- 1The Key Laboratory of Industrial Biotechnology, Ministry of Education, School of Biotechnology, Jiangnan University, Wuxi, China
- 2Ningxia Key Laboratory for Food Microbial-Applications Technology and Safety Control, Yinchuan, China
- 3School of Agriculture, Ningxia University, Yinchuan, China
- 4Department of Chemical and Biomolecular Engineering, The Ohio State University, Columbus, OH, United States
Prodigiosin (PG) is a typical secondary metabolite mainly produced by Serratia marcescens. CpxR protein is an OmpR family transcriptional regulator in Gram-negative bacteria. Firstly, it was found that insertion mutation of cpxR in S. marcescens JNB 5-1 by a transposon Tn5G increased the production of PG. Results from the electrophoretic mobility shift assay (EMSA) indicated that CpxR could bind to the promoter of the pig gene cluster and repress the transcription levels of genes involved in PG biosynthesis in S. marcescens JNB 5-1. In the ΔcpxR mutant strain, the transcription levels of the pig gene cluster and the genes involved in the pathways of PG precursors, such as proline, pyruvate, serine, methionine, and S-adenosyl methionine, were significantly increased, hence promoting the production of PG. Subsequently, a fusion segment composed of the genes proC, serC, and metH, responsible for proline, serine, and methionine, was inserted into the cpxR gene in S. marcescens JNB 5-1. On fermentation by the resultant engineered S. marcescens, the highest PG titer reached 5.83 g/L and increased by 41.9%, relative to the parental strain. In this study, we revealed the role of CpxR in PG biosynthesis and provided an alternative strategy for the engineering of S. marcescens to enhance PG production.
Introduction
Prodigiosin (PG), a red-colored tripyrrole, has gained increased interest because of its immunosuppressive, anticancer activity. It also has antifungal, antibacterial, antiprotozoal, and antimalarial properties; hence, it has a high potential for pharmaceutical application (Gastmeier, 2014; Gupta et al., 2014). Moreover, it is a good alternative to synthetic colorants and a promising source of food colorants. Prodigiosin is a typical secondary metabolite mainly produced by Serratia marcescens (Williamson et al., 2006; Papireddy et al., 2011), which usually appears in the later stages of bacterial growth (Williams, 1973). Incubation temperature plays an important role in PG synthesis in S. marcescens. PG could be efficiently produced at 28°C and sharply reduced at 37°C or higher (Williams et al., 1971; Tanaka et al., 2004).
The gene cluster for the biosynthesis of PG has been identified in Serratia, and most genes in the cluster have been functionally assigned. The bifurcated pathway culminates into two enzymatically condensed terminal products, 2-methyl-3-n-amyl-pyrrole (MAP) and 4-methoxy-2,2′-bipyrrole-5-carbaldehyde (MBC), and the pig clusters pigD, pigE, and pigB encode proteins that are involved in the production of MAP, while pigA and pigF–pigN encode proteins responsible for the synthesis of MBC (Grimont and Grimont, 1978; Von Graevenitz, 1980; Harris, 2004; Williamson et al., 2006). Proline is converted to 4-hydroxy-2,2′-bipyrrole-5-carbaldehyde (HBC) through a six-step reaction prior to methylation of the HBC hydroxyl group by S-adenosyl methionine-dependent PigF, resulting in the formation of MBC.
Previous studies have largely focused on quorum sensing (Matilla et al., 2015), signal transduction (Fineran et al., 2005), and two-component regulation systems (Horng et al., 2010) to determine the regulatory mechanisms which play an important role in the production of PG. But the thermoregulated mechanism of the production of this secondary metabolite is yet to be elucidated. Moreover, bacteria have evolved a wide variety of signal transduction systems in order to cope with changes in the external environment to ensure their survival and reproduction. This regulatory system has been divided into three categories according to the number of components involved: one-component regulatory system, two-component regulatory system (TCS), and three-component regulatory system (Marijuán et al., 2010). The cpx system is a canonical TCS, broadly conserved among Gram-negative bacteria, which consists of the sensor histidine kinase protein CpxA and the regulatory protein CpxR. Studies have suggested the role of CpxA in detecting changes in the external environment, including pH and the overexpression of envelope proteins (such as NlpE or pili subunits), as well as toxic concentrations of metal ions (Nakayama and Watanabe, 1995; Hunke et al., 2012; May et al., 2019). The detection of these stress signals activates CpxA, leading to autophosphorylation at the conserved 151 histidine residue. The histidine residue then carries a phosphate group and transfers it to the aspartic acid residue at position 51 of the response regulatory protein CpxR, hence activating CpxR (Yamamoto and Ishihama, 2006; MacRitchie et al., 2008). The phosphorylated CpxR finally binds to the specific sequence of the target gene promoter and initiates transcription. In this study, we firstly found that there was an increase in the proline, pyruvate, serine, and methionine biosynthetic pathway in the ΔcpxR mutant strain, which was beneficial to PG production. So, in this study, we discuss the effects of CpxR on prodigiosin production and further details of its mechanism function.
Materials and Methods
Bacterial Strains and Growth Conditions
S. marcescens JNB5-1 mutants derived from S. marcescens JNB 5-1 were grown in Luria–Bertani (LB) medium or fermentation medium. Escherichia coli BL21 (DE3) and E. coli DH5α, selected for the expression host, and the E. coli S17-1 λpir that pUT-Km replicated in were cultured in LB medium. The antibiotics used for selection in E. coli or S. marcescens JNB 5-1 were ampicillin (Amp), Kan, Chl, apramycin (Apm), clindamycin (Cli), and gentamicin (GM) at concentrations of 100, 50, 25, 50, 100, and 25 μg/ml, respectively. All incubations were performed either at 30 or 37°C. The strains in this work are listed in Supplementary Table S1.
Tn5G Transposon Mutagenesis
The method used for mutagenesis was the same as that in our previous research (Pan et al., 2019).
Measuring the Promoter of the pig Gene Cluster
Promoters of different lengths were amplified by PCR, and the plasmid pKK 232-8 (or pKKG) was linearized by double-enzyme digestion, followed by recombination of the linear DNA fragment obtained above by T4 ligase (Takara). Then, the recombinant plasmid pKK 232-8 (or pKKG) was transformed into E. coli DH5α competent cells, positive clones were selected, and successful construction was verified.
Measuring the Production of PG
The fermentation broth was dissolved in acidic ethanol (pH 3.0), moderately diluted (50-, 250-, and 1,000-fold), and the diluted sample was sealed and stored for 8 h (fully dissolved), centrifuged at 3,276 × g for 10 min, and the supernatant was taken. OD535 was determined. The blank control is acidic ethanol (Kalivoda et al., 2010).
Assay of PigF
A general procedure for measuring PigF activity was adopted from previous studies (Arnow, 1937; Dhar and Rosazza, 2000). The PigF assay buffer was made of 0.05 M Na2HPO4-KH2PO4 (pH 7.0). The assay mixture was composed of the assay buffer containing 1 mM dithiothreitol (DTT), 10 mM MgCl2, 2 mM S-adenosyl methionine, 1 mM 4-hydroxy-2,20-bipyrrole-5-carbaldehyde, and 2.5 mg of the enzyme in a final volume of 1 ml. The enzyme reaction mixtures were incubated at 30°C for 25 min before being terminated by the addition of 1 ml of 0.5 M HCl, followed by the addition of 1 ml each of 10% NaNO2, 10% NaMoO4, and 1 M NaOH. The absorbance at 510 nm of this solution was immediately measured to determine the amount of substrates consumed in the reaction.
CAT and GFP Fluorescence Experiments
The activity of the chloramphenicol acetyltransferase (CAT) enzyme was measured using Shaw's method, with slight modifications (Shaw, 1975). The reaction system was made of 250 mmol L−1 Tris-HCl (pH 7.8), 0.1 mmol L−1 acetyl-CoA, 0.4 mg mL−1 DTNB, and an appropriate amount of crude enzyme solution. The above reaction mixture was incubated for 2 min in 37°C water, and chloramphenicol was added to a final concentration of 0.1 mmol L−1. The light absorption value of A412 was measured immediately after mixing. The reaction solution without chloramphenicol was used as a control.
For the green fluorescent protein (GFP) fluorescence experiments, the BL21 (DE3)-containing plasmid with GFP was cultured in a 24-well plate at 37°C, 180 rpm, for 12 h. The fluorescence intensity (the excitation/emission wavelengths were 480/510) was measured and the BL21 (DE3)-containing plasmid without GFP was used as a control.
Construction of Mutants
Allelic replacement of the gene open reading frame (ORF) was performed by plasmid pUT-Km, which is a suicide plasmid with a π-protein-dependent origin of replication from the R6K plasmid, so the plasmid can only be replicated in strains expressing the π protein, such as λpir-lysogenic E. coli S17-1 λpir, SM10(λpir) (de Lorenzo et al., 1990).
For gene knockout, the upstream (containing 9 bp of gene to be knocked out) and downstream (containing 9 bp of gene to be knocked out) homology arms of the gene to be knocked out and the resistance marker gene were amplified by PCR (Supplementary Figure S1). The plasmid pUT-Km is linearized by double-enzyme digestion and recombination of the linear DNA fragment obtained above using the ClonExpress II One Step Cloning Kit (Vazyme, Nanjing, China). Then, the recombinant plasmid pUT-Km was transformed into E. coli S17-1 λpir competent cells, positive clones were selected, and successful construction was verified.
For gene knock-in, the knockout method was slightly modified, with harvest and amplification of the upstream homology arms, downstream homology arms, and resistance genes, but should amplify a gene to be replaced in the knock-in method and then homologous recombination of the upstream homology arm, replacement gene, resistance marker gene, and the downstream homology arm. The recombinant plasmid pUT-Km was mobilized into S. marcescens JNB 5-1 by conjugation. Transconjugants grown on the LB plates contain both Apm (50 μg/ml) and Cli (100 μg/ml) (Maseda et al., 2009). Mutant candidates were screened by colony PCR using primer pairs, which are listed in Supplementary Table S3 (Zhang et al., 2010). The construction of mutants that replaced the cpxR loci in S. marcescens JNB5-1 was done as described above, and the primers used are listed in Supplementary Table S3. The recombinant strain was obtained by screening on the agar plates containing Apm and Chl, with the results showing that the gene was successfully inserted into the gene of S. marcescens JNB 5-1.
RNA Extraction and Quantitative Real-Time PCR
RNA was prepared from cultures with stationary-phase cultures in LB medium at 30 or 37°C, respectively. Briefly, total RNA was treated with RNase-free pipette tips and RNA was purified using a FastPure Cell/Tissue Total RNA Isolation Kit (Vazyme, Nanjing, China). Of the total RNA, 1 μg was subjected to reverse transcription with 4 μl 5 × HiScript II qRT SuperMix II(Vazyme, Nanjing, China), and the cDNA was subjected to real-time quantitative PCR analysis using the AceQ qPCR SYBR Green Master Mix (Vazyme, Nanjing, China), with the indicated primer pairs listed in Supplementary Table S1. 16S rRNA was used as the internal reference gene and monitored in real time with the StepOnePlus PCR system (Applied Biosystems).
Electrophoretic Mobility Shift Assay
You's method was slightly modified (You et al., 2018). The cpxR gene was amplified with the indicated primers (listed in Supplementary Table S3), cloned into plasmid pET-28a for expression with 6 × His-tag, and then purified. The pig gene promoter and target gene ATG front sequences (possible promoter regions) were amplified with the indicated primers (listed in Supplementary Table S3) and ligated to the plasmid pMD19T (Simple) (listed in Supplementary Table S2). Subsequently, the promoter was amplified with the fluorescent-labeled primers M13-47-cy3 and RV-M-Cy3. The purified protein and the Cy3-labeled promoter were incubated in 2 × binding buffer (40 mM Tris-HCl, pH 7.5), 4 mM MgCl2, 100 mM NaCl, 10% glycerol, 2 mM DTT, 0.2 mg/ml bovine serum albumin (BSA), 0.02 mg/ml poly(dI-dC), and 1 mM EDTA for 30 min at 25°C. The incubated mixture was electrophoresed on a 5% native polyacrylamide gel electrophoresis (PAGE) for about 1 or 1.5 h. The gel was visualized with ImageQuant LAS 4000 (GE Healthcare Life Sciences, USA).
Shake Flask Fermentation Assay
The method with our previous research (Pan et al., 2019), with slight modifications. Briefly, JNB5-1 and SMCR were grown at 30°C and 37°C in a rotary shaker with fermentation medium (Sucrose 2%, Beef extract 1.5%, CaCl2 1%, L-proline 0.75%, MgSO4·7H2O 0.02%, and FeSO4·7H2O 0.006%) at 180 rpm for 120 h. The production of PG were determined at different fermentation time intervals (0, 12, 24, 36, 48, 60, 72, 84, 96, and 120 h) in triplicate.
Statistical Analysis
Experiments were done at least twice, with a minimum of three biological replicates. Origin software and TB tools were used for the drawing of the figures, with Student's t-tests, Mann–Whitney U-test, Fisher's exact test, and one-way ANOVA with Tukey's posttest. Significance was set at a P-value of < 0.05
Results
PG Production Could Be Enhanced by Disruption of CpxR in S. marcescens JNB 5-1
A Tn5G transposon was employed to identify genes, particularly those that influence PG production in S.marcescens JNB 5-1. The results from inverse PCR and sequencing of a mutant strain with a significantly increased PG production revealed that transposon insertion into the 38-bp position of a gene in JNB 5-1 could enhance PG production, and the selected gene was later mapped to the BVG90_20710 gene encoding a DNA-binding response regulator on the S. marcescens strain UMH8 genome (Figure 1A). Furthermore, bioinformatics analysis predicted that the selected gene from S. marcescens JNB 5-1 encodes the two-component regulatory system DNA-binding transcriptional regulator CpxR with a homology of 100% (699/699) to cpxR of the S. marcescens strain UMH8. At the protein level, the selected gene had a 91.38, 90.95, and 91.38% similarity to the cpxR of E. coli K12, Salmonella enterica, and Yersinia pestis, respectively (Supplementary Figure S2). Phylogenetic analysis suggests that the BVG90_20710-encoded protein is structurally intermediate in relatedness between Y. pestis and the highly similar Klebsiella pneumoniae (Figure 1B).
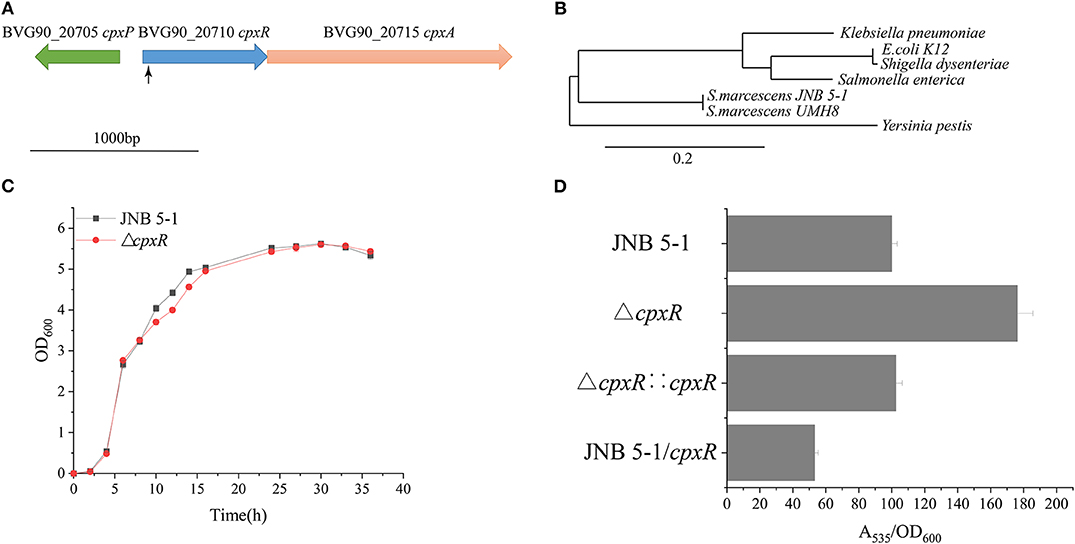
Figure 1. Isolation of increased prodigiosin (PG) production in the ΔcpxR mutant. (A) Relative positions of cpxA, cpxR, and cpxP in the CpxARP system in the Serratia marcescens UMH8 strain. The locus inserted by the transposon is marked with a black arrow. (B) Evolutionary relationship with several known proteins among the Enterobacteriaceae. The Phylogram (Dereeper et al., 2008) was drawn from www.Phylogeny.fr in the mode of “one-click” using default setting. The number of substitutions per site is proportional to the length of the branch. (C) Growth pattern of JNB 5-1 and the ΔcpxR mutant in Luria–Bertani (LB) medium. (D) PG yield in JNB 5-1, ΔcpxR mutant, complementary strain, and overexpression strain. All the yield displayed by A535/OD600 normalized to JNB 5-1 cultivated at 30°C; the mean ± SD from three independent experiments are shown.
The ΔcpxR mutant displayed a similar growth pattern to that of JNB 5-1 when cultivated in LB medium, indicating that the deletion of cpxR had no significant effect on the growth of JNB 5-1 (Figure 1C). Additionally, there was a 76% increase in PG production observed in the ΔcpxR mutant compared to S. marcescens JNB 5-1, and the complementary strain had restored the level of PG similar to that of JNB 5-1 (Figure 1D). The ΔcpxR mutant complementary analysis using the cpxR on the JNB 5-1 chromosome further supported the observed phenotypic changes that increased PG production. To confirm that the phenotypic change of JNB 5-1 was due to the loss of cpxR or effect on the adjacent genes, the in-frame knockout of cpxR was introduced into the JNB 5-1 chromosome by plasmid pUT mini-Km, and the resulting mutant had a similar phenotype to the transposon insertion mutant, resulting in increased PG production. These results indicated that the increase in PG production was due to the mutation of cpxR rather than to another mutation or effects on adjacent genes. The ΔcpxR mutant phenotypes were also complemented by cpxR from E. coli K12, S. enterica, and Y. pestis, with the resulting phenotype showing consistent PG production and growth to that complemented by cpxR from JNB 5-1. These results suggest that cpxR is highly conserved among the Enterobacteriaceae. Additionally, plasmid-borne overexpression of cpxR in the JNB 5-1 strain (with empty plasmid) showed a 46.8% decrease in PG production, indicating the inhibitory effect of cpxR (Figure 1D).
CpxR Binds to the pig Gene Cluster Promoter
To further investigate the effect of CpxR on PG production in JNB 5-1, we analyzed the transcription levels of the pig gene cluster genes from JNB 5-1 and the ΔcpxR mutant by quantitative real-time PCR (qRT-PCR). The results showed that the transcription levels of the pig gene cluster genes in the ΔcpxR mutant were upregulated (Figure 4). This was consistent with the previously discovered phenomenon, which further confirmed that the yield change of PG was influenced by CpxR. We hypothesized that CpxR affected the genes' expressions on the pig gene cluster by possibly binding to the pig gene cluster promoter.
A promoter plays a critical role in providing transcriptional capacity for nearly every natural and synthetic circuit or pathway (Redden and Alper, 2015). To gain more understanding of the mechanism that CpxR regulates prodigiosin biosynthesis, we constructed a series of recombinant plasmids with different promoter lengths in the putative promoter region of pigA. Based on the analysis of the CAT and GFP fluorescence experiments and the Virtual Footprint online software (Munch et al., 2005), the sequence between −263 and the start codon before pigA was identified as the promoter of the pig gene cluster used in subsequent experiments (Figure 2A and Supplementary Figure S3).
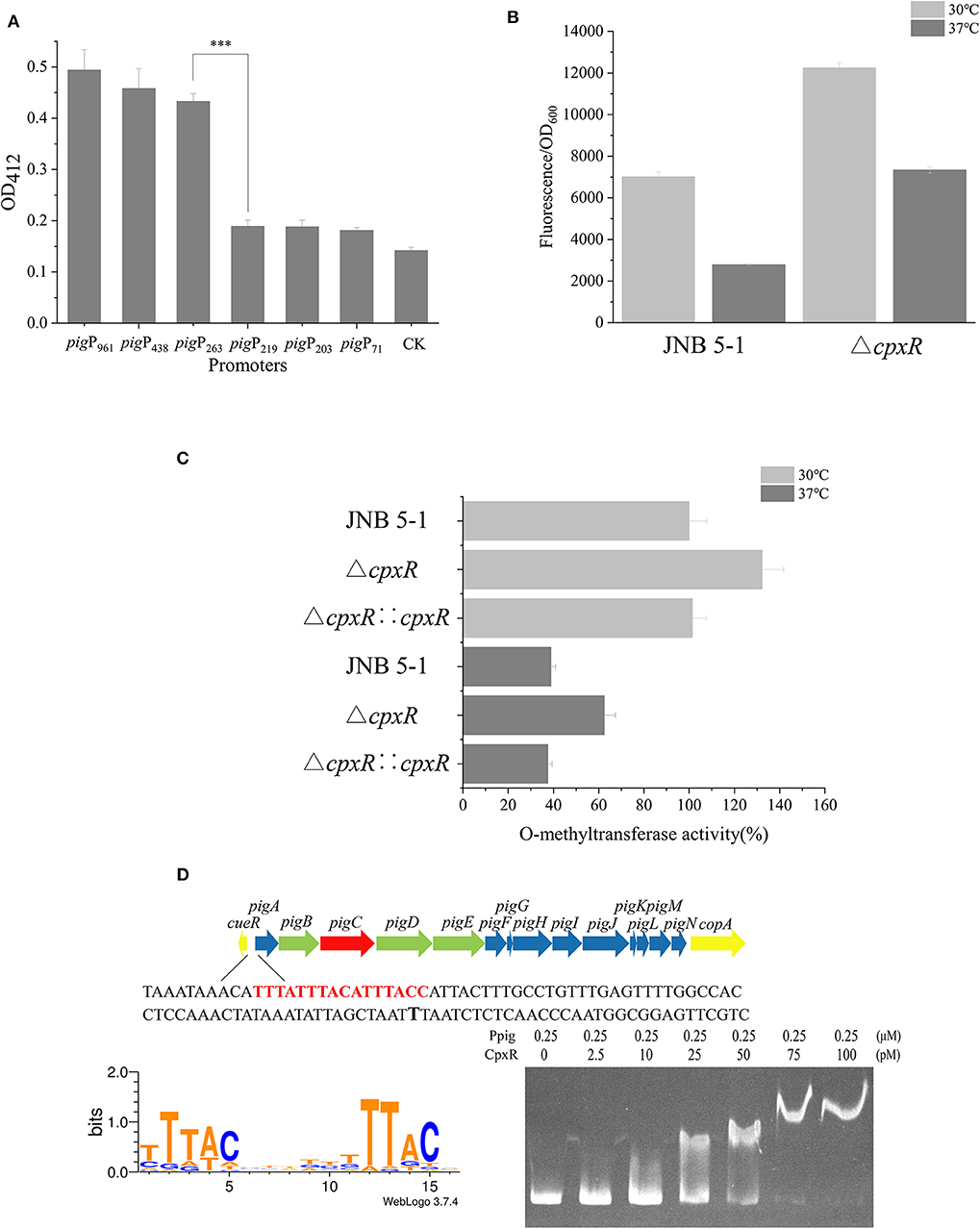
Figure 2. CpxR interaction with the pig gene cluster promoter region in JNB 5-1. (A) Characterization of promoter activity at different lengths in chloramphenicol acetyltransferase (CAT) activity. (B) Green fluorescent protein (GFP) fluorescence intensity of Ppig-gfp in JNB 5-1 and the ΔcpxR mutant at 30 and 37°C. All the results displayed as fluorescence intensity/OD600. (C) Whole O-methyltransferase activity from the crude cell extract derived from wild-type (with empty plasmid), cpxR mutant, and cpxR complement strain at 30 and 37°C. All the results displayed as percentage normalized to JNB 5-1 cultivated at 30°C. (D) CpxR bound to the pig promoter. The top picture is the pig gene cluster of the 16 genes in S. marcescens involved in the prodigiosin biosynthetic pathway. The yellow block arrow on both ends indicate the genes that are involved in copper ion transport. The 14 genes in the middle are involved in the biosynthesis of prodigiosin, in which the green block arrow indicates the genes encoding the protein that synthesizes MAP. Blue block arrow indicates the gene encoding the protein that synthesizes MBC, and red block arrow shows genes encoding the condensing enzyme in the condensation of MAP and MBC to form prodigiosin. The shown sequence is the front sequence of pigA (pig promoter), the red region is the CpxR binding region online, and the black bold T is the −10 region. The bottom left picture is the motif of CpxR harvested online, and bottom right picture is the result of the electrophoretic mobility shift assay (EMSA) wherein CpxR bound to the pig promoter. The mean ± SD from three independent experiments are shown. Significant differences in activity are calculated by one-way ANOVA. ***P < 0.001.
To investigate the effect of CpxR on the transcriptional level of the pig gene cluster in JNB 5-1, we constructed a recombinant plasmid carrying the fusion reporter gene (Ppig-gfp, a pig gene cluster promoter with gfp), as shown in Figure 2B. Evidently, the ΔcpxR mutant possessed a higher fluorescence intensity relative to JNB 5-1 at 30 and 37°C, respectively, while the fluorescence intensity of GFP at 30°C was higher than that at 37°C in both the ΔcpxR mutant and JNB 5-1, which is consistent with our hypothesis. Specifically, we found that the ΔcpxR mutant was 74.6 and 163.5% higher than JNB 5-1 at 30 and 37°C, respectively. However, in the JNB 5-1 and ΔcpxR mutants, the fluorescence intensity of GFP at 30°C was 151.9 and 66.9% higher than that at 37°C, respectively. Taken together, these results suggested that temperature had an effect on cpxR and modulated the pig gene cluster promoter (Figure 2B).
Furthermore, a key pigF gene from the pig gene cluster encoding O-methyltransferase (PigF) was carried out to verify the effect of CpxR on PG biosynthesis in JNB 5-1 at 30 and 37°C. The results showed that O-methyltransferase activity was the highest in the ΔcpxR mutant compared with JNB 5-1 and the cpxR complementary strain at 30 and 37°C (Figure 2C), implying the significant effect of CpxR on pigF or the pig gene cluster.
EMSA was performed to further gain insight into the regulation mechanism of CpxR on the pig cluster promoter. As shown in Figure 2D, the binding affinity increased with an increase in the amount of CpxR protein, suggesting that CpxR bound to the pig gene cluster promoter. Interestingly, there was an oblique upward gradient in the binding affinity of CpxR even when the amount of pig promoter added is maintained at 0.25 μM. Online analysis on Regulon DB (Santos-Zavaleta et al., 2019) predicted that the CpxR motif is “TTTACNNNNNTTTACN” and binding site was upstream of the pigA “TTTATTTACATTTACC” in JNB 5-1 (Figure 2D). The CpxR motif was highly conserved in the pig gene cluster promoter when mapping it to the annotated S. marcescens (Supplementary Figure S4).
Inhibition of PG Production by the cpx System Corresponds to Temperature in S. marcescens JNB 5-1
Considering the impact of temperature on PG biosynthesis, we investigated the relationship between the cpx system and temperature. A qRT-PCR assay was performed on cpx for further analysis. As shown in Figure 3A, cpxR and cpxA were upregulated, with 3.72- and 5.44-fold increases at the transcription level. Subsequently, dynamic transcription-level analysis of the cpx system between 30 and 37°C in JNB 5-1 showed a significantly increased transcription level of cpxA at 37°C (Figures 3A,B). Initially, there was no change in the transcription level of cpxR (0–6 h), but then an increase in the transcription level was observed at the exponential phase (6–12 h), as shown in Figure 3B. This may be attributed to the activation of cpxR by CpxA in response to environmental stress. Both cpxA and cpxR represented a downtrend when the temperature was lowered to 30°C. Subsequently, after 12 h of JNB 5-1 cultivation, the temperatures were altered from 30 to 37°C and vice versa, and then dynamic transcription-level tests on cpxA and cpxR were performed. The results indicated that the transcription level of cpxA increased 3.95- and 5.91-fold when cultivated at 37°C after 12 and 24 h, respectively, and decreased by 56.2 and 35.8% when cultivated at 30°C, respectively. A similar trend was observed in cpxR, where the transcription level increased by 2.69- and 4.46-fold when turned to 37°C, respectively, and decreased by 46.9 and 34.8% when lowered to 30°C, respectively (Figure 3B). These results suggested that the cpx system might affect PG production in the S. marcescens JNB 5-1 strain in response to temperature.
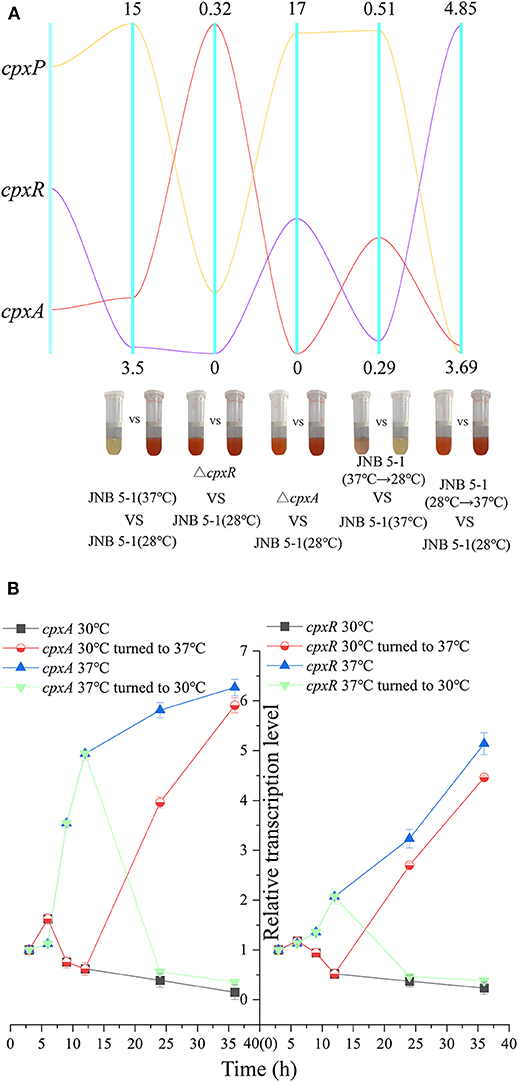
Figure 3. Dynamic analysis of the cpx system in response to temperature. (A) Dynamic analysis of the pig gene cluster and the cpx system. The five vertical line data represent the comparisons of JNB 5-1 between 30 and 37°C, of the cpxR mutant and JNB 5-1 at 30°C, between the cpxA mutant and JNB 5-1 at 30°C, between the JNB 5-1 transferred from 37 to 30°C and JNB 5-1 cultivated at 37°C, and between the JNB 5-1 transferred from 30 to 37°C and JNB 5-1 cultivated at 30°C. Yellow line represents gene cpxP, purple line represents gene cpxR, and red line represents gene cpxA. The tubes are the color change of the each strain (meaning change in PG yield). (B) Dynamic transcription level analysis of cpxA and cpxR. qPCR is performed for the characterization of dynamic change of transcription in cpxA and cpxR in JNB 5-1. The temperatures were altered from 30 to 37°C after 12 h of JNB 5-1 cultivation and vice versa. All the values of qRT-PCR were normalized to the first sample (3 h).
Subsequently, we examined the transcript level of the cpx system regulatory factor CpxP, which could be activated by CpxR transcription and also acts as an accessory protein to regulate the cpx system in the negative feedback loop, to further confirm the relationship between the cpx system and temperature. A significant difference in the transcription level of cpxP from JNB 5-1 was observed between 30 and 37°C, with the transcription level at 37°C 15-fold higher than that at 30°C. However, the transcription level of cpxP in the ΔcpxR mutant decreased to 11% when compared to JNB 5-1 cultivated at 30°C, and the transcription level of cpxP in the ΔcpxA mutant increased by 16.47-fold (Figure 3A). Furthermore, a similar trend to cpxR where the transcription level changed in response to temperature was observed, shown in Figure 3A. Taken together, these results suggested that CpxAR-dependent auxiliary protein CpxP expression is temperature-dependent, indicating that there is a substantial impact on CpxAR since the cpx system could be activated at 37°C (Figure 3A).
ΔcpxR Mutant Promotes the Supply of PG Synthesis Precursors
To gain insight on the effect of CpxR in JNB 5-1 and the changes in the transcription levels of genes on the metabolic pathways involved in PG synthesis in the ΔcpxR mutant, we performed qPCR analysis on the genes related to the known precursors for PG synthesis (proline, serine, and L-methionine synthesis) in S. marcescens. We found that the transcription levels of the precursors involved in PG synthesis and energy genes were improved. The results showed that the transcript levels of glutamate-5-semialdehyde dehydrogenase (proA), glutamate 5-kinase (proB), pyrroline-5-carboxylate reductase (proC), and proline iminopeptidase (pip) were upregulated 4.97-, 5.43-, 6.57-, and 3.23-fold, respectively, in the ΔcpxR mutant compared to JNB 5-1, indicating that proline biosynthesis was enhanced in the ΔcpxR mutant (Figure 4). Furthermore, the transcription levels of phosphoserine aminotransferase (serC) and phosphoserine phosphatase (serB), related to serine synthesis, were also upregulated 4.55- and 3.62-fold, respectively, indicating that serine biosynthesis was promoted in the ΔcpxR mutant. qRT-PCR analysis also showed that methionine synthase (metH) and S-adenosyl methionine decarboxylase (speD) were upregulated 4.81- and 2.74-fold, respectively, showing that S-adenosyl methionine, required for the transformation of HBC to MBC, increased in the ΔcpxR mutant (Figure 4).
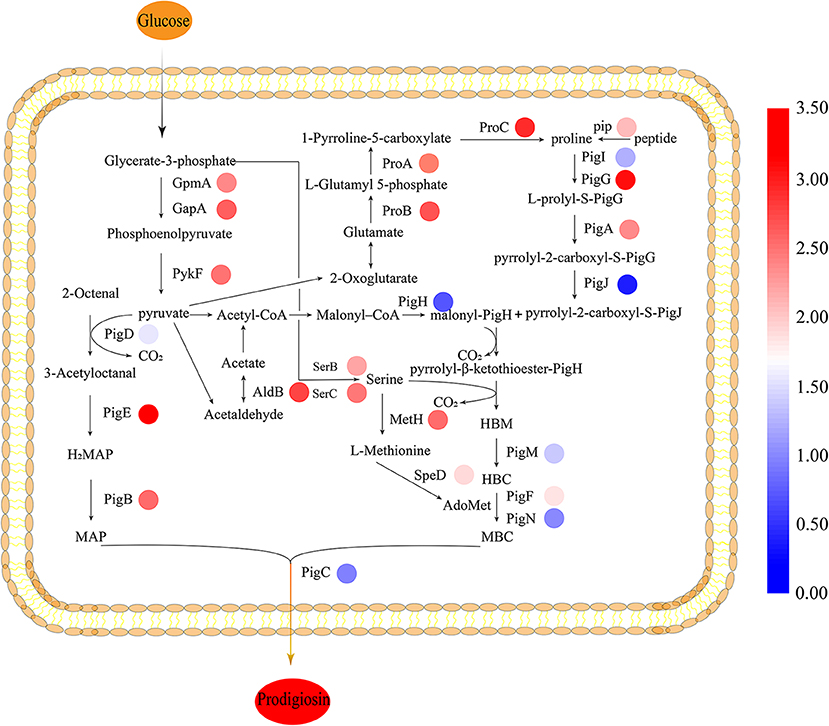
Figure 4. Biosynthesis pathway of prodigiosin in JNB 5-1. All the relative transcription-level values were log2-transformed. The biosynthesis of prodigiosin in Serratia marcescens is synthesized by a bifurcated pathway of MAP and MBC condensation to form prodigiosin by the condensing enzyme PigC. The colored rings beside the genes represent the transcription level, shown as log2 (fold change). MAP, 2-methyl-3-n-amyl-pyrrole; MBC, 4-methoxy-2,2′-bipyrrole-5-carbaldehyde; AdoMet, S-adenosyl methionine; HBC, 4-hydroxy-2,2′-bipyrrole-5-carbaldehyde; HBM, 4-hydroxy-2,2′-bipyrrole-5-methanol. JNB 5-1 and the cpxR mutant were cultivated at 30°C.
Moreover, the transcription levels of 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase (gpmA), glyceraldehyde 3-phosphate dehydrogenase (gapA), and pyruvate kinase (pykF) related to pyruvate synthesis were upregulated 4.21-, 5.14-, and 4.67-fold, respectively. The transcript level of aldehyde dehydrogenase gene (aldB), related to acetaldehyde synthesis, was also upregulated 5.83-fold. Acetaldehyde metabolism is essential for the supply of acetyl-CoA, which could be catalyzed to malonyl-CoA that is beneficial for PG synthesis (Figure 4).
Reconstruction of S. marcescens JNB 5-1 Strain for Efficient Production of Prodigiosin
Proline, serine, and methionine are important factors that promote PG production, considering thiamine and sodium acetate are also beneficial for PG biosynthesis. Therefore, proline, serine, methionine, thiamine, sodium acetate, and a proline–methionine combination were added into the medium for JNB 5-1 growth. The combination of proline–methionine showed the highest PG production relative to the control, and the use of proline or methionine alone also increased PG production (Figure 5B). Subsequently, we performed the plasmid-borne proA, proB, proC, serB, serC, and metH overexpression on strain JNB 5-1. As anticipated, plasmid-borne proC, serC, and metH overexpression in strain JNB 5-1 increased by 52.3%, 33.7 and 37.5%, respectively, compared with JNB 5-1 in LB medium harboring empty plasmid (Figure 5C).
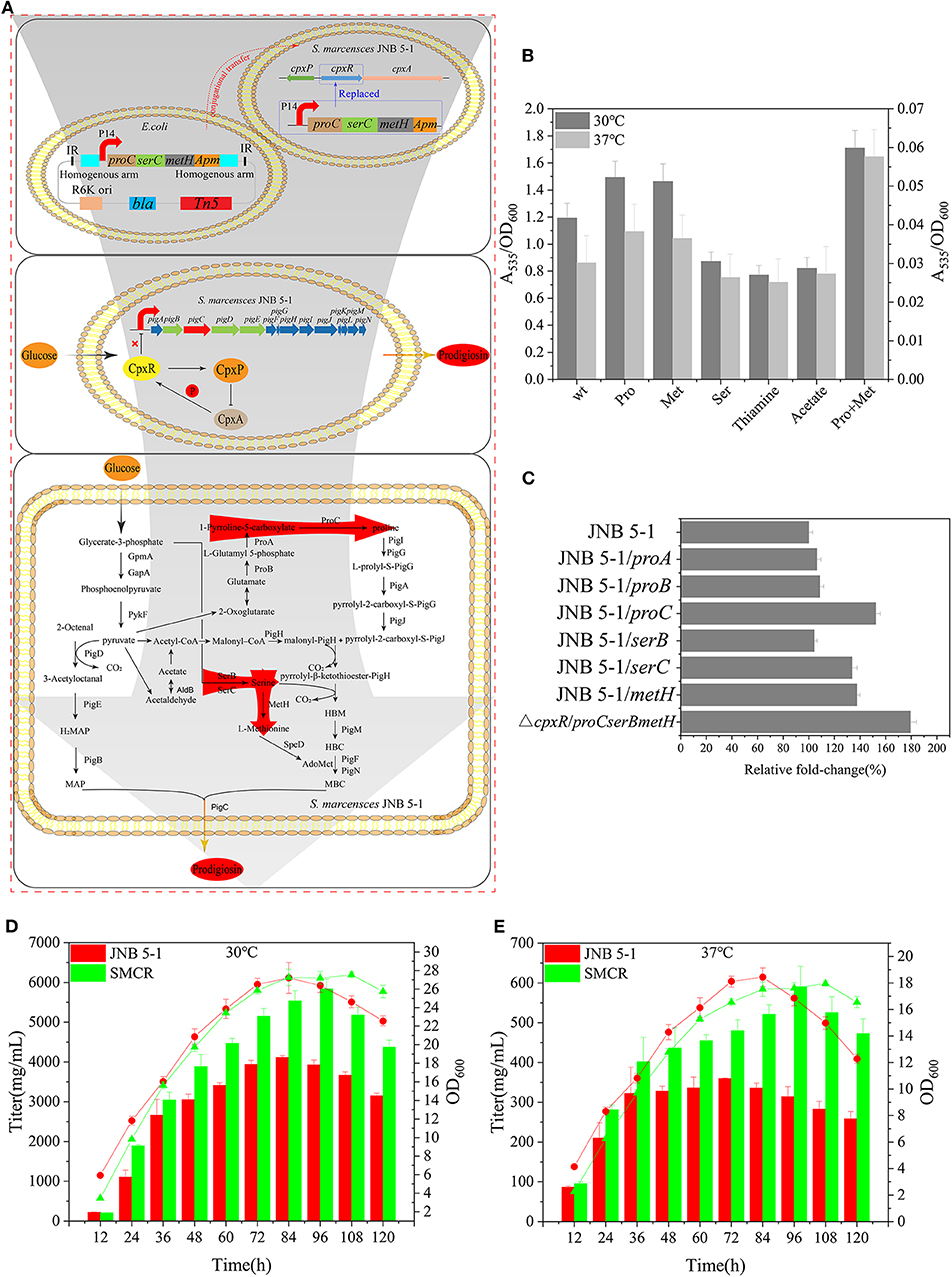
Figure 5. Reconstruction of Serratia marcescens JNB 5-1 and fermentation. (A) Workflow of the reconstruction of S. marcescens JNB 5-1. The top picture displays the reconstruction of S. marcescens JNB 5-1, replacing cpxR with proC, serC, and metH. The pUT plasmid was used as a knock-in plasmid to send the genes to the JNB 5-1 chromosome by conjugative transfer into JNB 5-1. Subsequently, homologous recombination after entering the cell (blue box). The middle picture represents the relationship between CpxA, CpxR, and CpxP and the regulation of CpxR on the pig gene cluster. The picture displays the biosynthesis of prodigiosin in the hyperproducing S. marcescens, in which cpxR was replaced by proC, serC, and metH. Red arrow region is the enhanced pathway by overexpression of proC, serC, and metH. (B) Addition of different precursors cultivated in Luria–Bertani (LB) medium at 30 and 37°C. All the yield displayed as A535/OD600. Left y-axils and right y-axils represent A535/OD600 from strains cultivated at 30 and 37°C, respectively. (C) Plasmid-borne overexpression of proA, proB, proC, serB, serC, and metH in JNB 5-1 in LB cultivated at 30°C. The last result displayed the promotion of mutant replacing the cpxR loci by proC, serC, and metH compared with JNB 5-1. All the yield displayed as A535/OD600 normalized to JNB 5-1 cultivated at 30°C. (D,E) Fermentation of mutants and JNB 5-1 cultivated at 30 and 37°C, respectively. Lines and columns represent OD600 and titer, respectively. The means ± SD from three independent experiments are shown.
Further, in order to overcome the low level of single-copy gene expression by homologous recombination, we integrated the proC, serC, and metH into the cpxR loci (Figure 5A). The titer of PG increased 79% in the recombinant strains with proC, serC, and metH integrated into the cpxR loci (the obtained recombinant strain was renamed SMCH) compared with JNB 5-1 in LB medium (Figure 5C). To fully substantiate the roles of proC, serC, and metH and cpxR, SMCH was subjected to shake flask fermentation for the production of PG in the fermentation medium. Overall, the PG production in SMCH was significantly higher than that of JNB 5-1 at both 30 and 37°C. Specifically, at 30°C, SMCH grew slowly compared to JNB 5-1 at the beginning of growth, and there was no difference between the two strains at the logarithmic phase, as shown in the Figures 5D,E. JNB 5-1 entered the death phase after 84 h, with the stationary period commencing in SMCH and the death phase later beginning at 120 h. Significantly, the highest yield of PG produced in SMCH was 5.83 g/L, which was 41.9% higher than that in JNB 5-1. When cultured at 37°C, there was no significant difference in the growth curve of SMCH and JNB 5-1 from 0 to 84 h, when JNB 5-1 entered the stationary phase. Specific to PG production, SMCH reached the highest yield of 590 mg/L, which was 64.1% higher than JNB 5-1. The results showed that the integration of proC, serC, and metH into the cpxR gene resulted in an increase in the expressions of proC, serC, and metH, thus improving the production of prodigiosin in S. marcescens JNB 5-1.
Discussion
We herein show a novel insight into the biosynthesis of PG with regulator response to temperature (RRT), in which a two-component regulatory system was thermoregulated and the regulatory protein can bind to the pig gene cluster promoter. The Cpx two-component regulatory system that inhibits PG production was screened by transposon insertion, which was thermoregulated and transferred this effect to PG biosynthesis. Subsequently, we reconstructed the key gene of the proline, serine, and methionine metabolic pathway by knock-in and knockout, in which the copy number between proC and the Cpx two-component regulatory system changed. This strategy can be used in metabolically engineered strains to improve the copy numbers of proC, serC, and metH. Lastly, we achieved a hyperproducing S. marcescens through the entire genome-modified metabolic strain.
There have been many reports on the effects of the production of PG. Kim et al. improved the prodigiosin production from 0.658 to 1.628 g/L by an antibiotic mutagenesis using chloramphenicol (Kim et al., 2008). Haddix and colleagues harvested the highest production of 1.314 g/L by modifying various fermentation parameters (Haddix and Shanks, 2020); however, the regulatory network may play an important role. The two-component regulation system is an expression system established by bacteria to cope with changes in the external environment (Cheung and Hendrickson, 2010). Previous reports on the Cpx two-component regulatory system have shown that CpxR could regulate the degP, dsbA, and ppiA genes that encode heat shock protease, disulfide oxidoreductase, and peptide prolyl isomerase, respectively (De Wulf et al., 1999). Our study correlated the Cpx two-component regulatory system to temperature regulation and PG production. The deletion of CpxR resulted in an increase of PG production in JNB 5-1, but did not affect JNB 5-1 growth. However, the overexpression of cpxR not only reduced the ability to synthesize PG in JNB 5-1 but also affected the growth of JNB 5-1. Spinola et al. (2010) found that the deletion of cpxA led to the accumulation of CpxR in Haemophilus ducreyi and weakened its survival (viability) in vivo. A similar result was observed, as shown in Figure 3A, where the transcription level of cpxR was upregulated in the ΔcpxA mutant. Thus, the overexpression of cpxR and the deletion of cpxA in JNB 5-1 showed that the growth of JNB 5-1 was influenced. García Véscovi Eleonora et al. also found that CpxR expression is temperature-dependent by changes in the cpxP transcriptional levels (Bruna et al., 2018). However, we observed that the cpx system changed with temperature change (Figure 3A). The transcription levels of the genes in the pig gene cluster also changed in S. marcescens JNB5-1 and in the ΔcpxR and ΔcpxA mutants. Furthermore, it was confirmed by EMSA that CpxR bound to the pig gene cluster promoter. However, we also noticed that PG production increased in the ΔcpxR mutant, but decreased in the ΔcpxA mutant, and this could be attributed to the influence of cpxA on the growth of S. marcescens JNB 5-1 (Delhaye et al., 2016). Moreover, since CpxA is a protein that responds to changes in the surrounding cell membrane environment, such as pH and salt ion concentration, deletion of cpxA probably resulted in the inability of cells to adapt to changes in the external environment. We found that S. marcescens JNB 5-1 had a highest titer (4.11 g/L) of PG after 84 h, while SMCH produced 5.83 g/L of PG after 96 h. We hypothesized that this was related to the relieved product feedback inhibition due to cpxR knockout, which was in agreement with previous reports that CpxR is related to cell membrane biosynthesis (May et al., 2019; Simpson and Trent, 2019).
Proline, synthesized via the glutamate pathway by a series of enzymes (ProA, ProB, and ProC), is involved in the first step of MBC synthesis catalyzed by PigG and PigI. It activates L-proline using ATP, resulting in the interaction of L-proline with the thiol group in PigG to form a complex L-prolyl-S-PigG under the action of Pig I, and then PigA oxidizes the L-prolyl-S-PCP to pyrrolyl-2-carboxyl-S-PCP (Williamson et al., 2006). Our research showed that the transcription levels of proA, proB, and proC involved in proline biosynthesis increased to varying degrees in the ΔcpxR mutant, resulting in increased proline synthesis, which was beneficial to PG biosynthesis. This result is in line with a study by Siva et al. that reported enhanced PG production in Serratia rubidaea after the addition of proline alone or in combination with methionine (Siva et al., 2012). Moreover, an increased PG production was achieved by plasmid overexpression of proC and by adding proline to the fermentation medium in our study. Subsequently, the expression of proC was increased by inserting proC into the cpxR loci, hence a reconstructed S. marcescens JNB 5-1 strain with efficient production of prodigiosin.
The transcription levels of the genes involved in pyruvate, serine, and methionine metabolism were also upregulated. Pyruvate is involved in the first step of the MAP branch in the PG biosynthesis bifurcated pathway, which forms 3-acetyloctanal with 2-octenal under the catalysis of PigD and then generates MAP via the catalysis of PigE and PigB (Williamson et al., 2006). Wasserman et al. also confirmed the derivation of C2 of MAP, and its attached methyl group is from pyruvate by 14C and 13C labeling (Wasserman et al., 1973). Initially, we added pyruvate and thiamine to the medium; however, there was no significant increase in PG production. We therefore suspected that pyruvate could be involved in many reactions in the bacteria and was diverted by other metabolic pathways. On the other hand, the maximum rate-limiting factor for PG production could be in the MBC branch. Also, it was predicted that serine involved in the biosynthesis of MBC was condensed with the pyrrolyl-β-ketothioester of PigH, forming 4-hydroxy-2,2′-bipyrrole-5-methanol (HBM) (Williamson et al., 2006), which is an intermediate of HBC required for the synthesis of MBC via the action of S-adenosyl methionine-dependent PigF and PigN (Williamson et al., 2005). The replication of the plasmid containing individual genes (proC, serC, and metH) was carried out by means of circular replication to achieve higher copy numbers. However, the replication generated a large amount of single DNAs, making the plasmid unstable (Aoki et al., 1987; Zhang et al., 2019), thus limiting its industrial application. Therefore, we used integrated expression strategy to insert the gene of the key synthetic enzyme for PG into a certain position on the chromosome of S. marcescens JNB5-1, allowing the inserted gene to replicate as the host chromosome replicates. Although the integrated expression level was lower than the plasmid expression level, by increasing the copy number of the integrated proC, we were able to achieve high levels of target protein production. In addition, we knocked out a suppressor gene of S. marcescens JNB 5-1, resulting in a decreased inhibitory effect on prodigiosin synthesis in S. marcescens JNB 5-1 (Zhang et al., 2019). As anticipated, the integration of proC, serC, and metH into the cpxR loci enhanced the transcription and expression of proC, serC, and metH in S. marcescens JNB5-1.
In conclusion, our study demonstrated that the two-component regulatory system cpx in S. marcescens JNB 5-1 is temperature-regulated and inhibited PG biosynthesis. The proposed RRT system hinted that temperature drives the expression of the cpx system, further regulating PG synthesis. The reconstructed high copy numbers of proC, serC, and metH in S. marcescens JNB 5-1 by homologous recombination increased the PG yield.
Data Availability Statement
All datasets generated for this study are included in the article/Supplementary Material.
Author Contributions
ZR and TY conceived and designed the study and critically revised the manuscript. TO critically revised the manuscript. YS carried out the experiments, analyzed the data, and drafted the manuscript. LW carried out the experiments. XP, HF, HZ, and S-TY contributed to the revision of the manuscript. All authors read and approved the final manuscript.
Funding
This work was supported by the National Key Research and Development Program of China (2018YFA0900300), the National Natural Science Foundation of China (31870066, 21778024, 31570085), National First-Class Discipline Program of Light Industry Technology and Engineering (LITE2018-06), the Program of Introducing Talents of Discipline to Universities (111-2-06), key research and development program of Ningxia hui autonomous region (2017BY069), the science and technology innovation team foundation of Ningxia hui autonomous region (KJT2017001), Top-notch Academic Programs Project of Jiangsu Higher Education Institutions and the Priority Academic Program Development of Jiangsu Higher Education Institution.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fbioe.2020.00344/full#supplementary-material
References
Aoki, T., Noguchi, N., Sasatsu, M., and Kono, M. (1987). Complete nucleotide sequence of pTZ12, a chloramphenicol-resistance plasmid of Bacillus subtilis. Gene 51, 107–111. doi: 10.1016/0378-1119(87)90481-1
Arnow, L. E. (1937). colorimetric determination of the components of 3,4-dihydroxyphenylalaninetyrosine mixtures. J. Biol. Chem. 118, 531–537.
Bruna, R. E., Molino, M. V., Lazzaro, M., Mariscotti, J. F., and García Véscovi, E. (2018). CpxR-dependent thermoregulation of Serratia marcescens PrtA metalloprotease expression and its contribution to bacterial biofilm formation. J. Bacteriol. 6:18. doi: 10.1128/JB.00006-18
Cheung, J., and Hendrickson, W. A. (2010). Sensor domains of two-component regulatory systems. Curr. Opin. Microbiol. 13, 116–123. doi: 10.1016/j.mib.2010.01.016
de Lorenzo, V., Herrero, M., Jakubzik, U., and Timmis, K. N. (1990). Mini-Tn5 transposon derivatives for insertion mutagenesis, promoter probing, and chromosomal insertion of cloned DNA in gram-negative eubacteria. J. Bacteriol. 172, 6568–6572. doi: 10.1128/JB.172.11.6568-6572.1990
De Wulf, P., Kwon, O., and Lin, E. C. (1999). The CpxRA signal transduction system of Escherichia coli: growth-related autoactivation and control of unanticipated target operons. J. Bacteriol. 181, 6772–6778. doi: 10.1128/JB.181.21.6772-6778.1999
Delhaye, A., Collet, J., and Laloux, G. (2016). Fine-tuning of the cpx envelope stress response is required for cell wall homeostasis in Escherichia coli. mBio 7, e16–e47. doi: 10.1128/mBio.00047-16
Dereeper, A., Guignon, V., Blanc, G., Audic, S., Buffet, S., Chevenet, F., et al. (2008). Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Res. 36, W465–W469. doi: 10.1093/nar/gkn180
Dhar, K., and Rosazza, J. P. (2000). Purification and characterization of Streptomyces griseus catechol O-methyltransferase. Appl. Environ. Microbiol. 66, 4877–4882. doi: 10.1128/AEM.66.11.4877-4882.2000
Fineran, P. C., Slater, H., Everson, L., Hughes, K., and Salmond, G. P. C. (2005). Biosynthesis of tripyrrole and β-lactam secondary metabolites in Serratia: integration of quorum sensing with multiple new regulatory components in the control of prodigiosin and carbapenem antibiotic production. Mol. Microbiol. 56, 1495–1517. doi: 10.1111/j.1365-2958.2005.04660.x
Gastmeier, P. (2014). Serratia marcescens: an outbreak experience. Front. Microbiol. 5:81. doi: 10.3389/fmicb.2014.00081
Grimont, P. A., and Grimont, F. (1978). The genus Serratia. Ann. Rev. Microbiol. 32:221. doi: 10.1146/annurev.mi.32.100178.001253
Gupta, N., Hocevar, S. N., Moultonmeissner, H. A., Stevens, K. M., Mcintyre, M. G., Jensen, B., et al. (2014). Outbreak of Serratia marcescens bloodstream infections in patients receiving parenteral nutrition prepared by a compounding pharmacy. Clin. Infect. Dis. 59, 1–8. doi: 10.1093/cid/ciu218
Haddix, P. L., and Shanks, R. (2020). Production of prodigiosin pigment by Serratia marcescens is negatively associated with cellular ATP levels during high-rate, low-cell-density growth. Can. J. Microbiol. 66, 243–255. doi: 10.1139/cjm-2019-0548
Harris, A. K. P. (2004). The Serratia gene cluster encoding biosynthesis of the red antibiotic, prodigiosin, shows species- and strain-dependent genome context variation. Microbiology 150, 3547–3560. doi: 10.1099/mic.0.27222-0
Horng, Y., Chang, K., Liu, Y., Lai, H., and Soo, P. (2010). The RssB/RssA two-component system regulates biosynthesis of the tripyrrole antibiotic, prodigiosin, in Serratia marcescens. Int. J. Med. Microbiol. 300, 304–312. doi: 10.1016/j.ijmm.2010.01.003
Hunke, S., Keller, R., and Müller, V. S. (2012). Signal integration by the Cpx-envelope stress system. FEMS Microbiol. Lett. 326, 12–22. doi: 10.1111/j.1574-6968.2011.02436.x
Kalivoda, E. J., Stella, N. A., Aston, M. A., Fender, J. E., Thompson, P. P., Kowalski, R. P., et al. (2010). Cyclic AMP negatively regulates prodigiosin production by Serratia marcescens. Res. Microbiol. 161, 158–167. doi: 10.1016/j.resmic.2009.12.004
Kim, S. J., Lee, H. K., Lee, Y. K., and Yim, J. H. (2008). Mutant selection of Hahella chejuensis KCTC 2396 and statistical optimization of medium components for prodigiosin yield-up. J. Microbiol. 46, 183–188. doi: 10.1007/s12275-008-0037-y
MacRitchie, D. M., Buelow, D. R., Price, N. L., and Raivio, T. L. (2008). Two-component signaling and gram negative envelope stress response systems. Adv. Exp. Med. Biol. 631, 80–110. doi: 10.1007/978-0-387-78885-2_6
Marijuán, P. C., Navarro, J., and Del Moral, R. (2010). On prokaryotic intelligence: strategies for sensing the environment. Biosystems 99, 94–103. doi: 10.1016/j.biosystems.2009.09.004
Maseda, H., Hashida, Y., Konaka, R., Shirai, A., and Kourai, H. (2009). Mutational upregulation of a resistance-nodulation-cell division-type multidrug efflux pump, SdeAB, upon exposure to a biocide, cetylpyridinium chloride, and antibiotic resistance in Serratia marcescens. Antimicrob. Agents Chemother. 53, 5230–5235. doi: 10.1128/AAC.00631-09
Matilla, M. A., Leeper, F. J., and Salmond, G. P. C. (2015). Biosynthesis of the antifungal haterumalide, oocydin A, in Serratia, and its regulation by quorum sensing, RpoS and Hfq. Environ. Microbiol. 17, 2993–3008. doi: 10.1111/1462-2920.12839
May, K. L., Lehman, K. M., Mitchell, A. M., and Grabowicz, M. (2019). A stress response monitoring lipoprotein trafficking to the outer membrane. mBio 10:e00618-19. doi: 10.1128/mBio.00618-19
Munch, R., Hiller, K., Grote, A., Scheer, M., Klein, J., Schobert, M., et al. (2005). Virtual footprint and PRODORIC: an integrative framework for regulon prediction in prokaryotes. Bioinformatics 21, 4187–4189. doi: 10.1093/bioinformatics/bti635
Nakayama, S., and Watanabe, H. (1995). Involvement of cpxA, a sensor of a two-component regulatory system, in the pH-dependent regulation of expression of Shigella sonnei virF gene. J. Bacteriol. 177, 5062–5069. doi: 10.1128/JB.177.17.5062-5069.1995
Pan, X., Sun, C., Tang, M., Liu, C., Zhang, J., You, J., et al. (2019). Loss of serine-type D-Ala-D-Ala carboxypeptidase daca enhances prodigiosin production in Serratia marcescens. Front. Bioeng. Biotechnol. 7:367. doi: 10.3389/fbioe.2019.00367
Papireddy, K., Smilkstein, M., Kelly, J. X., Shweta Salem, S. M., Alhamadsheh, M., et al. (2011). Antimalarial activity of natural and synthetic prodiginines. J. Med. Chem. 54, 5296–5306. doi: 10.1021/jm200543y
Redden, H., and Alper, H. S. (2015). The development and characterization of synthetic minimal yeast promoters. Nat. Commun. 6:7810. doi: 10.1038/ncomms8810
Santos-Zavaleta, A., Salgado, H., Gama-Castro, S., Sanchez-Perez, M., Gomez-Romero, L., Ledezma-Tejeida, D., et al. (2019). RegulonDB v 10.5: tackling challenges to unify classic and high throughput knowledge of gene regulation in E. coli K-12. Nucleic Acids Res. 47, D212–D220. doi: 10.1093/nar/gky1077
Shaw, W. V. (1975). Chloramphenicol acetyltransferase from chloramphenicol-resistant bacteria. Methods Enzymol. 43, 737–755. doi: 10.1016/0076-6879(75)43141-X
Simpson, B. W., and Trent, M. S. (2019). Emerging roles for NlpE as a sensor for lipoprotein maturation and transport to the outer membrane in Escherichia coli. MBio 10:e01302-19. doi: 10.1128/mBio.01302-19
Siva, R., Subha, K., Bhakta, D., Ghosh, A. R., and Babu, S. (2012). Characterization and enhanced production of prodigiosin from the spoiled coconut. Appl. Biochem. Biotechnol. 166, 187–196. doi: 10.1007/s12010-011-9415-8
Spinola, S. M., Fortney, K. R., Baker, B., Janowicz, D. M., Zwickl, B., Katz, B. P., et al. (2010). Activation of the CpxRA system by deletion of cpxA impairs the ability of Haemophilus ducreyi to infect humans. Infect. Immun. 78, 3898–3904. doi: 10.1128/IAI.00432-10
Tanaka, Y., Yuasa, J., Baba, M., Tanikawa, T., Nakagawa, Y., and Matsuyama, T. (2004). Temperature-dependent bacteriostatic activity of Serratia marcescens. Microbes Environ. 19, 236–240. doi: 10.1264/jsme2.19.236
Wasserman, H. H., Skles, R. J., Peverada, P., Shaw, C. K., Cushley, R. J., and Lipsky, C. R. (1973). Biosynthesis of prodigiosin. Incorporation patterns of C-labeled alanine, proline, glycine, and serine elucidated by fourier transform nuclear magnetic resonance. J. Am. Chem. Soc. 95, 6874–6875. doi: 10.1021/ja00801a080
Williams, R. P. (1973). Biosynthesis of prodigiosin, a secondary metabolite of Serratia marcescens. Appl. Microbiol. 25, 396–402. doi: 10.1128/AEM.25.3.396-402.1973
Williams, R. P., Gott, C. L., Qadri, S. M., and Scott, R. H. (1971). Influence of temperature of incubation and type of growth medium on pigmentation in Serratia marcescens. J. Bacteriol.106, 438–443. doi: 10.1128/JB.106.2.438-443.1971
Williamson, N. R., Fineran, P. C., Leeper, F. J., and Salmond, G. P. C. (2006). The biosynthesis and regulation of bacterial prodiginines. Nat. Rev. Microbiol. 4, 887–899. doi: 10.1038/nrmicro1531
Williamson, N. R., Simonsen, H. T., Ahmed, R. A. A., Goldet, G., Slater, H., Woodley, L., et al. (2005). Biosynthesis of the red antibiotic, prodigiosin, in Serratia: identification of a novel 2-methyl-3-n-amyl-pyrrole (MAP) assembly pathway, definition of the terminal condensing enzyme, and implications for undecylprodigiosin biosynthesis in Streptomyces. Mol. Microbiol. 56, 971–989. doi: 10.1111/j.1365-2958.2005.04602.x
Yamamoto, K., and Ishihama, A. (2006). Characterization of copper-inducible promoters regulated by CpxA/CpxR in Escherichia coli. Biosci. Biotechnol. Biochem.70, 1688–1695. doi: 10.1271/bbb.60024
You, J., Sun, L., Yang, X., Pan, X., Huang, Z., Zhang, X., et al. (2018). Regulatory protein SrpA controls phage infection and core cellular processes in Pseudomonas aeruginosa. Nat. Commun. 9:1846. doi: 10.1038/s41467-018-04232-6
Zhang, L., Sun, J. A., Hao, Y., Zhu, J., Chu, J., Wei, D., et al. (2010). Microbial production of 2,3-butanediol by a surfactant (serrawettin)-deficient mutant of Serratia marcescens H30. J. Indus. Microbiol. Biotechnol. 37, 857–862. doi: 10.1007/s10295-010-0733-6
Keywords: CpxR, Serratia marcescens, prodigiosin, temperature-sensitive, metabolic engineering
Citation: Sun Y, Wang L, Pan X, Osire T, Fang H, Zhang H, Yang S-T, Yang T and Rao Z (2020) Improved Prodigiosin Production by Relieving CpxR Temperature-Sensitive Inhibition. Front. Bioeng. Biotechnol. 8:344. doi: 10.3389/fbioe.2020.00344
Received: 25 November 2019; Accepted: 27 March 2020;
Published: 03 June 2020.
Edited by:
Jiazhang Lian, Zhejiang University, ChinaReviewed by:
Dae-Hee Lee, Korea Research Institute of Bioscience and Biotechnology (KRIBB), South KoreaJiabin Li, First Affiliated Hospital, Anhui Medical University, China
Copyright © 2020 Sun, Wang, Pan, Osire, Fang, Zhang, Yang, Yang and Rao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Taowei Yang, eXR3MTIyOCYjeDAwMDQwOzE2My5jb20=; Zhiming Rao, cmFvemhtJiN4MDAwNDA7amlhbmduYW4uZWR1LmNu
†Present address: Zhiming Rao, School of Biotechnology, Jiangnan University, Wuxi, China
 Yang Sun
Yang Sun Lijun Wang1
Lijun Wang1 Xuewei Pan
Xuewei Pan Shang-Tian Yang
Shang-Tian Yang Zhiming Rao
Zhiming Rao