- Department of Applied Chemistry, College of Science, Northeast Agricultural University, Harbin, China
4-Hydroxyphenylpyruvate dioxygenase (HPPD) is one of the most vital targets for herbicides discovery. In search for HPPD inhibitors with novel scaffolds, a series of aryl-naphthyl methanone derivatives have been designed and synthesized through alkylation and Friedel-Crafts acylation reactions. The bioassay indicated some of these compounds displayed preferable herbicidal activity at the rate of 0.75 mmol/m2 by post-emergence application, in which compound 3h displayed the best herbicidal activity. The molecular docking showed that compound 3h could bind well to the active site of the AtHPPD. This study shows that aryl-naphthyl methanone derivatives could be a potential lead structure for further development of novel herbicides.
Introduction
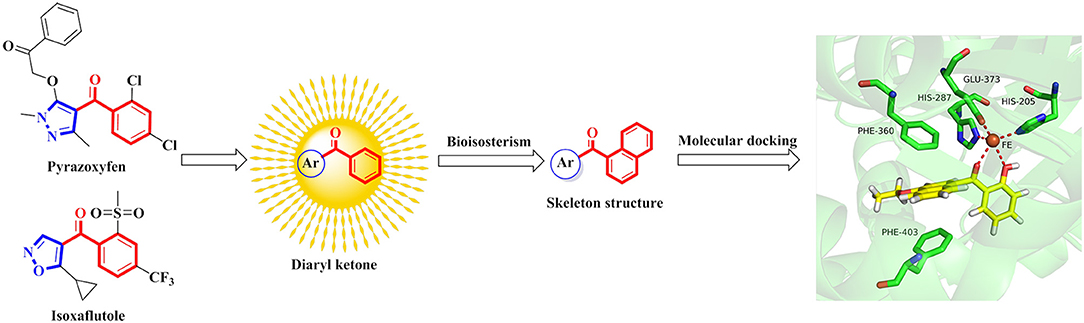
4-Hydroxyphenylpyruvate dioxygenase (EC 1.13.11.27, HPPD) is an iron-dependent, nonheme oxygenase present in most organisms. It is involved in the second step of the tyrosine degradation pathway, and catalyses the conversion of 4-hydroxyphenyl pyruvic acid (HPPA) into homogentisic acid (HGA) (Xu et al., 2014; Wang et al., 2015b). HGA is the intermediate of tyrosine catabolism and the precursor of the plastoquinone and tocopherols. The plastoquinone plays an important role in assisting phytoene desaturase to catalyse and synthesize the carotenoid. HPPD inhibiting-based herbicides cause low level of the carotenoid, which indirectly affects the photosynthesis. When exposed to sunlight, plants will develop into albinism followed by gangrene and death treated with HPPD inhibitors (Mitchell et al., 2001; Moran, 2005; Wojcik et al., 2014). There are many advantages of HPPD inhibitors, such as excellent crop selectivity, low application rate, low toxicity, broad-spectrum weed control, and benign environmental effects (Beaudegnies et al., 2009; Elmore et al., 2011; Laschi et al., 2016; Wang et al., 2016).
Up to now, approximately 15 HPPD inhibitors have been commercialized, which are primarily assorted into three classifications: triketones, pyrazoles, and isoxazoles (Zhu et al., 2005; Witschel, 2009; Wang et al., 2015b). Among the commercial HPPD herbicides, pyrazole and isoxazole derivatives have been studied diffusely due to their structural diversity. The first commercial pyrazole product, pyrazolynate, was launched by Sankyo in 1980 for controlling annual and perennial weeds in rice at a dose of 3–4 kg ai/ha (Kubo et al., 2012). Five years later, Ishihara Sangyo Kaisha commercialized the second pyrazole herbicide, pyrazoxyfen, which was applied in irrigated fields for broad-leaved weeds, was used at rates of 3 kg ai/ha (He et al., 2017). In 1990, the isoxaflutole was registered by BASF, using in corn field for selective pre-emergence herbicide at a dose of 120–150 g ai/ha (Luscombe and Pallett, 1996). Nearly, the ultra-efficient herbicide, topramezone, was commercialized by BASF as a postemergence herbicide with excellent selectivity to control all major grasses and broadleaf weeds in corn at a dose of 12–75 g ai/ha (Rene et al., 2003).
The commercialized herbicide pyrazoxyfen and isoxaflutole were common HPPD inhibitors, and it was found that the active substructure was mainly based on diaryl ketone (Figure 1). Many studies on HPPD inhibitors have proven that improved conjugated system of diaryl ketone will increase the π-π interaction, and enhance the chelation with Fe2+ to improve herbicidal activity (Lee et al., 1997; Ahrens et al., 2013; Santucci et al., 2017). For example, the HPPD inhibitor triketone-quinoline exhibited promising and broad-spectrum herbicidal activity at the rate of 150 g ai/ha with increased conjugated system by introducing quinoline group (Wang et al., 2015a). Furthermore, a series of pyrazole-quinoline compounds were included in the BASF patent, which exhibited well weed-inhibiting activity at the dosage of 63–125 g ai/ha (He et al., 2017). As a part of our job in designing and synthesizing novel HPPD inhibitors (Fu et al., 2017a,b, 2018), herein, the skeleton structure was obtained by replacing benzene with naphthalene based on bioisosterism. And the aromatic part was modified with different aromatic heterocycles. Through this strategy, we hoped that the better herbicidal activity would be attained. Twenty-three aryl-naphthyl methanone derivatives were synthesized through alkylation and Friedel-Crafts acylation reactions. The greenhouse experiments showed that compound 3h at a dose of 2.1 kg ai/ha has better herbicidal activity than pyrazoxyfen at rates of 3 kg ai/ha, and most of the synthesized compounds showed “good” to “excellent” herbicidal activity against barnyard grass at rates of 2–4 kg ai/ha.
Materials and Methods
Chemicals and Instruments
All the solvents and reactants were analytical pure and applied without purification. Analytical thin-layer chromatography (TLC) was exercised in silica gel GF254. Column chromatographic purification was carried out using silica gel. Yields were not optimized. The melting point was gauged on a Beijing Taike point instrument (X-4) and was uncorrected. The infrared (IR) spectra were collected on a Bruker ALPHA-T spectrometer (in KBr pallets). The nuclear magnetic resonance (NMR) spectrum were collected on a Bruker AV-400 spectrometer using CDCl3 as solvent and tetramethylsilane (TMS) as internal standard. The high-resolution mass spectrum was collected on a high resolution mass spectrometer (HRMS) of Bruker. Crystallographic data of the compound was measured on a Rigaku R-AXIS RAPID area-detector diffractometer.
General Procedure for Preparation of Intermediate Naphthyl Ether (2a and 2b) (Momeni, 2007)
The naphthol 1a or 1b (2 mmol) and anhydrous K2CO3 (14 mmol) were ground together into fine powder and stirred vigorously at 60°C in 100 mL flask. Alkylating agent (dimethyl sulfate or diethyl sulfate, 3 mmol) was added directly and heated with vigorous stirring for 2 h, and the TLC was used to monitor reaction progress. Water was added to the mixture after the reaction finished, then the solid was filtrated and washed with water. Collect the liquid, the products were extracted with diethyl ether, and the solvent was evaporated. The crude product was purified by column chromatography with petroleum ether and EtOAc (V:V=10:1) as eluent or by recrystallization in EtOAc-n-hexane (in the case of solid ethers). The spectral data for intermediate 2a and 2b are available in the Supporting Information.
General Procedure for Preparation of Aryl-Naphthyl Methanone 3a-w
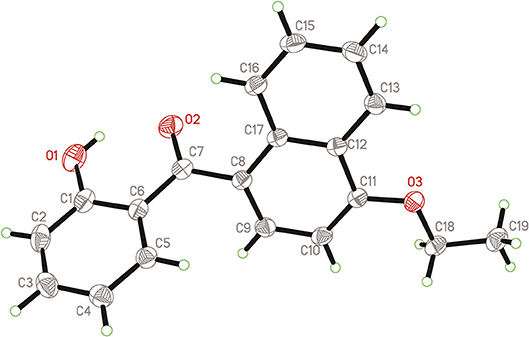
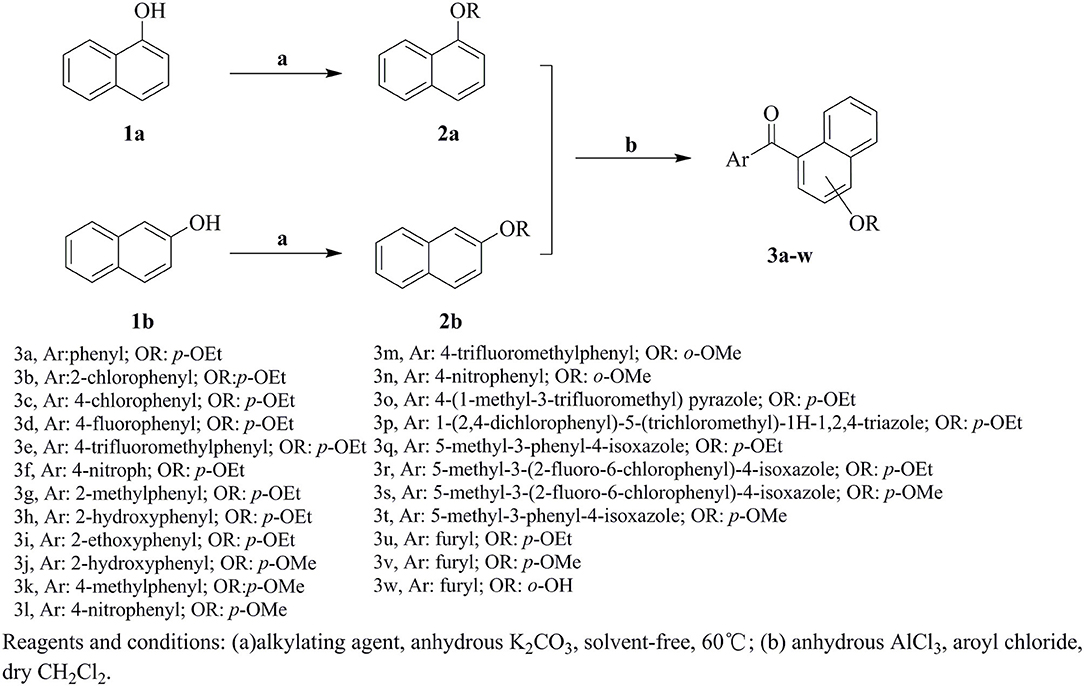
Naphthyl ether (10 mmol) and aroyl chloride (12 mmol) were mixed in dry CH2Cl2 (50 mL) at 0°C, and adding anhydrous AlCl3 (22 mmol) to the solution with stirring. Then the mixture was refluxed and stirred for additional 2 h. After the reaction was completed, the solvent was removed. Then 10% HCl (50 mL) were added to the mixture and stirred vigorously for 30 min, the crude product was obtained after filtration. The crude products were purified by column chromatography with petroleum ether-EtOAc (V:V = 15:1) as eluent or by recrystallization in EtOAc-n-hexane. In addition, compound 3h was recrystallized from a mixture of ethyl acetate and petroleum ether to afford a suitable crystal and its structure was determined by single crystal X-ray diffraction analysis. The molecular structure of compound 3h with atom-numbering is shown in Figure 2.
Compounds Data
(4-Ethoxynaphthalen-1-yl)(Phenyl)Methanone (3a)
White solid; yield, 65%; mp, 109–110°C; IR (KBr, cm−1) ν: 3073-2893 (C-H), 1651 (C = O), 1579-1445 (C=C); 1H NMR (400 MHz, CDCl3, ppm) δ: 8.37-8.47 (d, J = 9.8 Hz, 2H, Ar-H), 7.89-7.84 (d, J = 7.0 Hz, 2H, Ar-H), 7.63-7.45 (m, 6H, Ar-H), 6.84-6.74 (d, J = 8.1 Hz, 1H, Ar-H), 4.26-4.33 (m, 2H, O-CH2), 1.59-1.64 (t, J = 9.2 Hz, 3H, -CH3); 13C NMR (100 MHz, CDCl3, ppm) δ: 197.36, 157.75, 139.53, 132.71, 132.46, 131.63, 131.63, 130.30, 130.30, 128.26, 128.01, 128.01, 127.83, 125.86, 125.73, 122.38, 102.62, 64.11, 14.74. HRMS (ESI): m/z [M+H+] calculated for monoisotopic mass 277.1150, found 277.1225.
(2-Chlorophenyl)(4-ethoxynaphthalen-1-yl)methanone (3b)
Yellow solid; yield, 59%; mp, 143–144°C; IR (KBr, cm−1) ν: 3056-2894 (C-H), 1643 (C=O), 1578-1429 (C=C); 1H NMR (400 MHz, CDCl3, ppm) δ: 8.55-8.39 (m, 2H, Ar-H), 8.10-7.83 (m, 4H, Ar-H), 7.61-7.64 (m, 3H, Ar-H), 6.79-6.81 (d, J = 8.1 Hz, 1H, Ar-H), 4.27-4.34 (m, 2H, O-CH2), 1.59-1.64 (t, J = 9.2 Hz, 3H, -CH3); 13C NMR (100 MHz, CDCl3, ppm) δ: 196.03, 158.00, 138.91, 137.90, 132.64, 131.68, 131.68, 131.57, 128.62, 128.62, 128.16, 127.46, 125.93, 125.88, 125.63, 122.48, 102.66, 64.20, 14.75. HRMS (ESI): m/z [M+H+] calculated for monoisotopic mass 311.0761, found 311.0836.
(4-Chlorophenyl)(4-ethoxynaphthalen-1-yl)methanone (3c)
White solid; yield, 53%; mp, 138–140°C; IR (KBr, cm−1) ν: 3055-2893 (C-H), 1643 (C = O), 1578-1429 (C=C); 1H NMR (400 MHz, CDCl3, ppm) δ: 8.35-8.43 (m, 2H, Ar-H), 7.79-7.81 (d, J = 8.1 Hz, 2H, Ar-H), 7.44-7.60 (m, 5H, Ar-H), 6.79-6.81 (d, J = 8.1 Hz, 1H, Ar-H), 4.28-4.35 (m, 2H, O-CH2), 1.60-1.64 (t, J = 9.2 Hz, 3H, -CH3); 13C NMR (100 MHz, CDCl3, ppm) δ: 195.85, 158.34, 147.21, 141.20, 132.67, 132.56, 130.20, 130.20, 128.41, 128.41, 126.90, 125.97, 125.93, 125.62, 125.55, 122.49, 102.57, 64.23, 14.71. HRMS (ESI): m/z [M+H+] calculated for monoisotopic mass 311.0761, found 311.0836.
(4-Ethoxynaphthalen-1-yl)(4-fluorophenyl)methanone (3d)
Yellow solid; yield, 57%; mp, 117–119°C; IR (KBr, cm−1) ν: 3076-2893 (C-H), 1643(C=O), 1578-1430 (C = C); 1H NMR (400 MHz, CDCl3, ppm) δ: 7.86-8.43 (m, 4H, Ar-H), 7.56-7.59 (m, 3H, Ar-H), 7.09-7.17 (m, 2H, Ar-H), 6.78-6.80 (d, J = 8.1 Hz, 1H, Ar-H), 4.25-4.32 (m, 2H, O-CH2), 1.59-1.63 (t, J = 9.2 Hz, 3H, -C-CH3); 13C NMR (100 MHz, CDCl3, ppm) δ: 199.55, 158.77, 140.99, 137.06, 134.84, 132.73, 130.98, 130.13, 129.12, 128.75, 127.53, 126.01, 125.92, 125.87, 125.32, 122,43, 102.74, 64.19, 14.70. HRMS (ESI): m/z [M+H+] calculated for monoisotopic mass 295.1056, found 295.1126.
(4-Ethoxynaphthalen-1-yl)(4-trifluoromethylphenyl)methanone (3e)
White solid; yield, 56%; mp, 155–156°C; IR (KBr, cm-1) ν: 3053-2887 (C-H), 1634 (C=O), 1572-1429 (C = C); 1H NMR (400 MHz, CDCl3, ppm) δ: 7.43-8.43 (m, 9H, Ar-H), 6.75-6.80 (d, J = 8.1 Hz, 1H, Ar-H), 4.28-4.35 (m, 2H, O-CH2), 1.60-1.64 (t, J = 9.2 Hz, 3H, -C-CH3); 13C NMR (100 MHz, CDCl3, ppm) δ: 195.85, 158.34, 147.21, 141.20, 132.67, 132.56, 132.56, 130.39, 130.20, 128.41, 128.41, 126.90, 125.97, 125.93, 125.62, 125.55, 122.49, 102.57, 64.23, 14.71. HRMS (ESI): m/z [M+H+] calculated for monoisotopic mass 345.1024, found 345.1094.
(4-Ethoxynaphthalen-1-yl)(4-nitrophenyl)methanone (3f)
Yellow solid; yield, 61%; mp, 152–153°C; IR (KBr, cm−1) ν: 3053-2887 (C-H), 1634 (C=O), 1572-1429 (C=C); 1H NMR (400 MHz, CDCl3, ppm) δ: 8.31-8.57 (m, 4H, Ar-H), 7.95-7.98 (m, 2H, Ar-H), 7.56-7.67 (m, 3H, Ar-H), 6.78-6.81 (d, J = 8.2 Hz, 1H, Ar-H), 4.28-4.35 (m, 2H, O-CH2), 1.60-1.65 (t, J = 9.2 Hz, 3H, -C-CH3); 13C NMR (100 MHz, CDCl3, ppm) δ: 195.12, 158.88, 149.75, 145.08, 133.40, 132.64, 132.64, 130.91, 128.73, 128.73, 126.16, 126.12, 125.95, 125.54, 123.47, 122.60, 102.59, 64.35, 14.69. HRMS (ESI): m/z [M+H+] calculated for monoisotopic mass 322.1001, found 322.1070.
(4-Ethoxynaphthalen-1-yl)(2-methylphenyl)methanone (3g)
Yellow solid; yield, 50%; mp, 126–127°C; IR (KBr, cm−1) ν: 3067-2939 (C-H), 1639 (C=O), 1573-1429 (C=C); 1H NMR (400 MHz, CDCl3, ppm) δ: 8.99-9.02 (d, J = 8.5Hz, 1H, Ar-H), 8.42-8.45 (m, 1H, Ar-H), 7.25-7.69 (m, 7H, Ar-H), 6.78-6.71 (d, J = 8.2 Hz, 1H, Ar-H), 4.22-4.29 (m, 2H, O-CH2), 2.41 (s, 3H, -CH3), 1.57-1.62 (t, J = 9.2 Hz, 3H, -C-CH3); 13C NMR (100 MHz, CDCl3, ppm) δ: 199.55, 158.77, 140.99, 137.06, 134.84, 132.73, 130.98, 130.13, 129.12, 128.75, 127.53, 126.01, 125.92, 125.87, 125.32, 122,43, 102.74, 64.19, 20.20, 14.70. HRMS (ESI): m/z [M+H+] calculated for monoisotopic mass 291.1307, found 291.1375.
(4-Ethoxynaphthalen-1-yl)(2-hydroxyphenyl)methanone (3h)
Yellow solid; yield, 61%; mp, 125–127°C; IR (KBr, cm−1) ν: 3272 (-OH), 3051-2932 (C-H), 1624 (C = O), 1575-1443 (C = C); 1H NMR (400 MHz, CDCl3, ppm) δ: 12.43 (s, 1H, -OH), 7.99-8.54 (m, 2H, Ar-H), 7.47-7.57 (m, 5H, Ar-H), 6.79-7.13 (m, 3H, Ar-H), 4.28-4.34 (m, 2H, O-CH2), 1.61-1.65 (t, J = 6.8Hz, 3H, -C-CH3); 13C NMR (100 MHz, CDCl3, ppm) δ: 202.94, 163.42, 157.34, 136.43, 133.95, 132.03, 129.40, 127.87, 127.53, 125.85, 125.81, 125.23, 122.57, 120.78, 118.65, 118.31, 102.81, 64.15, 14.78. HRMS (ESI): m/z [M+H+] calculated for monoisotopic mass 293.1099, found 293.1175.
(4-Ethoxynaphthalen-1-yl)(2-ethoxyphenyl)methanone (3i)
Yellow solid; yield, 32%; mp, 146–147°C; IR (KBr, cm−1) ν: 3079-2865 (C-H), 1617 (C=O), 1583-1439 (C = C); 1H NMR (400 MHz, CDCl3, ppm) δ: 8.81-8.93 (d, J = 8.5 Hz, 1H, Ar-H), 8.30-8.43 (d, J = 8.5 Hz, 1H, Ar-H), 7.36-7.72 (m, 5H, Ar-H), 6.60-7.10 (m, 3H, Ar-H), 4.22-4.27 (m, 2H, O-CH2), 3.85-3.91 (m, 2H, O-CH2), 1.55-1.59 (t, J = 6.8Hz, 3H, -CH3), 0.92-0.96 (t, J = 6.8Hz, 3H, -CH3); 13C NMR (100 MHz, CDCl3, ppm) δ: 197.31, 158.27, 157.13, 133.51, 132.48, 131.87, 131.35, 130.09, 128.64, 128.34, 125.94, 125.69, 125.56, 122.17, 120.42, 112.88, 102.73, 64.18, 64.08, 14.71, 14.34. HRMS (ESI): m/z [M+H+] calculated for monoisotopic mass 321.1412, found 321.1490.
(2-Hydroxyphenyl)(4-ethoxynaphthalen-1-yl)methanone (3j)
Yellow solid; yield, 69%; mp, 108–110°C; IR (KBr, cm−1) ν: 3032-2917 (C-H), 1606(C=O), 1567-1431 (C=C); 1H NMR (400 MHz, CDCl3, ppm) δ: 12.41 (s, 1H, -OH), 8.35-8.41 (m, 1H, Ar-H), 8.03-8.09 (m, 1H, Ar-H), 7.45-7.59 (m, 5H, Ar-H), 7.10-7.14 (d, J = 0.8 Hz, 1H, Ar-H), 6.78-6.90 (m, 2H, Ar-H), 4.11 (s, 3H, O-CH3); 13C NMR (100 MHz, CDCl3, ppm) δ: 202.94, 163.42, 157.93, 136.47, 133.93, 131.97, 129.20, 127.91, 127.82, 125.97, 125.72, 125.26, 122.44, 120.75, 118.66, 118.32, 102.17, 55.85. HRMS (ESI): m/z [M+H+] calculated for monoisotopic mass 279.0943, found 279.1018.
(4-Methoxynaphthalen-1-yl)(4-methylphenyl)methanone (3k)
White solid; yield, 80%; mp, 122–123°C; IR (KBr, cm−1) ν: 3047-2819 (C-H), 1633 (C=O), 1565-1443 (C=C); 1H NMR (400 MHz, CDCl3, ppm) δ: 8.27-8.43 (m, 2H, Ar-H), 7.73-7.84 (d, J = 8.1 Hz, 2H, Ar-H), 7.53-7.62 (m, 3H, Ar-H), 7.25-7.32 (d, J = 8.0 Hz, 2H, Ar-H), 6.75-6.87 (d, J = 8.0 Hz, 1H, Ar-H), 4.09 (s, 3H, O-CH3), 2.46 (s, 3H, CH3); 13C NMR (100 MHz, CDCl3, ppm) δ: 197.15, 158.08, 143.44, 136.70, 132.59, 130.75, 130.54, 130.54, 129.01, 129.01, 128.58, 127.90, 125.80, 125.77, 122.24, 122.24, 102.01, 55.79, 21.72. HRMS (ESI): m/z [M+H+] calculated for monoisotopic mass 277.1150, found 277.1225.
(4-Methoxynaphthalen-1-yl)(4-nitrophenyl)methanone (3l)
Brown solid; yield, 52%; mp, 105–107°C; IR (KBr, cm−1) ν: 3060-2923 (C-H), 1629 (C=O), 1561-1431 (C=C); 1H NMR (400 MHz, CDCl3, ppm) δ: 8.26-8.63 (m, 4H, Ar-H), 7.89-8.03 (d, J = 8.8 Hz, 2H, Ar-H), 7.52-7.70 (m, 3H, Ar-H), 6.76-6.87 (d, J = 8.2 Hz, 1H, Ar-H), 4.11 (s, 3H, O-CH3); 13C NMR (100 MHz, CDCl3, ppm) δ: 195.12, 159.46, 149.78, 144.96, 133.22, 132.56, 130.93, 130.93, 128.76, 126.42, 126.28, 125.89, 125.56, 123.49, 123.49, 122.50, 102.01, 55.98. HRMS (ESI): m/z [M+H+] calculated for monoisotopic mass 308.0845, found 308.0919.
(4-Trifluoromethylphenyl)(2-methoxynaphthalen-1-yl)methanone(3m)
White solid; yield, 40%; mp, 119–120°C; IR (KBr, cm−1) ν: 3040-2820 (C-H), 1660 (C=O), 1566-1423 (C=C); 1H NMR (400 MHz, CDCl3, ppm) δ: 7.69-8.05 (m, 6H, Ar-H), 7.35-7.56 (m, 4H, Ar-H), 3.84(s, 3H, O-CH3); 13C NMR (100 MHz, CDCl3, ppm) δ: 196.75, 154.43, 148.01, 140.69, 131.89, 131.73, 131.61, 129.79, 129.61, 128.84, 128.30, 127.79, 125.93, 125.69, 125.65, 124.33, 123.74, 112.89, 56.48. HRMS (ESI): m/z [M+H+] calculated for monoisotopic mass 331.0868, found 331,0943.
(2-Methoxynaphthalen-1-yl)(4-nitrophenyl)methanone (3n)
Brown solid; yield, 59%; mp, 125–127°C; IR (KBr, cm−1) ν: 3060-2923 (C-H), 1629 (C=O), 1561-1431 (C=C); 1H NMR (400 MHz, CDCl3, ppm) δ: 8.31-8.57 (m, 4H, Ar-H), 7.95-7.98 (m, 2H, Ar-H), 7.56-7.67 (m, 3H, Ar-H), 6.78-6.81 (d, J = 8.2 Hz, 1H, Ar-H), 4.11 (s, 3H, O-CH3); 13C NMR (100 MHz, CDCl3, ppm) δ: 202.94, 163.42, 157.93, 136.47, 133.93, 131.97, 131.97, 129.19, 127.91, 125.97, 125.73, 125.26, 122.44, 120.76, 118.66, 118.32, 102.17, 55.84. HRMS (ESI): m/z [M+H+] calculated for monoisotopic mass 294.0688, found 294.0759.
(4-(1-Methyl-3-trifluoromethyl)pyrazole)(4-ethoxynaphthalen-1-yl)methanone (3o)
White solid; yield, 33%; mp, 109–110°C; IR (KBr, cm−1) ν: 3089-2941 (C-H), 1639 (C=O), 1573-1430 (C=C); 1H NMR (400 MHz, CDCl3, ppm) δ: 8.28-8.68 (m, 2H, Ar-H), 7.45-7.81 (m, 4H, Ar-H), 6.69-6.82 (d, J = 8.1 Hz, 1H, Ar-H), 4.25-4.32 (m, 2H, O-CH2), 3.97 (s, 3H, CH3), 1.58-1.63 (t, J = 9.2 Hz, 3H, -C-CH3); 13C NMR (100 MHz, CDCl3, ppm) δ: 188.42, 159.10, 136.79, 132.91, 132.19, 129.28, 129.14, 126.78, 126.65, 126.29, 123.56, 123.16, 119.66, 119.66, 103.28, 64.99, 40.53, 15.47. HRMS (ESI): m/z [M+H+] calculated for monoisotopic mass 349.1086, found 349.1162.
(3-(2,4-Dichlorophenyl)-5-(Trichloromethyl)-1,2,4-Triazole)(4-Ethoxynaphthalen-1-yl)Methanone (3p)
Brown solid; yield, 30%; mp, 166–167°C; IR (KBr, cm−1) ν: 3086-2887 (C-H), 1650 (C=O), 1572-1426 (C=C); 1H NMR (400 MHz, CDCl3, ppm) δ: 8.85-9.04 (d, J = 8.4 Hz, 1H, Ar-H), 8.18-8.46 (m, 2H, Ar-H), 7.37-7.77 (m, 5H, Ar-H), 6.78-6.91 (d, J = 8.4 Hz, 1H, Ar-H), 4.28-4.35 (m, 2H, O-CH2), 1.58-1.62 (t, J = 9.2 Hz, 3H, -C-CH3); 13C NMR (100 MHz, CDCl3, ppm) δ: 183.63, 158.99, 158.69, 154.62, 137.42, 135.17, 133.52, 132.99, 132.25, 130.19, 129.90, 128.40, 127.05, 125.34, 125.24, 125.03, 124.14, 121.78, 102.22, 84.78, 63.64, 14.00. HRMS (ESI): m/z [M+H+] calculated for monoisotopic mass 527.9529, found 527.9607.
(4-Ethoxynaphthalen-1-yl)(5-methyl-3-phenyl-4-isoxazole)methanone (3q)
White solid; yield, 71%; mp, 132–133°C; IR (KBr, cm−1) ν: 2987-2900 (C-H), 1638 (C=O), 1573-1443 (C=C); 1H NMR (400 MHz, CDCl3, ppm) δ: 8.13-8.96 (m, 2H, Ar-H), 7.12-7.74 (m, 8H, Ar-H), 6.48-6.67 (d, J = 8.2 Hz, 1H, Ar-H), 4.20-4.25 (m, 2H, O-CH2), 2.47 (s, 3H, CH3), 1.55-1.59 (t, J = 6.8 Hz, 3H, -C-CH3); 13C NMR (100 MHz, CDCl3, ppm) δ: 190.12, 172.64, 162.29, 159.02, 133.76, 132.21, 129.52, 128.95, 128.44, 128.44, 128.30, 128.30, 127.26, 126.03, 125.77, 125.29, 122.46, 117.58, 102.72, 102.72, 64.24, 14.61, 12.91. HRMS (ESI): m/z [M+H+] calculated for monoisotopic mass 358.1365, found 358.1443.
(5-Methyl-3-(2-fluoro-6-chlorophenyl)-4-isoxazole)(4-ethoxynaphthalen-1-yl)methanone (3r)
White solid; yield, 70%; mp, 179–180°C; IR (KBr, cm−1) ν: 3083-2883 (C-H), 1650 (C=O), 1576-1431 (C=C); 1H NMR (400 MHz, CDCl3, ppm) δ: 8.11-8.60 (m, 2H, Ar-H), 7.41-7.75 (m, 3H, Ar-H), 6.51-7.21 (m, 4H, Ar-H), 4.18-4.25 (m, 2H, O-CH2), 2.57 (s, 3H, CH3), 1.54-1.59 (t, J = 9.2 Hz, 3H, -C-CH3); 13C NMR (100 MHz, CDCl3, ppm) δ: 188.74, 173.34, 157.99, 155.20, 134.45, 134.40, 131.46, 131.33, 130.90, 130.78, 128.15, 127.05, 125.55, 125.27, 124.87, 121.91, 118.64, 113.79, 113.50, 102.13, 63.85, 14.32, 12.91. HRMS (ESI): m/z [M+H+] calculated for monoisotopic mass 410.0881, found 410.0961.
(5-Methyl-3-(2-fluoro-6-chlorophenyl)-4-isoxazole)(4-ethoxynaphthalen-1-yl)methanone (3s)
White solid; yield, 84%; mp, 140–142°C; IR (KBr, cm−1) ν: 3055-2821 (C-H), 1638 (C=O), 1562-1431 (C=C); 1H NMR (400 MHz, CDCl3, ppm) δ: 8.21-8.50 (m, 2H, Ar-H), 7.47-7.67 (m, 3H, Ar-H), 7.12-7.19 (m, 1H, Ar-H), 7.01-7.06 (d, J = 8.1 Hz, 1H, Ar-H), 6.59-6.87 (m, 2H, Ar-H), 4.01 (s, 3H, O-CH3), 2.58 (s, 3H, CH3); 13C NMR (100 MHz, CDCl3, ppm) δ: 189.10, 173.71, 161.68, 158.92, 155.53, 131.67, 131.54, 131.25, 131.16, 128.52, 127.59, 126.01, 125.46, 125.20, 125.17, 122.11, 118.91, 114.09, 113.87, 101.79, 55.83, 13.28. HRMS (ESI): m/z [M+H+] calculated for monoisotopic mass 396.0724, found 396.0800.
(4-Ethoxynaphthalen-1-yl)(5-methyl-3-phenyl-4-isoxazole)methanone (3t)
White solid; yield, 86%; mp, 155–156°C; IR (KBr, cm−1) ν: 3033-2919 (C-H), 1629 (C=O), 1562-1432 (C=C); 1H NMR (400 MHz, CDCl3, ppm) δ: 8.75-8.83 (d, J = 8.5 Hz, 1H, Ar-H), 8.26-8.35 (m, 1H, Ar-H), 7.45-7.73 (m, 5H, Ar-H), 7.25-7.30 (d, J = 4.1 Hz, 1H, Ar-H), 7.17-7.23 (d, J = 7.6 Hz, 2H, Ar-H), 6.56-6.66 (d, J = 8.2 Hz, 1H, Ar-H), 4.02 (s, 3H, O-CH3), 2.47 (s, 3H, CH3); 13C NMR (100 MHz, CDCl3, ppm) δ: 190.15, 172.73, 162.31, 159.61, 133.55, 132.13, 129.53, 129.53, 128.98, 128.44, 128.30, 127.58, 127.58, 126.15, 125.70, 125.31, 122.35, 117.56, 117.56, 102.09, 55.87, 12.93. HRMS (ESI): m/z [M+H+] calculated for monoisotopic mass 344.1208, found 344.1286.
(4-Ethoxynaphthalen-1-yl)(furan-2-yl)methanone (3u)
Yellow solid; yield, 61%; mp, 123–124°C; IR (KBr, cm−1) ν: 3126-2940 (C-H), 1631 (C=O), 1577-1428 (C=C); 1H NMR (400 MHz, CDCl3, ppm) δ: 8.31-8.47 (m, 2H, Ar-H), 7.47-7.89 (m, 4H, Ar-H), 7.02-7.13 (d, J = 3.3 Hz, 1H, Ar-H), 6.76-6.86 (d, J = 8.1 Hz, 1H, Ar-H), 6.54-6.62 (m, 1H, Ar-H), 4.26-4.33 (m, 2H, O-CH2), 1.58-1.63 (t, J = 9.2 Hz, 3H, -C-CH3); 13C NMR (100 MHz, CDCl3, ppm) δ: 183.66, 157.91, 153.40, 147.20, 132.29, 130.43, 128.09, 127.14, 125.87, 125.83, 125.28, 122.36, 120.59, 112.09, 102.61, 64.12, 14.74. HRMS (ESI): m/z [M+H+] calculated for monoisotopic mass 267.0943, found 267.1014.
(Furan-2-yl)(4-methoxynaphthalen-1-yl)methanone (3v)
Yellow solid; yield, 65%; mp, 133–134°C; IR (KBr, cm−1) ν: 3089-2916 (C-H), 1620 (C=O), 1567-1456 (C=C); 1H NMR (400 MHz, CDCl3, ppm) δ: 8.31-8.47 (m, 2H, Ar-H), 7.47-7.89 (m, 4H, Ar-H), 7.02-7.13 (d, J = 3.3 Hz, 1H, Ar-H), 6.76-6.86 (d, J = 8.1 Hz, 1H, Ar-H), 6.54-6.62 (m, 1H, Ar-H), 4.01 (s, 3H, O-CH3); 13C NMR (100 MHz, CDCl3, ppm) δ: 185.12, 157.24, 152.08, 147.65, 137.72, 134.55, 129.75, 129.73, 127.05, 126.32, 124.10, 121.68, 120.72, 112.66, 112.33. HRMS (ESI): m/z [M+H+] calculated for monoisotopic mass 253.0786, found 253.0861.
(Furan-2-yl)(2-hydroxylnaphthalen-1-yl)methanone (3w)
Yellow solid; yield, 43%; mp, 102–103°C; IR (KBr, cm−1) ν: 3220 (-OH), 3114-2904 (C-H), 1635 (C = O), 1566-1447 (C = C); 1H NMR (400 MHz, CDCl3, ppm) δ: 11.14 (s, 1H, -OH), 8.84 (s, 1H, Ar-H), 7.82-7.91 (m, 2H, Ar-H), 7.69-7.75 (d, J = 8.4 Hz, 1H, Ar-H), 7.46-7.59 (m, 2H, Ar-H), 7.33-7.41 (m, 2H, Ar-H), 6.68-6.75 (m, 1H, Ar-H); 13C NMR (100 MHz, CDCl3, ppm) δ: 185.12, 157.24, 152.08, 147.65, 137.72, 134.55, 129.75, 129.73, 127.05, 126.32, 124.10, 121.68, 120.72, 112.66, 112.33. HRMS (ESI): m/z [M+H+] calculated for monoisotopic mass 253.0786, found 253.1984.
X-Ray Diffraction Analysis
The cell dimensions and strengths of compound 3h (0.54 × 0.49 × 0.39 mm) were using a gauged at 298 K on Rigaku R-AXIS RAPID area detector diffractometer with graphite monochromated Mo-Kα radiation (λ = 0.71073 Å); θmax = 27.48; 13900 measured reflections, 3397 independent reflections (Rint = 0.0296). The structure was solved by direct method using SHELXS 97 and refined with SHELXL 97 (Sheldrick, 1997). Full-matrix least-squares refinement based on F2 using the weight of ω = 1/[σ2() + (0.0702P)2+0.0736P] gave final values of R = 0.0568, ωR = 0.1262, max/min residual electron density = 0.182 and−0.222 e.Å−3. Crystallographic data has been deposited at the Cambridge Crystallographic Data Center as supplementary publication number CCDC 1856302. Copies of the data can be obtained, free of charge, on application to CCDC, 12 Union Road, Cambridge CB21EZ, UK [www.ccdc.cam.ac.uk/data_request/cif].
As shown in Figure 2, compound 3h is compose of three rings, and the bond lengths and bond angles in the structure of 3h are in the usual ranges. It is obviously that there is π-π conjunctive effect between benzene ring, C7=O2 and naphthalene ring, which caused shorter bond length of C6-C7 [1.48(2) Å] and C7-C8 [1.49(2) Å] than the typical C-C bond length [1.54 Å]. The torsion angle of C11-O3-C18-C19 is 175.75(1)°, which indicated the ethoxy and the naphthalene ring are in a coplanar state. The molecule structure is almost at the same plane, due to the large conjugate system and torsion angle.
Biological Assays
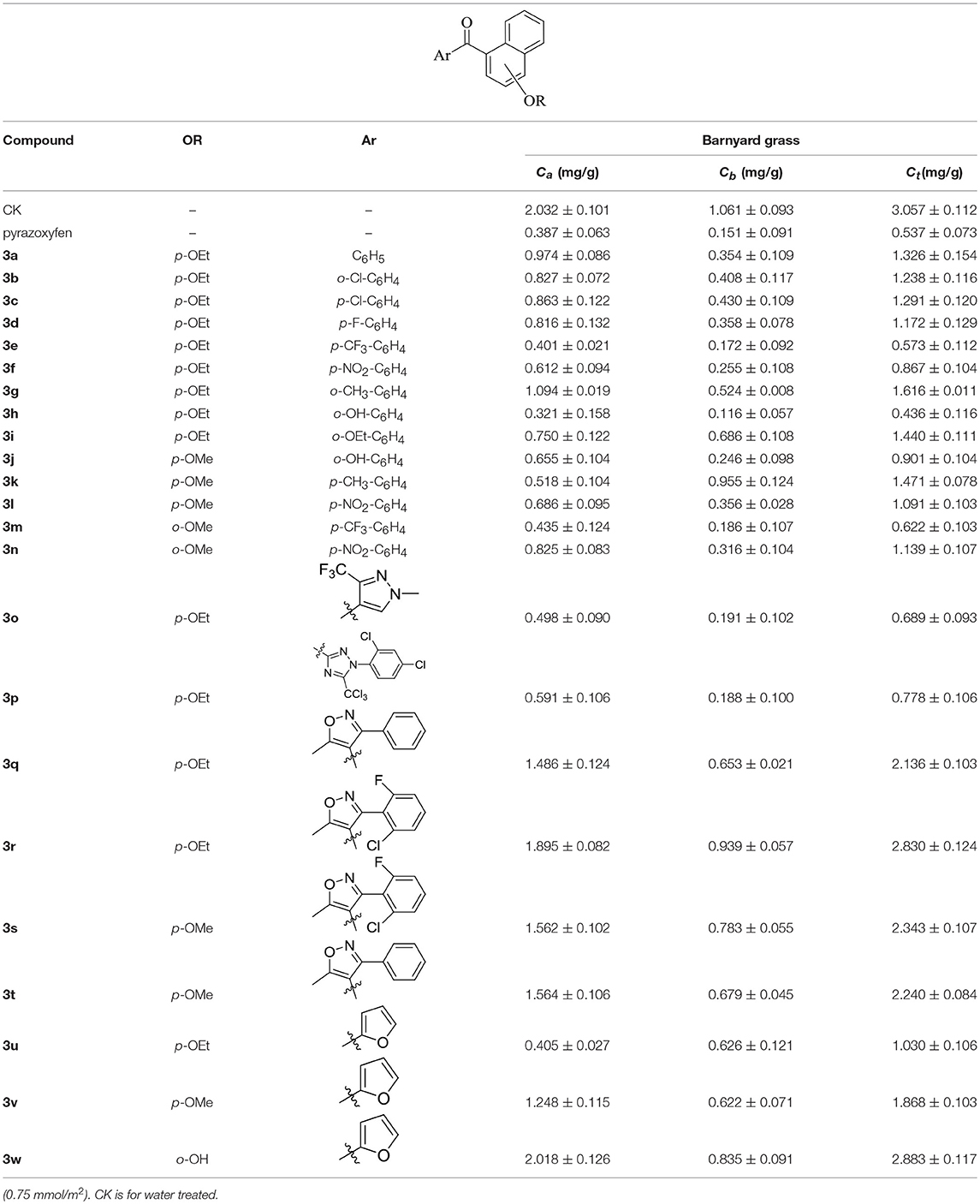
Herbicidal activities were determined using barnyard grass as indicating crops. The barnyard grass was planted in pots and grown in a greenhouse at around 25°C. The spraying treatment was conducted at rates of 2–4 kg ai/ha when the barnyard grass achieved the two-leaf period. After 7d, the chlorophyll content was surveyed and evaluated (Table 1). The commercial herbicide pyrazoxyfen was chosen as a positive control. The content of chlorophyll a (Ca), chlorophyll b (Cb), and total chlorophyll (Ct) were determined according to the literature (Wang et al., 2013).
Computational Methods
The structure of compounds 3h and inhibitor DAS869 were constructed by the sketch module of Tripos Associates, Inc. (2001). Subsequently, the Gasteiger-Huckel method was employed to calculate the partial atomic charges and the molecules were optimized. The crystal structure of Arabidopsis thaliana HPPD (AtHPPD, PDB ID 1TFZ) was obtained from the Protein Data Bank (PDB). Docking was modeled using the CDOCKER method in Accelrys Discovery Studio 2.5 (Catalyst, Version 4.10, 2005). In the preparation of protein structure, water and some other co-crystallized small molecules were removed, and a CHARMM force field was appended. After that, the binding site was limited in a 13.0 Å subset region from the center of the known ligand. The prepared protein and compounds were used to be docking receptor and ligand, respectively. In the process of docking, after the energy minimization using the smart minimize way, the top 10 conformations were saved for every ligand based on -CDOCKER ENERGY, and the remaining parameters were set to the default values.
Results and Discussion
Synthetic Chemistry
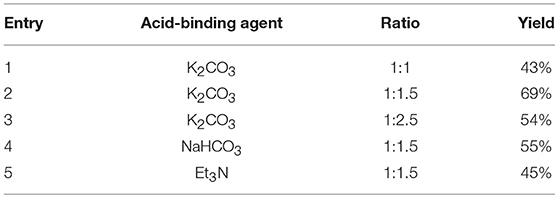
The synthetic route for the preparation of compounds 3a-w is depicted in Figure 3. In the process (a), the intermediate 2a was synthesized using naphthol and dimethyl sulfate as the starting materials, and anhydrous K2CO3 as the acid-binding agent. It should be noted that this process was solvent-free reaction, and the mixing rate of naphthol and the alkylating agents played a vital role. The insufficient mixing rate resulted in low yields. In addition, the effect of acid-binding agents was explored. As shown in Table 2, it was found that anhydrous K2CO3 was a better acid-binding agent than anhydrous NaHCO3 and Et3N. To further improve the yield, the ratios of naphthol and the alkylating agents were evaluated. The results indicated that naphthol-alkylating agent ratio in 1:1.5 provided better yields than ratio 1:1 or 1:2.5.
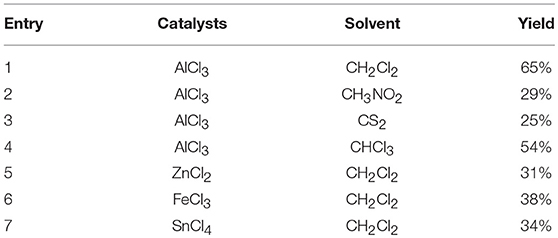
The target compounds 3a-w were synthesized via Friedel-Crafts acylation reaction. The synthesis of 3a was first carried out with ZnCl2 as catalysts, CHCl3 as solvent, at room temperature for 24 h, however, the target compound were not obtained. The different Lewis acids catalysts were evaluated. The results indicated that anhydrous AlCl3 was the best catalysts with excellent yield than SnCl4, ZnCl2, or FeCl3. To promote the yields, the solvent influences were investigated as well. The experiment results showed that CH2Cl2 possessed better yield than CH3NO2, CS2, and CHCl3. The detail exploration of the synthetic process to compound 3a is shown in Table 3.
The effects of substituent pattern on the yields were studied. It was found that different aromatic substitutents displayed diverse yields comparing compounds 3g, 3o-q, and 3u, and the order was as following: isoxazole>furan>benzene>pyrazole>triazole. The substituted position on the naphthalene ring was also explored. Generally the yield of α-substitution was better than β-substitution, and the yield with methoxy substituted was higher than that of ethoxy, for example, the yield of compound 3e with 56% was better than compound 3m. Moreover, the yields of compounds 3q and 3r with ethoxy substituted were lower than compounds 3s and 3t with methoxy substituted, which was mainly due to the steric hindrance of ethoxy was greater than methoxy.
Biological Activity and the Structure-Activity Relationships (SAR)
The herbicidal activities of compounds 3a-w (2–4 kg ai/ha) against barnyard grass in the greenhouse are list in Table 1. The chlorophyll content was gauged after spraying with compounds 3a-w for 7 days, and the commercial herbicide pyrazoxyfen was selected as positive control. As anticipated, the treated barnyard grass displayed unique bleaching symptoms, as typical HPPD herbicides. The biological activity evaluation indicated that most of the synthetic compounds showed good to excellent herbicidal activity. Compound 3h at a dose of 2.1 kg ai/ha exhibited better herbicidal activity than pyrazoxyfen at rates of 3 kg ai/ha.
As shown in Table 1, all of the compounds 3a-w decreased the content of Ca, Cb, and Ct in different degrees. Comparing the compounds 3a-n, all of them showed good herbicidal activity, especially compound 3h exhibited the best activity with a hydroxyl on the o-benzene ring, even better than pyrazoxyfen with o-PhCOCH2O, which may be attributed to the steric effects. It should be noted that the electron-withdrawing groups CF3 (3e, 3m), NO2 (3f, 3l, and 3n), F (3d), and Cl (3b, 3c) introduced on the phenyl were more beneficial for the herbicidal activity than electron-donating groups CH3 (3g, 3k) and OEt (3i). And it was observed that the stronger electron-withdrawing ability exhibited the better herbicidal activity. For example, compound 3e showed higher herbicidal activity than compounds 3b and 3c. In the same cases, the electron-donating substitution showed weak herbicidal activity. Thus, the SAR could be summarized as follow: CF3>NO2>F>Cl>H>OEt>CH3. The substituted position on the naphthalene ring was also compared. Generally the herbicidal activity of α-substitution was better than β-substitution. For instance, the herbicidal activity of compound 3l with p-OMe was better than compound 3m with o-OMe. Furthermore, the aromatic moiety also affected the inhibition. The results illustrated that benzene ring performed higher herbicidal activity than pyrazole. Compounds 3e exhibited better activity than compound 3o. In the same way, as the aromatic ring was triazole (3p), it displayed worse herbicidal activity than pyrazole (3o), and the isoxazole (3q) exhibited lower herbicidal activity than furan (3u). For the synthetic compounds, different aromatic subunits displayed diverse herbicidal activity, and the substituted benzene exhibited better herbicidal activity than others aromatic heterocycles.
Docking Study
During more than 30 years of investigation, knowledge about HPPD has advanced with technology, including X-ray crystallography and bioinformatics. Consequently, 40 HPPD structures have been determined, uploaded, and stored in PDBs (Ndikuryayo et al., 2017). In order to explain the interaction of the novel compounds with HPPD at the molecular level, the docking binding modes was compared with the reported crystal structure of AtHPPD in complex with pyrazoles inhibitor DAS869 (Pauly et al., 2013). As shown in Figure 4, both compounds 3h and DAS869 are well-matched to the active pocket of HPPD. The simulated complex of DAS869 and inhibitors 3h with the binding site of AtHPPD are displayed in Figures 5A,B. The inhibitor 3h was noticed to constitute a bidentate combination with the Fe2+, forming twisted square-pyramidal complex with a mainly five-coordinate. The naphthalene ring was “sandwiched” by the phenyl rings of Phe 360 and Phe 403, forming π-π interplay with the two amino acid residues, which was similar to DAS869 in complex with AtHPPD. Compared the distances of π-π interplay, it was observed that the distance of the naphthalene ring of inhibitor 3h with Phe 360 was 4.0 Å and that with Phe 403 was 3.4 Å; the distance of the benzene ring of DAS869 with Phe 360 was 4.5 Å and that with Phe 403 was 4.5 Å. The short distances of the naphthalene ring from Phe 360 and Phe 403 may be liable for the better AtHPPD inhibitory activity of inhibitor 3h.
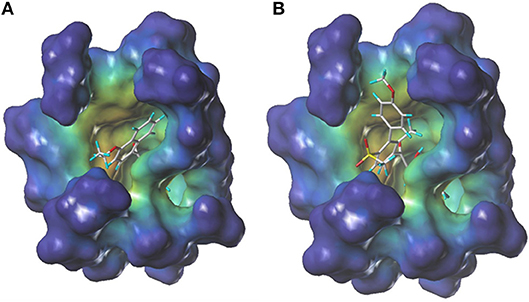
Figure 4. The docking modeling of 3h (A) and DAS869 (B) with AtHPPD at active site. The carbon atoms are shown in gray, the hydrogen atoms are shown in cyans, the oxygen atoms are shown in red, and the nitrogen atoms are shown in blue, the sulfur atom is shown in yellow.
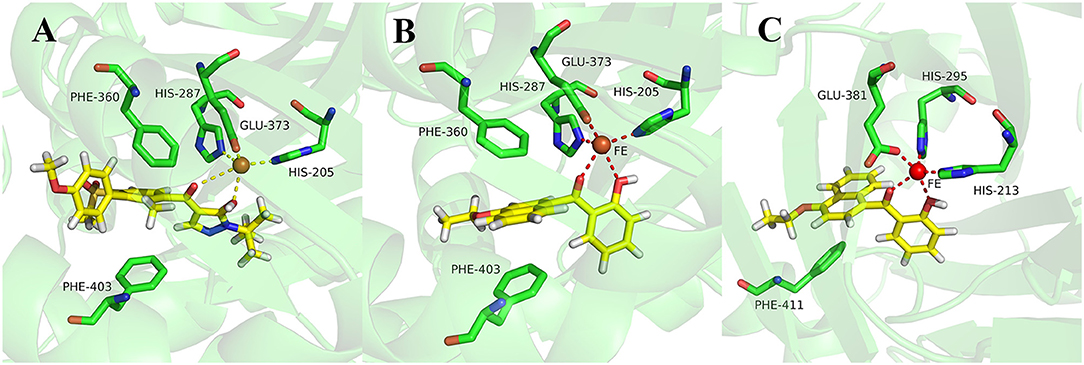
Figure 5. (A) Binding mode of DAS869 with 1TFZ. (B) Binding mode of compound 3h with 1TFZ. (C) Binding mode of compound 3h with Q9ARF9. Ligands DAS869 and compound 3h were shown in yellow.
So as to further explain the specificity interaction of the novel compounds with HPPD of others plants, six plants HPPD structures in PDB were selected for molecular docking including maize (PDB ID 1SP8), lectranthus scutellarioides (PDB ID Q9ARF9), camelina sativa (PDB ID 5YCK), brassica napus (PDB ID Q60Y65), daucus carota (PDB ID 3VLB) and hordeum vulgare (PDB ID 3TCM). The docking results showed that compound 3h could only bind to Q9ARF9, however, the naphthalene ring didn't form a favorable sandwich π-π interaction with the key residues of Q9ARF9 (Figure 5C). These consequences illustrated that compound 3h was safe for crops.
Conclusion
In summary, a series of new aryl-naphthyl methanone derivatives were designed and synthesized as effective HPPD inhibitors. The bioassay indicated that the target compounds showed excellent HPPD inhibitory activity. It is inspiring that compound 3h exhibits the best activity even better than the commercial herbicide pyrazoxyfen. This present work indicates that aryl-naphthyl methanone scaffold can be used as a potential lead structure to develop new HPPD inhibitors.
Author Contributions
YF and FY constructed the workflow. KW and J-XK synthesized and purified the compounds. PW provided the chemicals for all biological assays and helped in relevant literature. SG and L-XZ performed the mass spectrometric analysis. YF completed the paper.
Funding
This work was supported by the National Nature Science Foundation of China (No. 31772208), and Natural Science Foundation of Heilongjiang Province (ZD2017002), and the Research Science Foundation in Technology Innovation of Harbin (2015RAXXJ032).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fchem.2019.00002/full#supplementary-material
References
Ahrens, H., Lange, G., Müller, T., Rosinger, C., Willms, L., and Van, A. A. (2013). 4-Hydroxyphenylpyruvate dioxygenase inhibitors in combination with safeners: solutions for modern and sustainable agriculture. Angew. Chem. Int. Ed. 52, 9388–9398. doi: 10.1002/anie.201302365
Beaudegnies, R., Edmunds, A. J., Fraser, T. E., Hall, R. G., Hawkes, T. R., Mitchell, G., et al. (2009). Herbicidal 4-hydroxyphenylpyruvate dioxygenase inhibitors—a review of the triketone chemistry story from a syngenta perspective. Bioorg. Med. Chem. 17, 4134–4152. doi: 10.1016/j.bmc.2009.03.015
Elmore, M. T., Brosnan, J. T., Kopsell, D. A., Breeden, G. K., and Mueller, T. C. (2011). Response of hybrid bermudagrass (Cynodon dactylon × C. transvaalensis) to three HPPD-inhibitors. Weed Sci. 59, 458–463. doi: 10.1614/WS-D-11-00045.1
Fu, Y., Sun, Y. N., Yi, K. H., Li, M. Q., Cao, H. F., Li, J. Z., et al. (2017a). 3D Pharmacophore-based virtual screening and docking approaches toward the discovery of novel HPPD inhibitors. Molecules 22:959. doi: 10.3390/molecules22060959
Fu, Y., Sun, Y. N., Yi, K. H., Li, M. Q., Cao, H. F., Li, J. Z., et al. (2018). Combination of virtual screening protocol byin silicotoward the discovery of novel 4-hydroxyphenylpyruvate dioxygenase inhibitors. Front. Chem. 6:14. doi: 10.3389/fchem.2018.00014
Fu, Y., Wang, M. X., Zhang, D., Hou, Y. W., Gao, S., Zhao, L. X., et al. (2017b). Design, synthesis, and herbicidal activity of pyrazole benzophenone derivatives. RSC Adv. 7, 46858–46865. doi: 10.1039/c7ra09858h
He, B., Wang, D. W., Yang, W. C., Chen, Q., and Yang, G. F. (2017). Advances in research on HPPD structure and pyrazole-containing herbicides. Chin. J. Org. Chem. 37, 2895–2904. doi: 10.6023/cjoc201705031
Kubo, T., Ohno, M., Nagasawa, S., Kose, T., and Kawata, K. (2012). Behavior of herbicide pyrazolynate and its hydrolysate in paddy fields after application. Bull. Environ. Contam. Toxicol. 89, 985–989. doi: 10.1007/s00128-012-0793-6
Laschi, M., Bernardini, G., Dreassi, E., Millucci, L., Geminiani, M., Braconi, D., et al. (2016). Inhibition of para-hydroxyphenylpyruvate dioxygenase by analogues of the herbicide nitisinone as a strategy to decrease homogentisic acid levels, the causative agent of alkaptonuria. ChemMedChem 11, 674–678. doi: 10.1002/cmdc.201500578
Lee, D. L., Prisbylla, M. P., Cromartie, T. H., Dagarin, D. P., Howard, S. W., and Mutter, L. C. (1997). The discovery and structural requirements of inhibitors of p-hydroxyphenylpyruvate dioxygenase. Weed Sci. 45, 601–609.
Luscombe, B. M., and Pallett, K. E. (1996). Isoxaflutole for weed control in maize, Pestic. Outlook 7, 29–32.
Mitchell, G., Bartlett, D. W., Fraser, T. E., Hawkes, T. R., Holt, D. C., Townson, J. K., et al. (2001). Mesotrione: a new selective herbicide for use in maize. Pest Manag. Sci. 57, 120–128. doi: 10.1002/1526-4998(200102)57:2<120::AID-PS254>3.0.CO;2-E
Momeni, A. R. (2007). Solvent-free Williamson synthesis: an efficient, simple, and convenient method for chemoselective etherification of phenols and bisphenols. Synthet. Commun. 38, 1807–1815. doi: 10.1080/00397910701316268
Moran, G. R. (2005). 4-Hydroxyphenylpyruvate dioxygenase. Arch. Biochem. Biophys. 433, 117–128. doi: 10.1016/j.abb.2004.08.015
Ndikuryayo, F., Moosavi, B., Yang, W. C., and Yang, G. F. (2017). 4-Hydroxyphenylpyruvate dioxygenase inhibitors: from chemical biology to agrochemicals. J. Agric. Food Chem. 65, 8523–8537. doi: 10.1021/acs.jafc.7b03851
Pauly, J., Spiteller, D., Linz, J., Jacobs, J., Allen, C., Nett, M., et al. (2013). Ralfuranone thioether production by the plant pathogen ralstonia solanacearum. ChemBioChem. 14, 2169–2178. doi: 10.1002/cbic.201300364
Rene, L., Michael, K., Joachim, G., Michael, R., and Von, D. W. (2003). Verfahren zur Herstellung von 2-Alkyl-3-(4,5-Dihydroisoxazol-3-yl)-Acylbenzolen. U.S. Patent NO 20030149276. Washington, DC: U.S. Patent and Trademark Office.
Santucci, A., Bernardini, G., Braconi, D., Petricci, E., and Manetti, F. (2017). 4-Hydroxyphenylpyruvate dioxygenase and its inhibition in plants and animals: small molecules as herbicides and agents for the treatment of human inherited diseases. J. Med. Chem. 60, 4101–4125. doi: 10.1021/acs.jmedchem.6b01395
Wang, D. W., Lin, H. Y., Bo, H., Wu, F. X., Tao, C., Chen, Q., et al. (2016). An efficient one-pot synthesis of 2-(aryloxyacetyl)cyclohexane-1,3-diones as herbicidal 4-hydroxyphenylpyruvate dioxygenase inhibitors. J. Agric. Food Chem. 64, 8986–8993. doi: 10.1021/acs.jafc.6b04110
Wang, D. W., Lin, H. Y., Cao, R. J., Chen, T., Wu, F. X., Hao, G. F., et al. (2015a). Synthesis and herbicidal activity of triketone-quinoline hybrids as novel 4-hydroxyphenylpyruvate dioxygenase inhibitors. J. Agric. Food Chem. 63, 5587–5596. doi: 10.1021/acs.jafc.5b01530
Wang, D. W., Lin, H. Y., Cao, R. J., Ming, Z. Z., Chen, T., Hao, G. F., et al. (2015b). Design, synthesis and herbicidal activity of novel quinazoline-2,4-diones as 4-hydroxyphenylpyruvate dioxygenase inhibitors. Pest Manag. Sci. 71, 1122–1132. doi: 10.1002/ps.3894
Wang, R., Chen, Y. Z., Chen, L. S., Peng, S. F., Wang, X. N., Yang, X. H., et al. (2013). Correlation analysis of spad value and chlorophyll content in leaves of camellia oleifera. J. Cent. South. Univ. Forest. Tech. 33, 77–80. doi: 10.14067/j.cnki.1673-923x.2013.02.011
Witschel, M. (2009). Design, synthesis and herbicidal activity of new iron chelating motifs for HPPD-inhibitors. Bioorg. Med. Chem. 17, 4221–4229. doi: 10.1016/j.bmc.2008.11.006
Wojcik, A., Broclawik, E., Siegbahn, P. E. M., Lundberg, M., Moran, G., and Borowski, T. (2014). Role of substrate positioning in the catalytic reaction of 4-hydroxyphenylpyruvate dioxygenase-A QM/MM study. J. Am. Chem. Soc. 136, 14472–14485. doi: 10.1021/ja506378u
Xu, Y. L., Lin, H. Y., Cao, R. J., Ming, Z. Z., Yang, W. C., and Yang, G. F. (2014). Pyrazolone-quinazolone hybrids: a novel class of human 4-hydroxyphenylpyruvate dioxygenase inhibitors. Bioorg. Med. Chem. 22, 5194–5211. doi: 10.1016/j.bmc.2014.08.011
Keywords: 4-hydroxyphenylpyruvate dioxygenase, design, synthesis, aryl-naphthyl methanone, herbicidal activity
Citation: Fu Y, Wang K, Wang P, Kang J-X, Gao S, Zhao L-X and Ye F (2019) Design, Synthesis, and Herbicidal Activity Evaluation of Novel Aryl-Naphthyl Methanone Derivatives. Front. Chem. 7:2. doi: 10.3389/fchem.2019.00002
Received: 29 August 2018; Accepted: 03 January 2019;
Published: 22 January 2019.
Edited by:
Simone Brogi, Università Degli Studi di Siena, ItalyReviewed by:
Konstantinos M. Kasiotis, Benaki Phytopathological Institute, GreeceFrancisco Solano, University of Murcia, Spain
Ning Ma, Tianjin University, China
Copyright © 2019 Fu, Wang, Wang, Kang, Gao, Zhao and Ye. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Fei Ye, eWVmZWlAbmVhdS5lZHUuY24=
 Ying Fu
Ying Fu Kui Wang
Kui Wang Shuang Gao
Shuang Gao Fei Ye
Fei Ye