Introduction: Although messenger RNA (mRNA) has great potential to treat various diseases, its successful intracellular delivery remains challenging[1]. Lipid nanoparticles (LNPs) have been widely used over the past decade to deliver small interfering RNAs (siRNAs) to silence protein expression in the liver[2] but have only recently been reported to deliver mRNA to produce protein in vivo[3]. Because of the considerable size, stability, and charge differences between siRNA and mRNA, we hypothesized that the lipid nanoparticle formulation parameters of historically-used siRNA LNPs could be optimized to more efficiently deliver mRNA payloads rather than siRNA payloads (Fig. 1).
Materials and Methods: The original siRNA LNP formulation was made with ionizable lipid C12-200, first reported by Love et al[4]. in 2010, along with three additional excipients: helper phospholipid, cholesterol, and lipid-anchored polyethylene glycol (PEG). We encapsulated mRNA encoding for Erythropoietin (EPO), a secreted serum protein, into this original formulation to establish a baseline from which to improve. We then varied several parameters in the LNP formulation simultaneously: lipid:RNA weight ratio, molar composition of excipients, and phospholipid structure. To reduce the number of formulations required to deduce statistically significant trends in our large, multi-dimensional design space, we employed Design of Experiment (DOE) techniques[5] to design sequential nanoparticle libraries which were tested in mice. EPO mRNA-loaded LNPs were injected intravenously at 15 µg total mRNA per mouse, and blood was collected 6 hours post-injection; an ELISA assay was then used to quantify serum EPO levels. The first library used a Definitive Screening Design to parse the design space, the second library used a fractional factorial design to hone in on an optimized formulation, and the third library used a one-variable-at-a-time methodology to rigorously optimize what we determined to be the most important design factor.
Results and Discussion: After optimization with the three libraries of LNP formulations as described above, we significantly (p < 0.05) increased the efficiency of liver-targeting C12-200 LNPs seven-fold for EPO mRNA delivery relative to the formulation previously used for siRNA delivery (Fig. 2). We found that increased lipid:RNA weight ratios and the use of the phospholipid 1,2-dioleoyl-sn-gylcero-3-phosphoethanolamine (DOPE) were the most crucial factors in determining LNP efficacy (Fig. 3). Interestingly, when our mRNA-optimized LNP formulation was loaded with siRNA, it silenced protein expression in hepatocytes nearly identically to the original siRNA formulation, suggesting that siRNA-loaded LNPs may be more tolerant than mRNA-loaded LNP of formulation design space differences.
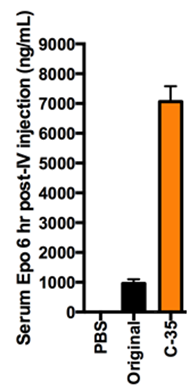
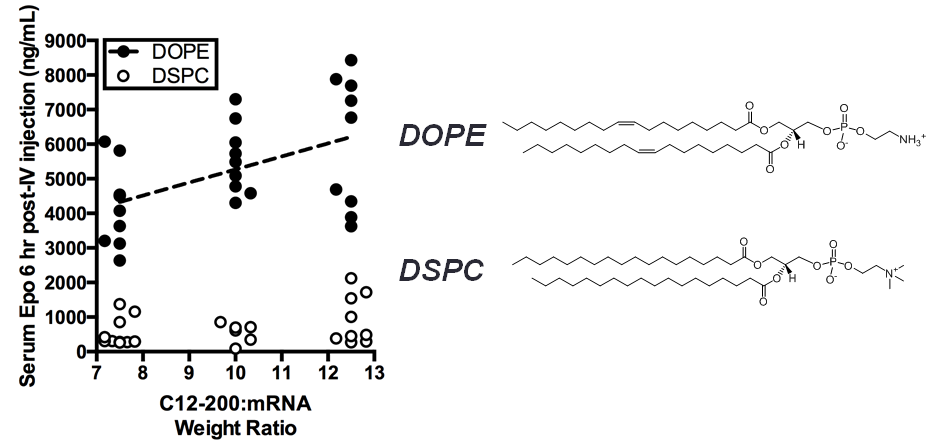
Conclusion: We significantly increased the potency of liver-targeting C12-200 LNPs for mRNA delivery using Design of Experiment methodologies, while showing that the same mRNA-optimized LNPs did not confer additional potency when delivering siRNA. This work provides evidence that some technologies previously used for siRNA delivery may require further optimization for better mRNA delivery. Additionally, the general strategy presented here could be used to screen multi-dimensional design spaces in vivo for optimizing other biomaterials and payloads.
Owen S. Fenton; Michael W. Heartlein; Frank DeRosa; Nicki Watson; Prof. Sumona Mondal; Shire Pharmaceuticals
References:
[1] Sahin, U.; Karikó, K.; Türeci, Ö. Nat. Rev. Drug Discov. 2014, 13 (10), 759–780.
[2] Whitehead, K. A.; Langer, R.; Anderson, D. G. Nat. Rev. Drug Discov. 2009, 8 (2), 129–138.
[3] Thess, A.; Grund, S.; Mui, B. L.; Hope, M. J.; Baumhof, P.; Fotin-Mleczek, M.; Schlake, T. Mol. Ther. 2015, 1–9.
[4] Love, K. T.; Mahon, K. P.; Levins, C. G.; Whitehead, K. a; Querbes, W.; Dorkin, J. R.; Qin, J.; Cantley, W.; Qin, L. L.; Racie, T.; Frank-Kamenetsky, M.; Yip, K. N.; Alvarez, R.; Sah, D. W. Y.; De Fougerolles, A.; Fitzgerald, K.; Koteliansky, V.; Akinc, A.; Langer, R.; Anderson, D. G. Proc. Natl. Acad. Sci. U. S. A. 2010, 107 (5), 1864–1869.
[5] Montgomery, D. C. Design and Analysis of Experiments, 7th ed.; John Wiley & Sons, 2008.