
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
MINI REVIEW article
Front. Chem. , 12 September 2018
Sec. Crop Biology and Sustainability
Volume 6 - 2018 | https://doi.org/10.3389/fchem.2018.00264
This article is part of the Research Topic Drought Tolerance in Crop Plants View all 12 articles
Drought induced stress is often a bottleneck of agricultural crop production. Invariably, field crops across all agro-ecological regions succumb to it with an yield penalty. Drought massively affects the growth and harvestable yield in crops and has become an imminent problem necessitating breeding of tolerant crops. It induces myriad changes of biochemical, molecular, and physiological nature that manifest into aberrant plant morphology. The response to drought in plants incites a signaling cascade that involves perception and translation of drought signal leading to concomitant modulation of gene expression and de novo osmolyte synthesis. The intricate patterns of expression of these genes vary from early induction to late responsive genes. While one class of genes codes for products imparting osmotolerance and protection to plants, the second class predominantly modulates target gene expression by an intricate signal transduction mechanism. This review summarizes both canonical and non-canonical cascades of drought stress response in plants, delineating the mechanism in rice (Oryza sativa) and emphasizes hydropenia induced lipid signaling.
Altered physiological conditions disrupt cellular homeostasis and orchestrate stress in plants. Invariably, plants during their growth period are exposed to multiple stresses such as drought, shading (low light intensity), low temperature, salinity, flooding, heat, oxidative stress, and heavy metal toxicity (Shivakumara et al., 2017; Shivaraj et al., 2017). All these stresses, individually or in combination eventually hamper productivity of the crops (Fang and Xiong, 2015; Joshi et al., 2016; Gupta et al., 2017). Amongst all, drought i.e., hydropenia is the most devastating environmental stress (Gaspar et al., 2002) and impacts multiple morphological changes that are visible in all “Phenological stages of plant/crop growth” (Zhang et al., 2017). It decreases crop stand in field (Lambers et al., 2008) and reduces harvestable yield and economic harvest in crops (Thirunavukkarasu et al., 2017; Van Gioi et al., 2017). It is estimated that drought will impact 30% global loss of crop yield by 2025 (Zhang, 2011). With climate change looming large, water deficit has become a cardinal issue of agriculture as climate models have predicted an increase in severity and frequency of drought (Walter et al., 2011; IPCC, 2012). Additionally, the growing water scarcity/mis-management of the available water is a major threat to sustainable domestic, industrial, and agricultural development (Hamdy et al., 2003).
Edaphologically, drought in crops results from short-fall in the required precipitation leading to reduced available water in the soil. Additionally, dry atmospheric condition increases water loss from plants by evapo-transpiration. Aside the precipitations, drought depends on evapo-transpiration, soil water holding capacity, crop water requirements, and ability of plants to efficiently utilize available water (Toker et al., 2007). Multiple molecular (Pornsiriwong et al., 2017) and cellular responses (Comas et al., 2013; Chen et al., 2017; Pornsiriwong et al., 2017) become operative with on-set of drought in plants. Nevertheless, tolerence to drought amongst crops shows variation between and/or within the crops. Depending upon the exhibited symptoms, effect of drought on plants are classified as slight, moderate, severe, and very severe on the basis of relative water content (RWC) (Gigon et al., 2004). While plants are able to withstand slight stress by evoking tolerance mechanism, mild drought induces regulation of water loss and uptake in plants allowing maintenance of relative water content (RWC) with minimally altered photosynthetic capacity and quantum yield.
Drought hinders plant growth/development with commensurate reduction in accumulation of biomass. Farooq et al. (2009) and Li et al. (2009) identified that the consequences of drought in crop plants range from reduced (i) cell division and expansion, (ii) leaf size and stem elongation, (iii) perturbed water/nutrient relations and stomatal oscillations, and (iv) diminished water use efficiency (WUE) (Farooq et al., 2009; Li et al., 2009). With on-set of water deficit, abscisic acid (ABA) biosynthesis is stimulated in plants which reduces stomatal conductance and transpirational losses (Yamaguchi-Shinozaki and Shinozaki, 2006). Cell division and cell enlargement in drought stressed plants is negatively affected as water potential/cellular turgor is lost, and photosynthesis decreases (Farooq et al., 2009; Taiz and Zeiger, 2010). These massive physiological changes in plants reduce root, shoot, and flower fresh/dry weight (Liu et al., 2011) with maximum reduction in total leaf area (Farooq et al., 2010). It also affects crop phenology and induces early transition from the vegetative to the reproductive phase (Desclaux and Roumet, 1996) leading to altered crop growth cycle.
Hydropenia has pronounced negative effect on crop yield. Particularly, drought during silking stage in maize reduces total biomass accumulation by 37%. It also reduces yield by negatively affecting at grain-filling stage (reduction by 34%) and at maturity (by 21%) (Kamara et al., 2003). In rice, drought-induced physiological changes such as stomatal closure decreases intake of CO2 and eventually decreases photosynthesis (Flexas et al., 2005) due to reduction in carbon capture that imbalances the source and sink partitioning of photosynthets, reduces the phloem loading, reduces assimilate translocation and dry matter partitioning (Farooq et al., 2009). Additionally, photorespiration becomes operative which leads to decline in the carbon fixation in rice. In C3 plants, such as rice, Rubisco is the key enzyme in CO2 assimilation and acts either as carboxylase or as an oxygenase depending upon the internal concentration of CO2/O2 in leaf. At moderate water stress with closed stomata, Rubisco acts as an oxygenase as prevailing cellular O2 concentration is higher than CO2 concentration. This increase in photorespiration due to drought at the “Expense of carbon-fixation” (Ghannoum, 2008) leads to yield reduction in rice.
Plants evoke myriad morphological and biochemical adaptations at whole-plant and cellular-levels to ward off stresses of drought. Noteworthy, among them are the three canonical mechanisms such as (i) drought escape, (ii) drought avoidance, and (iii) drought resistance (Yamaguchi-Shinozaki and Shinozaki, 2006). Drought escape is the mechanism that invigorates plants to complete their life cycle before drought sets in, so that the seeds enter in to dormancy before the dry conditions prevail e.g., desert plants saving themselves from extinction. However, “Drought avoidance mechanism in plants involve maintaining high water status/cellular hydration” either by absorbing more water from soil or by reducing loss of water by transpiration. In contrast, drought tolerance is the ability of plants to continue normal cellular metabolism and growth activity at low water potential despite prevailing stress condition and/or ability to recover fast after stress. A crop is considered tolerant, only if it survives drought with minimal yield penalty. These plants maintain the cellular turgor through osmotic adjustment and protoplasmic resistance (Mitra, 2001) by accumulation of free proline (Munns, 2005).
In response to drought, plants activate three main categories of genes that canonically modulate biochemical/physiological and/or molecular pathways (Dash et al., 2014). They are (1) genes involved in “protection of membranes; water and ion uptake/transport” imparting cellular tolerance (2) regulatory genes involved in signaling/transcriptional control, and (3) novel genes of unknown function reported to impart drought tolerence. Plants extrinsically perceive environmental stress and transfer the signal through cascades of molecules. These signaling molecules trigger the expression of specific genes leading to appropriate physiological/biochemical responses (Shinozaki and Yamaguchi-Shinozaki, 2007; Golldack et al., 2014; Hu and Xiong, 2014). A number of genes/transcription factors showing differential expression to drought have been identified in plants (Yamaguchi-Shinozaki and Shinozaki, 2006; Joshi et al., 2016). These are known to be involved in cellular responses such as “stress perception and transcriptional regulation” of drought responsive genes (Lata and Prasad, 2011). These genes code for “Protein kinases, phytohormones, transcription factors” (Lata et al., 2015), osmoprotectants and “late embryogenesis abundant (LEA)” proteins (Varshney et al., 2011; Golldack et al., 2014; Todaka et al., 2015; Sah et al., 2016) imparting tolerance to dehydration.
Perceived response to drought in plants is broadly categorized into ABA-dependent or ABA-independent pathway. In rice, ABA-Responsive cis-Elements (ABRE;PyACGTGG/TC) are enriched compared to Arabidsopsis and soybean (Maruyama et al., 2011) and in response to drought, ABA concentration dramatically increases in vegetative parts. Increased ABA triggers (i) stomatal closure, (ii) stress proteins and metabolites accumulation (protect cells during stress), and (iii) H2O2 accumulation in guard cells that signals reduction in water loss from the plant (Mustilli et al., 2002; Kwak et al., 2003; Wang and Song, 2008). The ABA independent pathway, elucidated earlier, involves H2O2 mediated stomatal closure in rice (Huang et al., 2009).
The signaling mechanism to drought in plants involves sensing and relaying of dehydration signal from plasma membrane to the nucleus (Sanders et al., 1999; Ramanjulu and Bartels, 2002). This is accomplished through several protein phosphorylation mechanisms involving kinases viz. the mitogen activated protein kinases (MAPKs) and receptor-like kinases (RLKs) (Das and Pandey, 2010; Tena et al., 2011; Seybold et al., 2014). In rice, “DROUGHT-HYPERSENSITIVE MUTANT1 (DSM1)”—a protein kinase—scavenges the reactive oxygen species (ROS) produced under drought stress. The dsm1 mutants are hypersensitive to drought during seedling and panicle development stage (Ning et al., 2010). The stress-responsive RLK genes such as stress induced protein kinase 1 (OsSIK1), growth under drought kinase (GUDK) were found to be induced by drought stress in rice. While, rice overexpressing OsSIK1 showed tolerance to drought (Ouyang et al., 2010); GUDK phosphorylates apetala 2/ERF37 (OsAP37) that activates stress-regulated genes (Ramegowda et al., 2014) in rice.
Several, transcription factors (TFs) regulating hydropenia signaling in rice has also been elucidated. Most of these TFs bind to cis-regulatory elements and belong to “AP2/ERF, bZIP, NAC, MYB, WRKY, bHLH, NF-Y, and CAMTA” families (Umezawa et al., 2006; Licausi et al., 2013; Castilhos et al., 2014; Shao et al., 2015). Over-expression of these TFs in rice showed increased ability of plant to withstand drought. Notably, DREB (dehydration-responsive element-binding protein) transcription factors act as key players in ABA independent pathway of drought tolerance. Among DREBs, DREB1/CBF, and DREB2 are involved in drought stress (Srivasta et al., 2010; Nakashima et al., 2014). The transgenic rice plants expressing DREB1A yield more compared to the non-transgenic plants (Datta et al., 2012) under drought condition. Similarly, over-expression of NAC transcription factors (NAP and ONAC022) “reduce rate of water loss and transpiration, decrease number of open stomata and increase proline content” in rice (Hong et al., 2016). Nevertheless, at vegetative stage, they impart “enhanced tolerance to high salinity, drought, and cold” while increase yield despite drought in flowering stage (Liang et al., 2014).
Besides drought responsive elements, proteins have been identifed in hydropenia (Goyal et al., 2005). Accumulation of LEA have been detected in seeds as well as in vegetative tissues (Ingram and Bartels, 1996; He et al., 2012; Liu et al., 2013). Expression of LEA protein encoding genes, OsEM1 and OsLEA3-1 enhances tolerance of rice under water deficit (Xiao et al., 2007; Yu et al., 2016). Similarly, high cuticular wax in many crops imparts tolerance to drought (Xue et al., 2017). Crops having more cuticular wax than reduced/ non-waxy crops show drought-tolerance and higher yield (Zhou L. et al., 2013; Guo et al., 2016) due to strong correlation between the wax content and WUE (Zhu and Xiong, 2013). Recently, waxy crystal-spare leaf 1 (OsWSL1) is reported to be involved in cuticular wax accumulation in rice (Yu et al., 2008) and grain lenght 1-6 (OsGL1-6) has been identified to synthesize fatty aldehyde decarbonylase required for formation of wax in epidermis and in vascular bundles (Zhou L. et al., 2013). Mutants defective in OsGL1 are sensitive to drought as they accumulate less cuticular wax (Mao et al., 2012). DEEPER ROOTING 1 mutant (DRO1) governing root architecture and drought stress have been identified in rice. DRO1 controls cell elongation at the root tip and changes the angle of root growth to downward direction (Uga et al., 2013) to fetch more water for growth.
Hydropenia induced abscisic acid (ABA) production also induces de novo expression of both structural and functional genes. Yamaguchi-Shinozaki and Shinozaki (2006) proposed operation of two pathways; (i) ABA-dependent pathway and (ii) ABA independent pathway. While the former pathway involves expression of genes “that may or may not require protein biosynthesis” the latter does not involve ABA for their induction. MYB and MYC transcription factors represent ABA dependent pathway while bZIP transcription factors don't require ABA synthesis and the target genes containing “abscisic acid response elements (ABREs) with core ACGT-containing G-box” (Chaves et al., 2003). The ABA independent pathway involves the “Water-deficit-inducible genes” that do not require ABA for their induction. The promoters of these genes contain a conserved “Dehydration responsive element (DRE)” and are induced by external stimuli (Yamaguchi-Shinozaki and Shinozaki, 2006).
Besides activation of TFs/proteins; lipids are also involved in abiotic stress tolerance in plants (Okazaki and Saito, 2014; Hou et al., 2016). Seminal discoveries have elucidated lipid signaling in response to drought (Darwish et al., 2009; Golldack et al., 2014) in plants. Lipids such as wax, cutin, and suberin directly contribute to the alleviation of drought (Samuels et al., 2008) by reduction of cellular dehydration (Okazaki and Saito, 2014) and lipid metabolism (Gigon et al., 2004). It is reported (Kosma et al., 2009; Seo et al., 2011) that ABA treatment increases layers of these hydrocarbons in plants. While, overexpression of wax biosynthetic genes increases tolerance to drought (Yang et al., 2011; Luo et al., 2013; Zhou L. et al., 2013; Zhou M. et al., 2013), plants depleted of wax are less tolerant to drought (Qin et al., 2011; Seo et al., 2011; Mao et al., 2012; Zhu and Xiong, 2013). Lipid signaling in plants includes generation of “Inositol phosphate, lysophospholipids (LPLs), phosphatidic acid (PA), oxylipins, sphingolipids, diacylglycerol (DAG), free fatty acids (FFA), and N-acylethnolamine” that are generated from phospholipids (Munnik and Testerink, 2009; Saucedo-García et al., 2015). The mechanism involves generation of phosphatidic acid (PA) by rapid activation of phospholipase C (PLC) and phospholipase D (PLD) enzymes (Munnik et al., 1998, 2000; Pical et al., 1999). The enzyme PA kinase, by a attenuation mechanism, reduces phosphatidic acid (Munnik et al., 1996, 2000; Pical et al., 1999) to produce diacyl-glycerol pyrophosphate (DAGPP). Similarly, hyperosmotically stimulated cells change concentration of phosphatidylinositol phosphate, phosphatidylinositol 4,5-bisphosphate (Einspahr et al., 1988; Cho et al., 1993; Pical et al., 1999) and/or their novel isomers (Dove et al., 1997) to adjust to hydropenia.
The action of phospholipases and lipid intermediates depicting their role during drought is summarized in Figure 1. It reveals DAG and Inositol-3-phosphate are produced by PLC. InP3 increases the Ca2+ concentration in cytosol (Staxen et al., 1999). Consequently, Ca2+ and PtInP2 stimulate PLD (Wang, 2000; Zheng et al., 2000). The activated PLD generates phosphatidic acid from phospholipids. Cellular homeostasis of PtdOH is maintained by the opposing actions of kinsases and phosphatases that interconvert DAG, phosphatidic acid, and DAG-PPi in plants. Thus, the pool of phosphatidic acid acts as an important hub of lipid signaling/biosynthesis (Liscovitch et al., 2000). However, PLD directly does not alter activity of PLA but oxylipin synthesis is inferred to be activated by PLD (Wang, 2000). On the contrary, LysoPL produced by PLA directly inhibits PLD activity (Ryu et al., 1997) that maintains phospholipid homeostasis in plants.
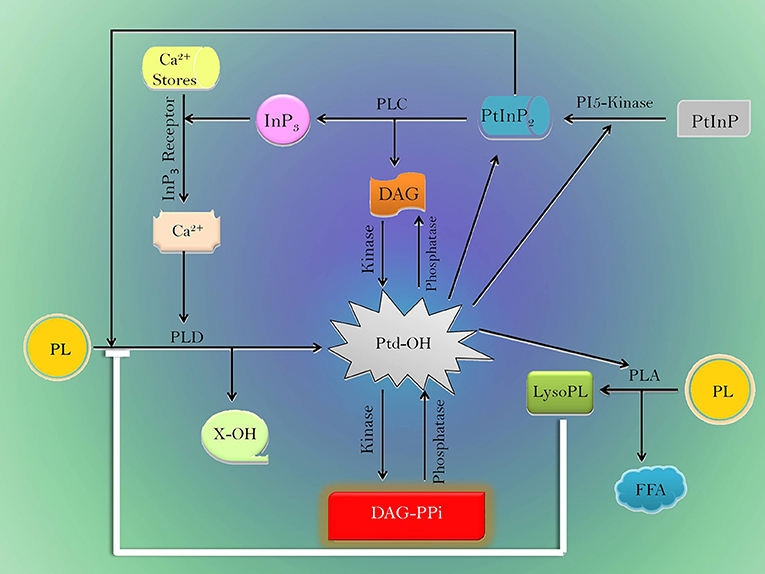
Figure 1. Model depicting the network of various phospholipases and lipid mediators during drought stress. Black line-induction, blue line-inhibition. PLA, Phospholipase A; PLC, Phospholipase C; PLD, Phospholipase D; DAG, Diacyl glycerol; DAG-PPi, Diacyl-glycerol pyrophosphate; PL, Phospholipid; Lyso PL, lysophospholipids; FFA, Free fatty acid; InP3, Inositol 1,4,5-trisphosphate; PI5-kinase, Phosphatidylinositol 4-phosophate 5-kinase; PtInP, Phosphatidylinositol monophosphate; PtInP2, Phosphatidylinositol 4,5-bisphosphate; PtdOH, Phosphatidic acid, and X-OH- Free head group.
With climate change looming large over modern intensive agriculture, frequency and severity of drought is predicted to increase. The erratic precipitation will cause large scale disruption in shallow rainfed rice agro-ecosystems leading to significant reduction in economic harvest. Thus, development of drought tolerant varieties by altering cellular homeostasis of lipids/proteins/carbohydrates is necessary for ensuring enhanced crop production in rainfed agro-ecological regions with unpredictable climatic conditions.
PD and RR planned, collected information, and organized the manuscript. VR and SP provided critical input and edited the manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The reviewer MK and handling Editor declared their shared affiliation.
We thank our laboratory members Dr. Payal Gupta, Dr. Shivraj SM, Ms. Varsha Jain, and Mr. Soumyadeep Mukherjee for providing input for the manuscript at initial stage. Work in PD's laboratory is supported by ICAR-NPTC and ICAR-NASF. Our sincere apology to those researchers whose work could not be cited due to space limitations.
Castilhos, G., Lazzarotto, F., Spagnolo-Fonini, L., Bodanese-Zanettini, M. H., and Margis-Pinheiro, M. (2014). Possible roles of basic helix-loop-helix transcription factors in adaptation to drought. Plant Sci. 223, 1–7. doi: 10.1016/j.plantsci.2014.02.010
Chaves, M. M., Maroco, J. P., and Pereira, J. S. (2003). Understanding plant responses to drought—from genes to the whole plant. Funct. Plant Biol. 30, 239–264. doi: 10.1071/FP02076
Chen, Z. H., Chen, G., Dai, F., Wang, Y., Hills, A., Ruan, Y. L., et al. (2017). Molecular evolution of grass stomata. Trends Plant Sci. 22, 124–139. doi: 10.1016/j.tplants.2016.09.005
Cho, M. H., Shears, S. B., and Boss, W. F. (1993). Changes in phosphatidylinositol metabolism in response to hyperosmotic stress in Daucus carota L. cells grown in suspension culture. Plant Physiol. 103, 637–647. doi: 10.1104/pp.103.2.637
Comas, L. H., Becker, S. R., Von Mark, V. C., Byrne, P. F., and Dierig, D. A. (2013). Root traits contributing to plant productivity under drought. Front. Plant Sci. 4:442. doi: 10.3389/fpls.2013.00442
Darwish, E., Testerink, C., Khalil, M., El-Shihy, O., and Munnik, T. (2009). Phospholipid signaling responses in salt-stressed rice leaves. Plant Cell Physiol. 50, 986–997. doi: 10.1093/pcp/pcp051
Das, R., and Pandey, G. K. (2010). Expressional analysis and role of calcium regulated kinases in abiotic stress signaling. Curr. Genomics 11, 2–13. doi: 10.2174/138920210790217981
Dash, P. K., Cao, Y., Jailani, A. K., Gupta, P., Venglat, P., Xiang, D., et al. (2014). Genome-wide analysis of drought induced gene expression changes in flax (Linum usitatissimum). GM Crops Food 5, 106–119. doi: 10.4161/gmcr.29742
Datta, K., Baisakh, N., Ganguly, M., Krishnan, S., Yamaguchi Shinozaki, K., et al. (2012). Overexpression of Arabidopsis and rice stress genes' inducible transcription factor confers drought and salinity tolerance to rice. Plant Biotechnol. J. 10, 579–586. doi: 10.1111/j.1467-7652.2012.00688.x
Desclaux, D., and Roumet, P. (1996). Impact of drought stress on the phenology of two soybean (Glycine max L. Merr) cultivars. Field Crops Res. 46, 61–70. doi: 10.1016/0378-4290(95)00086-0
Dove, S. K., Cooke, F. T., Douglas, M. R., and Sayers, L. G. (1997). Osmotic stress activates phosphatidylinositol-3, 5-bisphosphate synthesis. Nature 390, 187–192. doi: 10.1038/36613
Einspahr, K. J., Peeler, T. C., and Thompson, J. (1988). Rapid changes in polyphosphoinositide metabolism associated with the response of Dunaliella salina to hypoosmotic shock. J. Biol. Chem. 263, 5775–5779.
Fang, Y., and Xiong, L. (2015). General mechanisms of drought response and their application in drought resistance improvement in plants. Cell. Mol. Life Sci. 72, 673–689. doi: 10.1007/s00018-014-1767-0
Farooq, M., Basra, S., Wahid, A., Ahmad, N., and Saleem, B. (2009). Improving the drought tolerance in rice (Oryza sativa L.) by exogenous application of salicylic acid. J. Agron. Crop Sci. 195, 237–246. doi: 10.1111/j.1439-037X.2009.00365.x
Farooq, M., Kobayashi, N., Ito, O., Wahid, A., and Serraj, R. (2010). Broader leaves result in better performance of indica rice under drought stress. J. Plant Physiol. 167, 1066–1075. doi: 10.1016/j.jplph.2010.03.003
Flexas, J., Galmes, J., Ribas-Carbo, M., and Medrano, H. (2005). “The effects of water stress on plant respiration,” in Plant Respiration, eds H. Lambers and M. Ribas-Carbo (Dordrecht: Springer), 85–94.
Gaspar, T., Franck, T., Bisbis, B., Kevers, C., Jouve, L., Hausman, J.-F., et al. (2002). Concepts in plant stress physiology. Application to plant tissue cultures. Plant Growth Regul. 37, 263–285. doi: 10.1023/A:1020835304842
Ghannoum, O. (2008). C4 photosynthesis and water stress. Ann. Bot. 103, 635–644. doi: 10.1093/aob/mcn093
Gigon, A., Matos, A. R., Laffray, D., Zuily-Fodil, Y., and Pham-Thi, A.-T. (2004). Effect of drought stress on lipid metabolism in the leaves of Arabidopsis thaliana (ecotype Columbia). Ann. Bot. 94, 345–351. doi: 10.1093/aob/mch150
Golldack, D., Li, C., Mohan, H., and Probst, N. (2014). Tolerance to drought and salt stress in plants: unraveling the signaling networks. Front. Plant Sci. 5:151. doi: 10.3389/fpls.2014.00151
Goyal, K., Walton, L. J., and Tunnacliffe, A. (2005). LEA proteins prevent protein aggregation due to water stress. Biochem. J. 388, 151–157. doi: 10.1042/BJ20041931
Guo, J., Xu, W., Yu, X., Shen, H., Li, H., Cheng, D., et al. (2016). Cuticular wax accumulation is associated with drought tolerance in wheat near-isogenic lines. Front. Plant Sci. 7:1809. doi: 10.3389/fpls.2016.01809
Gupta, P., Saini, R., and Dash, P. K. (2017). Origin and evolution of group XI secretory phospholipase A2 from flax (Linum usitatissimum) based on phylogenetic analysis of conserved domains. 3 Biotech 7:216. doi: 10.1007/s13205-017-0790-x
Hamdy, A., Ragab, R., and Scarascia-Mugnozza, E. (2003). Coping with water scarcity: water saving and increasing water productivity. Irrigat. Drainage 52, 3–20. doi: 10.1002/ird.73
He, S., Tan, L., Hu, Z., Chen, G., Wang, G., and Hu, T. (2012). Molecular characterization and functional analysis by heterologous expression in E. coli under diverse abiotic stresses for OsLEA5, the atypical hydrophobic LEA protein from Oryza sativa L. Mol. Genet. Genomics. 287, 39–54. doi: 10.1007/s00438-011-0660-x
Hong, Y., Zhang, H., Huang, L., Li, D., and Song, F. (2016). Overexpression of a stress-responsive NAC transcription factor gene ONAC022 improves drought and salt tolerance in rice. Front. Plant Sci. 7:4. doi: 10.3389/fpls.2016.00004
Hou, Q., Ufer, G., and Bartels, D. (2016). Lipid signalling in plant responses to abiotic stress. Plant Cell Environ. 39, 1029–1048. doi: 10.1111/pce.12666
Hu, H., and Xiong, L. (2014). Genetic engineering and breeding of drought-resistant crops. Annu. Rev. Plant Biol. 65, 715–741. doi: 10.1146/annurev-arplant-050213-040000
Huang, X. Y., Chao, D. Y., Gao, J. P., Zhu, M. Z., Shi, M., and Lin, H. X. (2009). A previously unknown zinc finger protein, DST, regulates drought and salt tolerance in rice via stomatal aperture control. Genes Dev. 23, 1805–1817. doi: 10.1101/gad.1812409
Ingram, J., and Bartels, D. (1996). The molecular basis of dehydration tolerance in plants. Annu. Rev. Plant Biol. 47, 377–403. doi: 10.1146/annurev.arplant.47.1.377
IPCC (2012). Special Report on Renewable Energy Sources and Climate Change Mitigation. Nairobi: UNEP.
Joshi, R., Wani, S. H., Singh, B., Bohra, A., Dar, Z. A., Lone, A. A., et al. (2016). Transcription factors and plants response to drought stress: current understanding and future directions. Front. Plant Sci. 7:1029. doi: 10.3389/fpls.2016.01029
Kamara, A., Menkir, A., Badu-Apraku, B., and Ibikunle, O. (2003). The influence of drought stress on growth, yield and yield components of selected maize genotypes. J. Agric. Sci. 141, 43–50. doi: 10.1017/S0021859603003423
Kosma, D. K., Bourdenx, B., Bernard, A., Parsons, E. P., Lü, S., Joubès, J., et al. (2009). The impact of water deficiency on leaf cuticle lipids of Arabidopsis. Plant Physiol. 151, 1918–1929. doi: 10.1104/pp.109.141911
Kwak, J. M., Mori, I. C., Pei, Z. M., Leonhardt, N., Torres, M. A., Dangl, J. L., et al. (2003). NADPH oxidase AtrbohD and AtrbohF genes function in ROS-dependent ABA signaling in Arabidopsis. EMBO J. 22, 2623–2633. doi: 10.1093/emboj/cdg277
Lambers, H., Chapin, F. S., and Pons, T. L. (2008). Plant Physiological Ecology, 2nd Edn. New York, NY: Springer.
Lata, C., Muthamilarasan, M., and Prasad, M. (2015). “Drought stress responses and signal transduction in plants,” in Elucidation of Abiotic Stress Signaling in Plants, ed G. Pandey (New York, NY: Springer), 195–225.
Lata, C., and Prasad, M. (2011). Role of DREBs in regulation of abiotic stress responses in plants. J. Exp. Bot. 62, 4731–4748. doi: 10.1093/jxb/err210
Li, Y., Ye, W., Wang, M., and Yan, X. (2009). Climate change and drought: a risk assessment of crop-yield impacts. Clim. Res. 39, 31–46. doi: 10.3354/cr00797
Liang, C., Wang, Y., Zhu, Y., Tang, J., Hu, B., Liu, L., et al. (2014). OsNAP connects abscisic acid and leaf senescence by fine-tuning abscisic acid biosynthesis and directly targeting senescence-associated genes in rice. Proc. Natl. Acad. Sci. U.S.A. 111, 10013–10018. doi: 10.1073/pnas.1321568111
Licausi, F., Ohme-Takagi, M., and Perata, P. (2013). APETALA2/Ethylene Responsive Factor (AP2/ERF) transcription factors: mediators of stress responses and developmental programs. New Phytol. 199, 639–649. doi: 10.1111/nph.12291
Liscovitch, M., Czarny, M., Fiucci, G., and Xiaoqing, T. (2000). Phospholipase D: molecular and cell biology of a novel gene family. Biochem. J. 345, 401–415. doi: 10.1042/bj3450401
Liu, H., Wang, X., Wang, D., Zou, Z., and Liang, Z. (2011). Effect of drought stress on growth and accumulation of active constituents in Salvia miltiorrhiza bunge. Ind. Crops Prod. 33, 84–88. doi: 10.1016/j.indcrop.2010.09.006
Liu, Y., Wang, L., Xing, X., Sun, L., Pan, J., Kong, X., et al. (2013). ZmLEA3, a multifunctional group 3 LEA protein from maize (Zea mays L.), is involved in biotic and abiotic stresses. Plant Cell Physiol. 54, 944–959. doi: 10.1093/pcp/pct047
Luo, X., Bai, X., Sun, X., Zhu, D., Liu, B., Ji, W., et al. (2013). Expression of wild soybean WRKY20 in Arabidopsis enhances drought tolerance and regulates ABA signalling. J. Exp. Bot. 64, 2155–2169. doi: 10.1093/jxb/ert073
Mao, B., Cheng, Z., Lei, C., Xu, F., Gao, S., Ren, Y., et al. (2012). Wax crystal-sparse leaf2, a rice homologue of WAX2/GL1, is involved in synthesis of leaf cuticular wax. Planta 235, 39–52. doi: 10.1007/s00425-011-1481-1
Maruyama, K., Todaka, D., Mizoi, J., Yoshida, T., Kidokoro, S., Matsukura, S., et al. (2011). Identification of cis-acting promoter elements in cold-and dehydration-induced transcriptional pathways in Arabidopsis, rice, and soybean. DNA Res. 19, 37–49. doi: 10.1093/dnares/dsr040
Mitra, J. (2001). Genetics and genetic improvement of drought resistance in crop plants. Curr. Sci. 80, 758–763.
Munnik, T., de Vrije, T., Irvine, R. F., and Musgrave, A. (1996). Identification of diacylglycerol pyrophosphate as a novel metabolic product of phosphatidic acid during G-protein activation in plants. J. Biol. Chem. 271, 15708–15715. doi: 10.1074/jbc.271.26.15708
Munnik, T., Irvine, R. F., and Musgrave, A. (1998). Phospholipid signalling in plants. Biochim. Biophys. Acta 1389, 222–272. doi: 10.1016/S0005-2760(97)00158-6
Munnik, T., Meijer, H. J., Ter Riet, B., Hirt, H., Frank, W., Bartels, D., et al. (2000). Hyperosmotic stress stimulates phospholipase D activity and elevates the levels of phosphatidic acid and diacylglycerol pyrophosphate. Plant J. 22, 147–154. doi: 10.1046/j.1365-313x.2000.00725.x
Munnik, T., and Testerink, C. (2009). Plant phospholipid signaling: “in a nutshell.” J. Lipid Res. 50, S260–S265. doi: 10.1194/jlr.R800098-JLR200
Munns, R. (2005). Genes and salt tolerance: bringing them together. New Phytol. 167, 645–663. doi: 10.1111/j.1469-8137.2005.01487.x
Mustilli, A. C., Merlot, S., Vavasseur, A., Fenzi, F., and Giraudat, J. (2002). Arabidopsis OST1 protein kinase mediates the regulation of stomatal aperture by abscisic acid and acts upstream of reactive oxygen species production. Plant Cell 14, 3089–3099. doi: 10.1105/tpc.007906
Nakashima, K., Yamaguchi-Shinozaki, K., and Shinozaki, K. (2014). The transcriptional regulatory network in the drought response and its crosstalk in abiotic stress responses including drought, cold, and heat. Front. Plant Sci. 5:170. doi: 10.3389/fpls.2014.00170
Ning, J., Li, X., Hicks, L. M., and Xiong, L. (2010). A Raf-like MAPKKK gene DSM1 mediates drought resistance through reactive oxygen species scavenging in rice. Plant Physiol. 152, 876–890. doi: 10.1104/pp.109.149856
Okazaki, Y., and Saito, K. (2014). Roles of lipids as signaling molecules and mitigators during stress response in plants. Plant J. 79, 584–596. doi: 10.1111/tpj.12556
Ouyang, S. Q., Liu, Y. F., Liu, P., Lei, G., He, S. J., Ma, B., et al. (2010). Receptor-like kinase OsSIK1 improves drought and salt stress tolerance in rice (Oryza sativa) plants. Plant J. 62, 316–329. doi: 10.1111/j.1365-313X.2010.04146.x
Pical, C., Westergren, T., Dove, S. K., Larsson, C., and Sommarin, M. (1999). Salinity and hyperosmotic stress induce rapid increases in phosphatidylinositol 4, 5-bisphosphate, diacylglycerol pyrophosphate, and phosphatidylcholine in Arabidopsis thaliana cells. J. Biol. Chem. 274, 38232–38240. doi: 10.1074/jbc.274.53.38232
Pornsiriwong, W., Estavillo, G. M., Chan, K. X., Tee, E. E., Ganguly, D., Crisp, P. A., et al. (2017). A chloroplast retrograde signal, 3'-phosphoadenosine 5'-phosphate, acts as a secondary messenger in abscisic acid signaling in stomatal closure and germination. Elife 6:e23361. doi: 10.7554/eLife.23361
Qin, B. X., Tang, D., Huang, J., Li, M., Wu, X. R., Lu, L. L., et al. (2011). Rice OsGL1-1 is involved in leaf cuticular wax and cuticle membrane. Mol. Plant 4, 985–995. doi: 10.1093/mp/ssr028
Ramanjulu, S., and Bartels, D. (2002). Drought-and desiccation-induced modulation of gene expression in plants. Plant Cell Environ. 25, 141–151. doi: 10.1046/j.0016-8025.2001.00764.x
Ramegowda, V., Basu, S., Krishnan, A., and Pereira, A. (2014). Rice GROWTH UNDER DROUGHT KINASE is required for drought tolerance and grain yield under normal and drought stress conditions. Plant Physiol. 166, 1634–1645. doi: 10.1104/pp.114.248203
Ryu, S. B., Karlsson, B. H., Ozgen, M., and Palta, J. P. (1997). Inhibition of phospholipase D by lysophosphatidylethanolamine, a lipid-derived senescence retardant. Proc. Natl. Acad. Sci. U.S.A. 94, 12717–12721. doi: 10.1073/pnas.94.23.12717
Sah, S. K., Reddy, K. R., and Li, J. (2016). Abscisic acid and abiotic stress tolerance in crop plants. Front. Plant Sci. 7:571. doi: 10.3389/fpls.2016.00571
Samuels, L., Kunst, L., and Jetter, R. (2008). Sealing plant surfaces: cuticular wax formation by epidermal cells. Annu. Rev. Plant Biol. 59, 683–707. doi: 10.1146/annurev.arplant.59.103006.093219
Sanders, D., Brownlee, C., and Harper, J. F. (1999). Communicating with calcium. Plant Cell 11, 691–706. doi: 10.1105/tpc.11.4.691
Saucedo-García, M., Gavilanes-Ruíz, M., and Arce-Cervantes, O. (2015). Long-chain bases, phosphatidic acid, MAPKs, and reactive oxygen species as nodal signal transducers in stress responses in Arabidopsis. Front. Plant Sci. 6:55. doi: 10.3389/fpls.2015.00055
Seo, P. J., Lee, S. B., Suh, M. C., Park, M. J., Go, Y. S., and Park, C.-M. (2011). The MYB96 transcription factor regulates cuticular wax biosynthesis under drought conditions in Arabidopsis. Plant Cell 23, 1138–1152. doi: 10.1105/tpc.111.083485
Seybold, H., Trempel, F., Ranf, S., Scheel, D., Romeis, T., and Lee, J. (2014). Ca2+ signalling in plant immune response: from pattern recognition receptors to Ca2+ decoding mechanisms. New Phytol. 204, 782–790. doi: 10.1111/nph.13031
Shao, H., Wang, H., and Tang, X. (2015). NAC transcription factors in plant multiple abiotic stress responses: progress and prospects. Front. Plant Sci. 6:902. doi: 10.3389/fpls.2015.00902
Shinozaki, K., and Yamaguchi-Shinozaki, K. (2007). Gene networks involved in drought stress response and tolerance. J. Exp. Bot. 58, 221–227. doi: 10.1093/jxb/erl164
Shivakumara, T. N., Sreevathsa, R., Dash, P. K., Sheshshayee, M., Papolu, P. K., Rao, U., et al. (2017). Overexpression of Pea DNA Helicase 45 (PDH45) imparts tolerance to multiple abiotic stresses in chili (Capsicum annuum L.). Sci. Rep. 7:2760. doi: 10.1038/s41598-017-02589-0
Shivaraj, S. M., Deshmukh, R. K., Rai, R., Bélanger, R., Agrawal, P. K., and Dash, P. K. (2017). Genome-wide identification, characterization, and expression profile of aquaporin gene family in flax (Linum usitatissimum). Sci. Rep. 7:46137. doi: 10.1038/srep46137
Srivasta, A., Mehta, S., Lindlof, A., and Bhargava, S. (2010). Over-represented promoter motifs in abiotic stress-induced DREB genes of rice and sorghum and their probable role in regulation of gene expression. Plant Signal. Behav. 5, 775–784. doi: 10.4161/psb.5.7.11769
Staxen, I., Pical, C., Montgomery, L. T., Gray, J. E., Hetherington, A. M., and Mcainsh, M. R. (1999). Abscisic acid induces oscillations in guard-cell cytosolic free calcium that involve phosphoinositide-specific phospholipase C. Proc. Natl. Acad. Sci. U.S.A. 96, 1779–1784. doi: 10.1073/pnas.96.4.1779
Tena, G., Boudsocq, M., and Sheen, J. (2011). Protein kinase signaling networks in plant innate immunity. Curr. Opin. Plant Biol. 14, 519–529. doi: 10.1016/j.pbi.2011.05.006
Thirunavukkarasu, N., Sharma, R., Singh, N., Shiriga, K., Mohan, S., Mittal, S., et al. (2017). Genomewide expression and functional interactions of genes under drought stress in maize. Int. J. Genomics 2017:2568706. doi: 10.1155/2017/2568706
Todaka, D., Shinozaki, K., and Yamaguchi-Shinozaki, K. (2015). Recent advances in the dissection of drought-stress regulatory networks and strategies for development of drought-tolerant transgenic rice plants. Front. Plant Sci. 6:84. doi: 10.3389/fpls.2015.00084
Toker, C., Lluch, C., Tejera, N., Serraj, R., and Siddique, K. (2007). “23 Abiotic Stresses,” in Chickpea Breedingand Management, eds S. S. Yadav, R. J. Redden, W. Chen, and B. Sharma (London, UK: CABI), 474.
Uga, Y., Sugimoto, K., Ogawa, S., Rane, J., Ishitani, M., Hara, N., et al. (2013). Control of root system architecture by DEEPER ROOTING 1 increases rice yield under drought conditions. Nat. Genet. 45, 1097–1102. doi: 10.1038/ng.2725
Umezawa, T., Fujita, M., Fujita, Y., Yamaguchi-Shinozaki, K., and Shinozaki, K. (2006). Engineering drought tolerance in plants: discovering and tailoring genes to unlock the future. Curr. Opin. Biotechnol. 17, 113–122. doi: 10.1016/j.copbio.2006.02.002
Van Gioi, H., Mallikarjuna, M. G., Shikha, M., Pooja, B., Jha, S. K., Dash, P. K., et al. (2017). Variable level of dominance of candidate genes controlling drought functional traits in maize hybrids. Front. Plant Sci. 8:940. doi: 10.3389/fpls.2017.00940
Varshney, R. K., Bansal, K. C., Aggarwal, P. K., Datta, S. K., and Craufurd, P. Q. (2011). Agricultural biotechnology for crop improvement in a variable climate: hope or hype? Trends Plant Sci. 16, 363–371. doi: 10.1016/j.tplants.2011.03.004
Walter, J., Nagy, L., Hein, R., Rascher, U., Beierkuhnlein, C., Willner, E., et al. (2011). Do plants remember drought? Hints towards a drought-memory in grasses. Environ. Exp. Bot. 71, 34–40. doi: 10.1016/j.envexpbot.2010.10.020
Wang, P., and Song, C. P. (2008). Guard-cell signalling for hydrogen peroxide and abscisic acid. New Phytol. 178, 703–718. doi: 10.1111/j.1469-8137.2008.02431.x
Wang, X. (2000). Multiple forms of phospholipase D in plants: the gene family, catalytic and regulatory properties, and cellular functions. Prog. Lipid Res. 39, 109–149. doi: 10.1016/S0163-7827(00)00002-3
Xiao, B., Huang, Y., Tang, N., and Xiong, L. (2007). Over-expression of a LEA gene in rice improves drought resistance under the field conditions. Theor. Appl. Genet. 115, 35–46. doi: 10.1007/s00122-007-0538-9
Xue, D., Zhang, X., Lu, X., Chen, G., and Chen, Z. H. (2017). Molecular and evolutionary mechanisms of cuticular wax for plant drought tolerance. Front. Plant Sci. 8:621. doi: 10.3389/fpls.2017.00621
Yamaguchi-Shinozaki, K., and Shinozaki, K. (2006). Transcriptional regulatory networks in cellular responses and tolerance to dehydration and cold stresses. Annu. Rev. Plant Biol. 57, 781–803. doi: 10.1146/annurev.arplant.57.032905.105444
Yang, J., Ordiz, M. I., Jaworski, J. G., and Beachy, R. N. (2011). Induced accumulation of cuticular waxes enhances drought tolerance in Arabidopsis by changes in development of stomata. Plant Physiol. Biochem. 49, 1448–1455. doi: 10.1016/j.plaphy.2011.09.006
Yu, D., Ranathunge, K., Huang, H., Pei, Z., Franke, R., Schreiber, L., et al. (2008). Wax Crystal-Sparse Leaf1 encodes a β-ketoacyl CoA synthase involved in biosynthesis of cuticular waxes on rice leaf. Planta 228, 675–685. doi: 10.1007/s00425-008-0770-9
Yu, J., Lai, Y., Wu, X., Wu, G., and Guo, C. (2016). Overexpression of OsEm1 encoding a group I LEA protein confers enhanced drought tolerance in rice. Biochem. Biophys. Res. Commun. 478, 703–709. doi: 10.1016/j.bbrc.2016.08.010
Zhang, G., Zhang, M., Zhao, Z., Ren, Y., Li, Q., and Wang, W. (2017). Wheat TaPUB1 modulates plant drought stress resistance by improving antioxidant capability. Sci. Rep. 7:7549. doi: 10.1038/s41598-017-08181-w
Zhang, J. (2011). China's success in increasing per capita food production. J. Exp. Bot. 62, 3707–3711. doi: 10.1093/jxb/err132
Zheng, L., Krishnamoorthi, R., Zolkiewski, M., and Wang, X. (2000). Distinct Ca2+ binding properties of novel C2 domains of plant phospholipase Dα and β. J. Biol. Chem. 275, 19700–19706. doi: 10.1074/jbc.M001945200
Zhou, L., Ni, E., Yang, J., Zhou, H., Liang, H., Li, J., et al. (2013). Rice OsGL1-6 is involved in leaf cuticular wax accumulation and drought resistance. PLoS ONE 8:e65139. doi: 10.1371/journal.pone.0065139
Zhou, M., Li, D., Li, Z., Hu, Q., Yang, C., Zhu, L., et al. (2013). Constitutive expression of a miR319 gene alters plant development and enhances salt and drought tolerance in transgenic creeping bentgrass. Plant Physiol. 161, 1375–1391. doi: 10.1104/pp.112.208702
Keywords: drought, plant growth, crops, rice, abiotic stress, lipid signaling
Citation: Dash PK, Rai R, Rai V and Pasupalak S (2018) Drought Induced Signaling in Rice: Delineating Canonical and Non-canonical Pathways. Front. Chem. 6:264. doi: 10.3389/fchem.2018.00264
Received: 31 August 2017; Accepted: 12 June 2018;
Published: 12 September 2018.
Edited by:
Raju Datla, National Research Council Canada (NRC-CNRC), CanadaReviewed by:
Manoj Kulkarni, National Research Council Canada (NRC-CNRC), CanadaCopyright © 2018 Dash, Rai, Rai and Pasupalak. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Prasanta K. Dash, cGRhc0BucmNwYi5vcmc=
Surendranath Pasupalak, cGFzdXBhbGFrQHlhaG9vLmNvLmlu
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.