- Laboratory of Molecular Virology, International Center for Genetic Engineering and Biotechnology, Trieste, Italy
Classical antiviral therapy targets viral functions, mostly viral enzymes or receptors. Successful examples include precursor herpesvirus drugs, antiretroviral drugs that target reverse transcriptase and protease, influenza virus directed compounds as well as more recent direct antiviral agents (DAA) applied in the treatment of hepatitis C virus (HCV). However, from early times, the possibility of targeting the host cell to contain the infection has frequently re-emerged as an alternative and complementary antiviral strategy. Advantages of this approach include an increased threshold to the emergence of resistance and the possibility to target multiple viruses. Major pitfalls are related to important cellular side effects and cytotoxicity. In this mini-review, the concept of host directed antiviral therapy will be discussed with a focus on the most recent advances in the field of Flaviviruses, a family of important human pathogens for which we do not have antivirals available in the clinics.
Introduction
The discovery of acyclovir about 40 years ago marked a new era in the field of antivirals (Elion, 1993). Acyclovir acts as a prodrug being activated by the viral thymidine kinase, but targeting the viral DNA polymerase. Therefore, the drug is highly specific with a narrow range of action that includes herpes simplex, varicella zoster, and cytomegalovirus. High specificity leads to the emergence of viruses encoding a thymidine kinase resistant to acyclovir. Acute episodes of herpes infection are manageable with acyclovir, unless prolonged treatment, as in immune compromised patients where resistance usually emerges, is required (Palù et al., 1992). In such cases, drugs with alternative targets are needed. This scheme of high specificity, low toxicity and emergence of resistant viruses is a concern associated with every antiviral treatment of the last few decades. Therefore, there is a constant pressure to develop combined therapies against different viral targets to decrease viral fitness. This concept is particularly relevant for life-long therapies for chronic infections, such as the human immune deficiency virus (HIV) (Van Lint et al., 2013). Recently, a combination of novel inhibitors of viral functions achieved the remarkable objective of eradication of chronic HCV infection (Pawlotsky et al., 2015). Success stories like these certainly reinforce the validity of the approach and the importance of targeting viral functions for the development of successful antivirals. Nevertheless, other approaches targeting cellular functions, collectively called host directed (antiviral) therapies (HDT) are being considered either to enhance the activity of direct drugs, or to cover more infectious agents at the same time.
Before the advent of direct antiviral agents (DAA), the therapy for hepatitis C was based on a combination of recombinant interferon-α (IFNα) and ribavirin, a nucleoside analog with broad antiviral activity and poor specificity. Interferons are natural cellular proteins that trigger an innate antiviral response of the cell, as well as, stimulate the adaptive immune response. The combination of IFNα and Ribavirin results in higher sustained virological response rates (McHutchison et al., 1998). Hence, this is a perfect example of combination of a HDT, IFNα, with an antiviral agent, Ribavirin, although the latter may be targeting host factors as well, thus explaining its broad-spectrum antiviral activity. The treatment is not as effective as using DAAs and has some important side effects, but has been the only option for HCV treatment since decades. Paradoxically, the molecular mechanisms of antiviral activity of IFNα emerged only lately, after being used extensively in the clinics, not only as an antiviral, but also in the treatment of other diseases. IFNα is a signaling molecule that binds to its receptor and triggers a kinase cascade and transcription factor activation, leading to the induction of several interferon-stimulated genes (ISGs) with antiviral activity. ISG or a combination of ISGs, directly responsible for particular virus inhibition, is currently an area of intense investigation (Schoggins and Rice, 2011).
HCV belongs to the family Flaviviridae, genus Hepacivirus, while a large number of viruses of human interest belong to the genus Flavivirus (Lindenbach et al., 2007; Baud et al., 2017; Carletti et al., 2017). These include arthropod-borne viruses (arboviruses) delivered by mosquitoes and ticks such as Dengue virus (DENV), Yellow fever virus (YFV), West-Nile virus (WNV), Japanese encephalitis virus (JEV), Zika virus (ZIKV), and tick-borne encephalitis virus (TBEV). These viruses represent a threat to the population particularly in areas naïve to the infection and lately, due to climate change and increased movement of people, became responsible of sudden outbreaks outside endemic areas. Given their increasing importance for human health and the lack of treatment options, these viruses represent a challenge for the development of novel direct antivirals. Current approaches at the pre-clinical stage include targeting the viral enzymes such as: RNA-dependent RNA polymerase, methyltransferase, protease and helicase. However, Flavivirus is a large family and it is difficult to predict which of the virus would be responsible for the next epidemic. Therefore, HDT could represent a strategy for pan-Flavivirus agents, either blocking essential host cell pathways, required by the virus for replication, or activating cellular intrinsic antiviral programs that are common among family members. Only a handful of HDT antiviral drugs have been evaluated up to clinical trials so far and all of them are being used in the treatment of DENV as a result of repurposing. In this review we will focus exclusively on antiviral HDT as an alternative option in treating Flavivirus infection, leaving drugs aimed at viral targets to other readings.
Flavivirus Structure and Life Cycle
Flavivirus virions are composed of a nucleocapsid (C protein), protecting a single-stranded RNA genome (vRNA) of approximately 11 kb with positive polarity, surrounded by a lipid bilayer containing two envelope glycoproteins: E (envelope) and M (membrane) (Lindenbach et al., 2007). Flavivirus genomes are modified with a 5′ m7G cap structure but lack the 3′ polyadenylated tail that is characteristic of most cellular mRNAs. A single long open reading frame (ORF), flanked by 5'and 3' non-coding regions (NCRs), encodes a polyprotein, which is cleaved into different viral proteins by host and viral proteases. The structural proteins capsid (C), pre-membrane (prM), and envelope (E) precede the non-structural proteins NS1, NS2A, NS2B, NS3 (helicase and protease), NS4A, NS4B, and NS5 (RNA-dependent RNA polymerase and methyltransferase). A complete understanding of the process whereby Flaviviruses attach and enter into mammalian and mosquito cells has not been reached, although for certain members (i.e., DENV) more detailed information is available (Muñoz et al., 1998; Acosta et al., 2009; Piccini et al., 2015). It has been proposed that attachment factors on the cell surface are responsible for the first low affinity contact of the virus. Concentration of the virus on the cell surface serves to facilitate binding to specific receptors that eventually promote effective entry in the target cells (Navarro-Sanchez et al., 2003; Miller et al., 2008; Meertens et al., 2012). The uncoating of the nucleocapsid in the cytosol is followed by a Cap-dependent first-round of translation of viral RNA. The multi-transmembrane domain polyprotein precursor localizes on the endoplasmic reticulum (ER), where it is cleaved by cellular and viral enzymes. The NS5 polymerase synthesizes a complementary RNA strand, which then serves as a template for the asymmetric synthesis of additional vRNA. Flavivirus replication occurs in virus-induced vesicles that appear as spherical invagination of the endoplasmic reticulum (ER) and may serve to limit diffusion of viral/host proteins and to protect replication intermediates from the surveillance of cellular cytoplasmic receptors (Miorin et al., 2013; Romero-Brey and Bartenschlager, 2014). The genomic vRNA is believed to be extruded from these replication compartments and assembled with the C protein on the ER membrane, which then buds into the ER lumen. Newly synthesized virions are transported via the secretory pathway, where glycan modification of E and prM as well as cleavage of prME occurs, followed by release at the cell surface (Lindenbach et al., 2007).
OMICS Based Approach for HDT Target Selection
Viruses encode only few essential genes and heavily rely on the host cells to complete their replication cycle. The cell responds to infection by activating antiviral pathways. Therefore, hundreds of host factors are required for either supporting or limiting viral infection, and hence, their modulation could represent optimal targets for HDT. A number of loss-of-function genetic screens based on RNA interference (RNAi), on insertional mutagenesis in human haploid cells (HAP1) or on Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR) technology have been conducted over the years to provide an unbiased and comprehensive strategy to uncover host factors that promote or restrict virus replication (Krishnan et al., 2008; Sessions et al., 2009; Marceau et al., 2016; Zhang et al., 2016). A limited number of gain-of-function screens, based on overexpression of a set of putative antiviral effectors or of cellular microRNAs (miRNA), have also been performed (Schoggins et al., 2011; Smith et al., 2017). Other screens based on viral protein-protein or viral RNA-protein interaction provided information on the cellular pathways engaged by the infection, albeit only after proper functional validation (Ward et al., 2011; Heaton et al., 2016; Phillips et al., 2016). Genetic screens and interactome analysis are valuable instruments to understand the viral replication cycle, although often similar screens poorly overlap in the output, indicating high variability in their execution. Bottom-up approaches, such as those outlined above, are complemented by top-down strategies that take advantage of libraries of compounds of various sources, such as herbal drugs from traditional medicine or drugs approved for the treatment of other pathologies (Sood et al., 2015; Barrows et al., 2016; Wang et al., 2017). Repurposing strategies are particularly attractive because, if successful, may provide ready-to-use drugs that have already passed several steps in the approval process for human treatment. Drug repurposing, also known as drug re-profiling or drug repositioning, is the identification of new therapeutic indications for drugs that are in the clinics or compounds that failed at some stage of the clinical trials. Drug repurposing can be considered a first line approach for neglected infectious diseases primarily occurring in developing countries, where an effective treatment is urgently needed. This strategy reduce time and risks intrinsic to the drug discovery process to quickly advance a drug-candidate to late-stage development, therefore it is very attractive for pharmaceutical companies as it may open new markets for proprietary compounds.
Top-down approaches suffer from difficulties in the definition of the active principle and from the lack of knowledge of the target, except in the case of repurposing where the target has already been identified in most cases. To note, however, that the inhibitory concentration of a drug, repurposed for Flaviviruses infection, could be very different to that used therapeutically for the original disease. This has two important implications, first that the active target might be different, second that the clinical trials should be repeated at the new active concentrations.
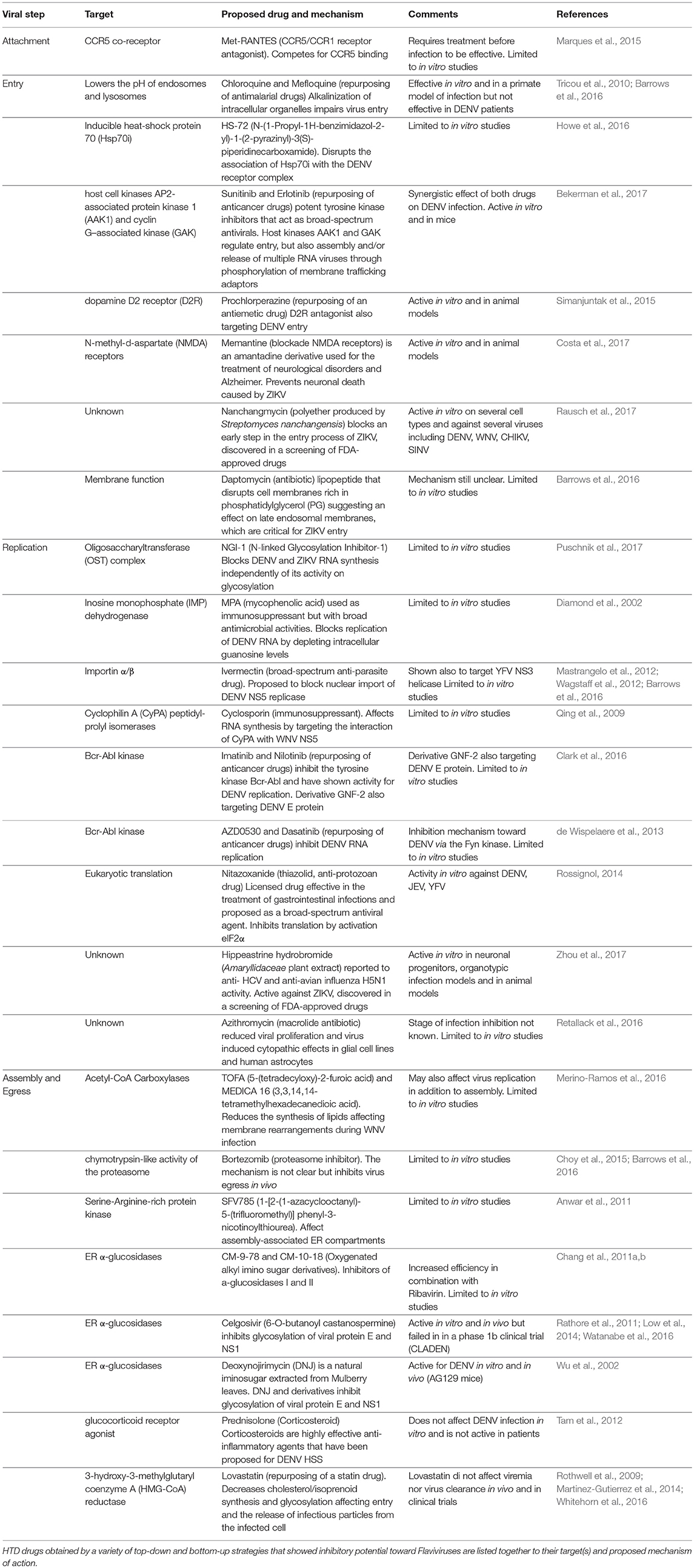
The results from such efforts and from “educated guess” approaches resulted in a number of compounds at various levels of testing. To note, however, that Flaviviruses have dual hosts: mosquitoes/ticks or vertebrates (birds, mammals). Therefore, the analysis that follows will be limited to mammalian cells in view of human treatment. To this end, the lifecycle of Flaviviruses has been divided in four steps: (i) attachment and entry, (ii) translation and polyprotein processing, (iii) replication, and (iv) egress. Attention has been put on druggable host factors at the various steps of the viral life cycle and on promising HTDs that have been discovered as a result of repurposing or screenings. A scheme illustrating the approaches to identify HTD for Flavivirus infection is shown in Figure 1, while Table 1 summarizes the antiviral HTD drugs for Flavivirus treatment proposed to date.
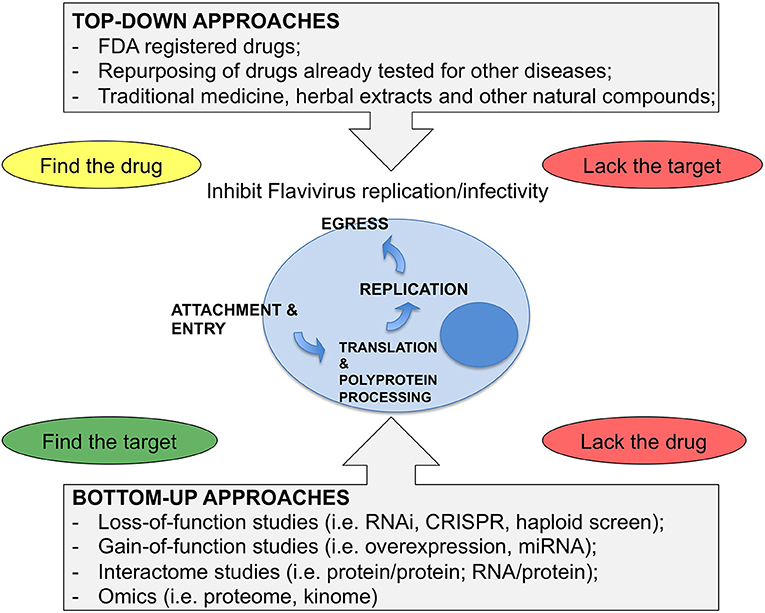
Figure 1. Strategies for antiviral HDT. (Top-Down) approaches consider drugs from various sources as starting material for a screen of antiviral activity. These approaches can identify drugs with both viral and cellular targets. However, when starting from complex mixtures, it is not easy to define the active compound, while screenings of Food and Drug Administration (FDA) registered drugs have the advantage of knowing the drug in advance and, in most cases, the target. Bottom-up approaches are based on unbiased screens by depleting or overexpressing host factors or miRNAs. Here the critical issue is to find the drug for the target that is identified by the screen. (Bottom-Up) approaches also include various approaches where the network of virus-host interactions is studied or where the response of the cell to the infection is analyzed as a whole. These methods require functional validation and only after that it is possible to proceed further and select the drugs.
Attachment and Entry
The E protein coating the virion is responsible for attachment to the cell surface through low-affinity receptors and co-receptors. It is likely that Flaviviruses require multiple co-receptors that define tissue tropism and pathogenesis (Perera et al., 2008; Perera-Lecoin et al., 2013). A number of receptors, for several members of the family, have been proposed in vitro. For example, antagonists of the chemokine co-receptor CCR5, developed for HIV-1 therapy, were effective in DENV treatment (Marques et al., 2015). However, a complete picture of (co)-receptors for Flavivirus entry is yet not available and identification of HDT targets of attachment awaits further studies. After attachment, the virus enters the cell by endocytosis, which could be either clathrin-dependent or independent based on the virus and cell type analyzed (Kalia et al., 2013). Nevertheless, acidification of the late endosome, mediated by the vacuolar-type H + ATPase (vATPase) complex, is a common essential step for all Flaviviruses that is enhanced by the transmembrane protein ribonuclease kappa (RNASEK) (Perreira et al., 2015). Acidification induces conformational changes in the E protein that expose hydrophobic peptides. This permits the fusion of the viral and vesicle membranes to allow nucleocapsid access to the cytoplasm (Kaufmann and Rossmann, 2011). As an example of repurposing, the anti-malaria drug Chloroquine is capable of alkalinizing intracellular organelles, such as endosomes and lysosomes. With such mechanisms, chloroquine is suitable to block entry of flaviviruses that require a pH-mediated fusion step in endosomes. In vitro, this drug inhibited DENV replication in a dose-dependent manner at non-toxic concentrations, with the additional benefit of lowering the activation of pro-inflammatory cytokines (Tricou et al., 2010). However, treatment of patients had no favorable effect on viremia or fever. The final step of the entry, that is uncoating, is least understood. However, several reports have demonstrated that it requires ubiquitin-mediated destabilization of the capsid, which allows ribosomes to access viral RNA (Krishnan et al., 2008; Byk et al., 2016).
Translation and Polyprotein Processing
The viral RNA is capped to allow initiation of translation by host factors. However, DENV has been shown to be able to switch from cap-dependent to cap-independent RNA translation when host cell translation is inhibited experimentally (Edgil et al., 2006). Lack of a poly(A) tail appears to be compensated by direct binding of the poly(A) binding protein (PABP) to the 5′-end of the viral genome (Polacek et al., 2009). In addition, recent findings showed that ZIKV and DENV suppress host cell translation, while translation of their RNA is maintained (Roth et al., 2017; Reid et al., 2018). Ribosomal proteins are required for core ribosome activities and for translation of specific subsets of mRNA and possibly vRNA. RPS25, RPL18, RPLP1/2 have been shown to be required for Flavivirus infectivity and likely required for translation of vRNA (Le Sommer et al., 2012; Cervantes-Salazar et al., 2015; Marceau et al., 2016; Campos et al., 2017). Flavivirus translation occurs at the ER membrane, most probably through the signal recognition particle (SRP) pathway. Indeed, several screens identified proviral components of the SRP pathway and translocon complex (Sessions et al., 2009; Le Sommer et al., 2012; Marceau et al., 2016; Zhang et al., 2016). Antiviral host factors include the TIA/R proteins that bind the TBEV genomes to inhibit translation (Albornoz et al., 2014). The viral polyprotein is cleaved by NS3 and its cofactor NS2B (NS2B/3 complex) and cellular proteases. Three subunits (SPCS1/2/3) of the signal peptidase complex (SPC) have been shown to be required for DENV and WNV infectivity (Zhang et al., 2016). Intriguingly, dependence on SPC appears to be a peculiarity of Flaviviruses not shared by any other RNA virus making it a promising target for therapy. Protein chaperons have been shown to be important for Flavivirus infection at multiple steps of the viral life cycle. HSP70 appears to be required for NS5 folding, but also associates with the capsid and is involved in assembly and possibly in entry (Ye et al., 2013; Taguwa et al., 2015). Protein chaperones, such as HSP70 or HSP90, are considered generally pro-viral as they promote protein folding. These could represent important targets for antiviral therapy, particularly in association with direct antiviral agents to decrease the threshold for mutagenesis leading to resistance (Geller et al., 2007; Anderson et al., 2014). The small molecule HS-72 has been shown to inhibit the stress-inducible form of HSP70 leading to a defect in DENV entry (Howe et al., 2016).
Replication
Replication of Flavivirus genomes occurs in replication vesicles spatially separated from sites of translation (Welsch et al., 2009; Gillespie et al., 2010; Miorin et al., 2013). This probably allows the switch from two incompatible processes targeting the same vRNA: translation and RNA synthesis as well as protecting viral dsRNA from innate sensing (Overby et al., 2010; Miorin et al., 2012). Several host RNA-binding proteins have been involved in vRNA synthesis: exoribonuclease family member 3 (ERI3) localizes to sites of viral replication and enhances RNA synthesis of DENV and YFV (Ward et al., 2016). The AU-rich binding factor 1, p45 isoform (AUF1 p45) and its cofactor arginine methyltransferase (PRMT1) have been shown to promote WNV vRNA cyclization, which is required for vRNA synthesis (Friedrich et al., 2014, 2016). Several other host factors have been shown to bind the viral RNA, but their mechanism of action still remains elusive. Replication vesicles contain both non-structural viral proteins and vRNA and are connected to the cytosol by a pore. The RNA-dependent RNA polymerase NS5 catalyzes the synthesis of the minus-strand vRNA that becomes the template for multiple rounds of positive-stranded vRNA, which then could either be translated, further replicated or assembled in new virions. ER membrane rearrangement is a characteristic hallmark of all Flaviviruses, but the molecular details of the process, which likely requires several host factors, are still not understood. The ER associated oligosaccharyltransferase (OST) complex was recently found to be associated with viral non-structural proteins and required for DENV RNA synthesis. The catalytic function of the OST complex is not required for DENV replication, suggesting that the complex may have a structural role in the formation of replication complex (Marceau et al., 2016). Both STT3A and STT3B subunits of the OST complex are required for DENV replication, but only STT3A is required for YFV, ZIKV, and WNV infectivity, highlighting differences in the requirement for OST complex variants among Flaviviruses. Interestingly, the OST inhibitor NGI-1 has pan-Flavivirus antiviral activity highlighting the critical importance of this cellular pathway, which could be exploited for antiviral HDT (Puschnik et al., 2017). Other processes that induce ER membranes rearrangements have been implicated in Flavivirus replication. DENV, ZIKV, and YFV strongly depend on the ER membrane complex (EMC) for their replication (Savidis et al., 2016). The EMC is involved in transmembrane proteins processing, maturation and is also associated to the OST complex. During Flavivirus replication, an abundant non-coding RNA fragment (termed sfRNA for subgenomic flaviviral RNA) has been detected in infected cells (Pijlman et al., 2008). The sfRNA is derived from incomplete degradation of the viral 3′ NCR by the cellular 5′-3′ exonuclease Xrn1 and has been proposed to regulate various cellular pathways to facilitate flaviviral pathogenicity and to inhibit the interferon/stress response (Bidet et al., 2014; Manokaran et al., 2015). Flaviviral RNA is also edited by host methyltransferases for N6-adenosine methylation (m6A), which modulates viral replication (Gokhale et al., 2016; Lichinchi et al., 2016). Generation of sfRNA and m6A-vRNA are peculiar aspects of the Flavivirus life cycle dependent on host factors that could be targeted for HDT.
Assembly and Egress
Assembly of virions initiate at the ER membrane by association between the capsid and vRNA, which results in the formation of nucleocapsid. Electron microscopy images show assembly occurring juxtaposed to the replication vesicles, possibly to facilitate the process of extruding the vRNA from the vesicles pore and binding to the capsid (Romero-Brey and Bartenschlager, 2014). Association of the nucleocapsid with E and prM heterodimers, inserted into the ER membrane, precedes budding of immature viral particles into the ER lumen. Viral particles are then transported along the secretory pathway to the Golgi where maturation and N-linked glycosylation of prM and E proteins takes place. A reduction in pH triggers a conformational change in prME spikes, which involves the same vATPase required for entry (Duan et al., 2008). The cellular protease Furin cleaves prM in this acidified compartment, converting the immature viral particle into a fully infectious virus that is subsequently released from the cell by vesicular fusion with the plasma membrane. Lipid droplets were shown to play a role in DENV assembly, either as storage for capsid protein or for nucleocapsid formation (Samsa et al., 2009). Several host factors have been involved in capsid localization to lipid droplets indicating the importance of this compartment (Iglesias et al., 2015). Cholesterol is also involved in the formation of lipid droplets required for DENV replication. Lovastatin inhibits DENV by reducing the release of infectious particles from the infected cell, but also by inhibiting virus entry (Rothwell et al., 2009; Martinez-Gutierrez et al., 2014). In vivo and in clinical trials, Lovastatin did not affect viremia nor virus clearance (Whitehorn et al., 2016). Corticosteroids may alleviate symptoms such as, capillary permeability and hemorrhage that are exacerbated in severe Dengue infection. Corticosteroids do not inhibit, neither enhance, Dengue infection in vitro and in patients these treatments were not efficacious (Tam et al., 2012). A key druggable enzyme for lipid synthesis is the acetyl-CoA carboxylase and its inhibitors have been shown to reduce WNV replication significantly (Merino-Ramos et al., 2016). However, targeting lipid metabolism could affect several steps of the virus life cycle. The host helicase DDX56 binds capsid and may facilitate transfer of viral RNA from the replication vesicles to the ER membrane, which is the site of WNV assembly (Xu and Hobman, 2012). Moreover, druggable targets for antiviral therapy could be the Src kinases that have been shown to be involved in the late stages of DENV life cycle and in promoting WNV trafficking through the secretory pathway (Hirsch et al., 2005). Proteins of the endosomal sorting complex required for transport (ESCRT) localize to the sites of DENV and JEV assembly, and their depletion inhibited the production of infectious virus (Tabata et al., 2016). Furthermore, the proteasome inhibitor Bortenzomib was also shown to inhibit DENV production in vitro and in vivo (Choy et al., 2015) as well as ZIKV. However, the mechanism for the latter was not described (Barrows et al., 2016). Other factors such as the Ras-related in brain protein Rab8b and the ADP-ribosylation proteins Arf4/5 were shown to promote Flavivirus egress (Kudelko et al., 2012; Kobayashi et al., 2016). Disruption of the KDEL-based recycling process was shown to be required for DENV egress by associating with prM (Li et al., 2015). The kinase inhibitor SFV785 targets the recruitment and assembly of the nucleocapsid during DENV assembly and reduces the production of infectious DENV (Anwar et al., 2011). An important process that occurs in the secretory pathway is the glycosylation of viral proteins. Within Flavivruses, the targets of glycosylation are the prM, E and NS1 proteins. Flaviviruses strongly depend on glycosylation for infectivity (Sessions et al., 2009). Several iminosugar derivatives, that inhibit glycosylases, demonstrated potent antiviral activity against Flaviviruses, either alone or in combination with Ribavirin (Chang et al., 2011a). Celgosivir (6-O-butanoyl castanospermine) is an inhibitor of ER α-glycosidases, active against DENV with in vitro EC50 in the sub-micromolar range and good selective index. Oral Celgosivir is active in a model of lethal Dengue in mice with reduction of viremia and protection against virus-induced mortality (Rathore et al., 2011; Watanabe et al., 2016). However, in a phase 1b clinical trial (CELADEN), celgosivir failed to show a significant antiviral effect, although modest antiviral trends were observed in patients with secondary infection (Low et al., 2014). Celgosivir antiviral activity against Dengue is both cell and virus-strain dependent and timing of treatment appears to be critical. Although Celgosivir failed in clinical trials, approaches targeting viral protein glycosylation appear very promising because, in addition to affecting directly virus assembly and egress, it also induce ER stress and the unfolded protein response, which could potentiate antiviral activity through apoptosis or the innate signaling pathway (Smith, 2014).
Concluding Remarks
The road toward HDT for treatment of Flavivirus infections is still long, but investing in a detailed understanding of the virus lifecycle at the molecular levels could eventually lead to novel and more potent strategies for treatment.
Current progress outlined in this review identified a panel of compounds with different cellular targets that have a potential inhibitory activity against Flaviviruses. In particular, two pathways are showing great promise: the ER associated OST complex required for RNA synthesis and the induction of ER stress that may induce innate signaling pathways. Studies need to move quickly to in vivo and to clinical trials when appropriate, but even if compounds result not as effective as expected, they remain very informative on the cellular pathways most relevant to Flavivirus infection. Concerning repurposing approaches, the most promising drugs are probably the kinase inhibitors that are already being used for the treatment of inflammation and cancer. These drugs could quickly move to the clinics for antiviral HDT.
The near future would hopefully witness broad-range treatments that will provide a first line pharmaceutical coverage in the case of new epidemics of members of this family of viruses.
Author Contributions
AM drafted the manuscript. TC and MKZ reviewed the manuscript, added references, and prepared the figure.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
Work on flaviviruses in AM's laboratory is supported by the Beneficientia Stiftung, Vaduz Lichtenstein, and by the FLAVIPOC, SEVARE, and ZIDECHIP projects from the Regionez FVG of Italy. We thank present and past members of the lab for fruitful discussions and excellent cooperation.
References
Acosta, E. G., Castilla, V., and Damonte, E. B. (2009). Alternative infectious entry pathways for dengue virus serotypes into mammalian cells. Cell. Microbiol. 11, 1533–1549. doi: 10.1111/j.1462-5822.2009.01345.x
Albornoz, A., Carletti, T., Corazza, G., and Marcello, A. (2014). The stress granule component TIA-1 binds tick-borne encephalitis virus RNA and is recruited to perinuclear sites of viral replication to inhibit viral translation. J. Virol. 88, 6611–6622. doi: 10.1128/JVI.03736-13
Anderson, I., Low, J. S., Weston, S., Weinberger, M., Zhyvoloup, A., Labokha, A. A., et al. (2014). Heat shock protein 90 controls HIV-1 reactivation from latency. Proc. Natl. Acad. Sci. U.S.A. 111, E1528–E1537. doi: 10.1073/pnas.1320178111
Anwar, A., Hosoya, T., Leong, K. M., Onogi, H., Okuno, Y., Hiramatsu, T., et al. (2011). The kinase inhibitor SFV785 dislocates dengue virus envelope protein from the replication complex and blocks virus assembly. PLoS ONE 6:e23246. doi: 10.1371/journal.pone.0023246
Barrows, N. J., Campos, R. K., Powell, S. T., Prasanth, K. R., Schott-Lerner, G., Soto-Acosta, R., et al. (2016). A Screen of FDA-Approved Drugs for Inhibitors of Zika Virus Infection. Cell Host Microbe 20, 259–270. doi: 10.1016/j.chom.2016.07.004
Baud, D., Gubler, D. J., Schaub, B., Lanteri, M. C., and Musso, D. (2017). An update on Zika virus infection. Lancet 390, 2099–2109. doi: 10.1016/S0140-6736(17)31450-2
Bekerman, E., Neveu, G., Shulla, A., Brannan, J., Pu, S. Y., Wang, S., et al. (2017). Anticancer kinase inhibitors impair intracellular viral trafficking and exert broad-spectrum antiviral effects. J. Clin. Invest. 127, 1338–1352. doi: 10.1172/JCI89857
Bidet, K., Dadlani, D., and Garcia-Blanco, M. A. (2014). G3BP1, G3BP2 and CAPRIN1 are required for translation of interferon stimulated mRNAs and are targeted by a dengue virus non-coding RNA. PLoS Pathog. 10:e1004242. doi: 10.1371/journal.ppat.1004242
Byk, L. A., Iglesias, N. G., De Maio, F. A., Gebhard, L. G., Rossi, M., and Gamarnik, A. V. (2016). Dengue virus genome uncoating requires ubiquitination. MBio 7:e00804–16. doi: 10.1128/mBio.00804-16
Campos, R. K., Wong, B., Xie, X., Lu, Y. F., Shi, P. Y., Pompon, J., et al. (2017). RPLP1 and RPLP2 are essential flavivirus host factors that promote early viral protein accumulation. J. Virol. 91:e01706–16. doi: 10.1128/JVI.01706-16
Carletti, T., Zakaria, M. K., and Marcello, A. (2017). The host cell response to tick-borne encephalitis virus. Biochem. Biophys. Res. Commun. 492, 533–540. doi: 10.1016/j.bbrc.2017.02.006
Cervantes-Salazar, M., Angel-Ambrocio, A. H., Soto-Acosta, R., Bautista-Carbajal, P., Hurtado-Monzon, A. M., Alcaraz-Estrada, S. L., et al. (2015). Dengue virus NS1 protein interacts with the ribosomal protein RPL18: this interaction is required for viral translation and replication in Huh-7 cells. Virology 484, 113–126. doi: 10.1016/j.virol.2015.05.017
Chang, J., Schul, W., Butters, T. D., Yip, A., Liu, B., Goh, A., et al. (2011a). Combination of alpha-glucosidase inhibitor and ribavirin for the treatment of dengue virus infection in vitro and in vivo. Antiviral Res. 89, 26–34. doi: 10.1016/j.antiviral.2010.11.002
Chang, J., Schul, W., Yip, A., Xu, X., Guo, J. T., and Block, T. M. (2011b). Competitive inhibitor of cellular alpha-glucosidases protects mice from lethal dengue virus infection. Antiviral Res. 92, 369–371. doi: 10.1016/j.antiviral.2011.08.003
Choy, M. M., Zhang, S. L., Costa, V. V., Tan, H. C., Horrevorts, S., and Ooi, E. E. (2015). Proteasome inhibition suppresses dengue virus egress in antibody dependent infection. PLoS Negl. Trop. Dis. 9:e0004058. doi: 10.1371/journal.pntd.0004058
Clark, M. J., Miduturu, C., Schmidt, A. G., Zhu, X., Pitts, J. D., Wang, J., et al. (2016). GNF-2 inhibits dengue virus by targeting Abl kinases and the viral E protein. Cell Chem. Biol. 23, 443–452. doi: 10.1016/j.chembiol.2016.03.010
Costa, V. V., Del Sarto, J. L., Rocha, R. F., Silva, F. R., Doria, J. G., Olmo, I. G., et al. (2017). N-Methyl-d-Aspartate (NMDA) receptor blockade prevents neuronal death induced by Zika virus infection. MBio 8:e00350–17. doi: 10.1128/mBio.00350-17
de Wispelaere, M., LaCroix, A. J., and Yang, P. L. (2013). The small molecules AZD0530 and dasatinib inhibit dengue virus RNA replication via Fyn kinase. J. Virol. 87, 7367–7381. doi: 10.1128/JVI.00632-13
Diamond, M. S., Zachariah, M., and Harris, E. (2002). Mycophenolic acid inhibits dengue virus infection by preventing replication of viral RNA. Virology 304, 211–221. doi: 10.1006/viro.2002.1685
Duan, X., Lu, X., Li, J., and Liu, Y. (2008). Novel binding between pre-membrane protein and vacuolar ATPase is required for efficient dengue virus secretion. Biochem. Biophys. Res. Commun. 373, 319–324. doi: 10.1016/j.bbrc.2008.06.041
Edgil, D., Polacek, C., and Harris, E. (2006). Dengue virus utilizes a novel strategy for translation initiation when cap-dependent translation is inhibited. J. Virol. 80, 2976–2986. doi: 10.1128/JVI.80.6.2976-2986.2006
Elion, G. B. (1993). Acyclovir: discovery, mechanism of action, and selectivity. J. Med. Virol. (Suppl. 1), 2–6. doi: 10.1002/jmv.1890410503
Friedrich, S., Schmidt, T., Geissler, R., Lilie, H., Chabierski, S., Ulbert, S., et al. (2014). AUF1 p45 promotes West Nile virus replication by an RNA chaperone activity that supports cyclization of the viral genome. J. Virol. 88, 11586–11599. doi: 10.1128/JVI.01283-14
Friedrich, S., Schmidt, T., Schierhorn, A., Lilie, H., Szczepankiewicz, G., Bergs, S., et al. (2016). Arginine methylation enhances the RNA chaperone activity of the West Nile virus host factor AUF1 p45. RNA 22, 1574–1591. doi: 10.1261/rna.055269.115
Geller, R., Vignuzzi, M., Andino, R., and Frydman, J. (2007). Evolutionary constraints on chaperone-mediated folding provide an antiviral approach refractory to development of drug resistance. Genes Dev. 21, 195–205. doi: 10.1101/gad.1505307
Gillespie, L. K., Hoenen, A., Morgan, G., and Mackenzie, J. M. (2010). The Endoplasmic reticulum provides the membrane platform for biogenesis of the flavivirus replication complex. J. Virol. 84, 10438–47. doi: 10.1128/JVI.00986-10
Gokhale, N. S., McIntyre, A. B. R., McFadden, M. J., Roder, A. E., Kennedy, E. M., Gandara, J. A., et al. (2016). N6-Methyladenosine in flaviviridae viral RNA genomes regulates infection. Cell Host Microbe 20, 654–665. doi: 10.1016/j.chom.2016.09.015
Heaton, N. S., Moshkina, N., Fenouil, R., Gardner, T. J., Aguirre, S., Shah, P. S., et al. (2016). Targeting viral proteostasis limits influenza virus, HIV, and dengue virus infection. Immunity 44, 46–58. doi: 10.1016/j.immuni.2015.12.017
Hirsch, A. J., Medigeshi, G. R., Meyers, H. L., DeFilippis, V., Fruh, K., Briese, T., et al. (2005). The Src family kinase c-Yes is required for maturation of West Nile virus particles. J. Virol. 79, 11943–11951. doi: 10.1128/JVI.79.18.11943-11951.2005
Howe, M. K., Speer, B. L., Hughes, P. F., Loiselle, D. R., Vasudevan, S., and Haystead, T. A. (2016). An inducible heat shock protein 70 small molecule inhibitor demonstrates anti-dengue virus activity, validating Hsp70 as a host antiviral target. Antiviral Res. 130, 81–92. doi: 10.1016/j.antiviral.2016.03.017
Iglesias, N. G., Mondotte, J. A., Byk, L. A., De Maio, F. A., Samsa, M. M., Alvarez, C., et al. (2015). Dengue virus uses a non-canonical function of the host GBF1-Arf-COPI system for capsid protein accumulation on lipid droplets. Traffic 16, 962–977. doi: 10.1111/tra.12305
Kalia, M., Khasa, R., Sharma, M., Nain, M., and Vrati, S. (2013). Japanese encephalitis virus infects neuronal cells through a clathrin-independent endocytic mechanism. J. Virol. 87, 148–162. doi: 10.1128/JVI.01399-12
Kaufmann, B., and Rossmann, M. G. (2011). Molecular mechanisms involved in the early steps of flavivirus cell entry. Microbes Infect. 13, 1–9. doi: 10.1016/j.micinf.2010.09.005
Kobayashi, S., Suzuki, T., Kawaguchi, A., Phongphaew, W., Yoshii, K., Iwano, T., et al. (2016). Rab8b regulates transport of west nile virus particles from recycling endosomes. J. Biol. Chem. 291, 6559–6568. doi: 10.1074/jbc.M115.712760
Krishnan, M. N., Ng, A., Sukumaran, B., Gilfoy, F. D., Uchil, P. D., Sultana, H., et al. (2008). RNA interference screen for human genes associated with West Nile virus infection. Nature 455, 242–245. doi: 10.1038/nature07207
Kudelko, M., Brault, J. B., Kwok, K., Li, M. Y., Pardigon, N., Peiris, J. S., et al. (2012). Class II ADP-ribosylation factors are required for efficient secretion of dengue viruses. J. Biol. Chem. 287, 767–777. doi: 10.1074/jbc.M111.270579
Le Sommer, C., Barrows, N. J., Bradrick, S. S., Pearson, J. L., and Garcia-Blanco, M. A. (2012). G protein-coupled receptor kinase 2 promotes flaviviridae entry and replication. PLoS Negl. Trop. Dis. 6:e1820. doi: 10.1371/journal.pntd.0001820
Li, M. Y., Grandadam, M., Kwok, K., Lagache, T., Siu, Y. L., Zhang, J. S., et al. (2015). KDEL receptors assist dengue virus exit from the endoplasmic reticulum. Cell Rep. 10, 1496–1507. doi: 10.1016/j.celrep.2015.02.021
Lichinchi, G., Zhao, B. S., Wu, Y., Lu, Z., Qin, Y., He, C., et al. (2016). Dynamics of human and viral RNA methylation during Zika Virus infection. Cell Host Microbe 20, 666–673. doi: 10.1016/j.chom.2016.10.002
Lindenbach, B. D., Thiel, H. J., and Rice, C. M. (2007). “Flaviviridae: the viruses and their replication,” in Fields Virology, eds D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, and M. A. Martin (Philadelphia, PA: Lippincott, Williams & Wilkins), 1101–1152.
Low, J. G., Sung, C., Wijaya, L., Wei, Y., Rathore, A. P. S., Watanabe, S., et al. (2014). Efficacy and safety of celgosivir in patients with dengue fever (CELADEN): a phase 1b, randomised, double-blind, placebo-controlled, proof-of-concept trial. Lancet Infect. Dis. 14, 706–715. doi: 10.1016/S1473-3099(14)70730-3
Manokaran, G., Finol, E., Wang, C., Gunaratne, J., Bahl, J., Ong, E. Z., et al. (2015). Dengue subgenomic RNA binds TRIM25 to inhibit interferon expression for epidemiological fitness. Science 350:217–221. doi: 10.1126/science.aab3369
Marceau, C. D., Puschnik, A. S., Majzoub, K., Ooi, Y. S., Brewer, S. M., Fuchs, G., et al. (2016). Genetic dissection of Flaviviridae host factors through genome-scale CRISPR screens. Nature 535, 159–163. doi: 10.1038/nature18631
Marques, R. E., Guabiraba, R., Del Sarto, J. L., Rocha, R. F., Queiroz, A. L., Cisalpino, D., et al. (2015). Dengue virus requires the CC-chemokine receptor CCR5 for replication and infection development. Immunology 145, 583–596. doi: 10.1111/imm.12476
Martinez-Gutierrez, M., Correa-Londono, L. A., Castellanos, J. E., Gallego-Gomez, J. C., and Osorio, J. E. (2014). Lovastatin delays infection and increases survival rates in AG129 mice infected with dengue virus serotype 2. PLoS ONE 9:e87412. doi: 10.1371/journal.pone.0087412
Mastrangelo, E., Pezzullo, M., De Burghgraeve, T., Kaptein, S., Pastorino, B., Dallmeier, K., et al. (2012). Ivermectin is a potent inhibitor of flavivirus replication specifically targeting NS3 helicase activity: new prospects for an old drug. J. Antimicrob. Chemother. 67, 1884–1894. doi: 10.1093/jac/dks147
McHutchison, J. G., Gordon, S. C., Schiff, E. R., Shiffman, M. L., Lee, W. M., Rustgi, V. K., et al. (1998). Interferon alfa-2b alone or in combination with ribavirin as initial treatment for chronic hepatitis C. Hepatitis Interventional Therapy Group. N. Engl. J. Med. 339, 1485–1492. doi: 10.1056/NEJM199811193392101
Meertens, L., Carnec, X., Lecoin, M. P., Ramdasi, R., Guivel-Benhassine, F., Lew, E., et al. (2012). The TIM and TAM families of phosphatidylserine receptors mediate dengue virus entry. Cell Host Microbe 12, 544–557. doi: 10.1016/j.chom.2012.08.009
Merino-Ramos, T., Vazquez-Calvo, A., Casas, J., Sobrino, F., Saiz, J. C., and Martin-Acebes, M. A. (2016). Modification of the host cell lipid metabolism induced by hypolipidemic drugs targeting the acetyl coenzyme A carboxylase impairs West Nile virus replication. Antimicrob. Agents Chemother. 60, 307–315. doi: 10.1128/AAC.01578-15
Miller, J. L., de Wet, B. J., Martinez-Pomares, L., Radcliffe, C. M., Dwek, R. A., Rudd, P. M., et al. (2008). The mannose receptor mediates dengue virus infection of macrophages. PLoS Pathog. 4:e17. doi: 10.1371/annotation/98b92fca-fa6e-4bf3-9b39-13b66b640476
Miorin, L., Albornoz, A., Baba, M. M., D'Agaro, P., and Marcello, A. (2012). Formation of membrane-defined compartments by tick-borne encephalitis virus contributes to the early delay in interferon signaling. Virus Res. 163, 660–666. doi: 10.1016/j.virusres.2011.11.020
Miorin, L., Romero-Brey, I., Maiuri, P., Hoppe, S., Krijnse-Locker, J., Bartenschlager, R., et al. (2013). Three-dimensional architecture of tick-borne encephalitis virus replication sites and trafficking of the replicated RNA. J. Virol. 87, 6469–6481. doi: 10.1128/JVI.03456-12
Muñoz, M. L., Cisneros, A., Cruz, J., Das, P., Tovar, R., and Ortega, A. (1998). Putative dengue virus receptors from mosquito cells. FEMS Microbiol. Lett. 168, 251–258. doi: 10.1016/S0378-1097(98)00434-0
Navarro-Sanchez, E., Altmeyer, R., Amara, A., Schwartz, O., Fieschi, F., Virelizier, J. L., et al. (2003). Dendritic-cell-specific ICAM3-grabbing non-integrin is essential for the productive infection of human dendritic cells by mosquito-cell-derived dengue viruses. EMBO Rep. 4, 723–728. doi: 10.1038/sj.embor.embor866
Overby, A. K., Popov, V. L., Niedrig, M., and Weber, F. (2010). Tick-borne encephalitis virus delays interferon induction and hides its double-stranded RNA in intracellular membrane vesicles. J. Virol. 84, 8470–8483. doi: 10.1128/JVI.00176-10
Palù, G., Gerna, G., Bevilacqua, F., and Marcello, A. (1992). A point mutation in the thymidine kinase gene is responsible for acyclovir-resistance in herpes simplex virus type 2 sequential isolates. Virus Res. 25, 133–144. doi: 10.1016/0168-1702(92)90105-I
Pawlotsky, J. M., Feld, J. J., Zeuzem, S., and Hoofnagle, J. H. (2015). From non-A, non-B hepatitis to hepatitis C virus cure. J. Hepatol. 62, S87–S99. doi: 10.1016/j.jhep.2015.02.006
Perera, R., Khaliq, M., and Kuhn, R. J. (2008). Closing the door on flaviviruses: entry as a target for antiviral drug design. Antiviral Res. 80, 11–22. doi: 10.1016/j.antiviral.2008.05.004
Perera-Lecoin, M., Meertens, L., Carnec, X., and Amara, A. (2013). Flavivirus entry receptors: an update. Viruses 6, 69–88. doi: 10.3390/v6010069
Perreira, J. M., Aker, A. M., Savidis, G., Chin, C. R., McDougall, W. M., Portmann, J. M., et al. (2015). RNASEK Is a V-ATPase-associated factor required for endocytosis and the replication of rhinovirus, influenza A virus, and dengue virus. Cell Rep. 12, 850–863. doi: 10.1016/j.celrep.2015.06.076
Phillips, S. L., Soderblom, E. J., Bradrick, S. S., and Garcia-Blanco, M. A. (2016). Identification of proteins bound to dengue viral RNA in vivo reveals new host proteins important for virus replication. MBio 7, e01865–15. doi: 10.1128/mBio.01865-15
Piccini, L. E., Castilla, V., and Damonte, E. B. (2015). Dengue-3 virus entry into vero cells: role of clathrin-mediated endocytosis in the outcome of infection. PLoS ONE 10:e0140824. doi: 10.1371/journal.pone.0140824
Pijlman, G. P., Funk, A., Kondratieva, N., Leung, J., Torres, S., van der Aa, L., et al. (2008). A highly structured, nuclease-resistant, noncoding RNA produced by flaviviruses is required for pathogenicity. Cell Host Microbe 4, 579–591. doi: 10.1016/j.chom.2008.10.007
Polacek, C., Friebe, P., and Harris, E. (2009). Poly(A)-binding protein binds to the non-polyadenylated 3' untranslated region of dengue virus and modulates translation efficiency. J. Gen. Virol. 90, 687–692. doi: 10.1099/vir.0.007021-0
Puschnik, A. S., Marceau, C. D., Ooi, Y. S., Majzoub, K., Rinis, N., Contessa, J. N., et al. (2017). A small-molecule oligosaccharyltransferase inhibitor with pan-flaviviral activity. Cell Rep. 21, 3032–3039. doi: 10.1016/j.celrep.2017.11.054
Qing, M., Yang, F., Zhang, B., Zou, G., Robida, J. M., Yuan, Z., et al. (2009). Cyclosporine inhibits flavivirus replication through blocking the interaction between host cyclophilins and viral NS5 protein. Antimicrob. Agents Chemother. 53, 3226–3235. doi: 10.1128/AAC.00189-09
Rathore, A. P., Paradkar, P. N., Watanabe, S., Tan, K. H., Sung, C., Connolly, J. E., et al. (2011). Celgosivir treatment misfolds dengue virus NS1 protein, induces cellular pro-survival genes and protects against lethal challenge mouse model. Antiviral Res. 92, 453–460. doi: 10.1016/j.antiviral.2011.10.002
Rausch, K., Hackett, B. A., Weinbren, N. L., Reeder, S. M., Sadovsky, Y., Hunter, C. A., et al. (2017). Screening bioactives reveals nanchangmycin as a broad spectrum antiviral active against Zika Virus. Cell Rep. 18, 804–815. doi: 10.1016/j.celrep.2016.12.068
Reid, D. W., Campos, R. K., Child, J. R., Zheng, T., Chan, K. W. K., Bradrick, S. S., et al. (2018). Dengue virus selectively annexes endoplasmic reticulum-associated translation machinery as a strategy for co-opting host cell protein synthesis. J. Virol. 92:e01766–17. doi: 10.1128/JVI.01766-17
Retallack, H., Di Lullo, E., Arias, C., Knopp, K. A., Laurie, M. T., Sandoval-Espinosa, C., et al. (2016). Zika virus cell tropism in the developing human brain and inhibition by azithromycin. Proc. Natl. Acad. Sci. U.S.A. 113, 14408–14413. doi: 10.1073/pnas.1618029113
Romero-Brey, I., and Bartenschlager, R. (2014). Membranous replication factories induced by plus-strand RNA viruses. Viruses 6, 2826–2857. doi: 10.3390/v6072826
Rossignol, J. F. (2014). Nitazoxanide: a first-in-class broad-spectrum antiviral agent. Antiviral Res. 110, 94–103. doi: 10.1016/j.antiviral.2014.07.014
Roth, H., Magg, V., Uch, F., Mutz, P., Klein, P., Haneke, K., et al. (2017). Flavivirus infection uncouples translation suppression from cellular stress responses. MBio 8:e02150–16. doi: 10.1128/mBio.02150-16
Rothwell, C., Lebreton, A., Young Ng, C., Lim, J. Y., Liu, W., Vasudevan, S., et al. (2009). Cholesterol biosynthesis modulation regulates dengue viral replication. Virology 389, 8–19. doi: 10.1016/j.virol.2009.03.025
Samsa, M. M., Mondotte, J. A., Iglesias, N. G., Assuncao-Miranda, I., Barbosa-Lima, G., Da Poian, A. T., et al. (2009). Dengue virus capsid protein usurps lipid droplets for viral particle formation. PLoS Pathog. 5:e1000632. doi: 10.1371/journal.ppat.1000632
Savidis, G., McDougall, W. M., Meraner, P., Perreira, J. M., Portmann, J. M., Trincucci, G., et al. (2016). Identification of Zika Virus and Dengue Virus dependency factors using functional genomics. Cell Rep. 16, 232–246. doi: 10.1016/j.celrep.2016.06.028
Schoggins, J. W., and Rice, C. M. (2011). Interferon-stimulated genes and their antiviral effector functions. Curr. Opin. Virol. 1, 519–525. doi: 10.1016/j.coviro.2011.10.008
Schoggins, J. W., Wilson, S. J., Panis, M., Murphy, M. Y., Jones, C. T., Bieniasz, P., et al. (2011). A diverse range of gene products are effectors of the type I interferon antiviral response. Nature 472, 481–485. doi: 10.1038/nature09907
Sessions, O. M., Barrows, N. J., Souza-Neto, J. A., Robinson, T. J., Hershey, C. L., Rodgers, M. A., et al. (2009). Discovery of insect and human dengue virus host factors. Nature 458, 1047–1050. doi: 10.1038/nature07967
Simanjuntak, Y., Liang, J. J., Lee, Y. L., and Lin, Y. L. (2015). Repurposing of prochlorperazine for use against dengue virus infection. J. Infect. Dis. 211, 394–404. doi: 10.1093/infdis/jiu377
Smith, J. A. (2014). A new paradigm: innate immune sensing of viruses via the unfolded protein response. Front. Microbiol. 5:222. doi: 10.3389/fmicb.2014.00222
Smith, J. L., Jeng, S., McWeeney, S. K., and Hirsch, A. J. (2017). A MicroRNA screen identifies the wnt signaling pathway as a regulator of the interferon response during flavivirus infection. J. Virol. 91:e02388-16. doi: 10.1128/JVI.02388-16
Sood, R., Raut, R., Tyagi, P., Pareek, P. K., Barman, T. K., Singhal, S., et al. (2015). Cissampelos pareira linn: natural source of potent antiviral activity against all four Dengue Virus serotypes. PLoS Negl. Trop. Dis. 9:e0004255. doi: 10.1371/journal.pntd.0004255
Tabata, K., Arimoto, M., Arakawa, M., Nara, A., Saito, K., Omori, H., et al. (2016). Unique requirement for ESCRT factors in flavivirus particle formation on the endoplasmic reticulum. Cell Rep. 16, 2339–2347. doi: 10.1016/j.celrep.2016.07.068
Taguwa, S., Maringer, K., Li, X., Bernal-Rubio, D., Rauch, J. N., Gestwicki, J. E., et al. (2015). Defining Hsp70 subnetworks in Dengue Virus replication reveals key vulnerability in flavivirus infection. Cell 163, 1108–1123. doi: 10.1016/j.cell.2015.10.046
Tam, D. T., Ngoc, T. V., Tien, N. T., Kieu, N. T., Thuy, T. T., Thanh, L. T., et al. (2012). Effects of short-course oral corticosteroid therapy in early dengue infection in Vietnamese patients: a randomized, placebo-controlled trial. Clin. Infect. Dis. 55, 1216–1224. doi: 10.1093/cid/cis655
Tricou, V., Minh, N. N., Van, T. P., Lee, S. J., Farrar, J., Wills, B., et al. (2010). A randomized controlled trial of chloroquine for the treatment of dengue in Vietnamese adults. PLoS Negl. Trop. Dis. 4:e785. doi: 10.1371/journal.pntd.0000785
Van Lint, C., Bouchat, S., and Marcello, A. (2013). HIV-1 transcription and latency: an update. Retrovirology 10:67. doi: 10.1186/1742-4690-10-67
Wagstaff, K. M., Sivakumaran, H., Heaton, S. M., Harrich, D., and Jans, D. A. (2012). Ivermectin is a specific inhibitor of importin alpha/beta-mediated nuclear import able to inhibit replication of HIV-1 and dengue virus. Biochem. J. 443, 851–856. doi: 10.1042/BJ20120150
Wang, S., Liu, Y., Guo, J., Wang, P., Zhang, L., Xiao, G., et al. (2017). Screening of FDA-approved drugs for inhibitors of japanese encephalitis virus infection. J. Virol. 91:e01055-17. doi: 10.1128/JVI.01055-17
Ward, A. M., Bidet, K., Yinglin, A., Ler, S. G., Hogue, K., Blackstock, W., et al. (2011). Quantitative mass spectrometry of DENV-2 RNA-interacting proteins reveals that the DEAD-box RNA helicase DDX6 binds the DB1 and DB2 3' UTR structures. RNA Biol. 8, 1173–1186. doi: 10.4161/rna.8.6.17836
Ward, A. M., Calvert, M. E., Read, L. R., Kang, S., Levitt, B. E., Dimopoulos, G., et al. (2016). The Golgi associated ERI3 is a Flavivirus host factor. Sci. Rep. 6:34379. doi: 10.1038/srep34379
Watanabe, S., Chan, K. W., Dow, G., Ooi, E. E., Low, J. G., and Vasudevan, S. G. (2016). Optimizing celgosivir therapy in mouse models of dengue virus infection of serotypes 1 and 2: the search for a window for potential therapeutic efficacy. Antiviral Res. 127, 10–19. doi: 10.1016/j.antiviral.2015.12.008
Welsch, S., Miller, S., Romero-Brey, I., Merz, A., Bleck, C. K., Walther, P., et al. (2009). Composition and three-dimensional architecture of the dengue virus replication and assembly sites. Cell Host Microbe 5, 365–375. doi: 10.1016/j.chom.2009.03.007
Whitehorn, J., Nguyen, C. V. V., Khanh, L. P., Kien, D. T. H., Quyen, N. T. H., Tran, N. T. T., Hang, N. T., Truong, N. T., Hue Tai, L. T., Cam Huong, N. T., et al. (2016). Lovastatin for the treatment of adult patients with dengue: a randomized, double-blind, placebo-controlled trial. Clin. Infect. Dis. 62, 468–476. doi: 10.1093/cid/civ949
Wu, S. F., Lee, C. J., Liao, C. L., Dwek, R. A., Zitzmann, N., and Lin, Y. L. (2002). Antiviral effects of an iminosugar derivative on flavivirus infections. J. Virol. 76, 3596–3604. doi: 10.1128/JVI.76.8.3596-3604.2002
Xu, Z., and Hobman, T. C. (2012). The helicase activity of DDX56 is required for its role in assembly of infectious West Nile virus particles. Virology 433, 226–235. doi: 10.1016/j.virol.2012.08.011
Ye, J., Chen, Z., Zhang, B., Miao, H., Zohaib, A., Xu, Q., et al. (2013). Heat shock protein 70 is associated with replicase complex of Japanese encephalitis virus and positively regulates viral genome replication. PLoS ONE 8:e75188. doi: 10.1371/journal.pone.0075188
Zhang, R., Miner, J. J., Gorman, M. J., Rausch, K., Ramage, H., White, J. P., et al. (2016). A CRISPR screen defines a signal peptide processing pathway required by flaviviruses. Nature 535, 164–168. doi: 10.1038/nature18625
Zhou, T., Tan, L., Cederquist, G. Y., Fan, Y., Hartley, B. J., Mukherjee, S., et al. (2017). High-content screening in hPSC-neural progenitors identifies drug candidates that inhibit Zika virus infection in fetal-like organoids and adult brain. Cell Stem Cell 21, 274.e5–283.e5. doi: 10.1016/j.stem.2017.06.017
Keywords: flavivirus, antiviral, host-directed therapy, screening tools, mechanism of action
Citation: Zakaria MK, Carletti T and Marcello A (2018) Cellular Targets for the Treatment of Flavivirus Infections. Front. Cell. Infect. Microbiol. 8:398. doi: 10.3389/fcimb.2018.00398
Received: 04 June 2018; Accepted: 23 October 2018;
Published: 12 November 2018.
Edited by:
Shelton S. Bradrick, The University of Texas Medical Branch at Galveston, United StatesReviewed by:
Penghua Wang, University of Connecticut Health Center, United StatesZhilong Yang, Kansas State University, United States
Copyright © 2018 Zakaria, Carletti and Marcello. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Alessandro Marcello, bWFyY2VsbG9AaWNnZWIub3Jn
†Present Address: Mohammad Khalid Zakaria, The Pirbright Institute, Surrey, United Kingdom
 Mohammad Khalid Zakaria
Mohammad Khalid Zakaria Tea Carletti
Tea Carletti Alessandro Marcello
Alessandro Marcello