- 1Department of Laboratory Medicine, Renji Hospital, School of Medicine, Shanghai Jiaotong University, Shanghai, China
- 2Department of Laboratory Medicine, Shanghai Children's Medical Center, Shanghai Jiaotong University School of Medicine, Shanghai, China
- 3Shanghai Institute for Veterinary Drug and Feeds Control, Shanghai, China
Staphylococcus aureus is highly pathogenic and can cause diseases in both humans and domestic animals. In animal species, including ruminants, S. aureus may cause severe or sub-clinical mastitis. This study aimed to investigate the molecular profile, antimicrobial resistance, and genotype/phenotype correlation of 212 S. aureus isolates recovered from cases of bovine mastitis from 2014 to 2015 in the Shanghai and Zhejiang areas of China. Nineteen sequence types (STs) were determined by multi-locus sequence typing, while the dominant ST was ST97, followed by ST520, ST188, ST398, ST7, and ST9. Within 14 methicillin-resistant S. aureus (MRSA) isolates and 198 methicillin-susceptible S. aureus (MSSA) isolates, ST97 was the predominant MSSA clone and ST9-MRSA-SCCmecXII-spa t899 was the most common MRSA clone. The MRSA strains showed much higher rates of resistance to multiple antibiotics than did MSSA strains. Compared with other MSSA strains, MSSA ST398 was more resistant to clindamycin, erythromycin, and ciprofloxacin. No isolates were resistant to vancomycin, teicoplanin, or linezolid. The molecular profiles of the virulence genes varied in different strains. ST520 strains carried seg-sei-sem-sen-seo genes, and ST9 and ST97 harbored sdrD-sdrE genes. Virulence phenotype analysis showed diversity in different clones. Biofilm formation ability was significantly enhanced in ST188 and ST7, and red blood cell lysis capacity was relatively strong in all S. aureus strains of animal origin except ST7. Our results indicate that MSSA was the predominant S. aureus strain causing bovine mastitis in eastern regions of China. However, the presence of multidrug resistant and toxigenic MRSA clone ST9 suggests that comprehensive surveillance of S. aureus infection should be implemented in the management of animal husbandry products.
Introduction
Staphylococcus aureus is a common facultative pathogenic bacterium that has long been recognized as a challenge in both human and veterinary medicine (Nemeghaire et al., 2014). In cattle, it is responsible for approximately one-third of cases of clinical and subclinical mastitis (Bradley et al., 2007; Botrel et al., 2010), a disease that causes major economic loss in the dairy industry worldwide. Since MRSA was first reported in the United Kingdom in 1961, it has become a global cause of hospital-associated (HA) and community-associated (CA) infections (Uhlemann et al., 2014). In the past few decades, a new subgroup of S. aureus, so-called livestock-associated S. aureus (LA-SA) has been described. There have been several reports of MRSA colonization and/or infections in dairy cattle since the first report of MRSA in mastitis in 1972 (Devriese et al., 1972; Monecke et al., 2007; Fessler et al., 2010; Huber et al., 2010). LA-SA ST398 was originally reported to have emerged in the Netherlands among pigs and pig farmers in 2003 (Voss et al., 2005), and was later found in Austria, Germany, and Denmark (Van Cleef et al., 2011; Bal et al., 2016). It developed into an overwhelmingly dominant lineage in Europe and North America (Vanderhaeghen et al., 2010; Cuny et al., 2013). Unlike those in Europe and North America, the epidemic LA MRSA clones in Asian countries have their own characteristics. LA MRSA ST9 has been predominantly isolated from pigs in China, Hong Kong, Taiwan, Thailand, and Malaysia. LA MRSA ST221 has been reported in Japan, and LA MRSA ST398 has emerged among pigs in Singapore and South Korea (Cui et al., 2009; Guardabassi et al., 2009; Neela et al., 2009; Baba et al., 2010; Ho J. et al., 2012; Larsen et al., 2012). Although the sequence type of LA MRSA isolates was mainly ST9 in most Asian countries, these isolates harbored different SCCmec elements in different areas, such as SCCmec III in China, SCCmec IVb, or V in Hong Kong, SCCmec V in Malaysia, SCCmec IX in Thailand and non-types I–VII in Taiwan (Larsen et al., 2012; Fang et al., 2014; Chuang and Huang, 2015). ST9 strains were usually associated with the MDR phenotype, with high resistance rates (>80%) to erythromycin, clindamycin, tetracycline, ciprofloxacin and gentamicin (Wan et al., 2013). To date, most studies on LA S. aureus have been performed in pigs, mainly with MRSA strains, even though the vast majority of S. aureus strains are host-specific. The data on molecular typing and antibiotic-related studies of bovine derived S. aureus are very limited. A relatively higher incidence of S. aureus mastitis has been reported in China than in other countries, which may cause a particular public threat (Li et al., 2011; He et al., 2014). According to previous studies performed in six Chinese provinces from March 2010 to August 2013, the most popular S. aureus clones causing bovine mastitis were primarily ST97 (51.9%), ST398 (13.6%) and ST2154 (8.6%), with ST97-MRSA-SCCmec IV representing the most common clone (50%; Wang et al., 2015). However, the characteristics of S. aureus from dairy cows in eastern regions of China have not yet been thoroughly discussed. To provide elementary evidence for developing the appropriate treatment and control measures for bovine mastitis, it is essential to understand the molecular epidemiology and the antibiotic resistance of S. aureus infections locally. Therefore, the present study was designed to analyze the molecular characteristics of S. aureus strains in bovine mastitis isolated from 2014 to 2015 in the Shanghai and Zhejiang areas.
Materials and Methods
Sample Collection and Bacterial Isolation
In total, 212 S. aureus isolates were isolated from dairy cows with mastitis from 2014 to 2015 in farms in the Shanghai and Zhejiang areas of China. Milk samples were taken from cows with clinical mastitis that had symptoms including color change in the milk, inflammation of the udder, and decreased milk production. For milk collection, the udders of the clinical mastitis cows were cleaned with water and dried. Cotton balls with 70% ethanol were used to disinfect the surfaces of the udder. The first few streams of milk were dropped. The collected milk was kept in a cooler with ice and transported to the laboratory within 8 h. The milk samples were cultured on 5% blood plate and inoculated at 37°C for 24 h. S. aureus identification was based on Gram staining, classical microbiological tests, including catalase and coagulase activity, and were further characterized using VITEK 2 Compact GP ID Card (bioMérieux, Marcy l'Etoile, France). S. aureus ATCC43300 was used as a quality control organism. All S. aureus isolates were stored at −80°C.
DNA Extraction, MRSA Identification, spa Typing, and SCCmec Typing
DNA was extracted as previously described (Hartmann et al., 1997). MRSA identification and mecA gene detection were performed using a previously published triplex PCR (Maes et al., 2002). The strains were further characterized by spa typing, as previously described (Harmsen et al., 2003). The resulting spa type was assigned by submitting the data to the S. aureus spa type database (http://www.spaserver.ridom.de). Staphylococcal cassette chromosome mec (SCCmec) types were determined by means of two multiplex PCRs (M-PCRs) designed for the detection of the mec-complex and the ccr-complex (Kondo et al., 2007; Yan et al., 2016). Specific primers, as previously described (Wu et al., 2015) was used in isolates with SCCmec types that could not be determined using the SCCmec typing strategy as previously described (Kondo et al., 2007).
MLST Typing and goeBURST Algorithm
Isolates were screened using a previously described method (Enright and Spratt, 1999) to detect the following seven housekeeping genes: carbamate kinase (arcC), shikimate dehydrogenase (aroE), glycerol kinase (glp), guanylate kinase (gmk), phosphate acetyltransferase (pta), triosephosphate isomerase (tpi), and acetyl coenzyme A acetyltransferase (yqiL). The sequences of the PCR products were compared with the existing sequences available from the MLST website (http://www.mlst.net) for S. aureus, and the allelic number was determined for each sequence. The goeBURST algorithm (http://goeBURST.phyloviz.net) was used to infer the evolutionary relatedness of the STs.
PFGE Typing
PFGE was used to compare the genetic diversity of the 10 isolates of ST9 (one isolate of MSSA ST9 and nine isolates of MRSA ST9). Briefly, Sma I-digested DNA embedded in agarose plugs was subjected to PFGE analysis at 14°C in a CHEF-MAPPER system (Bio-Rad) at 6 V/cm, in 0.5 × Tris-borate-EDTA buffer, in two stages: first stage, initial pulse, 5 s; final pulse, 15 s for 10 h; second stage, initial pulse, 15 s, final pulse, 60 s for 10 h; angle 120°.
Antimicrobial Susceptibility Testing
The standard disk diffusion method was used to test the antibiotic susceptibility of all isolates, and the results were interpreted in accordance with the Clinical and Laboratory Standards Institute (CLSI) guidelines (Ceriotti et al., 2012). Antibiogram classifications were made on the basis of susceptibility to 13 antimicrobials: gentamycin (CN), cefazolin (KZ), cefuroximesodium (CXM), oxacillin (OX), sulfamethoxazole + trimethoprim (SXT), penicillin (P), clindamycin (DA), erythromycin (E), cefoxitin (FOX), ciprofloxacin (CIP), teicoplanin (TEC), vancomycin (VA), and linezolid (LZD).
Detection of Virulence Genes
PCR amplification for virulence genes was performed in the dominant STs (the number was >10) for the following 30 staphylococcal virulence genes: the staphylococcal enterotoxin genes (sea, seb, sec, sed, see, seg, seh, sei, sem, sen, seo, seq, sek), the toxic shock syndrome toxin (tsst), the arginine catabolic mobile gene (arcA), the exfoliative toxin genes (eta, etb), leukotoxin (lukF/S, lukE, lukM; Lina and Etienne, 1999), the hemolysin genes (hla, hlb, hld, hlg), the serine protease (sspA) and the adhesion genes (clfA, icaA, fnbA, sdrC, and sdrE) as previously described (Arvidson and Tegmark, 2001; Peacock et al., 2002; Wardenburg et al., 2007). The amplification was carried out on a GeneAmp 9700 thermal cycler (Applied Biosystems, NY, USA) under the following conditions: an initial 5 min denaturation at 94°C, followed by 35 cycles of 30 s at 94°C, 30 s at 55°C, and 30 s at 72°C, with a final extension at 72°C for 7 min. In each PCR, a positive control and a negative control (distilled water) were included. The PCR fragments were visualized by agarose gel electrophoresis and ethidium bromide staining.
Semiquantitative Biofilm Assay
A semiquantitative biofilm assay was performed using 96-well tissue culture plates based on the method described by Wang et al. (Wang and Gao, 2008) with the following modification. After the cells were fixed in Bouin's fixative for 1 h, the cells were washed gently two times in phosphate-buffered saline and then stained with 0.1% crystal violet solution. The stain was washed off gently under slowly running water and the plates were dried at room temperature. The absorbance of the stained biofilm was measured at 560 nm using Synergy2 (Bio-Tek).
Human Erythrocyte Lysis Assay
Human blood was obtained from healthy people during physical examinations in Renji Hospital, Shanghai Jiao Tong University School of Medicine, Shanghai, China with ethical consent. Supernatants were collected from bacterial cultures grown for 8 h. Hemolytic activities were determined by incubating samples with human erythrocytes (2% v/v in Dulbecco's phosphate buffered saline, PBS) for 1 h at 37°C. Hemolysis was determined by measuring the optical density at 540 nm using an enzyme-linked immunosorbent assay reader.
Statistical Analysis
Statistical analyses were performed using Stata software (version10.1/SE, Stata Corp., College Station, TX, USA). We used the χ2 and Fisher's exact tests, as appropriate for the analysis of categorical data. Unpaired two-tailed Student's t-tests were performed to analyze the statistical significance of biofilm formation and human erythrocytes lysis in S. aureus strains. P < 0.05 was considered statistically significant.
Results
MLST, spa, SCCmec Types
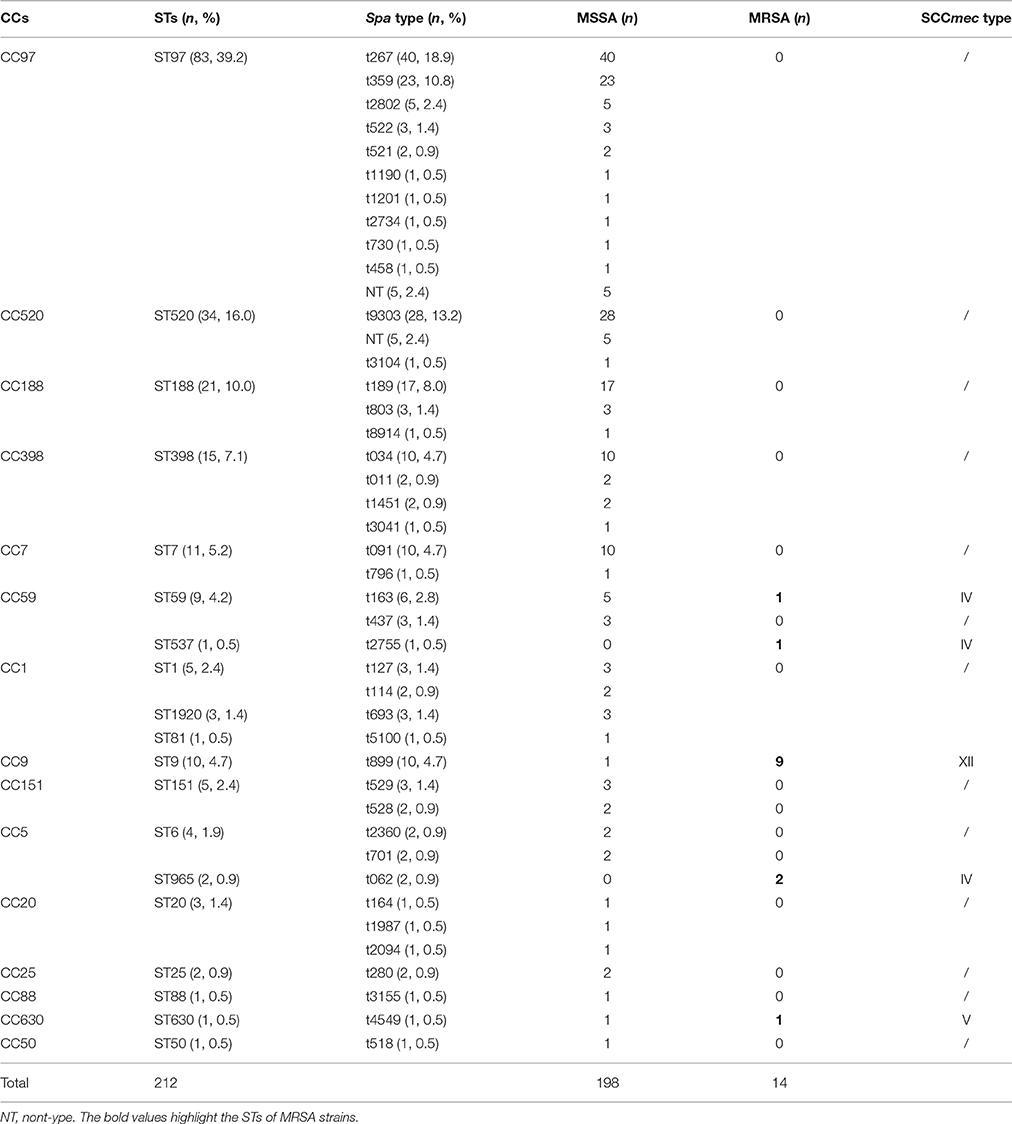
In total, 212 S. aureus isolates from bovine mastitis were collected in this study, including 198 (93.4%) MSSA and 14 (6.6%) MRSA isolates. Information associated with the isolation and identification of MSSA, MRSA, spa types, MLST, and SCCmec types is shown in Table 1. Fourteen isolates were confirmed as MRSA by the cefoxitin disc diffusion test and as mecA-positive by PCR. Nineteen distinct STs were identified within the 212 isolates, with ST97 as the most frequently represented isolate (39.2%, 83/212), accounting for more than one third of all S. aureus isolates, followed by ST520 (16%, 34/212), ST188 (10%, 21/212), ST398 (7.1%, 15/212), ST7 (5.2%, 11/212), and ST9 (4.7%, 10/212). The spa typing discriminated 212 S. aureus isolates into 42 spa types. Among these, spa t267 was the most common type (18.9%, 40/212), followed by t9303 (13.2%, 28/212), t359 (10.8%, 23/212), t189 (8.0%, 17/212), t034 (4.7%, 10/212), t091 (4.7%, 10/212), t899 (4.7%, 10/212), and t163 (2.8%, 6/212). Each of the remaining spa types had no more than five isolates. By SCCmec typing, only three types (types IV, V, and XII) were found among 14 MRSA isolates. Type XII was the most predominant type XII and was found in nine isolates (64.3%, 9/14). Type IV was found in four isolates (28.6%, 4/14), and type V was found in one isolate (7.1%, 1/14).
The eBURST analysis of bovine-associated S. aureus using all STs available in the MLST database is shown. A population scatter diagram of the bovine-associated isolates shows nine main primary groups (Figure 1). The eBURST algorithm clustered all 19 STs isolated from bovine mastitis into 14 clonal complexes, as follows, CC1, CC9, CC97, CC25, CC188, CC630, CC398, CC50, CC151, CC7, CC20, CC59, CC88, and CC5. Among these complexes, CC97 had 83 isolates, CC520 had 34 isolates, CC188 had 21 isolates, CC398 had 15 isolates, CC7 had 11 isolates, CC59 had 10 isolates, and CC9 had 10 isolates. Each of other CCs had <10 isolates.
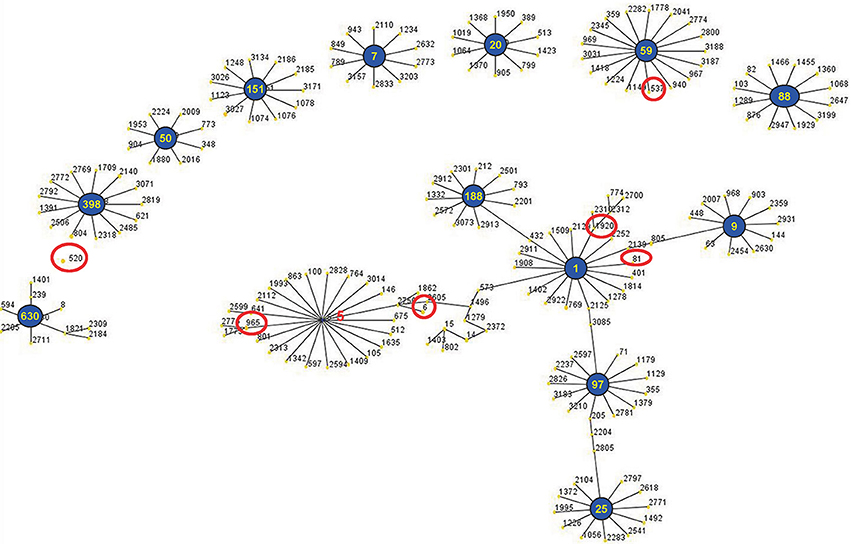
Figure 1. Application of the eBURST algorithm to MLST data for the collection of 212 S. aureus isolates. All 19 STs are represented by a filled circle. Blue and red circles represent STs that are group and sub-group founders, respectively. CC includes the groups of connected STs, considering that STs have at least six alleles in common with at least one other ST inside a CC.
Antimicrobial Susceptibility Profiles
The results of antibiotic susceptibility testing showed that all of the isolates were susceptible to vancomycin, teicoplanin, and linezolid. Although the majority of the strains were resistant to penicillin (73.1%), they were still susceptible to most of the antibiotics tested. The resistance rates to other antibiotics tested were 31.6% to gentamicin, 23.6% to erythromycin, 16.5% to clindamycin, 11.8% to ciprofloxacin, 6.6% to cefoxitin, or oxacillin, 4.7% to cefuroxime, 4.2% to sulfamethoxazole/trimethoprim, and 3.8% to cefazolin (Table 2).
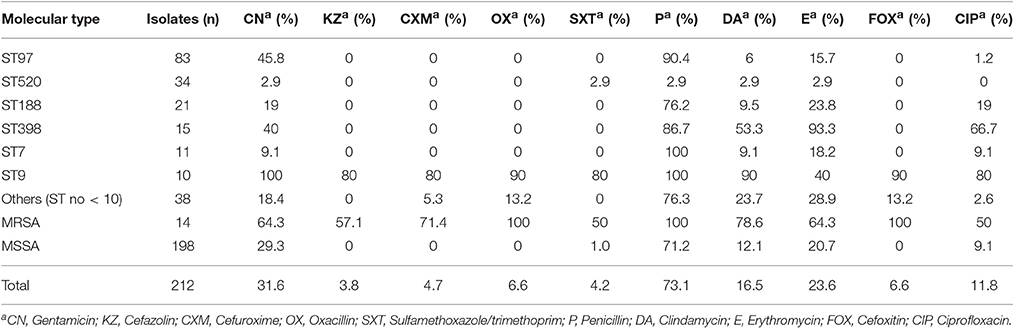
Table 2. Antimicrobial susceptibility profiles of S. aureus causing bovine mastitis in eastern regions of China arranged by STs.
Among the 212 S. aureus isolates, 50 (23.6%) strains were resistant to ≥3 antibiotics, including 14 (6.6%) MRSA and 36 (17.0%) MSSA strains. Nineteen (9.0%) isolates were resistant to five or more antibiotics, seventeen (8.0%) were resistant to four antibiotics and fourteen (6.6%) were resistant to three antibiotics. In the MSSA strains, 36 (17.0%) strains were resistant to ≥3 antibiotics, 22 (10.4%) strains showed resistance to ≥4 antibiotics, and 7 (3.3%) strains were resistant to ≥5 antibiotics. However, all the MRSA strains were found to be resistant to at least four antibiotics.
The resistance profiles of S. aureus isolates differed by their STs (Table 2). ST9 strains displayed the MDR phenotype, with high resistance (≥80%) to nine of the antibiotics tested except erythromycin (40%). ST398 isolates showed a higher resistance rate to erythromycin (93.3%), penicillin (86.7%), and ciprofloxacin (66.7%). ST97, ST188, and ST7 isolates were more resistant to penicillin (90.4, 76.2, and 100%, respectively) than to other antibiotics. Among 34 ST520 strains, only nine were resistant to one or two types of antibiotics, and the other ST520 isolates were susceptible to all antibiotics tested.
Detection of Virulence Genes
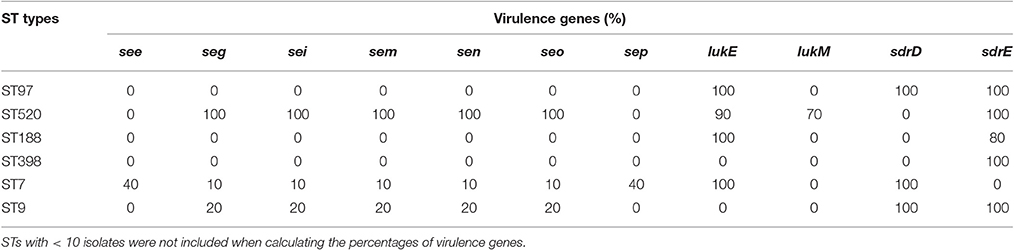
In the present study, 30 virulence genes were screened in six predominant STs. The distribution of virulence genes differed among the strains according to ST. Some genes were present in all of the strains, but some genes were not found in any strain. For example, all of the strains carried four hemolysin genes (hla, hlb, hld, hlg), one serine protease gene (sspA), and three adhesion genes (icaA, clfA, fnbA), but none of the strains harbored tst, arcA, eta, etb, or pvl. The carriage of staphylococcal enterotoxin genes was strongly influenced by MLST profiles. Thirteen staphylococcal enterotoxin genes (sea, seb, sec, sed, see, seg, seh, sei, sem, sen, seo, seq, sek) were detected among these strains. Some enterotoxin genes (sea, seb, sed, seh, sej, sek, seq) could not be found in any strain. No enterotoxin gene was found in ST97, ST188, or ST398 isolates. The see-sep genes were only present in the ST7 strain, but the seg-sei-sem-sen-seo genes were present in ST7, ST9, and ST520 strains. Compared to the other STs, ST520 had the highest carrying frequency. The seg-sei-sem-sen-seo genes were present in all 10 ST520 isolates (Table 3). Leucotoxins are regarded as virulence factors of S. aureus. Only ST520 (70%) carried both lukE and lukM. LukM could not be found in other STs, but lukE was detected in various other STs, including ST97, ST188, and ST7. The serine-aspartate repeat proteins (sdr) are members of a family of surface proteins that contribute to the pathogenicity of S. aureus. ST97 and ST9, which were the predominant STs of MSSA and MRSA, respectively, contained both sdrD and sdrE. Furthermore, sdrD only existed in ST7, but sdrE was present in various other STs, such as ST520, ST188, and ST398.
Biofilm Formation and Human Erythrocyte Lysis in S. aureus Strains
The ability of S. aureus to form biofilms is considered an important virulence factor because biofilms can tolerate antimicrobial agents, making the bacterium extremely difficult to eradicate. The biofilm assay was performed in the ST97 (randomly selected, n = 39), ST520 (randomly selected, n = 18), ST188 (randomly selected, n = 10), ST398 (randomly selected, n = 10), ST7 (randomly selected, n = 10), and ST9 (n = 10) strains. Compared to ST97 isolates, biofilm formation ability was enhanced significantly in ST188 and ST7 strains (Figure 2A). Moreover, ST7 strains had stronger biofilm formation than ST188 isolates (P < 0.05). There were no significant differences among the other STs, including ST97, ST9, ST520, and ST398.
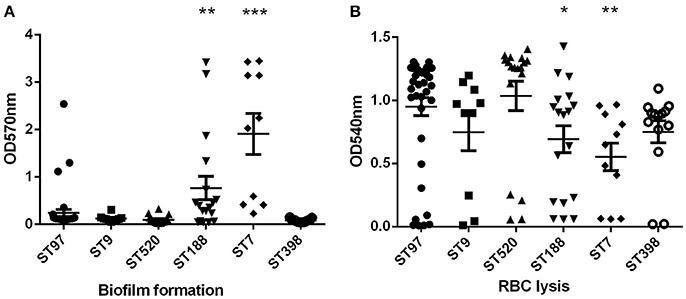
Figure 2. Biofilm formation and red blood cells lysis. (A) Semi-quantitative biofilm analysis of STs with more than 10 isolates. ***P < 0.001 (unpaired t-test, ST7 vs. ST97); **P < 0.01 (unpaired t-test, ST188 vs. ST97). (B) Hemolytic capacities of STs with more than 10 isolates. *P < 0.05 (unpaired t-test, ST188 vs. ST97) **P < 0.01 (unpaired t-test, ST7 vs. ST97).
Hemolysis (lysis of erythrocytes) is also a significant virulence determinant of S. aureus, representing a crucial means for the bacteria to acquire iron. Lysis of red blood cell capacity was relatively strong in all S. aureus strains of animal origin, except ST7 (Figure 2B). The hemolytic capacities of ST7 and ST188 strains were weaker than in ST97 isolates. Among these strains, ST7 had the strongest biofilm and the hemolytic capacity.
Discussion
S. aureus is the most common causative organism of bovine mastitis, which is considered a common, complicated and economically unbearable disease in dairy animals worldwide (Wyder et al., 2011). Dairy animals with mastitis frequently shed S. aureus into the milk supply, which can contribute to food poisoning in humans. This study intended to investigate the molecular profile and antimicrobial resistance of S. aureus isolates recovered from bovine mastitis from 2014 to 2015 in eastern regions of China.
In this study, we characterized 212 S. aureus isolates from bovine mastitis milk samples collected from farms in Shanghai and Zhejiang areas and identified 6.6% as MRSA strains, which was lower than western countries and Hong Kong (16–21.3%; Guardabassi et al., 2009), but higher than Korea (21/657, 3.2%; Lim et al., 2012), Malaysia (1.4%; Neela et al., 2009), and Japan (0.9%; Baba et al., 2010). These different frequencies may be due to the different animal populations studied or the implemented methodologies, among other factors.
Our study identified 14 bovine MRSA strains, including nine ST9-SCCmecXII-t899 strains, four belonging to SCCmec IV type (two ST965-t062, one ST59-t437, one ST537-t2755), and one SCCmecV strain (ST630-t4549). The MRSA isolates from eastern regions of China belonged to different spa and SCCmec types, suggesting their genetic diversity. In those previous studies, CC9 was identified as the predominant MRSA associated with pig farming and harbored various SCCmec types in different areas. For example, SCCmec was type III in mainland China, types IVb or V in Hong Kong, non-types I–VII in Taiwan, type V in Malaysia and type IX in Thailand (Cui et al., 2009; Ho J. et al., 2012; Larsen et al., 2012). In addition, SCCmec XII was a new SCCmec element that was first identified in two ST9-MRSA-t899 strains from pigs in China (Ho P. L. et al., 2012; Wang et al., 2015; Wu et al., 2015). It included a rare class C2 mec gene complex and a ccrC gene complex. The newly identified SCCmec was 48.575 kb in length and contained 45 predicted open reading frames. Therefore, the emerging ST9-MRSA-SCCmec XII strains can be isolated in both pigs and bovines in China, suggesting the potential possibility of its cross-species transmission. Notably, the CC9 MRSA isolates in the current study were shown to be multidrug-resistant, with high resistance rates to β-lactams and eight other antibiotics widely used in chemotherapy. The presence of multidrug resistant MRSA clone CC9 suggests an alarming situation and could be a serious challenge to therapy. The PFGE result in our study showed that the restriction profiles of 10 CC9 strains were identical with only one fragment difference between MRSA CC9 and MSSA CC9 (Supplementary Image 1). These observations support the notion that CC9 MSSA lineages may provide a stable genetic environment for the integration of SCCmec in favor of their infection and transmission in different areas.
Among the 212 S. aureus isolates, the prevalence of MSSA in this study was 93.4%. In addition, our study revealed more heterogeneous MSSA lineages, 16 distinct STs were identified in MSSA, but only five STs were found in MRSA. The most frequently represented lineage in MSSA was CC97 (39.2%, 83/212), which accounted for more than one third of all S. aureus isolates, followed by CC520 (16%, 34/212), CC188 (10%, 21/212), CC398 (7.1%, 15/212), and CC7 (5.2%, 11/212). CC97 was found to be the most predominant clone, which is consistent with a recent study on bovine mastitis infection that also identified CC97 as the dominant strain type among six provinces of China from March 2010 to August 2013 (Wang et al., 2015). However, the difference is that all CC97 isolates identified in our study were MSSA strains. In particular, CC97 strains have become the dominant lineage in Chile (Smith et al., 2005), Brazil (Aires-de-Sousa et al., 2007; Rabello et al., 2007), Japan (Hata et al., 2010), the Netherlands (Ikawaty et al., 2009), and the United States (Smith et al., 2005). We found that CC97 isolates were more resistant to gentamicin and penicillin than other MSSA isolates. This implies that appropriate drug selection based on different MSSA types may reduce the reservoir of drug-resistant bacteria.
The second most frequent MLST type shown here was CC520 (16%), which was only reported in a few studies and could indicate the emergence of a new epidemic clone. Compared to other lineages, CC520 carried more virulence elements but presented a lower drug resistance rate, which may facilitate its success as a pathogen spread in bovines. Moreover, in our study, 21 MSSA isolates belonged to CC188, which is a double locus variant (DLV) of CC1 that includes MW2/USA400, the highly virulent and first-known Panton-Valentine leukocidin (PVL)-positive MRSA strain (Centers for Disease Control and Prevention, 1999). In a previous study of human infection, CC188 was the predominant clone in community-onset MSSA infections (Chen et al., 2012). However, the contamination of bovine mastitis samples with S. aureus by humans cannot be excluded. We propose that these results represent a probable transient acquisition from human handling, which will require more attention in further studies.
LA MRSA was originally reported to have emerged in 2003 among healthy pigs and pig farmers in France and the Netherlands (Witte et al., 2007). In European countries and in North America, CC398 was the overwhelmingly dominant LA MRSA, whereas CC9 predominated in most Asian countries (Cortimiglia et al., 2016; Sharma et al., 2016). In our study, all CC398 strains were identified as MSSA strains. Additionally, the observed spa type distribution of MSSA CC398 mirrored the most frequently detected spa types for MRSA CC398: t034. In contrast to the large number of reports on MRSA CC398 (Fessler et al., 2010; Schijffelen et al., 2010; Smith and Pearson, 2010; Crombé et al., 2012; Price et al., 2012), limited attention has been given to the ecology of MSSA CC398 in livestock farms (Hasman et al., 2010). MSSA CC398 is a clone of clinical importance that affects humans and livestock in different geographic regions. Price et al. analyzed 89 isolates and concluded that LA-MRSA likely originates from humans as MSSA. They theorized that the recent emergence of LA-MRSA can be seen as a reintroduction of MSSA that acquired methicillin-resistance to the original host (Price et al., 2012). Therefore, CC398-MSSA strains in this study may be a risk factor for the transmission of MRSA to people in close contact with the infected animals. Additionally, the present study showed that CC398 was more resistant to clindamycin, erythromycin, and ciprofloxacin. Our results may contribute greatly to the understanding of the emergence and transmission of MRSA CC398 among livestock because they may indicate the existence of a reservoir from which MRSA CC398 strains can emerge.
S. aureus is the most common causative organism of bovine mastitis. However, the treatment and control of this infection often fails due to the complex nature of the S. aureus strains. The development of drug resistance is a significant feature of these organisms. The results of this research revealed that all MRSA isolates were resistant to oxacillin, penicillin and cefoxitin. All MSSA isolates were susceptible to cefazolin, cefuroxime, and oxacillin. In general, MRSA strains showed much higher resistance rates to all the tested antibiotics than MSSA strains (Table 3). All the MRSA strains were found to be resistant to at least four antibiotics. It was apparent that CC9 strains had significantly higher multiple antibiotic resistance profiles compared with other lineages. The widespread resistance of cattle-derived S. aureus will be a serious challenge to bovine mastitis therapy. The ability of bacterial pathogens to produce biofilms is regarded as a major cause of resistance to antibiotics and as responsible for persistent infections and innate immune defense for both animal and human infectious diseases (Cucarella et al., 2004; Bjarnsholt, 2013). The biofilm formed in multilayer growth of bacteria, avoiding bacterial clearance by the host immune system. In our study, we detected biofilm formation in different lineages. The results showed that the biofilm formation ability of CC7 and CC188 strains was significantly higher than other lineages. However, the resistance rates to different antibiotics in CC7 and CC188 were not higher than other lineages. This may indicate that the mechanism of drug resistance in bovine mastitis-associated S. aureus may depend on other factors than biofilm formation. While biofilm formation increases drug resistance and infection time, it can reduce bacterial virulence. Hemolysis (lysis of erythrocytes) is a significant virulence determinant of S. aureus, representing a crucial way for the bacteria to acquire iron. As shown in our study, lysis of red blood cell capacity was relatively strong in all S. aureus strains of animal origin, except CC7 and CC188. Therefore, different types of S. aureus have different potential risks to human beings.
In conclusion, the prevailing S. aureus strains in bovine mastitis were mainly MSSA strains in eastern regions of China. However, the presence of the multidrug-resistant and toxigenic MRSA clone ST9-t899 SCCmec XII suggests an alarming situation and might be a serious challenge to therapy, creating a public health concern. There were differences in the resistance genes, virulence genes and biological activities of the different molecular types of strains. As S. aureus from livestock may emerge as a threat to public health, comprehensive surveillance should be taken against S. aureus infection in the management of animal husbandry products.
Ethics Statement
Blood of healthy individuals for the lysis of erythrocytes by bacterial culture was collected using a standard method in accordance with a protocol approved by the ethics committee of Renji Hospital, School of Medicine, Shanghai Jiaotong University, Shanghai, China. All individuals provided written informed consent prior to donating blood.
Author Contributions
ML, JS, and XW planned and supervised the experiments and wrote the paper. TL, HL, XW, QG, and YD performed the experiments and/or analyzed the data.
Funding
This work was supported by the National Natural Science Foundation of China (grants 81371875, 81671975, and 81601737), the Shanghai Committee of Science and Technology, China (grant 15411960500) and the Foundation for Innovative Research Groups of the National Natural Science Foundation of China (grant 81421001).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors would like to thank the microbiologists and the technical staff of Shanghai Institute for Veterinary Drug and Feeds Control for collecting the bacterial isolates and for laboratory testing.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fcimb.2017.00127/full#supplementary-material
Supplementary Image 1. Pulsed-field gel electrophoresis (PFGE) of SmaI macro-restriction fragments of 10 bovine mastitis-associated isolates of ST9 isolated in this study. Lane M, XbaI-digested DNA of Salmonella enterica serovar as a reference molecular size marker. Lanes 1–10: 10 isolates of ST9 (Lane 1–5, 7–10 were MRSA ST9, Lane 6 was MSSA ST9).
References
Aires-de-Sousa, M., Parente, C. E., Vieira-da-Motta, O., Bonna, I. C., Silva, D. A., and de Lencastre, H. (2007). Characterization of Staphylococcus aureus isolates from buffalo, bovine, ovine, and caprine milk samples collected in Rio de Janeiro State, Brazil. Appl. Environ. Microbiol. 73, 3845–3849. doi: 10.1128/AEM.00019-07
Arvidson, S., and Tegmark, K. (2001). Regulation of virulence determinants in Staphylococcus aureus. Int. J. Med. Microbiol. 291, 159–170. doi: 10.1078/1438-4221-00112
Baba, K., Ishihara, K., Ozawa, M., Tamura, Y., and Asai, T. (2010). Isolation of meticillin-resistant Staphylococcus aureus (MRSA) from swine in Japan. Int. J. Antimicrob. Agents 36, 352–354. doi: 10.1016/j.ijantimicag.2010.06.040
Bal, A. M., Coombs, G. W., Holden, M. T., Lindsay, J. A., Nimmo, G. R., Tattevin, P., et al. (2016). Genomic insights into the emergence and spread of international clones of healthcare-, community- and livestock-associated meticillin-resistant Staphylococcus aureus: blurring of the traditional definitions. J. Glob. Antimicrob. Resist. 6, 95–101. doi: 10.1016/j.jgar.2016.04.004
Bjarnsholt, T. (2013). The role of bacterial biofilms in chronic infections. APMIS (Suppl. 136), 1–51. doi: 10.1111/apm.12099
Botrel, M. A., Haenni, M., Morignat, E., Sulpice, P., Madec, J. Y., and Calavas, D. (2010). Distribution and antimicrobial resistance of clinical and subclinical mastitis pathogens in dairy cows in Rhone-Alpes, France. Foodborne Pathog. Dis. 7, 479–487. doi: 10.1089/fpd.2009.0425
Bradley, A. J., Leach, K. A., Breen, J. E., Green, L. E., and Green, M. J. (2007). Survey of the incidence and aetiology of mastitis on dairy farms in England and Wales. Vet. Rec. 160, 253–257. doi: 10.1136/vr.160.8.253
Ceriotti, F., Zakowski, J., Sine, H., Altaie, S., Horowitz, G., Pesce, A. J., et al. (2012). Clinical and Laboratory Standards Institute (CLSI).
Chen, F. J., Siu, L. K., Lin, J. C., Wang, C. H., and Lu, P. L. (2012). Molecular typing and characterization of nasal carriage and community-onset infection methicillin-susceptible Staphylococcus aureus isolates in two Taiwan medical centers. BMC Infect. Dis. 12:343. doi: 10.1186/1471-2334-12-343
Chuang, Y. Y., and Huang, Y. C. (2015). Livestock-associated meticillin-resistant Staphylococcus aureus in Asia: an emerging issue? Int. J. Antimicrob. Agents 45, 334–340. doi: 10.1016/j.ijantimicag.2014.12.007
Cortimiglia, C., Luini, M., Bianchini, V., Marzagalli, L., Vezzoli, F., Avisani, D., et al. (2016). Prevalence of Staphylococcus aureus and of methicillin-resistant S. aureus clonal complexes in bulk tank milk from dairy cattle herds in Lombardy region (Northern Italy). Epidemiol. Infect. 144, 3046–3051. doi: 10.1017/S0950268816001576
Crombé, F., Willems, G., Dispas, M., Hallin, M., Denis, O., Suetens, C., et al. (2012). Prevalence and antimicrobial susceptibility of methicillin-resistant Staphylococcus aureus among pigs in Belgium. Microb. Drug Resist. 18, 125–131. doi: 10.1089/mdr.2011.0138
Cucarella, C., Tormo, M. A., Ubeda, C., Trotonda, M. P., Monzon, M., Peris, C., et al. (2004). Role of biofilm-associated protein bap in the pathogenesis of bovine Staphylococcus aureus. Infect. Immun. 72, 2177–2185. doi: 10.1128/IAI.72.4.2177-2185.2004
Cui, S., Li, J., Hu, C., Jin, S., Li, F., Guo, Y., et al. (2009). Isolation and characterization of methicillin-resistant Staphylococcus aureus from swine and workers in China. J. Antimicrob. Chemother. 64, 680–683. doi: 10.1093/jac/dkp275
Cuny, C., Kock, R., and Witte, W. (2013). Livestock associated MRSA (LA-MRSA) and its relevance for humans in Germany. Int. J. Med. Microbiol. 303, 331–337. doi: 10.1016/j.ijmm.2013.02.010
Devriese, L. A., Van Damme, L. R., and Fameree, L. (1972). Methicillin (cloxacillin)-resistant Staphylococcus aureus strains isolated from bovine mastitis cases. Zentralbl. Veterinrmed. B 19, 598–605. doi: 10.1111/j.1439-0450.1972.tb00439.x
Fang, H. W., Chiang, P. H., and Huang, Y. C. (2014). Livestock-associated methicillin-resistant Staphylococcus aureus ST9 in pigs and related personnel in Taiwan. PLoS ONE 9:e88826. doi: 10.1371/journal.pone.0088826
Fessler, A., Scott, C., Kadlec, K., Ehricht, R., Monecke, S., and Schwarz, S. (2010). Characterization of methicillin-resistant Staphylococcus aureus ST398 from cases of bovine mastitis. J. Antimicrob. Chemother. 65, 619–625. doi: 10.1093/jac/dkq021
Guardabassi, L., O'Donoghue, M., Moodley, A., Ho, J., and Boost, M. (2009). Novel lineage of methicillin-resistant Staphylococcus aureus, Hong Kong. Emerg. Infect. Dis. 15, 1998–2000. doi: 10.3201/eid1512.090378
Harmsen, D., Claus, H., Witte, W., Rothgänger, J., Claus, H., Turnwald, D., et al. (2003). Typing of methicillin-resistant Staphylococcus aureus in a university hospital setting by using novel software for spa repeat determination and database management. J. Clin. Microbiol. 41, 5442–5448. doi: 10.1128/JCM.41.12.5442-5448.2003
Hartmann, F. A., Trostle, S. S., and Klohnen, A. A. (1997). Isolation of methicillin-resistant Staphylococcus aureus from a postoperative wound infection in a horse. J. Am. Vet. Med. Assoc. 211, 590–592.
Hasman, H., Moodley, A., Guardabassi, L., Stegger, M., Skov, R. L., and Aarestrup, F. M. (2010). Spa type distribution in Staphylococcus aureus originating from pigs, cattle and poultry. Vet. Microbiol. 141, 326–331. doi: 10.1016/j.vetmic.2009.09.025
Hata, E., Katsuda, K., Kobayashi, H., Uchida, I., Tanaka, K., and Eguchi, M. (2010). Genetic variation among Staphylococcus aureus strains from bovine milk and their relevance to methicillin-resistant isolates from humans. J. Clin. Microbiol. 48, 2130–2139. doi: 10.1128/JCM.01940-09
He, J.-z., Wang, A.-q., Gang, L., Jian, G., Ali, T., and Bo, H. (2014). Biofilm formation and biofilm-associated genes assay of Staphylococcus aureus isolated from Bovine subclinical Mastitis in China. Pak. Vet. J. 34, 508–513.
Ho, J., O'Donoghue, M., Guardabassi, L., Moodley, A., and Boost, M. (2012). Characterization of methicillin-resistant Staphylococcus aureus isolates from pig carcasses in Hong Kong. Zoonoses Public Health 59, 416–423. doi: 10.1111/j.1863-2378.2012.01473.x
Ho, P. L., Chow, K. H., Lai, E. L., Law, P. Y., Chan, P. Y., Ho, A. Y., et al. (2012). Clonality and antimicrobial susceptibility of Staphylococcus aureus and methicillin-resistant S. aureus isolates from food animals and other animals. J. Clin. Microbiol. 50, 3735–3737. doi: 10.1128/JCM.02053-12
Huber, H., Koller, S., Giezendanner, N., Stephan, R., and Zweifel, C. (2010). Prevalence and characteristics of meticillin-resistant Staphylococcus aureus in humans in contact with farm animals, in livestock, and in food of animal origin, Switzerland, 2009. Euro Surveill. 15, 19542. doi: 10.5167/uzh-41122
Ikawaty, R., Brouwer, E. C., Jansen, M. D., Van, D. E., Mevius, D., Verhoef, J., et al. (2009). Characterization of Dutch Staphylococcus aureus from bovine mastitis using a multiple locus variable number tandem repeat analysis. Vet. Microbiol. 136, 277–284. doi: 10.1016/j.vetmic.2008.10.034
Kondo, Y., Ito, T., Ma, X. X., Watanabe, S., Kreiswirth, B. N., Etienne, J., et al. (2007). Combination of multiplex PCRs for staphylococcal cassette chromosome mec type assignment: rapid identification system for mec, ccr, and major differences in junkyard regions. Antimicrob. Agents Chemother. 51, 264–274. doi: 10.1128/AAC.00165-06
Larsen, J., Imanishi, M., Hinjoy, S., Tharavichitkul, P., Duangsong, K., Davis, M. F., et al. (2012). Methicillin-resistant Staphylococcus aureus ST9 in pigs in Thailand. PLoS ONE 7:e31245. doi: 10.1371/journal.pone.0031245
Li, L. I., Yang, H. J., Liu, D. C., Hong-Bin, H. E., Wang, C. F., Zhong, J. F., et al. (2011). Biofilm formation and analysis of associated genes involved in Staphylococcus isolates from Bovine Mastitis. Sci. Agric. Sin. 84, 439–449. doi: 10.1360/052011-114
Lim, S. K., Nam, H. M., Jang, G. C., Lee, H. S., Jung, S. C., and Kwak, H. S. (2012). The first detection of methicillin-resistant Staphylococcus aureus ST398 in pigs in Korea. Vet. Microbiol. 155, 88–92. doi: 10.1016/j.vetmic.2011.08.011
Lina, G., and Etienne, J. (1999). Involvement of panton-valentine leukocidin—producing Staphylococcus aureus in primary skin infections and pneumonia. Clin. Infect. Dis. 29, 1128–1132. doi: 10.1086/313461
Maes, N., Magdalena, J., Rottiers, S., De Gheldre, Y., and Struelens, M. J. (2002). Evaluation of a triplex PCR assay to discriminate Staphylococcus aureus from coagulase-negative Staphylococci and determine methicillin resistance from blood cultures. J. Clin. Microbiol. 40, 1514–1517. doi: 10.1128/JCM.40.4.1514-1517.2002
Monecke, S., Kuhnert, P., Hotzel, H., Slickers, P., and Ehricht, R. (2007). Microarray based study on virulence-associated genes and resistance determinants of Staphylococcus aureus isolates from cattle. Vet. Microbiol. 125, 128–140. doi: 10.1016/j.vetmic.2007.05.016
Neela, V., Zafrul, A. M., Mariana, N. S., Belkum, A. V., Yun, K. L., and Rad, E. G. (2009). Prevalence of ST9 methicillin-resistant Staphylococcus aureus among pigs and pig handlers in Malaysia. J. Clin. Microbiol. 47:4138. doi: 10.1128/jcm.01363-09
Nemeghaire, S., Argudin, M. A., Haesebrouck, F., and Butaye, P. (2014). Epidemiology and molecular characterization of methicillin-resistant Staphylococcus aureus nasal carriage isolates from bovines. BMC Vet. Res. 10:153. doi: 10.1186/1746-6148-10-153
Peacock, S. J., Moore, C. E., Justice, A., Kantzanou, M., Story, L., Mackie, K., et al. (2002). Virulent combinations of adhesin and toxin genes in natural populations of Staphylococcus aureus. Infect. Immun. 70, 4987–4996. doi: 10.1128/IAI.70.9.4987-4996.2002
Price, L. B., Stegger, M., Hasman, H., Aziz, M., Larsen, J., Andersen, P. S., et al. (2012). Staphylococcus aureus CC398: host adaptation and emergence of methicillin resistance in livestock. mBio 3:e00305-11. doi: 10.1128/mBio.00305-11
Rabello, R. F., Moreira, B. M., Lopes, R. M., Teixeira, L. M., Riley, L. W., and Castro, A. C. (2007). Multilocus sequence typing of Staphylococcus aureus isolates recovered from cows with mastitis in Brazilian dairy herds. J. Med. Microbiol. 56(Pt 11), 1505–1511. doi: 10.1099/jmm.0.47357-0
Schijffelen, M. J., Boel, C. H., van Strijp, J. A., and Fluit, A. C. (2010). Whole genome analysis of a livestock-associated methicillin-resistant Staphylococcus aureus ST398 isolate from a case of human endocarditis. BMC Genomics 11:376. doi: 10.1186/1471-2164-11-376
Sharma, M., Nunezgarcia, J., Kearns, A. M., Doumith, M., Butaye, P. R., Argudín, M. A., et al. (2016). Livestock-associated methicillin resistant Staphylococcus aureus (LA-MRSA) Clonal Complex (CC) 398 isolated from UK animals belong to European Lineages. Front. Microbiol. 7:1741. doi: 10.3389/fmicb.2016.01741
Smith, E. M., Green, L. E., Medley, G. F., Bird, H. E., Fox, L. K., Schukken, Y. H., et al. (2005). Multilocus sequence typing of intercontinental bovine Staphylococcus aureus isolates. J. Clin. Microbiol. 43, 4737–4743. doi: 10.1128/JCM.43.9.4737-4743.2005
Smith, T. C., and Pearson, N. (2010). The emergence of Staphylococcus aureus ST398. Vector Borne Zoonotic Dis. 11, 327–339. doi: 10.1089/vbz.2010.0072
Uhlemann, A. C., Otto, M., Lowy, F. D., and DeLeo, F. R. (2014). Evolution of community- and healthcare-associated methicillin-resistant Staphylococcus aureus. Infect. Genet. Evol. 21, 563–574. doi: 10.1016/j.meegid.2013.04.030
Centers for Disease Control and Prevention (CDC). (1999). From the centers for disease control and prevention. Four pediatric deaths from community-acquired methicillin-resistant Staphylococcus aureus — Minnesota and North Dakota, 1997–1999. JAMA 282, 1123–1125.
Van Cleef, B. A., Monnet, D. L., Voss, A., Krziwanek, K., Allerberger, F., Struelens, M., et al. (2011). Livestock-associated methicillin-resistant Staphylococcus aureus in humans, Europe. Emerg. Infect. Dis. 17, 502–505. doi: 10.3201/eid1703.101036
Vanderhaeghen, W., Cerpentier, T., Adriaensen, C., Vicca, J., Hermans, K., and Butaye, P. (2010). Methicillin-resistant Staphylococcus aureus (MRSA) ST398 associated with clinical and subclinical mastitis in Belgian cows. Vet. Microbiol. 144, 166–171. doi: 10.1016/j.vetmic.2009.12.044
Voss, A., Loeffen, F., Bakker, J., Klaassen, C., and Wulf, M. (2005). Methicillin-resistant Staphylococcus aureus in pig farming. Emerg. Infect. Dis. 11, 1965–1966. doi: 10.3201/eid1112.050428
Wan, M. T., Lauderdale, T. L., and Chou, C. C. (2013). Characteristics and virulence factors of livestock associated ST9 methicillin-resistant Staphylococcus aureus with a novel recombinant staphylocoagulase type. Vet. Microbiol. 162, 779–784. doi: 10.1016/j.vetmic.2012.10.003
Wang, D., Wang, Z., Yan, Z., Wu, J., Ali, T., Li, J., et al. (2015). Bovine mastitis Staphylococcus aureus: antibiotic susceptibility profile, resistance genes and molecular typing of methicillin-resistant and methicillin-sensitive strains in China. Infect. Genet. Evol. 31, 9–16. doi: 10.1016/j.meegid.2014.12.039
Wang, L., and Gao, Q. (2008). SarZ is a key regulator of biofilm formation and virulence in Staphylococcus epidermidis. J. Infect. Dis. 197, 1254–1262. doi: 10.1086/586714
Wardenburg, J. B., Patel, R. J., and Schneewind, O. (2007). Surface Proteins and exotoxins are required for the pathogenesis of Staphylococcus aureus Pneumonia. Infect. Immun. 75, 1040–1044. doi: 10.1128/IAI.01313-06
Witte, W., Strommenger, B., Stanek, C., and Cuny, C. (2007). Methicillin-resistant Staphylococcus aureus ST398 in humans and animals, Central Europe. Emerg. Infect. Dis. 13, 255–258. doi: 10.3201/eid1302.060924
Wu, Z., Li, F., Liu, D., Xue, H., and Zhao, X. (2015). Novel Type XII Staphylococcal cassette chromosome mec harboring a new cassette chromosome recombinase, CcrC2. Antimicrob. Agents Chemother. 59, 7597–7601. doi: 10.1128/AAC.01692-15
Wyder, A. B., Boss, R., Naskova, J., Kaufmann, T., Steiner, A., and Graber, H. U. (2011). Streptococcus spp. and related bacteria: their identification and their pathogenic potential for chronic mastitis – a molecular approach. Res. Vet. Sci. 91, 349–357. doi: 10.1016/j.rvsc.2010.09.006
Yan, X., Li, Z., Chlebowicz, M. A., Tao, X., Ni, M., Hu, Y., et al. (2016). Genetic features of livestock-associated Staphylococcus aureus ST9 isolates from Chinese pigs that carry the lsa (E) gene for quinupristin/dalfopristin resistance. Int. J. Med. Microbiol. 306, 722–729. doi: 10.1016/j.ijmm.2016.08.001
Keywords: bovine mastitis, S. aureus, MLST-genotyping, SCCmec typing, antimicrobial resistance
Citation: Li T, Lu H, Wang X, Gao Q, Dai Y, Shang J and Li M (2017) Molecular Characteristics of Staphylococcus aureus Causing Bovine Mastitis between 2014 and 2015. Front. Cell. Infect. Microbiol. 7:127. doi: 10.3389/fcimb.2017.00127
Received: 31 December 2016; Accepted: 28 March 2017;
Published: 19 April 2017.
Edited by:
Joan A. Geoghegan, Trinity College, Dublin, IrelandCopyright © 2017 Li, Lu, Wang, Gao, Dai, Shang and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jun Shang, 592568052@qq.com
Min Li, ruth_limin@126.com
†These authors have contributed equally to this work.
 Tianming Li
Tianming Li Huiying Lu1†
Huiying Lu1† Xing Wang
Xing Wang Qianqian Gao
Qianqian Gao Yingxin Dai
Yingxin Dai Min Li
Min Li