- 1Department of Microbiology and Medical Science, Chungnam National University School of Medicine, Daejeon, South Korea
- 2Department of Nanobiomedical Science, Dankook University, Cheonan, South Korea
Multidrug efflux systems contribute to antimicrobial resistance and pathogenicity in bacteria. Here, we report the identification and characterization of a transcriptional regulator AcrR controlling the yet uncharacterized multidrug efflux pump, AcrAB in Acinetobacter nosocomialis. In silico analysis revealed that the homologs of AcrR and AcrAB are reported in the genomes of many other bacterial species. We confirmed that the genes encoding the AcrAB efflux pump, acrA and acrB forms a polycistronic operon which is under the control of acrR gene upstream of acrA. Bioinformatic analysis indicated the presence of AcrR binding motif in the promoter region of acrAB operon and the specific binding of AcrR was confirmed by electrophoretic mobility shift assay (EMSA). The EMSA data showed that AcrR binds to −89 bp upstream of the start codon of acrA. The mRNA expression analysis depicted that the expression of acrA and acrB genes are elevated in the deletion mutant compared to that in the wild type confirming that AcrR acts as a repressor of acrAB operon in A. nosocomialis. The deletion of acrR resulted in increased motility, biofilm/pellicle formation and invasion in A. nosocomialis. We further analyzed the role of AcrR in A. nosocomialis pathogenesis in vivo using murine model and it was shown that acrR mutant is highly virulent inducing severe infection in mouse leading to host death. In addition, the intracellular survival rate of acrR mutant was higher compared to that of wild type. Our data demonstrates that AcrR functions as an important regulator of AcrAB efflux pump and is associated with several phenotypes such as motility, biofilm/pellicle formation and pathogenesis in A. nosocomialis.
Introduction
Acinetobacter spp. are important opportunistic bacterial pathogens primarily associated with nosocomial infections worldwide. These are the causative organisms for a number of pathological conditions including bacteremia, pneumonia, meningitis, urinary tract infections, and wound infections (Peleg et al., 2008; Lee et al., 2011; Nemec et al., 2011). The medically relevant Acinetobacter spp. predominantly come from a single group, designated as Acinetobacter baumannii (Ab) group which includes A. baumannii, Acinetobacter nosocomialis, Acinetobacter pittii, Acinetobacter seifertii, and Acinetobacter dijkshoorniae. All the members of the Ab group are opportunistic pathogens and have the capacity to acquire multidrug resistance phenotype (Nemec et al., 2015; Cosgaya et al., 2016; Marí-Almirall et al., 2017). A. nosocomialis is the most comprehensively studied strain in the Ab group second to A. baumannii. A. baumannii is reported to rapidly develop resistance to multiple antibiotics and is one of the most challenging pathogens for healthcare institutions (Fournier and Richet, 2006; Antunes et al., 2014; Doi et al., 2015). The increasing rate of multidrug resistance (MDR) phenotypes associated with Acinetobacter infections are so alarming that the Centers for Disease Control and Prevention (CDC) has listed Acinetobacter as a severe threat to mankind and last line therapeutic options like carbapenems, tigecycline, and colistin are now recommended to treat majority of Acinetobacter infections (Peleg and Hooper, 2010; CDC, 2013). Moreover, according to the recent epidemiological data, MDR strains acquire multidrug resistance phenotype faster than any of the other Gram-negative bacteria reported over the last decade (Giammanco et al., 2017).
Multidrug efflux pumps contribute to antibiotic resistance by extruding out drugs and are found in almost all bacterial species. These pumps have been categorized into five families on the basis of sequence similarity: resistance-nodulation-cell division (RND), the major facilitator, small multidrug resistance, multidrug and toxic compound extrusion, and ATP-binding cassette (ABC) families (Putman et al., 2000; Paulsen et al., 2001). In Gram negative bacteria, RND family efflux pumps are mostly effective in resistance to antibiotics (Nikaido, 1996; Murakami et al., 2002, 2006) and consist of an RND protein (the inner membrane component), membrane fusion protein (MFP: the periplasmic component), and the outer membrane protein (OMP: the outer membrane protein). The RND pumps were first reported in Escherichia coli and Pseudomonas aeruginosa independently and those include AcrAB-TolC (E. coli) (Ma et al., 1993, 1995), MexAB-OprM, MexCD-OprJ, MexEF-OprN etc. (P. aeruginosa) (Poole et al., 1993, 1996; Gotoh et al., 1995; Köhler et al., 1997). In E. coli, the increased level of antibiotic resistance is attributed majorly by this housekeeping efflux system AcrAB-TolC (Thanassi et al., 1997). The substrate profile of the AcrAB-TolC efflux pump in E. coli includes chloramphenicol, fluoroquinolone, tetracycline, novobiocin, rifampin, fusidic acid, nalidixic acid, and β-lactam antibiotics (Piddock, 2006). Apart from E. coli, the function of AcrAB efflux pump or its homologs on multidrug resistance has been reported in a number of bacterial strains including pseudomonads, Salmonella enterica serovar Typhimurium, Enterobacter aerogenes, Klebsiella pneumoniae, Enterobacter cloacae, Campylobacter spp., and Borrelia burgdorferi, etc. (Poole et al., 1993; Pumbwe et al., 2004; Buckley et al., 2006; Bunikis et al., 2008). Like that in E. coli, the AcrAB-TolC system in S. enterica is reported to actively expel different classes of antimicrobial agents such as quinolones, chloramphenicol, tetracycline, and nalidixic acid (Poole, 2007; Nishino et al., 2009).
The efflux pumps are almost exclusive to A. baumannii, A. pittii, and A. nosocomialis and their orthologues are distributed in other bacterial pathogens, such as Pseudomonas spp. The AdeABC efflux pump of A. baumannii confers resistance to various classes of antibiotics including aminoglycosides, tetracyclines, erythromycin, chloramphenicol, trimethoprim, fluoroquinolones, some β-lactams, tigecycline etc. (Magnet et al., 2001; Marchand et al., 2004).
The adeABC operon is found in 80% of the clinical isolates and its expression is tightly regulated by the two-component system, AdeRS (Marchand et al., 2004). A second RND efflux pump was reported in A. baumannii known as the AdeIJK which contributes resistance to β-lactams, chloramphenicol, tetracycline, tigecycline, erythromycin, lincosamides, fluoroquinolones, fusidic acid, novobiocin, rifampin, trimethoprim, acridine, safranin, pyronine, and sodium dodecyl sulfate (Laurence et al., 2007). The RND efflux pump, AdeFGH, encoded by adeFGH operon provides high-level resistance to fluoroquinolones, chloramphenicol, trimethoprim, clindamycin etc. and is under the control of a putative LysR-type transcriptional regulator, named adeL, which is located upstream of the adeFGH operon (Coyne et al., 2010a,b). Besides their role in antibiotic resistance, RND efflux pumps contribute to virulence and biofilm formation in A. baumannii (Richmond et al., 2016).
AnoR is a LuxR-type transcriptional regulator controlling the production of quorum sensing signals, N-acyl homoserine lactones (AHLs) in A. nosocomialis ATCC 17903 (Oh and Choi, 2015). Recently, we analyzed the whole transcriptome of A. nosocomialis wild-type (WT) and the anoR deletion mutant (unpublished data), and it revealed the downregulation of two uncharacterized efflux pump genes, NCTC8102_00280 and NCTC8102_00290. In silico analysis pointed out that these genes belong to the RND-type transport system and many homologs are found in the bacterial community. The NCTC8102_00280 and NCTC8102_00290 showed identity to the acrA and acrB genes of the acrAB operon, constituting the AcrAB-TolC efflux pump (Okusu et al., 1996) of E. coli. The regulation of expression of RND pumps are tightly regulated by an intricate balance between local regulators (generally repressors) and global regulators. It has been reported previously that acrAB is under the control of a local repressor, AcrR in E. coli (Ma et al., 1996). Further in silico analysis of the A. nosocomialis genome revealed an AcrR homolog upstream of acrA of the acrAB operon. In this study, we report the regulation of acrAB efflux pump by AcrR and the data indicate that AcrR plays a major role in motility, biofilm/pellicle formation and pathogenesis in A. nosocomialis by repressing the acrAB operon.
Materials and Methods
Bacterial Strains, Plasmids, and Culture Conditions
Bacterial strains and plasmids used in this study are shown in Table 1. Unless otherwise stated, E. coli strains were grown in Luria-Bertani (LB) medium (10 g/L tryptone, 5 g/L yeast extract, 10 g/L NaCl) at 37°C, while the A. nosocomialis strains were grown at 30 or 37°C in either LB or Mueller Hinton (MH) media (Difco). When appropriate, antibiotics were added to media for E. coli (kanamycin, 50 μg/ml; chloramphenicol, 20 μg/ml) as well as A. nosocomialis (kanamycin, 30 μg/ml; ampicillin, 100 μg/ml). Human aveolar epithelial cell line (A549) was cultured in Dulbecco's Modified Eagle's Medium (DMEM, WELGENE Inc., Korea) supplemented with 10% fetal bovine serum (FBS, WELGENE Inc., Korea) and penicillin G (100 U/ml)/streptomycin (100 ug/ml). The cells were maintained at 37°C in 5% (vol/vol) CO2 in 75-cm2 tissue culture flasks until used.
DNA Manipulations
Standard cloning procedures were followed in this study (Sambrook et al., 1989). Oligonucleotides were purchased from Macrogen Co., Ltd., Korea and all the oligonucleotides used in this study are given in Table 2. The genomic DNA from A. nosocomialis and the plasmid DNA from E. coli were isolated using the PureHelix™ Genomic DNA Prep Kit (Nanohelix Co. Ltd., Korea) and AccuPrep® Plasmid Extraction Kit (Bioneer Corp., Korea), respectively. The plasmids were introduced into E. coli and A. nosocomialis by heat shock method (Hanahan, 1983) and conjugation (Oh et al., 2015), respectively. PrimeSTAR GXL Taq DNA polymerase (TaKaRa, Japan) was used for polymerase chain reaction (PCR) amplification of DNA fragments. The DNA fragments were purified or eluted from gel using HiGene™ Gel & PCR Purification System (Biofact Co., Ltd., Korea). Restriction and DNA modifying enzymes were purchased from New England Biolabs, USA. The nucleotide sequence of all recombinant strains and mutants were confirmed by sequencing (Macrogen Co., Ltd., Korea).
In Silico Analysis
In silico analysis was performed to identify homologs of acrAB operon and acrR, using the A. nosocomialis ATCC 17903 (NCTC 8102) genome sequence retrieved from GenBank (GenBank accession no. CP029351). Phylogenetic analysis and molecular evolutionary analyzes were conducted to determine the relationship of acrR of A. nosocomialis with that of other related strains. The strains used for the construction of phylogenetic tree are given in Supplementary Table 1. Phylogenetic tree was constructed using MEGA 6.06 (Tamura et al., 2013). The sequences were aligned using MUSCLE (Edgar, 2004) and pairwise distances were estimated using the Maximum Likelihood approach. In order to find out the binding motif of AcrR in the promoter region of acrA/acrR (PacrA/acrR) of A. nosocomialis, multiple sequence alignment was carried out with the PacrA/acrR and the known AcrR binding motif from E. coli (Su et al., 2007).
Overexpression and Purification of His6-AcrR
For the overexpression of hexahistidyl-tagged AcrR (His6-AcrR), the complete coding region of acrR (624 bp) of A. nosocomialis was amplified by PCR and subcloned into His-tag expression vector, pET28a yielding pET-acrR. The recombinant plasmid was introduced into E. coli strain BL21(DE3) by transformation. The AcrR was overexpressed by adding 1 mM isopropyl β-D-1-thiogalactopyranoside (IPTG) at an OD600 of 0.5 and incubated the culture further for 5 h at 30°C. Induced cells were harvested by centrifugation and lysed by sonication (Vibra-Cell™, Sonics & Materials, Inc., USA) in lysis buffer [20 mM Tris-HCl (pH 8), 500 mM NaCl and 5 mM imidazole]. After centrifugation, the soluble supernatant was mixed with nickel–nitrilotriacetic acid–agarose solution (Qiagen, Germany) and loaded into polypropylene column (Thermo Fisher Scientific, USA). The column was washed with lysis buffer and the protein was eluted in elution buffer with varying imidazole concentration [20 mM Tris-HCl (pH 8), 500 mM NaCl, and 150/300 mM imidazole]. The purified AcrR was dialyzed overnight against phosphate buffered saline (PBS) to remove imidazole and directly used for the promoter binding assay.
Electrophoretic Mobility Shift Assay (EMSA)
EMSA was carried out to check the binding of AcrR to the promoter region of acrA/acrR according to the protocol described earlier (Alves and Cunha, 2012). A promoter fragment of 148 bp and 16s rDNA of 149 was amplified by PCR using the oligonucleotide primers listed in Table 2. In the binding assay, 200 ng of the PacrA/acrR fragment was incubated with varying amounts of purified His6-AcrR (0 to 0.5 μg) in 15 μl of binding buffer [250 mM phosphate buffer (pH 7.5), containing 50 mM NaCl, 500 mM KCl, 10 mM dithiothrietol, 10 mM ethylenediaminetetraacetic acid, 3 μg bovine serum albumin and 1 μg Poly[d(I-C)]]. To confirm the binding specificity of AcrR, 16s rDNA was included in the EMSA reaction mixture instead of the promoter fragment. The binding assay was carried out at room temperature (RT) for 30 min and the reaction mixtures were electrophoresed in 6% prerunned (20 min at 80 V) native polyacrylamide gel in 1x Tris/Borate/EDTA (TBE) buffer for 1 h at 80 V followed by staining with ECOred (MOSAICON, Korea) and visualization.
RNA Extraction and Real-Time Quantitative Reverse Transcription PCR (RT-PCR)
Cultures were inoculated into LB medium and incubated until the exponential phase. Total RNA was extracted with High Pure RNA Isolation Kit (Roche Diagnostics GmbH, Germany) according to the manufacturer's instructions. Complementary DNA (cDNA) was synthesized from 1 μg of DNase-treated total RNA with Reverse Transcription Master Premix (ELPIS Biotech. Inc., Korea) per manufacturer's instructions. Quantitative RT-PCR was performed in triplicates using StepOnePlus™ Real-Time PCR System (Applied Biosystems, USA) with Power SYBR® Green PCR Master Mix (Applied Biosystems, USA) and indicated primers (Table 2). Gene expression was normalized by the ΔΔCT method (Livak and Schmittgen, 2001) and 16S rRNA was used as a reference.
Construction of acrR Deletion Mutant and Complemented Strain
To construct the deletion mutant of acrR, markerless gene deletion method was followed as described previously (Oh et al., 2015). The complete open reading frame of the acrR (624 bp) was deleted by overlap extension PCR. For this, the upstream and downstream regions of the acrR were amplified using two sets of primer pair (AcrR01F and AcrR01R; AcrR02F and AcrR02R). nptI conferring kanamycin resistance was amplified from pUC4K using the primers, U1 and U2. Overlap extension PCR was carried out to anneal the three PCR products at their overlapping regions using the primers AcrR01F and U2. The fusion product was ligated into FspI-digested pHKD01 to result in pOH959. The recombinant plasmid pOH959 was introduced into A. nosocomialis by conjugation as described previously (Oh et al., 2015) and the integration of POH959 into the chromosome was confirmed by selection of colonies on LB plate containing ampicillin and kanamycin. To screen the colonies having mutated acrR gene, kanamycin-resistant colonies were grown overnight in LB plate containing 10% (w/v) sucrose. A sucrose-resistant and kanamycin sensitive cell (acrR deletion mutant) was selected and the double crossover event was confirmed by PCR. For single-copy acrR complementation in the acrR mutant, the acrR including its native promoter was inserted into the intergenic region that is located downstream of phaC encoding alpha/beta hydrolase by a modified markerless gene deletion method. Briefly, the acrR including its promoter region and the downstream regions (part I and part II) of phaC were amplified using the primer pairs AcrR03F/AcrR03R, PhaC01F/PhaC01R, and PhaC02F/PhaC02R, respectively. The nptI was amplified from the pUC4K plasmid using the primer pair U1/U2. The primers, AcrR03F and AcrR03R contained additional 20 nucleotides at their 5′ ends that are complementary to the regions of part I and part II of the downstream region of phaC, respectively, while PhaC02R carried additional 20 nucleotides at its 5′ end that is complementary to the part of kanamycin resistance cassette. All the four PCR products obtained were annealed together by overlap extension PCR using the primers PhaC01F and U2. The recombinant DNA fragment from the overlap extension PCR was ligated into the FspI-digested pHKD01 to generate pOH993. The recombinant plasmid, pOH993 was introduced into acrR deletion mutant by conjugation and the recombinants were screened as described previously (Oh et al., 2015).
Motility Test
Surface-associated motility was examined on MH plate containing 0.25% Eiken soft agar (Eiken chemical, Co., Ltd., Japan) as described previously (Skiebe et al., 2012). In order to minimize the variation between plates, 30 ml of the motility agar was poured to each plate in the laminar flow hood and the plates were dried for 30 min. A. nosocomialis WT, acrR deletion mutant and complemented mutant strains cultivated in MH broth overnight were diluted to an OD600 of 1 and 5 μl of the samples were spotted onto the motility plate. The plates were incubated at 30°C for 18 h and the motility on top of agar was observed. The plates were prepared fresh for each surface motility experiment and the test was repeated on at least three separate occasions.
Biofilm/Pellicle Formation Assay
Biofilm and pellicle formation assays were adapted from previously described protocols with slight modifications (Lee et al., 2008; Giles et al., 2015). For biofilm and pellicle formation assay, A. nosocomialis WT, acrR deletion mutant and complemented strains were cultivated in MH broth at 37°C. The overnight cultures were diluted to an OD600 of 1 and 25 μl of the diluted cultures were added to 5 ml of MH broth in polystyrene tubes (Ø 17 mm × H 100 mm) (SPL Life Sciences, Korea). The tubes were incubated at different time periods at 30°C without shaking. For biofilm assay, planktonic cells were removed post-incubation and washed thrice with sterile distilled water. For pellicle assay, 100% ethanol was carefully added to the bottom of the pellicle and the pellicle was removed and resuspended in 1 ml of PBS. The pellicle suspension was centrifuged at 13,000 rpm and the supernatant was carefully removed. Both biofilm and pellicle was stained with 0.1% (wt/vol) crystal violet (Sigma, USA) for 15 min at RT followed by washing with sterile distilled water thrice and air drying at RT. The stain absorbed to biofilm or pellicle was eluted with 30% acetic acid and quantitated by measuring the optical density at 595 nm. The pellicle/biofilm assay was performed in quintuplicates at three different occasions.
Adherence Assay
To check the adherence ability of acrR deletion mutant, A549 epithelial cells were seeded on 13 mm cell culture coverslips (Nunc™, Thermo Scientific, USA) in 24-well plates containing DMEM (WELGENE Inc., Korea) supplemented with 10% FBS (WELGENE Inc., Korea) and incubated at 37°C in 5% (vol/vol) CO2 for 24 h. A. nosocomialis WT, acrR deletion mutant and complemented strains were grown in LB broth overnight at 37°C, washed and diluted in DMEM containing FBS. The cell line was infected with different A. nosocomialis strains at a ratio of bacteria to host cells of 100:1 (MOI of 100) and incubated at 37°C in 5% CO2 for 5 h. After washing five times with Dulbecco's Phosphate Buffered Saline (DPBS), the cells were fixed with methanol for 20 min, and stained with Giemsa solution (Lee et al., 2006).
Invasion Assay
For the invasion assay, 1 × 105 A549 epithelial cells were seeded into 24-well plates containing DMEM (WELGENE Inc., Korea) supplemented with 10% FBS (WELGENE Inc., Korea) and incubated at 37°C in 5% (vol/vol) CO2 for 24 h. Cultures of A. nosocomialis WT, acrR deletion mutant and complemented strains were grown in LB broth overnight at 37°C, washed and diluted in DMEM supplemented with FBS. The cell line was washed with DPBS (WELGENE Inc., Korea) and infected with the bacterial cultures at an MOI of 100. After incubation for 5 h at 37°C in 5% (vol/vol) CO2, the wells were washed three times with DPBS before adding DMEM-FBS containing 300 μg/ml of gentamycin to kill all external bacteria. The microplates were incubated at 37°C in 5% CO2 for 2 h. The cells were washed with DPBS, lysed with Ultrapure water (WELGENE Inc., Korea) containing 0.25% Triton X-100 for 20 min at RT and appropriate dilutions were dropped onto LB agar plates. The plates were incubated overnight and CFUs were counted to quantify bacteria surviving intracellularly. To visualize the invasion of bacteria by fluorescence microscopy, cells were seeded on 13 mm glass coverslips and incubated for 24 h. After infection for 5 h, the cells were fixed with 3.7% paraformaldehyde for 10 min and permeabilized with 0.25% Triton X-100. Bacterial cells were labeled with polyclonal anti-rabbit AbOmpA antibody (1:1,000) followed by Alexa Fluor® 594-conjugated goat anti-rabbit IgG antibody (Invitrogen, USA). Actin was stained with Alexa Fluor® 488 phalloidin (Invitrogen, USA) and nucleus with DAPI (6-diamidino-2-phenylindole, Invitrogen, USA). The invasion of A. nosocomialis strains into epithelial cells was observed under Leica DMi8 confocal microscope (Leica Microsystems Ltd. Germany).
Animal Experiments
Seven weeks old female BALB/c mice were used to assess the virulence of the acrR deletion mutant. The mice were obtained from Nara Biotech, Korea and the animals were maintained under specific-pathogen-free conditions according to the Institutional Animal Care and Use Committee (IACUC) of Chungnam National University guidelines (permission number: CNU-00999). The animal experiments were performed in accordance with the guidelines of Korean Food and Drug Administration. For induction of neutropenic mouse model, 7-week-old female BALB/c mice were injected intraperitoneally with 100 mg of cyclophosphamide (CTX, Sigma. USA)/kg of body weight on day 1 and 4 prior to injection of bacterial cells, respectively. The mice were anesthetized and groups containing three were infected intratracheally with 1 × 108 CFU/ml of A. nosocomialis WT, acrR deletion mutant and complemented mutant. One hundred microliters of PBS was used to inject the control mice group. Mice were sacrificed 3 days post-infection, and lungs were harvested to assess the bacterial load. The lungs were homogenized and the cell suspension was serially diluted and spotted onto LB plates to count the bacteria. For histological analysis, tissues were subjected to hematoxylin/eosin, or immunohistochemical staining with OmpA (outer membrane protein A) antibody to visualize the bacteria.
Statistical Analysis
Whenever applicable, the experiments were repeated at least three times with consistent results. The significance of difference between two groups was determined by unpaired Student's t-test and that among more than three groups was evaluated with one-way ANOVA followed by Tukey's multiple comparison test using statistical software GraphPad Prism v5.01 (GraphPad Software Inc., USA). *P < 0.05, **P < 0.01, and ***P < 0.001 were considered as statistically significant.
Results
The A. nosocomialis Genome Encodes acrAB Operon Which Is Under the Control of acrR
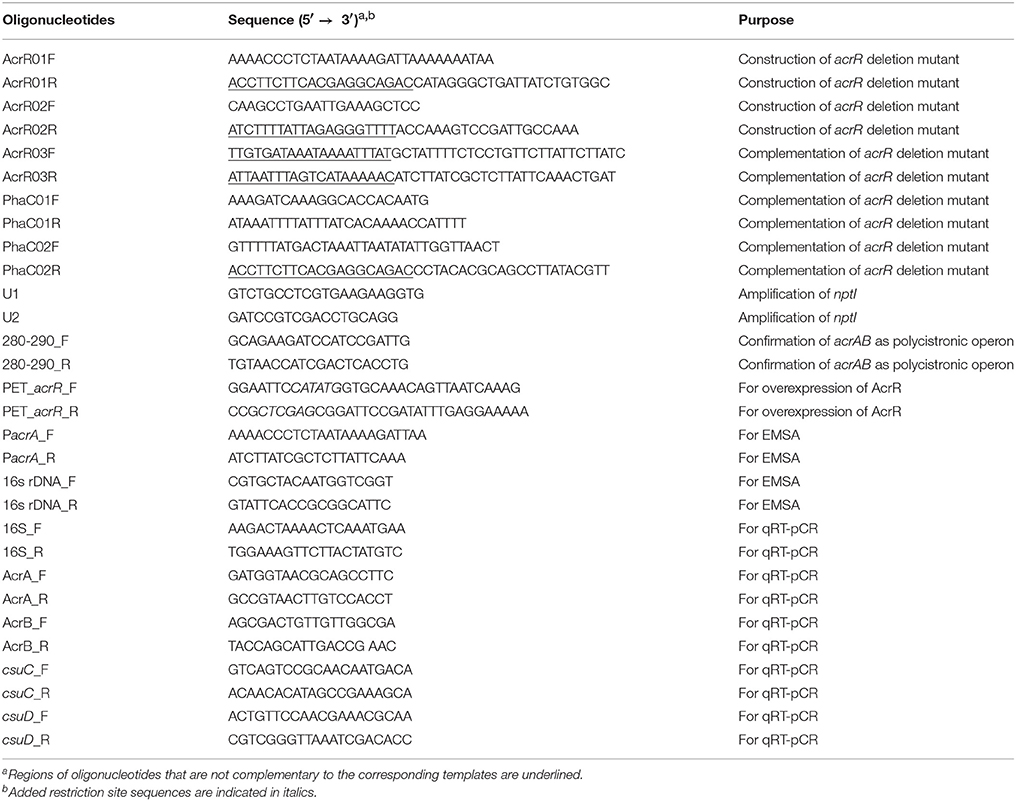
In this study, in silico analysis revealed the presence of a well-defined acrAB efflux pump operon in A. nosocomialis comprising of acrA, which encodes the membrane protein, and acrB, encoding acriflavine resistance protein (Figure 1A). The acrA and acrB genes of A. nosocomialis showed 24 and 25% identity to those of E. coli, respectively. The gene which encodes the outer membrane protein of the AcrAB efflux pump of A. nosocomialis is yet to the identified. The acrAB efflux pump is under the control of acrR gene upstream of it (Figure 1A) and the acrAB operon and acrR are divergently transcribed under the same promoter activity. We confirmed that acrAB is a polycistronic operon by amplifying the genomic region covering the 3′-end of acrA and 5′-end of acrB using cDNA prepared from DNAse-treated RNA (Figure 1B). The acrR is widely distributed among bacterial species and phylogenetic analysis was carried out to find out the relationships of acrR. The acrR from Acinetobacter strains showed close similarity with that from V. parahaemolyticus RIMD 2210633 and M. haemolytica PHL213 (Figure 1C). Also, the AcrR from A. nosocomialis shared high similarity of 85.38% and 88.41% with that of A. baumannii 17978 and A. baumannii 19606, respectively (Figure 1D). The well-studied AcrR from E. cloacae, E. coli K12 and S. enterica shared similarity of 25.9, 24.31, and 24.86%, respectively.
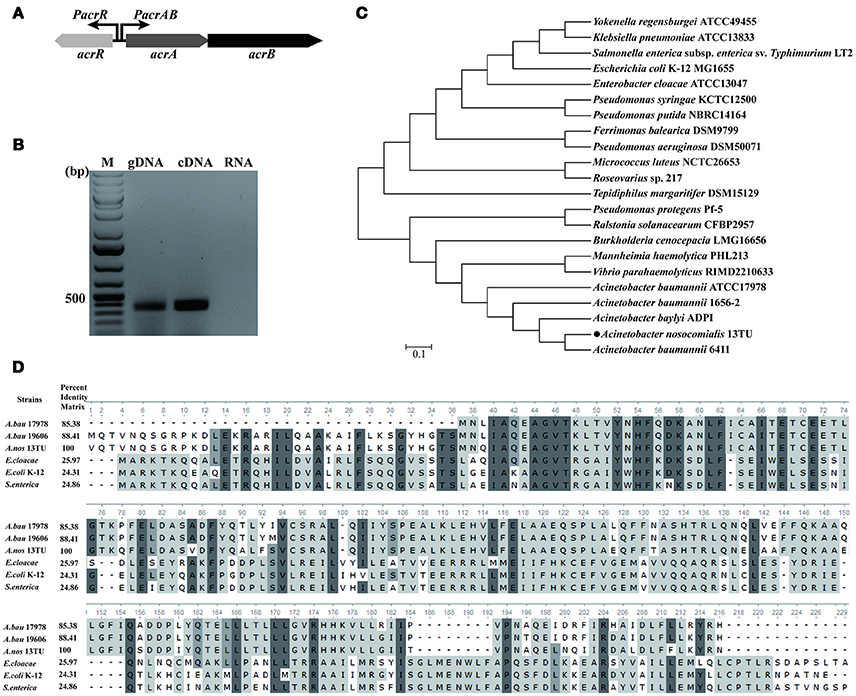
Figure 1. Genomic organization of acrAB operon and in silico analysis of acrR. (A) The schematic representation of the acrAB efflux pump operon in A. nosocomialis. The acrR gene which is divergently transcribed from the acrAB operon shares a common promoter region with the acrAB operon. (B) Confirmation of acrAB as polycistronic operon. RT-PCR analysis was performed using total RNA isolated from A. nosocomialis wild type strain grown in LB medium at 37°C. A negative control and positive control reactions were performed using DNase-treated RNA and genomic DNA as templates, respectively. The size of the amplified fragments was estimated by comparing it with the size marker DNAs (lane M). (C) Phylogenetic analysis of acrR from A. nosocomialis with that of other bacterial strains. The tree was constructed using MEGA 6.06 and the pairwise distance was calculated using Maximum Likelihood approach according to the aligned sequences. (D) Multiple sequence alignment of AcrR from A. nosocomialis with homologs from different bacterial strains. Residues that are conserved across all sequences are highlighted in dark gray and matches across the sequences by light gray. Pairwise sequence identity (%) is also shown. A.bau 17978, A. baumannii 17978; A.nos 13TU, A. nosocomialis; A.bau 19606; A. baumannii 19606.
AcrR Binds to the Promoter Region of acrA/acrR
In E. coli, AcrR functions as one of the regulators modulating the expression of acrAB operon (Ma et al., 1996). In this study, in silico analysis revealed the presence of an AcrR binding motif in the promoter region of acrA/acrR (Figure 2A). In order to examine the direct interaction between AcrR and PacrA/acrR, EMSA was performed with a 148 bp fragment of the promoter region and purified AcrR. With the addition of AcrR, the free DNA substrate disappeared and the DNA bands shifted (Figure 2B), indicating that AcrR could bind to the acrAB regulatory region. The observed interactions were specific because no AcrR binding of a 16S rDNA control fragment was observed (Figure 2B). The AcrR binding motif was located at position −87 (TTTATAAGTTTTTTTTAATTAATGTACC) relative to the translational start site (Figure 2A), according to the consensus sequences reported earlier in E. coli (Su et al., 2007). The consensus bases matching with those in the AcrR binding motif in E. coli are underlined. The EMSA data clearly shows that AcrR specifically binds to the promoter region of acrA/acrR and thus suggests an involvement in the regulation of expression of acrAB operon.
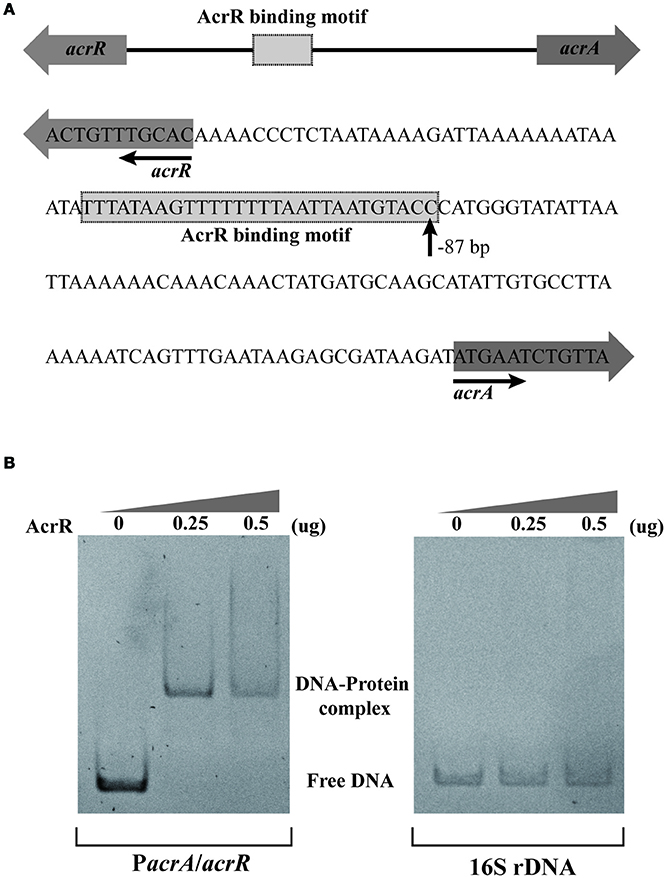
Figure 2. EMSA analysis for the AcrR interaction with the acrR-acrAB intergenic region. (A) Scheme and nucleotide sequence of the intergenic region of acrR and acrAB. A putative AcrR binding motif located in the promoter region between acrR and acrA is shown. The number beneath the indicated binding site of AcrR points the position of the first nucleotide in the binding motif relative to the translational start site of acrA. (B) EMSA result showing the interaction between AcrR and the promoter region of acrA/acrR. The promoter region and 16s rDNA (as control) was amplified by PCR and incubated without or with different concentrations of purified AcrR. 1 μg of poly[d(I-C)] was added to each reaction mixture in order to avoid non-specific binding.
AcrR Negatively Regulates acrAB Expression
Previous studies have pointed out that the expression of acrAB operon is downregulated by the local repressor AcrR in E. coli and S. Typhimurium (Olliver et al., 2004; Pourahmad Jaktaji and Jazayeri, 2013). In A. nosocomialis, the DNA-binding assay revealed an interaction of AcrR with the promoter region of acrA/acrR. To functionally analyze the effect of AcrR on acrAB expression in A. nosocomialis, RT-PCR transcription analyses were performed using wild type and acrR mutant strains. Deletion of acrR increased acrA and acrB expression in the mutant compared to the wild type (Figure 3). The data demonstrates that AcrR negatively regulates the expression of acrAB operon, similar to that in E. coli and S. Typhimurium.
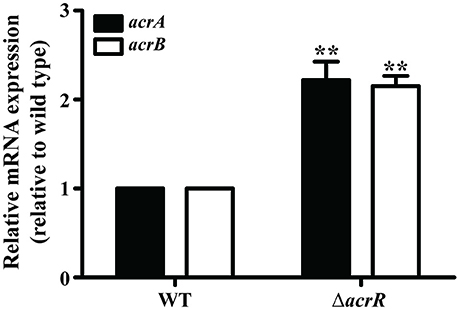
Figure 3. Transcription of acrA and acrB are significantly upregulated in the acrR deletion mutant. The relative acrA (black bars) and acrB (white bars) mRNA expression levels in the wild type (WT) and acrR mutant (ΔacrR) were determined as described in the section “Materials and Methods.” Values are mean ± SD (n = 3) and asterisks indicate a significant difference in mRNA expression between the mutant and the wild type at **P < 0.01.
Deleting acrR Enhances Motility
The acrR deletion mutant of A. nosocomialis ATCC 17903 was constructed using markerless gene deletion method based on double cross over. The detailed protocol for the construction of deletion mutant is given in the “Materials and Methods” section. The whole acrR gene was deleted and the deletion was confirmed by PCR (Figure 4A) followed by sequencing. For the complementation of acrR mutant, the acrR coding region along with its native promoter was inserted into intergenic region that is located downstream of phaC encoding alpha/beta hydrolase. The complementation of the acrR mutant was confirmed by PCR. To characterize the acrR mutant, A. nosocomialis wild type, acrR mutant and complemented mutant were cultivated in MH broth and the OD600 was measured until 24 h of incubation. It was noticed that the growth pattern of acrR mutant was not different from that of wild type.
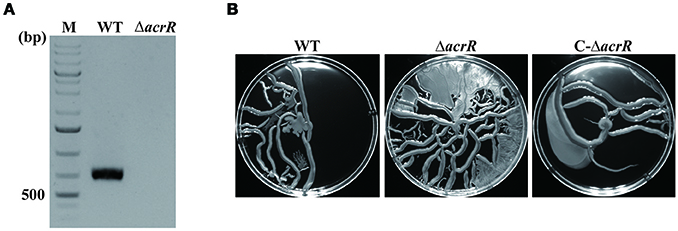
Figure 4. Confirmation of acrR deletion by PCR and motility assay. (A) Verification of acrR deletion mutant was carried out by PCR using genomic DNA as template. WT, wild type; ΔacrR, acrR mutant; M, DNA molecular weight marker. (B) Enhanced motility when acrR is deleted. Overnight cultures of A. nosocomialis wild type, acrR mutant and complemented mutant were cultivated in MH medium containing 0.25% Eiken agar and incubated at 30°C. The plates were observed after 18 h and photographed. The motility experiment was conducted in triplicates at three different occasions. Values are mean ± SD (n = 3).
It has been reported previously that AcrR modulates motility in E. coli (Kim et al., 2016). To check the effect of acrR deletion on motility in A. nosocomialis, overnight cultures of A. nosocomialis WT, acrR deletion mutant and complemented mutant were spotted on MH medium containing 0.25% Eiken soft agar. The data showed that motility of the acrR mutant is significantly increased compared to that of wild type, indicating that AcrR plays an important role in the regulation of motility in A. nosocomialis (Figure 4B). As expected, the acrR complemented strain restored the motility similar to that of the wild type strain.
Deletion of acrR Increases Biofilm/Pellicle Formation
Multidrug efflux pumps are known to be important constituent of biofilm formation (Lynch et al., 2007; Kvist et al., 2008; Matsumura et al., 2011; Baugh et al., 2012). In S. enterica serovar Typhimurium, the mutation in the acrAB operon led to impaired biofilm formation (Baugh et al., 2014; Schlisselberg et al., 2015). Similarly, the acrB mutant of E. coli also depicted decreased biofilm formation (Kvist et al., 2008; Matsumura et al., 2011). Therefore, we checked the effect of acrR deletion on biofilm/pellicle formation in A. nosocomialis by cultivating the strains in MH broth at 30°C over a period of time. Crystal violet staining was carried out to quantify separately the biofilm attached to the culture tubes and the pellicle formed at the air-liquid interface. acrR mutant exhibited increased biofilm formation even though the growth rate of the mutant was similar to that of wild type (Figure 5). The wild type A. nosocomialis displayed maximum biofilm formation at 48 h while the acrR mutant at 72 h (Figure 5A). There was a 2-fold increase in biofilm formation in the acrR mutant at 72 h compared to that of wild type. In the case of complemented mutant, the biofilm formation was almost similar to that of wild type (Figure 5B). As in the case of biofilm formation, the pellicle formation was also increased in the acrR mutant compared to that of wild type at different time periods (48 and 72 h) (Figure 5C). The highest pellicle formation was observed in the acrR mutant at 72 h. The above results suggest that AcrR is a negative regulator of biofilm/pellicle formation in A. nosocomialis.
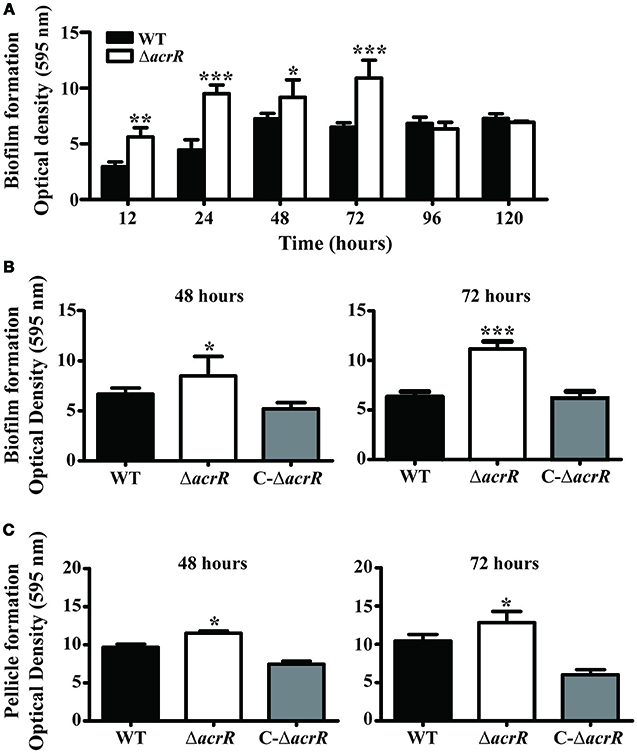
Figure 5. acrR mutant displays increased biofilm/pellicle formation. The strains were cultivated in MH medium at 30°C and biofilm/pellicle formation was quantified at different time periods based on crystal violet staining method. (A) Time-dependent biofilm formation by A. nosocomialis wild type and acrR mutant. (B) Crystal violet staining of the biofilm formation in wild type (WT), acrR mutant (ΔacrR) and complemented mutant (C-ΔacrR) at 48 and 72 h. Graphical representation of the biofilm formation is also shown. (C) The pellicle formation in wild type (WT), acrR mutant (ΔacrR) and complemented mutant (C-ΔacrR) at 48 h and 72 h are shown. Values are mean ± SD (n = 5) and asterisks indicate significantly different average biofilm or pellicle value when compared with wild type at *P < 0.05, **P < 0.01, and ***P < 0.001. The biofilm/pellicle assay was performed at three different occasions.
acrR Deletion Enhances Cell Adhesion and Invasion
Multidrug efflux pumps including AcrAB-TolC efflux system play an important role in the adherence and invasiveness of pathogenic bacteria to the host cells (Hirakata et al., 2002; Buckley et al., 2006). We determined the adherence and invasiveness of acrR mutant to A549 epithelial cells and noticed higher adherence and invasion ability compared to the wild type (Figure 6, Supplementary Figure 1). The adherence and invasiveness of the complemented mutant was similar to that of the wild type. The data depicts that AcrR plays a negative role in the adherence and invasiveness of A. nosocomialis. We checked the expression level of the genes, csuC and csuD which encodes components of the CsuA/BABCDE chaperone-usher pili assembly system important for the assembly and production of pili involved in adhesion in Acinetobacter by mRNA expression analysis. It was demonstrated that the expression of csuC and csuD genes are upregulated in the acrR deletion mutant compared to the wild type (Supplementary Figure 2). Thus, it can be concluded that the elevated expression of csuC and csuD might contribute to the increased adhesion and invasion in the acrR mutant.
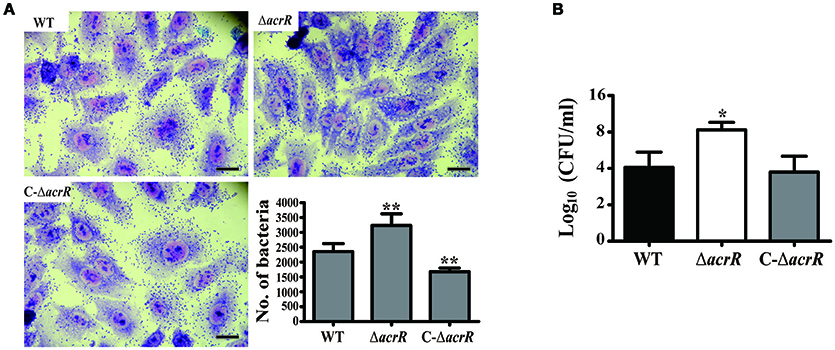
Figure 6. acrR deletion mutant displays increased cell adhesion and invasion. To check the effect of acrR deletion on the adherence and invasiveness of A. nosocomialis, A549 epithelial cells were infected with wild type (WT), acrR mutant (ΔacrR) and complemented mutant (C-ΔacrR) at MOI 100, and incubated at 37°C in 5% CO2 for 5 h. (A) For the adhesion assay, the infected cells were fixed with methanol post-infection and stained with Giemsa solution. The adherence ability of acrR mutant was higher than that of WT and C-ΔacrR. The number of bacteria adhered to the epithelial cells is graphically represented. Values are mean ± SD (n = 3) and asterisks indicate significantly different average bacterial count when compared with wild type at **P < 0.01. Scale bar, 50 μm. (B) To check the effect of acrR deletion on the invasiveness of A. nosocomialis, the extracellular bacteria were removed post-infection followed by cell lysis and CFU counting. Values are mean ± SD (n = 3) and asterisk indicate significantly different average CFU value when compared with wild type at *P < 0.05.
acrR Deletion Mutant Displays Increased Virulence
It has been reported previously that the AcrAB-TolC efflux system plays a direct role in the virulence of pathogenic bacteria. In S. Typhimurium, acrB insertional inactivated/deleted mutant displayed less virulence compared to the wild type strain (Buckley et al., 2006; Perrett et al., 2009; Webber et al., 2009). To investigate the role of AcrR on virulence, we infected BALB/c mice with wild type, acrR mutant and complemented mutant of A. nosocomialis intratracheally. Survival curve was plotted to assess the overall virulence of the mutant compared to the wild type. As shown in Figure 7A, the mice infected with the acrR mutant had a significantly lower survival rate. The CFUs of the mutant in the lungs of infected mice were 1.5 times higher than that of wild type in the infected lungs (Figure 7B). In addition, H&E staining and IHC of the lung tissues revealed that the acrR mutant severely damaged the lungs compared to the wild type (Figure 7C). The lung tissues from acrR mutant depicted increased cellular infiltration compared to that of the wild type. The IHC data showed higher number of bacterial cells in the damaged lung tissues from the acrR mutant than from the wild type tissues. The above results demonstrate that AcrR suppresses virulence in A. nosocomialis and thus the acrR mutant is highly pathogenic than the wild type.
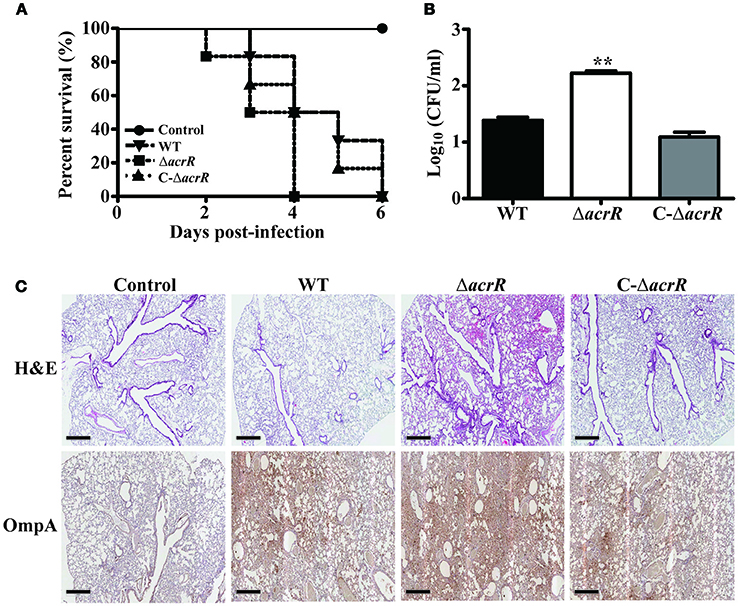
Figure 7. Deletion of acrR promotes virulence in A. nosocomialis. To investigate the effect of acrR deletion on the virulence of A. nosocomialis, mice were intratracheally infected with A. nosocomialis wild type (WT), acrR mutant (ΔacrR) and complemented mutant (C-ΔacrR). (A) Survival curve of mice infected with different bacterial strains. The lethality was higher in the case of mice infected with acrR mutant compared to the wild type. (B) Bacterial burden in lungs infected with A. nosocomialis strains. Lungs were homogenized 3 days post-infection and CFU counting was performed. (C) H&E, IHC staining results showing severe tissue damage in lungs of mice infected with acrR mutant compared to that with wild type. Values are mean ± SD (n = 3) and asterisks indicate a significant difference in CFU value compared with the wild type at **P < 0.01. Scale bar, 500 μm.
Discussion
Multidrug efflux pumps have been recognized as playing a pivotal role in drug resistance, cell division, pathogenicity and biofilm formation (Nikaido, 1996; Buckley et al., 2006; Kvist et al., 2008). Here, we identified an operon encoding AcrA and AcrB in A. nosocomialis, which shows significant homology to the AcrAB efflux pump of E. coli. The acrAB operon in A. nosocomialis depicted high similarity to the arpAB operon which plays a role in aminoglycoside resistance in A. baumannii 5075 (Tipton et al., 2017). In E. coli, the acrAB operon is regulated by AcrR, encoded by acrR located 140 bp upstream of the acrAB operon (Ma et al., 1996). The AcrR belongs to TetR family of transcriptional regulators and is composed of an N-terminal domain containing a DNA-binding helix-turn-helix (HTH) motif, and a C-terminal domain containing an unique ligand-binding sequence (Ramos et al., 2005; Li et al., 2007; Su et al., 2007). In silico analysis revealed that the DNA-binding motif of AcrR is highly conserved in the bacterial community (data not shown). Similar to that in E. coli, the acrR in A. nosocomialis is located upstream of acrAB operon and is divergently transcribed from the same promoter of acrA. Also, the AcrR binding motif identified in the PacrA/acrR of A. nosocomialis showed close similarity to that in E. coli.
In silico analysis and further confirmation by EMSA revealed that AcrR of A. nosocomialis binds to the promoter region of acrA/acrR and might control the expression of acrAB. The expression of acrAB was elevated in the acrR mutant, which was confirmed by RT-PCR. In E. coli, AcrR negatively modulates the expression of the AcrAB efflux pump and the absence of functional AcrR resulted in increased transcription of acrAB (Ma et al., 1996). In addition, AcrR acts as an autorepressor regulating its own gene expression in E. coli.
It was found that deletion of acrR leads to increased motility in A. nosocomialis, which is in accordance with the motility observed in the acrR mutant of E. coli (Kim et al., 2016). In E. coli, the deletion of acrR induces transcription of most motility genes, indicating that the toxic-compounds-response regulator AcrR participates in motility to escape toxic compounds. The inactivation of acrB in S. enterica negatively impacts survival in the host and motility (Webber et al., 2009) contributing to reduced pathogenicity (Khoramian-Falsafi et al., 1990). The data from our study reveal that AcrR has a negative role to play in motility by controlling the expression of AcrAB efflux pump.
The absence of the AcrAB efflux pump repressor, AcrR contributed to increased biofilm/pellicle formation, indicating that the AcrAB efflux pump is positively associated with biofilm formation in A. nosocomialis. It has been reported previously that a number of multidrug efflux systems, including AcrAB-TolC, play a role in biofilm formation in various Gram negative species, including E. coli, Klebsiella and S. Typhimurium (Kvist et al., 2008; Baugh et al., 2012). In addition, an upregulated expression of multidrug efflux genes, acrA and acrB has been noticed in biofilm cells of E. coli (Zou et al., 2012). The study that explores the functional link between multidrug efflux pump and biofilm formation in S. Typhimurium revealed that several multidrug efflux pumps, including AcrAB plays a role in the formation of extracellular matrix (Baugh et al., 2014). It has been known that the extracellular biofilm matrix of E. coli and Salmonella is majorly composed of structural curli proteins which help them in adhesion to inert surfaces as well as to host cells (Cegelski et al., 2009; Baugh et al., 2014). The curli proteins are constituted by the major and minor curli subunits which are synthesized by the csgBA operon (Grund and Weber, 1988; Andersson et al., 2013). It has been noticed previously that the acrB and tolC mutants of S. Typhimurium lack the production of curli due to the transcriptional repression of csgA and csgB genes (Baugh et al., 2014), indicating the importance of AcrAB efflux pump in curli production.
It has been known for decades that there is a significant correlation between biofilm formation and multidrug resistance (Anderl et al., 2000). The bacterial cells which are associated to biofilm can tolerate antibacterial agents far better than planktonic cells. The MexAB-OprM and MexCD-OprJ pumps of P. aeruginosa have been shown to be involved in biofilm-specific mechanisms for resistance (Gillis et al., 2005). In addition, the efflux pump operon, PA1874-1877 in P. aeruginosa was seen upregulated during biofilm formation and that appeared to be involved in biofilm-mediated antibiotic resistance (Zhang and Mah, 2008). Further studies on the expression level of AcrAB efflux pump and multidrug resistance under biofilm-forming conditions would shed light on the involvement of this pump in biofilm-mediated antibiotic resistance in A. nosocomialis.
The invasiveness and pathogenesis of A. nosocomialis was elevated when the acrR gene was deleted. Earlier studies have shown that the AcrAB-TolC efflux system and its homologs are crucial for the invasion/virulence of S. Typhimurium, Moraxella catarrhalis, P. aeruginosa, and Erwinia amylovora (Hirakata et al., 2002; Burse et al., 2004; Spaniol et al., 2015; Wang-Kan et al., 2017). Buckley et al. reported that functional AcrB is important for adhesion, invasion and survival of S. Typhimurium in eukaryotic cells, and the impaired virulence in the AcrB-defective mutant is due to the loss of efflux pump activity (Buckley et al., 2006). It was suggested that the poor invasion/intracellular survival of the acrB mutant might be accounted by the inability of the bacteria to export host antimicrobials, such as basic peptides or toxic compounds present in lysosomes as the natural function of the AcrAB-TolC efflux system is to expel such compounds from the bacterial cell (Shafer et al., 1998; Buckley et al., 2006). In addition, the decrease in adhesion and/or invasion of the acrB mutant could also be due to the lack of secretion of adhesins, which might be substrates of AcrAB-TolC efflux pump (Buckley et al., 2006). Furthermore, transcriptome analysis of the acrB mutant depicted downregulation of Salmonella pathogenicity island (SPI) genes that are essential for invasion, bacterial survival and persistence within the host (Webber et al., 2009). Also, it is known that multidrug efflux systems are required for the optimal production of virulence factors besides its role in the export of toxic chemicals (Burse et al., 2004; Bina et al., 2008). In this study, the deletion of acrR led to the upregulation of csuC and csuD genes which are important for the production and assembly of the virulence factor pili needed for adhesion in A. nosocomialis. Based on the fact that acrAB operon is induced in the acrR mutant, it could be suggested that the overexpression of the AcrAB efflux pump might play a role in the enhanced production of virulence factors contributing to increased biofilm formation, adhesion, invasion and pathogenesis in A. nosocomialis.
The involvement of multidrug efflux pumps in quorum sensing (QS) was first reported in P. aeruginosa in which the MexAB-OprM plays a role in the efflux of autoinducer N-(3-oxododecanoyl)-L-homoserine lactone (3-oxo-C12-HSL) (Pearson et al., 1999). The mexAB-oprM mutant displayed reduced diffusion of the 3-oxo-C12-HSL into the extracellular environment resulting in loss of virulence. The transcription of quorum sensing transporter genes lsrACDBK and the operon repressor lsrR was decreased in the acrB mutant of S. Typhimurium (Wang-Kan et al., 2017), indicating the role of AcrAB in the expression of virulence factors as lsrR controls the expression of SPI-related genes (Choi et al., 2012). In Burkholderia pseudomallei, the extracellular secretion of AHLs is absolutely dependent on the function of BpeAB-OprB efflux pump (Chan et al., 2007). Interestingly, in our study, RNA sequencing and mRNA expression analysis of the anoR mutant of A. nosocomialis depicted the downregulation of acrA and acrB genes (data unpublished). It has been reported earlier that the expression of mexAB-oprM of P. aeruginosa is limited by the intracellular concentration of the autoinducer molecules (Maseda et al., 2004). Thus, the decreased expression of acrA and acrB in the anoR mutant could be very well-associated with the absence of AHLs in the mutant background. It is very well-known that QS plays a crucial role in motility, biofilm formation and the production of virulence factors (Rutherford and Bassler, 2012). Thus, the enhanced AcrAB efflux pump activity in the acrR mutant might attribute well to the AHLs-mediated cell-to-cell signaling, contributing to increased motility, biofilm formation and virulence in the mutant.
In summary, we report a novel regulator, AcrR which negatively regulates the uncharacterized multidrug efflux operon, acrAB in A. nosocomialis. The bacterial motility, biofilm/pellicle formation and virulence were increased in the acrR mutant with the upregulated expression of acrAB operon. This supports the notion that besides contributing to antibiotic resistance, multidrug efflux pumps display a variety of functions in accordance to the environment and the bacterial behavior. Further studies are ongoing for in-depth characterization of the AcrAB efflux pump so that the knowledge could be used for developing efflux pump inhibitors as they are invaluable tools to restore the activity of agents to which the efflux pump confers resistance and also to reduce the ability of bacteria to infect their host.
Author Contributions
BS, JK, MO, and CC designed the research. BS, JK, DK, KW, and MO performed the research. BS, JK, DK, KW, MO, and CC analyzed the data. BS, MO, and CC wrote the paper.
Funding
This research was supported by the National Research Foundation of Korea (NRF) grant funded by the Korean government (MSIT) (No. 2016R1C1B1016370). This research was supported by Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education (NRF-2016R1D1A1B03931010).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2018.00270/full#supplementary-material
References
Alves, C., and Cunha, C. (2012). “Electrophoretic mobility shift assay: analyzing protein - nucleic acid interactions,” in Gel Electrophoresis, ed S. Magdeldin (IntechOpen), 205–228. doi: 10.5772/37619
Anderl, J. N., Franklin, M. J., and Stewart, P. S. (2000). Role of antibiotic penetration limitation in Klebsiella pneumoniae biofilm resistance to ampicillin and ciprofloxacin. Antimicrob. Agents Chemother. 44, 1818–1824. doi: 10.1128/AAC.44.7.1818-1824.2000
Andersson, E. K., Bengtsson, C., Evans, M. L., Chorell, E., Sellstedt, M., Lindgren, A. E., et al. (2013). Modulation of curli assembly and pellicle biofilm formation by chemical and protein chaperones. Chem. Biol. 20, 1245–1254. doi: 10.1016/j.chembiol.2013.07.017
Antunes, L. C., Visca, P., and Towner, K. J. (2014). Acinetobacter baumannii: evolution of a global pathogen. Pathog. Dis. 71, 292–301. doi: 10.1111/2049-632X.12125
Baugh, S., Ekanayaka, A. S., Piddock, L. J., and Webber, M. A. (2012). Loss of or inhibition of all multidrug resistance efflux pumps of Salmonella enterica serovar Typhimurium results in impaired ability to form a biofilm. J. Antimicrob. Chemother. 67, 2409–2417. doi: 10.1093/jac/dks228
Baugh, S., Phillips, C. R., Ekanayaka, A. S., Piddock, L. J., and Webber, M. A. (2014). Inhibition of multidrug efflux as a strategy to prevent biofilm formation. J. Antimicrob. Chemother. 69, 673–681. doi: 10.1093/jac/dkt420
Bina, X. R., Provenzano, D., Nguyen, N., and Bina, J. E. (2008). Vibrio cholerae RND family efflux systems are required for antimicrobial resistance, optimal virulence factor production, and colonization of the infant mouse small intestine. Infect. Immun. 76, 3595–3605. doi: 10.1128/IAI.01620-07
Buckley, A. M., Webber, M. A., Cooles, S., Randall, L. P., La Ragione, R. M., Woodward, M. J., et al. (2006). The AcrAB-TolC efflux system of Salmonella enterica serovar Typhimurium plays a role in pathogenesis. Cell. Microbiol. 8, 847–856. doi: 10.1111/j.1462-5822.2005.00671.x
Bunikis, I., Denker, K., Ostberg, Y., Andersen, C., Benz, R., and Bergström, S. (2008). An RND-type efflux system in Borrelia burgdorferi is involved in virulence and resistance to antimicrobial compounds. PLoS Pathog. 4:e1000009. doi: 10.1371/journal.ppat.1000009
Burse, A., Weingart, H., and Ullrich, M. S. (2004). The phytoalexin-inducible multidrug efflux pump AcrAB contributes to virulence in the fire blight pathogen, Erwinia amylovora. Mol. Plant. Microbe Interact. 17, 43–54. doi: 10.1094/MPMI.2004.17.1.43
Cegelski, L., Pinkner, J. S., Hammer, N. D., Cusumano, C. K., Hung, C. S., Chorell, E., et al. (2009). Small-molecule inhibitors target Escherichia coli amyloid biogenesis and biofilm formation. Nat. Chem. Biol. 5, 913–919. doi: 10.1038/nchembio.242
Chan, Y. Y., Bian, H. S., Tan, T. M., Mattmann, M. E., Geske, G. D., Igarashi, J., et al. (2007). Control of quorum sensing by a Burkholderia pseudomallei multidrug efflux pump. J. Bacteriol. 189, 4320–4324. doi: 10.1128/JB.00003-07
Choi, J., Shin, D., Kim, M., Park, J., Lim, S., and Ryu, S. (2012). LsrR-mediated quorum sensing controls invasiveness of Salmonella typhimurium by regulating SPI-1 and flagella genes. PLoS ONE 7:e37059. doi: 10.1371/journal.pone.0037059
Cosgaya, C., Marí-Almirall, M., Van Assche, A., Fernández-Orth, D., Mosqueda, N., Telli, M., et al. (2016). Acinetobacter dijkshoorniae sp. nov., a member of the Acinetobacter calcoaceticus-Acinetobacter baumannii complex mainly recovered from clinical samples in different countries. Int. J. Syst. Evol. Microbiol. 66, 4105–4111.doi: 10.1099/ijsem.0.001318
Coyne, S., Guigon, G., Courvalin, P., and Périchon, B. (2010a). Screening and quantification of the expression of antibiotic resistance genes in Acinetobacter baumannii with a microarray. Antimicrob. Agents Chemother. 54, 333–340. doi: 10.1128/AAC.01037-09
Coyne, S., Rosenfeld, N., Lambert, T., Courvalin, P., and Périchon, B. (2010b). Overexpression of resistance-nodulation-cell division pump AdeFGH confers multidrug resistance in Acinetobacter baumannii. Antimicrob. Agents Chemother. 54, 4389–4393. doi: 10.1128/AAC.00155-10
Doi, Y., Murray, G. L., and Peleg, A. Y. (2015). Acinetobacter baumannii: evolution of antimicrobial resistance-treatment options. Semin. Respir. Crit. Care Med. 36, 85–98. doi: 10.1055/s-0034-1398388
Edgar, R. C. (2004). MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797. doi: 10.1093/nar/gkh340
Fournier, P. E., and Richet, H. (2006). The epidemiology and control of Acinetobacter baumannii in health care facilities. Clin. Infect. Dis. 42, 692–699. doi: 10.1086/500202
Giammanco, A., Calà, C., Fasciana, T., and Dowzicky, M. J. (2017). Global assessment of the activity of tigecycline against multidrug-resistant Gram-Negative pathogens between 2004 and 2014 as part of the tigecycline evaluation and surveillance trial. mSphere 2:e00310-16. doi: 10.1128/mSphere.00310-16
Giles, S. K., Stroeher, U. H., Eijkelkamp, B. A., and Brown, M. H. (2015). Identification of genes essential for pellicle formation in Acinetobacter baumannii. BMC Microbiol. 15:116. doi: 10.1186/s12866-015-0440-6
Gillis, R. J., White, K. G., Choi, K. H., Wagner, V. E., Schweizer, H. P., and Iglewski, B. H. (2005). Molecular basis of azithromycin-resistant Pseudomonas aeruginosa biofilms. Antimicrob. Agents Chemother. 49, 3858–3867. doi: 10.1128/AAC.49.9.3858-3867.2005
Gotoh, N., Tsujimoto, H., Poole, K., Yamagishi, J., and Nishino, T. (1995). The outer membrane protein OprM of Pseudomonas aeruginosa is encoded by oprK of the mexA-mexB-oprK multidrug resistance operon. Antimicrob. Agents Chemother. 39, 2567–2569. doi: 10.1128/AAC.39.11.2567
Grund, S., and Weber, A. (1988). A new type of fimbriae on Salmonella typhimurium. J. Vet. Med. 35, 779–782. doi: 10.1111/j.1439-0450.1988.tb00560.x
Hanahan, D. (1983). Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166, 557–580. doi: 10.1016/S0022-2836(83)80284-8
Hirakata, Y., Srikumar, R., Poole, K., Gotoh, N., Suematsu, T., Kohno, S., et al. (2002). Multidrug efflux systems play an important role in the invasiveness of Pseudomonas aeruginosa. J. Exp. Med. 196, 109–118. doi: 10.1084/jem.20020005
Khoramian-Falsafi, T., Harayama, S., Kutsukake, K., and Pechère, J. C. (1990). Effect of motility and chemotaxis on the invasion of Salmonella typhimurium into HeLa cells. Microb. Pathog. 9, 47–53. doi: 10.1016/0882-4010(90)90039-S
Kim, Y. J., Im, S. Y., Lee, J. O., and Kim, O. B. (2016). Potential swimming motility variation by AcrR in Escherichia coli. J. Microbiol. Biotechnol. 26, 1824–1828. doi: 10.4014/jmb.1607.07058
Köhler, T., Michéa-Hamzehpour, M., Henze, U., Gotoh, N., Curty, L. K., and Pechère, J. C. (1997). Characterization of MexE-MexF-OprN, a positively regulated multidrug efflux system of Pseudomonas aeruginosa. Mol. Microbiol. 23, 345–354. doi: 10.1046/j.1365-2958.1997.2281594.x
Kvist, M., Hancock, V., and Klemm, P. (2008). Inactivation of efflux pumps abolishes bacterial biofilm formation. Appl. Environ. Microbiol. 74, 7376–7382. doi: 10.1128/AEM.01310-08
Laurence, D. P., Sophie, M., Sylvie, B., Thierry, L., and Patrice, C. (2007). AdeIJK, a resistance-nodulation-cell division pump effluxing multiple antibiotics in Acinetobacter baumannii. Antimicrob. Agents Chemother. 52, 557–562. doi: 10.1128/AAC.00732-07
Lee, H. W., Koh, Y. M., Kim, J., Lee, J. C., Lee, Y. C., Seol, S. Y., et al. (2008). Capacity of multidrug-resistant clinical isolates of Acinetobacter baumannii to form biofilm and adhere to epithelial cell surfaces. Clin. Microbiol. Infect. 14, 49–54. doi: 10.1111/j.1469-0691.2007.01842.x
Lee, J. C., Koerten, H., van den Broek, P., Beekhuizen, H., Wolterbeek, R., van den Barselaar, M., et al. (2006). Adherence of Acinetobacter baumannii strains to human bronchial epithelial cells. Res. Microbiol. 157, 360–366. doi: 10.1016/j.resmic.2005.09.011
Lee, Y. C., Huang, Y. T., Tan, C. K., Kuo, Y. W., Liao, C. H., Lee, P. I., et al. (2011). Acinetobacter baumannii and Acinetobacter genospecies 13TU and 3 bacteraemia: comparison of clinical features, prognostic factors and outcomes. J. Antimicrob. Chemother. 66, 1839–1846. doi: 10.1093/jac/dkr200
Li, M., Gu, R., Su, C. C., Routh, M. D., Harris, K. C., Jewell, E. S., et al. (2007). Crystal structure of the transcriptional regulator AcrR from Escherichia coli. J. Mol. Biol. 374, 591–603. doi: 10.1016/j.jmb.2007.09.064
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Lynch, S. V., Dixon, L., Benoit, M. R., Brodie, E. L., Keyhan, M., Hu, P., et al. (2007). Role of the rapA gene in controlling antibiotic resistance of Escherichia coli biofilms. Antimicrob. Agents Chemother. 51, 3650–3658. doi: 10.1128/AAC.00601-07
Ma, D., Alberti, M., Lynch, C., Nikaido, H., and Hearst, J. E. (1996). The local repressor AcrR plays a modulating role in the regulation of acrAB genes of Escherichia coli by global stress signals. Mol. Microbiol. 19, 101–112. doi: 10.1046/j.1365-2958.1996.357881.x
Ma, D., Cook, D. N., Alberti, M., Pon, N. G., Nikaido, H., and Hearst, J. E. (1993). Molecular cloning and characterization of acrA and acrE genes of Escherichia coli. J. Bacteriol. 175, 6299–6313. doi: 10.1128/jb.175.19.6299-6313.1993
Ma, D., Cook, D. N., Alberti, M., Pon, N. G., Nikaido, H., and Hearst, J. E. (1995). Genes acrA and acrB encode a stress-induced efflux system of Escherichia coli. Mol. Microbiol. 16, 45–55. doi: 10.1111/j.1365-2958.1995.tb02390.x
Magnet, S., Courvalin, P., and Lambert, T. (2001). Resistance-nodulation-cell division-type efflux pump involved in aminoglycoside resistance in Acinetobacter baumannii strain BM4454. Antimicrob. Agents Chemother. 45, 3375–3380. doi: 10.1128/AAC.45.12.3375-3380.2001
Marchand, I., Damier-Piolle, L., Courvalin, P., and Lambert, T. (2004). Expression of the RND-type efflux pump AdeABC in Acinetobacter baumannii is regulated by the AdeRS two-component system. Antimicrob. Agents Chemother. 48, 3298–3304. doi: 10.1128/AAC.48.9.3298-3304.2004
Marí-Almirall, M., Cosgaya, C., Higgins, P. G., Van Assche, A., Telli, M., Huys, G., et al. (2017). MALDI-TOF/MS identification of species from the Acinetobacter baumannii (Ab) group revisited: inclusion of the novel A. seifertii and A. dijkshoorniae species. Clin. Microbiol. Infect. 23, 210.e1–210.e9. doi: 10.1016/j.cmi.2016.11.020
Maseda, H., Sawada, I., Saito, K., Uchiyama, H., Nakae, T., and Nomura, N. (2004). Enhancement of the mexAB-oprM efflux pump expression by a quorum-sensing autoinducer and its cancellation by a regulator, MexT, of the mexEF-oprN efflux pump operon in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 48, 1320–1328. doi: 10.1128/AAC.48.4.1320-1328.2004
Matsumura, K., Furukawa, S., Ogihara, H., and Morinaga, Y. (2011). Roles of multidrug efflux pumps on the biofilm formation of Escherichia coli K-12. Biocontrol Sci. 16, 69–72. doi: 10.4265/bio.16.69
Murakami, S., Nakashima, R., Yamashita, E., Matsumoto, T., and Yamaguchi, A. (2006). Crystal structures of a multidrug transporter reveal a functionally rotating mechanism. Nature 443, 173–179. doi: 10.1038/nature05076
Murakami, S., Nakashima, R., Yamashita, E., and Yamaguchi, A. (2002). Crystal structure of bacterial multidrug efflux transporter AcrB. Nature 419, 587–593. doi: 10.1038/nature01050
Nemec, A., Krizova, L., Maixnerova, M., Sedo, O., Brisse, S., and Higgins, P. G. (2015). Acinetobacter seifertii sp. nov., a member of the Acinetobacter calcoaceticus-Acinetobacter baumannii complex isolated from human clinical specimens. Int. J. Syst. Evol. Microbiol. 65, 934–942. doi: 10.1099/ijs.0.000043
Nemec, A., Krizova, L., Maixnerova, M., van der Reijden, T. J., Deschaght, P., Passet, V., et al. (2011). Genotypic and phenotypic characterization of the Acinetobacter calcoaceticus-Acinetobacter baumannii complex with the proposal of Acinetobacter pittii sp. nov. (formerly Acinetobacter genomic species 3) and Acinetobacter nosocomialis sp. nov. (formerly Acinetobacter genomic species 13TU). Res. Microbiol. 162, 393–404. doi: 10.1016/j.resmic.2011.02.006
Nikaido, H. (1996). Multidrug efflux pumps of gram-negative bacteria. J. Bacteriol. 178, 5853–5859. doi: 10.1128/jb.178.20.5853-5859.1996
Nishino, K., Nikaido, E., and Yamaguchi, A. (2009). Regulation and physiological function of multidrug efflux pumps in Escherichia coli and Salmonella. Biochim. Biophys. Acta 1794, 834–843. doi: 10.1016/j.bbapap.2009.02.002
Oh, M. H., and Choi, C. H. (2015). Role of LuxIR homologue AnoIR in Acinetobacter nosocomialis and the effect of virstatin on the expression of anoR gene. J. Microbiol. Biotechnol. 25, 1390–1400. doi: 10.4014/jmb.1504.04069
Oh, M. H., Lee, J. C., Kim, J., Choi, C. H., and Han, K. (2015). Simple method for markerless gene deletion in multidrug-resistant Acinetobacter baumannii. Appl. Environ. Microbiol. 81, 3357–3368. doi: 10.1128/AEM.03975-14
Okusu, H., Ma, D., and Nikaido, H. (1996). AcrAB efflux pump plays a major role in the antibiotic resistance phenotype of Escherichia coli multipleantibiotic-resistance (Mar) mutants. J. Bacteriol. 178, 306–308. doi: 10.1128/jb.178.1.306-308.1996
Olliver, A., Valle, M., Chaslus-Dancla, E., and Cloeckaert, A. (2004). Role of an acrR mutation in multidrug resistance of in vitro-selected fluoroquinolone-resistant mutants of Salmonella enterica serovar Typhimurium. FEMS Microbiol. Lett. 238, 267–272. doi: 10.1111/j.1574-6968.2004.tb09766.x
Paulsen, I. T., Chen, J., Nelson, K. E., and Saier, M. H. J. (2001). Comparative genomics of microbial drug efflux systems. J. Mol. Microbiol. Biotechnol. 3, 145–150.
Pearson, J. P., Van Delden, C., and Iglewski, B. H. (1999). Active efflux and diffusion are involved in transport of Pseudomonas aeruginosa cell-to-cell signals. J. Bacteriol. 181, 1203–1210.
Peleg, A. Y., and Hooper, D. C. (2010). Hospital-acquired infections due to gram-negative bacteria. N. Engl. J. Med. 362, 1804–1813. doi: 10.1056/NEJMra0904124
Peleg, A. Y., Seifert, H., and Paterson, D. L. (2008). Acinetobacter baumannii: emergence of a successful pathogen. Clin. Microbiol. Rev. 21, 538–582. doi: 10.1128/CMR.00058-07
Perrett, C. A., Karavolos, M. H., Humphrey, S., Mastroeni, P., Martinez-Argudo, I., Spencer, H., et al. (2009). LuxS-based quorum sensing does not affect the ability of Salmonella enterica serovar Typhimurium to express the SPI-1 type 3 secretion system, induce membrane ruffles, or invade epithelial cells. J. Bacteriol. 191, 7253–7259. doi: 10.1128/JB.00727-09
Piddock, L. J. (2006). Multidrug-resistance efflux pumps - not just for resistance. Nat. Rev. Microbiol. 4, 629–636. doi: 10.1038/nrmicro1464
Poole, K. (2007). Efflux pumps as antimicrobial resistance mechanisms. Ann. Med. 39, 162–176. doi: 10.1080/07853890701195262
Poole, K., Gotoh, N., Tsujimoto, H., Zhao, Q., Wada, A., Yamasaki, T., et al. (1996). Overexpression of the mexC-mexD-oprJ efflux operon in nfxB-type multidrug-resistant strains of Pseudomonas aeruginosa. Mol. Microbiol. 21, 713–724. doi: 10.1046/j.1365-2958.1996.281397.x
Poole, K., Krebes, K., McNally, C., and Neshat, S. (1993). Multiple antibiotic resistance in Pseudomonas aeruginosa: evidence for involvement of an efflux operon. J. Bacteriol. 175, 7363–7372. doi: 10.1128/jb.175.22.7363-7372.1993
Pourahmad Jaktaji, R., and Jazayeri, N. (2013). Expression of acrA and acrB Genes in Esherichia coli Mutants with or without marR or acrR Mutations. Iran. J. Basic Med. Sci. 16, 1254–1258.
Pumbwe, L., Randall, L. P., Woodward, M. J., and Piddock, L. J. (2004). Expression of the efflux pump genes cmeB, cmeF and the porin gene porA in multiple-antibiotic-resistant Campylobacter jejuni. J. Antimicrob. Chemother. 54, 341–347. doi: 10.1093/jac/dkh331
Putman, M., van Veen, H. W., and konings, W. N. (2000). Molecular properties of bacterial multidrug transporters. Microbiol. Mol. Biol. Rev. 64, 672–693. doi: 10.1128/MMBR.64.4.672-693.2000
Ramos, J. L., Martínez-Bueno, M., Molina-Henares, A. J., Terán, W., Watanabe, K., Zhang, X., et al. (2005). The TetR family of transcriptional repressors. Microbiol. Mol. Biol. Rev. 69, 326–356. doi: 10.1128/MMBR.69.2.326-356.2005
Richmond, G. E., Evans, L. P., Anderson, M. J., Wand, M. E., Bonney, L. C., Ivens, A., et al. (2016). The Acinetobacter baumannii two-component system AdeRS regulates genes required for multidrug efflux, biofilm formation, and virulence in a strain-specific manner. MBio 7, e00430-16. doi: 10.1128/mBio.00430-16
Rutherford, S. T., and Bassler, B. L. (2012). Bacterial quorum sensing: its role in virulence and possibilities for its control. Cold Spring Harb. Perspect. Med. 2:a012427. doi: 10.1101/cshperspect.a012427
Sambrook, J., Fritsch, E. F., and Maniatis, T. (1989). Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory.
Schlisselberg, D. B., Kler, E., Kisluk, G., Shachar, D., and Yaron, S. (2015). Biofilm formation ability of Salmonella enterica serovar Typhimurium acrAB mutants. Int. J. Antimicrob. Agents 46, 456–459. doi: 10.1016/j.ijantimicag.2015.06.011
Shafer, W. M., Qu, X., Waring, A. J., and Lehrer, R. I. (1998). Modulation of Neisseria gonorrhoeae susceptibility to vertebrate antibacterial peptides due to a member of the resistance/nodulation/division efflux pump family. Proc. Natl. Acad. Sci. U.S.A. 95, 1829–1833. doi: 10.1073/pnas.95.4.1829
Simon, R., Priefer, U., and Puhler, A. (1983). A broad host range mobilization system for in vivo genetic engineering transposon mutagenesis in gram negative bacteria. Nat. Biotechnol. 1, 784–791. doi: 10.1038/nbt1183-784
Skiebe, E., de Berardinis, V., Morczinek, P., Kerrinnes, T., Faber, F., Lepka, D., et al. (2012). Surface-associated motility, a common trait of clinical isolates of Acinetobacter baumannii, depends on 1,3-diaminopropane. Int. J. Med. Microbiol. 302, 117–128. doi: 10.1016/j.ijmm.2012.03.003
Spaniol, V., Bernhard, S., and Aebi, C. (2015). Moraxella catarrhalis AcrAB-OprM efflux pump contributes to antimicrobial resistance and is enhanced during cold shock response. Antimicrob. Agents Chemother. 59, 1886–1894. doi: 10.1128/AAC.03727-14
Studier, F. W., and Moffatt, B. A. (1986). Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J. Mol. Biol. 189, 113–130. doi: 10.1016/0022-2836(86)90385-2
Su, C. C., Rutherford, D. J., and Yu, E. W. (2007). Characterization of the multidrug efflux regulator AcrR from Escherichia coli. Biochem. Biophys. Res. Commun. 361, 85–90. doi: 10.1016/j.bbrc.2007.06.175
Tamura, K., Stecher, G., Peterson, D., Filipski, A., and Kumar, S. (2013). MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729. doi: 10.1093/molbev/mst197
Thanassi, D. G., Cheng, L. W., and Nikaido, H. (1997). Active efflux of bile salts by Escherichia coli. J. Bacteriol. 179, 2512–2518. doi: 10.1128/jb.179.8.2512-2518.1997
Tipton, K. A., Farokhyfar, M., and Rather, P. N. (2017). Multiple roles for a novel RND-type efflux system in Acinetobacter baumannii AB5075. Microbiologyopen 6:e00418. doi: 10.1002/mbo3.418
Wang-Kan, X., Blair, J. M. A., Chirullo, B., Betts, J., La Ragione, R. M., Ivens, A., et al. (2017). Lack of AcrB efflux function confers loss of virulence on Salmonella enterica serovar Typhimurium. MBio 8:e00968-17. doi: 10.1128/mBio.00968-17
Webber, M. A., Bailey, A. M., Blair, J. M., Morgan, E., Stevens, M. P., Hinton, J. C., et al. (2009). The global consequence of disruption of the AcrAB-TolC efflux pump in Salmonella enterica includes reduced expression of SPI-1 and other attributes required to infect the host. J. Bacteriol. 191, 4276–4285. doi: 10.1128/JB.00363-09
Zhang, L., and Mah, T. F. (2008). Involvement of a novel efflux system in biofilm-specific resistance to antibiotics. J. Bacteriol. 190, 4447–4452. doi: 10.1128/JB.01655-07
Keywords: A. nosocomialis, efflux pump, AcrR, antibiotic resistance, quorum sensing, biofilm, virulence
Citation: Subhadra B, Kim J, Kim DH, Woo K, Oh MH and Choi CH (2018) Local Repressor AcrR Regulates AcrAB Efflux Pump Required for Biofilm Formation and Virulence in Acinetobacter nosocomialis. Front. Cell. Infect. Microbiol. 8:270. doi: 10.3389/fcimb.2018.00270
Received: 15 May 2018; Accepted: 19 July 2018;
Published: 07 August 2018.
Edited by:
Justin Merritt, Oregon Health & Science University, United StatesReviewed by:
Justin A. Thornton, Mississippi State University, United StatesEsther Orozco, Centro de Investigación y de Estudios Avanzados del Instituto Politécnico Nacional (CINVESTAV-IPN), Mexico
Copyright © 2018 Subhadra, Kim, Kim, Woo, Oh and Choi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Man Hwan Oh, eXkxMDkxQGRhbmtvb2suYWMua3I=
Chul Hee Choi, Y2hvaWNoQGNudS5hYy5rcg==
 Bindu Subhadra1
Bindu Subhadra1 Chul Hee Choi
Chul Hee Choi