- 1Department of Anatomical, Histological, Forensic and Orthopedic Sciences, Section of Histology and Medical Embryology, Sapienza University of Rome, Laboratory Affiliated to Istituto Pasteur Italia-Fondazione Cenci Bolognetti, Rome, Italy
- 2Department of Biology and Biotechnology ‘Charles Darwin’, Sapienza University of Rome, Rome, Italy
RNA chemical modifications in coding and non-coding RNAs have been known for decades. They are generally installed by specific enzymes and, in some cases, can be read and erased by other specific proteins. The impact of RNA chemical modifications on gene expression regulation and the reversible nature of some of these modifications led to the birth of the word epitranscriptomics, in analogy with the changes that occur on DNA and histones. Among more than 100 different modifications identified so far, most of the epitranscriptomics studies focused on the N6-methyladenosine (m6A), which is the more abundant internal modification in protein coding RNAs. m6A can control several pathways of gene expression, including spicing, export, stability, and translation. In this review, we describe the interplay between m6A and non-coding RNAs, in particular microRNAs and lncRNAs, with examples of its role in gene expression regulation. Finally, we discuss its relevance in cell development and disease.
Introduction
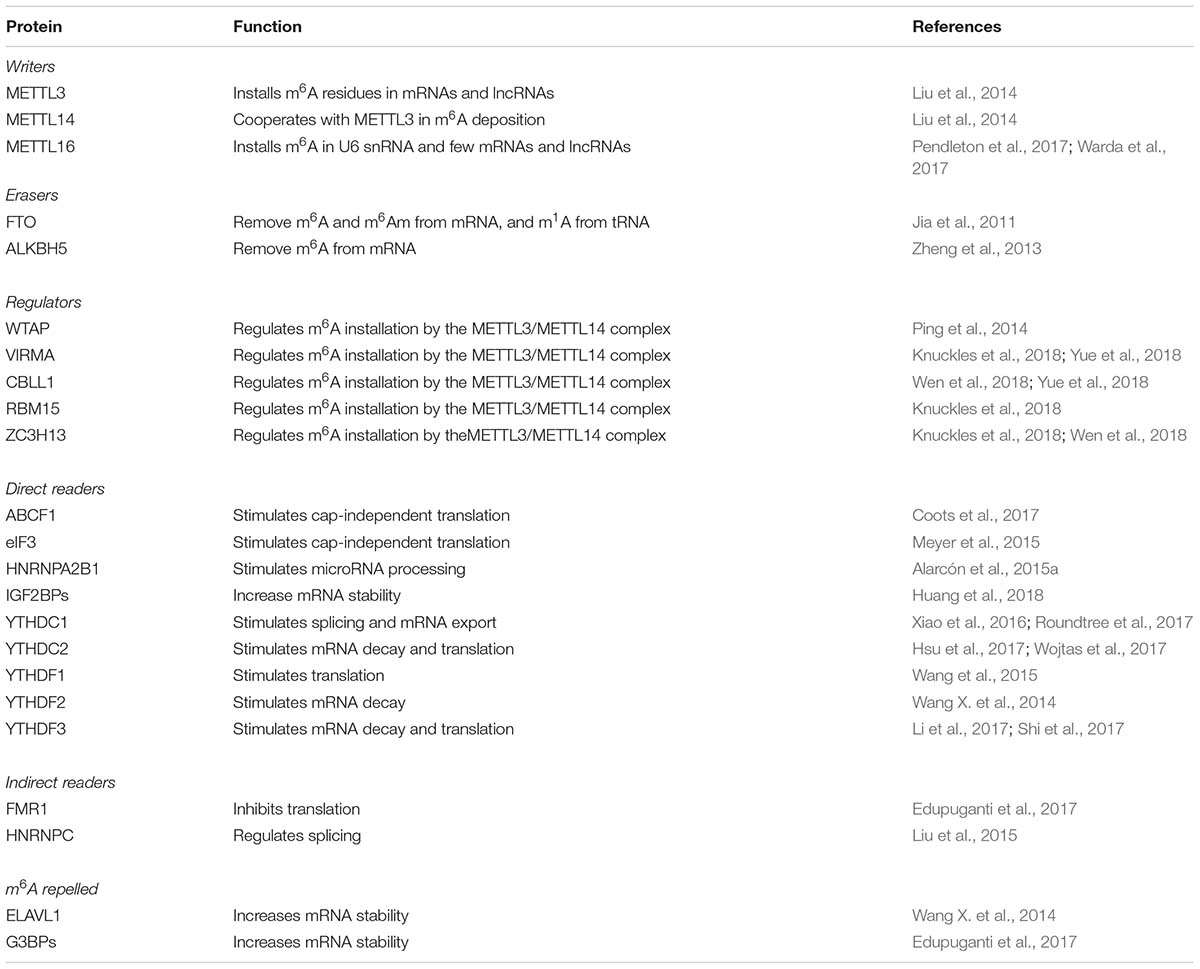
To date, more then 100 chemical modifications have been described in non-coding and protein coding RNAs (see The RNA Modification database1). The majority of them occur in transfer RNA (tRNA) and ribosomal RNA (rRNA), while a minority of them occur in messenger RNAs (mRNA) and long non-coding RNAs (lncRNAs). In all cases RNA modifications may play important role in RNA folding, stability and function; in view of the fact that, similarly to epigenetics, they can affect gene expression without changing the sequence of the RNA molecules, they are now referred to as “epitranscriptomics.” In metazoan, N6-methyladenosine (m6A) is the more abundant internal modification in mRNAs and lncRNAs and plays relevant roles in several steps of gene expression, including splicing, export, stability, and translation. Notably, m6A modification is present in all three different phylogenetic domains, Eukaya, Bacteria, and Archea (Carell et al., 2012), and is also present in viral RNAs (Lavi and Shatkin, 1975) where it has important regulatory functions (Meyer and Jaffrey, 2014). In analogy with DNA and histone modifications, m6A is a dynamic mark. It is installed by writers, removed by erasers and recognized by reader proteins. Despite its discovery in the early 1970s (Desrosiers et al., 1974; Perry and Kelley, 1974; Adams and Cory, 1975), the precise function of m6A residues in gene expression regulation remained elusive until recently with the development of high throughput methodologies for mapping of m6A residues in the whole transcriptome (Dominissini et al., 2012; Meyer et al., 2012; Linder et al., 2015). These methods used specific immunoprecipitation of m6A modified RNAs coupled to RNA sequencing. There are currently two different methodologies. The first one was developed by two independent groups (referred to as m6A-Seq or MeRIP-seq) and sequences immunoprecipitated m6A RNA fragment of about 200 nt, thus, does not allow for mapping of m6A residues at single-nucleotide resolution (Dominissini et al., 2012; Meyer et al., 2012). The second one (referred to as miCLIP), uses UV cross-linking to covalently bind the anti m6A antibody to modified RNAs, which induces mutation and truncation during reverse transcription, allowing for identification of single modified nucleotides within RNA species (Linder et al., 2015). m6A in mRNAs and lncRNAs can be installed by two independent complexes (Table 1): the heterodimeric complex of METTL3/METTL14 (methyltransferase-like protein 3 and 14), also referred to as MAC (m6A-METTL Complex), and the homodimeric complex of METTL16 (methyltransferase-like protein 16) (reviewed in Zhao et al., 2017; Lence et al., 2019). The MAC complex methylates adenosine during transcriptional elongation within the consensus motif RRACH (R = A/G; H = A/C/U), and is assisted in adenine selection by a multiprotein complex called MACOM (m6A-METTL-associated complex) composed of Wilms tumour 1-associated protein (WTAP), Vir-like m6A methyltransferase-associated (VIRMA), Cbl proto-oncogene like 1 (CBLL1, also known as Hakai), RNA-binding motif 15 (RBM15) or its paralog RBM15B, and zinc finger CCCH-type containing 13 (ZC3H13) proteins (Lence et al., 2019) (Table 1). The METTL3 is the catalytic component of the complex while METTL14 is required for RNA binding and stabilization. Recently, METTL14 was also found to interact with histone H3 trimethylation at Lys36 (H3K36me3), a marker for RNA polymerase II (RNA pol II) transcription elongation, thus ensuring modification of nascent RNAs in both intronic and exonic regions (Huang et al., 2019). However, m6A residues in mature mRNA molecules follow precise distribution and are enriched near the stop codon and untranslated regions. On the other hand, the METTL16 complex acts on a specific stem–loop structure of RNA containing the UACAGAGAA sequence. This complex acts on only a few percentages on methylated mRNAs and lncRNAs. However, between targeted RNAs, there is the human S-adenosylmethionine (SAM) synthetase MAT2A (Pendleton et al., 2017; Shima et al., 2017; Warda et al., 2017), which regulates cellular levels of the methyl donor SAM. Therein, METTL16 can regulate the activity of all cellular methyltransferases, including METTL3/MELL14. Moreover, it is also responsible for the methylation in the spliceosomal U6 small nuclear RNA (snRNA).
m6A modification can be removed by two demethylase enzymes belonging to the AlkB family of the Fe(II) and α-ketoglutarate-dependent dioxygenases: ALKBH5 (alkB homolog 5) and FTO (fat-mass and obesity associated protein) (Zhao et al., 2017) (Table 1). The first one is specific for m6A removal while the second can also demethylate N6, 2-O-dimethyladenosine (m6Am), which is installed in mRNA if the first transcribed nucleotide is adenosine, and N1-methyladenosine (m1A) in tRNA (Wei et al., 2018).
Although m6A modification does not prevent Watson-Crick base pairing of A-U nucleotides, m6A residues can affect tertiary interactions involving Hoogsteen base pairs that use the N6 atom of A for H-bond (Meyer and Jaffrey, 2014). Therein, m6A residues within RNA molecules can produce local changing in the RNA structure that can alter RNA folding and affect the interaction with proteins and RNAs (Meyer and Jaffrey, 2014; Liu et al., 2015; Edupuganti et al., 2017). However, the function of m6A modifications in gene expression regulation is mainly mediated by m6A readers (reviewed in Patil et al., 2018) (Table 1). Proteins of the YT521-B homology (YTH) domain family were the first to be identified. In humans, there are five members: the nuclear YTHDC1, and the cytoplasmic YTHDC2, YTHDF1, YTHDF2, and YTHDF3. Mechanistically, YTHDC1 binding in the nucleus regulates alternative splicing and promotes RNA export while YTHDF2 binding stimulates mRNA decay and YTHDF1 binding promotes translation. The YTHDF3 reader can cooperate with both YTHDF1 and YTFDF2 on modified mRNA while in circular RNAs it can promote translation independently from other YTH proteins. Additional readers, lacking the YTH domains, have been also identified, including the translational regulators eIF3 and ABCF1, which positively regulate translation of modified mRNA and the insulin-like growth factor mRNA-binding proteins IGFBP-1, -2 and- 3, which enhance RNA stability and translation (Huang et al., 2018). Therein, once installed, m6A modifications can produce several different outputs on the regulation of specific RNAs which are not easily predictable.
Notably, m6A plays an important role in embryonic stem cells (ESCs) by controlling cell fate transition and deletion of METTL3 and METTL16 in mouse results in embryonic lethality, indicating essential function for m6A modification in the regulation of gene expression programs required for embryo development (Batista et al., 2014; Wang Y. et al., 2014; Geula et al., 2015; Bertero et al., 2018; Mendel et al., 2018). Moreover, m6A plays also important role in mouse adult brain, by regulating synaptic function and stress-induced responses (Engel et al., 2018; Koranda et al., 2018), and in the hematopoietic system, by controlling stem cell differentiation and homeostasis (Lv et al., 2018; Wang et al., 2018; Yao et al., 2018). Even if most of the m6A studies focused on its direct role on mRNA function, recent evidences showed that m6A can also regulate the synthesis and function of microRNAs and lncRNAs. In addition, microRNAs and lncRNAs can also influence the function of m6A modification in mRNAs. Here, we review the impact of m6A on the regulation of these non-coding RNAs and we discuss the interplay between m6A, microRNAs and lncRNAs in cell development and disease.
Impact of Epitranscriptomics on MicroRNAs Biogenesis and Function
MicroRNAs (miRNAs) are endogenously encoded short RNAs (∼21 nucleotides, nt) that are produced from long primary transcripts (pri-miRNA) transcribed by RNA Pol II (reviewed in Bartel, 2018). More than 50% of human miRNAs are encoded in introns of coding and non-coding pre-mRNAs and are connected to the expression of their host genes. The pri-miRNA is cleaved co-transcriptionally by a protein complex, named Microprocessor, containing the nuclear RNase III-type endonuclease Drosha and the DGCR8 (DiGeorge syndrome critical region gene 8) protein. The recognition of the pri-miRNA by the Microprocessor requires a stem–loop structure formed by the mature miRNA and single-stranded regions flanking the stem–loop. The cleavage by Drosha produces a stem–loop pre-miRNA of about 70 nt that is then recognized and exported to the cytoplasm by the transport receptor Exportin 5. In the cytoplasm, the pre-miRNA is cleaved by the RNase III-type endonuclease Dicer, releasing the miRNA duplex. One strand is then incorporated into the silencing complex containing one Argonaute (AGO) protein (in human AGO1, AGO2, AGO3, and AGO4) and the TNRC6 protein (also called GW182). The miRNA directs the Ago complex to its target mRNAs through perfect complementarity between sequences in the 3′ì untranslated region (3ì-UTR) and a stretch of 6 nucleotides (from nucleotides 2 to 7) in the 5′ region of the miRNA, also referred to as “seed.” The Ago proteins recruit TNRC6 protein, which stimulates mRNA deadenylation by interacting with deadenylase complexes and, consequently, produces mRNA destabilization and translational repression (Jonas and Izaurralde, 2015; Bartel, 2018). In addition, TNRC6 also recruits DDX6, a helicase that enhances both the decay and translational repression of target mRNAs (Jonas and Izaurralde, 2015; Bartel, 2018). m6A modification may affect microRNA synthesis and function at multiple levels (Figure 1A). A strong correlation between m6A residues in the 3′-UTR and miRNA-binding sites has been identified (Meyer et al., 2012). This has suggested the existence of a functional interaction between m6A modification and miRNAs targeting. In particular, the presence of m6A residues within the complementary region between the 3′-UTR and the miRNA seed might destabilize A-U pairing, thus decreasing duplex stability and affecting miRNA interaction. However, even if alterations in miRNA binding might contribute to some of the observed effects of m6A modification, the impact of m6A modifications within mRNAs on miRNA targeting is still not clear. On the other hand, m6A can also affect miRNA synthesis. m6A marks are deposited co-transcriptionally on a set of pri-miRNA molecules and are read by the HNRNPA2B1 proteins that, in turn, stimulate nuclear miRNA processing by recruiting the Microprocessor component DGCR8 (Alarcón et al., 2015a, b). Therein, alteration in m6A deposition may unbalance cellular miRNA levels. Moreover, upon acute temperature stress, the METTL3/METL14 complex can co-transcriptionally recruit the DGCR8 protein on stem–loop structures present in heat-shock genes, independently from the presence of an embedded precursor microRNA. Thus, promoting their subsequent nuclear degradation by the microprocessor complex (Knuckles et al., 2017). In view of the recent discovery of the association of METTL14 with chromatin during transcriptional elongation (Huang et al., 2019), we can speculate that the METTL3/METTL14 complex may contribute to the co-transcriptional recruitment of the Microprocessor complex on pri-miRNA transcripts. In addition, it has also been shown that, in hepatocellular carcinoma (HCC), METTL14 can directly recruit DCGR8 on the m6A modified pri-miRNA encoding for oncosuppressor miR-126a (Ma et al., 2017). In particular, low levels of METTL14 in HCC are associated with low levels of miR-126a and increased metastatic capacity (Ma et al., 2017). More importantly, ectopic expression of miR-126a in HCC cells ameliorated the metastatic phenotype induced by METTL14 downregulation (Ma et al., 2017).
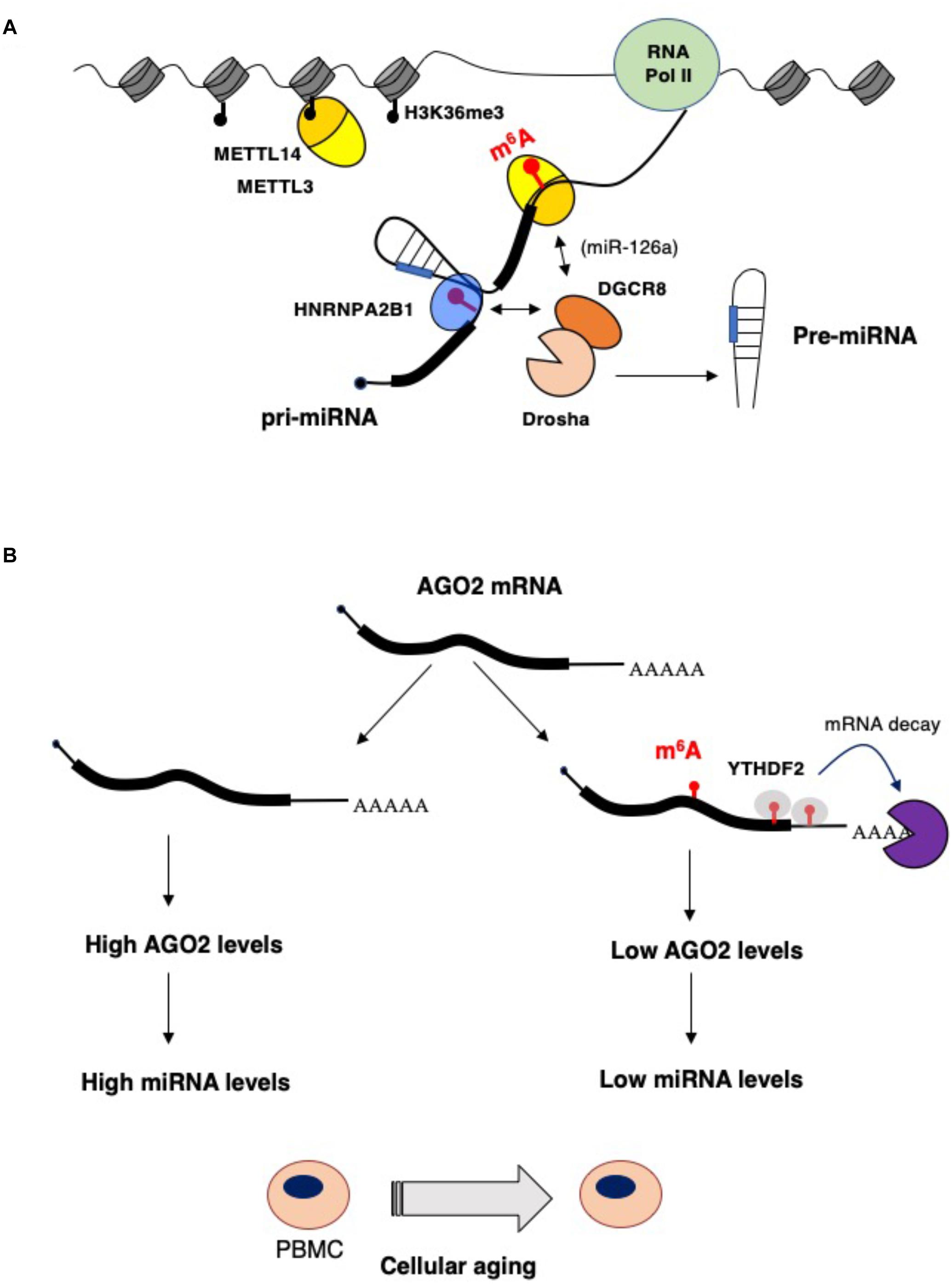
Figure 1. Impact of epitranscriptomics on microRNAs biogenesis and function. (A) m6A stimulates microRNA processing by recruiting the Drosha cofactor DGCR8 by the m6A reader HNBRPA2B1 (Alarcón et al., 2015a, b) or, in the case of the miR-126a, by direct interaction with METTL14 (Ma et al., 2017). (B) During aging of peripheral blood mononuclear cells (PBMCs), AGO2 and, eventually, miRNA levels are decreased by higher m6A modification of AGO2 mRNA. This results in enhanced mRNA decay that is very likely mediated by the YTHDF2 reader (Min et al., 2018).
Another example highlighting the role m6A modification on miRNAs processing has been recently reported for miR-25-3p. Zang and colleagues indeed described the impact of cigarette smoking on miR-25-3p maturation by m6A modification in pancreatic ductal adenocarcinoma (Zhang et al., 2019). In this manuscript, the authors described how the cigarette smoking induced the upregulation of METTL3 expression by affecting METTL3 promoter epigenetic regulation. This results in a METTL3-dependent modification of pri-miR-25-3p and an increase of miR-25-3p processing. The induction of miR-25-3p affects the expression of its target PH domain leucine-rich-repeats protein phosphatase 2 (PHLPP2), with an consequential impact on the AKT-p70S6K signaling pathway (Zhang et al., 2019). These results suggest that a METTL3-miR-25-3p-PHLPP2-AKT regulatory axis could be relevant for the transformation process induced by cigarette smoking in pancreatic tissue.
Interestingly, an additional mechanism leading to the modulation of m6A miRNAs modification is represented by the DDX3-dependent network. Indeed, DDX3, a member of the family of DEAD-box RNA helicases, has been shown to be able to interact with the RNA m6A demethylases, such as ALKBH5, resulting in m6A RNA demethylation. With specific regard to miRNAs, DDX3, thanks to its ability to also interact with AGO2 protein, may contribute to miRNAs demethylation. In summary, the functional contribution of DDX3 to the control of cell growth and proliferation may be at least in part mediated by its interaction with ALKBH5 and AGO2, relevant for the demethylation of mRNAs and miRNAs (Shah et al., 2017).
miRNA levels can also be controlled by m6A modification of AGO2 mRNA (Figure 1B). In a study performed on human peripheral blood mononuclear cells (PBMCs) from young and old donors, the AGO2 mRNA was found highly m6A methylated in young PBMCs and this correlated with a lower level of AGO2 mRNA in old PBMCs during aging (Min et al., 2018). AGO2 levels are important for both miRNA synthesis and function. Indeed, a lower level of AGO2 in old PBMCS resulted in an altered level of miRNA expression (Min et al., 2018), indicating that m6A modification on AGO2 mRNA contributes to cellular aging by regulating global miRNAs synthesis.
Impact of Epitranscriptomics on LncRNA Regulation and Functions
LncRNAs are generally defined as transcripts longer than 200 nt without coding potential (reviewed in Fatica and Bozzoni, 2014). The human genome contains 16,193 genes encoding for lncRNAs (Gencode v30), which can produce more than 30,000 lncRNA transcripts. The majority of lncRNAs, but not all, share several features with coding mRNAs; they are 5′ capped, spliced and, in most of the cases, polyadenylated. Similar to mRNAs, lncRNAs are also m6A methylated and the levels of m6A residues strongly depend on the cell line, tissue type and growth condition (Meyer et al., 2012; Han et al., 2019; Xiao et al., 2019). In cell lines, the enrichment score of m6A peaks within mRNAs and lncRNAs is very similar (Han et al., 2019). However, it has been shown recently that in human fetal tissues, a lower proportion of lncRNA is m6A modified compared to mRNA (Xiao et al., 2019). In contrast to mRNAs, m6A residues in lncRNAs are distributed along the whole body of the transcript and are more present in lncRNAs that undergo alternative splicing (Xiao et al., 2019). Thus, this indicates a possible function for m6A modification in regulation of lncRNA isoforms. Many lncRNAs are retained and function in the nucleus. Nuclear lncRNA may regulate gene expression by several mechanisms, such as modulating the activity of regulatory protein complexes, regulating chromosomal conformations and, more generally, nuclear organization (reviewed in Engreitz et al., 2016). In particular, different lncRNAs regulate gene expression by guiding regulatory complex to specific gene loci. This is generally achieved by lncRNA interaction with chromatin associated proteins, local chromosomal architecture or by forming an RNA-DNA triple helix (Engreitz et al., 2016; Li et al., 2016). LncRNA local structure and interaction with specific proteins plays an important role in lncRNA function. Therein, m6A modification might regulate lncRNA function by providing binding sites for m6A reader proteins or, alternatively, might regulate local RNA structure to allow access for specific RNA-binding proteins to nearby m6A residues. Furthermore, m6A modification may also influence RNA–DNA triple helix formation, in which a lncRNA binds with sequence specificity through Hoogsteen base pairs in the major groove of a Watson–Crick base-paired DNA duplex. Therein, m6A modification can potentially affect lncRNA interaction with specific DNA loci.
Metastasis-associated lung adenocarcinoma transcript 1 (MALAT1), also known as nuclear-enriched abundant transcript 2 (NEAT2), is a highly expressed nuclear lncRNA, frequently upregulated in cancer, that contains several m6A modifications (Dominissini et al., 2012). Even if MALAT1 is transcribed by RNA pol II, it lacks a canonical poly-A tail. The high stability observed for MALAT1 transcript is ensured by a triple-helix at its 3′-end that specifically binds METTL16 (Brown et al., 2014, 2016). MALAT1 accumulates in the nuclear speckels, which are nuclear domains enriched in splicing factors, and associates with different splicing regulators such as the serine/arginine-rich (SR) proteins and the protein heterogeneous nuclear ribonucleoprotein C (Tripathi et al., 2010; Zhou et al., 2016). In cell lines, MALAT1 silencing alters the alternative splicing of specific pre-mRNAs (Tripathi et al., 2010). However, MALAT1 knock-out in mouse has no effect on the alternative splicing nor on the formation of nuclear speckles (Nakagawa et al., 2012; Zhang et al., 2012). A further study showed that MALAT1 can also bind to the Polycomb 2 protein (Pc2), a component of the Polycomb Repressive Complex -1 (PRC1), and that it can act as scaffold in distinct subnuclear compartments required for coordinated regulation of gene transcription (Yang et al., 2011). Notably, m6A modifications identified in MALAT1 can alter the accessibility of the RNA motif to which proteins bind, through changing its local structure (Liu et al., 2015; Spitale et al., 2015; Zhou et al., 2016), a mechanism known as “m6A switch” (Liu et al., 2015). Therein, m6A might affect the function of MALAT1 in splicing and transcription by regulating RNA-protein interactions.
MALAT1 can also interact with microRNAs and act as a competing endogenous RNA (ceRNA), thus affecting microRNA binding to target mRNAs, in different cell types (Leucci et al., 2013; Han et al., 2015; Hirata et al., 2015; Xiao et al., 2015). Even in some cases, the interaction of MALAT1 with microRNAs has been reported to occur in the nucleus (Leucci et al., 2013), whereas in other cases, a specific translocation of MALAT1 in the cytoplasm is required (Han et al., 2015). However, the mechanism responsible for its translocation is still not known. Notably, m6A modification is directly involved in RNA nuclear export (Zheng et al., 2013; Roundtree et al., 2017; Lesbirel et al., 2018). Therein, the level of m6A modifications within MALAT1 might directly control its cellular localization and, eventually, its ceRNA activity. Alternatively, m6A modifications might directly regulate the RNA-RNA interactions between MALAT1 and targeted microRNAs, as recently reported for another ceRNA (Yang et al., 2018).
The X-inactive specific transcript (Xist), an important regulator of X-chromosome inactivation in mammals, is another highly m6A methylated lncRNA. An initial shRNA screening performed in mESCs identified three components of the MACOM complex, WTAP, VIRMA (also known as KIAA1429) and RBM15 proteins, which are all regulators of Xist activity (Moindrot et al., 2015). Moreover, knockdown of RBM15 and WTAP greatly impaired Xist mediated epigenetic silencing (Moindrot et al., 2015). In addition, WTAP protein was also identified as a stable interactor of Xist RNA (Chu et al., 2015). More recently, it has been confirmed that m6A modification is strictly required for Xist-mediated transcriptional repression and that knockdown of the METTL3 writer inhibits X chromosome silencing (Patil et al., 2016). The RBM15 protein was identified as the MACOM components that interacts with and guides the METTL3/METTL14 complex for the formation of the 78 m6A present in Xist RNA (Patil et al., 2016). Furthermore, it was also shown that the YTHDC1 reader recognized the m6A marks in mESCs and is required for Xist activity even if the mechanism has not yet been clarified (Patil et al., 2016). Notably, tethering of YTHDC1 on Xist in the absence of m6A residues is sufficient for its repressive function, indicating that m6A per se is not required for Xist activity. YTHDC1 is the only nuclear reader of the YTH family and is usually involved in the regulation of pre-mRNA spicing through recruiting splicing factors (Xiao et al., 2016). Similarly, YTHDC1 binding on Xist might function by bridging cis- acting regulatory elements on XIST RNA with trans-acting proteins required for transcriptional silencing.
Recently, it has also been found that the enhancer RNAs (eRNAs), non-coding transcripts produced from enhancer regions that act as regulators of transcription, are highly m6A modified (Xiao et al., 2019). This has suggested that m6A modification might contribute to the enhancer function of eRNAs during transcription.
In the cytoplasm, lncRNAs may regulate mRNA stability and translation by recruiting regulatory proteins to interacting mRNAs or by acting as ceRNAs (reviewed Fatica and Bozzoni, 2014). Therein, m6A residues might affect cytoplasmic lncRNA function with the same mechanisms described above. A recently identified cytoplasmic lncRNA whose function is regulated by m6A is lincRNA 1281 (linc1281) (Yang et al., 2018). Linc1281 is required for mESC differentiation and acts as a ceRNA by sequestering miRNAs of the let-7 family (Yang et al., 2018). Notably, linc1281 contains different m6A marks in its 3′-end region that are required for the binding of let-7 (Yang et al., 2018). It has been proposed that the presence of m6A in linc1281 can act as m6A-switch for specific RNA binding proteins, which will eventually regulate the interaction with let-7. However, the identity of such proteins has not yet been discovered. A similar mechanism has been already proposed for the binding of HuR (ELAVL1) protein and miRNAs to mRNAs encoding developmental regulators in mESCs (see below).
In view of the fact that many lncRNAs can be m6A modified and that m6A can affect their expression levels and functions, it very likely that other examples of lncRNAs regulated by m6A modification will follow soon.
Interplay Between Non-Coding RNAs and m6A Effector Proteins
As highlighted above, m6A is a relevant modification for non-coding RNA biogenesis and functional activity. However, it has been also reported that lncRNAs may control the function of the epitranscriptomics machinery. For example, the expression of the nuclear lncRNA FOXM1-AS allows the interaction between the FOXM1 nascent RNA and the m6A demethylase ALKBH5, that results in the demethylation of FOXM1 transcripts. This promotes the binding of HuR protein (also known as ELAVL1) with FOXM1 pre-mRNA, resulting in an elevated expression of FOXM1. Interestingly, it has also been shown that, in glioblastoma stem-like cells (GSCs), the m6A demethylase ALKBH5 is highly expressed, and the depletion of ALKBH5 and FOXM1-AS disrupts GSC tumorigenesis through the reduction FOXM1 expression (Zhang et al., 2017) (Figure 2A).
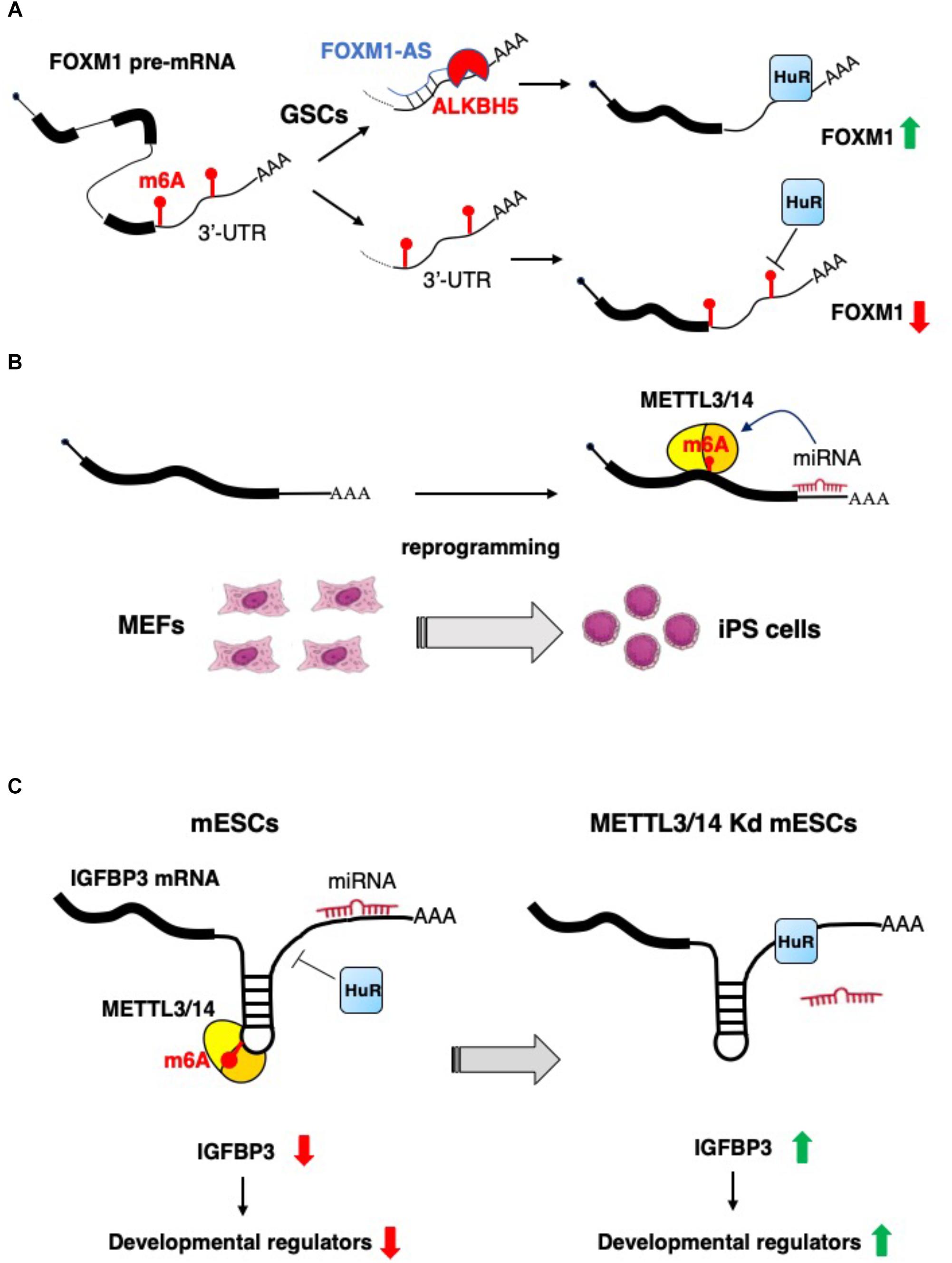
Figure 2. Examples of the interplay between non-coding RNAs and epitranscriptomics. (A) In glioblastoma stem-like cells (GSCs) the expression of FOXM1 is increased by the concomitant expression of the antisense transcript FOXM1-AS, which, in turn, promote m6A demethylation by recruiting ALKBH5 (Zhang et al., 2017). (B) m6A RNA methylation is positively regulated by microRNAs, which recruit METTL3 on specific mRNA and promotes reprogramming to pluripotency (Chen et al., 2015). (C) m6A modification decreases the IGFBP3 mRNA levels by inhibiting the binding of HuR and promoting the interaction with microRNAs. IGFBP3 protein positively regulates the stability of different developmental regulators. This mechanism ensures low level of IGFBP3 in mESCs (Wang Y. et al., 2014).
Recently, an additional function of non-coding RNAs in the control of m6A modification has emerged. In particular, it has been reported that the expression of epitranscriptomics machinery components may be controlled by miRNAs through the targeting of their corresponding mRNAs. Here, we include some representative examples of such modulation.
miR-145, broadly reported as a tumor suppressor miRNA, was shown to control the expression of the YTHDF2 reader. YTHDF2 is involved in the deadenylation and decay of m6A-containing RNAs through a direct interaction and recruitment of the CCR4-NOT deadenylase complex (Du et al., 2016).
Specifically, in liver cancer cells, miR-145 downregulates YTHDF2 mRNA expression with an increase on the overall levels of mRNAs containing m6A residues, as evaluated by dot-blot and immunofluorescence analyses with anti m6A antibodies, which do not allow m6A mapping in specific RNA species (Yang et al., 2017). Interestingly, this increase is inhibited by YTHDF2 overexpression, supporting the central role of this protein in this regulation (Yang et al., 2017). Accordingly, miR-145 expression levels are negatively correlated with those of YTHDF2 in HCC tissues. Functionally, miR-145 is able to suppresses the proliferation of HCC cells through the modulation of m6A-modified mRNA levels by targeting the 3′-UTR of YTHDF2 mRNA.
Another example is represented by miR-33a. In non-small-cell lung carcinoma (NSCLC) cells, it was recently described that miR-33a, by targeting the 3′-UTR of METTL3 mRNA, reduces the expression of METTL3 at both mRNA and protein levels and, eventually, global m6A mRNA methylation, with a functional reduction of cellular proliferation and anchorage-independent growth (Du et al., 2017).
Recently, it has been reported that the oncogenic properties of glioblastoma stem cells (GSCs) in terms of proliferation, migration, and invasion could be influenced by a novel miR-29a/QKI-6/WTAP molecular axis (Xi et al., 2017). Of note, the overexpression of miR-29a inhibits WTAP expression and the activation of the ERK and PI3K/AKT pathways by downregulating QKI-6 expression and impairing the oncogenic abilities of GSCs (Xi et al., 2017). The impact of this regulatory network on m6A levels have not been addressed in this study. However, considering the relevance of WTAP in regulating the methylation activity of the METTL3-METTL14 complex (Ping et al., 2014), it might be speculated that miR-29a, besides regulating DNA methylation during cell reprograming by targeting DNA-methyl-transferases (DNMTs) (Hysolli et al., 2016), could also have a crucial role in the modulation of RNA m6A methylation during neoplastic transformation processes.
Lately, a novel role of miRNAs in the regulation of m6A modification has emerged. Specifically, an enrichment of seed sequences for miRNAs has been observed in transcripts presenting m6A residues using next generation sequencing (NGS) approaches (Chen et al., 2015). This study evaluated the contribution of miRNAs to the ab initio induction of m6A methylation by depletion or overexpression of Dicer to modulate the overall miRNAs activity. In particular, Dicer expression favors the induction of m6A methylation on target mRNAs, highlighting the tight connection between miRNAs activity and m6A modification. Mechanistically, Dicer promotes the localization of METTL3 in nuclear speckles, enhancing its interaction with the transcript subjected to m6A modification. In the same study, by using m6A-Seq, the authors identified m6A levels specific to different degrees of pluripotency by analyzing various experimental models, such as ESCs, induced pluripotent stem cells (iPSCs), neural stem cells (NSCs), and testicular sertoli cells (SCs). They revealed the existence of both cell-common and cell-specific modified transcripts associated with biological processes such as stem cell maintenance and cell differentiation. Moreover, to explore the contribution of m6A modification in cell reprograming, the authors overexpressed human METTL3 into mouse embryonic fibroblasts (MEFs), expressing the reprograming factors Oct4, Sox2, Klf4, and c-Myc, and evidenced that the reprograming efficiency of MEFs was significantly improved by the increase METTL3-dependent m6A levels (Chen et al., 2015) (Figure 2B).
Of note, a role for m6A as a signal for miRNA-dependent degradation of transcripts encoding developmental regulators in mESCs has also recently emerged. Indeed, during mESCs normal development, the METTL3/METTL14-dependent m6A methylation of transcripts encoding developmental regulators blocks HuR protein binding and results in miRNAs-mediated transcript destabilization (Wang Y. et al., 2014). The loss of m6A modification, in METTL3 and METTL14 knocked-down cells, allows for HuR-mRNA interaction and reduction of miRNA functional activity, improving transcripts stability and promoting loss of the mESC ground state (Wang Y. et al., 2014) (Figure 2C).
On the contrary, it has also been shown that the presence of m6A modification may serve as a protective signal, which inhibits mRNA degradation through the binding of the IGF2BPs readers. In the case of SRF transcript, m6A modification allows interaction with IGF2BP inhibiting miRNA-mediated decay in cancer cells (Müller et al., 2019). SRF induction is associated with tumor cell phenotype and poor prognosis (Muller et al., 2019). In addition, IGF2BP1 can also stabilize different oncogenic mRNAs by inhibiting general mRNA degradation, as reported for MYC mRNA (Huang et al., 2018).
Conclusion
We are witnessing an impressive increase in the number of studies elucidating the role of m6A modification in cell development and cancer. However, the majority of these studies mainly focused on the impact of m6A marks on coding RNAs, while an important contribution of ncRNA molecules is emerging, such as lncRNAs and microRNAs, on the function of the epitranscriptome. Moreover, lncRNA and miRNA function can be itself regulated by m6A. Nevertheless, different questions still need to be answered. In particular, mapping of m6A modification in mRNAs and lncRNAs showed a different distribution of m6A marks within these two types of RNA pol II transcripts. However, it is still not known how this is achieved and, above all, if specific regulatory factors act differentially in controlling the m6A methylases activity on mRNA and lncRNA molecules. Moreover, these studies have mainly used polyA+ RNA for m6A mapping, therein excluding many regulatory ncRNAs missing a polyA tail. Other important remaining questions in the field will be to determine if small ncRNAs such as miRNAs or the recently functionally characterized Y RNAs contain m6A residues and if these are relevant for their regulatory function in normal and pathological conditions. Furthermore, another important issue concerns the impact of the interplay between m6A modification and non-coding RNA molecules on cell fate determination and development, and which are relevant for these biological processes. In this review, we have described some examples, but the general impact of this emerging molecular network will be clarified after extensive further investigation.
Author Contributions
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
Funding
The research leading to these results has received funding from the AIRC IG 2018 – ID. 21406 project, Istituto Pasteur Italia – Fondazione Cenci Bolognetti and “Progetti Ateneo” Sapienza University of Rome to FF and the AIRC IG 2015 – ID. 17352 project and “Progetti Ateneo” Sapienza University of Rome to AF.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We apologize for not directly citing many crucial references; these references can, however, be found in the cited manuscripts.
Footnotes
References
Adams, J. M., and Cory, S. (1975). Modified nucleosides and bizarre 5′-termini in mouse myeloma mRNA. Nature 255, 28–33. doi: 10.1038/255028a0
Alarcón, C. R., Goodarzi, H., Lee, H., Liu, X., Tavazoie, S., and Tavazoie, S. F. (2015a). HNRNPA2B1 is a mediator of m6A-dependent nuclear RNA processing events. Cell 162, 1299–1308. doi: 10.1016/j.cell.2015.08.011
Alarcón, C. R., Lee, H., Goodarzi, H., Halberg, N., and Tavazoie, S. F. (2015b). N6-methyladenosine marks primary microRNAs for processing. Nature 519, 482–485. doi: 10.1038/nature14281
Batista, P. J., Molinie, B., Wang, J., Qu, K., Zhang, J., Li, L., et al. (2014). m6A RNA modification controls cell fate transition in mammalian embryonic stem cells. Cell Stem Cell 15, 707–719. doi: 10.1016/j.stem.2014.09.019
Bertero, A., Brown, S., Madrigal, P., Osnato, A., Ortmann, D., Yiangou, L., et al. (2018). The SMAD2/3 interactome reveals that TGFβ controls m(6)A mRNA methylation in pluripotency. Nature 555, 256–259. doi: 10.1038/nature25784
Brown, J. A., Bulkley, D., Wang, J., Valenstein, M. L., Yario, T. A., Steitz, T. A., et al. (2014). Structural insights into the stabilization of MALAT1 noncoding RNA by a bipartite triple helix. Nat. Struct. Mol. Biol. 21, 633–640. doi: 10.1038/nsmb.2844
Brown, J. A., Kinzig, C. G., De Gregorio, S. J., and Steitz, J. A. (2016). Methyltransferase-like protein 16 binds the 3′-terminal triple helix of MALAT1 long noncoding RNA. Proc. Natl. Acad. Sci. U.S.A. 13, 14013–14018. doi: 10.1073/pnas.1614759113
Carell, T., Brandmayr, C., Hienzsch, A., Müller, M., Pearson, D., Reiter, V., et al. (2012). Structure and function of noncanonical nucleobases. Angew. Chem. Int. Ed. Engl. 51, 7110–7131. doi: 10.1002/anie.201201193
Chen, T., Hao, Y. J., Zhang, Y., Li, M. M., Wang, M., Han, W., et al. (2015). m(6)A RNA methylation is regulated by microRNAs and promotes reprogramming to pluripotency. Cell Stem Cell 16, 289–301. doi: 10.1016/j.stem.2015.01.016
Chu, C., Zhang, Q. C., da Rocha, S. T., Flynn, R. A., Bharadwaj, M., Calabrese, J. M., et al. (2015). Systematic discovery of Xist RNA binding proteins. Cell 161, 404–416. doi: 10.1016/j.cell.2015.03.025
Coots, R. A., Liu, X. M., Mao, Y., Dong, L., Zhou, J., Wan, J., et al. (2017). m(6)A facilitates eIF4F-independent mRNA translation. Mol. Cell 68, 504–514.e7. doi: 10.1016/j.molcel.2017.10.002
Desrosiers, R., Friderici, K., and Rottman, F. (1974). Identification of methylated nucleosides in messenger RNA from novikoff hepatoma cells. Proc. Natl. Acad. Sci. U.S.A. 71, 3971–3975. doi: 10.1073/pnas.71.10.3971
Dominissini, D., Moshitch-Moshkovitz, S., Schwartz, S., Salmon-Divon, M., Ungar, L., Osenberg, S., et al. (2012). Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature 485, 201–206. doi: 10.1038/nature11112
Du, H., Zhao, Y., He, J., Zhang, Y., Xi, H., Liu, M., et al. (2016). YTHDF2 destabilizes m(6)A-containing RNA through direct recruitment of the CCR4-NOT deadenylase complex. Nat. Commun. 7:12626. doi: 10.1038/ncomms12626
Du, M., Zhang, Y., Mao, Y., Mou, J., Zhao, J., Xue, Q., et al. (2017). MiR-33a suppresses proliferation of NSCLC cells via targeting METTL3 mRNA. Biochem. Biophys. Res. Commun. 482, 582–589. doi: 10.1016/j.bbrc.2016.11.077
Edupuganti, R. R., Geiger, S., Lindeboom, R. G. H., Shi, H., Hsu, P. J., Lu, Z., et al. (2017). N(6)-methyladenosine (m(6)A) recruits and repels proteins to regulate mRNA homeostasis. Nat. Struct. Mol. Biol. 24, 870–878. doi: 10.1038/nsmb.3462
Engel, M., Eggert, C., Kaplick, P. M., Eder, M., Röh, S., Tietze, L., et al. (2018). The role of m(6)A/m-RNA methylation in stress response regulation. Neuron 99, 389–403.e9. doi: 10.1016/j.neuron.2018.07.009
Engreitz, J. M., Ollikainen, N., and Guttman, M. (2016). Long non-coding RNAs: spatial amplifiers that control nuclear structure and gene expression. Nat. Rev. Mol. Cell Biol. 17, 756–770. doi: 10.1038/nrm.2016.126
Fatica, A., and Bozzoni, I. (2014). Long non-coding RNAs: new players in cell differentiation and development. Nat. Rev. Genet. 15, 7–21. doi: 10.1038/nrg3606
Geula, S., Moshitch-Moshkovitz, S., Dominissini, D., Mansour, A. A., Kol, N., Salmon-Divon, M., et al. (2015). Stem cells. m6A mRNA methylation facilitates resolution of naïve pluripotency toward differentiation. Science 347, 1002–1006. doi: 10.1126/science.1261417
Han, X., Yang, F., Cao, H., and Liang, Z. (2015). Malat1 regulates serum response factor through miR-133 as a competing endogenous RNA in myogenesis. FASEB J. 29, 3054–3064. doi: 10.1096/fj.14-259952
Han, Y., Feng, J., Xia, L., Dong, X., Zhang, X., Zhang, S., et al. (2019). CVm6A: a visualization and exploration database for m6as in cell lines. Cells 8:E168. doi: 10.3390/cells8020168
Hirata, H., Hinoda, Y., Shahryari, V., Deng, G., Nakajima, K., Tabatabai, Z. L., et al. (2015). Noncoding RNA MALAT1 promotes aggressive renal cell carcinoma through Ezh2 and interacts with miR-205. Cancer Res. 75, 1322–1331. doi: 10.1158/0008-5472.CAN-14-2931
Hsu, P. J., Zhu, Y., Ma, H., Guo, Y., Shi, X., Liu, Y., et al. (2017). Ythdc2 is an N6-methyladenosine binding protein that regulates mammalian spermatogenesis. Cell Res. 27, 1115–1127. doi: 10.1038/cr.2017.99
Huang, H., Weng, H., Sun, W., Qin, X., Shi, H., Wu, H., et al. (2018). Recognition of RNA N(6)-methyladenosine by IGF2BP proteins enhances mRNA stability and translation. Nat. Cell Biol. 20, 285–295. doi: 10.1038/s41556-018-0045-z
Huang, H., Weng, H., Zhou, K., Wu, T., Zhao, B. S., Sun, M., et al. (2019). Histone H3 trimethylation at lysine 36 guides m(6)A RNA modification co-transcriptionally. Nature 567, 414–419. doi: 10.1038/s41586-019-1016-7
Hysolli, E., Tanaka, Y., Su, J., Kim, K. Y., Zhong, T., Janknecht, R., et al. (2016). Regulation of the DNA methylation landscape in human somatic cell reprogramming by the miR-29 family. Stem Cell Rep. 7, 43–54. doi: 10.1016/j.stemcr.2016.05.014
Jia, G., Fu, Y., Zhao, X., Dai, Q., Zheng, G., Yang, Y., et al. (2011). N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat. Chem. Biol. 7, 885–887. doi: 10.1038/nchembio.687
Jonas, S., and Izaurralde, E. (2015). Towards a molecular understanding of microRNA-mediated gene silencing. Nat. Rev. Genet. 16, 421–433. doi: 10.1038/nrg3965
Knuckles, P., Carl, S. H., Musheev, M., Niehrs, C., Wenger, A., and Bühler, M. (2017). RNA fate determination through cotranscriptional adenosine methylation and microprocessor binding. Nat. Struct. Mol. Biol. 24, 561–569. doi: 10.1038/nsmb.3419
Knuckles, P., Lence, T., Haussmann, I. U., Jacob, D., Kreim, N., Carl, S. H., et al. (2018). Zc3h13/Flacc is required for adenosine methylation by bridging the mRNA-binding factor Rbm15/Spenito to the m(6)A machinery component Wtap/Fl(2)d. Genes Dev. 32, 415–429. doi: 10.1101/gad.309146.117
Koranda, J. L., Dore, L., Shi, H., Patel, M. J., Vaasjo, L. O., Rao, M. N., et al. (2018). Mettl14 is essential for epitranscriptomic regulation of striatal function and learning. Neuron 99, 283–292.e5. doi: 10.1016/j.neuron.2018.06.007
Lavi, S., and Shatkin, A. J. (1975). Methylated simian virus 40-specific RNA from nuclei and cytoplasm of infected BSC-1 cells. Proc. Natl. Acad. Sci. U.S.A. 72, 2012–2016. doi: 10.1073/pnas.72.6.2012
Lence, T., Paolantoni, C., Worpenberg, L., and Roignant, J. Y. (2019). Mechanistic insights into m6A RNA enzymes. Biochim. Biophys. Acta Gene Regul. Mech. 1862, 222–229. doi: 10.1016/j.bbagrm.2018.10.014
Lesbirel, S., Viphakone, N., Parker, M., Parker, J., Heath, C., Sudbery, I., et al. (2018). The m(6)A-methylase complex recruits TREX and regulates mRNA export. Sci. Rep. 8:13827. doi: 10.1038/s41598-018-32310-8
Leucci, E., Patella, F., Waage, J., Holmstrøm, K., Lindow, M., Porse, B., et al. (2013). microRNA-9 targets the long non-coding RNA MALAT1 for degradation in the nucleus. Sci. Rep. 3:2535. doi: 10.1038/srep02535
Li, A., Chen, Y. S., Ping, X. L., Yang, X., Xiao, W., Yang, Y., et al. (2017). Cytoplasmic m6A reader YTHDF3 promotes mRNA translation. Cell Res. 27, 444–447. doi: 10.1038/cr.2017.10
Li, Y., Syed, J., and Sugiyama, H. (2016). RNA-DNA triplex formation by long noncoding RNAs. (2016). Cell Chem. Biol. 23, 1325–1333. doi: 10.1016/j.chembiol.2016.09.011
Linder, B., Grozhik, A. V., Olarerin-George, A. O., Meydan, C., Mason, C. E., and Jaffrey, S. R. (2015). Single-nucleotide-resolution mapping of m6A and m6Am throughout the transcriptome. Nat. Methods 12, 767–772. doi: 10.1038/nmeth.3453
Liu, J., Yue, Y., Han, D., Wang, X., Fu, Y., Zhang, L., et al. (2014). A METTL3-METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation. Nat. Chem. Biol. 10, 93–95. doi: 10.1038/nchembio.1432
Liu, N., Dai, Q., Zheng, G., He, C., Parisien, M., and Pan, T. (2015). N(6)-methyladenosine-dependent RNA structural switches regulate RNA-protein interactions. Nature 518, 560–564. doi: 10.1038/nature14234
Lv, J., Zhang, Y., Gao, S., Zhang, C., Chen, Y., Li, W., et al. (2018). Endothelial-specific m(6)A modulates mouse hematopoietic stem and progenitor cell development via Notch signaling. Cell Res. 28, 249–252. doi: 10.1038/cr.2017.143
Ma, J. Z., Yang, F., Zhou, C. C., Liu, F., Yuan, J. H., Wang, F., et al. (2017). METTL14 suppresses the metastatic potential of hepatocellular carcinoma by modulating N(6) -methyladenosine-dependent primary MicroRNA processing. Hepatology 65, 529–543. doi: 10.1002/hep.28885
Mendel, M., Chen, K. M., Homolka, D., Gos, P., Pandey, R. R., McCarthy, A. A., et al. (2018). Methylation of structured RNA by the m(6)A Writer METTL16 Is Essential for Mouse Embryonic Development. Mol. Cell 71, 986–1000.e11. doi: 10.1016/j.molcel.2018.08.004
Meyer, K. D., and Jaffrey, S. R. (2014). The dynamic epitranscriptome: N6-methyladenosine and gene expression control. Nat. Rev. Mol. Cell Biol. 15, 313–326. doi: 10.1038/nrm3785
Meyer, K. D., Patil, D. P., Zhou, J., Zinoviev, A., Skabkin, M. A., Elemento, O., et al. (2015). 5′ UTR m(6)A promotes cap-independent translation. Cell 163, 999–1010. doi: 10.1016/j.cell.2015.10.012
Meyer, K. D., Saletore, Y., Zumbo, P., Elemento, O., Mason, C. E., and Jaffrey, S. R. (2012). Comprehensive analysis of mRNA methylation reveals enrichment in 3′ UTRs and near stop codons. Cell 149, 1635–1646. doi: 10.1016/j.cell.2012.05.003
Min, K. W., Zealy, R. W., Davila, S., Fomin, M., Cummings, J. C., Makowsky, D., et al. (2018). Profiling of m6A RNA modifications identified an age-associated regulation of AGO2 mRNA stability. Aging Cell 17:e12753. doi: 10.1111/acel.12753
Moindrot, B., Cerase, A., Coker, H., Masui, O., Grijzenhout, A., Pintacuda, G., et al. (2015). A pooled shRNA screen identifies Rbm15, Spen, and Wtap as factors required for Xist RNA-mediated silencing. Cell Rep. 12, 562–572. doi: 10.1016/j.celrep.2015.06.053
Müller, S., Glaß, M., Singh, A. K., Haase, J., Bley, N., Fuchs, T., et al. (2019). IGF2BP1 promotes SRF-dependent transcription in cancer in a m6A- and miRNA-dependent manner. Nucleic Acids Res. 47, 375–390. doi: 10.1093/nar/gky1012
Nakagawa, S., Ip, J. Y., Shioi, G., Tripathi, V., Zong, X., Hirose, T., et al. (2012). Malat1 is not an essential component of nuclear speckles in mice. RNA 18, 1487–1499. doi: 10.1261/rna.033217.112
Patil, D. P., Chen, C. K., Pickering, B. F., Chow, A., Jackson, C., Guttman, M., et al. (2016). m(6)A RNA methylation promotes XIST-mediated transcriptional repression. Nature 537, 369–373. doi: 10.1038/nature19342
Patil, D. P., Pickering, B. F., and Jaffrey, S. R. (2018). Reading m(6)A in the transcriptome: m(6)A-binding proteins. Trends Cell Biol. 28, 113–127. doi: 10.1016/j.tcb.2017.10.001
Pendleton, K. E., Chen, B., Liu, K., Hunter, O. V., Xie, Y., Tu, B. P., et al. (2017). The U6 snRNA m(6)A methyltransferase METTL16 regulates SAM synthetase intron retention. Cell 169, 824–835.e14. doi: 10.1016/j.cell.2017.05.003
Perry, R. P., and Kelley, D. E. (1974). Existence of methylated messenger RNA in mouse L cells. Cell 1, 37–42. doi: 10.1016/0092-8674(74)90153-6
Ping, X. L., Sun, B. F., Wang, L., Xiao, W., Yang, X., Wang, W. J., et al. (2014). Mammalian WTAP is a regulatory subunit of the RNA N6-methyladenosine methyltransferase. Cell Res. 24, 177–189. doi: 10.1038/cr.2014.3
Roundtree, I. A., Luo, G. Z., Zhang, Z., Wang, X., Zhou, T., Cui, Y., et al. (2017). YTHDC1 mediates nuclear export of N(6)-methyladenosine methylated mRNAs. eLife 6:e31311. doi: 10.7554/eLife.31311
Shah, A., Rashid, F., Awan, H. M., Hu, S., Wang, X., Chen, L., et al. (2017). The DEAD-Box RNA helicase DDX3 interacts with m6A RNA demethylase ALKBH5. Stem Cells Int. 2017:8596135. doi: 10.1155/2017/8596135
Shi, H., Wang, X., Lu, Z., Zhao, B. S., Ma, H., Hsu, P. J., et al. (2017). YTHDF3 facilitates translation and decay of N6-methyladenosine-modified RNA. Cell Res. 27, 315–328. doi: 10.1038/cr.2017
Shima, H., Matsumoto, M., Ishigami, Y., Ebina, M., Muto, A., Sato, Y., et al. (2017). S-Adenosylmethionine synthesis is regulated by selective N6-adenosine methylation and mRNA degradation involving METTL16 and YTHDC1. Cell Rep. 21, 3354–3363. doi: 10.1016/j.celrep.2017.11.092
Spitale, R. C., Flynn, R. A., Zhang, Q. C., Crisalli, P., Lee, B., Jung, J. W., et al. (2015). Structural imprints in vivo decode RNA regulatory mechanisms. Nature 519, 486–490. doi: 10.1038/nature14263
Tripathi, V., Ellis, J. D., Shen, Z., Song, D. Y., Pan, Q., Watt, A. T., et al. (2010). The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Mol. Cell 39, 925–938. doi: 10.1016/j.molcel.2010.08.011
Wang, H., Zuo, H., Liu, J., Wen, F., Gao, Y., Zhu, X., et al. (2018). Loss of YTHDF2-mediated m(6)A-dependent mRNA clearance facilitates hematopoietic stem cell regeneration. Cell Res. 28, 1035–1038. doi: 10.1038/s41422-018-0082-y
Wang, X., Zhao, B. S., Roundtree, I. A., Lu, Z., Han, D., Ma, H., et al. (2015). N(6)-methyladenosine modulates messenger RNA translation efficiency. Cell 161, 1388–1399. doi: 10.1016/j.cell.2015.05.014
Wang, Y., Li, Y., Toth, J. I., Petroski, M. D., Zhang, Z., and Zhao, J. C. (2014). N6-methyladenosine modification destabilizes developmental regulators in embryonic stem cells. Nat. Cell Biol. 16, 191–198. doi: 10.1038/ncb2902
Wang, X., Lu, Z., Gomez, A., Hon, G. C., Yue, Y., Han, D., et al. (2014). N6-methyladenosine-dependent regulation of messenger RNA stability. Nature 505, 117–120. doi: 10.1038/nature12730
Warda, A. S., Kretschmer, J., Hackert, P., Lenz, C., Urlaub, H., Höbartner, C., et al. (2017). Human METTL16 is a N(6)-methyladenosine (m(6)A) methyltransferase that targets pre-mRNAs and various non-coding RNAs. EMBO Rep. 18, 2004–2014. doi: 10.15252/embr.201744940
Wei, J., Liu, F., Lu, Z., Fei, Q., Ai, Y., He, P. C., et al. (2018). Differential m(6)A, m(6)A(m), and m(1)A demethylation mediated by FTO in the cell nucleus and cytoplasm. Mol. Cell 71, 973–985.e5. doi: 10.1016/j.molcel.2018.08.011
Wen, J., Lv, R., Ma, H., Shen, H., He, C., Wang, J., et al. (2018). Zc3h13 regulates nuclear RNA m(6)A methylation and mouse embryonic stem cell self-renewal. Mol. Cell 69, 1028–1038.e6. doi: 10.1016/j.molcel.2018.02.015
Wojtas, M. N., Pandey, R. R., Mendel, M., Homolka, D., Sachidanandam, R., and Pillai, R. S. (2017). Regulation of m6A transcripts by the 3′5′ RNA helicase YTHDC2 is essential for a successful meiotic program in the mammalian germline. Mol. Cell 68, 374–387.e12. doi: 10.1016/j.molcel.2017.09.021
Xi, Z., Wang, P., Xue, Y., Shang, C., Liu, X., Ma, J., et al. (2017). Overexpression of miR-29a reduces the oncogenic properties of glioblastoma stem cells by downregulating quaking gene isoform 6. Oncotarget 8, 24949–24963. doi: 10.18632/oncotarget.15327
Xiao, H., Tang, K., Liu, P., Chen, K., Hu, J., Zeng, J., et al. (2015). LncRNA MALAT1 functions as a competing endogenous RNA to regulate ZEB2 expression by sponging miR-200s in clear cell kidney carcinoma. Oncotarget 6, 38005–38015. doi: 10.18632/oncotarget.5357
Xiao, S., Cao, S., Huang, Q., Xia, L., Deng, M., Yang, M., et al. (2019). The RNA N(6)-methyladenosine modification landscape of human fetal tissues. Nat. Cell Biol. 21, 651–661. doi: 10.1038/s41556-019-0315-4
Xiao, W., Adhikari, S., Dahal, U., Chen, Y. S., Hao, Y. J., Sun, B. F., et al. (2016). Nuclear m(6)A reader YTHDC1 regulates mRNA splicing. Mol. Cell 61, 507–519. doi: 10.1016/j.molcel.2016.01.012
Yang, D., Qiao, J., Wang, G., Lan, Y., Li, G., Guo, X., et al. (2018). N6-methyladenosine modification of lincRNA 1281 is critically required for mESC differentiation potential. Nucleic Acids Res. 46, 3906–3920. doi: 10.1093/nar/gky130
Yang, L., Lin, C., Liu, W., Zhang, J., Ohgi, K. A., Grinstein, J. D., et al. (2011). ncRNA- and Pc2 methylation-dependent gene relocation between nuclear structures mediates gene activation programs. Cell 147, 773–788. doi: 10.1016/j.cell.2011.08.054
Yang, Z., Li, J., Feng, G., Gao, S., Wang, Y., Zhang, S., et al. (2017). MicroRNA-145 modulates N6-methyladenosine levels by targeting the 3′-untranslated mRNA region of the N6-methyladenosine binding YTH domain family 2 protein. J. Biol. Chem. 292, 3614–3623. doi: 10.1074/jbc.M116.749689
Yao, Q. J., Sang, L., Lin, M., Yin, X., Dong, W., Gong, Y., et al. (2018). Mettl3-Mettl14 methyltransferase complex regulates the quiescence of adult hematopoietic stem cells. Cell Res. 28, 952–954. doi: 10.1038/s41422-018-0062-2
Yue, Y., Liu, J., Cui, X., Cao, J., Luo, G., Zhang, Z., et al. (2018). VIRMA mediates preferential m(6)A mRNA methylation in 3′UTR and near stop codon and associates with alternative polyadenylation. Cell Discov. 4:10. doi: 10.1038/s41421-018-0019-0
Zhang, B., Arun, G., Mao, Y. S., Lazar, Z., Hung, G., Bhattacharjee, G., et al. (2012). The lncRNA Malat1 is dispensable for mouse development but its transcription plays a cis-regulatory role in the adult. Cell Rep. 2, 111–123. doi: 10.1016/j.celrep.2012.06.003
Zhang, J., Bai, R., Li, M., Ye, H., Wu, C., Wang, C., et al. (2019). Excessive miR-25-3p maturation via N6-methyladenosine stimulated by cigarette smoke promotes pancreatic cancer progression. Nat. Commun. 10:1858. doi: 10.1038/s41467-019-09712-x
Zhang, S., Zhao, B. S., Zhou, A., Lin, K., Zheng, S., Lu, Z., et al. (2017). m6A demethylase ALKBH5 maintains tumorigenicity of glioblastoma stem-like cells by sustaining FOXM1 expression and cell proliferation program. Cancer Cell 31, 591–606.e6. doi: 10.1016/j.ccell.2017.02.013
Zhao, B. S., Roundtree, I. A., and He, C. (2017). Post-transcriptional gene regulation by mRNA modifications. Nat. Rev. Mol. Cell. Biol. 1, 31–42. doi: 10.1038/nrm.2016.132
Zheng, G., Dahl, J. A., Niu, Y., Fedorcsak, P., Huang, C. M., Li, C. J., et al. (2013). ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol. Cell. 49, 18–29. doi: 10.1016/j.molcel.2012.10.015
Keywords: epitranscriptomics, m6A, RNA modifications, non-coding RNAs, microRNAs, lncRNAs, cell reprogramming, ESC development
Citation: Fazi F and Fatica A (2019) Interplay Between N6-Methyladenosine (m6A) and Non-coding RNAs in Cell Development and Cancer. Front. Cell Dev. Biol. 7:116. doi: 10.3389/fcell.2019.00116
Received: 27 March 2019; Accepted: 07 June 2019;
Published: 28 June 2019.
Edited by:
Karthikeyan Narayanan, West Virginia University, United StatesReviewed by:
Shizuka Uchida, University of Louisville, United StatesSaba Valadkhan, Case Western Reserve University, United States
Copyright © 2019 Fazi and Fatica. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Francesco Fazi, ZnJhbmNlc2NvLmZhemlAdW5pcm9tYTEuaXQ=; Alessandro Fatica, YWxlc3NhbmRyby5mYXRpY2FAdW5pcm9tYTEuaXQ=
 Francesco Fazi
Francesco Fazi Alessandro Fatica
Alessandro Fatica