Corrigendum: Genomic Characterization of mcr-1-carrying Salmonella enterica Serovar 4,[5],12:i:- ST 34 Clone Isolated From Pigs in China
- 1Institute of Preventive Veterinary Sciences, Department of Veterinary Medicine, College of Animal Sciences, Zhejiang University, Hangzhou, China
- 2Zhejiang Province Center for Disease Control and Prevention, Hangzhou, China
- 3College of Veterinary Medicine, Henan University of Animal Husbandry and Economy, Zhengzhou, China
- 4Zhejiang Provincial Key Laboratory of Preventive Veterinary Medicine, College of Animal Sciences, Zhejiang University, Hangzhou, China
Salmonella enterica serovar 4,[5],12:i:-, so-called Typhimurium monophasic variant, has become one of the most frequently isolated serovars both in humans and in animals all over the world. The increasing prevalence of mcr-1-carrying Salmonella poses significant global health concerns. However, the potential role of Salmonella 4,[5],12:i:- in mcr-1 gene migration through the food chain to the human remains obscure. Here, we investigated 337 Salmonella isolates from apparently healthy finishing pigs, which is rarely studied, obtained from pig farms and slaughterhouses in China. The mcr-1 gene was found in four colistin-resistant S. enterica 4,[5],12:i:- isolates. Notably, all four isolates belonged to sequence type 34 (ST34) with multidrug resistance phenotype. Further genomic sequencing and antimicrobial resistance characterization confirmed that mcr was responsible for the colistin resistance, and the conjugation assay demonstrated that three of four isolates carried mcr-1 in IncHI2 plasmid. Importantly, mcr-1 and class-1 integron were found to co-localize in two strains with IncHI2 plasmid. By collecting all the mcr-1-carrying Typhimurium and monophasic variant strains across the food chain (farm animals, animal-origin food, and humans), our phylogenomic analysis of available 66 genomes, including four strains in this study, demonstrated an independent phylogenetic cluster of all eight Chinese swine-originated isolates and one human isolate. Together, this study provides direct evidence for clonal and pork-borne transmission of mcr-1 by Salmonella 4,[5],12:i:- ST34 in China and highlighted a domestication pathway by acquisition of additional antimicrobial resistance determinants in Chinese ST34 isolates.
Introduction
Salmonella spp. are important zoonotic pathogen commonly identified in farmed livestock. A particular monophasic variant of Salmonella Typhimurium (Salmonella 4,[5],12:i:-) emerged in the mid-1990s and became one of the most widespread serovars (Hopkins et al., 2010; Wang et al., 2019). Recently, some studies proposed it as the global pandemic clone (Alicia et al., 2018; Mulvey et al., 2018; Biswas et al., 2019). Importantly, Salmonella 4,[5],12:i:- were suggested to be associated with swine (Linxian et al., 2017) and currently considered with high frequency of multidrug resistance potential among Salmonella serovars, posing significant public health concerns worldwide (Elnekave et al., 2018; Mather et al., 2018; Monte et al., 2019).
Salmonella serovar 4,[5],12:i:- is also commonly found in isolates from humans in China and has been linked to food animals and animal-borne products, particularly swine and pork (Linxian et al., 2017; Paudyal et al., 2018). Pork products are considered as one of the main sources of human Salmonella infections (Lu et al., 2019).
As a result of the extensive use of colistin for veterinary purposes especially for the control and prophylaxis of Enterobacteriaceae infections, the mobilized colistin-resistant (mcr) Salmonella 4,[5],12:i:- has been disseminated in humans, animals, as well as food products, including pork (Linxian et al., 2017; Alicia et al., 2018; Elbediwi et al., 2019). mcr carrying Salmonella 4,[5],12:i:- has also been reported from Asia, European countries, and North America (Monte et al., 2019). However, the study of Salmonella 4,[5],12:i:- in the finishing pigs is largely lacking and the direct evidence of the pork-borne transmission, particularly in certain critical antimicrobial resistance, i.e., mobile colistin resistance gene migration, remains obscure in China. Here, by focusing on the asymptomatic finishing pigs, an underappreciated modulator in the pork-borne transmission chain, we aimed to characterize the genomic features and evaluated the potential role of pork-borne transmission for mcr-1-carrying Salmonella 4,[5],12:i:-.
Materials and Methods
Sample Collection
Randomized sampling was done as a part of epidemiological surveillance for detection of colistin-resistant Salmonella isolates in finishing pigs. Between March 2017 and November 2017, a total of 1732 fecal samples were collected from randomly selected 45 pig farms, and two pig slaughtering facilities at Henan province in China.
Isolation and Identification of Salmonella Isolates
The isolation, identification, and serotyping of the organisms were done according to previous protocols (Jiang et al., 2019). The obtained isolates were then confirmed as Salmonella by polymerase chain reaction (PCR) using specific primers for amplification of the enterotoxin stn gene as recommended (Zhu et al., 2015). Monophasic S. Typhimurium isolates were serotyped by O-, H- antigens (Jiang et al., 2019) and confirmed by PCR (Bugarel et al., 2012), as described previously.
Antimicrobial Susceptibility Testing
Broth micro-dilution method was used to determine the minimum inhibitory concentrations (MICs) of 16 antimicrobial drugs for all isolates and results were interpreted according to CLSI protocols (Chattopadhyay et al., 2013). The antimicrobials used include colistin, ampicillin, amoxicillin–clavulanic acid, chloramphenicol, streptomycin, florfenicol, tetracycline, kanamycin, doxycycline, gentamicin, sulfamethoxazole, sulfamethoxazole/trimethoprim, ciprofloxacin, enrofloxacin, ceftriaxone, and cefotaxime.
PCR Screening of mcr Genes
The genomic DNA was extracted from the isolates and subjected for screening mcr genes of various types (mcr-1 to mcr-8), using a multiplex PCR as recommended (Wang et al., 2018). To elaborate, PCR amplification was performed using iTaq TM DNA polymerase with the following cycling conditions: 34 cycles of 94°C for 20 s, 50°C for 20 s, and 72°C for 30 s, followed by 1 cycle of 72°C for 5 min.
Bacterial Conjugation Assay
Bacterial conjugation for the mcr-1 positive Salmonella isolates was done in a liquid and solid mating-out assay (Lampkowska et al., 2008) using Escherichia coli J53 (streptomycin- and rifampicin-resistant) to detect the transferability of the gene. Trans-conjugants were selected on LB agar plates containing rifampicin (100 mg/L) and colistin (2 mg/L). The conjugation efficiency rate was estimated as a number of trans-conjugants per total recipients.
Genomic Sequencing and Data Analysis
The genomic DNA was first extracted and then sequenced on the Illumina MiSeq platform. The quality of sequencing and trimming was checked with FastQC toolkit (Bolger et al., 2014). The raw reads for each strain were assembled by using SPAdes 4.0.1. PLACNETw (Vielva et al., 2017) web tool was used to reconstruct the plasmid genome from the whole-genome sequence. The reconstructed plasmid contigs were aligned against the non-redundant database1 to find the best plasmid match. QUAST (Gurevich et al., 2013) was used to assess the assembled genomes through basic statistics generation, including the total number of contig, the length of contig, and N50. Prokka 1.14, with the “default” settings, was used to annotate the assembled genomes. The Genomic DNA library was constructed using Nextera XT DNA library construction kit (Illumina, United States, No. FC-131-1024), followed by genomic sequencing using Miseq Reagent Kit v2 300cycle kit (Illumina, United States, No. MS-102-2002). High-throughput genome sequencing was accomplished by the Illumina Miseq sequencing platform, as previously described (Paudyal et al., 2019; Biswas et al., 2020; Yu et al., 2020).
Salmonella monophasic in silico serotyping was done by SISTR (Yoshida et al., 2016) web tool. Multilocus sequence typing (MLST), detection of resistance genes, and plasmid replicon were conducted in the Center for Genomic Epidemiology (CGE)2 platform. Blastn3 and BRIG (Alikhan et al., 2011) tools were used to separate the plasmid contigs of the mcr-1-carrying plasmid and display circular comparison. Snapgene software4 was used to display the full annotated versions of the mcr-1-carrying plasmids (Supplementary Figures S1–S3). Pan-genome analysis was performed using the Roary pipeline (version 3.4.2) (Page et al., 2015).
Phylogenomic analysis was used Snippy v4.4.4 to obtain SNPs alignment and the phylogenetic tree was built by the maximum likelihood method with RAxML5 based on the recombination-free SNPs. To identify the phylogenetic relationship of mcr-1-carrying isolates, four strains of swine origin isolated in this study and all available mcr-1 positive S. Typhimurium and monophasic Typhimurium genomic sequences were retrieved from GenBank, including two environmental, 14 from swine (live pigs and pork), 17 from other animals, and 31 from humans, and were pooled together for further phylogenetic analysis.
Results
Characterization of Salmonella Isolates From Pigs
A total of 337 Salmonella strains were isolated from 1732 fecal samples of healthy finishing pigs randomly selected from 45 pig farms and two slaughterhouses in Henan province in China between March 2017 and November 2017. S. Derby was the most abundant serovar in both farms and slaughterhouses (Figure 1A). The prevalence of S. Typhimurium and monophasic variant serovars in the farms was higher than that in slaughterhouses. We also noticed that the isolation rate of the monophasic S. Typhimurium serovar was higher than other S. Typhimurium in slaughterhouses (Figure 1A). Most of the isolates exhibited multidrug resistance (resistant to three or more different antimicrobials classes) pattern by showing resistance to β-lactams, sulfonamides, phenicols, and tetracycline classes. Notably, the monophasic Typhimurium isolates (N = 7) from finishing pigs showed the highest resistance to all examined antimicrobials (Figure 1B). Importantly, four monophasic S. Typhimurium isolates (Sal_13.3, Sal_13.5, Sal_136, and Sal_15.5) obtained from pig farms showed resistance to colistin (MIC = 4 mg/L). These four isolates were isolated in 2017; three of them (Sal_13.3, Sal_13.5, and Sal_13.6) were obtained from the same farm located in Jiyuan city; however, Sal_15.5 was obtained from another farm located in Changhui city.
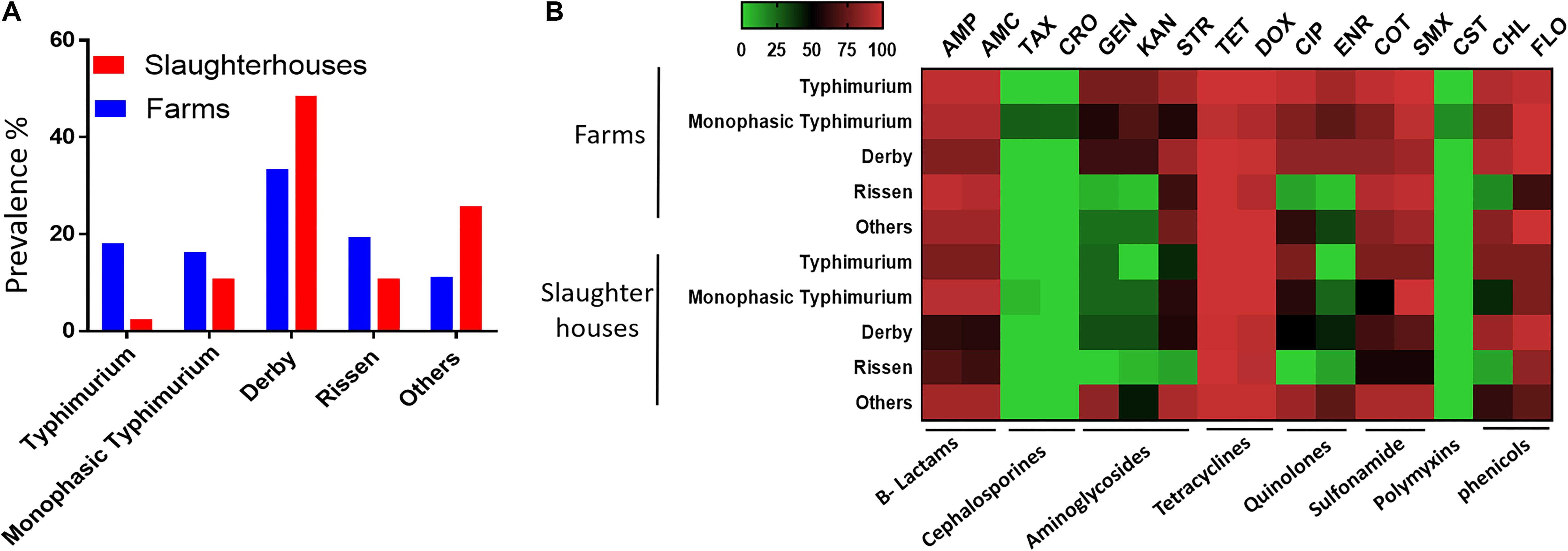
Figure 1. The serovar and antibiotic resistance diversity of Salmonella across farm and slaughterhouses. (A) Spatial variation of the distribution of multiple Salmonella serovars isolated from fecal samples analyzed in this study. (B) Distribution of the average resistance (in percent) of various serovars regardless of the source of isolation toward multiple antibiotics. The names of the antibiotics are abbreviated as ampicillin (AMP), amoxicillin (AMC), cefotaxime (TAX), ceftriaxone (CRO), gentamicin (GEN), kanamycin (KAN), streptomycin (STR), tetracycline (TET), doxycycline (DOX), ciprofloxacin (CIP), enrofloxacin (ENR), sulfa-trimethoprim (COT), sulfamethoxazole (SMX), colistin (CST), chloramphenicol (CHL), and florfenicol (FLO).
Polymerase chain reaction screening of all isolates also confirmed that only these four isolates harbored the mcr-1 gene. No positive isolates were identified from slaughterhouses. MLST subtyping and in silico serotyping, including the serum agglutination test and PCR assay, confirmed that the four strains were Salmonella enterica serovar 4,[5],12:i- and belonged to sequence type 34 (ST34) (Table 1).
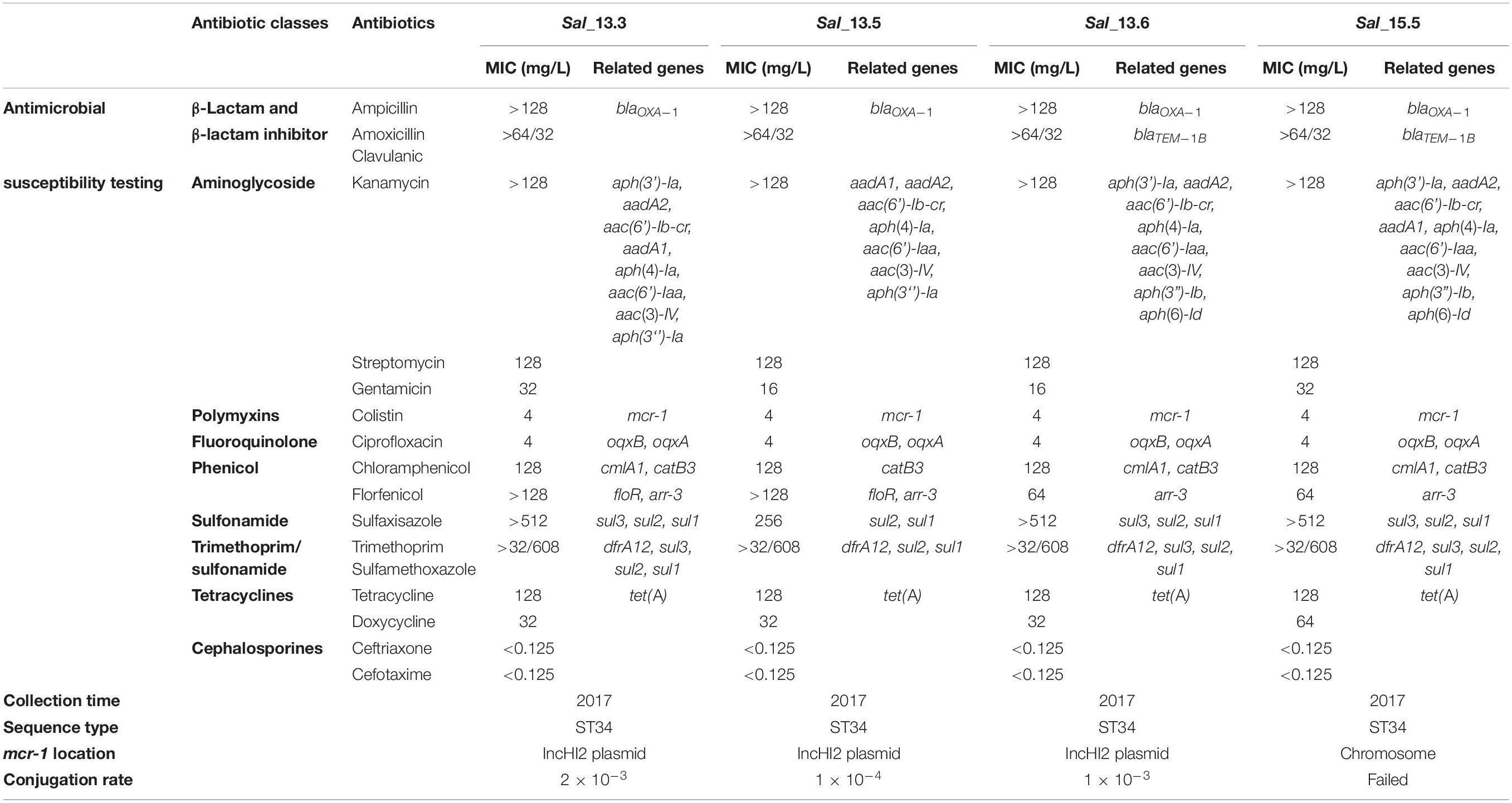
Table 1. Antimicrobial susceptibility, conjugation rate and whole genome analysis of mcr-1 positive strains isolated in this study.
Bacterial Conjugation Assay
The mcr-1 gene in Sal_13.3, Sal_13.5, and Sal_13.6 strains could be transmitted to E. coli J53 (streptomycin- and rifampicin-resistant) with a conjugation efficiency of 2 × 10–3, 1 × 10–4, and 1 × 10–3 per donor cell, respectively. However, conjugal transfer failed in Sal_15.5.
Antimicrobial Susceptibility and Related Genetic Determinants
All four mcr-carrying isolates showed multidrug resistance pattern but were sensitive to carbapenems and cephalosporines (Table 1). Phenotypic results correlated with the presence of the different antimicrobial resistance genes. As illustrated in Table 1, the four mcr-1-positive isolates exhibited MIC to colistin = 4 mg/L. They were phenotypically resistant to tetracyclines, which was confirmed by the presence of tet(A) gene. They also showed resistance to ampicillin and amoxicillin–clavulanic acid conferred by blaOXA–1 or blaTEM–1B (Table 1). Furthermore, co-resistance was observed for streptomycin, kanamycin, and gentamicin with an average of six different genes, which were responsible for aminoglycoside resistance. aac(6’)Ib-cr gene, responsible for resistance to the aminoglycosides in addition to quinolones, was also detected in all isolates. Sal_13.3 and Sal_13.5 were resistant to florfenicol (MIC > 128 mg/L) due to the presence of flor and arr-3 genes. However, Sal_13.6 and Sal_15.5 exhibited higher MIC value (64) and that may be due to the presence of the flor gene, which was specifically presented in these two isolates. All isolates showed resistance to chloramphenicol, which is encoded by cmlA1 and catB3, and ciprofloxacin, which is encoded by oqxB and oqxA. Importantly, mcr-1, along with one copy of Class-1 integron, has been identified in pSAL_13.3 and pSAL_13.6. Class-1 integron in both isolates have three copies of aph(4)-Ia and one copy of cmlA in their gene cassettes (Figure 2).
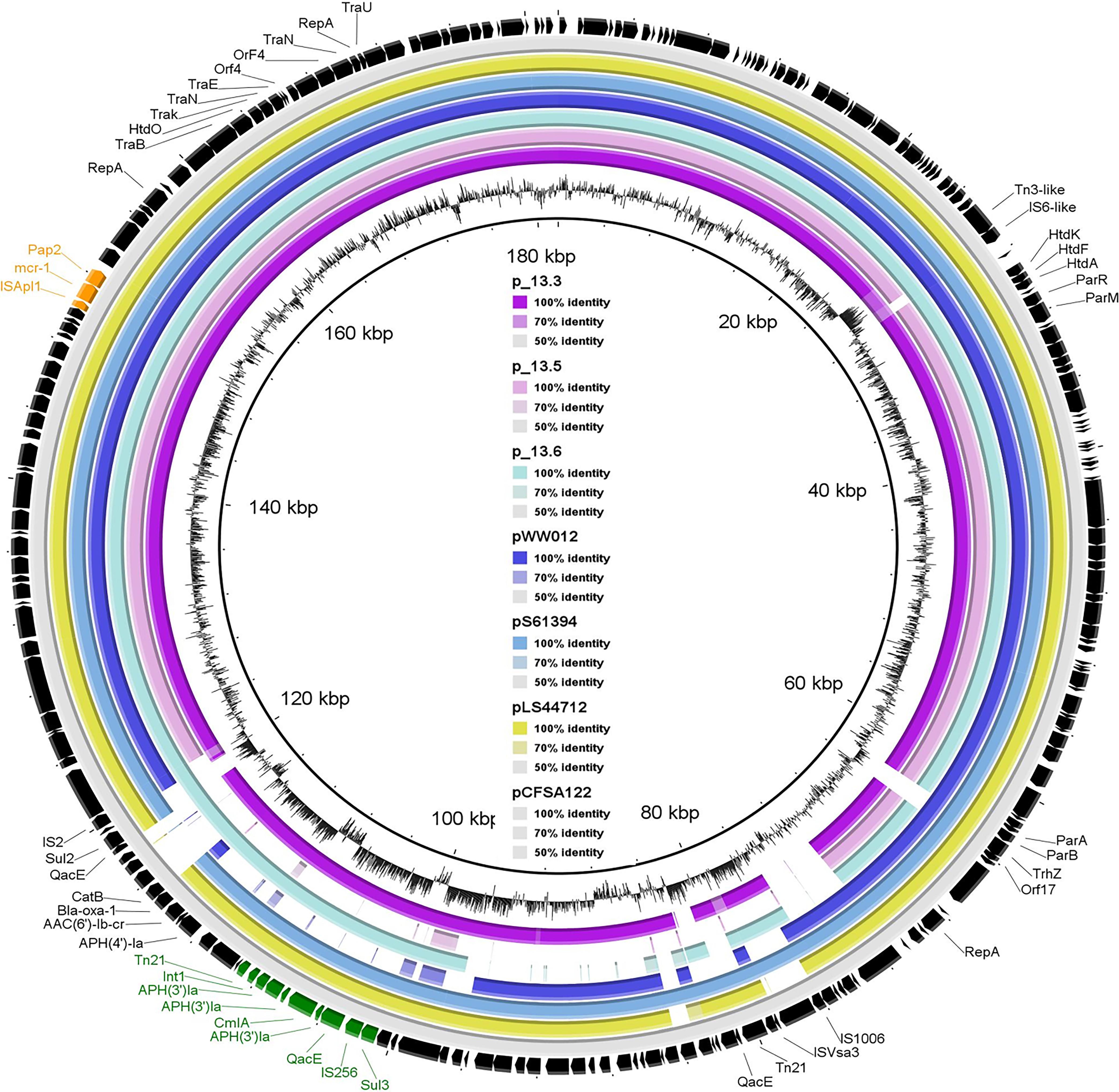
Figure 2. Sequence comparison of reconstructed mcr-1-positive plasmid from whole-genome sequence of SAL_13.3, SAL_13.5, SAL_13.6 with pLS44712 (NZ_CP035918), pS61394 (NZ_CP035916), pWW012 (NZ_CP022169), and pCFSA122-1 (NZ_CP033224.2) from a Chinese pork sample as a reference strain. Black arrows refer to pCFSA122-1, purple color refers to pSAL_13.3, pink color refers to pSAL_13.5, light blue color refers to SAL_13.6, dark blue color refers to pLS44712, blue color refers to pS61394, and yellow color refers to pWW012.
Genetic Background of the mcr-1 Gene
The mcr-1-coding sequence was located directly upstream of an open reading frame encoding PAP2 family protein, which is frequently associated with mcr-1. The complete version of ISApl1 located downstream of the mcr-1 cassette was detected in SAL_13.3, SAL_13.5, and SAL_13.6, and an incomplete version of ISApl1 was detected in SAL_15.5 (Figure 3).
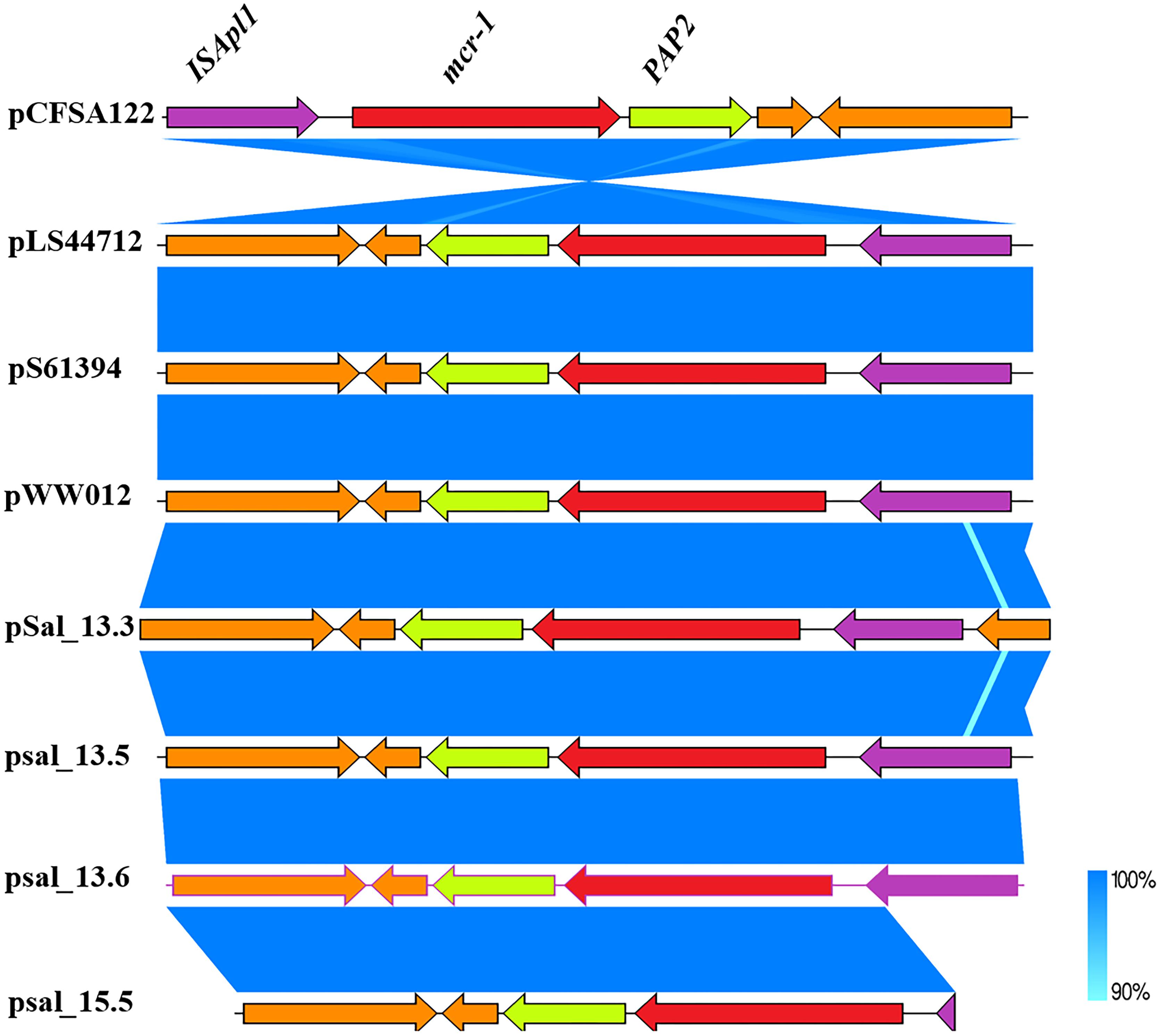
Figure 3. Genetic comparison of mcr-1 gene context between SAL_13.3, SAL_13.5, SAL_13.6 with pLS44712 (NZ_CP035918), pS61394 (NZ_CP035916), pWW012 (NZ_CP022169), and pCFSA122-1 (NZ_CP033224.2). N.B. Blue color in mcr-1 gene context refers to 90–100% identity and SAL_15.5 carries mcr-1 gene on chromosome.
The Plasmids Carrying mcr-1 Gene
The pSAL_13.3_mcr-1 (175,265 bp) (Supplementary Figure S1), pSAL_13.5_mcr-1 (163,786 bp) (Supplementary Figure S2), and pSAL_13.6_mcr-1 (171,333 bp) (Supplementary Figure S3) are IncHI2 plasmid, reconstructed from the whole genomic sequence by using PLANCETw. All mcr- plasmids were successfully transferred by conjugation to the recipient strain, signifying their ability to carry the mcr genes between isolates. For all isolates, genome sequencing allowed the detection of mcr-1 genes in the same contigs as replicons of the plasmid families. The plasmid sequence had a typical IncHI2-type backbone coding replication and conjugative transfer region, and also carried IncF replicons. In general, the structure of the IncHI2 plasmids seemed to present high similarity, with all of them having the conjugative transfer system, HigB-HigA toxin–antitoxin system for plasmid maintenance, and a tellurium resistant operon (Supplementary Figures S1–S3). Additionally, these plasmids also harbored antimicrobial resistance genes of different categories, including aminoglycosides, beta-lactams, tetracycline, phenicols, sulfonamides, and trimethoprim (Figure 2 and Supplementary Figure S4). They also showed >95% nucleotide sequence identity to the corresponding region of the mcr-1 positive InCHI2 plasmids obtained from Chinese S. enterica isolates pLS44712 (NZ_CP035918), pS61394 (NZ_CP035916), pWW012 (NZ_CP022169), and pCFSA122-1 (NZ_CP033224.2) (Figure 2). The plasmids harbored mcr-1 along with many other resistance genes.
Comparative Genomics and Clonal Nature of Chinese mcr-Positive Isolates
In order to evaluate the relationship between the four mcr-carrying isolates and all mcr-1 positive S. Typhimurium and monophasic variant isolates from different countries and sources, the phylogenetic tree of 62 mcr-1 positive isolates, including 34 monophasic S. Typhimurium and 28 S. Typhimurium isolates with genomes available in the NCBI database, was used to test the clonal feature (Figure 4). Except for SAMN10914547, all Chinese, including pig, pork, and human isolates, were clustered together, composed of two closely related independent subclades, and the whole-genome sequencing of the pork and human isolates showed that they were monophasic S. Typhimurium or 4,[5],12:i:-, also belonging to ST34, and all of them, except SAMN10290237, have the mcr-1 gene carried on IncHI2 plasmids. Additionally, Pan-genome analysis with Roary pipeline tool (Page et al., 2015) exhibited similar patterns of the genomes of all Chinese isolates. Further analysis revealed that 4597 (85.5%) out of 5663 genes were conserved among the completed genomes of all nine Chinese isolates (Figure 4 and Supplementary Figure S5). These results suggest the vital role of the food chain in the dissemination of mcr-carrying S. Typhimurium in China. We also found that there is a small difference in distance between phylogenetic branches of all mcr-1-positive InCHI2 plasmids obtained from Chinese S. enterica isolates pLS44712 (NZ_CP035918), pS61394 (NZ_CP035916), pWW012 (NZ_CP022169), and pCFSA122-1 (NZ_CP033224.2), indicating the close relation among these plasmids, which are from the same Inc type (Supplementary Figure S6). Salmonella isolate CFSA12 was reported as a mutant strain that has lost the mcr-1 gene from its wild strain WW012 of serovar Typhimurium (Hu et al., 2019).
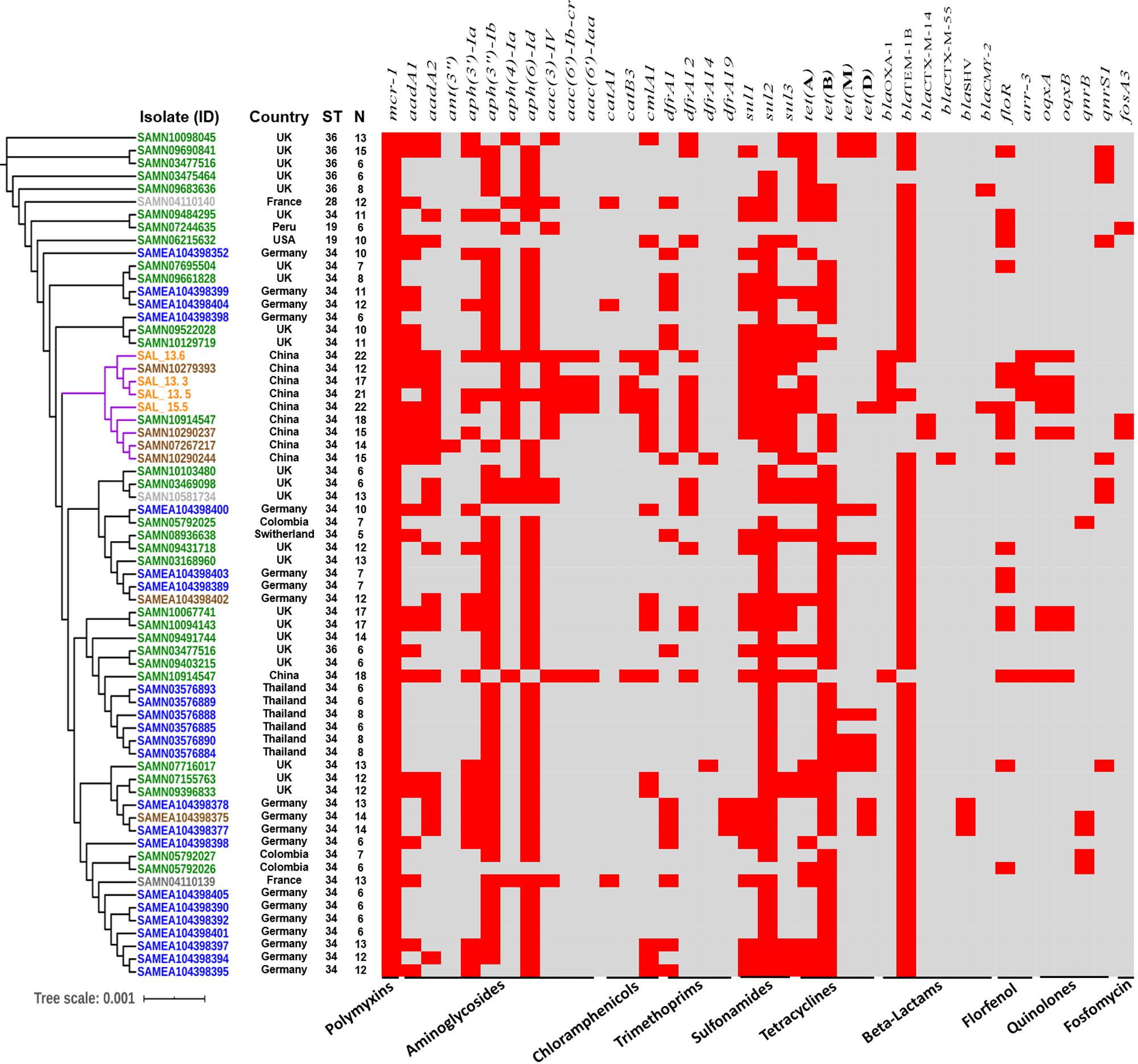
Figure 4. Global phylogenomic analysis of mcr-1-positive S. Typhimurium and monophasic isolates from different hosts retrieved from NCBI database (sample ID). Green color refers to human strains, blue color refers to animal strains, brown color refers to pork strains, gray color refers to environmental strains, and orange color refers to the isolated strains in this study. N refers to the numbers of resistant genes. ST refers to the sequence type.
Global Phylogenomic Analysis of mcr-1-Carrying S. Typhimurium
We retrieved all mcr-1-carrying Salmonella Typhimurium from the NCBI database in addition to the isolates obtained in this study to construct the phylogenomic tree (Figure 4). We noticed that mcr-1-carrying S. Typhimurium has been isolated from various sources like humans, animals, food products, and the environment. A small difference in distance between phylogenetic branches of the isolates was identified with a scale bar at 0.01, indicating a very close genetic relationship between the isolates obtained from different sources. The whole-genome analysis of all mcr-1-carrying isolates exhibited 58 of the 66 isolates, including those isolates reported in this study (Figure 4 and Supplementary Figure S7). These results highlighted an interesting host preference of mcr-1 gene and a worldwide prevalence of the mcr-positive Salmonella Typhimurium ST34. All isolates including the strains determined in this study carry antimicrobial resistance genes for aminoglycosides and β-lactams. aph(6)-Id (55 isolates, 83%) and blaTEM–1B (58 isolates, 87.8%) were the most detected genes among all the mcr-1-carrying isolates. BlaOXA–1 gene was only detected in Chinese isolates. We noticed that among all isolates, 47 (71%) were isolated between 2016 and 2018 (Supplementary Figure S7). Interestingly, the Chinese isolates have the highest number of antimicrobial resistance genes among all the examined isolates (Figure 3).
Discussion
Monophasic Salmonella 4,[5],12:i:- has become a global new epidemic multidrug-resistant clone associated with animal and human infections (Hopkins et al., 2010). For unknown reason, this particular clone was preferentially associated with swine, particularly in finishing herds, where the spillage of the intestinal contents during slaughter is a primary risk factor for the cross-contamination (Rodríguez and Suárez, 2014; Paudyal and Yue, 2019). As mentioned earlier, S. enterica 4,[5],12:i:- ST34 carrying mcr-1 have been reported in humans and pork (Li et al., 2016; Alicia et al., 2018). Here, S. enterica 4,[5],12:i:- isolated from asymptomatic finishing pigs were used to evaluate their role in mcr-1 gene transmission via the food chain.
We have investigated the prevalence of mcr-1 gene among Salmonella strains obtained from pigs in farms in Henan, China. Out of 337 Salmonella isolates, four (1.1%) isolates were positive for mcr-1. All other mcr genes (mcr-2 to mcr-8) have not been detected by the established multiplex PCR assay (Wang et al., 2018). The incidence rate of mcr-1 in Salmonella was much lower as compared to other Enterobacteriaceae (Liu et al., 2016; Malhotra-Kumar et al., 2016). The low prevalence of Salmonella harboring mcr-1 was also reported in other studies in China, England, and Wales (Doumith et al., 2016; Li et al., 2016).
All mcr-1-positive S. enterica 4,[5],12:i:- strains, belonging to ST34, were often related to an evolving multidrug-resistant S. enterica 4,[5],12:i:- clade in Australia, China, Italy, and United States (Li et al., 2016; Alicia et al., 2018; Elnekave et al., 2018). It is likely that the clonal dissemination of S. enterica 4,[5],12:i:- ST34 contributes to the spread of the mcr-1 gene among food animals in China (Lu et al., 2019) and may become a global significant public health concern (Elnekave et al., 2018; Mather et al., 2018; Monte et al., 2019).
It has been noticed that mcr-1 was carried also on IncHI2 plasmids in three of our strains, which is similar to that reported for S. Typhimurium ST34 (Linxian et al., 2017). Conjugation experiments confirmed the ability of all the isolates except SAL_15.5 to mobilize the antimicrobial-resistant gene to a recipient strain, and Genome sequencing data verified the presence of the conjugative determents. IncHI2/HI2A plasmids are typically large (García-Fernández and Carattoli, 2010), multidrug-resistant plasmids that have been accompanied by a range of antimicrobial and metal resistance genes in Salmonella species from humans and food-producing animals (Linxian et al., 2017; Elnekave et al., 2018; Mather et al., 2018; Paudyal et al., 2018; Biswas et al., 2019; Elbediwi et al., 2019; Lu et al., 2019; Monte et al., 2019). The presence of IncHI2 plasmids in Salmonella serovars indicates that horizontal transfer of mcr-1-harboring plasmids might have also contributed to the spread of mcr-1 and other resistant determinants in these bacteria (Liu et al., 2018). IncHI2 plasmids also carried a diversity of antimicrobial resistance genes from different categories, including aminoglycosides, beta-lactams, tetracycline, sulfonamides, and phenicols. Common antimicrobials were used to be administered during the rearing cycle in pig production and could persist for a long period in food-producing animals. These plasmids also contained genes encoding small multidrug resistance efflux transporter (QacE) conferring resistance to quaternary ammonium compounds (QACs). QAC has been commonly used as disinfectants with a wide application in the food industry (Quinn et al., 2015). Resistance to disinfectants presumably confers these clones the capacity to select and survive under available extreme conditions.
The mcr-1 flanking regions have also been reported in previous studies (Li et al., 2016; Malhotra-Kumar et al., 2016; Linxian et al., 2017). The ISApl1 flanking mcr-1 gene seems to play a crucial role in the dissemination of mcr-1 transposition between various incompatibility families of plasmids (Snesrud et al., 2016; Poirel et al., 2017), particularly in IncHI2 plasmids (Li et al., 2016; Linxian et al., 2017). In this study, we detected complete and incomplete copies of ISApl1 element downstream the mcr-1 gene, fixing the mcr-1 gene to the plasmid. These differences in the surrounding regions of mcr-1 probably indicate different stages in the evolution of the plasmid (Snesrud et al., 2016) or due to inadequate sequencing depth and coverage.
Salmonella serovars have a wide host range and can be transmitted to a broad diversity of animals, including mammals, birds, fish, and insects (Pan et al., 2019). Besides, Salmonella can grow in plants and can survive in protozoa, soil, and water (Silva et al., 2014; Pan et al., 2018). Hence, broad-host-range Salmonella can be transmitted via feces from wild animals, farm animals, and pets or by consumption of a wide variety of common foods: poultry, beef, pork, seafood, milk, fruit, and vegetables (Pan et al., 2019; Elbediwi et al., 2020). Phylogenomic analysis of four strains, determined in this study, with all available mcr-1-carrying S. Typhimurium and monophasic isolates from swine, poultry, humans, and environment, showed that these four strains were closely related and clustered together with four additional Chinese pork isolates and one human isolate (Figure 4). However, in silico serotyping of these isolates were monophasic S. Typhimurim (4,[5],12:i:-), and besides sharing the same sequence type (ST34), all, except for SAMN10290237, have the mcr-1 gene carried on IncHI2 plasmids. In addition, there is very limited genetic difference in the distance between the branches of the evolutionary tree of the genomes, indicating the consistency with the sequence type results. We could not prove the potential role of the transmission by performing in vivo experiments. These findings suggested that pork, pigs, and human monophasic S. Typhimurim (4,[5],12:i:-) isolates might be from the same source, and pork-borne transmission played a crucial role in the transmission of mcr-1-carrying S. enterica 4,[5],12:i:- ST34. Further enhanced surveillance should pay particular attention to the IncHI2-mediated mcr-1 transmission in monophasic S. enterica ST34.
Notably, the closely related Chinese swine-originated isolates were reported from Henan and Guangxi provinces (Supplementary Figure S4), top pig producers in China with the density exceeding 100 hogs per 100 acres (USDA, 2009; Gale et al., 2012). Additionally, Yang et al. (2019) reported that these two provinces were the highest antibiotics-consuming hot spots of pig production in China.
Conclusion
This study provided essential knowledge of the pig–pork chain in the transmission of mcr-1 by Salmonella 4,[5],12:i:- in China. In addition, it highlighted the importance of the occurrence of IncHI2 plasmids in S. enterica 4,[5],12:i:-, which may act as a vehicle for the mcr-1 gene and multiple antimicrobial-resistant genes during their dissemination through the food chain. Furthermore, the spread of similar IncHI2-like plasmids and Salmonella serovar 4,[5],12:i:- clones carrying mcr-1 emphasizes the requirements for internationally coordinated response strategies and continuing surveillance to mitigate mcr-carrying bacteria dissemination.
Data Availability Statement
The datasets generated for this study can be found in the NCBI Bioproject, accession number PRJNA573539.
Ethics Statement
No ethical approval was required for the current study. Fecal samples were obtained from farm pigs, with the permission of the farmers. Live animals were not handled directly. Oral agreement and permission was obtained from the farmers as well as the slaughterhouse manager before the sampling.
Author Contributions
ME designed the study and prepared the first draft, figures, and tables. HP, ME, and BW did the data analysis. ZJ collected the samples and did the microbiological isolation. SB and YL reviewed the manuscript. MY and ME finalized the manuscript and managed the project. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the National Program on Key Research Project of China (2017YFC1600103, 2018YFD0500501, and 2019YFE0103900) as well as the European Union’s Horizon 2020 Research and Innovation Program under Grant Agreement No. 861917–SAFFI, the Zhejiang Provincial Natural Science Foundation of China (LR19C180001), and the Zhejiang Provincial Key R&D Program of China (2020C02032).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fbioe.2020.00663/full#supplementary-material
FIGURE S1 | Genetic structure of mcr-1-carrying pSal_13.3.
FIGURE S2 | Genetic structure of mcr-1-carrying pSal_13.5.
FIGURE S3 | Genetic structure of mcr-1-carrying pSal_13.6.
FIGURE S4 | Sequence comparison of reconstructed mcr-1-positive plasmid from whole-genome sequence of SAL_13.3, SAL_13.5, and SAL_13.6 with highlighting the presence of antimicrobial resistance genes.
FIGURE S5 | Phylogenomic analysis, metadata, and comparative genomics analysis of Chinese mcr-1-positive monophasic S. Typhimurium (4,[5],12:i:-) in addition to mcr-1 location in these isolates. Green color refers to human isolates, brown color refers to pork, and orange color refers to the isolated strains in this study. ∗Salmonella isolate CFSA12 was reported as a mutant strain that has lost the mcr-1 gene from its wild strain Salmonella Typhimurium WW012.
FIGURE S6 | Phylogenomic analysis of reconstructed mcr-1-positive plasmid from whole-genome sequence of SAL_13.3, SAL_13.5, and SAL_13.6 genome sequence of SAL_13.3, SAL_13.5, SAL_13.6 with pLS44712 (NZ_CP035918), pS61394 (NZ_CP035916), pWW012 (NZ_CP022169), and pCFSA122-1 (NZ_CP033224.2) from a Chinese pork sample as a reference strain.
FIGURE S7 | Circular comparison and phylogenomic analysis a of global mcr-1-positive S. Typhimurium and monophasic variant according to year and source of isolation, sequence type, and place of isolation. Ring 1 refers to year of isolation, ring 2 refers to sequence type, ring 3 refers to isolation sources, and ring 4 refers to the place of isolation.
Footnotes
- ^ ftp://ftp.ncbi.nlm.nih.gov/blast/db/FASTA/nr.gz
- ^ https://cge.cbs.dtu.dk/services/
- ^ http://blast.ncbi.nlm.nih.gov/Blast.cgi
- ^ https://www.snapgene.com/
- ^ http://phylobench.vital-it.ch/raxml-bb/index.php
References
Alicia, A., Qinning, W., Nathan, B., Rosemarie, S., Chayanika, B., Cristina, S., et al. (2018). Multidrug-resistant Salmonella enterica 4,[5],12:i:- sequence type 34, new south wales, Australia, 2016–2017. Emerg. Infect. Dis. J. 24, 751–753. doi: 10.3201/eid2404.171619
Alikhan, N.-F., Petty, N. K., Ben Zakour, N. L., and Beatson, S. A. (2011). BLAST ring image generator (BRIG): simple prokaryote genome comparisons. BMC Genomics 12:402. doi: 10.1186/1471-2164-12-402
Biswas, S., Elbediwi, M., Gu, G., and Yue, M. (2020). Genomic characterization of new variant of hydrogen sulfide (H2S)-producing Escherichia coli with multidrug resistance properties carrying the mcr-1 gene in china dagger. Antibiotics 9:80. doi: 10.3390/antibiotics9020080
Biswas, S., Li, Y., Elbediwi, M., and Yue, M. (2019). Emergence and dissemination of mcr-carrying clinically relevant Salmonella typhimurium monophasic clone ST34. Microorganisms 7:298. doi: 10.3390/microorganisms7090298
Bolger, A. M., Lohse, M., and Usadel, B. (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. doi: 10.1093/bioinformatics/btu170
Bugarel, M., Vignaud, M. L., Moury, F., Fach, P., and Brisabois, A. (2012). Molecular identification in monophasic and nonmotile variants of Salmonella enterica serovar Typhimurium. Microbio. Open 1, 481–489. doi: 10.1002/mbo3.39
Chattopadhyay, S., Taub, F., Paul, S., Weissman, S. J., and Sokurenko, E. V. (2013). Microbial variome database: point mutations, adaptive or not, in bacterial core genomes. Mol. Biol. Evol. 30, 1465–1470. doi: 10.1093/molbev/mst048
Doumith, M., Godbole, G., Ashton, P., Larkin, L., Dallman, T., Day, M., et al. (2016). Detection of the plasmid-mediated mcr-1 gene conferring colistin resistance in human and food isolates of Salmonella enterica and Escherichia coli in England and Wales. J. Antimicrob. Chemother. 71, 2300–2305.
Elbediwi, M., Li, Y., Paudyal, N., Pan, H., Li, X., Xie, S., et al. (2019). Global burden of colistin-resistant bacteria: mobilized colistin resistance genes study (1980–2018). Microorganisms 7:461. doi: 10.3389/fmicb.2019.01513
Elbediwi, M., Pan, H., Biswas, S., Li, Y., and Yue, M. (2020). Emerging colistin resistance in Salmonella enterica serovar Newport isolates from human infections. Emerg. Microb. Infect. 9, 535–538. doi: 10.1080/22221751.2020.1733439
Elnekave, E., Hong, S., Mather, A. E., Boxrud, D., Taylor, A. J., Lappi, V., et al. (2018). Salmonella enterica serotype 4,[5],12:i:- in Swine in the United States midwest: an emerging multidrug-resistant clade. Clin. Infect. Dis. 66, 877–885. doi: 10.1093/cid/cix909
Gale, F., Marti, D., and Hu, D. (2012). China’s Volatile Pork Industry/LDP-M-211-01. Washington, DC: Economic Research Service.
García-Fernández, A., and Carattoli, A. (2010). Plasmid double locus sequence typing for IncHI2 plasmids, a subtyping scheme for the characterization of IncHI2 plasmids carrying extended-spectrum β-lactamase and quinolone resistance genes. J. Antimicrob. Chemother. 65, 1155–1161. doi: 10.1093/jac/dkq101
Gurevich, A., Saveliev, V., Vyahhi, N., and Tesler, G. (2013). QUAST: quality assessment tool for genome assemblies. Bioinformatics 29, 1072–1075. doi: 10.1093/bioinformatics/btt086
Hopkins, K. L., Kirchner, M., Guerra, B., Granier, S. A., Lucarelli, C., Porrero, M. C., et al. (2010). Multiresistant Salmonella enterica serovar 4,[5],12:i:- in Europe: a new pandemic strain? Eurosurveillance 15:19580.
Hu, Y., Fanning, S., Gan, X., Liu, C., Nguyen, S., Wang, M., et al. (2019). Salmonella harbouring the mcr-1 gene isolated from food in China between 2012 and 2016. J. Antimicrob. Chemother. 74, 826–828. doi: 10.1093/jac/dky496
Jiang, Z., Paudyal, N., Xu, Y., Deng, T., Li, F., Pan, H., et al. (2019). Antibiotic resistance profiles of Salmonella recovered from finishing pigs and slaughter facilities in Henan, China. Front. Microbiol. 10:1513. doi: 10.3389/fmicb.2019.01513
Lampkowska, J., Feld, L., Monaghan, A., Toomey, N., Schjørring, S., Jacobsen, B., et al. (2008). A standardized conjugation protocol to asses antibiotic resistance transfer between lactococcal species. Int. J. Food Microbiol. 127, 172–175. doi: 10.1016/j.ijfoodmicro.2008.06.017
Li, X. P., Fang, L. X., Song, J. Q., Xia, J., Huo, W., Fang, J. T., et al. (2016). Clonal spread of mcr-1 in PMQR-carrying ST34 Salmonella isolates from animals in China. Sci. Rep. 6:38511.
Linxian, Y., Jing, W., Yanling, G., Yiyun, L., Yohei, D., Renjie, W., et al. (2017). mcr-1-Harboring Salmonella enterica serovar typhimurium sequence type 34 in pigs. China. Emerg. Infect. Dis. J. 23, 291–295. doi: 10.3201/eid2302.161543
Liu, B. T., Song, F. J., and Zou, M. (2018). Characterization of Highly prevalent plasmids coharboring mcr-1, oqxAB, and blaCTX-M and plasmids harboring oqxAB and blaCTX-M in Escherichia coli Isolates from food-producing animals in China. Microb. Drug Resist. 25, 108–119. doi: 10.1089/mdr.2017.0391
Liu, Y.-Y., Wang, Y., Walsh, T. R., Yi, L.-X., Zhang, R., Spencer, J., et al. (2016). Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect. Dis. 16, 161–168. doi: 10.1016/s1473-3099(15)00424-7
Lu, X., Zeng, M., Xu, J., Zhou, H., Gu, B., Li, Z., et al. (2019). Epidemiologic and genomic insights on mcr-1-harbouring Salmonella from diarrhoeal outpatients in Shanghai, China, 2006-2016. EBioMedicine 4, 133–144. doi: 10.1016/j.ebiom.2019.03.006
Malhotra-Kumar, S., Xavier, B. B., Das, A. J., Lammens, C., Butaye, P., and Goossens, H. (2016). Colistin resistance gene mcr-1 harboured on a multidrug resistant plasmid. Lancet Infect. Dis. 16, 283–284. doi: 10.1016/s1473-3099(16)00012-8
Mather, A. E., Phuong, T. L. T., Gao, Y., Clare, S., Mukhopadhyay, S., Goulding, D. A., et al. (2018). New variant of multidrug-resistant Salmonella enterica serovar typhimurium associated with invasive disease in immunocompromised patients in vietnam. mBio 9:e01056-18.
Monte, D. F., Nelson, V., Cerdeira, L., Keelara, S., Greene, S., Griffin, D., et al. (2019). Multidrug- and colistin-resistant Salmonella enterica 4,[5],12:i:- sequence type 34 carrying the mcr-3.1 gene on the IncHI2 plasmid recovered from a human. J. Med. Microbiol. 68:1694. doi: 10.1099/jmm.0.001057
Mulvey, M. R., Bharat, A., Boyd, D. A., Irwin, R. J., and Wylie, J. (2018). Characterization of a colistin-resistant Salmonella enterica 4,[5],12:i:- harbouring mcr-3.2 on a variant IncHI-2 plasmid identified in Canada. J. Med. Microbiol. 67, 1673–1675. doi: 10.1099/jmm.0.000854
Page, A. J., Cummins, C. A., Hunt, M., Wong, V. K., Reuter, S., Holden, M. T., et al. (2015). Roary: rapid large-scale prokaryote pan genome analysis. Bioinformatics 31, 3691–3693. doi: 10.1093/bioinformatics/btv421
Pan, H., Paudyal, N., Li, X., Fang, W., and Yue, M. (2018). Multiple food-animal-borne route in transmission of antibiotic-resistant Salmonella newport to humans. Front. Microbiol. 9:23. doi: 10.3389/fmicb.2018.00023
Pan, H., Zhou, X., Chai, W., Paudyal, N., Li, S., Zhou, X., et al. (2019). Diversified sources for human infections by Salmonella enterica serovar newport. Transbound. Emerg. Dis. 66, 1044–1048. doi: 10.1111/tbed.13099
Paudyal, N., Pan, H., Elbediwi, M., Zhou, X., Peng, X., Li, X., et al. (2019). Characterization of Salmonella dublin isolated from bovine and human hosts. BMC Microbiol. 19:226.
Paudyal, N., Pan, H., Liao, X., Zhang, X., Li, X., Fang, W., et al. (2018). A meta-analysis of major foodborne pathogens in chinese food commodities between 2006 and 2016. Foodborne Pathog Dis. 15, 187–197. doi: 10.1089/fpd.2017.2417
Paudyal, N., and Yue, M. (2019). Antimicrobial resistance in the “Dark Matter”. Clin. Infect. Dis. 69, 379–380. doi: 10.1093/cid/ciz007
Poirel, L., Kieffer, N., and Nordmann, P. (2017). In vitro study of ISApl1-mediated mobilization of the colistin resistance Gene mcr-1. Antimicrob. Agents Chemother. 61:e00127-17. doi: 10.1128/AAC.00127-17
Quinn, M. M., Henneberger, P. K., Braun, B., Delclos, G. L., Fagan, K., Huang, V., et al. (2015). Cleaning and disinfecting environmental surfaces in health care: toward an integrated framework for infection and occupational illness prevention. Am. J. Infect. Control 43, 424–434. doi: 10.1016/j.ajic.2015.01.029
Rodríguez, D. M., and Suárez, M. C. (2014). Salmonella spp. in the pork supply chain: a risk approach. Rev. Colomb. Cien. Pecu. 27, 65–75.
Silva, C., Calva, E., and Maloy, S. (2014). One health and food-borne disease: Salmonella transmission between humans, animals, and plants. Microbiol. Spectr. 2:OH-0020-2013. doi: 10.1128/microbiolspec.OH-0020-2013
Snesrud, E., He, S., Chandler, M., Dekker, J. P., Hickman, A. B., McGann, P., et al. (2016). A model for transposition of the colistin resistance gene mcr-1 by ISApl1. Antimicrob. Agents Chemother. 60, 6973–6976. doi: 10.1128/aac.01457-16
USDA (2009). Economic Research Service Using Data From China National Development, and Reform Commission. (Data) Not Adjusted for Inflation. Washington, DC: USDA.
Vielva, L., de Toro, M., Lanza, V. F., and de la Cruz, F. (2017). PLACNETw: a web-based tool for plasmid reconstruction from bacterial genomes. Bioinformatics 33, 3796–3798.
Wang, X., Biswas, S., Paudyal, N., Pan, H., Li, X., Fang, W., et al. (2019). Antibiotic resistance in Salmonella typhimurium isolates recovered from the food chain through national antimicrobial resistance monitoring system between 1996 and 2016. Front. Microbiol. 10:985. doi: 10.3389/fmicb.2019.00985
Wang, X., Wang, Y., Zhou, Y., Li, J., Yin, W., Wang, S., et al. (2018). Emergence of a novel mobile colistin resistance gene, mcr-8, in NDM-producing Klebsiella pneumoniae. Emerg. Microb. Infect. 7:122.
Yang, H., Paruch, L., Chen, X., van Eerde, A., Skomedal, H., Wang, Y., et al. (2019). Antibiotic application and resistance in swine production in china: current situation and future perspectives. Front. Vet. Sci. 6:136. doi: 10.3389/fvets.2019.00136
Yoshida, C. E., Kruczkiewicz, P., Laing, C. R., Lingohr, E. J., Gannon, V. P. J., Nash, J. H. E., et al. (2016). The Salmonella in silico typing resource (SISTR): an open web-accessible tool for rapidly typing and subtyping draft Salmonella genome assemblies. PLoS One 11:e0147101. doi: 10.1371/journal.pone.0147101
Yu, H., Elbediwi, M., Zhou, X., Shuai, H., Lou, X., Wang, H., et al. (2020). Epidemiological and genomic characterization of campylobacter jejuni isolates from a foodborne outbreak at Hangzhou, China. Int. J. Mol. Sci. 21:3001. doi: 10.3390/ijms21083001
Keywords: Salmonella enterica serovar 4, [5], 12:i:-, ST34, Colistin, mcr-1, food chain
Citation: Elbediwi M, Wu B, Pan H, Jiang Z, Biswas S, Li Y and Yue M (2020) Genomic Characterization of mcr-1-carrying Salmonella enterica Serovar 4,[5],12:i:- ST 34 Clone Isolated From Pigs in China. Front. Bioeng. Biotechnol. 8:663. doi: 10.3389/fbioe.2020.00663
Received: 16 January 2020; Accepted: 28 May 2020;
Published: 30 June 2020.
Edited by:
Sandeep Tiwari, Federal University of Minas Gerais, BrazilReviewed by:
Junjie Yue, Institute of Biotechnology (CAAS), ChinaFlavia Figueira Aburjaile, Federal University of Pernambuco, Brazil
Copyright © 2020 Elbediwi, Wu, Pan, Jiang, Biswas, Li and Yue. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Min Yue, bXl1ZUB6anUuZWR1LmNu
†These authors have contributed equally to this work
 Mohammed Elbediwi
Mohammed Elbediwi Beibei Wu2†
Beibei Wu2† Silpak Biswas
Silpak Biswas Min Yue
Min Yue