- 1Centre for Infectious Diseases and Microbiology, The Westmead Institute for Medical Research, Sydney, NSW, Australia
- 2Sydney Medical School-Westmead, The University of Sydney, Sydney, NSW, Australia
- 3Marie Bashir Institute for Infectious Diseases and Biosecurity, University of Sydney, Sydney, NSW, Australia
- 4Centre for Infectious Diseases and Microbiology–Public Health, NSW Health Pathology, Westmead Hospital, Sydney, NSW, Australia
- 5MRC Laboratory for Molecular Cell Biology, University College London, London, United Kingdom
Invasive fungal pathogens cause more than 300 million serious human infections and 1.6 million deaths per year. A clearer understanding of the mechanisms by which these fungi cause disease is needed to identify novel targets for urgently needed therapies. Kinases are key components of the signaling and metabolic circuitry of eukaryotic cells, which include fungi, and kinase inhibition is currently being exploited for the treatment of human diseases. Inhibiting evolutionarily divergent kinases in fungal pathogens is a promising avenue for antifungal drug development. One such group of kinases is the phospholipase C1-dependent inositol polyphosphate kinases (IPKs), which act sequentially to transfer a phosphoryl group to a pre-phosphorylated inositol sugar (IP). This review focuses on the roles of fungal IPKs and their IP products in fungal pathogenicity, as determined predominantly from studies performed in the model fungal pathogen Cryptococcus neoformans, and compares them to what is known in non-pathogenic model fungi and mammalian cells to highlight potential drug targeting opportunities.
Invasive fungal diseases threaten human and animal health and food security. Invasive fungal diseases have caused recent major dieoffs and extinctions in wild animal species and plants, forcing overuse of antifungals to sustain our food supply (Fisher et al., 2012; Editorial Stop neglecting fungi, 2017). World-wide, invasive fungal pathogens cause more than 300 million serious human infections and 1.6 million deaths per year (Editorial Stop neglecting fungi, 2017). Infections caused by Cryptococcus neoformans, Candida albicans and Aspergillus fumigatus are common in immunocompromised individuals (e.g., those with HIV, blood cancers, and organ transplants) and a lack of effective antifungal drugs has compromised recent medical advances in the treatment of these conditions. Current antifungal agents are either toxic, lack broad-spectrum activity or are sub-optimally efficacious due to poor absorption, contributing to side effects and high rates of morbidity and mortality (Brown et al., 2012). Prolonged treatment courses breed drug resistance. Despite the urgent need, no new drug classes have been marketed since the introduction of echinocandins in 2002 (Denning, 2002).
Kinases are key components of signaling and metabolic pathways in all eukaryotes, including fungi. In silico predictions in C. neoformans identified 183 kinases, and experimental data show that 63 of them play a role in pathogenicity (Lee et al., 2016). The inhibition of kinases by small molecules is of major interest to pharmaceutical companies. With over 30 kinase inhibitors approved for treating non-infectious diseases, selective inhibition of fungal kinase activity is a potentially appealing antifungal therapeutic strategy. Kinases catalyze the transfer of phosphoryl groups from high energy molecules, predominantly ATP, to a variety of substrates. Eukaryotic protein kinases (where the substrate is a protein) are the most-well characterized. In contrast, kinases that target lipids and phosphorylated sugars are less well-studied.
Despite their reduced prevalence, these latter kinases provide a source of secondary messengers, which have crucial roles in signal transduction. Inhibitors targeting phosphoinositide kinases, which use the lipid phosphatidylinositol or its phosphorylated derivatives (e.g., PIP2) as substrates are under investigation as anticancer agents (Fabbro, 2015). They transfer a phosphoryl group to a specific position on the inositol ring of the phospholipid head group. In contrast, the inositol polyphosphate kinases (IPKs), which are often confused with the phosphoinositide kinases, are distinct in that they phosphorylate a variety of cytosolic, water-soluble inositol polyphosphates (IPs) originating from the phosphatidylinositol head group. In yeast, the first of these substrates is inositol trisphosphate (IP3). IP3 is generated by the hydrolytic action of the delta isoform of PLC (PLC1 in yeast) on PIP2. IPKs sequentially phosphorylate IP3 to create a combination of molecules with monophosphates at various positions on the inositol ring (IPs), or a combination of monophosphates and di- (or pyro-) phosphates (PP-IPs). The PP moiety harbors a “high-energy” phosphoanhydride bond, similar to ATP. Presumably due to their function as phosphate donors, PP-IPs undergo rapid turnover in eukaryotic cells (Menniti et al., 1993). Both the phosphorylated lipid products of phosphoinositide kinases and the soluble IPs/PP-IPs products of IPKs regulate diverse cellular processes in eukaryotes, including proliferation, survival, cytoskeletal arrangement, vesicle traffic, glucose transport, and platelet function (Fruman et al., 1998; Sarmah and Wente, 2010). Perturbed synthesis of IPs is also linked to apoptosis and insulin secretion (Nagata et al., 2005; Illies et al., 2007; Sarmah and Wente, 2010), and developmental defects in vertebrates (Frederick et al., 2005; Sarmah et al., 2005). IPs also regulate DNA repair, chromatin remodeling, transcription and gene expression, mRNA export, telomere length, exocytosis, RNA editing, translation, and Ca2+ channels (York et al., 1999; Shen et al., 2003; Steger et al., 2003; Macbeth et al., 2005; Saiardi et al., 2005; Alcazar-Roman and Wente, 2008; Bolger et al., 2008; Sarmah and Wente, 2009).
Identification of the PLC1-dependent IPK Pathway as a New Virulence-Related Signaling Pathway in the Model Fungal Pathogen C. neoformans
The IPK pathway has been characterized in mammalian cells and in model (non-pathogenic) yeast. The most substantial progress in characterizing the pathway in a fungal pathogen has come from studies performed in C. neoformans (Lev et al., 2013, 2015; Li et al., 2016; Li C. et al., 2017). To date, the only non-fungal pathogen in which kinases in the pathway have been studied is the protozoan parasite, Trypanosoma brucei, which causes sleeping sickness (Cordeiro et al., 2017).
C. neoformans is a respiratory pathogen that disseminates to the central nervous system. It causes fatal meningitis in about a quarter of a million people per year. The majority of these fatalities occur in individuals living in Sub-Saharan Africa and South East Asia, who have limited access to appropriate antifungal drugs (Rajasingham et al., 2017). The advantages of using C. neoformans as a working model are its genetically tractable and annotated haploid genome, and the availability of robust vertebrate and invertebrate infection models. Using S. cerevisiae IPK protein sequences in homology searches, we identified a series of putative IPK-encoding genes in the C. neoformans strain H99 genome and designated them Arg1, Arg2, Ipk1, Kcs1, and Vip1/Asp1 (Figure 1). Similar to the fission yeast, Schizosaccharomyces pombe, Vip1 is referred to as Asp1 in the cryptococcal database (Topolski et al., 2016; Pascual-Ortiz et al., 2018). The most striking difference between the yeast and mammalian IPK pathways is the linearity, and hence lack of redundancy in the yeast pathway. In mammalian cells, there is substantial redundancy, with 3 alternative enzymes able to convert IP3 to IP5 (Li et al., 2016).
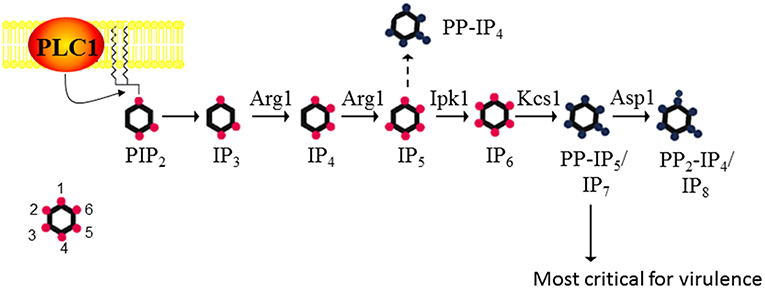
Figure 1. The inositol polyphosphate biosynthesis pathway in C. neoformans. PLC1 –derived IP3 is sequentially phosphorylated to IP4, IP5, and IP6 by Arg1 and Ipk1. Kcs1 generates PP-IP4 and PP-IP5/IP7 from IP5 and IP6, respectively. However, PP-IP4 is only detected in the absence of Ipk1 (hence the dashed line). PP2-IP4 is derived from Asp1. The insert represents the position of the phosphates on the inositol ring. Figure was adapted from Lev et al. (2015), Li et al. (2016).
Metabolic profiling of cryptococcal (Cn) IPK deletion mutants was used to determine the substrate specificity of each IPK. CnArg1 and CnArg2 are most similar to the S. cerevisiae (Sc) IP3/IP4 kinase, ScArg82/Ipk2/ArgIII/IPMK (Lev et al., 2013; Li C. et al., 2017). However, only CnArg1 plays a role in IP metabolism in C. neoformans: in the ARG1 deletion mutant (arg1Δ), but not the arg2Δ mutant, soluble IP3 accumulated and was not further phosphorylated to IP4−8 (Lev et al., 2015). Furthermore, enzymatic assays using recombinant CnArg1 confirmed that this kinase uses ATP to convert IP3 to IP4 and IP5 (Li C. et al., 2017), indicating that CnArg1 is the major IP3 kinase in C. neoformans. It was shown that the I(1,4,5)P3 substrate of Arg1 is derived from phosphatidylinositol 4,5-bisphosphate (PIP2) via the hydrolytic activity of phospholipase C1 (PLC1) (Chayakulkeeree et al., 2008; Lev et al., 2013). CnArg1 could restore defective phenotypes in the Scarg82Δ mutant, confirming that CnArg1 is the ortholog of ScArg82 (Li C. et al., 2017). CnArg2 has no identified cellular function but is unlikely to be a pseudo-kinase as it retains the conserved “P-x-x-x-D-x-K-x-G” catalytic motif essential for IP3 kinase activity in ScArg82 and CnArg1 (Saiardi et al., 1999; Bertsch et al., 2000). The reason for IP3 kinase gene duplication in this basidiomycetous yeast but not in the ascomycetes is unknown, but may reflect evolutionary diversification.
Metabolic profiling confirmed that CnIpk1 is the major IP5 kinase in C. neoformans, using IP5 as a substrate to produce a molecule with a fully phosphorylated inositol ring, namely, IP6 (Li et al., 2016). CnKcs1 and CnAsp1 were shown to be inositol pyrophosphate synthases: CnKcs1 phosphorylates IP6 at position 5 generating 5-PP-IP5 (IP7), which is further phosphorylated by CnAsp1 at position 1 to produce 1,5-PP2-IP4 (IP8)(Lev et al., 2015). In S. cerevisiae, the homolog of Asp1, Vip1, also phosphorylates IP6, producing 1-PP-IP5. However, this ScVip1 activity is very low as the 1-PP-IP5 product can only be detected by HPLC when both ScKCS1 and the inositol pyrophosphatase-encoding gene, ScDDP1, are deleted, the assumption being that any 1-PP-IP5 made by Asp1 is broken down by ScDdp1 (Mulugu et al., 2007). Whether Asp1 can phosphorylate IP6 in C. neoformans, to produce 1-PP-IP5, remains to be determined.
Cnarg1Δ, Cnkcs1Δ, and Cnipk1Δ were either avirulent or severely attenuated in virulence in a mouse inhalation model as discussed in the next section (Lev et al., 2013, 2015; Li et al., 2016; Li C. et al., 2017). This was in contrast to Cnarg2Δ and Cnasp1Δ, which retained a WT-like virulence profile. The importance of CnArg1 and CnKcs1 in cryptococcal cellular function was also highlighted by the fact that neither deletion mutant is represented in a signature-tagged kinome mutant library created by Lee et al in a genome-wide study of cryptococcal kinases (Lee et al., 2016). Only the Cnipk1Δ and Cnasp1Δ mutants are present in this library, with CnIpk1 also implicated in pathogenicity (Lee et al., 2016).
IP3 Kinase, Arg1, has the Most Significant Impact on Fungal Cellular Function and Pathogenicity, Acting via its IP Products
The impact of the loss of IP3 kinase function has been studied in C. neoformans and C. albicans. Compared to other IPKs, loss of CnArg1 had the most debilitating effect on cryptococcal phenotype (Li C. et al., 2017), while the phenotype and metabolic profile of the Cnarg2Δ mutant was similar to that of WT (Lev et al., 2013). Cnarg1Δ phenotypes included a significant reduction in growth at human body temperature (37°C) in either rich (YPD) or a more physiological cell culture medium (RPMI/5%CO2), a cell separation defect and enlarged vacuoles (Lev et al., 2013; Li C. et al., 2017). Additional defects included reduced mating filamentation and production of the virulence factors urease and melanin, and a cell wall integrity defect (Lev et al., 2013; Li C. et al., 2017). The latter was associated with a thickened cell wall and hypermannosylation of the cell wall-associated/secreted virulence determinant phospholipase B1 (PLB1) (Li C. et al., 2017). Although PLB1 release into the culture medium was attenuated, more cell-associated PLB1 was consistent with a secretion block. Capsules of the Cnarg1Δ mutant were smaller than those of WT, and this defect correlated with a higher rate of Cnarg1Δ phagocytosis by human peripheral blood monocytes and rapid clearance from lung in a mouse infection model (Li C. et al., 2017). The Cnarg1Δ mutant also exhibited reduced growth in the presence of non-glucose carbon sources and during phosphate starvation (Li C. et al., 2017). Cnarg1Δ phenotypes are summarized in Figure 2.
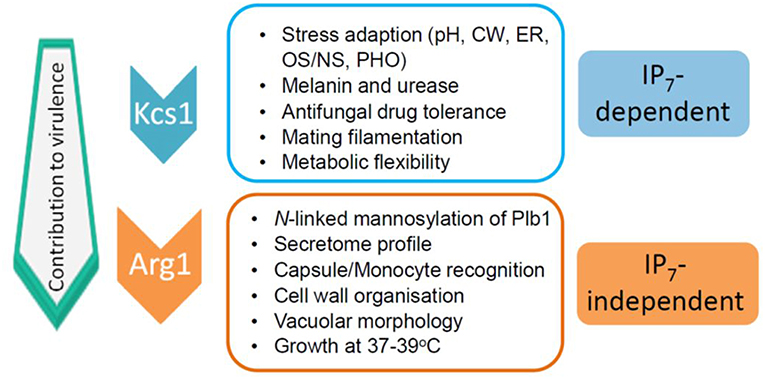
Figure 2. IP7-dependent and IP7-independent functions conveyed via CnArg1. Due to these combined functions, the contribution of CnArg1 to fungal virulence is greater than that of CnKcs1. pH—alkaline pH stress; CW—cell wall stress induced by calcofluor white, Congo red, SDS and caffeine; ER—endoplasmic reticulum stress caused by tunicamycin and DTT; OS/NS—oxidative and nitrosative stress; PHO—low phosphate.
In a recent study by Li J. et al. (2017), the impact of loss of the IP3 kinase gene (referred to as IPK2) was investigated in diploid C. albicans. The authors were unable to delete both copies of CaIPK2, consistent with an essential role for CaIPK2 in cellular function. Instead they created a heterozygous IPK2 deletion mutant and controlled expression of the remaining IPK2 allele using the inducible MET3 promoter. In contrast to the suppression of virulence-associated functions observed in C. neoformans following deletion of ARG1, suppression of CaIPK2 in C. albicans promoted virulence-related functions, such as hyphal development, by increasing expression of hyphal-specific genes and the transport of hypha-specific factors, and secretion of degradative enzymes such as proteases and lipases, which would include phospholipases (Li J. et al., 2017). Hyphal development and secretion of degradative enzymes allow C. albicans to invade the epithelial barrier and avoid detection by the innate immune response. The effect of diminished IPK2 gene expression on the virulence of C. albicans in animal infection models remains to be determined.
Although C. neoformans does not form hyphae during vegetative growth, it does alter its morphology during mating. However, loss of CnArg1 reduced mating-associated filamentation in C. neoformans (Lev et al., 2013). The functional significance of the differing roles of CnArg1 and CaIpk2 in regulating morphology in these two fungal pathogens remains to be determined. In contrast, overproduction of lipases, in particular PLB1, was a consistent phenotype in Arg1-deficient C. neoformans and C. albicans. Although increased PLB1 abundance on the cryptococcal cell surface could potentially lead to enhanced virulence, the opposite was found to be true (Li C. et al., 2017). Li J. et al. (2017) also found that loss of IPK in C. albicans increased damage to macrophages. Although macrophage damage by Cnarg1Δ was not assessed, increased uptake of the Cnarg1Δ mutant by blood monocytes was observed (Li C. et al., 2017). This may have resulted in a higher rate of macrophage death due to the oxidative burst directed at the internalized pathogen. Similar to the mammalian and S. cerevisiae Ipk2/Arg82 homologs, CaIpk2 was observed to be nuclear (Li J. et al., 2017). The cellular location of CnArg1 remains to be determined.
Potential Lipid Substrates of IP3 Kinases
Another potential metabolic function of Arg1 is to phosphorylate PIP2 and thus act as a PI3K. Both human and yeast IPMKs (i.e. ARG82/IPK2) display PI3K activity in vitro (Resnick et al., 2005). Using mammalian cell transfection studies, Resnick et al. (2005) showed that human IPMKs were exclusively localized in the nucleus and exhibited wortmannin-insensitive PI3K activity to regulate transcription. In contrast, canonical PI3Ks are predominantly cytosolic and wortmannin-sensitive. The importance of Arg82 PI3K activity in vivo has not been directly addressed. The cellular location of CnArg1 and whether CnArg1 functions as a PI3K to use membrane phosphoinositides as a substrate, also remains to be determined.
Functions of IP3 Kinases Which Are Independent of Catalytic Activity
In S. cerevisiae, Arg82 is nuclear-localized and possesses functions independent of its catalytic activity. Using a kinase-dead mutant strain in which key catalytic residues were mutated, ScArg82 was shown to be a regulatory component of the arginine-sensitive ArgR-Mcm1 transcriptional complex, which is required for regulating gene expression associated with arginine synthesis and breakdown (Bechet et al., 1970; Dubois et al., 1987; Bosch and Saiardi, 2012). These data led to the assignment of the “Arg” nomenclature. Given that the Arg1 homolog in C. albicans is also nuclei-localized, it too could potentially function to regulate gene expression (Li J. et al., 2017). Whether CnArg1 is nuclear and whether all of the attenuated phenotypes associated with the Cnarg1Δ mutant are due solely to the absence of one or more of its enzymatic products remains to be determined. However, due to the Cnarg1Δ mutant sharing several phenotypic defects with the Cnkcs1Δ and the CnKcs1 kinase-dead mutants (Lev et al., 2015), many of the key functions of Arg1 in pathogenicity are attributable to IP7 as discussed below.
Hierarchy of Importance of IPs in Fungal Pathogenesis Favors PP-IP4 and PP-IP5/IP7
Role of IP8
To dissect the importance of cryptococcal IPs downstream of IP3 in cellular function, the phenotypes of the WT, Cnipk1Δ, Cnipk1Δkcs1Δ, Cnkcs1Δ, and Cnasp1Δ deletion mutants, and their virulence profile in animal models, were compared. Taking this approach, the impact of absent metabolites on cellular function could be assessed. The asp1Δ mutant was found to accumulate IP7 and was deficient in IP8. No impact of absent IP8 on cellular function was observed as the Cnasp1Δ mutant exhibited a WT phenotype under all growth conditions tested. As expected from the phenotypic assessment, Cnasp1Δ virulence was similar to WT in a mouse inhalation model (Lev et al., 2015). In contrast, IP8 is required for cellular functions such as dimorphic switching, polarized growth and microtubule cytoskeletal regulation in S. pombe, Aspergillus nidulans and the basidiomycete, Ustilago maydis (Feoktistova et al., 1999; Pohlmann and Fleig, 2010; Pohlmann et al., 2014). In addition to IP8, Vip1 in S. cerevisiae produces 1-PP-IP5. It is interesting to note that the structure of ScVip1 is atypical, and differs significantly from other IPKs: it contains an N-terminal kinase domain and a C-terminal histidine acid phosphatase domain. This acid phosphatase domain functions to repress the activity of the kinase domain (Pascual-Ortiz et al., 2018).
Role of IP7
In contrast to IP8, the absence of its precursor, IP7 in the Cnkcs1Δ mutant had a profound impact on phenotype and virulence. However, the effect was not as marked as that in the Cnarg1Δ mutant, which is also deficient in IP7. A comparison of the phenotypes of the two mutants allowed IP7-dependent and IP7-independent functions to be distinguished as summarized in Figure 2 (Lev et al., 2015; Li C. et al., 2017). The Cnkcs1Δ mutant had a less severe 37°C growth defect than the Cnarg1Δ mutant in rich medium (although the temperature defect for both mutants was similar in the more physiological cell culture medium). The Cnkcs1Δ mutant also had a cell wall integrity defect but displayed neither the abnormal multi-layered, thickened cell walls nor the enlarged vacuoles observed for the Cnarg1Δ mutant. Cnarg1Δ cells were enlarged, while Cnkcs1Δ cells were WT-like. The Cnkcs1Δ mutant also produced lower amounts of the virulence factors, melanin, urease and PLB1 but had larger and more mucoid capsules compared to the Cnarg1Δ mutant. The PLB1 production defect in the kcs1Δ mutant was different to that of the Cnarg1Δ mutant: relative to WT and the Cnarg1Δ mutant, the Cnkcs1Δ mutant produced less total PLB1 (cell-associated and secreted), while only the release of cell-associated PLB1 was affected (i.e. blocked) in the Cnarg1Δ mutant. The secretome profile was also altered in the Cnarg1Δ mutant and this may reflect an altered cell wall proteome and the unusual cell wall architecture. The N-linked mannosylation defect was also absent in the Cnkcs1Δ mutant. The Cnkcs1Δ mutant was poorly recognized by phagocytes. This is in contrast to the Cnarg1Δ mutant which was readily phagocytosed. Both mutants were compromised in utilizing carbon sources other than glucose. This metabolic defect is predicted to impact negatively on their ability to survive in the low glucose environment of the host lung (Garnett et al., 2012). Both mutants were avirulent in a mouse inhalation model (no mice exhibited symptoms of infection up to 60 days post-infection). However, Cnarg1Δ infection was cleared rapidly while kcs1Δ infection resulted in lower lung burdens and no dissemination to the CNS. These results suggested that IP7 is an important metabolite in promoting fungal fitness and dissemination. Reduced recognition of Cnkcs1Δ cells by primary or immortalized monocytes and failure to elicit a strong immune response in vivo and in vitro correlated with reduced exposure of mannoproteins on the Cnkcs1Δ cell surface.
Taken together it was concluded that IP7 is the most crucial of the IPs for cryptococcal cellular function, fitness and virulence, being essential for fungal metabolic adaptation to the host environment, immunorecognition, dissemination, and pathogenicity (Lev et al., 2015). Functions for IP7 have also been identified in S. cerevisiae, and include the promotion of resistance to salt stress, cell wall integrity and vacuolar morphogenesis (Dubois et al., 2002) and autophagy (Taylor et al., 2012).
RNAseq analysis of the IPK deletion mutant set in C. neoformans (arg1Δ, ipk1Δ, and kcs1Δ) supports a role for IPs in the regulation of gene expression, with genes involved in mRNA translation and ribosomal biosynthesis being more highly expressed than in the WT strain (Lev et al., 2015). The RNAseq data also showed that the pattern of disregulated gene expression is mostly similar in the 3 CnIPK mutants (Li et al., 2016). Since IP7 is commonly absent in the 3 IPK mutants, the changes can therefore be attributed predominantly to the absence of IP7. IP7 may modulate gene expression via interaction with transcription and translation machinery or via pyrophosphorylation of its components as discussed below. Alternatively, altered gene expression in the absence of IP7 could be due to compensatory changes.
As mentioned above for IP3 kinases, IPKs can perform functions independent of their catalytic activity, such as acting as scaffolds to tether proteins into complexes as has been described for ScArg82 (see above). They also have roles in subcellular protein targeting and DNA binding (Rauch et al., 2011). To investigate whether this is true of CnKcs1, a “kinase-dead” mutant was created by altering critical catalytic residues using site-directed mutagenesis, and its phenotype compared with that of Cnkcs1Δ. The same attenuation in phenotype was observed in both mutants i.e., a cell wall integrity and high temperature growth defect, reduced LAC1 expression, laccase activity and melanin production and larger and more mucoid capsules (Lev et al., 2015) (summarized in Figure 2). The phenotypes observed for both the Cnkcs1Δ mutant and the kinase-dead mutant are therefore due solely to the absence of its enzymatic product IP7 as IP8 does not contribute to the virulence profile.
Kcs1 (presumably via IP7) also plays a role in inositol homeostasis in S. cerevisiae and C. neoformans (Ye et al., 2013; Liao et al., 2018). By comparing the phenotype of WT and the kcs1Δ mutants, Kcs1 was shown to negatively regulate inositol uptake and catabolism. However, in contrast to Kcs1 function in S. cerevisiae, Kcs1 did not regulate inositol biosynthesis in C. neoformans. Irrespective of the presence of glucose, inositol also repressed KCS1 gene expression in C. neoformans, potentially impacting PP-IP5/IP7 levels (El Alami et al., 2003).
Role of IP6 and 5-PP-IP4
IP6 is the most abundant IP species. However, by comparing the phenotypes of Cnipk1Δ, Cnipk1Δkcs1Δ, and Cnkcs1Δ, IP6 was shown to not play a significant role in cryptococcal virulence. This is presumably because other IP and PP-IP species can compensate for its absence, or for the absence of IP7 as discussed below. Thus, targeting IP6 production may not be the best approach for drug development.
Although Kcs1 is the major IP6 kinase in C. neoformans, phosphorylating IP6 to create IP7, it also acts as an IP5 kinase when IP6 is absent (i.e., in the Cnipk1Δ mutant). The IP5 that accumulates in Cnipk1Δ is converted by CnKcs1 to another pyrophosphate species, 5-PP-IP4, via the addition of a phosphoryl group to the existing phosphate at position five (see Figure 1) (Li et al., 2016). 5-PP-IP4 is therefore only detected when IPK1 function is disrupted and its physiological relevance remains to be determined. Relative to IP7, 5-PP-IP4 is produced by CnKcs1 in significant quantities when CnIpk1 function is abolished, and the presence of 5-PP-IP4 in Cnipk1Δ correlates with an increased virulence phenotype relative to that of Cnipk1Δkcs1Δ and Cnkcs1Δ (Li et al., 2016). This suggests that the “bypass pathway” responsible for production of PP-IP4 can partially compensate for the absence of IP7 (and hence its progenitor IP6) in the Cnipk1Δ mutant. This functional redundancy is most likely due to the structural similarity between the two PP-IP species (see Figure 1). Interestingly, unlike Cnipk1Δkcs1Δ and Cnkcs1Δ, Cnipk1Δ lung and brain burdens were high and resulted in a 20% mortality rate by 50 days post infection (Li et al., 2016). The Cnipk1Δ mutant is therefore well-tolerated in the mouse and slow to be detected by the host immune system. Elevated 5-PP-IP4 in Cnipk1Δ correlated with the rescue of alternative carbon source utilization but not with the rescue of laccase and urease production and ability to grow under oxidative/ nitrosative stress (Li et al., 2016).
In summary, a comparison of the phenotypes of CnIPK mutants confirmed that CnArg1 and IP7 are the most crucial IPK and IP species, respectively, for fungal cellular function and virulence, and allowed IP7-dependent and IP7-independent functions of CnArg1 to be distinguished (Figure 2). Possible reasons for IP7-deficent Cnarg1Δ having the most attenuated virulence phenotype of all of the IPK mutants, are the loss of IP4/5 and/or elevated levels of PIP2 and IP3. The potential impact of these changes in the IP profile is discussed below.
IP7 and the Regulation of Phosphate Homeostasis in Fungi
The pleiotropic phenotype of the IP7-deficient Cnkcs1Δ strain confirms that many of the important functions of CnArg1 are conveyed via IP7 (Lev et al., 2015). It was demonstrated that both the ability to synthesize IP7 and to activate the fungal phosphate acquisition (PHO) pathway, are essential for C. neoformans to grow in the blood and disseminate from the lung to the brain (Lev et al., 2015, 2017). This is consistent with the IPK and PHO pathways being connected in C. neoformans via IP7, as has been reported in S. cerevisae and S. pombe. No information is available on a possible connection between IP7 and the PHO pathway in other pathogenic fungi.
IP7 is extraordinarily rich in phosphate and its biosynthesis is directly tied to phosphate availability. Moreover, it was suggested that in S. cerevisiae, IP7 serves as a cellular energy sensor due to its high energy pyrophosphate (PP) bond. The Km of ScKcs1 for ATP is 1.1–1.4 mM, which is similar to the physiological concentration of ATP in the absence of nutritional stress (Wilson et al., 2013). Thus, when ATP is depleted, IP7 should also be depleted.
How the PHO Pathway Becomes Activated
The fungal PHO pathway is activated when phosphate becomes scarce in the environment or at alkaline pH. The cyclin-dependent kinase inhibitor Pho81, then inhibits the cyclin-dependent kinase Pho85, which ceases phosphorylation of the transcription factor Pho4. The helix-loop-helix transcription factor Pho4, is exported from the nucleus when it is phosphorylated, and imported into the nucleus when it is dephosphorylated. In S. cerevisiae and C. albicans, Pho4 associates with Pho2 to trigger expression of genes involved in phosphate acquisition. In C. neoformans, Pho4 is the sole transcription factor regulating PHO gene expression. One of the PHO gene products, secreted acid phosphatase, is widely used to monitor PHO pathway activation in variety of fungal species using a simple and robust colorimetric enzyme assay. The PHO pathway was recently implicated in cryptococcal pathogenesis following the demonstration that CnPho4, and therefore the expression of CnPho4-dependent genes, is essential for growth of C. neoformans in the blood and for cryptococcal dissemination to the brain (Lev et al., 2017). Ikeh et al also showed that CaPho4 is essential for disseminated candidiasis (Ikeh et al., 2016).
A number of earlier studies in S. cerevisiae reported perturbation of PHO gene expression in ScIPK mutants: Auesukaree et al (Auesukaree et al., 2005) demonstrated that the acid phosphatase encoding gene, PHO5, was constitutively expressed in the Scplc1Δ, Scarg82Δ, and Sckcs1Δ mutants and therefore not responsive to phosphate deprivation. Similarly, El Alami reported up-regulation of PHO5, and other PHO genes, in Scarg82Δ and Sckcs1Δ under non-inducing (phosphate replete) conditions (El Alami et al., 2003). There is some controversy as to which IP7 isoform is involved in PHO pathway activation: Lee et al demonstrated that ScVip1-derived IP7 (the 1-PP-IP5 isoform) interacts non-covalently with ScPho81 in the context of the ScPho80-Pho85-Pho81 complex, thus preventing Pho4 from accessing the kinase active site (Lee et al., 2007, 2008). Their data showed a corresponding increase in IP7 following phosphate deprivation. However, these findings are controversial: firstly, the level of ATP declines as a result of phosphate deprivation, rendering an increase in ATP-dependent IP7 production unlikely (Choi et al., 2017). In fact, Lonetti et al demonstrated a decrease in the abundance of ScKcs1-derived IP7 (5-PP-IP5) under similar conditions (Lonetti et al., 2011). It is possible that due to the low affinity of ScKcs1 for ATP, the activity of ScKcs1 is highly sensitive to ATP fluctuations, while the activity of ScVip1 is not affected, leading to an observed increase in IP7. Secondly, in a study assessing PHO pathway activation in IP6K mutants, it was shown that in the absence of ScKcs1, but not ScVip1, expression of PHO5 and PHO84 is constitutive, implicating ScKcs1-derived 5-PP-IP5, not Vip1-derived 1-PP-IP5, in PHO pathway regulation (Nishizawa et al., 2008; Choi et al., 2017).
IP7 Interacts With SPX Domains of PHO Pathway Components
SPX domains are found in eukaryotic proteins involved in phosphate homeostasis. In a recent breakthrough study, Wild et al demonstrated that SPX domains in S. cerevisiae and the model plant, Arabidopsis, provide a positively-charged binding module for various PP-IPs (Wild et al., 2016). They found that SPX domains bind IP6 and IP7 with high affinity (Kd in nM range) and other IPs including IP3/IP4/IP5 and free orthophosphate, with markedly lower affinity (Kd in μM range). They hypothesized that the abundance of IP7, which changes in response to phosphate deprivation, communicates cellular phosphate levels to SPX domain proteins to trigger an adaptive response (Wild et al., 2016). Whether IP7 interacts with SPX domains of PHO pathway components in fungal pathogens to fine-tune phosphate homeostasis, remains to be investigated.
The genome of S. cerevisiae encodes 10 SPX domain proteins, all containing the SPX domain at the N-terminus [(Desfougeres et al., 2016), reviewed in Azevedo and Saiardi (2017)]. Four of these proteins are components of the vacuolar transport complex (VTC). The others are Pho91, Pho87, Pho90, Gde1/Pho81, Syg1, and Xpr1. SPX is derived from Syg1, Pho81, and Xpr1, which are the proteins in which the SPX domain was first discovered. The VTC complex couples the production of inorganic polymers of phosphate (polyphosphates or polyP) with their transport to vacuoles and acidocalciosome-like vacuoles where they are stored. PolyP in these stores are promptly mobilized as a source of phosphate when the intracellular phosphate concentration is insufficient. IP6 and 5-PP-IP5 were shown to bind to the SPX domains of the VTC protein complex, stimulating polyP biosynthesis in isolated yeast vacuoles in vitro (Wild et al., 2016). Interestingly, IP6 and IP7 (the 5-PP-IP5 isoform) also bind to the SPX domain in the Na+/Pi symporter Pho91, triggering phosphate and sodium release from yeast vacuoles in vitro (Potapenko et al., 2018). Pho87 and Pho90 are low affinity plasma membrane phosphate transporters, which function when phosphate is available. In these proteins, the SPX domain performs an inhibitory function, limiting low-affinity phosphate uptake (Hurlimann et al., 2009). Pho81/GDE1 is the CDK inhibitor of the PHO pathway but also has glycerophosphodiesterase activity, which is thought to generate glycerol-phosphate during phosphate starvation (Fisher et al., 2005). Syg1 is predicted to be a phosphate transporter based on its similarity to the mammalian phosphate exporter Xpr1 (Giovannini et al., 2013). PP-IP interactions with the SPX domain of Syg1 have not been investigated. Of the SPX domain proteins in S. cerevisiae, only four have homologs in C. neoformans, including CnPho91, CnPho81, CnSyg1, and the CnVtc4 component of the VTC complex. Vtc4 and Pho81 have a similar function in C. neoformans and S. cerevisiae (Kretschmer et al., 2014; Toh-e et al., 2015), while the roles for CnPho91 and CnSyg1 have not been determined.
In summary, identification of SPX domain proteins as cellular IP7 receptors is a milestone in our understanding of how these molecules orchestrate numerous cellular functions. Given the different functions of each SPX domain protein, the impact of each SPX-IP7 interaction in fungal pathogenicity should be assessed on a case by case basis to gain a full understanding of this novel regulatory mechanism.
PP-IPs and the Regulation of Stress Response
In S. cerevisiae, all PP-IP species regulate chromatin remodeling, working in parallel with the target of rapamycin complex 1 (TORC1) pathway to control class I histone deacetylase Rpd3L activity and elicit global transcriptional changes in response to stress or starvation (Worley et al., 2013). Phenotypic cluster analysis of the cryptococcal kinome (Lee et al., 2016) identified the IP5 kinase Ipk1, and therefore potentially its distal PP-IP products, as working in the same pathway as the essential TORC1 kinase complex, rather than working in parallel as reported by Worley et al. (2013). In support of this, phosphate was identified as a nutrient monitored by TOR in Candida albicans (Liu et al., 2017). PP-IPs have also been reported to regulate apoptosis and telomere length by antagonizing the actions of PI3K-related protein kinases in the nucleus (Saiardi et al., 2005).
PP-IPs Exert Regulatory Effects by Pyrophosphorylation
Direct binding of IP7 to SPX proteins as described above, is only one mechanism by which PP-IPs exert pleiotropic effects in eukaryotic cells. However, PP-IPs are unique in that they can also modulate the activity of their target proteins by pyrophosphorylating them. This non-enzymatic reaction involves transferring the terminal phosphoryl group from their PP moiety to a pre-existing phosphate on the target, creating a new PP bond (Bhandari et al., 2007). For example, IP7 regulates yeast rRNA synthesis (a first step in ribosome biosynthesis) via pyrophosphorylation of RNA polymerase I (Thota et al., 2015). A recent study in mammalian cells by Chanduri et al demonstrated that IP7 regulates dynein-driven cytoplasmic vesicular transport along the microtubule. Ser51 in the dynein intermediate chain is pyrophosphorylated by IP7, and this modification promotes the interaction of dynein with the p150Glued subunit of dynactin, which recruits the motor to vesicles. This mechanism provides an explanation for how PP-IPs affect intracellular vesicular trafficking in mammalian cells (Chanduri et al., 2016; Saiardi, 2016). In a high throughput study in S. cerevisiae, multiple potential pyrophosphorylation targets (as well as IP6 -and IP7-interacting proteins) were identified using specially synthesized resins. These resins expose the non-hydrolysable IP7 analog, 5-PCP-IP5 (or IP6) to capture interacting proteins from whole yeast cell lysates (Wu et al., 2016). These studies have paved the way for investigating the consequences of such interactions in promoting fungal pathogenesis.
Mechanistic Insight into the Role of IP7 Precursors in Fungal Cellular Function and Pathogenicity
In fungal pathogens, it is still not clear whether IP3/4/5 serve predominantly as precursors for the synthesis of IP7, whether these metabolic precursors themselves modulate cellular function, and if so how? Evidence from fungal and animal model organisms suggests that IP3/4/5 evolved to have independent roles in cellular function. These studies provide a rich source of information for continuing investigation into the mechanisms by which these IP species exert their roles in fungal pathogenicity.
As previously mentioned, the metabolic profile of the Cnarg1Δ mutant is characterized by the absence of IP4−6 and PP-IP5 and the accumulation of IP3 and PIP2. In S. cerevisiae accumulation of PIP2 is responsible for defects in organization of actin cytoskeleton, cell wall, vacuolar morphology, endocytosis, and clathrin-mediated sorting between the Golgi and endosomes (Stefan et al., 2002). IP3 accumulation could also impact negatively on cellular function in the Cnarg1Δ mutant. For example, elevated IP3 could compete with PIP2 for the binding of cytoplasmic proteins with a pleckstrin homology (PH) domain (Varnai et al., 2002). Elevated IP3 could also potentially alter calcium homeostasis in C. neoformans, which has implications for virulence as altering calcium homeostasis by ablating the function of cryptococcal vacuolar calcium channels attenuates virulence in animal models (Squizani et al., 2018). In C. neoformans, the source of IP3 is PIP2, which is hydrolysed by PLC1 (Lev et al., 2013). In mammalian cells, PLC1-derived IP3 causes the release of Ca2+ from the endoplasmic reticulum (ER), where calcium is predominantly stored, following engagement of an IP3 receptor (Mikoshiba et al., 1994; Bergsma et al., 2001). This leads to activation of the serine/threonine phosphatase, calcineurin. However, the spike in IP3 in the Cnarg1Δ mutant did not lead to calcineurin activation in C. neoformans (Lev et al., 2013). Despite this observation, whether or not elevated IP3 has an impact on intracellular calcium levels in Cnarg1Δ, and the subcellular location of any calcium spikes, remains to be determined.
In C. albicans it was shown that suppressing IPK2 expression, which is predicted to cause IP3 accumulation, did in fact correlate with a spike in cytoplasmic calcium (Li J. et al., 2017). This is consistent with IP3 playing a role in regulating calcium homeostasis. The authors concluded that elevation in cytosolic calcium was due to calcium release from the vacuole due to IP3-mediated activation of the vacuolar calcium channel, Yvc1. In contrast to mammalian cells, the major calcium storage organelle in fungi is the vacuole, not the ER. Their results are consistent with those observed in the S. cerevisiae ARG82/IPK2 deletion mutant, where elevated IP3 caused constitutive activation of Yvc1, leading to an increase in cytoplasmic calcium (Tisi et al., 2004; Bouillet et al., 2012). Whether Yvc1 is a receptor for Arg1/Ipk2-derived IP3 in C. albicans and C. neoformans, remains to be determined.
In mammalian cells inositol 1,4,5,6-tetrakisphosphate (IP4) acts as intermolecular glue to facilitate binding of class 1 histone deacetylases to their cognate co-repressor to form transcription repression complexes. This mechanism of HDAC activation is thought to be conserved in class I HDACs from yeast to humans (Watson et al., 2012, 2016; Millard et al., 2013). A different IP4 isoform (inositol 1,3,4,5-tetrakisphosphate), inositol 1,3,4,5,6-pentakisphosphate (IP5), and inositol hexakisphosphate (IP6) have been reported to control protein phosphorylation by regulating the activity of the serine/threonine kinase casein kinase 2 (CK2), which phosphorylates a diverse array of proteins and is thought to be responsible for 20% of the eukaryotic phosphoproteome (Solyakov et al., 2004).
Numerous studies report a direct binding role for IP6 in the regulation of nuclear processes in eukaryotes. While IP6 does not have a dominant role in cryptococcal pathogenesis, it has been shown to bind to the mRNA export factor, Gle1, in S. cerevisiae to regulate mRNA export (Alcazar-Roman et al., 2010) and to the RNA editing enzyme, ADAR2, in humans to promote its stability and activity (Macbeth et al., 2005). IP6 binding also has a role in signaling, regulating oligomerization and cellular localization of Arrestin-2 (Milano et al., 2006). More recently, human host-derived IP6 has been reported to play a role in HIV-1 assembly and maturation (Dick et al., 2018).
Focus of Future Studies in Determining the Roles of IPs and PP-IPs in Fungal Pathogenicity
Future work in fungal pathogenesis needs to address whether, and which, transcription factors respond to IPs, and how IPs regulate virulence-related signaling pathways, including the PHO pathway. Crosstalk between IPs and Ras signaling has been reported in the basidiomycete, Schizophyllum commune, where lithium-induced activation of the IPK pathway, leading to increased levels of PP-IPs, was not observed in a Ras1 mutant (Murry et al., 2019).
IPKs as Novel Antifungal Drug Targets
The health and socioeconomic costs associated with the treatment of invasive fungal disease are enormous with estimates of ~2.6 billion per year in the US alone (Benedict et al., 2019). Reasons for this include the availability of only a small arsenal of antifungals with few targets. Some of the drawbacks of these drugs include poor bioavailability, lack of broad-spectrum efficacy, and toxicity. Furthermore, drug resistance is emerging. The gold standard for treating cryptococcal meningoencephalitis is a combination of the polyene amphotericin B and the uracil derivative 5-fluoro-uracil. 5-fluoro-uracil is not suitable for monotherapy because of the rapid development of resistance and hematologic toxicities. Similarly, amphotericin B is nephrotoxic and requires intravenous administration. Fluconazole is fungistatic and most commonly used for maintenance (suppressive) therapy and as an alternative to amphotericin B, especially in resource-limited regions (Sloan et al., 2009; Perfect et al., 2010). Although fluconazole inhibits the growth of Cryptococcus at low drug concentrations, it predisposes the patient to clinical relapse and the development of drug resistance (Sloan et al., 2009; Krysan, 2015). Echinocandins are the newest class of antifungal drug introduced into clinical practice in the early 2000s (Roemer and Krysan, 2014). Unfortunately, the echinocandins are not effective against Cryptococcus (Perfect et al., 2010). Given the limitations of our current antifungal drug arsenal, targeting essential pathways other than ergosterol and cell wall biosynthesis would be advantageous. The IPK pathway represents one such target particularly because it has a pleotropic role in virulence affecting the production of numerous virulence factors and is essential for promoting disseminated meningitis which is fatal without treatment. In the case of targeting IPKs, existing kinase inhibitors could be repurposed or entirely new inhibitors could be developed.
Another advantage of targeting the fungal IPK pathway is that there are no signaling redundancies. The pathway is linear and therefore non-redundant in fungi, while in humans there is a high degree of redundancy particularly in the steps converting IP3 to IP5. Thus, the chances of fungal IPK inhibitors blocking conversion of IP3 to IP5 in the host is minimal.
Several candidate molecules have been reported to inhibit IPKs, but not protein kinases, with different degrees of selectivity. The cell permeable purine-based compound, N2-(m-(trifluoromethyl)benzyl) N6-(p-nitrobenzyl) purine (TNP), selectively inhibits both human IP3K and IP6K enzymes by competing with ATP (Chang et al., 2002; Padmanabhan et al., 2009). The selectivity of TNP for IP6K is significantly higher than for IP3K. Similar to the effects of KCS1 deletion, TNP causes vacuolar fragmentation in S. cerevisiae (Padmanabhan et al., 2009). In addition to TNP, a number of natural and synthetic polyphenolic compounds were found to inhibit all three mammalian IP3K isoforms, as well as IPMK. One of these compounds, ellagic acid, is found in fruits and vegetables and has antiproliferative and antioxidant properties (Khanduja et al., 1999; Seeram et al., 2005). Chlorogenic acid, a major polyphenolic compound in coffee, selectively inhibited human IPMK at IC50 of 1.15 μM (Mayr et al., 2005). Chlorogenic acid, like a number of other natural polyphenols, was reported to suppress the growth of tumor cells in vitro (Feng et al., 2005; Yamagata et al., 2018). Furthermore, derivatives of chlorogenic acid exhibited antifungal activity against C. neoformans, C. albicans, and A. fumigatus (Ma et al., 2007). Polyphenolic compounds inhibit enzymes other than IPKs, in particular Topoisomerase II, but in many cases the IC50 for other enzymes is much higher than the IC50 for IPKs. Inhibition of IP3K and IPMK by polyphenolics is of a linear mixed type with respect to ATP binding, and of a linear non-competitive type with respect to IP3. This complex mechanism of binding is considered advantageous for developing highly selective inhibitors (Mayr et al., 2005). Thus, chemical derivatisation of polyphenolic compounds represents a promising strategy for developing novel antifungal molecules.
Human protein and lipid kinases have been widely investigated as targets for specific inhibitors as they are directly involved in a variety of disease-associated processes, especially cancerous growth. The key challenge is designing kinase inhibitors with the desired target specificity. The ideal inhibitors should be selective only for their target kinase (have minimal off target effects) and potent (only require a low dose) to reduce the potential for liver and kidney toxicity. However, this is usually almost impossible to achieve given that the human kinome is comprised of over 500 protein kinases. Despite this difficulty, as of 2014, 30 kinase inhibitors were approved for clinical use, mainly for oncological treatment purposes (Fabbro, 2015). The majority of these inhibitors target the ATP binding site. Conservation of the ATP binding site often causes inhibitors to cross-react with other kinases resulting in compounds with promiscuous profiles. Inhibitor specificity can be gained by designing inhibitors that bind to two different sites, one of which is the ATP-binding site. Alternatively, some inhibitors use structural features of the target kinase that do not include the ATP binding pocket (Fabbro, 2015). The methods used for developing human kinase inhibitors and the collections of compounds already available, provide opportunities for repurposing as well as creating new inhibitors with desired selectivity for fungal IPKs. Obtaining crystal structures for fungal IPKs will also facilitate drug discovery. So far the only IPK from a fungal pathogen that has been crystalized is Ipk1 (IP5 kinase) from C. neoformans (Oh et al., 2017).
Conclusions
Multiple studies demonstrate that IPs and PP-IPs play major roles in cellular function in all eukaryotes, from humans to fungi. C. neoformans has provided an essential model for elucidating the IPK pathway in fungal pathogens, and for demonstrating that IPKs play crucial roles in growth, fitness and virulence, predominantly via the more distal product IP7. In silico analysis of fungal genomes demonstrate that this pathway is also present in the major human fungal pathogens, C. albicans and A. fumigatus. Although inability to completely ablate IP3 kinase function in C. albicans has hindered progress in determining the impact of IPK function on the pathogenicity of C. albicans in animal infection models, the difficulties experienced point to this kinase having an essential role in fungal fitness. Furthermore, due to technical difficulties in detecting and quantifying IPs and PP-IPs, the mechanism of action of these metabolic messengers is only partially understood and efforts to study this in fungal and mammalian cells is ongoing. Despite the ubiquity of IPs and PP-IPs in nature, the IPK pathways that lead to their synthesis have become more complex and redundant in higher eukaryotes, while the pathway in fungi has remained linear. The lack of redundancy and low sequence conservation, coupled with what is now known about the importance of IPKs in cryptococcal pathogenicity, renders fungal IPKs, particularly the lead kinase Arg1, attractive targets for the development of an anticryptococcal drug. Pending further investigation these drugs could potentially have a broader antifungal specificity.
Author Contributions
JD and SL wrote the manuscript. CL, TS, AS, and DD proofread the manuscript and provided helpful suggestions.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We sincerely thank Dorothea Fiedler for proofreading this manuscript and for her helpful suggestions.
References
Alcazar-Roman, A. R., Bolger, T. A., and Wente, S. R. (2010). Control of mRNA export and translation termination by inositol hexakisphosphate requires specific interaction with Gle1. J. Biol. Chem. 285, 16683–16692. doi: 10.1074/jbc.M109.082370
Alcazar-Roman, A. R., and Wente, S. R. (2008). Inositol polyphosphates: a new frontier for regulating gene expression. Chromosoma 117, 1–13. doi: 10.1007/s00412-007-0126-4
Auesukaree, C., Tochio, H., Shirakawa, M., Kaneko, Y., and Harashima, S. (2005). Plc1p, Arg82p, and Kcs1p, enzymes involved in inositol pyrophosphate synthesis, are essential for phosphate regulation and polyphosphate accumulation in Saccharomyces cerevisiae. J. Biol. Chem. 280, 25127–25133. doi: 10.1074/jbc.M414579200
Azevedo, C., and Saiardi, A. (2017). Eukaryotic phosphate homeostasis: the inositol pyrophosphate perspective. Trends Biochem. Sci. 42, 219–231. doi: 10.1016/j.tibs.2016.10.008
Bechet, J., Greenson, M., and Wiame, J. M. (1970). Mutations affecting the repressibility of arginine biosynthetic enzymes in Saccharomyces cerevisiae. Eur. J. Biochem. FEBS 12, 31–39. doi: 10.1111/j.1432-1033.1970.tb00817.x
Benedict, K., Jackson, B. R., Chiller, T., and Beer, K. D. (2019). Estimation of direct healthcare costs of fungal diseases in the United States. Clin. Infect. Dis. 68, 1791–1797. doi: 10.1093/cid/ciy776
Bergsma, J. C., Kasri, N. N., Donaton, M. C., De Wever, V., Tisi, R., de Winde, J. H., et al. (2001). PtdIns(4,5)P(2) and phospholipase C-independent Ins(1,4,5)P(3) signals induced by a nitrogen source in nitrogen-starved yeast cells. Biochem. J. 359, 517–523. doi: 10.1042/bj3590517
Bertsch, U., Deschermeier, C., Fanick, W., Girkontaite, I., Hillemeier, K., Johnen, H., et al. (2000). The second messenger binding site of inositol 1,4,5-trisphosphate 3-kinase is centered in the catalytic domain and related to the inositol trisphosphate receptor site. J. Biol. Chem. 275, 1557–1564. doi: 10.1074/jbc.275.3.1557
Bhandari, R., Saiardi, A., Ahmadibeni, Y., Snowman, A. M., Resnick, A. C., Kristiansen, T. Z., et al. (2007). Protein pyrophosphorylation by inositol pyrophosphates is a posttranslational event. Proc. Natl. Acad. Sci. U.S.A. 104, 15305–15310. doi: 10.1073/pnas.0707338104
Bolger, T. A., Folkmann, A. W., Tran, E. J., and Wente, S. R. (2008). The mRNA export factor Gle1 and inositol hexakisphosphate regulate distinct stages of translation. Cell 134, 624–633. doi: 10.1016/j.cell.2008.06.027
Bosch, D., and Saiardi, A. (2012). Arginine transcriptional response does not require inositol phosphate synthesis. J. Biol. Chem. 287, 38347–38355. doi: 10.1074/jbc.M112.384255
Bouillet, L. E., Cardoso, A. S., Perovano, E., Pereira, R. R., Ribeiro, E. M., Tropia, M. J., et al. (2012). The involvement of calcium carriers and of the vacuole in the glucose-induced calcium signaling and activation of the plasma membrane H(+)-ATPase in Saccharomyces cerevisiae cells. Cell Calcium 51, 72–81. doi: 10.1016/j.ceca.2011.10.008
Brown, G. D., Denning, D. W., Gow, N. A., Levitz, S. M., Netea, M. G., and White, T. C. (2012). Hidden killers: human fungal infections. Sci. Transl. Med. 4:165rv13. doi: 10.1126/scitranslmed.3004404
Chanduri, M., Rai, A., Malla, A. B., Wu, M., Fiedler, D., Mallik, R., et al. (2016). Inositol hexakisphosphate kinase 1 (IP6K1) activity is required for cytoplasmic dynein-driven transport. Biochem. J. 473, 3031–3047. doi: 10.1042/BCJ20160610
Chang, Y. T., Choi, G., Bae, Y. S., Burdett, M., Moon, H. S., Lee, J. W., et al. (2002). Purine-based inhibitors of inositol-1,4,5-trisphosphate-3-kinase. Chembiochem 3, 897–901. doi: 10.1002/1439-7633(20020902)3:9<897::AID-CBIC897>3.0.CO;2-B
Chayakulkeeree, M., Sorrell, T. C., Siafakas, A. R., Wilson, C. F., Pantarat, N., Gerik, K. J., et al. (2008). Role and mechanism of phosphatidylinositol-specific phospholipase C in survival and virulence of Cryptococcus neoformans. Mol. Microbiol. 69, 809–826. doi: 10.1111/j.1365-2958.2008.06310.x
Choi, J., Rajagopal, A., Xu, Y. F., Rabinowitz, J. D., and O'Shea, E. K. (2017). A systematic genetic screen for genes involved in sensing inorganic phosphate availability in Saccharomyces cerevisiae. PLoS ONE. 12:e0176085. doi: 10.1371/journal.pone.0176085
Cordeiro, C. D., Saiardi, A., and Docampo, R. (2017). The inositol pyrophosphate synthesis pathway in Trypanosoma brucei is linked to polyphosphate synthesis in acidocalcisomes. Mol. Microbiol. 106, 319–333. doi: 10.1111/mmi.13766
Denning, D. W. (2002). Echinocandins: a new class of antifungal. J Antimicrobial Chemother. 49, 889–891. doi: 10.1093/jac/dkf045
Desfougeres, Y., Gerasimaite, R. U., Jessen, H. J., and Mayer, A. (2016). Vtc5, a novel subunit of the vacuolar transporter chaperone complex, regulates polyphosphate synthesis and phosphate homeostasis in yeast. J. Biol. Chem. 291, 22262–22275. doi: 10.1074/jbc.M116.746784
Dick, R. A., Zadrozny, K. K., Xu, C. F, Schur, K. M., Lyddon, T. D., et al. (2018). Inositol phosphates are assembly co-factors for HIV-1. Nature 560, 509–512. doi: 10.1038/s41586-018-0396-4
Dubois, E., Bercy, J., and Messenguy, F. (1987). Characterization of two genes, ARGRI and ARGRIII required for specific regulation of arginine metabolism in yeast. Mol. General Genetics MGG 207, 142–148. doi: 10.1007/BF00331501
Dubois, E., Scherens, B., Vierendeels, F., Ho, M. M., Messenguy, F., and Shears, S. B. (2002). In Saccharomyces cerevisiae, the inositol polyphosphate kinase activity of Kcs1p is required for resistance to salt stress, cell wall integrity, and vacuolar morphogenesis. J. Biol. Chem. 277, 23755–23763. doi: 10.1074/jbc.M202206200
El Alami, M., Messenguy, F., Scherens, B., and Dubois, E. (2003). Arg82p is a bifunctional protein whose inositol polyphosphate kinase activity is essential for nitrogen and PHO gene expression but not for Mcm1p chaperoning in yeast. Mol. Microbiol. 49, 457–468. doi: 10.1046/j.1365-2958.2003.03562.x
Fabbro, D. (2015). 25 years of small molecular weight kinase inhibitors: potentials and limitations. Mol. Pharmacol. 87, 766–775. doi: 10.1124/mol.114.095489
Feng, R., Lu, Y., Bowman, L. L., Qian, Y., Castranova, V., and Ding, M. (2005). Inhibition of activator protein-1, NF-kappaB, and MAPKs and induction of phase 2 detoxifying enzyme activity by chlorogenic acid. J. Biol. Chem. 280, 27888–27895. doi: 10.1074/jbc.M503347200
Feoktistova, A., McCollum, D., Ohi, R., and Gould, K. L. (1999). Identification and characterization of Schizosaccharomyces pombe asp1(+), a gene that interacts with mutations in the Arp2/3 complex and actin. Genetics 152, 895–908.
Fisher, E., Almaguer, C., Holic, R., Griac, P., and Patton-Vogt, J. (2005). Glycerophosphocholine-dependent growth requires Gde1p (YPL110c) and Git1p in Saccharomyces cerevisiae. J. Biol. Chem. 280, 36110–36117. doi: 10.1074/jbc.M507051200
Fisher, M. C., Henk, D. A., Briggs, C. J., Brownstein, J. S., Madoff, L. C., McCraw, S. L., et al. (2012). Emerging fungal threats to animal, plant and ecosystem health. Nature 484, 186–194. doi: 10.1038/nature10947
Frederick, J. P., Mattiske, D., Wofford, J. A., Megosh, L. C., Drake, L. Y., Chiou, S. T., et al. (2005). An essential role for an inositol polyphosphate multikinase, Ipk2, in mouse embryogenesis and second messenger production. Proc. Natl. Acad. Sci. U.S.A. 102, 8454–8459. doi: 10.1073/pnas.0503706102
Fruman, D. A., Meyers, R. E., and Cantley, L. C. (1998). Phosphoinositide kinases. Ann. Rev. Biochem. 67, 481–507. doi: 10.1146/annurev.biochem.67.1.481
Garnett, J. P., Baker, E. H., and Baines, D. L. (2012). Sweet talk: insights into the nature and importance of glucose transport in lung epithelium. Eur. Respir. J. 40, 1269–1276. doi: 10.1183/09031936.00052612
Giovannini, D., Touhami, J., Charnet, P., Sitbon, M., and Battini, J. L. (2013). Inorganic phosphate export by the retrovirus receptor XPR1 in metazoans. Cell Rep. 3, 1866–1873. doi: 10.1016/j.celrep.2013.05.035
Hurlimann, H. C., Pinson, B., Stadler-Waibel, M., Zeeman, S. C., and Freimoser, F. M. (2009). The SPX domain of the yeast low-affinity phosphate transporter Pho90 regulates transport activity. EMBO Rep. 10, 1003–1008. doi: 10.1038/embor.2009.105
Ikeh, M. A., Kastora, S. L., Day, A. M., Herrero-de-Dios, C. M., Tarrant, E., Waldron, K. J., et al. (2016). Pho4 mediates phosphate acquisition in Candida albicans and is vital for stress resistance and metal homeostasis. Mol. Biol. Cell. 27, 2784–2801. doi: 10.1091/mbc.e16-05-0266
Illies, C., Gromada, J., Fiume, R., Leibiger, B., Yu, J., Juhl, K., et al. (2007). Requirement of inositol pyrophosphates for full exocytotic capacity in pancreatic beta cells. Science 318, 1299–1302. doi: 10.1126/science.1146824
Khanduja, K. L., Gandhi, R. K., Pathania, V., and Syal, N. (1999). Prevention of N-nitrosodiethylamine-induced lung tumorigenesis by ellagic acid and quercetin in mice. Food Chem. Toxicol. 37, 313–318. doi: 10.1016/S0278-6915(99)00021-6
Kretschmer, M., Reiner, E., Hu, G., Tam, N., Oliveira, D. L., Caza, M., et al. (2014). Defects in phosphate acquisition and storage influence virulence of Cryptococcus neoformans. Infect. Immunity 82, 2697–2712. doi: 10.1128/IAI.01607-14
Krysan, D. J. (2015). Toward improved anti-cryptococcal drugs: novel molecules and repurposed drugs. Fungal Genetics Biol. 78, 93–98. doi: 10.1016/j.fgb.2014.12.001
Lee, K. T., So, Y. S., Yang, D. H., Jung, K. W., Choi, J., Lee, D. G., et al. (2016). Systematic functional analysis of kinases in the fungal pathogen Cryptococcus neoformans. Nat. Commun. 7:12766. doi: 10.1038/ncomms12766
Lee, Y. S., Huang, K., Quiocho, F. A., and O'Shea, E. K. (2008). Molecular basis of cyclin-CDK-CKI regulation by reversible binding of an inositol pyrophosphate. Nat. Chem. Biol. 4, 25–32. doi: 10.1038/nchembio.2007.52
Lee, Y. S., Mulugu, S., York, J. D., and O'Shea, E. K. (2007). Regulation of a cyclin-CDK-CDK inhibitor complex by inositol pyrophosphates. Science 316, 109–112. doi: 10.1126/science.1139080
Lev, S., Desmarini, D., Li, C., Chayakulkeeree, M., Traven, A., Sorrell, T. C., et al. (2013). Phospholipase C of Cryptococcus neoformans regulates homeostasis and virulence by providing inositol trisphosphate as a substrate for Arg1 kinase. Infect. Immunity 81, 1245–1255. doi: 10.1128/IAI.01421-12
Lev, S., Kaufman-Francis, K., Desmarini, D., Juillard, P. G., Li, C., Stifter, S. A., et al. (2017). Pho4 is essential for dissemination of Cryptococcus neoformans to the host brain by promoting phosphate uptake and growth at alkaline pH. mSphere 2:16. doi: 10.1128/mSphere.00381-16
Lev, S., Li, C., Desmarini, D., Saiardi, A., Fewings, N. L., Schibeci, S. D., et al. (2015). Inositol Pyrophosphate IP7 Is Crucial for Metabolic Adaptation to the Host Environment and Pathogenicity. mBio 6:15. doi: 10.1128/mBio.00531-15
Li, C., Lev, S., Desmarini, D., Kaufman-Francis, K., Saiardi, A., A., et al. (2017). IP3-4 kinase Arg1 regulates cell wall homeostasis and surface architecture to promote clearance of Cryptococcus neoformans infection in a mouse model. Virulence 8, 1833–1848. doi: 10.1080/21505594.2017.1385692
Li, C., Lev, S., Saiardi, A., Desmarini, D., Sorrell, T. C., and Djordjevic, J. T. (2016). Identification of a major IP5 kinase in Cryptococcus neoformans confirms that PP-IP5/IP7, not IP6, is essential for virulence. Sci. Rep. 6:23927. doi: 10.1038/srep23927
Li, J., Zhang, B., Ma, T., Wang, H., Zhang, B., Yu, Q., et al. (2017). Role of the inositol polyphosphate multikinase Ipk2 in regulation of hyphal development, calcium signaling and secretion in Candida albicans. Mycopathologia 182, 609–623. doi: 10.1007/s11046-017-0138-4
Liao, G., Wang, Y., Liu, T. B., Kohli, G., Qian, W., Shor, E., et al. (2018). Role of the inositol pyrophosphate multikinase Kcs1 in Cryptococcus inositol metabolism. Fungal Genetics Biol. 113, 42–51. doi: 10.1016/j.fgb.2018.01.006
Liu, N. N., Flanagan, P. R., Zeng, J., Jani, N. M., Cardenas, M. E., Moran, G. P., et al. (2017). Phosphate is the third nutrient monitored by TOR in Candida albicans and provides a target for fungal-specific indirect TOR inhibition. Proc. Natl. Acad. Sci. U.S.A. 114, 6346–6351. doi: 10.1073/pnas.1617799114
Lonetti, A., Szijgyarto, Z., Bosch, D., Loss, O., Azevedo, C., and Saiardi, A. (2011). Identification of an evolutionarily conserved family of inorganic polyphosphate endopolyphosphatases. J. Biol. Chem. 286, 31966–31974. doi: 10.1074/jbc.M111.266320
Ma, C. M., Kully, M., Khan, J. K., Hattori, M., and Daneshtalab, M. (2007). Synthesis of chlorogenic acid derivatives with promising antifungal activity. Bioorganic Med. Chem. 15, 6830–6833. doi: 10.1016/j.bmc.2007.07.038
Macbeth, M. R., Schubert, H. L., Vandemark, A. P., Lingam, A. T., Hill, C. P., and Bass, B. L. (2005). Inositol hexakisphosphate is bound in the ADAR2 core and required for RNA editing. Science 309, 1534–1539. doi: 10.1126/science.1113150
Mayr, G. W., Windhorst, S., and Hillemeier, K. (2005). Antiproliferative plant and synthetic polyphenolics are specific inhibitors of vertebrate inositol-1,4,5-trisphosphate 3-kinases and inositol polyphosphate multikinase. J. Biol. Chem. 280, 13229–13240. doi: 10.1074/jbc.M500545200
Menniti, F. S., Miller, R. N., Putney, J. W. Jr., and Shears, S. B. (1993). Turnover of inositol polyphosphate pyrophosphates in pancreatoma cells. J. Biol Chem. 268, 3850–3856.
Mikoshiba, K., Furuichi, T., and Miyawaki, A. (1994). Structure and function of IP3 receptors. Semin. Cell Biol. 5, 273–281. doi: 10.1006/scel.1994.1033
Milano, S. K., Kim, Y. M., Stefano, F. P., Benovic, J. L., and Brenner, C. (2006). Nonvisual arrestin oligomerization and cellular localization are regulated by inositol hexakisphosphate binding. J. Biol. Chem. 281, 9812–9823. doi: 10.1074/jbc.M512703200
Millard, C. J., Watson, P. J., Celardo, I., Gordiyenko, Y., Cowley, S. M., Robinson, C. V., et al. (2013). Class I HDACs share a common mechanism of regulation by inositol phosphates. Mol. Cell 51, 57–67. doi: 10.1016/j.molcel.2013.05.020
Mulugu, S., Bai, W., Fridy, P. C., Bastidas, R. J., Otto, J. C., Dollins, D. E., et al. (2007). A conserved family of enzymes that phosphorylate inositol hexakisphosphate. Science 316, 106–109. doi: 10.1126/science.1139099
Murry, R., Kniemeyer, O., Krause, K., Saiardi, A., and Kothe, E. (2019). Crosstalk between Ras and inositol phosphate signaling revealed by lithium action on inositol monophosphatase in Schizophyllum commune. Adv. Biol. Regul. 72, 78–88. doi: 10.1016/j.jbior.2019.01.001
Nagata, E., Luo, H. R., Saiardi, A., Bae, B. I., Suzuki, N., and Snyder, S. H. (2005). Inositol hexakisphosphate kinase-2, a physiologic mediator of cell death. J. Biol. Chem. 280, 1634–1640. doi: 10.1074/jbc.M409416200
Nishizawa, M., Komai, T., Katou, Y., Shirahige, K., Ito, T., and Toh, E. A. (2008). Nutrient-regulated antisense and intragenic RNAs modulate a signal transduction pathway in yeast. PLoS Biol. 6, 2817–2830. doi: 10.1371/journal.pbio.0060326
Oh, J., Lee, D. G., Bahn, Y. S., and Rhee, S. (2017). Crystal structure of inositol 1,3,4,5,6-pentakisphosphate 2-kinase from Cryptococcus neoformans. J. Struct. Biol. 200, 118–123. doi: 10.1016/j.jsb.2017.09.004
Padmanabhan, U., Dollins, D. E., Fridy, P. C., York, J. D., and Downes, C. P. (2009). Characterization of a selective inhibitor of inositol hexakisphosphate kinases: use in defining biological roles and metabolic relationships of inositol pyrophosphates. J. Biol. Chem. 284, 10571–10582. doi: 10.1074/jbc.M900752200
Pascual-Ortiz, M., Saiardi, A., Walla, E., Jakopec, V., Kunzel, N. A., Span, I., et al. (2018). Asp1 bifunctional activity modulates spindle function via controlling cellular inositol pyrophosphate levels in Schizosaccharomyces pombe. Mol. Cell. Biol. 38, e00047–18. doi: 10.1128/MCB.00047-18
Perfect, J. R., Dismukes, W. E., Dromer, F., Goldman, D. L., Graybill, J. R., Hamill, R. J., et al. (2010). Clinical practice guidelines for the management of cryptococcal disease: 2010 update by the infectious diseases society of america. Clin. Infect. Dis. 50, 291–322. doi: 10.1086/649858
Pohlmann, J., and Fleig, U. (2010). Asp1, a conserved 1/3 inositol polyphosphate kinase, regulates the dimorphic switch in Schizosaccharomyces pombe. Mol. Cell. Biol. 30, 4535–4547. doi: 10.1128/MCB.00472-10
Pohlmann, J., Risse, C., Seidel, C., Pohlmann, T., Jakopec, V., Walla, E., et al. (2014). The Vip1 inositol polyphosphate kinase family regulates polarized growth and modulates the microtubule cytoskeleton in fungi. PLoS Genet. 10:e1004586. doi: 10.1371/journal.pgen.1004586
Potapenko, E., Cordeiro, C. D., Huang, G., Storey, M., Wittwer, C., Dutta, A. K., et al. (2018). 5-Diphosphoinositol pentakisphosphate (5-IP7) regulates phosphate release from acidocalcisomes and yeast vacuoles. J. Biol. Chem. 293, 19101–19112. doi: 10.1074/jbc.RA118.005884
Rajasingham, R., Smith, R. M., Park, B. J., Jarvis, J. N., Govender, N. P., Chiller, T. M., et al. (2017). Global burden of disease of HIV-associated cryptococcal meningitis: an updated analysis. Lancet Infect. Dis. 17, 873–881. doi: 10.1016/S1473-3099(17)30243-8
Rauch, J., Volinsky, N., Romano, D., and Kolch, W. (2011). The secret life of kinases: functions beyond catalysis. Cell Commun. Signal. CCS 9:23. doi: 10.1186/1478-811X-9-23
Resnick, A. C., Snowman, A. M., Kang, B. N., Hurt, K. J., Snyder, S. H., and Saiardi, A. (2005). Inositol polyphosphate multikinase is a nuclear PI3-kinase with transcriptional regulatory activity. Proc. Natl. Acad. Sci. U.S.A. 102, 12783–12788. doi: 10.1073/pnas.0506184102
Roemer, T., and Krysan, D. J. (2014). Antifungal drug development: challenges, unmet clinical needs, and new approaches. Cold Spring Harb. Perspect. Med. 4:a019703. doi: 10.1101/cshperspect.a019703
Saiardi, A. (2016). Protein pyrophosphorylation: moving forward. Biochem. J. 473, 3765–3768. doi: 10.1042/BCJ20160710C
Saiardi, A., Erdjument-Bromage, H., Snowman, A. M., Tempst, P., and Snyder, S. H. (1999). Synthesis of diphosphoinositol pentakisphosphate by a newly identified family of higher inositol polyphosphate kinases. Curr. Biol. CB 9, 1323–1326. doi: 10.1016/S0960-9822(00)80055-X
Saiardi, A., Resnick, A. C., Snowman, A. M., Wendland, B., and Snyder, S. H. (2005). Inositol pyrophosphates regulate cell death and telomere length through phosphoinositide 3-kinase-related protein kinases. Proc. Natl. Acad. Sci. U.S.A. 102, 1911–1914. doi: 10.1073/pnas.0409322102
Sarmah, B., Latimer, A. J., Appel, B., and Wente, S. R. (2005). Inositol polyphosphates regulate zebrafish left-right asymmetry. Dev. Cell. 9, 133–145. doi: 10.1016/j.devcel.2005.05.002
Sarmah, B., and Wente, S. R. (2009). Dual functions for the Schizosaccharomyces pombe inositol kinase Ipk1 in nuclear mRNA export and polarized cell growth. Eukaryotic Cell 8, 134–146. doi: 10.1128/EC.00279-08
Sarmah, B., and Wente, S. R. (2010). Inositol hexakisphosphate kinase-2 acts as an effector of the vertebrate Hedgehog pathway. Proc. Natl. Acad. Sci. U.S.A. 107, 19921–19926. doi: 10.1073/pnas.1007256107
Seeram, N. P., Adams, L. S., Henning, S. M., Niu, Y., Zhang, Y., Nair, M. G., et al. (2005). In vitro antiproliferative, apoptotic and antioxidant activities of punicalagin, ellagic acid and a total pomegranate tannin extract are enhanced in combination with other polyphenols as found in pomegranate juice. J. Nutr. Biochem. 16, 360–367. doi: 10.1016/j.jnutbio.2005.01.006
Shen, X., Xiao, H., Ranallo, R., Wu, W. H., and Wu, C. (2003). Modulation of ATP-dependent chromatin-remodeling complexes by inositol polyphosphates. Science 299, 112–114. doi: 10.1126/science.1078068
Sloan, D. J., Dedicoat, M. J., and Lalloo, D. G. (2009). Treatment of cryptococcal meningitis in resource limited settings. Curr. Opin. Infect. Dis. 22, 455–463. doi: 10.1097/QCO.0b013e32832fa214
Solyakov, L., Cain, K., Tracey, B. M., Jukes, R., Riley, A. M., Potter, B. V., et al. (2004). Regulation of casein kinase-2 (CK2) activity by inositol phosphates. J. Biol. Chem. 279, 43403–43410. doi: 10.1074/jbc.M403239200
Squizani, E. D., Oliveira, N. K., J., Reuwsaat, C. V., Marques, B. M., Lopes, W., et al. (2018). Cryptococcal dissemination to the central nervous system requires the vacuolar calcium transporter Pmc1. Cell. Microbiol. 20:e12803. doi: 10.1111/cmi.12803
Stefan, C. J., Audhya, A., and Emr, S. D. (2002). The yeast synaptojanin-like proteins control the cellular distribution of phosphatidylinositol (4,5)-bisphosphate. Mol. Biol. Cell 13, 542–557. doi: 10.1091/mbc.01-10-0476
Steger, D. J., Haswell, E. S., Miller, A. L., Wente, S. R., and O'Shea, E. K. (2003). Regulation of chromatin remodeling by inositol polyphosphates. Science 299, 114–116. doi: 10.1126/science.1078062
Taylor, R. Jr., Chen, P. H., Chou, C. C., Patel, J., and Jin, S. V. (2012). KCS1 deletion in Saccharomyces cerevisiae leads to a defect in translocation of autophagic proteins and reduces autophagosome formation. Autophagy 8, 1300–1311. doi: 10.4161/auto.20681
Thota, S. G., Unnikannan, C. P., Thampatty, S. R., Manorama, R., and Bhandari, R. (2015). Inositol pyrophosphates regulate RNA polymerase I-mediated rRNA transcription in Saccharomyces cerevisiae. Biochem. J. 466, 105–114. doi: 10.1042/BJ20140798
Tisi, R., Belotti, F., Wera, S., Winderickx, J., Thevelein, J. M., and Martegani, E. (2004). Evidence for inositol triphosphate as a second messenger for glucose-induced calcium signalling in budding yeast. Curr. Genetics 45, 83–89. doi: 10.1007/s00294-003-0465-5
Toh-e, A., Ohkusu, M., Li, H. M., Shimizu, K., Takahashi-Nakaguchi, A., Gonoi, T., et al. (2015). Identification of genes involved in the phosphate metabolism in Cryptococcus neoformans. Fungal Genetics Biol. 80, 19–30. doi: 10.1016/j.fgb.2015.04.019
Topolski, B., Jakopec, V., Kunzel, N. A., and Fleig, U. (2016). Inositol pyrophosphate kinase Asp1 modulates chromosome segregation fidelity and spindle function in Schizosaccharomyces pombe. Mol. Cell. Biol. 36, 3128–3140. doi: 10.1128/MCB.00330-16
Varnai, P., Lin, X., Lee, S. B., Tuymetova, G., Bondeva, T., Spat, A., et al. (2002). Inositol lipid binding and membrane localization of isolated pleckstrin homology (PH) domains. Studies on the PH domains of phospholipase C delta 1 and p130. J. Biol. Chem. 277, 27412–22. doi: 10.1074/jbc.M109672200
Watson, P. J., Fairall, L., Santos, G. M., and Schwabe, J. W. (2012). Structure of HDAC3 bound to co-repressor and inositol tetraphosphate. Nature 481, 335–340. doi: 10.1038/nature10728
Watson, P. J., Millard, C. J., Riley, A. M., Robertson, N. S., Wright, L. C., Godage, H. Y., et al. (2016). Insights into the activation mechanism of class I HDAC complexes by inositol phosphates. Nat. Commun. 7:11262. doi: 10.1038/ncomms11262
Wild, R., Gerasimaite, R., Jung, J. Y., Truffault, V., Pavlovic, I., Schmidt, A., et al. (2016). Control of eukaryotic phosphate homeostasis by inositol polyphosphate sensor domains. Science 352, 986–990. doi: 10.1126/science.aad9858
Wilson, M. S., Livermore, T. M., and Saiardi, A. (2013). Inositol pyrophosphates: between signalling and metabolism. Biochem. J. 452, 369–379. doi: 10.1042/BJ20130118
Worley, J., Luo, X., and Capaldi, A. P. (2013). Inositol pyrophosphates regulate cell growth and the environmental stress response by activating the HDAC Rpd3L. Cell Rep. 3, 1476–1482. doi: 10.1016/j.celrep.2013.03.043
Wu, M., Chong, L. S., Perlman, D. H., Resnick, A. C., and Fiedler, D. (2016). Inositol polyphosphates intersect with signaling and metabolic networks via two distinct mechanisms. Proc. Natl. Acad. Sci. U.S.A. 113, E6757–E6765. doi: 10.1073/pnas.1606853113
Yamagata, K., Izawa, Y., Onodera, D., and Tagami, M. (2018). Chlorogenic acid regulates apoptosis and stem cell marker-related gene expression in A549 human lung cancer cells. Mol. Cell. Biochem. 441, 9–19. doi: 10.1007/s11010-017-3171-1
Ye, C., Bandara, W. M., and Greenberg, M. L. (2013). Regulation of inositol metabolism is fine-tuned by inositol pyrophosphates in Saccharomyces cerevisiae. J. Biol. Chem. 288, 24898–24908. doi: 10.1074/jbc.M113.493353
Keywords: inositol polyphosphate kinase, inositol pyrophosphate, IP7, IP, PP-IP, virulence, signaling, Cryptococcus neoformans
Citation: Lev S, Li C, Desmarini D, Sorrell TC, Saiardi A and Djordjevic JT (2019) Fungal Kinases With a Sweet Tooth: Pleiotropic Roles of Their Phosphorylated Inositol Sugar Products in the Pathogenicity of Cryptococcus neoformans Present Novel Drug Targeting Opportunities. Front. Cell. Infect. Microbiol. 9:248. doi: 10.3389/fcimb.2019.00248
Received: 15 April 2019; Accepted: 26 June 2019;
Published: 15 July 2019.
Edited by:
Yong-Sun Bahn, Yonsei University, South KoreaReviewed by:
James Kronstad, University of British Columbia, CanadaLukasz Kozubowski, Clemson University, United States
Copyright © 2019 Lev, Li, Desmarini, Sorrell, Saiardi and Djordjevic. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Julianne T. Djordjevic, anVsaWFubmUuZGpvcmRqZXZpY0BzeWRuZXkuZWR1LmF1
 Sophie Lev
Sophie Lev Cecilia Li
Cecilia Li Desmarini Desmarini
Desmarini Desmarini Tania C. Sorrell
Tania C. Sorrell Adolfo Saiardi
Adolfo Saiardi Julianne T. Djordjevic
Julianne T. Djordjevic