- 1Department of Otolaryngology, Peking Union Medical College Hospital, Chinese Academy of Medical Sciences and Peking Union Medical College, Beijing, China
- 2Department of Otolaryngology, China-Japan Friendship Hospital, Beijing, China
- 3Chinese Academy of Medical Sciences and Peking Union Medical College, Beijing, China
- 4School of Medicine, Tsinghua University, Beijing, China
- 5Department of Cardiology, Peking Union Medical College Hospital, Peking Union Medical College and Chinese Academy of Medical Sciences, Beijing, China
- 6Tsinghua-Peking Center for Life Sciences, Tsinghua University, Beijing, China
- 7Department of Otolaryngology, The Ohio State University, Columbus, OH, United States
- 8Key Laboratory of Cell Proliferation and Differentiation of the Ministry of Education, College of Life Sciences, Peking University, Beijing, China
- 9Department of Medical Genetics and National Laboratory of Medical Molecular Biology, Institute of Basic Medical Sciences, Chinese Academy of Medical Sciences and Peking Union Medical College, Beijing, China
- 10Department of Otolaryngology Head and Neck Surgery, Beijing Tiantan Hospital, Capital Medical University, Beijing, China
Background: Hemifacial microsomia (HFM) is a type of rare congenital syndrome caused by developmental disorders of the first and second pharyngeal arches that occurs in one out of 5,600 live births. There are significant gaps in our knowledge of the pathogenic genes underlying this syndrome.
Methods: Whole exome sequencing (WES) was performed on five patients, one asymptomatic carrier, and two marry-in members of a five-generation pedigree. Structure of WARP (product of VWA1) was predicted using the Phyre2 web portal. In situ hybridization and vwa1-knockdown/knockout studies in zebrafish using morpholino and CRISPR/Cas9 techniques were performed. Cartilage staining and immunofluorescence were carried out.
Results: Through WES and a set of filtration, we identified a c.G905A:p.R302Q point mutation in a novel candidate pathogenic gene, VWA1. The Phyre2 web portal predicted alterations in secondary and tertiary structures of WARP, indicating changes in its function as well. Predictions of protein-to-protein interactions in five pathways related to craniofacial development revealed possible interactions with four proteins in the FGF pathway. Knockdown/knockout studies of the zebrafish revealed deformities of pharyngeal cartilage. A decrease of the proliferation of cranial neural crest cells (CNCCs) and alteration of the structure of pharyngeal chondrocytes were observed in the morphants as well.
Conclusion: Our data suggest that a mutation in VWA1 is functionally linked to HFM through suppression of CNCC proliferation and disruption of the organization of pharyngeal chondrocytes.
Introduction
Hemifacial microsomia (HFM [MIM: 164210]), also known as otomandibular dysostosis and craniofacial microsomia, is a type of rare congenital genetic syndrome caused by developmental disorders of the first and second pharyngeal arches. The estimated occurrence rate is 1 in 5,600 live births, with no gender or laterality differences (Grabb, 1965; Xu et al., 2015). As one of the manifestations of the oculo-auriculo-vertebral spectrum (OAVS), recently re-estimated to show a prevalence of 3.8/100,000 live births, additional clinical findings are also frequently found in HFM patients, including other craniofacial anomalies and conditions affecting other organs and systems, such as urogenital anomalies, brain anomalies, microcephaly, heart defects, and short stature (Tasse et al., 2005). These variable clinical classifications have led to difficulties in classifying the severity of the disease. To date, several generations of classification systems have been developed, from the original Pruzansky-Kaban classification, which focuses mainly on the mandible and glenoid fossa (Kaban et al., 1988), to the SAT system, which takes skeletal malformations, auricular involvement and soft tissue defects into consideration, and finally to the widely accepted OMENS (Orbit, Mandible, Ear, Nerve, Soft tissue) and OMENS + system, which further includes additional disease manifestations (Tuin et al., 2015).
A complex etiology underlies this genetic syndrome. Environmental factors (e.g., retinoic acid exposure and vasoactive medications), maternal intrinsic factors (e.g., multiple pregnancies and maternal diabetes), and genetic factors, including both chromosomal variations (e.g., karyotype abnormalities of 14q22.3–q23.3) and mutations, can all lead to HFM (Ou et al., 2008; Northup et al., 2010). Most cases with documented pedigrees exhibit an autosomal-dominant inheritance pattern with incomplete penetrance and variable degrees of craniofacial deformities (Vendramini-Pittoli and Kokitsu-Nakata, 2009). To date, three possible pathogenic hypotheses have been proposed: (1) vascular abnormality and hemorrhage, which may interfere with the development of the first and second pharyngeal arches; (2) damage to Meckel’s cartilage, which may lead to apparent malformation; and (3) disruption of the development of cranial neural crest cells (CNCCs), which may disrupt the normal formation of maxillofacial bones (Chen et al., 2018). The three hypotheses are interconnected as CNCCs give rise to Meckel’s cartilage, and vascular abnormality can either interferes with CNCC development or cause damage to Meckel’s cartilage. Disruption of CNCC development can also in turn cause vascular malformation. Yet, there are significant gaps in our knowledge of the pathogenic genes underlying this syndrome.
In vertebrates, CNCCs origin from the ectoderm. They make up a mesenchymal core between the ectoderm and the endoderm along with the mesoderm cells and play important roles in the development of cartilage bones, peripheral nervous system and connective tissues of the head and neck (Schilling and Kimmel, 1994; Kimmel et al., 1998; Frisdal and Trainor, 2014). After gastrulation, CNCCs separate from the neural plate via epithelial-mesenchymal transition and migrate to the pharyngeal arches, where their development is further regulated by both intrinsic patterning information and endodermal signals such as RA (retinoic acid), FGF (fibroblast growth factor) and WNT (Frisdal and Trainor, 2014). In humans, the first pharyngeal arch gives rise to structures including the zygomatic arch, maxillary, mandible, malleus and incus, whereas the second pharyngeal arch gives rise to the stapes, styloid process and lesser horn of hyoid (Frisdal and Trainor, 2014). The patterning is conserved in zebrafish, where the first pharyngeal arch forms the jaw and the second forms jaw supporting structures (Schilling and Kimmel, 1994).
Here we analyzed an extended five-generation HFM pedigree. A novel candidate pathogenic gene, VWA1 (MIM: 611901), encoding von Willebrand factor A domain-containing protein 1, was identified by whole-exome sequencing (WES). Functional studies in zebrafish showed that vwa1 contributes to craniofacial cartilage development by supporting both the proliferation of CNCC and the organization of pharyngeal chondrocytes.
Materials and Methods
Participants
The study was conducted on an HFM pedigree at PUMCH (Peking Union Medical College Hospital). A comprehensive clinical history was taken and a complete physical examination was performed on all subjects. Ethical approval for this study was obtained from the institutional review board of PUMCH (JS-796). All participants or their legal guardians gave informed written consent to all clinical and genetic studies. All in vivo experiments were carried out in accordance with CALAS (Chinese Association for Laboratory Animal Science).
WES and Variants Identification
WES was performed on five patients, one asymptomatic carrier, and two marry-in members of the pedigree. Genomic DNA was extracted from peripheral blood samples using a TIANamp Blood DNA Kit (Tiangen, Beijing, China) according to the manufacturer’s protocol. Exome enrichment was performed using a Sure Select Human All Exon v6 kit (65 Mb) (Agilent, Santa Clara, CA, United States), which yielded an average sequencing depth of 100-fold and a coverage of 99%. Uncovered regions were checked for large genomic deletions and none was found. Enriched shotgun libraries were sequenced on a HiseqX platform (Illumina, San Diego, CA, United States). SNPs and indels were functionally annotated using ANNOVAR and categorized into missense, nonsense and splice-site mutations, and other genomic features.
To identify the most likely pathogenic mutations, we filtered out (1) synonymous and non-coding variants (with the exception of splicing site mutations that might create an ectopic splicing site); (2) variants with an allele frequency of 0.005 or higher in the 1000 Genomes (1KG) Project, Exome Sequencing Project (ESP6500) and the Genome Aggregation Database (gnomAD); (3) variants that were not detected in one or more of the affected participants; and (4) variants that were present in the marry-in members of the pedigree. After filtration, rare or novel mutations in patients of the pedigree were verified by polymerase chain reaction (PCR) amplification and Sanger sequencing.
Molecular Analysis of Variants
The webserver Phyre21 and the online software program PSIPRED (v3.3)2 were used to predict secondary and tertiary structural variations associated with potentially pathogenic mutations of candidate genes (Huang et al., 2014; Kelley et al., 2015). The web-based program String3 was used to predict functional protein associations with WNT, BMP (bone morphogenic protein), FGF, RA, and endothelin-1 pathways, which are closely related to craniofacial development (Frisdal and Trainor, 2014; Graf et al., 2016).
Zebrafish and Embryos
Adult zebrafish were maintained under standard conditions (Hans and Westerfield, 2007), and embryos were staged using standard methods (Kimmel et al., 1995). Tuebingen and the transgenic fish lines, Tg (sox10:EGFP) (ID:CZ156, ba2Tg/ +) and Tg (nkx2.3:EGFP) (Li et al., 2018), were used in the experiments.
Knockdown and Knockout of vwa1
The morpholino oligonucleotide, 5′-TTC GGA CCT CCA TGA CGG GAC TAA A-3′, targeting the translation start site of zebrafish vwa1 (vwa1-ATG MO) was designed and synthesized by Gene Tools (Gene Tools). To verify that the craniofacial deformities observed in these morphants were the result of reduced expression of vwa1 rather than off-target effects of the morpholino, we injected a 1.5-fold (w/w) greater amount of a morpholino targeting p53 (5′-GCG CCA TTG CTT TGC AAG AAT TG-3′) together with the vwa1-ATG MO (Robu et al., 2007). To validate the efficacy of vwa1-ATG MO, we constructed pCS2 + vwa1-ATG-MO-mCherry plasmid, inserting the morpholino binding sequence into the translation initiation site of mCherry. We injected the in vitro transcribed mRNA with/without vwa1-ATG morpholino into 1-cell embryos and observed the fluorescence at 1 dpf.
We also knocked out vwa1 in zebrafish using the CRISPR/Cas9 system. Detailed methods were in accordance with previous work (Wu et al., 2018). Four guide RNAs (gRNAs, Supplementary Table 1) were mixed and co-injected with Cas9 into 1-cell stage embryos. The gRNAs were designed prior to construction of the Genome-Scale Guide Set using the CRISPR Design Website4. Templates for gRNA production were generated by annealing and elongating a forward primer containing a T7 promoter and guide sequence and a reverse primer encoding the standard chimeric gRNA scaffold. The sequences used are presented in Supplementary Table 1. The DNA template was in vitro transcribed, and gRNA was microinjected into 1-cell embryos. The efficacy of gRNA was verified by extracting crude genomic DNA from whole vwa1-knockout zebrafish embryos, followed by PCR amplification and sequencing (Wu et al., 2018). A morpholino targeting upf3a was also injected to inhibit compensatory effects of DNA damage (Shum et al., 2016).
Whole-Mount in situ Hybridization
Whole-mount in situ hybridization (ISH) was performed on zebrafish as previously reported (Thisse and Thisse, 2008). A template for the vwa1 antisense probe was constructed by amplifying full-length zebrafish vwa1 using the forward primer 5′-ATG GAG GTC CGA AAG GCG CT-3′ and the reverse primer 5′-TAA TAC GAC TCA CTA TAG GGT CAA GCC GTG CAG CAG AGT C-3′ containing the T7 polymerase sequence. The antisense probe was synthesized using T7 polymerase. Probes against dlx2a (Akimenko et al., 1994), crestin (Xia et al., 2013), and sox9a (Chiang et al., 2001) were also used. Unless stated otherwise, these experiments were repeated for three times, and at each time 30 embryos were stained for each gene at each experimental setting.
Cartilage Staining
Embryos were fixed in 4% paraformaldehyde (PFA) at 4°C overnight and then washed in phosphate-buffered saline containing 0.1% Tween-20 (PBST) for 8 h. After staining cartilage with 0.015% Alcian blue overnight at 4°C, embryos were rehydrated with a graded series of alcohols to distilled water and then treated with 0.25% trypsin at room temperature until embryo tissues became transparent. The embryos were then transferred to a solution of 1% KOH/3% H2O2 for 10 min and washed twice in PBST for 5 min each (Li et al., 2018). Embryos were also fixed and stained with Wheat Germ Agglutinin (WGA) (Invitrogen) and DAPI (Sigma-Aldrich) as described by Dale and Topczewski (2011) to show the structure of pharyngeal chondrocytes (Sisson et al., 2015). Z-stack images of the embryos were taken using either a Zeiss LSM 510 or LSM 700 laser confocal microscope. Unless stated otherwise, these experiments were repeated for 3 times, and at each time 50 embryos were stained for each injection setting.
Immunofluorescence
Tg (sox10:EGFP) zebrafish embryos were fixed with 4% phosphate-buffered paraformaldehyde and washed with PBST for 20 min, then immunostained with PHH3 (1/400; sc-374669, Santa Cruz Biotechnology) antibodies. Terminal deoxynucleotidyl transferase dUTP nick-end labeling (TUNEL) assays were performed using the In Situ Cell Death Detection Kit, TMR red (12156792910, Roche) according to the manufacturer’s instructions. DAPI (4′,6-diamidino-2-phenylindole) was used to visualize nuclei. Anti-PHH3 staining was designed to indicate proliferation and TUNAL assays to indicate apoptosis. In either analysis, only cells that were double positive for anti-PHH3/TUNEL and EGFP were counted manually in order to make sure that they were CNCCs. All immunofluorescence images were captured using a Nikon A1R + confocal microscope using the same settings for all experiments. The experiments were repeated for 2 times.
Statistical Analysis
All experiments were performed in triplicate, and unpaired t-tests were used to analyze all data sets. Results were considered statistically significant at p < 0.05. Congruent results (>85% of the embryos observed) were reached in all phenotypic observational experiments and all selected images are representative.
Results
Clinical Presentation
8 of 11 direct relatives in this five-generation pedigree displayed HFM to varying degrees (Figure 1A). The proband showed grade III bilateral ear deformities and III:1 in the pedigree had grade II unilateral ear deformity according to the Marx classification, whereas other patients exhibited unilateral grade III ear deformities. All patients exhibited mandibular hypoplasia. Pure-tone audiometry testing showed moderate to severe conductive hearing loss. The family denied consanguinity. Detailed physical examination, along with cardiac, renal, and abdominal ultrasound was carried out on all 12 living members of the pedigree and no deformities of other systems were diagnosed. The disorder appeared to follow an autosomal-dominant segregation pattern with incomplete penetrance and variable expressivity. Individual IV:3 and V:3 showed no malformation of all the systems examined, and refused to give blood for sequencing. Individual IV:1 was predicted to be a carrier of the pathogenic gene.
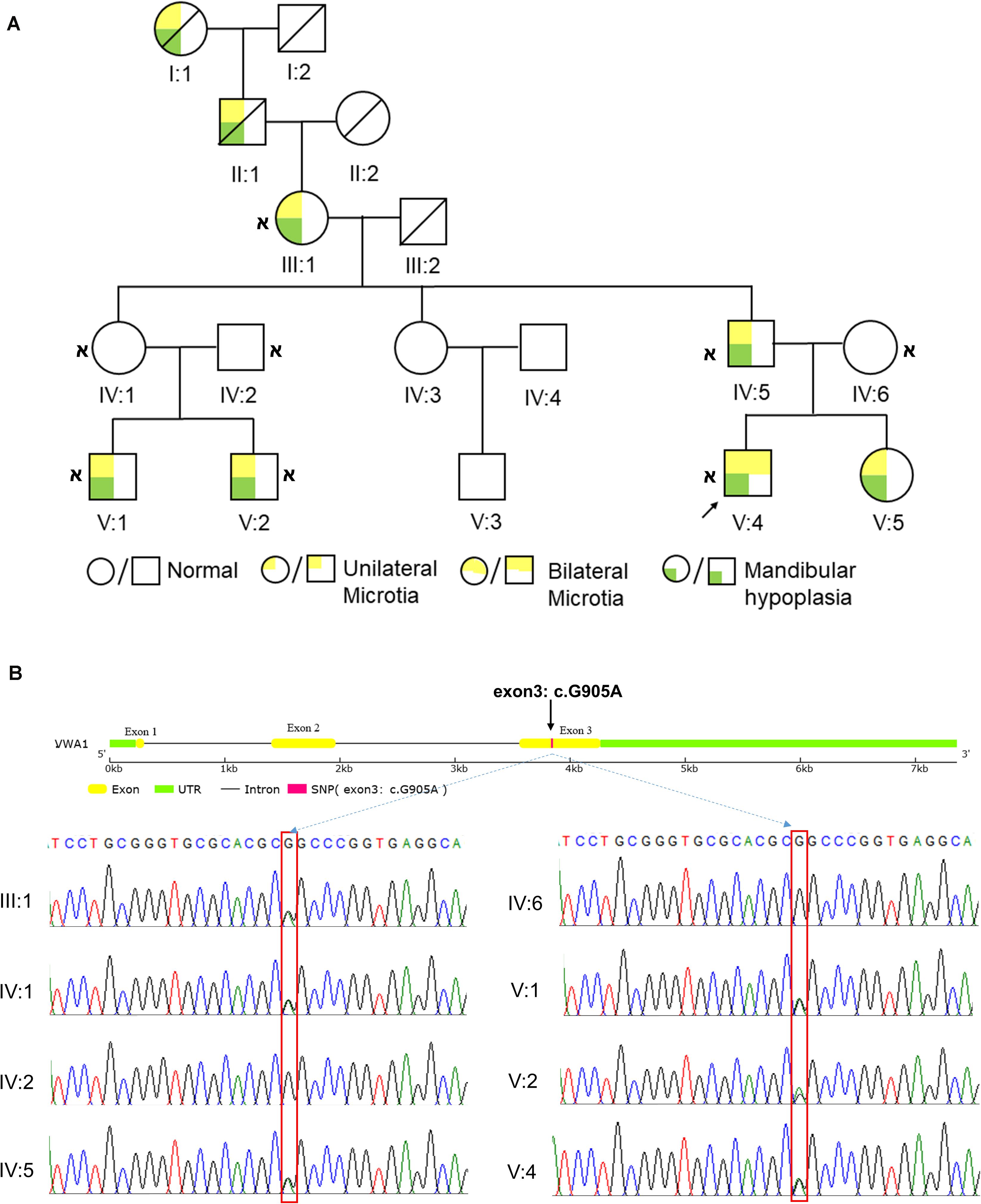
Figure 1. HFM pedigree and the identification of vwa1. (A) Diagram of the five-generation HFM pedigree. This is a five-generation Chinese kinship in which 8 of 11 direct relatives displayed HFM to varying degrees. The proband showed grade III bilateral ear deformities; individual III:1 in the pedigree had a grade II unilateral ear deformity, whereas other patients exhibited unilateral grade III ear deformities. All patients exhibited mandibular hypoplasia. Circles denote females and squares denote males. The text immediately below the symbols shows the ID of the corresponding family member;  indicates that whole-exome sequencing was done. (B) Sanger sequencing results of the VWA1 mutation in the pedigree. Patients III:1, IV:5, V:1, V:2 and V:4, and the asymptomatic carrier IV:1, all had the same non-synonymous point mutation (c.G905A:p.R302Q) in VWA1, whereas the two marry-in members of the family (IV:2 and IV:6) did not.
indicates that whole-exome sequencing was done. (B) Sanger sequencing results of the VWA1 mutation in the pedigree. Patients III:1, IV:5, V:1, V:2 and V:4, and the asymptomatic carrier IV:1, all had the same non-synonymous point mutation (c.G905A:p.R302Q) in VWA1, whereas the two marry-in members of the family (IV:2 and IV:6) did not.
Identification of VWA1 Mutation and Molecular Analysis Results
In order to identify the variants, we first collected peripheral blood from family members and extracted cDNA. Individual IV:1 was regarded as carrier and the two marry-in members (IV:2 and IV:6) were regarded as controls. After careful filtration as mentioned in the method section, WES revealed eight candidate genes: VWA1, SLC35G4, HRC, KLK1, ZBTB45, FBLN2, HECW1, and PGBD3. These eight genes were present in all six living affected individuals tested. All candidate genes went through secondary and tertiary structure predictions and expression profiles of all genes that have homologs in zebrafish were constructed (data not shown). Basing on: (1) VWA1 mutation caused the most apparent structural changes; (2) VWA1 associated variance had previously been reported in a patient presenting with mandibulofacial dysostosis and microtia (Zhang et al., 2010); (3) vwa1 showed specific expression in the mandible area of zebrafish, we suspected that VWA1, encoding WARP (von Willebrand factor A-domain-related protein 1), was most likely associated with HFM. The non-synonymous point mutation (c.G905A:p.R302Q) in the VWA1 gene was shared by all patients tested and the predicted carrier as well (Figure 1B).
Secondary and tertiary structures of sequences flanking the mutation were predicted (Figures 2A–C). The mutation introduced a R302Q amino acid substitution in the protein’s loop area, altering upstream and downstream secondary structures as well, including the possible disruption of a helix and introduction of several sheets (Figure 2A). The change from arginine, a basic amino acid, to glutamine, a neutral amino acid, altered the position of the amino acid’s side chain and brought changes to the local tertiary structural prediction, indicating changes in both the conformation and function of WARP (Figures 2B,C). Prediction of protein-to-protein interactions of WARP were carried out and five pathways: WNT, BMP (bone morphogenic protein), FGF, RA, and endothelin-1, which are closely related to craniofacial development, were selected for detailed analysis (Frisdal and Trainor, 2014; Graf et al., 2016). This analysis showed that WARP may interact with four proteins-FGF23, SPP1 (osteopontin), CDH2 (cadherin 2), and SDC2 (syndecan 2)-of the FGF pathway (Figure 2D). These taken together indicated that VWA1 is indeed a strong candidate pathogenic gene of this HFM pedigree.
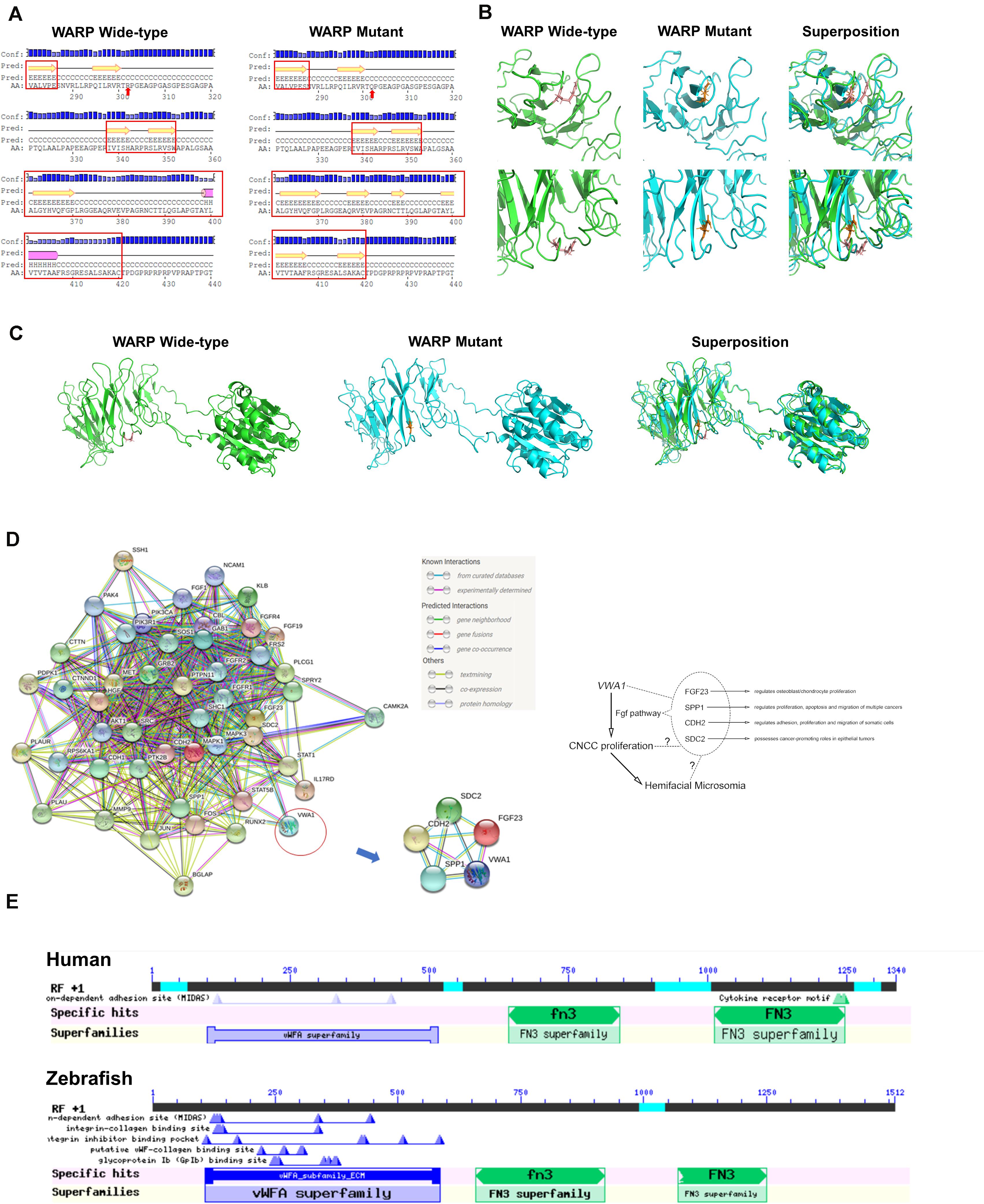
Figure 2. Predicted WARP structures, protein interactions and conserved domains. (A) Predicted secondary structures of wild-type and mutant protein sequences flanking the mutation. Secondary structural features are annotated as follows: pink cylinder, α-helix; yellow arrow, β-sheet; black line, coil; Conf, confidence; Pred, predicted; H in Pred line, helix; C in Pred line, coil; E in Pred line, sheet; AA, amino acid; red arrow, mutant amino acid. Local (B) and global (C) views of the predicted tertiary structures of wild-type (green) and mutant (blue) proteins. Structural superposition analyses demonstrating the change in protein structure brought about by the mutation are also displayed. (D) Protein-to-protein interactions of WARP with the FGF pathway. Predictions using the web-based program, String, indicated that WARP may functionally interact with four proteins of FGF pathway: FGF23, SPP1, CDH2, and SDC2. (E) Domains of WARP conserved in humans and zebrafish. Humans and zebrafish have the same WARP conserved domains: a von Willebrand factor A-domain, the first fibronectin type III repeat, and the second fibronectin type III repeat.
VWA1 Homolog in Zebrafish, vwa1, Is Specifically Expressed in the Mandible
The protein encoded by VWA1 in humans was shown to contain three conserved domains: a von Willebrand factor A-domain, the first fibronectin type III repeat and the second fibronectin type III repeat. There is one homologous gene, vwa1, in zebrafish which has the same conserved domains (Figure 2E).
To verify that vwa1 plays an important role in craniofacial development, we first examined its spatiotemporal expression during zebrafish embryonic development using in situ hybridization (n = 45). We found that, from 19 h post-fertilization (hpf) to 1 day post-fertilization (dpf), vwa1 expression was observed at the medial edge of the pharyngeal arch, with a slight concentration in the rostral region that gives rise to craniofacial structures (Figures 3A–D). At 19 hpf and 1 dpf (Figures 3B,D). From 2 to 4 dpf, expression of the vwa1 gene in the head became increasingly concentrated in the mandibular region (Figures 3E–J). Staining for vwa1 in the pharyngeal-pouch was more intense than that in the pharyngeal arch (Figures 3F–H). Expression of vwa1 was also found in the epidermis of the brain and somites (Figures 3C,D), which was in accordance with previous reports in mice (Brachvogel et al., 2008; Allen et al., 2009). These observations strongly suggest that zebrafish vwa1 is specifically expressed in the mandible during the period of pharyngeal arch development.
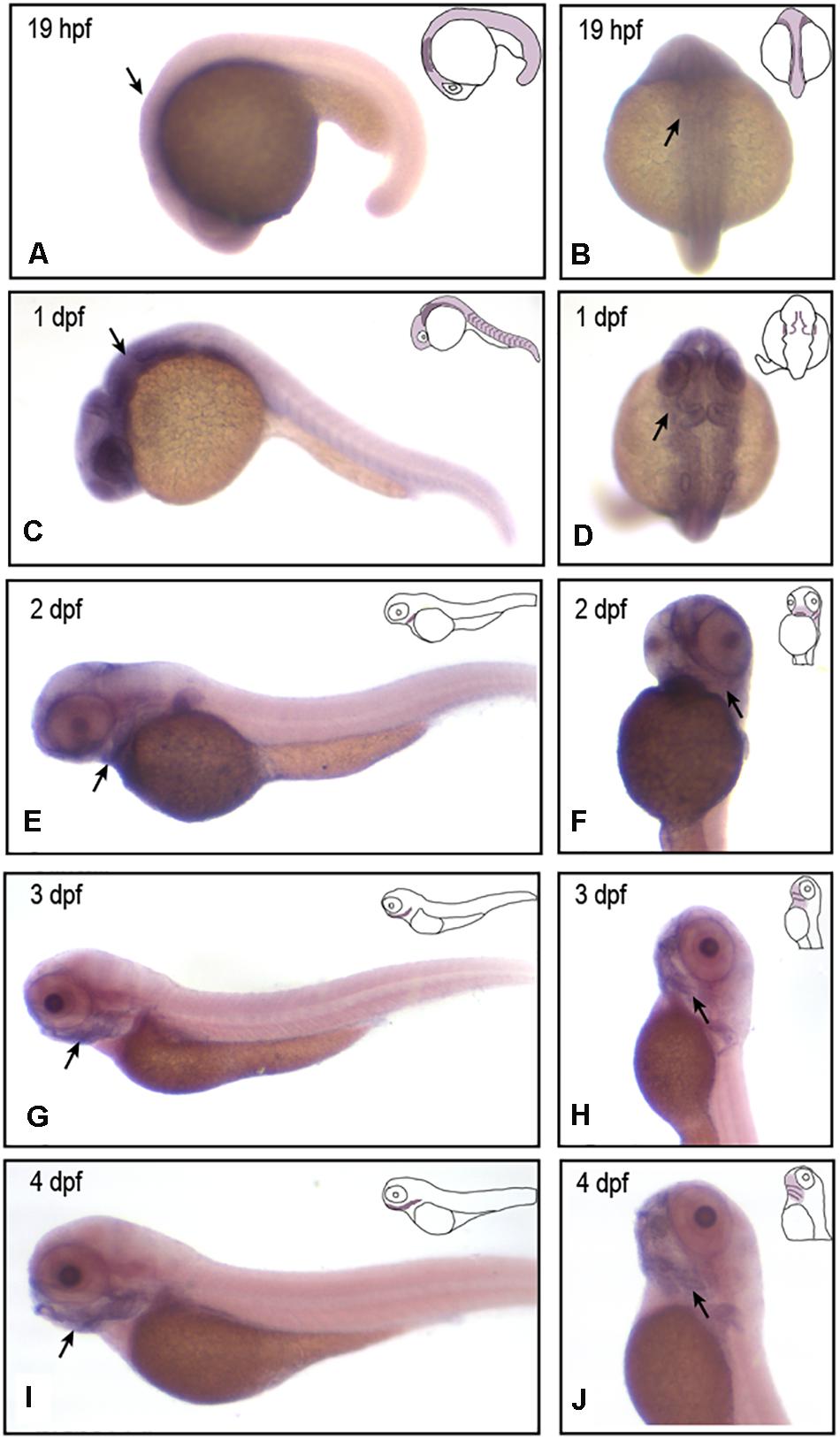
Figure 3. vwa1 expression in developing zebrafish embryos. In situ hybridization (ISH) results showing the expression of vwa1 at 19 hpf (A,B), 1 dpf (C,D), 2 dpf (E,F), 3 dpf (G,H) and 4 dpf (I,J) stages in lateral (A,C,E,G,I), dorsal (B,D) and lateral-ventral (F,H,J) views. At 19 hpf and 1 dpf, vwa1 was expressed throughout the body, with expression in the head being higher than that in other parts of the body (black arrows in A,C). At 19 hpf and 1 dpf, vwa1 expression was detected at the medial edge of the pharyngeal arch (black arrows in B,D). At 2 dpf, vwa1 staining in the head began to increase slightly, especially in the mandible (black arrow in E). At 3 dpf and 4 dpf, expression of vwa1 was restricted to the mandible (black arrows in G,I). Staining of vwa1 in the pharyngeal pouch was stronger than that in the pharyngeal arch (black arrows in F–H). vwa1 was also expressed in the epidermis of the brain (D) and in somites (C,E,G,I).
Reduced vwa1 Expression Causes Malformation of Pharyngeal Cartilage
To determine if vwa1 is required for the development of pharyngeal cartilages, we knocked down its function in zebrafish embryos using an antisense morpholino oligonucleotide targeting the translation start site of vwa1 mRNA (vwa1-ATG MO) at the 1-cell stage, which is proved effective. Higher amounts of vwa1-ATG MO caused more severe deformities (Supplementary Figure 2), and in subsequent experiments a dose of 1 ng was applied.
To reduce off-targeting effects of morpholinos, we co-injected a 1.5-fold (w/w) greater amount of p53 MO together with vwa1-ATG MO (Robu et al., 2007). Previous research has shown that p53 null mutation in mice can rescue the craniofacial damage caused by disruption of Brca1 (Robu et al., 2007; Kitami et al., 2018). We performed injection of p53 at different concentration without vwa1-ATG MO to rule out its potential effects and no visible malformation was observed at each concentration (Supplementary Figure 2). Co-injection of p53 MO together with vwa1-ATG MO induced observable phenotypic changes in 132/150 embryos. After injecting 1 ng vwa1-ATG MO and 1.5 ng p53 MO, there was a reduction in the overall size of jaw structures at 4 dpf compared with uninjected controls (Figure 4Aa,d). Alcian blue staining revealed that the first and second pharyngeal arches were severely hypoplastic, and the third to seventh pharyngeal arches (Cb1-5) were absent (Figure 4Ae,h). The joint junctions were also abnormal: the angle between cartilage derived from the first pharyngeal arch, Meckel’s (M) and palatoquadrate (Pq), became smaller, and the angle between cartilage derived from the second pharyngeal arch, ceratohyal (Ch), became larger (Figure 4Ae,i,h,l). Besides, injected embryos also demonstrated ventral swelling suggestive of cardiovascular dysfunction. Whether vwa1 had potential effects on cardiovascular development would require further study.
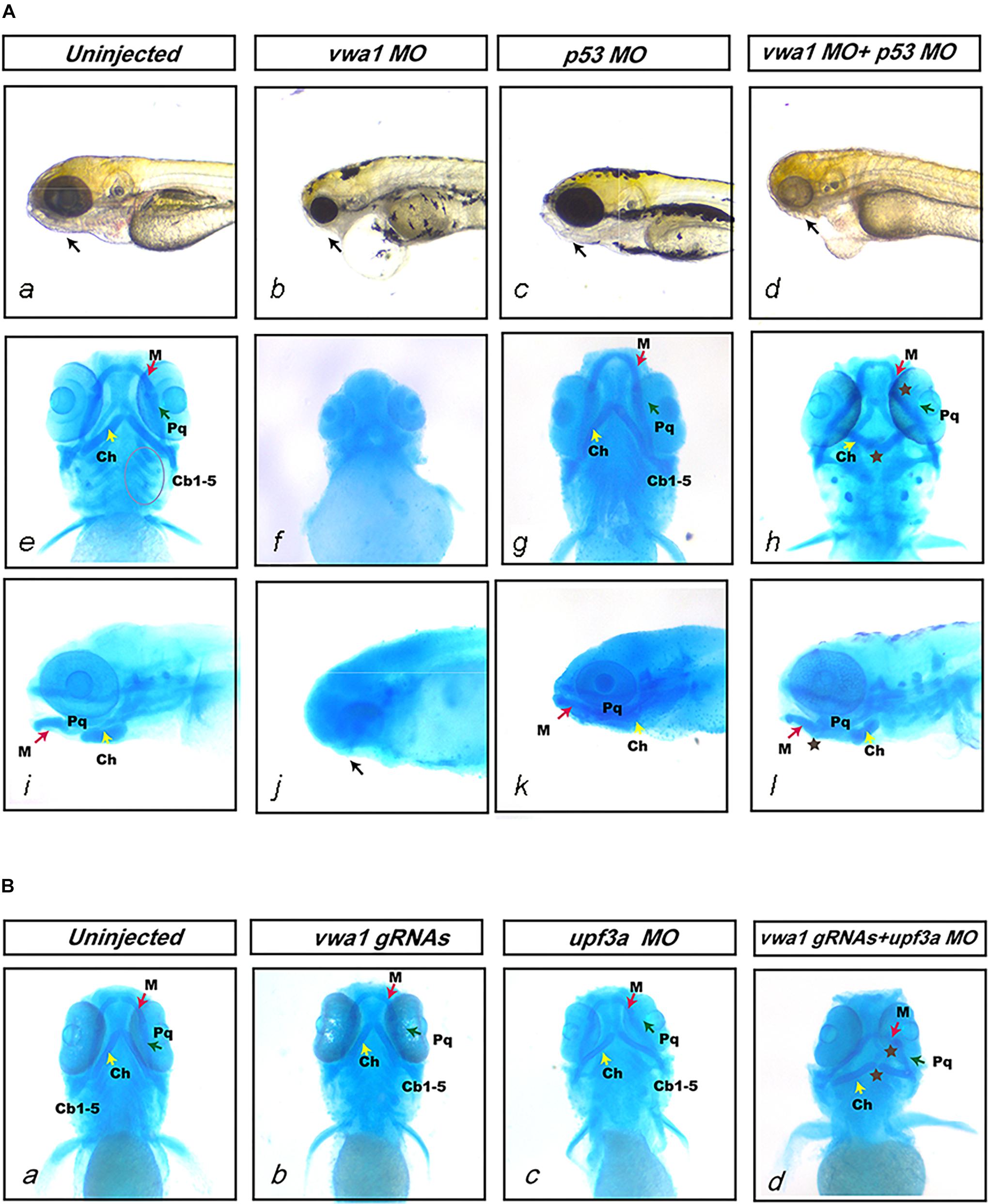
Figure 4. Deformities of pharyngeal cartilage in vwa1 morphants and mutants. (A) Deformities of pharyngeal cartilage in vwa1 morphants. The figure shows the jaws of uninjected and vwa1 morpholino (MO)-, p53 MO- and vwa1 + p53 MO-injected morphants under a stereoscope and Alcian blue staining of morphant embryos. Injection of 1 ng vwa1-ATG MO caused jaw structures to almost completely disappear (b, f, and g). Injection of 1 ng vwa1-ATG MO with p53 MO reduced the overall size of jaw structures at 4 dpf compared with uninjected controls and 1.5 ng p53 MO-injected morphants (a and d), whereas there was no significant difference compared with controls after injection of p53 MO (a–d and i–l). The first and second pharyngeal arches were severely hypoplastic (brown ⋆ in h and l), and the third to seventh pharyngeal arches (Cb1-5, circled in e) were absent (h). Joint junctions were abnormal: the angle between cartilage derived from the first pharyngeal arch, Meckel’s (M) and palatoquadrate (Pq) became smaller, whereas the angle between cartilage derived from the second pharyngeal arch ceratohyal (Ch) became larger (brown ⋆ in h and l). (B) Deformities of pharyngeal cartilage in vwa1-knockout mutants. a, b, c, and d shows Alcian blue staining results for uninjected embryos and embryos injected with vwa1 gRNAs, upf3a MO, and vwa1 gRNAs together with upf3a MO. Embryos injected with vwa1 gRNAs or upf3a MO showed no significant deformities in pharyngeal cartilage compared with uninjected embryos (b and c). Injection of vwa1 gRNAs together with upf3a MO resulted in pharyngeal cartilage deformities similar to those in vwa1 morphants, including hypoplasia of first and second pharyngeal cartilages, disappearance of the third to seventh pharyngeal arches, and abnormal joint junctions (brown ⋆ in d).
To further verify the causal relationship between vwa1 variants and the malformed pharyngeal cartilages observed in vwa1 zebrafish mutants, we knocked out vwa1 using four different gRNAs, all of which proved effective. Alcian blue staining revealed no deformities in the vwa1-knockout mutant (Figure 4Bb). Given the phenotype of vwa1-ATG morphants, this unexpected finding suggested potential compensatory effects. To mitigate potential genetic compensatory effects attributable to DNA damage, we co-injected vwa1-knockout mutants with morpholinos targeting upf3a to inhibit nonsense-mediated RNA decay (Shum et al., 2016). Embryos injected only with upf3a MO had no significant deformities in pharyngeal cartilages (Figure 4Bc). Embryos co-injected with upf3a MO showed deformities in pharyngeal cartilage similar to those of vwa1-ATG morphants, including hypoplasticity of the first and second pharyngeal cartilages, the disappearance of third to seventh pharyngeal arches and abnormal joint junctions (Figure 4Bd). Collectively, these data suggest that vwa1 plays a very important role in craniofacial development, such that its loss causes deformities of pharyngeal cartilage.
Migration and Differentiation of CNCCs Are Not Affected by vwa1
The higher expression of vwa1 in the pharyngeal endoderm led us to examine morphogenesis of the pharyngeal pouch in vwa1-ATG morphants. To this end, we knocked down the expression of the vwa1 in Tg (nkx2.3:EGFP) transgenic fish (Li et al., 2018), in which we could observe the pharyngeal endoderm cells. At approximately 38 hpf, the pharyngeal pouch in vwa1-ATG morphants appeared somewhat wider, but there were no striking differences compared with controls (Figure 5A).
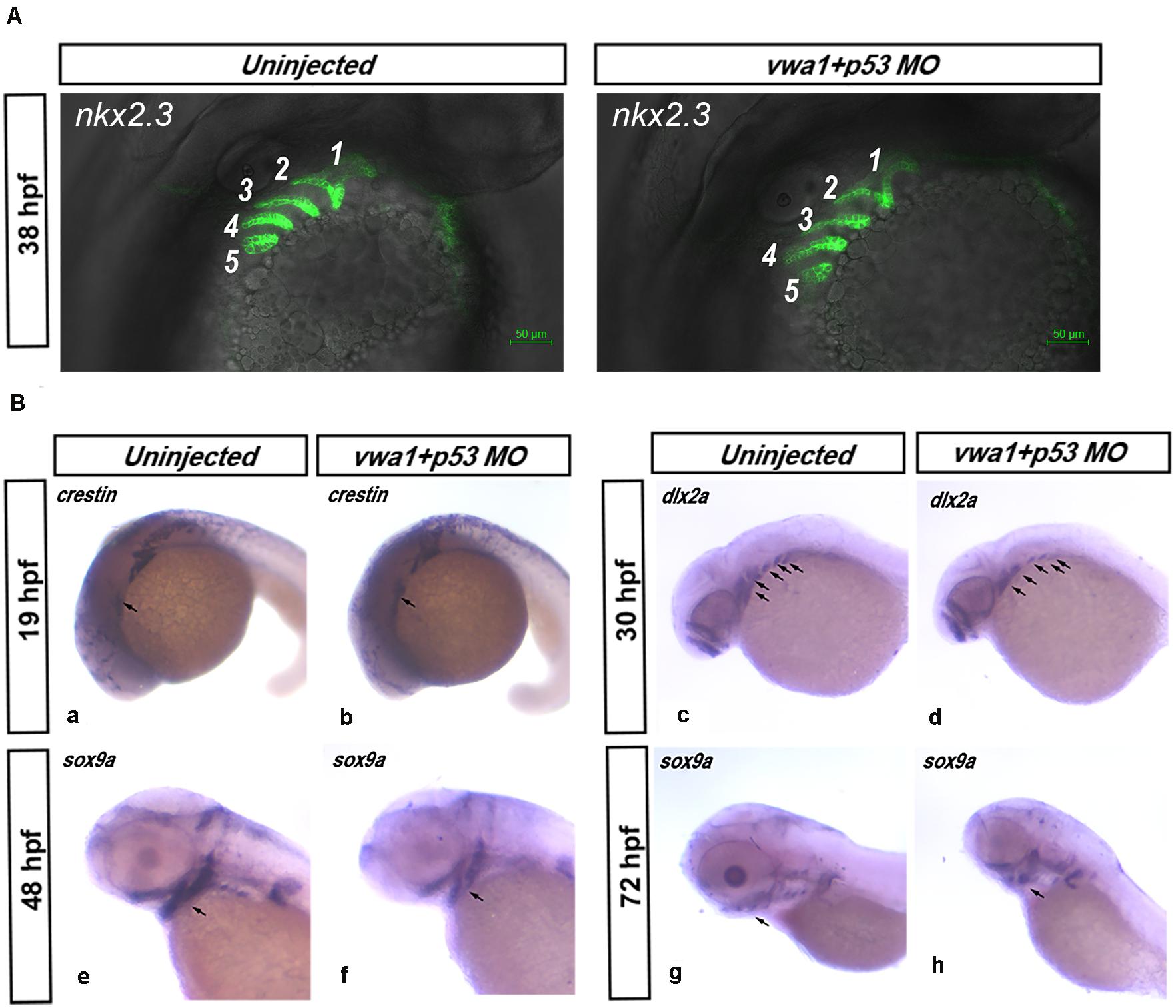
Figure 5. Effects of reduced vwa1 on the development of pharyngeal cartilage. (A) Endodermal pouch in Tg (nkx2.3:EGFP) embryos. At about 38 hpf, when the endodermal pouch is almost fully formed, no apparent difference was observed in the pharyngeal pouch in vwa1 morphants (right) compared with uninjected morphants (left). (B) vwa1 knockdown causes a reduction in cranial neural crest cell numbers. At about 19 hpf, when neural crest cells migrate to the cranial area, the expression of crestin in vwa1-knockdown embryos was not obviously different from that of uninjected control embryos (a and b). At 30 hpf, when migration of neural crest cells is complete, dlx2a was expressed in similar domains, but the area of these domains was reduced significantly in vwa1 morphants (c and d). At 48 and 72 hpf, when the cranial neural crest has differentiated into pharyngeal chondrocytes, sox9a was expressed in similar domains, but the areas of these domains were reduced significantly in vwa1 morphants (e–h). These data show that, in vwa1 morphants, neural crest cells generally could migrate to the pharyngeal arch and specify to chondrocytes, but their numbers were reduced.
We next used in situ hybridization to monitor the expression of the following marker genes involved in the migration and specification of the CNCCs: crestin, which is expressed in neural crest at about 19 hpf, a time when CNCCs migrate to the cranial area; dlx2a, which is expressed in all CNCCs in the pharyngeal arches at 30 hpf, when CNCCs have completed their migration; as well as sox9a, markers of cartilage differentiation (Yan et al., 2002; Johnson et al., 2011; Kim et al., 2015). At 19 hpf, there was no obvious difference in crestin expression between vwa1-ATG morphants and uninjected embryos, suggesting that CNCCs can migrate normally to the arches. At 30, 48 and 72 hpf, dlx2a and sox9a were expressed in similar domains, but the areas of these domains were significantly reduced in the vwa1-ATG morphants compared with uninjected embryos. These data show that, in the vwa1-ATG morphants, CNCCs were generally capable of migrating to the pharyngeal arches and differentiating into chondrocytes, but their number was reduced (Figure 5B).
Proliferation of CNCCs Requires vwa1
We hypothesized that the reduction in the number of CNCCs was attributable to changes in their proliferation and/or apoptosis. To identify proliferating and apoptotic cells, we immunostained for PHH3 and performed TUNEL assay, respectively, at 30 hpf. To confirm that the cells we observed in pharyngeal arches were indeed CNCCs, we used Tg (sox10:EGFP) embryos. Immunofluorescence images were captured (Figures 6A,B) and statistical analysis of 20 vwa1-ATG morphants vs. 20 uninjected embryos on cell proliferation, and 10 vwa1-ATG morphants vs. 10 uninjected embryos on cell apoptosis showed that proliferation of CNCCs was reduced significantly from an average of 41.30 ± 3.02 cells per fish (uninjected embryos) to 20.35 ± 3.45 cells per fish (vwa1-ATG morphants) (n = 20, p < 0.05), whereas apoptosis was not significantly affected, with that of that of uninjected embryos being 1.50 ± 0.81 cells per fish and vwa1-ATG morphants being 1.40 ± 1.11 cells per fish (n = 10). These data, combined with the bioinformatics prediction of protein-to-protein interactions of WARP (Figure 2D), suggest that vwa1 is required for the proliferation of CNCCs. It may regulate CNCC proliferation via crosstalk with FGF pathway and its knockdown leads to deformities of pharyngeal cartilages by reducting CNCC proliferation.
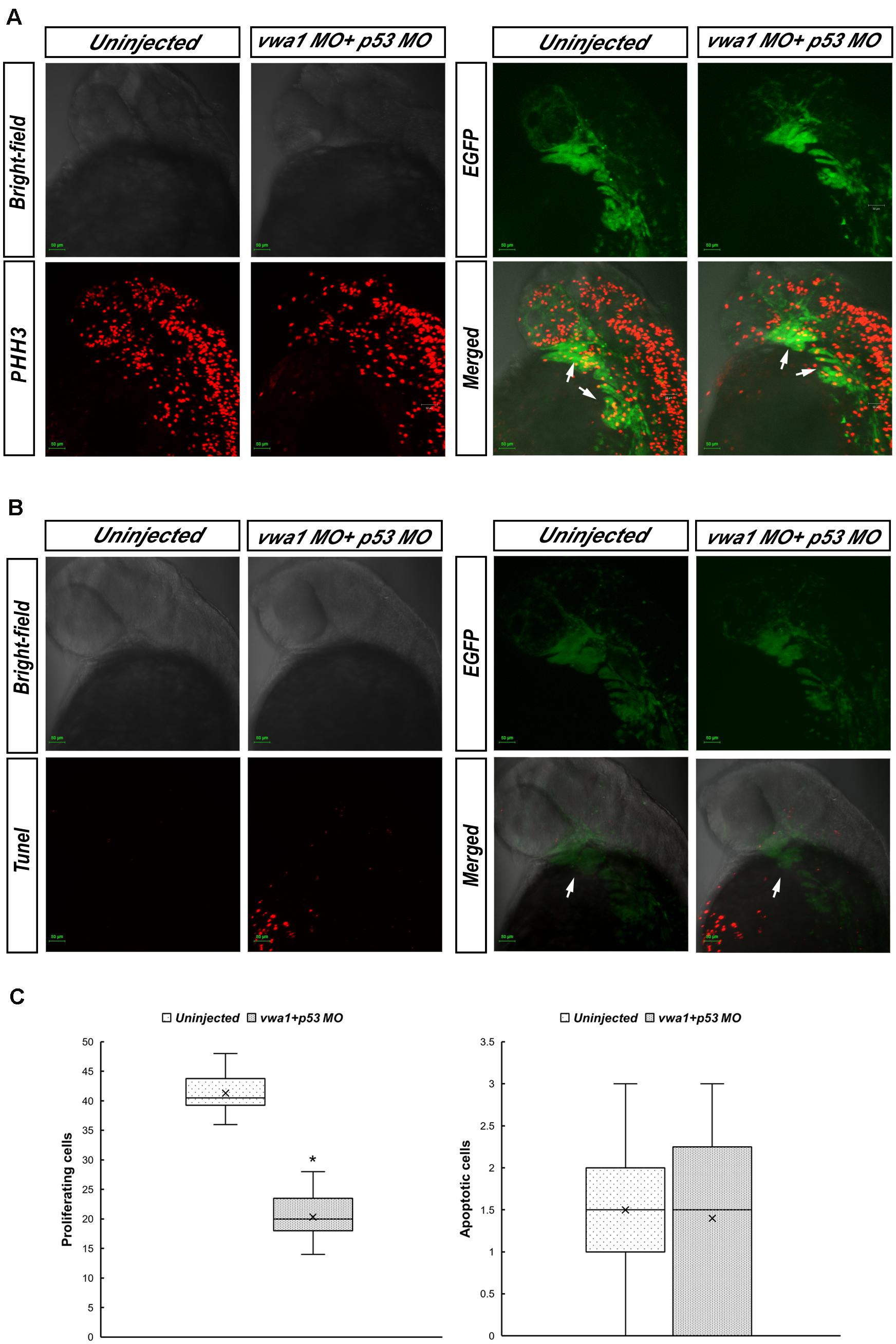
Figure 6. Proliferation and apoptosis of cranial neural crest cells at approximately 30 hpf. (A) Immunofluorescence results showing anti-phosphohistone H3 (PHH3) staining demonstrating cell proliferation. Merged image demonstrates higher anti-PHH3 signals in cranial neural crest cells compared with uninjected embryos. Arrows: pharyngeal arches. (B) Apoptosis assessed by immunofluorescence detection of TUNEL assay. The apoptosis signal in the vwa1 + p53 MO group did not coincide with EGFP fluorescence. Arrows: pharyngeal arches. (C) Comparison of apoptotic and proliferative numbers of neural crest cells in pharyngeal arches between vwa1 + p53 MO-injected embryos and uninjected control embryos. The difference in cell proliferation was statistically significant (p < 0.05, Student’s t-test). Lower/upper whiskers: 1st and 4th quartiles of the data; box: 2nd and 3rd quartiles of the data; mid-line: median of the data; ×: mean value of the data; *: significantly different.
vwa1-ATG Morphants Exhibit Disorganization of Pharyngeal Chondrocytes
As WARP is localized to the extracellular matrix of chondrocytes (Allen et al., 2006), we hypothesized that the arrangement of chondrocytes might be altered and could contribute to the pharyngeal cartilage deformities. To examine this, we used fluorescent wheat germ agglutinin (WGA) to stain glycoproteins in the chondrocyte membrane. The uninjected embryos exhibited a very stereotypic shape among individuals at 4 dpf, with thin and elongated chondrocytes assembled on top of one another, forming a characteristic “stack of pennies” organization (Figure 7A). In the vwa1-ATG morphants, many of the cartilage elements were not only smaller, but greatly deformed as well-compared to the uninjected ones (Figure 7B). These results show that knockdown of vwa1 leads to disorganization of pharyngeal chondrocytes and subsequent deformities in pharyngeal cartilage.
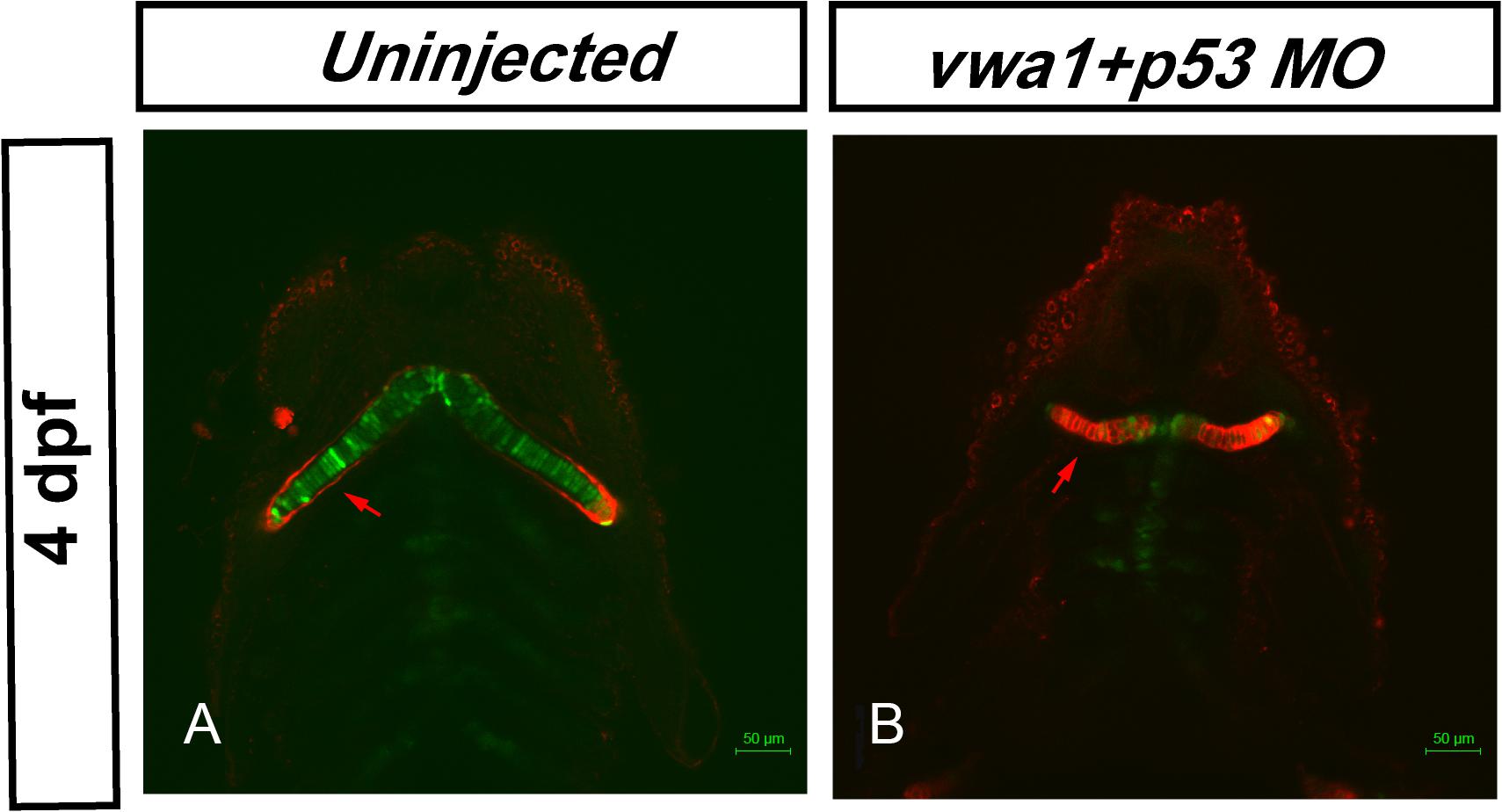
Figure 7. Knockdown of vwa1 leads to disorganization of pharyngeal chondrocytes at 4 dpf. (A) In uninjected control embryos at 4 dpf, craniofacial cartilage had formed and showed a very stereotypic shape between individuals, exhibiting the characteristic “stack of pennies” organization in which thin and elongated chondrocytes are assembled on top of one another to form their respective cartilage element. (B) In vwa1 morphants, the size and length-width ratio of many cartilage chondrocytes were smaller and the cartilage elements were greatly deformed compared with those of uninjected controls.
Discussion
In the present study, point mutations in 8 candidate genes shared by all affected individuals: VWA1, SLC35G4, HRC, KLK1, ZBTB45, FBLN2, HECW1 and PGBD3, were identified by WES in this five-generation HFM pedigree. Basing on structural prediction, literature review and functional study in zebrafish, we predicted the c.G905A:p.R302Q mutation in the VWA1 gene to be most likely associated with HFM. In the fifth generation, the mutation in V:1 and V:2 was inherited from a non-HFM mother (IV:1), indicating incomplete penetrance of the disease. Although only a single base is mutated, in silico prediction demonstrated that it may lead to significant changes in the secondary and tertiary structure of VWA1 protein, suggesting substantial changes in protein function.
VWA1 was primarily known for its expression in cartilage cells, in basement membranes of the peripheral nervous system and in skeletal and cardiac muscle (Fitzgerald et al., 2002; Allen et al., 2008; Fitzgerald, 2019). Subsequent research also revealed a distinct localization of VWA1 in the inner ear vasculature, suggesting a potential role in maintaining its integrity (Duong et al., 2011). Although VWA1 has surfaced in research on several diseases and has been suspected to be associated with neuro-musculoskeletal disorders, to date, no direct disease-causing variants have been directly identified (Dieppedale et al., 2013; Fitzgerald, 2019). Zhang et al. (2010) previously reported copy number variance of VWA1 and PYGO2 in a patient presenting with mandibulofacial dysostosis and microtia. At that time, VWA1 was suspected, but not confirmed, to be among the candidate genes responsible for the abnormal phenotype. Considering the low incidence of HFM, the fact that the mutation in was found in unrelated sporadic case and a five-generation pedigree reinforces the validity of the conclusion that the c.G905A:p.R302Q mutation in VWA1 is a likely pathogenic mutation for HFM.
Knockdown of vwa1 using vwa1-ATG MO resulted in hypoplasia of facial cartilage in zebrafish. In contrast, knockout of vwa1 using CRISPR/Cas9 (with four different gRNAs) did not cause deformities in pharyngeal cartilage. Notably, Vwa1 knockout in mice also did not induce observable craniofacial deformities as was reported in a previous study (Allen et al., 2009). The inconsistency in vwa1-ATG MO knockdown and CRISPR/Cas9 knockout led us to further inspect the mechanisms behind. We speculated that the lack of pharyngeal cartilage deformity in vwa1-knockout zebrafish was attributable to genetic compensation, which is considered a possible explanation for phenotypic differences between gene knockout and gene knockdown (Ma et al., 2019). mRNA bearing a premature termination codon promptly triggers a genetic compensation response that involves upf3a and components of the COMPASS complex (Ma et al., 2019). After we knocked down the function of upf3a, the vwa1-knockout zebrafish exhibited deformities similar to those in vwa1-knockdown embryos. These data show that vwa1 dysfunction genuinely affects craniofacial development. Notably, apart from craniofacial anomalies, knockdown of vwa1 also resulted in ventral swelling, which might indicate cardiovascular dysfunction. Neural crest cells also participate in the development of the heart and similar situation where a gene plays important roles in both ear and cardiovascular development has been reported in eya4 (Schönberger et al., 2005; Wentzel and Eriksson, 2011). Therefore, whether vwa1 holds potential effects on cardiac development requires further exploration. The phenotypic changes in zebrafish indicated potential haploinsufficiency mechanisms behind the potential pathogenic effects of vwa1. However, the possibility that truncated WARP produced by the point mutation in humans may interfere with normal WARP function and demonstrate a dominant negative pathogenic fashion was not excluded (Messina et al., 2016a, b).
In vwa1-ATG morphants, the chondrocytes of pharyngeal cartilage appeared smaller and disorganized compared with the normal characteristic “stack of pennies” organization. This observation is understandable as WARP serves as an important component of the extracellular matrix (ECM), whose composition is a complex mixture of collagens and non-collagenous proteins (Allen et al., 2006). Research has revealed a regulatory role for proteoglycans, glycosaminoglycans, proteases, and glycosidases in cell proliferation and differentiation (Roughley, 2006). Moreover, the special avascular nature of cartilage further highlights the important effects of the ECM on the diffusion, distribution, and binding capacity of signaling molecules (Esko and Selleck, 2002; Cortes et al., 2009). ECM proteins also have the capacity to bind to receptors and initiate signal transduction (Streuli, 1999). Therefore, a disturbance in the ECM microenvironment caused by knockdown of vwa1 could contribute to the overall phenotypic deformities in pharyngeal cartilage.
Upon reducing vwa1 function, the remaining cartilaginous elements in vwa1-ATG morphants seemed to adopt appropriate patterns, both the first and second pharyngeal arch derived structures were affected, and markers for craniofacial development were generally expressed in appropriate locations. These data suggest that vwa1 does not play a vital role in the migration, aggregation or specification of cranial neural crest cells, but may be very important in regulating their number. In support of this hypothesis, the size of both craniofacial cartilage and cranial neural crest cell marker domains were reduced in vwa1-ATG morphants, indicating a change in CNCC number. Reduced CNCC number could result from decreased proliferation and increased apoptosis, which have both been proven to be associated with different kinds of craniofacial anomalies. For example, reduced proliferation of CNCCs in the medial nasal process mesenchyme is required for upper lip closure and has been identified in the pathogenesis of cleft lip (Everson et al., 2017). Meanwhile, increased apoptosis of CNCCs could affect secondary palate fusion and has been proven to be associated with significant mid-facial anomalies such as cleft palate (Wilson et al., 2016; Welsh et al., 2018; Wang et al., 2019). Based on our data, we observed a decrease in cell proliferation, but no change in cell apoptosis, in vwa1-ATG morphants. Taken together, these data indicate that vwa1 is required for the proliferation of CNCCs.
CNCCs’ proliferation, apoptosis, migration and differentiation have been reported to be tightly regulated by a delicate and intricate extracellular signaling system, which includes SHH, WNT, RA, FGF, endothelin and BMP (Frisdal and Trainor, 2014; Graf et al., 2016; Everson et al., 2017). As an important component of the ECM, VWA1 is likely to regulate CNCC proliferation via extracellular signal transduction. Bioinformatics analyses predicted possible crosstalk of WARP with four proteins in the FGF pathway: FGF23, SPP1, CDH2, and SDC2. The FGF pathway is known to play crucial roles in the specification, patterning and migration of cranial neural crest cells (Diez Del Corral and Morales, 2017). Elements of this pathway have also been shown to regulate cranial neural crest cell proliferation (Sasaki et al., 2006). In particular, FGF23, a bone-derived hormone produced by osteoblasts and osteocytes that signals through FGFRs and α-Klotho, has a well-established association with renal phosphate wasting diseases (Erben, 2017). While research has demonstrated regulatory roles of FGF23 on osteoblast and chondrocyte proliferation in vitro and in vivo (Kawai et al., 2013), its effects on cranial neural crest cells await further study. CDH2 plays important roles in the adhesion, proliferation and migration of somatic cells (Marie et al., 2014), and has also been reported to function in the morphogenesis of zebrafish inner ear (Babb-Clendenon et al., 2006). SPP1 is an ECM protein closely associated with tumorigenesis and bone remodeling (Sousa et al., 2012; Saleh et al., 2016). Previous researches have reported its potential expression in cranial suture tissues of zebrafish (Topczewska et al., 2016). SDC2 has been reported to regulate angiogenic sprouting in zebrafish (Chen et al., 2004), and has distinct expression in embryonic zebrafish brain (Hofmeister et al., 2013). The overlap of spatiotemporal expression patterns and functional similarities between WARP and these proteins further support the possibility of their potential interactions. The crosstalk with these membrane proteins and secreted signaling molecules suggests complicated interactions among signaling pathways, especially the FGF pathway, which may underlie the suppressive effects of vwa1 knockdown on the proliferation of cranial neural crest cells (Figure 2D).
Conclusion
We reported that VWA1 is a strong candidate gene for HFM, showing that vwa1 variants influences the proliferation of CNCCs and the organization of pharyngeal chondrocytes. More patients with VWA1 mutations and corresponding genetic mouse models will be required to further confirm the causal role of this gene. The identification of this candidate gene for HFM can not only help to decipher the pathways involved in craniofacial development, but provide possible targets for genetic testing and therapeutic intervention as well.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Ethics Statement
The studies involving human participants were reviewed and approved by the institutional review board of PUMCH (JS-796). Written informed consent to participate in this study was provided by the participants’ legal guardian/next of kin. The animal study was reviewed and approved by the institutional review board of PUMCH (JS-796).
Author Contributions
XC and SZ obtained funding. YW, SZ, and XC conceived the study. YW and BZ designed the experiments. YW and LL conducted the experiments. YW and LP participated in the literature search, data collection, data analysis, and drafted the manuscript. XL conducted the bioinformatics protein structural prediction. YC, XF, and PW participated in the collection and treatment of the pedigree. YL participated in the genetic analysis of the pedigree. All authors have read and approved the final manuscript.
Funding
This work was supported by grant to XC from the General Programs of National Natural Science Foundation of China (81974143) and by grant to SZ from the National Key Research and Development Program of China (2016YFC0901500).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We are grateful to all the individuals in the pedigree for their participation. We thank Professor Xue Zhang for the guidance in the genetic analysis of the pedigree.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcell.2020.571004/full#supplementary-material
FIGURE S1 | Efficacy of vwa1-ATG Morpholino. (A) Structure of pCS2 + vwa1-ATG-MO-mCherry plasmid. (B) At 1 dpf, fluorescence could be observed in embryos injected with pCS2 + vwa1-ATG-MO-mCherry mRNA (a), showing that the mRNA could be translated into mCherry fluorescent protein. No fluorescence was observed in embryos co-injected with pCS2 + vwa1-ATG- MO-mCherry mRNA and vwa1-ATG MO (b), showing that mCherry was effectively knocked down. Fluorescence similar to that of pCS2 + vwa1-ATG-MO-mCherry injected embryos was observed in embryos co-injected with pCS2 + vwa1-ATG-MO-mCherry mRNA and p53 MO (c), showing that unspecific MO could not knock down mCherry.
FIGURE S2 | Deformities of pharyngeal cartilage in vwa1 morphants. The figure shows the jaws of uninjected and vwa1 morpholino (MO)-, p53 MO- and vwa1 + p53 MO-injected morphants Alcian blue staining of morphant embryos in different dose. Injection of vwa1-ATG MO and vwa1 + p53 MO caused deformities in pharyngeal cartilages, and higher amounts of MO caused more severe deformities. There was no significant difference compared with controls after injection of p53 MO.
TABLE S1 | Sequences of the primers of gRNA and the probes of the markers.
Footnotes
- ^ http://www.sbg.bio.ic.ac.uk/phyre2
- ^ http://bioinf.cs.ucl.ac.uk/psipred/
- ^ https://string-db.org
- ^ http://crispr.mit.edu
References
Akimenko, M. A., Ekker, M., Wegner, J., Lin, W., and Westerfield, M. (1994). Combinatorial expression of three zebrafish genes related to distal-less: part of a homeobox gene code for the head. J. Neurosci. 14, 3475–3486. doi: 10.1523/jneurosci.14-06-03475.1994
Allen, J. M., Bateman, J. F., Hansen, U., Wilson, R., Bruckner, P., Owens, R. T., et al. (2006). WARP is a novel multimeric component of the chondrocyte pericellular matrix that interacts with perlecan. J. Biol. Chem. 281, 7341–7349. doi: 10.1074/jbc.m513746200
Allen, J. M., Brachvogel, B., Farlie, P. G., Fitzgerald, J., and Bateman, J. F. (2008). The extracellular matrix protein WARP is a novel component of a distinct subset of basement membranes. Matrix Biol. 27, 295–305. doi: 10.1016/j.matbio.2008.01.005
Allen, J. M., Zamurs, L., Brachvogel, B., Schlötzer-Schrehardt, U., Hansen, U., Lamandé, S. R., et al. (2009). Mice lacking the extracellular matrix protein WARP develop normally but have compromised peripheral nerve structure and function. J. Biol. Chem. 284, 12020–12030. doi: 10.1074/jbc.M806968200
Babb-Clendenon, S., Shen, Y.-C., Liu, Q., Turner, K. E., Mills, M. S., Cook, G. W., et al. (2006). Cadherin-2 participates in the morphogenesis of the zebrafish inner ear. J. Cell Sci. 119(Pt 24), 5169–5177. doi: 10.1242/jcs.03299
Brachvogel, B., Farlie, P. G., Fitzgerald, J., and Bateman, J. F. (2008). The extracellular matrix protein WARP is a novel component of a distinct subset. Matrix Biol. 27, 295–305.
Chen, E., Hermanson, S., and Ekker, S. C. (2004). Syndecan-2 is essential for angiogenic sprouting during zebrafish development. Blood 103, 1710–1719. doi: 10.1182/blood-2003-06-1783
Chen, Q., Zhao, Y., Shen, G., and Dai, J. (2018). Etiology and pathogenesis of hemifacial microsomia. J. Dent. Res. 97, 1297–1305. doi: 10.1177/0022034518795609
Chiang, E. F., Pai, C. I., Wyatt, M., Yan, Y. L., Postlethwait, J., and Chung, B. (2001). Two sox9 genes on duplicated zebrafish chromosomes: expression of similar transcription activators in distinct sites. Dev. Biol. 231, 149–163. doi: 10.1006/dbio.2000.0129
Cortes, M., Baria, A. T., and Schwartz, N. B. (2009). Sulfation of chondroitin sulfate proteoglycans is necessary for proper Indian hedgehog signaling in the developing growth plate. Development 136, 1697–1706. doi: 10.1242/dev.030742
Dale, R. M., and Topczewski, J. (2011). Identification of an evolutionarily conserved regulatory element of the zebrafish col2a1a gene. Dev. Biol. 357, 518–531. doi: 10.1016/j.ydbio.2011.06.020
Dieppedale, J., Gesbert, G., Ramond, E., Chhuon, C., Dubail, I., Dupuis, M., et al. (2013). Possible links between stress defense and the tricarboxylic acid (TCA) cycle in Francisella pathogenesis. Mol. Cell. Proteom. 12, 2278–2292. doi: 10.1074/mcp.M112.024794
Diez Del Corral, R., and Morales, A. V. (2017). The multiple roles of FGF signaling in the developing spinal cord. Front. Cell Dev. Biol. 5:58. doi: 10.3389/fcell.2017.00058
Duong, T., Lopez, I. A., Ishiyama, A., and Ishiyama, G. (2011). Immunocytochemical distribution of WARP (von Willebrand A domain-related protein) in the inner ear. Brain Res. 1367, 50–61. doi: 10.1016/j.brainres.2010.10.056
Erben, R. G. (2017). Pleiotropic actions of FGF23. Toxicol. Pathol. 45, 904–910. doi: 10.1177/0192623317737469
Esko, J. D., and Selleck, S. B. (2002). Order out of chaos: assembly of ligand binding sites in heparan sulfate. Annu. Rev. Biochem. 71, 435–471. doi: 10.1146/annurev.biochem.71.110601.135458
Everson, J. L., Fink, D. M., Yoon, J. W., Leslie, E. J., Kietzman, H. W., Ansen-Wilson, L. J., et al. (2017). Sonic hedgehog regulation of promotes cranial neural crest mesenchyme proliferation and is disrupted in cleft lip morphogenesis. Development 144, 2082–2091. doi: 10.1242/dev.149930
Fitzgerald, J. (2019). WARP: a unique extracellular matrix component of cartilage, muscle, and endothelial cell basement membranes. Anat. Rec. 303, 1619–1623. doi: 10.1002/ar.24087
Fitzgerald, J., Tay Ting, S., and Bateman, J. F. (2002). WARP is a new member of the von Willebrand factor A-domain superfamily of extracellular matrix proteins. FEBS Lett. 517, 61–66. doi: 10.1016/s0014-5793(02)02579-6
Frisdal, A., and Trainor, P. A. (2014). Development and evolution of the pharyngeal apparatus. Wiley Interdiscip. Rev. Dev. Biol. 3, 403–418. doi: 10.1002/wdev.147
Grabb, W. C. (1965). The first and second branchial arch syndrome. Plast. Reconstruct. Surg. 36, 485–508. doi: 10.1097/00006534-196511000-00001
Graf, D., Malik, Z., Hayano, S., and Mishina, Y. (2016). Common mechanisms in development and disease: BMP signaling in craniofacial development. Cytokine Growth Fact. Rev. 27, 129–139. doi: 10.1016/j.cytogfr.2015.11.004
Hans, S., and Westerfield, M. (2007). Changes in retinoic acid signaling alter otic patterning. Development 134, 2449–2458. doi: 10.1242/dev.000448
Hofmeister, W., Devine, C. A., and Key, B. (2013). Distinct expression patterns of syndecans in the embryonic zebrafish brain. Gene Expr. Patterns 13, 126–132. doi: 10.1016/j.gep.2013.02.002
Huang, Y. J., Mao, B., Aramini, J. M., and Montelione, G. T. (2014). Assessment of template-based protein structure predictions in CASP10. Proteins 82(Suppl. 2), 43–56. doi: 10.1002/prot.24488
Johnson, C. W., Hernandez-Lagunas, L., Feng, W., Melvin, V. S., Williams, T., and Artinger, K. B. (2011). Vgll2a is required for neural crest cell survival during zebrafish craniofacial development. Dev. Biol. 357, 269–281. doi: 10.1016/j.ydbio.2011.06.034
Kaban, L. B., Moses, M. H., and Mulliken, J. B. (1988). Surgical correction of hemifacial microsomia in the growing child. Plast. Reconstr. Surg. 82, 9–19. doi: 10.1097/00006534-198807000-00003
Kawai, M., Kinoshita, S., Kimoto, A., Hasegawa, Y., Miyagawa, K., Yamazaki, M., et al. (2013). FGF23 suppresses chondrocyte proliferation in the presence of soluble alpha-Klotho both in vitro and in vivo. J. Biol. Chem. 288, 2414–2427. doi: 10.1074/jbc.M112.410043
Kelley, L. A., Mezulis, S., Yates, C. M., Wass, M. N., and Sternberg, M. J. (2015). The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 10, 845–858. doi: 10.1038/nprot.2015.053
Kim, Y.-I., No Lee, J., Bhandari, S., Nam, I.-K., Yoo, K.-W., Kim, S.-J., et al. (2015). Cartilage development requires the function of Estrogen-related receptor alpha that directly regulates sox9 expression in zebrafish. Sci. Rep. 5:18011. doi: 10.1038/srep18011
Kimmel, C. B., Ballard, W. W., Kimmel, S. R., Ullmann, B., and Schilling, T. F. (1995). Stages of embryonic development of the zebrafish. Dev. Dyn. 203, 253–310. doi: 10.1002/aja.1002030302
Kimmel, C. B., Miller, C. T., Kruze, G., Ullmann, B., BreMiller, R. A., Larison, K. D., et al. (1998). The shaping of pharyngeal cartilages during early development of the zebrafish. Dev. Biol. 203, 245–263. doi: 10.1006/dbio.1998.9016
Kitami, K., Kitami, M., Kaku, M., Wang, B., and Komatsu, Y. (2018). BRCA1 and BRCA2 tumor suppressors in neural crest cells are essential for craniofacial bone development. PLoS Genet. 14:e1007340. doi: 10.1371/journal.pgen.1007340
Li, L., Mao, A., Wang, P., Ning, G., Cao, Y., and Wang, Q. (2018). Endodermal pouch-expressed dmrt2b is important for pharyngeal cartilage formation. Biol. Open 7:bio.035444. doi: 10.1242/bio.035444
Ma, Z., Zhu, P., Shi, H., Guo, L., Zhang, Q., Chen, Y., et al. (2019). PTC-bearing mRNA elicits a genetic compensation response via Upf3a and COMPASS components. Nature 568, 259–263. doi: 10.1038/s41586-019-1057-y
Marie, P. J., Hay, E., Modrowski, D., Revollo, L., Mbalaviele, G., and Civitelli, R. (2014). Cadherin-mediated cell-cell adhesion and signaling in the skeleton. Calcif. Tissue Intern. 94, 46–54. doi: 10.1007/s00223-013-9733-7
Messina, G., Atterrato, M. T., and Dimitri, P. (2016a). When chromatin organisation floats astray: the srcap gene and floating-harbor syndrome. J. Med. Genet. 53, 793–797. doi: 10.1136/jmedgenet-2016-103842
Messina, G., Atterrato, M. T., Fanti, L., Giordano, E., and Dimitri, P. (2016b). Expression of human Cfdp1 gene in Drosophila reveals new insights into the function of the evolutionarily conserved BCNT protein family. Sci. Rep. 6:25511. doi: 10.1038/srep25511
Northup, J. K., Matalon, D., Hawkins, J. C., Matalon, R., and Velagaleti, G. V. (2010). Pericentric inversion, inv(14)(p11.2q22.3), in a 9-month old with features of Goldenhar syndrome. Clin. Dysmorphol. 19, 185–189. doi: 10.1097/MCD.0b013e3283359386
Ou, Z., Martin, D. M., Bedoyan, J. K., Cooper, M. L., Chinault, A. C., Stankiewicz, P., et al. (2008). Branchiootorenal syndrome and oculoauriculovertebral spectrum features associated with duplication of SIX1, SIX6, and OTX2 resulting from a complex chromosomal rearrangement. Am. J. Med. Genet. Part A 146a, 2480–2489. doi: 10.1002/ajmg.a.32398
Robu, M. E., Larson, J. D., Nasevicius, A., Beiraghi, S., Brenner, C., Farber, S. A., et al. (2007). p53 activation by knockdown technologies. PLoS Genet. 3:e78. doi: 10.1371/journal.pgen.0030078
Roughley, P. J. (2006). The structure and function of cartilage proteoglycans. Eur. Cells Mater. 12, 92–101. doi: 10.22203/ecm.v012a11
Saleh, S., Thompson, D. E., McConkey, J., Murray, P., and Moorehead, R. A. (2016). Osteopontin regulates proliferation, apoptosis, and migration of murine claudin-low mammary tumor cells. BMC Cancer 16:359. doi: 10.1186/s12885-016-2396-9
Sasaki, T., Ito, Y., Bringas, P. Jr., Chou, S., Urata, M. M., Slavkin, H., et al. (2006). TGFbeta-mediated FGF signaling is crucial for regulating cranial neural crest cell proliferation during frontal bone development. Development 133, 371–381. doi: 10.1242/dev.02200
Schilling, T. F., and Kimmel, C. B. (1994). Segment and cell type lineage restrictions during pharyngeal arch development in the zebrafish embryo. Development 120, 483–494.
Schönberger, J., Wang, L., Shin, J. T., Kim, S. D., Depreux, F. F., Zhu, H., et al. (2005). Mutation in the transcriptional coactivator EYA4 causes dilated cardiomyopathy and sensorineural hearing loss. Nat. Genet. 37, 418–422. doi: 10.1038/ng1527
Shum, E. Y., Jones, S. H., Shao, A., Dumdie, J., Krause, M. D., Chan, W. K., et al. (2016). The Antagonistic gene paralogs Upf3a and Upf3b govern nonsense-mediated RNA decay. Cell 165, 382–395. doi: 10.1016/j.cell.2016.02.046
Sisson, B. E., Dale, R. M., Mui, S. R., Topczewska, J. M., and Topczewski, J. (2015). A role of glypican4 and wnt5b in chondrocyte stacking underlying craniofacial cartilage morphogenesis. Mech. Dev. 138(Pt 3), 279–290. doi: 10.1016/j.mod.2015.10.001
Sousa, S., Valerio, F., and Jacinto, A. (2012). A new zebrafish bone crush injury model. Biol. Open 1, 915–921. doi: 10.1242/bio.2012877
Streuli, C. (1999). Extracellular matrix remodelling and cellular differentiation. Curr. Opin. Cell Biol. 11, 634–640. doi: 10.1016/s0955-0674(99)00026-5
Tasse, C., Bohringer, S., Fischer, S., Ludecke, H. J., Albrecht, B., and Horn, D. (2005). Oculo-auriculo-vertebral spectrum (OAVS): clinical evaluation and severity scoring of 53 patients and proposal for a new classification. Eur. J. Med. Genet. 48, 397–411. doi: 10.1016/j.ejmg.2005.04.015
Thisse, C., and Thisse, B. (2008). High-resolution in situ hybridization to whole-mount zebrafish embryos. Nat. Protoc. 3, 59–69. doi: 10.1038/nprot.2007.514
Topczewska, J. M., Shoela, R. A., Tomaszewski, J. P., Mirmira, R. B., and Gosain, A. K. (2016). The morphogenesis of cranial sutures in Zebrafish. PLoS One 11:e0165775. doi: 10.1371/journal.pone.0165775
Tuin, A. J., Tahiri, Y., Paine, K. M., Paliga, J. T., Taylor, J. A., and Bartlett, S. P. (2015). Clarifying the relationships among the different features of the OMENS+ classification in craniofacial microsomia. Plast. Reconstr. Surg. 135, 149e–156e. doi: 10.1097/prs.0000000000000843
Vendramini-Pittoli, S., and Kokitsu-Nakata, N. M. (2009). Oculoauriculovertebral spectrum: report of nine familial cases with evidence of autosomal dominant inheritance and review of the literature. Clin. Dysmorphol. 18, 67–77. doi: 10.1097/MCD.0b013e328323a7dd
Wang, Q., Kurosaka, H., Kikuchi, M., Nakaya, A., Trainor, P. A., and Yamashiro, T. (2019). Perturbed development of cranial neural crest cells in association with reduced sonic hedgehog signaling underlies the pathogenesis of retinoic-acid-induced cleft palate. Dis. Model. Mech. 12:dmm.040279. doi: 10.1242/dmm.040279
Welsh, I. C., Hart, J., Brown, J. M., Hansen, K., Rocha Marques, M., Aho, R. J., et al. (2018). Pbx loss in cranial neural crest, unlike in epithelium, results in cleft palate only and a broader midface. J. Anat. 233, 222–242. doi: 10.1111/joa.12821
Wentzel, P., and Eriksson, U. J. (2011). Altered gene expression in rat cranial neural crest cells exposed to a teratogenic glucose concentration in vitro: paradoxical downregulation of antioxidative defense genes. Birth Defects Res. B Dev. Reprod. Toxicol. 92, 487–497. doi: 10.1002/bdrb.20321
Wilson, N. R., Olm-Shipman, A. J., Acevedo, D. S., Palaniyandi, K., Hall, E. G., Kosa, E., et al. (2016). SPECC1L deficiency results in increased adherens junction stability and reduced cranial neural crest cell delamination. Sci. Rep. 6:17735. doi: 10.1038/srep17735
Wu, R. S., Lam, I. I., Clay, H., Duong, D. N., Deo, R. C., and Coughlin, S. R. (2018). A rapid method for directed gene knockout for screening in G0 zebrafish. Dev. Cell 46, 112–125.e4. doi: 10.1016/j.devcel.2018.06.003
Xia, Z., Tong, X., Liang, F., Zhang, Y., Kuok, C., Zhang, Y., et al. (2013). Eif3ba regulates cranial neural crest development by modulating p53 in zebrafish. Dev. Biol. 381, 83–96. doi: 10.1016/j.ydbio.2013.06.009
Xu, S., Zhang, Z., Tang, X., Yin, L., Liu, W., and Shi, L. (2015). The influence of gender and laterality on the incidence of hemifacial microsomia. J. Craniof. Surg. 26, 384–387. doi: 10.1097/scs.0000000000001336
Yan, Y.-L., Miller, C. T., Nissen, R. M., Singer, A., Liu, D., Kirn, A., et al. (2002). A zebrafish sox9 gene required for cartilage morphogenesis. Development 129, 5065–5079.
Keywords: hemifacial microsomia, VWA1, zebrafish, cranial neural crest cell, whole exome sequencing
Citation: Wang Y, Ping L, Luan X, Chen Y, Fan X, Li L, Liu Y, Wang P, Zhang S, Zhang B and Chen X (2020) A Mutation in VWA1, Encoding von Willebrand Factor A Domain-Containing Protein 1, Is Associated With Hemifacial Microsomia. Front. Cell Dev. Biol. 8:571004. doi: 10.3389/fcell.2020.571004
Received: 09 June 2020; Accepted: 19 August 2020;
Published: 09 September 2020.
Edited by:
Duan Ma, Fudan University, ChinaReviewed by:
Giovanni Messina, Sapienza University of Rome, ItalyGuang Wang, Jinan University, China
Xuesong Yang, Jinan University, China
Daniela Anne Braun, University Hospital Münster, Germany
Copyright © 2020 Wang, Ping, Luan, Chen, Fan, Li, Liu, Wang, Zhang, Zhang and Chen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Bo Zhang, YnpoYW5nQHBrdS5lZHUuY24=; Xiaowei Chen, Y2hlbnh3X3B1bWNoQDE2My5jb20=
†These authors have contributed equally to this work
 Yibei Wang1,2†
Yibei Wang1,2† Xiaodong Luan
Xiaodong Luan Shuyang Zhang
Shuyang Zhang Xiaowei Chen
Xiaowei Chen