- 1Department of Laboratory Medicine and Pathobiology, MaRS Centre, Faculty of Medicine, University of Toronto, Toronto, ON, Canada
- 2Canada Research Chairs Program, Faculty of Medicine, University of Toronto, Toronto, ON, Canada
Genetic loci are non-randomly arranged in the nucleus of the cell. This order, which is important to overall genome expression and stability, is maintained by a growing number of factors including the nuclear envelope, various genetic elements and dedicated protein complexes. Here, we review evidence supporting roles for non-coding RNAs (ncRNAs) in the regulation of spatial genome organization and its impact on gene expression and cell survival. Specifically, we discuss how ncRNAs from single-copy and repetitive DNA loci contribute to spatial genome organization by impacting perinuclear chromosome tethering, major nuclear compartments, chromatin looping, and various chromosomal structures. Overall, our analysis of the literature highlights central functions for ncRNAs and their transcription in the modulation of spatial genome organization with connections to human health and disease.
Introduction
Spatial genome organization involves the 3D structure, positioning, and interactions of chromatin within the nucleus. This is a non-random process that is characterized by the regulation of various nuclear domains, topological associations, and epigenetic signatures. For example, decondensed euchromatin domains, which include active enhancer elements and are generally conducive to transcription, are found preferentially within the nuclear interior. On the other hand, heterochromatin domains are densely packed chromosome regions that are occupied by gene-silencing histone marks, which include histone H3 methylated on Lysine 27 (H3K27me3) or Lysine 9 (H3K9me2 and H3K9me3) (Rice and Allis, 2001; Richards and Elgin, 2002). Such heterochromatin domains are preferentially located near the nuclear periphery or a major nuclear compartment called the nucleolus.
In fact, the nuclear genome is generally arranged within several cytologically distinct compartments. In addition to the prominent nucleolus, other nuclear compartments include the Cajal bodies, speckles, paraspeckles, and histone locus bodies. Nuclear compartments generally form via dynamic self-organization of their different constituents at sites of gene clusters (Mao et al., 2011; Sleeman and Trinkle-Mulcahy, 2014; Wang et al., 2016). For example, nucleoli encompass the tandem ribosomal DNA (rDNA) repeats while histone locus bodies form around the histone-encoding gene clusters. Early studies identified a role for molecular crowding in the formation of some nuclear compartments (Richter et al., 2007; Cho and Kim, 2012). High concentrations of macromolecules in a local environment creates crowding and promotes formation of weak non-covalent bonds between the macromolecules, thereby forming membrane-less nuclear compartments. Consistent with this notion, the formation of several nuclear compartments is driven by liquid-liquid phase separation (Zhu and Brangwynne, 2015; Hall et al., 2019). Importantly, the three-dimensional organization of chromatin into these nuclear compartments often underlies the expression and stability of the various genetic loci that are harbored within such nuclear bodies. For example, actively transcribed genes often associate with the periphery of nuclear speckles, which are sites of RNA processing (Hu et al., 2019). Disruption of nuclear speckles changes gene expression profiles by decreasing intrachromosomal interactions between active chromatin regions.
In addition to the formation of cytologically distinct nuclear compartments, the genome is organized into topologically associated domains (TADs) (Dixon et al., 2012), which can be viewed as three-dimensional building blocks of looped chromatin domains (Lieberman-Aiden et al., 2009; Dixon et al., 2012). TADs are present in the genomes of several eukaryotes including Drosophila (Sexton et al., 2012), mice (Krefting et al., 2018) and humans (Lieberman-Aiden et al., 2009), and are categorized into type A (active genes) and type B (inactive genes) compartments. TADs can regulate transcription by acting as insulators, preventing the spread of euchromatin or heterochromatin marks and regulating enhancer-promoter interactions.
Topologically associated domains are built or defined by their associated proteins, which include the cohesin complex, condensin complex and CCCTCF binding factor (CTCF) which binds DNA in a sequence-specific manner (Dixon et al., 2012; Zuin et al., 2014). Cohesin and condensin are ring-shaped protein complexes that bind chromatin independently of the DNA sequence and mediate chromatin looping, bringing distant DNA sequences along the linear genome into close proximity within the 3D space of the nucleus (Nuebler et al., 2018). The cohesin and condensin complexes, which are composed of structural maintenance of chromosome (SMC) proteins, extrude the DNA into loops through an ATP hydrolysis-dependent mechanism (Burmann et al., 2017; Diebold-Durand et al., 2017; Ganji et al., 2018). Cohesin loading onto chromatin is mediated by the loading factor NIPBL, the absence of which results in the loss of local TAD patterns (Schwarzer et al., 2017). The DNA is extruded until cohesin reaches a boundary element or insulator such as CTCF (Nuebler et al., 2018; Vian et al., 2018). CTCF is a DNA binding protein that mostly associates with TAD boundary regions, insulator sequences, and imprinting control regions (Rao et al., 2014; Sanborn et al., 2015). CTCF is responsible for the majority of chromatin loops across the human genome and is thus an important regulator of spatial genome organization.
Another regulator of spatial genome organization is the nuclear envelope, which harbors the inner nuclear membrane (INM) proteins and nuclear pore complexes (NPCs) and is lined by the nuclear lamina (NL), which is a meshwork of lamin and lamin-associated proteins. The nuclear lamins are important regulators of chromatin organization (Kind et al., 2015). Genes that are activated for transcription are commonly repositioned from the NL to either the nuclear interior or closer to NPCs. Regions of the chromatin that interact with the lamina are referred to as lamina associated domains (LADs), and this association is mediated by lamin-associated proteins. In mammals (Guelen et al., 2008), nematodes (Ikegami et al., 2010) and flies (Pickersgill et al., 2006; van Bemmel et al., 2010), LADs mostly harbor silent or weakly expressed genes, and contain heterochromatin marks such as H3K9me3 and H3K9me2 (Casolari et al., 2004; Wen et al., 2009), whereas budding yeast has no lamina or LADs and its genome is instead organized into gene crumples and directly tethered to INM or NPC proteins (Taddei et al., 2006; Mekhail et al., 2008; Hsieh et al., 2015). In Drosophila cells, NL disruption alters LAD composition such that there is more histone H3 acetylated on Lysine 9 (H3K9Ac) and less chromatin compaction (Ulianov et al., 2019). Furthermore, association of chromosomes with the nuclear lamina limits their mobility within the nucleus (Wang H. et al., 2018). In addition, studies in different organisms revealed that NPCs can regulate chromatin structure and function (Dilworth et al., 2005; Brown et al., 2008; Mekhail and Moazed, 2010). For example, the nucleoporins from which NPCs are built can associate with the promoters of active genes in yeast, thereby regulating gene expression (Schmid et al., 2006).
In addition to nuclear compartments, TADs/LADs, the nuclear envelope and their associated protein complexes, non-coding RNAs (ncRNAs) have emerged as major regulators of spatial genome organization. ncRNAs are RNA molecules that are not translated into proteins. ncRNAs are categorized based on their size – long (>200 bp) and short (<200 bp) – and are implicated in numerous cellular processes including transcription, mRNA splicing, and protein translation (Mortazavi et al., 2008; Khalil et al., 2009; Palazzo and Lee, 2015). ncRNAs emerging from within a given genetic locus can regulate transcription at the same locus (cis) or elsewhere in the genome (trans). Here we review ncRNAs that emerge from single-copy DNA loci or repetitive DNA loci and have diverse roles in spatial genome organization, thus impacting gene expression and stability. Collectively, ncRNAs impact spatial genome organization by modulating perinuclear chromosome tethering, the formation of major nuclear compartments, chromatin looping and various chromosomal structures. These roles of ncRNAs often intersect with various other regulators of genome structure and function.
Non-Coding Rna at Single Copy Loci
Single copy loci include genes required for cell function and survival and can give rise to ncRNAs that regulate higher order chromatin structure and positioning (Figure 1). ncRNAs and their active transcription can mediate chromatin looping to bring distant DNA regions into close proximity and reposition genetic loci to regulate their expression. Nuclear bodies, such as Cajal bodies and paraspeckles, are formed by ncRNA transcription and can regulate the localization or sequestration of transcriptional regulators. Furthermore, ncRNAs play roles in organismal development by regulating the subnuclear positioning and transcriptional status of the X chromosome, HOX genes and Kcnq1 genes. In this section we discuss roles for ncRNAs and their transcription in the control of spatial gene positioning, chromatin remodeling and nuclear compartmentalization.
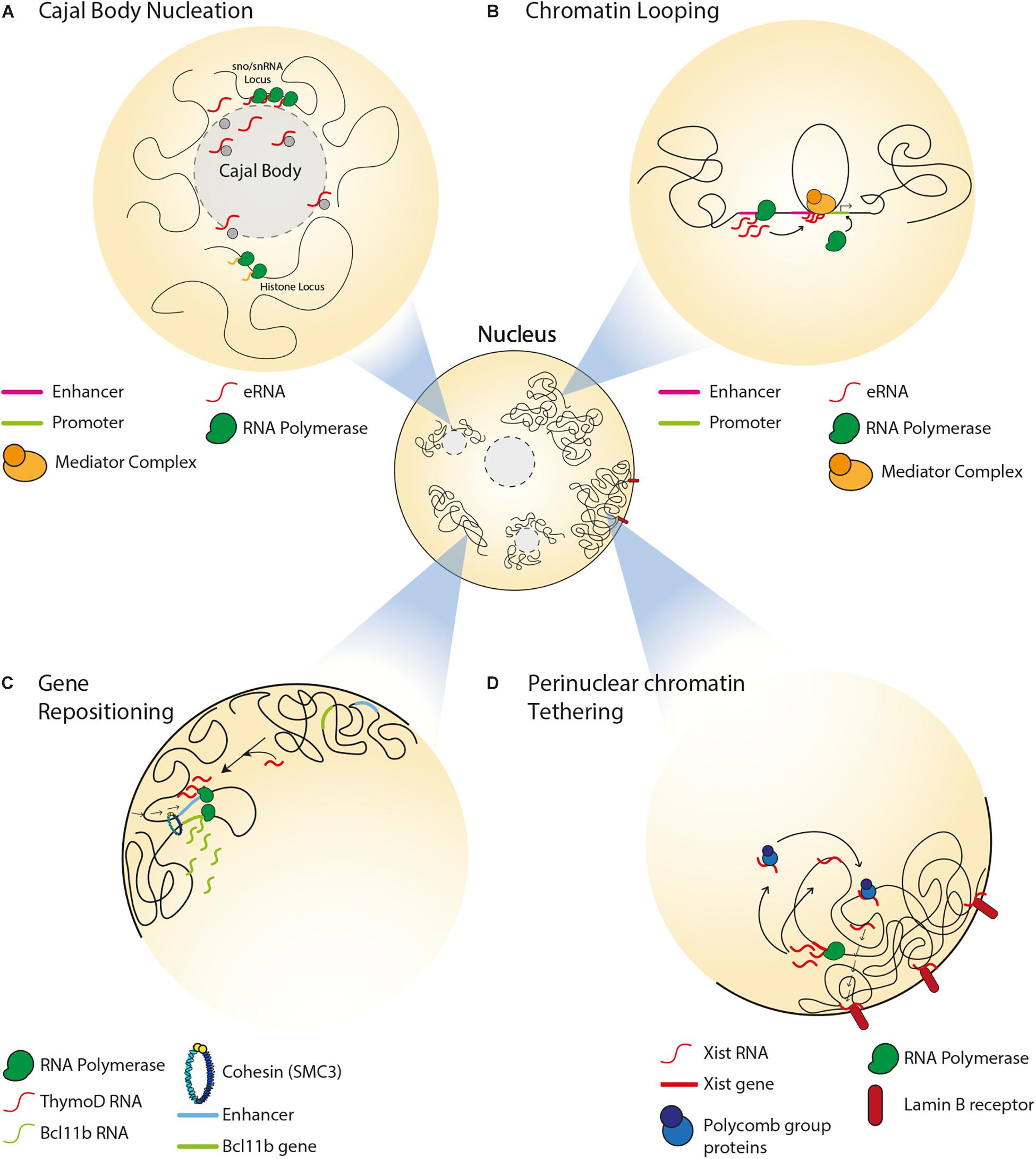
Figure 1. Spatial organization of single copy loci by ncRNAs. (A) Transcription of snRNA genes and interaction between intron-encoded snoRNA/snRNAs with coilin mediate Cajal body formation. Cajal bodies associate with sn/snoRNA and histone gene loci and regulate their gene expression. (B) Transcription of an enhancer can produce eRNAs, which associate with the mediator complex and enable chromatin looping, thereby driving enhancer-promoter interaction. (C) In the absence of the ThymoD ncRNA, the enhancer for the BCL11B locus is at nuclear periphery. ThymoD ncRNA mediates enhancer repositioning away from the nuclear periphery and drives chromatin looping of the enhancer bringing it in close proximity to the BCL11B locus, thereby allowing for the transcriptional activation of this locus. (D) In the somatic tissues of placental mammals, Xist lncRNA tethers the inactive X chromosome to the nuclear lamina by interacting with lamin B receptor. Xist interacts with polycomb proteins to establish the heterochromatin state of the inactive X chromosome. Xist also mediates relocation of active genes from the surface of the X chromosome to its interior.
ncRNAs in the Formation and Maintenance of Nuclear Compartments
Non-coding RNAs can impact the structure and function of nuclear compartments such as Cajal bodies. The latter are involved in various processes including telomerase biogenesis, 3′-end processing of histone pre-mRNAs, as well as the processing, assembly and maturation of spliceosomal small nuclear ribonucleoproteins (snRNPs) (Sawyer et al., 2016). Cajal bodies associate with small nuclear and nucleolar RNA (sn/snoRNA) gene loci, such that these genes form intra- and inter-chromosomal clusters around the bodies (Figure 1A) (Wang et al., 2016). In fact, the formation of Cajal bodies is itself mediated by the transcription of snRNA genes (Kaiser et al., 2008) and by interactions between intron-encoded snoRNA/snRNAs and a protein called coilin (Kaiser et al., 2008; Machyna et al., 2014). This is in accordance with studies reporting the loss of Cajal bodies during mitosis and their reformation during early G1 upon the resumption of transcription (Carmo-Fonseca et al., 1993; Strzelecka et al., 2010). Furthermore, these ncRNA-dependent Cajal bodies are responsible for the spatial organization and expression of other types of genes, including those encoding for histones or pre-mRNA splicing factors (Sawyer et al., 2016; Wang et al., 2016; Wang H. et al., 2018).
Paraspeckles are nuclear bodies that form in response to environmental stress at and around the NEAT1 gene (nuclear enriched abundant transcript 1), which is transcribed into the long ncRNAs (lncRNAs) NEAT1_1 (Men ε) and NEAT1_2 (Men β) (Sunwoo et al., 2009). These lncRNAs and their ongoing transcription are required for the nucleation and maintenance of these nuclear compartments (Shevtsov and Dundr, 2011). Transcriptional upregulation of NEAT1 increases paraspeckle size and sequestration of paraspeckle-associated transcriptional regulators, such as the splicing factor proline/glutamine-rich (SFPQ) (Hirose et al., 2014). In contrast, repression of NEAT1 disrupts paraspeckles, releases paraspeckle-associated proteins into the nucleoplasm and hyper-induces the transcription of various genes including ADARB2 (adenosine deaminase RNA-specific B2), which is involved in RNA editing (Clemson et al., 2009; Mao et al., 2011; Hirose et al., 2014; Imamura et al., 2014).
NEAT1 can also regulate the subnuclear localization of growth control genes by associating with Polycomb 2 protein (Pc2), a key subunit of the chromatin-repressive PRC1 complex (Yang et al., 2011). Methylation/demethylation cycles of Pc2 dictate its association with two ncRNAs, TUG1 (Taurine up-regulated 1) and NEAT1, which are found in two distinct nuclear bodies. Methylated Pc2 preferentially interacts with the TUG1 ncRNA within the transcriptionally repressive Polycomb nuclear bodies, thereby silencing the Pc2-associated growth control genes. On the other hand, demethylation of Pc2 results in its preferential interaction with NEAT1, which relocates Pc2 together with its associated growth control genes to inter-chromosomal granules within which the genes can be actively transcribed.
NEAT1 is commonly induced upon viral infection and can regulate the transcriptional activation of various antiviral genes (Ma et al., 2017). The splicing factor SFPQ is a transcriptional repressor of the antiviral gene IL-8. Recently, NEAT1 has been shown to mediate the relocation of SFPQ from the IL-8 promoter to paraspeckles, thereby activating IL-8 gene expression (Imamura et al., 2014). Paraspeckles and NEAT1 have also been linked to cancer biology, where they can have both oncogenic and tumor suppressive roles. In some cancers, the upregulation of NEAT1 and associated paraspeckles can be driven by tumor microenvironmental conditions and estrogen receptor stimulation, respectively (Chakravarty et al., 2014; Choudhry et al., 2015). This upregulation is associated with increases in active epigenetic marks and cellular proliferation. Surprisingly, in some types of cancer, upregulation of NEAT1 and paraspeckles prevented cellular transformation and tumorigenesis (Adriaens et al., 2016). Overall, these findings highlight functional connections between ncRNAs and nuclear compartments. These studies also underscore the importance of understanding the exact roles that ncRNAs can exert within different biological and clinical settings.
ncRNAs and Chromatin Looping
Non-coding RNAs can regulate gene expression by mediating chromatin remodeling between enhancers and promoters. Transcription of enhancers in mammalian cells can give rise to a type of ncRNA that is referred to as enhancer RNA (eRNA), which can bring an enhancer and promoter in close proximity by mediating the formation of a DNA loop, and associate with mediator complexes to drive the expression of target genes (Figure 1B) (Kim et al., 2010; Orom et al., 2010). For example, activation of estrogen receptor-α induces the transcription of eRNAs that mediate chromatin looping, thereby driving transcription-inducing enhancer-promoter interactions at target genes (Li W. et al., 2013). Another class of ncRNAs, which is known as ncRNA-activating (ncRNA-a), has a function similar to that of eRNA (Lai et al., 2013; Li W. et al., 2013). These ncRNA-a species activate their neighboring genes by associating with the mediator complex and enabling chromatin looping in cis. This 3D chromatin configuration and gene expression are reduced upon disruption of ncRNA-a species or mediator, suggesting the dependence of chromatin loop structure and function on interactions between ncRNA-a and mediator.
The active transcription of ncRNAs can also result in the looping of DNA, bringing gene loci in close proximity or blocking transcription of distant genes. In the plant Arabidopsis thaliana, transcription of the ncRNA APOLO forms a chromatin loop encompassing the promoter of its neighboring gene, PID (Ariel et al., 2014), which is the key regulator of polar auxin transport and root development (Benjamins et al., 2001). This APOLO-mediated 3D chromatin configuration, which is also influenced by PRC1 and PRC2 (polycomb repressive complex 1 and 2), limits the access of Pol II to the PID promoter, thereby regulating the transcriptional activity of this gene (Ariel et al., 2014). Disruption of the APOLO-dependent expression of PID results in defects in root development, highlighting the importance of ncRNA-mediated chromatin remodeling to plant growth and development (Benjamins et al., 2001).
Intergenic transcription-driven chromatin looping is also implicated in lymphocyte development. In developing B cells, V(D)J recombination is required for the assembly of antigen receptors (Alt et al., 1984; Sayegh et al., 2005). Importantly, V(D)J recombination requires changes to the 3D configuration of the immunoglobulin heavy locus (Igh) in order to bring the VH, DH and JH genes in close proximity, which in turn allows the genetic rearrangements to occur (Kosak et al., 2002; Medvedovic et al., 2013). Prior to rearrangement, non-coding transcription at this locus occurs at the VH intergenic region in the antisense orientation (Yancopoulos and Alt, 1985). This intergenic region contains Pax5-activated intergenic repeat (PAIR) elements (Fuxa et al., 2004), which are transcriptionally upregulated in the absence of CTCF (Degner et al., 2009). Antisense transcription of these PAIR elements in pro-B cells mediates long-distance interaction with the Eμ region on the Igh locus (Verma-Gaur et al., 2012). The resulting DNA looping brings the distal VH into close proximity with DJH and allows for VH to DJH recombination. These DNA loops are not observed in the absence of ncRNA transcription, highlighting the importance of active ncRNA transcription to V(D)J recombination and its role in B cell development.
In developing T cells, expression of BCL11B (BAF Chromatin Remodeling Complex Subunit BCL11B) promotes expression of T-lineage-specific genes and suppresses expression of the genes associated with alternative cell fates (Li et al., 2010). Activation of BCL11B expression is mediated by an enhancer that is located at the so-called intergenic control region (ICR) (Li L. et al., 2013). Repositioning of the enhancer from the nuclear lamina to the interior allows for the transcriptional activation of BCL11B (Figure 1C) (Isoda et al., 2017). Importantly, this relocation within nuclear space is mediated by transcription of the ncRNA ThymoD (thymocyte differentiation factor), which mediates DNA demethylation at CTCF binding sites and subsequent activation of CTCF/cohesin-dependent chromatin looping.
ncRNAs and X Chromosome Silencing and Positioning
One of the well-studied ncRNAs implicated in mammalian 3D genome organization is Xist (X inactive specific transcript), a 17 kb lncRNA that mediates inactivation of one of the X chromosomes during early female embryonic development (Brown et al., 1992). Xist is specifically transcribed from the inactive X chromosome. Xist occupies inactive regions of the X chromosome before spreading across transcriptionally active regions and initiating their inactivation. Subsequently, the inactive X chromosome forms a heterochromatic structure, which is referred to as Barr body and is found at the perinuclear and perinucleolar regions, where transcription silencing machineries are enriched (Zhang et al., 2007). Of note, tethering of the inactive X chromosome to the nuclear lamina is the result of interactions between Xist and the INM-embedded lamin B receptor (Figure 1D) (Chen et al., 2016). This interaction repositions transcriptionally active DNA regions of the X chromosome in close proximity with Xist and its transcriptional silencing domain, thereby promoting the spread of Xist across the chromosome. In female embryonic stem cells, the spreading of Xist along an X chromosome results in the establishment of polycomb group proteins-dependent heterochromatin and exclusion of transcription machineries (Figure 1D) (Plath et al., 2003; Okamoto et al., 2004; Chaumeil et al., 2006; Schoeftner et al., 2006). During this process, active genes that were once on the surface of the X chromosome relocate to the interior, forming Xist-containing transcriptionally silent domains (Chaumeil et al., 2006). Furthermore, Xist maintains this heterochromatic nuclear compartment by acting in cis to repel cohesin and other chromatin looping factors that typically facilitate gene expression (Minajigi et al., 2015). Consequently, compared to the active X chromosome, the inactive X chromosome is devoid of TADs, which can nonetheless be re-established upon depletion of Xist and restoration of cohesin loading (Nora et al., 2012). Taken together, these findings highlight how the Xist lncRNA mediates mammalian X chromosome inactivation through the formation of perinuclear heterochromatin domains and the exclusion of factors that can promote chromatin looping and gene expression.
The expression of Xist on the active X chromosome is regulated by another lncRNA, Tsix, which is transcribed antisense to Xist (Stavropoulos et al., 2001). Transcription of Tsix represses Xist expression in cis through epigenetic processes (Stavropoulos et al., 2001; Shibata and Lee, 2004). In mouse embryonic stem cells, the X chromosome lacking Tsix transcription was non-randomly inactivated (Lee and Lu, 1999; Luikenhuis et al., 2001), and induction of Tsix transcription resulted in targeted X chromosome activation (Luikenhuis et al., 2001). Therefore, Tsix and Xist play antagonistic roles in regulating X chromosome inactivation during embryonic stem cell differentiation.
In addition to Xist and Tsix, Firre (functional intergenic repeating element) is another lncRNA that is transcribed from a locus on the X chromosome (Hacisuleyman et al., 2014). Firre can maintain the silencing of the X chromosome by tethering it to the perinucleolar compartment (Yang et al., 2015). In addition, Firre interacts with the nuclear matrix factor hnRNPU and colocalizes with five distinct trans-chromosomal loci, which reside in spatial proximity to the Firre locus. This colocalization is lost in the absence of Firre, suggesting a role of this ncRNA in the establishment of higher order chromosomal architectures within nuclear space.
Typically, cells randomly choose whether the maternal or paternal X chromosome is inactivated. However, under certain circumstances, there can be bias toward one parental X chromosome. Such a bias is referred to as skewed X inactivation. In females, this can result in diseases such as Duchenne muscular dystrophy and hemophilia A (Yoshioka et al., 1998; Renault et al., 2007). Incomplete silencing of the X chromosome can also result in skewed X inactivation since some genes manage to evade silencing and remain therefore expressed. For example, escape of the steroid sulfatase locus from silencing can trigger X-linked ichthyosis, which is a group of diseases characterized by very dry skin (Hernandez-Martin et al., 1999). Thus, ncRNAs operate at the interface of spatial genome organization and epigenetic silencing to mediate X chromosome inactivation, the dysregulation of which underlies different human diseases.
ncRNAs and the Spatial Organization of Developmental Genes
During vertebrate development, ncRNAs can regulate the spatial organization of gene clusters, such as the HOX genes (Flyamer et al., 2017). HOX genes, which are homeotic genes involved in antero-posterior body axis development in vertebrates, are found on four spatially clustered chromosomal loci (HOXA, HOXB, HOXC, and HOXD). The genes are separated into distinct topological compartments based on their transcriptional profile, and during development, there exists a dynamic switch between these topological domains (Noordermeer et al., 2011). This higher order structure of the HOX gene clusters is regulated by intergenic ncRNAs (Wang et al., 2011). For example, the ncRNA HOTTIP (HOXA transcript at the distal tip) is transcribed from the 5′ edge of the HOXA locus and is required for maintaining the compartmentalization of the locus. HOTTIP can associate with and target the WD repeat mixed lineage leukemia (WDR-MLL) complex across the HOXA locus to yield active histone marks. This in return maintains the active state of some of the HOXA genes. HOTTIP also physically associates with CTCF, which can bind to six conserved binding sites at HOXA and serve as an insulator (Wang F. et al., 2018). This contributes to the discrete expression profile of genes across the HOXA locus. The dynamic 3D architecture of these gene clusters is important as it dictates the transcriptional profile of the HOX genes during development. Dysregulation of HOX gene expression can abrogate limb and skeletal development in murine and Drosophila embryos (Di-Poi et al., 2010; Andrey et al., 2013). Therefore, regulation of the spatial organization of HOX genes by ncRNAs is important for organismal development.
Another critical component of development is known as genetic imprinting, which consists of the silencing of one parental allele. Imprinted genes tend to spatially cluster and this allows for their coordinated regulation during development. lncRNAs have been shown to regulate the expression and large-scale chromatin structure of these genes through interaction with histone modifying proteins and chromatin looping (Umlauf et al., 2004; Terranova et al., 2008; Zhang et al., 2014). In early mammalian embryos, the Kcnq1 genes cluster into a compact subnuclear compartment, devoid of transcriptional activity (Verona et al., 2003; Lewis et al., 2006). This nuclear compartment is enriched with repressive histone marks and silencing protein complexes such as polycomb proteins (Umlauf et al., 2004; Terranova et al., 2008). Formation of this higher order repressive domain and its localization within the perinucleolar compartment is mediated by the Kcnq1ot1 ncRNA, which associates with the H3K9me3 repressive histone mark and polycomb proteins (Mohammad et al., 2008; Pandey et al., 2008; Terranova et al., 2008). Kcnq1ot1 is an antisense ncRNA (∼100 kb) that is transcribed from the intronic region of the Kcnq1 locus of one of the parental chromosomes. Deletion of Kcnq1ot1 results in expression of the parental allele that is normally silent (Mancini-Dinardo et al., 2006). More recently, this ncRNA has been shown to directly interact with the chromosome, through its 5′ terminal region, in order to mediate intrachromosomal looping between the Kcnq1 promoter and Kcnq1ot1 promoter KvDMR (Zhang et al., 2014). These promoters are 200 kb apart in the linear genome (Zhang et al., 2014). However, promoter looping results in the imprinting of the Kcnq1 cluster, or its allelic silencing. Deletion of KvDMR can result in biallelic expression of maternal-specific genes in the Kcnq1 cluster and growth deficiency in mice (Fitzpatrick et al., 2002; Shin et al., 2008). In humans, loss of imprinting can lead to Beckwith–Wiedemann syndrome, which is associated with cancer growth and progression (Lee et al., 1999; Fitzpatrick et al., 2002; Valente et al., 2019). Therefore, regulation of the spatial organization of the Kcnq1 gene cluster by Kcnq1ot1 is important for mammalian genetic imprinting and healthy development. Overall, these findings suggest that ncRNAs play a role in regulating gene expression during development via establishment of nuclear compartments and regulation of locus positioning within nuclear space.
Taken together, ncRNAs from single-copy loci contribute to spatial genome organization through chromatin remodeling, nuclear compartmentalization and the subnuclear positioning of various genes within nuclear space. These roles of ncRNA help mediate cellular processes that are central to the proper control of gene expression, genome stability, development, and health.
Non-Coding Rna at Repetitive Dna Loci
Eukaryotic genomes are largely composed of repetitive DNA sequences that can be generally classified as tandem or interspersed repeats. Tandem repeats include satellite and minisatellite repeats (e.g., centromeres) as well as microsatellite repeats (telomeres). Interspersed repeats include transposable elements that are either retrotransposons or DNA transposons. Retrotransposons include LTR-retrotransposons such as HERV and non-LTR retrotransposons such as SINEs (e.g., Alu), LINEs (e.g., LINE-1) or SVAs. It is also important to note that some types of repeats such as human ribosomal DNA (rDNA) can be arranged in tandem repeats that are interspersed throughout the linear genome. Regardless of their relative genomic location, DNA repeats are often clustered within nuclear space. This can facilitate their transcriptional co-regulation, minimize their potential deleterious interaction with the rest of the genome and control their exposure to potentially genome-destabilizing DNA recombination and repair machineries.
Repetitive DNA sequences are non-randomly arranged within the nucleus. For example, rDNA repeats are physically sequestered in the nucleolar compartment of the nucleus. This sequestration can be driven by inter- or intra-chromosomal interactions, or even direct tethering to the nuclear envelope in some organisms (O’Sullivan et al., 2009; Mekhail and Moazed, 2010; Chan et al., 2011; Hult et al., 2017). Telomeres, which are at the ends of linear chromosomes, often colocalize within PML bodies (Chang et al., 2013) at the nuclear interior or within telomeric clusters or bouquets at the nuclear periphery (Gotta et al., 1996). In budding yeast, the Transposons of Yeast 1 (Ty1) retrotransposons cluster within or near nucleoli (O’Sullivan et al., 2009), while centromeres cluster at the yeast spindle pole body (Jin et al., 2000). Importantly, several ncRNAs from repetitive DNA loci have emerged as major players that mediate crosstalk between spatial genome organization, expression and stability. Here were review such ncRNAs, which emerge from rDNA repeats, telomeric regions, transposable elements, and centromeres.
Non-coding RNAs in rDNA Structure and Function
Non-coding RNAs can play a role in the spatial organization and function of rDNA through the modulation of heterochromatin formation. Transcription of rDNA into ribosomal RNA (rRNA) molecules is dependent on the varying demand for protein synthesis that cells experience in response to intracellular signals or environmental stimuli. Therefore, despite the existence of 100s of rDNA repeats in eukaryotes, only a fraction of rDNA units are transcribed, while the remainder of the repeats is epigenetically silenced. Transcriptionally active rDNA units are marked by DNA hypomethylation, H4Ac and H3K4me2, whereas inactive rDNA units are marked by promoter hypermethylation, histone H4 hypoacetylation and methylation of H3K9, H3K27, and H4K20 (Santoro et al., 2002). Deposition of these marks is facilitated by the nucleolar remodeling complex (NoRC), which guides chromatin remodeling proteins to the rDNA and other loci. In mice, NoRC is recruited to nucleoli through interaction between the large subunit of NoRC (TIP5) and promoter-associated RNAs (pRNAs) that overlap the rDNA promoter (Mayer et al., 2006). A class of pRNAs termed PAPAS (promoter and pre-rRNA antisense) covers the rDNA promoter, and levels of PAPAS generally reflect the physiological state of the cell, such that there is an anti-correlation between PAPAS and pre-rRNA levels (Figure 2A) (Bierhoff et al., 2010). In quiescent mammalian cells, PAPAS is induced, binds to the histone methyltransferase Suv4-20h2, targets it to the rDNA promoter and downregulates rRNA transcription through enhanced H4K20me3 (Bierhoff et al., 2014). In addition, upon heat shock, upregulation of PAPAS attenuates pre-rRNA synthesis by recruiting another chromatin remodeling complex named CHD4/NuRD to the mammalian rDNA promoter (Zhao et al., 2018). On another front, in mammalian cells under hypotonic stress conditions, PAPAS upregulation recruits NuRD to reposition the rDNA promoter-bound nucleosome to the “off” position, thereby halting pre-rRNA synthesis (Zhao et al., 2016). Interestingly, pRNA-dependent heterochromatin formation at rDNA has also been shown to initiate the downstream establishment of heterochromatic structures at genomic regions that are in close proximity but lie outside of the murine nucleolus (Savic et al., 2014). Taken together, these studies reveal that under different environmental conditions, promoter-associated ncRNAs from repetitive loci can silence gene expression in cis through various processes. Future studies should explore how such ncRNAs are induced under different environmental conditions.
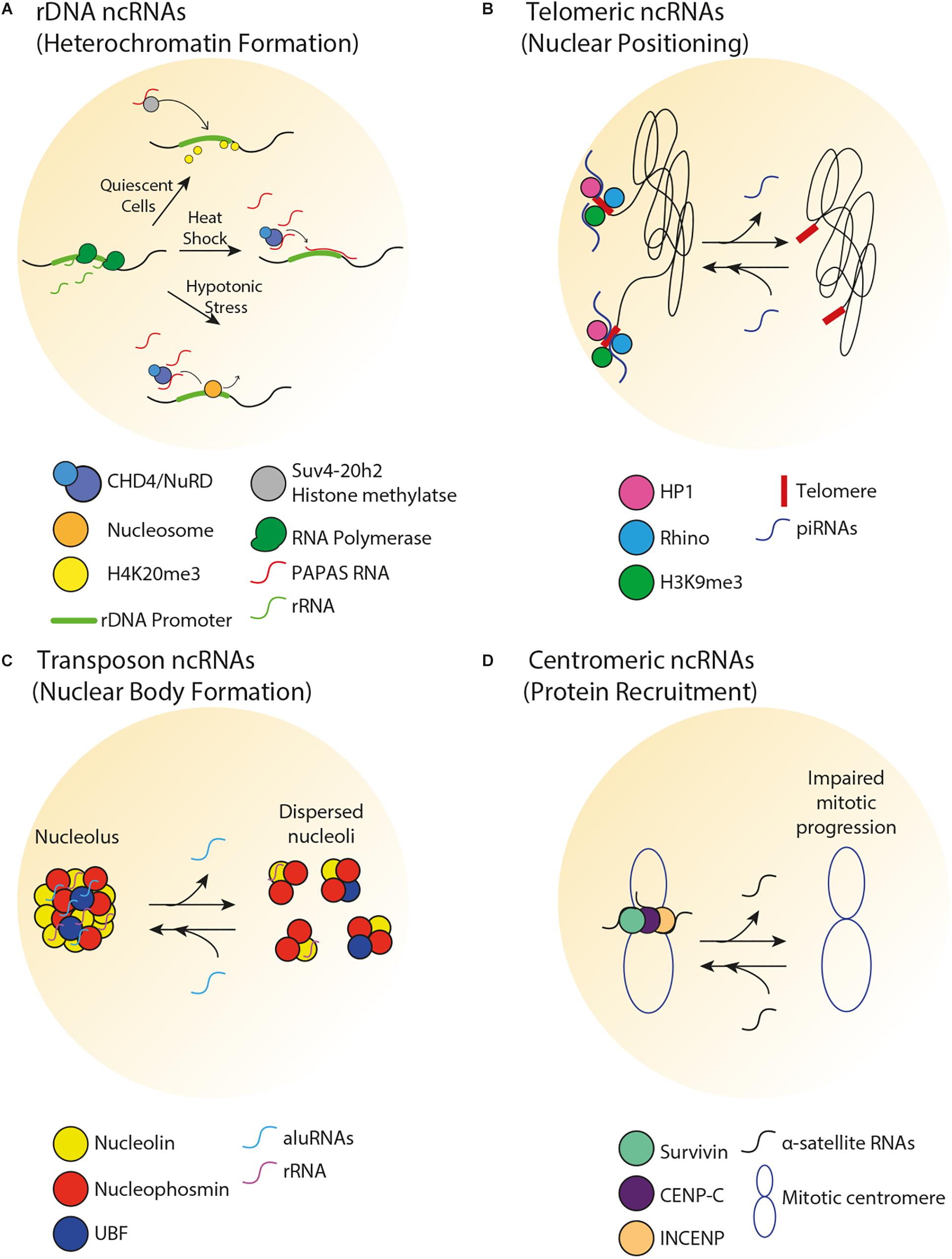
Figure 2. Spatial organization of repetitive DNA loci by ncRNA. (A) In human cells, there is an inverse correlation between PAPAS and pre-rRNA levels. In quiescent cells, PAPAS binds to the Suv4-20h2 histone methyltransferase and directs it to the rDNA promoter for H4K20me3-dependent repression. Upon heat shock, PAPAS hybridizes with the rDNA promoter and recruits the CHD4/NuRD complex, thereby preventing rDNA transcription. Upon hypotonic stress, upregulation of PAPAS recruits the CHD4/NuRD complex to reposition rDNA promoter-bound nucleosome to the off position, thereby halting pre-rRNA synthesis. (B) In germline tissues of flies, piRNAs transcribed from the telomeric region mediate perinuclear positioning of telomeres and promote HP1, Rhino, and H3K9me3 enrichments at telomeres. (C) In human cells, aluRNAs enriched in the nucleolus interact with nucleolin to maintain nucleolar structure and function. (D) In human cells, α-satellite RNAs associate with and promote the centromeric enrichment of Survivin, CENP-C, and INCENP in order to maintain centromere stability.
The nucleolus typically exhibits a phase separation-driven tripartite organization into a fibrillar centre (FC), dense fibrillar component (DFC), and granular component (GC) (Feric et al., 2016; Hall et al., 2019). Upon exposure to environmental stresses including heat shock or acidosis, a couple of ncRNAs induced from the mammalian rDNA intergenic spacer (IGS) dissolve this tripartite organization, structurally remodeling the nucleolus into a so-called “protein detention centre” (DC) (Mekhail et al., 2005; Jacob et al., 2013). The DC is suggested to be spatially, dynamically and biochemically distinct from the standard tripartite domains (Jacob et al., 2013). This structural remodeling of the mammalian nucleolus can arrest rRNA synthesis and create a hub for immobilized proteins, effectively mediating their nucleolar sequestration and functional inactivation (Audas et al., 2012). Upon removal of the environmental stressor, the ncRNAs are repressed, DC is dissolved and tripartite nucleolar organization is re-established (Jacob et al., 2013). Thus, ncRNAs spatially remodel the nucleolus during stress. Importantly, future studies should explore how cells control the generation and function of such intergenic ncRNAs under varying environmental conditions.
The organization of rDNA repeat regions into epigenetically silent chromatin structures is essential to proper cellular function and alterations in this organization may be associated with human disease. For example, rDNA hypermethylation is characteristic of early Alzheimer’s disease (Pietrzak et al., 2011), upregulation of rRNA expression is characteristic of tumor cells (White, 2005; Montanaro et al., 2008; Bywater et al., 2013) and rRNA dysfunction is linked to a group of genetic diseases known as ribosomopathies (Narla and Ebert, 2010; Nakhoul et al., 2014). In addition, in yeast, the dysregulation of IGS ncRNAs at rDNA repeats has been associated with premature aging through three distinct mechanisms. First, loss of IGS silencing leads to the upregulation of IGS ncRNAs, which displace cohesin complexes, triggering rDNA instability and premature aging (Saka et al., 2013). Second, IGS ncRNAs are prone to the formation of DNA replication-blocking RNA–DNA hybrid-containing structures called R-loops (Salvi et al., 2014). When these structures accumulate, as in some yeast mutants, unequal sister chromatid exchange events occur within the rDNA repeats, leading to chromosome instability and premature cellular aging (Salvi et al., 2014). Lastly, in yeast genetic models of neurodegenerative diseases, hyper-reductions in IGS ncRNA levels can lead to rDNA copy number instability and premature cellular aging (Ostrowski et al., 2018). Thus, ncRNAs that play important roles in the epigenetic silencing and organization of rDNA repeats can impact processes underlying organismal health span.
Crosstalk Between Telomeric ncRNAs, Heterochromatin, and Subnuclear Positioning
The telomeres at the end of linear chromosomes are often heterochromatic. In vertebrates, telomeres are composed of hexameric 5′-TTAGGG-3′ repeats that are flanked by repetitive subtelomeric regions. Telomeric and subtelomeric repeats exhibit heterochromatic marks (H3K9me3, H4K20me3, and hypoacetylation of H3 and H4). Loss of heterochromatin disrupts telomere length control, increases telomeric recombination and promotes premature cellular senescence (Garcia-Cao et al., 2004; Benetti et al., 2007). Interestingly, the establishment of telomeric heterochromatin is influenced by a type of ncRNA called telomeric repeat-containing RNA (TERRA), which is composed of UUAGGG repeats (Nergadze et al., 2009). TERRA transcription typically initiates from subtelomeric CpG islands and proceeds to chromosomal ends. Several lines of evidence support a role for TERRA in the regulation of heterochromatin and other structures near chromosome ends. First, TERRA associates with TIP5 and subsequently recruits NoRC and the histone-modifying enzymes Suv4-20h2 and SIRT6 to human telomeres (Postepska-Igielska et al., 2013). Second, loss of human TERRA decreases telomeric H3K9me3 and HP1 enrichments and induces the DNA damage response (Blasco, 2007; Deng et al., 2009). Third, TERRA facilitates heterochromatin-promoting interactions between the human Shelterin complex, HP1 and the origin recognition complex (Deng et al., 2009). Fourth, TERRA transcription initiates at subtelomeric CTCF-binding sites, tentatively suggesting that the transcription of TERRA is itself spatially regulated by chromosome looping (Beishline et al., 2017). ncRNAs also regulate telomeric heterochromatin formation in non-vertebrate species. For example, small ncRNAs are implicated in heterochromatin formation at fission yeast telomeres (Cao et al., 2009). Together, these studies highlight a role for telomeric ncRNAs in the promotion of local heterochromatin structures and consequent prevention of premature cellular senescence. Importantly, there is crosstalk between telomeric heterochromatin and the subnuclear positioning of telomeres. For example, in budding yeast, the constitutive co-localization of telomeres in a handful of clusters at the nuclear periphery increases the local concentration of chromatin silencing factors, reinforcing telomeric heterochromatin and limiting access to the potentially genome-destabilizing recombination machinery (Therizols et al., 2006; Schober et al., 2009; Chan et al., 2011). In the fly germline, loss of some PIWI-interacting RNAs (piRNAs) that are typically transcribed from telomeric regions decreased perinuclear telomere positioning and lowered the local enrichment of HP1, Rhino, and H3K9me3 (Figure 2B) (Radion et al., 2018).
Connections exist between telomere malfunction and disease. The aberrant loss of telomeric heterochromatin can trigger telomeric DNA damage responses and recombination events, which are associated with several diseases (Hagelstrom et al., 2010). The accumulation of TERRA-associated R-loops drives telomere instability in the rare autosomal recessive syndrome ICF (immunodeficiency, centromeric instability, and facial anomalies; Sagie et al., 2017). Similarly, in budding yeast, elevated TERRA levels can promote premature senescence (Wanat et al., 2018). On another front, various changes to TERRA levels are linked to cancer (Artandi and DePinho, 2010), dyskeratosis congenita (Armanios et al., 2009; Gu et al., 2009; Mason and Bessler, 2011) and aplastic anemia (Armanios et al., 2009; O’Sullivan and Karlseder, 2010; Armanios, 2012). We refer the reader elsewhere for a full review on the emerging connections between telomeric ncRNAs and disease (Maicher et al., 2012). Taken together, these studies suggest that telomeric ncRNAs modulate heterochromatin formation and subnuclear positioning at telomeres to promote health and longevity.
Transposable Elements
Similar to other repetitive DNA loci, transposable elements are often silenced by heterochromatin formation to limit the potentially deleterious effects of such elements (Slotkin and Martienssen, 2007). Transposable elements are silenced through a wide range of chromatin modifications, including DNA methylation, histone modifications (e.g., H3K9me and H4K20me) and chromatin condensation (Slotkin and Martienssen, 2007). Similar to PAPAS-dependent recruitment of Suv4-20h2 to the rDNA, in quiescent human cells, it was reported that ncRNAs from the transposable elements IAP and LINE-1 recruit Suv4-20h2 to mediate H4K20me3 enrichment and condense chromatin at transposable elements (Bierhoff et al., 2014). Such elements are also silenced through the action of small ncRNAs. For example, murine piRNAs generated from retroelements are bound to the PIWI-like protein MIWI2 and translocated into the nucleus to silence retroelements through de novo DNA methylation (Aravin et al., 2008; De Fazio et al., 2011). Additionally, small RNAs generated from LINE-1 and IAP retroelements can regulate their epigenetic state in mouse embryos (Fadloun et al., 2013). Taken together, these studies suggest that ncRNAs help establish the epigenetic states necessary to keep transposable elements in check.
In addition to regulating chromatin compaction at transposable elements, transposon-associated ncRNAs can modulate the spatial organization of the nucleolus. For example, in HeLa cells, transcripts originating from intronic Alu elements (aluRNAs) become enriched in the nucleolus, where they interact with the nucleolin (NCL) protein and contribute to the maintenance of nucleolar structure and function (Figure 2C) (Caudron-Herger et al., 2015). Similar processes were observed in human keratinocytes and fibroblasts for aluRNAs, and for the related B1 transcripts in mice. Interestingly, aluRNAs can somehow target other genomic loci to the nucleolus (Caudron-Herger et al., 2015), tentatively suggesting that these ncRNAs may impact spatial genome organization by establishing physical links within and outside of the nucleolus.
Given the high mutagenic potential of transposable element activity, it is perhaps not surprising that these elements have been linked to disease (Belancio et al., 2009). Transposons can promote disease through several processes including insertional mutations, deleterious non-allelic homologous recombination and the generation of cis-acting signals that modify gene expression (Belancio et al., 2009). It is estimated that ∼0.3% of human genetic diseases are caused by retroelements (Callinan and Batzer, 2006). For example, 15 human diseases are caused by Alu insertions while 18 germ-line diseases and 6 types of cancer are caused by unequal homologous recombination events between Alu repeats (Deininger and Batzer, 1999; Burns, 2017). In addition, LINE-1 and SVAs are causative agents in numerous other human diseases (Belancio et al., 2009). In fact, the increased activity of transposable elements is a known contributing factor to neurodegenerative diseases such as Alzheimer’s disease, Aicardi Goutières syndrome, multiple sclerosis, and amyotrophic lateral sclerosis (Guo et al., 2018; Tam et al., 2019). Elevated expression of transposable elements is also a potential mechanism underlying the pathogenic development of various mental disorders including schizophrenia, bipolar disorder, autism spectrum disorders, and major depression (Misiak et al., 2019). In the context of these various diseases, it is thought that loss of heterochromatin structures may be a major contributor to the increased transposable element activity and its deleterious impact. Together, the literature indicates that ncRNAs from transposable elements can positively contribute to spatial genome organization and stability, but that losing control of such elements can disrupt genome function and promote disease.
Centromeres
Centromeres are tandem repeats, which are largely assembled into heterochromatic structures and are important for kinetochore function and chromosome integrity. Centromeres are composed of centric and pericentric regions, which have different chromatin structures that are epigenetically established. CENP-A-containing centric chromatin is characterized by H3K4me3, while pericentric regions are enriched in H3K9me2, H3K9me3, H4K20, and HP1. Heterochromatin formation at centric and pericentric regions is mediated by NoRC, similar to heterochromatin formation at rDNA (Wong et al., 2007; Nemeth et al., 2010). In fact, the common positioning of centromeres near nucleoli may contribute to this dual role for NoRC at rDNA and centromeres (Wong et al., 2007; Nemeth et al., 2010).
Several classes of centromeric ncRNAs have been found to play a role in the establishment of centromeric heterochromatin and kinetochore function across a wide range of species. Importantly, centromeric heterochromatin is maintained by low levels of satellite repeat RNAs (Diaz et al., 1981; Trapitz et al., 1988; Rudert et al., 1995; Li and Kirby, 2003; Martens et al., 2005; Wong et al., 2007). In fission yeast, short-interfering RNAs produced by pericentromeric ‘otr’ ncRNAs help establish and maintain pericentric heterochromatin (Volpe et al., 2002), while in budding yeast, the expression of centromere-derived lncRNAs (cenRNAs) must be fine-tuned in order to maintain centromere function (Ling and Yuen, 2019). Increased cenRNA levels result in chromosome instability, aneuploidy and down-regulation of centromeric proteins while decreased cenRNA levels also result in chromosome instability. There is overwhelming evidence that centromeric- or pericentromeric-derived ncRNAs are important for the recruitment of centromeric proteins (Figure 2D) (Maison et al., 2002; Wong et al., 2007; Ferri et al., 2009; Chan et al., 2012). In Drosophila, centromeric SAT III ncRNAs act as a structural component of the kinetochore and are required for the recruitment of centromeric proteins (Rosic et al., 2014). In mice, lncRNAs produced from major pericentromeric satellite repeats recruit the SUMOylated form of HP1 through direct interaction with DNA at the site of their transcription (Maison et al., 2011). Murine major satellite-derived ncRNAs have also been shown to form RNA–DNA hybrids that promote the association of histone lysine methyltransferases Suv39h1 and Suv39h2 with polynucleosomes (Velazquez Camacho et al., 2017), suggesting a function for these ncRNAs in establishing heterochromatic structures. In human cells, single-stranded α-satellite RNAs are required for nucleolar localization of CENP-C and INCENP in interphase cells (Wong et al., 2007). Reducing or increasing centromeric transcription decreases the loading of several CENP proteins (Bergmann et al., 2011, 2012). In human cells and X. laevis egg extracts, loss of α-satellite ncRNAs reduces centromeric localization of the kinetochore protein Aurora-B and causes improper spindle attachment and chromosome misalignment (Ideue et al., 2014; Blower, 2016). Additionally, studies in maize, human cells and X. laevis suggest that centromeric ncRNAs stabilize CENP-C binding to DNA (Du et al., 2010; Grenfell et al., 2016; McNulty et al., 2017). Murine minor satellite repeat transcripts associate with CENP-A and regulate the structure and function of centromeres during stress and differentiation (Bouzinba-Segard et al., 2006; Ferri et al., 2009). Moreover, aberrant accumulation of these transcripts disrupts chromosome segregation, weakens sister chromatid cohesion, abrogates centromeric epigenetic signatures and results in the accumulation of micronuclei. Together, these studies reveal that the maintenance of an optimal level of centromeric ncRNAs may be important for centromeric function.
While mammalian centromeres can often co-localize with nucleoli in S phase cells, budding yeast centromeres cluster with each other at the spindle pole body, which is opposite the nucleolus (Mekhail and Moazed, 2010). Importantly, this co-localization may contribute to the cells’ ability to survive DNA double strand breaks (DSBs). Specifically, it was proposed that centromeres are released from the spindle pole body upon DNA damage induction to allow for increased chromosome flexibility and facilitate donor-acceptor locus contacts necessary for homology-directed repair (Strecker et al., 2016). The release of centromeres also drove the formation of intranuclear microtubule filaments onto which damaged DNA was mobilized by motor proteins to repair-conducive nuclear neighborhoods (Chung et al., 2015; Oshidari et al., 2018, 2019). It will be important to test whether endogenous transcription of centromeric ncRNAs contributes to this increased genome flexibility and formation of intranuclear filaments mediating DNA repair. Consistent with this possibility, the forced expression of an inducible gene integrated within a single centromere was sufficient to trigger the formation of the intranuclear microtubule filaments that are typically only observed upon DNA damage induction (Oshidari et al., 2018).
Changes to the epigenetic state of centromeres has been linked to disease (Black and Giunta, 2018). Tandemly arranged satellite repeats are prone to recombination events that can lead to chromosome rearrangements, genetic abnormalities and karyotypic abnormalities that are hallmarks of cancer (Kim et al., 2013). In addition, a study examining epigenetic signatures in ICF patients reported that, in all of the patients studied, juxtacentromeric satellite II repeats exhibited hypomethylation, tentatively suggesting that this altered epigenetic feature may underlie the chromosome fragility observed in ICF patients (Miniou et al., 1994). Centromeric repeat-associated ncRNAs have been implicated in chromatin-related changes in age and age-related diseases. There is a correlation between centromeric instability and senescence, which is potentially explained by an age-related loss of CENP-A at centromeres (Lee et al., 2010; Maehara et al., 2010; Hedouin et al., 2017). Senescence-related loss of CENP-A may be mediated by alterations to the levels of centromeric repeat transcripts, due to the fact that constitutive pericentromeric heterochromatin is decondensed in senescent cells (Swanson et al., 2013; Giunta and Funabiki, 2017). It has been directly shown that high rates of centromeric transcription can cause CENP-A translocation and mitotic arrest (Hedouin et al., 2017). Interestingly, some forms of cancer are characterized by elevated levels of α-satellite and pericentromeric satellite ncRNAs (Ting et al., 2011). These ncRNAs can form deleterious R-loop structures, which have been suggested to contribute to pericentromeric instability in several cancers (Bersani et al., 2015).
Taken together, the literature reveals numerous intersections between various types of ncRNAs and spatial genome organization in the modulation of repetitive DNA loci and their broader impact on the genome and health.
Concluding Remarks
In this review we have highlighted roles of ncRNAs and intergenic transcription from single copy and repetitive DNA loci in the regulation of spatial genome organization. Several ncRNAs participate in spatial genome organization through several common mechanisms of action, such as chromatin looping and heterochromatin formation, while others operate through distinct pathways such as perinuclear tethering. Deregulation of spatial genome organization is associated with developmental and age-related diseases including cancer. Although aberrant expression of ncRNAs has been implicated in disease, more direct or causal links between such ncRNAs, spatial genome organization and pathobiology should be established (Palazzo and Lee, 2015). Future studies should aim to identify the exact molecular switches that induce ncRNA-dependent changes to spatial genome organization, and whether these regulatory mechanisms are conserved across evolution. Furthermore, we expect future studies to identify novel processes through which ncRNAs can regulate the relationship between genome structure and function.
Author Contributions
NK and LO wrote the manuscript and prepared the figures. The manuscript was edited and updated by KM.
Funding
NK was supported by a scholarship from the Canadian Institutes of Health Research (CIHR). LO was supported by a scholarship from the Ontario Graduate Studies (OGS) Program. This work was supported by CIHR grants (#388041 and #399687) to KM, who holds the Canada Research Chair (CRC, #950-230661) in Spatial Genome Organization.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank members of the Mekhail lab for fruitful discussions.
References
Adriaens, C., Standaert, L., Barra, J., Latil, M., Verfaillie, A., Kalev, P., et al. (2016). p53 induces formation of NEAT1 lncRNA-containing paraspeckles that modulate replication stress response and chemosensitivity. Nat. Med. 22, 861–868. doi: 10.1038/nm.4135
Alt, F. W., Yancopoulos, G. D., Blackwell, T. K., Wood, C., Thomas, E., Boss, M., et al. (1984). Ordered rearrangement of immunoglobulin heavy chain variable region segments. EMBO J. 3, 1209–1219. doi: 10.1002/j.1460-2075.1984.tb01955.x
Andrey, G., Montavon, T., Mascrez, B., Gonzalez, F., Noordermeer, D., Leleu, M., et al. (2013). A switch between topological domains underlies HoxD genes collinearity in mouse limbs. Science 340:1234167. doi: 10.1126/science.1234167
Aravin, A. A., Sachidanandam, R., Bourc’his, D., Schaefer, C., Pezic, D., Toth, K. F., et al. (2008). A piRNA pathway primed by individual transposons is linked to de novo DNA methylation in mice. Mol. Cell. 31, 785–799. doi: 10.1016/j.molcel.2008.09.003
Ariel, F., Jegu, T., Latrasse, D., Romero-Barrios, N., Christ, A., Benhamed, M., et al. (2014). Noncoding transcription by alternative RNA polymerases dynamically regulates an auxin-driven chromatin loop. Mol. Cell. 55, 383–396. doi: 10.1016/j.molcel.2014.06.011
Armanios, M. (2012). Telomerase and idiopathic pulmonary fibrosis. Mutat. Res. 730, 52–58. doi: 10.1016/j.mrfmmm.2011.10.013
Armanios, M., Alder, J. K., Parry, E. M., Karim, B., Strong, M. A., and Greider, C. W. (2009). Short telomeres are sufficient to cause the degenerative defects associated with aging. Am. J. Hum. Genet. 85, 823–832. doi: 10.1016/j.ajhg.2009.10.028
Artandi, S. E., and DePinho, R. A. (2010). Telomeres and telomerase in cancer. Carcinogenesis 31, 9–18. doi: 10.1093/carcin/bgp268
Audas, T. E., Jacob, M. D., and Lee, S. (2012). Immobilization of proteins in the nucleolus by ribosomal intergenic spacer noncoding RNA. Mol. Cell. 45, 147–157. doi: 10.1016/j.molcel.2011.12.012
Beishline, K., Vladimirova, O., Tutton, S., Wang, Z., Deng, Z., and Lieberman, P. M. (2017). CTCF driven TERRA transcription facilitates completion of telomere DNA replication. Nat. Commun. 8:2114. doi: 10.1038/s41467-017-02212-w
Belancio, V. P., Deininger, P. L., and Roy-Engel, A. M. (2009). LINE dancing in the human genome: transposable elements and disease. Genome Med. 1:97. doi: 10.1186/gm97
Benetti, R., Gonzalo, S., Jaco, I., Schotta, G., Klatt, P., Jenuwein, T., et al. (2007). Suv4-20h deficiency results in telomere elongation and derepression of telomere recombination. J. Cell Biol. 178, 925–936. doi: 10.1083/jcb.200703081
Benjamins, R., Quint, A., Weijers, D., Hooykaas, P., and Offringa, R. (2001). The PINOID protein kinase regulates organ development in Arabidopsis by enhancing polar auxin transport. Development 128, 4057–4067.
Bergmann, J. H., Jakubsche, J. N., Martins, N. M., Kagansky, A., Nakano, M., Kimura, H., et al. (2012). Epigenetic engineering: histone H3K9 acetylation is compatible with kinetochore structure and function. J. Cell Sci. 125(Pt 2), 411–421. doi: 10.1242/jcs.090639
Bergmann, J. H., Rodriguez, M. G., Martins, N. M., Kimura, H., Kelly, D. A., Masumoto, H., et al. (2011). Epigenetic engineering shows H3K4me2 is required for HJURP targeting and CENP-A assembly on a synthetic human kinetochore. EMBO J. 30, 328–340. doi: 10.1038/emboj.2010.329
Bersani, F., Lee, E., Kharchenko, P. V., Xu, A. W., Liu, M., Xega, K., et al. (2015). Pericentromeric satellite repeat expansions through RNA-derived DNA intermediates in cancer. Proc. Natl. Acad. Sci. U.S.A. 112, 15148–15153. doi: 10.1073/pnas.1518008112
Bierhoff, H., Dammert, M. A., Brocks, D., Dambacher, S., Schotta, G., and Grummt, I. (2014). Quiescence-induced LncRNAs trigger H4K20 trimethylation and transcriptional silencing. Mol. Cell. 54, 675–682. doi: 10.1016/j.molcel.2014.03.032
Bierhoff, H., Schmitz, K., Maass, F., Ye, J., and Grummt, I. (2010). Noncoding transcripts in sense and antisense orientation regulate the epigenetic state of ribosomal RNA genes. Cold. Spring Harb. Symp. Quant. Biol 75, 357–364. doi: 10.1101/sqb.2010.75.060
Black, E. M., and Giunta, S. (2018). Repetitive fragile sites: centromere satellite DNA as a source of genome instability in human diseases. Genes 9:E615. doi: 10.3390/genes9120615
Blasco, M. A. (2007). The epigenetic regulation of mammalian telomeres. Nat. Rev. Genet. 8, 299–309. doi: 10.1038/nrg2047
Blower, M. D. (2016). Centromeric transcription regulates aurora-B localization and activation. Cell Rep. 15, 1624–1633. doi: 10.1016/j.celrep.2016.04.054
Bouzinba-Segard, H., Guais, A., and Francastel, C. (2006). Accumulation of small murine minor satellite transcripts leads to impaired centromeric architecture and function. Proc. Natl. Acad. Sci. U.S.A. 103, 8709–8714. doi: 10.1073/pnas.0508006103
Brown, C. J., Hendrich, B. D., Rupert, J. L., Lafreniere, R. G., Xing, Y., Lawrence, J., et al. (1992). The human XIST gene: analysis of a 17 kb inactive X-specific RNA that contains conserved repeats and is highly localized within the nucleus. Cell 71, 527–542. doi: 10.1016/0092-8674(92)90520-m
Brown, C. R., Kennedy, C. J., Delmar, V. A., Forbes, D. J., and Silver, P. A. (2008). Global histone acetylation induces functional genomic reorganization at mammalian nuclear pore complexes. Genes Dev. 22, 627–639. doi: 10.1101/gad.1632708
Burmann, F., Basfeld, A., Vazquez Nunez, R., Diebold-Durand, M. L., Wilhelm, L., and Gruber, S. (2017). Tuned SMC arms drive chromosomal loading of prokaryotic condensin. Mol. Cell. 65, 861.e–872.e. doi: 10.1016/j.molcel.2017.01.026
Burns, K. H. (2017). Transposable elements in cancer. Nat. Rev. Cancer 17, 415–424. doi: 10.1038/nrc.2017.35
Bywater, M. J., Pearson, R. B., McArthur, G. A., and Hannan, R. D. (2013). Dysregulation of the basal RNA polymerase transcription apparatus in cancer. Nat. Rev. Cancer 13, 299–314. doi: 10.1038/nrc3496
Callinan, P. A., and Batzer, M. A. (2006). Retrotransposable elements and human disease. Genome Dyn. 1, 104–115. doi: 10.1159/000092503
Cao, F., Li, X., Hiew, S., Brady, H., Liu, Y., and Dou, Y. (2009). Dicer independent small RNAs associate with telomeric heterochromatin. RNA 15, 1274–1281. doi: 10.1261/rna.1423309
Carmo-Fonseca, M., Ferreira, J., and Lamond, A. I. (1993). Assembly of snRNP-containing coiled bodies is regulated in interphase and mitosis–evidence that the coiled body is a kinetic nuclear structure. J. Cell Biol. 120, 841–852. doi: 10.1083/jcb.120.4.841
Casolari, J. M., Brown, C. R., Komili, S., West, J., Hieronymus, H., and Silver, P. A. (2004). Genome-wide localization of the nuclear transport machinery couples transcriptional status and nuclear organization. Cell 117, 427–439. doi: 10.1016/s0092-8674(04)00448-9
Caudron-Herger, M., Pankert, T., Seiler, J., Nemeth, A., Voit, R., Grummt, I., et al. (2015). Alu element-containing RNAs maintain nucleolar structure and function. EMBO J. 34, 2758–2774. doi: 10.15252/embj.201591458
Chakravarty, D., Sboner, A., Nair, S. S., Giannopoulou, E., Li, R., Hennig, S., et al. (2014). The oestrogen receptor alpha-regulated lncRNA NEAT1 is a critical modulator of prostate cancer. Nat. Commun. 5:5383. doi: 10.1038/ncomms6383
Chan, F. L., Marshall, O. J., Saffery, R., Kim, B. W., Earle, E., Choo, K. H., et al. (2012). Active transcription and essential role of RNA polymerase II at the centromere during mitosis. Proc. Natl. Acad. Sci. U.S.A. 109, 1979–1984. doi: 10.1073/pnas.1108705109
Chan, J. N., Poon, B. P., Salvi, J., Olsen, J. B., Emili, A., and Mekhail, K. (2011). Perinuclear cohibin complexes maintain replicative life span via roles at distinct silent chromatin domains. Dev. Cell 20, 867–879. doi: 10.1016/j.devcel.2011.05.014
Chang, F. T., McGhie, J. D., Chan, F. L., Tang, M. C., Anderson, M. A., Mann, J. R., et al. (2013). PML bodies provide an important platform for the maintenance of telomeric chromatin integrity in embryonic stem cells. Nucleic Acids Res. 41, 4447–4458. doi: 10.1093/nar/gkt114
Chaumeil, J., Le Baccon, P., Wutz, A., and Heard, E. (2006). A novel role for Xist RNA in the formation of a repressive nuclear compartment into which genes are recruited when silenced. Genes Dev. 20, 2223–2237. doi: 10.1101/gad.380906
Chen, C. K., Blanco, M., Jackson, C., Aznauryan, E., Ollikainen, N., Surka, C., et al. (2016). Xist recruits the X chromosome to the nuclear lamina to enable chromosome-wide silencing. Science 354, 468–472. doi: 10.1126/science.aae0047
Cho, E. J., and Kim, J. S. (2012). Crowding effects on the formation and maintenance of nuclear bodies: insights from molecular-dynamics simulations of simple spherical model particles. Biophys. J. 103, 424–433. doi: 10.1016/j.bpj.2012.07.007
Choudhry, H., Albukhari, A., Morotti, M., Haider, S., Moralli, D., Smythies, J., et al. (2015). Tumor hypoxia induces nuclear paraspeckle formation through HIF-2alpha dependent transcriptional activation of NEAT1 leading to cancer cell survival. Oncogene 34, 4482–4490. doi: 10.1038/onc.2014.378
Chung, D. K., Chan, J. N., Strecker, J., Zhang, W., Ebrahimi-Ardebili, S., Lu, T., et al. (2015). Perinuclear tethers license telomeric DSBs for a broad kinesin- and NPC-dependent DNA repair process. Nat. Commun. 6:7742. doi: 10.1038/ncomms8742
Clemson, C. M., Hutchinson, J. N., Sara, S. A., Ensminger, A. W., Fox, A. H., Chess, A., et al. (2009). An architectural role for a nuclear noncoding RNA: NEAT1 RNA is essential for the structure of paraspeckles. Mol. Cell. 33, 717–726. doi: 10.1016/j.molcel.2009.01.026
De Fazio, S., Bartonicek, N., Di Giacomo, M., Abreu-Goodger, C., Sankar, A., Funaya, C., et al. (2011). The endonuclease activity of Mili fuels piRNA amplification that silences LINE1 elements. Nature 480, 259–263. doi: 10.1038/nature10547
Degner, S. C., Wong, T. P., Jankevicius, G., and Feeney, A. J. (2009). Cutting edge: developmental stage-specific recruitment of cohesin to CTCF sites throughout immunoglobulin loci during B lymphocyte development. J. Immunol. 182, 44–48. doi: 10.4049/jimmunol.182.1.44
Deininger, P. L., and Batzer, M. A. (1999). Alu repeats and human disease. Mol. Genet. Metab. 67, 183–193. doi: 10.1006/mgme.1999.2864
Deng, Z., Norseen, J., Wiedmer, A., Riethman, H., and Lieberman, P. M. (2009). TERRA RNA binding to TRF2 facilitates heterochromatin formation and ORC recruitment at telomeres. Mol. Cell. 35, 403–413. doi: 10.1016/j.molcel.2009.06.025
Diaz, M. O., Barsacchi-Pilone, G., Mahon, K. A., and Gall, J. G. (1981). Transcripts from both strands of a satellite DNA occur on lampbrush chromosome loops of the newt Notophthalmus. Cell 24, 649–659. doi: 10.1016/0092-8674(81)90091-x
Diebold-Durand, M. L., Lee, H., Ruiz Avila, L. B., Noh, H., Shin, H. C., Im, H., et al. (2017). Structure of full-length SMC and rearrangements required for chromosome organization. Mol. Cell 67, 334.e5–347.e5. doi: 10.1016/j.molcel.2017.06.010
Dilworth, D. J., Tackett, A. J., Rogers, R. S., Yi, E. C., Christmas, R. H., Smith, J. J., et al. (2005). The mobile nucleoporin Nup2p and chromatin-bound Prp20p function in endogenous NPC-mediated transcriptional control. J. Cell Biol. 171, 955–965. doi: 10.1083/jcb.200509061
Di-Poi, N., Montoya-Burgos, J. I., Miller, H., Pourquie, O., Milinkovitch, M. C., and Duboule, D. (2010). Changes in Hox genes’ structure and function during the evolution of the squamate body plan. Nature 464, 99–103. doi: 10.1038/nature08789
Dixon, J. R., Selvaraj, S., Yue, F., Kim, A., Li, Y., Shen, Y., et al. (2012). Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature 485, 376–380. doi: 10.1038/nature11082
Du, Y., Topp, C. N., and Dawe, R. K. (2010). DNA binding of centromere protein C (CENPC) is stabilized by single-stranded RNA. PLoS Genet. 6:e1000835. doi: 10.1371/journal.pgen.1000835
Fadloun, A., Le Gras, S., Jost, B., Ziegler-Birling, C., Takahashi, H., Gorab, E., et al. (2013). Chromatin signatures and retrotransposon profiling in mouse embryos reveal regulation of LINE-1 by RNA. Nat. Struct. Mol. Biol. 20, 332–338. doi: 10.1038/nsmb.2495
Feric, M., Vaidya, N., Harmon, T. S., Mitrea, D. M., Zhu, L., Richardson, T. M., et al. (2016). Coexisting liquid phases underlie nucleolar subcompartments. Cell 165, 1686–1697. doi: 10.1016/j.cell.2016.04.047
Ferri, F., Bouzinba-Segard, H., Velasco, G., Hube, F., and Francastel, C. (2009). Non-coding murine centromeric transcripts associate with and potentiate Aurora B kinase. Nucleic Acids Res. 37, 5071–5080. doi: 10.1093/nar/gkp529
Fitzpatrick, G. V., Soloway, P. D., and Higgins, M. J. (2002). Regional loss of imprinting and growth deficiency in mice with a targeted deletion of KvDMR1. Nat. Genet. 32, 426–431. doi: 10.1038/ng988
Flyamer, I. M., Gassler, J., Imakaev, M., Brandao, H. B., Ulianov, S. V., Abdennur, N., et al. (2017). Single-nucleus Hi-C reveals unique chromatin reorganization at oocyte-to-zygote transition. Nature 544, 110–114. doi: 10.1038/nature21711
Fuxa, M., Skok, J., Souabni, A., Salvagiotto, G., Roldan, E., and Busslinger, M. (2004). Pax5 induces V-to-DJ rearrangements and locus contraction of the immunoglobulin heavy-chain gene. Genes Dev. 18, 411–422. doi: 10.1101/gad.291504
Ganji, M., Shaltiel, I. A., Bisht, S., Kim, E., Kalichava, A., Haering, C. H., et al. (2018). Real-time imaging of DNA loop extrusion by condensin. Science 360, 102–105. doi: 10.1126/science.aar7831
Garcia-Cao, M., O’Sullivan, R., Peters, A. H., Jenuwein, T., and Blasco, M. A. (2004). Epigenetic regulation of telomere length in mammalian cells by the Suv39h1 and Suv39h2 histone methyltransferases. Nat. Genet. 36, 94–99. doi: 10.1038/ng1278
Giunta, S., and Funabiki, H. (2017). Integrity of the human centromere DNA repeats is protected by CENP-A, CENP-C, and CENP-T. Proc. Natl. Acad. Sci. U.S.A. 114, 1928–1933. doi: 10.1073/pnas.1615133114
Gotta, M., Laroche, T., Formenton, A., Maillet, L., Scherthan, H., and Gasser, S. M. (1996). The clustering of telomeres and colocalization with Rap1, Sir3, and Sir4 proteins in wild-type Saccharomyces cerevisiae. J. Cell Biol. 134, 1349–1363. doi: 10.1083/jcb.134.6.1349
Grenfell, A. W., Heald, R., and Strzelecka, M. (2016). Mitotic noncoding RNA processing promotes kinetochore and spindle assembly in Xenopus. J. Cell Biol. 214, 133–141. doi: 10.1083/jcb.201604029
Gu, B., Bessler, M., and Mason, P. J. (2009). Dyskerin, telomerase and the DNA damage response. Cell Cycle 8, 6–10. doi: 10.4161/cc.8.1.7265
Guelen, L., Pagie, L., Brasset, E., Meuleman, W., Faza, M. B., Talhout, W., et al. (2008). Domain organization of human chromosomes revealed by mapping of nuclear lamina interactions. Nature 453, 948–951. doi: 10.1038/nature06947
Guo, C., Jeong, H. H., Hsieh, Y. C., Klein, H. U., Bennett, D. A., De Jager, P. L., et al. (2018). Tau activates transposable elements in Alzheimer’s Disease. Cell Rep. 23, 2874–2880. doi: 10.1016/j.celrep.2018.05.004
Hacisuleyman, E., Goff, L. A., Trapnell, C., Williams, A., Henao-Mejia, J., Sun, L., et al. (2014). Topological organization of multichromosomal regions by the long intergenic noncoding RNA Firre. Nat. Struct. Mol. Biol. 21, 198–206. doi: 10.1038/nsmb.2764
Hagelstrom, R. T., Blagoev, K. B., Niedernhofer, L. J., Goodwin, E. H., and Bailey, S. M. (2010). Hyper telomere recombination accelerates replicative senescence and may promote premature aging. Proc. Natl. Acad. Sci. U.S.A. 107, 15768–15773. doi: 10.1073/pnas.1006338107
Hall, A. C., Ostrowski, L. A., and Mekhail, K. (2019). Phase separation as a melting pot for DNA repeats. Trends Genet. 35, 589–600. doi: 10.1016/j.tig.2019.05.001
Hedouin, S., Grillo, G., Ivkovic, I., Velasco, G., and Francastel, C. (2017). CENP-A chromatin disassembly in stressed and senescent murine cells. Sci. Rep. 7:42520. doi: 10.1038/srep42520
Hernandez-Martin, A., Gonzalez-Sarmiento, R., and De Unamuno, P. (1999). X-linked ichthyosis: an update. Br. J. Dermatol. 141, 617–627. doi: 10.1046/j.1365-2133.1999.03098.x
Hirose, T., Virnicchi, G., Tanigawa, A., Naganuma, T., Li, R., Kimura, H., et al. (2014). NEAT1 long noncoding RNA regulates transcription via protein sequestration within subnuclear bodies. Mol. Biol. Cell 25, 169–183. doi: 10.1091/mbc.E13-09-0558
Hsieh, T. H., Weiner, A., Lajoie, B., Dekker, J., Friedman, N., and Rando, O. J. (2015). Mapping nucleosome resolution chromosome folding in yeast by micro-C. Cell 162, 108–119. doi: 10.1016/j.cell.2015.05.048
Hu, S., Lv, P., Yan, Z., and Wen, B. (2019). Disruption of nuclear speckles reduces chromatin interactions in active compartments. Epigenetics Chromatin 12:43. doi: 10.1186/s13072-019-0289-2
Hult, C., Adalsteinsson, D., Vasquez, P. A., Lawrimore, J., Bennett, M., York, A., et al. (2017). Enrichment of dynamic chromosomal crosslinks drive phase separation of the nucleolus. Nucleic Acids Res. 45, 11159–11173. doi: 10.1093/nar/gkx741
Ideue, T., Cho, Y., Nishimura, K., and Tani, T. (2014). Involvement of satellite I noncoding RNA in regulation of chromosome segregation. Genes Cells 19, 528–538. doi: 10.1111/gtc.12149
Ikegami, K., Egelhofer, T. A., Strome, S., and Lieb, J. D. (2010). Caenorhabditis elegans chromosome arms are anchored to the nuclear membrane via discontinuous association with LEM-2. Genome Biol. 11:R120. doi: 10.1186/gb-2010-11-12-r120
Imamura, K., Imamachi, N., Akizuki, G., Kumakura, M., Kawaguchi, A., Nagata, K., et al. (2014). Long noncoding RNA NEAT1-dependent SFPQ relocation from promoter region to paraspeckle mediates IL8 expression upon immune stimuli. Mol. Cell. 53, 393–406. doi: 10.1016/j.molcel.2014.01.009
Isoda, T., Moore, A. J., He, Z., Chandra, V., Aida, M., Denholtz, M., et al. (2017). Non-coding transcription instructs chromatin folding and compartmentalization to dictate enhancer-promoter communication and t cell fate. Cell 171, 103.e18–119.e18. doi: 10.1016/j.cell.2017.09.001
Jacob, M. D., Audas, T. E., Uniacke, J., Trinkle-Mulcahy, L., and Lee, S. (2013). Environmental cues induce a long noncoding RNA-dependent remodeling of the nucleolus. Mol. Biol. Cell 24, 2943–2953. doi: 10.1091/mbc.E13-04-0223
Jin, Q. W., Fuchs, J., and Loidl, J. (2000). Centromere clustering is a major determinant of yeast interphase nuclear organization. J. Cell Sci. 113(Pt 11), 1903–1912.
Kaiser, T. E., Intine, R. V., and Dundr, M. (2008). De novo formation of a subnuclear body. Science 322, 1713–1717. doi: 10.1126/science.1165216
Khalil, A. M., Guttman, M., Huarte, M., Garber, M., Raj, A., Rivea Morales, D., et al. (2009). Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc. Natl. Acad. Sci. U.S.A. 106, 11667–11672. doi: 10.1073/pnas.0904715106
Kim, T. K., Hemberg, M., Gray, J. M., Costa, A. M., Bear, D. M., Wu, J., et al. (2010). Widespread transcription at neuronal activity-regulated enhancers. Nature 465, 182–187. doi: 10.1038/nature09033
Kim, T. M., Xi, R., Luquette, L. J., Park, R. W., Johnson, M. D., and Park, P. J. (2013). Functional genomic analysis of chromosomal aberrations in a compendium of 8000 cancer genomes. Genome Res. 23, 217–227. doi: 10.1101/gr.140301.112
Kind, J., Pagie, L., de Vries, S. S., Nahidiazar, L., Dey, S. S., Bienko, M., et al. (2015). Genome-wide maps of nuclear lamina interactions in single human cells. Cell 163, 134–147. doi: 10.1016/j.cell.2015.08.040
Kosak, S. T., Skok, J. A., Medina, K. L., Riblet, R., Le Beau, M. M., Fisher, A. G., et al. (2002). Subnuclear compartmentalization of immunoglobulin loci during lymphocyte development. Science 296, 158–162. doi: 10.1126/science.1068768
Krefting, J., Andrade-Navarro, M. A., and Ibn-Salem, J. (2018). Evolutionary stability of topologically associating domains is associated with conserved gene regulation. BMC Biol. 16:87. doi: 10.1186/s12915-018-0556-x
Lai, F., Orom, U. A., Cesaroni, M., Beringer, M., Taatjes, D. J., Blobel, G. A., et al. (2013). Activating RNAs associate with Mediator to enhance chromatin architecture and transcription. Nature 494, 497–501. doi: 10.1038/nature11884
Lee, J. T., and Lu, N. (1999). Targeted mutagenesis of Tsix leads to nonrandom X inactivation. Cell 99, 47–57. doi: 10.1016/s0092-8674(00)80061-6
Lee, M. P., DeBaun, M. R., Mitsuya, K., Galonek, H. L., Brandenburg, S., Oshimura, M., et al. (1999). Loss of imprinting of a paternally expressed transcript, with antisense orientation to KVLQT1, occurs frequently in Beckwith-Wiedemann syndrome and is independent of insulin-like growth factor II imprinting. Proc. Natl. Acad. Sci. U.S.A. 96, 5203–5208. doi: 10.1073/pnas.96.9.5203
Lee, S. H., Itkin-Ansari, P., and Levine, F. (2010). CENP-A, a protein required for chromosome segregation in mitosis, declines with age in islet but not exocrine cells. Aging 2, 785–790. doi: 10.18632/aging.100220
Lewis, A., Green, K., Dawson, C., Redrup, L., Huynh, K. D., Lee, J. T., et al. (2006). Epigenetic dynamics of the Kcnq1 imprinted domain in the early embryo. Development 133, 4203–4210. doi: 10.1242/dev.02612
Li, L., Leid, M., and Rothenberg, E. V. (2010). An early T cell lineage commitment checkpoint dependent on the transcription factor Bcl11b. Science 329, 89–93. doi: 10.1126/science.1188989
Li, L., Zhang, J. A., Dose, M., Kueh, H. Y., Mosadeghi, R., Gounari, F., et al. (2013). A far downstream enhancer for murine Bcl11b controls its T-cell specific expression. Blood 122, 902–911. doi: 10.1182/blood-2012-08-447839
Li, W., Notani, D., Ma, Q., Tanasa, B., Nunez, E., Chen, A. Y., et al. (2013). Functional roles of enhancer RNAs for oestrogen-dependent transcriptional activation. Nature 498, 516–520. doi: 10.1038/nature12210
Li, Y. X., and Kirby, M. L. (2003). Coordinated and conserved expression of alphoid repeat and alphoid repeat-tagged coding sequences. Dev. Dyn. 228, 72–81. doi: 10.1002/dvdy.10355
Lieberman-Aiden, E., van Berkum, N. L., Williams, L., Imakaev, M., Ragoczy, T., Telling, A., et al. (2009). Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science 326, 289–293. doi: 10.1126/science.1181369
Ling, Y. H., and Yuen, K. W. Y. (2019). Point centromere activity requires an optimal level of centromeric noncoding RNA. Proc. Natl. Acad. Sci. U.S.A. 116, 6270–6279. doi: 10.1073/pnas.1821384116
Luikenhuis, S., Wutz, A., and Jaenisch, R. (2001). Antisense transcription through the Xist locus mediates Tsix function in embryonic stem cells. Mol. Cell. Biol. 21, 8512–8520. doi: 10.1128/MCB.21.24.8512-8520.2001
Ma, H., Han, P., Ye, W., Chen, H., Zheng, X., Cheng, L., et al. (2017). The long noncoding RNA NEAT1 exerts antihantaviral effects by acting as positive feedback for RIG-I signaling. J. Virol. 91:e2250-16. doi: 10.1128/JVI.02250-16
Machyna, M., Kehr, S., Straube, K., Kappei, D., Buchholz, F., Butter, F., et al. (2014). The coilin interactome identifies hundreds of small noncoding RNAs that traffic through Cajal bodies. Mol. Cell. 56, 389–399. doi: 10.1016/j.molcel.2014.10.004
Maehara, K., Takahashi, K., and Saitoh, S. (2010). CENP-A reduction induces a p53-dependent cellular senescence response to protect cells from executing defective mitoses. Mol. Cell. Biol. 30, 2090–2104. doi: 10.1128/MCB.01318-09
Maicher, A., Kastner, L., and Luke, B. (2012). Telomeres and disease: enter TERRA. RNA Biol. 9, 843–849. doi: 10.4161/rna.20330
Maison, C., Bailly, D., Peters, A. H., Quivy, J. P., Roche, D., Taddei, A., et al. (2002). Higher-order structure in pericentric heterochromatin involves a distinct pattern of histone modification and an RNA component. Nat. Genet. 30, 329–334. doi: 10.1038/ng843
Maison, C., Bailly, D., Roche, D., Montes de Oca, R., Probst, A. V., Vassias, I., et al. (2011). SUMOylation promotes de novo targeting of HP1alpha to pericentric heterochromatin. Nat. Genet. 43, 220–227. doi: 10.1038/ng.765
Mancini-Dinardo, D., Steele, S. J., Levorse, J. M., Ingram, R. S., and Tilghman, S. M. (2006). Elongation of the Kcnq1ot1 transcript is required for genomic imprinting of neighboring genes. Genes Dev. 20, 1268–1282. doi: 10.1101/gad.1416906
Mao, Y. S., Zhang, B., and Spector, D. L. (2011). Biogenesis and function of nuclear bodies. Trends Genet. 27, 295–306. doi: 10.1016/j.tig.2011.05.006
Martens, J. H., O’Sullivan, R. J., Braunschweig, U., Opravil, S., Radolf, M., Steinlein, P., et al. (2005). The profile of repeat-associated histone lysine methylation states in the mouse epigenome. EMBO J. 24, 800–812. doi: 10.1038/sj.emboj.7600545
Mason, P. J., and Bessler, M. (2011). The genetics of dyskeratosis congenita. Cancer Genet. 204, 635–645. doi: 10.1016/j.cancergen.2011.11.002
Mayer, C., Schmitz, K. M., Li, J., Grummt, I., and Santoro, R. (2006). Intergenic transcripts regulate the epigenetic state of rRNA genes. Mol. Cell. 22, 351–361. doi: 10.1016/j.molcel.2006.03.028
McNulty, S. M., Sullivan, L. L., and Sullivan, B. A. (2017). Human centromeres produce chromosome-specific and array-specific alpha satellite transcripts that are complexed with CENP-A and CENP-C. Dev. Cell 42, 226.e6–240.e6. doi: 10.1016/j.devcel.2017.07.001
Medvedovic, J., Ebert, A., Tagoh, H., Tamir, I. M., Schwickert, T. A., Novatchkova, M., et al. (2013). Flexible long-range loops in the VH gene region of the Igh locus facilitate the generation of a diverse antibody repertoire. Immunity 39, 229–244. doi: 10.1016/j.immuni.2013.08.011
Mekhail, K., Khacho, M., Carrigan, A., Hache, R. R., Gunaratnam, L., and Lee, S. (2005). Regulation of ubiquitin ligase dynamics by the nucleolus. J. Cell Biol. 170, 733–744. doi: 10.1083/jcb.200506030
Mekhail, K., and Moazed, D. (2010). The nuclear envelope in genome organization, expression and stability. Nat. Rev. Mol. Cell Biol. 11, 317–328. doi: 10.1038/nrm2894
Mekhail, K., Seebacher, J., Gygi, S. P., and Moazed, D. (2008). Role for perinuclear chromosome tethering in maintenance of genome stability. Nature 456, 667–670. doi: 10.1038/nature07460
Minajigi, A., Froberg, J., Wei, C., Sunwoo, H., Kesner, B., Colognori, D., et al. (2015). Chromosomes. A comprehensive Xist interactome reveals cohesin repulsion and an RNA-directed chromosome conformation. Science 349:aab2276. doi: 10.1126/science.aab2276
Miniou, P., Jeanpierre, M., Blanquet, V., Sibella, V., Bonneau, D., Herbelin, C., et al. (1994). Abnormal methylation pattern in constitutive and facultative (X inactive chromosome) heterochromatin of ICF patients. Hum. Mol. Genet. 3, 2093–2102. doi: 10.1093/hmg/3.12.2093
Misiak, B., Ricceri, L., and Sasiadek, M. M. (2019). Transposable elements and their epigenetic regulation in mental disorders: current evidence in the field. Front. Genet. 10:580. doi: 10.3389/fgene.2019.00580
Mohammad, F., Pandey, R. R., Nagano, T., Chakalova, L., Mondal, T., Fraser, P., et al. (2008). Kcnq1ot1/Lit1 noncoding RNA mediates transcriptional silencing by targeting to the perinucleolar region. Mol. Cell. Biol. 28, 3713–3728. doi: 10.1128/MCB.02263-07
Montanaro, L., Trere, D., and Derenzini, M. (2008). Nucleolus, ribosomes, and cancer. Am. J. Pathol. 173, 301–310. doi: 10.2353/ajpath.2008.070752
Mortazavi, A., Williams, B. A., McCue, K., Schaeffer, L., and Wold, B. (2008). Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods 5, 621–628. doi: 10.1038/nmeth.1226
Nakhoul, H., Ke, J., Zhou, X., Liao, W., Zeng, S. X., and Lu, H. (2014). Ribosomopathies: mechanisms of disease. Clin. Med. Insights Blood Disord. 7, 7–16. doi: 10.4137/CMBD.S16952
Narla, A., and Ebert, B. L. (2010). Ribosomopathies: human disorders of ribosome dysfunction. Blood 115, 3196–3205. doi: 10.1182/blood-2009-10-178129
Nemeth, A., Conesa, A., Santoyo-Lopez, J., Medina, I., Montaner, D., Peterfia, B., et al. (2010). Initial genomics of the human nucleolus. PLoS Genet. 6:e1000889. doi: 10.1371/journal.pgen.1000889
Nergadze, S. G., Farnung, B. O., Wischnewski, H., Khoriauli, L., Vitelli, V., Chawla, R., et al. (2009). CpG-island promoters drive transcription of human telomeres. RNA 15, 2186–2194. doi: 10.1261/rna.1748309
Noordermeer, D., Leleu, M., Splinter, E., Rougemont, J., De Laat, W., and Duboule, D. (2011). The dynamic architecture of Hox gene clusters. Science 334, 222–225. doi: 10.1126/science.1207194
Nora, E. P., Lajoie, B. R., Schulz, E. G., Giorgetti, L., Okamoto, I., Servant, N., et al. (2012). Spatial partitioning of the regulatory landscape of the X-inactivation centre. Nature 485, 381–385. doi: 10.1038/nature11049
Nuebler, J., Fudenberg, G., Imakaev, M., Abdennur, N., and Mirny, L. A. (2018). Chromatin organization by an interplay of loop extrusion and compartmental segregation. Proc. Natl. Acad. Sci. U.S.A. 115, E6697–E6706. doi: 10.1073/pnas.1717730115
Okamoto, I., Otte, A. P., Allis, C. D., Reinberg, D., and Heard, E. (2004). Epigenetic dynamics of imprinted X inactivation during early mouse development. Science 303, 644–649. doi: 10.1126/science.1092727
Orom, U. A., Derrien, T., Beringer, M., Gumireddy, K., Gardini, A., Bussotti, G., et al. (2010). Long noncoding RNAs with enhancer-like function in human cells. Cell 143, 46–58. doi: 10.1016/j.cell.2010.09.001
Oshidari, R., Huang, R., Medghalchi, M., Elizabeth, Y. W., Ashgriz, N., Lee, H. O., et al. (2019). DNA repair by Rad52 liquid droplets. bioRxiv [Preprint]. doi: 10.1101/768119
Oshidari, R., Strecker, J., Chung, D. K. C., Abraham, K. J., Chan, J. N. Y., Damaren, C. J., et al. (2018). Nuclear microtubule filaments mediate non-linear directional motion of chromatin and promote DNA repair. Nat. Commun. 9:2567. doi: 10.1038/s41467-018-05009-7
Ostrowski, L. A., Hall, A. C., Szafranski, K. J., Oshidari, R., Abraham, K. J., Chan, J. N. Y., et al. (2018). Conserved Pbp1/Ataxin-2 regulates retrotransposon activity and connects polyglutamine expansion-driven protein aggregation to lifespan-controlling rDNA repeats. Commun. Biol. 1:187. doi: 10.1038/s42003-018-0187-3
O’Sullivan, J. M., Sontam, D. M., Grierson, R., and Jones, B. (2009). Repeated elements coordinate the spatial organization of the yeast genome. Yeast 26, 125–138. doi: 10.1002/yea.1657
O’Sullivan, R. J., and Karlseder, J. (2010). Telomeres: protecting chromosomes against genome instability. Nat. Rev. Mol. Cell Biol. 11, 171–181. doi: 10.1038/nrm2848
Palazzo, A. F., and Lee, E. S. (2015). Non-coding RNA: what is functional and what is junk? Front. Genet. 6:2. doi: 10.3389/fgene.2015.00002
Pandey, R. R., Mondal, T., Mohammad, F., Enroth, S., Redrup, L., Komorowski, J., et al. (2008). Kcnq1ot1 antisense noncoding RNA mediates lineage-specific transcriptional silencing through chromatin-level regulation. Mol. Cell. 32, 232–246. doi: 10.1016/j.molcel.2008.08.022
Pickersgill, H., Kalverda, B., de Wit, E., Talhout, W., Fornerod, M., and van Steensel, B. (2006). Characterization of the Drosophila melanogaster genome at the nuclear lamina. Nat. Genet. 38, 1005–1014. doi: 10.1038/ng1852
Pietrzak, M., Rempala, G., Nelson, P. T., Zheng, J. J., and Hetman, M. (2011). Epigenetic silencing of nucleolar rRNA genes in Alzheimer’s disease. PLoS One 6:e22585. doi: 10.1371/journal.pone.0022585
Plath, K., Fang, J., Mlynarczyk-Evans, S. K., Cao, R., Worringer, K. A., Wang, H., et al. (2003). Role of histone H3 lysine 27 methylation in X inactivation. Science 300, 131–135. doi: 10.1126/science.1084274
Postepska-Igielska, A., Krunic, D., Schmitt, N., Greulich-Bode, K. M., Boukamp, P., and Grummt, I. (2013). The chromatin remodelling complex NoRC safeguards genome stability by heterochromatin formation at telomeres and centromeres. EMBO Rep. 14, 704–710. doi: 10.1038/embor.2013.87
Radion, E., Morgunova, V., Ryazansky, S., Akulenko, N., Lavrov, S., Abramov, Y., et al. (2018). Key role of piRNAs in telomeric chromatin maintenance and telomere nuclear positioning in Drosophila germline. Epigenetics Chromatin 11:40. doi: 10.1186/s13072-018-0210-4
Rao, S. S., Huntley, M. H., Durand, N. C., Stamenova, E. K., Bochkov, I. D., Robinson, J. T., et al. (2014). A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell 159, 1665–1680. doi: 10.1016/j.cell.2014.11.021
Renault, N. K., Dyack, S., Dobson, M. J., Costa, T., Lam, W. L., and Greer, W. L. (2007). Heritable skewed X-chromosome inactivation leads to haemophilia A expression in heterozygous females. Eur. J. Hum. Genet. 15, 628–637. doi: 10.1038/sj.ejhg.5201799
Rice, J. C., and Allis, C. D. (2001). Histone methylation versus histone acetylation: new insights into epigenetic regulation. Curr. Opin. Cell Biol. 13, 263–273. doi: 10.1016/s0955-0674(00)00208-8
Richards, E. J., and Elgin, S. C. (2002). Epigenetic codes for heterochromatin formation and silencing: rounding up the usual suspects. Cell 108, 489–500. doi: 10.1016/s0092-8674(02)00644-x
Richter, K., Nessling, M., and Lichter, P. (2007). Experimental evidence for the influence of molecular crowding on nuclear architecture. J. Cell Sci. 120(Pt 9), 1673–1680. doi: 10.1242/jcs.03440
Rosic, S., Kohler, F., and Erhardt, S. (2014). Repetitive centromeric satellite RNA is essential for kinetochore formation and cell division. J. Cell Biol. 207, 335–349. doi: 10.1083/jcb.201404097
Rudert, F., Bronner, S., Garnier, J. M., and Dolle, P. (1995). Transcripts from opposite strands of gamma satellite DNA are differentially expressed during mouse development. Mamm. Genome 6, 76–83. doi: 10.1007/bf00303248
Sagie, S., Toubiana, S., Hartono, S. R., Katzir, H., Tzur-Gilat, A., Havazelet, S., et al. (2017). Telomeres in ICF syndrome cells are vulnerable to DNA damage due to elevated DNA:RNA hybrids. Nat. Commun. 8:14015. doi: 10.1038/ncomms14015
Saka, K., Ide, S., Ganley, A. R., and Kobayashi, T. (2013). Cellular senescence in yeast is regulated by rDNA noncoding transcription. Curr. Biol. 23, 1794–1798. doi: 10.1016/j.cub.2013.07.048
Salvi, J. S., Chan, J. N., Szafranski, K., Liu, T. T., Wu, J. D., Olsen, J. B., et al. (2014). Roles for Pbp1 and caloric restriction in genome and lifespan maintenance via suppression of RNA-DNA hybrids. Dev. Cell 30, 177–191. doi: 10.1016/j.devcel.2014.05.013
Sanborn, A. L., Rao, S. S., Huang, S. C., Durand, N. C., Huntley, M. H., Jewett, A. I., et al. (2015). Chromatin extrusion explains key features of loop and domain formation in wild-type and engineered genomes. Proc. Natl. Acad. Sci. U.S.A. 112, E6456–E6465. doi: 10.1073/pnas.1518552112
Santoro, R., Li, J., and Grummt, I. (2002). The nucleolar remodeling complex NoRC mediates heterochromatin formation and silencing of ribosomal gene transcription. Nat. Genet. 32, 393–396. doi: 10.1038/ng1010
Savic, N., Bar, D., Leone, S., Frommel, S. C., Weber, F. A., Vollenweider, E., et al. (2014). lncRNA maturation to initiate heterochromatin formation in the nucleolus is required for exit from pluripotency in ESCs. Cell Stem Cell 15, 720–734. doi: 10.1016/j.stem.2014.10.005
Sawyer, I. A., Sturgill, D., Sung, M. H., Hager, G. L., and Dundr, M. (2016). Cajal body function in genome organization and transcriptome diversity. Bioessays 38, 1197–1208. doi: 10.1002/bies.201600144
Sayegh, C. E., Jhunjhunwala, S., Riblet, R., and Murre, C. (2005). Visualization of looping involving the immunoglobulin heavy-chain locus in developing B cells. Genes Dev. 19, 322–327. doi: 10.1101/gad.1254305
Schmid, M., Arib, G., Laemmli, C., Nishikawa, J., Durussel, T., and Laemmli, U. K. (2006). Nup-PI: the nucleopore-promoter interaction of genes in yeast. Mol. Cell. 21, 379–391. doi: 10.1016/j.molcel.2005.12.012
Schober, H., Ferreira, H., Kalck, V., Gehlen, L. R., and Gasser, S. M. (2009). Yeast telomerase and the SUN domain protein Mps3 anchor telomeres and repress subtelomeric recombination. Genes Dev. 23, 928–938. doi: 10.1101/gad.1787509
Schoeftner, S., Sengupta, A. K., Kubicek, S., Mechtler, K., Spahn, L., Koseki, H., et al. (2006). Recruitment of PRC1 function at the initiation of X inactivation independent of PRC2 and silencing. EMBO J. 25, 3110–3122. doi: 10.1038/sj.emboj.7601187
Schwarzer, W., Abdennur, N., Goloborodko, A., Pekowska, A., Fudenberg, G., Loe-Mie, Y., et al. (2017). Two independent modes of chromatin organization revealed by cohesin removal. Nature 551, 51–56. doi: 10.1038/nature24281
Sexton, T., Yaffe, E., Kenigsberg, E., Bantignies, F., Leblanc, B., Hoichman, M., et al. (2012). Three-dimensional folding and functional organization principles of the Drosophila genome. Cell 148, 458–472. doi: 10.1016/j.cell.2012.01.010
Shevtsov, S. P., and Dundr, M. (2011). Nucleation of nuclear bodies by RNA. Nat. Cell Biol. 13, 167–173. doi: 10.1038/ncb2157
Shibata, S., and Lee, J. T. (2004). Tsix transcription- versus RNA-based mechanisms in Xist repression and epigenetic choice. Curr. Biol. 14, 1747–1754. doi: 10.1016/j.cub.2004.09.053
Shin, J. Y., Fitzpatrick, G. V., and Higgins, M. J. (2008). Two distinct mechanisms of silencing by the KvDMR1 imprinting control region. EMBO J. 27, 168–178. doi: 10.1038/sj.emboj.7601960
Sleeman, J. E., and Trinkle-Mulcahy, L. (2014). Nuclear bodies: new insights into assembly/dynamics and disease relevance. Curr. Opin. Cell Biol. 28, 76–83. doi: 10.1016/j.ceb.2014.03.004
Slotkin, R. K., and Martienssen, R. (2007). Transposable elements and the epigenetic regulation of the genome. Nat. Rev. Genet. 8, 272–285. doi: 10.1038/nrg2072
Stavropoulos, N., Lu, N., and Lee, J. T. (2001). A functional role for Tsix transcription in blocking Xist RNA accumulation but not in X-chromosome choice. Proc. Natl. Acad. Sci. U.S.A. 98, 10232–10237. doi: 10.1073/pnas.171243598
Strecker, J., Gupta, G. D., Zhang, W., Bashkurov, M., Landry, M. C., Pelletier, L., et al. (2016). DNA damage signalling targets the kinetochore to promote chromatin mobility. Nat. Cell Biol. 18, 281–290. doi: 10.1038/ncb3308
Strzelecka, M., Oates, A. C., and Neugebauer, K. M. (2010). Dynamic control of Cajal body number during zebrafish embryogenesis. Nucleus 1, 96–108. doi: 10.4161/nucl.1.1.10680
Sunwoo, H., Dinger, M. E., Wilusz, J. E., Amaral, P. P., Mattick, J. S., and Spector, D. L. (2009). MEN epsilon/beta nuclear-retained non-coding RNAs are up-regulated upon muscle differentiation and are essential components of paraspeckles. Genome Res. 19, 347–359. doi: 10.1101/gr.087775.108
Swanson, E. C., Manning, B., Zhang, H., and Lawrence, J. B. (2013). Higher-order unfolding of satellite heterochromatin is a consistent and early event in cell senescence. J. Cell Biol. 203, 929–942. doi: 10.1083/jcb.201306073
Taddei, A., Van Houwe, G., Hediger, F., Kalck, V., Cubizolles, F., Schober, H., et al. (2006). Nuclear pore association confers optimal expression levels for an inducible yeast gene. Nature 441, 774–778. doi: 10.1038/nature04845
Tam, O. H., Ostrow, L. W., and Gale Hammell, M. (2019). Diseases of the nERVous system: retrotransposon activity in neurodegenerative disease. Mob DNA 10:32. doi: 10.1186/s13100-019-0176-1
Terranova, R., Yokobayashi, S., Stadler, M. B., Otte, A. P., van Lohuizen, M., Orkin, S. H., et al. (2008). Polycomb group proteins Ezh2 and Rnf2 direct genomic contraction and imprinted repression in early mouse embryos. Dev. Cell 15, 668–679. doi: 10.1016/j.devcel.2008.08.015
Therizols, P., Fairhead, C., Cabal, G. G., Genovesio, A., Olivo-Marin, J. C., Dujon, B., et al. (2006). Telomere tethering at the nuclear periphery is essential for efficient DNA double strand break repair in subtelomeric region. J. Cell Biol. 172, 189–199. doi: 10.1083/jcb.200505159
Ting, D. T., Lipson, D., Paul, S., Brannigan, B. W., Akhavanfard, S., Coffman, E. J., et al. (2011). Aberrant overexpression of satellite repeats in pancreatic and other epithelial cancers. Science 331, 593–596. doi: 10.1126/science.1200801
Trapitz, P., Wlaschek, M., and Bunemann, H. (1988). Structure and function of Y chromosomal DNA. II. Analysis of lampbrush loop associated transcripts in nuclei of primary spermatocytes of Drosophila hydei by in situ hybridization using asymmetric RNA probes of four different families of repetitive DNA. Chromosoma 96, 159–170. doi: 10.1007/bf00331048
Ulianov, S. V., Doronin, S. A., Khrameeva, E. E., Kos, P. I., Luzhin, A. V., Starikov, S. S., et al. (2019). Nuclear lamina integrity is required for proper spatial organization of chromatin in Drosophila. Nat. Commun. 10, 1176. doi: 10.1038/s41467-019-09185-y
Umlauf, D., Goto, Y., Cao, R., Cerqueira, F., Wagschal, A., Zhang, Y., et al. (2004). Imprinting along the Kcnq1 domain on mouse chromosome 7 involves repressive histone methylation and recruitment of Polycomb group complexes. Nat. Genet. 36, 1296–1300. doi: 10.1038/ng1467
Valente, F. M., Sparago, A., Freschi, A., Hill-Harfe, K., Maas, S. M., Frints, S. G. M., et al. (2019). Transcription alterations of KCNQ1 associated with imprinted methylation defects in the Beckwith-Wiedemann locus. Genet. Med. 21, 1808–1820. doi: 10.1038/s41436-018-0416-7
van Bemmel, J. G., Pagie, L., Braunschweig, U., Brugman, W., Meuleman, W., Kerkhoven, R. M., et al. (2010). The insulator protein SU(HW) fine-tunes nuclear lamina interactions of the Drosophila genome. PLoS One 5:e15013. doi: 10.1371/journal.pone.0015013
Velazquez Camacho, O., Galan, C., Swist-Rosowska, K., Ching, R., Gamalinda, M., Karabiber, F., et al. (2017). Major satellite repeat RNA stabilize heterochromatin retention of Suv39h enzymes by RNA-nucleosome association and RNA:DNA hybrid formation. eLife 6:e25293. doi: 10.7554/eLife.25293
Verma-Gaur, J., Torkamani, A., Schaffer, L., Head, S. R., Schork, N. J., and Feeney, A. J. (2012). Noncoding transcription within the Igh distal V(H) region at PAIR elements affects the 3D structure of the Igh locus in pro-B cells. Proc. Natl. Acad. Sci. U.S.A. 109, 17004–17009. doi: 10.1073/pnas.1208398109
Verona, R. I., Mann, M. R., and Bartolomei, M. S. (2003). Genomic imprinting: intricacies of epigenetic regulation in clusters. Annu. Rev. Cell Dev. Biol. 19, 237–259. doi: 10.1146/annurev.cellbio.19.111401.092717
Vian, L., Pekowska, A., Rao, S. S. P., Kieffer-Kwon, K. R., Jung, S., Baranello, L., et al. (2018). The energetics and physiological impact of cohesin extrusion. Cell 173, 1165.e20–1178.e20. doi: 10.1016/j.cell.2018.03.072
Volpe, T. A., Kidner, C., Hall, I. M., Teng, G., Grewal, S. I., and Martienssen, R. A. (2002). Regulation of heterochromatic silencing and histone H3 lysine-9 methylation by RNAi. Science 297, 1833–1837. doi: 10.1126/science.1074973
Wanat, J. J., Logsdon, G. A., Driskill, J. H., Deng, Z., Lieberman, P. M., and Johnson, F. B. (2018). TERRA and the histone methyltransferase Dot1 cooperate to regulate senescence in budding yeast. PLoS One 13:e0195698. doi: 10.1371/journal.pone.0195698
Wang, F., Tang, Z., Shao, H., Guo, J., Tan, T., Dong, Y., et al. (2018). Long noncoding RNA HOTTIP cooperates with CCCTC-binding factor to coordinate HOXA gene expression. Biochem. Biophys. Res. Commun. 500, 852–859. doi: 10.1016/j.bbrc.2018.04.173
Wang, H., Xu, X., Nguyen, C. M., Liu, Y., Gao, Y., Lin, X., et al. (2018). CRISPR-mediated programmable 3D genome positioning and nuclear organization. Cell 175, 1405.e14–1417.e14. doi: 10.1016/j.cell.2018.09.013
Wang, K. C., Yang, Y. W., Liu, B., Sanyal, A., Corces-Zimmerman, R., Chen, Y., et al. (2011). A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression. Nature 472, 120–124. doi: 10.1038/nature09819
Wang, Q., Sawyer, I. A., Sung, M. H., Sturgill, D., Shevtsov, S. P., Pegoraro, G., et al. (2016). Cajal bodies are linked to genome conformation. Nat. Commun. 7:10966. doi: 10.1038/ncomms10966
Wen, B., Wu, H., Shinkai, Y., Irizarry, R. A., and Feinberg, A. P. (2009). Large histone H3 lysine 9 dimethylated chromatin blocks distinguish differentiated from embryonic stem cells. Nat. Genet. 41, 246–250. doi: 10.1038/ng.297
White, R. J. (2005). RNA polymerases I and III, growth control and cancer. Nat. Rev. Mol. Cell Biol. 6, 69–78. doi: 10.1038/nrm1551
Wong, L. H., Brettingham-Moore, K. H., Chan, L., Quach, J. M., Anderson, M. A., Northrop, E. L., et al. (2007). Centromere RNA is a key component for the assembly of nucleoproteins at the nucleolus and centromere. Genome Res. 17, 1146–1160. doi: 10.1101/gr.6022807
Yancopoulos, G. D., and Alt, F. W. (1985). Developmentally controlled and tissue-specific expression of unrearranged VH gene segments. Cell 40, 271–281. doi: 10.1016/0092-8674(85)90141-2
Yang, F., Deng, X., Ma, W., Berletch, J. B., Rabaia, N., Wei, G., et al. (2015). The lncRNA Firre anchors the inactive X chromosome to the nucleolus by binding CTCF and maintains H3K27me3 methylation. Genome Biol. 16:52. doi: 10.1186/s13059-015-0618-0
Yang, L., Lin, C., Liu, W., Zhang, J., Ohgi, K. A., Grinstein, J. D., et al. (2011). ncRNA- and Pc2 methylation-dependent gene relocation between nuclear structures mediates gene activation programs. Cell 147, 773–788. doi: 10.1016/j.cell.2011.08.054
Yoshioka, M., Yorifuji, T., and Mituyoshi, I. (1998). Skewed X inactivation in manifesting carriers of Duchenne muscular dystrophy. Clin. Genet. 53, 102–107. doi: 10.1111/j.1399-0004.1998.tb02655.x
Zhang, H., Zeitz, M. J., Wang, H., Niu, B., Ge, S., Li, W., et al. (2014). Long noncoding RNA-mediated intrachromosomal interactions promote imprinting at the Kcnq1 locus. J. Cell Biol. 204, 61–75. doi: 10.1083/jcb.201304152
Zhang, L. F., Huynh, K. D., and Lee, J. T. (2007). Perinucleolar targeting of the inactive X during S phase: evidence for a role in the maintenance of silencing. Cell 129, 693–706. doi: 10.1016/j.cell.2007.03.036
Zhao, Z., Dammert, M. A., Grummt, I., and Bierhoff, H. (2016). lncRNA-induced nucleosome repositioning reinforces transcriptional repression of rRNA genes upon hypotonic stress. Cell Rep. 14, 1876–1882. doi: 10.1016/j.celrep.2016.01.073
Zhao, Z., Senturk, N., Song, C., and Grummt, I. (2018). lncRNA PAPAS tethered to the rDNA enhancer recruits hypophosphorylated CHD4/NuRD to repress rRNA synthesis at elevated temperatures. Genes Dev. 32, 836–848. doi: 10.1101/gad.311688.118
Zhu, L., and Brangwynne, C. P. (2015). Nuclear bodies: the emerging biophysics of nucleoplasmic phases. Curr. Opin. Cell Biol. 34, 23–30. doi: 10.1016/j.ceb.2015.04.003
Keywords: nuclear organization, non-coding RNA, DNA repeats, nucleolus, Cajal bodies (CBs)
Citation: Khosraviani N, Ostrowski LA and Mekhail K (2019) Roles for Non-coding RNAs in Spatial Genome Organization. Front. Cell Dev. Biol. 7:336. doi: 10.3389/fcell.2019.00336
Received: 10 October 2019; Accepted: 29 November 2019;
Published: 19 December 2019.
Edited by:
Patrizio Dimitri, Sapienza University of Rome, ItalyReviewed by:
Igor V. Sharakhov, Virginia Tech, United StatesMaurizio D’Esposito, Italian National Research Council (CNR), Italy
Copyright © 2019 Khosraviani, Ostrowski and Mekhail. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Karim Mekhail, a2FyaW0ubWVraGFpbEB1dG9yb250by5jYQ==
†ORCID: Negin Khosraviani, orcid.org/0000-0003-0588-3618; Lauren A. Ostrowski, orcid.org/0000-0001-6922-6196
 Negin Khosraviani
Negin Khosraviani Lauren A. Ostrowski
Lauren A. Ostrowski Karim Mekhail
Karim Mekhail