- 1Molecular Neurobiology Group, Institute of Physiological Chemistry, University of Marburg, Marburg, Germany
- 2DFG Research Training Group, Membrane Plasticity in Tissue Development and Remodeling, GRK 2213, University of Marburg, Marburg, Germany
- 3Center for Mind, Brain and Behavior, University of Marburg and Justus-Liebig-University Giessen, Giessen, Germany
- 4Department of Pharmacological and Biomolecular Sciences, University of Milan, Milan, Italy
Cyclase-associated protein (CAP) has been discovered three decades ago in budding yeast as a protein that associates with the cyclic adenosine monophosphate (cAMP)-producing adenylyl cyclase and that suppresses a hyperactive RAS2 variant. Since that time, CAP has been identified in all eukaryotic species examined and it became evident that the activity in RAS-cAMP signaling is restricted to a limited number of species. Instead, its actin binding activity is conserved among eukaryotes and actin cytoskeleton regulation emerged as its primary function. However, for many years, the molecular functions as well as the developmental and physiological relevance of CAP remained unknown. In the present article, we will compile important recent progress on its molecular functions that identified CAP as a novel key regulator of actin dynamics, i.e., the spatiotemporally controlled assembly and disassembly of actin filaments (F-actin). These studies unraveled a cooperation with ADF/Cofilin and Twinfilin in F-actin disassembly, a nucleotide exchange activity on globular actin monomers (G-actin) that is required for F-actin assembly and an inhibitory function towards the F-actin assembly factor INF2. Moreover, by focusing on selected model organisms, we will review current literature on its developmental and physiological functions, and we will present studies implicating CAP in human pathologies. Together, this review article summarizes and discusses recent achievements in understanding the molecular, developmental and physiological functions of CAP, which led this protein emerge as a novel CAPt’n of actin dynamics.
Introduction
Cyclase-associated protein (CAP) has been discovered three decades ago in the budding yeast Saccharomyces (S.) cerevisiae as a component of a complex involved in activation of RAS-family GTPase ras-like protein 2 (RAS2), which controls activity of the adenylyl cyclase (AC) and, hence, cyclic adenosine monophosphate (cAMP) signaling (Fedor-Chaiken et al., 1990; Field et al., 1990). Yeast expressing mutant CAP variants suppress a phenotype (heat shock and nitrogen starvation sensitivity) elicited by the hyperactive RAS2 variant RAS2-V19, thus explaining its alternate name suppressor of RAS2-V19 (SRV2; Fedor-Chaiken et al., 1990). However, CAP mutant yeast displayed cellular defects including impaired growth and altered morphology that could not be attributed to defective RAS2-cAMP signaling (Field et al., 1990; Zelicof et al., 1993; Hubberstey and Mottillo, 2002), thereby suggesting an implication in additional cellular processes. Indeed, subsequent studies identified CAP as a multifunctional protein that apart from regulating RAS2 activity is capable of actin binding (Lila and Drubin, 1997; Hubberstey and Mottillo, 2002; Ono, 2013; Zhou et al., 2014a). Since these initial studies, CAP has been detected in all eukaryotic species examined (Ono, 2013). Lower eukaryotic organisms and most invertebrates possess only one CAP, instead some invertebrates [e.g., Caenorhabditis (C.) elegans] as well as vertebrates have two CAP isoforms with tissue specific expression (Ono, 2013). Studies on model organisms led to the assumption that CAP’s activity in RAS-cAMP signaling is restricted to a limited number of species including some fungi and protists (Bahn and Sundstrom, 2001; Hubberstey and Mottillo, 2002; Ono, 2013). However, a recent study suggested an interaction of CAP-actin complexes with AC in human pancreatic cancer cells (Quinn et al., 2017). Instead, CAP’s actin binding activity is highly conserved and present in higher eukaryotic cells (Ono, 2013; Zhou et al., 2014a). Early studies suggested a rather passive role for CAP in actin cytoskeleton regulation, which was believed to act via sequestering globular actin monomers (G-actin; Hubberstey and Mottillo, 2002). This view has changed drastically in the last decade, because CAP has been implicated in almost all steps relevant for actin dynamics, the spatiotemporally controlled assembly and disassembly of actin filaments (F-actin). Specifically, these studies unraveled (i) a cooperation of CAP with key actin regulators such as ADF/Cofilin and Twinfilin in F-actin disassembly including dissociation of actin subunits from filaments’ barbed and pointed ends as well as F-actin severing, (ii) a nucleotide exchange activity on G-actin that is required for F-actin assembly, and (iii) an inhibitory function towards the F-actin assembly factor inverted formin 2 (INF2), and they linked each individual actin activity to specific protein domains (Chaudhry et al., 2013; Jansen et al., 2014; Johnston et al., 2015; Kotila et al., 2018, 2019; Mu et al., 2019, 2020; Shekhar et al., 2019). In this article, we will summarize and discuss important recent progress in CAP’s structure and molecular functions, focusing on those studies that have been published since Shoichiro Ono’s excellent review in 2013 (Ono, 2013). Moreover, we will review current literature on CAP’s manifold developmental and physiological functions, focusing on selected model organisms including Drosophila (D.) melanogaster, C. elegans and mouse, and we will present studies implicating CAP in the mechanisms of human diseases. Together, in this review article we will provide a comprehensive overview of CAP’s molecular, developmental and physiological functions.
Recent Achievements in Structure and Molecular Functions
Structure and Domain Organization
Cyclase-associated proteins are multifunctional proteins, composed of 526 amino acids (AA) in yeast and 474 (CAP1) or 476 (CAP2) AA in mouse, and they consist of several distinct motifs and domains, including an oligomerization domain (OD), a helical folded domain (HFD), two proline-rich motifs (P1, P2) separated by a Wiscott-Aldrich-Syndrome protein (WASP) homology 2 (WH2) domain and followed by a domain termed CAP and RP2 (CARP) domain that harbors a dimerization motif at the most C-terminal part of the protein (Figure 1). Crystallization of full length CAP was not possible due to its tendency to form high molecular weight aggregates, possession of autoproteolytic activity and possibly due to ‘unstable’ WH2/P2 domains (Hofmann et al., 2002; Yusof et al., 2005; Kotila et al., 2018). Consequently, only the crystal structures of HFD and β-sheets within the CARP domain have been determined to date (Ksiazek et al., 2003; Dodatko et al., 2004; Mavoungou et al., 2004; Kotila et al., 2018, 2019). In this section, we will provide an overview of the structure and function for each individual CAP domain and motif. The next section comprises a detailed description of CAP’s activities in actin dynamics regulation. Although most of the initial work has been carried out in yeast, we decided to avoid the yeast nomenclature in this section and will refer to SRV2 as yeast CAP to keep a better flow. Further, in both sections, we did not differentiate between individual CAP isoforms.
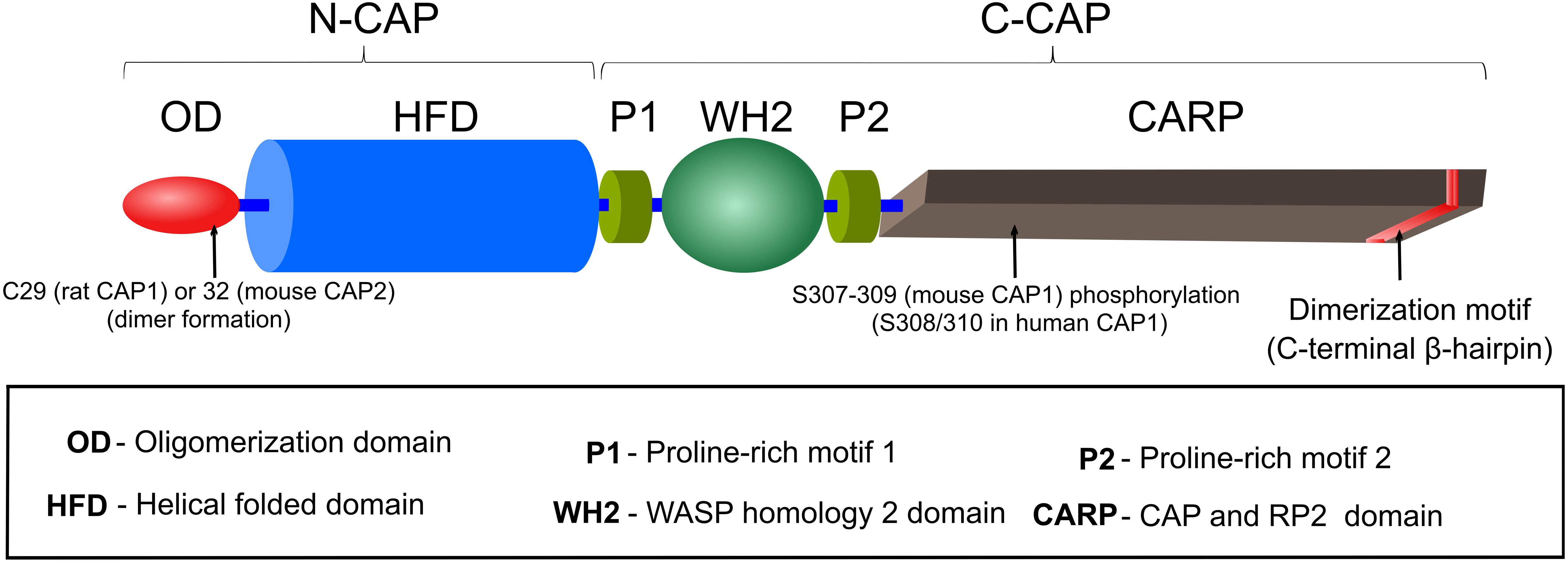
Figure 1. Domain organization of CAP. A detailed description of CAP’s motifs and domains is provided in the section ‘Structure and domain organization’.
Oligomerization Domain
Oligomerization domain (OD) is composed of the N-terminal ∼40 AA residues, which were predicted to form a coiled-coil structure due to presence of heptad repeat motifs (αXXαXXX), in which α represents a hydrophobic AA (Nishida et al., 1998). This region is sufficient for binding and activation of AC in yeast (Nishida et al., 1998), a function that is absent from CAP in higher eukaryotes (Hubberstey and Mottillo, 2002; Ono, 2013). In comparison to other CAP domains, OD is least conserved among species (Matviw et al., 1992). However, a requirement of this region for oligomerization is conserved from yeast to mammals, and oligomerization enhances CAP’s actin dynamics regulatory potential (Quintero-Monzon et al., 2009; Chaudhry et al., 2013; Jansen et al., 2014). Since in unbound protein OD is either unstructured or in random coil conformation, it has been hypothesized that this region could acquire coiled-coil structure upon AC binding or self-oligomerization (Ksiazek et al., 2003; Mavoungou et al., 2004). A specific role of this domain in oligomerization will be discussed below. Notably, this domain also includes a conserved cysteine residue, which recently was implicated in covalent dimer formation, Cofilin1 interaction and regulation of actin dynamics (Liu et al., 2018; Pelucchi et al., 2020a).
Helical Folded Domain
Helical folded domain encompasses AA residues ∼40–220 in (mouse) CAP, which forms an α-helix bundle composed of six antiparallel helices (15–25 AA residues each) that are connected by irregular loops of 5–12 AA residues (Ksiazek et al., 2003; Mavoungou et al., 2004). Although, earlier structural studies proposed that HFD could form dimers (Ksiazek et al., 2003; Yusof et al., 2005, 2006), later it was suggested that HFD/HFD interaction might be rather unspecific and transient and most likely does not contribute to stable dimer formation (Mavoungou et al., 2004; Yusof et al., 2006).
From a functional perspective, HFD binds to complexes composed of G-actin together with either ADF/Cofilin or Twinfilin (Moriyama and Yahara, 2002; Quintero-Monzon et al., 2009; Johnston et al., 2015), and it promotes F-actin depolymerization by interacting with ADF/Cofilin-bound pointed ends or Twinfilin-bound barbed ends (Johnston et al., 2015; Kotila et al., 2019; Shekhar et al., 2019). In addition, HFD interacts with ADF/Cofilin-bound F-actin to promote severing (Chaudhry et al., 2013), but this mechanism seems not to be the major contributor to CAP-dependent F-actin disassembly (Kotila et al., 2019; Shekhar et al., 2019). HFD’s structure is unique, not shared by any other ABP and differs from other actin-binding domains (ABD) in the mode of G-actin binding (Ksiazek et al., 2003; Kotila et al., 2019). Thus it binds efficiently to ADP-G-actin only in complex with ADF/Cofilin or Twinfilin (Moriyama and Yahara, 2002; Quintero-Monzon et al., 2009; Johnston et al., 2015). However, HFD’s interaction interface on G-actin does not overlap with that of ADF-H (actin depolymerizing factor homology domain), the ABD of ADF/Cofilin and Twinfilin, thereby allowing simultaneous binding of CAP, G-actin and ADF/Cofilin or Twinfilin (Kotila et al., 2019). HFD binds to the pointed end of G-actin, between subdomains (SD) 2 and 4. On the contrary, ADF-H binds to the barbed end between SD1 and SD3. It was proposed that binding of ADF/Cofilin induces a slight twist in the actin molecule (Kotila et al., 2019), thereby enabling HFD binding (Moriyama and Yahara, 2002).
Proline-Rich Motifs (P1, P2) and WH2 Domain
The central region comprising AA residues ∼220–320 in (mouse) CAP consists of two proline-rich motifs (P1, P2), which are separated by a WASP homology 2 (WH2) domain. Profilin, an ABP that promotes actin polymerization, binds to P1 of yeast CAP in vitro and in vivo (Bertling et al., 2007). Instead, Profilin can interact with both P1 and P2 domains of mouse CAP (Makkonen et al., 2013). Compared to P1, P2 contains fewer proline residues. It interacts with Src homology 3 (SH3) domain-containing proteins such as actin-binding protein 1 (ABP1) and is required for CAP localization to cortical actin patches in yeast (Freeman et al., 1996; Lila and Drubin, 1997; Balcer et al., 2003). Interestingly, human CAP also interacts with SH3 domain of the tyrosine kinase abelson murine leukemia viral oncogene homolog 1 (ABL) through its P1 region (Freeman et al., 1996). The functional relevance of CAP’s interaction with Profilin or ABL remained elusive, also because P1 mutations only caused very subtle effects in yeast (Bertling et al., 2007). However, genetic studies in D. melanogaster revealed at least functional interaction of CAP with Profilin or ABL, e.g., in determining cell morphology or in growth cone function (Wills et al., 1999, 2002; Benlali et al., 2000).
The function of the WH2 domain, located between P1 and P2 and encompassing AA residues ∼250–280, was deduced from its homology to other WH2 containing proteins including type I nucleation promoting factors (e.g., WASP and WAVE), Ena/VASP or β-Thymosin that participate in actin nucleation, F-actin elongation or G-actin sequestering, respectively (Paunola et al., 2002; Dominguez, 2016). Unlike most other WH2 domains that bind ATP-G-actin with higher affinity than ADP-G-actin (Dominguez, 2016), yeast CAP’s WH2 binds both ATP- and ADP-bound G-actin with similar affinity (Kd 1.5 μM) (Chaudhry et al., 2010). In comparison to yeast counterpart, mouse CAP’s WH2 possess slightly higher affinity towards ATP-bound G-actin (Kd 0.73 μM) (Makkonen et al., 2013). Whether this underlies species specific or experimental difference needs to be clarified. Yeast and mouse CAP’s WH2, together with the adjacent CARP domain (see below), are necessary for efficient catalysis of the ATP-for-ADP exchange on ADF/Cofilin-bound G-actin (Quintero-Monzon et al., 2009; Chaudhry et al., 2010; Jansen et al., 2014). Specific mutations disrupting G-actin-binding of WH2 caused actin disorganization associated with cell growth and cell morphogenesis defects in yeast (Chaudhry et al., 2010). WH2 domain of human CAP might bind to the ABP INF2, but additional CAP domains or interactions are necessary for this function (Mu et al., 2019, 2020). The functional relevance of this interaction is discussed below.
CAP and Retinitis Pigmentosa Protein 2 (CARP) Domain
Cyclase-associated protein’s C-terminus, between AA residues ∼320 and 474 possesses a β-sheet structure composed of six coils of right-handed parallel β-strands forming the core of the β-sheet and an additional C-terminal β-hairpin, which extends away from the core and participates in homodimer formation (Dodatko et al., 2004). β-hairpin interacts with the core β-sheet of a second CAP and forms a stable strand-exchanged dimer (Dodatko et al., 2004; Hliscs et al., 2010; Kotila et al., 2018). Since CAP’s β-sheet displays structural similarity to otherwise functionally unrelated proteins including X-linked retinitis pigmentosa protein 2 (RP2) this domain is referred to as CAP and RP2 (CARP) domain (Dodatko et al., 2004). CARP binds ADP-G-actin with high affinity (Kd 0.02–0.05 μM) and in 1:1 stoichiometry (Mattila et al., 2004; Makkonen et al., 2013). This interaction is unique in several ways (Iwase and Ono, 2017; Kotila et al., 2018). First, two CARP domains form a homodimer and bind simultaneously two ADP-G-actin, whereby each G-actin interacts with both CARP domains. Second, CARP binds to the pointed end of ADP-G-actin on SD1, SD2, and SD3 and forms the largest binding interface among known ABD (Kotila et al., 2018). These two observations explain the high affinity of CARP, however, only in dimeric form, towards ADP-G-actin (Iwase and Ono, 2016; Kotila et al., 2018). Thus, mutations disrupting CARP homodimer formation abolish interaction with ADP-G-actin and its recharging to ATP-G-actin (Iwase and Ono, 2016). Interestingly, structural analysis also revealed that CARP binding of G-actin could create a sterical clash with Profilin or ADF-H (Kotila et al., 2018). These findings provide an explanation why CARP competes with Profilin and ADF-H-containing ABP such as ADF/Cofilin and Twinfilin and enhances ADF/Cofilin dissociation from ADP-G-actin, thus priming it for nucleotide exchange (Mattila et al., 2004; Chaudhry et al., 2010; Ono, 2013; Johnston et al., 2015; Kotila et al., 2018). Notably, CARP’s binding interface on ADP-G-actin does not overlap with that of HFD, which is in line with the notion that both domains, presumably in cooperation with WH2, can work sequentially in ADF/Cofilin-mediated F-actin depolymerization and G-actin recharging (Kotila et al., 2019; Shekhar et al., 2019).
Function of CAP in Actin Dynamics
In order to maintain fast F-actin dynamics a tight balance between barbed end polymerization and pointed end depolymerization of F-actin is needed (Pollard, 2016). It has been demonstrated that ADF/Cofilin, in addition to F-actin severing, promotes pointed end depolymerization (Pollard, 2016; Shekhar and Carlier, 2017; Wioland et al., 2017), albeit with a relatively slow speed that cannot explain fast F-actin disassembly in cells (Miyoshi and Watanabe, 2013). Furthermore, upon ADF/Cofilin-mediated depolymerization ADP-G-actin must be removed from ADF/Cofilin and subsequently converted (recharged) to polymerization competent ATP-G-actin, a function which has been dedicated to Profilin (Goldschmidt-Clermont et al., 1991). However, compared to ADF/Cofilin (Kd 0.1 μM), the ADP-G-actin affinity of Profilin (Kd 1.5 μM) is rather low (Carlier et al., 1997; Vinson et al., 1998), suggesting that additional factors are required to mediate G-actin transition from ADF/Cofilin to Profilin and to convert ADP-G-actin into Profilin-binding favored form, i.e., ATP-G-actin. CAPs are ideal candidates for these functions, since they possess necessary protein domains to (i) efficiently interact with ADF/Cofilin-ADP-G-actin complexes and ADF/Cofilin-decorated filaments’ pointed ends (Moriyama and Yahara, 2002; Quintero-Monzon et al., 2009; Kotila et al., 2019; Shekhar et al., 2019), (ii) displace ADF/Cofilin from ADP-G-actin (Balcer et al., 2003; Mattila et al., 2004; Chaudhry et al., 2014; Kotila et al., 2018), and (iii) exhibit nucleotide exchanging activity on G-actin (Balcer et al., 2003; Chaudhry et al., 2007; Quintero-Monzon et al., 2009; Nomura et al., 2012). Recent studies from Pekka Lappalainen’s lab and from Bruce Goode’s lab (Kotila et al., 2018, 2019; Shekhar et al., 2019), together with previously generated data by the ADF/Cofilin and CAP research community (Ono, 2013), made a significant impact on our understanding how CAP can perform the aforementioned functions in vitro, and most likely in vivo. Based on this, we will summarize a proposed model for F-actin depolymerization and G-actin recharging carried out by CAP (Figure 2).
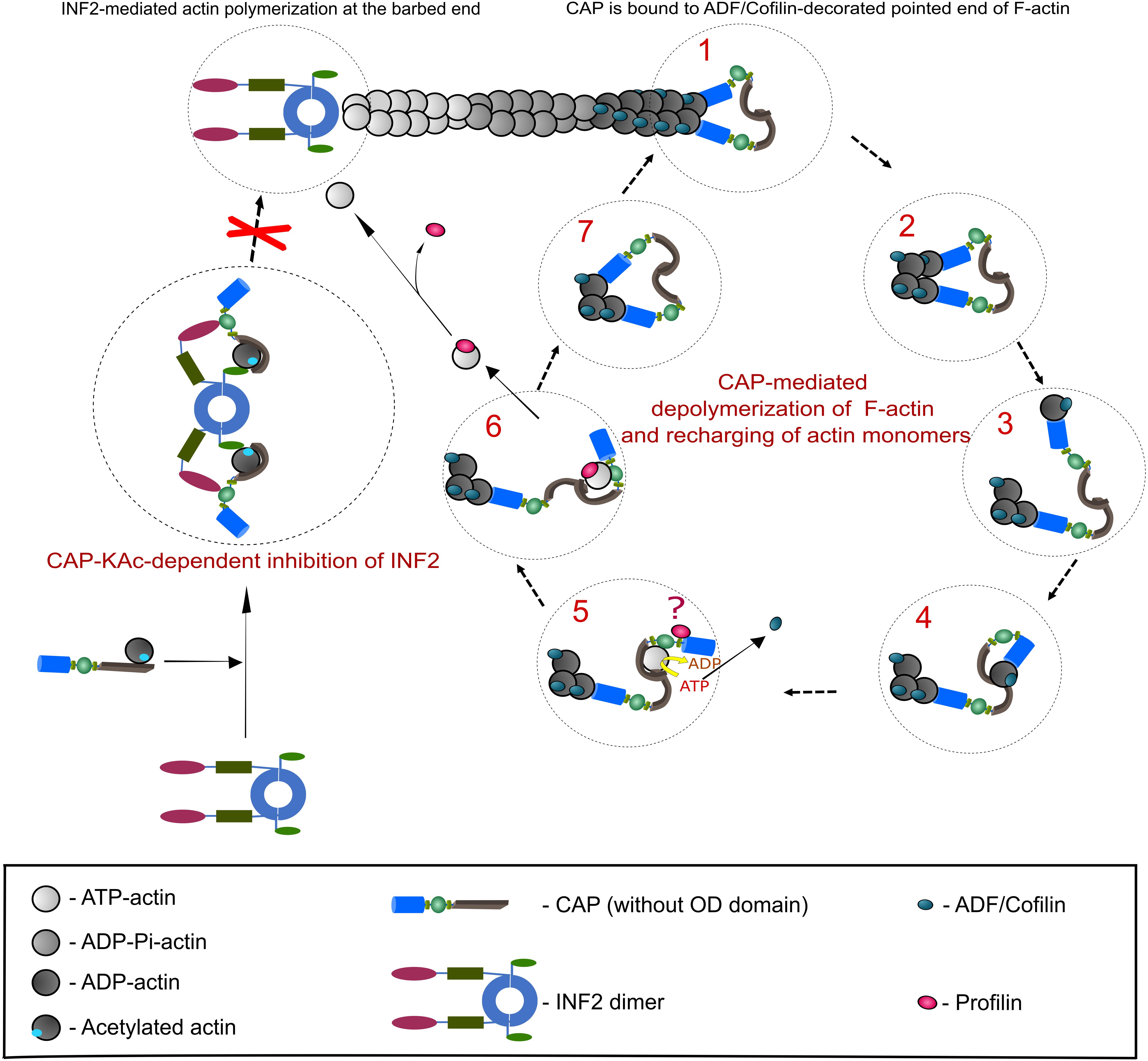
Figure 2. CAP-mediated regulation of actin dynamics. This scheme depicts the best studied CAP-dependent mechanisms relevant for actin dynamics. A detailed description of both mechanisms is provided in the section ‘Function of CAP in actin dynamics’, which also includes additional actin regulatory mechanisms that depend on CAP. Right panel presents a stepwise CAP-dependent depolymerization mechanism of ADF/Cofilin-decorated F-actin pointedends (based on Kotila et al., 2018, 2019). Although CAP forms hexamers, for the purpose of clarity we depicted them as dimers. Additionally, OD was removed. Two HFD (blue) of CAP dimer bind to ultimate and penultimate actin subunit at ADF/Cofilin-decorated pointed end (step 1). HFD thereby destabilizes intermolecular interaction of both actin subunits (step 2). HFD removes ADF/Cofilin-bound ultimate actin subunit from pointedend (step 3) and hands it over to CARPdomain (gray; step 4). Binding of CARP and WH2 (green) domains releases ADF/Cofilin and HFD from G-actin. Next, G-actin is recharged (ADP to ATP) and most likely transferred to Profilin (red) that binds CAP’s P1 motif (step 5 and 6). Question mark in step 5 indicates that the functional relationship between CAP and Profilin is still unclear. Polymerization competent, Profilin-bound ATP-G-actin can now be used for F-actin assembly. Step 7 indicates that a new round of CAP-dependent depolymerisation of ADF/Cofilin-bound actin starts. Left panel: in addition to actin depolymerisation at ADF/Cofilin-decorated pointed ends, CAP can impede F-actin assembly by inhibiting INF2 (depicted as dimer; based on Mu et al., 2019). Binding of a complex composed of lysine-acetylated (KAc-) actin and CAP to INF2’s DID (Diaphanous inhibitory domain, magenta) and DAD (Diaphanous autoregulatory domain, light green) keepINF2 in an inactive state, thereby inhibiting INF2-mediated actin polymerization.
F-Actin Depolymerizing and G-Actin Recharging
Cyclase-associated protein has two segments, N-CAP and C-CAP, which have specific biochemical activities and can function separately in controlling actin dynamics (Balcer et al., 2003; Mattila et al., 2004; Quintero-Monzon et al., 2009; Chaudhry et al., 2014). N-CAP is composed of OD and HFD (Ksiazek et al., 2003; Mavoungou et al., 2004), which have been implicated in oligomerization as well as F-actin depolymerization and severing (Quintero-Monzon et al., 2009; Chaudhry et al., 2013; Jansen et al., 2014; Kotila et al., 2019; Shekhar et al., 2019). C-CAP comprises P1, WH2 domain, P2 and CARP domain and catalyzes nucleotide exchange of G-actin (Moriyama and Yahara, 2002; Dodatko et al., 2004; Quintero-Monzon et al., 2009; Makkonen et al., 2013; Nomura and Ono, 2013; Jansen et al., 2014; Kotila et al., 2018). Structural analysis and modeling of a tripartite complex composed of a ADF-H, ADP-actin and HFD showed that ADF/Cofilin binding to ADP-actin at the filament’s pointed end changes the conformation of the actin subunits to favor simultaneous docking of two HFD to both the ultimate and the penultimate actin subunit (Tanaka et al., 2018; Kotila et al., 2019). HFD bound to the penultimate subunit destabilizes the interaction between both subunits, whereby the ADF/Cofilin-bound ultimate subunit dissociates from the pointed end while being bound to HFD (Kotila et al., 2019). As mentioned above, ADP-G-actin binding interfaces of ADF-H from ADF/Cofilin (or Twinfilin) and HFD from CAP do not overlap (Paavilainen et al., 2008). Therefore, ADF/Cofilin stays bound to HFD via ADP-G-actin, in line with previous biochemical data showing that N-CAP interacts efficiently only with ADF/Cofilin-ADP-G-actin complexes (Moriyama and Yahara, 2002; Quintero-Monzon et al., 2009). Furthermore, using dye labeled ADF/Cofilin-decorated F-actin, it was shown that both mouse and yeast N-CAP associate with pointed ends (Kotila et al., 2019; Shekhar et al., 2019), where the HFD binding interfaces of two terminal actin subunits might be exposed (Kotila et al., 2019). Importantly, treatment of either Gelsolin- or CAPZ-capped ADF/Cofilin-decorated F-actin with mouse N-CAP or yeast CAP enhanced pointed end depolymerization 30 or 100 fold, respectively (Kotila et al., 2019; Shekhar et al., 2019). Moreover, in comparison to the steady state (bare) F-actin depolymerization, presence of N-CAP or yeast CAP together with ADF/Cofilin increased the rate of pointed end depolymerization up to 100 or 330 fold, respectively (Kotila et al., 2019; Shekhar et al., 2019). Strikingly, although treatment of bare F-actin with mouse N-CAP increased pointed end depolymerization 2–3 folds (Kotila et al., 2019), the full length yeast CAP displayed 7-fold increase, twice as efficient as in ‘ADF/Cofilin only’ condition (Shekhar et al., 2019). Finally, full length mouse and yeast CAP accelerates pointed end F-actin depolymerization in conditions with high concentrations of Profilin and G-actin, thus mimicking physiological conditions (Kotila et al., 2019; Shekhar et al., 2019). Based on these observations, the depolymerization rate at ADF/Cofilin-bound pointed end by mouse N-CAP and yeast CAP was calculated as 13 and 44 subunits/second, respectively (Kotila et al., 2019; Shekhar et al., 2019). These values approach the estimated rates of actin turnover in vivo (Miyoshi and Watanabe, 2013), implying that synergistic interaction of ADF/Cofilin and CAP is the main driver of rapid F-actin depolymerization at pointed ends (Kotila et al., 2019; Shekhar et al., 2019). Twinfilin, another ADF-H-containing ABP involved in G-actin sequestering and barbed end capping, has been recently shown to enhance barbed end and pointed end F-actin depolymerization in presence of yeast CAP 3 and 17 fold, respectively (Goode et al., 1998; Paavilainen et al., 2007; Johnston et al., 2015). Despite the fact that mouse Twinfilin and CAP failed to jointly induce pointed end depolymerization, F-actin disassembly at barbed end was activated to the same extent (threefold) as with their yeast homologs (Hilton et al., 2018). Although, the pointed end disassembly mechanism might be similar to the one of ADF/Cofilin, how exactly CAP induces Twinfilin-mediated barbed end depolymerization needs further clarification.
In addition to CAP’s actin depolymerization activity inherent to N-CAP, C-CAP can promote dissociation of ADF/Cofilin from ADP-G-actin and catalyze nucleotide exchange on G-actin (Balcer et al., 2003; Mattila et al., 2004; Chaudhry et al., 2007; Quintero-Monzon et al., 2009; Nomura et al., 2012; Makkonen et al., 2013; Jansen et al., 2014). Mechanistically, this can be explained by the finding that ADP-G-actin binding interface of CARP does not overlap with that of HFD (Kotila et al., 2018, 2019). Thus, after removal of the ADF/Cofilin-bound terminal actin subunit from pointed ends by HFD, ADP-G-actin is transferred to the CARP domain, which has high affinity for ADP-G-actin (Moriyama and Yahara, 2002; Mattila et al., 2004; Kotila et al., 2018). Intriguingly, CARP binding to ADP-G-actin induces a sterical clash with ADF-H, and the WH2 binding of G-actin might further destabilize the association of ADF-H and ADP-G-actin (Balcer et al., 2003; Mattila et al., 2004; Chaudhry et al., 2010, 2014; Kotila et al., 2018). As a result, HFD as well as ADF/Cofilin dissociates from ADP-G-actin, which becomes subsequently recharged with ATP presumably by CARP in cooperation with WH2 (Chaudhry et al., 2014; Jansen et al., 2014; Kotila et al., 2018). Biochemical assays confirm the requirement of complete mouse and yeast C-CAP segment for nucleotide exchange of ADF/Cofilin-bound ADP-G-actin, implying that WH2 and CARP domains must be connected for efficient activity in G-actin recharging (Chaudhry et al., 2014; Jansen et al., 2014). In the next step, the only domain of CAP that has comparably higher affinity to ATP-G-actin, i.e., WH2 domain (Chaudhry et al., 2010), might release the polymerization competent G-actin to the surrounding or transfer it to ATP-G-actin-binding proteins such as Profilin (Bertling et al., 2007; Makkonen et al., 2013). Future studies will show which of those mechanisms occurs in vivo.
F-Actin Severing
F-actin severing by ADF/Cofilin enhances actin turnover, at least in part, by increasing the number of filaments’ pointed ends that can be depolymerized (Pollard, 2016). Based on real time assessment of immobilized F-actin sparsely labeled with biotin, it has been proposed that yeast and mouse CAP can enhance F-actin severing activity of ADF/Cofilin by direct interaction with ADF/Cofilin-decorated F-actin segments (Normoyle and Brieher, 2012; Chaudhry et al., 2013; Jansen et al., 2014). However, follow up studies using F-actin immobilized only via barbed end, thereby leaving the complete filament free in solution, showed that CAP effect on ADF/Cofilin-mediated F-actin severing is very modest when compared to F-actin depolymerization (Kotila et al., 2019; Shekhar et al., 2019). To which extent CAP accelerates ADF/Cofilin-mediated F-actin severing in vivo awaits further studies.
INF2 Inhibition
In addition to the aforementioned functions, CAP controls actin dynamics via its inhibitory function towards the F-actin assembly factor INF2 (Mu et al., 2019, 2020). Specifically, binding of lysine-acetylated actin (KAc-actin) containing CAP to INF2’s regulatory regions, i.e., DID (Diaphanous inhibitory domain) and DAD (Diaphanous autoregulatory domain), keeps INF2 in an inactive state (Mu et al., 2019, 2020). INF2 has been previously implicated in F-actin assembly at contact sites between the endoplasmic reticulum (ER) and mitochondria, which is relevant for mitochondrial recruitment of Dynamin-related protein 1 (DRP1), mitochondrial fission and calcium transfer from ER to mitochondria (Korobova et al., 2013; Chakrabarti et al., 2018). Interestingly, yeast CAP has been recently implicated in mitochondria morphology and function, too (Chen et al., 2019). This study revealed an interaction of yeast CAP with DRP1 and showed that CAP deletion caused elongated-hyperfused mitochondria associated with lower reserved respiration capacity. Hence, these data suggested a pro-fission activity for yeast CAP. CAP may control mitochondria morphology and function via regulating INF2 activity. However, apart from INF2, ADF/Cofilin has been implicated in mitochondrial DRP1-recruitment, mitochondrial dynamics and function, too (Chua et al., 2003; Klamt et al., 2009; Rehklau et al., 2012, 2017; Hoffmann et al., 2019), thereby offering an additional mode of CAP action on mitochondria.
G-Actin Sequestration
Based on earlier in vitro studies showing that CAP inhibits spontaneous actin polymerization, it was concluded that CAP acts as an actin sequestering protein (Gieselmann and Mann, 1992; Freeman et al., 1995; Mattila et al., 2004; Chaudhry et al., 2007; Peche et al., 2012; Nomura and Ono, 2013). Although CAP has high affinity to ADP-G-actin (Kd 0.02–0.05 μM; Mattila et al., 2004; Makkonen et al., 2013), it also possesses fast catalytic activity towards nucleotide exchange on G-actin (Chaudhry et al., 2010, 2013; Jansen et al., 2014). Taking into consideration that compared to Profilin (Kd 0.1 μM) or WH2 domain containing proteins CAP possesses relatively poor ATP-G-actin affinity (Kd 1.5 μM; Vinson et al., 1998; Chaudhry et al., 2010; Carlier and Shekhar, 2017), it is less likely that CAPs can efficiently sequester G-actin in a complex milieu of the cell. However, further investigations are needed to clarify CAP’s G-actin sequestering function and its implication in vivo.
Oligomerization as Necessity
Using different methods such as gel filtration or analytical ultracentrifugation, previous studies identified yeast, mouse and human CAP as part of a 600 kDa complex containing six CAP and six G-actin, thereby suggesting formation of CAP hexamers (Moriyama and Yahara, 2002; Balcer et al., 2003; Quintero-Monzon et al., 2009; Jansen et al., 2014; Mu et al., 2019). Furthermore, negative staining electron microscopy revealed that yeast and mouse N-CAP segments including OD and HFD form a wheel-like appearance with six symmetrical protrusions. Interestingly, within each of this protrusion one could dock only one HFD, thus forming again a hexameric structure. Additionally, other studies stated that human N-CAP form tetramers (Purde et al., 2019). Whether this discrepancy is due to different experimental conditions needs further clarification. The most N-terminal coiled-coil OD has been implicated in oligomerization, since yeast CAP lacking OD could only dimerize (Quintero-Monzon et al., 2009), most likely via CARP domain (see CARP domain section). Corroborating this result, human N-CAP lacking OD formed mostly monomers, albeit to a small extent also dimers (Purde et al., 2019). Importantly, deletion of OD in yeast CAP reduced ADF/Cofilin-mediated F-actin turnover in vitro and caused defects in cell growth, morphology and actin organization in vivo (Quintero-Monzon et al., 2009). Interestingly, a substitution of OD of human N-CAP with artificial ODs, capable of forming either dimers or trimers, potentiated cofilin mediated F-actin depolymerization and severing better with increasing level of oligomerization (Purde et al., 2019). Likewise, dimerizing HFD of human CAP with glutathione-S-transferase partially rescued a 20-fold reduction in pointed end depolymerization of ADF/Cofilin-decorated F-actin by OD lacking N-CAP (Kotila et al., 2019). In summary, these observations strongly suggest that higher-order oligomerization state is necessary for CAP’s function in actin dynamics.
Concluding Remarks on CAP Function in Actin Dynamics
Taken together, studies of the last decade drastically changed our view of CAP’s contribution to actin dynamics. Instead of being G-actin sequestering factor or passive player in actin turnover, these studies unraveled important functions for CAP in almost all steps of actin dynamics including F-actin depolymerization, F-actin severing, G-actin recharging and F-actin polymerization, making CAP an essential player in actin dynamics regulation (Chaudhry et al., 2013; Jansen et al., 2014; Johnston et al., 2015; Kotila et al., 2019; Mu et al., 2019; Shekhar et al., 2019).
Cellular, Developmental and Physiological Functions
Cellular Functions in Lower Eukaryotic Organisms
Cyclase-associated protein functions in budding yeast have been outlined above. Briefly, studies in S. cerevisiae revealed a role for CAP in RAS2-cAMP signaling (Fedor-Chaiken et al., 1990; Field et al., 1990), which may depend on CAP’s stimulatory effect on post-translational RAS2 modification (Lila and Drubin, 1997). Furthermore, an actin regulatory activity of CAP was first described in yeast (Vojtek et al., 1991; Balcer et al., 2003; Mattila et al., 2004; Bertling et al., 2007). Apart from S. cerevisiae, cellular CAP functions have been studied in the soil-dwelling amoeba Dictyostelium (D.) discoideum. CAP inactivation in D. discoideum caused defects in cell polarization, F-actin organization and phototaxis, thereby confirming the relevance of CAP for various cellular functions. In D. discoideum, CAP is relevant for both cAMP signaling and actin cytoskeleton regulation (Noegel et al., 2004; Sultana et al., 2012), similar to the yeast CAP. Furthermore, actin-regulating activity was located in CAP’s C-terminal region in D. discoideum, and it was inhibited by phosphatidylinositol 4,5-bisphosphate (PIP2) (Gottwald et al., 1996). In migrating amoeba CAP, was located at anterior and posterior plasma membrane regions and enriched in leading fronts upon chemotactic stimulation, thereby suggesting a function in PIP2-dependent actin cytoskeleton regulation. Moreover, these observations suggested important developmental functions for CAP in multicellular organisms.
Developmental and Physiological Functions in Invertebrates
Drosophila melanogaster only possesses a single CAP homolog (Ono, 2013), which has been identified simultaneously in two independent screens of mutant fly strains and termed Capulet and Act up, respectively (Baum et al., 2000; Benlali et al., 2000). In this review we stick to the term Capulet, which has been used more frequently in literature. Both screens unraveled Capulet as an important F-actin regulator that controls developmental patterning processes via actin-dependent mechanisms (Stevenson and Theurkauf, 2000). The first study found Capulet in a screen for oocyte polarity defects (Baum et al., 2000). During oogenesis, Capulet controls spatial F-actin assembly that is relevant for microtubule organization and, hence, for the asymmetric distribution of cell polarity determinants. The second study found antagonistic F-actin functions in the eye disk for Capulet and Chickadee, the fly homolog of Profilin. Although F-actin levels were increased in Capulet and reduced in Chickadee mutants (Benlali et al., 2000), both strains displayed similar defects in cell shape changes during eye development. These changes are relevant for the establishment of the eye disk morphogenetic furrow that restricts Sonic hedgehog (SHH) signaling and prevents premature photoreceptor differentiation. Consequently, SHH signaling and photoreceptor differentiation was less confined in mutant eyes (Benlali et al., 2000), demonstrating that actin-dependent cell morphological changes controlled by Capulet and Chickadee govern intercellular signaling cascades during development.
Since these pioneering, first genetic studies in multicellular organisms, several other important developmental functions have been unraveled for Capulet. It counterbalances F-actin assembly promoted by the Ena/VASP (vasodilator-stimulated phosphoprotein) protein Enabled at apical adherens junctions of follicular epithelia cells that cover oocytes during oogenesis (Baum and Perrimon, 2001). In this process, Capulet cooperated with the tyrosine kinase ABL, which interacts via its SH3 domain with Capulet’s proline-rich motif (Freeman et al., 1996). A function for Capulet in F-actin regulation at apical adherens junctions has been described also for wing epithelial cells (Major and Irvine, 2005). In these cells, Capulet acts downstream of Notch signaling in establishing a boundary between different cell populations and, hence, in wing compartmentalization. Boundary formation and wing compartmentalization was preserved in ABL mutants, demonstrating that ABL is not relevant for adherens junctions in wing epithelial cells. Hence, Capulet does not necessarily require ABL to be functional in epithelia cells. However, similar to follicular epithelia, a cooperation of Capulet and ABL has been reported in neuronal growth cones, which are dynamic and F-actin-enriched structures that navigate axons through the developing central nervous system (Wills et al., 2002). Specifically, Capulet and ABL interact in growth cone repulsion downstream of the secreted guidance cue Slit and its receptors of the Roundabout family, a pathway that spatially controls midline crossing of axons. Interestingly, Chickadee has been identified as another interaction partner of ABL in growth cones (Wills et al., 1999). Different from Capulet mutants, Chickadee mutants displayed growth cone arrest phenotype, suggesting opposing functions for both ABP in growth cones, similar to the eye disk (Benlali et al., 2000). While this study linked Capulet to F-actin regulation during neuron differentiation, another study described abnormal F-actin aggregates upon Capulet inactivation in differentiated neurons (Medina et al., 2008). In line with a biochemical function in releasing ADF/Cofilin from actin complexes (Johnston et al., 2015; Kotila et al., 2019; Shekhar et al., 2019), neurons from Capulet mutants displayed rod-like structures consisting of actin and ADF/Cofilin. Interestingly, similar ADF/Cofilin-actin rods have been found together with amyloid deposits and neurofibrillary tangles in brains from Alzheimer’s Disease (AD) patients as well as in abnormal actin aggregates termed Hirano bodies, which have been reported for AD and Parkinson’s disease (Minamide et al., 2000; Heredia et al., 2006; Gallo, 2007). Hence, defects in CAP-dependent neuronal actin dynamics may contribute to the pathology of human neurodegenerative diseases (see below).
Different from D. melanogaster and most other invertebrates, the nematode C. elegans expresses two CAP isoforms that have been termed CAS-1 and CAS-2, because the abbreviation CAP has been used already for actin capping proteins (Ono, 2013). CAS-1 and CAS-2 are encoded by distinct genes and differ in their expression pattern (Nomura et al., 2012; Nomura and Ono, 2013). While CAS-1 is abundant in muscle tissue, CAS-2 expression is restricted to non-muscle cells. In line with a conserved function in actin cytoskeleton regulation, CAS-1 binds G-actin and enhances exchange of actin-bound nucleotides (Nomura et al., 2012). Further, it promotes F-actin turnover in the presence of UNC-60B, the muscle-specific ADF/Cofilin homolog in C. elegans, which has been identified as an essential regulator of sarcomere F-actin organization in the larval body wall muscle (Ono et al., 1999, 2003). Genetic CAS-1 inactivation caused developmental arrest at larval stages, immobility as well as a severe F-actin disorganization in larval body wall muscles, while F-actin structures appeared normal in non-muscle tissues (Nomura et al., 2012). This study revealed a specific function for CAS-1 in striated muscles, and it strongly suggested a cooperative activity of CAS-1 and UNC-60B in sarcomere F-actin organization during myofibril differentiation, similar to a model that has been proposed for the mammalian homologs Cofilin2 and CAP2 (Kepser et al., 2019).
The second CAP homolog CAS-2 has been identified in C. elegans just a few years ago (Nomura and Ono, 2013). In vitro studies unraveled a primary function for CAS-2 in nucleotide exchange on G-actin and, hence, in F-actin assembly, which depends on β-sheets located in its CARP domain and on a C-terminal dimerization motif (Nomura and Ono, 2013; Iwase and Ono, 2016, 2017). CAS-2 has been shown to antagonize F-actin depolymerization and G-actin sequestration by the ADF/Cofilin homolog UNC-60A (Nomura and Ono, 2013). UNC-60A is widely expressed in non-muscle tissues and essential for embryonic cytokinesis and assembly of contractile actin networks in the somatic gonad (Ono et al., 2003, 2008), and CAS-2 may have similar important in vivo functions. However, the precise CAS-2 expression pattern has not been resolved to date and, due to the lack of CAS-2 mutant worms, its developmental and physiological functions remained unknown.
Developmental and Physiological Functions in Vertebrates
Unlike most invertebrates, vertebrates express two CAP isoforms with different expression patterns (Ono, 2013). In most vertebrate species investigated to date, one of these isoforms (CAP1) is broadly expressed, while expression of the second (CAP2) is restricted to a limited number of tissues, including heart, skeletal muscle and brain as well as - in lower amounts - in skin, testes and lung (Bertling et al., 2004; Peche et al., 2007). This led to the suggestion that vertebrate CAPs evolved cell type-specific functions (Ono, 2013), similar to the muscle-specific function of CAS-1 in C. elegans (Nomura et al., 2012). Although abundance in striated muscles has been reported for CAP2 in several species from frog (Xenopus laevis) and zebrafish (Danio rerio) to mammals (Bertling et al., 2004; Peche et al., 2007; Wolanski et al., 2009; Effendi et al., 2012), its function in striated muscles has been studied only in mice, in which systemic inactivation caused a dilated cardiomyopathy (DCM) together with impaired cardiac conduction (Peche et al., 2012; Stockigt et al., 2016). These defects might be caused by disturbed sarcomere organization and/or by reduced cooperativity of calcium-induced force generation, which have been both shown for isolated CAP2 mutant myofibrils (Peche et al., 2012). Impaired heart physiology in CAP2 mutant mice has been confirmed in an independent study that also included a mutant strain with specific CAP2 inactivation in cardiac muscle cells (Field et al., 2015). This study further reported that cardiac conduction defects can culminate in a complete heart block, which likely caused increased lethality of systemic and cardiomyocyte-specific mutants (Peche et al., 2012; Field et al., 2015). Of note, DCM and cardiac conduction defects have been recently associated with CAP2 mutations in humans (Aspit et al., 2019).
Gene expression analyses revealed an upregulation of fetal genes in hearts from CAP2 mutant mice prior to the manifestation of clinical symptoms (Xiong et al., 2019). Interestingly, target genes of the transcription factor serum response factor (SRF) including genes encoding for α-actin isoforms were overrepresented among upregulated genes. Pharmacological inhibition of SRF activity not only normalized expression of SRF downstream targets, but also prolonged normal cardiac function and survival in CAP2 mutant mice, thereby demonstrating that SRF dysregulation contributed to heart defects in mutant mice. Myocardin-related transcription factor (MRTF) is an important co-activator of SRF, which is sequestered by G-actin and promotes expression of cytoskeleton-related genes upon release from G-actin complexes (Olson and Nordheim, 2010; Esnault et al., 2014). CAP2 may control SRF-dependent gene expression in cardiomyocytes via regulating availability of G-actin, in line with the elevated nuclear MRTF levels in CAP2 mutant hearts. These findings support an interesting model, in which actin regulators such as CAP2 are developmentally and physiologically relevant not only by controlling local F-actin dynamics in subcellular structures, but also by governing gene expression.
While these studies unequivocally demonstrated the relevance of CAP2 for heart physiology in mice and human, CAP2 seems to be dispensable for heart development in embryogenesis (Peche et al., 2012; Field et al., 2015). Conversely, a recent study identified an important function for CAP2 in myofibril differentiation during skeletal muscle development (Kepser et al., 2019). Specifically, CAP2 inactivation delayed the sequential exchange of α-actin isoforms from smooth muscle and cardiac α-actin to skeletal muscle α-actin during early postnatal development. This delay coincided with the onset of motor function deficits and histopathological changes characterized by a high frequency of displaced myofibrils termed ring fibers (Kepser et al., 2019). A very similar delay in the ‘α-actin switch’ has been reported for mutant mice lacking Cofilin2 (Gurniak et al., 2014), suggesting that CAP2 and Cofilin2 cooperate in myofibril actin cytoskeleton differentiation, similar to CAS-1 and UNC-60B in C. elegans (Nomura et al., 2012). Notably, Cofilin2 mutant mice displayed skeletal muscle phenotypes similar to myopathies described for human patients with CFL2 mutations (Agrawal et al., 2007; Ockeloen et al., 2012; Ong et al., 2014). It is therefore tempting to speculate that mutations in the human CAP2 gene may cause skeletal muscle defects, too.
Deletions of the short arm of chromosome 6 that include the human CAP2 gene, have been described in a rare developmental disorder named 6p22 syndrome (Davies et al., 1999; Bremer et al., 2009; Celestino-Soper et al., 2012; Di Benedetto et al., 2013). This syndrome is characterized by developmental delays, heart defects as well as autism spectrum disorders (ASD) symptoms, which have been associated with synaptic defects (Bourgeron, 2015). Because CAP2 mutant mice displayed a delay in motor functions during postnatal development together with heart defects (Peche et al., 2012; Field et al., 2015; Kepser et al., 2019), a contribution of CAP2 loss to 6p22 syndrome has been suggested. Notably, apart from striated muscles, CAP2 is abundant in brain and present in different brain areas including cerebral cortex and hippocampus (Bertling et al., 2004; Peche et al., 2007; Kumar et al., 2016; Pelucchi et al., 2020a). In differentiated neurons, CAP2 is present in postsynaptic compartments (dendritic spines) of excitatory synapses and located in the F-actin enriched region underneath the postsynaptic density (Pelucchi et al., 2020a). CAP2 inactivation differently affected neuron structure and dendritic spine morphology in cerebral cortex and hippocampus. While primary cortical neurons from CAP2 mutant mice showed an increase in dendrite complexity and spine density (Kumar et al., 2016), CAP2 downregulation in primary hippocampal neurons reduced dendritic arborization and enlarged spines, which was associated with decreased synaptic excitatory transmission and impaired synaptic plasticity (Pelucchi et al., 2020a). Interestingly, CAP2 function in spine morphology and synaptic plasticity required its ability to form disulfide cross-linked homodimers, which were mediated by cysteine-residues at position 32 (C32). C32-dependent covalent dimerization of CAP2 was crucial for interaction with Cofilin1 (Pelucchi et al., 2020a), a key actin regulator in dendritic spines that controls spine morphology as well as different forms of synaptic plasticity, including long-term potentiation (LTP) and long-term depression (LTD; Rust, 2015). The mutation of the C32 of CAP2 significantly decreases, but not completely eliminates, CAP2 self-association and the binding to Cofilin1, suggesting that CAP2 can still form oligomers that are able to interact with Cofilin1. Yet, the lack of C32 covalent CAP2 dimers leads to a loss of function of CAP2/Cofilin1 complex on actin depolymerization, emphasizing the importance of C32 disulfide bond formation for the CAP2 function (Pelucchi et al., 2020a). In Cofilin1 mutant mice, mature spines were strongly enlarged in hippocampal and striatal neurons and LTD, which is associated with spine shrinkage or retraction, was not inducible (Rust et al., 2010; Wolf et al., 2015; Zimmermann et al., 2015), suggesting that Cofilin1 is relevant for F-actin disassembly in mature spines. Instead, during the early phase of LTP, Cofilin1 is recruited into dendritic spines where it may promote F-actin assembly that is required for spine expansion (Bosch et al., 2014). CAP2 may have a crucial role in the LTP-induced enrichment of Cofilin1 in spines: it has been shown that the C32-dependent CAP2 covalent dimerization and association to Cofilin1 are triggered by LTP and are required for LTP-induced Cofilin1 translocation into spines, spine remodeling and the potentiation of synaptic transmission (Pelucchi et al., 2020a). Redox-regulated disulfide bond formation represents an important post-translational control mechanism employed by several proteins, such as transcription factors, signaling proteins and cytoskeletal components to adjust their functional activity when reactive oxygen species (ROS) start to accumulate (Cremers and Jakob, 2013). Indeed, it is becoming clear that ROS are not simply toxic species, but are often transiently and locally produced as part of signaling pathways (Schippers et al., 2012; Cremers and Jakob, 2013). In neuronal cells, ROS generation has been also implicated in plasticity events (Bórquez et al., 2016), since it has been shown that the production of superoxide anion radical is required for the full expression of LTP and for memory tasks (Massaad and Klann, 2011). In light of these considerations, it might be possible that LTP-triggered changes in the redox balance could trigger the disulfide bond formation of CAP2 covalent dimers.
Together, these data implicated CAP2 in cellular processes that are believed to be fundamental for learning and memory (Holtmaat and Svoboda, 2009), and they suggested that CAP2 loss may contribute to ASD symptoms in 6p22 syndrome. Interestingly, a C29-dependent mechanism in covalent dimer formation relevant for F-actin and Cofilin1 binding has been also reported for CAP1 in rat mesangial cells (Liu et al., 2018). However, the physiological relevance of CAP1 covalent dimerization and Cofilin1 interaction has not been studied in neurons to date. Unlike C32 in CAP2, C29 in CAP1 is not conserved in humans, thereby limiting the relevance of CAP1 covalent dimerization for synaptic function.
Immunoblot analysis and in situ hybridization revealed broad CAP1 expression in mice, both during development and in adulthood (Bertling et al., 2004; Peche et al., 2007). However, only very little is known about its developmental and physiological functions, also because appropriate mouse models were missing. Transcription activator-like effector nuclease (TALEN)-engineered systemic CAP1 mutant mice have been reported just recently, but these mutants died at embryonic day 16.5, and their developmental defects have not been analyzed yet (Jang et al., 2019). Instead, heterozygous mutants with substantially reduced CAP1 protein levels were viable and showed defects in lipoprotein metabolism. Specifically, CAP1 was identified as an interaction partner of proprotein convertase subtilisin/kexin type-9 (PCSK9), which induces internalization and lysosomal degradation of low-density lipoprotein (LDL) receptor (LDLR). PCSK9 thereby enhances serum levels of LDL and LDL cholesterol, and it emerged as a valuable therapeutic target for atherosclerotic cardiovascular diseases (Burke et al., 2017). Mechanistically, PCSK9-binding of CAP1 promotes Caveolin-1-dependent endocytosis and lysosomal degradation of PCSK9-LDLR complexes (Jang et al., 2019). This pathway was impaired upon CAP1 inactivation, and heterozygous mice displayed increased LDLR levels and consequently reduced serum levels of LDL and LDL cholesterol. Hence, modulation of CAP1 activity may provide a novel therapeutic avenue for atherosclerosis and other cardiovascular diseases (Dron and Hegele, 2020). Apart from its function in lipoprotein metabolism, no other in vivo function has been reported for CAP1 to date, underlying the exigency of a conditional mouse model, which would also allow to test whether or not CAP1 is relevant for cytokine signaling, inflammation, adipose biology, coronary artery disease, chronic obstructive pulmonary disease or renal disease as suggested by recent studies (Lee et al., 2014; Xie et al., 2014; Munjas et al., 2017; Munjas et al., 2020).
Potential Implication in Human Diseases
As a major cytoskeletal component in eukaryotic cells, F-actin is involved in a variety of cellular processes. Together with actin motor proteins, it constitutes the primary machinery for the generation of protrusive and contractile forces (Castellano et al., 2001; Amberg et al., 2012). F-actin dynamic is relevant for cell migration, phagocytosis and membrane trafficking. In addition, actin is the target of executioner Caspases during apoptosis, and experiences oxidative damage when cellular stress occurs (Davidson and Wood, 2016). In this framework, ABP are critical for the precise control of the actin cytoskeleton, since they are responsible for forming the F-actin structures at the right place and time within the cell. In light of these considerations, the actin cytoskeleton and, thereby, ABP play key roles in many aspects of human health, ranging from embryonic development to aging, and are implicated in several diseases and pathological processes including cancer metastasis, wound repair, inflammation or neurodegenerative disorders (Davidson and Wood, 2016; Lai and Wong, 2020; Pelucchi et al., 2020b). As far as concern CAP1 and CAP2, potential roles have been described for various human pathologies apart from the already mentioned contributions of CAP1 to atherosclerosis or other cardiovascular diseases and of CAP2 to heart diseases and 6p22 syndrome.
CAP1 at the Crossroads of Metabolism and Cancer
Several publications reported altered CAP1 expression in a growing list of human cancers that include glioma, oral squamous cell carcinoma as well as breast, pancreatic, liver, lung and epithelial ovarian cancer. CAP1 is a protein relevant for cell migration and, thereby, presumably in metastasis formation. Indeed, dynamic actin cytoskeletal rearrangement, based on repeated cycles of F-actin turnover, is the primary driving force of cell migration and cancer cell invasiveness (Hall, 2009; Fife et al., 2014). Overexpression of CAP1 may have significant clinical implications as a diagnostic/prognostic factor for lung cancer (Tan et al., 2013), esophageal squamous cell carcinoma (Li et al., 2013), epithelial ovarian cancer (Hua et al., 2015) and glioma (Bao et al., 2016; Fan et al., 2016). The loss of CAP1 expression affects the breast cancer cell cycle (Yu et al., 2014), retards the glioma cells proliferation (Bao et al., 2016; Fan et al., 2016) and inhibits cell cycle progression in epithelial ovarian cancer cells (Hua et al., 2015).
CAP1 overexpression in hepatocellular carcinoma specimens correlates with tumor metastasis. Moreover, CAP1 co-localizes with actin in the leading edge of lamellipodia in hepatocellular carcinoma cells (Liu et al., 2014). CAP1 down-regulation impairs cells migration in hepatocellular carcinoma cells (Liu et al., 2014), in esophageal squamous cell carcinoma (Li et al., 2013), in breast cancer cells (Yu et al., 2014) and in glioma cells (Bao et al., 2016; Fan et al., 2016). However, the role for CAP1 in human cancers and in cell migration is still controversial, with mounting evidence suggesting a role that is dependent on the type or even subtype of cancer. CAP1 knockdown impaired F-actin dynamics, which in most cells leads to reduced cell motility. However, depletion of CAP1 in HeLa cells, while causing reduction in dynamics, actually caused increased cell motility through activation of cell adhesion signals (Zhou et al., 2014a). In metastatic breast cancer cells, depletion of CAP1 stimulated both the invasiveness and cell proliferation, while in non-metastatic MCF-7 cancer cells it actually had opposite effects (Zhang and Zhou, 2016).
Pancreatic cancer has the worst prognosis among cancers due to the difficulty of early diagnosis and its aggressive behavior. CAP1 was found upregulated in pancreatic cancer xenografts transplanted into immuno-deficient mice, and CAP1-positive tumor cells in clinical specimens correlated with the presence of lymph node metastasis and with the poor prognosis of patients (Yamazaki et al., 2009). CAP1 inactivation resulted in reduced lamellipodium formation, cell motility and invasion (Yamazaki et al., 2009). In another study, no changes in CAP1 expression in pancreatic cancer lines have been reported, but an increase in CAP1 phosphorylation at serine residues S308/S310 has been detected (Wu et al., 2019). CAP1 phosphorylation at this tandem phospho-site controls binding of Cofilin1 and actin and provide a mechanism that controls F-actin dynamics (Zhou et al., 2014b). The phosphorylation mutants showed defects in alleviating the elevated focal adhesion kinase (FAK) activity and enhanced focal adhesions in the CAP1 knockdown cells (Zhang et al., 2020). Overall, these results support the idea that transient CAP1 phosphorylation controls F-actin dynamics and cell adhesion. Interestingly, glycogen synthase kinase 3 (GSK3), which was reported to be hyper-activated in pancreatic cancer, can phosphorylate CAP1 (Zhou et al., 2014b). Disrupting CAP1 phospho-regulation via GSK3 inhibition or expressing phospho-site mutants compromised CAP1 functions in alleviating enhanced stress fibers and in rescuing invasiveness of CAP1-knockdown pancreatic cancer cells. These data suggest that transient CAP1 phosphorylation is relevant for the control of pancreatic cancer cell invasiveness (Wu et al., 2019). The involvement of this tandem phospho-site has been reported also in breast cancer cells, in which CAP1 has a role in the invasiveness and in regulating proliferative transformation of cancer cells, with ERK (extracellular signal-regulated kinase) signaling playing pivotal roles in mediating both cell functions (Zhang and Zhou, 2016).
In addition to the above-described mechanism related to cell migration and metastasis formation, CAP1 can contribute to cancer pathogenesis as Resistin receptor. Human Resistin is primarily expressed in and secreted from monocytes (Patel et al., 2003). Resistin-mediated chronic inflammation can lead to obesity, atherosclerosis, and other cardiometabolic diseases. In addition to the toll-like receptor 4 (TLR4; Tarkowski et al., 2010), CAP1 has been identified as receptor for human Resistin. Resistin binding of CAP1 via its SH3 domain upregulates cAMP concentration, protein kinase A (PKA) activity, and NF-κB-related transcription of inflammatory cytokines. Even though several biochemical binding assays have demonstrated the direct interaction between CAP1 and Resistin (Lee et al., 2014), the biological mechanism underlying CAP1/Resistin association at the plasma membrane requires further investigations considering that CAP1 lacks a transmembrane domain. Such concern should be taken into account also in relation to the binding of CAP1 to caveolin-1 and PCSK9, which is implicated in the caveolae-dependent endocytosis and lysosomal degradation of the LDLR. Further studies addressing how CAP1 is associated to the membrane are necessary to fully understand CAP1 cellular function.
CAP1 mediates the inflammatory response triggered by Resistin both in cultured human monocytes and in white adipose tissue in humanized Resistin mice in vivo (Lee et al., 2014). In addition, Resistin increases chemokine production by fibroblast-like synoviocytes via CAP1 in synovial tissue, thus contributing to the pathogenesis of rheumatoid arthritis (Sato et al., 2017).
Considering Resistin’s ability to stimulate lipid uptake and atherosclerotic plaque progression, CAP1 and Resistin levels have been assessed in patients affected by coronary artery disease. The results revealed a significant increase in plasma Resistin levels and in CAP1 expression in peripheral blood mononuclear cells of coronary artery disease patients, suggesting that Resistin is able to exert its effects stronger on cells with up-regulated CAP1 (Munjas et al., 2017).
An increase in Resistin plasma levels and CAP1 expression in peripheral blood mononuclear cells has been reported in colorectal cancer patients (Mihajlovic et al., 2019). Resistin can also contribute to pancreatic cancer pathogenesis, since its levels are increased in pancreatic cancer patients and correlate positively with tumor grades. Moreover, the Resistin receptors CAP1 and TLR4 mediate the effects of Resistin on cancer cells through activation of STAT3 (signal transducer and activator of transcription 3) and are implicated in the resistance to chemotherapy (Zhang et al., 2019).
Obesity represents a major risk for developing several types of cancer, including breast cancer (Calle and Kaaks, 2004). Resistin is among the top modulated adipokines secreted by adipocytes under obesity-associated metabolic conditions and therefore represents a plausible soluble mediator in the link between obesity, metabolic complications and breast cancer via the binding to CAP1 (Rosendahl et al., 2018). In a study, CAP1 gene and Resistin gene variants were associated with increased risk of breast cancer among Mexican women (Munoz-Palomeque et al., 2018). CAP1 is expressed across a large panel of breast cancer cell lines and primary human tumor and high CAP1 expression is associated with poor tumor characteristics and impaired prognosis among breast cancer patients (Rosendahl et al., 2018). Low CAP1 tumor expression was associated with higher body fatness and worse survival outcomes in breast cancer patients (Bergqvist et al., 2020). Moreover, Resistin increases breast cancer metastasis potential through induction epithelial to mesenchymal transition, a process in which cancer cells lose their epithelial characteristics and gain mesenchymal-like features, and that these effects may be associated with CAP1 (Avtanski et al., 2019).
CAP2 Role in AD, Wound Repair and Cancer
We have mentioned above the importance of CAP2 for synaptic plasticity that is relevant for learning and memory. F-actin alterations in dendritic spines have been described in AD, the most common form of dementia characterized by synaptic dysfunction in the early stages of the pathogenesis (Pelucchi et al., 2020b). CAP2 levels and synaptic localization are specifically reduced in the hippocampus, but not in the cortex of AD patients. Interestingly, CAP2 levels are increased in the cerebrospinal fluid of AD patients, but not in subjects affected by frontotemporal dementia, indicating the specificity of the alteration for this form of dementia. Furthermore, in AD hippocampal synapses CAP2 covalent dimer levels are decreased and Cofilin1 association to CAP2 covalent dimer/monomer is altered. These data suggested the presence of an ineffective CAP2-Cofilin1 complex in AD hippocampal synapses, which may contribute to impaired structural plasticity in AD (Pelucchi et al., 2020a).
CAP2 could be also involved in wound repair since CAP2 mutant mice showed an altered wound healing response (Kosmas et al., 2015). CAP2 in murine and human skin is present in the nucleus, in the cytosol and in the cell periphery. The keratinocytes from CAP2 mutant mice showed reduced velocity and a delay in scratch closure. Moreover, in human wounds, CAP2 is also expressed in hyper-proliferative epidermis. The fibroblasts of CAP2 mutant mice develop extended protrusions, increased focal adhesions and showed slower migration velocity, thereby suggesting a model in which a stabilization of focal adhesions as well as a disruption of cell polarity impaired motility of CAP2-deficient cells (Kosmas et al., 2015). The formation of a dense meshwork of peripheral F-actin and the disruption of the cell polarity may also contribute to reduced cell motility necessary to promote the wound healing process (Kosmas et al., 2015).
Similar to CAP1, CAP2 has been implicated in cancer pathogenesis and particularly in invasiveness. CAP2 is overexpressed in different cancer and is an unfavorable biomarker for prognostic prediction for patients affected by breast cancer (Xu et al., 2016), epithelial ovarian cancer (Adachi et al., 2020), malignante melanoma (Masugi et al., 2015), gastric cancer (Li et al., 2020), and glioma (Saker et al., 2020). Furthermore, oligonucleotides array technology revealed CAP2 as one of the genes upregulated in early hepatocellular carcinoma (Chuma et al., 2003). CAP2 overexpression is observed in a stepwise manner during the hepatocellular carcinoma progression and, in the early stages, the invading tumor cells were CAP2-positive (Shibata et al., 2006). Indeed, CAP2 overexpression is considered a poor prognostic biomarker for hepatocellular carcinoma patients (Fu et al., 2015). Interestingly, CAP2 and actin co-localized in the leading edge of lamellipodia from hepatocellular carcinoma cells. CAP2 knockdown inhibited lamellipodia extension upon serum stimulation and decreased cell motility. These data suggest a role for CAP2 in F-actin dynamics at the leading edge of lamellipodia, which is a characteristic feature of motile cells. Moreover, the overexpression of CAP2 correlates with portal vein invasion and intrahepatic metastasis, indicating CAP2 involvement in promoting the invasive behavior of hepatocellular carcinoma cells (Effendi et al., 2012).
Concluding Remarks
Actin dynamics is coordinated by multiple ABP and involved in a variety of cellular functions including muscle contraction, cell movement, intracellular transport, and transcriptional regulation within the nucleus. In this complex picture, CAP, an ABP conserved among eukaryotes, has acquired crucial roles. First described as a G-actin sequestering factor, more recently CAP has emerged as a molecular hub able to orchestrate F-actin depolymerization, G-actin recharging and F-actin severing. The existence of two CAP isoforms with relatively low homology (Yu et al., 1994) and different expression patterns in vertebrates, further increases the complexity. CAP1 is essential in most cell types, while CAP2 appears to have unique roles. Considering that most studies so far have been focused on CAP1, it will be important to investigate the function of CAP2 in specific cell types expressing high CAP2 levels.
In addition to the actin regulating activity, CAP1 has been identified as Resistin receptor and protein partner of PCSK9, thus highlighting the involvement of CAP1 in metabolic processes. Such novel biological function suggests that CAP could be not just an ABP, but it can be involved in multiple biological pathways. This hypothesis is supported by the involvement of CAP1 and CAP2 in the pathogenesis of different diseases.
How are CAP-mediated processes governed in cells? To address this issue, it will be crucial to integrate structural information and biological studies to identify post-translational modifications and signaling pathways controlling CAP activity. Such information will be relevant for a comprehensive characterization of CAP’s cellular functions and for understanding potential CAP dysregulation that may contribute to the pathogenesis of human diseases.
Author Contributions
MR, SK, SP, and EM wrote the article. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by grants from the Cariplo Foundation (project code 2018-0511) to MR and EM, by grants to MR [German Research Foundation (DFG) RU 1232/7-1 and Research Campus Mid-Hessen (FCMH) “Mitochondrial network initiative”], and by grants to EM [Italian Ministry of Education, University and Research (MIUR) PRIN 2017B9NCSX, MIUR Progetto Eccellenza, PON “Ricerca e Innovazione” PerMedNet Project (ARS01_01226), and Fondo di sviluppo unimi-linea2 – PSR2019].
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Abbreviations
AA, amino acid; ABD, actin-binding domain; ABL, abelson murine leukemia viral oncogene homolog 1; ABP, actin-binding protein; ABP1, actin-binding protein 1; AC, adenylyl cyclase; AD, Alzheimer’s disease; ADF, actin depolymerizing factor; ADF-H, actin depolymerizing factor homology domain; ASD, autism spectrum disorder; CAP, cyclase-associated protein; CAPZ, capping protein in Z band; CARP, CAP and RP2 domain; CAS-1/-2, CAP homologs in C. elegans; C-CAP, C-terminal part of CAP; DAD, diaphanous autoregulatory domain; DCM, dilated cardiomyopathy; DID, diaphanous inhibitory domain; DRP1, dynamin-related protein 1; ER, endoplasmic reticulum; ERK, extracellular signal-regulated kinase; F-actin, actin filament; FAK, focal adhesion kinase; G-actin, globular actin monomer; GSK3, glycogen synthase kinase 3; HFD, helical folded domain; INF2, inverted formin 2; KAc, lysine-acetylated; LDL, low-density lipoprotein; LDLR, LDL receptor; LTD, long-term depression; LTP, long-term potentiation; MRTF, myocardin-related transcription factor; N-CAP, N-terminal part of CAP; OD, oligomerization domain; P1, proline-rich motif 1; P2, proline-rich motif 2; PCSK9, proprotein convertase subtilisin/kexin type-9; PIP2, phosphatidylinositol 4,5-bisphosphate; PKA, protein kinase A; RAS2, ras-like protein 2; RP2, retinitis pigmentosa protein 2; SD, subdomain; SH3, Src homology 3 domain; SHH, sonic hedgehog; SRF, serum response factor; SRV2, suppressor of Ras2-Val19; STAT3, signal transducer and activator of transcription 3; TALEN, transcription activator-like effector nuclease; TLR4, toll-like receptor 4; UNC-60A/-60B, ADF/Cofilin homologs in C. elegans; VASP, vasodilator-stimulated phosphoprotein; WASP, Wiscott-Aldrich-Syndrome protein; WH2, WASP homology 2 domain.
References
Adachi, M., Masugi, Y., Yamazaki, K., Emoto, K., Kobayashi, Y., Tominaga, E., et al. (2020). Upregulation of cyclase-associated actin cytoskeleton regulatory protein 2 in epithelial ovarian cancer correlates with aggressive histologic types and worse outcomes. Jpn. J. Clin. Oncol. 50, 643–652. doi: 10.1093/jjco/hyaa026
Agrawal, P. B., Greenleaf, R. S., Tomczak, K. K., Lehtokari, V. L., Wallgren-Pettersson, C., Wallefeld, W., et al. (2007). Nemaline myopathy with minicores caused by mutation of the CFL2 gene encoding the skeletal muscle actin-binding protein, cofilin-2. Am. J. Hum. Genet. 80, 162–167. doi: 10.1086/510402
Amberg, D., Leadsham, J. E., Kotiadis, V., and Gourlay, C. W. (2012). Cellular ageing and the actin cytoskeleton. Subcell. Biochem. 57, 331–352. doi: 10.1007/978-94-007-2561-4_15
Aspit, L., Levitas, A., Etzion, S., Krymko, H., Slanovic, L., Zarivach, R., et al. (2019). CAP2 mutation leads to impaired actin dynamics and associates with supraventricular tachycardia and dilated cardiomyopathy. J. Med. Genet. 56, 228–235. doi: 10.1136/jmedgenet-2018-105498
Avtanski, D., Chen, K., and Poretsky, L. (2019). Resistin and adenylyl cyclase-associated protein 1 (CAP1) regulate the expression of genes related to insulin resistance in BNL CL.2 mouse liver cells. Data Brief 25:104112. doi: 10.1016/j.dib.2019.104112
Bahn, Y. S., and Sundstrom, P. (2001). CAP1, an adenylate cyclase-associated protein gene, regulates bud-hypha transitions, filamentous growth, and cyclic AMP levels and is required for virulence of Candida albicans. J. Bacteriol. 183, 3211–3223. doi: 10.1128/JB.183.10.3211-3223.2001
Balcer, H. I., Goodman, A. L., Rodal, A. A., Smith, E., Kugler, J., Heuser, J. E., et al. (2003). Coordinated regulation of actin filament turnover by a high-molecular-weight Srv2/CAP complex, cofilin, profilin, and Aip1. Curr. Biol. 13, 2159–2169. doi: 10.1016/j.cub.2003.11.051
Bao, Z., Qiu, X., Wang, D., Ban, N., Fan, S., Chen, W., et al. (2016). High expression of adenylate cyclase-associated protein 1 accelerates the proliferation, migration and invasion of neural glioma cells. Pathol. Res. Pract. 212, 264–273. doi: 10.1016/j.prp.2015.12.017
Baum, B., Li, W., and Perrimon, N. (2000). A cyclase-associated protein regulates actin and cell polarity during Drosophila oogenesis and in yeast. Curr. Biol. 10, 964–973. doi: 10.1016/S0960-9822(00)00640-0
Baum, B., and Perrimon, N. (2001). Spatial control of the actin cytoskeleton in Drosophila epithelial cells. Nat. Cell Biol. 3, 883–890. doi: 10.1038/ncb1001-883
Benlali, A., Draskovic, I., Hazelett, D. J., and Treisman, J. E. (2000). act up controls actin polymerization to alter cell shape and restrict Hedgehog signaling in the Drosophila eye disc. Cell 101, 271–281. doi: 10.1016/S0092-8674(00)80837-5
Bergqvist, M., Elebro, K., Sandsveden, M., Borgquist, S., and Rosendahl, A. H. (2020). Effects of tumor-specific CAP1 expression and body constitution on clinical outcomes in patients with early breast cancer. Breast Cancer Res. 22:67. doi: 10.1186/s13058-020-01307-5
Bertling, E., Hotulainen, P., Mattila, P. K., Matilainen, T., Salminen, M., and Lappalainen, P. (2004). Cyclase-associated protein 1 (CAP1) promotes cofilin-induced actin dynamics in mammalian nonmuscle cells. Mol. Biol. Cell 15, 2324–2334. doi: 10.1091/mbc.e04-01-0048
Bertling, E., Quintero-Monzon, O., Mattila, P. K., Goode, B. L., and Lappalainen, P. (2007). Mechanism and biological role of profilin-Srv2/CAP interaction. J. Cell Sci. 120, 1225–1234. doi: 10.1242/jcs.000158
Bórquez, D. A., Urrutia, P. J., Wilson, C., van Zundert, B., Núñez, M. T., and González-Billault, C. (2016). Dissecting the role of redox signaling in neuronal development. J. Neurochem. 137, 506–517. doi: 10.1111/jnc.13581
Bosch, M., Castro, J., Saneyoshi, T., Matsuno, H., Sur, M., and Hayashi, Y. (2014). Structural and molecular remodeling of dendritic spine substructures during long-term potentiation. Neuron 82, 444–459. doi: 10.1016/j.neuron.2014.03.021
Bourgeron, T. (2015). From the genetic architecture to synaptic plasticity in autism spectrum disorder. Nat. Rev. Neurosci. 16, 551–563. doi: 10.1038/nrn3992
Bremer, A., Schoumans, J., Nordenskjold, M., Anderlid, B. M., and Giacobini, M. (2009). An interstitial deletion of 7.1Mb in chromosome band 6p22.3 associated with developmental delay and dysmorphic features including heart defects, short neck, and eye abnormalities. Eur. J. Med. Genet. 52, 358–362. doi: 10.1016/j.ejmg.2009.06.002
Burke, A. C., Dron, J. S., Hegele, R. A., and Huff, M. W. (2017). PCSK9: regulation and target for drug development for dyslipidemia. Annu. Rev. Pharmacol. Toxicol. 57, 223–244. doi: 10.1146/annurev-pharmtox-010716-104944
Calle, E. E., and Kaaks, R. (2004). Overweight, obesity and cancer: epidemiological evidence and proposed mechanisms. Nat. Rev. Cancer 4, 579–591. doi: 10.1038/nrc1408
Carlier, M. F., Laurent, V., Santolini, J., Melki, R., Didry, D., Xia, G. X., et al. (1997). Actin depolymerizing factor (ADF/cofilin) enhances the rate of filament turnover: implication in actin-based motility. J. Cell Biol. 136, 1307–1322. doi: 10.1083/jcb.136.6.1307
Carlier, M. F., and Shekhar, S. (2017). Global treadmilling coordinates actin turnover and controls the size of actin networks. Nat. Rev. Mol. Cell Biol. 18, 389–401. doi: 10.1038/nrm.2016.172
Castellano, F., Chavrier, P., and Caron, E. (2001). Actin dynamics during phagocytosis. Semin. Immunol. 13, 347–355. doi: 10.1006/smim.2001.0331
Celestino-Soper, P. B., Skinner, C., Schroer, R., Eng, P., Shenai, J., Nowaczyk, M. M., et al. (2012). Deletions in chromosome 6p22.3-p24.3, including ATXN1, are associated with developmental delay and autism spectrum disorders. Mol. Cytogenet. 5:17. doi: 10.1186/1755-8166-5-17
Chakrabarti, R., Ji, W. K., Stan, R. V., de Juan Sanz, J., Ryan, T. A., and Higgs, H. N. (2018). INF2-mediated actin polymerization at the ER stimulates mitochondrial calcium uptake, inner membrane constriction, and division. J. Cell Biol. 217, 251–268. doi: 10.1083/jcb.201709111
Chaudhry, F., Breitsprecher, D., Little, K., Sharov, G., Sokolova, O., and Goode, B. L. (2013). Srv2/cyclase-associated protein forms hexameric shurikens that directly catalyze actin filament severing by cofilin. Mol. Biol. Cell 24, 31–41. doi: 10.1091/mbc.e12-08-0589
Chaudhry, F., Guerin, C., von Witsch, M., Blanchoin, L., and Staiger, C. J. (2007). Identification of Arabidopsis cyclase-associated protein 1 as the first nucleotide exchange factor for plant actin. Mol. Biol. Cell 18, 3002–3014. doi: 10.1091/mbc.e06-11-1041
Chaudhry, F., Jansen, S., Little, K., Suarez, C., Boujemaa-Paterski, R., Blanchoin, L., et al. (2014). Autonomous and in Trans functions for the two halves of Srv2/CAP in promoting actin turnover. Cytoskeleton 71, 351–360. doi: 10.1002/cm.21170
Chaudhry, F., Little, K., Talarico, L., Quintero-Monzon, O., and Goode, B. L. (2010). A central role for the WH2 domain of Srv2/CAP in recharging actin monomers to drive actin turnover in vitro and in vivo. Cytoskeleton 67, 120–133. doi: 10.1002/cm.20429
Chen, Y. C., Cheng, T. H., Lin, W. L., Chen, C. L., Yang, W. Y., Blackstone, C., et al. (2019). Srv2 is a pro-fission factor that modulates yeast mitochondrial morphology and respiration by regulating actin assembly. iScience 11, 305–317. doi: 10.1016/j.isci.2018.12.021
Chua, B. T., Volbracht, C., Tan, K. O., Li, R., Yu, V. C., and Li, P. (2003). Mitochondrial translocation of cofilin is an early step in apoptosis induction. Nat. Cell Biol. 5, 1083–1089. doi: 10.1038/ncb1070
Chuma, M., Sakamoto, M., Yamazaki, K., Ohta, T., Ohki, M., Asaka, M., et al. (2003). Expression profiling in multistage hepatocarcinogenesis: identification of HSP70 as a molecular marker of early hepatocellular carcinoma. Hepatology 37, 198–207. doi: 10.1053/jhep.2003.50022
Cremers, C. M., and Jakob, U. (2013). Oxidant sensing by reversible disulfide bond formation. J. Biol. Chem. 288, 26489–26496. doi: 10.1074/jbc.R113.462929
Davidson, A. J., and Wood, W. (2016). Unravelling the actin cytoskeleton: a new competitive edge? Trends Cell Biol. 26, 569–576. doi: 10.1016/j.tcb.2016.04.001
Davies, A. F., Mirza, G., Sekhon, G., Turnpenny, P., Leroy, F., Speleman, F., et al. (1999). Delineation of two distinct 6p deletion syndromes. Hum. Genet. 104, 64–72. doi: 10.1007/s004390050911
Di Benedetto, D., Di Vita, G., Romano, C., Giudice, M. L., Vitello, G. A., Zingale, M., et al. (2013). 6p22.3 deletion: report of a patient with autism, severe intellectual disability and electroencephalographic anomalies. Mol. Cytogenet. 6:4. doi: 10.1186/1755-8166-6-4
Dodatko, T., Fedorov, A. A., Grynberg, M., Patskovsky, Y., Rozwarski, D. A., Jaroszewski, L., et al. (2004). Crystal structure of the actin binding domain of the cyclase-associated protein. Biochemistry 43, 10628–10641. doi: 10.1021/bi049071r
Dominguez, R. (2016). The WH2 domain and actin nucleation: necessary but insufficient. Trends Biochem. Sci. 41, 478–490. doi: 10.1016/j.tibs.2016.03.004
Dron, J. S., and Hegele, R. A. (2020). A tip of the CAP1 to cholesterol metabolism. Eur. Heart J. 41, 253–254. doi: 10.1093/eurheartj/ehz771
Effendi, K., Yamazaki, K., Mori, T., Masugi, Y., Makino, S., and Sakamoto, M. (2012). Involvement of hepatocellular carcinoma biomarker, cyclase-associated protein 2 in zebrafish body development and cancer progression. Exp. Cell Res. 319, 35–44. doi: 10.1016/j.yexcr.2012.09.013
Esnault, C., Stewart, A., Gualdrini, F., East, P., Horswell, S., Matthews, N., et al. (2014). Rho-actin signaling to the MRTF coactivators dominates the immediate transcriptional response to serum in fibroblasts. Genes Dev. 28, 943–958. doi: 10.1101/gad.239327.114
Fan, Y. C., Cui, C. C., Zhu, Y. S., Zhang, L., Shi, M., Yu, J. S., et al. (2016). Overexpression of CAP1 and its significance in tumor cell proliferation, migration and invasion in glioma. Oncol. Rep. 36, 1619–1625. doi: 10.3892/or.2016.4936
Fedor-Chaiken, M., Deschenes, R. J., and Broach, J. R. (1990). SRV2, a gene required for RAS activation of adenylate cyclase in yeast. Cell 61, 329–340. doi: 10.1016/0092-8674(90)90813-T
Field, J., Vojtek, A., Ballester, R., Bolger, G., Colicelli, J., Ferguson, K., et al. (1990). Cloning and characterization of CAP, the S. cerevisiae gene encoding the 70 kd adenylyl cyclase-associated protein. Cell 61, 319–327. doi: 10.1016/0092-8674(90)90812-S
Field, J., Ye, D. Z., Shinde, M., Liu, F., Schillinger, K. J., Lu, M., et al. (2015). CAP2 in cardiac conduction, sudden cardiac death and eye development. Sci. Rep. 5:17256. doi: 10.1038/srep17256
Fife, C. M., McCarroll, J. A., and Kavallaris, M. (2014). Movers and shakers: cell cytoskeleton in cancer metastasis. Br. J. Pharmacol. 171, 5507–5523. doi: 10.1111/bph.12704
Freeman, N. L., Chen, Z., Horenstein, J., Weber, A., and Field, J. (1995). An actin monomer binding activity localizes to the carboxyl-terminal half of the Saccharomyces cerevisiae cyclase-associated protein. J. Biol. Chem. 270, 5680–5685. doi: 10.1074/jbc.270.10.5680
Freeman, N. L., Lila, T., Mintzer, K. A., Chen, Z., Pahk, A. J., Ren, R., et al. (1996). A conserved proline-rich region of the Saccharomyces cerevisiae cyclase-associated protein binds SH3 domains and modulates cytoskeletal localization. Mol. Cell. Biol. 16, 548–556. doi: 10.1128/MCB.16.2.548
Fu, J., Li, M., Wu, D. C., Liu, L. L., Chen, S. L., and Yun, J. P. (2015). Increased expression of CAP2 indicates poor prognosis in hepatocellular carcinoma. Transl. Oncol. 8, 400–406. doi: 10.1016/j.tranon.2015.08.003
Gallo, G. (2007). Tau is actin up in Alzheimer’s disease. Nat. Cell Biol. 9, 133–134. doi: 10.1038/ncb0207-133
Gieselmann, R., and Mann, K. (1992). ASP-56, a new actin sequestering protein from pig platelets with homology to CAP, an adenylate cyclase-associated protein from yeast. FEBS Lett. 298, 149–153. doi: 10.1016/0014-5793(92)80043-G
Goldschmidt-Clermont, P. J., Machesky, L. M., Doberstein, S. K., and Pollard, T. D. (1991). Mechanism of the interaction of human platelet profilin with actin. J. Cell Biol. 113, 1081–1089. doi: 10.1083/jcb.113.5.1081
Goode, B. L., Drubin, D. G., and Lappalainen, P. (1998). Regulation of the cortical actin cytoskeleton in budding yeast by twinfilin, a ubiquitous actin monomer-sequestering protein. J. Cell Biol. 142, 723–733. doi: 10.1083/jcb.142.3.723
Gottwald, U., Brokamp, R., Karakesisoglou, I., Schleicher, M., and Noegel, A. A. (1996). Identification of a cyclase-associated protein (CAP) homologue in Dictyostelium discoideum and characterization of its interaction with actin. Mol. Biol. Cell 7, 261–272. doi: 10.1091/mbc.7.2.261
Gurniak, C. B., Chevessier, F., Jokwitz, M., Jonsson, F., Perlas, E., Richter, H., et al. (2014). Severe protein aggregate myopathy in a knockout mouse model points to an essential role of cofilin2 in sarcomeric actin exchange and muscle maintenance. Eur. J. Cell Biol. 93, 252–266. doi: 10.1016/j.ejcb.2014.01.007
Hall, A. (2009). The cytoskeleton and cancer. Cancer Metastasis Rev 28, 5–14. doi: 10.1007/s10555-008-9166-3
Heredia, L., Helguera, P., de Olmos, S., Kedikian, G., Sola Vigo, F., LaFerla, F., et al. (2006). Phosphorylation of actin-depolymerizing factor/cofilin by LIM-kinase mediates amyloid beta-induced degeneration: a potential mechanism of neuronal dystrophy in Alzheimer’s disease. J. Neurosci. 26, 6533–6542. doi: 10.1523/JNEUROSCI.5567-05.2006
Hilton, D. M., Aguilar, R. M., Johnston, A. B., and Goode, B. L. (2018). Species-Specific Functions of Twinfilin in Actin Filament Depolymerization. J. Mol. Biol. 430, 3323–3336. doi: 10.1016/j.jmb.2018.06.025
Hliscs, M., Sattler, J. M., Tempel, W., Artz, J. D., Dong, A., Hui, R., et al. (2010). Structure and function of a G-actin sequestering protein with a vital role in malaria oocyst development inside the mosquito vector. J. Biol. Chem. 285, 11572–11583. doi: 10.1074/jbc.M109.054916
Hoffmann, L., Rust, M. B., and Culmsee, C. (2019). Actin(g) on mitochondria – a role for cofilin1 in neuronal cell death pathways. Biol. Chem. 400, 1089–1097. doi: 10.1515/hsz-2019-0120
Hofmann, A., Hess, S., Noegel, A. A., Schleicher, M., and Wlodawer, A. (2002). Crystallization of cyclase-associated protein from Dictyostelium discoideum. Acta Crystallogr. D Biol. Crystallogr. 58, 1858–1861. doi: 10.1107/S0907444902013306
Holtmaat, A., and Svoboda, K. (2009). Experience-dependent structural synaptic plasticity in the mammalian brain. Nat. Rev. Neurosci. 10, 647–658. doi: 10.1038/nrn2699
Hua, M., Yan, S., Deng, Y., Xi, Q., Liu, R., Yang, S., et al. (2015). CAP1 is overexpressed in human epithelial ovarian cancer and promotes cell proliferation. Int. J. Mol. Med. 35, 941–949. doi: 10.3892/ijmm.2015.2089
Hubberstey, A. V., and Mottillo, E. P. (2002). Cyclase-associated proteins: CAPacity for linking signal transduction and actin polymerization. FASEB J. 16, 487–499. doi: 10.1096/fj.01-0659rev
Iwase, S., and Ono, S. (2016). The C-terminal dimerization motif of cyclase-associated protein is essential for actin monomer regulation. Biochem. J. 473, 4427–4441. doi: 10.1042/BCJ20160329
Iwase, S., and Ono, S. (2017). Conserved hydrophobic residues in the CARP/beta-sheet domain of cyclase-associated protein are involved in actin monomer regulation. Cytoskeleton 74, 343–355. doi: 10.1002/cm.21385
Jang, H. D., Lee, S. E., Yang, J., Lee, H. C., Shin, D., Lee, H., et al. (2019). Cyclase-associated protein 1 is a binding partner of proprotein convertase subtilisin/kexin type-9 and is required for the degradation of low-density lipoprotein receptors by proprotein convertase subtilisin/kexin type-9. Eur. Heart J. 71, 239–252. doi: 10.1093/eurheartj/ehz566
Jansen, S., Collins, A., Golden, L., Sokolova, O., and Goode, B. L. (2014). Structure and mechanism of mouse cyclase-associated protein (CAP1) in regulating actin dynamics. J. Biol. Chem. 289, 30732–30742. doi: 10.1074/jbc.M114.601765
Johnston, A. B., Collins, A., and Goode, B. L. (2015). High-speed depolymerization at actin filament ends jointly catalysed by Twinfilin and Srv2/CAP. Nat. Cell Biol. 17, 1504–1511. doi: 10.1038/ncb3252
Kepser, L. J., Damar, F., De Cicco, T., Chaponnier, C., Proszynski, T. J., Pagenstecher, A., et al. (2019). CAP2 deficiency delays myofibril actin cytoskeleton differentiation and disturbs skeletal muscle architecture and function. Proc. Natl. Acad. Sci. U.S.A. 116, 8397–8402. doi: 10.1073/pnas.1813351116
Klamt, F., Zdanov, S., Levine, R. L., Pariser, A., Zhang, Y., Zhang, B., et al. (2009). Oxidant-induced apoptosis is mediated by oxidation of the actin-regulatory protein cofilin. Nat. Cell Biol. 11, 1241–1246. doi: 10.1038/ncb1968
Korobova, F., Ramabhadran, V., and Higgs, H. N. (2013). An actin-dependent step in mitochondrial fission mediated by the ER-associated formin INF2. Science 339, 464–467. doi: 10.1126/science.1228360
Kosmas, K., Eskandarnaz, A., Khorsandi, A. B., Kumar, A., Ranjan, R., Eming, S. A., et al. (2015). CAP2 is a regulator of the actin cytoskeleton and its absence changes infiltration of inflammatory cells and contraction of wounds. Eur. J. Cell Biol. 94, 32–45. doi: 10.1016/j.ejcb.2014.10.004
Kotila, T., Kogan, K., Enkavi, G., Guo, S., Vattulainen, I., Goode, B. L., et al. (2018). Structural basis of actin monomer re-charging by cyclase-associated protein. Nat. Commun. 9:1892. doi: 10.1038/s41467-018-04231-7
Kotila, T., Wioland, H., Enkavi, G., Kogan, K., Vattulainen, I., Jegou, A., et al. (2019). Mechanism of synergistic actin filament pointed end depolymerization by cyclase-associated protein and cofilin. Nat. Commun. 10:5320. doi: 10.1038/s41467-019-13213-2
Ksiazek, D., Brandstetter, H., Israel, L., Bourenkov, G. P., Katchalova, G., Janssen, K. P., et al. (2003). Structure of the N-terminal domain of the adenylyl cyclase-associated protein (CAP) from Dictyostelium discoideum. Structure 1, 1171–1178. doi: 10.1016/S0969-2126(03)00180-1
Kumar, A., Paeger, L., Kosmas, K., Kloppenburg, P., Noegel, A. A., and Peche, V. S. (2016). Neuronal actin dynamics, spine density and neuronal dendritic complexity are regulated by CAP2. Front. Cell. Neurosci. 10:180. doi: 10.3389/fncel.2016.00180
Lai, W. F., and Wong, W. T. (2020). Roles of the actin cytoskeleton in aging and age-associated diseases. Ageing Res. Rev. 58:101021. doi: 10.1016/j.arr.2020.101021
Lee, S., Lee, H. C., Kwon, Y. W., Lee, S. E., Cho, Y., Kim, J., et al. (2014). Adenylyl cyclase-associated protein 1 is a receptor for human resistin and mediates inflammatory actions of human monocytes. Cell Metab. 19, 484–497. doi: 10.1016/j.cmet.2014.01.013
Li, L., Fu, L. Q., Wang, H. J., and Wang, Y. Y. (2020). CAP2 is a valuable biomarker for diagnosis and prognostic in patients with gastric cancer. Pathol. Oncol. Res. 26, 273–279. doi: 10.1007/s12253-018-0450-4
Li, M., Yang, X., Shi, H., Ren, H., Chen, X., Zhang, S., et al. (2013). Downregulated expression of the cyclase-associated protein 1 (CAP1) reduces migration in esophageal squamous cell carcinoma. Jpn. J. Clin. Oncol. 43, 856–864. doi: 10.1093/jjco/hyt093
Lila, T., and Drubin, D. G. (1997). Evidence for physical and functional interactions among two Saccharomyces cerevisiae SH3 domain proteins, an adenylyl cyclase-associated protein and the actin cytoskeleton. Mol. Biol. Cell 8, 367–385. doi: 10.1091/mbc.8.2.367
Liu, Y., Cui, X., Hu, B., Lu, C., Huang, X., Cai, J., et al. (2014). Upregulated expression of CAP1 is associated with tumor migration and metastasis in hepatocellular carcinoma. Pathol. Res. Pract. 210, 169–175. doi: 10.1016/j.prp.2013.11.011
Liu, Y., Xiao, W., Shinde, M., Field, J., and Templeton, D. M. (2018). Cadmium favors F-actin depolymerization in rat renal mesangial cells by site-specific, disulfide-based dimerization of the CAP1 protein. Arch. Toxicol. 92, 1049–1064. doi: 10.1007/s00204-017-2142-3
Major, R. J., and Irvine, K. D. (2005). Influence of Notch on dorsoventral compartmentalization and actin organization in the Drosophila wing. Development 132, 3823–3833. doi: 10.1242/dev.01957
Makkonen, M., Bertling, E., Chebotareva, N. A., Baum, J., and Lappalainen, P. (2013). Mammalian and malaria parasite cyclase-associated proteins catalyze nucleotide exchange on G-actin through a conserved mechanism. J. Biol. Chem. 288, 984–994. doi: 10.1074/jbc.M112.435719
Massaad, C. A., and Klann, E. (2011). Reactive oxygen species in the regulation of synaptic plasticity and memory. Antioxid. Redox Signal. 14, 2013–2054. doi: 10.1089/ars.2010.3208
Masugi, Y., Tanese, K., Emoto, K., Yamazaki, K., Effendi, K., Funakoshi, T., et al. (2015). Overexpression of adenylate cyclase-associated protein 2 is a novel prognostic marker in malignant melanoma. Pathol. Int. 65, 627–634. doi: 10.1111/pin.12351
Mattila, P. K., Quintero-Monzon, O., Kugler, J., Moseley, J. B., Almo, S. C., Lappalainen, P., et al. (2004). A high-affinity interaction with ADP-actin monomers underlies the mechanism and in vivo function of Srv2/cyclase-associated protein. Mol. Biol. Cell 15, 5158–5171. doi: 10.1091/mbc.e04-06-0444
Matviw, H., Yu, G., and Young, D. (1992). Identification of a human cDNA encoding a protein that is structurally and functionally related to the yeast adenylyl cyclase-associated CAP proteins. Mol. Cell. Biol. 12, 5033–5040. doi: 10.1128/MCB.12.11.5033
Mavoungou, C., Israel, L., Rehm, T., Ksiazek, D., Krajewski, M., Popowicz, G., et al. (2004). NMR structural characterization of the N-terminal domain of the adenylyl cyclase-associated protein (CAP) from Dictyostelium discoideum. J. Biomol. NMR 29, 73–84. doi: 10.1023/B:JNMR.0000019513.86120.98
Medina, P. M., Worthen, R. J., Forsberg, L. J., and Brenman, J. E. (2008). The actin-binding protein capulet genetically interacts with the microtubule motor kinesin to maintain neuronal dendrite homeostasis. PLoS One 3:e3054. doi: 10.1371/journal.pone.0003054
Mihajlovic, M., Ninic, A., Sopic, M., Miljkovic, M., Stefanovic, A., Vekic, J., et al. (2019). Association among resistin, adenylate cyclase-associated protein 1 and high-density lipoprotein cholesterol in patients with colorectal cancer: a multi-marker approach, as a hallmark of innovative predictive, preventive, and personalized medicine. EPMA J. 10, 307–316. doi: 10.1007/s13167-019-00178-x
Minamide, L. S., Striegl, A. M., Boyle, J. A., Meberg, P. J., and Bamburg, J. R. (2000). Neurodegenerative stimuli induce persistent ADF/cofilin-actin rods that disrupt distal neurite function. Nat. Cell Biol. 2, 628–636. doi: 10.1038/35023579
Miyoshi, T., and Watanabe, N. (2013). Can filament treadmilling alone account for the F-actin turnover in lamellipodia? Cytoskeleton 70, 179–190. doi: 10.1002/cm.21098
Moriyama, K., and Yahara, I. (2002). Human CAP1 is a key factor in the recycling of cofilin and actin for rapid actin turnover. J. Cell Sci. 115, 1591–1601.
Mu, A. M., Fung, T. S., Francomacaro, L. M., Huynh, T., Kotila, T., Svindrych, Z., et al. (2020). Regulation of INF2-mediated actin polymerization through site-specific lysine acetylation of actin itself. Proc. Natl. Acad. Sci. U.S.A. 117, 439–447. doi: 10.1073/pnas.1914072117
Mu, A. M., Fung, T. S., Kettenbach, A. N., Chakrabarti, R., and Higgs, H. N. (2019). A complex containing lysine-acetylated actin inhibits the formin INF2. Nat. Cell Biol. 21, 592–602. doi: 10.1038/s41556-019-0307-4
Munjas, J., Sopic, M., Bogavac-Stanojevic, N., Kravljaca, M., Miljkovic, M., Simic-Ogrizovic, S., et al. (2020). Serum resistin, adenylate cyclase-associated protein 1 gene expression, and carotid intima-media thickness in patients with end-stage renal disease and healthy controls. Cardiorenal Med. 10, 51–60. doi: 10.1159/000503416
Munjas, J., Sopic, M., Spasojevic-Kalimanovska, V., Kalimanovska-Ostric, D., Andelkovic, K., and Jelic-Ivanovic, Z. (2017). Association of adenylate cyclase-associated protein 1 with coronary artery disease. Eur. J. Clin. Invest. 47, 659–666. doi: 10.1111/eci.12787
Munoz-Palomeque, A., Guerrero-Ramirez, M. A., Rubio-Chavez, L. A., Rosales-Gomez, R. C., Lopez-Cardona, M. G., Barajas-Avila, V. H., et al. (2018). Association of RETN and CAP1 SNPs, expression and serum resistin levels with breast cancer in Mexican women. Genet. Test. Mol. Biomarkers 22, 209–217. doi: 10.1089/gtmb.2017.0212
Nishida, Y., Shima, F., Sen, H., Tanaka, Y., Yanagihara, C., Yamawaki-Kataoka, Y., et al. (1998). Coiled-coil interaction of N-terminal 36 residues of cyclase-associated protein with adenylyl cyclase is sufficient for its function in Saccharomyces cerevisiae Ras pathway. J. Biol. Chem. 273, 28019–28024. doi: 10.1074/jbc.273.43.28019
Noegel, A. A., Blau-Wasser, R., Sultana, H., Muller, R., Israel, L., Schleicher, M., et al. (2004). The cyclase-associated protein CAP as regulator of cell polarity and cAMP signaling in Dictyostelium. Mol. Biol. Cell 15, 934–945. doi: 10.1091/mbc.e03-05-0269
Nomura, K., Ono, K., and Ono, S. (2012). CAS-1, a C. elegans cyclase-associated protein, is required for sarcomeric actin assembly in striated muscle. J. Cell Sci. 125, 4077–4089. doi: 10.1242/jcs.104950
Nomura, K., and Ono, S. (2013). ATP-dependent regulation of actin monomer-filament equilibrium by cyclase-associated protein and ADF/cofilin. Biochem. J. 453, 249–259. doi: 10.1042/BJ20130491
Normoyle, K. P., and Brieher, W. M. (2012). Cyclase-associated protein (CAP) acts directly on F-actin to accelerate cofilin-mediated actin severing across the range of physiological pH. J. Biol. Chem. 287, 35722–35732. doi: 10.1074/jbc.M112.396051
Ockeloen, C. W., Gilhuis, H. J., Pfundt, R., Kamsteeg, E. J., Agrawal, P. B., Beggs, A. H., et al. (2012). Congenital myopathy caused by a novel missense mutation in the CFL2 gene. Neuromuscul. Disord. 22, 632–639. doi: 10.1016/j.nmd.2012.03.008
Olson, E. N., and Nordheim, A. (2010). Linking actin dynamics and gene transcription to drive cellular motile functions. Nat. Rev. Mol. Cell Biol. 11, 353–365. doi: 10.1038/nrm2890
Ong, R. W., AlSaman, A., Selcen, D., Arabshahi, A., Yau, K. S., Ravenscroft, G., et al. (2014). Novel cofilin-2 (CFL2) four base pair deletion causing nemaline myopathy. J. Neurol. Neurosurg. Psychiatry 85, 1058–1060. doi: 10.1136/jnnp-2014-307608
Ono, K., Parast, M., Alberico, C., Benian, G. M., and Ono, S. (2003). Specific requirement for two ADF/cofilin isoforms in distinct actin-dependent processes in Caenorhabditis elegans. J. Cell Sci. 116, 2073–2085. doi: 10.1242/jcs.00421
Ono, K., Yamashiro, S., and Ono, S. (2008). Essential role of ADF/cofilin for assembly of contractile actin networks in the C. elegans somatic gonad. J Cell Sci. 121, 2662–2670. doi: 10.1242/jcs.034215
Ono, S. (2013). The role of cyclase-associated protein in regulating actin filament dynamics – more than a monomer-sequestration factor. J. Cell Sci. 126, 3249–3258. doi: 10.1242/jcs.128231
Ono, S., Baillie, D. L., and Benian, G. M. (1999). UNC-60B, an ADF/cofilin family protein, is required for proper assembly of actin into myofibrils in Caenorhabditis elegans body wall muscle. J. Cell Biol. 145, 491–502. doi: 10.1083/jcb.145.3.491
Paavilainen, V. O., Hellman, M., Helfer, E., Bovellan, M., Annila, A., Carlier, M. F., et al. (2007). Structural basis and evolutionary origin of actin filament capping by twinfilin. Proc. Natl. Acad. Sci. U.S.A. 104, 3113–3118. doi: 10.1073/pnas.0608725104
Paavilainen, V. O., Oksanen, E., Goldman, A., and Lappalainen, P. (2008). Structure of the actin-depolymerizing factor homology domain in complex with actin. J. Cell Biol. 182, 51–59. doi: 10.1083/jcb.200803100
Patel, L., Buckels, A. C., Kinghorn, I. J., Murdock, P. R., Holbrook, J. D., Plumpton, C., et al. (2003). Resistin is expressed in human macrophages and directly regulated by PPAR gamma activators. Biochem. Biophys. Res. Commun. 300, 472–476. doi: 10.1016/S0006-291X(02)02841-3
Paunola, E., Mattila, P. K., and Lappalainen, P. (2002). WH2 domain: a small, versatile adapter for actin monomers. FEBS Lett. 513, 92–97. doi: 10.1016/S0014-5793(01)03242-2
Peche, V., Shekar, S., Leichter, M., Korte, H., Schroder, R., Schleicher, M., et al. (2007). CAP2, cyclase-associated protein 2, is a dual compartment protein. Cell. Mol. Life Sci. 64, 2702–2715. doi: 10.1007/s00018-007-7316-3
Peche, V. S., Holak, T. A., Burgute, B. D., Kosmas, K., Kale, S. P., Wunderlich, F. T., et al. (2012). Ablation of cyclase-associated protein 2 (CAP2) leads to cardiomyopathy. Cell. Mol. Life Sci. 70, 527–543. doi: 10.1007/s00018-012-1142-y
Pelucchi, S., Stringhi, R., and Marcello, E. (2020a). Dendritic spines in Alzheimer’s disease: how the actin cytoskeleton contributes to synaptic failure. Int. J. Mol. Sci. 21:908. doi: 10.3390/ijms21030908
Pelucchi, S., Vandermeulen, L., Pizzamiglio, L., Aksan, B., Yan, J., Konietzny, A., et al. (2020b). CAP2 dimerization regulates Cofilin in synaptic plasticity and Alzheimer’s disease. Brain Commun. doi: 10.1093/braincomms/fcaa086 [Epub ahead of print].
Pollard, T. D. (2016). Actin and actin-binding proteins. Cold Spring Harb. Perspect. Biol. 8:a018226. doi: 10.1101/cshperspect.a018226
Purde, V., Busch, F., Kudryashova, E., Wysocki, V. H., and Kudryashov, D. S. (2019). Oligomerization affects the ability of human cyclase-associated proteins 1 and 2 to promote actin severing by cofilins. Int. J. Mol. Sci. 20:5647. doi: 10.3390/ijms20225647
Quinn, S. N., Graves, S. H., Dains-McGahee, C., Friedman, E. M., Hassan, H., Witkowski, P., et al. (2017). Adenylyl cyclase 3/adenylyl cyclase-associated protein 1 (CAP1) complex mediates the anti-migratory effect of forskolin in pancreatic cancer cells. Mol. Carcinog. 56, 1344–1360. doi: 10.1002/mc.22598
Quintero-Monzon, O., Jonasson, E. M., Bertling, E., Talarico, L., Chaudhry, F., Sihvo, M., et al. (2009). Reconstitution and dissection of the 600-kDa Srv2/CAP complex: roles for oligomerization and cofilin-actin binding in driving actin turnover. J. Biol. Chem. 284, 10923–10934. doi: 10.1074/jbc.M808760200
Rehklau, K., Gurniak, C. B., Conrad, M., Friauf, E., Ott, M., and Rust, M. B. (2012). ADF/cofilin proteins translocate to mitochondria during apoptosis but are not generally required for cell death signaling. Cell Death Differ. 19, 958–967. doi: 10.1038/cdd.2011.180
Rehklau, K., Hoffmann, L., Gurniak, C. B., Ott, M., Witke, W., Scorrano, L., et al. (2017). Cofilin1-dependent actin dynamics control DRP1-mediated mitochondrial fission. Cell Death Dis. 8:e3063. doi: 10.1038/cddis.2017.448
Rosendahl, A. H., Bergqvist, M., Lettiero, B., Kimbung, S., and Borgquist, S. (2018). Adipocytes and obesity-related conditions jointly promote breast cancer cell growth and motility: associations with CAP1 for prognosis. Front. Endocrinol. 9:689. doi: 10.3389/fendo.2018.00689
Rust, M. B. (2015). ADF/cofilin: a crucial regulator of synapse physiology and behavior. Cell. Mol. Life Sci. 72, 3521–3529. doi: 10.1007/s00018-015-1941-z
Rust, M. B., Gurniak, C. B., Renner, M., Vara, H., Morando, L., Gorlich, A., et al. (2010). Learning, AMPA receptor mobility and synaptic plasticity depend on n-cofilin-mediated actin dynamics. EMBO J. 29, 1889–1902. doi: 10.1038/emboj.2010.72
Saker, Z., Bahmad, H. F., Fares, Y., Al Najjar, Z., Saad, M., Harati, H., et al. (2020). Prognostic impact of adenylyl cyclase-associated protein 2 (CAP2) in glioma: a clinicopathological study. Heliyon 6:e03236. doi: 10.1016/j.heliyon.2020.e03236
Sato, H., Muraoka, S., Kusunoki, N., Masuoka, S., Yamada, S., Ogasawara, H., et al. (2017). Resistin upregulates chemokine production by fibroblast-like synoviocytes from patients with rheumatoid arthritis. Arthritis Res. Ther. 19:263. doi: 10.1186/s13075-017-1472-0
Schippers, J. H. M., Nguyen, H. M., Lu, D., Schmidt, R., and Mueller-Roeber, B. (2012). ROS homeostasis during development: an evolutionary conserved strategy. Cell. Mol. Life Sci. 69, 3245–3257. doi: 10.1007/s00018-012-1092-4
Shekhar, S., and Carlier, M. F. (2017). Enhanced depolymerization of actin filaments by ADF/cofilin and monomer funneling by capping protein cooperate to accelerate barbed-end growth. Curr. Biol. 27, 1990–1998. doi: 10.1016/j.cub.2017.05.036
Shekhar, S., Chung, J., Kondev, J., Gelles, J., and Goode, B. L. (2019). Synergy between cyclase-associated protein and cofilin accelerates actin filament depolymerization by two orders of magnitude. Nat. Commun. 10:5319. doi: 10.1038/s41467-019-13268-1
Shibata, R., Mori, T., Du, W., Chuma, M., Gotoh, M., Shimazu, M., et al. (2006). Overexpression of cyclase-associated protein 2 in multistage hepatocarcinogenesis. Clin. Cancer Res. 12, 5363–5368. doi: 10.1158/1078-0432.CCR-05-2245
Stevenson, V. A., and Theurkauf, W. E. (2000). Actin cytoskeleton: putting a CAP on actin polymerization. Curr. Biol. 10, R695–R697. doi: 10.1016/S0960-9822(00)00712-0
Stockigt, F., Peche, V. S., Linhart, M., Nickenig, G., Noegel, A. A., and Schrickel, J. W. (2016). Deficiency of cyclase-associated protein 2 promotes arrhythmias associated with connexin43 maldistribution and fibrosis. Arch. Med. Sci. 12, 188–198. doi: 10.5114/aoms.2015.54146
Sultana, H., Neelakanta, G., Rivero, F., Blau-Wasser, R., Schleicher, M., and Noegel, A. A. (2012). Ectopic expression of cyclase associated protein CAP restores the streaming and aggregation defects of adenylyl cyclase a deficient Dictyostelium discoideum cells. BMC Dev. Biol. 12:3. doi: 10.1186/1471-213X-12-3
Tan, M., Song, X., Zhang, G., Peng, A., Li, X., Li, M., et al. (2013). Overexpression of adenylate cyclase-associated protein 1 is associated with metastasis of lung cancer. Oncol. Rep. 30, 1639–1644. doi: 10.3892/or.2013.2607
Tanaka, K., Takeda, S., Mitsuoka, K., Oda, T., Kimura-Sakiyama, C., Maéda, Y., et al. (2018). Structural basis for cofilin binding and actin filament disassembly. Nat. Commun. 9:1860. doi: 10.1038/s41467-018-04290-w
Tarkowski, A., Bjersing, J., Shestakov, A., and Bokarewa, M. I. (2010). Resistin competes with lipopolysaccharide for binding to toll-like receptor 4. J. Cell. Mol. Med. 14, 1419–1431. doi: 10.1111/j.1582-4934.2009.00899.x
Vinson, V. K., De La Cruz, E. M., Higgs, H. N., and Pollard, T. D. (1998). Interactions of Acanthamoeba profilin with actin and nucleotides bound to actin. Biochemistry 37, 10871–10880. doi: 10.1021/bi980093l
Vojtek, A., Haarer, B., Field, J., Gerst, J., Pollard, T. D., Brown, S., et al. (1991). Evidence for a functional link between profilin and CAP in the yeast S. cerevisiae. Cell 66, 497–505. doi: 10.1016/0092-8674(81)90013-1
Wills, Z., Emerson, M., Rusch, J., Bikoff, J., Baum, B., Perrimon, N., et al. (2002). A Drosophila homolog of cyclase-associated proteins collaborates with the Abl tyrosine kinase to control midline axon pathfinding. Neuron 36, 611–622. doi: 10.1016/S0896-6273(02)01022-X
Wills, Z., Marr, L., Zinn, K., Goodman, C. S., and Van Vactor, D. (1999). Profilin and the Abl tyrosine kinase are required for motor axon outgrowth in the Drosophila embryo. Neuron 22, 291–299. doi: 10.1016/S0896-6273(00)81090-9
Wioland, H., Guichard, B., Senju, Y., Myram, S., Lappalainen, P., Jégou, A., et al. (2017). ADF/cofilin accelerates actin dynamics by severing filaments and promoting their depolymerization at both ends. Curr. Biol. 27, 1956–1967. doi: 10.1016/j.cub.2017.05.048
Wolanski, M., Khosrowshahian, F., Jerant, L., Jap, I. S., Brockman, J., and Crawford, M. J. (2009). Expression of CAP2 during early Xenopus embryogenesis. Int. J. Dev. Biol. 53, 1063–1067. doi: 10.1387/ijdb.062158mw
Wolf, M., Zimmermann, A. M., Gorlich, A., Gurniak, C. B., Sassoe-Pognetto, M., Friauf, E., et al. (2015). ADF/cofilin controls synaptic actin dynamics and regulates synaptic vesicle mobilization and exocytosis. Cereb. Cortex 25, 2863–2875. doi: 10.1093/cercor/bhu081
Wu, H., Hasan, R., Zhang, H., Gray, J., Williams, D., Miller, M., et al. (2019). Phosphorylation regulates CAP1 (cyclase-associated protein 1) functions in the motility and invasion of pancreatic cancer cells. Sci. Rep. 9:4925. doi: 10.1038/s41598-019-41346-3
Xie, S. S., Hu, F., Tan, M., Duan, Y. X., Song, X. L., and Wang, C. H. (2014). Relationship between expression of matrix metalloproteinase-9 and adenylyl cyclase-associated protein 1 in chronic obstructive pulmonary disease. J. Int. Med. Res. 42, 1272–1284. doi: 10.1177/0300060514548290
Xiong, Y., Bedi, K., Berritt, S., Attipoe, B. K., Brooks, T. G., Wang, K., et al. (2019). Targeting MRTF/SRF in CAP2-dependent dilated cardiomyopathy delays disease onset. JCI Insight 4:e124629. doi: 10.1172/jci.insight.124629
Xu, L., Peng, S., Huang, Q., Liu, Y., Jiang, H., Li, X., et al. (2016). Expression status of cyclaseassociated protein 2 as a prognostic marker for human breast cancer. Oncol. Rep. 36, 1981–1988. doi: 10.3892/or.2016.5051
Yamazaki, K., Takamura, M., Masugi, Y., Mori, T., Du, W., Hibi, T., et al. (2009). Adenylate cyclase-associated protein 1 overexpressed in pancreatic cancers is involved in cancer cell motility. Lab. Invest. 89, 425–432. doi: 10.1038/labinvest.2009.5
Yu, G., Swiston, J., and Young, D. (1994). Comparison of human CAP and CAP2, homologs of the yeast adenylyl cyclase-associated proteins. J. Cell Sci. 107, 1671–1678.
Yu, X. F., Ni, Q. C., Chen, J. P., Xu, J. F., Jiang, Y., Yang, S. Y., et al. (2014). Knocking down the expression of adenylate cyclase-associated protein 1 inhibits the proliferation and migration of breast cancer cells. Exp. Mol. Pathol. 96, 188–194. doi: 10.1016/j.yexmp.2014.02.002
Yusof, A. M., Hu, N. J., Wlodawer, A., and Hofmann, A. (2005). Structural evidence for variable oligomerization of the N-terminal domain of cyclase-associated protein (CAP). Proteins 58, 255–262. doi: 10.1002/prot.20314
Yusof, A. M., Jaenicke, E., Pedersen, J. S., Noegel, A. A., Schleicher, M., and Hofmann, A. (2006). Mechanism of oligomerisation of cyclase-associated protein from Dictyostelium discoideum in solution. J. Mol. Biol. 362, 1072–1081. doi: 10.1016/j.jmb.2006.08.008
Zelicof, A., Gatica, J., and Gerst, J. E. (1993). Molecular cloning and characterization of a rat homolog of CAP, the adenylyl cyclase-associated protein from Saccharomyces cerevisiae. J. Biol. Chem. 268, 13448–13453.
Zhang, H., Ramsey, A., Xiao, Y., Karki, U., Xie, J. Y., Xu, J., et al. (2020). Dynamic phosphorylation and dephosphorylation of cyclase-associated protein 1 by antagonistic signaling through cyclin-dependent kinase 5 and cAMP are critical for the protein functions in actin filament disassembly and cell adhesion. Mol. Cell. Biol. 40:e00282-19. doi: 10.1128/MCB.00282-19
Zhang, H., and Zhou, G. L. (2016). CAP1 (cyclase-associated protein 1) exerts distinct functions in the proliferation and metastatic potential of breast cancer cells mediated by ERK. Sci. Rep. 6:25933. doi: 10.1038/srep25933
Zhang, M., Yan, L., Wang, G. J., and Jin, R. (2019). Resistin effects on pancreatic cancer progression and chemoresistance are mediated through its receptors CAP1 and TLR4. J. Cell. Physiol. 234, 9457–9466. doi: 10.1002/jcp.27631
Zhou, G. L., Zhang, H., and Field, J. (2014a). Mammalian CAP (Cyclase-associated protein) in the world of cell migration: roles in actin filament dynamics and beyond. Cell Adh. Migr. 8, 55–59. doi: 10.4161/cam.27479
Zhou, G. L., Zhang, H., Wu, H., Ghai, P., and Field, J. (2014b). Phosphorylation of the cytoskeletal protein CAP1 controls its association with cofilin and actin. J. Cell Sci. 127, 5052–5065. doi: 10.1242/jcs.156059
Keywords: cyclase-associated protein, CAP, SRV2, Cofilin, F-actin, G-actin
Citation: Rust MB, Khudayberdiev S, Pelucchi S and Marcello E (2020) CAPt’n of Actin Dynamics: Recent Advances in the Molecular, Developmental and Physiological Functions of Cyclase-Associated Protein (CAP). Front. Cell Dev. Biol. 8:586631. doi: 10.3389/fcell.2020.586631
Received: 23 July 2020; Accepted: 26 August 2020;
Published: 24 September 2020.
Edited by:
Lei-Miao Yin, Shanghai University of Traditional Chinese Medicine, ChinaReviewed by:
Shoichiro Ono, Emory University, United StatesDmitri Kudryashov, The Ohio State University, United States
Copyright © 2020 Rust, Khudayberdiev, Pelucchi and Marcello. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Marco B. Rust, bWFyY28ucnVzdEBzdGFmZi51bmktbWFyYnVyZy5kZQ==; Elena Marcello, ZWxlbmEubWFyY2VsbG9AdW5pbWkuaXQ=
 Marco B. Rust
Marco B. Rust Sharof Khudayberdiev
Sharof Khudayberdiev Silvia Pelucchi
Silvia Pelucchi Elena Marcello
Elena Marcello