- 1Biology and Evolution of Marine Organisms, Stazione Zoologica Anton Dohrn Napoli, Naples, Italy
- 2School of Biological Sciences, Georgia Institute of Technology, Atlanta, GA, United States
The tunicate Ciona robusta is an emerging model system to study the evolution of the nervous system. Due to their small embryos and compact genomes, tunicates, like Ciona robusta, have great potential to comprehend genetic circuitry underlying cell specific gene repertoire, among different neuronal cells. Their simple larvae possess a sensory vesicle comprising two pigmented sensory organs, the ocellus and the otolith. We focused here on Klhl21/30, a gene belonging to Kelch family, that, in Ciona robusta, starts to be expressed in pigmented cell precursors, becoming specifically maintained in the otolith precursor during embryogenesis. Evolutionary analyses demonstrated the conservation of Klhl21/30 in all the chordates. Cis-regulatory analyses and CRISPR/Cas9 mutagenesis of potential upstream factors, revealed that Klhl21/30 expression is controlled by the combined action of three transcription factors, Mitf, Dmrt, and Msx, which are downstream of FGF signaling. The central role of Mitf is consistent with its function as a fundamental regulator of vertebrate pigment cell development. Moreover, our results unraveled a new function for Dmrt and Msx as transcriptional co-activators in the context of the Ciona otolith.
Introduction
Understanding the developmental logics that orchestrate specific gene expression inside the nervous system represents a fascinating challenge in cell and developmental biology. However, the identification of the molecular processes underlying cell specific expression among different neuronal cells is very difficult using vertebrate models, due to the relative complexity of vertebrate embryos and genomes, as well as the numbers of genes involved.
Within the chordates, the tunicate subphylum is the sister group of vertebrates, forming with them the clade Olfactores (Delsuc et al., 2006). Due to their small, invariant embryos and compact genomes (Berná and Alvarez-Valin, 2014), tunicates have great potential to help to uncover the genetic circuitry regulating chordate-specific mechanisms of neural development. The larva of the tunicate Ciona robusta possesses two distinct pigmented sensory organs, the otolith and the ocellus, contained in the anterior sensory vesicle, a structure evolutionarily related to the forebrain of vertebrates (Moret et al., 2005; Dufour et al., 2006). The ocellus is formed by 30 photoreceptors, three lens cells and one melanized cup-shaped cell (Horie et al., 2005). Due to its association with photoreceptors, the ocellus has been compared to the vertebrate eye (Kusakabe et al., 2001; Nakagawa et al., 2002; Nakashima et al., 2003; D’Aniello et al., 2006). In contrast, the otolith is a single spherical cell containing melanin granules and attached to the sensory vesicle floor through a tight stalk (Dilly, 1962, 1969). It has been proposed that the displacement of the otolith within the fluid-filled sensory vesicle lumen stimulates a putative mechanosensory antenna cell in the adjacent brain, thus playing a key role in negative geotropism and gravitational orientation (Tsuda et al., 2003; Sakurai et al., 2004). Supporting this model, an ammonium channel regulates the sensory vesicle fluid composition and otolith functioning (Marino et al., 2007). In addition, the pigmentation of these sensory organs is fundamental for normal photo- and geotactic behaviors in the close sibling species C. savignyi (Jiang, 2005).
During embryonic development, ocellus and otolith pigment cells derive from a pair of left/right pigment cell precursors (PCP) in the neural plate that intercalate at the dorsal midline of the neural tube during neurulation (Cole and Meinertzhagen, 2004). Later, during neural tube closure, PCPs divide twice and give rise to a total of 8 cells at early tailbud stage, which express melanogenic genes belonging to the Tyrosinase (Tyr) family (Tief et al., 1996; Caracciolo et al., 1997; Esposito et al., 2012; Haupaix et al., 2014; Racioppi et al., 2014, 2017). The loss of Tyr function in Ciona results in pigment-free larvae (Sordino et al., 2008; Crocetta et al., 2015), whereas some tunicate species (e.g., Molgula occulta) have lost Tyr family genes and lack melanin pigmentation altogether (Racioppi et al., 2017).
The majority of the transcription factors and cell signaling molecules implicated in otolith and ocellus differentiation are expressed in the whole PCP lineage, even though only two cells will become pigmented. A FGF-dependent transcriptional code for the formation of Ciona PCPs was recently partially elucidated (Racioppi et al., 2014). Among the two pigmented cells the anterior-posterior order of intercalation orchestrates ocellus versus otolith pigment cell determination: the anterior cell always becomes the otolith while the posterior cell always gives rise to the ocellus pigment cell. These mechanisms, which are still not well studied, involve a Wnt and FoxD-mediated suppression of Pax3/7-dependent activation of the Microphthalmia-associated transcription factor gene (Mitf) (Abitua et al., 2012).
Here, we focused on the transcriptional control of Klhl21/30, a gene that we found to be specifically expressed in the ocellus and otolith precursors, during embryogenesis. Klhl21/30 belongs to the Kelch-like (Klhl) family of genes, encoding for proteins characterized by the presence of multiple Kelch motifs, which are evolutionarily conserved, but poorly characterized, short domains implicated in protein-protein interactions (Adams et al., 2000). In mammalian cells, Klhl21 is thought to be involved in E3-ubiquitination during cytokinesis and regulation of cortical dynamics (Maerki et al., 2009; Courtheoux et al., 2016) and is implicated in diverse types of carcinoma (Shi et al., 2016; Chen et al., 2018). We provide the most updated evolutionary reconstruction of Klhl family, demonstrating that Klhl21/30 is ultra-conserved in chordates. This gene has dynamic expression pattern in Ciona PCPs, becoming restricted to the otolith during embryogenesis. Klhl21/30 transcription is governed by a complex regulatory code, in fact, we have identified the minimal key cis-regulatory element able to drive Klhl21/30 expression in the C. robusta otolith, containing functional binding sites for the transcription factors Mitf, Msx, and Dmrt. Using tissue-specific CRISPR/Cas9-mediated gene knockouts, we found that Mitf is central to Klhl21/30 expression in the otolith, as it is for many pigmentation markers in the pigment cells of vertebrates (Levy et al., 2006), with Msx and Dmrt acting as co-activators.
Results
Kelch-like Gene Family Evolution in Chordates
A lineage-specific transcription profiling of genes downstream of fibroblast growth factor signaling (FGF), was used to find novel players involved in pigment cell formation (Racioppi et al., 2014). Among these, we found a Kelch-like (Klhl) gene family member (Kyoto Hoya gene model KH.L84.23), which exhibited similar expression values of known PCP markers, including Tyr, Tyrp.a and Rab32/38 and that, by reciprocal BLASTs, is similar to vertebrate Kelch-like 21 (Klhl21) and Kelch-like 30 (Klhl30).
To shed light on the evolutionary origins of the Klhl21/30 gene and to gain insights into the poorly studied Klhl family in metazoans, we performed an evolutionary analysis of these proteins in chordates. First, we analyzed their domain organization in PROSITE (de Castro et al., 2006) and Ensembl databases. This revealed the presence of one BTB/POZ domain, one BACK domain and five Kelch repeats (Supplementary Figure S1) in both C. robusta Klhl21/30 and Homo sapiens KLHL21 (Dhanoa et al., 2013). To confirm the orthology of C. robusta Klhl21/30 (KH.L84.23) and study the evolution of Kelch-like genes in chordates, we performed a phylogenetic survey employing a manually curated database (Supplementary Table S1). Our phylogenetic reconstruction included 118 Kelch-like protein sequences from the cephalochordate amphioxus Branchiostoma belcheri, tunicates Oikopleura dioica and Ciona robusta, and human Homo sapiens as representative of vertebrates (Figure 1). Sequences with high degree of molecular divergence have been excluded from phylogeny and listed in the Supplementary Table S2. In total, we identified 25 Klhl subfamilies, conserved throughout chordate evolution from cephalochordates to vertebrates, with a unique exception (Klhl5L) that is tunicate-specific (Figure 1). Genome search and phylogenetic tree defined the complete Kelch-like toolkit of B. belcheri (37), C. robusta (32), O. dioica (14), H. sapiens (42): this confirms the previous survey performed in human (Dhanoa et al., 2013) and represents the first step into understanding Kelch-like evolution in all three chordate subphyla. Though the number of Kelch-like genes in the amphioxus genome has been shaped by various amphioxus-specific duplications (as in the case of Klhl42 duplications), our survey is coherent with relative genomic stasis of this slow-evolving branch (Putnam et al., 2008). In contrast, C. robusta retained 32 Klhl genes and 16 subfamilies, whilst O. dioica maintained 14 genes and 10 subfamilies. This is coherent to the tendency of the fast-evolving Oikopleura dioica to lose large portions of gene families (Berná and Alvarez-Valin, 2014; Albalat and Cañestro, 2016; Martí-Solans et al., 2016; Coppola et al., 2019). Otherwise, we also detected different tunicate-specific duplications of certain Kelch-like genes in either species (Figure 1).
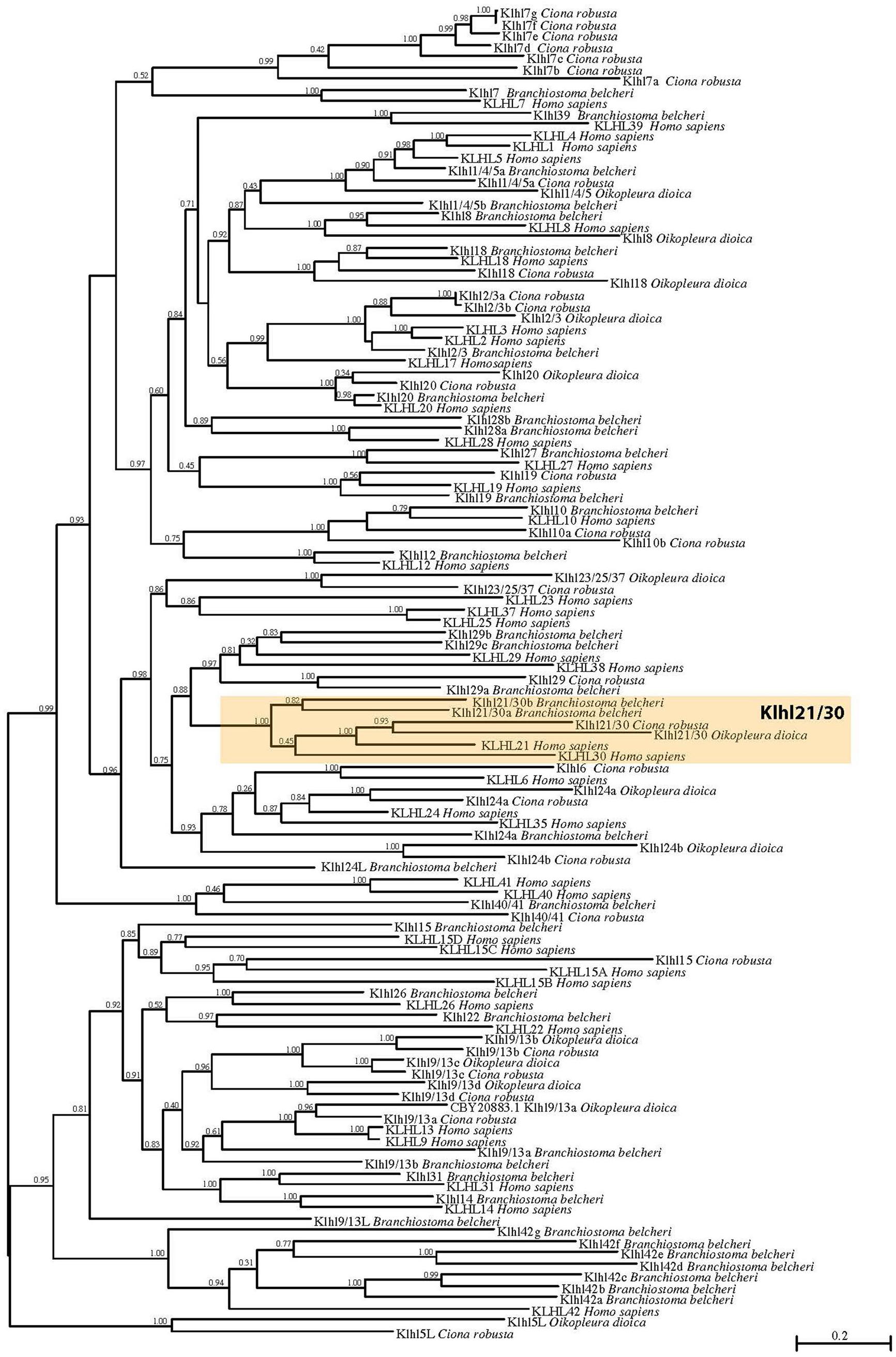
Figure 1. Phylogenetic tree of Kelch-like proteins in chordates. Maximum likelihood (ML) reconstruction of 118 Kelch-like members from four key chordate species: amphioxus Branchiostoma belcheri, larvacean Oikopleura dioica, ascidian Ciona robusta, human Homo sapiens. The phylogeny demonstrates the existence of a robust Klhl21/30 subfamily (orange box). Numbers at the branches indicate replicates obtained using the ML estimation method.
In particular, our phylogenetic tree clustered Ciona KH.L84.23 with one O. dioica protein, two B. belcheri proteins (deriving from a local duplication) and H. sapiens KLHL21 and KLHL30, forming a protein class with robust support that we named Klhl21/30 (orange box; Figure 1). The presence of two related genes in human speaks in favor of a common origin for vertebrate Klhl21 and Klhl30, which could have derived from a local duplication or a whole-genome duplication event in the vertebrate ancestor (Abi-Rached et al., 2002; Dehal and Boore, 2005). In contrast, invertebrate chordates possess a single Klhl21/30 gene, similar to other invertebrates such as the nematode Caenorhabditis elegans and the sea urchin Strongylocentrotus purpuratus (data not shown). Remarkably, Klhl21/30 is one of the few Kelch-like genes not lost by O. dioica and therefore could be functionally relevant for conserved developmental processes. Moreover, we analyzed the Klhl21/30 locus in several available tunicate genomes, finding high degree of local synteny surrounding Klhl21/30 in C. robusta, C. savignyi, Phallusia mammillata, and Halocynthia roretzi (Supplementary Figure S2). Our survey prompted us to describe an ancestral cluster of 11 genes in tunicates, nearly all conserved between C. robusta and P. mammillata. Interestingly, all surveyed species exhibited linkage between Klhl21/30 and Nph4 (nephrocystin-4) genes, which is also observed in human between KLHL21 and NPH4. Conversely, this linkage has been lost for KLHL30 (Supplementary Figure S3). The comparison of Klhl21 and Klhl30 genome environment of species as coelacanth Latimeria chalumnae, spotted gar Lepisosteus oculatus, frog Xenopus tropicalis and human, demonstrated the conservation of flanking genes during gnathostome evolution (Supplementary Figure S3). Moreover, the presence of orthologous genes on both the surveyed chromosomal regions, strongly indicated Klhl21 and Klhl30 as paralogs deriving from one of the events of whole-genome duplication (WGDs) occurred at the root of vertebrate radiation (Dehal and Boore, 2005).
In sum, we have reported here the first evolutionary study dedicated to Kelch-like family in chordates, focused on the seemingly indispensable Klhl21/30 subfamily, which was present in all chordate species examined.
Klhl21/30 Dynamic Expression in Otolith
The analysis of the spatio-temporal expression pattern of Klhl21/30, by whole mount in situ hybridization (WISH) in embryos at different developmental stages (Figures 2A–D), showed that during C. robusta embryogenesis, Klhl21/30 is expressed from initial tailbud stage onward in the pigment cell lineage (Figures 2A–D). In particular, at initial tailbud stage it is expressed in two cells (Figures 2A,B), possibly the ones originating the otolith and ocellus pigmented cells of the larva. Later in development, from the middle tailbud stage onward, Klhl21/30 transcript became restricted specifically to one cell (Figures 2C,D). Double WISH using the pigment cell marker, Tyrp1/2a, was performed to better define PCP-specific expression. Detailed and thorough analysis of the expression domain proved that Klhl21/30 is expressed specifically in the a11.193 pair, i.e. the most posterior cells marked by Tyrp1/2a (Haupaix et al., 2014; Racioppi et al., 2014) (white arrow in Figures 2E–F”’) later, starting from middle tailbud stage, the Klhl21/30 expression became restricted to the otolith precursor (white arrow in Figures 2G–G”’), while it disappears from the ocellus pigment cell precursor. Thus, Klhl21/30 represents the earliest-expressed otolith specific gene described so far. Its expression is maintained in the otolith pigment cell precursor while it is downregulated in the ocellus pigment cell precursor, even before expression of the βγ-crystallin, which is expressed specifically in the otolith starting from the larval stage (Shimeld et al., 2005). Klhl21/30 expression in PCPs is consistent with microarray data (Racioppi et al., 2014), in which Klhl21/30 expression was observed to decrease between 8 and 12 hpf. Furthermore, its expression was shown to be dependent on the FGF signaling, which is required for PCPs specification. Coherently, in situ hybridizations performed using Klhl21/30 probe on tailbud embryos electroporated with a construct, Tyrp > dnFGFr, able to drive a dominant-negative form of the FGF receptor in the PCPs (Squarzoni et al., 2011), showed a sharp decrease in the number of tailbud embryos expressing Klhl21/30 in PCPs compared to control (Supplementary Figure S4). Altogether, these data identified Klhl21/30 as the earliest marker of the Ciona otolith and the first Klhl member described in tunicates.
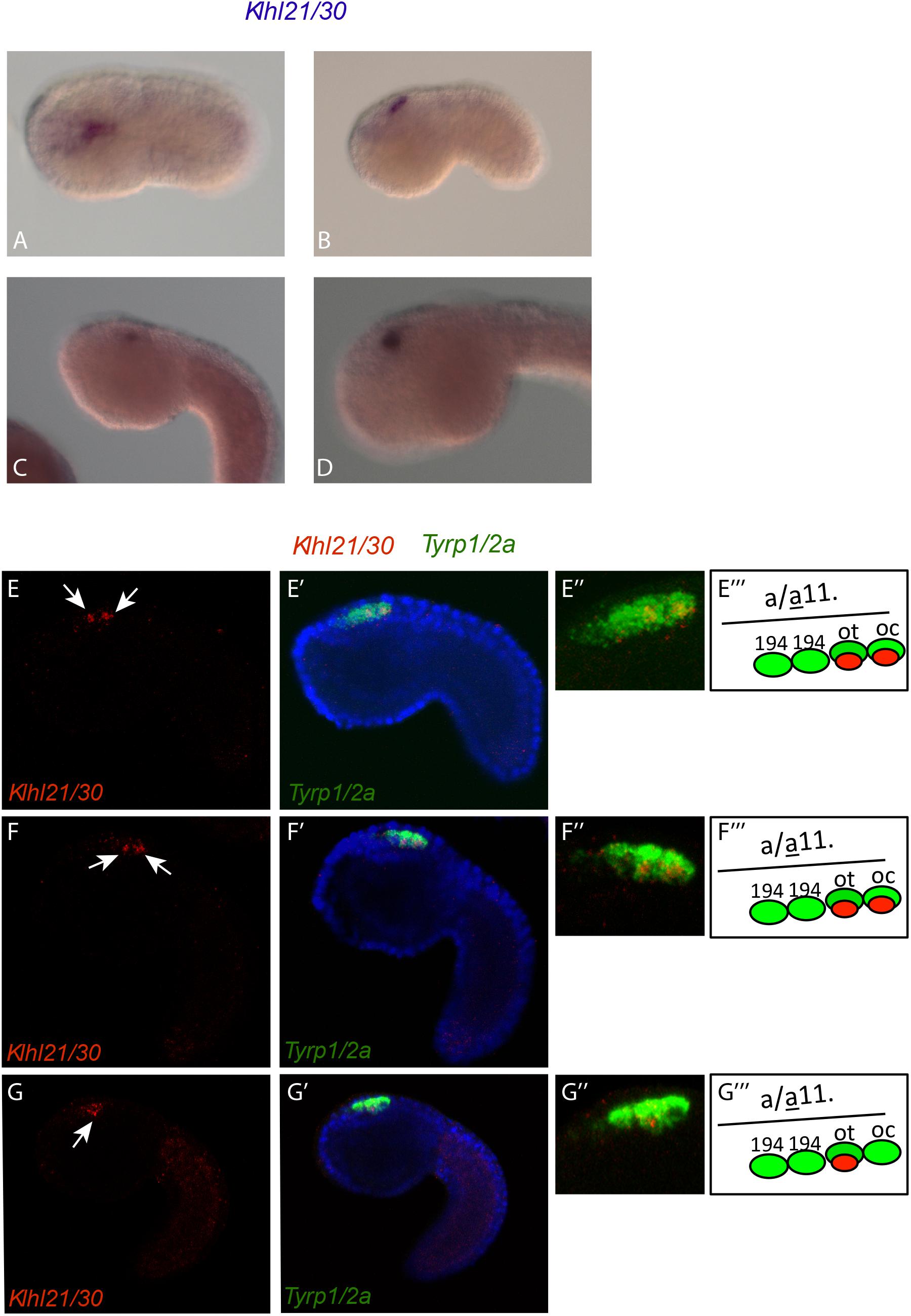
Figure 2. Expression of Klhl21/30 during Ciona robusta embryogenesis. Klhl21/30 expression was detected by in situ hybridization in the C. robusta pigment cell lineage at initial tailbud (A), early tailbud (B), middle tailbud (C), late tailbud (D). Double WISH with Tyrp1/2a that colocalizes precisely Klhl21/30 among PCPs. (E–F”’) Early tailbud I (E–E”) and early tailbud II (F–F”) stages, white arrow indicates the two cells expressing Klhl21/30 that colocalizes with the posterior two Tyrp1/2a positive cells. (G–G”’) Middle tailbud stage. One white arrow indicates the sole cell expressing Klhl21/30 at this stage that colocalizes with otolith precursor as shown in (G–G”’).
Cis-Regulatory Analysis of Klhl21/30 Expression
Since Klhl21/30 was identified as a specific marker for PCPs as well as the earliest otolith marker in Ciona robusta, we sought to study its transcriptional regulation to understand the regulatory logics underlying differentiation of these structures (Figure 3). In order to discover the cis-regulatory element involved in the control of Klhl21/30 expression, we took advantage of the genome conservation tracks in the ANISEED genome browser (Brozovic et al., 2018) to select a 949-bp non-coding region upstream Klhl21/30 (“KlA”, −1044 to −95 from the start codon), a region presenting conserved peaks with the sibling species C. savignyi (Figure 3A). This KlA fragment was cloned in a vector containing a GFP reporter gene downstream of a human β-globin minimal promoter (Zeller et al., 2006). Once electroporated the KlA > GFP reporter plasmid into Ciona embryos (Corbo et al., 1997), we detected otolith-specific GFP fluorescence in 70% of larvae at stage 26 (Hotta et al., 2007; Figures 3B,C). By WISH using GFP probe, we registered transcription of KlA > GFP at the middle tailbud stage in one cell of the sensory vesicle, possibly corresponding otolith precursor (Supplementary Figure S5). This indicates that the absence of GFP fluorescence until the larval stage is due to a delay in GFP maturation and accumulation. Our reporter plasmid thus recapitulates the endogenous expression of Klhl21/30 at least in the otolith, confirming the restriction of its expression to the otolith precursor in the late embryogenesis. To dissect the regulatory logics underlying the expression of Klhl21/30, we focused on two smaller, highly conserved regions within the KlA sequence: KlB (385 bp long, −918 to −553) and KlC (441 bp long, −536 to −95). While KlC did not drive GFP expression, 43% of larvae electroporated with KlB > GFP recapitulated the strong GFP signal in otolith, suggesting that KlB fragment retains the minimal regulatory information necessary to drive Klhl21/30 expression (Figure 3C). When we divided KlB roughly into two halves, KlD (199 bp long, −918 to −719) and KlE (166 bp long, −719 to −553), neither was sufficient to drive GFP expression (Figure 3C). However, we found that two smaller fragments centered around KlB, which are KlF (257 bp long, −810 to −553) and KlG (321 bp long, −874 to −553), drove GFP expression in 10 and 22% of larvae, respectively (Figure 3C). These fragments therefore represent the minimal cis-regulatory elements sufficient to recapitulate Klhl21/30 expression. We hypothesized that the KlB region contains transcription factors binding sites (TFBS) crucial for the sustained activation of Klhl21/30 in pigment cell precursors. Therefore, we searched in this fragment putative TFBS using Genomatix software and CIS-BP database with C. intestinalis motifs (Weirauch et al., 2014) and these results have been compared to JASPAR database (Khan et al., 2018; Figure 4). Among several predicted binding sites, we focused our attention on two bHLH-binding motifs. In Ciona 44 bHLH genes have been found (Satou et al., 2003) and, for most of them, expression pattern during development has been described (ANISEED database). We selected Mitf as possible factor binding the bHLH sites in the Klhl21/30 regulatory region (yellow in Figure 4A) because of involvement of Mitf in eye development and in the activation of melanogenic markers Tyr and Tyrp/DCT in vertebrates (Goding, 2000; Curran et al., 2010) and its specific expression in the pigment cell lineage of Ciona (Curran et al., 2010; Abitua et al., 2012) and Halocynthia roretzi (Yajima et al., 2003). We demonstrated that Mitf is co-expressed with Klhl21/30 in both otolith and ocellus pigmented cells precursors at early tailbud stage (Figures 4B–B”’) and that both became restricted to otolith precursor starting from middle tailbud stage (Figures 4C–C”’). Previous experiments showed also that Mitf is downregulated upon perturbation of FGF signaling (Racioppi et al., 2014). To test the relevance of these two putative sites for Mitf (CACGTG), we mutated them individually in the KlB reporter, naming these mutant constructs K_Mitf_mut1 and K_Mitf_mut2 (Figure 4E). When each of these constructs was electroporated, the percentage of larvae showing GFP signal in the otolith was close to zero (2.5 and 3%, respectively, Figure 4E). These data strongly support these binding sites for Mitf as involved in KlB activity, leading to hypothesize that Mitf activates Klhl21/30 in Ciona robusta.
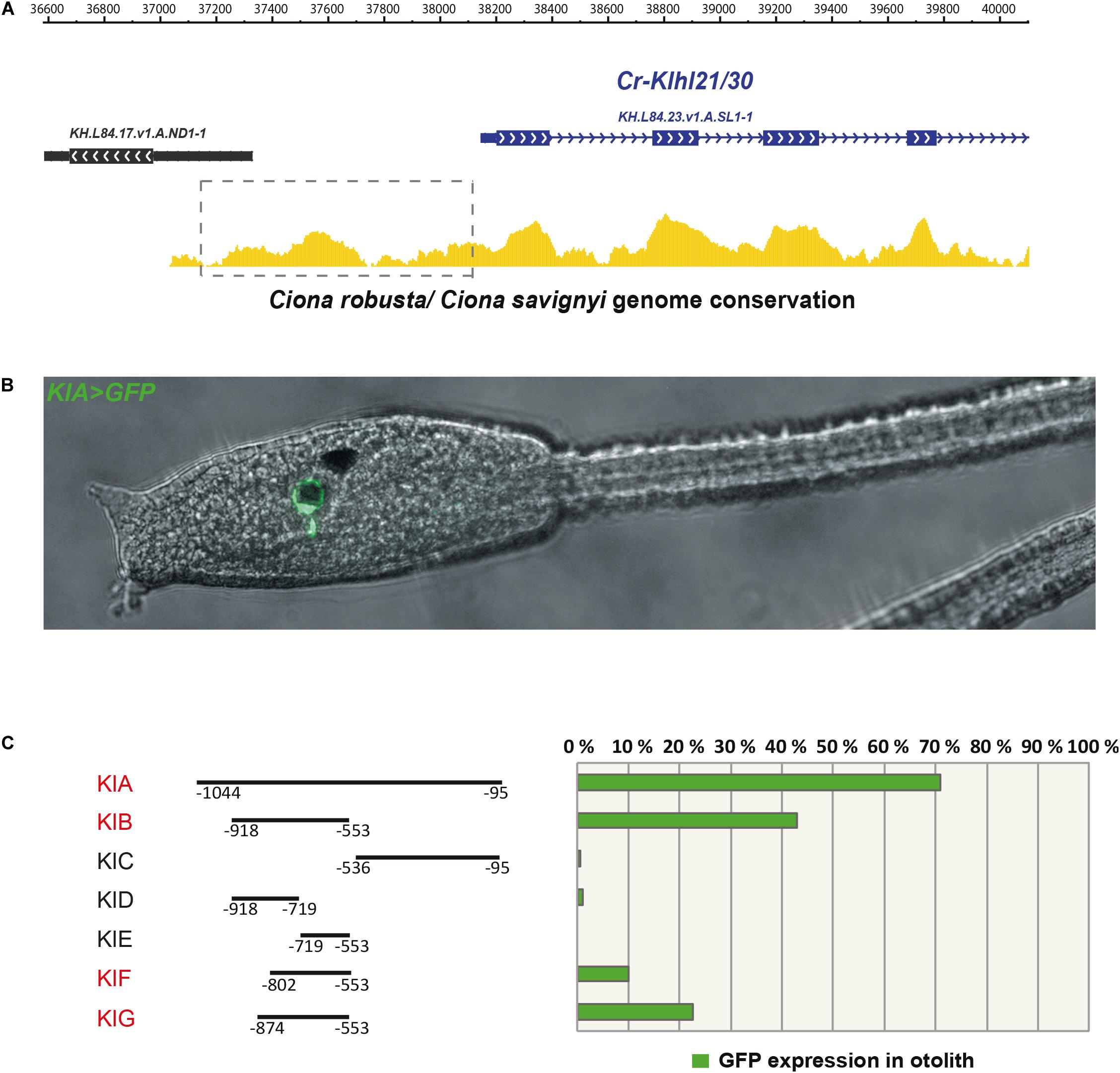
Figure 3. The Klhl21/30 cis-regulatory region. (A) Alignment of Klhl21/30 locus between Ciona robusta and Ciona savignyi employing WASHU browser, with a dashed rectangle indicating the surveyed intergenic region. (B) Side view of larva embryo electroporated with KlA > GFP construct. (C) Percentage of larvae expressing GFP reporter in otolith with schematic representations of the dissected fragments and analysis by gene reporter assay (the fragments driving expression in otolith are highlighted in red); each bar represents the combined number of larvae counted during at least three trials; n > 150 embryos scored for transgene expression.
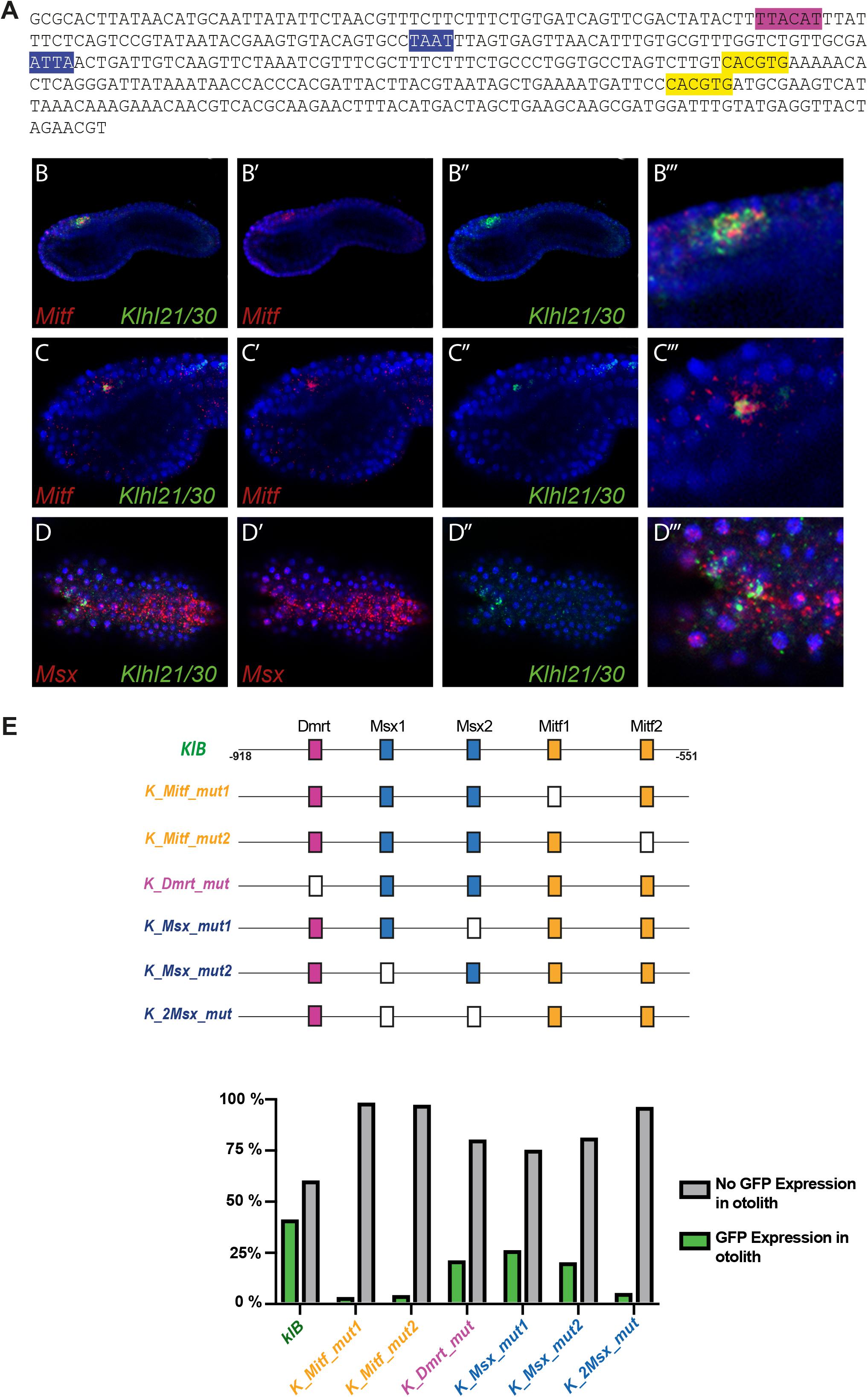
Figure 4. Regulatory logic underlying Klhl21/30 expression. (A) KlB region with the transcription factor binding sites (TFBS) analyzed: Mitf (yellow-orange), Dmrt (violet), Msx (blue). (B) Co-expression in pigment cell precursors of Klhl21/30 with Mitf at early tailbud (B–B”’) and middle tailbud (C–C”’), and Msx (D–D”’), respectively. (E) Mutational analysis with percentages of larvae expressing GFP, employing KlB > GFP as control; the TFBS are shown using rectangles with the same color code reported for the sequence, while the mutated sites are represented with white rectangles. Experiments were repeated at least three times and 200 embryos were counted each time; all the mutated constructs were significant versus the control KlB (Fisher exact test, p < 0.00001).
We also identified a well-supported binding site in KlB for Dmrt (TTACAT, violet in Figure 4A), a transcription factor expressed in the early a-line neural plate (Wagner and Levine, 2012). C. savignyi Dmrt mutants present abnormalities in the development of the larval sensory vesicle (Tresser et al., 2010) and the expression of the C. robusta ortholog is under early FGF control (Imai et al., 2006). The KlB fragment harboring a mutated Dmrt site (K_Dmrt_mut) drives GFP expression only in 20% of electroporated larvae, roughly half of the activity of wild-type KlB (Figure 4E). Additionally, two binding sites attributed to the homeodomain transcription factors caught our attention (ATTA, blue in Figure 4A). Among TFs able to bind these sites we focused on Msx because of its early expression in C. robusta pigment cell precursors (Aniello et al., 1999; Russo et al., 2004), and given that it is sharply down-regulated when FGF is blocked (Racioppi et al., 2014). We detected co-expression of Msx and Klhl21/30 in the PCPs at early tailbud stage (Figures 4D–D”’) As development proceeds, the expression of Msx in PCP decrease becoming excluded from most of the a9.49 derivatives in 12 hpf tailbud embryos as already described (Racioppi et al., 2014). This early expression suggests that Msx might be activating early Klhl21/30 transcription, even though Msx factors normally have been described to act as repressors in Ciona (Roure and Darras, 2016). We individually mutated each putative Msx binding site in KlB, resulting in two mutant constructs, which we termed K_Msx_mut1 and K_Msx_mut2. The mutated reporters caused a decrease in the percentage of GFP-expressing larvae of 24 and 19%, respectively, with respect to the wild-type (Figure 4E). Mutating both putative Msx sites simultaneously caused a strong reduction in GFP expression, observed in only 4% of larvae (Figure 4E, K_2xMsx_mut). Taken together, our data suggest that also Dmrt and Msx could be involved in the transcriptional activation of Klhl21/30 with a novel role in Ciona.
Functional Analysis of Trans-Acting Factors by CRISPR/Cas9
To test the potential role of Mitf, Dmrt and Msx as transcriptional activators of Klhl21/30, we used CRISPR/Cas9-mediated mutagenesis to knock out these factors in C. robusta embryos (Stolfi et al., 2014; Gandhi et al., 2017, 2018). To design primers to generate sgRNAs for Mitf, Dmrt, and Msx, we used CRISPOR v4.0 portal (Haeussler et al., 2016), which takes in account the potential single-nucleotide polymorphisms (SNPs) and the specificity of each sgRNA and provides their predicted “efficacy” (predicted efficiency of their ability to induce specific DSBs) by the Fusi/Doench algorithm (Doench et al., 2016). Using a series of candidate sgRNAs, we targeted different parts of the coding regions of the selected genes (Supplementary Table S3), synthesized U6 > sgRNA cassettes through One-Step Overlap PCR (OSO-PCR) and validated them by peakshift analysis (Gandhi et al., 2017; see section “Materials and Methods” for details; Supplementary Table S3).
We co-electroporated each gene-specific U6 > sgRNA cassettes, (targeting either Mitf, Dmrt, or Msx) together with Fog > Cas9, which drives Cas9 expression in the a-line blastomeres through the Fog promoter (Rothbächer et al., 2007), and the KlB > GFP reporter plasmid to verify the effect of the experiments on Klhl21/30 isolated regulatory region. As a control, we co-electroporated Fog > Cas9 and KlB > GFP with an sgRNA cassette targeting the mesoderm-specific transcription factor Mesp.5 (Davidson et al., 2005), which is not expressed in the pigment cell lineage. While the control sample had a proportion of GFP-expressing larvae comparable to electroporation with KlB > GFP alone (43% Figure 5A, compared with Figure 2C), we detected a loss of GFP fluorescence in larvae electroporated with sgRNA cassettes targeting the three putative regulators of Klhl21/30 (Figure 5A). When Mitf was knocked out, KlB > GFP was expressed in only 3% of larvae, a sharp decrease compared to the control condition. Moreover, the knockout of Dmrt and Msx also reduced GFP expression, with only 10 and 4% of larvae showing the KlB > GFP expression, respectively (Figure 5A). Because of the strong effect of Mitf loss-of-function on KlB > GFP activity in C. robusta, and the evolutionary conservation of Mitf function in pigment cell development (Goding, 2000; Levy et al., 2006), we performed a complementary Mitf gain-of-function experiment. We co-electroporated KlB > GFP along with Ebf −2.6 kb > H2B:mCherry and Ebf −2.6 kb/ + 15 STOP > Mitf to overexpress Mitf in Ebf + neural progenitors during development. These resulted in ectopic KlB > GFP expression in Ebf + cells in the central nervous system (Figures 5B–B”), suggesting that Mitf drive the transcription of Klhl21/30 in these cells.
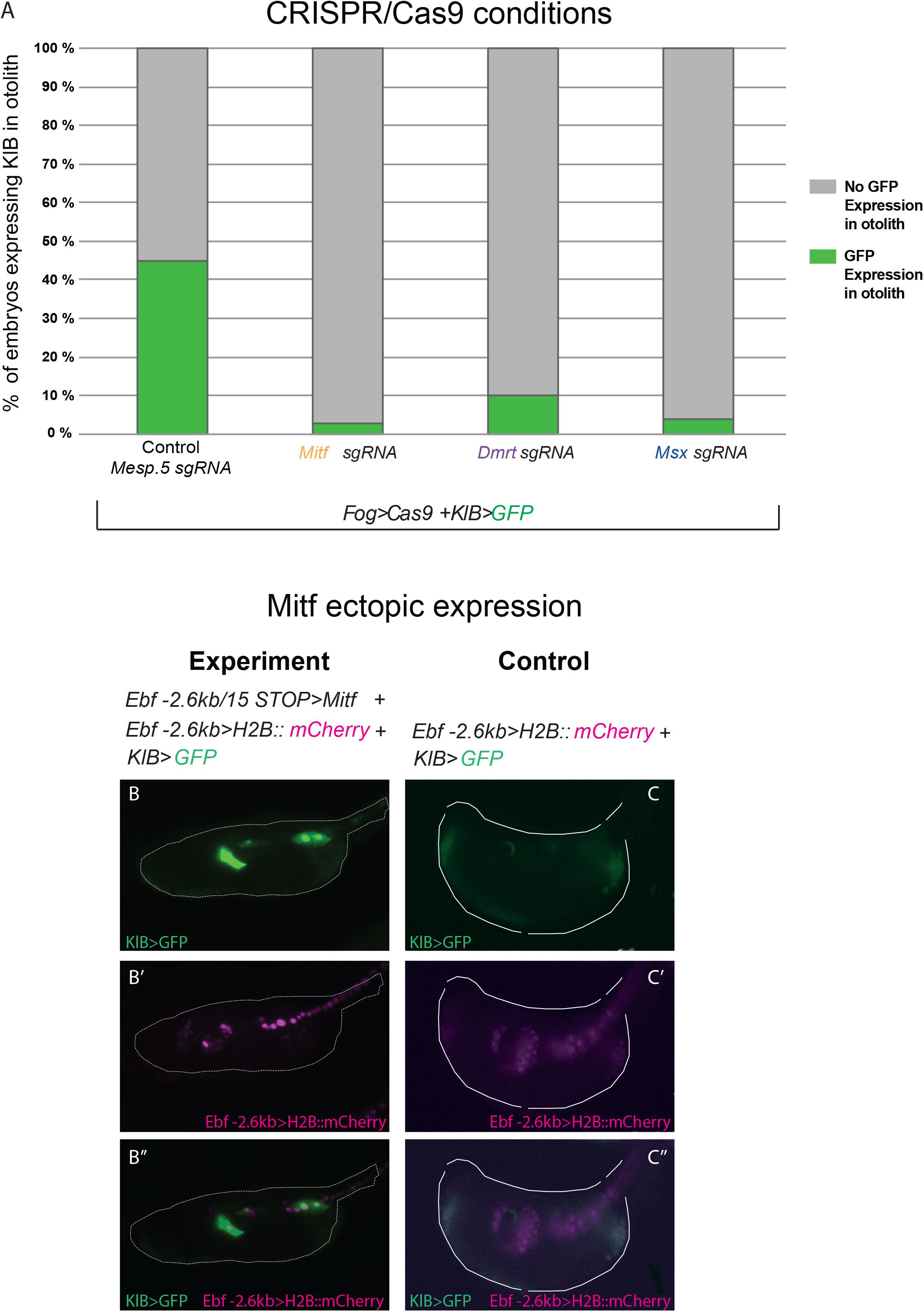
Figure 5. Functional approach on Klhl21/30 regulators. (A) Histogram with phenotypic assays for otolith-specific loss of KlB > GFP in F0 embryos. Larvae were electroporated with 35 μg Fog > Cas9, 60 μg KlB > GFP plus 30 μl of OSO-PCR-based sgRNA cassettes U6 > Mesp.5 (control), U6 > Mitf-ex3 106, U6 > Dmrt-ex2 76, and U6 > Msx-ex3 209, respectively. (B,C) Mitf overexpression driving ectopic KlB > GFP expression: control (60 μg KlB > GFP, 10 μg Ebf –2.6 kb > H2B:mCherry), overexpression experiment (60 μg KlB > GFP, 10 μg Ebf –2.6 kb > H2B:mCherry, 50 μg Ebf –2.6 kb/ + 15 STOP > Mitf). (B–B”) Ectopic expression of KlB > GFP in EBF + cells expressing Mitf ectopically. (C–C”) Control larva showing KlB > GFP expression in the otolith (C,C”) but no ectopic expression in EBF + cells (C”). Experiments were performed at least three times and 100 embryos were scored each time; each site-directed mutagenesis was significant versus the control Mesp.5 (Fisher exact test, p < 0.00001).
Thus, these data strongly suggest that, downstream the FGF signaling, Klhl21/30 is under the control of Mitf, with Dmrt and Msx acting as important co-activators.
Discussion
Our work sheds light on a novel Ciona robusta marker of pigment cell development, the gene Klhl21/30, which represents the earliest gene specifically restricted to the otolith pigmented cell during development. We show that Klhl21/30 is a highly conserved member of the poorly studied Klhl family, whose components have been implicated in various protein-protein interaction and ubiquitination functions. Although human genome encompasses 42 Kelch-like genes (Dhanoa et al., 2013; Figure 1), our phylogenetic analysis revealed the basic chordate toolkit of Klhl genes (Figure 1), whose roles, during development, have yet to be fully explored. We described the Klhl toolkit of C. robusta (32), the larvacean tunicate O. dioica (14), and the cephalochordate (amphioxus) B. belcheri (37). Thus, we deduced that 25 ancestral subfamilies were present at the stem of chordates and are conserved in human, plus one (Klhl5L) tunicate-specific. Despite the tendency of tunicates to lose genes and the relative genomic stasis of cephalochordates (Putnam et al., 2008), we detected several local duplications in these groups. For instance, in tunicates the subfamily Klhl9/13 has undergone tremendous increase, while in amphioxus we found Klhl21/30 duplication and a dramatic expansion of Klhl42 genes. With respect to the ancestral chordate Klhl toolkit, larvaceans lost more than 50% of subfamilies, while we registered an expansion of the family in amphioxus, Ciona and human. Interestingly, the Klhl increase in invertebrate chordates has been driven by different events of gene duplication, whilst WGDs (Abi-Rached et al., 2002; Dehal and Boore, 2005) could have been a major role in modeling human (and vertebrate) Klhl repertoire. Further investigations in other species might be important to understand the exact role exerted by gene duplications, WGDs and losses in shaping the Klhl toolkit, especially in vertebrates. Taken together, our data represent the most detailed analysis of the evolutionary landscape of Klhl family in Chordata so far. Strikingly, Klhl21/30 is one of the most conserved subfamilies and our syntenic survey demonstrates its conservation in all the tunicates (Supplementary Figure S2). Furthermore, Klhl21/30 represents one of few subfamilies retained by the gene-loser O. dioica, hinting at its relative importance. Moreover, we detected a WGD-origin of human (and vertebrates) KLHL21 and KLHL30 genes (Supplementary Figure S3).
There is scarcity of information about the function and expression of genes of the Klhl21/30 subfamily. Klhl21 is known to bind E3 ubiquitin ligase through Cul3 (Maerki et al., 2009; Courtheoux et al., 2016) and Klhl30 has been identified as a circadian pathway gene involved in the onset of glioma (Madden et al., 2014). Like many other Klhl members, Klhl21 is implicated in diverse types of carcinoma, probably due to its role in cell division (Shi et al., 2016; Huang et al., 2017; Chen et al., 2018). Our findings represent the first expression data for Klhl21/30 subfamily in the chordate nervous system. In C. robusta, Klhl21/30 is initially transcribed in two cells of the pigment cell lineage alongside well-known markers of pigmented cells such as Tyr, Tyrp, and Rab32/38 (Coppola et al., 2016; Racioppi et al., 2019), which are conserved in chordate pigment cell evolution (Esposito et al., 2012; Coppola et al., 2016). Interestingly, Klhl21/30 became restricted to the otolith precursor from middle tailbud stage onward. This renders Klhl21/30 the earliest marker of Ciona otolith so far identified, well before the activation of βγ-crystallin in the otolith at the larval stage (Shimeld et al., 2005). Expression data from invertebrates and vertebrates could be crucial to understand the potential functional conservation during evolution of poorly studied Klhl21/30 genes.
The sustained expression of Klhl21/30 in the otolith prompted us to investigate the transcriptional regulation of this gene. Taking advantage of genome conservation with the sibling species C. savignyi, we identified the smallest region containing the TFBS crucial for Klhl21/30 expression in the otolith. Our cis-regulatory mutational analyses suggested the involvement of three transcription factors (Mitf, Dmrt, Msx) in the activation of Klhl21/30 (Figure 3). The tissue-specific, CRISPR/Cas9-mediated loss-of-function of Mitf, Dmrt and Msx resulted in highly penetrant downregulation of the Klhl21/30 reporter in electroporated larvae, suggesting that Klhl21/30 expression depends on these trans-acting factors (Figure 5A). Moreover, a gain-of-function experiment confirmed Mitf as the main regulator for Klhl21/30 reporter expression in other cells of the nervous system. Due to the fact that some Ebf + cells also express Msx, it may be that also Msx collaborates in the ectopic activation of Klhl21/30 reporter. On the other hand, the low efficiency of ectopic expression of Klhl21/30 in Ebf > Mitf ectopic experiment could reflect lack of the Msx or Dmrt in these cells or low penetrance of CRISPR/Cas9 for these genes. We therefore conclude that Mitf, a key regulator of melanocyte development and melanoma in vertebrates (Levy et al., 2006), seems to be an indispensable transcription factor for Klhl21/30 expression in Ciona, with Dmrt and Msx acting as potential co-factors. However, these transcription factors could also work without a direct interaction with the regulatory region responsible for Klh21/30 expression in pigment cell lineage.
Interestingly, Mitf shows sustained expression in the otolith at later developmental stages (Figure 4; Abitua et al., 2012) confirming that this factor could be the final node of the regulatory network underlying the fate determination between otolith and ocellus pigment cells in Ciona as already suggested by Abitua et al. (2012). Besides, Mitf controls also the expression of Rab32/38, a conserved melanogenic marker (Racioppi et al., 2019). The role of Mitf in pigment cell development has been observed also in Drosophila melanogaster (Hallsson et al., 2004), and the function of Mitf as master regulator of genes related to pigment cells and pigmentation is conserved in vertebrates (Levy et al., 2006; Vachtenheim and Borovanský, 2010). However, in this latter Mitf plays a crucial role also in other processes as mast cell development (Morii, 2004; Putnam et al., 2008) and osteoclast biogenesis (Hershey and Fisher, 2004), while in tunicates this bHLH factor seems to be involved only in pigment cell development, and otolith specification, as suggested also by its specific expression during embryogenesis (Figures 4B–C”’; Yajima et al., 2003). If we consider also the specific expression of Mitf in the first pigmented spot of the early-branching amphioxus (Yu et al., 2008), we can speculate that the ancestral Mitf function in early development of chordates was related only to pigment cells and, in vertebrates, it acquired new specializations. However, it is not known if tunicates possess cell types homologous to mast cells or osteoclasts, especially in the poorly studied adult phase. Therefore, a more thorough understanding of Mitf functions (and the existence of other PCPs-specific genes regulated by Mitf) throughout the entire life cycle of invertebrate chordates will be necessary to corroborate this hypothesis.
Our data hinted at Dmrt and Msx as positive regulators of Klhl21/30 (Figures 4, 5). Activated very early by FGF signaling (Imai et al., 2006), the Dmrt gene is crucial for otolith development in the sister species C. savignyi, in which Dmrt mutants typically possess a single ocellus pigment cell but lack an otolith (Tresser et al., 2010). Our results further suggest that Dmrt is a co-activator of an otolith-specific gene, Klhl21/30. With respect to Msx, this transcription factor is traditionally considered a repressor in Ciona (Roure and Darras, 2016) and in different vertebrates (Takeda et al., 2000; Takahashi et al., 2001; Xie et al., 2013). However, in mouse, Msx1 and Msx2 are the transcriptional activators of the proneural gene Atoh1 during spinal cord patterning (Duval et al., 2014). Our findings reveal that the function as activator for Msx might be present also in tunicates.
Importantly, while Mitf has clearly a role in Klhl21/30 activation in Ciona, the fragment containing solely Mitf binding sites (KlE) is not sufficient to drive expression of Klhl21/30 in otolith. In contrast, fragments that include Dmrt and Msx binding sites in addition to Mitf binding sites are sufficient for reporter gene activity and one of them (KlB > GFP) has been utilized for Mitf gain-of-function experiment (Figures 5B–C”). Together with our cis-regulatory mutational and CRISPR/Cas9-mediated knockout data, this suggests that all three factors could be key activators. Alternatively, the much earlier expression of Dmrt and Msx in PCPs evokes that their early binding to the Klhl21/30 cis-regulatory region might be required for later transcriptional activation by otolith specifier Mitf, acting as a so-called “pioneer factors” (Zaret and Carroll, 2011), involved in keeping the chromatin accessible to recruit other late transcriptional activators. Novel functional data regarding Dmrt and Msx in Ciona could unveil their function as activators and/or pioneer factors, and if this role is restricted to pigment cells or if it extends to more broadly anterior neural plate derivatives.
In sum, we provided the first description of a Klhl member in Ciona robusta, identifying it as the earliest otolith pigment cell marker and we elucidated the gene regulatory network (GRN) controlling its expression.
Materials and Methods
Evolutionary Analyses and Transcription Factor Binding Site Analysis
Kelch-like protein sequences from vertebrate Homo sapiens were used as queries in BLASTp and tBLASTn searches in ANISEED (Brozovic et al., 2018), NCBI or Ensembl genome databases of selected species. Orthology was initially assessed by reciprocal best blast hit (RBBH) approach and supported by phylogenetic analyses. Evolutionary reconstructions were performed using ML inferences calculated with PhyML v3.0 and automatic modality of selection of substitution model (Guindon et al., 2010) using protein alignments generated with MUSCLE (Edgar, 2004) and ClustalX (Larkin et al., 2007) programs. Alignment of Supplementary Figure S1 has been carried out employing ClustalX. Full protein sequences were used in our analysis. Accession numbers and names for phylogenetic tree of Figure 1 are listed in Supplementary Table S1, while those excluded are encompassed in Supplementary Table S2. The analysis of synteny conservation (shown in Supplementary Figures S2, S3) was performed by employing ANISEED, Ensembl and Genomicus databases (Louis et al., 2015). To predict putative transcription factor binding sites (TFBS) in the surveyed cis-regulatory regions, we utilized the MatInspector module of the Genomatix Software Suite and CIS-BP employing a Ciona intestinalis (former name for Ciona robusta) DNA-binding-domain classes database (Weirauch et al., 2014). We also used the JASPAR database (Khan et al., 2018) to recognize the potential binding sites.
Animals and Embryo Electroporation
Adults of Ciona robusta were collected from the Gulf of Naples, or from San Diego, CA, United States, by M-REP. Gametes from many animals were gathered separately for in vitro cross-fertilization followed by dechorionation and electroporation as previously illustrated (Christiaen et al., 2009a; Racioppi et al., 2014). Electroporated plasmid amounts (e.g., 10 μg) were per 700 μl of total volume. Embryos were staged according to the developmental timeline shown in Hotta et al. (2007). To visualize GFP, embryos were fixed in MEM-FA (3.7% methanol-free formaldehyde, 0.1 M MOPS pH 7.4, 0.5 M NaCl, 2 mM MgSO4, 1 mM EGTA) for 30 min and washed several times in PBS-NH4Cl and in PBS containing 0.05% Triton X-100. Each electroporation of Figures 3, 4 was carried out using 60 μg of plasmid. The statistical significance of electroporations associated to Figure 4 was validated using Fisher exact test. The statistical significance of electroporations of Supplementary Figure S4 was evaluated employing Chi-square test for trend.
In situ Hybridization
Single and double in situ hybridization experiments were performed out essentially as described previously (Christiaen et al., 2009b; Racioppi et al., 2014), using DIG- and FLUO-labeled riboprobes, anti-DIG-POD and anti-FLUO-POD Fab fragments (Roche, Indianapolis, IN), and Tyramide Amplification Signal with Fluorescein (Perkin Elmer, MA). The antisense riboprobes were obtained from plasmids contained in the C. intestinalis gene collection release I: Klhl21/30 (KH2012:KH.L84.23, GC17e22), Tyrp1/2a (KH2012:KH.C8.537, GC31h05), Mitf (KH2012:KH.C10.106, GC28k08), Dmrt (KH2012:KH.S544.3, GC02f18), and Msx (KH2012:KH.C2.957, GC42h24) (Satou et al., 2002).
Molecular Cloning
The cis-regulatory elements upstream Klhl21/30 were PCR-amplified from genomic DNA and their localization on upstream sequence is shown in Supplementary Figure S6. Insertion of the products into pSP72 vector containing GFP (Zeller et al., 2006) was carried out using TOPO-TA Cloning kit (Invitrogen). The QuickChange Site-Directed Mutagenesis Kit (Agilent) was employed to generate the mutations inside the putative binding sites (Mitf, Dmrt, Msx) identified in the sequence of the KlB element (Figure 4). All the oligos used for cloning the putative regulatory regions and for mutational experiments are listed in Supplementary Table S4.
Functional Experiments by CRISPR/Cas9 and Gain-of-Function
Cas9 and sgRNA expression vectors were constructed or used as previously described (Stolfi et al., 2014; Gandhi et al., 2017). The predictive algorithm used for designing in vivo-transcribed sgRNAs has been the Fusi/Doench (Doench et al., 2016), available on CRISPOR portal (Haeussler et al., 2016). The target sequences have been selected not too close to the translational start, to have the higher impact on the function of protein of interest (Gandhi et al., 2017). DNA oligos used to generate sgRNAs are listed in Supplementary Table S3. One-step overlap PCR (OSO-PCR) was employed for the fast synthesis of a U6 > sgRNA cassettes through a single PCR reaction, performed using Platinum Pfx Polymerase (Invitrogen). The products were cloned into pCESA plasmid using Gibson Assembly Cloning Kit (NEB). Before the electroporation, the products were purified utilizing AMPure XP (Agencourt).
Mitf, Msx, and Dmrt sgRNAs were validated by PCR amplification and Sanger sequencing of the targeted region as previously described (Gandhi et al., 2018). Pooled larvae were lysed for 30 min in 180 μl buffer + 5 μl of Proteinase K from QIAamp DNA Micro Kit (Qiagen) and eluted in 20 μl of water. Approximately 200 ng/μl of genomic DNA extracted from hatched larvae was used for “touchdown” PCRs with Platinum Pfx Polymerase (Invitrogen), as described in Gandhi et al. (2017). The genomic oligos (“peakshift oligos”) employed for “touchdown” PCRs were selected at 150–500 bp away from the primer (Supplementary Table S3), to ensure a proper peakshift. Electroporations were performed as single biological replicates. Electroporation mix recipes can be found in the Supplementary Figure S7. The statistical significance of electroporations associated to Figure 5 was validated using Fisher exact test. Mitf overexpression construct (Ebf-2.6kb/ + 15 STOP > Mitf) was designed as described in Supplementary Figure S7: STOP indicates the presence of stop codon between the ATG of Ebf and the NotI site where Mitf was cloned (no amino acid sequence of Ebf is included in Mitf protein). Images were captured using Confocal ZEISS LSM 700 or ZEISS Apotome.2 compound microscopes.
Data Availability Statement
All datasets presented in this study are included in the article/Supplementary Material.
Author Contributions
UC and FR: conceptualization. UC and AK: investigation. UC, AS, and FR: data curation. AS and FR: supervision. UC: writing. AS and FR: review and editing. All authors contributed to the article and approved the submitted version.
Funding
UC has been supported by SZN OU Ph.D. fellowship and by short-term fellowships from EMBO (7534) and the Company of Biologists (DEVTF-171108). AK has been supported by SZN OU Ph.D. fellowship. AS was supported by NIH award (R00 HD084814).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Susanne Gibboney for helping validate sgRNAs for CRISPR/Cas9.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcell.2020.569601/full#supplementary-material
FIGURE S1 | Domain conservation among Klhl21/30 proteins of Ciona robusta and Homo sapiens. BTB/POZ (bold), BACK (underlined), Kelch-repeat 1 (green), Kelch-repeat 2 (yellow), Kelch-repeat 3 (turquoise), Kelch-repeat 4 (magenta), Kelch-repeat 5 (gray). Domains mapped using Smart classification available on Ensembl database (H. sapiens KLHL30: ENSP00000386389.1; H. sapiens KLHL21: ENSP00000366886.4; C. robusta Klhl21/30: ENSCINP00000014893.3). Ciona Klhl21/30 shares the same domain architecture of human KLHL (according to Dhanoa et al., 2013).
FIGURE S2 | Klhl21/30 synteny analysis in Olfactores. Identification of an ancestral cluster in ascidians, comprising Klhl21/30 gene (orange boxes). Ascidians and human share the gene duplet formed by Klhl21/30 and Nphp4 (red rectangle). Each orthologous gene has been represented using the same color code and black arrows for transcription direction.
FIGURE S3 | Klhl21/30 synteny analysis in Gnathostomes. Conservation of genes flanking gnathostome Klhl21 and Klhl30 (orange boxes) comparing Latimeria chalumnae, Lepisosteus oculatus, Xenopus tropicalis and Homo sapiens. Each orthologous gene has been represented using the same color code and black arrows for transcription direction.
FIGURE S4 | Expression of Klhl21/30 in embryos electroporated with dnFGFR. Evaluation of percentage of embryos showing expression in the otolith respect to wild-type (wt). The experiment was repeated three times, showing a significant trend respect to control (Chi-square test for trend, p < 0.0001; n = 200).
FIGURE S5 | GFP transcription in tailbud embryos electroporated with KlA >GFP. In situ hybridization experiment using GFP probe on tailbud embryos electroporated with KlA >GFP construct (A,B).
FIGURE S6 | Localization of regions used for cis-regulatory analysis (Figure 3) inside upstream region of Ciona robusta Klhl21/30 (KH.L84.23). Different colors indicate the selected regions and the oligos used for cloning, with respective combinations.
FIGURE S7 | Electroporation mixes and constructs used for functional experiments.
TABLE S1 | List of Klhl protein sequences employed for phylogeny of Figure 1.
TABLE S2 | Klhl protein sequences excluded from phylogeny for their high degree of molecular divergence. Their names were assessed by genome search and reciprocal BLASTs.
TABLE S3 | (A) List of oligonucleotides used for functional assays. Names contain indication on the position of the target sequence inside the selected exon (e.g., Mitf-ex3 106 indicates that the target sequence is within exon3 starting from its nucleotide 106). In bold and capital letters the sgRNA N (19) target sequences. In lowercase the protospacer sequence (for OSO PCR) appended 3′ to a forward primer and, in reverse complement, appended 3′ to the reverse primer (Gandhi et al., 2017). In red are highlighted the oligos whose sgRNAs were able to cause deletions. “Specificity” indicates the capability of the synthetized sgRNA to target a specific sequence (0–100); “efficacy” represents a prediction of sgRNA ability to induce Cas9-mediated DSBs (0–100); “SNPs” is the number of potential single-nucleotide polimorphisms associated to each sgRNA, which can possibly affect its pairing to the target. All these values were retrieved from CRISPOR portal. (B) List of “peakshift” genomic oligos (in blue) used to verify the effect of deletions on specific exons (ex i.e., exon).
TABLE S4 | List of oligos used for cloning and mutational experiments (in red are shown the mutated nucleotides, with respect to wt sequence).
References
Abi-Rached, L., Gilles, A., Shiina, T., Pontarotti, P., and Inoko, H. (2002). Evidence of en bloc duplication in vertebrate genomes. Nat. Genet. 31, 100–105. doi: 10.1038/ng855
Abitua, P. B., Wagner, E., Navarrete, I. A., and Levine, M. (2012). Identification of a rudimentary neural crest in a non-vertebrate chordate. Nature 492, 104–107. doi: 10.1038/nature11589
Adams, J., Kelso, R., and Cooley, L. (2000). The kelch repeat superfamily of proteins: propellers of cell function. Trends Cell Biol. 10, 17–24. doi: 10.1016/s0962-8924(99)01673-6
Aniello, F., Locascio, A., Villani, M. G., Di Gregorio, A., Fucci, L., and Branno, M. (1999). Identification and developmental expression of Ci-msxb: a novel homologue of Drosophila msh gene in Ciona intestinalis. Mech. Dev. 88, 123–126. doi: 10.1016/s0925-4773(99)00178-1
Berná, L., and Alvarez-Valin, F. (2014). Evolutionary genomics of fast evolving tunicates. Genome Biol. Evol. 6, 1724–1738. doi: 10.1093/gbe/evu122
Brozovic, M., Dantec, C., Dardaillon, J., Dauga, D., Faure, E., Gineste, M., et al. (2018). ANISEED 2017: extending the integrated ascidian database to the exploration and evolutionary comparison of genome-scale datasets. Nucleic Acids Res. 46, D718–D725.
Caracciolo, A., Gesualdo, I., Branno, M., Aniello, F., Di Lauro, R., and Palumbo, A. (1997). Specific cellular localization of tyrosinase mRNA during Ciona intestinalis larval development. Dev. Growth Differ. 39, 437–444. doi: 10.1046/j.1440-169x.1997.t01-3-00004.x
Chen, J., Song, W., Du, Y., Li, Z., Xuan, Z., Zhao, L., et al. (2018). Inhibition of KLHL21 prevents cholangiocarcinoma progression through regulating cell proliferation and motility, arresting cell cycle and reducing Erk activation. Biochem. Biophys. Res. Commun. 499, 433–440. doi: 10.1016/j.bbrc.2018.03.152
Christiaen, L., Wagner, E., Shi, W., and Levine, M. (2009a). Isolation of sea squirt (Ciona) gametes, fertilization, dechorionation, and development. Cold Spring Harb. Protoc. 2009:db.rot5344.
Christiaen, L., Wagner, E., Shi, W., and Levine, M. (2009b). Whole-mount in situ hybridization on sea squirt (Ciona intestinalis) embryos. Cold Spring Harb. Protoc. 2009:db.rot5348.
Cole, A. G., and Meinertzhagen, I. A. (2004). The central nervous system of the ascidian larva: mitotic history of cells forming the neural tube in late embryonic Ciona intestinalis. Dev. Biol. 271, 239–262. doi: 10.1016/j.ydbio.2004.04.001
Coppola, U., Annona, G., D’Aniello, S., and Ristoratore, F. (2016). Rab32 and Rab38 genes in chordate pigmentation: an evolutionary perspective. BMC Evol. Biol. 16:26. doi: 10.1186/s12862-016-0596-1
Coppola, U., Ristoratore, F., Albalat, R., and D’Aniello, S. (2019). The evolutionary landscape of the Rab family in chordates. Cell. Mol. Life Sci. 76, 4117–4130. doi: 10.1007/s00018-019-03103-7
Corbo, J. C., Levine, M., and Zeller, R. W. (1997). Characterization of a notochord-specific enhancer from the Brachyury promoter region of the ascidian, Ciona intestinalis. Development 124, 589–602.
Courtheoux, T., Enchev, R. I., Lampert, F., Gerez, J., Beck, J., Picotti, P., et al. (2016). Cortical dynamics during cell motility are regulated by CRL3(KLHL21) E3 ubiquitin ligase. Nat. Commun. 7:12810.
Crocetta, F., Marino, R., Cirino, P., Macina, A., Staiano, L., Esposito, R., et al. (2015). Mutation studies in ascidians: a review. Genesis 53, 160–169.
Curran, K., Lister, J. A., Kunkel, G. R., Prendergast, A., Parichy, D. M., and Raible, D. W. (2010). Interplay between Foxd3 and Mitf regulates cell fate plasticity in the zebrafish neural crest. Dev. Biol. 344, 107–118. doi: 10.1016/j.ydbio.2010.04.023
D’Aniello, S., D’Aniello, E., Locascio, A., Memoli, A., Corrado, M., Russo, M. T., et al. (2006). The ascidian homolog of the vertebrate homeobox gene Rx is essential for ocellus development and function. Differentiation 74, 222–234. doi: 10.1111/j.1432-0436.2006.00071.x
Davidson, B., Shi, W., and Levine, M. (2005). Uncoupling heart cell specification and migration in the simple chordate Ciona intestinalis. Development 132, 4811–4818. doi: 10.1242/dev.02051
de Castro, E., Sigrist, C. J. A., Gattiker, A., Bulliard, V., Langendijk-Genevaux, P. S., Gasteiger, E., et al. (2006). ScanProsite: detection of PROSITE signature matches and ProRule-associated functional and structural residues in proteins. Nucleic Acids Res. 34, W362–W365.
Dehal, P., and Boore, J. L. (2005). Two rounds of whole genome duplication in the ancestral vertebrate. PLoS Biol. 3:e314. doi: 10.1371/journal.pbio.0030314
Delsuc, F., Brinkmann, H., Chourrout, D., and Philippe, H. (2006). Tunicates and not cephalochordates are the closest living relatives of vertebrates. Nature 439, 965–968. doi: 10.1038/nature04336
Dhanoa, B. S., Cogliati, T., Satish, A. G., Bruford, E. A., and Friedman, J. S. (2013). Update on the Kelch-like (KLHL) gene family. Hum. Genomics 7:13. doi: 10.1186/1479-7364-7-13
Dilly, P. N. (1962). Studies on the receptors in the cerebral vesicle of ascidian tadpole. Q. J. Microsc. Sci. 105, 13–20.
Dilly, P. N. (1969). Studies on the receptors in Ciona intestinalis. Z. Zellforsch. Mikrosk. Anat. 96, 63–65. doi: 10.1007/bf00321477
Doench, J. G., Fusi, N., Sullender, M., Hegde, M., Vaimberg, E. W., Donovan, K. F., et al. (2016). Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9. Nat. Biotechnol. 34, 184–191. doi: 10.1038/nbt.3437
Dufour, H. D., Chettouh, Z., Deyts, C., de Rosa, R., Goridis, C., Joly, J.-S., et al. (2006). Precraniate origin of cranial motoneurons. Proc. Natl. Acad. Sci. U.S.A. 103, 8727–8732. doi: 10.1073/pnas.0600805103
Duval, N., Daubas, P., Bourcier de Carbon, C., St Cloment, C., Tinevez, J.-Y., Lopes, M., et al. (2014). Msx1 and Msx2 act as essential activators of Atoh1 expression in the murine spinal cord. Development 141, 1726–1736. doi: 10.1242/dev.099002
Edgar, R. C. (2004). MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics 5:113. doi: 10.1186/1471-2105-5-113
Esposito, R., D’Aniello, S., Squarzoni, P., Pezzotti, M. R., Ristoratore, F., and Spagnuolo, A. (2012). New insights into the evolution of metazoan tyrosinase gene family. PLoS One 7:e35731. doi: 10.1371/journal.pone.0035731
Gandhi, S., Haeussler, M., Razy-Krajka, F., Christiaen, L., and Stolfi, A. (2017). Evaluation and rational design of guide RNAs for efficient CRISPR/Cas9-mediated mutagenesis in Ciona. Dev. Biol. 425, 8–20. doi: 10.1016/j.ydbio.2017.03.003
Gandhi, S., Razy-Krajka, F., Christiaen, L., and Stolfi, A. (2018). CRISPR knockouts in ciona embryos. Adv. Exp. Med. Biol. 1029, 141–152. doi: 10.1007/978-981-10-7545-2_13
Goding, C. R. (2000). Mitf from neural crest to melanoma: signal transduction and transcription in the melanocyte lineage. Genes Dev. 14, 1712–1728.
Guindon, S., Dufayard, J.-F., Lefort, V., Anisimova, M., Hordijk, W., and Gascuel, O. (2010). New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst. Biol. 59, 307–321. doi: 10.1093/sysbio/syq010
Haeussler, M., Schönig, K., Eckert, H., Eschstruth, A., Mianné, J., Renaud, J.-B., et al. (2016). Evaluation of off-target and on-target scoring algorithms and integration into the guide RNA selection tool CRISPOR. Genome Biol. 17:148.
Hallsson, J. H., Haflidadóttir, B. S., Stivers, C., Odenwald, W., Arnheiter, H., Pignoni, F., et al. (2004). The basic helix-loop-helix leucine zipper transcription factor Mitf is conserved in Drosophila and functions in eye development. Genetics 167, 233–241. doi: 10.1534/genetics.167.1.233
Haupaix, N., Abitua, P. B., Sirour, C., Yasuo, H., Levine, M., and Hudson, C. (2014). Ephrin-mediated restriction of ERK1/2 activity delimits the number of pigment cells in the Ciona CNS. Dev. Biol. 394, 170–180.
Hershey, C. L., and Fisher, D. E. (2004). Mitf and Tfe3: members of a b-HLH-ZIP transcription factor family essential for osteoclast development and function. Bone 34, 689–696. doi: 10.1016/j.bone.2003.08.014
Horie, T., Orii, H., and Nakagawa, M. (2005). Structure of ocellus photoreceptors in the ascidian Ciona intestinalis larva as revealed by an anti-arrestin antibody. J. Neurobiol. 65, 241–250. doi: 10.1002/neu.20197
Hotta, K., Mitsuhara, K., Takahashi, H., Inaba, K., Oka, K., Gojobori, T., et al. (2007). A web-based interactive developmental table for the ascidian Ciona intestinalis, including 3D real-image embryo reconstructions: I. From fertilized egg to hatching larva. Dev. Dyn. 236, 1790–1805. doi: 10.1002/dvdy.21188
Huang, G., Kaufman, A. J., Xu, K., Manova, K., and Singh, B. (2017). Squamous cell carcinoma-related oncogene (SCCRO) neddylates Cul3 protein to selectively promote midbody localization and activity of Cul3KLHL21 protein complex during abscission. J. Biol. Chem. 292, 15254–15265. doi: 10.1074/jbc.m117.778530
Imai, K. S., Levine, M., Satoh, N., and Satou, Y. (2006). Regulatory blueprint for a chordate embryo. Science 312, 1183–1187. doi: 10.1126/science.1123404
Jiang, D. (2005). Pigmentation in the sensory organs of the ascidian larva is essential for normal behavior. J. Exp. Biol. 208, 433–438. doi: 10.1242/jeb.01420
Khan, A., Fornes, O., Stigliani, A., Gheorghe, M., Castro-Mondragon, J. A., van der Lee, R., et al. (2018). JASPAR 2018: update of the open-access database of transcription factor binding profiles and its web framework. Nucleic Acids Res. 46, D260–D266.
Kusakabe, T., Kusakabe, R., Kawakami, I., Satou, Y., Satoh, N., and Tsuda, M. (2001). Ci-opsin1, a vertebrate-type opsin gene, expressed in the larval ocellus of the ascidian Ciona intestinalis. FEBS Lett. 506, 69–72. doi: 10.1016/s0014-5793(01)02877-0
Larkin, M. A., Blackshields, G., Brown, N. P., Chenna, R., McGettigan, P. A., McWilliam, H., et al. (2007). Clustal W and Clustal X version 2.0. Bioinformatics 23, 2947–2948. doi: 10.1093/bioinformatics/btm404
Levy, C., Khaled, M., and Fisher, D. E. (2006). MITF: master regulator of melanocyte development and melanoma oncogene. Trends Mol. Med. 12, 406–414. doi: 10.1016/j.molmed.2006.07.008
Louis, A., Nguyen, N. T. T., Muffato, M., and Roest Crollius, H. (2015). Genomicus update 2015: karyoView and matrixView provide a genome-wide perspective to multispecies comparative genomics. Nucleic Acids Res. 43, D682–D689.
Madden, M. H., Anic, G. M., Thompson, R. C., Nabors, L. B., Olson, J. J., Browning, J. E., et al. (2014). Circadian pathway genes in relation to glioma risk and outcome. Cancer Causes Control 25, 25–32. doi: 10.1007/s10552-013-0305-y
Maerki, S., Olma, M. H., Staubli, T., Steigemann, P., Gerlich, D. W., Quadroni, M., et al. (2009). The Cul3–KLHL21 E3 ubiquitin ligase targets Aurora B to midzone microtubules in anaphase and is required for cytokinesis. J. Cell Biol. 187, 791–800. doi: 10.1083/jcb.200906117
Marino, R., Melillo, D., Di Filippo, M., Yamada, A., Pinto, M. R., De Santis, R., et al. (2007). Ammonium channel expression is essential for brain development and function in the larva of Ciona intestinalis. J. Comp. Neurol. 503, 135–147. doi: 10.1002/cne.21370
Martí-Solans, J., Belyaeva, O. V., Torres-Aguila, N. P., Kedishvili, N. Y., Albalat, R., and Cañestro, C. (2016). Coelimination and survival in gene network evolution: dismantling the RA-signaling in a chordate. Mol. Biol. Evol. 33, 2401–2416. doi: 10.1093/molbev/msw118
Moret, F., Christiaen, L., Deyts, C., Blin, M., Vernier, P., and Joly, J.-S. (2005). Regulatory gene expressions in the ascidian ventral sensory vesicle: evolutionary relationships with the vertebrate hypothalamus. Dev. Biol. 277, 567–579. doi: 10.1016/j.ydbio.2004.11.004
Morii, E. (2004). Roles of MITF for development of mast cells in mice: effects on both precursors and tissue environments. Blood 104, 1656–1661. doi: 10.1182/blood-2004-01-0247
Nakagawa, M., Orii, H., Yoshida, N., Jojima, E., Horie, T., Yoshida, R., et al. (2002). Ascidian arrestin (Ci-arr), the origin of the visual and nonvisual arrestins of vertebrate. Eur. J. Biochem. 269, 5112–5118. doi: 10.1046/j.1432-1033.2002.03240.x
Nakashima, Y., Kusakabe, T., Kusakabe, R., Terakita, A., Shichida, Y., and Tsuda, M. (2003). Origin of the vertebrate visual cycle: genes encoding retinal photoisomerase and two putative visual cycle proteins are expressed in whole brain of a primitive chordate. J. Comp. Neurol. 460, 180–190. doi: 10.1002/cne.10645
Putnam, N. H., Butts, T., Ferrier, D. E. K., Furlong, R. F., Hellsten, U., Kawashima, T., et al. (2008). The amphioxus genome and the evolution of the chordate karyotype. Nature 453, 1064–1071.
Racioppi, C., Coppola, U., Christiaen, L., and Ristoratore, F. (2019). Transcriptional regulation of Rab32/38, a specific marker of pigment cell formation in Ciona robusta. Dev. Biol. 448, 111–118. doi: 10.1016/j.ydbio.2018.11.013
Racioppi, C., Kamal, A. K., Razy-Krajka, F., Gambardella, G., Zanetti, L., di Bernardo, D., et al. (2014). Fibroblast growth factor signalling controls nervous system patterning and pigment cell formation in Ciona intestinalis. Nat. Commun. 5:4830.
Racioppi, C., Valoroso, M. C., Coppola, U., Lowe, E. K., Titus Brown, C., Swalla, B. J., et al. (2017). Evolutionary loss of melanogenesis in the tunicate Molgula occulta. EvoDevo 8:11.
Rothbächer, U., Bertrand, V., Lamy, C., and Lemaire, P. (2007). A combinatorial code of maternal GATA, Ets and beta-catenin-TCF transcription factors specifies and patterns the early ascidian ectoderm. Development 134, 4023–4032. doi: 10.1242/dev.010850
Roure, A., and Darras, S. (2016). Msxb is a core component of the genetic circuitry specifying the dorsal and ventral neurogenic midlines in the ascidian embryo. Dev. Biol. 409, 277–287. doi: 10.1016/j.ydbio.2015.11.009
Russo, M. T., Donizetti, A., Locascio, A., D’Aniello, S., Amoroso, A., Aniello, F., et al. (2004). Regulatory elements controlling Ci-msxb tissue-specific expression during Ciona intestinalis embryonic development. Dev. Biol. 267, 517–528. doi: 10.1016/j.ydbio.2003.11.005
Sakurai, D., Goda, M., Kohmura, Y., Horie, T., Iwamoto, H., Ohtsuki, H., et al. (2004). The role of pigment cells in the brain of ascidian larva. J. Comp. Neurol. 475, 70–82. doi: 10.1002/cne.20142
Satou, Y., Imai, K. S., Levine, M., Kohara, Y., Rokhsar, D., and Satoh, N. (2003). A genome wide survey of developmentally relevant genes in Ciona intestinalis. I. Genes for bHLH transcription factors. Dev. Genes Evol. 213, 213–221. doi: 10.1007/s00427-003-0319-7
Satou, Y., Yamada, L., Mochizuki, Y., Takatori, N., Kawashima, T., Sasaki, A., et al. (2002). A cDNA resource from the basal chordate Ciona intestinalis. Genesis 33, 153–154.
Shi, L., Zhang, W., Zou, F., Mei, L., Wu, G., and Teng, Y. (2016). KLHL21, a novel gene that contributes to the progression of hepatocellular carcinoma. BMC Cancer 16:815. doi: 10.1186/s12885-016-2851-7
Shimeld, S. M., Purkiss, A. G., Dirks, R. P. H., Bateman, O. A., Slingsby, C., and Lubsen, N. H. (2005). Urochordate betagamma-crystallin and the evolutionary origin of the vertebrate eye lens. Curr. Biol. 15, 1684–1689. doi: 10.1016/j.cub.2005.08.046
Sordino, P., Andreakis, N., Brown, E. R., Leccia, N. I., Squarzoni, P., Tarallo, R., et al. (2008). Natural variation of model mutant phenotypes in Ciona intestinalis. PLoS One 3:e2344. doi: 10.1371/journal.pone.0002344
Squarzoni, P., Parveen, F., Zanetti, L., Ristoratore, F., and Spagnuolo, A. (2011). FGF/MAPK/Ets signaling renders pigment cell precursors competent to respond to Wnt signal by directly controlling Ci-Tcf transcription. Development 138, 1421–1432. doi: 10.1242/dev.057323
Stolfi, A., Gandhi, S., Salek, F., and Christiaen, L. (2014). Tissue-specific genome editing in Ciona embryos by CRISPR/Cas9. Development 141, 4115–4120. doi: 10.1242/dev.114488
Takahashi, K., Nuckolls, G. H., Takahashi, I., Nonaka, K., Nagata, M., Ikura, T., et al. (2001). Msx2 is a repressor of chondrogenic differentiation in migratory cranial neural crest cells. Dev. Dyn. 222, 252–262. doi: 10.1002/dvdy.1185
Takeda, M., Saito, Y., Sekine, R., Onitsuka, I., Maeda, R., and Maéno, M. (2000). Xenopus msx-1 regulates dorso-ventral axis formation by suppressing the expression of organizer genes. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 126, 157–168. doi: 10.1016/s0305-0491(00)00194-2
Tief, K., Schmidt, A., Aguzzi, A., and Beermann, F. (1996). Tyrosinase is a new marker for cell populations in the mouse neural tube. Dev. Dyn. 205, 445–456. doi: 10.1002/(sici)1097-0177(199604)205:4<445::aid-aja8>3.0.co;2-i
Tresser, J., Chiba, S., Veeman, M., El-Nachef, D., Newman-Smith, E., Horie, T., et al. (2010). doublesex/mab3 related-1 (dmrt1) is essential for development of anterior neural plate derivatives in Ciona. Development 137, 2197–2203. doi: 10.1242/dev.045302
Tsuda, M., Sakurai, D., and Goda, M. (2003). Direct evidence for the role of pigment cells in the brain of ascidian larvae by laser ablation. J. Exp. Biol. 206, 1409–1417. doi: 10.1242/jeb.00235
Vachtenheim, J., and Borovanský, J. (2010). “Transcription physiology” of pigment formation in melanocytes: central role of MITF. Exp. Dermatol. 19, 617–627. doi: 10.1111/j.1600-0625.2009.01053.x
Wagner, E., and Levine, M. (2012). FGF signaling establishes the anterior border of the Ciona neural tube. Development 139, 2351–2359. doi: 10.1242/dev.078485
Weirauch, M. T., Yang, A., Albu, M., Cote, A. G., Montenegro-Montero, A., Drewe, P., et al. (2014). Determination and inference of eukaryotic transcription factor sequence specificity. Cell 158, 1431–1443. doi: 10.1016/j.cell.2014.08.009
Xie, H., Cherrington, B. D., Meadows, J. D., Witham, E. A., and Mellon, P. L. (2013). Msx1 homeodomain protein represses the αGSU and GnRH receptor genes during gonadotrope development. Mol. Endocrinol. 27, 422–436. doi: 10.1210/me.2012-1289
Yajima, I., Endo, K., Sato, S., Toyoda, R., Wada, H., Shibahara, S., et al. (2003). Cloning and functional analysis of ascidian Mitf in vivo: insights into the origin of vertebrate pigment cells. Mech. Dev. 120, 1489–1504. doi: 10.1016/j.mod.2003.08.009
Yu, J.-K., Yu, J., Meulemans, D., McKeown, S. J., and Bronner-Fraser, M. (2008). Insights from the amphioxus genome on the origin of vertebrate neural crest. Genome Res. 18, 1127–1132. doi: 10.1101/gr.076208.108
Zaret, K. S., and Carroll, J. S. (2011). Pioneer transcription factors: establishing competence for gene expression. Genes Dev. 25, 2227–2241. doi: 10.1101/gad.176826.111
Keywords: tunicates, otolith, Mitf, Klhl family evolution, pigment cells, CRISPR/Cas9, cis-regulatory regions
Citation: Coppola U, Kamal AK, Stolfi A and Ristoratore F (2020) The Cis-Regulatory Code for Kelch-like 21/30 Specific Expression in Ciona robusta Sensory Organs. Front. Cell Dev. Biol. 8:569601. doi: 10.3389/fcell.2020.569601
Received: 04 June 2020; Accepted: 17 August 2020;
Published: 11 September 2020.
Edited by:
David Ellard Keith Ferrier, University of St Andrews, United KingdomReviewed by:
Takehiro Kusakabe, Konan University, JapanIldiko M. L. Somorjai, University of St Andrews, United Kingdom
Copyright © 2020 Coppola, Kamal, Stolfi and Ristoratore. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Filomena Ristoratore, Zmlsb21lbmEucmlzdG9yYXRvcmVAc3puLml0
 Ugo Coppola
Ugo Coppola Ashwani Kumar Kamal1
Ashwani Kumar Kamal1 Alberto Stolfi
Alberto Stolfi Filomena Ristoratore
Filomena Ristoratore