- 1School of Basic Medicine Sciences, Chengdu University of Traditional Chinese Medicine, Chengdu, China
- 2Department of Respiratory, Affiliated Hospital of Chengdu University of Traditional Chinese Medicine, Chengdu, China
Chronic obstructive pulmonary disease (COPD) is a serious public health concern worldwide. By 2040, 4.41 million people are estimated to expire annually due to COPD. However, till date, it has remained difficult to alter the activity or progress of the disease through treatment. In order to address this issue, the best way would be to find biomarkers and new therapeutic targets for COPD. DNA methylation (DNAm) may be a potential biomarker for disease prevention, diagnosis, and prognosis, and its reversibility further makes it a potential drug design target in COPD. In this review, we aimed to explore the role of DNAm as biomarkers and disease mediators in different tissue samples from patients with COPD.
Introduction
Chronic obstructive pulmonary disease (COPD) is a chronic progressive disease (Rabe and Watz, 2017). Its pathological features mainly include irreversible airway obstruction, mucus secretion, and inflammation (Celli and MacNee, 2004). Fifty years ago, it had prompted the establishment of the Department of Pulmonary Diseases. Unfortunately, with the world’s population aging rapidly, COPD has become an increasingly serious problem (Crapo, 2019). The Global Burden of Disease Study (GBDS) had estimated ∼299 million patients to have COPD, worldwide, in 2017 (GBD 2017 Disease and Injury Incidence and Prevalence Collaborators, 2018). In 2016, ∼2.93 million patients across the world died of the disease due to its high incidence rate. By 2040, the number of deaths due to COPD is expected to be double of that in 2016, which implies ∼4.41 million deaths due to COPD (Foreman et al., 2018). The above data, however, does not include the patients with COPD who died of cardiovascular disease (Calverley et al., 2007), and those with some cardiovascular diseases along with airflow restriction (Franssen et al., 2016). Therefore, the global mortality caused by COPD is remarkably underestimated, and in near future, it could pose a serious public health concern. For many years, the clinical treatment of COPD has focused on relieving patients’ symptoms, and improving their health and quality of life. However, till date, it has remained very difficult to alter the progress of this disease through treatment. COPD is a heterogeneous disease, which may need precise and personalized therapeutic approach (Heaney and McGarvey, 2017). Moreover, early diagnosis of the disease would be necessary, before the occurrence of disability or irreversible lung structural changes (Lowe et al., 2019). The best way to solve this problem would be to explore the biomarkers of COPD and identify new therapeutic targets.
COPD is a complex disease affected by environmental factors (smoking or drugs); therefore, its pathological mechanism is driven by genetic and epigenetic variation (Qiu et al., 2012; Vucic et al., 2014). Through genome-wide genetic association studies (GWAS), many COPD-susceptible gene loci, such as FAM13A (Cho et al., 2010; Chen et al., 2015), CHRNA 3/5 (Pillai et al., 2009; Hardin et al., 2012) and HHIP (Pillai et al., 2009), had been identified previously (Young et al., 2008, 2010, 2011; Hancock et al., 2010; Lambrechts et al., 2010; Repapi et al., 2010; Van Durme et al., 2010; Chappell et al., 2011). Interestingly, many single nucleotide polymorphisms related to COPD were found in the non-coding region and intron regulatory regions of the gene; this result was consistent with many complex diseases (Hindorff et al., 2009; Maurano et al., 2012). These single nucleotide polymorphisms may be speculated to be involved in the regulation of epigenetic modification. Some differential methylated sites (CPGs), unrelated to GWAS but closely related to those regarding smoking (the main exposure factor of COPD), have potential significance in COPD susceptibility (Qiu et al., 2012, 2015; Vucic et al., 2014). Therefore, genetic variation and environmental exposure mediated by epigenetic modification may enhance the risk of COPD (Qiu et al., 2012).
Epigenetic modification mainly includes DNA methylation and various post-translational modifications of histones (histone PTM, including histone acetylation, methylation, phosphorylation, ubiquitination and sulfonation). Among them, COPD is mainly related to histone acetylation. Histone deacetylase is the key enzyme to control the proinflammatory cytokines related to COPD (Bowman et al., 2009). The balance of histone acetylation/deacetylation in COPD patients will shifted toward acetylation (Szulakowski et al., 2006). The imbalance of histone acetylation and deacetylation changes the nucleosome structure in the transcription of inflammatory cytokines, affects the expression of inflammatory genes (Mroz et al., 2007, 2008). This leads to the change of gene expression profile in COPD patients (Szulakowski et al., 2006).
DNA methylation (DNAm) is a tissue-specific epigenetic modification involved in the regulation of gene expression (Vucic et al., 2014). It plays an important role in many chronic complex diseases, like cancer (Luo et al., 2020) and aging (Horvath and Raj, 2018), and is the basis of normal physiological development. Many studies, till date, have confirmed DNA methylation, related to COPD, to be a potential biomarker for disease prevention, diagnosis, and prognostic evaluation (Qiu et al., 2012; Sundar et al., 2017). Furthermore, its reversibility makes it an attractive candidate for designing disease biomarkers and drugs. In this review, we focused on the role of DNAm as biomarkers and disease mediators in different tissue samples from patients with COPD.
Biomarker in Blood
Blood samples from patients with COPD had previously been reported to show hypomethylation of DNA, which is related to severity of the disease (Zinellu et al., 2017c). Some studies had suggested DNA hypomethylation to be related to oxidative stress (OS) via various mechanisms (Fomenko et al., 2007; Hitchler and Domann, 2007; Franco et al., 2008). The redox state of cells, with higher degree of oxidation, may lead to the reduction of DNAm through redox regulation of related enzymes, such as SAM-dependent methyltransferase (Fomenko et al., 2007). In an environment with higher level of oxidation, the activity of methionine adenosyltransferase is decreased, which in turn catalyzes the addition of methionine to adenosine to synthesize SAM (Pajares et al., 1992). The important role of OS in COPD has now been confirmed (Figure 1; Zinellu et al., 2016a, b, 2016); the characteristics of hypoxia in COPD can enhance OS by increasing ROS production at the level of mitochondrial respiratory chain (Chandel and Schumacker, 2000; Hoppeler et al., 2003). In contrast, Angelo Zinellu et al. had reported DNA hypomethylation in the blood of patients with COPD to be related to disease severity, although there was no significant correlation between DNAm and OS index (Zinellu et al., 2017c). This may be due to the relatively small sample size of the study or the lack of severely unstable patients with COPD (particularly with high oxidative stress). At the same time, Qiu et al. (2012) had screened 27578 CpG loci in subjects with COPD, by using the HumanMethylation27 array (Illumina, San Diego, CA, United States). A total of 349 CpG loci was found to be significantly related to the susceptibility and severity of COPD. Gene ontology analysis, based on these CpGs, suggested the involvement of immune and inflammatory system pathways, stress response, and external stimulation. Hypomethylation of SERPINA1 and fucosyltransferase-7 (FUT7) genes was found to be closely related to COPD and pulmonary dysfunction. Recently, the haplotypes of two major alleles of SERPINA1 were found to be related to reduced risk of COPD (Ponce-Gallegos et al., 2019). SERPINA1 gene encodes a sialic acid protein called α-1 antitrypsin (AAT), whose main function is to protect the lower respiratory tract from degradation by neutrophil elastase (Salahuddin, 2010). AAT deficiency is generally believed to be the most important genetic risk factor of COPD. Decrease of α-1 antitrypsin level in circulation leads to the increase of elastase activity in pulmonary neutrophils; it is the imbalance of protease–antiprotease that leads to lung remodeling (Brode et al., 2012). FUT7 encodes sialyl Lewis X (SLeX), which can promote leukocyte migration to inflammatory tissue (Miyashiro et al., 2004). Furthermore, the SLeX is a ligand of E-selectin (ES), which is increased in patients with COPD (Di Stefano et al., 1994). Therefore, future research should focus on whether the relative hypomethylation of FUT7 could change the peripheral expression of SLex and eventually lead to the transfer of neutrophils to the lung. If this hypothesis could be confirmed, it will be another exciting and biologically credible way. Some new differential methylation sites related to COPD had been identified in the DNA of Korean blood samples, including cg03559389 (DIP2C) and a rare variant (rs140198372) of cg19904425 (SERPINA 12) (Lee et al., 2017). Among them, the DIP2C mutation was found in lung cancer samples (Gao et al., 2013) and SERPINA12 was reported to be related to airflow restriction (Jackson et al., 2016). For the systematic evaluation of COPD as well as DNAm of peripheral blood in a population-based study, no consistent CpG locus was found in the study of COPD status or that of lung function value (Machin et al., 2017). Large-scale research on vertical design to solve reverse causality may prove to be a more fruitful research approach.
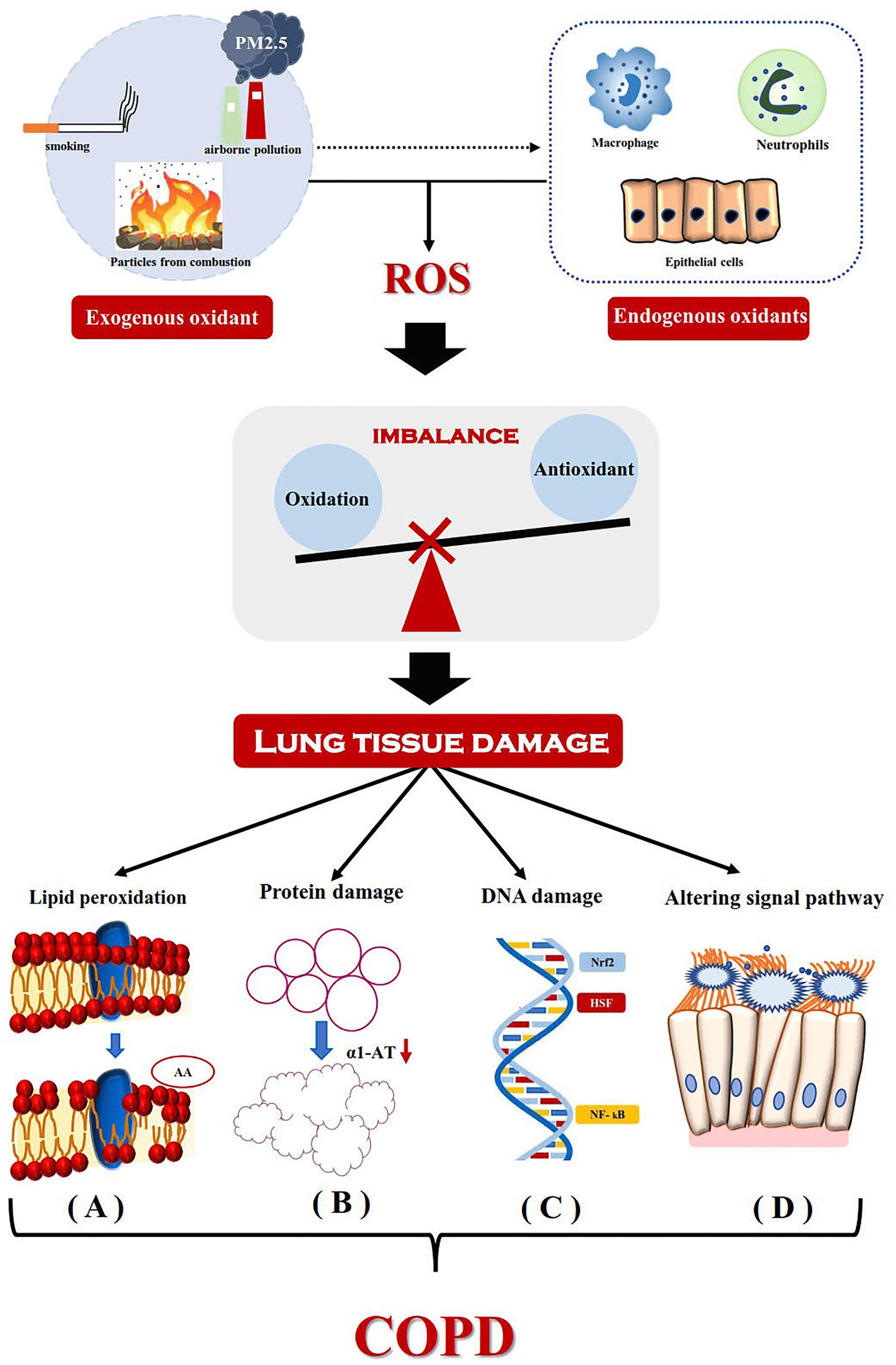
Figure 1. Mechanism for the development of COPD driven by oxidative stress. Exogenous oxidants from the environment and endogenous oxidants (released by activated neutrophils, alveolar macrophages and epithelial cells) lead to the production of reactive oxygen species and cause lung tissue damage. It mainly includes lipid peroxidation, Protein damage, DNA damage and Altering signal pathway. (A) Lipid peroxidation results in the hydrolysis of phospholipids in the cell membrane and changes in the structure and permeability of the cell membrane. Arachidonic acid produced by hydrolysis of phosphoric acid molecules, its metabolites thromboxane, prostaglandin E and leukotriene C4 are involved in the inflammatory process. (B) The inactivation of α1-AT breaks the balance between protease and anti-protease, which do great damage to elastin and eventually leads to emphysema. (C) Gene expression of inflammatory mediators induced by DNA damage. (D) Oxidative stress changes TNF-α-mediated signaling pathway, which leads to increased mucus secretion and ciliated cell dysfunction. ROS, reactive oxygen species; AA, arachidonic acid; α1-AT, alpha-1-antitrypsin; Nrf2, nuclear factor erythroid-2-related factor2; HSF, heat shock transcription factor; NF- kB, Nuclear Factor-κB.
Blood biomarkers are useful for the assessment of systemic properties of COPD (Faner et al., 2014). Compared to lung tissue samples, the risk in blood sampling is lower. In addition, sampling of blood samples can be easily repeated, which is beneficial for longitudinal monitoring of disease progression. Compared to other minimally invasive biological samples (such as urine or sputum), blood samples have a wider range of application, more repeatability, and are less time-consuming. These characteristics make blood sampling more practical in large-scale research (Regan et al., 2019).
Biomarker in Sputum
Methylation of P16 and GATA4 promoter in sputum samples was assumed to be an early biomarker of COPD (Sood et al., 2010; Meek et al., 2015). GATA4 is a transcription regulator for many cell cycle genes (Rojas et al., 2008). In humans, it is a crucial transcription factor for normal lung development (Ackerman et al., 2007). Moreover, methylation of GATA4 gene has been found to regulate oxidative stress and/or airway inflammation, which is related to the health status of patients with COPD (Yao and Rahman, 2011). Therefore, methylation of GATA4 gene in sputum may independently predict the health status of patients with COPD (Meek et al., 2015). P16 mediated cell cycle arrest and senescence (Takahashi et al., 2006), The absence of p16 activated the accumulation of Akt and cyclin D, upregulated IGF1 pathway, promoted the proliferation and regeneration of aecii, and finally effectively prevented the cigarette induced emphysema (Cottage et al., 2019). Of course, there are also some researchers who are opposed to the protective effect of p16, but the sample size of relevant studies is very small (only 3 cases), and the reliability of the results needs further study (Sundar et al., 2018).
The methylation pattern of tumor suppressor gene (TSG) promoter was studied in sputum samples of patients with COPD; they included cyclin dependent kinase inhibitor 2A (Berger and Bardeesy, 2007), MGMT (Belinsky et al., 2002), and CDH1 (Elloul et al., 2006). The extent of methylation in CDKN2A and MGMT was significantly higher. Induction of abnormal methylation in TSG in sputum samples could be a useful tool for early diagnosis of COPD (Guzmán et al., 2012).
Biomarker in Tracheal Epithelium and Lung Tissue
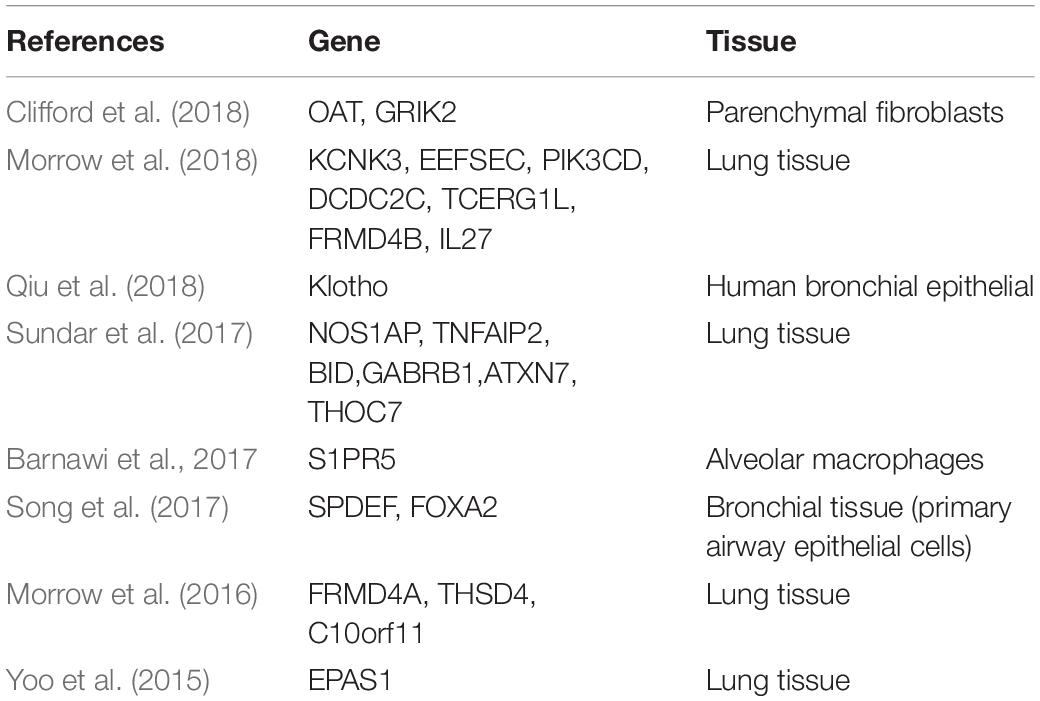
COPD is a heterogeneous inflammatory disease of the airways, alveolus, and microvasculature (Rabe and Watz, 2017). Since DNAm in lung tissue (Cheng et al., 2016; Morrow et al., 2016) and small airway tissue (Vucic et al., 2014) is closely related to COPD, tissue-specific DNAm may be considered to occur in COPD. Unlike the hypomethylation pattern of DNAm in blood (Qiu et al., 2012), most differentially methylated genes in COPD-associated small airway epithelia (SAE) are hypermethylated. Although collection of samples of airway and lung tissues is difficult, a large volume of literature exists regarding DNAm markers in tracheal epithelium and lung tissues (Table 1).
In view of the common genetic and epigenetic variation of COPD, some researchers follow the results of GWAS as a guide to prioritize the identified loci related to COPD (Morrow et al., 2018) with epigenetic annotation (Kundaje et al., 2015), and highlight the mechanisms related to complex traits. The genetic control of DNAm has been observed in a variety of complex traits and diseases (Olsson et al., 2014; Hannon et al., 2016; Bonder et al., 2017). Morrow et al. (2016) had analyzed the whole genome DNAm of homogenous lung tissue samples from patients with COPD. The differential methylation sites were integrated with previous genome-wide studies and four of them, namely CHRM1, DTX1, GLT1D1, and C10orf11, were focused upon in the study (Morrow et al., 2016). The CHRM1 is known to be associated with airway constriction, and a small candidate gene study had shown it to be associated with asthma (Maeda et al., 2006) and nicotine dependence (Lou et al., 2006). In addition, the role of other muscarinic receptor genes had also been previously reported in COPD (Cherubini et al., 2016).
In the lung parenchyma of patients with COPD, researchers had found differentially methylated genes to be closely related to the top typical pathways (such as β-γ signal or cancer mechanism), diseases and disorders (tissue damage and abnormality, cancer and respiratory system diseases), and molecular and cellular functions (Sundar et al., 2017). Genomic DNAm analysis had confirmed the changes in DNAm status of suggestive genes such as NOS1AP, TNFAIP2, BID, GABRB1, ATXN7, and THOC7 in COPD-lung tissue, and further verified the changes by pyrosequencing. DNAm was suggested to possibly play a key role in the regulation of gene expression related to molecular pathways and cellular processes in COPD (Yao and Rahman, 2011). Small airways are the main sites of airflow obstruction in patients with COPD; therefore, detection of markers in small-airway epithelium of patients with COPD could have important biological and clinical significance. Three pathways in SAE were found to have potential significance in the pathogenesis of COPD, namely phosphatase and tensin homolog (PTEN) signal, NF-E2-related factor 2 (Nrf2) mediated oxidative stress, and interleukin-17F(IL-17F) inflammatory response pathway (Vucic et al., 2014).
DNAm has a high level of cell-type specificity, there are some studies on DNAm of individual cell types in patients with COPD. Clifford et al. (2018) had studied the genomic DNAm of fibroblasts isolated from lung parenchyma and airway, and found DNAm to be related to differential gene expression only in the parenchymal fibroblasts. In addition, 359 individual differential CpG loci were found in the parenchymal fibroblasts. The methylation sites of OAT and GRIK2 genes significantly increased their expression in COPD cells (Clifford et al., 2018). Barnawi et al. (2017) had found phagocytosis of apoptotic cells by alveolar macrophages in patients with COPD to be controlled by epigenetic regulation, and decrease of methylation to possibly be the cause of increased S1PR5 gene expression in alveolar macrophages and of the defect in COPD-related efferent cells. Notch-mediated hypermethylation of Klotho in alveolar macrophages and airway epithelial cells inhibited the expression of Klotho and promoted inflammatory response and apoptosis in COPD (Qiu et al., 2018). Demethylation of NF-κ B-mediated pathway gene is related to the deterioration of COPD. However, TET1/2 plays an important role in the regulation of DNAm and production of cytokine/chemokine by NF-κ B, STAT3, IKK, and NIk genes in A549 cells (Kaur and Batra, 2020).
Goblet cell metaplasia is a common feature of COPD, which is related to mucus hypersecretion. SAM-pointed domain, a transcription factors containing ETS like factor (SPDEF) and forkhead box protein A2 (FOXA2), regulates the differentiation of goblet cells. Airway mucin 5AC, secreted by SPDEF, is aggravated the differentiation of goblet cells and mucin production (Park et al., 2007; Chen et al., 2009; Rajavelu et al., 2015), while FOXA2 effectively inhibited the differentiation of pulmonary goblet cells (Tang et al., 2013). Song et al. (2017) had found hypermethylation of CPG-8 in the promoter of SPDEF and hypomethylation of CpG-14 and CpG-15 in FOXA2, was identified the abnormal methylation of SPDEF and FOXA2 during the differentiation of goblet cells is the basic factor of mucus hypersecretion in COPD, thus providing a new approach to understand mucus hypersecretion from the perspective of epigenetics.
Some researchers had identified key regulatory factors in lung samples of patients with COPD by integrating functional genome, epigenetic data, and higher-order phenotype data (Yoo et al., 2015). Endothelial PAS domain protein 1 (EPAS1) was found to be the only key regulatory factor with a significant overlap of multiple gene sets related to COPD, and the protein level of EPAS1 was low in the lung tissue of patients with COPD (Yoo et al., 2015). EPAS1 is a hypoxia responsive transcription factor, also known as hypoxia inducible factor 2 α (HIF2A) (Ema et al., 1997; Tian et al., 1997). Its expression in lung and endothelial cells was higher than that of hypoxia inducible factor 1 α (HIF1A). The decrease of EPAS1 expression observed in COPD actually lead to inadaptability of hypoxia response (Kent et al., 2011). Therefore, it would be highly significant to understand the role and mechanism of EPAS1 in the treatment of diseases.
The above study found significant levels of abnormal DNAm to be different across lung tissues and cell types. It depends on the number of samples (Sundar et al., 2017), the integration method of gene expression (Yoo et al., 2015), and the integration scheme with GWAS results (Morrow et al., 2016).
Discussion
DNAm is obviously a potential biomarker for disease prevention, diagnosis, and prognosis. Owing to its reversibility, it has been widely considered as a biomarker and drug design target in COPD. While it has huge prospects in research, there are some limitations in the studies as well. First, the studies were limited by the type of sample: either they were conducted in blood, related to blood biomarkers of the disease (although the transformation of lung pathology was limited), or they are carried out in the lung tissue, where the cell type-specific methylation spectrum could hide the disease-related changes. Stueve et al. (2017) confirmed that DNA methylation from peripheral blood can serve as a surrogate marker for DNA methylation in lung tissue, which shows that it is feasible to search for cross tissue DNA methylation markers (especially in blood and lung tissue) in COPD patients. Moreover, mixed cell population in the whole tissue might complicate the study of the mechanism of cell type-specificity in subsequent diseases. Second, cross-sectional studies hindered the causal relationship between methylation changes and COPD status or lung function level. We are not sure whether these are prior to COPD or the result of COPD. Finally, the studies focused on the European population, with few methylation and transcriptome data from other populations, such as the Asians.
Nevertheless, DNAm as a potential biomarker of COPD, has a broad research prospect. First of all, COPD, as a non-neoplastic lesion, often occurs simultaneously with lung cancer (Tockman et al., 1987; Mannino et al., 2003). Hypermethylation of IL-12RBETA2 and WIF-1 has been found frequently in the transition from COPD to lung cancer (Suzuki et al., 2010). Therefore, COPD and lung cancer are suggested to be an epigenetic continuum characterized by the accumulation of methylation markers over time. In addition, large-scale methylation studies may help to reveal the details of epigenetic development of COPD into lung cancer, which would be of great significance to public health. Second, the discovery of CRISPR-Cas9 has promoted the development of epigenetic editing (Wu et al., 2018). Its advantages in targeting, safety, and preventing non-targeting effect make epigenetic editing, based on epigenetic markers, feasible. In addition, unlike genome editing, epigenetic editing does not lead to permanent changes in the genome. The core of its function is to correct abnormal epigenetic markers to affect the expression or function of specific genes. This could provide a very effective method for the treatment of COPD in future. It is also urgent to obtain dynamic and persistent information of epigenetic markers for different cell types.
Author Contributions
L-XH, Q-SH, and W-HL designed the manuscript. L-XH wrote the first draft of the manuscript. Z-HT, Q-SH, W-HL, and L-XH wrote specific sections of the manuscript. All authors contributed to the article and approved the submitted version.
Funding
We wish to thank the National Natural Science Foundation of China (Grant No. 81873204) and the National Key Research and Development Program of China (Grant No. 2017YFC1703304) for financial support.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We sincerely thank Prof. Chen Wei for his valuable comments on the manuscript. We would also like to thank Editage for English language editing too.
References
Ackerman, K. G., Wang, J., Luo, L., Fujiwara, Y., Orkin, S. H., and Beier, D. R. (2007). Gata4 is necessary for normal pulmonary lobar development. Am. J. Respir. Cell Mol. Biol. 36, 391–397. doi: 10.1165/rcmb.2006-0211RC
Barnawi, J., Jersmann, H., Haberberger, R., Hodge, S., and Meech, R. (2017). Reduced DNA methylation of sphingosine-1 phosphate receptor 5 in alveolar macrophages in COPD: a potential link to failed efferocytosis. Respirology 22, 315–321. doi: 10.1111/resp.12949
Belinsky, S. A., Palmisano, W. A., Gilliland, F. D., Crooks, L. A., Divine, K. K., Winters, S. A., et al. (2002). Aberrant promoter methylation in bronchial epithelium and sputum from current and former smokers. Cancer Res. 62, 2370–2377.
Berger, J. H., and Bardeesy, N. (2007). Modeling INK4/ARF tumor suppression in the mouse. Curr. Mol. Med. 7, 63–75. doi: 10.2174/156652407779940477
Bonder, M. J., Luijk, R., Zhernakova, D. V., Moed, M., Deelen, P., Vermaat, M., et al. (2017). Disease variants alter transcription factor levels and methylation of their binding sites. Nat. Genet. 49, 131–138. doi: 10.1038/ng.3721
Bowman, R. V., Wright, C. M., Davidson, M. R., Francis, S. M., Yang, I. A., and Fong, K. M. (2009). Epigenomic targets for the treatment of respiratory disease. Expert Opin. Ther. Targets 13, 625–640. doi: 10.1517/14728220902926119
Brode, S. K., Ling, S. C., and Chapman, K. R. (2012). Alpha-1 antitrypsin deficiency: a commonly overlooked cause of lung disease. CMAJ 184, 1365–1371. doi: 10.1503/cmaj.111749
Calverley, P. M., Anderson, J. A., Celli, B., Ferguson, G. T., Jenkins, C., Jones, P. W., et al. (2007). Salmeterol and fluticasone propionate and survival in chronic obstructive pulmonary disease. N. Engl. J. Med. 356, 775–789. doi: 10.1056/NEJMoa063070
Celli, B. R., and MacNee, W. (2004). Standards for the diagnosis and treatment of patients with COPD: a summary of the ATS/ERS position paper. Eur. Respir. J. 23, 932–946. doi: 10.1183/09031936.04.00014304
Chandel, N. S., and Schumacker, P. T. (2000). Cellular oxygen sensing by mitochondria: old questions, new insight. J. Appl. Physiol. 88, 1880–1889. doi: 10.1152/jappl.2000.88.5.1880
Chappell, S. L., Daly, L., Lotya, J., Alsaegh, A., Guetta-Baranes, T., Roca, J., et al. (2011). The role of IREB2 and transforming growth factor beta-1 genetic variants in COPD: a replication case-control study. BMC Med. Genet. 12:24. doi: 10.1186/1471-2350-12-24
Chen, G., Korfhagen, T. R., Xu, Y., Kitzmiller, J., Wert, S. E., Maeda, Y., et al. (2009). SPDEF is required for mouse pulmonary goblet cell differentiation and regulates a network of genes associated with mucus production. J. Clin. Invest. 119, 2914–2924. doi: 10.1172/jci39731
Chen, W., Brehm, J. M., Manichaikul, A., Cho, M. H., Boutaoui, N., Yan, Q., et al. (2015). A genome-wide association study of chronic obstructive pulmonary disease in Hispanics. Ann. Am. Thorac. Soc. 12, 340–348. doi: 10.1513/AnnalsATS.201408-380OC
Cheng, L., Liu, J., Li, B., Liu, S., Li, X., and Tu, H. (2016). cigarette smoke-induced hypermethylation of the GCLC gene is associated with COPD. Chest 149, 474–482. doi: 10.1378/chest.14-2309
Cherubini, E., Esposito, M. C., Scozzi, D., Terzo, F., Osman, G. A., Mariotta, S., et al. (2016). Genetic polymorphism of CHRM2 in COPD: clinical significance and therapeutic implications. J. Cell. Physiol. 231, 1745–1751. doi: 10.1002/jcp.25277
Cho, M. H., Boutaoui, N., Klanderman, B. J., Sylvia, J. S., Ziniti, J. P., Hersh, C. P., et al. (2010). Variants in FAM13A are associated with chronic obstructive pulmonary disease. Nat. Genet. 42, 200–202. doi: 10.1038/ng.535
Clifford, R. L., Fishbane, N., Patel, J., MacIsaac, J. L., McEwen, L. M., Fisher, A. J., et al. (2018). Altered DNA methylation is associated with aberrant gene expression in parenchymal but not airway fibroblasts isolated from individuals with COPD. Clin. Epigenet. 10:32. doi: 10.1186/s13148-018-0464-5
Cottage, C. T., Peterson, N., Kearley, J., Berlin, A., Xiong, X., Huntley, A., et al. (2019). Targeting p16-induced senescence prevents cigarette smoke-induced emphysema by promoting IGF1/Akt1 signaling in mice. Commun. Biol. 2:307. doi: 10.1038/s42003-019-0532-1
Crapo, J. D. (2019). Fifty years of the division of lung diseases and the evolution of pulmonary research and medicine. Chronic Obstr. Pulm. Dis. 6, 292–296. doi: 10.15326/jcopdf.6.4.2019.0160
Di Stefano, A., Maestrelli, P., Roggeri, A., Turato, G., Calabro, S., Potena, A., et al. (1994). Upregulation of adhesion molecules in the bronchial mucosa of subjects with chronic obstructive bronchitis. Am. J. Respir. Crit. Care Med. 149(3 Pt 1), 803–810. doi: 10.1164/ajrccm.149.3.7509705
Elloul, S., Silins, I., Tropé, C. G., Benshushan, A., Davidson, B., and Reich, R. (2006). Expression of E-cadherin transcriptional regulators in ovarian carcinoma. Virchows Archiv. 449, 520–528. doi: 10.1007/s00428-006-0274-6
Ema, M., Taya, S., Yokotani, N., Sogawa, K., Matsuda, Y., and Fujii-Kuriyama, Y. (1997). A novel bHLH-PAS factor with close sequence similarity to hypoxia-inducible factor 1alpha regulates the VEGF expression and is potentially involved in lung and vascular development. Proc. Natl. Acad. Sci. U. S. A. 94, 4273–4278. doi: 10.1073/pnas.94.9.4273
Faner, R., Tal-Singer, R., Riley, J. H., Celli, B., Vestbo, J., MacNee, W., et al. (2014). Lessons from ECLIPSE: a review of COPD biomarkers. Thorax 69, 666–672. doi: 10.1136/thoraxjnl-2013-204778
Fomenko, D. E., Xing, W., Adair, B. M., Thomas, D. J., and Gladyshev, V. N. (2007). High-throughput identification of catalytic redox-active cysteine residues. Science 315, 387–389. doi: 10.1126/science.1133114
Foreman, K. J., Marquez, N., Dolgert, A., Fukutaki, K., Fullman, N., McGaughey, M., et al. (2018). Forecasting life expectancy, years of life lost, and all-cause and cause-specific mortality for 250 causes of death: reference and alternative scenarios for 2016-40 for 195 countries and territories. Lancet 392, 2052–2090. doi: 10.1016/s0140-6736(18)31694-5
Franco, R., Schoneveld, O., Georgakilas, A. G., and Panayiotidis, M. I. (2008). Oxidative stress, DNA methylation and carcinogenesis. Cancer Lett. 266, 6–11. doi: 10.1016/j.canlet.2008.02.026
Franssen, F. M., Soriano, J. B., Roche, N., Bloomfield, P. H., Brusselle, G., Fabbri, L. M., et al. (2016). Lung function abnormalities in smokers with ischemic heart disease. Am. J. Respir. Crit. Care Med. 194, 568–576. doi: 10.1164/rccm.201512-2480OC
Gao, J., Aksoy, B. A., Dogrusoz, U., Dresdner, G., Gross, B., Sumer, S. O., et al. (2013). Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. signal. 6:pI1. doi: 10.1126/scisignal.2004088
GBD 2017 Disease and Injury Incidence and Prevalence Collaborators (2018). Global, regional, and national incidence, prevalence, and years lived with disability for 354 diseases and injuries for 195 countries and territories, 1990-2017: a systematic analysis for the global burden of disease study 2017. Lancet 392, 1789–1858. doi: 10.1016/s0140-6736(18)32279-7
Guzmán, L., Depix, M. S., Salinas, A. M., Roldán, R., Aguayo, F., Silva, A., et al. (2012). Analysis of aberrant methylation on promoter sequences of tumor suppressor genes and total DNA in sputum samples: a promising tool for early detection of COPD and lung cancer in smokers. Diagn. Pathol. 7:87. doi: 10.1186/1746-1596-7-87
Hancock, D. B., Eijgelsheim, M., Wilk, J. B., Gharib, S. A., Loehr, L. R., Marciante, K. D., et al. (2010). Meta-analyses of genome-wide association studies identify multiple loci associated with pulmonary function. Nat. Genet. 42, 45–52. doi: 10.1038/ng.500
Hannon, E., Dempster, E., Viana, J., Burrage, J., Smith, A. R., Macdonald, R., et al. (2016). An integrated genetic-epigenetic analysis of schizophrenia: evidence for co-localization of genetic associations and differential DNA methylation. Genome Biol. 17:176. doi: 10.1186/s13059-016-1041-x
Hardin, M., Zielinski, J., Wan, E. S., Hersh, C. P., Castaldi, P. J., Schwinder, E., et al. (2012). CHRNA3/5, IREB2, and ADCY2 are associated with severe chronic obstructive pulmonary disease in Poland. Am. J. Respir. Cell Mol. Biol. 47, 203–208. doi: 10.1165/rcmb.2012-0011OC
Heaney, L. G., and McGarvey, L. P. (2017). Personalised medicine for asthma and chronic obstructive pulmonary disease. Respiration 93, 153–161. doi: 10.1159/000455395
Hindorff, L. A., Sethupathy, P., Junkins, H. A., Ramos, E. M., Mehta, J. P., Collins, F. S., et al. (2009). Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc. Natl. Acad. Sci. U. S. A. 106, 9362–9367. doi: 10.1073/pnas.0903103106
Hitchler, M. J., and Domann, F. E. (2007). An epigenetic perspective on the free radical theory of development. Free Radic. Biol. Med. 43, 1023–1036. doi: 10.1016/j.freeradbiomed.2007.06.027
Hoppeler, H., Vogt, M., Weibel, E. R., and Flück, M. (2003). Response of skeletal muscle mitochondria to hypoxia. Exp. Physiol. 88, 109–119. doi: 10.1113/eph8802513
Horvath, S., and Raj, K. (2018). DNA methylation-based biomarkers and the epigenetic clock theory of ageing. Nat. Rev. Genet. 19, 371–384. doi: 10.1038/s41576-018-0004-3
Jackson, V. E., Ntalla, I., Sayers, I., Morris, R., Whincup, P., Casas, J. P., et al. (2016). Exome-wide analysis of rare coding variation identifies novel associations with COPD and airflow limitation in MOCS3, IFIT3 and SERPINA12. Thorax 71, 501–509. doi: 10.1136/thoraxjnl-2015-207876
Kaur, G., and Batra, S. (2020). Regulation of DNA methylation signatures on NF-κB and STAT3 pathway genes and TET activity in cigarette smoke extract-challenged cells/COPD exacerbation model in vitro. Cell biol. Toxicol. Online ahead of print doi: 10.1007/s10565-020-09522-8
Kent, B. D., Mitchell, P. D., and McNicholas, W. T. (2011). Hypoxemia in patients with COPD: cause, effects, and disease progression. Int. J. Chronic Obstr. Pulm. Dis. 6, 199–208. doi: 10.2147/copd.s10611
Kundaje, A., Meuleman, W., Ernst, J., Bilenky, M., Yen, A., Heravi-Moussavi, A., et al. (2015). Integrative analysis of 111 reference human epigenomes. Nature 518, 317–330. doi: 10.1038/nature14248
Lambrechts, D., Buysschaert, I., Zanen, P., Coolen, J., Lays, N., Cuppens, H., et al. (2010). The 15q24/25 susceptibility variant for lung cancer and chronic obstructive pulmonary disease is associated with emphysema. Am. J. Respir. Crit. Care Med. 181, 486–493. doi: 10.1164/rccm.200909-1364OC
Lee, M. K., Hong, Y., Kim, S. Y., Kim, W. J., and London, S. J. (2017). Epigenome-wide association study of chronic obstructive pulmonary disease and lung function in Koreans. Epigenomics 9, 971–984. doi: 10.2217/epi-2017-0002
Lou, X. Y., Ma, J. Z., Payne, T. J., Beuten, J., Crew, K. M., and Li, M. D. (2006). Gene-based analysis suggests association of the nicotinic acetylcholine receptor beta1 subunit (CHRNB1) and M1 muscarinic acetylcholine receptor (CHRM1) with vulnerability for nicotine dependence. Hum. Genet. 120, 381–389. doi: 10.1007/s00439-006-0229-7
Lowe, K. E., Regan, E. A., Anzueto, A., Austin, E., Austin, J. H. M., Beaty, T. H., et al. (2019). COPDGene(®) 2019: redefining the diagnosis of chronic obstructive pulmonary disease. Chronic Obstr. Pulm. Dis. 6, 384–399. doi: 10.15326/jcopdf.6.5.2019.0149
Luo, H., Zhao, Q., Wei, W., Zheng, L., Yi, S., Li, G., et al. (2020). Circulating tumor DNA methylation profiles enable early diagnosis, prognosis prediction, and screening for colorectal cancer. Sci. Transl. Med. 12:eaax7533. doi: 10.1126/scitranslmed.aax7533
Machin, M., Amaral, A. F., Wielscher, M., Rezwan, F. I., Imboden, M., Jarvelin, M. R., et al. (2017). Systematic review of lung function and COPD with peripheral blood DNA methylation in population based studies. BMC Pulm. Med. 17:54. doi: 10.1186/s12890-017-0397-3
Maeda, Y., Hizawa, N., Jinushi, E., Honda, A., Takahashi, D., Fukui, Y., et al. (2006). Polymorphisms in the muscarinic receptor 1 gene confer susceptibility to asthma in Japanese subjects. Am. J. Respir. Crit. Care Med. 174, 1119–1124. doi: 10.1164/rccm.200601-081OC
Mannino, D. M., Aguayo, S. M., Petty, T. L., and Redd, S. C. (2003). Low lung function and incident lung cancer in the united states: data from the first national health and nutrition examination survey follow-up. Arch. Intern. Med. 163, 1475–1480. doi: 10.1001/archinte.163.12.1475
Maurano, M. T., Humbert, R., Rynes, E., Thurman, R. E., Haugen, E., Wang, H., et al. (2012). Systematic localization of common disease-associated variation in regulatory DNA. Science 337, 1190–1195. doi: 10.1126/science.1222794
Meek, P. M., Sood, A., Petersen, H., Belinsky, S. A., and Tesfaigzi, Y. (2015). Epigenetic change (GATA-4 gene methylation) is associated with health status in chronic obstructive pulmonary disease. Biol. Res. Nurs. 17, 191–198. doi: 10.1177/1099800414538113
Miyashiro, M., Furuya, S., Fujishige, K., and Sugita, T. (2004). Highly sensitive cell-based assay system to monitor the sialyl Lewis X biosynthesis mediated by alpha1-3 fucosyltransferase-VII. Biochem. Biophys. Res. commun. 324, 98–107. doi: 10.1016/j.bbrc.2004.09.025
Morrow, J. D., Cho, M. H., Hersh, C. P., Pinto-Plata, V., Celli, B., Marchetti, N., et al. (2016). DNA methylation profiling in human lung tissue identifies genes associated with COPD. Epigenetics 11, 730–739. doi: 10.1080/15592294.2016.1226451
Morrow, J. D., Glass, K., Cho, M. H., Hersh, C. P., Pinto-Plata, V., Celli, B., et al. (2018). Human lung DNA methylation quantitative trait loci colocalize with chronic obstructive pulmonary disease genome-wide association loci. Am. J. Respir. Crit. Care Med. 197, 1275–1284. doi: 10.1164/rccm.201707-1434OC
Mroz, R. M., Holownia, A., Chyczewska, E., and Braszko, J. J. (2008). Chronic obstructive pulmonary disease: an update on nuclear signaling related to inflammation and anti-inflammatory treatment. J. Physiol. Pharm. 59(Suppl. 6), 35–42.
Mroz, R. M., Noparlik, J., Chyczewska, E., Braszko, J. J., and Holownia, A. (2007). Molecular basis of chronic inflammation in lung diseases: new therapeutic approach. J. Physiol. Pharm. 58(Suppl. 5), 453–460.
Olsson, A. H., Volkov, P., Bacos, K., Dayeh, T., Hall, E., Nilsson, E. A., et al. (2014). Genome-wide associations between genetic and epigenetic variation influence mRNA expression and insulin secretion in human pancreatic islets. PLoS Genet 10:e1004735. doi: 10.1371/journal.pgen.1004735
Pajares, M. A., Durán, C., Corrales, F., Pliego, M. M., and Mato, J. M. (1992). Modulation of rat liver S-adenosylmethionine synthetase activity by glutathione. J. Biol. Chem. 267, 17598–17605.
Park, K. S., Korfhagen, T. R., Bruno, M. D., Kitzmiller, J. A., Wan, H., Wert, S. E., et al. (2007). SPDEF regulates goblet cell hyperplasia in the airway epithelium. J. Clin. Invest. 117, 978–988. doi: 10.1172/jci29176
Pillai, S. G., Ge, D., Zhu, G., Kong, X., Shianna, K. V., Need, A. C., et al. (2009). A genome-wide association study in chronic obstructive pulmonary disease (COPD): identification of two major susceptibility loci. PLoS Genet. 5:e1000421. doi: 10.1371/journal.pgen.1000421
Ponce-Gallegos, M. A., Pérez-Rubio, G., García-Carmona, A., García-Gómez, J., Hernández-Zenteno, R., Ramírez-Venegas, A., et al. (2019). Haplotype in SERPINA1 (AAT) is associated with reduced risk for COPD in a mexican mestizo population. Int. J. Mol. Sci. 21:195 doi: 10.3390/ijms21010195
Qiu, J., Zhang, Y. N., Zheng, X., Zhang, P., Ma, G., and Tan, H. (2018). Notch promotes DNMT-mediated hypermethylation of Klotho leads to COPD-related inflammation. Exp. Lung Res. 44, 368–377. doi: 10.1080/01902148.2018.1556749
Qiu, W., Baccarelli, A., Carey, V. J., Boutaoui, N., Bacherman, H., Klanderman, B., et al. (2012). Variable DNA methylation is associated with chronic obstructive pulmonary disease and lung function. Am. J. Respir. Crit. Care Med. 185, 373–381. doi: 10.1164/rccm.201108-1382OC
Qiu, W., Wan, E., Morrow, J., Cho, M. H., Crapo, J. D., Silverman, E. K., et al. (2015). The impact of genetic variation and cigarette smoke on DNA methylation in current and former smokers from the COPDGene study. Epigenetics 10, 1064–1073. doi: 10.1080/15592294.2015.1106672
Rabe, K. F., and Watz, H. (2017). Chronic obstructive pulmonary disease. Lancet 389, 1931–1940. doi: 10.1016/s0140-6736(17)31222-9
Rajavelu, P., Chen, G., Xu, Y., Kitzmiller, J. A., Korfhagen, T. R., and Whitsett, J. A. (2015). Airway epithelial SPDEF integrates goblet cell differentiation and pulmonary Th2 inflammation. J. Clin. Invest. 125, 2021–2031. doi: 10.1172/jci79422
Regan, E. A., Hersh, C. P., Castaldi, P. J., DeMeo, D. L., Silverman, E. K., Crapo, J. D., et al. (2019). Omics and the search for blood biomarkers in chronic obstructive pulmonary disease. insights from COPDGene. Am. J. Respir. Cell Mol. Biol. 61, 143–149. doi: 10.1165/rcmb.2018-0245PS
Repapi, E., Sayers, I., Wain, L. V., Burton, P. R., Johnson, T., Obeidat, M., et al. (2010). Genome-wide association study identifies five loci associated with lung function. Nat. Genet. 42, 36–44. doi: 10.1038/ng.501
Rojas, A., Kong, S. W., Agarwal, P., Gilliss, B., Pu, W. T., and Black, B. L. (2008). GATA4 is a direct transcriptional activator of cyclin D2 and Cdk4 and is required for cardiomyocyte proliferation in anterior heart field-derived myocardium. Mol. Cell. Biol. 28, 5420–5431. doi: 10.1128/mcb.00717-08
Salahuddin, P. (2010). Genetic variants of alpha1-antitrypsin. Curr. Prot. Pept. Sci. 11, 101–117. doi: 10.2174/138920310790848368
Song, J., Heijink, I. H., Kistemaker, L. E. M., Reinders-Luinge, M., Kooistra, W., Noordhoek, J. A., et al. (2017). Aberrant DNA methylation and expression of SPDEF and FOXA2 in airway epithelium of patients with COPD. Clin. Epigenet. 9:42. doi: 10.1186/s13148-017-0341-7
Sood, A., Petersen, H., Blanchette, C. M., Meek, P., Picchi, M. A., Belinsky, S. A., et al. (2010). Wood smoke exposure and gene promoter methylation are associated with increased risk for COPD in smokers. Am. J. Respir. Crit. Care Med. 182, 1098–1104. doi: 10.1164/rccm.201002-0222OC
Stueve, T. R., Li, W. Q., Shi, J., Marconett, C. N., Zhang, T., Yang, C., et al. (2017). Epigenome-wide analysis of DNA methylation in lung tissue shows concordance with blood studies and identifies tobacco smoke-inducible enhancers. Hum. Mol. Genet. 26, 3014–3027. doi: 10.1093/hmg/ddx188
Sundar, I. K., Rashid, K., Gerloff, J., Li, D., and Rahman, I. (2018). Genetic ablation of p16(INK4a) does not protect against cellular senescence in mouse models of chronic obstructive pulmonary disease/emphysema. Am. J. Respir. Cell Mol. Biol. 59, 189–199. doi: 10.1165/rcmb.2017-0390OC
Sundar, I. K., Yin, Q., Baier, B. S., Yan, L., Mazur, W., Li, D., et al. (2017). DNA methylation profiling in peripheral lung tissues of smokers and patients with COPD. Clin. Epigenetics 9:38. doi: 10.1186/s13148-017-0335-5
Suzuki, M., Wada, H., Yoshino, M., Tian, L., Shigematsu, H., Suzuki, H., et al. (2010). Molecular characterization of chronic obstructive pulmonary disease-related non-small cell lung cancer through aberrant methylation and alterations of EGFR signaling. Ann. Surg. Oncol. 17, 878–888. doi: 10.1245/s10434-009-0739-3
Szulakowski, P., Crowther, A. J., Jiménez, L. A., Donaldson, K., Mayer, R., Leonard, T. B., et al. (2006). The effect of smoking on the transcriptional regulation of lung inflammation in patients with chronic obstructive pulmonary disease. Am. J. Respir. Crit. Care Med. 174, 41–50. doi: 10.1164/rccm.200505-725OC
Takahashi, A., Ohtani, N., Yamakoshi, K., Iida, S., Tahara, H., Nakayama, K., et al. (2006). Mitogenic signalling and the p16INK4a-Rb pathway cooperate to enforce irreversible cellular senescence. Nat. Cell Biol. 8, 1291–1297. doi: 10.1038/ncb1491
Tang, X., Liu, X. J., Tian, C., Su, Q., Lei, Y., Wu, Q., et al. (2013). Foxa2 regulates leukotrienes to inhibit Th2-mediated pulmonary inflammation. Am. J. Respir. Cell Mol. Biol. 49, 960–970. doi: 10.1165/rcmb.2013-0122OC
Tian, H., McKnight, S. L., and Russell, D. W. (1997). Endothelial PAS domain protein 1 (EPAS1), a transcription factor selectively expressed in endothelial cells. Genes Dev. 11, 72–82. doi: 10.1101/gad.11.1.72
Tockman, M. S., Anthonisen, N. R., Wright, E. C., and Donithan, M. G. (1987). Airways obstruction and the risk for lung cancer. Ann. Intern. Med. 106, 512–518. doi: 10.7326/0003-4819-106-4-512
Van Durme, Y. M., Eijgelsheim, M., Joos, G. F., Hofman, A., Uitterlinden, A. G., Brusselle, G. G., et al. (2010). Hedgehog-interacting protein is a COPD susceptibility gene: the rotterdam study. Eur. Respir. J. 36, 89–95. doi: 10.1183/09031936.00129509
Vucic, E. A., Chari, R., Thu, K. L., Wilson, I. M., Cotton, A. M., Kennett, J. Y., et al. (2014). DNA methylation is globally disrupted and associated with expression changes in chronic obstructive pulmonary disease small airways. Am. J. Respir. Cell Mol. Biol. 50, 912–922. doi: 10.1165/rcmb.2013-0304OC
Wu, D. D., Song, J., Bartel, S., Krauss-Etschmann, S., Rots, M. G., and Hylkema, M. N. (2018). The potential for targeted rewriting of epigenetic marks in COPD as a new therapeutic approach. Pharmacol. Ther. 182, 1–14. doi: 10.1016/j.pharmthera.2017.08.007
Yao, H., and Rahman, I. (2011). Current concepts on oxidative/carbonyl stress, inflammation and epigenetics in pathogenesis of chronic obstructive pulmonary disease. Toxicol. Appl. Pharmacol. 254, 72–85. doi: 10.1016/j.taap.2009.10.022
Yoo, S., Takikawa, S., Geraghty, P., Argmann, C., Campbell, J., Lin, L., et al. (2015). Integrative analysis of DNA methylation and gene expression data identifies EPAS1 as a key regulator of COPD. PLoS Genet. 11:e1004898. doi: 10.1371/journal.pgen.1004898
Young, R. P., Hopkins, R. J., Hay, B. A., Epton, M. J., Black, P. N., and Gamble, G. D. (2008). Lung cancer gene associated with COPD: triple whammy or possible confounding effect? Eur. Respir. J. 32, 1158–1164. doi: 10.1183/09031936.00093908
Young, R. P., Hopkins, R. J., Whittington, C. F., Hay, B. A., Epton, M. J., and Gamble, G. D. (2011). Individual and cumulative effects of GWAS susceptibility loci in lung cancer: associations after sub-phenotyping for COPD. PLoS One 6:e16476. doi: 10.1371/journal.pone.0016476
Young, R. P., Whittington, C. F., Hopkins, R. J., Hay, B. A., Epton, M. J., Black, P. N., et al. (2010). Chromosome 4q31 locus in COPD is also associated with lung cancer. Eur. Respir. J. 36, 1375–1382. doi: 10.1183/09031936.00033310
Zinellu, A., Fois, A. G., Sotgia, S., Sotgiu, E., Zinellu, E., Bifulco, F., et al. (2016a). Arginines plasma concentration and oxidative stress in mild to moderate COPD. PLoS One 11:e0160237. doi: 10.1371/journal.pone.0160237
Zinellu, A., Fois, A. G., Sotgia, S., Zinellu, E., Bifulco, F., Pintus, G., et al. (2016b). Plasma protein thiols: an early marker of oxidative stress in asthma and chronic obstructive pulmonary disease. Eur. J. Clin. Invest. 46, 181–188. doi: 10.1111/eci.12582
Zinellu, A., Sotgiu, E., Fois, A. G., Zinellu, E., Sotgia, S., Ena, S., et al. (2017c). Blood global DNA methylation is decreased in non-severe chronic obstructive pulmonary disease (COPD) patients. Pulm. Pharmacol. Ther. 46, 11–15. doi: 10.1016/j.pupt.2017.08.006
Keywords: DNA methylation, COPD, biomarker, tissue specificity, lung cancer
Citation: He L-X, Tang Z-H, Huang Q-S and Li W-H (2020) DNA Methylation: A Potential Biomarker of Chronic Obstructive Pulmonary Disease. Front. Cell Dev. Biol. 8:585. doi: 10.3389/fcell.2020.00585
Received: 08 May 2020; Accepted: 16 June 2020;
Published: 07 July 2020.
Edited by:
Liang Cheng, Harbin Medical University, ChinaReviewed by:
Caixia Li, Chengdu Institute of Biology (CAS), ChinaBin Wu, Chongqing Hospital of Traditional Chinese Medicine, China
Ming Liu, West China Fourth Hospital of Sichuan University, China
Copyright © 2020 He, Tang, Huang and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wei-Hong Li, bHdoQGNkdXRjbS5lZHUuY24=; Qing-Song Huang, aHFzMTI2MUAxMjYuY29t
 Lin-Xi He
Lin-Xi He Zhao-Hui Tang1
Zhao-Hui Tang1 Wei-Hong Li
Wei-Hong Li