
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Bioeng. Biotechnol. , 20 March 2020
Sec. Computational Genomics
Volume 8 - 2020 | https://doi.org/10.3389/fbioe.2020.00201
This article is part of the Research Topic Computational Epigenetics in Human Diseases, Cell Differentiation, and Cell Reprogramming, Volume I View all 42 articles
 Ni Kou1†
Ni Kou1† Wenyang Zhou2†
Wenyang Zhou2† Yuzhu He1
Yuzhu He1 Xiaoxia Ying1
Xiaoxia Ying1 Songling Chai1
Songling Chai1 Tao Fei1
Tao Fei1 Wenqi Fu1
Wenqi Fu1 Jiaqian Huang1
Jiaqian Huang1 Huiying Liu1*
Huiying Liu1*Accumulating evidence showed that Interleukin (IL) level is associated with Osteoporosis. Whereas, most of these associations are based on observational studies. Thus, their causality was still unclear. Mendelian randomization (MR) is a widely used statistical framework that uses genetic instrumental variables (IVs) to explore the causality of intermediate phenotype with disease. To classify their causality, we conducted a MR analysis to investigate the effect of IL-18 level on the risk of Osteoporosis. First, based on summarized genome-wide association study (GWAS) data, 8 independent IL-18 SNPs reaching genome-wide significance were deemed as IVs. Next, Simple median method was used to calculate the pooled odds ratio (OR) of these 8 SNPs for the assessment of IL-8 on the risk of Osteoporosis. Then, MR-Egger regression was utilized to detect potential bias due to the horizontal pleiotropy of these IVs. As a result of simple median method, we get the SE (−0.001; 95% CI−0.002 to 0; P = 0.042), which means low IL-18 level could increases the risk of the development of Osteoporosis. The low intercept (0; 95% CI −0.001 to 0; P = 0.59) shows there is no bias due to the horizontal pleiotropy of the IVs.
Osteoporosis is a chronic disease with a variety of causals to bone mineral density and bone loss of quality (Sambrook, 2006). Since the bone weakening in Osteoporosis patients, it increases the risk of a broken bone and other diseases among the elderly (Tu et al., 2018). Bone is a type of living tissue, which is constantly being broken down and replaced. When the creation of new bone doesn't keep up with the loss of old one, Osteoporosis occurs (Seo et al., 2018; Papaleontiou et al., 2019; Wang et al., 2019). Osteoporosis affects all the countries including men and women. For example, about 90,000,000 Osteoporosis patients in china, which covers the 7.1% of the total population. Since lots of complications affected by Osteoporosis and its incurability, it is very important for preventing the Osteoporosis. Whereas, there are no typical symptoms in the early stages of bone loss according to the current knowledge (Hennemann, 2002). This raises the difficulties for prevention. To solve this problem, it is urgent to expose the causal clinical phenotypes of Osteoporosis.
Interleukin (IL) is a class of cytokines, which is produced by a variety of cells and also functions on a variety of cells (Kato and Perl, 2018; Zhang et al., 2018). Current, about forty types of IL was discoveries in human body. IL-18 is one of them, which locates at 11q22.2-22.3. IL-18 is powerful inflammatory cytokines, the most characteristic feature of which is the regulation of cellular proliferation and differentiation (Weiss et al., 2018; Youssef et al., 2018; Prencipe et al., 2019; Valero et al., 2019). Recent studies show that IL-18 plays important roles in immune regulation, resistance to infection and anti-tumor. Furthermore, IL-18 has been identified as the causal of multiple chronic diseases, such as type 2 diabetes (Zou et al., 2018).
The relationship between IL and Osteoporosis has been investigated in observational studies decades of years. Early in 1993, Lewis et al. investigated a transgenic mice with disorder in bone homeostasis that inappropriately express the cytokine IL-4 (Lewis et al., 1993). And then they observed that Osteoporosis was associated with the IL-4. In 2005, Rusinska et al. evaluated the relationship between multiple ILs and the etiopathogenesis of idiopathic osteoporosis in children (Rusinska and Chlebnasokół, 2005). In 2010, Edwards et al. exposed the relationship between IL-6 and rheumatoid arthritis-associated osteoporosis (Edwards and Williams, 2010).
Although current advantages on investigating the relationship between IL-18 and Osteoporosis, it is still not clear whether IL-18 is the consequent or causal effect of the Osteoporosis. This is the common problem for many associations between phenotypes and diseases. With the development of Genome-Wide Association Studies (GWAS) and identification of molecular mechanism in recent years (Li et al., 2015; Zhou et al., 2017a,b; Tang et al., 2018; Tan et al., 2019), Mendelian randomization (MR) analysis is widely used to expose the causal effect of phenotypes on the development of diseases. For example, body mass index and C-reaction protein are identified as the causal effect of the development of type 2 diabetes (Cheng et al., 2018b, 2019c). Meanwhile, some negative associations are also exposed, such as associations between infant length and type 2 diabetes (Zhuang et al., 2019a). As other statistical analysis and machine learning methods (Du et al., 2018; Liao et al., 2018; Wang B. et al., 2018; Wang L. et al., 2018; Cheng et al., 2019b; Han et al., 2019; Lv et al., 2019; Yang et al., 2019; Zeng et al., 2019; Zou and Ma, 2019; Zhao et al., 2020), MR is an instrumental variable (IV) based method for inferring associations between phenotypes and diseases. As shown in Figure 1, genetic variants as SNPs are often used as IVs. This is because that SNPs are genetic characterize and occurred before phenotypes and diseases, which can avoid reverse causality. Here Z (e.g., SNPs) represents IVs, X is phenotype (e.g., IL-18), and Y is the disease (Osteoporosis). To conduct MR analysis, the IVs should meet two assumptions. One is that SNPs should be robustly associated with phenotype (IL-18), and the other is that SNPs can influence the disease only through the phenotype.
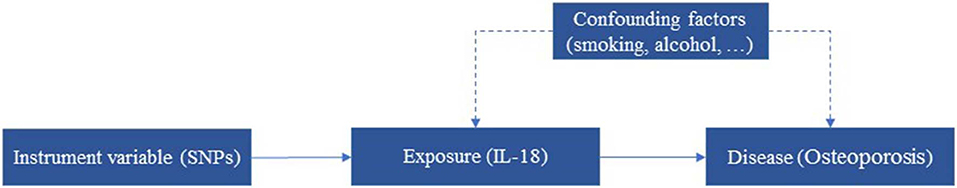
Figure 1. MR analysis using SNPs as instrumental variables for estimating the influence of IL-18 on the risk of Osteoporosis.
To explore the causal effect of IL-18 on the development of Osteoporosis, we conducted a MR analysis in this study. First, we defined a framework for processing data to establish IVs for MR analysis. Next, simple median method was used for calculating the pooled result based on IVs. Then, to avoid bias and analysis the limitation of our method, MR Egger analysis and leave-one-out validation was conducted.
Summary-level data of GWAS dataset was the fundamental for MR analysis. IVs should be extracted from IL-8 related GWAS dataset. And the further analysis need Osteoporosis related GWAS dataset. To meet the MR assumptions and reduce the bias, the summarized GWAS data was processed. Subsequently, MR analysis involving simple median method, leave-one-out validation, MR-Egger analysis was conducted to comprehensively assess the causal effect of IL-8 on the risk of the development of Osteoporosis.
In 2014, Matteini et al. (2014) performed a genome-wide association study (GWAS) on Cardiovascular Health Study and InCHIANTI cohorts. Totally, it contains 1200 InCHIANTI cohorts and 3200 CHS population. After genotyping, they used GWAPower software to assess the difference in power of the combined InCHIANTI-CHS meta-analysis compared to single study analyses. Then, significant SNPs of IL-18 were identified and used for analyzing causal relationship of IL-18 on the risk of Type 2 Diabetes (Zhuang et al., 2019a). Here, we extracted SNPs, effect allele (EA), allele frequencies, beta coefficients, and standard errors (SEs) as summarized data.
In 2018, Bycroft et al. published their prospective cohort study (Bycroft et al., 2018), which contains approximately 500,000 individuals from across the United Kingdom, aged between 40 and 69 at recruitment. It also provided detailed description and summarized data of deep genetic and phenotypic data. We downloaded summarized GWAS data of Osteoporosis from UK Biobank, which involves susceptibility loci together with other SNPs, beta coefficient, EA, SEs and their P-values, and etc. Totally, it contains 933 cases and 360,261 controls.
We process the summarized GWAS data for constructing IVs of MR analysis. Here IVs are genetic variants. Each SNP should be significantly associated with IL-18, and should be not associated with Osteoporosis. Thus, we extracted SNPs significant associated with IL-18, and then removed these SNPs associated with Osteoporosis. We defined P < 5*10−8 as significant associated SNPs of IL-18, and we defined P-value more than 0.05 as not associated SNPs of Osteoporosis. To avoid over-precise estimates due to genetic pleiotropy, we should remove these SNPs with potential linkage disequilibrium (LD) relationships. The analogous method has been used in the MR analysis of causal effect of phenotype on T2DM (Cheng et al., 2019c; Zhuang et al., 2019b). To remove SNPs with LDs, we ranked significant SNPs of IL-18 based on P-values. For each SNP, we removed those SNPs in LD with it (R2 threshold of 0.001) or within 10,000 kb physical distance based on a reference dataset (Devuyst, 2015). This process was iterated for each of significant SNPs of IL-18.
MR analysis contains simple median method, leave-one-out validation, MR-Egger regression analysis (Bowden et al., 2015). Simple median method was used for assessing the pooled influence of IL-18 on the risk of Osteoporosis. Leave-one-out validation was performed for assessing sensitivity of each of IVs. Egger regression analysis was used to evaluate pleiotropy bias of IVs.
• Simple median method
Simple median method was described in the previous study (Burgess et al., 2016) for evaluating the influence of clinical phenotype on the risk of disease, which is defined as following equation.
where X, Y, and Z are IL-18, Osteoporosis, and IVs, respectively, Wald ratio (betaXY) of IL-18 to Osteoporosis through specified IV, betaZY is the per-allele log(OR) of Osteoporosis from summarized GWAS data of Osteoporosis. betaZX is the per-allele log(OR) of IL-18 from summarized GWAS data of IL-18. Then, we calculated SE of association between IL-18 and Osteoporosis for each Wald ratio, which is defined as Equation 2.
where SEZY and SEZX represent the SE of the IV-Osteoporosis and IV-IL-18 associations from corresponding summarized GWAS data, respectively. Next, we calculated 95% confidence intervals (CIs) from the SE of each Wald ratio. To get the pooled influence of these IVs, simple median method was used as meta-analysis for estimating comprehensive influence of IVs.
• Leave-one-out validation
We used leave-one-out validation for evaluating the sensitivity of each of IVs as following. To assess the influence of a SNP of IVs to the pooled result, we remove this SNP from IVs to get the result using simple median method. Thus, the corresponding result is obtained without considering this SNP. The fluctuation of the pooled results before and after removing the SNP could reflect the sensitivity of this SNP. This process was iterated on each of these IVs to get the influence for each of IVs.
• MR-Egger analysis
We conducted a MR-Egger regression analysis about asymmetry test to measure bias based on potential pleiotropic effect of IVs (Bowden et al., 2015). The MR-Egger regression is source from Egger regression, which is designed for detecting bias due to small study and pleiotropy in meta-analysis. Here, MR-Egger used intercept as an estimated value for evaluating the average pleiotropic effect of IVs. For example, the larger or smaller an intercept, the more of pleiotropy effect should be. All statistical tests for this study were undertaken using the R Package of MRBase (Hemani et al., 2018).
Totally, 8 SNPs were extracted as significant associated SNPs of IL-18. Those SNPs were not associated with Osteoporosis and have no LD associations. As a result, those 8 significant SNPs of IL-18 were eventually selected as IVs for the MR analysis, which were shown in the Table 1. Each line of the table documents 10 items involving the SNP, EA, chromosome position, beta coefficients and SE of the SNP on the risk of IL-18 and Osteoporosis, and so on.
After using 8 individual SNPs as IVs for MR analysis based on two summary-level GWAS data, we used simple median method for pooled analysis. Figure 2 shows that there is no evidence of heterogeneity between variants of the summarized data. As a result, we get the SE (−0.001; 95% CI −0.002 to 0; P = 0.042), which means low IL-18 level could increase the risk of the development of Osteoporosis.
Figure 3 shows estimate result of the leave-one-out analysis. After removing rs6760105, rs6748621, rs7577696, or rs2250417 from 8 IVs, the estimate value shows small fluctuation. And the result is consistent with using all the IVs. This means that this four SNPs activate weak influence to the estimate result. In comparison, after removing rs212713, rs2300702, rs2268797, or rs212745, estimate value shows large fluctuation. This means that this four SNPs activate strong influence to the estimate result. The detailed information about leave-one-out validation result is shown in Table 2.
Figure 4 shows the effect estimate based on MR-Egger regression. The low intercept (0; 95% CI −0.001 to 0; P = 0.59) shows there is no bias due to the horizontal pleiotropy of the IVs.
Till now, it is not clear that the level of IL-18 is the causal or consequence of the development of Osteoporosis. To expose the relationship, we conducted an MR analysis based on two summarized GWAS datasets in this study. As a result of simple median method, we get the SE (−0.001; 95% CI −0.002 to 0; P = 0.042). This means that low IL-18 level could increase the risk of the development of Osteoporosis.
Observational study is a widely used way to reveal the associations between phenotypes and diseases. Whereas, it couldn't be used for exposing the causal effect. With the increase of GWAS data and abundance of molecular characterize (Cheng, 2019; Cheng et al., 2019a; Dao et al., 2020; Zhang et al., 2020), more and more researchers choose MR analysis for this purpose. MR analysis is an IV-based framework, which requires summarized GWAS data. In recent years, MR analysis has helped us to identify lots of causal effects, such as body mass index and C-reactive protein increase the risk of type 2 diabetes (Cheng et al., 2019c; Zhuang et al., 2019b). Here, the number of the case and control for GWAS data is very important for the estimation. In the previous study, the summarized data of IL-18 GWAS data has been applied in exposing the relationship between IL-18 and T2DM (Zhuang et al., 2019a). And the number of case and control is over 5,000. In addition, the UK biobank provided 933 Osteoporosis and 360,261 controls. The number of the samples for IL-18 and Osteoporosis GWAS data is the baseline for our MR analysis.
To make the MR analysis reliably, the summarized GWAS data was processed strictly to choose suitable IVs. First, significant associated SNPs of IL-18 (P < 5*10−8) was extracted. Then, these Osteoporosis (P < 5*10−2) associated SNPs were removed. This process is to make sure the IVs meet the requirement of MR's assumption. In addition, to reduce the bias of IVs due to the pleiotropic effect, SNPs in IVs with LDs were removed. In actually, the comprehensive effect of IVs are used for estimating the causal effect. The SNPs with LDs should be deemed as a SNP to reduce the bias based on the replication. To detect the potential bias due to the horizontal pleiotropy of IVs, MR egger method was conducted. As a result, we got the low intercept (0; 95% CI −0.001 to 0; P = 0.59), which shows there is no bias due to the horizontal pleiotropy of the IVs. All the data process of IVs is to reduce the bias, and make the result more reliably.
Although there is no direct associations between IVs and Osteoporosis, we also validated their potential linkages. We downloaded associated genes of Osteoporosis from a widely used functional annotation database OAHG (Cheng et al., 2016, 2018a), and compared them with genes that IVs located at (Table 1). Up to 440 genes of Osteoporosis were documented in OAHG, and no intersection between them and genes of IVs. This validate further that no potential bias of IVs based on current knowledge.
Although MR analysis show the advantages in distinguishing causal effect from general associations. It also has the limitations. With the incensement of samples, the summarized data of phenotype and disease should be a little alteration. This could lead to the changes in fluctuations about summarized data of GWAS data and even in IVs. This would influence the estimated result. Thus, to avoid this problem, the huge number of samples is very important. In this study, we do a leave-one-out analysis to judge which SNPs could influence our results. As a result, we find out SNPs rs6760105, rs6748621, rs7577696, or rs2250417 shows a little capability in influencing the result. In comparison, SNPs rs212713, rs2300702, rs2268797, or rs212745 could largely influence the result, all of which are located at genes SRD5A2, SLC30A6, and NLRC4. With the increase of the GWAS data, the impact of these SNPs could be decreased.
In addition to simple median method, IVW method is another frequently used method for MR analysis. As a result of IVW method, we get SE (0; 95% CI −0.001 to 0.001; P = 0.625), which is inconsistent with the result of simple median method. Currently, most of current MR analysis based on different methods often show inconsistent results. Each method has its advantages and limitations. In general, the causal effect could be validated when the relationship is supported by one of these methods. To provide more reliable validation, it needs randomized controlled trial (RCT). Whereas, it is very hard to conduct RCT. Thus, most of current causal effect between clinical phenotypes and diseases are validated based on MR analysis. Although current success in applying MR analysis, it couldn't substitute for RCT.
In summary, 8 SNPs were used as IVs for estimating the causal effect of IL-18 on the development of Osteoporosis. Results show that low IL-18 level could increase the risk of the development of Osteoporosis based on simple median method. In considering the limitation of MR method and current samples of GWAS data, further experiment for the conclusion is expected.
Publicly available datasets were analyzed in this study. This data can be found here: http://biobank.ctsu.ox.ac.uk/crystal/field.cgi?id=20002.
HL conceived and designed the experiments. NK and WZ analyzed the data. NK, WZ, and HL wrote the manuscript. All authors read and approved the final manuscript.
This work was supported by the National Natural Science Foundation of China (Grant No. 61871068).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Bowden, J., Davey Smith, G., and Burgess, S. (2015). Mendelian randomization with invalid instruments: effect estimation and bias detection through Egger regression. Int. J. Epidemiol. 44, 512–525. doi: 10.1093/ije/dyv080
Burgess, S., Bowden, J., Dudbridge, F., and Thompson, S. G. (2016). Robust instrumental variable methods using multiple candidate instruments with application to Mendelian randomization. Methodology. arXiv.org [Preprint]. arXiv:1606.03729.
Bycroft, C., Freeman, C., Petkova, D., Elliott, L. T., Sharp, K., Motyer, A., et al. (2018). The UK Biobank resource with deep phenotyping and genomic data. Nature 562, 203–209. doi: 10.1038/s41586-018-0579-z
Cheng, L. (2019). Computational and biological methods for gene therapy. Curr. Gene Ther. 19, 210–210. doi: 10.2174/156652321904191022113307
Cheng, L., Hu, Y., Sun, J., Zhou, M., and Jiang, Q. (2018a). DincRNA: a comprehensive web-based bioinformatics toolkit for exploring disease associations and ncRNA function. Bioinformatics 34, 1953–1956. doi: 10.1093/bioinformatics/bty002
Cheng, L., Qi, C., Zhuang, H., Fu, T., and Zhang, X. (2019a). gutMDisorder: a comprehensive database for dysbiosis of the gut microbiota in disorders and interventions. Nucl. Acids Res. 48, D554–D560. doi: 10.1093/nar/gkz843
Cheng, L., Sun, J., Xu, W. Y., Dong, L. X., Hu, Y., and Zhou, M. (2016). OAHG: an integrated resource for annotating human genes with multi-level ontologies. Sci. Rep. 6:34820. doi: 10.1038/srep34820
Cheng, L., Zhao, H., Wang, P., Zhou, W., Luo, M., Li, T., et al. (2019b). Computational methods for identifying similar diseases. Mol. Ther. Nucl. Acids 18, 590–604. doi: 10.1016/j.omtn.2019.09.019
Cheng, L., Zhuang, H., Ju, H., Yang, S., Han, J., Tan, R., et al. (2019c). Exposing the causal effect of body mass index on the risk of type 2 diabetes mellitus: a Mendelian randomization study. Front Genet 10:94. doi: 10.3389/fgene.2019.00094
Cheng, L., Zhuang, H., Yang, S., Jiang, H., Wang, S., and Zhang, J. (2018b). Exposing the causal effect of C-reactive protein on the risk of type 2 diabetes mellitus: a Mendelian randomization study. Front. Genet. 9:657. doi: 10.3389/fgene.2018.00657
Dao, F. Y., Lv, H., Zulfiqar, H., Yang, H., Su, W., Gao, H., et al. (2020). A computational platform to identify origins of replication sites in eukaryotes. Brief. Bioinform. doi: 10.1093/bib/bbaa017. [Epub ahead of print].
Devuyst, O. (2015). The 1000 Genomes project: welcome to a new world. Perit. Dial. Int. 35, 676–677. doi: 10.3747/pdi.2015.00261
Du, X. Q., Li, X. R., Li, W., Yan, Y. T., and Zhang, Y. P. (2018). Identification and analysis of cancer diagnosis using probabilistic classification vector machines with feature selection. Curr. Bioinform. 13, 625–632. doi: 10.2174/1574893612666170405125637
Edwards, C. J., and Williams, E. (2010). The role of interleukin-6 in rheumatoid arthritis-associated osteoporosis. Osteoporos. Int. 21, 1287–1293. doi: 10.1007/s00198-010-1192-7
Han, J., Han, X., Kong, Q., and Cheng, L. (2019). psSubpathway: a software package for flexible identification of phenotype-specific subpathways in cancer progression. Bioinformatics. doi: 10.1093/bioinformatics/btz894. [Epub ahead of print].
Hemani, G., Zheng, J., Elsworth, B., Wade, K. H., Haberland, V., Baird, D., et al. (2018). The MR-Base platform supports systematic causal inference across the human phenome. eLife 7:e34408. doi: 10.7554/eLife.34408
Hennemann, A. (2002). Osteoporosis: prevention, diagnosis and therapy. Medizinische Monatsschrift Für Pharmazeuten 25, 164–167.
Kato, H., and Perl, A. (2018). Blockade of treg cell differentiation and function by the interleukin-21–mechanistic target of rapamycin axis via suppression of autophagy in patients with systemic lupus erythematosus. Arthritis Rheumatol. 70, 427–438. doi: 10.1002/art.40380
Lewis, D. B., Liggitt, H. D., Effmann, E. L., Motley, S. T., Teitelbaum, S. L., Jepsen, K. J., et al. (1993). Osteoporosis induced in mice by overproduction of interleukin 4. Proc. Natl. Acad. Sci. U.S.A. 90, 11618–11622. doi: 10.1073/pnas.90.24.11618
Li, P., Guo, M., Wang, C., Liu, X., and Zou, Q. (2015). An overview of SNP interactions in genome-wide association studies. Brief. Funct. Genomics 14, 143–155. doi: 10.1093/bfgp/elu036
Liao, Z. J., Li, D. P., Wang, X. R., Li, L. S., and Zou, Q. (2018). Cancer diagnosis through IsomiR expression with machine learning method. Curr. Bioinform. 13, 57–63. doi: 10.2174/1574893611666160609081155
Lv, H., Zhang, Z. M., Li, S. H., Tan, J. X., Chen, W., and Lin, H. (2019). Evaluation of different computational methods on 5-methylcytosine sites identification. Brief. Bioinform. doi: 10.1093/bib/bbz048. [Epub ahead of print].
Matteini, A. M., Li, J., Lange, E. M., Tanaka, T., Lange, L. A., Tracy, R. P., et al. (2014). Novel gene variants predict serum levels of the cytokines IL-18 and IL-1ra in older adults. Cytokine 65, 10–16. doi: 10.1016/j.cyto.2013.10.002
Papaleontiou, M., Banerjee, M., Reyes-Gastelum, D., Hawley, S. T., and Haymart, M. R. (2019). Risk of osteoporosis and fractures in patients with thyroid cancer: a case–control study in U.S. Veterans. Oncologist 24, 1166–1173. doi: 10.1634/theoncologist.2019-0234
Prencipe, G., Bracaglia, C., and Benedetti, F. D. (2019). Interleukin-18 in pediatric rheumatic diseases. Curr. Opin. Rheumatol. 31, 421–427. doi: 10.1097/BOR.0000000000000634
Rusinska, A., and Chlebnasokół, D. (2005). Evaluation of interleukin-1 and -6 in the etiopathogenesis of idiopathic osteoporosis and osteopenia in children. Arch Immunol Ther Exp 53, 257–265.
Seo, G. H., Kang, H. Y., and Choe, E. K. (2018). Osteoporosis and fracture after gastrectomy for stomach cancer: a nationwide claims study. Medicine 97:e0532. doi: 10.1097/MD.0000000000010532
Tan, J. X., Li, S. H., Zhang, Z. M., Chen, C. X., Chen, W., Tang, H., et al. (2019). Identification of hormone binding proteins based on machine learning methods. Math. Biosci. Eng. 16, 2466–2480. doi: 10.3934/mbe.2019123
Tang, H., Zhao, Y. W., Zou, P., Zhang, C. M., Chen, R., Huang, P., et al. (2018). HBPred: a tool to identify growth hormone-binding proteins. Int. J. Biol. Sci. 14, 957–964. doi: 10.7150/ijbs.24174
Tu, K. N., Lie, J. D., C, Wan, K. V., Cameron, M., and Hyun, D. (2018). Osteoporosis: a review of treatment options. P T 43, 92–104.
Valero, N., Mosquera, J., Torres, M., Duran, A., and Veliz, T. (2019). Increased serum ferritin and interleukin-18 levels in children with dengue. Braz. J. Microbiol. 50, 649–656. doi: 10.1007/s42770-019-00105-2
Wang, B., Lu, K., Zheng, X., Su, B. Y., Zhou, Y. M., Chen, P., et al. (2018). Early stage identification of Alzheimer's disease using a two-stage ensemble classifier. Curr. Bioinform. 13, 529–535. doi: 10.2174/1574893613666180328093114
Wang, J., Chen, L., Zhang, Y., Li, C.-G., and Shu, B. (2019). Association between serum vitamin B 6 concentration and risk of osteoporosis in the middle-aged and older people in China: a cross-sectional study. BMJ Open 9:e028129. doi: 10.1136/bmjopen-2018-028129
Wang, L., Ping, P. Y., Kuang, L. N., Ye, S. T., Lqbal, F. M. B., and Pei, T. R. (2018). A novel approach based on Bipartite network to predict human microbe-disease associations. Curr. Bioinform. 13, 141–148. doi: 10.2174/1574893612666170911143601
Weiss, E. S., Girard-Guyonvarc'h, C., Holzinger, D., de Jesus, A. A., Tariq, Z., Picarsic, J., et al. (2018). Interleukin-18 diagnostically distinguishes and pathogenically promotes human and murine macrophage activation syndrome. Blood 131, 1442–1455. doi: 10.1182/blood-2017-12-820852
Yang, H., Yang, W., Dao, F. Y., Lv, H., Ding, H., Chen, W., et al. (2019). A comparison and assessment of computational method for identifying recombination hotspots in Saccharomyces cerevisiae. Brief. Bioinform. bbz123. doi: 10.1093/bib/bbz123. [Epub ahead of print].
Youssef, D. M., Al-Atif, A. M. A., El-Khateeb, S. S. H., and Elshal, A. S. (2018). Evaluation of interleukin-18 in children with steroid-sensitive nephrotic syndrome before and after using levamisole. Saudi J. Kidney Dis. Transpl. 29, 591–597. doi: 10.4103/1319-2442.235173
Zeng, X. X., Wang, W., Deng, G. S., Bing, J. X., and Zou, Q. (2019). Prediction of potential disease-associated microRNAs by using neural networks. Mol. Ther. Nucl. Acids 16, 566–575. doi: 10.1016/j.omtn.2019.04.010
Zhang, J., Li, Z., Liu, L., Wang, Q., Li, S., Chen, D., et al. (2018). Long noncoding RNA TSLNC8 is a tumor suppressor that inactivates the interleukin-6/STAT3 signaling pathway. Hepatology 67, 171–187. doi: 10.1002/hep.29405
Zhang, Z. Y., Yang, Y. H., Ding, H., Wang, D., Chen, W., and Lin, H. (2020). Design powerful predictor for mRNA subcellular location prediction in Homo sapiens. Brief. Bioinform. doi: 10.1093/bib/bbz177. [Epub ahead of print].
Zhao, X., Jiao, Q., Li, H., Wu, Y., Wang, H., Huang, S., et al. (2020). ECFS-DEA: an ensemble classifier-based feature selection for differential expression analysis on expression profiles. BMC Bioinform. 21:43. doi: 10.1186/s12859-020-3388-y
Zhou, M., Zhang, Z., Zhao, H., Bao, S., Cheng, L., and Sun, J. (2017b). An immune-related six-lncRNA signature to improve prognosis prediction of glioblastoma multiforme. Mol. Neurobiol. 55, 3684–3697. doi: 10.1007/s12035-017-0572-9
Zhou, M., Zhao, H. Q., Xu, W. Y., Bao, S. Q., Cheng, L., and Sun, J. (2017a). Discovery and validation of immune-associated long non-coding RNA biomarkers associated with clinically molecular subtype and prognosis in diffuse large B cell lymphoma. Mol. Cancer 2017:16. doi: 10.1186/s12943-017-0580-4
Zhuang, H., Han, J., Cheng, L., and Liu, S. (2019a). A positive causal influence of IL-18 levels on the risk of T2DM: a mendelian randomization study. Front. Genet. 10:295. doi: 10.3389/fgene.2019.00295
Zhuang, H., Zhang, Y., Yang, S., Cheng, L., and Liu, S. L. (2019b). A Mendelian randomization study on infant length and type 2 diabetes mellitus risk. Curr Gene Ther 19, 224–231. doi: 10.2174/1566523219666190925115535
Zou, Q., and Ma, Q. (2019). The application of machine learning to disease diagnosis and treatment. Math. Biosci. 320:108305. doi: 10.1016/j.mbs.2019.108305
Keywords: genome-wide association studies, Interleukin-18, Osteoporosis, casual effect, Mendelian randomization
Citation: Kou N, Zhou W, He Y, Ying X, Chai S, Fei T, Fu W, Huang J and Liu H (2020) A Mendelian Randomization Analysis to Expose the Causal Effect of IL-18 on Osteoporosis Based on Genome-Wide Association Study Data. Front. Bioeng. Biotechnol. 8:201. doi: 10.3389/fbioe.2020.00201
Received: 29 January 2020; Accepted: 28 February 2020;
Published: 20 March 2020.
Edited by:
Meng Zhou, Wenzhou Medical University, ChinaReviewed by:
Lei Deng, Central South University, ChinaCopyright © 2020 Kou, Zhou, He, Ying, Chai, Fei, Fu, Huang and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Huiying Liu, bGh5MDQ1MTIwMDBAZG11LmVkdS5jb20=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.