- Department of Oral Biology, University of Florida, Gainesville, FL, United States
Streptococci, including the dental pathogen Streptococcus mutans, undergo cell-to-cell signaling that is mediated by small peptides to control critical physiological functions such as adaptation to the environment, control of subpopulation behaviors and regulation of virulence factors. One such model pathway is the regulation of genetic competence, controlled by the ComRS signaling system and the peptide XIP. However, recent research in the characterization of this pathway has uncovered novel operons and peptides that are intertwined into its regulation. These discoveries, such as cell lysis playing a critical role in XIP release and importance of bacterial self-sensing during the signaling process, have caused us to reevaluate previous paradigms and shift our views on the true purpose of these signaling systems. The finding of new peptides such as the ComRS inhibitor XrpA and the peptides of the RcrRPQ operon also suggests there may be more peptides hidden in the genomes of streptococci that could play critical roles in the physiology of these organisms. In this review, we summarize the recent findings in S. mutans regarding the integration of other circuits into the ComRS signaling pathway, the true mode of XIP export, and how the RcrRPQ operon controls competence activation. We also look at how new technologies can be used to re-annotate the genome to find new open reading frames that encode peptide signals. Together, this summary of research will allow us to reconsider how we perceive these systems to behave and lead us to expand our vocabulary of peptide signals within the genus Streptococcus.
Introduction
The idea that bacterial self-produced extracellular mediators could influence phenotypic behaviors first originated in the 1960s when Dr. Alexander Tomasz published on an environmental factor that was both heat-labile and sensitive to peptidases and could synchronize a Streptococcus population into a transformable state (Tomasz, 1965). At that time, it was the traditional view that bacterial populations were lacking mechanisms for specific intercellular communication and only cells of “higher organisms” coded for means to induce or reinforce the expression of many physiological properties. Coupled with the findings in Vibrio that bioluminescence required a self-produced factor in a term referred to at that time as autoinduction (Nealson et al., 1970), paradigms began to shift and it was recognized that bacterial populations could behave as a biological unit with considerable, although temporary, coordination among its members. Since that time, the study of bacterial communication or quorum sensing as it is now termed has rapidly expanded (Fuqua et al., 1994; Whiteley et al., 2017). We now know that the factor that Tomasz described is a small peptide or “pheromone” (Havarstein et al., 1997) and the phenomenon is widespread in Gram-positive controlling not only transformation, but also sporulation (Perego and Hoch, 1996) and production of extracellular toxins that impact virulence potential (Ji et al., 1995).
Today, the study of genetic competence (the transient phenotypic state leading to transformation as Tomasz described) remains a model pathway to dissect and explore cell-to-cell communication in Gram-positive bacteria. This is especially true within the genus Streptococcus, where extensive research progress has identified two distinct competence-activating signaling systems that divides the phylogenetic groups within the genus (Håvarstein, 2010). The well-studied ComCDE system reigns supreme in competence regulation among the Mitis and Anginosus groups that includes the popular model organism Streptococcus pneumoniae. ComCDE consists of a traditional two-component system signal transduction cascade composed of a histidine kinase and response regulator and is a model form of extracellular peptide signaling. The Pyogenic, Salivarius, and Bovis groups contain an intracellular signaling system, termed ComRS, comprised of a cytosolic transcriptional regulator that recognizes specific DNA sequences within promoter regions upon binding of its cognate peptide that is imported into the cells to regulate their competence activation. Finally there is the Mutans group of Streptococcus that contain both systems and regulate genetic competence dependent on the environmental conditions (Son et al., 2012). Being the oddball, Streptococcus mutans and its genetic competence circuit has served as an attractive model to sort out the interplay between the extracellular and intracellular signaling systems. This review will focus solely on the research and findings within S. mutans; for broader reviews on genetic competence within streptococci as a whole, readers are encouraged to view excellent reviews elsewhere that cover this topic (Jimenez and Federle, 2014; Johnston et al., 2014; Fontaine et al., 2015; Straume et al., 2015; Lin et al., 2016; Shanker and Federle, 2017).
Natural transformation in S. mutans was first described by Perry and Kuramitsu (1981). While not all recovered S. mutans isolates are transformable due to mutations in critical competence genes (Cornejo et al., 2013; Palmer et al., 2013), S. mutans has gained favorability for use as a model organism due to its ease of genetic manipulation and low biosafety level required for working with the organism (Lemos et al., 2013). The strain UA159 (early designations referred to this isolate as UAB577), was isolated in 1982 by P.W. Caufield at the University of Alabama Birmingham from a child with active cavities and was selected over the years for study due to its high degree of transformability over other S. mutans isolates (Murchison et al., 1986; Tao et al., 1993). Therefore, the wild-type and lab-adapted S. mutans has been selected for its peptide signaling and high degree of natural competence. In the last decade alone, numerous studies have enlightened our understanding of mechanisms by which these systems function as well as how these systems are in-tune with environmental sensing and stress response on a global scale to both habitat and foe (Figure 1). Along with these benchmark studies, more questions than answers have been raised with discoveries of novel peptides that appear to be associated with genetic competence leading to the mudding of once clear pathways by which these regulators operate.
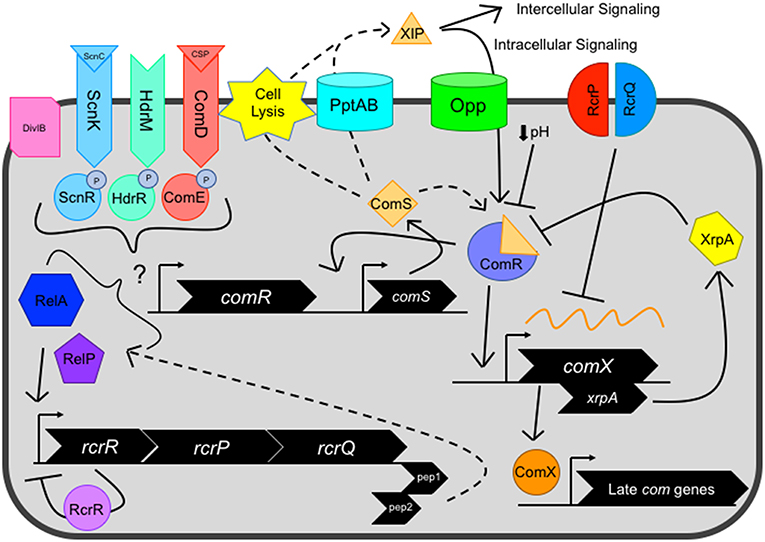
Figure 1. Current view of the genetic competence pathway in S. mutans. Competence activation is centrally mediated by the ComRS system, consisting of the Rgg-like transcriptional regulator ComR, and ComS, a 17-aa precursor of the 7-aa XIP peptide. ComS is processed into XIP and exported by unknown mechanism(s), potentially facilitated by dedicated transport systems such as PptAB or through cell lysis. Externalized XIP can activate nearby cells in an intercellular signaling mechanism or is re-imported by oligopeptide permeases (Opp) in an intracellular signaling manner. Cytosolic ComS or XIP interacts with the transcriptional regulator ComR, forming the ComR-XIP complex. The complex recognizes a select palindromic sequence termed the ComR-box, located within the promoter of both comX and comS, the latter creating a positive feedback loop for the system. Several negative regulators can also impact ComRS signaling, including decreased environmental pH and XrpA, located intragenically within the comX coding sequence on a separate reading frame that inhibits ComRS signaling through direct interaction with ComR. After competence activation, the alternative sigma factor ComX directs the RNA polymerase to a regulon consisting of late competence genes whose products make up the machinery needed for DNA uptake and potential homologous recombination of the single stranded DNA into the genome. However, the RcrRPQ operon, that is negatively regulated by RcrR, can shut down ComX production through overexpression of the ABC transporters RcrP and RcrQ that results in the processing/degradation of comX mRNA. Additionally, two peptides located at the 3′ end of the rcrQ coding region termed Pep1 and Pep2 are believed to modulate competence signaling through interactions with the (p)ppGpp enzymes RelA and RelP. Several distal regulators, including two component systems ComDE, ScnKR, and HdrMR, membrane protein DivIB and (p)ppGpp concentration mediated by the dual synthetase/hydrolase enzyme RelA can impact ComRS signaling.
Bridging ComCDE and ComRS—A Missed Connection
Early work on the genetic competence circuit within streptococci was completed in the Mitis and Anginosus groups, where a secreted peptide termed competence stimulating peptide (CSP) was found to activate the system in a density-dependent manner (Håvarstein et al., 1995; Havarstein et al., 1997). A similar peptide was identified in S. mutans, and addition of a synthetic version of the peptide (sCSP) both to planktonic and biofilm-grown cells greatly increased the number of S. mutans transformants when donor DNA was present (Li et al., 2001). For the next decade, the CSP peptide and ComDE signaling system was seen as the activator for genetic competence in S. mutans. The system follows the classical extracellular peptide signaling mechanism of other Gram-positive bacteria, in which a ribosomally produced precursor (in this example ComC), is processed during export by the ComAB secretion system (Hui and Morrison, 1991). The proteolytic cleavage site arises immediately after a double-glycine motif, commonly observed at the end of leader peptides for non-lantibiotic peptide bacteriocins and CSPs produced from other streptococci (Havarstein et al., 1997). This cleavage results in a mature peptide that is 21 residues long. An 18 amino acid (aa) version of CSP that lacks the three C-terminal amino acid residues was purified from S. mutans strain GS5 culture supernatants that had been pre-treated with the 21-aa version of CSP to identify the resulting antimicrobial peptides and lantibiotics that would be produced (Petersen et al., 2006). It was later realized that the 21-aa version of CSP undergoes a second processing step by the SepM extracellular membrane-associated protease where the final three amino acids are removed to generate the functional 18-aa form. The 21-aa version alone is non-functional and is not able to activate competence or bacteriocin production (Hossain and Biswas, 2012; Biswas et al., 2016). Several sequenced clinical isolates of S. mutans do not encode the final three C-terminal amino acids of comC and generate the functional 18-aa upon secretion (Palmer et al., 2013). Once the extracellular concentration of functional CSP reaches a critical threshold level, it is bound and activates the transmembrane histidine kinase ComD which autophosphorylates and subsequently transfers the phosphate group to ComE, a cytoplasmic response regulator (Kreth et al., 2007; Xie et al., 2010; Martin et al., 2013). Recent study into the structure-function relationship between CSP and ComD binding have shown that hydrophobic residues in the central region of the CSP 18-aa are critical for ComD receptor binding (Bikash et al., 2018), that the first few N-terminal residues are dispensable for signaling (Syvitski et al., 2007), and that two of the three extracellular loops of ComD are required for CSP recognition while the third plays little role (Dong et al., 2016). Early studies of CSP and the ComCDE operon suggested the presence of a second CSP receptor, as addition of CSP to comCDE mutant biofilms partially restored architecture similar to that of wild-type biofilms (Li et al., 2002).
Beginning in 2009, evidence for another peptide signaling system that regulated genetic competence in Streptococcus began to emerge (Fontaine et al., 2009; Gardan et al., 2009; Monnet et al., 2014). Using S. mutans as the model for this pathway (Mashburn-Warren et al., 2010), this second route for competence induction was found to include a cytosolic Rgg-like transcriptional regulator termed ComR and a 17-aa peptide precursor encoded immediately downstream in ComS. ComS is processed into an active 7-aa peptide derived from the C-terminus of ComS called XIP (comX Inducing Peptide). In other streptococci, the ComRS pathway follows the traditional intercellular signaling paradigm in that the ribosomally translated ComS is exported into the extracellular space and cleaved by a protease(s) to yield XIP (Mashburn-Warren et al., 2010; Shanker and Federle, 2017). After XIP is produced and released, it is transported back into the cell by the oligopeptide ABC transporter Opp (Mashburn-Warren et al., 2010; Son et al., 2012). The re-imported XIP can then be specifically bound by ComR to form a dimeric ComR-XIP complex that functions as a transcriptional activator for the promoters of comX and comS (Fontaine et al., 2013; Khan et al., 2016; Talagas et al., 2016). The ComR-XIP complex recognizes a ComR-box, consisting of a 20-bp palindromic motif, whose core GACA/TGTC inverted repeat is conserved across ComRS-containing streptococci (Fontaine et al., 2009; Mashburn-Warren et al., 2010). The activation of comS creates a positive feedback loop that amplifies ComS and possibly XIP production (Son et al., 2012; Fontaine et al., 2013). The model for XIP release and processing may differ in S. mutans. While three independent research groups have detected XIP in supernatant fluids of S. mutans (Desai et al., 2012; Khan et al., 2012; Wenderska et al., 2012), the mechanisms for secretion or processing have not been identified in this organism to date (Kaspar et al., 2017). Notably, in Streptococcus thermophilus (Gardan et al., 2013), the Eep protease processes ComS to XIP, but an equivalent function for proteases in S. mutans with characteristics similar to Eep has not been demonstrated (Khan et al., 2012). Chang and Federle (Chang and Federle, 2016) have recently shown the ability of the ABC transporter PptAB to export small hydrophobic peptides used in Rgg-signaling systems in Firmicutes, however removal of pptAB in S. mutans only partially disrupted XIP signaling suggesting still other XIP export mechanisms are present.
The Mutans group of Streptococcus is unique in that most strains encode both ComCDE and ComRS pathways (Håvarstein, 2010). Differing environmental conditions, which include pH, redox and growth phase, influence the activity of each pathway (Hagen and Son, 2017). Addition of sCSP to growing cultures of S. mutans in a peptide-rich medium, such as BHI, results in activation within the entire cell population of transcription for the biogenesis of bacteriocins via direct binding of phosphorylated ComE to a conserved sequence in the promoter regions of these genes and operons (Kreth et al., 2007; Lemme et al., 2011). Transcription of comX can also be induced by CSP, but this generally occurs in only a subset of organisms in a population (Son et al., 2012). It does not involve direct binding of ComE to the comX promoter, and the underlying mechanism for CSP-dependent activation of comX is not well-understood (Kreth et al., 2007; Hung et al., 2011). Consistent with this observation, the ComCDE system of S. mutans appears to have evolved from a common ancestor of the BlpCHR system in S. pneumoniae, which does not regulate competence but does induce bacteriocins in the pneumococcus and some related organisms (Johnston et al., 2014; Shanker et al., 2016; Mignolet et al., 2018). For this reason, the ComCDE system has begun being referred to as BlpCHR in S. mutans or MutCDE (Reck et al., 2015; Shanker and Federle, 2017). The proximal regulator for direct activation of competence is the ComRS signaling system as removal of comR results in a non-transformable strain (Mashburn-Warren et al., 2010).
Several intriguing hypotheses have been put forth to connect the ComCDE and ComRS pathways and to explain how activation of ComCDE may result in comX upregulation. An early hypothesis suggested that a ComE-regulated bacteriocin, CipB, may have a moonlighting function in regulating ComRS expression as cipB mutants showed decreased transformation efficiency (Dufour et al., 2011). Further adding complexity to the system are the ScnCRK and HdrRM systems whose genetic alterations seem to impact ComR production and/or stability leading to decreases in comX expression (Okinaga et al., 2010b; Kim et al., 2013). ScnRK comprise a two-component system and 61-aa peptide in ScnC. Unlike other two component systems, ScnC is believed to block the activity of ScnRK when bound rather than activate it (Kim et al., 2013). HdrRM consists of a membrane bound inhibitor protein (HdrM) that antagonizes the activity of an associated LytTR family transcription activator (HdrR) (Okinaga et al., 2010a). Competence has been shown to be strongly activated either when HdrR is overexpressed or when the HdrM membrane-bound protein is inactivated via mutation (Okinaga et al., 2010b). Finally, a recent genome-wide transposon screen to uncover novel regulators of genetic competence in S. mutans highlighted DivIB, a protein proposed to participate in the regulation of cell division (Shields et al., 2017). Loss of divIB resulted in decreased PcomX activity when stimulated with sCSP, however PcomX remained active when sXIP was supplied. Importantly, bacteriocin production by the ΔdivIB strain appeared not to be altered, suggesting that the ComCDE pathway in this strain was still functional despite a lack of comX activation by CSP. These data highlight DivIB as focus for future research into crosstalk between ComCDE and ComRS systems in S. mutans.
While it is unclear how ComCDE may affect ComRS expression, a feedback loop from ComRS to ComCDE was recently uncovered by two independent studies (Reck et al., 2015; Son et al., 2015b). The ComRS system was found to be required for comE activation by either CSP or XIP through ComX production. A ComX binding site (termed a cin-box motif) was found within the promoter of comE. PcomE was unresponsive to addition of XIP to cell cultures after disruption of the cin-box (Son et al., 2015b). A second finding in this study was that bacteriocin activation via ComE required a low threshold of CSP (only 100 nM) and activated the entire population (Son et al., 2015b). This is in contrast to comX activation by CSP, where a higher threshold is required and only activates a subpopulation (Son et al., 2012). This finding raises the question of how increasing CSP concentrations can provide greater stimulation of comX if the activity of cipB is already saturated. One potential explanation is that the higher CSP concentrations activate an additional, unidentified signaling pathway in parallel to cipB that further stimulates the ComRS system and comX. One thing is clear: ComCDE signaling is much less sensitive to certain environmental signals compared to ComRS signaling. Thus, the two signaling systems may collect different information about the environment for different purposes. Bacteriocin production relies on the simpler ComCDE system, depending more on a population density input (true quorum-sensing behavior), whereas competence development under ComRS is sensitive to fine changes in environmental signals such as media type, carbohydrate source (Moye et al., 2016), oxidative stress (De Furio et al., 2017) and pH levels (Guo et al., 2014; Son et al., 2015a). The bidirectional flow of information between the two systems may therefore allow regulation of additional behaviors in response to combinations of inputs.
Into the Milieu—Are Peptide Signals Meant to be Shared?
The activation of genetic competence in Streptococcus spp. has traditionally been viewed as a quorum sensing-like process. Secreted oligopeptides of the ComCDE or ComRS systems are excreted into the extracellular milieu, where they accumulate until a critical threshold concentration is overcome that activates their respective systems, usually via a positive feedback loop. This process is seen as a community-wide behavior, such that bacterial subpopulations are able to coordinate a multicellular-type response. Perhaps the best example of this behavior is the colonization of Vibrio fischeri in the light organ of the squid Euprymna scolopes, where high cell density of V. fischeri leads to bioluminescence that is activated by a cell-to-cell signaling process (Waters and Bassler, 2005). In streptococci, the peptides that activate genetic competence have been shown to be potentially induced by environmental stressors such as antibiotics, oxidative stress and nutrient/carbohydrate sources (Claverys et al., 2006; Ahn et al., 2007; Perry et al., 2009; Håvarstein, 2010; Moye et al., 2016; De Furio et al., 2017). The activation of competence can then be seen as a response by the population to challenging conditions in order to increase genome plasticity and fitness for the survival of a few.
For S. mutans, it was not clear whether ComRS signaling would be active within biofilms due to strict environmental requirements including a growth medium devoid of peptides and a pH > 6.5 left (Son et al., 2012, 2015a; Guo et al., 2014). Two separate studies of S. mutans grown in biofilm populations indeed found that ComRS signaling could be active under these conditions. In the first, sXIP peptide was supplied to growing biofilms at different time points (Shields and Burne, 2016). As the biofilms matured (20–23 h), increasing concentrations of sXIP were required to induce measurable comX activation, and only a fraction of the cells responded to the signal. This was in contrast to early biofilms (5–7 h), where robust comX activity was measured and the majority of the population responded. In a second study, a co-culturing system was designed such that one S. mutans strain (a “sender”) would overproduce the XIP peptide precursor ComS, exporting large amounts of XIP to the extracellular space so that a receiver strain, which was unable to produce XIP (ΔcomS), would internalize the XIP via the Opp transport system and a response could be measured (Kaspar et al., 2017). This study concluded that XIP had the potential to act as a diffusible signal and that the XIP released by a sender could activate a nearby responder in biofilm populations. However, as more exopolysaccharide material was produced due to an increase in sucrose concentrations, signaling dramatically decreased showing that the intercellular signaling would be limited in range. Furthermore, signaling could not be successfully observed when the sender/receiver co-culture was embedded in an agarose matrix that viewed single cell populations over a limited time. The inability of the system to activate broader cell populations leaves open the question the efficiency of XIP as a true intercellular signaling system.
The importance of bacterial “self-sensing” or intracellular signaling in an autocrine-like mechanism has gained renewed focus in the last several years. In Bacillus subtilis, a co-culture system similar to the sender/responder experiment discussed above showed that the peptide-secreting cells displayed a stronger response to the signal than non-secreting cells due to self-sensing (Bareia et al., 2018). A similar finding was recently made in S. mutans, in that a ΔcomS strain activated comX to a lower degree in response to exogenously-provided XIP compared to a wild-type strain (Underhill et al., 2018). A comS-overproducing strain has also been shown to self-activate independent to changes in the rate of medium replacement in a microfluidic device, suggesting that external XIP accumulation played little role in this strain's ability to activate competence (Underhill et al., 2018). The basis that XIP serves as intercellular signal has always been built on previous paradigms, detection of XIP by LC-MS/MS in supernates of S. mutans cultures that were grown to high cell densities (Khan et al., 2012; Wenderska et al., 2012), and the fact that filtrates of S. mutans cultures grown to high density induced PcomX activity in reporter strains (Desai et al., 2012). However, the substantial data against this paradigm includes (i) requirement for an intact comS gene in order for CSP to elicit any comX response in a peptide-rich medium (Son et al., 2012), (ii) the XIP importer Opp not being required for the bimodal PcomX activation seen in peptide-rich medium (Son et al., 2012), and (iii) fluid replacement not altering the induction of comX in a strain that overexpresses comS (Underhill et al., 2018). These findings must force us to reconsider the importance of extracellular XIP accumulated in the milieu and whether genetic competence is truly a cell-to-cell signaling behavior. Several other recent findings in other organisms support this anti-social behavior in signaling molecules. In Pseudomonas aeruginosa, an aggregate model that recapitulates the biogeographical properties of infection in the cystic fibrosis lung found that cell-to-cell signaling only occurs within but not between aggregates or cell clusters that are spaced some distance apart (Darch et al., 2018). Autocrine-like signaling via the ComCDE pathway was observed in S. pneumoniae due to cell chaining from exposure to the antibiotics aztreonam and clavulanic acid that extended the length of the limited competence window (Domenech et al., 2018). Specifically for XIP, it is known in Streptococcus thermophilus that XIP is not excreted into the extracellular medium and may be limited to just the bacterial cell surface, favoring a self-sensing activation model (Gardan et al., 2013).
A final piece to the puzzle in determining if S. mutans XIP is a real extracellular signal is finding (or not finding) a dedicated XIP exporter. As previously mentioned, there have been several attempts to find a XIP exporter that went unsuccessful including deletion of Eep and PptAB (Khan et al., 2012; Chang and Federle, 2016). This leaves room to postulate that either S. mutans XIP relies on a suite of non-specific exporters in its delivery to the external milieu, or XIP is released by a mechanism different from that in other streptococci. One hypothesis is that S. mutans cell lysis is the dominant pathway for release of XIP. After competence activation in S. mutans, a subpopulation of cells will undergo autolysis (Perry et al., 2009; Wenderska et al., 2012). Autolysis is mediated in S. mutans through the activities of encoded cell wall hydrolases that are a part of the ComX regulon (Khan et al., 2016), and possibly by intracellular bacteriocins (Perry et al., 2009). Release of XIP to bacterial kin could also be a part of this process—release of both a source of genetic material (eDNA) and activating peptide at the same time is a logical approach so that DNA is available as cells become fully competent. Support for this new idea arises from failed competence activation of a reporter strain grown in supernatant of a S. mutans strain lacking the major autolysin AtlA (Kaspar et al., 2017). Why would S. mutans lack a dedicated peptide export system that seems common in other streptococci? It may come down to lifestyle. S. mutans is an obligate biofilm organism, where regulated cell death and lysis is critical for biofilm maturation and stability through release of extracellular matrix components such as eDNA (Liao et al., 2014). S. mutans may have simply evolved in such a way that a dedicated XIP exporter was dispensable, and autolysis of cells within the biofilm community was sufficient. The explanation of cell lysis being responsible for XIP release is not out of line with previous data where a high optical density is needed for XIP to be identified within cellular supernatants and competence autoactivation in reporter strains (Khan et al., 2012; Wenderska et al., 2012). ComRS signaling in S. mutans may just be more reliant on timing (i.e., specific points in biofilm maturation) rather than true cellular density commonly observed in other quorum sensing systems.
The Curious Case Of The Rcrrpq Operon
In 2011, Seaton et al. (2011) targeted and explored an operon in S. mutans that was linked to stress tolerance, (p)ppGpp accumulation and genetic competence and later termed RcrRPQ for Rel and Competence Related (Figure 2A). RcrR is a MarR-type negative transcriptional regulator that binds to the promoter region for the operon (Seaton et al., 2015). Downstream of rcrR are two ABC transporters, rcrP and rcrQ. Much of the research detailed by Seaton et al. discusses the phenotypes displayed by two separate mutants of rcrR: ΔrcrR-P contains a polar antibiotic resistant cassette in the replacement of the rcrR coding region (a strong transcriptional terminator is included after the antibiotic resistance gene), and ΔrcrR-NP contains a non-polar antibiotic resistant cassette (no transcriptional terminator is included). The ABC transporters rcrP and rcrQ in ΔrcrR-NP are upregulated 100-fold due to the loss of the negative regulator rcrR that controls the expression of the operon. Meanwhile, the presence of the transcriptional terminator greatly reduces the upregulation in ΔrcrR-P, resulting in the ABC transporters being expressed similarly to wild-type levels. Thus, we can assume that the phenotypes observed in ΔrcrR-P are from only the loss of rcrR, while those seen in ΔrcrR-NP may be due to both loss of rcrR as well as upregulation of rcrP and rcrQ.
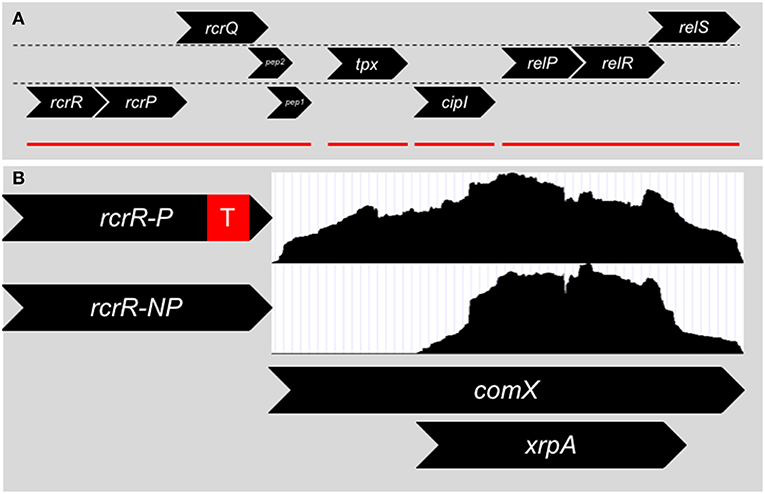
Figure 2. (A) Genomic organization surrounding the rcrRPQ operon of S. mutans. Genes are divided into their respective reading frames. rcrR encodes for a MarR-like transcriptional regulator that negatively regulates the transcription of the operon. rcrP and rcrQ are ABC-type multidrug/protein/lipid transporters. At the end of rcrQ are two small peptides encoded in separate reading frames. The start codon of Pep 1 overlaps with the stop codon of rcrQ. Pep 2's reading frame begins inside the rcrQ coding sequence and overlaps with the beginning of Pep 1. Downstream of the rcrRPQ operon is tpx, a thiol peroxidase, and cipI, a bacteriocin immunity protein. Finally relP, a small alarmone (p)ppGpp synthetase, along with the two component system relRS are further downstream. Mutations within rcrRPQ effect the expression of the relPRS operon and vice versa. Red lines denote operon structures within the region. (B) Two different mutants of rcrR are mainly used to investigate the operon that differ only by the presence of a transcriptional terminator (red box) within the antibiotic resistance cassette used to replace the gene. The ΔrcrR-P strain expresses the rcrP and rcrQ transporters at wild-type levels due to the presence of the strong transcriptional terminator and is hyper-transformable in terms of genetic competence, while the ΔrcrR-NP strain produces the transporters at 100x higher than wild-type background, leading to a non-transformable state. Differences in transformability between ΔrcrR-P and ΔrcrR-NP is the result of an altered comX transcript between the two strains. RNA-Seq read counts (white boxes) between the 5′ and 3′ ends of comX are even in the ΔrcrR-P background and a full transcript is produced. However, in the ΔrcrR-NP background, reads only map to the 3′ end and a smaller transcript is produced that corresponds to the intragenic coding region of xrpA that led to its discovery.
Changes in the frequency of genetic transformation, both at basal levels as well as with addition of either S. mutans competence peptides, CSP or XIP, distinguish the ΔrcrR-P and ΔrcrR-NP mutants: ΔrcrR-P is hyper-transformable while ΔrcrR-NP cannot be transformed under any growth condition tested. This discrepancy between the two strains was made even more confusing by the early evidence that both comS and comX were upregulated in either strain (Seaton et al., 2011). It was clear that the competence phenotypes between the two strains could not be from defects in signaling, but rather from some type of post-activation inhibition. The answer was found when read accumulation between the two transcriptomes of either strain was compared using RNA-Seq (Kaspar et al., 2015). While it had appeared that comX was upregulated in ΔrcrR-NP via relative real-time PCR, only the 3′ half of the comX full-length transcript was present—the 5′ half was missing (Figure 2B). In fact, ΔrcrR-NP did not make a functional ComX protein at all (Kaspar et al., 2015). This could answer how ΔrcrR-NP was non-transformable, but the why is still left to ponder. Does RcrRPQ function as a kill switch for the competence circuit for S. mutans? One suggestion is that in a stressful condition that is sensed by the MarR-like regulator RcrR, the ABC transporters become upregulated, leading to the degradation of the 5′ comX mRNA transcript. Unfortunately, the signal for RcrR and the substrate for the transporters RcrP and RcrQ is unknown. The genomic location of the operon, however, may give us a clue. The (p)ppGpp synthetase relP and a two-component system termed relRS are encoded immediately downstream of RcrRPQ, with mutations of rcrR affecting relP transcription suggesting interplay between the two operons (Seaton et al., 2011). It has been shown that (p)ppGpp levels can affect competence activation through ComRS signaling, and subsequently deletion of the dual (p)ppGpp synthetase/hydrolase enzyme RelA can revert the non-transformable ΔrcrR-NP strain into a phenotype similar to its hyper-transformable counterpart ΔrcrR-P (Kaspar et al., 2016), further linking the rcrRPQ operon to (p)ppGpp. The operon could directly respond to (p)ppGpp accumulation or stressors that lead to changes in (p)ppGpp concentration within the cells.
The degradation of the 5′ comX transcript still left an abundance of the 3′ transcript remaining. This transcript does serve a purpose. Encoded within just a few bases of the transcript start was a 69 amino acid open reading frame (ORF). On the chance that this ORF coded for a functional protein, the start codon was mutated and evaluated for phenotypes (Kaspar et al., 2015). While the start codon mutant had minimal phenotype differences in an S. mutans wild-type background, the mutation completely restored the accumulation of full-length comX transcript as well as transformability in the ΔrcrR-NP strain. The ORF was named XrpA for comX Regulatory Peptide. Further research into the function of XrpA identified XrpA as a novel inhibitor of competence development through direct interaction with ComR, detailed through fluorescent polarization assays (Kaspar et al., 2018). While the exact mechanism by which XrpA binding to ComR inhibits ComRS signaling remains unknown, one hypothesis is that ComR is unable to form a dimeric complex when XrpA is bound (Talagas et al., 2016; Kaspar et al., 2018). XrpA does not appear to directly compete with XIP for the ComR peptide binding pocket (Kaspar et al., 2018). Several intriguing questions remain, including the translational mechanism by which the internal ORF XrpA is produced from the comX mRNA transcript, and whether XrpA is continuously active in the cell or is triggered by an environmental response that switches off ComRS signaling.
XrpA was not the only peptide that has been discovered that is connected to the RcrRPQ operon. Ahn et al. (2014) uncovered two peptides serendipitously at the end of the rcrQ ORF when two separately constructed rcrQ mutants, that differed only in the 3′ endpoint of the antibiotic cassette replacement, displayed altered phenotypes. Upon further review of the genome, two ORFs encoded on separate frames from rcrQ was noticed that could account for the phenotypic differences. One ORF's (rcrQ Pep1) start codon overlapped with the stop codon of rcrQ, while the other (rcrQ Pep2) overlaps with both the rcrQ ORF and the Pep1 ORF. When both peptides were removed in ΔrcrR-NP, the strain became hyper-transformable similar to what was observed when xrpA and relA was removed (Ahn et al., 2014; Kaspar et al., 2015, 2016). There was no change in the phenotypes of ΔrcrR-P, suggesting that de-repression of the operon is needed for the peptides to exert their effect. Individual deletion of either peptide through mutation of the start codon had marginal effects on growth rate in response to the competence activating peptide CSP as well as transformation efficiency—the best observable phenotypes were always when both peptides were deleted. Determining the function of the rcrQ peptides is currently ongoing. Interestingly, the activities of this operon appear limited to just S. mutans. Deletion of the operon in Streptococcus gordonii resulted in no changes to transformation efficiency but increased sensitivity to oxygen and reactive oxygen species in a similar manner to phenotypes observed in S. mutans (Shields and Burne, 2015). Perhaps further connecting the rcrRPQ operon with (p)ppGpp will shed new insights into the mechanisms by which the transporters and peptides integrate stress tolerance signals into the governance of competence regulation.
More Peptide to be Discovered?
The ecological niche for the genus Streptococcus is narrow in scope, as almost all members are intimately associated and do not survive outside of a host, although some have been isolated in waste waters, on plants, and in milk and cheese (Mundt, 1982). Among them, the streptococci that inhabit the human oral cavity are uniquely adapted to the continuum of environmental insults as well as microbial antagonism within biofilm populations (Bowen et al., 2017). These challenges include rapid fluctuations in carbohydrate concentrations and other nutrient availability, exposure to environmental oxygen, and reactive oxygen species produced by competitors, reductions of environmental pH and exposure to the metabolic by-products of the other organisms that comprise oral biofilms (Lemos et al., 2013). To adapt to such constantly changing conditions, the microbiota employ numerous systems to optimize their gene expression patterns, cellular physiology, and fitness. For example, the S. mutans genome contains 14 two component systems and a single alternative sigma factor (comX) that together help to integrate sensory inputs from the environment or cell envelope into appropriate genetic and physiologic responses. Isolates of S. mutans have genomes that are about 2 megabases (Cornejo et al., 2013). This is comparatively small and in contrast to other host-associated and free-living organisms that have substantially larger genomes and a much greater repertoire of two component systems and alternative sigma factors to govern gene expression in response to specific environmental stimuli. Thus, S. mutans has evolved in a way that, instead of using specific sensing proteins, uses a set of global transcriptional regulators, conserved stress-response circuits (molecular chaperones, protease chaperones), monitoring of metabolic status (phosphosugar pools, HPr, acetyl phosphate levels), and sugar-specific permeases to interpret the environment and integrate this information with the physiologic status of the cells to fine-tune gene expression patterns and virulence capacities. Additionally, these bacteria must also coordinate mechanisms that allow for their establishment and growth among a consortia of other microbes in competitive interactions (Bowen et al., 2017). Therefore, it would not be surprising to find that these bacteria have encoded numerous small peptide/protein mediators to optimally monitor and adapt cell physiology to these numerous affronts to account for their small genome size.
After uncovering the unusual xrpA and the rcrQ peptides, one must ask if we are underestimating the number of small peptides/proteins found within the genomes of streptococci that impact the fitness and virulence potential of these organisms. Simple analysis of the S. mutans UA159 genome shows that there are over 18,000 ORFs that have the potential to code for proteins >100 nucleotides in length. At the time of the S. mutans genome sequencing in 2002, 1,963 ORFs were assigned using the ORF prediction software GLIMMER 2.0 with the resulting annotations still in use today (Delcher et al., 1999; Ajdić et al., 2002). Through about two decades of research after the publication of the original genome, we know some of the resulting annotations are incorrect and ORFs containing peptide precursors such as ComS were not identified. This leaves open the real possibility that some protein/peptides encoded within small ORFs are overlooked that either serve as signaling molecules or act as small protein effectors. There are over 4,000 small ORFs that overlap with another ORF similar to the rcrQ peptides, 12,000+ ORFs that are intragenic to another larger ORF such as the relationship between comX and xrpA, and greater than a 1,000 ORFs that are intergenic that have no assigned coding function (Figure 3). These numbers highlight the potential that we may be still unaware of critical factors that shape bacterial metabolism, responses, and overall physiology.
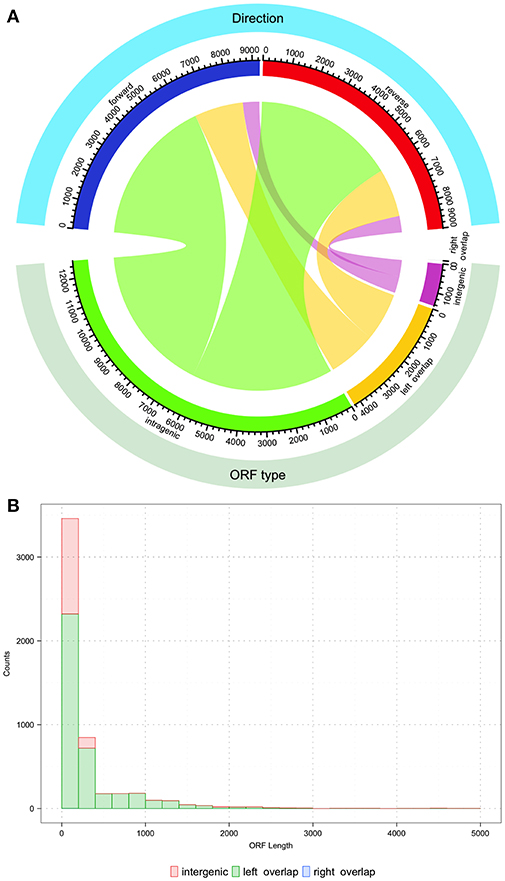
Figure 3. Description of potential coding open reading frames (ORFs) within the S. mutans UA159 genome. The S. mutans UA159 genome was downloaded from NCBI (Biosample SAMN02604090, Accession PRJNA333) with all annotations and loaded into Geneious (v 11.1.4). All ORFs (>100 nucleotides) were predicted with Geneious. (A) Chord plot showing the relationship between the type of predicted ORF and the direction (forward or reverse strand). From the total of 18,276 predicted ORFs, around 69% (12,599 ORFs) are completely intragenic or within another coding sequence. The count of ORFs that overlap with another on its 5′ end is 23% (4231 ORFs) and < 0.001% (1 ORF of length 123 bp) that overlap on its 3′ end. Finally, 8% (1445 ORFs) are located entirely within intergenic regions. Regarding the orientation of the ORFs, from the plot it can be concluded that roughly half of the total ORF count are oriented in opposite directions. Chord plot was generated with the function “chordDiagram” from the R package “circlize” (Gu et al., 2014). (B) Histogram of the nucleotide length distribution for intergenic, left, and right overlapping ORFs. Histogram was generated with the R package “ggplot2” (Wickham, 2016).
Several advancements in technologies are notable toward the future pursuit of these unidentified factors. One popular technique for detection of low molecular weight proteins and peptides is liquid chromatography-tandem mass spectrometry (LS-MS/MS). The power of this technique was recently shown by a study of peptidic small molecules produced both in monocultures of oral bacteria isolates as well as in in vitro multispecies biofilms (Edlund et al., 2017). Only 2.2% of the peptides produced in these cultures could be putatively annotated, and the diversity of secreted peptides changed over growth of the biofilm cultures. This study reinforces the perception that we are severely lacking in our understanding of secreted factors, especially in the context of complex communities where cell-to-cell signaling, cooperation and antagonism interactions shape microbiome composition and persistence. While LS-MS/MS can determine the nature of small peptides present in a given sample, it's a greater challenge to resolve where in the genome these peptides originated. Terminomics, or the study of protein terminal sequences, is able to assist in determining additional non-annotated ORFs as well as ORF boundary adjustments from the original computational prediction (Dandekar et al., 2000; Jaffe et al., 2004; Hartmann and Armengaud, 2014; Berry et al., 2016). These strategies aim to identify the N-termini within an entire proteome through both N-termini enrichment strategies, such as in vivo acetylation (Chen et al., 2013), as well as bioinformatic data analysis (Hartmann and Armengaud, 2014). To date, proteogenomics has been used to re-evaluate the genomes of over 20 bacterial species, including relevant human pathogens Helicobacter pylori (Müller et al., 2013), Mycobacterium tuberculosis (Kelkar et al., 2011), Salmonella Typhimurium (Ansong et al., 2011), and Yersinia pestis (Payne et al., 2010). Specific selection of the N-termini is more beneficial toward the discovery of small peptides rather than traditional mass spectrometry as the technique reduces the high complexity and dynamic range of peptide fragments present in the sample, decreases the amount of peptides that may go undetected from poor ionization and those that lack charged residues, and eases biases present in traditional data analysis pipelines due to poor or low spectrum present from each peptide (Duncan et al., 2010; Berry et al., 2016).
The emergence of next-generation genomics and emerging transcriptomics technologies also offers opportunities to re-characterize a genome. These techniques are more user-friendly and do not come at a high cost burden compared to proteogenomics. Whole transcriptome shotgun sequencing, commonly referred to as RNA-Seq, allows for the annotation of transcriptional features at single-nucleotide precision in a strand-specific manner (Croucher and Thomson, 2010; Creecy and Conway, 2015). While RNA-Seq is commonly used in bacterial studies to determine differential expression of transcripts between strains and/or conditions, RNA-Seq can also be used to define operon architecture, identify promoters, transcriptional start sites, terminators, and antisense RNAs (Conway et al., 2014; Sharma and Vogel, 2014; Thomason et al., 2015). Proper identification and annotation of these features within a genome may reveal novel transcripts not previously recognized, such as the case with S. mutans xrpA (Kaspar et al., 2015). One pitfall of this analysis is pervasive transcription in non-canonical locations, leading to false interpretations about potential coding regions (Wade and Grainger, 2014).
RNA-Seq has also led to the development of derivative technologies that follow a similar protocol of RNA sequencing but with a specific focus rather than the entire transcriptome. One of these newer technologies is ribosomal profiling (Ribo-Seq) that maps the exact positions of ribosomes on transcripts by nuclease footprinting and subsequent RNA sequencing (Ingolia et al., 2009, 2012). Prior to cell harvest, a drug such as chloramphenicol is added to cultures to pause actively translating ribosomes. Purified mRNAs are then treated with nucleases to degrade regions that are not protected by the ribosome, leaving only 20–40 bp footprints that are mapped to the original genome or mRNA such that the location of a ribosome can be determined (Ingolia et al., 2011). Ribo-Seq takes transcriptome profiling a step further to determine which transcripts are actively being translated at the time of cell harvest. Ribo-Seq can be used to answer several key questions—it can be used as a quantitative proteomics tool to monitor known and new protein synthesis, measurement of protein synthesis rates which may be clues of posttranscriptional regulation, determination of ORF start codons and locations of translational pausing (Brar and Weissman, 2015). In the hunt for small proteins and peptides, the location of ribosome density along a given mRNA can be a critical sign that a specific ORF is actually translated, rather than being just pervasive translation. Two key studies in Escherichia coli have shown this proof of concept. In the first, ribosomes were paused with tetracycline that led to an accumulation of ribosomes at start codons (Nakahigashi et al., 2016). This technique was used to reannotate the N-termini of many known ORFs while also leading to the discovery of several unannotated ORFs in intergenic regions. In the second, ribosome elongation and initiation data was combined using both Onc112 and retapamulin which appear to be better alternatives than tetracycline for initiation site determination (Seefeldt et al., 2015; Meydan et al., 2019; Weaver et al., 2019). Predicted small proteins were then tagged, validating 38 of the 41 predicted (Weaver et al., 2019). Included in the validation were several proteins that contained substantial overlap with another ORF but encoded on a separate frame. While this technique offers several clear advantages for new ORF discovery and reannotation of the genome, users should be cognizant that this is still a relatively new technology that can be significantly impacted by the drug choice for stalling of ribosomes and the method chosen for rapid freezing of samples (Mohammad et al., 2019). Ribo-Seq protocols continue to evolve.
While there are several methods available for the discovery for new small peptides/proteins, the best approach in the future may actually be combining technologies. For instance, ribosome profiling may be first used for prediction of new start site initiations that is followed up by confirmation using proteogenomics and N-terminal mapping of peptide samples (Koch et al., 2014). While these high-throughput techniques offer speed and ability to re-map entire genomes at a time, validation of new peptides and proteins may still come down to tagging a protein of interest and visualizing a band on a gel. As these techniques become more popular and the bar for usage becomes lower in terms of cost and simplicity to the user, a mass re-evaluation of genomes to find hidden gems we may have missed could be on the horizon.
Conclusion and Prospects
The study of cell-to-cell signaling in Streptococcus has provided a valuable model system to understand complex processes such as gene regulation, stress response, signal perception and transduction, and subpopulation and community behaviors. We are now approaching 25 years since the first characterization of CSP in Streptococcus pneumoniae (Håvarstein et al., 1995), yet we continue to make new discoveries that lead us to re-evaluate our understanding and realize the complexity by which these systems are controlled. This is especially true in S. mutans, where the findings of XrpA and inclusion of RcrRPQ into the story has only complicated matters. While there are many lingering questions, they should serve as examples to broaden our search for novel regulators that critically impact the physiology of key pathogens. We should be excited by the prospect that there may still be more out there currently hidden in the genomes of Gram-positive bacteria. One recent example is the finding of a leaderless secreted peptide consisting of only 8 amino acids in Streptococcus pyogenes that controls the production of the major virulence factor SpeB (Do et al., 2017). This determination of a critical molecular mechanism in a previously undescribed regulatory pathway offers hope for new therapeutic intervention that was previously not recognized. As we move closer to a post-antibiotic world and where manipulation of one's microbiome may prove to be a key curative strategy, these new research methods and findings should offer new approaches that move us closer to the selective targeting, disruption and removal of major pathogenic organisms.
Author Contributions
JK contributed to conception, design, drafted, and critically revised the review. AW contributed to the figures, design, and critically revised the review. All authors gave final approval to the review and agree to be accountable for all aspects of the work.
Funding
The authors are supported by the National Institute of Dental and Craniofacial Research of the National Institutes of Health under Award Number R01 DE013239. JK is supported by F32 DE028479 from the National Institute of Dental and Craniofacial Research.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Dr. Robert Burne for this critical reading of the review and providing thoughtful suggestions.
References
Ahn, S. J., Kaspar, J., Kim, J. N., Seaton, K., and Burne, R. A. (2014). Discovery of novel peptides regulating competence development in Streptococcus mutans. J. Bacteriol. 196, 3735–3745. doi: 10.1128/JB.01942-14
Ahn, S. J., Wen, Z. T., and Burne, R. A. (2007). Effects of oxygen on virulence traits of Streptococcus mutans. J. Bacteriol. 189, 8519–8527. doi: 10.1128/JB.01180-07
Ajdić, D., McShan, W. M., McLaughlin, R. E., Savić, G., Chang, J., Carson, M. B., et al. (2002). Genome sequence of Streptococcus mutans UA159, a cariogenic dental pathogen. Proc. Natl. Acad. Sci. U.S.A. 99, 14434–14439. doi: 10.1073/pnas.172501299
Ansong, C., Tolić, N., Purvine, S. O., Porwollik, S., Jones, M., Yoon, H., et al. (2011). Experimental annotation of post-translational features and translated coding regions in the pathogen Salmonella Typhimurium. BMC Genom. 12:433. doi: 10.1186/1471-2164-12-433
Bareia, T., Pollak, S., and Eldar, A. (2018). Self-sensing in Bacillus subtilis quorum-sensing systems. Nat. Microbiol. 3, 83–89. doi: 10.1038/s41564-017-0044-z
Berry, I. J., Steele, J. R., Padula, M. P., and Djordjevic, S. P. (2016). The application of terminomics for the identification of protein start sites and proteoforms in bacteria. Proteomics 16, 257–272. doi: 10.1002/pmic.201500319
Bikash, C. R., Hamry, S. R., and Tal-Gan, Y. (2018). Structure–activity relationships of the competence stimulating peptide in Streptococcus mutans reveal motifs critical for membrane protease SepM recognition and ComD receptor activation. ACS Infect. Dis. 9, 1385–1394. doi: 10.1021/acsinfecdis.8b00115
Biswas, S., Cao, L., Kim, A., and Biswas, I. (2016). SepM, a streptococcal protease involved in quorum sensing, displays strict substrate specificity. J. Bacteriol. 198, 436–447. doi: 10.1128/JB.00708-15
Bowen, W. H., Burne, R. A., Wu, H., and Koo, H. (2017). Oral biofilms: pathogens, matrix, and polymicrobial interactions in microenvironments. Trends Microbiol. 26, 229–242. doi: 10.1016/j.tim.2017.09.008
Brar, G. A., and Weissman, J. S. (2015). Ribosome profiling reveals the what, when, where and how of protein synthesis. Nat. Rev. Mol. Cell Biol. 16, 651–664. doi: 10.1038/nrm4069
Chang, J. C., and Federle, M. J. (2016). PptAB exports Rgg quorum-sensing peptides in Streptococcus. PLoS ONE 11:e0168461. doi: 10.1371/journal.pone.0168461
Chen, S.-H., Chen, C.-R., Chen, S.-H., Li, D.-T., and Hsu, J.-L. (2013). Improved N α -acetylated peptide enrichment following dimethyl labeling and SCX. J. Proteome Res. 12, 3277–3287. doi: 10.1021/pr400127j
Claverys, J.-P., Prudhomme, M., and Martin, B. (2006). Induction of competence regulons as a general response to stress in gram-positive bacteria. Ann. Rev. 60, 451–475. doi: 10.1146/annurev.micro.60.080805.142139
Conway, T., Creecy, J. P., Maddox, S. M., Grissom, J. E., Conkle, T. L., Shadid, T. M., et al. (2014). Unprecedented high-resolution view of bacterial operon architecture revealed by RNA sequencing. MBio 5:e01442–e01414. doi: 10.1128/mBio.01442-14
Cornejo, O. E., Lefébure, T., Bitar, P. D., Lang, P., Richards, V. P., Eilertson, K., et al. (2013). Evolutionary and population genomics of the cavity causing bacteria Streptococcus mutans. Mol. Biol. Evol. 30, 881–893. doi: 10.1093/molbev/mss278
Creecy, J. P., and Conway, T. (2015). Quantitative bacterial transcriptomics with RNA-seq. Curr. Opin. Microbiol. 23, 133–140. doi: 10.1016/j.mib.2014.11.011
Croucher, N. J., and Thomson, N. R. (2010). Studying bacterial transcriptomes using RNA-seq. Curr. Opin. Microbiol. 13, 619–624. doi: 10.1016/j.mib.2010.09.009
Dandekar, T., Huynen, M., Regula, J. T., Ueberle, B., Zimmermann, C. U., Andrade, M. A., et al. (2000). Re-annotating the Mycoplasma pneumoniae genome sequence: adding value, function and reading frames. Nucleic Acids Res. 28, 3278–3288. doi: 10.1093/nar/28.17.3278
Darch, S. E., Simoska, O., Fitzpatrick, M., Barraza, J. P., Stevenson, K. J., Bonnecaze, R. T., et al. (2018). Spatial determinants of quorum signaling in a Pseudomonas aeruginosa infection model. Proc. Natl. Acad. Sci. U.S.A. 115, 4779–4784. doi: 10.1073/pnas.1719317115
De Furio, M., Ahn, S. J., Burne, R. A., and Hagen, S. J. (2017). Oxidative stressors modify the response of Streptococcus mutans to its competence signal peptides. Appl. Environ. Microbiol. 83, e01345–e01317. doi: 10.1128/AEM.01345-17
Delcher, A. L., Harmon, D., Kasif, S., White, O., and Salzberg, S. L. (1999). Improved microbial gene identification with GLIMMER. Nucleic Acids Res. 27, 4636–4641. doi: 10.1093/nar/27.23.4636
Desai, K., Mashburn-Warren, L., Federle, M. J., and Morrison, D. A. (2012). Development of competence for genetic transformation of Streptococcus mutans in a chemically defined medium. J. Bacteriol. 194, 3774–3780. doi: 10.1128/JB.00337-12
Do, H., Makthal, N., VanderWal, A. R., Rettel, M., Savitski, M. M., Peschek, N., et al. (2017). Leaderless secreted peptide signaling molecule alters global gene expression and increases virulence of a human bacterial pathogen. Proc. Natl. Acad. Sci. U.S.A. 114, E8498–E8507. doi: 10.1073/pnas.1705972114
Domenech, A., Slager, J., and Veening, J.-W. (2018). Antibiotic-induced cell chaining triggers pneumococcal competence by reshaping quorum sensing to autocrine-like signaling. Cell Rep. 25:e3. doi: 10.1016/j.celrep.2018.11.007
Dong, G., Tian, X.-L., Cyr, K., Liu, T., Lin, W., Tziolas, G., et al. (2016). Membrane topology and structural insights into the peptide pheromone receptor ComD, A quorum-sensing histidine protein kinase of Streptococcus mutans. Sci. Rep. 6:26502. doi: 10.1038/srep26502
Dufour, D., Cordova, M., Cvitkovitch, D. G., and Lévesque, C. M. (2011). Regulation of the competence pathway as a novel role associated with a streptococcal bacteriocin. J. Bacteriol. 193, 6552–6559. doi: 10.1128/JB.05968-11
Duncan, M. W., Aebersold, R., and Caprioli, R. M. (2010). The pros and cons of peptide-centric proteomics. Nat. Biotechnol. 28, 659–664. doi: 10.1038/nbt0710-659
Edlund, A., Garg, N., Mohimani, H., Gurevich, A., He, X., Shi, W., et al. (2017). Metabolic fingerprints from the human oral microbiome reveal a vast knowledge gap of secreted small peptidic molecules. mSystems 2:e00058–e00017. doi: 10.1128/mSystems.00058-17
Fontaine, L., Boutry, C., de Frahan, M. H., Delplace, B., Fremaux, C., Horvath, P., et al. (2009). A novel pheromone quorum-sensing system controls the development of natural competence in Streptococcus thermophilus and Streptococcus salivarius. J. Bacteriol. 192, 1444–1454. doi: 10.1128/JB.01251-09
Fontaine, L., Goffin, P., Dubout, H., Delplace, B., Baulard, A., Lecat-Guillet, N., et al. (2013). Mechanism of competence activation by the ComRS signalling system in streptococci. Mol. Microbiol. 87, 1113–1132. doi: 10.1111/mmi.12157
Fontaine, L., Wahl, A., Fléchard, M., Mignolet, J., and Hols, P. (2015). Regulation of competence for natural transformation in streptococci. Infect. Genet. Evol. 33, 343–360. doi: 10.1016/j.meegid.2014.09.010
Fuqua, W. C., Winans, S. C., and Greenberg, E. P. (1994). Quorum sensing in bacteria: the LuxR-LuxI family of cell density-responsive transcriptional regulators. J. Bacteriol. 176, 269–275. doi: 10.1128/jb.176.2.269-275.1994
Gardan, R., Besset, C., Gitton, C., Guillot, A., Fontaine, L., Hols, P., et al. (2013). Extracellular life cycle of ComS, the competence-stimulating peptide of Streptococcus thermophilus. J. Bacteriol. 195, 1845–1855. doi: 10.1128/JB.02196-12
Gardan, R., Besset, C., Guillot, A., Gitton, C., and Monnet, V. (2009). The oligopeptide transport system is essential for the development of natural competence in Streptococcus thermophilus strain LMD-9. J. Bacteriol. 191, 4647–4655. doi: 10.1128/JB.00257-09
Gu, Z., Gu, L., Eils, R., Schlesner, M., and Brors, B. (2014). circlize implements and enhances circular visualization in R. Bioinformatics 30, 2811–2812. doi: 10.1093/bioinformatics/btu393
Guo, Q., Ahn, S.-J., Kaspar, J., Zhou, X., and Burne, R. A. (2014). Growth phase and pH influence peptide signaling for competence development in Streptococcus mutans. J. Bacteriol. 196, 227–236. doi: 10.1128/JB.00995-13
Håvarstein, L. S. (2010). Increasing competence in the genus Streptococcus. Mol. Microbiol. 78, 541–544. doi: 10.1111/j.1365-2958.2010.07380.x
Håvarstein, L. S., Coomaraswamy, G., and Morrison, D. A. (1995). An unmodified heptadecapeptide pheromone induces competence for genetic transformation in Streptococcus pneumoniae. Proc. Natl. Acad. Sci. U.S.A. 92, 11140–11144. doi: 10.1073/pnas.92.24.11140
Hagen, S. J., and Son, M. (2017). Origins of heterogeneity in Streptococcus mutans competence: interpreting an environment-sensitive signaling pathway. Phys. Biol. 14:015001. doi: 10.1088/1478-3975/aa546c
Hartmann, E. M., and Armengaud, J. (2014). N-terminomics and proteogenomics, getting off to a good start. Proteomics 14, 2637–2646. doi: 10.1002/pmic.201400157
Havarstein, L. S., Hakenbeck, R., and Gaustad, P. (1997). Natural competence in the genus Streptococcus: evidence that streptococci can change pherotype by interspecies recombinational exchanges. J. Bacteriol. 179, 6589–6594. doi: 10.1128/jb.179.21.6589-6594.1997
Hossain, M. S., and Biswas, I. (2012). An extracelluar protease, SepM, generates functional competence-stimulating peptide in Streptococcus mutans UA159. J. Bacteriol. 194, 5886–5896. doi: 10.1128/JB.01381-12
Hui, F. M., and Morrison, D. A. (1991). Genetic transformation in Streptococcus pneumoniae: nucleotide sequence analysis shows comA, a gene required for competence induction, to be a member of the bacterial ATP-dependent transport protein family. J. Bacteriol. 173, 372–381. doi: 10.1128/jb.173.1.372-381.1991
Hung, D. C. I., Downey, J. S., Ayala, E. A., Kreth, J., Mair, R., Senadheera, D. B., et al. (2011). Characterization of DNA binding sites of the ComE response regulator from Streptococcus mutans. J. Bacteriol. 193, 3642–3652. doi: 10.1128/JB.00155-11
Ingolia, N. T., Brar, G. A., Rouskin, S., McGeachy, A. M., and Weissman, J. S. (2012). The ribosome profiling strategy for monitoring translation in vivo by deep sequencing of ribosome-protected mRNA fragments. Nat. Protoc. 7, 1534–1550. doi: 10.1038/nprot.2012.086
Ingolia, N. T., Ghaemmaghami, S., Newman, J. R. S., and Weissman, J. S. (2009). Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science 324, 218–223. doi: 10.1126/science.1168978
Ingolia, N. T., Lareau, L. F., and Weissman, J. S. (2011). Ribosome profiling of mouse embryonic stem cells reveals the complexity and dynamics of mammalian proteomes. Cell 147, 789–802. doi: 10.1016/j.cell.2011.10.002
Jaffe, J. D., Berg, H. C., and Church, G. M. (2004). Proteogenomic mapping as a complementary method to perform genome annotation. Proteomics 4, 59–77. doi: 10.1002/pmic.200300511
Ji, G., Beavis, R. C., and Novick, R. P. (1995). Cell density control of staphylococcal virulence mediated by an octapeptide pheromone. Proc. Natl. Acad. Sci. U.S.A. 92, 12055–12059. doi: 10.1073/pnas.92.26.12055
Jimenez, J. C., and Federle, M. J. (2014). Quorum sensing in group A Streptococcus. Front. Cell. Infect. Microbiol. 4:127. doi: 10.3389/fcimb.2014.00127
Johnston, C., Martin, B., Fichant, G., Polard, P., and Claverys, J.-P. (2014). Bacterial transformation: distribution, shared mechanisms and divergent control. Nat. Rev. Microbiol. 12, 181–196. doi: 10.1038/nrmicro3199
Kaspar, J., Ahn, S.-J., Palmer, S. R., Choi, S. C., Stanhope, M. J., and Burne, R. A. (2015). A Unique ORF within the comX gene of Streptococcus mutans regulates genetic competence and oxidative stress tolerance. Mol. Microbiol. 96, 463–482. doi: 10.1111/mmi.12948
Kaspar, J., Kim, J. N., Ahn, S.-J., and Burne, R. A. (2016). An essential role for (p)ppGpp in the integration of stress tolerance, peptide signaling, and competence development in Streptococcus mutans. Front. Microbiol. 7:1162. doi: 10.3389/fmicb.2016.01162
Kaspar, J., Shields, R. C., and Burne, R. A. (2018). Competence inhibition by the XrpA peptide encoded within the comX gene of Streptococcus mutans. Mol. Microbiol. 109, 345–364. doi: 10.1111/mmi.13989
Kaspar, J., Underhill, S. A. M., Shields, R. C., Reyes, A., Rosenzweig, S., Hagen, S. J., et al. (2017). Intercellular communication via the comX-Inducing Peptide (XIP) of Streptococcus mutans. J. Bacteriol. 00404–17. doi: 10.1101/148320
Kelkar, D. S., Kumar, D., Kumar, P., Balakrishnan, L., Muthusamy, B., Yadav, A. K., et al. (2011). Proteogenomic analysis of Mycobacterium tuberculosis by high resolution mass spectrometry. Mol. Cell. Proteomics 10:M111.011627. doi: 10.1074/mcp.M111.011627
Khan, R., Rukke, H. V., Høvik, H., Åmdal, H. A., Chen, T., Morrison, D. A., et al. (2016). Comprehensive Transcriptome profiles of Streptococcus mutans UA159 Map core streptococcal competence genes. mSystems 1:e00038–e00015. doi: 10.1128/mSystems.00038-15
Khan, R., Rukke, H. V., Ricomini Filho, A. P., Fimland, G., Arntzen, M. Ø., Thiede, B., et al. (2012). Extracellular identification of a processed type II ComR/ComS pheromone of Streptococcus mutans. J. Bacteriol. 194, 3781–3788. doi: 10.1128/JB.00624-12
Kim, J. N., Stanhope, M. J., and Burne, R. A. (2013). Core-gene-encoded peptide regulating virulence-associated traits in Streptococcus mutans. J. Bacteriol. 195, 2912–2920. doi: 10.1128/JB.00189-13
Koch, A., Gawron, D., Steyaert, S., Ndah, E., Crappé, J., De Keulenaer, S., et al. (2014). A proteogenomics approach integrating proteomics and ribosome profiling increases the efficiency of protein identification and enables the discovery of alternative translation start sites. Proteomics 14, 2688–2698. doi: 10.1002/pmic.201400180
Kreth, J., Hung, D. C., Merritt, J., Perry, J., Zhu, L., Goodman, S. D., et al. (2007). The response regulator ComE in Streptococcus mutans functions both as a transcription activator of mutacin production and repressor of CSP biosynthesis. Microbiology 153, 1799–1807. doi: 10.1099/mic.0.2007/005975-0
Lemme, A., Gröbe, L., Reck, M., Tomasch, J., and Wagner-Döbler, I. (2011). Subpopulation-specific transcriptome analysis of competence-stimulating-peptide-induced Streptococcus mutans. J. Bacteriol. 193, 1863–1877. doi: 10.1128/JB.01363-10
Lemos, J. A., Quivey, R. G., Koo, H., and Abranches, J. (2013). Streptococcus mutans: a new gram-positive paradigm? Microbiology 159, 436–445. doi: 10.1099/mic.0.066134-0
Li, Y.-H., Tang, N., Aspiras, M. B., Lau, P. C., Lee, J. H., Ellen, R. P., et al. (2002). A quorum-sensing signaling system essential for genetic competence in Streptococcus mutans is involved in biofilm formation. J. Bacteriol. 184, 2699–2708. doi: 10.1128/JB.184.10.2699-2708.2002
Li, Y. H., Lau, P. C., Lee, J. H., Ellen, R. P., and Cvitkovitch, D. G. (2001). Natural genetic transformation of Streptococcus mutans growing in biofilms. J. Bacteriol. 183, 897–908. doi: 10.1128/JB.183.3.897-908.2001
Liao, S., Klein, M. I., Heim, K. P., Fan, Y., Bitoun, J. P., Ahn, S.-J., et al. (2014). Streptococcus mutans extracellular DNA is upregulated during growth in biofilms, actively released via membrane vesicles, and influenced by components of the protein secretion machinery. J. Bacteriol. 196, 2355–2366. doi: 10.1128/JB.01493-14
Lin, J., Zhu, L., and Lau, G. W. (2016). Disentangling competence for genetic transformation and virulence in Streptococcus pneumoniae. Curr. Genet. 62, 97–103. doi: 10.1007/s00294-015-0520-z
Martin, B., Soulet, A.-L., Mirouze, N., Prudhomme, M., Mortier-Barrière, I., Granadel, C., et al. (2013). ComE/ComE~P interplay dictates activation or extinction status of pneumococcal X-state (competence). Mol. Microbiol. 87, 394–411. doi: 10.1111/mmi.12104
Mashburn-Warren, L., Morrison, D. A., and Federle, M. J. (2010). A novel double-tryptophan peptide pheromone controls competence in Streptococcus spp. via an Rgg regulator. Mol. Microbiol. 78, 589–606. doi: 10.1111/j.1365-2958.2010.07361.x
Meydan, S., Marks, J., Klepacki, D., Sharma, V., Baranov, P. V., Firth, A. E., et al. (2019). Retapamulin-assisted ribosome profiling reveals the alternative bacterial proteome. Molcul. Cell 74, 481–493.e6. doi: 10.1016/j.molcel.2019.02.017
Mignolet, J., Fontaine, L., Sass, A., Nannan, C., Mahillon, J., Coenye, T., et al. (2018). Circuitry rewiring directly couples competence to predation in the gut dweller Streptococcus salivarius. Cell Rep. 22, 1627–1638. doi: 10.1016/j.celrep.2018.01.055
Mohammad, F., Green, R., and Buskirk, A. R. (2019). A systematically-revised ribosome profiling method for bacteria reveals pauses at single-codon resolution. Elife 8:e42591. doi: 10.7554/eLife.42591
Monnet, V., Juillard, V., and Gardan, R. (2014). Peptide conversations in Gram-positive bacteria. Crit. Rev. Microbiol. 42:339–351. doi: 10.3109/1040841X.2014.948804
Moye, Z. D., Son, M., Rosa-Alberty, A. E., Zeng, L., Ahn, S.-J., Hagen, S. J., et al. (2016). Effects of carbohydrate source on genetic competence in Streptococcus mutans. Appl. Environ. Microbiol. 82:4821–4834. doi: 10.1128/AEM.01205-16
Müller, S. A., Findei,ß, S., Pernitzsch, S. R., Wissenbach, D. K., Stadler, P. F., Hofacker, I. L., et al. (2013). Identification of new protein coding sequences and signal peptidase cleavage sites of Helicobacter pylori strain 26695 by proteogenomics. J. Proteom. 86, 27–42. doi: 10.1016/j.jprot.2013.04.036
Mundt, J. O. (1982). The ecology of the streptococci. Microb. Ecol. 8, 355–369. doi: 10.1007/BF02010675
Murchison, H. H., Barrett, J. F., Cardineau, G. A., and Curtiss, R. (1986). Transformation of Streptococcus mutans with chromosomal and shuttle plasmid (pYA629) DNAs. Infect. Immun. 54, 273–282.
Nakahigashi, K., Takai, Y., Kimura, M., Abe, N., Nakayashiki, T., Shiwa, Y., et al. (2016). Comprehensive identification of translation start sites by tetracycline-inhibited ribosome profiling. DNA Res. 23, 193–201. doi: 10.1093/dnares/dsw008
Nealson, K. H., Platt, T., and Hastings, J. W. (1970). Cellular control of the synthesis and activity of the bacterial luminescent system. J. Bacteriol. 104, 313–322.
Okinaga, T., Niu, G., Xie, Z., Qi, F., and Merritt, J. (2010a). The hdrRM operon of Streptococcus mutans encodes a novel regulatory system for coordinated competence development and bacteriocin production. J. Bacteriol. 192, 1844–1852. doi: 10.1128/JB.01667-09
Okinaga, T., Xie, Z., Niu, G., Qi, F., and Merritt, J. (2010b). Examination of the hdrRM regulon yields insight into the competence system of Streptococcus mutans. Mol. Oral Microbiol. 25, 165–177. doi: 10.1111/j.2041-1014.2010.00574.x
Palmer, S. R., Miller, J. H., Abranches, J., Zeng, L., Lefebure, T., Richards, V. P., et al. (2013). Phenotypic heterogeneity of genomically-diverse isolates of Streptococcus mutans. PLoS ONE 8:e61358. doi: 10.1371/annotation/ffff8cd5-b8fa-4d3c-a993-e5169198f1e6
Payne, S. H., Huang, S.-T., and Pieper, R. (2010). A proteogenomic update to Yersinia: enhancing genome annotation. BMC Genomics 11:460. doi: 10.1186/1471-2164-11-460
Perego, M., and Hoch, J. A. (1996). Cell-cell communication regulates the effects of protein aspartate phosphatases on the phosphorelay controlling development in Bacillus subtilis. Proc. Natl. Acad. Sci. U.S.A. 93, 1549–1553. doi: 10.1073/pnas.93.4.1549
Perry, D., and Kuramitsu, H. K. (1981). Genetic transformation of Streptococcus mutans. Infect. Immun. 32, 1295–1297.
Perry, J. A., Jones, M. B., Peterson, S. N., Cvitkovitch, D. G., and Lévesque, C. M. (2009). Peptide alarmone signalling triggers an auto-active bacteriocin necessary for genetic competence. Mol. Microbiol. 72, 905–917. doi: 10.1111/j.1365-2958.2009.06693.x
Petersen, F. C., Fimland, G., and Scheie, A. A. (2006). Purification and functional studies of a potent modified quorum-sensing peptide and a two-peptide bacteriocin in Streptococcus mutans. Mol. Microbiol. 61, 1322–1334. doi: 10.1111/j.1365-2958.2006.05312.x
Reck, M., Tomasch, J., and Wagner-Döbler, I. (2015). The alternative sigma factor SigX controls bacteriocin synthesis and competence, the two quorum sensing regulated traits in Streptococcus mutans. PLoS Genet. 11:e1005353. doi: 10.1371/journal.pgen.1005353
Seaton, K., Ahn, S.-J., and Burne, R. A. (2015). Regulation of competence and gene expression in Streptococcus mutans by the RcrR transcriptional regulator. Mol. Oral Microbiol. 30, 147–159. doi: 10.1111/omi.12079
Seaton, K., Ahn, S.-J., Sagstetter, A. M., and Burne, R. A. (2011). A transcriptional regulator and ABC transporters link stress tolerance, (p)ppGpp, and genetic competence in Streptococcus mutans. J. Bacteriol. 193, 862–874. doi: 10.1128/JB.01257-10
Seefeldt, A. C., Nguyen, F., Antunes, S., Pérébaskine, N., Graf, M., Arenz, S., et al. (2015). The proline-rich antimicrobial peptide Onc112 inhibits translation by blocking and destabilizing the initiation complex. Nat. Struct. Mol. Biol. 22, 470–475. doi: 10.1038/nsmb.3034
Shanker, E., and Federle, M. (2017). Quorum sensing regulation of competence and bacteriocins in Streptococcus pneumoniae and mutans. Genes 8:15. doi: 10.3390/genes8010015
Shanker, E., Morrison, D. A., Talagas, A., Nessler, S., Federle, M. J., and Prehna, G. (2016). Pheromone Recognition and selectivity by ComR proteins among Streptococcus Species. PLOS Pathog. 12:e1005979. doi: 10.1371/journal.ppat.1005979
Sharma, C. M., and Vogel, J. (2014). Differential RNA-seq: the approach behind and the biological insight gained. Curr. Opin. Microbiol. 19, 97–105. doi: 10.1016/j.mib.2014.06.010
Shields, R. C., and Burne, R. A. (2015). Conserved and divergent functions of RcrRPQ in Streptococcus gordonii and S. mutans. FEMS Microbiol. Lett. 362:fnv119. doi: 10.1093/femsle/fnv119
Shields, R. C., and Burne, R. A. (2016). Growth of Streptococcus mutans in biofilms alters peptide signaling at the sub-population level. Front. Microbiol. 7:1075. doi: 10.3389/fmicb.2016.01075
Shields, R. C., O'Brien, G., Maricic, N., Kesterson, A., Grace, M., Hagen, S. J., et al. (2017). Genome-wide screens reveal new gene products that influence genetic competence in Streptococcus mutans. J. Bacteriol. 200:e00508–17. doi: 10.1128/JB.00508-17
Son, M., Ahn, S.-J., Guo, Q., Burne, R. A., and Hagen, S. J. (2012). Microfluidic study of competence regulation in Streptococcus mutans: environmental inputs modulate bimodal and unimodal expression of comX. Mol. Microbiol. 86, 258–272. doi: 10.1111/j.1365-2958.2012.08187.x
Son, M., Ghoreishi, D., Ahn, S.-J., Burne, R. A., and Hagen, S. J. (2015a). Sharply tuned pH response of genetic competence regulation in Streptococcus mutans: a microfluidic study of the environmental sensitivity of comX. Appl. Environ. Microbiol. 81, 5622–5631. doi: 10.1128/AEM.01421-15
Son, M., Shields, R. C., Ahn, S.-J., Burne, R. A., and Hagen, S. J. (2015b). Bidirectional signaling in the competence regulatory pathway of Streptococcus mutans. FEMS Microbiol. Lett. 362:fnv159. doi: 10.1093/femsle/fnv159
Straume, D., Stamsås, G. A., and Håvarstein, L. S. (2015). Natural transformation and genome evolution in Streptococcus pneumoniae. Infect. Genet. Evol. 33, 371–380. doi: 10.1016/j.meegid.2014.10.020
Syvitski, R. T., Tian, X.-L., Sampara, K., Salman, A., Lee, S. F., Jakeman, D. L., et al. (2007). Structure-activity analysis of quorum-sensing signaling peptides from Streptococcus mutans. J. Bacteriol. 189, 1441–1450. doi: 10.1128/JB.00832-06
Talagas, A., Fontaine, L., Ledesma-Garca, L., Mignolet, J., Li de la Sierra-Gallay, I., Lazar, N., et al. (2016). Structural insights into streptococcal competence regulation by the cell-to-cell communication system ComRS. PLOS Pathog. 12:e1005980. doi: 10.1371/journal.ppat.1005980
Tao, L., MacAlister, T. J., and Tanzer, J. M. (1993). Transformation Efficiency of EMS-induced mutants of Streptococcus mutans of altered cell shape. J. Dent. Res. 72, 1032–1039. doi: 10.1177/00220345930720060701
Thomason, M. K., Bischler, T., Eisenbart, S. K., Förstner, K. U., Zhang, A., Herbig, A., et al. (2015). Global transcriptional start site mapping using differential RNA sequencing reveals novel antisense RNAs in Escherichia coli. J. Bacteriol. 197, 18–28. doi: 10.1128/JB.02096-14
Tomasz, A. (1965). Control of the competent state in pneumococcus by a hormone-like cell product: an example for a new type of regulatory mechanism in bacteria. Nature 208, 155–159. doi: 10.1038/208155a0
Underhill, S. A. M., Shields, R. C., Kaspar, J. R., Haider, M., Burne, R. A., and Hagen, S. J. (2018). Intracellular signaling by the comRS system in Streptococcus mutans genetic competence. mSphere 3:e00444–e00418. doi: 10.1128/mSphere.00444-18
Wade, J. T., and Grainger, D. C. (2014). Pervasive transcription: illuminating the dark matter of bacterial transcriptomes. Nat. Rev. Microbiol. 12, 647–653. doi: 10.1038/nrmicro3316
Waters, C. M., and Bassler, B. L. (2005). QUORUM SENSING: cell-to-cell communication in bacteria. Annu. Rev. Cell Dev. Biol. 21, 319–346. doi: 10.1146/annurev.cellbio.21.012704.131001
Weaver, J., Mohammad, F., Buskirk, A. R., and Storz, G. (2019). Identifying small proteins by ribosome profiling with stalled initiation complexes. MBio 10:e02819–e02818. doi: 10.1128/mBio.02819-18
Wenderska, I. B., Lukenda, N., Cordova, M., Magarvey, N., Cvitkovitch, D. G., and Senadheera, D. B. (2012).A novel function for the competence inducing peptide, XIP, as a cell death effector of Streptococcus mutans. FEMS Microbiol. Lett. 336, 104–112. doi: 10.1111/j.1574-6968.2012.02660.x
Whiteley, M., Diggle, S. P., and Greenberg, E. P. (2017). Progress in and promise of bacterial quorum sensing research. Nature 551, 313–320. doi: 10.1038/nature24624
Wickham, H. (2016). ggplot2: Elegant Graphics for Data Analysis, 2nd Edn. New York, NY: Springer-Verlag.
Keywords: peptides, bacterial communication, cell-to-cell signaling, genetic competence, transformation, LC-MS/MS, RNA-Seq, Ribo-Seq
Citation: Kaspar JR and Walker AR (2019) Expanding the Vocabulary of Peptide Signals in Streptococcus mutans. Front. Cell. Infect. Microbiol. 9:194. doi: 10.3389/fcimb.2019.00194
Received: 01 April 2019; Accepted: 21 May 2019;
Published: 06 June 2019.
Edited by:
N. Luisa Hiller, Carnegie Mellon University, United StatesReviewed by:
Laura Ledesma-García, Catholic University of Louvain, BelgiumTodd Kitten, Virginia Commonwealth University, United States
Celine M. Levesque, University of Toronto, Canada
Copyright © 2019 Kaspar and Walker. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Justin R. Kaspar, jkaspar@dental.ufl.edu
 Justin R. Kaspar
Justin R. Kaspar Alejandro R. Walker
Alejandro R. Walker