
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Cell. Infect. Microbiol. , 13 September 2019
Sec. Molecular Bacterial Pathogenesis
Volume 9 - 2019 | https://doi.org/10.3389/fcimb.2019.00326
This article is part of the Research Topic Decoding the Streptococcal Language View all 6 articles
 Anfal Shakir Motib1,2
Anfal Shakir Motib1,2 Firas A. Y. Al-Bayati1,3
Firas A. Y. Al-Bayati1,3 Irfan Manzoor4
Irfan Manzoor4 Sulman Shafeeq4
Sulman Shafeeq4 Anagha Kadam5
Anagha Kadam5 Oscar P. Kuipers4
Oscar P. Kuipers4 N. Luisa Hiller5
N. Luisa Hiller5 Peter W. Andrew1
Peter W. Andrew1 Hasan Yesilkaya1*
Hasan Yesilkaya1*Streptococcus pneumoniae is able to cause deadly diseases by infecting different tissues, each with distinct environmental and nutritional compositions. We hypothesize that the adaptive capabilities of the microbe is an important facet of pneumococcal survival in fluctuating host environments. Quorum-sensing (QS) mechanisms are pivotal for microbial host adaptation. We previously demonstrated that the TprA/PhrA QS system is required for pneumococcal utilization of galactose and mannose, neuraminidase activity, and virulence. We also showed that the system can be modulated by using linear molecularly imprinted polymers. Due to being a drugable target, we further studied the operation of this QS system in S. pneumoniae. We found that TprA controls the expression of nine different operons on galactose and mannose. Our data revealed that TprA expression is modulated by a complex regulatory network, where the master regulators CcpA and GlnR are involved in a sugar dependent manner. Mutants in the TprA/PhrA system are highly attenuated in their survival in nasopharynx and lungs after intranasal infection, and growth in blood after intravenous infection.
Streptococcus pneumoniae causes an array of diseases with high morbidity and mortality including pneumonia, bacteraemia, meningitis, and otitis media (Weiser et al., 2018), especially among children and elderly. The high incidence of pneumococcal diseases is compounded by a rising trend of antimicrobial resistance among pneumococcal strains. This requires new approaches to develop anti-infectives against this microbe. Adaptation to changing environmental conditions has been linked to pneumococcal virulence (Sandrini et al., 2014; Kadam et al., 2017; Zhi et al., 2018) hence, pneumococcal proteins involved in detection and processing of environmental signals may be targeted to develop new anti-infectives.
Upon detection of environmental alterations, bacteria adjust their gene expression in order to obtain a phenotype that is suitable for the environment to which they are exposed (Federle, 2012). Transcriptional regulators enable the microbes to sense and respond to environmental stimuli, by altering gene expression. While individual cells have inherent adaptive capability, in certain cases, the adaptation to a given stimulus occurs at the population level via detection and processing of chemical signals. Community level coordination of gene expression is known as quorum sensing (QS). QS systems have been implicated in wide range of biological functions, including surface attachment, oxidative stress resistance, microbial competition, and virulence (Federle, 2012; Cuevas et al., 2017; Zhi et al., 2018). These systems have received considerable attention not least because of their far-reaching impact on the fundamental biology of microbes but also from the more applied perspective for their utility as anti-infective targets (Brooks and Brooks, 2014).
In the pneumococcus, the competence (com) locus and LuxS-mediated AI-2 are the best-studied QS systems (Alloing et al., 1996; Trappetti et al., 2017). However, functional genomic studies have revealed several other QS systems in S. pneumoniae (Hoover et al., 2015; Cuevas et al., 2017; Junges et al., 2017; Kadam et al., 2017; Motib et al., 2017; Aggarwal et al., 2018; Zhi et al., 2018). Prominent among these is the RRNPP (Rap, Rgg, NprR, PlcR, PrgX) family of transcriptional regulators, which are characterized at the structural level by the presence of tetratricopeptide repeats that mediate regulator-peptide interactions (Cook and Federle, 2014). Recently, we and others characterized several Rgg (Rgg144, Rgg939) and TprA (TprA, TprA2) family regulators in S. pneumoniae (Hoover et al., 2015; Cuevas et al., 2017; Junges et al., 2017; Kadam et al., 2017; Motib et al., 2017; Zhi et al., 2018). We found that Rgg144 and Rgg939 QS circuits are induced by short hydrophobic peptides (Shp) upon detection of galactose or mannose (Zhi et al., 2018). Microarray analyses revealed that the expression of a large number of genes involved in essential physiological functions and virulence is controlled by Rgg/Shp QS systems. Interestingly, our data revealed evidence for cross-talk between these systems, and showed the involvement of Rgg systems in colonization and virulence (Zhi et al., 2018). Additionally, we studied a novel streptococcal cell-cell communication peptide, known as virulence peptide 1 (VP1), regulated by Rgg/Shp144, which promotes pneumococcal virulence and biofilm formation (Cuevas et al., 2017).
TprA/PhrA is another member of the RRNPP family found in pneumococci. Hoover et al. showed PhrA/TprA QS system's role in galactose metabolism and the modulation of a predicted lantibiotic gene cluster (Hoover et al., 2015). TprA is a PlcR ortholog (Hoover et al., 2015), first characterized in Bacillus, and reported to control expression of genes for most of their extracellular virulence factors (Agaisse et al., 1999; Gohar et al., 2008). The function of this regulator relies on the existence of a signaling peptide (PapR), the gene of which is located about 70 bp downstream from plcR (Gominet et al., 2001). Once imported, PapR binds PlcR, modifying its interaction with the PlcR box, located in the promoter regions of PlcR regulated genes (Rocha-Estrada et al., 2010), and controlling their transcription. The number of TprA/PhrA homologs varies across pneumococcal lineages (Kadam et al., 2017). Specifically, we have shown that strains in the PMEN1 lineage (also known as Spain23F-1 and SPN23F) encode not only TprA/PhrA, but also the homologous TprA2/PhrA2 signaling systems. We showed that TprA2/PhrA2 is a negative regulator of a genomic locus encoding a lanthionine-containing peptide (lcpA), and the presence of this QS system restrains the virulence of PMEN1 through the control exerted over lcpA expression. Interestingly, PhrA2 can induce the expression of the TprA/PhrA system not only in PMEN1 strains, but also in type 2 strain D39 from a divergent lineage. This suggests that this feature may allow PMEN1 strains to modify the transcriptomic and functional attributes of a multi-strain pneumococcal community. Given that an estimated 20–40% of pneumococcal infections are multi-strains, the implication of this finding is considerable for understanding pneumococcal virulence and anti-infective design (Everett et al., 2012; Saha et al., 2015).
While the TprA2/PhrA2 system restrains virulence in a mouse colonization model, the TprA/PhrA strain is essential for virulence in the type 2 D39 strain, as we demonstrated in our recent study using the models of mouse pneumococcal pneumonia and chinchilla otitis media (Kadam et al., 2017; Motib et al., 2017). It was shown by us, and others, that PhrA levels are repressed in glucose and raised in galactose and that the TprA/PhrA system controls the expression of a predicted lantibiotic gene cluster that encodes members of the bacteriocin family of antimicrobial peptides (Hoover et al., 2015). Our data also indicated that TprA/PhrA controls a neuraminidase gene expression and synthesis when the pneumococcus is grown on galactose (Motib et al., 2017). Moreover, we were able to modulate the function of TprA/PhrA system using linear molecularly imprinted polymers (LMIP), which interferes with the interaction of TprA/PhrA, thus, abrogating phenotypic manifestation of the system both in vitro and in vivo (Motib et al., 2017). This clearly showed that TprA/PhrA is a drugable anti-infective target. Therefore, to extend our knowledge of this QS system, we studied the regulatory interactions between TprA and PhrA, and assigned a role to CcpA and GlnR in regulation of TprA/PhrA. In addition, we have further evaluated TprA/PhrA's role in pneumococcal growth in the nasopharynx and lungs, and in septicaemia.
The strains used in this study are listed in Table S1. S. pneumoniae D39 was grown microaerobically at 37°C either in brain heart infusion broth (BHI), blood agar base (BAB) (Oxoid, UK) supplemented with 5% (v/v) defibrinated horse blood, or in chemically defined medium (CDM) supplemented with 55 mM of the selected sugar (Yesilkaya et al., 2006, 2008). For anaerobic growth, an anaerobic cabinet was used. Escherichia coli was grown in Luria broth with shaking at 37°C, or in Luria agar (Oxoid). Growth was monitored by measuring the OD600, or by determining colony counts. Growth rates (μ) were calculated through linear regressions of the plots of ln(OD600) vs. time during the exponential growth phase. Spectinomycin, tetracycline, and kanamycin were added at 100, 15, and 250 μg/mL, respectively, for pneumococcal cultures, and for E. coli ampicillin and kanamycin were used at 100 and 50 μg/mL, respectively.
Insertion-deletion mutants were constructed by transforming an in vitro mutagenized SOEing construct as described before (Al-Bayati et al., 2017). Briefly, a spectinomycin (SpecR) resistance gene cassette was amplified using the primes Spec-F and Spec-R (Table S2). The primers LF-SOEX-F/LF-SOEX-R and RF-SOEX-F/RF-SOEX-R (where X indicates the gene code) were used to amplify the left and right flanking regions of each target gene, generating PCR products of ~600 bp. The amplified antibiotic resistance cassette and the PCR products flanking the target gene were then fused using LF-SOEX-F and RF-SOEX-R primers. The fused product was gel-purified after electrophoresis (Qiagen) and transformed into S. pneumoniae (Bricker and Camilli, 1999). The transformants were selected on BAB plates containing the appropriate antibiotic and the mutations were confirmed by PCR and sequencing. To rule out the polar effect of mutation, an intact copy of tprA with its putative promoter was introduced into ΔtprA using the pCEP plasmid (Guiral et al., 2006; Table S1).
Chromosomal transcriptional lacZ fusions to the target promoters were constructed by using the integration plasmid pPP1 (Halfmann et al., 2007). The putative promoter regions were amplified using the primers modified to incorporate SphI and BamHI sites (indicated with Fusion-F/R in Table S2, and then ligated into similarly digested pPP1. All plasmid constructs were confirmed by sequencing, and integrated into pneumococcal genome via double crossover. β-galactosidase activity was measured as described before using cells grown anaerobically in CDM at 37°C supplemented with 55 mM of the selected sugars (Kadam et al., 2017; Motib et al., 2017).
The Trizol reagent kit (Invitrogen, UK) was used to isolate the total RNA. First strand cDNA synthesis was done at 42°C for 50 min using SuperScript III reverse transcriptase and random hexamers, according to the manufacturer's instructions (Invitrogen, UK). cDNA (15 ng) was amplified in a 20 μL reaction volume containing 1X SensiMix SYBR Master mix (Bioline, UK) and 3 pmol of each primer (Table S2). The transcription level of specific genes was normalized to gyrB transcription, which was amplified in parallel with SPD0709F and SP0709R primers. The results were analyzed by the comparative CT method (Livak and Schmittgen, 2001).
Streptococcus pneumoniae D39 and its isogenic mutant strain ΔtprA were grown microaerobically in CDM supplemented either with 55 mM galactose or mannose as the main carbon source (Gaspar et al., 2014), or in M17 medium supplemented with 0.5% glucose (Difco) The experiments were repeated with four biological replicates. The MicroPrep software package was used to obtain the microarray data from the slides. CyberT implementation of a variant of the t-test (http://wiki.gcc.rug.nl/wiki/GccStart) was performed and false discovery rates (FDRs) were calculated (Gominet et al., 2001). For differentially expressed genes, p < 0.001 and FDR < 0.05 were taken as standard. For the identification of differentially expressed genes, a Bayesian p-value of < 0.001 and a fold change cut-off 2-fold was applied. All other procedures for the DNA microarray experiments and data analysis were performed as described before (Shafeeq et al., 2015). Microarray data have been submitted to GEO (Gene Expression Omnibus) database under the accession numbers GSE 67668 and GSE133010.
ccpA, tprA, and glnR coding regions were amplified by PCR, and the amplicons were cloned into pLEICS-01 (Al-Bayati et al., 2017). The recombinant plasmid was transformed into E. coli BL21 (DE3) and gene expression was induced with 0.5 mM IPTG at 25°C. Recombinant proteins were then purified using Talon® Metal Affinity resin (Clontech Inc., UK) as previously described (Zhi et al., 2018). The identity of the purified recombinant proteins was achieved by matrix-assisted laser desorption ionization—time of flight (MALDI-TOF) done by PNACL (University of Leicester).
Neuraminidase activity was assayed using chromogenic 2-O-(p-nitrophenyl) -α-D-N-acetylneuraminic acid (pNP- NANA) (Sigma, United Kingdom), as previously described (Yesilkaya et al., 2006). The absorbance of released p-nitrophenol was measured photometrically at 405 nm. The protein concentration was measured according to the method described by Bradford (1976). One unit of enzyme activity was defined as the amount of enzyme that produced 1 μmol p-nitrophenol per min per milligram of protein under standard assay conditions.
Putative promoter regions and the presence of regulatory elements of target genes were identified using the bacterial promoter prediction tool (BPROM) (Solovyev et al., 2010) and the motif-based sequence analysis tools (MEME). The putative promoter regions were amplified using EMSA-F/R primers (Table S2).
The binding reaction was set up by mixing a constant amount of target promoter probes (~10 nmol), and increasing amounts (0.1–0.5 μM) of purified and dialyzed His-tagged proteins, in the presence of 5X binding buffer (20 mM Tris-HCl pH 7.5, 30 mM KCl, 1 mM DTT, 1 mM EDTA pH 8.0 and 10% v/v glycerol) at room temperature for 20 min in a total volume of 20 μL. The binding reaction mixture was run on an 8% w/v non-denaturing polyacrylamide gel. The detection of DNA-protein complexes relied on SYBR green staining (Molecular Probes fluorescence-based EMSA kit, Invitrogen), or the use of 5′-FAM labeled DNA probe, obtained with primers that incorporate fluorescently labeled tags. Typhoon Trio+ scanner (GE Healthcare Life Sciences) with a 526 nm short-pass wavelength filter was used for visualizing DNA-protein complexes.
Female, 8–10 week old MF1 mice weighing ~30–35 g (Charles River, UK), were anesthetized with 2.5% v/v isoflurane (Astra Zeneca, Macclesfield, UK) over oxygen (2–2.5 liter/min). For the pneumonia model, ~1.5 × 106 pneumococci, in 50 μL PBS (Gaspar et al., 2014; Glanville et al., 2018) were administered dropwise into the nostrils of the anesthetized mouse, held vertically. After infections, CFU were determined to ensure that the intended number of pneumococci had been administered. At 24 and 48 h post-infection, a pre-selected group of mice was culled under terminal anesthesia. Nasopharyngeal tissues and the lungs were removed, homogenized, and pneumococcal numbers determined by plating out the serial dilutions of the homogenates.
For intravenous infection, mice were infected with ~1 × 106 CFU in 100 μL PBS, pH 7.0, through a dorsal tail vein. Mice were observed for disease signs over 168 h. In addition, at 24 and 48 h post-infection, a sample of blood was taken from a tail vein and the CFU/ml was determined by plating serial dilutions.
Mouse experiments at the University of Leicester were performed under appropriate project (permit no. 60/4327) and personal (permit no. 80/10279) licenses according to the United Kingdom Home Office guidelines under the Animals Scientific Procedures Act 1986, and the University of Leicester ethics committee approval. The protocol was approved by both the U.K. Home Office and the University of Leicester ethics committee. Where indicated, the procedures were carried out under anesthesia with isoflurone. Animals were housed in individually ventilated cages in a controlled environment and were frequently monitored after infection. In bacterial infection experiments mice were humanely culled immediately if they became lethargic.
The TprA/PhrA system is induced by host-derived sugars, so we studied the influence of TprA on phrA in different sugar sources. To this end we utilized a LacZ reporter assay and isogenic mutants. For this, PtprA and PphrA driven ß-galactosidase activity was measured in wild type or in ΔtprA and ΔphrA. The results showed that in the condition where the tprA promoter activity is high, that is in presence of mannose and galactose, TprA reduces PphrA-driven ß-galactosidase activity relative to its expression in the wild type D39 strain, indicating that TprA is a repressor of phrA (p < 0.01 and p < 0.001 in mannose and galactose, respectively) (Figure 1). In glucose or N-acetylglucosamine, where the PtprA activity is low, TprA does not have any effect on phrA expression (Figure 1). Moreover, it was also found that phrA activates its own expression on mannose and galactose, when its promoter activity is up, but this regulatory effect could not be observed on glucose and N-acetylglucosamine (Figure 1).
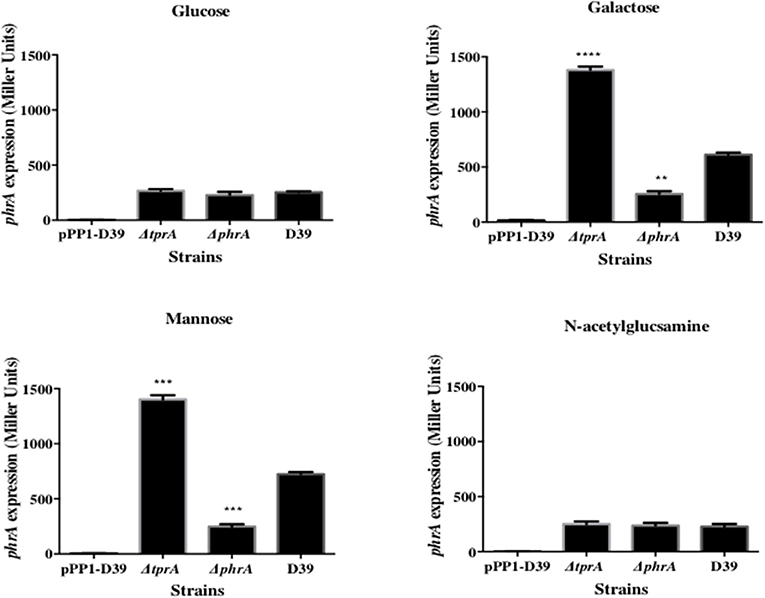
Figure 1. Analysis of phrA regulation in different sugars using a LacZ reporter assay. The activity of ß-glactosidase under regulation of PphrA is expressed in nmol p-nitrophenol/min/mL (Miller Units). A strain containing promoterless pPP1 plasmid (pPP1-D39) was included as a control in the assay. Values are average of three independent experiments each with three replicates. Bars indicate standard error of mean (SEM). ** p < 0.01, *** p < 0.001, **** p < 0.0001.
TprA may also positively regulate its own expression (Figure 2). In glucose or N-acetylglucosamine, where TprA levels are low, the absence of tprA or phrA does not influence tprA gene expression levels. In contrast, in galactose and mannose where TprA levels are high, deletion of tprA leads to a decrease in tprA promoter expression, consistent with autoinduction of TprA. Deletion of phrA does not have a significant influence on tprA gene expression consistent with TprA also having PhrA-independent activity. Thus, expression of the TprA regulon must depend on the balance between PhrA bound and unbound forms (Figure 2).
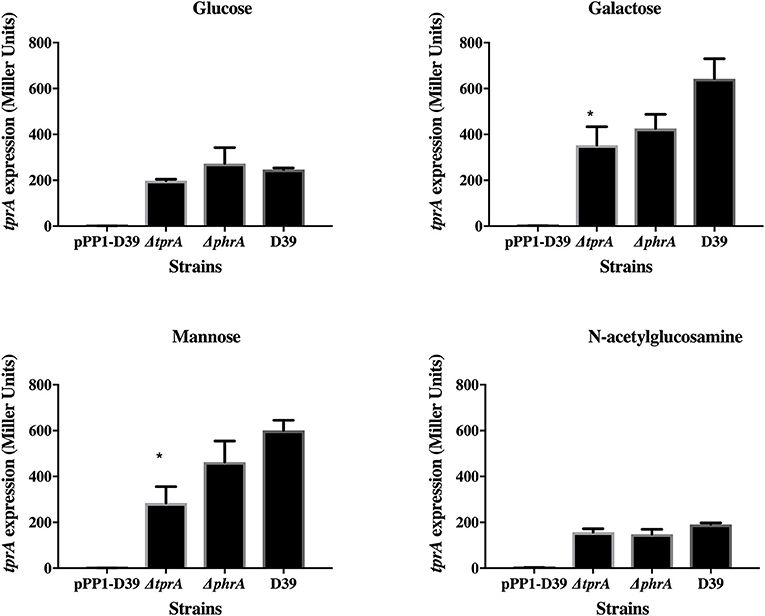
Figure 2. Analysis of tprA regulation in different sugars using a LacZ reporter assay. The activity of ß-glactosidase under regulation of PtprA is expressed in nmol p-nitrophenol/min/mL (Miller Units). A strain containing promoterless pPP1 plasmid (pPP1-D39) was included as a control in the assay. Values are average of three independent experiments each with three replicates. Bars indicate standard error of mean (SEM). * p < 0.05.
To analyse the regulon of TprA we selected galactose and mannose growth conditions, where TprA is predicted to be playing an inhibitory role, and where both the TprA promoter and PhrA promoter activities are high. We compared gene expression in wild type and ΔtprA mutants using a microarray chip. The results showed that in galactose, 61 genes were differentially expressed, of these 46 were up-regulated, and 15 were down regulated in the mutant (Tables S3–S6). The up-regulated genes could be divided into eight putative operons. The most up-regulated was 11 genes, SPD_1747-SPD_1756, downstream of phrA, including a bacteriocin synthesis cluster, that showed between 5- and 73-fold change. This locus is predicted to synthesize lantibiotics and its change in expression in the tprA mutant was also reported by Hoover et al. (2015). Lantibiotics are secreted cyclic peptides produced by various bacterial species, and many function as bacteriocins (Willey and van der Donk, 2007). Pneumococci encode a large number of predicted bacteriocins, which have been linked to inter and intra species competitions (Dawid et al., 2007; Rezaei et al., 2018; Wholey et al., 2019). PTS and ABC transporters involved in galactose and mannose uptake, and mannose metabolism were also up-regulated by 2- to 4.5 fold. Additionally, a cluster of genes, SPD_1944-SPD_1948, was up-regulated by 3–8 fold in the mutant. This locus contains a gene for synthesis of a putative CAAX amino terminal protease, for a hypothetical protein, a membrane protein and a putative transcriptional regulator. Previously we demonstrated the expression of this locus was also raised in a mutant of pflB, which codes for pyruvate formate lyase, indicating that this locus has a role in control of the mixed acid fermentation seen when the pneumococcus is propagated on galactose or mannose (Al-Bayati et al., 2017). The notably downregulated genes included eight tRNA genes and a GlnR family transcriptional regulator.
On mannose, 29 genes were differentially regulated. Off these 21 were up-regulated and eight were down-regulated. The upregulated genes include three operons: a locus containing putative genes possibly involved in membrane protein synthesis (SPD_1514-SPD_1519), the downstream bacteriocin loci, and the hypothetical set with a CAAX protease. The two latter sets were also observed in the galactose, but the fold changes observed between the presence and absence of TprA, are less accentuated in mannose than galactose. The notably downregulated genes on mannose included SPD_0093, SPD_0094, and SPD_0095. These genes code for hypothetical proteins of unknown function, and are highly conserved in the fully sequenced pneumococcal genomes (Liu et al., 2017). Haas et al. (2005) reported that exposure to a lethal concentration of vancomycin led to significant alteration in the transcription of these genes in multiple S. pneumoniae strains.
Direct interaction of TprA with the differentially expressed genes was analyzed by EMSA. To do this, fluorescently labeled DNA probes representing the putative promoters of phrA (SPD_1746), a hypothetical protein upregulated in mannose (SPD_1517), and a L-fucose phosphate aldolase upregulated in galactose (SPD_1994) were prepared and mixed with different concentration of recombinant TprA. The result showed that in the presence of TprA, the mobility of labeled probes has changed compared to that of DNA alone, indicative of a direct interaction between TprA and these DNA probes (Figure S1). TprA binding was specific, as the recombinant protein could not bind to the putative promoter of gyrB, which is not within TprA regulon.
As TprA/PhrA expression is influenced by different sugars, we reasoned that the system may be controlled by other transcriptional regulators involved in regulation of sugar metabolism, such as CcpA, which is the main transcriptional regulator of carbon catabolite control in S. pneumoniae and functions by binding to DNA at catabolite-responsive element (cre) sites. Further support for this analysis originated from the fact that a cre-like sequence is located in the putative promoter region of tprA (Hoover et al., 2015; Figure 3). In addition to CcpA, GlnR's (encoded by SPD_0447) role in regulation of tprA, was also investigated as glnR was found to be differentially expressed in ΔtprA relative to wild type (Table S3). We demonstrated by electrophoretic mobility shift assay (EMSA) analysis that both CcpA and GlnR bind to the putative promoter region of tprA (Figure 3), indicating direct interaction of CcpA and GlnR with the putative promoter of tprA (PtprA). This binding was specific as CcpA and GlnR could not bind to upstream region of gyrB (data not shown).
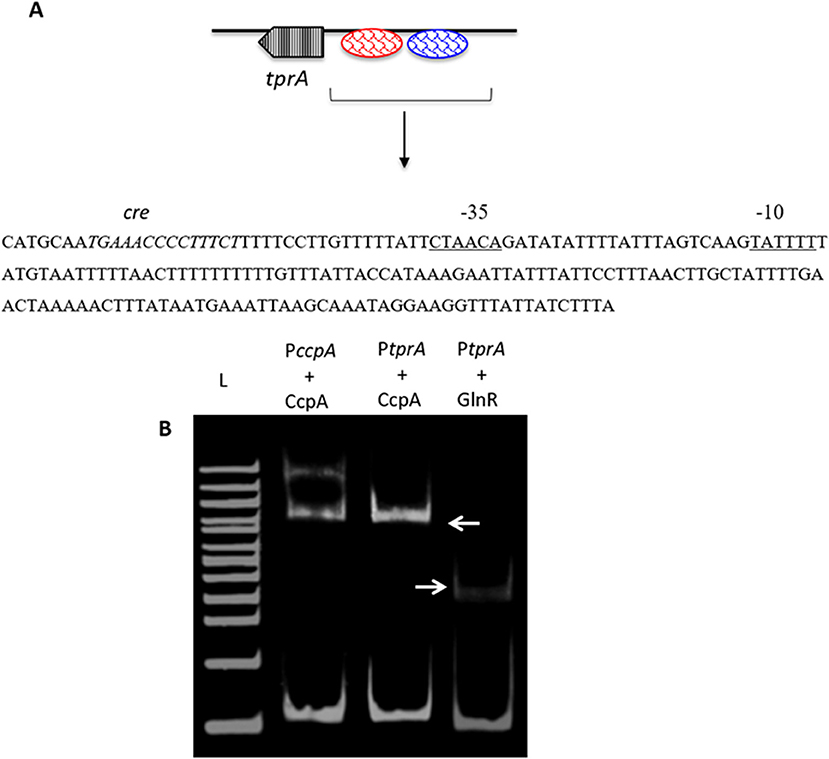
Figure 3. Electrophoretic mobility shift assay showing the direct interaction of His-CcpA and His-GlnR with the putative promoter region of tprA. (A) Illustration showing the analysis of the predicted promoter region of PtprA used in EMSA. The core promoter regions, −10 and −35 sequences, are underlined, and the putative transcriptional factor binding sites are shown in blue, and the putative cre sequence is in red. (B) Direct interaction of 0.5 μM CcpA and GlnR with the putative promoter sequence of tprA. CcpA binding to its own promoter containing cre sequence was used as a control (lane L- 500 ng of 100 bp DNA ladder (NEB). Gels were stained with SYBR Green EMSA for visualizing DNA. The arrows indicate the mobility shift.
To test the influence of CcpA and GlnR on tprA regulation further, we used PtprA reporter strains. To this end, PtprA was fused to a promoterless lacZ and reporter strains were created in wild type (PtprA::lacZ-wt), ΔccpA (PtprA::lacZ-ΔccpA), and ΔglnR (PtprA::lacZ-ΔglnR). We found that the PtprA activity in PtprA-lacZ-ΔglnR was lower than in PtprA-lacZ-wt in both glucose and galactose in the presence or absence of oxygen. This finding suggests that GlnR is a positive regulator of PtprA activity and active in multiple sugars and oxygen conditions (Table 1).
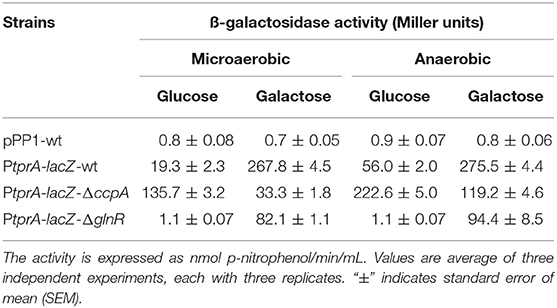
Table 1. Expression levels (in Miller units) of pneumococcal transcriptional lacZ-fusions to the promoter of tprA in different genetic backgrounds grown microaerobically and anaerobically in CDM supplemented with 55 mM of glucose or galactose.
On the other hand, PtprA activity in PtprA-lacZ-ΔccpA was lower than PtprA-lacZ-wt in galactose, and higher than PtprA-lacZ-wt in glucose. CcpA is a known repressor, and these findings suggest that it represses PtprA expression on glucose and de-represses on galactose (Table 1). These data show that TprA regulation is controlled by multiple regulators, in that regulation via CcpA is highly dependent on carbohydrate sources and promotes tprA expression on galactose.
Previously, we reported the essential role of TprA/PhrA system in pneumococcal virulence after intranasal infection but did not determine the growth profiles of pneumococcal strains in the nasopharynx, lungs and blood during the course of infection (Motib et al., 2017). In this study, we evaluated TprA/PhrA's role in growth of S. pneumoniae in the respiratory tract after intranasal infection, and in blood after intravenous infection by following growth of pneumococci in these tissues. The growth profiles of pneumococcal strains in the nasopharynx and lung, were determined after intranasal infection. In the nasopharynx, the numbers of wild-type D39 bacteria increased gradually by 24 (Log10 3.52 ± 0.12 CFU/mg, n = 5) and 48 h (Log10 4.02 ± 0.08 CFU/mg, n = 5) post-infection (Figure 4A). In contrast, the numbers of ΔtprA and ΔphrA did not change at 24 and 48 h post infection (Figure 4A). The difference between the number of bacteria recovered from the wild type and the mutant strains was highly significant (p < 0.0001). In the lungs, the numbers of wild type increased approximately two log over 48 h, while the numbers of ΔphrA did not change significantly at 24 and 48 h post infection relative to the numbers determined immediately after post-infection (p > 0.05). Moreover, the ΔtprA was cleared 24 h post infection. These data show that both TprA and PhrA are required for pneumococcal growth in the respiratory tract, however the relative contributions of the individual components of this QS system in pneumococcal survival differ.
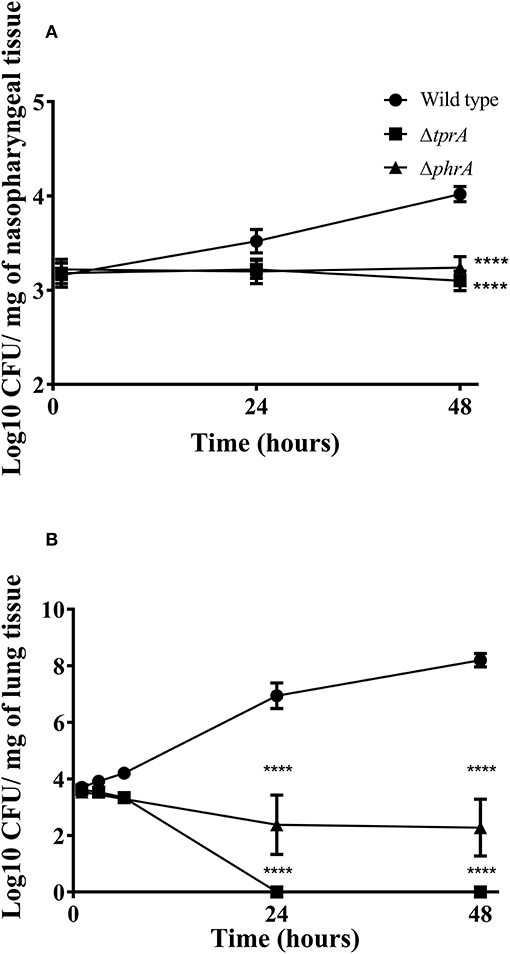
Figure 4. Survival of pneumococcal strains in nasopharynx (A) and lungs (B). Mice were infected intranasally with ~1 × 106 CFU/mouse. Each datum point was collected from five animals. Bars indicate standard error of mean (SEM). ****p < 0.0001.
The in vitro data suggest that TprA/PhrA should not play a key role in after intravenous inoculation, given the higher levels of glucose in the blood. To test this, we compared the wild type, ΔtprA and ΔphrA in a systemic infection model by administering the strains through the dorsal tail vein. The results showed that all the mice infected either with ΔtprA or ΔphrA survived the challenge (168 h), whereas five out of six mice infected with D39 reached the assay end point (median survival: 58 ± 48.8 h, n = 6). Moreover, the CFU/ml of D39 at 24- and 48 h post infection was Log10 3.25 ± 0.56 (n = 6) and Log10 5.95 ± 1.36 (n = 6), respectively, whereas ΔtprA and ΔphrA could not be detected in the blood at these time points. These data demonstrate that the role of the TprA/PhrA extends beyond galactose metabolism contributing to metabolic function in diverse conditions.
In order to explore the reasons behind the growth attenuation of the mutant in blood, we determined, by microarray analysis, the transcriptome of ΔtprA on glucose, because the microbe can be exposed to up to 10 mM glucose in blood (Philips et al., 2003). The analysis showed that 53 genes were differentially expressed in ΔtprA relative to the wild type: 25 down-regulated and 28 up-regulated in the mutant compared to the wild type, affecting five putative operons, and several other genes with various metabolic functions (Tables S7, S8). In these five putative operons, the expression was up in four affecting the expression of genes involved in the synthesis of dihydroorotate dehydrogenase, endo-beta-N-acetylglucosaminidase (lytB) (SPD_0852-SPD_0853), iron transporters (SPD_0916-SPD_0917), aminotransferase (SPD_0978-SPD_0979), and carbamoyl-phosphate synthase (SPD_1132- SPD_1133), while it was down in one operon affecting a locus responsible for the synthesis of heat shock proteins (SPD_0458-SPD_0460). Other notable differentially expressed genes included transcriptional regulators (comX1, SPD_0144, spxA, hrcA), the genes encoding for several choline binding proteins including cbpE and pcpA, and 18 hypothetical genes with unknown function. These data demonstrate that the TprA/PhrA system has a role in wider metabolic functions beyond galactose metabolism.
The mechanism of pneumococcal adaptation in tissues with different sugar composition is not fully understood. We demonstrate that the TprA/PhrA system is one of the regulatory systems involved in metabolism of galactose and mannose, and is essential for pneumococcal survival in tissues. This system can be targeted by linear molecularly imprinted polymers to modulate the consumption of host-derived sugars, the activity of neuraminidase, and pneumococcal survival in the host (Motib et al., 2017).
Previously we demonstrated that the TprA/PhrA system is essential for virulence in a mouse model of pneumonia, as well as in a chinchilla otitis media model (Motib et al., 2017). The results obtained in this study clearly demonstrate that in the absence of this QS system, the pneumococcus cannot grow in the nasopharynx and lungs. The effect of TprA mutation was severe on pneumococcal survival particularly in the lungs because it was cleared off after 24 h post infection. While ΔphrA could be detected in the lungs, these pneumococci could not successfully disseminate into the blood (Motib et al., 2017). This shows that TrpA requires PhrA binding for its full function. Growth attenuation of phrA mutant is probably due to the lack of activation of TprA/PhrA QS system. However, TprA independent functions of PhrA cannot be ruled out, and this needs to be further investigated. Cross regulatory interaction among the members of RRNPP family are well-documented (Cook and Federle, 2014; Zhi et al., 2018).
The TprA/PhrA system is induced on galactose and mannose, sugars found predominantly in the structure of host glycans in respiratory secretions (King et al., 2006). However, this does not exclude TprA/PhrA system's involvement in other metabolic functions. Both ΔtprA and ΔphrA were avirulent in septicaemia model, and thus, given that glucose concentration is higher than galactose and mannose in blood, abrogation of virulence is unlikely due only to effects on galactose and mannose metabolism. Thus, we conclude that TprA has functions other than its role in galactose and mannose metabolism. Support for this postulation comes from TprA regulon analysis on these two sugars, which showed that in addition to its involvement in sugar metabolism, TprA controls the expression of genes involved in other pathways including iron metabolism and synthesis of predicted bacteriocins. Our microarray analysis of ΔtprA on glucose also supports the view that TprA/PhrA systems regulatory function extends beyond its control over galactose and mannose metabolism.
Attenuation of colonization and virulence of TprA/PhrA mutants in the respiratory tract is very likely due to the inability to cleave, transport and metabolize host sugars. We showed previously by EMSA and by reporter assays that the expression of nanA, which encodes for the main pneumococcal neuraminidase activity, is upregulated by TprA on galactose but not on glucose (Motib et al., 2017). The involvement of nanA in pneumococcal colonization and virulence has been linked to the removal of terminal sialic acid from host glycoconjugates. The initial cleavage is critical as this process exposes other internal sugar linkages to the activity of pneumococcal glycosidases, such as galactosidases and mannosidases, which cleave galactose and mannose, respectively (King, 2010). The reduction in neuraminidase activity in the ΔtprA is consistent with growth attenuation of this mutant on mucin, very likely due to reduced concentration of free sugars and subsequent deprivation of nutrients.
Our results suggest the involvement of the TrpA/PhrA system to the wider metabolic network and show the complexity of regulation of sugar metabolism in S. pneumoniae as illustrated in Figure 5. Here we found that tprA expression is regulated not only by autoinduction on mannose and galactose, but also by CcpA, the master regulator of sugar metabolism, and by GlnR, a regulator of glutamine and glutamate metabolism (Hendriksen et al., 2008). CcpA is the activator of TprA on galactose and a repressor on glucose. GlnR is an activator independent of sugar type. The presence of multiple regulator-binding sites highlights the intertwined nature of different aspects of central metabolic pathways. It is very likely that these proteins bind to cre like sequence located proximal to tprA, which is known to be the binding site for CcpA.
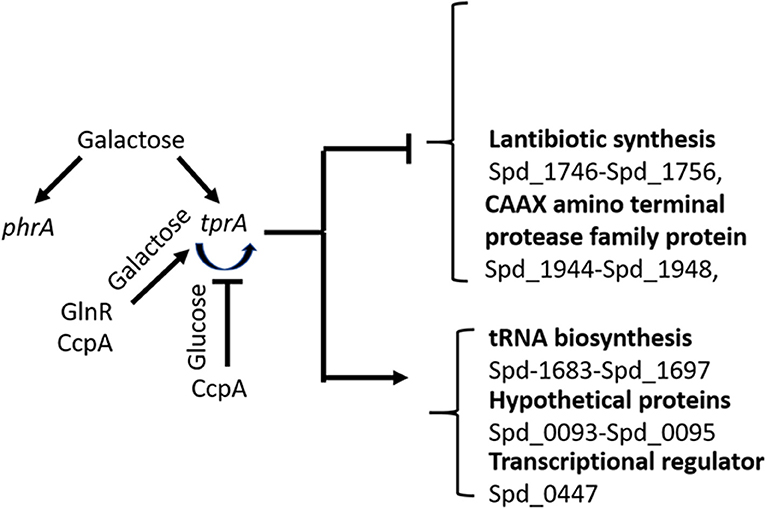
Figure 5. Schematic diagram of TprA/PhrA mediated regulation. Blunt arrows indicate down- and pointed arrows show upregulation. The curve arrows show self-induction. Some of the genes that are up- or down regulated by TprA/PhrA system on mannose and galactose are also shown.
Interfering with bacterial communication systems is considered to be an effective strategy to develop novel anti-infectives (Clatworthy et al., 2007; Brooks and Brooks, 2014). QS system homologs are not present in mammals hosts and conserved among pathogenic bacteria, meaning that the possibility of host-toxicity would be low (Clatworthy et al., 2007). We previously demonstrated that TprA/PhrA system is a drugable target. This study adds to our understanding of how this system affects the pneumococcal gene regulation, survival, and virulence.
The datasets generated for this study can be found in the GEO/GSE 67668.
Mouse experiments at the University of Leicester were performed under appropriate project (permit no. 60/4327) and personal (permit no. 80/10279) licenses according to the United Kingdom Home Office guidelines under the Animals Scientific Procedures Act 1986, and the University of Leicester ethics committee approval. The protocol was approved by both the U.K. Home Office and the University of Leicester ethics committee. Where indicated, the procedures were carried out under anesthesia with isoflurone. Animals were housed in individually ventilated cages in a controlled environment, and were frequently monitored after infection. In bacterial infection experiments mice were humanely culled immediately if they became lethargic.
AM, FA-B, IM, SS, AK, OK, NH, PA, and HY made substantial contributions to the conception or design of the work, the acquisition, analysis, and interpretation of data for the work, gave the final approval of the manuscript to be published, and agreed to be accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. AM, OK, NH, PA, and HY contributed to the drafting the work and critically revising the manuscript for important intellectual content.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2019.00326/full#supplementary-material
Agaisse, H., Gominet, M., Okstad, O. A., Kolsto, A. B., and Lereclus, D. (1999). PlcR is a pleiotropic regulator of extracellular virulence factor gene expression in Bacillus thuringiensis. Mol. Microbiol. 32, 1043–1053. doi: 10.1046/j.1365-2958.1999.01419.x
Aggarwal, S. D., Eutsey, R., West-Roberts, J., Domenech, A., Xu, W., Abdullah, I. T., et al. (2018). Function of BriC peptide in the pneumococcal competence and virulence portfolio. PLoS Pathog. 14:e1007328. doi: 10.1371/journal.ppat.1007328
Al-Bayati, F. A., Kahya, H. F., Damianou, A., Shafeeq, S., Kuipers, O. P., Andrew, P. W., et al. (2017). Pneumococcal galactose catabolism is controlled by multiple regulators acting on pyruvate formate lyase. Sci. Rep. 7:43587. doi: 10.1038/srep43587
Alloing, G., Granadel, C., Morrison, D. A., and Claverys, J. P. (1996). Competence pheromone, oligopeptide permease, and induction of competence in Streptococcus pneumoniae. Mol. Microbiol. 21, 471–478. doi: 10.1111/j.1365-2958.1996.tb02556.x
Bradford, M. M. (1976). A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72, 248–254. doi: 10.1016/0003-2697(76)90527-3
Bricker, A. L., and Camilli, A. (1999). Transformation of a type 4 encapsulated strain of Streptococcus pneumoniae. FEMS Microbiol. Lett. 172, 131–135. doi: 10.1111/j.1574-6968.1999.tb13460.x
Brooks, B. D., and Brooks, A. E. (2014). Therapeutic strategies to combat antibiotic resistance. Adv. Drug Deliv. Rev. 78, 14–27. doi: 10.1016/j.addr.2014.10.027
Clatworthy, A. E., Pierson, E., and Hung, D. T. (2007). Targeting virulence: a new paradigm for antimicrobial therapy. Nat. Chem. Biol. 3, 541–548. doi: 10.1038/nchembio.2007.24
Cook, L. C., and Federle, M. J. (2014). Peptide pheromone signaling in Streptococcus and Enterococcus. FEMS Microbiol. Rev. 38, 473–492. doi: 10.1111/1574-6976.12046
Cuevas, R. A., Eutsey, R., Kadam, A., West-Roberts, J. A., Woolford, C. A., Mitchell, A. P., et al. (2017). A novel streptococcal cell-cell communication peptide promotes pneumococcal virulence and biofilm formation. Mol. Microbiol. 105, 554–571. doi: 10.1111/mmi.13721
Dawid, S., Roche, A. M., and Weiser, J. N. (2007). The blp bacteriocins of Streptococcus pneumoniae mediate intraspecies competition both in vitro and in vivo. Infect. Immun. 75, 443–451. doi: 10.1128/IAI.01775-05
Everett, D. B., Cornick, J., Denis, B., Chewapreecha, C., Croucher, N., Harris, S., et al. (2012). Genetic characterisation of Malawian pneumococci prior to the roll-out of the PCV13 vaccine using a high-throughput whole genome sequencing approach. PLoS ONE 7:e44250. doi: 10.1371/journal.pone.0044250
Federle, M. (2012). Pathogenic streptococci speak, but what are they saying? Virulence 3, 92–94. doi: 10.4161/viru.3.1.18652
Gaspar, P., Al-Bayati, F. A., Andrew, P. W., Neves, A. R., and Yesilkaya, H. (2014). Lactate dehydrogenase is the key enzyme for pneumococcal pyruvate metabolism and pneumococcal survival in blood. Infect. Immun. 82, 5099–5109. doi: 10.1128/IAI.02005-14
Glanville, D. G., Han, L., Maule, A. F., Woodacre, A., Thanki, D., Abdullah, I. T., et al. (2018). RitR is an archetype for a novel family of redox sensors in the streptococci that has evolved from two-component response regulators and is required for pneumococcal colonization. PLoS Pathog. 14:e1007052. doi: 10.1371/journal.ppat.1007052
Gohar, M., Faegri, K., Perchat, S., Ravnum, S., Okstad, O. A., Gominet, M., et al. (2008). The PlcR virulence regulon of Bacillus cereus. PLoS ONE 3:e2793. doi: 10.1371/journal.pone.0002793
Gominet, M., Slamti, L., Gilois, N., Rose, M., and Lereclus, D. (2001). Oligopeptide permease is required for expression of the Bacillus thuringiensis plcR regulon and for virulence. Mol. Microbiol. 40, 963–975. doi: 10.1046/j.1365-2958.2001.02440.x
Guiral, S., Henard, V., Laaberki, M. H., Granadel, C., Prudhomme, M., Martin, B., et al. (2006). Construction and evaluation of a chromosomal expression platform (CEP) for ectopic, maltose-driven gene expression in Streptococcus pneumoniae. Microbiology 152, 343–349. doi: 10.1099/mic.0.28433-0
Haas, W., Kaushal, D., Sublett, J., Obert, C., and Tuomanen, E. I. (2005). Vancomycin stress responsein a sensitive and a tolerant strain of Streptococcus pneumoniae. J. Bacteriol. 187, 8205–8210. doi: 10.1128/JB.187.23.8205-8210.2005
Halfmann, A., Hakenbeck, R., and Bruckner, R. (2007). A new integrative reporter plasmid for Streptococcus pneumoniae. FEMS Microbiol. Lett. 268, 217–224. doi: 10.1111/j.1574-6968.2006.00584.x
Hendriksen, W. T., Bootsma, H. J., Estevao, S., Hoogenboezem, T., De Jong, A., De Groot, R., et al. (2008). CodY of Streptococcus pneumoniae: link between nutritional gene regulation and colonization. J. Bacteriol. 190, 590–601. doi: 10.1128/JB.00917-07
Hoover, S. E., Perez, A. J., Tsui, H. C. T., Sinha, D., Smiley, D. L., Dimarchi, R. D., et al. (2015). A new quorum-sensing system (TprA/PhrA) for Streptococcus pneumoniae D39 that regulates a lantibiotic biosynthesis gene cluster. Mol. Microbiol. 97, 229–243. doi: 10.1111/mmi.13029
Junges, R., Salvadori, G., Shekhar, S., Amdal, H. A., Periselneris, J. N., Chen, T., et al. (2017). A quorum-sensing system that regulates Streptococcus pneumoniae biofilm formation and surface polysaccharide production. mSphere 2:e00324-17. doi: 10.1128/mSphere.00324-17
Kadam, A., Eutsey, R. A., Rosch, J., Miao, X., Longwell, M., Xu, W., et al. (2017). Promiscuous signaling by a regulatory system unique to the pandemic PMEN1 pneumococcal lineage. PLoS Pathog. 13:e1006339. doi: 10.1371/journal.ppat.1006339
King, S. J. (2010). Pneumococcal modification of host sugars: a major contributor to colonization of the human airway? Mol. Oral Microbiol. 25, 15–24. doi: 10.1111/j.2041-1014.2009.00564.x
King, S. J., Hippe, K. R., and Weiser, J. N. (2006). Deglycosylation of human glycoconjugates by the sequential activities of exoglycosidases expressed by Streptococcus pneumoniae. Mol Microbiol 59, 961–974. doi: 10.1111/j.1365-2958.2005.04984.x
Liu, X., Li, J. W., Feng, Z., Luo, Y., Veening, J. W., and Zhang, J. R. (2017). Transcriptional repressor PtvR regulates phenotypic tolerance to vancomycin in Streptococcus pneumoniae. J. Bacteriol. 199, e00054–e00017. doi: 10.1128/JB.00054-17
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta CT) method. Methods. 25, 402–408. doi: 10.1006/meth.2001.1262
Motib, A., Guerreiro, A., Al-Bayati, F., Piletska, E., Manzoor, I., Shafeeq, S., et al. (2017). Modulation of quorum sensing in a Gram-positive pathogen by linear molecularly imprinted polymers with anti-infective properties. Angew. Chem. Int. Ed. Engl. 56, 16555–16558. doi: 10.1002/anie.201709313
Philips, B. J., Meguer, J. X., Redman, J., and Baker, E. H. (2003). Factors determining the appearance of glucose in upper and lower respiratory tract secretions. Intens. Care Med. 29, 2204–2210. doi: 10.1007/s00134-003-1961-2
Rezaei, J. R., van Tonder, A. J., King, J. P., Harrold, C. L., and Brueggemann, A. B. (2018). Genome sequencing reveals a large and diverse repertoire of antimicrobial peptides. Front. Microbiol. 9:2012. doi: 10.3389/fmicb.2018.02012
Rocha-Estrada, J., Aceves-Diez, A. E., Guarneros, G., and De La Torre, M. (2010). The RNPP family of quorum-sensing proteins in Gram-positive bacteria. Appl. Microbiol. Biotechnol. 87, 913–923. doi: 10.1007/s00253-010-2651-y
Saha, S., Modak, J. K., Naziat, H., Al-Emran, H. M., Chowdury, M., Islam, M., et al. (2015). Detection of co-colonization with Streptococcus pneumoniae by algorithmic use of conventional and molecular methods. Vaccine 33, 713–718. doi: 10.1016/j.vaccine.2014.11.040
Sandrini, S., Alghofaili, F., Freestone, P., and Yesilkaya, H. (2014). Host stress hormone norepinephrine stimulates pneumococcal growth, biofilm formation and virulence gene expression. BMC Microbiol. 14:180. doi: 10.1186/1471-2180-14-180
Shafeeq, S., Afzal, M., Henriques-Normark, B., and Kuipers, O. P. (2015). Transcriptional profiling of UlaR-regulated genes in Streptococcus pneumoniae. Genom Data 4, 57–59. doi: 10.1016/j.gdata.2015.02.004
Solovyev, V. V., Shahmuradov, I. A., and Salamov, A. A. (2010). Identification of promoter regions and regulatory sites. Methods Mol. Biol. 674, 57–83. doi: 10.1007/978-1-60761-854-6_5
Trappetti, C., Mcallister, L. J., Chen, A., Wang, H., Paton, A. W., Oggioni, M. R., et al. (2017). Autoinducer 2 signaling via the phosphotransferase FruA drives galactose utilization by Streptococcus pneumoniae, resulting in hypervirulence. MBio 8:e02269–16. doi: 10.1128/mBio.02269-16
Weiser, J. N., Ferreira, D. M., and Paton, J. C. (2018). Streptococcus pneumoniae: transmission, colonization and invasion. Nat Rev Microbiol. 16, 355–367. doi: 10.1038/s41579-018-0001-8
Wholey, W. Y., Abu-Khdeir, M., Yu, E. A., Siddiqui, S., Esimai, O., and Dawid, S. (2019). Characterization of the competitive pneumocin peptides of Streptococcus pneumoniae. Front. Cell Infect. Microbiol. 9:55. doi: 10.3389/fcimb.2019.00055
Willey, J. M., and van der Donk, W. A. (2007). Lantibiotics: peptides of diverse structure and function. Annu. Rev. Microbiol. 61, 477–501. doi: 10.1146/annurev.micro.61.080706.093501
Yesilkaya, H., Manco, S., Kadioglu, A., Terra, V. S., and Andrew, P. W. (2008). The ability to utilize mucin affects the regulation of virulence gene expression in Streptococcus pneumoniae. FEMS Microbiol. Lett. 278, 231–235. doi: 10.1111/j.1574-6968.2007.01003.x
Yesilkaya, H., Soma-Haddrick, S., Crennell, S. J., and Andrew, P. W. (2006). Identification of amino acids essential for catalytic activity of pneumococcal neuraminidase A. Res. Microbiol. 157, 569–574. doi: 10.1016/j.resmic.2005.12.009
Keywords: Streptococcus pneumoniae, transcriptional regulation, TprA/PhrA, sugar metabolism, virulence
Citation: Motib AS, Al-Bayati FAY, Manzoor I, Shafeeq S, Kadam A, Kuipers OP, Hiller NL, Andrew PW and Yesilkaya H (2019) TprA/PhrA Quorum Sensing System Has a Major Effect on Pneumococcal Survival in Respiratory Tract and Blood, and Its Activity Is Controlled by CcpA and GlnR. Front. Cell. Infect. Microbiol. 9:326. doi: 10.3389/fcimb.2019.00326
Received: 15 May 2019; Accepted: 29 August 2019;
Published: 13 September 2019.
Edited by:
Jorge Eugenio Vidal, University of Mississippi Medical Center, United StatesReviewed by:
Anders P. Hakansson, Lund University, SwedenCopyright © 2019 Motib, Al-Bayati, Manzoor, Shafeeq, Kadam, Kuipers, Hiller, Andrew and Yesilkaya. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hasan Yesilkaya, aHkzQGxlLmFjLnVr
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.