
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
MINI REVIEW article
Front. Cell Dev. Biol. , 20 February 2020
Sec. Signaling
Volume 8 - 2020 | https://doi.org/10.3389/fcell.2020.00095
This article is part of the Research Topic Resident and Ectopic FGF Signaling in Development and Disease View all 9 articles
Fibroblast growth factor (FGF) receptor 4 (FGFR4) belongs to a family of tyrosine kinase receptor. FGFR4 is highly activated in certain types of cancer and its activation is closely associated with its specific ligand, FGF19. Indeed, FGF19-FGFR4 signaling is implicated in many cellular processes including cell proliferation, migration, metabolism, and differentiation. Since active FGF19-FGFR4 signaling acts as an oncogenic pathway in certain types of cancer, the development and therapeutic evaluation of FGFR4-specific inhibitors in cancer patients is a topic of significant interest. In this review, we aim to provide an updated overview of currently-available FGFR4 inhibitors and their ongoing clinical trials, as well as upcoming potential therapeutics. Further, we examined the possibility of enhancing the therapeutic efficiency of FGFR4 inhibitors in cancer patients. We also discussed the underlying molecular mechanisms of oncogenic activation of FGFR4 by FGF19.
The fibroblast growth factor receptors (FGFRs) consist of four highly conserved transmembrane receptor tyrosine kinases (FGFR1-4) (Lin and Desnoyers, 2012). When FGFs bind to their specific FGFRs, they subsequently activate downstream signaling and regulate many biological processes including embryonic development, cell proliferation, differentiation, and tissue repair (Wilkie et al., 1995). FGF-FGFR signaling is dysregulated in many diseases, including cancers. Compared to the other three FGFR family members, the function of the FGFR4 signaling pathway in cancer is less well-characterized. Recently, it has been reported that the activation of FGF19-FGFR4 signaling is closely associated with cancer development and progression (Zhang et al., 2011; Chen et al., 2019), suggesting that FGF19-FGFR4 might be an attractive target for effective anticancer therapeutics (Schadt et al., 2018; Gao et al., 2019).
FGFR4 is a member of the FGFR family, contains tyrosine kinase domains, and plays a critical role in embryonic development, tissue repair, tumor angiogenesis, and tumor progression (Wu et al., 2011). The human FGFR4 gene is located on the long arm of chromosome 5 (5q35.1) and spans a genomic region of about 11 kb. The FGFR4 gene has five transcript variants with three of them encoding the FGFR4 isoform 1 with 802 amino acids. The estimated molecular weight of FGFR4 isoform 1 is around 95–110 kDa, depending on its glycosylation levels (Raja et al., 2019). The basic protein domain structure of the FGFR4 is composed of four parts including a signaling peptide, three extracellular immunoglobulin (Ig) domains, a transmembrane domain, and an intracellular tyrosine kinase domain (Figure 1A). Similar with FGFR1-3, the extracellular region of FGFR4 has Ig-like domains (IgI, IgII, and IgIII), which support specific ligand-binding, including FGF19. Unlike FGFR1-3, FGFR4 does not have splice variants in IgIII, which generates the IIIb and IIIc transcript variants encoding different receptor isoforms. Therefore, single ligand binding domain of FGFR4 enables high-affinity bindings of its specific ligands, suggesting that it would be possible to develop highly specific inhibitors (Touat et al., 2015). Genetic alterations of FGFRs were found in 7.1% of cancers. Compared with the other three members of the FGFR family (about 7%), genetic alternations of FGFR4 (0.5%) were relatively infrequent. Gene amplification was most frequent type of alteration in all FGFR4 gene alterations with frequency of 69% (Presta et al., 2017). Overexpression of FGFR4 has been reported in several solid tumors including hepatocellular carcinoma (HCC), breast cancer, oropharyngeal squamous cell carcinoma (OPSCC), and pancreatic cancer (Lang and Teng, 2019). High expression of FGFR4 was reported to be associated with poor overall survival of NSCLC (Huang et al., 2015) and malignant pleural mesothelioma (Vlacic et al., 2019). Particularly, FGFR4 expression was significantly upregulated in most liver cancer cases, and enhanced FGF19-FGFR4 signaling is linked to HCC progression, metastasis, and poor survival (Sawey et al., 2011). Although high expression of FGFR4 was observed in OPSCC and pancreatic cancer, FGFR4 does not show prognostic significance in these cancers (Motoda et al., 2011; Koole et al., 2015).
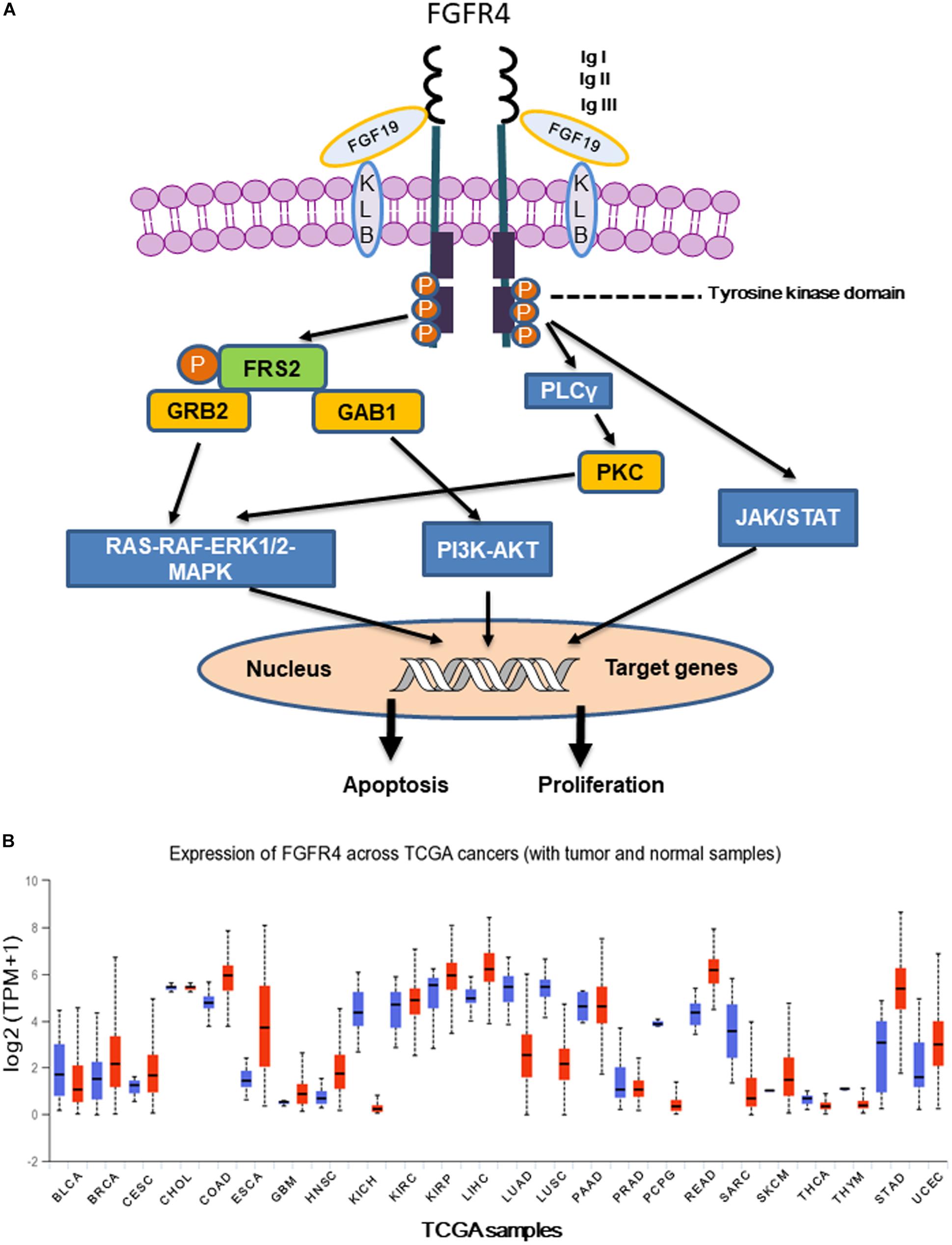
Figure 1. (A) Following FGFR4 and Klotho (KLB) bind to FGF19, activated FGFR4 forms homo- or heterodimer and subsequently activates multiple downstream signaling pathways including Ras-Raf- MAPK, PI3K-AKT, EMT, and JAK/STAT. (B) Expression of FGFR4 in various The Cancer Genome Atlas (TCGA) cancer types by http://ualcan.path.uab.edu/cgi-bin/Pan-cancer.pl?genenam=FGFR4, X axis represents 33 TCGA different cancer types (ACC, adrenocortical carcinoma; BLCA, bladder urothelial carcinoma; BRCA, breast invasive carcinoma; CESC, cervical squamous cell carcinoma and endocervical adenocarcinoma; CHOL, cholangio carcinoma; COAD, colon adenocarcinoma; DLBC, lymphoid neoplasm diffuse large B-cell lymphoma; ESCA, esophageal carcinoma; GBM, glioblastoma multiforme; HNSC, head and neck squamous cell carcinoma; KICH, kidney chromophobe; KIRC, kidney renal clear cell carcinoma; KIRP, kidney renal papillary cell carcinoma; LAML, acute myeloid leukemia; LGG, brain lower grade glioma; LIHC, liver hepatocellular carcinoma; LUAD, lung adenocarcinoma; LUSC, lung squamous cell carcinoma; MESO, mesothelioma; OV, ovarian serous cystadenocarcinoma; PAAD, pancreatic adenocarcinoma; PCPG, pheochromocytoma and paraganglioma; PRAD, prostate adenocarcinoma; READ, rectum adenocarcinoma; SARC, sarcoma; SKCM, skin cutaneous melanoma; STAD, stomach adenocarcinoma; TGCT, testicular germ cell tumors; THCA, thyroid carcinoma; THYM, thymoma; UCEC, uterine corpus endometrial carcinoma; UCS, uterine carcinosarcoma; UVM, uveal melanoma). Y axis represents log2 (TPM + 1) transformed expression data for plotting (TPM, transcripts per million).
The human FGF family is comprised of 22 members that are divided into seven subfamilies according to their sequences and biochemical functions. FGFs are involved in broad biological processes including embryogenesis, angiogenesis, tissue homeostasis, and cancer progression (Presta et al., 2017). The FGF19 subfamily is called hormone-like FGFs and includes FGF19, FGF21, and FGF23. Unlike other FGF families that bind to heparin-sulfate proteoglycan for paracrine signaling by FGFRs, FGF19 subfamily binds to FGFRs and its co-receptor, Klotho, to transfer endocrine signaling (Katoh, 2016). β-Klotho (KLB) forms a complex with FGFR4, enhancing the affinity between FGF19 and FGFR4 (Repana and Ross, 2015). Promoter hypermethylation induced downregulation of KLB was frequently seen in breast, colon, pancreatic and stomach cancers (Dallol et al., 2015). In contrast, elevated expression of KLB and FGFR4 was reported in HCC (Poh et al., 2012), suggesting dual functions of KLB in cancer development and progression. FGF19 subfamily functions as a secreted endocrine signal that regulates various metabolic process. FGF19 promotes protein synthesis by activating mTOR signaling, and enhances glycogen synthesis through inhibition of GSK3α and GSK3β. In contrast, FGF19 reduces gluconeogenesis by inhibiting cAMP regulatory element binding protein/PGC1A axis (Prieto-Dominguez et al., 2018). The FGF19 genomic locus is on the chromosome 11q13.3, and is frequently amplified in human cancers. For example, amplification of the FGF19 genomic locus including the well-established proto-oncogene CCND1, was found in liver cancer, breast cancer, lung cancer, bladder cancer and esophageal cancer (Sawey et al., 2011; Hoover et al., 2015; Tiong et al., 2016; Zhang et al., 2017). Hyperactivation of FGFR4 by FGF19 was reported in colon cancer cells and hepatocellar carcinoma. An animal model showed that a neutralizing antibody against FGF19 was able to abrogate liver cancer development in transgenic mice and inhibited xenograft tumors from HCC and colon cancer, suggesting feasibility of FGF19 as a therapeutic target (Desnoyers et al., 2008; French et al., 2012). Particularly, the functions of FGF19 are well-characterized in HCC. FGF19 gene expression is significantly upregulated in HCC tissues compared to normal adjacent liver tissues, which suggests that the amplified FGF19 genomic locus might be directly linked to upregulated FGF19 gene expression (Zhao H. et al., 2016). In addition, serum FGF19 levels are significantly higher in preoperative HCC patients than post-operative HCC patients, suggesting that FGF19 may serve as a non-invasive biomarker of disease (Miura et al., 2012). Recently, Hyeon et al. (2013) analyzed 281 patients who underwent curative resection and found that 48% of the HCC tissues had overexpressed FGF19 protein. FGF19 expression was positively associated with a larger tumor size, more advanced Barcelona Clinic Liver Cancer (BCLC) stage, early recurrence, and poor prognosis.
Accumulating evidence indicates that FGFR4 plays a critical role in cancer progression and metastasis, particularly in those cancer patients who have FGF19 amplification. Klotho interacts with FGFR4 forming 1:1 heterocomplex to facilitate binding between FGF19 and FGFR4. Later FGF19 binds to heterocomplex of Klotho and FGFR4 to form homodimer receptor complex, activating downstream signaling pathways (Gao et al., 2017; Lang and Teng, 2019; Raja et al., 2019). Here, we summarized and discussed four main signaling pathways downstream of FGF19-FGFR4 signaling including Ras-Raf-MAPK, PI3K-AKT, epithelial–mesenchymal transition (EMT), and the signal transducer and activator of transcription (STAT) pathway (Figure 1A).
Ras/Raf/MAPK and PI3K-AKT pathways are two major pathways that are frequently dysregulated in cancer. The Ras/Raf/MAPK pathway is aberrantly activated in a number of cancers, and is involved in the regulation of cell proliferation, migration, survival, and apoptosis (Cary et al., 1999; Stacey, 2003; Boonstra et al., 2013). When FGFR4 is activated, phosphorylated FGFR4 recruits and phosphorylates intracellular specific adaptor protein FGFR-substrate-2 (FRS2), which in turn recruits the adaptor proteins GRB2 and SOS, subsequently activating the Ras-Raf-MAPK pathway. Meanwhile, GRB2 binds to GAB1, leading to activation of the PI3K-AKT pathway (Touat et al., 2015). PI3K-AKT signaling leads to decreased apoptosis and increased cell growth/proliferation. High activation of the PI3K-AKT pathway has been noted in a number of cancers including prostate cancer (Toren and Zoubeidi, 2014), lung cancer (Qian et al., 2006), and HCC (Zhou et al., 2011). On the other hand, once activated, FGFR4 phosphorylates phospholipase Cγ (PLCγ) which in turn activates protein kinase C (PKC), and subsequently stimulates the Ras/Raf/MAPK pathway, contributing to cell proliferation and migration (Goetz and Mohammadi, 2013).
The FGF19-FGFR4 axis has been reported to promote the metastatic potential of HCC. When activated, the FGF19-FGFR4 pathway enhances GSK3β-βcatenin signaling, consequently inducing EMT and resulting in increased HCC metastasis (Goetz and Mohammadi, 2013; Zhao H. et al., 2016). In addition, activated FGFR4 stimulates AKT, ERK1/2, and Src, promoting the invasive activity of colon cancer (CRC) cells. Inversely, knockdown of FGFR4 reduces the migratory and invasive ability of CRC cells by upregulating epithelial marker E-cadherin and downregulating mesenchymal marker Snail, suggesting a pivotal role of FGFR4 in CRC metastasis (Peláez-García et al., 2013). Moreover, increased activation of the FGFR4-GSK3β-βcatenin axis was also reported in CRC metastasis. FOXC1 was found to directly bind to the promoter region of FGFR4 and enhance FGFR4 expression. High levels of FOXC1 have been reported to be correlated with CRC metastasis, recurrence, and poor prognosis (Liu et al., 2018).
The JAKs-STATs pathway is an efficient and well-organized system which is principally dedicated to the regulation of gene expression (Zhao C. G. et al., 2016). Basically, activation of JAKs-STATs pathway requires cytokine-mediated phosphorylation and activation of non-receptor tyrosine kinases, JAKs (Nicolas et al., 2013). Once FGF19 binds to FGFR4/Klotho complex, FGF19-dependent FGFR4 dimerization leads to a conformational change in its structure, which facilitates intermolecular transphosphorylation and recruits downstream adaptor proteins for further activation of signaling cascades (Touat et al., 2015). It has been reported that FGF19 acts through the receptor complex FGFR4-KLB to activate JAKs/STATs signaling. FGF19-FGFR4 dimerization induces JAKs phosphorylation and activation, and activated JAKs phosphorylate STATs (Zhou et al., 2014). Activated STATs then form homo- or heterodimers to translocate to the nucleus where they carry out their function as a transcription factor (Nicolas et al., 2013). In addition, a single nucleotide polymorphism, rs351855 (G/A) results in an amino acid change in the FGFR4 protein, producing the Arg388-variant. The FGFR4 Arg388-variant enhances STAT3 signaling, promotes tumor progression, and is associated with poor prognosis of multiple cancers (Ulaganathan et al., 2015; Kogan et al., 2018). In the Rhabdomyosarcoma (RMS), mutations in FGFR4 tyrosine kinase domain (K535 and E550) might activate STAT3 signaling, increasing tumor growth and metastatic potential in mice model (Taylor et al., 2009).
Increasing evidence indicates that the FGF19-FGFR4 pathway plays a pivotal role in multiprocess of cancer initiation and progression (Wu et al., 2010, 2011). Recombinant FGF19 promotes proliferation and the invasive ability of HCC cells, however, knockdown of FGF19 or FGFR4 by siRNA inhibits proliferation and invasion and induces apoptosis of HCC cells. FGF19 is highly expressed in HCC, and patients with low FGF19 expression have prolonged 5-year survival rates compared to those patients with high FGF19 expression (Miura, 2012). Overexpression of FGF19 in transgenic mice has been shown to promote liver tumor formation by enhancing the expression of α-fetoprotein (AFP), an oncofetal protein used as a marker for neoplastic transformation of hepatocytes (Nicholes et al., 2002). Meanwhile, FGFR4 has been reported to contribute to HCC progression by regulating AFP secretion, proliferation, and preventing apoptosis of HCC cells (Han et al., 2009).
In breast cancer, upregulation of FGFR4 is partly due to gene amplification. Co-expression of FGFR4 and FGF19 was observed in about 30% of primary breast cancers. Furthermore, FGFR4 and FGF19 expressions were significantly associated with phosphorylated AKT expression, suggesting that FGFR4 might be a therapeutic target for these patients (Jaakkola et al., 1993; Penault-Llorca et al., 1995; Tiong et al., 2016). In addition, FGFR4 overexpression is closely associated with advanced tumor stage and poor prognosis in astrocytomas. FGFR4 was also reported to be a prognostic marker in advanced stage, high grade serous ovarian cancer. Inhibiting FGFR4 and its ligand markedly reduces ovarian tumor growth both in vitro and in vivo, indicating that FGFR4 could also be a promising therapeutic target in ovarian cancer that has high expression of FGFR4 (Zaid et al., 2013). Similar results were also observed in prostate cancer. The expression of FGFR4 was markedly increased in prostate cancer and was associated with higher Gleason score and worse outcomes (Sahadevan et al., 2007; Sugiyama et al., 2010). FGFR4 transmembrane domain polymorphism (FGFR4 Gly388Arg) has been reported to be associated with increased risks of breast and prostate cancer in Asian population (Xu et al., 2010). FGFR4-Arg388 could activate ERK, c-scr and STAT3 signaling, contributing to tumor progression. Increased risk for developing the malignant tumor was reported in prostate, however, decreased risk was observed in head and neck squamous cell carcinoma (HNSCC) and soft tissue sarcomas for FGFR4-Arg388 allele. Accumulating evidences also suggested that FGFR4-Arg388 allele was frequently seen in rapid tumor progression, increased metastasis and advanced stage of patients with breast, prostate, colon and lung cancer (Heinzle et al., 2014).
Interaction between FGF19 and FGFR4 plays an essential role in colorectal tumorigenesis. Blocking either FGF19 or FGFR4 significantly reduces in vivo xenograft tumor growth of colon cancer cells (Pai et al., 2008). In vitro, knockdown of FGFR4 suppresses colon cancer cell proliferation and migration, again indicating that FGFR4 is a promising therapeutic target in colon cancer (Heinzle et al., 2012).
The main obstacle of targeted cancer therapy is the development of drug resistance. Significantly increased FGFR4 expression was observed in doxorubicin-resistant breast cancer cells, and knockdown of FGFR4 could increase sensitivity to doxorubicin. Mechanistically, expression of the anti-apoptotic protein BCL-XL was induced by increased FGFR4 through the MAPK signaling cascade (Roidl et al., 2009). Furthermore, activated FGF19-FGFR4 signaling has been reported to increase resistance against doxorubicin in basal-like breast cancer. Treatment of high-expressing FGF19 or FGFR4 positive breast cancer cells with an antibody against FGF19 or siRNA to FGF19 sensitizes the cells to doxorubicin (Tiong et al., 2016). Silencing FGFR4 sensitizes CRC cells to 5-fluorouracil (5-FU) or oxaliplatin treatment (Zhao H. et al., 2016).
The importance of FGF19-FGFR4 signaling is well-characterized in HCC. It has been reported that the activation of FGF19-FGFR4 signaling is important in multiple tyrosine kinase inhibitor (TKI), sorafenib resistance for the treatment of HCC. Overexpression of FGF19 could suppress sorafenib-induced, ROS-dependent apoptosis (Gao et al., 2017). Elevated FGF19-FGFR4 signaling is also able to reduce ROS-mediated apoptosis through inhibition of the caspase-3 pathway in colonic epithelial cells (Valastyan and Weinberg, 2011).
Currently, several FGFRs inhibitors are under investigation for the treatment of various cancers including breast, bladder, lung, liver, cholangiocarcinoma, and glioblastoma (Zaid et al., 2013). So far, some of these pan-FGFRs inhibitors have been reported to have efficient inhibitory effects for FGFRs including FIIN-2 (Masaru, 2016), JNJ-42756493 (Tabernero et al., 2015), LY2874455 (Tie et al., 2014), and ponatinib (Cortes et al., 2012). Infigratinib (BGJ398) selectively targets FGFR (1–3) with high affinity, but also targets FGFR4 with less affinity (Huynh et al., 2019). Meanwhile, accumulating evidence suggests that there could be more variability between FGFR4 and other FGFRs than was initially expected. It has been reported that there are a number of amino-acid substitutions in the tyrosine kinase domains of FGFR4 (Dutra et al., 2015; Katoh, 2016). FGF401 (Weiss et al., 2019; Zhou et al., 2019), BLU9931 (Hagel et al., 2015), and BLU-554 (Hatlen et al., 2019; Kim et al., 2019) have been reported to be the most highly potent and selective FGFR4 inhibitors. Representative FGFR4 inhibitors that are currently undergoing clinical trials were shown in Table 1. A phase I study of fisogatinib (BLU-554) validated that aberrant FGF19/FGFR4 signaling is a targetable oncogenic driver in HCC; fisogatinib was well-tolerated and most adverse events were manageable grade 1/2 gastrointestinal event – primarily diarrhea, nausea, and vomiting (Kim et al., 2019). Compared to the other three members of FGFRs, FGFR4 has one notably distinct amino acid change. Cys552 on FGFR4 is unique among the four paralogs of FGFRs and thus provides a selectivity handle to specifically target FGFR4. Cys552 in FGFR4 is required for the covalent binding with BLU9931 (Hoeflich et al., 2015). Selective FGFR4 targeting has been shown to reduce adverse effects, and the simultaneous targeting of FGFR4 and other receptor tyrosine kinases (RTKs) has been shown to indirectly enhance anti-tumor activity through normalization of the tumor microenvironment (Repana and Ross, 2015; Katoh, 2016).
Another approach to targeting the FGF19-FGFR4 signaling pathway is the use of monoclonal antibodies against FGF19 or FGFR4. Desnoyers et al. (2008) produced a neutralizing anti-FGF19 monoclonal antibody that prevented liver cancer formation in FGF19 transgenic (FGF-TG) mice treated with diethylnitrosamine, a potent hepatocarcinogen which accelerate the development of HCC in FGF19-TG mice (French et al., 2012). The safety of this antibody was tested in cynomolgus monkeys, an appropriate model because there is similar binding affinity of anti-FGF19 to human and cynomolgus monkey FGF19. Treatment of monkeys with anti-FGF19 antibody increased bile acid synthesis and changed the expression of bile transporters in the liver (Pai et al., 2012). LD1 is a neutralizing antibody against FGFR4 that has been shown to inhibit FGFR4 mediated signaling, colony formation, and cell proliferation. LD1 is also able to suppress tumor growth in a preclinical model of liver cancer in vivo (Loilome et al., 2009). U3-1784, a novel FGFR4 targeting antibody, is a high-affinity fully humanized antibody. U3-1784 could compete with various FGFs for their FGFR4 binding sites, therefore inhibits receptor activation and downstream signaling, including FRS2 and ERK. Animal studies showed that U3-1784 exerts strong antitumor effects specific to liver cancer cells overexpressing FGF19 (Bartz et al., 2019). Strong antitumor effects and the lower toxicity of anti-FGFR4 antibody suggests a general strategy for avoiding adverse events with FGFR4 inhibitors for liver cancer therapy.
Aberrant FGFR4 expression was observed in a number of human cancers, especially in HCC (Figure 1B) (Fischbach et al., 2015). Importantly, selective inhibitors of FGFR4 have demonstrated clinical benefit in HCC patients with high FGF19 expression (Kim et al., 2019). FGF19 binds to FGFR4 and activates GSK3β-Nrf2 signaling cascade to exert cytoprotective role against ER stress in HCC (Teng et al., 2017). It has been reported that both rearranged distal enhancement regions and point mutations in the proximal promoter region might cause FGFR4 overexpression (Koole et al., 2015). Moreover, inhibition of miRNAs targeting FGFR4 or aberrant long non-coding RNA (lncRNA) expression leads to FGFR4 overexpression (Masaru, 2016). FGFR4 inhibitors directly or indirectly suppress tumor growth by influencing the tumor microenvironment, particularly paracrine signaling, angiogenesis, and immune evasion (Masaru, 2016).
FGFR4 has been reported as an oncogenic driver in FGF19-positive advanced HCC. A therapeutic strategy against targeting FGF19/FGFR4 is being actively investigated (Dutra et al., 2015; Huynh et al., 2019). Meanwhile, inhibition of FGF19-FGFR4 signaling has been reported to be associated with increased risk of hepatotoxicity and cardiac toxicity (Lin and Desnoyers, 2012). Thus, careful preclinical studies evaluating the potential risks of treatment with FGFR4 inhibitors are critical to further development (Wang and Jacob, 2011; Foo et al., 2012).
Immune checkpoint blockade therapy has demonstrated several advantages in clinical oncology. PD-1 ligand (PD-L1) is presented in cancer cells and stromal/immune cells, while PD-1 and CTLA-4 are expressed in CD8+ T cells or Treg cells (Wu et al., 2014). Since both PD-1 and CTLA-4 signaling pathways are directly or indirectly involved in the inhibition of cytotoxic T cell functions; anti-PD-1, anti-PDL1, and anti-CTLA-4 monoclonal antibodies have been clinically used for cancer immunotherapy (Das et al., 2015). Interestingly, FGFR4 inhibitors have been shown to target immune cells in the tumor microenvironment, such as myeloid-derived suppressor cells (MDSCs) and M2-type tumor-associating macrophages (m2-tams), and are expected to indirectly inhibit PD-L1 expression in tumor cells and stromal/immune cells through influencing the tumor microenvironment (Masaru, 2016). Therefore, combination therapy with FGFR4 inhibitors and immune checkpoint blockers (anti-PD-1 or anti-CTLA-4 mAb) might be a promising therapeutic method for certain cancer patients.
In conclusion, the positive results of phase-I study for fisogatinib (BLU-554) in advanced HCC expressing FGF19 indicate that the development of covalently binding FGFR4 specific inhibitors is an important research direction. Meanwhile, searching for biomarkers that could predict the responses to FGFR4-targeting therapy are urgently needed. Another approach to target FGF19-FGFR4 signaling is monoclonal antibodies and FGF ligand traps which can block downstream signal of FGFR4 by interfering with ligand binding or receptor dimerization. Based on promising preliminary results from clinical trials of FGFR4 inhibitors, more efforts are needed to recognize patients who most likely benefit from treatment, and to design and implement effective combination therapies.
YL and MC collected the articles and made Figures. YC and CZ guided YL and MC to collect articles and make figures. XL prerevised the manuscript. CZ and RC wrote the manuscript and made a table.
This work was supported by grants from the National Natural Science Foundation of China (81672305) to RC.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Bartz, R., Fukuchi, K., Ohtsuka, T., Lange, T., Gruner, K., Watanabe, I., et al. (2019). Preclinical development of U3-1784, a Novel FGFR4 antibody against cancer, and avoidance of its on-target toxicity. Mol. Cancer Ther. 18, 1832–1843. doi: 10.1158/1535-7163.MCT-18-0048
Boonstra, J., Rijken, P., Humbel, B., Cremers, F., Verkleij, A., and Van Be, H. P. (2013). The epidermal growth factor. Cell Biol. Int. 19, 413–430.
Cary, L. A., Han, D. C., and Guan, J. L. (1999). Integrin-mediated signal transduction pathways. Histol. Histopathol. 14, 1001–1009.
Chen, J., Du, F., Dang, Y., Li, X., Qian, M., Feng, W., et al. (2019). FGF19-mediated upregulation of SOX18 promotes hepatocellular carcinoma metastasis by transactivating FGFR4 and FLT4. Hepatology (in press). doi: 10.1002/hep.30951
Cortes, J. E., Kantarjian, H., Shah, N. P., Bixby, D., Mauro, M. J., Flinn, I., et al. (2012). Ponatinib in refractory philadelphia chromosome-positive leukemias. N. Engl. J. Med. 367, 2075–2088. doi: 10.1056/NEJMoa1205127
Dallol, A., Buhmeida, A., Merdad, A., Al-Maghrabi, J., Gari, M. A., Abu-Elmagd, M. M., et al. (2015). Frequent methylation of the KLOTHO gene and overexpression of the FGFR4 receptor in invasive ductal carcinoma of the breast. Tumour Biol. 36, 9677–9683. doi: 10.1007/s13277-015-3733-3
Das, R., Verma, R., Sznol, M., Boddupalli, C. S., Gettinger, S. N., Kluger, H., et al. (2015). Combination therapy with anti-CTLA-4 and anti-PD-1 leads to distinct immunologic changes in vivo. J. Immunol. 194, 950–959. doi: 10.4049/jimmunol.1401686
Desnoyers, L. R., Pai, R., Ferrando, R. E., Hotzel, K., Le, T., Ross, J., et al. (2008). Targeting FGF19 inhibits tumor growth in colon cancer xenograft and FGF19 transgenic hepatocellular carcinoma models. Oncogene 27, 85–97. doi: 10.1038/sj.onc.1210623
Dutra, R. L., Santos, M. D., Mendes, S. O., Peterle, G. T., Louro, I. D., and da Silva Conforti, A. M. Á (2015). “FGFR4 as a biomarker in squamous cell cancers of mouth and oropharynx,” in Biomarkers in Cancer. Biomarkers in Disease: Methods, Discoveries and Applications, eds V. Preedy and V. Patel (Dordrecht: Springer).
Fischbach, A., Rogler, A., Erber, R., Stoehr, R., Poulsom, R., Heidenreich, A., et al. (2015). Fibroblast growth factor receptor (FGFR) gene amplifications are rare events in bladder cancer. Histopathology 66, 639–649. doi: 10.1111/his.12473
Foo, J. N., Liu, J. J., and Tan, E. K. (2012). Whole-genome and whole-exome sequencing in neurological diseases. Nat. Rev. Neurol. 8, 508–517. doi: 10.1038/nrneurol.2012.148
French, D. M., Lin, B. C., Wang, M., Adams, C., Shek, T., Hotzel, K., et al. (2012). Targeting FGFR4 inhibits hepatocellular carcinoma in preclinical mouse models. PLoS One 7:e36713. doi: 10.1371/journal.pone.0036713
Gao, L., Wang, X., Tang, Y., Huang, S., Hu, C. A., and Teng, Y. (2017). FGF19/FGFR4 signaling contributes to the resistance of hepatocellular carcinoma to sorafenib. J. Exp. Clin. Cancer Res. 36:8. doi: 10.1186/s13046-016-0478-9
Gao, L. X., Lang, L. W., Zhao, X. D., Shay, C., Shull, A. Y., and Teng, Y. (2019). FGF19 amplification reveals an oncogenic dependency upon autocrine FGF19/FGFR4 signaling in head and neck squamous cell carcinoma. Oncogene 38, 2394–2404. doi: 10.1038/s41388-018-0591-7
Goetz, R., and Mohammadi, M. (2013). Exploring mechanisms of FGF signalling through the lens of structural biology. Nat. Rev. Mol. Cell Biol. 14, 166–180. doi: 10.1038/nrm3528
Hagel, M., Miduturu, C., Sheets, M., Rubin, N., Weng, W., Stransky, N., et al. (2015). First selective small molecule inhibitor of FGFR4 for the treatment of hepatocellular carcinomas with an activated FGFR4 signaling pathway. Cancer Dis. 5, 424–437. doi: 10.1158/2159-8290.CD-14-1029
Han, K. H., Pok, S., Streit, S., Ruhe, J. E., Hart, S., Lim, K. S., et al. (2009). Fibroblast growth factor receptor 4 regulates proliferation, anti-apoptosis and alpha-fetoprotein secretion during hepatocellular carcinoma progression and represents a potential target for therapeutic intervention. J. Hepatol. 50, 118–127. doi: 10.1016/j.jhep.2008.08.015
Hatlen, M. A., Schmidt-Kittler, O., Sherwin, C. A., Rozsahegyi, E., Rubin, N., Sheets, M. P., et al. (2019). Acquired on-target clinical resistance validates FGFR4 as a driver of hepatocellular carcinoma. Cancer Dis. 9, 1686–1695. doi: 10.1158/2159-8290.CD-19-0367
Heinzle, C., Erdem, Z., Paur, J., Grasl-Kraupp, B., Holzmann, K., Grusch, M., et al. (2014). Is fibroblast growth factor receptor 4 a suitable target of cancer therapy? Curr. Pharm. Des. 20, 2881–2898. doi: 10.2174/13816128113199990594
Heinzle, C., Gsur, A., Hunjadi, M., Erdem, Z., Gauglhofer, C., Stattner, S., et al. (2012). Differential effects of polymorphic alleles of FGF receptor 4 on colon cancer growth and metastasis. Cancer Res. 72, 5767–5777. doi: 10.1158/0008-5472.CAN-11-3654
Hoeflich, K., Hagel, M., Miduturu, C., Sheets, M., Rubin, N., Weng, W., et al. (2015). O048 : first selective small molecule inhibitor of FGFR4 for the treatment of hepatocellular carcinomas with an activated FGFR4 signaling pathway. Cancer Dis. 5, 424–437. doi: 10.1158/2159-8290.cd-14-1029
Hoover, H., Li, J., Marchese, J., Rothwell, C., Borawoski, J., Jeffery, D. A., et al. (2015). Quantitative proteomic verification of membrane proteins as potential therapeutic targets located in the 11q13 amplicon in cancers. J. Proteome Res. 14, 3670–3679. doi: 10.1021/acs.jproteome.5b00508
Huang, H. P., Feng, H., Qiao, H. B., Ren, Z. X., and Zhu, G. D. (2015). The prognostic significance of fibroblast growth factor receptor 4 in non-small-cell lung cancer. Onco Targets Ther. 8, 1157–1164. doi: 10.2147/OTT.S81659
Huynh, H., Lee, L. Y., Goh, K. Y., Ong, R., Hao, H. X., Huang, A., et al. (2019). Infigratinib mediates vascular normalization, impairs metastasis, and improves chemotherapy in hepatocellular carcinoma. Hepatology 69, 943–958. doi: 10.1002/hep.30481
Hyeon, J., Ahn, S., Lee, J. J., Song, D. H., and Park, C. K. (2013). Expression of fibroblast growth factor 19 is associated with recurrence;and poor prognosis of hepatocellular carcinoma. Dig. Dis. Sci. 58, 1916–1922. doi: 10.1007/s10620-013-2609-x
Jaakkola, S., Salmikangas, P., Nylund, S., Partanen, J., Armstrong, E., Pyrhonen, S., et al. (1993). Amplification of fgfr4 gene in human breast and gynecological cancers. Int. J. Cancer 54, 378–382. doi: 10.1002/ijc.2910540305
Katoh, M. (2016). FGFR inhibitors: effects on cancer cells, tumor microenvironment and whole-body homeostasis (Review). Int. J. Mol. Med. 38, 3–15. doi: 10.3892/ijmm.2016.2620
Kim, R. D., Sarker, D., Meyer, T., Yau, T., Macarulla, T., Park, J. W., et al. (2019). First-in-human phase l study of fisogatinib (BLU-554) validates aberrant fibroblast growth factor 19 signaling as a driver event in hepatocellular carcinoma. Cancer Dis. 9, 1696–1707. doi: 10.1158/2159-8290.cd-19-0555
Kogan, D., Grabner, A., Yanucil, C., Faul, C., and Ulaganathan, V. K. (2018). STAT3-enhancing germline mutations contribute to tumor-extrinsic immune evasion. J. Clin. Investigat. 128, 1867–1872. doi: 10.1172/JCI96708
Koole, K., van Kempen, P. M., van Bockel, L. W., Smets, T., van der Klooster, Z., Dutman, A. C., et al. (2015). FGFR4 Is a potential predictive biomarker in oral and oropharyngeal squamous cell carcinoma. Pathobiology 82, 280–289. doi: 10.1159/000439536
Lang, L., and Teng, Y. (2019). Fibroblast growth factor receptor 4 targeting in cancer: new insights into mechanisms and therapeutic strategies. Cells 8:31. doi: 10.3390/cells8010031
Lin, B. C., and Desnoyers, L. R. (2012). FGF19 and Cancer. Adv. Exp. Med. Biol. 728, 183–194. doi: 10.1007/978-1-4614-0887-1_12
Liu, J., Zhang, Z., Li, X., Chen, J., Wang, G., Tian, Z., et al. (2018). Forkhead box C1 promotes colorectal cancer metastasis through transactivating ITGA7 and FGFR4 expression. Oncogene 37, 5477–5491. doi: 10.1038/s41388-018-0355-4
Loilome, W., Joshi, A. D., Rhys, C. M. J. A., Piccirillo, S., Angelo, V. L., Gallia, G. L., et al. (2009). Glioblastoma cell growth is suppressed by disruption of fibroblast growth factor pathway signaling. J. Neurooncol. 94, 359–366. doi: 10.1007/s11060-009-9885-5
Masaru, K. (2016). FGFR inhibitors: effects on cancer cells, tumor microenvironment and whole-body homeostasis (Review). Int. J. Mol. Med. 38, 3–15. doi: 10.3892/ijmm.2016.2620
Miura, S. (2012). Fibroblast growth factor 19 expression correlates with tumor progression and poorer prognosis of hepatocellular carcinoma. BMC Cancer 12:56. doi: 10.1186/1471-2407-12-56
Miura, S., Mitsuhashi, N., Shimizu, H., Kimura, F., Yoshidome, H., Otsuka, M., et al. (2012). Fibroblast growth factor 19 expression correlates with tumor progression and poorer prognosis of hepatocellular carcinoma. BMC Cancer 12:56. doi: 10.1186/1471-2407-12-56
Motoda, N., Matsuda, Y., Onda, M., Ishiwata, T., Uchida, E., and Naito, Z. (2011). Overexpression of fibroblast growth factor receptor 4 in high-grade pancreatic intraepithelial neoplasia and pancreatic ductal adenocarcinoma. Int. J. Oncol. 38, 133–143.
Nicholes, K., Guillet, S., Tomlinson, E., Hillan, K., Wright, B., Frantz, G. D., et al. (2002). A mouse model of hepatocellular carcinoma: ectopic expression of fibroblast growth factor 19 in skeletal muscle of transgenic mice. Am. J. Pathol. 160, 2295–2307.
Nicolas, C. S., Amici, M., Bortolotto, Z. A., Doherty, A., Csaba, Z., Fafouri, A., et al. (2013). The role of JAK-STAT signaling within the CNS. JAKSTAT 2:e22925. doi: 10.4161/jkst.22925
Pai, R., Dunlap, D., Qing, J., Mohtashemi, I., Hotzel, K., and French, D. M. (2008). Inhibition of fibroblast growth factor 19 reduces tumor growth by modulating beta-catenin signaling. Cancer Res. 68, 5086–5095. doi: 10.1158/0008-5472.CAN-07-2325
Pai, R., French, D., Ning, M., Hotzel, K., Plise, E., Salphati, L., et al. (2012). Antibody-mediated inhibition of fibroblast growth factor 19 results in increased bile acids synthesis and ileal malabsorption of bile acids in cynomolgus monkeys. Toxicol. Sci. 126, 446–456. doi: 10.1093/toxsci/kfs011
Peláez-García, A., Barderas, R., Torres, S., Hernández-Varas, P., Teixidó, J., Bonilla, F., et al. (2013). FGFR4 role in epithelial-mesenchymal transition and its therapeutic value in colorectal cancer. PLoS One 8:e63695. doi: 10.1371/journal.pone.0063695
Penault-Llorca, F., Bertucci, F., Adelaide, J., Parc, P., Coulier, F., Jacquemier, J., et al. (1995). Expression of FGF and FGF receptor genes in human breast cancer. Int. J. Cancer 61, 170–176.
Poh, W., Wong, W., Ong, H., Aung, M. O., Lim, S. G., Chua, B. T., et al. (2012). Klotho-beta overexpression as a novel target for suppressing proliferation and fibroblast growth factor receptor-4 signaling in hepatocellular carcinoma. Mol. Cancer 11:14. doi: 10.1186/1476-4598-11-14
Presta, M., Chiodelli, P., Giacomini, A., Rusnati, M., and Ronca, R. (2017). Fibroblast growth factors (FGFs) in cancer: FGF traps as a new therapeutic approach. Pharmacol. Ther. 179, 171–187. doi: 10.1016/j.pharmthera.2017.05.013
Prieto-Dominguez, N., Shull, A. Y., and Teng, Y. (2018). Making way for suppressing the FGF19/FGFR4 axis in cancer. Future Med. Chem. 10, 2457–2470. doi: 10.4155/fmc-2018-0099
Qian, J., Yong, Z., Rahman, S. M., and Massion, P. P. (2006). Implications of different mechanisms of PI3K/AKT activation in non-small cell lung cancer. Cancer Res. 66, 305–306. doi: 10.1097/JTO.0b013e3181a1084f
Raja, A., Park, I., Haq, F., and Ahn, S. M. (2019). FGF19-FGFR4 signaling in hepatocellular carcinoma. Cells 8:536. doi: 10.3390/cells8060536
Repana, D., and Ross, P. (2015). Targeting FGF19/FGFR4 pathway: a novel therapeutic Strategy for hepatocellular carcinoma. Diseases 3, 294–305. doi: 10.3390/diseases3040294
Roidl, A., Berger, H. J., Kumar, S., Bange, J., Knyazev, P., and Ullrich, A. (2009). Resistance to chemotherapy is associated with fibroblast growth factor receptor 4 up-regulation. Clin. Cancer Res. 15, 2058–2066. doi: 10.1158/1078-0432.CCR-08-0890
Sahadevan, K., Darby, S., Leung, H. Y., Mathers, M. E., Robson, C. N., and Gnanapragasam, V. J. (2007). Selective over-expression of fibroblast growth factor receptors 1 and 4 in clinical prostate cancer. J. Pathol. 213, 82–90. doi: 10.1002/path.2205
Sawey, E. T., Chanrion, M., Cai, C., Wu, G., Zhang, J., Zender, L., et al. (2011). Identification of a therapeutic strategy targeting amplified FGF19 in liver cancer by oncogenomic screening. Cancer Cell 19, 347–358. doi: 10.1016/j.ccr.2011.01.040
Schadt, H. S., Wolf, A., Mahl, J. A., Wuersch, K., Couttet, P., Schwald, M., et al. (2018). Bile acid sequestration by cholestyramine mitigates FGFR4 inhibition-induced ALT elevation. Toxicol. Sci. 163, 265–278. doi: 10.1093/toxsci/kfy031
Stacey, D. W. (2003). Cyclin D1 serves as a cell cycle regulatory switch in actively proliferating cells. Curr. Opin. Cell Biol. 15, 158–163. doi: 10.1016/s0955-0674(03)00008-5
Sugiyama, N., Varjosalo, M., Meller, P., Lohi, J., Hyytiainen, M., Kilpinen, S., et al. (2010). Fibroblast growth factor receptor 4 regulates tumor invasion by coupling fibroblast growth factor signaling to extracellular matrix degradation. Cancer Res. 70, 7851–7861. doi: 10.1158/0008-5472.CAN-10-1223
Tabernero, J., Bahleda, R., Dienstmann, R., Infante, J. R., Mita, A., Italiano, A., et al. (2015). Phase I dose-escalation study of JNJ-42756493, an oral pan-fibroblast growth factor receptor inhibitor, in patients with advanced solid tumors. J. Clin. Oncol. 33, 3401–3408. doi: 10.1200/JCO.2014.60.7341
Taylor, J. G., Cheuk, A. T., Tsang, P. S., Chung, J. Y., Song, Y. K., Desai, K., et al. (2009). Identification of FGFR4-activating mutations in human rhabdomyosarcomas that promote metastasis in xenotransplanted models. J. Clin. Investigat. 119, 3395–3407. doi: 10.1172/JCI39703
Teng, Y., Zhao, H., Gao, L., Zhang, W., Shull, A. Y., and Shay, C. (2017). FGF19 Protects hepatocellular carcinoma cells against endoplasmic reticulum stress via activation of FGFR4-GSK3beta-Nrf2 signaling. Cancer Res. 77, 6215–6225. doi: 10.1158/0008-5472.CAN-17-2039
Tie, J., Bang, Y. J., Park, Y. S., Kang, Y. K., Monteith, D., Hartsock, K., et al. (2014). Abstract CT215: A phase I trial of LY2874455, a fibroblast growth factor receptor inhibitor, in patients with advanced cance. Cancer Res. 74(Suppl. 19):CT215.
Tiong, K. H., Tan, B. S., Choo, H. L., Chung, F. F., Hii, L. W., Tan, S. H., et al. (2016). Fibroblast growth factor receptor 4 (FGFR4) and fibroblast growth factor 19 (FGF19) autocrine enhance breast cancer cells survival. Oncotarget 7, 57633–57650. doi: 10.18632/oncotarget.9328
Toren, P., and Zoubeidi, A. (2014). Targeting the PI3K/Akt pathway in prostate cancer: challenges and opportunities (review). Int. J. Oncol. 45, 1793–1801. doi: 10.3892/ijo.2014.2601
Touat, M., Ileana, E., Postel-Vinay, S., Andre, F., and Soria, J. C. (2015). Targeting FGFR signaling in cancer. Clin. Cancer Res. 21, 2684–2694. doi: 10.1158/1078-0432.ccr-14-2329
Ulaganathan, V. K., Sperl, B., Rapp, U. R., and Ullrich, A. (2015). Germline variant FGFR4 p.G388R exposes a membrane-proximal STAT3 binding site. Nature 528, 570–574. doi: 10.1038/nature16449
Valastyan, S., and Weinberg, R. A. (2011). Tumor metastasis: molecular insights and evolving paradigms. Cell 147, 275–292. doi: 10.1016/j.cell.2011.09.024
Vlacic, G., Hoda, M. A., Klikovits, T., Sinn, K., Gschwandtner, E., Mohorcic, K., et al. (2019). Expression of FGFR1-4 in malignant pleural mesothelioma tissue and corresponding cell lines and its relationship to patient survival and fgfr inhibitor sensitivity. Cells 8:1091. doi: 10.3390/cells8091091
Wang, B., and Jacob, S. T. (2011). Role of cancer stem cells in hepatocarcinogenesis. Genome Med. 3:11. doi: 10.1186/gm225
Weiss, A., Adler, F., Buhles, A., Stamm, C., Fairhurst, R. A., Kiffe, M., et al. (2019). FGF401, a first-in-class highly selective and potent FGFR4 inhibitor for the treatment of FGF19-driven hepatocellular cancer. Mol. Cancer Ther. 18, 2194–2206. doi: 10.1158/1535-7163.MCT-18-1291
Wilkie, A. O., Morriss-Kay, G. M., Jones, E. Y., and Heath, J. K. (1995). Functions of fibroblast growth factors and their receptors. Curr. Biol. 5, 500–507.
Wu, A. L., Coulter, S., Liddle, C., Wong, A., Easthamanderson, J., French, D. M., et al. (2011). FGF19 Regulates cell proliferation, glucose and bile acid metabolism via fgfr4-dependent and independent pathways. PLoS One 6:e17868. doi: 10.1371/journal.pone.0017868
Wu, X., Ge, H., Lemon, B., Vonderfecht, S., Weiszmann, J., Hecht, R., et al. (2010). FGF19-induced hepatocyte proliferation is mediated through fgfr4 activation. J. Biol. Chem. 285, 5165–5170. doi: 10.1074/jbc.M109.068783
Wu, X., Zhang, H., Xing, Q., Cui, J., Li, J., Li, Y., et al. (2014). PD-1+ CD8+ T cells are exhausted in tumours and functional in draining lymph nodes of colorectal cancer patients. Br. J. Cancer 111, 1391–1399. doi: 10.1038/bjc.2014.416
Xu, W., Li, Y., Wang, X., Chen, B., Wang, Y., Liu, S., et al. (2010). FGFR4 transmembrane domain polymorphism and cancer risk: a meta-analysis including 8555 subjects. Eur. J. Cancer 46, 3332–3338. doi: 10.1016/j.ejca.2010.06.017
Zaid, T. M., Yeung, T. L., Thompson, M. S., Leung, C. S., Harding, T., Co, N. N., et al. (2013). Identification of FGFR4 as a potential therapeutic target for advanced-stage, high-grade serous ovarian cancer. Clin. Cancer Res. 19, 809–820. doi: 10.1158/1078-0432.CCR-12-2736
Zhang, X., Kong, M., Zhang, Z., Xu, S., Yan, F., Wei, L., et al. (2017). FGF19 genetic amplification as a potential therapeutic target in lung squamous cell carcinomas. Thorac Cancer 8, 655–665. doi: 10.1111/1759-7714.12504
Zhang, Z. D., Wang, Z. J., Hua, L. X., Chen, S. Q., Tong, N., Xu, B., et al. (2011). FGFR4 Gly388Arg polymorphism contributes to prostate cancer development and progression: a meta-analysis of 2618 cases and 2305 controls. BMC Cancer 11:84. doi: 10.1186/1471-2407-11-84
Zhao, C. G., Li, H. M., Lin, H. J., Yang, S. L., Lin, J. Y., and Liang, G. (2016). Feedback activation of STAT3 as a cancer drug-resistance mechanism. Trends Pharmacol. Sci. 37, 47–61. doi: 10.1016/j.tips.2015.10.001
Zhao, H., Lv, F., Liang, G., Huang, X., Wu, G., Zhang, W., et al. (2016). FGF19 promotes epithelial-mesenchymal transition in hepatocellular carcinoma cells by modulating the GSK3beta/beta- catenin signaling cascade via FGFR4 activation. Oncotarget 7, 13575–13586. doi: 10.18632/oncotarget.6185
Zhou, M., Wang, X., Phung, V., Lindhout, D. A., Mondal, K., Hsu, J. Y., et al. (2014). Separating tumorigenicity from bile acid regulatory activity for endocrine hormone FGF19. Cancer Res. 74, 3306–3316. doi: 10.1158/0008-5472.CAN-14-0208
Zhou, Q., Lui, V. W., and Yeo, W. (2011). Targeting the PI3K/Akt/mTOR pathway in hepatocellular carcinoma. Future Oncol. 7, 1149–1167. doi: 10.2217/fon.11.95
Keywords: FGF19, FGFR4, inhibitors, cancer, targeted therapy
Citation: Liu Y, Cao M, Cai Y, Li X, Zhao C and Cui R (2020) Dissecting the Role of the FGF19-FGFR4 Signaling Pathway in Cancer Development and Progression. Front. Cell Dev. Biol. 8:95. doi: 10.3389/fcell.2020.00095
Received: 25 November 2019; Accepted: 04 February 2020;
Published: 20 February 2020.
Edited by:
Ana Cuenda, Spanish National Research Council, SpainReviewed by:
Cynthia L. Bristow, Alpha-1 Biologics, United StatesCopyright © 2020 Liu, Cao, Cai, Li, Zhao and Cui. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chengguang Zhao, emhhb2NoZW5nZ3VhbmdAd211LmVkdS5jbg==; Ri Cui, d3ptdWN1aXJpQDE2My5jb20=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.