- 1Division of Biological Sciences, University of Montana, Missoula, MT, United States
- 2Center for Integrative Bioinformatics Vienna, Max F. Perutz Laboratories, Medical University of Vienna, Vienna, Austria
- 3Department of Biology, University of Colorado, Colorado Springs, CO, United States
- 4Center for Biomolecular Structure and Dynamics, University of Montana, Missoula, MT, United States
The Lyme disease spirochete Borrelia (Borreliella) burgdorferi must tolerate nutrient stress to persist in the tick phase of its enzootic life cycle. We previously found that the stringent response mediated by RelBbu globally regulates gene expression to facilitate persistence in the tick vector. Here, we show that RelBbu regulates the expression of a swath of small RNAs (sRNA), affecting 36% of previously identified sRNAs in B. burgdorferi. This is the first sRNA regulatory mechanism identified in any spirochete. Threefold more sRNAs were RelBbu-upregulated than downregulated during nutrient stress and included antisense, intergenic and 5′ untranslated region sRNAs. RelBbu-regulated sRNAs associated with genes known to be important for host infection (bosR and dhhp) as well as persistence in the tick (glpF and hk1) were identified, suggesting potential mechanisms for post-transcriptional regulation of gene expression.
Introduction
Borrelia (Borreliella) burgdorferi, the causative agent of Lyme disease (Burgdorfer et al., 1982; Benach et al., 1983; Steere et al., 1983), exists in nature in an enzootic cycle alternating between Ixodes ticks and vertebrate hosts (Lane et al., 1991; Radolf et al., 2012; Caimano et al., 2016). Tick larvae acquire B. burgdorferi by feeding on an infected host, typically a small mammal (Piesman and Schwan, 2010). B. burgdorferi must adapt to persist in the tick midgut during the molt into nymphs and then transmit back to a vertebrate during nymphal feeding (Radolf et al., 2012; Corona and Schwartz, 2015; Caimano et al., 2016). In order to survive these disparate environments, B. burgdorferi alters its pattern of gene expression in response to environmental signals, including temperature and nutrients (Radolf et al., 2012; Troxell and Yang, 2013; Iyer and Schwartz, 2016; Samuels and Samuels, 2016; Stevenson and Seshu, 2017). Central to this response is the RpoN-RpoS alternative sigma factor cascade that transcriptionally controls expression of numerous genes required for the transmission from the tick and the establishment of infection in the mammal (Hübner et al., 2001; Fisher et al., 2005; Burtnick et al., 2007; Caimano et al., 2007; Dunham-Ems et al., 2012; Ouyang et al., 2012; Grove et al., 2017). Additionally, the enhancer-binding protein and response regulator Rrp2 (Yang et al., 2003; Burtnick et al., 2007), the transcriptional regulators BosR (Boylan et al., 2003; Hyde et al., 2009; Ouyang et al., 2009, 2011; Katona, 2015) and BadR (Miller et al., 2013; Ouyang and Zhou, 2015), as well as the intracellular second messengers cyclic dimeric GMP (c-di-GMP) (Rogers et al., 2009; Sultan et al., 2010; He et al., 2011; Caimano et al., 2015) and guanosine pentaphosphate and tetraphosphate [(p)ppGpp] (Bugrysheva et al., 2015; Drecktrah et al., 2015) act in concert with RpoN and RpoS to regulate genes required for either host infection or tick persistence (Samuels, 2011; Caimano et al., 2016). However, post-transcriptional regulation of gene expression remains mostly unexplored in B. burgdorferi (Lybecker and Samuels, 2007; Jutras et al., 2013; Dulebohn et al., 2014).
Small, noncoding RNAs (sRNAs) have emerged as major regulators of gene expression, although many of the mechanisms are not well understood (Waters and Storz, 2009; Storz et al., 2011; Caldelari et al., 2013; Quereda and Cossart, 2017; Hör et al., 2018). The sRNAs that affect mRNA expression directly are divided into two classes: cis-acting, which function in close proximity to their genomic location, and trans-acting, which target more distal elements (Georg and Hess, 2011). Both act by binding target mRNAs via complementary base pairing, albeit sometimes with imperfect complementarity (Thomason and Storz, 2010). Antisense RNAs (asRNAs) are a class of sRNAs that are transcribed opposite to the protein-coding sense strand and have complete complementarity to the corresponding sense RNA. Antisense transcripts regulate gene expression through a variety of different mechanisms: transcriptional interference, transcriptional attenuation, translational stimulation or inhibition, and RNA stability, but detailed mechanisms remain elusive despite their ubiquity (Thomason and Storz, 2010; Lybecker et al., 2014). The double-stranded RNA (dsRNA) created by sense-antisense pairing can be cleaved by RNase III (Anacker et al., 2018), which can destabilize or stabilize the RNA, leading to degradation or translation, respectively (Lioliou et al., 2012; Lybecker et al., 2014; Gordon et al., 2017). Thus, levels of asRNA and the associated mRNA may be coordinately upregulated, which suggests the asRNA stabilizes the mRNA, or reciprocally regulated, which suggests destabilization of the mRNA. Riboswitches are another cis-acting regulatory RNA. They are usually found in the 5′ untranslated region (5′ UTR) of the mRNA they regulate and function by altering their secondary structure in response to environmental signals and binding intracellular metabolites (Breaker, 2012; Serganov and Nudler, 2013; Sherwood and Henkin, 2016; Nelson and Breaker, 2017). sRNAs transcribed from the intergenic regions between genes (IG RNAs) are typically trans-acting sRNAs that have imperfect pairing with their target(s) and regulate the translation and/or degradation of the target RNA (Storz et al., 2011). Thus, a vast transcriptomic armada of noncoding sRNAs exists that provide an additional layer of gene regulation with important implications for post-transcriptional control and, ultimately, protein expression.
The stringent response is invoked by bacteria to adapt and survive nutrient stresses, including decreased levels of amino acids, carbon, phosphate, and iron, by increasing the levels of the intracellular second messenger (p)ppGpp (Chatterji and Ojha, 2001; Potrykus and Cashel, 2008; Liu et al., 2015). This alarmone causes a sea change in physiology, metabolism and gene expression by altering RNA polymerase (RNAP) sigma factor selectivity and associated expression from RpoD-dependent and RpoS-dependent promoters (Dalebroux and Swanson, 2012; Hauryliuk et al., 2015). The enzymes RelA and SpoT, as well as their homologs, control the levels of (p)ppGpp (Potrykus and Cashel, 2008; Atkinson et al., 2011; Dalebroux and Swanson, 2012; Hauryliuk et al., 2015). RelA synthesizes (p)ppGpp and SpoT either synthesizes or hydrolyzes (p)ppGpp in a number of bacteria; however, these two activities are combined in a single bifunctional enzyme, RelBbu, which mediates the stringent response in B. burgdorferi (Concepcion and Nelson, 2003; Bugrysheva et al., 2005; Drecktrah et al., 2015). RelBbu globally regulates gene expression to facilitate persistence in the tick during the transition from unfed to fed nymphs when B. burgdorferi experiences nutrient deprivation as the blood meal is absorbed in the tick midgut (Drecktrah et al., 2015). Previously, we and others found that expression of the glycerol metabolism (glp) operon, which is important for persistence in the tick (He et al., 2011; Pappas et al., 2011), is RelBbu-dependent (Bugrysheva et al., 2015; Drecktrah et al., 2015). Additionally, the expression of host-associated virulence genes, including bosR as well as dbpBA, encoding decorin-binding proteins, and vlsE, encoding an antigenic variation locus, are regulated by RelBbu (Drecktrah et al., 2015).
DsrA, the first regulatory sRNA identified and characterized in B. burgdorferi, regulates temperature-dependent translation of the alternative sigma factor RpoS (Lybecker and Samuels, 2007). More recently, small RNA-sequencing analyses of B. burgdorferi revealed a vast sRNA landscape containing over 1000 sRNAs, including asRNAs, IG RNAs, and 5′ UTR sRNAs, as well as intraRNAs, which are partial transcripts from the same strand encoding the ORF (Popitsch et al., 2017). Popitsch et al. (2017) described temperature-regulated sRNA expression under conditions that mimic the tick vector (23°C) and mammalian host (37°C) environments, which suggested a complex and dynamic role for sRNAs in the enzootic cycle. Another study identified 351 growth-phase-dependent sRNAs in B. burgdorferi (Arnold et al., 2016). Together, these studies have illuminated an impressive tally of sRNAs and have laid the foundation to investigate the regulatory mechanisms for sRNA expression in the Lyme disease spirochete (Lybecker and Samuels, 2017).
In this study, we discovered that the RelBbu-mediated stringent response is a major regulator of sRNA expression in B. burgdorferi, affecting the levels of a third (241 out of 666) of all identified sRNAs (not including intraRNAs). sRNA-regulated gene expression may play a larger regulatory role in this bacterium compared to others as many of the protein-mediated transcriptional regulators and signaling pathways are absent in B. burgdorferi (Fraser et al., 1997; Samuels and Radolf, 2009). Herein, we identify sRNAs associated with known virulence factors and metabolic pathways, and we integrate the RelBbu-dependent sRNA transcriptome and mRNA transcriptome (Drecktrah et al., 2015), evincing implications for sRNA-regulated gene expression in regard to host infectivity and persistence in the vector.
Materials and Methods
B. burgdorferi Strains and Growth Conditions
Low-passage B. burgdorferi strain B31-5A4 and relBbu mutant (ΔrelBbu) strains (Drecktrah et al., 2015) were cultured at 35°C in Barbour-Stoenner-Kelly II (BSK) liquid medium, pH 7.6, containing 6% rabbit serum (RS) (Pel-Freez Biologicals) without gelatin (Barbour, 1984; Samuels et al., 2018). For nutrient stress experiments, 40-ml cultures of wild-type and ΔrelBbu B. burgdorferi strains were grown as described above until reaching stationary phase (~1 × 108 cells ml−1) and collected by centrifugation at 8,000 × g for 10 min at 4°C; the supernatant was discarded and cells were resuspended in RPMI (without glutamate) (Mediatech) for 6 h for the nutrient stress samples. Cell density was determined by enumeration using a Petroff-Hausser cell counting chamber (Samuels et al., 2018).
RNA Isolation
RNA was isolated form B. burgdorferi cultures using hot phenol extraction as previously described (Jahn et al., 2008; Drecktrah et al., 2015; Popitsch et al., 2017). RNA was then treated with Turbo DNase according to the manufacturer's instructions. The RNase inhibitor SUPERase-In (Invitrogen) was diluted 1:25 in the reaction mixture and samples were incubated at 37°C for 30 min. DEPC-water was added to bring the total volume up to 300 μl and transferred to a 2.0 ml phase-lock gel heavy tube (5Prime). Three hundred microliter of ultrapure phenol:chloroform:isoamyl was added and samples were inverted 10 times. Samples were centrifuged at 20,800 × g for 7 min at 4°C. Three hundred microliters of chloroform, pH 4.3 (Fisher) was added to the top layer in the phase lock tube and samples inverted 10 times. Samples were centrifuged at 20,800 × g for 7 min at 4°C. The aqueous phase was transferred to a new RNase-free tube and 1 μl of 20 mg ml−1 glycogen, 1/10 volume of 3 M sodium acetate, pH 5.2 and, finally, three volumes of ice-cold 100% ethanol were added. Samples were inverted gently to mix and incubated at −20°C overnight. Precipitated RNA was pelleted by centrifugation at 20,800 × g for 45 min at 4°C. The pellet was washed with 75% ethanol and centrifuged at 20,800 × g for 20 min at 4°C. Pellets were dried for 1 min and resuspended in 20 μl of DEPC-water.
RNA Sequencing
RNA sequencing cDNA libraries were generated from two independent biological replicates as previously described (Drecktrah et al., 2015). Importantly, the ribosomal-depleted (Ribo-Zero rRNA removal kit, Illumina) total RNA was fragmented and size-selected (75–300 nucleotides) on an 8% TBE-Urea gel to include both sRNAs and fragmented mRNAs. Libraries were sequenced on an Illumina HiSeq 2000 with single-end 50-base-pair reads at the Vienna Biocenter Core Facilities Next Generation Sequencing unit. As previously described (Popitsch et al., 2017), reads were demultiplexed and adapters were clipped with cutadapt. After quality control, the reads were mapped to the B. burgdorferi B31 reference genome with NextGenMap 0.4.10 (Sedlazeck et al., 2013) using standard parameters and a minimum identity threshold of 90%; multireads and reads with a mapping quality smaller than 20 were pruned. FeatureCounts (Liao et al., 2014) was used to calculate raw read counts for all datasets. Annotation of the RelBbu-dependent sRNAs was based on Popitsch et al. (2017) and gene ontogeny of associated ORFs was guided by the Schutzer annotation (Di et al., 2014). Since the isolated RNA included both small RNAs and fragmented mRNAs, intraRNAs, which are located within annotated genes, could not be distinguished from mRNAs and were not included in the differential sRNA expression analyses. From the read counts, we calculated differential expression between wild type and the relBbu mutant during nutrient stress using edgeR and filtered the results by adjusted P-value ≤ 0.05. To enable inspection in a genome browser, we used CODOC (Popitsch, 2014) to compute strand-specific, normalized depth-of-coverage signals.
Northern Blot Analysis
RNA was analyzed by Northern blotting as previously described (Popitsch et al., 2017). Five to fifteen micrograms of RNA was combined with an equal volume of 2 × RNA loading dye and denatured by heating at 65°C for 10 min. Samples were separated on an 8% TBE-Urea gel before transfer to HybondXL membranes (Amersham) by electrotransfer. Membranes were UV-crosslinked and blocked with ULTRAhyb Oligo Hybridization Buffer (Invitrogen) for 1 h at 40°C. Membranes were probed with 32P-labeled oligonucleotides overnight at 40°C in a rotating hybridization oven. ffs, which encodes the 4.5S RNA of the signal recognition particle, was used as a loading control because 4.5S RNA levels are not dependent on RelBbu (Drecktrah et al., 2015). Membranes were washed with 2 × SSC + 0.5% SDS, wrapped in plastic, exposed to an imaging plate for 12–48 h, and analyzed using a Fujifilm FLA-3000G phosphorimager. 32P-labeled oligonucleotides (Table S1) were end-labeled with gamma 32P-ATP (Perkin-Elmer) and T4 PNK (New England Biolabs) according to the manufacturer's instructions.
Results and Discussion
In this study, we identify the RelBbu-dependent sRNA transcriptome under conditions of nutrient stress to better understand gene regulation during persistence of B. burgdorferi in its tick vector. We found that RelBbu regulates approximately one third of the 666 sRNAs (not including intraRNAs) identified in B. burgdorferi by Popitsch et al. (2017), with greater than threefold more sRNAs upregulated than downregulated: 187 sRNAs upregulated (Table S2) and 54 sRNAs downregulated (Table S3) during nutrient stress (Figure 1). sRNAs from each category, IG RNA, asRNA and 5′ UTR sRNA, were RelBbu-regulated, with asRNAs being the majority of upregulated sRNAs (Figure 2A) and IG RNAs as the largest category of downregulated sRNAs (Figure 3A). RelBbu-regulated sRNAs were found throughout the genome, including the linear chromosome and the numerous linear and circular plasmids (Fraser et al., 1997; Brisson et al., 2012). sRNAs from all categories with significant and greater than twofold changes in sRNA levels from deep sequencing were validated by Northern blot analyses (Figures 4, 5 and Figures S2–S4). To gain insight into the cellular processes regulated by the RelBbu via sRNAs, we categorized the function of the ORFs associated with asRNA and the 5′ UTR sRNAs using biological function gene ontology. Not surprisingly, genes of unknown function were the most common ORF associated with both the upregulated asRNA (Figure 2B) and downregulated 5′ UTR sRNAs (Figure 3C). Interestingly, many asRNAs that were downregulated were opposite genes encoding RNA modification enzymes and other translational machinery (Figure 3B, TL category) and downregulated 5′ UTR sRNAs were associated with rRNA (Figure 3C). Many upregulated 5′ UTR sRNAs were associated with genes encoding translational machinery (Figure 2C, TL category). sRNAs associated with pseudogenes may have important trans-acting functions that provide selection for the maintenance of both the sRNA and pseudogene. These results suggest that sRNAs play a previously unrecognized role in the regulation of rRNA expression or stability, which is the original hallmark of the stringent response and the namesake of the relA gene (relaxed rRNA synthesis). The implications of these initial observations await further investigation.
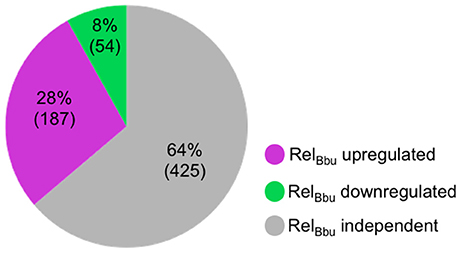
Figure 1. RelBbu-dependent sRNAs identified by RNA-seq comparing wild-type and ΔrelBbu strains shifted from BSKII + RS to RPMI (nutrient stress). sRNA with greater than twofold upregulation (magenta) or downregulation (green) that were significant (EdgeR p-adjusted of <0.05) were enumerated. The number of sRNAs not affected by RelBbu are in gray (RelBbu independent). Analyses are based on the annotated sRNAs identified by Popitsch et al. (2017), excluding intraRNAs.
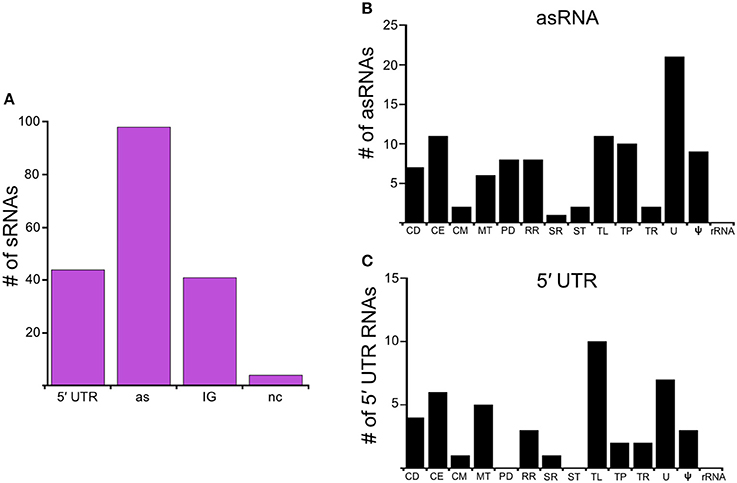
Figure 2. RelBbu-upregulated sRNAs. (A) sRNAs are classified according to their genomic context and include: 5′ untranslated region (5′ UTR), antisense (as), intergenic (IG), and non-coding (nc) RNAs. Functional categories of genes with RelBbu-upregulated asRNAs (B) and 5′ UTR sRNAs (C). Functional categories are abbreviated: cell division (CD), cell envelope (CE), cell motility (CM), metabolism (MT), protein degradation (PD), DNA recombination and repair (RR), stress response (SR), signal transduction (ST), translation (TL), transporter (TP), transcription (TR), unknown (U), pseudogene (ψ), and ribosomal RNA (rRNA).
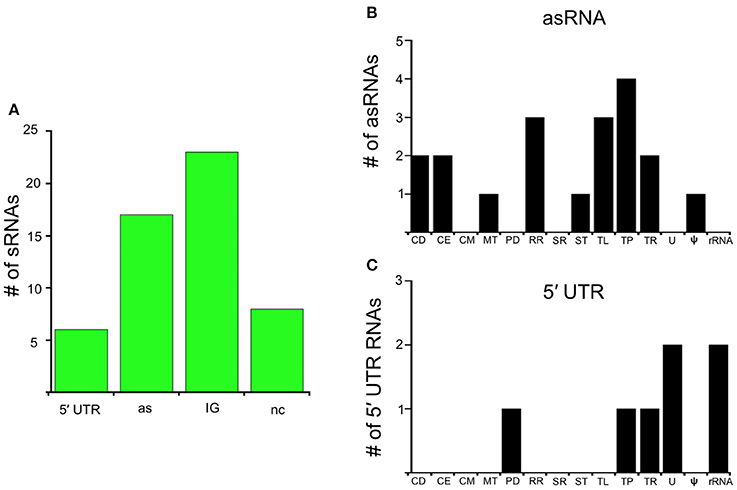
Figure 3. RelBbu-downregulated sRNAs. (A) sRNAs are classified according to their genomic context and include: 5′ untranslated region (5′ UTR), antisense (as), intergenic (IG), and non-coding (nc) RNAs. Functional categories of genes with RelBbu-downregulated asRNAs (B) and 5′ UTR sRNAs (C). Functional categories are abbreviated: cell division (CD), cell envelope (CE), cell motility (CM), metabolism (MT), protein degradation (PD), DNA recombination and repair (RR), stress response (SR), signal transduction (ST), translation (TL), transporter (TP), transcription (TR), unknown (U), pseudogene (ψ), and ribosomal RNA (rRNA).
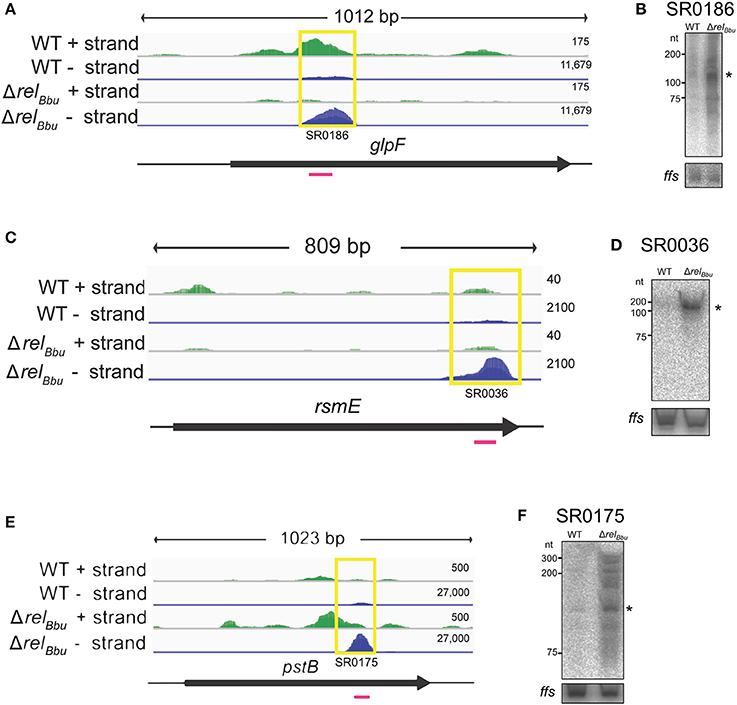
Figure 4. RelBbu-dependent asRNAs. The deep sequencing reads for glpF and glpF asRNA (SR0186) (A), rsmE and rsmE asRNA (SR0036) (C), and pstB and pstB asRNA (SR0175) (E) from both the wild-type and ΔrelBbu strains during nutrient stress are shown in coverage maps with both biological replicates overlaid and displayed in dark and light green (plus strand) and dark and light blue (negative strand). The number associated with each strand represents the normalized number of reads mapped and varies depending on the strand. The corresponding ORFs are shown in black below the RNA-seq coverage maps. The yellow boxes define the position of the called sRNA with the small RNA (SR) number and the magenta lines represent the position of the 32P-labeled oligonucleotide probe used in the Northern blot analyses. Northern blot analyses of total RNA from wild-type and ΔrelBbu strains using 32P-labeled oligonucleotide probes (Table S1) to the asRNA SR0186 (B), asRNA SR0036 (D), and asRNA SR0175 (F). For each Northern blot, ffs (4.5S RNA) was used as a loading control. Asterisks denote the asRNA sizes predicted from RNA-seq analyses.
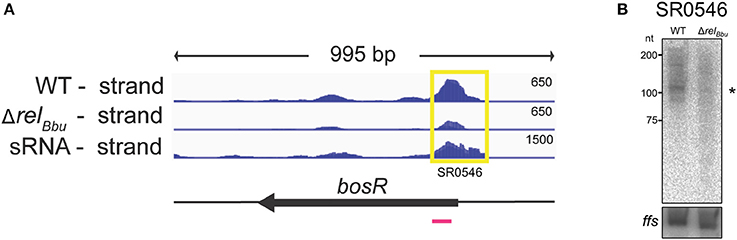
Figure 5. The bosR 5′ UTR sRNA (SR0546) is RelBbu-dependent. (A) The deep sequencing reads from both the wild-type and ΔrelBbu strains during nutrient stress are shown in coverage maps with both biological replicates overlaid in dark and light blue (negative strand); the deep sequencing reads from the sRNA coverage map are from Popitsch et al. (2017). The number associated with each strand represents the normalized number of reads and varies depending on the library. The corresponding ORF is shown in black below the RNA-seq coverage maps. The yellow box defines the position of the called sRNA with the SR number and the magenta line represents the position of the 32P-labeled oligonucleotide probe used in the Northern blot analysis. (B) Northern blot analysis of total RNA from wild-type and ΔrelBbu strains using 32P-labeled oligonucleotide probes (Table S1) to the asRNA SR0546 and ffs (4.5S RNA) as a loading control. Asterisk denotes the sRNA size predicted from RNA-seq analyses.
cis-Encoded 5′ UTR sRNAs and asRNAs
5′ UTR sRNAs and asRNAs likely regulate their associated mRNAs, and the mechanism of regulation can be gleaned from comparing their respective RelBbu-dependent regulation patterns. The regulatory response, either upregulated or downregulated, of RelBbu -dependent 5′ UTR sRNAs and asRNAs was compared to the regulatory response of their cognate mRNAs we previously reported (Drecktrah et al., 2015). The number of sRNAs whose cognate mRNA were affected in the same direction, either both upregulated or both downregulated, were designated co-regulated (Figure S1, blue bars), those oppositely affected were termed inversely regulated (Figure S1, orange bars), and the cognate mRNAs that were not RelBbu-dependent were categorized as not changed (Figure S1, gray bars). Only a single 5′ UTR sRNA was inversely regulated with its cognate mRNA while 19 5′ UTR sRNAs were co-regulated with their associated mRNAs (Figure S1). An equal number of asRNA were co-regulated (15/116) and inversely regulated (15/116) with their associated mRNAs (Figure S1). Most mRNAs associated with the RelBbu -regulated 5′ UTR sRNAs and asRNAs were unchanged by RelBbu (Figure S1, gray bars); these sRNAs may act in trans or may regulate the translation rather than the stability of their mRNA targets. Some 5′ UTR sRNAs may be the termination products of transcriptional riboswitches (Breaker, 2012; Serganov and Nudler, 2013; Sherwood and Henkin, 2016) and, in fact, a ppGpp-sensing riboswitch was recently discovered (Sherlock et al., 2018), suggesting that B. burgdorferi may possess a direct RNA-mediated mechanism for regulating gene expression via the stringent response.
RelBbu Regulates glpF asRNA
RelBbu regulates an asRNA associated with the glycerol metabolic (glp) operon. Previously, we found that RelBbu regulates the glp operon, upregulating expression of the glycerol uptake facilitator (glpF) and glycerol kinase (glpK), while downregulating glycerol 3-phosphate dehydrogenase (glpD) (Drecktrah et al., 2015). Our analyses of the RelBbu-regulated sRNA transcriptome revealed the asRNA SR0186 opposite to glpF was downregulated more than threefold by RelBbu during nutrient stress (Figure 4A). These results were confirmed by Northern blot analyses (Figure 4B). The glp operon encodes gene products responsible for the import and metabolism of glycerol and has been shown to be important for persistence in the tick, where glycerol is thought to be a key carbon source for B. burgdorferi (He et al., 2011; Pappas et al., 2011; Corona and Schwartz, 2015; Caimano et al., 2016). Notably, the glp operon is a focal point of several B. burgdorferi signaling systems, including c-di-GMP, the alternative sigma factor RpoS and the transcriptional activator BosR, which all regulate glp transcription in response to environmental signals (Hyde et al., 2006; He et al., 2011; Grove et al., 2017). glpF asRNA is a highly expressed sRNA whose levels greatly exceed that of glpF mRNA (Figure 4A). The levels of SR0186 asRNA and glpF transcript are reciprocally regulated by RelBbu, suggesting a mechanism in which sense-antisense pairing destabilizes the glpF transcript and controls glycerol utilization during the tick phase of the enzootic cycle.
RelBbu Regulates sRNAs Associated With rRNA Methyltransferase Genes
rRNAs and tRNAs in the cell are extensively post-transcriptionally modified. RelBbu regulates multiple sRNAs associated with genes encoding RNA modifying enzymes. The asRNA SR0036 is opposite rsmE (bb0062), which encodes a methyltransferase, and the asRNA SR0081 is associated with bb0129, which is predicted to encode an rRNA methyltransferase. SR0036 was downregulated by RelBbu during nutrient stress (Figures 4C,D), while rsmE mRNA was upregulated at stationary phase (Drecktrah et al., 2015), suggesting that the antisense-sense pairing destabilizes the rsmE transcript. SR0081 was RelBbu-upregulated threefold during nutrient stress, although its cognate mRNA (bb0129) levels were not significantly changed (Drecktrah et al., 2015). In addition, 5′ UTR sRNAs associated with rRNA methyltransferases were targeted by RelBbu: the sRNA SR0033 is associated with bb0052, which encodes an rRNA/tRNA methyltransferase, and was RelBbu-upregulated (more than eightfold) during nutrient stress as was the bb0052 mRNA (more than twofold) (Drecktrah et al., 2015), suggesting that the antisense-sense pairing stabilizes bb0052 mRNA. The 5′ UTR sRNA SR0226 was RelBbu-upregulated about fourfold, but there was no significant change in mRNA levels of the associated gene rrmJ (bb0313), which is predicted to encode a large subunit rRNA methyltransferase (Fraser et al., 1997). Studies in other organisms have revealed a role for rRNA methylation in ribosome biogenesis and ribosome stability/function (Nachtergaele and He, 2017; Sergiev et al., 2018). rRNA methylation in B. burgdorferi has not been studied, but our results identifying numerous RelBbu-regulated sRNAs associated with the genes encoding these modifying enzymes suggest the stringent response controls rRNA modification along with the canonical reduction in rRNA expression.
RelBbu Regulates pstB asRNA
The stringent response in B. burgdorferi also targets two asRNAs linked to genes predicted to encode the high-affinity inorganic phosphate transport (pst) operon (Fraser et al., 1997). The first gene, pstS (bb0215), encodes a substrate-binding protein that was shown to specifically bind inorganic phosphate (Brautigam et al., 2014). The next genes are pstC (bb0216) and pstA (bb0217), predicted to encode the permeases, followed by pstB (bb0218), predicted to encode the nucleotide-binding protein of the phosphate ABC transporter (Fraser et al., 1997). The pstB asRNA SR0175 was RelBbu-downregulated (more than sixfold) under nutrient stress (Figures 4E,F) and pstB mRNA followed suit (Drecktrah et al., 2015). Another sRNA, pstA asRNA SR0171, was RelBbu-upregulated while pstA transcript was RelBbu-downregulated (Drecktrah et al., 2015). Thus, our results suggest the pstB antisense-sense pairing stabilizes pstB mRNA while the pstA pairing destabilizes its mRNA. In E. coli, expression of the pst operon is regulated by the two-component PhoR/PhoB system, as part of the Pho regulon, and is induced in response to inorganic phosphate starvation. The B. burgdorferi genome does not contain an annotated PhoR/PhoB two-component regulatory system (Fraser et al., 1997; Galperin, 2010). We propose that the B. burgdorferi pst operon, and potentially phosphate import, is regulated by the RelBbu-dependent sense-antisense pairing, at least under our nutrient stress conditions. At first, downregulation of the phosphate transport system during nutrient stress seems counterintuitive, but our nutrient stress conditions shift B. burgdorferi from normal growth medium (BSK + RS) to RPMI, which actually increases inorganic phosphate concentrations more than sixfold. Thus, in our model, nutrient stress conditions, which have increased inorganic phosphate levels, signal RelBbu-dependent downregulation of pstB and pstA transcript, potentially via asRNA regulation, presumably resulting in lower PstB and PstA protein levels and less phosphate transport. pstB asRNA and pstB mRNA are not regulated by temperature (Popitsch et al., 2017), c-di-GMP (Caimano et al., 2015), the transcriptional regulator BadR (Miller et al., 2013), or growth phase (Arnold et al., 2016).
RelBbu Regulates the 5′ UTR sRNA of bosR
The 5′ UTR sRNA SR0546, which is associated with the gene encoding the transcriptional regulator BosR, was RelBbu-upregulated (greater than fourfold) during nutrient stress in the RNA-seq data set and validated by Northern blot analyses (Figures 5A,B). BosR is a Fur/PerR homolog that serves as a transcriptional regulator activating the RpoN-RpoS sigma factor cascade that controls the synthesis of a number of host-induced lipoproteins (Boylan et al., 2003; Hyde et al., 2009; Ouyang et al., 2009; Samuels and Radolf, 2009; Katona, 2015). Additionally, BosR has been shown to directly repress expression of genes encoding the tick-phase-specific lipoproteins OspA and OspD during mammalian infection (Wang et al., 2013; Shi et al., 2014). bosR expression is regulated not only by RelBbu (Drecktrah et al., 2015), but also by the XylR/NagC homolog BadR (Miller et al., 2013; Ouyang and Zhou, 2015) and by its own protein product (Ouyang et al., 2016). The RelBbu-mediated upregulation of both the bosR 5′ UTR sRNA and bosR mRNA (each greater than fourfold) during nutrient stress suggests yet another layer of regulation enhancing bosR transcription or stability. The mechanistic relationship between the bosR 5′ UTR sRNA and mRNA, and how it may affect BosR function, await further investigation.
RelBbu Regulates DsrA and 6S RNA
The stringent response also upregulated two annotated sRNAs in B. burgdorferi: DsrA (2.6-fold) and 6S RNA (3.3-fold). DsrA, the first regulatory sRNA identified in B. burgdorferi, post-transcriptionally regulates the temperature-dependent production of RpoS, the sigma factor that activates genes required for mammalian infection (Lybecker and Samuels, 2007). Interestingly, rpoS mRNA levels were upregulated by RelBbu in stationary phase, but not during nutrient stress (Drecktrah et al., 2015). Note that DsrA, like other trans-encoded sRNAs, may have more than one target RNA.
6S RNA is a widely conserved noncoding RNA that forms a secondary structure mimicking an open promoter complex and binds RNAP, resulting in global transcriptional changes (Wassarman and Storz, 2000; Trotochaud and Wassarman, 2005). 6S RNAs from many bacteria accumulate in stationary growth phase and facilitate the shift from RpoD-dependent to RpoS-dependent promoter gene expression to adapt cells to stationary-phase stresses (Wassarman and Storz, 2000). The predicted B. burgdorferi 6S RNA (Bb6S) was identified in a bioinformatic analysis (Barrick et al., 2005) and visualized in RNA-seq studies (Arnold et al., 2016; Popitsch et al., 2017), but its function and regulation were unknown. We found that Bb6S was RelBbu-upregulated during nutrient stress. This is consistent with the observation that the stringent response increases 6S RNA levels in the soil bacterium Rhizobium etli (Vercruysse et al., 2011). Additionally, 6S RNA was found to regulate expression of relA and (p)ppGpp levels in E. coli (Cavanagh et al., 2010). Thus, our findings augment evidence that the two major transcriptional regulators targeting sigma factor selectivity coordinate their activities to influence global gene expression. The mechanism by which the stringent response alters Bb6S levels remains to be determined, but likely affects transcription, stability or processing/degradation.
RelBbu Regulates asRNAs Associated With Signaling Systems
Nucleotide second messengers have central roles in bacterial signaling systems (Pesavento and Hengge, 2009; Kalia et al., 2013; Fahmi et al., 2017; Nelson and Breaker, 2017; Hall and Lee, 2018), and B. burgdorferi is no exception (Novak et al., 2014; Caimano et al., 2016; Samuels and Samuels, 2016). Besides for (p)ppGpp and c-di-GMP, cyclic dimeric AMP (c-di-AMP) was recently recognized as an intracellular signal in B. burgdorferi (Ye et al., 2014; Savage et al., 2015). DhhP is the essential phosphodiesterase that degrades c-di-AMP, and is required for activation of RpoS (Ye et al., 2014), and CdaA putatively synthesizes c-di-AMP (Savage et al., 2015). SR0008, an asRNA opposite of cdaA, is RelBbu-upregulated sixfold (Table S2) and SR0500, an asRNA opposite of dhhP, is RelBbu-downregulated sixfold (Table S3), which implies a role for the stringent response in regulating c-di-AMP levels via an antisense-sense mechanism in B. burgdorferi, although the specific function of c-di-AMP in the spirochete is as of now obscure.
c-di-GMP, which regulates genes required in the tick vector and during enzootic cycle transitions (Novak et al., 2014; Caimano et al., 2016), is synthesized by the diguanlyate cyclase Rrp1(Rogers et al., 2009; He et al., 2011; Sze et al., 2013; Caimano et al., 2015). Rrp1, a two-component response regulator, is activated by its cognate histidine kinase Hk1 (Caimano et al., 2011; Bauer et al., 2015). SR0315 is an asRNA opposite of hk1 and up-regulated sixfold by RelBbu (Table S2). Although the regulatory mechanism has not yet been deciphered, this finding suggests an interaction between the two guanine nucleotide signaling systems as previously hypothesized (Drecktrah et al., 2015).
Perspectives and Conclusions
Only two other (p)ppGpp-dependent (relA/spoT or rel mutant) bacterial sRNA transcriptomes have been published to our knowledge: Salmonella enterica serovar Typhimurium (Ramachandran et al., 2012, 2014; Amin et al., 2016) and the soil bacterium Rhizobium etli (Vercruysse et al., 2011). In Salmonella, there appear to be more sRNAs downregulated by (p)ppGpp than upregulated, in contrast to B. burgdorferi where there are three times more sRNAs upregulated than downregulated by (p)ppGpp. However, these results may depend on the type of environmental conditions inducing the stringent response. Stringent response-regulated sRNAs in Salmonella were associated with genes involved in porin expression and resistance to antibiotics (Ramachandran et al., 2014). In R. etli, 28 sRNAs were found to be (p)ppGpp-upregulated and five (p)ppGpp-downregulated, with the majority being novel, uncharacterized sRNAs (Vercruysse et al., 2011). Notably, 6S RNA levels were targeted by the stringent response. 6S RNA levels were (p)ppGpp-upregulated in R. etli as we found in B. burgdorferi, but 6S RNA was downregulated in Salmonella (Ramachandran et al., 2014).
The stringent response may not only modulate sRNA levels, but also target RNA-binding proteins, such as the RNA chaperone Hfq, to affect mRNA levels. In E. coli, the RelA-mediated stringent response stimulated oligomerization of Hfq monomers to affect the function of sRNAs (Argaman et al., 2012). An unusual homolog of Hfq was identified in B. burgdorferi and shown to play a role in host infectivity (Lybecker et al., 2010), but the effect of RelBbu on Hfq activity has not yet been examined.
Post-transcriptional sRNA-mediated regulation of mRNA is now established as an important facet of control of gene expression. B. burgdorferi has an impressive number of sRNAs that may be paramount in gene regulation to compensate for the dearth of transcriptional activators and repressors as well as two-component systems (Arnold et al., 2016; Adams et al., 2017; Lybecker and Samuels, 2017; Popitsch et al., 2017). Our results demonstrate that the RelBbu-induced stringent response controls the levels of a third of all known sRNAs (not including intraRNAs). The mechanisms of sRNA regulation and the functional significance of RelBbu-regulated sRNAs in vertebrate infectivity and persistence in the tick provide an exciting and potentially fruitful avenue for dissecting gene regulation.
Data Availability Statement
All sequence files are available from the National Center for Biotechnology Information Sequence Read Archive database (accession numbers SRX971832 and SRX971835).
Author Contributions
DD, ML, and DSS conceived and designed the study. DD, LH, and ML performed the experiments. PR performed the bioinformatics analyses. DD, PR, ML, and DSS interpreted the results. DD wrote the first draft of the manuscript. DD, PR, ML, and DSS wrote sections of the manuscript. All authors read and approved the submitted version.
Funding
This work was supported by the National Institute of Allergy and Infectious Disease grant R01 AI051486 to DSS.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Melissa Caimano and Chris Li for the invitation to contribute our work and for their patience.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2018.00231/full#supplementary-material
Figure S1. Correlation of RelBbu-dependent sRNAs and cognate mRNAs. The mRNAs associated with RelBbu-dependent 5′ UTR sRNAs or asRNAs were classified as co-regulated (blue bar), inversely regulated (orange bar), or not changed (gray bars) in relation to the sRNA. The correlation was performed using the RelBbu-regulated mRNA transcriptome from Drecktrah et al. (2015).
Figure S2. The murJ asRNA (SR0688) is RelBbu-dependent. (A) The deep sequencing reads from both the wild-type and ΔrelBbu strains during nutrient stress are shown in coverage maps with both biological replicates overlaid in dark and light green (plus strand) and dark and light blue (negative strand). The number associated with each strand represents the normalized number of reads mapped and varies depending on the strand. The corresponding ORFs is shown in black below the RNA-seq mapped reads. The yellow box defines the position of the called sRNA with the SR number and the magenta line represents the position of the 32P-labeled oligonucleotide probe used in the Northern blot analyses. (B) Northern blot analyses of total RNA from wild-type and ΔrelBbu strains using a 32P-labeled oligonucleotide probe (Table S1) to the asRNA SR0688 and ffs as a loading control. Asterisk denotes the sRNA size predicted from RNA-seq analyses.
Figure S3. The IG sRNAs SR0725 and SR0735 are RelBbu-dependent. (A,C) The deep sequencing reads from both the wild-type and ΔrelBbu strains during nutrient stress are shown in coverage maps with both biological replicates overlaid in dark and light blue (negative strand). The number associated with each strand represents the normalized number of reads. The corresponding ORFs is shown in black below the RNA-seq coverage maps. The yellow box defines the position of the called sRNA with the SR number and the magenta line represents the position of the 32P-labeled oligonucleotide probe used in the Northern blot analyses. (B,D) Northern blot analyses of total RNA from wild-type and ΔrelBbu strains using a 32P-labeled oligonucleotide probe (Table S1) to the asRNAs SR0725 (B) and SR0735 (D) and ffs as a loading control.
Figure S4. The IG sRNAs SR0961, SR0891, and SR0793 are RelBbu-dependent. (A,C,E) The deep sequencing reads from both the wild-type and ΔrelBbu strains during nutrient stress are shown in coverage maps with both biological replicates overlaid in dark and light green (plus strand). The number associated with each strand represents the normalized number of reads. The corresponding ORFs are shown in black below the RNA-seq coverage maps. The yellow box defines the position of the called sRNA with the SR number and the magenta line represents the position of the 32P-labeled oligonucleotide probes used in the Northern blot analyses. (B,D,F) Northern blot analyses of total RNA from wild-type and ΔrelBbu strains using a 32P-labeled oligonucleotide probe (Table S1) to the asRNAs SR0961 (B), SR0891 (D), and SR0793 (F) and ffs as a loading control. Asterisks denote the sRNA sizes predicted from RNA-seq analyses.
Table S1. List of sRNAs and oligonucleotides used in Northern blot analyses.
Table S2. List of RelBbu-upregulated sRNAs.
Table S3. List of RelBbu-downregulated sRNAs.
References
Adams, P. P., Flores Avile, C., Popitsch, N., Bilusic, I., Schroeder, R., Lybecker, M., et al. (2017). In vivo expression technology and 5′ end mapping of the Borrelia burgdorferi transcriptome identify novel RNAs expressed during mammalian infection. Nucleic Acids Res. 45, 775–792. doi: 10.1093/nar/gkw1180
Amin, S. V., Roberts, J. T., Patterson, D. G., Coley, A. B., Allred, J. A., Denner, J. M., et al. (2016). Novel small RNA (sRNA) landscape of the starvation-stress response transcriptome of Salmonella enterica serovar typhimurium. RNA Biol. 13, 331–342. doi: 10.1080/15476286.2016.1144010
Anacker, M. L., Drecktrah, D., LeCoultre, R. D., Lybecker, M., and Samuels, D. S. (2018). RNase III processing of rRNA in the Lyme disease spirochete Borrelia burgdorferi. J. Bacteriol. 200:e00035-18. doi: 10.1128/JB.00035-18
Argaman, L., Elgrably-Weiss, M., Hershko, T., Vogel, J., and Altuvia, S. (2012). RelA protein stimulates the activity of RyhB small RNA by acting on RNA-binding protein Hfq. Proc. Natl. Acad. Sci. U.S.A. 109, 4621–4626. doi: 10.1073/pnas.1113113109
Arnold, W. K., Savage, C. R., Brissette, C. A., Seshu, J., Livny, J., and Stevenson, B. (2016). RNA-seq of Borrelia burgdorferi in multiple phases of growth reveals insights into the dynamics of gene expression, transcriptome architecture, and noncoding RNAs. PLoS ONE 11:e0164165. doi: 10.1371/journal.pone.0164165
Atkinson, G. C., Tenson, T., and Hauryliuk, V. (2011). The RelA/SpoT homolog (RSH) superfamily: distribution and functional evolution of ppGpp synthetases and hydrolases across the tree of life. PLoS ONE 6:e23479. doi: 10.1371/journal.pone.0023479
Barbour, A. G. (1984). Isolation and cultivation of Lyme disease spirochetes. Yale J. Biol. Med. 57, 521–525.
Barrick, J. E., Sudarsan, N., Weinberg, Z., Ruzzo, W. L., and Breaker, R. R. (2005). 6S RNA is a widespread regulator of eubacterial RNA polymerase that resembles an open promoter. RNA 11, 774–784. doi: 10.1261/rna.7286705
Bauer, W. J., Luthra, A., Zhu, G., Radolf, J. D., Malkowski, M. G., and Caimano, M. J. (2015). Structural characterization and modeling of the Borrelia burgdorferi hybrid histidine kinase Hk1 periplasmic sensor: a system for sensing small molecules associated with tick feeding. J. Struct. Biol. 192, 48–58. doi: 10.1016/j.jsb.2015.08.013
Benach, J. L., Bosler, E. M., Hanrahan, J. P., Coleman, J. L., Bast, T. F., Habicht, G. S., et al. (1983). Spirochetes isolated from the blood of two patients with Lyme disease. N. Engl. J. Med. 308, 740–742. doi: 10.1056/NEJM198303313081302
Boylan, J. A., Posey, J. E., and Gherardini, F. C. (2003). Borrelia oxidative stress response regulator, BosR: a distinctive Zn-dependent transcriptional activator. Proc. Natl. Acad. Sci. U.S.A. 100, 11684–11689. doi: 10.1073/pnas.2032956100
Brautigam, C. A., Ouyang, Z., Deka, R. K., and Norgard, M. V. (2014). Sequence, biophysical, and structural analyses of the PstS lipoprotein (BB0215) from Borrelia burgdorferi reveal a likely binding component of an ABC-type phosphate transporter. Protein Sci. 23, 200–212. doi: 10.1002/pro.2406
Breaker, R. R. (2012). Riboswitches and the RNA world. Cold Spring Harb. Perspect. Biol. 4:a003566. doi: 10.1101/cshperspect.a003566
Brisson, D., Drecktrah, D., Eggers, C. H., and Samuels, D. S. (2012). Genetics of Borrelia burgdorferi. Annu. Rev. Genet. 46, 515–536. doi: 10.1146/annurev-genet-011112-112140
Bugrysheva, J. V., Bryksin, A. V., Godfrey, H. P., and Cabello, F. C. (2005). Borrelia burgdorferi rel is responsible for generation of guanosine-3′-diphosphate-5′-triphosphate and growth control. Infect. Immun. 73, 4972–4981. doi: 10.1128/IAI.73.8.4972-4981.2005
Bugrysheva, J. V., Pappas, C. J., Terekhova, D. A., Iyer, R., Godfrey, H. P., Schwartz, I., et al. (2015). Characterization of the RelBbu regulon in Borrelia burgdorferi reveals modulation of glycerol metabolism by (p)ppGpp. PLoS ONE 10:e0118063. doi: 10.1371/journal.pone.0118063
Burgdorfer, W., Barbour, A. G., Hayes, S. F., Benach, J. L., Grunwaldt, E., and Davis, J. P. (1982). Lyme disease–a tick-borne spirochetosis? Science 216, 1317–1319.
Burtnick, M. N., Downey, J. S., Brett, P. J., Boylan, J. A., Frye, J. G., Hoover, T. R., et al. (2007). Insights into the complex regulation of rpoS in Borrelia burgdorferi. Mol. Microbiol. 65, 277–293. doi: 10.1111/j.1365-2958.2007.05813.x
Caimano, M. J., Drecktrah, D., Kung, F., and Samuels, D. S. (2016). Interaction of the Lyme disease spirochete with its tick vector. Cell. Microbiol. 18, 919–927. doi: 10.1111/cmi.12609
Caimano, M. J., Dunham-Ems, S., Allard, A. M., Cassera, M. B., Kenedy, M., and Radolf, J. D. (2015). Cyclic di-GMP modulates gene expression in Lyme disease spirochetes at the tick-mammal interface to promote spirochete survival during the blood meal and tick-to-mammal transmission. Infect. Immun. 83, 3043–3060. doi: 10.1128/IAI.00315-15
Caimano, M. J., Iyer, R., Eggers, C. H., Gonzalez, C., Morton, E. A., Gilbert, M. A., et al. (2007). Analysis of the RpoS regulon in Borrelia burgdorferi in response to mammalian host signals provides insight into RpoS function during the enzootic cycle. Mol. Microbiol. 65, 1193–1217. doi: 10.1111/j.1365-2958.2007.05860.x
Caimano, M. J., Kenedy, M. R., Kairu, T., Desrosiers, D. C., Harman, M., Dunham-Ems, S., et al. (2011). The hybrid histidine kinase Hk1 is part of a two-component system that is essential for survival of Borrelia burgdorferi in feeding Ixodes scapularis ticks. Infect. Immun. 79, 3117–3130. doi: 10.1128/IAI.05136-11
Caldelari, I., Chao, Y., Romby, P., and Vogel, J. (2013). RNA-mediated regulation in pathogenic bacteria. Cold Spring Harb. Perspect. Med. 3:a010298. doi: 10.1101/cshperspect.a010298
Cavanagh, A. T., Chandrangsu, P., and Wassarman, K. M. (2010). 6S RNA regulation of relA alters ppGpp levels in early stationary phase. Microbiology 156, 3791–3800. doi: 10.1099/mic.0.043992-0
Chatterji, D., and Ojha, A. K. (2001). Revisiting the stringent response, ppGpp and starvation signaling. Curr. Opin. Microbiol. 4, 160–165. doi: 10.1016/S1369-5274(00)00182-X
Concepcion, M. B., and Nelson, D. R. (2003). Expression of spoT in Borrelia burgdorferi during serum starvation. J. Bacteriol. 185, 444–452. doi: 10.1128/JB.185.2.444-452.2003
Corona, A., and Schwartz, I. (2015). Borrelia burgdorferi: carbon metabolism and the tick-mammal enzootic cycle. Microbiol. Spectr. 3:MBP-0011-2014. doi: 10.1128/microbiolspec.MBP-0011-2014
Dalebroux, Z. D., and Swanson, M. S. (2012). ppGpp: magic beyond RNA polymerase. Nat. Rev. Microbiol. 10, 203–212. doi: 10.1038/nrmicro2720
Di, L., Pagan, P. E., Packer, D., Martin, C. L., Akther, S., Ramrattan, G., et al. (2014). BorreliaBase: a phylogeny-centered browser of Borrelia genomes. BMC Bioinformatics 15:233. doi: 10.1186/1471-2105-15-233
Drecktrah, D., Lybecker, M., Popitsch, N., Rescheneder, P., Hall, L. S., and Samuels, D. S. (2015). The Borrelia burgdorferi RelA/SpoT homolog and stringent response regulate survival in the tick vector and global gene expression during starvation. PLoS Pathog. 11:e1005160. doi: 10.1371/journal.ppat.1005160
Dulebohn, D. P., Hayes, B. M., and Rosa, P. A. (2014). Global repression of host-associated genes of the Lyme disease spirochete through post-transcriptional modulation of the alternative sigma factor RpoS. PLoS ONE 9:e93141. doi: 10.1371/journal.pone.0093141
Dunham-Ems, S. M., Caimano, M. J., Eggers, C. H., and Radolf, J. D. (2012). Borrelia burgdorferi requires the alternative sigma factor RpoS for dissemination within the vector during tick-to-mammal transmission. PLoS Pathog. 8:e1002532. doi: 10.1371/journal.ppat.1002532
Fahmi, T., Port, G. C., and Cho, K. H. (2017). c-di-AMP: an essential molecule in the signaling pathways that regulate the viability and virulence of gram-positive bacteria. Genes 8:197. doi: 10.3390/genes8080197
Fisher, M. A., Grimm, D., Henion, A. K., Elias, A. F., Stewart, P. E., Rosa, P. A., et al. (2005). Borrelia burgdorferi σ54 is required for mammalian infection and vector transmission but not for tick colonization. Proc. Natl. Acad. Sci. U.S.A. 102, 5162–5167. doi: 10.1073/pnas.0408536102
Fraser, C. M., Casjens, S., Huang, W. M., Sutton, G. G., Clayton, R., Lathigra, R., et al. (1997). Genomic sequence of a Lyme disease spirochete, Borrelia burgdorferi. Nature 390, 580–586. doi: 10.1038/37551
Galperin, M. Y. (2010). Diversity of structure and function of response regulator output domains. Curr. Opin. Microbiol. 13, 150–159. doi: 10.1016/j.mib.2010.01.005
Georg, J., and Hess, W. R. (2011). cis-antisense RNA, another level of gene regulation in bacteria. Microbiol. Mol. Biol. Rev. 75, 286–300. doi: 10.1128/MMBR.00032-10
Gordon, G. C., Cameron, J. C., and Pfleger, B. F. (2017). RNA sequencing identifies new RNase III cleavage sites in Escherichia coli and reveals increased regulation of mRNA. MBio 8:e00128-17. doi: 10.1128/mBio.00128-17
Grove, A. P., Liveris, D., Iyer, R., Petzke, M., Rudman, J., Caimano, M. J., et al. (2017). Two distinct mechanisms govern RpoS-mediated repression of tick-phase genes during mammalian host adaptation by Borrelia burgdorferi, the Lyme disease spirochete. MBio 8:e01204-17. doi: 10.1128/mBio.01204-17
Hall, C. L., and Lee, V. T. (2018). Cyclic-di-GMP regulation of virulence in bacterial pathogens. Wiley Interdiscip. Rev. RNA 9:e1454. doi: 10.1002/wrna.1454
Hauryliuk, V., Atkinson, G. C., Murakami, K. S., Tenson, T., and Gerdes, K. (2015). Recent functional insights into the role of (p)ppGpp in bacterial physiology. Nat. Rev. Microbiol. 13, 298–309. doi: 10.1038/nrmicro3448
He, M., Ouyang, Z., Troxell, B., Xu, H., Moh, A., Piesman, J., et al. (2011). Cyclic di-GMP is essential for the survival of the Lyme disease spirochete in ticks. PLoS Pathog. 7:e1002133. doi: 10.1371/journal.ppat.1002133
Hör, J., Gorski, S. A., and Vogel, J. (2018). Bacterial RNA biology on a genome scale. Mol. Cell 70, 785–799. doi: 10.1016/j.molcel.2017.12.023
Hübner, A., Yang, X., Nolen, D. M., Popova, T. G., Cabello, F. C., and Norgard, M. V. (2001). Expression of Borrelia burgdorferi OspC and DbpA is controlled by a RpoN-RpoS regulatory pathway. Proc. Natl. Acad. Sci. U.S.A. 98, 12724–12729. doi: 10.1073/pnas.231442498
Hyde, J. A., Seshu, J., and Skare, J. T. (2006). Transcriptional profiling of Borrelia burgdorferi containing a unique bosR allele identifies a putative oxidative stress regulon. Microbiology 152, 2599–2609. doi: 10.1099/mic.0.28996-0
Hyde, J. A., Shaw, D. K., Smith, R. III., Trzeciakowski, J. P., and Skare, J. T. (2009). The BosR regulatory protein of Borrelia burgdorferi interfaces with the RpoS regulatory pathway and modulates both homeostatic and pathogenic properties of the Lyme disease spirochete. Mol. Microbiol. 74, 1344–1355. doi: 10.1111/j.1365-2958.2009.06951.x
Iyer, R., and Schwartz, I. (2016). Microarray-based comparative genomic and transcriptome analysis of Borrelia burgdorferi. Microarrays 5:9. doi: 10.3390/microarrays5020009
Jahn, C. E., Charkowski, A. O., and Willis, D. K. (2008). Evaluation of isolation methods and RNA integrity for bacterial RNA quantitation. J. Microbiol. Methods 75, 318–324. doi: 10.1016/j.mimet.2008.07.004
Jutras, B. L., Jones, G. S., Verma, A., Brown, N. A., Antonicello, A. D., Chenail, A. M., et al. (2013). Posttranscriptional self-regulation by the Lyme disease bacterium's BpuR DNA/RNA-binding protein. J. Bacteriol. 195, 4915–4923. doi: 10.1128/JB.00819-13
Kalia, D., Merey, G., Nakayama, S., Zheng, Y., Zhou, J., Luo, Y., et al. (2013). Nucleotide, c-di-GMP, c-di-AMP, cGMP, cAMP, (p)ppGpp signaling in bacteria and implications in pathogenesis. Chem. Soc. Rev. 42, 305–341. doi: 10.1039/C2CS35206K
Katona, L. I. (2015). The Fur homologue BosR requires Arg39 to activate rpoS transcription in Borrelia burgdorferi and thereby direct spirochaete infection in mice. Microbiology 161, 2243–2255. doi: 10.1099/mic.0.000166
Lane, R. S., Piesman, J., and Burgdorfer, W. (1991). Lyme borreliosis: relation of its causative agent to its vectors and hosts in North America and Europe. Annu. Rev. Entomol. 36, 587–609. doi: 10.1146/annurev.en.36.010191.003103
Liao, Y., Smyth, G. K., and Shi, W. (2014). featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930. doi: 10.1093/bioinformatics/btt656
Lioliou, E., Sharma, C. M., Caldelari, I., Helfer, A. C., Fechter, P., Vandenesch, F., et al. (2012). Global regulatory functions of the Staphylococcus aureus endoribonuclease III in gene expression. PLoS Genet. 8:e1002782. doi: 10.1371/journal.pgen.1002782
Liu, K., Bittner, A. N., and Wang, J. D. (2015). Diversity in (p)ppGpp metabolism and effectors. Curr. Opin. Microbiol. 24, 72–79. doi: 10.1016/j.mib.2015.01.012
Lybecker, M. C., Abel, C. A., Feig, A. L., and Samuels, D. S. (2010). Identification and function of the RNA chaperone Hfq in the Lyme disease spirochete Borrelia burgdorferi. Mol. Microbiol. 78, 622–635. doi: 10.1111/j.1365-2958.2010.07374.x
Lybecker, M. C., and Samuels, D. S. (2007). Temperature-induced regulation of RpoS by a small RNA in Borrelia burgdorferi. Mol. Microbiol. 64, 1075–1089. doi: 10.1111/j.1365-2958.2007.05716.x
Lybecker, M. C., and Samuels, D. S. (2017). Small RNAs of Borrelia burgdorferi: characterizing functional regulators in a sea of sRNAs. Yale J. Biol. Med. 90, 317–323.
Lybecker, M., Bilusic, I., and Raghavan, R. (2014). Pervasive transcription: detecting functional RNAs in bacteria. Transcription 5:e944039. doi: 10.4161/21541272.2014.944039
Miller, C. L., Karna, S. L., and Seshu, J. (2013). Borrelia host adaptation Regulator (BadR) regulates rpoS to modulate host adaptation and virulence factors in Borrelia burgdorferi. Mol. Microbiol. 88, 105–124. doi: 10.1111/mmi.12171
Nachtergaele, S., and He, C. (2017). The emerging biology of RNA post-transcriptional modifications. RNA Biol. 14, 156–163. doi: 10.1080/15476286.2016.1267096
Nelson, J. W., and Breaker, R. R. (2017). The lost language of the RNA world. Sci. Signal. 10:eaam8812. doi: 10.1126/scisignal.aam8812
Novak, E. A., Sultan, S. Z., and Motaleb, M. A. (2014). The cyclic-di-GMP signaling pathway in the Lyme disease spirochete, Borrelia burgdorferi. Front. Cell. Infect. Microbiol. 4:56. doi: 10.3389/fcimb.2014.00056
Ouyang, Z., and Zhou, J. (2015). BadR (BB0693) controls growth phase-dependent induction of rpoS and bosR in Borrelia burgdorferi via recognizing TAAAATAT motifs. Mol. Microbiol. 98, 1147–1167. doi: 10.1111/mmi.13206
Ouyang, Z., Deka, R. K., and Norgard, M. V. (2011). BosR (BB0647) controls the RpoN-RpoS regulatory pathway and virulence expression in Borrelia burgdorferi by a novel DNA-binding mechanism. PLoS Pathog. 7:e1001272. doi: 10.1371/journal.ppat.1001272
Ouyang, Z., Kumar, M., Kariu, T., Haq, S., Goldberg, M., Pal, U., et al. (2009). BosR (BB0647) governs virulence expression in Borrelia burgdorferi. Mol. Microbiol. 74, 1331–1343. doi: 10.1111/j.1365-2958.2009.06945.x
Ouyang, Z., Narasimhan, S., Neelakanta, G., Kumar, M., Pal, U., Fikrig, E., et al. (2012). Activation of the RpoN-RpoS regulatory pathway during the enzootic life cycle of Borrelia burgdorferi. BMC Microbiol. 12:44. doi: 10.1186/1471-2180-12-44
Ouyang, Z., Zhou, J., and Norgard, M. V. (2016). Evidence that BosR (BB0647) Is a positive autoregulator in Borrelia burgdorferi. Infect. Immun. 84, 2566–2574. doi: 10.1128/IAI.00297-16
Pappas, C. J., Iyer, R., Petzke, M. M., Caimano, M. J., Radolf, J. D., and Schwartz, I. (2011). Borrelia burgdorferi requires glycerol for maximum fitness during the tick phase of the enzootic cycle. PLoS Pathog. 7:e1002102. doi: 10.1371/journal.ppat.1002102
Pesavento, C., and Hengge, R. (2009). Bacterial nucleotide-based second messengers. Curr. Opin. Microbiol. 12, 170–176. doi: 10.1016/j.mib.2009.01.007
Piesman, J., and Schwan, T. G. (2010). “Ecology of borreliae and their arthropod vectors,” in Borrelia: Molecular Biology, Host Interaction and Pathogenesis, eds D. S. Samuels and J. D. Radolf (Norfolk: Caister Academic Press), 251–278.
Popitsch, N. (2014). CODOC: efficient access, analysis and compression of depth of coverage signals. Bioinformatics 30, 2676–2677. doi: 10.1093/bioinformatics/btu362
Popitsch, N., Bilusic, I., Rescheneder, P., Schroeder, R., and Lybecker, M. (2017). Temperature-dependent sRNA transcriptome of the Lyme disease spirochete. BMC Genomics 18:28. doi: 10.1186/s12864-016-3398-3
Potrykus, K., and Cashel, M. (2008). (p)ppGpp: still magical? Annu. Rev. Microbiol. 62, 35–51. doi: 10.1146/annurev.micro.62.081307.162903
Quereda, J. J., and Cossart, P. (2017). Regulating bacterial virulence with RNA. Annu. Rev. Microbiol. 71, 263–280. doi: 10.1146/annurev-micro-030117-020335
Radolf, J. D., Caimano, M. J., Stevenson, B., and Hu, L. T. (2012). Of ticks, mice and men: understanding the dual-host lifestyle of Lyme disease spirochaetes. Nat. Rev. Microbiol. 10, 87–99. doi: 10.1038/nrmicro2714
Ramachandran, V. K., Shearer, N., and Thompson, A. (2014). The primary transcriptome of Salmonella enterica serovar Typhimurium and its dependence on ppGpp during late stationary phase. PLoS ONE 9:e92690. doi: 10.1371/journal.pone.0092690
Ramachandran, V. K., Shearer, N., Jacob, J. J., Sharma, C. M., and Thompson, A. (2012). The architecture and ppGpp-dependent expression of the primary transcriptome of Salmonella Typhimurium during invasion gene expression. BMC Genomics 13:25. doi: 10.1186/1471-2164-13-25
Rogers, E. A., Terekhova, D., Zhang, H. M., Hovis, K. M., Schwartz, I., and Marconi, R. T. (2009). Rrp1, a cyclic-di-GMP-producing response regulator, is an important regulator of Borrelia burgdorferi core cellular functions. Mol. Microbiol. 71, 1551–1573. doi: 10.1111/j.1365-2958.2009.06621.x
Samuels, D. S. (2011). Gene regulation in Borrelia burgdorferi. Annu. Rev. Microbiol. 65, 479–499. doi: 10.1146/annurev.micro.112408.134040
Samuels, D. S., and Radolf, J. D. (2009). Who is the BosR around here anyway? Mol. Microbiol. 74, 1295–1299. doi: 10.1111/j.1365-2958.2009.06971.x
Samuels, D. S., and Samuels, L. R. N. (2016). Gene regulation during the enzootic cycle of the Lyme disease spirochete. For. Immunopathol. Dis. Therap. 7, 205–212. doi: 10.1615/ForumImmunDisTher.2017019469
Samuels, D. S., Drecktrah, D., and Hall, L. S. (2018). “Genetic transformation and complementation,” in Borrelia burgdorferi: Methods and Protocols, eds U. Pal and O. Buyuktanir (New York, NY: Humana Press), 183–200. doi: 10.1007/978-1-4939-7383-5_15
Savage, C. R., Arnold, W. K., Gjevre-Nail, A., Koestler, B. J., Bruger, E. L., Barker, J. R., et al. (2015). Intracellular concentrations of Borrelia burgdorferi cyclic di-AMP are not changed by altered expression of the CdaA synthase. PLoS ONE 10:e0125440. doi: 10.1371/journal.pone.0125440
Sedlazeck, F. J., Rescheneder, P., and von Haeseler, A. (2013). NextGenMap: fast and accurate read mapping in highly polymorphic genomes. Bioinformatics 29, 2790–2791. doi: 10.1093/bioinformatics/btt468
Serganov, A., and Nudler, E. (2013). A decade of riboswitches. Cell 152, 17–24. doi: 10.1016/j.cell.2012.12.024
Sergiev, P. V., Aleksashin, N. A., Chugunova, A. A., Polikanov, Y. S., and Dontsova, O. A. (2018). Structural and evolutionary insights into ribosomal RNA methylation. Nat. Chem. Biol. 14, 226–235. doi: 10.1038/nchembio.2569
Sherlock, M. E., Sudarsan, N., and Breaker, R. R. (2018). Riboswitches for the alarmone ppGpp expand the collection of RNA-based signaling systems. Proc. Natl. Acad. Sci. U.S.A. 115, 6052–6057. doi: 10.1073/pnas.1720406115
Sherwood, A. V., and Henkin, T. M. (2016). Riboswitch-mediated gene regulation: novel RNA architectures dictate gene expression responses. Annu. Rev. Microbiol. 70, 361–374. doi: 10.1146/annurev-micro-091014-104306
Shi, Y., Dadhwal, P., Li, X., and Liang, F. T. (2014). BosR functions as a repressor of the ospAB operon in Borrelia burgdorferi. PLoS ONE 9:e109307. doi: 10.1371/journal.pone.0109307
Steere, A. C., Grodzicki, R. L., Kornblatt, A. N., Craft, J. E., Barbour, A. G., Burgdorfer, W., et al. (1983). The spirochetal etiology of Lyme disease. N. Engl. J. Med. 308, 733–740. doi: 10.1056/NEJM198303313081301
Stevenson, B., and Seshu, J. (2017). “Regulation of gene and protein expression in the Lyme disease spirochete,” in Current Topics in Microbiology and Immunology, ed B. Adler (Berlin; Heidelberg: Springer). doi: 10.1007/82_2017_49
Storz, G., Vogel, J., and Wassarman, K. M. (2011). Regulation by small RNAs in bacteria: expanding frontiers. Mol. Cell 43, 880–891. doi: 10.1016/j.molcel.2011.08.022
Sultan, S. Z., Pitzer, J. E., Miller, M. R., and Motaleb, M. A. (2010). Analysis of a Borrelia burgdorferi phosphodiesterase demonstrates a role for cyclic-di-guanosine monophosphate in motility and virulence. Mol. Microbiol. 77, 128–142. doi: 10.1111/j.1365-2958.2010.07191.x
Sze, C. W., Smith, A., Choi, Y. H., Yang, X., Pal, U., Yu, A., et al. (2013). Study of the response regulator Rrp1 reveals its regulatory role in chitobiose utilization and virulence of Borrelia burgdorferi. Infect. Immun. 81, 1775–1787. doi: 10.1128/IAI.00050-13
Thomason, M. K., and Storz, G. (2010). Bacterial antisense RNAs: how many are there, and what are they doing? Annu. Rev. Genet. 44, 167–188. doi: 10.1146/annurev-genet-102209-163523
Trotochaud, A. E., and Wassarman, K. M. (2005). A highly conserved 6S RNA structure is required for regulation of transcription. Nat. Struct. Mol. Biol. 12, 313–319. doi: 10.1038/nsmb917
Troxell, B., and Yang, X. F. (2013). Metal-dependent gene regulation in the causative agent of Lyme disease. Front. Cell. Infect. Microbiol. 3:79. doi: 10.3389/fcimb.2013.00079
Vercruysse, M., Fauvart, M., Jans, A., Beullens, S., Braeken, K., Cloots, L., et al. (2011). Stress response regulators identified through genome-wide transcriptome analysis of the (p)ppGpp-dependent response in Rhizobium etli. Genome Biol. 12:R17. doi: 10.1186/gb-2011-12-2-r17
Wang, P., Dadhwal, P., Cheng, Z., Zianni, M. R., Rikihisa, Y., Liang, F. T., et al. (2013). Borrelia burgdorferi oxidative stress regulator BosR directly represses lipoproteins primarily expressed in the tick during mammalian infection. Mol. Microbiol. 89, 1140–1153. doi: 10.1111/mmi.12337
Wassarman, K. M., and Storz, G. (2000). 6S RNA regulates E. coli RNA polymerase activity. Cell 101, 613–623. doi: 10.1016/S0092-8674(00)80873-9
Waters, L. S., and Storz, G. (2009). Regulatory RNAs in bacteria. Cell 136, 615–628. doi: 10.1016/j.cell.2009.01.043
Yang, X. F., Alani, S. M., and Norgard, M. V. (2003). The response regulator Rrp2 is essential for the expression of major membrane lipoproteins in Borrelia burgdorferi. Proc. Natl. Acad. Sci. U.S.A. 100, 11001–11006. doi: 10.1073/pnas.1834315100
Keywords: Lyme disease, Borrelia burgdorferi, stringent response, small RNA, gene regulation, RNA sequencing, guanosine tetraphosphate and pentaphosphate, spirochetes
Citation: Drecktrah D, Hall LS, Rescheneder P, Lybecker M and Samuels DS (2018) The Stringent Response-Regulated sRNA Transcriptome of Borrelia burgdorferi. Front. Cell. Infect. Microbiol. 8:231. doi: 10.3389/fcimb.2018.00231
Received: 19 April 2018; Accepted: 19 June 2018;
Published: 05 July 2018.
Edited by:
Melissa Jo Caimano, University of Connecticut Health Center, United StatesReviewed by:
Philip E. Stewart, National Institute of Allergy and Infectious Diseases (NIAID), United StatesZhiming Ouyang, University of South Florida, United States
Copyright © 2018 Drecktrah, Hall, Rescheneder, Lybecker and Samuels. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Dan Drecktrah, ZGFuLmRyZWNrdHJhaEBtc28udW10LmVkdQ==
 Dan Drecktrah
Dan Drecktrah Laura S. Hall1
Laura S. Hall1 Meghan Lybecker
Meghan Lybecker D. Scott Samuels
D. Scott Samuels