Table 2). Compared with healthy group, Chao, ace and Simpson indices were higher, and the Shannon index was lower for diarrhea samples, which signified the abundance for diarrhea samples was higher than healthy samples, but the reverse was diversity. After treating diarrhea samples with L. casei ZX633, microbial richness and diversity reduced slightly, as indicated by a lower ace index, and a higher Simpson index.
Bacterial community differences and similarities in fecal groups were analyzed based on genus levels (Figure 4A). We observed 89, 228, and 244 genera in healthy, diarrhea and treated samples, respectively, of which 65 were common genera to all groups, accounting for 20.44% of total observed genera (318). Moreover, 4, 9 and 100 genera were shared by healthy and diarrhea samples, healthy and treatment samples, and diarrhea and treated samples, respectively, whereas 140 unique genera were found in healthy, diarrhea and treatment groups (11, 59, and 70, respectively). Fecal microbiota from the three groups are shown (Figure 4B). The relative genus abundance in all samples was Bifidobacterium (51.3%), Escherichia-Shigella (13.8%), Streptococcus (10.62%), and Bacteroides (6.84%). Sequences unclassified into any known groups were specified as “others.”
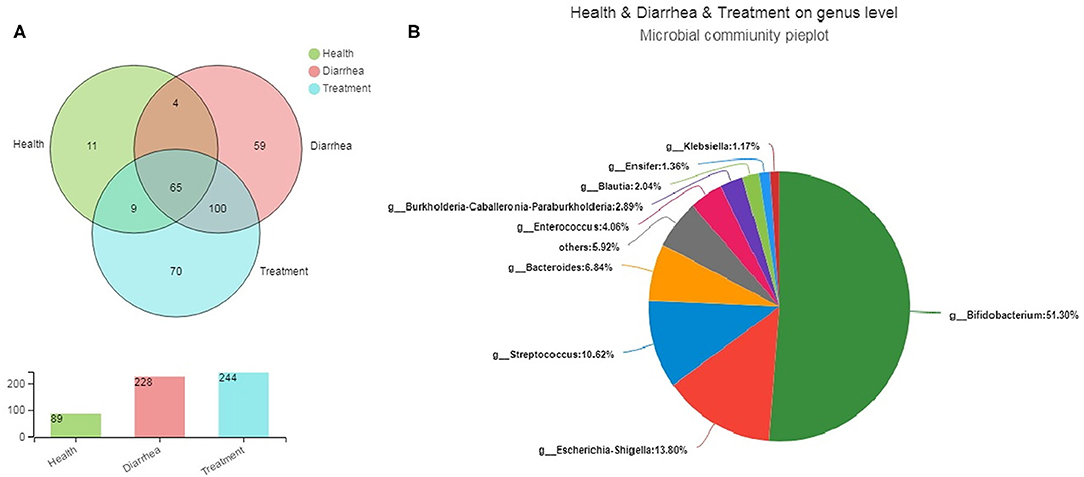
Figure 4. The core microbiota of infant feces at genus levels. (A) A Venn diagram showing bacterial numbers shared within and between sample groups. (B) Bacterial communities in fecal samples. Less than 1% unclassified genus abundance was merged into “others”.
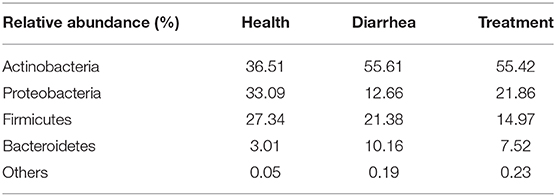
In fecal samples, sequences were classified into 23 different bacterial phyla, 38 classes, 85 orders, 163 families, 318 genera and 427 species, based on Illumina platform analyses. At the phylum level (Table 3), Actinobacteria recorded the highest abundance, and phyla that had relatively high abundance were followed by Proteobacteria, Firmicutes, and Bacteroidetes. As observed, Proteobacteria abundance was largely increased while Bacteroidetes were decreased in the treatment group, when compared to the healthy group.
Bacterial communities based on order and genus level classifications are shown (Figure 5). Microbial composition structures in diarrhea samples differed from healthy samples, which trended toward the healthy group after treatment with L. casei ZX633. At the order level (Figure 5A), when compared with healthy group, Lactobacillales and Bacteroidales abundance was higher and Enterobacteriales and Clostridiales abundance were lower in the diarrhea group, however these improved after treatment. At the genus level (Figure 5B), Escherichia-Shigella and Clostridioides abundance increased, and Streptococcus, Bacteroides, Enterococcus, and Veillonella richness decreased slightly.
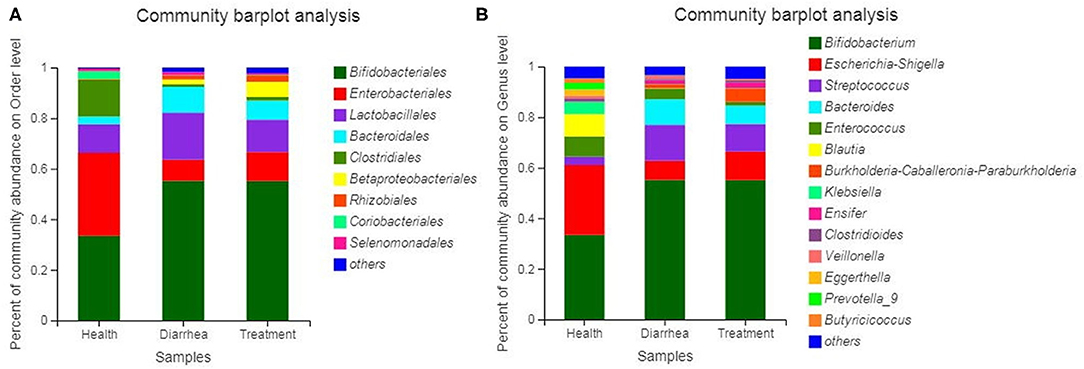
Figure 5. Bacterial community and relative abundance, at the order (A) and genus (B) levels for all infant feces. Taxa with an abundance <1% are included in “others”.
Microbial composition and relationships between species and samples showed that predominant bacteria from different groups were similar (Figure 6A). Bifidobacterium, Escherichia-Shigella, Streptococcus, Bacteroides, and Enterococcus were the five most dominant genera in all fecal samples. The in vitro recovery of gut microbial composition after L. casei ZX633 treatment is shown (Figure 6B). After treating diarrhea fecal samples with the probiotic, Escherichia-Shigella abundance increased from 7.56 to 11.29%, while Streptococcus and Bacteroides abundance separately decreased, from 14.21 to 10.86% and 10.05 to 7.29%, respectively, which trended toward healthy levels.
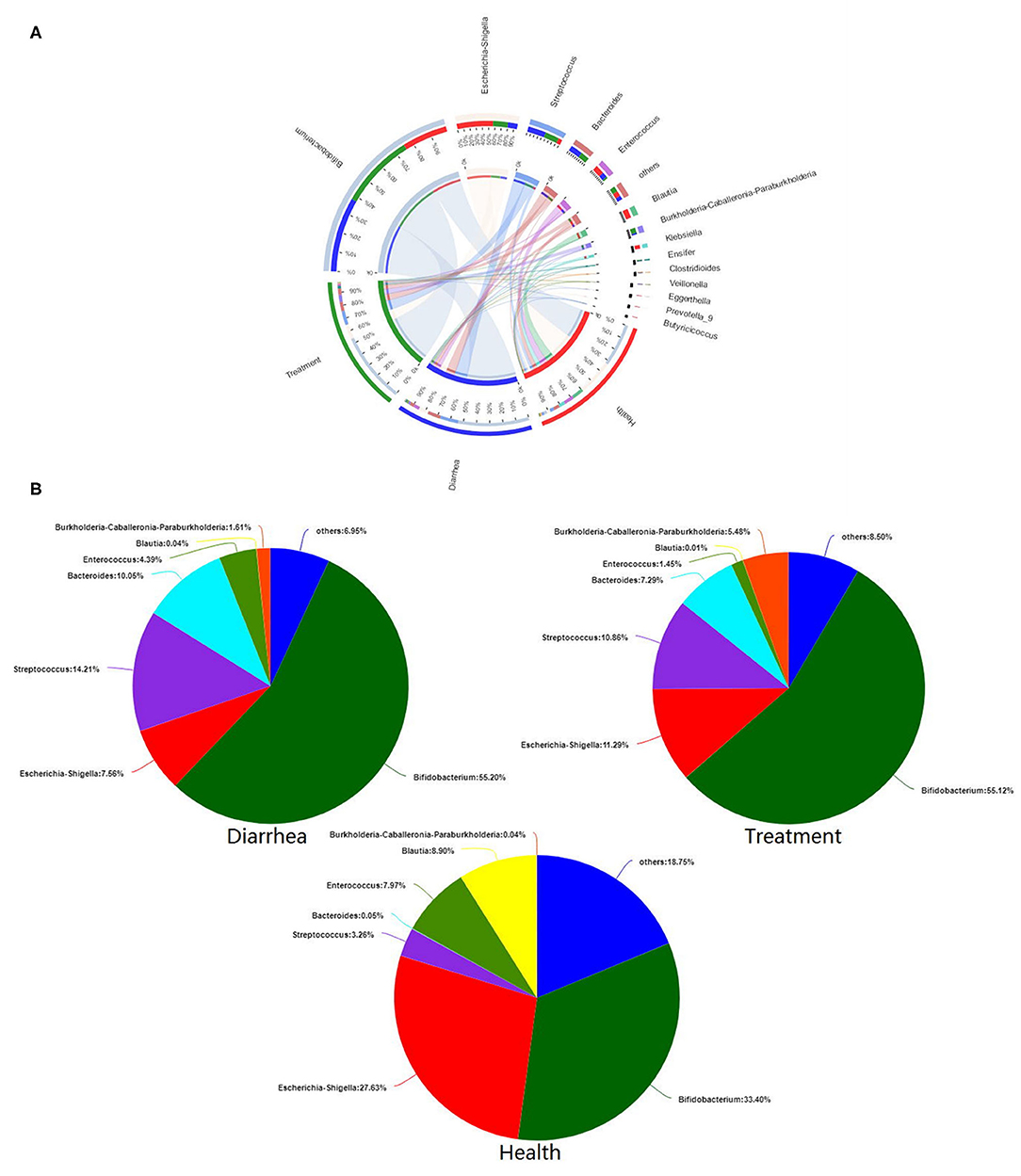
Figure 6. The effects of L. casei ZX633 on the gut microbiota of infants with diarrhea. (A) A circos plot shows the linkage between samples and bacterial genera. (B) Pie charts show gut microbial composition recovery after L. casei ZX633 treatment.
The Relationship Between SCFA Levels and Fecal Bacterial Communities
Our correlation heat map showed that the relationship between SCFA levels and microbial composition at the genus level in different infant fecal samples were varied (Figure 7). In the healthy group, butyric acid was significantly (P < 0.05) and positively related to Eggerthella. In the diarrhea group, formic acid was significantly (P < 0.001) and positive correlated with Bradyrhizobium, Burkholderia-Caballeronia-Paraburkholderia, and Ensifer, and obviously (P < 0.05) negatively correlated with Streptococcus and Clostridium. Acetic acid was positively (P < 0.05) correlated with Paracoccus, whereas butyric acid was significantly (P < 0.001) and positively correlated with Enterobacter, negatively (P < 0.05) correlated with Escherichia-Shigella, and extremely (P < 0.001) negatively correlated with Streptococcus. In treatment group, formic acid was significantly (P < 0.05) and positively correlated with Bradyrhizobium, Burkholderia-Caballeronia-Paraburkholderia, and Beggiatoaceae, propionic acid showed a notable (P < 0.05) negative correlation to Bradyrhizobium, and was positively (P < 0.05) correlated to Veillonella, butyric acid demonstrated a positive (P < 0.05) correlation with Clostridioides, and a negative (P < 0.05) correlation with Bacteroides and Lactobacillus.
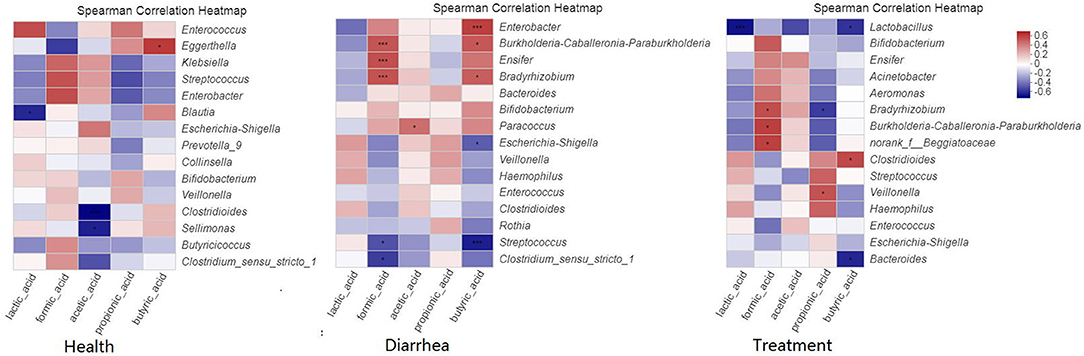
Figure 7. Spearman's correlation heatmaps of the top 15 genera and SCFAs. The X and Y axes are environmental factors and genus, respectively. *P < 0.05, **P < 0.01, ***P < 0.001.
Discussion
The gut microbiota is a large and complicated ecosystem, with a mutualistic relationship with its human or animal host. The system plays an important role in maintaining gut health and preventing gut infection (Yan et al., 2018). Moreover, there is evidence that microbes at one site can affect the host at other sites, and interesting discoveries on gut microbial community linked with the milieu of the brain, respiratory and urogenital tracts, heart and skin, resulting in that microbial community might be new approaches to health maintenance, disease prevention and even treatment (Reid et al., 2017). Diarrhea is related to an imbalance in the gut microbiota, such as the presence of specific pathogens like Clostridium (Mallina et al., 2018), Salmonella (Tadesse, 2014), and Escherichia (Chingwaru and Vidmar, 2017; Mekonnen et al., 2019). Given the potential resistance effects of some antibiotic therapies for bacterial diarrhea, studies have suggested that probiotics could be an effective strategies in combatting these diarrheal diseases (Szajewska et al., 2011; Hojsak et al., 2018). Hence, we strategically selected L. casei ZX633 from healthy feces, and examined its in vitro effects on the composition of intestinal microbiota. Since the cultural properties of LAB in vitro are crucial to select probiotic strains for treating gut microbiota modulation (Bubnov et al., 2018), we have previously studied the probiotic properties of L. casei ZX633 (Wang et al., 2020), including antibiotic sensitivity, aggregation, gastrointestinal survival rate, and so on. Generally, LAB isolated from the human beings are considered to be safe, however, specific research on its internal safety issues will be taken into consideration in further experiments.
LAB are widely distributed in nature. Currently, at least 40 genera are known, mainly comprising Lactococcus, Lactobacillus, Leuconostoc, Enterococcus, Pediococcus, and Streptococcus. Dairy products contain several genera, including Lactococcus (Smit et al., 2005), S. acidophilus (Ott et al., 2000), and Leuconostoc (Sanchez et al., 2005), whereas Lactobacillus (Bernardeau et al., 2008) mainly occurs in plants, animals and silage. Enterococcus are widespread in the digestive system of humans and animals (Giraffa, 2002). A previous study showed that Lactobacillus was the most prevalent genus in infant feces, followed by Enterococcus (Rubio et al., 2014). In this study, E. faecium was the most frequently isolated species from infant feces, either healthy or diarrheal, whereas Lactobacillus was the second. These observations agree with a previous study showing that Enterococcus was the main species isolated from infant feces (Solis et al., 2010).
We also investigated changes in microbiota viability and SCFAs. Some studies have suggested that LAB species contribute to the prevention and treatment of numerous gastrointestinal tract disorders, including diarrhea (Wanke and Szajewska, 2014; Xu et al., 2019). Similarly, Clostridium is a commensal gut microbe, and is the foremost cause of nosocomial bacterial infections, making it the most vicious of the enteric pathogens (Kalakuntla et al., 2019). When compared with the diarrhea group, some microbiota species were modulated in the treatment group, such as reductions in Clostridium and increased LAB, which could be linked to diarrhea remission. The most abundant microbial metabolites were acetate, propionate and butyrate, each of which acts as a microbial regulatory factor and major modulators of host-microbiome interactions, affecting gut homeostasis and defenses against pathogen invasion (He et al., 2019). L. casei ZX633 appeared to exert different effects on SCFA levels, showing reductions in formic acid and increased propionic acid in the treatment group. These findings indicate that L. casei ZX633 treatment could improve SCFA levels, and promote gut health in infants with diarrhea, which are closely correlated with the development of microbiota community in diarrhea group toward healthy group.
Soltan et al. reported that LAB were capable of secreting acids, bacteriocins and other by-products that could neutralize infectious pathogens (Soltan Dallal et al., 2017). As a corollary, we observed decreased microbial community richness and diversity after processing feces samples, which may have been attributable to L. casei ZX633. In accordance with our data, Lai et al. showed a lower abundance and diversity in the gut microbiota of children, when supplemented with a probiotic L. casei (Lai et al., 2019).
According to previous reports, the gut microbiota in infants is enriched with Bifidobacterium, Escherichia-Shigella, Streptococcus, and Bacteroides, and high levels of Bifidobacterium (Yan et al., 2018; Derrien et al., 2019), consistent with our results. Furthermore, we observed a higher abundance of Escherichia-Shigella and Enterococcus, and a lower abundance of Bacteroides in healthy feces, when compared with the diarrhea group. This observation agreed with a recent study showing a decreased abundance of Escherichia-Shigella and Enterococcus, and an increase abundance in Bacteroides, in community-acquired pneumonia in children (Ren et al., 2020). Bacteroides can alter gut permeability, increasing immune infiltration, and weakening intestinal epithelial repair (Hua et al., 2016), which eventually promotes the migration of enteric pathogens through the intestinal barrier (Ren et al., 2017). Studies have shown that Shigella normally colonizes the human gut and is associated with intestinal infections, such as bacillary dysentery (Anderson et al., 2016), and Escherichia causes a broad range of diseases, from diarrheal diseases to extra-intestinal diseases (Walsham et al., 2016; Gallardo et al., 2017). Nevertheless, Escherichia is the most common commensal aerobic bacterium in the gut microbiota of humans which had dualism between commensalism and pathogenicity (Massot et al., 2016). Accordingly, an increase in Escherichia-Shigella abundance in the treatment group was normal and beneficial. Additionally, Streptococcus is frequently isolated from infected abdominal cavities and urogenital systems; the rate of Streptococcal infections is gradually increasing (Wang et al., 2019). Higher Streptococcus abundance was found in the diarrhea group, which may possibly indicate infection. After treatment with L. casei ZX633, diarrhea was improved as manifested by a reduction in the relative abundance of Streptococcus.
The composition and activity of intestinal microbiota influences intestinal environments, and therefore fecal SCFA profiles (Hemalatha et al., 2017), which exert vital influences on intestinal health. SCFAs are important for gut integrity, glucose homeostasis and immune function (Shibata et al., 2017; Liu et al., 2018). Furthermore, the increase in SCFA levels may contribute to the repairs intestinal mucosa, ameliorates permeability, and alleviates diarrhea (Liu et al., 2019). For example, Lee et al. (2017) reported SCFAs mitigate pro-inflammatory responses by inhibiting the NF-κB pathway and may promoted anti-inflammatory cytokine (i.e., IL-10) production. SCFAs are produced by colonic anaerobic microbial communities via the fermentation of indigestible fibrous matter, and some luminal amino acids (Kong et al., 2016). Butyric acid production has been shown to affect the intestines, inhibit pathogen growth, and promote healthy intestinal flora (Liu et al., 2014). In our study, due to changes in SCFA levels and bacterial abundance, correlations between bacterial abundance and SCFAs in different infant fecal conditions were varied.
Conclusions
In summary, we compared changes in intestinal microbiota in diarrhea samples treated with the probiotic, L. casei ZX633. The intestinal flora of diarrhea samples was different to healthy samples, including an increased abundance of some pathogenic bacteria. When supplemented with L. casei ZX633, this reduced gut microbiota destruction and promoted the healthy development of intestinal microbial diversity in diarrhea. Our in vitro study provides a basis on which to further explore L. casei ZX633 as a probiotic for gut microbiota modulation in infants with diarrhea. Further, more comprehensive studies should be performed before these strains are used in clinical practice including the role of inflammation, immunity, intestinal barrier function, and so on.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/sra/PRJNA642333, PRJNA642333.
Author Contributions
XW, MZ, WW, and ZT designed the study and wrote the manuscript. XW, MZ, and YL performed the experiments. XW conducted the statistical and bioinformatics analysis. HL, HZ, and ZT were involved in the revision of the manuscript. All the authors reviewed and approved the final manuscript.
Funding
This work was supported by the Qinghai Province Key R&D and Transformation Plan of China (No. 2020-NK-127), Foundation of Key Technology Research Project of Henan Province (Grant No. 192102110082), and Major collaborative innovation project of Zhengzhou (Key discipline construction project of Zhengzhou University) (No. xkzdjc201905).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Anderson, M., Sansonetti, P. J., and Marteyn, B. S. (2016). Shigella diversity and changing landscape: insights for the twenty-first century. Front. Cell. Infect. Microbiol. 6:45. doi: 10.3389/fcimb.2016.00045
Bautista-Gallego, J., Ferrocino, I., Botta, C., Ercolini, D., Cocolin, L., and Rantsiou, K. (2019). Probiotic potential of a Lactobacillus rhamnosus cheese isolate and its effect on the fecal microbiota of healthy volunteers. Food Res. Int. 119, 305–314. doi: 10.1016/j.foodres.2019.02.004
Bernardeau, M., Vernoux, J. P., Henri-Dubernet, S., and Gueguen, M. (2008). Safety assessment of dairy microorganisms: the Lactobacillus genus. Int. J. Food Microbiol. 126, 278–285. doi: 10.1016/j.ijfoodmicro.2007.08.015
Bubnov, R. V., Babenko, L. P., Lazarenko, L. M., Mokrozub, V. V., and Spivak, M. Y. (2018). Specific properties of probiotic strains: relevance and benefits for the host. EPMA J. 9, 205–223. doi: 10.1007/s13167-018-0132-z
Bubnov, R. V., Spivak, M. Y., Lazarenko, L. M., Bomba, A., and Boyko, N. V. (2015). Probiotics and immunity: provisional role for personalized diets and disease prevention. EPMA J. 6:14. doi: 10.1186/s13167-015-0036-0
Chingwaru, W., and Vidmar, J. (2017). Potential of Zimbabwean commercial probiotic products and strains of Lactobacillus plantarum as prophylaxis and therapy against diarrhoea caused by Escherichia coli in children. Asian Pac. J. Trop. Med. 10, 57–63. doi: 10.1016/j.apjtm.2016.12.009
Derrien, M., Alvarez, A. S., and de Vos, W. M. (2019). The gut microbiota in the first decade of life. Trends Microbiol. 27, 997–1010. doi: 10.1016/j.tim.2019.08.001
Gallardo, P., Izquierdo, M., Vidal, R. M., Chamorro-Veloso, N., Rossello-Mora, R., O'Ryan, M., et al. (2017). Distinctive gut microbiota is associated with diarrheagenic Escherichia coli infections in chilean children. Front. Cell. Infect. Microbiol. 7:424. doi: 10.3389/fcimb.2017.00424
Gheziel, C., Russo, P., Arena, M. P., Spano, G., Ouzari, H. I., Kheroua, O., et al. (2019). Evaluating the probiotic potential of Lactobacillus plantarum strains from algerian infant feces: towards the design of probiotic starter cultures tailored for developing countries. Probiotics Antimicrob. Proteins 11, 113–123. doi: 10.1007/s12602-018-9396-9
Giraffa, G. (2002). Enterococci from foods. FEMS Microbiol. Rev. 26, 163–171. doi: 10.1111/j.1574-6976.2002.tb00608.x
He, T., Zhu, Y. H., Yu, J., Xia, B., Liu, X., Yang, G. Y., et al. (2019). Lactobacillus johnsonii L531 reduces pathogen load and helps maintain short-chain fatty acid levels in the intestines of pigs challenged with Salmonella enterica Infantis. Vet. Microbiol. 230, 187–194. doi: 10.1016/j.vetmic.2019.02.003
Hemalatha, R., Ouwehand, A. C., Saarinen, M. T., Prasad, U. V., Swetha, K., and Bhaskar, V. (2017). Effect of probiotic supplementation on total lactobacilli, bifidobacteria and short chain fatty acids in 2-5-year-old children. Microb. Ecol. Health Dis. 28:1298340. doi: 10.1080/16512235.2017.1298340
Hill, C., Guarner, F., Reid, G., Gibson, G. R., Merenstein, D. J., Pot, B., et al. (2014). Expert consensus document. The International Scientific Association for probiotics and prebiotics consensus statement on the scope and appropriate use of the term probiotic. Nat. Rev. Gastroenterol. Hepatol. 11, 506–514. doi: 10.1038/nrgastro.2014.66
Hojsak, I., Szajewska, H., Canani, R. B., Guarino, A., Indrio, F., Kolacek, S., et al. (2018). Probiotics for the prevention of nosocomial diarrhea in children. J. Pediatr. Gastroenterol. Nutr. 66, 3–9. doi: 10.1097/MPG.0000000000001637
Hua, X., Goedert, J. J., Pu, A., Yu, G., and Shi, J. (2016). Allergy associations with the adult fecal microbiota: analysis of the American gut project. EBioMedicine 3, 172–179. doi: 10.1016/j.ebiom.2015.11.038
Kalakuntla, A. S., Nalakonda, G., Nalakonda, K., Pidikiti, C. V., and Aasim, S. A. (2019). Probiotics and Clostridium difficile: a review of dysbiosis and the rehabilitation of gut microbiota. Cureus 11:e5063. doi: 10.7759/cureus.5063
Kong, X. F., Ji, Y. J., Li, H. W., Zhu, Q., Blachier, F., Geng, M. M., et al. (2016). Colonic luminal microbiota and bacterial metabolite composition in pregnant Huanjiang mini-pigs: effects of food composition at different times of pregnancy. Sci. Rep. 6:37224. doi: 10.1038/srep37224
Kwok, L. Y., Guo, Z., Zhang, J., Wang, L., Qiao, J., Hou, Q., et al. (2015). The impact of oral consumption of Lactobacillus plantarum P-8 on faecal bacteria revealed by pyrosequencing. Benef. Microbes 6, 405–413. doi: 10.3920/BM2014.0063
Kwok, L. Y., Wang, L., Zhang, J., Guo, Z., and Zhang, H. (2014). A pilot study on the effect of Lactobacillus casei Zhang on intestinal microbiota parameters in Chinese subjects of different age. Benef. Microbes 5, 295–304. doi: 10.3920/BM2013.0047
Lai, H. H., Chiu, C. H., Kong, M. S., Chang, C. J., and Chen, C. C. (2019). Probiotic Lactobacillus casei: effective for managing childhood diarrhea by altering gut microbiota and attenuating fecal inflammatory markers. Nutrients 11:1150. doi: 10.3390/nu11051150
Lee, C., Kim, B. G., Kim, J. H., Chun, J., Im, J. P., and Kim, J. S. (2017). Sodium butyrate inhibits the NF-kappa B signaling pathway and histone deacetylation, and attenuates experimental colitis in an IL-10 independent manner. Int. Immunopharmacol. 51, 47–56. doi: 10.1016/j.intimp.2017.07.023
Li, Y. T., Xu, H., Ye, J. Z., Wu, W. R., Shi, D., Fang, D. Q., et al. (2019). Efficacy of Lactobacillus rhamnosus GG in treatment of acute pediatric diarrhea: a systematic review with meta-analysis. World J. Gastroenterol. 25, 4999–5016. doi: 10.3748/wjg.v25.i33.4999
Liu, B., Qian, J., Wang, Q., Wang, F., Ma, Z., and Qiao, Y. (2014). Butyrate protects rat liver against total hepatic ischemia reperfusion injury with bowel congestion. PLoS ONE 9:e106184. doi: 10.1371/journal.pone.0106184
Liu, B., Wang, W., Zhu, X., Sun, X., Xiao, J., Li, D., et al. (2018). Response of gut microbiota to dietary fiber and metabolic interaction with SCFAs in piglets. Front. Microbiol. 9:2344. doi: 10.3389/fmicb.2018.02344
Liu, C. S., Liang, X., Wei, X. H., Jin, Z., Chen, F. L., Tang, Q. F., et al. (2019). Gegen qinlian decoction treats diarrhea in piglets by modulating gut microbiota and short-chain fatty acids. Front. Microbiol. 10:825. doi: 10.3389/fmicb.2019.00825
Mallina, R., Craik, J., Briffa, N., Ahluwalia, V., Clarke, J., and Cobb, A. G. (2018). Probiotic containing Lactobacillus casei, Lactobacillus bulgaricus, and Streptococcus thermophiles (ACTIMEL) for the prevention of Clostridium difficile associated diarrhoea in the elderly with proximal femur fractures. J. Infect. Public Health 11, 85–88. doi: 10.1016/j.jiph.2017.04.001
Massot, M., Picard, B., and Denamur, E. (2016). Diversity and variability in populations of E. coli over time within the gut microbiota. Rev. Francoph. Lab. 2016, 35–43. doi: 10.1016/S1773-035X(16)30326-4
Mekonnen, G. K., Mengistie, B., Sahilu, G., Kloos, H., and Mulat, W. (2019). Etiologies of diarrhea and drug susceptibility patterns of bacterial isolates among under-five year children in refugee camps in Gambella region, Ethiopia: a case control study. BMC Infect. Dis. 19:1008. doi: 10.1186/s12879-019-4599-6
Ott, A., Germond, J. E., and Chaintreau, A. (2000). Origin of acetaldehyde during milk fermentation using (13)C-labeled precursors. J. Agric. Food Chem. 48, 1512–1517. doi: 10.1021/jf9904867
Pilla, R., Guard, B. C., Steiner, J. M., Gaschen, F. P., Olson, E., Werling, D., et al. (2019). Administration of a synbiotic containing Enterococcus faecium does not significantly alter fecal microbiota richness or diversity in dogs with and without food-responsive chronic enteropathy. Front. Vet. Sci. 6:277. doi: 10.3389/fvets.2019.00277
Reid, G., Abrahamsson, T., Bailey, M., Bindels, L. B., Bubnov, R., Ganguli, K., et al. (2017). How do probiotics and prebiotics function at distant sites? Benef. Microbes 8, 521–533. doi: 10.3920/BM2016.0222
Ren, X., Gamallat, Y., Liu, D., Zhu, Y., Meyiah, A., Yan, C., et al. (2020). The distribution characteristics of intestinal microbiota in children with community-acquired pneumonia under five years of age. Microb. Pathog. 142:104062. doi: 10.1016/j.micpath.2020.104062
Ren, X., Zhu, Y., Gamallat, Y., Ma, S., Chiwala, G., Meyiah, A., et al. (2017). E. coli O124 K72 alters the intestinal barrier and the tight junctions proteins of guinea pig intestine. Biomed. Pharmacother. 94, 468–473. doi: 10.1016/j.biopha.2017.07.123
Rubio, R., Jofre, A., Martin, B., Aymerich, T., and Garriga, M. (2014). Characterization of lactic acid bacteria isolated from infant faeces as potential probiotic starter cultures for fermented sausages. Food Microbiol. 38, 303–311. doi: 10.1016/j.fm.2013.07.015
Sanchez, J. I., Martinez, B., and Rodriguez, A. (2005). Rational selection of Leuconostoc strains for mixed starters based on the physiological biodiversity found in raw milk fermentations. Int. J. Food Microbiol. 105, 377–387. doi: 10.1016/j.ijfoodmicro.2005.04.025
Shibata, N., Kunisawa, J., and Kiyono, H. (2017). Dietary and microbial metabolites in the regulation of host immunity. Front. Microbiol. 8:2171. doi: 10.3389/fmicb.2017.02171
Shin, S.-H., and Shin, H.-S. (2018). The comparison of clinical characteristics in three types of viral acute diarrhea in infants and toddlers and the effect of Lactobacillus acidophilus on rotaviral diarrhea. J. Bacteriol. Virol. 48:166. doi: 10.4167/jbv.2018.48.4.166
Smit, G., Smit, B. A., and Engels, W. J. (2005). Flavour formation by lactic acid bacteria and biochemical flavour profiling of cheese products. FEMS Microbiol. Rev. 29, 591–610. doi: 10.1016/j.fmrre.2005.04.002
Solis, G., de Los Reyes-Gavilan, C. G., Fernandez, N., Margolles, A., and Gueimonde, M. (2010). Establishment and development of lactic acid bacteria and bifidobacteria microbiota in breast-milk and the infant gut. Anaerobe 16, 307–310. doi: 10.1016/j.anaerobe.2010.02.004
Soltan Dallal, M. M., Davoodabadi, A., Abdi, M., Hajiabdolbaghi, M., Sharifi Yazdi, M. K., Douraghi, M., et al. (2017). Inhibitory effect of Lactobacillus plantarum and Lb. fermentum isolated from the faeces of healthy infants against nonfermentative bacteria causing nosocomial infections. New Microbe New Infect. 15, 9–13. doi: 10.1016/j.nmni.2016.09.003
Szajewska, H., Wanke, M., and Patro, B. (2011). Meta-analysis: the effects of Lactobacillus rhamnosus GG supplementation for the prevention of healthcare-associated diarrhoea in children. Aliment. Pharmacol. Ther. 34, 1079–1087. doi: 10.1111/j.1365-2036.2011.04837.x
Tadesse, G. (2014). Prevalence of human Salmonellosis in Ethiopia: a systematic review and meta-analysis. BMC Infect. Dis. 14:88. doi: 10.1186/1471-2334-14-88
Walker, C. L. F., Rudan, I., Liu, L., Nair, H., Theodoratou, E., Bhutta, Z. A., et al. (2013). Global burden of childhood pneumonia and diarrhoea. Lancet 381, 1405–1416. doi: 10.1016/S0140-6736(13)60222-6
Walsham, A. D., MacKenzie, D. A., Cook, V., Wemyss-Holden, S., Hews, C. L., Juge, N., et al. (2016). Lactobacillus reuteri inhibition of enteropathogenic Escherichia coli adherence to human intestinal epithelium. Front. Microbiol. 7:244. doi: 10.3389/fmicb.2016.00244
Wang, X., Wang, W., Lv, H., Zhang, H., Liu, Y., Zhang, M., et al. (2020). Probiotic potential and wide-spectrum antimicrobial activity of lactic acid bacteria isolated from infant feces. Probiot. Antimicrob. Prot. doi: 10.1007/s12602-020-09658-3. [Epub ahead of print].
Wang, Y., Li, A., Jiang, X., Zhang, H., Mehmood, K., Zhang, L., et al. (2018). Probiotic potential of leuconostoc pseudomesenteroides and lactobacillus strains isolated from yaks. Front. Microbiol. 9:2987. doi: 10.3389/fmicb.2018.02987
Wang, Y., Li, A., Liu, J., Mehmood, K., Wangdui, B., Shi, H., et al. (2019). L. pseudomesenteroides and L. johnsonii isolated from yaks in Tibet modulate gut microbiota in mice to ameliorate enteroinvasive Escherichia coli-induced diarrhea. Microb. Pathog. 132, 1–9. doi: 10.1016/j.micpath.2019.04.020
Wanke, M., and Szajewska, H. (2014). Probiotics for preventing healthcare-associated diarrhea in children: a meta-analysis of randomized controlled trials. Pediatr. Pol. 89, 8–16. doi: 10.1016/j.pepo.2013.12.003
Wen, H., Yin, X., Yuan, Z., Wang, X., and Su, S. (2018). Comparative analysis of gut microbial communities in children under 5 years old with diarrhea. J. Microbiol. Biotechnol. 28, 652–662. doi: 10.4014/jmb.1711.11065
Wongsen, S., Werawatganon, D., and Tumwasorn, S. (2019). Lactobacillus plantarum B7 attenuates Salmonella typhimurium infection in mice: preclinical study in vitro and in vivo. Asian Biomed. 12, 211–218. doi: 10.1515/abm-2019-0022
Xu, H., Zhao, F., Hou, Q., Huang, W., Liu, Y., Zhang, H., et al. (2019). Metagenomic analysis revealed beneficial effects of probiotics in improving the composition and function of the gut microbiota in dogs with diarrhoea. Food Funct. 10, 2618–2629. doi: 10.1039/C9FO00087A
Yan, M., Chen, W., Li, N., Ren, J., Chen, C., You, C., et al. (2018). Impact of Lactobacillus plantarum ST-III on the composition of infant gut microbiota and its potential synergism with breast milk and infant formula as revealed by an in vitro study. Int. Dairy J. 85, 66–72. doi: 10.1016/j.idairyj.2018.05.014
Yang, L., Yuan, X., Li, J., Dong, Z., and Shao, T. (2019). Dynamics of microbial community and fermentation quality during ensiling of sterile and nonsterile alfalfa with or without Lactobacillus plantarum inoculant. Bioresour. Technol. 275, 280–287. doi: 10.1016/j.biortech.2018.12.067
Zhang, B., Wang, Y. P., Tan, Z. F., Li, Z. W., Jiao, Z., and Huang, Q. C. (2016). Screening of probiotic activities of Lactobacilli strains isolated from traditional Tibetan Qula, a raw Yak milk cheese. Asian Australas. J. Anim. Sci. 29, 1490–1499. doi: 10.5713/ajas.15.0849
Zhang, M., Wang, X., Cui, M., Wang, Y., Jiao, Z., and Tan, Z. (2018). Ensilage of oats and wheatgrass under natural alpine climatic conditions by indigenous lactic acid bacteria species isolated from high-cold areas. PLoS ONE 13:e0192368. doi: 10.1371/journal.pone.0192368
Keywords: Lactobacillus casei, infant, diarrhea, gut microbiota, high-throughput sequencing
Citation: Wang X, Zhang M, Wang W, Lv H, Zhang H, Liu Y and Tan Z (2020) The in vitro Effects of the Probiotic Strain, Lactobacillus casei ZX633 on Gut Microbiota Composition in Infants With Diarrhea. Front. Cell. Infect. Microbiol. 10:576185. doi: 10.3389/fcimb.2020.576185
Received: 27 June 2020; Accepted: 13 August 2020;
Published: 10 September 2020.
Edited by:
Frederic Antonio Carvalho, INSERM U1107 Douleur et Biophysique Neurosensorielle (Neuro-Dol), FranceReviewed by:
Rostyslav V. Bubnov, National Academy of Sciences of Ukraine, UkraineJorge Reinheimer, National University of Littoral, Argentina
Diego Javier Mercanti, Instituto de Lactología Industrial (UNL-CONICET), Argentina
Roxana Beatriz Medina, CONICET Centro de Referencia para Lactobacilos (CERELA), Argentina
Copyright © 2020 Wang, Zhang, Wang, Lv, Zhang, Liu and Tan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhongfang Tan, tzhongfang@zzu.edu.cn
†These authors share first authorship
 Xing Wang
Xing Wang Miao Zhang1†
Miao Zhang1† Zhongfang Tan
Zhongfang Tan