- 1Stem Cell and Tissue Engineering Laboratory, Department of Orthopedics, West Virginia University, Morgantown, WV, United States
- 2Department of Plastic and Reconstructive Surgery, Shanghai Ninth People’s Hospital Affiliated with Shanghai Jiao Tong University School of Medicine, Shanghai, China
- 3Department of Biochemistry and Molecular Genetics, University of Colorado Denver, Aurora, CO, United States
- 4Bioinformatics Core Facility, West Virginia University, Morgantown, WV, United States
- 5Department of Microbiology, Immunology and Cell Biology, School of Medicine, West Virginia University, Morgantown, WV, United States
- 6WVU Cancer Institute, Robert C. Byrd Health Sciences Center, West Virginia University, Morgantown, WV, United States
Adult stem cells have unique properties in both proliferation and differentiation preference. In this study, we hypothesized that adipose stem cells have a depot-dependent lineage preference. Four rabbits were used to provide donor-matched adipose stem cells from either subcutaneous adipose tissue (ScAT) or infrapatellar fat pad (IPFP). Proliferation and multi-lineage differentiation were evaluated in adipose stem cells from donor-matched ScAT and IPFP. RNA sequencing (RNA-seq) and proteomics were conducted to uncover potential molecular discrepancy in adipose stem cells and their corresponding matrix microenvironments. We found that stem cells from ScAT exhibited significantly higher proliferation and adipogenic capacity compared to those from donor-matched IPFP while stem cells from IPFP displayed significantly higher chondrogenic potential compared to those from donor-matched ScAT. Our findings are strongly endorsed by supportive data from transcriptome and proteomics analyses, indicating a site-dependent lineage preference of adipose stem cells.
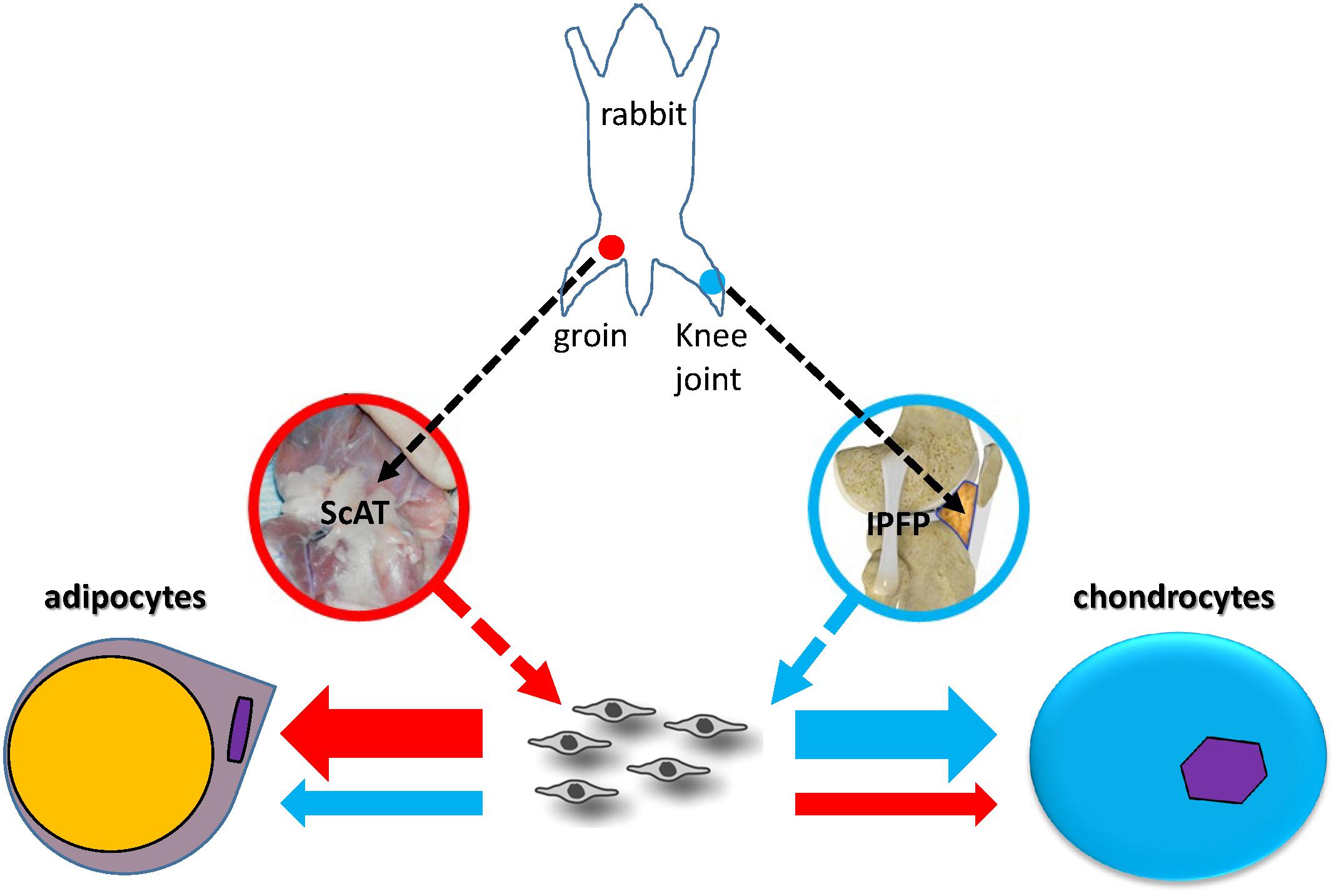
Graphical Abstract. Lineage preference of adult stem cells might be influenced by local matrix microenvironment. In this study, stem cells from subcutaneous adipose tissue (ScAT) had more adipogenic capacity and those from infrapatellar fat pad (IPFP) possessed advantageous chondrogenic potential; these findings are fully endorsed by molecular evidence from RNA Seq and proteomic analyses.
Introduction
Adult stem cells, found in varied tissues such as bone marrow, adipose, and synovium, have the capacity for proliferation and differentiation and are important cell sources for clinical therapy (Brown et al., 2019). However, adult stem cells from different sources have different capacities for proliferation and tissue-specific differentiation (Pizzute et al., 2015). This discrepancy even exists in a single tissue, for example, adipose tissue. Recent evidence indicates that, compared with subcutaneous adipose tissue (ScAT), infrapatellar fat pad (IPFP) is a unique adipose tissue with some distinct features (Sun et al., 2018). ScAT, a fat pad beneath the skin, is becoming an attractive stem cell source for tissue regeneration due to its ease of harvest in large quantities with a minimally invasive procedure (Chu et al., 2019). IPFP, located inside the knee joint and outside the synovial membrane, is becoming a promising stem cell source for cartilage engineering and regeneration (Sun et al., 2018).
In osteoarthritis, IPFP exhibits both anti-inflammatory and pro-inflammatory characteristics (Alegre-Aguarón et al., 2012). Increasing evidence indicates the superiority of IPFP-derived stem cells (IPFSCs) over subcutaneous adipose stem cells (ScASCs) in chondrogenic differentiation capacity (Alegre-Aguarón et al., 2012; Lopa et al., 2014); however, most reports are not donor-matched studies, leading to an uncertain conclusion unless large-scale samples are used. In this study, donor-matched IPFSCs and ScASCs isolated from four New Zealand white rabbits were used for comparison in cell proliferation and tri-lineage differentiation capacities. We hypothesized that adipose stem cells have a depot-dependent lineage preference. Transcriptome and proteome analyses were used to facilitate an in-depth understanding of site-dependent variations of lineage preference.
Materials and Methods
Cell Culture and Proliferation and Surface Phenotype Evaluation
Donor-matched inguinal ScAT and IPFP were obtained from four 4-month-old male New Zealand white rabbits. This project was approved by our Institutional Animal Care and Use Committee (IACUC). The harvested tissues were finely minced and digested using 0.1% collagenase P (Roche, Indianapolis, IN, United States) for 90 min at 37°C in a shaking water bath. The collected cells were cultured in growth medium containing alpha-minimum essential medium (αMEM), 10% fetal bovine serum, 100 U/mL penicillin, 100 μg/mL streptomycin, and 0.25 μg/mL fungizone (Invitrogen, Carlsbad, CA, United States) in a 37°C, 5% CO2, humidified incubator. After reaching 80% confluence, ScASCs and IPFSCs were detached and counted (n = 4) using a hemocytometer, then seeded on T175 flasks at a density of 3000/cm2 for expansion. The cells were counted for three passages after harvesting. Cell population doubling time (PD time) was then calculated based on the formula “PD time = T∗log (2) / (log (N1) – log (N0))”. T stands for incubation time, N1 stands for harvesting cell number, and N0 stands for seeding cell number.
Passage 1 ScASCs and IPFSCs from four donors were used to perform EdU (5-ethynyl-2′-deoxyuridine) cell proliferation assay (cat no. C1024; Invitrogen) according to manufacturer’s protocols. Briefly, the cells were seeded at a density of 3000/cm2, grew to 40% confluence, and then were incubated with 10 μM EdU solution for 3 h before detaching, fixing, permeabilizing, and staining the cells. EdU fluorescence was detected and analyzed by FACSCalibur (BD Biosciences, San Jose, CA, United States) using FCS Express 5 software (De Novo Software, Los Angeles, CA, United States).
A CD146 assay was performed on passage 1 ScASCs and IPFSCs from four donors. Briefly, 3 × 105 cells (n = 4) were incubated with CD146 antibody conjugated with phycoerythrin (cat no. 12-1469-42; Thermo Fisher Scientific, Milford, MA, United States) in darkness for 30 min. The fluorescence was analyzed by FACSCalibur using FCS Express 5 software (De Novo Software, Los Angeles, CA, United States).
Stemness and Senescence Gene Expression
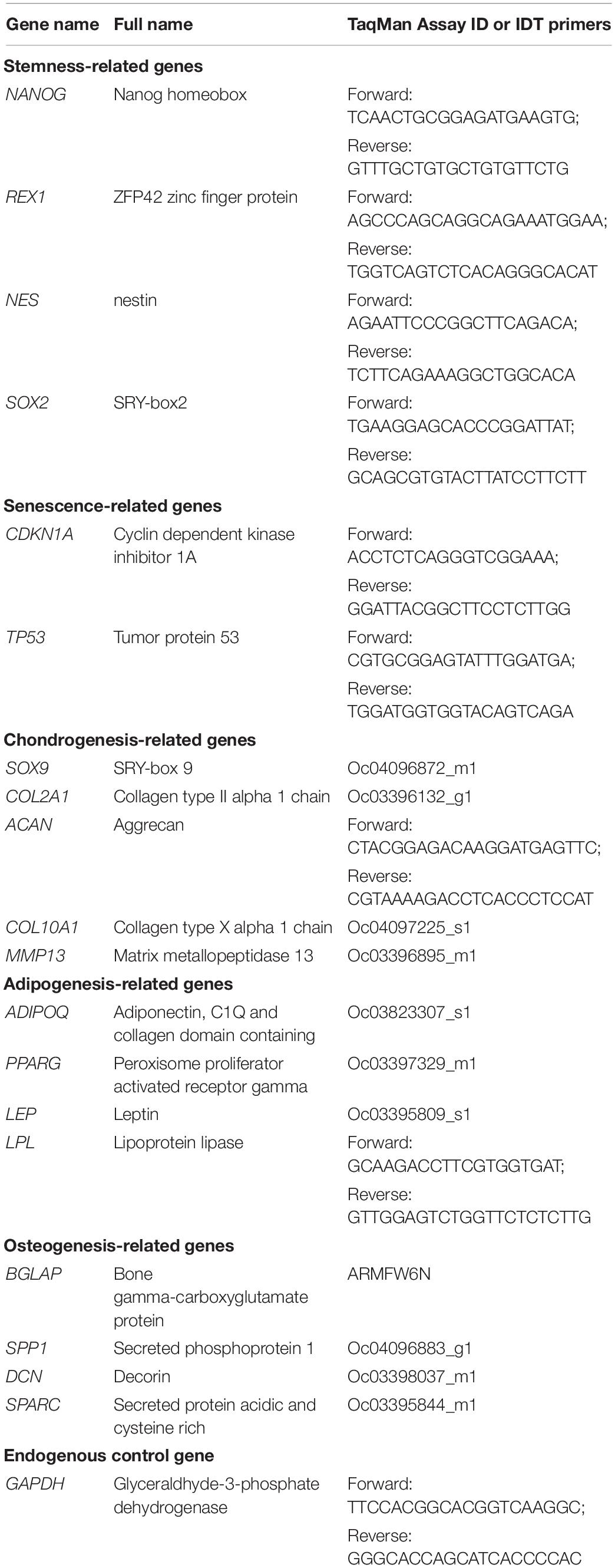
Expanded cells were evaluated using real-time quantitative polymerase chain reaction (qPCR) for differences between ScASCs and IPFSCs in stemness and senescence-related gene expression. Total RNA was extracted from passage 1 cells of all four donors using TRIzol (Invitrogen). Then cDNA was synthesized from mRNA by reverse transcriptase using a High-Capacity cDNA Archive Kit (Thermo Fisher Scientific). Primers of stemness-related genes, senescence-related genes, and an endogenous reference gene (Table 1) were customized from Integrated DNA Technologies (IDT, Coralville, IA, United States) as Sybr green gene expression assay using their qPCR tool. qPCR was performed with iCycler iQTM Multicolor RT-PCR Detection and calculated by computer software (PerkinElmer, Wellesley, MA, United States).
Chondrogenic Induction and Evaluation
Passage 2 expanded cells (4 × 105) were centrifuged at 300 g for 7 min in a 15-ml polypropylene tube to form a pellet. After 24 h incubation (day 0), the pellets were cultured with serum free medium containing high-glucose Dulbecco’s modified Eagle’s medium, 40 μg/mL proline, 0.1 μM ascorbic acid-2-phosphate, 100 nM dexamethasone, 1 × ITS premix (BD Biosciences),100 U/mL penicillin, 100 μg/mL streptomycin, 0.25 μg/mL fungizone, and 10 ng/mL recombinant human transforming growth factor beta3 (TGFβ3, PeproTech Inc., Rocky Hill, NJ, United States) in a 37°C, 5% CO2, humidified incubator for up to 18 days.
Total RNA from ScASCs and IPFSCs (n = 4) was extracted from pellets using an RNase-free pestle in TRIzol, and other qPCR procedures were the same as described above. Primers for chondrogenic-related genes and the endogenous control gene (Table 1) were customized as TaqMan® gene expression assay from Applied Biosystems (Foster City, CA, United States). ACAN primer was customized as Sybr green gene expression assay from IDT (Table 1).
Adipogenic Induction and Evaluation
When passage 2 cells grew to 90% confluence, they were cultured in adipogenic medium consisting of growth medium supplemented with 1 μM dexamethasone, 10 μM insulin (Biovendor, Asheville, NC, United States), 0.5 mM 3-isobutyl-1-methylxanthine, and 200 μM indomethacin in a 37°C, 5% CO2, humidified incubator for as long as 21 days. Primers for qPCR analysis (n = 4) of adipogenic-related genes (Table 1) were customized as TaqMan® gene expression assay from Applied Biosystems. LPL primer was customized as Sybr green gene expression assay from IDT (Table 1).
Adipogenic induced cell samples were dissolved in lysis buffer with protease inhibitors. Total protein was quantified by a NanoDrop Spectrophotometer. Twenty micrograms of proteins (n = 4) were separated on a 12% polyacrylamide gel and transferred to nitrocellulose membrane (30 V, overnight at 4°C). Membranes were blocked with 5% non-fat milk in TBS (Tris-buffered saline) and probed with primary adiponectin (ADIPOQ) monoclonal antibody (cat no. MA1-054; Thermo Fisher Scientific) followed by secondary antibody conjugated with HRP (horseradish peroxidase) (cat no. RK244131; Thermo Fisher Scientific). Next, a chemiluminescence kit (GE Healthcare, Chicago, IL, United States) was used for developing; the blot was imaged using a GE gel documentation. Anti-β actin antibody (Invitrogen) was used to normalize the loading amounts.
Osteogenic Induction and Evaluation
When passage 2 cells grew to 90% confluence, they were cultured with osteogenic medium consisting of growth medium supplemented with 0.01 μM dexamethasone, 50 mg/L ascorbic acid-2-phosphate, and 10 mM β-glycerophosphate in a 37°C, 5% CO2, humidified incubator for as long as 21 days. Primers for qPCR analysis (n = 4) of osteogenic-related genes (Table 1) were customized as TaqMan® gene expression assays from Applied Biosystems.
Protein extracts were assayed for osteocalcin using Osteocalcin (OCN) assay kit (cat no. MBS2019743; MyBioSource, San Diego, CA, United States) following manufacturer’s instructions. Briefly, protein samples (n = 4) were added to wells with pre-coated osteocalcin antibody and incubated at room temperature for 1 h. Upon incubation, 100 uL of detection reagent A was added to the microwells and incubated at 37°C for 1 h. Microwells were washed and incubated with 100 μL of detection reagent B at 37°C for 30 min. Upon washing, 90 μL of 3,3’,5,5’- tetramethylbenzidine substrate solution was added and the sample was incubated at 37°C for 30 min. The reaction was stopped by the addition of 50 μL of stop solution. This enzyme-linked immunosorbent assay (ELISA) was quantitatively measured using a microplate reader at 450 nm.
RNA-Seq Analysis
The total RNA samples collected from donor-matched passage 1 IPFSCs (n = 4) and ScASCs (n = 4) underwent an initial quality control (QC) check. When samples were deemed high quality [RIN (RNA Integrity Number) > 8], a Next Generation Library was built followed by a final QC check, which involved Qubit quantification and size estimation on the Agilent Bioanalyzer using a High Sensitivity DNA chip. The completed libraries were sequenced on their HiSeq 2500 (Illumina, Inc., San Diego, CA, United States). FastQC1 and multiQC (Ewels et al., 2016) were used to confirm that the read quality was good and no trimming was required. We mapped the pair-end RNA-Seq reads to the reference genome of Oryctolagus cuniculus (OryCun2.0) from Ensembl (Zerbino et al., 2018) using the Subread aligner (Liao et al., 2013). The number of read counts corresponding to gene transcripts annotated by Ensembl (OryCun2.0.98) were summarized by the featureCounts function from the Rsubread package (Liao et al., 2019). Gene expression levels were quantified by Reads Per Kilobase of transcript, per Million mapped reads (RPKM) with in-house script. We applied Edger3 (McCarthy et al., 2012) to call differentially expressed genes with the following criteria: fold change (FC) > 2, false discovery rate (FDR) < 0.05, and an average expression (RPKM) higher than one in at least one condition.
Proteomics Analysis of Donor-Matched Cells and Their Deposited Matrices
Donor-matched passage 1 IPFSCs (n = 4; C-IPFSC) and ScASCs (n = 4; C-ScASC) as well as the decellularized extracellular matrices (dECM) deposited by passage 1 IPFSCs (n = 4; E-IPFSC) and ScASCs (n = 4; E-ScASC) were used for this analysis. dECM was prepared following our previous protocol (Li and Pei, 2018). Briefly, 0.2% gelatin solution was used to coat a tissue culture plastic (TCP) flask for 1 h, followed by treatment with 1% glutaraldehyde and 1M ethanolamine for 30 min each. Both cells were cultured in the above-treated TCP flasks using growth medium until 100% confluence followed by supplementation with 250 μM L-ascorbic acid phosphate magnesium salt for 10 days and then treatment with Extraction Buffer (0.5% Triton X-100 and 20 mM NH4OH in phosphate buffered saline) (Pizzute et al., 2016). Cell lysates (50 μg) were diluted in 8 M urea in 100 mM ammonium bicarbonate (ABC) pH 8.5 for digestion according to the Filter Aided Sample Preparation (FASP) protocol. dECMs were processed as previously described (Barrett et al., 2017). Briefly, samples were lyophilized, and digested with 100 mM cyanogen bromide (CNBr). The resultant digest was solubilized in 8 M urea and FASP digested with sequencing grade trypsin (Wiśniewski et al., 2009). Liquid chromatography tandem mass spectrometry (LC-MS/MS) was performed on an Eksigent 2D nano LC coupled to an LTQ-Orbitrap Velos mass spectrometer. MS acquisition parameters were detailed previously (Hill et al., 2015). Raw files were directly loaded into Proteome Discoverer 2.2 3 (Thermo Fisher Scientific) and searched against mouse SwissProt database using an in-house MascotTM server (Version 2.5, Matrix Science). Mass tolerances were +/- 10ppm for MS peaks, and +/- 0.6 Da for MS/MS fragment ions. CNBr/Trypsin specificity was used allowing for 1 missed cleavage. Met oxidation, proline hydroxylation, protein N-terminal acetylation, and peptide N-terminal pyroglutamic acid formation were allowed for variable modifications while carbamidomethyl of Cys was set as a fixed modification. Label Free Quantifications were done using the Minora Feature detector for peak intensity based abundance. Protein thresholds of 1% FDR were used to filter for high confidence identifications. Results were directly exported into Microsoft Excel. Statistical enumeration of the data was achieved through Metaboanalyst (v3.6) (Xia and Wishart, 2016) and pathway analysis was done using the Reactome (Fabregat et al., 2018).
Statistical Analyses
For the data from flow analysis, ELISA, and qPCR, results are presented as the mean and the standard deviation of the mean; the t-test was used to assess data between two groups. All statistical analyses were performed with SPSS 13.0 statistical software (SPSS, Inc., Chicago, IL, United States). p < 0.05 was considered statistically significant.
Results
Stem Cell Proliferation and Surface Marker Expression
ScASCs and donor-matched IPFSCs showed similar fibroblast-like morphology (Figure 1A). Population doubling (PD) time of ScASCs was significantly shorter than IPFSCs (Figure 1B) and relative EdU incorporation of ScASCs was higher than IPFSCs (Figure 1C), indicating that ScASCs were superior to donor-matched IPFSCs in cell proliferation. Surface phenotype assay showed that CD146 expression was negligible in ScASCs with an average of 0.39% (n = 4) while in donor-matched IPFSCs, the average CD146 expression was 4.52% (n = 4), significantly higher than ScASCs (Figure 1D). Due to the commercial unavailability of surface marker antibodies targeting rabbit CD44, CD73, CD90, and CD105, RNA-seq was used to measure these surface marker expressions at mRNA levels including CD44 (Log2FC from IPFSCs to ScASCs was 1.20, FDR = 9.22E-09), NT5E (5′-Nucleotidase Ecto for CD73) (Log2FC from IPFSCs to ScASCs was −0.99, FDR = 1.69E-05), THY1 (Thy-1 Cell Surface Antigen for CD90) (Log2FC from IPFSCs to ScASCs was 0.39, FDR = 1.00E+00), and ENG (Endoglin for CD105) (Log2FC from IPFSCs to ScASCs was −0.67, FDR = 1.32E-01) (Figure 1E). Interestingly, qPCR data showed that there were no significant differences in stemness-related genes NANOG, REX1, NES, and SOX2, and senescence-related genes CDKN1A and TP53 in donor-matched adipose stem cells (Figure 1F).
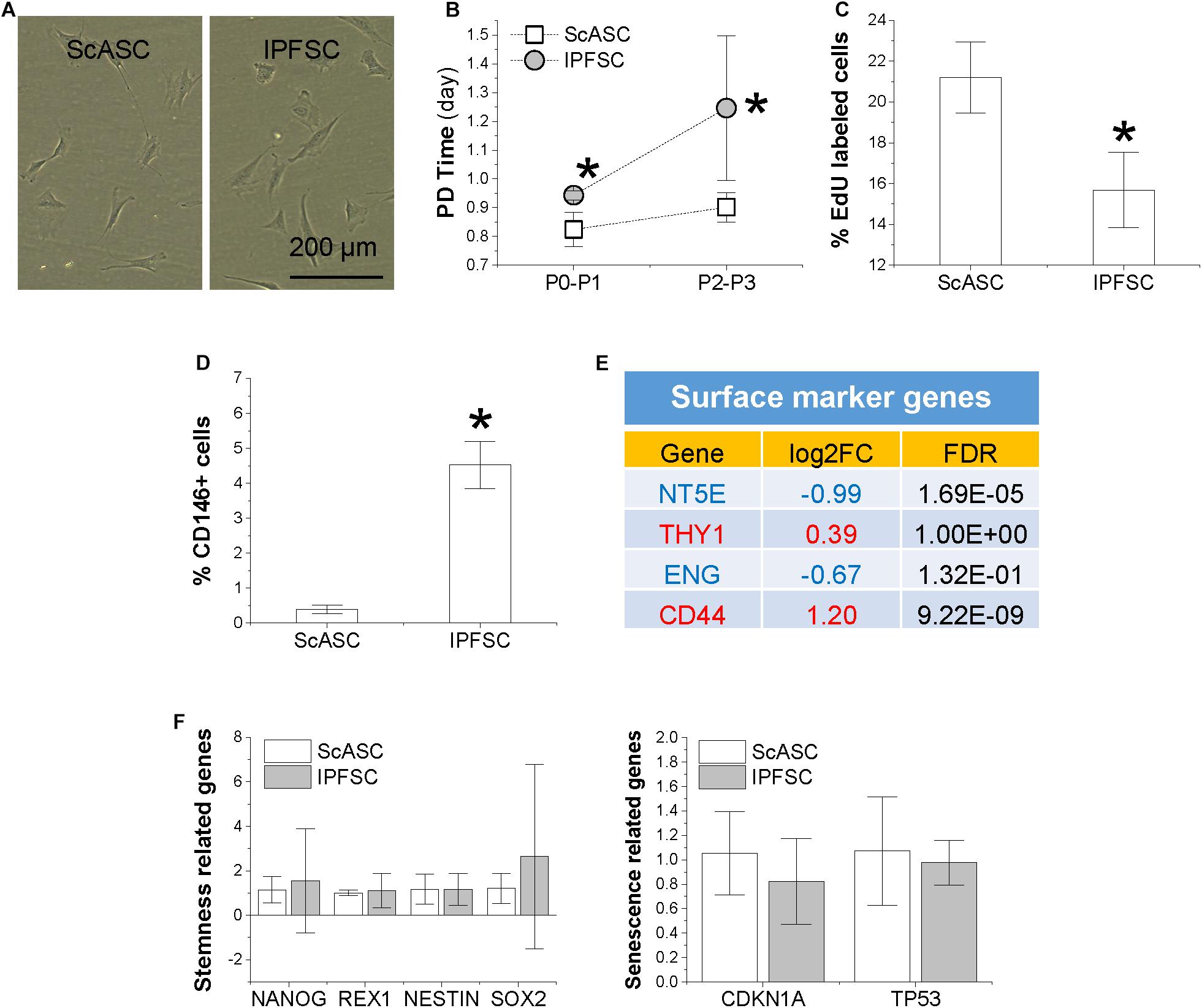
Figure 1. Comparison of proliferation capacity between ScASCs (n = 4) and donor-matched IPFSCs (n = 4) in (A) cell morphology; (B) cell population doubling (PD) time calculated by cell counting; (C) relative EdU incorporation; (D) CD146 surface marker expression level evaluated by flow cytometry analysis; (E) other surface marker gene expression levels evaluated by RNA-seq, in which a positive value in log2FC means higher expression in IPFSCs (in red) and a negative value means higher expression in ScASCs (in blue); and (F) stemness-related genes NANOG, REX1, NESTIN, and SOX2 and senescence-related genes CDKN1A and TP53 measured by qPCR. Data are shown as bar charts. * indicates a significant difference compared to the corresponding control group (p < 0.05).
Adaptation of Transcription Factors During Differentiation Induction
To determine the adaptation of transcription factors in ScASCs and donor-matched IPFSCs, qPCR was used to evaluate SOX9 for chondrogenesis, PPARG for adipogenesis, and RUNX2 for osteogenesis during differentiation induction. Before induction, only SOX9 expression was significantly higher in IPFSCs compared with ScASCs (Figure 2A). After an 18-day chondrogenic induction, SOX9 expression was maintained at a higher level in IPFSCs similar to day 0 while PPARG expression was significantly lower than ScASCs despite a remarkable increase of PPARG in both cells (Figures 2B,C). After 21-day adipogenic induction, the difference in SOX9 expression between groups was diminished; interestingly, both PPARG and RUNX2 expression in IPFSCs were significantly lower than donor-matched ScASCs (Figure 2D). After a 21-day osteogenic induction, the difference in SOX9 expression decreased but IPFSCs still had a higher level than ScASCs; intriguingly, IPFSCs exhibited a higher expression level in PPARG than ScASCs (Figure 2E).
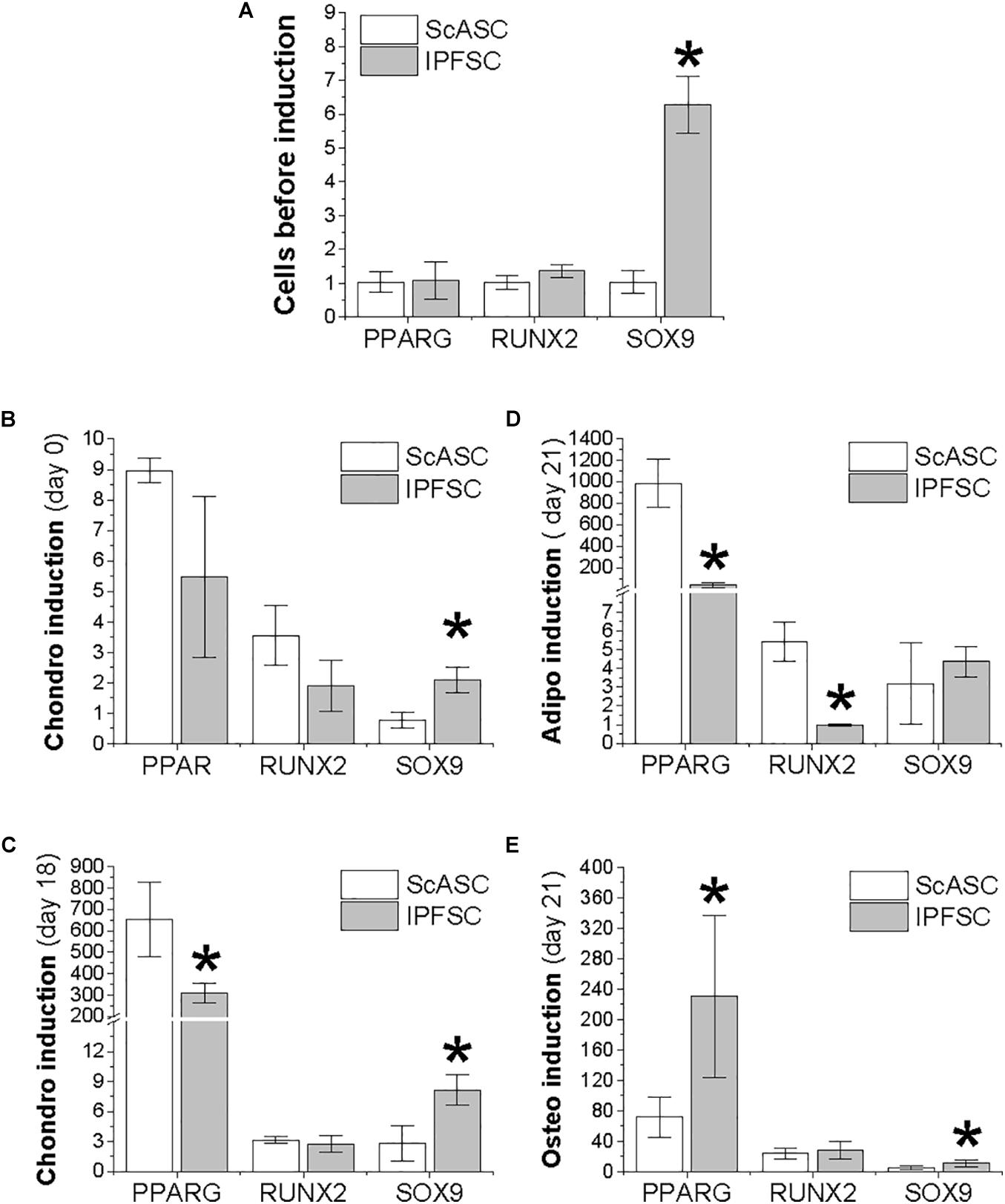
Figure 2. Expression of transcription factors in ScASCs (n = 4) and donor-matched IPFSCs (n = 4) measured by qPCR. SOX9, PPARG, and RUNX2 were used to represent chondrogenesis, adipogenesis, and osteogenesis, respectively. Expanded cells were evaluated before induction (A) and after induction (B–E): (B) 24 h after centrifugation to form a pellet but before chondrogenic induction; (C) 18 days after chondrogenic induction; (D) 21 days after adipogenic induction; and (E) 21 days after osteogenic induction. Data are shown as bar charts. Expression of each target gene in undifferentiated IPFSCs (A) and differentiated ScASCs/IPFSCs (B–E) is plotted against undifferentiated ScASCs (A), which is set as “1”. * indicates a significant difference compared to the corresponding control group (p < 0.05).
Development of Differentiation Factors During Induction
To further explore the differentiation preference of donor-matched adipose stem cells, qPCR was used to evaluate COL2A1 and ACAN as markers of chondrogenesis, ADIPOQ and LPL for adipogenesis, and BGLAP and SPP1 for osteogenesis during induction. Before induction, IPFSCs had significantly higher expression of COL2A1 and ACAN than ScASCs (Figure 3A), in line with SOX9 expression (Figure 2A). The advantageous expression of COL2A1 and ACAN was further strengthened during chondrogenic induction (Figure 3B). No significant difference of COL10A1 expression was observed between groups; however, we found that MMP13 expression was more than 1000 times higher in IPFSCs compared with ScASCs before induction, which became comparable between groups after chondrogenic induction (Figure 3C).
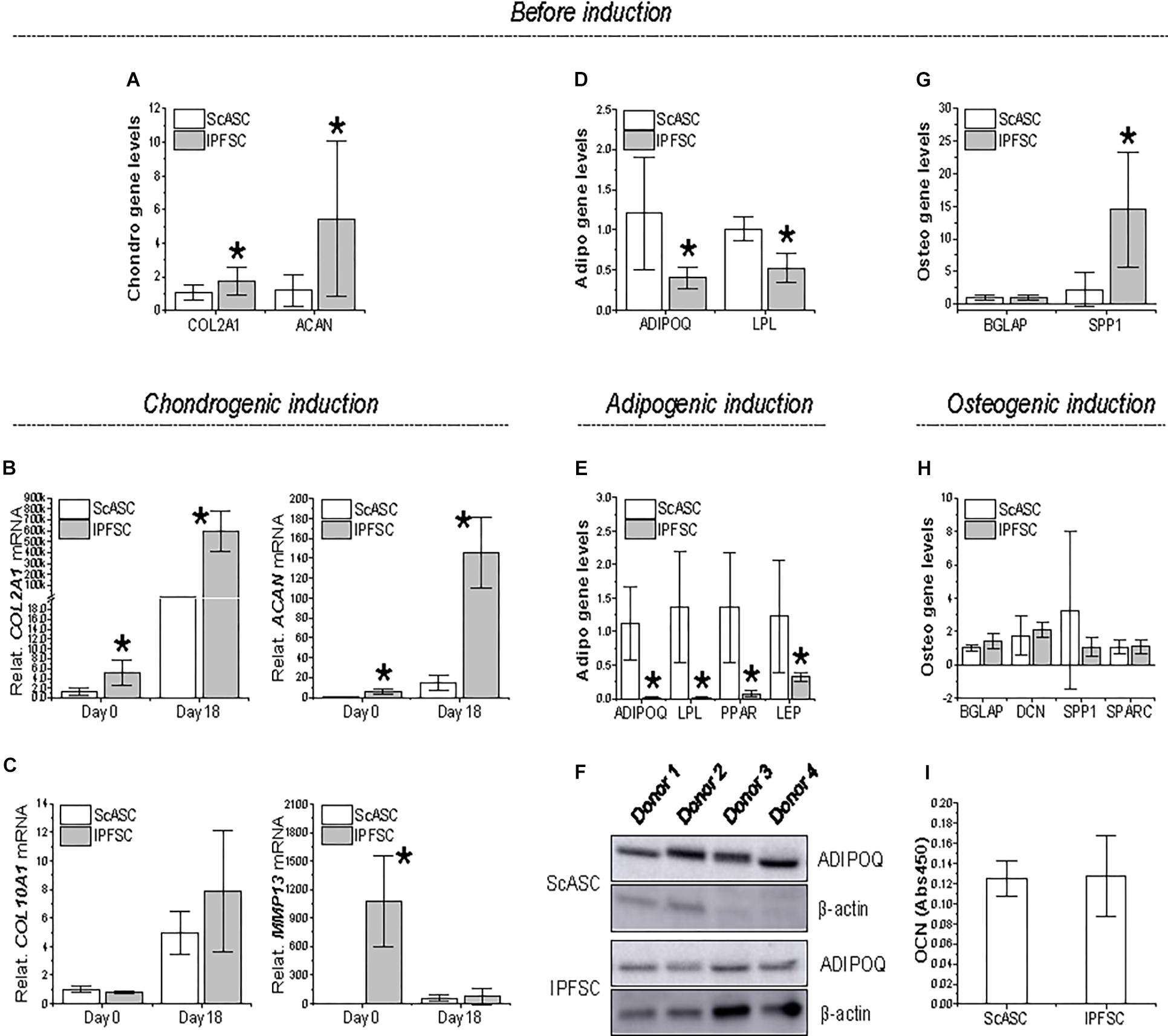
Figure 3. Expression of differentiation genes in ScASCs (n = 4) and donor-matched IPFSCs (n = 4) before and after induction. For chondrogenic differentiation, typical markers, COL2A1 and ACAN, were evaluated using qPCR before (A) and after chondrogenic induction (B). Hypertrophic markers COL10A1 and MMP13 were also measured after chondrogenic induction (C). For adipogenic differentiation, typical markers, ADIPOQ and LPL, were evaluated using qPCR before (D) and after adipogenic induction (E). LEP expression was also measured after induction (E). Adiponectin (ADIPOQ) expression was confirmed using Western blot analysis with β-actin as an internal control (F). For osteogenic differentiation, typical markers, BGLAP and SPP1, were evaluated using qPCR before (G) and after osteogenic induction (H). Expression of DCN and SPARC were also measured after induction (H). Osteocalcin (OCN) expression was confirmed using ELISA analysis (I). Expression of each target gene in IPFSCs is plotted against the corresponding ScASCs, which is set as “1”, but for those in chondrogenic induction (B/C), expression in day 0 is plotted as “1”. Data are shown as bar charts. * indicates a significant difference compared to the corresponding control group (p < 0.05).
Before induction, ScASCs exhibited higher expression of ADIPOQ and LPL than IPFSCs (Figure 3D), despite no significant difference in transcription factor PPARG expression (Figure 2A). The dominant expression of ADIPOQ and LPL became dramatically increased during adipogenic induction along with differentiation gene LEP and transcription factor PPARG expression (Figure 3E). Moreover, higher expression of adiponectin (ADIPOQ) in ScASCs than donor-matched IPFSCs was also confirmed in Western blot analysis (Figure 3F).
For osteogenic differentiation, despite no difference in BGLAP expression before induction, IPFSCs displayed a higher expression of SPP1 than ScASCs (Figure 3G), which became comparable after 21-day osteogenic induction along with other differentiation genes BGLAP, DCN, and SPARC expression (Figure 3H). Furthermore, comparable expression of OCN between ScASCs and IPFSCs was also confirmed in ELISA analysis (Figure 3I).
Transcriptome Difference Between ScASCs and Donor-Matched IPFSCs
We applied RNA-seq data analysis to explore the difference in gene expression levels between ScASCs and IPFSCs. Principal coordinate analysis on gene expression across all genes revealed that samples from ScASCs and IPFSCs were from two distinctive clusters on the first dimension (Figure 4A). By applying EdgeR (McCarthy et al., 2012) with stringent thresholds [FC > 2 and FDR < 0.05], we identified 359 and 277 protein-coding genes upregulated and downregulated from the ScASC samples to the IPFSC samples, respectively (Figure 4B). Intriguingly, differentially expressed genes were enriched in those encoding transcription factors (Figure 4C), including the Homeobox (HOX) gene family related to stem cell differentiation (Seifert et al., 2015; Figure 4D).
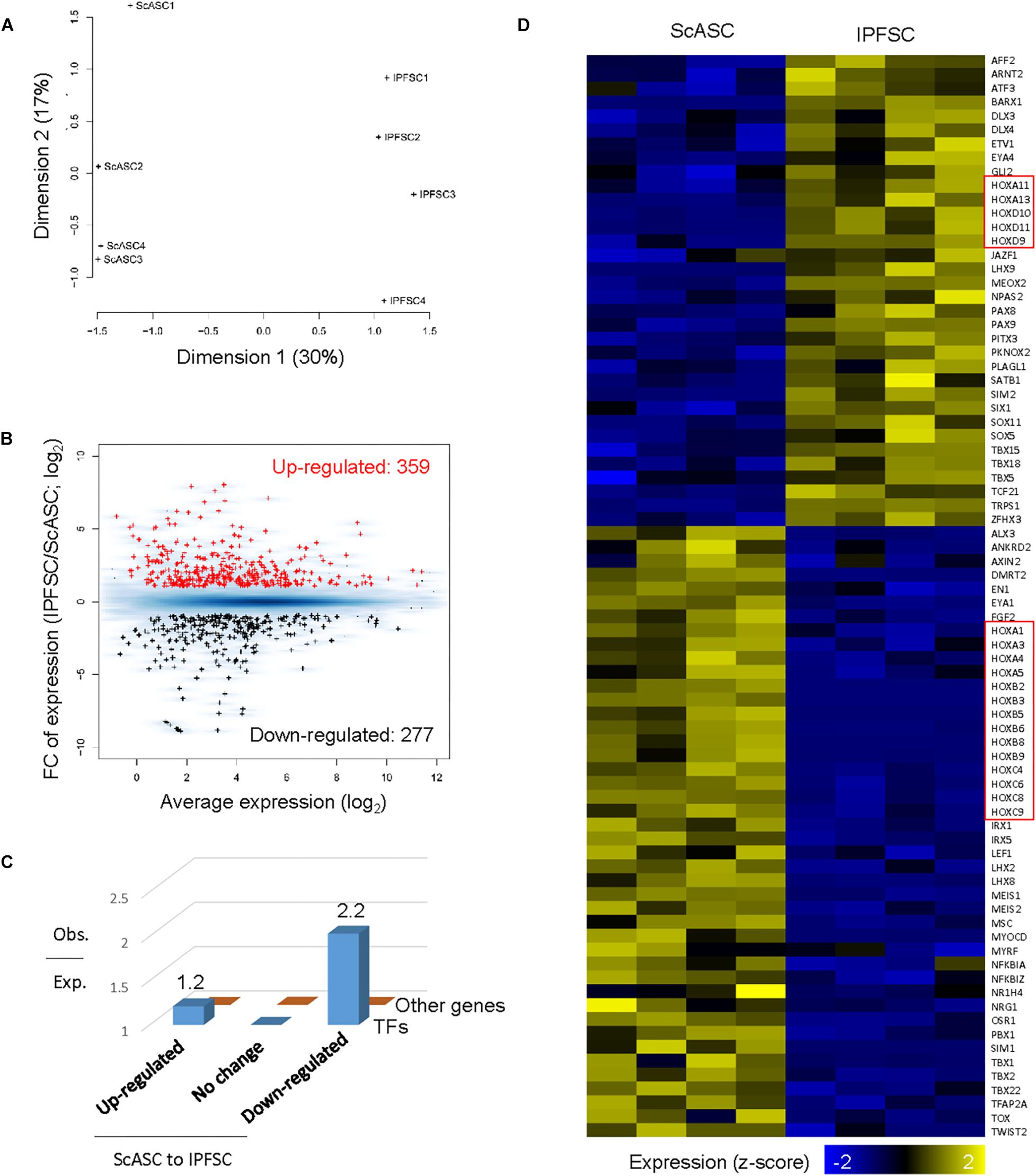
Figure 4. Differentially expressed gene analysis of ScASCs and donor-matched IPFSCs from four rabbits (n = 4). (A) Principal coordinates analysis on gene expression for RNA-Seq samples from ScASCs and IPFSCs. (B) MA plot showing the average and fold change (FC) of gene expression for differentially expressed genes between ScASCs and IPFSCs. Red: genes upregulated from ScASCs to IPFSCs; Black: genes downregulated from ScASC to IPFSCs; Blue background: all expressed genes. (C) Bar plot showing the ratio of observed number to the expected number of genes for transcription factor genes and for other genes grouped based on their expression changes between ScASCs and IPFSCs. (D) Heat map showing the expression changes between ScASCs and IPFSCs for differentially expressed transcription factors. Genes sorted first by their expression changes and then by names. Red rectangles: Homeobox (HOX) genes.
We next examined the expression changes on particular groups of genes known to function in chondrogenesis and/or adipogenesis. The data were presented as Log2FC of expression and FDR. Functional annotation clustering was categorized (Figure 5) including (A) HOX gene family; (B) T (the brachyury gene)-box (TBX) gene family; (C) TGFβ (TGFB) gene family; (D) SOX gene family; (E) Bone morphogenetic protein (BMP) gene family; (F) Wingless/integrase-1 (WNT) gene signals; (G) Collagen (COL) gene family; (H) Lysyl oxidase (LOX) gene family; (I) Genes coding for Basement membrane proteins; (J) Genes coding Matrix turnover enzymes; (K) Integrin (ITG) gene family; (L) Aquaporin (AQP) gene family; (M) Chondrogenic-related genes; and (N) Adipogenic-related genes.
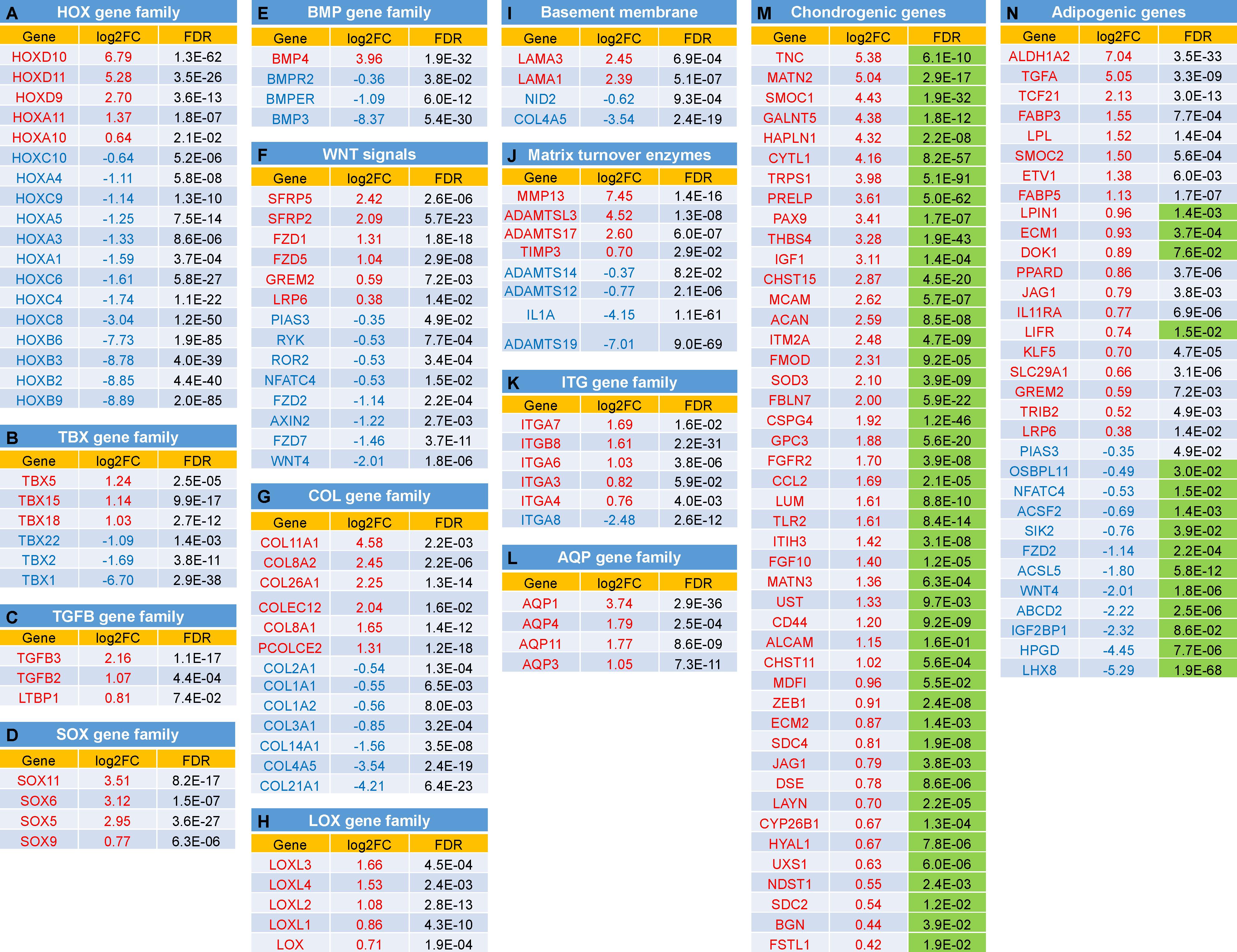
Figure 5. Expression changes between IPFSCs and ScASCs for selected groups of genes with function in chondrogenesis and/or adipogenesis: (A) Homeobox (HOX) gene family; (B) T (the brachyury gene)-box (TBX) gene family; (C) TGFβ (TGFB) gene family (D) SOX gene family; (E) Bone morphogenetic protein (BMP) gene family; (F) Wingless/integrase-1 (WNT) gene signals; (G) Collagen (COL) gene family; (H) Lysyl oxidase (LOX) gene family; (I) Genes coding for Basement membrane proteins; (J) Genes coding Matrix turnover enzymes; (K) Integrin (ITG) gene family; (L) Aquaporin (AQP) gene family; (M) Chondrogenic-related genes; and (N) Adipogenic-related genes. Green shaded FDR indicates favorable effect of the gene on chondrogenesis (M) or adipogenesis (N). A positive value in log2FC means higher expression in IPFSCs (in red), and a negative value means higher expression in ScASCs (in blue). Orange shaded FDR: > 0.05.
Proteome Difference Between ScASCs and Donor-Matched IPFSCs
Both cells and deposited proteins were interrogated with bottom-up mass spectrometry-based proteomics to monitor differences in protein abundance (Figure 6A). The pathway analysis highlighted that the matrix proteins deposited by ScASCs mapped to more cell cycle regulators than traditional ECM proteins (Figures 6B,C), such as the uptick in fibril associated collagens with interrupted triple helices (FACIT collagens) including COL12A1 (FC = 2.26, p = 0.010) and COL14A1 (FC = 4.73, p = 0.002), Thrombospondin 1 (THBS1) (FC = 4.60, p = 0.008), THBS2 (FC = 8.37, p = 0.009), Asporin (ASPN, FC = 9.70, p = 0.002), Podocan (PODN, FC = 1.70, p = 0.009), and Versican (VCAN, FC = 3.10, p = 0.001). These proteins did not seem to be laying down as much ECM in general as those deposited by IPFSCs as shown by both the pathway analysis and the relatively lower amount of structural ECM.
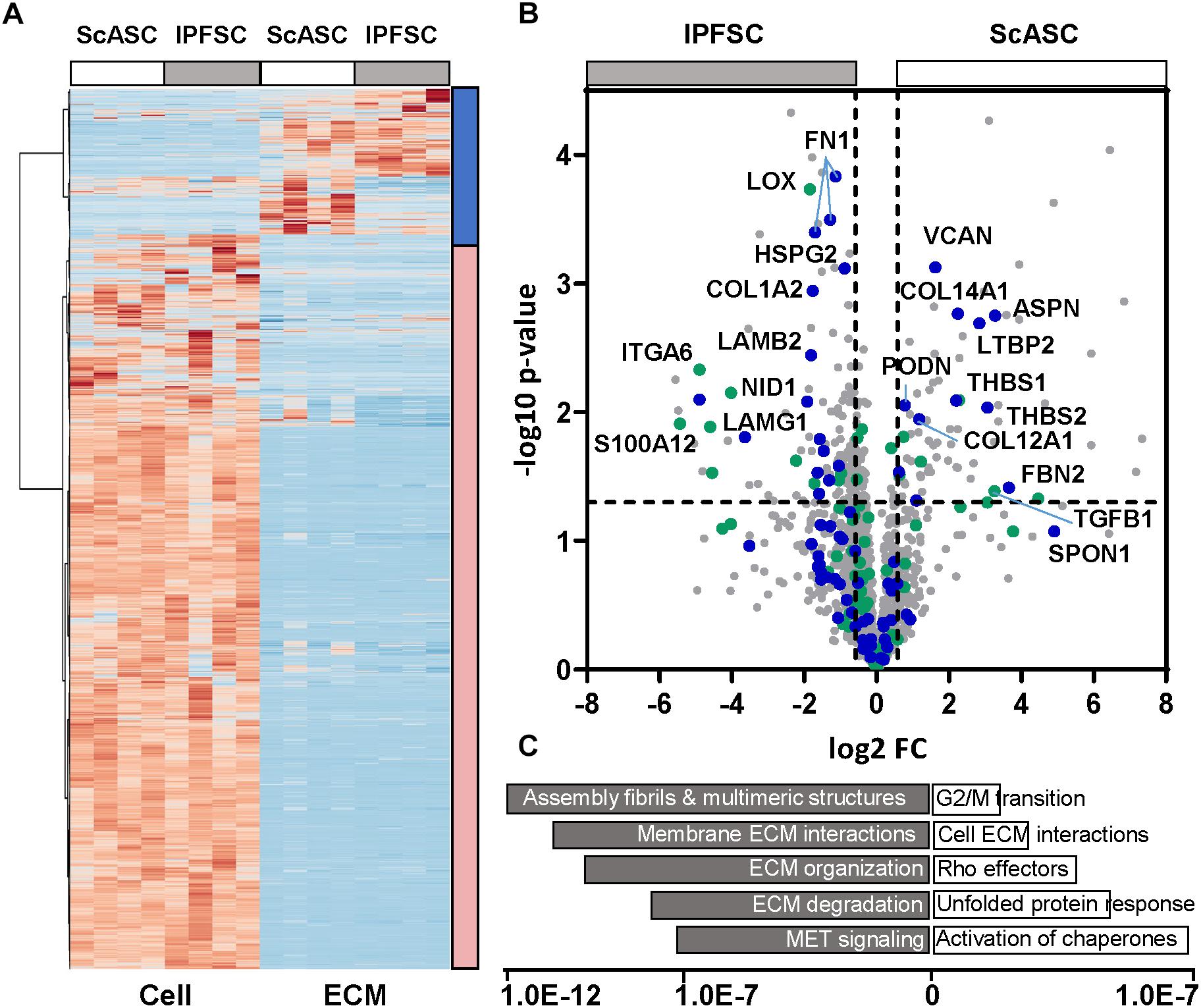
Figure 6. Proteomic results of cells and ECMs from ScASCs versus IPFSCs of four donor-matched rabbits. (A) Heatmap for global representation of protein differences (Z score used). The blue bar on the left indicates the region highly enriched in ECM components, the red bar indicates the region that contains primarily intracellular proteins. (B) Volcano plot with the Y-axis indicating -log10 p-value and the X-axis of the fold change (FC) in log2. Core ECM proteins are indicated in blue and Matrisome associated proteins are in green. (C) Gene enrichment analysis (top pathways, Reactome) is shown with the X-axis indicating the statistical significance based on false discovery rates using the Benjamani_hochberg method.
In contrast, the matrix proteins enriched from IPFSCs include numerous basement membrane components (Figures 6B,C), such as nidogen 1 (NID1) (FC = 0.27, p = 0.008), heparin sulfate proteoglycan 2 (HSPG2)/perlecan (FC = 0.54, p = 0.001), laminin subunits including LAMA5 (FC = 0.10, p = 0.030), LAMB1 (FC = 0.40, p = 0.033), LAMB2 (FC = 0.28, p = 0.004), and LAMG1 (FC = 0.34, p = 0.005). Of relevance, and consistent with the mRNA level, the integrin A6 subunit (ITGA6) that forms a laminin receptor was also significantly increased (FC = 0.03, p = 0.005). In addition, various isoforms of fibronectin (FN) were significantly higher. Interestingly, compared to those secreted by ScASCs, collagen modifying enzymes, such as LOX and tissue transglutaminase (TG2, FC = 0.49, p = 0.050), accompanying fibrillar collagens, such as COL5A1 (FC = 0.36, p = 0.020) and COL5A3 (FC = 0.48, p = 0.030), were significantly upregulated in the matrix proteins deposited by IPFSCs. Consistent with the measured mRNA levels, LOX was significantly increased (FC = 0.27, p = 0.000) in matrix proteins deposited by IPFSCs, with similar trends in LOX-like 1 (LOXL1) (FC = 0.30, p = 0.036) and LOXL4 (FC = 0.48, p = 0.030).
Discussion
Adult stem cells have the ability to differentiate into varied lineages. Increasing evidence indicates that adult stem cells from different tissues do not have equivalent ability toward a specific lineage differentiation (Jones and Pei, 2012; Pizzute et al., 2015). However, there is a lack of comparable studies using donor-matched adult stem cells and their matrix microenvironment. In this study, we found that, despite both deriving from adipose tissue, ScASCs and IPFSCs isolated from different depots of four donor-matched rabbits exhibited significant divergence in both proliferation and differentiation potential. Transcriptome and proteomics data provide robust evidence to support our hypothesis that there are site-dependent variations in lineage potential and preference of stem cells from adipose tissue.
Our proliferation data showed that ScASCs grew faster than donor-matched IPFSCs, which might result from higher levels of COL12A1, COL14A1, THBS1, THBS2, and VCAN expression in the matrix proteins deposited by ScASCs. These matrix proteins trend higher in tumor microenvironments and tissues to support proliferative or structurally rigid phenotypes (Wight et al., 2014; Subramanian and Schilling, 2015; Belotti et al., 2016). We also found that IPFSCs expressed significantly higher CD146 than donor-matched ScASCs, which was in line with the gene expression level in RNA-seq data (Log2FC from IPFSCs to ScASCs in CD146/MCAM was 2.62, FDR = 5.7E-07). Regardless of the role as a perivascular marker for adult stem cells (Crisan et al., 2008), a higher level of CD146 expression in adult stem cells might be associated with an advantage in chondrogenic differentiation (Hagmann et al., 2014; Su et al., 2015). Despite the failure to measure expression of surface markers CD90 and CD105 using flow cytometry on both ScASCs and IPFSCs (data not shown), our RNA-seq data indicated that the genes (NT5E, THY1, ENG, and CD44) encoding surface marker proteins (CD73, CD90, CD105, and CD44) were expressed in both cells with NT5E preferentially expressed in ScASCs and CD44 advantageously expressed in IPFSCs. We also found no statistical difference in measured stemness-related genes (NANOG, REX1, NESTIN, and SOX2), indicating that these genes expressed in the donor-matched IPFSCs and ScASCs might not contribute to the discrepancy of a specific lineage differentiation found in this study.
Our differentiation data indicated that IPFSCs exhibited a significantly higher level of chondrogenic markers compared with donor-matched ScASCs. RNA-seq data showed that advantageous genes in IPFSCs can be categorized into several clusters based on functional annotation clustering. For example, the HOX gene, an evolutionarily highly conserved gene family, determines lineage specific differentiation of adult stem cells once tissue damage occurs (Seifert et al., 2015). In this study, HOXD genes, including HOXD9-11 and HOXA10/11, were only dominantly expressed in IPFSCs over donor-matched ScASCs. HOXD genes, one of four grouped HOX clusters (HOXA, HOXB, HOXC, and HOXD) in vertebrates, are suggested to play a role in stem cell based chondrogenesis (Jung and Tsonis, 1998; González-Martín et al., 2014). HOXA11 and HOXD11 double mutant mice had shorter limb formation (Boulet and Capecchi, 2004). Compared with IPFSCs, however, donor-matched ScASCs exhibited robust expression of other HOX genes, such as HOXA4, HOXA5, HOXC4, and HOXC8, which are closely associated with adipogenesis rather than osteogenesis (Levi et al., 2010). An exception is HOXC8 which does not inhibit but retards chondrogenesis, resulting in a boost of proliferating precursor cells and a negative influence on chondrogenesis (Yueh et al., 1998; Cormier et al., 2003).
Another group of conservative transcription factors, TBX family genes, also plays crucial roles throughout development. Our RNA-seq data showed that TBX5, TBX15, and TBX18 were highly expressed in IPFSCs compared to donor-matched ScASCs. TBX5 was reported to activate WNT and fibroblast growth factor (FGF) signaling in upper limb development (Takeuchi et al., 2003). Overexpression of TBX15 in 3T3-L1 preadipocytes impeded adipocyte differentiation and reduced triglyceride content (Gesta et al., 2011). TBX15 and TBX18 co-expressed in the limb mesenchyme indicate their functions during limb development (Tanaka and Tickle, 2004; Singh et al., 2005). Recently, TBX18 was reported to co-express with SOX9 in chondrogenic progenitors and differentiating chondrocytes (Haraguchi et al., 2015). In contrast, TBX1 was reported to be preferentially expressed in the inducible pool of brown adipocytes (beige cells) present in white fat of the inguinal depot (Wu et al., 2012). The above findings indicate that two fat depots, ScAT and IPFP, exhibit distinct HOX and TBX conservative gene expression profiles that likely influence their differentiation preference.
The TGFβ superfamily is composed of two subfamilies; the TGFβ subfamily mainly includes TGFβs (1, 2, and 3) and the BMP subfamily mainly includes BMPs (2, 4-10) (Wang et al., 2014). A considerable amount of in vitro data indicate that TGFβ signaling pathways promote mesenchymal condensation (Barry et al., 2001; Tuli et al., 2003) and the early stages of chondrocyte differentiation, but restrain terminal hypertrophy (Mueller and Tuan, 2008; Shintani et al., 2013). In this study, we found that all TGFβ isotypes (TGFB1, TGFB2, and TGFB3) and latent-TGFB-binding protein 1 (LTBP1) were preferentially expressed in IPFSCs rather than donor-matched ScASCs. However, the protein level showed an opposite trend for TGFB1 and LTBP2, indicating that, while ScASCs have more available bound ligand, IPFSCs may have more free ligand, which needs to be further confirmed. This finding is in line with the advantageous expression in IPFSCs of the combination of SOX5, SOX6, and SOX9 (the SOX trio) that supplies signals sufficient to induce permanent cartilage (Ikeda et al., 2004) and of SOX11 that contributes to the regulation of growth/differentiation factor 5 (GDF5) in joint maintenance (Kan et al., 2013). Our data also showed that only BMP4 exhibited a statistically higher level of expression in IPFSCs compared with donor-matched ScASCs. BMP4 plays an important role in retaining chondrogenic phenotype, through both increasing matrix production and suppressing the production of collagen type X (Reddi, 2001; Steinert et al., 2003). Another important BMP isotype in chondrogenesis, BMP2, was expressed at a higher level in IPFSCs than in donor-matched ScASCs despite no significant difference (FDR = 5.9E-01). Interestingly, BMP3, which was reported to inhibit bone formation by hindering BMP2 activity (Bahamonde and Lyons, 2001), was significantly expressed in ScASCs compared with donor-matched IPFSCs. These findings indicate that IPFSCs have an expression profile consistent with endochondral bone formation over donor-matched ScASCs. The above data support that TGFβ signaling cooperated with BMP signaling in stimulating articular cartilage regeneration (Wang et al., 2014).
Increasing evidence indicates that the canonical Wnt signaling pathway inhibits adipogenesis while the noncanonical Wnt pathway promotes adipogenesis by antagonizing the Wnt/β-catenin pathway (Christodoulides et al., 2009). In accordance with the above findings, we found that preferential expression favoring Wnt/β-catenin signaling genes such as SPARC-related modular calcium binding protein 2 (SMOC2) (Nie and Sage, 2009), low-density lipoprotein receptor-related protein 6 (LRP6) (Peröbner et al., 2012), and Gremlin 2 (GREM2) (Wu et al., 2015) in IFPSCs might be responsible for the inhibition of adipogenesis and the enhancement of chondrogenesis. In contrast, expression inhibiting Wnt/β-catenin signaling genes such as WNT4 (Liu et al., 2016) and nuclear factor of activated T-cells 4 (NFATC4) (Yang et al., 2006) in ScASCs might be responsible for promoting adipogenesis and blocking chondrogenesis (Church et al., 2002).
Interestingly, we found FACIT collagen, such as COL14A1 and COL21A1, was expressed at a higher level in ScASCs rather than IPFSCs. We also found that, despite collagen modifying enzymes detectable in both cells, IPFSCs exhibited significantly higher expression of LOX isoforms and tissue transglutaminase (Log2FC from IPFSCs to ScASCs for TGM2 was 3.68, FDR = 4.9E-06) at both mRNA and protein levels than ScASCs, along with fibrillar collagens COL5A1 and COL5A3, which participate in the formation of a fibrillar collagen network and the regulation of fibrillogenesis (Imamura et al., 2000; Malfait et al., 2005; Lincoln et al., 2006). These findings indicate that IPFSCs are more suitable adipose stem cells for chondrogenesis compared to donor-matched ScASCs. During chondrogenesis, the contents of collagen type II and of non-reducible collagen cross-links were positively linked with tensile biomechanical properties and the functional integrity of cartilage tissues (Williamson et al., 2003). LOX family enzyme activities catalyze the eventual enzymatic conversion that is needed for the generation of biosynthetic collagen cross-links. For example, knockdown studies demonstrated that LOXL2 expression is required for mouse ATDC5 chondroprogenitors’ differentiation toward chondrocytes through regulation of SNAIL and SOX9, two important transcription factors for chondrogenesis (Iftikhar et al., 2011). Interestingly, hypoxia increased LOX gene expression, which promoted pyridinoline cross-links in engineered cartilage (Makris et al., 2013).
Regarding the matrix components, we found that IPFSCs deposited more basement membrane proteins compared to ScASCs, including LAMA1, LAMA3, and HSPG2 at an mRNA level and LAMA5, LAMB1, LAMB2, LAMG1, NID1, and HSPG2 at a protein level. In contrast, NID2 and COL4A5 were significantly more highly expressed in ScASCs compared to donor-matched IPFSCs. Given the function of HSPG2 encoding perlecan as the “factotum” in cartilage (Melrose et al., 2008) and the role of laminins in cartilage regeneration (Sun et al., 2017), NID2 and COL4A5 expression might be associated with adipogenic differentiation, which remains to be elucidated.
Interestingly, matrix turnover enzymes were uniquely preferentially expressed in IPFSCs or ScASCs. For example, matrix metallopeptidase 13 (MMP13), known as collagenase 3, was found exclusively in IPFSCs. The enzyme’s initial role is in tissue remodeling, especially during embryonic development, but it is also highly active in some pathological processes such as osteoarthritis and cancer (Inada et al., 2004; Tardif et al., 2004). In osteoarthritis, MMP13 was highly expressed in chondrocytes and synovial cells responsible for the degradation of cartilage matrix (Inada et al., 2004; Davidson et al., 2006; van den Berg, 2011). However, little is known about MMP13 expression in IPFP, either in healthy donors or in the pathological process. In this study, qPCR data showed that MMP13 was expressed more than 1000-fold higher in IPFSCs than donor-matched ScASCs during cell condensation (day 0 pellet), in accordance with RNA-seq data of cell samples (Log2FC from IPFSCs to ScASCs in MMP13 was 7.45, FDR = 1.4E-16). Interestingly, after an 18-day chondrogenic induction, IPFSCs exhibited a comparable expression of MMP13 to donor-matched ScASCs. From the proteomics analysis, A disintegrin and metalloproteinase with thrombospondin motifs 5 (ADAMTS5) and ADAMTS1 were identified as significantly higher levels in IPFSCs versus a much higher level of ADAMTSL1 (ADAMTS like 1) in ScASCs. The protease profile is likely important for key processing events associated with the physiological role of IPFP in support of the remodeling of articular cartilage, which requires further investigation.
Integrin, a transmembrane receptor, facilitates cell-ECM adhesion. Interestingly, more integrins were advantageously expressed in IPFSCs including ITGA3, ITGA4, ITGA6, ITGA7, and ITGB8 compared to ITGA8, which was superiorly expressed in donor-matched ScASCs. Another transmembrane protein, aquaporin, called the “the plumbing system for cells”, was found to serve as a water channel to rapidly move water through cells. Chondrocytes from articular cartilage are unique due to their sensitivity to water transfer and ionic and osmotic adjustments from the extracellular environment and are responsible for the creation of synovial fluid. A recent report showed that the expression of AQP1 and AQP3 were upregulated during chondrogenic induction of human adipose stem cells, indicative of physiological modification of functionally mature chondrocytes to the local environment (Graziano et al., 2018). This finding was consistent with our data, in which all statistically significant AQP gene expression was advantageously found in IPFSCs rather than donor-matched ScASCs, not only AQP1 and AQP3 but also AQP4 and AQP11.
Furthermore, we found that important clusters associated with cartilage matrix were preferentially expressed in IPFSCs over donor-matched ScASCs. For instance, one of the clusters is glycosaminoglycan (GAG) metabolic and biosynthetic process, including N-Deacetylase And N-Sulfotransferase 1 (NDST1), Polypeptide N-Acetylgalactosaminyltransferase 5 (GALNT5), Biglycan (BGN), Carbohydrate Sulfotransferase 11 (CHST11), Dermatan-sulfate epimerase (DSE), Hyaluronidase-1 (HYAL1), and Inter-alpha-trypsin inhibitor heavy chain H3 (ITIH3), along with GAG binding function cluster, genes of CD44, ACAN, BGN, BMP4, Chemokine (C-C motif) ligand 2 (CCL2), Extracellular matrix protein 2 (ECM2), FGF10, FGFR2, Fibulin 7 (FBLN7), Follistatin-like 1 (FSTL1), Hyaluronan and proteoglycan link protein 1 (HAPLN1), Layilin (LAYN), LPL, procollagen C-endopeptidase enhancer 2 (PCOLCE2), Proline/arginine-rich end leucine-rich repeat protein (PRELP), Superoxide dismutase 3 (SOD3), THBS4, and Toll-like receoptor 2 (TLR2). The CD44 receptor transduces signals on chondrocytes, which is an important mediator of cell-matrix interactions, especially the homeostasis maintained by hyaluronan-CD44 interactions (Knudson and Loeser, 2002). Another crucial cluster is chondroitin sulfate proteoglycan and its metabolic process of which the advantageously expressed genes in IPFSCs include ACAN, BGN, Fibromodulin (FMOD), HSPG2, Syndecan 2 (SDC2), SDC4, IGFI, Cytokine-like 1 (CYTL1), CHST11, CHST15, Chondroitin sulfate N-acetylgalactosaminyltransferase 2 (CSGALNACT2), DSE, and Uronyl 2-sulfotransferase (UST). IGFI encodes insulin-like growth factor-I, which is the predominant anabolic growth factor and decreases matrix catabolism for articular cartilage (Jenniskens et al., 2006). Apparently, IPFSCs naturally express abundant genes involved in the chondrogenesis pathway by modulating cartilage development and cartilage ECM while ScASCs show no significant correlation with chondrogenesis.
Both ScASCs and donor-matched IPFSCs expressed plenty of genes related to lipid and fatty acid in their biosynthetic, catabolic, binding, and homeostasis processes. Among these differentially expressed genes, none of them showed a distinct tendency toward the brown adipose tissue due to either nondetectable expression of Uncoupling protein-1 (UCP1), Peroxisome proliferator-activated receptor-γ coactivator 1-α (PGC1A), and Forkhead box protein C2 (FOXC2), or comparable expression of Cell death inducing DFFA-like effector A (CIDEA) (Log2FC from IPFSCs to ScASCs was −1.45, FDR = 7.1E-01), a core set of brown adipose genes (Harms and Seale, 2013). Given the preferential expression of PPARG, a master regulator of adipogenesis (Kawai and Rosen, 2010), in ScASCs, most privileged associated genes favored adipogenesis, including but not limited to ATP Binding Cassette Subfamily G Member 1 (ABCG1) (Frisdal et al., 2015), Fibrillin-1 (FBN1) (Davis et al., 2016), NFATC4 (Yang et al., 2006), Oxysterol-binding protein-related protein 11 (OSBPL11) (Zhou et al., 2012), Paternally expressed gene-10 (PEG10) (Hishida et al., 2007), Salt inducible kinase 2 (SIK2) (Henriksson et al., 2015), and WNT4 (Christodoulides et al., 2009). Protein inhibitor of activated STAT 3 (PIAS3) did not favor adipogenesis (Deng et al., 2006). In contrast, excluding a few genes favoring adipogenesis, such as Docking protein 1 (DOK1) (Hosooka et al., 2008), ECM1 (Challa et al., 2015), and Leukemia inhibitor factor receptor alpha (LIFR) (Aubert et al., 1999), most related genes inhibited adiogenic differentiation in IPFSCs, including but not limited to Fatty acid binding protein 4 (FABP4) (Garin-Shkolnik et al., 2014), GREM2 (Wu et al., 2015), Jagged canonical NOTCH ligand 1 (JAG1) (Ugarte et al., 2009), LRP6 (Peröbner et al., 2012), SMOC2 (Nie and Sage, 2009), and Tribbles homolog 2 (TRIB2) (Naiki et al., 2007). The above findings indicate that, compared with ScASCs, donor-matched IPFSCs exhibit less potential to differentiate toward adipose tissue.
Our study did not find a significant difference in expression levels of osteogenic markers between ScASCs and donor-matched IPFSCs after induction. It has long been speculated that IPFSCs have a higher chondrogenic potential than other fat depots in the body (Sun et al., 2018), which might be due to the IPFP’s anatomic site that is closer to synovium and cartilage; it is regarded as a special form of fibro-adipose tissue (Macchi et al., 2016), or a continuation of synovium. IPFSCs may share some common characteristics with synovium-derived stem cells, a tissue-specific stem cell for chondrogenesis (Jones and Pei, 2012). A study comparing IPFSCs with near knee joint ScASCs from donor-matched osteoarthritis patients found that IPFSCs had a superior chondrogenic potential; ScASCs were superior in osteogenesis while both were similar in adipogenic capacity (Lopa et al., 2014). In this study with donor-matched normal adipose stem cells, we observed clear superiority of IPFSCs for chondrogenesis. However, we obtained divergent results on adipogenic and osteogenic potential. There were two possible explanations for the discrepancy; first, species differences between humans and rabbits can lead to considerable diversity, and secondly, Lopa et al. harvested IPFSCs from aged patients affected by osteoarthritis (Lopa et al., 2014) while we harvested from young healthy rabbits. The adult stem cell profile could be greatly affected by disease and aging (Li and Pei, 2012; Lynch and Pei, 2014).
Taken together, by donor-matched comparison and high throughput assay of transcriptome and proteomics, this study investigated the variance of depot-specific adult stem cells from adipose tissues. The findings might shed some light on the hypothesis that adipose stem cells have a depot-dependent lineage preference. Here, we have profiled RNA and protein levels to identify unique transcriptional programs, signaling components, and matrix environments that reveal novel insights into the higher chondrogenic potential of IPFSCs.
Data Availability Statement
The RNA-Seq datasets generated for this study can be found in the GEO, under the accession number GSE142626, https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE142626. The data that support the findings of this study are available from the corresponding author upon reasonable request.
Ethics Statement
The animal study was reviewed and approved by IACUC of West Virginia University, protocol number 1608003840.
Author Contributions
TW contributed to the conception and design, collection and/assembly of data, data analysis and interpretation, manuscript writing, and final approval of the manuscript. RH and MD contributed to the collection and/assembly of data, data analysis and interpretation, manuscript writing, and final approval of the manuscript. AI, GH, LZ, and KH contributed to the data analysis and interpretation, manuscript writing, and final approval of the manuscript. MP contributed to the conception and design, financial support, data analysis and interpretation, manuscript writing, and final approval of the manuscript.
Funding
This work was supported by Research Grants from the Musculoskeletal Transplant Foundation (MTF) and the National Institutes of Health (1R01AR067747-01A1) to MP. We also would like to acknowledge the WVU Flow Cytometry and Single Cell Core Facility and the grants that support the facility, TME CoBRE grant P20GM131322 and the WV CTSI grant GM104942. The work was also partially supported by the National Institute of General Medical Sciences Grant 5U54GM104942-04 to G.H. and P20 GM103434 to AI.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Suzanne Danley for editing the manuscript.
Footnotes
References
Alegre-Aguarón, E., Desportes, P., García-Álvarez, F., Castiella, T., Larrad, L., and Martínez-Lorenzo, M. J. (2012). Differences in surface marker expression and chondrogenic potential among various tissue-derived mesenchymal cells from elderly patients with osteoarthritis. Cells Tissues Organs 196, 231–240. doi: 10.1159/000334400
Aubert, J., Dessolin, S., Belmonte, N., Li, M., McKenzie, F. R., Staccini, L., et al. (1999). Leukemia inhibitory factor and its receptor promote adipocyte differentiation via the mitogen-activated protein kinase cascade. J. Biol. Chem. 274, 24965–24972. doi: 10.1074/jbc.274.35.24965
Bahamonde, M. E., and Lyons, K. M. (2001). BMP3: to be or not to be a BMP. J. Bone Joint Surg. Am. 83A(Pt 1), S56–S62.
Barrett, A. S., Wither, M. J., Hill, R. C., Dzieciatkowska, M., D’Alessandro, A., Reisz, J. A., et al. (2017). Hydroxylamine chemical digestion for insoluble extracellular matrix characterization. J. Proteome Res. 16, 4177–4184. doi: 10.1021/acs.jproteome.7b00527
Barry, F., Boynton, R. E., Liu, B., and Murphy, J. M. (2001). Chondrogenic differentiation of mesenchymal stem cells from bone marrow: differentiation-dependent gene expression of matrix components. Exp. Cell Res. 268, 189–200. doi: 10.1006/excr.2001.5278
Belotti, D., Capelli, C., Resovi, A., Introna, M., and Taraboletti, G. (2016). Thrombospondin-1 promotes mesenchymal stromal cell functions via TGFβ and in cooperation with PDGF. Matrix Biol. 55, 106–116. doi: 10.1016/j.matbio.2016.03.003
Boulet, A. M., and Capecchi, M. R. (2004). Multiple roles of Hoxa11 and Hoxd11 in the formation of the mammalian forelimb zeugopod. Development 131, 299–309. doi: 10.1242/dev.00936
Brown, C., McKee, C., Bakshi, S., Walker, K., Hakman, E., Halassy, S., et al. (2019). Mesenchymal stem cells: cell therapy and regeneration potential. J. Tissue Eng. Regen. Med. 13, 1738–1755. doi: 10.1002/term.2914
Challa, T. D., Straub, L. G., Balaz, M., Kiehlmann, E., Donze, O., Rudofsky, G., et al. (2015). Regulation of de novo adipocyte differentiation through cross talk between adipocytes and preadipocytes. Diabetes 64, 4075–4087. doi: 10.2337/db14-1932
Christodoulides, C., Lagathu, C., Sethi, J. K., and Vidal-Puig, A. (2009). Adipogenesis and WNT signalling. Trends Endocrinol. Metab. 20, 16–24. doi: 10.1016/j.tem.2008.09.002
Chu, D. T., Nguyen Thi Phuong, T., Tien, N. L. B., Tran, D. K., Minh, L. B., Thanh, V. V., et al. (2019). Adipose tissue stem cells for therapy: an update on the progress of isolation, culture, storage, and clinical application. J. Clin. Med. 8:E917. doi: 10.3390/jcm8070917
Church, V., Nohno, T., Linker, C., Marcelle, C., and Francis-West, P. (2002). Wnt regulation of chondrocyte differentiation. J. Cell Sci. 115(Pt 24), 4809–4818. doi: 10.1242/jcs.00152
Cormier, S. A., Mello, M. A., and Kappen, C. (2003). Normal proliferation and differentiation of Hoxc-8 transgenic chondrocytes in vitro. BMC Dev. Biol. 3:4. doi: 10.1186/1471-213x-3-4
Crisan, M., Yap, S., Casteilla, L., Chen, C. W., Corselli, M., Park, T. S., et al. (2008). A perivascular origin for mesenchymal stem cells in multiple human organs. Cell Stem Cell 3, 301–313. doi: 10.1016/j.stem.2008.07.003
Davidson, R. K., Waters, J. G., Kevorkian, L., Darrah, C., Cooper, A., Donell, S. T., et al. (2006). Expression profiling of metalloproteinases and their inhibitors in synovium and cartilage. Arthritis Res. Ther. 8:R124.
Davis, M. R., Arner, E., Duffy, C. R., De Sousa, P. A., Dahlman, I., Arner, P., et al. (2016). Expression of FBN1 during adipogenesis: relevance to the lipodystrophy phenotype in Marfan syndrome and related conditions. Mol. Genet. Metab. 119, 174–185. doi: 10.1016/j.ymgme.2016.06.009
Deng, J., Hua, K., Caveney, E. J., Takahashi, N., and Harp, J. B. (2006). Protein inhibitor of activated STAT3 inhibits adipogenic gene expression. Biochem. Biophys. Res. Commun. 339, 923–931. doi: 10.1016/j.bbrc.2005.10.217
Ewels, P., Magnusson, M., Lundin, S., and Käller, M. (2016). MultiQC: summarize analysis results for multiple tools and samples in a single report. Bioinformatics 32, 3047–3048. doi: 10.1093/bioinformatics/btw354
Fabregat, A., Jupe, S., Matthews, L., Sidiropoulos, K., Gillespie, M., and Garapati, P. (2018). The reactome pathway knowledgebase. Nucleic Acids Res. 46, D649–D655. doi: 10.1093/nar/gkx1132
Frisdal, E., Le Lay, S., Hooton, H., Poupel, L., Olivier, M., Alili, R., et al. (2015). Adipocyte ATP-binding cassette G1 promotes triglyceride storage, fat mass growth, and human obesity. Diabetes 64, 840–855. doi: 10.2337/db14-0245
Garin-Shkolnik, T., Rudich, A., Hotamisligil, G. S., and Rubinstein, M. (2014). FABP4 attenuates PPARgamma and adipogenesis and is inversely correlated with PPARgamma in adipose tissues. Diabetes 63, 9009–9011. doi: 10.2337/db13-0436
Gesta, S., Bezy, O., Mori, M. A., Macotela, Y., Lee, K. Y., and Kahn, C. R. (2011). Mesodermal developmental gene Tbx15 impairs adipocyte differentiation and mitochondrial respiration. Proc. Natl. Acad. Sci. U.S.A. 108, 2771–2776. doi: 10.1073/pnas.1019704108
González-Martín, M. C., Mallo, M., and Ros, M. A. (2014). Long bone development requires a threshold of Hox function. Dev. Biol. 392, 454–465. doi: 10.1016/j.ydbio.2014.06.004
Graziano, A. C. E., Avola, R., Pannuzzo, G., and Cardile, V. (2018). Aquaporin1 and 3 modification as a result of chondrogenic differentiation of human mesenchymal stem cell. J. Cell Physiol. 233, 2279–2291. doi: 10.1002/jcp.26100
Hagmann, S., Frank, S., Gotterbarm, T., Dreher, T., Eckstein, V., and Moradi, B. (2014). Fluorescence activated enrichment of CD146+ cells during expansion of human bone-marrow derived mesenchymal stromal cells augments proliferation and GAG/DNA content in chondrogenic media. BMC Musculoskelet. Disord. 15:322. doi: 10.1186/1471-2474-15-322
Haraguchi, R., Kitazawa, R., and Kitazawa, S. (2015). Epigenetic regulation of Tbx18 gene expression during endochondral bone formation. Cell Tissue Res. 359, 503–512. doi: 10.1007/s00441-014-2028-0
Harms, M., and Seale, P. (2013). Brown and beige fat: development, function and therapeutic potential. Nat. Med. 19, 1252–1263. doi: 10.1038/nm.3361
Henriksson, E., Säll, J., Gormand, A., Wasserstrom, S., Morrice, N. A., Fritzen, A. M., et al. (2015). SIK2 regulates CRTCs, HDAC4 and glucose uptake in adipocytes. J. Cell Sci. 128, 472–486. doi: 10.1242/jcs.153932
Hill, R. C., Calle, E. A., Dzieciatkowska, M., Niklason, L. E., and Hansen, K. C. (2015). Quantification of extracellular matrix proteins from a rat lung scaffold to provide a molecular readout for tissue engineering. Mol. Cell Proteomics 14, 961–973. doi: 10.1074/mcp.M114.045260
Hishida, T., Naito, K., Osada, S., Nishizuka, M., and Imagawa, M. (2007). peg10, an imprinted gene, plays a crucial role in adipocyte differentiation. FEBS Lett. 581, 4272–4278. doi: 10.1016/j.febslet.2007.07.074
Hosooka, T., Noguchi, T., Kotani, K., Nakamura, T., Sakaue, H., Inoue, H., et al. (2008). Dok1 mediates high-fat diet-induced adipocyte hypertrophy and obesity through modulation of PPAR-gamma phosphorylation. Nat. Med. 14, 188–193. doi: 10.1038/nm1706
Iftikhar, M., Hurtado, P., Bais, M. V., Wigner, N., Stephens, D. N., Gerstenfeld, L. C., et al. (2011). Lysyl oxidase-like-2 (LOXL2) is a major isoform in chondrocytes and is critically required for differentiation. J. Biol. Chem. 286, 909–918. doi: 10.1074/jbc.M110.155622
Ikeda, T., Kamekura, S., Mabuchi, A., Kou, I., Seki, S., Takato, T., et al. (2004). The combination of SOX5, SOX6, and SOX9 (the SOX trio) provides signals sufficient for induction of permanent cartilage. Arthritis Rheum. 50, 3561–3573. doi: 10.1002/art.20611
Imamura, Y., Scott, I. C., and Greenspan, D. S. (2000). The pro-alpha3(V) collagen chain. Complete primary structure, expression domains in adult and developing tissues, and comparison to the structures and expression domains of the other types V and XI procollagen chains. J. Biol. Chem. 275, 8749–8759. doi: 10.1074/jbc.275.12.8749
Inada, M., Wang, Y., Byrne, M. H., Rahman, M. U., Miyaura, C., López-Otín, C., et al. (2004). Critical roles for collagenase-3 (Mmp13) in development of growth plate cartilage and in endochondral ossification. Proc. Natl. Acad. Sci. U.S.A. 101, 17192–17197. doi: 10.1073/pnas.0407788101
Jenniskens, Y. M., Koevoet, W., de Bart, A. C., Weinans, H., Jahr, H., Verhaar, J. A., et al. (2006). Biochemical and functional modulation of the cartilage collagen network by IGF1. TGFbeta2 and FGF2. Osteoarthritis Cartilage 14, 1136–1146. doi: 10.1016/j.joca.2006.04.002
Jones, B., and Pei, M. (2012). Synovium-derived stem cells: a tissue-specific stem cell for cartilage tissue engineering and regeneration. Tissue Eng. Part B Rev. 18, 301–311. doi: 10.1089/ten.teb.2012.0002
Jung, J. C., and Tsonis, P. A. (1998). Role of 5’ HoxD genes in chondrogenesis in vitro. Int. J. Dev. Biol. 42, 609–615.
Kan, A., Ikeda, T., Fukai, A., Nakagawa, T., Nakamura, K., Chung, U. I., et al. (2013). SOX11 contributes to the regulation of GDF5 in joint maintenance. BMC Dev. Biol. 13:4. doi: 10.1186/1471-213X-13-4
Kawai, M., and Rosen, C. J. (2010). PPARgamma: a circadian transcription factor in adipogenesis and osteogenesis. Nat. Rev. Endocrinol. 6, 629–636. doi: 10.1038/nrendo.2010.155
Knudson, W., and Loeser, R. F. (2002). CD44 and integrin matrix receptors participate in cartilage homeostasis. Cell Mol. Life Sci. 59, 36–44. doi: 10.1007/s00018-002-8403-0
Levi, B., James, A. W., Glotzbach, J. P., Wan, D. C., Commons, G. W., and Longaker, M. T. (2010). Depot-specific variation in the osteogenic and adipogenic potential of human adipose-derived stromal cells. Plast. Reconstr. Surg. 126, 822–834. doi: 10.1097/PRS.0b013e3181e5f892
Li, J., and Pei, M. (2012). Cell senescence: a challenge in cartilage engineering and regeneration. Tissue Eng. Part B Rev. 18, 270–287. doi: 10.1089/ten.TEB.2011.0583
Li, J., and Pei, M. (2018). A protocol to prepare decellularized stem cell matrix for rejuvenation of cell expansion and cartilage regeneration. Methods Mol. Biol. 1577, 147–154. doi: 10.1007/7651_2017_27
Liao, Y., Smyth, G. K., and Shi, W. (2013). The Subread aligner: fast, accurate and scalable read mapping by seed-and-vote. Nucleic Acids Res. 41:e108. doi: 10.1093/nar/gkt214
Liao, Y., Smyth, G. K., and Shi, W. (2019). The R package Rsubread is easier, faster, cheaper and better for alignment and quantification of RNA sequencing reads. Nucleic Acids Res. 47:e47. doi: 10.1093/nar/gkz114
Lincoln, J., Florer, J. B., Deutsch, G. H., Wenstrup, R. J., and Yutzey, K. E. (2006). ColVa1 and ColXIa1 are required for myocardial morphogenesis and heart valve development. Dev. Dyn. 235, 3295–3305. doi: 10.1002/dvdy.20980
Liu, R., Li, N., Lin, Y., Wang, M., Peng, Y., Lewi, K., et al. (2016). Glucagon like peptide-1 promotes adipocyte differentiation via the Wnt4 mediated sequestering of beta-catenin. PLoS One 11:e0160212. doi: 10.1371/journal.pone.0160212
Lopa, S., Colombini, A., Stanco, D., de Girolamo, L., Sansone, V., and Moretti, M. (2014). Donor-matched mesenchymal stem cells from knee infrapatellar and subcutaneous adipose tissue of osteoarthritic donors display differential chondrogenic and osteogenic commitment. Eur. Cell Mater. 27, 298–311. doi: 10.22203/ecm.v027a21
Lynch, K., and Pei, M. (2014). Age associated communication between cells and matrix: a potential impact on stem cell-based tissue regeneration strategies. Organogenesis 10, 289–298. doi: 10.4161/15476278.2014.970089
Macchi, V., Porzionato, A., Sarasin, G., Petrelli, L., Guidolin, D., Rossato, M., et al. (2016). The infrapatellar adipose body: a histotopographic study. Cells Tissues Organs 201, 220–231. doi: 10.1159/000442876
Makris, E. A., Hu, J. C., and Athanasiou, K. A. (2013). Hypoxia-induced collagen crosslinking as a mechanism for enhancing mechanical properties of engineered articular cartilage. Osteoarthritis Cartilage 21, 634–641. doi: 10.1016/j.joca.2013.01.007
Malfait, F., Coucke, P., Symoens, S., Loeys, B., Nuytinck, L., and De Paepe, A. (2005). The molecular basis of classic Ehlers-Danlos syndrome: a comprehensive study of biochemical and molecular findings in 48 unrelated patients. Hum. Mutat. 25, 28–37. doi: 10.1002/humu.20107
McCarthy, D. J., Chen, Y., and Smyth, G. K. (2012). Differential expression analysis of multifactor RNA-Seq experiments with respect to biological variation. Nucleic Acids Res. 40, 4288–4297. doi: 10.1093/nar/gks042
Melrose, J., Hayes, A. J., Whitelock, J. M., and Little, C. B. (2008). Perlecan, the “jack of all trades” proteoglycan of cartilaginous weight-bearing connective tissues. Bioessays 20, 457–469. doi: 10.1002/bies.20748
Mueller, M. B., and Tuan, R. S. (2008). Functional characterization of hypertrophy in chondrogenesis of human mesenchymal stem cells. Arthritis Rheum. 58, 1377–1388. doi: 10.1002/art.23370
Naiki, T., Saijou, E., Miyaoka, Y., Sekine, K., and Miyajima, A. (2007). TRB2, a mouse Tribbles ortholog, suppresses adipocyte differentiation by inhibiting AKT and C/EBPbeta. J. Biol. Chem. 282, 24075–24082. doi: 10.1074/jbc.m701409200
Nie, J., and Sage, E. H. (2009). SPARC inhibits adipogenesis by its enhancement of beta-catenin signaling. J. Biol. Chem. 284, 1279–1290. doi: 10.1074/jbc.M808285200
Peröbner, I., Karow, M., Jochum, M., and Neth, P. (2012). LRP6 mediates Wnt/β-catenin signaling and regulates adipogenic differentiation in human mesenchymal stem cells. Int. J. Biochem. Cell Biol. 44, 1970–1982. doi: 10.1016/j.biocel.2012.07.025
Pizzute, T., Lynch, K., and Pei, M. (2015). Impact of tissue-specific stem cells on lineage specific differentiation: a focus on musculoskeletal system. Stem Cell Rev. Rep. 11, 119–132. doi: 10.1007/s12015-014-9546-8
Pizzute, T., Zhang, Y., He, F., and Pei, M. (2016). Ascorbate-dependent impact on cell-derived matrix in modulation of stiffness and rejuvenation of infrapatellar fat derived stem cells toward chondrogenesis. Biomed. Mater. 11:045009. doi: 10.1088/1748-6041/11/4/045009
Reddi, A. H. (2001). Bone morphogenetic proteins: from basic science to clinical applications. J. Bone Joint Surg. Am. 83A Suppl 1(Pt 1), S1–S6.
Seifert, A., Werheid, D. F., Knapp, S. M., and Tobiasch, E. (2015). Role of Hox genes in stem cell differentiation. World J. Stem Cells 7, 583–595. doi: 10.4252/wjsc.v7.i3.583
Shintani, N., Siebenrock, K. A., and Hunziker, E. B. (2013). TGF-ß1 enhances the BMP-2-induced chondrogenesis of bovine synovial explants and arrests downstream differentiation at an early stage of hypertrophy. PLoS One 8:e53086. doi: 10.1371/journal.pone.0053086
Singh, M. K., Petry, M., Haenig, B., Lescher, B., Leitges, M., and Kispert, A. (2005). The T-box transcription factor Tbx15 is required for skeletal development. Mech. Dev. 122, 131–144. doi: 10.1016/j.mod.2004.10.011
Steinert, A., Weber, M., Dimmler, A., Julius, C., Schütze, N., Nöth, U., et al. (2003). Chondrogenic differentiation of mesenchymal progenitor cells encapsulated in ultrahigh-viscosity alginate. J. Orthop. Res. 21, 1090–1097. doi: 10.1016/s0736-0266(03)00100-1
Su, X., Zuo, W., Wu, Z., Chen, J., Wu, N., Ma, P., et al. (2015). CD146 as a new marker for an increased chondroprogenitor cell sub-population in the later stages of osteoarthritis. J. Orthop. Res. 33, 84–91. doi: 10.1002/jor.22731
Subramanian, A., and Schilling, T. F. (2015). Tendon development and musculoskeletal assembly: emerging roles for the extracellular matrix. Development 142, 4191–4204. doi: 10.1242/dev.114777
Sun, Y., Chen, S., and Pei, M. (2018). Comparative advantages of infrapatellar fat pad: an emerging stem cell source for regenerative medicine. Rheumatology 57, 2072–2086. doi: 10.1093/rheumatology/kex487
Sun, Y., Wang, T. L., Toh, W. S., and Pei, M. (2017). The role of laminins in cartilaginous tissues: from development to regeneration. Eur. Cell Mater. 34, 40–54. doi: 10.22203/eCM.v034a03
Takeuchi, J. K., Koshiba-Takeuchi, K., Suzuki, T., Kamimura, M., Ogura, K., and Ogura, T. (2003). Tbx5 and Tbx4 trigger limb initiation through activation of the Wnt/Fgf signaling cascade. Development 130, 2729–2739. doi: 10.1242/dev.00474
Tanaka, M., and Tickle, C. (2004). Tbx18 and boundary formation in chick somite and wing development. Dev. Biol. 268, 470–480. doi: 10.1016/j.ydbio.2003.12.036
Tardif, G., Reboul, P., Pelletier, J. P., and Martel-Pelletier, J. (2004). Ten years in the life of an enzyme: the story of the human MMP-13 (collagenase-3). Mod. Rheumatol. 14, 197–204. doi: 10.3109/s10165-004-0292-7
Tuli, R., Tuli, S., Nandi, S., Huang, X., Manner, P. A., Hozack, W. J., et al. (2003). Transforming growth factor-beta-mediated chondrogenesis of human mesenchymal progenitor cells involves N-cadherin and mitogen-activated protein kinase and Wnt signaling cross-talk. J. Biol. Chem. 278, 41227–41236. doi: 10.1074/jbc.m305312200
Ugarte, F., Ryser, M., Thieme, S., Fierro, F. A., Navratiel, K., Bornhäuser, M., et al. (2009). Notch signaling enhances osteogenic differentiation while inhibiting adipogenesis in primary human bone marrow stromal cells. Exp. Hematol. 37, 867–875. doi: 10.1016/j.exphem.2009.03.007
van den Berg, W. B. (2011). Osteoarthritis year 2010 in review: pathomechanisms. Osteoarthritis Cartilage 19, 338–341. doi: 10.1016/j.joca.2011.01.022
Wang, R. N., Green, J., Wang, Z., Deng, Y., Qiao, M., Peabody, M., et al. (2014). Bone Morphogenetic Protein (BMP) signaling in development and human diseases. Genes Dis. 1, 87–105.
Wight, T. N., Kinsella, M. G., Evanko, S. P., Potter-Perigo, S., and Merrilees, M. J. (2014). Versican and the regulation of cell phenotype in disease. Biochim. Biophys. Acta 1840, 2441–2451. doi: 10.1016/j.bbagen.2013.12.028
Williamson, A. K., Chen, A. C., Masuda, K., Thonar, E. J., and Sah, R. L. (2003). Tensile mechanical properties of bovine articular cartilage: variations with growth and relationships to collagen network components. J. Orthop. Res. 21, 872–880. doi: 10.1016/s0736-0266(03)00030-5
Wiśniewski, J. R., Zougman, A., Nagaraj, N., and Mann, M. (2009). Universal sample preparation method for proteome analysis. Nat. Methods 6, 359–362. doi: 10.1038/nmeth.1322
Wu, J., Boström, P., Sparks, L. M., Ye, L., Choi, J. H., Giang, A. H., et al. (2012). Beige adipocytes are a distinct type of thermogenic fat cell in mouse and human. Cell 150, 366–376. doi: 10.1016/j.cell.2012.05.016
Wu, Q., Tang, S. G., and Yuan, Z. M. (2015). Gremlin 2 inhibits adipocyte differentiation through activation of Wnt/β-catenin signaling. Mol. Med. Rep. 12, 5891–5896. doi: 10.3892/mmr.2015.4117
Xia, J., and Wishart, D. S. (2016). Using MetaboAnalyst 3.0 for comprehensive metabolomics data analysis. Curr. Protoc. Bioinformatics 55, 14.10.1–14.10.91.
Yang, T. T., Suk, H. Y., Yang, X., Olabisi, O., Yu, R. Y., Durand, J., et al. (2006). Role of transcription factor NFAT in glucose and insulin homeostasis. Mol. Cell Biol. 26, 7372–7387. doi: 10.1128/mcb.00580-06
Yueh, Y. G., Gardner, D. P., and Kappen, C. (1998). Evidence for regulation of cartilage differentiation by the homeobox gene Hoxc-8. Proc. Natl. Acad. Sci. U.S.A. 95, 9956–9961. doi: 10.1073/pnas.95.17.9956
Zerbino, D. R., Achuthan, P., Akanni, W., Amode, M. R., Barrell, D., Bhai, J., et al. (2018). Ensembl 2018. Nucleic Acids Res. 46, D754–D761. doi: 10.1093/nar/gkx1098
Keywords: adipogenesis, adipose stem cell, chondrogenesis, infrapatellar fat pad, osteogenesis, subcutaneous adipose tissue
Citation: Wang T, Hill RC, Dzieciatkowska M, Zhu L, Infante AM, Hu G, Hansen KC and Pei M (2020) Site-Dependent Lineage Preference of Adipose Stem Cells. Front. Cell Dev. Biol. 8:237. doi: 10.3389/fcell.2020.00237
Received: 13 December 2019; Accepted: 20 March 2020;
Published: 15 April 2020.
Edited by:
Pavel Makarevich, Lomonosov Moscow State University, RussiaReviewed by:
Egor Plotnikov, Lomonosov Moscow State University, RussiaElena Andreeva, Russian Academy of Sciences, Russia
Dmitry Traktuev, University of Florida, United States
Copyright © 2020 Wang, Hill, Dzieciatkowska, Zhu, Infante, Hu, Hansen and Pei. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ming Pei, bXBlaUBoc2Mud3Z1LmVkdQ==
 Tingliang Wang
Tingliang Wang Ryan C. Hill3
Ryan C. Hill3 Aniello M. Infante
Aniello M. Infante Gangqing Hu
Gangqing Hu Kirk C. Hansen
Kirk C. Hansen Ming Pei
Ming Pei