- 1Department of Biology, Trent University, Peterborough, ON, Canada
- 2Institut Lumière Matière, CNRS UMR 5306, Université Claude Bernard Lyon 1, Université de Lyon, Lyon, France
Cytokinins (CKs) are a diverse group of evolutionarily significant growth-regulating molecules. While the CK biosynthesis and signal transduction pathways are the most well-understood in plant systems, these molecules have been identified in all kingdoms of life. This review follows the recent discovery of an expanded CK profile in the social amoeba, Dictyostelium discoideum. A comprehensive review on the present knowledge of CK biosynthesis, signal transduction, and CK-small molecule interactions within members of Dictyostelia will be summarized. In doing so, the utility of social amoebae will be highlighted as a model system for studying the evolution of these hormone-like signaling agents, which will set the stage for future research in this area.
Introduction
Cytokinin Roles in Plants and Beyond
The cytokinins (CKs) encompass a group of evolutionarily significant molecules, most well known for their roles in signaling, where they orchestrate all levels of plant growth and development (Mok and Mok, 2001). Chemically, these molecules are N6 adenine derivatives, and they exhibit a broad phylogenetic occurrence from bacteria to humans sharing both common and diverging roles, such as growth promotion and virulence, among others (Figure 1A; Akiyoshi et al., 1984; Golovko et al., 2000; see reviews by Spíchal, 2012 and Kabbara et al., 2018). Recent genetic and molecular analyses have led to an explosion of research on the pleiotropic effects of these hormones in plant species (see reviews Kieber and Schaller, 2014; Durán-Medina et al., 2017). The early and simple documented roles of CKs at the single-cell level, such as the promotion of cell growth and differentiation, have now expanded to more complex roles involving whole-plant organization and beyond, such as: shoot initiation, leaf senescence, vascular and embryonic development, and nutrient uptake, among others (Kieber and Schaller, 2014). In concert, the elucidation of key elements in CK biosynthesis, perception, and signal transduction pathways have been pivotal to the understanding of these hormones on a molecular level.
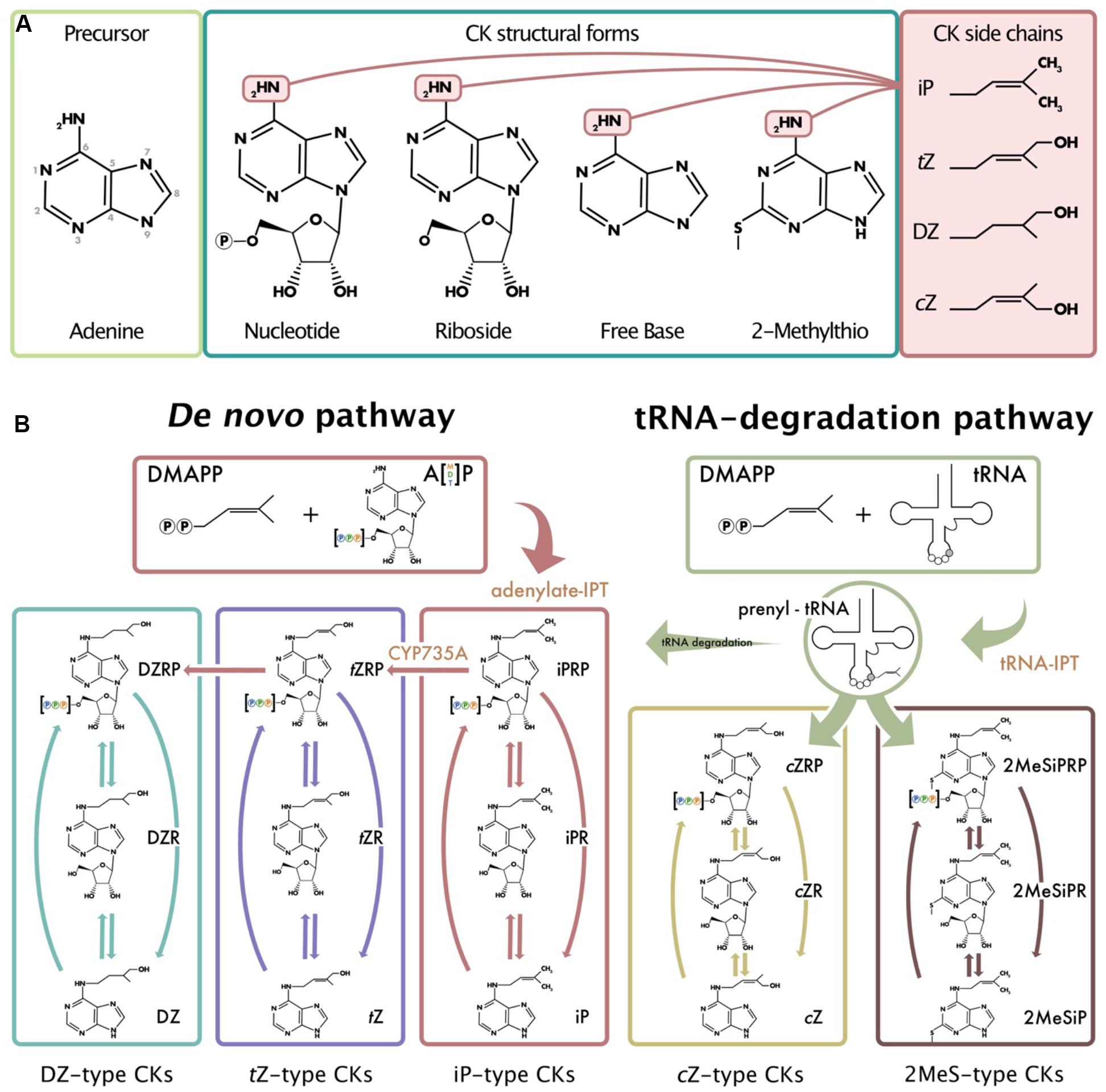
Figure 1. Common isoprenoid cytokinin (CK) structures and model of isoprenoid CK biosynthesis derived from plant, fungi, and bacterial models. (A) Depicts the numbering system used in CK nomenclature, the common CK structural forms, and the various isoprenoid side chains (iP, tZ, DZ, and cZ) that can be attached to the N6 position of the adenine. (B) Depicts the two different pathways from which isoprenoid CKs can be formed: the methylerythritol phosphate pathway (de novo pathway) and the mevalonate pathway (tRNA-degradation pathway). Arrows are representative of the various enzymes that convert between the different CK types and structural derivatives. Orange font indicates specific enzymes. The 2MeS-type CKs shown as a product of tRNA degradation all possess 2MeSiP-type side chains; however, 2MeStZ- and 2MeScZ-type are also possible. Note that not all CK structural forms and enzymes responsible for CK biosynthesis and metabolism are depicted (e.g., glucosides or cytokinin oxidase, etc.). Adapted from Kamada-Nobusada and Sakakibara (2009), Spíchal (2012), Morrison et al. (2015, 2017).
In plants, biosynthesis of the most abundant CK type, the isoprenoid-type CKs, occurs through two different pathways: the methylerythritol phosphate pathway (MEP; de novo pathway) and the mevalonate pathway (MVA; tRNA-degradation pathway) (Figure 1B). Isoprenoid- (iP), trans-zeatin- (tZ), and dihydrozeatin-type CKs (DZ) are predominately derived from the de novo biosynthesis pathway through adenylate-isopentenyl transferases (IPTs) (Sakakibara, 2006). In contrast to the de novo pathway, the MVA or tRNA-degradation pathway is responsible for the production of cis-zeatin-type CKs (cZ) (Figure 1; Miyawaki et al., 2006). In fungi and mammals, there is evidence that iP- and methyl-thiolated-type (2MeS-) CKs are also derived from the tRNA degradation pathway (Morrison et al., 2017; Seegobin et al., 2018). In both pathways, IPTs facilitate N-prenylation of the adenosine molecule at the N6 terminus (Figure 1B) (Kakimoto, 2001; Takei et al., 2001; Sakakibara, 2006). When the isoprenoid donor dimethylallyl pyrophosphate (DMAPP) acts with IPT, isopentenyladenine-type (iP-type) CKs are formed. From iP-type CKs, other CKs can be generated (e.g., tZ, DZ) through modifications of the side chain. Specifically, adenylate-IPTs catalyze the transfer of the isoprenoid moiety to adenine through the rate limiting reaction to form iP nucleotides (either mono-, di-, or tri-phosphate; iPRPs). Therefore, the production of all other isoprenoid CK types is dependent upon the initial presence of IPT to facilitate iPRP production. In the case of tRNA-IPTs, prenylation occurs on tRNA molecules at position A37, and upon degradation, the tRNA-derived CKs contribute to the pool of unbound CKs in the organism (Gajdosová et al., 2011). From plant CK biosynthesis pathways, it is well-known that there are various CK types, and they are distinguished by their characteristic side-chain attachments at the N6 position of the adenine (e.g., iP or tZ) (Figure 1A; Kamada-Nobusada and Sakakibara, 2009). Within each CK-type, there are various structural derivatives or forms that exist which determine the level of biological activity within the organism, such as: free bases, ribosides and nucleotides, and conjugates with glucose, xylose, or amino acid residues (Figure 1A). Generally, the nucleotide forms are considered inactive precursors from which the more biologically active riboside and free base forms can be created.
Recently, an increasing amount of attention has been placed on uncovering the roles of CKs outside of the plant kingdom. In fact, these signaling molecules and or components of their biosynthesis pathways are present in organisms of all kingdoms. There is a possibility that CKs have roles in the growth and development of all organisms that produce them. Owing to their ubiquitous presence in all kingdoms, CKs are viewed as primary candidates for the study of interkingdom, hormone-like, signaling molecules (Kabbara et al., 2018). Recent evidence involving CKs opens many new doors for research beyond the plant kingdom. The human pathogen, Mycobacterium tuberculosis, secretes CKs that induce transcriptional changes affecting both the metabolome and staining properties of M. tuberculosis (Samanovic et al., 2018). These findings clearly demonstrate strong evidence of the involvement of CKs as signaling molecules on an interkingdom level. A phytopathogenic bacterial receptor was identified revealing a conserved mechanism by which bacteria can respond to CK in order to defend themselves against their host innate immune response (Wang et al., 2017; Chen et al., 2019). Moreover, a possible role for CKs in animal-microbiota relationships with implications for human health was explored in a recent review (Chanclud and Lacombe, 2017). In the apicomplexan parasite, Toxoplasma gondii, CKs regulate growth, the cell cycle, and apicoplast proliferation (which affects the viability of the parasite) (Andrabi et al., 2018). The thermophilic archaeon, Sulfolobus islandicus REY15A, was the first archaeal species identified to possess a CK-activating enzyme, lonely guy (LOG) (Mayaka et al., 2019). Furthermore, the detection of six additional CK forms, beyond the one previously identified (N6-isopentenyladenine-9-riboside), were reported in mammalian tissues (Seegobin et al., 2018). Among these various examples of CK production or activity in non-plant biota, there is a common theme of CKs exhibiting biological functions in many kingdoms of life. Exogenous application of CKs on mammalian cancer cell lines show that CKs have anticancer activity (Voller et al., 2010). Moreover, CKs affect growth and germination in the social amoeba, Dictyostelium discoideum and in the biotrophic fungus, Claviceps purpurea (Hinsch et al., 2015; Aoki et al., 2019). In the coming years, it is likely that we will continue to see increasing roles of CKs beyond the plant kingdom as research continues to use organisms from all kingdoms of life.
The Dictyostelia
The Dictyostelids are soil-dwelling, amoeboid protozoans belonging to the Amoebozoa phylum (Raper, 1984; Romeralo et al., 2011). These eukaryotes are often referred to as social amoebae, owing to their unique life cycle, which consists of two mutually exclusive states: vegetative growth (single-celled amoebae) and development (multicellular organism) (Figure 2). Individual amoeboid cells grow asexually and divide mitotically, feeding upon soil bacteria and/or decaying leaf litter until resources are depleted. Starvation triggers the developmental program through the secretion of a chemical messenger, cAMP, which acts as a chemoattractant initiating the migration of neighboring amoebae (i.e., chemotaxis) to form an aggregate of cells (Konijn et al., 1967). These aggregates are collectively known as mounds. Tens of thousands of aggregated cells then undergo morphogenesis to form a multicellular pseudoplasmodium (slug), which migrates toward light and warmth (see review by Schaap, 2011). Cells within the slug terminally differentiate into either stalk or spores to form the final life cycle stage, a fruiting body, which consists of specialized stalk cells and a droplet of spores that sits atop the stalk (Schilde and Schaap, 2013; Loomis, 2014).
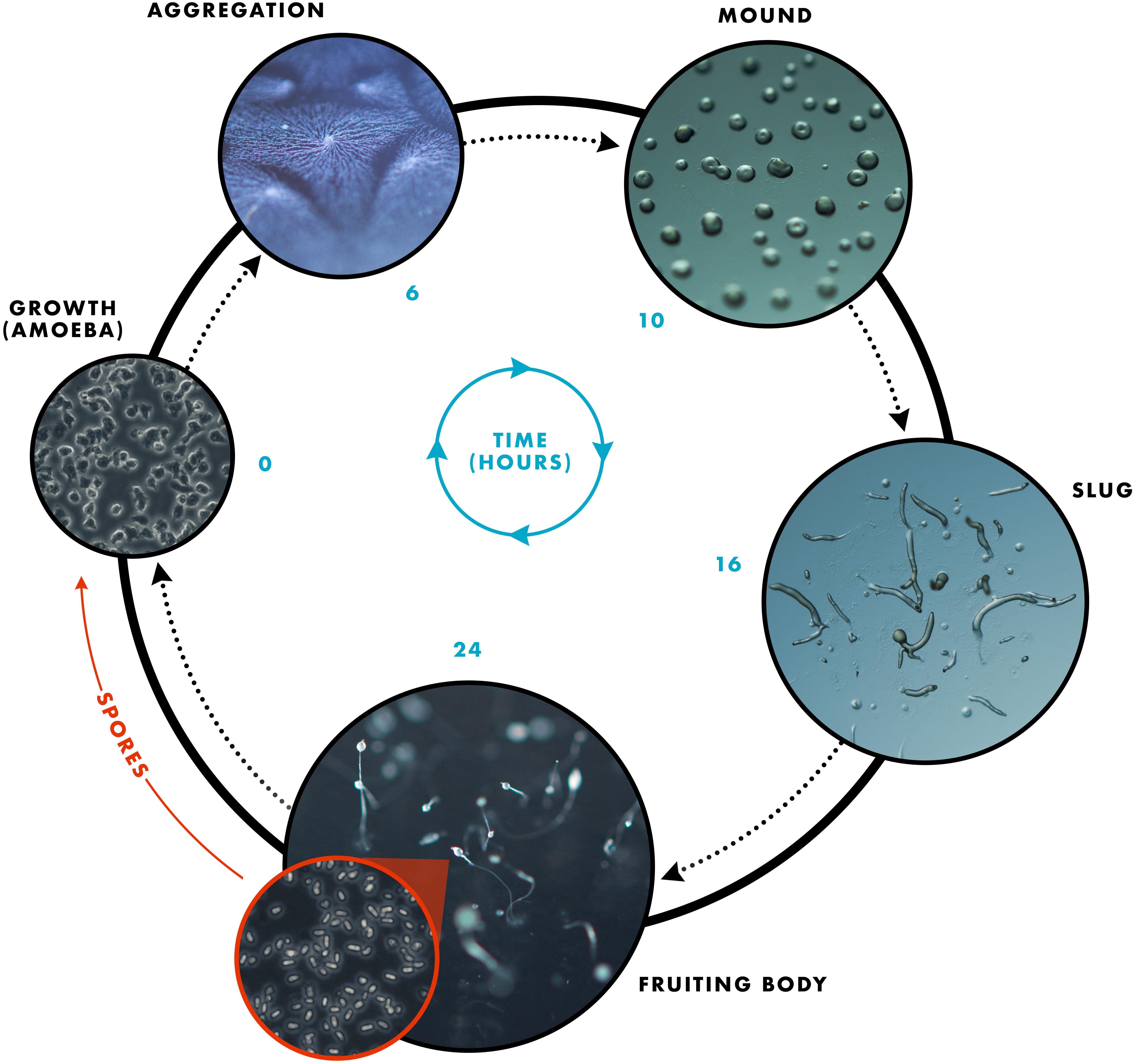
Figure 2. The D. discoideum life cycle showing the transition from a single-celled amoeba to a multicellular organism. Upon starvation, the developmental program is initiated in which single-celled amoeba aggregate toward a central location, as depicted in the aggregation image. The cells continue to aggregate to form the mound structure, which occurs after approximately 10 h starvation. Transition from the mound to the slug life cycle stage takes approximately 6–8 h. Finally, culmination generates a fruiting body consisting of a slender stalk and mass of spores that forms 24 h after the onset of starvation. Each spore gives rise to a single amoeba upon germination when food resources are available.
The most well-known organism of the Amoebozoa phylum is D. discoideum, hereafter referred to as D. discoideum. The genome of D. discoideum was the first free-living protozoan to be sequenced (Eichinger et al., 2005). Prior to sequencing, over five decades of intensive research on this social amoeba led to increased understanding of various cellular processes, such as chemotaxis and differentiation. Uniquely situated at the juncture of plants and animals, sharing many traits between the two kingdoms, the Dictyostelids offer a matchless platform to assess a wide variety of cellular and developmental processes. In light of the unique life cycle of Dictyostelid species, paired with its position in phylogeny and possession of CK biosynthetic and signal transduction components, this review will highlight how Dictyostelids can be used to study the role of CKs beyond the plant kingdom from an evolutionary perspective.
Cytokinins in Dictyostelia
Overview
The first articles published on the presence of CK in Dictyostelids involved the discovery of a novel CK in D. discoideum, known as discadenine (DA) (Obata et al., 1973; Tanaka et al., 1975; Abe et al., 1976). DA was discovered in the spore mass and was characterized as a potent inhibitor of spore germination. A notable discovery involving CKs in Dictyostelids by Taya et al. (1978a) revealed the existence of a CK biosynthetic pathway unrelated to the tRNA CK degradation pathway. Moreover, 5′-AMP was shown to be the acceptor molecule for the isopentenyl group forming the precursor molecule, isopentenyl adenine nucleotide (iPRP). Collectively, this early work in Dictyostelids laid the foundation for the CK biosynthesis pathway to be mapped in Arabidopsis thaliana (see reviews by Sakakibara, 2006; Kamada-Nobusada and Sakakibara, 2009). Following the initial discovery of an alternate biosynthetic pathway in Dictyostelids, the presence of N6-isopentenyladenine (iP) was identified at the start of fruiting body formation, also known as culmination, and iP was shown to be the precursor molecule to DA (Tanaka et al., 1978). Further analysis revealed a developmental regulation of CK in D. discoideum, as both iP and DA were detected following the onset of culmination (Ihara et al., 1980). This early evidence of the developmental role of CK in D. discoideum was later confirmed and expanded upon by Anjard and Loomis (2008). More recently, a comprehensive scan of 30 potential CKs in D. discoideum revealed that 6 different CKs are synthesized and secreted during growth, development, and germination (Aoki et al., 2019). Total levels of CK production were highest in the fruiting body and during germination, followed by aggregation and single-celled growth. Interestingly, iP-type CKs were the dominant CK forms during single-celled growth and aggregation, and DA was not present during these early life cycle stages. We detected high levels of DA following the onset of culmination, which is consistent with previous research (Ihara et al., 1980; Aoki et al., 2019). Exogenous application of iP affected single-celled growth by prolonging the stationary phase of cultures (Aoki et al., 2019). Together, these results indicate a greater role for CKs during the D. discoideum life cycle. The focus of this review will be on CK biosynthesis, metabolism, signal transduction, and CK-small molecule interactions in D. discoideum. We will also compare this information to what is known in other CK-producing organisms to provide an evolutionary context for CK function.
Cytokinin Biosynthesis and Metabolism
iptA – Adenylate Isopentenyltransferase
In D. discoideum, there are three identified IPT genes, only one of which has been functionally characterized (Anjard and Loomis, 2008). A phylogenic analysis of the corresponding proteins revealed that the D. discoideum genome encodes one adenylate-IPT and two tRNA-IPTs (Anjard and Loomis, 2008). The adenylate-IPT gene, denoted iptA, encodes a 283 amino acid, 32 kDa protein (IptA, DDB0233672) (Anjard and Loomis, 2008). IptA is a developmentally regulated protein, and its activity peaks during the late culmination stage of D. discoideum development (Figure 3; Ihara et al., 1980, 1984; Rot et al., 2009). Purification of IptA reveals that it is highly unstable and loses most of its activity after one day (Ihara et al., 1984). In terms of substrate specificity, 5′-AMP is the most effective substrate for IptA followed by ADP, which is 60–80% as effective. ATP, adenine, and adenosine are not substrates for IptA in D. discoideum (Ihara et al., 1984). The activity of D. discoideum IptA is dependent upon divalent metal cations (Zn2+, Mg2+, Mn2+), which is consistent with other prenyl-transfer reactions (Durbecq et al., 2001). The Km values of D. discoideum IptA, under optimum conditions (pH 7.0, 1 mM Zn2+, and 25°C) for both 5′-AMP and isopentenylpyrophosphate are 1.0 × 10–7 M and 2.2 × 10–6 M, respectively (Ihara et al., 1984). While the role of IptA in D. discoideum is consistent with plants and other CK-producing biota, the aforementioned properties of the enzyme (Km, substrate specificity, etc.) are variable among CK-producing organisms, such as A. thaliana, and the plant-pathogenic bacterium, Agrobacterium tumefaciens (Kakimoto, 2001; Sugawara et al., 2008). For instance, the adenylate-IPT from D. discoideum and A. tumefaciens use AMP as the prenyl acceptor molecule, while A. thaliana adenylate-IPTs preferentially use ADP or ATP (Taya et al., 1978a; Kakimoto, 2001).
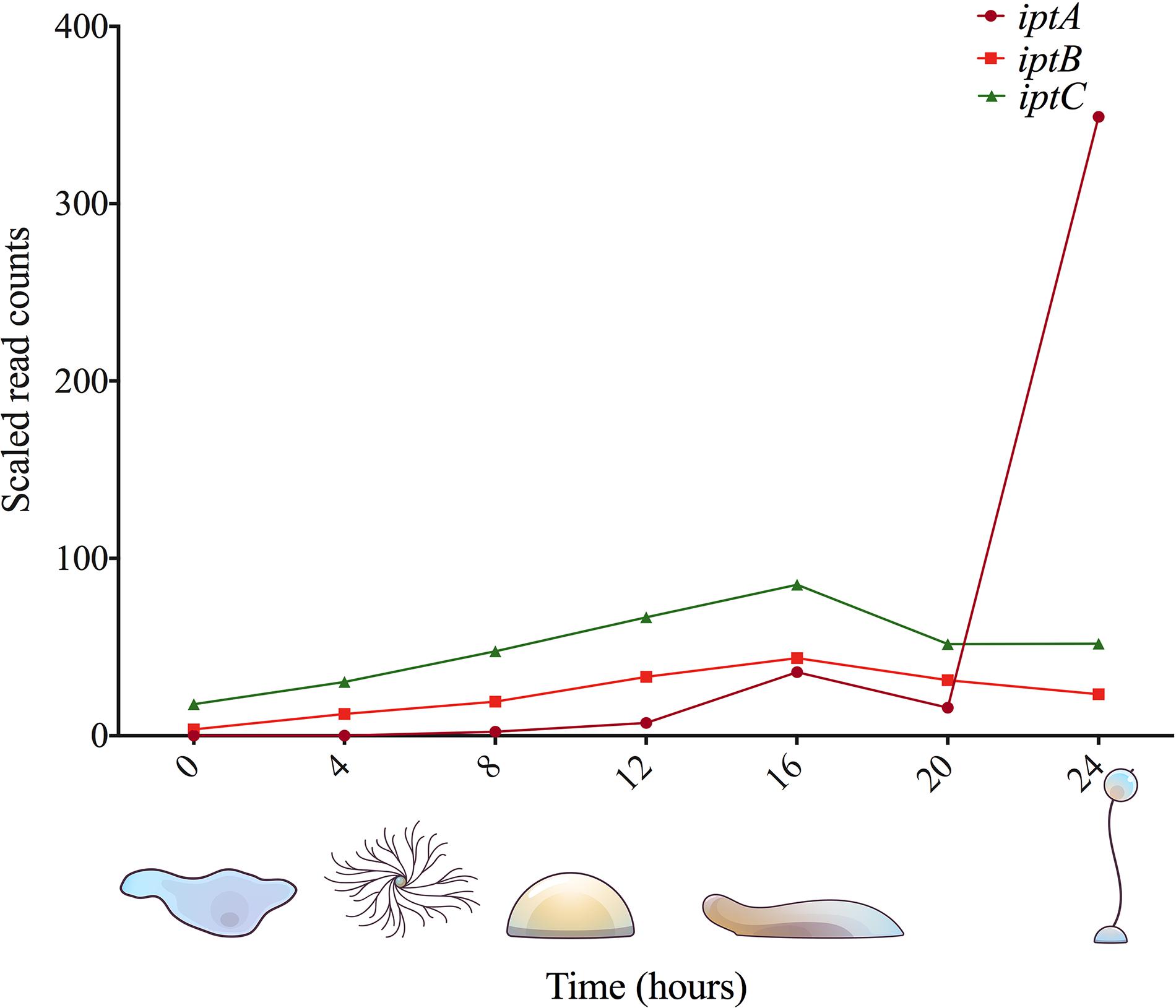
Figure 3. Gene expression analysis of known and putative CK biosynthesis genes in D. discoideum. RNA-Seq data was obtained from dictyExpress (www.dictyexpress.biolab.si) (Rot et al., 2009).
Disruption of iptA, through homologous recombination in the AX4 parental strain of D. discoideum, reduced total CK production and impaired spore viability (Anjard and Loomis, 2008). The rate of CK accumulation in iptA– cells was 90% less than that observed in wild-type cells when developed from vegetative cells on filters over a 30-h period (CK production was assessed every 2 h starting from 20 to 30 h). Developing iptA– cells with wild-type cells resulted in improved sporulation, indicating a non-cell autonomous phenotype (Anjard and Loomis, 2008). A threshold concentration of CK was calculated to determine the amount of endogenous CK necessary to fully induce spore formation, which was 10 nM. iptA– cells took 30 h to reach this concentration, roughly 6–8 h longer than wild-type cells. As a result, these findings support the conclusion that CKs play a significant role in sporulation in D. discoideum (Anjard and Loomis, 2008).
Although IptA is responsible for catalyzing the reaction for the synthesis of isopentenyl adenine-type (iP-type) CKs, organisms do not usually display a full complement of CK forms and this likely reflects the different pathways present and relative enzyme activities therein. Thus far, six CK forms have been identified in D. discoideum: cis-Zeatin (cZ), DA, iP, N6-isopentenyladenine-9-riboside (iPR), N6-isopentenyladenine-9-riboside-5′ phosphate (iPRP), and 2-methylthio-N6-isopentenyladenine (2MeSiP) (Figure 4; Abe et al., 1976; Tanaka et al., 1978; Taya et al., 1978a; Aoki et al., 2019). Of these identified CK forms, the most well-studied are iP and DA; DA will be discussed at greater lengths in a following section entitled discadenine sythase. The biosynthesis of iP-type CKs in D. discoideum appears to be similar to other CK-producing organisms, as the traditional CK forms of iP (nucleotide, riboside, and free base) have been identified throughout the life cycle (Aoki et al., 2019). Of the iP-type CKs identified in D. discoideum, the free base fraction, iP, is the most prevalent CK form detected (Anjard and Loomis, 2008; Aoki et al., 2019). Other than higher plants, only a handful of other CK-producing species contain adenylate-IPT genes: the plant pathogenic bacteria, A. tumefaciens (Akiyoshi et al., 1984) and Rhodococcus fascians (Crespi et al., 1992); the land moss, Physcomitrella patens (Lindner et al., 2014); the cyanobacterium, Nostoc sp. (Frébortová et al., 2015); and the social amoebae, D. discoideum and Dictyostelium purpureum (Anjard and Loomis, 2008; Sucgang et al., 20111).
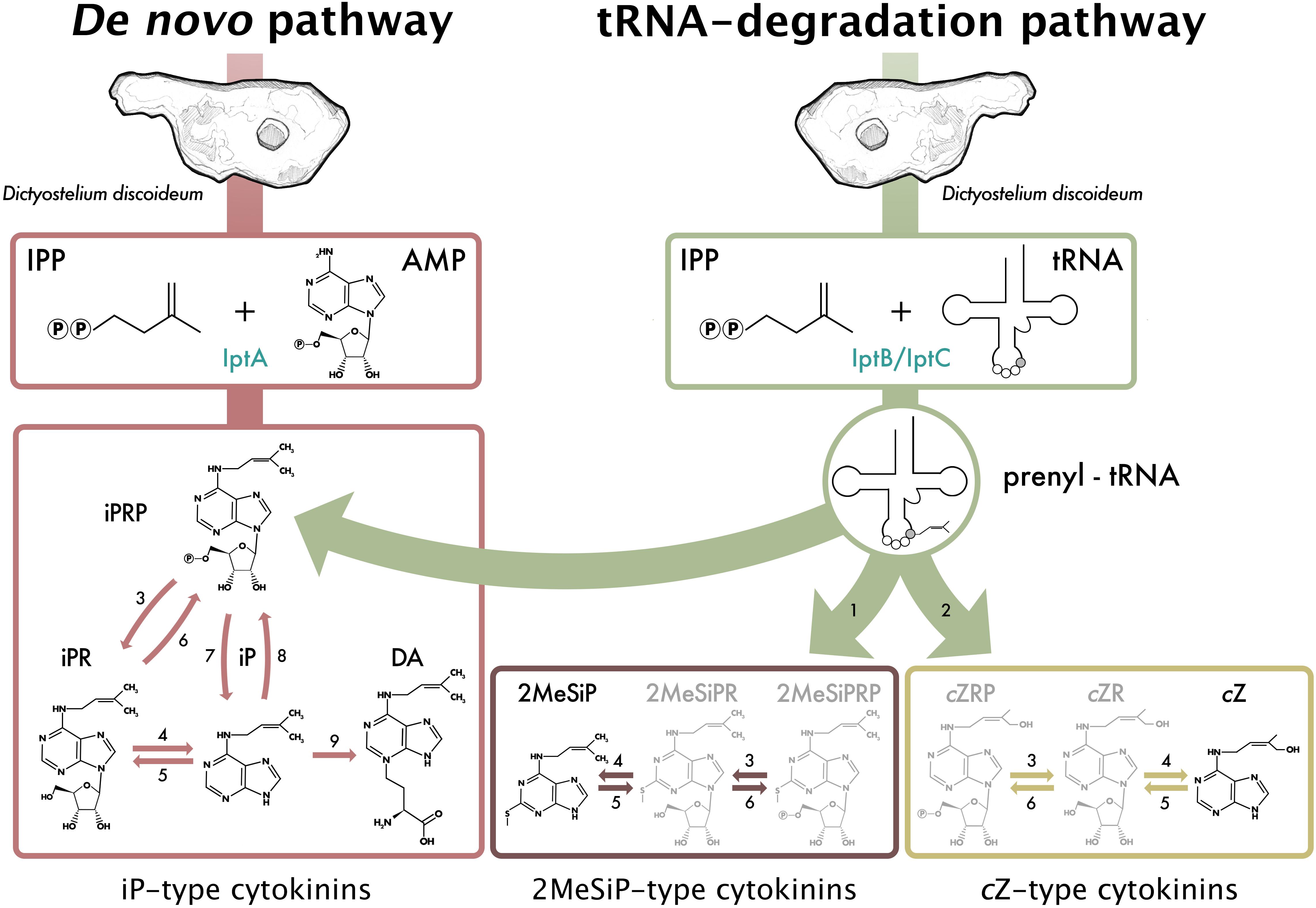
Figure 4. Proposed cytokinin (CK) biosynthesis pathway based on the CKs detected throughout all stages of the D. discoideum life cycle. Black CK structures denote CKs that have been previously detected in D. discoideum, and grayed CK structures are indicative of common CK forms detected in other CK-producing biosynthetic pathways. Numbers represent known or inferred enzymes from previous D. discoideum, plant, fungi, and bacteria studies and are as follows: 1. cdk5rap1-like ortholog (DDB_G0287079); 2. cis-hydroxylase; 3. 50-ribonucleotide phosphohydrolase; 4. adenosine nucleosidase; 5. purine nucleoside phosphorylase; 6. adenosine kinase; 7. CK phosphoribohydrolase (LOG-like ortholog, DDB_G0281309); 8. adenine phosphoribosyltransferase; 9. discadenine synthase (Taya et al., 1978a, b; Anjard and Loomis, 2008; Kamada-Nobusada and Sakakibara, 2009; Spíchal, 2012; Morrison et al., 2017; Nishii et al., 2018). Reproduced with permission from Aoki et al. (2019).
iptB and iptC – Putative tRNA Isopentenyltransferases
In D. discoideum, there are two putative tRNA-IPTs: iptB and iptC. In the commonly used AX3 and AX4 strains of D. discoideum, there is a large duplication on chromosome 2, which affects iptC resulting in two copies of this gene, denoted iptC-1 and iptC-2 (Bloomfield et al., 2008). From phylogenic analyses, IptB and IptC cluster with tRNA-IPTs that are closely related to bacterial and eukaryotic tRNA-IPTs, respectively; however, there has been no functional characterization to date (Anjard and Loomis, 2008; Lindner et al., 2014; Nishii et al., 2018). Despite tRNA-IPTs being the most abundant IPT-type conserved across kingdoms of life, much less is known about it compared to adenylate-IPTs in terms of CK biosynthesis (Schäfer et al., 2015). iptB encodes a predicted 522 amino acid, 61 kDa protein (IptB, DDB0233673), and iptC encodes a predicted 413 amino acid, 48 kDa protein (IptC, DDB0233671). While these proteins have not been previously studied, it is likely that IptB and IptC contribute to the collective pool of CKs in D. discoideum, as the disruption of iptA leads to a drastic reduction of CK levels, but not a complete elimination of CK biosynthesis (Anjard and Loomis, 2008). In support of this idea, the recent CK profiles of D. discoideum follow a similar trend to what is found in CK-producing organisms with dominant tRNA-IPT CK pathways, possessing mostly iP-type CKs (Figure 4; Aoki et al., 2019). Therefore, the peak in expression at the 16-h stage of development (slug stage) could coincide with preparing the cells for the encapsulation of spores, where tRNA-bound CKs could be degraded to contribute to the pool of free CKs required for encapsulation at the 20-h time point (Figure 31). In D. discoideum, there is little known about tRNA modification. Total tRNA transcript abundance does not increase during development, but tRNA modifications do (Palatnik et al., 1977; Rosengarten et al., 2017). The peak in expression of iptB and iptC throughout development aligns with these studies, which further support the putative roles of tRNA modification assigned to both of these gene products (Figure 3).
Beyond the role of CK biosynthesis, tRNA-IPTs are involved in many other functions, such as translation fidelity (in yeast and humans), in vitro growth and gene expression (in bacteria), and tRNA-gene mediated silencing (tgm) and drug resistance under environmental stress (in yeast), among others (Ericson and Bjork, 1986; Tolerico et al., 1999; Spinola et al., 2005; Suzuki et al., 2012; Pratt-Hyatt et al., 2013; Yu et al., 2017; see review by Dabravolski, 2020). Dabravolski (2020) summarizes that the roles of tRNA-IPTs can be broken up into different categories: (1) tRNA-isopentenylation-related, (2) tRNA-isopentenylation-unrelated, and (3) CK production upon tRNA degradation. Interestingly, a secondary zinc finger domain in addition to the IPP transferase domain (Pfam: IPPT) is required for the tgm role of certain tRNA-IPTs (Dabravolski, 2020 and references therein). IptB contains both of these domains, but IptC does not. Therefore, perhaps IptB has a secondary role involving tgm. With this in mind, more work with iptB and iptC knockout cell lines is necessary to determine the extent to which tRNA-IPTs contribute to CK production in D. discoideum and to other roles.
Discadenine Synthase
Unlike both adenylate-IPTs and tRNA-IPTs, discadenine synthase (or synthetase) is unique to members of Dictyostelia (Taya et al., 1980; Abe et al., 1981). DA synthase is responsible for catalyzing the reaction that creates the novel CK and potent spore germination inhibitor, DA (Taya et al., 1978b; Ihara et al., 1980, 1986; Abe et al., 1981). Specifically, DA is synthesized through direct transfer of the 3-amino-3-carboxy-propyl moiety of S-adenosylmethionine (SAM) to iP (Figure 5; Taya et al., 1978b). While the gene encoding DA synthase remains unidentified, various properties of the protein have been studied in D. discoideum (Taya et al., 1978b; Ihara et al., 1980, 1986). Protein BLAST and HMMER searches in D. discoideum using known 3-amino-3-carboxypropyl transferases from other organisms, such as tRNA-uridine aminocarboxylpropyl transferase in Escherichia coli, to identify potential DA synthase gene candidates reveal no promising candidates.
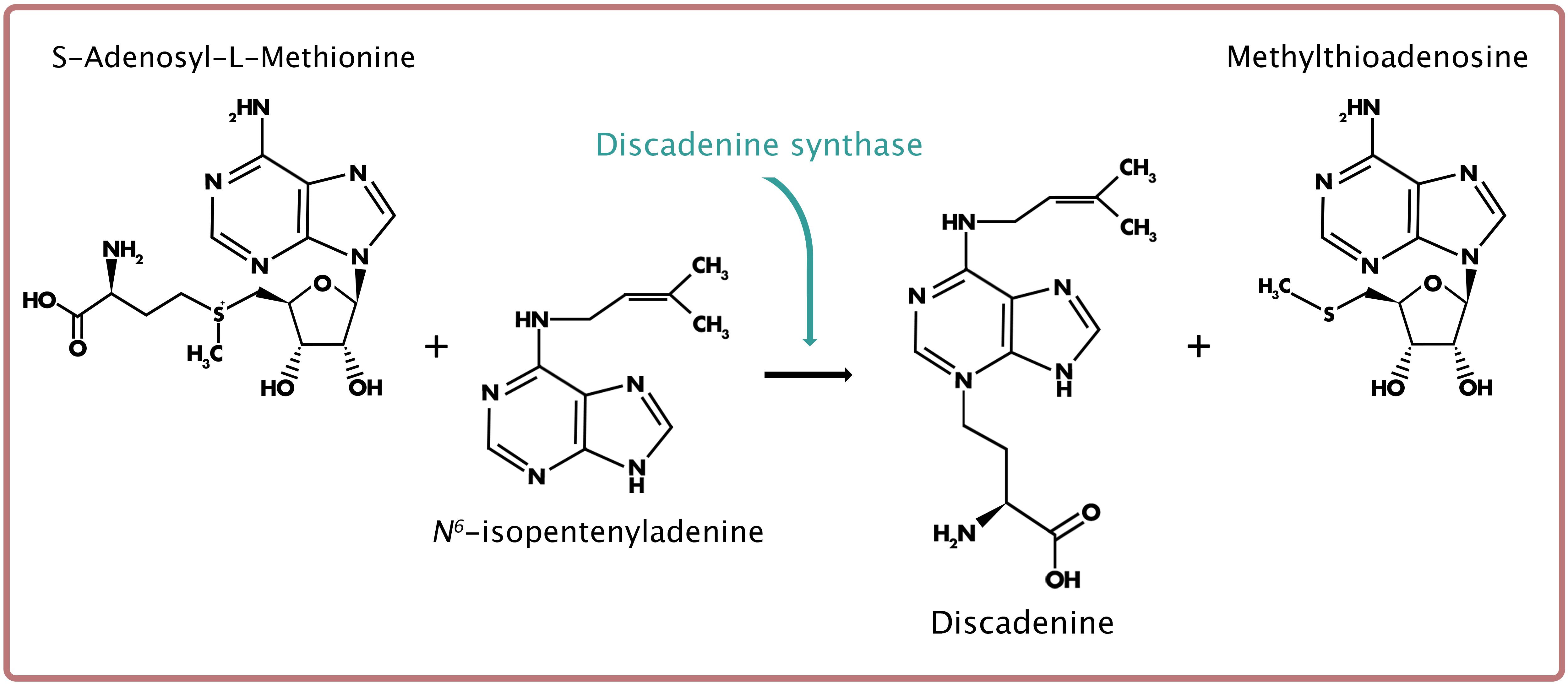
Figure 5. Reaction scheme for discadenine (DA) biosynthesis, derived from N6- isopentenyladenine (iP) and catalyzed by discadenine synthase.
The activity of DA synthase increases during D. discoideum development (Ihara et al., 1980). The protein is active earlier than IptA and continually increases in activity following aggregation, which includes a brief hold in activity after 16-h of development (slug stage), followed by a major peak at 26 h (post-fruiting body development) and a final smaller peak at 31 h. Considering that iP is the precursor to DA, it is puzzling that the activity of IptA peaks 10 h later than DA synthase (Ihara et al., 1980). The sharp rise in IptA activity near the onset of sporulation may indicate that large amounts of iP are rapidly converted to DA allowing for the precipitous encapsulation and dormancy of spores. In support of the incongruent enzymatic activities of IptA and DA synthase, similar findings were observed in D. discoideum between the developmentally regulated enzymes UDP-glucose epimerase and UDP-galactose polysaccharide transferase (Telser and Sussman, 1971).
The CK produced through DA synthase is unlike all other known naturally occurring CKs. Structurally, DA is an adenine derivative and is recognized as the first natural purine to possess an α-amino acid residue on theN3 position of the adenine (Figure 5; Nomura et al., 1977). Unlike its precursor, iP, DA does not promote cell proliferation (at concentrations between 1 nM and 1 μM), nor does it prolong the stationary phase of single-celled growth (Aoki et al., 2019). However, when tested using the classical cytokinin assay (i.e., tobacco callus cell division bioassay), DA exhibits CK activity at concentrations tested between 0.5 and 16 μM (Nomura et al., 1977). More rigorous testing of DA as a CK was performed using several classical cytokinin and ligand-binding assays designed to assess CK activity in other organisms (Mik et al., 2017). The CK-like activity of DA was confirmed through the tobacco callus (cell division), Amaranthus (anthocyanin production), and detached wheat leaf (senescence) bioassays, and the activities were compared to that of a known potent, aromatic CK, benzyladenine (BA). The tobacco callus bioassay demonstrated most clearly the CK activity of DA, as the proliferation of CK-dependent callus cells was stimulated in a dose-dependent manner (at concentrations between 1 nM and 100 μM). Interestingly, the same concentration at which BA exerted cytotoxicity (10 μM) did not affect callus cell growth for cells treated with 10 μM DA. In fact, DA-treated callus cells continued to proliferate at this concentration. While N3-substituted CK derivatives (i.e., N3-methylated BA) have been shown to reduce CK activity, the N3-α-amino acid residue in DA was shown to decrease the cytotoxicity of the molecule compared to BA. As further confirmation of the CK-like activity, DA reduced the binding of isotopically labeled tZ in the A. thaliana receptors, AHK3 and CRE1/AHK4 (Mik et al., 2017). Of the two receptors, a higher affinity of DA occurred for CRE1/AHK4. Moreover, in a bacterial receptor assay, where the activation of the A. thaliana CK receptors results in the expression of the β-galactosidase reporter gene, DA elicited expression in both receptors, also in a dose-dependent manner (Mik et al., 2017). Therefore, DA possesses growth-promoting properties as a CK (in other organisms), as well as growth-inhibiting properties as an inhibitor of spore germination (in D. discoideum).
The proposed activities for DA may reflect a specific functionality within certain members of the Dictyostelium genus. A correlation between acrasins, or chemotactic agents, and spore germination inhibitors was noted between various Dictyostelid species (Taya et al., 1980; Abe et al., 1981). Of the 6 Dictyostelid species tested in these studies, DA and DA synthase were detected in only three species (namely D. discoideum, D. purpureum, and Dictyostelium mucoroides). These three species were the only species tested that use cAMP as their chemotactic agent for aggregation. Furthermore, inhibition of spore germination was achieved through treatment with 2 μM DA in these same three species, but not in the other three tested Dictyostelid species that utilize other chemotactic agents besides cAMP for aggregation (Dictyostelium minutum, Dictyostelium lacteum, and Polysphondylium violaceum). Based on these findings, it was postulated that spore germination inhibitors, including DA, may be both biochemically and evolutionarily linked with the chemotactic agent (Taya et al., 1980). This link with cAMP and DA will be discussed further in the CK-small molecule functional interactions section.
LOG – CK Phosphoribohydrolase or Lonely Guy
Cytokinin phosphoribohydrolase (LOG) is responsible for the direct conversion of CK nucleotides into their most biologically active forms, the free bases (Kurakawa et al., 2007). This enzyme was first discovered in rice (Oryza sativa) where a loss of function mutant resulted in decreased floral organs and flowers with only one stamen; accordingly, it was wryly named “lonely guy” (LOG). Since this discovery, LOG homologs have been characterized in other plant species as well as bacteria, fungi, and archaea (Kuroha et al., 2009; Hinsch et al., 2015; Samanovic and Darwin, 2015; Seo and Kim, 2017; Mayaka et al., 2019; Moramarco et al., 2019). A Protein BLAST search revealed a putative LOG homolog (DDB_G0281309) in D. discoideum that presents strong homology with known LOG genes from plants and bacteria (Figure 6A). Interestingly, there are two peaks in the expression of the putative LOG gene: during vegetative growth and late development, both of which coincide with a time when CK accumulation and biological effects have been observed in D. discoideum (Figure 6B; Anjard and Loomis, 2008; Rot et al., 2009; Aoki et al., 2019). LOG knockout and overexpression analyses in A. thaliana reveal pleiotropic effects on plant growth and development, and ultimately conclude that LOG plays a fundamental role in the regulation of CK across all developmental stages of A. thaliana (Kuroha et al., 2009). Considering the widespread occurrence of LOG genes among various kingdoms, it is highly likely this uncharacterized gene has phosphoribohydrolase activity in D. discoideum.
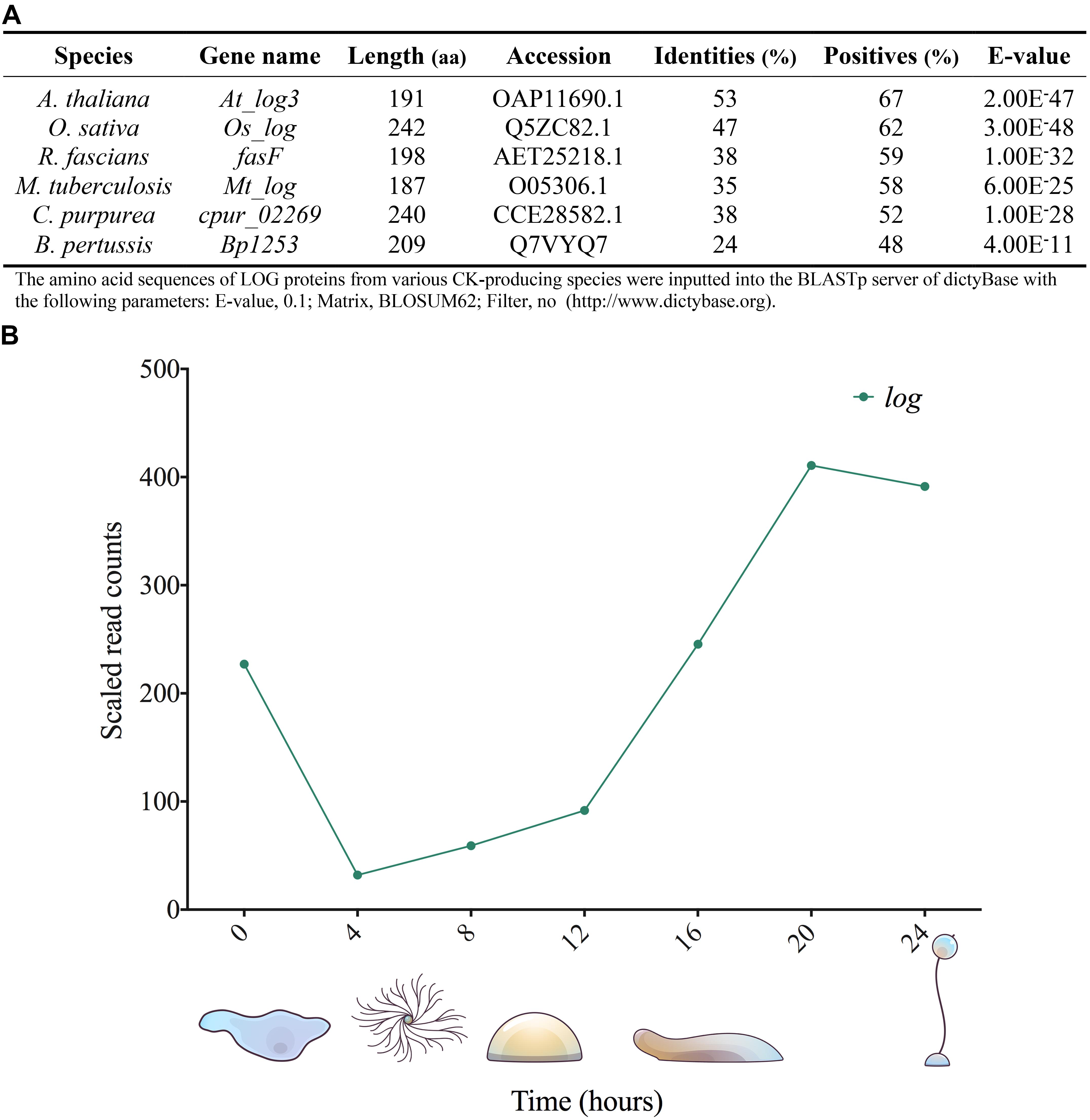
Figure 6. Dictyostelium discoideum has a putative log gene (DDB_G0281309). (A) presents LOG homologs from several organisms. Protein BLASTs were performed using the dictyBase BLAST server to assess sequence similarity of LOG homologs to the protein product of the putative log gene, DDB_G0281309 (http://www.dictybase.org). (B) Gene expression profile of the putative log gene using RNA-Seq data obtained from dictyExpress (www.dictyexpress.biolab.si) (Rot et al., 2009).
CKX – Cytokinin Oxidase/Dehydrogenase
Cytokinin oxidase/dehydrogenase (CKX) inactivates CKs through oxidative cleavage of the N6 side chain from the adenine ring (Frébort et al., 2011). While CKX-activity has been identified primarily in plant species, similar enzymatic activities have been demonstrated in both D. discoideum and Saccharomyces cerevisiae (Van Kast and Laten, 1987; Armstrong and Firtel, 1989). An enzyme with CKX-like activity was assayed by Armstrong and Firtel (1989) throughout all stages of D. discoideum growth and development. There was a peak in enzyme activity from growth to aggregation and then a steady decline throughout the remainder of D. discoideum development. The purified enzyme catalyzed the cleavage of the N6 side chain from iP to adenine, and for this reason, was discussed as being similar to CKX in higher plants. However, Protein BLAST searches for both D. discoideum and S. cerevisiae do not reveal CKX-related sequences. Considering this point, Schmülling et al. (2003) speculates that CK breakdown via CKX is not conserved across all CK-producing organisms. In fact, the radish plant, Raphanus sativus, inactivates CKs solely through N-conjugation (Parker and Letham, 1973). The regulatory enzymes responsible for CK deactivation through N- or O-conjugation with a sugar moiety, commonly glucose, are known as uridine diphosphate glycosyltransferases (Šmehilová et al., 2016). A brief search on dictyBase revealed five characterized glucosyltransferases, four of which contained annotations for N-linked glucosylation. In our previous CK profiling experiments, we did not detect CK-glucosides at any stage of the D. discoideum life cycle; however, we did not search all possible conjugate alternatives (Aoki et al., 2019). It is likely that the observed CKX-like activity in D. discoideum occurs through a non-CK specific degrading enzyme, such as one of the mentioned D. discoideum glucosyltransferases. Further work is necessary to determine (1) if D. discoideum produces any CK-conjugates and (2) if so, what enzyme is responsible for CK deactivation.
CK Secretion and Translocation
Like most hormones, CKs are synthesized intracellularly before being secreted into the extracellular space. Transporter proteins play a key role in the inter- and intracellular distribution of CKs. This topic has yet to be explored in D. discoideum, so we will draw on what is known in plant systems. In A. thaliana, the transmembrane ABC transporter, ABCG14, is responsible for the active transport of CKs from the roots to the shoots via the xylem (Ko et al., 2014; Zhang et al., 2014). This ATPase transporter localizes to the plasma membrane of root cells, and its inactivation prevents the translocation of CKs. Experiments using radioactively labeled CKs show that AtABCG14 acts as a CK exporter rather than an importer (Zhang et al., 2014). Potential regulation of CK secretion at the level of the ABC transporter is possible but has not been investigated. The AtABCG14 protein is characterized as a half-transporter, composed of both an ATPase binding domain and a transmembrane domain. To be functional, half-transporters must associate with another polypeptide containing both a binding domain and a transmembrane domain to form a homo- or heterodimer. Alternatively, genes encoding full ABCG transporters, with all four domains (two binding domains and two transmembrane domains) residing on a single polypeptide, are found in plants but not animals. Clear homologs of AtABCG14 are found across the plant kingdom and likely play a similar role in CK transport. The D. discoideum genome possesses a wide variety of ABC transporters encompassing 71 different genes, which includes 24 for the G family (8 half transporters and 16 full transporters) (Anjard et al., 2002; see also update in http://www.dictybase.org).
A sequence comparison shows that AtABCG14 presents strong homology with members of the D. discoideum ABCG family, especially DdABCG22. A null mutant of DdABCG22 was generated during a systematic study of ABC transporters (Miranda et al., 2013). This null strain presented delayed development and reduced spore viability – a phenotype that would be expected in the case of impaired CK secretion. However, the CK production of this strain was not evaluated. Furthermore, DdABCG22 was also found to influence vegetative cell dispersion during a screening for mutants reverting the dispersive phenotype of the ami8-mutant (Nagasaki and Uyeda, 2008). As CKs were recently shown to have a role during vegetative cell stage, such results are not unexpected (Aoki et al., 2019). Therefore, assessing CK production and secretion in the DdABCG22 null strain would give insight into whether this protein is involved in active transport of CKs or if its phenotype is unrelated to CK.
Two other protein families play a dominant role in CK transport in plants – the purine uptake permease (PUP) transporter family and the equilibrative nucleoside transporter (ENT) family. Certain members of the PUP transporter family allow for CK import into plant cells, specifically CK-nucleotides (Liu et al., 2019). The CK-specific PUP permeases appear to have evolved during terrestrial plant colonization between the bryophytes and the lycophytes from the pre-existing nucleotide sugar permease precursor (Jelesko, 2012). Thus, it is not surprising that homologs of PUP permease cannot be found in the D. discoideum genome. However, we cannot exclude the existence of a permease with similar function that would have evolved independently within Dictyostelids. The ENT family of transporters are also responsible for CK import into plant cells – specifically CK-ribosides (Liu et al., 2019). Out of the four ENT gene products in rice, one, OsENT2 was shown to transport CK-ribosides, as well as adenosine (Hirose et al., 2005). D. discoideum possesses three uncharacterized ENT genes similar to OsENT2 but also to other ENTs, making it impossible to predict their substrate specificity without further experimentation (Table 1).

Table 1. Sequence similarity of three uncharacterized equilibrative nucleoside transporter (ENT) proteins in D. discoideum to the known cytokinin transporter protein, OsENT2, in Oryza sativa.
CK Signal Transduction
The canonical CK signal transduction pathway in plants involves a multi-step phosphorelay system that interacts through a complex form of the two-component signaling (TCS) pathway (Hwang and Sheen, 2001). TCS pathways are dominant in prokaryotes, especially bacteria, where they comprise the basic stimulus-response regulatory network allowing organisms to sense and respond to nearly all environmental stimuli (Stock et al., 2000). It is important to note that not all CK-producing organisms that respond to CK possess TCS elements. This is the case in several mammalian studies where CKs act as agonists to the adenosine receptors A2A and A3 (Blad et al., 2011; Lee et al., 2012; Lappas, 2015). However, the remainder of this section will focus on components of the TCS CK signaling pathway, as are present in D. discoideum. Classically, the TCS pathway involves a histidine kinase (HK), which acts as the receptor, and a response regulator protein (RR), which, once activated, elicits a specific response through downstream effectors. This is slightly modified in land plants, where the CK signal transduction pathway involves a series of sequential phosphorylation events that alternate between His and Asp residues initiated by HKs, are perpetuated by histidine phosphotransfer proteins (HPs), and finished by RRs (Kieber and Schaller, 2018). A conserved extracellular loop of the HK transmembrane receptors belonging to the cyclase/histidine kinase-associated sensory extracellular (CHASE) domain-containing HK family is responsible for the initiation of CK signaling (Anantharaman and Aravind, 2001). Interestingly, the number of reported CK-producing organisms that possess conserved CHASE domains in recent years has expanded and will be described below (Kabbara et al., 2018).
DhkA and AcgA
In D. discoideum, there are two CHASE-domain containing proteins that are both involved in encystation and sporulation – the HK, DhkA, and the adenylyl cyclase of germination stage protein, AcgA (Alvarez-Curto et al., 2007; Anjard and Loomis, 2008). Neither of these proteins appear to be a CK receptor in D. discoideum, as null mutants for both proteins have a normal response to CK treatment (Anjard and Loomis, 2008). However, a dhkA and acgA double knockout has yet to be tested for definitive confirmation that neither of these proteins function as the CK receptor in D. discoideum. While CK was shown to act independently of DhkA, DhkA is still thought to regulate spore germination (Anjard and Loomis, 2008). In pre-spore cells, the peptide, spore differentiation factor 2 (SDF-2), binds to the CHASE domain in DhkA, which leads to the dephosphorylation of the cAMP phosphodiesterase, RegA, resulting in its inactivation. In this same time frame, the CK signaling pathway in D. discoideum (discussed below) converges to facilitate spore formation by inducing cAMP production (Figure 7; Anjard and Loomis, 2008).
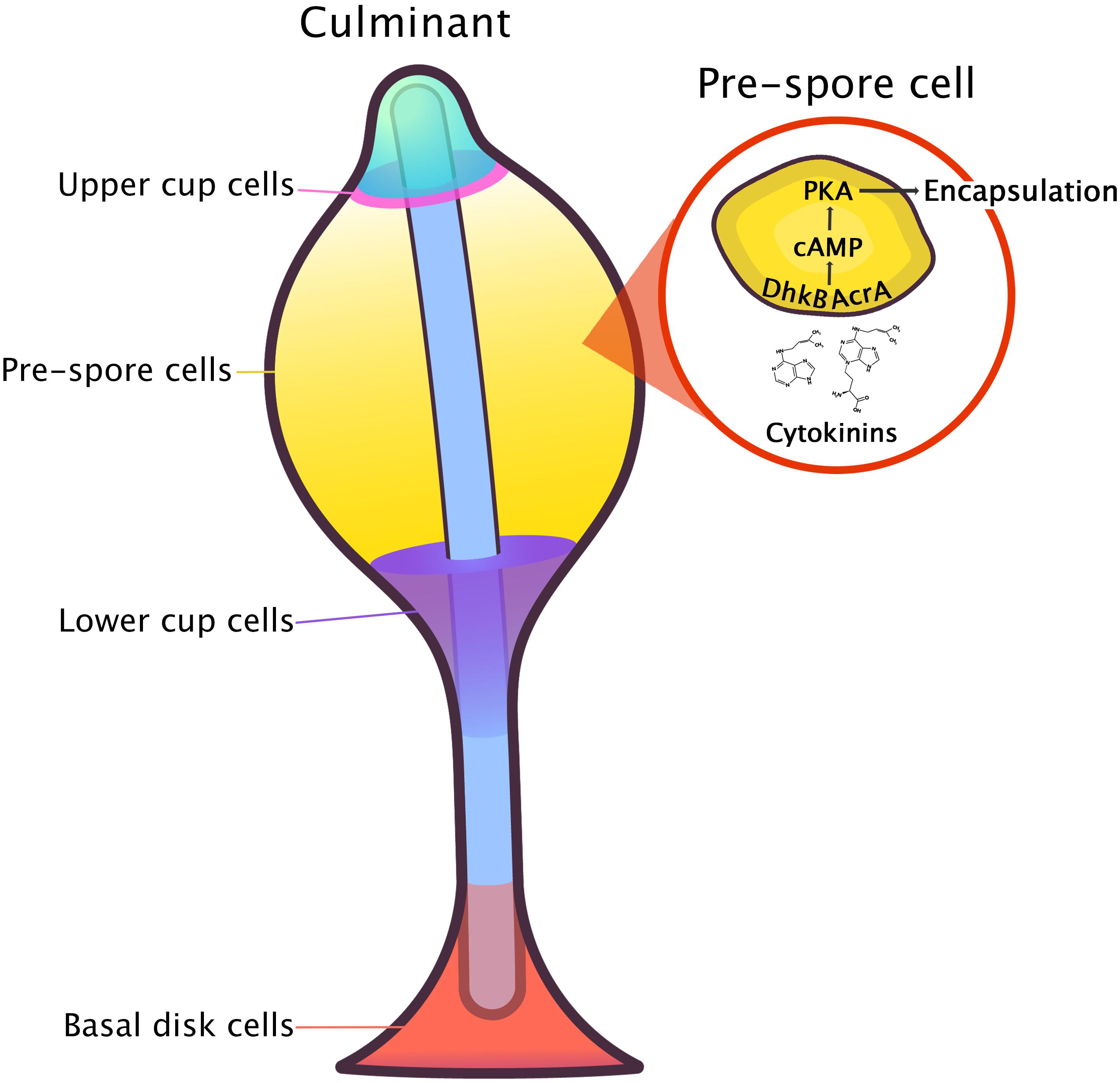
Figure 7. A model for the induction of sporulation by CK proposed by Anjard and Loomis (2008). Cytokinin is perceived by an unknown receptor and initiates a downstream phosphorelay through interaction of the histidine kinase, DhkB, and the adenylyl cyclase, AcrA This, in turn, leads to an increase in intracellular cAMP where the CK pathway and the SDF-2 signaling pathway converge at the level of PKA activation. An increase in PKA activity then leads to rapid encapsulation of spores.
DhkB and AcrA
The CK signal transduction pathway leading to the induction of sporulation is dependent on the HK, DhkB, and the adenylate cyclase with response regulator domain, AcrA (Anjard and Loomis, 2008). dhkB encodes a 1,969 amino acid, 219 kDA protein (DhkB, DDB0215358), whereas acrA encodes a 2,123 amino acid, 243 kDA protein (AcrA, DDB0191294). dhkB is one of 13 functional HK genes in the D. discoideum genome that encodes a HK protein possessing several potential transmembrane domains and extracellular loops (Anjard and Loomis, 2003). Because of the large size of both of these transmembrane proteins, they were tested in a CK binding assay to determine if they were CK receptors (Anjard and Loomis, 2008). Wild-type cells (22-h) undergoing development were tested for their ability to bind 3H-labeled iP in the presence of 1 mM adenine to minimize non-specific background binding. Wild-type cells had a binding affinity for 3H-iP of 6 nM. Furthermore, specific binding of 3H-iP in vegetative and 22-h developed wild-type cells and 22-h developed dhkB– and acrA– cells was determined for each of the respective cell types. Developed 22-h wild-type, dhkB–, and acrA– cells all bound similar levels of 3H-iP indicating that DhkB and AcrA are unlikely to be CK receptors. Vegetative wild-type cells bound 8-times less CK in comparison to the three tested developed cell types suggesting that the receptor is developmentally regulated. Zinda and Singleton (1998) reported that dhkB– null cells develop into fruiting bodies within 22–24 h. However, precocious germination of spores quickly ensues as the mutant dhkB– spores fail to respond to the spore germination inhibitor, DA. The germinated amoebae within the spore head quickly dehydrate owing to the high osmolarity maintained in the spore mass, and the majority of the cells are non-viable. dhkB– cells with partially constitutive PKA activity (dhkB–/K), developed at low densities and failed to differentiate into spores when treated with DA or other CKs; however, they could differentiate when treated with the spore inducers SDF-1, SDF-2, and GABA at levels similar to those found in wild-type cells (Anjard and Loomis, 2008). AcrA carries two response regulatory regions, which were suggested as potential targets for the interaction between DhkB and AcrA (Anjard et al., 2001; see reviews by Kriebel and Parent, 2004 and Loomis, 2014). Like dhkB– cells, acrA– spores germinated precociously resulting in almost no viable cells (Söderbom et al., 1999). An abnormal phenotype was observed in acrA– cells, in which the knockout strain formed fruiting bodies with abnormally thin stalks and glassy, translucent spore masses. Interestingly, this phenotype was also observed in the dhkB– and dhkA– knockout strains (Zinda and Singleton, 1998; Wang et al., 1999). Consistent with dhkB– cells, acrA– cells also failed to differentiate into spores following treatment with DA, iP, or tZ, but differentiated normally in response to other sporulation inducers (Anjard and Loomis, 2008).
AcaA, RdeA, and RegA
Several other known proteins affecting D. discoideum development were studied to determine whether they were involved in the induction of sporulation by CKs (Anjard and Loomis, 2008). The adenylyl cyclase of aggregation protein, AcaA, was identified as a possible candidate for interaction with CK owing to its similarity to AcrA. acaA– cells responded well to CKs, SDF-2, and GABA, indicating that the SDF-2 induction of spore encapsulation is not dependent upon a specific source of cAMP derived from this adenylyl cyclase (Anjard and Loomis, 2008). The rapid development protein, RdeA, and the previously mentioned cAMP phosphodiesterase, RegA, were tested to rule out any indirect effects of CK sporulation induction through stimulation of these proteins in the SDF-2 pathway. As mentioned earlier, spore induction through both the SDF-2 and CK signaling pathways occurs at precisely the same time during development. Previous work on spore induction pathways indicated that the peptide, SDF-2, generated from the Acyl-CoA binding protein, AcbA, acts on the receptor HK DhkA, which is present on both pre-spore and pre-stalk cells (Anjard and Loomis, 2005). The binding of SDF-2 to DhkA inhibits phosphorelay between RdeA and RegA. As such, the unphosphorylated response regulation region of RegA results in an accumulation of cAMP through AcrA, which increases the activity of PKA triggering rapid encapsulation of spores (Wang et al., 1999). Cells lacking RdeA and RegA responded to CK normally but were unable to respond to SDF-2 or GABA, thus leading to the conclusion that these proteins are not required for the induction of sporulation by CKs (Anjard and Loomis, 2008). While there have been no direct interactions observed between these three mentioned proteins and CKs, their combined roles lead to increased cAMP production, which is essential for encapsulation (Schaap and Schilde, 2018).
In summary, the signal transduction pathway for the induction of sporulation by CK is dependent upon DhkB and AcrA. CK signaling acts indirectly with the proteins involved in the SDF-2 signal transduction pathway, DhkA, RdeA, and RegA resulting in increased cAMP signaling. Furthermore, the other adenylyl cyclases, AcaA and AcgA, do not appear to directly interact with the CK pathway, unlike AcrA. However, there is an underlying connection with CK and cAMP in D. discoideum that will be discussed in the CK-small molecule functional interaction section. Potential cross-talk with CKs and other signaling pathways is likely and is common in other systems (e.g., CKs and auxin in plants); therefore, this would be an area of interest for future studies. Currently, there are no recognized receptors in D. discoideum that respond to CK, so further investigation is necessary to identify how CKs enact their effects. A candidate gene was found during systematic screening for developmental mutants after REMI mutagenesis. The mutant, DGG1110, presented a strikingly similar phenotype to the acrA null strain. Both strains develop normally but form spores that fail to remain dormant in the spore mass, leading to the death of the emerging amoebae. The gene inactivated in the DGG1110 strain (DDB_G0269128), encodes a predicted 544 amino acid protein with no clear homology to a known gene. However, this sequence presents a potential extracellular domain flanked by two transmembrane regions with a similar size and topology to the CHASE domain. While the homology with such domain is low, the mere presence of a potential extracellular loop in addition to the lack of response to CK in the null mutant make this gene a strong candidate for being the unidentified CK receptor in D. discoideum.
Future Directions – CK-Small Molecule Functional Interactions
CK-cAMP Interactions
cAMP is considered a universal second messenger controlling a wide variety of biological processes in all kingdoms of life (McDonough and Rodriguez, 2012). Among Dictyostelia, cAMP plays a dominant role in developmental signaling (Schaap, 2011). In the evolution of Dictyostelids, the roles of cAMP progressed from those involved in biological processes relating to stress to those regulating chemotactic aggregation, morphogenesis, and differentiation (Schaap, 2016). While the links between cAMP and the developmental program of D. discoideum have been extensively studied, intriguing suggestions of interactions between cAMP and CK have appeared over the last half century and remain largely unexplored.
Early studies on the role of CK in D. discoideum report an interesting correspondence between chemoattractants and spore germination inhibitors (Taya et al., 1980; Abe et al., 1981). As mentioned above, only the species that use cAMP as their acrasin also produce and respond to the spore germination inhibitor, DA. Moreover, during CK synthesis, the acceptor molecule of the isopentenyl side chain, 5′-AMP, which is catalyzed by IPT to create iP nucleotides, can be derived from cAMP through hydrolysis from cAMP phosphodiesterase (Taya et al., 1980). This evolutionary link between cAMP and CK among Dictyostelia was further confirmed through recent molecular phylogenetic analyses where all members of the most recently diverged species of Dictyostelia, group 4, use cAMP as their acrasin despite the use of various other chemoattractants among members belonging to the earlier diverged species of Dictyostelia in groups 1–3 (Schaap et al., 2006; Schaap, 2011). Therefore, it is likely that the earlier diverged species of Dictyostelia in groups 1–3 do not possess the enzyme to synthesize DA unlike the species belonging to group 4 who use cAMP as their chemoattractant.
Among other CK-producing organisms, links between cAMP and CK have been established. In A. thaliana, Plakidou-Dymock et al. (1998) identified seven potential seven transmembrane-spanning G-protein coupled receptors candidates. One of the candidates, GCR1, was isolated owing to its high similarity to other characterized 7-transmembrane proteins. Interestingly, the GCR1 protein is most closely related to the D. discoideum cAMP receptors, CarA-D. Transcriptome analysis of GCR1 in A. thaliana revealed new evidence that this protein is involved in hormonal responses, including CK biosynthesis and response, among many other functions (Chakraborty et al., 2015). Therefore, it would be of high interest to study the Car receptors in D. discoideum as potential CK receptors. In E. coli, CKs have a growth-promoting effect by modulating the activity of enzymes responsible for cAMP biosynthesis and degradation (Judewicz et al., 1973; Coppola et al., 1976). A similar effect is seen in mammalian cells where exogenously added CKs act as competitive inhibitors of cAMP phosphodiesterase (Hecht et al., 1974). While a link between cAMP and CK is suggested through the reported studies, much remains to be understood between the functional interplay between these two molecules. Considering the dominant role cAMP plays throughout the D. discoideum life cycle, D. discoideum provides an excellent system to further unravel the interaction between CKs and cAMP.
CK-NO Interactions
Like cAMP, nitric oxide (NO) is prevalent among all kingdoms and is one of the most common signaling molecules among living organisms (Schmidt and Walter, 1994; Lamattina et al., 2003; Moncada and Higgs, 2006). In D. discoideum, endogenous production of NO acts as a regulator of differentiation during development (Tao et al., 1997). NO-releasing compounds, such as sodium nitroprusside, inhibit D. discoideum aggregation by affecting the ability of cells to generate cAMP pulses (Tao et al., 1992, 1996). On the contrary, treatment with NO-scavengers, such as oxyhemoglobin (oxyHb), stimulates D. discoideum aggregation (Tao et al., 1997). Combining these respective results, a working model for the action of NO was proposed in D. discoideum: NO inhibits cAMP production in vegetative cells; upon starvation, the cells overcome the NO-inhibiting effects through cAMP pulses initiating the developmental program of D. discoideum. Total endogenous CK concentrations increase as cells transition from growth to aggregation (Aoki et al., 2019). This increase in endogenous CKs coincides with the decreases in NO that occurs during aggregation. Perhaps CKs have a secondary role in regulating NO levels in D. discoideum, specifically in the initiation of the developmental program.
Looking at other CK-producing organisms, it is clear that interactions between NO and CKs exist. A protective role of CKs against reactive nitrogen species was shown, in which CKs were shown to act as NO-scavengers (Liu et al., 2013). This same study reported that NO can chemically modify the CK structure to produce novel reaction products both in vitro and in vivo. Considering this evidence, future research looking into interactions between CK and NO in D. discoideum would be insightful to see if CK has a secondary role in scavenging NO during aggregation. Furthermore, the strong interactions between CK and NO, as demonstrated in plants and other CK-producing biota, could be used to elucidate candidate genes in D. discoideum involving NO synthesis and regulation (Husain et al., 2010; Feng et al., 2013; Liu et al., 2013; Samanovic and Darwin, 2015).
Conclusion
The Dictyostelid system is unlike any other model organism utilized to assess the evolution of CKs as signaling agents among and between kingdoms of life. In fact, many new emerging areas of D. discoideum research encompass interactions with organisms in other kingdoms. These emerging areas include topics such as, D. discoideum as farmers of symbiotic bacteria, host defense against pathogenic bacteria and nematodes, and interactions with spore-dispersing organisms – all of which are influenced by interkingdom signaling, and perhaps CKs (Bozzaro and Eichinger, 2011; Brock et al., 2013; Smith et al., 2014; Novohradská et al., 2017). While there is accumulating evidence documenting the conservation of CK biosynthetic and signal transduction pathways among Dictyostelia species, much remains to be understood about the role of CK within this clade. Considering the widespread occurrence of CKs in both single-celled and multicellular organisms, Dictyostelids offer a unique opportunity to assess how CKs have evolved from roles at the cellular level to roles in controlling complex events during multicellular development. Furthermore, the dominant role that cAMP and NO play in various aspects of D. discoideum development expand the utility of this organism for studying not only the evolution of CKs beyond the plant kingdom, but also CK interactions with other signaling molecules. An additional promising small molecule to be investigated in the future for CK interactions is inorganic polyphosphate (poly P). Several studies highlight the drastic accumulation of poly P throughout the course of D. discoideum development, and ppk1– cells have developmental defects in germination similar to those described for the CK-dependent dhkB– and acrA– null cells (Zhang et al., 2005; Livermore et al., 2016). Utilizing this model system, specific functions of CKs can be observed at the single cell level and beyond into multicellular organization and development, which may offer insight into how CKs have evolved as molecules, facilitating physiological interactions among and between various kingdoms.
Author Contributions
MA and NE wrote the first draft of the manuscript. CA wrote sections of the manuscript. All authors contributed to manuscript revision, read and approved the submitted version.
Funding
MA is supported by the Queen Elizabeth II Graduate Scholarship in Science and Technology (QEII-GSST). NE, CB, and RH are supported by Discovery Grants from the Natural Sciences and Engineering Research Council of Canada (RGPIN-05436 to NE, and RGPIN-2020-04843 to CB, RGPIN-2018-04855 to RH).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
References
Abe, H., Hashimoto, K., and Uchiyama, M. (1981). Discadenine distribution in cellular slime molds and its inhibitory activity on spore germination. Agric. Biol. Chem. 45, 1295–1296. doi: 10.1080/00021369.1981.10864699
Abe, H., Uchiyama, M., Tanaka, Y., and Saito, H. (1976). Structure of discadenine, a spore germination inhibitor from the cellular slime mold Dictyostelium discoideum. Tetrahedron Lett. 17, 3807–3810. doi: 10.1016/S0040-4039(00)93115-0
Akiyoshi, D. E., Klee, H., Amasino, R. M., Nester, E. W., and Gordon, M. P. (1984). T-DNA of Agrobacterium tumefaciens encodes an enzyme of cytokinin biosynthesis. Proc. Nati. Acad. Sci. U.S.A. 81, 5994–5998. doi: 10.1073/pnas.81.19.5994
Alvarez-Curto, E., Saran, S., Meima, M., Zobel, J., Scott, C., and Schaap, P. (2007). cAMP production by adenylyl cyclase G induces differentiation in Dictyostelium slugs. Development 134, 959–966. doi: 10.1242/dev.02775
Anantharaman, V., and Aravind, L. (2001). The CHASE domain: a predicted ligand-binding module in plant cytokinin receptors and other eukaryotic and bacterial receptors. Trends Biochem. Sci. 26, 579–582. doi: 10.1016/S0968-0004(01)01968-5
Andrabi, S. B. A., Tahara, M., Matsubara, R., Toyama, T., Aonuma, H., Sakakibara, H., et al. (2018). Plant hormone cytokinins control cell cycle progression and plastid replication in apicomplexan parasites. Parasitol. Int. 67, 47–58. doi: 10.1016/j.parint.2017.03.003
Anjard, C., and Loomis, W. F. (2003). “Histidine Kinases of Dictyostelium,” in Histidine Kinases in Signal Transduction, eds M. Inouye and R. Dutta (San Diego: Academic Press), 421–438. doi: 10.1016/B978-012372484-7/50021-7
Anjard, C., and Loomis, W. F. (2005). Peptide signaling during terminal differentiation of Dictyostelium. Proc. Natl. Acad. Sci. U.S.A. 102, 7607–7611. doi: 10.1073/pnas.0501820102
Anjard, C., and Loomis, W. F. (2008). Cytokinins induce sporulation in Dictyostelium. Development 135, 819–827. doi: 10.1242/dev.018051
Anjard, C., and Loomis, W. F., Dictyostelium Sequencing Consortium (2002). Evolutionary analyses of ABC transporters of Dictyostelium discoideum. Eukaryot. Cell 1, 643–652. doi: 10.1128/ec.1.4.643-652.2002
Anjard, C., Söderbom, F., and Loomis, W. F. (2001). Requirements for the adenylyl cyclases in the development of Dictyostelium. Development 128, 3649–3654.
Aoki, M. M., Kisiala, A. B., Li, S., Stock, N. L., Brunetti, C. R., Huber, R. J., et al. (2019). Cytokinin detection during the Dictyostelium discoideum life cycle: profiles are dynamic and affect cell growth and spore germination. Biomolecules 9:702. doi: 10.3390/biom9110702
Armstrong, D. J., and Firtel, R. A. (1989). Cytokinin oxidase activity in the cellular slime mold, Dictyostelium discoideum. Dev. Biol. 136, 491–499. doi: 10.1016/0012-1606(89)90274-1
Blad, C. C., von Frijtag Drabbe Künzel, J. K., de Vries, H., Mulder-Krieger, T., Bar-Yehuda, S., Fishman, P., et al. (2011). Putative role of the adenosine A3 receptor in the antiproliferative action of N6-(2-isopentenyl)adenosine. Purinergic Signal. 7, 453–462. doi: 10.1007/s11302-011-9244-9
Bloomfield, G., Tanaka, Y., Skelton, J., Ivens, A., and Kay, R. R. (2008). Widespread duplications in the genomes of laboratory stocks of Dictyostelium discoideum. Genome Biol. 9:R75. doi: 10.1186/gb-2008-9-4-r75
Bozzaro, S., and Eichinger, L. (2011). The professional phagocyte Dictyostelium discoideum as a model host for bacterial pathogens. Curr. Drug Targets 12, 942–954. doi: 10.2174/138945011795677782
Brock, D. A., Read, S., Bozhchenko, A., Queller, D. C., and Strassmann, J. E. (2013). Social amoeba farmers carry defensive symbionts to protect and privatize their crops. Nat. Commun. 4:2385. doi: 10.1038/ncomms3385
Chakraborty, N., Sharma, P., Kanyuka, K., Pathak, R. R., Choudhury, D., Hooley, R. A., et al. (2015). Transcriptome analysis of Arabidopsis GCR1 mutant reveals its roles in stress, hormones, secondary metabolism and phosphate starvation. PLoS One 10:e0122423. doi: 10.1371/journal.pone.0117819
Chanclud, E., and Lacombe, B. (2017). Plant hormones: key players in gut microbiota and human diseases? Trends Plant Sci. 22, 754–758. doi: 10.1016/j.tplants.2017.07.003
Chen, P., Jiao, X., Zhang, Y., Wu, L., Tang, D. J., Li, P., et al. (2019). The crystal structure of the phytopathogenic bacterial sensor PcrK reveals different cytokinin recognition mechanism from the plant sensor AHK4. J. Struct. Biol. 208, 69–76. doi: 10.1016/j.jsb.2019.08.001
Coppola, S., Marino, P., and Zoina, A. (1976). Cytokinin contents and cAMP metabolism during growth of Escherichia coli. Experientia 32, 566–567. doi: 10.1007/BF01990164
Crespi, M., Messens, E., Caplan, A. B., van Montagu, M., and Desomer, J. (1992). Fasciation induction by the phytopathogen Rhodococcus fascians depends upon a linear plasmid encoding a cytokinin synthase gene. EMBO J. 11, 795–804. doi: 10.1002/j.1460-2075.1992.tb05116.x
Dabravolski, S. (2020). Multi-faceted nature of the tRNA isopentenyltransferase. Funct. Plant Biol. 47, 475–485. doi: 10.1071/FP19255
Durán-Medina, Y., Díaz-Ramírez, D., and Marsch-Martínez, N. (2017). Cytokinins on the Move. Front. Plant Sci. 8:146. doi: 10.3389/fpls.2017.00146
Durbecq, V., Sainz, G., Oudjama, Y., Clantin, B., Bompard-Gilles, C., Tricot, C., et al. (2001). Crystal structure of isopentenyl diphosphate:dimethylallyl diphosphate isomerase. EMBO J. 20, 1530–1537. doi: 10.1093/emboj/20.7.1530
Eichinger, L., Pachebat, J. A., Glöckner, G., Rajandream, M.-A., Sucgang, R., Berriman, M., et al. (2005). The genome of the social amoeba Dictyostelium discoideum. Nature 435, 43–57. doi: 10.1038/nature03481
Ericson, J. U., and Bjork, G. R. (1986). Pleiotropic effects induced by modification deficiency next to the anticodon of tRNA from Salmonella typhimurium LT2. J. Bacteriol. 166, 1013–1021. doi: 10.1128/jb.166.3.1013-1021.1986
Feng, J., Wang, C., Chen, Q., Chen, H., Ren, B., Li, X., et al. (2013). S-nitrosylation of phosphotransfer proteins represses cytokinin signaling. Nat. Commun. 4:1529. doi: 10.1038/ncomms2541
Frébort, I., Kowalska, M., Hluska, T., Frébortová, J., and Galuszka, P. (2011). Evolution of cytokinin biosynthesis and degradation. J. Exp. Bot. 62, 2431–2452. doi: 10.1093/jxb/err004
Frébortová, J., Greplová, M., Seidl, M. F., Heyl, A., and Frébort, I. (2015). Biochemical characterization of putative adenylate dimethylallyltransferase and cytokinin dehydrogenase from Nostoc sp. PCC 7120. PLoS One 10:e0138468. doi: 10.1371/journal.pone.0138468
Gajdosová, S., Spíchal, L., Kamínek, M., Hoyerová, K., Novák, O., Dobrev, P. I., et al. (2011). Distribution, biological activities, metabolism, and the conceivable function of cis-Zeatin-type cytokinins in plants. J. Exp. Bot. 62, 2827–2840. doi: 10.1093/jxb/erq457
Golovko, A., Hjälm, G., Sitbon, F., and Nicander, B. (2000). Cloning of a human tRNA isopentenyl transferase. Gene 258, 85–93. doi: 10.1016/S0378-1119(00)00421-2
Hecht, S. M., Faulkner, R. D., and Hawrelak, S. D. (1974). Competitive inhibition of beef heart cyclic AMP phosphodiesterase by cytokinins and related compounds. Proc. Natl. Acad. Sci. U.S.A. 71, 4670–4674. doi: 10.1073/pnas.71.12.4670
Hinsch, J., Vrabka, J., Oeser, B., Novák, O., Galuszka, P., and Tudzynski, P. (2015). De novo biosynthesis of cytokinins in the biotrophic fungus Claviceps purpurea. Environ. Microbiol. 17, 2935–2951. doi: 10.1111/1462-2920.12838
Hirose, N., Makita, N., Yamaya, T., and Sakakibara, H. (2005). Functional characterization and expression analysis of a gene, OsENT2, encoding an equilibrative nucleoside transporter in rice suggest a function in cytokinin transport. Plant Physiol. 138, 196–206. doi: 10.1104/pp.105.060137
Husain, M., Jones-Carson, J., Song, M., McCollister, B. D., Bourret, T. J., and Vázquez-Torres, A. (2010). Redox sensor SsrB Cys203 enhances Salmonella fitness against nitric oxide generated in the host immune response to oral infection. Proc. Natl. Acad. Sci. U.S.A. 107, 14396–14401. doi: 10.1073/pnas.1005299107
Hwang, I., and Sheen, J. (2001). Two-component circuitry in Arabidopsis cytokinin signal transduction. Nature 413, 383–389. doi: 10.1038/35096500
Ihara, M., Tanaka, Y., Yanagisawa, K., Taya, Y., and Nishimura, S. (1986). Purification and some properties of discadenine synthase from Dictyostelium discoideum. BBA 881, 135–140. doi: 10.1016/0304-4165(86)90106-6
Ihara, M., Taya, Y., and Nishimura, S. (1980). Developmental regulation of cytokinin, spore germination inhibitor discadenine and related enzymes in Dictyostelium discoideum. Exp. Cell Res. 126, 273–278. doi: 10.1016/0014-4827(80)90265-7
Ihara, M., Taya, Y., Nishimura, S., and Tanaka, Y. (1984). Purification and some properties of Δ2-isopentenylpyrophosphate:5’AMP Δ2-isopentenyltransferase from the cellular slime mold Dictyostelium discoideum. Arch. Biochem. Biophys. 230, 652–660. doi: 10.1016/0003-9861(84)90446-6
Jelesko, J. G. (2012). An expanding role for purine uptake permease-like transporters in plant secondary metabolism. Front. Plant Sci. 3:78. doi: 10.3389/fpls.2012.00078
Judewicz, N. D., De Robertis, E. M., and Torres, H. N. (1973). Inhibition of Escherichia coli growth by cyclic adenosine 3’, 5’-monophosphate. Biochem. Biophys. Res. Commun. 52, 1257–1262. doi: 10.1016/0006-291X(73)90636-0
Kabbara, S., Schmülling, T., and Papon, N. (2018). CHASEing cytokinin receptors in plants, bacteria, fungi, and beyond. Trends Plant Sci. 23, 179–181. doi: 10.1016/j.tplants.2018.01.001
Kakimoto, T. (2001). Identification of plant cytokinin biosynthetic enzymes as dimethylallyl diphosphate:ATP/ADP isopentenyltransferases. Plant Cell Physiol. 42, 677–685. doi: 10.1093/pcp/pce112
Kamada-Nobusada, T., and Sakakibara, H. (2009). Molecular basis for cytokinin biosynthesis. Phytochemistry 70, 444–449. doi: 10.1016/j.phytochem.2009.02.007
Kieber, J. J., and Schaller, G. E. (2018). Cytokinin signaling in plant development. Development 145:dev149344. doi: 10.1242/dev.149344
Ko, D., Kang, J., Kiba, T., Park, J., Kojima, M., Do, J., et al. (2014). Arabidopsis ABCG14 is essential for the root-to-shoot translocation of cytokinin. Proc. Natl. Acad. Sci. U.S.A. 111, 7150–7155. doi: 10.1073/pnas.1321519111
Konijn, T. M., Van De Meene, J. G., Bonner, J. T., and Barkley, D. S. (1967). The acrasin activity of adenosine-3’,5’-cyclic phosphate. Proc. Natl. Acad. Sci. U.S.A. 58, 1152–1154. doi: 10.1073/pnas.58.3.1152
Kriebel, P. W., and Parent, C. A. (2004). Adenylyl cyclase expression and regulation during the differentiation of Dictyostelium discoideum. IUBMB Life 56, 541–546. doi: 10.1080/15216540400013887
Kurakawa, T., Ueda, N., Maekawa, M., Kobayashi, K., Kojima, M., Nagato, Y., et al. (2007). Direct control of shoot meristem activity by a cytokinin-activating enzyme. Nature 445, 652–655. doi: 10.1038/nature05504
Kuroha, T., Tokunaga, H., Kojima, M., Ueda, N., Ishida, T., Nagawa, S., et al. (2009). Functional analyses of LONELY GUY cytokinin-activating enzymes reveal the importance of the direct activation pathway in Arabidopsis. Plant Cell 21, 3152–3169. doi: 10.1105/tpc.109.068676
Lamattina, L., García-Mata, C., Graziano, M., and Pagnussat, G. (2003). Nitric oxide: the versatility of an extensive signal molecule. Annu. Rev. Plant Biol. 54, 109–136. doi: 10.1146/annurev.arplant.54.031902.134752
Lappas, C. M. (2015). The plant hormone zeatin riboside inhibits T lymphocyte activity via adenosine A2A receptor activation. Cell Mol. Immunol. 12, 107–112. doi: 10.1038/cmi.2014.33
Lee, Y.-C., Yang, Y.-C., Huang, C.-L., Kuo, T.-Y., Lin, J.-H., Yang, D.-M., et al. (2012). When cytokinin, a plant hormone, meets the adenosine A2A receptor: a novel neuroprotectant and lead for treating neurodeenerative disorders? PLoS One 7:e38865. doi: 10.1371/journal.pone.0038865
Lindner, A. C., Lang, D., Seifert, M., Podlešáková, K., Novák, O., Strnad, M., et al. (2014). Isopentenyltransferase-1 (IPT1) knockout in Physcomitrella together with phylogenetic analyses of IPTs provide insights into evolution of plant cytokinin biosynthesis. J. Exp. Bot. 65, 2533–2543. doi: 10.1093/jxb/eru142
Liu, C. J., Zhao, Y., and Zhang, K. (2019). Cytokinin transporters: multisite players in cytokinin homeostasis and signal distribution. Front. Plant Sci. 10:693. doi: 10.3389/fpls.2019.00693
Liu, W. Z., Kong, D. D., Gu, X. X., Gao, H. B., Wang, J. Z., Xia, M., et al. (2013). Cytokinins can act as suppressors of nitric oxide in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 110, 1548–1553. doi: 10.1073/pnas.1213235110
Livermore, T. M., Chubb, J. R., and Saiardi, A. (2016). Developmental accumulation of inorganic pholyphosphate affects germination and energetic metabolism in Dictyostelium discoideum. Proc. Natl. Acad. Sci. U.S.A. 113, 996–1001. doi: 10.1073/pnas.1519440113
Loomis, W. F. (2014). Cell signaling during development of Dictyostelium. Dev. Biol. 391, 1–16. doi: 10.1016/j.ydbio.2014.04.001
Mayaka, J. B., Huang, Q., Xiao, Y., Zhong, Q., Ni, J., and Shen, Y. (2019). The Lonely Guy (LOG) homologue SiRe_0427 from the thermophilic archaeon Sulfolobus islandicus REY15A is a phosphoribohydrolase representing a novel group. Appl. Environ. Microbiol 85:e01739-19. doi: 10.1128/AEM.01739-19
McDonough, K. A., and Rodriguez, A. (2012). The myriad roles of cyclic AMP in microbial pathogens: from signal to sword. Nat. Rev. Microbiol. 10, 27–38. doi: 10.1038/nrmicro2688
Mik, V., Mièková, Z., Doležal, K., Frébort, I., and Pospíšil, T. (2017). Activity of (+)-discadenine as a plant cytokinin. J. Nat. Prod. 80, 2136–2140. doi: 10.1021/acs.jnatprod.6b01165
Miranda, E. R., Zhuchenko, O., Toplak, M., Santhanam, B., Zupan, B., Kuspa, A., et al. (2013). ABC transporters in Dictyostelium discoideum development. PLoS One 8:e70040. doi: 10.1371/journal.pone.0070040
Miyawaki, K., Tarkowski, P., Matsumoto-Kitano, M., Kato, T., Sato, S., Tarkowska, D., et al. (2006). Roles of Arabidopsis ATP/ADP isopentenyltransferases and tRNA isopentenyltransferases in cytokinin biosynthesis. Proc. Natl. Acad. Sci. U.S.A. 103, 16598–16603. doi: 10.1073/pnas.0603522103
Mok, D. W., and Mok, M. C. (2001). Cytokinin metabolism and action. Annu. Rev. Plant Physiol. Plant Mol. Biol. 52, 89–118. doi: 10.1146/annurev.arplant.52.1.89
Moncada, S., and Higgs, E. A. (2006). The discovery of nitric oxide and its role in vascular biology. Br. J. Pharmacol. 147, S193–S201. doi: 10.1038/sj.bjp.0706458
Moramarco, F., Pezzicoli, A., Salvini, L., Leuzzi, R., Pansegrau, W., and Balducci, E. (2019). A LONELY GUY protein of Bordetella pertussis with unique features is related to oxidative stress. Sci. Rep. 9, 1–12. doi: 10.1038/s41598-019-53171-9
Morrison, E. N., Emery, R. J. N., and Saville, B. J. (2017). Fungal derived cytokinins are necessary for normal Ustilago maydis infection of maize. Plant Pathol. 66, 726–742. doi: 10.1111/ppa.12629
Morrison, E. N., Knowles, S., Hayward, A., Thorn, R. G., Saville, B. J., and Emery, R. J. N. (2015). Detection of phytohormones in temperate forest fungi predicts consistent abscisic acid production and a common pathway for cytokinin biosynthesis. Mycologia 107, 245–257. doi: 10.3852/14-157
Nagasaki, A., and Uyeda, T. Q. P. (2008). Screening of genes involved in cell migration in Dictyostelium. Exp. Cell Res. 314, 1136–1146. doi: 10.1016/j.yexcr.2007.12.002
Nishii, K., Wright, F., Chen, Y. Y., and Möller, M. (2018). Tangled history of a multigene family: the evolution of ISOPENTENYLTRANSFERASE genes. PLoS One 13:e0201198. doi: 10.1371/journal.pone.0201198
Nomura, T., Tanaka, Y., Abe, H., and Uchiyama, M. (1977). Cytokinin activity of discadenine: a spore germination inhibitor of Dictyostelium discoideum. Phytochemistry 16, 1819–1820. doi: 10.1016/0031-9422(71)85097-5
Novohradská, S., Ferling, I., and Hillmann, F. (2017). Exploring virulence determinants of filamentous fungal pathogens through interactions with soil amoebae. Front. Cell. Infect. Microbiol. 7:497. doi: 10.3389/fcimb.2017.00497
Obata, Y., Abe, H., Tanaka, Y., Yanagisawa, K., and Uchiyama, M. (1973). Isolation of a spore germination inhibitor from a cellular slime mold Dictyostelium discoideum. Agric. Biol. Chem. 37, 1989–1990. doi: 10.1271/bbb1961.37.1989
Palatnik, C. M., Katz, E. R., and Brenner, M. (1977). Isolation and characterization of transfer RNAs from Dictyostelium discoideum during growth and development. J. Biol. Chem. 252, 694–703.
Parker, C. W., and Letham, D. S. (1973). Regulators of cell division in plant tissues. XVI - Metabolism of zeatin by radish cotyledons and hypocotyls. Planta 114, 199–218. doi: 10.1007/BF00389036
Plakidou-Dymock, S., Dymock, D., and Hooley, R. (1998). A higher plant seven-transmembrane receptor that influences sensitivity to cytokinins. Curr. Biol. 8, 315–324. doi: 10.1016/S0960-9822(98)70131-9
Pratt-Hyatt, M., Pai, D. A., Haeusler, R. A., Wozniak, G. G., Good, P. D., Miller, E. L., et al. (2013). Mod5 protein binds to tRNA gene complexes and affects local transcriptional silencing. Proc. Natl. Acad. Sci. U.S.A. 110, E3081–E3089. doi: 10.1073/pnas.1219946110
Romeralo, M., Cavender, J. C., Landolt, J. C., Stephenson, S. L., and Baldauf, S. L. (2011). An expanded phylogeny of social amoebas (Dictyostelia) shows increasing diversity and new morphological patterns. BMC Evol. Biol. 11:84. doi: 10.1186/1471-2148-11-84
Rosengarten, R. D., Santhanam, B., Kokosar, J., and Shaulsky, G. (2017). The long noncoding RNA transcriptome of Dictyostelium discoideum development. G3 7, 387–398. doi: 10.1534/g3.116.037150
Rot, G., Parikh, A., Curk, T., Kuspa, A., Shaulsky, G., and Zupan, B. (2009). dictyExpress: a Dictyostelium discoideum gene expression database with an explorative data analysis web-based interface. BMC Bioinform. 10:265. doi: 10.1186/1471-2105-10-265
Sakakibara, H. (2006). Cytokinins: activity, biosynthesis, and translocation. Annu. Rev. Plant Biol. 57, 431–449. doi: 10.1146/annurev.arplant.57.032905.105231
Samanovic, M. I., and Darwin, K. H. (2015). Cytokinins beyond plants: synthesis by Mycobacterium tuberculosis. Microb. Cell 2, 168–170. doi: 10.15698/mic2015.05.203
Samanovic, M. I., Hsu, H. C., Jones, M. B., Jones, V., McNeil, M. R., Becker, S. H., et al. (2018). Cytokinin signaling in Mycobacterium tuberculosis. mBio 9:e00989-18. doi: 10.1128/mBio.00989-18
Schaap, P. (2011). Evolution of developmental cyclic adenosine monophosphate signaling in the Dictyostelia from an amoebozoan stress response. Dev. Growth Differ. 53, 452–462. doi: 10.1111/j.1440-169X.2011.01263.x
Schaap, P. (2016). Evolution of developmental signalling in Dictyostelid social amoebas. Curr. Opin. Genet. Dev. 39, 29–34. doi: 10.1016/j.gde.2016.05.014
Schaap, P., and Schilde, C. (2018). Encystation: the most prevalent and underinvestigated differentiation pathway of eukaryotes. Microbiology 164, 727–739. doi: 10.1099/mic.0.000653
Schaap, P., Winckler, T., Nelson, M., Alvarez-Curto, E., Elgie, B., Hagiwara, H., et al. (2006). Molecular phylogeny and evolution of morphology in the social amoebas. Science 314, 661–663. doi: 10.1126/science.1130670
Schäfer, M., Brütting, C., Canales, I. M., Großkinsky, D. K., Vankova, R., Baldwin, I. T., et al. (2015). The role of cis-zeatin-type cytokinins in plant growth regulation and mediating responses to environmental interactions. J. Exp. Bot. 66, 4873–4884. doi: 10.1093/jxb/erv214
Schilde, C., and Schaap, P. (2013). The Amoebozoa. Totowa, NJ: Humana Press, 1–15. doi: 10.1007/978-1-62703-302-2_1
Schmidt, H. H., and Walter, U. (1994). NO at work. Cell 78, 919–925. doi: 10.1016/0092-8674(94)90267-4
Schmülling, T., Werner, T., Riefler, M., Krupková, E., and Bartrina y Manns, I. (2003). Structure and function of cytokinin oxidase/dehydrogenase genes of maize, rice, Arabidopsis and other species. J. Plant Res. 116, 241–252. doi: 10.1007/s10265-003-0096-4
Seegobin, M., Kisiala, A., Noble, A., Kaplan, D., Brunetti, C., and Emery, R. J. N. (2018). Canis familiaris tissues are characterized by different profiles of cytokinins typical of the tRNA degradation pathway. FASEB J. 32, 6575–6581. doi: 10.1096/fj.201800347
Seo, H., and Kim, K. J. (2017). Structural basis for a novel type of cytokinin-activating protein. Sci. Rep. 7:45985. doi: 10.1038/srep45985
Šmehilová, M., Dobrůšková, J., Novák, O., Takáč, T., and Galuszka, P. (2016). Cytokinin-specific glycosyltransferases possess different roles in cytokinin homeostasis maintenance. Front. Plant Sci. 7:1264. doi: 10.3389/fpls.2016.01264
Smith, J., Queller, D. C., and Strassmann, J. E. (2014). Fruiting bodies of the social amoeba Dictyostelium discoideum increase spore transport by Drosophila. BMC Evol. Biol. 14:105. doi: 10.1186/1471-2148-14-105
Söderbom, F., Anjard, C., Iranfar, N., Fuller, D., and Loomis, W. F. (1999). An adenylyl cyclase that functions during late development of Dictyostelium. Development 126, 5463–5471.
Spíchal, L. (2012). Cytokinins – recent news and views of evolutionally old molecules. Funct. Plant Biol. 39, 267. doi: 10.1071/FP11276
Spinola, M., Galvan, A., Pignatiello, C., Conti, B., Pastorino, U., Nicander, B., et al. (2005). Identification and functional characterization of the candidate tumor suppressor gene TRIT1 in human lung cancer. Oncogene 24, 5502–5509. doi: 10.1038/sj.onc.1208687
Stock, A. M., Robinson, V. L., and Goudreau, P. N. (2000). Two-component signal transduction. Annu. Rev. Biochem. 69, 183–215. doi: 10.1146/annurev.biochem.69.1.183
Sucgang, R., Kuo, A., Tian, X., Salerno, W., Parikh, A., Feasley, C. L., et al. (2011). Comparative genomics of the social amoebae Dictyostelium discoideum and Dictyostelium purpureum. Genome Biol. 12:R20. doi: 10.1186/gb-2011-12-2-r20
Sugawara, H., Ueda, N., Kojima, M., Makita, N., Yamaya, T., and Sakakibara, H. (2008). Structural insight into the reaction mechanism and evolution of cytokinin biosynthesis. Proc. Natl. Acad. Sci. U.S.A. 105, 2734–2739. doi: 10.1073/pnas.0707374105
Suzuki, G., Shimazu, N., and Tanaka, M. (2012). A yeast prion, Mod5, promotes acquired drug resistance and cell survival under environmental stress. Science 336, 355–359. doi: 10.1126/science.1219491
Takei, K., Sakakibara, H., and Sugiyama, T. (2001). Identification of genes encoding adenylate isopentenyltransferase, a cytokinin biosynthesis enzyme, in Arabidopsis thaliana. J. Biol. Chem. 276, 26405–26410. doi: 10.1074/jbc.M102130200
Tanaka, Y., Abe, H., Uchiyama, M., Taya, Y., and Nishimura, S. (1978). Isopentenyladenine from Dictyostelium discoideum. Phytochemistry 17, 543–544. doi: 10.1016/s0031-9422(00)89356-5
Tanaka, Y., Hashimoto, Y., Yanagisawa, K., Abe, H., and Uchiyama, M. (1975). Partial structure of a spore germination inhibitor from a cellular slime mold, Dictyostelium discoideum. Agric. Biol. Chem. 39, 1929–1932. doi: 10.1080/00021369.1975.10861883
Tao, Y., Howlett, A., and Klein, C. (1992). Nitric oxide stimulates the ADP-ribosylation of a 41-kDa cytosolic protein in Dictyostelium discoideum. Proc. Natl. Acad. Sci. U.S.A. 89, 5902–5906. doi: 10.1073/pnas.89.13.5902
Tao, Y., Howlett, A., and Klein, C. (1996). Nitric oxide inhibits the initiation of cAMP pulsing in Dictyostelium discoideum without altering receptor-activated adenylate cyclase. Cell. Signal. 8, 26–34. doi: 10.1016/0898-6568(95)02011-x
Tao, Y. P., Misko, T. P., Howlett, A. C., and Klein, C. (1997). Nitric oxide, an endogenous regulator of Dictyostelium discoideum differentiation. Development 124, 3587–3595.
Taya, Y., Tanaka, Y., and Nishimura, S. (1978a). 5’-AMP is a direct precursor of cytokinin in Dictyostelium discoideum. Nature 271, 545–547. doi: 10.1038/271545a0
Taya, Y., Tanaka, Y., and Nishimura, S. (1978b). Cell-free biosynthesis of discadenine, a spore germination inhibitor of Dictyostelium discoideum. FEBS Lett. 89, 326–328. doi: 10.1016/0014-5793(78)80247-6
Taya, Y., Yamada, T., and Nishimura, S. (1980). Correlation between acrasins and spore germination inhibitors in cellular slime molds. J. Bacteriol. 143, 715–719. doi: 10.1128/jb.143.2.715-719.1980
Telser, A., and Sussman, M. (1971). Uridine diphosphate galactose-4-epimerase, a developmentally regulated enzyme in the cellular slime mold Dictyostelium discoideum. J. Biol. Chem. 246, 2252–2257.
Tolerico, L. H., Benko, A. L., Aris, J. P., Stanford, D. R., Martin, N. C., and Hopper, A. K. (1999). Saccharomyces cerevisiae Mod5p-II contains sequences antagonistic for nuclear and cytosolic locations. Genetics 151, 57–75.
Van Kast, C. A., and Laten, H. M. (1987). Cytokinin utilization by adenine-requiring mutants of the yeast Saccharomyces cerevisiae. Plant Physiol. 83, 726–727. doi: 10.1104/pp.83.4.726
Voller, J., Zatloukal, M., Lenobel, R., Doleal, K., Bére, T., Krytof, V., et al. (2010). Anticancer activity of natural cytokinins: a structure-activity relationship study. Phytochemistry 71, 1350–1359. doi: 10.1016/j.phytochem.2010.04.018
Wang, F. F., Cheng, S. T., Wu, Y., Ren, B. Z., and Qian, W. (2017). A bacterial receptor PcrK senses the plant hormone cytokinin to promote adaptation to oxidative stress. Cell Rep. 21, 2940–2951. doi: 10.1016/j.celrep.2017.11.017
Wang, N., Söderbom, F., Anjard, C., Shaulsky, G., and Loomis, W. F. (1999). SDF-2 induction of terminal differentiation in Dictyostelium discoideum is mediated by the membrane-spanning sensor kinase DhkA. Mol. Cell. Biol. 19, 4750–4756. doi: 10.1128/mcb.19.7.4750
Yu, J. M., Wang, D., Pierson, L. S., and Pierson, E. A. (2017). Disruption of MiaA provides insights into the regulation of phenazine biosynthesis under suboptimal growth conditions in Pseudomonas chlororaphis 30-84. Microbiology 163, 94–108. doi: 10.1099/mic.0.000409
Zhang, H., Gómez-García, M. R., Brown, M. R. W., and Kornber, A. (2005). Inorganic polyphosphate in Dicytostelium discoideum: influence on development, sporulation, and predation. Proc. Natl. Acad. Sci. U.S.A. 102, 2731–2735. doi: 10.1073/pnas.0500023102
Zhang, K., Novák, O., Wei, Z., Gou, M., Zhang, X., Yu, Y., et al. (2014). Arabidopsis ABCG14 protein controls the acropetal translocation of root-synthesized cytokinins. Nat. Commun. 5:3274. doi: 10.1038/ncomms4274
Keywords: cytokinins, Dictyostelia, Dictyostelium discoideum, cytokinin biosynthesis, cytokinin signaling, development
Citation: Aoki MM, Emery RJN, Anjard C, Brunetti CR and Huber RJ (2020) Cytokinins in Dictyostelia – A Unique Model for Studying the Functions of Signaling Agents From Species to Kingdoms. Front. Cell Dev. Biol. 8:511. doi: 10.3389/fcell.2020.00511
Received: 11 April 2020; Accepted: 28 May 2020;
Published: 19 June 2020.
Edited by:
Venkaiah Betapudi, Chemical and Biological Defense Division (USDHS), United StatesReviewed by:
Richard H. Gomer, Texas A&M University, United StatesShweta Saran, Jawaharlal Nehru University, India
Francisco P. Chávez, University of Chile, Chile
Pascale G. Charest, University of Arizona, United States
Copyright © 2020 Aoki, Emery, Anjard, Brunetti and Huber. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Megan M. Aoki, bWVnYW5hb2tpQHRyZW50dS5jYQ==
 Megan M. Aoki1*
Megan M. Aoki1* R. J. Neil Emery
R. J. Neil Emery Christophe Anjard
Christophe Anjard