- 1College of Life and Environmental Sciences—Biosciences, University of Exeter, Exeter, United Kingdom
- 2School of Life Sciences, University of Warwick, Coventry, United Kingdom
The formation of persister cells is one mechanism by which bacteria can survive exposure to environmental stresses. We show that Campylobacter jejuni 11168H forms persister cells at a frequency of 10−3 after exposure to 100 × MIC of penicillin G for 24 h. Staining the cell population with a redox sensitive fluorescent dye revealed that penicillin G treatment resulted in the appearance of a population of cells with increased fluorescence. We present evidence, to show this could be a consequence of increased redox protein activity in, or associated with, the electron transport chain. These data suggest that a population of penicillin G treated C. jejuni cells could undergo a remodeling of the electron transport chain in order to moderate membrane hyperpolarization and intracellular alkalization; thus reducing the antibiotic efficacy and potentially assisting in persister cell formation.
Introduction
Campylobacter jejuni is the leading bacterial cause of gastroenteritis in the world estimated be causing almost 100 million cases worldwide (Asuming-Bediako et al., 2019) with 1 million cases a year in the US, and over 250,000 cases in the European Union (Chlebicz and Slizewska, 2018). In many areas of the world the reported incidence of disease appears to be increasing (Chlebicz and Slizewska, 2018). The avian gut is the normal site of carriage of the bacterium and most cases in humans arise from the ingestion of contaminated poultry (Chlebicz and Slizewska, 2018; Igwaran and Okoh, 2019; Rossler et al., 2019). In developing countries, Campylobacter is hyper-endemic, a leading bacterial cause of diarrhoeal disease and a major cause of infant mortality (Asuming-Bediako et al., 2019; Igwaran and Okoh, 2019). The bacterium is notoriously fastidious and survives poorly under laboratory conditions, yet it appears to be ubiquitous in the environment, forming reservoirs of infection (Murphy et al., 2006; Bronowski et al., 2014). Environmental survival of the bacterium is essential for transmission to new hosts. Survival outside of the host is also evidenced in the ability of the bacterium to tolerate abiotic stresses encountered during food processing (Ligowska et al., 2011; Garcia-Sanchez et al., 2017).
One mechanism employed by bacteria to survive otherwise lethal stresses is the formation of persister cells (Lewis, 2005, 2007). Persistence has been defined as the ability of a subset of the population (perisister cells) to survive exposure to a bactericidal drug concentration (Balaban et al., 2019). It is an example of phenotypic switching; the ability of a genetically identical population of organisms to display diverse phenotypes under a given environment (Balaban et al., 2004; Lewis, 2007). There is good evidence that persister cells are present in the bacterial population before exposure to the stress. They can be revealed by exposing the population to a supra-lethal dose of a bactericidal antibiotic (Lewis, 2007; Balaban et al., 2019), which kills most of the bacterial population, except for the persister cells. The ability of persister cells to survive exposure to abiotic stresses, including multiples of the minimum inhibitory concentration (MIC) of antibiotics, biocides and killing by toxic metals, is ascribed to their low growth rates or dormancy (Harrison et al., 2005; Lewis, 2008; Harms et al., 2016). There is also increasing evidence that persister cells make up a subpopulation of antibiotic-resistant cells in biofilms (Lewis, 2007, 2008; Jayaraman, 2008; Harrison et al., 2009; Kim et al., 2009).
The mechanisms by which persister cells form and their molecular makeup have been studied intensively over the past few years and reveal some common mechanisms but have also revealed that a diverse range of molecular events are associated with persister cell formation (Wilmaerts et al., 2019). Toxin-antitoxin systems may play a role by regulating metabolism and targeting functions such as transcription, translation and DNA replication (Harms et al., 2016). Another mechanism involves energy metabolism: a number of reports linking the electron transport chain to persister cell formation (Harms et al., 2016). Drug efflux pumps may contribute to their formation too (Harms et al., 2016). We have previously shown that oxygen availability can influence persister cell formation (Hemsley et al., 2014). At the time of submitting this manuscript there were no reports of the formation of persister cells by Campylobacter.
Here, we set out to investigate whether C. jejuni 11168H is able to form persister cells and to investigate the molecular makeup of these cells. C. jejuni strain 11168H is a strain, selected as representative, for the first Campylobacter genome sequencing project (Parkhill et al., 2000). This should provide new insight into the mechanisms by which C. jejuni 11168H survives exposure to antibiotic stresses and could reveal mechanisms that it uses to survive in the environment.
Materials and Methods
Growth of C. jejuni
C. jejuni 11168H was cultured on Columbia agar plates (CBA) supplemented with either 5–9% (v/v) horse blood or with Skirrow selective supplement (Oxoid Ltd., Basingstoke, UK) and 5–9% (v/v) horse blood in a variable atmosphere incubator (VAIN) (Don Whitley Scientific, Bingley, UK) under microaerobic conditions (5% O2, 85% N2, 10% CO2) at 37°C for 24 or 48 h. The bacteria were sub-cultured into 25 ml of Mueller-Hinton broth (Oxoid) and grown under microaerobic conditions as before (Champion et al., 2010).
Staining With BacLight™
Bacteria were enumerated using BacLight™ Live/Dead Bacterial Viability Kit (Life Technologies, Paisley, UK) according to the manufacturers' instructions. Previous studies have demonstrated the utility of BacLight™ for the differentiation of live and dead C. jejuni (Alonso et al., 2002; He and Chen, 2010; Kim et al., 2014). Briefly, aliquots of bacterial cells were treated with premixed stain, mixed and incubated for 15 min in the dark. Two μl of the resulting bacterial suspension was then placed on poly-L-lysine coated glass slides (Sigma, Dorset, UK) along with 2 μl of 1 μm polystyrene latex beads (Sigma), and fluorescence viewed using a Zeiss Fluorescence Microscope with an FITC filter. The total number of cells and the number of specifically stained cells was calculated using Image J software (Schneider et al., 2012).
Staining With Redox Sensor Green
Aliquots of the bacterial cell suspension in 1 ml PBS were stained with 4 μl of BacLight™ Redox Sensor Green reagent (RSG) (Life Technologies) for 10 min in the dark at room temperature and then visualized after excitation at 490 nm by microscopy as described above or flow cytometry as detailed below.
Exposure to Penicillin G
The MIC of penicillin G toward C. jejuni 11168H was measured using ETEST® antimicrobial susceptibility test strips (Biomerieux, Basingstoke, UK). Overnight broth cultures were diluted to the equivalent of a McFarland turbidity standard of 0.5 and used to create a lawn on Mueller-Hinton agar plates. The ETEST® strips were added and the plates were incubated at 37°C for 24 h under microaerobic conditions and zones of growth inhibition recorded. We found that the MIC of penicillin G was 11.43 μg/ml. For studies where C. jejuni 11168H was exposed to penicillin G, bacteria were first grown in MHB broth for 14 h and the cell harvested by centrifugation and re-suspended in fresh MHB or in MHB containing 100 × the MIC of penicillin G (1,143 μg/ml).
Bacterial Killing by Penicillin G
Broth cultured bacteria were incubated for 24 h with either 100 × the MIC of penicillin G (Sigma) in Mueller Hinton broth or in Mueller Hinton broth alone. At intervals, samples of the cultures were taken, centrifuged and the cell pellet re-suspended in phosphate buffered saline (PBS; pH 7.2). Bacteria were then enumerated as described above. To calculate the number of colony forming units (CFU), bacteria were serially diluted in PBS, plated onto Mueller Hinton agar plates (Oxoid) and incubated at 37°C under microaerobic conditions. The frequency of persister cell formation was calculated as the number of C. jejuni cells cultured after 24 h exposure to 100 × MIC of penicillin/number of C. jejuni cells cultured before exposure to antibiotic. Assays were carried out in triplicate.
Flow Cytometry
Aliquots of untreated and penicillin G-treated C. jejuni 11168H were stained with Redox Sensor Green as described above, and analyzed in a BD FACS Aria III (Becton Dickenson fluorescence-activated cell sorting (FACS) cytometer using a 488 nm laser. Emission of fluorescence was detected at 530 ± 30 nm. Three distinct RSG stained populations (bright, dim and unstained, 3 × 106 cells) were sorted into 10 ml falcon tubes and kept on ice.
The populations of bacterial cells were collected as described above, concentrated by centrifugation at 11,337 × g for 30 min at 4°C and then pooled and lysed in 70 μl of Bugbuster reagent (Merck Millipore, Dorset, UK). After shaking at room temperature for 20 min the bacterial lysates were stored at −80°C.
Mass Spectrometry
Proteomics was performed as described previously (Goggs et al., 2013). Briefly, an UltiMate™3000 nano HPLC system in line with an LTQ-Orbitrap Velos mass spectrometer (Thermo Scientific) was used. The raw data files were processed and quantified using Proteome Discoverer software v1.2 (Thermo Scientific) and searched against UniProt C. jejuni strain 11168H database using the SEQUEST algorithm. The reverse database search option was enabled and all peptide data was filtered to satisfy false discovery rate (FDR) of 5%. Abundance of each protein in each sample was calculated using the average area measurements of the three most abundant peptides matching to each protein (Top3 method) (Ahrne et al., 2013). Normalization of the mass spectrometric data (protein abundances) was performed globally at protein level. Abundance of each protein was expressed as the fraction of the signal derived from the total abundance detected in each sample (Ting et al., 2009). This value was then compared for each protein in the penicillin G treated and control samples.
Proteins with significantly differential abundance were identified using the R packages limma and q-value (Smyth, 2004), when proteins with constant abundance across replicates were removed from the analysis. The former package was used for significance analysis, calculating p-values. The latter package was used for false discovery rate control, calculating q-values. Proteins with a q < 0.05 and more than 2-fold change difference in abundance were considered significant.
Online Tools
Cellular localization of the proteins encoded in the C. jejuni strain 11168H genome was predicted using PSORTb v.3.0.2 (https://www.psort.org/psortb/) (Yu et al., 2010). C. jejuni strain 11168H proteins were classified into functional categories based on clusters of orthologous gene (COG) designations; COG categories were assigned to each protein using eggNOG-mapper (http://eggnog-mapper.embl.de/) (Huerta-Cepas et al., 2017, 2019).
Results and Discussion
C. jejuni 11168H Forms Persister Cells
We exposed C. jejuni 11168H to penicillin G, because beta-lactam antibiotics were previously used successfully to reveal persister cells in other species (Balaban et al., 2019) and globally the penicillins are widely used in poultry farming, as growth promoters and to treat disease (Allen and Stanton, 2014; Manyi-Loh et al., 2018). Therefore, C. jejuni in the avian gut could be exposed to high doses of penicillin.
We grew C. jejuni 11168H in broth for 14 h (mid-log phase), and then exposed cultures to 100 × the MIC (11.43 μg/ml). At intervals the numbers of total cells, culturable cells, cells strained by BacLight™ and cells stained using RSG were determined for up to 48 h post dosing (Figure 1). Staining with RSG reveals metabolic activity because in metabolically active cells redox sensor green is reduced as a consequence of electron transport chain function, forming a green fluorescent dye (Jaen et al., 2019).
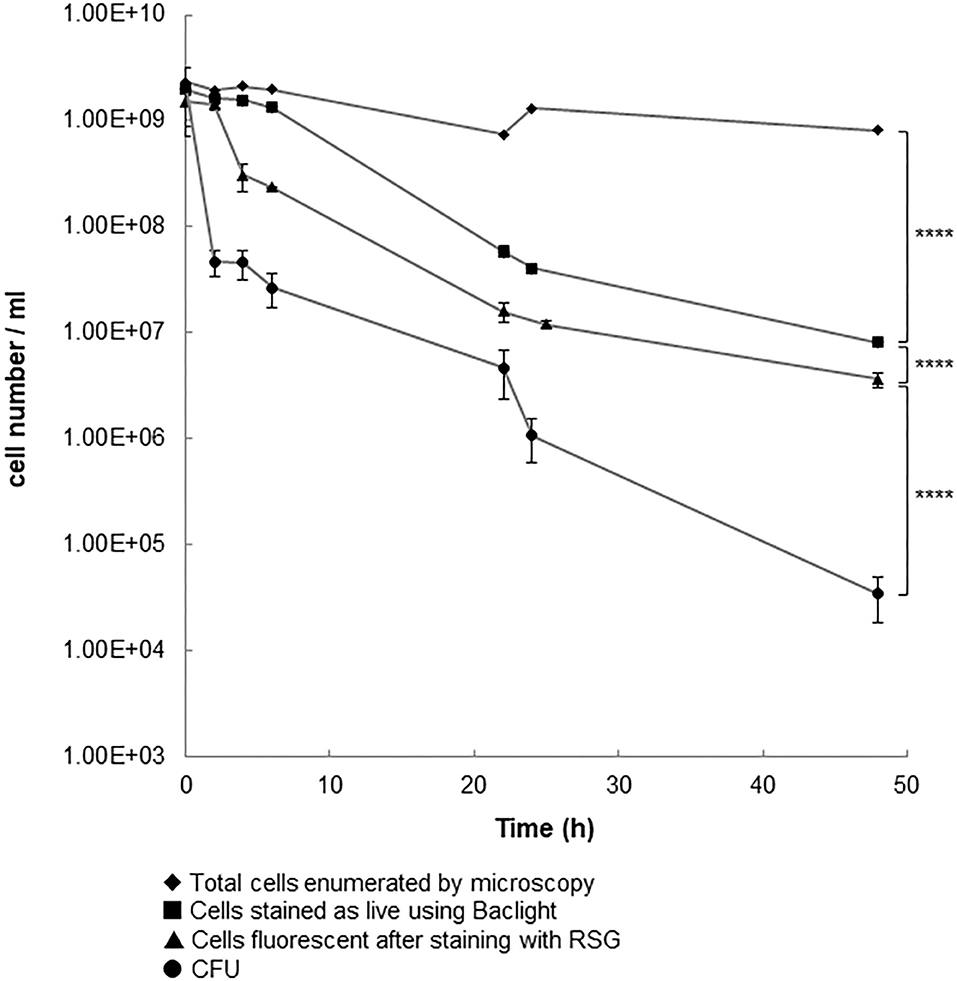
Figure 1. Cell populations following the addition of penicillin G to 14 h old cultures of C. jejuni 11168H. A log phase culture (14 h) of C. jejuni was incubated for 24 h with 100 × the MIC of penicillin G. Results shown are the mean of three replicates with bars corresponding to the standard error of the mean shown at each data point. ****p < 0.0001 (unpaired t-test).
The numbers of culturable cells (CFU) and of cells that fluoresced after staining with RSG showed bi-phasic reductions, which is characteristic of the killing of drug-sensitive cells followed by the much slower decline in the number of drug tolerant persister cells (Lewis, 2005). The numbers of cells that stained green with BacLight™ declined over the course of the experiment but did not show a biphasic reduction. When we used older cultures (24 or 48 h instead of than 14 h) for this experiment, we saw similar patterns of changes in total cells numbers, culturable cells, or BacLight™ stained cells (data not shown). The total number of cells did not show the same pattern of decline. When the population that survived exposure to antibiotic was re-cultured in fresh broth, we found that the MIC of penicillin G (11 μg/ml) was similar to that of the population at the start of the experiment. This is consistent with the non-inherited and antibiotic-tolerant phenotype of persister cells. Based on the number of cells that could be cultured after exposure of the population to 100 × MIC of penicillin G for 24 h, we calculate the persister cell frequency to be 5.25 × 10−4. It is reported (Lewis, 2007) that the frequency of persister cell formation in other bacterial species is typically 10−3 to 10−6 (Keren et al., 2004; Lewis, 2007). The high frequency with which C. jejuni 11168H forms persister cells might explain the ability of this bacterium to survive a wide range of environmental insults when exposed to antibiotics. Since our manuscript was submitted Ovsepian et al. (2020) reported that C. jejuni strains 81–176 and RM1221 form persister cells which are revealed by exposure to ciprofloxacin at frequencies of 10−5 to 10−7 after exposure to this drug for 22 h. This finding confirms that a range of strains of C. jejuni can form persister cells. However, these authors were unable to demonstrate persister cells resistant to 100 × MIC of ampicillin. This could indicate strain-specific differences in persister cell formation or differences in the responses to ampicillin or penicillin G exposure. Strain-specific differences in persister cell formation and differences in survival after exposure to different drugs is well-established (Fisher et al., 2017). Further work is required to investigate these possibilities in C. jejuni.
Flow Cytometry Reveals Three Populations After Exposure to Penicillin G
We next compared the flow cytometry histograms of cultures of C. jejuni 11168H that had been cultured for 14 h in MHB (T0 culture) with the histograms of cells that were subsequently incubated at 37°C under micro-aerobic conditions in either MHB or in MHB containing 100 × MIC of penicillin G. The histograms showed cells grown in MHB, had a normally distributed range of fluorescence signal intensities, ranging from dim to bright (Figure 2; T0). A small sub-population of cells were unstained.
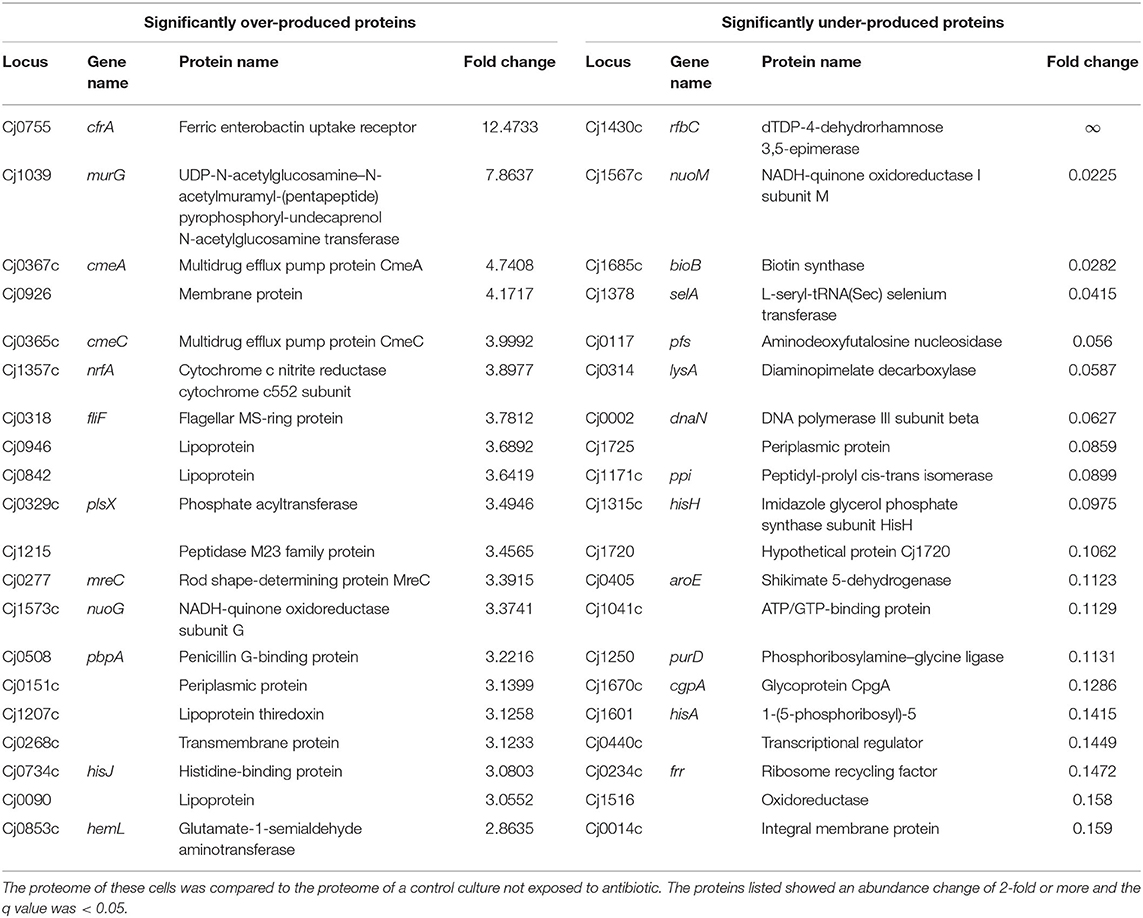
Table 1. Top 20 proteins significantly over-produced (left hand side) or under-produced (right hand side) in C. jejuni cells that strained brightly with RSG after exposure to 100 × MIC of penicillin G for 24 h.
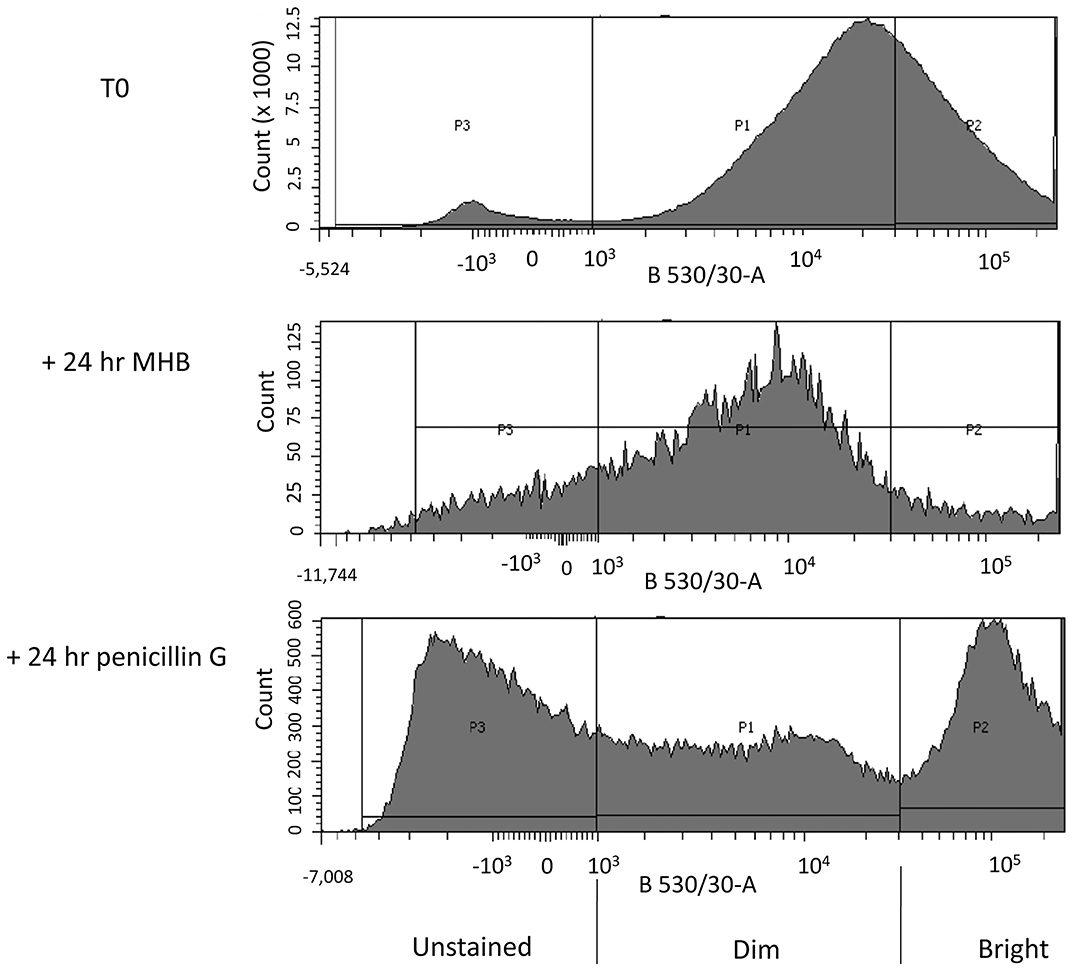
Figure 2. FACS profiles of C. jejuni 11168H after staining with RSG. Three biological replicates of each of the following: A log phase (14 h growth) C. jejuni culture (T0), a log phase culture incubated for a further 24 h in MHB (control + 24 h MHB) and a log phase culture incubated for a further 24 h in MHB containing 100 × MIC of penicillin G (+ 24 h Penicillin G), were stained with Redox Sensor Green (RSG). The stained cultures were then analyzed by FACS. Fluorescence intensity of RSG stained C. jejuni cells at 530 ± 30 nm (x-axis) after excitation with a 488 nm laser is plotted against bacterial cell count (y-axis). Different populations of cells were gated according to the intensity of the RSG fluorescence of each population (P1–stained, P2–brightly stained, and P3–unstained).
A parallel culture was not stained or analyzed by flow cytometry, and instead these cells were collected by centrifugation and re-suspended in fresh MHB and incubated for 24 h before staining with RSG and then analyzed using flow cytometry (Figure 2; +24 h MHB). In this antibiotic-free control we found a population of cells with a normally distributed range of fluorescence signal intensities from dim to bright. We found that the median signal intensity was reduced compared to the T0 culture.
In the test culture, we processed cells as detailed above but after centrifugation we re-suspended them in MHB containing 100 × MIC of penicillin G and then incubated for 24 h before staining with RSG. After flow cytometry analysis the penicillin G treated culture separated into three populations, labeled P1-P3 (Figure 2; +24 h penicillin G).
Proteomic Analysis of C. jejuni With Different RSG Fluorescence Signals
We next analyzed the proteomic makeup of three biological replicates of cells incubated for 24 h in MHB (control) or three biological replicates of cells incubated for 24 h in MHB containing 100 × MIC of penicillin G. The control or penicillin-treated cultures were stained with RSG and sorted using FACS into cells that did not fluoresce, cells that fluoresced weakly or cells that fluoresced brightly (Figure 2). We collected broadly similar numbers of cells (1–3 × 106) events in each of these groups and the collected cells were lysed and subjected to tryptic digests to release proteins and analyzed by mass spectrometry. Our results were also normalized according to the abundance of each protein relative to the total protein detected by mass spectrometry in each sample. The C. jejuni 11168H genome encodes 1,572 proteins and 95% (1,493) of these were detected in the combined untreated and penicillin G treated samples.
In the control cultures, we first compared the proteomes of the dimly stained cells and brightly stained cells. We did not identify differentially produced proteins, confirming that these were essentially the same population of cells. Considering that the dimly stained cells were the predominant population in the control culture, we next carried out a detailed comparison of the proteome of these cells with the proteome of the brightly stained cells in the penicillin G treated culture (Supplementary Table 1).
We identified 1,331 proteins in the dimly stained cells from the control culture and 1,217 proteins in the brightly stained cells from the penicillin G treated culture. A comparison of these two datasets (Table 1) revealed 44 significantly more abundant and 87 significantly less abundant proteins in the brightly stained cells from the penicillin G treated culture. These proteins were assigned to COG functional categories (Figure 3). The cell membrane/envelope biogenesis and the energy production and conversion groups had the greatest number of proteins with increased abundance. These proteins included the CmeA and CmeC components of the CmeABC efflux pump, which has previously been associated with antibiotic, bile, heavy metals and other antimicrobials in C. jejuni (Iovine, 2013). The upregulation of this efflux pump has also been reported after phage infection (Sacher et al., 2018). Our proteome analysis also highlighted MurG (Cj1039) to be over-produced, which catalysis the final intracellular step of peptidoglycan synthesis (Shaku et al., 2020), indicating a possible role of this enzyme in restoring cell wall integrity after exposure to penicillin G. The majority of the proteins with increased abundance were associated with translation and amino acid metabolism and transport.
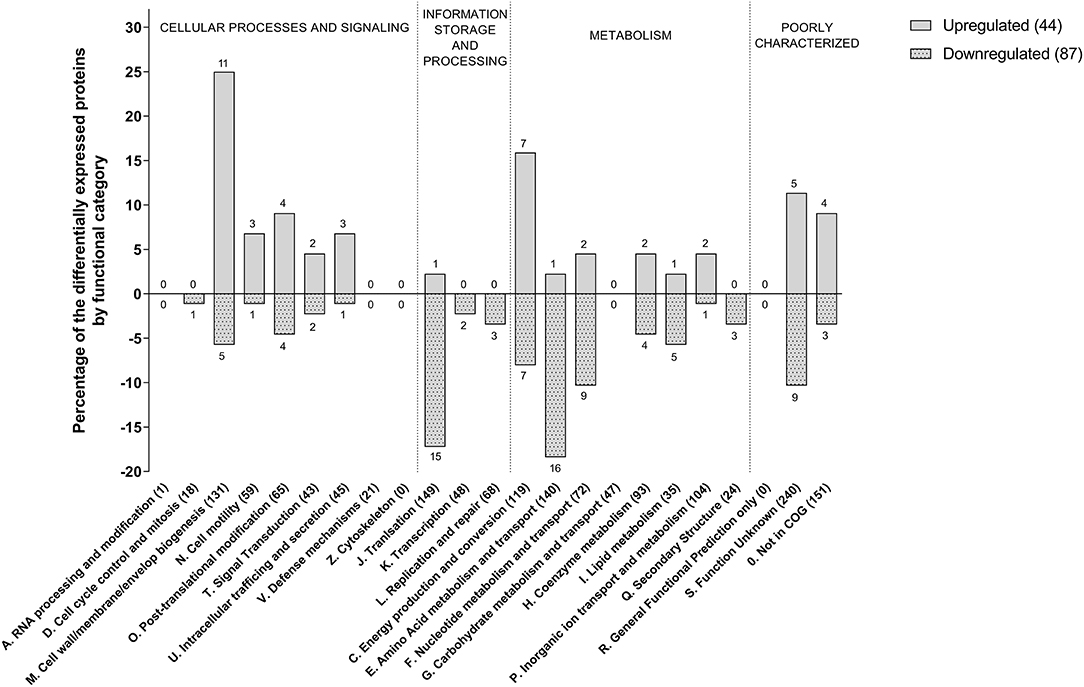
Figure 3. Functional COG categories of differentially produced proteins in penicillin G treated C. jejuni cells compared to untreated cells. Only significant produced proteins (q < 0.05) with abundance changes of 2-fold or more are shown.
The increase in RSG fluorescence signal from bright cells after treatment with penicillin G indicates reduction of the dye within the cell. RSG stain will fluoresce when modified by bacterial reductases (Jaen et al., 2019) which are predominantly located in, or associated with, the electron transport chain of the inner membrane.
Analysis of the proteome of the penicillin G treated cells reveals that a component (NuoG, Cj1573c) of an unusual flavodoxin driven membrane-bound quinone-reductase Complex I (an NADH-quinone oxidoreductase but lacking the NADH dehydrogenase module subunits nuoE and nuoF) (Weerakoon and Olson, 2008) and a component (SdhA Cj0437) of a fumarate reductase (Cj0437-0439) (Taylor and Kelly, 2019) (previously mis-annotated as a succinate dehydrogenase) are significantly over-produced. These two key respiratory complexes are responsible for electron transfer to/from the menaquinone pool during respiration. The β (AtpD Cj1355) and δ (AtpH Cj0104) subunits of the F1 ATP synthase are also significantly over-produced.
In addition, the periplasmic facing cytochrome c nitrite reductase (NrfA, Cj 1357c) (Baymukhametov et al., 2018) was over-produced in penicillin treated cells. It is a periplasmic respiratory enzyme that couples to the formate dehydrogenase (Cj1508-1511), via menaquinol oxidation, in order to generate a membrane potential (Δp). NrfA is a pentaheme containing c-type cytochrome that catalysis the six-electron reduction of nitrite to ammonium by receiving electrons from menaquinol (Sellars et al., 2002). In E. coli, the NrfA nitrite reductase is normally produced during both anoxic and micro-oxic conditions, and in addition to reducing nitrite has also been shown to play a defensive role in NO detoxification (Poock et al., 2002). Our results show that NrfA in C. jejuni 11168H is significantly over-produced in penicillin G treated cells with a fold change of 3.9, and could provide a route to an alternative electron acceptor and protect against nitrosative stress (Pittman et al., 2007). The combined effects of these over-produced redox proteins undoubtedly contributes to the enhanced RSG intensity seen in the antibiotic treated cells and indicate a remodeling of the electron transfer chain toward a less electrogenic process. A potential shift from hydrogen or formate oxidation (which provides the highest measurable values of membrane potential of the available electron donors) to utilizing the flavodoxin driven membrane-bound quinone-reductase (Complex I) could help to dissipate the high membrane potential. Similarly, using periplasmic facing electroneutral terminal reductases such as NrfA or fumarate reductase, that accept electrons directly from the menaquinol pool rather than a QCR (Cj1184c−1186c), will also function to reduce the Δp as their H+/e− = 0 (Taylor and Kelly, 2019). The net effect of these changes would modify the bioenergetics of the system and reduce the number of protons translocated per electron transfer, and thus aid to moderate Δp, ATP production and intracellular pH homeostasis. The reduction in the hyperpolarization of the cytoplasmic membrane could lead indirectly to restrain the ROS formation and reduce intracellular alkalization (Voskuil et al., 2018).
The efficacy of antibiotics being linked to changes in cellular respiration has been reported previously (Lobritz et al., 2015). In E. coli and S. aureus exposure to that treatment with bactericidal antibiotics gave rise to acceleration in respiration rate, and that inhibition of cellular respiration by creating a knockout mutant deficient in cytochrome c oxidase was sufficient to attenuate drug bactericidal activity. Furthermore, it was demonstrated that when the basal rate of electron transfer was accelerated, by uncoupling the electron transfer chain from ATP synthesis, the effectiveness of the antibiotic was increased. There is a general consensus that, for aerobes, the generation of reactive oxygen species (ROS) and the subsequent oxidative damage of many macromolecules is a known secondary effect of many bactericidal antibiotics including fluoroquinolones, beta-lactams and aminoglycosides (Dwyer et al., 2014).
The reason that potentiation of beta-lactam activity is less pronounced in bacteria grown at low oxygen levels may be because ROS levels are much lower under low oxygen aerobic conditions (Oh et al., 2015). Consistent with this suggestion, we (Hemsley et al., 2014) and others (Hamad et al., 2011) have shown that beta-lactam antibiotics are less effective in killing the Gram-negative bacterium Burkolderia pseudomallei under anaerobic compared to aerobic conditions. We would expect low levels of ROS in C. jejuni grown under microaerobic conditions. Also, the bacterium has a number of ROS detoxification enzymes (Taylor and Kelly, 2019) including; superoxide dismutase SodB (Cj0169), alkyl hydroxide reductase AhpC (Cj0334), catalase KatA (Cj1385), thiol peroxidases Tpx (Cj0779), bacterioferritin comigratory protein Bcp (Cj0271), two cytochrome c peroxidases (Cj0020c and Cj0358) and methionine sulfoxide reductases MsrA and MsrB (Cj0637 and Cj1112), none of which are significantly regulated upon treatment with penicillin G. Therefore, in summary we believe that unlike most bacteria studies to date, ROS do not play a major role in antibiotic killing in C. jejuni 11168H.
Persister cell formation in bacteria has previously been associated with reduced levels of metabolic activity and reduced membrane potential (Δp) and can be induced by perturbing the intracellular ATP levels. Although, recently it has been shown that the addition of salicylate can induce persister cell formation in E. coli via a mechanism that generates ROS (Wang et al., 2017). It has been suggested that the salicylate induces ROS generation and causes a decrease in membrane potential, which in turn leads to reduced metabolism and an increase in persistence. Further studies with E. coli using moderate levels of hydrogen peroxide (300–600 μM H2O2) as a direct source of ROS showed a protection against a lethal dose of ofloxacin by increasing persister cell formation by an order of magnitude (Vega et al., 2012). The metabolic burst we saw in penicillin G treated C. jejuni cells under microaerobic conditions could result from a remodeling of the electron transport chain to prevent hyperpolarization of the inner membrane and potentially curb intracellular alkalization and limit ROS formation. Therefore, the combination of these changes might play a broader role in supporting persister cell formation.
Conclusion
In this study we set out to investigate how antibiotics affect C. jejuni 11168H and how this in turn might influence environmental survival. We report that C. jejuni 11168H forms persister cells after exposure to penicillin G. Another important finding from our study is the appearance of a population of cells with increased levels of redox proteins in cells exposed to penicillin G, resulting in a greater signal from cells stained with RSG. It is not clear if cells with increased levels of redox proteins are a feature of persister cells and further work would be required to explore this possibility.
Data Availability Statement
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD021418.
Author Contributions
The project was conceived by DS, OC, OS, and RWT. Experimental work was carried out by HM, OC, and RKT. Flow cytometry was enabled and supported by JL and RKT. Data analysis was carried out by AK-S, CB, HM, RKT, RWT, and ZY. Manuscript writing was carried out AK-S, CB, DS, HM, OS, RKT, RWT, SW, and ZY. All authors have read and approved this manuscript.
Funding
HM, AK-S, and SW were supported in part by a BBSRC grant (BB/N016513/1) to SW, RWT, and CB.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank the University of Exeter BioCatalysis center for support with flow cytometry analysis. We thank Kate Heesom at the University of Bristol Proteomics Facility for carrying out Mass spectrophotometry.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2020.565975/full#supplementary-material
References
Ahrne, E., Molzahn, L., Glatter, T., and Schmidt, A. (2013). Critical assessment of proteome-wide label-free absolute abundance estimation strategies. Proteomics 13, 2567–2578. doi: 10.1002/pmic.201300135
Allen, H. K., and Stanton, T. B. (2014). Altered egos: antibiotic effects on food animal microbiomes. Annu. Rev. Microbiol. 68, 297–315. doi: 10.1146/annurev-micro-091213-113052
Alonso, J. L., Mascellaro, S., Moreno, Y., Ferrús, M. A., and Hernández, J. (2002). Double-staining method for differentiation of morphological changes and membrane integrity of Campylobacter coli cells. Appl. Environ. Microbiol. 68 5151–5154. doi: 10.1128/AEM.68.10.5151-5154.2002
Asuming-Bediako, N., Parry-Hanson Kunadu, A., Abraham, S., and Habib, I. (2019). Campylobacter at the human-food interface: the african perspective. Pathogens 8:87. doi: 10.3390/pathogens8020087
Balaban, N. Q., Helaine, S., Lewis, K., Ackermann, M., Aldridge, B., Andersson, D. I., et al. (2019). Definitions and guidelines for research on antibiotic persistence. Nat. Rev. Microbiol. 17, 441–448. doi: 10.1038/s41579-019-0196-3
Balaban, N. Q., Merrin, J., Chait, R., Kowalik, L., and Leibler, S. (2004). Bacterial persistence as a phenotypic switch. Science (New York, N.Y.) 305, 1622–1625. doi: 10.1126/science.1099390
Baymukhametov, T. N., Chesnokov, Y. M., Pichkur, E. B., Boyko, K. M., Tikhonova, T. V., Myasnikov, A. G., et al. (2018). Three-dimensional structure of cytochrome c nitrite reductase as determined by cryo-electron microscopy. Acta Naturae 10, 48–56. doi: 10.32607/20758251-2018-10-3-48-56
Bronowski, C., James, C. E., and Winstanley, C. (2014). Role of environmental survival in transmission of Campylobacter jejuni. FEMS Microbiol. Lett. 356, 8–19. doi: 10.1111/1574-6968.12488
Champion, O. L., Karlyshev, A. V., Senior, N. J., Woodward, M., La Ragione, R., Howard, S. L., et al. (2010). Insect infection model for Campylobacter jejuni reveals that O-methyl phosphoramidate has insecticidal activity. J. Infect. Dis. 201, 776–782. doi: 10.1086/650494
Chlebicz, A., and Slizewska, K. (2018). Campylobacteriosis, salmonellosis, yersiniosis, and listeriosis as zoonotic foodborne diseases: a review. Int. J. Environ. Res. Public Health 15:863. doi: 10.3390/ijerph15050863
Dwyer, D. J., Belenky, P. A., Yang, J. H., MacDonald, I. C., Martell, J. D., Takahashi, N., et al. (2014). Antibiotics induce redox-related physiological alterations as part of their lethality. Proc. Natl. Acad. Sci. U.S.A 111, E2100–E2109. doi: 10.1073/pnas.1401876111
Fisher, R. A., Gollan, B., and Helaine, S. (2017). Persistent bacterial infections and persister cells. Nat. Rev. Microbiol. 15, 453–464. doi: 10.1038/nrmicro.2017.42
Garcia-Sanchez, L., Melero, B., Jaime, I., Hanninen, M. L., Rossi, M., and Rovira, J. (2017). Campylobacter jejuni survival in a poultry processing plant environment. Food microbiology 65, 185–192. doi: 10.1016/j.fm.2017.02.009
Goggs, R., Harper, M. T., Pope, R. J., Savage, J. S., Williams, C. M., Mundell, S. J., et al. (2013). RhoG protein regulates platelet granule secretion and thrombus formation in mice. J. Biol. Chem. 288, 34217–34229. doi: 10.1074/jbc.M113.504100
Hamad, M. A., Austin, C. R., Stewart, A. L., Higgins, M., Vazquez-Torres, A., and Voskuil, M. I. (2011). Adaptation and antibiotic tolerance of anaerobic Burkholderia pseudomallei. Antimicrob. Agents Chemother. 55, 3313–3323. doi: 10.1128/AAC.00953-10
Harms, A., Maisonneuve, E., and Gerdes, K. (2016). Mechanisms of bacterial persistence during stress and antibiotic exposure. Science (New York, N.Y.) 354:aaf4268. doi: 10.1126/science.aaf4268
Harrison, J. J., Ceri, H., Roper, N. J., Badry, E. A., Sproule, K. M., and Turner, R. J. (2005). Persister cells mediate tolerance to metal oxyanions in Escherichia coli. Microbiology (Reading, England) 151, 3181–3195. doi: 10.1099/mic.0.27794-0
Harrison, J. J., Wade, W. D., Akierman, S., Vacchi-Suzzi, C., Stremick, C. A., Turner, R. J., et al. (2009). The chromosomal toxin gene yafQ is a determinant of multidrug tolerance for Escherichia coli growing in a biofilm. Antimicrob. Agents Chemother. 53, 2253–2258. doi: 10.1128/AAC.00043-09
He, Y., and Chen, C. Y. (2010). Quantitative analysis of viable, stressed and dead cells of Campylobacter jejuni strain 81-176. Food Microbiol. 27, 439–446. doi: 10.1016/j.fm.2009.11.017
Hemsley, C. M., Luo, J. X., Andreae, C. A., Butler, C. S., Soyer, O. S., and Titball, R. W. (2014). Bacterial drug tolerance under clinical conditions is governed by anaerobic adaptation but not anaerobic respiration. Antimicrob Agents Chemother. 58, 5775–5783. doi: 10.1128/AAC.02793-14
Huerta-Cepas, J., Forslund, K., Coelho, L. P., Szklarczyk, D., Jensen, L. J., von Mering, C., et al. (2017). Fast genome-wide functional annotation through orthology assignment by eggNOG-mapper. Mol. Biol. Evol. 34, 2115–2122. doi: 10.1093/molbev/msx148
Huerta-Cepas, J., Szklarczyk, D., Heller, D., Hernandez-Plaza, A., Forslund, S. K., Cook, H., et al. (2019). eggNOG 5.0: a hierarchical, functionally and phylogenetically annotated orthology resource based on 5090 organisms and 2502 viruses. Nucleic Acids Res. 47, D309–D314. doi: 10.1093/nar/gky1085
Igwaran, A., and Okoh, A. I. (2019). Human campylobacteriosis: a public health concern of global importance. Heliyon 5:e02814. doi: 10.1016/j.heliyon.2019.e02814
Iovine, N. M. (2013). Resistance mechanisms in Campylobacter jejuni. Virulence 4, 230–240. doi: 10.4161/viru.23753
Jaen, K. E., Velazquez, D., Delvigne, F., Sigala, J. C., and Lara, A. R. (2019). Engineering E. coli for improved microaerobic pDNA production. Bioprocess Biosyst. Eng. 42, 1457–1466. doi: 10.1007/s00449-019-02142-5
Jayaraman, R. (2008). Bacterial persistence: some new insights into an old phenomenon. J. Biosci. 33, 795–805. doi: 10.1007/s12038-008-0099-3
Keren, I., Kaldalu, N., Spoering, A., Wang, Y., and Lewis, K. (2004). Persister cells and tolerance to antimicrobials. FEMS Microbiol. Lett. 230, 13–18. doi: 10.1016/S0378-1097(03)00856-5
Kim, J. S., Lee, E. J., and Kim, Y. J. (2014). Inactivation of Campylobacter jejuni with dielectric barrier discharge plasma using air and nitrogen gases. Foodborne Pathog. Dis. 11, 645–651. doi: 10.1089/fpd.2013.1732
Kim, Y., Wang, X., Ma, Q., Zhang, X. S., and Wood, T. K. (2009). Toxin-antitoxin systems in Escherichia coli influence biofilm formation through YjgK (TabA) and fimbriae. J. Bacteriol. 191, 1258–1267. doi: 10.1128/JB.01465-08
Lewis, K. (2005). Persister cells and the riddle of biofilm survival. Biochemistry (Mosc) 70, 267–274. doi: 10.1007/s10541-005-0111-6
Lewis, K. (2007). Persister cells, dormancy and infectious disease. Nat. Rev. Microbiol. 5, 48–56. doi: 10.1038/nrmicro1557
Lewis, K. (2008). Multidrug tolerance of biofilms and persister cells. Curr. Topics Microbiol. Immunol. 322, 107–131. doi: 10.1007/978-3-540-75418-3_6
Ligowska, M., Cohn, M. T., Stabler, R. A., Wren, B. W., and Brondsted, L. (2011). Effect of chicken meat environment on gene expression of Campylobacter jejuni and its relevance to survival in food. Int. J. Food Microbiol. 145 (Suppl. 1), S111–S115. doi: 10.1016/j.ijfoodmicro.2010.08.027
Lobritz, M. A., Belenky, P., Porter, C. B., Gutierrez, A., Yang, J. H., Schwarz, E. G., et al. (2015). Antibiotic efficacy is linked to bacterial cellular respiration. Proc. Natl. Acad. Sci. U.S.A. 112, 8173–8180. doi: 10.1073/pnas.1509743112
Manyi-Loh, C., Mamphweli, S., Meyer, E., and Okoh, A. (2018). Antibiotic use in agriculture and its consequential resistance in environmental sources: potential public health implications. Molecules 23:795. doi: 10.3390/molecules23040795
Murphy, C., Carroll, C., and Jordan, K. N. (2006). Environmental survival mechanisms of the foodborne pathogen Campylobacter jejuni. J. Appl. Microbiol. 100, 623–632. doi: 10.1111/j.1365-2672.2006.02903.x
Oh, E., McMullen, L., and Jeon, B. (2015). Impact of oxidative stress defense on bacterial survival and morphological change in Campylobacter jejuni under aerobic conditions. Front. Microbiol. 6:295. doi: 10.3389/fmicb.2015.00295
Ovsepian, A., Larsen, M. H., Vegge, C. S., and Ingmer, H. (2020). Ciprofloxacin-induced persister-cells in Campylobacter jejuni. Microbiology. 166, 849–853. doi: 10.1099/mic.0.000953
Parkhill, J., Wren, B. W., Mungall, K., Ketley, J. M., Churcher, C., Basham, D., et al. (2000). The genome sequence of the food-borne pathogen Campylobacter jejuni reveals hypervariable sequences. Nature 403, 665–668. doi: 10.1038/35001088
Pittman, M. S., Elvers, K. T., Lee, L., Jones, M. A., Poole, R. K., Park, S. F., et al. (2007). Growth of Campylobacter jejuni on nitrate and nitrite: electron transport to NapA and NrfA via NrfH and distinct roles for NrfA and the globin Cgb in protection against nitrosative stress. Mol. Microbiol. 63, 575–590. doi: 10.1111/j.1365-2958.2006.05532.x
Poock, S. R., Leach, E. R., Moir, J. W., Cole, J. A., and Richardson, D. J. (2002). Respiratory detoxification of nitric oxide by the cytochrome c nitrite reductase of Escherichia coli. J. Biol. Chem. 277, 23664–23669. doi: 10.1074/jbc.M200731200
Rossler, E., Signorini, M. L., Romero-Scharpen, A., Soto, L. P., Berisvil, A., Zimmermann, J. A., et al. (2019). Meta-analysis of the prevalence of thermotolerant Campylobacter in food-producing animals worldwide. Zoonoses Public Health 66, 359–369. doi: 10.1111/zph.12558
Sacher, J. C., Flint, A., Butcher, J., Blasdel, B., Reynolds, H. M., Lavigne, R., et al. (2018). Transcriptomic analysis of the Campylobacter jejuni response to T4-like phage NCTC 12673 infection. Viruses 10:332. doi: 10.3390/v10060332
Schneider, C. A., Rasband, W. S., and Eliceiri, K. W. (2012). NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9, 671–675. doi: 10.1038/nmeth.2089
Sellars, M. J., Hall, S. J., and Kelly, D. J. (2002). Growth of Campylobacter jejuni supported by respiration of fumarate, nitrate, nitrite, trimethylamine-N-oxide, or dimethyl sulfoxide requires oxygen. J. Bacteriol. 184, 4187–4196. doi: 10.1128/JB.184.15.4187-4196.2002
Shaku, M., Ealand, C., Matlhabe, O., Lala, R., and Kana, B. D. (2020). Peptidoglycan biosynthesis and remodeling revisited. Adv. Appl. Microbiol. 112, 67–103. doi: 10.1016/bs.aambs.2020.04.001
Smyth, G. K. (2004). Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol. 3:3. doi: 10.2202/1544-6115.1027
Taylor, A. J., and Kelly, D. J. (2019). The function, biogenesis and regulation of the electron transport chains in Campylobacter jejuni: new insights into the bioenergetics of a major food-borne pathogen. Adv. Microb. Physiol. 74, 239–329. doi: 10.1016/bs.ampbs.2019.02.003
Ting, L., Cowley, M. J., Hoon, S. L., Guilhaus, M., Raftery, M. J., and Cavicchioli, R. (2009). Normalization and statistical analysis of quantitative proteomics data generated by metabolic labeling. Mol. Cell Proteomics 8, 2227–2242. doi: 10.1074/mcp.M800462-MCP200
Vega, N. M., Allison, K. R., Khalil, A. S., and Collins, J. J. (2012). Signaling-mediated bacterial persister formation. Nat. Chem. Biol. 8, 431–433. doi: 10.1038/nchembio.915
Voskuil, M. I., Covey, C. R., and Walter, N. D. (2018). Antibiotic lethality and membrane bioenergetics. Adv. Microb. Physiol. 73, 77–122. doi: 10.1016/bs.ampbs.2018.06.002
Wang, T., El Meouche, I., and Dunlop, M. J. (2017). Bacterial persistence induced by salicylate via reactive oxygen species. Sci. Rep. 7:43839. doi: 10.1038/srep43839
Weerakoon, D. R., and Olson, J. W. (2008). The Campylobacter jejuni NADH:ubiquinone oxidoreductase (complex I) utilizes flavodoxin rather than NADH. J. Bacteriol. 190, 915–925. doi: 10.1128/JB.01647-07
Wilmaerts, D., Windels, E. M., Verstraeten, N., and Michiels, J. (2019). General mechanisms leading to persister formation and awakening. Trends Genet 35, 401–411. doi: 10.1016/j.tig.2019.03.007
Yu, N. Y., Wagner, J. R., Laird, M. R., Melli, G., Rey, S., Lo, R., et al. (2010). PSORTb 3.0: improved protein subcellular localization prediction with refined localization subcategories and predictive capabilities for all prokaryotes. Bioinformatics 26, 1608–1615. doi: 10.1093/bioinformatics/btq249
Keywords: Campylobacter jejuni, proteomics, persister cell, antibiotic, electron transport
Citation: Morcrette H, Kovacs-Simon A, Tennant RK, Love J, Wagley S, Yang ZR, Studholme DJ, Soyer OS, Champion OL, Butler CS and Titball RW (2020) Campylobacter jejuni 11168H Exposed to Penicillin Forms Persister Cells and Cells With Altered Redox Protein Activity. Front. Cell. Infect. Microbiol. 10:565975. doi: 10.3389/fcimb.2020.565975
Received: 26 May 2020; Accepted: 11 September 2020;
Published: 19 October 2020.
Edited by:
Alessandra Piccirillo, University of Padua, ItalyReviewed by:
Martin I. Voskuil, University of Colorado Denver, United StatesJens Kreth, Oregon Health and Science University, United States
Daise Aparecida Rossi, Federal University of Uberlandia, Brazil
Roberta Torres Melo, Federal University of Uberlandia, Brazil
Copyright © 2020 Morcrette, Kovacs-Simon, Tennant, Love, Wagley, Yang, Studholme, Soyer, Champion, Butler and Titball. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Richard W. Titball, r.w.titball@exeter.ac.uk
 Helen Morcrette
Helen Morcrette Andrea Kovacs-Simon
Andrea Kovacs-Simon Richard K. Tennant
Richard K. Tennant John Love
John Love Sariqa Wagley
Sariqa Wagley Zheng R. Yang1
Zheng R. Yang1 David J. Studholme
David J. Studholme Clive S. Butler
Clive S. Butler Richard W. Titball
Richard W. Titball