
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Cell. Infect. Microbiol. , 14 May 2019
Sec. Clinical Microbiology
Volume 9 - 2019 | https://doi.org/10.3389/fcimb.2019.00162
This article is part of the Research Topic Novel Methods for Rapid Identification and Susceptibility Testing of Bacteria View all 19 articles
 Menglan Zhou1,2,3
Menglan Zhou1,2,3 Timothy Kudinha4
Timothy Kudinha4 Bin Du5
Bin Du5 Jinmin Peng5
Jinmin Peng5 Xiaojun Ma6
Xiaojun Ma6 Yang Yang1,3
Yang Yang1,3 Ge Zhang1,3
Ge Zhang1,3 Jingjia Zhang1,3
Jingjia Zhang1,3 Qiwen Yang1,3*
Qiwen Yang1,3* Ying-Chun Xu1,3*
Ying-Chun Xu1,3*Background: Rapid screening of patients for colonization with carbapenemase-producing organisms (CPO), coupled with implementation of infection prevention strategies, has the potential to contain the spread of CPO.
Methods: We first evaluated the performance of Xpert Carba-R assay (in comparison with other phenotypic methods) for carbapenemase detection using clinical isolates, and then used it to determine the intestinal CPO colonization in hospitalized patients. We then assessed the effectiveness of patient isolation in controlling the spread of CPO in a medical intensive care unit.
Results: The Xpert Carba-R assay required the least processing time to reveal results and showed a 94.5% sensitivity and specificity in carbapenemase detection, except for IMP-8 (n = 4). During a 6-month study period, 134 patients in one ward were studied for CPO colonization and infection. Fifteen patients (11.2%) were colonized by CPO as detected by Xpert Carba-R assay, including three NDM, three IMP, and nine KPC possessing strains. The overall colonization and CPO infection rates were both 11.2% each. Isolation of patients with CPO led to a reduction in both colonization (from 28.6 to 5.6%) and infection rates (from 35.7 to 2.8%) during the study period (p < 0.05).
Conclusion: Active surveillance of CPO utilizing the Xpert Carba-R assay supplemented with immediate patient isolation, proved to be an effective strategy to limit the spread of CPO in a health care setting.
The rapid dissemination of carbapenemase-resistant organisms (CROs) has been reported worldwide, and is predominantly attributed to the production of carbapenemases (Tzouvelekis et al., 2012). Nosocomial transmission of carbapenemase-producing organisms (CPO) is a serious public health concern as invasive infections by these organisms are associated with a very high mortality rate (Lasserre et al., 2015; Rotova et al., 2017). Treatment options for CPO are limited as the organisms often exhibit widespread resistance to multiple antibiotics (Pannala et al., 2018). Colonization by CPO may promote infection, and carriers, particularly asymptomatic ones, may act as an important reservoir for CPO dissemination within the hospital setting. Furthermore, carriers of CPO are at higher risk of acquiring a subsequent infection (Borer et al., 2012). Rapid and accurate screening for CPO colonization among hospitalized patients, coupled with implementation of infection prevention strategies, including strict isolation practices, has the potential to interrupt the spread of CPO in hospitals (Wiener-Well et al., 2010; Singh et al., 2012).
Nevertheless, such interventions are critically dependent on accurate and rapid identification of patients colonized by CPO through laboratory testing. In this regard, the introduction of rapid and sensitive methodologies for the detection of CPO is of utmost importance. Unfortunately, detection of CPO still remains a challenge for most clinical microbiology laboratories in China. Currently, several phenotypic methods are available for CPO screening, including growth-based assays (modified Hodge test [MHT]) (Akhi et al., 2017), rapid colorimetric-based assays (manual and commercial versions of the Carba NP test) (Gauthier et al., 2017), matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS)-based meropenem hydrolysis assays (MHA) (Papagiannitsis et al., 2015), immune-chromatographic lateral flow assays (Wareham and Abdul Momin, 2017), and most recently, the modified carbapenem inactivation method (mCIM) (Tamma et al., 2017). All of these assays are culture-based and thus limited to detection of bacterial strains.
The Food and Drug Administration (FDA)-cleared Xpert Carba-R (Cepheid, Sunnydale, CA) assay, on the other hand, is a multiplex PCR that detects the genes encoding for the 5 most common carbapenemases, including KPC, NDM, VIM, IMP, and OXA-48-like enzymes, both directly from bacterial colonies and from rectal swabs, in 2 easy steps that can be completed within 1 h (Moore et al., 2017). Several studies have been conducted in the United States and Europe to assess the accuracy of the Xpert Carba-R assay compared to conventional culture and DNA sequencing, in the detection of CPOs, with reported sensitivity and specificity ranging from 96.6 −100% to 94.2–100%, respectively (Genc et al., 2016; Oviano et al., 2016; Tato et al., 2016; Hoyos-Mallecot et al., 2017). To date, only one study has assessed the performance of Xpert Carba-R assay in determining the intestinal CPO colonization rates, and this was performed in an unselected intensive care unit cohort of patients in Korea. In that study, the assay exhibited good sensitivity, sufficient enough to detect CPO even when culture was negative (Kim et al., 2016). However, none of the previous studies have assessed the utility of combining CPO colonization detection (by Xpert Carba-R assay) with implementation of intervention measures, for CPO infection control.
Thus, in the present study, we conducted a two-stage study to assess the utility of the Xpert Carba-R assay in controlling the transmission of CPO in China. The first stage evaluated the performance of Xpert Carba-R assay (compared with other phenotypic methods) for carbapenemase detection using clinically obtained carbapenem resistant Enterobacteriaceae (CRE) isolates. In the second stage, we conducted an active CPO surveillance study by determining the intestinal CPO colonization rates among hospitalized patients using Xpert Carba-R assay (by directly testing rectal swabs of hospitalized patients), and coupled this with patient isolation in a medical intensive care unit (MICU) ward, in order to contain the spread of CPO.
A total of 100 clinical non-duplicate CRE isolates from blood (n = 24), sputum (n = 21), midstream urine (n = 20), ascitic fluid (n = 6), abscess (n = 6), drainage fluid (n = 5), tissue (n = 5), wound (n = 3), bile (n = 2), catheter (n = 2), and various sterile and non-sterile sites (n = 6), referred by 34 teaching hospitals in China, to Peking Union Medical College Hospital (PUMCH), from 2004 to 2014, were included. All the Enterobacteriaceae isolates were identified by MALDI-TOF MS (Bruker Biotyper Bruker Daltonik, Bremen, Germany). CRE was defined as resistance to one of the carbapenems, imipenem (IPM), meropenem (MEM) or ertapenem (ETP), by broth microdilution method (BMD) according to the latest CLSI 2018 breakpoints criteria (Clinical Laboratory Standards Institute, 2018).
blaIMP, blaVIM, blaNDM, blaAIM, blaSPM, blaKPC, blaDIM, blaBIC, blaGIM, blaSIM, and blaOXA−48 genes were detected by multiplex PCR as previously described (Poirel et al., 2011). blaGES, blaNMC, blaSME, and blaIMI genes were each detected by a single primer set (Queenan and Bush, 2007). All positive isolates were sent for subsequent DNA sequencing. The obtained gene sequences were compared with those in the database located at NCBI blast server (http://blast.ncbi.nlm.nih.gov). A minimum of 99% sequence identity and 100% coverage threshold was deemed sufficient enough for confirmation of each gene (Pierce et al., 2017).
All the CRE isolates were subcultured from frozen stocks onto 5% sheep blood agar (BAP) plate at 35°C for an overnight incubation, followed by a second subculture prior to phenotypic testing including MHT, Carba NP, mCIM and MHA, as previously described (Zhou et al., 2018). As for the Xpert Carba-R assay (Cepheid, Sunnyvale, USA), which detects the blaKPC, blaNDM, blaVIM, blaOXA−48, and blaIMP−1 carbapenem resistance genes in a GeneXpert cartridge, isolates to be tested were inoculated into the sample reagent and loaded into the cartridge following the manufacturer's instructions. The assay has a run time of 47 min in the instrument. Quality control was performed according to the Xpert package insert. Results were analyzed using GeneXpert Software Version 4.3 (AlTamimi et al., 2017; Miller et al., 2017). All the results generated by phenotypic tests and Xpert Carba-R assay were compared to reference DNA sequencing results.
The investigators performing phenotypic testing were blinded to the identity of the isolates. Agreement and validity values were calculated with a 95% confidence interval (CI) based on an exact binomial distribution. Data were analyzed using trend Chi-square tests, SPSS, version 15.0 (SPSS Inc, Chicago, USA).
We performed a single-center prospective study from January 1st to June 30th 2017 in a Medical ICU (MICU) ward in PUMCH. The study was approved by the Human Research Ethics Committee of PUMCH and written informed consent was obtained from each patient or their relatives.
On the very first day (January 1) of the study, rectal swabs were collected from all the patients in the MICU ward and screened for CPO by Xpert Carba-R assay. After that, follow-up tests were performed for each patient upon admission, and at weekly intervals until discharge from MICU. Patients with a positive result were isolated in a designated room immediately. CPO colonization rates were calculated monthly based on Xpert Carba-R assay results. For each patient admitted in the ward at the time of the study, medical records were checked to determine if any CPO infection occurred during hospitalization. Xpert Carba-R assay was used for carbapenemase detection if any CRO was isolated from sterile sites.
A double swab set (Venturi Transystem; Copan, CA) and transport medium (liquid Stuart transport swab; Copan) were used to collect and transport rectal swab specimens from eligible subjects. One swab was inoculated onto a BAP and a China Blue Agar (CBA) plate (screen for gram-negative bacteria), and streaked for isolation followed by an overnight incubation at 35°C. Colonies growing on the CBA plate were identified by MALDI-TOF MS. Susceptibility testing to confirm carbapenem resistance was performed by standard disk diffusion method with ETP, IPM, and MEM, following the CLSI guidelines (Clinical Laboratory Standards Institute, 2018). If the presence of a carbapenem-non-susceptible organism was confirmed (intermediate or resistant) to ETP, IPM or MEM using the CLSI interpretive criteria in document M100-S28 (Clinical Laboratory Standards Institute, 2018), DNA was extracted and then purified, amplified and sequenced using primers as previously described (Moore et al., 2017). The other swab was used for the Xpert Carba-R assay as per the manufacturer's instructions. To minimize the potential bias of sampling differences, the two swabs were gently rolled against one another before starting the Cepheid Xpert Carba-R assay and culture procedure (Tato et al., 2016).
Ten bacterial species were identified among 100 CRE, including 38 Klebsiella pneumoniae, 22 Enterobacter cloacae, 20 Escherichia coli, seven K. oxytoca, four Citrobacter freundii, three Enterobacter aerogenes, two each of Serratia marcescens and K. planticola, one each of Proteus mirabilis and Providencia rettgeri. A total of 73 (73%) isolates were positive for carbapenemase genes, among which 30 Klebsiella pneumoniae carbapenemase (KPC), 20 New Delhi metallo-β-lactamase (NDM), 19 Imipenem metallo-β-lactamase (IMP), two each of IMP & Verona integron-encoded metallo-β-lactamase (VIM) and KPC & IMP carbapenemase genes, were identified (Supplementary Table S1).
The results showed a notable variation between phenotypic assays and molecular-based Xpert Carba-R assay in terms of rapidity, sensitivity, specificity, positive predictive value (PPV), and negative predictive value (NPV), in detecting carbapenemase production among all CRE (Table 1). In the phenotypic assays, Carba NP, and MHA assays take 2–3 h to get results whilst MHT and mCIM take 16–24 h. The molecular-based Xpert Carba-R assay takes less than an hour to complete. The overall sensitivity and specificity for Xpert Carba-R assay was 94.5 and 100%, respectively. Using this method, a 100% sensitivity and specificity was observed for all the carbapenemase genes except IMP (sensitivity: 79.0%), for which four isolates possessing IMP-8 were not detected. However, this gene is not within the detection range of the Xpert Carba-R assay. The 2 h-incubation MHA exhibited the highest overall sensitivity, specificity, PPV and NPV of 100% each, followed by Carba NP and mCIM-MEM with a sensitivity of 98.6% each. In contrast, the 1 h-incubation MHA performed relatively poorly, with an overall sensitivity of 91.8%, mainly due to failure to detect KPC (Supplementary Table S1). Among three carbapenem disks using mCIM, MEM performed the best with the highest sensitivity (98.6%) and specificity (100%) compared to IPM (sensitivity: 97.3%; specificity: 96.3%) and ETP (sensitivity: 95.9%; specificity: 100%). The MHT assay performed the worst in CPO detection, with the lowest sensitivity (87.7%) and specificity (85.2%).
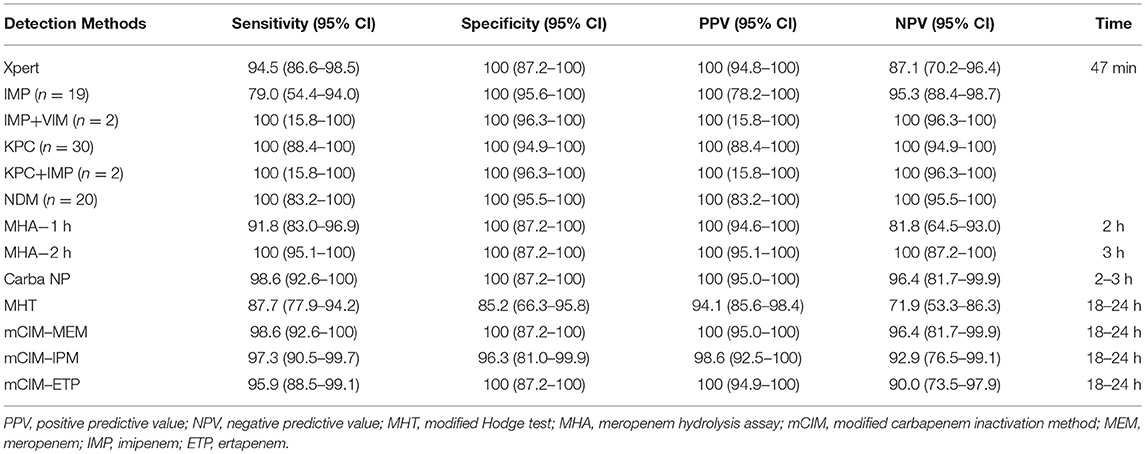
Table 1. Comparison of the performance of Xpert Carba-R assay vs. other phenotypic methods for carbapenemase detection using prospectively collected CRE isolates.
A total of 134 patients were included in the study, and 350 rectal swabs were collected and screened for CPO with the Xpert Carba-R assay during the 6-month study period. Eighty-three (61.9%) of the patients were screened for CPO colonization at least twice during hospitalization (Supplementary Table S2). Of the 134 patients tested, 15 patients (corresponding to 51 specimens) were CPO positive by Xpert Carba-R assay, including three NDM, three IMP, and nine KPC. Conventional culture and DNA sequencing confirmed one NDM-producing and nine KPC-producing K. pneumoniae strains. For three cases involving IMPs, one of NDM, and one of KPC, no organisms were detected from swab culture. In another NDM positive rectal swab, no carbapenemase gene was detected from the carbapenem-sensitive but ESBL-positive E. coli.
Monthly CPO rectal colonization rates and infection rates in the MICU ward are shown in Tables 2, 3. The overall rectal colonization and CPO infection rates were both 11.2% each. At the beginning of the study, four out of 14 patients in the ward were CPO positive by Xpert Carba-R assay, contributing to a colonization rate of 28.6%. The four patients were isolated immediately. All the patients, including those newly admitted, were continually monitored for CPO colonization at weekly intervals, and all positive patients were isolated. Subsequent surveillance showed a significant decreasing trend in the colonization rate from 21.7% in January to 5.6% in June (p = 0.011) (Table 2).
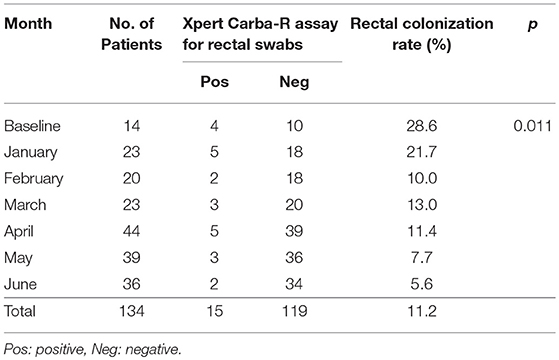
Table 2. Monthly CPO rectal colonization rates based on Xpert Carba-R assay during the 6-month study period.
In 13 out of the 15 patients who tested positive for CPO colonization by Xpert Carba-R assay, follow-up medical record review revealed that CRO was isolated from at least one sterile site (blood, abscess, etc) of the patients during hospitalization (Figure 1). Two different CPOs were identified in three patients (P001, P021, P027), one of which was positive for KPC (K. pneumoniae) and the other was negative (Acinetobacter baumannii) by Xpert Carba-R assay. Noticeably, for one of these three patients (P001), initial colonization screening was positive for IMP instead of KPC, and swab culture was negative for the organism. Similarly, in one patient who developed KPC-producing K. pneumoniae infection (P052), the initial colonization screening was positive for NDM rather than KPC.
Overall, a total of 17 patients including 15 male and two female with an average age of 59 years old were found to be colonized or infected with CPO. Sixteen patients had a history of prior hospitalization before admitted to MICU. Invasive operations were performed in 15 out of the 17 patients and five of them had steroid use within 3 months. Thirteen patients had a previous exposure to antibiotics, nine of which had used carbapenems before (Supplementary Table S3). Three of them were infected with CRP with the same carbapenemase as that detected at initial colonization screening by Xpert Carba-R assay, including one whose initial swab culture was negative for the organism, all of which were KPC-producing K. pneumoniae (Supplementary Table S2). In six other patients with CR A. baumannii infection which was negative by Xpert Carba-R assay, OXA-23 or OXA-24 plus OXA-51 carbapenemases were identified in subsequent DNA sequencing (Figure 1). For these patients, initial colonization screening and swab culture revealed diverse results, including three KPC-producing K. pneumoniae (P021, P027, P047), two IMP (P001, P003), and one NDM (P098) which was negative in the swab culture. No CRO infection was observed in the remaining two patients (P012, P081) despite a positive colonization result. On the contrary, in two patients whose colonization screening and swab culture were both negative (P038, P039), KPC carbapenemase was identified in the K. pneumoniae isolates (Supplementary Table S2). Overall, a significant decreasing trend in the infection rate was shown from 35.7% of baseline to 2.8% in June (p = 0.000) (Table 3).
Active surveillance for potential CPO carriers has been recommended as an aid to infection control by CDC to contain the spread of these strains, and has already been adopted as a routine clinical practice in many parts of the world (Albiger et al., 2015). However, limited data is available on the intestinal carriage of CPO in patients in China. So far, only two studies have been published on carbapenemase-producing Enterobacteriaceae (CPE) colonization rates in China, with rates of 2.6% and 0.5% reported in mainland China and Hong Kong, respectively. However, both studies utilized conventional culture-based methods for colonization screening, which was considered time-consuming and laborious, and also had low sensitivity (Kim et al., 2016). To the best of our knowledge, this is the first study to determine the prevalence of CPO intestinal colonization among inpatients in China using Xpert Carba-R assay.
Most studies conducted to date mainly focused on the prevalence of CRE colonization using MacConkey agar to screen for lactose-fermenting colonies (Ben-David et al., 2011; Gijon et al., 2012; Swaminathan et al., 2013; Banach et al., 2014). Subsequent carbapenemase detection was usually based on culture-dependent phenotypic methods, with reported carriage rates varying by geographic locale (Ben-David et al., 2011; Gijon et al., 2012; Swaminathan et al., 2013; Banach et al., 2014). We evaluated the performance of Xpert Carba-R assay using prospectively collected CRE isolates in the first stage of our study. Compared to all four phenotypic methods studied, Xpert Carba-R assay provided more information about specific carbapenemase genes present in each isolate, and accurately identified all the carbapenemase genes tested except for IMP-8 (n = 4), which is actually beyond the detection limit of the assay. MHT assay required an overnight incubation and still showed poor sensitivity (87.7%) and specificity (85.2%), as it produced nine false-negative (FN) results for NDM-producing isolates and four false-positive (FP) results, as previously reported (Supplementary Table S1) (Miller et al., 2017). Similarly, the mCIM test also needed an overnight incubation but demonstrated a much better sensitivity (95.9–98.6%) and reported fewer FN results associated with one, two, and three NDM-producing isolates for mCIM-MEM, IPM and ETP, respectively (Supplementary Table S1). For Carba-NP and MHA assays, results can be available within 3 h but both require cumbersome preparation for dedicated reagents, with associated costs and training needs. FN results were observed in five KPC and one IMP isolates for MHA-1h and in one KPC for Carba-NP test (Supplementary Table S1). From the above findings, Xpert Carba-R assay appears to be a good alternative in carbapenemase detection when accuracy and rapidity are taken into consideration. Most importantly, it can provide specific carbapenemase information for each isolate even when multiple resistance genes are present, an outcome also reported in previous studies (Tato et al., 2016), and which cannot be achieved by any of the other phenotypic methods.
In the second stage of our study, we studied the utility of the Xpert Carba-R assay in clinical practice by determining the intestinal CPO colonization rates in patients (by directly testing rectal swabs of hospitalized patients), and coupled this with patient isolation in a MICU ward, in order to contain the spread of CPO. Based on our data, a much higher average CPO colonization rate (11.2%) was observed in our study compared to previous rates of 2.6 and 0.5% reported in mainland China and Hong Kong, respectively (Zhao et al., 2014; Cheng et al., 2016). While geographic locale and time variation may be a possible explanation for the differences in rectal colonization rates, another possible reason is that we calculated the colonization rate based on Xpert Carba-R assay results, whereas the two previous studies utilized conventional culture-based methods for colonization screening, which is considered to be of low sensitivity (Zhao et al., 2014; Cheng et al., 2016; Kim et al., 2016). As our results show, for three IMP, one KPC, and one NDM cases, as detected by Xpert Carba-R assay, no organisms were obtained from swab culture. In another patient whose rectal specimen was positive for NDM by Xpert Carba-R assay, only ESBL was confirmed in subsequent culture of E. coli. One possibility is that these patients may have been receiving antimicrobial agents which inhibited the growth of the organisms (Tenover et al., 2013; Tato et al., 2016). Furthermore, it is also possible that the bacterial load in the rectal swab specimens was too low for successful culture on agar plates (Tato et al., 2016). Another possibility is that some or all carbapenemase-producing organisms have fastidious growth requirements, or carry a modified sequence of the target gene that was not expressed or was expressed at low levels (Tenover et al., 2013; Tato et al., 2016).
Despite the high baseline CPO colonization rates, a significant decreasing trend was observed in both the CPO colonization and infection rates during the course of the study period (p < 0.05). A total of 15 patients tested positive for CPO colonization initially and 13 of them developed subsequent CRO infection, eight of which had the same carbapenemase (KPC) detected in both the colonizing and infecting bacterial strain, suggesting a pathway from colonization to infection. Two other patients whose initial colonization was with IMP and NDM possessing strains, also developed KPC-producing K. pneumoniae infection afterwards. Six patients who were initially CPO colonized (three KPC, two IMP, and one NDM) developed CR A. baumannii infection but tested negative for carbapenemase by Xpert Carba-R assay. Subsequent DNA sequencing of these isolates confirmed the presence of OXA-23/24 plus OXA-51 carbapenemases, which are beyond the detection range of Xpert Carba-R assay. Two patients who were negative for CPO colonization also developed CPO infection, with KPC-producing K. pneumoniae isolated from the respiratory tract. The route of infection acquisition in these patients was uncertain and requires to be further investigated. Nevertheless, generalizing from the significant decline in CPO colonization and infection rates, it is reasonable to speculate that active surveillance plus patient isolation was an effective measure in the containment of CPO spread.
Our study has several limitations. First, the number of organisms used to establish the sensitivity and specificity of the Xpert Carba-R assay did not cover the entire range of the CP genes that could be detected. No OXA-positive isolate was detected in our study, both in the evaluation and clinical stages due to the scarcity of isolates with this genotype in our setting. At the time of writing, <50 OXA-positive CROs have been reported in previous studies from China (Liu et al., 2015; Guo et al., 2016; Yin et al., 2017). Secondly, we did not use nutrient broth for bacteria enrichment on collected swabs in our study. Consequently, for those swabs that tested positive by Xpert Carba-R assay but negative by direct plate culture, we were unable to trace the root cause. Third, despite the multiple measures recommended by CDC for control of CPE (Centers for Disease Control and Prevention, 2015), we only evaluated the effect of active surveillance plus patient isolation in this tentative study; more preventive strategies should be implemented as soon as possible to better contain the spread of CPE.
In conclusion, our study suggests that the Xpert CARBA-R assay is an easy-to-use and accurate assay for the detection of carbapenemase genes, which allows for rapid identification of patients colonized with CPO directly from rectal swabs. However, the assay is limited in the range of genes detected as it can only detect five carbapenemase genes. Active CPO surveillance using this assay, supplemented with immediate patient isolation, proved to be an effective measure to limit the spread of CPO in a health care setting. The higher CPO colonization rate (11.2%) in the MICU ward of a tertiary care hospital in China compared to previous similar settings (Zhao et al., 2014; Cheng et al., 2016; Kim et al., 2016), is a cause for concern, and calls for immediate action to tackle CPO spread.
The study was approved by the Human Research Ethics Committee of PUMCH and written informed consent was obtained from each patient or their relatives.
MZ, QY, and Y-CX conceived and designed the experiments, performed the experiments, analyzed the data, and wrote the paper. YY helped perform the experiments. MZ, TK, BD, JP, XM, YY, GZ, JZ, QY, and Y-CX approved the final version of the manuscript.
This work was supported by The National Key Research and Development Program of China (2018YFC1200100, 2018YFC1200105), Outstanding Talents Training Funding Project of Dongcheng District, Beijing (2017), Graduate Innovation Fund of Peking Union Medical College (grant no. 2017-1002-1-21), CAMS Innovation Fund for Medical Sciences (grant no. 2016-I2M-1-014), CAMS Initiative for Innovative Medicine (grant no. 2016-I2M-3-014), National Natural Science Foundation of China (grant no. 81101287) and Youth Foundation of Peking Union Medical College Hospital (2013) and the Cepheid Company. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2019.00162/full#supplementary-material
Akhi, M.T., Khalili, Y., Ghotaslou, R., Kafil, H.S., Yousefi, S., Nagili, B., et al. (2017). Carbapenem inactivation: a very affordable and highly specific method for phenotypic detection of carbapenemase-producing Pseudomonas aeruginosa isolates compared with other methods. J. Chemother. 29, 144–149. doi: 10.1080/1120009X.2016.1199506
Albiger, B., Glasner, C., Struelens, M. J., Grundmann, H., Monnet, D. L., and European Survey of carbapenemase-producing enterobacteriaceae working, g. (2015). Carbapenemase-producing Enterobacteriaceae in Europe: assessment by national experts from 38 countries, May 2015. Euro. Surveill. 20:45. doi: 10.2807/1560-7917.ES.2015.20.45.30062
AlTamimi, M., AlSalamah, A., AlKhulaifi, M., and AlAjlan, H. (2017). Comparison of phenotypic and PCR methods for detection of carbapenemases production by enterobacteriaceae. Saudi J. Biol. Sci. 24, 155–161. doi: 10.1016/j.sjbs.2016.07.004
Banach, D. B., Francois, J., Blash, S., Patel, G., Jenkins, S. G., LaBombardi, V., et al. (2014). Active surveillance for carbapenem-resistant Enterobacteriaceae using stool specimens submitted for testing for Clostridium difficile. Infect. Control Hosp. Epidemiol. 35, 82–84. doi: 10.1086/674391
Ben-David, D., Masarwa, S., Navon-Venezia, S., Mishali, H., Fridental, I., Rubinovitch, B., et al. (2011). Carbapenem-resistant Klebsiella pneumoniae in post-acute-care facilities in Israel. Infect. Control Hosp. Epidemiol. 32, 845–853. doi: 10.1086/661279
Borer, A., Saidel-Odes, L., Eskira, S., Nativ, R., Riesenberg, K., Livshiz-Riven, I., et al. (2012). Risk factors for developing clinical infection with carbapenem-resistant Klebsiella pneumoniae in hospital patients initially only colonized with carbapenem-resistant K. pneumoniae. Am. J. Infect. Control 40, 421–425. doi: 10.1016/j.ajic.2011.05.022
Centers for Disease Control and Prevention (2015). Guidance for Control of Carbapenem-Resistant Enterobacteriaceae (CRE). 2015 Toolkit. Atlanta, GA: Centers for Disease Control and Prevention.
Cheng, V. C., Chen, J. H., So, S. Y., Wong, S. C., Chau, P. H., Wong, L. M., et al. (2016). A novel risk factor associated with colonization by carbapenemase-producing enterobacteriaceae: use of proton pump inhibitors in addition to antimicrobial treatment. Infect. Control Hosp. Epidemiol. 37, 1418–1425. doi: 10.1017/ice.2016.202
Clinical and Laboratory Standards Institute (2018). Performance Standards for Antimicrobial Susceptibility Testing: Twenty-Eighth Informational Supplement, M100-S28. Wayne, PA: Clinical and Laboratory Standards Institute.
Gauthier, L., Bonnin, R. A., Dortet, L., and Naas, T. (2017). Retrospective and prospective evaluation of the Carbapenem inactivation method for the detection of carbapenemase-producing enterobacteriaceae. PLoS ONE. 12:e0170769. doi: 10.1371/journal.pone.0170769
Genc, O., Aksu, E., and Gulcan, A. (2016). The identification of carbapenemase types in Enterobacteriaceae by using molecular assay and phenotyping confirmation tests. J. Microbiol. Methods 125, 8–11. doi: 10.1016/j.mimet.2016.03.010
Gijon, D., Curiao, T., Baquero, F., Coque, T. M., and Canton, R. (2012). Fecal carriage of carbapenemase-producing Enterobacteriaceae: a hidden reservoir in hospitalized and nonhospitalized patients. J. Clin. Microbiol. 50, 1558–1563. doi: 10.1128/JCM.00020-12
Guo, L., An, J., Ma, Y., Ye, L., Luo, Y., Tao, C., et al. (2016). Nosocomial outbreak of OXA-48-producing Klebsiella pneumoniae in a Chinese hospital: clonal transmission of ST147 and ST383. PLoS ONE. 11:e0160754. doi: 10.1371/journal.pone.0160754
Hoyos-Mallecot, Y., Ouzani, S., Dortet, L., Fortineau, N., and Naas, T. (2017). Performance of the Xpert((R)) Carba-R v2 in the daily workflow of a hygiene unit in a country with a low prevalence of carbapenemase-producing Enterobacteriaceae. Int. J. Antimicrob. Agents 49, 774–777. doi: 10.1016/j.ijantimicag.2017.01.025
Kim, D. K., Kim, H. S., Pinto, N., Jeon, J., D'Souza, R., Kim, M. S., et al. (2016). Xpert CARBA-R assay for the detection of carbapenemase-producing organisms in intensive care unit patients of a Korean tertiary care hospital. Ann. Lab. Med. 36, 162–165. doi: 10.3343/alm.2016.36.2.162
Lasserre, C., De Saint Martin, L., Cuzon, G., Bogaerts, P., Lamar, E., Glupczynski, Y., et al. (2015). Efficient detection of carbapenemase activity in enterobacteriaceae by matrix-assisted laser desorption ionization-time of flight mass spectrometry in less than 30 minutes. J. Clin. Microbiol. 53, 2163–2171. doi: 10.1128/JCM.03467-14
Liu, Y., Feng, Y., Wu, W., Xie, Y., Wang, X., Zhang, X., et al. (2015). First Report of OXA-181-Producing Escherichia coli in China and characterization of the isolate using whole-genome sequencing. Antimicrob. Agents Chemother. 59, 5022–5025. doi: 10.1128/AAC.00442-15
Miller, S. A., Hindler, J. A., Chengcuenca, A., and Humphries, R.M. (2017). Use of ancillary carbapenemase tests to improve specificity of phenotypic definitions for carbapenemase-producing enterobacteriaceae. J. Clin. Microbiol. 55, 1827–1836. doi: 10.1128/JCM.00157-17
Moore, N. M., Canton, R., Carretto, E., Peterson, L. R., Sautter, R. L., Traczewski, M. M., et al. (2017). Rapid identification of five classes of carbapenem resistance genes directly from rectal swabs by use of the Xpert carba-R assay. J. Clin. Microbiol. 55, 2268–2275. doi: 10.1128/JCM.00137-17
Oviano, M., Torres, I., Gonzalez, M., and Bou, G. (2016). Evaluation of a novel procedure for rapid detection of carbapenemase-producing Enterobacteriaceae (CPE) using the LightMix(R) modular carbapenemase kits. J. Antimicrob. Chemother. 71, 3420–3423. doi: 10.1093/jac/dkw356
Pannala, R., Baldwin, B., Aluru, V., Grys, T.E., Holmes, J., Miller, L.J., et al. (2018). Prospective study of the feasibility of point-of-care testing strategy for carbapenem-resistant organism detection. Endosc. Int. Open. 6, E58–E63. doi: 10.1055/s-0043-122141
Papagiannitsis, C. C., Studentova, V., Izdebski, R., Oikonomou, O., Pfeifer, Y., Petinaki, E., et al. (2015). Matrix-assisted laser desorption ionization-time of flight mass spectrometry meropenem hydrolysis assay with NH4HCO3, a reliable tool for direct detection of carbapenemase activity. J. Clin. Microbiol. 53, 1731–1735. doi: 10.1128/JCM.03094-14
Pierce, V. M., Simner, P. J., Lonsway, D. R., Roe-Carpenter, D. E., Johnson, J. K., Brasso, W.B., et al. (2017). Modified carbapenem inactivation method for phenotypic detection of carbapenemase production among enterobacteriaceae. J. Clin. Microbiol. 55, 2321–2333. doi: 10.1128/JCM.00193-17
Poirel, L., Walsh, T. R., Cuvillier, V., and Nordmann, P. (2011). Multiplex PCR for detection of acquired carbapenemase genes. Diagn. Microbiol. Infect. Dis. 70, 119–123. doi: 10.1016/j.diagmicrobio.2010.12.002
Queenan, A. M., and Bush, K. (2007). Carbapenemases: the versatile beta-lactamases. Clin. Microbiol. Rev. 20, 440–458. doi: 10.1128/CMR.00001-07
Rotova, V., Papagiannitsis, C. C., Skalova, A., Chudejova, K., and Hrabak, J. (2017). Comparison of imipenem and meropenem antibiotics for the MALDI-TOF MS detection of carbapenemase activity. J Microbiol. Methods 137, 30–33. doi: 10.1016/j.mimet.2017.04.003
Singh, K., Mangold, K. A., Wyant, K., Schora, D.M., Voss, B., Kaul, K. L., et al. (2012). Rectal screening for Klebsiella pneumoniae carbapenemases: comparison of real-time PCR and culture using two selective screening agar plates. J. Clin. Microbiol. 50, 2596–2600. doi: 10.1128/JCM.00654-12
Swaminathan, M., Sharma, S., Poliansky Blash, S., Patel, G., Banach, D. B., Phillips, M., et al. (2013). Prevalence and risk factors for acquisition of carbapenem-resistant enterobacteriaceae in the setting of endemicity. Infect. Control Hosp. Epidemiol. 34, 809–817. doi: 10.1086/671270
Tamma, P. D., Opene, B. N., Gluck, A., Chambers, K. K., Carroll, K. C., and Simner, P. J. (2017). Comparison of 11 phenotypic assays for accurate detection of carbapenemase-producing enterobacteriaceae. J. Clin. Microbiol. 55, 1046–1055. doi: 10.1128/JCM.02338-16
Tato, M., Ruiz-Garbajosa, P., Traczewski, M., Dodgson, A., McEwan, A., Humphries, R., et al. (2016). Multisite evaluation of cepheid Xpert Carba-R assay for detection of carbapenemase-producing Organisms in rectal swabs. J. Clin. Microbiol. 54, 1814–1819. doi: 10.1128/JCM.00341-16
Tenover, F. C., Canton, R., Kop, J., Chan, R., Ryan, J., Weir, F., et al. (2013). Detection of colonization by carbapenemase-producing gram-negative Bacilli in patients by use of the Xpert MDRO assay. J. Clin. Microbiol. 51, 3780–3787. doi: 10.1128/JCM.01092-13
Tzouvelekis, L. S., Markogiannakis, A., Psichogiou, M., Tassios, P. T., and Daikos, G. L. (2012). Carbapenemases in Klebsiella pneumoniae and other enterobacteriaceae: an evolving crisis of global dimensions. Clin. Microbiol. Rev. 25, 682–707. doi: 10.1128/CMR.05035-11
Wareham, D. W., and Abdul Momin, M. H. (2017). Rapid detection of carbapenemases in enterobacteriaceae: evaluation of the resist-3 O.K.N. (OXA-48, KPC, NDM) lateral flow multiplexed assay. J. Clin. Microbiol. 55, 1223–1225. doi: 10.1128/JCM.02471-16
Wiener-Well, Y., Rudensky, B., Yinnon, A. M., Kopuit, P., Schlesinger, Y., Broide, E., et al. (2010). Carriage rate of carbapenem-resistant Klebsiella pneumoniae in hospitalised patients during a national outbreak. J. Hosp. Infect. 74, 344–349. doi: 10.1016/j.jhin.2009.07.022
Yin, D., Dong, D., Li, K., Zhang, L., Liang, J., Yang, Y., et al. (2017). Clonal dissemination of OXA-232 carbapenemase-producing Klebsiella pneumoniae in neonates. Antimicrob. Agents Chemother. 61:8. doi: 10.1128/AAC.00385-17
Zhao, Z. C., Xu, X. H., Liu, M. B., Wu, J., Lin, J., and Li, B. (2014). Fecal carriage of carbapenem-resistant enterobacteriaceae in a Chinese university hospital. Am. J. Infect. Control 42, e61–64. doi: 10.1016/j.ajic.2014.01.024
Keywords: Xpert Carba-R assay, colonization, carbapenemase-producing organisms, infection control, screening
Citation: Zhou M, Kudinha T, Du B, Peng J, Ma X, Yang Y, Zhang G, Zhang J, Yang Q and Xu Y-C (2019) Active Surveillance of Carbapenemase-Producing Organisms (CPO) Colonization With Xpert Carba-R Assay Plus Positive Patient Isolation Proves to Be Effective in CPO Containment. Front. Cell. Infect. Microbiol. 9:162. doi: 10.3389/fcimb.2019.00162
Received: 28 October 2018; Accepted: 29 April 2019;
Published: 14 May 2019.
Edited by:
Nahed Ismail, University of Illinois at Chicago, United StatesReviewed by:
Yamuna Devi Bakthavatchalam, Christian Medical College and Hospital, IndiaCopyright © 2019 Zhou, Kudinha, Du, Peng, Ma, Yang, Zhang, Zhang, Yang and Xu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Qiwen Yang, eWFuZ3Fpd2VuODFAdmlwLjE2My5jb20=
Ying-Chun Xu, eHljcHVtY2hAMTM5LmNvbQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.