
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Cell. Infect. Microbiol. , 30 July 2019
Sec. Clinical Microbiology
Volume 9 - 2019 | https://doi.org/10.3389/fcimb.2019.00264
 Amir Arastehfar1†
Amir Arastehfar1† Sadegh Khodavaisy2,3†
Sadegh Khodavaisy2,3† Farnaz Daneshnia1
Farnaz Daneshnia1 Mohammad-Javad Najafzadeh4
Mohammad-Javad Najafzadeh4 Shahram Mahmoudi3,5
Shahram Mahmoudi3,5 Arezoo Charsizadeh6
Arezoo Charsizadeh6 Mohammad-Reza Salehi7
Mohammad-Reza Salehi7 Hossein Zarrinfar8
Hossein Zarrinfar8 Abbas Raeisabadi4
Abbas Raeisabadi4 Somayeh Dolatabadi9
Somayeh Dolatabadi9 Zahra Zare Shahrabadi10
Zahra Zare Shahrabadi10 Kamiar Zomorodian10*
Kamiar Zomorodian10* Weihua Pan11*
Weihua Pan11* Ferry Hagen1,12,13
Ferry Hagen1,12,13 Teun Boekhout1,11,14
Teun Boekhout1,11,14Despite the increasing occurrence of Candida orthopsilosis and Candida metapsilosis in clinical settings, little is known about their microbiological and clinical properties. Herein, we conducted a national retrospective study (2014–2019) from multiple centers in Iran. Among the 1,770 Candida isolates collected, we identified 600 Candida parapsilosis species complex isolates. Isolate identification was performed by 9-plex PCR, matrix-assisted laser desorption-time of flight mass spectrometry (MALDI-TOF MS), and rDNA sequencing, and antifungal susceptibility testing (AFST) followed CLSI M27-A3/S4; genotyping was performed by amplified fragment length polymorphism (AFLP) analysis; and clinical information was mined. Thirty-one isolates of C. orthopsilosis from various clinical sources, one mixed sample (blood) concurrently containing C. orthopsilosis and C. parapsilosis and one isolate of C. metapsilosis from a nail sample were identified. Although both 9-plex PCR and MALDI-TOF successfully identified all isolates, only 9-plex PCR could identify the agents in a mixed sample. For the C. orthopsilosis isolates, resistance (non-wild type) was noted only for itraconazole (n = 4; 12.5%). Anidulafungin and fluconazole showed the highest and voriconazole had the lowest geometric mean values. AFLP analysis showed three main and four minor genotypes. Interestingly, 90% of nail isolates clustered with 80% of the blood isolates within two clusters, and four blood isolates recovered from four patients admitted to a hospital clustered into two genotypes and showed a high degree of similarity (>99.2%), which suggests that C. orthopsilosis disseminates horizontally. Supported by our data and published case studies, C. orthopsilosis and C. metapsilosis can be linked to challenging clinical failures, and successful outcomes are not always mirrored by in vitro susceptibility. Accordingly, conducting nationwide studies may provide more comprehensive data, which is required for a better prognosis and clinical management of patients.
With advancements in identification tools and changes in clinical practices, a distinct trend of an increasing prevalence of non-Candida albicans Candida (NCAC) species in clinical settings has been revealed (Lamoth et al., 2018). The recent arrival and increase in the amount of azole-resistant Candida parapsilosis isolates (Grossman et al., 2015; Govender et al., 2016; Asadzadeh et al., 2017a; Choi et al., 2018; Thomaz et al., 2018; Singh et al., 2019), and the ability of this species to be horizontally transmitted from the hands of healthcare workers (HCWs) (Thomaz et al., 2018) emphasize the importance of surveillance studies to limit its spread in healthcare settings. Additionally, if left undetected, this yeast can be the source of fatal candidemia outbreaks, and it can persist in the hospital environment for a long period of time (Wang et al., 2016). Phylogenetic analysis performed by Tavanti et al. (2005) showed that C. parapsilosis is a species complex comprising C. parapsilosis sensu stricto, C. orthopsilosis, and C. metapsilosis (Tavanti et al., 2005). Although C. orthopsilosis and C. metapsilosis are less virulent than C. parapsilosis, they have the ability to cause a wide range of clinical manifestations ranging from superficial (Feng et al., 2012) to fatal invasive bloodstream infections (Barbedo et al., 2015). Besides, clinical failure for infections caused by C. orthopsilosis and C. metapsilosis have been reported in some studies, while infected patients underwent prolonged administration of antifungals (Choi et al., 2010; Wessel et al., 2013; Oliveira et al., 2014; Heslop et al., 2015; Charsizadeh et al., 2018). On the other hand, a survey conducted in Italy (Pisa and Rome) showed that 40% of C. orthopsilosis isolates were resistant to fluconazole (FLZ), and among them, 100 and 68.7% of FLZ-resistant C. orthopsilosis isolates were cross-resistant to two and three most commonly used azoles (Rizzato et al., 2018). Lines of evidence show that azole resistance in the C. parapsilosis complex species is mainly mediated by a specific mutation in ERG11 (A395T) (Choi et al., 2018; Rizzato et al., 2018), and unlike other Candida species, efflux pumps might not play a main role in azole resistance (Mello et al., 2017). Moreover, it has been shown that mutations in hotspot 1 (HS1) and HS2 of FKS1 are linked to echinocandin resistance in the C. parapsilosis species complex (Garcia-Effron et al., 2008).
Variability in virulence factors and antifungal susceptibility patterns among members of the C. parapsilosis species complex points to the importance of correct species-level identification (Neji et al., 2017a). Phenotypic assays, such as biochemical assays, are unable to differentiate species within the C. parapsilosis species complex (Neji et al., 2017b), while PCR-based molecular assays (Tavanti et al., 2007; Mirhendi et al., 2010; Arastehfar et al., 2018), matrix-assisted laser desorption-time of flight mass spectrometry (MALDI-TOF MS) (De Carolis et al., 2014), and sequencing of so-called barcoding genes (Tavanti et al., 2005) allow correct species-level identification.
Genomic studies have led to the discovery that C. orthopsilosis and C. metapsilosis were derived from the hybridization of species with non-pathogenic lineages (Pryszcz et al., 2014, 2016). As a result, genotyping techniques may provide a better understanding of the evolution of the mechanism of pathogenicity in this complex. Moreover, the application of typing techniques may not only aid in detecting the source of infection but may also broaden our knowledge of the biological niches of a species of interest. Amplified fragment length polymorphism (AFLP) analysis is regarded as the preferred typing choice for members of the C. parapsilosis species complex (Tavanti et al., 2007), Candida albicans (Asadzadeh et al., 2017b), Candida auris (Schelenz et al., 2016), and Aspergillus terreus (Kathuria et al., 2015).
Herein, we conducted a multicenter study and collected all presumptively identified isolates of C. parapsilosis from three main metropolitan cities of Iran (Tehran, Shiraz, and Mashhad) from 2014 to 2019. Isolates were identified by MALDI-TOF MS, a previously described 9-plex PCR (Arastehfar et al., 2018) and sequencing of rDNA. Moreover, isolates were genotyped by AFLP, their antifungal susceptibility pattern was determined, and genes conferring resistance to FLZ (ERG11) and echinocandins (HS1 and HS2 of FKS1) were sequenced.
In total, we collected 600 presumptively identified C. parapsilosis species complex isolates among 1,770 isolates of Candida species isolates recovered (2014–2019) from three major clinical centers in Iran, namely, Tehran, Shiraz, and Mashhad (Table 1). C. parapsilosis species complex isolates constituted 33.8% of all Candida species isolates recovered from the aforementioned centers. C. parapsilosis species complex strains were mainly isolated from blood (n = 167) and other non-sterile sites (n = 433; Table 1). Studies undertaken by the centers included in this study were individually reviewed and approved by ethical committee members in each center (IR.SUMS.REC.1397.365, IR MUMS fm REC.1397.268, IR. TUMS.SPH.REC.1396.4195). To ensure anonymity, patients were assigned numerical codes. All patients gave written informed consent in accordance with the ethical permit of the centers involved in this study. Strains were grown on Sabouraud dextrose agar (SDA) and incubated at 37°C for 24–48 h. To ensure that samples with mixed species were identified, all clinical samples were struck on CHROMagar (Candiselect, Bio-Rad, USA), and incubated at 37°C for 48 h.
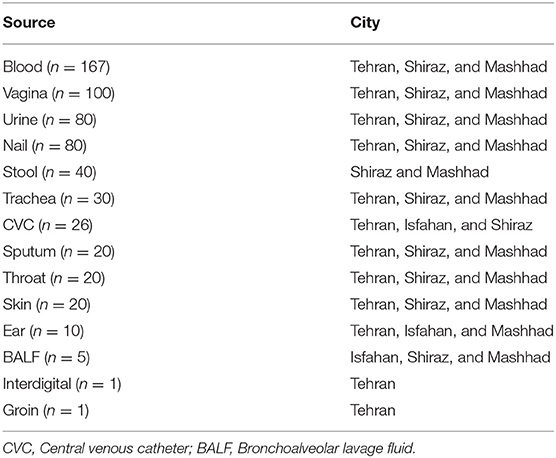
Table 1. Presumptively identified C. parapsilosis species complex isolates collected from clinical centers.
A previously CTAB-based DNA extraction protocol was used to extract DNA samples (Theelen et al., 2001). Primarily, isolated strains were identified by MALDI-TOF MS (MicroFlex LTD, Bruker, Bremen, Germany) using a full-extraction method (Cendejas-Bueno et al., 2012) and a 9-plex PCR differentiating nine species within the C. albicans, Candida glabrata, and C. parapsilosis species complexes (Arastehfar et al., 2018). Strains identified as C. orthopsilosis and C. metapsilosis were further identified by sequencing of the large subunit (LSU) and internal transcribed spacer sequences (ITS) of the rDNA domain using LR5 and ITS5 primers (Stielow et al., 2015).
To assess the genotypic diversity of C. orthopsilosis and C. metapsilosis, a previously defined AFLP protocol was used (Marchetta et al., 2018). In brief, 5 μl of a DNA sample was mixed with restriction-ligation reactions containing HpyCH4 IV and MseI adapters and restriction enzymes and T4 ligase and incubated at room temperature for 90 min. Subsequently, the ligation-restriction reactions were stopped by the addition of 80 μl of 10 mM Tris-HCl (pH 8.3), and diluted products were added to PCRs containing HpyCH4 IV and MseI primers. In the next stage, PCR products were purified using Sephadex (Sigma Aldrich, St. Louis, Missouri, USA) and diluted 50 times with Milli-Q water; 1 μl of PCR product was mixed with master mixes containing standard ladder size, incubated for 1 min at 100°C, and finally subjected to an ABI 3730XL DNA analyzer (Thermo Fisher Scientific, Waltham, Massachusetts, USA). BioNumerics software V7.6 (Applied Math Inc., Austin, Texas, USA) was used to analyze the AFLP data. Reference and type strains of C. metapsilosis (CBS 2315, CBS 2916, and CBS 10907) and C. orthopsilosis (CBS 10906) were used for comparative purposes.
CLSI M27-A3/S4 broth micro dilution (BMD) was used for the AFST of the C. orthopsilosis and C. metapsilosis isolates (Clinical Laboratory Standards Institute, 2008, 2012). AFST included the following drugs: fluconazole (FLZ) (Pfizer, New York, USA), voriconazole (VRZ) (Pfizer, New York, USA), itraconazole (ITZ) (Santa Cruz Biotech, Dallas, USA), amphotericin B (AMB) (Sigma Chemical Corporation, St. Louis, MO), micafungin (MFG) (Astellas Pharma Inc., Japan), and anidulafungin (AFG) (Pfizer A/S, Ballerup, Denmark). Reference strains of C. parapsilosis (CBS 604) and Candida krusei (CBS 5147) were used for quality control purposes. The MIC values were visually determined after incubating the plates for 24 h at 37°C. Due to the lack of a species-specific clinical breakpoint and epidemiological cut-off values for C. orthopsilosis and C. metapsilosis, the obtained MIC values were compared with those of C. parapsilosis. Moreover, due to the inter laboratory variation and unreliability of caspofungin (Espinel-Ingroff et al., 2013), this drug was not investigated in the current study. Isolates showing MIC values ≥8 μg/ml for FLZ, MFG, and ANF and those showing MIC values ≥1 for VRZ were regarded as resistant (Pfaller and Diekema, 2012). Due to the lack of clinical breakpoints for AMB and ITZ, their corresponding MIC values were interpreted based on epidemiological cut-off values (ECV) and non-wild type (NWT) values when the MIC values were >2 and >0.5 μg/ml, respectively (Pfaller and Diekema, 2012).
As resistance to azoles in C. orthopsilosis is mainly mediated by a specific point mutation (A395T) that resulted in a missense mutation of Y132F (Mello et al., 2017; Rizzato et al., 2018), primers targeting this region were used (Table 2). Moreover, C. parapsilosis species complex universal primers (from unpublished data) targeting HS1 and HS2 of FKS1 (Table 2) were included to explore the potential non-synonymous mutations conferring resistance to echinocandins.
PCRs contained the following ingredients: 5 μl of PCR buffer (10X NH4 without MgCl2), 2 mM MgCl2, 10 picomole target primers (ERG11F/R and HS1F/R and HS2F/R), 0.2 mM mixed dNTPs (dNTP mix, 100 mM, Bioline), and 1.25 units of Taq polymerase (BioTaq DNA Polymerase, Bioline). Milli-Q water was used to adjust the volume to 50 μl. PCR reactions were subjected to Applied Biosystem 2720 Thermal Cycler (Thermo Fisher Scientific, Waltham, Massachusetts, USA) with the following program: one cycle of 95°C for 5 min; followed by 35 cycles of 95°C for 30 s, 52°C for 30 s, and 72°C for 30 s; and finally, one cycle of 72°C for 8 min.
The dideoxy-chain termination sequencing protocol was used for sequencing of target genes, and the generated contigs were curated, assembled and edited by SeqMan Pro (DNASTAR, Madison, USA). Curated sequences were aligned using MEGA v7.0 (Temple University, Philadelphia, USA). The obtained sequences of ERG11 were compared with the corresponding reference sequences of XM_003870254.1 (Riccombeni et al., 2012; Rizzato et al., 2018), and the sequences of HS1 and HS2 were compared with those presented by Garcia-Effron et al. (2008).
The C. orthopsilosis and C. metapsilosis strains obtained from this study were deposited in the culture collection of Westerdijk Fungal Biodiversity Institute, and their corresponding sequences of ITS and LSU rDNA, HS1 and HS2 of FKS1, and ERG11 were deposited in GenBank (https://www.ncbi.nlm.nih.gov/genbank/) (Supplementary Table 1).
In total, 32 C. orthopsilosis isolates were recovered from 31 patients, with a median age of 39 years old (1 month-90 years) and one C. metapsilosis isolate from a 30-year-old man (Table 3). Women constituted the vast majority of patients (n = 22; 68.7%). Tehran had the highest number of C. orthopsilosis isolates (n = 19; 57.6%), followed by Mashhad (n = 13; 39.4%), and Shiraz (n = 1; 3%). C. orthopsilosis isolates were mainly from blood (n = 10; 31.2%) and nail (n = 10; 31.2%) samples, followed by urine (n = 5; 15.6%), vaginal (n = 3; 9.3%), tracheal (n = 2; 6.2%), and groin and interdigital (each n = 1; 3.2%) samples (Table 3). The only isolate of C. metapsilosis was recovered from a nail sample. Diabetes (n = 5), hematological malignancies (n = 4), and pneumonia (n = 2) were the most encountered underlying conditions (considering that the majority of samples were obtained from outpatients and the underlying conditions were not available for some patients). All patients with invasive candidiasis due to C. orthopsilosis were treated with broad-spectrum antibiotics. In total, 10 patients were treated with antifungals and AMB was the most widely used antifungal (n = 7; 70%), followed by the combination of CSP, FLZ, and AMB (each n = 3; 30%; Table 3). Four patients infected with C. orthopsilosis died and the corresponding isolates were recovered from blood (n = 2), vagina (n = 1), and trachea (n = 1). For comparison purposes, case report studies describing microbiological and clinical outcomes are presented in Supplementary Table 2.
C. orthopsilosis and C. metapsilosis comprised 1.8 and 0.05% of all Candida species isolates and 5.3 and 0.17% of all C. parapsilosis species complex isolates, respectively. Both 9-plex PCR and MALDI-TOF MS consistent with ITS and LSU rDNA sequencing successfully identified all C. orthopsilosis, C. metapsilosis, and C. parapsilosis isolates. One of the blood samples concurrently harbored both C. orthopsilosis and C. parapsilosis, which were identified based on colony morphology (wrinkled colonies for C. parapsilosis and round colonies for C. orthopsilosis). Sequencing and MALDI-TOF MS identified this mixed sample as C. parapsilosis, while the 9-plex PCR successfully identified both C. parapsilosis and C. orthopsilosis (Figure 1).
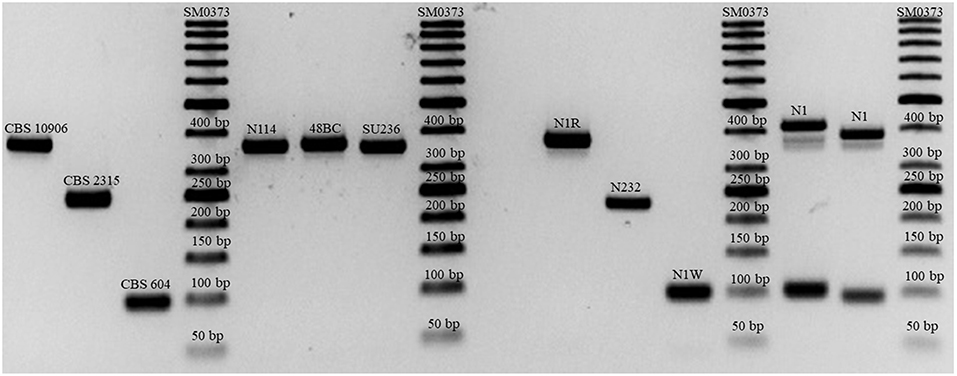
Figure 1. Successful differentiation of the C. parapsilosis species complex and mixed isolates of C. parapsilosis and C. orthopsilosis (N1 with double bands representing both species).
AFLP was employed to explore the genotypic diversity of C. orthopsilosis and C. metapsilosis isolates included in this study (Figure 2). In total, three major genotypes, namely, G1 (n = 12), G2 (n = 6), and G3 (n = 10), along with four minor genotypes, each containing one strain, were detected. Isolates of G1 were mainly obtained from nail samples (66.6%), and 80% of blood isolates (n = 8) belonged to G1 and G2 clustered with isolates recovered from nail samples (n = 10; 90%; Figure 2). Isolates grouped in G3 were from a diverse range of clinical sources, including nails, blood, urine, vagina, trachea, and interdigital. A geographical trend was observed for the clustering of some genotypes, where G1 and G3 isolates came mainly from Tehran and Mashhad, respectively. Moreover, four blood isolates distributed in G1 and G2 (each containing two isolates) recovered from a neonatal ICU ward in Tehran (Children's Medical Center) showed a clonal pattern with a similarity of >99.2% (Figure 2).
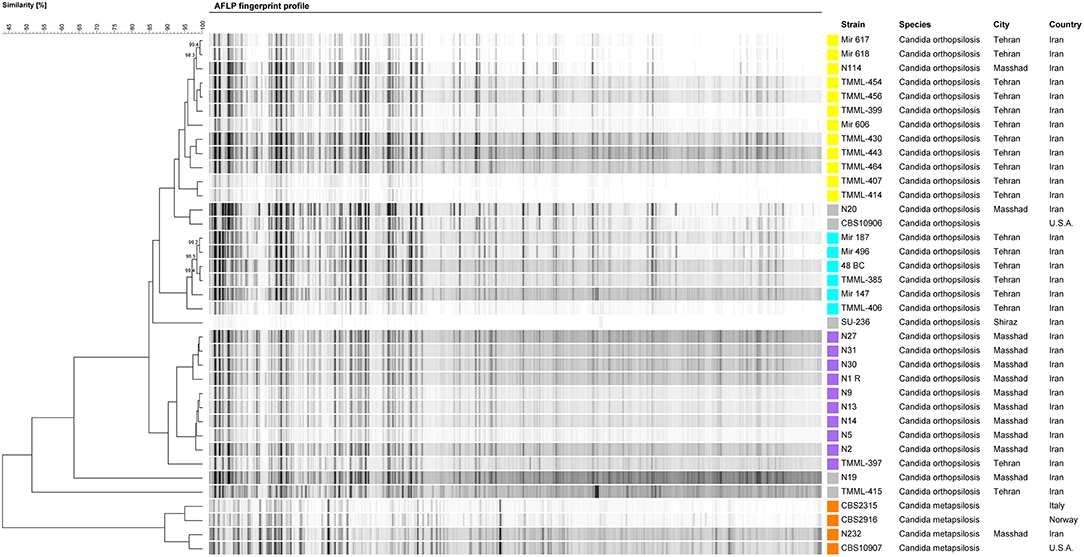
Figure 2. AFLP fingerprint profile of C. orthopsilosis and C. metapsilosis isolates included in this study. Each genotype is assigned a distinct color.
Antifungal susceptibility data for all isolates of C. orthopsilosis and C. metapsilosis are presented in Tables 4, 5. All isolates were susceptible to ANF (≤8 μg/ml) and MFG (≤8 μg/ml) and had a wild-type (WT) phenotype in the presence of AMB (<2 μg/ml). FLZ-susceptible dose-dependent (SDD) (= 4 μg/ml) and VRZ-intermediate (I) (0.25–0.5 μg/ml) were noted in 3.12 and 6.25% of isolates, respectively. For ITZ, 12.5% of isolates showed a NWT phenotype against this drug (>0.5 μg/ml). ANF and FLZ showed the highest geometric mean values (~1.0), followed by MFG (0.68), ITZ and AMB (0.31), and VRZ (0.02) (Table 4).
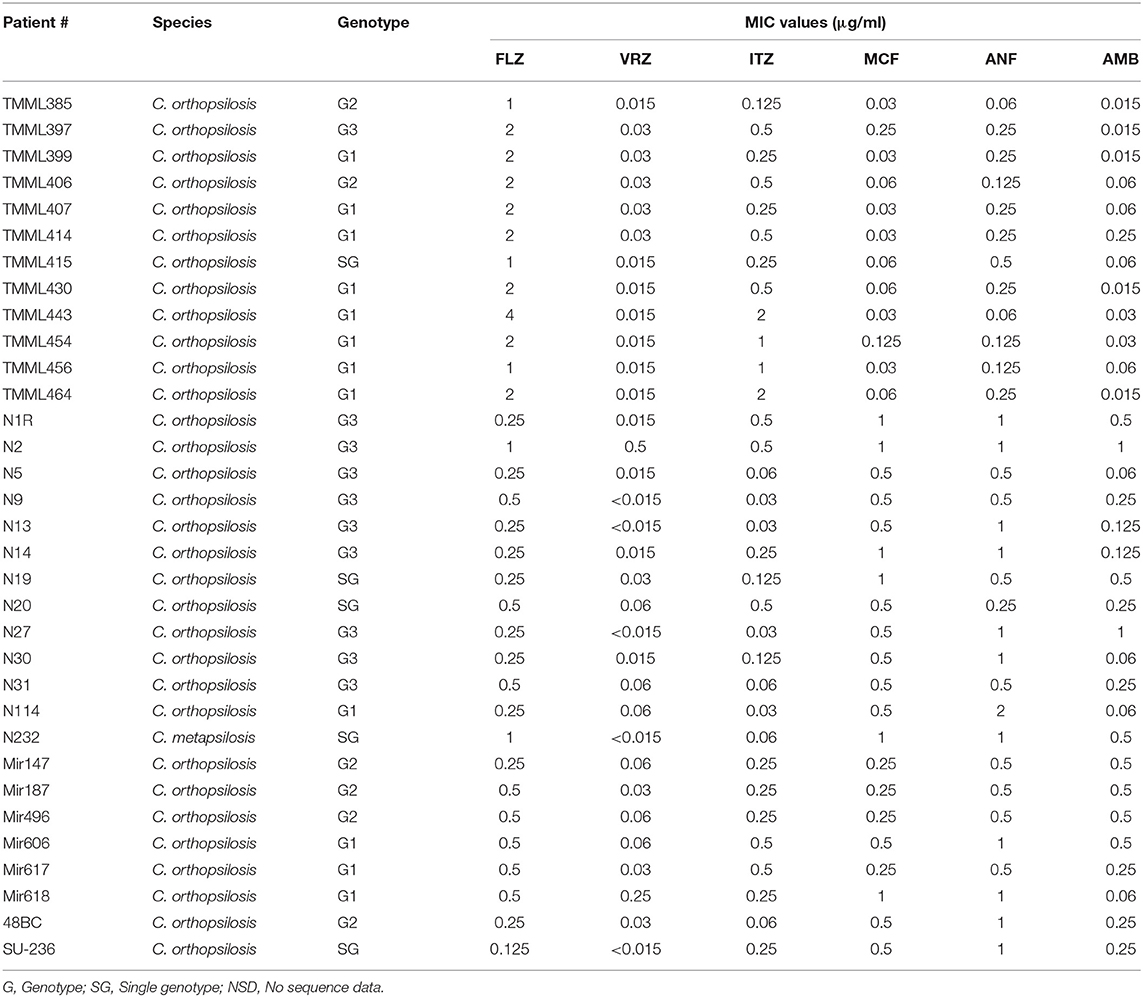
Table 5. Antifungal susceptibility testing data and sequencing of genes conferring resistance to echinocandins (HS1 and HS2 of FKS1) and azoles (ERG11).
Although successful PCR amplification and sequencing results were obtained for all target genes of C. orthopsilosis, sequences of acceptable quality were not obtained for ERG11 of C. metapsilosis. All isolates harbored WT ERG11 and HS1 and HS2 of FKS1 (Table 5).
In this study, we present the largest collection of C. orthopsilosis (n = 32) and the first case of C. metapsilosis recovered from Iranian patients. In a previous study, Mohammadi et al. (2017) explored the antifungal susceptibility of different and smaller sets of Iranian C. orthopsilosis (n = 18) isolates, but the association of genotypic diversity and clinical data, mechanism of resistance via sequencing of ERG11 and HS1 and HS2 of FKS1, and comparison of MALDI-TOF and 9-plex PCR in the context of sequencing were not assessed.
In our study, C. orthopsilosis and C. metapsilosis were responsible for 5.3 and 0.17% of C. parapsilosis species complex infections, respectively. The extremely low prevalence of C. metapsilosis in this study is similar to observations from other studies conducted in Iran (Mohammadi et al., 2017), Italy (Romeo et al., 2012; Lovero et al., 2016), Venezuela (Moreno et al., 2017), and Kuwait (Asadzadeh et al., 2009) but, contrasts the observations reported for East China (Ge et al., 2012). In contrast, other studies from Africa (Neji et al., 2017b), Latin America (Goncalves et al., 2010), Europe (Gomez-Lopez et al., 2008), and other Asian countries (Tay et al., 2009; Chen et al., 2010) isolated both C. orthopsilosis and C. metapsilosis from blood samples, although with varying prevalences. The low prevalence of C. metapsilosis could be related to the reduced virulence and biofilm-production ability of this emerging pathogen (Gago et al., 2014), but this substantial variability might be indicative of the involvement of other factors, such as variation in the microbiome structure observed in different populations and environments. For instance, in East China, authors noted that C. metapsilosis was responsible for 60% of the C. parapsilosis species complex infections in one of the centers included in the study, and these isolates were mainly obtained from cutaneous samples of dermatological outpatients (Ge et al., 2012). The authors attributed this unusual C. metapsilosis prevalence to a different microbiome population of infected patients who might have shared the same working environment (Ge et al., 2012). This might be a plausible explanation, as C. metapsilosis has been found in the commensal (Ghannoum et al., 2010) and environmental (Trofa et al., 2008) microbiomes. Additionally, it has been shown that drinking water (Willis et al., 2018) and specific lifestyle (Valles et al., 2018) might have an impact on the microbiome structure, and this finding may further justify this observed marked difference in the epidemiology of this species complex.
Although C. parapsilosis is one of the most prominent Candida species to cause clonal outbreaks (Wang et al., 2016; Singh et al., 2019), this phenomenon has not been observed for C. orthopsilosis and C. metapsilosis. Interestingly, we noted that four isolates obtained from four patients in a neonatal ICU ward (Tehran) clustered in two genotypes with a high degree of genetic similarity (>99.2%), which is in contrast to the observation that clinical C. orthopsilosis isolates showed a high level of genetic diversity (Tavanti et al., 2007). The hybrid nature of C. orthopsilosis isolates (Pryszcz et al., 2014) and the fact that those isolates were recovered from various health care settings located in different countries (Tavanti et al., 2007) might explain the high level of genetic diversity observed in that study. On the other hand, we noticed that 80% of C. orthopsilosis blood isolates clustered with 90% of C. orthopsilosis isolates obtained from nail samples. This finding, along with the possible clonality of C. orthopsilosis isolates and the simultaneous isolation of this species from both central venous catheter (CVC) and blood samples reported previously (Barbedo et al., 2015), might imply that C. orthopsilosis, similar to C. parapsilosis, could be horizontally transferred from the hands of healthcare workers.
MALDI-TOF MS and Sanger sequencing are the most accurate means of identification in clinical settings. However, in this study, we observed that both MALDI-TOF MS and sequencing of ITS and LSU rDNA failed to identify C. parapsilosis and C. orthopsilosis from a mixed isolate obtained from blood, while the 9-plex PCR yielded two bands representing both species. A study from Portugal showed that 9.5% of C. parapsilosis blood isolates were a mixture of C. parapsilosis and C. orthopsilosis (Barbedo et al., 2015). Because polyfungal infections are associated with a high rate of mortality (Kim et al., 2013), it seems relevant to utilize sensitive and specific assays to identify the causative agents of mixed samples. Moreover, the application of such techniques can reveal a possible mixed sample to technicians and, as a result, might prevent the underestimation of these emerging yeast species; consequently, this may lead to a better epidemiological, microbiological, and clinical understanding.
Except for 3.12% FLZ-SDD, 6.25% VRZ-I, and 12.5% ITZ-NWT phenotypes, our C. orthopsilosis isolates together with a single C. metapsilosis isolate were susceptible to all major antifungal drugs tested. The lack of FLZ and echinocandin resistance was further proven by sequencing ERG11 and HS1 and HS2 of FKS1. Although antifungal resistance for C. orthopsilosis (Mohammadi et al., 2017; Brilhante et al., 2018) and C. metapsilosis (Chen et al., 2010) is considered a rare phenomenon, a study conducted in Italy revealed that almost 40% of C. orthopsilosis isolates were resistant to FLZ, and among them, almost 100% of isolates were cross-resistant to at least two azole drugs (Rizzato et al., 2018). Given that some FLZ-R genotypes of C. parapsilosis can persist in hospital settings for several years (Choi et al., 2018) in addition to the possible clonality of C. orthopsilosis presented in this study, this finding emphasizes the paramount importance of typing studies to limit the spread and to find the source of a given C. orthopsilosis FLZ-R genotype.
C. orthopsilosis followed by C. metapsilosis are considered the least virulent and benign species within the C. parapsilosis species complex, while studies dealing with clinical cases proved otherwise and showed that these two species can be linked to challenging septic arthritis (Heslop et al., 2015), keratitis (Wessel et al., 2013), and blood-borne infections (Choi et al., 2010; Oliveira et al., 2014; Charsizadeh et al., 2018). In our study, almost 33% of patients admitted to the ICU (n = 4) died, despite three of them received AMB or FLZ. Surprisingly, the MIC values of those C. orthopsilosis isolates derived from treated patients were susceptible to all antifungals used (except for one ITZ-R isolate). This discrepancy between clinical outcome and in vitro AFST has been noted in a keratitis case caused by C. orthopsilosis (Wessel et al., 2013). In that study, the recovered C. orthopsilosis isolate was susceptible to FLZ, VRZ, and AMB, and despite prolonged treatment with topical or systemic VRZ along with AMB, the patient manifested clinical failure, and surgical intervention finally alleviated the symptoms (Wessel et al., 2013). Surprisingly, apart from one study that showed the efficacy of FLZ (Alencar et al., 2017), the remaining studies unanimously showed the fatality of C. orthopsilosis infection (Choi et al., 2010; Oliveira et al., 2014; Charsizadeh et al., 2018) along with the lack of efficacy of FLZ and CSP (Choi et al., 2010), FLZ and AMB (Oliveira et al., 2014), FLZ (Heslop et al., 2015), and AMB (Charsizadeh et al., 2018). This variability in clinical outcome is shown even for the two C. metapsilosis fungemia cases, where one study showed successful treatment via only CVC removal without antifungal drug intervention (Asadzadeh et al., 2016), while the other study showed FLZ and AMB treatment failure (Oliveira et al., 2014). In addition to host-related underlying conditions and variability in tissue penetration of antifungal drugs (Zhao et al., 2017), these discrepancies between in vitro AFST and clinical outcome and the relatively high rate of clinical failure in case studies could be a strain-dependent phenomenon and may be explained by variation in microbiological factors, such as biofilm formation. Moreover, a recent study disclosed that this discrepancy between clinical outcome and MIC data might be due to the presence of a distinguished category of cells called tolerant cells that typically are miscategorized as susceptible via in vitro susceptibility protocols, while these cells can slowly grow in the presence of antifungal protocols (Rosenberg et al., 2018).
The discrepancy between in vitro AFST and the clinical failure of infections caused by both C. orthopsilosis and C. metapsilosis underscores the importance of the implementation of appropriate identification tools. Although MALDI-TOF MS and Sanger sequencing are the most accurate means of identification currently used in medical mycology, the application of molecular assays for laboratories lacking these tools is recommended to broaden our knowledge about the epidemiology, clinical profile, and microbiological features of these two underrated Candida species. However, the application of molecular assays, such as 9-plex PCR, can be a supplementary tool to guide the identification of causative agents of mixed samples that are not identifiable via CHROMagar and even MALDI-TOF MS and rDNA sequencing. Moreover, the possible clonal transmission of C. orthopsilosis noted in this study warrants further analysis to reinforce our findings and may reveal that employing resolutive typing techniques may have infection control implications in the case of outbreaks caused by C. orthopsilosis. Unfortunately, the lack of isolates derived from environmental samples and hands of healthcare workers from hospitals where C. orthopsilosis blood isolates were obtained and the lack of assessment of the biofilm-production ability of C. orthopsilosis isolates are the main limitations of this study.
The raw data supporting the conclusions of this manuscript will be made available by the authors, without undue reservation, to any qualified researcher.
Studies undertaken by involved centers were individually reviewed and approved by ethical committee members in each center (IR.SUMS.REC.1397.365, IR MUMS fm REC.1397.268, IR. TUMS.SPH.REC.1396.4195). In order to ensure the anonymity, patients were assigned with numerical codes. All patients gave written informed consent as in accordance with the ethical committee of centers involved.
AA, SK, and FD designed the study, collected the data, drafted the manuscript, and performed part of the experimental studies. M-JN, SM, AC, M-RS, HZ, AR, SD, ZZ, and FH participated in the experimental studies, data collection, and revising of the manuscript. SK, M-JN, AC, and KZ provided the clinical isolates. WP, KZ, and TB supervised the study and revised the manuscript.
This work was supported by the Major National R&D Projects of the National Health Department (2018ZX10101003), European Union's Horizon 2020 Research and Innovation Program under the Marie Sklodowska-Curie Grant Agreement No. 642095, Shiraz University of Medical Sciences (98-01-43-19754), Tehran University of Medical Sciences & Health Services (grant No. 97-02-211-38592), National Natural Science Foundation of China (31770161), Shanghai Science and Technology Committee (Grant Nos. 14DZ2272900 and 14495800500).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2019.00264/full#supplementary-material
Alencar, D. S. O., Tsujisaki, R. A. S., Spositto, F. L. E., Nunes, M. O., Almeida, A. A., Martins, M. D. A., et al. (2017). Candidaemia due to Candida parapsilosis species complex at a hospital in Brazil: clinical characteristics and antifungal susceptibility profile. Rev. Iberoam Micol. 34, 106–108. doi: 10.1016/j.riam.2016.06.008
Arastehfar, A., Fang, W., Pan, W., Liao, W., Yan, L., and Boekhout, T. (2018). Identification of nine cryptic species of Candida albicans, C. glabrata, and C. parapsilosis complexes using one-step multiplex PCR. BMC Infect. Dis. 18:480. doi: 10.1186/s12879-018-3381-5
Asadzadeh, M., Ahmad, S., Al-Sweih, N., Gulati, R. R., and Khan, Z. (2016). First isolation of Candida metapsilosis in Kuwait, an emerging global opportunistic pathogen. J. Mycol. Med. 26, 46–50. doi: 10.1016/j.mycmed.2015.11.001
Asadzadeh, M., Ahmad, S., Al-Sweih, N., and Khan, Z. (2017a). Epidemiology and molecular basis of Resistance to fluconazole among clinical Candida parapsilosis isolates in Kuwait. Microb. Drug Resist. 23, 966–972. doi: 10.1089/mdr.2016.0336
Asadzadeh, M., Ahmad, S., Al-Sweih, N., and Khan, Z. (2017b). Molecular fingerprinting studies do not support intrahospital transmission of Candida albicans among candidemia Patients in Kuwait. Front. Microbiol. 8:247. doi: 10.3389/fmicb.2017.00247
Asadzadeh, M., Ahmad, S., Al-Sweih, N., and Khan, Z. U. (2009). Rapid molecular differentiation and genotypic heterogeneity among Candida parapsilosis and Candida orthopsilosis strains isolated from clinical specimens in Kuwait. J. Med. Microbiol. 58, 745–752. doi: 10.1099/jmm.0.008235-0
Barbedo, L. S., Vaz, C., Pais, C., Figueiredo-Carvalho, M. H. G., de Muniz, M. M., et al. (2015). Different scenarios for Candida parapsilosis fungaemia reveal high numbers of mixed C. parapsilosis and Candida orthopsilosis infections. J. Med. Microbiol. 64, 7–17. doi: 10.1099/jmm.0.080655-0
Brilhante, R. S. N., Sales, J. A., da Silva, M. L. Q., de Oliveira, J. S., Pereira, L., Pereira-Neto, W. A., et al. (2018). Antifungal susceptibility and virulence of Candida parapsilosis species complex: an overview of their pathogenic potential. J. Med. Microbiol. 67, 903–914. doi: 10.1099/jmm.0.000756
Cendejas-Bueno, E., Kolecka, A., Alastruey-Izquierdo, A., Theelen, B., Groenewald, M., Kostrzewa, M., et al. (2012). Reclassification of the Candida haemulonii complex as Candida haemulonii (C. haemulonii group I), C. duobushaemulonii sp. nov. (C. haemulonii group II), and C. haemulonii var. vulnera var. nov.: three multiresistant human pathogenic yeasts. J. Clin. Microbiol. 50, 3641–3651. doi: 10.1128/JCM.02248-12
Charsizadeh, A., Mirhendi, H., Nikmanesh, B., Eshaghi, H., Rahmani, M., Farhang, A., et al. (2018). Candidemia in children caused by uncommon species of Candida. Arc. Pediatr. Infect. Dis. 6:e11895. doi: 10.5812/pedinfect.11895
Chen, Y.-C., Lin, Y.-H., Chen, K.-W., Lii, J., Teng, H.-J., and Li, S.-Y. (2010). Molecular epidemiology and antifungal susceptibility of Candida parapsilosis sensu stricto, Candida orthopsilosis, and Candida metapsilosis in Taiwan. Diagn. Microbiol. Infect. Dis. 68, 284–292. doi: 10.1016/j.diagmicrobio.2010.07.004
Choi, H. J., Shin, J. H., Park, K. H., Shin, M. G., Suh, S. P., and Ryang, D. W. (2010). A fatal case of Candida orthopsilosis fungemia. Korean J Clin Microbiol. 13, 140–143
Choi, Y. J., Kim, Y.-J., Yong, D., Byun, J.-H., Kim, T. S., Chang, Y. S., et al. (2018). Fluconazole-resistant Candida parapsilosis bloodstream isolates with Y132F mutation in ERG11 gene, South Korea. Emerg. Infect. Dis. 24, 1768–1770. doi: 10.3201/eid2409.180625
Clinical and Laboratory Standards Institute (2008). Reference Method for Broth Dilution Antifungal Susceptibility Testing of Yeasts; Approved Standard—Third Edition. Wayne, PA: M27-A3 CLSI.
Clinical and Laboratory Standards Institute (2012). Reference Method for Broth Dilution Antifungal Susceptibility Testing of Yeasts: Fourth Informational Supplement. Wayne, PA: M27-S4 CLSI.
De Carolis, E., Hensgens, L. A. M., Vella, A., Posteraro, B., Sanguinetti, M., Senesi, S., et al. (2014). Identification and typing of the Candida parapsilosis complex: MALDI-TOF MS vs. AFLP. Med. Mycol. 52, 123–130. doi: 10.1093/mmy/myt009
Espinel-Ingroff, A., Arendrup, M. C., Pfaller, M. A., Bonfietti, L. X., Bustamante, B., Canton, E., et al. (2013). Interlaboratory variability of Caspofungin MICs for Candida spp. Using CLSI and EUCAST methods: should the clinical laboratory be testing this agent? Antimicrob Agents Chemother. 57, 5836–5842. doi: 10.1128/AAC.01519-13
Feng, X., Ling, B., Yang, G., Yu, X., Ren, D., and Yao, Z. (2012). Prevalence and distribution profiles of Candida parapsilosis, Candida orthopsilosis and Candida metapsilosis responsible for superficial candidiasis in a Chinese university hospital. Mycopathologia 173, 229–234. doi: 10.1007/s11046-011-9496-5
Gago, S., Garcia-Rodas, R., Cuesta, I., Mellado, E., and Alastruey-Izquierdo, A. (2014). Candida parapsilosis, Candida orthopsilosis, and Candida metapsilosis virulence in the non-conventional host Galleria mellonella. Virulence 5, 278–285. doi: 10.4161/viru.26973
Garcia-Effron, G., Katiyar, S. K., Park, S., Edlind, T. D., and Perlin, D. S. (2008). A naturally occurring proline-to-alanine amino acid change in Fks1p in Candida parapsilosis, Candida orthopsilosis, and Candida metapsilosis accounts for reduced echinocandin susceptibility. Antimicrob. Agents Chemother. 52, 2305–2312. doi: 10.1128/AAC.00262-08
Ge, Y. P., Boekhout, T., Zhan, P., Lu, G. X., Shen, Y. N., Li, M., et al. (2012). Characterization of the Candida parapsilosis complex in East China: species distribution differs among cities. Med. Mycol. 50, 56–66. doi: 10.3109/13693786.2011.591440
Ghannoum, M. A., Jurevic, R. J., Mukherjee, P. K., Cui, F., Sikaroodi, M., Naqvi, A., et al. (2010). Characterization of the oral fungal microbiome (mycobiome) in healthy individuals. PLoS Pathog. 6:e1000713. doi: 10.1371/journal.ppat.1000713
Gomez-Lopez, A., Alastruey-Izquierdo, A., Rodriguez, D., Almirante, B., Pahissa, A., Rodriguez-Tudela, J. L., et al. (2008). Prevalence and susceptibility profile of Candida metapsilosis and Candida orthopsilosis: results from population-based surveillance of candidemia in Spain. Antimicrob. Agents Chemother. 52, 1506–1509. doi: 10.1128/AAC.01595-07
Goncalves, S. S., Amorim, C. S., Nucci, M., Padovan, A. C. B., Briones, M. R. S., Melo, A. S. A., et al. (2010). Prevalence rates and antifungal susceptibility profiles of the Candida parapsilosis species complex: results from a nationwide surveillance of candidaemia in Brazil. Clin. Microbiol. Infect. 16, 885–887. doi: 10.1111/j.1469-0691.2009.03020.x
Govender, N. P., Patel, J., Magobo, R. E., Naicker, S., Wadula, J., Whitelaw, A., et al. (2016). Emergence of azole-resistant Candida parapsilosis causing bloodstream infection: results from laboratory-based sentinel surveillance in South Africa. J. Antimicrob. Chemother. 71, 1994–2004. doi: 10.1093/jac/dkw091
Grossman, N. T., Pham, C. D., Cleveland, A. A., and Lockhart, S. R. (2015). Molecular mechanisms of fluconazole resistance in Candida parapsilosis isolates from a U.S. surveillance system. Antimicrob. Agents Chemother. 59:1030 LP-1037. doi: 10.1128/AAC.04613-14
Heslop, O. D., De Ceulaer, K., Rainford, L., and Nicholson, A. (2015). A case of Candida orthopsilosis associated septic arthritis in a patient with Systemic Lupus Erythematosus (SLE). Med. Mycol. Case Rep. 7, 1–3. doi: 10.1016/j.mmcr.2014.11.001
Kathuria, S., Sharma, C., Singh, P. K., Agarwal, P., Agarwal, K., Hagen, F., et al. (2015). Molecular epidemiology and in-vitro antifungal susceptibility of Aspergillus terreus species complex isolates in Delhi, India: evidence of genetic diversity by amplified fragment length polymorphism and microsatellite typing. PLoS ONE 10:e0118997. doi: 10.1371/journal.pone.0118997
Kim, H. K., Park, Y. K., Wang, H.-J., Kim, B. W., Shin, S. Y., Lim, S.-K., et al. (2013). Epidemiology and clinical features of post-transplant bloodstream infection: an analysis of 222 consecutive liver transplant recipients. Infect. Chemother. 45, 315–324. doi: 10.3947/ic.2013.45.3.315
Lamoth, F., Lockhart, S. R., Berkow, E. L., and Calandra, T. (2018). Changes in the epidemiological landscape of invasive candidiasis. J. Antimicrob. Chemother. 73(suppl_1):i4-i13. doi: 10.1093/jac/dkx444
Lovero, G., Borghi, E., Balbino, S., Cirasola, D., De Giglio, O., Perdoni, F., et al. (2016). Molecular identification and echinocandin susceptibility of Candida parapsilosis complex bloodstream isolates in Italy, 2007-2014. PLoS ONE 11:e0150218. doi: 10.1371/journal.pone.0150218
Marchetta, A., Gerrits van den Ende, B., Al-Hatmi, A. M. S., Hagen, F., Zalar, P., Sudhadham, M., et al. (2018). Global molecular diversity of the halotolerant fungus Hortaea werneckii. Life 8:E31. doi: 10.3390/life8030031
Mello, E., Vella, A., Carolis, E., de Sanguinetti, M., Cuore, C. S., and Health, P. (2017). “Antifungal resistance and susceptibility testing Molecular mechanisms of azole resistance in clinical isolates of Candida orthopsilosis,” in ESCMID Conference (Vienna), 6–7.
Mirhendi, H., Bruun, B., Schonheyder, H. C., Christensen, J. J., Fuursted, K., Gahrn-Hansen, B., et al. (2010). Molecular screening for Candida orthopsilosis and Candida metapsilosis among Danish Candida parapsilosis group blood culture isolates: proposal of a new RFLP profile for differentiation. J. Med. Microbiol. 59, 414–420. doi: 10.1099/jmm.0.017293-0
Mohammadi, R., Mirhendi, H., Hedayati, M. T., and Badali, H. (2017). Caspofungin-non-susceptible Candida orthopsilosis isolated from onychomycosis in Iran. Iran J. Public Health. 46, 235–241.
Moreno, X., Reviákina, V., Panizo, M. M., Ferrara, G., García, N., Alarcón, V., et al. (2017). Molecular identification and in vitro antifungal susceptibility of blood isolates of the Candida parapsilosis species complex in Venezuela. Rev. Iberoam Micol. 34, 165–170. doi: 10.1016/j.riam.2016.11.005
Neji, S., Hadrich, I., Trabelsi, H., Abbes, S., Cheikhrouhou, F., Sellami, H., et al. (2017a). Virulence factors, antifungal susceptibility and molecular mechanisms of azole resistance among Candida parapsilosis complex isolates recovered from clinical specimens. J. Biomed. Sci. 24:67. doi: 10.1186/s12929-017-0376-2
Neji, S., Trabelsi, H., Hadrich, I., Cheikhrouhou, F., Sellami, H., Makni, F., et al. (2017b). Molecular study of the Candida parapsilosis complex in Sfax, Tunisia. Med. Mycol. 55, 137–144. doi: 10.1093/mmy/myw063
Oliveira, V. K. P., Paula, C. R., Colombo, A. L., Merseguel, K. B., Nishikaku, A. S., Moreira, D., et al. (2014). Candidemia and death by Candida orthopsilosis and Candida metapsilosis in neonates and children. Pediatr. Neonatol. 55, 75–76. doi: 10.1016/j.pedneo.2013.07.006
Pfaller, M. A., and Diekema, D. J. (2012). Progress in antifungal susceptibility testing of Candida spp. by use of clinical and laboratory standards institute broth microdilution methods, 2010 to 2012. J. Clin. Microbiol. 50, 2846–2856. doi: 10.1128/JCM.00937-12
Pryszcz, L. P., Nemeth, T., Gacser, A., and Gabaldon, T. (2014). Genome comparison of Candida orthopsilosis clinical strains reveals the existence of hybrids between two distinct subspecies. Genome Biol. Evol. 6, 1069–1078. doi: 10.1093/gbe/evu082
Pryszcz, L. P., Nemeth, T., Saus, E., Ksiezopolska, E., Hegedusova, E., Nosek, J., et al. (2016). Correction: the genomic aftermath of hybridization in the opportunistic pathogen Candida metapsilosis. PLoS Genet. 12:e1006202. doi: 10.1371/journal.pgen.1006202
Riccombeni, A., Vidanes, G., Proux-Wera, E., Wolfe, K. H., and Butler, G. (2012). Sequence and analysis of the genome of the pathogenic yeast Candida orthopsilosis. PLoS ONE 7:e35750. doi: 10.1371/journal.pone.0035750
Rizzato, C., Poma, N., Zoppo, M., Posteraro, B., Mello, E., Bottai, D., et al. (2018). CoERG11 A395T mutation confers azole resistance in Candida orthopsilosis clinical isolates. J. Antimicrob Chemother. 73, 1815–1822. doi: 10.1093/jac/dky122
Romeo, O., Delfino, D., Costanzo, B., Cascio, A., and Criseo, G. (2012). Molecular characterization of Italian Candida parapsilosis isolates reveals the cryptic presence of the newly described species Candida orthopsilosis in blood cultures from newborns. Diagn. Microbiol. Infect. Dis. 72, 234–238. doi: 10.1016/j.diagmicrobio.2011.12.002
Rosenberg, A., Ene, I. V., Bibi, M., Zakin, S., Segal, E. S., Ziv, N., et al. (2018). Antifungal tolerance is a subpopulation effect distinct from resistance and is associated with persistent candidemia. Nat. Commun. 9:2470. doi: 10.1038/s41467-018-04926-x
Schelenz, S., Hagen, F., Rhodes, J. L., Abdolrasouli, A., Chowdhary, A., Hall, A., et al. (2016). First hospital outbreak of the globally emerging Candida auris in a European hospital. Antimicrob. Resist. Infect. Control 5:35. doi: 10.1186/s13756-016-0132-5
Singh, A., Singh, P. K., de Groot, T., Kumar, A., Mathur, P., Tarai, B., et al. (2019). Emergence of clonal fluconazole-resistant Candida parapsilosis clinical isolates in a multicentre laboratory-based surveillance study in India. J. Antimicrob. Chemother. 74, 1260–1268. doi: 10.1093/jac/dkz029
Stielow, J. B., Levesque, C. A., Seifert, K. A., Meyer, W., Iriny, L., Smits, D., et al. (2015). One fungus, which genes? Development and assessment of universal primers for potential secondary fungal DNA barcodes. Persoonia 35, 242–263. doi: 10.3767/003158515X689135
Tavanti, A., Davidson, A. D., Gow, N., a, R., Maiden, M. C. J., and Odds, F. C. (2005). Candida orthopsilosis and Candida metapsilosis spp. nov. to replace Candida parapsilosis Groups II and III. J. Clin. Microbiol. 43, 284–292. doi: 10.1128/JCM.43.1.284
Tavanti, A., Hensgens, L. A. M., Ghelardi, E., Campa, M., and Senesi, S. (2007). Genotyping of Candida orthopsilosis clinical isolates by amplification fragment length polymorphism reveals genetic diversity among independent isolates and strain maintenance within patients. J. Clin. Microbiol. 45, 1455–1462. doi: 10.1128/JCM.00243-07
Tay, S. T., Na, S. L., and Chong, J. (2009). Molecular differentiation and antifungal susceptibilities of Candida parapsilosis isolated from patients with bloodstream infections. J. Med. Microbiol. 58, 185–191. doi: 10.1099/jmm.0.004242-0
Theelen, B., Silvestri, M., Gueho, E., van Belkum, A., and Boekhout, T. (2001). Identification and typing of Malassezia yeasts using amplified fragment length polymorphism (AFLP™), random amplified polymorphic DNA (RAPD) and denaturing gradient gel electrophoresis (DGGE). FEMS Yeast Res. 1, 79–86. doi: 10.1111/j.1567-1364.2001.tb00018.x
Thomaz, D. Y., de Almeida, J. N. J., Lima, G. M. E., Nunes, M., de, O., Camargo, C. H., et al. (2018). An azole-resistant Candida parapsilosis outbreak: clonal persistence in the intensive care unit of a Brazilian teaching hospital. Front. Microbiol. 9:2997. doi: 10.3389/fmicb.2018.02997
Trofa, D., Gacser, A., and Nosanchuk, J. D. (2008). Candida parapsilosis, an emerging fungal pathogen. Clin. Microbiol. Rev. 21, 606–625. doi: 10.1128/CMR.00013-08
Valles, Y., Inman, C. K., Peters, B. A., Ali, R., Wareth, L. A., Abdulle, A., et al. (2018). Types of tobacco consumption and the oral microbiome in the United Arab Emirates Healthy Future (UAEHFS) Pilot Study. Sci. Rep. 8:11327. doi: 10.1038/s41598-018-29730-x
Wang, H., Zhang, L., Kudinha, T., Kong, F., Ma, X.-J., Chu, Y.-Z., et al. (2016). Investigation of an unrecognized large-scale outbreak of Candida parapsilosis sensu stricto fungaemia in a tertiary-care hospital in China. Sci. Rep. 6:27099. doi: 10.1038/srep27099
Wessel, J. M., Bachmann, B. O., Meiller, R., and Kruse, F. E. (2013). Fungal interface keratitis by Candida orthopsilosis following deep anterior lamellar keratoplasty. BMJ Case Rep. 2013:8361. doi: 10.1136/bcr-2012-008361
Willis, J. R., Gonzalez-Torres, P., Pittis, A. A., Bejarano, L. A., Cozzuto, L., Andreu-Somavilla, N., et al. (2018). Citizen science charts two major “stomatotypes” in the oral microbiome of adolescents and reveals links with habits and drinking water composition. Microbiome 6:218. doi: 10.1186/s40168-018-0592-3
Keywords: C. parapsilosis species complex, Iran, AFLP genotyping, AFST, C. orthopsilosis, C. metapsilosis, clonality
Citation: Arastehfar A, Khodavaisy S, Daneshnia F, Najafzadeh M-J, Mahmoudi S, Charsizadeh A, Salehi M-R, Zarrinfar H, Raeisabadi A, Dolatabadi S, Zare Shahrabadi Z, Zomorodian K, Pan W, Hagen F and Boekhout T (2019) Molecular Identification, Genotypic Diversity, Antifungal Susceptibility, and Clinical Outcomes of Infections Caused by Clinically Underrated Yeasts, Candida orthopsilosis, and Candida metapsilosis: An Iranian Multicenter Study (2014–2019). Front. Cell. Infect. Microbiol. 9:264. doi: 10.3389/fcimb.2019.00264
Received: 11 March 2019; Accepted: 08 July 2019;
Published: 30 July 2019.
Edited by:
Max Maurin, Université Grenoble Alpes, FranceReviewed by:
Elisa Borghi, University of Milan, ItalyCopyright © 2019 Arastehfar, Khodavaisy, Daneshnia, Najafzadeh, Mahmoudi, Charsizadeh, Salehi, Zarrinfar, Raeisabadi, Dolatabadi, Zare Shahrabadi, Zomorodian, Pan, Hagen and Boekhout. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Kamiar Zomorodian, em9tb3JvZGlhbkBzdW1zLmFjLmly; Weihua Pan, cGFud2VpaHVhQHNtbXUuZWR1LmNu
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.