
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
BRIEF RESEARCH REPORT article
Front. Cell. Infect. Microbiol. , 14 August 2020
Sec. Bacteria and Host
Volume 10 - 2020 | https://doi.org/10.3389/fcimb.2020.00416
The obligate intracellular bacterium Chlamydia trachomatis is the leading cause of bacterial sexually transmitted infections. Once internalized in host cells, C. trachomatis undergoes a biphasic developmental cycle within a membrane-bound compartment, known as the inclusion. Successful establishment of the intracellular niche relies on bacterial Type III effector proteins, such as Inc proteins. In vitro and in vivo systems have contributed to elucidating the intracellular lifestyle of C. trachomatis, but additional models combining the archetypal environment of infection with the advantages of in vitro systems are needed. Organoids are three-dimensional structures that recapitulate the microanatomy of an organ's epithelial layer, bridging the gap between in vitro and in vivo systems. Organoids are emerging as relevant model systems to study interactions between bacterial pathogens and their hosts. Here, we took advantage of recently developed murine endometrial organoids (EMOs) and present a C. trachomatis-murine EMO infection model system. Confocal microscopy of EMOs infected with fluorescent protein-expressing bacteria revealed that inclusions are formed within the cytosol of epithelial cells. Moreover, infection with a C. trachomatis strain that allows for the tracking of RB to EB transition indicated that the bacteria undergo a full developmental cycle, which was confirmed by harvesting infectious bacteria from infected EMOs. Finally, the inducible gene expression and cellular localization of a Chlamydia Inc protein within infected EMOs further demonstrated that this model is compatible with the study of Type III secreted effectors. Altogether, we describe a novel and relevant system for the study of Chlamydia-host interactions.
Chlamydia is the most commonly reported bacterial sexually transmitted disease in the world with rates continuing to increase annually (Centers for Disease Control and Prevention, 2019). The etiological agent, Chlamydia trachomatis, is an obligate intracellular bacterium that, after colonization of the lower female reproductive tract, ascends to infect the uterine endometrium and fallopian tubes. Currently, there is no vaccine available against Chlamydia (Phillips et al., 2019). Although antibiotics can be effective against C. trachomatis infections, 70–80% of infections in women are asymptomatic and thus left untreated (Geisler, 2010; Malhotra et al., 2013). Untreated infections can lead to pelvic inflammatory disease, ectopic pregnancy, and infertility (Haggerty et al., 2010). Following entry into a host epithelial cell, Chlamydia resides within a membrane-bound compartment, known as the inclusion (Gitsels et al., 2019). Within the inclusion, Chlamydia undergoes a biphasic developmental cycle alternating between two distinct bacterial forms (Moulder, 1991; Abdelrahman and Belland, 2005). Infection begins with the infectious, but non-replicative elementary body (EB). The EB-containing inclusion traffics toward the nucleus, and EBs transition to the replicative, but non-infectious form (reticulate body, RB). After multiple rounds of replication and ~24–36 h post-bacterial entry, the RBs asynchronously transition back to the EB form. After 2–3 days of infection, EBs are released from the host cell following cell lysis or by extrusion (Hybiske and Stephens, 2007; Zuck et al., 2016).
As an intracellular bacterium, Chlamydia must remodel the membrane of the inclusion and manipulate its host cell to establish its replication niche. This process is mediated via a number of bacterially-encoded Type III effector proteins (Ferrell and Fields, 2016; Bugalhão and Mota, 2019). Some of these effector proteins, known as inclusion membrane proteins (Inc), are translocated to and embedded within the inclusion membrane to mediate interaction between the inclusion and the host (Dehoux et al., 2011; Lutter et al., 2012; Moore and Ouellette, 2014; Bugalhão and Mota, 2019; Gitsels et al., 2019).
Much of what is known regarding Chlamydia's mechanisms to manipulate host cells comes from in vitro studies using cancer-derived cell lines, such as HeLa cells, which provide ease of maintenance and genetic manipulation, but lack much of the relevance of an in vivo environment (Dolat and Valdivia, 2019). Beyond cancer-derived cell lines, immortalized and polarized cell lines of reproductive tissues have broadened the relevance of in vitro systems (Igietseme et al., 1994; Guseva et al., 2005, 2007; Moore et al., 2008; King et al., 2009), and primary cells have provided systems with minimal artificial manipulation of cells (Prozialeck et al., 2002; Roth et al., 2010; Zadora et al., 2019; Maffei et al., 2020). Further, the use of three-dimensional (3D) and multi-cell type models have allowed for incorporating aspects of cell-cell interactions and tissue architecture (Nogueira et al., 2017; Edwards et al., 2019). Finally, whole tissue or ex vivo models are among the most relevant established systems, as they exhibit a more complete environment of infection (Hutchinson et al., 1979; Cooper et al., 1990; Roth et al., 2010; Jerchel et al., 2012; Kessler et al., 2012). However, they cannot be maintained long-term and are not easily accessible. Murine models to study pathogenesis and host immune response associated with Chlamydia infection are also commonly used, but the study of specific molecular mechanisms involved is limited (De Clercq et al., 2013). While each of the above-mentioned systems has its benefits, additional model systems that better combine the advantages of in vitro systems and in vivo models would be valuable to gain a better understanding of the molecular mechanisms underlying the interaction of C. trachomatis with its host cell.
Organoids present an ex vivo model system that bridges the gap between artificial in vitro studies and less tractable in vivo systems. Under niche-mimicking culture conditions, adult stem cells self-organize into 3D near-physiological structures termed organoids (Shamir and Ewald, 2014). The stemness of organoids results in a self-renewing system that can be cultivated indefinitely. Organoids have been generated from various tissue types, most notably the intestinal epithelium, and were originally used as a tool to study mammalian tissue biology and diseases (Dutta et al., 2017). In the last 6 years, the use of intestinal organoids was extended to the study of bacterial infection with Helicobacter pylori, Escherichia coli, Salmonella, and Clostridium difficile (Zhang et al., 2014; Forbester et al., 2015; Leslie et al., 2015; Bartfeld, 2016; In et al., 2016; Tao et al., 2016; Dutta and Clevers, 2017; Dutta et al., 2017; Pompaiah and Bartfeld, 2017; Pleguezuelos-Manzano et al., 2020).
Two female reproductive tract organoid models were recently developed and show promise for the study of sexually transmitted infections (STIs). First, human fallopian tube organoids were used to model chronic C. trachomatis infection (Kessler et al., 2015, 2019). Second, endometrial organoids (EMOs), which recapitulate the epithelial layer of the endometrium, defined as the innermost layer of the uterus, were established from both murine and human tissues (Boretto et al., 2017, 2019; Turco et al., 2017). EMOs are generated by culturing stem cells found within endometrial glands, which are proposed to be responsible for regenerating the epithelial cell layer of the endometrium (Gargett et al., 2016; Boretto et al., 2017; Turco et al., 2017; Cousins et al., 2018). EMOs contain both the ciliated epithelial cells and secretory cells present in the endometrial epithelial layer. Additionally, they maintain accurate cell polarity, cell-cell junctions, mucus production, and sex hormone response (Boretto et al., 2017; Turco et al., 2017). Despite the great potential of EMOs to increase the relevance of the study of STIs, they have not been used in the context of infections with pathogens.
Here, we describe a novel murine EMO model system to study Chlamydia infection. We demonstrate that C. trachomatis successfully infects and completes its developmental cycle within murine EMOs. In addition, we show that this system is compatible with inducible gene expression and detection of tagged Chlamydia Inc proteins by immunofluorescence microscopy. Together, our data show that the C. trachomatis-EMO model system has the potential to preserve the tools of in vitro models while contributing aspects of the cellular environment of in vivo systems.
All genetic manipulations and containment work were approved by the UVA Biosafety Committee and are in compliance with the section III-D-1-a of the National Institutes of Health guidelines for research involving recombinant DNA molecules. Mice were not bred for the sole purpose of harvesting endometrial tissues. Endometrial tissues for EMO generation was sourced from otherwise discarded murine reproductive tracts.
HeLa cells were obtained from the ATCC (CCL-2) and cultured at 37°C with 5% CO2 in high-glucose Dulbecco's modified Eagle's medium (DMEM; Invitrogen) supplemented with 10% heat-inactivated fetal bovine serum (FBS; Invitrogen). C. trachomatis Lymphogranuloma venereum, Type II were obtained from ATCC (L2/434/Bu VR-902B). C. trachomatis strain expressing mCherry alone and the RB-to-EB transition fluorescent reporter strain were described previously (Cortina et al., 2019). The C. trachomatis strain expressing mCherry constitutively and IncV-3xFLAG under the control of the anhydrotetracycline inducible promoter was described previously (Stanhope et al., 2017). C. trachomatis propagation and HeLa cell infection was performed as previously described (Derré et al., 2007).
The method for generating murine endometrial organoids was adapted from Boretto et al. as follows (Boretto et al., 2017). Uterine horns were isolated from otherwise discarded genital tract tissue from naïve 6–8 week-old female C57BL/6J mice, bisected, and minced. Tissue fragments were incubated in 30 mM ethylenediaminetetraacetic acid (EDTA) in PBS for 1 h at 37°C followed by mechanical dissociation in cold PBS. Dissociated tissue and supernatant were passed through a 70 μm cell strainer (Corning) to isolate endometrial glands and supplemented with 10% heat inactivated FBS. Isolated glands in PBS/FBS were centrifuged at 230 g for 5 min, and the pellet was resuspended in DMEM. The sample was centrifuged at 230 g for 5 min, and the pellet was resuspended in mixture of 70% growth factor reduced Matrigel (Corning) in DMEM. Ten to twenty microliters drops of the suspension were plated onto pre-warmed non-treated 24 well plates (1–2 drops per well) and incubated at 37°C for ~20–30 min to allow for Matrigel solidification. Mouse IntestiCultTM organoid growth medium (STEMCELL Technologies) supplemented with 1X penicillin-streptomycin-glutamine (PSQ; Gibco) was added to cover the Matrigel drops. Cultures were maintained at 37°C with 5% CO2, and culture medium was changed every 2 days. EMOs were visible within 2–5 days.
EMOs were passaged every 7–14 days. Culture medium was removed from the wells, and TrypLETM Express Enzyme (1X) without phenol red (Gibco) was added for 15 min at 37°C. TrypLETM was inactivated by addition of DMEM. EMOs were collected and centrifuged at 230 g for 5 min at 4°C. The pellet was resuspended in DMEM and subjected to mechanical disruption to fully dissociate EMOs into single cells. Dissociated cells were centrifuged at 230 g for 5 min at 4°C, and the pellet was resuspended in 70% Matrigel in DMEM and plated as described above. Culture medium supplemented with 1X PSQ and 10 μM Y-27632 (ROCK inhibitor; Millipore Sigma) was added. Culture medium containing ROCK inhibitor was used for the first subsequent medium change, but removed for additional medium changes.
One day prior to infection, culture medium was replaced with medium free of PSQ. At the time of infection, culture medium was removed, and cold PBS was added to dissolve Matrigel, followed by vigorous pipetting to break apart EMOs. The fragmented EMOs were incubated in DMEM for 15 min at 37°C for 5% CO2, followed by centrifugation at 290 g for 5 min. The pellet of ~50–100 fragmented EMOs was incubated with ~5 × 109 inclusion forming units (IFU) of the indicated strain of C. trachomatis in 100 μl of DMEM for 1 h at 37°C to allow for infection. Matrigel was added to generate a 70% Matrigel mixture which was plated as described above. IntestiCultTM without PSQ or Rock inhibitor was used.
Uninfected or infected EMOs embedded in Matrigel were plated in eight well-chambered coverglass wells (Thermo Scientific Nunc). At the indicated times, EMOs were fixed in 1X PBS containing 4% paraformaldehyde for 1 h at room temperature (RT). Antibodies and fluorescent dyes were diluted in PBS containing 0.5% Triton X-100 and 5% bovine serum albumin (BSA). Incubation with primary and secondary antibodies was conducted overnight at 4°C and 1–2 h at RT, respectively. Primary and secondary staining of fixed samples was followed by three washes with 1X PBS.
For Figure 3B, HeLa cells seeded onto glass coverslips were fixed for 20 min in PBS containing 4% paraformaldehyde. Coverslips were stained with Hoechst dye diluted in PBS containing 0.1% Triton X-100 for 1 h at RT, followed by three washes with 1X PBS. Coverslips were mounted with DABCO antifade-containing mounting media.
Samples were imaged with a Nikon epifluorescence microscope or a Leica DMi8 confocal microscope equipped with the Andor iXon ULTRA 888BV EMCCD camera and driven by the IQ software. Where indicated, live samples were imaged in a humidified live cell environmental chamber set at 37°C and 5% CO2. Images were processed using the Imaris software (Bitplane, Belfast, United Kingdom).
The following primary antibodies and dyes were used: goat polyclonal anti-Chlamydia major outer membrane protein (MOMP; 1:200, Virostat) and mouse monoclonal anti-FLAG (1:1,000, Sigma). The following secondary antibodies were used: Alexa FluorTM 488-conjugated donkey anti-goat IgG (1:500, Molecular Probes) and Alexa FluorTM 488- or 594-conjugated goat anti-mouse IgG (1:500, Molecular Probes). The following dyes were used: CellMaskTM Green (1:200, Invitrogen), SYTOXTM Green (1:10,000, Invitrogen), SYTOXTM Blue (1:5,000, Invitrogen), Alexa FluorTM 594 Phalloidin (1:200, Molecular Probes), Hoechst (1:1,000, Molecular Probes), and TO-PROTM-3 Iodide (1:1,000, Invitrogen).
EMOs were infected with mCherry-expressing C. trachomatis as described above. At 48 hpi (hours post-infection), the culture medium was removed and replaced with cold water to dissolve the Matrigel, followed by vigorous pipetting to break apart and lyse epithelial cells of the EMOs and release the bacteria. Bacteria and unlysed cells were collected and centrifuged at 8,000 rpm for 20 min at 4°C. Pelleted sample was resuspended in cold water, followed by additional vigorous pipetting and centrifugation with the same conditions to ensure that all infected cells were lysed. Pelleted sample was resuspended in RT DMEM supplemented with 10% FBS and plated onto a confluent HeLa cell monolayer, followed by centrifugation at 1,200 rpm for 20 min at RT. HeLa cells were incubated with harvested EBs for 32 h at 37°C in the presence of 5% CO2, at which point samples were fixed, stained, and imaged as described above.
EMOs were infected with C. trachomatis expressing mCherry constitutively and IncV-3xFLAG under the control of the anhydrotetracycline inducible promoter, as described above. At 24 hpi, the culture medium was replaced with culture medium supplemented with 20 ng/ml aTc. Infected EMOs were incubated in the presence of aTc at 37°C with 5% CO2 for 6 h, at which point samples were fixed, stained, and imaged as described above.
Images presented in this study are representative of multiple infected EMOs analyzed from at least three independent experiments performed with independent EMO preparations.
Murine endometrial organoids (EMOs) were generated by adaptation of a previously established protocol (Boretto et al., 2017). Uterine horns excised from otherwise discarded genital tract tissues of naïve mice were bisected and minced to expose the endometrium (Figure 1A). Endometrial glands, which contain the stem and progenitor cells that replenish the endometrium during the estrous cycle (Gargett et al., 2016; Cousins et al., 2018), were isolated by chemical dissociation of the tissue, embedded within a 3D extracellular matrix, Matrigel, and cultured in the presence of growth factors. Within ~7 days, mature EMOs were observed (Figure 1B). EMOs were then passaged and expanded every 7–14 days by dissociation into individual cells by trypsin digest and re-embedding in Matrigel. Live confocal imaging of EMOs, stained with CellMaskTM to visualize the plasma membrane of individual epithelial cells, revealed the previously observed 3D oblate spheroid structure composed of a single cellular layer surrounding a hollow lumen (Figures 1C–E).
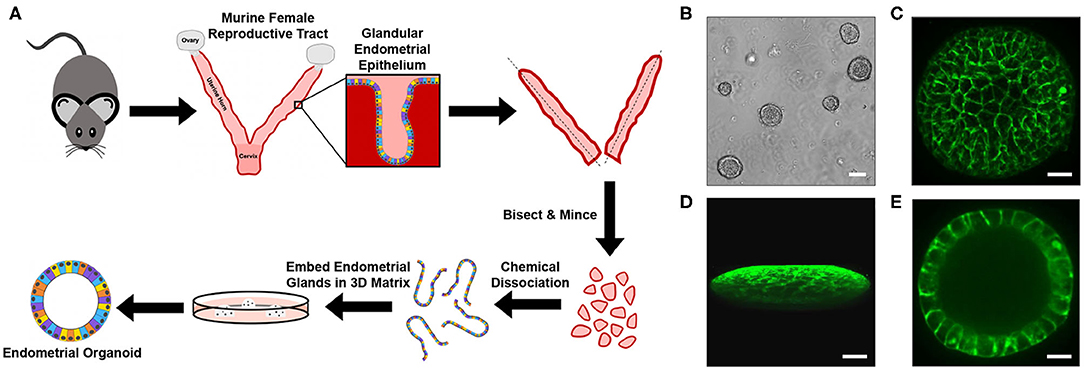
Figure 1. Generation of murine EMOs. (A) Cartoon diagram depicting the method used to generate EMOs from female C57BL/6J mice. (B) Brightfield image of mature murine EMOs embedded in Matrigel following passage and expansion. Scale bar: 80 μm. (C–E) Confocal micrographs of a live EMO stained with CellMaskTM Green to mark plasma membranes. A top view of a 3D reconstruction of an EMO (C), a side view of a 3D reconstruction of an EMO (D), and a single plane across the middle of an EMO (E) are shown. Scale bar: 50 μm.
As Chlamydia encounters the apical surface of the endometrial epithelium during natural infection, one obstacle to EMO infection is the polarization of the epithelial cells with the apical side facing the lumen (Boretto et al., 2017). To circumvent this problem, 3D EMOs were mechanically fragmented prior to incubation with mCherry-expressing C. trachomatis for 1 h (Figure 2A). Infected EMO fragments were then embedded in Matrigel and incubated for 32 h to allow for 3D EMOs to reform and the bacteria to replicate. Immunostaining of C. trachomatis major outer membrane protein (MOMP) revealed the presence of bacteria-filled inclusions within the cellular layer of EMOs (Figure 2B). To determine the specific localization of inclusions, infected EMOs were fixed and stained with SYTOXTM Green to visualize nuclei (Figure 2C, green) and red fluorescent Phalloidin to visualize cortical actin and microvilli, thereby allowing detection of cell boundaries (Figure 2C, red). Inclusions, which were indicated by mCherry fluorescence and SYTOXTM Green staining, were detected within the cytosol of individual cells. Together, these data indicate that C. trachomatis successfully infects murine EMOs and forms inclusions within the cytosol of epithelial cells.
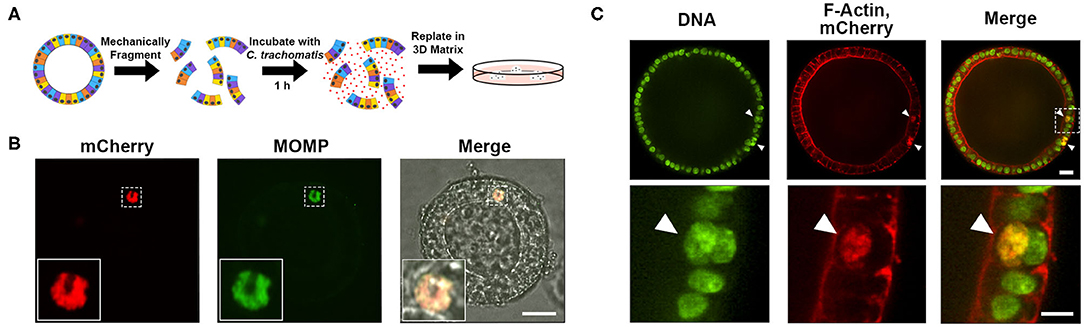
Figure 2. Infection of murine EMOs with C. trachomatis. (A) Cartoon diagram depicting the method of infecting EMOs with C. trachomatis by mechanical fragmentation. (B) Confocal micrographs of an EMO infected with mCherry-expressing C. trachomatis (red, left panel, mCherry). Infected EMOs were fixed at 32 hpi and immunostained with anti-C. trachomatis MOMP (green, middle panel, MOMP). The merge, including a brightfield image, is shown on the right. Higher magnification of the indicated boxed area is shown in the inset of each panel. A single plane across the middle of the inclusion is shown. Scale bar: 40 μm. (C) Confocal micrographs of an EMO infected with mCherry-expressing C. trachomatis (red, middle panels, mCherry) fixed at 32 hpi and stained with SYTOXTM Green (green, left panels, DNA) and Phalloidin (red, middle panels, F-Actin). The merge is shown on the right. A single plane across the middle of the EMO and inclusions is shown. Top panels: whole EMO. Scale bar: 40 μm. Bottom panels: higher magnification of indicated boxed area. Inclusions are indicated by arrow heads. Scale bar: 20 μm.
To determine if C. trachomatis underwent a complete developmental cycle within EMOs, we used our fluorescent reporter strain of C. trachomatis that allows for tracking of the RB to EB transition (Cortina et al., 2019). This strain expresses the mCherry fluorescent protein under the control of the groESL promoter, resulting in constitutive expression throughout the developmental cycle. It also expresses the green fluorescent protein (GFP) under the control of the late omcA promoter, which is expressed in the middle of the developmental cycle as bacteria transition from the RB to EB form (Shaw et al., 2000; Belland et al., 2003). At 24 h post-infection (hpi) murine EMOs infected with the RB-to-EB reporter strain displayed inclusions that were positive for mCherry with very few bacteria expressing GFP (Figure 3A, 24 hpi). At 48 hpi, inclusions had grown in size and were filled with GFP-expressing bacteria, indicating that RBs had successfully transitioned to EBs (Figure 3A, 48 hpi). To independently confirm the production of infectious EBs, EMOs were infected with mCherry-expressing C. trachomatis, and bacteria harvested from EMOs 48 hpi were used to infect HeLa cells (Figure 3B). The presence of mCherry-positive inclusions within the HeLa cell monolayer indicated that infectious EBs were indeed produced within EMOs. Altogether, these data demonstrate that C. trachomatis undergoes a full developmental cycle and generates infectious progeny within murine EMOs.
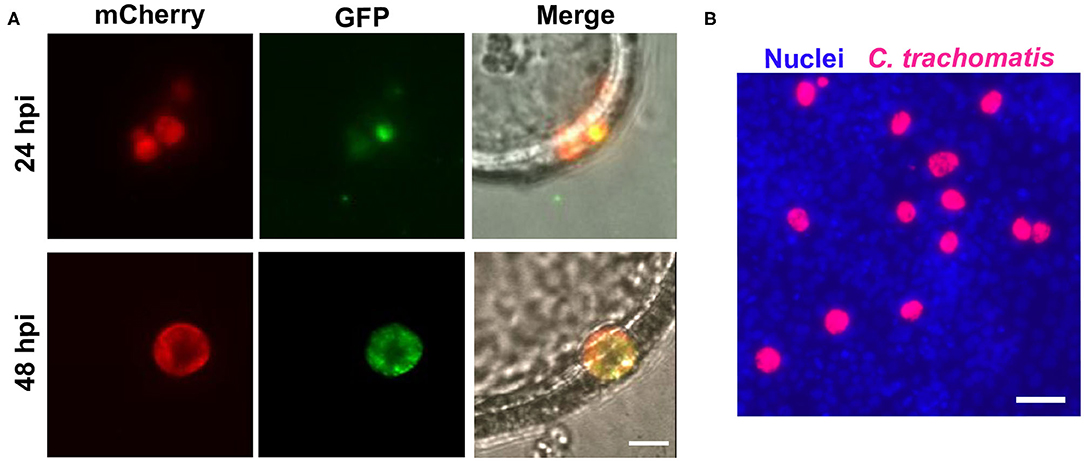
Figure 3. C. trachomatis completes a full developmental cycle in murine EMOs. (A) Live epifluorescence micrographs of two different EMOs infected with the RB-to-EB reporter strain of C. trachomatis for 24 h (top panels) or 48 h (bottom panels). Bacteria at the RB stage expressing mCherry alone are shown in red (left panels, mCherry), and bacteria that have transitioned to EB stage expressing GFP are shown in green (middle panels, GFP). The merge, including a brightfield image, is shown on the right. A single plane across the middle of the EMO and inclusions is shown. Scale bar: 10 μm. (B) Immunofluorescence image of a HeLa cell monolayer infected with mCherry-expressing C. trachomatis (red) collected from infected EMOs at 48 hpi. At 32 hpi, the infected HeLa cells were fixed and stained with the Hoechst DNA dye (blue). Scale bar: 40 μm.
Chlamydia relies on inclusion-localized Inc proteins to facilitate direct interaction between host cell proteins and the inclusion membrane (Dehoux et al., 2011; Lutter et al., 2012; Moore and Ouellette, 2014; Bugalhão and Mota, 2019; Gitsels et al., 2019). Strains of C. trachomatis expressing tagged Inc proteins under an inducible promoter have become a valuable tool to assess the localization and function of Inc proteins during infection (Agaisse and Derré, 2014; Bauler and Hackstadt, 2014; Mirrashidi et al., 2015; Weber et al., 2015; Han and Derré, 2017; Nguyen et al., 2018). With this system, the expression of the inc gene of interest is induced by the addition of anhydrotetracycline (aTc), and the corresponding protein is detected by immunofluorescence microscopy. To test if this system was applicable to EMOs, EMOs were infected with C. trachomatis expressing mCherry constitutively and IncV-3xFLAG under the control of the aTc inducible promoter. At 24 hpi, IncV-3xFLAG expression was induced for 6 h followed by fixation and immunostaining with anti-FLAG antibodies. Confocal microscopy revealed that IncV-3xFLAG expression was successfully induced within EMOs. As exemplified by Figure 4A, multiple inclusions within an infected EMO were positive for the IncV-3xFLAG signal. This result was also confirmed by imaging a single Z plane across an inclusion (Figure 4B). Moreover, we note the crescent shape pattern of the IncV signal on one side of the inclusion, which is in agreement with the previously described inclusion localization of IncV in HeLa cells (Stanhope et al., 2017). The ability to induce and detect the expression of IncV-3xFLAG in infected EMOs demonstrates that the EMO model system is compatible with the study of the cellular localization of Chlamydia inclusion membrane proteins and, most likely, effector proteins in general.
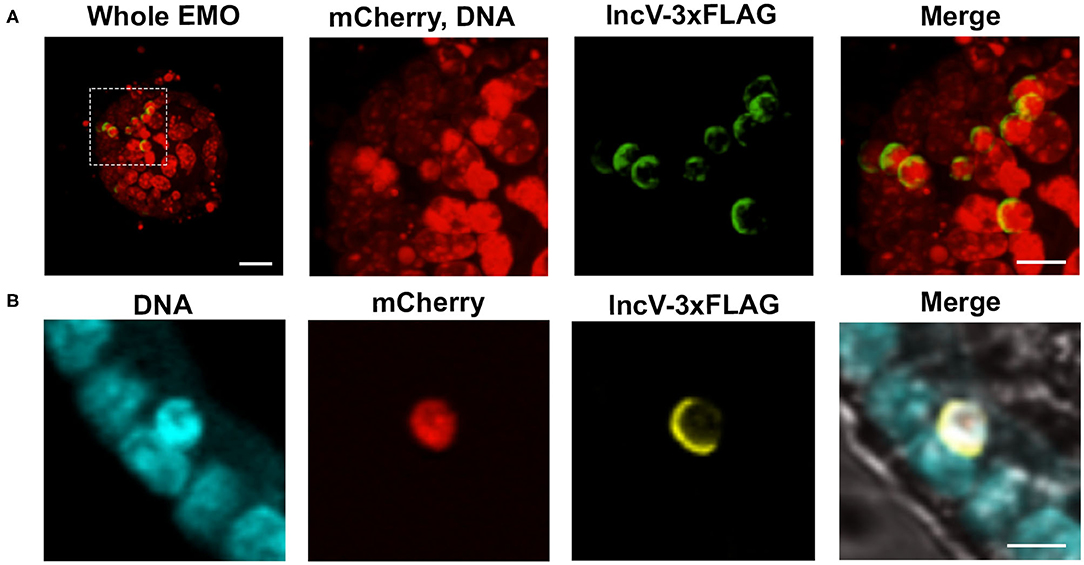
Figure 4. Induction of gene expression and inclusion localization of an Inc protein within infected murine EMOs. (A) 3D confocal micrographs of an EMO infected for 30 h with mCherry-expressing C. trachomatis constitutively (red) and IncV-3xFLAG under the control of an aTc inducible promoter (green), and stained with TO-PROTM-3 (red, DNA). A low magnification merge image corresponding to the whole EMO is shown on the left (Whole EMO). Scale bar: 40 μm. A high magnification of individual channels and the merge image of the indicated boxed area is shown in the three panels to the right. Scale bar: 20 μm (B) Confocal micrographs of an EMO infected for 30 h with C. trachomatis expressing mCherry constitutively (red) and IncV-3xFLAG under the control of an aTc inducible promoter (yellow), stained with SYTOXTM Blue (blue, DNA) and antibodies against FLAG (yellow, IncV-3xFLAG). The merge, including a brightfield image, is shown on the right. A single plane across the middle of the EMO and inclusion is shown. Scale bar: 20 μm.
Much of the current understanding of the molecular processes used by Chlamydia to interact with its host cell comes from in vitro systems (Dolat and Valdivia, 2019). Although these models are more easily maintained and manipulated than in vivo models, they do not fully recapitulate the cell-cell interactions and 3D nature present in the natural environment of the infected tissues. EMOs present an ex vivo model system that bridges the gap between less relevant in vitro studies and less tractable in vivo systems. Our data demonstrate that murine EMOs can serve as a novel model system in which to study Chlamydia host-pathogen interactions.
Our methods described here for generating and infecting murine EMOs were developed to be accessible and without the need for specialized equipment. The largest barrier to infection of EMOs with C. trachomatis is access to the cellular apical surface. A number of different methods have been developed for bacterial infection of organoids, including generation of 2D organoid monolayers and microinjection of bacteria into organoid lumens (Dutta et al., 2017; Yin and Zhou, 2018). Microinjection is especially appropriate for invasion or early infection studies and offers the advantage of minimally disrupting the integrity of the EMO at the time of infection. It also allows for a tight control of the number of bacteria delivered in the EMO lumen. Another method, which was used here, is infection of mechanically fragmented organoids. This approach leads to a temporary disruption of the 3D architecture of the EMOs and does not allow for a precise control of the MOI, which would preclude it from invasion studies. Nevertheless, the cellular organization of each EMO fragment is preserved and post-invasion stages of the infection can be monitored in the 3D environment of the primary cell epithelium of the resealed EMO. This method is also less technically challenging and does not require any special equipment.
Our analysis of C. trachomatis infected EMOs indicated that Chlamydia undergoes a productive developmental cycle in EMOs. Beyond 48 h of infection, we observed ruptured inclusions (not shown), however this phenomenon was not accompanied by the formation of secondary inclusions 72 h post-infection. Further studies will be required to determine if this phenomenon is due to technical limitations of keeping infected EMOs alive for a long period of time or reflective of the response of EMOs to infection and/or inclusion lysis.
The model presented here complements a recently described human fallopian tube organoid model, which has been used to address tubal pathology during chronic Chlamydia infection (Kessler et al., 2015, 2019). The fallopian tube organoid and EMO models together provide the ability to observe mechanisms of chlamydial infection in the relevant upper female genital tract tissue. Moreover, generation of human EMOs to model Chlamydia infection would further enhance the relevance of the EMO model. Human EMOs have been described (Boretto et al., 2017, 2019; Turco et al., 2017), and the methods developed here for infecting murine EMOs could be easily translatable.
Genetic methods can further develop this model and expand its use in studying pathogenesis of C. trachomatis. Genetic manipulation of EMOs using lentiviral vector systems would incorporate the genetic tractability of traditional cancer-derived cell lines. Such systems have already been developed with intestinal organoids and could be adapted for EMOs, as techniques for lentiviral infection are similar to the methods described here for C. trachomatis infection (Maru et al., 2016). Another method to improve the genetic tractability of EMOs is the use of genetically modified mice for primary endometrial tissue. Boretto et al. successfully generated EMOs from TdTomato knock-in mice, demonstrating the possibility of using the power of mouse genetics with this system (Boretto et al., 2017).
Together, our results indicate that murine EMOs can be used as a novel system with which to study C. trachomatis infection. This model offers a simple system to study direct interactions between C. trachomatis and the epithelial cells of the endometrium, providing the relative ease of maintenance and microscopy methods of in vitro cell culture, as well as the tissue architecture and relevant cell types of an in vivo model.
The datasets generated for this study are available upon request to the corresponding author.
RB performed the experiment. ID supervised the study. RB and ID conceived the experiments, analyzed the data, and wrote the manuscript. MR provided the endometrial tissue. MR, MB, and HV provided expertise and edited the manuscript. All authors contributed to the article and approved the submitted version.
This work was supported by National Institute of Allergy and Infectious Diseases grants AI007046 to RB and AI146649 and AI101441 to ID. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We thank Dr. Laura Shankman and Samantha Fleury (University of Virginia) for assistance with organoid culturing techniques and Francesca Azar (University of Virginia) for assistance in providing otherwise discarded murine endometrium tissues. We also thank Dr. Hervé Agaisse, Dr. Marìa-Eugenia Cortina, and Rachel Ende (University of Virginia) for critical reading of the manuscript.
Abdelrahman, Y. M., and Belland, R. J. (2005). The chlamydial developmental cycle. FEMS Microbiol. Rev. 29, 949–959. doi: 10.1016/j.femsre.2005.03.002
Agaisse, H., and Derré, I. (2014). Expression of the effector protein IncD in Chlamydia trachomatis mediates recruitment of the lipid transfer protein CERT and the endoplasmic reticulum-resident protein VAPB to the inclusion membrane. Infect. Immun. 82, 2037–2047. doi: 10.1128/IAI.01530-14
Bartfeld, S. (2016). Modeling infectious diseases and host-microbe interactions in gastrointestinal organoids. Dev. Biol. 420, 262–270. doi: 10.1016/j.ydbio.2016.09.014
Bauler, L. D., and Hackstadt, T. (2014). Expression and targeting of secreted proteins from Chlamydia trachomatis. J. Bacteriol. 196, 1325–1334. doi: 10.1128/JB.01290-13
Belland, R. J., Zhong, G., Crane, D. D., Hogan, D., Sturdevant, D., Sharma, J., et al. (2003). Genomic transcriptional profiling of the developmental cycle of Chlamydia trachomatis. Proc. Natl. Acad. Sci. U.S.A. 100, 8478–8483. doi: 10.1073/pnas.1331135100
Boretto, M., Cox, B., Noben, M., Hendriks, N., Fassbender, A., Roose, H., et al. (2017). Development of organoids from mouse and human endometrium showing endometrial epithelium physiology and long-term expandability. Development 144, 1775–1786. doi: 10.1242/dev.148478
Boretto, M., Maenhoudt, N., Luo, X., Hennes, A., Boeckx, B., Bui, B., et al. (2019). Patient-derived organoids from endometrial disease capture clinical heterogeneity and are amenable to drug screening. Nat. Cell Biol. 21, 1041–1051. doi: 10.1038/s41556-019-0360-z
Bugalhão, J. N., and Mota, L. J. (2019). The multiple functions of the numerous Chlamydia trachomatis secreted proteins: the tip of the iceberg. Microb. Cell 6, 414–449. doi: 10.15698/mic2019.09.691
Centers for Disease Control and Prevention (2019). Sexually Transmitted Disease Surveillance 2018. Atlanta, GA: U.S. Department of Health and Human Services. doi: 10.15620/cdc.79370
Cooper, M. D., Rapp, J., Jeffery-Wiseman, C., Barnes, R. C., and Stephens, D. S. (1990). Chlamydia trachomatis infection of human fallopian tube organ cultures. J. Gen. Microbiol. 136, 1109–1115. doi: 10.1099/00221287-136-6-1109
Cortina, M. E., Ende, R. J., Bishop, R. C., Bayne, C., and Derré, I. (2019). Chlamydia trachomatis and Chlamydia muridarum spectinomycin resistant vectors and a transcriptional fluorescent reporter to monitor conversion from replicative to infectious bacteria. PLoS One 14:e0217753. doi: 10.1371/journal.pone.0217753
Cousins, F. L., Dorien, F. O., and Gargett, C. E. (2018). Endometrial stem/progenitor cells and their role in the pathogenesis of endometriosis. Best Pract. Res. Clin. Obstet. Gynaecol. 50, 27–38. doi: 10.1016/j.bpobgyn.2018.01.011
De Clercq, E., Kalmar, I., and Vanrompay, D. (2013). Animal models for studying female genital tract infection with Chlamydia trachomatis. Infect. Immun. 81, 3060–3067. doi: 10.1128/IAI.00357-13
Dehoux, P., Flores, R., Dauga, C., Zhong, G., and Subtil, A. (2011). Multi-genome identification and characterization of chlamydiae-specific type III secretion substrates: the Inc proteins. BMC Genomics 12:109. doi: 10.1186/1471-2164-12-109
Derré, I., Pypaert, M., Dautry-Varsat, A., and Agaisse, H. (2007). RNAi screen in drosophila cells reveals the involvement of the Tom complex in Chlamydia infection. PLoS Pathog. 3:e155. doi: 10.1371/journal.ppat.0030155
Dolat, L., and Valdivia, R. H. (2019). A renewed tool kit to explore Chlamydia pathogenesis: from molecular genetics to new infection models. F1000 Res. 8:F1000. doi: 10.12688/f1000research.18832.1
Dutta, D., and Clevers, H. (2017). Organoid culture systems to study host–pathogen interactions. Curr. Opin. Immunol. 48, 15–22. doi: 10.1016/j.coi.2017.07.012
Dutta, D., Heo, I., and Clevers, H. (2017). Disease modeling in stem cell-derived 3D organoid systems. Curr. Opin. Immunol. 23, 393–410. doi: 10.1016/j.molmed.2017.02.007
Edwards, V. L., Smith, S. B., McComb, E. J., Tamarelle, J., Ma, B., Humphrys, M. S., et al. (2019). The cervicovaginal microbiota-host interaction modulates Chlamydia trachomatis infection. mBio 10:e01548–19. doi: 10.1128/mBio.01548-19
Ferrell, J. C., and Fields, K. A. (2016). A working model for the type III secretion mechanism in Chlamydia. Microbes Infect. 18, 84–92. doi: 10.1016/j.micinf.2015.10.006
Forbester, J. L., Goulding, D., Vallier, L., Hannan, N., Hale, C., Pickard, D., et al. (2015). Interaction of Salmonella enterica serovar Typhimurium with intestinal organoids derived from human induced pluripotent stem cells. Infect. Immun. 83, 2926–2934. doi: 10.1128/IAI.00161-15
Gargett, C. E., Schwab, K. E., and Deane, J. A. (2016). Endometrial stem/progenitor cells: the first 10 years. Hum. Reprod. Update 22, 137–163. doi: 10.1093/humupd/dmv051
Geisler, W. M. (2010). Duration of untreated, uncomplicated Chlamydia trachomatis genital infection and factors associated with Chlamydia resolution: a review of human studies. J. Infect. Dis. 201(Suppl. 2), S104–S113. doi: 10.1086/652402
Gitsels, A., Sanders, N., and Vanrompay, D. (2019). Chlamydial infection from outside to inside. Front. Microbiol. 10:2329. doi: 10.3389/fmicb.2019.02329
Guseva, N. V., Dessus-Babus, S., Moore, C. G., Whittimore, J. D., and Wyrick, P. B. (2007). Differences in Chlamydia trachomatis serovar E growth rate in polarized endometrial and endocervical epithelial cells grown in three-dimensional culture. Infect. Immun. 75, 553–564. doi: 10.1128/IAI.01517-06
Guseva, N. V., Dessus-Babus, S. C., Whittimore, J. D., Moore, C. G., and Wyrick, P. B. (2005). Characterization of estrogen-responsive epithelial cell lines and their infectivity by genital Chlamydia trachomatis. Microbes Infect. 7, 1469–1481. doi: 10.1016/j.micinf.2005.05.004
Haggerty, C. L., Gottlieb, S. L., Taylor, B. D., Low, N., Xu, F., and Ness, R. B. (2010). Risk of sequelae after Chlamydia trachomatis genital infection in women. J. Infect. Dis. 201, 134–155. doi: 10.1086/652395
Han, Y., and Derré, I. (2017). A co-infection model system and the use of chimeric proteins to study Chlamydia inclusion proteins interaction. Front. Cell. Infect. Microbiol. 7:79. doi: 10.3389/fcimb.2017.00079
Hutchinson, G. R., Taylor-Robinson, D., and Dourmashkin, R. R. (1979). Growth and effect of chlamydiae in human and bovine oviduct organ cultures. Br. J. Vener. Dis. 55, 194–202. doi: 10.1136/sti.55.3.194
Hybiske, K., and Stephens, R. S. (2007). Mechanisms of host cell exit by the intracellular bacterium Chlamydia. Proc. Natl. Acad. Sci. U.S.A. 104, 11430–11435. doi: 10.1073/pnas.0703218104
Igietseme, J. U., Wyrick, P. B., Goyeau, D., and Rank, R. G. (1994). An in vitro model for immune control of chlamydial growth in polarized epithelial cells. Infect. Immun. 62, 3528–3535. doi: 10.1128/IAI.62.8.3528-3535.1994
In, J., Foulke-Abe, J., Zachos, N. C., Hansen, A. M., Kaper, J. B., Bernstein, H. D., et al. (2016). Enterohemorrhagic Escherichia coli reduces mucus and intermicrovillar bridges in human stem cell-derived colonoids. Cell. Mol. Gastroenterol. Hepatol. 2, 48–62.e3. doi: 10.1016/j.jcmgh.2015.10.001
Jerchel, S., Knebel, G., König, P., Bohlmann, M. K., and Rupp, J. (2012). A human fallopian tube model for investigation of C. trachomatis infections. J. Vis. Exp. 66, 1–6. doi: 10.3791/4036
Kessler, M., Hoffmann, K., Brinkmann, V., Thieck, O., Jackisch, S., Toelle, B., et al. (2015). The Notch and Wnt pathways regulate stemness and differentiation in human fallopian tube organoids. Nat. Commun. 6:8989. doi: 10.1038/ncomms9989
Kessler, M., Hoffmann, K., Fritsche, K., Brinkmann, V., Mollenkopf, H. J., Thieck, O., et al. (2019). Chronic Chlamydia infection in human organoids increases stemness and promotes age-dependent CpG methylation. Nat. Commun. 10:1194. doi: 10.1038/s41467-019-09144-7
Kessler, M., Zielecki, J., Thieck, O., Mollenkopf, H. J., Fotopoulou, C., and Meyer, T. F. (2012). Chlamydia trachomatis disturbs epithelial tissue homeostasis in fallopian tubes via paracrine Wnt signaling. Am. J. Pathol. 180, 186–198. doi: 10.1016/j.ajpath.2011.09.015
King, A. E., Wheelhouse, N., Cameron, S., McDonald, S. E., Lee, K.-F., Entrican, G., et al. (2009). Expression of secretory leukocyte protease inhibitor and elafin in human fallopian tube and in an in-vitro model of Chlamydia trachomatis infection. Hum. Reprod. 24, 679–686. doi: 10.1093/humrep/den452
Leslie, J. L., Huang, S., Opp, J. S., Nagy, M. S., Kobayashi, M., Young, V. B., et al. (2015). Persistence and toxin production by Clostridium difficile within human intestinal organoids result in disruption of epithelial paracellular barrier function. Infect. Immun. 83, 138–145. doi: 10.1128/IAI.02561-14
Lutter, E. I., Martens, C., and Hackstadt, T. (2012). Evolution and conservation of predicted inclusion membrane proteins in chlamydiae. Comp. Funct. Genomics 2012:362104. doi: 10.1155/2012/362104
Maffei, B., Laverriere, M., Wu, Y., Triboulet, S., Perrinet, S., Duchateau, M., et al. (2020). Infection-driven activation of transglutaminase 2 boosts glucose uptake and hexosamine biosynthesis in epithelial cells. EMBO J. 39:e102166. doi: 10.15252/embj.2019102166
Malhotra, M., Sood, S., Mukherjee, A., Muralidhar, S., and Bala, M. (2013). Genital Chlamydia trachomatis: an update. Indian J. Med. Res. 138, 303–316.
Maru, Y., Orihashi, K., and Hippo, Y. (2016). Lentivirus-based stable gene delivery into intestinal organoids. Methods Mol. Biol. 1422, 13–21. doi: 10.1007/978-1-4939-3603-8_2
Mirrashidi, K. M., Elwell, C. A., Verschueren, E., Johnson, J. R., Frando, A., Von Dollen, J., et al. (2015). Global mapping of the inc-human interactome reveals that retromer restricts Chlamydia infection. Cell Host Microbe 18, 109–121. doi: 10.1016/j.chom.2015.06.004
Moore, E. R., Fischer, E. R., Mead, D. J., and Hackstadt, T. (2008). The chlamydial inclusion preferentially intercepts basolaterally directed sphingomyelin-containing exocytic vacuoles. Traffic 9, 2130–2140. doi: 10.1111/j.1600-0854.2008.00828.x
Moore, E. R., and Ouellette, S. P. (2014). Reconceptualizing the chlamydial inclusion as a pathogen-specified parasitic organelle: an expanded role for Inc proteins. Front. Cell. Infect. Microbiol. 4:157. doi: 10.3389/fcimb.2014.00157
Moulder, J. W. (1991). Interaction of Chlamydiae and host cells in vitro. Microbiol. Rev. 55, 143–190. doi: 10.1128/MMBR.55.1.143-190.1991
Nguyen, P. H., Lutter, E. I., and Hackstadt, T. (2018). Chlamydia trachomatis inclusion membrane protein MrcA interacts with the inositol 1,4,5-trisphosphate receptor type 3 (ITPR3) to regulate extrusion formation. PLoS Pathog. 14:e1006911. doi: 10.1371/journal.ppat.1006911
Nogueira, A. T., Braun, K. M., and Carabeo, R. A. (2017). Characterization of the growth of Chlamydia trachomatis in in vitro-generated stratified epithelium. Front. Cell. Infect. Microbiol. 7:438. doi: 10.3389/fcimb.2017.00438
Phillips, S., Quigley, B. L., and Timms, P. (2019). Seventy years of Chlamydia vaccine research–limitations of the past and directions for the future. Front. Microbiol. 10:70. doi: 10.3389/fmicb.2019.00070
Pleguezuelos-Manzano, C., Puschhof, J., Huber, A. R., van Hoeck, A., Wood, H. M., Nomburg, J., et al. (2020). Mutational signature in colorectal cancer caused by genotoxic pks(+) E. coli. Nature 580, 269–273. doi: 10.1038/s41586-020-2080-8
Pompaiah, M., and Bartfeld, S. (2017). Gastric organoids: an emerging model system to study Helicobacter pylori pathogenesis. Curr. Top. Microbiol. Immunol. 400, 149–168. doi: 10.1007/978-3-319-50520-6_7
Prozialeck, W. C., Fay, M. J., Lamar, P. C., Pearson, C. A., Sigar, I., and Ramsey, K. H. (2002). Chlamydia trachomatis disrupts N-cadherin-dependent cell-cell junctions and sequesters β-catenin in human cervical epithelial cells. Infect. Immun. 70, 2605–2613. doi: 10.1128/IAI.70.5.2605-2613.2002
Roth, A., Konig, P., van Zandbergen, G., Klinger, M., Hellwig-Burgel, T., Daubener, W., et al. (2010). Hypoxia abrogates antichlamydial properties of IFN-gamma in human fallopian tube cells in vitro and ex vivo. Proc. Natl. Acad. Sci. U.S.A. 107, 19502–19507. doi: 10.1073/pnas.1008178107
Shamir, E. R., and Ewald, A. J. (2014). Three-dimensional organotypic culture: experimental models of mammalian biology and disease. Nat. Rev. Mol. Cell Biol. 15, 647–664. doi: 10.1038/nrm3873
Shaw, E. I., Dooley, C. A., Fischer, E. R., Scidmore, M. A., Fields, K. A., and Hackstadt, T. (2000). Three temporal classes of gene expression during the Chlamydia trachomatis developmental cycle. Mol. Microbiol. 37, 913–925. doi: 10.1046/j.1365-2958.2000.02057.x
Stanhope, R., Flora, E., Bayne, C., and Derré, I. (2017). IncV, a FFAT motif-containing Chlamydia protein, tethers the endoplasmic reticulum to the pathogen-containing vacuole. Proc. Natl. Acad. Sci. U.S.A. 114, 12039–12044. doi: 10.1073/pnas.1709060114
Tao, L., Zhang, J., Meraner, P., Tovaglieri, A., Wu, X., Gerhard, R., et al. (2016). Frizzled proteins are colonic epithelial receptors for C. difficile toxin B. Nature 538, 350–355. doi: 10.1038/nature19799
Turco, M. Y., Gardner, L., Hughes, J., Cindrova-Davies, T., Gomez, M. J., Farrell, L., et al. (2017). Long-term, hormone-responsive organoid cultures of human endometrium in a chemically defined medium. Nat. Cell Biol. 19, 568–577. doi: 10.1038/ncb3516
Weber, M. M., Bauler, L. D., Lam, J., and Hackstadt, T. (2015). Expression and localization of predicted inclusion membrane proteins in Chlamydia trachomatis. Infect. Immun. 83, 4710–4718. doi: 10.1128/IAI.01075-15
Yin, Y., and Zhou, D. (2018). Organoid and enteroid modeling of Salmonella infection. Front. Cell. Infect. Microbiol. 8:102. doi: 10.3389/fcimb.2018.00102
Zadora, P. K., Chumduri, C., Imami, K., Berger, H., Mi, Y., Selbach, M., et al. (2019). Integrated phosphoproteome and transcriptome analysis reveals Chlamydia-induced epithelial-to-mesenchymal transition in host cells. Cell Rep. 26, 1286–1302.e1288. doi: 10.1016/j.celrep.2019.01.006
Zhang, Y.-G., Wu, S., Xia, Y., and Sun, J. (2014). Salmonella-infected crypt-derived intestinal organoid culture system for host-bacterial interactions. Physiol. Rep. 2:e12147. doi: 10.14814/phy2.12147
Keywords: Chlamydia, endometrial organoids, 3D culture, developmental cycle, inclusion membrane proteins
Citation: Bishop RC, Boretto M, Rutkowski MR, Vankelecom H and Derré I (2020) Murine Endometrial Organoids to Model Chlamydia Infection. Front. Cell. Infect. Microbiol. 10:416. doi: 10.3389/fcimb.2020.00416
Received: 04 May 2020; Accepted: 07 July 2020;
Published: 14 August 2020.
Edited by:
Daniel E. Voth, University of Arkansas for Medical Sciences, United StatesReviewed by:
Mary M. Weber, The University of Iowa, United StatesCopyright © 2020 Bishop, Boretto, Rutkowski, Vankelecom and Derré. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Isabelle Derré, aWQ4bUB2aXJnaW5pYS5lZHU=
†Present address: Matteo Boretto, Hubrecht Institute, Royal Netherlands Academy of Arts and Sciences, Utrecht, Netherlands
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.