- 1Department of Internal Medicine, Faculty of Medicine, King Abdulaziz University, Jeddah, Saudi Arabia
- 2Department of Biology, Faculty of Biological Sciences, University of Jeddah, Jeddah, Saudi Arabia
- 3Department of Clinical Laboratory Sciences, College of Applied Medical Sciences, Taif University, Makkah, Saudi Arabia
- 4Department of Medical Laboratory Technology, Faculty of Applied Medical Sciences, King Abdulaziz University, Jeddah, Saudi Arabia
- 5Center of Excellence in Genomic Medicine Research, Faculty of Applied Medical Sciences, King Abdulaziz University, Jeddah, Saudi Arabia
Sphingosine-1-phosphate (S1P) is a pleiotropic sphingolipid derived by the phosphorylation of sphingosine either by sphingosine kinase 1 (SPHK1) or SPHK2. Importantly, S1P acts through five different types of G-protein coupled S1P receptors (S1PRs) in immune cells to elicit inflammation and other immunological processes by enhancing the production of various cytokines, chemokines, and growth factors. The airway inflammation in asthma and other respiratory diseases is augmented by the activation of immune cells and the induction of T-helper cell type 2 (Th2)-associated cytokines and chemokines. Therefore, studying the S1P mediated signaling in airway inflammation is crucial to formulate effective treatment and management strategies for asthma and other respiratory diseases. The central aim of this study is to characterize the molecular targets induced through the S1P/S1PR axis and dissect the therapeutic importance of this key axis in asthma, airway inflammation, and other related respiratory diseases. To achieve this, we have adopted both high throughput next-generation knowledge discovery platforms such as SwissTargetPrediction, WebGestalt, Open Targets Platform, and Ingenuity Pathway Analysis (Qiagen, United States) to delineate the molecular targets of S1P and further validated the upstream regulators of S1P signaling using cutting edge multiple analyte profiling (xMAP) technology (Luminex Corporation, United States) to define the importance of S1P signaling in asthma and other respiratory diseases in humans.
Introduction
Sphingosine-1-phosphate (S1P), a sphingolipid, is one of the essential modulators of various cellular processes such as differentiation, survival, growth, etc. (Aarthi et al., 2011). Sphingolipids are recently termed as “morphogenetic lipids” for their crucial role in embryonic stem cell differentiation, survival, and postnatal development (Wang et al., 2018). It was recently demonstrated that S1P plays an important role in allergic anaphylaxis and asthma (Schauberger et al., 2016; Kim, 2019). Asthma is a heterogeneous and complex disease typically characterized by bronchial hyperactivity, remodeling of lung tissues, and chronic bronchial inflammation (Mims, 2015). Importantly, asthma is predisposed by both genetic and environmental factors and the incidence of Asthma has been ever increasing in both children and adults around the globe (Beran et al., 2015; McGeachie et al., 2015). Approximately 1000 people die each day due to Asthma and there were 339.4 million people affected by asthma globally in 2016. This represents a 3.6% increase in age-standardized prevalence since 2006 (Global Asthma Network, 2018; Global Initiative for Asthma, 2018). Asthma affects individuals of all ages and all ethnicity and the economic burden of respiratory diseases to governments, healthcare systems, families, and patients are on the rise globally (Singh, 2005; Ellwood et al., 2017). Hence, creating new applied scientific knowledge in the area of asthma and related respiratory diseases has become a key priority around the globe (Ellwood et al., 2017; Koshak et al., 2017).
S1P is the ligand for G-protein coupled receptors (GPCRs) that belong to the Endothelial Differentiation Gene (EDG) family of proteins termed as S1P receptors (S1PRs) (van Koppen et al., 1996; Hla, 2001; Spiegel and Milstien, 2003; Aarthi et al., 2011). S1P can specifically stimulate distinct genetic events depending on the relative expression of S1PRs as well as downstream G-proteins (An et al., 1998; Aarthi et al., 2011; Manikandan et al., 2012). The intracellular S1P is transported through ATP binding cassette transporter, ABCC1 to the extracellular milieu (Mitra et al., 2006; Nieuwenhuis et al., 2009) and causes the chemotaxis of inflammatory cells that is a pre-requisite for T-lymphocyte exit from primary and secondary lymph organs (Aarthi et al., 2011). It was shown that the S1P/S1PR axis is important for the initiation of various pathophysiological processes that possibly leading to critical diseases such as cancer, inflammatory diseases, and disorders in humans and other higher eukaryotes (Aarthi et al., 2011). Recently, FDA has approved the first oral drug, named Gilenya (FTY720 or Fingolimod), a sphingosine analog for the treatment of relapse in Multiple Sclerosis (MS) (Aarthi et al., 2011). The sphingolipid metabolism was significantly affected in asthma and the S1P levels were increased and correlated with the severity of the disease (Mohammed and Harikumar, 2017; Saluja et al., 2017). The clinical phenotypes of asthma with distinct disease mechanisms are critically influenced by levels of sphingolipid metabolites such as S1P (McGeachie et al., 2015). It was shown that the changes in sphingolipid metabolism and airway hyperresponsiveness were positively correlated in patients suffering from dust mite allergy and the intravascular levels of both sphinganine-1-phosphate (SA1P) and S1P, were significantly associated with the severity of bronchial hyperreactivity (Kowal et al., 2019). The phenotypes of asthma may be distinct based on the levels of sphingolipid metabolites and the change in sphingolipids could represent a pathophysiological variation in bronchial hypersensitivity to allergens in asthmatics (McGeachie et al., 2015).
To address this important problem, we have designed this study to delineate the functions of S1P in the development of asthma and other respiratory diseases. Here, we have utilized both next-generation knowledge discovery platforms such as SwissTargetPrediction, WebGestalt, Open Targets Platform, and Ingenuity Pathway Analysis (Qiagen, United States) to delineate the molecular targets of S1P and validated the upstream regulators of S1P signaling using cutting edge multiple analyte profiling (xMAP) technology (Luminex Corporation, United States) (Bahlas et al., 2019; Pushparaj, 2019; Harakeh et al., 2020; Jafri et al., 2020).
Materials and Methods
SwissTargetPrediction Analysis
The in silico prediction of the molecular targets of S1P was performed using Swiss Target Prediction, a virtual screening web tool (Gfeller et al., 2013). Both canonical and isomeric SMILES of S1P were used as an input sequence as described before (Daina et al., 2019). In the SwissTargetPrediction web tool, the similarity principle is used to predict the targets by reverse screening strategy. In the SwissTargetPrediction web tool, the predictions are performed from analogous molecules in 2D and 3D, from 376342 experimentally active compounds that are significantly interacting with 3068 known macromolecular targets (Daina and Zoete, 2019).
WebGestalt Analysis of S1P Targets
The Over Representation Analysis (ORA) of the molecular targets of S1P derived using Swiss Target Prediction was used in the WebGestalt tool (wGSEA) (Zhang et al., 2005; Wang et al., 2013, 2017; Liao et al., 2019). Using wGSEA gene lists obtained from large scale -omics studies were categorized based on biological, molecular, and cellular functions. wGSEA is a freely available open-source platform1 that enables a more broad, effective, flexible, and interactive functional enrichment study (Liao et al., 2019). The latest version of the wGSEA recognizes 155175 functional categories, 342 gene identifiers, and 12 organisms including many user-defined functional databases (Liao et al., 2019).
To functionally characterize the S1P-induced molecular profile, we analyzed the molecular targets of S1P obtained by Swiss Target Prediction using ORA (Liao et al., 209). Here, the enrichment method was chosen as ORA, the selected organism was Homo sapiens, and gene ontology (Molecular Function, Biological Function, and Cellular Function) was selected for each type of analysis. The reference list for each analysis included all mapped Entrez gene IDs from the selected platform genome. The parameters for the enrichment analysis included the minimum number of IDs in the category (5), the maximum number of IDs in the category (2000), the Benjamini Hochberg (BH) method (P < 0.05) for computing the False Discovery Rate (FDR) (P < 0.05), and the significance Level (Top 10).
Open Targets Platforms Analysis
The Open Targets Platform was used to find the S1P molecular targets associated with respiratory diseases (Koscielny et al., 2017; Carvalho-Silva et al., 2019; Zhang et al., 2019). The evidence from scientific literature, animal models, genomics, transcriptomics, genetics, and drugs are used in the Open Targets Platform to score and rank target-disease associations and aid target prioritization (Carvalho-Silva et al., 2019; Zhang et al., 2019). The query list with 93 molecular targets of S1P was used to find the respiratory diseases significantly (P < 0.05) regulated by the S1P signaling and its associated molecular networks.
Ingenuity Pathway Analysis
Ingenuity pathway analysis (IPA) software has a cutting edge up to date next generation knowledge base consists of clarified scientific information from publications, databases, and other relevant resources (Jafri et al., 2020). Here, we applied the IPA software (Qiagen, United States) to functionally annotate the gene clusters and identified biologically significant pathways regulated by S1P. The molecular target of S1P was subjected to Core Analysis in the IPA to delineate biologically relevant canonical pathways as well as novel molecular signatures, using the right-tailed Fisher Exact Test and Benjamini Hochberg Correction (BHC) for multiple testing (P < 0.05), affecting the respiratory diseases through S1P/SPHK pathway to deduce unique disease-causing gene clusters.
Luminex xMAP Assay for Biomarkers of Asthma
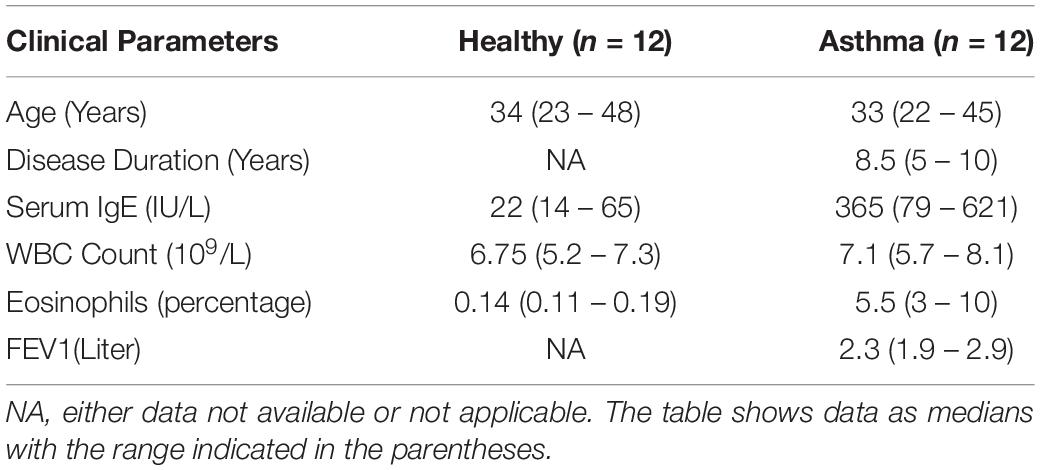
The blood samples were collected from healthy volunteers and asthma patients and both groups were age, and sex-matched. Healthy non-atopic individuals (n = 12) were used as control and the asthma patients (n = 12) were with moderate to severe disease state, non-smokers, and without any other co-morbid conditions such as chronic obstructive pulmonary disease (COPD), other types of pulmonary diseases and disorders, diabetes, cancer, autoimmune diseases, etc. The institutional ethical approval was obtained before collecting the samples and the study was approved by the Scientific Research Committee, Deanship of Scientific Research (DSR), King Abdulaziz University (KAU), Jeddah. The peripheral blood isolated was kept at room temperature for clotting and then centrifuged at 3000 rpm for 10 min for the separation of serum and the aliquots were stored at −80°C till the xMAP assays were done (Bahlas et al., 2019). The evaluation of cytokines and growth factors filtered using the next-generation knowledge discovery platforms was done in both healthy controls and asthma patients using a multi-cytokine/chemokine (30-plex) magnetic-bead based fluorescence assay and the results were obtained based on the standard curves generated using the 30plex standards (16 plex and 14 plex vials) supplied with the xMAP kit (Catalog No: LHC6003M) (Novex, Invitrogen, United States) using MAGPIX multiplex platform as described before (Bahlas et al., 2019; Pushparaj, 2019; Harakeh et al., 2020; Jafri et al., 2020; Pushparaj, 2020). Serum samples were also used to evaluate the concentration of S1P in both healthy controls and asthma patients using a competitive ELISA method as described before (Milara et al., 2012). The absorbance was measured at 450 nm using SpectraMax i3 Multi-Mode Reader with MiniMax Imaging Cytometer (Molecular Devices, United States).
Statistical Analyses
The raw xMAP data was analyzed by the xPONENT analysis software (Version 4.2) (Luminex Corporation, Austin, TX, United States) to determine the absolute concentration of cytokines and growth factors in both healthy control and asthma groups (Bahlas et al., 2019; Harakeh et al., 2020; Jafri et al., 2020). The statistical significance was further computed by using GraphPad Prism Version 8.3 (GraphPad Software, San Diego, CA, United States). P ≤ 0.05 was considered to be statistically significant based on the Student’s t-test (Two Tail). The values were represented as mean ± SD. Furthermore, an F-test to compare the variances and simple correlation analysis of S1P levels against cytokines and chemokines in the serum of asthma patients was also performed using GraphPad Prism (Version 8.3) software.
Results
In silico Prediction of the Molecular Targets of S1P Using SwissTargetPrediction
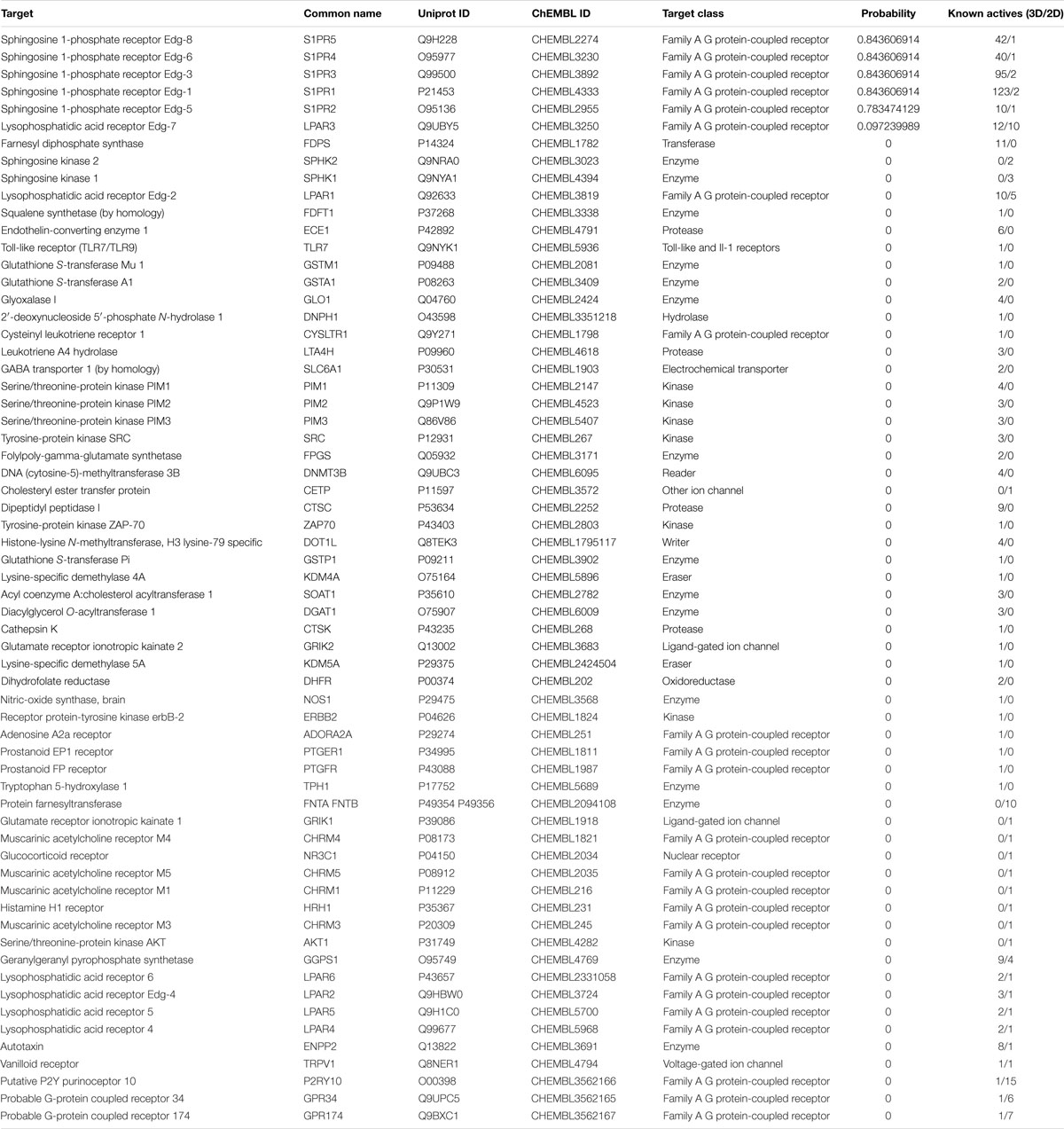
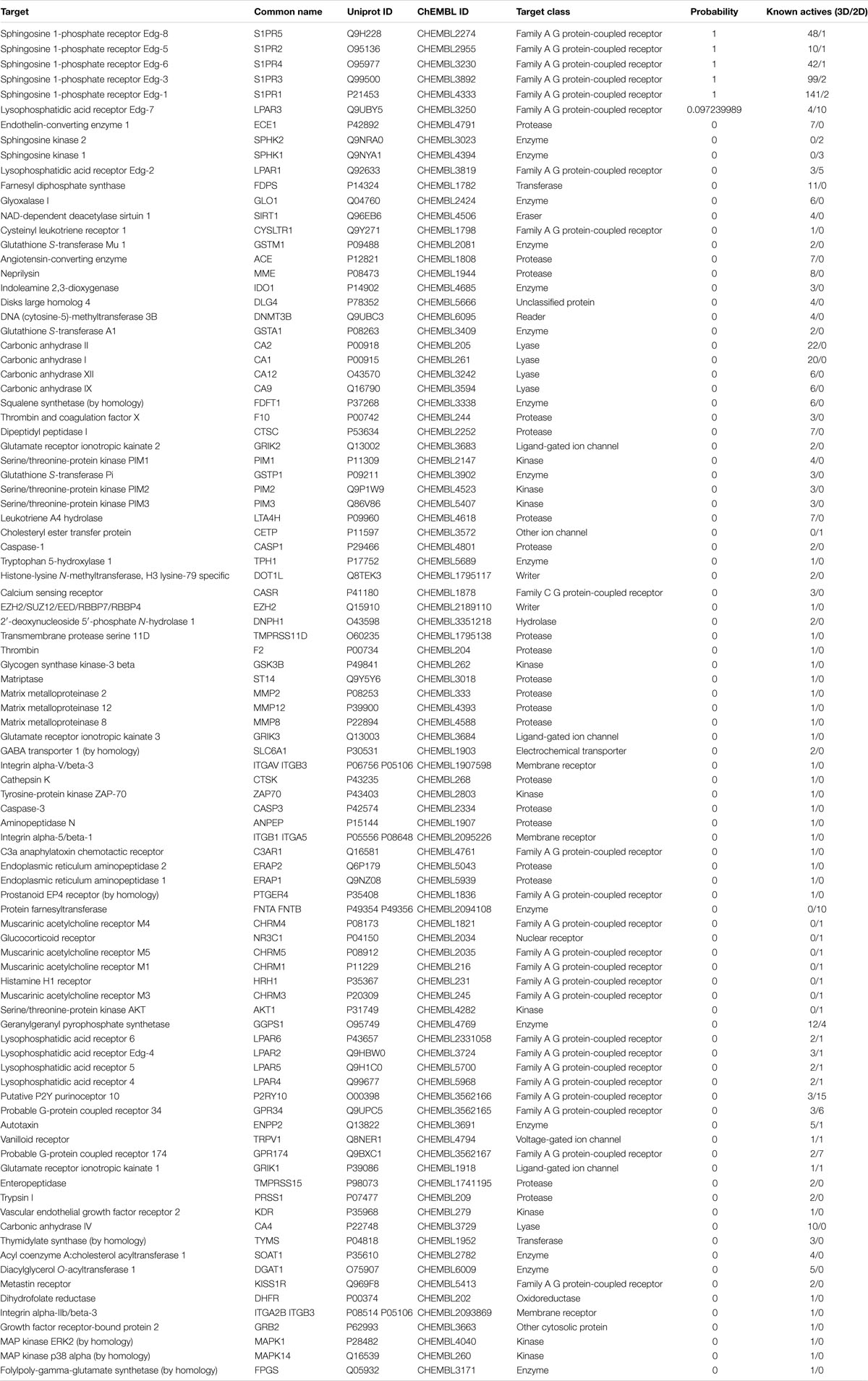
In the present study, the SwissTargetPrediction was performed for S1P (C18H38NO5P) using both Canonical [CCCCCCCCCCCCCC = CC(C(COP(= O)(O)O)N)O] and Isomeric (CCCCCCCCCCCCC/C = C/[C@H]([C@H](COP(= O) (O)O)N)O) Simplified Molecular Input Line Entry System (SMILES) codes (Supplementary Figure S1) computed by OEChem (Version 2.1.5), have shown that it interacts with 64 and 93 molecules respectively (Tables 1A,B) with the highest percentage of binding (46.7 and 53.3%, respectively) with Family A GPCRs (Figure 1).
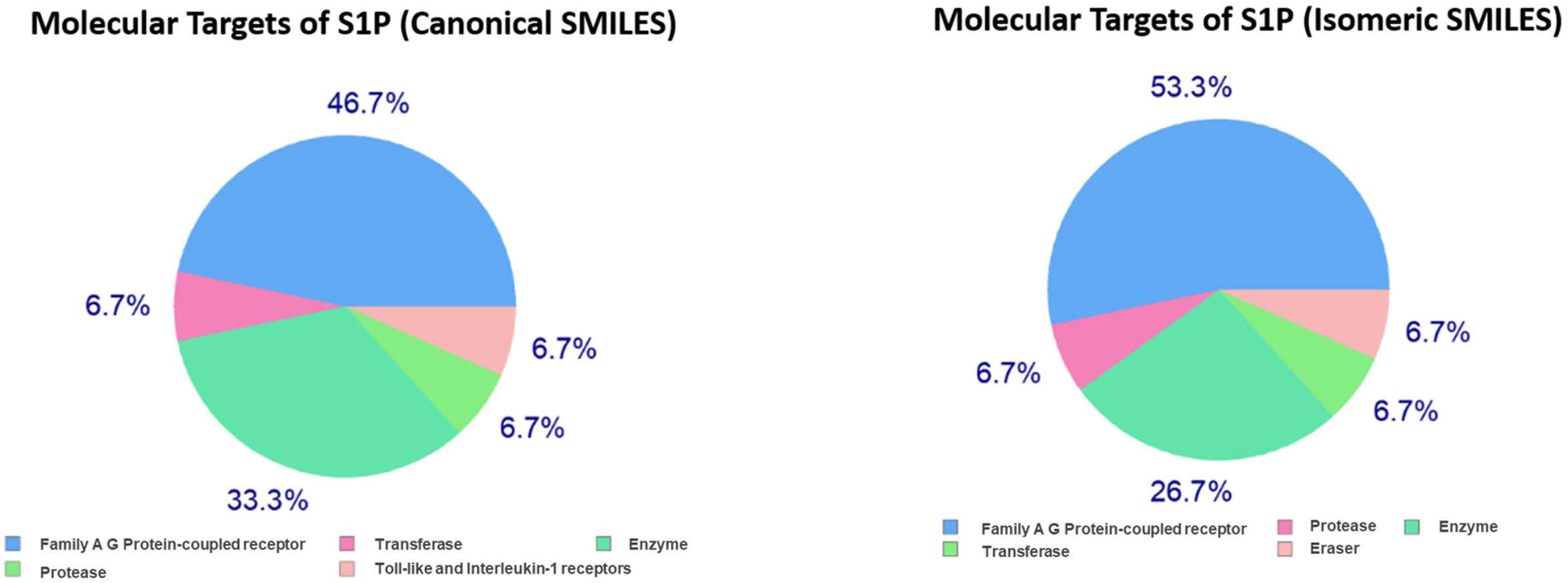
Figure 1. Swiss Target Prediction for sphingosine-1-phosphate (S1P). Top 15 percent of the molecular targets (Homo sapiens) of S1P obtained using both canonical and isomeric Simplified Molecular Input Line Entry System (SMILES) codes in Swiss Target Prediction server.
Over Representation Analysis (ORA) of the Molecular Targets of S1P Using WebGestalt
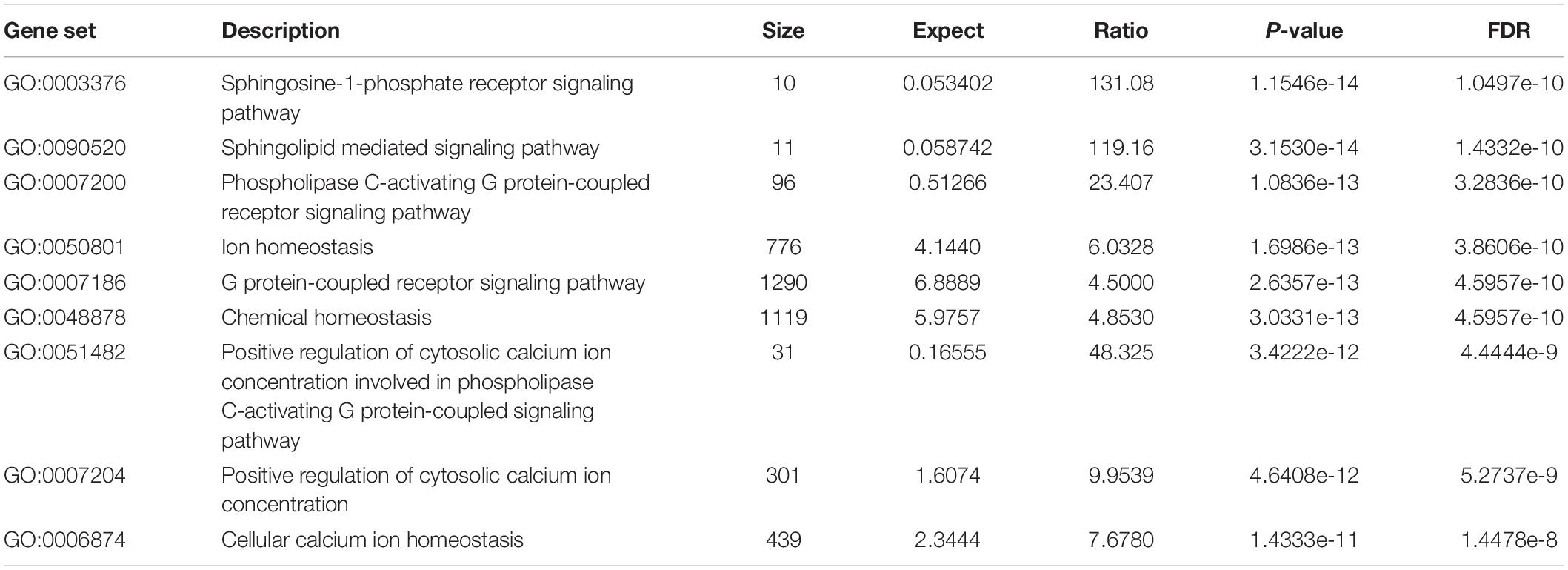
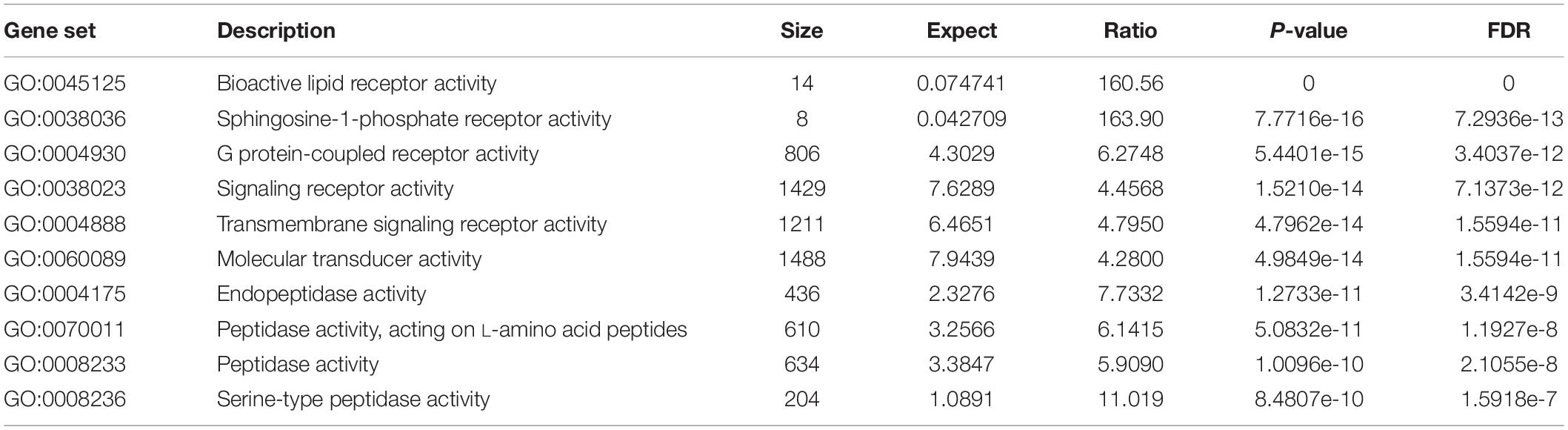
All the 93 molecular targets of S1P obtained using isomeric SMILES were used as input molecules in WebGestalt Open Source Tool to perform the ORA. GO Slim Summary for S1P Molecular Targets in Humans showing Biological Process, Cellular Component, and Molecular Function category in the red, blue, and green bar, respectively. The height of the bar represents the number of IDs in the user list and also in the category (Figure 2). The query list had 93 targets of S1P in which 89 were mapped to 89 unique Entrez gene IDs unambiguously, and the remaining 4 could not be mapped to any Entrez gene ID. Therefore, the GO Slim summary was established upon the 89 distinctive Entrez gene IDs (Figure 2). The reference list was mapped to 61506 Entrez gene IDs and 16671 IDs were annotated to the selected functional categories that were used as the reference for the enrichment analysis. The GO Biological Processes such as sphingolipid mediated signaling pathway, S1P receptor signaling pathway, chemical homeostasis, positive regulation of cytosolic calcium ion concentration, and cellular calcium ion homeostasis, and phospholipase C-activating G protein-coupled receptor signaling pathway were significantly regulated (P < 0.01; Q < 0.01) in ORA (Table 2). The GO Molecular Functions such as the bioactive lipid receptor activity, S1P receptor activity, transmembrane signaling receptor activity, molecular transducer activity, G protein-coupled receptor activity, etc. were significantly regulated (P < 0.01; Q < 0.01) in the ORA (Table 3). The molecules involved in the bioactive receptor activity (GO:0045125) comprise of GPR174, LPAR1, LPAR2, LPAR3, LPAR4, S1PR1, S1PR2, S1PR3, S1PR4, S1PR5, SPHK1, and SPHK2 (Table 4).
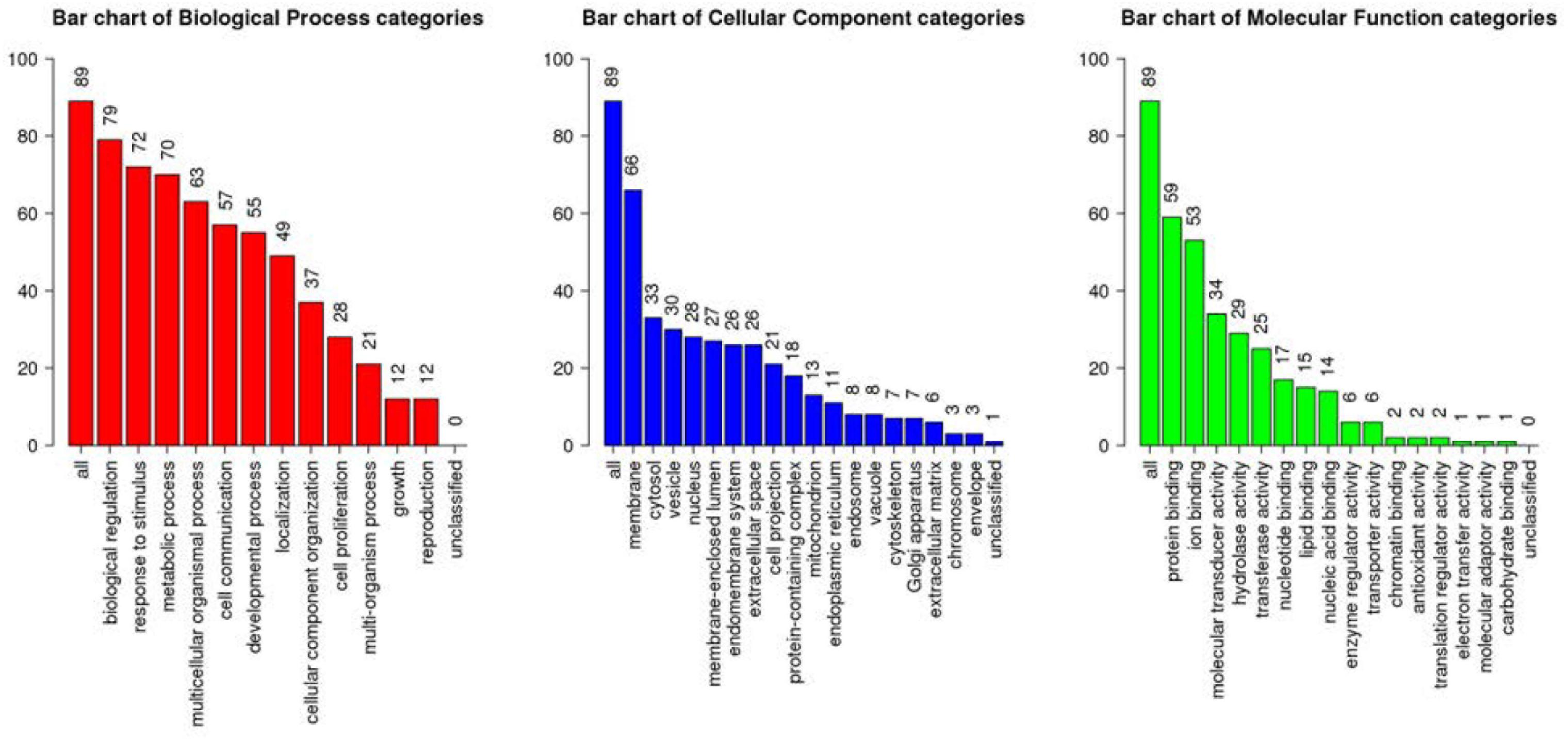
Figure 2. Over Representation Analysis (ORA) using Web-based Gene Set Analysis Toolkit (WebGestalt). GO Slim Summary for S1P molecular targets in humans showing Biological Process, Cellular Component, and Molecular Function categories in the red, blue, and green bar, respectively. The height of the bar represents the number of IDs in the user list and also in the GO category.
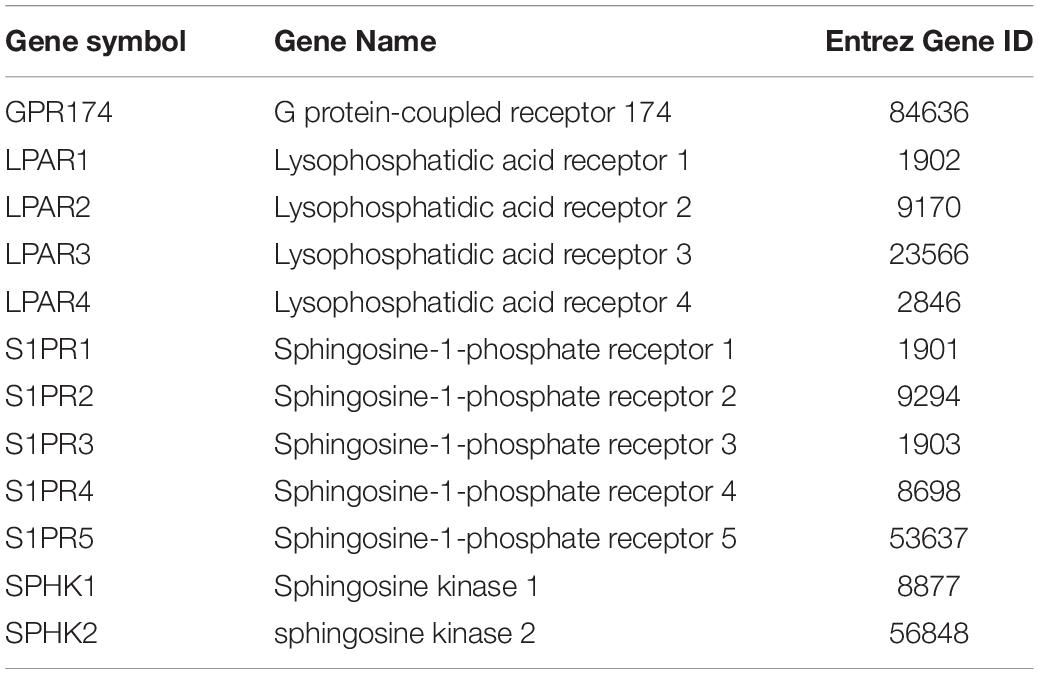
Table 4. The genes involved in the bioactive receptor activity (GO:0045125) regulated by S1P in Homo sapiens.
Identification of S1P-Induced Molecular Targets in Asthma and Other Respiratory Diseases
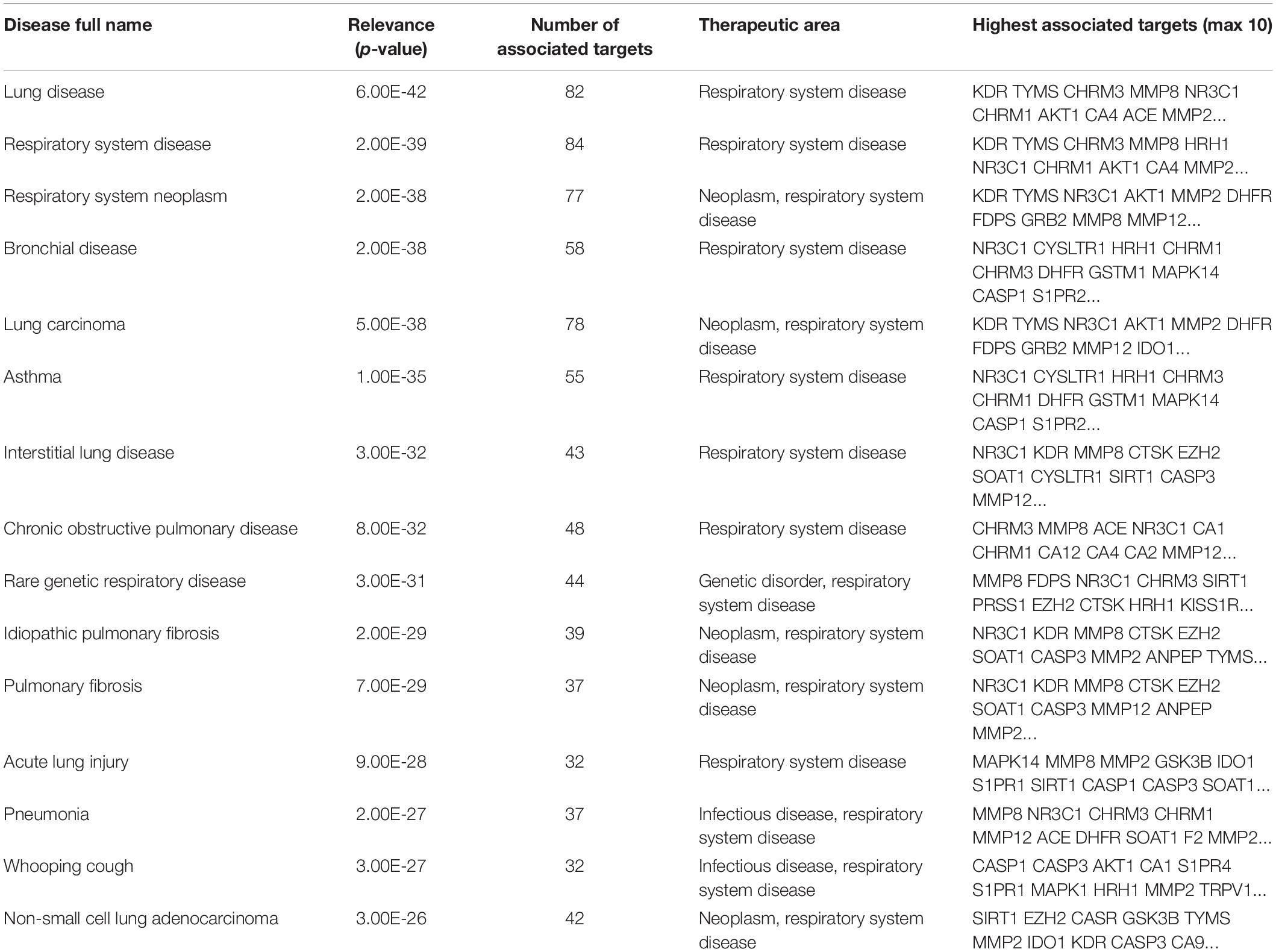
Then, we used the Open Targets Platform to find the S1P molecular targets associated with respiratory diseases. Our findings showed that about 109 types of respiratory diseases were significantly (P < 0.05) affected by the molecular targets of S1P. The top 15 respiratory diseases significantly (P < 0.01; Q < 0.01) induced by S1P and its molecular targets were lung disease, respiratory system disease, respiratory system neoplasm, bronchial disease, lung carcinoma, asthma, interstitial lung disease, COPD, Rare genetic respiratory disease, idiopathic pulmonary fibrosis, pulmonary fibrosis, acute lung injury, pneumonia, whooping cough, and non-small cell lung adenocarcinoma (Table 5).
Ingenuity Pathway Analysis of the Differentially Regulated Gene Networks by S1P/S1PR Axis in Respiratory Diseases
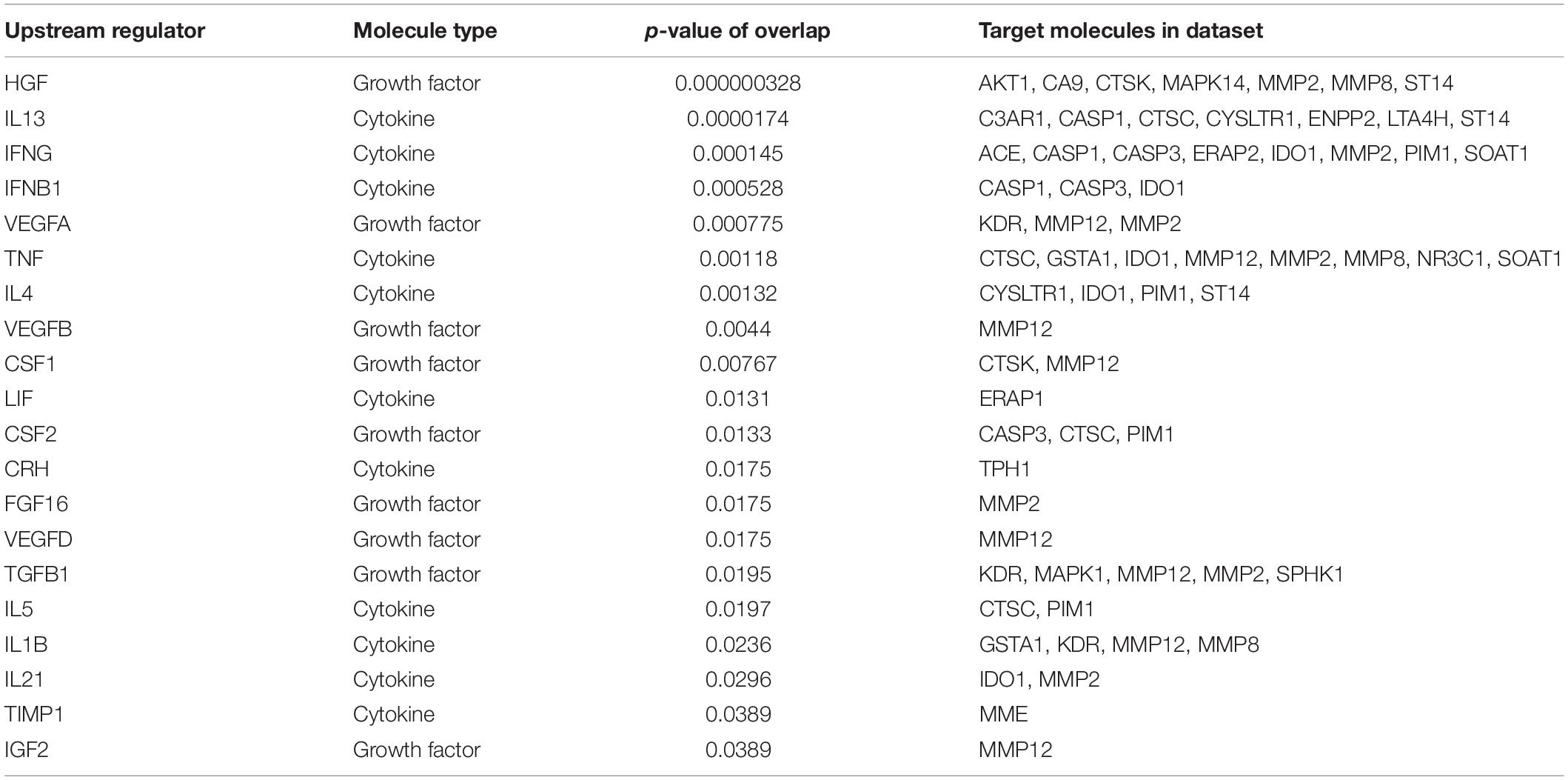
Next, we utilized the IPA to deduce the canonical pathways, upstream regulators, causal functions, diseases, and bio functions, and non-directional unique networks significantly impacted by the S1P mediated signaling molecules. The IPA core analysis of the molecular targets of S1P revealed that the canonical pathways such as eNOS signaling, S1P signaling, GPCRs signaling, ceramide signaling, G a12/13 signaling, human embryonic stem cell pluripotency, and endocannabinoid cancer inhibition pathway (Figure 3) were potentially regulated (P < 0.05). About 263 molecules play a significant role (P < 0.05) as upstream regulators of the molecular targets of S1P (Supplementary Table S1). Furthermore, we have filtered the upstream regulators based on the molecule types such as cytokines, and growth factors using the IPA. The cytokines such as Interleukin-13 (IL-13), Interferon-gamma (IFN-γ), IFN-β1, Tumor Necrosis Factor-alpha (TNF-α), Interleukin-4 (IL-4), Interleukin-5 (IL-5), Interleukin-1 (IL-1β), Interleukin-21 (IL-21), Tissue Inhibitor of Metalloproteinases-1 (TIMP-1) that has cytokine-like activity, and cytokine-inducing Corticotropin-Releasing Hormone (CRH) were found to be the upstream regulators of the S1P signaling. The growth factors such as Hepatocyte Growth Factor (HGF), Vascular Endothelial Growth Factors (VEGF), Colony Stimulating Factor 1 (CSF1) or Macrophage Colony-Stimulating Factor (M-CSF), Colony Stimulating Factor-2 (CSF2) or Granulocyte-Macrophage Colony-Stimulating Factor (GM-CSF), VEGF-A, VEGF-B, VEGF-D, Fibroblast Growth Factor 16 (FGF16), Insulin-like Growth Factor (IGF), and Transforming Growth Factor-beta 1 (TGF-β1) were identified as the upstream regulators of S1P signaling (Table 6).
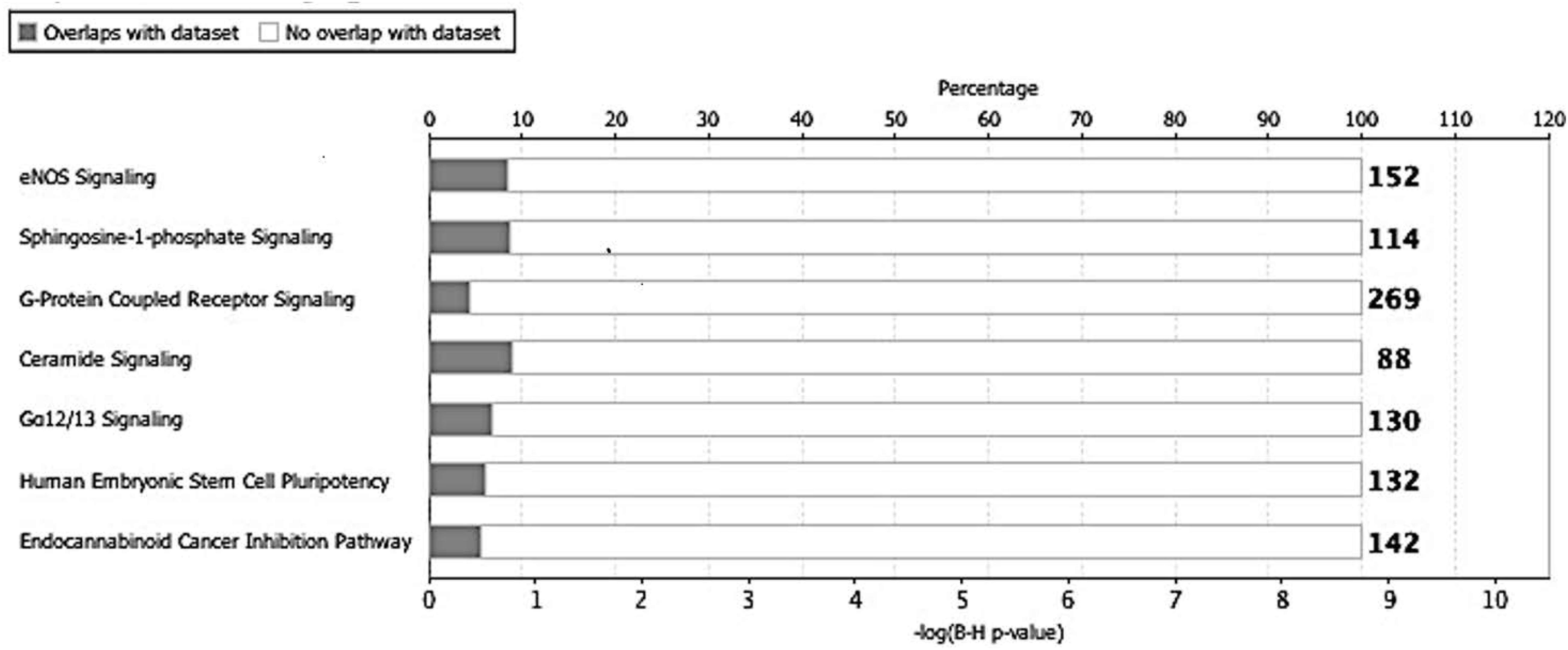
Figure 3. Canonical pathways regulated by the molecular targets of S1P. The IPA core analysis of the molecular targets of S1P revealed that the canonical pathways such as eNOS signaling, sphingosine-1-phosphate signaling, G-protein coupled receptor signaling, ceramide signaling, G a12/13 signaling, human embryonic stem cell pluripotency, and endocannabinoid cancer inhibition pathway were potentially regulated (P < 0.05).
Validation of Biomarkers in Asthma Using Luminex xMAP Technology
Besides, we have used the Luminex xMAP assay to validate the cytokines and growth factors that were upstream of the S1P signaling in asthma. Here, we have analyzed the levels of cytokines such as IL-13, IFN-γ, TNF-α, IL-4, IL-5, and IL-1β (Figure 4) and growth factors such as HGF, VEGF, and CSF2 or GM-CSF along with the estimation of serum S1P concentration in asthma patients compared with the healthy controls (Figure 5). The clinical and laboratory characteristics of asthma patients and healthy controls were given in Table 7. The upstream regulator cytokines of S1P signaling such as IL-13, IFN-γ, TNF-α, IL-4, IL-5, and IL-1β were significantly (P < 0.01) increased in the serum of asthma patients compared the healthy controls. Similarly, the growth factors that were found to be upstream of S1P signaling such as HGF, VEGF, and CSF2 were significantly (P < 0.01) increased in the serum of asthma patients compared to the healthy controls. The S1P levels were significantly increased in the serum of asthma patients compared to the healthy controls. All the cytokines and growth factors analyzed by the Luminex xMAP assay were positively correlated with the S1P levels in the serum of asthmatics (Supplementary Table S2).
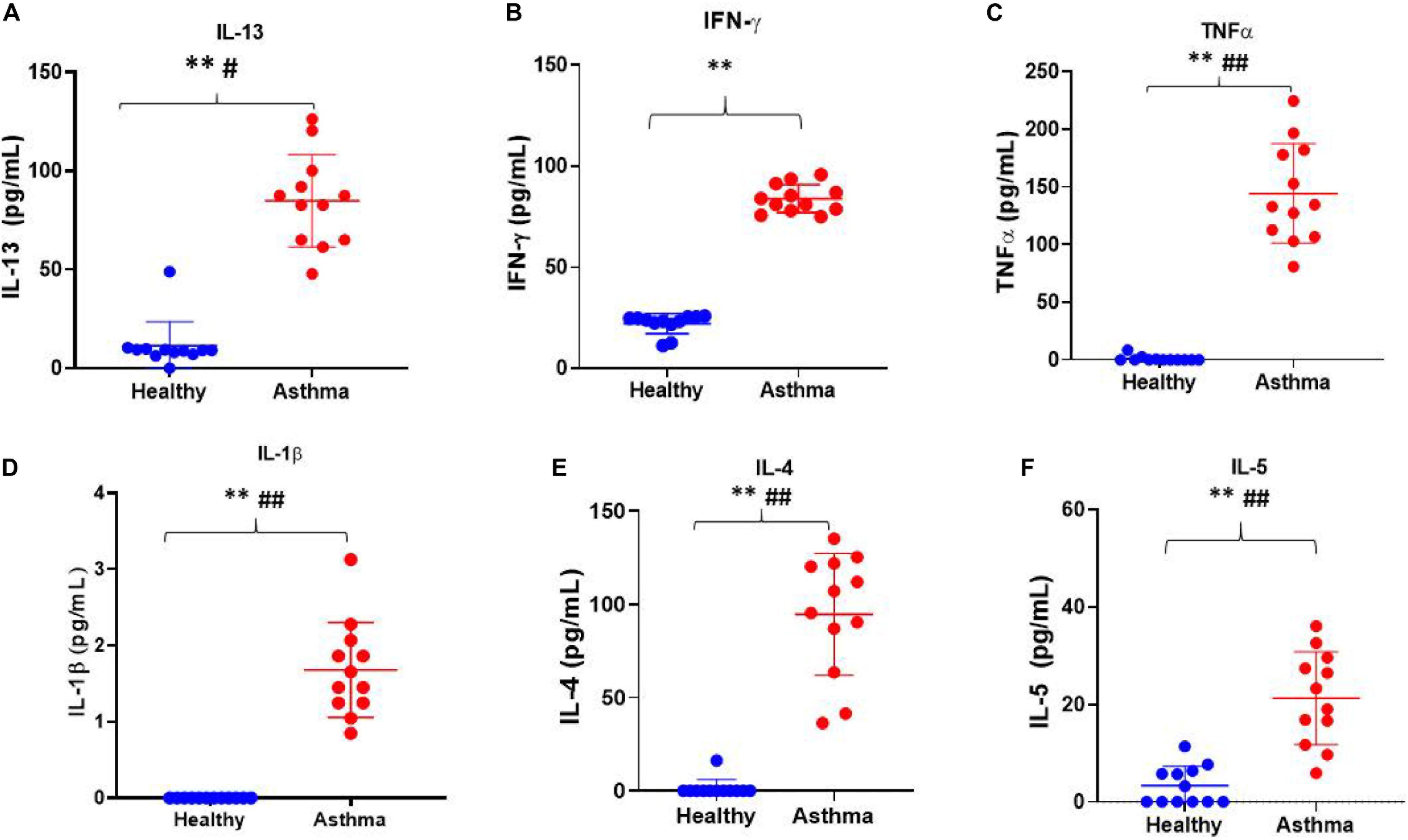
Figure 4. Luminex xMAP assay to validate the cytokines that were upstream of the S1P signaling in asthma. The upstream regulator cytokines of S1P signaling such as IL-13 (A), IFN-γ (B), TNF-α (C), IL-1β (D), IL-4 (E), and IL-5 (F) were significantly (T-test ∗∗P < 0.01, F-test #P < 0.05, and ##P < 0.01) elevated in the serum of asthma patients compared the healthy controls.
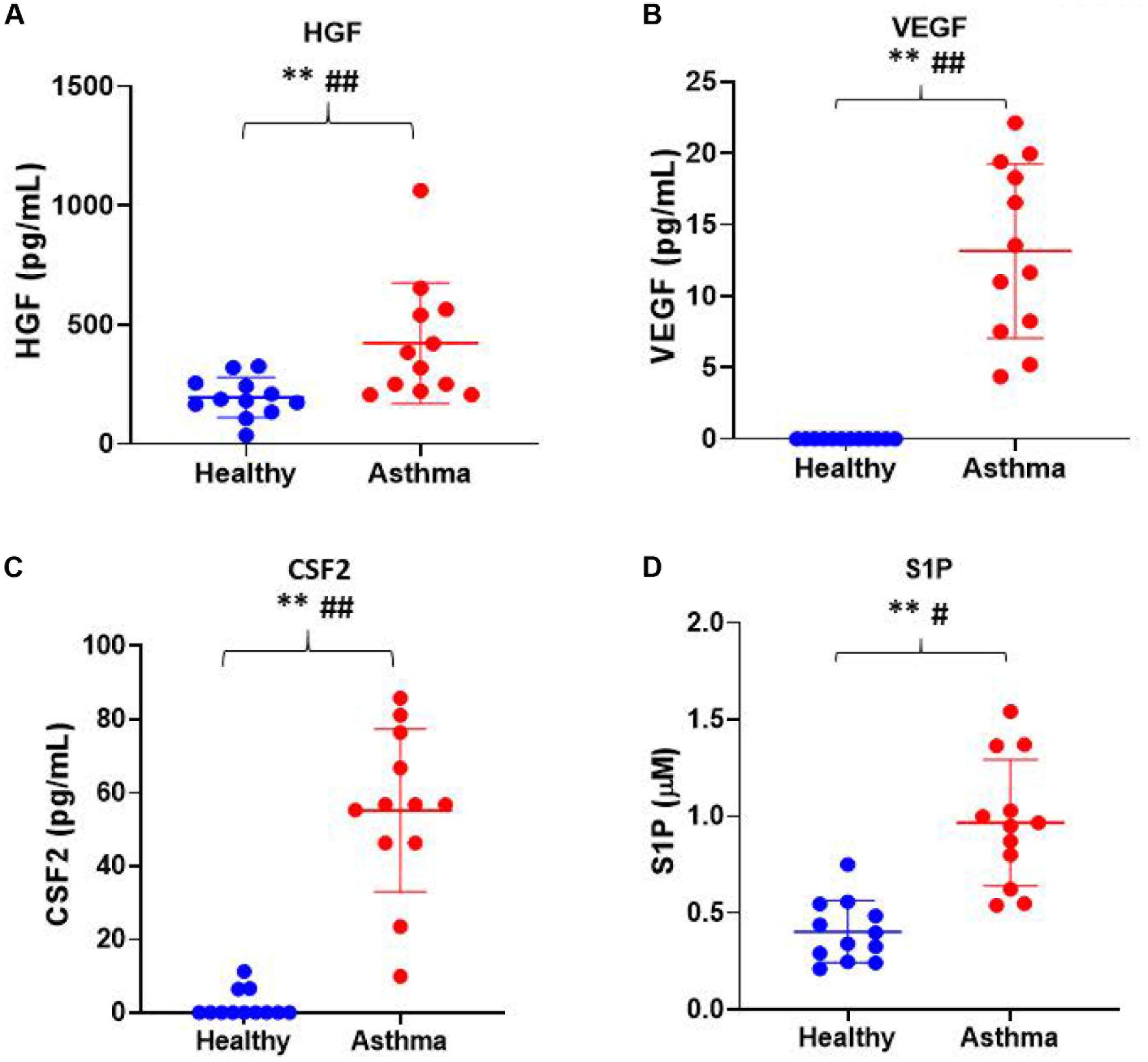
Figure 5. Luminex xMAP assay to validate the growth factors that were upstream of the S1P signaling in asthma. The growth factors that were found to be upstream of S1P signaling such as (A) HGF, (B) VEGF, (C) CSF2, and (D) S1P were significantly (T-test ∗∗P < 0.01, F-test #P < 0.05, and ##P < 0.01) elevated in the serum of asthma patients compared to the healthy controls.
Discussion
In the present study, to decode the biological and molecular functions of bioactive lipid molecule, S1P in the development of asthma and other respiratory diseases, we have applied both next-generation knowledge discovery platforms such as SwissTargetPrediction, WebGestalt, Open Targets Platform, and IPA and validated the key upstream regulators of S1P signaling using Luminex xMAP technology. Currently, knowledge discovery and big data analytical platforms are swiftly transforming the Biomedical Research landscape (Cirillo and Valencia, 2019; Paczkowska et al., 2020). All the chemical or biochemical compounds with pharmacological or pathological actions influencing diagnosis, treatment, or recuperation, and prevention of a disease can be used in next-generation knowledge discovery platforms using a unique SMILES code (Weininger, 1988; Ratnawati et al., 2018). SMILES, postulated by David Weininger, is a chemical annotation method that symbolizes a simple molecule structure (Weininger, 1988; Ratnawati et al., 2018). Here, we have used the isomeric SMILES of S1P to deduce its downstream molecular targets using the SwissTargetPrediction server (Gfeller et al., 2013; Daina and Zoete, 2019; Daina et al., 2019).
Several studies have shown that S1P augments airway hyperactivity, bronchoconstriction, and airway remodeling in asthma and other related respiratory diseases (Roviezzo et al., 2010; Petrache and Berdyshev, 2016; Liu et al., 2018) and the S1P receptors are differentially expressed in the lymphoid tissues, dendritic cells, and the lung (Aarthi et al., 2011). S1P is an effective biologically active paracrine mediator that regulates various cellular functions such as apoptosis, cell growth, cell proliferation, immune regulation, etc. (Le Stunff et al., 2004; Aarthi et al., 2011). Here, we have further identified that biological processes such as S1PR signaling pathway, sphingolipid mediated signaling pathway, chemical homeostasis, positive regulation of cytosolic calcium ion concentration, and cellular calcium ion homeostasis were regulated by S1P. Importantly, the interaction of S1P with the five types of S1PRs and four types of LPARs on the plasma membrane triggers an intracellular cascade of reactions leading to the biosynthesis of various pro-inflammatory mediators that contribute to the pathogenicity in asthma. Also, the S1P regulates various molecular functions that are essential for the pathogenesis of asthma and respiratory diseases such as the bioactive lipid receptor activity, S1P receptor activity, G protein-coupled receptor activity, signaling receptor activity, transmembrane signaling receptor activity, molecular transducer activity, endopeptidase activity, peptidase activity acting on L-amino acid peptides, peptidase activity, and serine-type peptidase activity. We found that the S1P signaling was associated with more than 100 types of respiratory diseases such as lung disease, respiratory system disease, respiratory system neoplasm, bronchial disease, lung carcinoma, asthma, interstitial lung disease, COPD, rare genetic respiratory disease, idiopathic pulmonary fibrosis, pulmonary fibrosis, acute lung injury, pneumonia, whooping cough, non-small cell lung adenocarcinoma, etc., Besides, the S1P levels were found to be increased in the bronchoalveolar lavage fluids of allergic asthma patients and were also positively associated with augmented airway inflammation (Ammit et al., 2001; Kim, 2019). Altered sphingolipid metabolism according to the phenotype and genotype of asthma was deduced using metabolic studies in asthma patients (McGeachie et al., 2015; Kim, 2019). Similarly, the sphingolipid metabolism was found to be disturbed in aspirin-exacerbated respiratory disease (AERD), a serious type of adult-onset eosinophilic asthma (Trinh et al., 2016; Reinke et al., 2017).
In the present study, we have found several upstream regulators of S1P signaling including various cytokines such as IL-13, IFN-γ, IFN-β1, TNF-α, IL-4, IL-5, IL-1β, IL-21, TIMP-1, and CRH, and growth factors such as HGF, VEGF, CSF1, CSF2, VEGF-A, VEGF-B, VEGF-D, FGF16, IGF, and TGFβ1. Using multiplex xMAP technology, we have further validated that the levels of the upstream cytokines of S1P signaling such as IL-13, IFN-γ, TNF-α, IL-4, IL-5, and IL-1β were increased in the serum of asthma patients. Similarly, the growth factors such as HGF, VEGF, and CSF2 were increased in the serum of asthmatic patients. To further decode the association of the upstream cytokines and growth factors with S1P, we have estimated the S1P concentration in the serum of asthmatics. The levels of cytokines and growth factors analyzed using xMAP technology positively correlated with the S1P levels in the serum of asthmatics which further vouch for the significance of S1P signaling in the pathogenicity of asthma.
Asthma is a chronic disease and the incidence is ever-increasing around the globe (Al-Moamary et al., 2019). Most of the asthmatics have disease exacerbation caused by the Th2 type of immune cells compared to other types such as T-helper type 1 (Th1) and T-helper type 17 (Th17) and other immune cells such as mast cells, eosinophils, and bronchial smooth muscles, myofibroblasts and epithelial cells (Manikandan et al., 2012). The Th2 associated cytokines play a vital role in the development of allergy and airway remodeling in asthma (Komai-Koma et al., 2012). The asthmatics studied here had more eosinophils in their blood and the levels of Th2 serum cytokines such as IL-4, IL-5, and IL-13 were increased compared to the healthy controls. Here, most of the cytokines and growth factors examined were associated positively with an increase in the serum S1P. The sphingosine kinases regulate the expression of Th2, Th1, Th17, chemokines, and an array of other pro-inflammatory factors by catalyzing the conversion of sphingosine into S1P (Nayak et al., 2010; Aarthi et al., 2011). Many asthmatics have an uncontrolled disease state even though they are under a treatment regimen which could be due to the increased production of S1P (Aarthi et al., 2011) and a recent study found that ceramide/S1P rheostat was indeed dysregulated in uncontrolled asthma (Kim et al., 2020). Hence, it is prudent that the ceramide/S1P production may be targeted in controlling the airway inflammation in asthma (Kim et al., 2020). Importantly, the in silico results obtained using our next generation knowledge discovery pipeline coupled with xMAP assay also showed a potential association of S1P in asthma and other respiratory diseases which is detrimental for the patients due to its ability to trigger an array of cytokines, chemokines, and growth factors. More importantly, an increase in the upstream regulator Th2 cytokines such as IL-4, IL-5, and IL-13 is potentially harmful to asthmatics and play an underlying role in the disease pathogenesis. Similarly, the growth factors such as VEGF, CSF2, and HGF were found to be increased in asthmatics and may also be responsible for making the disease chronically worse. A recent study showed that the inhibition of S1P receptors potentially augments the inhibition of VEGF (Fischl et al., 2019). The S1P/S1PR signaling was regulated through the activation of SPHK1 and SPHK 2 to produce S1P by the phosphorylation of sphingosine (Nayak et al., 2010; Aarthi et al., 2011) and the airway hyperresponsiveness and inflammation was potentially reduced by SPHK inhibitors and FTY720, a synthetic analog of S1P and S1PR agonist (Aarthi et al., 2011) and other S1PR modulators (Marciniak et al., 2018), in animal models of allergic asthma (Sawicka et al., 2003; Idzko et al., 2006; Price et al., 2013).
We conclude that the S1P and its associated signaling molecules, either upstream or downstream, are pharmacological targets of great significance for the development of novel drugs for asthma and other related respiratory diseases in humans. However, one of the critical limitations of this study is the less number of samples used for the validation of biomarkers in the serum associated with S1P signaling in asthma. In future, we will be collecting more samples from patients suffering from atopic and non-atopic asthma and other respiratory diseases and disorders with varying severity that undergo different treatment regimens, for further validation of upstream regulator molecules associated with S1P signaling along with the key cytokines, chemokines, and growth factors that play a crucial role in the pathogenesis of an array of respiratory diseases and disorders.
Data Availability Statement
The datasets generated for this study are available on request to the corresponding author.
Ethics Statement
The studies involving human participants were reviewed and approved by our institutional ethical committee, and the study was accepted by the Scientific Research Committee, Deanship of Scientific Research (DSR), King Abdulaziz University (KAU), Jeddah. The patients/participants provided their written informed consent to participate in this study.
Author Contributions
All authors designed and conducted the experiments, analyzed the data, wrote the manuscript, and contributed to the editing of the manuscript and the scientific discussions. SB, LD, and PP proposed the research idea.
Funding
This work was approved and funded by the Deanship of Scientific Research (DSR), the King Abdulaziz University, Jeddah, the Kingdom of Saudi Arabia (KSA) through Grant Number G-608-140-37.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The handling editor declared a past co-authorship with one of the authors PP.
Acknowledgments
We acknowledge with thanks the Deanship of Scientific Research (DSR), King Abdulaziz University, for their excellent technical support.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcell.2020.00444/full#supplementary-material
FIGURE S1 | The flow diagram illustrates the essential steps in the in silico experiments using next-generation knowledge discovery platforms to uncover the molecular targets of S1P and their association with various respiratory diseases and disorders and the subsequent validation of disease-specific biomarkers and their relationship with S1P in Asthma.
TABLE S1 | The list of 263 upstream regulators identified based on the IPA core analysis of the molecular targets of S1P.
TABLE S2 | Simple linear regression analysis of S1P with upstream regulatory cytokines and growth factors in asthma patients.
Footnotes
References
Aarthi, J. J., Darendeliler, M. A., and Pushparaj, P. N. (2011). Dissecting the role of the S1P/S1PR axis in health and disease. J. Dent. Res. 90, 841–854. doi: 10.1177/0022034510389178
Al-Moamary, M., Alhaider, S., Alangari, A., Al Ghobain, M., Zeitouni, M., Idrees, M., et al. (2019). The Saudi initiative for asthma - 2019 update: guidelines for the diagnosis and management of asthma in adults and children. Ann. Thorac. Med. 14, 3–48. doi: 10.4103/atm.ATM-327-18
Ammit, A. J., Hastie, A. T., Edsall, L. C., Hoffman, R. K., Amrani, Y., Krymskaya, V. P., et al. (2001). Sphingosine 1-phosphate modulates human airway smooth muscle cell functions that promote inflammation and airway remodeling in asthma. FASEB J. 15, 1212–1214. doi: 10.1096/fj.00-0742fje
An, S., Goetzl, E. J., and Lee, H. (1998). Signaling mechanisms and molecular characteristics of G protein-coupled receptors for lysophosphatidic acid and sphingosine 1-phosphate. J. Cell. Biochem. 72, 147–157. doi: 10.1002/(sici)1097-4644(1998)72:30/31%2B<147::aid-jcb19>3.0.co;2-f
Bahlas, S., Damiati, L., Dandachi, N., Sait, H., Alsefri, M., and Pushparaj, P. N. (2019). Rapid immunoprofiling of cytokines, chemokines, and growth factors in patients with active rheumatoid arthritis using Luminex Multiple Analyte Profiling technology for precision medicine. Clin. Exp. Rheumatol. 37, 112–119.
Beran, D., Zar, H. J., Perrin, C., Menezes, A. M., and Burney, P. (2015). Burden of asthma and chronic obstructive pulmonary disease and access to essential medicines in low-income and middle-income countries. Lancet Respir. Med. 3, 159–170. doi: 10.1016/S2213-2600(15)00004-1
Carvalho-Silva, D., Pierleoni, A., Pignatelli, M., Ong, C. K., Fumis, L., Karamanis, N., et al. (2019). Open targets platform: new developments and updates two years on. Nucleic Acids Res. 47, D1056–D1065. doi: 10.1093/nar/gky1133
Cirillo, D., and Valencia, A. (2019). Big data analytics for personalized medicine. Curr. Opin. Biotechnol. 58, 161–167. doi: 10.1016/j.copbio.2019.03.004
Daina, A., Michielin, O., and Zoete, V. (2019). SwissTargetPrediction: updated data and new features for efficient prediction of protein targets of small molecules. Nucleic Acids Res. 47, W357–W364. doi: 10.1093/nar/gkz382
Daina, A., and Zoete, V. (2019). Application of the swissdrugdesign online resources in virtual screening. Int. J. Mol. Sci. 20:4612. doi: 10.3390/ijms20184612
Ellwood, P., Asher, M. I., Billo, N. E., Bissell, K., Chiang, C. Y., Ellwood, E. M., et al. (2017). The Global Asthma Network rationale and methods for Phase I global surveillance: prevalence, severity, management and risk factors. Eur. Respir. J. 49:1601605. doi: 10.1183/13993003.01605-2016
Fischl, A. S., Wang, X., Falcon, B. L., Almonte-Baldonado, R., Bodenmiller, D., Evans, G., et al. (2019). Inhibition of Sphingosine phosphate receptor 1 signaling enhances the efficacy of VEGF Receptor Inhibition. Mol. Cancer. Ther. 18, 856–867. doi: 10.1158/1535-7163
Gfeller, D., Michielin, O., and Zoete, V. (2013). Shaping the interaction landscape of bioactive molecules. Bioinformatics 29, 3073–3079. doi: 10.1093/bioinformatics/btt540
Global Asthma Network (2018). The Global Asthma Report 2018. Available online at: http://www.globalasthmareport.org/Global%20Asthma%20Report%202018.pdf (accessed January 18, 2020).
Global Initiative for Asthma (2018). 2018 GINA Report: Global Strategy for Asthma Management and Prevention 2018. Available online at: https://ginasthma.org/wp-content/uploads/2018/04/wms-GINA-2018-report-tracked_v1.3.pdf (accessed January 18, 2020).
Harakeh, S., Kalamegam, G., Pushparaj, P. N., Al-Hejin, A., Alfadul, S. M., Al Amri, T., et al. (2020). Chemokines and their association with body mass index among healthy Saudis. Saudi J. Biol. Sci. 27, 6–11. doi: 10.1016/j.sjbs.2019.03.00
Hla, T. (2001). Sphingosine 1-phosphate receptors. Prostaglandins Other Lipid Mediat. 64, 135–142. doi: 10.1016/S0090-6980(01)00109-5
Idzko, M., Hammad, H., Van Nimwegen, M., Kool, M., Müller, T., Soullié, T., et al. (2006). Local application of FTY720 to the lung abrogates experimental asthma by altering dendritic cell function. J. Clin. Invest. 116, 2935–2944. doi: 10.1172/JCI28295
Jafri, M. A., Kalamegam, G., Abbas, M., Al-Kaff, M., Ahmed, F., Bakhashab, S., et al. (2020). Deciphering the association of cytokines, chemokines, and growth factors in chondrogenic differentiation of human bone marrow mesenchymal stem cells using an ex vivo osteochondral culture system. Front. Cell Dev. Biol. 7:380. doi: 10.3389/fcell.2019.00380
Kim, S. H. (2019). Sphingosine-1-phosphate: biomarker, contributor, or target for asthma? Allergy Asthma Immunol. Res. 11, 299–301. doi: 10.4168/aair.2019.11.3.299
Kim, S. H., Jung, H. W., Kim, M., Moon, J. Y., Ban, G. Y., Kim, S. J., et al. (2020). Ceramide/sphingosine-1-phosphate imbalance is associated with distinct inflammatory phenotypes of uncontrolled asthma. Allergy. doi: 10.1111/all.14236 [Epub ahead of print].
Komai-Koma, M., Brombacher, F., Pushparaj, P. N., Arendse, B., McSharry, C., Alexander, J., et al. (2012). Interleukin-33 amplifies IgE synthesis and triggers mast cell degranulation via interleukin-4 in naïve mice. Allergy Eur. J. Allergy Clin. Immunol. 67, 1118–1126. doi: 10.1111/j.1398-9995.2012.02859.x
Koscielny, G., An, P., Carvalho-Silva, D., Cham, J. A., Fumis, L., Gasparyan, R., et al. (2017). Open targets: a platform for therapeutic target identification and validation. Nucleic Acids Res. 45, D985–D994. doi: 10.1093/nar/gkw1055
Koshak, A., Wei, L., Koshak, E., Wali, S., Alamoudi, O., Demerdash, A., et al. (2017). Nigella sativa supplementation improves asthma control and biomarkers: a randomized, double-blind, placebo-controlled trial. Phyther. Res. 31, 403–409. doi: 10.1002/ptr.5761
Kowal, K., Żebrowska, E., and Chabowski, A. (2019). Altered sphingolipid metabolism is associated with asthma phenotype in house dust mite-allergic patients. Allergy Asthma Immunol. Res. 11, 330–342. doi: 10.4168/aair.2019.11.3.330
Le Stunff, H., Milstien, S., and Spiegel, S. (2004). Generation and metabolism of bioactive sphingosine-1-phosphate. J. Cell. Biochem. 92, 882–899. doi: 10.1002/jcb.20097
Liao, Y., Wang, J., Jaehnig, E. J., Shi, Z., and Zhang, B. (2019). WebGestalt 2019: gene set analysis toolkit with revamped UIs and APIs. Nucleic Acids Res. 47, W199–W205. doi: 10.1093/nar/gkz401
Liu, L., Zhai, C., Pan, Y., Zhu, Y., Shi, W., Wang, J., et al. (2018). Sphingosine-1-phosphate induces airway smooth muscle cell proliferation, migration, and contraction by modulating hippo signaling effector yap. Am. J. Physiol. Lung Cell. Mol. Physiol. 315, L609–L621. doi: 10.1152/ajplung.00554.2017
Manikandan, J., Kothandaraman, N., Hande, M. P., and Pushparaj, P. N. (2012). Deciphering the structure and function of FcεRI/mast cell axis in the regulation of allergy and anaphylaxis: a functional genomics paradigm. Cell. Mol. Life Sci. 69, 1917–1929. doi: 10.1007/s00018-011-0886-0
Marciniak, A., Camp, S. M., Garcia, J. G. N., and Polt, R. (2018). An update on sphingosine-1-phosphate receptor 1 modulators. Bioorg. Med. Chem. Lett. 28, 3585–3591. doi: 10.1016/j.bmcl.2018.10.042
McGeachie, M. J., Dahlin, A., Qiu, W., Croteau-Chonka, D. C., Savage, J., Wu, A. C., et al. (2015). The metabolomics of asthma control: a promising link between genetics and disease. Immun. Inflamm. Dis. 3, 224–238. doi: 10.1002/iid3.61
Milara, J., Navarro, R., Juan, G., Peiró, T., Serrano, A., Ramón, M., et al. (2012). Sphingosine-1-phosphate is increased in patients with idiopathic pulmonary fibrosis and mediates epithelial to mesenchymal transition. Thorax 67, 147–156. doi: 10.1136/thoraxjnl-2011-200026
Mims, J. W. (2015). Asthma: definitions and pathophysiology. Int. Forum Allergy Rhinol. 5, S2–S6. doi: 10.1002/alr.21609
Mitra, P., Oskeritzian, C. A., Payne, S. G., Beaven, M. A., Milstien, S., and Spiegel, S. (2006). Role of ABCC1 in export of sphingosine-1-phosphate from mast cells. Proc. Natl. Acad. Sci. U.S.A. 103, 16394–16399. doi: 10.1073/pnas.0603734103
Mohammed, S., and Harikumar, K. B. (2017). Sphingosine 1-phosphate: a novel target for lung disorders. Front. Immunol. 8:296. doi: 10.3389/fimmu.2017.00296
Nayak, D., Huo, Y., Kwang, W. X. T., Pushparaj, P. N., Kumar, S. D., Ling, E. A., et al. (2010). Sphingosine kinase 1 regulates the expression of proinflammatory cytokines and nitric oxide in activated microglia. Neuroscience 166, 132–144. doi: 10.1016/j.neuroscience.2009.12.020
Nieuwenhuis, B., Lüth, A., Chun, J., Huwiler, A., Pfeilschifter, J., Schäfer-Korting, M., et al. (2009). Involvement of the ABC-transporter ABCC1 and the sphingosine 1-phosphate receptor subtype S1P3 in the cytoprotection of human fibroblasts by the glucocorticoid dexamethasone. J. Mol. Med. 87, 645–657. doi: 10.1007/s00109-009-0468-x
Paczkowska, M., Barenboim, J., Sintupisut, N., Fox, N. S., Zhu, H., Abd-Rabbo, D., et al. (2020). Integrative pathway enrichment analysis of multivariate omics data. Nat. Commun. 11:735. doi: 10.1038/s41467-019-13983-9
Petrache, I., and Berdyshev, E. V. (2016). Ceramide signaling and metabolism in pathophysiological states of the lung. Annu. Rev. Physiol. 78, 463–480. doi: 10.1146/annurev-physiol-021115-105221
Price, M. M., Oskeritzian, C. A., Falanga, Y. T., Harikumar, K. B., Allegood, J. C., Alvarez, S. E., et al. (2013). A specific sphingosine kinase 1 inhibitor attenuates airway hyperresponsiveness and inflammation in a mast cell-dependent murine model of allergic asthma. J. Allergy Clin. Immunol. 131, 501–511.e1. doi: 10.1016/j.jaci.2012.07.014
Pushparaj, P. N. (2019). “Multiple analyte profiling (xMAP) technology coupled with functional bioinformatics strategies: potential applications in protein biomarker profiling in autoimmune inflammatory diseases,” in Essentials of Bioinformatics, Vol. II, eds N. A. Shaik et al. (Cham: Springer), 151–165. doi: 10.1007/978-3-030-18375-2_9
Pushparaj, P. N. (2020). “Translational interest of immune profiling,” in Precision Medicine for Investigators, Practitioners and Providers, eds J. Faintuch and S. Faintuch (Cambridge, MA: Academic Press), 105–122. doi: 10.1016/b978-0-12-819178-1.00011-3
Ratnawati, D. E., Marjono, M., and Anam, S. (2018). Prediction of active compounds from SMILES codes using backpropagation algorithm. AIP Conf. Proc. 2021:060009. doi: 10.1063/1.5062773
Reinke, S. N., Gallart-Ayala, H., Gómez, C., Checa, A., Fauland, A., Naz, S., et al. (2017). Metabolomics analysis identifies different metabotypes of asthma severity. Eur. Respir. J. 49:1601740. doi: 10.1183/13993003.01740-2016
Roviezzo, F., D’Agostino, B., Brancaleone, V., De Gruttola, L., Bucci, M., De Dominicis, G., et al. (2010). Systemic administration of sphingosine-1-phosphate increases bronchial hyperresponsiveness in the mouse. Am. J. Respir. Cell Mol. Biol. 42, 572–577. doi: 10.1165/rcmb.2009-0108OC
Saluja, R., Kumar, A., Jain, M., Goel, S. K., and Jain, A. (2017). Role of sphingosine-1-phosphate in mast cell functions and asthma and its regulation by non-coding RNA. Front. Immunol. 8:587. doi: 10.3389/fimmu.2017.00587
Sawicka, E., Zuany-Amorim, C., Manlius, C., Trifilieff, A., Brinkmann, V., Kemeny, D. M., et al. (2003). Inhibition of Th1- and Th2-mediated airway inflammation by the sphingosine 1-phosphate receptor agonist FTY720. J. Immunol. 171, 6206–6214. doi: 10.4049/jimmunol.171.11.6206
Schauberger, E., Peinhaupt, M., Cazares, T., and Lindsley, A. W. (2016). Lipid mediators of allergic disease: pathways, treatments, and emerging therapeutic targets. Curr. Allergy Asthma Rep. 16:48. doi: 10.1007/s11882-016-0628-3
Singh, M. (2005). The burden of asthma in children: an Asian perspective. Paediatr. Respir. Rev. 6, 14–19. doi: 10.1016/j.prrv.2004.11.003
Spiegel, S., and Milstien, S. (2003). Sphingosine-1-phosphate: an enigmatic signalling lipid. Nat. Rev. Mol. Cell Biol. 4, 397–407. doi: 10.1038/nrm1103
Trinh, H. K. T., Kim, S. C., Cho, K., Kim, S. J., Ban, G. Y., Yoo, H. J., et al. (2016). Exploration of the sphingolipid metabolite, sphingosine-1-phosphate and sphingosine, as novel biomarkers for aspirin-exacerbated respiratory disease. Sci. Rep. 6:36599. doi: 10.1038/srep36599
van Koppen, C. J., Meyer Zu Heringdorf, D., Laser, K. T., Zhang, C., Jakobs, K. H., Bünemann, M., et al. (1996). Activation of a high affinity Gi protein-coupled plasma membrane receptor by sphingosine-1-phosphate. J. Biol. Chem. 271, 2082–2087. doi: 10.1074/jbc.271.4.2082
Wang, G., Spassieva, S. D., and Bieberich, E. (2018). Ceramide and S1P signaling in embryonic stem cell differentiation. Methods Mol. Biol. 1697, 153–171. doi: 10.1007/7651_2017_43
Wang, J., Duncan, D., Shi, Z., and Zhang, B. (2013). WEB-based GEne SeT AnaLysis Toolkit (WebGestalt): update 2013. Nucleic Acids Res. 41, W77–W83. doi: 10.1093/nar/gkt439
Wang, J., Vasaikar, S., Shi, Z., Greer, M., and Zhang, B. (2017). WebGestalt 2017: a more comprehensive, powerful, flexible and interactive gene set enrichment analysis toolkit. Nucleic Acids Res. 45, W130–W137. doi: 10.1093/nar/gkx356
Weininger, D. (1988). SMILES, a chemical language and information system: 1: introduction to methodology and encoding rules. J. Chem. Inf. Comput. Sci. 28, 31–36. doi: 10.1021/ci00057a005
Zhang, B., Kirov, S., and Snoddy, J. (2005). WebGestalt: an integrated system for exploring gene sets in various biological contexts. Nucleic Acids Res. 33, W741–W748. doi: 10.1093/nar/gki475
Keywords: asthma, respiratory diseases, sphingosine-1-phosphate, SwissTargetPrediction, WebGestalt, open targets platform, ingenuity pathway analysis, Luminex xMAP technology
Citation: Bahlas S, Damiati LA, Al-Hazmi AS and Pushparaj PN (2020) Decoding the Role of Sphingosine-1-Phosphate in Asthma and Other Respiratory System Diseases Using Next Generation Knowledge Discovery Platforms Coupled With Luminex Multiple Analyte Profiling Technology. Front. Cell Dev. Biol. 8:444. doi: 10.3389/fcell.2020.00444
Received: 12 February 2020; Accepted: 12 May 2020;
Published: 19 June 2020.
Edited by:
Ahmed Rebai, Centre of Biotechnology of Sfax, TunisiaReviewed by:
Shafiq Ur Rehman, Gyeongsang National University, South KoreaIkram Ullah, International Islamic University, Islamabad, Pakistan
Copyright © 2020 Bahlas, Damiati, Al-Hazmi and Pushparaj. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sami Bahlas, ZHJiYWhsYXNAZ21haWwuY29t
 Sami Bahlas1*
Sami Bahlas1* Ayman S. Al-Hazmi
Ayman S. Al-Hazmi Peter Natesan Pushparaj
Peter Natesan Pushparaj