- Department of Molecular, Cell and Cancer Biology, University of Massachusetts Medical School, Worcester, MA, United States
RLIM/Rnf12 is an E3 ubiquitin ligase that has originally been identified as a transcriptional cofactor associated with LIM domain transcription factors. Indeed, this protein modulates transcriptional activities and multiprotein complexes recruited by several classes of transcription factors thereby enhancing or repressing transcription. Around 10 years ago, RLIM/Rnf12 has been identified as a major regulator for the process of X chromosome inactivation (XCI), the transcriptional silencing of one of the two X chromosomes in female mice and ESCs. However, the precise roles of RLIM during XCI have been controversial. Here, we discuss the cellular and developmental functions of RLIM as an E3 ubiquitin ligase and its roles during XCI in conjunction with its target protein Rex1.
Introduction
X dosage compensation in female mice occurs early during embryogenesis in two waves. An early, imprinted form of XCI (iXCI), which silences exclusively the paternally inherited X (Xp), directly follows zygotic genome activation (ZGA) at the end of the 2-cell stage. While this pattern of XCI is maintained in extraembryonic tissues including trophoblast and primitive endoderm, epiblast cells which give rise to the embryo proper reactivate the Xp (XCR) and undergo a random form of XCI (rXCI) around implantation (Payer, 2016). The long non-coding (lnc) RNA Xist plays crucial roles during both forms of XCI and paints the X from which it is expressed (Brown et al., 1992; Penny et al., 1996). Initiation of Xist transcription is considered the onset of XCI and an early phase of continued Xist expression is required for the maintenance of the XCI state (Wutz and Jaenisch, 2000). Prior to upregulation of Xist during rXCI, which is mostly investigated in female ESC models, both X chromosomes transiently move into spatial proximity, a process known as X pairing (Bacher et al., 2006; Xu et al., 2006; Augui et al., 2007). Xist transcription is inhibited in trans by pluripotency transcription factors including Rex1 (Navarro et al., 2010) and in cis by the lnc RNA Tsix, which is transcribed antisense to Xist (Lee et al., 1999; Lee, 2000).
The Rlim gene (also known as Rnf12) encodes a RING finger ubiquitin ligase (E3) (Ostendorff et al., 2002). During mouse development Rlim mRNA is widely expressed, while RLIM protein expression is more restricted in cell types and tissues (Ostendorff et al., 2006). In cells, RLIM protein shuttles between the nucleus and cytoplasm in a phosphorylation-dependent manner (Jiao et al., 2013) but in most cell types, RLIM protein is detected in the nucleus, where many of its substrate proteins reside that include transcription factors and transcriptional co-regulators. Indeed, RLIM is involved in regulating the dynamics of DNA-bound multiprotein complexes in promoters/enhancers
(Ostendorff et al., 2002; Güngör et al., 2007; Johnsen et al., 2009). RLIM can self-ubiquitinate and mutations of the RING finger results in gain-of function activities and stabilization of the mutated protein (Ostendorff et al., 2002).
Important in vivo functions of Rlim have been discovered in female mice. In mammary glands of pregnant and lactating females, RLIM serves as a survival factor specifically for milk-producing alveolar cells (Jiao et al., 2012, 2013). Moreover, Rlim/Rnf12 has been identified as a major activator of XCI in female ESCs (Jonkers et al., 2009) and required for iXCI in female mice (Shin et al., 2010). In an ESC model RLIM interacts with Rex1/Zfp42, a transcriptional repressor of Xist, leading to proteasomal degradation and initiation of XCI (Gontan et al., 2012). However, Rlim was found dispensable for rXCI in epiblast tissues and in other ESC model systems (Shin et al., 2010, 2014), and thus over the last years there was much confusion on the roles and importance of Rlim during rXCI. Recent work has identified Rex1 as the critical Rlim target during iXCI and directly compared XCI in various ESC model systems (Wang et al., 2017; Gontan et al., 2018). Results illuminate major roles of Rlim in conjunction with Rex1 during XCI in nuclei of cells, thereby partially clarifying the existing controversy.
RLIM as a RING Finger E3 Ubiquitin Ligase
RLIM/Rnf12 was first identified as an antigen recognized by autologous antibodies of renal cancer patients (Scanlan et al., 1999) and as a cofactor negatively affecting the transcriptional and developmental activity of LIM homeodomain transcription factors (Bach et al., 1999). The gene Rlim maps to the X chromosome and is conserved from humans to chick (Ostendorff et al., 2000). RLIM protein in mice encompasses 600 amino acids (Figure 1A) and contains several conserved domains, including nuclear localization and export sequences (NLS and NES, respectively), a centrally located basic domain (BD) and a C-terminal RING-H2 zinc finger domain. Indeed, both the NLS and the NES are functional and rapid nucleocytoplasmic shuttling of the RLIM protein in a phosphorylation-dependent manner has been demonstrated (Jiao et al., 2013), even though in most cell types RLIM protein is detected predominantly in the nuclear compartment. The C-terminal RING H2 zinc finger motif identifies RLIM as an E3 ubiquitin protein ligase. RING finger E3 ligases are part of the ubiquitin proteasome system (UPS) involving an E1 activating enzyme, E2 ubiquitin-conjugating enzymes and E3 ubiquitin ligases which, by recognizing the substrate proteins, confer the specificity of the ubiquitination reaction (Metzger et al., 2014). Ubiquitinated proteins are often targeted for degradation by the 26S proteasome but this depends on mono-, multi- or poly-ubiquitination chains and the chain linkage type (Hicke, 2001; Pickart, 2001; Ikeda and Dikic, 2008; Metzger et al., 2014). Unlike the cullin subclass of RING finger E3s (Petroski and Deshaies, 2005), RLIM belongs to a subclass of RING finger E3s in which substrate recognition and ubiquitination ligase functions are mediated by the same protein (Metzger et al., 2012), and many substrate proteins of RLIM are recognized and bound by the Basic Domain (Figure 1A). As RLIM locates mostly to the nucleus, it is not surprising that substrates comprise mainly nuclear proteins, including LIM-only (LMO) and CLIM/Ldb cofactors (Ostendorff et al., 2002; Hiratani et al., 2003), the histone de-acetylase HDAC2 (Kramer et al., 2003), the telomeric protein TRF1 (Her and Chung, 2009), the oncoprotein Stathmin (Chen et al., 2014), estrogen receptor alpha (ERalpha) (Johnsen et al., 2009), the TFIIIB subunit BRF1 (Wang et al., 2019) and SMAD7 involved in TGFbeta signaling (Zhang et al., 2012). Due to its E3 ligase activity, RLIM regulates cellular levels of transcriptional co-regulators and is involved in the dynamics of multiprotein complexes on DNA in promoters/enhancers (Ostendorff et al., 2002; Güngör et al., 2007; Johnsen et al., 2009). Importantly, RLIM also targets Rex1 for proteasomal degradation (Gontan et al., 2012), a pluripotency factor expressed in early embryos (Climent et al., 2013) that acts as a transcriptional repressor of Xist. RLIM is able to self-ubiquitinate (Ostendorff et al., 2002), a hallmark of RING finger E3s, which likely contributes to its reported short half-life (Becker et al., 2003). Moreover, RLIM targets another RING E3 ligase Mdm2 for degradation (Gao et al., 2016) and is the target of the RING E3 ligase SIAH1/2 (Kramer et al., 2008). Indeed, as part of regulatory networks, both self-ubiquitination of RING finger E3s and ubiquitination of RING E3s by heterologous E3s frequently occurs in vivo (Weissman et al., 2011). These activities likely contribute to the observed differences in the widespread developmental Rlim mRNA expression, and the more restricted RLIM protein expression (Ostendorff et al., 2006).
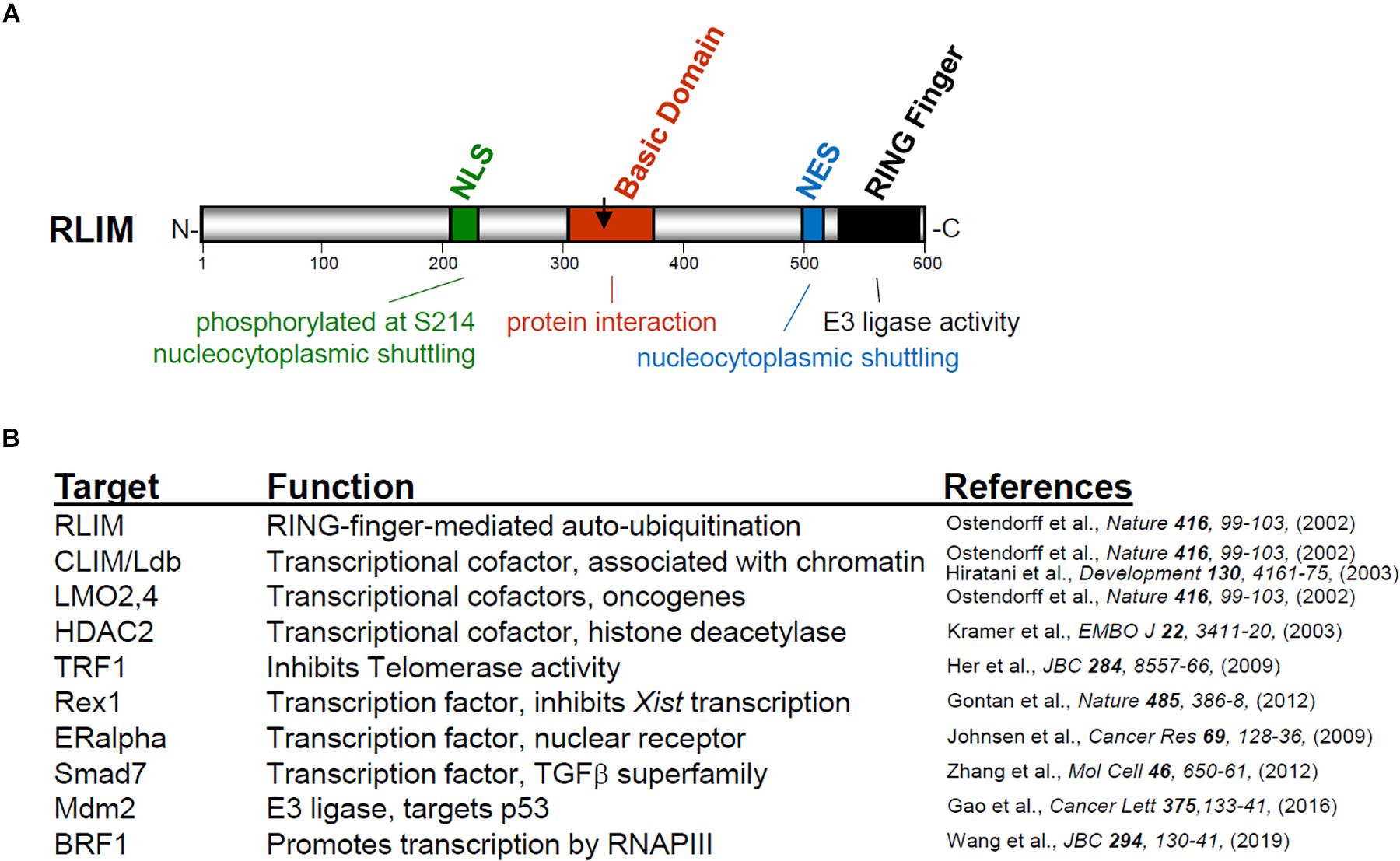
Figure 1. (A) Structure of RLIM protein indicating conserved domains, their functions and amino acid position. The arrow indicates the C-terminus of RLIM333. (B) Reported Rlim target proteins for ubiquitination. Note that most proteins are nuclear.
The activity of E3 ligases toward a specific substrate protein is influenced by many factors including the correct cellular location and enzyme stability (Weissman et al., 2011). Moreover, E3 activity is regulated via post-translational modifications both at the level of substrate or E3 enzyme (Metzger et al., 2012, 2014). Thus, mutations in many different domains within an E3 enzyme may ultimately influence levels of target proteins. Because instability domains in substrate proteins are generally recognized by multiple E3 ligases, mutations in the RING finger often lead to dominant-negative or gain-of-function effects, inhibiting the actions of the full lengths enzyme and/or other E3s, depending on the cellular context (Bach and Ostendorff, 2003). Indeed, forced expression of RING finger mutated RLIM in cells inhibits the functions of endogenous RLIM and likely other E3 ligases, leading to dramatically increased cellular levels of target proteins CLIM and HDAC2 (Ostendorff et al., 2002; Kramer et al., 2003), while levels of these proteins in RlimKO cells only increase marginally (Shin et al., 2010).
Not surprisingly, mutations in genes encoding E3 ligases are often associated with human diseases mostly due to mis-regulation of substrate protein levels (Ciechanover and Schwartz, 2004), and mutations in Rlim have been associated with intellectual disability and autism (Tonne et al., 2015; Hu et al., 2016). However, while evidence suggests altered neuronal differentiation in mouse male ESCs lacking Rlim/Rnf12 (Bustos et al., 2018), CNS development in mice lacking Rlim appears normal and thus, it is unclear at this stage whether human disability phenotypes are caused by the lack of functional RLIM protein, gain-of-function activity acquired by the mutated RLIM or a combination of both. Male mice systemically lacking Rlim are born at Mendelian ratios, appear healthy and grow up to be fertile (Shin et al., 2010, 2014; Wang et al., 2016) and even though minor functions cannot be excluded, these results indicate no general, major developmental function of Rlim in male mice. However, during female embryogenesis, major roles of Rlim have been identified in conjunction with Rex1 during X dosage compensation.
Rlim/Rnf12, Rex1 and rXCI
In an attempt to validate a stochastic model of rXCI that postulates the existence of an X-linked activator of XCI (Monkhorst et al., 2008), Rlim/Rnf12 was identified as an activator of XCI (Jonkers et al., 2009). Using the established F121 female ESC line, a KO ESC line was generated (Jonkers et al., 2009; Barakat et al., 2011), hereafter referred to as Rnf12KO. Indeed, upon differentiation, Rnf12KO ESCs failed to activate Xist transcription and initiate XCI, and in corresponding heterozygous ESCs rXCI was skewed toward inactivation of the KO allele (Jonkers et al., 2009; Barakat et al., 2011). Moreover, data from an Rnf12KO derived ESC line indicated that X-pairing was not needed for XCI (Barakat et al., 2014). Based on these results a model was proposed in which the dose of RLIM expressed from two alleles was required to trigger rXCI (Barakat et al., 2011). However, ESCs lacking Rlim, which were isolated from an independent mouse model containing a conditional Rlim KO allele (referred to as RlimKO) developed Xist clouds and initiated XCI upon differentiation in vitro and in vivo, and in tetraploid complementation experiments these ESCs were able to undergo rXCI and form post-gastrulation embryos (Shin et al., 2010, 2014). rXCI in the absence of Rlim in mice was supported by targeting the cKO of Rlim specifically in epiblasts via Sox2-Cre. Moreover, RLIM protein levels in WT embryos are downregulated specifically in differentiating epiblast cells at the timepoint of X-reactivation, shortly before the onset of rXCI (Shin et al., 2014). However, in contrast to differentiating epiblast cells, RLIM downregulation does not occur in ESCs differentiated in vitro (Wang et al., 2017). Thus, while Rlim is dispensable for rXCI in vivo and in female ESCs freshly isolated from embryos, it was required in the female F121 ESCs.
The identification of the pluripotency factor Rex1/Zfp42 as RLIM target (Gontan et al., 2012) has proven a major step in XCI regulation. Indeed, RLIM strongly interacts with and polyubiquitinates Rex1 in female ESCs leading to targeted proteasomal degradation. Rex1 acts as a potent repressor of Xist transcription both by repressing Xist directly in trans and in cis by activating the lncTsix (Gontan et al., 2012). Thus, the proteasomal targeting of Rex1 has provided a mechanism for Rlim’s positive action on XCI.
Two recent papers have provided much evidence not only that the dosage of nuclear Rex1 is critical in regulating the XCI process but have also shed light on the ESC controversy. One study generated various CRISPR/Cas9 mediated ESCs carrying an Rlim deletion in a Pgk12.1 (Norris et al., 1994) female ESC background (RlimKOp) (Wang et al., 2017). These cells were able to undergo XCI at much higher rates of efficiency when directly compared with Rnf12KO ESCs. It was discovered that the Rnf12KO did not represent a full knockout but that a 45kD truncated RLIM protein consisting of 333 amino acids (referred to as RLIM333) is expressed from the “KO” Rlim locus (Wang et al., 2017). As RLIM333 contains the NLS and a partial BD but lacks the NES and the RING finger (see Figure 1A), this protein can no longer shuttle but is trapped in the nucleus and consistent with the fact that Rex1 interacts with the BD, forced expression of RLIM333 in ESCs (RlimKO or WT) leads to nuclear accumulation of Rex1, indicating gain-of-function activity. Moreover, while Rnf12KO ESCs (Rlim333f) express Rex1 in the nucleus, cell derivatives with a CRISPR/Cas9-mediated deletion of RLIM333 (Rlim0f, in F121 background) are XCI competent at least to some degree and displayed mostly cytoplasmic Rex1 (Supplementary Figure 1). Together, these results indicate a direct connection between nuclear accumulation of Rex1 and the presence of RLIM333 and show that lack of Rlim in ESCs does not necessarily lead to nuclear accumulation of Rex1 (Wang et al., 2017). However, in a newly generated ESC line (in F121) that lacks the entire Rlim ORF (referred to as Rnf12CR–/CR–) and is unable to undergo XCI, XCI activity was restored upon additional deletion of Rex1 (Gontan et al., 2018). In contrast to ESCs examined in the first study, however, Rex1 accumulated in nuclei of Rnf12CR–/CR– cells, presumably due to lack of E3 ligase activity (Gontan et al., 2018). These results are important as they demonstrate that in Rnf12KO it is Rex1 activity that prevents these cells from undergoing XCI. Thus, in various female ESC lines nuclear Rex1 accumulation is caused by the presence of RLIM333 or the lack of Rlim, while the latter does not affect Rex1 in all ESC lines.
The observed differences in Rex1 distribution in various ESCs models suggests the existence of additional pathways regulating both levels and subcellular localization of Rex1 that might include mechanisms affecting RLIM’s E3 ligase activity and/or differences in cellular competence factor repertoire. In this context it is important to mention that various ESC lines WT for Rlim display very different endogenous levels of Rex1, with Pgk12.1 ESCs expressing low but F121 and the male E14 (Hooper et al., 1987) lines much higher levels of Rex1, and at least endogenous Rex1 levels and cellular localization in Pgk12.1 are Rlim-independent (Wang et al., 2017). Moreover, ESC culture conditions may play an important role as physiological O2 levels improved XCI efficiency in RlimKO ESC lines (Wang et al., 2017). A powerful Rlim-independent cellular pathway regulating Rex1 is induced upon ESC differentiation, when Rex1 protein rapidly drops to undetectable levels even in the absence of Rlim (Gontan et al., 2012; Wang et al., 2017). Thus, one common feature among various ESC lines lacking Rlim is that nuclear exclusion of Rex1 strongly correlates with their ability to undergo XCI in vitro.
Roles of Rlim/Rnf12 During iXCI
In contrast to rXCI, there is little controversy about the major role Rlim plays during iXCI. Indeed, female mice receiving a maternally transmitted Rlim KO allele (KOm) die (Shin et al., 2010) and not a single female receiving a KOm allele has been born in over 10 years of breeding. In contrast, females receiving a paternally transmitted KO allele (KOp) are born at Mendelian ratios and appear to undergo normal XCI. This parent-of-origin effect is a consequence of the fact that iXCI silences exclusively the Xp and the Rlim gene is X-linked. KOm females die around implantation due to defective development of trophoblast tissues caused by iXCI failure (Shin et al., 2010).
Concerning the precise function during iXCI, a crucial role of Rlim in the maintenance of iXCI at blastocyst stages appears clear. This role was genetically established by inducing the Rlim cKO in embryos after ZGA via a paternally transmitted Rosa26-Cre transgene as well as by data obtained from RNA-seq experiments following expression profiles of X-linked genes and Xist transcription in single embryos across pre-implantation stages comparing RlimKO with WT embryos (Figures 2A,B; Wang et al., 2016). Another mouse model that targets the Rlim gene has recently been generated based on Rnf12KO ESCs. Like RlimKO mice, females receiving a maternal Rnf12KO allele do not undergo iXCI leading to defective trophoblast development (Gontan et al., 2018). Strikingly, iXCI is rescued in female mice containing a Rnf12/Rex1 double KO, establishing a genetic link between these two genes during iXCI. This result is exciting as it identifies Rex1 as the critical target of Rlim during iXCI in vivo. Moreover, Rex1 protein accumulates in nuclei of Rnf12KO blastocysts (Gontan et al., 2018). Indeed, in WT animals, Rex1 mRNA levels increase dramatically from 4 cell staged embryos to early blastocyst stages (Figure 2C; Wang et al., 2016), and Rex1 protein is detected in the cytoplasm of cells throughout mouse pre-implantation development, while it is undetectable in nuclei of cells in WT embryos up to morula stages (Climent et al., 2013). These data emphasize the critical role that Rlim plays in the nuclear exclusion of Rex1 during iXCI.
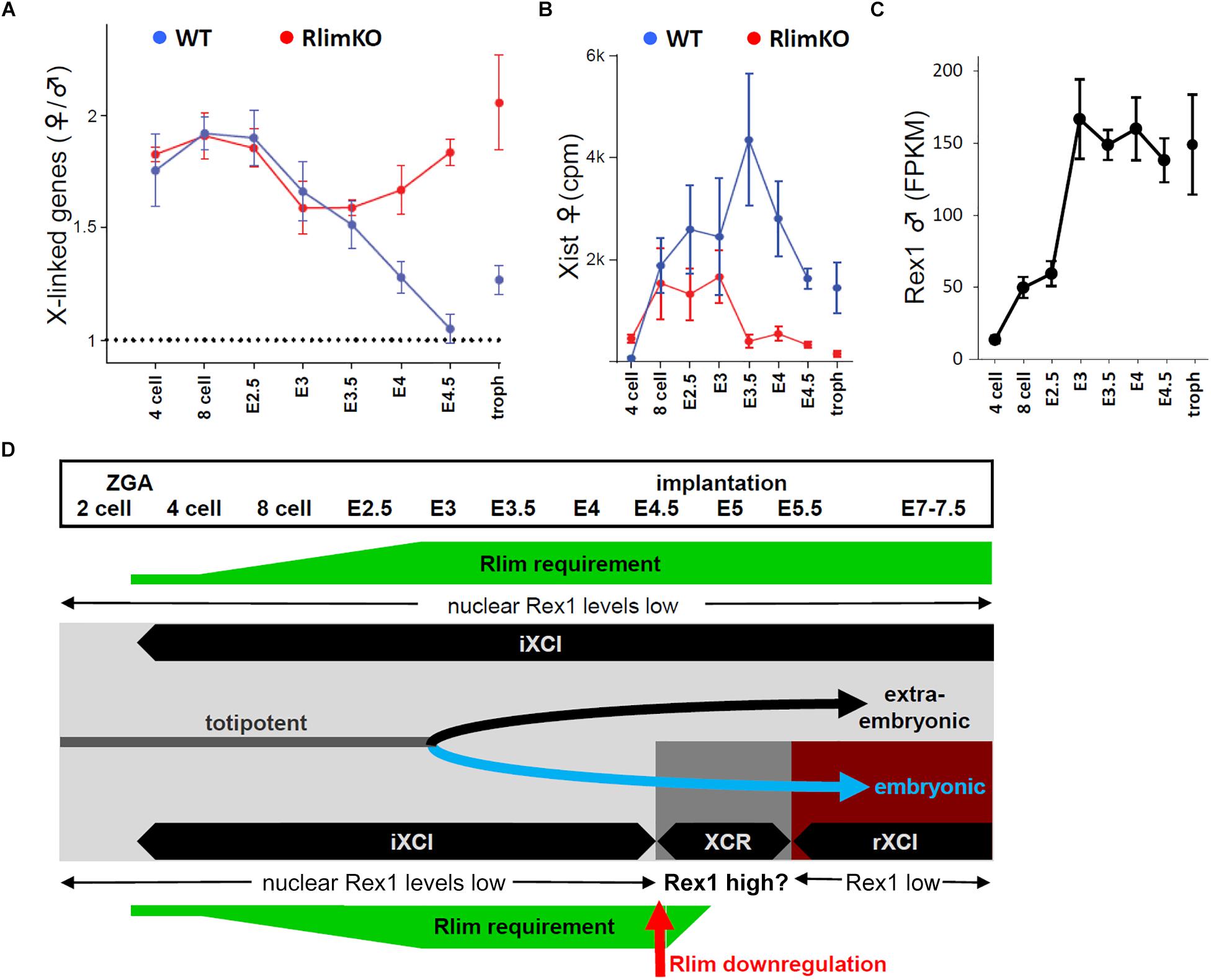
Figure 2. (A–C) Comparisons of gene expression profiles during preimplantation development as elucidated by single embryo RNA-seq. All data shown originate from the same dataset (RlimKO; Wang et al., 2017). Shown are comparisons of total X-linked gene expression in female RlimKO and WT embryos, normalized to males (A), Xist RNA expression in RlimKO and WT females (B), and Rex1 expression in males (C). Note that once Rex1 reaches high levels at embryonic stage E3, Xist levels in RlimKO females drop dramatically at E3.5 halting/reversing the process of X dosage compensation. (D) Summary of XCI during female mouse embryogenesis. The requirement of Rlim (green) and its effect on nuclear Rex 1 protein levels are shown. Embryonic stages, iXCI, rXCI and XCR as well as cell lineages are indicated (gray, totipotent cell lineage; black, extraembryonic cell lineages; blue, embryonic/epiblast cell lineage).
As for the role of Rlim in initiating Xist expression and iXCI, the results are less clear-cut. At early pre-implantation stages single embryo RNA-seq data indicate that the process of X dosage compensation initiates at least to some degree in females lacking Rlim but is then interrupted at early blastocyst stages (Figures 2A–C; Wang et al., 2016), when Rex1 levels increase. Formation of Xist clouds in a significant number of cells up to E2.5 was confirmed by RNA-FISH in RlimKO females. Importantly, the RlimKO embryos were generated using mothers systemically lacking Rlim including in their germline (Wang et al., 2016). Because the profiles of X-linked gene expression in RlimKO embryos mirror those observed in WT females at early pre-implantation stages (Figure 2A), this result argues against influences of maternally transmitted Rex1 which, like Rlim is expressed in oocytes (Climent et al., 2013), on initiation of iXCI in next generation embryos. However, when compared to WT, the number of cells lacking a Xist cloud is diminished in RlimKO females at all timepoints investigated (Shin et al., 2010; Wang et al., 2016). Therefore, while iXCI initiation is not absolutely dependent on Rlim at least in the C57Bl/6 genetic background, its presence likely promotes this process.
It is important to point out here that in the Rnf12KO system, embryos are expected to express the RLIM333 protein in all cells at pre-implantation stages, but this has not been examined. Because effects of RLIM333 expression and Rlim deletion on Rex1 are partially overlapping, additional repressive contributions of the truncated Rlim protein on the iXCI process cannot be excluded, and iXCI in Rnf12KO animals appears more severely inhibited when compared to RlimKO animals, especially during iXCI initiation (Gontan et al., 2018). Thus, while the Rex1KO in Rnf12KO mice removes the effects on XCI induced by both lack of Rlim and the presence of RLIM333, this model cannot distinguish what caused the nuclear accumulation of Rex1 protein.
Discussion
The discovery of Rex1 as the crucial RLIM target during iXCI represents a major finding, assigning critical roles for the interplay of both genes in controlling the process of X dosage compensation in mice, in particular for iXCI and possibly XCR in epiblast cells. A general theme that emerges is that the major role of Rlim during XCI is to regulate nuclear Rex1 levels in order to enable Xist transcription. This is supported by genetic data in mice, Rex1 mRNA and protein expression during pre-implantation development and some data from ESC model systems. Incorporating these data in a previously formulated model (Wang et al., 2017) (see also Figure 2D), X dosage compensation in mice might be carried out in a scenario similar to that outlined below: Initiation of Xist transcription at the end of 2-/beginning of 4 cell-stage likely occurs under low levels of embryonic Rex1 (Figure 2C) and at least in part independent of Rlim. As Rex1 mRNA levels increase during pre-implantation development, the function/requirement of Rlim becomes more important to keep Rex1 protein excluded from nuclei of cells to ensure the continuation/maintenance of Xist transcription and the iXCI process. Thus, when Rlim is downregulated specifically in epiblasts of late blastocysts, it is likely that high Rex1 mRNA levels translate into rapid nuclear accumulation of Rex1 protein. Because Rex1 represses Xist but activates Tsix transcription, this spike in nuclear Rex1 likely facilitates the XCR process and/or contributes to an introduction/establishment of Xist regulation in cis via Tsix during rXCI in vivo. However, to ensure rXCI, Rex1 needs to be eliminated from nuclei of differentiating epiblast cells in a Rlim-independent manner, to allow for initiation of Xist transcription. The mechanisms for this are unclear but it is tempting to speculate the involvement of a pathway that is activated upon epiblast differentiation, partially mimicked in ESCs by LIF removal/embryoid body formation. Xist activation and rXCI in differentiating epiblast cells would then occur in the absence of both RLIM and Rex1. Conceptually, such a model shifts the critical dose dependency of the XCI process from Rlim to nuclear Rex1, as during iXCI Rlim is transcribed mostly from a single allele.
Concerning initiation of iXCI, it is important to point out that both RLIM and Rex1 proteins are detectable in the female germline (Shin et al., 2010; Climent et al., 2013, Wang et al., 2016). However, while initiation of iXCI in embryos occurs at least to some degree in the C57Bl/6 genetic background, this likely depends on maternal Rex1 levels, which might vary in individual females or in females of other genetic backgrounds. Thus, in early female embryos RLIM may serve as a safeguard of iXCI, protecting against maternal Rex1. Indeed, the ability of RLIM to target DNA-bound proteins (Ostendorff et al., 2002) is predicted to efficiently counteract repressive functions of both maternally transmitted and embryonic Rex1 on Xist regulatory sequences.
Concerning XCI in various female ESC models, the ability of female ESCs to undergo XCI correlates with low nuclear levels of Rex1. However, nuclear exclusion and overall cellular levels of Rex1 is controlled by RLIM in some ESC lines, but not in others, indicating the existence of alternative pathway(s) that may be sensitive to culture and/or differentiation conditions and possibly also involving multiple mechanisms on RLIM regulation such as E3 ligase activity and differences in the epigenetic make-up selected via clonal line selection. Moreover, various ESC lines might represent different developmental states with possibly leaky suppression of Rlim-independent pathway(s) controlling Rex1, thereby explaining different baseline levels and localization of Rex1 protein in cells. Thus, while RLIM appears the only factor controlling nuclear exclusion of Rex1 at early pre-implantation stages, in ESCs Rlim represents only one of multiple pathways. These data suggest that the dynamics of both Rlim and Rex1 regulation during mouse development is not always paralleled in ESCs in culture, and more work is needed to elucidate the cellular regulation of Rex1 protein.
Concerning RLIM333, the presence of this truncated protein in Rnf12KO-derived systems is based on a cloning artifact. RLIM333 causes nuclear accumulation of Rex1, an effect overlapping with that of the RlimKO and thus, this gain-of-function activity will be eliminated by an additional Rex1KO. However, because RLIM333 lacks specific protein domains, its activity is no longer associated with pathways regulating the full-length protein in terms of expression, stability and E3 ligase activity. In Rnf12KO-derived model systems including mice, there is likely enhanced effects on nuclear Rex1 due to the lack of RLIM and the presence of RLIM333, potentially influencing various aspects of the iXCI process in a temporary, qualitative and quantitative manner, and the effects of RLIM333 on XCI cannot be distinguished from those induced by the lack of Rlim. Outside the Rex1-Rlim axis, RLIM has other E3 ligase targets most of which are also targeted by other E3s. While the KO has little/no effect on such targets, the presence of RLIM333 might, and thus other phenotypes are likely. This is why the reported lethality in male Rnf12KO (Gontan et al., 2018) is concerning as in RlimKO models including germline RlimKO models (Cre-recombinase negative) no lethality has been observed in males lacking RLIM over more than 10 years. Therefore, data obtained by the Rnf12KO model need to be interpreted not only from the KO perspective, but also from a gain-of-function perspective. Because the terms “KO” or “−/−” are incorrect and cause confusion, derivatives of the Rnf12KO ESCs should be renamed to account for the presence of RLIM333.
In summary, recent results have assigned a crucial importance to the Rlim-Rex1 axis in controlling XCI both in vivo and in vitro. Going forward, it will be important to elucidate the functional evolutionary conservation of this module in other species, the dynamics between both factors specifically in epiblast cells during implantation including effects on Xist/Tsix, and to identify factors that mediate rXCI in vivo.
Data Availability Statement
All datasets generated for this study are included in the article/Supplementary Material.
Ethics Statement
The animal study was reviewed and approved by UMMS Institute of Animal Care and Usage Committee.
Author Contributions
Both authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
Funding
This work was supported by NIH grant R01GM128168.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcell.2019.00258/full#supplementary-material
References
Augui, S., Filion, G. J., Huart, S., Nora, E., Guggiari, M., Maresca, M., et al. (2007). Sensing X chromosome pairs before X inactivation via a novel X-pairing region of the Xic. Science 318, 1632–1636. doi: 10.1126/science.1149420
Bach, I., and Ostendorff, H. P. (2003). Orchestrating nuclear functions: ubiquitin sets the rhythm. Trends Biochem. Sci. 28, 189–195. doi: 10.1016/s0968-0004(03)00055-0
Bach, I., Rodriguez-Esteban, C., Carriere, C., Bhushan, A., Krones, A., Rose, D. W., et al. (1999). RLIM inhibits functional activity of LIM homeodomain transcription factors via recruitment of the histone deacetylase complex. Nat. Genet. 22, 394–399. doi: 10.1038/11970
Bacher, C. P., Guggiari, M., Brors, B., Augui, S., Clerc, P., Avner, P., et al. (2006). Transient colocalization of X-inactivation centres accompanies the initiation of X inactivation. Nat. Cell Biol. 8, 293–299. doi: 10.1038/ncb1365
Barakat, T. S., Gunhanlar, N., Pardo, C. G., Achame, E. M., Ghazvini, M., Boers, R., et al. (2011). RNF12 activates Xist and is essential for X chromosome inactivation. PLoS Genet. 7:e1002001. doi: 10.1371/journal.pgen.1002001
Barakat, T. S., Loos, F., van Staveren, S., Myronova, E., Ghazvini, M., Grootegoed, J. A., et al. (2014). The trans-activator RNF12 and cis-acting elements effectuate X chromosome inactivation independent of x-pairing. Mol. Cell. 53, 965–978. doi: 10.1016/j.molcel.2014.02.006
Becker, T., Bossenz, M., Tursun, B., Schluter, A., Peters, M. A., Becker, C. G., et al. (2003). Comparing protein stabilities during zebrafish embryogenesis. Methods Cell Sci. 25, 85–89. doi: 10.1023/b:mics.0000006895.03556.9b
Brown, C. J., Hendrich, B. D., Rupert, J. L., Lafreniere, R. G., Xing, Y., Lawrence, J., et al. (1992). The human XIST gene: analysis of a 17 kb inactive X-specific RNA that contains conserved repeats and is highly localized within the nucleus. Cell 71, 527–542. doi: 10.1016/0092-8674(92)90520-m
Bustos, F., Segarra-Fas, A., Chaugule, V. K., Brandenburg, L., Branigan, E., Toth, R., et al. (2018). RNF12 X-Linked intellectual disability mutations disrupt E3 ligase activity and neural differentiation. Cell Rep. 23, 1599–1611. doi: 10.1016/j.celrep.2018.04.022
Chen, X., Shen, J., Li, X., Wang, X., Long, M., Lin, F., et al. (2014). Rlim, an E3 ubiquitin ligase, influences the stability of stathmin protein in human osteosarcoma cells. Cell. Signal. 26, 1532–1538. doi: 10.1016/j.cellsig.2014.03.018
Ciechanover, A., and Schwartz, A. L. (2004). The ubiquitin system: pathogenesis of human diseases and drug targeting. Biochim. Biophys. Acta 1695, 3–17. doi: 10.1016/j.bbamcr.2004.09.018
Climent, M., Alonso-Martin, S., Perez-Palacios, R., Guallar, D., Benito, A. A., Larraga, A., et al. (2013). Functional analysis of rex1 during preimplantation development. Stem Cells Dev. 22, 459–472. doi: 10.1089/scd.2012.0211
Gao, K., Wang, C., Jin, X., Xiao, J., Zhang, E., Yang, X., et al. (2016). RNF12 promotes p53-dependent cell growth suppression and apoptosis by targeting MDM2 for destruction. Cancer Lett. 375, 133–141. doi: 10.1016/j.canlet.2016.02.013
Gontan, C., Achame, E. M., Demmers, J., Barakat, T. S., Rentmeester, E., van, I. W., et al. (2012). RNF12 initiates X-chromosome inactivation by targeting REX1 for degradation. Nature 485, 386–390. doi: 10.1038/nature11070
Gontan, C., Mira-Bontenbal, H., Magaraki, A., Dupont, C., Barakat, T. S., Rentmeester, E., et al. (2018). REX1 is the critical target of RNF12 in imprinted X chromosome inactivation in mice. Nat. Commun. 9:4752. doi: 10.1038/s41467-018-07060-w
Güngör, C., Taniguchi-Ishigaki, N., Ma, H., Drung, A., Tursun, B., Ostendorff, H. P., et al. (2007). Proteasomal selection of multiprotein complexes recruited by LIM homeodomain transcription factors. Proc. Natl. Acad. Sci. U.S.A. 104, 15000–15005. doi: 10.1073/pnas.0703738104
Her, Y. R., and Chung, I. K. (2009). Ubiquitin ligase RLIM modulates telomere length homeostasis through a proteolysis of TRF1. J. Biol. Chem. 284, 8557–8566. doi: 10.1074/jbc.M806702200
Hicke, L. (2001). Protein regulation by monoubiquitin. Nat. Rev. Mol. Cell Biol. 2, 195–201. doi: 10.1038/35056583
Hiratani, I., Yamamoto, N., Mochizuki, T., Ohmori, S. Y., and Taira, M. (2003). Selective degradation of excess Ldb1 by Rnf12/RLIM confers proper Ldb1 expression levels and Xlim-1/Ldb1 stoichiometry in Xenopus organizer functions. Development 130, 4161–4175. doi: 10.1242/dev.00621
Hooper, M., Hardy, K., Handyside, A., Hunter, S., and Monk, M. (1987). HPRT-deficient (Lesch-Nyhan) mouse embryos derived from germline colonization by cultured cells. Nature 326, 292–295. doi: 10.1038/326292a0
Hu, H., Haas, S. A., Chelly, J., Van Esch, H., Raynaud, M., de Brouwer, A. P., et al. (2016). X-exome sequencing of 405 unresolved families identifies seven novel intellectual disability genes. Mol. Psychiatry. 21, 133–48. doi: 10.1038/mp.2014
Ikeda, F., and Dikic, I. (2008). Atypical ubiquitin chains: new molecular signals. protein modifications: beyond the usual suspects’ review series. EMBO Rep. 9, 536–542. doi: 10.1038/embor.2008.93
Jiao, B., Ma, H., Shokhirev, M. N., Drung, A., Yang, Q., Shin, J., et al. (2012). Paternal RLIM/Rnf12 is a survival factor for milk-producing alveolar cells. Cell 149, 630–641. doi: 10.1016/j.cell.2012.02.056
Jiao, B., Taniguchi-Ishigaki, N., Gungor, C., Peters, M. A., Chen, Y. W., Riethdorf, S., et al. (2013). Functional activity of RLIM/Rnf12 is regulated by phosphorylation-dependent nucleocytoplasmic shuttling. Mol. Biol. Cell 24, 3085–3096. doi: 10.1091/mbc.E13-05-0239
Johnsen, S. A., Gungor, C., Prenzel, T., Riethdorf, S., Riethdorf, L., Taniguchi-Ishigaki, N., et al. (2009). Regulation of estrogen-dependent transcription by the LIM cofactors CLIM and RLIM in breast cancer. Cancer Res. 69, 128–136. doi: 10.1158/0008-5472.CAN-08-1630
Jonkers, I., Barakat, T. S., Achame, E. M., Monkhorst, K., Kenter, A., Rentmeester, E., et al. (2009). RNF12 is an X-Encoded dose-dependent activator of X chromosome inactivation. Cell 139, 999–1011. doi: 10.1016/j.cell.2009.10.034
Kramer, O. H., Muller, S., Buchwald, M., Reichardt, S., and Heinzel, T. (2008). Mechanism for ubiquitylation of the leukemia fusion proteins AML1-ETO and PML-RARalpha. FASEB J. 22, 1369–1379. doi: 10.1096/fj.06-8050com
Kramer, O. H., Zhu, P., Ostendorff, H. P., Golebiewski, M., Tiefenbach, J., Peters, M. A., et al. (2003). The histone deacetylase inhibitor valproic acid selectively induces proteasomal degradation of HDAC2. EMBO J. 22, 3411–3420. doi: 10.1093/emboj/cdg315
Lee, J. T. (2000). Disruption of imprinted X inactivation by parent-of-origin effects at tsix. Cell 103, 17–27. doi: 10.1016/s0092-8674(00)00101-x
Lee, J. T., Davidow, L. S., and Warshawsky, D. (1999). Tsix, a gene antisense to Xist at the X-inactivation centre. Nat. Genet. 21, 400–404. doi: 10.1038/7734
Metzger, M. B., Hristova, V. A., and Weissman, A. M. (2012). HECT and RING finger families of E3 ubiquitin ligases at a glance. J. Cell Sci. 125, 531–537. doi: 10.1242/jcs.091777
Metzger, M. B., Pruneda, J. N., Klevit, R. E., and Weissman, A. M. (2014). RING-type E3 ligases: master manipulators of E2 ubiquitin-conjugating enzymes and ubiquitination. Biochim. Biophys. Acta 1843, 47–60. doi: 10.1016/j.bbamcr.2013.05.026
Monkhorst, K., Jonkers, I., Rentmeester, E., Grosveld, F., and Gribnau, J. (2008). X inactivation counting and choice is a stochastic process: evidence for involvement of an X-linked activator. Cell 132, 410–421. doi: 10.1016/j.cell.2007.12.036
Navarro, P., Oldfield, A., Legoupi, J., Festuccia, N., Dubois, A., Attia, M., et al. (2010). Molecular coupling of tsix regulation and pluripotency. Nature 468, 457–460. doi: 10.1038/nature09496
Norris, D. P., Patel, D., Kay, G. F., Penny, G. D., Brockdorff, N., Sheardown, S. A., et al. (1994). Evidence that random and imprinted Xist expression is controlled by preemptive methylation. Cell 77, 41–51. doi: 10.1016/0092-8674(94)90233-x
Ostendorff, H. P., Bossenz, M., Mincheva, A., Copeland, N. G., Gilbert, D. J., Jenkins, N. A., et al. (2000). Functional characterization of the gene encoding RLIM, the corepressor of LIM homeodomain factors. Genomics 69, 120–130. doi: 10.1006/geno.2000.6311
Ostendorff, H. P., Peirano, R. I., Peters, M. A., Schluter, A., Bossenz, M., Scheffner, M., et al. (2002). Ubiquitination-dependent cofactor exchange on LIM homeodomain transcription factors. Nature 416, 99–103. doi: 10.1038/416099a
Ostendorff, H. P., Tursun, B., Cornils, K., Schluter, A., Drung, A., Gungor, C., et al. (2006). Dynamic expression of LIM cofactors in the developing mouse neural tube. Dev. Dyn. 235, 786–791. doi: 10.1002/dvdy.20669
Payer, B. (2016). Developmental regulation of X-chromosome inactivation. Semin. Cell Dev. Biol. 56, 88–99. doi: 10.1016/j.semcdb.2016.04.014
Penny, G. D., Kay, G. F., Sheardown, S. A., Rastan, S., and Brockdorff, N. (1996). Requirement for Xist in X chromosome inactivation. Nature 379, 131–137. doi: 10.1038/379131a0
Petroski, M. D., and Deshaies, R. J. (2005). Function and regulation of cullin-RING ubiquitin ligases. Nat. Rev. Mol. Cell Biol. 6, 9–20. doi: 10.1038/nrm1547
Pickart, C. M. (2001). Mechanisms underlying ubiquitination. Annu. Rev. Biochem. 70, 503–533. doi: 10.1146/annurev.biochem.70.1.503
Scanlan, M. J., Gordan, J. D., Williamson, B., Stockert, E., Bander, N. H., Jongeneel, V., et al. (1999). Antigens recognized by autologous antibody in patients with renal-cell carcinoma. Int. J. Cancer 83, 456–464. doi: 10.1002/(sici)1097-0215(19991112)83:4<456::aid-ijc4>3.0.co;2-5
Shin, J., Bossenz, M., Chung, Y., Ma, H., Byron, M., Taniguchi-Ishigaki, N., et al. (2010). Maternal Rnf12/RLIM is required for imprinted X-chromosome inactivation in mice. Nature 467, 977–981. doi: 10.1038/nature09457
Shin, J., Wallingford, M. C., Gallant, J., Marcho, C., Jiao, B., Byron, M., et al. (2014). RLIM is dispensable for X-chromosome inactivation in the mouse embryonic epiblast. Nature 511, 86–89. doi: 10.1038/nature13286
Tonne, E., Holdhus, R., Stansberg, C., Stray-Pedersen, A., Petersen, K., Brunner, H. G., et al. (2015). Syndromic X-linked intellectual disability segregating with a missense variant in RLIM. Eur. J. Hum. Genet. 23, 1652–1656. doi: 10.1038/ejhg.2015.30
Wang, F., McCannell, K. N., Boskovic, A., Zhu, X., Shin, J., Yu, J., et al. (2017). Rlim-dependent and -independent pathways for X chromosome inactivation in female ESCs. Cell Rep. 21, 3691–3699. doi: 10.1016/j.celrep.2017.12.004
Wang, F., Shin, J., Shea, J. M., Yu, J., Boskovic, A., Byron, M., et al. (2016). Regulation of X-linked gene expression during early mouse development by rlim. eLife 5:e19127. doi: 10.7554/eLife.19127
Wang, F., Zhao, K., Yu, S., Xu, A., Han, W., and Mei, Y. (2019). RNF12 catalyzes BRF1 ubiquitination and regulates RNA polymerase III-dependent transcription. J. Biol. Chem. 294, 130–141. doi: 10.1074/jbc.RA118.004524
Weissman, A. M., Shabek, N., and Ciechanover, A. (2011). The predator becomes the prey: regulating the ubiquitin system by ubiquitylation and degradation. Nat. Rev. Mol. Cell Biol. 12, 605–620. doi: 10.1038/nrm3173
Wutz, A., and Jaenisch, R. (2000). A shift from reversible to irreversible X inactivation is triggered during ES cell differentiation. Mol. Cell. 5, 695–705. doi: 10.1016/s1097-2765(00)80248-8
Xu, N., Tsai, C. L., and Lee, J. T. (2006). Transient homologous chromosome pairing marks the onset of X inactivation. Science 311, 1149–1152. doi: 10.1126/science.1122984
Keywords: mouse genetics, XCI, RLIM, Rnf12, Rex1, Xist regulation
Citation: Wang F and Bach I (2019) Rlim/Rnf12, Rex1, and X Chromosome Inactivation. Front. Cell Dev. Biol. 7:258. doi: 10.3389/fcell.2019.00258
Received: 22 August 2019; Accepted: 16 October 2019;
Published: 31 October 2019.
Edited by:
Celine Morey, UMR 7216 Epigénétique et Destin Cellulaire, FranceReviewed by:
Atsushi Fukuda, Tokai University, JapanSha Sun, University of California, Irvine, United States
Copyright © 2019 Wang and Bach. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ingolf Bach, aW5nb2xmLmJhY2hAdW1hc3NtZWQuZWR1
 Feng Wang
Feng Wang Ingolf Bach
Ingolf Bach