- 1Dipartimento di Farmacia, Scienze della Salute e della Nutrizione – Universitá della Calabria (DFSSN-UNICAL), Ed. Polifunzionale, Arcavacata di Rende (CS), Rende, Italy
- 2Centro di Ricerca Olivicoltura, Frutticoltura, Agrumicoltura, Consiglio per la Ricerca in Agricoltura e L’analisi dell’Economia Agraria (CREA-OFA), Rende, Italy
- 3Dipartimento di Biologia, Ecologia e Scienza della Terra, Università della Calabria, Arcavacata di Rende, Italy
Olive leaf extract is characterized by a high content of phenols and flavonoids (oleuropein, luteolin, and their derivatives). These compounds are defined as secondary metabolites and exert such as anti-inflammatory, antioxidant, and antimicrobial activities. We investigated the in vitro antifungal activity of two olive leaf extracts (named EF1 and EF2) against a Fusarium proliferatum (AACC0215) strain that causes diseases to many economically important plants and synthesizing diverse mycotoxins. In this work, we aimed to identify the most appropriate concentration between the tested two olive leaf extracts to develop a safe, stable and efficient drug delivery system. Qualitative and quantitative analyses of the two olive leaf extracts by (HPLC) were performed. Furthermore, we also evaluated the antifungal effects of the two leaf extracts when encapsulated in chitosan-tripolyphosphate nanoparticles. The major compound in both EF1 and EF2 was oleuropein, with 336 and 603 mg/g, respectively, however, high concentrations of flavonoid were also present. EF1 and EF2 showed a concentration depended effect on F. proliferatum (AACC0215) viability. Our results showed a great efficacy of EF1/nanoparticles at the higher concentration tested (12X) against the target species. In this case, we observed an inhibition rate to both germination and growth of 87.96 and 58.13%, respectively. We suggest that EF1 olive leaf extracts, as free or encapsulated in chitosan-tripolyphosphate nanoparticles, could be used as fungicides to control plant diseases. Finally, future application of these findings may allow to reduce the dosage of fungicides potentially harmful to human health.
Introduction
Nanoparticle formulation is beneficial in different fields including electronics, textiles, mobile phones, food, paper, robotics, fertilizers, pesticides, and agrochemical industries. In recent years, an increased interest has been developed for natural polymers which have a versatility due to their chemical, physical and functional properties. The wide range of potential applications has led to their use in various fields of research, mainly in the biomedical, cosmetics, food and pharmaceuticals (Agnihotri et al., 2004; Manna and Patil, 2009).
Chitosan (CS) has emerged as one of the most promising polymers for the formation of nanoparticles (NPs) (Kashyap et al., 2015), mainly due to its biodegradable and biocompatible properties, its moderate or lack of toxicity to animals and humans, and for its antimicrobial and antifungal activity (López-León et al., 2005; Zhou and Chen, 2008; Akamatsu et al., 2010). Chitosan nanoparticles have gained considerable popularity as a carrier for the active ingredient delivery for various applications owing to their biocompatibility, biodegradability, high permeability, cost-effectiveness, and non-toxicity (Shukla et al., 2013). Various procedures can be employed to synthesize CSNPs, such as emulsion formation, coacervation, spray drying, ionotropic gelation. The method selected is mainly dependent on the substances encapsulated, and the route of administration. So, by varying the concentration and the molecular weight of the polymer and by using copolymers and crosslinking agents, efficient delivery systems for the pharmaceutical, biomedical and agricultural industry could be obtained (Höhne et al., 2007; Nasti et al., 2009).
In agriculture, NPs could be used as vectors to control release of agrochemicals, such as fertilizers, pesticides, herbicides and plant growth regulators (Cota-Arriola et al., 2013). Plants are continuously exposed to a series of pathogenic microorganisms such as fungi, oomycetes and bacteria, which can attack the plant both above and below ground (Buhtz et al., 2015) and cause the evolution of devastating epidemics and significant yield losses of annual crops, seriously affecting the economy. Several fungal species belonging to the genus Fusarium are known for their ability to colonize a wide variety of host plants, such as tomatoes, potatoes, cereal and tobacco (Desjardins, 2003; Schweigkofler et al., 2004; Alves-Santos et al., 2007; Nguyen et al., 2016). The most common symptoms of the disease are wilting, yellow leaves, dry collar, chlorosis, premature leaf drop, browning of the vascular system and growth arrest. When the disease spreads to the whole plant, necrosis and death occurs (Trapero-Casas and Jiménes-Dìaz, 1985). Fusarium produces mycotoxins which can have an important role in pathogen virulence during infection of the plant (Nguyen et al., 2016). The control of these fungi, responsible for pre- and post-harvested diseases of agricultural products, is an issue that remains unresolved, along with the excessive environmental impacts of chemicals to tackle this problem. Current efforts are focused to search new strategies and effective alternatives for microbial control and to reduce the excessive use of synthetic fungicides which negatively impact the environment and human and animal health (Cota-Arriola et al., 2013; Rodriguez-Maturino et al., 2015).
Plants have been a rich source of bioactive compounds for millennia, while the use of plant derivatives to produce nanobiotechnological formulations has gained scientific and technological importance in recent years (Joanitti and Silva, 2014). Olea europaea belongs to the Oleaceae family and it is native of the Mediterranean region. Olive oil, fruit and leaves have been recognized as important components of medicine and of a healthy diet. The extract from olive leaves were reported to have anticancer, antioxidative and anti-inflammatory properties (Le Toutour and Guedon, 1992; Anter et al., 2011). In addition to the health benefits described above, it is claimed that extracts from olive leaves may aid in the treatment of a broad range of infectious diseases. They have important pharmacological properties attributable primarily to the phenolic content (Omar, 2010). The main phenolic compound present in the leaves and fruits of olive tree is oleuropein (Ole) (Bianco et al., 1999; Goldsmith et al., 2015) and the detectable amount ranges from 17% to 23%, depending on the harvesting period (Le Toutour and Guedon, 1992). Olive leaves extract is characterized by a high content of phenolic compounds and flavonoids such as Ole, hydroxytyrosol and their derivatives (Zorić et al., 2016) and luteolin 7-glucoside and their derivatives (Sudjana et al., 2009). The antimicrobial activity of Ole and leaf extracts has been examined previously (Markin et al., 2003; Sudjana et al., 2009).
In this work, we aimed to identify the most appropriate concentration between the tested two olive leaf extracts to develop a safe, stable and efficient drug delivery system. These CSNPs were synthesized by a chemical route and displayed certain characteristics defined by preparation conditions. The physical and chemical characterization of the nanoformulation such as mean particle size, zeta potential values, polydispersity index (PDI), and EE were evaluated. In this study, the CSNPs antifungal effect was evaluated against F. proliferatum (AACC0215) strain through an in vitro assay, looking at different concentrations and preparations of the olive leaf extracts.
Experimental
Sample Preparation
The plant material used for the extraction is represented by fresh leaves of Carolea cultivar collected in November 2015 from plants grown in the Botanical Gardens of the University of Calabria, Arcavacata di Rende (CS) (GPS coordinates: latitude 39.357548; longitude 16.228990). The plants were identified by Dr. Nicodemo Passalacqua curator of the Botanical Gardens of University of Calabria.
Extraction and Characterization of the Olive Leaf Extracts
The method used for the extraction of olive leaf extracts is that described in Muzzalupo et al. (2011) with some modifications. Olive leaves (20 grams corresponding to about 100 leaves) were homogenized in 100 mL of a mixture of acetone and methanol in a ratio of 1:1 (v/v). The homogenization was carried out using an Ultra-TURRAX® (IKA, Seneco Science, Milan, Italy) for 5 min at room temperature. The homogenized mixture was vacuum filtered, and the liquid portion was recovered. The pellet was re-homogenized with the previous mixture and the process was repeated a further two times. The filtrate obtained was evaporated to dryness with a rotavapor (Strike 202 Rotary Evaporator, Steroglass, Perugia, Italy) and resuspended with 80 mL of distilled water. The filtrate was washed, in the separating funnel, with different solvents with increasing polarity: n-hexane, ethyl ether, chloroform and ethyl acetate. All solvents used are pure (ACS grade solvents, Sigma-Aldrich, Milano, Italy). The washings with n-hexane and ethyl ether were discarded, instead those from the chloroform and ethyl acetate phases were recovered and kept separate.
The extract obtained from the chloroform was referred to as “leaf extract 1” (EF1), while that derived from ethyl acetate as “leaf extract 2” (EF2). The two extracts were made anhydrous with sodium sulfate, filtered and evaporated to dryness and stored at −20°C in the dark.
Determination of Total Phenolic Compounds
The total phenolic content of each extract was determined spectrophotometrically at 750 nm using Folin-Ciocalteu reagent (Fuentes et al., 2012). To 1 mL of the sample to be tested were added 0.5 mL of Folin-Ciocolteu (Sigma-Aldrich) and left in the dark for 5 min. Subsequently 3 mL of Na2CO3 (Sigma-Aldrich) at 20% and 5.5 mL of distilled water were added. After 20 min, spent in the dark and at room temperature, samples were centrifuged at 3,500 rpm for 10 min. A calibration curve was calculated using pure Ole (Extrasynthèse, ZI Lyon-Nord, Genay, France). The total phenolic compounds are expressed as Ole milligrams per grams of extract.
Identification of Phenolic Compounds Contained in Each Extract by HPLC
Both extracts (EF1 and EF2), solubilized in methanol, were characterized performing HPLC analysis. The procedure used is that reported by Montedoro et al. (1992): HPLC JASCO LC-2000 plus equipped with a pump PU-2080 and UV-2075 detector (JASCO), with a RP-18 column Spherisorb ODS-2 (160 mm x 4.6 mm, Waters, Vimodrone, Italy) and injection volume of 20 μL; the flow rate was 1 mL/min at room temperature; the mobile phase used was 2% acetic acid in water (A) and methanol (B) for a total running time of 45 min, and the gradient conditions were as follows: 95% A-5% B for 2 min, 75% A-25% B for 8 min, 60% A-40% B for 10 min, 60% A-50% B for 10 min and 0% A-100% B for 10 min, until it stops; the eluents were detected at 280 nm. As phenolic standards were used: Ole, verbascoside, luteolin-4′-O-glucoside, luteoloside, luteolin, apigenin-7-O-glucoside and apigenin, all purchased from Extrasynthése.
Preparation and Characterization of the CSNPs
The CSNPs were prepared by ionotropic gelation method, reported by Rampino et al. (2013), with some modifications. Dispersions of chitosan were prepared, at a concentration of 1 mg/mL, by dissolving the medium molecular weight chitosan (50,000–190,000 Da, 75–85% deacetylated, Sigma Aldrich) in a solution of hydrochloric acid to 0.04% (v/v) and then stirring for 1 h. The pH of the CS solution was adjusted to 5.5 by NaOH. 1 mL of Ole or EF1 or EF2 water solution was added to 5 mL of the chitosan solution leaving it under stirring for a few minutes and adjusting the pH to 5.5. Tripolyphosphate (Sigma Aldrich) was dissolved in distilled water to a final concentration of 2 mg/mL and was added dropwise to the chitosan solution in a volumetric ratio of 1:5. The resulting solution was stirred for 30 min at room temperature. Moreover, CSNPs without the leaf extract were prepared. All formulations assayed (1X, solutions and carried CSNPs) were prepared at a final concentration of 100 mg/L of Ole (Table 1).
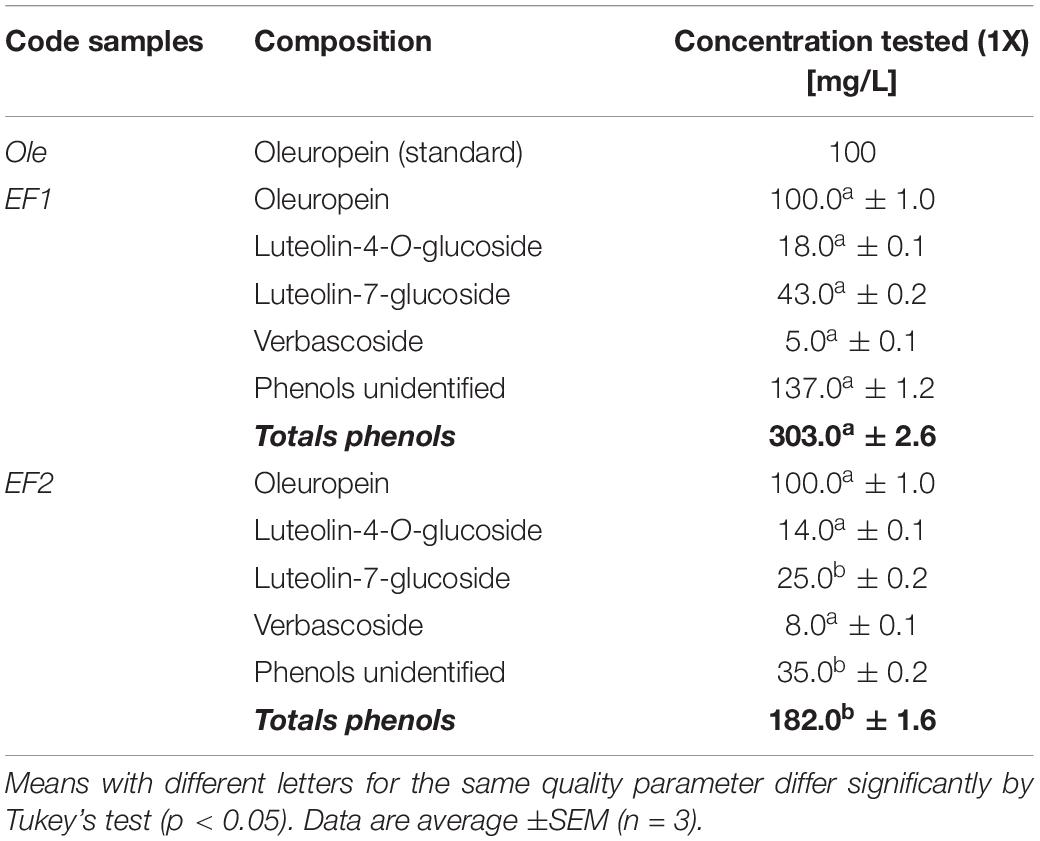
Table 1. Phenolic compounds (Oleuropein – Ole; leaf extract 1 – EF1; leaf extract 2 – EF2) used as free or encapsulated in CSNPs, for in vitro assays against Fusarium proliferatum (AACC0215).
All nanoformulations were characterized in terms of particle size, size distribution, PDI and zeta potential using a Zetasizer ZS (Malvern Instrument Ltd., Malvern), based on the DLS technique. DLS measurements of the samples were performed at 25°C with a detection angle of 90°.
Evaluation of Drug Loaded Efficiency
The extracts UV spectrum had a single maximum of absorption at 280 nm and this aspect allowed us to treat the extracts as a single component. The EE was calculated using the following formula (Shi et al., 2014):
Where drug means the phenolic compounds under study (EF1, EF2 and Ole). Each preparation was filtered using the syringe filters with a porosity equal to 0.2 μm (Millipore, Italy). 100 μL of filtrate are taken and brought to a final volume of 5 mL with distilled water. The amount of free and total drug was calculated by using the V-530 spectrophotometer (JASCO) at 280 nm (Mazzotta et al., 2020).
In vitro Olive Leaf Extracts Release
The release of Ole and leaf extracts from CSNPs was estimated using the method reported in Varuna Kumara and Basavaraj (2015), with some modifications. 2 mL of CSNPs/Ole or CSNPs/EF2 were taking and placing in pre-treated dialysis tubes Spectra/Por 4 (MWCO: 12–14 kD, Spectrum Laboratories, Inc., Canada). These were dipped into 50 mL of PBS solution (pH = 5.9) and left to stir at room temperature. At predetermining time points, 2 mL of the medium were taken and replaced with the same volumetric amount of fresh PBS. The solution was analyzed by UV-VIS spectrophotometry to evaluate the drug content.
Assessment of Antifungal Activity
Used Strains
In order to evaluate the antifungal activity of prepared extracts, the F. proliferatum (AACC0215) strain were used. Isolation and identification of the strain was described in a previous study (Muto et al., 2014). In brief, F. proliferatum (AACC0215) was isolated from colonized cloves of garlic (Allium sativum) collected at Altomonte, Cosenza, Italy in the year 2013, and taxonomically characterized as described in Muto et al. (2014). This strain was sub-cultured on potato dextrose broth (PDB) and incubated in darkness at 24°C. The suspension was diluted to a concentration of 1 × 105 spores/mL. Afterward it was divided into 1.5 mL aliquots and stored at −80°C in 25% glycerol (Steinkellner and Mammerler, 2007).
In vitro Test for the Evaluation of Germination
To verify the ability and success of F. proliferatum (AACC0215) to germinate in the presence of EF1, EF2, and Ole, different tests were performed using multiwell plates for cell cultures (Sigma-Aldrich). Inside of each well 200 μL of test preparation and 10 μL of conidial suspension at a concentration of 1 × 105 conidia/mL were added. Table 1 shows the composition of the analyzed individual preparations, respectively, containing Ole, EF1 and EF2 in solution or carrier to the CSNPs. The concentration used for each treatment was 3, 6, 9, and 12 times the initial one. As a control, the fungus was inoculated into the wells containing sterile water. The F. proliferatum (AACC0215) was incubated at 25–27°C in the dark and under aerobic conditions. After 24 h, 20 μL of each solution were taken to prepare slides by using a Malassez cell. The samples were observed under an optical microscope (DMRB Leica Microsystems, Milan, Italy) at × 400 magnification, equipped with a digital camera (DFC490 Leica Microsystems). The evaluation of germinated conidia and index of germination were performed according to Benslim et al., 2016. The conidia were considered as germinated when the germ tube length exceeded the diameter of the conidium (Figure 1; Boch et al., 1999; Rosengaus et al., 2000).
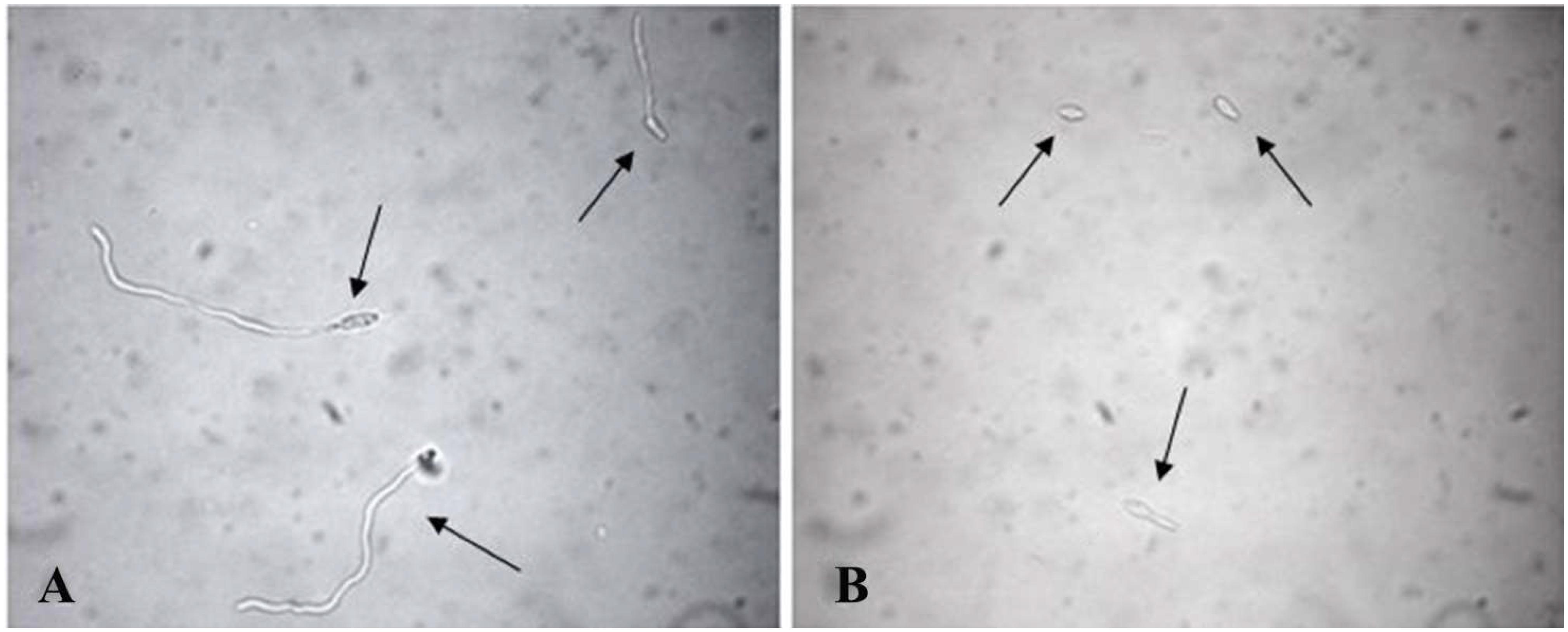
Figure 1. Conidia of Fusarium proliferatum (AACC0215) germinated in distilled water (A) and in presence of Ole (B) after 24 h. The samples were observed under an optical microscope at X400 magnification. The “arrow heads” indicate the germinated (A) and the non-germinated conidia (B).
For each slide, a total of 500 conidia were counted, by determining the percentage of inhibition rate (% IRg) by using the following formula:
In vitro Inhibition to Growth
The essays were conducted as described in Taskeen-Un-Nisa et al. (2011) with small modifications. Assays were performed in Petri plates containing 25 mL of PDA supplemented with streptomycin and ampicillin, at a final concentration of 6 mg/L each. At the center of the plates, a sterile polycarbonate filter with 0.8 μM porosity (Isopore Membrane Filters, Millipore) was placed on the PDA and 50 μL of the solutions to be tested were added on it.
In order to test the in vitro activity of the individual preparations at increasing concentrations, volumes of 3, 6, 9, and 12 times more than the starting solution described in Table 1 were loaded on the polycarbonate filter in the Petri capsule. In this way a thin and uniform film was formed on the surface.
PDA plates, to which a filter of 50 μL of ethanol had been added, were used as controls. Subsequently, the dry filters were inoculated at the center with 4 μL of conidial suspension at a concentration of 1 × 105 spores/mL and they were incubated in the dark at 24°C under aerobic conditions for 6 days. At the end of incubation, the capsule was photographed, and the image was analyzed by using the ImageJ software (vers. 1.49v National Institutes of Health, United States) to calculate the area, expressed in square millimeters, occupied by the mycelium.
The percentage of growth inhibition (I%) was calculated using the following formula (Chin Ming et al., 2015):
Where AC represents the average value of the area of mycelium used as a control and AT the average area value of the mycelium inoculated on plates treated with the individual preparations (Ole, EF1 and EF2 free and carrier to the CSNPs) (Kaiser et al., 2005; Taskeen-Un-Nisa et al., 2011).
Statistical Analysis
Statistical analysis was performed with XLSTAT v.2016. All data obtained from in vitro tests were compared by using One-way ANOVA, with Tukey’s multiple comparison test. All results are the mean of at least three individual experiments. All the values obtained from chemical analysis and biological tests are calculated from triplicate data were expressed as means ±standard error.
Results and Discussion
Analysis of Olive Leaf Extracts
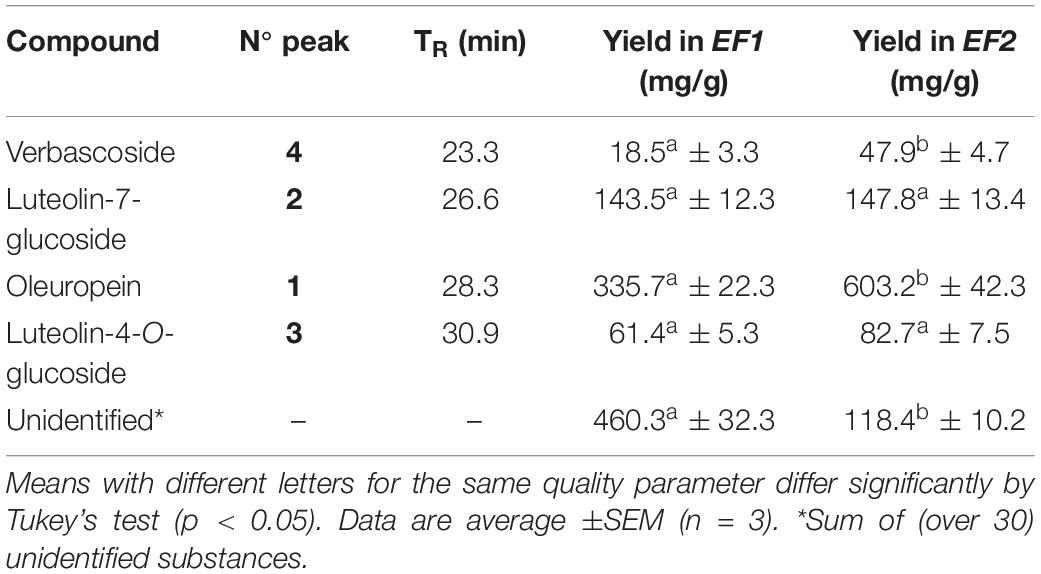
The identification of the phenolic compounds was carried out by comparing the retention times obtained from the HPLC analysis of the olive leaf extracts and those of the available standards. The results of the HLPC analysis of EF1 and EF2 showed a different content of phenols, in relation to the extraction procedure followed and their hydrophilicity (Table 2). In chloroform (EF1) and ethyl acetate (EF2) olive leaf extracts four phenol compounds were identified and quantified: Ole, verbascoside, luteolin-4′-O-glucoside, and luteolin-7-glucoside (Figure 2). Phenol compounds apigenin, luteolin, and apigenin-7-O-glucoside were not detected.
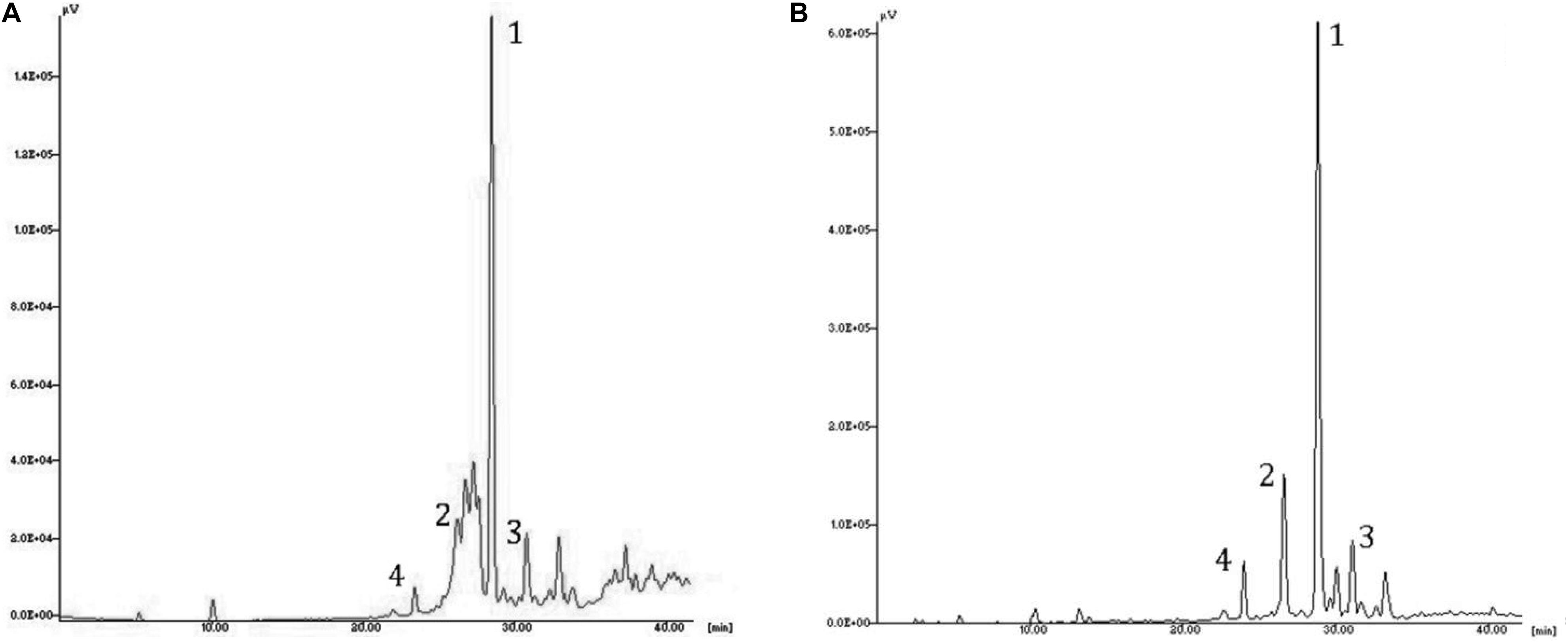
Figure 2. HPLC chromatogram of the leaf extracts EF1 (A) and EF2 (B). Assignment of the compounds to the identified peaks: (1) oleuropein, (2) luteolin-7-glucoside, (3) luteolin-4′-O-glucoside; (4) verbascoside.
The most abundant compound of chloroform and ethyl acetate olive leaf extracts is Ole, with 336 mg/g and 603 mg/g of extract, respectively. Furthermore, in both olive leaf extracts, there are numerous (over 30) unidentified phenolic compounds, which represented 460 and 118 mg/g of extract, respectively (Table 2).
The EF2 extract had a lower level of total phenols when compared to EF1 extract, 0.75 g and 1.04 g of extract, respectively. This suggests that most of the compounds present in the olive leaves were less hydrophilic phenols (Tan et al., 2014). Consistent with this, chloroform is less polar than ethyl acetate. The obtained results are in line with previously published data for O. europaea leaves, where Ole was identified as the major phenol compound extract (Yateem et al., 2014).
Nanoparticles Preparation and Characterization
Chitosan nanoparticles were prepared by ionotropic gelation with the dropwise addition of tripolyphosphate to a chitosan solution. Formation of NPs occurs quickly upon mixing tripolyphosphate and chitosan solutions and this is due to the electrostatic interactions between the positively charged primary amino groups of chitosan and the negatively charged groups of tripolyphosphate (Servat-Medina et al., 2015). The different formulations of CSNPs, containing different concentrations of Ole, showed good stability over time. They were stored in the dark at 4°C. When monitored after 30–40 days, they did not show sedimentation, creaming or flocculation. The particle size, PDI, zeta potential and EE are displayed in Table 3 for all nanoformulations.
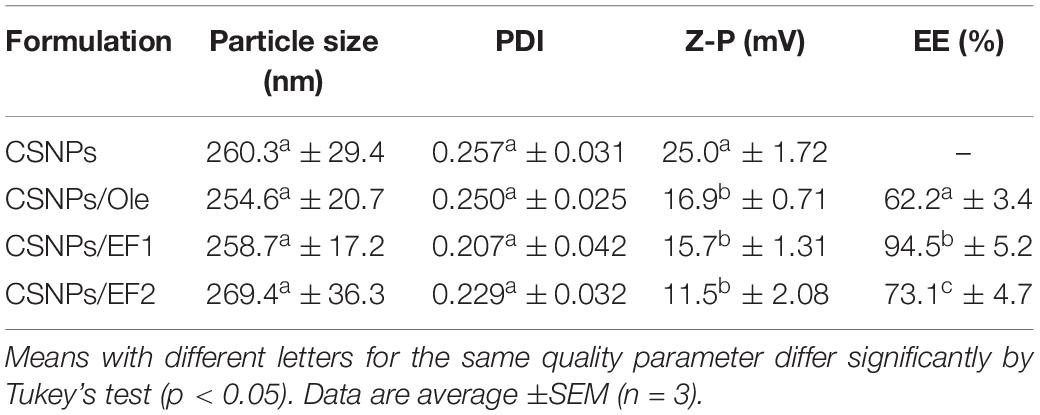
Table 3. The particle size, polydispersity index (PDI), zeta potential (Z-P), and encapsulation efficiency (EE%).
The NPs have always shown dimensions between 250 to 270 nm. As for the PDI values, these are always lower than 0.3 and this indicates a clear homogeneity of the system. No significant differences were seen in the Z-potential except for CSNPs/EF2 whose values are lower than those of CSNPs, indicating a greater presence of negative charge density. This result could possibly be related to the chemical nature of the unidentified compounds present in different percentages in the two leaf extracts.
During the formation of NPs, bioactive molecules are trapped both inside and on the surface of such particles. However, there is an initial burst release probably due to the drug on the surface, followed by a prolonged release (Bahreini et al., 2014).
Commercial Ole is completely released after 2 h of dialysis, while Ole in CSNPs is released more slowly, reaching the maximum value of 77% after 6 h (Figure 3A). EF1 after 45 h of dialysis against water has a negligible release, probably due to a lower hydrophilia. EF2 extract, both in solution and in NPs, is released in a lower percentage and more slowly than Ole (Figure 3B). Specifically, the free solution EF2 reaches a release value equal to 62% after 5 h, instead the release values of EF2 encapsulated in the NPs, reach the maximum value, equal to 38%, only after 30 h. This behavior may, probably, depend on the presence of lipophilic compounds in leaves, including lipophilic phenols, that are not allowed to pass into PBS.
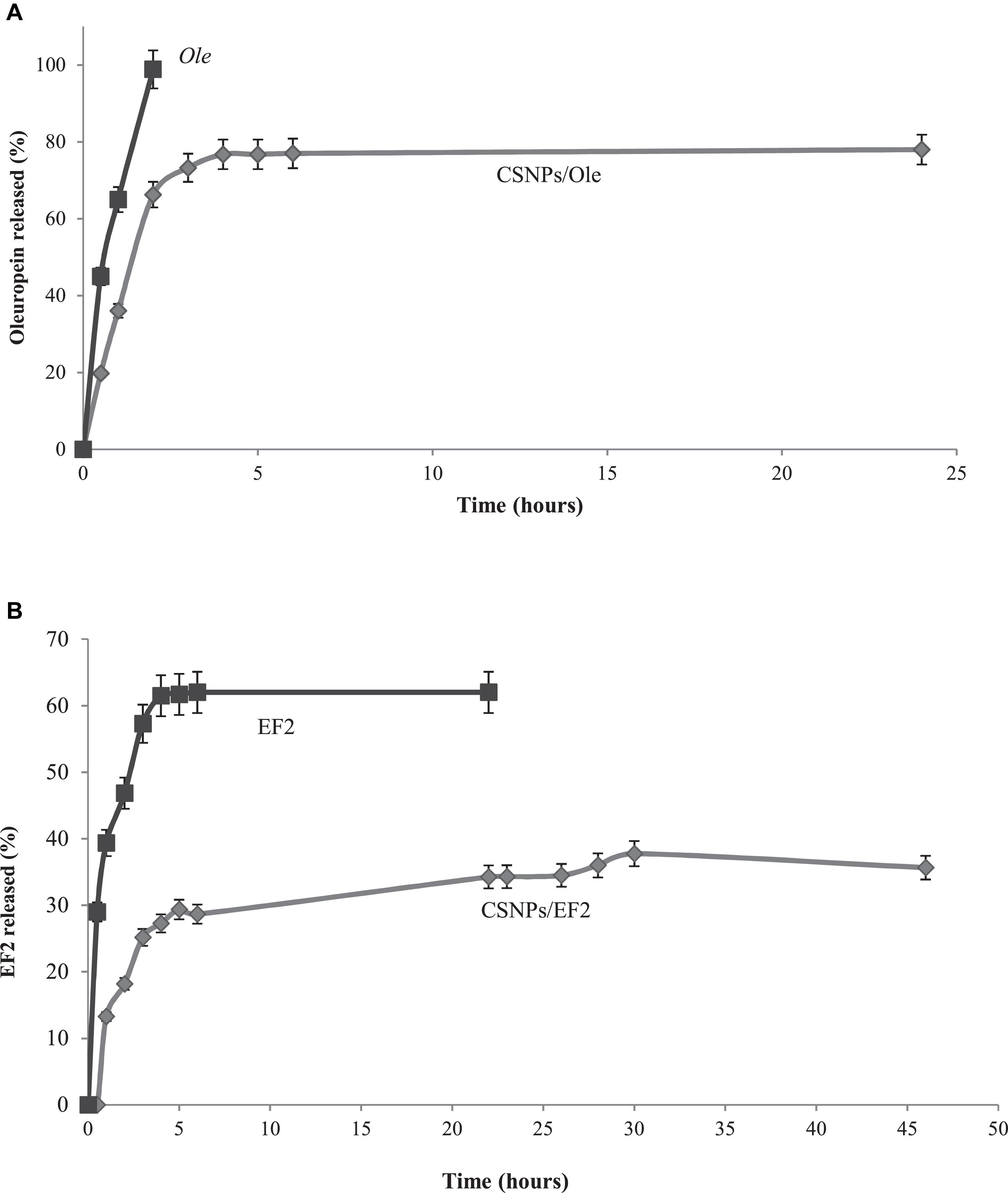
Figure 3. (A) In vitro release profile of the commercial oleuropein in solution (Ole) (■) and in chitosan nanoparticles (CSNPs/Ole) (•). Data are average ±S.E.M. (n = 3). (B) In vitro release profile of EF2 leaf extract in solution (■) and in chitosan nanoparticles (CSNPs/EF2) (•). Data are average ± SEM (n = 3).
Assessment of Antifungal Activity
In vitro Test for the Evaluation of Germination
The antifungal activity of Ole, EF1, and EF2 in solution or carrier on the CSNPs on conidial germination of F. proliferatum (AACC0215) is shown in Table 4. The obtained results were compared with distilled water. All formulations show an inhibition percentage of conidial germination versus control.
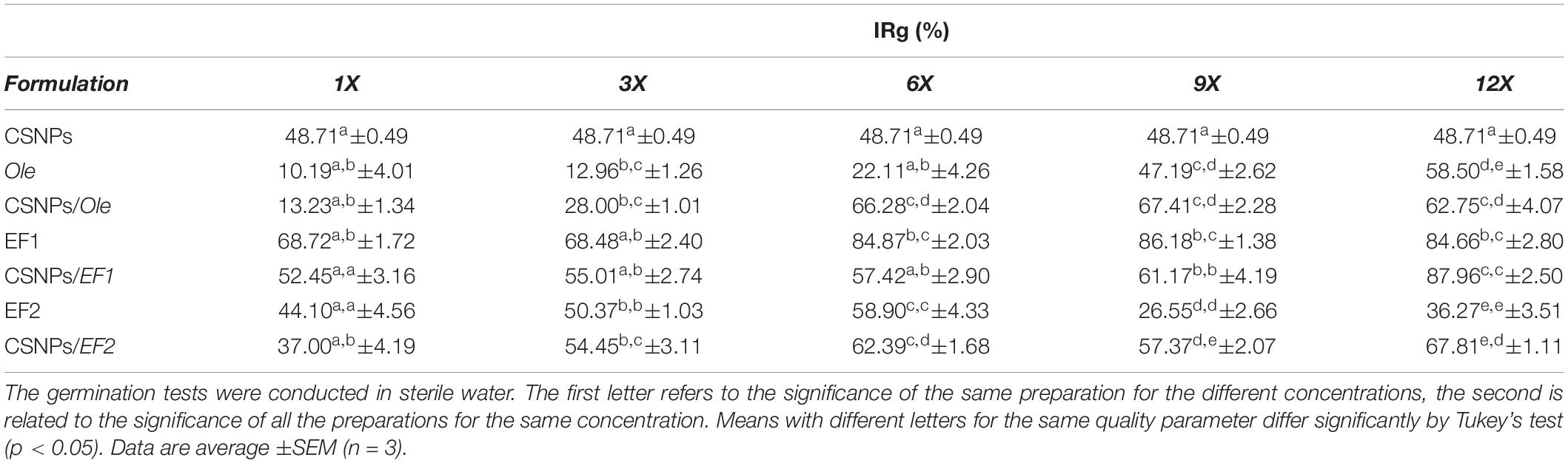
Table 4. In vitro Fusarium proliferatum (AACC0215) percentage of inhibition rate (% IRg) in the presence of pure chitosan nanoparticles (CSNPs), oleuropein (Ole), leaf extract 1 (EF1) and leaf extract 2 (EF2) free in solution or encapsulated into NPs.
The maximum inhibition percentage of conidial germination was obtained at the 12X concentration (87.96%), in CSNPs/EF1 formulation, while the lowest one was obtained at the 1X concentration (10.19%) of Ole solutions. The empty CSNPs markedly reduced conidial germination of Fusarium compared to the control, equal to 48.71%. The mechanism of action could be based on the electrostatic interaction between the amine group of chitosan and the negatively charged compounds (phospholipids, proteins, amino acids) of the cell membrane of fungi (Rabea et al., 2003; Liu et al., 2004).
The results obtained have shown a reduction in the germination capacity of the conidia, depending on the concentrations employed, except for the solution prepared with EF2 (Table 4). A reduction in the germination capacity of conidia was recorded as the concentration increased. In fact, in the presence of Ole the highest percentage of inhibition of germination was obtained at the highest concentration used 12 mg/L (12X) with a percentage of 58.50%, while with the CSNPs/Ole complex the maximum (67.41%) was obtained at the concentration of 900 mg/L (9X).
Regarding the leaf extract EF1, the germination capacity of the conidia is very limited compared to Ole, reaching a maximum value of 68.72% at the lowest concentration (1X). However, in the CSNPs/EF1 system, the inhibition of germination increases to between 52.45% (1X) and 61.17% (9X) (Table 4). The formulation that records the greatest reduction in germination capacity of F. proliferatum (AACC0215) conidia, reaching 87.96% (12X), is CSNPs/EF1 at the highest concentration assayed. Considering the solution containing only CSNPs, it produced an IRg% equal to 48.71. This result is the same in all treatments because there is no drug inside the CSNPs, so the concentration of each component of the NPs is the same in all the concentrations tested.
The CSNPs/Ole and CSNPs/EF1 complex results suggest that these systems act with an enhanced effect at the highest concentrations. It is the behavior of the CSNP/Ole formulations obtained with a low Ole content (1X and 3X) that have not shown the expected performance with respected to IRg%. The IRg% decreases significantly compared to the control, this result can be attributed to a decrease of the CSNPs chelated positive charges by the hydroxyl groups of the Ole. With respect to EF2, the obtained results showed a decrease in the inhibition of germination at the highest concentrations tested with a value of 26.55% at the concentration 9X. The increased germination capacity may depend on the presence of impurity within the extract that is likely to be used as a source of energy from the fungus and which could stimulate germination of spores.
In vitro Inhibition to Growth
The results obtained have shown an antifungal activity for almost all the analyzed samples, with the exception of six formulations (CSNPs, Ole 1X, and 3X; CSNPs/OLE 1x; CSNPs/EF1 1x; EF2 9x, Table 5) that induce a stimulation in growth.
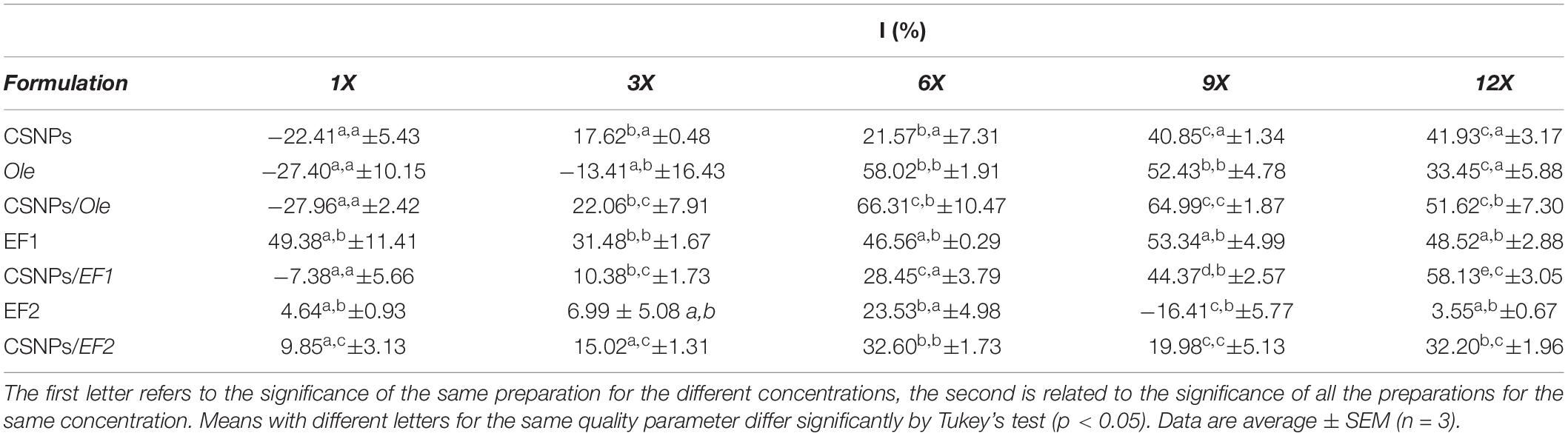
Table 5. In vitro Fusarium proliferatum (AACC0215) percentage of growth inhibition (% I) in the presence of pure chitosan nanoparticles (CSNPs), oleuropein (Ole), leaf extract 1 (EF1) and leaf extract 2 (EF2) free in solution or encapsulated into NPs.
In almost all the concentrations tested, the empty CSNPs inhibited F. proliferatum (AACC0215) growth by 17.62, 21.57, 40.85, and 41.93%, respectively. Pure Ole and EF1 at higher concentrations, both in solution and in CSNPs, exhibit a higher inhibition rate of growth than EF2 (Table 5). Ole showed good antifungal activity at concentrations of 600 mg/L and 900 mg/L (6X and 9X) with a I% equal, respectively, at 58.02% and 52.43% when administered directly and at 66.31 and 64.99% if encapsulated in CSNPs. EF1 fungicidal effect was quite similar to all concentrations used. EF1 solution has shown inhibitory activity already at the lowest concentration (1X) with a I% equal to 49.38%. When EF1 was combined with CSNPs, F. proliferatum (AACC0215) growth inhibition was found to be directly proportional to the concentration used, reaching a value of 58.13% at the highest concentration (12X). EF2 pure exhibited lower antifungal activity with a maximum inhibition value of 23.53% at concentration 6X. However, the maximum of growth inhibition percentage was obtained at 6X and 12x concentration by using the complex CSNPs/EF2 (32.60 and 32.20%, respectively).
All formulations containing CSNPs have an activity of I% greater or not significant compared to the control. Only in the case of CSNPs/EF2 the higher concentrations appear to have significantly lower values than the control. These results, as mentioned above, is probably due to the presence of impurities of the extract EF2 that the mushroom uses as a source of energy and which stimulate its growth.
In this study, it was observed that the tested phenolic compounds exert a cytotoxic activity in vitro against F. proliferatum (AACC0215) and this activity increase when they are complexed with CSNPs. These results are in line with other studies in which phenols extract from olive leaves show an antifungal activity (Markin et al., 2003; Steinkellner and Mammerler, 2007; Goldsmith et al., 2015; Rodriguez-Maturino et al., 2015; Zorić et al., 2016). As reported by Ansari et al. (2013) the phenols antimicrobial activity could be due to a synergistic action of the antioxidant and chelating power of the hydroxyl groups of the phenolic ring that form hydrogen bonds with cell wall proteins of microorganisms. Chitosan also exhibits antimicrobial activity based on the electrostatic interaction between the amine group of chitosan and the negatively charged compounds (phospholipids, proteins, amino acids) of the cell membrane of fungi (Rabea et al., 2003; Liu et al., 2004). Therefore, the interaction between phenolic compounds complexed with CSNPs cause an alteration of the integrity and permeability of the microbial cell.
Conclusion
Olive leaf extract-encapsulated CSNPs were obtained by ionotropic gelation method. The characterization of synthesized NPs showed that the size of leaf extract/CSNPs was 254.6–269.4 nm, the EE ranged from 62.2 at 94.5% and Zeta potential varied from 11.5 at 25.0 mV. As for the polydispersity index values, the lower was 0.207 and this indicates a clear homogeneity of the system. The nanoformulation thus achieved may be explored for the target delivery of phenols for disease control.
Considering the highest concentration (12X) tested, leaf extract/CSNPs showed greater efficacy than pure extracts (EF1 and EF2) and the commercial formulation (Ole) against F. proliferatum (AACC0215). We suggest that the EF1 olive leaf extracts, as free or encapsulated in chitosan-tripolyphosphate nanoparticles, could be used as fungicides to control plant diseases. Finally, future application of these findings may allow to reduce the dosage of fungicides potentially harmful to human health.
Data Availability Statement
All datasets generated for this study are included in the article/supplementary material.
Author Contributions
IM, AC, NP, and RM designed the research, analyzed the data, and discussed the results. GB performed the research and discussed the results. All authors contributed to improving the manuscript and approved the final manuscript.
Funding
The projects “Olio extra vergine d’oliva digital ID management – ODIN,” POR Calabria 2014–2020; “L’aglio Testa Rossa Cardinale della valle dell’Esaro e del Fullone (nuova selezione varietale)”, PSR Calabria 2007–2013; Salvaguardia e valorizzazione del patrimonio olivicolo italiano con azioni di ricerca nel settore della difesa fitosanitaria – SALVAOLIVI” MIPAAF D.M. 33437/2017; “Characterization and enhancement of table and dual-purpose olives – ALIVE” MIPAAF D.M. 93880/2017 financially supported this study.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
Dr. Lorena Tavano is kindly thanked for her assistance in nanoparticle preparations. The authors would like to thank Dr. Tiziana Belfiore and Dr. Veronica Vizzarri for their counseling on phytopathogenic fungi.
Abbreviations
CS, chitosan; CSNPs, chitosan nanoparticles; DLS, dinamic light scattering; EE, encapsulation efficiency; EF1, leaf extract 1; EF2, leaf extract 2; HPLC, high performance liquid chromatography; NPs, nanoparticles; Ole, oleuropein; PBS, phosphate-buffered saline; PDA, potato dextrose agar; PDI, polydispersityindex.
References
Agnihotri, S. A., Mallikarjuna, N. N., and Aminabhavi, T. M. (2004). Recent advances on chitosan-based micro and nanoparticles in drug delivery. J. Control Release 100, 5–28. doi: 10.1016/j.jconrel.2004.08.010
Akamatsu, K., Kaneko, D., Sugawara, T., Kikuchi, R., and Nakoo, S. I. (2010). Three preparation methods for monodispersed chitosan microspheres using the SPG membrane emulsification technique and mechanisms of microsphere formation. Ind. Eng. Chem. Res. 49, 3236–3241. doi: 10.1021/ie901821s
Alves-Santos, F. M., Martínez-Bermejo, D., Rodríguez-Molina, M. C., and Diez, J. J. (2007). Cultural characteristics, pathogenicity and genetic diversity of Fusarium oxysporum isolates from tobacco fields in Spain. Physiol. Mol. Plant. Pathol. 71, 26–32. doi: 10.1016/j.pmpp.2007.09.007
Ansari, M., Anurag, A., Fatima, Z., and Hameed, S. (2013). Natural phenolic compounds: a potential antifungal agent. Microb. Pathog. Strateg. Combat. them Sci. Technol. Educ. 1, 1189–1195.
Anter, J., Fernandez-Bedmar, Z., Villatoro-Pulido, M., Demyda-Peyras, S., Moreno-Millán, M., Alonso-Moraga, A., et al. (2011). A pilot study on the DNA-protective, cytotoxic, and apoptosis-inducing properties of olive-leaf-extracts. Mutat. Res. 723, 165–170. doi: 10.1016/j.mrgentox.2011.05.005
Bahreini, E., Aghaiypour, K., Abbasalipourkabir, R., Mokarram, A. R., Goodarzi, M. T., and Saidijam, M. (2014). Preparation and nanoencapsulation of L-asparaginase II in chitosan-tripolyphosphate nanoparticles and in vitro release study. Nanoscale Res. Let. 9, 340–353. doi: 10.1186/1556-276X-9-340
Benslim, A., Mezaache-Aichour, S., Haichour, N., Chebel, S., and Mihoub Zerroug, M. (2016). Evaluation of inhibition of fungal spore germination by rhizospheric bacterial extracts. ARRB 11, 1–7. doi: 10.9734/arrb/2016/31228
Bianco, A. D., Muzzalupo, I., Piperno, A., Romeo, G., and Uccella, N. (1999). Bioactive derivatives of oleuropein from olive fruits. J. Agric. Food Chem. 47, 3531–3534. doi: 10.1021/jf981240p
Boch, C. H., Jeger, M. J., Mughogho, L. K., Cardwell, K. F., and Mtisi, E. (1999). Effect of dew point temperature and conidium age on germination, germ tube growth and infection of maize and sorghum by Peronosclerospora sorghi. Mycol. Res. 103, 859–864. doi: 10.1017/S0953756298007886
Buhtz, A., Witzel, K., Strehmel, N., Ziegler, J., Abel, S., and Grosch, R. (2015). Perturbations in the primary metabolism of Tomato and Arabidopsis thalianaplants infected with the soil-borne fungus Verticillium dahliae. PLoS One 10:e0138242. doi: 10.1371/journal.pone.0138242
Chin Ming, E., Sunar, N. M., Leman, A. M., and Othman, N. (2015). Direct growth inhibition assay of total airborne fungi with application of biocide-treated malt extract agar. MethodsX 2, 340–344. doi: 10.1016/j.mex.2015.07.002
Cota-Arriola, O., Cortez-Rocha, M., Burgos-Hernandez, A., Ezquerra-Brauer, J., and Plascencia-Jatomea, M. (2013). Controlled release matrices and micro/nanoparticles of chitosan with antimicrobial potential: development of new strategies for microbial control in agriculture. J. Sci. Food Agric. 93, 1525–1536. doi: 10.1002/jsfa.6060
Desjardins, A. E. (2003). Gibberella from A (venaceae) to Z (eae). Annu. Rev. Phytopathol. 41, 177–198. doi: 10.1146/annurev.phyto.41.011703.115501
Fuentes, E., Báez, M. E., Bravo, M., Cid, C., and Labra, F. (2012). Determination of total phenolic content in olive oil Samples by UV–visible spectrometry and multivariate calibration. Food Anal. Methods 5, 1311–1319. doi: 10.1007/s12161-012-9379-5
Goldsmith, C. D., Vuong, Q. V., Sadeqzadeh, E., Stathopoulos, C. E., Roach, P. D., and Scarlett, C. J. (2015). Phytochemical properties and anti-proliferative activity of Olea europaea L. leaf extracts against pancreatic cancer cells. Molecules 20, 12992–13004. doi: 10.3390/molecules200712992
Höhne, S., Frenzel, R., Heppe, A., and Simon, F. (2007). Hydrophobic chitosan microparticles: heterogeneous phase-reaction of chitosan with hydrophobic carbonyl reagents. Biomacromolecules 8, 2051–2058. doi: 10.1021/bm0702354
Joanitti, G. A., and Silva, L. P. (2014). The emerging potential of by-products as platforms for drug delivery systems. Curr. Drug Targets 15, 478–485. doi: 10.2174/13894501113149990171
Kaiser, C., van der Merwe, R., Bekker, T. F., and Labuschagne, N. (2005). In-vitro inhibition of mycelial growth of several phytopathogenic fungi, including Phytophthora cinnamomic by soluble silicon. South African Avocado Growers’. Assoc. Yearb. 28:74.
Kashyap, P. L., Xiang, X., and Heiden, P. (2015). Chitosan nanoparticle based delivery systems for sustainable agriculture. Int. J. Biol. Macromol. 77, 36–51. doi: 10.1016/j.ijbiomac.2015.02.039
Le Toutour, B., and Guedon, D. (1992). Antioxidative activities of Olea europaea leaves and related phenolic compounds. Phytochemistry 31, 1173–1178. doi: 10.1016/0031-9422(92)80255-D
Liu, H., Du, Y., Wang, X., and Sun, L. (2004). Chitosan kills bacteria through cell membrane damage. Int. J. Food Microbiol. 95, 147–155. doi: 10.1016/j.ijfoodmicro.2004.01.022
López-León, T., Carvalho, E. L. S., Seijo, B., Ortega-Vinuesa, J. L., and Bastos-González, D. (2005). Physicochemical characterization of chitosan nanoparticles: electrokinetic and stability behavior. J. Colloid Interface Sci. 283, 344–351. doi: 10.1016/j.jcis.2004.08.186
Manna, V., and Patil, S. (2009). Borax mediated layer-by-layer self-assembly of neutral poly(vinyl-alcohol) and chitosan. J. Phys. Chem. B. 113, 9137–9142. doi: 10.1021/jp9025333
Markin, D., Duek, L., and Berdicevsky, I. (2003). In vitro antimicrobial activity of olive leaves. Mycoses 46, 132–136. doi: 10.1046/j.1439-0507.2003.00859.x
Mazzotta, E., De Benedittis, S., Qualtieri, A., and Muzzalupo, R. (2020). Actively targeted and redox responsive delivery of anticancer drug by chitosan nanoparticles. Pharmaceutics 12:26. doi: 10.3390/pharmaceutics12010026
Montedoro, G., Servili, M., Baldioli, M., and Miniati, E. (1992). Simple and hydrolyzable phenolic compounds in virgin olive oil. I: their extraction, separation, and quantitative and semiquantitative evaluation by HPLC. J. Agric. Food Chem. 40, 1571–1576. doi: 10.1021/jf00021a019
Muto, A., Pisani, F., Pacenza, M., Greco, F., Tavano, L., Gagliardi, O., et al. (2014). Isolation and characterization of Fusarium species associated with cloves of garlic (Allium sativa). Final report of project “L’aglio Testa Rossa Cardinale della valle dell’Esaro e del Fullone (nuova selezione varietale)”, PSR Calabria 2007-2013 Misura 124 “Cooperazione per lo sviluppo di nuovi prodotti, processi e tecnologie nei settori agricolo e alimentare, e in quello forestale. Calabria region, Italy, 15–55.
Muzzalupo, I., Stefanizzi, F., Perri, E., and Chiappetta, A. A. (2011). Transcript levels of CHL P gene, antioxidants and chlorophylls contents in olive (Olea europaea L.) pericarps: a comparative study on eleven olive cultivars harvested in two ripening stages. Plant Foods Hum. Nutr. 66, 1–10. doi: 10.1007/s11130-011-0208-6
Nasti, A., Zaki, N. M., de Leonardis, P., Ungphaiboon, S., Sansongsak, P., Rimoli, M. G., et al. (2009). Chitosan/TPP and chitosan/TPP-hyaluronic acid nanoparticles: systematic optimization of the preparative process and preliminary biological evaluation. Pharm. Res. 26, 1918–1930. doi: 10.1007/s11095-009-9908-0
Nguyen, T. T. X., Dehne, H.-W., and Steiner, U. (2016). Histopathological assessment of the infection of maize leaves by Fusarium graminearum. F. proliferatum, and F. verticillioides. Fungal Biol. 120, 1094–1104. doi: 10.1016/j.funbio.2016.05.013
Omar, S. H. (2010). Oleuropein in olive and its pharmacological effects. Sci. Pharm 78, 133–154. doi: 10.3797/scipharm.0912-18
Rabea, E. I., Mohamed, E. T., Steven, C. V., Smagghe, G., and Steurbaut, W. (2003). Chitosan as antimicrobial agent: applications and mode of action. Biomacromolecules 4, 1457–1465. doi: 10.1021/bm034130m
Rampino, A., Borgogna, M., and Blasi, P. (2013). Chitosan nanoparticles: preparation, size evolution and stability. Int. J. Pharm. 455, 219–228. doi: 10.1016/j.ijpharm.2013.07.034
Rodriguez-Maturino, A., Troncoso-Rojas, R., Sánchez-Estrada, A., González-Mendoza, D., Ruiz-Sanchez, E., Zamora-Bustillos, R., et al. (2015). Antifungal effect of phenolic and carotenoids extracts from chiltepin (Capsicum annum var. glabriusculum) on Alternaria alternata and Fusarium oxysporum. Rev. Argent. Microbiol. 47, 72–77. doi: 10.1016/j.ram.2014.12.005
Rosengaus, R. B., Lefebvre, M. L., and Traniello, J. F. A. (2000). Inhibition of fungal spore germination by nasutitermes: evidence for a possible antiseptic role of soldier defensive secretions. J. Chem. Ecol. 26, 21–39. doi: 10.1023/A:1005481209579
Schweigkofler, W., O’Donnell, K., and Garbelotto, M. (2004). Detection and quantification of airborne conidia of Fusarium circinatum, the causal agent of pine pitch canker, from two California sites by using a real-time PCR approach combined with a simple spore trapping method. Appl. Environ. Microbiol. 70, 3512–3520. doi: 10.1128/AEM.70.6.3512-3520.2004
Servat-Medina, L., González-Gómez, A., Reyes-Ortega, F., Sousa, I. M. O., Queiroz, N. D. C. A., Zago, P. M. W., et al. (2015). Chitosan–tripolyphosphate nanoparticles as Arrabidaeachica standardized extract carrier: synthesis, characterization, biocompatibility, and antiulcerogenic activity. Int. J. Nanomed. 10, 3897–3907. doi: 10.2147/IJN.S83705
Shi, Y., Wan, A., Shi, Y., Zhang, Y., and Chen, Y. (2014). Experimental and mathematical studies on the drug release properties of aspirin loaded chitosan nanoparticles. BioMed Res. Int. 2014, 1–8. doi: 10.1155/2014/613619
Shukla, S. K., Mishra, A. K., Arotiba, O. A., and Mamba, B. B. (2013). Chitosan-based nanomaterials: a state-of-the-art review. Int. J. Biol. Macromol. 59, 6–58. doi: 10.1016/j.ijbiomac.2013.04.043
Steinkellner, S., and Mammerler, R. (2007). Effect of flavonoids on the development of Fusarium oxysporum f. sp. Lycopersici. J. Plant. Interact 2, 17–23. doi: 10.1080/17429140701409352
Sudjana, A. N., D’Orazio, C., Ryan, V., Rasool, N., Ng, J., Islam, N., et al. (2009). Antimicrobial activity of commercial Olea europaea (olive) leaf extract. Int. J. Antimicrob. Agents 33, 461–463. doi: 10.1016/j.ijantimicag.2008.10.026
Tan, S. P., Parks, S. E., Stathopoulos, C. E., and Roach, P. D. (2014). Extraction of flavanoids form bitter melon. Food Nutr. Sci. 5, 458–465.
Taskeen-Un-Nisa, A. H., Yaqub Bhat, M., Pala, S. A., and Mir, R. A. (2011). In vitro inhibitory effect of fungicides and botanicals on mycelial growth and spore germination of Fusarium oxysporum. J. Biopest 4, 53–56.
Trapero-Casas, A., and Jiménes-Dìaz, R. M. (1985). Fungal wilt and root rot diseases of chickpea in southern Spain. Phytopathology 75, 1146–1551.
Varuna Kumara, J. B., and Basavaraj, M. (2015). Synthesis, characterization and hemocompatibility evaluation of curcumin encapsulated chitosan nanoparticles for oral delivery. IJAR 3, 604–611.
Yateem, H., Afaneh, I., and Al-Rimawi, F. (2014). Optimum conditions for oleuropein extraction from olive leaves. IJAST 4, 153–157.
Zhou, H. Y., and Chen, X. G. (2008). Biocompatibility and characteristics of chitosan/cellulose acetate microspheres for drug delivery. Front. Mater. Sci. China 5:367–378. doi: 10.1007/s11706-011-0146-0
Keywords: antifungal activity, olive leaf extracts, oleuropein, biofungicides, nanoformulates
Citation: Muzzalupo I, Badolati G, Chiappetta A, Picci N and Muzzalupo R (2020) In vitro Antifungal Activity of Olive (Olea europaea) Leaf Extracts Loaded in Chitosan Nanoparticles. Front. Bioeng. Biotechnol. 8:151. doi: 10.3389/fbioe.2020.00151
Received: 13 September 2019; Accepted: 13 February 2020;
Published: 03 March 2020.
Edited by:
Haifeng Zhao, South China University of Technology, ChinaReviewed by:
Selin Şahin Sevgili, Istanbul University, TurkeyLukasz Stepien, Institute of Plant Genetics (PAN), Poland
Copyright © 2020 Muzzalupo, Chiappetta, Badolati, Picci and Muzzalupo. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Innocenzo Muzzalupo, aW5ub2NlbnpvLm11enphbHVwb0BjcmVhLmdvdi5pdA==; bXV6emFsdXBvaUBob3RtYWlsLmNvbQ==; Adriana Chiappetta, YWRyaWFuYS5jaGlhcHBldHRhQHVuaWNhbC5pdA==
 Innocenzo Muzzalupo
Innocenzo Muzzalupo Giuliana Badolati1
Giuliana Badolati1 Adriana Chiappetta
Adriana Chiappetta Rita Muzzalupo
Rita Muzzalupo