- Department of Biotechnology, Panjab University, Chandigarh, India
Rv1288, a conserved hypothetical protein of M. tuberculosis (M.tb), was recently characterized as two-domain esterase enzyme by in silico study. In the present study, Rv1288 and its domains (Est and Lyt) were cloned individually from M.tb into E. coli for expression and purification. The purified rRv1288 and rEst proteins exhibited lipolytic activity with medium chain length esters as optimum substrates, while Lyt domain did not show enzymatic activity. However, presence of Lyt domain resulted in enhanced rate of protein aggregation at higher temperature. Both rRv1288 and rEst followed the similar patterns of substrate specificity, temperature and pH activity. Site directed mutagenesis confirmed the Ser-294, Asp-391 and His-425 as catalytic site residues. Rv1288 was found to be present in cell wall fraction of M.tb H37Ra. Peptidoglycan binding activity of Rv1288 and its domains demonstrated that the Lyt domain is essential for anchoring protein to the cell wall. Expression of rv1288 was up regulated in M.tb under nutrient starved condition. Over expression of rv1288 in surrogate host M. smegmatis led to change in colony morphology, enhanced pellicle and aggregate formation that might be linked with the changed lipid composition of bacterial cell wall. Cell wall of M. smegmatis expressing rv1288 had higher amount of lipids, with a significant increase in trehalose dimycolate content. Rv1288 also leads to increase in drug resistance of M. smegmatis. Rv1288 also enhanced the intracellular survival of M. smegmatis in Raw264.7 cell line. Overall, this study suggested that Rv1288, a cell wall localized carboxyl hydrolase with mycolyl-transferase activity, modulated the cell wall lipids to favor the survival of bacteria under stress condition.
Introduction
Tuberculosis (TB) plagued humankind since the beginning of medical history. Even today, it is the leading killer of human beings globally from any infectious disease. According to WHO global tuberculosis report 2017, approximately 6.3 million new cases and 1.6 million deaths from TB were reported in 2016, across the world (WHO, 2016). The gravity of the situation had worsened due to poorly understood biology, pathogenesis and long dormancy stage of causative organism Mycobacterium tuberculosis (M.tb), undiagnosed primary and re-infection, reasonably asymptomatic condition, limited anti-TB drugs, long duration treatment resulting in poor compliance by patients, the emergence of drug-resistant strains and co-infection with HIV (Ford et al., 2015; Petersen et al., 2017; Queiroz and Riley, 2017). Hence new, effective and cheap drugs active against active as well as latent TB, compatible with antiretroviral therapy, are the need of the hour (Mahamed et al., 2017). Under these circumstances identification of new drug targets, which might be explored to develop new anti TB drugs, are urgently needed.
Lipids and lipid metabolism played significant role in infection, intracellular survival and pathogenicity of M.tb. Cell wall lipids were reported to interact with host resulting in modulation of metabolism and immune response, regulation of transport of drugs, nutrients and other molecules. During infection, M.tb stored lipids in the form of triacylglycerol, and hydrolyzed them for nutrition and energy requirements during latent and reactivation stage (Lovewell et al., 2016; Barisch and Soldati, 2017; Queiroz and Riley, 2017; Gago et al., 2018). Approximately 6% of M.tb genome encoded for the lipid metabolism enzymes (Lew et al., 2011). M.tb had 26 genes, encoding for putative lipases (Lip A to Lip Z), classified as lip gene family (Cole et al., 1998). However, other than these, many other proteins, possessing the typical characteristics of lipase/esterase enzymes (α/β serine hydrolase fold and conserved penta-peptide sequence) were identified bioinformatically. Though α/β serine hydrolase fold is possessed by many other enzyme families such as acetylcholineesterase, thioesterase, proline iminopeptidase/oligopeptidase, serine carboxylpeptidase, eposide hydrolase and many more (Holmquist, 2000), several of these proteins demonstrated lipolytic activity contrary to their annotated function (Lun and Bishai, 2007; Anand et al., 2011; Kumar et al., 2017c). The possible role of several lipases/esterases in the life of M.tb had been investigated extensively till date. These were involved in the various functions such as synthesis and maintenance of M.tb unique cell envelop, degradation of host cell lipids, xenobiotic degradation, toxic epoxide degradation, evasion and modulation of host immune system, elicit humoral response, dentritic cell maturation and antigen presentation, adaptation during adverse conditions like dormancy and acidification (Deb et al., 2006; Xu et al., 2010; Madan-Lala et al., 2014; Singh et al., 2014c, 2016b; Jadeja et al., 2016; Barisch and Soldati, 2017; Chownk et al., 2017; Johnson, 2017; Kaur et al., 2017a; Kumar et al., 2017b; Vemula et al., 2018). Such variety of functions were possible due to the fact that, these mycobacterial lipolytic enzymes possess many additional activities also like β lactamase, Mycolyltransferase, TDM hydrolase, Eposide Hydrolase, Phospholipase, etc. (Zhang et al., 2005; Singh et al., 2016a; Chownk et al., 2017; Queiroz and Riley, 2017). One such activity is mycolyltransferase and TDM hydrolase activity. They are the most well-characterized member of mycobacterial alpha/beta hydrolases, both biochemically and genetically. Yang et al. (2014) reported Rv3451 as TDM hydrolase, which remodels the M.tb cell wall to enhance the nutrient flux, and increased its intracellular survival in MyD88-/- mice (Yang et al., 2014). A cutinase like serine carboxylesterase Msmeg_1529 acts as TDM hydrolase in M. smegmatis (Ojha et al., 2010). Therefore, these enzymes could be used as drug targets for developing new anti-mycobacterial drugs (Gago et al., 2018).
Approximately 4,000 genes were identified in M.tb genome, but still around 1,100 genes had unknown function and were classified under unknown or conserved hypothetical proteins category. Some of these hypothetical genes were also presumed to encode for lipolytic enzymes (Kumar et al., 2017d). Recently several studies were carried out to elucidate the role of such hypothetical proteins such as Rv2224c (Lun and Bishai, 2007; Rengarajan et al., 2008; Naffin-Olivos et al., 2017) and Rv0774c (Kumar et al., 2017c,e). However, lots of information was still buried in these hypothetical proteins. Therefore, to completely understand the physiology of M. tb, there is a need to find out the role of these unknown/uncharacterized gene products.
One such protein Rv1288, a 456 amino acid protein, was categorized as conserved hypothetical protein with probable protease activity in tuberculist database. This protein shared 26–31% sequence identity with the Antigen 85 complex (FbpA, FbpB, and FbpC) (Takayama et al., 2005). Homologous proteins of Rv1288 were found in other mycobacterium species such as M. smegmatis, M. bovis, M. avium, and M. marinum based on sequence alignment analysis. Rv1288 was composed of two domains, first LytE domain at N-terminal, consisted of three consecutive LysM motifs and second esterase domain at C- terminal belongs to esterase D family (Doerks et al., 2012). Kumar et al., performed the bioinformatics analysis by homology modeling and molecular docking to study the probable role of this protein in M.tb. It was suggested as a probable lipolytic enzyme by in silico experiments (Takayama et al., 2005; Kumar et al., 2017a). The LytE domain was predicted to be responsible for the binding of this protein with mycobacterial cell wall. Esterase domain showed the presence of HGGG tetrapeptide motif, GxSxG pentapeptide sequence and S294, D391 and H425 as catalytic triad. Further docking studies predicted the high binding affinity of serine hydrolase inhibitors such as THL and PMSF with this enzyme (Kumar et al., 2017a). However, all these properties were annotated on the basis of bioinformatics and prediction methods and needed to be validated by wet lab experiments. Therefore, in the present study, full length Rv1288 and its domains were cloned and expressed in E. coli in order to purify and characterized them. We analyzed the expression of Rv1288 under different stress conditions. Effect of Rv1288 on different cell surface properties was also explored to elucidate the functional role of Rv1288 in physiology of M. tb.
Materials and Methods
Reagents, Chemicals, Bacterial Strains, and Vectors
All reagents and chemicals were purchased from Hi-Media laboratories (India), and Sigma- Aldrich (USA). All the bacterial strains were procured from IMTECH-MTCC, Chandigarh. E. coli DH5α and E. coli BL21 DE3 strains were grown in Luria Broth (LB media) at 37°C and 180 rpm. LB media supplemented with kanamycin (30 μg/ml) and chloramphenicol (35 μg/ml) antibiotics was used for growth of recombinant strains of E. coli. M.tb H37Ra and M. smegmatis mc2155 were grown in M7H9 media (Sigma Aldrich, USA) supplemented with 1% glycerol, 0.05% Tween-80 and 10% Oleic acid- albumin-dextrose-catalase (OADC) (BD Bioscience), at 37°C and 180 rpm. For growth of M. smegmatis recombinant strains, kanamycin and hygromycin were added to M7H9 media at a concentration of 50 μg/ml and 100 μg/ml.
Expression of rv1288 in M. tuberculosis H37Ra Under Various Stress Conditions
M.tb H37Ra culture was inoculated in M7H9 media and grown at 37°C, 180 rpm up to mid log phase. Culture was centrifuged at 4,000 rpm for 10 min. Cell pellet was washed twice with 1X PBS (pH 7.4), re-suspended in stress media, such as acidic stress (M7H9 media, pH adjusted to 4.5 with HCl), nutrient stress (1X PBS, pH 7.4) and iron stress (using iron deficient media) (Kumar et al., 2017e) along with control culture (M7H9 media). All the samples were incubated at 37°C for 6 h. Total RNA was extracted from the samples using TRIzol Reagent (Sigma Aldrich, USA) according to manufacturer's guidelines along with cryogrinding. cDNA was prepared from equal amount of RNA of all samples by using Revert Aid first strand synthesis kit (Thermofisher, USA). Quantitative real time PCR (qRT-PCR) (Applied Biosystem® Step OneTM real time PCR) was performed to quantify the expression of rv1288 in each sample using SYBR Green/ROX qPCR master mix (Fermantas). 16S rRNA was used as an internal control in each set of reactions. Fold change in expression in different samples were calculated by 2−ΔΔCt. rv3203, rv3097c, and rv560c were used as positive control for acidic, nutritive and iron stress conditions, respectively (Rodriguez et al., 2002; Deb et al., 2006; Singh et al., 2014b). The specificity of results was validated by melting curve analysis.
Cloning, Expression, and Purification of rv1288, est, and lyt Domain
For cloning of rv1288, its domains est and lyt gene sequences were amplified from genomic DNA of M.tb H37Rv (kind gift from Dr. U.D. Gupta, JALMA, Agra, India) using designated set of primers (Table S1). Amplified products were excised and eluted from the gel. PCR product and pET28a expression vector were double digested with restriction enzyme (BamHI and HindIII. Digested gene fragments were ligated with vector by T4 DNA ligase (Invitrogen, USA). Ligated products were used to transform E. coli DH5α competent cells. Transformed cells were confirmed by sequencing (Xcelris Labs Ltd., India). Positive recombinant plasmids were transformed into the E. coli BL21 DE3 cells. All the cultures were grown overnight in 10 ml LB medium containing kanamycin. These overnight grown cultures were used to inoculate 250 ml of LB media. Bacterial cells were grown at 37°C, 180 rpm, until absorbance (600 nm) reached 0.4–0.6 i.e., mid log phase. All the cultures were induced with IPTG (0.05 mM) at 15°C, 180 rpm for 16–20 h. Cultures were centrifuged at 10,000 rpm for 20 min at 4°C. Cell pellet were suspended in 30 ml of cell lysis buffer (50 mM Sodium phosphate buffer, pH 8.0, 150 mM NaCl, 0.2% (v/v) triton-X 100). Cell lysates were further subjected to sonication (21% amplitude, 10 sec on and off pulse) for 10 min with intermittent cooling by Misonix Sonicator (Qsonia, USA). Cell debris were removed by centrifugation at 7,000 rpm for 20 min. Ni NTA column was equilibrated with equilibration buffer [sodium phosphate buffer (50 mM, pH 8.0), NaCl (300 mM), imidazole (20 mM)]. After equilibration, protein (supernatant collected by centrifugation) was passed through the column. Unbound or loosely bound proteins were washed from the column by passing 10 volume wash buffer [sodium phosphate buffer (50 mM, pH 8.0), NaCl (50 mM), imidazole (50 mM)]. Elution of bound protein was carried out by elution buffer [sodium phosphate buffer (50 mM, pH 8.0), NaCl (300 mM), imidazole (300 mM)] in 1 ml protein fractions. Absorbance at 280 nm was measured to find out the amount of protein in each fraction. All the fractions containing protein were pooled and dialyzed overnight in 50 mM phosphate buffer. The integrity and purity of protein was analyzed on 10% SDS-PAGE. Protein concentration was estimated by Bicinchoninic acid (BCA) method (kit by Banglore Genei, India).
Skimmed Milk Plate Assay
Skimmed milk plate assay were used for the qualitative determination of protease activity.
Skimmed milk (2%) agar plates were prepared in 50 mM sodium phosphate buffer (pH 8.0) and wells were punched (Brown and Foster, 1970; Drake and Montie, 1988). Hundred microliter of enzyme (50 μg/ml) and control buffer (100 μl) were added to the wells. Plates were incubated at 37°C for overnight. Trypsin (50 μg/ml) was used as positive control. Zone of hydrolysis around the well would indicate the protease activity of test protein.
Tributyrin Plate Assay
The esterase activity of proteins was investigated by tributyrin plate assay method. Tributyrin emulsion (1%) agar plates were prepared in 50 mM phosphate buffer (pH 8.0). Assay was performed by adding 100 μl each of enzymes (50 μg/ml) and buffer as control in the wells. Plates were incubated at 37°C for overnight for developing zone of clearance.
Enzyme Assay
Enzymatic assays were carried out using pNP (para nitro phenyl)- derivative esters with little modifications. Reaction mixture was prepared by adding 700 μl sodium phosphate buffer (50 mM, pH 8.0), 100 μl protein (50 μg/ml), 100 μl sodium deoxycholate (10 mM), and 100 μl substrate (2 mM, pNP ester). Blank reaction mixture contained all components except for the protein. The reaction mix was incubated for 15 min at optimum temperature. After incubation 250 μl of Na2CO3 (0.1 M) was added to inhibit the reaction. Hydrolysis product was estimated by measuring absorbance at 420 nm was measured using in UV/Vis spectrophotometer (GE Healthcare, UK). One unit of enzyme activity was defined as the amount of enzyme, required to release 1 μmole of para-nitrophenol from substrate in 1 min under standard lipolytic enzyme assay conditions. Specific activity was expressed in U/mg of protein.
Substrate Specificity
The substrate specificity of rRv1288 and rEst proteins was determined by using different carbon chain length (C2, C4, C8, C10, C12, C14, C16, and C18) pNP esters as substrate in standard enzyme assay conditions. Stock solutions of substrates were prepared in absolute ethanol (10 mM stock).
Biochemical Characterization of rRv1288 and rEst
To determine the optimum temperature for enzyme activity, reaction mixture was incubated at temperature ranging from 20 to 80°C. To study the effect of pH, enzyme assay was performed in different pH buffers (pH 4.0 to 12.0) (Kumar et al., 2017e) and reaction mix was incubated at optimum temperature. Relative enzyme activity at various temperature/ pH was calculated by taking the maximum enzyme activity as 100%.
To find out the effect of temperature on stability of proteins, purified proteins (50 μg/ml) were incubated at different temperatures (20–80°C) for 1 h, and the cooled on ice for 10 min. Similarly, purified proteins were incubated in different pH buffer (pH 4.0 to 12.0) for 1 h at room temperature (RT). After incubation time, standard lipolytic enzyme assay was performed at optimum temperature and ph. Residual activity of enzymes was calculated by taking activity of un-incubated enzyme as 100%.
Enzyme assay was performed with different concentrations of pNP- caprylate (0.1–1 mM) to study the effect of substrate on rate of enzyme reaction. Lineweaver-Burk plot was drawn to calculate the Michaelis–Menten constant (Km), maximum velocity for the reaction (Vmax), kcat and kcat/Km.
Effect of Phenylmethanesulfonylfluride (PMSF)
The purified rRv1288 and rEst proteins were incubated with PMSF (5 mM) at RT for 1 h. Standard enzyme assay was performed and residual enzyme activity of incubated proteins was calculated as compared to control (untreated proteins). The reaction mix without enzyme, but with PMSF, served as blank.
Site Direct Mutagenesis
To confirm the predicted catalytic residues, Ser-294-Ala, Asp-391-Ala, and His-425-Ala mutants of Rv1288 were created by overlapping extension PCR. Primers used for the mutagenesis were mentioned in Table S1. Mutations were confirmed by gene sequencing. All the mutants were cloned, expressed, and purified according to the protocol used for wild type Rv1288 protein. Enzyme activities of mutants and wild type proteins were determined by standard lipolytic enzyme assay. Residual activity was calculated by taking enzyme activity of wild type protein as 100%.
Biophysical Characterization
For biophysical studies, CD spectra of both the proteins were obtained (JASCO J-815 spectrophotometer, Japan) at 190–260 nm wavelength using 2 mM path length cuvette at RT. The negative ellipticity was expressed as [θ], degcm2 dmol−1. Secondary structure elements were estimated quantitatively by spectra software using Yang's reference. Purified proteins (100 μg) were incubated at different temperature (20 to 90°C) for 10 min and spectra were collected at 190–260 nm wavelengths at 1 nm band width. Buffer served as blank for each spectrum.
To determine the intrinsic fluorescence, fluorescence spectra of both the proteins were obtained (JASCO J-815 spectro fluorometer, Japan) using 10 mm path length quartz cuvette at excitation wavelength 295 nm (5 nm slit width). Emission was recorded at wavelength 310–400 nm (10 nm slit width). The spectra of proteins pre-incubated (10 min) at different temperature (20 to 90°C) were also collected. Buffer served as blank for each spectrum.
Subcellular Localization of Rv1288
The mid log phase culture of M. tb H37Ra was harvested and fractionated. Bacterial cells were centrifuged at 10,000 g for 30 min at 4°C. Extracellular proteins present in culture supernatant were precipitated with 80% ammonium sulfate saturation and dialyzed against 1 X PBS and used as culture filtrate fraction. Cell pellet was washed twice with PBS, and further lysed by sonication in PBS (10 × of 60 s cycle with 2 min cooling period). Cell lysate were centrifuged twice at 4,000 × g for 10 min to remove the unlysed cells. Supernatent was further centrifuged at 27,000 × g for 1 h at 4°C to separate the cytosolic fractions (supernatant) and cell wall fractions (pellet) (Rezwan et al., 2007).
Raising Antibody Against rRv1288
Purified rRv1288 was used to raise polyclonal antibodies in rabbit against the protein. Appropriate ethical clearance was taken from Institutional Ethics Committee, Panjab University, Chandigarh. Protein (200 μg) was emulsified with Freund's complete antigen (1:1 ratio) for first dose and with Freund's incomplete antigen (1:1 ratio) for two booster doses. Two weeks old rabbit were acclimatized in animal house conditions for 2 weeks. At day 1, blood was obtained from rabbit via orbital bleeding method (puncturing marginal ear vein) for control sample. At day 1, first dose was administered by subcutaneous injection. At day 14 and day 28, first and second booster doses were given, respectively. At day 33, blood sample was obtained and kept at 37°C for 1 h for the blood clotting. Blood was centrifuged at 4,000 rpm at 4°C for 15 min. The upper transparent layer of serum was carefully separated and stored at −20°C. Serum was checked for the presence of antibodies by dot blot experiment. Cross reactivity of Rv1288 IgG antibody was checked with the bacillus lipase, LipV and Lip N of M. tuberculosis.
The presence of Rv1288 protein in different fractions was identified by western blot using rabbit generated anti Rv1288 IgG (1:2,000) primary antibody, followed by alkaline phosphate conjugated anti rabbit IgG secondary antibody (1: 5,000) with intermittent washing. Anti-GroEL IgG (1:5,000) and Anti-antigen 85 complex (1:5,000) generated in rabbit were used as marker to check the purity of fractions. BCIP-NBT substrate (Chromus Biotech, India) was used to develop the blot.
Peptidoglycan Binding Assay
Peptidoglycan binding assay was performed according to the modified protocol from (Yoshida et al., 1996). Briefly, 1 mg of Bacillus peptidoglycans (Sigma Co., USA) were dispersed in 1 ml of Tris binding buffer (20 mM Tris-Cl buffer, pH 6.8, and 150 mM NaCl). Peptidoglycans were washed twice with 1 ml of Tris binding buffer by centrifugation and sedimentation at 12,000 g for 5 min. The sedimented peptidoglycans were resuspended in 500 μl of Tris binding buffer. 100 μl of each protein (250 μg/ml) was mixed with 60 μl of Tris binding buffer and 160 μl of peptidoglycan suspension. Mixture was incubated at 4°C for 30 min and centrifuged for 5 min at 12,000 g. The peptidoglycan pellets were transferred to fresh microcentrifuge tubes and washed with 300 μl of Tris binding buffer. The pellet of purified rRv1288 without peptodoglycan was used as control to rule out the possibility of getting some denatured or aggregated protein in pellet. The supernatant and pellet fraction were subjected to SDS-PAGE.
Cloning and Expression of rv1288, est, and lyt in M. smegmatis
For cloning in M. smegmatis, all the recombinant pET28a plasmid (rv1288, est, lyt, and S294A mutant) and pVV16 E. coli-Mycobacterium shuttle vector were double digested with restriction enzymes (BamHI and HindIII). The digested gene fragments from pET28a were ligated with the digested pVV16 vector and used to transform E. coli DH5α cells. Transformants were screened for the presence of insert by colony PCR and plasmid migration, followed by confirmation by sequence analysis. Recombinant plasmids were electroporated into competent M. smegmatis mc2155 cells at 25 μF and 2.5 kV for 5 ms in Gene pulser, (BioRad, USA) followed by immediate addition of M7H9 media. After electroporation, cells were grown at 37°C, 180 rpm for 6 h, followed by spreading on M7H10 agar plates supplemented with kanamycin and hygromycin. Plates were incubated at 37°C for 5–6 days.
For localization, MS_Est and MS_Lyt (recombinant M. smegmatis strains cloned with Est and Lyt domain, respectively) were fractionated using same protocol as mentioned above. Western blot was performed using mice anti-His IgG (1:5,000) as primary antibody and alkaline phosphate conjugated anti mice IgG (1:5,000) as secondary antibody with intermittent washing.
Colony Morphology and Growth Kinetics of MS_Vec and MS_1288
M. smegmatis expressing rv1288 (MS_1288) and vector alone (MS_Vec) were spread on M7H10 agar plates supplemented with 1% glycerol, 10% OADC, and 0.05% Tween-80. Plates were incubated at 37°C for 5–6 days. For the high resolution, microphotography of the colonies was carried out by Nikon Camera. For growth kinetics MS_Vec and MS_1288 were inoculated (from 1% overnight grown cultures) in 100 ml of M7H9 supplemented with 1% glycerol, 0.05% Tween-80, and 10% OADC and grown at 37°C, 180 rpm. Absorbance at 600 nm was measured and colony forming units (cfu) were determined at regular interval of 24 h.
Mycobacterial Cell Surface Properties: Pellicle and Aggregate Formation
For pellicle formation all the cultures were grown in 10 ml M7H9 medium with 1% glycerol, 10% OADC (without Tween-80) at 37°C without shaking. Cultures were photographed at interval of 24 h (Singh et al., 2016b). For aggregate formation all the cultures were grown in 10 ml M7H9 medium with 1% glycerol, 10% OADC (without Tween-80) at 37°C, 180 rpm for 24 and 48 h. Cell aggregates formed in both the cultures were allowed to settle down by placing the tubes at static conditions at RT and then photographs were captured.
Effect on Drug Resistance
MS_Vec and MS_1288 were also checked for the antibiotic sensitivity using resazurin colorimetric redox indicator test (Kumar et al., 2017c). Both the cultures were grown to mid log phase. Equal number of cells (2 × 105) were suspended in M7H9 media devoid of Tween-80 and added to the 48 well plates. Drug were diluted and added to the wells at different concentrations of rifampicin (1, 2, 4, 6, 8, 10 μg/ml), streptomycin (0.5, 1, 2, 4, 8, 16 μg/ml) and isoniazid (0.5, 1, 2, 4, 8, 16 μg/ml). Plates were incubated for 2 h at 37°C. Fresh working solution of resazurin was prepared by diluting the 10X stock (in 20% Tween-80) in 1:1 dilution. Eight microliterof working resazurin solution was added to each well. Plates were incubated at 37°C for 2 days. Plates were observed for change of blue to pink color at regular interval of time.
In another experiment, equal number of cells (2 × 105) were suspended in M7H9 media devoid of Tween-80 and added to the 48 well plates. Drug were diluted and added to the wells at different concentrations of rifampicin (1, 2, 4, 6, 8, 10 μg/ml), streptomycin (0.5, 1, 2, 4, 8, 16 μg/ml) and, isoniazid (0.5, 1, 2, 4, 8, 16 μg/ml). Plates were incubated for 2 h at 37°C. Cells were appropriately diluted and spread on the M7H10 plates supplemented with kanamycin and hygromycin for cfu count.
Lipid Composition of Cell Wall of MS_Vec and MS_1288
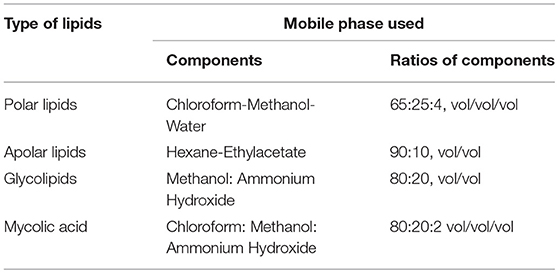
Both the recombinant M. smegmatis strains were grown to mid log phase. Cells were harvested by centrifugation at 4,000 rpm for 10 min. Total lipids were extracted from equal weights of mycobacterial wet pellets, by continuous stirring in 50 ml chloroform-methanol (2:1, v/v) at RT for overnight. This organic phase was evaporated in rotary evaporator to obtain the whole cell lipids in dried form. Amount of lipid content for both the cultures was measured by weighing and equal amount of both the lipids were resuspended in 100 μl of chloroform. Two microliters of each lipids were loaded on Silica gel 60 plates (Merck, USA) for separation of different types of lipids including polar, apolar lipids, glycolipids and mycolic acid containing glycolipids by thin layer chromatography (TLC) analysis on using different mobile phases as mentioned in Table 1. TDM (Sigma, 10 mg/ml) was used as control in mycolic acid TLC plates. Iodine staining was used to visualize the TLC plates.
Effect of Nutrition Stress on Survival of MS_Vec and MS_1288
MS_1288 and MS_Vec were grown in M7H9 medium till mid log phase. Cells were centrifuged at 2,000 g for 5 min and washed twice with 1X PBS (pH 7.4). Cell pellets were resuspended in 1X PBS and were incubated at 37°C for 24 h. For determination of cfu count, cells were serial diluted and plated onto M7H10 agar plates supplemented with kanamycin and hygromycin.
Effect on Growth of MS_1288 in Presence of Different Lipids
Equal number of cells of MS_1288 and MS_Vec were inoculated in 5 ml of different media[M7H9 medium supplemented with 1% glycerol, or 1% tributyrin (C4) or 1 mg/ml trilaurin (C12)]. All the tubes were incubated at 37°C, 180 rpm for 24 h. The cells growing on M7H9 media alone served as control. For determination of cfu count, cells were serial diluted and plated onto M7H10 agar plates supplemented with kanamycin and hygromycin.
Effect on Intracellular Survival
Murine macrophage cell line RAW 264.7 were maintained in Dulbecco's Modified Eagle's Medium (DMEM) containing 10% FBS, streptomycin (100 μg/ml), penicillin (100 IU/ml) and 1% L- Glutamine at 37°C in CO2 incubator (5% CO2). For intracellular survival, cells were seeded in 12 well (2 × 105 cell per well) surface treated culture plates in DMEM medium (without antibiotics). MS_Vec and MS_1288 were internalized into macrophage cell line with MOI values 10:1 ratio for 3 h at 37°C in 5% CO2. Afterwards, media was removed from the wells. Cells were washed twice with PBS. DMEM media with antibiotics was added to kill the extracellular bacteria and then cells were incubated for 1, 6, 12, 24, 48, and 72 h. At each time interval, cells were again washed twice with PBS. Cells were lysed with 100 μl lysis buffer (0.1% triton X 100 in PBS). The numbers of intracellular mycobacteria were enumerated by spreading suitable dilutions onto the M7H10 agar plates containing Kan+/Hyg+ antibiotics, to compare the ability of cultures to infect macrophages.
Results
Up-Regulation of rv1288 Expression in Nutritive Stress
Once inside the host, mycobacterium encountered various stress conditions. Expression of genes of mycobacteria changed during stress conditions (acidic, nutrient, and iron starvation)might help the adaptation and survival of bacilli inside host cells. Therefore, we investigated the expression of rv1288 in M.tb H37Ra grown under different in vitro stress environment and it was found to be up-regulated by 2.5 fold under nutrient depletion condition with respect to 16S rRNA reference gene. However, no significant change in expression of rv1288 was observed under acidic and iron stress conditions (Figure 1).
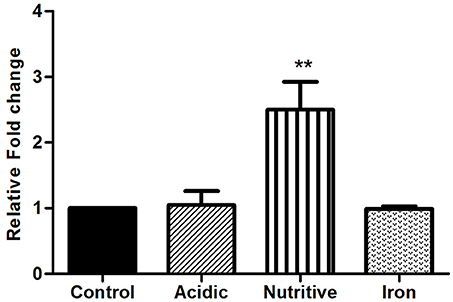
Figure 1. Expression analysis of rv1288 under different stress conditions by real time PCR. The mid log phase culture of M. tb H37Ra culture was exposed to acidic, nutritive and iron stress for 6 h. Relative mRNA expression of rv1288 was calculated with housekeeping gene 16S rRNA as internal control. All experiments were carried out three times in triplicate (n = 3) and error bars indicated standard deviation. Student's t-test was performed to show the statistical significance (**p < 0.01).
Comparative Esterase Activity of rRv1288 and its Domains
Expressions of all the three proteins (rRv1288, rEst domain, and rLyt domain) were optimized to get the expressed proteins in the soluble fraction and were purified by affinity chromatography (Figure 2A). All the proteins were purified to homogeneity. The purified rRv1288 did not show any proteolytic activity as no zone of clearance was observed on skimmed milk plate (Figure 2B). However, purified rRv1288 and rEst proteins showed clearance zone on emulsified tributyrin plate (Figure 2C), while no zone was observed with rLyt domain. The lipolytic activity of enzyme was further investigated with synthetic lipolytic substrates, pNP esters of different carbon chain length. Both the rRv1288 and rEst proteins exhibited hydrolytic activity, with pNP-caprylate (C8) as optimum substrate followed by pNP- capricate (C10) while no lipolytic activity was observed with rLyt protein. Increasing carbon chain length beyond C14 drastically decreased the enzyme activity of both the proteins (Figure 2D). Specific activity of rRv1288 and rEst were 96 ± 8 and 92 ± 5 U/mg, respectively with pNP-caprylate as substrate.
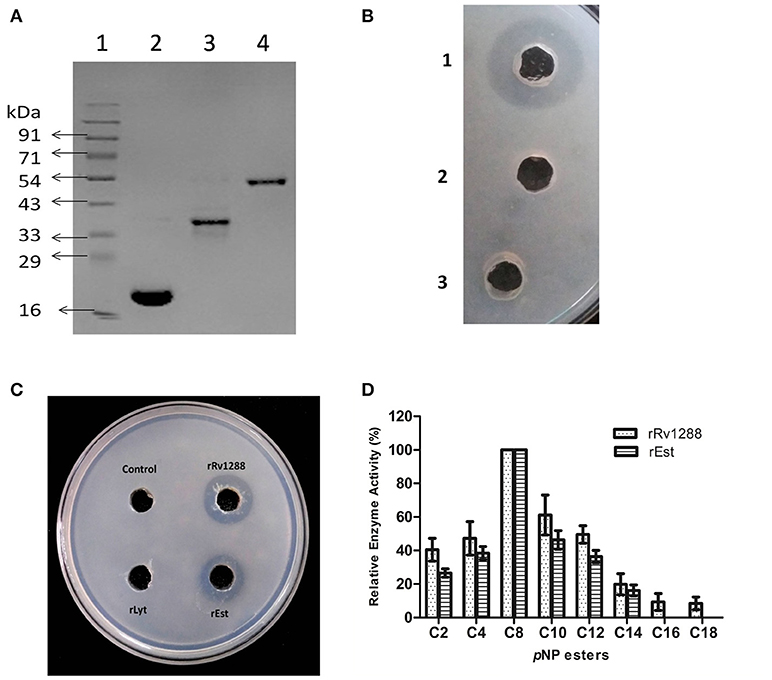
Figure 2. (A) SDS–PAGE analysis of purified rRv1288 and its domains. 1: Protein Marker, 2: Purified rLytE domain, 3: Purified esterase domain (rEst), 4: Purified full length Rv1288. (B) Skimmed milk plate for protease activity. 1 = Trypsin, 2 = 50 mM phosphate buffer and 3 = purified rRv1288. Trypsin was used as positive control. (C) Tributyrin plate assay of rRv1288 and its domains. 50 mM phosphate buffer was taken as control. (D) Substrate specificity of Rv1288 and Est domain toward different p-NP esters. C2: pNP-acetate, C4: pNP-butyrate, C8: pNP-caprylate, C10: pNP-capricate, C12: pNP-laurate, C14: pNP- myristate, C16: pNP-palmitate, C18: pNP- stearate. The enzyme activity with pNP-caprylate was taken as 100%. The values represent the means ± SD of three independent experiments.
Effect of Temperature and pH on Activity and Stability of rRv1288 and rEst Proteins
Optimum temperature and pH for activity of rRv1288 and rEst protein was observed to be 45°C and pH 9.0 (Figures 3A,C). Both the proteins displayed negligible activity and stability at higher temperature (70 and 80°C) and extreme pH (4.0, 5.0, 11.0, and 12.0) (Figures 3B,D). Thus, both the full length Rv1288 and esterase domain displayed similar biochemical characteristics. All parameters were almost similar for both the proteins suggesting that LytE domain did not alter the optimum temperature/pH for enzyme activity as well as stability of esterase domain.
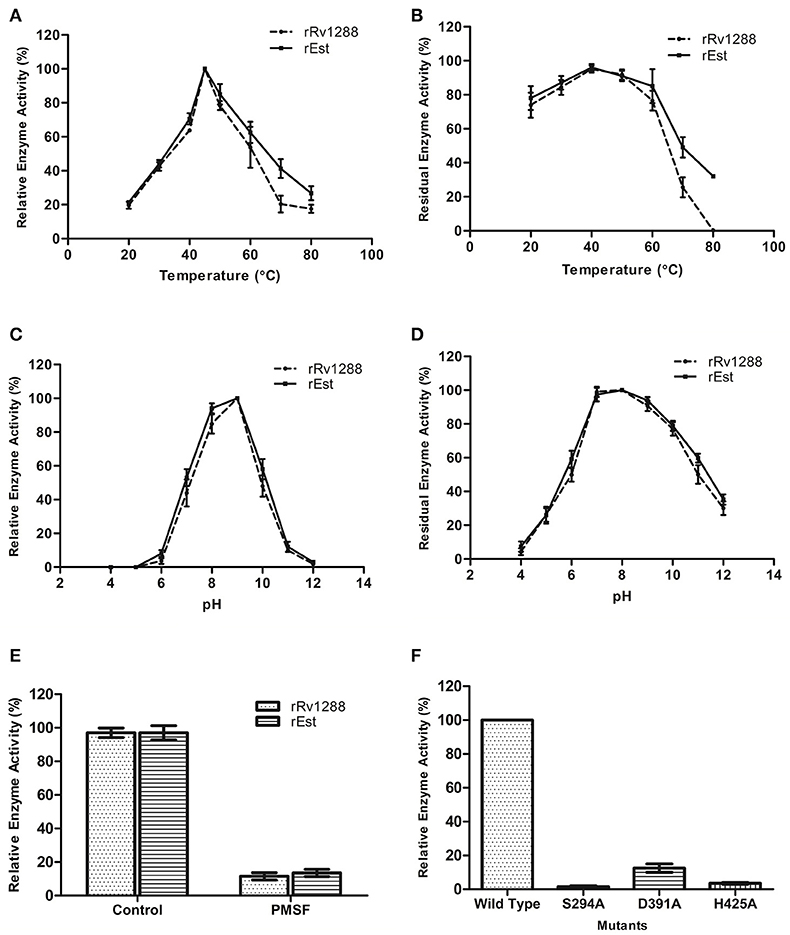
Figure 3. Biochemical characterization of rRv1288 and Est Domain. Effect of temperature (A) and pH (C) on enzyme activity. Effect of temperature and pH on enzyme stability (B,D) of rRv1288 (dashed line), and its Est domain (solid line). The relative enzyme activity were expressed as a percentage activity at pH 9.0 and 45°C. For checking stability, enzymes were incubated at respective pH and residual activity were calculated by taking the activity of purified enzymes (without incubation) as 100% in standard conditions. (E) Effect of PMSF on enzyme activity of rRv1288 and rEst proteins. (F) Relative enzyme activity of mutants (Ser294Ala, Asp391Ala, and His425Ala) in comparison to wild type Rv1288 protein. Results are representative of three independent biological replicates and shown as mean ± SD.
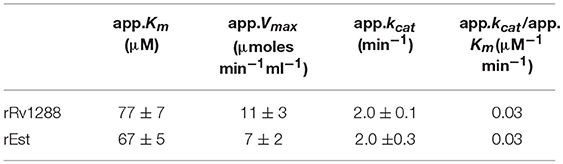
Kinetic Studies
Kinetic studies of rRv1288 and rEst were performed at different concentration of substrate ranging from 0.1 to 1 mM. The apparent Michaelis- Menten constant (Km) and maximum velocity (Vmax), turnover number (kcat) and catalytic efficiency (kcat / Km) were calculated from Lineweaver-Burk plot (Figure S1) and mentioned in Table 2. All parameters were almost similar for both the proteins suggesting that LytE domain did not alter the catalytic activity of full length protein.
Catalytic Residues for Enzyme Activity
Significant loss in esterase enzyme activity (90%) was observed with Ser modifier, PMSF for both rRv1288 and rEst proteins (Figure 3E). To validate the predicted catalytic residues S294A, D391A and H425A mutants of Rv1288 were generated. Complete loss of enzyme activity was observed in S294A and D425A mutants. However, D391A mutant lost nearly 85% the total enzyme activity as compared to wild type rRv1288 protein (Figure 3F).
Biophysical Studies
Presence of α/β hydrolase fold in rRv1288 and rEst proteins was confirmed by Far UV CD spectra (Figure 4A). The relative amount of secondary structure of rRv1288 was calculated to be 25% α-helix, 20.8% β-sheets, 25.4% turns, and 28.7% random coil, while, 21.4% α-helices, 25.4% β-sheets, 36.4% turns, and 16.8% random coil for rEst respectively. Both recombinant proteins demonstrated similar CD spectra pattern, with higher minima at 208 nm than at 222 nm. During the thermal unfolding studies of both the proteins, negative molar ellipticity reduced gradually with rise in temperature (Figure 4B). The helical conformation and minima at 222 nm of proteins were stable up to 50°C. Beyond this the minima was abruptly shifted, indicating the unfolding/ denaturation of protein.
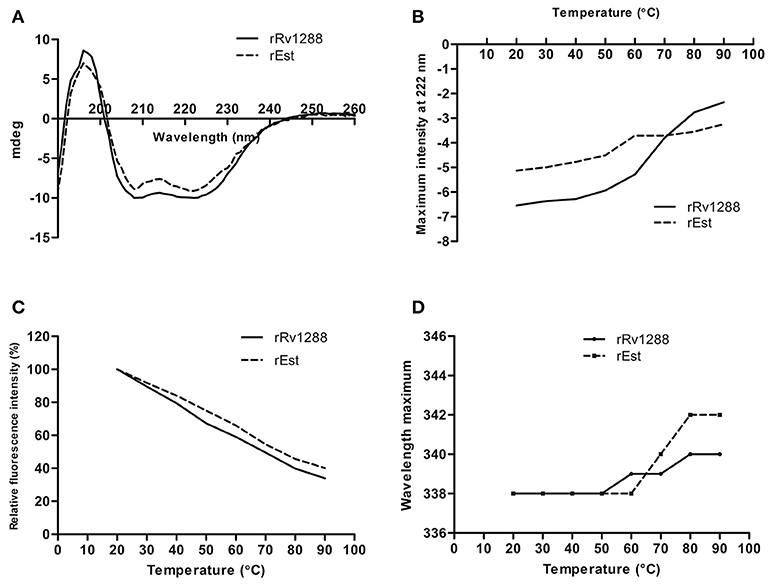
Figure 4. Biophysical Characterization of rRv1288 and rEst. (A) Far—UV CD spectra of recombinant purified rRv1288 and rEst proteins measured in 10 mM phosphate buffer (pH 8.0) at 25°C at wavelength 190–280 nm. (B) Thermal unfolding of rRv1288 and rEst proteins recorded at far UV-CD spectra at 222 nm wavelength with temperture increasing from 20 to 90°C at rate of 1°C/min. (C) Effect of temperature on maximum fluorescence intensity. Relative maximum fluorescence intensity of rRv1288 and rEst proteins with temperature increasing from 20 to 90°C. (D) Wavelength peak maxima for rRv1288 and rEst proteins at different temperatures (20 to 90°C). Each spectra represented the average data of three independent scans.
Effect of temperature on conformation change in tertiary structure was also monitored by measuring the intrinsic tryptophan fluorescence intensity. The rRv1288 and rEst protein has 9 and 8 tryptophan amino acid, respectively. The maximum intensity was recorded at 338 nm for both enzymes. With rise in temperature, fluorescence intensity was gradually decreased. Rate of reduction in maximum peak intensity is slightly more in rRv1288 as compared to rEst. Peak maxima were also shifted from 338 to 340 nm and 342 nm for rRv1288 and rEst respectively, with increase in temperature from 50 to 90°C (Figures 4C,D). Shifting of peak maxima toward higher wavelength is known as red shift. So, higher red shift was observed in case of rEst. Buffer was used as blank for each spectrum.
Localization
For detection of localization of Rv1288, western blot analysis of cytosolic, membrane and secreted fractions of M.tb H37Ra was carried out using anti-Rv1288 polyclonal antibody raised in rabbit. Control marker GroEL and antigen 85 complex were observed in cytoplasmic fraction and membrane fraction, respectively, which indicated the purity of fractions. A 49 kDa band was observed only in the cell wall fraction, but not in the cytosolic and culture filtrate fractions (Figure 5A). This showed that Rv1288 is mainly present in cell wall fraction.
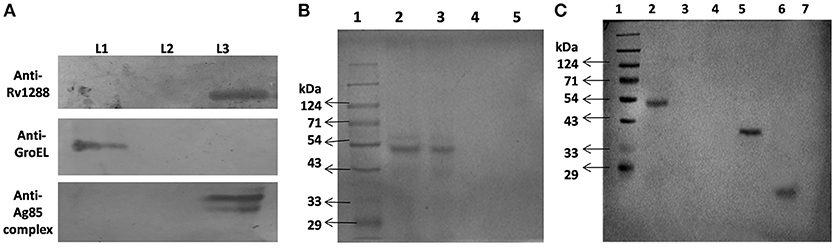
Figure 5. (A) Immunolocalization of Rv1288, GroEL and antigen 85 complex in M. tb H37Ra. L1 = Cytosolic fraction, L2 = Culture filtrate protein fraction, L3 = Cell wall fraction. (B) Peptidoglycan Binding Assay of rRv1288. 1: Protein marker, 2: Purified rRv1288 protein, 3: Pellet fraction after incubation of rRv1288 and peptidoglycan, 4: Supernatant fraction after incubation of rRv1288 and peptidoglycan, 5: Pellet fraction of control (only protein, no peptidoglycan). (C) Peptidoglycan Binding Assay of rRv1288 and its domains. Bacillus peptidoglycans were incubated with the purified rRv1288, Est and Lyt proteins for 30 min, centrifuged, washed and loaded on the SDS-PAGE gel. 1: Protein Marker, 2: Pellet fraction after incubation with rRv1288, 3: Supernatant fraction after incubation with rRv1288, 4: Pellet fraction after incubation with rEst protein, 5: Supernatant fraction after incubation with rEst protein, 6: Pellet fraction after incubation with rLyt protein, 7: Supernatant fraction after incubation with rLyt protein.
Peptidoglycan Binding Assay
Full length rRv1288, rEst and rLyt domain were examined for their peptidoglycan binding capability. After incubating all the three proteins with peptidoglycan, only the full length rRv1288 and rLyt domain were detected in the pellet fraction containing peptidoglycans, while rEst domain was present in the supernatant fraction (Figures 5B,C). This indicated the rRv1288 could bind to peptidoglycan of cell wall and Lyt domain mediated this binding, while rEst domain did not have the binding affinity for bacillus peptidoglycans. However, no band was observed in the control, proving that pelting of proteins were peptidoglycan dependent and not due to insoluble proteins (Figure 5B).
Effect of Recombinant rv1288 Over-Expression in M. smegmatis
To study the effect of over-expression of Rv1288 in M. smegmatis, rv1288 gene was cloned in E. coli- Mycobacterium shuttle vector pVV16 and expressed in the surrogate host M. smegmatis mc2155. The colony morphology of MS_1288 was different from the control MS_Vec. The colony surface of MS_Vec was dry, irregular, rough with bulging while for recombinant it was spherical, smooth and wet (Figures 6A,B). However, over-expression of rv1288 did not alter the growth pattern of M. smegmatis mc2155 significantly (Figures 6C,D).
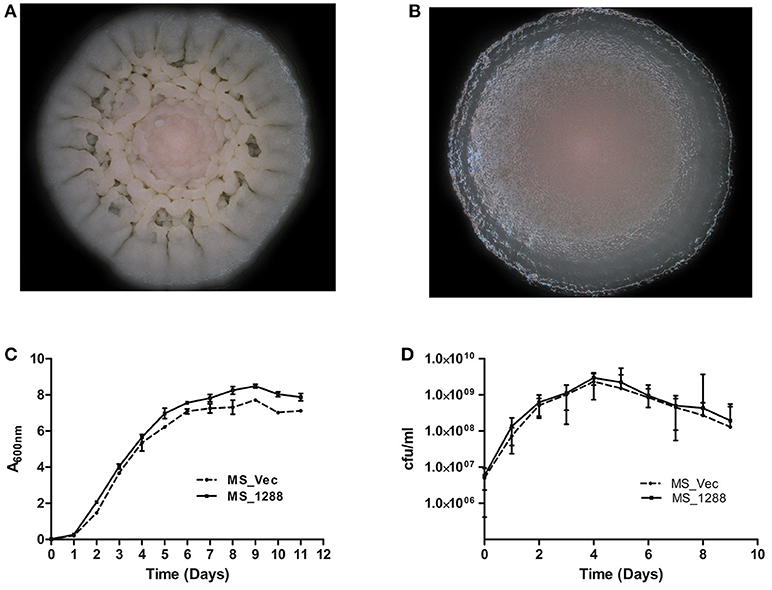
Figure 6. Effect of rv1288 on Colony morphology: Enlarged view of single colony of (A) MS_Vec and (B) MS_1288. Graphs showing growth curve in (C) absorbance at 600 nm and (D) cfu count of MS_1288 with MS_Vec as control. The data is representative of 3 independent experiments (mean ± SD).
Effect on Membrane Hydrophobicity
We also investigated the cell surface properties such as pellicle formation and cellular aggregation. Pellicles are biofilm like layer that formed at the air liquid interface in static cultures. MS_1288 and formed pellicle earlier and thicker as compared to MS_Vec and MS_S294A _1288 (Figure 7A) under similar experimental conditions. Aggregate formation in cultures was also examined in the absence of Tween-80. Tween-80 is a detergent that prevented clumping of mycobacterial cells. In absence of Tween-80, cellular aggregation resulted in reduced turbidity of cultures at 48 h. More aggregation was observed in MS_1288 and MS_S294A _1288 cells as compared to MS_Vec (Figure 7B).
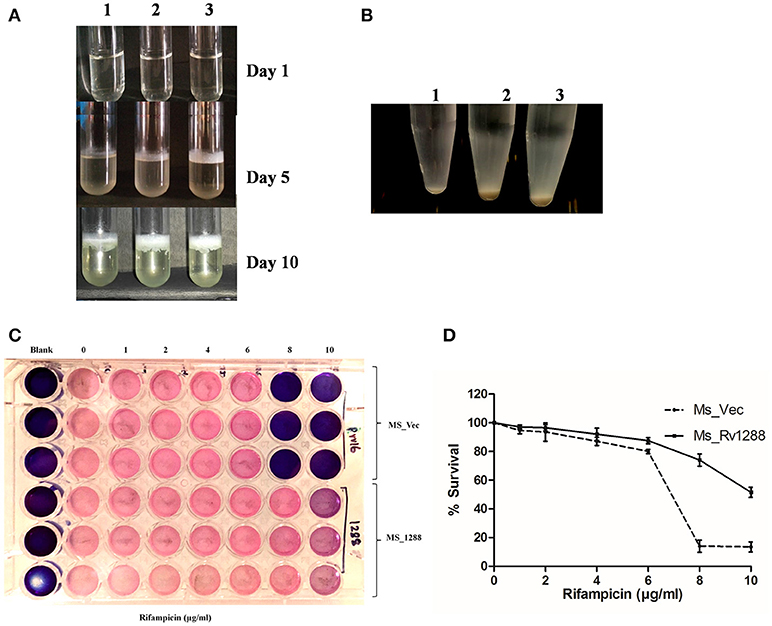
Figure 7. (A) Pellicle formation in standing cultures of MS_Vec (1), MS_S294A_1288 (2) and MS_1288 (3) (3) in M7H9 medium, in absence of Tween-80 at 37°C for various time points. (B) Aggregate formation by MS_Vec (1), MS_1288 (2) and MS_S294A_1288 (3) strains in M7H9 medium, absence of Tween-80 at 37°C with shaking for 48 h. Cultures were allowed to settle at room temperature for 30 min before taking photographs. (C) Effect of Rv1288 on drug susceptibility of M. smegmatis. Mid log phase culture of MS_Vec and MS_1288 were diluted in M7H9 medium without Tween-80 and were treated with different concentrations of Rifampicin (1–10 μg/ml). After 2 h incubation resazurin dye was added and plates were observed for the change in color. Blue color represents dead bacterial cells and pink represents live cells. (D) MS_Vec and MS_1288 were cultured in the presence of rifampicin at 37°C for 48 h. Survival of both the culture was measured by cfu count. Graph represent the % survival (Cfu count in the absence of rifampicin was considered as 100% survival). Experiment was performed in triplicate and results were shown as mean ± SD.
Effect on Drug Resistance
The effect of over expression of rv1288 on drug sensitivity of M. smegmatis was studied in the presence of anti-TB drugs. MS_1288 showed significant increase in drug resistance as compared to control MS_Vec culture in case of rifampicin. MS_1288 exhibited resistance at 8 μg/ml of rifampicin, while control cells were killed at this concentration (Figures 7C,D). However, no significant change in drug sensitivity was observed in case of streptomycin and isoniazid (Data not shown). Enhanced drug resistance might be due to modulation of cell wall lipids/increased hydrophobicity of membrane and hence increase in permeability for rifampicin (hydrophobic molecule) but reduced permeability for hydrophilic isoniazid.
Lipid Content of MS_Vec and MS_1288 Cultures
Since the change in morphology, pellicle and aggregate formation as well as drug sensitivity was observed in previous experiments, we investigated the lipid content of MS_Vec and MS_1288. MS_1288 had higher total lipid content as compared to control (Figure 8A). However, no difference was observed in polar and apolar lipid content (Figure 8B). Since mycolic acid is the main component of cell wall and Rv1288 was predicted to be a mycolyl acyl transferase, we further examined the mycolic acid rich glycolipids. TLC results showed the increased amount of TDM (Trehalose dimycolate) in MS_1288 in comparison to MS_Vec (Figure 8C).
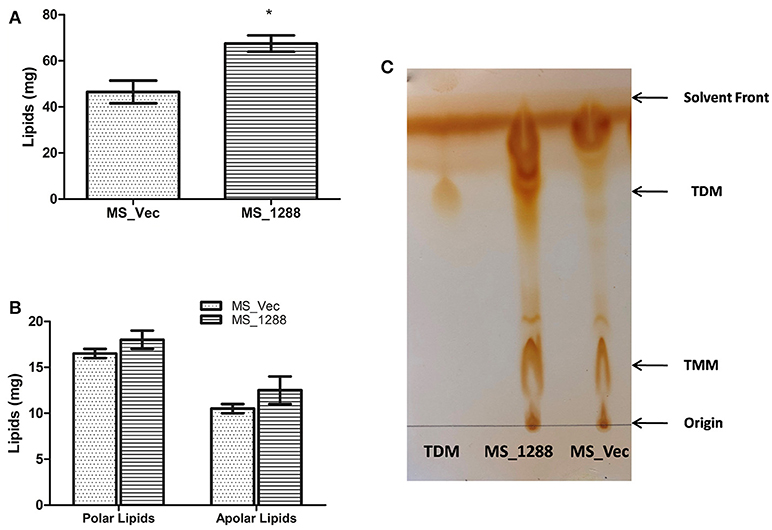
Figure 8. (A) Quantitative estimation of total lipids extracted from the equal weight of MS_Vec and MS_1288. (B) Polar and apolar lipids extracted from the equal weight of MS_Vec and MS_1288. (C) Mycolic acid containing glycolipids were separated from the pool of total polar lipids on TLC plate using chloroform: methanol: ammonium hydroxide (80:20:2) as mobile phase. TLC results showed the increased amount of TDM content in MS_1288 in comparison to MS_Vec. Results are expressed as mean ± SD from three independent experiments. All experiments were carried out three times in triplicate (n = 3) and error bars indicated standard deviation. Student's t-test was performed to show the statistical significance (*p < 0.05).
Effect of Nutrition Stress on Survival of MS_Vec and MS_1288
Since rv1288 was found to be upregulated under nutrition starvation conditions in M. tuberculosis H37Ra, we also investigated the effect of same on survival of MS_Vec and MS_1288 under nutrition starvation conditions. MS_1288 exhibited nearly 3 folds higher cfu count (Figure 9A) under nutrition depletion condition than MS_Vec.
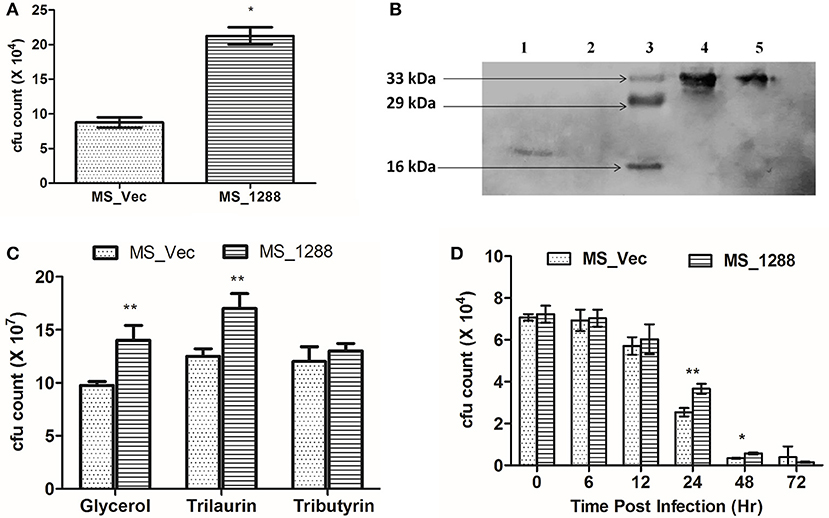
Figure 9. (A) Effect of expression of rv1288 on survival of M. smegmatis in nutrition stress. Graph showing cfu count of MS_Vec with MS_1288 under nutrition stressed conditions. The data is representative of 3 independent experiments. All experiments were carried out three times in triplicate (n = 3) and error bars indicated standard deviation. Student's t-test was performed to show the statistical significance (*p < 0.05). (B) Immunolocalization of Est and Lyt in recombinant M. smegmatis. 1 = Cell wall fraction of MS_Lyt, 2 = Cytosolic fraction MS_Lyt, 3 = Protein Marker, 4 = Cytosolic fraction MS_est, 5 = Cell wall fraction MS_est. (C) Cfu count of MS_Vec and MS_1288 in presence of M7H9 medium supplemented with Glycerol, Trilaurin and tributyrin, at 37°C, 180 rpm for 24 h. (D) Intracellular survival of MS_Vec and MS_1288. Cfu count of intracellular MS_Vec and MS_1288 were measured after 6,12, 24, 48, and 72 h post infection. The data is representive of three independent experiments.
Intracellular Localization of Est and Lyt Domain in M. smegmatis
Since, we found that Rv1288 protein was present in cell wall fraction of M.tb H37Ra, attempts were made to find out the localization of individual domain in recombinant M. smegmatis (MS_Est and MS_Lyt) using Anti-His antibody. Est domain was localized in both the cell wall and cytoplasmic fraction, while Lyt domain was found only in the cell wall fraction (Figure 9B).
Effect on Growth of MS_1288 in Presence of Different Lipids
Since Rv1288 was found to be membrane bound esterase, we tested the hydrolyzing efficiency of Rv1288 protein on lipids present in extracellular environment. MS_Vec and MS_1288 culture were grown in the presence of glycerol, tributyrin and trilaurin instead of usual Tween-80 in the medium. MS_1288 clearly showed the enhanced growth in the presence of glycerol and trilaurin as compared to control (Figure 9C). However, no significant change in growth was observed in case of tributyrin.
Effect on Intracellular Survival
Inside the host, M. tb resides mostly in macrophage cells and they are commonly used as model system to study the intracellular survival. Therefore, we tested the effect of MS_Vec and MS_1288 in intracellular survival of M. smegmatis in Raw 264.7 murine macrophage cell lines. Expression of Rv1288 enhanced the survival of M. smegmatis inside the macrophage cell line at 24 h and 48 h (Figure 9D).
Discussion
The mycobacterial proteins categorized as conserved, hypothetical with unknown functions restricted our understanding of physiology of this pathogen. Several hypothetical proteins shared characteristic feature with the other functionally known proteins. Many of these proteins including Rv1288 consisted of α/β hydrolase fold, i.e., characteristic features of lipase/esterase/protease enzymes and predicted to have hydrolase activity (Kumar et al., 2017d). Other than Rv1288, only one protein, Rv2719, a cell wall hydrolase and potential regulator of cell division in M.tb, was reported to have Lyt domain (Chauhan et al., 2006). Earlier reports suggested that proteins having LysM domain act as virulence factor in several human pathogens like Staphylococcus aureus, Listeria monocytogenes, Neisseria meningitides, Francisella tularensis, Streptococcus agalactiae etc. (Martin et al., 2002; Lenz et al., 2003; Kajimura et al., 2005; Melillo et al., 2006; Oldfield et al., 2007; Buist et al., 2008). Several lipases/esterases from mycobacteria, when expressed in E. coli, were reported to be expressed as inactive protein and formed inclusion bodies. However, absence of cysteine residue and disulfide bond along with aliphatic and instability index (< 40), predicted the expression of all the three proteins (Rv1288, Est, and Lyt) in soluble form (Idicula-Thomas and Balaji, 2005). Our study was in line with the predictions, as we were able to purify these proteins from the soluble fraction at low IPTG concentration and induction temperature. This strategy was successfully used for expression of proteins in soluble fraction in several other studies (Sinha et al., 2005; Zhang et al., 2005; Deb et al., 2006; Kaur et al., 2017b; Lin et al., 2017).
Conserved domain analysis revealed that Rv1288 belongs to “α/β hydrolase family,” which was further validated by CD spectroscopy. In contrast to the prediction by tuberculist, the protein demonstrated esterase activity and did not show protease activity. Both the full length and Est domain proteins showed similar specific activity and hydrolysis pattern, suggesting that Lyt domain did not affect the catalytic ability of Est domain. Our data was consistent with previous studies that demonstrated that several mycobacterium lipolytic enzymes prefer medium chain length esters (Guo et al., 2010; Sultana et al., 2013; Singh et al., 2014b) and short chain length esters (Zhang et al., 2005; Rengarajan et al., 2008; Shen et al., 2012; Jadeja et al., 2016; Singh et al., 2016a; Kumar et al., 2017e; Lin et al., 2017). The aggregate formation at higher temperature by rRv1288 only followed by loss in activity suggested the role of highly hydrophobic Lyt domain in protein aggregation and decreased stability of Rv1288. The protein aggregation during thermal unfolding was reported to contribute toward the smaller magnitude of red shift (Duy and Fitter, 2006). This study was in contrast to Rv0774c, where C terminal lipolytic domain is comparatively less stable than the full length protein (Kumar et al., 2017e). Previously, some mycobacterial lipases were reported to be stable at higher temperatures i.e., LipD (Singh et al., 2014a), LipV (Singh et al., 2014b),LipL (Singh et al., 2016a), LipN (Jadeja et al., 2016), and LipU (Kaur et al., 2017a).
Lyt domain is known to interact with the peptidoglycan moiety of cell wall (Buist et al., 2008). Molecular docking studies also demonstrated that it could bind to the N-acetyl glucosamine moiety of peptidoglycan (Kumar et al., 2017a). The peptidoglycan binding assay suggested that Rv1288 was attached to the cell wall of M.tb by Lyt domain. In line to our prediction, sub-cellular localization studies confirmed the presence of Rv1288 in cell wall fraction of M.tb H37Ra. This is consistent with mass spectrometry analysis that found the Rv1288 in membrane fraction of M.tb H37Rv (De Souza et al., 2011). Similar to our finding, Rv2719 protein of M.tb, containing Lyt domain was also localized on cell wall (Chauhan et al., 2006). Interestingly, in M.tb there are several cell wall anchored lipase/esterase such as LipY (Singh et al., 2014c), Rv0183 (Xu et al., 2010), and Rv2224c (Lun and Bishai, 2007) that were reported to play an important role in pathogenesis and virulence.
The proteome of M.tb is dynamic and kept changing in order to facilitate the survival of pathogen under stress conditions (Geiman et al., 2006; Saviola, 2010). Rv1288 was previously reported in hypoxia sample by mass spectrometry (James et al., 2013) and in dormant phase (Hasan et al., 2006; Murphy and Brown, 2007; Agüero et al., 2008). Current regimens are not much effective against the bacterium in latent phase, especially in hypoxic granulomas present in the lung (Murphy and Brown, 2007). It was earlier reported that survival of the mycobacteria depend on their potential to metabolize the stored lipids, as they are the major energy source during latency (Barisch and Soldati, 2017). In this study, rv1288 was up-regulated under nutrient starved condition and expression of rv1288 gene in M. smegmatis resulted in enhanced survival under nutrient starved condition. As this is a membrane bound enzyme and it showed enhanced growth in presence of glycerol and trilaurin as carbon source, the probable role of this protein in host lipid utilization cannot be ruled out. Thus, it was suggested that under nutrient starved condition, Rv1288 might hydrolyze intracellular or extracellular lipids in order to facilitate its survival. Previously, other lipases LipY and LipX were also reported to be up regulated in nutrition starvation (Betts et al., 2002; Deb et al., 2006).
To mimic the over expression of rv1288 in M.tb under stress condition, we over-expressed rv1288 in M. smegmatis. Over expression of this protein resulted in altered colony morphology and slight increase in growth rate of bacterium. Similar to these results, expression of genes such as rv1818c (Delogu et al., 2004), rv1169c (Singh et al., 2016b) and rv0774c (Kumar et al., 2017c), changed the colony morphology and growth rate of M. smegmatis. It has been reported that colony morphology of mycobacterium species is associated with their virulence, drug resistance, infection, intracellular survival, immune modulation, and altered signaling pathways (Kuze and Uchihira, 1984; Shiratsuchi et al., 1993; Hubert et al., 2002). TLC results suggested that the Rv1288 might be responsible for increased amount of TDM content. The change in cell surface properties of M. smegmatis expressing rv1288, like colony morphology, pellicle formation, aggregation, and change in drug sensitivity pointed toward the remodeling of cell envelop resulting in change in permeability that might help the intracellular survival of mycobacteria. MS_1288 also showed enhanced intracellular survival in Raw 264.7 cells as compared to the control. Various report showed that altered colony morphology, increase pellicle formation, and aggregative nature were attributed to the change in composition and remodeling of cell wall lipids (Singh et al., 2016b; Kumar et al., 2017c). In M.tb, mycolyl transferases are differentially expressed under different conditions to enable the bacteria to evade host immune system and survive (Ojha et al., 2010; Queiroz and Riley, 2017). Earlier, Takayama et al. (2005) reported Rv1288 as a probable mycolyl transferase II on the basis of its similarity to Ag85 complex proteins. Ag85 complex proteins catalyze the synthesis of TDM or cell wall arabinogalactan-mycolate from TMM by mycoyltransferase activity (Queiroz and Riley, 2017). Lipid profiling studies showed increase in total lipid and TDM content in rv1288 expressing M. smegmatis strain. Virulent M.tb strains are usually rich in TDM (Singh et al., 2017). Similarly the expression of mycolyl transferase Ag85A protein in M. smegmatis resulted in increase concentration of the glycolipids TDM and TMM in the cell (Elamin et al., 2011). Almost all mycolyl transferases possessed carboxylesterase domain, and their ability to transfer the mycolyl moiety is usually dependent on this domain (Dautin et al., 2017). Recently, Rv0774c was also reported as mycolyl transferase II, that altered the lipid composition by synthesizing TDM (Kumar et al., 2017c). Rv1169c was also demonstrated to modulate the cell wall lipids (Singh et al., 2016b). Similar to our study, Rv3451 in M.tb was reported as TDM hydrolase, which was overexpressing under nutrition starvation conditions. Rv3451 was also reported to increase the growth rate and reactivation of M.tb in an immunocompromised animal model (Yang et al., 2014). These findings, strongly suggested that Rv1288 might possess mycolyl transferase activity.
Conclusion
In summary, Rv1288 was confirmed as a cell wall anchored esterase of M.tb that was up regulated under nutrition depletion conditions and observed to support the bacterial growth under nutritive starvation condition. The Lyt domain was essential for placing the protein at cell wall. The expression of protein altered the colony morphology, cell surface properties, drug resistance, lipid content of the cell wall of M. smegmatis and its intracellular survival inside the macrophage cell line. The mycolyl transferase activity of Rv1288 could be correlated with increased concentration of the glycolipid TDM. These results pointed toward its important role in intracellular survival of mycobacteria. Further studies with knockout of rv1288 in M.tbH37Rv are required to confirm the precise role of Rv1288 in lipid modulation.
Authors Contributions
PM performed the experiments and wrote manuscript. AK analyzed the data and manuscript writing. JasK performed experiments related to lipids. JagK conceived and designed the whole study.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We acknowledge UGC-SAP and DST PURSE grant to JagK at Department of Biotechnology, and DBT, New Delhi, India and CSIR, New Delhi, India for providing fellowship to PM and JasK, respectively. We would also like to acknowledge Dr. Vinay K. Nandicoori (Scientist, NII, New Delhi, India) and Dr. Garima Khare (DU, New Delhi, India) for providing control antibodies for subcellular localization experiment.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2018.00421/full#supplementary-material
References
Agüero, F., Al-Lazikani, B., Aslett, M., Berriman, M., Buckner, F. S., Campbell, R. K., et al. (2008). Genomic-scale prioritization of drug targets: the TDR Targets database. Nature reviews. Drug discovery 7:900. doi: 10.1038/nrd2684
Anand, P., Sankaran, S., Mukherjee, S., Yeturu, K., Laskowski, R., Bhardwaj, A., et al. (2011). Structural Annotation of Mycobacterium tuberculosis Proteome. PLoS ONE 6:e27044. doi: 10.1371/journal.pone.0027044
Barisch, C., and Soldati, T. (2017). Mycobacterium marinum degrades both triacylglycerols and phospholipids from its Dictyostelium host to synthesise its own triacylglycerols and generate lipid inclusions. PLoS Pathog. 13:e1006095. doi: 10.1371/journal.ppat.1006095
Betts, J. C., Lukey, P. T., Robb, L. C., McAdam, R. A., and Duncan, K. (2002). Evaluation of a nutrient starvation model of Mycobacterium tuberculosis persistence by gene and protein expression profiling. Mol. Microbiol. 43, 717–731. doi: 10.1046/j.1365-2958.2002.02779.x
Brown, M. R., and Foster, J. H. (1970). A simple diagnostic milk medium for Pseudomonas aeruginosa. J. Clin. Pathol. 23, 172–177. doi: 10.1136/jcp.23.2.172
Buist, G., Steen, A., Kok, J., and Kuipers, O. P. (2008). LysM, a widely distributed protein motif for binding to (peptido) glycans. Mol. Microbiol. 68, 838–847. doi: 10.1111/j.1365-2958.2008.06211.x
Chauhan, A., Maloney, E., Moore, J., Fol, M., Madiraju, M. V., et al. (2006). Interference of Mycobacterium tuberculosis cell division by Rv2719c, a cell wall hydrolase. Mol. Microbiol. 62, 132–147. doi: 10.1111/j.1365-2958.2006.05333.x
Chownk, M., Sharma, A., Singh, K., and Kaur, J. (2017). mesT, a unique epoxide hydrolase, is essential for optimal growth of Mycobacterium tuberculosis in the presence of styrene oxide. Future Microbiol. 12, 527–546 doi: 10.2217/fmb-2016-0206
Cole, S., Brosch, R., Parkhill, J., Garnier, T., Churcher, C., Harris, D., et al. (1998). Erratum: Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 396:190. doi: 10.1038/24206
Dautin, N., de Sousa-d'Auria, C., Constantinesco-Becker, F., Labarre, C., Oberto, J., Li de la Sierra-Gallay, I., et al. (2017). Mycoloyltransferases: a large and major family of enzymes shaping the cell envelope of Corynebacteriales. Biochim. Biophys. Acta 1861, 3581–3592. doi: 10.1016/j.bbagen.2016.06.020
De Souza, G. A., Arntzen, M. Ø., Fortuin, S., Schürch, A. C., Målen, H., McEvoy, C. R., et al. (2011). Proteogenomic analysis of polymorphisms and gene annotation divergences in prokaryotes using a clustered mass spectrometry-friendly database. Mol. Cell. Proteomics 10:M110.002527. doi: 10.1074/mcp.M110.002527
Deb, C., Daniel, J., Sirakova, T. D., Abomoelak, B., Dubey, V. S., and Kolattukudy, P. E. (2006). A novel lipase belonging to the hormone-sensitive lipase family induced under starvation to utilize stored triacylglycerol in Mycobacterium tuberculosis. J. Biol. Chem. 281, 3866–3875. doi: 10.1074/jbc.M505556200
Delogu, G., Pusceddu, C., Bua, A., Fadda, G., Brennan, M. J., and Zanetti, S. (2004). Rv1818c-encoded PE_PGRS protein of Mycobacterium tuberculosis is surface exposed and influences bacterial cell structure. Mol. Microbiol. 52, 725–733. doi: 10.1111/j.1365-2958.2004.04007.x
Doerks, T., Van Noort, V., Minguez, P., and Bork, P. (2012). Annotation of the M. tuberculosis hypothetical orfeome: adding functional information to more than half of the uncharacterized proteins. PLoS ONE 7:e34302. doi: 10.1371/journal.pone.0034302
Drake, D., and Montie, T. C. (1988). Flagella, motility and invasive virulence of Pseudomonas aeruginosa. Microbiology 134, 43–52. doi: 10.1099/00221287-134-1-43
Duy, C., and Fitter, J. (2006). How aggregation and conformational scrambling of unfolded states govern fluorescence emission spectra. Biophys. J. 90, 3704–3711. doi: 10.1529/biophysj.105.078980.
Elamin, A. A., Stehr, M., Spallek, R., Rohde, M., and Singh, M. (2011). The Mycobacterium tuberculosis Ag85A is a novel diacylglycerol acyltransferase involved in lipid body formation. Mol. Microbiol. 81, 1577–1592. doi: 10.1111/j.1365-2958.2011.07792.x
Ford, N., Shubber, Z., Meintjes, G., Grinsztejn, B., Eholie, S., Mills, E. J., et al. (2015). Causes of hospital admission among people living with HIV worldwide: a systematic review and meta-analysis. Lancet HIV 2, e438–e444. doi: 10.1016/S2352-3018(15)00137-X
Gago, G., Diacovich, L., and Gramajo, H. (2018). Lipid metabolism and its implication in mycobacteria–host interaction. Curr. Opin. Microbiol. 41, 36–42. doi: 10.1016/j.mib.2017.11.020
Geiman, D. E., Raghunand, T. R., Agarwal, N., and Bishai, W. R. (2006). Differential gene expression in response to exposure to antimycobacterial agents and other stress conditions among seven Mycobacterium tuberculosis whiB-like genes. Antimicrob. Agents Chemother. 50, 2836–2841. doi: 10.1128/AAC.00295-06
Guo, J., Zheng, X., Xu, L., Liu, Z., Xu, K., Li, S., et al. (2010). Characterization of a novel esterase Rv0045c from Mycobacterium tuberculosis. PLoS ONE 5:e13143. doi: 10.1371/journal.pone.0013143
Hasan, S., Daugelat, S., Rao, P. S., and Schreiber, M. (2006). Prioritizing genomic drug targets in pathogens: application to Mycobacterium tuberculosis. PLoS Comput. Biol. 2:e61. doi: 10.1371/journal.pcbi.0020061
Holmquist, M. (2000). Alpha beta-hydrolase fold enzymes structures, functions and mechanisms. Curr. Protein Peptide Sci. 1, 209–235. doi: 10.2174/1389203003381405
Hubert, M. T., Josephy, S. I., Chan, E. D., Fouts, D., and Cooper, A. M. (2002). Activation of the mitogen-activated protein kinase signaling pathway is instrumental in determining the ability of Mycobacterium avium to grow in murine macrophages. J. Immunol. 168, 825–833. doi: 10.4049/jimmunol.168.2.825
Idicula-Thomas, S., and Balaji, P. V. (2005). Understanding the relationship between the primary structure of proteins and its propensity to be soluble on overexpression in Escherichia coli. Protein Sci. 14, 582–592. doi: 10.1110/ps.041009005.
Jadeja, D., Dogra, N., Arya, S., Singh, G., Singh, G., and Kaur, J. (2016). Characterization of LipN (Rv2970c) of Mycobacterium tuberculosis H37Rv and its probable role in xenobiotic degradation. J. Cell. Biochem. 117, 390–401. doi: 10.1002/jcb.25285
James, B. W., Bacon, J., and Marsh, P. (2013). Mycobacterial Antigens Expressed Under Low Oxygen Tension. United States Patent 8404826.
Johnson, G. (2017). The α/β hydrolase fold proteins of Mycobacterium tuberculosis, with reference to their contribution to virulence. Curr. Protein Peptide Sci. 18, 190–210. doi: 10.2174/1389203717666160729093515
Kajimura, J., Fujiwara, T., Yamada, S., Suzawa, Y., Nishida, T., Oyamada, Y., et al. (2005). Identification and molecular characterization of an N-acetylmuramyl-l-alanine amidase Sle1 involved in cell separation of Staphylococcus aureus. Mol. Microbiol. 58, 1087–1101. doi: 10.1111/j.1365-2958.2005.04881.x
Kaur, G., Saini, v., Kumari, B., and Kaur, J. (2017a). Characterization of an extracellular protein, Rv1076 from M. tuberculosis with a potential role in humoral response. Int. J. Biol. Macromol. 101, 621–629. doi: 10.1016/j.ijbiomac.2017.03.096
Kaur, J., Kumar, A., and Kaur, J. (2017b). Strategies for optimization of heterologous protein expression in E. coli: Roadblocks and reinforcements. Int. J. Biol. Macromol. 106, 803–822. doi: 10.1016/j.ijbiomac.2017.08.080
Kumar, A., Maan, P., Singh, G., and Kaur, J. (2017a). In-silico characterization of a hypothetical protein, Rv1288 of Mycobacterium tuberculosis Containing an esterase signature and an uncommon LytE domain. Curr. Comput. Aided Drug Des. 13, 101–111. doi: 10.2174/1573409912666161124144725
Kumar, A., Manisha Sangha, G. K., Shrivastava, A., and Kaur, J. (2017b). The immunosuppressive effects of a novel recombinant LipQ (Rv2485c) protein of Mycobacterium tuberculosis on human macrophage cell lines. Microb. Pathog. 107, 361–367. doi: 10.1016/j.micpath.2017.04.015
Kumar, A., Saini, V., Kumar, A., Kaur, J., and Kaur, J. (2017c). Modulation of trehalose dimycolate and immune system by Rv0774c protein enhanced the intracellular survival of Mycobacterium smegmatis in human macrophages cell line. Front. Cell. Infect. Microbiol. 7:289. doi: 10.3389/fcimb.2017.00289
Kumar, A., Sharma, A., Kaur, G., Makkar, P., and Kaur, J. (2017d). Functional characterization of hypothetical proteins of Mycobacterium tuberculosis with possible esterase/lipase signature: a cumulative in silico and in vitro approach. J. Biomol. Struct. Dyn. 35, 1226–1243. doi: 10.1080/07391102.2016.1174738
Kumar, A., Singh, S. M., Singh, R., and Kaur, J. (2017e). Rv0774c, an iron stress inducible, extracellular esterase is involved in immune-suppression associated with altered cytokine and TLR2 expression. Int. J. Med. Microbiol. 307, 126–138. doi: 10.1016/j.ijmm.2017.01.003
Kuze, F., and Uchihira, F. (1984). Various colony-formers of Mycobacterium avium-intracellulare. Eur. J. Respir. Dis. 65, 402–410.
Lenz, L. L., Mohammadi, S., Geissler, A., and Portnoy, D. A. (2003). SecA2-dependent secretion of autolytic enzymes promotes Listeria monocytogenes pathogenesis. Proc. Natl. Acad. Sci.U.S.A. 100, 12432–12437. doi: 10.1073/pnas.2133653100
Lew, J. M., Kapopoulou, A., Jones, L. M., and Cole, S. T. (2011). TubercuList−10 years after. Tuberculosis 91, 1–7. doi: 10.1016/j.tube.2010.09.008
Lin, Y., Li, Q., Xie, L., and Xie, J. (2017). Mycobacterium tuberculosis rv1400c encodes functional lipase/esterase. Protein Expr. Purif. 129, 143–149. doi: 10.1016/j.pep.2016.04.013
Lovewell, R. R., Sassetti, C. M., and VanderVen, B. C. (2016). Chewing the fat: lipid metabolism and homeostasis during M. tuberculosis infection. Curr. Opin. Microbiol. 29, 30–36. doi: 10.1016/j.mib.2015.10.002
Lun, S., and Bishai, W. R. (2007). Characterization of a novel cell wall-anchored protein with carboxylesterase activity required for virulence in Mycobacterium tuberculosis. J. Biol. Chem. 282, 18348–18356. doi: 10.1074/jbc.M700035200
Madan-Lala, R., Sia, J. K., King, R., Adekambi, T., Monin, L., Khader, S. A., et al. (2014). Mycobacterium tuberculosis impairs dendritic cell functions through the serine hydrolase Hip1. J. Immunol. 192, 4263–4272. doi: 10.4049/jimmunol.1303185
Mahamed, D., Boulle, M., Ganga, Y., Mc Arthur, C., Skroch, S., Oom, L., et al. (2017). Intracellular growth of Mycobacterium tuberculosis after macrophage cell death leads to serial killing of host cells. Elife 6:e22028. doi: 10.7554/eLife.22028
Martin, D., Rioux, S., Gagnon, E., Boyer, M., Hamel, J., Charland, N., et al. (2002). Protection from group B streptococcal infection in neonatal mice by maternal immunization with recombinant Sip protein. Infect. Immun. 70, 4897–4901. doi: 10.1128/IAI.70.9.4897-4901.2002
Melillo, A., Sledjeski, D. D., Lipski, S., Wooten, R. M., Basrur, V., and Lafontaine, E. R. (2006). Identification of a Francisella tularensis LVS outer membrane protein that confers adherence to A549 human lung cells. FEMS Microbiol. Lett. 263, 102–108. doi: 10.1111/j.1574-6968.2006.00413.x
Murphy, D. J., and Brown, J. R. (2007). Identification of gene targets against dormant phase Mycobacterium tuberculosis infections. BMC Infect. Dis. 7:84. doi: 10.1186/1471-2334-7-84
Naffin-Olivos, J. L., Daab, A., White, A., Goldfarb, N. E., Milne, A. C., Liu, D., et al. (2017). Structure determination of Mycobacterium tuberculosis serine protease Hip1 (Rv2224c). Biochemistry 56, 2304–2314 doi: 10.1021/acs.biochem.6b01066
Ojha, A. K., Trivelli, X., Guerardel, Y., Kremer, L., and Hatfull, G. F. (2010). Enzymatic hydrolysis of trehalose dimycolate releases free mycolic acids during mycobacterial growth in biofilms. J. Biol. Chem. 285, 17380–17389. doi: 10.1074/jbc.M110.112813
Oldfield, N. J., Bland, S. J., Taraktsoglou, M., Ramos, F. J. D., Robinson, K., Wooldridge, K. G., et al. (2007). T-cell stimulating protein A (TspA) of Neisseria meningitidis is required for optimal adhesion to human cells. Cell. Microbiol. 9, 463–478. doi: 10.1111/j.1462-5822.2006.00803.x
Petersen, E., Maeurer, M., Marais, B., Migliori, G. B., Mwaba, P., Ntoumi, F., et al. (2017). World TB day 2017: advances, challenges and opportunities in the “End-TB” Era. Int. J. Infect. Dis.56, 1–5. doi: 10.1016/j.ijid.2017.02.012
Queiroz, A., and Riley, L. W. (2017). Bacterial immunostat: Mycobacterium tuberculosis lipids and their role in the host immune response. Rev. Soc. Bras. Med. Trop. 50, 9–18. doi: 10.1590/0037-8682-0230-2016
Rengarajan, J., Murphy, E., Park, A., Krone, C. L., Hett, E. C., Bloom, B. R., et al. (2008). Mycobacterium tuberculosis Rv2224c modulates innate immune responses. Proc. Natl. Acad. Sci.U.S.A. 105, 264–269. doi: 10.1073/pnas.0710601105
Rezwan, M., Lanéelle, M.-A., Sander, P., and Daff,é, M. (2007). Breaking down the wall: fractionation of mycobacteria. J. Microbiol. Methods 68, 32–39. doi: 10.1016/j.mimet.2006.05.016
Rodriguez, G. M., Voskuil, M. I., Gold, B., Schoolnik, G. K., and Smith, I. (2002). ideR, an essential gene in Mycobacterium tuberculosis: role of IdeR in iron-dependent gene expression, iron metabolism, and oxidative stress response. Infect. Immun. 70, 3371–3381. doi: 10.1128/IAI.70.7.3371-3381.2002
Saviola, B. (2010). “All stressed out: mycobacterial responses to stress,”in Current Research, Technology, and Education Topics in Applied Microbiology and Microbial Biotechnology. Microbiology Book Series Edition, ed. A. Mendez-Vilas (Formatex Research Center), 545–549.
Shen, G., Singh, K., Chandra, D., Serveau-Avesque, C., Maurin, D., Canaan, S., et al. (2012). LipC (Rv0220) is an immunogenic cell surface esterase of Mycobacterium tuberculosis. Infect. Immun. 80, 243–253. doi: 10.1128/IAI.05541-11
Shiratsuchi, H., Toossi, Z., Mettler, M. A., and Ellner, J. J. (1993). Colonial morphotype as a determinant of cytokine expression by human monocytes infected with Mycobacterium avium. J. Immunol. 150, 2945–2954.
Singh, G., Arya, S., Kumar, A., Narang, D., and Kaur, J. (2014a). Molecular characterization of oxidative stress-inducible LipD of Mycobacterium tuberculosis H37Rv. Curr. Microbiol. 68, 387–396. doi: 10.1007/s00284-013-0486-3
Singh, G., Arya, S., Narang, D., Jadeja, D., Singh, G., Gupta, U., et al. (2014b). Characterization of an acid inducible lipase Rv3203 from Mycobacterium tuberculosis H37Rv. Mol. Biol. Rep. 41, 285–296. doi: 10.1007/s11033-013-2861-3
Singh, G., Kumar, A., Arya, S., Gupta, U. D., Singh, K., and Kaur, J. (2016a). Characterization of a novel esterase Rv1497 of Mycobacterium tuberculosis H37Rv demonstrating β-lactamase activity. Enzyme Microb. Technol. 82, 180–190. doi: 10.1016/j.enzmictec.2015.10.007
Singh, G., Kumar, A., Maan, P., and Kaur, J. (2017). Cell wall associated factors of Mycobacterium tuberculosis as major virulence determinants: current perspectives in drugs discovery and design. Curr. Drug Targets 18, 1904–1918. doi: 10.2174/1389450118666170711150034
Singh, P., Rao, R. N., Reddy, J. R., Prasad, R. B., Kotturu, S. K., Ghosh, S., et al. (2016b). PE11, a PE/PPE family protein of Mycobacterium tuberculosis is involved in cell wall remodeling and virulence. Sci. Rep. 6:21624. doi: 10.1038/srep21624
Singh, V. K., Srivastava, M., Dasgupta, A., Singh, M. P., Srivastava, R., and Srivastava, B. S. (2014c). Increased virulence of Mycobacterium tuberculosis H37Rv overexpressing LipY in a murine model. Tuberculosis 94, 252–261. doi: 10.1016/j.tube.2014.02.001
Sinha, S. C., Wetterer, M., Sprang, S. R., Schultz, J. E., and Linder, J. U. (2005). Origin of asymmetry in adenylyl cyclases: structures of Mycobacterium tuberculosis Rv1900c. EMBO J. 24, 663–673. doi: 10.1038/sj.emboj.7600573
Sultana, R., Vemula, M. H., Banerjee, S., and Guruprasad, L. (2013). The PE16 (Rv1430) of Mycobacterium tuberculosis is an esterase belonging to serine hydrolase superfamily of proteins. PLoS ONE 8:e55320. doi: 10.1371/journal.pone.0055320
Takayama, K., Wang, C., and Besra, G. S. (2005). Pathway to synthesis and processing of mycolic acids in Mycobacterium tuberculosis. Clin. Microbiol. Rev. 18, 81–101. doi: 10.1128/CMR.18.1.81-101.2005
Vemula, M., Balakrishnan, K., Banerjee, S., and Guruprasad, L. (2018). Mycobacterium tuberculosis PE1 and PE2 proteins carrying conserved α/β-serine hydrolase domain are esterases hydrolyzing short to medium chain p-nitrophenyl esters. Progr. Biophys. Mol. Biol. doi: 10.1016/j.pbiomolbio.2018.04.012
Xu, G., Jia, H., Li, Y., Liu, X., Li, M., and Wang, Y. (2010). Hemolytic phospholipase Rv0183 of Mycobacterium tuberculosis induces inflammatory response and apoptosis in alveolar macrophage RAW264. 7 cells. Can. J. Microbiol. 56, 916–924. doi: 10.1139/W10-079
Yang, Y., Kulka, K., Montelaro, R. C., Reinhart, T. A., Sissons, J., Aderem, A., et al. (2014). A hydrolase of trehalose dimycolate induces nutrient influx and stress sensitivity to balance intracellular growth of Mycobacterium tuberculosis. Cell Host Microbe 15, 153–163. doi: 10.1016/j.chom.2014.01.008
Yoshida, H., and Ashida, M. (1996). Purification of a peptidoglycan recognition protein from hemolymph of the silkworm, Bombyx mori. (0021-9258 (Print)). doi: 10.1074/jbc.271.23.13854
Keywords: Mycobacterium tuberculosis, esterase, cell wall, nutrient starvation, TDM content, peptidoglycan binding
Citation: Maan P, Kumar A, Kaur J and Kaur J (2018) Rv1288, a Two Domain, Cell Wall Anchored, Nutrient Stress Inducible Carboxyl-Esterase of Mycobacterium tuberculosis, Modulates Cell Wall Lipid. Front. Cell. Infect. Microbiol. 8:421. doi: 10.3389/fcimb.2018.00421
Received: 09 July 2018; Accepted: 16 November 2018;
Published: 03 December 2018.
Edited by:
Stephane Canaan, Centre National de la Recherche Scientifique (CNRS), FranceReviewed by:
Kyle Rohde, University of Central Florida, United StatesLautaro Diacovich, CONICET Instituto de Biología Molecular y Celular de Rosario (IBR), Argentina
Copyright © 2018 Maan, Kumar, Kaur and Kaur. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jagdeep Kaur, amFnc2VraG9uQHlhaG9vLmNvbQ==;amFnc2VraG9uQHB1LmFjLmlu
 Pratibha Maan
Pratibha Maan Arbind Kumar
Arbind Kumar Jashandeep Kaur
Jashandeep Kaur Jagdeep Kaur
Jagdeep Kaur