- Department of Immunology, St. Jude Children's Research Hospital, Memphis, TN, United States
Programmed cell death is regulated by evolutionarily conserved pathways that play critical roles in development and the immune response. A newly recognized pathway for proinflammatory programmed cell death called PANoptosis is controlled by a recently identified cytoplasmic multimeric protein complex named the PANoptosome. The PANoptosome can engage, in parallel, three key modes of programmed cell death—pyroptosis, apoptosis, and necroptosis. The PANoptosome components have been implicated in a wide array of human diseases including autoinflammatory diseases, neurodegenerative diseases, cancer, microbial infections, and metabolic diseases. Here, we review putative components of the PANoptosome and present a phylogenetic analysis of their molecular domains and interaction motifs that support complex assembly. We also discuss genetic data that suggest PANoptosis is coordinated by scaffolding and catalytic functions of the complex components and propose mechanistic models for PANoptosome assembly. Overall, this review presents potential mechanisms governing PANoptosis based on evolutionary analysis of the PANoptosome components.
Introduction
Programmed cell death (PCD) is an evolutionarily conserved process that plays central roles in maintaining organismal homeostasis. Three key PCD pathways have been studied in great detail—pyroptosis (inflammasome-dependent PCD executed by gasdermin family members), apoptosis (PCD mediated by the apoptosome and executioner caspases), and necroptosis (PCD mediated by RIPK3 and the downstream effector MLKL). Previous studies have provided a foundation for us to understand the active and extensive crosstalk between the inflammasome/pyroptosis and apoptosis and necroptosis (Lamkanfi et al., 2008; Malireddi et al., 2010, 2018, 2020; Gurung et al., 2014, 2016a; Lukens et al., 2014; Kuriakose et al., 2016; Zheng et al., 2020). Some pathogenic challenges, such as influenza A virus (IAV) infection (Kuriakose et al., 2016), inhibition of the homeostasis-regulating transforming growth factor β-activated kinase 1 (TAK1) (Malireddi et al., 2018, 2020), and functional alterations in receptor-interacting serine/threonine-protein kinase (RIPK) 1 (Dannappel et al., 2014; Dillon et al., 2014; Kaiser et al., 2014; Rickard et al., 2014a; Dondelinger et al., 2019; Newton et al., 2019a; Lalaoui et al., 2020), can induce pyroptosis, apoptosis, and necroptosis together. These observations raised a question about regulation of PCD in these cases—are the three pathways activated independently of each other, or is a single cell death-inducing complex controlling all three? Studies using mice lacking Tak1 in myeloid cells and small molecule inhibitors of TAK1 kinase activity suggested the latter (Malireddi et al., 2019, 2020). This evidence led to the identification of a single cell death-inducing complex that is assembled to induce a PCD phenomenon called PANoptosis (Malireddi et al., 2019, 2020). The complex has subsequently been named the PANoptosome (Figure 1A) (Christgen et al., 2020). The PANoptosome was initially shown to contain RIPK1, apoptosis-associated speck-like protein containing a caspase recruitment domain (ASC), nucleotide-binding oligomerization domain (NOD)-like receptor pyrin domain-containing 3 (NLRP3), and caspase (CASP) 8 (Malireddi et al., 2020). A subsequent study determined that RIPK3, CASP6, Z-DNA-binding protein 1 (ZBP1), and CASP1 are also components of the PANoptosome that is assembled in response to IAV infection (Zheng et al., 2020). Collectively, these studies showed that the PANoptosome contains molecules critical for pyroptosis, apoptosis, and necroptosis and is able to activate all three to execute proinflammatory cell death (Christgen et al., 2020). Therefore, PANoptosis provides a mechanism for the host to activate alternative cell death defense mechanisms if one or more of the PCD pathways are compromised by a pathogen or other blockade. In this review, we will discuss PANoptosis and the molecular components of the PANoptosome complex, including their domain structures and functions, and use evolutionary analyses to contextualize the interactions between these molecules.
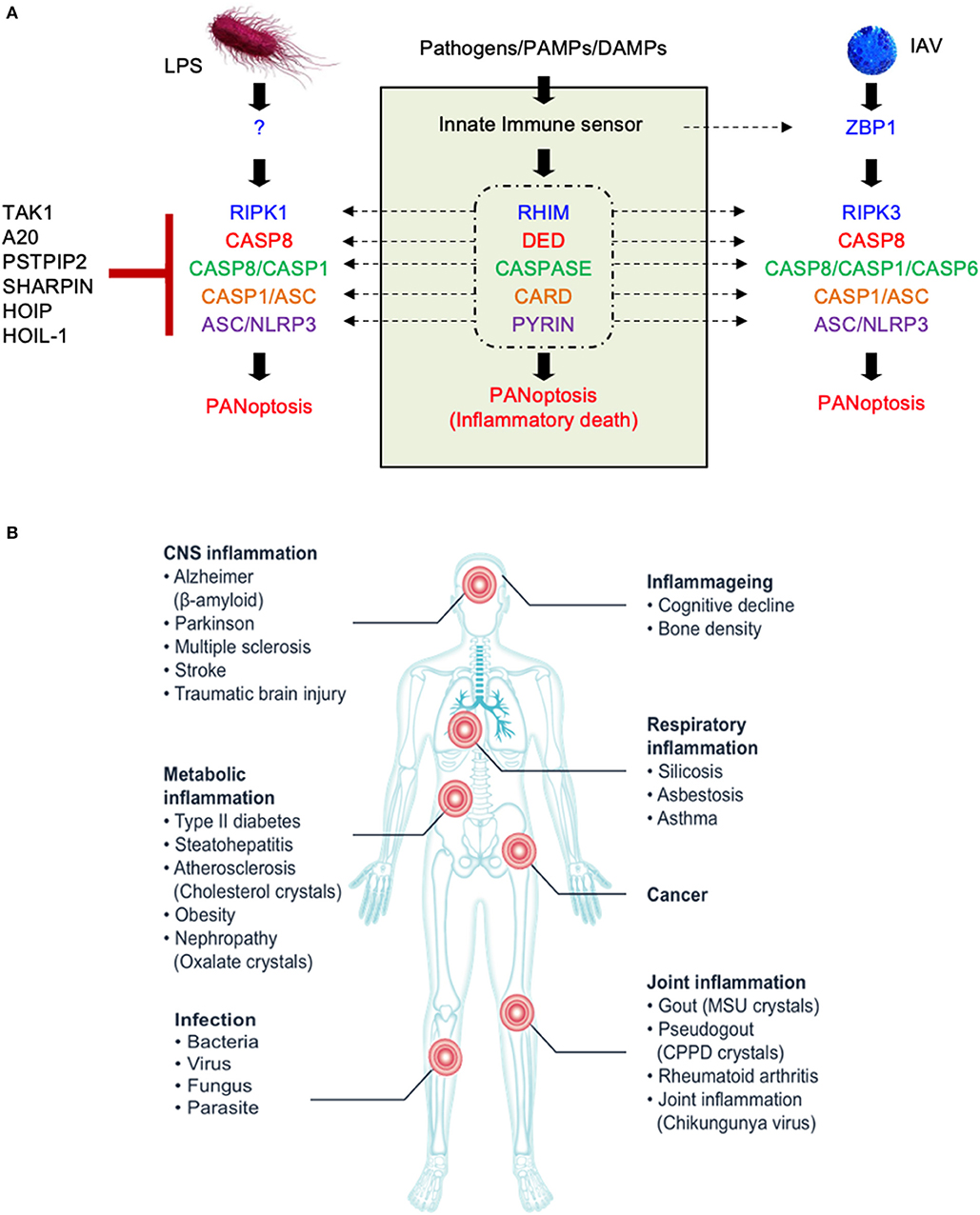
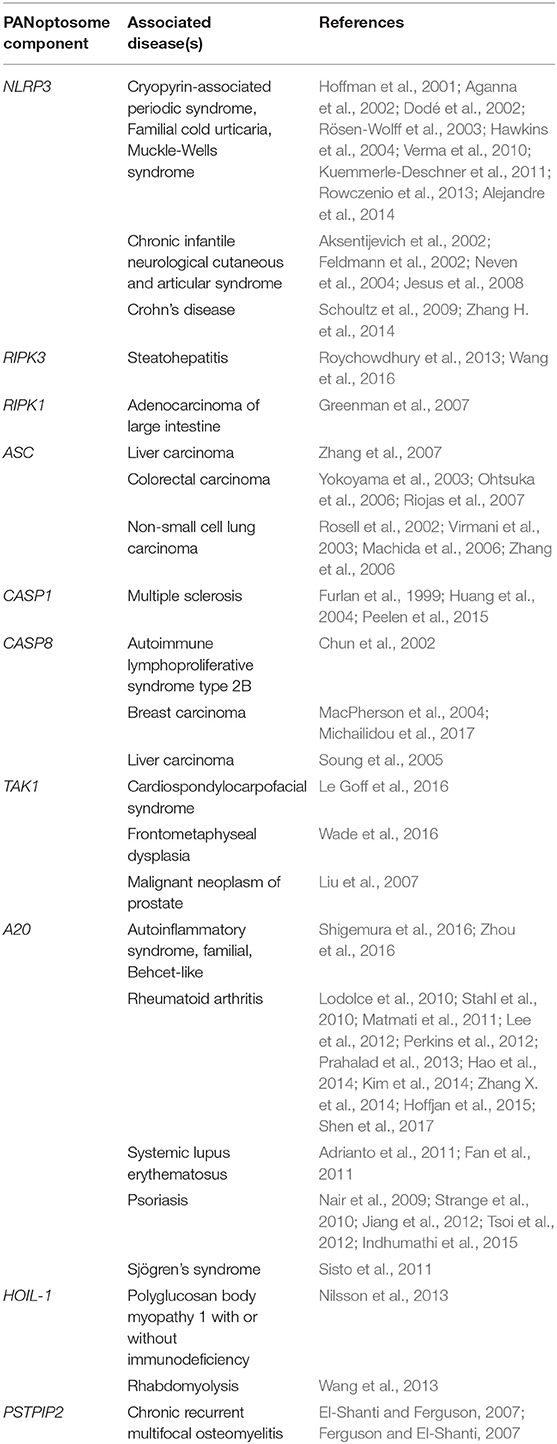
Figure 1. The molecular components of the PANoptosome and the human health significance of PANoptosis. (A) Proposed domains of PANoptosome components required for complex assembly in response to various stimuli. PANoptosome assembly is mediated by key molecular motifs collectively called death fold domains. Genetic evidence suggests that PANoptosome activation is inhibited by TAK1, PSTPIP2, SHARPIN, HOIP, HOIL-1, and A20. (B) Defects in the PANoptosome components have been implicated in a range of human diseases.
Established Mechanisms of PANoptosis
Pathogen sensing by the innate immune system activates intracellular signaling cascades to induce a proinflammatory immune response. Many innate sensors also trigger PCD either by directly interacting with mediators of cell death or through the downstream effects of secreted cytokines. One pathogen that elicits proinflammatory immune responses and PCD is IAV. Yearly seasonal flu outbreaks are caused by IAV strains, but hypervirulent strains sporadically emerge and can result in catastrophic pandemics, such as the 1918 Spanish flu, which killed approximately 50 million people worldwide (Johnson and Mueller, 2002). One feature that made the Spanish flu so deadly was its ability to initiate a dramatic host inflammatory response to the virus, culminating in a rapid “cytokine storm,” severe lung tissue damage, and death. The NLRP3 inflammasome was shown to be a major antiviral host defense mechanism controlling lung inflammation during IAV infection (Kanneganti et al., 2006a,b; Thomas et al., 2009). However, the PCD induced by IAV is not entirely dependent on the NLRP3 inflammasome. The loss of a component essential for just one PCD pathway does not protect the cells from IAV-induced cell death (Kuriakose et al., 2016). Cells lacking the ability to induce pyroptosis, apoptosis, and necroptosis collectively due to genetic loss of Ripk3 and Casp8 (or Fadd) are protected (Kuriakose et al., 2016). Combinatorial pharmacological inhibition of these three PCD pathways also protects the cells. IAV-induced PCD is also dependent on type I interferon signaling, as cells lacking interferon alpha and beta receptor subunit 1 (IFNAR1) are resistant to IAV-induced cell death (Kuriakose et al., 2016). These findings suggest the existence of upstream regulator(s) controlled by type I interferon signaling. The search for the upstream molecules responsible for NLRP3 inflammasome activation and PCD identified ZBP1—cells lacking Zbp1 are completely protected from PCD (Kuriakose et al., 2016). Later studies provided additional confirmation that ZBP1 is responsible for RIPK3-dependent cell death during IAV infection (Thapa et al., 2016; Kesavardhana et al., 2017). These observations led to the first description of PANoptosis (Malireddi et al., 2019). Mechanistically, ZBP1 drives PANoptosis by recruiting RIPK3 and CASP8 to the PANoptosome upon sensing IAV infection (Kuriakose et al., 2016; Kesavardhana et al., 2017; Kuriakose and Kanneganti, 2017; Zheng et al., 2020).
Although the ZBP1 PANoptosome promotes PANoptosis to shape the immune response against viral infections, its role in maintaining organismal homeostasis is increasingly being appreciated. Lethality in mice caused by the Ripk1RHIM mutation can be rescued by concomitantly deleting ZBP1 or mutating the RHIM domain in ZBP1 (Lin et al., 2016; Newton et al., 2016; Jiao et al., 2020), suggesting that RIPK1 inhibits the aberrant induction of PANoptosis by ZBP1. This evidence extends the concept of PANoptosis beyond the context of infection and shows that it can also regulate fundamental organismal processes such as development.
Further evidence for the role of PANoptosis in organismal processes can be seen from studies with RIPK1. The regulation of PANoptosis by RIPK1 is essential for homeostasis, cell death, and inflammatory immune responses. The scaffolding function of RIPK1 promotes cell survival signaling via the assembly of complex-I, which blocks both early and late cell death checkpoints downstream of TNFR1 signaling (Gerlach et al., 2011; Vince et al., 2012; Dondelinger et al., 2013; Peltzer et al., 2016, 2018; Ting and Bertrand, 2016; Taraborrelli et al., 2018), while RIPK1's kinase activity or receptor-interacting protein homotypic interaction motif (RHIM) function enables PCD (Dillon et al., 2014; Kaiser et al., 2014; Rickard et al., 2014b; Dowling et al., 2015; Newton et al., 2019a; Lalaoui et al., 2020; Tao et al., 2020). Deletion of RIPK1 in mice is embryonically lethal, which may be caused by the activation of RIPK3-regulated PANoptosis-like cell death through CASP8 and Fas-associated protein with a death domain (FADD) leading to systemic inflammation (Kelliher et al., 1998; Zhang et al., 2011; Dillon et al., 2014). In addition, mutations inactivating CASP8 catalytic activity lead to embryonic lethality in mice mediated by PANoptosis through the activation of RIPK1, RIPK3-MLKL, and CASP1 (Fritsch et al., 2019; Newton et al., 2019b). Mutating the CASP8 cleavage site in RIPK1, which is essential to regulate RIPK1 activity, also causes embryonic lethality and inflammation (Newton et al., 2019a; Lalaoui et al., 2020; Tao et al., 2020). Collectively, these studies provide evidence for the regulation of PANoptosis by RIPK1 and CASP8.
Another key regulator of PANoptosis was identified through a study of the complex-I component TAK1 (Malireddi et al., 2018). TAK1 was found to play an important role in NLRP3 inflammasome activation and to also modulate RIPK1 activity to regulate PANoptosis (Malireddi et al., 2019). In the absence of TAK1 activity due to pharmacological inhibition or genetic loss, RIPK1 activates PANoptosis and inflammation through its kinase activity (Malireddi et al., 2018; Orning et al., 2018; Sarhan et al., 2018). A follow up study discovered RIPK1 kinase-independent activation of PANoptosis, questioning the dogma that RIPK1 kinase function is required for PCD (Malireddi et al., 2020). These studies suggest that loss of function or deregulation of RIPK1 homeostasis can engage PANoptosis and affect organismal development and immune responses.
The discovery of PANoptosis in the contexts of IAV infection and organismal homeostasis led us to revisit autoinflammatory diseases in which components of the PCD machinery have been implicated to determine whether PANoptosis also plays a role here. A mutation in the Pstpip2 gene (Pstpip2cmo) causes autoinflammation via CASP8- and NLRP3/CASP1-regulated PCD. Combined deletion of Ripk3, Casp1, and Casp8, which blocks all three arms of PANoptosis (pyroptosis, apoptosis, and necroptosis), rescued the mice from the autoinflammatory condition, while inhibition of a single arm was not sufficient for rescue (Lukens et al., 2014; Gurung et al., 2016a), suggesting the involvement of PANoptosis in mediating the autoinflammatory condition. Another example of PANoptosis in autoinflammation can be seen in mice with a mutation in Sharpin (Sharpincpdm). The inflammation in Sharpincpdm mice can be rescued by deletion of TNF or expression of kinase-dead RIPK1 (Gerlach et al., 2011; Berger et al., 2014). It can also be rescued by hemizygous deletion of CASP8 along with the loss of RIPK3 (Rickard et al., 2014b). Skin inflammation can also be delayed by the loss of Nlrp3 or the inflammasome-dependent cytokine IL-1β (Berger et al., 2014; Douglas et al., 2015; Gurung et al., 2016b). Involvement of the three arms of PANoptosis suggests a role for PCD and the inflammatory cytokines released by PANoptotic cells in driving inflammation in Sharpincpdm mice.
Several other proteins associated with autoinflammation with dual roles in cell survival and inflammatory immune signaling pathways have been implicated in PANoptosis. In addition to RIPK1, deletion of other complex-I molecules, including HOIP, HOIL-1, and A20, also leads to embryonic lethality and autoinflammatory diseases, suggesting they may have a role in regulating PANoptosis (Gerlach et al., 2011; Vince et al., 2012; Dondelinger et al., 2013; Vande Walle et al., 2014; Peltzer et al., 2016; Ting and Bertrand, 2016; Taraborrelli et al., 2018; Zhang et al., 2019).
Whether it is induced in response to infection, during organismal homeostasis, or in the context of autoinflammation, PANoptosis is executed by a molecular complex called the PANoptosome that integrates apoptotic, necroptotic, and inflammasome components. Downstream cytokines such as IL-1β and IL-18 and damage-associated molecular patterns (DAMPs) released by PANoptotic cells act as alarmins initiating and amplifying the inflammatory response. Overall, the PANoptosome is responsible for driving this inflammatory form of cell death, and defects in PANoptosome components have been implicated in a host of human diseases including autoinflammatory and neurodegenerative diseases, cancer, and susceptibility to pathogenic infections (Figure 1B, Table 1).
Components of the PANoptosome
Genetic and biochemical data suggest that proteins that are part of the PANoptosome can be generally categorized into three classes—(1) ZBP1 and NLRP3 as putative PAMP and DAMP sensors, (2) ASC and FADD as adaptors, and (3) RIPK1, RIPK3, CASP1, and CASP8 as catalytic effectors. A recent study identified CASP6 as a critical component of the PANoptosome that promotes its assembly by reinforcing the interaction between RIPK3 and ZBP1 in the complex, suggesting a novel scaffold function for this presumed apoptotic executioner caspase (Zheng et al., 2020). Whether the catalytic activity of CASP6 is required for PANoptosome assembly remains to be fully understood. Additionally, classification of RIPK1 presents a challenge. Earlier studies had shown that RIPK1 kinase activity was required for the induction of necroptosis. However, a more recent study showed that RIPK1 is recruited to the PANoptosome, and the kinase-dead mutant of RIPK1 is capable of inducing PANoptosis, suggesting that RIPK1 may act as an adaptor in some cases and a catalytic effector in others (Malireddi et al., 2020). These observations suggest that PANoptosome components might have multiple biochemical functions involved in the execution of PANoptosis.
The Inflammasome, Apoptosome, and Necroptosome as Templates for Panoptosome Formation
PANoptosome assembly likely requires domain-specific homotypic and heterotypic interactions between the molecular components. A closer look at the molecular mechanisms of PCD makes it clear that multimeric protein interactions and catalytic activities constitute two major functionally important aspects to induce cell death. In the case of pyroptosis, cell death is executed by a hetero-multimeric complex called the inflammasome (Martinon et al., 2002; Kanneganti et al., 2006a; Mariathasan et al., 2006; Sutterwala et al., 2007; Bürckstümmer et al., 2009; Fernandes-Alnemri et al., 2009; Hornung et al., 2009; Broz and Dixit, 2016). Inflammasome assembly involves homotypic interactions among the caspase recruitment domains (CARDs) and PYRIN domains (PYDs) of inflammasome sensors, the adaptor ASC, and the catalytic actuator CASP1 (Man and Kanneganti, 2015; Broz and Dixit, 2016). Intrinsic apoptosis is driven by another hetero-multimeric complex known as the apoptosome. Apoptosome assembly exploits the homotypic interactions of CARDs for assembly and activation of the catalytic actuator CASP9 for cell death execution (Zou et al., 1999; Acehan et al., 2002; Bao and Shi, 2007; Riedl and Salvesen, 2007; Zmasek and Godzik, 2013). Extrinsic apoptosis induced by death receptors exploits the homotypic interactions mediated by death effector domains (DEDs) and death domains (DEATH) to assemble the death-inducing signaling complex (DISC), which activates the cysteine protease CASP8 (Lo et al., 2015; Yang, 2015). In necroptosis, the molecular mechanism of assembly and the molecular composition of the necroptosome are not entirely clear, but available data suggest that assembly might require the homotypic interactions of RHIMs and the kinase activity of RIPK3 for cell death execution (Wallach et al., 2011; Li et al., 2012; Vanden Berghe et al., 2016; Zhang et al., 2016; Mompeán et al., 2018; Weber et al., 2018). The PANoptosome likely combines many of the same interactions as the inflammasome, DISC, and necroptosome and acts as a versatile PCD complex utilizing necroptosome-associated kinase, as well as apoptosis- and pyroptosis-associated protease, activities for cell death execution.
Evolution of the PANoptosome Components
Genetic studies have established that PANoptosome components have evolved to play key roles in cellular homeostasis, development, and inflammatory immune responses (Lin et al., 2016; Newton et al., 2016, 2019a,b; Malireddi et al., 2018; Fritsch et al., 2019; Kesavardhana et al., 2020; Lalaoui et al., 2020). Although some of the conserved molecular motifs present in cell death-executing complexes exist in all branches and kingdoms of life, others are present only in higher eukaryotes and viruses. The key molecules involved in pyroptosis, apoptosis, and necroptosis share these domains, which indicates that the three pathways employ similar assembly mechanisms for large hetero-multimeric cell death-inducing complexes.
To develop a framework to understand the interactions between these molecules, we performed an evolutionary analysis to help identify important domains for PANoptosome assembly because the critical domains are more likely to coevolve. We separated the domains of the PANoptosome components into three classes—(1) assembly domains, (2) catalytic domains, and (3) sensing domains. Assembly domains include CARD, DEATH, DED, PYD, and RHIM. Catalytic domains are Peptidase_C14 (caspase domain), Pkinase (kinase domain in RIPK3), Pkinase_Tyr (kinase domain in RIPK1), and NACHT (ATPase domain in NLRP3). Sensing domains include z-alpha and LRR_6. We used the Pfam database to look at the evolution of these domains (Sonnhammer et al., 1997). Four of the assembly domains (CARD, DEATH, DED, and PYD) are also called death fold domains due to their critical role in assembling PCD-executing complexes. The death fold domains seem to be present only in multicellular organisms and a few viruses that infect them (Figure 2A). Although Pkinase_Tyr, RHIM, z-alpha, and LRR_6 domains do exist in prokaryotes, most of the proteins containing these domains are present in eukaryotes with a few exceptions in viruses (Figures 2A–C). Pkinase, Peptidase_C14, and NACHT domains are more widely distributed among the kingdoms of life (Figure 2B). These data suggest that some of the molecular motifs responsible for catalytic functions might have been present in the common ancestors of eukaryotes and prokaryotes. However, a functional PANoptosome might have coevolved with the development of multicellularity because the death fold domains that seem to be essential for its assembly are present mainly in metazoans. This is in line with the role of PANoptosis in modulating organismal homeostasis (Dillon et al., 2014; Malireddi et al., 2018, 2019; Newton et al., 2019a; Lalaoui et al., 2020).
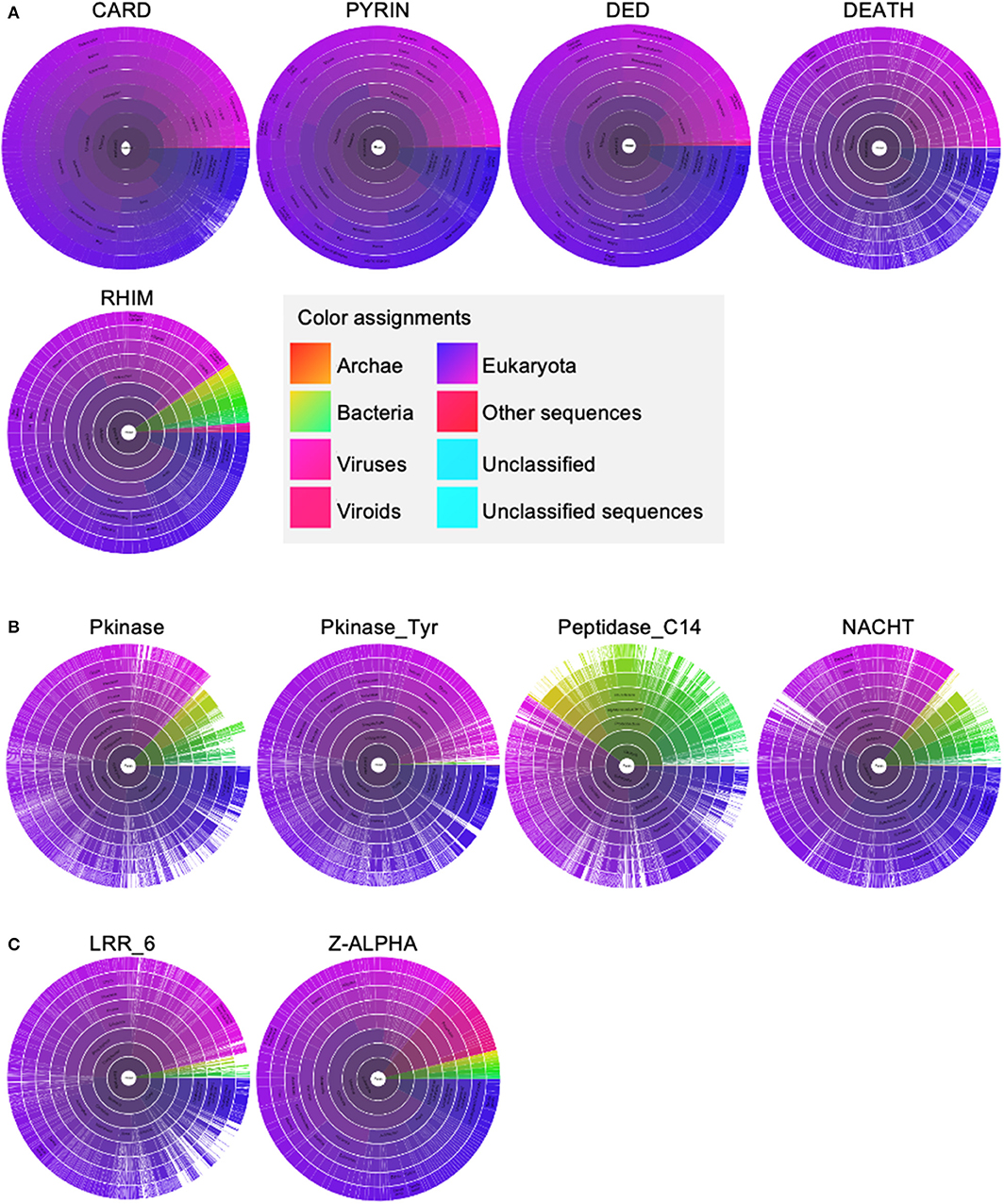
Figure 2. The distribution of molecular motifs present in PANoptosome components. (A) Distribution of assembly domains. (B) Distribution of catalytic domains. (C) Distribution of sensing domains. All domain names and their distributions were obtained from the Pfam database (Sonnhammer et al., 1997). Sunburst visualizations and color assignment legends were exported from the Pfam database.
The Mechanism of PANoptosome Assembly Through Phylogenetic Analysis
PANoptosome components contain a wide range of molecular motifs that can be utilized for assembly of the active complex. However, a clear domain-domain homotypic interaction-based assembly mechanism has not yet been deciphered. Phylogenetic analysis of the assembly domains of the components using Maximum Likelihood Analysis allows a testable model for the mechanism of PANoptosome assembly to be built (Figure 3). Since the homotypic interactions between the assembly domains are associated with sequence similarity between them, the domains capable of heterotypic interactions would hypothetically be closer together in a phylogenetic tree. Phylogenetic analysis revealed that one of the DED motifs in CASP8 (DED2CASP8) is in the same branch as the PYD from NLRP3 and ASC. Previous studies have revealed that the PYD of ASC (PYDASC) is required for CASP8 polymerization potentially through a heterotypic interaction between DED2CASP8 and PYDASC (Vajjhala et al., 2015), corroborating the phylogenetic data. The CASP8 DED1 motif (DED1CASP8) was close to the FADD DED motif (DEDFADD). The homotypic interaction mediated by DEDFADD and DED1CASP8 is known to be important for DISC assembly during extrinsic apoptosis (Kischkel et al., 1995; Muzio et al., 1996; Fu et al., 2016). Another feature of the phylogenetic tree was that the CARD domain of ASC (CARDASC) and one of the RHIM domains of ZBP1 (RHIM2ZBP1) were closer to each other than to CARDASC and CARDCASP1 or RHIM1ZBP1 and RHIM2ZBP1, respectively. This result suggests that upon ligand sensing, ZBP1 may be able to induce assembly of the PANoptosome by recruiting ASC through a heterotypic interaction between RHIM2ZBP1 and CARDASC.
Figure 3. Using phylogenetic analysis to understand the molecular mechanism governing PANoptosome assembly. Phylogenetic analysis of assembly domains present in PANoptosome components. Amino acid sequences corresponding to the domains were obtained from the Uniprot database (The UniProt Consortium, 2015). Multiple sequence alignment and phylogenetic analysis were performed using the Mega-X software package (Kumar et al., 2018). ClustalW was used for multiple sequence alignment. Maximum likelihood analysis with bootstrap 500 iterations was used to generate the phylogenetic tree. The consensus phylogenetic tree is shown. If a protein contains multiple copies of a domain, the domains are numbered beginning at the N-terminus. For example, the first occurrence of the RHIM domain in ZBP1 is named RHIM 1, and the second occurrence is named RHIM 2.
These observations suggest a putative assembly mechanism for the ZBP1 PANoptosome involving IAV vRNP sensing by ZBP1 (Figure 3). In this model, activated ZBP1 would bind to ASC through RHIM2ZBP1 and CARDASC heterotypic interactions and induce ASC polymerization. PYDASC can then be engaged to recruit NLRP3 into the complex through PYDNLRP3. Since CASP1 activation in IAV-infected cells is strictly dependent on NLRP3, NLRP3 recruitment to the PANoptosome must precede recruitment of CASP1 (Kanneganti et al., 2006b). In cells lacking TAK1 kinase activity, CASP1 activation is again dependent on NLRP3, but the identity of the upstream sensor that induces assembly of the PANoptosome in these cells remains unclear (Malireddi et al., 2018). This sequence of events suggests that there may be a conformational change in CARDASC after PYDNLRP3 binding to PYDASC that would allow CASP1 binding. The change in binding capacity could also be due to an increase in valency because the NACHT domain of NLRP3 (NACHTNLRP3) increases PYDASC polymerization (Cai et al., 2014; Lu et al., 2014; Samir and Kanneganti, 2019). While the RHIM2ZBP1 valency is occupied to build the ASC-mediated portion of the complex, the free RHIM valency (RHIM1ZBP1) can recruit RIPK1 and/or RIPK3. CASP8 can be recruited to the complex in two ways—(1) through the DED domain of RIPK1 (DEDRIPK1) interacting with one of the DEDCASP8 domains and/or (2) through PYDASC interacting with DED2CASP8. Future experiments will help define the specific molecular interactions facilitating PANoptosome assembly.
Alternative Assembly Strategies
Two recent studies have suggested a role for interactions mediated by intrinsically disordered regions (IDR) in inducing PCD (Samir et al., 2019; Zheng et al., 2020). In one study, the interaction between NLRP3 and DDX3X was proposed to be IDR-mediated and was critical for pyroptosis downstream of NLRP3 inflammasome activation (Samir et al., 2019). In another study, CASP6 was found to promote IAV-induced PANoptosis dependent on an IDR-mediated interaction between CASP6 and RIPK3. This interaction increased the efficiency of RIPK3 recruitment to the ZBP1-induced PANoptosome (Zheng et al., 2020). The possibility of IDR involvement in the assembly of the PANoptosome increases the range of available interaction modalities because all known PANoptosome components have predicted IDRs. Additionally, some of the assembly domains have the ability to undergo prionoid phase transitions (Cai et al., 2014; Lu et al., 2014; Li et al., 2018). Therefore, it is likely that assembly of the PANoptosome can lead to signal amplification through polymerization and prionoid phase transitions, resulting in the assembly of large cytoplasmic supramolecular structures that can be visualized using immunofluorescence microscopy.
The PANoptosome contains molecules that can induce PCD by multiple mechanisms. This means that a full complex may not need to be assembled before cells undergo cell death. This provides opportunities, and difficulties, for the molecular dissection of PANoptosis. For example, inhibition of one arm of PANoptosis will not be sufficient to prevent cells from dying. This can be exploited to study the ways in which different PCD pathways crosstalk with each other. Additionally, identification of apical sensors that control PANoptosis as a whole will be critical for the modulation of this process. So far ZBP1 is the only apical sensor that has been identified to induce PANoptosis in response to IAV infection (Kuriakose et al., 2016; Kesavardhana et al., 2017). However, TAK1 inhibition-induced PANoptosis provides compelling evidence for the existence of alternative modes of PANoptosis activation that do not depend on the ZBP1 PANoptosome but instead result from pathogen-induced cellular perturbations being detected as danger signals, triggering the activation of this highly inflammatory form of cell death (Malireddi et al., 2018, 2020; Orning et al., 2018; Sarhan et al., 2018). These studies indicate that PANoptosis has evolved as a robust fail-safe mechanism to induce inflammatory cell death and promote protective antimicrobial immune responses, especially in response to pathogens that target key inflammatory signaling cascades or cell death pathways (Philip et al., 2014; Weng et al., 2014; Malireddi et al., 2018, 2019, 2020; Orning et al., 2018; Sarhan et al., 2018). It remains to be seen if there are additional sensors that are activated in response to other pathogenic challenges to induce PANoptosis. It is possible that other apical sensors for PANoptosis do not engage exactly the same molecular machinery as ZBP1 or TAK1 inhibition, allowing PANoptosomes with differing compositions to form. This would be in line with the lessons learned from pyroptosis and inflammasomes, as the sensors NLRP1b, NLRP3, and NLRC4 have differential requirements for the adaptor ASC when assembling the inflammasome (Case et al., 2009; Abdelaziz et al., 2011; De Jong et al., 2014; Opdenbosch et al., 2014; Patankar et al., 2015; Broz and Dixit, 2016; Li et al., 2018). The ZBP1 PANoptosome can be thought of as the founding member of the PANoptosome class of PCD-inducing complexes, and evidence suggests there are additional forms of the PANoptosome (Malireddi et al., 2018, 2020; Christgen et al., 2020).
Summary
The recent progress in our understanding of the extensive crosstalk between different cell death and signaling cascades unequivocally establishes the existence of multifaceted signaling platforms. The PANoptosome represents one such platform which can engage multiple cell death modalities including pyroptosis, apoptosis, and necroptosis to form the collective inflammatory cell death pathway of PANoptosis. The PANoptosome appears to have evolved to efficiently utilize diverse modes of cellular communication that include domain-based scaffolds and enzyme-based catalytic modifications to engage PANoptosis. Using these diverse cellular mechanisms might be an important way in which the PANoptosome incorporates fundamentally different modes of cell death to orchestrate homeostasis during developmental programs while driving a robust inflammatory form of cell death for immune-mediated protection against microbial infections. Because many of the PANoptosome components have been associated with disease, including autoinflammatory, infectious, and neurodegenerative diseases and cancer, improved understanding of the molecular underpinnings of the PANoptosome will be able to inform the development of targeted inhibitors and activators of inflammatory cell death for therapeutic modulation of inflammation and the immune response.
Author Contributions
PS performed the phylogenetic and evolutionary analyses. All authors contributed to writing and editing the manuscript.
Funding
Research studies from the Kanneganti lab are supported by the US National Institutes of Health (AR056296, CA163507, AI124346, and AI101935 to T-DK) and the American Lebanese Syrian Associated Charities (to T-DK).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We apologize to our colleagues whose work was not cited due to space limitations. We thank Rebecca Tweedell, Ph.D., for scientific editing and Emily VanGilder of the Biomedical Communications Department at St. Jude Children's Research Hospital for assistance with Figure 1A.
References
Abdelaziz, D. H. A., Gavrilin, M. A., Akhter, A., Caution, K., Kotrange, S., Khweek, A. A., et al. (2011). Asc-dependent and independent mechanisms contribute to restriction of legionella pneumophila infection in murine macrophages. Front. Microbiol. 2:18. doi: 10.3389/fmicb.2011.00018
Acehan, D., Jiang, X., Morgan, D. G., Heuser, J. E., Wang, X., and Akey, C. W. (2002). Three-dimensional structure of the apoptosome: implications for assembly, procaspase-9 binding, and activation. Mol. Cell 9, 423–432. doi: 10.1016/S1097-2765(02)00442-2
Adrianto, I., Wen, F., Templeton, A., Wiley, G., King, J. B., Lessard, C. J., et al. (2011). Association of a functional variant downstream of TNFAIP3 with systemic lupus erythematosus. Nat. Genet. 43, 253–258. doi: 10.1038/ng.766
Aganna, E., Martinon, F., Hawkins, P. N., Ross, J. B., Swan, D. C., Booth, D. R., et al. (2002). Association of mutations in the NALP3/CIAS1/PYPAF1 gene with a broad phenotype including recurrent fever, cold sensitivity, sensorineural deafness, and AA amyloidosis. Arthritis Rheum. 46, 2445–2452. doi: 10.1002/art.10509
Aksentijevich, I., Nowak, M., Mallah, M., Chae, J. J., Watford, W. T., Hofmann, S. R., et al. (2002). De novo CIAS1 mutations, cytokine activation, and evidence for genetic heterogeneity in patients with neonatal-onset multisystem inflammatory disease (NOMID): a new member of the expanding family of pyrin-associated autoinflammatory diseases. Arthritis Rheum. 46, 3340–3348. doi: 10.1002/art.10688
Alejandre, N., Ruiz-Palacios, A., García-Aparicio, A. M., Blanco-Kelly, F., Bermúdez, S., Fernández-Sanz, G., et al. (2014). Description of a new family with cryopyrin-associated periodic syndrome: risk of visual loss in patients bearing the R260W mutation. Rheumatol. Oxf. Engl. 53, 1095–1099. doi: 10.1093/rheumatology/ket486
Bao, Q., and Shi, Y. (2007). Apoptosome: a platform for the activation of initiator caspases. Cell Death Differ. 14, 56–65. doi: 10.1038/sj.cdd.4402028
Berger, S. B., Kasparcova, V., Hoffman, S., Swift, B., Dare, L., Schaeffer, M., et al. (2014). Cutting edge: RIP1 kinase activity is dispensable for normal development but is a key regulator of inflammation in SHARPIN-deficient mice. J. Immunol. 192, 5476–5480. doi: 10.4049/jimmunol.1400499
Broz, P., and Dixit, V. M. (2016). Inflammasomes: mechanism of assembly, regulation and signalling. Nat. Rev. Immunol. 16, 407–420. doi: 10.1038/nri.2016.58
Bürckstümmer, T., Baumann, C., Blüml, S., Dixit, E., Dürnberger, G., Jahn, H., et al. (2009). An orthogonal proteomic-genomic screen identifies AIM2 as a cytoplasmic DNA sensor for the inflammasome. Nat. Immunol. 10, 266–272. doi: 10.1038/ni.1702
Cai, X., Chen, J., Xu, H., Liu, S., Jiang, Q.-X., Halfmann, R., et al. (2014). Prion-like polymerization underlies signal transduction in antiviral immune defense and inflammasome activation. Cell 156, 1207–1222. doi: 10.1016/j.cell.2014.01.063
Case, C. L., Shin, S., and Roy, C. R. (2009). Asc and ipaf inflammasomes direct distinct pathways for caspase-1 activation in response to legionella pneumophila. Infect. Immun. 77, 1981–1991. doi: 10.1128/IAI.01382-08
Christgen, S., Zheng, M., Kesavardhana, S., Karki, R., Malireddi, R. K. S., Banoth, B., et al. (2020). Identification of the PANoptosome: a molecular platform triggering pyroptosis, apoptosis, and necroptosis (PANoptosis). Front. Cell. Infect. Microbiol.
Chun, H. J., Zheng, L., Ahmad, M., Wang, J., Speirs, C. K., Siegel, R. M., et al. (2002). Pleiotropic defects in lymphocyte activation caused by caspase-8 mutations lead to human immunodeficiency. Nature 419, 395–399. doi: 10.1038/nature01063
Dannappel, M., Vlantis, K., Kumari, S., Polykratis, A., Kim, C., Wachsmuth, L., et al. (2014). RIPK1 maintains epithelial homeostasis by inhibiting apoptosis and necroptosis. Nature 513, 90–94. doi: 10.1038/nature13608
De Jong, H. K., Koh, G. C. K. W., van Lieshout, M. H. P., Roelofs, J. J. T. H., van Dissel, J. T., van der Poll, T., et al. (2014). Limited role for ASC and NLRP3 during in vivo Salmonella Typhimurium infection. BMC Immunol. 15:30. doi: 10.1186/s12865-014-0030-7
Dillon, C. P., Weinlich, R., Rodriguez, D. A., Cripps, J. G., Quarato, G., Gurung, P., et al. (2014). RIPK1 blocks early postnatal lethality mediated by caspase-8 and RIPK3. Cell 157, 1189–1202. doi: 10.1016/j.cell.2014.04.018
Dodé, C., Le Dû, N., Cuisset, L., Letourneur, F., Berthelot, J.-M., Vaudour, G., et al. (2002). New mutations of CIAS1 that are responsible for Muckle-Wells syndrome and familial cold urticaria: a novel mutation underlies both syndromes. Am. J. Hum. Genet. 70, 1498–1506. doi: 10.1086/340786
Dondelinger, Y., Aguileta, M. A., Goossens, V., Dubuisson, C., Grootjans, S., Dejardin, E., et al. (2013). RIPK3 contributes to TNFR1-mediated RIPK1 kinase-dependent apoptosis in conditions of cIAP1/2 depletion or TAK1 kinase inhibition. Cell Death Differ. 20, 1381–1392. doi: 10.1038/cdd.2013.94
Dondelinger, Y., Delanghe, T., Priem, D., Wynosky-Dolfi, M. A., Sorobetea, D., Rojas-Rivera, D., et al. (2019). Serine 25 phosphorylation inhibits RIPK1 kinase-dependent cell death in models of infection and inflammation. Nat. Commun. 10, 1–16. doi: 10.1038/s41467-019-09690-0
Douglas, T., Champagne, C., Morizot, A., Lapointe, J.-M., and Saleh, M. (2015). The inflammatory caspases-1 and−11 mediate the pathogenesis of dermatitis in sharpin-deficient mice. J. Immunol. 195, 2365–2373. doi: 10.4049/jimmunol.1500542
Dowling, J. P., Nair, A., and Zhang, J. (2015). A novel function of RIP1 in postnatal development and immune homeostasis by protecting against RIP3-dependent necroptosis and FADD-mediated apoptosis. Front. Cell Dev. Biol. 3:12. doi: 10.3389/fcell.2015.00012
El-Shanti, H. I., and Ferguson, P. J. (2007). Chronic recurrent multifocal osteomyelitis: a concise review and genetic update. Clin. Orthop. 462, 11–19. doi: 10.1097/BLO.0b013e3180986d73
Fan, Y., Tao, J.-H., Zhang, L.-P., Li, L.-H., and Ye, D.-Q. (2011). The association between BANK1 and TNFAIP3 gene polymorphisms and systemic lupus erythematosus: a meta-analysis. Int. J. Immunogenet. 38, 151–159. doi: 10.1111/j.1744-313X.2010.00990.x
Feldmann, J., Prieur, A.-M., Quartier, P., Berquin, P., Certain, S., Cortis, E., et al. (2002). Chronic infantile neurological cutaneous and articular syndrome is caused by mutations in CIAS1, a gene highly expressed in polymorphonuclear cells and chondrocytes. Am. J. Hum. Genet. 71, 198–203. doi: 10.1086/341357
Ferguson, P. J., and El-Shanti, H. I. (2007). Autoinflammatory bone disorders. Curr. Opin. Rheumatol. 19, 492–498. doi: 10.1097/BOR.0b013e32825f5492
Fernandes-Alnemri, T., Yu, J.-W., Datta, P., Wu, J., and Alnemri, E. S. (2009). AIM2 activates the inflammasome and cell death in response to cytoplasmic DNA. Nature 458, 509–513. doi: 10.1038/nature07710
Fritsch, M., Günther, S. D., Schwarzer, R., Albert, M.-C., Schorn, F., Werthenbach, J. P., et al. (2019). Caspase-8 is the molecular switch for apoptosis, necroptosis and pyroptosis. Nature 575, 683–687. doi: 10.1038/s41586-019-1770-6
Fu, T.-M., Li, Y., Lu, A., Li, Z., Vajjhala, P. R., Cruz, A. C., et al. (2016). Cryo-EM structure of caspase-8 tandem DED filament reveals assembly and regulation mechanisms of the death-inducing signaling complex. Mol. Cell 64, 236–250. doi: 10.1016/j.molcel.2016.09.009
Furlan, R., Filippi, M., Bergami, A., Rocca, M. A., Martinelli, V., Poliani, P. L., et al. (1999). Peripheral levels of caspase-1 mRNA correlate with disease activity in patients with multiple sclerosis; a preliminary study. J. Neurol. Neurosurg. Psychiatry 67, 785–788. doi: 10.1136/jnnp.67.6.785
Gerlach, B., Cordier, S. M., Schmukle, A. C., Emmerich, C. H., Rieser, E., Haas, T. L., et al. (2011). Linear ubiquitination prevents inflammation and regulates immune signalling. Nature 471, 591–596. doi: 10.1038/nature09816
Greenman, C., Stephens, P., Smith, R., Dalgliesh, G. L., Hunter, C., Bignell, G., et al. (2007). Patterns of somatic mutation in human cancer genomes. Nature 446, 153–158. doi: 10.1038/nature05610
Gurung, P., Anand, P. K., Malireddi, R. K. S., Walle, L. V., Opdenbosch, N. V., Dillon, C. P., et al. (2014). FADD and caspase-8 mediate priming and activation of the canonical and noncanonical Nlrp3 inflammasomes. J. Immunol. 192, 1835–1846. doi: 10.4049/jimmunol.1302839
Gurung, P., Burton, A., and Kanneganti, T.-D. (2016a). NLRP3 inflammasome plays a redundant role with caspase 8 to promote IL-1β-mediated osteomyelitis. Proc. Natl. Acad. Sci. 113, 4452–4457. doi: 10.1073/pnas.1601636113
Gurung, P., Sharma, B. R., and Kanneganti, T.-D. (2016b). Distinct role of IL-1β in instigating disease in Sharpin cpdm mice. Sci. Rep. 6, 1–11. doi: 10.1038/srep36634
Hao, G., Li, Y., Liu, J., and Wo, M. (2014). TNFAIP3 rs2230926 polymorphisms in rheumatoid arthritis of southern Chinese Han population: a case-control study. Int. J. Clin. Exp. Pathol. 7, 8958–8961.
Hawkins, P. N., Lachmann, H. J., Aganna, E., and McDermott, M. F. (2004). Spectrum of clinical features in muckle-wells syndrome and response to anakinra. Arthritis Rheum. 50, 607–612. doi: 10.1002/art.20033
Hoffjan, S., Okur, A., Epplen, J. T., Wieczorek, S., Chan, A., and Akkad, D. A. (2015). Association of TNFAIP3 and TNFRSF1A variation with multiple sclerosis in a German case-control cohort. Int. J. Immunogenet. 42, 106–110. doi: 10.1111/iji.12183
Hoffman, H. M., Mueller, J. L., Broide, D. H., Wanderer, A. A., and Kolodner, R. D. (2001). Mutation of a new gene encoding a putative pyrin-like protein causes familial cold autoinflammatory syndrome and muckle-wells syndrome. Nat. Genet. 29, 301–305. doi: 10.1038/ng756
Hornung, V., Ablasser, A., Charrel-Dennis, M., Bauernfeind, F., Horvath, G., Caffrey, D. R., et al. (2009). AIM2 recognizes cytosolic dsDNA and forms a caspase-1-activating inflammasome with ASC. Nature 458, 514–518. doi: 10.1038/nature07725
Huang, W.-X., Huang, P., and Hillert, J. (2004). Increased expression of caspase-1 and interleukin-18 in peripheral blood mononuclear cells in patients with multiple sclerosis. Mult. Scler. Houndmills Basingstoke Engl. 10, 482–487. doi: 10.1191/1352458504ms1071oa
Indhumathi, S., Rajappa, M., Chandrashekar, L., Ananthanarayanan, P. H., Thappa, D. M., and Negi, V. S. (2015). TNFAIP3 and TNIP1 polymorphisms confer psoriasis risk in South Indian Tamils. Br. J. Biomed. Sci. 72, 168–173. doi: 10.1080/09674845.2015.11665748
Jesus, A. A., Silva, C. A., Segundo, G. R., Aksentijevich, I., Fujihira, E., Watanabe, M., et al. (2008). Phenotype-genotype analysis of cryopyrin-associated periodic syndromes (CAPS): description of a rare non-exon 3 and a novel CIAS1 missense mutation. J. Clin. Immunol. 28, 134–138. doi: 10.1007/s10875-007-9150-7
Jiang, X., Tian, H., Fan, Y., Chen, J., Song, Y., Wang, S., et al. (2012). Expression of tumor necrosis factor alpha-induced protein 3 mRNA in peripheral blood mononuclear cells negatively correlates with disease severity in psoriasis vulgaris. Clin. Vaccine Immunol. CVI 19, 1938–1942. doi: 10.1128/CVI.00500-12
Jiao, H., Wachsmuth, L., Kumari, S., Schwarzer, R., Lin, J., Eren, R. O., et al. (2020). Z-nucleic-acid sensing triggers ZBP1-dependent necroptosis and inflammation. Nature 580, 391–395. doi: 10.1038/s41586-020-2129-8
Johnson, N. P. A. S., and Mueller, J. (2002). Updating the accounts: global mortality of the 1918-1920 “Spanish” influenza pandemic. Bull. Hist. Med. 76, 105–115. doi: 10.1353/bhm.2002.0022
Kaiser, W. J., Daley-Bauer, L. P., Thapa, R. J., Mandal, P., Berger, S. B., Huang, C., et al. (2014). RIP1 suppresses innate immune necrotic as well as apoptotic cell death during mammalian parturition. Proc. Natl. Acad. Sci. U.S.A. 111, 7753–7758. doi: 10.1073/pnas.1401857111
Kanneganti, T.-D., Body-Malapel, M., Amer, A., Park, J.-H., Whitfield, J., Franchi, L., et al. (2006b). Critical Role for cryopyrin/Nalp3 in activation of caspase-1 in response to viral infection and double-stranded RNA. J. Biol. Chem. 281, 36560–36568. doi: 10.1074/jbc.M607594200
Kanneganti, T.-D., Özören, N., Body-Malapel, M., Amer, A., Park, J.-H., Franchi, L., et al. (2006a). Bacterial RNA and small antiviral compounds activate caspase-1 through cryopyrin/Nalp3. Nature 440, 233–236. doi: 10.1038/nature04517
Kelliher, M. A., Grimm, S., Ishida, Y., Kuo, F., Stanger, B. Z., and Leder, P. (1998). The death domain kinase RIP mediates the TNF-induced NF-κB signal. Immunity 8, 297–303. doi: 10.1016/S1074-7613(00)80535-X
Kesavardhana, S., Kuriakose, T., Guy, C. S., Samir, P., Malireddi, R. K. S., Mishra, A., et al. (2017). ZBP1/DAI ubiquitination and sensing of influenza vRNPs activate programmed cell death. J. Exp. Med. jem.20170550. doi: 10.1084/jem.20170550
Kesavardhana, S., Malireddi, R. K. S., and Kanneganti, T.-D. (2020). Caspases in cell death, inflammation, and gasdermin-induced pyroptosis. Annu. Rev. Immunol. 38, 567–95. doi: 10.1146/annurev-immunol-073119-095439
Kim, S.-K., Choe, J.-Y., Bae, J., Chae, S.-C., Park, D.-J., Kwak, S. G., et al. (2014). TNFAIP3 gene polymorphisms associated with differential susceptibility to rheumatoid arthritis and systemic lupus erythematosus in the Korean population. Rheumatol. Oxf. Engl. 53, 1009–1013. doi: 10.1093/rheumatology/ket473
Kischkel, F. C., Hellbardt, S., Behrmann, I., Germer, M., Pawlita, M., Krammer, P. H., et al. (1995). Cytotoxicity-dependent APO-1 (Fas/CD95)-associated proteins form a death-inducing signaling complex (DISC) with the receptor. EMBO J. 14, 5579–5588. doi: 10.1002/j.1460-2075.1995.tb00245.x
Kuemmerle-Deschner, J. B., Lohse, P., Koetter, I., Dannecker, G. E., Reess, F., Ummenhofer, K., et al. (2011). NLRP3 E311K mutation in a large family with Muckle-Wells syndrome–description of a heterogeneous phenotype and response to treatment. Arthritis Res. Ther. 13:R196. doi: 10.1186/ar3526
Kumar, S., Stecher, G., Li, M., Knyaz, C., and Tamura, K. (2018). MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35, 1547–1549. doi: 10.1093/molbev/msy096
Kuriakose, T., and Kanneganti, T.-D. (2017). ZBP1: innate sensor regulating cell death and inflammation. Trends Immunol. 39, 123–134. doi: 10.1016/j.it.2017.11.002
Kuriakose, T., Man, S. M., Malireddi, R. K. S., Karki, R., Kesavardhana, S., Place, D. E., et al. (2016). ZBP1/DAI is an innate sensor of influenza virus triggering the NLRP3 inflammasome and programmed cell death pathways. Sci. Immunol. 1, aag2045-aag2045. doi: 10.1126/sciimmunol.aag2045
Lalaoui, N., Boyden, S. E., Oda, H., Wood, G. M., Stone, D. L., Chau, D., et al. (2020). Mutations that prevent caspase cleavage of RIPK1 cause autoinflammatory disease. Nature 577, 103–108. doi: 10.1038/s41586-019-1828-5
Lamkanfi, M., Kanneganti, T.-D., Van Damme, P., Vanden Berghe, T., Vanoverberghe, I., Vandekerckhove, J., et al. (2008). Targeted peptidecentric proteomics reveals caspase-7 as a substrate of the caspase-1 inflammasomes. Mol. Cell Proteomics 7, 2350–2363. doi: 10.1074/mcp.M800132-MCP200
Le Goff, C., Rogers, C., Le Goff, W., Pinto, G., Bonnet, D., Chrabieh, M., et al. (2016). Heterozygous Mutations in MAP3K7, Encoding TGF-β-Activated Kinase 1, Cause Cardiospondylocarpofacial Syndrome. Am. J. Hum. Genet. 99, 407–413. doi: 10.1016/j.ajhg.2016.06.005
Lee, Y. H., Bae, S.-C., Choi, S. J., Ji, J. D., and Song, G. G. (2012). Associations between TNFAIP3 gene polymorphisms and rheumatoid arthritis: a meta-analysis. Inflamm. Res. Off. J. Eur. Histamine Res. Soc. Al 61, 635–641. doi: 10.1007/s00011-012-0455-5
Li, J., McQuade, T., Siemer, A. B., Napetschnig, J., Moriwaki, K., Hsiao, Y.-S., et al. (2012). The RIP1/RIP3 necrosome forms a functional amyloid signaling complex required for programmed necrosis. Cell 150, 339–350. doi: 10.1016/j.cell.2012.06.019
Li, Y., Fu, T.-M., Lu, A., Witt, K., Ruan, J., Shen, C., et al. (2018). Cryo-EM structures of ASC and NLRC4 CARD filaments reveal a unified mechanism of nucleation and activation of caspase-1. Proc. Natl. Acad. Sci. U.S.A. 115, 10845–10852. doi: 10.1073/pnas.1810524115
Lin, J., Kumari, S., Kim, C., Van, T.-M., Wachsmuth, L., Polykratis, A., et al. (2016). RIPK1 counteracts ZBP1-mediated necroptosis to inhibit inflammation. Nature 540, 124–128. doi: 10.1038/nature20558
Liu, W., Chang, B.-L., Cramer, S., Koty, P. P., Li, T., Sun, J., et al. (2007). Deletion of a small consensus region at 6q15, including the MAP3K7 gene, is significantly associated with high-grade prostate cancers. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 13, 5028–5033. doi: 10.1158/1078-0432.CCR-07-0300
Lo, Y.-C., Lin, S.-C., Yang, C.-Y., and Tung, J.-Y. (2015). Tandem DEDs and CARDs suggest novel mechanisms of signaling complex assembly. Apoptosis 20, 124–135. doi: 10.1007/s10495-014-1054-4
Lodolce, J. P., Kolodziej, L. E., Rhee, L., Kariuki, S. N., Franek, B. S., McGreal, N. M., et al. (2010). African-derived genetic polymorphisms in TNFAIP3 mediate risk for autoimmunity. J. Immunol. Baltim. Md 184, 7001–7009. doi: 10.4049/jimmunol.1000324
Lu, A., Magupalli, V. G., Ruan, J., Yin, Q., Atianand, M. K., Vos, M. R., et al. (2014). Unified polymerization mechanism for the assembly of ASC-dependent inflammasomes. Cell 156, 1193–1206. doi: 10.1016/j.cell.2014.02.008
Lukens, J. R., Gurung, P., Vogel, P., Johnson, G. R., Carter, R. A., McGoldrick, D. J., et al. (2014). Dietary modulation of the microbiome affects autoinflammatory disease. Nature 516, 246–249. doi: 10.1038/nature13788
Machida, E. O., Brock, M. V., Hooker, C. M., Nakayama, J., Ishida, A., Amano, J., et al. (2006). Hypermethylation of ASC/TMS1 is a sputum marker for late-stage lung cancer. Cancer Res. 66, 6210–6218. doi: 10.1158/0008-5472.CAN-05-4447
MacPherson, G., Healey, C. S., Teare, M. D., Balasubramanian, S. P., Reed, M. W. R., Pharoah, P. D. P., et al. (2004). Association of a common variant of the CASP8 gene with reduced risk of breast cancer. J. Natl. Cancer Inst. 96, 1866–1869. doi: 10.1093/jnci/dji001
Malireddi, R. K., Ippagunta, S., Lamkanfi, M., and Kanneganti, T. -D. (2010). Cutting edge: proteolytic inactivation of poly(ADP-ribose) polymerase 1 by the Nlpr3 and Nlrc4 inflammasomes. J. Immunol. 185, 3127–3130. doi: 10.4049/jimmunol.1001512
Malireddi, R. K. S., Gurung, P., Kesavardhana, S., Samir, P., Burton, A., Mummareddy, H., et al. (2020). Innate immune priming in the absence of TAK1 drives RIPK1 kinase activity-independent pyroptosis, apoptosis, necroptosis, and inflammatory disease. J. Exp. Med. 217:jem.20191644. doi: 10.1084/jem.20191644
Malireddi, R. K. S., Gurung, P., Mavuluri, J., Dasari, T. K., Klco, J. M., Chi, H., et al. (2018). TAK1 restricts spontaneous NLRP3 activation and cell death to control myeloid proliferation. J. Exp. Med. jem.20171922. doi: 10.1084/jem.20171922
Malireddi, R. K. S., Kesavardhana, S., and Kanneganti, T.-D. (2019). ZBP1 and TAK1: master regulators of NLRP3 inflammasome/pyroptosis, apoptosis, and necroptosis (PAN-optosis). Front. Cell. Infect. Microbiol. 9:406. doi: 10.3389/fcimb.2019.00406
Man, S. M., and Kanneganti, T.-D. (2015). Regulation of inflammasome activation. Immunol. Rev. 265, 6–21. doi: 10.1111/imr.12296
Mariathasan, S., Weiss, D. S., Newton, K., McBride, J., O'Rourke, K., Roose-Girma, M., et al. (2006). Cryopyrin activates the inflammasome in response to toxins and ATP. Nature 440, 228–232. doi: 10.1038/nature04515
Martinon, F., Burns, K., and Tschopp, J. (2002). The inflammasome: a molecular platform triggering activation of inflammatory caspases and processing of proIL-β. Mol. Cell 10, 417–426. doi: 10.1016/S1097-2765(02)00599-3
Matmati, M., Jacques, P., Maelfait, J., Verheugen, E., Kool, M., Sze, M., et al. (2011). A20 (TNFAIP3) deficiency in myeloid cells triggers erosive polyarthritis resembling rheumatoid arthritis. Nat. Genet. 43, 908–912. doi: 10.1038/ng.874
Michailidou, K., Lindström, S., Dennis, J., Beesley, J., Hui, S., Kar, S., et al. (2017). Association analysis identifies 65 new breast cancer risk loci. Nature 551, 92–94. doi: 10.1038/nature24284
Mompeán, M., Li, W., Li, J., Laage, S., Siemer, A. B., Bozkurt, G., et al. (2018). The structure of the necrosome RIPK1-RIPK3 core, a human hetero-amyloid signaling complex. Cell 173, 1244-1253.e10. doi: 10.1016/j.cell.2018.03.032
Muzio, M., Chinnaiyan, A. M., Kischkel, F. C., O'Rourke, K., Shevchenko, A., Ni, J., et al. (1996). FLICE, a novel FADD-homologous ICE/CED-3-like protease, is recruited to the CD95 (Fas/APO-1) death-inducing signaling complex. Cell 85, 817–827. doi: 10.1016/S0092-8674(00)81266-0
Nair, R. P., Duffin, K. C., Helms, C., Ding, J., Stuart, P. E., Goldgar, D., et al. (2009). genomewide scan reveals association of psoriasis with IL-23 and NF-κB pathways. Nat. Genet. 41, 199–204. doi: 10.1038/ng.311
Neven, B., Callebaut, I., Prieur, A.-M., Feldmann, J., Bodemer, C., Lepore, L., et al. (2004). Molecular basis of the spectral expression of CIAS1 mutations associated with phagocytic cell-mediated autoinflammatory disorders CINCA/NOMID, MWS, and FCU. Blood 103, 2809–2815. doi: 10.1182/blood-2003-07-2531
Newton, K., Wickliffe, K. E., Dugger, D. L., Maltzman, A., Roose-Girma, M., Dohse, M., et al. (2019a). Cleavage of RIPK1 by caspase-8 is crucial for limiting apoptosis and necroptosis. Nature 574, 428–431. doi: 10.1038/s41586-019-1548-x
Newton, K., Wickliffe, K. E., Maltzman, A., Dugger, D. L., Reja, R., Zhang, Y., et al. (2019b). Activity of caspase-8 determines plasticity between cell death pathways. Nature 575, 679–682. doi: 10.1038/s41586-019-1752-8
Newton, K., Wickliffe, K. E., Maltzman, A., Dugger, D. L., Strasser, A., Pham, V. C., et al. (2016). RIPK1 inhibits ZBP1-driven necroptosis during development. Nature 540, 129–133. doi: 10.1038/nature20559
Nilsson, J., Schoser, B., Laforet, P., Kalev, O., Lindberg, C., Romero, N. B., et al. (2013). Polyglucosan body myopathy caused by defective ubiquitin ligase RBCK1. Ann. Neurol. 74, 914–919. doi: 10.1002/ana.23963
Ohtsuka, T., Liu, X.-F., Koga, Y., Kitajima, Y., Nakafusa, Y., Ha, C.-W., et al. (2006). Methylation-induced silencing of ASC and the effect of expressed ASC on p53-mediated chemosensitivity in colorectal cancer. Oncogene 25, 1807–1811. doi: 10.1038/sj.onc.1209204
Opdenbosch, N. V., Gurung, P., Walle, L. V., Fossoul, A., Kanneganti, T.-D., and Lamkanfi, M. (2014). Activation of the NLRP1b inflammasome independently of ASC-mediated caspase-1 autoproteolysis and speck formation. Nat. Commun. 5:4209. doi: 10.1038/ncomms4209
Orning, P., Weng, D., Starheim, K., Ratner, D., Best, Z., Lee, B., et al. (2018). Pathogen blockade of TAK1 triggers caspase-8-dependent cleavage of gasdermin D and cell death. Science 362:1064–1069. doi: 10.1126/science.aau2818
Patankar, Y. R., Mabaera, R., and Berwin, B. (2015). Differential ASC requirements reveal a key role for neutrophils and a noncanonical IL-1β response to Pseudomonas aeruginosa. Am. J. Physiol. Lung Cell. Mol. Physiol. 309, L902–913. doi: 10.1152/ajplung.00228.2015
Peelen, E., Damoiseaux, J., Muris, A.-H., Knippenberg, S., Smolders, J., Hupperts, R., et al. (2015). Increased inflammasome related gene expression profile in PBMC may facilitate T helper 17 cell induction in multiple sclerosis. Mol. Immunol. 63, 521–529. doi: 10.1016/j.molimm.2014.10.008
Peltzer, N., Darding, M., Montinaro, A., Draber, P., Draberova, H., Kupka, S., et al. (2018). LUBAC is essential for embryogenesis by preventing cell death and enabling haematopoiesis. Nature 557, 112–117. doi: 10.1038/s41586-018-0064-8
Peltzer, N., Darding, M., and Walczak, H. (2016). Holding RIPK1 on the ubiquitin leash in TNFR1 signaling. Trends Cell Biol. 26, 445–461. doi: 10.1016/j.tcb.2016.01.006
Perkins, E. A., Landis, D., Causey, Z. L., Edberg, Y., Reynolds, R. J., Hughes, L. B., et al. (2012). Association of single-nucleotide polymorphisms in CCR6, TAGAP, and TNFAIP3 with rheumatoid arthritis in African Americans. Arthritis Rheum. 64, 1355–1358. doi: 10.1002/art.33464
Philip, N. H., Dillon, C. P., Snyder, A. G., Fitzgerald, P., Wynosky-Dolfi, M. A., Zwack, E. E., et al. (2014). Caspase-8 mediates caspase-1 processing and innate immune defense in response to bacterial blockade of NF-κB and MAPK signaling. Proc. Natl. Acad. Sci. U.S.A. 111, 7385–7390. doi: 10.1073/pnas.1403252111
Prahalad, S., Conneely, K. N., Jiang, Y., Sudman, M., Wallace, C. A., Brown, M. R., et al. (2013). Susceptibility to childhood-onset rheumatoid arthritis: investigation of a weighted genetic risk score that integrates cumulative effects of variants at five genetic loci. Arthritis Rheum. 65, 1663–1667. doi: 10.1002/art.37913
Rickard, J. A., Anderton, H., Etemadi, N., Nachbur, U., Darding, M., Peltzer, N., et al. (2014b). TNFR1-dependent cell death drives inflammation in Sharpin-deficient mice. ELife 3:e03464. doi: 10.7554/eLife.03464
Rickard, J. A., O'Donnell, J. A., Evans, J. M., Lalaoui, N., Poh, A. R., Rogers, T., et al. (2014a). RIPK1 regulates RIPK3-MLKL-driven systemic inflammation and emergency hematopoiesis. Cell 157, 1175–1188. doi: 10.1016/j.cell.2014.04.019
Riedl, S. J., and Salvesen, G. S. (2007). The apoptosome: signalling platform of cell death. Nat. Rev. Mol. Cell Biol. 8, 405–413. doi: 10.1038/nrm2153
Riojas, M. A., Guo, M., Glöckner, S. C., Machida, E. O., Baylin, S. B., and Ahuja, N. (2007). Methylation-induced silencing of ASC/TMS1, a pro-apoptotic gene, is a late-stage event in colorectal cancer. Cancer Biol. Ther. 6, 1710–1716. doi: 10.4161/cbt.6.11.4829
Rosell, R., Monzo, M., O'Brate, A., and Taron, M. (2002). Translational oncogenomics: toward rational therapeutic decision-making. Curr. Opin. Oncol. 14, 171–179. doi: 10.1097/00001622-200203000-00006
Rösen-Wolff, A., Quietzsch, J., Schröder, H., Lehmann, R., Gahr, M., and Roesler, J. (2003). Two German CINCA (NOMID) patients with different clinical severity and response to anti-inflammatory treatment. Eur. J. Haematol. 71, 215–219. doi: 10.1034/j.1600-0609.2003.00109.x
Rowczenio, D. M., Trojer, H., Russell, T., Baginska, A., Lane, T., Stewart, N. M., et al. (2013). Clinical characteristics in subjects with NLRP3 V198M diagnosed at a single UK center and a review of the literature. Arthritis Res. Ther. 15:R30. doi: 10.1186/ar4171
Roychowdhury, S., McMullen, M. R., Pisano, S. G., Liu, X., and Nagy, L. E. (2013). Absence of receptor interacting protein kinase 3 prevents ethanol-induced liver injury. Hepatol. Baltim. Md 57, 1773–1783. doi: 10.1002/hep.26200
Samir, P., and Kanneganti, T.-D. (2019). Hidden aspects of valency in immune system regulation. Trends Immunol. 40, 1082–1094. doi: 10.1016/j.it.2019.10.005
Samir, P., Kesavardhana, S., Patmore, D. M., Gingras, S., Malireddi, R. K. S., Karki, R., et al. (2019). DDX3X acts as a live-or-die checkpoint in stressed cells by regulating NLRP3 inflammasome. Nature 573, 590–594. doi: 10.1038/s41586-019-1551-2
Sarhan, J., Liu, B. C., Muendlein, H. I., Li, P., Nilson, R., Tang, A. Y., et al. (2018). Caspase-8 induces cleavage of gasdermin D to elicit pyroptosis during Yersinia infection. Proc. Natl. Acad. Sci. U.S.A. 115, E10888–E10897. doi: 10.1073/pnas.1809548115
Schoultz, I., Verma, D., Halfvarsson, J., Törkvist, L., Fredrikson, M., Sjöqvist, U., et al. (2009). Combined polymorphisms in genes encoding the inflammasome components NALP3 and CARD8 confer susceptibility to Crohn's disease in Swedish men. Am. J. Gastroenterol. 104, 1180–1188. doi: 10.1038/ajg.2009.29
Shen, N., Ruan, Y., Lu, Y., Jiang, X., Sun, H., Gao, G., et al. (2017). Three single nucleotide polymorphisms of TNFAIP3 gene increase the risk of rheumatoid arthritis. Oncotarget 8, 20784–20793. doi: 10.18632/oncotarget.15265
Shigemura, T., Kaneko, N., Kobayashi, N., Kobayashi, K., Takeuchi, Y., Nakano, N., et al. (2016). Novel heterozygous C243Y A20/TNFAIP3 gene mutation is responsible for chronic inflammation in autosomal-dominant Behçet's disease. RMD Open 2, e000223. doi: 10.1136/rmdopen-2015-000223
Sisto, M., Lisi, S., Lofrumento, D. D., Ingravallo, G., Maiorano, E., and D'Amore, M. (2011). A failure of TNFAIP3 negative regulation maintains sustained NF-κB activation in Sjögren's syndrome. Histochem. Cell Biol. 135, 615–625. doi: 10.1007/s00418-011-0821-3
Sonnhammer, E. L., Eddy, S. R., and Durbin, R. (1997). Pfam: a comprehensive database of protein domain families based on seed alignments. Proteins 28, 405–420. doi: 10.1002/(SICI)1097-0134(199707)28:3<405::AID-PROT10>3.0.CO;2-L
Soung, Y. H., Lee, J. W., Kim, S. Y., Sung, Y. J., Park, W. S., Nam, S. W., et al. (2005). Caspase-8 gene is frequently inactivated by the frameshift somatic mutation 1225_1226delTG in hepatocellular carcinomas. Oncogene 24, 141–147. doi: 10.1038/sj.onc.1208244
Stahl, E. A., Raychaudhuri, S., Remmers, E. F., Xie, G., Eyre, S., Thomson, B. P., et al. (2010). Genome-wide association study meta-analysis identifies seven new rheumatoid arthritis risk loci. Nat. Genet. 42, 508–514. doi: 10.1038/ng.582
Strange, A., Capon, F., Spencer, C. C., Knight, J., Weale, M. E., Allen, M. H., et al. (2010). Genome-wide association study identifies new psoriasis susceptibility loci and an interaction between HLA-C and ERAP1. Nat. Genet. 42, 985–990. doi: 10.1038/ng.694
Sutterwala, F. S., Mijares, L. A., Li, L., Ogura, Y., Kazmierczak, B. I., and Flavell, R. A. (2007). Immune recognition of pseudomonas aeruginosa mediated by the IPAF/NLRC4 inflammasome. J. Exp. Med. 204, 3235–3245. doi: 10.1084/jem.20071239
Tao, P., Sun, J., Wu, Z., Wang, S., Wang, J., Li, W., et al. (2020). A dominant autoinflammatory disease caused by non-cleavable variants of RIPK1. Nature 577, 109–114. doi: 10.1038/s41586-019-1830-y
Taraborrelli, L., Peltzer, N., Montinaro, A., Kupka, S., Rieser, E., Hartwig, T., et al. (2018). LUBAC prevents lethal dermatitis by inhibiting cell death induced by TNF, TRAIL and CD95L. Nat. Commun. 9, 1–12. doi: 10.1038/s41467-018-06155-8
Thapa, R. J., Ingram, J. P., Ragan, K. B., Nogusa, S., Boyd, D. F., Benitez, A. A., et al. (2016). DAI senses influenza a virus genomic RNA and activates RIPK3-dependent cell death. Cell Host Microbe 20, 674–681. doi: 10.1016/j.chom.2016.09.014
The UniProt Consortium (2015). UniProt: a hub for protein information. Nucleic Acids Res. 43, D204–D212. doi: 10.1093/nar/gku989
Thomas, P. G., Dash, P., Aldridge, J. R., Ellebedy, A. H., Reynolds, C., Funk, A. J., et al. (2009). The intracellular sensor NLRP3 mediates key innate and healing responses to influenza A virus via the regulation of caspase-1. Immunity 30, 566–575. doi: 10.1016/j.immuni.2009.02.006
Ting, A. T., and Bertrand, M. J. M. (2016). More to life than NF-κB in TNFR1 signaling. Trends Immunol. 37, 535–545. doi: 10.1016/j.it.2016.06.002
Tsoi, L. C., Spain, S. L., Knight, J., Ellinghaus, E., Stuart, P. E., Capon, F., et al. (2012). Identification of fifteen new psoriasis susceptibility loci highlights the role of innate immunity. Nat. Genet. 44, 1341–1348. doi: 10.1038/ng.2467
Vajjhala, P. R., Lu, A., Brown, D. L., Pang, S. W., Sagulenko, V., Sester, D. P., et al. (2015). The inflammasome adaptor ASC induces procaspase-8 death effector domain filaments. J. Biol. Chem. 290, 29217–29230. doi: 10.1074/jbc.M115.687731
Vande Walle, L., Van Opdenbosch, N., Jacques, P., Fossoul, A., Verheugen, E., Vogel, P., et al. (2014). Negative regulation of the NLRP3 inflammasome by A20 protects against arthritis. Nature 512, 69–73. doi: 10.1038/nature13322
Vanden Berghe, T., Hassannia, B., and Vandenabeele, P. (2016). An outline of necrosome triggers. Cell. Mol. Life Sci. 73, 2137–2152. doi: 10.1007/s00018-016-2189-y
Verma, D., Eriksson, P., Sahdo, B., Persson, A., Ejdebäck, M., Särndahl, E., et al. (2010). Two adult siblings with atypical cryopyrin-associated periodic syndrome due to a novel M299V mutation in NLRP3. Arthritis Rheum. 62, 2138–2143. doi: 10.1002/art.27489
Vince, J. E., Wong, W. W.-L., Gentle, I., Lawlor, K. E., Allam, R., O'Reilly, L., et al. (2012). Inhibitor of apoptosis proteins limit RIP3 kinase-dependent interleukin-1 activation. Immunity 36, 215–227. doi: 10.1016/j.immuni.2012.01.012
Virmani, A., Rathi, A., Sugio, K., Sathyanarayana, U. G., Toyooka, S., Kischel, F. C., et al. (2003). Aberrant methylation of TMS1 in small cell, non small cell lung cancer and breast cancer. Int. J. Cancer 106, 198–204. doi: 10.1002/ijc.11206
Wade, E. M., Daniel, P. B., Jenkins, Z. A., McInerney-Leo, A., Leo, P., Morgan, T., et al. (2016). Mutations in MAP3K7 that alter the activity of the TAK1 signaling complex cause frontometaphyseal dysplasia. Am. J. Hum. Genet. 99, 392–406. doi: 10.1016/j.ajhg.2016.05.024
Wallach, D., Kovalenko, A., and Kang, T.-B. (2011). 'Necrosome'-induced inflammation: must cells die for it? Trends Immunol. 32, 505–509. doi: 10.1016/j.it.2011.07.004
Wang, K., Kim, C., Bradfield, J., Guo, Y., Toskala, E., Otieno, F. G., et al. (2013). Whole-genome DNA/RNA sequencing identifies truncating mutations in RBCK1 in a novel Mendelian disease with neuromuscular and cardiac involvement. Genome Med. 5:67. doi: 10.1186/gm471
Wang, S., Ni, H.-M., Dorko, K., Kumer, S. C., Schmitt, T. M., Nawabi, A., et al. (2016). Increased hepatic receptor interacting protein kinase 3 expression due to impaired proteasomal functions contributes to alcohol-induced steatosis and liver injury. Oncotarget 7, 17681–17698. doi: 10.18632/oncotarget.6893
Weber, K., Roelandt, R., Bruggeman, I., Estornes, Y., and Vandenabeele, P. (2018). Nuclear RIPK3 and MLKL contribute to cytosolic necrosome formation and necroptosis. Commun. Biol. 1:6. doi: 10.1038/s42003-017-0007-1
Weng, D., Marty-Roix, R., Ganesan, S., Proulx, M. K., Vladimer, G. I., Kaiser, W. J., et al. (2014). Caspase-8 and RIP kinases regulate bacteria-induced innate immune responses and cell death. Proc. Natl. Acad. Sci. 111, 7391–7396. doi: 10.1073/pnas.1403477111
Yang, J. K. (2015). Death effecter domain for the assembly of death-inducing signaling complex. Apoptosis 20, 235–239. doi: 10.1007/s10495-014-1060-6
Yokoyama, T., Sagara, J., Guan, X., Masumoto, J., Takeoka, M., Komiyama, Y., et al. (2003). Methylation of ASC/TMS1, a proapoptotic gene responsible for activating procaspase-1, in human colorectal cancer. Cancer Lett. 202, 101–108. doi: 10.1016/j.canlet.2003.08.027
Zhang, C., Li, H., Zhou, G., Zhang, Q., Zhang, T., Li, J., et al. (2007). Transcriptional silencing of the TMS1/ASC tumour suppressor gene by an epigenetic mechanism in hepatocellular carcinoma cells. J. Pathol. 212, 134–142. doi: 10.1002/path.2173
Zhang, H., Wang, Z., Lu, X., Wang, Y., Zhong, J., and Liu, J. (2014). NLRP3 gene is associated with ulcerative colitis (UC), but not Crohn's Disease (CD), in Chinese han population. Inflamm. Res. Off. J. Eur. Histamine Res. Soc. Al 63, 979–985. doi: 10.1007/s00011-014-0774-9
Zhang, H., Zhou, X., McQuade, T., Li, J., Chan, F. K.-M., and Zhang, J. (2011). Functional complementation between FADD and RIP1 in embryos and lymphocytes. Nature 471, 373–376. doi: 10.1038/nature09878
Zhang, J., Webster, J. D., Dugger, D. L., Goncharov, T., Roose-Girma, M., Hung, J., et al. (2019). Ubiquitin ligases cIAP1 and cIAP2 limit cell death to prevent inflammation. Cell Rep. 27, 2679–2689.e3. doi: 10.1016/j.celrep.2019.04.111
Zhang, J., Yang, Y., He, W., and Sun, L. (2016). Necrosome core machinery: MLKL. Cell. Mol. Life Sci. 73, 2153–2163. doi: 10.1007/s00018-016-2190-5
Zhang, X., Li, W., Zhang, X., Zhao, L., Zhang, X., Jiang, L., et al. (2014). Single nucleotide polymorphisms in TNFAIP3 were associated with the risks of rheumatoid arthritis in northern Chinese Han population. BMC Med. Genet. 15:56. doi: 10.1186/1471-2350-15-56
Zhang, Z., Tan, S., and Zhang, L. (2006). Prognostic value of apoptosis-associated speck-like protein containing a CARD gene promoter methylation in resectable non-small-cell lung cancer. Clin. Lung Cancer 8, 62–65. doi: 10.3816/CLC.2006.n.035
Zheng, M., Karki, R., Vogel, P., and Kanneganti, T.-D. (2020). Caspase-6 is a key regulator of innate immunity, inflammasome activation, and host defense against influenza a virus. Cell. doi: 10.1016/j.cell.2020.03.040
Zhou, Q., Wang, H., Schwartz, D. M., Stoffels, M., Park, Y. H., Zhang, Y., et al. (2016). Loss-of-function mutations in TNFAIP3 leading to A20 haploinsufficiency cause an early-onset autoinflammatory disease. Nat. Genet. 48, 67–73. doi: 10.1038/ng.3459
Zmasek, C. M., and Godzik, A. (2013). Evolution of the animal apoptosis network. Cold Spring Harb. Perspect. Biol. 5, a008649. doi: 10.1101/cshperspect.a008649
Keywords: PANoptosome, PANoptosis, ASC, caspase-1, RIPK1, RIPK3, ZBP1, caspase-8
Citation: Samir P, Malireddi RKS and Kanneganti T-D (2020) The PANoptosome: A Deadly Protein Complex Driving Pyroptosis, Apoptosis, and Necroptosis (PANoptosis). Front. Cell. Infect. Microbiol. 10:238. doi: 10.3389/fcimb.2020.00238
Received: 02 April 2020; Accepted: 24 April 2020;
Published: 03 June 2020.
Edited by:
Yousef Abu Kwaik, University of Louisville, United StatesReviewed by:
Bodduluri Haribabu, University of Louisville, United StatesRama Vankayalapati, University of Texas at Tyler, United States
Copyright © 2020 Samir, Malireddi and Kanneganti. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Thirumala-Devi Kanneganti, dGhpcnVtYWxhLWRldmkua2FubmVnYW50aUBzdGp1ZGUub3Jn
 Parimal Samir
Parimal Samir R. K. Subbarao Malireddi
R. K. Subbarao Malireddi Thirumala-Devi Kanneganti
Thirumala-Devi Kanneganti