- 1Women's Hospital, School of Medicine, Zhejiang University, Hangzhou, China
- 2State Key Laboratory for Diagnosis and Treatment of Infectious Diseases, Collaborative Innovation Center for Diagnosis and Treatment of Infectious Diseases, The First Affiliated Hospital, Zhejiang University School of Medicine, Hangzhou, China
- 3School of Medicine, Zhejiang University Hangzhou, Hangzhou, China
- 4Department of Neurosurgery, The Second Affiliated Hospital, Zhejiang University School of Medicine, Hangzhou, China
MicroRNAs (miRNAs) and long non-coding RNAs (lncRNAs) are components of many signaling pathways associated with tumor aggressiveness and cancer metastasis. Some lncRNAs are classified as competitive endogenous RNAs (ceRNAs) that bind to specific miRNAs to prevent interaction with target mRNAs. Studies have shown that the hepatocyte growth factor/mesenchymal-epithelial transition factor (HGF/c-Met) pathway is involved in physiological and pathological processes such as cell growth, angiogenesis, and embryogenesis. Overexpression of c-Met can lead to sustained activation of downstream signals, resulting in carcinogenesis, metastasis, and resistance to targeted therapies. In this review, we evaluated the effects of anti-oncogenic and oncogenic non-coding RNAs (ncRNAs) on c-Met, and the interactions among lncRNAs, miRNAs, and c-Met in cancer using clinical and tissue chromatin immunoprecipition (ChIP) analysis data. We summarized current knowledge of the mechanisms and effects of the lncRNAs/miR-34a/c-Met axis in various tumor types, and evaluated the potential therapeutic value of lncRNAs and/or miRNAs targeted to c-Met on drug-resistance. Furthermore, we discussed the functions of lncRNAs and miRNAs in c-Met-related carcinogenesis and potential therapeutic strategies.
Introduction
Mesenchymal-epithelial transition factor (c-Met) is a receptor tyrosine kinase that belongs to the MET (MNNG HOS transforming gene) family (Salgia, 2017). C-Met is encoded by the human MET gene, located on chromosome 7 (bands q21–q31) (Liu, 1998). As a kinase receptor, c-Met is a 190 kDa glycoprotein heterodimer localized on the surface of epithelial and endothelial cells, and it has two binding sites for its specific ligand, HGF/SF (hepatocyte growth factor or scatter factor). The first binding site contains the IPT3 and IPT4 domains, which have high affinity for the N domain of HGF. The second binding site is the SEMA domain, which has low affinity for the SPH domain of activated HGF (Stamos et al., 2004; Basilico et al., 2008). Binding of HGF can initiate stable c-Met homodimerization, which is mediated by adaptor proteins (GAB1 and GRB2), and then activates a number of key signaling pathways, including PI3K/Akt, Erk1/2, JAK/STAT, Src, Ras/MAPK, and Wnt/β-catenin (Imura et al., 2016; Pilotto et al., 2017), to induce cell proliferation, migration, invasion, and other biological effects (Ponzetto et al., 1994; Johnson and Lapadat, 2002) (Figure 1A). The HGF/c-Met axis is involved in biological and pathological processes such as embryogenesis, wound healing, and hepatic renal and epidermis regeneration (Parikh et al., 2014). The oncogenicity of c-Met is rarely caused by genetic alteration, and is more often due to upregulation of the wild-type gene (Trusolino et al., 2010; Gherardi et al., 2012). Amplification of c-Met was detected in patients with advanced solid cancers (Jardim et al., 2014). Furthermore, mutations, overexpression, or amplification of the MET gene in some tumor types resulted in aberrant HGF/c-Met axis activity, which induced cell motility and proliferation, promoted tumor development, and led to resistance to radiotherapy and targeted drug therapy in multiple cancers (Minuti et al., 2012; Barrow-Mcgee et al., 2016; Bahrami et al., 2017). Clinical trials of drug monotherapies targeted to c-Met have shown promising outcomes against multiple cancer types (Spigel et al., 2013; Solomon et al., 2014; Kogita et al., 2015). However, these medications cause significant side effects. Treatments that target the HGF/c-Met axis require further development.
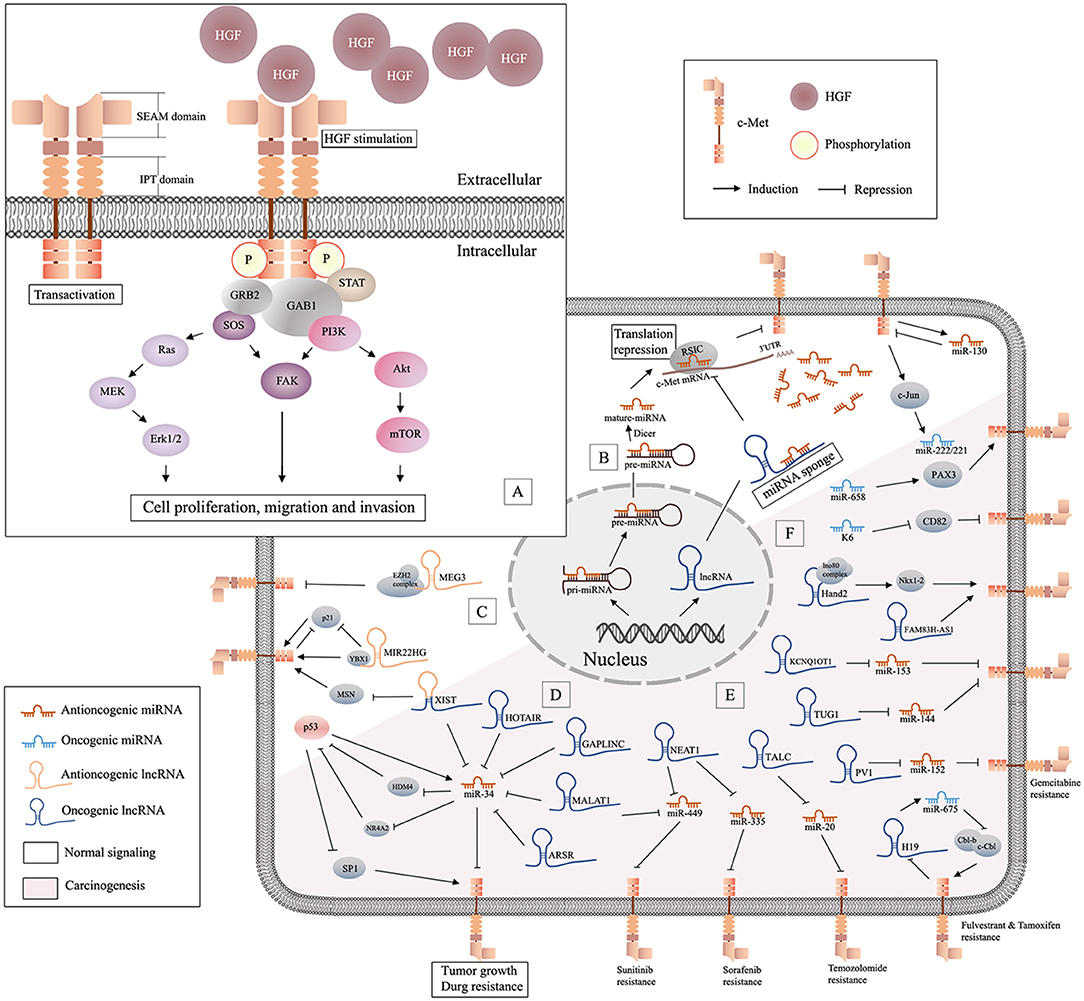
Figure 1. Schematic of HGF/c-Met signaling and lncRNA/miRNA/c-Met interaction. (A) Binding of HGF initiates stable c-Met homodimerization, which activates various downstream signaling pathways; (B) miRNAs are generated from the nucleus and can be decoyed by specific lncRNAs in cytoplasm, and antioncogenic miRNAs may suppress tumor progression by repressing the translation of c-Met mRNA; (C), illustration of antioncogenic lncRNAs (MEG3 and MIR22HG) in the regulation of c-Met; (D) the graphical representation of p53/miR-34 and lncRNA/miR-34/c-Met pathway; (E) oncogenic lncRNAs/miRNAs/c-Met in drug resistance; (F) oncogenic miRNAs in c-Met related carcinogenesis.
Early studies have proved that besides proteins, there are lots of unique RNAs encoded by the genome are functional. Subsequent researches discovered a variety of non-coding RNAs (ncRNAs), including microRNAs (miRNAs) and long non-coding RNAs (lncRNAs) (Bejerano et al., 2004; Johnsson et al., 2014). Non-coding RNAs (ncRNAs) are involved in many signaling pathways associated with tumor aggressiveness and metastasis. MiRNAs are 20-22 nucleotide (nt) non-coding, highly conserved RNA molecules present in all human cells. MiRNAs can regulate 20–30% of all transcripts (Krol et al., 2010). The effect of specific miRNAs can be found in normal and cancer tissues, and in different cancer subtypes (Iorio and Croce, 2012). In the last 20 years, the role of miRNAs in oncogenesis has received increased attention. MiRNAs are encoded and transcribed as initial miRNA transcripts (pri-miRNAs), and processed to the precursor miRNAs (pre-miRNA) that harbor a stem-loop structure in nucleus. Then pre-miRNAs are processed into dsRNAs by RNase III enzyme DICER1, and merged into RNA-induced silencing complex (RISC) in cytoplasm. Only one strand of the dsRNA is preserved in RISC as miRNAs, while the other undergoes fast degradation (De Los Santos et al., 2019). MiRNAs can downregulate specific genes at the post-transcriptional level by interaction with the 3' UTR) of target messenger RNA (mRNA). They function in different ways via sequence complementarity: imperfect base pair match results in translational repression or mRNA cleavage, while perfect pairing leads to deadenylation (Ruvkun, 2001) (Figure 1B). Studies have suggested that miRNAs may be potential diagnostic and prognostic cancer biomarkers.
NcRNAs >200 nt in length are referred to as lncRNAs (Quinn and Chang, 2016). LncRNAs regulate various cellular processes including chromatin, transcriptional and post-transcriptional modification, and scaffolding. LncRNAs are ubiquitous in subnuclear domains and in the cytoplasm. MiRNAs regulate mRNA expression through degradation or silencing of mRNAs, while lncRNAs activate and repress genes via diverse mechanisms at the transcriptional or post-translational levels. MiRNAs and lncRNAs can interact to modulate the transcriptome (Yoon et al., 2014). Prevalent mechanism of action of lncRNAs in cancer is as miRNA decoys. LncRNAs can act as competitive endogenous RNAs (ceRNAs), decoys, or sponges that bind to specific miRNAs to prevent interactions with target mRNAs (Figure 1B). Some lncRNAs act as precursors of miRNAs to repress the downstream gene. Also, there are other lncRNAs can modulate mRNA through the complementary sequences (Salmena et al., 2011). Although lncRNAs are relatively stable, they are dysregulated in many diseases (Du et al., 2013). Increasing numbers of studies have focused on the functions of miRNAs and lncRNAs in homeostasis and maintenance of cell identity, pathogenesis, and carcinogenesis. Studies have shown that lncRNAs and miRNAs may act as independent master regulators of carcinogenesis, and interactions among lncRNAs, miRNAs, and specific mRNAs may be important in tumor-related processes.
A number of studies have correlated aberrant expression of lncRNAs, miRNAs, and c-Met with various cancers. In this review, we summarized the latest studies of the interplay between ncRNA (especially miRNAs and lncRNA) and the HGF/c-Met axis. In addition, we showed through this literature review that miRNAs and lncRNAs are critical regulators of c-Met in multiple cancers. Furthermore, the lncRNAs/miRNAs/c-Met axis may have potential as a target for treatment of cancer.
The miRNAs/c-Met Axis in Cancer
Studies have shown that c-Met is a target gene of multiple miRNAs (Tables 1, 2) that play critical roles in controlling HGF/c-Met activity. This section summarizes recent studies of miRNAs that regulating c-Met.
Antioncogenic miRNAs Target c-Met
miRNA-34 Family and c-Met
The miR-34 family, which includes miR-34a, miR-34b, and miR-34c, has been shown to inhibit cell cycle progression, epithelial-mesenchymal transition (EMT), metastasis, and stemness, and was shown to promote apoptosis, which resulted in suppression of tumor growth and carcinogenesis (Rokavec et al., 2014). All members of the miR-34 family negatively regulate c-Met (Figure 1D). Some studies have shown that miR-34 may be the primary miRNA regulator of c-Met. Mature miR-34, which lacks a 5'-phosphate, is inactive in cells. Phosphorylation of miR-34 resulting from DNA damage leads to activation of miR-34 (Salzman et al., 2016). MiR-34a and miR-34b levels are downregulated in non-small cell lung cancer (NSCLC) tissue compared to those in paired normal tissue. In addition, decreased levels of miR-34a have been associated with a higher risk of relapse, and lower levels of miR-34b expression have been associated with a higher frequency of lymph node metastasis (Gallardo et al., 2009). The miR-34 family inhibits the expression of c-Met, which is part of the p53 tumor suppressor network (Hwang et al., 2011). Wild p53 negatively regulates c-Met expression by transactivation of miR-34 or inhibition of SP1 binding to MET promoter (Hwang et al., 2011). On the other hand, HDM4, a potent inhibitor of p53, is repressed by miR-34 directly. Therefore, miRNA-34 and p53 form a positive feedback loop, resulting in inhibition of c-Met (Migliore and Giordano, 2009). Nuclear receptor subfamily 4 group A member 2 (NR4A2) is an orphan nuclear receptor that is upregulated in cancer and is involved in malignant biological properties. A study showed that overexpression of miR-34 or activation of p53 inhibited the expression of NR4A2. On the other hand, NR4A2 can suppress p53 and transcriptional targets, including miR-34, which suggested another feedback mechanism of p53/miR-34 (Hwang et al., 2011). The levels of miR-34a are significantly lower in hepatocellular carcinoma (HCC) than those in non-tumorous tissues, and are further reduced in HCC with metastasis or portal vein tumor embolus. Studies have shown that miR-34a suppressed cell growth, migration, and invasion, and increased cellular apoptosis and caspase activity by targeting c-Met in HCC cells (Dang et al., 2013). In addition, miR-34a downregulated Snail directly or through inhibition of HGF/c-Met, which promoted EMT in gastric cancer (Liu Y. W. et al., 2015). Furthermore, miR-34a attenuated proliferation and invasion in gastric cancer (Wei et al., 2015) and colorectal cancer (Luo et al., 2018). MiR-34a might also suppress brain tumors by targeting c-Met and Notch, which resulted in inhibition of cell proliferation, reduced cell survival, and decreased cell invasion of glioma and medulloblastoma cells, but not astrocytes (Li et al., 2009). A stable and efficient cationic carrier (dendritic polyglycerolamine) of mature miR-34a has been shown to inhibit cell cycle progression, proliferation, and migration of glioblastoma cells through modulation of c-Met (Ofek et al., 2016). Another study showed that overexpression of miR-34a suppressed tumor growth and metastasis of osteosarcoma (OS), possibly through downregulation of c-Met (Yan et al., 2012). Bioengineered miR-34a has showed to decrease the expression of c-Met and inhibited OS cell growth and invasion by inducing apoptosis and cell cycle arrest. This study confirmed the effectiveness and safety of bioengineered miR-34a using an orthotopic OS xenograft tumor mouse model (Zhao et al., 2016). Another study showed an inverse relationship between the expression of miR-34a-5p and c-Met in bone metastasis, but not in non-metastatic or metastatic breast carcinomas (Maroni et al., 2017). The expression level of miR-34a in triple-negative (MDA-MB-231) and HER2-overexpressing (SK-BR-3) cells was lower than that in normal breast cells (MCF-10A). MiR-34a has been shown to target c-Met and AXL in triple-negative breast cancer (Hajalirezay Yazdi et al., 2018). In addition, miR-34 has been increasingly studied because of its role in tumor growth, metastasis (Maroni et al., 2017), and drug resistance (Ghandadi and Sahebkar, 2016). Studies have also focused on miR-34a as a therapeutic agent and prognostic biomarker (Chen et al., 2017a).
miR-206 and c-Met
Recent evidence suggests that miR-206 inhibited HGF-induced EMT and angiogenesis in NSCLC via the c-Met/PI3k/Akt/mTOR pathway (Chen et al., 2016b). In other studies, miRNA-206, an inhibitor of c-Met and Bcl-2, was shown to promote apoptosis and inhibit tumor cell proliferation, migration, invasion, and colony formation in NSCLC (Sun et al., 2015a). MiRNA-206 has been shown to regulate cisplatin resistance and EMT in human lung adenocarcinoma cells by targeting c-Met (Chen et al., 2016a). In addition, miR-206 has been shown to sensitize HGF-induced gefitinib-resistant human lung cancer cells through inhibition of c-Met signaling and the EMT (Jiao et al., 2018). MiR-206 also has been shown to prevent the pathogenesis of hepatocellular carcinoma by modulating the expression of the MET proto-oncogene and cyclin-dependent kinase 6 in mice (Wu et al., 2017). Another study showed that miR-206 inhibited hepatocellular carcinoma cell proliferation and migration, but promoted apoptosis, by modulating the expression of c-Met (Wang et al., 2019). Furthermore, miR-206 expression was downregulated in GC cells, particularly in metastatic lesions, and loss of miR-206 promoted gastric cancer metastasis through activation of the PAX3/c-Met pathway (Zhang et al., 2015). Similarly, miR-206 was also significantly downregulated in CRC tissues, and a series of loss-of-function and gain-of-function assays were performed to demonstrate that miR-206 suppressed CRC cell proliferation and invasion by targeting FMNL2 and c-Met (Ren et al., 2016).
miR-1 and c-Met
MiR-1, which is abundant in cardiac and smooth muscles, is expressed in the lung and is downregulated in primary lung cancer tissues and cell lines. A previous study has reported that exogenous miR-1 significantly reduced the expression of c-Met, and enhanced sensitivity to doxorubicin-induced apoptosis (Nasser et al., 2008). The seed sequences of miR-1 and miR-206 are identical, and both inhibited c-Met and the downstream Akt and Erk pathways, and HGF-induced EMT, and increased gefitinib sensitivity in lung cancers (Koshizuka et al., 2017; Jiao et al., 2018). Downregulation of miR-1 and upregulation of c-Met have also been reported in gastric, prostate, cervical, and ovarian cancers (Han et al., 2015; Qu et al., 2017; Cheng et al., 2019; Gao et al., 2019). MiR-1 directly inhibits the expression of c-Met/Akt/mTOR and suppresses cell proliferation, migration, and EMT.
Other Antioncogenic miRNAs and c-Met
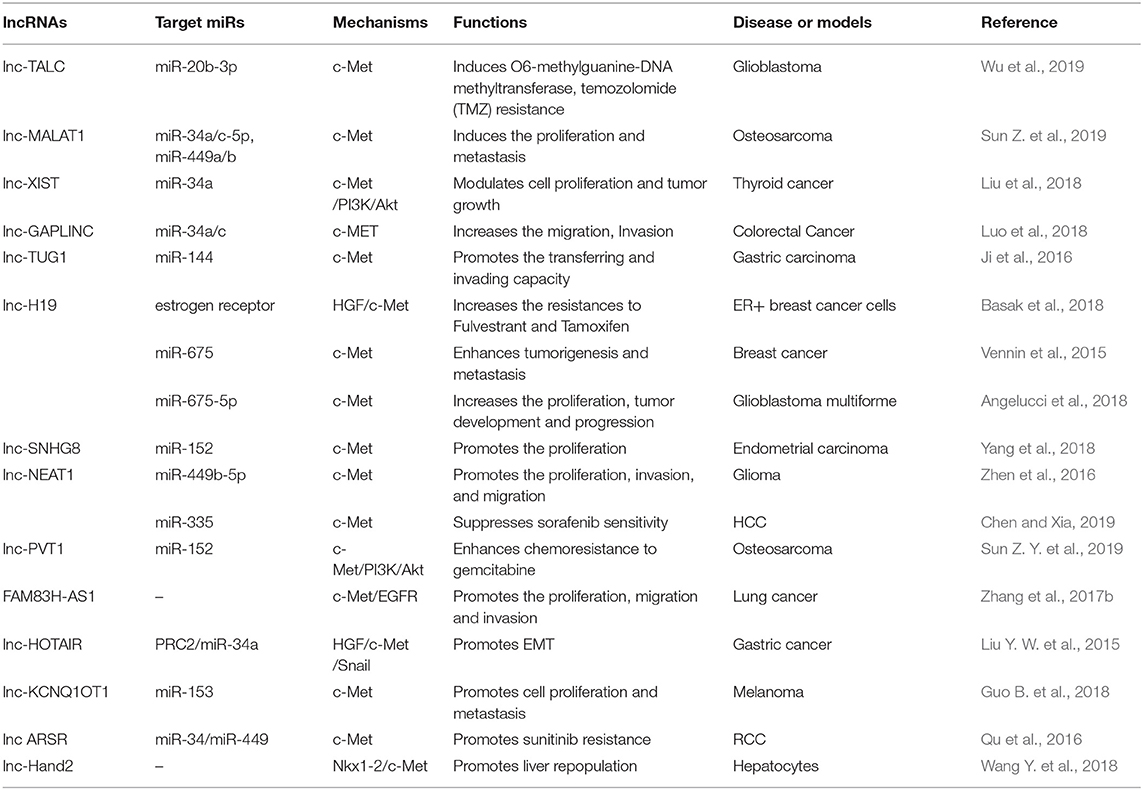
Most miRNAs in c-Met signaling are antioncogenic. Besides above, there are antioncogenic miRNAs include miR-410, -598, -182, and -144-3p in nervous system (Chen et al., 2012; Kouri et al., 2015; Lan et al., 2015; Wang N. et al., 2018); miR-140-5p in retinoblastoma (Liao et al., 2018) and miR-144 in uveal melanoma (Sun et al., 2015c); miR-365-3p, -152, -23b, -27b, and -143 in oral squamous cell carcinoma (OSCC) (Xu et al., 2015; Fukumoto et al., 2016; Li et al., 2018a; Huang et al., 2019). MiR-449b and -3666 are downregulated in thyroid cancer (Chen et al., 2015a; Wang et al., 2016); and miR-133b, -340, -335, and -185 are inhibit tumor progression in breast cancer (Wu et al., 2011; Fu et al., 2014; Gao et al., 2015; Wang Q. Y. et al., 2018). On account of the attention, a lot of miRNAs are found deficiency in lung cancer, including miR-200a, 168-198, -203, -182, -27b, -19a, -329, -16, -128, -31, -335, -1026, -139-5p, -105-3p338-3p, and miR-7515 in lung cancer (Kim et al., 2008; Lee et al., 2013; Wan et al., 2014; Sun et al., 2015b, 2016; Zhen et al., 2015; Zhu et al., 2015, 2018b; Hou et al., 2016; Jiang et al., 2016a; Cao et al., 2017; Pazzaglia et al., 2017; Zhou et al., 2017; Andriani et al., 2018; Li et al., 2018b; Zhang et al., 2018; Du et al., 2019). The digestive system cancer are also found absence of some miRNAs, including miR-449c, -26a/b, -16, and -144 in gastric cancer (Huang et al., 2015; Liu J. et al., 2015; Wu Z. et al., 2015; Li et al., 2016b; Zhang et al., 2017a); miR-198, -449a, -181-5p, and -122 in HCC (Tan et al., 2011; Korhan et al., 2014; Cheng et al., 2018); miR-320 in cholangiocarcinoma (Zhu et al., 2018a); miR-335-5p in pancreatic cancer (Cao et al., 2016); miR-146a, -141, and -137 in colorectal cancer (Chen et al., 2017b; Long et al., 2017; Bleau et al., 2018). While miR-32-5p, -23b, -27b, -323-3p and -433 are reported inhibit c-Met signaling in urinary cancer (Hu et al., 2013; Chiyomaru et al., 2015; Xu et al., 2016; Li et al., 2017a; Wang M. et al., 2018). Moreover, miR-148-3p, -119a-5p, -485, -454-3p, -23b, -138, -130 and -195 are relevant to the malignancy of reproductive system (Mariani et al., 2014; Campos-Viguri et al., 2015; Kinose et al., 2015; Wu J. et al., 2015; Li et al., 2016a; Nara et al., 2016; Yeung et al., 2017; Guo Y. et al., 2018; Wang S. et al., 2018; Wang W. et al., 2018); and miR-198, -206, -454, and -489-3p are relevant to the skeletal system tumor (Niu et al., 2015; Liu et al., 2017; Georges et al., 2018); miR-340, -137 and -27b in hematological malignancy (Zhang et al., 2016a; Umezu et al., 2017; Jia et al., 2018).
Oncogenic miRNAs and c-Met
Some miRNAs exert oncogenic effects through modulation of the HGF/c-Met axis (Figure 1F). Studies have shown that miR-29b-1-5p was upregulated in OSCC compared with normal oral epithelium, and this upregulation correlated with OSCC histological grade and poor prognosis. In addition, up-regulation of miR-29b-1-5p showed a synergistic effect with c-Met, resulting in induction of the EMT in OSCC cells (Kurihara-Shimomura et al., 2019). MiR-658 may have an opposite effects on the metastasis of gastric cancer (MGC) compared to that induced by miR-206. Serum levels of miR-658 in patients with distant MGC were significantly higher than those with no MGC, and overexpression of miR-658 has been shown to activate the PAX3/c-Met pathway to promote metastasis of GC cells (Wu et al., 2018). Interestingly, viral miRNAs may also be oncogenic. MiR-K6 is a Kaposi's sarcoma-associated herpes virus (KSHV)-encoded microRNA, and miR-K6-5p has been shown to specifically inhibit the expression of endogenous CD82, a metastasis suppressor, and directly interact with c-Met to inhibit its activation, resulting in endothelial cell invasion and angiogenesis (Li et al., 2017b). Tumor necrosis factor (TNF)-related apoptosis inducing ligand (TRAIL) is a member of the TNF-α family that induces apoptosis by binding to death receptors on the cell surface. Some scholars noted that NSCLC cells that overexpressed miR-221/222 were TRAIL-resistant and showed increased migration and invasion (Garofalo et al., 2008). They also found that c-Met conferred resistance to TRAIL-induced cell death and enhanced tumorigenicity of lung cancer cells by activation of miR-221 and miR-222 through the c-Jun transcription factor (Garofalo et al., 2009). In another study, low levels of miR-130a were detected in NSCLC, and miR-130a targeted c-Met and reduced TRAIL resistance through c-Jun-mediated down-regulation of miR-221 and miR-222 (Acunzo et al., 2012). Evidence is also proposed that c-Met activation increased miR-130b levels, inhibited androgen receptor expression, and promoted cancer spreading and resistance to hormone ablation therapy. They also showed that c-Met/miR-130b axis expression in exosomes isolated from peripheral blood was a non-invasive tool for active surveillance and therapy monitoring (Cannistraci et al., 2017).
LncRNAs/c-Met Axis in Cancer
Oncogenic lncRNAs Target c-Met
Most lncRNAs that target c-Met act as ceRNAs, decoys, or sponges, and bind to specific miRNAs to prevent suppression of c-Met (Salmena et al., 2011) (Table 3).
LncRNA-H19 and c-Met
Lnc-H19, a precursor of miR-675, has been associated with carcinogenesis. Investigators reported that overexpression of lnc-H19/miR-675 enhanced the oncogenic action of breast cancer cells, and increased cell proliferation and migration in vitro and in vivo. A study showed that lnc-H19/miR-675 enhanced activation of c-Met and EGFR, which resulted in sustained activation of Akt and Erk, and increased proliferation and migration through direct binding to c-Cbl and Cbl-b mRNA (Vennin et al., 2015). In contrast, in endocrine therapy resistant (ETR) cells, lnc-H19 regulated ERα transcript and protein levels, and maintained levels of ERα following treatment with fulvestrant. Pharmacological inhibitors of Notch and HGF/c-Met signaling decreased lnc-H19 and ERα expression, which resulted in decreased resistance to Tam and fulvestrant in ETR cells (Basak et al., 2018), which may represent a novel therapeutic strategy for ETR. A recent study showed that lnc-H19/miR-675-5p and c-Met levels were elevated in cancer stem cells (CSCs) of glioblastoma multiforme compared to those in peritumoral tissue (Angelucci et al., 2018).
Lnc-X-Inactive Specific Transcript (XIST) and c-Met
A study showed elevated levels of XIST and reduced levels of miR-34a in thyroid cancer tissues and cell lines. XIST serves as an oncogenic lncRNA, and a ceRNA (sponging) for miR-34a as well. XIST may compete with c-Met for miR-34a binding, which promotes thyroid cancer cell proliferation and tumor growth (Liu et al., 2018).
Lnc-Nuclear Enriched Abundant Transcript 1 (NEAT1) and c-Met
NEAT1, a highly abundant lncRNA in the nucleus, was found to be present at higher levels in glioma than in non-cancerous brain tissues. NEAT1 acts as a molecular sponge for miR-449b-5p, which leads to upregulation of c-Met, resulting in glioma pathogenesis (Zhen et al., 2016). Recently, in vivo and in vitro studies have shown the potential treatments for sorafenib resistance in HCC (Chen and Xia, 2019). NEAT1 enhanced c-Met/Akt pathway signaling by negatively regulating miR-335, which resulted in sorafenib resistance.
Other Oncogenic lncRNAs and c-Met
Other oncogenic lncRNAs include lnc-TALC in glioblastoma (Wu et al., 2019); lnc-FAM83H in lung cancer (Zhang et al., 2017b); lnc-TUG1 and lnc-HOTAIR in gastric cancer (Liu Y. W. et al., 2015); lnc-Hand2 in liver cancer (Wang Y. et al., 2018); lnc-ARSR in renal cell cancer (Qu et al., 2016); lnc-GAPLINC in colorectal cancer (Luo et al., 2018); lnc-SNHG8 in endometrial cancer (Yang et al., 2018); lnc-MALAT1 and LNC-PV1 in osteosarcoma (Sun Z. Y. et al., 2019); and lnc-KCNQ1OT1 in melanoma (Guo B. et al., 2018).
Antioncogenic lncRNAs That Target c-Met
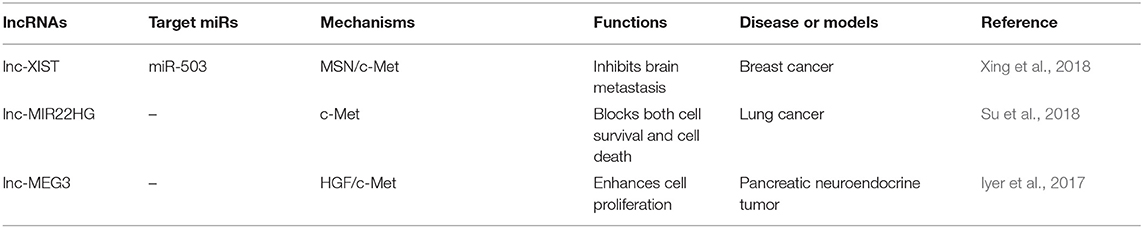
In addition to most lncRNAs act as miRNAs sponges to upregulate the expression of c-Met, there are also some lncRNAs suppress the cancer progression by targeting c-Met through other pathways (Figure 1C). Reduced expression of XIST was observed in patients with breast cancer. Interestingly, XIST plays an antioncogenic role in breast cancer. Levels of XIST were inversely correlated with brain metastasis. Downregulation of XIST resulted in activation of c-Met and induction of the EMT, which resulted in increased secretion of exosomal miRNA-503 and suppression of T-cell proliferation, and promoted stemness of tumor cells (Xing et al., 2018). A previous study has attempted to explain that lncRNA-MIR22HG played a tumor suppressive role through dysregulation of the oncogenes YBX1, MET, and p21, which resulted in reduced cell survival and increased cell death signaling in human primary lung tumors (Su et al., 2018). Another study showed that loss of lncRNA-MEG3 was associated with increased expression of c-Met in pancreatic neuroendocrine tumors (PNETs). They also showed that MEG3 bound to unique genomic regions in and around the c-Met gene, which may be a potential therapeutic target to increase pancreatic islet beta-cell expansion to ameliorate beta-cell loss in diabetes (Iyer et al., 2017) (Table 4).
LncRNAs/miRNAs/c-Met Pathway
Oncogenic lncRNAs/miR-34a/c-Met Pathway
As mentioned above, miRNA-34 is crucial in HGF/c-Met signaling. Many studies have focused on lncRNAs, including lnc-XIST, lnc-MALAT1, lnc-GAPLINC, lnc-HOTAIR, and lnc-ARSR, as ceRNAs targeting to miR-34 (Figure 1D). Recently, scholars explored up-regulation of lnc-MALAT1 (lung adenocarcinoma transcript 1) in osteosarcoma tissues and cell lines, which was significantly associated with poor prognosis (Sun Z. et al., 2019). The results of this study suggested that lnc-MALAT1 increased osteosarcoma proliferation and metastasis by competitively binding to miR-34a/c-5p and miR-449a/b. Increased expression of lnc-GAPLINC was detected in CRC compared to that in non-cancerous tissues, and identified a GAPLINC/miR-34a/c-Met axis involved in cell migration and invasion in CRC tissues and cells (Luo et al., 2018). Another study showed that lncRNA-HOX antisense intergenic RNA (HOTAIR) was highly expressed in gastric cancer, particularly in the diffuse type. This study showed that HOTAIR epigenetically repressed miR-34a by binding to PRC2, and then activated the HGF/C-Met/Snail pathway, which resulted in EMT and accelerated GC tumor metastasis (Liu Y. W. et al., 2015). It has been demonstrated that lncRNA activated in RCC with sunitinib resistance (lnc-ARSR) correlated with poor sunitinib response (Qu et al., 2016). Lnc-ARSR might modulate AXL and c-Met by competitively binding to miR-34/miR-449, which may be a potential therapeutic target for mitigation of sunitinib resistance.
LncRNAs/miRNAs/c-Met in Drug Resistance
In this section, resistance to current anticancer strategies used for treatment of lung, kidney, and pancreatic cancer is discussed (Figure 1E). Lnc-ARSR causes sunitinib resistance in RCC, lnc-H19 promotes resistance to fulvestrant and tamoxifen in breast cancer, and lnc-NEAT1 enhances resistance to sorafenib in HCC. Other lncRNAs may promote drug resistance, and may be predictors and potential targets for cancer treatment. It is interesting to observe that lnc-TALC (temozolomide-associated lncRNA in glioblastoma recurrence) was required for temozolomide (TMZ) resistance and glioblastoma recurrence. Lnc-TALC, which was regulated by the Akt/FOXO3 axis, elevated c-Met expression by competitively binding to miR-20b-3p (Wu et al., 2019). Similarly, lncRNA-PVT1 (Plasmacytoma Variant Translocation 1) played a vital role in chemoresistance of osteosarcoma cells. PVT1 has been shown to target and downregulate miR-152, which results in c-Met/PI3K/AKT pathway activation, and contributes to resistance to gemcitabine (Sun Z. Y. et al., 2019).
miRNA, lncRNA-Based Therapeutics in c-Met-Related Cancer
Events such as c-Met gene mutation, overexpression, and amplification may be closely related to aberrant activation of HGF/c-Met signaling in human cancers (Barrow-Mcgee et al., 2016; Bahrami et al., 2017). Increasing preclinical evidence has allowed for development of specific molecular inhibitors, including anti-c-Met monoclonal antibodies and small molecule tyrosine kinase inhibitors (TKIs). Anti-c-Met monoclonal antibodies block binding to HGF, which prevents dimerization and induces degradation. Small molecules TKIs inhibit catalytic activity. Besides blocking monoclonal antibodies against c-Met or against the HGF ligand, c-Met inhibitors can be classified into two groups of TKIs: multi-kinase inhibitors and selective c-Met inhibitors. A number of clinical trials have been performed to evaluate drugs that target the HGF/c-Met axis, but the results of these trials have been unsatisfactory (Hughes and Siemann, 2018). The poor results of these studies suggested that HGF/c-Met signaling may be more complex than originally thought. Upstream regulation and additional functions of c-Met in progression of metastasis require further investigation. Many studies have evaluated the role of c-Met in cancer, but relationships among lncRNAs, miRNAs, and c-Met have not been characterized. Abundant clinical and ChIP analysis data has been generated to evaluate lncRNAs/miRNAs/c-Met, but few in vivo studies have been performed. Preclinical models and successful clinical trials are of critical importance. MiR-34a has been extensively studied and is a promising miRNA therapeutic candidate (Figure 2). Like miR-34, most miRNAs involved in HGF/c-Met signaling are anti-oncogenic, and the therapeutic approach via miRNAs mimics is considered as the main miRNA-based therapy. One of the biggest challenges is to ensure the delivery efficiency of miRNAs mimics to target cells. While RNase is abundant in serum and cytoplasm, miRNAs is short-lived in plasma (Sun et al., 2017). Moreover, chemically modify nucleotides can increase miRNAs expression and stability, but they are limited due to the low membrane penetrability. Hence, the main strategies are based on the nano-scaled carriers, which are divided to virus and non-virus. Viral vectors, including lentivirus, retrovirus and adenovirus are commonly used. The encapsulated genes can be transferred to the genome of target cells after virus infection. For instance, lentiviral miR-34a delivery system induced apoptosis in mammalian (MM) cells, reduced tumor size in MM xenografts (Di Martino et al., 2012), and prolonged prostate cancer animal survival (Liu et al., 2011). Among non-viral nanocarriers, lipid-based vector are frequently applied because of the high transfection efficiency, but they are limited on account of the poor stability in serum and the toxicity (Xue et al., 2015). To solve this, stable nucleic acid lipid particles (SNALPs) was synthesized, and SNALPs encapsulating miR-34a showed inhibition of MM xenograft growth with low toxicity (Di Martino et al., 2014). Besides, polymeric vector, another non-viral delivery system with the low immunogenicity and cytotoxicity is also used to inhibit the tumor via integrating miR-34a (Lin et al., 2019). The first miRNA-associated therapeutic drug, MRX34, was a lipid nanoparticle loaded with miR-34 mimics, and was tested in clinical trials (NCT01829971 and NCT02862145) (Beg et al., 2017). However, MRX34 was quickly withdrawn because of immune-related adverse events. Therefore, it is important to evaluate the safety of nanoparticles. Other non-viral nano particles are also used as delivery systems for miRNA therapeutics, like EnGeneIC Delivery Vehicle (EDV) nanocells (Taylor et al., 2015), synthetic polyethylenimine (PEI) (Akhtar and Benter, 2007), dendrimers (Duncan and Izzo, 2005), cyclodextrin (Gonzalez et al., 1999), chitosan (Ragelle et al., 2013), N-acetyl-D-galactosamine (Nair et al., 2014). Furthermore, it is important to develop reliable and effective approaches to development of lncRNA-based therapeutics. As discussed above, lots of known lncRNAs in the c-Met signaling are oncogenic. In order to silence oncogenic lncRNAs, three approaches might be used to promote their selective degradation of the lncRNAs, including: RNA interference (RNAi), Rnase H active antisense oligonucleotides (ASOs) and Clustered Regularly Interspaced Short Palindromic Repeats/CRISPR-Associated Protein 9 (CRISPR/Cas9) system; RNAi utilizes the RISC containing a specific siRNA to degrade the targeted lncRNA in cytoplasm (Dorsett and Tuschl, 2004). Rnase H ASOs use endogenous RNase H1, which cleaves the RNA in an RNA/DNA heteroduplex, to reduce the accumulation of lncRNA transcripts or block the functional domains (Ulitsky et al., 2011; Kurian et al., 2015) and CRISPR/Cas9 platform is a powerful genome-editing tool, which is already successfully applied to facilitate targeted genetic mutations. The endonuclease Cas9 is recruited to specific genomic sites via a sequence-dependent CRISPR RNA (crRNA) and a sequence-independent trans-activating CRISPRRNA (tracrRNA) (Jinek et al., 2012; Doudna and Charpentier, 2014). LncRNA-targeting efficiency and knockout confirmation have been done in vivo, which is a promising therapeutic technology (Hansmeier et al., 2019). Advances in delivery strategies may allow for development of novel lncRNA- and miRNA-based therapies, and the lncRNAs/miRNAs/c-Met axis may contain biomarkers for early detection and clinical management of patients with different types of cancers.
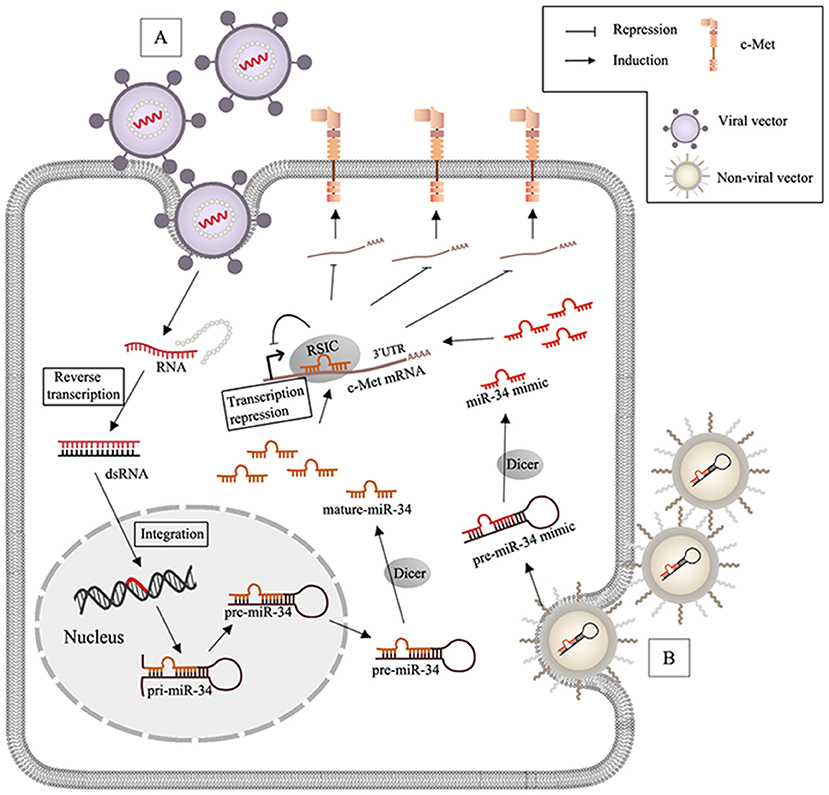
Figure 2. MiR-34 based therapy and delivery systems in c-Met-regulated cancers. (A) After virus (lentivirus or adenovirus) infects the target cells, RNA packaged by viral vectors is liberated with the wrapped protein and undergoes the reverse transcription to synthesize double strands RNA (dsRNA), and the genome of the host cell can integrate with dsRNA and generate miR-34; (B) non-viral nanoparticles including lipid- and polymeric carriers release the encapsulated miR-34 mimics after transfection.
Conclusions and Perspectives
Studies have shown that c-Met is associated with tumor formation, progression, and metastasis. Recent studies have highlighted the HGF/c-Met axis as a promising therapeutic target for treatment of metastatic tumors. Metastatic dissemination represents a critical problem in the treatment of malignancy, and results in poor prognosis and increased mortality in patients with cancer. The metastatic process includes growth and proliferation of the primary tumor, and migration, adhesion, invasion, and survival at distal sites. A number of lncRNAs and miRNAs that target c-Met may play different roles in carcinogenesis through various molecular mechanisms, including up-regulation or mutation of the MET gene. The relationships among lncRNAs, miRNAs, and c-Met in cancer require further characterization. Studies that evaluate the effect of specific lncRNAs or miRNAs on c-Met in different types of cancer are required to determine the therapeutic potential of targeting this signaling pathway.
Studies have shown that ncRNAs/HGF/c-Met signaling is involved in many physiological and pathological processes, and most miRNAs are anti-oncogenic and most lncRNAs are oncogenic. LncRNAs act as ceRNA, decoys, or sponges and bind to specific miRNAs, to prevent interactions with c-Met, which results in malignancy. Studies have shown that c-Met may drive oncogenesis and promote resistance to targeted therapies such as EGFR, VEGFR, HER2, and BRAF inhibitors (Engelman et al., 2007; Shojaei et al., 2010; Chapman et al., 2011; Saito et al., 2015). We reviewed the role of the lncRNA/miRNA/c-Met axis in drug resistance in specific cancers. Translation of molecular mechanisms of cancer resistance to clinical practice is necessary to develop therapeutic strategies to prevent or overcome resistance to targeted therapies, and to identify patient populations who are more likely to benefit from treatment with ncRNAs/HGF/c-Met targeted therapies.
In conclusion, overexpression of c-Met can be induced by upstream lncRNAs and/or miRNAs in various types of tumors (Figure 3). Clinical trials of ncRNAs/c-Met for mitigation of specific drug resistance have shown promising results. In this review, we discussed interactions among lncRNAs, miRNAs, and c-Met, and the relationships of these interactions with tumorigenesis and progression of various cancers. We also reviewed molecular mechanisms associated with the onset and development of cancer, and discussed novel diagnostic therapeutic approaches. Further identification of tissue-specific lncRNAs and miRNAs that directly or indirectly regulate c-Met, and characterization of the associated molecular mechanisms, are necessary.
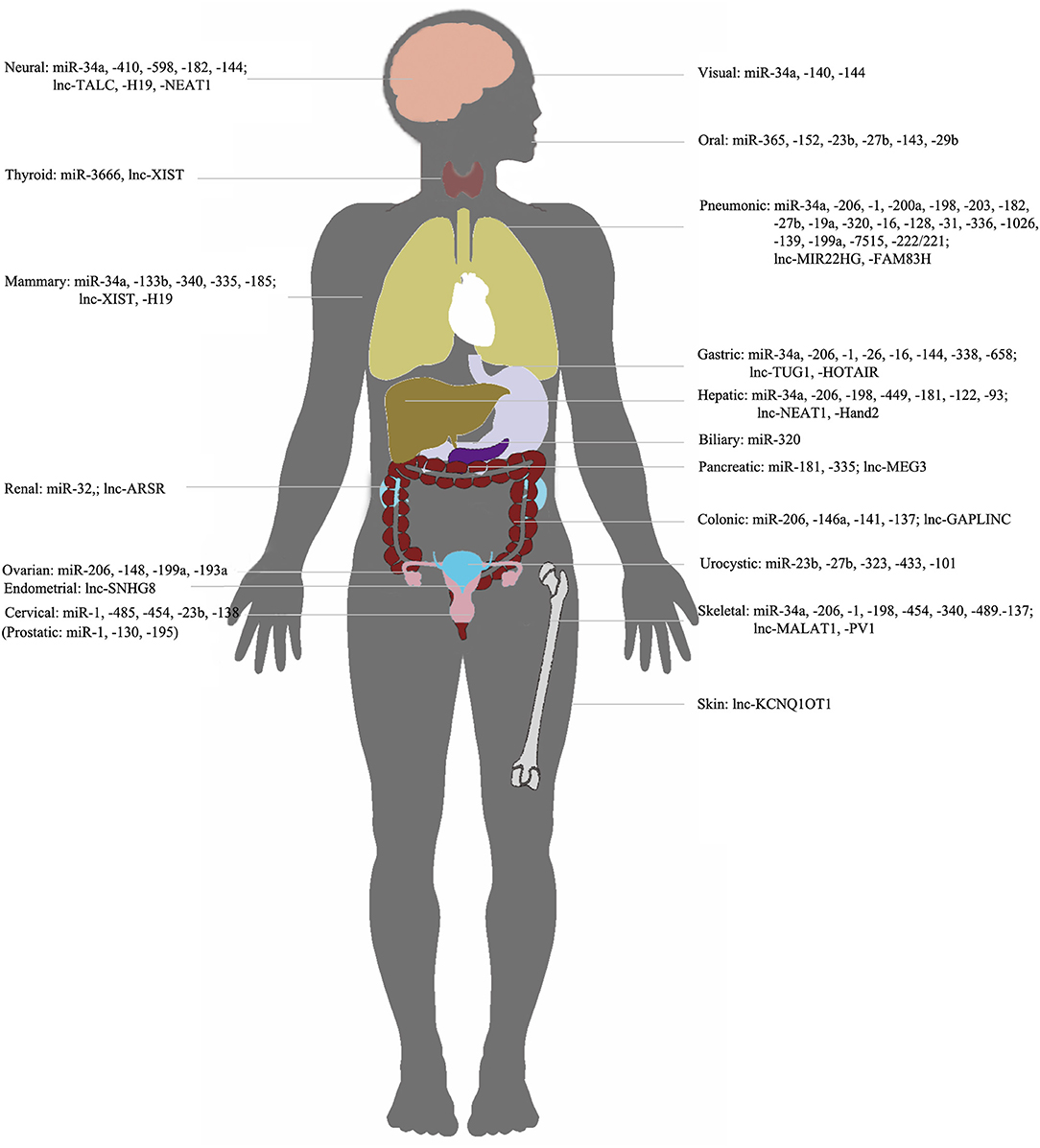
Figure 3. NcRNAs/c-met in different organs. Representation of organ-specific lncRNAs or miRNAs in c-Met regulation in cancer.
Author Contributions
HZ and ST drafted the manuscript. FZ revised the manuscript. AS and JL reviewed and modified the manuscript. All authors agreed on the final version.
Funding
This work was funded by China Postdoctoral Science Foundation (2017M612010) and National Natural Science Foundation of China (81701144, 81571413).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The handling editor and the authors are affiliated with Zhejiang University, Hangzhou, but do not have any collaborations.
References
Acunzo, M., Visone, R., Romano, G., Veronese, A., Lovat, F., Palmieri, D., et al. (2012). miR-130a targets MET and induces TRAIL-sensitivity in NSCLC by downregulating miR-221 and 222. Oncogene 31, 634–642. doi: 10.1038/onc.2011.260
Akhtar, S., and Benter, I. F. (2007). Nonviral delivery of synthetic siRNAs in vivo. J. Clin. Invest. 117, 3623–3632. doi: 10.1172/JCI33494
Andriani, F., Majorini, M. T., Mano, M., Landoni, E., Miceli, R., Facchinetti, F., et al. (2018). MiR-16 regulates the pro-tumorigenic potential of lung fibroblasts through the inhibition of HGF production in an FGFR-1- and MEK1-dependent manner. J. Hematol. Oncol. 11:45. doi: 10.1186/s13045-018-0594-4
Angelucci, C., D'alessio, A., Lama, G., Binda, E., Mangiola, A., Vescovi, A. L., et al. (2018). Cancer stem cells from peritumoral tissue of glioblastoma multiforme: the possible missing link between tumor development and progression. Oncotarget 9, 28116–28130. doi: 10.18632/oncotarget.25565
Bahrami, A., Shahidsales, S., Khazaei, M., Ghayour-Mobarhan, M., Maftouh, M., Hassanian, S. M., et al. (2017). C-Met as a potential target for the treatment of gastrointestinal cancer: current status and future perspectives. J. Cell. Physiol. 232, 2657–2673. doi: 10.1002/jcp.25794
Barrow-Mcgee, R., Kishi, N., Joffre, C., Menard, L., Hervieu, A., Bakhouche, B. A., et al. (2016). Beta 1-integrin-c-Met cooperation reveals an inside-in survival signalling on autophagy-related endomembranes. Nat. Commun. 7:11942. doi: 10.1038/ncomms12392
Basak, P., Chatterjee, S., Bhat, V., Su, A., Jin, H., Lee-Wing, V., et al. (2018). Long non-coding RNA H19 acts as an estrogen receptor modulator that is required for endocrine therapy resistance in ER+ breast cancer cells. Cell Physiol. Biochem. 51, 1518–1532. doi: 10.1159/000495643
Basilico, C., Arnesano, A., Galluzzo, M., Comoglio, P. M., and Michieli, P. (2008). A high affinity hepatocyte growth factor-binding site in the immunoglobulin-like region of Met. J. Biol. Chem. 283, 21267–21277. doi: 10.1074/jbc.M800727200
Beg, M. S., Brenner, A. J., Sachdev, J., Borad, M., Kang, Y. K., Stoudemire, J., et al. (2017). Phase I study of MRX34, a liposomal miR-34a mimic, administered twice weekly in patients with advanced solid tumors. Invest. New Drugs 35, 180–188. doi: 10.1007/s10637-016-0407-y
Bejerano, G., Pheasant, M., Makunin, I., Stephen, S., Kent, W. J., Mattick, J. S., et al. (2004). Ultraconserved elements in the human genome. Science 304, 1321–1325. doi: 10.1126/science.1098119
Bleau, A. M., Redrado, M., Nistal-Villan, E., Villalba, M., Exposito, F., Redin, E., et al. (2018). miR-146a targets c-met and abolishes colorectal cancer liver metastasis. Cancer Lett. 414, 257–267. doi: 10.1016/j.canlet.2017.11.008
Campos-Viguri, G. E., Jimenez-Wences, H., Peralta-Zaragoza, O., Torres-Altamirano, G., Soto-Flores, D. G., Hernandez-Sotelo, D., et al. (2015). miR-23b as a potential tumor suppressor and its regulation by DNA methylation in cervical cancer. Infect. Agent Cancer 10, 42. doi: 10.1186/s13027-015-0037-6
Cannistraci, A., Federici, G., Addario, A., Di Pace, A. L., Grassi, L., Muto, G., et al. (2017). C-Met/miR-130b axis as novel mechanism and biomarker for castration resistance state acquisition. Oncogene 36, 3718–3728. doi: 10.1038/onc.2016.505
Cao, J., Zhang, Y., Yang, J., He, S., Li, M., Yan, S., et al. (2016). NEAT1 regulates pancreatic cancer cell growth, invasion and migration though mircroRNA-335–5p/c-met axis. Am. J. Cancer Res. 6, 2361–2374.
Cao, X., Lai, S., Hu, F., Li, G., Wang, G., Luo, X., et al. (2017). miR-19a contributes to gefitinib resistance and epithelial mesenchymal transition in non-small cell lung cancer cells by targeting c-Met. Sci. Rep. 7:2939. doi: 10.1038/s41598-017-01153-0
Chapman, P. B., Hauschild, A., Robert, C., Haanen, J. B., Ascierto, P., Larkin, J., et al. (2011). Improved survival with vemurafenib in melanoma with BRAF V600E mutation. N. Engl. J. Med. 364, 2507–2516. doi: 10.1056/NEJMoa1103782
Chen, A. H., Qin, Y. E., Tang, W. F., Tao, J., Song, H. M., and Zuo, M. (2017a). MiR-34a and miR-206 act as novel prognostic and therapy biomarkers in cervical cancer. Cancer Cell Int. 17:63. doi: 10.1186/s12935-017-0431-9
Chen, L., Xu, L., and Wang, G. (2015a). Regulation of MET-mediated proliferation of thyroid carcinoma cells by miR-449b. Tumour Biol. 36, 8653–8660. doi: 10.1007/s13277-015-3619-4
Chen, L., Zhang, J., Feng, Y., Li, R., Sun, X., Du, W., et al. (2012). MiR-410 regulates MET to influence the proliferation and invasion of glioma. Int. J. Biochem Cell Biol. 44, 1711–1717. doi: 10.1016/j.biocel.2012.06.027
Chen, Q. Y., Jiao, D. M., Wang, J., Hu, H., Tang, X., Chen, J., et al. (2016a). miR-206 regulates cisplatin resistance and EMT in human lung adenocarcinoma cells partly by targeting MET. Oncotarget 7, 24510–24526. doi: 10.18632/oncotarget.8229
Chen, Q. Y., Jiao, D. M., Wu, Y. Q., Chen, J., Wang, J., Tang, X. L., et al. (2016b). MiR-206 inhibits HGF-induced epithelial-mesenchymal transition and angiogenesis in non-small cell lung cancer via c-Met /PI3k/Akt/mTOR pathway. Oncotarget 7, 18247–18261. doi: 10.18632/oncotarget.7570
Chen, Q. Y., Jiao, D. M., Yan, L., Wu, Y. Q., Hu, H. Z., Song, J., et al. (2015b). Comprehensive gene and microRNA expression profiling reveals miR-206 inhibits MET in lung cancer metastasis. Mol. Biosyst. 11, 2290–2302. doi: 10.1039/C4MB00734D
Chen, S., and Xia, X. (2019). Long noncoding RNA NEAT1 suppresses sorafenib sensitivity of hepatocellular carcinoma cells via regulating miR-335-c-Met. J. Cell Physiol. 234, 14999–15009. doi: 10.1002/jcp.27567
Chen, T., Cai, S. L., Li, J., Qi, Z. P., Li, X. Q., Ye, L. C., et al. (2017b). Mecp2-mediated Epigenetic Silencing of miR-137 contributes to colorectal adenoma-carcinoma sequence and tumor progression via relieving the suppression of c-Met. Sci. Rep. 7:44543. doi: 10.1038/srep44543
Cheng, J., Wu, L. M., Deng, X. S., Wu, J., Lv, Z., Zhao, H. F., et al. (2018). MicroRNA-449a suppresses hepatocellular carcinoma cell growth via G1 phase arrest and the HGF/MET c-Met pathway. Hepatobiliary Pancreat Dis. Int. 17, 336–344. doi: 10.1016/j.hbpd.2018.07.006
Cheng, Y., Yang, M., and Peng, J. (2019). Correlation the between the regulation of miRNA-1 in c-Met-induced EMT and cervical cancer progression. Oncol. Lett. 17, 3341–3349. doi: 10.3892/ol.2019.9971
Chiyomaru, T., Seki, N., Inoguchi, S., Ishihara, T., Mataki, H., Matsushita, R., et al. (2015). Dual regulation of receptor tyrosine kinase genes EGFR and c-Met by the tumor-suppressive microRNA-23b/27b cluster in bladder cancer. Int. J. Oncol. 46, 487–496. doi: 10.3892/ijo.2014.2752
Dai, C., Xie, Y., Zhuang, X., and Yuan, Z. (2018). MiR-206 inhibits epithelial ovarian cancer cells growth and invasion via blocking c-Met/AKT/mTOR signaling pathway. Biomed. Pharmacother. 104, 763–770. doi: 10.1016/j.biopha.2018.05.077
Dang, Y., Luo, D., Rong, M., and Chen, G. (2013). Underexpression of miR-34a in hepatocellular carcinoma and its contribution towards enhancement of proliferating inhibitory effects of agents targeting c-MET. PLoS ONE 8:e61054. doi: 10.1371/journal.pone.0061054
De Los Santos, M. C., Dragomir, M. P., and Calin, G. A. (2019). The role of exosomal long non-coding RNAs in cancer drug resistance. Cancer Drug Resist. 2, 1178–1192. doi: 10.20517/cdr.2019.74
Di Martino, M. T., Campani, V., Misso, G., Gallo Cantafio, M. E., Gulla, A., Foresta, U., et al. (2014). In vivo activity of miR-34a mimics delivered by stable nucleic acid lipid particles (SNALPs) against multiple myeloma. PLoS ONE 9:e90005. doi: 10.1371/journal.pone.0090005
Di Martino, M. T., Leone, E., Amodio, N., Foresta, U., Lionetti, M., Pitari, M. R., et al. (2012). Synthetic miR-34a mimics as a novel therapeutic agent for multiple myeloma: in vitro and in vivo evidence. Clin. Cancer Res. 18, 6260–6270. doi: 10.1158/1078-0432.CCR-12-1708
Dorsett, Y., and Tuschl, T. (2004). siRNAs: applications in functional genomics and potential as therapeutics. Nat. Rev. Drug Discov. 3, 318–329. doi: 10.1038/nrd1345
Doudna, J. A., and Charpentier, E. (2014). Genome editing. The new frontier of genome engineering with CRISPR-Cas9. Science 346:1258096. doi: 10.1126/science.1258096
Du, M., Wang, J., Chen, H., Wang, S., Chen, L., Xu, Y., et al. (2019). MicroRNA200a suppresses migration and invasion and enhances the radiosensitivity of NSCLC cells by inhibiting the HGF/cMet signaling pathway. Oncol. Rep. 41, 1497–1508. doi: 10.3892/or.2018.6925
Du, Z., Fei, T., Verhaak, R. G., Su, Z., Zhang, Y., Brown, M., et al. (2013). Integrative genomic analyses reveal clinically relevant long noncoding RNAs in human cancer. Nat. Struct. Mol. Biol. 20, 908–913. doi: 10.1038/nsmb.2591
Duncan, R., and Izzo, L. (2005). Dendrimer biocompatibility and toxicity. Adv. Drug. Deliv. Rev. 57, 2215–2237. doi: 10.1016/j.addr.2005.09.019
El Bezawy, R., De Cesare, M., Pennati, M., Deraco, M., Gandellini, P., Zuco, V., et al. (2017). Antitumor activity of miR-34a in peritoneal mesothelioma relies on c-MET and AXL inhibition: persistent activation of ERK and AKT signaling as a possible cytoprotective mechanism. J. Hematol. Oncol. 10:19. doi: 10.1186/s13045-016-0387-6
Engelman, J. A., Zejnullahu, K., Mitsudomi, T., Song, Y., Hyland, C., Park, J. O., et al. (2007). MET amplification leads to gefitinib resistance in lung cancer by activating ERBB3 signaling. Science 316, 1039–1043. doi: 10.1126/science.1141478
Feng, D., Zhu, N., Yu, C., and Lou, D. (2019). MicroRNA-34a suppresses human lens epithelial cell proliferation and migration via downregulation of c-Met. Clin. Chim. Acta. 495, 326–330. doi: 10.1016/j.cca.2019.04.060
Fu, P., Du, F., Yao, M., Lv, K., and Liu, Y. (2014). MicroRNA-185 inhibits proliferation by targeting c-Met in human breast cancer cells. Exp. Ther. Med. 8, 1879–1883. doi: 10.3892/etm.2014.1999
Fukumoto, I., Koshizuka, K., Hanazawa, T., Kikkawa, N., Matsushita, R., Kurozumi, A., et al. (2016). The tumor-suppressive microRNA-23b/27b cluster regulates the MET oncogene in oral squamous cell carcinoma. Int. J. Oncol. 49, 1119–1129. doi: 10.3892/ijo.2016.3602
Gallardo, E., Navarro, A., Vinolas, N., Marrades, R. M., Diaz, T., Gel, B., et al. (2009). miR-34a as a prognostic marker of relapse in surgically resected non-small-cell lung cancer. Carcinogenesis 30, 1903–1909. doi: 10.1093/carcin/bgp219
Gao, S., Zhao, Z., Wu, R., Wu, L., Tian, X., and Zhang, Z. (2019). MiR-1 inhibits prostate cancer PC3 cells proliferation through the Akt/mTOR signaling pathway by binding to c-Met. Biomed. Pharmacother. 109, 1406–1410. doi: 10.1016/j.biopha.2018.10.098
Gao, Y., Zeng, F., Wu, J. Y., Li, H. Y., Fan, J. J., Mai, L., et al. (2015). MiR-335 inhibits migration of breast cancer cells through targeting oncoprotein c-Met. Tumour Biol. 36, 2875–2883. doi: 10.1007/s13277-014-2917-6
Garofalo, M., Di Leva, G., Romano, G., Nuovo, G., Suh, S. S., Ngankeu, A., et al. (2009). miR-221&222 regulate TRAIL resistance and enhance tumorigenicity through PTEN and TIMP3 downregulation. Cancer Cell 16, 498–509. doi: 10.1016/j.ccr.2009.10.014
Garofalo, M., Quintavalle, C., Di Leva, G., Zanca, C., Romano, G., Taccioli, C., et al. (2008). MicroRNA signatures of TRAIL resistance in human non-small cell lung cancer. Oncogene 27, 3845–3855. doi: 10.1038/onc.2008.6
Georges, S., Calleja, L. R., Jacques, C., Lavaud, M., Moukengue, B., Lecanda, F., et al. (2018). Loss of miR-198 and−206 during primary tumor progression enables metastatic dissemination in human osteosarcoma. Oncotarget 9, 35726–35741. doi: 10.18632/oncotarget.26284
Ghandadi, M., and Sahebkar, A. (2016). MicroRNA-34a and its target genes: Key factors in cancer multidrug resistance. Curr. Pharm. Des. 22, 933–939. doi: 10.2174/1381612822666151209153729
Gherardi, E., Birchmeier, W., Birchmeier, C., and Vande Woude, G. (2012). Targeting MET in cancer: rationale and progress. Nat. Rev. Cancer 12, 89–103. doi: 10.1038/nrc3205
Gonzalez, H., Hwang, S. J., and Davis, M. E. (1999). New class of polymers for the delivery of macromolecular therapeutics. Bioconjug. Chem. 10, 1068–1074. doi: 10.1021/bc990072j
Guo, B., Zhang, Q., Wang, H., Chang, P., and Tao, K. (2018). KCNQ1OT1 promotes melanoma growth and metastasis. Aging 10, 632–644. doi: 10.18632/aging.101418
Guo, Y., Tao, M., and Jiang, M. (2018). MicroRNA-454–3p inhibits cervical cancer cell invasion and migration by targeting c-Met. Exp. Ther. Med. 15, 2301–2306. doi: 10.3892/etm.2018.5714
Hajalirezay Yazdi, S., Paryan, M., and Mohammadi-Yeganeh, S. (2018). An integrated approach of bioinformatic prediction and in vitro analysis identified that miR-34a targets MET and AXL in triple-negative breast cancer. Cell. Mol. Biol. Lett. 23:51. doi: 10.1186/s11658-018-0116-y
Han, C., Zhou, Y., An, Q., Li, F., Li, D., Zhang, X., et al. (2015). MicroRNA-1 (miR-1) inhibits gastric cancer cell proliferation and migration by targeting MET. Tumour Biol. 36, 6715–6723. doi: 10.1007/s13277-015-3358-6
Hansmeier, N. R., Widdershooven, P. J. M., Khani, S., and Kornfeld, J. W. (2019). Rapid generation of long noncoding RNA knockout mice using CRISPR/Cas9 technology. Noncoding RNA 5:E12. doi: 10.3390/ncrna5010012
Hong, J. H., Roh, K. S., Suh, S. S., Lee, S., Sung, S. W., Park, J. K., et al. (2015). The expression of microRNA-34a is inversely correlated with c-MET and CDK6 and has a prognostic significance in lung adenocarcinoma patients. Tumour Biol. 36, 9327–9337. doi: 10.1007/s13277-015-3428-9
Hou, C., Sun, B., Jiang, Y., Zheng, J., Yang, N., Ji, C., et al. (2016). MicroRNA-31 inhibits lung adenocarcinoma stem-like cells via down-regulation of MET-PI3K-Akt signaling pathway. Anticancer Agents Med. Chem. 16, 501–518. doi: 10.2174/1871520615666150824152353
Hou, Q., Tang, J., Wang, Z., Wang, C., Chen, X., Hou, L., et al. (2013). Inhibitory effect of microRNA-34a on retinal pigment epithelial cell proliferation and migration. Invest. Ophthalmol. Vis. Sci. 54, 6481–6488. doi: 10.1167/iovs.13-11873
Hu, Z., Lin, Y., Chen, H., Mao, Y., Wu, J., Zhu, Y., et al. (2013). MicroRNA-101 suppresses motility of bladder cancer cells by targeting c-Met. Biochem. Biophys. Res. Commun. 435, 82–87. doi: 10.1016/j.bbrc.2013.04.042
Huang, N., Wu, Z., Lin, L., Zhou, M., Wang, L., Ma, H., et al. (2015). MiR-338–3p inhibits epithelial-mesenchymal transition in gastric cancer cells by targeting ZEB2 and MACC1/Met/Akt signaling. Oncotarget 6, 15222–15234. doi: 10.18632/oncotarget.3835
Huang, W. C., Jang, T. H., Tung, S. L., Yen, T. C., Chan, S. H., and Wang, L. H. (2019). A novel miR-365-3p/EHF/keratin 16 axis promotes oral squamous cell carcinoma metastasis, cancer stemness and drug resistance via enhancing β5-integrin/c-met signaling pathway. J. Exp. Clin. Cancer. Res. 38:89. doi: 10.1186/s13046-019-1091-5
Hughes, V. S., and Siemann, D. W. (2018). Have clinical trials properly assessed c-Met inhibitors? Trends Cancer 4, 94–97. doi: 10.1016/j.trecan.2017.11.009
Hwang, C. I., Matoso, A., Corney, D. C., Flesken-Nikitin, A., Korner, S., Wang, W., et al. (2011). Wild-type p53 controls cell motility and invasion by dual regulation of MET expression. Proc. Natl. Acad. Sci. U.S.A. 108, 14240–14245. doi: 10.1073/pnas.1017536108
Imura, Y., Nakai, T., Yamada, S., Outani, H., Takenaka, S., Hamada, K., et al. (2016). Functional and therapeutic relevance of hepatocyte growth factor/c-MET signaling in synovial sarcoma. Cancer Sci. 107, 1867–1876. doi: 10.1111/cas.13092
Iorio, M. V., and Croce, C. M. (2012). microRNA involvement in human cancer. Carcinogenesis 33, 1126–1133. doi: 10.1093/carcin/bgs140
Iyer, S., Modali, S. D., and Agarwal, S. K. (2017). Long Noncoding RNA MEG3 is an epigenetic determinant of oncogenic signaling in functional pancreatic neuroendocrine tumor cells. Mol. Cell Biol. 37:e00278–e00217. doi: 10.1128/MCB.00278-17
Jardim, D. L., Tang, C., Gagliato Dde, M., Falchook, G. S., Hess, K., Janku, F., et al. (2014). Analysis of 1,115 patients tested for MET amplification and therapy response in the MD Anderson Phase I Clinic. Clin. Cancer. Res. 20, 6336–6345. doi: 10.1158/1078-0432.CCR-14-1293
Ji, T. T., Huang, X., Jin, J., Pan, S. H., and Zhuge, X. J. (2016). Inhibition of long non-coding RNA TUG1 on gastric cancer cell transference and invasion through regulating and controlling the expression of miR-144/c-Met axis. Asian Pac. J. Trop. Med. 9, 508–512. doi: 10.1016/j.apjtm.2016.03.026
Jia, Y. J., Liu, Z. B., Wang, W. G., Sun, C. B., Wei, P., Yang, Y. L., et al. (2018). HDAC6 regulates microRNA-27b that suppresses proliferation, promotes apoptosis and target MET in diffuse large B-cell lymphoma. Leukemia 32, 703–711. doi: 10.1038/leu.2017.299
Jiang, J., Feng, X., Zhou, W., Wu, Y., and Yang, Y. (2016a). MiR-128 reverses the gefitinib resistance of the lung cancer stem cells by inhibiting the c-met/PI3K/AKT pathway. Oncotarget 7, 73188–73199. doi: 10.18632/oncotarget.12283
Jiang, S., Zhao, C., Yang, X., Li, X., Pan, Q., Huang, H., et al. (2016b). miR-1 suppresses the growth of esophageal squamous cell carcinoma in vivo and in vitro through the downregulation of MET, cyclin D1 and CDK4 expression. Int. J. Mol. Med. 38, 113–122. doi: 10.3892/ijmm.2016.2619
Jiao, D., Chen, J., Li, Y., Tang, X., Wang, J., Xu, W., et al. (2018). miR-1–3p and miR-206 sensitizes HGF-induced gefitinib-resistant human lung cancer cells through inhibition of c-Met signalling and EMT. J. Cell Mol. Med. 22, 3526–3536. doi: 10.1111/jcmm.13629
Jinek, M., Chylinski, K., Fonfara, I., Hauer, M., Doudna, J. A., and Charpentier, E. (2012). A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337, 816–821. doi: 10.1126/science.1225829
Johnson, G. L., and Lapadat, R. (2002). Mitogen-activated protein kinase pathways mediated by ERK, JNK, and p38 protein kinases. Science 298, 1911–1912. doi: 10.1126/science.1072682
Johnsson, P., Lipovich, L., Grander, D., and Morris, K. V. (2014). Evolutionary conservation of long non-coding RNAs; sequence, structure, function. Biochim. Biophys. Acta. 1840, 1063–1071. doi: 10.1016/j.bbagen.2013.10.035
Kim, S., Lee, U. J., Kim, M. N., Lee, E. J., Kim, J. Y., Lee, M. Y., et al. (2008). MicroRNA miR-199a* regulates the MET proto-oncogene and the downstream extracellular signal-regulated kinase 2 (ERK2). J. Biol. Chem. 283, 18158–18166. doi: 10.1074/jbc.M800186200
Kinose, Y., Sawada, K., Nakamura, K., Sawada, I., Toda, A., Nakatsuka, E., et al. (2015). The hypoxia-related microRNA miR-199a-3p displays tumor suppressor functions in ovarian carcinoma. Oncotarget 6, 11342–11356. doi: 10.18632/oncotarget.3604
Kogita, A., Togashi, Y., Hayashi, H., Banno, E., Terashima, M., De Velasco, M. A., et al. (2015). Activated MET acts as a salvage signal after treatment with alectinib, a selective ALK inhibitor, in ALK-positive non-small cell lung cancer. Int. J. Oncol. 46, 1025–1030. doi: 10.3892/ijo.2014.2797
Korhan, P., Erdal, E., and Atabey, N. (2014). MiR-181a-5p is downregulated in hepatocellular carcinoma and suppresses motility, invasion and branching-morphogenesis by directly targeting c-Met. Biochem. Biophys. Res. Commun. 450, 1304–1312. doi: 10.1016/j.bbrc.2014.06.142
Koshizuka, K., Hanazawa, T., Fukumoto, I., Kikkawa, N., Matsushita, R., Mataki, H., et al. (2017). Dual-receptor (EGFR and c-MET) inhibition by tumor-suppressive miR-1 and miR-206 in head and neck squamous cell carcinoma. J. Hum. Genet. 62, 113–121. doi: 10.1038/jhg.2016.47
Kouri, F. M., Ritner, C., and Stegh, A. H. (2015). miRNA-182 and the regulation of the glioblastoma phenotype - toward miRNA-based precision therapeutics. Cell Cycle 14, 3794–3800. doi: 10.1080/15384101.2015.1093711
Krol, J., Loedige, I., and Filipowicz, W. (2010). The widespread regulation of microRNA biogenesis, function and decay. Nat. Rev. Genet. 11, 597–610. doi: 10.1038/nrg2843
Kurian, L., Aguirre, A., Sancho-Martinez, I., Benner, C., Hishida, T., Nguyen, T. B., et al. (2015). Identification of novel long noncoding RNAs underlying vertebrate cardiovascular development. Circulation 131, 1278–1290. doi: 10.1161/CIRCULATIONAHA.114.013303
Kurihara-Shimomura, M., Sasahira, T., Shimomura, H., Nakashima, C., and Kirita, T. (2019). The oncogenic activity of miR-29b-1-5p induces the epithelial-mesenchymal transition in oral squamous cell carcinoma. J. Clin. Med. 8:273. doi: 10.3390/jcm8020273
Lan, F., Yu, H., Hu, M., Xia, T., and Yue, X. (2015). miR-144-3p exerts anti-tumor effects in glioblastoma by targeting c-Met. J. Neurochem. 135, 274–286. doi: 10.1111/jnc.13272
Lee, J. M., Yoo, J. K., Yoo, H., Jung, H. Y., Lee, D. R., Jeong, H. C., et al. (2013). The novel miR-7515 decreases the proliferation and migration of human lung cancer cells by targeting c-Met. Mol. Cancer. Res. 11, 43–53. doi: 10.1158/1541-7786.MCR-12-0355
Li, B., Yang, X. X., Wang, D., and Ji, H. K. (2016a). MicroRNA-138 inhibits proliferation of cervical cancer cells by targeting c-Met. Eur. Rev. Med. Pharmacol. Sci. 20, 1109–1114.
Li, J., Xu, X., Meng, S., Liang, Z., Wang, X., Xu, M., et al. (2017a). MET/SMAD3/SNAIL circuit mediated by miR-323a-3p is involved in regulating epithelial–mesenchymal transition progression in bladder cancer. Cell Death Dis. 8:e3010. doi: 10.1038/cddis.2017.331
Li, M., Li, Z., Wang, X., Wang, Y., Zhao, C., and Wang, L. (2018a). Function of miR152 as tumor suppressor in oral squamous cell carcinoma cells by targeting cMET. Oncol Rep. 39, 1173–1180. doi: 10.3892/or.2017.6157
Li, S., Zhang, H., Wang, X., Qu, Y., Duan, J., Liu, R., et al. (2016b). Direct targeting of HGF by miR-16 regulates proliferation and migration in gastric cancer. Tumour Biol. 37, 15175–15183. doi: 10.1007/s13277-016-5390-6
Li, W., Hu, M., Wang, C., Lu, H., Chen, F., Xu, J., et al. (2017b). A viral microRNA downregulates metastasis suppressor CD82 and induces cell invasion and angiogenesis by activating the c-Met signaling. Oncogene 36, 5407–5420. doi: 10.1038/onc.2017.139
Li, Y., Guessous, F., Zhang, Y., Dipierro, C., Kefas, B., Johnson, E., et al. (2009). MicroRNA-34a inhibits glioblastoma growth by targeting multiple oncogenes. Cancer Res. 69, 7569–7576. doi: 10.1158/0008-5472.CAN-09-0529
Li, Y., Zhang, H., Li, Y., Zhao, C., Fan, Y., Liu, J., et al. (2018b). MiR-182 inhibits the epithelial to mesenchymal transition and metastasis of lung cancer cells by targeting the Met gene. Mol. Carcinog. 57, 125–136. doi: 10.1002/mc.22741
Li, Y. Q., Ren, X. Y., He, Q. M., Xu, Y. F., Tang, X. R., Sun, Y., et al. (2015). MiR-34c suppresses tumor growth and metastasis in nasopharyngeal carcinoma by targeting MET. Cell Death Dis. 6:e1618. doi: 10.1038/cddis.2014.582
Liao, Y., Yin, X., Deng, Y., and Peng, X. (2018). MiR-140-5p suppresses retinoblastoma cell growth via inhibiting c-Met/AKT/mTOR pathway. Biosci. Rep. 38:BSR20180776. doi: 10.1042/BSR20180776
Lin, F., Wen, D., Wang, X., and Mahato, R. I. (2019). Dual responsive micelles capable of modulating miRNA-34a to combat taxane resistance in prostate cancer. Biomaterials 192, 95–108. doi: 10.1016/j.biomaterials.2018.10.036
Liu, C., Kelnar, K., Liu, B., Chen, X., Calhoun-Davis, T., Li, H., et al. (2011). The microRNA miR-34a inhibits prostate cancer stem cells and metastasis by directly repressing CD44. Nat. Med. 17, 211–215. doi: 10.1038/nm.2284
Liu, H., Deng, H., Zhao, Y., Li, C., and Liang, Y. (2018). LncRNA XIST/miR-34a axis modulates the cell proliferation and tumor growth of thyroid cancer through MET-PI3K-AKT signaling. J. Exp. Clin. Cancer Res. 37, 279. doi: 10.1186/s13046-018-0950-9
Liu, J., Xue, H., Zhang, J., Suo, T., Xiang, Y., Zhang, W., et al. (2015). MicroRNA-144 inhibits the metastasis of gastric cancer by targeting MET expression. J. Exp. Clin. Cancer Res. 34:35. doi: 10.1186/s13046-015-0154-5
Liu, Q., Yang, G., and Qian, Y. (2017). Loss of MicroRNA-489-3p promotes osteosarcoma metastasis by activating PAX3-MET pathway. Mol. Carcinog. 56, 1312–1321. doi: 10.1002/mc.22593
Liu, Y. (1998). The human hepatocyte growth factor receptor gene: complete structural organization and promoter characterization. Gene 215, 159–169. doi: 10.1016/S0378-1119(98)00264-9
Liu, Y. W., Sun, M., Xia, R., Zhang, E. B., Liu, X. H., Zhang, Z. H., et al. (2015). LincHOTAIR epigenetically silences miR34a by binding to PRC2 to promote the epithelial-to-mesenchymal transition in human gastric cancer. Cell. Death Dis. 6:e1802. doi: 10.1038/cddis.2015.150
Long, Z. H., Bai, Z. G., Song, J. N., Zheng, Z., Li, J., Zhang, J., et al. (2017). miR-141 inhibits proliferation and migration of colorectal cancer SW480 cells. Anticancer Res. 37, 4345–4352. doi: 10.21873/anticanres.11828
Luo, Y., Ouyang, J., Zhou, D., Zhong, S., Wen, M., Ou, W., et al. (2018). Long noncoding RNA GAPLINC promotes cells migration and invasion in colorectal cancer cell by regulating miR-34a/c-MET signal pathway. Dig. Dis. Sci. 63, 890–899. doi: 10.1007/s10620-018-4915-9
Mariani, M., Mchugh, M., Petrillo, M., Sieber, S., He, S., Andreoli, M., et al. (2014). HGF/c-Met axis drives cancer aggressiveness in the neo-adjuvant setting of ovarian cancer. Oncotarget 5, 4855–4867. doi: 10.18632/oncotarget.2049
Maroni, P., Puglisi, R., Mattia, G., Care, A., Matteucci, E., Bendinelli, P., et al. (2017). In bone metastasis miR-34a-5p absence inversely correlates with Met expression, while Met oncogene is unaffected by miR-34a-5p in non-metastatic and metastatic breast carcinomas. Carcinogenesis 38, 492–503. doi: 10.1093/carcin/bgx027
Mataki, H., Seki, N., Chiyomaru, T., Enokida, H., Goto, Y., Kumamoto, T., et al. (2015). Tumor-suppressive microRNA-206 as a dual inhibitor of MET and EGFR oncogenic signaling in lung squamous cell carcinoma. Int. J. Oncol. 46, 1039–1050. doi: 10.3892/ijo.2014.2802
Migliore, C., and Giordano, S. (2009). MiRNAs as new master players. Cell Cycle 8, 2185–2186. doi: 10.4161/cc.8.14.9113
Minuti, G., Cappuzzo, F., Duchnowska, R., Jassem, J., Fabi, A., O'brien, T., et al. (2012). Increased MET and HGF gene copy numbers are associated with trastuzumab failure in HER2-positive metastatic breast cancer. Br. J. Cancer 107, 793–799. doi: 10.1038/bjc.2012.335
Nair, J. K., Willoughby, J. L., Chan, A., Charisse, K., Alam, M. R., Wang, Q., et al. (2014). Multivalent N-acetylgalactosamine-conjugated siRNA localizes in hepatocytes and elicits robust RNAi-mediated gene silencing. J. Am. Chem. Soc. 136, 16958–16961. doi: 10.1021/ja505986a
Nara, T., Narita, S., Mingguo, H., Yoshioka, T., Koizumi, A., Numakura, K., et al. (2016). Altered miRNA expression in high-fat diet-induced prostate cancer progression. Carcinogenesis 37, 1129–1137. doi: 10.1093/carcin/bgw108
Nasser, M. W., Datta, J., Nuovo, G., Kutay, H., Motiwala, T., Majumder, S., et al. (2008). Down-regulation of micro-RNA-1 (miR-1) in lung cancer. Suppression of tumorigenic property of lung cancer cells and their sensitization to doxorubicin-induced apoptosis by miR-1. J. Biol. Chem. 283, 33394–33405. doi: 10.1074/jbc.M804788200
Niu, G., Li, B., Sun, J., and Sun, L. (2015). miR-454 is down-regulated in osteosarcomas and suppresses cell proliferation and invasion by directly targeting c-Met. Cell Prolif. 48, 348–355. doi: 10.1111/cpr.12187
Ofek, P., Calderon, M., Mehrabadi, F. S., Krivitsky, A., Ferber, S., Tiram, G., et al. (2016). Restoring the oncosuppressor activity of microRNA-34a in glioblastoma using a polyglycerol-based polyplex. Nanomedicine 12, 2201–2214. doi: 10.1016/j.nano.2016.05.016
Ohta, K., Hoshino, H., Wang, J., Ono, S., Iida, Y., Hata, K., et al. (2015). MicroRNA-93 activates c-Met/PI3K/Akt pathway activity in hepatocellular carcinoma by directly inhibiting PTEN and CDKN1A. Oncotarget 6, 3211–3224. doi: 10.18632/oncotarget.3085
Parikh, R. A., Wang, P., Beumer, J. H., Chu, E., and Appleman, L. J. (2014). The potential roles of hepatocyte growth factor (HGF)-MET pathway inhibitors in cancer treatment. Onco. Targets Ther. 7, 969–983. doi: 10.2147/OTT.S40241
Pazzaglia, L., Novello, C., Conti, A., Pollino, S., Picci, P., and Benassi, M. S. (2017). miR-152 down-regulation is associated with MET up-regulation in leiomyosarcoma and undifferentiated pleomorphic sarcoma. Cell Oncol. 40, 77–88. doi: 10.1007/s13402-016-0306-4
Peng, Y., Guo, J. J., Liu, Y. M., and Wu, X. L. (2014). MicroRNA-34A inhibits the growth, invasion and metastasis of gastric cancer by targeting PDGFR and MET expression. Biosci. Rep. 34:e00112. doi: 10.1042/BSR20140020
Pilotto, S., Carbognin, L., Karachaliou, N., Ma, P. C., Rosell, R., Tortora, G., et al. (2017). Tracking MET de-addiction in lung cancer: a road towards the oncogenic target. Cancer Treat Rev. 60, 1–11. doi: 10.1016/j.ctrv.2017.08.002
Ponzetto, C., Bardelli, A., Zhen, Z., Maina, F., Dalla Zonca, P., Giordano, S., et al. (1994). A multifunctional docking site mediates signaling and transformation by the hepatocyte growth factor/scatter factor receptor family. Cell 77, 261–271. doi: 10.1016/0092-8674(94)90318-2
Qu, L., Ding, J., Chen, C., Wu, Z. J., Liu, B., Gao, Y., et al. (2016). Exosome-transmitted IncARSR promotes sunitinib resistance in renal cancer by acting as a competing endogenous RNA. Cancer Cell 29, 653–668. doi: 10.1016/j.ccell.2016.03.004
Qu, W., Chen, X., Wang, J., Lv, J., and Yan, D. (2017). MicroRNA-1 inhibits ovarian cancer cell proliferation and migration through c-Met pathway. Clin. Chim. Acta. 473, 237–244. doi: 10.1016/j.cca.2017.07.008
Quinn, J. J., and Chang, H. Y. (2016). Unique features of long non-coding RNA biogenesis and function. Nat. Rev. Genet. 17, 47–62. doi: 10.1038/nrg.2015.10
Ragelle, H., Vandermeulen, G., and Preat, V. (2013). Chitosan-based siRNA delivery systems. J. Control Release 172, 207–218. doi: 10.1016/j.jconrel.2013.08.005
Ren, X. L., He, G. Y., Li, X. M., Men, H., Yi, L. Z., Lu, G. F., et al. (2016). MicroRNA-206 functions as a tumor suppressor in colorectal cancer by targeting FMNL2. J. Cancer Res. Clin. Oncol. 142, 581–592. doi: 10.1007/s00432-015-2053-8
Rokavec, M., Li, H., Jiang, L., and Hermeking, H. (2014). The p53/miR-34 axis in development and disease. J. Mol. Cell Biol. 6, 214–230. doi: 10.1093/jmcb/mju003
Ruvkun, G. (2001). Molecular biology. Glimpses of a tiny RNA world. Science 294, 797–799. doi: 10.1126/science.1066315
Saito, S., Morishima, K., Ui, T., Hoshino, H., Matsubara, D., Ishikawa, S., et al. (2015). The role of HGF/MET and FGF/FGFR in fibroblast-derived growth stimulation and lapatinib-resistance of esophageal squamous cell carcinoma. BMC Cancer 15:82. doi: 10.1186/s12885-015-1065-8
Salgia, R. (2017). MET in lung cancer: biomarker selection based on scientific rationale. Mol. Cancer Ther. 16, 555–565. doi: 10.1158/1535-7163.MCT-16-0472
Salmena, L., Poliseno, L., Tay, Y., Kats, L., and Pandolfi, P. P. (2011). A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell 146, 353–358. doi: 10.1016/j.cell.2011.07.014
Salzman, D. W., Nakamura, K., Nallur, S., Dookwah, M. T., Metheetrairut, C., Slack, F. J., et al. (2016). miR-34 activity is modulated through 5'-end phosphorylation in response to DNA damage. Nat. Commun. 7:10954. doi: 10.1038/ncomms10954
Shojaei, F., Lee, J. H., Simmons, B. H., Wong, A., Esparza, C. O., Plumlee, P. A., et al. (2010). HGF/c-Met acts as an alternative angiogenic pathway in sunitinib-resistant tumors. Cancer Res. 70, 10090–10100. doi: 10.1158/0008-5472.CAN-10-0489
Simone, B. A., Ly, D., Savage, J. E., Hewitt, S. M., Dan, T. D., Ylaya, K., et al. (2014). microRNA alterations driving acute and late stages of radiation-induced fibrosis in a murine skin model. Int. J. Radiat. Oncol. Biol. Phys. 90, 44–52. doi: 10.1016/j.ijrobp.2014.05.003
Solomon, B. J., Mok, T., Kim, D. W., Wu, Y. L., Nakagawa, K., Mekhail, T., et al. (2014). First-line crizotinib versus chemotherapy in ALK-positive lung cancer. N. Engl. J. Med. 371, 2167–2177. doi: 10.1056/NEJMoa1408440
Spigel, D. R., Ervin, T. J., Ramlau, R. A., Daniel, D. B., Goldschmidt, J. H. Jr., Blumenschein, G. R. Jr., et al. (2013). Randomized phase II trial of Onartuzumab in combination with erlotinib in patients with advanced non-small-cell lung cancer. J. Clin. Oncol. 31, 4105–4114. doi: 10.1200/JCO.2012.47.4189
Stamos, J., Lazarus, R. A., Yao, X., Kirchhofer, D., and Wiesmann, C. (2004). Crystal structure of the HGF beta-chain in complex with the Sema domain of the Met receptor. EMBO J. 23, 2325–2335. doi: 10.1038/sj.emboj.7600243
Su, W., Feng, S., Chen, X., Yang, X., Mao, R., Guo, C., et al. (2018). Silencing of long noncoding RNA MIR22HG triggers cell survival/death signaling via oncogenes YBX1, MET, and p21 in lung cancer. Cancer Res. 78, 3207–3219. doi: 10.1158/0008-5472.CAN-18-0222
Sun, C., Liu, Z., Li, S., Yang, C., Xue, R., Xi, Y., et al. (2015a). Down-regulation of c-Met and Bcl2 by microRNA-206, activates apoptosis, and inhibits tumor cell proliferation, migration and colony formation. Oncotarget 6, 25533–25574. doi: 10.18632/oncotarget.4575
Sun, C., Sang, M., Li, S., Sun, X., Yang, C., Xi, Y., et al. (2015b). Hsa-miR-139–5p inhibits proliferation and causes apoptosis associated with down-regulation of c-Met. Oncotarget 6, 39756–39792. doi: 10.18632/oncotarget.5476
Sun, C. C., Li, S. J., Zhang, F., Pan, J. Y., Wang, L., Yang, C. L., et al. (2016). Hsa-miR-329 exerts tumor suppressor function through down-regulation of MET in non-small cell lung cancer. Oncotarget 7, 21510–21526. doi: 10.18632/oncotarget.7517
Sun, L., Bian, G., Meng, Z., Dang, G., Shi, D., and Mi, S. (2015c). MiR-144 inhibits uveal melanoma cell proliferation and invasion by regulating c-Met expression. PLoS ONE 10:e0124428. doi: 10.1371/journal.pone.0124428
Sun, Y., Zhao, Y., Zhao, X., Lee, R. J., Teng, L., and Zhou, C. (2017). Enhancing the therapeutic delivery of oligonucleotides by chemical modification and nanoparticle encapsulation. Molecules 22:E1724. doi: 10.3390/molecules22101724
Sun, Z., Zhang, T., and Chen, B. (2019). Long non-coding RNA metastasis-associated lung adenocarcinoma transcript 1 (MALAT1) promotes proliferation and metastasis of osteosarcoma cells by targeting c-Met and SOX4 via miR-34a/c-5p and miR-449a/b. Med. Sci. Monit. 25, 1410–1422. doi: 10.12659/MSM.912703
Sun, Z. Y., Jian, Y. K., Zhu, H. Y., and Li, B. (2019). lncRNAPVT1 targets miR-152 to enhance chemoresistance of osteosarcoma to gemcitabine through activating c-MET/PI3K/AKT pathway. Pathol. Res. Pract. 215, 555–563. doi: 10.1016/j.prp.2018.12.013
Tan, S., Li, R., Ding, K., Lobie, P. E., and Zhu, T. (2011). miR-198 inhibits migration and invasion of hepatocellular carcinoma cells by targeting the HGF/c-MET pathway. FEBS Lett. 585, 2229–2234. doi: 10.1016/j.febslet.2011.05.042
Taylor, K., Howard, C. B., Jones, M. L., Sedliarou, I., Macdiarmid, J., Brahmbhatt, H., et al. (2015). Nanocell targeting using engineered bispecific antibodies. MAbs 7, 53–65. doi: 10.4161/19420862.2014.985952
Tomihara, H., Yamada, D., Eguchi, H., Iwagami, Y., Noda, T., Asaoka, T., et al. (2017). MicroRNA-181b-5p, ETS1, and the c-Met pathway exacerbate the prognosis of pancreatic ductal adenocarcinoma after radiation therapy. Cancer Sci. 108, 398–407. doi: 10.1111/cas.13159
Trusolino, L., Bertotti, A., and Comoglio, P. M. (2010). MET signalling: principles and functions in development, organ regeneration and cancer. Nat. Rev. Mol. Cell Biol. 11, 834–848. doi: 10.1038/nrm3012
Ulitsky, I., Shkumatava, A., Jan, C. H., Sive, H., and Bartel, D. P. (2011). Conserved function of lincRNAs in vertebrate embryonic development despite rapid sequence evolution. Cell 147, 1537–1550. doi: 10.1016/j.cell.2011.11.055
Umezu, T., Imanishi, S., Azuma, K., Kobayashi, C., Yoshizawa, S., Ohyashiki, K., et al. (2017). Replenishing exosomes from older bone marrow stromal cells with miR-340 inhibits myeloma-related angiogenesis. Blood Adv. 1, 812–823. doi: 10.1182/bloodadvances.2016003251
Vennin, C., Spruyt, N., Dahmani, F., Julien, S., Bertucci, F., Finetti, P., et al. (2015). H19 non coding RNA-derived miR-675 enhances tumorigenesis and metastasis of breast cancer cells by downregulating c-Cbl and Cbl-b. Oncotarget 6, 29209–29223. doi: 10.18632/oncotarget.4976
Wan, L., Zhu, L., Xu, J., Lu, B., Yang, Y., Liu, F., et al. (2014). MicroRNA-409-3p functions as a tumor suppressor in human lung adenocarcinoma by targeting c-Met. Cell Physiol. Biochem. 34, 1273–1290. doi: 10.1159/000366337
Wang, G., Cai, C., and Chen, L. (2016). MicroRNA-3666 regulates thyroid carcinoma cell proliferation via MET. Cell Physiol. Biochem. 38, 1030–1039. doi: 10.1159/000443054
Wang, M., Sun, Y., Xu, J., Lu, J., Wang, K., Yang, D. R., et al. (2018). Preclinical studies using miR-32–5p to suppress clear cell renal cell carcinoma metastasis via altering the miR-32–5p/TR4/HGF/Met signaling. Int. J. Cancer 143, 100–112. doi: 10.1002/ijc.31289
Wang, N., Zhang, Y., and Liang, H. (2018). MicroRNA-598 inhibits cell proliferation and invasion of glioblastoma by directly targeting metastasis associated in colon cancer-1 (MACC1). Oncol. Res. 26, 1275–1283. doi: 10.3727/096504018X15185735627746
Wang, Q. Y., Zhou, C. X., Zhan, M. N., Tang, J., Wang, C. L., Ma, C. N., et al. (2018). MiR-133b targets Sox9 to control pathogenesis and metastasis of breast cancer. Cell Death Dis. 9:752. doi: 10.1038/s41419-018-0715-6
Wang, S., Zhang, Y., Yuan, S., and Ji, X. (2018). MicroRNA485 targets MACC1 and inhibits cervical cancer cell proliferation and invasion. Mol. Med. Rep. 18, 2407–2416. doi: 10.3892/mmr.2018.9186
Wang, W., Dong, J., Wang, M., Yao, S., Tian, X., Cui, X., et al. (2018). miR-148a-3p suppresses epithelial ovarian cancer progression primarily by targeting c-Met. Oncol. Lett. 15, 6131–6136. doi: 10.3892/ol.2018.8110
Wang, Y., Tai, Q., Zhang, J., Kang, J., Gao, F., Zhong, F., et al. (2019). MiRNA-206 inhibits hepatocellular carcinoma cell proliferation and migration but promotes apoptosis by modulating cMET expression. Acta. Biochim. Biophys. Sin. 51, 243–253. doi: 10.1093/abbs/gmy119
Wang, Y., Zhu, P., Wang, J., Zhu, X., Luo, J., Meng, S., et al. (2018). Long noncoding RNA lncHand2 promotes liver repopulation via c-Met signaling. J. Hepatol. 69, 861–872. doi: 10.1016/j.jhep.2018.03.029
Wei, B., Huang, Q. Y., Huang, S. R., Mai, W., and Zhong, X. G. (2015). MicroRNA34a attenuates the proliferation, invasion and metastasis of gastric cancer cells via downregulation of MET. Mol. Med. Rep. 12, 5255–5261. doi: 10.3892/mmr.2015.4110
Wu, H., Tao, J., Li, X., Zhang, T., Zhao, L., Wang, Y., et al. (2017). MicroRNA-206 prevents the pathogenesis of hepatocellular carcinoma by modulating expression of met proto-oncogene and cyclin-dependent kinase 6 in mice. Hepatology 66, 1952–1967. doi: 10.1002/hep.29374
Wu, J., Ji, A., Wang, X., Zhu, Y., Yu, Y., Lin, Y., et al. (2015). MicroRNA-195-5p, a new regulator of Fra-1, suppresses the migration and invasion of prostate cancer cells. J. Transl. Med. 13:289. doi: 10.1186/s12967-015-0650-6
Wu, P., Cai, J., Chen, Q., Han, B., Meng, X., Li, Y., et al. (2019). Lnc-TALC promotes O6-methylguanine-DNA methyltransferase expression via regulating the c-Met pathway by competitively binding with miR-20b-3p. Nat. Commun. 10:2045. doi: 10.1038/s41467-019-10025-2
Wu, Y., Wan, X., Ji, F., Song, Z., and Fang, X. (2018). Serum miR-658 induces metastasis of gastric cancer by activating PAX3-MET pathway: a population-based study. Cancer Biomark. 22, 111–118. doi: 10.3233/CBM-171045
Wu, Z., Wang, H., Fang, S., and Xu, C. (2015). MiR-449c inhibits gastric carcinoma growth. Life Sci. 137, 14–19. doi: 10.1016/j.lfs.2015.06.020
Wu, Z. S., Wu, Q., Wang, C. Q., Wang, X. N., Huang, J., Zhao, J. J., et al. (2011). miR-340 inhibition of breast cancer cell migration and invasion through targeting of oncoprotein c-Met. Cancer 117, 2842–2852. doi: 10.1002/cncr.25860
Xing, F., Liu, Y., Wu, S. Y., Wu, K., Sharma, S., Mo, Y. Y., et al. (2018). Loss of XIST in breast cancer activates MSN-c-Met and reprograms microglia via exosomal miRNA to promote brain metastasis. Cancer Res. 78, 4316–4330. doi: 10.1158/0008-5472.CAN-18-1102
Xu, P., Li, Y., Yang, S., Yang, H., Tang, J., and Li, M. (2015). Micro-ribonucleic acid 143 (MiR-143) inhibits oral squamous cell carcinoma (OSCC) cell migration and invasion by downregulation of phospho-c-Met through targeting CD44 v3. Oral Surg. Oral Med. Oral Pathol. Oral Radiol. 120, 43–51. doi: 10.1016/j.oooo.2015.02.486
Xu, X., Zhu, Y., Liang, Z., Li, S., Xu, X., Wang, X., et al. (2016). c-Met and CREB1 are involved in miR-433-mediated inhibition of the epithelial-mesenchymal transition in bladder cancer by regulating Akt/GSK-3beta/Snail signaling. Cell Death Dis. 7:e2088. doi: 10.1038/cddis.2015.274
Xue, H. Y., Guo, P., Wen, W. C., and Wong, H. L. (2015). Lipid-based nanocarriers for RNA delivery. Curr. Pharm. Des. 21, 3140–3147. doi: 10.2174/1381612821666150531164540
Yan, K., Gao, J., Yang, T., Ma, Q., Qiu, X., Fan, Q., et al. (2012). MicroRNA-34a inhibits the proliferation and metastasis of osteosarcoma cells both in vitro and in vivo. PLoS ONE 7:e33778. doi: 10.1371/journal.pone.0033778
Yang, C. H., Zhang, X. Y., Zhou, L. N., Wan, Y., Song, L. L., Gu, W. L., et al. (2018). LncRNA SNHG8 participates in the development of endometrial carcinoma through regulating c-MET expression by miR-152. Eur. Rev. Med. Pharmacol. Sci. 22, 1629–1637. doi: 10.26355/eurrev_201803_14698
Yang, Y. M., Lee, C. G., Koo, J. H., Kim, T. H., Lee, J. M., An, J., et al. (2015). Galpha12 overexpressed in hepatocellular carcinoma reduces microRNA-122 expression via HNF4alpha inactivation, which causes c-Met induction. Oncotarget 6, 19055–19069. doi: 10.18632/oncotarget.3957
Yeung, C. L., Tsang, T. Y., Yau, P. L., and Kwok, T. T. (2017). Human papillomavirus type 16 E6 suppresses microRNA-23b expression in human cervical cancer cells through DNA methylation of the host gene C9orf3. Oncotarget 8, 12158–12173. doi: 10.18632/oncotarget.14555
Yoon, J. H., Abdelmohsen, K., and Gorospe, M. (2014). Functional interactions among microRNAs and long noncoding RNAs. Semin. Cell Dev. Biol. 34, 9–14. doi: 10.1016/j.semcdb.2014.05.015
Yu, K. (2017). The Rs4938723 Polymorphism Reduces Expression of MicroRNA-34b and Increases the Risk of Recurrence after Endoscopic Dissection in Early Gastric Cancer. Cell Physiol. Biochem. 43, 1235–1246. doi: 10.1159/000481764
Zhan, F. B., Zhang, X. W., Feng, S. L., Cheng, J., Zhang, Y., Li, B., et al. (2019). MicroRNA-206 reduces osteosarcoma cell malignancy in vitro by targeting the PAX3-MET axis. Yonsei Med. J. 60, 163–173. doi: 10.3349/ymj.2019.60.2.163
Zhang, B., Ma, L., Wei, J., Hu, J., Zhao, Z., Wang, Y., et al. (2016a). miR-137 suppresses the phosphorylation of AKT and improves the dexamethasone sensitivity in multiple myeloma cells via targeting MITF. Curr. Cancer Drug Targets 16, 807–817. doi: 10.2174/1568009616666160203114140
Zhang, H., Deng, T., Liu, R., Bai, M., Zhou, L., Wang, X., et al. (2017a). Exosome-delivered EGFR regulates liver microenvironment to promote gastric cancer liver metastasis. Nat. Commun. 8:15016. doi: 10.1038/ncomms15016
Zhang, J., Fa, X., and Zhang, Q. (2019). MicroRNA206 exerts antioncogenic functions in esophageal squamous cell carcinoma by suppressing the cMet/AKT/mTOR pathway. Mol. Med. Rep. 19, 1491–1500. doi: 10.3892/mmr.2018.9775
Zhang, J., Feng, S., Su, W., Bai, S., Xiao, L., Wang, L., et al. (2017b). Overexpression of FAM83H-AS1 indicates poor patient survival and knockdown impairs cell proliferation and invasion via MET/EGFR signaling in lung cancer. Sci. Rep. 7:42819. doi: 10.1038/srep42819
Zhang, L., Xia, L., Zhao, L., Chen, Z., Shang, X., Xin, J., et al. (2015). Activation of PAX3-MET pathways due to miR-206 loss promotes gastric cancer metastasis. Carcinogenesis 36, 390–399. doi: 10.1093/carcin/bgv009
Zhang, P., Li, S., Lv, C., Si, J., Xiong, Y., Ding, L., et al. (2018). BPI-9016M, a c-Met inhibitor, suppresses tumor cell growth, migration and invasion of lung adenocarcinoma via miR203-DKK1. Theranostics 8, 5890–5902. doi: 10.7150/thno.27667
Zhang, Y., Schiff, D., Park, D., and Abounader, R. (2014). MicroRNA-608 and microRNA-34a regulate chordoma malignancy by targeting EGFR, Bcl-xL and MET. PLoS ONE 9:e91546. doi: 10.1371/journal.pone.0091546
Zhang, Z., Kong, Y., Yang, W., Ma, F., Zhang, Y., Ji, S., et al. (2016b). Upregulation of microRNA-34a enhances the DDP sensitivity of gastric cancer cells by modulating proliferation and apoptosis via targeting MET. Oncol. Rep. 36, 2391–2397. doi: 10.3892/or.2016.5016
Zhao, Y., Tu, M. J., Wang, W. P., Qiu, J. X., Yu, A. X., and Yu, A. M. (2016). Genetically engineered pre-microRNA-34a prodrug suppresses orthotopic osteosarcoma xenograft tumor growth via the induction of apoptosis and cell cycle arrest. Sci. Rep. 6:26611. doi: 10.1038/srep26611
Zhen, L., Yun-Hui, L., Hong-Yu, D., Jun, M., and Yi-Long, Y. (2016). Long noncoding RNA NEAT1 promotes glioma pathogenesis by regulating miR-449b-5p/c-Met axis. Tumour Biol. 37, 673–683. doi: 10.1007/s13277-015-3843-y
Zhen, Q., Liu, J., Gao, L., Liu, J., Wang, R., Chu, W., et al. (2015). MicroRNA-200a targets EGFR and c-Met to inhibit migration, invasion, and gefitinib resistance in non-small cell lung cancer. Cytogenet. Genome. Res. 146, 1–8. doi: 10.1159/000434741
Zheng, Z., Yan, D., Chen, X., Huang, H., Chen, K., Li, G., et al. (2015). MicroRNA-206: effective inhibition of gastric cancer progression through the c-Met pathway. PLoS ONE 10, e0128751. doi: 10.1371/journal.pone.0128751
Zhou, H., Liu, Y., Xiao, L., Hu, Z., and Xia, K. (2017). Overexpression of MicroRNA-27b inhibits proliferation, migration, and invasion via suppression of MET expression. Oncol. Res. 25, 147–154. doi: 10.3727/096504016X14732772150505
Zhou, J. Y., Chen, X., Zhao, J., Bao, Z., Chen, X., Zhang, P., et al. (2014). MicroRNA-34a overcomes HGF-mediated gefitinib resistance in EGFR mutant lung cancer cells partly by targeting MET. Cancer Lett. 351, 265–271. doi: 10.1016/j.canlet.2014.06.010
Zhu, H., Jiang, X., Zhou, X., Dong, X., Xie, K., Yang, C., et al. (2018a). Neuropilin-1 regulated by miR-320 contributes to the growth and metastasis of cholangiocarcinoma cells. Liver Int. 38, 125–135. doi: 10.1111/liv.13495
Zhu, K., and Wang, W. (2016). Green tea polyphenol EGCG suppresses osteosarcoma cell growth through upregulating miR-1. Tumour Biol. 37, 4373–4382. doi: 10.1007/s13277-015-4187-3
Zhu, L., Xue, F., Xu, X., Xu, J., Hu, S., Liu, S., et al. (2018b). MicroRNA-198 inhibition of HGF/c-MET signaling pathway overcomes resistance to radiotherapy and induces apoptosis in human non-small-cell lung cancer. J. Cell Biochem. 119, 7873–7886. doi: 10.1002/jcb.27204
Keywords: microRNAs, long non-coding RNAs, hepatocyte growth factor, c-Met, cancer, drug-resistance
Citation: Zhan H, Tu S, Zhang F, Shao A and Lin J (2020) MicroRNAs and Long Non-coding RNAs in c-Met-Regulated Cancers. Front. Cell Dev. Biol. 8:145. doi: 10.3389/fcell.2020.00145
Received: 19 November 2019; Accepted: 21 February 2020;
Published: 11 March 2020.
Edited by:
Xin Xu, College of Medicine, Zhejiang University, ChinaReviewed by:
Hai-long Piao, Dalian Institute of Chemical Physics (CAS), ChinaRizaldy Paz Scott, Northwestern University, United States
Copyright © 2020 Zhan, Tu, Zhang, Shao and Lin. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Anwen Shao, 21118116@zju.edu.cn; anwenshao@sina.com; Jun Lin, linjun@zju.edu.cn
 Hong Zhan
Hong Zhan Sheng Tu2
Sheng Tu2 Anwen Shao
Anwen Shao