- 1Department of General Surgery, Clinical Research Center of Geriatric Diseases in Hubei Province, Tongji Hospital, Tongji Medical College, Huazhong University of Science and Technology, Wuhan, China
- 2Department of Pediatrics, Union Hospital, Tongji Medical College, Huazhong University of Science and Technology, Wuhan, China
- 3Department of Hepatobiliary and Pancreatic Surgery, The First Affiliated Hospital of Zhengzhou University, Zhengzhou, China
- 4Hepatic Surgery Center, Key Laboratory of Organ Transplantation, Ministry of Education and Ministry of Public Health, Tongji Hospital, Tongji Medical College, Huazhong University of Science and Technology, Wuhan, China
Hepatocellular carcinoma (HCC) is one of the most common malignancies with poor outcomes. The main causes of HCC-related deaths are recurrence and metastasis. Long noncoding RNAs (lncRNAs) are recently identified as critical regulators in cancers. However, the lncRNAs involved in HCC recurrence and metastasis are poorly understood. In this study, via analyzing The Cancer Genome Atlas Liver Hepatocellular Carcinoma dataset, we identified a novel lncRNA LINC01134, which is highly expressed in HCC tissues and correlated with microvascular invasion, macrovascular invasion, recurrence, and poor overall survival of HCC patients. Functional experiments revealed that ectopic expression of LINC01134 promotes HCC cell migration and invasion in vitro and HCC liver metastasis and lung metastasis in vivo. Knockdown of LINC01134 represses HCC cell migration and invasion in vitro and HCC liver metastasis and lung metastasis in vivo. Mechanistically, we found that LINC01134 directly binds the promoter of AKT1S1 and activates AKT1S1 expression. Via activating AKT1S1, LINC01134 further activates NF-κB signaling. The expression of LINC01134 is significantly positively correlated with that of AKT1S1 in HCC tissues. In line with LINC01134, AKT1S1 is also highly expressed in HCC tissues and correlated with poor survival of HCC patients. Functional rescue experiments showed that repressing AKT1S1 or NF-κB signaling abrogates the roles of LINC01134 in HCC. Taken together, these findings recognized LINC01134 as a novel oncogenic lncRNA, which indicates vascular invasion, recurrence, and poor overall survival of HCC patients. LINC01134 promotes HCC metastasis via activating AKT1S1 expression and subsequently activating NF-κB signaling. This study suggested LINC01134 as a potential prognostic biomarker and therapeutic target for HCC.
Introduction
Liver cancer is one of the most common malignances worldwide (Bray et al., 2018). It is ranked sixth for incidence and fourth for mortality, with 841,080 estimated new cases and 781,631 estimated deaths in 2018 globally (Bray et al., 2018). Although the overall cancer mortality has fallen since 1991, the mortality for liver cancer is still increasing until now (Siegel et al., 2020). Hepatocellular carcinoma (HCC) is the major subtype and accounts for 90% of liver cancer. Very limited treatment options are available for HCC (Wen et al., 2019). Therefore, the outcomes of most HCC patients are still very poor with a 5-year survival rate of only about 18% (Siegel et al., 2020). Identification of specific molecular changes underlying HCC progression will facilitate the development of novel therapeutic strategies against HCC (Auger et al., 2015; Nasr et al., 2019).
Recent advances in human genome and transcriptome profiling have surprisingly found that most human genomes encode for noncoding RNAs (ncRNAs), but not for proteins (Iyer et al., 2015). Among these ncRNAs, long noncoding RNAs (lncRNAs) have been intensively studied in the last decade (Wang et al., 2018; Li et al., 2020). lncRNAs are a class of ncRNAs with more than 200 nucleotides (nt) in length (Ponting et al., 2009). Increasing evidences have demonstrated that lncRNAs are implicated in almost all physiological and pathological processes, including cancers (Esposito et al., 2019; Sweta et al., 2019; Zhang et al., 2019). Many lncRNAs are revealed to be dysregulated in various cancers, including HCC (Li et al., 2017; Berger et al., 2018). The lncRNAs reported to be upregulated in HCC include lncRNA-ATB, HULC, GIHCG, AWPPH, GPC3-AS1, CASC9, MCM3AP-AS1, and BZRAP1-AS1 (Yuan et al., 2014; Sui et al., 2016; Zhu et al., 2016; Zhao et al., 2017; Klingenberg et al., 2018; Xin et al., 2018; Wang W. et al., 2019; Wang Y. et al., 2019). The lncRNAs reported to be downregulated in HCC include CASC2, PSTAR, GAS8-AS1, and LINC000607 (Wang et al., 2017; Pan et al., 2018; Sun et al., 2018; Qin et al., 2019). Furthermore, many lncRNAs are reported to have critical roles in cancers, such as the regulation of cell proliferation, cell cycle, apoptosis, migration, invasion, metastasis, angiogenesis, epithelial–mesenchymal transition (EMT), autophagy, chemoresistance, and self-renewal (Cui et al., 2018; Hu et al., 2018, 2019; Kim et al., 2018; Mondal et al., 2018; Wang et al., 2018; Derderian et al., 2019; Keshavarz and Asadi, 2019). lncRNAs CASC9, PXN-AS1-L, PXN-AS1-S, RAB5IF, and MCM3AP-AS1 modulate HCC cell viability and growth via various mechanisms (Yuan et al., 2017; Klingenberg et al., 2018; Koo et al., 2019; Wang Y. et al., 2019). lncRNA-ATB, MITA1, MIR31HG, and LINC01093 were revealed to regulate HCC metastasis (Yuan et al., 2014; Yan et al., 2018; He et al., 2019; Ma et al., 2019). Thus, lncRNAs are gradually revealed to be important regulators in HCC. Although some of these lncRNAs have been studied in HCC, the total number of lncRNAs identified in human cells is more than 58,000, compared to the 21,000 protein-coding genes in human cells (Iyer et al., 2015). Therefore, other lncRNAs may also be linked to the initiation and progression of HCC.
Metastasis and recurrence are the main causes of HCC-induced deaths even with curative resection (Huang et al., 2019; Sakamoto et al., 2019). Therefore, we focused on the lncRNAs implicated in the metastasis and recurrence of HCC. Via analyzing The Cancer Genome Atlas (TCGA) Liver Hepatocellular Carcinoma (LIHC) dataset, we identified a novel lncRNA LINC01134, which is associated with poor survival of HCC patients. The expression and function of LINC01134 in human cancers have not been investigated. In this study, we further detected the expression and clinical significances of LINC01134 in HCC. Using in vitro and in vivo gain- and loss-of-function experiments, we found that LINC01134 promotes HCC cell migration and invasion and HCC liver metastasis and lung metastasis. Mechanistically, we found that LINC01134 directly binds the promoter of AKT1S1 and activates AKT1S1 expression. Via activating AKT1S1, LINC01134 further activates NF-κB signaling. Our findings unveiled that LINC01134 may be a potential therapeutic target against HCC metastasis.
Materials and Methods
Tissue Specimens
Eighty-four pairs of HCC tissues and paired adjacent noncancerous liver tissues and 20 portal vein tumor thrombus (PVTT) tissues were obtained from HCC patients who received surgery at Tongji Hospital (Wuhan, China) with written informed consent. None of the patients received chemotherapy and/or radiotherapy before surgery. The clinical parameters of these 84 HCC patients were obtained retrospectively from pathology reports and listed in Table 1. All tissue specimens were confirmed by pathological examination. Tissue specimens were acquired during surgery and immediately snap-frozen in liquid nitrogen and stored at −80°C until use. The Ethics Committee of Tongji Hospital (Wuhan, China) reviewed and approved this study.
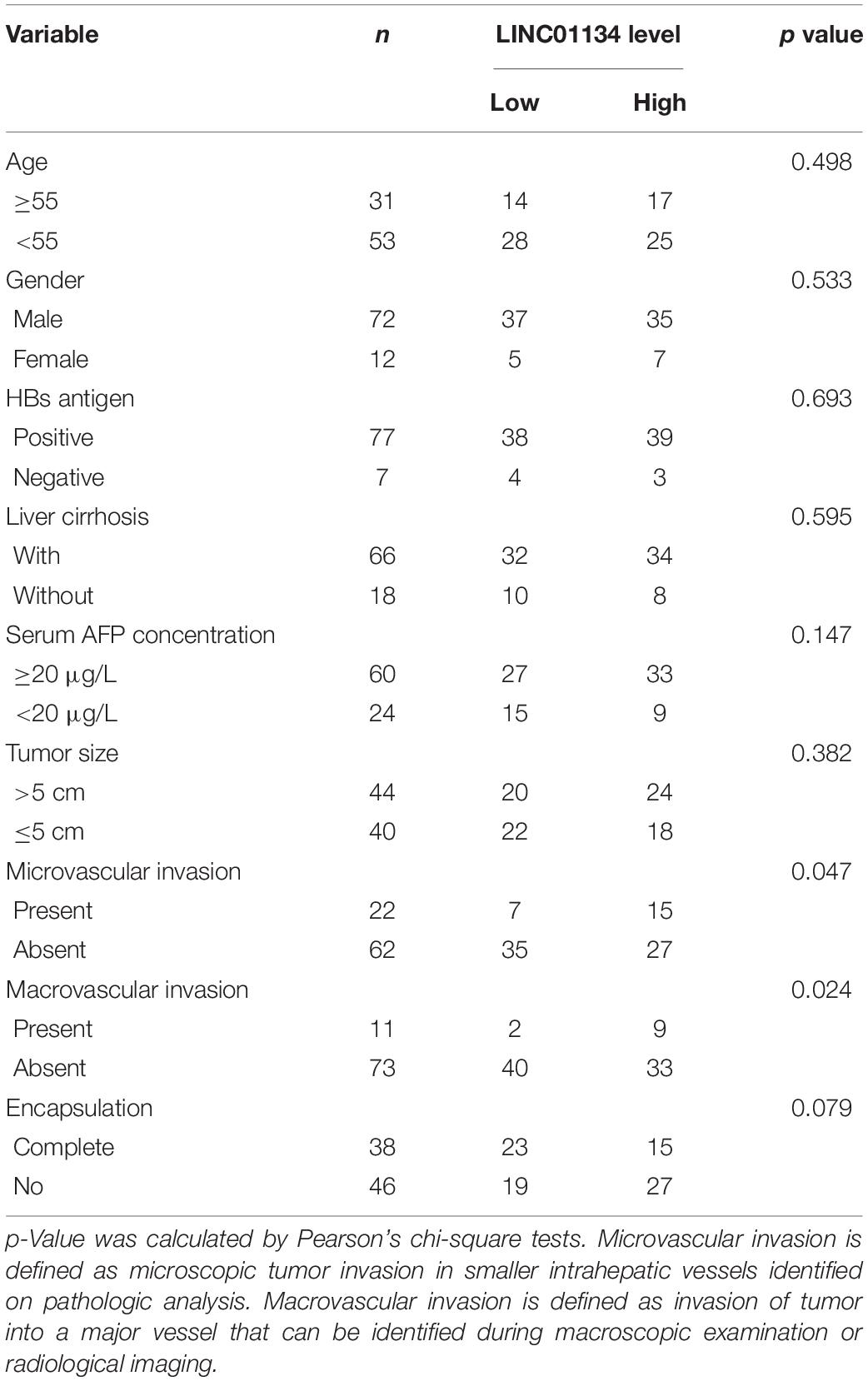
Table 1. Relationship between the LINC01134 levels and clinicopathological features of 84 HCC patients.
Cell Culture and Treatment
Human HCC cell lines SK-HEP-1, HCCLM3, and Huh7 were acquired from the Cell Bank of Type Culture Collection (Chinese Academy of Sciences, Shanghai, China). The cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM, Invitrogen, Carlsbad, CA, United States) supplemented with 10% fetal bovine serum (FBS, Invitrogen) and incubated at 37°C in a humidified incubator with 5% CO2. Where indicated, the HCC cells were treated with 20 nM rapamycin (Selleck, Houston, TX, United States) or 5 μM JSH-23 (Selleck) for 48 h.
RNA Extraction and Quantitative RT-PCR
Total RNA was extracted from indicated tissues and cells using the TRIzol reagent (Cat# 15596-026, Invitrogen) strictly following the manufacturer’s instruction. RNA concentration and quality were detected by the NanoDrop spectrophotometry (Thermo Fisher Scientific) with the A260/A280 ratios being allowed between 1.8 and 2.2. Next, cDNA was synthesized using the PrimeScriptTM II First-Strand cDNA Synthesis Kit (Cat# 6210A, Takara, Dalian, China) strictly following the manufacturer’s instruction. Quantitative RT-PCR (qRT-PCR) was carried out using the TB Green® Premix Ex TaqTM (Tli RNaseH Plus), ROX plus (Cat# RR42LR, Takara) on an ABI StepOnePlus real-time PCR system (Applied Biosystems, Irvine, CA, United States). The amplification conditions were as follows: 95°C for 30 s, followed by 40 cycles of 95°C for 5 s, 60°C for 30 s, and 72°C for 30 s. Amplification efficiencies are allowed to be between 95 and 105%. The primers used were as follows: for LINC01134 (LINC01134-202), 5′-CATATTTGAAAGGGGCAGAC-3′ (forward) and 5′-GGCAACATTAGCCAAACCTA-3′ (reverse); for LINC01134-201, 5′-CCGTTTCTCCGGTGCTATC-3′ (forward) and 5′-TTCTCCGCTGTGTCCTTCAT-3′ (reverse); for LINC01134-203, 5′-GCCATGTTTGAGCGAGGAA-3′ (forward) and 5′-AAGGAGGAGGATGGGAGTC-3′ (reverse); for LINC01134-204, 5′-TCACAGGCCTTGGGCCAGT-3′ (forward) and 5′-GCGTGGGAAACATAGCGGC-3′ (reverse); for AKT1S1, 5′-GAGGGCTCTTTGTGATGGATG-3′ (forward) and 5′-TGCTGTGTGGGTAGGGCTGA-3′ (reverse); for E-cadherin, 5′-GCCCCATCAGGCCTCCGTTT-3′ (forward) and 5′-ACCTTGCCTTCTTTGTCTTTGTTGGA-3′ (reverse); for vimentin, 5′-CCTGAACCTGAGGGAAACTAA-3′ (forward) and 5′-GCAGAAAGGCACTTGAAAGC-3′ (reverse); for NEAT1, 5′-GTCTTTCCATCCACTCACGTCT-3′ (forward) and 5′-GGACAACTAGATGCCGAGGTAG-3′ (reverse); for MALAT1, 5′-GGATCCTAGACCAGCATGCC-3′ (forward) and 5′-AAAGGTTACCATAAGTAAGTTCCAGAAAA-3′ (reverse); and for GAPDH, 5′-GGTCTCCTCTGACTTCAACA-3′ (forward) and 5′-GTGAGGGTCTCTCTCTTCCT-3′ (reverse). GAPDH was selected as endogenous control. The relative expression of RNAs was calculated using the −2ΔΔCt method.
Subcellular Fractionation
Subcellular fractionation was carried out as described before (Gagnon et al., 2014). The RNA in different subcellular components was extracted and detected by qRT-PCR as described above.
Vector Construction and Transfection
LINC01134 full-length sequences were generated by PCR with the primers 5′-GGAATTCACACTGGAGCAGGAAGTC-3′ (forward) and 5′-GCTCTAGACCATATGAGAATGAAGGTTTT-3′ (reverse). Next, the LINC01134 sequences were cloned into the EcoRI and XbaI sites of pcDNATM3.1(+) vector (Invitrogen) to generate the LINC01134 overexpression vector. LINC01134 full-length sequences with the deletion of 464 to 753 nt were synthesized by GenScript (Nanjing, China) and cloned into the pcDNATM3.1(+) vector to generate the mutated LINC01134 overexpression vector. Two independent cDNA oligonucleotides repressing LINC01134 (shLINC-1 and shLINC-2) and one cDNA oligonucleotide repressing AKT1S1 (shAKT1S1) were synthesized by GenePharma (Shanghai, China) and cloned into the GenePharma SuperSilencingTM shRNA expression vector pGPU6/Hygro. The sequences of shLINC-1 were 5′-CACCGGACAGGTTTGAGCTAGAAACTTCAAGAGAGTTT CTAGCTCAAACCTGTCCTTTTTTG-3′ (forward) and 5′-GATCCAAAAAAGGACAGGTTTGAGCTAGAAACTCTCTT GAAGTTTCTAGCTCAAACCTGTCC-3′ (reverse). The sequences of shLINC-2 were 5′-CACCGCGCATCCACTCAT TCACTCATTCAAGAGATGAGTGAATGAGTGGATGCGCTT TTTTG-3′ (forward) and 5′-GATCCAAAAAAGCGCATCCA CTCATTCACTCATCTCTTGAATGAGTGAATGAGTGGATG CGC-3′ (reverse). The sequences of shAKT1S1 were 5′-CACCGGAAACAGGACCTCCTCTAGATTCAAGAGATCTAG AGGAGGTCCTGTTTCCTTTTTTG-3′ (forward) and 5′-GA TCCAAAAAAGGAAACAGGACCTCCTCTAGATCTCTTGAA TCTAGAGGAGGTCCTGTTTCC-3′ (reverse). The sequences of negative control shRNA (shNC) were 5′-CAC CGTTCTCCGAACGTGTCACGTCAAGAGATTACGTGACAC GTTCGGAGAATTTTTTG-3′ (forward) and 5′-GATCCAAA AAATTCTCCGAACGTGTCACGTAATCTCTTGACGTGACA CGTTCGGAGAAC-3′ (reverse). AKT1S1 promoter sequences were generated by PCR with the primers 5′-GGGGTACCCTCCAGCATCACCTCTTCC-3′ (forward) and 5′-CCCAAGCTTGCCTACTCACCCACTTCGT-3′ (reverse) and then cloned into the KpnI and HindIII sites of pGL3-basic Luciferase Reporter Vector (Promega, Madison, WI, United States) to generate the AKT1S1 promoter reporter pGL3-AKT1S1. Transfection of vectors was undertaken using Lipofectamine 3000 (Invitrogen) according to the manufacturer’s instruction.
Stable Cell Line Construction
To construct LINC01134-stably-overexpressed HCC cells, LINC01134 overexpression vector and control pcDNATM3.1(+) vector were transfected into SK-HEP-1 and HCCLM3 cells. Forty-eight hours after transfection, the cells were treated with 800 μg/ml neomycin for 4 weeks to select LINC01134-overexpressed SK-HEP-1 and HCCLM3 cells. To construct LINC01134-stably-silenced HCC cells, shLINC-1, shLINC-2, and shNC were transfected into HCCLM3 and Huh7 cells. Seventy-two hours after transfection, the cells were treated with 800 μg/ml hygromycin for 4 weeks to select LINC01134-silenced HCCLM3 and Huh7 cells. To construct LINC01134-overexpressed and concurrently AKT1S1-silenced HCC cells, shAKT1S1 and shNC were transfected into LINC01134-stably-overexpressed SK-HEP-1 and HCCLM3 cells. Seventy-two hours after transfection, the cells were treated with 800 μg/ml neomycin and 800 μg/ml hygromycin for 4 weeks to select LINC01134-overexpressed and concurrently AKT1S1-silenced SK-HEP-1 and HCCLM3 cells. To construct luciferase-labeled cells, indicated HCC cells were infected with luciferase-expressing lentivirus (Ubi-MCS-firefly_Luciferase-IRES-Puromycin) (Cat# LVCON101, GeneChem, Shanghai, China) and selected with 2 μg/ml puromycin for 4 weeks to construct luciferase stably labeled HCC cells.
Transwell Migration and Invasion Assays
Cell migration and invasion abilities were assessed by Transwell migration and invasion assays, respectively. Transwell chambers (Corning, NY, United States) coated with Matrigel were used for Transwell invasion assay. The same Transwell chambers coated without Matrigel were used for Transwell migration assay. A total of 50,000 indicated HCC cells resuspended in FBS-free medium were plated into the upper chambers pre-coated with or without Matrigel. Complete medium was added to the lower chambers. After 48 h of incubation, the cells remaining on the upper chambers were removed. The migratory and invasive cells were fixed with paraformaldehyde and stained with crystal violet. At least five random fields were photographed under a photomicroscope (Zeiss, Oberkochen, Germany) to count the cell numbers.
Cell Viability Assay
Cell viability was measured using the Cell Counting Kit-8 (CCK-8) assay. Plated into 96-well plate per well were 3,000 indicated HCC cells. The cells were concurrently treated with 20 nM rapamycin (Cat# S1039, Selleck). At the indicated time, cell viability was measured using the CCK-8 (Cat# CK04, Dojindo Laboratories) following the manufacturer’s protocol.
Mouse Xenograft Models
Six-week-old male athymic BALB/c nude mice were used in this study, and the Ethics Committee of Tongji Hospital (Wuhan, China) reviewed and approved the mouse xenograft assays. For liver metastasis experiments, 3 × 106 indicated HCC cells were intrasplenically injected into nude mice. The mice were allowed to grow for another 4 weeks. Then, the mice were sacrificed, and the liver were resected, embedded in paraffin, and used to carry out routine hematoxylin–eosin (HE) staining. The number and size of liver metastatic nodules were assessed under a photomicroscope. For lung metastasis experiments, 2 × 106 luciferase-labeled indicated HCC cells were injected into tail veins of nude mice. The mice were allowed to grow for another 4 weeks. Lung metastases were monitored using the IVIS@ Lumina II system (Caliper Life Sciences, Hopkinton, MA, United States) 10 min after intraperitoneal injection of 4.0 mg of luciferin in 50 μl of saline.
Western Blot
Total protein was isolated from indicated HCC cells using RIPA lysis buffer (Beyotime, Shanghai, China) supplemented with protease inhibitor PMSF (Beyotime). After quantitation of protein concentration with Enhanced BCA Protein Assay Kit (Beyotime), an equal amount of protein was separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, followed by being transferred to a polyvinylidene fluoride membrane (Beyotime). After blocking, the membrane was incubated with primary antibodies against AKT1S1 (ab151719, 1:1,000, Abcam, Cambridge, MA, United States), phospho-AKT1S1 (ab226994, 1:1,000, Abcam), p65 (#8242, 1;1,000, Cell Signaling Technology), histone H3 (ab1791, 1:1,000, Abcam), or GAPDH (ab8245, 1:10,000, Abcam). After three washes, the membrane was further incubated with goat anti-rabbit IgG H&L (IRDye® 800CW) preadsorbed (ab216773, 1:10,000, Abcam) or goat anti-mouse IgG H&L (IRDye® 680RD) preadsorbed (ab216776, 1:10,000, Abcam). Lastly, the membrane was detected on the Odyssey infrared scanner (Li-Cor, Lincoln, NE, United States).
Chromatin Isolation by RNA Purification
Chromatin isolation by RNA purification (ChIRP) was carried out using the Magna ChIRPTM RNA Interactome Kit (Millipore, Bedford, MA, United States) following the manufacturer’s instruction. The sequences of the antisense DNA probes targeting LINC01134 were as follows: 1, 5′-gaaacatagcggctacaggg-3′; 2, 5′-aagcgccgttcacgaacatg-3′; 3, 5′-attttactttcaggcctttc-3′; 4, 5′-ttcaagtggtttctagctca-3′; 5, 5′-ataggtcttggctggttctc-3′; 6, 5′-gcaaacagcgaggtcaacac-3′; 7, 5′-cgagggaggacactgacatg-3′; 8, 5′-cgcagaatggcaggaatcaa-3′; 9, 5′-gaatgttcaggagagagggc-3′; 10, 5′-aaggtaggttctgggacatg-3′; and 11, 5′-gagtgggggtggtcactaac-3′. The enriched DNA was detected by qRT-PCR with the primers 5′-AGAGCGAGCCAGGACTTG-3′ (forward) and 5′-GGTATCTTATTGTGGTTTTGG-3′ (reverse) for the AKT1S1 promoter and 5′-TGTCACTACCGCAGAGCCT-3′ (forward) and 5′-GAGGACTTTGGGAACGACTG-3′ (reverse) for the GAPDH promoter.
Dual Luciferase Reporter Assays
To assess the effects of LINC01134 on AKT1S1 promoter activity, the AKT1S1 promoter reporter pGL3-AKT1S1 was co-transfected with pRL-TK and LINC01134 overexpression or silencing vectors into HCCLM3 cells. pRL-TK encodes renilla luciferase and was used as an endogenous reference. Forty-eight hours after transfection, the firefly luciferase and renilla luciferase activities were measured using the Dual-Luciferase Reporter Assay System (Promega). To assess the effects of LINC01134 on NF-κB transcriptional activity, firefly luciferase reporter containing NF-κB binding sites (pNFκB-luc) (Beyotime) was co-transfected with pRL-TK and LINC01134 overexpression or silencing vectors into HCCLM3 cells. Forty-eight hours after transfection, the firefly luciferase and renilla luciferase activities were measured using the Dual-Luciferase Reporter Assay System (Promega).
p65-DNA Binding Activity Assay
Forty-eight hours after transfection of LINC01134 overexpression or silencing vectors into HCCLM3 cells, the nuclear extracts from transfected cells were used to carry out p65-DNA binding activity with an NF-κB p65 Transcription Factor Assay Kit (ab133112, Abcam) following the manufacturer’s instruction.
Statistical Analysis
GraphPad Prism v6.0 (GraphPad Software, La Jolla, CA, United States) was used to undergo all statistical analyses. For comparison, Wilcoxon matched-pairs signed-rank test, log-rank test, Pearson’s chi-square test, unpaired two-sided Student’s t-test, one-way ANOVA followed by Dunnett’s multiple comparisons test, one-way ANOVA followed by Tukey’s multiple comparisons test, Mann–Whitney test, Kruskal–Wallis test followed by Dunn’s multiple comparisons test, and Spearman correlation analysis were performed as indicated. P < 0.05 was regarded as statistically significant.
Results
LINC01134 Is Highly Expressed and Correlated With Poor Survival of HCC Patients
To identify the genes correlated with survival of HCC patients, we analyzed TCGA LIHC dataset using the online tool GEPIA.1 The top 100 genes most significantly correlated with overall survival of HCC patients are shown in Supplementary Table 1. Although most of these genes are protein-coding genes, we noted a gene (ID: ENSG00000236423.5) which encodes an lncRNA LINC01134. Reanalyzing TCGA LIHC dataset, we found that LINC01134 is significantly highly expressed in HCC tissues (n = 369) compared with normal liver tissues (n = 50) (Figure 1A). Reanalyzing TCGA LIHC dataset with available survival data, we found that high levels of LINC01134 indicate shorter overall survival and disease-free survival times (Figures 1B,C). Searching Ensembl,2 we found that the LINC01134 has five exons with four isoforms (Supplementary Figure 1A). Quantitative RT-PCR with isoform-specific primers indicated that LINC01134-202 is the most abundant isoform in HCCLM3 cells and HCC tissues (Supplementary Figures 1B,C). Therefore, we focused on the most abundant isoform LINC01134-202. Three in silico tools, CPC, CPAT, and PhyloCSF, all predicted that LINC01134 as a ncRNA (Supplementary Figures 1D–F). To further explore the clinical significance of LINC01134, we collected 84 pairs of HCC tissues and paired adjacent noncancerous liver tissues. Quantitative RT-PCR results revealed that LINC01134 is significantly highly expressed in HCC tissues compared with noncancerous liver tissues (Figure 1D). In addition, we collected 20 PVTT tissues, which are intrahepatic metastases of HCC. qRT-PCR results revealed that LINC01134 is further highly expressed in PVTT tissues compared with HCC tissues (Figure 1D). Analyzing the correlation between LINC01134 expression levels and the clinical parameters revealed that LINC01134-highly-expressed HCC tissues showed more microvascular invasion and macrovascular invasion compared to LINC01134-lowly-expressed HCC tissues (Table 1). LINC01134-highly-expressed HCC patients had significantly shorter overall survival and disease-free survival compared to LINC01134-lowly-expressed HCC patients (Figures 1E,F).
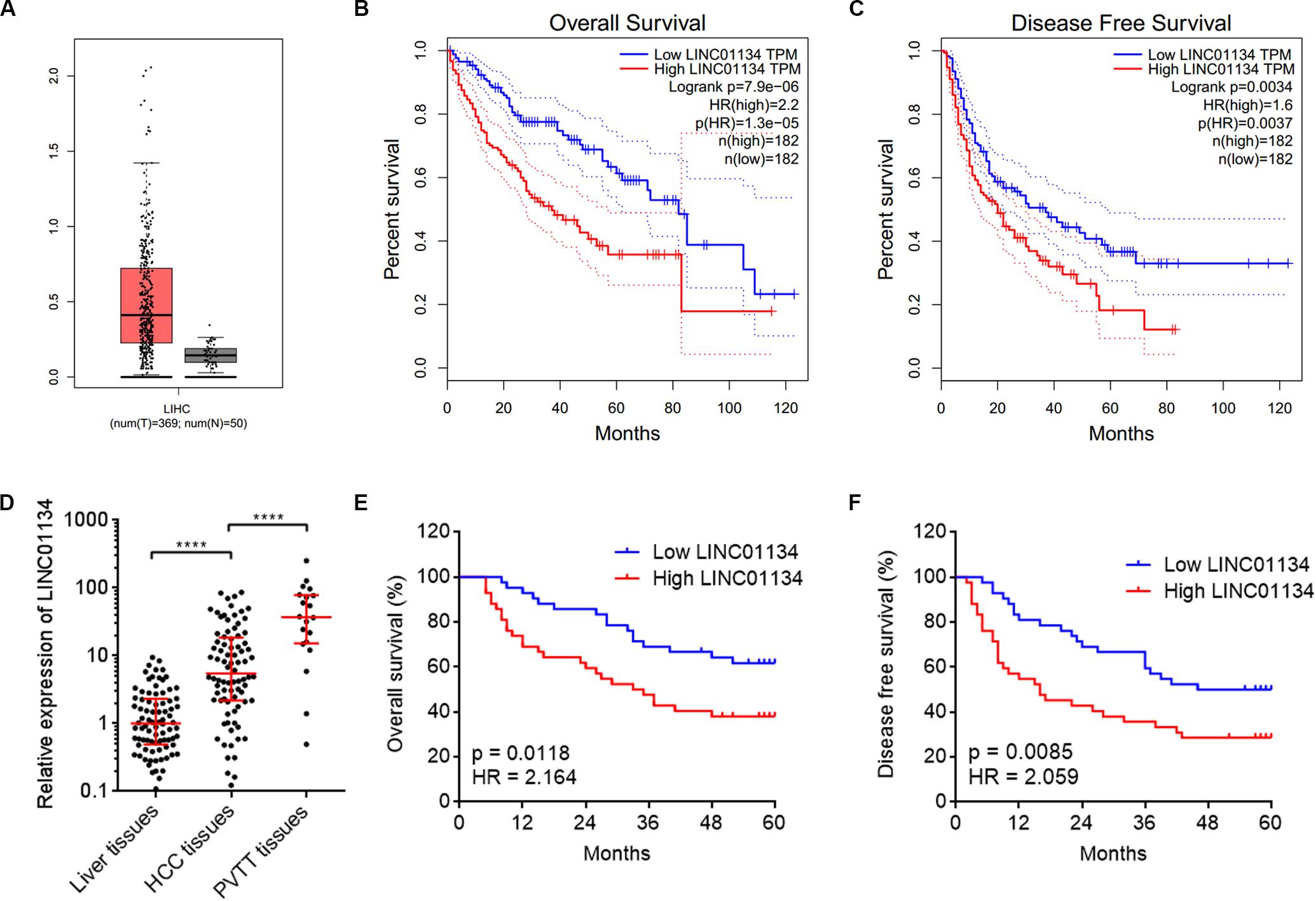
Figure 1. LINC01134 is highly expressed and correlated with poor prognosis of HCC patients. (A) The expression of LINC01134 in HCC tissues (n = 369) versus normal liver tissues (n = 50) from TCGA LIHC dataset. (B) Kaplan–Meier survival analysis of the correlation between LINC01134 expression levels and overall survival of HCC patients from TCGA LIHC dataset. (C) Kaplan–Meier survival analysis of the correlation between LINC01134 expression levels and disease-free survival of HCC patients from TCGA LIHC dataset. (D) The expression of LINC01134 in 84 pairs of HCC tissues and paired adjacent noncancerous liver tissues and 20 PVTT tissues was detected by qRT-PCR. ****p < 0.0001. The comparison between liver tissues and HCC tissues was calculated by Wilcoxon matched-pairs signed-rank test. The comparison between HCC tissues and PVTT tissues was calculated by Mann–Whitney test. (E) Kaplan–Meier survival analysis of the correlation between LINC01134 expression level and overall survival of these 84 HCC patients. p = 0.0118, HR = 2.164 by log-rank test. (F) Kaplan–Meier survival analysis of the correlation between LINC01134 expression level and disease-free survival of these 84 HCC patients. p = 0.0085, HR = 2.059 by log-rank test.
LINC01134 Promotes Migration and Invasion of HCC Cells
Due to LINC01134 being positively correlated with microvascular invasion, macrovascular invasion, and poor survival of HCC patients, we next investigated the roles of LINC01134 in HCC cell migration and invasion. LINC01134-stably-overexpressed HCC cells were constructed via stable transfection of LINC01134 overexpression plasmids into SK-HEP-1 and HCCLM3 cells. The overexpression efficiencies were confirmed by qRT-PCR (Figure 2A). Transwell migration assays showed that LINC01134-overexpressed SK-HEP-1 and HCCLM3 cells have stronger migratory ability compared to control SK-HEP-1 and HCCLM3 cells, respectively (Figure 2B). Transwell invasion assays showed that LINC01134-overexpressed SK-HEP-1 and HCCLM3 cells have stronger invasive ability compared to control SK-HEP-1 and HCCLM3 cells, respectively (Figure 2C). Furthermore, LINC01134-stably-silenced HCC cells were constructed via stable transfection of two independent LINC01134-specific shRNAs into HCCLM3 and Huh7 cells. The knockdown efficiencies were confirmed by qRT-PCR (Figure 2D). Transwell migration assays showed that LINC01134-silenced HCCLM3 and Huh7 cells have less migratory cell number compared to control HCCLM3 and Huh7 cells, respectively (Figure 2E). Transwell invasion assays showed that LINC01134-silenced HCCLM3 and Huh7 cells have less invasive cell number compared to control HCCLM3 and Huh7 cells, respectively (Figure 2F). Due to relationships between EMT, cell migration, and invasion, we next investigated whether LINC01134 regulates EMT in HCC cells. Epithelial and mesenchymal markers E-cadherin and vimentin, respectively, were assayed in LINC01134-overexpressing SK-HEP-1 and LINC01134-silenced HCCLM3 cells, by qRT-PCR. The results showed that neither LINC01134 overexpression nor LINC01134 silencing modulates EMT marker expression (Supplementary Figures 2A,B), which suggested that LINC01134 did not modulate EMT in HCC cells. Collectively, these data demonstrated that LINC01134 promotes HCC cell migration and invasion.
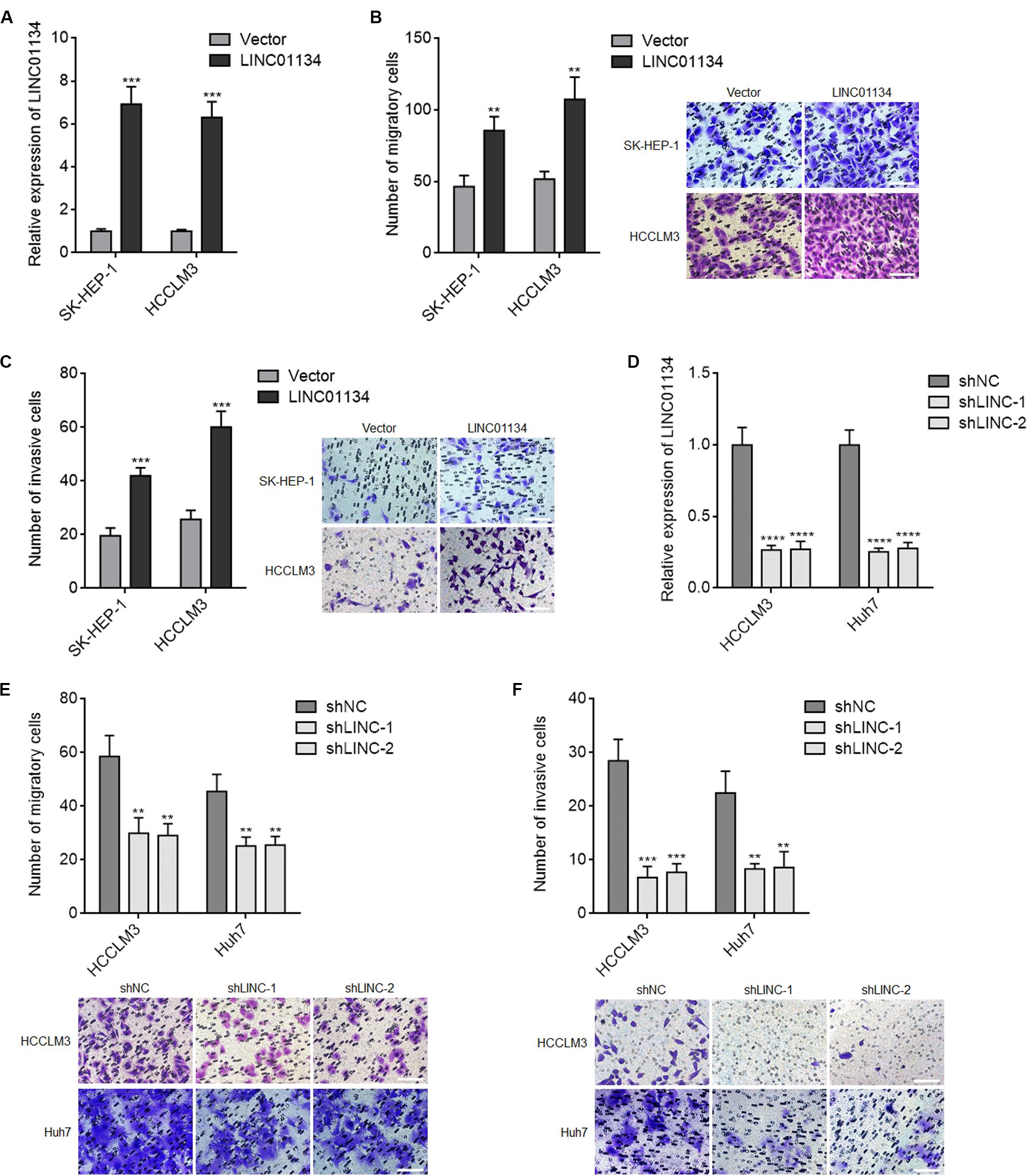
Figure 2. LINC01134 promotes migration and invasion of HCC cells. (A) The expression of LINC01134 in LINC01134-stably-overexpressed and control SK-HEP-1 and HCCLM3 cells was detected by qRT-PCR. (B) Transwell migration assays were conducted to assess the migration ability of LINC01134-stably-overexpressed and control SK-HEP-1 and HCCLM3 cells. Representative images of migratory cells were shown. Scale bar = 100 μm. (C) Transwell invasion assays were conducted to assess the invasion ability of LINC01134-stably-overexpressed and control SK-HEP-1 and HCCLM3 cells. Representative images of invasive cells were shown. Scale bar = 100 μm. (D) The expression of LINC01134 in LINC01134-stably-silenced and control HCCLM3 and Huh7 cells was detected by qRT-PCR. (E) Transwell migration assays were conducted to assess the migration ability of LINC01134-stably-silenced and control HCCLM3 and Huh7 cells. Representative images of migratory cells were shown. Scale bar = 100 μm. (F) Transwell invasion assays were conducted to assess the invasion ability of LINC01134-stably-silenced and control HCCLM3 and Huh7 cells. Representative images of invasive cells were shown. Scale bar = 100 μm. Results are shown as the mean ± SD of three independent experiments. **p < 0.01, ***p < 0.001, ****p < 0.0001 by unpaired two-sided Student’s t-test (A–C) or one-way ANOVA followed by Dunnett’s multiple comparisons test (D–F).
LINC01134 Promotes HCC Liver Metastasis and Lung Metastasis in vivo
To investigate the effects of LINC01134 in HCC liver metastasis in vivo, LINC01134-stably-overexpressed and control HCCLM3 cells were intrasplenically injected into nude mice. At the 28th day after injection, the liver metastatic nodules were detected by HE staining. As shown in Figures 3A–C, LINC01134-overexpressed HCCLM3 cells form more and larger liver metastatic nodules compared to control cells. LINC01134-stably-silenced and control HCCLM3 cells were also intrasplenically injected into nude mice. At the 28th day after injection, the liver metastatic nodules were detected by HE staining. As shown in Figures 3D–F, LINC01134-silenced HCCLM3 cells form less and smaller liver metastatic nodules compared to control cells. Next, the effects of LINC01134 on HCC lung metastasis were explored. Luciferase-labeled LINC01134-stably-overexpressed and control HCCLM3 cells were injected into the tail veins of nude mice. At the 28th day after injection, LINC01134-overexpressed HCCLM3 cells form more lung metastasis compared to control cells (Figure 3G). Luciferase-labeled LINC01134-stably-silenced and control HCCLM3 cells were also injected into the tail veins of nude mice. At the 28th day after injection, LINC01134-silenced HCCLM3 cells form less lung metastasis compared to control cells (Figure 3H). Therefore, these data suggested that LINC01134 promotes HCC liver metastasis and lung metastasis in vivo.
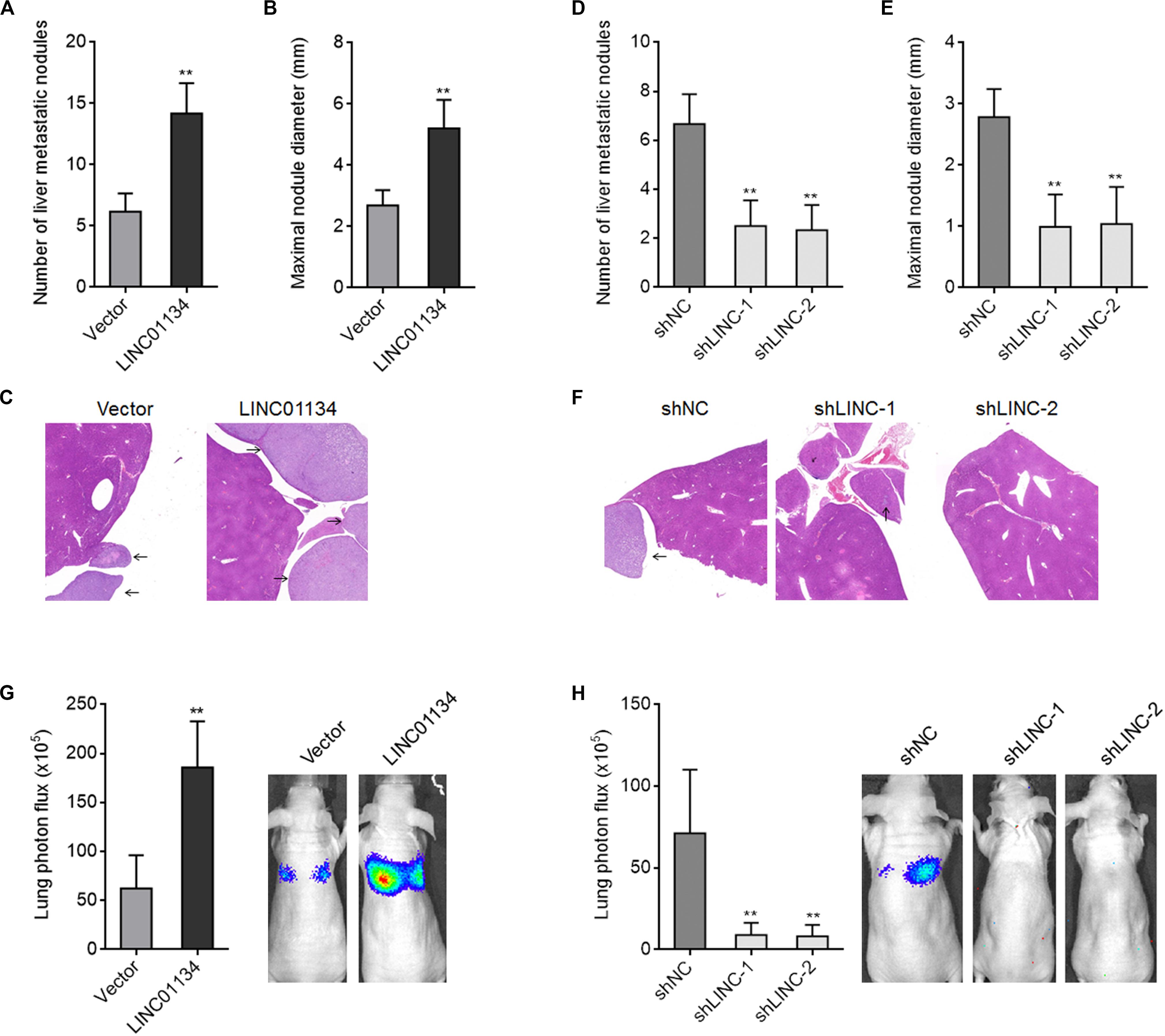
Figure 3. LINC01134 promotes HCC liver metastasis and lung metastasis in vivo. (A–C) LINC01134-stably-overexpressed and control HCCLM3 cells were intrasplenically injected into nude mice. At the 28th day after injection, the number (A) and size (B) of liver metastatic nodules were detected by HE staining (C). (D–F) LINC01134-stably-silenced and control HCCLM3 cells were intrasplenically injected into nude mice. At the 28th day after injection, the number (D) and size (E) of liver metastatic nodules were detected by HE staining (F). (G) Luciferase-labeled LINC01134-stably-overexpressed and control HCCLM3 cells were injected into the tail veins of nude mice. At the 28th day after injection, lung metastasis was assessed via detecting luciferase signal intensities. (H) Luciferase-labeled LINC01134-stably-silenced and control HCCLM3 cells were injected into the tail veins of nude mice. At the 28th day after injection, lung metastasis was assessed via detecting luciferase signal intensities. Results are shown as the mean ± SD of n = 6 mice in each group. **p < 0.01 by Mann–Whitney test (A–C,G) or Kruskal–Wallis test followed by Dunn’s multiple comparisons test (D–F,H).
The Expression of AKT1S1 Is Positively Correlated With That of LINC01134 in HCC Tissues
To explore the potential molecular mechanisms mediating the roles of LINC01134 in HCC, we searched the genes whose expressions are correlated with those of LINC01134 in TCGA LIHC dataset using the online tool R23 (Supplementary Table 2). Among the genes most significantly correlated with LINC01134, we noted AKT1S1, which has been reported to be a critical oncogene in several cancers, including HCC (Malla et al., 2015; Lv et al., 2017; Qi et al., 2020). The correlation between LINC01134 and AKT1S1 expression intensities based on TCGA LIHC dataset is shown in Figure 4A. Analyzing TCGA LIHC dataset, we found that in line with LINC01134, AKT1S1 is also significantly highly expressed in HCC tissues (n = 369) compared with normal liver tissues (n = 50) (Figure 4B). Reanalyzing TCGA LIHC dataset with available survival data, we found that in line with LINC01134, high levels of AKT1S1 indicate shorter overall survival time (Figure 4C). In our cohort containing 84 pairs of HCC tissues and paired adjacent noncancerous liver tissues and additional 20 PVTT tissues, we further confirmed that AKT1S1 is highly expressed in HCC tissues compared with noncancerous liver tissues and further highly expressed in PVTT tissues compared with HCC tissues (Figure 4D). The expression of AKT1S1 is also significantly positively correlated with that of LINC01134 in these 84 HCC tissues (Figure 4E). AKT1S1-highly-expressed HCC patients had significantly shorter overall survival compared to AKT1S1-lowly-expressed HCC patients (Figure 4F). The correlation between the expressions of LINC01134, AKT1S1, and EMT markers in TCGA LIHC dataset was further analyzed using R2. The results showed that the expressions of LINC01134 and AKT1S1 are not consistently correlated with EMT markers in TCGA LIHC dataset (Supplementary Figures 2C–F), which supported that LINC01134 and AKT1S1 did not modulate EMT in HCC.
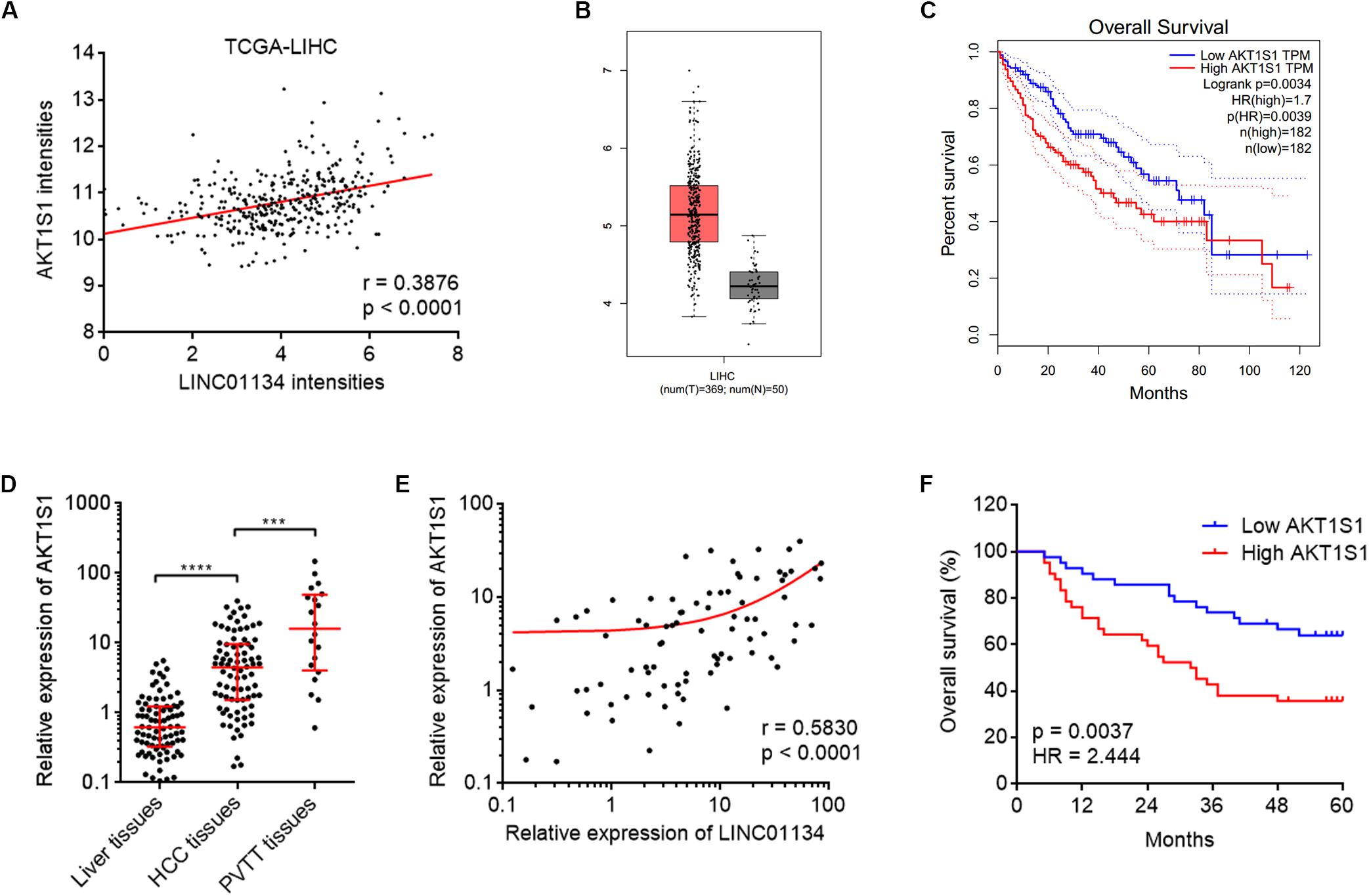
Figure 4. The positive correlation between AKT1S1 and LINC01134 expressions in HCC tissues. (A) The correlation between AKT1S1 and LINC01134 expression intensities from TCGA LIHC dataset. r = 0.3876, p < 0.0001 by Spearman correlation analysis. (B) The expression of AKT1S1 in HCC tissues (n = 369) versus normal liver tissues (n = 50) from TCGA LIHC dataset. (C) Kaplan–Meier survival analysis of the correlation between AKT1S1 expression levels and overall survival of HCC patients from TCGA LIHC dataset. (D) The expression of AKT1S1 in 84 pairs of HCC tissues and paired adjacent noncancerous liver tissues and 20 PVTT tissues was detected by qRT-PCR. ****p < 0.0001. The comparison between liver tissues and HCC tissues was calculated by Wilcoxon matched-pairs signed-rank test. The comparison between HCC tissues and PVTT tissues was calculated by Mann–Whitney test. (E) The correlation between LINC01134 expression level and AKT1S1 expression level in these 84 HCC tissues. r = 0.583, p < 0.0001 by Spearman correlation analysis. (F) Kaplan–Meier analyses of the correlation between AKT1S1 expression level and overall survival of these 84 HCC patients. p = 0.0037, HR = 2.444 by log-rank test.
LINC01134 Activates AKT1S1 Transcription via Directly Binding the Promoter of AKT1S1
Due to the significantly positive correlation between LINC01134 and AKT1S1 expressions in HCC tissues, we next investigated whether LINC01134 regulates AKT1S1 in HCC. AKT1S1 mRNA and protein levels in LINC01134-stably-overexpressed and LINC01134-silenced HCCLM3 cells were detected by qRT-PCR and western blot. As shown in Figures 5A–D, LINC01134-overexpressed HCCLM3 cells have significantly higher mRNA and protein expression of AKT1S1 compared to control cells, while LINC01134-silenced HCCLM3 cells have significantly lower mRNA and protein expression of AKT1S1 compared to control cells. These data suggested that LINC01134 activates AKT1S1 expression. To investigate the potential mechanisms underlying the modulation of AKT1S1 by LINC01134, we first assessed the subcellular localization of LINC01134 via biochemical fractionation. The results revealed that LINC01134 is mainly localized in the chromatin (Figure 5E), suggesting a potential role of LINC01134 in regulating gene transcription. Intriguingly, we identified a highly adversely complementary region between LINC01134 (464–753 nt) and the promoter of AKT1S1 (−1,802 to −1,498 bp) (Figure 5F). To explore whether LINC01134 directly binds the AKT1S1 promoter via this complementary region, ChIRP assays were conducted with biotinylated LINC01134 capture probes. As shown in Figure 5G, the AKT1S1 promoter is specifically enriched in the LINC01134 probe group, but not in the control probe group (LacZ probe). To investigate whether the binding of LINC01134 to the AKT1S1 promoter modulates AKT1S1 transcription, the promoter of AKT1S1 containing the LINC01134 binding site was cloned into the firefly luciferase reporter. Dual luciferase reporter assays showed that concurrent overexpression of LINC01134 significantly increased the luciferase activity of the AKT1S1 promoter, which was abrogated by the mutation of the complementary sequences on LINC01134 (Figure 5H). Conversely, LINC01134 knockdown significantly decreased the luciferase activity of the AKT1S1 promoter (Figure 5I). Collectively, these data suggested that LINC01134 activates AKT1S1 transcription via directly binding the AKT1S1 promoter.
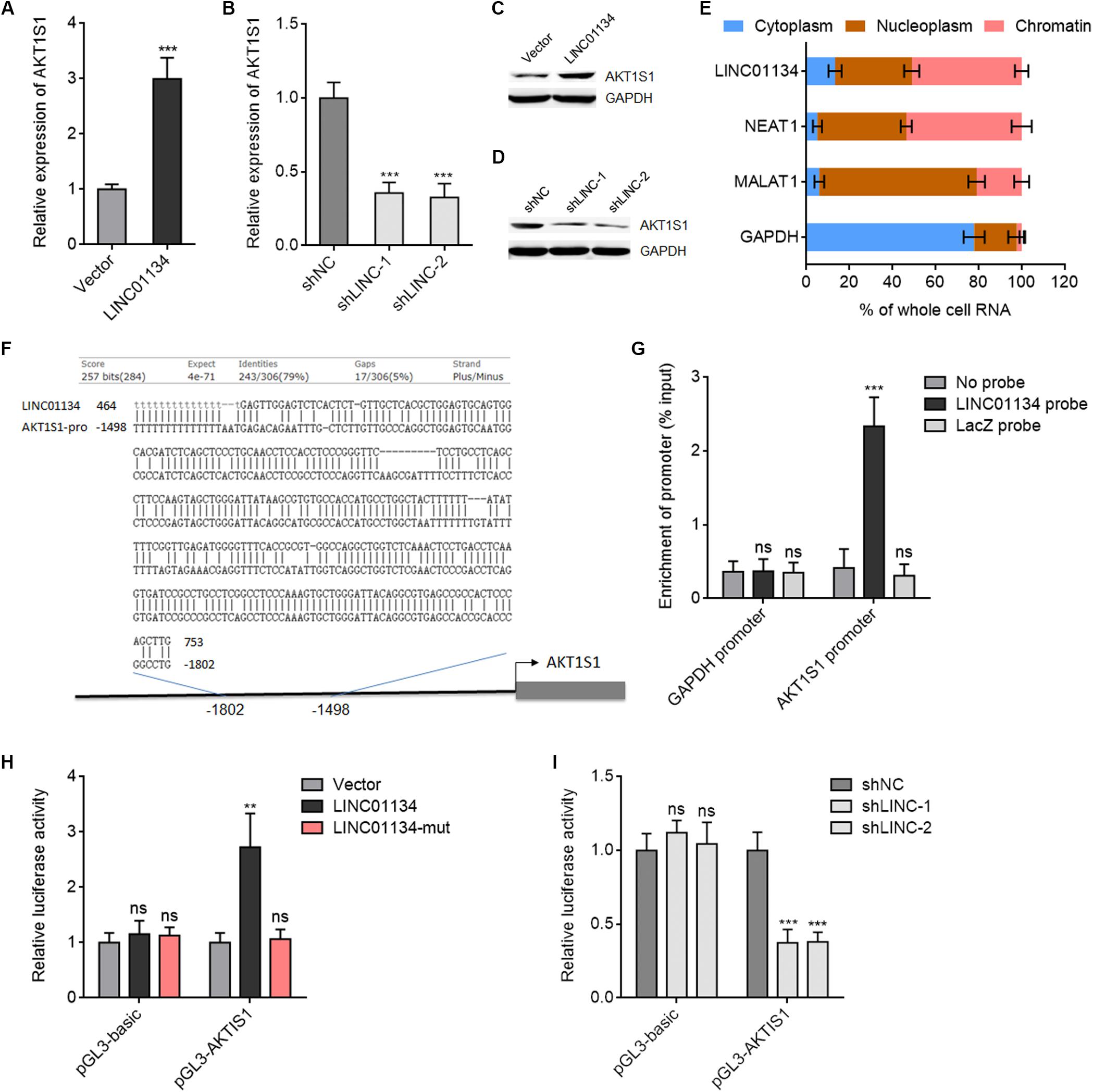
Figure 5. LINC01134 directly binds the AKT1S1 promoter and activates AKT1S1 expression. (A) AKT1S1 mRNA levels in LINC01134-stably-overexpressed and control HCCLM3 cells were detected by qRT-PCR. (B) AKT1S1 mRNA levels in LINC01134-stably-silenced and control HCCLM3 cells were detected by qRT-PCR. (C) AKT1S1 protein levels in LINC01134-stably-overexpressed and control HCCLM3 cells were detected by western blot. (D) AKT1S1 protein levels in LINC01134-stably-silenced and control HCCLM3 cells were detected by western blot. (E) Subcellular localization of LINC01134 and control transcripts was analyzed by qRT-PCR in biochemically fractionated HCCLM3 cells. NEAT1 was used as a chromatin-associated lncRNA control. MALAT1 was used as a nucleoplasm-localized lncRNA control. GAPDH mRNA was used as a cytoplasmic control. (F) The highly adversely complementary region between LINC01134 (464–753 nt) and AKT1S1 promoter (−1,802 to −1,498 bp). (G) ChIRP assays with LINC01134 capture probes were conducted in HCCLM3 cells. The enrichment of the AKT1S1 promoter and GAPDH promoter was detected by qRT-PCR. (H) After transient co-transfection of wild-type or complementary region mutated LINC01134 overexpression plasmids with firefly luciferase reporter containing the AKT1S1 promoter and pRL-TK (encoding renilla luciferase) into HCCLM3 cells, dual luciferase reporter assays were conducted to assess AKT1S1 promoter activity. Results are shown as the relative ratio of firefly luciferase activity to renilla luciferase activity. (I) After transient co-transfection of LINC01134-specific shRNAs with firefly luciferase reporter containing the AKT1S1 promoter and pRL-TK into HCCLM3 cells, dual luciferase reporter assays were conducted to assess AKT1S1 promoter activity. Results are shown as the relative ratio of firefly luciferase activity to renilla luciferase activity. Results are shown as the mean ± SD of three independent experiments. **p < 0.01, ***p < 0.001, ns, not significant, by unpaired two-sided Student’s t-test (A) or one-way ANOVA followed by Dunnett’s multiple comparisons test (B,G–I).
AKT1S1 Silencing Reverses the Roles of LINC01134 in Promoting HCC Migration, Invasion, and Metastasis
To explore whether the activation of AKT1S1 mediates the roles of LINC01134 in promoting HCC migration, invasion, and metastasis, AKT1S1 was stably silenced in LINC01134-stably-overexpressed SK-HEP-1 and HCCLM3 cells via stable transfection of AKT1S1-specific shRNAs (Figures 6A,B). Transwell migration assays revealed that the pro-migratory roles of LINC01134 are abolished by AKT1S1 silencing (Figure 6C). Transwell invasion assays revealed that the pro-invasive roles of LINC01134 are also abolished by AKT1S1 silencing (Figure 6D). LINC01134-stably-overexpressed and AKT1S1-stably-silenced HCCLM3 cells were intrasplenically injected into nude mice. At the 28th day after injection, the liver metastatic nodules were detected. As shown in Figures 6E–G, the increased number and size of liver metastatic nodules caused by LINC01134 overexpression are abolished by AKT1S1 silencing. Luciferase-labeled LINC01134-stably-overexpressed and AKT1S1-stably-silenced HCCLM3 cells were also injected into the tail veins of nude mice. At the 28th day after injection, the increased lung metastasis caused by LINC01134 overexpression is abolished by AKT1S1 silencing (Figure 6H). Rapamycin, a known mammalian target of rapamycin (mTOR) pathway inhibitor, is reported to inhibit AKT1S1 (Hu et al., 2015). We then treated LINC01134-stably-overexpressed SK-HEP-1 and HCCLM3 cells with rapamycin to repress AKT1S1 (Supplementary Figures 3A,B). Rapamycin treatment suppressed cell viability of HCC cells (Supplementary Figures 3C,D). Transwell migration assays showed that the pro-migratory roles of LINC01134 are abolished by rapamycin (Supplementary Figure 3E). Transwell invasion assays showed that the pro-invasive roles of LINC01134 are also abolished by rapamycin (Supplementary Figure 3F). These data supported the concept that LINC01134 promotes HCC migration and invasion via activation of AKT1S1.
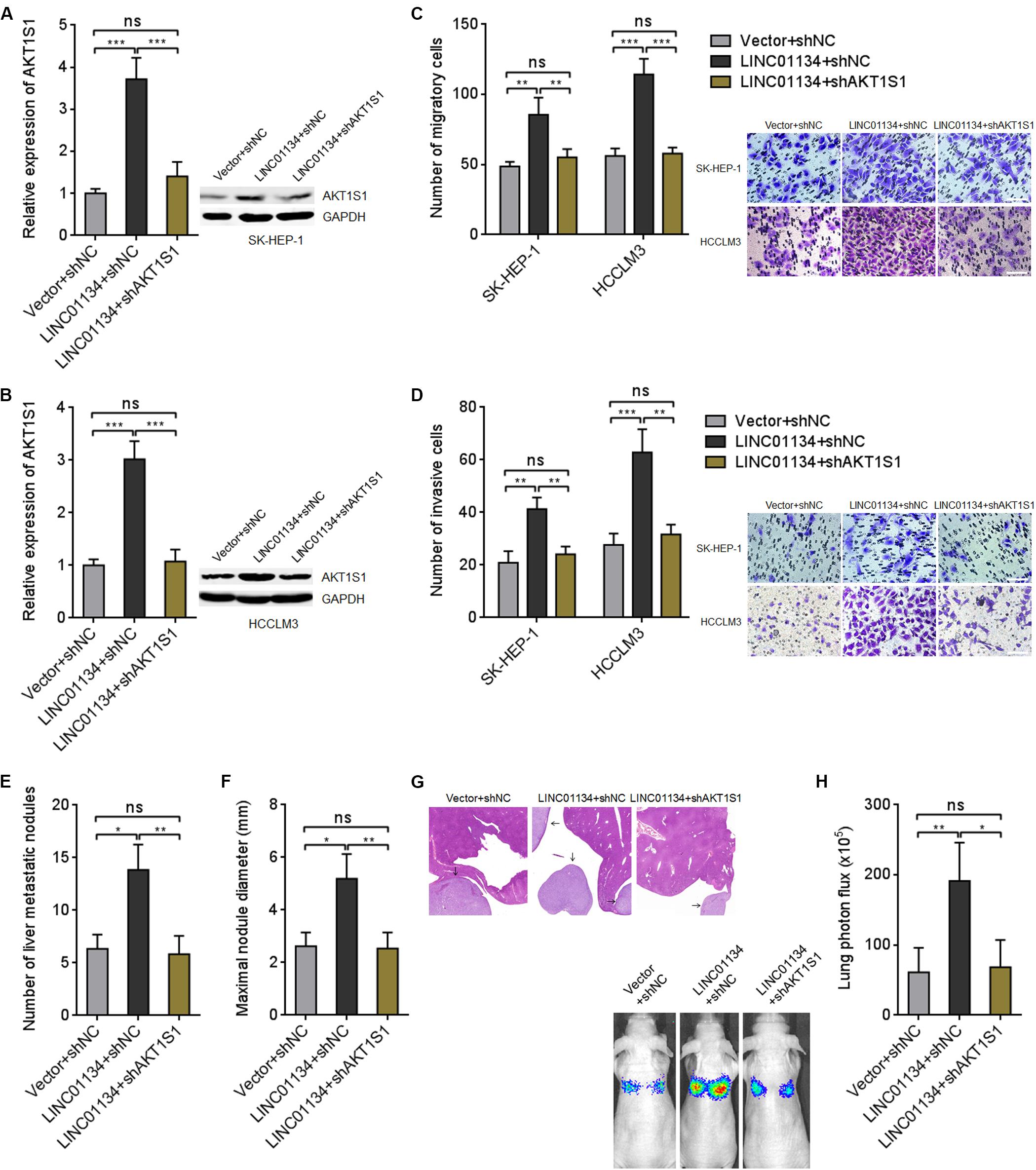
Figure 6. Inhibition of AKT1S1 reverses the roles of LINC01134 in HCC. (A) AKT1S1 mRNA and protein levels in LINC01134-stably-overexpressed and concurrently AKT1S1-stably-silenced SK-HEP-1 cells were detected by qRT-PCR and western blot. (B) AKT1S1 mRNA and protein levels in LINC01134-stably-overexpressed and concurrently AKT1S1-stably-silenced HCCLM3 cells were detected by qRT-PCR and western blot. (C) Transwell migration assays were conducted to assess the migration ability of LINC01134-stably-overexpressed and concurrently AKT1S1-stably-silenced SK-HEP-1 and HCCLM3 cells. Representative images of migratory cells were shown. Scale bar = 100 μm. (D) Transwell invasion assays were conducted to assess invasion ability of LINC01134-stably-overexpressed and concurrently AKT1S1-stably-silenced SK-HEP-1 and HCCLM3 cells. Representative images of invasive cells were shown. Scale bar = 100 μm. (E–G) LINC01134-stably-overexpressed and concurrently AKT1S1-stably-silenced HCCLM3 cells were intrasplenically injected into nude mice. At the 28th day after injection, the number (E) and size (F) of liver metastatic nodules were detected by HE staining (G). (H) Luciferase-labeled LINC01134-stably-overexpressed and concurrently AKT1S1-stably-silenced HCCLM3 cells were injected into the tail veins of nude mice. At the 28th day after injection, lung metastasis was assessed via detecting luciferase signal intensities. Results are shown as the mean ± SD of three independent experiments (A–D) or n = 6 mice in each group (E–H). *p < 0.05, **p < 0.01, ***p < 0.001, ns, not significant, by one-way ANOVA followed by Tukey’s multiple comparisons test (A–D) or Kruskal–Wallis test followed by Dunn’s multiple comparisons test (E,F,H).
LINC01134 Enhances NF-κB Transcriptional Activity in HCC
Previous report has shown that AKT1S1 enhances NF-κB transcriptional activity via associating with p65 (Zhu et al., 2017). Thus, we further investigated whether LINC01134 modulates NF-κB signaling via the activation of AKT1S1. Dual luciferase reporter assays showed that overexpression of LINC01134 significantly increases NF-κB transcriptional activity, which is abrogated by the mutation of the complementary sequences on LINC01134 (Figure 7A). Conversely, knockdown of LINC01134 significantly reduces NF-κB transcriptional activity (Figure 7B). The effects of LINC01134 on p65-DNA binding activity were further investigated. As shown in Figure 7C, overexpression of LINC01134 significantly increases p65-DNA binding activity, which is abrogated by the mutation of the complementary sequences on LINC01134. Conversely, knockdown of LINC01134 reduces p65-DNA binding activity (Figure 7D). To further investigate whether the roles of LINC01134 in promoting HCC cell migration and invasion are dependent on the regulation of AKT1S1-NF-κB signaling, LINC01134-stably-overexpressed SK-HEP-1 and HCCLM3 cells were treated with NF-κB signaling inhibitor JSH-23, which repressed p65 nuclear translocation (Supplementary Figure 4). Transwell migration assays showed that treatment with JSH-23 abrogated the pro-migratory roles of LINC01134 (Figure 7E). Transwell invasion assays showed that treatment with JSH-23 abrogated the pro-invasive roles of LINC01134 (Figure 7F). Thus, these data suggested that the activation of AKT1S1-NF-κB transcriptional activity at least partially mediates the roles of LINC01134 in promoting HCC cell migration and invasion.
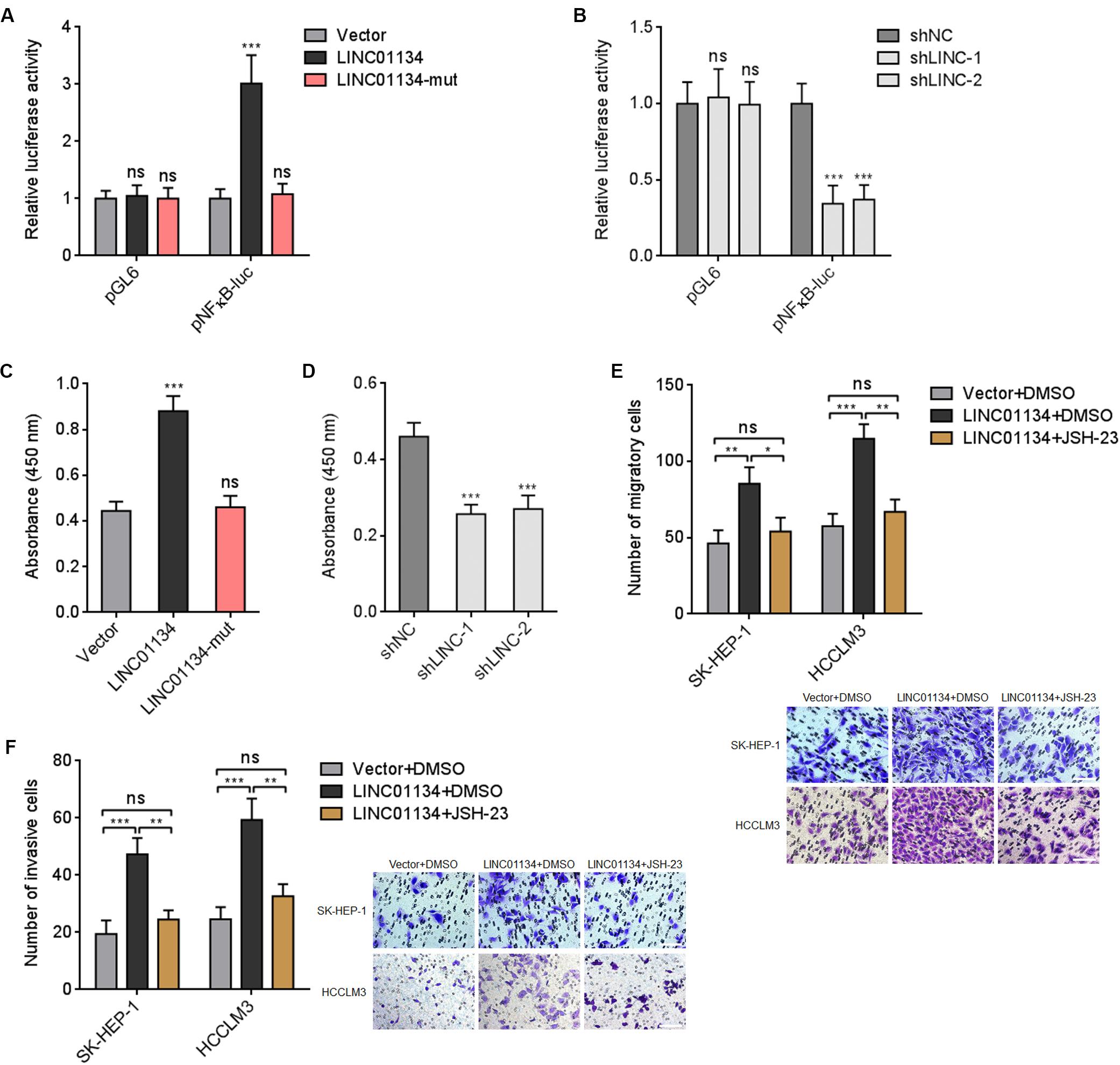
Figure 7. LINC01134 enhances NF-κB transcriptional activity. (A) After transient co-transfection of wild-type or complementary region mutated LINC01134 overexpression plasmids with firefly luciferase reporter containing NF-κB binding sites (pNFκB-luc) and pRL-TK into HCCLM3 cells, dual luciferase reporter assays were conducted to assess NF-κB transcriptional activity. Results are shown as the relative ratio of firefly luciferase activity to renilla luciferase activity. (B) After transient co-transfection of LINC01134-specific shRNAs with firefly luciferase reporter containing NF-κB binding sites (pNFκB-luc) and pRL-TK into HCCLM3 cells, dual luciferase reporter assays were conducted to assess NF-κB transcriptional activity. Results are shown as the relative ratio of firefly luciferase activity to renilla luciferase activity. (C) After transient transfection of wild-type or complementary region mutated LINC01134 overexpression plasmids into HCCLM3 cells, the p65-DNA binding activity was detected using an NF-κB p65 Transcription Factor Assay Kit. (D) After transient transfection of LINC01134-specific shRNAs into HCCLM3 cells, the p65-DNA binding activity was detected using an NF-κB p65 Transcription Factor Assay Kit. (E) LINC01134-stably-overexpressed and control SK-HEP-1 and HCCLM3 cells were treated with 5 μM JSH-23. Then, the migration ability of treated cells was assessed by Transwell migration assays. Representative images of migratory cells were shown. Scale bar = 100 μm. (F) LINC01134-stably-overexpressed and control SK-HEP-1 and HCCLM3 cells were treated with 5 μM JSH-23. Then, the invasion ability of treated cells was assessed by Transwell invasion assays. Representative images of invasive cells were shown. Scale bar = 100 μm. Results are shown as the mean ± SD of three independent experiments. **p < 0.01, ***p < 0.001, ns, not significant, by one-way ANOVA followed by Dunnett’s multiple comparisons test (A–D) or one-way ANOVA followed by Tukey’s multiple comparisons test (E,F).
Discussion
Recent advances in high-throughput sequencings have discovered more and more lncRNAs in diverse types of cancers (Iyer et al., 2015; Berger et al., 2018). Growing studies suggest the important clinical significances and roles of lncRNAs in many cancers, including HCC (Yuan et al., 2014). In this study, we provide another evidence for the implication of lncRNA in HCC.
In this study, we identified a relative novel lncRNA LINC01134. Although this lncRNA has been included in the National Center for Biotechnology Information (NCBI), the expression and roles of LINC01134 in human diseases have not been reported. To our knowledge, we first found that LINC01134 is significantly highly expressed in HCC tissues compared to noncancerous liver tissues. Increased expression of LINC01134 is positively correlated with microvascular invasion and macrovascular invasion but not correlated with age, gender, hepatitis B surface antigen (HBs antigen), liver cirrhosis, alpha-fetoprotein (AFP), tumor size, and encapsulation. Furthermore, high expression of LINC01134 is correlated with poor disease-free survival and overall survival. Our findings identified LINC01134 as an indicator of HCC recurrence and prognosis. The best cutoff values of LINC01134 could be obtained from larger and multicenter cohorts to predict patients’ survival. LINC01134 is 1,965 nt in length, and the gene encoding LINC01134 is located at chromosome 1p36.32. Analyzing TCGA dataset, we found that LINC01134 is also highly expressed in stomach cancer, prostate cancer, lung cancer, esophageal cancer, and bladder cancer and correlated with poor survival of adrenocortical carcinoma, kidney cancer, lung cancer, and glioma patients. These data suggest that LINC01134 may be a cancer-correlated lncRNA. The expressions and clinical significances of LINC01134 in other cancers need further detection.
Gain- and loss-of-function experiments showed that enhanced expression of LINC01134 promotes HCC cell migration and invasion in vitro and HCC liver metastasis and lung metastasis in vivo. Silencing of LINC01134 represses HCC cell migration and invasion in vitro and HCC liver metastasis and lung metastasis in vivo. Our findings identified LINC01134 as a lncRNA regulating HCC metastasis and suggested LINC01134 as a potential therapeutic target for HCC metastasis. Whether LINC01134 regulates the metastasis of other cancers need further investigation.
Different subcellular distributions of lncRNAs influence the different mechanisms of action of lncRNAs. For cytoplasmic lncRNAs, they could directly bind microRNAs and relieve the repressive roles of microRNAs on their genuine targets (Yuan et al., 2014). Furthermore, the cytoplasmic lncRNAs could bind proteins and change the posttranslational modification and/or stability of the interacted proteins (Wang et al., 2014). For nuclear lncRNAs, they often directly bind epigenetic modification enzymes, such as EZH2, and epigenetically regulate the transcription of target genes (Sun et al., 2016). Nuclear lncRNAs may also directly bind DNA, change the architecture of chromatin, and further modulate the transcription of target genes (Ding et al., 2016). In this study, we identified LINC01134 as a chromatin-binding lncRNA and found that LINC01134 directly binds the promoter of AKT1S1. AKT1S1, also known as PRAS40, is a substrate of Akt and a component of the mTOR complex 1 (Volkers and Sussman, 2013). The oncogenic roles of AKT1S1 have been reported in colon cancer, liver cancer, lung cancer, prostate cancer, breast cancer, and so on via regulating PI3K/Akt, mTOR, and/or NF-κB signaling pathways (Malla et al., 2015; Lv et al., 2017). In this study, we found a significantly positive correlation between LINC01134 and AKT1S1 expressions in HCC tissues. In line with LINC01134, AKT1S1 is also highly expressed in HCC and correlated with poor survival of HCC patients. Via binding the promoter of AKT1S1, LINC01134 activates AKT1S1 expression and further activates NF-κB signaling. Functional rescue experiments revealed that silencing AKT1S1 or blocking NF-κB signaling reversed the roles of LINC01134. Thus, our findings identified the activation of AKT1S1-NF-κB signaling as the critical mediators of the roles of LINC01134 in HCC. AKT1S1 could be phosphorylated by Akt and mTOR (Lv et al., 2017). Phosphorylated AKT1S1 was reported to promote HCC metastasis (Hu et al., 2015). In this study, we found that LINC01134 upregulates the total AKT1S1 protein level and also phospho-AKT1S1 level. The further downstream mechanisms after AKT1S1 phosphorylation in HCC metastasis need further investigation. Nevertheless, our findings identified AKT1S1 as a critical mediator of the roles of LINC01134 in promoting HCC metastasis. The effects of LINC01134–AKT1S1 promoter binding on the chromatin architecture of the AKT1S1 promoter need further investigation to elucidate AKT1S1 activation mechanisms. NF-κB signaling pathways are involved in various cancers, including HCC. NF-κB signaling modulates many biological behaviors of cancer cells, including proliferation, apoptosis, migration, invasion, and metastasis (Xu et al., 2013; Wang et al., 2015; Duan et al., 2018; Rodrigues et al., 2018). In this study, we mainly focused on their roles in HCC metastasis. Through regulating NF-κB signaling, the effects of LINC01134 and AKT1S1 on HCC cell proliferation and apoptosis need further investigation. During our revision, another group reported that LINC01134 promotes EMT and metastasis of HCC through the LINC01134/miR-324-5p/IGF3BP1/YY1 axis (Rong et al., 2020). We both reported the pro-metastatic roles of LINC01134 in HCC. But we identified different molecular mechanisms mediating the roles of LINC01134 in HCC, which reflect the diversity and complexity of lncRNAs’ roles and mechanisms in different cellular contexts. Nonetheless, we both identified LINC01134 as a potential therapeutic target for HCC.
Conclusion
Taken together, in this study, we identified a novel lncRNA, LINC01134, which is highly expressed in HCC tissues compared to noncancerous liver tissues. The expression of LINC01134 is positively correlated with microvascular invasion and macrovascular invasion. Furthermore, LINC01134 indicates recurrence and poor overall survival of HCC patients. LINC01134 was identified as a chromatin-binding lncRNA and directly binds the promoter of AKT1S1 and subsequently activates AKT1S1 transcription. Via activating AKT1S1, LINC01134 further activates NF-κB signaling. Via activating AKT1S1-NF-κB signaling, LINC01134 promotes HCC cell migration and invasion in vitro and metastasis in vivo (Figure 8). Our findings suggested LINC01134 as a potential prognostic biomarker and therapeutic target for HCC.
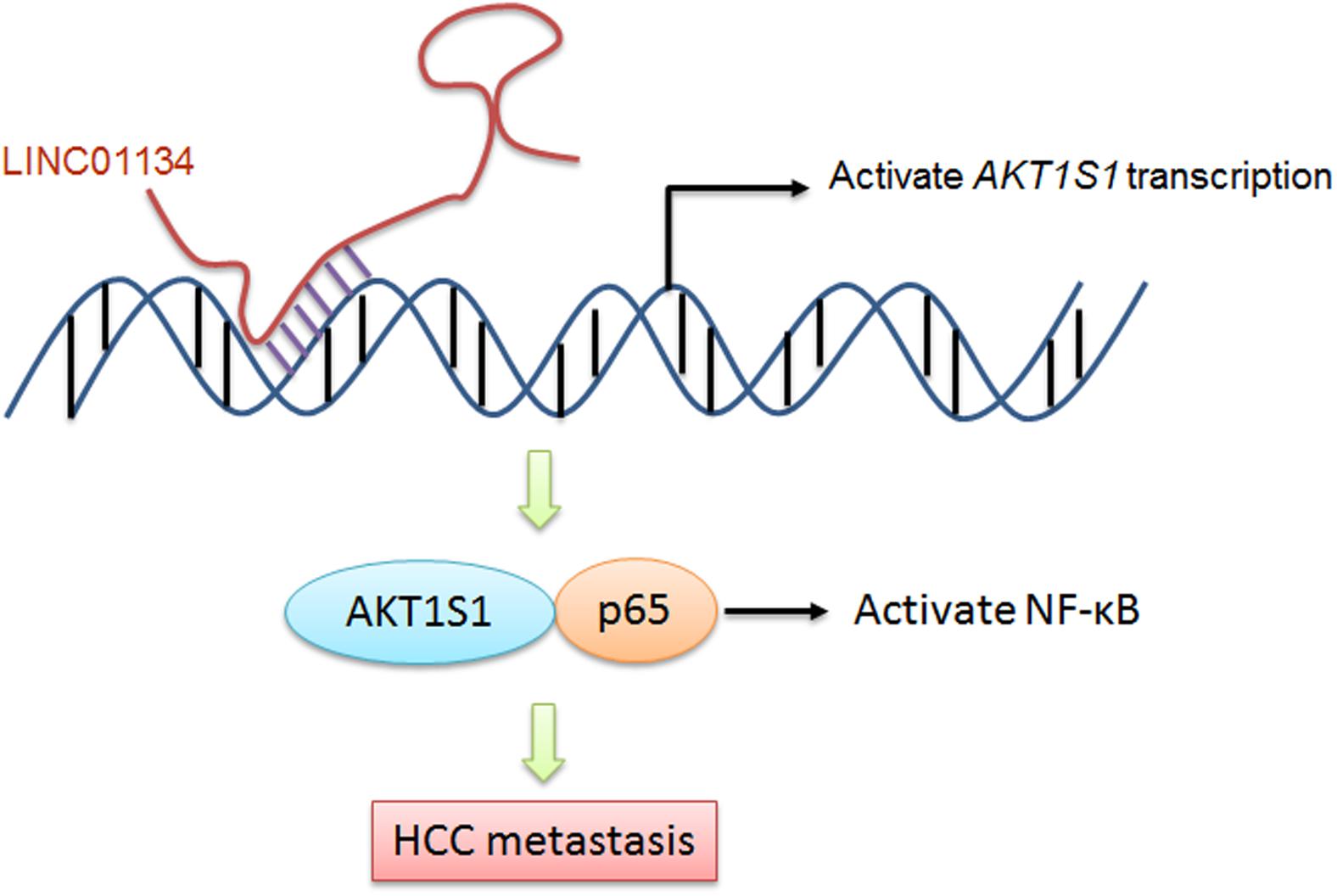
Figure 8. A schematic model of LINC01134 in promoting HCC metastasis via activating AKT1S1-NF-κB signaling.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation, to any qualified researcher.
Ethics Statement
The studies involving human participants were reviewed and approved by the Ethics Committee of Tongji Hospital (Wuhan, China). The patients/participants provided their written informed consent to participate in this study.
Author Contributions
LZ and KC designed this study. CW, YC, and LZ carried out the experiments. LZ, KC, and CW analyzed the data. LZ and CW wrote the manuscript. All authors have read and approved the final manuscript.
Funding
This work was supported by the National Natural Science Foundation of China (no. 81201554), Research Fund for Hepatobiliary and Pancreatic Cancer from Hubei Chen Xiaoping Foundation for Scientific and Technological Development (no. CXPJJH11900001-2019346), and Start Up Fund for Returnees from Tongji Hospital (no. 2019HGRY002).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcell.2020.00429/full#supplementary-material
Footnotes
- ^ http://gepia.cancer-pku.cn/detail.php
- ^ http://asia.ensembl.org/index.html
- ^ https://hgserver1.amc.nl/cgi-bin/r2/main.cgi
References
Auger, C., Alhasawi, A., Contavadoo, M., and Appanna, V. D. (2015). Dysfunctional mitochondrial bioenergetics and the pathogenesis of hepatic disorders. Front. Cell Dev. Biol. 3:40. doi: 10.3389/fcell.2015.00040
Berger, A. C., Korkut, A., Kanchi, R. S., Hegde, A. M., Lenoir, W., Liu, W., et al. (2018). A comprehensive pan-cancer molecular study of gynecologic and breast cancers. Cancer Cell 33:690-705.e9. doi: 10.1016/j.ccell.2018.03.014
Bray, F., Ferlay, J., Soerjomataram, I., Siegel, R. L., Torre, L. A., and Jemal, A. (2018). Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 68, 394–424. doi: 10.3322/caac.21492
Cui, Z., An, X., Li, J., Liu, Q., and Liu, W. (2018). LncRNA MIR22HG negatively regulates miR-141-3p to enhance DAPK1 expression and inhibits endometrial carcinoma cells proliferation. Biomed. Pharmacother. 104, 223–228. doi: 10.1016/j.biopha.2018.05.046
Derderian, C., Orunmuyi, A. T., Olapade-Olaopa, E. O., and Ogunwobi, O. O. (2019). PVT1 signaling is a mediator of cancer progression. Front. Oncol. 9:502. doi: 10.3389/fonc.2019.00502
Ding, L. J., Li, Y., Wang, S. D., Wang, X. S., Fang, F., Wang, W. Y., et al. (2016). Long noncoding RNA lncCAMTA1 promotes proliferation and cancer stem cell-like properties of liver cancer by inhibiting CAMTA1. Int. J. Mol. Sci. 17:1617. doi: 10.3390/ijms17101617
Duan, L., Wu, R., Zhang, X., Wang, D., You, Y., Zhang, Y., et al. (2018). HBx-induced S100A9 in NF-kappaB dependent manner promotes growth and metastasis of hepatocellular carcinoma cells. Cell Death Dis. 9:629. doi: 10.1038/s41419-018-0512-2
Esposito, R., Bosch, N., Lanzos, A., Polidori, T., Pulido-Quetglas, C., and Johnson, R. (2019). Hacking the cancer genome: profiling therapeutically actionable long non-coding RNAs using CRISPR-Cas9 screening. Cancer Cell 35, 545–557. doi: 10.1016/j.ccell.2019.01.019
Gagnon, K. T., Li, L., Janowski, B. A., and Corey, D. R. (2014). Analysis of nuclear RNA interference in human cells by subcellular fractionation and Argonaute loading. Nat. Protoc. 9, 2045–2060. doi: 10.1038/nprot.2014.135
He, J., Zuo, Q., Hu, B., Jin, H., Wang, C., Cheng, Z., et al. (2019). A novel, liver-specific long noncoding RNA LINC01093 suppresses HCC progression by interaction with IGF2BP1 to facilitate decay of GLI1 mRNA. Cancer Lett. 450, 98–109. doi: 10.1016/j.canlet.2019.02.033
Hu, F., Deng, X., Yang, X., Jin, H., Gu, D., Lv, X., et al. (2015). Hypoxia upregulates Rab11-family interacting protein 4 through HIF-1alpha to promote the metastasis of hepatocellular carcinoma. Oncogene 34, 6007–6017. doi: 10.1038/onc.2015.49
Hu, Q., Ye, Y., Chan, L. C., Li, Y., Liang, K., Lin, A., et al. (2019). Oncogenic lncRNA downregulates cancer cell antigen presentation and intrinsic tumor suppression. Nat. Immunol. 20, 835–851. doi: 10.1038/s41590-019-0400-7
Hu, W. L., Jin, L., Xu, A., Wang, Y. F., Thorne, R. F., Zhang, X. D., et al. (2018). GUARDIN is a p53-responsive long non-coding RNA that is essential for genomic stability. Nat. Cell Biol. 20, 492–502. doi: 10.1038/s41556-018-0066-7
Huang, Z., Zhou, J. K., Wang, K., Chen, H., Qin, S., Liu, J., et al. (2019). PDLIM1 inhibits tumor metastasis through activating Hippo signaling in hepatocellular carcinoma. Hepatology 71, 1643–1659. doi: 10.1002/hep.30930
Iyer, M. K., Niknafs, Y. S., Malik, R., Singhal, U., Sahu, A., Hosono, Y., et al. (2015). The landscape of long noncoding RNAs in the human transcriptome. Nat. Genet. 47, 199–208. doi: 10.1038/ng.3192
Keshavarz, M., and Asadi, M. H. (2019). Long non-coding RNA ES1 controls the proliferation of breast cancer cells by regulating the Oct4/Sox2/miR-302 axis. FEBS J. 286, 2611–2623. doi: 10.1111/febs.14825
Kim, J., Piao, H. L., Kim, B. J., Yao, F., Han, Z., Wang, Y., et al. (2018). Long noncoding RNA MALAT1 suppresses breast cancer metastasis. Nat. Genet. 50, 1705–1715. doi: 10.1038/s41588-018-0252-3
Klingenberg, M., Gross, M., Goyal, A., Polycarpou-Schwarz, M., Miersch, T., Ernst, A. S., et al. (2018). The long noncoding RNA cancer susceptibility 9 and RNA binding protein heterogeneous nuclear Ribonucleoprotein L form a complex and coregulate genes linked to AKT signaling. Hepatology 68, 1817–1832. doi: 10.1002/hep.30102
Koo, J. I., Lee, H. J., Jung, J. H., Im, E., Kim, J. H., Shin, N., et al. (2019). The Pivotal role of long noncoding RNA RAB5IF in the Proliferation of Hepatocellular Carcinoma Via LGR5 Mediated beta-Catenin and c-Myc signaling. Biomolecules 9:718. doi: 10.3390/biom9110718
Li, J. K., Chen, C., Liu, J. Y., Shi, J. Z., Liu, S. P., Liu, B., et al. (2017). Long noncoding RNA MRCCAT1 promotes metastasis of clear cell renal cell carcinoma via inhibiting NPR3 and activating p38-MAPK signaling. Mol. Cancer 16:111. doi: 10.1186/s12943-017-0681-0
Li, Y., Yan, G., Zhang, J., Chen, W., Ding, T., Yin, Y., et al. (2020). LncRNA HOXA11-AS regulates calcium oxalate crystal-induced renal inflammation via miR-124-3p/MCP-1. J. Cell Mol. Med. 24, 238–249. doi: 10.1111/jcmm.14706
Lv, D., Guo, L., Zhang, T., and Huang, L. (2017). PRAS40 signaling in tumor. Oncotarget 8, 69076–69085. doi: 10.18632/oncotarget.17299
Ma, M., Xu, H., Liu, G., Wu, J., Li, C., Wang, X., et al. (2019). Metabolism-induced tumor activator 1 (MITA1), an energy stress-inducible long noncoding RNA, Promotes Hepatocellular Carcinoma metastasis. Hepatology 70, 215–230. doi: 10.1002/hep.30602
Malla, R., Ashby, C. R. Jr., Narayanan, N. K., Narayanan, B., Faridi, J. S., et al. (2015). Proline-rich AKT substrate of 40-kDa (PRAS40) in the pathophysiology of cancer. Biochem. Biophys. Res. Commun. 463, 161–166. doi: 10.1016/j.bbrc.2015.05.041
Mondal, T., Juvvuna, P. K., Kirkeby, A., Mitra, S., Kosalai, S. T., Traxler, L., et al. (2018). Sense-Antisense lncRNA Pair Encoded by Locus 6p22.3 determines Neuroblastoma susceptibility via the USP36-CHD7-SOX9 Regulatory Axis. Cancer Cell 33:417-434.e7. doi: 10.1016/j.ccell.2018.01.020
Nasr, M. A., Salah, R. A., Abd Elkodous, M., Elshenawy, S. E., and El-Badri, N. (2019). Dysregulated MicroRNA Fingerprints and Methylation patterns in hepatocellular carcinoma, cancer stem cells, and Mesenchymal stem cells. Front. Cell Dev. Biol. 7:229. doi: 10.3389/fcell.2019.00229
Pan, W., Zhang, N., Liu, W., Liu, J., Zhou, L., Liu, Y., et al. (2018). The long noncoding RNA GAS8-AS1 suppresses hepatocarcinogenesis by epigenetically activating the tumor suppressor GAS8. J. Biol. Chem. 293, 17154–17165. doi: 10.1074/jbc.RA118.003055
Ponting, C. P., Oliver, P. L., and Reik, W. (2009). Evolution and functions of long noncoding RNAs. Cell 136, 629–641. doi: 10.1016/j.cell.2009.02.006
Qi, Z., Zhang, T., Song, L., Fu, H., Luo, H., Wu, J., et al. (2020). PRAS40 hyperexpression promotes hepatocarcinogenesis. EBiomedicine 51:102604. doi: 10.1016/j.ebiom.2019.102604
Qin, G., Tu, X., Li, H., Cao, P., Chen, X., Song, J., et al. (2019). Long noncoding RNA p53-Stabilizing and Activating RNA Promotes p53 signaling by inhibiting heterogeneous nuclear Ribonucleoprotein K deSUMOylation and suppresses hepatocellular carcinoma. Hepatology 71, 112–129. doi: 10.1002/hep.30793
Rodrigues, P., Patel, S. A., Harewood, L., Olan, I., Vojtasova, E., Syafruddin, S. E., et al. (2018). NF-kappaB-dependent Lymphoid Enhancer Co-option Promotes Renal Carcinoma Metastasis. Cancer Discov. 8, 850–865. doi: 10.1158/2159-8290.CD-17-1211
Rong, Z., Wang, Z., Wang, X., Qin, C., and Geng, W. (2020). Molecular interplay between linc01134 and YY1 dictates hepatocellular carcinoma progression. J. Exp. Clin. Cancer Res. 39:61. doi: 10.1186/s13046-020-01551-9
Sakamoto, K., Ogawa, K., Tohyama, T., Ueno, Y., Tamura, K., Inoue, H., et al. (2019). Serosal invasion is a strong prognostic factor for hepatocellular carcinoma after hepatectomy. Hepatol. Res. 49, 419–431. doi: 10.1111/hepr.13285
Siegel, R. L., Miller, K. D., and Jemal, A. (2020). Cancer statistics, 2020. CA Cancer J. Clin. 70, 7–30. doi: 10.3322/caac.21590
Sui, C. J., Zhou, Y. M., Shen, W. F., Dai, B. H., Lu, J. J., Zhang, M. F., et al. (2016). Long noncoding RNA GIHCG promotes hepatocellular carcinoma progression through epigenetically regulating miR-200b/a/429. J. Mol. Med. 94, 1281–1296. doi: 10.1007/s00109-016-1442-z
Sun, C. C., Li, S. J., Li, G., Hua, R. X., Zhou, X. H., and Li, D. J. (2016). Long Intergenic Noncoding RNA 00511 Acts as an Oncogene in Non-small-cell lung cancer by binding to EZH2 and Suppressing p57. Mol. Ther. Nucleic Acids 5:e385. doi: 10.1038/mtna.2016.94
Sun, Q. M., Hu, B., Fu, P. Y., Tang, W. G., Zhang, X., Zhan, H., et al. (2018). Long non-coding RNA 00607 as a tumor suppressor by modulating NF-kappaB p65/p53 signaling axis in hepatocellular carcinoma. Carcinogenesis 39, 1438–1446. doi: 10.1093/carcin/bgy113
Sweta, S., Dudnakova, T., Sudheer, S., Baker, A. H., and Bhushan, R. (2019). Importance of Long Non-coding RNAs in the development and disease of Skeletal Muscle and Cardiovascular Lineages. Front. Cell Dev. Biol. 7:228. doi: 10.3389/fcell.2019.00228
Volkers, M., and Sussman, M. (2013). mTOR/PRAS40 interaction: hypertrophy or proliferation. Cell Cycle 12, 3579–3580. doi: 10.4161/cc.26822
Wang, F., Yang, J. L., Yu, K. K., Xu, M., Xu, Y. Z., Chen, L., et al. (2015). Activation of the NF-kappaB pathway as a mechanism of alcohol enhanced progression and metastasis of human hepatocellular carcinoma. Mol. Cancer 14:10. doi: 10.1186/s12943-014-0274-0
Wang, F., Yuan, J. H., Wang, S. B., Yang, F., Yuan, S. X., Ye, C., et al. (2014). Oncofetal long noncoding RNA PVT1 promotes proliferation and stem cell-like property of hepatocellular carcinoma cells by stabilizing NOP2. Hepatology 60, 1278–1290. doi: 10.1002/hep.27239
Wang, W., Chen, G., Wang, B., Yuan, Z., Liu, G., Niu, B., et al. (2019). Long non-coding RNA BZRAP1-AS1 silencing suppresses tumor angiogenesis in hepatocellular carcinoma by mediating THBS1 methylation. J. Transl. Med. 17:421. doi: 10.1186/s12967-019-02145-6
Wang, Y., Yang, L., Chen, T., Liu, X., Guo, Y., Zhu, Q., et al. (2019). A novel lncRNA MCM3AP-AS1 promotes the growth of hepatocellular carcinoma by targeting miR-194-5p/FOXA1 axis. Mol. Cancer 18:28. doi: 10.1186/s12943-019-0957-7
Wang, Y., Liu, Z., Yao, B., Li, Q., Wang, L., Wang, C., et al. (2017). Long non-coding RNA CASC2 suppresses epithelial-mesenchymal transition of hepatocellular carcinoma cells through CASC2/miR-367/FBXW7 axis. Mol. Cancer 16:123. doi: 10.1186/s12943-017-0702-z
Wang, Z., Yang, B., Zhang, M., Guo, W., Wu, Z., Wang, Y., et al. (2018). lncRNA Epigenetic Landscape analysis identifies EPIC1 as an Oncogenic lncRNA that Interacts with MYC and promotes cell-cycle progression in cancer. Cancer Cell 33:706-720.e9. doi: 10.1016/j.ccell.2018.03.006
Wen, L., Xin, B., Wu, P., Lin, C. H., Peng, C., Wang, G., et al. (2019). An Efficient Combination Immunotherapy for Primary Liver Cancer by Harmonized Activation of Innate and Adaptive Immunity in Mice. Hepatology 69, 2518–2532. doi: 10.1002/hep.30528
Xin, X., Wu, M., Meng, Q., Wang, C., Lu, Y., Yang, Y., et al. (2018). Long noncoding RNA HULC accelerates liver cancer by inhibiting PTEN via autophagy cooperation to miR15a. Mol. Cancer 17:94. doi: 10.1186/s12943-018-0843-8
Xu, W. P., Yi, M., Li, Q. Q., Zhou, W. P., Cong, W. M., Yang, Y., et al. (2013). Perturbation of MicroRNA-370/Lin-28 homolog A/nuclear factor kappa B regulatory circuit contributes to the development of hepatocellular carcinoma. Hepatology 58, 1977–1991. doi: 10.1002/hep.26541
Yan, S., Tang, Z., Chen, K., Liu, Y., Yu, G., Chen, Q., et al. (2018). Long noncoding RNA MIR31HG inhibits hepatocellular carcinoma proliferation and metastasis by sponging microRNA-575 to modulate ST7L expression. J. Exp. Clin. Cancer Res. 37:214. doi: 10.1186/s13046-018-0853-9
Yuan, J. H., Liu, X. N., Wang, T. T., Pan, W., Tao, Q. F., Zhou, W. P., et al. (2017). The MBNL3 splicing factor promotes hepatocellular carcinoma by increasing PXN expression through the alternative splicing of lncRNA-PXN-AS1. Nat. Cell Biol. 19, 820–832. doi: 10.1038/ncb3538
Yuan, J. H., Yang, F., Wang, F., Ma, J. Z., Guo, Y. J., Tao, Q. F., et al. (2014). A long noncoding RNA activated by TGF-beta promotes the invasion-metastasis cascade in hepatocellular carcinoma. Cancer Cell 25, 666–681. doi: 10.1016/j.ccr.2014.03.010
Zhang, H., Liao, Z., Liu, F., Su, C., Zhu, H., Li, Y., et al. (2019). Long noncoding RNA HULC promotes hepatocellular carcinoma progression. Aging 11, 9111–9127. doi: 10.18632/aging.102378
Zhao, X., Liu, Y., and Yu, S. (2017). Long noncoding RNA AWPPH promotes hepatocellular carcinoma progression through YBX1 and serves as a prognostic biomarker. Biochim. Biophys. Acta Mol. Basis Dis. 1863, 1805–1816. doi: 10.1016/j.bbadis.2017.04.014
Zhu, G., Qi, Q., Havel, J. J., Li, Z., Du, Y., Zhang, X., et al. (2017). PRAS40 promotes NF-kappaB transcriptional activity through association with p65. Oncogenesis 6:e381. doi: 10.1038/oncsis.2017.80
Keywords: long noncoding RNA, hepatocellular carcinoma, metastasis, AKT1S1, NF-κB signaling
Citation: Wang C, Chen Y, Chen K and Zhang L (2020) Long Noncoding RNA LINC01134 Promotes Hepatocellular Carcinoma Metastasis via Activating AKT1S1 and NF-κB Signaling. Front. Cell Dev. Biol. 8:429. doi: 10.3389/fcell.2020.00429
Received: 12 February 2020; Accepted: 07 May 2020;
Published: 12 June 2020.
Edited by:
Jaime J. Carvajal, Andalusian Center for Development Biology (CABD), SpainReviewed by:
Ander Matheu, Biodonostia Health Research Institute (IIS Biodonostia), SpainFrederick Charles Campbell, Queen’s University Belfast, United Kingdom
Copyright © 2020 Wang, Chen, Chen and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lei Zhang, emhhbmdsQHRqaC50am11LmVkdS5jbg==; Kunlun Chen, Y2hlbmtsZm9yZnV0dXJlQDE2My5jb20=
 Chao Wang1
Chao Wang1 Kunlun Chen
Kunlun Chen