
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Cell. Infect. Microbiol. , 31 July 2020
Sec. Molecular Bacterial Pathogenesis
Volume 10 - 2020 | https://doi.org/10.3389/fcimb.2020.00369
 Peizhen Li1,2,3*†
Peizhen Li1,2,3*† Tingyuan Zhu1,2,3†
Tingyuan Zhu1,2,3† Danying Zhou1,2
Danying Zhou1,2 Wei Lu1,2
Wei Lu1,2 Hongmao Liu1,2
Hongmao Liu1,2 Zhewei Sun1,2
Zhewei Sun1,2 Jun Ying1,2
Jun Ying1,2 Junwan Lu1,2
Junwan Lu1,2 Xi Lin1,2,3
Xi Lin1,2,3 Kewei Li1,2,3
Kewei Li1,2,3 Jianchao Ying1,2,3
Jianchao Ying1,2,3 Qiyu Bao1,2,3*
Qiyu Bao1,2,3* Teng Xu4,5*
Teng Xu4,5*Bacterial resistance to antibiotics has become an important concern for public health. This study was aimed to investigate the characteristics and the distribution of the florfenicol-related resistance genes in bacteria isolated from four farms. A total of 106 florfenicol-resistant Gram-negative bacilli were examined for florfenicol-related resistance genes, and the positive isolates were further characterized. The antimicrobial sensitivity results showed that most of them (100, 94.33%) belonged to multidrug resistance Enterobacteriaceae. About 91.51% of the strains carried floR gene, while 4.72% carried cfr gene. According to the pulsed-field gel electrophoresis results, 34 Escherichia coli were subdivided into 22 profiles, the genetic similarity coefficient of which ranged from 80.3 to 98.0%. The multilocus sequence typing (MLST) results revealed 17 sequence types (STs), with ST10 being the most prevalent. The genome sequencing result showed that the Proteus vulgaris G32 genome consists of a 4.06-Mb chromosome, a 177,911-bp plasmid (pG32-177), and a 51,686-bp plasmid (pG32-51). A floR located in a drug-resistant region on the chromosome of P. vulgaris G32 was with IS91 family transposase, and the other floR gene on the plasmid pG32-177 was with an ISCR2 insertion sequence. The cfr gene was located on the pG32-51 flanked by IS26 element and TnpA26. This study suggested that the mobile genetic elements played an important role in the replication of resistance genes and the horizontal resistance gene transfer.
Florfenicol is a new type of broad-spectrum antibiotics of chloramphenicol for veterinary use, which was successfully developed in the late 1980's. It can also be recalled that thiamphenicol has the chemical formula C12H14Cl2FNO4S. CH3SO4 replaced the -NO2 group of chloramphenicol and gave florfenicol an obvious advantage in safety and efficacy compared with chloramphenicol. The antimicrobial spectrum of florfenicol is extremely wide. It has an inhibitory effect not only on the majority of Gram-negative and Gram-positive bacteria, as well as part of chlamydia and rickettsia, but also on chloramphenicol-resistant bacteria (Sams, 1995; Fang et al., 2020). Thus, florfenicol has been applied gradually across the world since it was launched in Japan in 1990. It is widely used in veterinary clinics, as a veterinary drug and feed additive, for its good therapeutic efficacy to bacterial diseases of pigs, cattle, poultry, and fish. However, with the irrational usage of florfenicol in clinical veterinary, serious resistance problems have emerged. Recently, many florfenicol-resistant bacteria and florfenicol-related resistance genes have been isolated from various animals. In 1996, florfenicol-resistant gene pp-flo was isolated from the multidrug-resistant plasmid R of the fish Pasteurella in Japan (Kim and Aoki, 1996). Then, floR was detected in the plasmids and the chromosomes of Escherichia coli from cattle, poultry, and pigs (Arcangioli et al., 1999; Bolton et al., 1999). In addition, it was also found in the IncC plasmid R55 of Klebsiella pneumoniae (Cloeckaert et al., 2001), a closely related plasmid of Salmonella newport (Meunier et al., 2003), and the chromosomes of Vibrio cholerae (Hochhut et al., 2001). Resistance to florfenicol in clinical settings has also been observed in human-originated E. coli isolates (Fernández-Alarcón et al., 2011). According to the study, the FloR protein contains 12 hydrophobic transmembrane regions belonging to the main susceptibility factor superfamily, which is located on the bacterial membrane and plays a significant role in the resistance to antibacterials (Hayes et al., 2006). In 2000, cfr gene was cloned from Staphylococcus sciuri by S. Schwarz, mediating the resistance of S. sciuri to florfenicol. The sequence analysis results suggested that cfr is a new type of florfenicol resistance gene (Schwarz et al., 2000). There was no homology between cfr and floR (floR-like), and it also does not share any homology with the amino acid sequence of chloramphenicol acetyltransferase. A novel gene fexA which encoded an efflux pump in Gram-positive coccus Staphylococcus lentus was located on a 34-kb plasmid PSPCFS2 that conferred resistance to florfenicol and chloramphenicol (Fessler et al., 2010). With in-depth studies, many florfenicol-associated resistance genes have been discovered constantly, such as the phenicol-specific exporter genes fexB, pexA (Liu et al., 2012), AcrAB-Tok multidrug efflux system tolC gene, acrB gene (Lee et al., 2000), and novel ATP-binding cassette (ABC) transporter gene optrA (Wang et al., 2015). Most of the genes co-existed with the bacterial mobile genetic elements, including plasmids, transposons, or integrons, which contributed to the rapid spread of florfenicol resistance genes to numerous bacterial species through horizontal gene transfer (HGT).
Therefore, the objective of this study was to investigate the prevalence and the distribution of florfenicol-related resistance genes in bacteria isolated from chicken farms and goose farms in Zhejiang Province. We also focused on the characterization of the potential novel mobile genetic elements associated with the dissemination of florfenicol-related resistance genes through HGT. It will be helpful to identify the major resistance determinants responsible for this resistance and provide evidence for the rational use of drugs in veterinary medicine.
A total of 32 samples were collected from the feces of chickens and geese from three different chicken farms and one goose farm in three regions of South China in 2015. The samples were processed as previously described (Miranda and Zemelman, 2002; Miranda and Rojas, 2007), and florfenicol-resistant strains were recovered by a spread plate method using Tryptic soy agar (Difco, Franklin Lakes, NJ, USA) containing 30 μg/ml of florfenicol (Schering-Plough, Kenilworth, NJ, USA), (Schwarz et al., 2004). The strains were identified by pathogen isolation, morphologic observation, biochemical reaction, and sequencing of 16S rDNA amplified by polymerase chain reaction (PCR). The purified strains were stored at −80°C in Tryptic soy broth (Difco) supplemented with 20% glycerol and florfenicol (30 μg/ml), (Fernández-Alarcón et al., 2010).
The minimum inhibitory concentrations (MICs) of florfenicol, tetracycline, chloramphenicol, cefuroxime sodium, enrofloxacin, levofloxacin, gentamicin, ampicillin, imipenem, ceftazidime, ciprofloxacin, colistin, and nalidixic acid were determined using the standard agar dilution method. The susceptibility results were categorized according to the guidelines from the Clinical and Laboratory Standards Institute (CLSI) 2017 (CLSI document M100-S27, 2017). E. coli ATCC25922 and one floR gene-positive K. pneumoniae strain1341 from our collection were used as controls.
The genes were detected by PCR. Genomic template was extracted using a bacterial genomic DNA extraction kit (Takara), following the manufacturer's instructions. The final 30 μl of PCR mixture containing ExTaq Premix (Takara), DNA template, and a pair of primers of the florfenicol resistance genes (floR, fexA, fexB, cfr, and optrA) was prepared for PCR (Cloeckaert et al., 2000; Zhang et al., 2011; Gómez-Sanz et al., 2013). The mixture was initially denatured at 94°C for 5 min, followed by 30 cycles of 94°C for 40 s, 55°C for 45 s, and 72°C for 50 s and was then elongated at 72°C for 10 min (Table 1). The PCR products were detected by electrophoresis in 1.5% agarose gels.
The chromosomal DNA of E. coli strains carrying the floR gene was digested with the restriction enzyme XbaI and then subjected to pulsed-field gel electrophoresis (PFGE) analysis (Xia et al., 2010). Salmonella enterica serovar Braenderup H9812 genome was used as a size standard. The gels were then electrophoresed in a CHEF-Mapper system (Bio-Rad, USA) in 1% pulsed-field certified agarose (Bio-Rad, USA) with 0.5× tris-borate and ethylene diamine tetraacetic acid as the running buffer at 14°C and 6 V/cm for 20 h. The pulse time ramped up from 5 to 20 s. Images were captured with Gel Doc system (Bio-Rad, USA).
E. coli DNA was extracted using a bacterial genomic DNA extraction kit (Takara, Dalian, China) following the manufacturer's instructions. Multilocus sequence typing (MLST) was performed using seven conserved housekeeping genes (adk, purA, fumC, mdh, icd, gyrB, and recA; https://pubmlst.org). The mixture was initially denatured at 94°C for 5 min, followed by 30 cycles of 94°C for 40 s, 54°C for 45 s, and 72°C for 50 s and was then elongated at 72°C for 10 min. The annealing temperatures were set at 54°C for adk, fumC, purA, and icd, 58°C for recA, and 60°C for gyrB and mdh. The amplified fragments for all loci were sequenced. The allelic profiles and sequence type determinations were performed according to the E. coli MLST website scheme. The MLST data were analyzed by using the eBURST algorithm (http://eburst.mlst.net), which assesses the relationship within clonal complexes.
Genomic DNA was isolated from bacterial cells grown overnight in Luria–Bertani broth at 37°C. The DNA was isolated by using QIAmp DNA mini kit (Qiagen, Valencia, CA, USA). The DNA concentrations and purity were determined by measuring the absorbance at 260 and 280 nm in a Nanodrop 2000 spectrophotometer. The genomic DNA of Proteus vulgaris G32 was sequenced by Pacific sequencing technology. The assembly of the sequence was performed with the help of SOAPdenovo v2.04, Celera Assembler 8.0 (Zhao et al., 2010), and Gap Closer v1.12 (Mendes et al., 2008). We used the Glimmer software to predict protein-coding genes with potential open reading frames (ORFs), (Delcher et al., 2007). RNAmmer and tRNAscan-SE were utilized to identify rRNA and tRNA genes, respectively (Lowe and Eddy, 1997; Lagesen et al., 2007). Gview was used to construct the basic genomic features (Petkau et al., 2010). BLASTX was used to annotate predicted protein-coding genes against the public protein database with an e-value threshold of 1e-5. Genome-wide identification of restriction–modification systems was conducted by using BLASTP searching against Rebase with >50% amino acid identity and >50% query coverage after all ORFs were theoretically translated (Roberts et al., 2015). The complete nucleotide sequences of the chromosome and the two plasmids of P. vulgaris G32 have been submitted to GenBank, and the accession numbers of the chromosome, pG32-177 and pG32-51, are CP053371, CP053372, and CP053373, respectively.
The DNA fragments containing the floR and the cfr genes were amplified by PCR using the primers (Table 1). The complete ORF fragment of the PCR product was cloned into a pMD19 vector (TaKaRa, Dalian, China). The recombinant clones were picked and sequenced. The recombinant plasmid was digested with restriction endonucleases, and the ORF fragment was recovered and further cloned into a pET28a vector (TaKaRa, Dalian, China). Finally, the recombinant plasmids were transformed into the host strain BL21. The minimal inhibitory concentrations of antibiotics were determined by the agar dilution method for the recipient strains BL21, BL21[pET28a-floR], and BL21[pET28a-cfr] in accordance with the guidelines of the Clinical and Laboratory Standards Institute (CLSI, 2017).
A total of 106 florfenicol-resistant Gram-negative bacilli were analyzed in this study. Most of the isolates were demonstrated to be E. coli (91 strains). The remaining isolates belonged to genera or species of Bacillus proteus (six strains), Shigella (five strains), K. pneumoniae (two strains), and Salmonella (two strains). The antibiotic susceptibility tests showed that most of them (100, 94.33%) were resistant to three commonly used veterinary antibiotics, and more than a half of the strains were resistant to more than seven antibiotics (55, 51.89%, Table 2). The percentages of strains resistant to florfenicol, ampicillin, enrofloxacin, tetracycline, chloramphenicol, cefuroxime sodium, gentamicin, levofloxacin, nalidixic acid, ceftazidime, and ciprofloxacin were 100% (106/106), 98.11% (104/106), 93.39% (99/106), 91.51% (97/106), 88.68% (94/106), 84.91% (90/106), 61.32% (65/106), 52.83% (56/106), 48.11% (51/106), 41.15% (44/106), and 36.68% (39/106), respectively. The lowest levels of resistance were found for colistin (3.77%, 4/106) and imipenem (0.94%, 1/106), (Table 3).
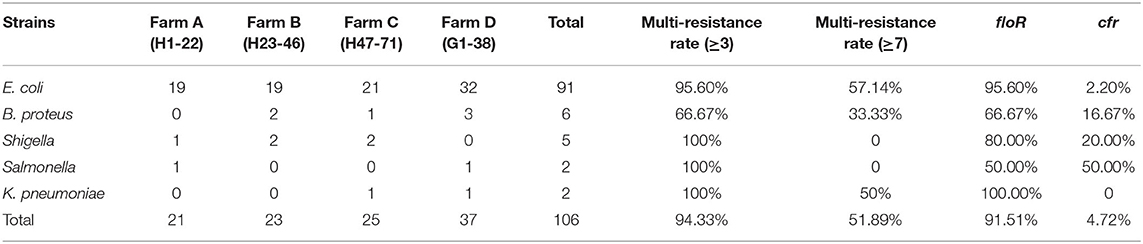
Table 2. Number, multi-resistance rate of resistant strains, and distribution of antimicrobial resistance genes.
We screened the florfenicol resistance genes from above 109 strains via PCR. About 91.51% (97/106) of the strains showed floR gene-positive results, 4.72% (5/106) showed cfr gene-positive results, while no other known florfenicol-resistant genes (fexA, fexB, and optrA) were detected (Table 2). Among them, only one isolate was positive for both floR and cfr, which was P. vulgaris G32 from the goose farm.
We conducted a further epidemiological study on E. coli, the most common florfenicol-resistant bacteria in this region. A total of 34 floR-positive E. coli strains which are resistant to more than seven drugs from four farms were typed by PFGE. Bands with sizes ranging from 20 to 700 kb were obtained. The number of electrophoretic bands was between 15 and 28. A restriction map containing 22 bands was obtained when a cutoff of identity was set at 100% (Figure 1). By UPGMA cluster analysis, the genetic similarity coefficient ranged from 80.3 to 98.0%; each band type included one to two strains. The similarity coefficient of E. coli H20 from chicken farm B and E. coli G23 from the geese farm was 93%. In addition, the similarity coefficient among E. coli H2, H7, H9 (farm A), and H18 (farm B) was 98%. These results indicated that the dissemination of floR in this district was most possibly mediated by HGT.
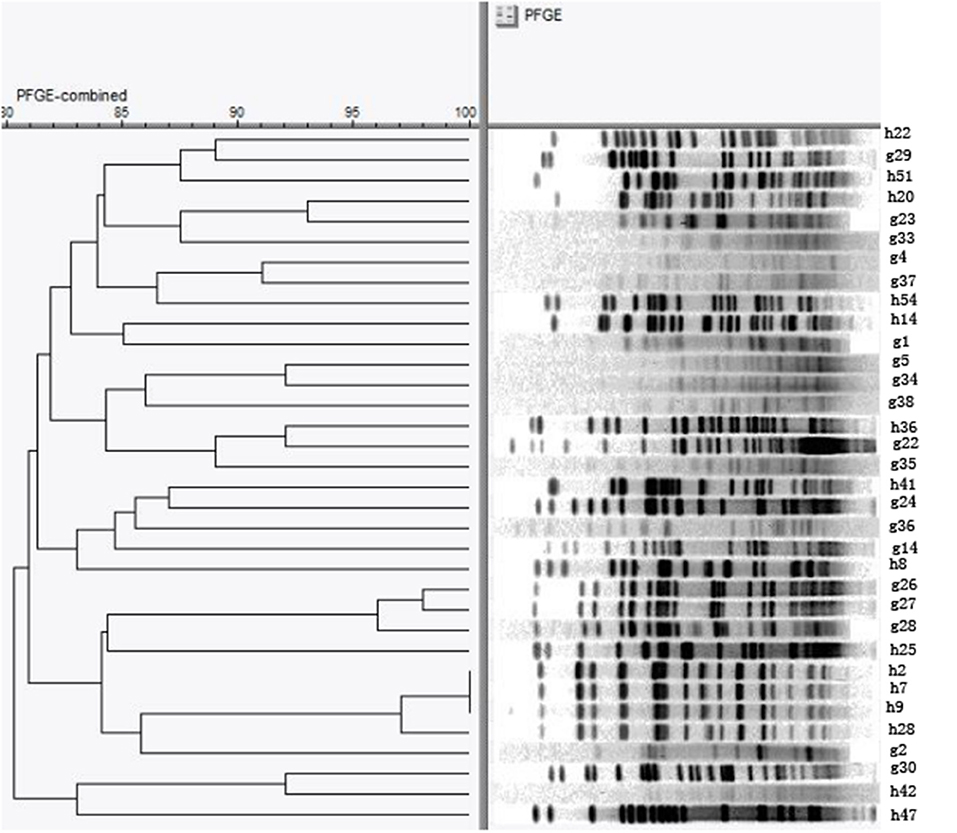
Figure 1. Pulsed-field gel electrophoresis (PFGE) pattern of 34 E. coli resistant to more than seven drugs. The chromosomal DNA of 34 E. coli isolates carrying the floR genes were digested with the restriction enzyme XbaI and then subjected to PFGE analysis.
Based on the abovementioned PFGE results, a total of 18 strains from the dominant clusters were selected for MLST analysis. As a result, 17 sequence types were identified, including six new STs (ST01n−06n), (Table 4 and Figure 2). ST10 and its single locus variants (SLVs) belonged to clonal complex 10 (CC10), which was the most commonly observed one. ST3544, ST10, and ST48 belonged to CC10, while ST155, ST01n, and ST02n were classified into CC155 (Figure 2). ST01n and ST02n were double-loci variants of ST155 because they had two different alleles. ST162 and ST03n belonged to CC469. ST03n was a SLV of ST469. ST165, ST05n, and ST06n belonged to CC65, CC206, and CC23, respectively. Furthermore, there were six singleton STs, including ST4417, ST1276, ST542, ST746, ST117, and ST04n, which could not be categorized according to any of the clonal complexes based on the MLST.
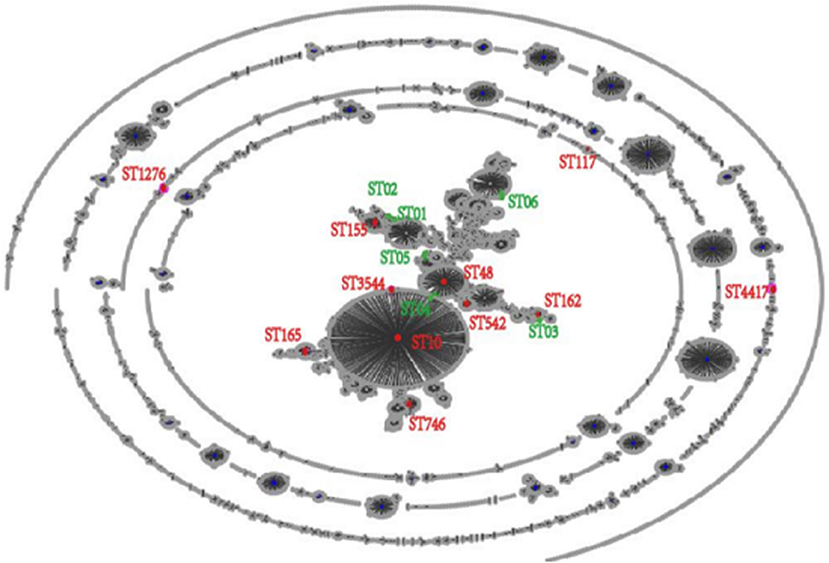
Figure 2. EBURST result of 6,475 sequence types (STs) in Pubmlst/E. coli database. All the 6,475 STs were clustered into 17 STs.
The results of PCR screening showed that the P. vulgaris G32 also carried floR and cfr resistance genes. It is rarely reported in B. proteus. The genome sequencing result showed that the P. vulgaris G32 genome consists of a 4.06-Mb chromosome with an average GC content of 38.1% encoding 3,590 open reading frames (Table 5), a 177,911-bp plasmid (pG32-177) encoding 251 ORFs, and a 51,686-bp plasmid (pG32-51) encoding 80 ORFs (Figure 3). A total of 34 antibiotic resistance genes were identified, of which eight were encoded on the chromosome, 24 (including cfr, floR, sul2, aminoglycoside resistance genes, β-lactamase genes, and so on) on pG32-177, and three on pG32-51. The co-linear analysis of the genomes among sequenced P. vulgaris G32, Proteus mirabilis CYPV1, and P. vulgaris FDAARGOS-366 in GenBank showed a high collinearity among the three chromosomes, except that partial inversion, insertion, and deletion were observed (Figure 4).
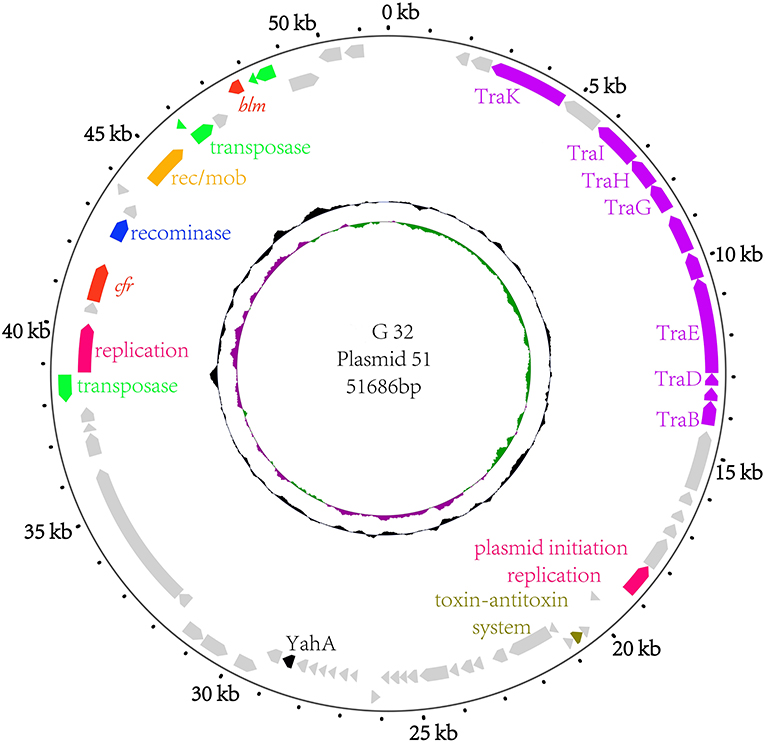
Figure 3. The circular map of the pG32-51 genome. Counting from outside toward the center, the first circle refers to the position in base pairs. The second circle consists of two-direction arrows which indicate the position of the gene and the marked genes encoded on the leading strand (outwards) or lagging strand (inwards). The different function genes are shown in different colors: red, drug-resistance related genes; purple, conjugation and transfer; green and yellow, transposase/insertion sequences; rose red, replication; blue, recominase; silver, unknown function genes. The third circle shows the GC skew (G–C/G+C), with a positive GC skew toward the outside and a negative GC skew toward the inside. The fourth circle shows the GC content with an average of 50%, whereby a G+C content of more than 50% is shown toward the outside and a G+C content of <50% toward the inside.
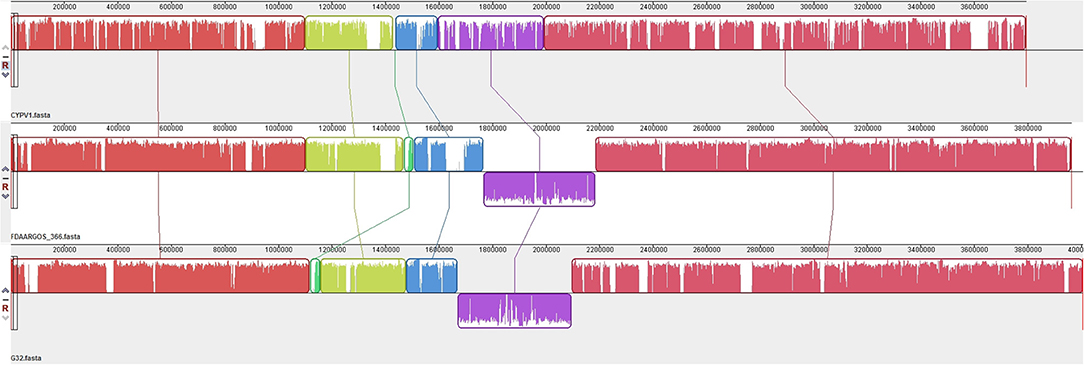
Figure 4. Genome structure comparison of P. vulgaris G32 with CYPV1 and FDAARGOS_366. The corresponding blocks from these three strains are shown according to the gene content or sequence similarities. The blocks below the line in strains CYPV1 and FDAARGOS_366 genome indicated that the sequence in the block was in a reverse direction compared to the corresponding region in P. vulgaris G32. The denser and higher lines represented more genes and higher similarities between the sequences.
A florfenicol-related resistance gene floR is located in a drug-resistant region on the chromosome of P. vulgaris G32, which is highly homologous to the Proteus chromosomes from Proteus CYPV1 and Proteus FDAARGOS-366 in GenBank. The upstream of regions all contained transcriptional regulators, ABC super family, regulatory proteins, and so on, and the downstream region of a 13,425-bp segment of P. vulgaris G32 is homologous with the corresponding segment of Proteus FDAARGOS-366 and is different from Proteus CYPV1 slightly. The 19,966-bp region containing floR in the middle is a specific fragment of P. vulgaris G32. The sequence displayed that there is a pair of IS91 family transposase locating upstream and downstream of the floR gene. Besides that, there are also TrbL/VirB6 associated with conjugation, transduction, and phage integrase family proteins existing in its downstream region. This gives reasonable explanations for the existence of this resistance segment. In other words, this segment was inserted into the chromosome with the help of integrase and transposase, and the insertion site was located between the repressor and the GMP synthetase. Transcriptional regulatory factor AlpA, regulatory protein RepA, and the stabilizing protein, which locates between TrbL/VirB6 and phage integrase family protein, also provide help for stable replication, transcription, and heredity to some extent. In addition, aminoglycoside-resistant genes strA and strB are also found in this region. Compared with the sequence of V. cholerae HC1037 (positions 95,522–102,832) from the clinical patients in the database, we can find that only part of the LysR open reading frame between IS91 family transposase and floR is missing in the chromosome, and a strA gene between sul2 and strB is different (Figure 5).
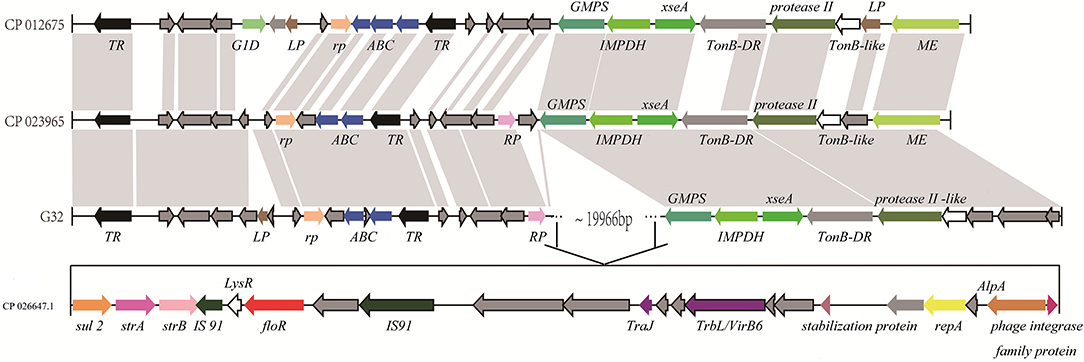
Figure 5. Comparison of the floR gene regions on the chromosome of P. vulgaris G32, Proteus CYPV1 (accession number CP012675), Proteus FDAARGOS-366 (accession number CP023965), and V. cholerae HC1037 (accession number CP026647.1). The homolog genes are marked with the same color and lined together, respectively.
In addition to the chromosome-encoded floR, P. vulgaris G32 also harbored another copy of floR gene on the plasmid pG32-177 with an ISCR2 insertion sequence. A series of transposition-related genes including tnpM, tnpR, and tnpA was also observed near ISCR2, equipping this region with the autonomous transposition. This 13,177-bp segment is highly homologous with those of chicken-derived E. coli YJMC8 from Guangzhou, China, in 2017 and of porcine enteropathogenic E. coli SHP45 from Guangzhou, China, in 2016 (Figure 6).
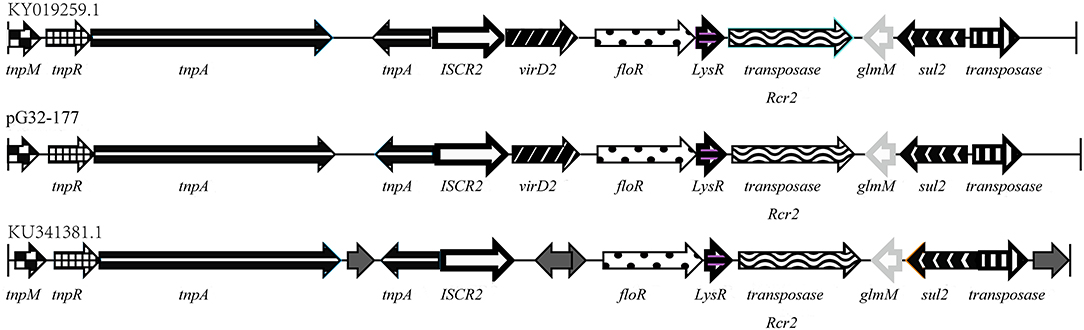
Figure 6. Comparison of the floR gene regions on the pG32-177, the sequenced plasmids of E. coli YJMC8 (accession number KY019259.1) and E. coli SHP45 (accession number KU341381.1).
The cfr gene is located on pG32-51, flanked by two or three identical copies of the 820-bp IS26 element encoding TnpA26 in the same orientation. Similar resistance gene regions have been detected in other sequenced plasmids of E. coli 8ZG12D, SH21G, and the sequences of P. vulgaris PV-01 (Figure 7). Compared with IS26 insertion sequence, pG32-51 lacks a 1,030-bp segment whose function has to be further studied but has a segment of 4,150 bp which cannot be found in E. coli 8ZG12D. Rep, which exists at 3,927 bp downstream of cfr between rec/mob and tnpA26 in P. vulgaris PV-01, was located in the upstream of cfr immediately of tnpA26 of pG32-51. In addition, the resistant insertion segment of pG32-51 containing cfr follows a segment of 1,568 bp containing tnpA26. So, the main structure of the drug-resistant region is IS26-cfr-recombinase-rec/mob-tnpA26.
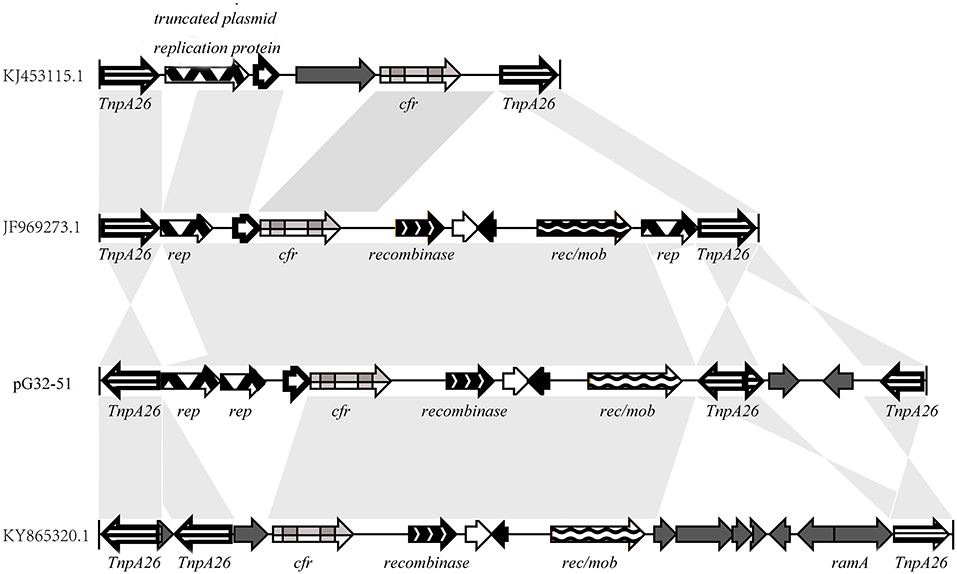
Figure 7. Comparison of the cfr gene regions on pG32-51, E. coli 8ZG12D (accession number KY865320.1), E. coli SH21G (accession number KJ453115.1), and the sequences of P. vulgaris strain PV-01 (accession number JF969273.1).
We cloned the complete ORFs of both floR and cfr into pET-28a vectors and transformed them separately into E. coli BL21. The MICs of the antibiotics and the recipient controls against a group of antimicrobial drugs were detected (Table 3). BL21[pET28a-floR] and BL21[pET28a-cfr] showed resistance to florfenicol and chloramphenicol.
Man has developed new anti-bacterials to inhibit and eliminate the increasingly widespread and complex drug resistance of bacteria. However, new resistant strains are also appearing along with the clinical application of new antibiotics. The problem of florfenicol resistance is becoming serious; more and more drug-resistant bacteria have been found in the breeding environment and in animals (Bossé et al., 2015a). In 2000, White et al. studied the resistance of E. coli isolated from diarrhea cattle to chloramphenicol and florfenicol and got the result that 42 out of 44 strains (florfenicol MICs ≥ 16 μg/ml) carried floR gene (White et al., 2000). Kuo found that the resistance of E. coli isolated from pigs in Taiwan to florfenicol increased from 39.2% in 2003 to 78.3% in 2007, and the prevalence rate of floR gene showed a rising trend (Kuo et al., 2014). From a breeding farm in Heilongjiang Province, China, 60 strains of E. coli from swine were detected with positive rate as 50% of cmlA and 80% of floR (Zhao et al., 2017). In this study, we have examined the prevalence of florfenicol-related resistance genes in bacteria of animal origin. The results suggest that floR was the main epidemic resistance gene of florfenicol (91.74%); only five strains were positive to cfr (4.59%), and the rest of the resistance genes were not detected. In addition, the positive rate of floR gene was similar to the proportion of strains (MICs ≥ 32 μg/ml), agreeing with the results of Claudia et al. (2011). A PFGE analysis suggested that the degree of similarity of the bacterial diversity was low, and clone transmissions might exist in the multidrug-resistant E. coli strains carrying floR genes. In addition, the MLST analysis indicated that E. coli sequence types ST10, ST3544, and ST48 all belonged to clone complex 10 (CC10), the largest and most popular clone complex in the world (Shabana et al., 2013; Maluta et al., 2014). The detection of the common sequence type (ST10) in E. coli isolated from patients, farm workers, pigs, wild birds, and river water was reported, suggesting a possible transmission among animals, humans, and the surrounding environments (Fischer et al., 2017; Gomi et al., 2017). In other words, the popular strains in our research shared a high degree of homology with other major epidemic clones in the world.
There are many reasons for the emergence of resistance. Among them, the most important and widespread resistance mechanism is the acquisition and the transmission of various resistance genes. Most of the florfenicol resistance genes are located in the mobile plasmids and the transposons. The florfenicol resistance gene pp-flo (Kim and Aoki, 1996) was located in the multidrug-resistant R plasmid. An analysis of the plasmid pMBSFl with floR gene of the E. coli isolates from pigs (Blickwede and Schwarz, 2004) indicated that the flanking regions of floR gene are composed of three sequences from different sources. They are highly homologous to transposon Tn5393, plasmid with floR from E. coli 10660, and transposon Tn1721, respectively. The floR in pM3446F of these Pasteurellaceae plasmids was described as a transposable element encoding floR, transcriptional regulator lysR, and transposase tnpA genes initially (Bossé et al., 2015b). Zhang et al. confirmed that an IncA/C plasmid carrying the multiresistance gene cfr in a porcine E. coli strain was flanked by two copies of IS256 (Zhang et al., 2014). Our study indicated that florfenicol resistance genes not only existed in the plasmids but also in the chromosomes of a bacterium. The floR gene in the chromosome of P. vulgaris G32 was mediated by the transposon IS91, which is different from the usual insertion sequence (Mataseje et al., 2014). Meunier et al. found the floR gene from bovine E. coli, which was shown to be associated with the insertion sequence ISCR2 (Meunier et al., 2010). Similarly, our floR in pG32-177 is also next to an ISCR2 insertion sequence, mediated by Tn21, indicating that the gene can be transferred with the composite transposons. The cfr gene in pG32-51 formed the structure of the composite transposon, recombinase-rec/mob-tnpA26, which can mediate the resistance genes mobile by homologous recombination or transferred by conjugation. That is consistent with the report of Doublet et al. (2005). Obviously, the molecular genetic background of florfenicol resistance genes is very complex. The transference of resistance plasmids in bacteria strengthens the resistance of drug-resistant bacteria and accelerates the spreading of resistance, and transposition links the resistance genes among chromosomes, plasmids, and phages and enriches the source of resistance plasmids.
The resistance problem of florfenicol has become a major problem in the development of animal husbandry, which needs to be tackled urgently. Resistance encoding genes determine the genetic complexity of the strains. The study on the prevalence and the environment of florfenicol resistance genes will contribute to the understanding of its origin, expression, and metastasis in molecular biology. It can also provide references for the rational usage of antibiotics and the further prevention of drug resistance in veterinary medicine.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/supplementary material.
PL, QB, and TX designed the experiment. PL, TZ, DZ, WL, HL, and JuY performed experiments. XL, JiY, and TX contributed to analysis the experimental data. PL, TZ, and QB wrote the manuscript. PL, ZS, and JL supported and designed the project. KL, QB, and TX critically revised the manuscript. All authors read and approved the manuscript.
This work was funded by grants from the National Natural Science Foundation of China (81501808, 81960381, and 80215049), the Science and Technology Foundation of Wenzhou City (Y20170205), and the Science & Technology Project of Inner Mongolia Autonomous Region, China (201802125).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Arcangioli, M. A., Leroy-Sétrin, S., Martel, J. L., and Chaslus-Dancla, E. (1999). A new chloramphenicol and florfenicol resistance gene flanked by two integron structures in Salmonella typhimurium DT104. FEMS Microbiol. Lett. 174, 327–332. doi: 10.1111/j.1574-6968.1999.tb13586.x
Blickwede, M., and Schwarz, S. (2004). Molecular analysis of florfenicol-resistant Escherichia coli isolates from pigs. J. Antimicrob. Chemother. 53, 58–64. doi: 10.1093/jac/dkh007
Bolton, L. F., Kelley, L. C., Lee, M. D., Fedorka-Cray, P. J., and Maurer, J. J. (1999). Detection of multidrug-resistant Salmonella enterica serotype typhimurium DT104 based on a gene which confers cross-resistance to florfenicol and chloramphenicol. J. Clin. Microbiol. 37, 1348–1351. doi: 10.1128/JCM.37.5.1348-1351.1999
Bossé, J. T., Li, Y., Atherton, T. G., Walker, S., Williamson, S. M., Rogers, J., et al. (2015b). Characterisation of a mobilisable plasmid conferring florfenicol and chloramphenicol resistance in actinobacillus pleuropneumoniae. Vet. Microbiol. 178, 279–282. doi: 10.1016/j.vetmic.2015.07.002
Bossé, J. T., Li, Y., Walker, S., Atherton, T., Fernandez Crespo, R., Williamson, S. M., et al. (2015a). Identification of dfrA14 in two distinct plasmids conferring trimethoprim resistance in actinobacillus pleuropneumoniae. J. Antimicrob. Chemother. 70, 2217–2222. doi: 10.1093/jac/dkv121
Cloeckaert, A., Baucheron, S., and Chaslus-Dancla, E. (2001). Nonenzymatic chloramphenicol resistance mediated by IncC plasmid R55 is encoded by a floR gene variant. Antimicrob. Agents Chemother. 45, 2381–2382. doi: 10.1128/AAC.45.8.2381-2382.2001
Cloeckaert, A., Baucheron, S., Flaujac, G., Schwarz, S., Kehrenberg, C., Martel, J. L., et al. (2000). Plasmid-mediated florfenicol resistance encoded by the floR gene in Escherichia coli isolated from cattle. Antimicrob. Agents Chemother. 44, 2858–2860. doi: 10.1128/AAC.44.10.2858-2860.2000
Delcher, A. L., Bratke, K. A., Powers, E. C., and Salzberg, S. L. (2007). Identifying bacterial genes and endosymbiont DNA with glimmer. Bioinformatics 23, 673–679. doi: 10.1093/bioinformatics/btm009
Doublet, B., Schwarz, S., Kehrenberg, C., and Cloeckaert, A. (2005). Florfenicol resistance gene floR is part of a novel transposon. Antimicrob. Agents Chemother. 49, 2106–2108. doi: 10.1128/AAC.49.5.2106-2108.2005
Fang, Y. Q., Li, S. Q., Ye, L. J., Yi, J., Li, X. F., Gao, C. K., et al. (2020). Increased bioaffinity and anti-inflammatory activity of florfenicol nanocrystals by wet grinding method. J. Microencapsul. 37, 109–120. doi: 10.1080/02652048.2019.1701115
Fernández-Alarcón, C., Miranda, C. D., Singer, R. S., López, Y., Rojas, R., Bello, H., et al. (2010). Detection of the floR gene in a diversity of florfenicol resistant gram-negative bacilli from freshwater salmon farms in Chile. Zoonoses Public Health 57, 181–188. doi: 10.1111/j.1863-2378.2009.01243.x
Fernández-Alarcón, C., Singer, R. S., and Johnson, T. J. (2011). Comparative genomics of multidrug resistance-encoding IncA/C plasmids from commensal and pathogenic Escherichia coli from multiple animal sources. PLoS ONE 6:e23415. doi: 10.1371/journal.pone.0023415
Fessler, A., Scott, C., Kadlec, K., Ehricht, R., Monecke, S., and Schwarz, S. (2010). Characterization of methicillin-resistant Staphylococcus aureus ST398 from cases of bovine mastitis. J. Antimicrob. Chemother. 65, 619–625. doi: 10.1093/jac/dkq021
Fischer, J., Hille, K., Ruddat, I., Mellmann, A., Köck, R., and Kreienbrock, L. (2017). Simultaneous Occurrence of MRSA and ESBL-producing Enterobacteriaceae on pig farms and in nasal and stool samples from farmers. Vet. Microbiol. 200, 107–113. doi: 10.1016/j.vetmic.2016.05.021
Gómez-Sanz, E., Kadlec, K., Fessler, A. T., Billerbeck, C., Zarazaga, M., Schwarz, S., et al. (2013). Analysis of a novel erm(T)- and cadDX-carrying plasmid from methicillin-susceptible Staphylococcus aureus ST398-t571 of human origin. J. Antimicrob. Chemother. 68, 471–473. doi: 10.1093/jac/dks411
Gomi, R., Matsuda, T., Matsumura, Y., Yamamoto, M., Tanaka, M., Ichiyama, S., et al. (2017). Whole-genome analysis of antimicrobial-resistant and extraintestinal pathogenic Escherichia coli in river water. Appl. Environ. Microbiol. 83, e02703–e02716. doi: 10.1128/AEM.02703-16
Hayes, E. T., Wilks, J. C., Sanfilippo, P., Yohannes, E., Tate, D. P., Jones, B. D., et al. (2006). Oxygen limitation modulates pH regulation of catabolism and hydrogenases, multidrug transporters, and envelope composition in Escherichia coli K-12. BMC Microbiol. 6:89. doi: 10.1186/1471-2180-6-89
Hochhut, B., Lotfi, Y., Mazel, D., Faruque, S. M., Woodgate, R., and Waldor, M. K. (2001). Molecular analysis of antibiotic resistance gene clusters in Vibrio cholerae O139 and O1 SXT constins. Antimicrob. Agents Chemother. 45, 2991–3000. doi: 10.1128/AAC.45.11.2991-3000.2001
Kim, E., and Aoki, T. (1996). Sequence analysis of the florfenicol resistance gene encoded in the transferable R-plasmid of a fish pathogen, Pasteurella piscicida. Microbiol. Immunol. 40, 665–669. doi: 10.1111/j.1348-0421.1996.tb01125.x
Kuo, H. C., Lauderdale, T. L., Lo, D. Y., Chen, C. L., Chen, P. C., Liang, S. Y., et al. (2014). An association of genotypes and antimicrobial resistance patterns among Salmonella isolates from pigs and humans in Taiwan. PLoS ONE 9:e95772. doi: 10.1371/journal.pone.0095772
Lagesen, K., Hallin, P., Rødland, E. A., Staerfeldt, H. H., Rognes, T., and Ussery, D. W. (2007). RNAmmer: consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Res. 35, 3100–3108. doi: 10.1093/nar/gkm160
Lee, A., Mao, W., Warren, M. S., Mistry, A., Hoshino, K., Okumura, R., et al. (2000). Interplay between efflux pumps may provide either additive or multiplicative effects on drug resistance. J. Bacteriol. 182, 3142–3150. doi: 10.1128/JB.182.11.3142-3150.2000
Liu, H., Wang, Y., Wu, C., Schwarz, S., Shen, Z., Jeon, B., et al. (2012). A novel phenicol exporter gene, fexB, found in enterococci of animal origin. J. Antimicrob. Chemother. 67, 322–325. doi: 10.1093/jac/dkr481
Lowe, T. M., and Eddy, S. R. (1997). tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 25, 955–964. doi: 10.1093/nar/25.5.955
Maluta, R. P., Logue, C. M., Casas, M. R., Meng, T., Guastalli, E. A., Rojas, T. C., et al. (2014). Overlapped sequence types (STs) and serogroups of avian pathogenic (APEC) and human extra-intestinal pathogenic (ExPEC) Escherichia coli isolated in Brazil. PLoS ONE 9:e105016. doi: 10.1371/journal.pone.0105016
Mataseje, L. F., Boyd, D. A., Lefebvre, B., Bryce, E., Embree, J., Gravel, D., et al. (2014). Complete sequences of a novel blaNDM-1-harbouring plasmid from Providencia rettgeri and an FII-type plasmid from Klebsiella pneumoniae identified in Canada. J. Antimicrob. Chemother. 69, 637–642. doi: 10.1093/jac/dkt445
Mendes, R. E., Bell, J. M., Turnidge, J. D., Yang, Q., Yu, Y., Sun, Z., et al. (2008). Carbapenem-resistant isolates of Klebsiella pneumoniae in China and detection of a conjugative plasmid (blaKPC-2 plus qnrB4) and a blaIMP-4 gene. Antimicrob. Agents Chemother. 52, 798–799. doi: 10.1128/AAC.01185-07
Meunier, D., Baucheron, S., Chaslus-Dancla, E., Martel, J. L., and Cloeckaert, A. (2003). Florfenicol resistance in Salmonella enterica serovar newport mediated by a plasmid related to R55 from Klebsiella pneumoniae. J. Antimicrob. Chemother. 51, 1007–1009. doi: 10.1093/jac/dkg141
Meunier, D., Jouy, E., Lazizzera, C., Doublet, B., Kobisch, M., Cloeckaert, A., et al. (2010). Plasmid-borne florfenicol and ceftiofur resistance encoded by the floR and blaCMY-2 genes in Escherichia coli isolates from diseased cattle in France. J. Med. Microbiol. 59, 467–471. doi: 10.1099/jmm.0.016162-0
Miranda, C. D., and Rojas, R. J. A. (2007). Occurrence of florfenicol resistance in bacteria associated with two Chilean salmon farms with different history of antibacterial usage. Aquaculture 266, 39–46. doi: 10.1016/j.aquaculture.2007.02.007
Miranda, C. D., and Zemelman, R. (2002). Antimicrobial multiresistance in bacteria isolated from freshwater Chilean salmon farms. Sci. Total Environ. 293, 207–218. doi: 10.1016/S0048-9697(02)00022-0
Petkau, A., Stuart-Edwards, M., Stothard, P., and Van Domselaar, G. (2010). Interactive microbial genome visualization with GView. Bioinformatics 26, 3125–3126. doi: 10.1093/bioinformatics/btq588
Roberts, R. J., Vincze, T., Posfai, J., and Macelis, D. (2015). REBASE–a database for DNA restriction and modification: enzymes, genes and genomes. Nucleic Acids Res. 43, D298–D299. doi: 10.1093/nar/gku1046
Sams, R. A. (1995). Florfenicol: chemistry and metabolism of a novel broad-spectrum antbiotic. Tierarzt Umschau 50, 703–707.
Schwarz, S., Kehrenberg, C., Doublet, B., and Cloeckaert, A. (2004). Molecular basis of bacterial resistance to chloramphenicol and florfenicol. FEMS Microbiol. Rev. 28, 519–542. doi: 10.1016/j.femsre.2004.04.001
Schwarz, S., Werckenthin, C., and Kehrenberg, C. (2000). Identification of a plasmid-borne chloramphenicol-florfenicol resistance gene in Staphylococcus sciuri. Antimicrob. Agents Chemother. 44, 2530–2533. doi: 10.1128/AAC.44.9.2530-2533.2000
Shabana, I. I., Zaraket, H., and Suzuki, H. (2013). Molecular studies on diarrhea-associated Escherichia coli isolated from humans and animals in Egypt. Vet. Microbiol. 167, 532–539. doi: 10.1016/j.vetmic.2013.08.014
Wang, Y., Lv, Y., Cai, J., Schwarz, S., Cui, L., Hu, Z., et al. (2015). A novel gene, optrA, that confers transferable resistance to oxazolidinones and phenicols and its presence in Enterococcus faecalis and Enterococcus faecium of human and animal origin. J. Antimicrob. Chemother. 70, 2182–2190. doi: 10.1093/jac/dkv116
White, D. G., Hudson, C., Maurer, J. J., Ayers, S., Zhao, S., Lee, M. D., et al. (2000). Characterization of chloramphenicol and florfenicol resistance in Escherichia coli associated with bovine diarrhea. J. Clin. Microbiol. 38, 4593–4598. doi: 10.1128/JCM.38.12.4593-4598.2000
Xia, L. N., Li, L., Wu, C. M., Liu, Y. Q., Tao, X. Q., Dai, L., et al. (2010). A survey of plasmid-mediated fluoroquinolone resistance genes from Escherichia coli isolates and their dissemination in Shandong, China. Foodborne Pathog. Dis. 7, 207–215. doi: 10.1089/fpd.2009.0378
Zhang, W. J., Wu, C. M., Wang, Y., Shen, Z. Q., Dai, L., Han, J., et al. (2011). The new genetic environment of cfr on plasmid pBS-02 in a Bacillus strain. J. Antimicrob. Chemother. 66, 1174–1175. doi: 10.1093/jac/dkr037
Zhang, W. J., Xu, X. R., Schwarz, S., Wang, X. M., Dai, L., Zheng, H. J., et al. (2014). Characterization of the IncA/C plasmid pSCEC2 from Escherichia coli of swine origin that harbours the multiresistance gene cfr. J. Antimicrob. Chemother. 69, 385–389. doi: 10.1093/jac/dkt355
Zhao, F., Bai, J., Wu, J., Liu, J., Zhou, M., Xia, S., et al. (2010). Sequencing and genetic variation of multidrug resistance plasmids in Klebsiella pneumoniae. PLoS ONE 5:e10141. doi: 10.1371/journal.pone.0010141
Keywords: flofenicol, floR, cfr, PFGE, MLST, genomics
Citation: Li P, Zhu T, Zhou D, Lu W, Liu H, Sun Z, Ying J, Lu J, Lin X, Li K, Ying J, Bao Q and Xu T (2020) Analysis of Resistance to Florfenicol and the Related Mechanism of Dissemination in Different Animal-Derived Bacteria. Front. Cell. Infect. Microbiol. 10:369. doi: 10.3389/fcimb.2020.00369
Received: 19 February 2020; Accepted: 16 June 2020;
Published: 31 July 2020.
Edited by:
Brian Ahmer, The Ohio State University, United StatesReviewed by:
Naouel Klibi, Tunis El Manar University, TunisiaCopyright © 2020 Li, Zhu, Zhou, Lu, Liu, Sun, Ying, Lu, Lin, Li, Ying, Bao and Xu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Peizhen Li, lpz0522@126.com; Qiyu Bao, baoqy@genomics.cn; Teng Xu, xuteng@wmu.edu.cn
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.