- 1Center for Quantitative Biology, Academy for Advanced Interdisciplinary Studies, College of Chemistry and Molecular Engineering, Beijing National Laboratory for Molecular Sciences (BNLMS), Peking University, Beijing, China
- 2Department of Pharmacology, School of Pharmacy, Fudan University, Shanghai, China
The aberrant activation of hedgehog (Hh) signaling pathway is closely related to human diseases. The upstream protein, N-terminal product of sonic hedgehog (ShhN) is overexpressed in many cancers and considered as a promising antitumor target. Inhibitors that bind to ShhN and break its interaction with the 12-transmembrane glycoprotein patched (Ptch) protein are highly wanted to tune down the abnormal Hh pathway activation. However, research of ShhN inhibitors remains lacking. In this paper, we computationally screened potential inhibitors against the ShhN-Ptch interaction interface, and tested their activities by experimental studies. Seven compounds (1-7) with diverse scaffolds, showed inhibition in cellular assays and directly bound to ShhN in vitro. The compounds were verified to modulate the Hh pathway activity. Furthermore, we studied the structure-activity relationship of the pyrimidine (7) derivatives and identified a potent compound (7_3d3) with IC50 of 0.4 ± 0.1 μM in cellular assays. These small molecule inhibitors of ShhN provide novel chemical probes for future investigations of Hh signaling.
Introduction
The hedgehog (Hh) signaling pathway is highly conserved in organisms ranging from insects to mammals, and plays an important role in embryogenesis and tissue homeostasis (Ingham et al., 2011; Briscoe and Therond, 2013). In mammalian cells, Hh family proteins bind to the 12-transmembrane glycoprotein patched (Ptch) protein to activate the 7-transmembrane protein smoothened (Smo). Then, Smo activates transcription factors in the Gli family via formation of a cytosolic complex. Activated Gli proteins then translocate into the nucleus to induce transcription of target genes (Hu et al., 2015).
Aberrant activation of the Hh pathway is associated with the initiation and progression of various cancers (Xie et al., 1998; Berman et al., 2003; Lum and Beachy, 2004; Merchant and Matsui, 2010; Francesca et al., 2018; Galperin et al., 2019). Blocking the Hh pathway has been reported to inhibit cancer cell growth and tumor development (Beauchamp et al., 2011), so the Hh signaling pathway has critical therapeutic potential. Sonic hedgehog (Shh), the key upstream component in the Hh pathway, is one of the Hh homologs that are overexpressed in many cancers (Oro et al., 1997; Rubin and de Sauvage, 2006). Full-length Shh undergoes an autocleavage process to generate Shh N-terminal product (ShhN) (Ingham et al., 2011), which cooperates with Ptch for signaling through a direct protein-protein interaction (PPI). As a secreted protein, ShhN is a promising target for drug development.
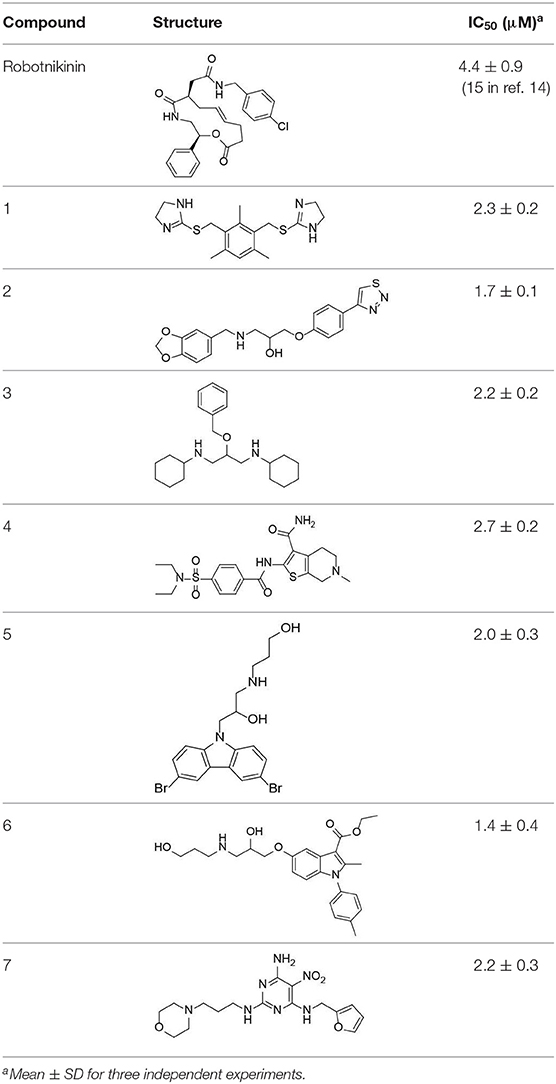
Despite the availability of neutralizing anti-Hh antibodies, discovery of potent inhibitors that disrupt the ShhN-Ptch interaction has been challenging. So far, there are only two related literature reports. Stanton et al. reported that robotnikinin (Stanton et al., 2009), the first small-molecular-weight ShhN inhibitor, has moderate Shh inhibitory activity (IC50 ≈ 15 μM) in cellular assays. The second small-molecular-weight ShhN inhibitor(HL2-m5) was a macrocyclic peptide with an IC50 of 0.25 μM in cellular assays, reported by Owens et al. (2017). Novel Hh small molecule inhibitors targeting Shh/Ptch interface need to be explored.
We present the discovery of organic small molecule inhibitors of PPI with ShhN. We defined the PPI of ShhN, used virtual screening to select potential inhibitors, and performed experimental validation preliminary. The structure-activity relationship (SAR) of one of the most potent compounds was studied.
Materials and Methods
Virtual Screening Targeting ShhN
The crystal structure of human ShhN (PDB ID: 3M1N) (Pepinsky et al., 2000) was used for a docking study with the defined PPI residues. Molecular docking-based virtual screening was performed using Glide. To increase the success rate of PPI inhibitor discovery, several chemical libraries were used, namely, the SPECS compound library (November 2014 version, ~200,000 compounds), Maybridge screening library (May 2015 version, ~50,000 compounds), and the specially designed PPI inhibitor library known as Life chemicals PPI inhibitors 3D similarity library (February 2015 version, ~17,000 compounds). We screened the libraries using the Glide SP mode (Friesner et al., 2004, 2006; Halgren et al., 2004) and selected 5% of the top-ranking compounds of each library for further study. Then, we used the Glide XP mode to make further selections. A total of 318 compounds with XP GScores of less than −6.0 kcal/mol were selected to filter by PAINS-Remover (Baell and Holloway, 2010). The remaining 263 compounds were evaluated manually according to the following criteria: (a) formatting hydrogen bond with ShhN, (b) occupying >80% of the pocket, (c) not a peptide, and (d) containing no metal atoms. Finally, 209 compounds were selected and purchased for further experimental tests.
Similarity Searching
The SciFinder website was used to conduct a 2D similarity search of compound 7. Forty-five compounds with >65% similarity from Life Chemicals Library were manual evaluated. Finally, 11 compounds were chosen and purchased.
Shape Screening in the Schrödinger software package was used for 3D similarity search (Sastry et al., 2011). Life chemicals PPI inhibitors library and SPECS library were screened separately. Forty-four compounds (Shape Sim > 0.45) from Life Chemicals and 26 compounds (Shape Sim > 0.60) from SPECS were manually selected. Finally, six compounds from Life Chemicals and five compounds from SPECS were purchased.
Chemicals
All identified hit compounds that were purchased claimed purities of >95%. 4, 5, 6, and 7 were confirmed using 1H NMR and high resolution mass spectra (HRMS); 10 derivatives of the 7 were confirmed using 1H NMR (Supporting Information III. Characterizations of the Compounds). The 1H NMR spectra were recorded at 400 MHz and are shown in Supplementary Materials. HRMS were recorded on a Bruker Apex IV FTMS mass spectrometer using electrospray ionization. Robotnikinin with >97% purity was purchased from Biovision (Milpitas, CA).
Expression and Purification of ShhN and ShhN-EGFP
PET-28(a)-human ShhN (Shh 24-197) plasmid was from Prof. Chuanmao Zhang. EGFP gene was subcloned into pET-28(a)-human ShhN vector to build ShhN-EGFP plasmids. All mutations were introduced by PCR and confirmed by sequencing with GENEWIZ (China).
His-ShhN plasmid or His-ShhN-EGFP plasmid was transformed into Escherichia coli strain Rosetta (DE3). The cells were cultivated at 37°C in Luria-Bertani (LB) medium containing 50 μg/mL kanamycin until the OD600 reached 1.0. Then, IPTG (final concentration 0.4 mM) was added to induce target protein expression. The cells were grown overnight at 20°C. Cells pellets were first resuspended and then lysed by sonication in a solution containing 20 mM HEPES, pH 7.4, 300 mM NaCl, 0.1 mM PMSF, and 1 mM DDT. The cell lysates were clarified by centrifugation at 35,000 g for 30 min at 4°C. The supernatants were filtered by the Millex-GV Filter Unit (Merck Millipore Ltd.) and then applied to the nickel column (HisTrap HP; GE Healthcare Biacore, Uppsala, Sweden). After the column was equilibrated with buffer A (20 mM HEPES, pH 7.4, 300 mM NaCl, 20 mM imidazole), proteins were eluted with a 0–100% gradient of buffer B (20 mM HEPES, pH 7.4, 300 mM NaCl, 500 mM imidazole). The target protein was centrifuged and applied to an S200 gel-filtration column (Sephacryl S-200 HR, GE Healthcare) that had been equilibrated with buffer C (20 mM HEPES, pH 7.4, 300 mM NaCl). The concentration of target protein was measured with a Nanodrop 2000 (Thermo Fisher Scientific, Waltham, MA).
SPR Assays
Biacore T200 (GE Healthcare, Uppsala, Sweden) was used to perform the experiments. ShhN was diluted to 10 ng/μL in 10 mM sodium acetate (pH 5.5) and immobilized on a CM5 sensor chip via standard EDC/NHS amine coupling to ~1,000 RU at 25°C. The control surface was activated in the absence of ShhN and used as a reference. Running buffer and sample buffer were the same and consisted of 10 mM HEPES, pH 7.4, 150 mM NaCl, 0.05% P20, and 5% DMSO. The samples were subsequently injected. All results were evaluated with Biacore T200 Evaluation Software. The reported small molecule inhibitor, robotnikinin (Stanton et al., 2009), was used as the positive reference.
Dual-Luciferase Reporter Assays
A dual-luciferase reporter assay was performed as previously described (Zhan et al., 2014; Kong et al., 2015). Shh-LIGHT2 cells, an Hh activity reporter cell line derived from NIH-3T3 cells by stably expressing 8 × Gli-binding site luciferase reporter (8 × GBS-luciferase) plasmid provided by Sasaki et al. (1999) and pRL-Renilla luciferase plasmid, were seeded into 96-well plates. After 24 h, ShhN-conditioned medium (ShhN CM) was added, and cells were exposed to molecules. The luciferase activities of cells were measured 36 h later with a Dual-Luciferase Reporter Assay System (E1960, Promega), according to the manufacturer's instructions, in a luminometer (Molecular Devices, Sunnyvale, CA). The firefly luciferase values were normalized to the Renilla luciferase activities, and IC50 values were fitted with the Hill equation with OriginPro 9.
Real-Time PCR
Ptch+/−p53−/− medulloblastoma cells were cultivated from spontaneous medulloblastoma developed by ptch+/−; p53−/− mice. All procedures were preapproved by the Animal Care and Use Committee of Fudan University and performed according to institutional policies. Briefly, medulloblastoma tissues were mechanically minced and digested by collagenase. The cells were routinely cultured using Neurobasal A medium (Invitrogen) plus B-27 supplement (Invitrogen), EGF 20 ng/mL (Invitrogen), bFGF 20 ng/mL (Invitrogen), non-essential amino acids, N-acetylcysteine 60 μg/mL. GDC-0449 was purchased from Biovision (Milpitas, CA). Cells in logarithmic growth were vaccinated in six-well plate with inoculum density of about 106. After 24 h, cells were exposed to molecules. Thirty-six hours later, cells were harvested.
Total RNA was extracted from cells using an RNAiso Plus Kit (TaKaRa, Dalian, China) according to the manufacturer's instructions and processed directly to cDNA by reverse transcription with a SuperScript III Kit (TaKaRa, Dalian, China). Semi-quantitative PCR amplification was conducted using Stratagene mx3005p (Agilent). All quantitative PCR amplifications were performed in triplicate with a SYBR Green Kit (TaKaRa, Dalian, China) in an iCycleriQ system (Bio-Rad, Hercules, CA). The mRNA expression level of Gli1 was normalized to that of GUSB. The primers for Gli1 were provided by Invitrogen (Shanghai, China): Gli1: 5′-GCAGTGGGTAACATGAGTGTCT-3′, 5′-AGGCACTAGAGTTGAGGAATTGT-3′.
Microscale Thermophoresis (MST) Analysis
Measurements were performed using a Monolith NT. 115 instrument (NanoTemper Inc., Germany). MST-optimized buffer (50 mM Tris·HCl, pH 7.4, 150 mM NaCl, 10 mM MgCl2, 0.1% Tween-20, 0.1% Pluronic-F127, 5% DMSO) was applied. Measurements were carried out using blue LED color with 20% LED power and 40% MST power. The concentration of ShhN-EGFP was adjusted according to the fluorescence. Mean values and standard deviations were calculated from experiments performed at least in triplicate. Nanotemper analysis software V1.5.41 was applied to analyze data.
NanoDSF Analysis
Measurements were performed using a Prometheus NT.48 instrument (NanoTemper Inc., Germany). ShhN was diluted to 0.2 mg/mL in buffer contained 10 mM HEPES, pH 7.4, 150 mM NaCl, and 5% DMSO. Measurements were carried out using 70% power with standard Prometheus NT.48 capillaries. The heating was 1.0°C/min from 20 to 95°C.
Results and Discussion
Discovery of ShhN–Ptch PPI Inhibitors
Our inhibitor discovery workflows include: determine the interface between ShhN and Ptch; molecular docking-based virtual screening; PAINS-Remover; manual evaluation.
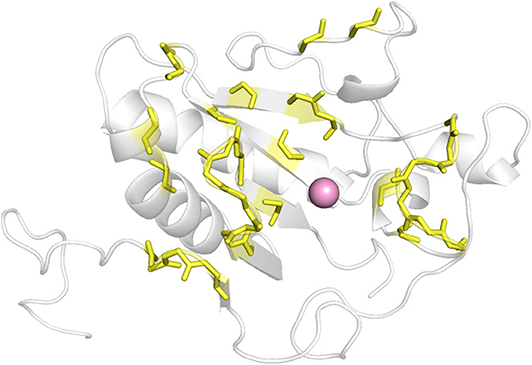
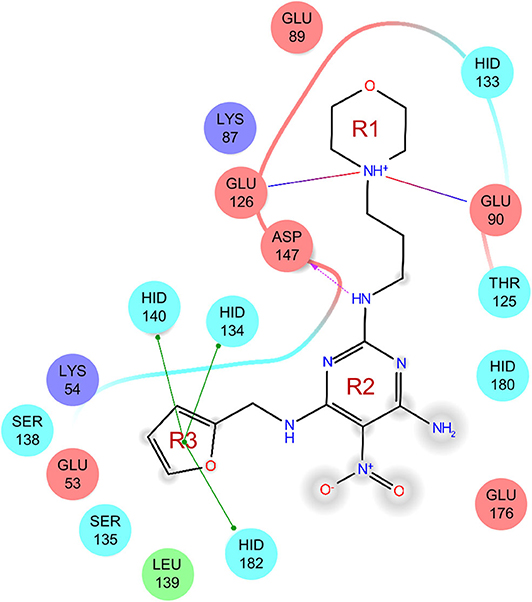
As there was no structural information of Shh and Ptch complex available at the beginning of this work, we determined which ShhN residues interact with Ptch based on the complex structures of ShhN with an antibody and a peptide. The antibody 5E1 has been shown to inhibit the interaction between Hh and Ptch, and the complex structure of Shh bound to the 5E1 fab fragment has been reported (PDB ID:3MXW) (Maun et al., 2010). The Hh-interacting protein (HHIP) L2 peptide competes with a Ptch1 L2-like peptide for Shh binding (Bosanac et al., 2009), and the complex structure of HHIP and Shh (PDB ID: 3HO5) has also been solved (Bosanac et al., 2009). We analyzed the interaction surfaces of these proteins with ShhN and found that interacting residues on ShhN (within 5 Å of the 5E1 fab fragment or HHIP) were similar. We then defined the following residues as potential PPI interface residues for docking study: L42, A43, Y44, K45, K87, E89, R123, T125, E126, D131, G132, H133, H134, S135, E136, S138, H140, D147, T149, D152, R153, R155, Y158, Y174, Y175, E176, S177, K178, A179, H180, and H182 (Figure 1).
The crystal structure 3M1N was used as the model to identify potential inhibitors of ShhN. Compounds from SPECS, Maybridge, and Life Chemicals PPI Inhibitors library were prepared by Ligprep to build their 3D structures. First, Glide SP mode was applied to a virtual screen. Second, the compounds with Glide SP GScores in the top 5% of each library were chosen to perform by Glide XP mode. Third, 318 compounds with XP GScore less than −6.0 kcal/mol were selected to filter by PAINS-Remover. With our effective strategy, the potential small molecule ShhN inhibitors were selected by virtual screening.
Compounds Identified by Initial Binding Screen Bind to ShhN in vitro
After manual evaluation, 209 compounds were bought and tested their binding to ShhN at 50 μM using SPR (Tables S1–S3). Of these selected compounds with diverse chemical structures, 45 showed binding to ShhN (RU > 5.0 at concentration of 50 μM). More than three quarters of the binding compounds have an imino group, and more than half of the compounds include fused heterocyclic structures like purine and 3,6-dibromo-9H-carbazol-9-yl.
Shh-Binding Compounds Inhibit Hh Signaling Pathway
We further tested the 45 Shh-binding compounds to assess their effects on the Hh signaling pathway. Among the tested compounds, 10 showed strong inhibition of Hh signaling (>85%). Compounds with similar framework were merged, resulting in seven hits. Ranks of the final seven identified hits are given in Table S4. The IC50 values of these compounds ranged from 1.4 to 2.7 μM, the reported ShhN inhibitor robotnikinin was measured with IC50 value 4.4 μM under the same conditions (Table 1).
Quantitative Measurement of Binding Constants Between ShhN and Active Compounds
We used microscale thermophoresis (MST) to quantitatively measure binding constants between the compounds and ShhN (Table S5 and Figure S1). A ShhN-EGFP fusion protein was constructed, expressed, and purified from bacterial cells. In MST binding assays, ShhN-EGFP was kept at a constant concentration according to the fluorescence signal. Different concentrations of the molecules were tested and dissociation constants (Kd) were derived. All above seven compounds showed significant binding signals. Compound 4 did not reach saturation in the MST dose-response curve due to low solubility. Dissociation constants of the other compounds ranged from 3.1 to 8.4 μM.
Target Identification ex vivo
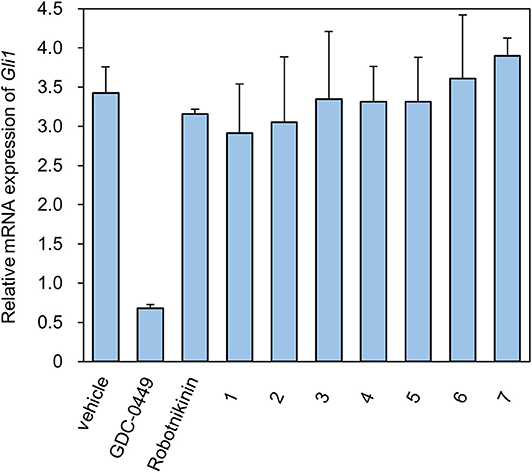
We designed ex vivo target identification studies, to show that the putative target of these compounds might locate upstream of ptch, acting like robotnikinin.
Levels of the Shh-induced transcription factor Gli1 are higher in Ptch+/−p53−/− medulloblastoma from mice than in control tissues as a result of increased Hh signaling activity (Wetmore et al., 2001). As a reliable indicator, Gli1 mRNA levels are used to monitor the activity of the Hh signaling pathway (Scales and de Sauvage, 2009). We measured the expression of Gli1 mRNA using RT-qPCR (Figure 2). As a compound targeting Smo, GDC-0449 was a positive control for inhibition of the expression of Gli1 mRNA. In contrast, ShhN binding robotnikinin did not inhibit Gli1 mRNA expression. In the literature (Stanton et al., 2009), the effect of robotnikinin on Gli1 expression was obtained from ShhN-treated primary human keratinocytes in Stanton's article, which harbor wild type ptch gene. Our verification is different from the report.
The seven tested compounds showed behavior similar to that of robotnikinin, indicating that they inhibited the Hh signaling pathway by targeting ShhN and not Smo.
The active compounds have diverse scaffolds, including derivatives of bis-imidazole (1), 1,3-benzodioxole (2), cyclohexane (3), sulfamide (4), carbazole (5), indole (6), and pyrimidine (7). The inhibitors target ShhN and show submicromolar to micromolar inhibition activities at the cellular level.
Binding Modes of the Active Compounds
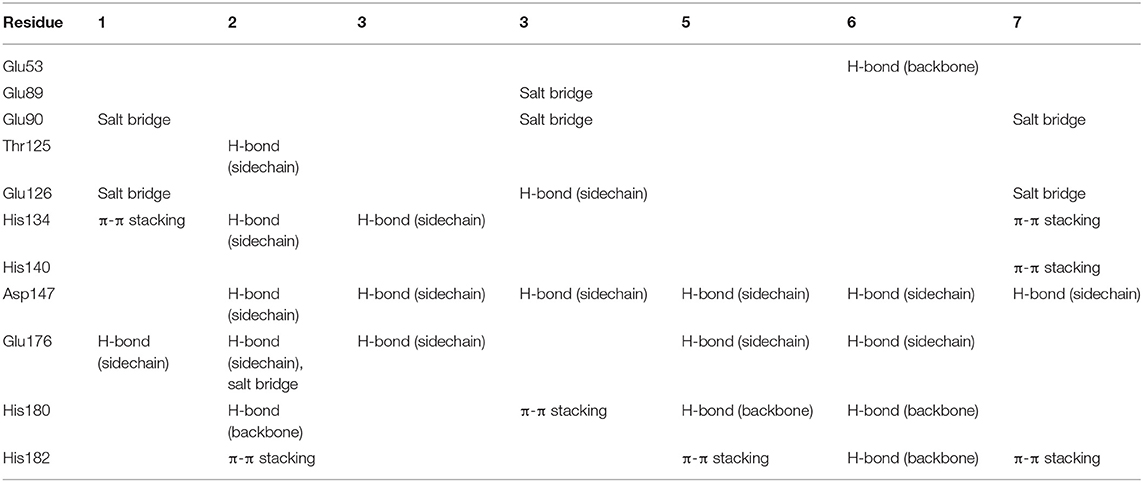
To gain more information at the molecular level, we analyzed the docking positions of the seven compounds. Although the seven compounds formed different interactions with ShhN, they shared similar interactions with key residues, including His134, Asp147, Glu176, His180, and His182 (Table 2).
SAR Study of Pyrimidine (7)
The uses of pyrimidine (7) have not been reported in the literature. Based on the docking structure, 7 can be divided into three parts (Figure 3): the 4-substitute-morpholine part (R1), the 5-nitropyrimidine-2,4,6-triamine part (R2) in the center of compound, and the furan ring part (R3).
We performed a 2D similarity search of 7 using SciFinder to explore the contributions of the R1 and R3 regions, and 11 compounds (7_2d1–7_2d11) with > 65% similarity were selected for further study. We also performed a 3D similarity search using Shape Screening (Sastry et al., 2011) to look for new frameworks of the R2 region, and another 11 compounds (7_3d1–7_3d11) were identified.
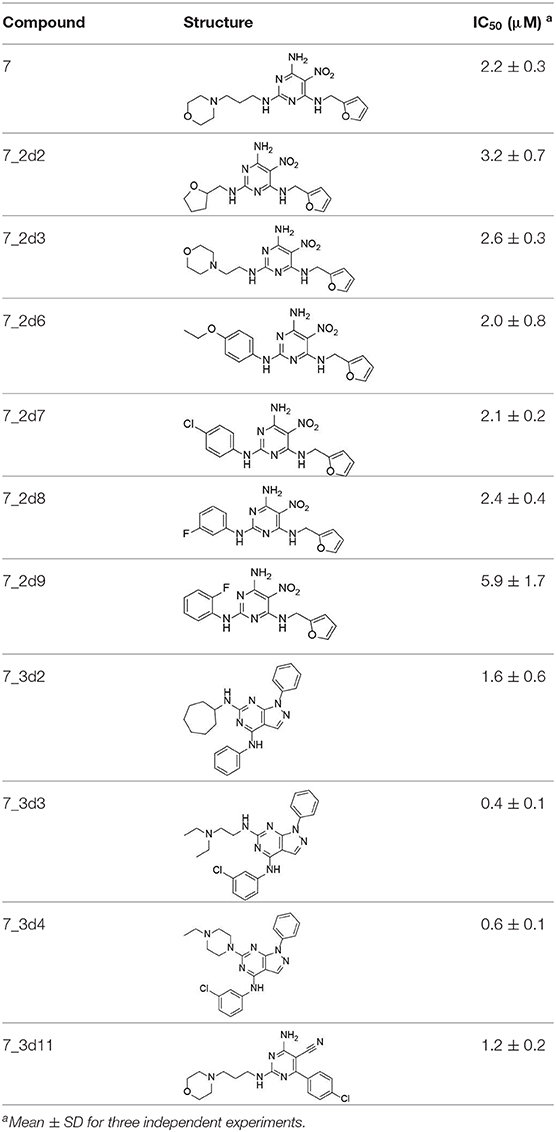
All 22 derivatives of 7 were tested at 5 μM with the dual-luciferase reporter cell assay (Table S6). Results showed some compounds inhibited Hh signaling. The IC50 values for the potent compounds are as shown in Table 3.
In the R1 region, changing 3-morpholinopropan-1-azyl to hydrogen (from 7 to 7_2d1) resulted in a significant decrease in inhibitory activity while keep R2 and R3 unchanged. The number of carbons in the linker appears to be of great importance, based on the decreasing trend of potency from 7 to 7_2d2 and 7_2d3. Replacement of 3-morpholinopropan-1-azyl by 4-substitute-benzene (7_2d6 and 7_2d7) did not alter inhibitory activity, which was maintained at a level similar to that of 7. Replacement of 3-morpholinopropan-1-azyl by 3-fluorobenzene (7_2d8) or 2-fluorobenzene (7_2d9) reduced potency, two times lower potency was observed when 2-position fluorobenzene was used as the replacement (7_2d9).
In the R2 region, when the central framework was changed to fused 1H-pyrazolo[3,4-d]pyrimidine-4,6-diamine (from 7_3d1 to 7_3d6), the inhibitory activities were greatly increased. Strikingly, the IC50 value of 7_3d3 reached at about 0.4 μM, which is currently the most active small molecule. In 7_3d3, the hydrogen atom of the amino-group was replaced by a large hydrophobic halogenated benzene (Figure S2). The fused ring, 1H-pyrazolo[3,4-d]pyrimidine presents a novel framework of the inhibitors. The other five compounds identified through 3D similarity search have central frameworks that are similar to that of 5-nitropyrimidine-2, 4, 6-triamine, among which only 7_3d11 showed better potency.
In the R3 region, changing furan in 7 to tetrahydrofuran (7_2d10) decreased the potency. In addition, changing furan in 7 to phenyl (7_2d11) caused toxicity.
Compared to the reported small-molecular-weight ShhN inhibitors, the IC50 of compound 7_3d3 is about 10 times stronger than robotnikinin (IC50 measured using the same assay in the present study); HL2-m5, a macrocycle peptide with a molecular weight of 1633 and an IC50 of 0.25 μM (determined using a similar cell-based assay as robotnikinin), 7_3d3 is much smaller with a molecular weight <500, and the non-cyclic structure makes it more suitable for further optimization.
To gain further insight into the effects of inhibitors against ShhN, a NanoDSF experiment was conducted. The results indicated that 7_3d3 improved the thermal stability of ShhN better than 7 (Figure S3), indicating an interaction of 7_3d3 toward ShhN in vitro.
Peng et al. (2009) reported that in different jobs, robotnikinin caused a significant decrease in mRNA Gli1 expression. If it is verified by a similar method, the selectivity of our compound to sonic hedgehog would be illustrated.
Ptch is attracting more attention by its association with the most common cancer in humans. The structure of Ptch1 was reported recently (Gong et al., 2018; Qi et al., 2018; Zhang et al., 2018; Qian et al., 2019) and our small molecule inhibitors of Shh/Ptch provide important clues for future investigation of Hh signaling.
Conclusion
In summary, using computational docking screen, SPR, MST, NanoDSF experiment and cellular assays, we discovered seven novel ShhN inhibitors with diverse scaffolds. The inhibitors bind ShhN with micromole affinity. At the cellular level, the compounds showed submicromolar to micromolar inhibition activities. In addition, all of these compounds passed the PAINS filter. We also established the structure-activity relationship for the pyrimidine series of compounds and identified the most potent compound with an IC50 of 0.4 μM in cellular assays.
Our study demonstrates the feasibility of developing novel inhibitors in the upstream of Hh signaling pathway, and presents an example of discovering and developing PPI inhibitors.
Data Availability Statement
All datasets generated for this study are included in the article/Supplementary Material.
Author Contributions
TY, WT, and YL searched literature, designed the research, and wrote the manuscript. TY, JW, JY, and WH performed the experiments, collected data, and analyzed the data. LL provided scientific suggestions and contributed to the manuscript revision. WT and YL supervised the project. All authors contributed to the article and approved the submitted version.
Funding
This work was supported in part by the National Natural Science Foundation of China (Grant Nos. 21877003, 21573012, 21633001, 81573452, 81773767) and the Ministry of Science and Technology of China (2016YFA0502303, 2015CB910300).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Prof. Chuanmao Zhang for providing plasmids and engaging in scientific discussions during the course of this project, and NanoTemper Inc. for providing a demo machine.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fchem.2020.00498/full#supplementary-material
References
Baell, J. B., and Holloway, G. A. (2010). New substructure filters for removal of pan assay interference compounds (PAINS) from screening libraries and for their exclusion in bioassays. J. Med. Chem. 53, 2719–2740. doi: 10.1021/jm901137j
Beauchamp, E. M., Ringer, L., Bulut, G., Sajwan, K. P., Hall, M. D., Lee, Y. C., et al. (2011). Arsenic trioxide inhibits human cancer cell growth and tumor development in mice by blocking Hedgehog/GLI pathway. J. Clin. Invest. 121, 148–160. doi: 10.1172/JCI42874
Berman, D. M., Karhadkar, S. S., Maitra, A., Oca, R. M. D., Gerstenblith, M. R., Briggs, K., et al. (2003). Widespread requirement for hedgehog ligand stimulation in growth of digestive tract tumors. Nature 425, 846–851. doi: 10.1038/nature01972
Bosanac, I., Maun, H. R., Scales, S. J., Wen, X. H., Lingel, A., Bazan, J. F., et al. (2009). The structure of SHH in complex with HHIP reveals a recognition role for the shh pseudo active site in signaling. Nat. Struct. Mol. Biol. 16, 691–U628. doi: 10.1038/nsmb.1632
Briscoe, J., and Therond, P. P. (2013). The mechanisms of hedgehog signalling and its roles in development and disease. Nat. Rev. Mol. Cell Biol. 14, 416–429. doi: 10.1038/nrm3598
Francesca, G., Mattia, M., and Paola, I. (2018). Current trends in Hedgehog signaling pathway inhibition by small molecules. Bioorg. Med. Chem. Lett. 28, 3131–3140. doi: 10.1016/j.bmcl.2018.08.033
Friesner, R. A., Banks, J. L., Murphy, R. B., Halgren, T. A., Klicic, J. J., Mainz, D. T., et al. (2004). Glide: a new approach for rapid, accurate docking and scoring. 1. Method and assessment of docking accuracy. J. Med. Chem. 47, 1739–1749. doi: 10.1021/jm0306430
Friesner, R. A., Murphy, R. B., Repasky, M. P., Frye, L. L., Greenwood, J. R., Halgren, T. A., et al. (2006). Extra precision glide: docking and scoring incorporating a model of hydrophobic enclosure for protein-ligand complexes. J. Med. Chem. 49, 6177–6196. doi: 10.1021/jm051256o
Galperin, I., Dempwolff, L., Diederich, W., and Lauth, M. (2019). Inhibiting hedgehog - an update on pharmacological compounds and targeting strategies. J. Med. Chem. 62, 8392–8411. doi: 10.1021/acs.jmedchem.9b00188
Gong, X., Qian, H., Cao, P., Zhao, X., Zhou, Q., Lei, J., et al. (2018). Structural basis for the recognition of sonic hedgehog by human Patchedl. Science 361:aas8935. doi: 10.1126/science.aas8935
Halgren, T. A., Murphy, R. B., Friesner, R. A., Beard, H. S., Frye, L. L., Pollard, W. T., et al. (2004). Glide: a new approach for rapid, accurate docking and scoring. 2. enrichment factors in database screening. J. Med. Chem. 47, 1750–1759. doi: 10.1021/jm030644s
Hu, L. P., Lin, X. Y., Lu, H., Chen, B. C., and Bai, Y. H. (2015). An overview of hedgehog signaling in fibrosis. Mol. Pharmacol. 87, 174–182. doi: 10.1124/mol.114.095141
Ingham, P. W., Nakano, Y., and Seger, C. (2011). Mechanisms and functions of Hedgehog signalling across the metazoa. Nat. Rev. Genet. 12, 393–406. doi: 10.1038/nrg2984
Kong, Y., Peng, Y. Q., Liu, Y., Xin, H., Zhan, X., and Tan, W. F. (2015). Twist1 and snail link hedgehog signaling to tumor-initiating cell-like properties and acquired chemoresistance independently of ABC transporters. Stem Cells 33, 1063–1074. doi: 10.1002/stem.1955
Lum, L., and Beachy, P. A. (2004). The Hedgehog response network: sensors, switches, and routers. Science 304, 1755–1759. doi: 10.1126/science.1098020
Maun, H. R., Wen, X. H., Lingel, A., de Sauvage, F. J., Lazarus, R. A., Scales, S. J., et al. (2010). Hedgehog pathway antagonist 5e1 binds hedgehog at the pseudo-active site. J. Biol. Chem. 285, 26570–26580. doi: 10.1074/jbc.M110.112284
Merchant, A. A., and Matsui, W. (2010). Targeting hedgehog - a cancer stem cell pathway. Clin. Cancer Res. 16, 3130–3140. doi: 10.1158/1078-0432.CCR-09-2846
Oro, A. E., Higgins, K. M., Hu, Z. L., Bonifas, J. M., Epstein, E. H., and Scott, M. P. (1997). Basal cell carcinomas in mice overexpressing sonic hedgehog. Science 276, 817–821. doi: 10.1126/science.276.5313.817
Owens, A. E., Paola, I. D., Hansen, W. A., Liu, Y. W., Khare, S. D., and Fasan, R. (2017). Design and evolution of a macrocyclic peptide inhibitor of the sonic hedgehog/patched interaction. J. Am. Chem. Soc. 139, 12559–12568. doi: 10.1021/jacs.7b06087
Peng, L. F., Stanton, B. Z., Maloof, N., Wang, X., and Schreiber, S.L. (2009). Syntheses of aminoalcohol-derived macrocycles leading to a small-molecule binder to and inhibitor of Sonic Hedgehog. Bioorg. Med. Chem. Lett. 19, 6319–6325, doi: 10.1016/j.bmcl.2009.09.089
Pepinsky, R. B., Rayhorn, P., Day, E. S., Dergay, A., Williams, K. P., Galdes, A., et al. (2000). Mapping sonic hedgehog-receptor interactions by steric interference. J. Biol. Chem. 275, 10995–11001. doi: 10.1074/jbc.275.15.10995
Qi, X., Schmiege, P., Coutavas, E., Wang, J., and Li, X. (2018). Structures of human patched and its complex with native palmitoylated sonic hedgehog. Nature 560, 128–132. doi: 10.1038/s41586-018-0308-7
Qian, H., Cao, P., Hu, M., Gao, S., Yan, N., and Gong, X. (2019). Inhibition of tetrameric patched1 by sonic hedgehog through an asymmetric paradigm. Nat. Commun. 10:2320. doi: 10.1038/s41467-019-10234-9
Rubin, L. L., and de Sauvage, F. J. (2006). Targeting the hedgehog pathway in cancer. Nat. Rev. Drug Discov. 5, 1026–1033. doi: 10.1038/nrd2086
Sasaki, H., Nishizaki, Y., Hui, C. C., Nakafuku, M., and Kondoh, H. (1999). Regulation of Gli2 and Gli3 activities by an amino-terminal repression domain: implication of Gli2 and Gli3 as primary mediators of Shh signaling. Development 126, 3915–3924.
Sastry, G. M., Dixon, S. L., and Sherman, W. (2011). Rapid shape-based ligand alignment and virtual screening method based on atom/feature-pair similarities and volume overlap scoring. J. Chem. Inform. Model. 51, 2455–2466. doi: 10.1021/ci2002704
Scales, S. J., and de Sauvage, F. J. (2009). Mechanisms of hedgehog pathway activation in cancer and implications for therapy. Trends Pharmacol. Sci. 30, 303–312. doi: 10.1016/j.tips.2009.03.007
Stanton, B. Z., Peng, L. F., Maloof, N., Nakai, K., Wang, X., Duffner, J. L., et al. (2009). A small molecule that binds Hedgehog and blocks its signaling in human cells. Nat. Chem. Biol. 5, 154–156. doi: 10.1038/nchembio.142
Wetmore, C., Eberhart, D. E., and Curran, T. (2001). Loss of p53 but not ARF Accelerates Medulloblastoma in mice heterozygous for patched. Cancer Res. 61, 513–516.
Xie, J. W., Murone, M., Luoh, S. M., Ryan, A., Gu, Q. M., Zhang, C. H., et al. (1998). Activating smoothened mutations in sporadic basal-cell carcinoma. Nature 391, 90–92. doi: 10.1038/34201
Zhan, X., Wang, J., Liu, Y., Peng, Y. Q., and Tan, W. F. (2014). GPCR-like signaling mediated by smoothened contributes to acquired chemoresistance through activating Gli. Mol. Cancer 13:4. doi: 10.1186/1476-4598-13-4
Keywords: hedgehog signaling pathway, N-terminal product of sonic hedgehog, Shh/Ptch interface, virtual screening, small molecule inhibitor, structure activity analysis
Citation: Yun T, Wang J, Yang J, Huang W, Lai L, Tan W and Liu Y (2020) Discovery of Small Molecule Inhibitors Targeting the Sonic Hedgehog. Front. Chem. 8:498. doi: 10.3389/fchem.2020.00498
Received: 12 December 2019; Accepted: 14 May 2020;
Published: 16 June 2020.
Edited by:
Giulia Morra, Istituto di Scienze e Tecnologie Chimiche Giulio Natta (SCITEC), ItalyReviewed by:
Jun-Seok Lee, Korea Institute of Science and Technology (KIST), South KoreaSalvatore Di Maro, Second University of Naples, Italy
Copyright © 2020 Yun, Wang, Yang, Huang, Lai, Tan and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wenfu Tan, d2Z0YW4mI3gwMDA0MDtmdWRhbi5lZHUuY24=; Ying Liu, bGl1eWluZyYjeDAwMDQwO3BrdS5lZHUuY24=
 Taikangxiang Yun1
Taikangxiang Yun1 Juan Wang
Juan Wang Jun Yang
Jun Yang Luhua Lai
Luhua Lai Wenfu Tan
Wenfu Tan Ying Liu
Ying Liu