
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Cell. Infect. Microbiol. , 02 April 2019
Sec. Clinical Microbiology
Volume 9 - 2019 | https://doi.org/10.3389/fcimb.2019.00074
This article is part of the Research Topic Beyond Antimicrobials: Non-Traditional Approaches to Combating Multidrug-Resistant Bacteria View all 24 articles
Antimicrobial resistance constitutes one of the major challenges facing humanity in the Twenty-First century. The spread of resistant pathogens has been such that the possibility of returning to a pre-antibiotic era is real. In this scenario, innovative therapeutic strategies must be employed to restrict resistance. Among the innovative proposed strategies, anti-virulence therapy has been envisioned as a promising alternative for effective control of the emergence and spread of resistant pathogens. This review presents some of the anti-virulence strategies that are currently being developed, it will cover strategies focused on quench pathogen quorum sensing (QS) systems, disassemble of bacterial functional membrane microdomains (FMMs), disruption of biofilm formation and bacterial toxin neutralization.
Antimicrobial resistance has turned a serious concern to the human health, because in addition to the death caused by drug-resistant pathogens (~700,000 death annually and it is estimated ~10 million for the year 2050), important medical procedures such as organ transplantation, cancer chemotherapy and surgery are also compromised (O'Neill, 2016). Antimicrobial resistance is a multifactorial phenomenon. Therefore, to circumvent it, a range of actions are needed (WHO, 2018). According that, the innovative antimicrobial compounds development that operate under different principles to those of conventional antibiotics constitutes an important element in the battle against resistance (Munguia and Nizet, 2017). Among the new therapeutic strategies, anti-virulence therapy has emerged as a promising alternative since instead of killing the pathogens; it tries to deprive them from their virulence factors. Accordingly, the selective pressure exerted over pathogens should be lower than that exerted by conventional antibiotics and the emergence and spread of resistant mutants could be less frequent (Sully et al., 2014; Daly et al., 2015; Quave et al., 2015; Vale et al., 2016; Munguia and Nizet, 2017). However, Pseudomonas aeruginosa has developed resistance to anti-virulence drugs (Maeda et al., 2012; García-Contreras et al., 2013, 2015).
Virulence factors are microbial components (biomolecules and structures) used by pathogens to colonize, invade and persist in a susceptible host (Peterson, 1996; Defoirdt, 2017). The production of these factors is under the control of regulatory mechanisms; therefore, in principle interference with these regulatory mechanisms could affect the production of several virulence factors (Defoirdt, 2017). In this regard, quorum-sensing systems (QS) are involved in the regulation of the production of several virulence factors and consequently constitute one of the most exploited targets for the development of anti-virulence drugs (Defoirdt, 2017; Schütz and Empting, 2018). Moreover, the proper folding and/or oligomerization of virulence factors are pivotal for their biological activities. Therefore, the bacterial machinery involved in the virulence factors assembly is also a suitable target for disturbing pathogen virulence via anti-virulence drugs (Heras et al., 2015; Kahler et al., 2018). Recently, it has been described that bacterial functional membrane microdomains (FMMs) play a significant role in the assembly of several virulence factors, hence turning FMMs in an attractive target for drug development (García-Fernández et al., 2017; Koch et al., 2017; Mielich-Süss et al., 2017). In addition to disrupting the production and assembly of virulence factors; anti-virulence drugs have also been focused on interfering with the virulence factor functions (Mühlen and Dersch, 2016; Dickey et al., 2017). In that view, toxin neutralization constitutes a useful strategy to diminish the virulence of pathogens, as secretion of toxins is used by pathogens to colonize the host as well as to evade host immune system response (Heras et al., 2015; Kong et al., 2016; Rudkin et al., 2017). In addition, biofilm growing is a strategy used by pathogens to overcome the host immune system response (Gunn et al., 2016; Watters et al., 2016). Several anti-virulence strategies have been directed to disturb biofilm via interference with bacterial adhesion, extracellular matrix production or disintegration of existing biofilm (Feng et al., 2018; Liu et al., 2018; Puga et al., 2018).
Given the significance attributed to anti-virulence therapy in the scientific community, and especially regarding antimicrobial resistance, this review is directed toward some recent findings in this area. It will uncover innovative strategies that are being implemented to quench pathogen quorum sensing (QS) systems, disassemble functional membrane microdomains (FMMs), disrupt biofilm formation and neutralize toxins (Figure 1 and Table 1). Some of the challenges that anti-virulence therapy faces as an emerging treatment in overcoming multidrug resistant pathogens will also be highlighted.
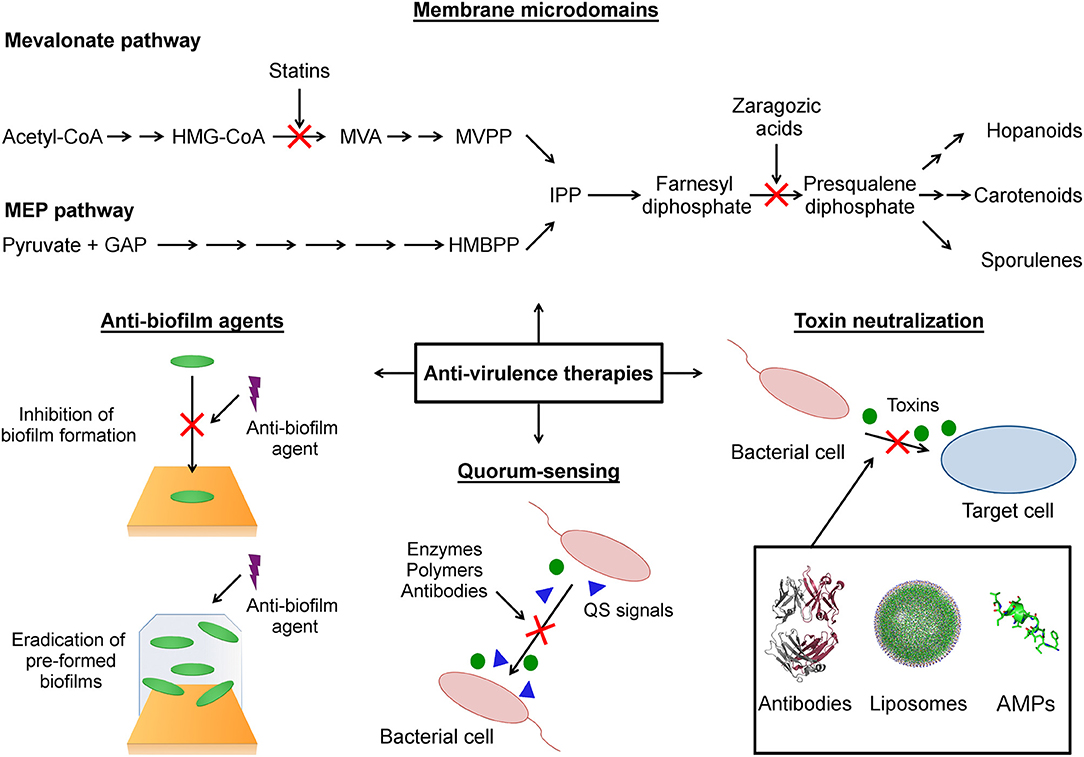
Figure 1. Schematic representation of anti-virulence strategies covered in this review. Membrane microdomains: The functional membrane microdomains (FMMs) are targeted by small molecules (statins, zaragozic acid) that inhibit the biosynthesis of their major constituent lipids (hopanoids, carotenoids). Anti-biofilm agents: This strategy focused on the use of agents that block the initial bacterial attachment to surface during biofilm formation and agents that destroy preformed biofilm. Quorum-sensing: The anti-virulence strategy that seeks modulate the production of virulence factors through interference with the quorum-sensing networks. Toxin neutralization: A strategy focused on block the action of toxins on host target cells. HMG-CoA (3-hydroxy-3-methylglutaryl-CoA), MVA (mevalonic acid), MVPP (5-diphosphomevalonate), GAP (D-glyceraldehyde-3-phosphate), HMBPP (4-hydroxy-3-methylbut-2-enyl-diphosphate), IPP (isopentenyl diphosphate), QS (quorum sensing), AMPs (antimicrobial peptides).
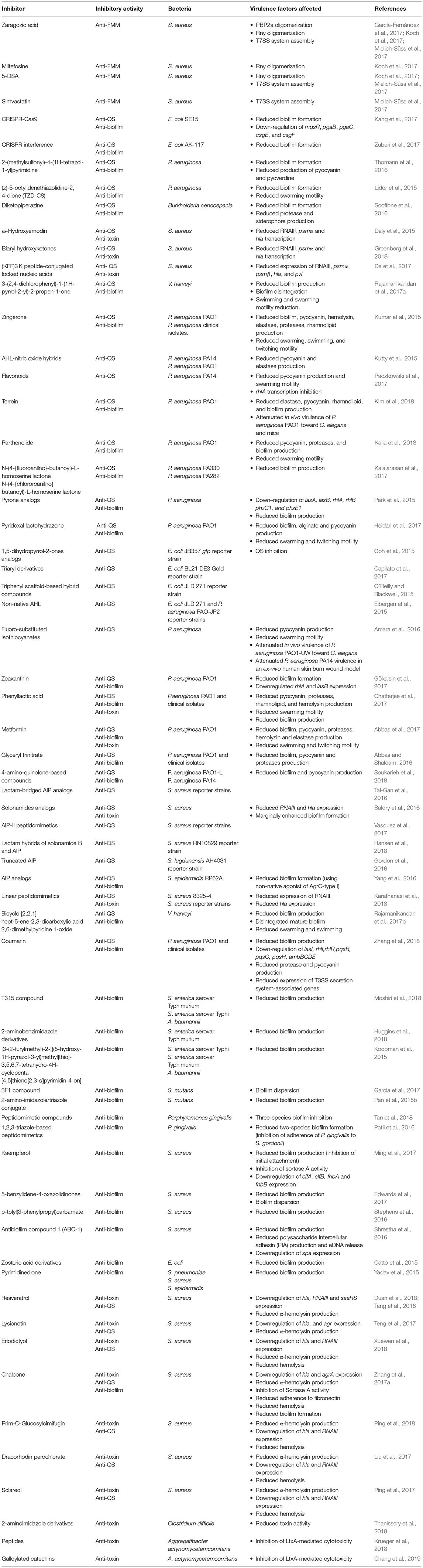
Table 1. Inhibitors of functional membrane microdomains assembly, quorum-sensing systems, biofilm formation, and toxin production and function.
To develop new therapeutic strategies against multidrug-resistant pathogens, a suitable approach could be designing antimicrobial compounds that target bacterial structures other than the targets of the major conventional antibiotics. In this respect, the bacterial cytoplasmic membrane constitutes an attractive target as it functions as a barrier that maintains favorable intracellular physicochemical conditions for the correct development of bacterial metabolism. In addition, the cytoplasmic membrane regulates the exchange of information and substances with the extracellular medium (Poolman et al., 2004; Strahl and Errington, 2017). Structural changes in membranes to resist antimicrobial compounds could involve an elevate fitness cost and consequently could be less likely to occur (Zasloff, 2002). However, it has been reported membrane modifications linked to resistance toward antimicrobial compounds (Nuri et al., 2015; Joo et al., 2016). Therefore, compounds that interfere with structural organization and/or functions associated with membranes without affecting bacterial growth could be desirable.
In particular, bacterial membranes contain FMMs which are eukaryotic lipid-raft–like domains that enclose a characteristic lipid and protein composition. Specifically, they appear be rich in polyisoprenoid lipids like hopanoids and carotenoids, conferring compact, rigid, and hydrophobic features that could limit the diffusion of FMM-associated proteins away from them (Bramkamp and Lopez, 2015). As regards FMM-associated protein composition, bacterial protein flotillins are essential FMM components. These proteins are closely associated with microdomains and are involved in the membrane fluidity regulation as well as promoting and stabilizing the assembly of specific protein complexes via their scaffold activity (Lopez and Koch, 2017). Other FMM-associated proteins are involved in signaling networks (e.g., sensor kinase KinC), protein secretion machinery (e.g., Sec Y and Sec A) and proteolytic complexes (e.g., FtsH protease complex) (Bramkamp and Lopez, 2015; Lopez and Koch, 2017). Most of these FMM-associated proteins are functionally active when they form multimeric complexes. Therefore, FMMs could be seen as protein complexes assembly platforms with punctuating distribution along bacterial cytoplasmic membrane, where the recruited proteins undergo efficient oligomerization and consequently become functionally active (Bramkamp and Lopez, 2015; Lopez and Koch, 2017).
Experimental evidence has shown that mutant strains in genes involved in the production of FMM structural components could be hampered in establishing virulence determinants. A Bacillus subtilis mutant in the yisP gene was ineffective in biofilm formation. The gene yisP encodes for phosphatase YisP, which produces farnesol from farnesyl diphosphate (López and Kolter, 2010; Feng et al., 2014). Moreover, B. subtilis double mutant (ΔfloT ΔyqfA) in genes that encode for the flotillin-like proteins FloT and YqfA (FloA) was defective in the formation of biofilms and sporulation (López and Kolter, 2010; Yepes et al., 2012). Bacillus subtilis mutants Δ yisP, Δ yuaG, Δ yqfA, and Δ yuaG Δ yqfA showed diminished Sec-dependent secretion efficiency (Bach and Bramkamp, 2013). Furthermore, a Campylobacter jejuni mutant in the gene cj0268c displayed reduced adherence to Caco2 cells. The gene cj0268c encodes for a protein that contains a SPFH domain, which is typical for flotillin-like proteins (Tareen et al., 2013). In addition, gnotobiotic IL-10−/− mice infected with C. jejuni cj0268c mutant developed reduced intestinal immunopathology in comparison with ones infected with parental strain and complemented strain (Heimesaat et al., 2014). Recently, it was observed that lack of the gene floA in Staphylococcus aureus may impact the function of the type VII secretion system (T7SS). Consequently, the ΔfloA mutant exhibited a reduced virulence in a murine model (Mielich-Süss et al., 2017). Similarly, another study showed that the S. aureus ΔfloA mutant exhibited diminished virulence in both an invertebrate infection model (Galleria mellonella) and a murine infection model. Specifically, a perturbed degradosome activity was observed; probably by the defective FloA-assisted oligomerization of RNase Rny in the ΔfloA mutant (Koch et al., 2017). All these experimental items of evidence suggest that FMMs could be an attractive target to develop anti-virulence therapy.
In accordance with the above described experimental evidence, several studies have demonstrated that small molecules, which interfere with polyisoprenoid lipid biosynthesis metabolic pathways and with FMM′ physicochemical properties, attenuated the virulence of pathogens in vitro and in vivo (Table 1) (García-Fernández et al., 2017; Koch et al., 2017; Mielich-Süss et al., 2017). Isoprenoids are organic molecules that involve significant diversity of chemical structures and functions; however, they all derive from isopentyl diphosphate (IPP) or its isomer dimethylallyl diphosphate (DMAPP) (Heuston et al., 2012; Pérez-Gil and Rodríguez-Concepción, 2013). These precursor molecules are synthesized in bacteria through two metabolic pathways including the mevalonate pathway and the 2C-methyl-D-erythritol 4-phosphate (MEP) pathway. The MEP pathway is used by most bacteria. However, the mevalonate pathway is used exclusively by pathogens like Borrelia burgdorferi, S. aureus, Streptococcus pneumoniae, Enterococcus faecalis, and is also present in animals (Wilding et al., 2000a; Pérez-Gil and Rodríguez-Concepción, 2013). Moreover, Listeria monocytogenes and some Streptomyces strains contain both pathways. Bacteria such as Rickettsia and Mycoplasma do not contain genes that encode for the enzymes involved in the mevalonate and MEP pathways, and therefore they lack both isoprenoid pathways (Pérez-Gil and Rodríguez-Concepción, 2013).
Because the mevalonate pathway is shared between humans and bacteria, in principle, hypercholesterolemia-treating drugs in humans could exert an inhibitory effect on the bacterial mevalonate pathway (Bramkamp and Lopez, 2015). Statins are a class of drugs that target the enzyme class I HMG-CoA reductase (3-hydroxy-3-methylglutaryl-Coenzyme A reductase), which catalyzes the conversion of 3-hydroxy-3-methylglutaryl-Coenzyme A (HMG-CoA) to coenzyme A (CoA) and mevalonate via reductive deacylation, in the human mevalonate pathway (Istvan and Deisenhofer, 2001; Tobert, 2003). Statins bind to the class I HMG-CoA reductase active site, affecting the binding of biological substrate HMG-CoA, and therefore act as competitive inhibitors (Istvan and Deisenhofer, 2001). Similarly, as takes place for human HMG-CoA reductase, statins are also class II HMG-CoA reductase (bacteria and archaea) inhibitors. However, statins exert a diminished inhibitory activity toward class II HMG-CoA reductases [inhibition constant (Ki) in the micro and millimolar range] than class I (Ki in the nanomolar range), which appear to be associated with structural differences between these enzyme classes (Alberts et al., 1980; Kim et al., 2000; Wilding et al., 2000b; Tabernero et al., 2003; Hedl and Rodwell, 2004; Haines et al., 2013). It is important to highlight that some statins with antibacterial activity against Gram-positive and Gram-negative bacteria have been reported. Such capability matched to their wide clinical use, potentially favoring the statins resistance emergence (Ko et al., 2017).
In addition to statins, another inhibitors of bacterial HMG-CoA reductase are the plant-derived compounds annonaceous acetogenins. Feng et al. (2011) showed that squamostating A, squamostating B, squamocin C, and asimicin A were more potent S. pneumoniae HMG-CoA reductase inhibitors than lovastatin (Feng et al., 2011). Moreover, Li et al. (2012) using a structure-based screening approach identified several potential S. pneumoniae HMG-CoA reductase inhibitors with IC50 values in the μM range (Li et al., 2012).
Other enzymes that have been targeted to disrupt the production of polyisoprenoids are the bacterial “head-to-head” terpene synthases. This class of enzymes catalyzes the formation of a cyclopropylcarbinyl diphosphate intermediate (e.g., presqualene diphosphate) via C1′-2, 3 condensation of two isoprenoid diphosphate molecules (e.g., farnesyl diphosphate) (Lin et al., 2010). Subsequently, the presqualene diphosphate undergoes rearrangement that involves ring opening catalyzed by the same “head-to-head” terpene synthase [e.g., S. aureus dehydrosqualene synthase (CrtM)] or by partner enzymes (e.g., Zymomonas mobilis HpnC) rendering dehydrosqualene or hydroxysqualene (Lin et al., 2010; Pan et al., 2015a; Schwalen et al., 2017). Because the “head-to-head” terpene synthases act on IPP synthesis downstream, inhibitors as zaragozic acid family compounds, which are potent inhibitors of “head-to-head” terpene synthases, could hinder the production of polyisoprenoid lipids in many bacterial species and, therefore, disturb the structural assembly and function of the FMMs (Bramkamp and Lopez, 2015).
Supporting the potential use of these sterol synthesis inhibitory drugs as effective anti-FMM drugs, García-Fernández et al. (2017) showed that the penicillin-binding protein PBP2a, which mediates resistance to β-lactam antibiotics in the pathogen methicillin-resistant Staphylococcus aureus (MRSA), was associated with FMMs. The treatment of the MRSA culture with 50 μM of zaragozic acid did not affect the bacterial growth but affected FMMs formation with the consequent disturbance of PBP2a oligomerization, which could imply a non-optimal functionality of PBP2a. In support of this, zaragozic acid-treated MRSA showed a β-lactam-sensitive phenotype in comparison with the non-treated MRSA. Likewise, MRSA treatment with simvastatin, lovastatin, mevastatin and pravastatin enhanced the antimicrobial activity of oxacillin toward the resistant pathogen. Moreover, oxacillin-treated MRSA-infected mice showed a significantly lower survival rate than oxacillin/zaragozic acid-treated MRSA-infected mice. Mice infected with a MRSA strain isolated from a pneumonia patient and treated with oxacillin exhibited a significantly higher bacterial load in lungs than oxacillin/zaragozic acid-treated mice (García-Fernández et al., 2017). This study show the potential of anti-FMM drugs to revert an antibiotic-resistant phenotype into an antibiotic-sensitive phenotype. Besides, it has been demonstrated that treatment of S. aureus with zaragozic acid, miltefosine and 5-doxyl-stearic acid (5-DSA) (miltefosine and 5-DSA alter physicochemical properties of lipid-rafts) disturbed FMMs assembly and consequently inhibited FloA oligomerization and scaffold activity. Impaired functional FloA could yield non-proper oligomerization of the RNase Rny, therefore affecting the S. aureus degradosome machinery, which is important for the regulation of virulence factors-coding genes expression (Koch et al., 2017). Using a murine infection model it was observed that S. aureus- infected mice treated with zaragozic acid, miltefosine, or 5-DSA displayed a significantly higher survival rate than non-treated infected mice. Additionally, in another infection experiment, it was demonstrated that in the treated animals' lungs, small RNAs rsaA and sau63 had significantly higher expression in comparison to untreated controls. This suggested defective degradosome function mediated by the inhibition of FloA oligomerization in vivo (Koch et al., 2017). Other evidence of anti-FMM drugs' potential in anti-virulence therapy was recently revealed. It was observed that S. aureus treatment with zaragozic acid, simvastatin, or 5-DSA disturbed the correct assembly of T7SS and consequently affected the secretion of T7SS-associated virulence factors. It was suggested that this effect of the anti-FMM compounds on T7SS functionality could involve impaired FloA scaffold activity, since FloA interacted with the component EssB of T7SS and assisted in the complex assembly. Additionally, it was verified that in BALB/c mice infected with S. aureus and treated with zaragozic acid the levels of IgM antibodies against T7SS substrates such as Esx A Esx B, Esx C, and Esx D were inferior to those in non-treated controls (Mielich-Süss et al., 2017).
For the establishment of successful host infection by pathogenic bacteria it is necessary to have coordinated actions among the population members of the infecting pathogen. These synchronized actions may be achieved through communication systems between bacteria. Therefore, bacterial communication systems are important players in the establishment of a successful host-infection process and consequently are attractive targets for developing anti-virulence therapeutic strategies (Defoirdt, 2017; Munguia and Nizet, 2017). One of the bacterial communication systems that is most studied and distributed among bacteria is the quorum-sensing (QS) network. Independently of the diversity of bacterial QS systems' architecture and functional components, these communication systems are commonly based on a sequence of events that consist of the production of chemical signaling molecules (autoinducers), which are secreted to the external medium and accumulate until reaching a threshold of concentration that is detected by bacteria. Subsequently, a change in the gene-expression patterns take place in response to the detected signaling molecules (Waters and Bassler, 2005; Papenfort and Bassler, 2016). The strategies for disturbing and manipulating QS networks aim to interfere with these basic events (Table 1).
However, these quorum quenching strategies have to face several challenges to become feasible therapeutic options. The expression of certain virulence factor could be subjected to the control of several regulatory mechanisms other than the targeted QS system (Arya and Princy, 2016). Depending on the environment factors find by the pathogen; some of these regulatory mechanisms could influence the virulence factor expression more predominantly than others (Goerke et al., 2001; Xiong et al., 2006; Zurek et al., 2014; Liu et al., 2016). Another element that could represent a challenge is the diversity of QS systems that could be present in a pathogen; and that these could form complex interconnected networks (Lee and Zhang, 2015; Koul et al., 2016). Furthermore, in some cases, the interference with the QS systems could promote the virulence instead of attenuated it (Köhler et al., 2010; García-Contreras, 2016). In addition, interference with QS systems could affect the pathogen growth, which could exert selective pressure and facilitate the emergence of resistant pathogens (García-Contreras, 2016). Moreover, in a polymicrobial infection, because the interaction between pathogens could be mediate by QS-controlled factors, the interference with QS systems in a pathogen potentially could facilitate the pathogenicity and antibiotic resistance of the co-infecting pathogens (O'brien and Fothergill, 2017; Radlinski et al., 2017). Also, it is important to have diagnostic tools sensitive enough that allow the detection of the infecting pathogen at low cellular densities; so that quorum quenching strategies could be implemented before the pathogen reach the quorum necessary to trigger their pathogenic potential (Kalia et al., 2019).
One of the approaches to disrupting QS systems is based on the interference in signal production. This strategy is centered on inhibiting the activity of the autoinducer-producing enzymes via small inhibitory molecules, which are mainly substrate structural analogs or transition state analogs and do not affect bacterial growth (LaSarre and Federle, 2013). The advantages of this strategy are that enzymes involved in the production of autoinducers are not present in mammalian cells and are encoded in the genome of several bacterial species (LaSarre and Federle, 2013; Pereira et al., 2013). Moreover, it is possible that an effect on the activity of a particular enzyme [e.g., 5′-methylthioadenosine/S adenosylhomocysteine nucleosidase (MTAN nucleosidase)] could in turn affect the production of more than one type of autoinducer (Gutierrez et al., 2009). However, the fact that target enzymes have intracellular localization raises several challenges for the implementation of this strategy. Primarily, the inhibitory compounds should overcome the diffusion barriers imposed by bacterial surface structures, particularly difficult in Gram-negative bacteria, which contain a double membrane system. Furthermore, once inside the cell, the inhibitors could be expelled to extracellular space by efflux pumps. In addition, the inhibitor compounds could inhibit enzymes involved in important metabolic processes, compromising the cellular viability, and therefore creating selective pressure on the bacteria (Hinsberger et al., 2014; Sahner et al., 2015; Ji et al., 2016).
Despite all these possible limitations, several studies using in cellulo and in vivo approaches suggested the feasibility of the use of autoinducer-producing enzyme inhibitors. Gutierrez et al. (2009) showed that transition state analogs of the enzyme MTAN nucleosidase suppressed the production of autoinducers by Vibrio cholerae and enterohemorrhagic Escherichia coli O157:H7. Specifically, the inhibitors 5′-methylthio-DADMe-Immucillin-A, 5′-ethylthio-DADMe-Immucillin-A, and 5′-butylthio-DADMe-Immucillin-A inhibited the MTAN activity in a dose-dependent fashion, and consequently the production of autoinducers, without disturbing bacterial growth in V. cholerae N1696 culture. Similar behavior was observed in Escherichia coli O157:H7 culture, where 5′-methylthio-DADMe-Immucillin-A and 5′-butylthio-DADMe-Immucillin-A inhibited the production of AI-2 in a dose-dependent mode, without affecting bacterial growth. Importantly, both bacterial strains maintained the 5′-butylthio-DADMe-Immucillin-A sensitive phenotype after growth for several generations when challenged with the inhibitor, suggesting that there was no emergence of resistance. In agreement with the reduction of AI-2 production, reduced biofilm-forming capacity was observed in both bacterial species when they were treated with the 5′-butylthio-DADMe-Immucillin-A inhibitor (Gutierrez et al., 2009).
Another bacterial pathogen for which QS signal-producing enzyme inhibitors have been developed is Pseudomonas aeruginosa. This pathogen contains a particular QS system [Pseudomonas quinolone system (pqs)] which uses PQS (3,4-dihydroxy-2-heptylquinoline, Pseudomonas quinolone signal) and HHQ (2-heptyl-4-hydroxyquinoline) as QS signal molecules (LaSarre and Federle, 2013; Papenfort and Bassler, 2016). The production of HHQ and PQS starts with activity of the enzyme anthranilyl-CoA ligase (PqsA) which catalyzes the activation of anthranilate to anthraniloyl-coenzyme A via an anthranilyl-AMP reaction intermediate. Sulfonyladenosine compounds (anthranilyl-MAS and anthranilyl-AMSN) that mimic the anthranilyl-AMP intermediate were PqsA inhibitors that reduced the production of HHQ and PQS by P. aeruginosa PA14 strain (Ji et al., 2016). In addition to intermediate-mimic compounds, other PqsA inhibitors are substrate structural analogs. P. aeruginosa PAO1strain challenged with the anthranilate analog methyl-anthranilate reduced in a dose-dependent fashion PQS production as well as elastase activity. Elastase is a virulence factor that is under the control of the pqs QS system (Calfee et al., 2001). However, in other study, the effect of methyl-anthranilate in reducing P. aeruginosa PA14-produced PQS and HHQ levels was less potent, and the most powerful compounds were halogenated anthranilate analogs. In addition, some of these halogenated analogs showed effectivity in vivo in reducing the virulence and limiting P. aeruginosa systemic dissemination in infected mice (Lesic et al., 2007). In addition to PqsA, another enzyme linked to the PQS biosynthesis pathway that has been targeted is PqsD. This enzyme catalyzes the formation of the 2-aminobenzoylacetyl-CoA by the condensation of anthraniloyl-CoA with malonyl-CoA via a tetrahedral transition state. The treatment of P. aeruginosa PA14 with the potent PqsD inhibitor (2-nitrophenyl) phenyl methanol disturbed the production of HHQ and PQS as well as reducing the biofilm volume (Storz et al., 2012). Moreover, some catechol-derivative compounds that act as PqsD inhibitors by blocking the access of the natural substrate to the active site of PqsD reduced the production of HHQ by P. aeruginosa (Allegretta et al., 2015).
Furthermore, the enzyme PqsBC, which catalyzes the condensation of octanoyl-CoA and 2-aminobenzoylacetate rendering HHQ is also a QS inhibitor target. In this regard, it was observed that the PqsBC competitive inhibitor 2-aminoacetophenone (2-AA) affected HHQ production by the recombinant P. putida KT2440 strain (Drees et al., 2016). However, strategies focused on PqsBC inhibition could carry unwanted effects. The PqsBC substrate 2-aminobenzoylacetate could transform in 2-AA or 2,4-dihydroxyquinoline (DHQ) (Dulcey et al., 2013). The 2-AA promoted the emergence of persister cells in pathogens as P. aeruginosa, A. baumannii and Burkholderia thailandensis, whereas DHQ has been linked to P. aeruginosa pathogenicity (Que et al., 2013; Gruber et al., 2016). In principle, the inhibition PqsBC could provoke the accumulation of 2-aminobenzoylacetate, favoring the formation of 2-AA and DHQ, which could promote the emergence of antibiotic tolerant pathogens and/or increase in the pathogenicity (Allegretta et al., 2017; Maura et al., 2017). In this regard, it has been demonstrated that the treatment of a P. aeruginosa mvfR mutant (constitutively expressed the pqs ABCDE operon) with some benzamide-benzimidazole compounds (PqsBC inhibitors) may provoked the accumulation of 2-AA and DHQ (Maura et al., 2017). In addition, in the P. aeruginosa PA14 parental strain, the treatment with some of these PqsBC inhibitors (specifically those also contain a low anti-MvfR (PqsR) activity) did affect only partially the production of 2-AA and DHQ and did not inhibit the tolerance to meropenem (Maura et al., 2017). In another study performed by Allegretta et al. (2017), the treatment of P. aeruginosa PA14 and a PA 14 pqsH mutant with PqsBC inhibitors, produced an increase at 2-AA and DHQ levels. One of these PqsBC inhibitors increased the subpopulation of persister P. aeruginosa PA14 cells to levels similar to a pqsBC mutant strain (Allegretta et al., 2017). Interestingly, in this study it was observed that treatment with PqsBC inhibitors increased the 4-hydroxy-2-heptylquinoline-N-oxide (HQNO) levels. This molecular specie has been also linked to the emergence of antibiotic tolerance in P. aeruginosa (Hazan et al., 2016; Allegretta et al., 2017). HQNO appears to boost bacterial autolysis with the subsequent DNA release, which facilitates biofilm formation making the pathogen more tolerant to antibiotics (Hazan et al., 2016). Moreover, the HQNO produced by P. aeruginosa also influence the S. aureus susceptibility to antibiotics (Orazi and O'Toole, 2017; Radlinski et al., 2017). In another study, it was shown that P. aeruginosa ΔpqsB and ΔpqsC mutants that only produce DHQ, were more virulent in C. elegans than a quinolone-null mutant ΔpqsAB. An increased colonization capacity of C. elegans was observed for the ΔpqsB mutant in comparison with the ΔpqsAB mutant (Gruber et al., 2016).
Additional support for the feasibility of using QS signal biosynthesis inhibitors in vivo was demonstrated by the use of ambuic acid as an anti-virulence compound in a murine model of intradermal MRSA challenge. The treatment with ambuic acid impaired the virulence exerted by S. aureus in the infected animals, as it attenuated skin ulcer formation and the signs of infection-induced morbidity. The anti-virulence effect of ambuic acid was mediated by inhibition of the agr quorum sensing system (Todd et al., 2017).
Among the quorum quenching strategies one of the most exploited is the inactivation of QS signals (LaSarre and Federle, 2013). This is a strategy that exists in the natural interactions between microbial populations, and it has been extrapolated as an approach to modulating the virulence of bacterial pathogens. This virulence modulation through interference with QS signal is centered mainly on the use of QS signal-degrading enzymes (LaSarre and Federle, 2013; Fetzner, 2015). The advantage of this strategy is that it targets the QS signal after it is secreted to extracellular medium. Therefore, there is greater access to the target and challenges associated with penetrating bacterial cells are avoided. Moreover, as an extracellular factor is targeted, the emergence and spread of resistance could be less probable, but potential resistance mechanisms have been envisioned (Defoirdt et al., 2010; Fetzner, 2015; Vale et al., 2016).
The most thoroughly characterized quorum quenching enzymes are acyl-homoserine lactone (acyl-HSL) lactonases, acyl-HSL acylases, and acyl-HSL oxidoreductases, which target the QS signal acyl-homoserine lactones (acyl-HSLs) (Fetzner, 2015). The acyl-HSL lactonases and acyl-HSL acylases destroy acyl-HSL molecules via homoserine lactone ring hydrolysis (specifically the ester bond) or amide bond hydrolysis between the acyl tail and the homoserine lactone ring, respectively. Otherwise, the acyl-HSL oxidoreductases modify acyl-HSLs molecules chemically via oxidation or reduction of the acyl chain instead of degrading them (LaSarre and Federle, 2013; Fetzner, 2015). Other types of quorum quenching enzymes that have been characterized include E. coli LsrK kinase that targets the AI-2, and dioxygenases Hod from Arthrobacter sp. Rue61a, AqdC1 and AqdC2 from Mycobacterium abscessus subsp abscessus and Rhodococcus erythropolis BG43 that target alkylquinolone-type molecules (Pustelny et al., 2009; Roy et al., 2010; Müller et al., 2015; Birmes et al., 2017). LsrK catalyzes the phosphorylation of AI-2 molecules, rendering phospho-AI-2, whereas dioxygenases mediate a dioxygenolytic cleavage of PQS, rendering N-octanoylanthranilic acid and carbon monoxide (Pustelny et al., 2009; Roy et al., 2010).
The potential for using quorum quenching enzymes in clinical infection treatment is supported by several studies. Recently, Utari et al. (2018) used mouse models of pulmonary P. aeruginosa infection and showed the efficacy of intranasally administered PvdQ acylase in hindering P. aeruginosa virulence. In a lethal infection model, PvdQ-treated animals presented a 5-fold lower bacterial load than non-treated animals, as well as a longer survival time. Moreover, PvdQ-treated mice showed lower lung inflammation, CXCL2 and TNF-α levels than non-treated animals in a sub-lethal infection model. It is noteworthy that intranasally supplied PvdQ acylase was shown to be safe as it was well tolerated by animals (Utari et al., 2018). Previously, using a Caenorhabditis elegans infection model, the potential of PvdQ acylase as an anti-virulence agent had been shown (Papaioannou et al., 2009). Penicillin V acylases PaPVA and AtPVA from the Gram-negative bacteria Pectobacterium atrosepticum and Agrobacterium tumefaciens also exerted quorum quenching activity on P. aeruginosa. The supplementation of these two acylases to P. aeruginosa PAO1 provoked a reduction in 3-oxo-C12-HSL levels, elastase activity, pyocyanin and biofilm production. In addition, the survival rates of G. mellonella infected with P. aeruginosa PAO1 pre-treated with acylases were higher than G. mellonella larvae infected with P. aeruginosa PAO1 without acylases pre-treatment (Sunder et al., 2017).
Another quorum quenching enzyme that has been tested in vivo is the engineered lactonase SsoPox-W263I. Using an acute lethal model of P. aeruginosa pneumonia in rats, Hraiech et al. (2014) demonstrated that intra-tracheally delivered SsoPox-W263I immediately after infection with P. aeruginosa PAO1 significantly reduced the mortality rate and the lung damage. SsoPox-W263I was well tolerated by rats (Hraiech et al., 2014). Recently, SsoPox-W263I showed anti-virulence activity against clinical P. aeruginosa isolates. Interestingly, SsoPox-W263I immobilization did not affect the anti-virulence activity against P. aeruginosa PAO1 (Guendouze et al., 2017). Moreover, AiiM lactonase attenuated P. aeruginosa PAO1 virulence in an acute pneumonia murine model. Mice infected via intratracheal with an AiiM-expressing P. aeruginosa PAO1 strain showed less lung injury, lower pro-inflammatory cytokines levels, and lower mortality than animals infected with an AiiM-nonexpressing P. aeruginosa PAO1 strain. In addition, in AiiM-expressing P. aeruginosa PAO1 infected mice there was a reduced systemic dissemination of the infection in comparison with the AiiM-nonexpressing P. aeruginosa PAO1 infected ones (Migiyama et al., 2013). Based on a C. elegans infection model it was demonstrated that lactonase MomL from Muricauda olearia increased the survival of P. aeruginosa PAO1-infected nematodes without showing toxic effects. However, a protective effect was not observed in A. baumannii-infected nematodes (Tang et al., 2015; Zhang et al., 2017b).
Furthermore, recently discovered quorum-quenching enzymes show potential as anti-virulence agents. Lactonases AaL isolated from Alicyclobacillus acidoterrestris, AiiK from Kurthia huakui LAM0618T and Aii810 from Mao-tofu metagenome, inhibited virulence factors production and biofilm formation by A. baumannii and P. aeruginosa PAO1 without affecting bacterial growth (Fan et al., 2017; Bergonzi et al., 2018; Dong et al., 2018). Another newly described quorum-quenching enzyme is AidA, which was identified in A. baumannii clinical isolates (López et al., 2017).
In addition to quorum-quenching enzymes, QS signal inactivation is also reached by the action of anti-QS signal antibodies and synthetic polymers that sequester it (Piletska et al., 2010; LaSarre and Federle, 2013). The use of antibodies with therapeutic aims offers desirable effects, such as high specificity to the target coupled with low off-target cytotoxicity (Palliyil et al., 2014). However, developing anti-QS signal antibodies is a challenging task, because these signals are small size molecules and generally not structurally complex, making them poor antigens (Palliyil et al., 2014). Despite this, several studies support the potential of antibodies in disturbing quorum-sensing networks. Recently, centered on the agr type I quorum-sensing system of S. aureus, a virus-like particle (VLP)-based agr type I vaccine was developed using a P. aeruginosa RNA bacteriophage PP7 coat protein inserted with a S. aureus sequence-modified autoinducer peptide-1 (AIP1S, cysteine was substituted by serine, YSTSDFIM). In this vaccinal candidate (PP7-AIP1S) the AIP1S peptide was exposed on the surface of the VLP, and immunized mice with PP7-AIP1S developed antibodies that specifically recognized the original S. aureus AIP1 in vitro. In addition, using a murine model of S. aureus SSTI (skin and soft tissue infection), it was observed that after a challenge with a virulent S. aureus USA300 isolate LAC (agr-type I), in PP7-AIP1S immunized mice reduced agr-type I-mediated pathogenesis was developed, compared to the non-immunized animals. Reduced alpha-hemolysin levels, as well as RNAIII transcription at the infection site in the immunized animals, together with the in vitro antibodies binding (from immunized animals) to AIP1 suggested the occurrence of immune suppression of agr-signaling during the infection (Daly et al., 2017). Using a peptide library displayed on VLP, O'Rourke et al. (2014) identified eight peptides (VLP-peptides) that bind specifically to the antigen-binding site of the monoclonal antibody AP4-24H11. This monoclonal antibody specifically bound and neutralized the autoinducer peptide-4 (AIP4) from S. aureus and protected animals from S. aureus pathogenicity, as was shown previously by Park et al. (2007). From the eight AP4-24H11-bonding VLP-peptides, two of them, when administered alone, apparently induced a protective response (reduced abscess and dermonecrosis) against S. aureus agr-type IV isolate AH1872 infection in immunized mice. Additionally, the immunization of mice with a combination of these two VLP-peptides protected those from S. aureus AH1872 infection via inhibition of agr-signaling (O'Rourke et al., 2014). Furthermore, it was observed that sheep-mouse chimeric monoclonal antibodies with affinity in the nanomolar range against HSL molecules protected C. elegans nematodes and mice infected with P. aeruginosa PA058. In infected mice, this protection appears to be associated with the antibody-mediated scavenging of HSL molecules and not by effects on the bacterial load (Palliyil et al., 2014). Moreover, immunized mice with 3-oxo- dodecanoyl homoserine lactone conjugated to BSA (3-oxo-c12-HSL-BSA) developed specific antibodies against the HSL and intermediate protection was observed after intranasal infection with P. aeruginosa PAO1. Interestingly, the lung bacterial burden was not affected in the immunized mice, and the levels of TNF-α (lung) and 3-oxo-c12-HSL (lung and serum) were lower than in non-immunized animals (Miyairi et al., 2006).
In addition to interference with QS signaling, monoclonal antibodies against QS signal molecules also protect from cytotoxic effects exerted by these molecules on host cells. Kaufmann et al. (2008) observed in vitro that the monoclonal antibody RS2-1G9 protected murine bone marrow-derived macrophages in a concentration-dependent fashion from the cytotoxicity associated with 3-oxo-C12-HSL (Kaufmann et al., 2008). Previously, it was demonstrated that serum from immunized animals with 3-oxo-C12-HSL-BSA inhibited the autoinducer-dependent apoptosis of the macrophage cell line P388D1 (Miyairi et al., 2006).
Synthetic polymers constitute another alternative for interference with QS signal. These polymers bind and sequester the QS signal without affecting bacterial growth; therefore, they should not exert selective pressure. A pioneering work by Piletska et al. (2010) demonstrated that signal-sequestering polymers interfered with the Vibrio fischeri QS network-based on 3-oxo-C6-HSL. The sequestering of 3-oxo-C6-HSL by the polymers impaired the bioluminescence production as well as biofilm formation (Piletska et al., 2010). In a subsequent work by this group, it was showed that an itaconic acid (IA)-based-molecular imprinted polymer (MIP) impaired P. aeruginosa biofilm formation by sequestering the 3-oxo-C12-HSL QS signal (Piletska et al., 2011). Moreover, linear polymers (IA-based polymers and methacrylic acid-based polymers) reduced V. fischeri bioluminescence and Aeromonas hydrophila biofilm production through lactones sequestering. The IA-based polymers were more effective than methacrylic acid-based polymers regarding the quorum-quenching activity. Importantly, the polymers did not show cytotoxic effects on mammalian cells and did not affect bacterial growth (Cavaleiro et al., 2015). Recently, it was observed that 2-hydroxyethyl methacrylate (HEMA)-based MIPs suppressed the biofilm formation by P. aeruginosa; however, IA-based MIPs were not effective in the biofilm attenuation (Ma et al., 2018).
Interference with signal detection is another of the most exploited strategies for disrupting QS systems. Some of these QS signal detection inhibitors are signal structural analogs that compete with the signal molecule by binding at the ligand-binding site in the receptor (Stevens et al., 2010). Moreover, other inhibitors could act in a non-competitive fashion (e.g., halogenate furanones, isothiocyanate-based covalent inhibitors, and flavonoids) (Koch et al., 2005; Amara et al., 2016; Paczkowski et al., 2017). The inhibitors binding to the receptors directly affect the signaling cascade by different pathways, including block signal binding, structural destabilization of the receptor, impaired receptor dimerization, impaired DNA binding, or impaired interaction with RNA polymerase (Stevens et al., 2010; Paczkowski et al., 2017; Suneby et al., 2017). Moreover, it has been shown that agonists of the QS signal can also exert inhibitory activity on QS systems. Some QS circuits are arranged in hierarchically cross-regulated networks, e.g., in P. aeruginosa the las-QS system positively regulates rhl- and pqs-QS systems, in addition the activated pqs-QS system also positively regulates the rhl-QS system, whereas this exerts negative regulation on the pqs-QS system (Lee and Zhang, 2015). Therefore, by modulating the activity of one QS system, it is possible to influence the activity of the other QS systems. In this respect, Welsh et al. (2015) observed that some agonists of RhlR attenuated the expression of virulence factors controlled by the pqs-QS system in P. aeruginosa. Disruption of cross-regulation of rhl-pqs systems was proposed as a novel mechanism of QS inhibition (Welsh et al., 2015).
The use of small molecules to disturb QS signal detection has to face several challenges. In this respect, structural stability of the inhibitors is a very important issue; some structural analogs of HSLs and AIPs are prone to hydrolysis, depending on the characteristics of the media (Glansdorp et al., 2004; Vasquez et al., 2017). In addition, inhibitors could be potentially degraded by enzymes as well as targeted by efflux pumps (Maeda et al., 2012; Grandclément et al., 2015). Moreover, the inhibitory effect observed could be strain-dependent, and it is therefore important to include several strains in the studies (García-Contreras et al., 2015). Despite the challenges, the feasibility of QS signal detection inhibition as a quorum-quenching strategy is supported by several in vivo studies.
Meta-bromo-thiolactone (mBTL) is a partial agonist/ partial antagonist of both RhlR and LasR receptors in the HSL-guided QS systems of P. aeruginosa, and RhlR inhibition is its main mechanism of action in vivo. This compound potently inhibited P. aeruginosa PA14 pyocyanin and biofilm production without affecting bacterial growth. In addition, the treatment of P. aeruginosa PA14 with mBTL down-regulated the expression of several LasR- and RhlR-controlled virulence factor genes. The treatment of wild-type and P. aeruginosa PA14 lasR mutant strains with mBTL reduced the pathogenesis exerted by these strains in C. elegans and human lung carcinoma cell line A549 (O'Loughlin et al., 2013). Another inhibitor of P. aeruginosa HSL-based QS systems that has been tested in vivo is the fungal metabolite terrein. The treatment of P. aeruginosa PAO1 with terrein provoked a reduction in a dose-dependent manner in the production of virulence factors elastase, pyocyanin, and rhamnolipid as well as in biofilm formation without affecting bacterial growth. In addition, terrein showed to be more stable than the QS inhibitor furanone C-30 and enhanced the anti-biofilm activity of ciprofloxacin when used in combination. Importantly, terrein mediated protection of C. elegans and mice against P. aeruginosa PAO1infection in a fast killing infection assay and murine airway infection model, respectively. Interestingly, it was observed that the QS system and c-di-GMP signaling pathway could be interconnected, and that terrein could act as a dual inhibitor of these systems (Kim et al., 2018). Moreover, HSL analogs that act as covalent inhibitors of LasR receptor were seen to be promising in vivo tests. Specifically, the isothiocyanate- and fluoroisothiocyanate-based covalent inhibitors (ITC-12 and ITC-F, respectively) attenuated the virulence of P. aeruginosa PAO1-UW and consequently increased the survival of C. elegans worms during an infection assay with this pathogen. The ITC-F treated group showed a significant survival rate in comparison with the control group. Moreover, using an ex-vivo human skin burn wound model it was observed that ITC-F and ITC-12 treatment impaired the establishment of infection by P. aeruginosa PA14 (Amara et al., 2016). In addition to HSL-based QS systems in P. aeruginosa, the PQS-based QS system is also involved in the regulation of virulence factor production. In this regard, maybe inhibitors that could affect these two QS system types would be desirable. Among them, 3-Phenyllactic acid (PLA) is an organic compound produced by Lactobacillus spp that acts as a QS sensing inhibitor that potentially could bind to RhlR and PqsR receptors with high affinity (Chatterjee et al., 2017). Recently, Chatterjee et al. (2017) showed that PLA impaired the attachment of P. aeruginosa PAO1 on a catheter tube, using a Medaka fish intraperitoneal catheter-associated infection models (Chatterjee et al., 2017).
Furthermore, agr-QS system inhibition has been shown to be an achievable strategy for controlling the virulence of pathogens like S. aureus. In this regard, atopic dermatitis is a chronic inflammatory skin disease where S. aureus triggers an immunopathology response through mast cell degranulation. This mast cell degranulation could be induced by the bacterial δ-toxin, which is encoded by the hld gene that is under control of the agr-QS system (Baldry et al., 2018; Geoghegan et al., 2018). Recently, the effectiveness of the agr-QS inhibitor solonamide B in suppressing the S. aureus δ-toxin-induced-inflammatory response was tested using a modified epicutaneous colonization mouse model. Animals infected with S. aureus and treated with solonamide B showed a reduced skin inflammatory cell infiltrate, less skin damage, reduced RNAIII expression and production of pro-inflammatory cytokines in comparison to non-treated animals, suggesting that agr-QS inhibitors could effectively attenuate S. aureus pathogenesis in vivo (Baldry et al., 2018).
In addition to the use of quorum quenching enzymes and quorum sensing inhibitors, some innovative therapeutic strategies to interfere with quorum sensing networks are being developed. One of the challenges in disrupting quorum sensing networks is the fact that a pathogen may possess several QS systems of the same class, for example P. aeruginosa contains the AHL-based systems LasRI and RhlRI (Lee and Zhang, 2015). Therefore, acquiring complete inhibition of the QS systems using quorum-quenching enzymes or quorum-quenching inhibitors in a monotherapy-based scheme could be difficult (Fong et al., 2018). Based on this challenge, Fong et al. (2018) tested the capacity of a combinatory therapy using the quorum-quenching enzyme AiiA and the quorum-sensing inhibitor G1 (LuxR-type receptor inhibitor) to suppress the QS systems in P. aeruginosa. It was observed that combinatory therapy inhibited the expression of lasB-gfp, pqsA-gfp, and rhlA-gfp in P. aeruginosa PAO1 bioreporter strains more potently than single treatments. The rhlA gene is involved in the biosynthesis of rhamnolipid and is under RhlR-transcriptional regulation. In accordance with this, the inhibitory effect on rhamnolipid biosynthesis was verified in a P. aeruginosa PAO1 strain. The level of synthetized rhamnolipid in the combinatory therapy-treated bacteria was nearly to the rhamnolipid level in a ΔlasIΔrhlI mutant strain (Fong et al., 2018). All this evidence suggests that using combinatory therapy with different types of quorum-quenching agents it is possible to disturb diverse quorum-sensing systems existing in pathogens.
In the combinatory therapy described above, a multi-target effect is achieved by joining two therapeutic agents that target different components in the QS networks. However, it is possible to get the same multi-target effect using a single compound (Thomann et al., 2016; Maura et al., 2017). Recently a drug with dual inhibitory activity toward the PqsR and PqsD components of the P. aeruginosa pqs QS system was developed. This dual inhibitor [2-(methylsulfonyl)-4-(1H-tetrazol-1-yl)pyrimidine] was developed from a common molecular scaffold existing in single PqsR- antagonist and PqsD-inhibitor. In vitro analysis showed that the dual inhibitor disturbed the production of the virulence factors pyocyanin and pyoverdine as well as the biofilm production by P. aeruginosa PA14. In addition, the dual inhibitory compound increased the survival rate of G. mellonella larvae infected with P. aeruginosa PA14 (Thomann et al., 2016). Moreover, Maura et al. (2017) observed that some benzamide-benzimidazole-based compounds also act as dual inhibitors of the pqs-QS system in P. aeruginosa. These dual inhibitory compounds targeted the proteins PqsR (MvfR) and PqsBC and could be grouped depending on their inhibition patterns in: PqsR-PqsBC dual inhibitors with high anti-PqsR and high anti-PqsBC activity or PqsR-PqsBC dual inhibitors with low anti-PqsR activity and high anti-PqsBC activity. The treatment with some of these dual inhibitors increased the survival rate of human lung epithelial cells and RAW264.7 macrophages when infected with P. aeruginosa PA14 (Maura et al., 2017). Among the dual inhibitors, those exert a high anti-PqsR activity constitute an attractive therapeutic option because interfere with the production of 2-AA and consequently limit the emergence of antibiotic-tolerant bacteria as was previously discussed.
Recently it has been demonstrated that it is possible to manipulate the AI-2 levels through “controller cells” (Quan et al., 2017). These “controller cells” are based on a subset of “consumer cells” and another of “supplier cells.” The “consumer cells” were engineered to overexpress genes involved in the uptake and processing of AI-2 in E. coli (e.g., lsrACDBK and lsrACDBFGK) while the “supplier cells” in genes involved in the biosynthesis of AI-2 (luxS and mtn). Because these “controller cells” influence the environmental AI-2 levels they will have a direct impact on biofilm formation. In line with this, it was observed that “consumer cells” decreased biofilm formation by E. coli reporter strain, whereas “supplier cells” enhanced biofilm formation (Quan et al., 2017). This suggests a route toward future therapeutic strategies based on engineered cells that act as “controller cells.” In addition, it is possible to modulate microbial behavior through AI-2 levels via a synthetic mammalian cell-based microbial-control device, as was demonstrated by Sedlmayer et al. (2018). This microbial-control device consisted of engineered mammalian cells with a formyl peptide sensor module coupled to AI-2 production and release module. Essentially, the engineered mammalian cells detect formyl peptides released by pathogens (peptides produced by a broad range of bacterial species) and trigger the production and release of AI-2. It was showed that biofilm formation by Candida albicans was reduced when this pathogen was co-cultured with microbial-control-engineered cells. This system appears to be a promising anti-virulence strategy, as ubiquitous pathogen signals are detected with high sensitivity (nM range), and robust production of the autoinducer takes place (without being toxic for the host) influencing bacterial communication without exerting selective pressure. In addition, the fact that autoinducer production is coupled to signal detection allows a synchronized response in accordance with the infection dynamic (Sedlmayer et al., 2018). Moreover, it possible to engineer bacteria that will sense the presence of pathogenic bacteria via the quorum sensing system and, once detected, will release anti-pathogens agents. Hwang et al. (2017) engineered a probiotic E. coli Nissle 1917 strain for sense 3-oxo-C12 HSL from P. aeruginosa and respond by autolysing itself via lysin E7 with the consequent release of the bacteriocin pyocin S5 and the anti-biofilm enzyme DspB, which exerted an anti-P. aeruginosa activity. The feasibility of this approach was demonstrated in vivo using C. elegans and murine infection models, where the engineered strain showed prophylactic and therapeutic effects (Hwang et al., 2017).
Although the strategy of interference with autoinducer biosynthesis has been based on the discovery and design of small molecules that inhibit the enzymatic activity, it has been envisioned that engineered bacterial strains could be an alternative. Recently, it was showed that it is possible to disrupt biofilm production in clinical isolates through the manipulation of the expression levels of the enzyme LuxS. Specifically, using the Clustered Regularly Interspaced Short Palindromic Repeats-Cas 9 interference (CRISPRi) system, the expression of LuxS enzyme was suppressed in clinical E. coli isolates. It was suggested that CRISPRi edited cells could be an alternative strategy for controlling biofilm production in nosocomial settings through CRISPRi system delivery via nucleic acid conjugation (Zuberi et al., 2017). However, delivery of CRISPRi system in nosocomial setting via nucleic acid conjugation could be a very challenging task. Given the fact that nucleic acid conjugation could occur between different bacterial species (Musovic et al., 2006; Goren et al., 2010; Crémet et al., 2012; Van Meervenne et al., 2012); could be possible the transfer of the CRISPRi system from the edited cells to co-existing bacteria other than target bacteria. In this regard, maybe the utilization of narrow host range plasmids as vectors for CRISPRi delivery could limit such potential off-target effect. In addition, the nucleic acid conjugation effectivity in established biofilms could be compromised (Merkey et al., 2011; Stalder and Top, 2016). This could limit the use of CRISPRi edited cells for the treatment of formed biofilms. In this sense, maybe the utilization of engineered phages as vehicles for CRISPRi system delivery could be an attractive alternative. Phages have shown be a CRISPR delivery system with specificity to pathogenic bacteria as well as with capacity to removing established biofilms (Lu and Collins, 2007; Bikard et al., 2014; Citorik et al., 2014; Alves et al., 2016; Fong et al., 2017).
Bacteria can live in a community called biofilm, a structure that can be formed by extracellular polymeric substances, such as DNA, protein, and polysaccharides (Flemming et al., 2016). After forming biofilms, bacteria can disperse and colonize other environments (Fleming and Rumbaugh, 2017). This lifestyle can protect bacteria against potential environmental stress, such as antibiotics and host defense components (Hall and Mah, 2017; Tseng et al., 2018). In the clinical situation, biofilm formed by pathogenic bacteria can establish themselves on human surfaces or medical devices, including implants, catheter, endotracheal tubes and others (Rieger et al., 2016; Konstantinović et al., 2017; Kenaley et al., 2018; Silva et al., 2018). Biofilm formation on these surfaces can serve as a source of infection. The successful establishment of pathogenic biofilm on human surfaces can cause chronic infections and limit the success of antibiotic therapy (Rybtke et al., 2015; Li et al., 2017). In general, combating biofilms may require high antibiotic doses and a combination of strategies (Ribeiro et al., 2016). Unfortunately, many of the marketed antibiotics fail to affect biofilm, especially if they are formed by resistant bacteria. To overcome this problem, researchers have prospected compounds from the natural world (from animals, plants, fungi, viruses, and even bacteria) or synthetics (synthesized through the chemical process and/or screened from chemical libraries) (Rajput et al., 2017). In both situations, anti-biofilm agents can be represented by a variety of organic and inorganic chemical compounds (Rajput et al., 2017).
Anti-virulence compounds against biofilms could be used to limit bacterial adhesion on surfaces (Liu et al., 2018; Ranfaing et al., 2018) to affect the production of an extracellular matrix (Feng et al., 2018) and to disturb the existing biofilm (Puga et al., 2018; Table 1). Some examples of anti-virulence compounds cited here work against non-pathogenic bacteria to humans and animals. However, the approaches using these bacteria may serve as proof of principle to study anti-virulence compounds against biofilms in a general way.
In the prevention scenario, one possibility consists of interfering with structures associated with the successful establishment of biofilms such as flagella (that favor the bacterial motility and interaction with surfaces) and fimbriae (with structures that facilitate bacterial adhesion). Higrocin C (a compound isolated from marine-derived Streptomyces sp. SCSGAA 0027) for example, suppressed swimming motility of Bacillus amyloliquefaciens SCSGAB0082, which could explain the biofilm inhibition (Wang et al., 2018a). Transcriptome studies showed downregulation (more than twofold) of genes associated with bacterial chemotaxis and flagellar motor (Wang et al., 2018a). Coumarin, for example, presents the ability to prevent bacteria biofilm without affecting bacterial growth (Lee et al., 2014). This compound repressed curli genes and motility genes in E. coli O157:H7 and reduced fimbriae production, swarming motility, and biofilm formation (Lee et al., 2014).
Another way to prevent biofilm formation consists of affecting the extracellular matrix production. A chemical compound named TCC (3, 3′, 4′, 5-tetrachlorosalicylanilide) for example, inhibited B. subtilis biofilm formation by reducing extracellular matrix production. This was associated with the repression of SinR protein negative regulated genes (involved in extracellular matrix production) (Feng et al., 2018). Other studies have shown that compounds that prevent biofilm formation can potentially affect bacterial cell communication by degrading quorum-sensing molecules (Ivanova et al., 2015; Passos da Silva et al., 2017).
In the context of combating existing biofilms, compounds can be used to destroy components of extracellular matrix, such DNA, proteins and carbohydrates (Puga et al., 2018). In this context, enzymes have been used as potential agents to disrupt mono and polymicrobial biofilm (Puga et al., 2018). DNAse I, for example, presents the ability to degrade extracellular DNA of Campylobacter jejuni, promoting biofilm removal without affecting bacterial viability (Brown et al., 2015).
The understanding of mechanisms involved in the formation of bacterial biofilm, as well as the understanding of their cells and biofilm structures, could indicate possible targets to develop compounds that affect biofilm without killing bacteria. In addition to potential anti-biofilm therapy, agents that can prevent or disperse biofilm could potentially combine with anti-virulence compounds. For example, anti-virulence agents could be used to neutralize endotoxins from bacterial cells that disperse from biofilms and thus prevent or minimize the harmful effects of the host inflammatory response against bacterial infection.
It is known that pathogenic bacteria may produce diverse virulence determinants in order to successfully survive host system responses, as well as colonizing a host (Kong et al., 2016). Among them, toxins comprise proteins expressed by bacteria during post-exponential and early stationary phases that have been divided into different classes, including hemolysin (Powers et al., 2015), leukotoxin (Zivkovic et al., 2011), exfoliative toxins (Bukowski et al., 2010), endotoxin (Heinbockel et al., 2018), among others. These protein-based toxins are intrinsically related to physical damage, biochemical degradation and signaling interruption in the host cells, resulting in immune system evasion and characterizing pathogen-to-host interactions (Wei et al., 2017). Bacterial toxin neutralization, for instance, has been shown to compromise bacterial proliferation and survival in the host (Ortines et al., 2018). More importantly, unlike antibiotic-based treatments, anti-toxin or anti-virulence therapies do not affect bacterial viability directly and, as a consequence, could impose reduced selective pressure, probably decreasing the frequency of resistance events (Rasko and Sperandio, 2010). In addition, anti-virulence compounds are also known to preserve the host's endogenous microbiome as they target virulent factors secreted exclusively by pathogenic bacteria (Clatworthy et al., 2007). In this context, here we described compounds, including antibodies, nanoparticles, small molecules, and bioactive peptides (Figure 1 and Table 1), which have been studied recently as promising candidates for anti-virulence therapies that aim to treat and prevent bacterial infections.
The α-toxin (AT), also known as α-hemolysin, is a key virulence factor expressed by S. aureus that has been investigated in different animal infection models, including bacteremia, pneumonia and skin/soft tissue infections (Surewaard et al., 2018). This toxin is capable of lysing red blood cells, and also targets monocytes, macrophages and neutrophils (Bubeck Wardenburg et al., 2007). Moreover, in the clinic, AT levels in patients are often correlated with disease severity (Jenkins et al., 2015). Studies have shown that rabbits with acute bacterial skin and skin structure infections (ABSSSI) caused by AT-expressing methicillin-resistant S. aureus (MRSA) develop severe infections similar to those observed in humans, including the presence of large dermonecrotic lesions. In contrast, rabbits infected with a mutant deficient AT strain developed only small dermonecrotic lesions (Le et al., 2016). One major anti-virulence strategy to neutralize AT consists of using antibodies. The study cited above also reported a significant decrease in the disease severity thought AT neutralization by treating the rabbits with an anti-AT human monoclonal antibody (mAb) (MEDI4893*) (Le et al., 2016). Similarly, Ortines et al. (2018) observed in non-diabetic and diabetic mice that S. aureus-infected animals passively immunized with anti-AT mAb (MEDI4893*) showed decreased wound size and bacterial counts when compared to the untreated controls. Moreover, those authors also showed the differential host immune response effects, revealing different patterns of macrophage, monocyte and neutrophil infiltrates, as well as neutrophil extracellular traps (NETs) in non-diabetic and diabetic mice (Ortines et al., 2018).
In addition to skin infections, S. aureus strains are often associated with respiratory mono-infections and co-infections with Gram-negative strains, including P. aeruginosa and Klebsiella pneumoniae. In a study by Cohen et al. (2016), it was shown that S. aureus AT, in a mixed pathogen-lung infection model, could potentiate Gram-negative bacterial dissemination and lethality. This situation, however, could be circumvented by the passive immunization of mice with an anti-AT mAb, leading to S. aureus and co-pathogens (Gram-negative bacteria) clearance in the lungs (Cohen et al., 2016). Additionally to mAb, the intravenous immunoglobulin (IVIG), which consists of a polyclonal human antibody pool, has been investigated regarding its protective effects against necrotizing pneumonia caused by different epidemic community-associated and hospital-associated MRSA strains (Diep et al., 2016). As reported by Diep et al. (2016), two IVIG antibodies specific to an AT (α-hemolysin, HTa) and a Panton-Valentine leukocidin (PVL) conferred protection on immunized rabbits against MRSA, leading to improved survival outcomes (Diep et al., 2016).
In the clinic, patients affected by bacteremia, including S. aureus, may present occlusion of small blood vessels by the formation of large platelet aggregates (van der Poll and Opal, 2008). In a recent study, it was reported that AT induces rapid platelet aggregation and liver injury, causing multi-organ dysfunction during S. aureus sepsis (Surewaard et al., 2018). Interestingly, however, all these damaging effects could be prevented in mice treated with the anti-AT mAb (MEDI4893*) (Surewaard et al., 2018), thus reinforcing the importance of monoclonal antibodies as bacterial toxin neutralizing agents in anti-virulence therapies. More recently, Wang et al. (2018b), reported a novel vaccine platform based on extracellular vesicles (EVs) from S. aureus. In that work, the authors purified EVs from a genetically engineered S. aureus capable of overexpressing detoxified cytolysins (HlaH35L and LuKE), which were non-toxic, immunogenic and protected mice from lethal sepsis caused by S. aureus (Wang et al., 2018b). Also in the field of S. aureus toxin neutralization, the monoclonal antibody, ASN100 (Arsanis Inc.), which consists of the combination of two human IgG1k monoclonal antibodies, ASN1 and ASN2, has shown promising results in the neutralization of six S. aureus toxins (Rouha et al., 2015; Badarau et al., 2016). Despite the advances in the usage ASN100 in the clinic, the company Arsanis Inc. has discontinued a phase II clinical trial for ASN100 as it failed to prove its effectiveness in high-risk, mechanically ventilated patients with S. aureus pneumonia.
Antibodies have also been applied as anti-virulent therapies involving Clostridium difficile, which represents a primary cause of nosocomial antibiotic-related diarrhea. This bacterium produces two main virulence factors, toxin A (TcdA) and toxin B (TcdB), responsible for gastrointestinal epithelial damage and colonic inflammation. In this matter, the engineering and use of TcdA/B-neutralizing antibodies appears as a promising approach to counter diarrhea episodes caused by C. difficile infections. With that in mind, Andersen et al. (2016) developed an antitoxin strategy to express TcdB-neutralizing antibody fragments in Lactobacillus strains in the gastrointestinal tract of hamsters infected with a TcdA−/ TcdB+ C. difficile strain. Initially, in vitro studies were carried out to confirm the ability of the expressed fragments in neutralizing the cytotoxic effect of TcdB. Moreover, in vivo assays revealed that Lactobacillus strains expressing two TcdB-neutralizing antibodies led to improved survival rates in the treated group. Furthermore, the protection with TcdB-neutralizing antibodies also preserved the gastrointestinal tract of the animals as no damages or limited inflammation were observed (Andersen et al., 2016). In addition to antibody-based therapies, studies have also explored the potential of small molecules as inhibitors of C. difficile TcdB. Tam et al. (2015) reported a high-throughput phenotypic method for screening small molecules capable of protecting human cells from TcdB. As a result, the authors reported a series of small molecules with diverse mechanisms of action on TcdB, including direct binding, sequestration of TcdB, non-competitive inhibition of the glucosyl-transferase activity of TcdB, as well as endosomal maturation inhibition (Tam et al., 2015). However, in vivo studies are still underway to confirm the effectiveness of these small molecules in C. difficile infections.
Apart from the application of anti-toxin antibodies and small molecules in anti-virulence therapies, studies have also highlighted the importance of engineered nanoparticle mimicking cell membranes (e.g., liposomes) in sequestering cytotoxic bacterial toxins both in vitro and in vivo (Fang et al., 2015). Artificial liposomes are constituted exclusively of natural lipids and therefore are not active against bacteria, thus allowing their usage in combination with antibiotics for bacterial infection treatment. Henry et al. (2015), for instance, showed the potential of artificial liposomes in sequestering bacterial toxins in vitro, along with the preservation of the integrity of mammalian cells. The authors also observed that, during in vivo experiments, the administration of artificial liposomes resulted in mice recovering from septicemia caused by S. aureus and Streptococcus pneumoniae, as well as mice being protected against pneumonia (Henry et al., 2015). Moreover, combining the artificial liposomes with conventional antibiotics, including vancomycin and penicillin, improved survival rates were observed when compared to mono-therapies (Henry et al., 2015).
Bacteria secrete a wide variety of toxins during host colonization and infection, which represents a bottleneck when it comes to vaccine development aiming at anti-virulence therapies. Indeed, vaccine strategies based on multiple targets (bacterial toxins) have already been reported; however, the identification and further confirmation of virulence factors secreted by bacteria is considered a costly and time-consuming method (Fujita and Taguchi, 2011). As an alternative, studies have proposed the use of multiantigenic nanotoxoids based on naturally occurring bacterial proteins to develop vaccines against pathogenic bacteria. Wei et al. (2017) reported a feasible approach for entrapping diverse toxins from bacterial protein preparations using a membrane-coated nanosponge construct capable of delivering these virulence factors in the organism and, consequently, combating bacterial infections. As for the other anti-virulence therapies here described, the nanoparticle-based neutralization and delivery not only usefully prevent severe bacterial infections but also decrease the risk of antibiotic resistance events (Wei et al., 2017).
Besides the secretion of protein-based toxins, the bacterial LPS in the host's blood stream is known to cause severe immune system stimulation, resulting in septic shock and sepsis (Rietschel et al., 1996). Among the strategies to neutralize LPS, the application of antimicrobial peptides (AMPs) has shown promising results. Moreover, the mechanisms of action and structural arrangements of some AMPs in contact with LPS have already been investigated, including polymyxins (Pristovsek and Kidric, 1999), temporins (Bhunia et al., 2011), and melittins (Bhunia et al., 2007). This class of antimicrobials is well known for its multifunctionality and structural diversity. Studies have shown that AMPs with extended activities, including immunomodulatory, are capable of binding to LPS and, consequently, decreasing the production of nitric oxide and tumor necrosis factor-α (TNF-α), which are commonly related to tissue damage (Pulido et al., 2012). Chih et al. (2015), for instance, have reported the antiendotoxin effects of two antimicrobial peptides, S1 and KWWK. Interestingly, the authors observed that LPS-neutralizing activities were directly related to the addition of β-naphthylalanine end-tags in both peptides, which was also reflected in the dose-dependent inhibition of nitrite oxide production and TNF-α release in vitro and in vivo (Chih et al., 2015). In addition, other AMPs, including members from the Pep19-2.5 family (Heinbockel et al., 2018) and retrocyclins (Kudryashova et al., 2015), have revealed the ability to unfold bacterial toxins, as well as causing conformational changes such as toxin aggregation and fluidity (Heinbockel et al., 2018).
The antimicrobial resistance threat has driven the global scientific community to search for effective solutions. Given the fact that antimicrobial resistance is a multifactorial phenomenon, the solution for this problem involves a range of approaches focused on controlling the factors that facilitate the emergence and spread of resistance. One of these approaches consists of developing new therapeutic agents that operate under different principles to the currently available antibiotics. In this respect, anti-virulence therapy has been envisioned as a promising alternative with the aim of controlling pathogen virulence in a pathogen-specific fashion, without exerting strong selective pressure on the pathogens.
However, as an emerging therapeutic strategy, anti-virulence therapy has to face several challenges. The selection of the targeted virulence factor(s) is of critical importance for the effectiveness of the strategy in terms of evolutionary robustness. In line with this, a suitable target should be a virulence factor whose disruption does not imply (or imply minimal) fitness consequences for the pathogen (Vale et al., 2016). Moreover, a virulence factor that is conserved between different pathogens could be ideal, because in principle it would be possible to treat polymicrobial infections with a single anti-virulence drug (Maura et al., 2016). It is necessary to understand the detailed dynamics of action of the targeted virulence factor as well as the dynamics of production (Dickey et al., 2017). For example, during P. aeruginosa infection of cystic fibrosis patients take place an acute to chronic infection transition. This shift involves down-regulation of virulence factors as the flagellum, T3SS secretion system, proteases, and others; while virulence factors as exopolysaccharides are up-regulated (Hogardt and Heesemann, 2013; Sousa and Pereira, 2014). Therefore, the anti-virulence agent that target some of these virulence factors should be supplemented in accordance with this dynamic of expression. In addition to knowing the targeted virulence factor production dynamics, it is important to know if this virulence factor undergoes chemical modifications that modulate its activity. Furthermore, as anti-virulence therapy works in a pathogen-specific fashion, it is important to have diagnostic methods like matrix-assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF), microarray-based nucleic acid test, magnetic resonance-based diagnostic, fluorescence in situ hybridization (FISH) test, next generation sequencing (NGS) and multiplex PCR-based diagnostic test, which permit rapid and precise identification of the infection-causing pathogen (Dickey et al., 2017; Messacar et al., 2017). It is also mandatory to define which parameters will be taken into account for measuring the effectivity of the anti-virulence therapy and for which type of infection the therapy is most suitable (Maura et al., 2016; Dickey et al., 2017). For example, in certain types of S. aureus infections (e.g., chronic and bacteremia), a dysfunctional agr-QS system appears to be beneficial for the pathogen (Khan et al., 2015). Moreover, recently it has been reported that defective agr-QS system could mediate the tolerance to certain antibiotics (gentamicin and ciprofloxacin; Kumar et al., 2017). In addition, it has been suggested that phenol-soluble modulin toxins (PSMs) are involved in the control of S. aureus persister cells population (Bojer et al., 2018). Because the PSMs production is under the control of the agr-QS system, it is probably that a defective agr-QS system down-regulate the expression of PSMs which could favor the emergence of persister cells to certain antibiotics. Therefore, in the above-pointed situations maybe anti-virulence therapies based on agr-QS system inhibition could be not a feasible strategy. Although anti-virulence therapy is an emerging field, several potential anti-virulence drugs have already been identified, and existing chemical libraries for antibiotic discovery could be a valuable source for rapid identification of novel anti-virulence drugs (Maura et al., 2016; Dickey et al., 2017). At this point, it is necessary to direct these potential anti-virulence candidates toward pre-clinical and clinical trials.
OF, MC, and SR wrote the manuscript. MC performed the figures. OLF designed and revised the manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
This work was supported by CAPES, CNPq, FAPDF, FUNDECT, UCB, and UCDB.
Abbas, H. A., Elsherbini, A. M., and Shaldam, M. A. (2017). Repurposing metformin as a quorum sensing inhibitor in Pseudomonas aeruginosa. Afr. Health Sci. 17, 808–819. doi: 10.4314/ahs.v17i3.24
Abbas, H. A., and Shaldam, M. A. (2016). Glyceryl trinitrate is a novel inhibitor of quorum sensing in Pseudomonas aeruginosa. Afr. Health Sci. 16, 1109–1117. doi: 10.4314/ahs.v16i4.29
Alberts, A., Chen, J., Kuron, G., Hunt, V., Huff, J., Hoffman, C., et al. (1980). Mevinolin: a highly potent competitive inhibitor of hydroxymethylglutaryl-coenzyme A reductase and a cholesterol-lowering agent. Proc. Natl. Acad. Sci. U.S.A. 77, 3957–3961. doi: 10.1073/pnas.77.7.3957
Allegretta, G., Maurer, C. K., Eberhard, J., Maura, D., Hartmann, R. W., Rahme, L., et al. (2017). In-depth profiling of MvfR-regulated small molecules in Pseudomonas aeruginosa after quorum sensing inhibitor treatment. Front. Microbiol. 8:924. doi: 10.3389/fmicb.2017.00924
Allegretta, G., Weidel, E., Empting, M., and Hartmann, R. W. (2015). Catechol-based substrates of chalcone synthase as a scaffold for novel inhibitors of PqsD. Eur. J. Med. Chem. 90, 351–359. doi: 10.1016/j.ejmech.2014.11.055
Alves, D. R., Perez-Esteban, P., Kot, W., Bean, J., Arnot, T., Hansen, L. H., et al. (2016). A novel bacteriophage cocktail reduces and disperses Pseudomonas aeruginosa biofilms under static and flow conditions. Microb. Biotechnol. 9, 61–74. doi: 10.1111/1751-7915.12316
Amara, N., Gregor, R., Rayo, J., Dandela, R., Daniel, E., Liubin, N., et al. (2016). Fine-tuning covalent inhibition of bacterial quorum sensing. Chembiochem 17, 825–835. doi: 10.1002/cbic.201500676
Andersen, K. K., Strokappe, N. M., Hultberg, A., Truusalu, K., Smidt, I., Mikelsaar, R. H., et al. (2016). Neutralization of clostridium difficile toxin B mediated by engineered lactobacilli that produce single-domain antibodies. Infect. Immun. 84, 395–406. doi: 10.1128/IAI.00870-15
Arya, R., and Princy, S. A. (2016). Exploration of modulated genetic circuits governing virulence determinants in Staphylococcus aureus. Indian J. Microbiol. 56, 19–27. doi: 10.1007/s12088-015-0555-3
Bach, J. N., and Bramkamp, M. (2013). Flotillins functionally organize the bacterial membrane. Mol. Microbiol. 88, 1205–1217. doi: 10.1111/mmi.12252
Badarau, A., Rouha, H., Malafa, S., Battles, M. B., Walker, L., Nielson, N., et al. (2016). Context matters: The importance of dimerization-induced conformation of the LukGH leukocidin of Staphylococcus aureus for the generation of neutralizing antibodies. MAbs 8, 1347–1360. doi: 10.1080/19420862.2016.1215791
Baldry, M., Kitir, B., Frøkiær, H., Christensen, S. B., Taverne, N., Meijerink, M., et al. (2016). The agr inhibitors solonamide B and analogues alter immune responses to Staphylococccus aureus but do not exhibit adverse effects on immune cell functions. PLoS ONE 11:e0145618. doi: 10.1371/journal.pone.0145618
Baldry, M., Nakamura, Y., Nakagawa, S., Frees, D., Matsue, H., Núñez, G., et al. (2018). Application of an agr-specific anti-virulence compound as therapy for Staphylococcus aureus-induced inflammatory skin disease. J. Infect. Dis. 218, 1009–1013. doi: 10.1093/infdis/jiy259
Bergonzi, C., Schwab, M., Naik, T., Daudé, D., Chabrière, E., and Elias, M. (2018). Structural and biochemical characterization of AaL, a quorum quenching lactonase with unusual kinetic properties. Sci. Rep. 8:11262. doi: 10.1038/s41598-018-28988-5
Bhunia, A., Domadia, P. N., and Bhattacharjya, S. (2007). Structural and thermodynamic analyses of the interaction between melittin and lipopolysaccharide. Biochim. Biophys. Acta 1768, 3282–3291. doi: 10.1016/j.bbamem.2007.07.017
Bhunia, A., Saravanan, R., Mohanram, H., Mangoni, M. L., and Bhattacharjya, S. (2011). NMR structures and interactions of temporin-1Tl and temporin-1Tb with lipopolysaccharide micelles: mechanistic insights into outer membrane permeabilization and synergistic activity. J. Biol. Chem. 286, 24394–24406. doi: 10.1074/jbc.M110.189662
Bikard, D., Euler, C. W., Jiang, W., Nussenzweig, P. M., Goldberg, G. W., Duportet, X., et al. (2014). Exploiting CRISPR-Cas nucleases to produce sequence-specific antimicrobials. Nat. Biotechnol. 32:1146. doi: 10.1038/nbt.3043
Birmes, F. S., Wolf, T., Kohl, T. A., Rüger, K., Bange, F., Kalinowski, J., et al. (2017). Mycobacterium abscessus subsp. abscessus is capable of degrading Pseudomonas aeruginosa quinolone signals. Front. Microbiol. 8:339. doi: 10.3389/fmicb.2017.00339
Bojer, M. S., Lindemose, S., Vestergaard, M., and Ingmer, H. (2018). Quorum sensing-regulated phenol-soluble modulins limit persister cell populations in Staphylococcus aureus. Front. Microbiol. 9:255. doi: 10.3389/fmicb.2018.00255
Bramkamp, M., and Lopez, D. (2015). Exploring the existence of lipid rafts in bacteria. Microbiol. Mol. Biol. Rev. 79, 81–100. doi: 10.1128/MMBR.00036-14
Brown, H. L., Hanman, K., Reuter, M., Betts, R. P., and Van Vliet, A. H. (2015). Campylobacter jejuni biofilms contain extracellular DNA and are sensitive to DNase I treatment. Front. Microbiol. 6:699. doi: 10.3389/fmicb.2015.00699
Bubeck Wardenburg, J., Bae, T., Otto, M., Deleo, F. R., and Schneewind, O. (2007). Poring over pores: alpha-hemolysin and Panton-Valentine leukocidin in Staphylococcus aureus pneumonia. Nat. Med. 13, 1405–1406. doi: 10.1038/nm1207-1405
Bukowski, M., Wladyka, B., and Dubin, G. (2010). Exfoliative toxins of Staphylococcus aureus. Toxins 2, 1148–1165. doi: 10.3390/toxins2051148
Calfee, M. W., Coleman, J. P., and Pesci, E. C. (2001). Interference with Pseudomonas quinolone signal synthesis inhibits virulence factor expression by Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. U.S.A. 98, 11633–11637. doi: 10.1073/pnas.201328498
Capilato, J. N., Philippi, S. V., Reardon, T., McConnell, A., Oliver, D. C., Warren, A., et al. (2017). Development of a novel series of non-natural triaryl agonists and antagonists of the Pseudomonas aeruginosa LasR quorum sensing receptor. Bioorg. Med. Chem. 25, 153–165. doi: 10.1016/j.bmc.2016.10.021
Cattò, C., Dell'Orto, S., Villa, F., Villa, S., Gelain, A., Vitali, A., et al. (2015). Unravelling the structural and molecular basis responsible for the anti-biofilm activity of zosteric acid. PLoS ONE 10:e0131519. doi: 10.1371/journal.pone.013151
Cavaleiro, E., Duarte, A. S., Esteves, A. C., Correia, A., Whitcombe, M. J., Piletska, E. V., et al. (2015). Novel linear polymers able to inhibit bacterial quorum sensing. Macromol. Biosci. 15, 647–656. doi: 10.1002/mabi.201400447
Chang, E. H., Huang, J., Lin, Z., and Brown, A. C. (2019). Catechin-mediated restructuring of a bacterial toxin inhibits activity. Biochim. Biophys. Acta General Subjects 1863, 191–198. doi: 10.1016/j.bbagen.2018.10.011
Chatterjee, M., D'Morris, S., Paul, V., Warrier, S., Vasudevan, A. K., Vanuopadath, M., et al. (2017). Mechanistic understanding of Phenyllactic acid mediated inhibition of quorum sensing and biofilm development in Pseudomonas aeruginosa. Appl. Microbiol. Biotechnol. 101, 8223–8236. doi: 10.1007/s00253-017-8546-4
Chih, Y. H., Lin, Y. S., Yip, B. S., Wei, H. J., Chu, H. L., Yu, H. Y., et al. (2015). Ultrashort antimicrobial peptides with antiendotoxin properties. Antimicrob. Agents Chemother. 59, 5052–5056. doi: 10.1128/AAC.00519-15
Citorik, R. J., Mimee, M., and Lu, T. K. (2014). Sequence-specific antimicrobials using efficiently delivered RNA-guided nucleases. Nat. Biotechnol. 32, 1141–1145. doi: 10.1038/nbt.3011
Clatworthy, A. E., Pierson, E., and Hung, D. T. (2007). Targeting virulence: a new paradigm for antimicrobial therapy. Nat. Chem. Biol. 3, 541–548. doi: 10.1038/nchembio.2007.24
Cohen, T. S., Hilliard, J. J., Jones-Nelson, O., Keller, A. E., O'Day, T., Tkaczyk, C., et al. (2016). Staphylococcus aureus alpha toxin potentiates opportunistic bacterial lung infections. Sci. Transl. Med. 8:329ra331. doi: 10.1126/scitranslmed.aad9922
Crémet, L., Bourigault, C., Lepelletier, D., Guillouzouic, A., Juvin, M.-E., Reynaud, A., et al. (2012). Nosocomial outbreak of carbapenem-resistant Enterobacter cloacae highlighting the interspecies transferability of the bla OXA-48 gene in the gut flora. J. Antimicrob. Chemother. 67, 1041–1043. doi: 10.1093/jac/dkr547
Da, F., Yao, L., Su, Z., Hou, Z., Li, Z., Xue, X., et al. (2017). Antisense locked nucleic acids targeting agrA inhibit quorum sensing and pathogenesis of community-associated methicillin-resistant Staphylococcus aureus. J. Appl. Microbiol. 122, 257–267. doi: 10.1111/jam.13321
Daly, S. M., Elmore, B. O., Kavanaugh, J. S., Triplett, K. D., Figueroa, M., Raja, H. A., et al. (2015). ω-Hydroxyemodin limits Staphylococcus aureus quorum sensing-mediated pathogenesis and inflammation. Antimicrob. Agents Chemother. 59, 2223–2235. doi: 10.1128/AAC.04564-14
Daly, S. M., Joyner, J. A., Triplett, K. D., Elmore, B. O., Pokhrel, S., Frietze, K. M., et al. (2017). VLP-based vaccine induces immune control of Staphylococcus aureus virulence regulation. Sci. Rep. 7:637. doi: 10.1038/s41598-017-00753-0
Defoirdt, T. (2017). Quorum-sensing systems as targets for antivirulence therapy. Trends Microbiol. 26, 313–328. doi: 10.1016/j.tim.2017.10.005
Defoirdt, T., Boon, N., and Bossier, P. (2010). Can bacteria evolve resistance to quorum sensing disruption? PLoS Pathog. 6:e1000989. doi: 10.1371/journal.ppat.1000989
Dickey, S. W., Cheung, G. Y., and Otto, M. (2017). Different drugs for bad bugs: antivirulence strategies in the age of antibiotic resistance. Nat. Rev. Drug Discovery 16:457. doi: 10.1038/nrd.2017.23
Diep, B. A., Le, V. T., Badiou, C., Le, H. N., Pinheiro, M. G., Duong, A. H., et al. (2016). IVIG-mediated protection against necrotizing pneumonia caused by MRSA. Sci. Transl. Med. 8:357ra124. doi: 10.1126/scitranslmed.aag1153
Dong, W., Zhu, J., Guo, X., Kong, D., Zhang, Q., Zhou, Y., et al. (2018). Characterization of AiiK, an AHL lactonase, from Kurthia huakui LAM0618 T and its application in quorum quenching on Pseudomonas aeruginosa PAO1. Sci. Rep. 8:6013. doi: 10.1038/s41598-018-24507-8
Drees, S. L., Li, C., Prasetya, F., Saleem, M., Dreveny, I., Williams, P., et al. (2016). PqsBC, a condensing enzyme in the biosynthesis of the Pseudomonas aeruginosa quinolone signal crystal structure, inhibition, and reaction mechanism. J. Biol. Chem. 291, 6610–6624. doi: 10.1074/jbc.M115.708453
Duan, J., Li, M., Hao, Z., Shen, X., Liu, L., Jin, Y., et al. (2018). Subinhibitory concentrations of resveratrol reduce alpha-hemolysin production in Staphylococcus aureus isolates by downregulating saeRS. Emerg. Microb. Infect. 7:136. doi: 10.1038/s41426-018-0142-x
Dulcey, C. E., Dekimpe, V., Fauvelle, D.-A., Milot, S., Groleau, M.-C., Doucet, N., et al. (2013). The end of an old hypothesis: the Pseudomonas signaling molecules 4-hydroxy-2-alkylquinolines derive from fatty acids, not 3-ketofatty acids. Chem. Biol. 20, 1481–1491. doi: 10.1016/j.chembiol.2013.09.021
Edwards, G. A., Shymanska, N. V., and Pierce, J. G. (2017). 5-Benzylidene-4-oxazolidinones potently inhibit biofilm formation in Methicillin-resistant Staphylococcus aureus. Chem. Commun. 53, 7353–7356. doi: 10.1039/C7CC03626D
Eibergen, N. R., Moore, J. D., Mattmann, M. E., and Blackwell, H. E. (2015). Potent and selective modulation of the RhlR quorum sensing receptor by using non-native ligands: an emerging target for virulence control in Pseudomonas aeruginosa. Chembiochem 16, 2348–2356. doi: 10.1002/cbic.201500357
Fan, X., Liang, M., Wang, L., Chen, R., Li, H., and Liu, X. (2017). Aii810, a novel cold-adapted N-acylhomoserine lactonase discovered in a metagenome, can strongly attenuate Pseudomonas aeruginosa virulence factors and biofilm formation. Front. Microbiol. 8:1950. doi: 10.3389/fmicb.2017.01950
Fang, R. H., Luk, B. T., Hu, C. M., and Zhang, L. (2015). Engineered nanoparticles mimicking cell membranes for toxin neutralization. Adv. Drug Deliv. Rev. 90, 69–80. doi: 10.1016/j.addr.2015.04.001
Feng, L., Zhou, L., Sun, Y., Gui, J., Wang, X., Wu, P., et al. (2011). Specific inhibitions of annonaceous acetogenins on class II 3-hydroxy-3-methylglutaryl coenzyme A reductase from Streptococcus pneumoniae. Bioorg. Med. Chem. 19, 3512–3519. doi: 10.1016/j.bmc.2011.04.019
Feng, X., Guo, W., Zheng, H., Du, J., Luo, H., Wu, Q., et al. (2018). Inhibition of biofilm formation by chemical uncoupler, 3, 3′, 4′, 5-tetrachlorosalicylanilide (TCS): from the perspective of quorum sensing and biofilm related genes. Biochem. Eng. J. 137, 95–99. doi: 10.1016/j.bej.2018.05.010
Feng, X., Hu, Y., Zheng, Y., Zhu, W., Li, K., Huang, C.-H., et al. (2014). Structural and functional analysis of Bacillus subtilis YisP reveals a role of its product in biofilm production. Chem. Biol. 21, 1557–1563. doi: 10.1016/j.chembiol.2014.08.018
Fetzner, S. (2015). Quorum quenching enzymes. J. Biotechnol. 201, 2–14. doi: 10.1016/j.jbiotec.2014.09.001
Fleming, D., and Rumbaugh, K. P. (2017). Approaches to dispersing medical biofilms. Microorganisms 5:15. doi: 10.3390/microorganisms5020015
Flemming, H.-C., Wingender, J., Szewzyk, U., Steinberg, P., Rice, S. A., and Kjelleberg, S. (2016). Biofilms: an emergent form of bacterial life. Nat. Rev. Microbiol. 14:563. doi: 10.1038/nrmicro.2016.94
Fong, J., Zhang, C., Yang, R., Boo, Z. Z., Tan, S. K., Nielsen, T. E., et al. (2018). Combination therapy strategy of quorum quenching enzyme and quorum sensing inhibitor in suppressing multiple quorum sensing pathways of P. aeruginosa. Sci. Rep. 8:1155. doi: 10.1038/s41598-018-19504-w
Fong, S. A., Drilling, A., Morales, S., Cornet, M. E., Woodworth, B. A., Fokkens, W. J., et al. (2017). Activity of bacteriophages in removing biofilms of Pseudomonas aeruginosa isolates from chronic rhinosinusitis patients. Front. Cell. Infect. Microbiol. 7:418. doi: 10.3389/fcimb.2017.00418
Fujita, Y., and Taguchi, H. (2011). Current status of multiple antigen-presenting peptide vaccine systems: application of organic and inorganic nanoparticles. Chem. Cent. J. 5:48. doi: 10.1186/1752-153X-5-48
Garcia, S., Blackledge, M., Michalek, S., Su, L., Ptacek, T., Eipers, P., et al. (2017). Targeting of Streptococcus mutans biofilms by a novel small molecule prevents dental caries and preserves the oral microbiome. J. Dent. Res. 96, 807–814. doi: 10.1177/0022034517698096
García-Contreras, R. (2016). Is quorum sensing interference a viable alternative to treat Pseudomonas aeruginosa infections? Front. Microbiol. 7:1454. doi: 10.3389/fmicb.2016.01454
García-Contreras, R., Martínez-Vázquez, M., Velázquez Guadarrama, N., Villegas Pañeda, A. G., Hashimoto, T., Maeda, T., et al. (2013). Resistance to the quorum-quenching compounds brominated furanone C-30 and 5-fluorouracil in Pseudomonas aeruginosa clinical isolates. Pathog. Dis. 68, 8–11. doi: 10.1111/2049-632X.12039
García-Contreras, R., Peréz-Eretza, B., Jasso-Chávez, R., Lira-Silva, E., Roldán-Sánchez, J. A., González-Valdez, A., et al. (2015). High variability in quorum quenching and growth inhibition by furanone C-30 in Pseudomonas aeruginosa clinical isolates from cystic fibrosis patients. Pathog. Dis. 73:ftv040. doi: 10.1093/femspd/ftv040
García-Fernández, E., Koch, G., Wagner, R. M., Fekete, A., Stengel, S. T., Schneider, J., et al. (2017). Membrane microdomain disassembly inhibits MRSA antibiotic resistance. Cell 171, 1354–1367.e20. doi: 10.1016/j.cell.2017.10.012
Geoghegan, J. A., Irvine, A. D., and Foster, T. J. (2018). Staphylococcus aureus and atopic dermatitis: a complex and evolving relationship. Trends Microbiol. 26, 484–497. doi: 10.1016/j.tim.2017.11.008
Glansdorp, F. G., Thomas, G. L., Lee, J. K., Dutton, J. M., Salmond, G. P., Welch, M., et al. (2004). Synthesis and stability of small molecule probes for Pseudomonas aeruginosa quorum sensing modulation. Org. Biomol. Chem. 2, 3329–3336. doi: 10.1039/b412802h
Goerke, C., Fluckiger, U., Steinhuber, A., Zimmerli, W., and Wolz, C. (2001). Impact of the regulatory loci agr, sarA and sae of Staphylococcus aureus on the induction of α-toxin during device-related infection resolved by direct quantitative transcript analysis. Mol. Microbiol. 40, 1439–1447. doi: 10.1046/j.1365-2958.2001.02494.x
Goh, W.-K., Gardner, C. R., Sekhar, K. V. C., Biswas, N. N., Nizalapur, S., Rice, S. A., et al. (2015). Synthesis, quorum sensing inhibition and docking studies of 1, 5-dihydropyrrol-2-ones. Bioorg. Med. Chem. 23, 7366–7377. doi: 10.1016/j.bmc.2015.10.025
Gökalsin, B., Aksoydan, B., Erman, B., and Sesal, N. C. (2017). Reducing virulence and biofilm of Pseudomonas aeruginosa by potential quorum sensing inhibitor carotenoid: zeaxanthin. Microb. Ecol. 74, 466–473. doi: 10.1007/s00248-017-0949-3
Gordon, C. P., Olson, S. D., Lister, J. L., Kavanaugh, J. S., and Horswill, A. R. (2016). Truncated autoinducing peptides as antagonists of Staphylococcus lugdunensis quorum sensing. J. Med. Chem. 59, 8879–8888. doi: 10.1021/acs.jmedchem.6b00727
Goren, M. G., Carmeli, Y., Schwaber, M. J., Chmelnitsky, I., Schechner, V., and Navon-Venezia, S. (2010). Transfer of carbapenem-resistant plasmid from Klebsiella pneumoniae ST258 to Escherichia coli in patient. Emerging Infect. Dis. 16, 1014–1017. doi: 10.3201/eid1606.091671
Grandclément, C., Tannières, M., Moréra, S., Dessaux, Y., and Faure, D. (2015). Quorum quenching: role in nature and applied developments. FEMS Microbiol. Rev. 40, 86–116. doi: 10.1093/femsre/fuv038
Greenberg, M., Kuo, D., Jankowsky, E., Long, L., Hager, C., Bandi, K., et al. (2018). Small-molecule AgrA inhibitors F12 and F19 act as antivirulence agents against Gram-positive pathogens. Sci. Rep. 8:14578. doi: 10.1038/s41598-018-32829-w
Gruber, J. D., Chen, W., Parnham, S., Beauchesne, K., Moeller, P., Flume, P. A., et al. (2016). The role of 2, 4-dihydroxyquinoline (DHQ) in Pseudomonas aeruginosa pathogenicity. PeerJ 4:e1495. doi: 10.7717/peerj.1495
Guendouze, A., Plener, L., Bzdrenga, J., Jacquet, P., Rémy, B., Elias, M., et al. (2017). Effect of quorum quenching lactonase in clinical isolates of Pseudomonas aeruginosa and comparison with quorum sensing inhibitors. Front. Microbiol. 8:227. doi: 10.3389/fmicb.2017.00227
Gunn, J. S., Bakaletz, L. O., and Wozniak, D. J. (2016). What is on the outside matters: the role of the extracellular polymeric substance of gram-negative biofilms in evading host immunity and as a target for therapeutic intervention. J. Biol. Chem. 291, 12538–12546. doi: 10.1074/jbc.R115.707547
Gutierrez, J. A., Crowder, T., Rinaldo-Matthis, A., Ho, M.-C., Almo, S. C., and Schramm, V. L. (2009). Transition state analogs of 5′-methylthioadenosine nucleosidase disrupt quorum sensing. Nat. Chem. Biol. 5:251. doi: 10.1038/nchembio.153
Haines, B. E., Wiest, O., and Stauffacher, C. V. (2013). The increasingly complex mechanism of HMG-CoA reductase. Acc. Chem. Res. 46, 2416–2426. doi: 10.1021/ar3003267
Hall, C. W., and Mah, T.-F. (2017). Molecular mechanisms of biofilm-based antibiotic resistance and tolerance in pathogenic bacteria. FEMS Microbiol. Rev. 41, 276–301. doi: 10.1093/femsre/fux010
Hansen, A. M., Peng, P., Baldry, M., Perez-Gassol, I., Christensen, S. B., Vinther, J. M. O., et al. (2018). Lactam hybrid analogues of solonamide B and autoinducing peptides as potent S. aureus AgrC antagonists. Eur. J. Med. Chem. 152, 370–376. doi: 10.1016/j.ejmech.2018.04.053
Hazan, R., Que, Y. A., Maura, D., Strobel, B., Majcherczyk, P. A., Hopper, L. R., et al. (2016). Auto poisoning of the respiratory chain by a quorum-sensing-regulated molecule favors biofilm formation and antibiotic tolerance. Curr. Biol. 26, 195–206. doi: 10.1016/j.cub.2015.11.056
Hedl, M., and Rodwell, V. W. (2004). Inhibition of the Class II HMG–CoA reductase of Pseudomonas mevalonii. Protein Sci. 13, 1693–1697. doi: 10.1110/ps.03597504
Heidari, A., Noshiranzadeh, N., Haghi, F., and Bikas, R. (2017). Inhibition of quorum sensing related virulence factors of Pseudomonas aeruginosa by pyridoxal lactohydrazone. Microb. Pathog. 112, 103–110. doi: 10.1016/j.micpath.2017.09.043
Heimesaat, M. M., Lugert, R., Fischer, A., Alutis, M., Kühl, A. A., Zautner, A. E., et al. (2014). Impact of Campylobacter jejuni cj0268c knockout mutation on intestinal colonization, translocation, and induction of immunopathology in gnotobiotic IL-10 deficient mice. PLoS ONE 9:e90148. doi: 10.1371/journal.pone.0090148
Heinbockel, L., Weindl, G., Martinez-de-Tejada, G., Correa, W., Sanchez-Gomez, S., Barcena-Varela, S., et al. (2018). Inhibition of lipopolysaccharide- and lipoprotein-induced inflammation by antitoxin peptide Pep19-2.5. Front. Immunol. 9:1704. doi: 10.3389/fimmu.2018.01704
Henry, B. D., Neill, D. R., Becker, K. A., Gore, S., Bricio-Moreno, L., Ziobro, R., et al. (2015). Engineered liposomes sequester bacterial exotoxins and protect from severe invasive infections in mice. Nat. Biotechnol. 33, 81–88. doi: 10.1038/nbt.3037
Heras, B., Scanlon, M. J., and Martin, J. L. (2015). Targeting virulence not viability in the search for future antibacterials. Br. J. Clin. Pharmacol. 79, 208–215. doi: 10.1111/bcp.12356
Heuston, S., Begley, M., Gahan, C. G., and Hill, C. (2012). Isoprenoid biosynthesis in bacterial pathogens. Microbiology 158, 1389–1401. doi: 10.1099/mic.0.051599-0
Hinsberger, S., de Jong, J. C., Groh, M., Haupenthal, J., and Hartmann, R. W. (2014). Benzamidobenzoic acids as potent PqsD inhibitors for the treatment of Pseudomonas aeruginosa infections. Eur. J. Med. Chem. 76, 343–351. doi: 10.1016/j.ejmech.2014.02.014
Hogardt, M., and Heesemann, J. (2013). Microevolution of Pseudomonas aeruginosa to a chronic pathogen of the cystic fibrosis lung. Curr Top Microbiol Immunol. 358, 91–118. doi: 10.1007/82_2011_199
Hraiech, S., Hiblot, J., Lafleur, J., Lepidi, H., Papazian, L., Rolain, J.-M., et al. (2014). Inhaled lactonase reduces Pseudomonas aeruginosa quorum sensing and mortality in rat pneumonia. PLoS ONE 9:e107125. doi: 10.1371/journal.pone.0107125
Huggins, W. M., Nguyen, T. V., Hahn, N. A., Baker, J. T., Kuo, L. G., Kaur, D., et al. (2018). 2-Aminobenzimidazoles as antibiofilm agents against Salmonella enterica serovar typhimurium. Medchemcomm 9, 1547–1552. doi: 10.1039/C8MD00298C
Hwang, I. Y., Koh, E., Wong, A., March, J. C., Bentley, W. E., Lee, Y. S., et al. (2017). Engineered probiotic Escherichia coli can eliminate and prevent Pseudomonas aeruginosa gut infection in animal models. Nat. Commun. 8:15028. doi: 10.1038/ncomms15028
Istvan, E. S., and Deisenhofer, J. (2001). Structural mechanism for statin inhibition of HMG-CoA reductase. Science 292, 1160–1164. doi: 10.1126/science.1059344
Ivanova, K., Fernandes, M. M., Francesko, A., Mendoza, E., Guezguez, J., Burnet, M., et al. (2015). Quorum-quenching and matrix-degrading enzymes in multilayer coatings synergistically prevent bacterial biofilm formation on urinary catheters. ACS Appl. Mater. Interfaces 7, 27066–27077. doi: 10.1021/acsami.5b09489
Jenkins, A., Diep, B. A., Mai, T. T., Vo, N. H., Warrener, P., Suzich, J., et al. (2015). Differential expression and roles of Staphylococcus aureus virulence determinants during colonization and disease. MBio. 6, e02272–e02214. doi: 10.1128/mBio.02272-14
Ji, C., Sharma, I., Pratihar, D., Hudson, L. L., Maura, D., Guney, T., et al. (2016). Designed small-molecule inhibitors of the anthranilyl-CoA synthetase PqsA block quinolone biosynthesis in Pseudomonas aeruginosa. ACS Chem. Biol. 11, 3061–3067. doi: 10.1021/acschembio.6b00575
Joo, H.-S., Fu, C.-I., and Otto, M. (2016). Bacterial strategies of resistance to antimicrobial peptides. Phil. Trans. R. Soc. B 371:20150292. doi: 10.1098/rstb.2015.0292
Kahler, C. M., Sarkar-Tyson, M., Kibble, E. A., Stubbs, K. A., and Vrielink, A. (2018). Enzyme targets for drug design of new anti-virulence therapeutics. Curr. Opin. Struct. Biol. 53, 140–150. doi: 10.1016/j.sbi.2018.08.010
Kalaiarasan, E., Thirumalaswamy, K., Harish, B. N., Gnanasambandam, V., Sali, V. K., and John, J. (2017). Inhibition of quorum sensing-controlled biofilm formation in Pseudomonas aeruginosa by quorum-sensing inhibitors. Microb. Pathog. 111, 99–107. doi: 10.1016/j.micpath.2017.08.017
Kalia, M., Yadav, V. K., Singh, P. K., Sharma, D., Narvi, S. S., and Agarwal, V. (2018). Exploring the impact of parthenolide as anti-quorum sensing and anti-biofilm agent against Pseudomonas aeruginosa. Life Sci. 199, 96–103. doi: 10.1016/j.lfs.2018.03.013
Kalia, V. C., Patel, S. K., Kang, Y. C., and Lee, J.-K. (2019). Quorum sensing inhibitors as antipathogens: biotechnological applications. Biotechnol. Adv. 37, 68–90. doi: 10.1016/j.biotechadv.2018.11.006
Kang, S., Kim, J., Hur, J. K., and Lee, S.-S. (2017). CRISPR-based genome editing of clinically important Escherichia coli SE15 isolated from indwelling urinary catheters of patients. J. Med. Microbiol. 66, 18–25. doi: 10.1099/jmm.0.000406
Karathanasi, G., Bojer, M. S., Baldry, M., Johannessen, B. A., Wolff, S., Greco, I., et al. (2018). Linear peptidomimetics as potent antagonists of Staphylococcus aureus agr quorum sensing. Sci. Rep. 8:3562. doi: 10.1038/s41598-018-21951-4
Kaufmann, G. F., Park, J., Mee, J. M., Ulevitch, R. J., and Janda, K. D. (2008). The quorum quenching antibody RS2-1G9 protects macrophages from the cytotoxic effects of the Pseudomonas aeruginosa quorum sensing signalling molecule N-3-oxo-dodecanoyl-homoserine lactone. Mol. Immunol. 45, 2710–2714. doi: 10.1016/j.molimm.2008.01.010
Kenaley, K. M., Blackson, T., Boylan, L., Ciarlo, J., Antunes, M., Shaffer, T. H., et al. (2018). Impact of endotracheal tube biofilm and respiratory secretions on airway resistance and mechanics of breathing in a neonatal lung model. J. Appl. Physiol. 125, 1227–1231. doi: 10.1152/japplphysiol.00083.2018
Khan, B. A., Yeh, A. J., Cheung, G. Y., and Otto, M. (2015). Investigational therapies targeting quorum-sensing for the treatment of Staphylococcus aureus infections. Expert Opin. Investig. Drugs 24, 689–704. doi: 10.1517/13543784.2015.1019062
Kim, B., Park, J.-S., Choi, H.-Y., Yoon, S. S., and Kim, W.-G. (2018). Terrein is an inhibitor of quorum sensing and c-di-GMP in Pseudomonas aeruginosa: a connection between quorum sensing and c-di-GMP. Sci. Rep. 8:8617. doi: 10.1038/s41598-018-26974-5
Kim, D.-Y., Stauffacher, C. V., and Rodwell, V. W. (2000). Dual coenzyme specificity of Archaeoglobus fulgidus HMG-CoA reductase. Protein Sci. 9, 1226–1234. doi: 10.1110/ps.9.6.1226
Ko, H. H., Lareu, R. R., Dix, B. R., and Hughes, J. D. (2017). Statins: antimicrobial resistance breakers or makers? PeerJ 5:e3952. doi: 10.7717/peerj.3952
Koch, B., Liljefors, T., Persson, T., Nielsen, J., Kjelleberg, S., and Givskov, M. (2005). The LuxR receptor: the sites of interaction with quorum-sensing signals and inhibitors. Microbiology 151, 3589–3602. doi: 10.1099/mic.0.27954-0
Koch, G., Wermser, C., Acosta, I. C., Kricks, L., Stengel, S. T., Yepes, A., et al. (2017). Attenuating Staphylococcus aureus virulence by targeting flotillin protein scaffold activity. Cell Chem. Biol. 24, 845–857.e46. doi: 10.1016/j.chembiol.2017.05.027
Köhler, T., Perron, G. G., Buckling, A., and Van Delden, C. (2010). Quorum sensing inhibition selects for virulence and cooperation in Pseudomonas aeruginosa. PLoS Pathog. 6:e1000883. doi: 10.1371/journal.ppat.1000883
Kong, C., Neoh, H. M., and Nathan, S. (2016). Targeting Staphylococcus aureus toxins: a potential form of anti-virulence therapy. Toxins 8:72. doi: 10.3390/toxins8030072
Konstantinović, N., Cirković, I., ÐDukić, S., Marić, V., and BoŽić, D. D. (2017). Biofilm formation of Achromobacter xylosoxidans on contact lens. Acta Microbiol. Immunol. Hung. 64, 293–300. doi: 10.1556/030.64.2017.005
Koopman, J. A., Marshall, J. M., Bhatiya, A., Eguale, T., Kwiek, J. J., and Gunn, J. S. (2015). Inhibition of Salmonella enterica biofilm formation using small molecule adenosine mimetics. Antimicrob. Agents Chemother. 59, 76–84. doi: 10.1128/AAC.03407-14
Koul, S., Prakash, J., Mishra, A., and Kalia, V. C. (2016). Potential emergence of multi-quorum sensing inhibitor resistant (MQSIR) bacteria. Indian J. Microbiol. 56, 1–18. doi: 10.1007/s12088-015-0558-0
Krueger, E., Hayes, S., Chang, E. H., Yutuc, S., and Brown, A. C. (2018). Receptor-based peptides for inhibition of RTX leukotoxin (LtxA) activity. ACS Infect. Dis. 4, 1073–1081. doi: 10.1021/acsinfecdis.7b00230
Kudryashova, E., Seveau, S., Lu, W., and Kudryashov, D. S. (2015). Retrocyclins neutralize bacterial toxins by potentiating their unfolding. Biochem. J. 467, 311–320. doi: 10.1042/BJ20150049
Kumar, K., Chen, J., Drlica, K., and Shopsin, B. (2017). Tuning of the lethal response to multiple stressors with a single-site mutation during clinical infection by Staphylococcus aureus. MBio. 8, e01476–e01417. doi: 10.1128/mBio.01476-17
Kumar, L., Chhibber, S., Kumar, R., Kumar, M., and Harjai, K. (2015). Zingerone silences quorum sensing and attenuates virulence of Pseudomonas aeruginosa. Fitoterapia 102, 84–95. doi: 10.1016/j.fitote.2015.02.002
Kutty, S. K., Barraud, N., Ho, K. K., Iskander, G. M., Griffith, R., Rice, S. A., et al. (2015). Hybrids of acylated homoserine lactone and nitric oxide donors as inhibitors of quorum sensing and virulence factors in Pseudomonas aeruginosa. Org. Biomol. Chem. 13, 9850–9861. doi: 10.1039/C5OB01373A
LaSarre, B., and Federle, M. J. (2013). Exploiting quorum sensing to confuse bacterial pathogens. Microbiol. Mol. Biol. Rev. 77, 73–111. doi: 10.1128/MMBR.00046-12
Le, V. T., Tkaczyk, C., Chau, S., Rao, R. L., Dip, E. C., Pereira-Franchi, E. P., et al. (2016). Critical role of alpha-toxin and protective effects of its neutralization by a human antibody in acute bacterial skin and skin structure infections. Antimicrob. Agents Chemother. 60, 5640–5648. doi: 10.1128/AAC.00710-16
Lee, J., and Zhang, L. (2015). The hierarchy quorum sensing network in Pseudomonas aeruginosa. Protein Cell 6, 26–41. doi: 10.1007/s13238-014-0100-x
Lee, J.-H., Kim, Y.-G., Cho, H. S., Ryu, S. Y., Cho, M. H., and Lee, J. (2014). Coumarins reduce biofilm formation and the virulence of Escherichia coli O157: H7. Phytomedicine 21, 1037–1042. doi: 10.1016/j.phymed.2014.04.008
Lesic, B., Lépine, F., Déziel, E., Zhang, J., Zhang, Q., Padfield, K., et al. (2007). Inhibitors of pathogen intercellular signals as selective anti-infective compounds. PLoS Pathog. 3:e126. doi: 10.1371/journal.ppat.0030126
Li, C., Wu, Y., Riehle, A., Ma, J., Kamler, M., Gulbins, E., et al. (2017). Staphylococcus aureus survives in cystic fibrosis macrophages forming a reservoir for chronic pneumonia. Infect. Immun. 85:e00883–16. doi: 10.1128/IAI.00883-16
Li, D., Gui, J., Li, Y., Feng, L., Han, X., Sun, Y., et al. (2012). Structure-based design and screen of novel inhibitors for class II 3-hydroxy-3-methylglutaryl coenzyme A reductase from Streptococcus Pneumoniae. J. Chem. Inf. Model. 52, 1833–1841. doi: 10.1021/ci300163v
Lidor, O., Al-Quntar, A., Pesci, E., and Steinberg, D. (2015). Mechanistic analysis of a synthetic inhibitor of the Pseudomonas aeruginosa LasI quorum-sensing signal synthase. Sci. Rep. 5:16569. doi: 10.1038/srep16569
Lin, F.-Y., Liu, C.-I., Liu, Y.-L., Zhang, Y., Wang, K., Jeng, W.-Y., et al. (2010). Mechanism of action and inhibition of dehydrosqualene synthase. Proc. Natl. Acad. Sci. U.S.A. 107, 21337–21342. doi: 10.1073/pnas.1010907107
Liu, Q., Yeo, W. S., and Bae, T. (2016). The SaeRS two-component system of Staphylococcus aureus. Genes 7:81. doi: 10.3390/genes7100081
Liu, X., Peng, L., Meng, J., Zhu, Z., Han, B., and Wang, S. (2018). Protein-mediated anti-adhesion surface against oral bacteria. Nanoscale 10, 2711–2714. doi: 10.1039/C7NR08844B
Liu, Y., Shi, D., Guo, Y., Li, M., Zha, Y., Wang, Q., et al. (2017). Dracorhodin perochlorate attenuates Staphylococcus aureus USA300 virulence by decreasing α-toxin expression. World J. Microbiol. Biotechnol. 33:17. doi: 10.1007/s11274-016-2129-x
Lopez, D., and Koch, G. (2017). Exploring functional membrane microdomains in bacteria: an overview. Curr. Opin. Microbiol. 36, 76–84. doi: 10.1016/j.mib.2017.02.001
López, D., and Kolter, R. (2010). Functional microdomains in bacterial membranes. Genes Dev. 24, 1893–1902. doi: 10.1101/gad.1945010
López, M., Mayer, C., Fernández-García, L., Blasco, L., Muras, A., Ruiz, F. M., et al. (2017). Quorum sensing network in clinical strains of A. baumannii: AidA is a new quorum quenching enzyme. PLoS ONE 12:e0174454. doi: 10.1371/journal.pone.0174454
Lu, T. K., and Collins, J. J. (2007). Dispersing biofilms with engineered enzymatic bacteriophage. Proc. Natl. Acad. Sci. U.S.A. 104, 11197–11202. doi: 10.1073/pnas.0704624104
Ma, L., Feng, S., de la Fuente-Núñez, C., Hancock, R. E., and Lu, X. (2018). Development of molecularly imprinted polymers to block quorum sensing and inhibit bacterial biofilm formation. ACS Appl. Mater. Interfaces. 10, 18450–18457. doi: 10.1021/acsami.8b01584
Maeda, T., García-Contreras, R., Pu, M., Sheng, L., Garcia, L. R., Tomás, M., et al. (2012). Quorum quenching quandary: resistance to antivirulence compounds. ISME J. 6, 493–501. doi: 10.1038/ismej.2011.122
Maura, D., Ballok, A. E., and Rahme, L. G. (2016). Considerations and caveats in anti-virulence drug development. Curr. Opin. Microbiol. 33, 41–46. doi: 10.1016/j.mib.2016.06.001
Maura, D., Drees, S. L., Bandyopadhaya, A., Kitao, T., Negri, M., Starkey, M., et al. (2017). Polypharmacology approaches against the Pseudomonas aeruginosa MvfR regulon and their application in blocking virulence and antibiotic tolerance. ACS Chem. Biol. 12, 1435–1443. doi: 10.1021/acschembio.6b01139
Merkey, B. V., Lardon, L. A., Seoane, J. M., Kreft, J. U., and Smets, B. F. (2011). Growth dependence of conjugation explains limited plasmid invasion in biofilms: an individual-based modelling study. Environ. Microbiol. 13, 2435–2452. doi: 10.1111/j.1462-2920.2011.02535.x
Messacar, K., Parker, S. K., Todd, J. K., and Dominguez, S. R. (2017). Implementation of rapid molecular infectious disease diagnostics: the role of diagnostic and antimicrobial stewardship. J. Clin. Microbiol. 57, 715–723. doi: 10.1128/JCM.02264-16
Mielich-Süss, B., Wagner, R. M., Mietrach, N., Hertlein, T., Marincola, G., Ohlsen, K., et al. (2017). Flotillin scaffold activity contributes to type VII secretion system assembly in Staphylococcus aureus. PLoS Pathog. 13:e1006728. doi: 10.1371/journal.ppat.1006728
Migiyama, Y., Kaneko, Y., Yanagihara, K., Morohoshi, T., Morinaga, Y., Nakamura, S., et al. (2013). Efficacy of AiiM, an N-acylhomoserine lactonase, against Pseudomonas aeruginosa in a mouse model of acute pneumonia. Antimicrob. Agents Chemother. 57, 3653–3658. doi: 10.1128/AAC.00456-13
Ming, D., Wang, D., Cao, F., Xiang, H., Mu, D., Cao, J., et al. (2017). Kaempferol inhibits the primary attachment phase of biofilm formation in Staphylococcus aureus. Front. Microbiol. 8:2263. doi: 10.3389/fmicb.2017.02263
Miyairi, S., Tateda, K., Fuse, E. T., Ueda, C., Saito, H., Takabatake, T., et al. (2006). Immunization with 3-oxododecanoyl-L-homoserine lactone–protein conjugate protects mice from lethal Pseudomonas aeruginosa lung infection. J. Med. Microbiol. 55, 1381–1387. doi: 10.1099/jmm.0.46658-0
Moshiri, J., Kaur, D., Hambira, C. M., Sandala, J. L., Koopman, J. A., Fuchs, J. R., et al. (2018). Identification of a small molecule anti-biofilm agent against Salmonella enterica. Front. Microbiol. 9:2804. doi: 10.3389/fmicb.2018.02804
Mühlen, S., and Dersch, P. (2016). Anti-virulence strategies to target bacterial infections. Curr. Top. Microbiol. Immunol. 398, 147–183. doi: 10.1007/82_2015_490
Müller, C., Birmes, F. S., Rückert, C., Kalinowski, J., and Fetzner, S. (2015). Rhodococcus erythropolis BG43 genes mediating Pseudomonas aeruginosa quinolone signal degradation and virulence factor attenuation. Appl. Environ. Microbiol. 22, 7720–7729. doi: 10.1128/AEM.02145-15
Munguia, J., and Nizet, V. (2017). Pharmacological targeting of the host–pathogen interaction: alternatives to classical antibiotics to combat drug-resistant superbugs. Trends Pharmacol. Sci. 38, 473–488. doi: 10.1016/j.tips.2017.02.003
Musovic, S., Oregaard, G., Kroer, N., and Sørensen, S. J. (2006). Cultivation-independent examination of horizontal transfer and host range of an IncP-1 plasmid among gram-positive and gram-negative bacteria indigenous to the barley rhizosphere. Appl. Environ. Microbiol. 72, 6687–6692. doi: 10.1128/AEM.00013-06
Nuri, R., Shprung, T., and Shai, Y. (2015). Defensive remodeling: how bacterial surface properties and biofilm formation promote resistance to antimicrobial peptides. Biochim. Biophys. Acta 1848, 3089–3100. doi: 10.1016/j.bbamem.2015.05.022
O'brien, S., and Fothergill, J. L. (2017). The role of multispecies social interactions in shaping Pseudomonas aeruginosa pathogenicity in the cystic fibrosis lung. FEMS Microbiol. Lett. 364:fnx128. doi: 10.1093/femsle/fnx128
O'Loughlin, C. T., Miller, L. C., Siryaporn, A., Drescher, K., Semmelhack, M. F., and Bassler, B. L. (2013). A quorum-sensing inhibitor blocks Pseudomonas aeruginosa virulence and biofilm formation. Proc. Natl. Acad. Sci. U.S.A. 110, 17981–17986. doi: 10.1073/pnas.1316981110
O'Neill, J. (2016). Tackling Drug-Resistant Infections Globally: Final Report and Recommendations. HM Government and Wellcome Trust.
Orazi, G., and O'Toole, G. A. (2017). Pseudomonas aeruginosa alters Staphylococcus aureus sensitivity to vancomycin in a biofilm model of cystic fibrosis infection. MBio. 8:e00873–17. doi: 10.1128/mBio.00873-17
O'Reilly, M. C., and Blackwell, H. E. (2015). Structure-based design and biological evaluation of triphenyl scaffold-based hybrid compounds as hydrolytically stable modulators of a LuxR-Type quorum sensing receptor. ACS Infect. Dis. 2, 32–38. doi: 10.1021/acsinfecdis.5b00112
O'Rourke, J. P., Daly, S. M., Triplett, K. D., Peabody, D., Chackerian, B., and Hall, P. R. (2014). Development of a mimotope vaccine targeting the Staphylococcus aureus quorum sensing pathway. PLoS ONE 9:e111198. doi: 10.1371/journal.pone.0111198
Ortines, R. V., Liu, H., Cheng, L. I., Cohen, T. S., Lawlor, H., Gami, A., et al. (2018). Neutralizing alpha-toxin accelerates healing of Staphylococcus aureus-infected wounds in nondiabetic and diabetic mice. Antimicrob. Agents Chemother. 62:e02288–17. doi: 10.1128/AAC.02288-17
Paczkowski, J. E., Mukherjee, S., McCready, A. R., Cong, J.-P., Aquino, C. J., Kim, H., et al. (2017). Flavonoids suppress Pseudomonas aeruginosa virulence through allosteric inhibition of quorum-sensing receptors. J. Biol. Chem. 292, 4064–4076. doi: 10.1074/jbc.M116.770552
Palliyil, S., Downham, C., Broadbent, I., Charlton, K., and Porter, A. J. (2014). High-sensitivity monoclonal antibodies specific for homoserine lactones protect mice from lethal Pseudomonas aeruginosa infections. Appl. Environ. Microbiol. 80, 462–469. doi: 10.1128/AEM.02912-13
Pan, J.-J., Solbiati, J. O., Ramamoorthy, G., Hillerich, B. S., Seidel, R. D., Cronan, J. E., et al. (2015a). Biosynthesis of squalene from farnesyl diphosphate in bacteria: three steps catalyzed by three enzymes. ACS Central Sci. 1, 77–82. doi: 10.1021/acscentsci.5b00115
Pan, W., Fan, M., Wu, H., Melander, C., and Liu, C. (2015b). A new small molecule inhibits Streptococcus mutans biofilms in vitro and in vivo. J. Appl. Microbiol. 119, 1403–1411. doi: 10.1111/jam.12940
Papaioannou, E., Wahjudi, M., Nadal-Jimenez, P., Koch, G., Setroikromo, R., and Quax, W. J. (2009). Quorum-quenching acylase reduces the virulence of Pseudomonas aeruginosa in a Caenorhabditis elegans infection model. Antimicrob. Agents Chemother. 53, 4891–4897. doi: 10.1128/AAC.00380-09
Papenfort, K., and Bassler, B. L. (2016). Quorum sensing signal–response systems in gram-negative bacteria. Nat. Rev. Microbiol. 14:576. doi: 10.1038/nrmicro.2016.89
Park, J., Jagasia, R., Kaufmann, G. F., Mathison, J. C., Ruiz, D. I., Moss, J. A., et al. (2007). Infection control by antibody disruption of bacterial quorum sensing signaling. Chem. Biol. 14, 1119–1127. doi: 10.1016/j.chembiol.2007.08.013
Park, S., Kim, H.-S., Ok, K., Kim, Y., Park, H.-D., and Byun, Y. (2015). Design, synthesis and biological evaluation of 4-(alkyloxy)-6-methyl-2H-pyran-2-one derivatives as quorum sensing inhibitors. Bioorg. Med. Chem. Lett. 25, 2913–2917. doi: 10.1016/j.bmcl.2015.05.054
Passos da Silva, D., Schofield, M. C., Parsek, M. R., and Tseng, B. S. (2017). An update on the sociomicrobiology of quorum sensing in gram-negative biofilm development. Pathogens 6:51. doi: 10.3390/pathogens6040051
Patil, P. C., Tan, J., Demuth, D. R., and Luzzio, F. A. (2016). 1, 2, 3-Triazole-based inhibitors of Porphyromonas gingivalis adherence to oral streptococci and biofilm formation. Bioorg. Med. Chem. 24, 5410–5417. doi: 10.1016/j.bmc.2016.08.059
Pereira, C. S., Thompson, J. A., and Xavier, K. B. (2013). AI-2-mediated signalling in bacteria. FEMS Microbiol. Rev. 37, 156–181. doi: 10.1111/j.1574-6976.2012.00345.x
Pérez-Gil, J., and Rodríguez-Concepción, M. (2013). Metabolic plasticity for isoprenoid biosynthesis in bacteria. Biochem. J. 452, 19–25. doi: 10.1042/BJ20121899
Peterson, J. W. (1996). “Bacterial pathogenesis,” in Medical Microbiology. 4th Edn. (University of Texas Medical Branch at Galveston).
Piletska, E. V., Stavroulakis, G., Karim, K., Whitcombe, M. J., Chianella, I., Sharma, A., et al. (2010). Attenuation of Vibrio fischeri quorum sensing using rationally designed polymers. Biomacromolecules 11, 975–980. doi: 10.1021/bm901451j
Piletska, E. V., Stavroulakis, G., Larcombe, L. D., Whitcombe, M. J., Sharma, A., Primrose, S., et al. (2011). Passive control of quorum sensing: prevention of Pseudomonas aeruginosa biofilm formation by imprinted polymers. Biomacromolecules 12, 1067–1071. doi: 10.1021/bm101410q
Ping, O., Mao, S., Xuewen, H., Kaiyu, W., Zhongqiong, Y., Hualin, F., et al. (2017). Sclareol protects Staphylococcus aureus-induced lung cell injury via inhibiting alpha-hemolysin expression. J. Microbiol. Biotechnol. 27, 19–25. doi: 10.4014/jmb.1606.06039
Ping, O., Ruixue, Y., Jiaqiang, D., Kaiyu, W., Jing, F., Yi, G., et al. (2018). Subinhibitory concentrations of prim-o-glucosylcimifugin decrease the expression of alpha-hemolysin in Staphylococcus aureus (USA300). Evid. Based Complement. Alternat. Med. 2018:7579808. doi: 10.1155/2018/7579808
Poolman, B., Spitzer, J. J., and Wood, J. M. (2004). Bacterial osmosensing: roles of membrane structure and electrostatics in lipid–protein and protein–protein interactions. Biochim. Biophys. Acta 1666, 88–104. doi: 10.1016/j.bbamem.2004.06.013
Powers, M. E., Becker, R. E., Sailer, A., Turner, J. R., and Bubeck Wardenburg, J. (2015). Synergistic action of Staphylococcus aureus alpha-toxin on platelets and myeloid lineage cells contributes to lethal sepsis. Cell Host Microbe 17, 775–787. doi: 10.1016/j.chom.2015.05.011
Pristovsek, P., and Kidric, J. (1999). Solution structure of polymyxins B and E and effect of binding to lipopolysaccharide: an NMR and molecular modeling study. J. Med. Chem. 42, 4604–4613. doi: 10.1021/jm991031b
Puga, C., Rodríguez-López, P., Cabo, M., SanJose, C., and Orgaz, B. (2018). Enzymatic dispersal of dual-species biofilms carrying Listeria monocytogenes and other associated food industry bacteria. Food Control 94, 222–228. doi: 10.1016/j.foodcont.2018.07.017
Pulido, D., Nogues, M. V., Boix, E., and Torrent, M. (2012). Lipopolysaccharide neutralization by antimicrobial peptides: a gambit in the innate host defense strategy. J. Innate Immun. 4, 327–336. doi: 10.1159/000336713
Pustelny, C., Albers, A., Büldt-Karentzopoulos, K., Parschat, K., Chhabra, S. R., Cámara, M., et al. (2009). Dioxygenase-mediated quenching of quinolone-dependent quorum sensing in Pseudomonas aeruginosa. Chem. Biol. 16, 1259–1267. doi: 10.1016/j.chembiol.2009.11.013
Quan, Y., Meng, F., Ma, X., Song, X., Liu, X., Gao, W., et al. (2017). Regulation of bacteria population behaviors by AI-2 “consumer cells” and “supplier cells”. BMC Microbiol. 17:198. doi: 10.1186/s12866-017-1107-2
Quave, C. L., Lyles, J. T., Kavanaugh, J. S., Nelson, K., Parlet, C. P., Crosby, H. A., et al. (2015). Castanea sativa (European Chestnut) leaf extracts rich in ursene and oleanene derivatives block Staphylococcus aureus virulence and pathogenesis without detectable resistance. PLoS ONE 10:e0136486. doi: 10.1371/journal.pone.0136486
Que, Y.-A., Hazan, R., Strobel, B., Maura, D., He, J., Kesarwani, M., et al. (2013). A quorum sensing small volatile molecule promotes antibiotic tolerance in bacteria. PLoS ONE 8:e80140. doi: 10.1371/journal.pone.0080140
Radlinski, L., Rowe, S. E., Kartchner, L. B., Maile, R., Cairns, B. A., Vitko, N. P., et al. (2017). Pseudomonas aeruginosa exoproducts determine antibiotic efficacy against Staphylococcus aureus. PLoS Biol. 15:e2003981. doi: 10.1371/journal.pbio.2003981
Rajamanikandan, S., Jeyakanthan, J., and Srinivasan, P. (2017a). Discovery of potent inhibitors targeting Vibrio harveyi LuxR through shape and e-pharmacophore based virtual screening and its biological evaluation. Microb. Pathog. 103, 40–56. doi: 10.1016/j.micpath.2016.12.003
Rajamanikandan, S., Jeyakanthan, J., and Srinivasan, P. (2017b). Molecular docking, molecular dynamics simulations, computational screening to design quorum sensing inhibitors targeting LuxP of Vibrio harveyi and its biological evaluation. Appl. Biochem. Biotechnol. 181, 192–218. doi: 10.1007/s12010-016-2207-4
Rajput, A., Thakur, A., Sharma, S., and Kumar, M. (2017). aBiofilm: a resource of anti-biofilm agents and their potential implications in targeting antibiotic drug resistance. Nucleic Acids Res. 46, D894–D900. doi: 10.1093/nar/gkx1157
Ranfaing, J., Dunyach-Remy, C., Lavigne, J.-P., and Sotto, A. (2018). Propolis potentiates the effect of cranberry (Vaccinium macrocarpon) in reducing the motility and the biofilm formation of uropathogenic Escherichia coli. PLoS ONE 13:e0202609. doi: 10.1371/journal.pone.0202609
Rasko, D. A., and Sperandio, V. (2010). Anti-virulence strategies to combat bacteria-mediated disease. Nat. Rev. Drug Discov. 9, 117–128. doi: 10.1038/nrd3013
Ribeiro, S. M., Felicio, M. R., Boas, E. V., Gonçalves, S., Costa, F. F., Samy, R. P., et al. (2016). New frontiers for anti-biofilm drug development. Pharmacol. Ther. 160, 133–144. doi: 10.1016/j.pharmthera.2016.02.006
Rieger, U. M., Djedovic, G., Pattiss, A., Raschke, G. F., Frei, R., Pierer, G., et al. (2016). Presence of biofilms on polyurethane-coated breast implants: preliminary results. J. Long Term Eff. Med. Implants 26, 237–243. doi: 10.1615/JLongTermEffMedImplants.2016016851
Rietschel, E. T., Brade, H., Holst, O., Brade, L., Muller-Loennies, S., Mamat, U., et al. (1996). Bacterial endotoxin: chemical constitution, biological recognition, host response, and immunological detoxification. Curr. Top. Microbiol. Immunol. 216, 39–81. doi: 10.1007/978-3-642-80186-0_3
Rouha, H., Badarau, A., Visram, Z. C., Battles, M. B., Prinz, B., Magyarics, Z., et al. (2015). Five birds, one stone: neutralization of α-hemolysin and 4 bi-component leukocidins of Staphylococcus aureus with a single human monoclonal antibody. MAbs 7, 243–254. doi: 10.4161/19420862.2014.985132
Roy, V., Fernandes, R., Tsao, C.-Y., and Bentley, W. E. (2010). Cross species quorum quenching using a native AI-2 processing enzyme. ACS Chem. Biol. 5, 223–232. doi: 10.1021/cb9002738
Rudkin, J. K., McLoughlin, R. M., Preston, A., and Massey, R. C. (2017). Bacterial toxins: offensive, defensive, or something else altogether? PLoS Pathog. 13:e1006452. doi: 10.1371/journal.ppat.1006452
Rybtke, M., Hultqvist, L. D., Givskov, M., and Tolker-Nielsen, T. (2015). Pseudomonas aeruginosa biofilm infections: community structure, antimicrobial tolerance and immune response. J. Mol. Biol. 427, 3628–3645. doi: 10.1016/j.jmb.2015.08.016
Sahner, J. H., Empting, M., Kamal, A., Weidel, E., Groh, M., Börger, C., et al. (2015). Exploring the chemical space of ureidothiophene-2-carboxylic acids as inhibitors of the quorum sensing enzyme PqsD from Pseudomonas aeruginosa. Eur. J. Med. Chem. 96, 14–21. doi: 10.1016/j.ejmech.2015.04.007
Schütz, C., and Empting, M. (2018). Targeting the Pseudomonas quinolone signal quorum sensing system for the discovery of novel anti-infective pathoblockers. Beilstein J. Org. Chem. 14, 2627–2645. doi: 10.3762/bjoc.14.241
Schwalen, C. J., Feng, X., Liu, W., Bing, O., Ko, T. P., Shin, C. J., et al. (2017). Head-to-head prenyl synthases in pathogenic bacteria. Chembiochem 18, 985–991. doi: 10.1002/cbic.201700099
Scoffone, V. C., Chiarelli, L. R., Makarov, V., Brackman, G., Israyilova, A., Azzalin, A., et al. (2016). Discovery of new diketopiperazines inhibiting Burkholderia cenocepacia quorum sensing in vitro and in vivo. Sci. Rep. 6:32487. doi: 10.1038/srep32487
Sedlmayer, F., Hell, D., Müller, M., Ausländer, D., and Fussenegger, M. (2018). Designer cells programming quorum-sensing interference with microbes. Nat. Commun. 9:1822. doi: 10.1038/s41467-018-04223-7
Shrestha, L., Kayama, S., Sasaki, M., Kato, F., Hisatsune, J., Tsuruda, K., et al. (2016). Inhibitory effects of antibiofilm compound 1 against Staphylococcus aureus biofilms. Microbiol. Immunol. 60, 148–159. doi: 10.1111/1348-0421.12359
Silva, T., Freitas, A., Pinheiro, M., Watanabe, E., and Albuquerque, R. (2018). Oral biofilm formation on different materials for dental implants. J. Visual. Exp. 136:e57756. doi: 10.3791/57756
Soukarieh, F., Vico Oton, E., Dubern, J.-F., Gomes, J., Halliday, N., de Pilar Crespo, M., et al. (2018). In silico and in vitro-guided identification of inhibitors of alkylquinolone-dependent quorum sensing in pseudomonas aeruginosa. Molecules 23:257. doi: 10.3390/molecules23020257
Sousa, A. M., and Pereira, M. O. (2014). Pseudomonas aeruginosa diversification during infection development in cystic fibrosis lungs—a review. Pathogens 3, 680–703. doi: 10.3390/pathogens3030680
Stalder, T., and Top, E. (2016). Plasmid transfer in biofilms: a perspective on limitations and opportunities. NPJ Biofilms Microbiomes 2:16022. doi: 10.1038/npjbiofilms.2016.22
Stephens, M. D., Yodsanit, N., and Melander, C. (2016). Evaluation of ethyl N-(2-phenethyl) carbamate analogues as biofilm inhibitors of methicillin resistant Staphylococcus aureus. Org. Biomol. Chem. 14, 6853–6856. doi: 10.1039/C6OB00706F
Stevens, A. M., Queneau, Y., Soulere, L., Bodman, S. V., and Doutheau, A. (2010). Mechanisms and synthetic modulators of AHL-dependent gene regulation. Chem. Rev. 111, 4–27. doi: 10.1021/cr100064s
Storz, M. P., Maurer, C. K., Zimmer, C., Wagner, N., Brengel, C., de Jong, J. C., et al. (2012). Validation of PqsD as an anti-biofilm target in Pseudomonas aeruginosa by development of small-molecule inhibitors. J. Am. Chem. Soc. 134, 16143–16146. doi: 10.1021/ja3072397
Strahl, H., and Errington, J. (2017). Bacterial membranes: structure, domains, and function. Annu. Rev. Microbiol. 71, 519–538. doi: 10.1146/annurev-micro-102215-095630
Sully, E. K., Malachowa, N., Elmore, B. O., Alexander, S. M., Femling, J. K., Gray, B. M., et al. (2014). Selective chemical inhibition of agr quorum sensing in Staphylococcus aureus promotes host defense with minimal impact on resistance. PLoS Pathog. 10:e1004174. doi: 10.1371/journal.ppat.1004174
Sunder, A. V., Utari, P. D., Ramasamy, S., van Merkerk, R., Quax, W., and Pundle, A. (2017). Penicillin V acylases from gram-negative bacteria degrade N-acylhomoserine lactones and attenuate virulence in Pseudomonas aeruginosa. Appl. Microbiol. Biotechnol. 101, 2383–2395. doi: 10.1007/s00253-016-8031-5
Suneby, E. G., Herndon, L. R., and Schneider, T. L. (2017). Pseudomonas aeruginosa LasR· DNA binding is directly inhibited by quorum sensing antagonists. ACS Infect. Dis. 3, 183–189. doi: 10.1021/acsinfecdis.6b00163
Surewaard, B. G. J., Thanabalasuriar, A., Zeng, Z., Tkaczyk, C., Cohen, T. S., Bardoel, B. W., et al. (2018). alpha-toxin induces platelet aggregation and liver injury during Staphylococcus aureus sepsis. Cell Host Microbe 24, 271–284.e73. doi: 10.1016/j.chom.2018.06.017
Tabernero, L., Rodwell, V. W., and Stauffacher, C. V. (2003). Crystal structure of a statin bound to a class II HMG-CoA reductase. J. Biol. Chem. 278, 19933–19938. doi: 10.1074/jbc.M213006200
Tal-Gan, Y., Ivancic, M., Cornilescu, G., Yang, T., and Blackwell, H. E. (2016). Highly stable, amide-bridged autoinducing peptide analogues that strongly inhibit the AgrC quorum sensing receptor in Staphylococcus aureus. Angewandte Chem. 128, 9059–9063. doi: 10.1002/ange.201602974
Tam, J., Beilhartz, G. L., Auger, A., Gupta, P., Therien, A. G., and Melnyk, R. A. (2015). Small molecule inhibitors of clostridium difficile toxin B-induced cellular damage. Chem. Biol. 22, 175–185. doi: 10.1016/j.chembiol.2014.12.010
Tan, J., Patil, P. C., Luzzio, F. A., and Demuth, D. R. (2018). In vitro and in vivo activity of peptidomimetic compounds that target the periodontal pathogen Porphyromonas gingivalis. Antimicrob. Agents Chemother. 62:e00400–18. doi: 10.1128/AAC.00400-18
Tang, F., Li, L., Meng, X., Li, B., Wang, C.-Q., Wang, S.-Q., et al. (2018). Inhibition of alpha-hemolysin expression by resveratrol attenuates Staphylococcus aureus virulence. Microb. Pathog. 127, 85–90. doi: 10.1016/j.micpath.2018.11.027
Tang, K., Su, Y., Brackman, G., Cui, F., Zhang, Y., Shi, X., et al. (2015). MomL, a novel marine-derived N-acyl homoserine lactonase from Muricauda olearia. Appl. Environ. Microbiol. 81, 774–782. doi: 10.1128/AEM.02805-14
Tareen, A. M., Lüder, C. G., Zautner, A. E., Groß, U., Heimesaat, M. M., Bereswill, S., et al. (2013). The Campylobacter jejuni Cj0268c protein is required for adhesion and invasion in vitro. PLoS ONE 8:e81069. doi: 10.1371/journal.pone.0081069
Teng, Z., Shi, D., Liu, H., Shen, Z., Zha, Y., Li, W., et al. (2017). Lysionotin attenuates Staphylococcus aureus pathogenicity by inhibiting α-toxin expression. Appl. Microbiol. Biotechnol. 101, 6697–6703. doi: 10.1007/s00253-017-8417-z
Thanissery, R., Zeng, D., Doyle, R. G., and Theriot, C. M. (2018). A small molecule-screening pipeline to evaluate the therapeutic potential of 2-aminoimidazole molecules against clostridium difficile. Front. Microbiol. 9:1206. doi: 10.3389/fmicb.2018.01206
Thomann, A., de Mello Martins, A. G., Brengel, C., Empting, M., and Hartmann, R. W. (2016). Application of dual inhibition concept within looped autoregulatory systems toward antivirulence agents against Pseudomonas aeruginosa infections. ACS Chem. Biol. 11, 1279–1286. doi: 10.1021/acschembio.6b00117
Tobert, J. A. (2003). Lovastatin and beyond: the history of the HMG-CoA reductase inhibitors. Nat. Rev. Drug Discov. 2:517. doi: 10.1038/nrd1112
Todd, D. A., Parlet, C. P., Crosby, H. A., Malone, C. L., Heilmann, K. P., Horswill, A. R., et al. (2017). Signal biosynthesis inhibition with ambuic acid as a strategy to target antibiotic-resistant infections. Antimicrob. Agents Chemother. 61:e00263–17. doi: 10.1128/AAC.00263-17
Tseng, B. S., Reichhardt, C., Merrihew, G. E., Araujo-Hernandez, S. A., Harrison, J. J., MacCoss, M. J., et al. (2018). A biofilm matrix-associated protease inhibitor protects Pseudomonas aeruginosa from proteolytic attack. MBio. 9:e00543–18. doi: 10.1128/mBio.00543-18
Utari, P. D., Setroikromo, R., Melgert, B. N., and Quax, W. J. (2018). PvdQ quorum quenching acylase attenuates Pseudomonas aeruginosa virulence in a mouse model of pulmonary infection. Front. Cell. Infection Microbiol. 8:119. doi: 10.3389/fcimb.2018.00119
Vale, P. F., McNally, L., Doeschl-Wilson, A., King, K. C., Popat, R., Domingo-Sananes, M. R., et al. (2016). Beyond killing can we find new ways to manage infection? Evol. Med. Public Health 2016, 148–157. doi: 10.1093/emph/eow012
van der Poll, T., and Opal, S. M. (2008). Host-pathogen interactions in sepsis. Lancet Infect. Dis. 8, 32–43. doi: 10.1016/S1473-3099(07)70265-7
Van Meervenne, E., Van Coillie, E., Kerckhof, F.-M., Devlieghere, F., Herman, L., De Gelder, L. S., et al. (2012). Strain-specific transfer of antibiotic resistance from an environmental plasmid to foodborne pathogens. Biomed Res. Int. 2012:834598. doi: 10.1155/2012/834598
Vasquez, J. K., Tal-Gan, Y., Cornilescu, G., Tyler, K. A., and Blackwell, H. E. (2017). Simplified AIP-II peptidomimetics are potent inhibitors of Staphylococcus aureus AgrC quorum sensing receptors. Chembiochem 18, 413–423. doi: 10.1002/cbic.201600516
Wang, J., Nong, X.-H., Amin, M., and Qi, S.-H. (2018a). Hygrocin C from marine-derived Streptomyces sp. SCSGAA 0027 inhibits biofilm formation in Bacillus amyloliquefaciens SCSGAB0082 isolated from South China Sea gorgonian. Appl. Microbiol. Biotechnol. 102, 1417–1427. doi: 10.1007/s00253-017-8672-z
Wang, X., Thompson, C. D., Weidenmaier, C., and Lee, J. C. (2018b). Release of Staphylococcus aureus extracellular vesicles and their application as a vaccine platform. Nat. Commun. 9:1379. doi: 10.1038/s41467-018-03847-z
Waters, C. M., and Bassler, B. L. (2005). Quorum sensing: cell-to-cell communication in bacteria. Annu. Rev. Cell Dev. Biol. 21, 319–346. doi: 10.1146/annurev.cellbio.21.012704.131001
Watters, C., Fleming, D., Bishop, D., and Rumbaugh, K. (2016). “Host responses to biofilm,” in Progress in Molecular Biology and Translational Science, eds M, San Francisco, B. San Francisco (Elsevier), 193–239.
Wei, X., Gao, J., Wang, F., Ying, M., Angsantikul, P., Kroll, A. V., et al. (2017). In situ capture of bacterial toxins for antivirulence vaccination. Adv. Mater. 29:1701644. doi: 10.1002/adma.201701644
Welsh, M. A., Eibergen, N. R., Moore, J. D., and Blackwell, H. E. (2015). Small molecule disruption of quorum sensing cross-regulation in Pseudomonas aeruginosa causes major and unexpected alterations to virulence phenotypes. J. Am. Chem. Soc. 137, 1510–1519. doi: 10.1021/ja5110798
WHO (2018). Antibiotic Resistance [Online]. Available: online at http://www.who.int/news-room/fact-sheets/detail/antibiotic-resistance (Accessed 16 september 2018).
Wilding, E. I., Brown, J. R., Bryant, A. P., Chalker, A. F., Holmes, D. J., Ingraham, K. A., et al. (2000a). Identification, evolution, and essentiality of the mevalonate pathway for isopentenyl diphosphate biosynthesis in gram-positive cocci. J. Bacteriol. 182, 4319–4327. doi: 10.1128/JB.182.15.4319-4327.2000
Wilding, E. I., Kim, D.-Y., Bryant, A. P., Gwynn, M. N., Lunsford, R. D., McDevitt, D., et al. (2000b). Essentiality, expression, and characterization of the class II 3-hydroxy-3-methylglutaryl coenzyme A reductase of Staphylococcus aureus. J. Bacteriol. 182, 5147–5152. doi: 10.1128/JB.182.18.5147-5152.2000
Xiong, Y. Q., Willard, J., Yeaman, M. R., Cheung, A. L., and Bayer, A. S. (2006). Regulation of Staphylococcus aureus α-toxin gene (hla) expression by agr, sarA and sae in vitro and in experimental infective endocarditis. J. Infect. Dis. 194, 1267–1275. doi: 10.1086/508210
Xuewen, H., Ping, O., Zhongwei, Y., Zhongqiong, Y., Hualin, F., Juchun, L., et al. (2018). Eriodictyol protects against Staphylococcus aureus-induced lung cell injury by inhibiting alpha-hemolysin expression. World J. Microbiol. Biotechnol. 34:64. doi: 10.1007/s11274-018-2446-3
Yadav, M. K., Go, Y. Y., Chae, S.-W., and Song, J.-J. (2015). The small molecule dam inhibitor, pyrimidinedione, disrupts Streptococcus pneumoniae biofilm growth in vitro. PLoS ONE 10:e0139238. doi: 10.1371/journal.pone.0139238
Yang, T., Tal-Gan, Y., Paharik, A. E., Horswill, A. R., and Blackwell, H. E. (2016). Structure–function analyses of a Staphylococcus epidermidis autoinducing peptide reveals motifs critical for AgrC-type receptor modulation. ACS Chem. Biol. 11, 1982–1991. doi: 10.1021/acschembio.6b00120
Yepes, A., Schneider, J., Mielich, B., Koch, G., García-Betancur, J. C., Ramamurthi, K. S., et al. (2012). The biofilm formation defect of a B acillus subtilis flotillin-defective mutant involves the protease FtsH. Mol. Microbiol. 86, 457–471. doi: 10.1111/j.1365-2958.2012.08205.x
Zasloff, M. (2002). Antimicrobial peptides of multicellular organisms. Nature 415, 389–395. doi: 10.1038/415389a
Zhang, B., Teng, Z., Li, X., Lu, G., Deng, X., Niu, X., et al. (2017a). Chalcone attenuates Staphylococcus aureus virulence by targeting sortase A and alpha-hemolysin. Front. Microbiol. 8:1715. doi: 10.3389/fmicb.2017.01715
Zhang, Y., Brackman, G., and Coenye, T. (2017b). Pitfalls associated with evaluating enzymatic quorum quenching activity: the case of MomL and its effect on Pseudomonas aeruginosa and Acinetobacter baumannii biofilms. PeerJ 5:e3251. doi: 10.7717/peerj.3251
Zhang, Y., Sass, A., Van Acker, H., Wille, J., Verhasselt, B., Van Nieuwerburgh, F., et al. (2018). Coumarin reduces virulence and biofilm formation in Pseudomonas aeruginosa by affecting quorum sensing, type III secretion and c-di-GMP levels. Front. Microbiol. 9:1952. doi: 10.3389/fmicb.2018.01952
Zivkovic, A., Sharif, O., Stich, K., Doninger, B., Biaggio, M., Colinge, J., et al. (2011). TLR 2 and CD14 mediate innate immunity and lung inflammation to staphylococcal Panton-Valentine leukocidin in vivo. J. Immunol. 186, 1608–1617. doi: 10.4049/jimmunol.1001665
Zuberi, A., Misba, L., and Khan, A. U. (2017). CRISPR interference (CRISPRi) inhibition of luxS gene expression in E. coli: an approach to inhibit biofilm. Front. Cell. Infection Microbiol. 7:214. doi: 10.3389/fcimb.2017.00214
Keywords: anti-virulence therapy, antibiotic resistance, bacterial membrane microdomains, quorum sensing, biofilms, bacterial toxins
Citation: Fleitas Martínez O, Cardoso MH, Ribeiro SM and Franco OL (2019) Recent Advances in Anti-virulence Therapeutic Strategies With a Focus on Dismantling Bacterial Membrane Microdomains, Toxin Neutralization, Quorum-Sensing Interference and Biofilm Inhibition. Front. Cell. Infect. Microbiol. 9:74. doi: 10.3389/fcimb.2019.00074
Received: 11 October 2018; Accepted: 05 March 2019;
Published: 02 April 2019.
Edited by:
Natalia V. Kirienko, Rice University, United StatesReviewed by:
Michael L. Vasil, School of Medicine, University of Colorado Denver, United StatesCopyright © 2019 Fleitas Martínez, Cardoso, Ribeiro and Franco. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Octavio Luiz Franco, b2NmcmFuY29AZ21haWwuY29t
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.