Corrigendum: Investigating the Effect of Selected Non-Saccharomyces Species on Wine Ecosystem Function and Major Volatiles
- 1Department of Viticulture and Oenology, Institute for Wine Biotechnology, Stellenbosch University, Stellenbosch, South Africa
- 2Department of Food, Environmental and Nutritional Sciences, University Degli Studi di Milano, Milan, Italy
Natural alcoholic fermentation is initiated by a diverse population of several non-Saccharomyces yeast species. However, most of the species progressively die off, leaving only a few strongly fermentative species, mainly Saccharomyces cerevisiae. The relative performance of each yeast species is dependent on its fermentation capacity, initial cell density, ecological interactions as well as tolerance to environmental factors. However, the fundamental rules underlying the working of the wine ecosystem are not fully understood. Here we use variation in cell density as a tool to evaluate the impact of individual non-Saccharomyces wine yeast species on fermentation kinetics and population dynamics of a multi-species yeast consortium in synthetic grape juice fermentation. Furthermore, the impact of individual species on aromatic properties of wine was investigated, using Gas Chromatography-Flame Ionization Detector. Fermentation kinetics was affected by the inoculation treatment. The results show that some non-Saccharomyces species support or inhibit the growth of other non-Saccharomyces species in the multi-species consortium. Overall, the fermentation inoculated with a high cell density of Starmerella bacillaris displayed the fastest fermentation kinetics while fermentation inoculated with Hanseniaspora vineae showed the slowest kinetics. The production of major volatiles was strongly affected by the treatments, and the aromatic signature could in some cases be linked to specific non-Saccharomyces species. In particular, Wickerhamomyces anomalus at high cell density contributed to elevated levels of 2-Phenylethan-1-ol whereas Starm. bacillaris at high cell density resulted in the high production of 2-methylpropanoic acid and 3-Hydroxybutanone. The data revealed possible direct and indirect influences of individual non-Saccharomyces species within a complex consortium, on wine chemical composition.
Introduction
Yeasts are the key components of the wine fermentation ecosystem and responsible for the conversion of grape sugars to CO2, ethanol and a kaleidoscope of volatile and non-volatile compounds (Setati et al., 2012; Alonso-del-Real et al., 2017; Mezzasalma et al., 2018). While the initial species composition of this ecosystem in a freshly pressed grape juice will be specific and unique to each juice, several species are found in most musts, independent of grape variety or region of origin (Sun et al., 2009; Bezzera-Bussoli et al., 2013; Milanović et al., 2013; Tristezza et al., 2013; Vigentini et al., 2016; Alonso-del-Real et al., 2017; Mezzasalma et al., 2017). Such common species can be considered as the core of the wine fermentation ecosystem. These species interact and compete with one another, and species with a higher relative fitness will persist longer and significantly influence the chemical composition and sensorial features of the final product (Tristezza et al., 2013; Wang et al., 2015; García et al., 2017).
The performance of each species is affected by abiotic factors such as the availability of nutrients, pH and oxygen, and biotic factors such as initial cell density and the presence of other species (Sadoudi et al., 2012; Medina et al., 2013; Taillandier et al., 2014; Alonso-del-Real et al., 2017; Shekhawat et al., 2017; González et al., 2018). While the impact of abiotic factors on wine fermentations has been investigated in many studies, relatively limited information regarding the impact of biotic factors is available. In particular, it remains unclear whether species-specific ecological interactions will have predictable consequences on the persistence of species, independently of the environmental conditions within individual grape musts.
Data thus far have shown that species such as Hanseniaspora uvarum, Starm. bacillaris, Lachancea thermotolerans, and Torulaspora delbrueckii, tend to persist longer throughout spontaneous fermentation when present at high initial cell densities (104−6) in grape must or inoculated at higher cell ratios than S. cerevisiae (e.g., 1000:1 and 10000:1) in simultaneous or sequential fermentations (Sun et al., 2009; Comitini et al., 2011; Gobbi et al., 2013; Medina et al., 2013; Bagheri et al., 2015; Liu et al., 2016; García et al., 2017). In contrast, the presence of some non-Saccharomyces species such as Metchnikowia pulcherrima and Williopsis saturnus at high cell densities only has a marginal effect on their competitiveness and persistence throughout fermentation (Moreira et al., 2005; Lee et al., 2013). Follow-up studies of several combinations of mixed-culture fermentations using one non-Saccharomyces species and S. cerevisiae showed that inoculation of some non-Saccharomyces species such as Hanseniaspora guilliermondii, H. vineae, M. pulcherrima, W. anomalus, and Starm. bacillaris at high cell densities generated wines with a distinctive aromatic profile, different from those produced by single cultures of S. cerevisiae (Gobbi et al., 2013; Medina et al., 2013; Izquierdo Cañas et al., 2014; Sun et al., 2014; González et al., 2018; Nisiotou et al., 2018). This can either be due to simple addition of metabolites produced by each yeast or specific metabolic activities that may be induced by the presence of other species (Gobbi et al., 2013; Ciani and Comitini, 2015). For instance, wines generated by fermentations with H. vineae and S. cerevisiae were characterized by higher concentrations of 2-phenylethyl acetate whereas those of L. thermotolerans and S. cerevisiae produced wines with higher concentrations of 2-phenylethanol (2-Phenylethan-1-ol) and glycerol (Gobbi et al., 2013; Medina et al., 2013). These particular contributions are related to the persistence and competitiveness of the non-Saccharomyces strains (Gobbi et al., 2013; Ciani and Comitini, 2015). On the other hand, some non-Saccharomyces species have been shown to contribute to wine aromatic profile, despite their lack of persistence in fermentation, which suggests that the contribution to aroma compounds is due to specific enzymatic activities such as β-glucosidase and esterase (Viana et al., 2011; Ciani and Comitini, 2015; De Ovalle et al., 2018). For instance, Sauvignon blanc fermentations inoculated with S. cerevisiae and M. pulcherrima were reported to generate wines with increased levels of ethyl 2-phenylacetate, 3-methylbutyl acetate and terpenols such as hotrienol, terpiniol, and linalool, despite the early rapid decline of M. pulcherrima (Sadoudi et al., 2012). Undoubtedly, each species interacts differently with S. cerevisiae and with other yeast species present in a wine ecosystem and such interactions influence the dominance and persistence of the yeasts as well as the analytical profiles of the wines (Gobbi et al., 2013; Medina et al., 2013; Ma et al., 2017). Indeed, some studies have reported antagonistic interactions between S. cerevisiae and L. thermotolerans (Gobbi et al., 2013), Starm. bacillaris (Sun et al., 2014), T. delbrueckii (Taillandier et al., 2014), H. guilliermondii (Albergaria et al., 2010), Starm. bacillaris (Englezos et al., 2016a, 2018a), and Brettanomyces bruxellensis (Branco et al., 2014). Such interactions have been attributed to cell-cell contact (Gobbi et al., 2013; Englezos et al., 2016a), restricted oxygen availability (Shekhawat et al., 2017; Englezos et al., 2018a), and production of antimicrobial peptides by S. cerevisiae (Branco et al., 2017). Conversely, other studies have demonstrated positive metabolic interaction between S. cerevisiae and Pichia fermentans (Ma et al., 2017). However, the results indicate that these interactions are strain-specific.
Taken together, most of the previous studies have focused on two-species mixed-culture fermentations (Medina et al., 2013; Taillandier et al., 2014; Branco et al., 2017; De Ovalle et al., 2018). Thus, we have a limited understanding of the effect of individual species in a multi-species yeast community. Furthermore, no information is available on potential interactions among non-Saccharomyces species and how such interactions may affect wine ecosystem and wine aroma. Previously, we reported on the establishment of a multi-species yeast consortium that proved to be a suitable approximation of a grape must ecosystem (Bagheri et al., 2017). The population dynamics of the multi-species consortium was evaluated by viable counts and Automated Ribosomal Intergenic Spacer Analysis (ARISA) methods. Similar to other methods, ARISA can introduce bias since it is unable to differentiate between live and dead cells. However, our data confirmed that ARISA and viable counts generated similar growth patterns for individual yeast species in the consortium throughout the fermentation. Furthermore, the population dynamics of the consortium closely resembled that of natural fermentations, making it a suitable model to assess yeast-yeast interactions in complex communities (Bagheri et al., 2017). The current study sought to understand the effect of individual yeast species on a wine yeast consortium. For this purpose, variation in cell density was used as a tool to understand how presence of individual species affects fermentation kinetics, population dynamics of the yeast consortium and the analytical profile of wines.
Materials and Methods
Yeast Consortium and Culture Conditions
A yeast consortium comprising seven yeast strains obtained from the culture collection of the Institute for Wine Biotechnology (IWBT) and a commercial yeast S. cerevisiae Lalvin EC1118 (Lallemand, Canada) were constructed as described in Bagheri et al. (2017). The yeast species used in the constructed yeast consortium are presented in Table 1. The yeast stock cultures were maintained in 20% (v/v) glycerol at −80°C and were streaked out on Wallerstein Laboratory Nutrient agar (WLN) (Sigma-Aldrich, Spain), when required. The plates were incubated at 30°C for 3–5 days.
Microfermentations
Fermentations were performed in synthetic grape juice medium (SGJM) at pH 3.5 (adapted from Henschke and Jiranek (1993) and Bely et al. (1990). The medium contained 200 g/L sugars (100 g/L glucose and 100 g/L fructose) and 300 mg/L assimilable nitrogen (460 mg/L NH4Cl and 180 mg/L amino acids). SGJM comprises major constituents including carbon, nitrogen, vitamins, minerals, organic acids and anaerobic factors necessary for yeast growth, formulated to closely mimic natural grape juice. However, lacks polyphenols, grape proteins, varietal thiols, and terpenes that provide the precursors to allow determination of yeast contribution to varietal aroma. Nonetheless, this medium provides a simple matrix in which reproducible data on yeast fermentation performance, gene expression patterns, and microbial interactions can be generated (Riou et al., 1997; Viana et al., 2014). Such reproducibility cannot be achieved in natural grape juice which can vary considerably between vintages and varietals.
The effect of cell density on the dynamics of yeast consortium was evaluated in presence and in absence of S. cerevisiae (Figures 1A,B). In the presence of S. cerevisiae, in each treatment, one non-Saccharomyces species was inoculated at approximately 106 cells/mL while the rest of yeast species in the consortium (6 non-Saccharomyces species and S. cerevisiae) was inoculated at approximately 104 cells/mL. In the absence of S. cerevisiae, the same inoculation strategy was applied. However, S. cerevisiae was not included in the yeast consortium. The inoculation treatments are labeled as X-dose, where (X) represents the respective yeast species that is inoculated at higher concentration. The control fermentations were performed in the presence of S. cerevisiae (NS-SC consortium) where each non-Saccharomyces species was inoculated at approximately 106 cells/mL and Sc at approximately 104 cells/mL. In contrast, in the absence of S. cerevisiae (NS consortium), 7 non-Saccharomyces was each inoculated at approximately 106 cells/mL.
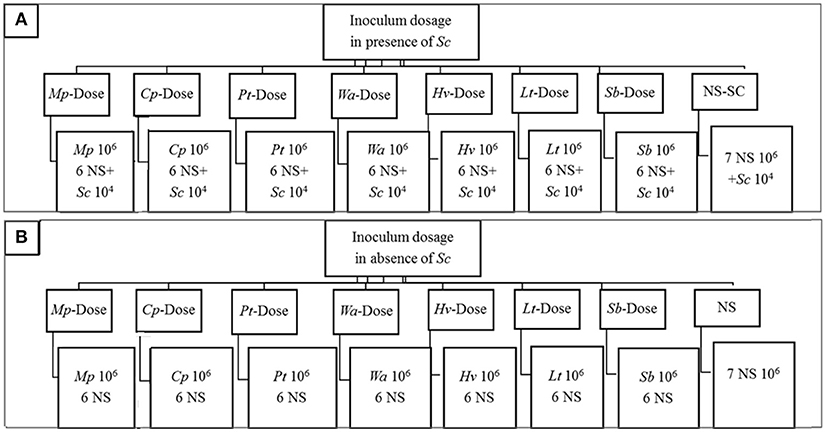
Figure 1. Outline of the experimental plan showing the dosage treatments with S. cerevisiae included in the inoculum (A) and in the absence of S. cerevisiae (B) Treatments are defined based on the strain codes. The following abbreviations were used in this figure. Mp, M. pulcherrima; Cp, C. parapsilosis; Pt, P. terricola; Wa, W. anomalus; Hv, H. vineae; Lt, L. thermotolerans; Sb, Starm. Bacillaris; NS, Non-Saccharomyces species. In the presence of S. cerevisiae, in each treatment, one non-Saccharomyces species was inoculated at approximately 106 while the rest of yeast species in the consortium (7 NS and Sc) was inoculated at approximately 104. In the absence of S. cerevisiae, the same inoculation strategy was applied. However, S. cerevisiae was not included in the yeast consortium. In control fermentations, in the presence of S. cerevisiae (NS-SC consortium), each NS species was inoculated at approximately 106 and Sc at approximately 104 whereas in the absence of Sc (NS consortium), 7 NS was each inoculated at approximately 106.
The fermentations were performed in 500 mL synthetic grape juice dispensed in 500 mL Erlenmeyer flasks fitted with fermentation locks. Static fermentations were carried out in triplicate, at 25°C. The samples were withdrawn at 2-day intervals to monitor the fermentation kinetics. Glucose and fructose were measured, using enzymatic kits, Enzytec™Fluid D-glucose (E5140), fructose (E5120) (Boehringer Mannheim, R-biopharm, Darmstadt, Germany). The fermentations were considered finished when the residual sugar (glucose and fructose) was less than 2 g/L.
ARISA
Previously, we have proved that ARISA and viable counts generated similar growth patterns for individual yeast species in the yeast consortium in wine fermentation (Bagheri et al., 2017). Thus, in the current study, ARISA was used to monitor the population dynamics throughout the fermentations. For this purpose, two-milliliter samples were withdrawn from fermentation flasks. Samples were centrifuged at 5630 × g for 10 min to collect the cells. Genomic DNA was extracted using the method described by Sambrook and Russell (2006). Concentrations of DNA samples were determined spectrophotometrically using the NanoDrop®ND-1000 (NanoDrop Technologies Inc., Wilmington, DE, USA). The concentration of all DNA samples was adjusted to 100 ng/μL. ITS1-5.8S rRNA-ITS2 gene was amplified using the carboxy-fluorescein labeled ITS1 primer (5′-6-FAM- TCC GTA GGT GAA CCT TGC GG-3′) and ITS4 (5′- TCC GTA GGT GAA CCTTGC GG-3′) as described in Slabbert et al. (2010).
ARISA fragments were separated by capillary electrophoresis on an ABI 3010x Genetic Analyser (Applied Biosystems) with a ROX 1.1 labeled size standard (75–1121 base pairs). ARISA profiles were analyzed using Genemapper software version 4.1 (Applied Biosystems). Only fragments with peak area larger than 0.5% of the total fluorescence were considered for further analysis. A bin size of 3 bp for species with ITS region below 700 bp and 5 bp for species with ITS region above 700 bp, was used to minimize the inaccuracies in the ARISA analysis (Slabbert et al., 2010). The abundance of each peak was calculated, dividing individual peak area by the total peak areas for the respective sample.
Analytical Methods
The volatile compounds of wines from different treatments were analyzed by liquid-liquid extraction method, using GC-FID as described by Louw et al. (2010). In brief, the extraction was performed by the addition of 4-methyl-2-pentanol as the internal standard (final concentration 5 mg/L) and 1 mL diethyl ether to each sample. The samples were sonicated for 5 min followed by centrifugation at 4000 × g for 5 min. The ether layer (supernatant) was removed and dried over Na2SO4. Separation of compounds was done, using a DB-FFAP capillary column (Agilent, Little Falls, Wilmington, USA) with dimensions 60 m length × 0.32 mm i.d. × 0.5 μm film thickness. Furthermore, a Hewlett Packard 6890 Plus GC instrument (Little Falls, USA) equipped with a split/splitless injector and a flame ionization detector (FID) were used for gas chromatography (GC). The gas chromatography was performed under the following conditions: an initial oven temperature of 33°C for 17 min, followed by an increase in temperature up to 240°C, for 5 min (12°C/min). Finally, three microliters of the diethyl-ether extract were injected at 200°C in split mode, with the split ratio of 15:1 and the split flow rate of 49.5 mL/min. The column flow rate was 3.3 mL/min, using hydrogen as carrier gas. The detector temperature was 250°C.
Statistical Analysis
All the fermentations and the chemical analysis were performed in at least three repeats. The values were presented as means ± SD. The differences between treatments were determined using analysis of variance (ANOVA) using the statistical software, Statistica version 13.0 (StatSoft Inc., Tulsa, Oklahoma, USA). The differences were considered significant should the p-values be equal to or less than 0.05. For multivariate data analysis, the principal component analysis was performed, using XLSTAT in Microsoft® Excel (2016).
Results
Fermentation Kinetics
The effect of individual yeast species on the wine yeast consortium was evaluated by conducting fermentations in which one of the 8 species was inoculated at a dosage 100 times higher than the rest of the species.
Overall, fermentations conducted in the absence of S. cerevisiae were sluggish and did not ferment to dryness. Residual sugar ranging between 88 and 107 g/L was detected after 28–32 days when the fermentations were terminated (data not shown). In contrast, all the fermentations in which S. cerevisiae was included in the consortium fermented to dryness, albeit at different rates. The fermentations inoculated with a high dosage of M. pulcherrima, C. parapsilosis, and P. terricola, displayed similar fermentation kinetics, taking 18 days to reach dryness (Figures 2A–C). H. vineae and L. thermotolerans fermentations displayed the slowest fermentation kinetics, taking 28 and 22 days to reach dryness (Figures 2E,F). In contrast, the fermentation inoculated with a higher concentration of W. anomalus and Starm. bacillaris (Figures 2D,G) displayed the fastest fermentation kinetics (14 and 12 days) compared to the rest of dosage treatments and the NS-SC control (Figure 2H). The Starm. bacillaris high inoculum fermentation displayed rapid consumption of fructose from the onset of fermentation whereas the rest of the treatments showed a similar consumption rate for both glucose and fructose in the early stages of fermentations and a faster consumption of glucose toward the middle of fermentations. Based on the fermentations slope calculated for the first 8 days of fermentations, the P. terricola and W. anomalus high inoculum fermentations exhibited the fastest glucose consumption.
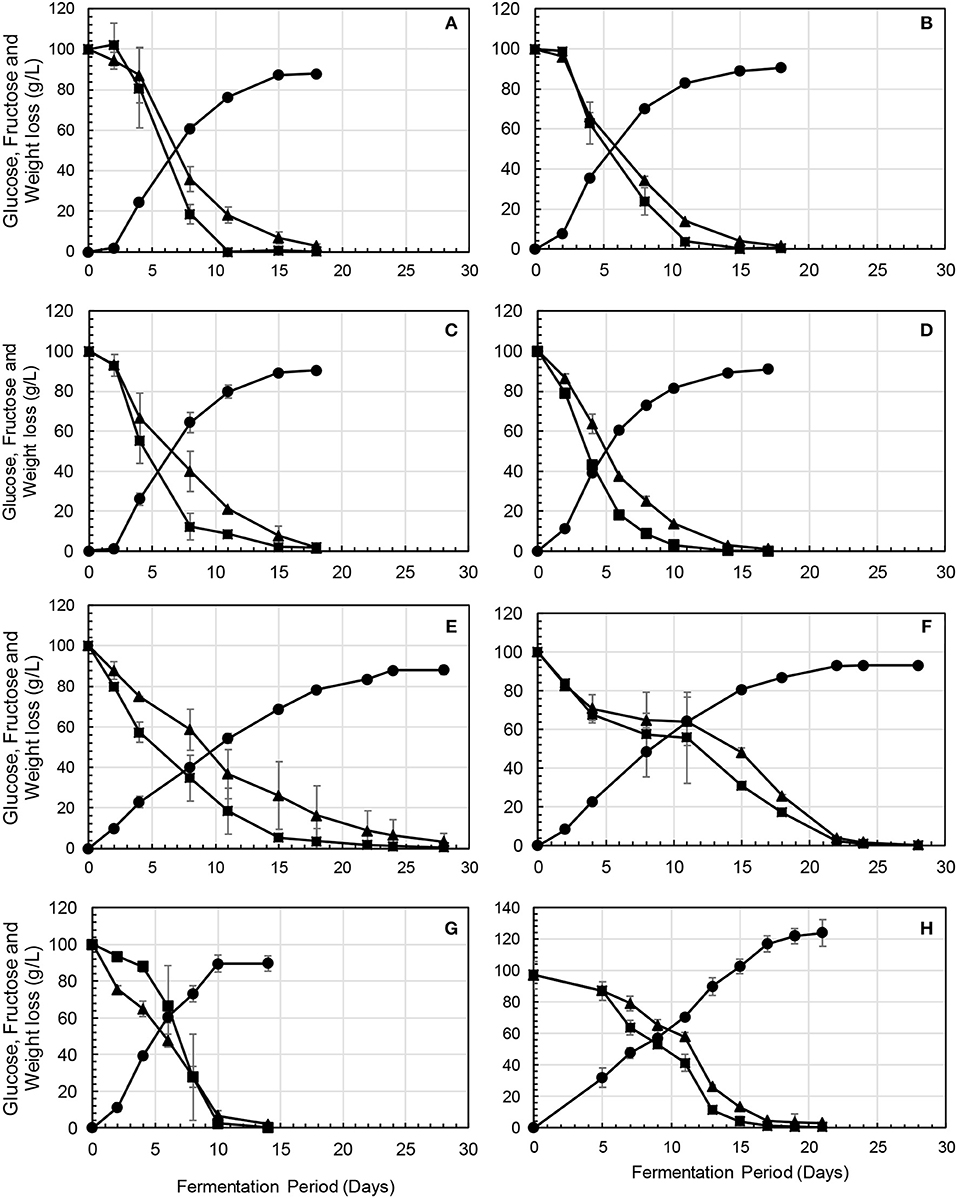
Figure 2. Fermentation profiles showing the kinetics of sugar consumption fructose (▴) and glucose (■), in (A) Mp-dose, (B) Cp-dose, (C) Pt-dose, (D), Wa-dose, (E) Hv-dose, (F) Lt-dose, (G) Sb-dose, and (H) NS-SC.
Population Dynamics
The growth of all species was evaluated during three stages of fermentation, beginning (10–20% sugar consumption), middle (40–60% consumption), and end of fermentation (over 95% sugar consumption).
Overall, a higher initial density of each species allowed the individual species to persist at a slightly higher relative abundance compared to the NS-SC fermentation where all species were inoculated at equal concentrations. However, the dynamics of the individual species were affected differently with regard to their length of persistence. For instance, M. pulcherrima and C. parapsilosis accounted for less than 10% of the population at the beginning of fermentation and were undetectable by the middle of fermentation (Figure 3). In contrast, W. anomalus and L. thermotolerans persisted until the middle of fermentation, while P. terricola, H. vineae, and Starm. bacillaris persisted until the end of fermentation in the respective fermentations where they were inoculated at higher dosages. Starm. bacillaris was the only species that persisted until the end of fermentation at a considerable level (12.7%). A high inoculation of some non-Saccharomyces species such as P. terricola and C. parapsilosis supported or inhibited the growth of other non-Saccharomyces species in the consortium. For instance, H. vineae was detected at higher relative abundance by the middle of fermentation in the Cp-dose and Pt-dose. Similarly, W. anomalus persisted until the middle of fermentation in the Pt-dose while it was below detection by the beginning of Lt-dose fermentation.
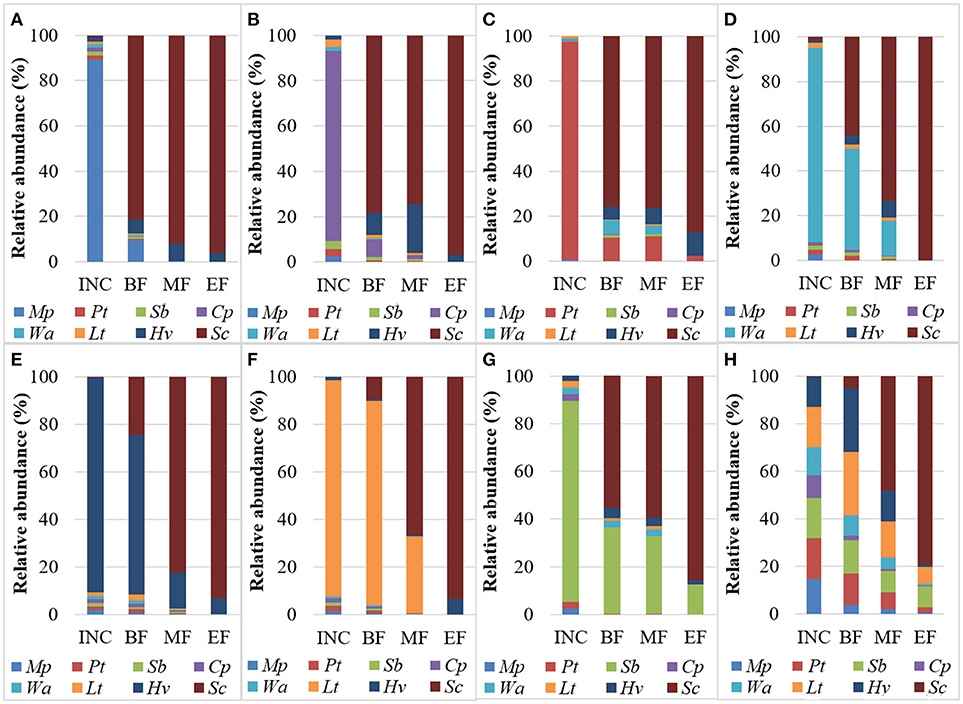
Figure 3. Distribution of yeast species (%) during fermentations at the inoculation time (INC), the beginning (BF), middle (MF), and end of fermentation (EF) in (A) Mp-dose, (B) Cp-dose, (C) Pt-dose, (D), Wa-dose, (E) Hv-dose, (F) Lt-dose, (G) Sb-dose, and (H) NS-SC.
Production of Major Volatiles
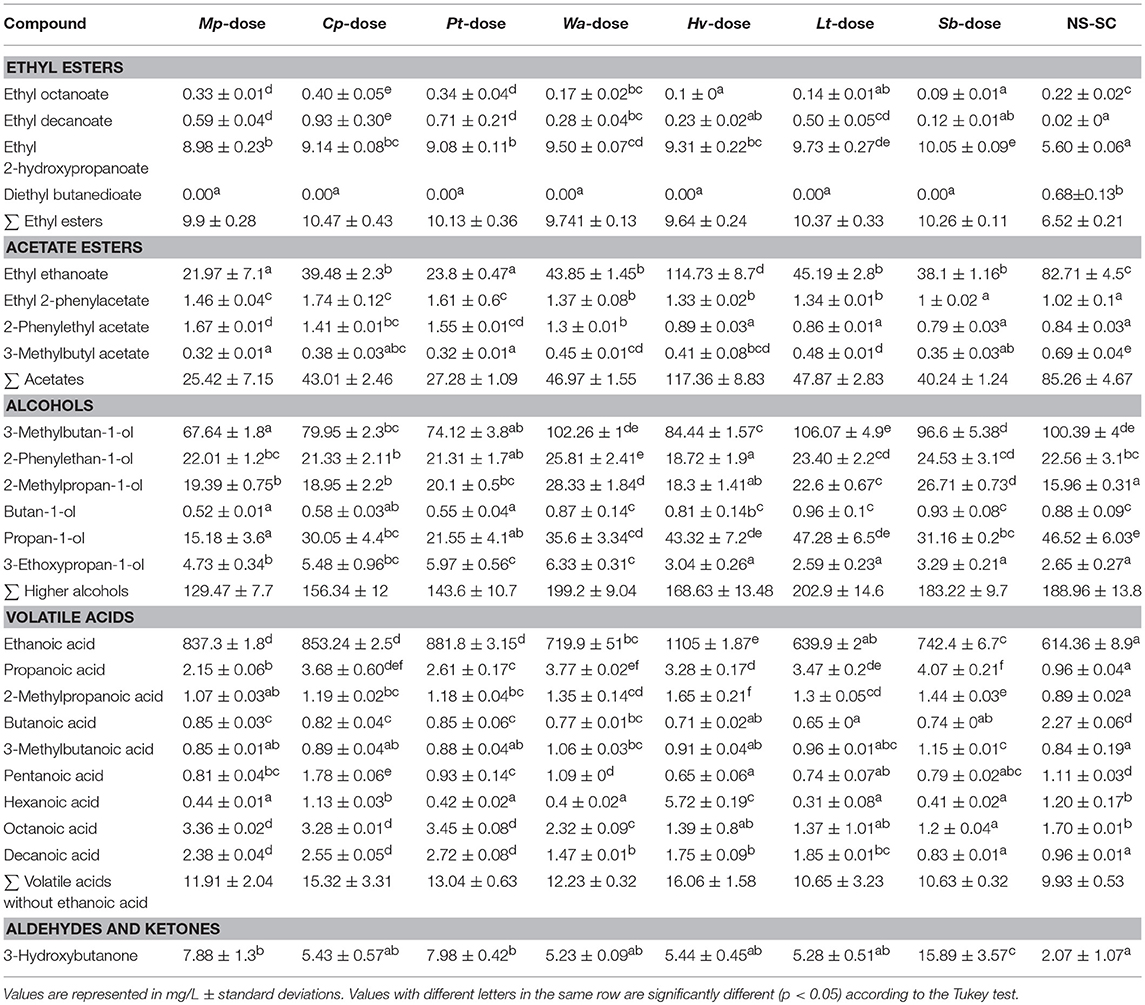
Production of major volatiles was evaluated for all the wines, in order to reveal a potential contribution of individual non-Saccharomyces species in the production of specific major volatiles. The NS-SC produced wine with the lowest total esters (6.52 mg/L) and volatiles acids (9.93 mg/L). Mp-dose, Cp-dose, and Pt-dose wines contained the lowest levels of higher alcohols while they generated the highest concentration of ethyl octanoate, ethyl decanoate, octanoic acid, and decanoic acid (Table 2). In contrast, the Hv-dose generated the highest total acetates (117.36 mg/L) and volatile acid (16.06 mg/L), which could be attributed to high ethyl ethanoate and hexanoic acid, respectively. Furthermore, the Hv-dose produced 1105 mg/L ethanoic acid, which was the highest amongst all the inoculation regimes. Conversely, the highest concentration of 3-Hydroxybutanone (15.89 mg/L) was recorded in the Sb-dose derived wine whereas the highest concentration of 2-Phenylethan-1-ol (25.81 mg/L) was generated in Wa-dose wine.
Principal component analysis (PCA) was applied to all quantifiable major volatiles to decipher the compounds which would drive the differentiation between the wines. PC1 and PC2 explained 66.21% of the variance. PC1 accounts for 40.18% of the variance and mainly shows separation of the wines into three major groups based on rapid (Cp-dose, Mp-dose, and Pt-dose), intermediary (Wa-dose, Hv-dose, Lt-dose, and Sb-dose) and slow (NS-SC) establishment of S. cerevisiae within the ecosystem. The NS-SC derived wine was clearly separated from the rest of treatments in the lower right quadrant of the PC1 and it was mainly associated with butanoic acid, ethyl ethanoate, 3-methylbutyl acetate, and diethyl butanedioate. Conversely, the Cp-dose, Pt-dose, and Mp-dose wines formed one group and were mainly associated with ethyl decanoate, ethyl octanoate, 2-phenylethyl acetate, ethyl 2-phenylacetate as well as octanoic acid and decanoic acid. PC2 accounts for 26.03% of the variance and further separates the Sb-dose wine from the intermediary group. Within this group, the Sb-dose wine was associated with 3-methylbutanoic acid, 2-methylpropanoic acid, and 2-methylpropan-1-ol. In contrast, the Wa-dose, Hv-dose, and Lt-dose were associated with hexanoic acid, 3-Methylbutan-1-ol, butan-1-ol, and propan-1-ol (Figure 4).
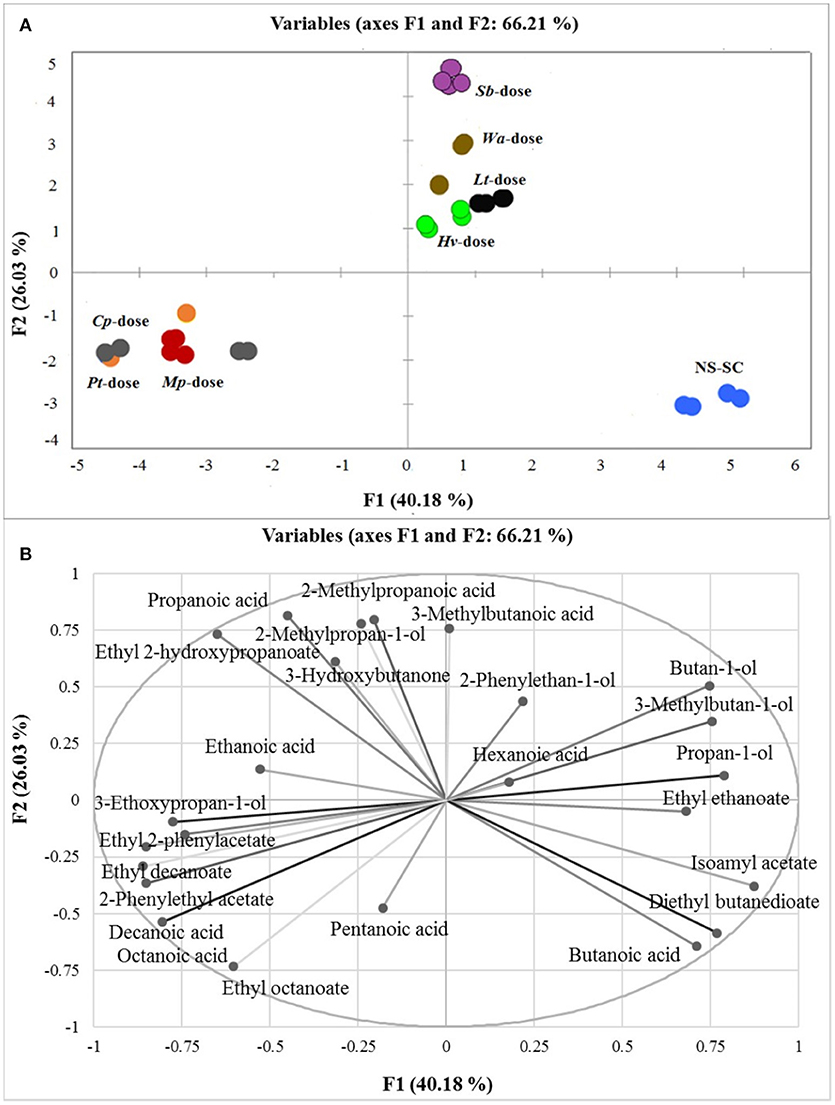
Figure 4. PCA score plot (A) and corresponding loading plot (B) of first and second principal components (PC) derived from PC analysis of the major volatile compounds produced in synthetic must fermentations. The major volatile compounds are represented by light grays whereas the treatments are as follows: Mp-dose (red), Cp-dose (gray), Pt-dose (orange), Wa-dose (brown), Hv-dose (green), Lt-dose (black), Sb-dose (purple), and NS-SC (blue). Some replicates were overlapped in each treatment (e.g. Pt-dose).
Discussion
The current study used the variation in cell densities as a tool to understand the influence of individual species on population dynamics in a complex community and on the chemicals responsible for wine aroma. For this purpose, one out of 8 species was inoculated at a dosage 100 times higher than the rest of the species.
The data showed that the persistence of some species, for instance, L. thermotolerans, Starm. bacillaris, W. anomalus, and H. vineae were enhanced considerably when they were inoculated at high concentration. The persistence of these species until the final stages of fermentation have been reported previously (Viana et al., 2011; Gobbi et al., 2013; Bagheri et al., 2015). These species have some common metabolic traits such as moderate (Starm. bacillaris, W. anomalus, and H. vineae) and strong fermentative capabilities (L. thermotolerans), high tolerance to different levels of ethanol (6–12%) and osmotic pressure (Sabel et al., 2014; Englezos et al., 2016a; Hranilovic et al., 2018; Martin et al., 2018) that can explain their persistence in wine fermentations. Despite their common metabolic traits, the persistence of each species in wine fermentation can be attributed to their species-specific characteristics. For instance, L. thermotolerans has shown to tolerate very high concentration of ethanol (12% v/v) and total SO2 (>200 mg/L) (Hranilovic et al., 2018; Nally et al., 2018) whereas W. anomalus has displayed high tolerance to low pH (Sabel et al., 2014). Persistence of Starm. bacillaris throughout fermentation can be attributed to its strong fructophilic characteristics, limited nitrogen requirements and the ability of this species to excrete some branched amino acids (Englezos et al., 2018b).
Despite the similar pattern observed for the persistence of L. thermotolerans, Starm. bacillaris, W. anomalus, and H. vineae throughout fermentations, the growth dynamics, as well as the kinetics of the respective fermentation processes, were remarkably divergent. Three patterns of fermentation kinetics comprising fast (Sb-dose and Wa-dose), intermediate (Cp-dose, Pt-dose, and Mp-dose) and slow (Lt-dose and Hv-dose) were evident. These trends revealed possible yeast-yeast interactions. For instance, inoculation of Starm. bacillaris at a high concentration resulted in fast fermentation kinetics which can be attributed to rapid simultaneous utilization of both fructose and glucose. We can, therefore, infer that there is a co-operative interaction between the two dominant yeasts (i.e., Starm. bacillaris and S. cerevisiae) in this fermentation. Indeed, several studies have shown that most strains of Starm. bacillaris are fructophilic while S. cerevisiae is known to exhibit preferential consumption of glucose. This co-operative interaction has been observed amongst other strains of these two species (Magyar and Tóth, 2011; Suzzi et al., 2012; Masneuf-Pomarede et al., 2015; Tofalo et al., 2016). These findings suggest that the interaction between the two could be more species-specific rather than strain specific. Despite the positive interaction observed in Sb-dose fermentation between these two species, a gradual decrease in the population of Starm. bacillaris was observed. This decline in the population of Starm. bacillaris can be due to several parameters such as lower fermentative capability of this species compared to S. cerevisiae due to several parameters such as cell to cell contact (Englezos et al., 2019), limited oxygen (Englezos et al., 2018b) and nutrient availability as suggested by Englezos et al. (2016b). In the case of Wa-dose fermentation, the sugar consumption rate displayed a faster utilization of glucose than fructose, however, this does not seem to affect the implantation of S. cerevisiae.
Within the intermediate fermentations, M. pulcherrima, C. parapsilosis, P. terricola were rapidly replaced by S. cerevisiae at an early stage of fermentations, despite the high initial density. We can infer that the growth of these species may be curtailed by other parameters. For instance, several strains of M. pulcherrima were reported to have lower growth rates than S. cerevisiae and required significantly higher oxygen input to grow in high sugar environments (Contreras et al., 2014; Shekhawat et al., 2017). Similar correlations for the growth of C. parapsilosis and oxygen availability were reported by Oh et al. (1998) and Holland et al. (2014), while the rapid decline in the population of P. terricola in Pt-dose could be due to weak fermentative capability of this species as previously suggested by Clemente-Jimenez et al. (2005) and Di Maro et al. (2007). Even though the rapid decline in the population of M. pulcherrima, C. parapsilosis, P. terricola can be attributed to their low fermentative capability and limited nutrient and oxygen availability, the distribution of yeast species in the consortium is strongly affected by the growth of individual species and the interaction among these species. It was evident in the current study that a high inoculum of C. parapsilosis created conducive conditions for enhanced persistence of H. vineae, while P. terricola promoted the persistence of both H. vineae and W. anomalus. This could suggest that at high dosage C. parapsilosis and P. terricola either produce compounds that retard the establishment of the other yeasts or compete strongly for nutrients. However, due to their sensitivity to oxygen limitation and increasing ethanol, the two yeasts still decline rapidly.
Our data revealed potential competition between H. vineae, L. thermotolerans, and S. cerevisiae. Indeed, the fermentations inoculated with high concentrations of L. thermotolerans and/or H. vineae were the slowest, probably due to the moderate (H. vineae) and strong (L. thermotolerans) fermentation capacity of these species accompanied by a delay in the implantation of S. cerevisiae. Medina et al. (2013), demonstrated that H. vineae competes with S. cerevisiae for the consumption of nitrogen and vitamins. This lack of nutrients retards the growth of S. cerevisiae while H. vineae is a moderate fermenter and cannot ferment to dryness. On the other hand, previous studies have confirmed that L. thermotolerans shows a high degree of competitiveness against S. cerevisiae which can retard the fermentation kinetics and delay the implantation of S. cerevisiae. Decline in the population of L. thermotolerans could be due to competition for oxygen between S. cerevisiae and L. thermotolerans (Shekhawat et al., 2017), a slow metabolic activity of L. thermotolerans under anoxic conditions (Nissen and Arneborg, 2003), cell to cell contact (Gobbi et al., 2013) as well as production of anti-microbial peptides by S. cerevisiae (Branco et al., 2017).
Concerning the major volatile production, a clear separation was observed between NS-SC and the dosage treatments, which confirmed that the production of major volatiles was strongly affected by the treatments. Further analysis of chemical compounds underlined that aromatic signature of some non-Saccharomyces species could be detected in wine where these species were inoculated at a high concentration. Our study for the first time highlighted that presence of W. anomalus at a high cell density could result in an elevated level of 2-Phenylethan-1-ol in wine whereas C. parapsilosis at a high cell density could generate the wine with high concentrations of valeric acid, ethyl decanoate, and ethyl octanoate. This result suggests that the presence of C. parapsilosis and W. anomalus at a high cell density in grape must can affect the quality of wine, despite the rapid decline of these species at the early stage of fermentation. Furthermore, the aromatic signature of H. vineae (high concentrations of ethyl ethanoate and ethanoic acid) and L. thermotolerans (high concentration of 3-Methylbutan-1-ol and low concentration of ethanoic acid) could be detected in wine where these species were inoculated at a high concentration. Similar results have been reported for wines obtained from the mixed culture fermentations of S. cerevisiae and H. vineae in other grape matrices such as Macabeo, Merlot, and Chardonnay (Viana et al., 2011; Medina et al., 2013; Lleixà et al., 2016). Similarly, the production of 3-Methylbutan-1-ol and ethanoic acid in Lt-dose wine followed a similar pattern in wine obtained from the mixed culture fermentation of L. thermotolerans and S. cerevisiae in pasteurized commercial white grape juice (Gobbi et al., 2013) and Airén grape juice (Benito et al., 2016). On the other hand, our study for the first time demonstrated that presence of W. anomalus at a high cell density could result in an elevated level of 2-Phenylethan-1-ol in wine whereas C. parapsilosis at a high cell density could generate the wine with high concentrations of valeric acid, ethyl decanoate, and ethyl octanoate. This result suggests that the presence of C. parapsilosis and W. anomalus at a high cell density in grape must can affect the quality of wine, despite the rapid decline of these species at the early stage of fermentation.
In conclusion, the data confirmed that persistence of non-Saccharomyces species throughout fermentation does not necessarily depend on their initial cell density. Our results reveal that the presence of individual non-Saccharomyces species will positively or negatively affect the growth of other species in the consortium. Therefore, the growth of non-Saccharomyces species in a multi-species community is affected by both the interactions between non-Saccharomyces and Saccharomyces species as well as the interactions among non-Saccharomyces species in the yeast consortium. Consequently, the production of major volatiles and the quality of wine is affected by these yeast-yeast interactions. In our data, we can link the production of certain compounds to the presence of specific non-Saccharomyces species, but only for those that were inoculated at a higher cell density. However, the production of these compounds did not necessarily depend on their persistence through the fermentation. Indeed, presence of some species at a high cell density was sufficient to affect the quality of wine. Additional experiments with adjusted relative inoculation cell densities will be required to better evaluate how the strain- or species-specific interactions within this ecosystem impact on fermentation outcomes. The results also highlight the usefulness of a consortium-based approach to better understand the dynamics of multi-species interactions and their impact on wine character. The result of current study is a first step to untangling the interactions within the wine ecosystem. Future work will focus to unravel mechanisms underlying yeast-yeast interactions observed in this study (metabolically and physically) and to evaluate how presence of two or three non-Saccharomyces species at high densities in the consortium affect the population dynamics and wine aroma.
Author Contributions
MS and FB conceptualized the study, Supervised the design of experimental layout. BB and PZ performed the experiments and analyzed data. BB wrote first draft of the manuscript. MS, FB, BB, PZ, and IV edited draft manuscript and approved final manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank the National Research Foundation (NRF) for funding through SARCHI (Grant number UID 83471 and grant number 101998), NRF-Technology and Human Resources for Industry Programme (grant number TP14080184824). We also thank the Wine Industry Network for Expertise and Technology (Winetech) SU IWBT 16/02. The authors gratefully acknowledge the YeSVitE consortium (FP7-IRSES-2013-GA no. 612441) for helpful discussion and Prof. Concetta Compagno for precious collaboration. Paolo Zambelli was supported by the YeSViTE project in his secondment to the Stellenbosch University.
References
Albergaria, H., Francisco, D., Gori, K., Arneborg, N., and Gírio, F. (2010). Saccharomyces cerevisiae CCMI 885 secretes peptides that inhibit the growth of some non-Saccharomyces wine-related strains. Appl. Microbiol. Biotechnol. 86, 965–972. doi: 10.1007/s00253-009-2409-6
Alonso-del-Real, J., Lairón-Peris, M., Barrio, E., and Querol, A. (2017). Effect of temperature on the prevalence of Saccharomyces non Cerevisiae species against a S. cerevisiae wine strain in wine fermentation, competition, physiological fitness, and influence in final wine composition. Front. Microbiol. 8:150. doi: 10.3389/fmicb.2017.00150
Bagheri, B., Bauer, F. F., and Setati, M. E. (2015). The diversity and dynamics of indigenous yeast communities in grape must from vineyards employing different agronomic practices and their influence on wine fermentation. S. Afr. J. Enol. Vitic. 36, 243–251. doi: 10.21548/36-2-957
Bagheri, B., Bauer, F. F., and Setati, M. E. (2017). The impact of Saccharomyces cerevisiae on a wine yeast consortium in natural and inoculated fermentations. Front. Microbiol. 8:1988. doi: 10.3389/fmicb.2017.01988
Bely, M., Sablayrolles, J. M., and Barre, P. (1990). Automatic detection of assimilable nitrogen deficiencies during alcoholic fermentation in oenological conditions. J. Ferment. Bioeng. 70, 246–252. doi: 10.1016/0922-338X(90)90057-4
Benito, A., Calderon, F., Palomere, F., and Benito, S. (2016). Quality and composition of airén wines fermented by sequential inoculation of Lachancea thermotolerans and Saccharomyces cerevisiae. Food Technol. Biotechnol. 54, 135–144. doi: 10.17113/ftb.54.02.16.4220
Bezzera-Bussoli, C., Baffi, M. A., Gomes, E., and Da-Silva, R. (2013). Yeast diversity isolated from Grape musts during spontaneous fermentation from a Brazilian winery. Curr. Microbiol. 67, 356–361. doi: 10.1007/s00284-013-0375-9
Branco, P., Francisco, D., Chambon, C., Hébraud, M., Arneborg, N., Almeida, M. G., et al. (2014). Identification of novel GAPDH-derived antimicrobial peptides secreted by Saccharomyces cerevisiae and involved in wine microbial interactions. Appl. Microbiol. Biotechnol. 98, 843–853. doi: 10.1007/s00253-013-5411-y
Branco, P., Francisco, D., Monteiro, M., Almeida, M. G., Caldeira, J., Arneborg, N., et al. (2017). Antimicrobial properties and death-inducing mechanisms of saccharomycin, a biocide secreted by Saccharomyces cerevisiae.Appl. Microbiol. Biotechnol. 101, 159–171. doi: 10.1007/s00253-016-7755-6
Ciani, M., and Comitini, F. (2015). Yeast interactions in multi-starter wine fermentation. Curr. Opin. Food Sci. 1, 1–6. doi: 10.1016/j.cofs.2014.07.001
Clemente-Jimenez, J. M., Mingorance-Cazorla, L., Martínez-Rodríguez, S., Las Heras-Vázquez, F. J., and Rodríguez-Vico, F. (2005). Influence of sequential yeast mixtures on wine fermentation. Int. J. Food Microbiol. 98, 301–308. doi: 10.1016/j.ijfoodmicro.2004.06.007
Comitini, F., Gobbi, M., Domizio, P., Romani, C., Lencioni, L., Mannazzu, I., et al. (2011). Selected non-Saccharomyces wine yeasts in controlled multi-starter fermentations with Saccharomyces cerevisiae. Food Microbiol. 28, 873–882. doi: 10.1016/j.fm.2010.12.001
Contreras, A., Hidalgo, C., Henschke, P. A., Chambers, P. J., Curtin, C., and Varela, C. (2014). Evaluation of non-Saccharomyces yeasts for the reduction of alcohol content in wine. Appl. Environ. Microbiol. 80, 1670–1678. doi: 10.1128/AEM.03780-13
De Ovalle, S., Cavello, I., Brena, B. M., Cavalitto, S., and González-Pombo, P. (2018). Production and characterization of a β-glucosidase from Issatchenkia terricola and its use for hydrolysis of aromatic precursors in Cabernet Sauvignon wine. LWT Food Sci. Technol. 87, 515–522. doi: 10.1016/j.lwt.2017.09.026
Di Maro, E., Ercolini, D., and Coppola, S. (2007). Yeast dynamics during spontaneous wine fermentation of the Catalanesca grape. Int. J. Food Microbiol. 117, 201–210. doi: 10.1016/j.ijfoodmicro.2007.04.007
Englezos, V., Cocolin, L., Rantsiou, K., Ortiz-Julien, A., Bloem, A., Dequin, S., et al. (2018a). Specific phenotypic traits of Starmerella bacillaris related to nitrogen source consumption and central carbon metabolite production during wine fermentation. Appl. Environ. Microbiol. 84, e00797–e00718. doi: 10.1128/AEM.00797-18
Englezos, V., Cravero, F., Torchio, F., Rantsiou, K., Ortiz-Julien, A., Lambri, M., et al. (2018b). Oxygen availability and strain combination modulate yeast growth dynamics in mixed culture fermentations of grape must with Starmerella bacillaris and Saccharomyces cerevisiae. Food Microbiol. 69, 179–188. doi: 10.1016/j.fm.2017.08.007
Englezos, V., Rantsiou, K., Cravero, F., Torchio, F., Ortiz-Julien, A., Gerbi, V., et al. (2016a). Starmerella bacillaris and Saccharomyces cerevisiae mixed fermentations to reduce ethanol content in wine. Appl. Microbiol. Biotechnol. 100, 5515–5526. doi: 10.1007/s00253-016-7413-z
Englezos, V., Rantsiou, K., Giacosa, S, Segade, G. R., Rolle, L., and Cocolin, L. (2019). Cell-to-cell contact mechanism modulates Starmerella bacillaris death in mixed culture fermentations with Saccharomyces cerevisiae. Int. J. Food Microbiol. 289, 106–114. doi: 10.1016/j.ijfoodmicro.2018.09.009
Englezos, V., Torchio, F., Cravero, F., Marengo, F., Giacosa, S., Gerbi, V., et al. (2016b). Aroma profile and composition of Barbera wines obtained by mixed fermentations of Starmerella bacillaris (synonym Candida zemplinina) and Saccharomyces cerevisiae. LWT Food Sci. Technol. 73, 567–575. doi: 10.1016/j.lwt.2016.06.063
García, M., Esteve-Zarzoso, B., Crespo, J., Cabello, J. M., and Arroyo, T. (2017). Yeast monitoring of wine mixed or sequential made by native strains from D.O “Vinos de Madrid” using real-time quantitative PCR. Front. Microbiol. 8:2520. doi: 10.3389/fmicb.2017.02520
Gobbi, M., Comitini, F., Domizio, P., Romani, C., Lencioni, L., Mannazzu, I., et al. (2013). Lachancea thermotolerans and Saccharomyces cerevisiae in simultaneous and sequential co-fermentation: a strategy to enhance acidity and improve the overall quality of wine. Food Microbiol. 33, 271–281. doi: 10.1016/j.fm.2012.10.004
González, B., Vázquez, J., Morcillo-Parra, M. A., Mas, A., Torija, M. J., and Beltran, G. (2018). The production of aromatic alcohols in non-Saccharomyces wine yeast is modulated by nutrient availability. Food Microbiol. 74, 64–74. doi: 10.1016/j.fm.2018.03.003
Henschke, P. A., and Jiranek, V. (1993). “Yeasts metabolism of nitrogen compounds,” in Wine Microbiology and Biotechnology, ed G. H. Fleet (Chur: Harwood Academic), 77–164.
Holland, L. M., Schröder, M. S., Turner, S. A., Taff, H., Andes, D., Grózer, Z., et al. (2014). Comparative phenotypic analysis of the major fungal pathogens Candida parapsilosis and Candida albicans. PLoS Pathog. 10:e1004365. doi: 10.1371/journal.ppat.1004365
Hranilovic, A. M., Gambetta, J., Schmidtke, L., Boss, P. R., Grbin, P., and Jiranek, V. (2018). Oenological traits of Lachancea thermotolerans show signs of domestication and allopatric differentiation. Sci. Rep. 8:14812. doi: 10.1038/s41598-018-33105-7
Izquierdo Cañas, P. M., Esteban, C., Romero, G., Heras, J. M., Mónica, M., and González, F. (2014). Influence of sequential inoculation of Wickerhamomyces anomalus and Saccharomyces cerevisiae in the quality of red wines. Eur. Food Res. Technol. 239, 279–286. doi: 10.1007/s00217-014-2220-1
Lee, P. R., Kho, S. H., Yu, B., Curran, P., and Liu, S. Q. (2013). Yeast ratio is a critical factor for sequential fermentation of papaya wine by Williopsis saturnus and Saccharomyces cerevisiae. Microb. Biotechnol. 6, 385–393. doi: 10.1111/1751-7915.12008
Liu, P.-T., Lu, L., Duan, C.-Q., and Yan, G.-L. (2016). The contribution of indigenous non-Saccharomyces wine yeast to the improved aromatic quality of cabernet sauvignon wines by spontaneous fermentation. Food Sci. Technol. 71, 356–363. doi: 10.1016/j.lwt.2016.04.031
Lleixà, J., Martín, V., Portillo, M. C., Carrau, F., Beltran, G., and Mas, A. (2016). Comparison of fermentation and wines produced by inoculation of Hanseniaspora vineae and Saccharomyces cerevisiae. Front. Microbiol. 7:338. doi: 10.3389/fmicb.2016.00338
Louw, L., Tredoux, A. G. J., van Rensburg, P., Kidd, M., Naes, T., and Nieuwoudt, H. H. (2010). Fermentation-derived aroma compounds in varietal young wines from South Africa. S. Afr. J. Enol. Vitic. 31, 213–225. doi: 10.21548/31-2-1418
Ma, D., Yan, X., Wang, Q., Zhang, Y., and Tao, Y. (2017). Performance of selected P. fermentans and its excellular enzyme in co-inoculation with S. cerevisiae for wine aroma enhancement. LWT 86, 361–370. doi: 10.1016/j.lwt.2017.08.018
Magyar, I., and Tóth, T. (2011). Comparative evaluation of some oenological properties in wine strains of Candida stellata, Candida zemplinina, Saccharomyces uvarum and Saccharomyces cerevisiae. Food Microbiol. 28, 94–100. doi: 10.1016/j.fm.2010.08.011
Martin, V., Valera, M. J., Medina, K., Boido, E., and Carrau, F. (2018). Oenological impact of the Hanseniaspora/Kloeckera yeast genus on wines—a review. Fermentation 4:76. doi: 10.3390/fermentation4030076
Masneuf-Pomarede, I., Juquin, E., Miot-Sertier, C., Renault, P., Laizet, Y., Salin, F., et al. (2015). The yeast Starmerella bacillaris (synonym Candida zemplinina) shows high genetic diversity in winemaking environments. FEMS Yeast Res. 15:fov045. doi: 10.1093/femsyr/fov045
Medina, K., Boido, E., Fariña, L., Gioia, O., Gomez, M. E., Barquet, M., et al. (2013). Increased flavour diversity of Chardonnay wines by spontaneous fermentation and co-fermentation with Hanseniaspora vineae. Food Chem. 141, 2513–2521. doi: 10.1016/j.foodchem.2013.04.056
Mezzasalma, V., Sandionigi, A., Bruni, I., Bruno, A., Lovicu, G., Casiraghi, M., et al. (2017). Grape microbiome as a reliable and persistent signature of field origin and environmental conditions in Cannonau wine production. PLoS ONE 12:e0184615. doi: 10.1371/journal.pone.0184615
Mezzasalma, V., Sandionigi, A., Guzzetti, L., Galimberti, A., Grando, M. S., Tardaguila, J., et al. (2018). Geographical and cultivar features differentiate grape microbiota in Northern Italy and Spain vineyards. Front. Microbiol. 9:946. doi: 10.3389/fmicb.2018.00946
Milanović, V., Comitini, F., and Ciani, M. (2013). Grape berry yeast communities: influence of fungicide treatments. Int. J. Food Microbiol. 161, 240–246. doi: 10.1016/j.ijfoodmicro.2012.12.019
Moreira, N., Mendes, F., Hogg, T., and Vasconcelos, I. (2005). Alcohols, esters and heavy sulphur compound production by pure and mixed cultures of apiculate wine yeasts. Int. J. Food Microbiol. 103, 285–294. doi: 10.1016/j.ijfoodmicro.2004.12.029
Nally, M. C., Ponsone, M. L., Pesce, V. M., Toro, M. E., Vazquez, F., and Chulze, S. (2018). Evaluation of behaviour of Lachancea thermotolerans biocontrol agents on grape fermentations. Lett. Appl. Microbiol. 67, 89–96. doi: 10.1111/lam.13001
Nisiotou, A., Sgouros, G., Mallouchos, A., Nisiotis, C. S., Michaelidis, C., Tassou, C., et al. (2018). The use of indigenous Saccharomyces cerevisiae and Starmerella bacillaris strains as a tool to create chemical complexity in local wines. Food Res. Int. 111, 498–508. doi: 10.1016/j.foodres.2018.05.035
Nissen, P., and Arneborg, N. (2003). Characterization of early death of non-Saccharomyces yeasts in mixed cultures with Saccharomyces cerevisiae. Arch. Microbiol. 180, 257–263. doi: 10.1007/s00203-003-0585-9
Oh, D., Kim, S., and Kim, J. (1998). Increase of xylitol production rate by controlling redox potential in Candida parapsilosis. Biotechnol. Bioeng. 58, 440–444.
Riou, C., Nicaud, J., Barre, P., and Gaillardin, C. (1997). Stationary-phase gene expression in Saccharomyces cerevisiae during wine fermentation. Yeast 13, 90–95. doi: 10.1002/(SICI)1097-0061(199708)13:10<903::AID-YEA145>3.0.CO;2-1
Sabel, A., Martens, S., Petri, A., König, H., and Claus, H. (2014). Wickerhamomyces anomalus AS1: a new strain with potential to improve wine aroma. Ann. Microbiol. 64, 483–491. doi: 10.1007/s13213-013-0678-x
Sadoudi, M., Tourdot-Maréchal, R., Rousseaux, S., Steyer, D., Gallardo Chacón, J. J., Ballester, J., et al. (2012). Yeast-yeast interactions revealed by aromatic profile analysis of Sauvignon Blanc wine fermented by single or co-culture of non- Saccharomyces and Saccharomyces yeasts. Food Microbiol. 32, 243–253. doi: 10.1016/j.fm.2012.06.006
Sambrook, J., and Russell, W.D (2006). Rapid isolation of yeast DNA. Cold Spring Harbour Protocols 2018:pdb.prot093542. doi: 10.1101/pdb.prot4039
Setati, M. E., Jacobson, D., Andong, U. C., and Bauer, F. (2012). The vineyard yeast microbiome, a mixed model microbial map. PLoS ONE 7:e52609. doi: 10.1371/journal.pone.0052609
Shekhawat, K., Bauer, F. F., and Setati, M. E. (2017). Impact of oxygenation on the performance of three non-Saccharomyces yeasts in co-fermentation with Saccharomyces cerevisiae. Appl. Microbiol. Biotechnol. 101, 2479–2491. doi: 10.1007/s00253-016-8001-y
Slabbert, E., Van Heerden, C. J., and Jacobs, K. (2010). Optimisation of automated ribosomal intergenic spacer analysis for the estimation of microbial diversity in fynbos soil. S. Afr. J. Sci. 106, 8–11. doi: 10.4102/sajs.v106i7/8.329
Sun, H., Ma, H., Hao, M., Pretorius, I. S., and Chen, S. (2009). Identification of yeast population dynamics of spontaneous fermentation in Beijing wine region, China. Ann. Microbiol. 59, 69–76. doi: 10.1007/BF03175601
Sun, S. Y., Gong, H. S., Jiang, X. M., and Zhao, Y. P. (2014). Selected non-Saccharomyces wine yeasts in controlled multi-starter fermentations with Saccharomyces cerevisiae on alcoholic fermentation behaviour and wine aroma of cherry wines. Food Microbiol. 44, 15–23. doi: 10.1016/j.fm.2014.05.007
Suzzi, G., Schirone, M., Sergi, M., Marianella, R. M., Fasoli, G., Aguzzi, I., et al. (2012). Multistarter from organic viticulture for red wine Montepulciano d'Abruzzo production. Front. Microbiol. 3:135. doi: 10.3389/fmicb.2012.00135
Taillandier, P., Lai, Q. P., Julien-Ortiz, A., and Brandam, C. (2014). Interaction between Torulaspora delbrueckii and Saccharomyces cerevisiae in wine fermentation: influence of inoculation and nitrogen content. World J.Microbiol. Biotechnol. 30, 1959–1967. doi: 10.1007/s11274-014-1618-z
Tofalo, R., Patrignani, F., Lanciotti, R., Perpetuini, G., Schirone, M., di Gianvito, et al. (2016). Aroma profile of montepulciano d'abruzzo wine fermented by single and co-culture starters of autochthonous Saccharomyces and non-Saccharomyces yeasts. Front Microbiol. 7:610. doi: 10.3389/fmicb.2016.00610
Tristezza, M., Vetrano, C., Bleve, G., Spano, G., Capozzi, V., Logrieco, A., et al. (2013). Biosafety aspects of yeast strains characterized from vineyards and spontaneous fermentations in the Apulia region, Italy. Food Microbiol. 36, 335–342. doi: 10.1016/j.fm.2013.07.001
Viana, F., Belloch, C., Vallés, S., and Manzanares, P. (2011). Monitoring a mixed starter of Hanseniaspora vineae–Saccharomyces cerevisiae in natural must: impact on 2-phenylethyl acetate production. Int. J. Food Microbiol. 151, 235–240. doi: 10.1016/j.ijfoodmicro.2011.09.005
Viana, T., Loureiro-Dias, M. C., and Prista, C. (2014). Efficient fermentation of an improved synthetic grape must by enological and laboratory strains of Saccharomyces cerevisiae. AMB Express 4:16. doi: 10.1186/s13568-014-0016-0
Vigentini, I., Maghradze, D., Petrozziello, M., Bonello, F., Mezzapelle, V., Valdetara, F., et al. (2016). Indigenous Georgian wine-associated yeasts and grape cultivars to edit the wine quality in a precision oenology perspective. Front. Microbiol. 7:352. doi: 10.3389/fmicb.2016.00352
Keywords: wine fermentation, population dynamics, yeast-yeast interactions, multi-starter fermentation, yeast consortium
Citation: Bagheri B, Zambelli P, Vigentini I, Bauer FF and Setati ME (2018) Investigating the Effect of Selected Non-Saccharomyces Species on Wine Ecosystem Function and Major Volatiles. Front. Bioeng. Biotechnol. 6:169. doi: 10.3389/fbioe.2018.00169
Received: 31 August 2018; Accepted: 25 October 2018;
Published: 13 November 2018.
Edited by:
Mingfeng Cao, University of Illinois at Urbana-Champaign, United StatesReviewed by:
Wenjing Cui, Jiangnan University, ChinaRajendran Velmurugan, Chulalongkorn University, Thailand
Pasquale Stano, University of Salento, Italy
Copyright © 2018 Bagheri, Zambelli, Vigentini, Bauer and Setati. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Mathabatha Evodia Setati, c2V0YXRpQHN1bi5hYy56YQ==
 Bahareh Bagheri
Bahareh Bagheri Paolo Zambelli2
Paolo Zambelli2 Ileana Vigentini
Ileana Vigentini Florian Franz Bauer
Florian Franz Bauer Mathabatha Evodia Setati
Mathabatha Evodia Setati