
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Cell. Infect. Microbiol. , 28 July 2020
Sec. Fungal Pathogenesis
Volume 10 - 2020 | https://doi.org/10.3389/fcimb.2020.00370
This article is part of the Research Topic Malassezia: A Skin Commensal Yeast Impacting Both Health And Disease View all 14 articles
Malassezia spp. are lipid-dependent yeasts, inhabiting the skin and mucosa of humans and animals. They are involved in a variety of skin disorders in humans and animals and may cause bloodstream infections in severely immunocompromised patients. Despite a tremendous increase in scientific knowledge of these yeasts during the last two decades, the epidemiology of Malassezia spp. related to fungemia remains largely underestimated most likely due to the difficulty in the isolation of these yeasts species due to their lipid-dependence. This review summarizes and discusses the most recent literature on Malassezia spp. infection and fungemia, its occurrence, pathogenicity mechanisms, diagnostic methods, in vitro susceptibility testing and therapeutic approaches.
Malassezia are lipid-dependent yeasts inhabiting the skin of healthy humans and other warm blooded animals. However, these yeasts may also act as opportunistic pathogens, causing dermatitis and otitis in animals, and dermatitis with (i.e., atopic dermatitis, folliculitis, and psoriasis) and without inflammation (Pityriasis versicolor) in humans (Guillot and Bond, 2020; Saunte et al., 2020). Besides their involvement in skin diseases, Malassezia spp. have increasingly been reported to cause severe systemic infections, especially among premature neonates and immunocompromised patients receiving parenteral nutrition (Miceli et al., 2011; Iatta et al., 2014a, 2018; Ilahi et al., 2017; Pedrosa et al., 2018). While fungemia caused by Candida species has been recognized as a cause of morbidity and mortality in hospitalized patients worldwide (Mellinghoff et al., 2018), the epidemiology of Malassezia-related fungemia remains largely underestimated most likely due to the difficulties in the isolation of these yeasts species due to their lipid-dependent growth (Iatta et al., 2018). Currently, the genus comprises 18 lipid-dependent species with a variable distribution on different hosts and pathologies (reviewed in Lorch et al., 2018; Guillot and Bond, 2020). Additionally, by using both fingerprinting methods and multigene sequence analysis, different Malassezia genotypes were identified as strictly related to the host, geographical origin, and/or clinical manifestations (Cafarchia et al., 2008, 2011b; Theelen et al., 2018). Several hypotheses have been proposed to explain the pathogenic mechanisms of these fungi, but the role of single species and genotypes in clinical manifestations remains to be elucidated (Theelen et al., 2018). In addition, scientific data suggest that Malassezia antifungal susceptibility profiles against azoles, amphotericin B (AmB) and terbinafine (TER) largely vary between Malassezia species or genotypes, thus influencing clinical management of patients (Theelen et al., 2018). This review summarizes and discusses the most recent literature on Malassezia fungemia, its occurrence, pathogenic mechanisms of the involved species, diagnostic methods, in vitro susceptibility testing and therapeutic approaches.
Since the designation of the genus Malassezia by Baillon in 1889, the taxonomy has been updated and currently the genus comprises 18 lipid-dependent species with different genotypes showing variable pathology and distribution on different hosts (reviewed in Lorch et al., 2018; Guillot and Bond, 2020). All Malassezia species are lipid-dependent and, with the exception of Malassezia pachydermatis, do not grow on Sabouraud Dextrose Agar (SDA), which is most commonly used for culturing fungi in clinical labs. The genus Malassezia occurs on the skin of humans and animals but some species have only been observed on either humans or animals (reviewed in Guillot and Bond, 2020). Interestingly, Malassezia yeasts are the major component of the healthy human skin mycobiome (Findley et al., 2013; Oh et al., 2014; Wu et al., 2015). A recent phylogenetic study evaluating six genes suggested that the genus is deeply rooted in the Ustilaginomycotina and has a sister relationship with the Ustilaginomycetes and Exobasidiomycetes, assigning the genus its own class, Malasseziomycetes (Wang et al., 2014). Based on a phylogenomics study using 164 core eukaryotic genes, three main clusters were identified: Cluster A consisting of M. furfur, M. japonica, M. obtusa, and M. yamatoensis; Sub-cluster B1, with the most abundant human skin inhabitants M. globosa and M. restricta; Sub-cluster B2 consisting of M. sympodialis, M. dermatis, M. caprae, M. equina, M. nana, and M. pachydermatis; and Cluster C forming a basal lineage with M. cuniculi and M. slooffiae (Wu et al., 2015). Four more species have been described since then: M. brasiliensis and M. psittaci from parrot (Cabañes et al., 2016), M. arunalokei from human scalp (Honnavar et al., 2017), and M. vespertilionis from bat (Lorch et al., 2018). Multiple genotypes of a species can colonize the same patient (Cafarchia et al., 2008; Machado et al., 2010; Ilahi et al., 2017) but some genetic types might be linked to a particular body site or pathology thus indicating an affiliation of Malassezia genotypes with host, geographical origin and/or clinical manifestations (Cafarchia et al., 2008, 2011a,b; Ilahi et al., 2017). In particular, amplified fragment length polymorphism (AFLP) patterns of M. furfur skin isolates from Ontario, Canada clustered separately from mainly European references from other body sites, suggesting geographical or ecological/clinical variability in the species (Gupta et al., 2004). Sequence analysis of the intergenic spacer (IGS1) distinguished specific M. globosa, M. restricta, and M. pachydermatis variants in seborrheic dermatitis, atopic eczema, and on healthy skin of humans and animals (Sugita et al., 2003, 2004; Kobayashi et al., 2011). Moreover, sequence analyses of the LSU rDNA showed distinct Malassezia spp. subtypes on different host species (Gaitanis et al., 2012). Multilocus sequence analysis that included the D1/D2 domains of LSU rDNA, the chs2 gene, and the ITS1 region grouped M. pachydermatis strains from skin of healthy dogs and from skin lesions in three main genotypes (A, B, and C) with eight ITS1 subtypes. Genotype B included isolates from dogs of European origin and appeared to be present on healthy dog skin, without producing phospholipase. The A and C genotypes and their subtypes seemed to be predominantly associated with skin lesions and their isolates showed high phospholipase production (Cafarchia et al., 2008; Machado et al., 2010). Similarly, IGS1 subtypes 3C and 3D displayed high phospholipase production and were more frequently isolated from skin lesions of dogs with atopic dermatitis (Kobayashi et al., 2011). Only three Malassezia species have been described to cause bloodstream infections: M. furfur, M. pachydermatis, and M. sympodialis. Interestingly, AFLP analysis or ITS sequences showed that only one main M. furfur or M. pachydermatis genotype seems to be involved in blood stream infections in immunocompromised hosts (Theelen et al., 2001; Kaneko et al., 2012; Ilahi et al., 2017). All these Malassezia genotypes might also colonize the skin of patients or of hospital staff which might represent the driver for these systemic infections (Theelen et al., 2001; Gupta et al., 2004; Kaneko et al., 2012). With respect to the assessment of antifungal microbiological profiles, a reference method has not yet been developed for these yeasts and the culture media, inoculum sizes, incubation times, and the criteria used to determine MIC endpoints differ among studies. However, evidence suggested that Malassezia antifungal susceptibility profiles against azoles, AmB and TER vary according to the Malassezia species, regardless of culture medium or other conditions employed. M. sympodialis and M. pachydermatis are the most susceptible and M. furfur and M. globosa the least susceptible species to azoles, AmB and TER (Rojas et al., 2014; Cafarchia et al., 2015; Pedrosa et al., 2019b). Itraconazole (ITZ) and ketoconazole (KTZ) were the most active drugs for all Malassezia species, and fluconazole (FLZ), voriconazole (VOR) and AmB the least active (Rojas et al., 2014; Cafarchia et al., 2015; Pedrosa et al., 2019b). Interestingly, antifungal profiles may also vary in relation to genotype (Sugita et al., 2005; Cafarchia et al., 2012). In particular, isolates derived from animal skin lesions and belonging to a unique genotype, showed reduced susceptibility to azoles when compared to genotypes associated with healthy skin (Cafarchia et al., 2008).
The epidemiology of Malassezia fungemia has not been well-investigated until now due to the scant surveillance studies with this focus (see Table 1). Malassezia furfur, M. sympodialis, and M. pachydermatis are the only Malassezia species isolated from bloodstream infections to date (Table 1). Some authors propose that the fungal density on the skin as well the host immunological competence might be driving factors influencing their pathogenic role (Wheeler et al., 2017; partially reviewed in Theelen et al., 2018). Malassezia bloodstream infections have been described in adult and child immunocompromised patients, and neonates (Table 1). According to general knowledge, the main ecological niche of Malassezia yeasts is human and animal skin, and they represent 50 to 80% of the total human skin mycobiome (Nagata et al., 2012; Findley et al., 2013; Gupta et al., 2014). Therefore, it would make sense to consider the skin mycobiome as a potential reservoir and point d'entrée for bloodstream infections. More is currently known about the adult human skin mycobiome but little information is available about the child and neonatal skin mycobiome. Even if Malassezia skin colonization may be the result of both maternal and environmental sources, data suggests that colonization of human skin begins immediately after birth and Malassezia species distribution varies with age (Jo et al., 2016; Ward et al., 2018). The limited data currently available about the cutaneous mycobiomes in preterm and term neonates shows that Malassezia species distribution on skin of neonates and children varies between studies, but M. globosa, M. furfur, M. sympodialis, and M. restricta seem to be the most prevalent species described (Bernier et al., 2002; Zomorodain et al., 2008; Jang et al., 2009; Gupta et al., 2014; Prohic et al., 2014; Jo et al., 2016; Paul et al., 2019). Variation between studies could be the result of methodological differences such as number of subjects studied, DNA extraction, target region selection and PCR-conditions, and data processing and interpretation; but also geographical and host-specific differences may explain study discrepancies. The study of Paul et al. determined M. restricta to be the most abundant Malassezia species in both term and preterm neonates (n = 30), similar to adult skin (Paul et al., 2019). Interestingly, this species has thus far not been linked to bloodstream infections. Malassezia colonization increases quickly after birth but a very important shift, making Malassezia the most predominant genus of the human skin mycobiome, takes place during adolescence and has been linked to lipid composition shifts of the skin (Grice and Segre, 2011; Findley et al., 2013; Jo et al., 2016). In adults, M. restricta and M. globosa seem to be the dominating Malassezia species on the skin, followed by M. sympodialis, depending on body site (Findley et al., 2013; Wu et al., 2015). Geographical factors may influence Malassezia species distribution on healthy human skin (Leong et al., 2019; Saunte et al., 2020). In addition, the proportion of Malassezia species isolated from the skin varies considerably among different medical conditions (Saunte et al., 2020). As the two most abundant Malassezia species on healthy human adult skin have not been linked to Malassezia bloodstream infections, studies focusing on genetic and functional differences between cells of Malassezia species involved in bloodstream infections and ones that are not, in addition to potentially selective host parameters, could help explain this. Recent molecular studies highlighted the genetic variability among single species implicated in both skin and systemic infections (Theelen et al., 2001; Kaneko et al., 2012; Gupta et al., 2014), suggesting pathogenicity variation at species and sub-species levels. Malassezia pachydermatis is mainly known from animals and is not commonly associated with human skin colonization yet in multiple cases it was linked to Malassezia bloodstream infections (Table 1).
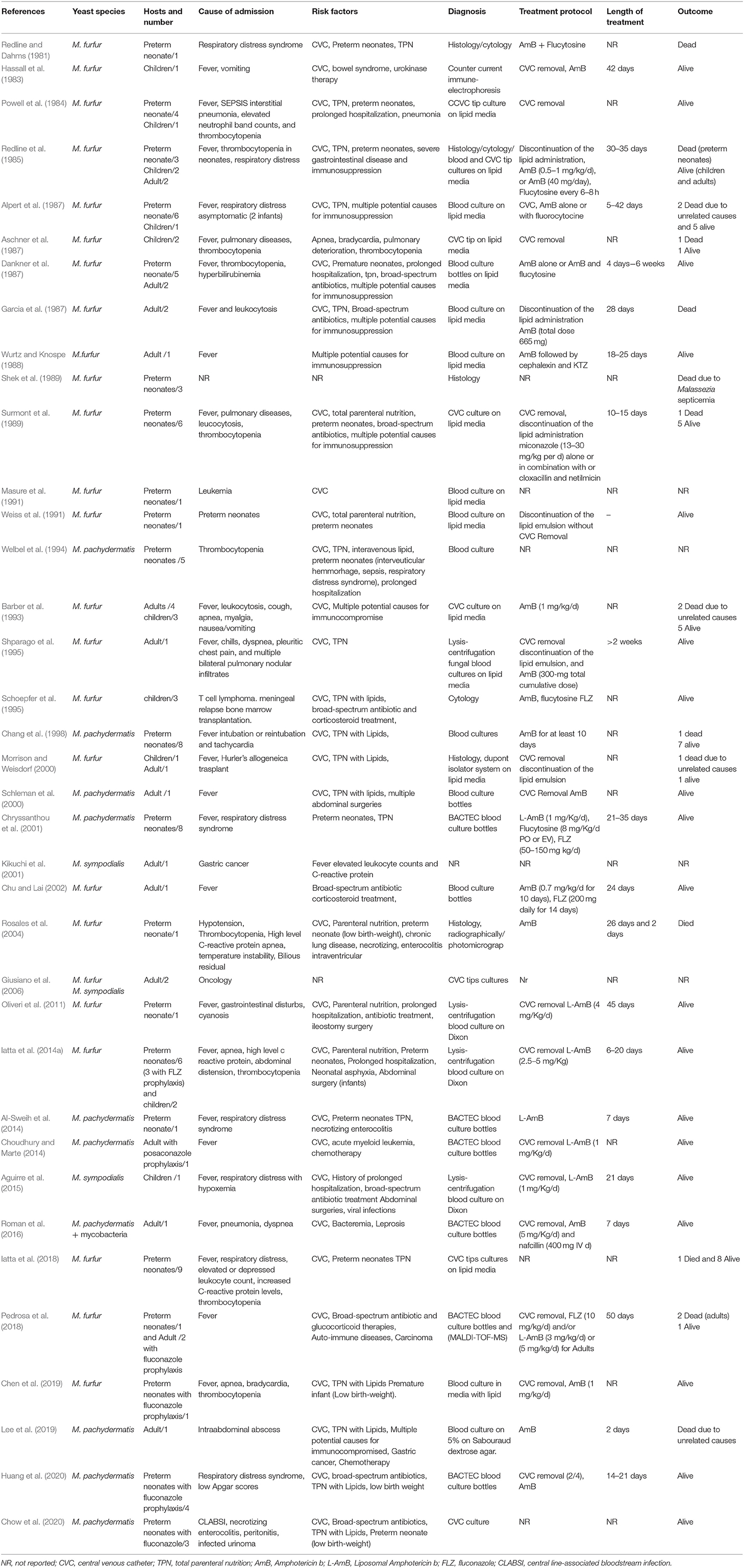
Table 1. Malassezia yeasts fungemia: yeast species, risk factors, diagnosis, treatment, and outcome.
A study evaluating cats and dogs (107 healthy and 123 with chronic otitis externa), showed that occurrence and population size of M. pachydermatis increased according to the presence in skin lesions, not only in affected areas of the skin but also at other sites without detectable skin lesions, suggesting that the yeasts could be easily transmitted from site to site because of scratching induced by pruritus (Cafarchia et al., 2005). This finding is also relevant from a zoonotic point of view since the yeasts may be mechanically transmitted from dogs to their owners (Morris et al., 2005) and subsequently can cause health problems as previously reported from an intensive care nursery (Chang et al., 1998). A similar transmission route—animal to human or human to human—from health care worker or family member for M. furfur or M. sympodialis may also occur and should be further explored in future studies. Even when these species are part of the stable mycobiomes of Malassezia bloodstream infection patients, isolates causing the infection could belong to different genotypes, transmitted from external sources. The first case of Malassezia spp. as a pathogen in bloodstream infection and sepsis was reported in 1981 by Redline and colleagues (Table 1). Until now, a total of 118 cases were published, but only three surveillance studies (Table 1). Malassezia furfur was the most encountered species with 82 cases, followed by M. pachydermatis (33 cases), and only three cases of M. sympodialis fungemia have been described. To date, Malassezia fungemia cases were observed with highest incidence in neonates (82 cases), followed by 23 in adults and 17 in children. Numerous cases were reported in the past two decades, particularly in neonates and infants receiving intravenous lipids. In our 1-year survey on yeast fungemia involving 290 neonatal and 17 pediatric patients with intravascular catheters, lipid parenteral nutrition, prolonged ward stay, and surgery, were evaluated. This diagnostic survey on bloodstream infections (BSIs) resulted in a higher prevalence of M. furfur (2.1%) than Candida spp. (1.4%) and suggested that Malassezia BSIs might be underestimated, due to improper diagnosis (Iatta et al., 2014a). The surveillance study was repeated 4 years later, and a total of 202 neonatal patients with intravascular catheters were enrolled (Iatta et al., 2017). A total of 10 cases of BSIs were registered, thus suggesting the relevance of these yeasts in catheter mediated fungemia (Iatta et al., 2018). Incubator, sheets and the skin of patient or hospital staff may represent potential sources of Malassezia infection. Recently, lipid infusion type, phototherapy light sources, central venous catheter placement, and prophylactic fluconazole were proposed as risk factors affecting Malassezia colonization and /or fungemia more than candidemia (Chen et al., 2019).
Almost all-available research to date focuses on Malassezia virulence factors linked to skin and pathogenesis for Malassezia BSIs has hardly been studied, likely due to the relatively low number of cases when compared to skin diseases. With an increasing number of cases in recent years and the likely underestimation of Malassezia BSIs as a result of the use of standard culture media without lipid supplementation in the clinic, future studies will hopefully address some of the missing knowledge. Here, we focus on known virulence factors of the three fungemia causing Malassezia species (i.e., M. pachydermatis, M. furfur, and M. sympodialis) and discuss findings, even if they were derived from skin. Virulence factors, to some extent, may be of a more general nature and could potentially also be relevant for bloodstream infections. Several hypotheses have been proposed to explain the pathogenic behavior of these fungi. The relationship between host and Malassezia metabolism seems to be key for the understanding the pathogenesis of infection (Cafarchia and Otranto, 2004; Velegraki et al., 2015; Theelen et al., 2018).
Various Malassezia cell characteristics may be involved in BSI pathogenesis. Malassezia spp. have cell walls with a very thick multi-layered structure that may protect them from different environmental stresses and have been described to help evade phagocytosis (Celis et al., 2017a). Biofilm formation has been linked to increased drug resistance and virulence. M. furfur, M. sympodialis, and M. pachydermatis are able to form biofilms, a process that seems to be strain dependent (Figueredo et al., 2013; Angiolella et al., 2017; Pedrosa et al., 2019a). In particular, M. pachydermatis strains isolated from dogs with and without skin lesions were able to form biofilms with variable extracellular matrix (ECM) quantity and structure depending on the sources (Figueredo et al., 2013). Accordingly, a structural heterogeneity of biofilm was found between those formed by M. furfur and M. sympodialis isolates, with both species exhibiting yeast aggregates in multilayer clusters but with a denser entrapment by a more gelatinous ECM in case of M. furfur biofilms (Pedrosa et al., 2019a). Biofilm formation and the extracellular matrix generation were responsible for the emergence of antifungal resistance (Figueredo et al., 2013; Angiolella et al., 2017). The biofilm formation was well-correlated with hydrophobicity, adherence, and phospholipase production of pathogenic M. pachydermatis and M. furfur cells, which may help explaining the change from a commensal to a pathogenic phenotype of these organisms (Figueredo et al., 2013; Angiolella et al., 2017). Malassezia furfur colonization of central venous catheters was already observed in 1994 (Sizun et al., 1994) and the ability M. pachydermatis to form biofilms on catheter surfaces in the laboratory was confirmed in another study (Cannizzo et al., 2007). Though not observed frequently, Malassezia yeasts are able to produce mycelium, a feature first suggested from an observation of hyphae in the scales of PV. Later, researchers managed to also obtain a mycelial phase for M. furfur in vitro (Saadatzadeh et al., 2001). In a recent review, the importance of morphological switching for fungal pathogens was highlighted: shape-shifting between different morphologies allows fungi to adapt to different host environments. The authors suggest that to understand pathogenesis mechanisms, it is crucial to establish how fungal morphology impacts virulence strategies (Min et al., 2020). It would be useful to further investigate Malassezia morphology switching and its potential relevance in bloodstream infections.
It has been shown that μ-opioid receptors (MORs) are present on M. pachydermatis cell membranes, having a role in modulating the phospholipase activities (Cafarchia et al., 2010). In animals without lesion MORs were expressed as dimers with other opioids thus resulting in an inactive form (Cafarchia et al., 2010). The high concentration of beta-endorphin normally present on lesioned skin of hosts (Pan, 2005; Honnavar et al., 2017) influenced the expression of MOR in their active form, thus favoring phospholipase production (Cafarchia et al., 2010). Increased phospholipase activity may be linked to the appearance of skin lesions and in some cases septicaemia (Cafarchia and Otranto, 2004; Vlachos et al., 2013).
Malassezia yeasts also produce esterases, lipases, and proteases, of which the latter have a crucial role in interactions with the host and microbial community such as Staphylococcus aureus thus making it unfavorable for colonization (Chen and Hill, 2005; Li et al., 2018; Tee et al., 2019). Additionally, M. furfur secretes aspartyl proteases, capable of degrading a wide range of human skin associated extracellular matrix (ECM) protein, and might also be able to modify the skin environment potentially interfering with wound re-epithelization (Poh et al., 2020). These enzymes could on one hand contribute to pathogenesis but on the other hand have a protective function, leading to a potentially very complex role for Malassezia on and in the human host. Lipases particularly have been considered virulence factors of Malassezia yeasts since they may be involved in the invasion, colonization, persistence and proliferation within host tissues (Petrokilidou et al., 2019). The first Malassezia gene encoding an extracellular lipase was identified in M. furfur and was designated as LIP1. Subsequently, orthologs were identified in M. pachydermatis, M. globosa, and M. restricta. Malassezia species possess multiple genes encoding putative lipases (from 9 to 14 depending on the species) that are differently involved in various skin disorders (Park et al., 2017; Tee et al., 2019). Interestingly, the lipase and phospholipase activities of Malassezia yeasts vary according to the species and they are implicated in both skin diseases and fungemia. In particular, M. furfur strains causing fungemia showed very high lipolytic enzyme activity thus suggesting that parenteral lipid emulsions may play an important role in modulating the growth and pathogenicity of Malassezia-yeasts in sepsis (Kaneko et al., 2012). Malassezia strains from skin disorders produce metabolites, such as indirubin and indolo[3,2-b] carbazole (ICZ) which are associated with carcinogenesis, immune regulation and mediation of ultraviolet radiation (UVR) damage (Gaitanis et al., 2011; Theelen et al., 2018). Recently, it has been shown that both M. furfur and M. sympodialis are able to produce nanovesicles enriched with allergens and/or proteins that interact with keratinocytes and monocytes, thus causing and maintaining the inflammation (Johansson et al., 2018; Zhang et al., 2019). In particular, nanovesicles produced by M. sympodialis (MalaEx) are also able to activate human keratinocytes causing an enhanced intercellular adhesion molecule-1 (ICAM-1) expression, which can cause an attraction of immune competent cells, thus causing host cutaneous defense to M. sympodialis (Vallhov et al., 2020). So far, the function of these extracellular vesicles for Malassezia spp. has only been studied in relation to skin but various studies in other fields related to similar kinds of nanovesicles show that their function in cell-to-cell communication may be much broader than that. For example, a study on human pathogen Cryptococcus gattii, showed that vesicles were taken up by macrophages of the infected host, allowing long distance pathogen-to-pathogen communication resulting in virulence enhancement (Bielska et al., 2018). It would be interesting to explore whether extracellular vesicles might also play a role in Malassezia BSIs.
In order to gain better understanding of Malassezia pathogenesis in general, and in BSIs in particular, it is important to assess known Malassezia virulence factors, as well as investigate potentially unknown factors, and perform comparative studies between BSI-derived isolates and skin isolates. In recent years, many useful new tools have been developed for this purpose. Although to date, no model systems for studying systemic Malassezia infections have been described, recent advances for other Malassezia-affected areas of the human body may offer useful insights for future application to studying host-pathogen interactions in BSIs. Sparber et al., reported of a murine epicutaneous infection model that allowed studying the interaction of Malassezia with mammalian skin in vivo (Sparber and LeibundGut-Landmann, 2019; Sparber et al., 2019) and in the same year the association of Malassezia with Crohn's disease was reported using mouse models (Limon et al., 2019). Performing Malassezia host-pathogen interaction studies in model systems such as the mouse may sometimes be difficult, but with the recent establishment of an in vivo infection model using Galleria mellonella, host-pathogen interaction studies are becoming more accessible. The G. mellonella model has multiple advantages, such as the absence of ethical hurdles, low cost, ease of use, yet the immune response has similarities with the human system (Torres et al., 2020). In addition, recently developed Malassezia transformation systems can aid in studying the role of specific genes and virulence factors in Malassezia pathogenicity. Agrobacterium tumefaciens mediated transformation systems for the BSI-relevant Malassezia species were developed, allowing direct gene manipulation to better understand gene function (Ianiri et al., 2016; Celis et al., 2017b). With the recent improvements to the CRISPR/Cas9 strategy, this gene editing system has already contributed to various fungal virulence studies (Malavia et al., 2020), and also its first application for research in functional Malassezia genetics has recently been reported (Ianiri et al., 2019).
Since multiple Malassezia species and/or genotypes with varying antifungal susceptibility profiles may cause unique or similar pathologies, serious concern about the diagnostic procedures and antifungal treatment has been raised. Isolation and enumeration of Malassezia cells from clinical specimens remain a challenge because of their lipid dependency. Although microscopy of swab specimens is useful for diagnosing animal and human dermatitis, a more accurate etiological diagnosis is needed in high-risk patients (Iatta et al., 2018). Clinical features, laboratory markers, strategies of patient management, and outcomes of Candida and Malassezia fungemia do not differ. In some studies, M. furfur fungemia appeared earlier than candidaemia (average day 26 vs. day 42), most likely due to its exogenous origin. In addition, duration of Malassezia fungemia is longer than candidaemia, most likely due to the late removal of the central venous catheters (CVC) and also as a result of the lower efficacy of the antifungal therapy (Iatta et al., 2014a, 2018). The first signs causing suspicion of Malassezia fungemia usually are bloodstream infections manifested in fever of unknown origin in hospitalized and severely immunocompromised patients with CVC (i.e., preterm neonates or cancer patients) or in patients with pulmonary distress (Table 1) (Morrison and Weisdorf, 2000; Iatta et al., 2015, 2018). Malassezia fungemia should however be confirmed with the laboratory isolation of the responsible agent and identification by molecular means (Morrison and Weisdorf, 2000; Iatta et al., 2015, 2018; Pedrosa et al., 2018). In literature Malassezia yeasts were mainly isolated by culturing blood or CVC-tip directly on specific media and mainly by using the Isolator system (Table 1). It has been shown that the automated blood culture system BacT/Alert is not suitable for detecting M. furfur fungemia (Campigotto et al., 2016; Iatta et al., 2018). Out of 9 M. furfur fungemia cases reported by using BacT/Alert bottles only 1 case of M. furfur fungemia was detected whereas all the M. pachydermatis cases were easily detected using the above mentioned method, since this species grows on the media included in the bottles (Iatta et al., 2018, Table 1). Interestingly, it has been shown that human blood has a toxic effect on yeast growth and the addition of 3% of palmitic acid in the bottle might be able to overcome the inhibitory effect of both small (0.5 ml) and larger (3 ml) volumes of blood, thus favoring Malassezia growth (Nelson et al., 1995). Finally, although molecular tools have been used to detect Malassezia yeasts from biological samples no studies were performed to molecularly diagnose Malassezia directly from blood, thus culture remains the gold standard to isolate and identify the yeasts. This method is also suitable for yeast quantification, viability assessment, and genotyping and eventually to test the antifungal susceptibility profile of the isolated species (Peker et al., 2018). Generally, clinics use standard culture media without lipid supplementation, likely leading to underdiagnosis of both M. furfur and M. sympodialis in BSIs. It is recommended to carefully assess Malassezia BSI risk factors and apply the use of lipid-rich culture media when Malassezia spp. may be the causative agent.
For treatment of Malassezia-related infections, azoles, and the polyene AmB are frequently employed, both in humans and animals. Topical antifungal agents (mainly azoles) are adequate for the management of localized skin lesions, while systemic ITZ or FLZ for severe skin diseases (Bond et al., 2020; Saunte et al., 2020). For catheter-related Malassezia infections, there are only recommendations, and patients were usually treated with catheter removal, discontinuation of lipid infusion and administration of antifungal drugs such as FLZ, AmB, and or VOR (Arendrup et al., 2014; Table 1). Amphotericin B is effective in the treatment of Malassezia systemic infections, both in preterm infants and adults, but both FLZ and posaconazole (POS) fail to prevent Malassezia fungemia (Table 1). Usually, about 24 days of AmB treatment might be useful for a positive outcome of Malassezia fungemia but the length of the treatment might be different depending on the Malassezia species (Table 1). M. pachydermatis fungemia resolve more quickly than M. furfur fungemia (about 14 vs. 24 days, Table 1).
Despite attempts to treat these fungal infections, a trend toward recurrence is often observed in humans and animals with dermatitis (Negre et al., 2009; Bond et al., 2010, 2020; Saunte et al., 2020). Moreover, the induction of in vitro FLZ resistance in M. pachydermatis as well as the clinical evidence of treatment failure with TER in patients with pityriasis versicolor and, with KTZ in dogs with otitis (Kim and Pandya, 1998; Gupta et al., 2014; Kim et al., 2018) or with FLZ or POS in preventing M. furfur fungemia in humans, suggested the occurrence of drug resistance phenomena in these yeast species (Choudhury and Marte, 2014; Iatta et al., 2014a; Pedrosa et al., 2018; Chen et al., 2019). Antifungal susceptibility test methods have not yet been standardized, neither by the Clinical and Laboratory Standards Institute (CLSI) nor by the European Committee on Antimicrobial Susceptibility Testing (EUCAST) (Arendrup et al., 2014), resulting in the absence of clinical breakpoints for these yeasts species. A recent study showed that drug efflux pumps (EPMs) are involved as defense mechanisms to azole drugs in Malassezia yeasts (Iatta et al., 2017). By using a broth microdilution chequerboard analysis, the in vitro efficacy of azoles in combination with EPMs (i.e., haloperidol-HAL, promethazine-PTZ, and cyclosporine) was evaluated. The MICs of FLZ and VOR of Malassezia spp. decreased in presence of sub-inhibitory concentrations of HAL and/or PTZ, and a synergistic effect was observed only in strains with FLZ MIC ≥ 128 μg/mL for M. furfur, FLZ MIC ≥ 64 μg/mL for M. pachydermatis, and VOR MIC ≥ 4 μg/mL in both Malassezia spp., suggesting that the above FLZ and VOR MIC values might be considered the cut-off to discriminate susceptible and resistant strains (Iatta et al., 2017). Finally, the in vitro susceptibility of Malassezia for echinocandins suggests that this genus is intrinsically resistant to these drugs. Indeed, MIC > 32 μg/mL were usually recorded for M. pachydermatis and M. furfur regardless of the employed CLSI protocol for testing drug efficacy (Prado et al., 2008; Yurayart et al., 2013; Al-Sweih et al., 2014; Leong et al., 2017).
Our knowledge on Malassezia yeasts has increased tremendously during the last two decades. Many questions remain ambiguous however, such as their role in pathology, diagnostic procedures, azole susceptibility profiles and clinical implications. Malassezia is the major component of the healthy human skin mycobiome but data on the constitution of the skin mycobiome and the abundance of Malassezia yeasts directly after birth and during the first years of life are highly inconclusive. Additional studies are needed as a better understanding of early (skin) mycobiome development may aid in understanding transmission routes, and may subsequently aid in infection containment. For example, in cases where M. pachydermatis was involved, transmission from pets via healthcare workers to patients was suggested which warrants better hygiene measures for hospital staff (Chang et al., 1998; Morris et al., 2005). Interpreting available literature on this topic should be done with caution as variation between studies may arise from methodological differences for factors such as study group size, identification methods and geography.
Concerning the causative species of bloodstream infection, epidemiological surveys suggest that M. furfur is the most common species involved. Although further studies are needed to understand the exact mechanisms of this finding; the colonization of skin of patients, as well as the higher virulence of Malassezia strains involved in fungemia, might be the driving factors influencing BSI epidemiology.
The occurrence of these infections seems lower among adult patients, and higher among neonatal patients, yet a slanted view exists as cases are likely underestimated, given the special lipid requirement of these yeast species (Iatta et al., 2018). Interestingly neonatal patients seem more frequently colonized with M. furfur strains than adults, but more data is needed to support this trend. Although lipid infusion and/or total parenteral nutrition seems to be one of the major risk factors in causing Malassezia fungemia due to the lipolytic properties of Malassezia yeasts, cases unrelated to intravenous nutrition were also observed (Table 1; Pedrosa et al., 2018; Chen et al., 2019). Interestingly, these infections are usually confined to severely immunocompromised hosts with CVCs, thus confirming the importance of this portal of entry (Iatta et al., 2018; Pedrosa et al., 2018; Chen et al., 2019). Several potential virulence factors have been described and may be involved in Malassezia pathogenesis of BSIs. In particular, the ability to produce enzymes such as lipases and/or phospholipases, production of various indolic compounds, the ability to form biofilms, and allergen enriched nanovesicle production were described for M. furfur and M. sympodialis which are frequently related to fungemia. The properties of these microorganisms to adhere to the skin and to medical indwelling devices by forming biofilms influence the antifungal profile of cells, which might represent another virulence factor in fungemia of severely immunocompromised hosts (Figueredo et al., 2013; Pedrosa et al., 2019a). Interestingly the structure of Malassezia biofilms is strain dependent and those of M. furfur and M. pachydermatis from skin lesions are composed of a more gelatinous extracellular matrix (Figueredo et al., 2013; Angiolella et al., 2018; Pedrosa et al., 2019a). Since the extracellular matrix is directly linked to the virulence of these yeasts, M. furfur and M.pachydermatis should be more virulent than M. sympodialis, which may explain the lower observed incidence of M. sympodialis fungemia (Figueredo et al., 2013; Angiolella et al., 2018; Pedrosa et al., 2019a). In recent years, useful new tools for studying virulence and host-pathogen interactions have been developed. Galleria mellonella has proven to be a promising in vivo infection model (Torres et al., 2020) and two studies showed the potential of a Agrobacterium tumefaciens mediated transformation system for studying the role of specific virulence related genes (Ianiri et al., 2016; Celis et al., 2017b). As the clinical outcome of Malassezia fungemia does not differ from that of Candida yeasts (Iatta et al., 2014a), clinical guidelines need to be organized for an early diagnosis of these yeasts infections. In particular, fever of unknown origin and very high values of C-reactive protein (CRP) should alert the clinicians to suspect Malassezia related systemic infections. Diagnostic procedures to recover Malassezia organisms from blood are not routinely available, and the most common system used in many laboratories for the detection of bacterial and fungal pathogens (i.e., BacT/Alert system), is ineffective for diagnosing M. furfur fungemia (Iatta et al., 2018). Recently, CVC culturing on lipid-supplemented media, has been proposed as a routine procedure to diagnose M. furfur fungemia in severely immunocompromised patients (Iatta et al., 2018).
Guidelines for the treatment of Malassezia spp. skin disorders of pet animals and humans have been assessed, but those related to systemic infections still need to be addressed. Clinical evidence indicated efficacy of azole drugs for the control of skin infections and of AmB for systemic ones (Table 1). However, common recurrences of skin disorders (Negre et al., 2009; Theelen et al., 2018; Bond et al., 2020) as well as the clinical evidence of treatment failure with TER in patients with pityriasis versicolor, with KTZ in dogs with otitis (Kim and Pandya, 1998; Gupta et al., 2014) or with FLZ or POS in preventing M. furfur fungemia in humans, suggested the occurrence of drug resistance phenomena in these yeast species.
Additionally, the high level of inter- and intraspecies differences of Malassezia antifungal profiles might explain the differences in mycological cure rates when an antifungal agent is used to treat what appears to be clinically the same disease state. Different Malassezia species might be involved in the same clinical diseases, and/or different genetic types with different antifungal profiles might colonize the same host (Prohic et al., 2015; Velegraki et al., 2015). However, even if the MIC data of Malassezia species vary according to the protocol used for susceptibility testing, there are evidences of a very low susceptibility of these yeasts to FLZ, VOR and echinocandins (reviewed in Theelen et al., 2018; Bond et al., 2020). Although Malassezia species show differences in their antifungal susceptibility in vitro, the in vivo efficacy of antifungal agents needs to be further evaluated by comparing in vitro MICs and clinical outcomes. Correlation of high azole MIC values for Malassezia spp. with unsuccessful treatment has been reported in some studies (Velegraki et al., 2005; Iatta et al., 2014a; Rojas et al., 2014). The above results need to be further validated with multicentre studies in order to develop therapeutic guidelines. For the moment, it is important to be aware that the genus Malassezia comprises a heterogeneous group of species and genotypes that may cause the same pathology, but may vary in their susceptibility to different antifungal agents. Species identification is of paramount importance not only for epidemiological surveillance and outbreak investigation, but also when therapy failure is registered. Interestingly, AmB is among the preferred therapeutic options for the first-line approach, mainly in patients under FLZ prophylaxis (Iatta et al., 2014b). Liposomal-AmB seems to be the most active drug due to the lipophilic nature of these yeasts even if in vitro resistance phenomena were registered for L–AmB. The favorable outcome of patients after therapy with micafungin and L-AmB followed by FLZ, might suggest in vivo synergism between these drugs as has been previously reported for Candida spp. (Rosato et al., 2012; Iatta et al., 2015). Although the observed frequency of systemic Malassezia infections is not very high, this may in part be due to underdiagnosis. Use of FLZ prophylaxis with reduced susceptibility for this drug, and the increase of immunocompromised patients, may lead to a higher number of observed Malassezia BSIs in the future. Regardless, emerging infections need to be timely diagnosed to aid clinicians in better patient management. The persistence of Malassezia yeasts on incubator surfaces and on the hands of health care workers or parents suggested the need for punctilious hygienic measures (Chang et al., 1998; Iatta et al., 2014a,b). The role of lipid infusion in aiding the spread of different Malassezia species infections should be also explored. The lack of sufficient literature showing the prevalence of Malassezia fungemia, low specificity and sensitivity of different blood culture systems used for the diagnosis of this fungal sepsis, the lack of standardized methods for in vitro antifungal susceptibility testing, as well as the lack of studies investigating drug resistance phenomena in these yeasts, call for further studies drafting guidelines for the diagnosis and correct management of Malassezia related diseases. Until specific guidelines for diagnosis and treatment of Malassezia bloodstream infections are available, clinicians must be aware of the patient population at risk for these infections and they must communicate to the laboratory the need to include special procedures to recover the organisms. Importantly, commonly used culture media in the clinic do not include lipid supplementation, which is needed for Malassezia yeasts to grow due to their lipid dependence. Finally, the very low susceptibility of some of these yeasts to azole drugs (i.e., FLZ and VOR) and echinocandins should be considered when a long term or prophylactic therapy is expected to be used. As stated before, Malassezia are the major fungal component of the human skin microbiome but for a long time the vast majority of scientific research focused on the role of bacterial microbiota in health and disease. In recent years, the role of fungi attracted more attention regarding their interplay with the human host, but also with other members of the microbiome. Any alteration in either host or microbiota can result in infections and some recent studies emphasized a role for Malassezia spp. in other parts of the body, and linked to other diseases known until then (Kong and Segre, 2020). One study reported a role for M. restricta in Crohn's disease, observing higher relative abundances of intestinal Malassezia compared with healthy controls, evoking inflammatory responses through CARD9 signaling (Limon et al., 2019). Though a few recent studies reported dysbiosis signatures of mycobiota in colorectal cancer (CRC) with enrichment of Malassezia in CRC compared with controls (Gao et al., 2017; Coker et al., 2019); a new study, for the first time, showed direct proof for the involvement of Malassezia in the pathogenesis of cancer. A much increased fungal community in pancreatic ductal adenocarcinomas (PDAs) was significantly enriched for Malassezia. Removal of the mycobiome reduced tumor growth, and only repopulation with Malassezia accelerated oncogenesis. PDA progression was dependent on mannose-binding lectin (MBL), which binds to glycans of the fungal cell wall to activate a part of the immune system called the complement cascade (Aykut et al., 2019). The relationship between Malassezia and the host is complex and more research is needed to understand the various roles that Malassezia may express in/on the human host, and the conditions that trigger them. However, the above mentioned recent advancements have paved the way with new tools and insights that may also benefit a better understanding of Malassezia bloodstream infections.
CC planned, wrote, and contributed to the critical review of the manuscript. WR and BT performed an initial electronic search and drafted and edited the manuscript. CC, BT, TB, and DO performed data cleaning and reviewed the manuscript. CC and BT approved the manuscript for submission. All authors contributed to the article and approved the submitted version.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Aguirre, C., Euliarte, C., Finquelievich, J., de los Ángeles Sosa, M., and Giusiano, G. (2015). Fungemia and interstitial lung compromise caused by Malassezia sympodialis in a pediatric patient. Rev. Iberoam. Micol. 32, 118–121. doi: 10.1016/j.riam.2014.01.002
Alpert, G., Bell, L. M., and Campos, J. M. (1987). Malassezia furfur fungemia in infancy. Clin. Pediatr. 26, 528–531. doi: 10.1177/000992288702601007
Al-Sweih, N., Ahmad, S., Joseph, L., Khan, S., and Khan, Z. (2014). Malassezia pachydermatis fungemia in a preterm neonate resistant to fluconazole and flucytosine. Med. Mycol. Case Rep. 5, 9–11. doi: 10.1016/j.mmcr.2014.04.004
Angiolella, L., Carradori, S., Maccallini, C., Giusiano, G., and Supuran, C. T. (2017). Targeting Malassezia species for novel synthetic and natural antidandruff agents. Curr. Med. Chem. 24, 2392–2412. doi: 10.2174/0929867324666170404110631
Angiolella, L., Leone, C., Rojas, F., Mussin, J., de Los Angeles Sosa, M., and Giusiano, G. (2018). Biofilm, adherence, and hydrophobicity as virulence factors in Malassezia furfur. Med. Mycol. 56, 110–116. doi: 10.1093/mmy/myx014
Arendrup, M. C., Boekhout, T., Akova, M., Meis, J. F., Cornely, O. A., Lortholary, O., and ESCMID EFISG study group ECMM. (2014). ESCMID and ECMM joint clinical guidelines for the diagnosis and management of rare invasive yeast infections. Clin. Microbiol. Infect. 20, 76–98. doi: 10.1111/1469-0691.12360
Aschner, J. L., Punsalang, A., Maniscalco, W. M., and Menegus, M. A. (1987). Percutaneous central venous catheter colonization with Malassezia furfur: incidence and clinical significance. Pediatrics 80, 535–539.
Aykut, B., Pushalkar, S., Chen, R., Li, Q., Abengozar, R., Kim, J. I., et al. (2019). The fungal mycobiome promotes pancreatic oncogenesis via activation of MBL. Nature 574, 264–267. doi: 10.1038/s41586-019-1608-2
Barber, G. R., Brown, A. E., Kiehn, T. E., Edwards, F. F., and Armstrong, D. (1993). Catheter-related Malassezia furfur fungemia in immunocompromised patients. Am. J. Med. 95, 365–370. doi: 10.1016/0002-9343(93)90304-8
Bernier, V., Weill, F. X., Hirigoyen, V., Elleau, C., Feyler, A., Labrèze, C., et al. (2002). Skin Colonization by Malassezia in Neonates and Infants. Arch. Dermatol. 138, 215–218. doi: 10.1001/archderm.138.2.215
Bielska, E., Sisquella, M. A., Aldeieg, M., Birch, C., O'Donoghue, E. J., and May, R. C. (2018). Pathogen-derived extracellular vesicles mediate virulence in the fatal human pathogen Cryptococcus gattii. Nat. Commun. 9:1556. doi: 10.1038/s41467-018-03991-6
Bond, R., Guillot, J., and Cabañes, F. J. (2010). “Malassezia yeasts in animal disease,” in Malassezia and the Skin: Science and Clinical Practice, eds T. Boekhout, E. Gueho, P. Mayser, and A. Velegraki (Berlin; Heidelberg: Springer), 271–299. doi: 10.1007/978-3-642-03616-3_10
Bond, R., Morris, D. O., Guillot, J., Bensignor, E. J., Robson, D., Mason, K. V., et al. (2020). Biology, diagnosis and treatment of Malassezia dermatitis in dogs and cats Clinical Consensus Guidelines of the World Association for Veterinary Dermatology. Vet. Dermatol. 31, 28–74. doi: 10.1111/vde.12809
Cabañes, F. J., Coutinho, S. D. A., Puig, L., Bragulat, M. R., and Castellá, G. (2016). New lipid-dependent Malassezia species from parrots. Annu. Rev. Microbiol. 33, 92–99. doi: 10.1016/j.riam.2016.03.003
Cafarchia, C., Dell'Aquila, M. E., Traversa, D., Albrizio, M., Guaricci, A. C., De Santis, T., et al. (2010). Expression of the μ-opioid receptor on Malassezia pachydermatis and its effect in modulating phospholipase production. Med. Mycol. 48, 73–78. doi: 10.3109/13693780902718347
Cafarchia, C., Figueredo, L. A., Iatta, R., Colao, V., Montagna, M. T., and Otranto, D. (2012). In vitro evaluation of Malassezia pachydermatis susceptibility to azole compounds using E-test and CLSI microdilution methods. Med. Mycol. 50, 795–801. doi: 10.3109/13693786.2012.674219
Cafarchia, C., Gallo, S., Capelli, G., and Otranto, D. (2005). Occurrence and population size of Malassezia spp. in the external ear canal of dogs and cats both healthy and with otitis. Mycopathologia 160, 143–149. doi: 10.1007/s11046-005-0151-x
Cafarchia, C., Gasser, R. B., Figueredo, L. A., Latrofa, M. S., and Otranto, D. (2011b). Advances in the identification of Malassezia. Mol. Cell. Probes 25, 1–7. doi: 10.1016/j.mcp.2010.12.003
Cafarchia, C., Gasser, R. B., Latrofa, M. S., Parisi, A., Campbell, B. E., and Otranto, D. (2008). Genetic variants of Malassezia pachydermatis from canine skin: body distribution and phospholipase activity. FEMS Yeast Res. 8, 451–459. doi: 10.1111/j.1567-1364.2008.00358.x
Cafarchia, C., Iatta, R., Immediato, D., Puttilli, M. R., and Otranto, D. (2015). Azole susceptibility of Malassezia pachydermatis and Malassezia furfur and tentative epidemiological cut-off values. Med. Mycol. 53, 743–748. doi: 10.1093/mmy/myv049
Cafarchia, C., Latrofa, M. S., Figueredo, L. A., Da Silva Machado, M. L., Ferreiro, L., Guillot, J., et al. (2011a). Physiological and molecular characterization of atypical lipid-dependent Malassezia yeasts from a dog with skin lesions: adaptation to a new host? Med. Mycol. 49, 365–374. doi: 10.3109/13693786.2010.531487
Cafarchia, C., and Otranto, D. (2004). Association between phospholipase production by Malassezia pachydermatis and skin lesions. J. Clin. Microbiol. 42, 4868–4869. doi: 10.1128/JCM.42.10.4868-4869.2004
Campigotto, A., Richardson, S. E., Sebert, M., TeKippe, E. M., Chakravarty, A., and Doern, C. D. (2016). Low utility of pediatric isolator blood culture system for detection of fungemia in children: a 10-year review. J. Clin. Microbiol. 54, 2284–2287. doi: 10.1128/JCM.00578-16
Cannizzo, F. T., Eraso, E., Ezkurra, P. A., Villar-Vidal, M., Bollo, E., Castellá, G., et al. (2007). Biofilm development by clinical isolates of Malassezia pachydermatis. Med. Mycol. 45,357–361. doi: 10.1080/13693780701225767
Celis, A. M., Vos, A. M., Triana, S., Medina, C. A., Escobar, N., Restrepo, S., et al. (2017b). Highly efficient transformation system for Malassezia furfur and Malassezia pachydermatis using Agrobacterium tumefaciens-mediated transformation. J. Microbiol. Methods 134, 1–6. doi: 10.1016/j.mimet.2017.01.001
Celis, A. M., Wösten, H. A. B., Triana, S., Restrepo, S., and de Cock, H. (2017a). Malassezia spp. beyond the mycobiota. SM Dermatol. J. 3, 1019-1–1019-10. doi: 10.36876/smdj.1019
Chang, H. J., Miller, H. L., Watkins, N., Arduino, M. J., Ashford, D. A., Midgley, G., et al. (1998). An epidemic of Malassezia pachydermatis in an intensive care nursery associated with colonization of health care workers' pet dogs. N. Engl. J. Med. 338, 706–711. doi: 10.1056/NEJM199803123381102
Chen, I. T., Chen, C. C., Huang, H. C., and Kuo, K. C. (2019). Malassezia furfur emergence and candidemia trends in a neonatal intensive care unit during 10 years: the experience of fluconazole prophylaxis in a single hospital. Adv. Neonatal. 20, E3–E8. doi: 10.1097/ANC.0000000000000640
Chen, T. A., and Hill, P. B. (2005). The biology of Malassezia organisms and their ability to induce immune responses and skin disease. Vet. Dermatol. 16, 4–26. doi: 10.1111/j.1365-3164.2005.00424.x
Choudhury, S., and Marte, R. L. (2014). Malassezia pachydermatis fungaemia in an adult on posaconazole prophylaxis for acute myeloid leukaemia. Pathology J. RCPA 46, 466–467. doi: 10.1097/PAT.0000000000000139
Chow, N. A., Chinn, R., Pong, A., Schultz, K., Kim, J., Gade, L., et al. (2020). Use of whole-genome sequencing to detect an outbreak of Malassezia pachydermatis infection and colonization in a neonatal intensive care unit—California, 2015–2016. Infect. Control Hosp. Epidemiol. 41, 1–3. doi: 10.1017/ice.2020.73
Chryssanthou, E., Broberger, U., and Petrini, B. (2001). Malassezia pachydermatis fungaemia in a neonatal intensive care unit. Acta Paediatr. 90, 323–327. doi: 10.1080/080352501300067712
Chu, C. M., and Lai, R. W. (2002). Malassezia furfur fungaemia in a ventilator-dependent patient without known risk factors. Hong Kong Med. J. 8, 212–215.
Coker, O. O., Nakatsu, G., Dai, R. Z., Wu, W. K. K., Wong, S. H., Ng, S. C., et al. (2019). Enteric fungal microbiota dysbiosis and ecological alterations in colorectal cancer. Gut 68, 654–662. doi: 10.1136/gutjnl-2018-317178
Dankner, W. M., Spector, S. A., Fierer, J., and Davis, C. E. (1987). Malassezia fungemia in neonates and adults: complication of hyperalimentation. Rev. Infect. Dis. 9, 743–753. doi: 10.1093/clinids/9.4.743
Figueredo, L. A., Cafarchia, C., Desantis, S., and Otranto, D. (2013). Biofilm formation of Malassezia pachydermatis from dogs. Vet. Microbiol. 160, 126–131. doi: 10.1016/j.vetmic.2012.05.012
Findley, K., Oh, J., Yang, J., Conlan, S., Deming, C., Meyer, J. A., et al. (2013). Topographic diversity of fungal and bacterial communities in human skin. Nature 498, 367–370. doi: 10.1038/nature12171
Gaitanis, G., Magiatis, P., Hantschke, M., Bassukas, I. D., and Velegraki, A. (2012). The Malassezia genus in skin and systemic diseases. Clin. Microbiol. Rev. 25, 106–141. doi: 10.1128/CMR.00021-11
Gaitanis, G., Velegraki, A., Magiatis, P., Pappas, P., and Bassukas, I. D. (2011). Could Malassezia yeasts be implicated in skin carcinogenesis through the production of aryl-hydrocarbon receptor ligands? Med. Hypotheses 77, 47–51. doi: 10.1016/j.mehy.2011.03.020
Gao, R., Kong, C., Li, H., Huang, L., Qu, X., Qin, N., et al. (2017). Dysbiosis signature of mycobiota in colon polyp and colorectal cancer. Eur. J. Clin. Microbiol Infect. Dis. 36, 2457–2468. doi: 10.1007/s10096-017-3085-6
Garcia, C. R., Johnston, B. L., Corvi, G., Walker, L. J., and George, W. L. (1987). Intravenous catheter-associated Malassezia furfur fungemia. Am. J. Med. 83, 790–792. doi: 10.1016/0002-9343(87)90917-X
Giusiano, G., Mangiaterra, M., Saito, V. G., Rojas, F., Gómez, V., and Díaz, M. C. (2006). Fluconazole and itraconazole resistance of yeasts isolated from the bloodstream and catheters of hospitalized pediatric patients. Chemotherapy 52, 254–259. doi: 10.1159/000094867
Grice, E. A., and Segre, J. A. (2011). The skin microbiome. Nat. Rev. Microbiol. 9, 244–253. doi: 10.1038/nrmicro2537
Guillot, J., and Bond, R. (2020). Malassezia yeasts in veterinary dermatology: an pdated overview. Front Cell Infect. Microbiol. 10:79. doi: 10.3389/fcimb.2020.00079
Gupta, A. K., Boekhout, T., Theelen, B., Summerbell, R., and Batra, R. (2004). Identification and typing of Malassezia species by amplified fragment length polymorphism and sequence analyses of the internal transcribed spacer and large-subunit regions of ribosomal DNA. J. Clin. Microbiol. 42, 4253–4260. doi: 10.1128/JCM.42.9.4253-4260.2004
Gupta, P., Chakrabarti, A., Singhi, S., Kumar, P., Honnavar, P., and Rudramurthy, S. M. (2014). Skin colonization by Malassezia spp. in hospitalized neonates and infants in a tertiary care centre in North India. Mycopathologia 178, 267–272. doi: 10.1007/s11046-014-9788-7
Hassall, E., Ulich, T., and Ament, M. E. (1983). Pulmonary embolus and Malassezia pulmonary infection related to urokinase therapy. J. Pediatr. 102, 722–725. doi: 10.1016/S0022-3476(83)80244-3
Honnavar, P., Chakrabarti, A., Prasad, G. S., Singh, P., Dogra, S., and Rudramurthy, S. M. (2017). β-Endorphin enhances the phospholipase activity of the dandruff causing fungi Malassezia globosa and Malassezia restricta. Med. Mycol. 55, 150–154. doi: 10.1093/mmy/myw058
Huang, C. Y., Peng, C. C., Hsu, C. H., Chang, J. H., Chiu, N. C., and Chi, H. (2020). Systemic infection caused by Malassezia pachydermatis in infants. Pediatr. Infect. Dis. J. 39, 444–448. doi: 10.1097/INF.0000000000002591
Ianiri, G., Averette, A. F., Kingsbury, J. M., Heitman, J., and Idnurm, A. (2016). Gene function analysis in the ubiquitous human commensal and pathogen Malassezia genus. mBio 7:e01853-16. doi: 10.1128/mBio.01853-16
Ianiri, G., Dagotto, G., Sun, S., and Heitman, J. (2019). Advancing functional genetics through Agrobacterium-mediated insertional mutagenesis and CRISPR/Cas9 in the commensal and pathogenic yeast Malassezia. Genetics 212, 1163–1179. doi: 10.1534/genetics.119.302329
Iatta, R., Battista, M., Miragliotta, G., Boekhout, T., Otranto, D., and Cafarchia, C. (2018). Blood culture procedures and diagnosis of Malassezia furfur bloodstream infections: strength and weakness. Med. Mycol. 56, 828–833. doi: 10.1093/mmy/myx122
Iatta, R., Cafarchia, C., Cuna, T., Montagna, O., Laforgia, N., Gentile, O., et al. (2014a). Bloodstream infections by Malassezia and Candida species in critical care patients. Med. Mycol. 52, 264–269. doi: 10.1093/mmy/myt004
Iatta, R., Figueredo, L. A., Montagna, M. T., Otranto, D., and Cafarchia, C. (2014b). In vitro antifungal susceptibility of Malassezia furfur from bloodstream infections. J. Med. Microbiol. 63, 1467–1473. doi: 10.1099/jmm.0.078709-0
Iatta, R., Immediato, D., Montagna, M. T., Otranto, D., and Cafarchia, C. (2015). In vitro activity of two amphotericin B formulations against Malassezia furfur strains recovered from patients with bloodstream infections. Med. Mycol.53, 269–274. doi: 10.1093/mmy/myu089
Iatta, R., Puttilli, M. R., Immediato, D., Otranto, D., and Cafarchia, C. (2017). The role of drug efflux pumps in Malassezia pachydermatis and Malassezia furfur defence against azoles. Mycoses 60, 178–182. doi: 10.1111/myc.12577
Ilahi, A., Hadrich, I., Goudjil, S., Kongolo, G., Chazal, C., Léké, A., et al. (2017). Molecular epidemiology of a Malassezia pachydermatis neonatal unit outbreak. Med. Mycol. 56, 69–77. doi: 10.1093/mmy/myx022
Jang, S. J., Lim, S. H., Ko, J. H., Oh, B. H., Kim, S. M., Song, Y. C., et al. (2009). The investigation on the distribution of Malassezia yeasts on the normal Korean skin by 26S rDNA PCR-RFLP. Ann. Dermatol. 21, 18–26. doi: 10.5021/ad.2009.21.1.18
Jo, J. H., Deming, C., Kennedy, E. A., Kennedy, E. A., Conlan, S., Polley, E. C., et al. (2016). Diverse human skin fungal communities in children converge in adulthood. J. Invest. Dermatol. 136, 2356–2363. doi: 10.1016/j.jid.2016.05.130
Johansson, H. J., Vallhov, H., Holm, T., Gehrmann, U., Andersson, A., and Johansson, C. (2018). Extracellular nanovesicles released from the commensal yeast Malassezia sympodialis are enriched in allergens and interact with cells in human skin. Sci. Rep. 8, 1–11. doi: 10.1038/s41598-018-27451-9
Kaneko, T., Murotani, M., Ohkusu, K., Sugita, T., and Makimura, K. (2012). Genetic and biological features of catheter-associated Malassezia furfur from hospitalized adults. Med. Mycol. 50, 74–80. doi: 10.3109/13693786.2011.584913
Kikuchi, K., Fujishiro, Y., Totsuka, K., Seshimo, A., Kameoka, S., Makimura, K., et al. (2001). A case of central venous catheter-related infection with Malassezia sympodialis. Nippon Ishinkin Gakkai Zasshi 42, 220–222. doi: 10.3314/jjmm.42.220
Kim, M., Cho, Y. J., Park, M., Choi, Y., Hwang, S. Y., and Jung, W. H. (2018). Genomic tandem quadruplication is associated with ketoconazole resistance in Malassezia pachydermatis. J. Microbiol Biotechnol. 28, 1937–1945. doi: 10.4014/jmb.1810.10019
Kim, N. Y., and Pandya, A. G. (1998). Pigmentary diseases. Med. Clin. North Am. 82, 1185–1207. doi: 10.1016/S0025-7125(05)70410-7
Kobayashi, T., Kano, R., Nagata, M., and Hasegawa, A. (2011). Genotyping of Malassezia pachydermatis isolates from canine healthy skin and atopic dermatitis by internal spacer 1 (IGS1) region analysis. Vet Dermatol. 22, 401–405. doi: 10.1111/j.1365-3164.2011.00961.x
Kong, H. H., and Segre, J. A. (2020). Cultivating fungal research. Science. 368, 365–366. doi: 10.1126/science.aaz8086
Lee, J., Cho, Y. G., Kim, D. S., Choi, S. I., and Lee, H. S. (2019). First case of catheter-related Malassezia pachydermatis fungemia in an adult. Ann. Lab. Med. 39, 99–101. doi: 10.3343/alm.2019.39.1.99
Leong, C., Buttafuoco, A., Glatz, M., and Bosshard, P. P. (2017). Antifungal susceptibility testing of Malassezia spp. with an optimized colorimetric broth microdilution method. J. Clin. Microbiol. 55, 1883–1893. doi: 10.1128/JCM.00338-17
Leong, C., Schmid, B., Toi, M. J., Wang, J., Irudayaswamy, A. S., Goh, J. P. Z., et al. (2019). Geographical and ethnic differences influence culturable commensal yeast diversity on healthy skin. Front. Microbiol. 10:1891. doi: 10.3389/fmicb.2019.01891
Li, H., Goh, B. N., Teh, W. K., Jiang, Z., Goh, J. P. Z., Goh, A., et al. (2018). Skin commensal Malassezia globosa secreted protease attenuates Staphylococcus aureus biofilm formation. J. Investig. Dermatol. 138, 1137–1145 doi: 10.1016/j.jid.2017.11.034
Limon, J. J., Tang, J., Li, D., Wolf, A. J., Michelsen, K. S., Funari, V., et al. (2019). Malassezia is associated with Crohn's disease and exacerbates colitis in mouse models. Cell Host Microbe 25, 377–388. doi: 10.1016/j.chom.2019.01.007
Lorch, J. M., Palmer, J. M., Vanderwolf, K. J., Schmidt, K. Z., Verant, M. L., Weller, T. J., et al. (2018). Malassezia vespertilionis sp. nov.: a new cold-tolerant species of yeast isolated from bats. Persoonia: Mol. Phylo. Evol. Fungi 41, 56–70. doi: 10.3767/persoonia.2018.41.04
Machado, M. L., Cafarchia, C., Otranto, D., Ferreira, R. R., Bianchi, S. P., Latrofa, M. S., et al. (2010). Genetic variability and phospholipase production of Malassezia pachydermatis isolated from dogs with diverse grades of skin lesions. Med. Mycol. 48, 889–892. doi: 10.3109/13693780903532080
Malavia, D., Gow, N. A. R., and Usher, J. (2020). Advances in molecular tools and in vivo models for the study of human fungal pathogenesis. Microorganisms 8:803. doi: 10.3390/microorganisms8060803
Masure, O., Leostic, C., Abalain, M. L., Chastel, C., Yakoub-Agha, I., Berthou, C., et al. (1991). Malassezia furfur septicaemia in a child with leukaemia. J. Infect. 23, 335–336. doi: 10.1016/0163-4453(91)93296-O
Mellinghoff, S. C., Cornely, O. A., and Jung, N. (2018). Essentials in Candida bloodstream infection. Infection 46, 897–899. doi: 10.1007/s15010-018-1218-1
Miceli, M. H., Díaz, J. A., and Lee, S. A. (2011). Emerging opportunistic yeast infections. Lancet Infect. Dis. 11, 142–151. doi: 10.1016/S1473-3099(10)70218-8
Min, K., Neiman, A. M., and Konopka, J. B. (2020). Fungal pathogens: shape-shifting invaders. Trends Microbiol. doi: 10.1016/j.tim.2020.05.001. [Epub ahead of print].
Morris, D. O., O'Shea, K., Shofer, F. S., and Rankin, S. (2005). Malassezia pachydermatis carriage in dog owners. Emerg. Infect. Dis. 11, 83–88. doi: 10.3201/eid1101.040882
Morrison, V. A., and Weisdorf, D. J. (2000). The spectrum of Malassezia infections in the bone marrow transplant population. Bone Marrow Transplant. 26, 645–648. doi: 10.1038/sj.bmt.1702566
Nagata, R., Nagano, H., Ogishima, D., Nakamura, Y., Hiruma, M., and Sugita, T. (2012). Transmission of the major skin microbiota, Malassezia, from mother to neonate. Pediatr. Int. 54, 350–355. doi: 10.1111/j.1442-200X.2012.03563.x
Negre, A., Bensignor, E., and Guillot, J. (2009). Evidence-based veterinary dermatology: a systematic review of interventions for Malassezia dermatitis in dogs. Vet. Dermatol. 20, 1–12. doi: 10.1111/j.1365-3164.2008.00721.x
Nelson, S. C., Yau, Y. C., Richardson, S. E., and Matlow, A. G. (1995). Improved detection of Malassezia species in lipid-supplemented Peds Plus blood culture bottles. J. Clin. Microbiol. 33, 1005–1007. doi: 10.1128/JCM.33.4.1005-1007.1995
Oh, J., Byrd, A., Deming, C., Conlan, S., NISC Comparative Sequencing Program, Kong, H. K., et al. (2014). Biogeography and individuality shape function in the human skin metagenome. Nature 514, 59–64. doi: 10.1038/nature13786
Oliveri, S., Trovato, L., Betta, P., Romeo, M., and Nicoletti, G. (2011). Malassezia furfur fungaemia in a neonatal patient detected by lysis-centrifugation blood culture method: first case reported in Italy. Mycoses 54, e638–e640. doi: 10.1111/j.1439-0507.2010.01955.x
Pan, Y. X. (2005). Diversity and complexity of the mu opioid receptor gene: alternative pre-mRNA splicing and promoters. DNA Cell Biol. 24, 736–750. doi: 10.1089/dna.2005.24.736
Park, M., Cho, Y. J., Lee, Y. W., and Jung, W. H. (2017). Whole genome sequencing analysis of the cutaneous pathogenic yeast Malassezia restricta and identification of the major lipase expressed on the scalp of patients with dandruff. Mycoses 60, 188–197. doi: 10.1111/myc.12586
Paul, A. A., Hoffman, K. L., Hagan, J. L., Sampath, V., Petrosino, J. F., and Pammi, M. (2019). Fungal cutaneous microbiome and host determinants in preterm and term neonates. Pediatr. Res. doi: 10.1038/s41390-019-0719-7. [Epub ahead of print]
Pedrosa, A. F., Lisboa, C., Branco, J., Almeida, A. C., Mendes, C., Pellevoisin, C., et al. (2019a). Malassezia colonisation on a reconstructed human epidermis: Imaging studies. Mycoses 62, 1194–1201. doi: 10.1111/myc.13011
Pedrosa, A. F., Lisboa, C., Faria-Ramos, I., Silva, R., Ricardo, E., Teixeira-Santos, R., et al. (2019b). Epidemiology and susceptibility profile to classic antifungals and over-the-counter products of Malassezia clinical isolates from a Portuguese University Hospital: a prospective study. J. Med. Microbiol. 68, 778–784. doi: 10.1099/jmm.0.000966
Pedrosa, A. F., Lisboa, C., and Rodrigues, A. G. (2018). Malassezia infections with systemic involvement: figures and facts. J. Dermatol. 45, 1278–1282. doi: 10.1111/1346-8138.14653
Peker, N., Couto, N., Sinha, B., and Rossen, J. W. (2018). Diagnosis of bloodstream infections from positive blood cultures and directly from blood samples: recent developments in molecular approaches. Clin. Microbiol. Infect. 24, 944–955. doi: 10.1016/j.cmi.2018.05.007
Petrokilidou, C., Pavlou, E., Gaitanis, G., Bassukas, I. D., Saridomichelakis, M. N., Velegraki, A., et al. (2019). The lipid profile of three Malassezia species assessed by Raman spectroscopy and discriminant analysis. Mol. Cell. Probes. 46:101416. doi: 10.1016/j.mcp.2019.06.006
Poh, S. E., Goh, J. P. Z., Fan, C., Chua, W., Gan, S. Q., Lim, P. L. K., et al. (2020). Identification of Malassezia furfur secreted aspartyl protease 1 (MfSAP1) and its role in extracellular matrix degradation. Front. Cell Infect. Microbiol. 9:148. doi: 10.3389/fcimb.2020.00148
Powell, D. A., Aungst, J., Snedden, S., Hansen, N., and Brady, M. (1984). Broviac catheter-related Malassezia furfur sepsis in five infants receiving intravenous fat emulsions. J. Pediatr. 105, 987–990. doi: 10.1016/S0022-3476(84)80096-7
Prado, M. R., Brito, É. H., Brilhante, R. S., Cordeiro, R. A., Leite, J. J., Sidrim, J. J., et al. (2008). Subculture on potato dextrose agar as a complement to the broth microdilution assay for Malassezia pachydermatis. J. Microbiol. Methods 75, 341–343. doi: 10.1016/j.mimet.2008.05.022
Prohic, A., Kuskunovic-Vlahovljak, S., Sadikovic, T. J., and Cavaljuga, S. (2015). The prevalence and species composition of Malassezia yeasts in patients with clinically suspected onychomycosis. Med. Arh. 69, 81–84. doi: 10.5455/medarh.2015.69.81-84
Prohic, A., Simic, D., Sadikovic, T. J., and Krupalija-fazlic, M. (2014). Distribution of Malassezia species on healthy human skin in Bosnia and Herzegovina: correlation with body part, age and gender. Iran. J. Microbiol. 6, 253–262.
Redline, R. W., and Dahms, B. B. (1981). Malassezia pulmonary vasculitis in an infant on long-term intralipid therapy. N. Engl. J. Med. 305, 1395–1398. doi: 10.1056/NEJM198112033052307
Redline, R. W., Redline, S. S., Boxerbaum, B., and Dahms, B. B. (1985). Systemic Malassezia furfur infections in patients receiving intralipid therapy. Hum. Pathol. 16, 815–822. doi: 10.1016/S0046-8177(85)80253-7
Rojas, F. D., Sosa, M. D. L. A., Fernandez, M. S., Cattana, M. E., Cordoba, S. B., and Giusiano, G. E. (2014). Antifungal susceptibility of Malassezia furfur, Malassezia sympodialis, and Malassezia globosa to azole drugs and amphotericin B evaluated using a broth microdilution method. Sabouraudia 52, 641–646. doi: 10.1093/mmy/myu010
Roman, J., Bagla, P., Ren, P., Blanton, L. S., and Berman, M. A. (2016). Malassezia pachydermatis fungemia in an adult with multibacillary leprosy. Med. Mycol. Case Rep. 12, 1–3. doi: 10.1016/j.mmcr.2016.05.002
Rosales, C. M., Jackson, M. A., and Zwick, D. (2004). Malassezia furfur meningitis associated with total parenteral nutrition subdural effuion. Pediatr. Dev. Pathol. 7, 86–90. doi: 10.1007/s10024-003-4030-5
Rosato, A., Piarulli, M., Immacolata Pia Schiavone, B., Teresa Montagna, M., Caggiano, G., Muraglia, M., et al. (2012). In vitro synergy testing of anidulafungin with fluconazole, tioconazole, 5-flucytosine and amphotericin B against some Candida spp. Med. Chem. 8, 690–698. doi: 10.2174/157340612801216184
Saadatzadeh, M. R., Ashbee, H. R., Holland, K. T., and Ingham, E. (2001). Production of the mycelial phase of Malassezia in vitro. Med. Mycol. 39, 487–493. doi: 10.1080/mmy.39.6.487.493
Saunte, D. M. L., Gaitanis, G., and Hay, R. J. (2020). Malassezia-associated skin diseases, the use of diagnostics and treatment. Front. Cell Infect. Microbiol. 20:112. doi: 10.3389/fcimb.2020.00112
Schleman, K. A., Tullis, G., and Blum, R. (2000). Intracardiac mass complicating Malassezia furfur fungemia. Chest 118, 1828–1829. doi: 10.1378/chest.118.6.1828
Schoepfer, C., Carla, H., Bezou, M. J., Cambon, M., Girault, D., Deméocq, F., et al. (1995). Septicémie à Malassezia furfur au décours d'une greffe de moelle. Arch de Pédiatrie 2, 245–248. doi: 10.1016/0929-693X(96)81136-5
Shek, Y. H., Tucker, M. C., Viciana, A. L., Manz, H. J., and Connor, D. H. (1989). Malassezia furfur disseminated infection in premature infants. Am. J. Clin. Pathol. 92, 595–603. doi: 10.1093/ajcp/92.5.595
Shparago, N. I., Bruno, P. P., and Bennett, J. (1995). Systemic Malassezia furfur infection in an adult receiving total parenteral nutrition. J. Am. Osteopath. Assoc. 95, 375–377. doi: 10.7556/jaoa.1995.95.6.375
Sizun, J., Karangwa, A., Giroux, J. D., Masure, O., Simitzis, A. M., Alix, D., et al. (1994). Malassezia furfur-related colonization and infection of central venous catheters. Intensive Care Med. 20, 496–499. doi: 10.1007/BF01711902
Sparber, F., de Gregorio, C., Steckholzer, S., Ferreira, F. M., Dolowschiak, T., Ruchti, F., et al. (2019). The skin commensal yeast Malassezia triggers a type 17 response that coordinates anti-fungal immunity and exacerbates skin inflammation. Cell Host Microbe. 25, 389–403. doi: 10.1016/j.chom.2019.02.002
Sparber, F., and LeibundGut-Landmann, S. (2019). Infecting mice with Malassezia spp. to study the fungus-host interaction. J. Vis. Exp. 153:e60175. doi: 10.3791/60175
Sugita, T., Kodama, M., Saito, M., Ito, T., Kato, Y., Tsuboi, R., et al. (2003). Sequence diversity of the intergenic spacer region of the rRNA gene of Malassezia globosa colonizing the skin of patients with atopic dermatitis and healthy individuals. J. Clin. Microbiol. 41, 3022–3027. doi: 10.1128/JCM.41.7.3022-3027.2003
Sugita, T., Tajima, M., Amaya, M., Tsuboi, R., and Nishikawa, A. (2004). Genotype analysis of Malassezia restricta as the major cutaneous flora in patients with atopic dermatitis and healthy subjects. Microbiol. Immunol. 48, 755–759. doi: 10.1111/j.1348-0421.2004.tb03601.x
Sugita, T., Tajima, M., Ito, T., Saito, M., Tsuboi, R., and Nishikawa, A. (2005). Antifungal activities of tacrolimus and azole agents against the eleven currently accepted Malassezia species. J. Clin. Microbiol. 43, 2824–2829. doi: 10.1128/JCM.43.6.2824-2829.2005
Surmont, I., Gavilanes, A., Vandepitte, J., Devlieger, H., and Eggermont, E. (1989). Malassezia furfur fungaemia in infants receiving intravenous lipid emulsions. A rarity or just underestimated? Eur. J. Pediatr. 148, 435–438. doi: 10.1007/BF00595906
Tee, C. B., Sei, Y., and Kajiwara, S. (2019). Secreted hydrolytic and haemolytic activities of Malassezia clinical strains. Mycopathologia 184, 227–238. doi: 10.1007/s11046-019-00330-1
Theelen, B., Cafarchia, C., Gaitanis, G., Bassukas, I. D., Boekhout, T., and Dawson, T. L. Jr. (2018). Malassezia ecology, pathophysiology, and treatment. Med. Mycol. 56, 10–25. doi: 10.1093/mmy/myx134
Theelen, B., Silvestri, M., Guého, E., Belkum, A., and Boekhout, T. (2001). Identification and typing of Malassezia yeasts using amplified polymorphic DNA (RAPD) and denaturing gradient gel electrophoresis (DGGE). FEMS Yeast Res. 1, 79–86. doi: 10.1111/j.1567-1364.2001.tb00018.x
Torres, M., Pinzón, E. N., Re, F. M., Heydys, M., Parra, G. C. M., Celis, A. M., et al. (2020). Galleria mellonella as a novelty in vivo model of host-pathogen interaction for Malassezia furfur CBS 1878 and Malassezia pachydermatis CBS 1879. Front. Cell. Infect. Microbiol. 10:199. doi: 10.3389/fcimb.2020.00199
Vallhov, H., Johansson, C., Veerman, R. E., and Scheynius, A. (2020). Extracellular vesicles released from the skin commensal yeast Malassezia sympodialis activate human primary keratinocytes. Front. Cell Infect. Microbiol. 24:6. doi: 10.3389/fcimb.2020.00006
Velegraki, A., Alexopoulos, E. C., Kritikou, S., and Gaitanis, G. (2005). Use of fatty acid RPMI 1640 media for testing susceptibilities of eight Malassezia species to the new triazole posaconazole and to six established antifungal agents by a aodified NCCLS M27-A2 microdilution method and etest. J. Clin. Microbiol.43:1014. doi: 10.1128/JCM.43.2.1014.2005
Velegraki, A., Cafarchia, C., Gaitanis, G., Iatta, R., and Boekhout, T. (2015). Malassezia infections in humans and animals: pathophysiology, detection, and treatment. PLoS Pathog. 11:e1004523. doi: 10.1371/journal.ppat.1004523
Vlachos, C., Gaitanis, G., Alexopoulos, E. C., Papadopoulou, C., and Bassukas, I. D. (2013). Phospholipase activity after β-endorphin exposure discriminates Malassezia strains isolated from healthy and seborrhoeic dermatitis skin. J. Eur. Acad. Dermatol. Venereol. 27, 1575–1578. doi: 10.1111/j.1468-3083.2012.04638.x
Wang, Q. M., Theelen, B., Groenewald, M., Bai, F. Y., and Boekhout, T. (2014). Moniliellomycetes and Malasseziomycetes, two new classes in Ustilaginomycotina. Persoonia 33, 41–47. doi: 10.3767/003158514X682313
Ward, T. L., Dominguez-Bello, M. G., Heisel, T., Al-Ghalith, G., Knights, D., and Gale, C. A. (2018). Development of the human mycobiome over the first month of life and across body sites. mSystems 3, 140–157. doi: 10.1128/mSystems.00140-17
Weiss, S. J., Schoch, P. E., and Cunha, B. A. (1991). Malassezia furfur fungemia associated with central venous catheter lipid emulsion infusion. Heart Lung: J. Critic. Care 20, 87–90.
Welbel, S. F., McNeil, M. M., Pramanik, A., Silberman, R., Oberle, A. D., Midgley, G., et al. (1994). Nosocomial Malassezia pachydermatis bloodstream infections in a neonatal intensive care unit. Pediatr. Infect. Dis. J. 13, 104–108. doi: 10.1097/00006454-199402000-00005
Wheeler, M. L., Limon, J. J., and Underhill, D. M. (2017). Immunity to commensal fungi: detente and disease. Annu. Rev. Pathol. 12, 359–385. doi: 10.1146/annurev-pathol-052016-100342
Wu, G., Zhao, H., Li, C., Rajapakse, M. P., Wong, W. C., Xu, J., et al. (2015). Genus-wide comparative genomics of Malassezia delineates its phylogeny, physiology, and niche adaptation on human skin. PLoS Genet. 11:e1005614. doi: 10.1371/journal.pgen.1005614
Wurtz, R. M., and Knospe, W. N. (1988). Malassezia furfur fungemia in a patient without the usual risk factors. Ann. Intern. Med. 109, 432–433. doi: 10.7326/0003-4819-109-5-432
Yurayart, C., Nuchnoul, N., Moolkum, P., Jirasuksiri, S., Niyomtham, W., Chindamporn, A., et al. (2013). Antifungal agent susceptibilities and interpretation of Malassezia pachydermatis and Candida parapsilosis isolated from dogs with and without seborrheic dermatitis skin. Med. Mycol. 51, 721–730. doi: 10.3109/13693786.2013.777165
Zhang, Y. J., Han, Y., Sun, Y. Z., Jiang, H. H., Liu, M., Qi, R. Q., et al. (2019). Extracellular vesicles derived from Malassezia furfur stimulate IL-6 production in keratinocytes as demonstrated in in vitro and in vivo models. J. Dermatol. Sci. 93, 168–175. doi: 10.1016/j.jdermsci.2019.03.001
Keywords: Malassezia spp., epidemiology, pathogenesis, fungemia, diagnosis, therapy, antifungal profile
Citation: Rhimi W, Theelen B, Boekhout T, Otranto D and Cafarchia C (2020) Malassezia spp. Yeasts of Emerging Concern in Fungemia. Front. Cell. Infect. Microbiol. 10:370. doi: 10.3389/fcimb.2020.00370
Received: 04 March 2020; Accepted: 16 June 2020;
Published: 28 July 2020.
Edited by:
Salomé LeibundGut-Landmann, University of Zurich, SwitzerlandReviewed by:
Adnane Sellam, Université de Montréal, CanadaCopyright © 2020 Rhimi, Theelen, Boekhout, Otranto and Cafarchia. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Claudia Cafarchia, Y2xhdWRpYS5jYWZhcmNoaWFAdW5pYmEuaXQ=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.