- 1Department of Endocrinology, The Affiliated Hospital of Qingdao University, Qingdao, China
- 2Department of Clinical Laboratory, The Affiliated Hospital of Qingdao University, Qingdao, China
Infectious diseases are the second most important cause of human death worldwide; Staphylococcus aureus (S. aureus) is a very common human pathogenic microorganism that can trigger a variety of infectious diseases, such as skin and soft tissue infections, endocarditis, osteomyelitis, bacteremia, and lethal pneumonia. Moreover, according to the sensitivity to antibiotic drugs, S. aureus can be divided into methicillin-sensitive Staphylococcus aureus (MSSA) and methicillin-resistant Staphylococcus aureus (MRSA). In recent decades, due to the evolution of bacteria and the abuse of antibiotics, the drug resistance of S. aureus has gradually increased, the infection rate of MRSA has increased worldwide, and the clinical anti-infective treatment for MRSA has become more difficult. Accumulating evidence has demonstrated that the resistance mechanisms of S. aureus are very complex, especially for MRSA, which is resistant to many kinds of antibiotics. Therefore, understanding the drug resistance of MRSA in a timely manner and elucidating its drug resistance mechanism at the molecular level are of great significance for the treatment of S. aureus infection. A large number of researchers believe that analyzing the molecular characteristics of S. aureus can help provide a basis for designing effective prevention and treatment measures against hospital infections caused by S. aureus and further monitor the evolution of S. aureus. This paper reviews the research status of MSSA and MRSA, the detailed mechanisms of the intrinsic antibiotic resistance and the acquired antibiotic resistance, the advanced research on anti-MRSA antibiotics and novel therapeutic strategies for MRSA treatment.
Introduction
Staphylococcus Aureus
Staphylococcus aureus (S. aureus) is one of main pathogens in hospital and community infections and can cause many infectious diseases, such as mild skin and soft tissue infections infective endocarditis, osteomyelitis, bacteremia, and fatal pneumonia (Lowy, 1998; Humphreys, 2012). Staphylococcus aureus was first discovered in 1880 in Aberdeen, Scotland, by surgeon Alexander Ogston from patients with ulcerated sores. Staphylococcus aureus belongs to the genus Staphylococcus, Firmicutes; is positive for Gram stain, ~0.8 μm in diameter, arranged in a “string of grapes” under a microscope, an aerobic or anaerobic; and grows optimally at 37°C, and at pH7.4 (Edwards and Massey, 2011; Gardete and Tomasz, 2014). The colonies on blood agar plate are thick, shiny, and round with a diameter of 1~2 mm (Gonzalez-Perez et al., 2019; Sato et al., 2019). Most of them are hemolytic, forming a transparent hemolytic ring around the colonies on blood agar plates (Sato et al., 2019). Moreover, S. aureus does not form spores or flagella, but possesses a capsule, can produce golden yellow pigment, and decompose mannitol (Tayeb-Fligelman et al., 2017). Additionally, it has also been found that tests of plasma coagulase, lactose fermentation and deoxyribonuclease are positive in S. aureus (Chino et al., 2017; Tayeb-Fligelman et al., 2017).
Methicillin-Resistant Staphylococcus Aureus (MRSA)
Fleming discovered penicillin in the 1940s and pioneered the era of antibiotics for infection treatment (Klevens et al., 2007; Klein et al., 2017). At the time, the infectious diseases caused by S. aureus were well-controlled, but with the widespread use of penicillin in the 1950s, penicillin-resistant S. aureus appeared in the clinic (Rayner and Munckhof, 2005; Pichereau and Rose, 2010). Penicillin-resistant S. aureus can produce penicillinase, which can hydrolyze the penicillin β-lactam ring, leading to resistance to penicillin. Later, scientists developed a new penicillinase-resistant semisynthetic penicillin named methicillin, which is resistant to the hydrolysis of β-lactamase (Rayner and Munckhof, 2005; Khoshnood et al., 2019). After being applied to the clinic in 1959, methicillin effectively controlled the infection of penicillin-resistant S. aureus (Chambers and Deleo, 2009; Jokinen et al., 2017). However, only 2 years after methicillin was applied, in 1961, British scientist Jevons reported the isolation of an MRSA strain; this resistance was produced by a gene encoding the penicillin-binding protein 2a or 2′ (PBP2a or PBP2′) (mecA) which was integrated into the chromosomal element (SCCmec) of methicillin-sensitive S. aureus (Schulte and Munson, 2019). Moreover, MRSA has rapidly become the most frequently occurring resistant pathogen identified in many parts of the world, including Europe, the United States, North Africa, the Middle East and East Asia (Mediavilla et al., 2012; Lakhundi and Zhang, 2018). According to its original source, MRSA is classified into hospital-acquired MRSA (HA-MRSA) and community-acquired MRSA (CA-MRSA) (Lindsay, 2013; Otto, 2013). In China, the proportion of hospital-acquired MRSA has reached 50.4% (Shang et al., 2016). Additionally, based on the Centers for Disease Control (CDC) in the US, the mortality rate of MRSA infection has exceeded that of acquired immune deficiency syndrome (AIDS), Parkinson's disease and murder (Lessa et al., 2012). Thus, the analysis of the molecular characteristics of S. aureus, which has become the focus of global public health concerns, can help us understand the prevalence of S. aureus, monitor the evolution of S. aureus, discover new molecular features of S. aureus, and provide information for developing novel drugs to against S. aureus (Figure 1A).
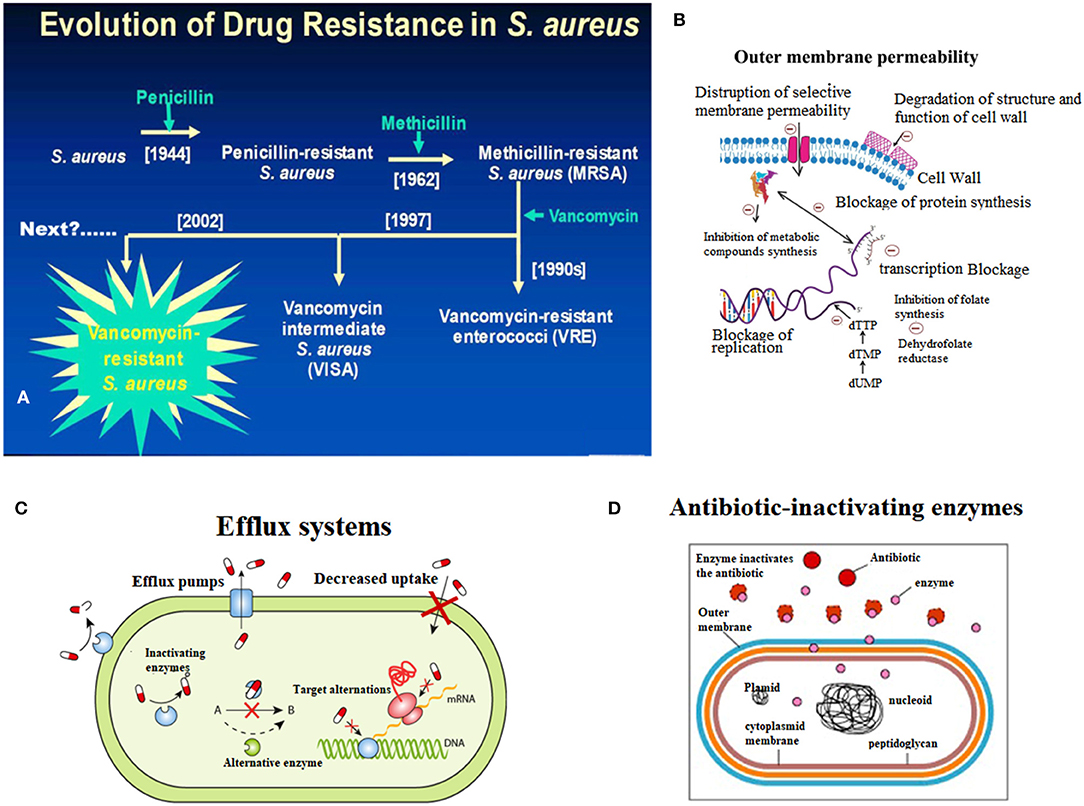
Figure 1. Endogenous resistance mechanism of Staphylococcus aureus. (A) Breif introduction of evolution of drug resistance in S. aureus. (B) The scheme of decreased outmembrane permeability caused drug resistance of S. aureus. (C) The role of active efflux systems in MRSA resistance. (D) The role of cellular enzymes in drug resistance of S. aureus.
Intrinsic Antibiotic Resistance
The resistance rates of S. aureus infection and multidrug resistant strains are increasing, making the clinical anti-infective treatment more difficult. The endogenous resistance mechanism mainly includes three aspects (Figure 1).
Outer Membrane Permeability
When the cell membrane permeability is lowered, the energy metabolism of the bacteria is affected, and therefore, drug absorption is reduced, leading to drug resistance (Li et al., 2013; Anuj et al., 2019). For example, the resistance of S. aureus to aminoglycosides is caused by a decrease in membrane permeability and finally results in a decrease in drug intake (Figure 1B) (Hori and Hiramatsu, 1994; Andrade et al., 2014).
Efflux Systems
The active efflux system of bacteria was discovered in 1980 by Ball and McMurry when studying the resistance of Escherichia coli to tetracycline (Seifi and Khoshbakht, 2016). Afterwards, the scholars conducted many experiments on the active efflux system, which confirmed that the active efflux system is a normal physiological structure of bacteria, and exists in sensitive strains (Lekshmi et al., 2018). When induced by substrates in the environment for a long time, efflux system-encoding genes are activated and expressed, and the ability to efflux drugs is greatly enhanced, thus leading to drug resistance (Zarate et al., 2019). Active drug efflux systems play a role in resistance to multiple drugs (Costa et al., 2018). There are three types of multidrug-pumping proteins present on the Staphylococcus aureus cell membrane: QacA, NorA, and Smr (Foster, 2016; Jang, 2016). Noguchi et al. considered QacA to be an important factor in MRSA (Noguchi et al., 2005; Nakaminami et al., 2019). Multidrug pumping proteins are all proton kinesins (Foster, 2016). That is, instead of relying on ATP hydrolysis to release energy, material exchange is performed by an electrochemical gradient formed by H+ on both sides of the cell membrane (Matano et al., 2017). Usually, it is a reversible process, that is, H+ moves from extracellular to intracellular, while intracellular harmful substances such as dyes and antibacterial drugs flow from the inside of the cell to the outside (Lowy, 1998). Experiments by Kristiansen et al. also demonstrated the role of active efflux systems in MRSA resistance (Mo et al., 2007) (Figure 1C).
Excessive Production of β-Lactamase
β-lactamase is an enzyme that catalyzes the hydrolysis of various β-lactam antibiotics (including antibiotics such as carbapenem broad-spectrum antibiotics), is encoded by bacterial chromosomal genes, and is transferable (Lee and Park, 2016). At present, the research shows that β-lactam antibiotics have a lethal effect on bacteria mainly through two mechanisms: first, by binding to penicillin-binding protein (PBPs, i.e., cell wall mucin synthase), which represses cell wall mucin synthesis, disrupts the cell wall, and leads to bacterial expansion and lysis; second, by triggering the autolytic enzyme activity of the bacteria, which resulted in autolysis and death (Matono et al., 2018). Excessive secretion of β-lactamase by MRSA mainly reduces the effect of antibiotics through two mechanisms, which lead to MRSA resistant (Khan et al., 2014). The first is the hydrolysis mechanism, that is, β-lactamase hydrolzes and inactivates β-lactam antibiotics; the second is the mechanism of pinching, that is, a large amount of β-lactamase binds quickly and firmly to extracellular antibiotics, preventing the antibiotics from reaching the intracellular space and therefore the antibiotics are not able to reach the target site, ultimately leading to MRSA resistance to antibiotics (Figure 1D) (Harada et al., 2014; Hashizume et al., 2017).
Acquired Antibiotic Resistance
Resistance by Mutations
Staphylococcus aureus can become drug-resistant by genetic mutations that alter the target DNA gyrase or reduce outer membrane proteins, thereby reducing drug accumulation (Kime et al., 2019; Yang et al., 2019). For example, the principle of resistance to clindamycin and erythromycin is caused by a modification in ribosomal RNA methylase (Heelan et al., 2004; Martinez et al., 2018).
Acquisition of Resistance Genes
Acquired resistance is a type of plasmid-mediated resistance (Foster, 2017). Through plasmid-mediated transduction, transformation, and insertion of drug-resistant genes, excessive β-lactamase can be produced, leading to bacteria resistance (Foster, 2017; Haaber et al., 2017). The mechanism of MRSA resistance is mainly because plasmids, or drug-resistant gene transmission mediated by plasmids, which can expand the genome and resistance genes can be transferred between S. aureus and other bacteria (Vestergaard et al., 2019). For example, MRSA can obtain drug-resistant plasmids from Enterococcus, further expanding and enhancing its resistance (Lazaris et al., 2017).
Biofilm-Mediated Resistance
Bacterial biofilm is an extracellular complex structure composed of a microbial population attached to the surface of the substrate, and its internal microorganisms are surrounded by a highly hydrated extracellular polymer matrix produced by itself, which is a protective way of survival for bacteria to adapt to their surroundings (Craft et al., 2019; Saxena et al., 2019). Moreover, the vast majority of bacteria in nature exist in the form of biofilms, and the most prominent feature of bacterial biofilms is their strong adhesion and drug resistance, which allowing bacteria to resist host immune responses and evade antibiotic killing (Kanwar et al., 2017). Their resistance to antibacterial drugs can increase to 1,000 times that of plankton. At present, domestic and foreign anti-biofilm treatments mainly focus on the continuous development of new antibacterial drugs, but antibiotics and chemical synthetic drugs used in clinical medicine have certain toxic effects (Saxena et al., 2019). Biofilm bacteria are prone to resistance to these conventional drugs, and resistant strains presented an increasing trend (Craft et al., 2019). Studies have shown that the combined application of traditional Chinese medicine and antibiotics has the advantage of reducing efficiency (Kanwar et al., 2017).
Persister Cells in Antibiotic Resistance
Persister cells are a small subset of cells that are genetically homologous but phenotypically heterogeneous in a microbial population, grow slowly, or dormant and survive high concentrations of antibiotics (Fisher et al., 2017). Early studies suggest that, unlike antibiotic resistance, bacterial retention is a physiological state of bacteria that temporarily resists antibiotic stress, and does not result in a change in the genotype (Kester and Fortune, 2014). However, this statement is being challenged because of the rapid development of high-throughput sequencing technology. When bacteria encounter external stimuli such as antibiotics, most of the bacteria are killed immediately, but a small percentage of the bacteria will resist this pressure by arresting growth and remaining inactive (Michiels et al., 2016). When the external pressure disappears, this small amount of bacteria can return to normal growth; we call these bacteria persister cell (Fisher et al., 2017). The presence of persister cells poses many obstacles to the complete elimination of bacterial infections and the prevention of recurrent infections (Fisher et al., 2017; Foster, 2017). Bacterial persister cells exhibit antibiotic tolerance, slow growth, and the ability to restart infection after antibiotic treatment. Antibiotics can cause fatal damage to bacteria, but persister cells can resist this killing by reducing cell growth and metabolism, and even by becoming dormant (Lewis, 2008). Bacterial persistence refers to a state of reduced metabolic activity that endows a subpopulation of isogenic bacteria with multidrug tolerance (Fisher et al., 2017). Persisters are phenotypic variants but not mutants (Kester and Fortune, 2014). Existing research results showed that the mechanism of bacterial persistence is complex and the related signaling pathways include toxins-antitoxin systems, cell physiological reduction of energy metabolism and protein and nucleic acid synthesis, DNA protection and repair systems, protease systems, trans-translation, external pumping system, and so on (Michiels et al., 2016; Fisher et al., 2017).
Research Advances in Commonly Used Antibiotics
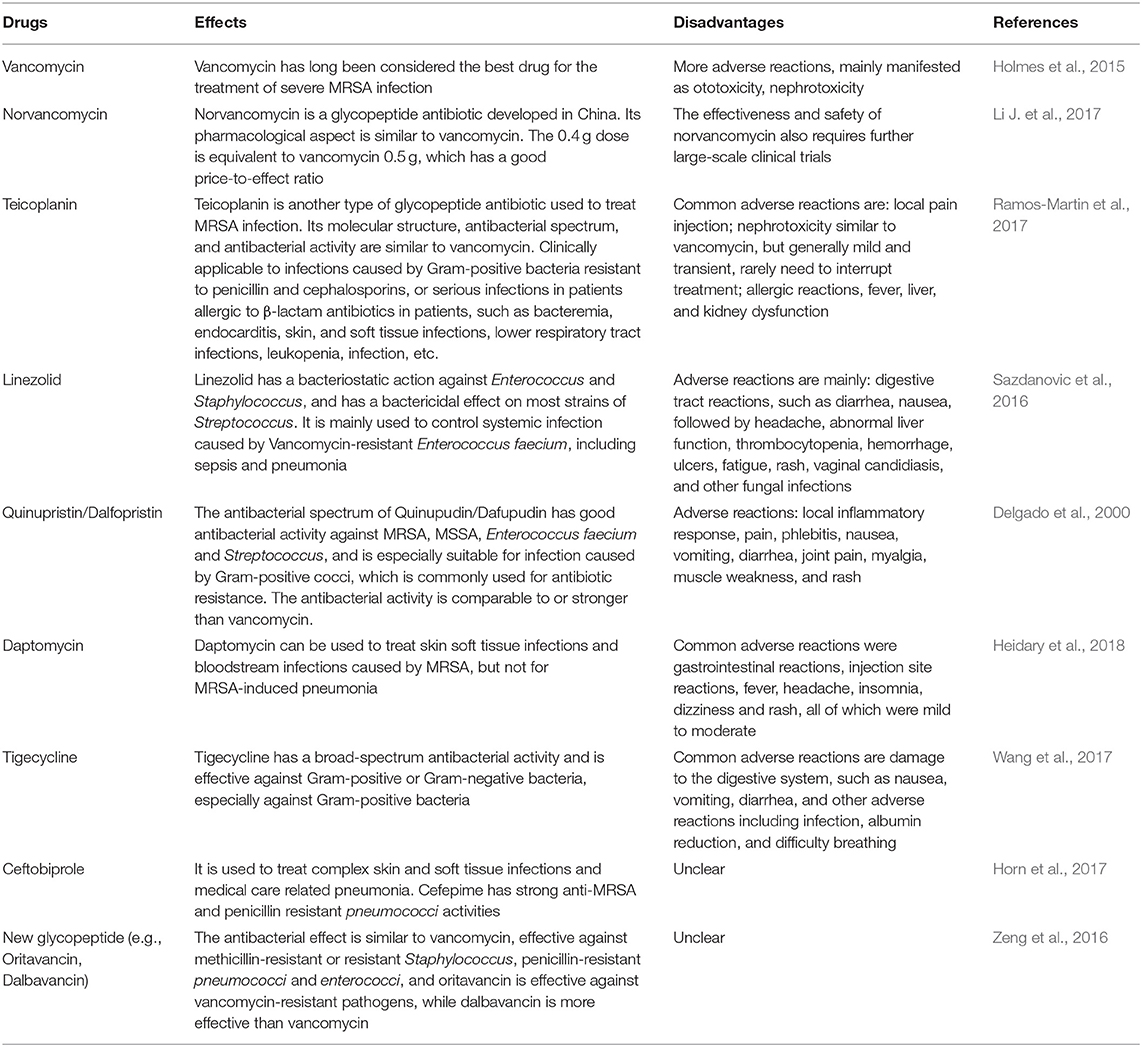
MRSA is a kind of multidrug resistant “super bacteria” resistant to penicillins, cephalosporins, chloramphenicol, lincomycin, aminoglycosides, tetracyclines, macrolides, quinophthalones, sulfonamides and rifampicin, which is a very difficult problem in clinical treatment (2016). Moreover, it has been reported that MRSA infection is one of the world's major infectious diseases due to its high rates of morbidity and mortality, which seriously threaten human health and have attracted the attention of the global medical community (Hassoun et al., 2017). Therefore, it is urgent to find effective drugs to treat multidrug resistant bacterial infections. We have listed some of the drugs used against anti-MRSA infections, that have been applied to in the clinic in Table 1, and three of them (i.e.,: vancomycin, daptomycin, and linezolid) have been highlighted.
Vancomycin
Vancomycin has long been considered the best drug for the treatment of severe MRSA infection, including both HA-MRSA and CA-MRSA which can cause serious, invasive infections such as pneumonia and sepsis (Holmes et al., 2015). Vancomycin has been known as the last line of defense line of against gram-positive cocci infection (Micek, 2007). It was found that the resistance mechanism of vancomycin is mainly specific binding of vancomycinto the bacterial cell wall via peptidoglycan precursor small peptides, which are terminated with D-alanyl-D-alanine; this binding inhibits the elongation and cross-linking of bacterial cell wall peptidoglycans, thereby repressing cell wall synthesis and ultimately leading to bacterial death (Micek, 2007; Haseeb et al., 2019). However, the resistance of Staphylococcus aureus to vancomycin is increasing daily, causing widespread concern in the medical community (Haseeb et al., 2019). Currently, a large number of researchers generally divide vancomycin-resistant Staphylococcus aureus into three types: vancomycin-resistant Staphylococcus aureus (VRSA), vancomycin-intermediate Staphylococcus aureus (VISA) and heterologous vancomycin resistant Staphylococcus aureus (hetero-VRSA) (Amberpet et al., 2019). VRSA refers to a minimal inhibitory concentration (MIC) of clinically isolated Staphylococcus aureus to vancomycin <32 mg/L, and was first reported in the United States in 2002. VISA means that the MIC of Staphylococcus aureus to vancomycin is 8–16 mg/L, the first strain was isolated in 1997 in Japan and has attracted the attention of medical community (Baseri et al., 2018). Subsequently, the United States, China and other places have successively discovered multiple VISA (Howden et al., 2010). Hetero-VRSA refers to the primary culture of Staphylococcus aureus isolated from clinical specimens. VRSA can be detected by the MH microbroth dilution method or agar dilution method and the MIC of vancomycin is ≤4 mg/L (Severin et al., 2004).
Daptomycin
Daptomycin is a cyclized lipopeptide drug that is extracted from the fermentation broth of Streptomyces roseosporus (Heidary et al., 2018). Its mechanism of action is to destroy the electric potential of the plasma membranes in the presence of calcium ions, but daptomycin does not inhibit the lipoteichoic acid (Taylor and Palmer, 2016). Due to its unique mechanism of action, daptomycin has no cross-resistance with other antibiotics and can be used to treat skin soft tissue infections and bloodstream infections caused by MRSA, but not MRSA-induced pneumonia because its activity can be suppressed by alveolar surfactant (Gomez Casanova et al., 2017). A large amount of evidence has reported that daptomycin has a faster bactericidal effect than vancomycin, linezolid or quinupristin/dalofopine (Stefani et al., 2015; Gomez Casanova et al., 2017). Furthermore, daptomycin has the effect of resisting most clinical gram-positive bacteria in vitro; therefore, daptomycin is mainly applied to treat infections of many drug-resistant bacteria, such as vancomycin-resistant enterococci, MRSA, glycopeptide-sensitive Staphylococcus aureus, coagulase-negative Staphylococci, and penicillin-resistant Streptococcus pneumoniae (Chuang et al., 2016). The United States has approved intravenous injections of daptomycin for the treatment of complex skin and soft tissue infections (Heidary et al., 2018). For the dosage form, daptomycin is currently only available in the form of an injection, and its oral dosage form is under study (Mediavilla et al., 2012).
Linezolid
Linezolid is a synthetic, new class of oxazolidinone antibacterial agents that inhibits enterococcistaphylococci, and most strains of Streptococcus (Sazdanovic et al., 2016). It is mainly used to control systemic infections caused by vancomycin-resistant Enterobacter faecium, such as sepsis, and pneumonia (Krueger and Unertl, 2002). Linezolid can bind to the 23S site of ribosomal RNA on the 50S subunit in bacteria inhibiting the 50S and 30S ribosomal subunits and preventing the formation of the 70S initiation complex, thereby interfering with protein synthesis (Livermore, 2003). This unique mechanism of action eliminates cross-resistance between linezolid and other antibiotics (Livermore, 2003; Hashemian et al., 2018). It has been reported that the survival rate and clinical cure rate of patients with MRSA infection treated with linezolid were significantly higher than those treated with vancomycin. Based on large-scale clinical studies, the oral and injection dosage forms of linezolid are equally effective in the treatment of MRSA, and are also effective against infections such as vancomycin-resistant enterococci, penicillin-resistant pneumococcal, and macrolide-resistant bacteriostatic streptococci (Hashemian et al., 2018). In view of the good therapeutic effect of the drug on multidrug resistant bacteria, it has been used clinically after being approved by the US Food and Drug Administration (FDA) in 2000 (Tyson et al., 2018). In 2007, linezolid also entered the Chinese market. Because it has a strong antibacterial effect on most gram-positive bacteria, it is considered to be an important choice for the treatment of MRSA (Hashemian et al., 2018).
Novel Therapeutic Strategies for MRSA Treatment
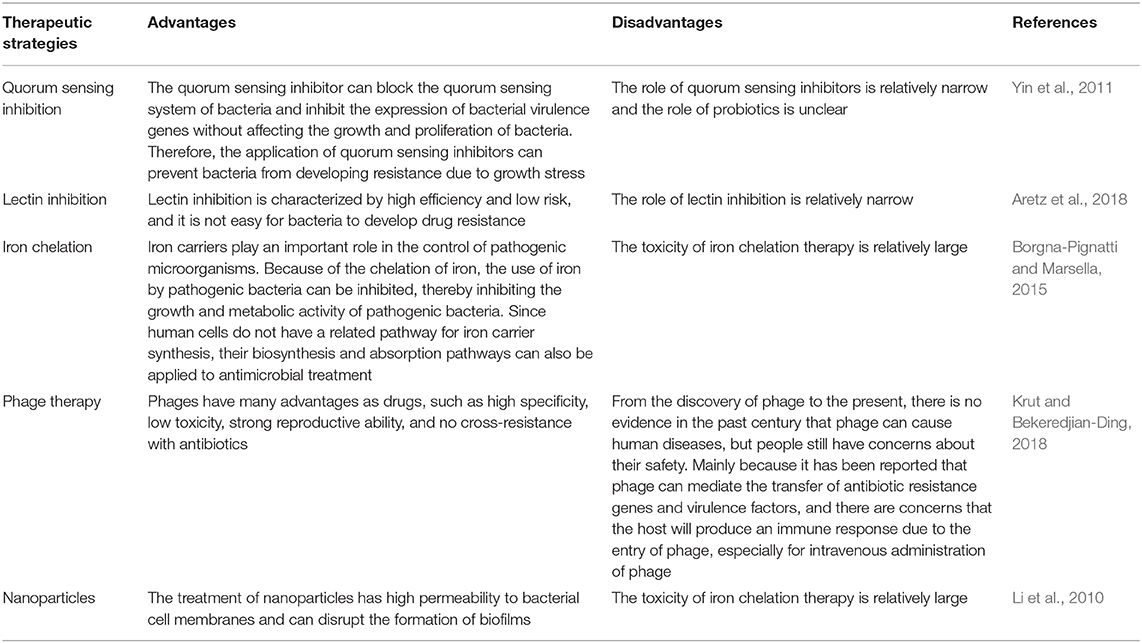
MRSA is multidrug resistant, not only resistant to β-lactam antibiotics, but also resistant to antimicrobial agents such as aminoglycosides, quinolones, and macrolides (Vestergaard et al., 2019). The mortality rate of systemic infection is more than 50%, which has become a worldwide problem in clinical and community anti-infective treatment, and it is difficult to treat (Lindsay, 2013; Vestergaard et al., 2019). At the same time, the hospital ICU is the main site for MRSA, which is likely to cause an outbreak (Lindsay, 2013). Therefore, many new drugs against MRSA are urgently needed. Table 2 lists the newly studied anti-MRSA drugs in recent years.
Quorum Sensing Inhibition
Quorum sensing is a phenomenon in which bacterial cells regulate the behavior of bacterial populations by sensing self-inducers. Bacteria secrete signal molecules called auto-inducing substances (Perez-Perez et al., 2017). When the extracellular concentration of these substances increases to a certain threshold with the concentration of the population, the bacteria turn on the expression of specific genes, thereby regulating the group behavior of the bacteria (Yin et al., 2011). This is an effective means of information exchange between bacteria, including bioluminescence, biofilm, and toxic gene expression and many other behaviors are regulated by the quorum sensing system (Kalia and Purohit, 2011). The microbial pathogenic properties of S. aureus are very complex, and are mainly related to virulence factors (Yin et al., 2011). These virulence factors are chiefly exotoxins that disrupt host cells, interfere with immune responses, and some proteins involved in adhesion and defense against host defenses (Kalia and Purohit, 2011; Yin et al., 2011). The expression of virulence factors is regulated by a complex network composed of multiple genes, of which agr a global regulatory factor H1, is the most important gene regulated by quorum sensing mechanisms (Haseeb et al., 2019). However, the inhibition of this quorum sensing mechanism in bacteria could result in the obstruction of biofilm formation, reduction in bacterial virulence and decreased bacterial resistance (Wang et al., 2017; Haseeb et al., 2019).
Lectin Inhibition
Lectin is a non-immune-derived sugar-binding protein that enables cell agglutination or precipitation of glycoconjugates (Aretz et al., 2018). It has been reported that lectins can not only agglutinate red blood cells, but also agglutinate with a variety of cells, such as pathogens, immune cells, and germ cells (Aretz et al., 2018; Alghadban et al., 2019). Presently, the application of lectin in the medical field is mainly the specific recognition and adhesion of lectins, which allows various pathogenic microorganisms to bind and infect their recipient cells (Alghadban et al., 2019). For example, certain mannose lectins can significantly affect the toxicity of HIV, which enables the development of antiviral drugs (Barre et al., 2019). Therefore, it is possible to use the characteristics of lectin to design and develop new clinical drugs, and fundamentally prevent the binding of pathogenic microorganisms to recipient cells, thus preventing most infectious diseases (Aretz et al., 2018).
Iron Chelation
Iron ions are essential nutrients for most organisms, including bacteria (Carver, 2018). Studies have shown that iron ions constitute the catalytic center of important biological enzymes such as oxidoreductase, and participate in various life activities such as electron transport, antioxidant reactions, and nucleic acid synthesis (Nuti et al., 2017). The antibiotic resistance of pathogenic bacteria is continuously increasing; therefore new antibacterial drugs urgently needed (Borgna-Pignatti and Marsella, 2015). One of the important mechanisms of bacterial resistance is to reduce the permeability of the outer membrane and thus hinder the entry of drug molecules into the cells (Carver, 2018). To circumvent the drug resistance mediated by this mechanism, one method is to attach the antibiotic molecule to an iron carrier, forming an iron carrier-antibiotic conjugate, and this iron carrier-antibiotic conjugate can selectively interact with the surface of the bacterial cell membrane (Rayner and Munckhof, 2005). The iron carrier outer membrane receptors interacts with this conjugate; the conjugate then crosses the outer cell membrane by active transport through an iron ion transport system (Bogdan et al., 2016). In this case, the iron carrier bound to the antibiotic can be bound to Fe3+, and the resulting complex (antibiotic-iron carrier-Fe3+) enters the cell. Finally, the drug is released inside the cell, thereby exerting antibacterial action (Bogdan et al., 2016).
Phage Therapy
At the beginning of their discovery, phage were used by the former Soviet Union and Eastern European medical communities to treat bacterial infections (Cisek et al., 2017). However, with the introduction of the antibiotic era, people gradually neglected in-depth research on phages. In recent years, due to the increasing global infection rate of drug-resistant bacteria, the use of antibiotics to treat bacterial infections has faced unprecedented challenges (Lin et al., 2017). The emergence of a series of drug-resistant pathogens such as Staphylococcus aureus, Pseudomonas aeruginosa, Acinetobacter baumannii, tuberculosis, Enterococcus faecalis, and especially MRSA, has led some scientists and clinicians to refocus their attention on phage research, leading to great progress in this area (Lin et al., 2017; Shlezinger et al., 2017). A large number of experiments have proven that phages can effectively improve the survival rate of animals infected with bacteria (Shlezinger et al., 2017). Compared with antibiotics, phage preparations have the advantages of high specificity, fast self-proliferation, and short development time (Krut and Bekeredjian-Ding, 2018). Phage therapy is considered to be one of the most promising therapies against human pathogens, including antibiotic resistant strains (Shlezinger et al., 2017). As early as 1921, phage were used to treat skin infections caused by staphylococci (Wang et al., 2017). In 2007, Italian researchers demonstrated that the phage Msa can effectively control lethal infections caused by S. aureus by establishing an intravenous injection model of in mice (Delgado et al., 2000). With the increase of drug-resistant bacteria, the advantages of phages have been recognized by more scholars. However, the biological characteristics of S. aureus phages and related animal studies over the years show that there are many limitations in the preparation, storage, and conditions of phages (Lin et al., 2017). Similar to antibiotics, bacteria may also be resistant to phages. However, the diversity and variability of phages in nature also provide an inexhaustible resource pool for phage-controlled bacteria. In addition, phage lysing enzymes that have a destructive effect on the basic framework of bacteria can make up for the lack of phage resistance (Shlezinger et al., 2017). Moreover, at present, phage therapy is still immature in clinical application. The main problems are as follows (Lin et al., 2017; Shlezinger et al., 2017): (1) most phages are highly specific and can only kill one or several subgroups of bacteria; (2) phage therapy in specific in vitro test presents effective, but it does not mean that it is equally effective in vivo; (3) Phages only begin to proliferate when the bacteria reach a certain density. Phages may be inoculated prematurely or at inappropriate doses, and may be eliminated by the body before they begin to proliferate. Therefore, determining the optimal inoculation time and dose will become a major difficulty in phage treatment. The above are common problems in phage therapy. Therefore, these problems also exist in the course of Staphylococcus aureus treatment.
Nanoparticles
Nanotechnology refers to the preparation, research and industrialization of substances at the nanoscale, as well as comprehensive technical systems for cross-research and industrialization using nanoscale materials (Wu et al., 2020). Studies have shown that nanotechnology can be applied in the fields of medicine, medicine, biology, chemistry, and information technology; therefore, it can play an important role in non-invasive minimally invasive medicine (Barbero et al., 2017). In the medical field, nanoparticles enhance the ability to deliver a drugin the human body (Li et al., 2010). After several layers of nanoparticle-encapsulated smart drugs enter the human body, they can actively search for and attack cancer cells or repair damaged tissues (Wang et al., 2020). China has successfully developed a new generation of nanoscale antibacterial drugs. The powdered nanoparticles are only 25 nanometers in diameter and have strong inhibition and killing effects on pathogenic microorganisms such as Escherichia coli and S. aureus (Li M. et al., 2017). Nanoscale antibacterial drugs have many properties such as spectrum, hydrophilicity and environmental protection, and they does not produce resistance due to the use of natural minerals (Howden et al., 2010).
Concluding Remark
Staphylococcus aureus is a bacterium that is ubiquitous in the environment and is common in the human body on the surface of the skin and in the upper respiratory tract mucosa (Lowy, 1998). Approximately 20% of human population are a long-term carriers of S. aureus, and most people do not show clinical symptoms; however S. aureus is still an important pathogen of humans (Humphreys, 2012). S. aureus can cause infections in hospitals and communities, and has become the leading pathogen in hospitals worldwide (Edwards and Massey, 2011). In the 1840s, penicillin was discovered by the British bacteriologist Fleming and was used in the clinic to control the S. aureus infection (Gardete and Tomasz, 2014). Later, various antibacterial drugs continued to emerge. However, this major research has also laid a hidden danger for human society. The widespread use of antibiotics has led to an increasing incidence of bacterial resistance, beginning with the emergence of multidrug resistant strains such as MRSA, which has been regarded as clinically important problem and has also attracted extensive attention from domestic and foreign research experts (Pichereau and Rose, 2010; Klein et al., 2017). Although the mortality rate of MRSA infection has declined in some European countries in recent years, MRSA is still a serious public health challenge worldwide (Rayner and Munckhof, 2005). Due to its characteristics of easy infection, high mortality and multidrug resistance, MRSA has become a stumbling block in clinical treatment (Khoshnood et al., 2019). Hence, how to effectively prevent and control MRSA has become a hot topic in modern research. Over the years, science and technology have progressed, and medicine has continued to develop. Humans have made outstanding achievements in the research of pathogenic factors of MRSA. Presently, vancomycin is likely still the best drug for curing MRSA infection (Micek, 2007). However, the multidrug resistance of MRSA has greatly increased the difficulty of human research (Vestergaard et al., 2019). Further research is needed to continuously study the ability of MRSA to cause infection and the antibiotic resistance pathways of MRSA, and to promote the development of new drugs against MRSA infection. The development of new drugs has given doctors more options to treat MRSA infections, providing greater protection to human health. However, the efficacy and safety of drugs require further clinical research.
Author Contributions
YW designed the concept. YG and GS drafted the manuscript. MS collected and analyzed the literature. JW edited the manuscript and revised the language.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
(2016). 36th International Symposium on Intensive Care and Emergency Medicine: Brussels, Belgium. Crit Care 20(Suppl 2):94. doi: 10.1186/s13054-016-1208-6
Alghadban, S., Kenawy, H. I., Dudler, T., Schwaeble, W. J., and Brunskill, N. J. (2019). Absence of the lectin activation pathway of complement ameliorates proteinuria-induced renal injury. Front. Immunol. 10:2238. doi: 10.3389/fimmu.2019.02238
Amberpet, R., Sistla, S., Sugumar, M., Nagasundaram, N., Manoharan, M., and Parija, S. C. (2019). Detection of heterogeneous vancomycin-intermediate Staphylococcus aureus: a preliminary report from south India. Indian J. Med. Res. 150, 194–198. doi: 10.4103/ijmr.IJMR_1976_17
Andrade, J. C., Morais-Braga, M. F., Guedes, G. M., Tintino, S. R., Freitas, M. A., Menezes, I. R., et al. (2014). Enhancement of the antibiotic activity of aminoglycosides by alpha-tocopherol and other cholesterol derivates. Biomed. Pharmacother. 68, 1065–1069. doi: 10.1016/j.biopha.2014.10.011
Anuj, S. A., Gajera, H. P., Hirpara, D. G., and Golakiya, B. A. (2019). Interruption in membrane permeability of drug-resistant Staphylococcus aureus with cationic particles of nanosilver. Eur. J. Pharm. Sci. 127, 208–216. doi: 10.1016/j.ejps.2018.11.005
Aretz, J., Anumala, U. R., Fuchsberger, F. F., Molavi, N., Ziebart, N., Zhang, H., et al. (2018). Allosteric inhibition of a mammalian lectin. J. Am. Chem. Soc. 140, 14915–14925. doi: 10.1021/jacs.8b08644
Barbero, F., Russo, L., Vitali, M., Piella, J., Salvo, I., Borrajo, M. L., et al. (2017). Formation of the protein corona: the interface between nanoparticles and the immune system. Semin Immunol. 34, 52–60. doi: 10.1016/j.smim.2017.10.001
Barre, A., Simplicien, M., Benoist, H., Van Damme, E. J. M., and Rouge, P. (2019). Mannose-specific lectins from marine algae: diverse structural scaffolds associated to common virucidal and anti-cancer properties. Mar. Drugs 17:440. doi: 10.3390/md17080440
Baseri, N., Najar-Peerayeh, S., and Bagheri Amiri, F. (2018). Prevalence of vancomycin-intermediate Staphylococcus aureus among clinical isolates in Iran: a systematic review and meta-analysis. J. Glob. Antimicrob. Resist. 15, 178–187. doi: 10.1016/j.jgar.2018.06.018
Bogdan, A. R., Miyazawa, M., Hashimoto, K., and Tsuji, Y. (2016). Regulators of iron homeostasis: new players in metabolism, cell death, and disease. Trends Biochem. Sci. 41, 274–286. doi: 10.1016/j.tibs.2015.11.012
Borgna-Pignatti, C., and Marsella, M. (2015). Iron chelation in thalassemia major. Clin. Ther. 37, 2866–2877. doi: 10.1016/j.clinthera.2015.10.001
Carver, P. L. (2018). The battle for iron between humans and microbes. Curr. Med. Chem. 25, 85–96. doi: 10.2174/0929867324666170720110049
Chambers, H. F., and Deleo, F. R. (2009). Waves of resistance: Staphylococcus aureus in the antibiotic era. Nat. Rev. Microbiol. 7, 629–641. doi: 10.1038/nrmicro2200
Chino, T., Nukui, Y., Morishita, Y., and Moriya, K. (2017). Morphological bactericidal fast-acting effects of peracetic acid, a high-level disinfectant, against Staphylococcus aureus and Pseudomonas aeruginosa biofilms in tubing. Antimicrob. Resist. Infect. Control 6:122. doi: 10.1186/s13756-017-0281-1
Chuang, Y. C., Lin, H. Y., Chen, P. Y., Lin, C. Y., Wang, J. T., and Chang, S. C. (2016). Daptomycin versus linezolid for the treatment of vancomycin-resistant enterococcal bacteraemia: implications of daptomycin dose. Clin. Microbiol. Infect. 22, 890.e1–890.e7 doi: 10.1016/j.cmi.2016.07.018
Cisek, A. A., Dabrowska, I., Gregorczyk, K. P., and Wyzewski, Z. (2017). Phage therapy in bacterial infections treatment: one hundred years after the discovery of bacteriophages. Curr. Microbiol. 74, 277–283. doi: 10.1007/s00284-016-1166-x
Costa, S. S., Sobkowiak, B., Parreira, R., Edgeworth, J. D., Viveiros, M., Clark, T. G., et al. (2018). Genetic diversity of norA, coding for a main efflux pump of Staphylococcus aureus. Front. Genet. 9:710. doi: 10.3389/fgene.2018.00710
Craft, K. M., Nguyen, J. M., Berg, L. J., and Townsend, S. D. (2019). Methicillin-resistant Staphylococcus aureus (MRSA): antibiotic-resistance and the biofilm phenotype. Medchemcomm 10, 1231–1241. doi: 10.1039/C9MD00044E
Delgado, G. Jr., Neuhauser, M. M., Bearden, D. T., and Danziger, L. H. (2000). Quinupristin-dalfopristin: an overview. Pharmacotherapy 20, 1469–1485. doi: 10.1592/phco.20.19.1469.34858
Edwards, A. M., and Massey, R. C. (2011). How does Staphylococcus aureus escape the bloodstream? Trends Microbiol. 19, 184–190 doi: 10.1016/j.tim.2010.12.005
Fisher, R. A., Gollan, B., and Helaine, S. (2017). Persistent bacterial infections and persister cells. Nat. Rev. Microbiol. 15, 453–464. doi: 10.1038/nrmicro.2017.42
Foster, T. J. (2016). The remarkably multifunctional fibronectin binding proteins of Staphylococcus aureus. Eur. J. Clin. Microbiol. Infect. Dis. 35, 1923–1931. doi: 10.1007/s10096-016-2763-0
Foster, T. J. (2017). Antibiotic resistance in Staphylococcus aureus. Current status and future prospects. FEMS Microb. Rev. 41, 430–449. doi: 10.1093/femsre/fux007
Gardete, S., and Tomasz, A. (2014). Mechanisms of vancomycin resistance in Staphylococcus aureus. J. Clin. Invest. 124, 2836–2840. doi: 10.1172/JCI68834
Gomez Casanova, N., Siller Ruiz, M., and Munoz Bellido, J. L. (2017). Mechanisms of resistance to daptomycin in Staphylococcus aureus. Rev. Esp. Quimioter. 30, 391–396.
Gonzalez-Perez, C. J., Tanori-Cordova, J., Aispuro-Hernandez, E., Vargas-Arispuro, I., and Martinez-Tellez, M. A. (2019). Morphometric parameters of foodborne related-pathogens estimated by transmission electron microscopy and their relation to optical density and colony forming units. J. Microbiol. Methods 165:105691 doi: 10.1016/j.mimet.2019.105691
Haaber, J., Penades, J. R., and Ingmer, H. (2017). Transfer of antibiotic resistance in Staphylococcus aureus. Trends Microbiol. 25, 893–905. doi: 10.1016/j.tim.2017.05.011
Harada, Y., Chong, Y., Shimono, N., Miyake, N., Uchida, Y., Kadowaki, M., et al. (2014). Nosocomial spread of meticillin-resistant Staphylococcus aureus with beta-lactam-inducible arbekacin resistance. J. Med. Microbiol. 63(Pt 5): 710–714. doi: 10.1099/jmm.0.065276-0
Haseeb, A., Ajit Singh, V., Teh, C. S. J., and Loke, M. F. (2019). Addition of ceftaroline fosamil or vancomycin to PMMA: an in vitro comparison of biomechanical properties and anti-MRSA efficacy. J. Orthop. Surg. 27:2309499019850324. doi: 10.1177/2309499019850324
Hashemian, S. M. R., Farhadi, T., and Ganjparvar, M. (2018). Linezolid: a review of its properties, function, and use in critical care. Drug Des. Devel. Ther. 12, 1759–1767. doi: 10.2147/DDDT.S164515
Hashizume, H., Takahashi, Y., Masuda, T., Ohba, S. I., Ohishi, T., Kawada, M., et al. (2017). In vivo efficacy of beta-lactam/tripropeptin C in a mouse septicemia model and the mechanism of reverse beta-lactam resistance in methicillin-resistant Staphylococcus aureus mediated by tripropeptin C. J. Antibiot. 71, 79–85. doi: 10.1038/ja.2017.88
Hassoun, A., Linden, P. K., and Friedman, B. (2017). Incidence, prevalence, and management of MRSA bacteremia across patient populations-a review of recent developments in MRSA management and treatment. Crit. Care 21:211. doi: 10.1186/s13054-017-1801-3
Heelan, J. S., Hasenbein, M. E., and McAdam, A. J. (2004). Resistance of group B streptococcus to selected antibiotics, including erythromycin and clindamycin. J. Clin. Microbiol. 42, 1263–1264. doi: 10.1128/JCM.42.3.1263-1264.2004
Heidary, M., Khosravi, A. D., Khoshnood, S., Nasiri, M. J., Soleimani, S., and Goudarzi, M. (2018). Daptomycin. J. Antimicrob. Chemother. 73, 1–11. doi: 10.1093/jac/dkx349
Holmes, N. E., Tong, S. Y., Davis, J. S., and van Hal, S. J. (2015). Treatment of methicillin-resistant Staphylococcus aureus: vancomycin and beyond. Semin. Respir. Crit. Care Med. 36, 17–30. doi: 10.1055/s-0034-1397040
Hori, S., and Hiramatsu, K. (1994). Mechanisms of antimicrobial resistance. Nihon Rinsho 52, 327–331.
Horn, K. S., Danziger, L. H., Rodvold, K. A., and Glowacki, R. C. (2017). Pharmacokinetic drug evaluation of ceftobiprole for the treatment of MRSA. Exp. Opin. Drug. Metab. Toxicol. 13, 463–472. doi: 10.1080/17425255.2017.1303481
Howden, B. P., Davies, J. K., Johnson, P. D., Stinear, T. P., and Grayson, M. L. (2010). Reduced vancomycin susceptibility in Staphylococcus aureus, including vancomycin-intermediate and heterogeneous vancomycin-intermediate strains: resistance mechanisms, laboratory detection, and clinical implications. Clin. Microbiol. Rev. 23, 99–139. doi: 10.1128/CMR.00042-09
Humphreys, H. (2012). Staphylococcus aureus: the enduring pathogen in surgery. Surgeon 10, 357–360. doi: 10.1016/j.surge.2012.05.003
Jang, S. (2016). Multidrug efflux pumps in Staphylococcus aureus and their clinical implications. J. Microbiol. 54, 1–8. doi: 10.1007/s12275-016-5159-z
Jokinen, E., Laine, J., Huttunen, R., Rahikka, P., Huhtala, H., Vuento, R., et al. (2017). Comparison of outcome and clinical characteristics of bacteremia caused by methicillin-resistant, penicillin-resistant and penicillin-susceptible Staphylococcus aureus strains. Infect. Dis. 49, 493–500. doi: 10.1080/23744235.2017.1292046
Kalia, V. C., and Purohit, H. J. (2011). Quenching the quorum sensing system: potential antibacterial drug targets. Crit. Rev. Microbiol. 37, 121–140. doi: 10.3109/1040841X.2010.532479
Kanwar, I., Sah, A. K., and Suresh, P. K. (2017). Biofilm-mediated antibiotic-resistant oral bacterial infections: mechanism and combat strategies. Curr. Pharm. Des. 23, 2084–2095 doi: 10.2174/1381612822666161124154549
Kester, J. C., and Fortune, S. M. (2014). Persisters and beyond: mechanisms of phenotypic drug resistance and drug tolerance in bacteria. Crit. Rev. Biochem. Mol. Biol. 49, 91–101. doi: 10.3109/10409238.2013.869543
Khan, S., Sallum, U. W., Zheng, X., Nau, G. J., and Hasan, T. (2014). Rapid optical determination of beta-lactamase and antibiotic activity. BMC Microbiol. 14:84. doi: 10.1186/1471-2180-14-84
Khoshnood, S., Heidary, M., Asadi, A., Soleimani, S., Motahar, M., Savari, M., et al. (2019). A review on mechanism of action, resistance, synergism, and clinical implications of mupirocin against Staphylococcus aureus. Biomed. Pharmacother. 109, 1809–1818. doi: 10.1016/j.biopha.2018.10.131
Kime, L., Randall, C. P., Banda, F. I., Coll, F., Wright, J., Richardson, J., et al. (2019). Transient silencing of antibiotic resistance by mutation represents a significant potential source of unanticipated therapeutic failure. MBio 10:e01755-19. doi: 10.1128/mBio.01755-19
Klein, E. Y., Mojica, N., Jiang, W., Cosgrove, S. E., Septimus, E., Morgan, D. J., et al. (2017). Trends in methicillin-resistant Staphylococcus aureus hospitalizations in the United States, 2010–2014. Clin. Infect. Dis. 65, 1921–1923. doi: 10.1093/cid/cix640
Klevens, R. M., Morrison, M. A., Nadle, J., Petit, S., Gershman, K., Ray, S., et al. (2007). Invasive methicillin-resistant Staphylococcus aureus infections in the United States. JAMA 298, 1763–1771. doi: 10.1001/jama.298.15.1763
Krueger, W. A., and Unertl, K. E. (2002). [New treatment option for gram-positive infections in critically ill patients - overview over linezolid]. Anasthesiol Intensivmed Notfallmed Schmerzther 37, 199–204. doi: 10.1055/s-2002-25077
Krut, O., and Bekeredjian-Ding, I. (2018). Contribution of the immune response to phage therapy. J. Immunol. 200, 3037–3044. doi: 10.4049/jimmunol.1701745
Lakhundi, S., and Zhang, K. (2018). Methicillin-Resistant Staphylococcus aureus: molecular characterization, evolution, and epidemiology. Clin. Microbiol. Rev. 31:e00020–18. doi: 10.1128/CMR.00020-18
Lazaris, A., Coleman, D. C., Kearns, A. M., Pichon, B., Kinnevey, P. M., Earls, M. R., et al. (2017). Novel multiresistance cfr plasmids in linezolid-resistant methicillin-resistant Staphylococcus epidermidis and vancomycin-resistant Enterococcus faecium (VRE) from a hospital outbreak: co-location of cfr and optrA in Vre. J. Antimicrob. Chemother. 72, 3252–3257. doi: 10.1093/jac/dkx292
Lee, Y. D., and Park, J. H. (2016). Phage conversion for beta-lactam antibiotic resistance of Staphylococcus aureus from foods. J. Microbiol. Biotechnol. 26, 263–269. doi: 10.4014/jmb.1508.08042
Lekshmi, M., Ammini, P., Adjei, J., Sanford, L. M., Shrestha, U., Kumar, S., et al. (2018). Modulation of antimicrobial efflux pumps of the major facilitator superfamily in Staphylococcus aureus. AIMS Microbiol. 4, 1–18. doi: 10.3934/microbiol.2018.1.1
Lessa, F. C., Mu, Y., Ray, S. M., Dumyati, G., Bulens, S., Gorwitz, R. J., et al. (2012). Impact of USA300 methicillin-resistant Staphylococcus aureus on clinical outcomes of patients with pneumonia or central line-associated bloodstream infections. Clin. Infect. Dis. 55, 232–241. doi: 10.1093/cid/cis408
Lewis, K. (2008). Multidrug tolerance of biofilms and persister cells. Curr. Top. Microbiol. Immunol. 322, 107–131. doi: 10.1007/978-3-540-75418-3_6
Li, J., He, S., Yang, Z., and Lu, C. (2017). Pharmacokinetics and cerebrospinal fluid penetration of norvancomycin in Chinese adult patients. Int. J. Antimicrob. Agents 49, 603–608. doi: 10.1016/j.ijantimicag.2017.01.014
Li, M., Al-Jamal, K. T., Kostarelos, K., and Reineke, J. (2010). Physiologically based pharmacokinetic modeling of nanoparticles. ACS Nano 4, 6303–6317 doi: 10.1021/nn1018818
Li, M., Zou, P., Tyner, K., and Lee, S. (2017). Physiologically based pharmacokinetic (PBPK) modeling of pharmaceutical nanoparticles. AAPS J. 19, 26–42. doi: 10.1208/s12248-016-0010-3
Li, N., Luo, M., Fu, Y. J., Zu, Y. G., Wang, W., Zhang, L., et al. (2013). Effect of corilagin on membrane permeability of Escherichia coli, Staphylococcus aureus and Candida albicans. Phytother. Res. 27, 1517–1523. doi: 10.1002/ptr.4891
Lin, D. M., Koskella, B., and Lin, H. C. (2017). Phage therapy: an alternative to antibiotics in the age of multi-drug resistance. World J. Gastrointest. Pharmacol. Ther. 8, 162–173. doi: 10.4292/wjgpt.v8.i3.162
Lindsay, J. A. (2013). Hospital-associated MRSA and antibiotic resistance-what have we learned from genomics? Int. J. Med. Microbiol. 303, 318–323. doi: 10.1016/j.ijmm.2013.02.005
Livermore, D. M. (2003). Linezolid in vitro: mechanism and antibacterial spectrum. J. Antimicrob. Chemother. 51(Suppl 2): ii9–16. doi: 10.1093/jac/dkg249
Lowy, F. D. (1998). Staphylococcus aureus infections. N. Engl. J. Med. 339, 520–532. doi: 10.1056/NEJM199808203390806
Martinez, N., Luque, R., Milani, C., Ventura, M., Banuelos, O., and Margolles, A. (2018). A gene homologous to rRNA methylase genes confers erythromycin and clindamycin resistance in bifidobacterium breve. Appl. Environ. Microbiol. 84, 166–176. doi: 10.1128/AEM.02888-17
Matano, L. M., Morris, H. G., Hesser, A. R., Martin, S. E. S., Lee, W., Owens, T. W., et al. (2017). Antibiotic that inhibits the ATPase activity of an ATP-binding cassette transporter by binding to a remote extracellular site. J. Am. Chem. Soc. 139, 10597–10600. doi: 10.1021/jacs.7b04726
Matono, T., Nagashima, M., Mezaki, K., Motohashi, A., Kutsuna, S., Hayakawa, K., et al. (2018). Molecular epidemiology of beta-lactamase production in penicillin-susceptible Staphylococcus aureus under high-susceptibility conditions. J. Infect. Chemother. 24, 153–155. doi: 10.1016/j.jiac.2017.10.014
Mediavilla, J. R., Chen, L., Mathema, B., and Kreiswirth, B. N. (2012). Global epidemiology of community-associated methicillin resistant Staphylococcus aureus (CA-MRSA). Curr. Opin. Microbiol. 15, 588–595. doi: 10.1016/j.mib.2012.08.003
Micek, S. T. (2007). Alternatives to vancomycin for the treatment of methicillin-resistant Staphylococcus aureus infections. Clin. Infect. Dis. 45(Suppl 3): S184–90. doi: 10.1086/519471
Michiels, J. E., Van den Bergh, B., Verstraeten, N., and Michiels, J. (2016). Molecular mechanisms and clinical implications of bacterial persistence. Drug. Resist. Updat. 29, 76–89. doi: 10.1016/j.drup.2016.10.002
Mo, X. N., Li, J. G., Tang, Y. C., Zhang, Y., Zhu, J. X., Tan, S. Q., et al. (2007). [The action of active efflux system on multi-drug resistance in methicillin resistant Staphylococcus aureus]. Zhonghua Jie He He Hu Xi Za Zhi 30, 40–43.
Nakaminami, H., Takadama, S., Okita, M., Sasaki, M., and Noguchi, N. (2019). Fast-acting bactericidal activity of olanexidine gluconate against qacA/B-positive methicillin-resistant Staphylococcus aureus. J. Med. Microbiol. 68, 957–960. doi: 10.1099/jmm.0.000979
Noguchi, N., Suwa, J., Narui, K., Sasatsu, M., Ito, T., Hiramatsu, K., et al. (2005). Susceptibilities to antiseptic agents and distribution of antiseptic-resistance genes qacA/B and smr of methicillin-resistant Staphylococcus aureus isolated in Asia during 1998 and 1999. J. Med. Microbiol. 54(Pt 6): 557–565. doi: 10.1099/jmm.0.45902-0
Nuti, R., Goud, N. S., Saraswati, A. P., Alvala, R., and Alvala, M. (2017). Antimicrobial peptides: a promising therapeutic strategy in tackling antimicrobial resistance. Curr. Med. Chem. 24, 4303–4314. doi: 10.2174/0929867324666170815102441
Otto, M. (2013). Community-associated MRSA: what makes them special? Int. J. Med. Microbiol. 303:324–330. doi: 10.1016/j.ijmm.2013.02.007
Perez-Perez, M., Jorge, P., Perez Rodriguez, G., Pereira, M. O., and Lourenco, A. (2017). Quorum sensing inhibition in Pseudomonas aeruginosa biofilms: new insights through network mining. Biofouling 33, 128–142. doi: 10.1080/08927014.2016.1272104
Pichereau, S., and Rose, W. E. (2010). Invasive community-associated MRSA infections: epidemiology and antimicrobial management. Expert Opin. Pharmacother. 11, 3009–3025. doi: 10.1517/14656566.2010.511614
Ramos-Martin, V., Johnson, A., McEntee, L., Farrington, N., Padmore, K., Cojutti, P., et al. (2017). Pharmacodynamics of teicoplanin against MRSA. J. Antimicrob. Chemother. 72, 3382–3389. doi: 10.1093/jac/dkx289
Rayner, C., and Munckhof, W. J. (2005). Antibiotics currently used in the treatment of infections caused by Staphylococcus aureus. Intern. Med. J. 35(Suppl 2):S3–16. doi: 10.1111/j.1444-0903.2005.00976.x
Sato, A., Yamaguchi, T., Hamada, M., Ono, D., Sonoda, S., Oshiro, T., et al. (2019). Morphological and biological characteristics of Staphylococcus aureus biofilm formed in the presence of plasma. Microb. Drug. Resist. 25, 668–676. doi: 10.1089/mdr.2019.0068
Saxena, P., Joshi, Y., Rawat, K., and Bisht, R. (2019). Biofilms: architecture, resistance, quorum sensing and control mechanisms. Indian J. Microbiol. 59, 3–12. doi: 10.1007/s12088-018-0757-6
Sazdanovic, P., Jankovic, S. M., Kostic, M., Dimitrijevic, A., and Stefanovic, S. (2016). Pharmacokinetics of linezolid in critically ill patients. Expert Opin. Drug. Metab. Toxicol. 12, 595–600. doi: 10.1517/17425255.2016.1170807
Schulte, R. H., and Munson, E. (2019). Staphylococcus aureus resistance patterns in wisconsin: 2018 surveillance of wisconsin organisms for trends in antimicrobial resistance and epidemiology (SWOTARE) program report. Clin. Med. Res. 17, 72–81. doi: 10.3121/cmr.2019.1503
Seifi, S., and Khoshbakht, R. (2016). Prevalence of tetracycline resistance determinants in broiler isolated Escherichia coli in Iran. Br. Poult. Sci. 57, 729–733. doi: 10.1080/00071668.2016.1232478
Severin, A., Tabei, K., Tenover, F., Chung, M., Clarke, N., and Tomasz, A. (2004). High level oxacillin and vancomycin resistance and altered cell wall composition in Staphylococcus aureus carrying the staphylococcal mecA and the enterococcal vanA gene complex. J. Biol. Chem. 279, 3398–3407. doi: 10.1074/jbc.M309593200
Shang, W., Hu, Q., Yuan, W., Cheng, H., Yang, J., Hu, Z., et al. (2016). Comparative fitness and determinants for the characteristic drug resistance of ST239-MRSA-III-t030 and ST239-MRSA-III-t037 strains isolated in China. Microb. Drug. Resist. 22, 185–192. doi: 10.1089/mdr.2015.0226
Shlezinger, M., Khalifa, L., Houri-Haddad, Y., Coppenhagen-Glazer, S., Resch, G., Que, Y. A., et al. (2017). Phage therapy: a new horizon in the antibacterial treatment of oral pathogens. Curr. Top. Med. Chem. 17, 1199–1211. doi: 10.2174/1568026616666160930145649
Stefani, S., Campanile, F., Santagati, M., Mezzatesta, M. L., Cafiso, V., and Pacini, G. (2015). Insights and clinical perspectives of daptomycin resistance in Staphylococcus aureus: a review of the available evidence. Int. J. Antimicrob. Agents 46, 278–289. doi: 10.1016/j.ijantimicag.2015.05.008
Tayeb-Fligelman, E., Tabachnikov, O., Moshe, A., Goldshmidt-Tran, O., Sawaya, M. R., Coquelle, N., et al. (2017). The cytotoxic Staphylococcus aureus PSMalpha3 reveals a cross-alpha amyloid-like fibril. Science 355, 831–833. doi: 10.1126/science.aaf4901
Taylor, S. D., and Palmer, M. (2016). The action mechanism of daptomycin. Bioorg. Med. Chem. 24, 6253–6268. doi: 10.1016/j.bmc.2016.05.052
Tyson, G. H., Sabo, J. L., Hoffmann, M., Hsu, C. H., Mukherjee, S., Hernandez, J., et al. (2018). Novel linezolid resistance plasmids in Enterococcus from food animals in the USA. J. Antimicrob. Chemother. 73, 3254–3258. doi: 10.1093/jac/dky369
Vestergaard, M., Frees, D., and Ingmer, H. (2019). Antibiotic resistance and the MRSA problem. Microbiol. Spectr. 7:GPP3-0057-2018. doi: 10.1128/microbiolspec.GPP3-0057-2018
Wang, J., Pan, Y., Shen, J., and Xu, Y. (2017). The efficacy and safety of tigecycline for the treatment of bloodstream infections: a systematic review and meta-analysis. Ann. Clin. Microbiol. Antimicrob. 16:24. doi: 10.1186/s12941-017-0199-8
Wang, Q., Yang, Q., Wu, W. (2020). Graphene-based steganographic aptasensor for information computing and monitoring toxins of biofilm in food. Front. Microbiol. 10:3139. doi: 10.3389/fmicb.2019.03139
Wu, W., Yu, C., Chen, J., Yang, Q. (2020). Fluorometric detection of copper ions using click chemistry and the target-induced conjunction of split DNAzyme fragments. Int. J. Environ. Analyt. Chem. 100, 324–332. doi: 10.1080/03067319.2019.1636977
Yang, J. J., Cheng, A., Tai, H. M., Chang, L. W., Hsu, M. C., and Sheng, W. H. (2019). Selected mutations by nemonoxacin and fluoroquinolone exposure among relevant gram-positive bacterial strains in Taiwan. Microb. Drug Resist. 26, 110–117. doi: 10.1089/mdr.2019.0048
Yin, S. L., Chang, Y. J., Deng, S. P., Wang, Q. C., Yu, W. G., and Gong, Q. H. (2011). [Research progress of new antibacterial drugs that target bacterial quorum sensing systems]. Yao Xue Xue Bao 46, 613–621.
Zarate, S. G., Morales, P., Swiderek, K., Bolanos-Garcia, V. M., and Bastida, A. (2019). A molecular modeling approach to identify novel inhibitors of the major facilitator superfamily of efflux pump transporters. Antibiotics 8:25. doi: 10.3390/antibiotics8010025
Keywords: Staphylococcus aureus, molecular mechanisms, acquired antibiotic resistance, antibiotic resistance therapy, cell membrane
Citation: Guo Y, Song G, Sun M, Wang J and Wang Y (2020) Prevalence and Therapies of Antibiotic-Resistance in Staphylococcus aureus. Front. Cell. Infect. Microbiol. 10:107. doi: 10.3389/fcimb.2020.00107
Received: 11 November 2019; Accepted: 27 February 2020;
Published: 17 March 2020.
Edited by:
Ren-You Gan, Institute of Urban Agriculture (CAAS), ChinaReviewed by:
Tian Ding, Zhejiang University, ChinaWei Wu, Qingdao Agricultural University, China
Juan M. Pericas, University Hospital Arnau de Vilanova, Spain
Copyright © 2020 Guo, Song, Sun, Wang and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yi Wang, YXVzdGlud3lAMTYzLmNvbQ==
 Yunlei Guo1
Yunlei Guo1 Yi Wang
Yi Wang