
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
PERSPECTIVE article
Front. Cell. Infect. Microbiol., 12 February 2020
Sec. Microbiome in Health and Disease
Volume 10 - 2020 | https://doi.org/10.3389/fcimb.2020.00022
This article is part of the Research TopicInteractions of the Nervous System with BacteriaView all 14 articles
 Walter J. Lukiw1,2,3*
Walter J. Lukiw1,2,3*The microbiome of the human gastrointestinal (GI)-tract is a rich and dynamic source of microorganisms that together possess a staggering complexity and diversity. Collectively these microbes are capable of secreting what are amongst the most neurotoxic and pro-inflammatory biopolymers known. These include lipopolysaccharide (LPS), enterotoxins, microbial-derived amyloids and small non-coding RNA (sncRNA). One of the major microbial species in the human GI-tract microbiome, about ~100-fold more abundant than Escherichia coli, is Bacteroides fragilis, an anaerobic, rod-shaped Gram-negative bacterium that secretes: (i) a particularly potent, pro-inflammatory LPS glycolipid subtype (BF-LPS); and (ii) a hydrolytic, extracellular zinc metalloproteinase known as B. fragilis toxin (BFT) or fragilysin. Ongoing studies support multiple observations that BF-LPS and BFT (fragilysin) disrupt paracellular barriers by cleavage of intercellular proteins, such as E-cadherin, between epithelial cells, resulting in ‘leaky’ barriers. These defective barriers, which also become more penetrable with age, in turn permit entry of microbiome-derived neurotoxic biopolymers into the systemic circulation from which they can next transit the blood-brain barrier (BBB) and gain access into the brain. This short communication will highlight some recent advances in this extraordinary research area that links the pro-inflammatory exudates of the GI-tract microbiome with innate-immune disturbances and inflammatory signaling within the human central nervous system (CNS) with reference to Alzheimer's disease (AD) wherever possible.
The gastrointestinal (GI)-tract microbiome, the largest reservoir of microbes in the human body, containing about 1014 microbial cells, is a complex, dynamic and abundant source of bacteria, methanogenic archaea, fungi, microbial eukaryotes, protozoa, viruses, and other microorganisms. Together the GI-tract microbiome possesses a remarkable microbiological diversity and staggering genetic complexity of at least 1000 major bacterial species. In the most recent estimate analyses of the GI-tract metagenomes of ~2100 donors well over 22.3 million non-redundant prokaryotic genes were detected, and at least half of all the genes identified were unique to an individual (Tierney et al., 2019). When compared to the established human genome content of 26.6 thousand protein-encoding transcripts of the human genome sequencing project obtained about ~18 years ago (Fields et al., 1994; Venter et al., 2001; Hicks et al., 2019) the number of microbial genes in the human GI-tract microbiome alone outnumbers human genes by about 837 to 1 (Tierney et al., 2019). Another interesting fact is that of the 52 major divisions of bacteria identified to date, only 2 phyla are known to predominate in the human GI-tract microbiome—the Gram-negative Bacteroidetes (representing about ~20–30% of all GI-tract resident bacteria) and the Gram-positive Firmicutes (representing ~70–80% of the total) with relatively minor contributions by Actinobacteria (~3%), Proteobacteria (~1%), Fusobacteria (~0.1%) and Verrucomicrobia (0.1%). Collectively these microorganisms represent: (i) “the microbial core” of the human GI-tract microbiome (Sarkar and Banerjee, 2019; Ticinesi et al., 2019); (ii) an extremely active, dynamic, and changing ecosystem dependent on the host's age, diet, environment, ethnicity, and health and/or disease status (Sender et al., 2016; Zhao and Lukiw, 2018a,b; Rinninella et al., 2019); (iii) a rich source of commensal bacteria usually beneficial to human health because of their abilities to metabolize and/or biosynthesize complex sugars, polysaccharides, and dietary fiber into volatile short chain fatty acids (SCFAs; including acetate, propionate, butyrate, valerate and lactate and other nutrients). SCFAs (i) normally function in the development, maintenance, and homeostasis of the host immune, neuro-endrocrine and digestive systems; and (ii) play important regulatory roles in glucose homeostasis, lipid metabolism and anti-inflammatory signaling in endothelial cells of the lining of the GI-tract, sometimes known as the intestinal endothelium. Interestingly, there is recent evidence that SCFAs can signal through G-protein coupled receptors (GPCRs) at the cell surface, including GPCR41, GPCR43, and GPCR109a and these activate signaling cascades that control multiple immune functions. Recent transgenic mouse studies support a key role of these GPCRs in the regulation of intestinal inflammation (Sears, 2009; Fathi and Wu, 2016; Lukiw, 2016a,b; Castillo-Álvarez and Marzo-Sola, 2019; Fox et al., 2019; Parada Venegas et al., 2019).
Over 99% of the microbes in the human GI-tract are facultative and obligate anaerobic bacteria; the most abundant Gram-negative bacterial Phylum in the human GI-tract microbiome are the Bacteroidetes, with a major Genus-species being represented by the obligate Gram-negative anaerobe Bacteroides fragilis. In some intestinal tract regions B. fragilis: (i) are present at about ~100-fold the abundance of the Proteobacteria Escherichia coli; (ii) colonize the human GI-tract at densities up to 8 × 1010 CFU per cm3, the highest density of any microbial colonization known in nature (Sears, 2009; Fathi and Wu, 2016; Rios-Covian et al., 2017; Patrick et al., 2019; Rinninella et al., 2019); and (iii) reside and proliferate exclusively in the GI-tract of mammals, suggesting a strong adaptation to the pH, biophysical and microbial composition of the gut environment (Bhattacharjee and Lukiw, 2013; Wexler and Goodman, 2017; Poeker et al., 2018; Castillo-Álvarez and Marzo-Sola, 2019).
In the human GI-tract there are 2 predominant strains of Bacteroides fragilis (B. fragilis) distinguished in part by their biosynthetic capabilities to synthesize and secrete a zinc-dependent metalloprotease toxin known as B. fragilis toxin (BFT) or fragilysin. Strains of Bacteroides that do not secrete BFT are called non-toxigenic B. fragilis while those that do secrete are called enterotoxigenic B. fragilis (ETBF; Allen et al., 2019). Relatively recently it has been established that enterotoxigenic strains of B. fragilis (ETBF) can rapidly proliferate in the mammalian GI-tract both in the absence of adequate dietary fiber and in the presence of high-fat cholesterol diets (Heinritz et al., 2016; Wexler and Goodman, 2017; Poeker et al., 2018; Zhao and Lukiw, 2018a,b). This proliferation enhances the intestinal abundance of B. fragilis and hence the potential of this Gram negative obligate anaerobe to secrete its formidable array of neurotoxic exudates. These primarily include: (i) the lipoglycan lipopolysaccharide (LPS), a particularly potent, pro-inflammatory LPS glycolipid subtype (BF-LPS); and (ii) the hydrolytic, extracellular zinc metalloproteinase known as ETBF-secreted Bacteroides fragilis toxin (BFT), also known as fragilysin. Recent characterization of BF-LPS and fragilysin have shown them to be amongst the most pro-inflammatory lipoglycans and enterotoxins known (Vines et al., 2000; Sears, 2009; Lukiw, 2016a,b; Zhao and Lukiw, 2018a,b; Batista et al., 2019; Sheppard et al., 2019). Both BF-LPS and fragilysin can leak through the normally protective mucosal barriers of the GI-tract intestinal endothelium to induce substantial inflammatory pathology both systemically and after BBB transit into vulnerable CNS compartments, including the neocortical parenchyma of the brain (Fathi and Wu, 2016; Lukiw, 2016a,b; Zhao and Lukiw, 2018a,b; Barton et al., 2019; Batista et al., 2019; Fox et al., 2019; Sheppard et al., 2019; Zhao et al., 2019). Indeed, while Bacteroides fragilis is an anaerobic, Gram-negative, rod-shaped bacillus, and part of the normal microbiota of the human colon and is generally commensal, this microbe can cause a “smoldering” systemic infection if displaced into the bloodstream or surrounding tissue following disease, trauma or surgery (Hill et al., 2014a,b; Montagne et al., 2017; Tulkens et al., 2018; Erdo and Krajcsi, 2019; Fox et al., 2019; Patrick et al., 2019; Sarkar and Banerjee, 2019; Sweeney et al., 2019). When the highly toxic exudates of enterotoxigenic strains of B. fragilis escape the microbial-dense environment of the human GI-tract they can produce substantial systemic inflammatory pathology with significant mortality and morbidity. B. fragilis proliferation is associated with, and causative for, bacteremia, brain and intra-abdominal abscess, cellulitis, colitis, diabetic ulcer, diarrhea, necrotizing fasciitis, sepsis, peritonitis, septicemia, association with and the development of multiple pro-inflammatory bowel cancers, systemic infection and systemic inflammation, the development of neurological diseases involving inflammatory neurodegeneration, and those neurological disorders that display a significantly elevated incidence of atypical developmental programming against a background of aging (Leshchyns'ka and Sytnyk, 2016; Agrawal et al., 2017; Shivaji, 2017; Zhao et al., 2019). Very recently LPS-induced systemic inflammation has been associated with synaptic loss and cognitive decline in multiple human neurological disorders and in animal models, and a role for LPS-mediated microglial release of pro-inflammatory cytokines (such as IL-1β) based on both in vivo and primary culture studies in vitro (Sheppard et al., 2019; Zhao et al., 2019).
Two anatomical gateways, including the gastrointestinal mucosa that includes the “GI-tract barrier” and the “blood-brain barrier (BBB),” each formed essentially by vascular epithelial and/or endothelial cells and epithelial-endothelial-derived basement membranes provide both a biophysical interface and a biological compartmentalization of the GI-tract microbiome, the systemic circulation, the brain parenchyma and distinct anatomical regions of the brain such as the neocortex (Figure 1). These barriers are a requisite for the essential maintenance of homeostasis and the physiological environment of each compartment; microorganisms of the GI-tract microbiome and their neurotoxins that are able to transit the single layer of epithelial cells have virtually unimpeded access into the systemic circulation (Varatharaj and Galea, 2017; Logsdon et al., 2018; Sweeney et al., 2019; Tulkens et al., 2018). Probably the most important structural components of these barriers are the multiple tight junctions between adjacent cells of vascular capillaries (Sweeney et al., 2018, 2019; Tulkens et al., 2018). For example, the blood-brain barrier of the CNS can selectively regulate its intracellular compartments and thereby isolate itself from rapid biochemical or biophysical changes that may occur in the systemic circulation.
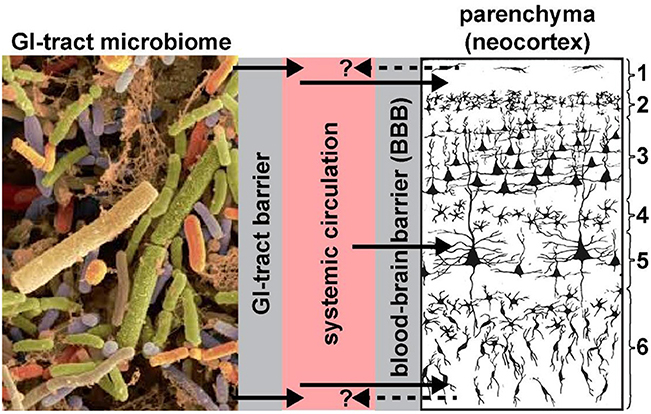
Figure 1. Highly schematicized depiction of the potential transfer of GI-tract microbiome-derived pro-inflammatory neurotoxins across the GI-tract barrier into the systemic circulation, followed by translocation across the BBB into the brain parenchyma (see Hill et al., 2014a,b; Hill and Lukiw, 2015; Zhao and Lukiw, 2018a; neocortical region shown; solid black arrows). Neurotoxins identified to date include Bacteroides fragilis lipopolysaccharide (BF-LPS) and enterotoxins such as the Bacteroides fragilis-derived toxin (BFT) also known as fragilysin. The contribution of other pro-inflammatory neurotoxins such as GI-tract-derived amyloids and bacterial sncRNAs are not well-understood and currently very little is known concerning their neurotoxicity and CNS-effects. BF-LPS, BFT (fragilysin), age, dietary toxins, traumatic brain injury (TBI) and vascular disease are known to effectively disrupt endothelial cell-based biophysical barriers in part through the cleavage, disruption and/or degeneration of cell-cell adhesion proteins (Wu et al., 1998; Clement et al., 2016; Zhao and Lukiw, 2018a,b; Sweeney et al., 2019). We speculate that all of these neurotoxins together have potential to constitute a highly neurotoxic pro-inflammatory GI-tract microbiome-derived cocktail greatly detrimental to the cytoarchitecture and signaling functions of neuronal and glial cells. Gram-negative bacterial-derived LPS and related neurotoxins have been recently observed within the systemic circulation and brain parenchyma (Zhan et al., 2016, 2018; Zhao et al., 2017a,c, 2019; Zhao and Lukiw, 2018a), within and around neurons, and in the later stages of AD completely encapsulating neuronal neocortical nuclei (Hill et al., 2014b,a; Hill and Lukiw, 2015; Zhao and Lukiw, 2018a,b; unpublished observations); this later action appears to impair the exit of neuron-specific transcripts such as the neurofilament-light (NF-L) chain and synapsin-2 (SYN2) messenger RNA (mRNA) from the neuronal nuclei; both NF-L and SYN2 mRNA abundance and expression are down-regulated in LPS-treated human neuronal-glial (HNG) cells in primary culture and in AD brain (Lukiw et al., 2018; Zhao et al., 2019). Whether neurotoxins from the brain parenchyma of the neocortex can cross the BBB back into the systemic circulation (dashed black arrows with question mark) is currently not well-understood; if so, these species may be useful serum biomarkers for both the diagnosis and prognosis of AD and other types of inflammatory neurodegeneration; left panel = GI-tract microbiome magnification x ~3000 (source: http://www.Injnbio.com; http://www.lnjnbio.com/nd.jsp?id=20; permission to reproduce granted; last accessed 26 November 2019); right panel = 6-layered structure of the human association neocortex; layers 3 and 5 are the pyramidal cell layers targeted by the AD process; other brain regions may also be affected; magnification x ~20 (source: adapted and redrawn from Martinez-Conde, 2018; last accessed 26 November 2019).
The surface area of the human GI-tract barrier and the BBB is remarkably large; for example although the interior of the small intestine is only about ~3.5 cm in diameter and ~7 m in length (due in large part to a concentrically folded mucosa) it has a total absorptive surface area of ~32 m2 (Helander and Fändriks, 2014; Sweeney et al., 2019). Similarly the surface area of the 600 km of the human brain's 5 um diameter microvessels, representing the majority of the BBB, corresponds to a total surface area of ~25 m2 (Wong et al., 2013). Hence the maintenance of these large and formidable biophysical barriers is a very active and energy-intense biological process (Wong et al., 2013; Varatharaj and Galea, 2017; Barton et al., 2019; Erdo and Krajcsi, 2019; Sweeney et al., 2019). BF-LPS, BFT (fragilysin) and other GI-tract microbiome-derived exudates are remarkable in their capabilities: (i) of breaking down the intercellular junctions of these barriers via their disruptive actions on cadherins and other cell-cell adhesion molecules (Wu et al., 1998; Wong et al., 2013; Sweeney et al., 2018); (ii) of altering BBB integrity and permeability (Varatharaj and Galea, 2017; Tulkens et al., 2018; Sweeney et al., 2019); (iii) of changing BBB transport rates (Wong et al., 2013; Logsdon et al., 2018); (iv) of modulating neuroimmune signaling or the transport of immune-regulatory molecules (Erdo and Krajcsi, 2019); (v) of trafficking dietary neurotoxins and pathogens into the brain (Sweeney et al., 2019; Wong et al., 2013); and/or (vi) of inducing the release of inflammatory and neuro-immune substances from the barrier cells (Hill et al., 2014a,b; Köhler et al., 2016; Lukiw, 2016a,b; Montagne et al., 2017; Varatharaj and Galea, 2017; Tulkens et al., 2018; Sweeney et al., 2018, 2019; Erdo and Krajcsi, 2019; Fox et al., 2019; Patrick et al., 2019; Sarkar and Banerjee, 2019).
Indeed, the most recent research evidence continues to strengthen the idea that one major, and virtually unlimited, source of pro-inflammatory neurotoxic signals in inflammatory neurodegeneration such as those typified by the AD process may originate from internally derived noxious exudates such as those supplied via the diet and metabolized by the GI-tract microbiome (Sweeney et al., 2018, 2019; Erdo and Krajcsi, 2019). Because of aging, traumatic brain injury (TBI), cerebrovascular deficits (some of which may be genetic), neurovascular pathology or neuroinflammatory brain degeneration, neurotoxic molecules can “leak” into the systemic circulation, prompting some investigators to propose that progressive neurodegenerative diseases such as AD supplied via the reflect a malfunction of key biophysical barriers including those of the GI-tract and BBB, and that AD is in fact a “defective barrier” disease (Bhattacharjee and Lukiw, 2013; Montagne et al., 2017; Sweeney et al., 2018, 2019; Erdo and Krajcsi, 2019). The abundance of blood-borne bacterial components including LPS, for example, represents a variable component of the human blood serum that can elicit variable systemic pro-inflammatory and innate-immune-modulatory responses in the host, resulting in systemic-immune activation by pathogen-associated molecular patterns (PAMPs), a process sometimes referred to as “microbial translocation” (Zhao et al., 2017a,b; Tulkens et al., 2018; Di Lorenzo et al., 2019; Logsdon et al., 2018; Patrick et al., 2019). Multiple recent reports further suggest that GI-tract dysbiosis and “leaky gut syndrome” constitute a vastly under-appreciated, under-studied and critical pathophysiological passageway for transport of GI-tract microbiome-derived neurotoxins across GI-tract and blood–brain biological barriers resulting in an age-related progression from systemic inflammation to neurovascular disease to CNS inflammation and degeneration that progressively contribute to critical aspects of neuropathology associated with age-related neurodegenerative disorders. These include neuropathological disorders such as AD, anxiety, autism spectrum disorder (ASD), depression, epilepsy, multiple sclerosis, Parkinson's disease (PD), prion disease, systemic inflammatory response syndrome, and other incapacitating and/or ultimately lethal neurological diseases of the human CNS (Hill et al., 2014a,b; Köhler et al., 2016; Li and Yu, 2017; Varatharaj and Galea, 2017; Zhao et al., 2017a,b, 2019; Griffiths and Mazmanian, 2018; Di Lorenzo et al., 2019; Fox et al., 2019; Patrick et al., 2019; Sarkar and Banerjee, 2019).
The extruded lipopolysaccharide shed from the human GI-tract microbiome-abundant Bacteroides fragilis (BF-LPS): (i) is one of the most pro-inflammatory and neurotoxic lipoglycans known (Sears, 2009; Fathi and Wu, 2016; Lukiw, 2016a,b; Allen et al., 2019); (ii) is linked to synaptic loss and cognitive decline in human patients and in animal models (Sheppard et al., 2019); and (iii) is recognized by the Toll receptors TLR2, TLR4, and/or CD14 microglial cell receptors, as are the pro-inflammatory and hydrophobic 42 amino acid amyloid-beta (Aβ42) peptides whose accumulation are a characteristic feature of AD brain (Sears, 2009; Zhao et al., 2015; Zhao and Lukiw, 2015; Lukiw, 2016a,b; Batista et al., 2019; Sheppard et al., 2019). Additional LPS-mediated pro-inflammatory actions, pathogenic mechanisms and neurodegeneration-promoting activities remain incompletely understood but remarkable progress is being made both in transgenic animal models and in human patient studies (Zhan et al., 2018; Zhao and Lukiw, 2018a,b; Barton et al., 2019; Sarkar and Banerjee, 2019; Sheppard et al., 2019; Wu et al., 2007). For example, LPS-induced synaptic loss and the impairment of cognition appear to be in part the result of a modified microglial activation, reactive oxidative species (ROS) or cytokine generation and oxidative stress damage, disruption of the intercellular adhesion proteins associated with the GI-tract or blood-brain barriers, the ROS mediated oxidation, atrophy, destruction and loss of synapse-related proteins, elevations in neuroinflammatory signaling or any combination of these events (Barton et al., 2019; Batista et al., 2019; Sheppard et al., 2019; Wu et al., 2007). One recently described BF-LPS mediated pathogenic and AD-relevant pathway is the robust activation of NF-κB (p50/p65) in human brain cells in primary culture and induction of a pro-inflammatory signaling pathway involving an NF-kB-regulated microRNA-146a, and the subsequent chronic and pathogenic over-stimulation of innate-immune and neuro-inflammatory pathways (Zhao and Lukiw, 2018b). These include deficits in the innate-immune system modulator complement factor H (CFH), decreases in the expression of the essential presynaptic neuronal phosphoprotein synapsin-2 (SYN2) and down-regulation of the neuron-specific neurofilament light chain (NF-L) cytoskeletal protein (Lukiw et al., 2018; Zhan et al., 2018; Zhao et al., 2019). These pathological signaling pathways appear to strongly contribute to synaptic disorganization and decline, neuronal atrophy and inflammation-mediated amyloidogenic neuropathology which are all characteristic attributes of the AD-affected brain.
The neurobiological signaling connections between the GI-tract microbiome and CNS disease remain incompletely understood. Eighteen years after the elucidation and characterization of the genes expressed in the human genome (Venter et al., 2001), the staggering genetic complexity of the human GI-tract and oral microbiomes have been analyzed with remarkable and unexpected results (Tierney et al., 2019). It is truly extraordinary that the potential contribution to human health and disease by the GI-tract microbiome with a total mass, complexity and diversity, and number of genes exceeding that of the liver could have been almost completely overlooked as recent as just ~10 years ago.
Several fundamental questions remain concerning nature of the microorganisms of the GI-tract microbiome, their compartmentalization within the GI-tract and their potential effects on the neuropathology, neurobiology, and the pathogenetics of inflammatory neurodegeneration and neuropsychiatric disease. It will be further interesting to discover: (i) the evolutionary history of the GI-tract microbiome and for example, why just 2 of 52 bacterial phyla were selected and evolved to be both dominant and symbiotic within the entire human metagenome; (ii) what patterns of microbial abundance, speciation, complexity, stoichiometry, dysbiosis and GI-tract-derived mixtures of neurotoxins are the most effective in promoting pathogenic inflammatory neuro-degeneration; (iii) if the incidence of blood-borne GI-tract-derived toxic elements in the systemic inflammation could be used as a pathological biomarker or be of prognostic value for AD and other progressive, age-related, neurodegenerative diseases; (iv) what would be the contribution of combinations of the microbial constituents including archaebacteria, fungi, protozoa, viruses, and other GI-tract resident microbes of the GI-tract microbiome to enhance neurological health; (v) the intriguing possibility that the composition of the GI-tract microbiome could be altered through diet, probiotics and/or prebiotics to optimize human neurological health; (vi) if the penetration of epithelial barriers by bacterial products occurs in the oral cavity in periodontal disease with similar systemic effects; (vii) the mechanism of the duality of GI-tract abundant Gram-negative anaerobic bacteria such as B. fragilis in behaving in both pro- and anti-inflammatory capacities, the role of capsular polysaccharides and IL-10 secreting B and T cells in this transition, and how the role of B. fragilis can switch from an abundant beneficial microbe and commensal microorganism to a highly neurotoxic one (Ramakrishna et al., 2019); and (viii) perhaps most importantly, if medical researchers along with neurologists and dieticians could devise a strategy, perhaps through “personalized medicine,” that promotes the lowering of noxious GI-tract microbes and their secretions that would optimize life-long GI-tract microbiome function and CNS health. This approach might minimize the risk of developing AD and other highly incapacitating human diseases as we age. Furthering our molecular-genetic and mechanistic understanding of how different secreted components of the GI-tract microbiome negatively affect the CNS may uncover potential and novel strategic approaches for the GI-tract microbiome-based modulation of neurological function, and the more effective clinical management of terminal, age-related neurological disorders.
The appreciation of a potential contribution from the GI-tract microbiome to human neurological health and devastating behavioral, amnestic and cognitive disorders such as AD is a relatively recent one (Bhattacharjee and Lukiw, 2013), and gathering recent evidence continues to strengthen this association (Johnson and Foster, 2018; Patrick et al., 2019; Sarkar and Banerjee, 2019; Sheppard et al., 2019; Strandwitz et al., 2019; Sweeney et al., 2019; Ticinesi et al., 2019; Tierney et al., 2019; Zhao et al., 2019). Dietary manipulations of the GI-tract microbiome including diets enriched in biologically soluble and insoluble fiber, that seem to neutralize the potentially neurotoxic secretions from Gram-negative bacilli such as Bacteroides fragilis might provide a life-long resolution to defer the development of human neuro-inflammatory degenerative disease (Heinritz et al., 2016; Chen et al., 2017; Poeker et al., 2018). It is becoming increasingly established that the contribution of the GI-tract microbiome and GI-tract microbiome-derived neurotoxins to pathogenic signaling associated with inflammatory neurodegeneration is: (i) age-related and progressive; (ii) contains multiple neurotoxic components with capability to breach biophysical barriers; (iii) constitute a virtually unlimited supply of BF-LPS, BFT (fragilysin) and other neurotoxins; and (iv) that the GI-tract microbiome comprises a “staggering” microbial genetic complexity, and the recent finding that at least half of all the genes identified are unique to each individual further underscores the interesting parallel in the heterogeneity between GI-tract microbiome composition and AD risk, onset and development (Patrick et al., 2019; Strandwitz et al., 2019; Ticinesi et al., 2019; Tierney et al., 2019). Of further recent interest is the potential involvement of the GI-tract microbiota-brain axis with the mental status of the host in that certain “psychotropic bacteria” and their secreted array of “psychobiotics” appear to influence the mental health of the host (Beck et al., 2019; Cheng et al., 2019; Kelly et al., 2019). Given that AD was originally referred to as a progressive and dementing “senile psychosis,” efficacious manipulation of the GI-tract microbiome might not only attenuate inflammatory neurodegeneration, synaptic disorganization and cognitive decline but also optimize healthy neuroimmune, neuroendocrine, humoral and brain signaling pathways that also promote well-being, anti-depressive and anxiolytic behaviors in patients affected by the AD process.
The datasets generated for this study are available on request to the corresponding author.
The studies involving human participants were reviewed and approved by Louisiana State University Institutional Review Board—only post-mortem human tissues were used in these studies. The ethics committee waived the requirement of written informed consent for participation.
WL distilled the results from all laboratory experiments at the LSU laboratories and performed literature searches of recent peer-reviewed publications in this research area, compiled all data, and wrote this manuscript.
Research on microRNAs, ethnobiology, botanical neurotoxins, pro-inflammatory and pathogenic signaling in the Lukiw laboratory involving the microbiome, the innate-immune response, amyloidogenesis, synaptogenesis, and neuro-inflammation in AD, prion and in other human neurological- and plant-viroid-based diseases was supported through an unrestricted grant to the LSU Eye Center from Research to Prevent Blindness (RPB); the Louisiana Biotechnology Research Network (LBRN) and NIH grants NEI EY006311, NIA AG18031, and NIA AG038834 (WL). The content of this manuscript was solely the responsibility of the authors and does not necessarily represent the official views of the National Institute on Aging, the National Center for Research Resources, or the National Institutes of Health.
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The research in this Perspectives article was presented in part at the Vavilov Institute of General Genetics Autumn 2018 Seminar Series (Институт общей генетики имени Вавилова Осень 2018 Семинар серии) in Moscow, Russia, October 2018, at the Society for Neuroscience (SFN) Annual Meeting, Chicago IL USA, November 2019. Sincere thanks are extended to Drs. L. Cong, F. Culicchia, C. Eicken, K. Navel, A. I. Pogue, W. Poon, E. Head, and the late Drs. J. M. Hill and P. N. Alexandrov for helpful discussions in this research area, for short postmortem interval (PMI) human brain and retinal tissues or extracts, for initial bioinformatics and data interpretation, and to A. I. Pogue and D. Guillot for expert technical assistance and medical artwork.
Agrawal, M., Ajazuddin Tripathi, D. K., Saraf, S., Saraf, S., Antimisiaris, S. G., et al. (2017). Recent advancements in liposomes targeting strategies to cross blood-brain barrier (BBB) for the treatment of Alzheimer's disease. J. Control Release 260, 61–77. doi: 10.1016/j.jconrel.2017.05.019
Allen, J., Hao, S., Sears, C. L., and Timp, W. (2019). Epigenetic changes induced by Bacteroides fragilis toxin. Infect. Immun. 87:e00447-18. doi: 10.1128/IAI.00447-18
Barton, S. M., Janve, V. A., McClure, R., Anderson, A., Matsubara, J. A., Gore, J. C., et al. (2019). Lipopolysaccharide induced opening of the blood brain barrier on aging 5XFAD mouse model. J. Alzheimers Dis. 67, 503–513. doi: 10.3233/JAD-180755
Batista, C. R. A., Gomes, G. F., Candelario-Jalil, E., Fiebich, B. L., and de Oliveira, A. C. P. (2019). Lipopolysaccharide-induced neuroinflammation as a bridge to understand neurodegeneration. Int. J. Mol. Sci. 20:E2293. doi: 10.3390/ijms20092293
Beck, B. R., Park, G. S., Jeong, D. Y., Lee, Y. H., Im, S., Song, W. H., et al. (2019). Multidisciplinary and comparative investigations of potential psychobiotic effects of Lactobacillus strains isolated from newborns and their impact on gut microbiota and ileal transcriptome in a healthy murine model. Front. Cell Infect. Microbiol. 9:269. doi: 10.3389/fcimb.2019.00269
Bhattacharjee, S., and Lukiw, W. J. (2013). Alzheimer's disease and the microbiome. Front. Cell. Neurosci. 7:153. doi: 10.3389/fncel.2013.00153
Castillo-Álvarez, F., and Marzo-Sola, M. E. (2019). Role of the gut microbiota in the development of various neurological diseases. Neurologia. doi: 10.1016/j.nrl.2019.03.017. [Epub ahead of print].
Chen, T., Long, W., Zhang, C., Liu, S., Zhao, L., and Hamaker, B. R. (2017). Fiber-utilizing capacity varies in Prevotella- versus Bacteroides-dominated gut microbiota. Sci. Rep. 7:2594. doi: 10.1038/s41598-017-02995-4
Cheng, L. H., Liu, Y. W., Wu, C. C., Wang, S., and Tsai, Y. C. (2019). Psychobiotics in mental health, neurodegenerative and neurodevelopmental disorders. J. Food Drug Anal. 27, 632–648. doi: 10.1016/j.jfda.2019.01.002
Clement, C., Hill, J. M., Dua, P., Culicchia, F., and Lukiw, W. J. (2016). Analysis of RNA from Alzheimer's disease post-mortem brain tissues. Mol. Neurobiol. 53, 1322–1328. doi: 10.1007/s12035-015-9105-6
Di Lorenzo, F., De Castro, C., Silipo, A., and Molinaro, A. (2019). Lipopolysaccharide structures of Gram-negative populations in the gut microbiota and effects on host interactions. FEMS Microbiol. Rev. 43, 257–272. doi: 10.1093/femsre/fuz002
Erdo, F., and Krajcsi, P. (2019). Age-related functional and expressional changes in efflux pathways at the blood-brain barrier. Front. Aging Neurosci. 11:196. doi: 10.3389/fnagi.2019.00196
Fathi, P., and Wu, S. (2016). Isolation, detection, and characterization of enterotoxigenic Bacteroides fragilis in clinical samples. Open Microbiol. J. 10, 57–63. doi: 10.2174/1874285801610010057
Fields, C., Adams, M. D., White, O., and Venter, J. C. (1994). How many genes in the human genome? Nat. Genet. 7, 345–346.
Fox, M., Knorr, D. A., and Haptonstall, K. M. (2019). Alzheimer's disease and symbiotic microbiota: an evolutionary medicine perspective. Ann. N Y. Acad. Sci. 1449, 3–24. doi: 10.1111/nyas.14129
Griffiths, J. A., and Mazmanian, S. K. (2018). Emerging evidence linking the gut microbiome to neurologic disorders. Genome Med. 10:98. doi: 10.1186/s13073-018-0609-3
Heinritz, S. N., Weiss, E., Eklund, M., Aumiller, T., Heyer, C. M., Messner, S., et al. (2016). Impact of a high-fat or high-fiber diet on intestinal microbiota and metabolic markers in a pig model. Nutrients 8:E317. doi: 10.3390/nu8050317
Helander, H. F., and Fändriks, L. (2014). Surface area of the digestive tract - revisited. Scand. J. Gastroenterol. 49, 681–689. doi: 10.3109/00365521.2014.898326
Hicks, M., Bartha, I., di Iulio, J., Venter, J. C., and Telenti, A. (2019). Functional characterization of 3D protein structures informed by human genetic diversity. Proc. Natl. Acad. Sci. U.S.A. 116, 8960–8965. doi: 10.1073/pnas.1820813116
Hill, J. M., Bhattacharjee, S., Pogue, A. I., and Lukiw, W. J. (2014b). The gastrointestinal tract microbiome and potential link to Alzheimer's disease. Front. Neurol. 5:43. doi: 10.3389/fneur.2014.00043
Hill, J. M., Clement, C., Pogue, A. I., Bhattacharjee, S., Zhao, Y., and Lukiw, W. J. (2014a). Pathogenic microbes, the microbiome, and Alzheimer's disease (AD). Front. Aging Neurosci. 6:127. doi: 10.3389/fnagi.2014.00127
Hill, J. M., and Lukiw, W. J. (2015). Microbial-generated amyloids and Alzheimer's disease (AD). Front. Aging Neurosci. 7:9. doi: 10.3389/fnagi.2015.00009
Johnson, K. V.-A., and Foster, K. R. (2018). Why does the microbiome affect behaviour? Nat. Rev. Microbiol. 16, 647–655. doi: 10.1038/s41579-018-0014-3
Kelly, J. R., Keane, V. O., Cryan, J. F., Clarke, G., and Dinan, T. G. (2019). Mood and microbes: gut to brain communication in depression. Gastroenterol Clin. North Am. 48, 389–405. doi: 10.1016/j.gtc.2019.04.006
Köhler, C. A., Maes, M., Slyepchenko, A., Berk, M., Solmi, M., Lanctôt, K. L., et al. (2016). The gut-brain axis, including the microbiome, leaky gut and bacterial translocation: mechanisms and pathophysiological role in Alzheimer's disease. Curr. Pharm. Des. 22, 6152–6166. doi: 10.2174/1381612822666160907093807
Leshchyns'ka, I., and Sytnyk, V. (2016). Synaptic cell adhesion molecules in Alzheimer's disease. Neural Plast. 2016:6427537. doi: 10.1155/2016/6427537
Li, D., and Yu, F. (2017). Peripheral inflammatory biomarkers and cognitive decline in older adults with and without Alzheimer's disease: a systematic review. J. Gerontol. Nurs. 2017, 1–7. doi: 10.3928/00989134-20170519-01
Logsdon, A. F., Erickson, M. A., Rhea, E. M., Salameh, T. S., and Banks, W. A. (2018). Gut reactions: how the blood-brain barrier connects the microbiome and the brain. Exp. Biol. Med. 243, 159–165. doi: 10.1177/1535370217743766
Lukiw, W. J. (2016a). Bacteroides fragilis lipopolysaccharide and inflammatory signaling in Alzheimer's disease. Front. Microbiol. 7:1544. doi: 10.3389/fmicb.2016.01544
Lukiw, W. J. (2016b). The microbiome, microbial-generated pro-inflammatory neurotoxins, and Alzheimer's disease. J. Sport Health Sci. 5, 393–396. doi: 10.1016/j.jshs.2016.08.008
Lukiw, W. J., Cong, L., Jaber, V., and Zhao, Y. (2018). Microbiome-derived lipopolysaccharide (LPS) selectively inhibits neurofilament light chain (NF-L) gene expression in human neuronal-glial (HNG) cells in primary culture. Front. Neurosci. 12:896. doi: 10.3389/fnins.2018.00896
Martinez-Conde, S. (2018). Cajal Institute (CSIC), Madrid Santiago Ramón y Cajal, the Young Artist Who Grew Up to Invent Neuroscience. Scientific American.
Montagne, A., Zhao, Z., and Zlokovic, B. V. (2017). Alzheimer's disease: a matter of blood-brain barrier dysfunction? J. Exp. Med. 214, 3151–3169. doi: 10.1084/jem.20171406
Parada Venegas, D., De la Fuente, M. K., Landskron, G., González, M. J., Quera, R., Dijkstra, G., et al. (2019). Short chain fatty acids (SCFAs)-mediated gut epithelial and immune regulation and its relevance for inflammatory bowel diseases. Front. Immunol. 10:277. doi: 10.3389/fimmu.2019.00277
Patrick, K. L., Bell, S. L., Weindel, C. G., and Watson, R. O. (2019). Exploring the “Multiple-hit Hypothesis” of neurodegenerative disease: bacterial infection comes up to bat. Front. Cell Infect. Microbiol. 9:138. doi: 10.3389/fcimb.2019.00138
Poeker, S. A., Geirnaert, A., Berchtold, L., Greppi, A., Krych, L., Steinert, R. E., et al. (2018). Understanding the prebiotic potential of different dietary fibers using an in vitro continuous adult fermentation model. Sci. Rep. 8:4318. doi: 10.1038/s41598-018-22438-y
Ramakrishna, C., Kujawski, M., Chu, H., Li, L., Mazmanian, S. K., and Cantin, E. M. (2019). Bacteroides fragilis polysaccharide A induces IL-10 secreting B and T cells that prevent viral encephalitis. Nat Commun. 10:2153. doi: 10.1038/s41467-019-09884-6
Rinninella, E., Raoul, P., Cintoni, M., Franceschi, F., Miggiano, G. A. D., Gasbarrini, A., et al. (2019). What is the healthy gut microbiota composition? A changing ecosystem across age, environment, diet, and diseases. Microorganisms. 7:E14. doi: 10.3390/microorganisms7010014
Rios-Covian, D., Salazar, N., Gueimonde, M., and de Los Reyes-Gavilan, C. G. (2017). Shaping the metabolism of intestinal Bacteroides population through diet to improve human health. Front. Microbiol. 8:376. doi: 10.3389/fmicb.2017.00376
Sarkar, R. S., and Banerjee, S. (2019). Gut microbiota in neurodegenerative disorders. J. Neuroimmunol. 328, 98–104. doi: 10.1016/j.jneuroim.2019.01.004
Sears, C. L. (2009). Enterotoxigenic Bacteroides fragilis: a rogue among symbiotes. Clin. Microbiol. Rev. 22, 349–369. doi: 10.1128/CMR.00053-08
Sender, R., Fuchs, S., and Milo, R. (2016). Revised estimates for the number of human and bacteria cells in the body. PLoS Biol. 14:e1002533. doi: 10.1371/journal.pbio.1002533
Sheppard, O., Coleman, M. P., and Durrant, C. S. (2019). Lipopolysaccharide-induced neuroinflammation induces presynaptic disruption through a direct action on brain tissue involving microglia-derived interleukin 1 beta. J. Neuroinflam. 16:106. doi: 10.1186/s12974-019-1490-8
Shivaji, S. (2017). We are not alone: a case for the human microbiome in extra intestinal diseases. Gut Pathog 9:13. doi: 10.1186/s13099-017-0163-3
Strandwitz, P., Kim, K. H., Terekhova, D., Liu, J. K., Sharma, A., Levering, J., et al. (2019). GABA-modulating bacteria of the human gut microbiota. Nat. Microbiol. 4, 396–403. doi: 10.1038/s41564-018-0307-3
Sweeney, M. D., Sagare, A. P., and Zlokovic, B. V. (2018). Blood-brain barrier breakdown in Alzheimer disease and other neurodegenerative disorders. Nat. Rev. Neurol. 14, 133–150. doi: 10.1038/nrneurol.2017.188
Sweeney, M. D., Zhao, Z., Montagne, A., Nelson, A. R., and Zlokovic, B. V. (2019). Blood-brain barrier: from physiology to disease and back. Physiol. Rev. 99, 21–78. doi: 10.1152/physrev.00050.2017
Ticinesi, A., Tana, C., and Nouvenne, A. (2019). The intestinal microbiome and its relevance for functionality in older persons. Curr. Opin. Clin. Nutr. Metab. Care 22, 4–12. doi: 10.1097/MCO.0000000000000521
Tierney, B. T., Yang, Z., Luber, J. M., Beaudin, M., Wibowo, M. C., et al. (2019). The landscape of genetic content in the gut and oral human microbiome. Cell Host Microbe 26, 283–295.e8. doi: 10.1016/j.chom.2019.07.008
Tulkens, J., Vergauwen, G., Van Deun, J., Geeurickx, E., Dhondt, B., Lippens, L., et al. (2018). Increased levels of systemic LPS-positive bacterial extracellular vesicles in patients with intestinal barrier dysfunction. Gut 69, 191–193. doi: 10.1136/gutjnl-2018-317726
Varatharaj, A., and Galea, I. (2017). The blood-brain barrier in systemic inflammation. Brain Behav. Immun. 60, 1–12. doi: 10.1016/j.bbi.2016.03.010
Venter, J. C., Adams, M. D., Myers, E. W., Li, P. W., Mural, R. J., Sutton, G. G., et al. (2001). The sequence of the human genome. Science 291, 1304–1351. doi: 10.1126/science.1058040
Vines, R. R., Perdue, S. S., Moncrief, J. S., Sentz, D. R., Barroso, L. A., Wright, R. L., et al. (2000). Fragilysin, the enterotoxin from Bacteroides fragilis, enhances the serum antibody response to antigen co-administered by the intranasal route. Vaccine 19, 655–660. doi: 10.1016/S0264-410X(00)00254-1
Wexler, A. G., and Goodman, A. L. (2017). An insider's perspective: Bacteroides as a window into the microbiome. Nat. Microbiol. 2:17026. doi: 10.1038/nmicrobiol.2017.26
Wong, A. D., Ye, M., Levy, A. F., Rothstein, J. D., Bergles, D. E, and Searson, P. C. (2013). The blood-brain barrier: an engineering perspective. Front. Neuroeng. 6:7. doi: 10.3389/fneng.2013.00007
Wu, S., Lim, K. C., Huang, J., Saidi, R. F., and Sears, C. L. (1998). Bacteroides fragilis enterotoxin cleaves the zonula adherens protein, E-cadherin. Proc. Natl. Acad. Sci. U.S.A. 95, 14979–14984. doi: 10.1073/pnas.95.25.14979
Wu, S., Rhee, K. J., Zhang, M., Franco, A., and Sears, C. L. (2007). Bacteroides fragilis toxin stimulates intestinal epithelial cell shedding and gamma-secretase-dependent E-cadherin cleavage. J Cell Sci. 120(Pt 11), 1944–52. doi: 10.1242/jcs.03455
Zhan, X., Stamova, B., Jin, L. W., DeCarli, C., Phinney, B., and Sharp, F. R. (2016). Gram-negative bacterial molecules associate with Alzheimer disease pathology. Neurology 87, 2324–2332. doi: 10.1212/WNL.0000000000003391
Zhan, X., Stamova, B., and Sharp, F. R. (2018). Lipopolysaccharide associates with amyloid plaques, neurons and oligodendrocytes in Alzheimer's disease brain: a review. Front. Aging Neurosci. 10:42. doi: 10.3389/fnagi.2018.00042
Zhao, Y., Cong, L., Jaber, V., and Lukiw, W. J. (2017c). Microbiome-derived lipopolysaccharide enriched in the perinuclear region of Alzheimer's disease brain. Front. Immunol. 8:1064. doi: 10.3389/fimmu.2017.01064
Zhao, Y., Cong, L., and Lukiw, W. J. (2017a). Lipopolysaccharide (LPS) accumulates in neocortical neurons of Alzheimer's disease (AD) brain and impairs transcription in human neuronal-glial primary co-cultures. Front. Aging Neurosci. 9:407. doi: 10.3389/fnagi.2017.00407
Zhao, Y., Dua, P., and Lukiw, W. J. (2015). Microbial sources of amyloid and relevance to amyloidogenesis and Alzheimer's disease (AD). J Alzheimers Dis Parkinsonism. 5:177.
Zhao, Y., Jaber, V., and Lukiw, W. J. (2017b). Secretory products of the human GI tract microbiome and their potential impact on Alzheimer's disease (AD): detection of lipopolysaccharide (LPS) in AD hippocampus. Front. Cell Infect. Microbiol. 7:318. doi: 10.3389/fcimb.2017.00318
Zhao, Y., and Lukiw, W. J. (2015). Microbiome-generated amyloid and potential impact on amyloidogenesis in Alzheimer's disease (AD). J. Nat. Sci. 1:e138.
Zhao, Y., and Lukiw, W. J. (2018a). Bacteroidetes neurotoxins and inflammatory neurodegeneration. Mol. Neurobiol. 55, 9100–9107. doi: 10.1007/s12035-018-1015-y
Zhao, Y., and Lukiw, W. J. (2018b). Microbiome-mediated upregulation of microRNA-146a in sporadic Alzheimer's disease. Front. Neurol. 9:145. doi: 10.3389/fneur.2018.00145
Zhao, Y., Sharfman, N. M., Jaber, V. R., and Lukiw, W. J. (2019). Down-regulation of essential synaptic components by GI-tract microbiome-derived lipopolysaccharide (LPS) in LPS-treated human neuronal-glial (HNG) cells in primary culture: relevance to Alzheimer's disease (AD). Front. Cell Neurosci. 13:314. doi: 10.3389/fncel.2019.00314
Keywords: Alzheimer's disease (AD), Bacteroides fragilis and BFT (fragilysin), dysbiosis, lipopolysaccharide (LPS), microbiome and microbial genetics, neurofilament light (NF-L), neuroinflammation, synapsin-2 (SYN2)
Citation: Lukiw WJ (2020) Gastrointestinal (GI) Tract Microbiome-Derived Neurotoxins—Potent Neuro-Inflammatory Signals From the GI Tract via the Systemic Circulation Into the Brain. Front. Cell. Infect. Microbiol. 10:22. doi: 10.3389/fcimb.2020.00022
Received: 20 August 2019; Accepted: 15 January 2020;
Published: 12 February 2020.
Edited by:
Ruth Ann Luna, Baylor College of Medicine, United StatesReviewed by:
J. Christopher Fenno, University of Michigan, United StatesCopyright © 2020 Lukiw. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Walter J. Lukiw, d2x1a2l3QGxzdWhzYy5lZHU=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.