- 1Department of Biology and Biotechnology Charles Darwin, Sapienza University of Rome, Rome, Italy
- 2Center for Human Technologies, Italian Institute of Technology, Genoa, Italy
In 1957, Francis Crick speculated that RNA, beyond its protein-coding capacity, could have its own function. Decade after decade, this theory was dramatically boosted by the discovery of new classes of non-coding RNAs (ncRNAs), including long ncRNAs (lncRNAs) and circular RNAs (circRNAs), which play a fundamental role in the fine spatio-temporal control of multiple layers of gene expression. Recently, many of these molecules have been identified in a plethora of different tissues, and they have emerged to be more cell-type specific than protein-coding genes. These findings shed light on how ncRNAs are involved in the precise tuning of gene regulatory mechanisms governing tissues homeostasis. In this review, we discuss the recent findings on the mechanisms used by lncRNAs and circRNAs to sustain skeletal and cardiac muscle formation, paying particular attention to the technological developments that, over the last few years, have aided their genome-wide identification and study. Together with lncRNAs and circRNAs, the emerging contribution of Piwi-interacting RNAs and transfer RNA-derived fragments to myogenesis will be also discussed, with a glimpse on the impact of their dysregulation in muscle disorders, such as myopathies, muscle atrophy, and rhabdomyosarcoma degeneration.
Introduction
In the last 50 years, RNA has been subjected to an unprecedented revaluation that gradually shifted the perspective on how gene expression is regulated from a coding to a non-coding point of view. With the discovery of messenger RNA (mRNA) codon–transfer RNA (tRNA) anticodon interaction, tRNAs have become forerunners of non-coding functionality (Hoagland et al., 1958; Steitz and Jakes, 1975). In roughly 30 years of experiments and observations, novel classes of non-coding RNAs (ncRNAs), such as ribosomal RNAs (rRNAs) (Scherrer and Darnell, 1962), small nuclear RNAs (Weinberg and Penman, 1968), small nucleolar RNAs (Reddy and Busch, 1988), and microRNAs (miRNAs) (Lee et al., 1993; Lagos-Quintana et al., 2001; Lau et al., 2001), attracted the attention of the scientific community by showing their participation to several biological processes. A significant contribution to these discoveries was made by the advances in both sequencing technologies and data open sharing, which brought the new millennium into the “omics” revolution. In this context, the identification and the extraordinary cell-type-specific expression of new classes of ncRNAs, including long ncRNAs (lncRNAs) and circular RNAs (circRNAs), provided new clues into the understanding of tissue homeostasis (Kapranov et al., 2007; Mattick, 2011; Nagano and Fraser, 2011; Hon et al., 2017).
LncRNAs are arbitrarily defined as transcripts longer than 200 nucleotides, which regulate gene expression at transcriptional and post-transcriptional level (Rinn and Chang, 2012; Ulitsky and Bartel, 2013; Fatica and Bozzoni, 2014). The function of these transcripts is in close connection with their specific subcellular localization, ranging from an almost exclusive nuclear (Heard and Disteche, 2006; Tripathi et al., 2010; Chen et al., 2016) or cytoplasmic (Cesana et al., 2011; van Heesch et al., 2014) enrichment, up to a more uniform and less defined distribution (Ballarino et al., 2015; Alessio et al., 2019). Together with lncRNAs, it is nowadays well-established that cells also express circRNAs. These are endogenously expressed and covalently closed single-stranded RNA species that derive from back-splicing circularization events (Jeck et al., 2013; Chen, 2016). Differently from linear lncRNA molecules, the lack of 5′- and 3′-ends confers to circRNAs greater stability and amplifies their chance to exert various biological tasks. As a common aspect, the function of both lncRNA and circRNA is determined by their ability to recognize nucleic acids by base pairing with their high versatility to interact with proteins (Guttman and Rinn, 2012; Rinn and Chang, 2012; Batista and Chang, 2013). For this reason, the post-genomic epoch has been marked by the establishment of innovative technologies, which have been extremely helpful for identifying the ncRNA interactome, thus providing crucial clues on their mechanisms of action (Ule et al., 2003; Licatalosi et al., 2008; Engreitz et al., 2014; McHugh and Guttman, 2018). The RNA antisense purification (RAP) is one of the most RNA-centric leading-edge approach able to purify and identify the interacting partners of a specific RNA (McHugh and Guttman, 2018). The protein-centric mirror technique is represented by the cross-linking and immunoprecipitation (CLIP) assay (Ule et al., 2003; Licatalosi et al., 2008). Both the systems exploit the ultraviolet (UV) cross-linking to create covalent linkages between directly interacting RNAs and proteins, purifying the molecule of interest under stringent conditions to reduce background signals. In addition, a method to systematically map RNA–RNA interactions, based on psoralen cross-linking, has been also developed (Engreitz et al., 2014).
To date, the vast majority of studies have focused on the role of miRNAs in muscle development (Ballarino et al., 2016; Wang J. et al., 2018; Colpaert and Calore, 2019). In this article, we highlight the importance of some less studied classes of ncRNAs, such as lncRNAs and circRNAs, focusing on their function in skeletal and cardiac muscles biology. We discuss paradigmatic examples that support their nuclear and cytoplasmic activities and report the latest findings of lncRNAs and circRNAs containing functional open reading frames (ORFs) (Chekulaeva and Rajewsky, 2019). In the final section, we also provide a broad overview on other classes of ncRNAs [i.e., Piwi-interacting RNAs (piRNAs) and tRNA fragments], which are well-known for their canonical functions but only recently emerged as functional in muscle physiology (Liapi et al., 2019).
lncRNAs in Skeletal and Cardiac Myogenesis
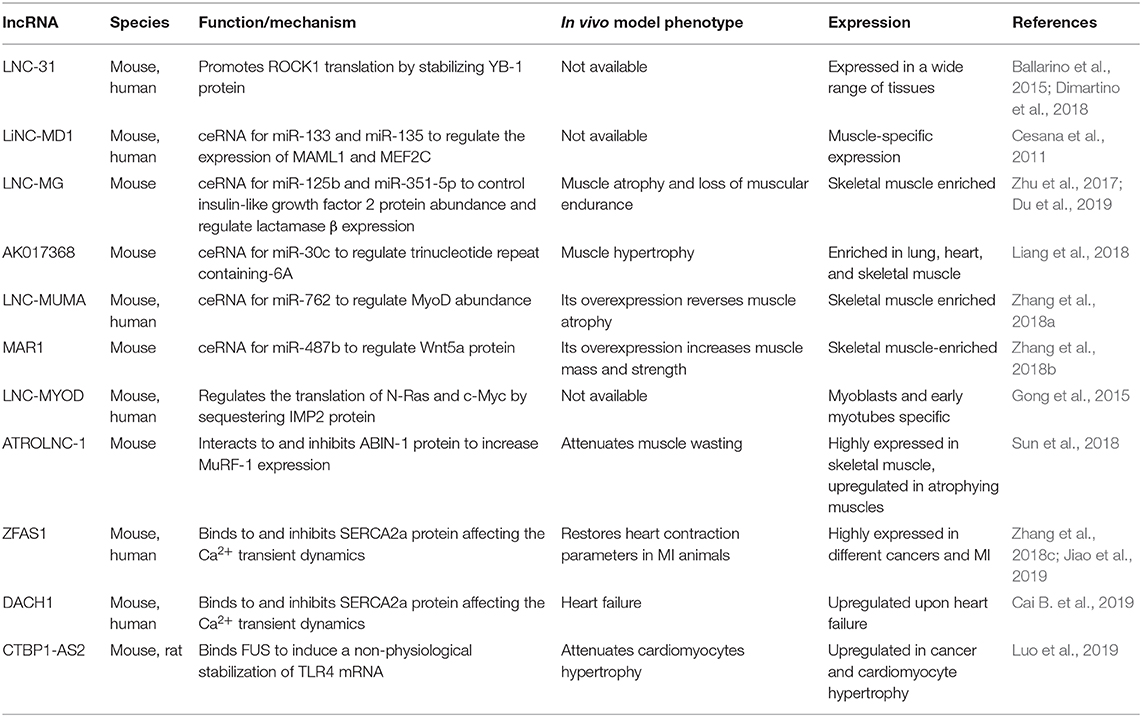
Since the establishment of the murine myoblast C2C12 cell line, which allowed to reproduce in vitro the different stages of myogenic differentiation (Yaffe and Saxel, 1977), and the discovery of MYOD1 as the “master gene” for myogenesis (Davis et al., 1987), a wealth of knowledge has been accumulated regarding the ncRNA-mediated regulatory networks governing muscle biology. Indeed, multiple examples of nuclear (Table 1) and cytoplasmic (Table 2) non-coding transcripts involved in the acquisition of both skeletal and cardiac muscle identity have been detailed over the last decade, with an increasing degree of emphasis on large species (Neguembor et al., 2014; Rotini et al., 2018).
Nuclear-Enriched lncRNAs
Aside from the “finished” non-coding transcript, the act of transcription has been proposed to be functional in myogenesis. Few years ago, Anderson et al. (2016) reported that the transcription of Hand2-associated lncRNA, named Upperhand (Uph) is required to establish a permissive chromatin environment at the Hand2 enhancer locus. Indeed, blockade of Uph transcription, but not the knockdown of the mature transcript, abolished Hand2 expression and caused heart failure and embryonic lethality in mice. In a very recent paper, Ritter et al. identified a novel lncRNA locus named Handsdown (Hdn) that is fundamental for the transcriptional regulation of Hand2 gene (Ritter et al., 2019). In situ hybridization of both Hdn and Hand2 demonstrates that the transcripts are coexpressed in the same cells or at least in the same parts of the tissue at different time points of embryonic heart specification. Interestingly, Hdn knockout shows that the locus is essential for embryonic development and uterine implantation, while its reduced expression in the heterozygous mice is haploinsufficient for proper heart formation. Chromosome conformation capture (Hi-C and 4C) analyses demonstrate that Hdn expression is crucial for structuring the genomic region and regulating in cis Hand2 production in a transcript-independent, transcription-based manner.
Upon their transcription, most of the nuclear-enriched lncRNAs regulate myogenesis by recruiting chromatin-modifying complexes to nearby (in cis) or distant (in trans) genomic loci. The Polycomb repressive complex 2 (PRC2), which catalyzes the methylation of lysine 27 on histone H3, is one of the most studied chromatin-modifying complexes found to be recruited by myogenic lncRNAs. Earlier studies from 2013 described two paradigmatic examples, which include Braveheart (Klattenhoff et al., 2013) and Fendrr (Grote et al., 2013). Overall, these findings highlighted the functional importance of lncRNA-based epigenetic mechanisms in the regulation of cell fate and greatly contributed to advance our understanding of the regulatory networks driving cardiac lineage commitment. In skeletal muscle, a more recent study (Jin et al., 2018) reported the identification of SYISL (SYNPO2-intron sense-overlapping), an abundant and intron-encoded lncRNA whose expression increases with C2C12 myoblasts differentiation. Mechanistically, SYISL promotes proliferation (and inhibits differentiation) by silencing the muscle-specific expression of myogenin, muscle creatine kinase, and myosin heavy chain (Myh4) through the recruitment of the PRC2 subunit enhancer of Zeste homolog 2 (EZH2), to their promoters. Similarly, Wang S. et al. (2019) described a functional interaction between EZH2 and Neat1 in myogenic cells. Nuclear Enriched Abundant Transcript 1 (Neat1) is a nuclear lncRNA essential for paraspeckles formation, stability, and integrity (Souquere et al., 2010), which exerts critical roles in several biological processes as well as in tumorigenesis and non-cancerous diseases (Ghafouri-Fard and Taheri, 2019; Prinz et al., 2019). During C2C12 differentiation, Neat1 sustains proliferation and blocks differentiation by recruiting EZH2 to p21 (cyclin-dependent kinase inhibitor 1 A) and to muscle specific promoters (i.e., Myog, Myh4, and Tnni2). To note, Neat1 depletion in vivo was shown to delay muscle regeneration induced by cardiotoxin treatment.
Besides the recruitment of PRC2, several nuclear-enriched lncRNAs regulate the binding of transcription factors and transcriptional coactivators to specific myogenic loci. Several of these lncRNAs, such as SRA (Caretti et al., 2006; Hubé et al., 2011), the CE and DRR endogenous RNAs (eRNAs) (Mousavi et al., 2013), and Linc-RAM (Yu et al., 2017) regulate the activity of the master transcription factor MyoD. Others impinge myogenesis either through the binding of specific myogenic factors or transcription regulators (Dum, Wang et al., 2015; Linc YY1, Zhou et al., 2015). Irm, a lncRNA that regulates myogenic genes expression in C2C12 cells by directly binding to MEF2D (Sui et al., 2019), belongs to the first category. A recent example from the second group includes Myolinc (AK142388), a lncRNA that promotes myoblasts fusion by the recruitment of the TAR DNA-binding protein 43 (TDP-43) in cis, on the neighboring Filip1 promoter, and in trans, on muscle-specific targets (Militello et al., 2018). As for Neat1, Myolinc knockdown causes a delayed regeneration of skeletal muscle in adult mice. Hitachi et al. recently described Myoparr, a promoter-associated lncRNA derived from the upstream region of the myogenin gene, which has a dual role in myogenesis: on the one hand, it is involved in myoblasts cell cycle withdrawal by the transactivation of myo-miRNAs expression; on the other hand, it is responsible for the activation in cis of myogenin transcription, which allows the entrance of myoblasts into myogenic differentiation. The latter is mediated by the interaction between Myoparr and the MyoD-coactivator Ddx17, which results in the RNA-Pol II recruitment on myogenin promoter and in transcriptional activation (Hitachi et al., 2019a). In a subsequent paper, the same authors also examined the role of Myoparr in skeletal muscle atrophy. They found that, in denervated muscles, Myoparr knockdown prevents atrophy by activating the bone morphogenetic protein signaling and by increasing the expression of GDF5, a muscle atrophy inhibitor. The existence of a Myoparr human counterpart makes the lncRNA potentially eligible as therapeutic target for neurogenic atrophy (Hitachi et al., 2019b).
Like Myoparr, it is not uncommon to find association between dysregulation of lncRNA expression and pathological conditions that induce changes in muscle mass, such as atrophy, hypertrophy, and cardiac remodeling. Another example is Pvt1, an evolutionary conserved lncRNA whose upregulation in cancer cells influences positively the stability of the oncoprotein transcription factor MYC (Tseng and Bagchi, 2015). A similar Pvt1/MYC interplay also plays a role during muscle atrophy, in which the Pvt1-mediated c-Myc stabilization impacts on the activity of Bcl-2, a crucial regulator of apoptosis and autophagy (Alessio et al., 2019). First evidence on the involvement of lncRNAs in hypertrophy came in 2014 from Chang's lab, which described a reduced hypertrophic growth and improved cardiac functions upon restoration of myosin heavy-chain-associated RNA transcripts (Mhrt) levels in transverse aortic constriction (TAC)-stressed hearts (Han et al., 2014). Analogously to Mhrt, in skeletal muscle cells, the Gm17281 lncRNA, known as Chronos, inhibits hypertrophy through the Bmp7 signaling. Mechanistically, Chronos acts as an epigenetic repressor of Bmp7 cascade through the recruitment of EZH2 on the Bmp7 promoter (Neppl et al., 2017). In murine cardiac fibroblasts, a microarray analysis led to the identification of several lncRNAs dysregulated after pressure overload upon TAC. Among the most abundant nuclear species, Piccoli et al. found the lncRNA maternally expressed gene 3 (Meg3). In cardiac fibroblasts, Meg3 controls the production of the matrix metalloproteinase-2 and, consequently, cardiac fibrosis (Piccoli et al., 2017). Indeed, Meg3 silencing was shown to reduce diastolic dysfunction and fibrosis by preventing the pathological induction of the matrix metalloproteinase-2 protein after pressure overload. Prospectively, the inhibition of Meg3 expression might represent a conceivable strategy to prevent the development of fibrosis and cardiomyocytes hypertrophic growth observed in heart diseases. The finding that Meg3 expression is upregulated in clinical heart failure samples and controls the apoptosis of human cardiomyocites (Wu et al., 2018) makes Meg3 particularly attracting as a therapeutic target. Charme (chromatin architect of muscle expression) is another nuclear-enriched lncRNA (Ballarino et al., 2018), whose ablation in vivo leads to a pronounced phenotype of cardiac remodeling at developmental onset. In myotubes, Charme contributes to the chromatin proximity between important myogenic loci, thus controlling their expression at transcriptional level. This epigenetic control is in line with emerging studies indicating that lncRNAs can act as modular scaffolds to shape the formation of chromosome territories where coregulated gene expression occurs (Clemson et al., 2009; Tripathi et al., 2010; Engreitz et al., 2013; Hacisuleyman et al., 2014; Ribeiro et al., 2018). Moreover, the existence of a Charme ortholog transcript produced by the human syntenic locus makes this lncRNA an appealing target for cardiovascular diseases.
Cytoplasmic-Enriched lncRNAs
In the cytoplasm (Table 2), lncRNAs mainly act as regulators of mRNA stability and translation. These activities depend on two major lncRNA peculiarities: (i) the capacity to base pair with RNA counterparts and (ii) the ability to act as flexible scaffolds for tethering RNAs and proteins and ensure their concerted action. Paradigmatic examples include the antisense lncRNAs known as half-STAU1-binding site RNAs (1/2-sbsRNAs) that induce Staufen1-mediated (STAU-mediated) mRNA decay (Wang J. et al., 2013). A new identified mechanism by which lncRNA–mRNA–protein interplay exerts a role in skeletal myogenesis is represented by lnc-31, a lncRNA controlling the maintenance of myoblast proliferation both in murine and human myoblasts (Ballarino et al., 2015). A key determinant for lnc-31 function is the presence of a 22nt-long region, which binds the 5′ untranslated region of ROCK1 mRNA and positively controls protein synthesis. The concomitant recruitment and the consequent stabilization of the translational activator Y-box protein 1 (YBX1) by lnc-31 concur to this translational control, as demonstrated by the decrease in ROCK1 protein upon Y-box protein 1 (YBX1) knockdown in proliferating myoblasts (Dimartino et al., 2018).
Another mechanism which underlies the capacity of lncRNAs to influence mRNA stability is based on their ability to act as competing endogenous RNAs (ceRNAs) for miRNAs (Cesana et al., 2011; Kallen et al., 2013; Han et al., 2015; Yan et al., 2015) or proteins (Gong et al., 2015). Since the discovery of linc-MD1, which was one of the first miRNA sponges to be identified, additional muscle relevant ceRNAs have been described. Recent examples include lnc-mg (Zhu et al., 2017), AK017386 (Liang et al., 2018), LncMUMA (Zhang et al., 2018a), and MAR1 (Zhang et al., 2018b) lncRNAs. Lnc-mg is a 5′-capped and polyadenylated ncRNA whose ablation (i) in vitro, reduces the capacity of murine satellite cells (MuSCs) to differentiate and (ii) in vivo, results in muscular atrophy associated to reduced muscle endurance. Mechanistically, lnc-mg acts as a miRNA-125b sponge to increase the abundance of insulin-like growth factor 2, an already known miRNA-125b target (Ge et al., 2011). More recently, lnc-mg was shown to act as a molecular sponge for miR-351-5p, which functions in skeletal myogenesis by targeting lactamase β (Du et al., 2019). AK017368 is a muscle highly enriched lincRNA able, to induce proliferation and inhibit myoblasts differentiation in vitro. AK017368 depletion in murine muscles induce fibers hypertrophy. Liang et al. (2018) demonstrated that AK017368 acts as a sponge for miR-30c by competing with its known mRNA target Tnrc6a (trinucleotide repeat containing-6A), already involved in control of myogenic differentiation by guiding Ago protein into the nucleus to lead miRNA-mediated gene silencing (Nishi et al., 2013). Another example of ceRNA promoting myogenesis is LncMUMA, a mechanical unloading-induced muscle atrophy-related lncRNA functioning as a sponge for miR-762 to regulate in vitro MyoD abundances (Zhang et al., 2018a). As LncMUMA enforced expression reverses the established muscle atrophy in hindlimb suspension mice, the above studies provide a novel therapeutic targeting for treating muscle atrophy following mechanical unloading. The same authors identified muscle anabolic regulator 1 (MAR1), a lncRNA that acts as miR-487b sponge to regulate Wnt5a protein, an important regulator of myogenesis (Tajbakhsh et al., 1998) and a known target of mir-487b in other cellular systems (Xi et al., 2013). MAR1 resulted to be positively correlated with muscle differentiation and growth, both in vitro and in vivo. Moreover, muscle mass and strength were increased in MAR1 overexpressing condition, suggesting a putative therapeutic role for muscle atrophy treatment.
Among the lncRNAs that are able to modulate the function of cytoplasmic proteins, good examples are lncMyoD (Gong et al., 2015) and Atrolnc-1 (Sun et al., 2018). LncMyoD is directly activated by MyoD at the onset of differentiation and negatively regulates the translation of N-Ras and c-Myc by directly binding and sequestering the IMP2 (insulin-like growth factor 2–mRNA-binding protein 2) factor. Atrolnc-1 was recently identified to be highly expressed in atrophying muscle from mice with cachexia. The authors demonstrated that Atrolnc-1 predominantly interacts with cytoplasmic proteins, and, in particular, they focused on A20 binding inhibitor of nuclear factor kappa B-1 (NF-κB-1), an inhibitor of IκB degradation and NF-κB activation (Mauro et al., 2006; Hooper et al., 2014). The consequence of this cytoplasmic interaction is the inhibition of A20 binding inhibitor of NF-κB-1 function, leading to an increased activity of NF-κB that in turn increases the expression of the ubiquitin E3-ligase MuRF-1 (Rom and Reznick, 2016). Moreover, the overexpression of Atrolnc-1 in wild-type muscles causes increased MuRF-1 expression, which results in myofiber atrophy.
In the heart, cytoplasmic lncRNAs have also been proposed as regulators of the calcium reuptake occurring after cardiac muscle contraction. ZFAS1 lncRNA is markedly upregulated in both mouse and human cardiac tissues subjected to myocardial infarction (MI). While its knockdown restores heart contraction parameters to normal levels in a MI mouse model, its overexpression in wild-type mice induces a MI-like phenotype. Mechanistically, ZFAS1 directly binds the SERCA2a protein, which is responsible for calcium reuptake in the sarcoplasmic reticulum, and it is responsible for the downregulation of the protein levels, thus affecting the Ca2+ transient dynamics (Zhang et al., 2018c). More recently, the same group described a similar mechanism for DACH1 lncRNA. This RNA molecule is upregulated in failing hearts of mice and humans and directly interacts with SERCA2a; its overexpression is associated with an augmented ubiquitination of the protein, driving it to proteasome degradation (Cai B. et al., 2019).
As described for nuclear lncRNAs, also the dysregulation of cytoplasmic lncRNAs expression can result in cardiac hypertrophy. For instance, the murine CTBP1-AS2 lncRNA is selectively upregulated in hypertrophic hearts and in Ang-II treated neonatal rat ventricular myocytes. In the cytoplasm, CTBP1-AS2 interacts with the RNA-binding protein fused in sarcoma (FUS) forming a ribonucleoparticle that includes also TLR4 mRNA. Both CTBP1-AS2 and FUS participate in the anomalous stabilization of TLR4 mRNA, which encodes for a well-known driver of the inflammation process at the base of cardiac hypertrophy (Luo et al., 2019).
lncRNA-Derived Micropeptides
Recently, the functional role of lncRNAs has expanded due to their ability to generate bioactive micropeptides (Table 3), which have been identified mainly in muscle-related functions and cancer development (Ji et al., 2015). The identification of small ORFs that are translatable and encode short peptides remains a major challenge. Two complementary approaches are normally used to discover functional small ORFs: (i) the computational one, based on bioinformatic predictions and (ii) the experimental one, that uses transcriptional and translational data. Several computational tools have been developed to estimate the coding potential of novel identified transcripts. Among the most utilized CPC (Kong et al., 2007) and its updated version CPC2 (Kang et al., 2017), CPAT (Wang L. et al., 2013), COME (Hu et al., 2017), LncRNApred (Pian et al., 2016), PORTRAIT (Arrial et al., 2009), CONC, and others can be cited. The experimental approach is mainly based on the genome-wide translatome that has been studied by ribosome footprinting, a technique introduced by Ingolia in 2009 (Ingolia et al., 2009). The analyses of ribosome dynamics during translation allows the identification of actively translated ORFs (Bazzini et al., 2014) including the ones deriving from previously non-protein coding annotated transcripts. Recently, by applying this technique to human hearts, 169 lncRNAs and 40 circRNAs encoding for previously unknown microproteins have been identified and in vivo validated (van Heesch et al., 2019); among them, already known microproteins such as DWORF (Nelson et al., 2016) and SPAR (Matsumoto et al., 2017) were detected (see below). Interestingly, the identified microproteins were associated to cellular processes mainly related to mitochondria. The identification of micropeptides led to hypothesize a dual function for those lncRNAs that were previously characterized for their non-coding functions. In the van Heesch et al. paper, the authors identified 27 human and 5 murine small ORFs in lncRNAs whose non-coding function was already assessed. Among the 169 identified micropeptides deriving from lncRNAs, 16 were cardiac or skeletal muscle specific, suggesting that the list of lncRNA-derived micropeptides, important for muscle physiology, will grow in the coming years. Among the ones that have been already characterized, myoregulin (MLN) was the first transmembrane microprotein identified; it takes origin from a previously annotated muscle-specific lncRNA (NR_038041). The small ORF, located in the third exon of the lncRNA, encodes for a 46-amino-acid long micropeptide. Olson's group showed that this single transmembrane alpha helix peptide interacts with the sarcoplasmic reticulum Ca2+-ATPase (SERCA) and impedes Ca2+ uptake into the sarcoplasmic reticulum, thus influencing muscle relaxation. According to this observation, MLN showed a strong structural resemblance with the sarcolipin (SLN) and phospholamban (PLN) micropeptides, already known to regulate Ca2+ pump activity by directly interacting with SERCA. MLN is the most abundant micropeptide in adult skeletal muscles. For this reason, the authors suggested MLN as a dominant regulator of SERCA activity. Overexpression of MLN peptide caused a reduction in Ca2+ uptake while MLN-null mice significantly enhanced Ca2+ handling and improved exercise performance (Anderson et al., 2015). A couple of years later, Zhou's lab described a different nuclear non-coding function for the same transcript (NR_038041) that was named Linc-RAM (see above) (Yu et al., 2017). More recently, the same lab extended this work by showing that the fibroblast growth factor FGF2 mediates self-renewal and differentiation of satellite cells by inhibiting Linc-RAM expression in a MyoD-dependent manner (Zhao et al., 2018). Linc-RAM represents a clear case of a non-coding RNA with a well-characterized dual role: one in the nucleus, as non-coding transcript, and the other in the cytoplasm as protein. DWORF (Dwarf Open Reading Frame) is another example of microprotein transcribed from a previously annotated muscle-specific lncRNA. It is highly expressed in the heart and is the third smallest full-length protein annotated in the mouse genome (34 amino acids long). In their paper, Nelson et al. (2016) showed how this peptide, conserved in vertebrates, is able to increase in mouse SERCA pump activity by displacing the already cited inhibitory peptides phospholamban, sarcolipin, and MLN. Hearts from DWORF-overexpressing mice exhibited an increased activity of SERCA, indicating a higher affinity for Ca2+, while slow skeletal muscles from DWORF-KO mice showed a reduced SERCA activity with a delayed Ca2+ clearance and muscle relaxation.
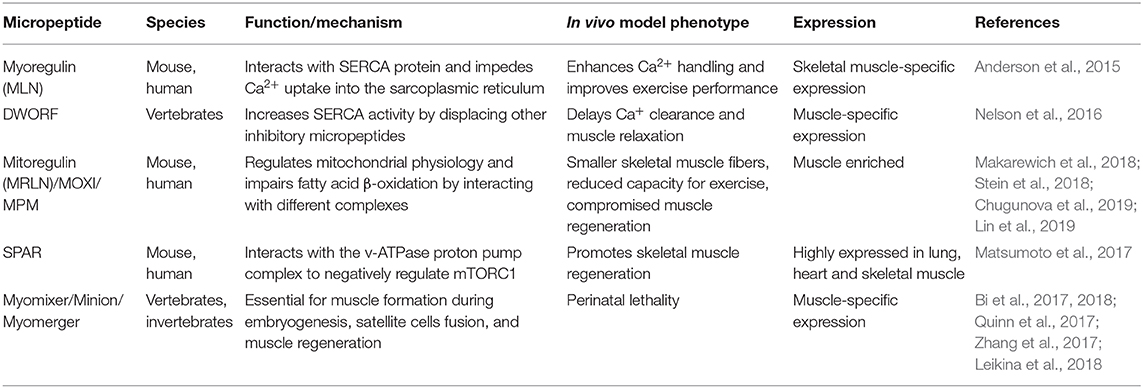
Table 3. Functional long non-coding RNA (lncRNA)-derived micropeptides in skeletal and cardiac myogenesis.
In mouse, 1500011K16Rik (LINC00116 in human) is a recently identified lncRNA enriched in heart and skeletal muscle tissues and highly coexpressed with mitochondrial genes. Two papers, published at the same time by different groups (Makarewich et al., 2018; Stein et al., 2018), demonstrated that this lncRNA contains, within its first exon, a nucleotide region that encodes for a predicted conserved single-pass transmembrane microprotein (56 amino acids long) named as mitoregulin (Mtln) because of its inner mitochondrial membranes' localization (Stein et al., 2018) or micropeptide regulator of β-oxidation (MOXI) because of its role in fatty acid β-oxidation (Makarewich et al., 2018). Stein et al. revealed its involvement in supporting mitochondrial high-molecular-weight supercomplexes assembly and/or stability. They demonstrated the high affinity between Mtln and cardiolipin, a phospholipid important for the maintenance of the integrity on the inner mitochondrial membrane. Makarewich et al. showed the ability of the MOXI micropeptide to interact with the mitochondrial trifunctional protein (MTP), an enzymatic complex that catalyzes the last three steps of long-chain fatty acids oxidation, and both groups suggested that Mtnl/MOXI-KO mice present an impaired fatty acid β-oxidation upon increased metabolic demand, probably due to inefficient mitochondrial complex (ri) organization after metabolic switches. Overexpression in HeLa cells and Mtln-KO mice observation allowed to unravel its ability to influence other mitochondrial processes such as membrane potential, Ca2+ retention capacity, and oxidative stress (Stein et al., 2018). Moreover, Makarewich et al. observed mitochondrial abnormalities in the KO mice. The role of this micropeptide in mitochondrial respiration was confirmed in a work that was under review when the previous two were published (Chugunova et al., 2019). In this paper, the authors demonstrated that the interaction of Mtln with NADH-dependent cytochrome b5 reductase stimulates the activity of mitochondrial respiratory chain complex I by providing a favorable lipid composition of the membrane. The role of Mtln in promoting myogenic differentiation, muscle growth, and regeneration was further studied by Lin et al., which refers to LINC00116 with the name of micropeptide in mitochondria (MPM, Lin et al., 2019).
The small regulatory polypeptide of amino acid response (SPAR) is another polypeptide translated from a lncRNA (LINC00961) that is conserved among species and highly expressed in heart, skeletal muscle, and lung (Matsumoto et al., 2017). SPAR is characterized by a conserved transmembrane domain that allows its localization at the membranes of late endosomes and lysosomes; in the lysosomal membrane, it interacts with two subunits of the v-ATPase proton pump complex to exert a negative regulation on the amino-acid-dependent mTORC1 activation. In the same paper, the authors demonstrated that loss of SPAR promotes post-injury skeletal-muscle regeneration by increasing mTORC1 activation.
It is noteworthy that all the null mice (Mtln-KO, MLN-KO, SPAR-KO, DWORF-KO) analyzed so far did not show any overt phenotype. The only exception is represented by the Myomixer micropeptide (84 amino acids long), which is generated by a previously annotated lncRNA (Gm7325); germline Myomixer null mice are characterized by perinatal lethality caused by the absence of multinucleated myofibers. This lncRNA has been identified in a loss-of-function screen aimed at the identification of essential genes involved in myoblasts fusion (Bi et al., 2017). Myomixer, also known as minion and myomerger, is essential for muscle formation during embryogenesis, satellite cells fusion, and muscle regeneration (Bi et al., 2017, 2018; Quinn et al., 2017; Zhang et al., 2017); it is embedded in the plasma membrane, and it was suggested to interact with the fusogenic protein Myomaker, thus promoting myoblasts fusion. A more recent paper, in contrast with the previous one, indicate that Myomaker does not need Myomixer to promote hemifusion and that the two proteins are involved in different steps of the fusion pathway (Leikina et al., 2018). Even if the mechanism of action remains to be elucidated, Myomixer role in promoting myoblasts fusion seems to be evolutionary conserved in vertebrates: the Zebrafish ortholog was able to induce heterologous cell fusion when overexpressed with the murine Myomaker (Bi et al., 2017).
CircRNAs in Skeletal and Cardiac Myogenesis
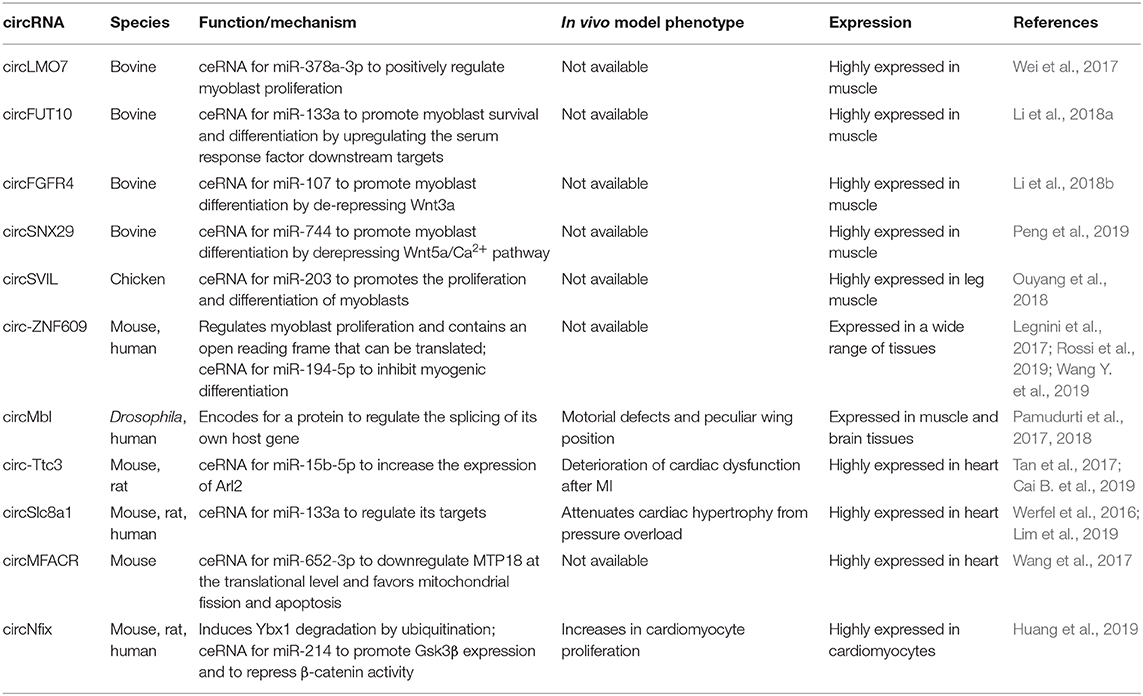
CircRNAs are originated from a non-canonical splicing reaction, called back splicing, in which a 3′ donor site is unconventionally fused to an upstream 5′ acceptor site. This mechanism generates highly stable and covalently closed molecules lacking a 5′ cap and a poly-A tail and thus inaccessible to the action of cellular exonucleases. Even though the first evidence of their existence goes back to the 1970s, circRNAs have been widely studied only after the advent of next generation sequencing technologies, being recognized as a common element of all metazoan transcriptomes (Wilusz, 2018). De novo discovery of circRNAs is based on the presence of exon-junction spanning reads in RNA-seq datasets; the effective existence of a newly identified circRNA then requires PCR validation with divergent primers and RNase R digestion assay to prove exonuclease resistance. This experimental pipeline has allowed the identification and annotation of hundreds of circRNAs, which are expressed and modulated during cultured myoblasts differentiation (Chen et al., 2018) or in developing and aging skeletal muscle of various mammalian species (Li et al., 2017; Wei et al., 2017) (Table 4). The high-throughput discovery of this large amount of circular RNAs has boosted the characterization of their biological significance and the molecular mechanisms of post-transcriptional regulation in which they are involved. A large part of circRNAs show an enrichment for miRNA binding sites in their nucleotide sequence, suggesting a role in competing endogenous RNA networks, which is also corroborated by their high stability. The research of differentially expressed circRNAs between C2C12 myoblasts and myotubes has identified 581 candidates; according to in silico prediction, the top 30 upregulated circRNAs could be part of ceRNA networks involving 91 miRNAs and core myogenic factors like myogenin, Myocyte enhancer factor 2a, myosin heavy chain (Myh)-1, Myh7, and Myh7b (Chen et al., 2018). Similarly, many differentially modulated circRNAs have been identified during several developmental stages of the bovine longissimus dorsi skeletal muscle (Wei et al., 2017). Among them, circLMO7, the most downregulated circRNA between adult and embryonic muscles, was shown to positively regulate myoblast proliferation while reducing myoblast apoptosis and differentiation. The biological function of circLMO7 was correlated with its ability to act as a competing endogenous RNA for miR-378a-3p. Starting from the same dataset, the authors demonstrated that other two candidates impact the development of bovine skeletal muscle through a miRNA-binding activity. Specifically, CircFUT10 was observed to promote myoblasts survival and differentiation and to reduce cell proliferation rate by directly binding to and inhibiting miR-133a; in this way, the circRNA upregulates the serum response factor downstream targets, resulting in a positive impact on myogenesis (Li et al., 2018a). On the other side, CircFGFR4 promotes bovine primary myoblasts differentiation by sequestering miR-107 and thus derepressing its known target Wnt3a, a key upstream factor of the Wnt/β-catenin signaling pathway (Li et al., 2018b). Recently, circSNX29 was identified as another player in the regulation of bovine skeletal muscle development (Peng et al., 2019). This molecule facilitates myoblast differentiation by sponging miR-744 to attenuate its inhibitory effect on the Wnt5a/Ca2+ signaling pathway.
Moreover, the analysis of circRNA expression during chicken skeletal muscle development identified circSVIL. circSVIL promotes the proliferation and differentiation of myoblasts by binding and antagonizing the function of miR-203, a post-transcriptional repressor of c-JUN and MEF2C (Ouyang et al., 2018). Furthermore, a massive high-throughput screening of both proliferating and differentiated human primary myoblasts and mouse C2C12 cells revealed an abundant population of cytoplasmic circRNAs, which showed a global accumulation during differentiation (Legnini et al., 2017). Interestingly, a large part of them was found deregulated in human myoblasts derived from Duchenne muscular dystrophy patients. A subset of highly conserved, strongly modulated circRNAs was selected for a wide phenotypic screening, from which circ-ZNF609 emerged as a regulator of myoblast proliferation, while Circ-QKI and circ-BNC2 showed a positive and negative effect on myogenesis, respectively. Of them, Circ-ZNF609 contains an ORF that can be translated in a cap-independent manner due to the IRES activity of the untranslated region (Legnini et al., 2017). More recently, it has been shown that CircZNF609 is upregulated in rhabdomyosarcoma biopsies and that its knockdown induces a specific arrest of G1/S transition rhabdomyosarcoma-derived cells. The cell cycle block is the result of a strong decrease in phosphorylated Akt protein level and an alteration of the p-Rb/Rb ratio (Rossi et al., 2019). The role of this molecule in tumor progression has been validated also in breast cancer, where high levels of Circ-ZNF609 are associated with poor prognosis (Wang S. et al., 2018). Moreover, the mouse homolog of CircZNF609, named circZfp609, has been shown to inhibit myogenic differentiation by sponging miR-194-5p (Wang Y. et al., 2019).
Circ-ZNF609 is only one of the examples of a protein-coding circRNA in eukaryotes (Chekulaeva and Rajewsky, 2019). The Muscleblind locus encodes for a splicing factor with a critical function in muscle development; its sequestration from aberrant CUG repeats is causative of myotonic dystrophy (Miller et al., 2000). In Drosophila, this locus produces also circMbl, an abundant protein-coding circRNA that regulates splicing of its own host gene (Pamudurti et al., 2017). Interestingly, circMbl knockdown in the whole organism has a specific effect on muscle development and function, as confirmed from gene expression analysis, motorial defects, and a peculiar wing position (Pamudurti et al., 2018). In their latest work, Chen et al. identified a subset of 224 circRNAs modulated during C2C12 differentiation with bona fide coding potential according to the number of open reading frames and N6-methyladenosine motifs, which are known to work as promoting factors for cap-independent translation (Chen et al., 2018). It was recently suggested that circular RNAs expression can be affected by splicing alterations related to myotonic dystrophy type 1 (DM1), a multisystemic disorder in which expanded CTG repeats in the DMPK gene leads to splicing abnormalities. CircRNAs expressed in DM1 skeletal muscles biopsies were identified by analyzing RNA-sequencing datasets, demonstrating the upregulation of CDYL, HIPK3, RTN4_03, and ZNF609 compared to healthy controls (Voellenkle et al., 2019). The circular fraction values were positively correlated with splicing biomarkers of disease severity, reaching higher values in more severely affected patients. Measurement of circular-to-linear ratios for these candidates resulted to be a good prediction method to discriminate DM1 patients from controls, suggesting a possible future use as biomarkers.
The miRNA-binding paradigmatic mechanism has been validated also in cardiac muscle. In a recent study, Tan et al. performed a deep RNA-sequencing from human and mouse hearts and across a differentiation time-course of human embryonic stem-cell-derived cardiomyocytes. They identified a total of 15,318 and 3,017 cardiac circRNAs within human and mouse, respectively. Among them, circ-Ttc3 resulted in one of the top highest expressed circRNAs in mouse heart (Tan et al., 2017). Cai L. et al. (2019) found that circTtc3 was markedly upregulated in the ischemic myocardium, whereas there was no significant change in linear Ttc3 RNA. Overexpression of circ-Ttc3 in cardiomyocytes counteracted hypoxia-induced ATP depletion and apoptotic death. Mechanistically, this circRNA sponges and inhibits miR-15b-5p leading to the increased expression of its target Arl2. Consistently, Arl2 knockdown partially abolished the beneficial effects of circ-Ttc3 overexpression on ATP production and apoptosis of cardiomyocytes. Thus, the Circ-Ttc3-miR-15b-Arl2 regulatory cascade may have a cardioprotective role in myocardial infarction (Cai L. et al., 2019). The same screening allowed the identification of circSlc8a1, which is the most abundant single-exon cardiac expressed. A new study demonstrated that circSlc8a1 inhibition attenuated cardiac hypertrophy, while its forced overexpression resulted in heart failure. As reported for the majority of the circRNAs identified so far, also this circular molecule act in the cytoplasm by sponging a miRNA, in particular the cardiac-enriched mir-133a. Considering that the overexpression of miR-133a attenuates cardiac hypertrophy both in vitro and in vivo, inhibiting circSlc8a1 may result as a novel therapeutic target for cardiac hypertrophy (Lim et al., 2019). An increasing number of studies have indicated that mitochondrial fission dysfunction occurs in many cardiac diseases, such as MI and heart failure. Recently, it has been suggested that circRNAs could play a role also in this context. In this regard, CircMFACR favors mitochondrial fission and apoptosis in the heart by sequestering miR-652-3p, which in turn downregulates MTP18 expression at the translational level. MTP18 is a nuclear-encoded mitochondrial membrane protein whose deficiency reduces mitochondrial fission and suppresses cardiomyocyte apoptosis and MI. Thus, the silencing of MFACR expression leads to a reduction in mitochondrial fragmentation and cell apoptosis. This study revealed that circRNAs can play an active role in the regulation of mitochondrial dynamics, cardiomyocyte apoptosis, and myocardial infarction (Wang et al., 2017). CircRNAs were found to regulate cardiac regeneration. One of the most recent example is circNfix, a superenhancer-regulated circular RNA whose depletion produces cardiomyocyte proliferation and angiogenesis. RNA pulldown and luciferase reporter assays revealed that circNfix acts both as a miR-214 sponge and as a destabilizing partner of Ybx1 via ubiquitin-dependent degradation (Huang et al., 2019).
piRNAs and tRFs in Skeletal and Cardiac Myogenesis
The regulatory potential of the non-protein-coding transcriptome is not limited to the better characterized lncRNAs but includes also less known and more recently discovered RNA species. The piRNAs are a class of small RNAs widely conserved in all the metazoans. Following transcription and processing, the mature piRNA form associates with different members of the PIWI proteins clade, thus creating silencing molecular complexes that defend the germline genome from transposon expression by acting at both transcriptional and post-transcriptional level (Ozata et al., 2019). The common perception of the PIWI–piRNA pathway as a germline-specific feature is nowadays questioned by the evidence of expression of this molecular machinery in a variety of somatic tissues (Lee et al., 2011; Perera et al., 2019). While the functional role of somatic piRNAs is still debated in other tissues, recent works suggest that these small RNAs could regulate heart regeneration due to their effect on the Akt pathway, a fundamental network in heart physiopathology. Microarray profiling of different populations of cardiac progenitors identified a consistent set of piRNAs, which are differentially modulated between cardiospheres, cardiosphere-derived cells, and cardiac fibroblasts, thus creating a specific molecular signature of these cell populations (Vella et al., 2016). In particular, piRNAs which are specifically upregulated in cardiosphere-derived cells act as negative regulators of L1 retrotransposon transcripts, whose inhibition has been shown to decrease upon post-ischemic myocardial damage by the activation of the Akt signaling (Lucchinetti et al., 2006).
tRNAs represent another class of small RNAs which are emerging as regulators of heart and skeletal muscle physiology (Figure 1). They are well-known for their canonical function as adaptor molecules during protein synthesis; however, it has been recently discovered that their controlled endonucleolitic cleavage could represent a source of small regulatory non-coding RNAs. These small molecules were first identified during miRNA library cloning experiments (Fu et al., 2005) and initially classified as random tRNA degradation misproducts; however, the conserved length and the recurrent position of cleavage sites suggested the existence of a dedicated enzymatic activity responsible for their biogenesis. tRNA fragments can be grouped in three categories (Liapi et al., 2019), based on their derivation from the 5′ or 3′ end of parental tRNA (5′/3′ tRFs), from the 3′ portion of the immature tRNA precursor (tRF1s), or from a cleavage event within the anticodon loop which splits the parental molecules in two 30–35 nucleotide long (5′ and 3′) halves. tRNA halves biogenesis is an evolutionary widespread molecular event of particular interest, since they tend to be produced as a response to several cellular stresses by the activity of the angiogenin (ANG) endonuclease. The first report of such mechanism in mammals has been made in tissues of various origin, including ex vivo starved mouse hearts, which showed a specific accumulation of the 5′ portion of Val-AAC tRNA (Fu et al., 2009). The accumulation of a small subset of tRNA halves compared to the complexity of parental tRNA pools corroborates the hypothesis of a controlled processing machinery and of a functional role for tRNA fragments, other than a simple mechanism of stress-induced translational shutdown. Moreover, the expression of specific tRNA fragments was found to be strongly upregulated in a rat model of induced myocardial hypertrophy (Shen et al., 2018); interestingly, transfection of a synthetic version of the two most upregulated tRFs in cultured cardiomyocites increases both hypertrophy marker expression and cell surface area. In particular, Kassiri et al. (2009) show that tRFs1, which derives from tRNA-Gly-GCC cleavage, targets the 3′ untranslated region of Timp3, a well-known cardiac hypertrophy regulatory factor in a miRNA-like manner. Remarkably, mice with induced myocardial hypertrophy had high levels of the identified tRFs also in sperm; a similar pattern of expression of these fragments was found in offspring hearts, which showed an intermediate phenotype with extended heart muscle fiber breakage and fibrosis. Taken together, these findings not only indicate a role for tRFs in the physiopathology of heart but also their involvement on the transgenerational inheritance of a non-genetic stress condition, as already reported for sperm-contained miRNAs (Rodgers et al., 2015).
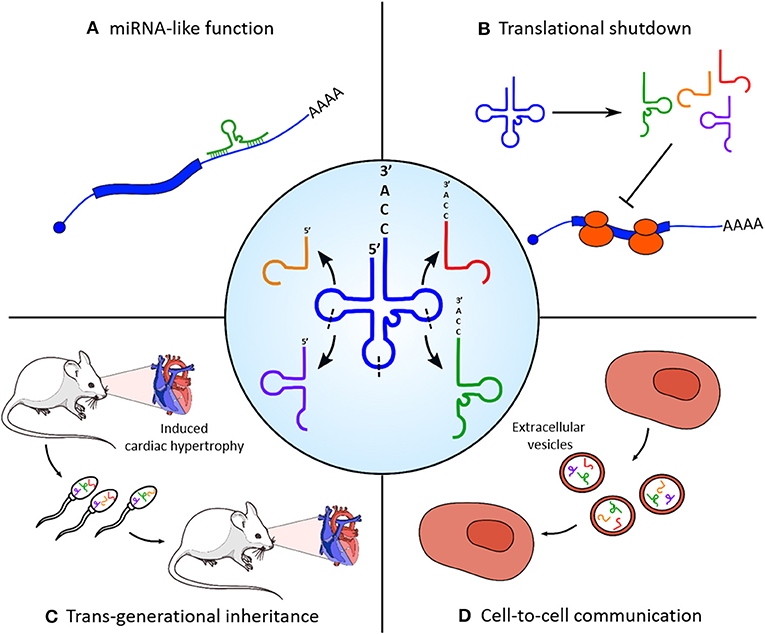
Figure 1. Functional roles of tRNA-derived fragments (tRFs) in skeletal and cardiac muscle homeostasis. Central panel: tRNAs are processed into different categories of tRFs by enzymatic cleavage on specific sites (indicated with dashed lines). (A) tRNA fragments can target the 3′ untranslated region (UTR) of protein-coding transcripts (e.g., Kassiri et al., 2009) and regulate their stability and translation efficiency with a microRNA (miRNA)-like mechanism. (B) Massive cleavage of specific tRNA species in stress conditions induces a rapid translational blockade. (C) Accumulation of tRFs in the sperm is responsible for the non-genetic transmission of myocardial hypertrophy phenotypic traits in the offspring. (D) tRFs produced from myoblasts are selectively loaded in extracellular vesicles and could act as mediators of cell-to-cell communication.
Recent works have shown that small RNAs are actively loaded into extracellular vesicles (EVs) and released in almost all kind of biofluids to act as mediators of cell-to-cell communication (Tkach and Théry, 2016). Even if this phenomenon has been widely studied for miRNAs, there are also evidences of exosome-mediated secretion of other small RNA species, such as piRNAs and tRNA fragments, from different tissue sources (Chiou et al., 2018; Tosar et al., 2018). Of note, the small RNA transcriptome from both human rhabdomyosarcoma RD4 and murine C2C12 myoblasts and of EVs produced by these cells has been recently analyzed (Sork et al., 2018). It emerged that small RNAs are underrepresented in EVs compared to their parental cells, suggesting a loading mechanism that is not explicable with simple diffusion. Moreover, C2C12-produced EVs present a strong enrichment of piRNAs and tRNA fragments compared to intracellular content, including the tRNA-Gly-GCC fragment cited above. Taken together, these evidence suggest that these small RNAs can be selectively included in secretory vesicles, with a potential role in long distance cell-to-cell communication events.
High-throughput small RNA sequencing from four types of wild-type and dystrophic (mdx) muscles and from sera allowed to discover that many piRNAs and tRFs are expressed in skeletal muscle tissue and differentially released in the circulation in dystrophic conditions. Furthermore, the high level of piR_000620 and piR_000935 detected in mdx serum was restored to nearly wild-type levels in response to DMD exon skipping with injected modified oligonucleotides (Coenen-Stass et al., 2018). The differential release of piRNAs has been also observed in serum vesicles isolated from heart failure patients compared to healthy donors, suggesting a possible use of these small RNAs as clinically relevant biomarkers (Yang et al., 2018).
Author Contributions
JM wrote the manuscript and selected the literature. DM and FD reviewed the manuscript and prepared tables and figures. MB proposed the topic, wrote the manuscript, and reviewed the text.
Funding
This work was partially supported by grants from Sapienza University (prot. RM11715C7C8176C1 and RM11916B7A39DCE5) and FFABR Anvur (2017) to MB and from Sapienza University (prot. RM118164363B1D21) to JM.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Alessio, E., Buson, L., Chemello, F., Peggion, C., Grespi, F., Martini, P., et al. (2019). Single cell analysis reveals the involvement of the long non-coding RNA Pvt1 in the modulation of muscle atrophy and mitochondrial network. Nucleic Acids Res. 47, 1653–1670. doi: 10.1093/nar/gkz007
Anderson, D. M., Anderson, K. M., Chang, C. L., Makarewich, C. A., Nelson, B. R., McAnally, J. R., et al. (2015). A micropeptide encoded by a putative long noncoding RNA regulates muscle performance. Cell 160, 595–606. doi: 10.1016/j.cell.2015.01.009
Anderson, K. M., Anderson, D. M., McAnally, J. R., Shelton, J. M., Bassel-Duby, R., and Olson, E. N. (2016). Transcription of the non-coding RNA upperhand controls Hand2 expression and heart development. Nature 539, 433–436. doi: 10.1038/nature20128
Arrial, R. T., Togawa, R. C., and Brigido Mde, M. (2009). Screening non-coding RNAs in transcriptomes from neglected species using PORTRAIT: case study of the pathogenic fungus Paracoccidioides brasiliensis. BMC Bioinformatics 10:239. doi: 10.1186/1471-2105-10-239
Ballarino, M., Cazzella, V., D'Andrea, D., Grassi, L., Bisceglie, L., Cipriano, A., et al. (2015). Novel long noncoding RNAs (lncRNAs) in myogenesis: a miR-31 overlapping lncRNA transcript controls myoblast differentiation. Mol. Cell Biol. 35, 728–736. doi: 10.1128/MCB.01394-14
Ballarino, M., Cipriano, A., Tita, R., Santini, T., Desideri, F., Morlando, M., et al. (2018). Deficiency in the nuclear long noncoding RNA Charme causes myogenic defects and heart remodeling in mice. EMBO J. 37:18. doi: 10.15252/embj.201899697
Ballarino, M., Morlando, M., Fatica, A., and Bozzoni, I. (2016). Non-coding RNAs in muscle differentiation and musculoskeletal disease. J Clin Invest. 126, 2021–2030. doi: 10.1172/JCI84419.
Batista, P. J., and Chang, H. Y. (2013). Long noncoding RNAs: cellular address codes in development and disease. Cell 152, 1298–1307. doi: 10.1016/j.cell.2013.02.012
Bazzini, A. A., Johnstone, T. G., Christiano, R., Mackowiak, S. D., Obermayer, B., Fleming, E. S., et al. (2014). Identification of small ORFs in vertebrates using ribosome footprinting and evolutionary conservation. EMBO J. 33, 981–993. doi: 10.1002/embj.201488411
Bi, P., McAnally, J. R., Shelton, J. M., Sánchez-Ortiz, E., Bassel-Duby, R., and Olson, E. N. (2018). Fusogenic micropeptide Myomixer is essential for satellite cell fusion and muscle regeneration. Proc. Natl. Acad. Sci. U.S.A. 115, 3864–3869. doi: 10.1073/pnas.1800052115
Bi, P., Ramirez-Martinez, A., Li, H., Cannavino, J., McAnally, J. R., Shelton, J. M., et al. (2017). Control of muscle formation by the fusogenic micropeptide myomixer. Science 356, 323–327. doi: 10.1126/science.aam9361
Cai, B., Zhang, Y., Zhao, Y., Wang, J., Li, T., Zhang, Y., et al. (2019). Long noncoding RNA-DACH1 (Dachshund Homolog 1) regulates cardiac function by inhibiting SERCA2a (sarcoplasmic reticulum calcium ATPase 2a). Hypertension 74, 833–842. doi: 10.1161/HYPERTENSIONAHA.119.12998
Cai, L., Qi, B., Wu, X., Peng, S., Zhou, G., Wei, Y., et al. (2019). Circular RNA Ttc3 regulates cardiac function after myocardial infarction by sponging miR-15b. J. Mol. Cell Cardiol. 130, 10–22. doi: 10.1016/j.yjmcc.2019.03.007
Caretti, G., Schiltz, R. L., Dilworth, F. J., Di Padova, M., Zhao, P., Ogryzko, V., et al. (2006). The RNA helicases p68/p72 and the noncoding RNA SRA are coregulators of MyoD and skeletal muscle differentiation. Dev. Cell. 11, 547–560. doi: 10.1016/j.devcel.2006.08.003
Cesana, M., Cacchiarelli, D., Legnini, I., Santini, T., Sthandier, O., Chinappi, M., et al. (2011). A long noncoding RNA controls muscle differentiation by functioning as a competing endogenous RNA. Cell 147, 358–369. doi: 10.1016/j.cell.2011.09.028. Erratum in: Cell. (2011) 147,947.
Chekulaeva, M., and Rajewsky, N. (2019). Roles of long noncoding RNAs and circular RNAs in translation. Cold Spring Harb Perspect Biol. 11:6. doi: 10.1101/cshperspect.a032680
Chen, C. K., Blanco, M., Jackson, C., Aznauryan, E., Ollikainen, N., Surka, C., et al. (2016). Xist recruits the X chromosome to the nuclear lamina to enable chromosome-wide silencing. Science 354, 468–472. doi: 10.1126/science.aae0047
Chen, L. L. (2016). The biogenesis and emerging roles of circular RNAs. Nat. Rev. Mol. Cell Biol. 17, 205–211. doi: 10.1038/nrm.2015.32
Chen, R., Jiang, T., Lei, S., She, Y., Shi, H., Zhou, S., et al. (2018). Expression of circular RNAs during C2C12 myoblast differentiation and prediction of coding potential based on the number of open reading frames and N6-methyladenosine motifs. Cell Cycle 17, 1832–1845. doi: 10.1080/15384101.2018.1502575
Chiou, N. T., Kageyama, R., and Ansel, K. M. (2018). Selective export into extracellular vesicles and function of tRNA fragments during T cell activation. Cell Rep. 25, 3356–3370. doi: 10.1016/j.celrep.2018.11.073
Chugunova, A., Loseva, E., Mazin, P., Mitina, A., Navalayeu, T., Bilan, D., et al. (2019). LINC00116 codes for a mitochondrial peptide linking respiration and lipid metabolism. Proc. Natl. Acad. Sci. U.S.A. 116, 4940–4945. doi: 10.1073/pnas.1809105116
Clemson, C. M., Hutchinson, J. N., Sara, S. A., Ensminger, A. W., Fox, A. H., Chess, A., et al. (2009). An architectural role for a nuclear noncoding RNA: NEAT1 RNA is essential for the structure of paraspeckles. Mol Cell. 33, 717–726. doi: 10.1016/j.molcel.2009.01.026
Coenen-Stass, A. M. L., Sork, H., Gatto, S., Godfrey, C., Bhomra, A., Krjutškov, K., et al. (2018). Comprehensive RNA-sequencing analysis in serum and muscle reveals novel small RNA signatures with biomarker potential for DMD. Mol. Ther. Nucleic Acids 13, 1–15. doi: 10.1016/j.omtn.2018.08.005
Colpaert, R. M. W., and Calore, M. (2019). microRNAs in cardiac diseases. Cells 8:737. doi: 10.3390/cells8070737
Davis, R. L., Weintraub, H., and Lassar, A. B. (1987). Expression of a single transfected cDNA converts fibroblasts to myoblasts. Cell 51, 987–1000. doi: 10.1016/0092-8674(87)90585-x
Dimartino, D., Colantoni, A., Ballarino, M., Martone, J., Mariani, D., Danner, J., et al. (2018). The long non-coding RNA lnc-31 interacts with Rock1 mRNA and mediates its YB-1-dependent translation. Cell Rep. 23, 733–740. doi: 10.1016/j.celrep.2018.03.101
Du, J., Zhang, P., Zhao, X., He, J., Xu, Y., Zou, Q., et al. (2019). MicroRNA-351-5p mediates skeletal myogenesis by directly targeting lactamase-β and is regulated by lnc-mg. FASEB J. 33, 1911–1926. doi: 10.1096/fj.201701394RRR
Engreitz, J. M., Pandya-Jones, A., McDonel, P., Shishkin, A., Sirokman, K., Surka, C., et al. (2013). The Xist lncRNA exploits three-dimensional genome architecture to spread across the X chromosome. Science 341:1237973. doi: 10.1126/science.1237973
Engreitz, J. M., Sirokman, K., McDonel, P., Shishkin, A. A., Surka, C., Russell, P., et al. (2014). RNA-RNA interactions enable specific targeting of noncoding RNAs to nascent Pre-mRNAs and chromatin sites. Cell 159, 188–199. doi: 10.1016/j.cell.2014.08.018
Fatica, A., and Bozzoni, I. (2014). Long non-coding RNAs: new players in cell differentiation and development. Nat. Rev. Genet. 15, 7–21. doi: 10.1038/nrg3606
Fu, H., Feng, J., Liu, Q., Sun, F., Tie, Y., Zhu, J., et al. (2009). Stress induces tRNA cleavage by angiogenin in mammalian cells. FEBS Lett. 583, 437–442. doi: 10.1016/j.febslet.2008.12.043
Fu, H., Tie, Y., Xu, C., Zhang, Z., Zhu, J., Shi, Y., et al. (2005). Identification of human fetal liver miRNAs by a novel method. FEBS Lett. 579, 3849–3854. doi: 10.1016/j.febslet.2005.05.064
Ge, Y., Sun, Y., and Chen, J. (2011). IGF-II is regulated by microRNA-125b in skeletal myogenesis. J. Cell Biol. 192, 69–81. doi: 10.1083/jcb.201007165
Ghafouri-Fard, S., and Taheri, M. (2019). Nuclear enriched abundant transcript 1 (NEAT1): a long non-coding RNA with diverse functions in tumorigenesis. Biomed. Pharmacother. 111, 51–59. doi: 10.1016/j.biopha.2018.12.070
Gong, C., Li, Z., Ramanujan, K., Clay, I., Zhang, Y., Lemire-Brachat, S., et al. (2015). A long non-coding RNA, LncMyoD, regulates skeletal muscle differentiation by blocking IMP2-mediated mRNA translation. Dev. Cell 34, 181–191. doi: 10.1016/j.devcel.2015.05.009
Grote, P., Wittler, L., Hendrix, D., Koch, F., Währisch, S., Beisaw, A., et al. (2013). The tissue-specific lncRNA Fendrr is an essential regulator of heart and body wall development in the mouse. Dev. Cell 24, 206–214.
Guttman, M., and Rinn, J. L. (2012). Modular regulatory principles of large non-coding RNAs. Nature 482,339–346. doi: 10.1038/nature10887
Hacisuleyman, E., Goff, L. A., Trapnell, C., Williams, A., Henao-Mejia, J., Sun, L., et al. (2014). Topological organization of multichromosomal regions by the long intergenic noncoding RNA Firre. Nat. Struct. Mol. Biol. 21, 198–206. doi: 10.1038/nsmb.2764
Han, P., Li, W., Lin, C. H., Yang, J., Shang, C., Nurnberg, S. T., et al. (2014). A long noncoding RNA protects the heart from pathological hypertrophy. Nature 514, 102–106. doi: 10.1038/nature13596
Han, X., Yang, F., Cao, H., and Liang, Z. (2015). Malat1 regulates serum response factor through miR-133 as a competing endogenous RNA in myogenesis. FASEB J. 29, 3054–3064. doi: 10.1096/fj.14-259952
Heard, E., and Disteche, C. M. (2006). Dosage compensation in mammals: fine-tuning the expression of the X chromosome. Genes Dev. 20, 1848–1867. doi: 10.1101/gad.1422906
Hitachi, K., Nakatani, M., Takasaki, A., Ouchi, Y., Uezumi, A., Ageta, H., et al. (2019b). Myogenin promoter-associated lncRNA Myoparr is essential for myogenic differentiation. EMBO Rep. 20:3. doi: 10.15252/embr.201847468
Hitachi, K., Nakatani, M., and Tsuchida, K. (2019a). Long non-coding RNA myoparr regulates GDF5 expression in denervated mouse skeletal muscle. Noncoding RNA 5:2. doi: 10.3390/ncrna5020033
Hoagland, M. B., Stephenson, M. L., Scott, J. F., Hecht, L. I., and Zamecnik, P. C. (1958). A soluble ribonucleic acid intermediate in protein synthesis. J. Biol. Chem. 231, 241–257.
Hon, C. C., Ramilowski, J. A., Harshbarger, J., Bertin, N., Rackham, O. J., Gough, J., et al. (2017). An atlas of human long non-coding RNAs with accurate 5' ends. Nature 543, 199–204. doi: 10.1038/nature21374
Hooper, C., Jackson, S. S., Coughlin, E. E., Coon, J. J., and Miyamoto, S. (2014). Covalent modification of the NF-κB essential modulator (NEMO) by a chemical compound can regulate its ubiquitin binding properties in vitro. J. Biol. Chem. 289, 33161–33174. doi: 10.1074/jbc.M114.582478
Hu, L., Xu, Z., Hu, B., and Lu, Z. J. (2017). COME: a robust coding potential calculation tool for lncRNA identification and characterization based on multiple features. Nucleic Acids Res. 45:e2. doi: 10.1093/nar/gkw79
Huang, S., Li, X., Zheng, H., Si, X., Li, B., Wei, G., et al. (2019). Loss of super-enhancer-regulated circRNA Nfix induces cardiac regeneration after myocardial infarction in adult mice. Circulation. 139, 2857–2876. doi: 10.1161/CIRCULATIONAHA.118.038361
Hubé, F., Velasco, G., Rollin, J., Furling, D., and Francastel, C. (2011). Steroid receptor RNA activator protein binds to and counteracts SRA RNA-mediated activation of MyoD and muscle differentiation. Nucleic Acids Res. 39, 513–525. doi: 10.1093/nar/gkq833
Ingolia, N. T., Ghaemmaghami, S., Newman, J. R., and Weissman, J. S. (2009). Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science 324, 218–223. doi: 10.1126/science.1168978
Jeck, W. R., Sorrentino, J. A., Wang, K., Slevn, M. K., Burd, C. E., Liu, J., et al. (2013). Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA 19, 141–157. doi: 10.1261/rna.035667.112
Ji, Z., Song, R., Regev, A., and Struhl, K. (2015). Many lncRNAs, 5'UTRs, and pseudogenes are translated and some are likely to express functional proteins. Elife 4:e08890. doi: 10.7554/eLife.08890
Jiao, L., Li, M., Shao, Y., Zhang, Y., Gong, M., Yang, X., et al. (2019). lncRNA-ZFAS1 induces mitochondria-mediated apoptosis by causing cytosolic Ca2+ overload in myocardial infarction mice model. Cell Death Dis. 10:942. doi: 10.1038/s41419-019-2136-6
Jin, J. J., Lv, W., Xia, P., Xu, Z. Y., Zheng, A. D., Wang, X. J., et al. (2018). Long noncoding RNA SYISL regulates myogenesis by interacting with polycomb repressive complex 2. Proc. Natl. Acad. Sci. U.S.A. 115, E9802–E9811. doi: 10.1073/pnas.1801471115
Kallen, A. N., Zhou, X. B., Xu, J., Qiao, C., Ma, J., Yan, L., et al. (2013). The imprinted H19 lncRNA antagonizes let-7 microRNAs. Mol. Cell. 52, 101–112. doi: 10.1016/j.molcel.2013.08.027
Kang, Y. J., Yang, D. C., Kong, L., Hou, M., Meng, Y. Q., Wei, L., et al. (2017). CPC2: a fast and accurate coding potential calculator based on sequence intrinsic features. Nucleic Acids Res. 41, W12–W16. doi: 10.1093/nar/gkx428
Kapranov, P., Cheng, J., Dike, S., Nix, D. A., Duttagupta, R., Willingham, A. T., et al. (2007). RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science 316, 1484–1488. doi: 10.1126/science.1138341
Kassiri, Z., Defamie, V., Hariri, M., Oudit, G. Y., Anthwal, S., Dawood, F., et al. (2009). Simultaneous transforming growth factor beta-tumor necrosis factor activation and cross-talk cause aberrant remodeling response and myocardial fibrosis in Timp3-deficient heart. J. Biol. Chem. 284, 29893–29904. doi: 10.1074/jbc.M109.028449
Klattenhoff, C. A., Scheuermann, J. C., Surface, L. E., Bradley, R. K., Fields, P. A., Steinhauser, M. L., et al. (2013). Braveheart, a long noncoding RNA required for cardiovascular lineage commitment. Cell 152, 570–583. doi: 10.1016/j.cell.2013.01.003
Kong, L., Zhang, Y., Ye, Z. Q., Liu, X. Q., Zhao, S. Q., Wei, L., et al. (2007). CPC: assess the protein-coding potential of transcripts using sequence features and support vector machine. Nucleic Acids Res. 35, W345–W349. doi: 10.1093/nar/gkm391
Lagos-Quintana, M., Rauhut, R., Lendeckel, W., and Tuschl, T. (2001). Identification of novel genes coding for small expressed RNAs. Science 294, 853–858. doi: 10.1126/science.1064921
Lau, N. C., Lim, L. P., Weinstein, E. G., and Bartel, D. P. (2001). An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science 294, 858–862. doi: 10.1126/science.1065062
Lee, E. J., Banerjee, S., Zhou, H., Jammalamadaka, A., Arcila, M., Manjunath, B. S., et al. (2011). Identification of piRNAs in the central nervous system. RNA 17, 1090–1099. doi: 10.1261/rna.2565011
Lee, R. C., Feinbaum, R. L., and Ambros, V. (1993). The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 75, 843–54, doi: 10.1016/0092-8674(93)90529-Y
Legnini, I., Di Timoteo, G., Rossi, F., Morlando, M., Briganti, F., Sthandier, O., et al. (2017). Circ-ZNF609 is a circular rna that can be translated and functions in myogenesis. Mol. Cell 66, 22–37.e9. doi: 10.1016/j.molcel.2017.02.017
Leikina, E., Gamage, D. G., Prasad, V., Goykhberg, J., Crowe, M., Diao, J., et al. (2018). Myomaker and Myomerger work independently to control distinct steps of membrane remodeling during myoblast fusion. Dev. Cell 46, 767–780. doi: 10.1016/j.devcel.2018.08.006
Li, C., Li, X., Yao, Y., Ma, Q., Ni, W., Zhang, X., et al. (2017). Genome-wide analysis of circular RNAs in prenatal and postnatal muscle of sheep. Oncotarget 8, 97165–97177. doi: 10.18632/oncotarget.21835
Li, H., Wei, X., Yang, J., Dong, D., Hao, D., Huang, Y., et al. (2018a). circFGFR4 promotes differentiation of myoblasts via binding miR-107 to relieve its inhibition of Wnt3a. Mol. Ther. Nucleic Acids 11, 272–283. doi: 10.1016/j.omtn.2018.02.012
Li, H., Yang, J., Wei, X., Song, C., Dong, D., Huang, Y., et al. (2018b). CircFUT10 reduces proliferation and facilitates differentiation of myoblasts by sponging miR-133a. J. Cell. Physiol. 233, 4643–4651. doi: 10.1002/jcp.26230
Liang, T., Zhou, B., Shi, L., Wang, H., Chu, Q., Xu, F., et al. (2018). lncRNA AK017368 promotes proliferation and suppresses differentiation of myoblasts in skeletal muscle development by attenuating the function of miR-30c. FASEB J. 32, 377–389. doi: 10.1096/fj.201700560RR
Liapi, E., van Bilsen, M., Verjans, R., and Schroen, B. (2019). tRNAs and tRNA fragments as modulators of cardiac and skeletal muscle function. Biochim. Biophys. Acta Mol. Cell Res. doi: 10.1016/j.bbamcr.2019.03.012. [Epub ahead of print].
Licatalosi, D. D., Mele, A., Fak, J. J., Ule, J., Kayikci, M., Chi, S. W., et al. (2008). HITS-CLIP yields genome-wide insights into brain alternative RNA processing. Nature 456, 464–469. doi: 10.1038/nature07488
Lim, T. B., Aliwarga, E., Luu, T. D. A., Li, Y. P., Ng, S. L., Annadoray, L., et al. (2019). Targeting the highly abundant circular RNA circSlc8a1 in cardiomyocytes attenuates pressure overload induced hypertrophy. Cardiovasc. Res. 115, 1998–2007. doi: 10.1093/cvr/cvz130
Lin, Y. F., Xiao, M. H., Chen, H. X., Meng, Y., Zhao, N., Yang, L., et al. (2019). A novel mitochondrial micropeptide MPM enhances mitochondrial respiratory activity and promotes myogenic differentiation. Cell Death Dis. 10:528. doi: 10.1038/s41419-019-1767-y
Lucchinetti, E., Feng, J., Silva, R.d., Tolstonog, G. V., Schaub, M. C., Schumann, G. G., et al. (2006). Inhibition of LINE-1 expression in the heart decreases ischemic damage by activation of Akt/PKB signaling. Physiol. Genomics 25, 314–324. doi: 10.1152/physiolgenomics.00251.2005
Luo, X., He, S., Hu, Y., Liu, J., and Chen, X. (2019). Sp1-induced LncRNA CTBP1-AS2 is a novel regulator in cardiomyocyte hypertrophy by interacting with FUS to stabilize TLR4. Cardiovasc. Pathol. 42, 21–29. doi: 10.1016/j.carpath.2019.04.005
Makarewich, C. A., Baskin, K. K., Munir, A. Z., Bezprozvannaya, S., Sharma, G., Khemtong, C., et al. (2018). MOXI is a mitochondrial micropeptide that enhances fatty acid β-oxidation. Cell Rep. 23, 3701–3709. doi: 10.1016/j.celrep.2018.05.058
Matsumoto, A., Pasut, A., Matsumoto, M., Yamashita, R., Fung, J., Monteleone, E., et al. (2017). mTORC1 and muscle regeneration are regulated by the LINC00961-encoded SPAR polypeptide. Nature 541, 228–232. doi: 10.1038/nature21034
Mattick, J. S. (2011). The central role of RNA in human development and cognition. FEBS Lett. 585, 1600–1616. doi: 10.1016/j.febslet.2011.05.001
Mauro, C., Pacifico, F., Lavorgna, A., Mellone, S., Iannetti, A., Acquaviva, R., et al. (2006). ABIN-1 binds to NEMO/IKKgamma and co-operates with A20 in inhibiting NF-kappaB. J. Biol. Chem. 281, 18482–18488. doi: 10.1074/jbc.M601502200
McHugh, C. A., and Guttman, M. (2018). RAP-MS: a method to identify proteins that interact directly with a specific RNA molecule in cells. Methods Mol. Biol. 1649, 473–488. doi: 10.1007/978-1-4939-7213-5_31
Militello, G., Hosen, M. R., Ponomareva, Y., Gellert, P., Weirick, T., John, D., et al. (2018). A novel long non-coding RNA Myolinc regulates myogenesis through TDP-43 and Filip1. J. Mol. Cell Biol. 10, 102–117. doi: 10.1093/jmcb/mjy025
Miller, J. W., Urbinati, C. R., Teng-Umnuay, P., Stenberg, M. G., Byrne, B. J., Thornton, C. A., et al. (2000). Recruitment of human muscleblind proteins to (CUG)n expansions associated with myotonic dystrophy. EMBO J. 19, 4439–4448. doi: 10.1093/Emboj/19.17.4439
Mousavi, K., Zare, H., Dell'orso, S., Grontved, L., Gutierrez-Cruz, G., Derfoul, A., et al. (2013). eRNAs promote transcription by establishing chromatin accessibility at defined genomic loci. Mol. Cell 51, 606–617. doi: 10.1016/j.molcel.2013.07.022
Nagano, T., and Fraser, P. (2011). No-nonsense functions for long noncoding RNAs. Cell 145, 178–181. doi: 10.1016/j.cell.2011.03.014;
Neguembor, M. V., Jothi, M., and Gabellini, D. (2014). Long noncoding RNAs, emerging players in muscle differentiation and disease. Skelet. Muscle 4:8. doi: 10.1186/2044-5040-4-8
Nelson, B. R., Makarewich, C. A., Anderson, D. M., Winders, B. R., Troupes, C. D., Wu, F., et al. (2016). A peptide encoded by a transcript annotated as long noncoding RNA enhances SERCA activity in muscle. Science 351, 271–275. doi: 10.1126/science.aad4076
Neppl, R. L., Wu, C. L., and Walsh, K. (2017). lncRNA Chronos is an aging-induced inhibitor of muscle hypertrophy. J. Cell. Biol. 216, 3497–3507. doi: 10.1083/jcb.201612100
Nishi, K., Nishi, A., Nagasawa, T., and Ui-Tei, K. (2013). Human TNRC6A is an Argonaute-navigator protein for microRNA-mediated gene silencing in the nucleus. RNA 19, 17–35. doi: 10.1261/rna.034769.112
Ouyang, H., Chen, X., Li, W., Li, Z., Nie, Q., and Zhang, X. (2018). Circular RNA circSVIL promotes myoblast proliferation and differentiation by sponging miR-203 in chicken. Front. Genet. 9:172. doi: 10.3389/fgene.2018.00172
Ozata, D. M., Gainetdinov, I., Zoch, A., O'Carroll, D., and Zamore, P. D. (2019). PIWI-interacting RNAs: small RNAs with big functions. Nat. Rev. Genet. 20, 89–108. doi: 10.1038/s41576-018-0073-3
Pamudurti, N. R., Bartok, O., Jens, M., Ashwal-Fluss, R., Stottmeister, C., Ruhe, L., et al. (2017). Translation of CircRNAs. Mol. Cell. 66, 9–21.e7. doi: 10.1016/j.molcel.2017.02.021
Pamudurti, N. R., Konakondla-Jacob, V. V., Krishnamoorthy, A., Ashwal-Fluss, R., Bartok, O., Wüst, S., et al. (2018). An in vivo knockdown strategy reveals multiple functions for circMbl. bioRxiv. doi: 10.1101/483271
Peng, S., Song, C., Li, H., Cao, X., Ma, Y., Wang, X., et al. (2019). Circular RNA SNX29 sponges miR-744 to regulate proliferation and differentiation of Myoblasts by activating the Wnt5a/Ca(2+) signaling pathway. Mol. Ther. Nucleic Acids. 16, 481–493. doi: 10.1016/j.omtn.2019.03.009
Perera, B. P. U., Tsai, Z. T., Colwell, M. L., Jones, T. R., Goodrich, J. M., Wang, K., et al. (2019). Somatic expression of piRNA and associated machinery in the mouse identifies short, tissue-specific piRNA. Epigenetics 14, 504–521. doi: 10.1080/15592294.2019.1600389
Pian, C., Zhang, G., Chen, Z., Chen, Y., Zhang, J., Yang, T., et al. (2016). LncRNApred: classification of long non-coding RNAs and protein-coding transcripts by the ensemble algorithm with a new hybrid feature. PLoS ONE 11:e0154567. doi: 10.1371/journal.pone.0154567
Piccoli, M. T., Gupta, S. K., Viereck, J., Foinquinos, A., Samolovac, S., Kramer, F. L., et al. (2017). Inhibition of the cardiac fibroblast-enriched lncRNA Meg3 prevents cardiac fibrosis and diastolic dysfunction. Circ. Res. 121, 575–583. doi: 10.1161/CIRCRESAHA.117.310624
Prinz, F., Kapeller, A., Pichler, M., and Klec, C. (2019). The implications of the long non-coding RNA NEAT1 in non-cancerous diseases. Int. J. Mol. Sci. 20:3. doi: 10.3390/ijms20030627
Quinn, M. E., Goh, Q., Kurosaka, M., Gamage, D. G., Petrany, M. J., Prasad, V., et al. (2017). Myomerger induces fusion of non-fusogenic cells and is required for skeletal muscle development. Nat. Commun. 8:15665. doi: 10.1038/ncomms15665
Reddy, R., and Busch, H. (1988). “Small nuclear RNAs: RNA sequences, structure, and modifications,” in Structure and Function of Major and Minor Small NuclearRibonucleoprotein Particles, ed Birnstiel ML (New York, NY: Springer-Verlag), 1–37.
Ribeiro, D. M., Zanzoni, A., Cipriano, A., Delli Ponti, R., Spinelli, L., Ballarino, M., et al. (2018). Protein complex scaffolding predicted as a prevalent function of long non-coding RNAs. Nucleic Acids Res. 46, 917–928. doi: 10.1093/nar/gkx1169
Rinn, J. L., and Chang, H. Y. (2012). Genome regulation by long noncoding RNAs. Ann. Rev. Biochem. 81, 145–166. doi: 10.1146/annurev-biochem-051410-092902
Ritter, N., Ali, T., Kopitchinski, N., Schuster, P., Beisaw, A., Hendrix, D. A., et al. (2019). The lncRNA locus handsdown regulates cardiac gene programs and is essential for early mouse development. Dev. Cell. 50, 644–657. doi: 10.1016/j.devcel.2019.07.013
Rodgers, A. B., Morgan, C. P., Leu, N. A., and Bale, T. L. (2015). Transgenerational epigenetic programming via sperm microRNA recapitulates effects of paternal stress. Proc. Natl. Acad. Sci. U.S.A. 112, 13699–13704. doi: 10.1073/pnas.1508347112
Rom, O., and Reznick, A. Z. (2016). The role of E3 ubiquitin-ligases MuRF-1 and MAFbx in loss of skeletal muscle mass. Free Rad. Biol. Med. 98, 218–230. doi: 10.1016/j.freeradbiomed.2015.12.031
Rossi, F., Legnini, I., Megiorni, F., Colantoni, A., Santini, T., Morlando, M., et al. (2019). Circ-ZNF609 regulates G1-S progression in rhabdomyosarcoma. Oncogene 38, 3843–3854. doi: 10.1038/s41388-019-0699-4
Rotini, A., Martínez-Sarr,à, E., Pozzo, E., and Sampaolesi, M. (2018). Interactions between microRNAs and long non-coding RNAs in cardiac development and repair. Pharmacol. Res. 127, 58–66. doi: 10.1016/j.phrs.2017.05.029
Scherrer, K., and Darnell, J. E. (1962). Sedimentation characteristics of rapidly labelled RNA from HeLa cells. Biochem. Biophys. Res. Commun. 7, 486–490. doi: 10.1016/0006-291x(62)90341-8
Shen, L., Gan, M., Tan, Z., Jiang, D., Jiang, Y., Li, M., et al. (2018). A novel class of tRNA-derived small non-coding RNAs respond to myocardial hypertrophy and contribute to intergenerational inheritance. Biomolecules 8:3. doi: 10.3390/biom8030054
Sork, H., Corso, G., Krjutskov, K., Johansson, H. J., Nordin, J. Z., Wiklander, O. P. B., et al. (2018). Heterogeneity and interplay of the extracellular vesicle small RNA transcriptome and proteome. Sci. Rep. 8:10813. doi: 10.1038/s41598-018-28485-9
Souquere, S., Beauclair, G., Harper, F., Fox, A., and Pierron, G. (2010). Highly ordered spatial organization of the structural long noncoding NEAT1 RNAs within paraspeckle nuclear bodies. Mol. Biol. Cell 21, 4020–4027. doi: 10.1091/mbc.E10-08-0690
Stein, C. S., Jadiya, P., Zhang, X., McLendon, J. M., Abouassaly, G. M., Witmer, N. H., et al. (2018). Mitoregulin: A lncRNA-encoded microprotein that supports mitochondrial supercomplexes and respiratory efficiency. Cell Rep. 23, 3710–3720.e8. doi: 10.1016/j.celrep.2018.06.002
Steitz, J. A., and Jakes, K. (1975). How ribosomes select initiator regions in mRNA: base pair formation between the 3' terminus of 16S rRNA and the mRNA during initiation of protein synthesis in Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 72, 4734–4738.
Sui, Y., Han, Y., Zhao, X., Li, D., and Li, G. (2019). Long non-coding RNA Irm enhances myogenic differentiation by interacting with MEF2D. Cell Death Dis. 10:181. doi: 10.1038/s41419-019-1399-2
Sun, L., Si, M., Liu, X., Choi, J. M., Wang, Y., Thomas, S. S., et al. (2018). Long-noncoding RNA Atrolnc-1 promotes muscle wasting in mice with chronic kidney disease. J. Cachexia Sarcopenia Muscle 9, 962–974. doi: 10.1002/jcsm.12321
Tajbakhsh, S., Borello, U., Vivarelli, E., Kelly, R., Papkoff, J., Duprez, D., et al. (1998). Differential activation of Myf5 and MyoD by different Wnts in explants of mouse paraxial mesoderm and the later activation of myogenesis in the absence of Myf5. Development. 125, 4155–4162.
Tan, W. L., Lim, B. T., Anene-Nzelu, C. G., Ackers-Johnson, M., Dashi, A., See, K., et al. (2017). A landscape of circular RNA expression in the human heart. Cardiovasc. Res. 113, 298–309. doi: 10.1093/cvr/cvw250
Tkach, M., and Théry, C. (2016). Communication by extracellular vesicles: where we are and where we need to go. Cell 164, 1226–1232. doi: 10.1016/j.cell.2016.01.043
Tosar, J. P., Rovira, C., and Cayota, A. (2018). Non-coding RNA fragments account for the majority of annotated piRNAs expressed in somatic non-gonadal tissues. Commun. Biol. 1:2. doi: 10.1038/s42003-017-0001-7
Tripathi, V., Ellis, J. D., Shen, Z., Song, D. Y., Pan, Q., Watt, A. T., et al. (2010). The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Mol. Cell. 39, 925–938. doi: 10.1016/j.molcel.2010.08.011
Tseng, Y. Y., and Bagchi, A. (2015). The PVT1-MYC duet in cancer. Mol. Cell Oncol. 2:e974467. doi: 10.4161/23723556.2014.974467
Ule, J., Jensen, K. B., Ruggiu, M., Mele, A., Ule, A., and Darnell, R. B. (2003). CLIP identifies Nova-regulated RNA networks in the brain. Science. 302, 1212–1215. doi: 10.1126/science.1090095
Ulitsky, I., and Bartel, D. P. (2013). lincRNAs: genomics, evolution, and mechanisms. Cell 154, 26–46. doi: 10.1016/j.cell.2013.06.020
van Heesch, S., van Iterson, M., Jacobi, J., Boymans, S., Essers, P. B., de Bruijn, E., et al. (2014). Extensive localization of long noncoding RNAs to the cytosol and mono- and polyribosomal complexes. Genome Biol. 15:R6. doi: 10.1186/gb-2014-15-1-r6
van Heesch, S., Witte, F., Schneider-Lunitz, V., Schulz, J. F., Adami, E., and Faber, A. B. (2019). The translational landscape of the human heart. Cell 178, 242–260. doi: 10.1016/j.cell.2019.05.010
Vella, S., Gallo, A., Lo Nigro, A., Galvagno, D., Raffa, G. M., Pilato, M., et al. (2016). PIWI-interacting RNA (piRNA) signatures in human cardiac progenitor cells. Int. J. Biochem. Cell Biol. 76, 1–11. doi: 10.1016/j.biocel.2016.04.012
Voellenkle, C., Perfetti, A., Carrara, M., Fuschi, P., Renna, L. V., Longo, M., et al. (2019). Dysregulation of circular RNAs in myotonic dystrophy type 1. Int. J. Mol. Sci. 20:8. doi: 10.3390/ijms20081938
Wang, J., Gong, C., and Maquat, L. (2013). Control of myogenesis by rodent SINE-containing lncRNAs. Genes Dev. 27, 793–804. doi: 10.1101/gad.212639.112
Wang, J., Yang, L. Z., Zhang, J. S., Gong, J. X., Wang, Y. H., Zhang, C. L., et al. (2018). Effects of microRNAs on skeletal muscle development. Gene. 668, 107–113. doi: 10.1016/j.gene.2018.05.039
Wang, K., Gan, T. Y., Li, N., Liu, C. Y., Zhou, L. Y., Gao, J. N., et al. (2017). Circular RNA mediates cardiomyocyte death via miRNA-dependent upregulation of MTP18 expression. Cell Death Differ. 24, 1111–1120. doi: 10.1038/cdd.2017.61
Wang, L., Park, H. J., Dasari, S., Wang, S., Kocher, J. P., and Li, W. (2013). CPAT: Coding-Potential Assessment Tool using an alignment-free logistic regression model. Nucleic Acids Res. 41:e74. doi: 10.1093/nar/gkt006
Wang, L., Zhao, Y., Bao, X., Zhu, X., Kwok, Y. K., Sun, K., et al. (2015). LncRNA Dum interacts with Dnmts to regulate Dppa2 expression during myogenic differentiation and muscle regeneration. Cell Res. 25, 335–350. doi: 10.1038/cr.2015.21
Wang, S., Xue, X., Wang, R., Li, X., Li, Q., Wang, Y., et al. (2018). CircZNF609 promotes breast cancer cell growth, migration, and invasion by elevating p70S6K1 via sponging miR-145-5p. Cancer Manag. Res. 10, 3881–3890. doi: 10.2147/CMAR.S174778
Wang, S., Zuo, H., Jin, J., Lv, W., Xu, Z., and Fan, Y. (2019). Long noncoding RNA Neat1 modulates myogenesis by recruiting Ezh2. Cell Death Dis. 10:505. doi: 10.1038/s41419-019-1742-7
Wang, Y., Li, M., Wang, Y., Liu, J., Zhang, M., Fang, X., et al. (2019). A Zfp609 circular RNA regulates myoblast differentiation by sponging miR-194-5p. Int. J. Biol. Macromol. 121, 1308–1313. doi: 10.1016/j.ijbiomac.2018.09.039
Wei, X., Li, H., Yang, J., Hao, D., Dong, D., Huang, Y., et al. (2017). Circular RNA profiling reveals an abundant circLMO7 that regulates myoblasts differentiation and survival by sponging miR-378a-3p. Cell Death Dis. 8:e3153. doi: 10.1038/cddis.2017.541
Weinberg, R. A., and Penman, S. (1968). Small molecular weight monodisperse nuclear RNA. J. Mol. Biol. 38, 289–304. doi: 10.1016/0022-2836(68)90387-2
Werfel, S., Nothjunge, S., Schwarzmayr, T., Strom, T., Meitinger, T., Engelhardt, S., et al. (2016). Characterization of circular RNAs in human, mouse and rat hearts. J. Mol. Cell Cardiol. 98, 103–107. doi: 10.1016/j.yjmcc.2016.07.007
Wilusz, J. E. (2018). A 360° view of circular RNAs: From biogenesis to functions. Wiley Interdiscip. Rev. RNA 9:e1478. doi: 10.1002/wrna.1478
Wu, H., Zhao, Z. A., Liu, J., Hao, K., Yu, Y., Han, X., et al. (2018). Long noncoding RNA Meg3 regulates cardiomyocyte apoptosis in myocardial infarction. Gene Ther. 25, 511–523. doi: 10.1038/s41434-018-0045-4
Xi, S., Xu, H., Shan, J., Tao, Y., Hong, J. A., Inchauste, S., et al. (2013). Cigarette smoke mediates epigenetic repression of miR-487b during pulmonary carcinogenesis. J. Clin. Invest. 123, 1241–1261. doi: 10.1172/JCI61271
Yaffe, D., and Saxel, O. (1977). A myogenic cell line with altered serum requirements for differentiation. Differentiation 7, 159–166.
Yan, B., Yao, J., Liu, J. Y., Li, X. M., Wang, X. Q., Li, Y. J., et al. (2015). lncRNA-MIAT regulates microvascular dysfunction by functioning as a competing endogenous RNA. Circ. Res. 116, 1143–1156. doi: 10.1161/CIRCRESAHA.116.305510
Yang, J., Xue, F. T., Li, Y. Y., Liu, W., and Zhang, S. (2018). Exosomal piRNA sequencing reveals differences between heart failure and healthy patients. Eur. Rev. Med. Pharmacol. Sci. 22, 7952–7961. doi: 10.26355/eurrev_201811_16423
Yu, X., Zhang, Y., Li, T., Ma, Z., Jia, H., and Chen, Q. (2017). Long non-coding RNA Linc-RAM enhances myogenic differentiation by interacting with MyoD. Nat. Commun. 8:14016. doi: 10.1038/ncomms14016
Zhang, Q., Vashisht, A. A., O'Rourke, J., Corbel, S. Y., Moran, R., Romero, A., et al. (2017). The microprotein Minion controls cell fusion and muscle formation. Nat. Commun. 8:15664. doi: 10.1038/ncomms15664
Zhang, Y., Jiao, L., Sun, L., Li, Y., Gao, Y., Xu, C., et al. (2018c). LncRNA ZFAS1 as a SERCA2a inhibitor to cause intracellular Ca2+ overload and contractiledysfunction in a mouse model of myocardial infarction. Circ. Res. 122, 1354–1368. doi: 10.1161/CIRCRESAHA.117.312117
Zhang, Z. K., Li, J., Guan, D., Liang, C., Zhuo, Z., Liu, J., et al. (2018a). Long noncoding RNA lncMUMA reverses established skeletal muscle atrophy following mechanical unloading. Mol. Ther. 26, 2669–2680. doi: 10.1016/j.ymthe.2018.09.014
Zhang, Z. K., Li, J., Guan, D., Liang, C., Zhuo, Z., Liu, J., et al. (2018b). A newly identified lncRNA MAR1 acts as a miR-487b sponge to promote skeletal muscle differentiation and regeneration. J. Cachexia Sarcopenia Muscle 9, 613–626. doi: 10.1002/jcsm.12281
Zhao, Y., Cao, F., Yu, X., Chen, C., Meng, J., Zhong, R., et al. (2018). Linc-RAM is required for FGF2 function in regulating myogenic cell differentiation. RNA Biol. 15, 404–412. doi: 10.1080/15476286.2018.1431494
Zhou, L., Sun, K., Zhao, Y., Zhang, S., Wang, X., Li, Y., et al. (2015). Linc-YY1 promotes myogenic differentiation and muscle regeneration through an interaction with the transcription factor YY1. Nat. Commun. 6:10026. doi: 10.1038/ncomms10026
Keywords: myogenesis, non-coding RNAs, long non-coding RNAs, lncRNAs, circular RNAs, circRNAs, Piwi-interacting RNAs, transfer RNA-derived fragments
Citation: Martone J, Mariani D, Desideri F and Ballarino M (2020) Non-coding RNAs Shaping Muscle. Front. Cell Dev. Biol. 7:394. doi: 10.3389/fcell.2019.00394
Received: 29 November 2019; Accepted: 26 December 2019;
Published: 07 February 2020.
Edited by:
Pavel Sumazin, Baylor College of Medicine, United StatesReviewed by:
Lucia Lorenzi, Ghent University, BelgiumAlessandra Perfetti, IRCCS Policlinico San Donato, Italy
Copyright © 2020 Martone, Mariani, Desideri and Ballarino. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Monica Ballarino, bW9uaWNhLmJhbGxhcmlub0B1bmlyb21hMS5pdA==
 Julie Martone
Julie Martone Davide Mariani
Davide Mariani Fabio Desideri
Fabio Desideri Monica Ballarino
Monica Ballarino